Highlights
-
•
2H6-antigen binding fragment forms a multimeric complex with non-structural 1 protein.
-
•
2H6 antibody binds tightly to non-structural 1 protein.
-
•
The binding affinity reduces significantly when threonine 49 is substituted with Alanine.
-
•
Intracellular expression of 2H6-scFv in mammalian cells reduces viral replication and release of progeny virus.
Abbreviations: IAV, influenza A virus; NS1, non-structural 1; RBD, RNA binding domain; CPSF30, cleavage and polyadenylation and specificity factor 30; PI3K, phosphoinositide 3-kinase; IFN, interferon; PKR, protein kinase R; OAS, 2′5′-oligoadenylate synthetase; dsRNA, double-stranded RNA; mAbs, monoclonal antibodies
Keywords: Non-structural 1 protein, Monoclonal antibody, Influenza A virus
Abstract
The emergence of resistant influenza A viruses highlights the continuous requirement of new antiviral drugs that can treat the viral infection. Non-structural 1 (NS1) protein, an indispensable component for efficient virus replication, can be used as a potential target for generating new antiviral agents. Here, we study the interaction of 2H6 monoclonal antibody with NS1 protein and also determine whether influenza virus replication can be inhibited by blocking NS1. The 2H6-antigen binding fragment (Fab) forms a multimeric complex with the NS1 RNA-binding domain (RBD). T49, a residue which forms a direct hydrogen bond with double stranded RNA, in NS1 protein was found to be critical for its interaction with 2H6 antibody. NS1(RBD) has high affinity to 2H6 with KD of 43.5 ± 4.24 nM whereas NS1(RBD)-T49A has more than 250 times lower affinity towards 2H6. Interestingly, the intracellular expression of 2H6-single-chain variable fragment (scFv) in mammalian cells caused a reduction in viral growth and the M1 viral protein level was significantly reduced in 2H6-scFv transfected cells in comparison to vector transfected cells at 12 h post infection. These results indicate that the tight binding of 2H6 to NS1 could lead to reduction in viral replication and release of progeny virus. In future, 2H6 antibody in combination with other neutralizing antibodies can be used to increase the potency of viral inhibition.
1. Introduction
Influenza A virus (IAV), a member of Orthomyxoviridae family, is still a threat to human health and a burden on the health services (Salomon and Webster, 2009). Despite many advances, IAVs are still a challenge for the scientists. IAVs are highly contagious and causative agents of seasonal flu epidemics resulting in morbidity, mortality and huge economic losses. Based on the circulating strains, seasonal influenza vaccines are developed annually or biannually, if needed, by WHO but the immunity provided is short-lived due to continuous change in the virus strains. Therefore, vaccination is usually required every year to be protected from seasonal flu that leads to increase in vaccine cost along with shortage of vaccines in developing countries. But the two main problems with vaccination are the time required to select, manufacture and deliver vaccine and the variable annual immunization rates (Couch, 2008). Besides vaccination, the antiviral agents are the therapeutic options to treat the infection. Antivirals against M2 protein and neuraminidase are available but their irrational use has led to the emergence of resistant strains (Agrawal et al., 2010, Hayden and Hay, 1992, Poland et al., 2009). Thus, there is a continued requirement of new antiviral agents against IAV.
The non-structural protein NS1 of IAV is a multifunctional protein associated with various viral functions including mRNA processing regulation via interactions with the cleavage and polyadenylation and specificity factor 30 (CPSF30), inhibition of cellular apoptosis by interaction with the p85β regulatory subunit of phosphoinositide 3-kinase (PI3K), limitation of interferon (IFN) production and the IFN-induced proteins, such as protein kinase R (PKR) and 2′5′-oligoadenylate synthetase (OAS)/RNase L by binding to double-stranded RNA (dsRNA) (Hale et al., 2008, Min and Krug, 2006), and inhibition of mRNA splicing by binding to U6 snRNA (Qiu et al., 1995, Wang and Krug, 1998). NS1 has two functionally distinct domains: the N-terminal RNA binding domain (RBD), consisting of three α-helices, and the C-terminal effector domain, consisting of seven β-strands and three α-helices. The RBD domain binds with low affinity to several RNA species in a sequence independent manner (Chien et al., 2004, Hatada and Fukuda, 1992, Qian et al., 1995), and effector domain predominantly interacts with host-cell proteins and also functionally stabilizes RBD domain (Wang et al., 2002).
NS1, a well conserved protein, is expressed at very high levels in infected cells (Krug and Etkind, 1973, Palese and Shaw, 2007). Therefore, NS1 protein is a good target for therapeutics development and several small molecules have been found to inhibit NS1 function resulting in reduced viral replication (Engel, 2013, Nayak et al., 2014, Woo et al., 2013). In our previous study, we generated a panel of new monoclonal antibodies (mAbs) against the RNA binding domain of NS1 (NS1(RBD)) (Tan et al., 2010). Here we report the biophysical characterization of one of these mAbs, named as 2H6, and NS1(RBD) protein interaction, and inhibition of viral replication by targeting NS1 protein in infected cells. 2H6 has been shown to bind to NS1 of different IAV subtypes, namely H5N1, H3N2 and H1N1 (Tan et al., 2010).
2. Materials and methods
2.1. Cell lines and virus
A549, 293T and MDCK cells were purchased from American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured at 37 °C in 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum. A/Puerto Rico/8/1934(H1N1) (PR8) virus was obtained from the American Type Culture Collection and grown in embryonated chicken eggs as previously described (Narasaraju et al., 2011).
2.2. Generation of anti-M1 monoclonal antibody
The cDNA encoding for the M1 gene from a H5N1 isolate (A/chicken/Hatay/2004(H5N1)), GenBank accession number AM040045) was cloned into the pGEX-6P1 vector (GE Healthcare, Uppsala, Sweden). Glutathione S-transferase (GST)-fused M1 protein was then expressed in Escherichia coli BL21(DE3) (Novagen, EMD Chemicals, Inc., Madison, WI, USA) and purified as previously described (Tan et al., 2010). The GST-fusion protein was then used to immunize mice and generate hybridomas as previously described (Oh et al., 2010). The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the Biological Resource Centre, A*STAR, Singapore (Protocol Number: 110693). All the procedures were carried out in strict accordance with the recommendations of the National Advisory Committee for Laboratory Animal Research (NACLAR) guidelines in Singapore. All efforts were made to minimize suffering and euthanasia was performed using carbon dioxide.
2.3. Ascites production
Ascites was produced by injecting hybridoma cells into the peritoneal cavities of pristine-primed BALB/c mice. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the Biological Resource Centre, A*STAR, Singapore (Protocol Number: 110694). All the procedures were carried out in strict accordance with the recommendations of the NACLAR guidelines in Singapore. All efforts were made to minimize suffering and euthanasia was performed using carbon dioxide.
2.4. Expression and purification of NS1 in bacteria
The gene encoding N-terminus of NS1 (1–73 aa) of A/chicken/Hatay/2004(H5N1) was cloned into modified pET-28a expression vector (Novagen) with N-terminal His-SUMO tag. The wild-type NS1(RBD) and its mutants (R38AK41A, S42A and T49A) were expressed in E . coli BL21(DE3)-RILP overnight at 20 °C, and protein expression was induced using 0.4 mM isopropyl β-d-thiogalactoside (IPTG). Cells were harvested by centrifugation and the cell pellet was resuspended in lysis buffer which was followed by passing it through a cell disruptor (www.avestin.com) for five times. After ultracentrifugation at 40,000 rpm for 1 h, the supernatants were loaded onto Ni–NTA affinity column for purification. Pooled eluted fractions were dialysed overnight against dialysis buffer (20 mM Tris (pH 7.4), 100 mM NaCl) supplemented with Ulp1 protease for His-SUMO cleavage. The fractions were reloaded onto Ni–NTA affinity column to remove His-SUMO tag. The flow through was then loaded onto HiLoad 26/60 Superdex 75 column (GE Healthcare), equilibrated in 25 mM Tris (pH 7.4), 500 mM NaCl and 10 mM DTT for further purification. The purified proteins were dialysed against dialysis buffer again and concentrated to 3 mg/mL in a Centriprep-10 (Amicon). Purified NS1 fragments were then subjected to Tricine–SDS–PAGE on a 15% gel which was stained using coomassie blue to visualize the purity of proteins.
2.5. Purification of 2H6 whole antibody and preparation of 2H6-antigen-binding fragment (Fab)
Antibody was purified from ascites by HiTrap Protein A column (GE Healthcare) according to manufacturer’s instructions. Briefly, ascites was diluted in 1× phosphate buffer saline (PBS) and injected in pre-equilibrated Protein A column. The whole antibody was eluted from the column using 25 mM glycine (pH 2.2) elution buffer and then subjected to papain cleavage according to manufacturer’s instructions. In brief, whole antibody was incubated with immobilized papain (Thermo Scientific) in digestion buffer (20 mM sodium phosphate (pH 7), 10 mM EDTA, 20 mM cysteine) overnight at 37 °C which was followed by Fab purification by HiTrap Protein L column (GE Healthcare) according to manufacturer’s instructions.
2.6. Circular dichroism (CD) spectroscopy
Far-UV CD spectra (260–190 nm) were recorded using a Jasco J-810 spectropolarimeter (Jasco Corporation, Tokyo, Japan). The cuvette chamber and instrument optics were continuously purged with 30 L of nitrogen/min before and during the measurements. The spectra were recorded using a scanning speed of 50 nm/min, a resolution of 0.1 nm and a bandwidth of 1 nm. NS1(RBD) (10 μM) and 2H6-Fab (5 μM) were dissolved in 1 mM phosphate buffer (pH 7.4) and placed in a cuvette with 0.1 cm path length. An average of three scans was taken to increase the signal to noise ratio and the baseline was subtracted.
2.7. Gel filtration chromatography
The NS1(RBD) and 2H6-Fab were mixed in 1:1 M ratio and incubated overnight at 4 °C. The NS1(RBD) and 2H6-Fab complex formation was examined by gel filtration chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare). The sample was injected in the column equilibrated with 50 mM Tris (pH 7.4) buffer containing 150 mM NaCl and eluted with the same buffer using AKTA purifier system (GE Healthcare Life Sciences, Piscataway, NJ, USA) at a flow rate of 1 mL/min. The gel filtration fractions were analyzed by SDS/PAGE on 4–20% ready gel (Bio-Rad).
2.8. Surface plasmon resonance (SPR)
The affinities for NS1(RBD) and NS1(RBD)-T49A binding to 2H6 antibody were determined using Biacore™ 3000 (GE Healthcare Life Sciences, Piscataway, NJ, USA) at 25 °C. Briefly, 2H6 antibody (20 μg/mL) diluted in 10 mM sodium acetate buffer (pH 5) was immobilized at a flow rate of 10 μL/min for 7 min on the surface of a CM5 chip (GE Healthcare) following the standard 1-ethyl-3-(3-dimethylpropyl)-carbodiimide (EDC) plus N-hydroxysuccinimide (NHS) (GE Healthcare) coupling chemistry. Kinetic measurements were carried out by 3 min injection at a flow rate of 20 μL/min of serial dilutions of NS1(RBD) and NS1(RBD)-T49A in HBS–EP buffer (GE Healthcare) from 400–25 nM and 12.5–0.78 μM, respectively and dissociation for 3 min. The 2H6 immobilized chip surface was regenerated by injection of 10 mM glycine (pH 2) for 45 s at a flow rate of 30 μL/min after each cycle. The data were collected and processed using the BIAevaluation software (GE Healthcare). Binding of 2H6 to all concentration series of NS1(RBD) and NS1(RBD)-T49A were analyzed using a 1:1 binding model. The data were corrected by subtraction of a “buffer only” control as well as zero 2H6 antibody flow cell.
2.9. Enzyme-linked immunosorbent assay (ELISA)
Purified wild-type and mutant NS1(RBD) proteins were diluted at different concentrations into 0.05 M carbonate–bicarbonate buffer (pH 9.6). Proteins (50 μL) were then coated onto 96-well ELISA plates (Nunc) overnight at 4 °C. The wells were blocked in 5% milk in PBS with 0.1% Tween 20 (PBST) for 1 h at 37 °C followed by addition of 100 μL of 2H6 whole antibody (5 μg/mL) as primary antibody to each well and incubated at 37 °C for 2–3 h. The wells were then washed in PBST followed by the addition of goat anti-mouse horse-radish peroxidase (HRP)-conjugated antibody (Pierce) as secondary antibody and incubated at 37 °C for 1 h. Tetramethylbenzidine substrate (Pierce) was added and reaction was stopped using 0.2 M sulfuric acid. Absorbance at 450 nm was recorded using an absorbance reader (Tecan Infinite M200).
2.10. Transient transfection of 2H6-single-chain variable fragment (scFv) and western blot analysis
Variable heavy (VH) and variable light (VL) genes were obtained from RNA extracted from the 2H6 or 7G12 hybridoma as described previously (Dang et al., 2013). 7G12 is a monoclonal antibody targeting the spike protein of severe acute respiratory syndrome (SARS) coronavirus (Lip et al., 2006). Subsequently, the 2H6-scFv and 7G12-scFv were constructed by using overlap PCR to link them via a GGGSGGGSGGGS linker. Finally, the 2H6-scFv and 7G12-scFv were cloned into the pXJ40-FLAG or pXJ40-Myc CMV expression vector so that a FLAG or Myc epitope is fused to its N terminus. The plasmids were then transfected into 80% confluent 293T or A549 cells by using X-treme GENE HP (Roche) according to the manufacturer’s instructions. After 24 h (293T) or 48 h (A549) post-transfection, the cells were rinsed using PBS and inoculated with PR8 for 1 h. The viral media were collected for titration by plaque assay. The transfected cells were lysed in lysis buffer (50 mM Tris (pH 8.0), 0.1% NP-40 and 150 mM NaCl) at 4 °C for 2 h, and the cell lysates were subsequently collected. The cell lysates (50 μg) were resolved using electrophoresis on an SDS–polyacrylamide gel and transferred to a nitrocellulose membrane (Bio-Rad). Antibodies against FLAG (Sigma), Myc (Santa Cruz), M1 (as described above), β-actin (Sigma) were used. After washing, the membrane was incubated with a HRP-conjugated secondary antibody (Pierce). The membranes were then washed and enhanced chemiluminescence substrate (Pierce) was added for detection using ChemiDoc™ MP Imaging System (Bio-Rad).
2.11. Plaque assay in MDCK cells
MDCK cells were seeded in 6-well plates. After 24 h incubation, plates were rinsed using PBS, and subsequently adsorbed with serially diluted supernatants containing viruses for 1 h at 37 °C. The medium was discarded and the cells were rinsed using PBS. The cells were overlaid with 2 mL of DMEM supplemented by 0.3% agar and 2 μg/mL TPCK-trypsin (Thermo Scientific). After incubation at 37 °C for 2 days, the cells were fixed using 10% formalin for 1 h and stained using a 0.1% crystal violet solution.
2.12. Statistical analysis
The two-tailed Student’s t test was applied to evaluate the statistical significance of differences measured from the data sets obtained in 3 independent experiments. p < 0.05 was considered statistically significant.
3. Results and discussion
3.1. The 2H6-Fab and NS1(RBD) protein forms a multimeric complex
In order to characterize the interaction between mAb 2H6 and NS1 protein, the NS1(RBD) protein was expressed in E. coli and purified while the 2H6-Fab was obtained from ascites fluid after papain cleavage (see Supplementary Fig. S1). Far-UV CD spectroscopy was then used to analyze the secondary structures of NS1(RBD) and 2H6-Fab. The spectrum showed minima at 208 and 220 nm for NS1(RBD) protein (Fig. 1 A), which indicates that it is rich in α-helices and is consistent with the three-dimensional structures of NS1(RBD) protein (Hale, 2014). The spectrum for 2H6-Fab showed minimum at 216 nm and maximum at 202 nm (Fig. 1B), which indicates that it is composed of β-sheeted structure (Satow et al., 1986, Tetin et al., 2003).
Fig. 1.
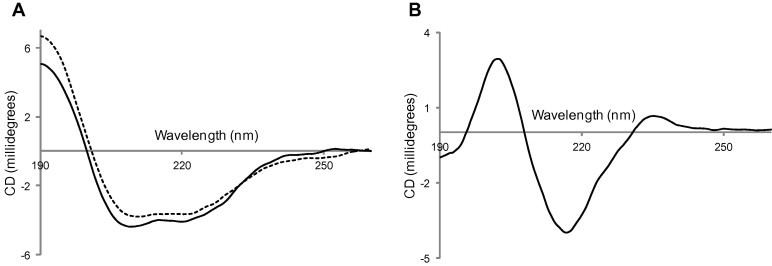
Secondary structure characterization of NS1(RBD) and 2H6 Fab. Far-UV CD spectra of (A) NS1(RBD) (solid line) and NS1(RBD)-T49A (dotted line), and (B) 2H6-Fab in 1 mM sodium phosphate buffer pH 7 were recorded using 0.1 cm pathlength cuvette.
Next, NS1(RBD) and 2H6-Fab were mixed in 1:1 M ratio, incubated overnight at 4 °C and then subjected to size exclusion chromatography. Three peaks were obtained after separation on a Superdex 200 column (Fig. 2 A). The 2H6-Fab and NS1(RBD) protein co-eluted in the first peak and the free 2H6-Fab eluted out as the major peak (Fig. 2B). The 7G12 antibody, which binds to the spike protein of SARS coronavirus (Lip et al., 2006), was used as a negative control for this experiment. In contrast to the results for 2H6-Fab, 7G12-Fab and NS1(RBD) eluted out of the column separately when the mixture was run on a Superdex 200 column (see Supplementary Fig. S2).
Fig. 2.
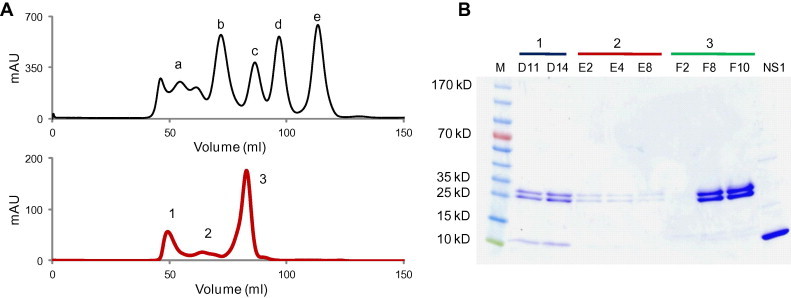
2H6 Fab–NS1(RBD) complex co-elution. (A) Gel filtration standard (Bio-Rad) profile on Superdex 200 column (top). a: Thyroglobulin (670 kDa); b: γ-Globulin (158 kDa); c: Ovalbumin (44 kDa); d: Myoglobin (17 kDa); e: Vitamin B12 (1.35 kDa). Gel filtration profile of the pre-incubated 2H6 Fab and NS1(RBD) protein (bottom). The complex was eluted out with 50 mM Tris–HCl buffer (pH 7.4) containing 150 mM NaCl. (B) SDS–PAGE analysis of the peaks eluted from the gel filtration column. Samples were resolved on 4–20% ready gel and stained with Coomassie Brilliant Blue-R250. M: PageRuler Prestained Protein Ladder; D11 and D14: Peak 1 fractions; E2, E4 and E8: Peak 2 fractions; F2, F8 and F10: Peak 3 fractions; NS1: Purified NS1(RBD) protein.
This result clearly indicates that NS1(RBD) interacts with 2H6-Fab and the elution time suggests that the antibody–antigen complex has a molecular weight greater than 158 kDa (Fig. 2A). Dynamic light scattering (DLS) was performed on the fraction containing this complex and the results reveal that it is homogenous and has an estimated molecular weight of 400 kDa (see Supplementary Fig. S3). Given that NS1(RBD) exists as a dimer of ∼18 kDa, binding to 2H6-Fab of ∼50 kDa will yield an expected size of ∼120 kDa assuming the complex has a globular structure. This may imply that the antigen–antibody complex is actually multimeric in nature. The occurrence of multimeric antigen–antibody complex has been reported in several studies (Sanny and Price, 1997, Santora et al., 2001). However, it cannot be ruled out that the complex does not have a globular structure and hence, show non-ideal chromatographic behavior. In-depth studies using more sophisticated biophysical techniques like nuclear magnetic resonance will be needed to get a better insight.
3.2. NS1(RBD)-T49A substitution mutant exhibited less affinity towards 2H6 antibody
Residues 42–53 in NS1 were previously shown to be required for interaction with 2H6 (Tan et al., 2010). Since the side-chains of residues S42 and T49 make direct hydrogen bond with the double stranded RNA (dsRNA) backbone and dsRNA binding affinity is reduced by up to 10-fold when either of them is mutated (Cheng et al., 2009), two substitution mutants, NS1(RBD)-S42A and NS1(RBD)-T49A, were generated. To determine if these residues in NS1 are critical for interaction with 2H6, we performed a comparative ELISA (Fig. 3 A). The results show that NS1(RBD) and NS1(RBD)-S42A bound comparably to 2H6 at all the antigen concentrations tested, while the binding of NS1(RBD)-T49A was significantly decreased when 0.125 or 0.031 μg/mL of protein was used for coating. No binding to Bovine serum albumin (BSA), which was used as a negative control, was observed. A double substitution mutant NS1(RBD)-R38AK41A was also tested because these two residues, which lie upstream of the 2H6 binding domain, have previously been shown to be important for RNA binding (Wang et al., 1999). The NS1(RBD)-R38AK41A mutant has similar binding to 2H6 as NS1(RBD) confirming that residues upstream of S42 are not likely to be involved in the interaction.
Fig. 3.
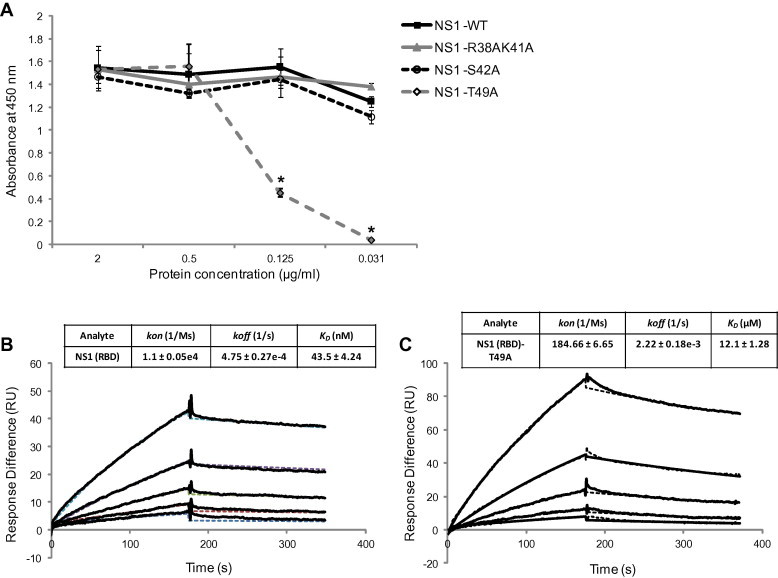
In vitro binding affinity analysis. (A) Comparative ELISA was performed to determine the NS1(RBD) residues critical for its interaction with 2H6. Wells coated with different concentrations of antigens were probed with 5 μg/mL of 2H6. All experiments were performed in triplicate, and the average values with standard deviations are plotted. Differences in absorbance readings between NS1(RBD) and NS1(RBD)-T49A were evaluated by unpaired t-test (∗, p < 0.05). (B) and (C) Representative SPR sensorgrams of the kinetics of association and dissociation of a range of concentrations from (B) 25–400 nM of NS1(RBD) or (C) 0.78–12.5 μM of NS1(RBD)-T49A to immobilized 2H6. Data was fitted as a 1:1 interaction model and the dotted lines represent the fitting of the original sensorgrams (solid line). Three independent analyses were carried out, and kinetic constants are reported ± SD.
Since T49 appears to play a key role in the NS1(RBD)–2H6 interaction, SPR was next used to compare the 2H6 antibody-binding affinity between NS1(RBD) and NS1(RBD)-T49A. 2H6 antibody was immobilized on a biosensor chip and various concentrations of NS1(RBD) or NS1(RBD)-T49A were passed through the chip. The kinetics of association and dissociation were recorded in SPR sensorgram. The kinetic rate constants kon and koff were determined from the ascending rate of resonance units during association and the descending rate during dissociation. The results show that NS1(RBD) has high affinity to 2H6 with KD of 43.5 ± 4.24 nM (Fig. 3B) whereas NS1(RBD)-T49A has a significantly lower affinity (KD = 12.1 ± 1.28 μM) (Fig. 3C), indicating that the T49 residue is important for strong binding between NS1 and 2H6. The CD spectrum of NS1(RBD)-T49A reveals that its secondary structure is similar to that of NS1(RBD) (Fig. 1A), indicating the substitution does not cause gross structural change.
3.3. Intracellular expression of 2H6 caused a reduction in viral replication
Since 2H6 binds to the RBD of NS1, it may have the ability to inhibit the function of NS1 in the infected cells. To express 2H6 intracellularly, the 2H6-scFv gene was cloned in a mammalian expression plasmid and transiently transfected into A549 cells. After 48 h, the cells were inoculated with 0.01 MOI of PR8 virus and the amount of virus secreted was determined by plaque quantitation at 12, 24, and 48 h post infection (hpi). As shown in Fig. 4 A, the viral growth in the 2H6-scFv transfected A549 cells was reduced by approximately 0.5 log PFU/mL at 24 and 48 hpi when compared to cells transfected with vector only. To determine if viral inhibition by 2H6-scFv is specific, the scFv of an irrelevant antibody 7G12, which binds to the spike protein of SARS coronavirus (Lip et al., 2006), was also used in this experiment. The viral growth in the 7G12-scFv transfected A549 cells did not show reduction when compared to cells transfected with vector only.
Fig. 4.
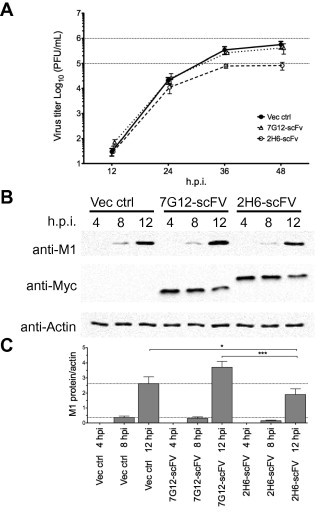
Effects of intracellular 2H6-scFv expression on the replication of influenza A virus. (A) Multiple-step growth curve of PR8 (0.01 MOI) in FLAG-tagged 2H6-scFv or 7G12-scFv expressing A549 cells was determined by plaque forming unit at 12, 24, and 48 hpi. (B) 293T cells expressing Myc-tagged 2H6-scFv or 7G12-scFv were infected by PR8 (2 MOI) and harvested at 4, 8, and 12 hpi. Western blot analysis was performed by using anti-M1, anti-Myc and anti-actin antibodies. (C) The expression levels of M1 were showed in graphs after normalization with actin. All experiments were performed in triplicate, and the average values with standard deviations are plotted. Differences in M1 expression between 2H6-scFv and vector transfected cells or between 2H6-scFv and 7G12-scFv transfected cells were evaluated by unpaired t-test (∗, p < 0.05; ∗∗∗, p < 0.001).
As only ∼50% of the A549 cells were transfected (data not shown), the experiment was repeated using the highly transfectable 293T cells. Following that, the cells were infected with 2 MOI of PR8 and harvested at 4, 8, and 12 hpi for western blot analysis. Consistent with the results obtained in A549 cells, the level of the M1 viral protein in the infected cells was significantly reduced in 2H6-scFv transfected cells when compared to the vector or 7G12-scFv transfected cells at 12 hpi (Fig. 4B and C). The average reduction in normalized M1 expression in 2H6-scFv transfected cells when compared to vector and 7G12-scFv transfected cells at 12 hpi is 27% and 49% respectively. These results suggest that the binding of 2H6 to NS1 could hinder the role of NS1 in infection and cause a reduction in viral replication and release.
4. Conclusion
Our results show that the murine mAb 2H6 binds tightly to the NS1 protein and the affinity of this interaction is significantly reduced when T49 is substituted by A. When the 2H6-scFv is expressed intracellularly, it binds to NS1 expressed in the infected cells and caused a significant reduction in viral protein expression and release of progeny virus. Since the side-chains of T49 are involved in RNA binding (Cheng et al., 2009), it is likely that the binding of 2H6 has a negative impact on the ability of NS1 to bind to RNA and hence, resulting in a reduction of viral replication.
NS1 is a viable drug target as it is involved in various processes that are crucial for viral replication and propagation (Hale et al., 2008). In the past, few small chemical compounds have been shown to inhibit both NS1 function and virus replication (Basu et al., 2009, Walkiewicz et al., 2011). But the precise mechanisms of action of these chemical compounds are still unknown and remain to be elucidated. Earlier, the function of CPSF30 binding site of the NS1 has been shown to be inhibited by the second and third zinc fingers (F2F3), a 61 residues fragment of CPSF30, resulting in the inhibition of IAV replication without detectable effects on cellular functions (Twu et al., 2006). Our approach of expressing 2H6-scFv in the cells to block IAV replication during infection is similar to their method of expressing F2F3 intracellularly to bind NS1 and inhibit its interaction with CPSF30. Interestingly, a recent publication has also reported the abilities of 3 NS1 specific-human scFvs to inhibit replication of influenza A virus (Yodsheewan et al., 2013). However, the mechanisms of binding are different as they bound to residues 7–18, 75–90 and 179–190 of NS1, respectively.
These studies open up a new opportunity to restrict viral replication by targeting different NS1 domains via different modes of inhibition. In future studies, it will be important to use a combination of different NS1 inhibiting molecules to determine if this can increase the potency of viral inhibition and/or minimize the chance of viral escape both in vitro and in vivo. Delivery of antibody or scFv into the cells is an important aspect of antiviral therapy. Thus, viral expression vectors such as adenovirus or lentivirus can be used for 2H6-scFv delivery. Moreover, cytoplasmic transduction peptide (CTP), a stretch of basic residues, has been extensively documented for its efficient delivery of biomolecules (Gump and Dowdy, 2007, Kim et al., 2006). Recently, hepatitis B virus replication has been reported to be inhibited by intracellular delivery of scFv against core protein via CTP (Xun et al., 2013). As an alternative to viral expression vectors, 2H6-scFv can be fused with CTP for the efficient cytoplasmic translocation of the scFv into the cells.
Acknowledgments
We thank Q. Duong Hong, M. Nguyen Tien, H. Le Thanh, B. Le Tran, K. DinhDuy (Institute of Biotechnology, Hanoi, Vietnam), and Lal S.K. (International Centre for Genetic Engineering and Biotechnology, New Delhi, India) for providing H5N1 virus-derived DNA constructs. This work is supported by the Singapore Ministry of Health’s National Medical Research Council under its NMRC-CBRG scheme [Grant No. NMRC/CBRG/0017/2012]. Initial work was supported by intramural funding at the Institute of Molecular and Cell Biology, A*STAR (Agency for Science, Technology and Research), Singapore.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.antiviral.2015.01.015.
Appendix A. Supplementary data
Purification of 2H6-Fab and NS1(RBD). (A) SDS–PAGE analysis of 2H6 antibody purified from ascites fluid using Protein A column chromatography. (B) SDS–PAGE analysis of the 2H6-Fab purification by Protein L column chromatography following papain cleavage. NS1(RBD) protein was expressed as a fusion protein in BL21 (DE3) cells. (C) SDS–PAGE analysis of the purified NS1(RBD) proteins. All the gels were stained with Coomassie Brilliant Blue-R250. M: PageRuler Prestained Protein Ladder; P: PageRuler Unstained Protein Ladder; FT: Flow through; W: Wash; E: Elution.
7G12-Fab and NS1(RBD) elute out separately. (A) Gel filtration profile of the pre-incubated 7G12-Fab and NS1(RBD) protein. The proteins were eluted out with 50 mM Tris–HCl buffer (pH 7.4) containing 150 mM NaCl. (B) SDS–PAGE analysis of the peaks eluted from the gel filtration column. Samples were resolved on 4–20% ready gel and stained with Coomassie Brilliant Blue-R250. M: PageRuler Prestained Protein Ladder; E10: Peak 1 fraction; F15, F13, F11, F8, F5 and F3: Peak 2 fractions; G2: Peak 3 fraction; NS1: Purified NS1(RBD) protein.
Dynamic light scattering. About 20 μl of the fraction containing 2H6-Fab and NS1(RBD) complex (peak 1) was used to perform DLS on DynaPro (Wyatt Technology, Santa Barbara, CA, USA).
References
- Agrawal A.S., Sarkar M., Ghosh S., Roy T., Chakrabarti S., Lal R., Mishra A.C., Chadha M.S., Chawla-Sarkar M. Genetic characterization of circulating seasonal influenza A viruses (2005–2009) revealed introduction of oseltamivir resistant H1N1 strains during 2009 in eastern India. Infect. Genet. Evol.: J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2010;10:1188–1198. doi: 10.1016/j.meegid.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Basu D., Walkiewicz M.P., Frieman M., Baric R.S., Auble D.T., Engel D.A. Novel influenza virus NS1 antagonists block replication and restore innate immune function. J. Virol. 2009;83:1881–1891. doi: 10.1128/JVI.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Wong S.M., Yuan Y.A. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009;19:187–195. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- Chien C.Y., Xu Y., Xiao R., Aramini J.M., Sahasrabudhe P.V., Krug R.M., Montelione G.T. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry. 2004;43:1950–1962. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- Couch R.B. Seasonal inactivated influenza virus vaccines. Vaccine. 2008;26(Suppl. 4):D5–D9. doi: 10.1016/j.vaccine.2008.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang V.T., Mandakhalikar K.D., Ng O.W., Tan Y.J. A simple methodology for conversion of mouse monoclonal antibody to human–mouse chimeric form. Clin. Dev. Immunol. 2013;2013:716961. doi: 10.1155/2013/716961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel D.A. The influenza virus NS1 protein as a therapeutic target. Antivir. Res. 2013;99:409–416. doi: 10.1016/j.antiviral.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gump J.M., Dowdy S.F. TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol. Med. 2007;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Hale B.G. Conformational plasticity of the influenza A virus NS1 protein. J. Gen. Virol. 2014 doi: 10.1099/vir.0.066282-0. [DOI] [PubMed] [Google Scholar]
- Hale B.G., Randall R.E., Ortin J., Jackson D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Hatada E., Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 1992;73(Pt. 12):3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- Hayden F.G., Hay A.J. Emergence and transmission of influenza A viruses resistant to amantadine and rimantadine. Curr. Top. Microbiol. Immunol. 1992;176:119–130. doi: 10.1007/978-3-642-77011-1_8. [DOI] [PubMed] [Google Scholar]
- Kim D., Jeon C., Kim J.H., Kim M.S., Yoon C.H., Choi I.S., Kim S.H., Bae Y.S. Cytoplasmic transduction peptide (CTP): new approach for the delivery of biomolecules into cytoplasm in vitro and in vivo. Exp. Cell Res. 2006;312:1277–1288. doi: 10.1016/j.yexcr.2005.12.029. [DOI] [PubMed] [Google Scholar]
- Krug R.M., Etkind P.R. Cytoplasmic and nuclear virus-specific proteins in influenza virus-infected MDCK cells. Virology. 1973;56:334–348. doi: 10.1016/0042-6822(73)90310-3. [DOI] [PubMed] [Google Scholar]
- Lip K.M., Shen S., Yang X., Keng C.T., Zhang A., Oh H.L., Li Z.H., Hwang L.A., Chou C.F., Fielding B.C., Tan T.H., Mayrhofer J., Falkner F.G., Fu J., Lim S.G., Hong W., Tan Y.J. Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J. Virol. 2006;80:941–950. doi: 10.1128/JVI.80.2.941-950.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.Y., Krug R.M. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′–5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasaraju T., Yang E., Samy R.P., Ng H.H., Poh W.P., Liew A.A., Phoon M.C., van Rooijen N., Chow V.T. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am. J. Pathol. 2011;179:199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak M.K., Agrawal A.S., Bose S., Naskar S., Bhowmick R., Chakrabarti S., Sarkar S., Chawla-Sarkar M. Antiviral activity of baicalin against influenza virus H1N1-pdm09 is due to modulation of NS1-mediated cellular innate immune responses. J. Antimicrob. Chemother. 2014;69:1298–1310. doi: 10.1093/jac/dkt534. [DOI] [PubMed] [Google Scholar]
- Oh H.L., Akerstrom S., Shen S., Bereczky S., Karlberg H., Klingstrom J., Lal S.K., Mirazimi A., Tan Y.J. An antibody against a novel and conserved epitope in the hemagglutinin 1 subunit neutralizes numerous H5N1 influenza viruses. J. Virol. 2010;84:8275–8286. doi: 10.1128/JVI.02593-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P., Shaw M.L. Orthomyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1647–1689. [Google Scholar]
- Poland G.A., Jacobson R.M., Ovsyannikova I.G. Influenza virus resistance to antiviral agents: a plea for rational use. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2009;48:1254–1256. doi: 10.1086/598989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X.Y., Chien C.Y., Lu Y., Montelione G.T., Krug R.M. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA. 1995;1:948–956. [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Nemeroff M., Krug R.M. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6–U2 and U6–U4 snRNA interactions during splicing. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- Salomon R., Webster R.G. The influenza virus enigma. Cell. 2009;136:402–410. doi: 10.1016/j.cell.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanny C.G., Price J.A. Analysis of antibody–antigen interactions using size-exclusion high-performance (pressure) liquid chromatography. Anal. Biochem. 1997;246:7–14. doi: 10.1006/abio.1996.9995. [DOI] [PubMed] [Google Scholar]
- Santora L.C., Kaymakcalan Z., Sakorafas P., Krull I.S., Grant K. Characterization of noncovalent complexes of recombinant human monoclonal antibody and antigen using cation exchange, size exclusion chromatography, and BIAcore. Anal. Biochem. 2001;299:119–129. doi: 10.1006/abio.2001.5380. [DOI] [PubMed] [Google Scholar]
- Satow Y., Cohen G.H., Padlan E.A., Davies D.R. Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A. J. Mol. Biol. 1986;190:593–604. doi: 10.1016/0022-2836(86)90245-7. [DOI] [PubMed] [Google Scholar]
- Tan Z., Akerstrom S., Wee B.Y., Lal S.K., Mirazimi A., Tan Y.J. A new panel of NS1 antibodies for easy detection and titration of influenza A virus. J. Med. Virol. 2010;82:467–475. doi: 10.1002/jmv.21709. [DOI] [PubMed] [Google Scholar]
- Tetin S.Y., Prendergast F.G., Venyaminov S.Y. Accuracy of protein secondary structure determination from circular dichroism spectra based on immunoglobulin examples. Anal. Biochem. 2003;321:183–187. doi: 10.1016/s0003-2697(03)00458-5. [DOI] [PubMed] [Google Scholar]
- Twu K.Y., Noah D.L., Rao P., Kuo R.L., Krug R.M. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J. Virol. 2006;80:3957–3965. doi: 10.1128/JVI.80.8.3957-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkiewicz M.P., Basu D., Jablonski J.J., Geysen H.M., Engel D.A. Novel inhibitor of influenza non-structural protein 1 blocks multi-cycle replication in an RNase L-dependent manner. J. Gen. Virol. 2011;92:60–70. doi: 10.1099/vir.0.025015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Krug R.M. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT–AC splicing by the influenza virus NS1 protein. RNA. 1998;4:55–64. [PMC free article] [PubMed] [Google Scholar]
- Wang W., Riedel K., Lynch P., Chien C.Y., Montelione G.T., Krug R.M. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Basler C.F., Williams B.R., Silverman R.H., Palese P., Garcia-Sastre A. Functional replacement of the carboxy-terminal two-thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J. Virol. 2002;76:12951–12962. doi: 10.1128/JVI.76.24.12951-12962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.M., Kim K.S., Lee J.M., Shim H.S., Cho S.J., Lee W.K., Ko H.W., Keum Y.S., Kim S.Y., Pathinayake P., Kim C.J., Jeong Y.J. Single-stranded DNA aptamer that specifically binds to the influenza virus NS1 protein suppresses interferon antagonism. Antivir. Res. 2013;100:337–345. doi: 10.1016/j.antiviral.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Xun Y., Pan Q., Tang Z., Chen X., Yu Y., Xi M., Zang G. Intracellular-delivery of a single-chain antibody against hepatitis B core protein via cell-penetrating peptide inhibits hepatitis B virus replication in vitro. Int. J. Mol. Med. 2013;31:369–376. doi: 10.3892/ijmm.2012.1210. [DOI] [PubMed] [Google Scholar]
- Yodsheewan R., Maneewatch S., Srimanote P., Thueng-In K., Songserm T., Dong-Din-On F., Bangphoomi K., Sookrung N., Choowongkomon K., Chaicumpa W. Human monoclonal ScFv specific to NS1 protein inhibits replication of influenza viruses across types and subtypes. Antivir. Res. 2013;100:226–237. doi: 10.1016/j.antiviral.2013.07.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purification of 2H6-Fab and NS1(RBD). (A) SDS–PAGE analysis of 2H6 antibody purified from ascites fluid using Protein A column chromatography. (B) SDS–PAGE analysis of the 2H6-Fab purification by Protein L column chromatography following papain cleavage. NS1(RBD) protein was expressed as a fusion protein in BL21 (DE3) cells. (C) SDS–PAGE analysis of the purified NS1(RBD) proteins. All the gels were stained with Coomassie Brilliant Blue-R250. M: PageRuler Prestained Protein Ladder; P: PageRuler Unstained Protein Ladder; FT: Flow through; W: Wash; E: Elution.
7G12-Fab and NS1(RBD) elute out separately. (A) Gel filtration profile of the pre-incubated 7G12-Fab and NS1(RBD) protein. The proteins were eluted out with 50 mM Tris–HCl buffer (pH 7.4) containing 150 mM NaCl. (B) SDS–PAGE analysis of the peaks eluted from the gel filtration column. Samples were resolved on 4–20% ready gel and stained with Coomassie Brilliant Blue-R250. M: PageRuler Prestained Protein Ladder; E10: Peak 1 fraction; F15, F13, F11, F8, F5 and F3: Peak 2 fractions; G2: Peak 3 fraction; NS1: Purified NS1(RBD) protein.
Dynamic light scattering. About 20 μl of the fraction containing 2H6-Fab and NS1(RBD) complex (peak 1) was used to perform DLS on DynaPro (Wyatt Technology, Santa Barbara, CA, USA).


