Abstract
The Million Veteran Program (MVP), initiated by the Department of Veterans Affairs (VA), aims to collect biosamples with consent from at least one million veterans. Presently, blood samples have been collected from over 800,000 enrolled participants. The size and diversity of the MVP cohort, as well as the availability of extensive VA electronic health records, make it a promising resource for precision medicine. MVP is conducting array-based genotyping to provide a genome-wide scan of the entire cohort, in parallel with whole-genome sequencing, methylation, and other ‘omics assays. Here, we present the design and performance of the MVP 1.0 custom Axiom array, which was designed and developed as a single assay to be used across the multi-ethnic MVP cohort. A unified genetic quality-control analysis was developed and conducted on an initial tranche of 485,856 individuals, leading to a high-quality dataset of 459,777 unique individuals. 668,418 genetic markers passed quality control and showed high-quality genotypes not only on common variants but also on rare variants. We confirmed that, with non-European individuals making up nearly 30%, MVP’s substantial ancestral diversity surpasses that of other large biobanks. We also demonstrated the quality of the MVP dataset by replicating established genetic associations with height in European Americans and African Americans ancestries. This current dataset has been made available to approved MVP researchers for genome-wide association studies and other downstream analyses. Further data releases will be available for analysis as recruitment at the VA continues and the cohort expands both in size and diversity.
Keywords: VA, Million Veteran Program, genotype data, quality control, genetic ancestry, genetic relatedness, SNP array design, GWAS, clinical variants, biobank
Introduction
The United States Department of Veterans Affairs (VA) initiated the Million Veteran Program (MVP) in 2011 to create a mega-biobank of at least one million samples with genetic data linked to nationally consolidated longitudinal clinical records.1 The initial and continuing goal of MVP is to create a national resource for research to improve the health of United States veterans and, more generally, to contribute to our understanding of human health. MVP has currently collected samples from over 800,000 Veteran participants and expects to exceed a total of 1 million participants in the next 2 to 3 years.
Although MVP is similar in some respects to other large biobank projects, such as the UK Biobank; the Kaiser Permanente Research Program on Genes, Environment, and Health (RPGEH); the China Kadoorie Biobank (CKB); and the DiscovEHR initiative,2, 3, 4 it is unique in several ways. As one of the largest single biobanking efforts to date, MVP satisfies the need for larger genetic datasets while also benefiting from a very rich, nationally integrated longitudinal clinical database housed in the largest consolidated healthcare network in the United States. This feature allows for enhanced clinical phenotyping capabilities. The availability of additional self-reported health and lifestyle survey information augments clinical data from the Veterans Information Systems and Technology Architecture (VistA)—the VA’s electronic health record (EHR).
Furthermore, with over 29% of participants self-reporting non-white ethnicity, MVP has substantial diversity in genetic ancestry and thus meets a pressing need for greater diversity in genome-wide association analyses so that researchers can discover novel associations, reduce false positives, and increase research equity.5, 6, 7, 8 As such, the MVP cohort provides an unprecedented opportunity for increasing the power of genome-wide association studies (GWASs) and will enable association discoveries regarding clinically important low-frequency and rare variants; such discoveries would only be possible in larger sample sizes. Reliable typing of these variants could provide explanations of missing heritability in complex or non-Mendelian diseases. However, the genetic diversity of MVP also poses challenges in genotype quality control.
In this report, we introduce the first installment of MVP genotype data consisting of 459,777 samples surveyed at 668,418 markers. In brief, we (1) describe the design of a research genotyping array with emphasis on clinically useful and/or rare variants applicable to multi-ethnic backgrounds; (2) describe the generation and quality control of genotyping data; (3) highlight some of the current MVP dataset’s unique features, including exploratory analyses of genetic ancestry; and (4) replicate effect sizes of previously reported variants associated with height in European Americans and African Americans. Overall, we find that the MVP genetic dataset, linked to deep phenotypic data, is a high-quality and diverse resource for performing genetic analyses.
Material and Methods
Human Subjects and Data and Sample Collection
The VA Central IRB, as well as the local IRBs at the VA Boston Healthcare System and the VA Connecticut Healthcare System, approved this project. An overview of the recruitment strategies and protocols is given in a previous publication.1 In brief, participants were recruited from approximately 60 VA healthcare facilities across the United States on a rolling basis. Informed consent was obtained from all participants. Participants consented to a blood draw and to have their DNA analyzed, as well as to linking their genetic information with their full clinical, survey, and other health data. Participants were also invited to answer two separate surveys about basic demographic information and lifestyle characteristics.
Blood drawn from consenting participants was shipped to the central biorepository in Boston, Massachusetts, where DNA was extracted and later shipped to two external vendors for genotyping on a custom Axiom array designed specifically for MVP (MVP 1.0). A description of the MVP 1.0 array design features is detailed in the Supplemental Data.
Thermo Fisher Scientific (formally Affymetrix) Axiom Genotyping Platform
The MVP 1.0 custom Axiom array is based on the Axiom Genotyping Platform. The Axiom genotyping platform utilizes a two-color, ligation-based assay using 30-mer oligonucleotide probes synthesized in situ onto a microarray substrate. Each single-nucleotide polymorphism (SNP) feature contains a unique oligomeric sequence complementary to the genomic sequence flanking the polymorphic site on either the forward or the reverse strand. Solution probes bearing attachment sites for one of two dyes, depending on the 3′ (SNP-site) base (A or T, versus C or G), are hybridized to the target complex, followed by ligation for specificity. Oligonucleotide sequences complementary to the forward or reverse strands are referred to as probesets. A marker (SNP or indel) can be interrogated by the probeset for the forward and/or reverse strand.
For additional details of the Axiom Genotyping Platform, see the Supplemental Materials and Methods.
Genotype Calling
We received unprocessed Axiom genotype data for 485,856 unique samples assayed by two vendors, referred to as vendor 1 and vendor 2, and performed genotype calling in batches grouped by vendor and sample processing date. By using data provided by the vendors and generated from our internal genotype calling process (see Supplemental Materials and Methods for details), we first analyzed the standard Axiom genotype quality metrics and compared these metrics between the two vendors.
After calling genotypes, we applied an advanced normalization procedure for mitigating plate-to-plate variation developed in collaboration with Thermo Fisher Scientific. The procedure was applied selectively on a per-batch basis to probesets exhibiting high plate-to-plate variance. After plate normalization, we applied standard marker quality-control procedures to clean and harmonize genotype calls across all the batches (Supplemental Materials and Methods), followed by advanced sample quality control (QC).
Advanced Sample QC
Sample Contamination
To detect and mitigate sample contamination, we assessed heterozygosity with PLINK, version 1.9, by calculating the F coefficient and quarantining samples with an F coefficient of less than −0.1. We assessed excess relatedness by using the relatedness inference software KING, version 2.0, and quarantined samples having a kinship coefficient of at least 0.1 with seven or more other samples within MVP. These samples had high dish QC (DQC) and low call rates and were outliers in comparison to the majority of samples in the MVP dataset (Figure S5D). Because a call rate below 98.5% correlated with excess sample heterozygosity or relatedness, we removed samples (15,436, or 3.00%) with call rates below this threshold.9 All samples that were removed or quarantined from the current release of MVP data will be re-genotyped and included in the future data releases.
Sample Mislabeling
We identified samples and plates demonstrating potential mislabeling issues by analyzing genotype concordance between intentional duplicate samples that were sent blinded to the vendors as new samples for genotyping. Of the 25,867 intentional duplicate pairs, only 211 (0.82%) pairs were highly discordant (greater than 1% discordance). Samples on plates with discordant intentional duplicate pairs were quarantined for further analysis and re-genotyping. We also removed both samples and plates if the duplicate pair had a relatedness coefficient of less than 0.45. These precautions were taken out of concern about potential plate swaps and led to 9,975 samples’ being quarantined.
Sample Misidentification
To discriminate between misidentified intentional duplicates (same samples intentionally genotyped twice), technical duplicates (controls repeatedly genotyped by vendors), and monozygotic twins, we calculated sample relatedness with the KING software, version 2.1.10 Monozygotic twins were confirmed by cross-referencing EHR data. Pairs with birth dates differing by no more than one day and having unique participant identifiers and first names were considered verified monozygotic twin pairs. Unverified samples were quarantined as potentially mislabeled and will be re-genotyped.
Sex Check
To confirm sample gender, we extracted markers genotyped on the X chromosome while excluding the pseudoautosomal region, used the sex-check command from PLINK, and compared the expected F coefficient on the X chromosome to the gender recorded in the sample’s EHR for all samples.11 Participants whose reported gender differed from that inferred by PLINK were quarantined from subsequent analysis. We also removed remaining samples on plates with four or more gender mismatches to account for potential plate swaps. The threshold is relatively low because of the low percentage of females in our dataset.
Advanced Marker QC
Advanced Marker QC Pipeline
We implemented three main approaches to create the advanced marker QC pipeline: (1) exclude probeset calls from all batches for probesets that failed advanced QC tests; (2) exclude probeset calls in a given batch for which the probeset is not recommended; and (3) choose the best probeset per marker for markers interrogated by multiple probesets and exclude probeset calls from all batches for the “not-best” probesets. Details of each steps of the advanced marker QC are available in Supplemental Materials and Methods and in Figures S4, S6A, and S7A.
The advanced marker QC pipeline produced an inclusion list of probesets that met quality standards across the entire MVP dataset. For each batch, we included a probeset in the dataset if it met all three of the following criteria: (1) it was included in the inclusion list; (2) it was recommended in that batch; and (3) it was the best probeset for a marker interrogated by multiple probesets. We then generated a list of probesets per batch, created PLINK marker list binary files for each batch, and merged all batches together by using the PLINK merge command.
Reproducibility of Genotype Calling
To assess the consistency of genotype calls across time and vendors, we analyzed the discordance between 25,867 intentional duplicate samples that were sent blinded to the vendors. After confirming that these sample pairs were genetically identical through KING relatedness inference, we determined the number of minor-allele pairs (MAPs) for each marker. A MAP is any pair of genotypes for a marker where both genotypes are called in the sample pair and where the pair contains at least one minor allele. We then calculated the number of discordant genotyping pairs per MAP for each marker. Normalizing by the number of MAPs renders different minor-allele frequency (MAF) bins comparable in the discordance calculation. Otherwise, rare markers will always have extremely low discordance rates because most samples carry the homozygous major genotype.
Additionally, within the 485,856 samples genotyped in the MVP cohort, we included 2,064 positive control samples. We called the genotypes of the positive controls along with other MVP samples across 112 batches organized by genotyping scan date for 668,418 markers passing advanced marker quality control. These genotypes were compared to the consensus positive-control genotype.
To construct the consensus genotype sequence, we calculated the frequency of each marker across the panel of 2,064 positive-control samples. Markers with MAF of less than 1% were set to homozygous in the consensus sequence, and markers with a MAF of greater than 49% were set to heterozygous in the consensus sequence. For markers with a MAF greater than or equal to 1% and less than or equal to 49% (536, or 0.082% of markers) or that had no observed calls (18,158, or 2.76%), we set the consensus genotype to missing.
We calculated concordance across all common (MAF ≥ 5%) and low-frequency (MAF < 5%) markers by assessing MAFs over the entire MVP sample. We then calculated concordance between the consensus sequence and each positive control. Concordance was defined as the number of matching called genotypes over the total number of called genotypes. Uncalled markers in either the positive control or the consensus sequence were not included in either the numerator or the denominator of the concordance calculation. We then plotted the concordance distribution for each batch’s positive controls across time.
Comparing MVP Allele Frequencies to Those from gnomAD and the UK Biobank
Genome Aggregation Database (gnomAD) version 2.1 data were downloaded online (see Web Resources). Markers in both gnomAD and MVP were matched on chromosome, start position, end position, reference allele, and alternative allele. For any mismatch, we checked strands and indel notations. Reference and alternative alleles were corrected, and their frequencies were recomputed when strands were flipped. Indels had their genomic coordinates and alleles recoded and harmonized.
UK Biobank summary data were downloaded online (see Web Resources). Markers shared between the UK Biobank and MVP were matched through the use of SNP rsIDs. Because information on marker chromosome, genomic positions, reference alleles, and alternate alleles were not provided in the summary statistics, we were unable to explicitly check for strand flips. However, as we expected, variant annotation in MVP and the UK Biobank tended to be well harmonized because both were genotyped on Axiom arrays and followed the same standard Axiom marker QC workflow; thus, we compared allele frequencies as is without excluding any variants.
For this analysis, European Americans (EAs) were defined as samples with a GBR (British in England and Scotland) proportion greater than 0.9 on the basis of ADMIXTURE results (described below), resulting in a sample size of 311,365. We used PLINK to compute allele frequencies by genetic ancestry subgroup via the “–freq” command with default filters and quality control parameters.
Genetic Relatedness
We performed additional preprocessing of the MVP dataset before analyzing genetic relatedness. We applied standard PLINK 1.9 filters for genotype missingness (> 5% removed), MAF (< 1% removed), and sample missingness (> 5% removed).11 We then conducted pairwise relatedness inference by using KING 2.1 to identify related pairs.10 KING explicitly accounts for population structure and is therefore an appropriate algorithm for our sample, which contains diverse genetic ancestry. However, KING is also known to overestimate relatedness in the presence of recent admixture. Therefore, we selected SNPs with a low load in principal components (PCs) 1–3 for a second round of KING, as was done in the UK Biobank.12
We ran the first round of KING with the command “–related–degree 3” to identify all potential pairs of individuals with closer than third-degree relatedness. From this result, we excluded all individuals with more than 200 third-degree relatives and also families with more than 100 members because we suspected they were artifacts of sample-processing errors such as low-level sample contamination. Then, a set of unrelated individuals was defined via the largest_independent_vertext_sets() function in the Python version of the igraph tool. Principal-component analysis (PCA) was then conducted with the unrelated samples. Only SNPs with a MAF greater than 0.01 and missingness less than 0.015 were considered for this PCA. 23 regions defined as having high linkage disequilibrium (LD) in the UK Biobank13 were also excluded, and then SNPs were pruned according to an r2 threshold of 0.1, a window of 1000 markers, and a step size of 80. In the end, 90,288 SNPs were selected for PCA, which was conducted with PLINK v2.00a2LM and the command “–pca var-wts approx” so that variant weights and fast PCA approximation could be obtained. We selected low-weight SNPs in PC1, PC2, and PC3 by adjusting the absolute weight threshold to keep at least two-thirds of the input SNPs, which led to 60,118 SNPs’ being put forward for the next round of KING.
The second round of KING was again conducted with the command “–related–degree 3.” The effect of using SNPs with low weights in PCs 1–3 on the distribution of the number of relatives per individual is shown in Figures S10A and S10B. We flagged 35 individuals with more than 200 third-degree relatives (UK Biobank reported nine individuals with more than 200 third-degree relatives), as well as all members of two clusters that were tightly interconnected with each other (Supplemental Materials and Methods and Figures S10C, S10D, and S11).
We defined genetically identical pairs as those having a kinship coefficient of 0.45 or greater (the maximum kinship coefficient output by KING is 0.5). However, given the large number of intentional duplicates samples in our dataset, we only considered genetically identical pairs as monozygotic twin pairs after cross-referencing EHR data as above. Parent-child pairs were defined as those having a kinship coefficient of greater than or equal to 0.19 and less than 0.45 and having less than 0.0025 percent of the genome held with zero alleles identical-by-state (IBS0). Sample pairs with a kinship coefficient greater than or equal to 0.19 and less than 0.45 and IBS0 greater than or equal to 0.0025 were designated as full siblings. Any pairs of participants with a kinship coefficient between 0.0884 and 0.19 were inferred to be second-degree or third-degree relatives. To identify potential trios in our sample, we extracted parent-child pairs in which a sample appeared twice. We then assessed the kinship coefficient between the other two participants. If the other two participants were not a related pair and consisted of one male and one female, we identified these three samples as a trio.
Genetic Ancestry
For genetic-ancestry analysis, we used the same set of markers used for relatedness analysis and applied LD pruning with PLINK (–indep-pairwise 1000 50 0.05), which left us with 50,000 markers.
Principal-Component Analysis
For the 1000 Genomes Project projection PCA, we merged the MVP dataset with the 1000 Genomes Project Phase 3 reference panel.14 We first filtered the 1000 Genomes Project dataset to ensure scalable merging with the MVP dataset. Markers with MAF less than 1% and any samples constituting related pairs were removed prior to LD pruning via PLINK according to the same parameters as above. We then calculated PCs by using the 1000 Genomes Project dataset and projected the MVP samples onto them with EIGENSOFT, version 6.0.1.15
We also calculated the PCs on the filtered MVP dataset alone by using the FastPCA method from the EIGENSOFT package for within-cohort PCA. For this PCA, we excluded all related individuals, whereas we kept all related individuals in the 1000 Genomes project PCA.
ADMIXTURE Analysis
In order to quantify ancestry proportions in MVP, we ran the program ADMIXTURE, version 1.3, on the MVP samples in supervised mode with five reference populations from the 1000 Genomes Project dataset as training data.16 We chose the five reference populations on the basis of their global geographic location to ensure global representativeness. The Yoruba in Ibadan, Nigeria (YRI) samples serve as a proxy for West African ancestry, the Luhya in Webuye, Kenya (LWK) for East African ancestry, the British in England and Scotland (GBR) for European ancestry, the Han Chinese in Beijing, China (CHB) for East Asian ancestry, and the Peruvians from Lima, Peru (PEL) for Native American ancestry (Figure S8C). Participants with more than 80% of their genetic ancestry attributed to one reference population were assigned to that reference. Remaining participants who had greater than 90% of their genetic ancestry derived from two reference populations were assigned to that pair of populations. Any participants not meeting the above two criteria were assigned to a separate subgroup (MVP_OTHER) and were assumed to contain admixture from three or more reference populations.
UMAP Analysis
We used Uniform Manifold Approximation Projection (UMAP), a dimensionality-reduction method that is useful for visualizing both global and local structure in data, to further visualize the genetic ancestry of the MVP cohort. A UMAP embedding was calculated on the basis of the first 10 principal components of unrelated samples with hyperparameters n_neighbors of 15 and min_distance of 0.1, which were suggested by a previous study on UK Biobank data.17 We then visualized the population structure by projecting subpopulations identified by our ADMIXTURE analysis onto the UMAP embedding.
GWAS of Height
Height measurements, dates of measurement, and dates of birth for each participant were extracted from the VA healthcare system’s EHR. Any height measurement outside the range of 48 to 84 inches was excluded,18 and inches were converted to meters. Age at measurement was calculated by subtracting the date of birth from the date of height measurement. Individuals younger than 18 or older than 120 years old were excluded. Sex was genetically determined by PLINK.
Markers whose genotype missingness was greater than 1%, as well as non-autosomal markers, were removed. Samples whose missingness was over 5% were also excluded. By using the results of the relatedness analysis described below, we also removed all closely related pairs.
After marker and sample filtering, we ran association tests by using BOLT-LMM13 with sex, age, age-squared, and the first 10 PCs as covariates. LD scores were calculated from the 1000 Genomes Project population subsets with ldsc 1.0.19 We generated model SNPs with PLINK 2.0 by pruning unrelated samples with an R-squared threshold of 0.2 (–pairwise-indep 1000 50 0.2). We also generated PCs by using PLINK 2.0 (–pca approx) on the cohorts that had model SNPs extracted.
We extracted the effect size, direction of effect, and allele for each previously associated marker from the GWAS catalog on March 21, 2019 and then extracted the effects for the markers present in the MVP association analysis. We then scaled the effect values within each study to between 0 and 1 to account for different height units and plotted the previously derived effects against those inferred in MVP.
Results
The MVP 1.0 Array
Array Design and Content
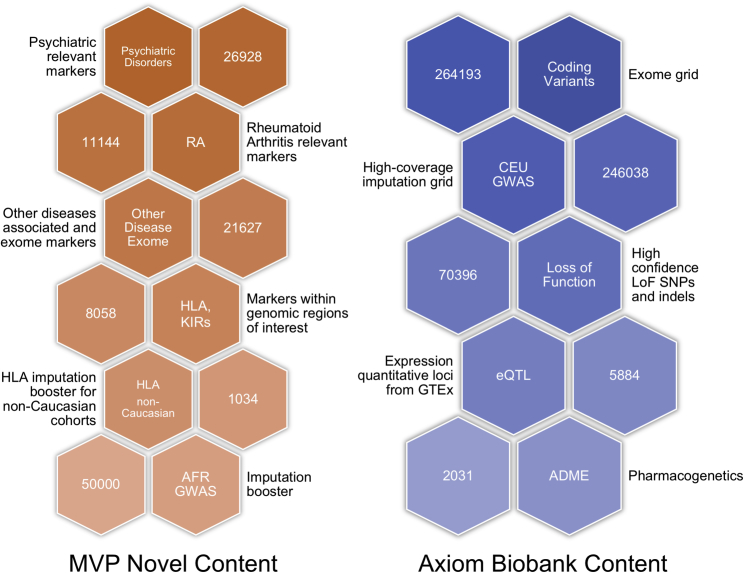
The MVP 1.0 array was based on the Applied Biosystems Axiom Biobank Genotyping Array with additional custom content developed for MVP (Figure 1). The Axiom Biobank Genotyping Array incorporates multiple content categories that are important for translational medicine research and discovery; such categories include modules for genome-wide coverage of common European variants, rare coding SNPs and indels, pharmacogenomics markers, expression quantitative trait loci (eQTLs), and loss-of-function markers (further described in Supplemental Materials and Methods). The MVP-1.0-specific modules were mainly SNPs and indels known to be associated with diseases and traits of interest to MVP (especially psychiatric disorders and rheumatoid arthritis), as well as a set of SNPs selected to improve African American imputation performance (Supplemental Materials). In total, 723,305 probesets interrogating 686,682 unique bi-allelic markers (SNPs and indels) based on the GRCh37 genome build were tiled onto the MVP 1.0 array. Among these, 270 are mitochondrial markers, 142 are in the non-pseudoautosomal regions of the Y chromosome, 1,139 are in the pseudoautosomal regions (PAR1 and PAR2) of the X and Y chromosomes, 18,026 are in the non-pseudoautosomal regions of the X chromosome, and the remaining 667,105 markers are autosomal markers (Table S1).
Figure 1.
Key MVP 1.0 Genotyping Array Modules
The modules are divided into those shared with the Axiom Biobank Genotyping Array and those unique to the MVP 1.0 array, along with descriptions and counts of unique markers in each module. Counts represent the number of markers in the module, and markers can be in more than one module.
MVP 1.0 Genotyping Quality Control and Assessment
Assessment of Overall Genotyping Performance
Figure S3 is an overview of the steps taken to ensure high-quality genotype data for the MVP cohort. Advanced genotype and sample QC were conducted in addition to the standard Affymetrix good-practice guidelines and are described in the Materials and Methods and Supplemental Materials and Methods. In addition, we further devised a batch variation correction step to apply to markers that showed significant allele frequency differences between releases (Supplemental Methods and Figures S4 and S6A).
We investigated multiple quality-control metrics for across and within the two assay vendors. Median Axiom DQC values for all genotyping batches were greater than 95 for both vendors (Figure S5A). Median QC call rate was also high, exceeding 99% for each genotyping batch (Figures S5B and S5C). Overall, sample call rates and other genotype quality-control metrics demonstrated high-quality genotype calls for MVP regardless of genotyping vendor (more detail is available in the Supplemental Materials and Methods).
Marker and Sample QC and Selection
The MVP 1.0 array contains a large amount of novel, custom marker content that has not been validated on other arrays. These markers were assayed with more than one probeset, so determining which probesets for a given marker performed best across all genotyped batches and removing systematically poor-quality probesets required advanced marker QC. Ultimately, we retained 668,418 markers representing 97.34% of the original markers and included 459,777 samples from a total of 485,856 unique genotyped samples in this data release. As expected, almost 98% of the markers that were previously tested on the Axiom biobank array were associated with a probeset that passed quality control, whereas 77% of the markers in the MVP 1.0 custom modules were associated with a probeset that remained after quality control. Additionally, although sample missingness (the fraction of missing genotype calls per individual; see Supplemental Materials and Methods) was slightly higher for vendor 1 than for vendor 2, almost all genotyped samples from both vendors exhibit missingness of less than 5% (Figure S6A).
We also either excluded or quarantined samples that did not meet sample QC criteria. Excluded samples include those expected to be removed by design or for known logistical or data errors. These samples include positive controls, samples with no or multiple unique participant identifiers, and samples in intentional duplicate pairs with the lower call rate. Quarantined samples are those that are temporarily removed from the dataset as a result of quality concerns. For instance, we investigated 1,149 pairs of samples with high relatedness to discriminate between misidentified intentional duplicates, technical duplicates (controls repeatedly genotyped by vendors), and monozygotic twins. Although we confirmed 49 monozygotic twins by cross-referencing with EHR data, the remaining 1,100 unintentional duplicate pairs could not be verified through independent means and were quarantined from data release as potentially mislabeled and will be re-genotyped. We also cross-checked genetically determined sample sex with EHR-reported gender information. Among the 485,856 unique genotyped samples, 2,000 (0.41%) did not have any reported gender information from either the EHR or self-reporting, and 2,073 (0.43%) of the remaining samples had a genetic sex that was opposite of the reported gender. We quarantined these samples for further analysis and potential re-genotyping (Table S2). The total number of samples that were excluded or quarantined from the current release of MVP genotype data and the reasons for exclusion are summarized in Table 1. All quarantined samples removed from the current data release will undergo further quality-control validation, be sent back to the vendors for re-genotyping, or will be otherwise verified before being included in subsequent data releases.
Table 1.
Quarantine and Exclusion Criteria for MVP Samples and Sample Count per Category
| Category | Number of Samples | Percentage of Samples |
|---|---|---|
| Starting MVP sample set for analysis | 514,383 | − |
| Intentionally duplicated samples | 25,291 | − |
| Uniquely genotyped individuals | 485,856 | 100.00% |
| Samples with call rates below 98.5% | 15,436 | 3.18% |
| Positive-control samples | 3,236 | 0.66% |
| Samples with sex misclassification | 1,450 | 0.29% |
| Samples on plates containing 4 or more sex misclassifications | 2,619 | 0.53% |
| Unintentionally duplicated samples | 1,149 | 0.23% |
| Samples on plates containing an intentional duplicate with high discordance | 9,975 | 2.05% |
| Samples with high heterozygosity | 248 | 0.05% |
| Samples with no or multiple unique participant identifiers | 71 | 0.01% |
| Intentionally duplicated samples with high discordance | 413 | 0.08% |
| Samples with 7 or more “relatives” | 466 | 0.09% |
| Samples excluded from the dataset | 28,527 | 5.87% |
| Samples quarantined from the dataset | 31,836 | 6.55% |
| Sample set in current data release | 459,777 | − |
Percentages are calculated from the total number of uniquely genotyped individuals (485,856). Categories are not mutually exclusive (i.e., a sample can be removed as a result of more than one category and is counted in each applicable category in the table).
Marker Missingness and Discordance by MAF
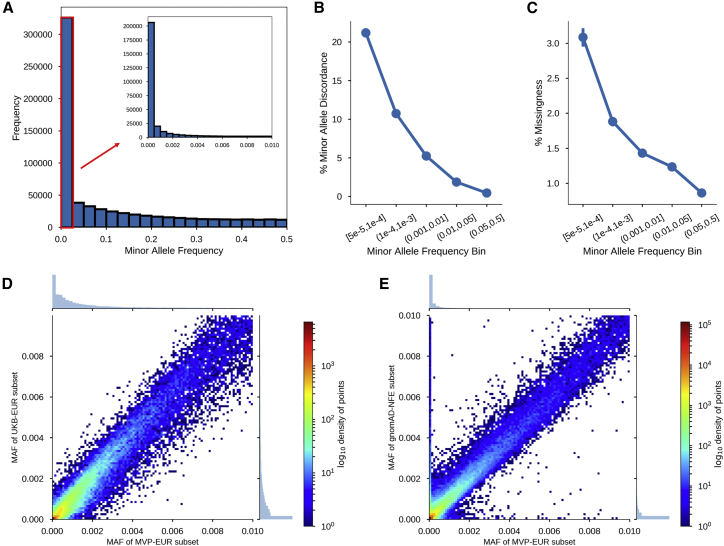
We assessed marker missingness in correlation with MAF. Overall, the MAF distribution of MVP 1.0 is highly skewed toward rare variants; 42.89% of markers have a MAF below 1%, and 33.89% have a MAF below 0.1% (Figure 2A). This result is by design: the content of the MVP array focuses on markers associated with potential disease phenotypes. We find that MAF is correlated with marker missingness, as shown in Figures 2C and S6B, and lower frequency variants are missing in a larger fraction of samples. Despite this trend, genotyping call rate among low-frequency markers is still relatively high. For example, 87.29% of rare markers (MAF < 0.1%) have a genotyping call rate of greater than 95%.
Figure 2.
Quality-Control Assessments on the MVP Dataset after Performance of the Advanced Marker Quality Control Procedures
(A) MAF distribution after sample QC filtering. The inset diagram shows the distribution for markers with a MAF below 1%.
(B) Minor-allele discordance rates per MAF bin, based on intentionally duplicated samples.
(C) Marker missingness rates per MAF bin, after sample QC filtering.
(D) Comparison of MAFs between the EA subset of MVP (MVP-EUR) and the UK Biobank European subset (UKB-EUR).
(E) Comparison of MAFs between MVP-EUR and the non-Finnish European subset of gnomAD (gnomAD-NFE).
Additionally, we examined marker genotype discordance rates across intentional duplicate sample pairs with respect to MAF. Discordance is calculated per MAP for each marker, and markers are binned by MAF. We found a correlation between MAF and discordance rate, such that lower-frequency variants had a higher rate of minor-allele discordance (Figures 2B and S6C).
Duplicate and Positive Control Samples for Continuous Quality Assessment
Importantly, because we employed two separate vendors for genotyping, we intentionally included 25,291 duplicate samples that were blinded to the vendors for independent assessment of genotype quality. This amounts to a target of 5% of all genotyped samples and is an effort to accurately assess genotyping quality on a continuous basis. Sample concordance among intentional duplicates or positive controls was very high; the median concordance rate was greater than 99.8% across all comparisons (Figure S7A).
Assessing concordance in positive-control samples also provides valuable information about the consistency and reproducibility of the MVP 1.0 array’s genotypes over time. Along with the MVP samples, 2,064 positive-control samples were genotyped on the MVP 1.0 array. As discussed in the Materials and Methods section, we constructed a consensus genotype sequence across 657,459 markers by using this panel of positive controls. For markers in the consensus sequence, 543,691 (82.70%) were homozygous, 95,079 (14.46%) were heterozygous, and 18,689 (2.84%) were uncalled. Concordance for each of the 2,064 positive-control samples is defined as the number of markers that agree with the consensus sequence divided by the number of called markers in the consensus sequence.
Overall positive-control concordance is shown in Figure S7A, and the distributions by batch of concordance values across all positive controls are shown in Figures S7B–S7D. The median concordance rate between each positive-control sample and the consensus sequence was 99.93% for all markers, 99.89% for common (MAF ≥ 5%) markers, and 100.00% for low-frequency (MAF < 5%) markers. The minimum observed concordance rate between a positive control and the consensus occurs during analysis of common markers, but this concordance rate is still high at 99.05%.
Concordance with HapMap Samples
To further test concordance and genotyping quality, we genotyped 96 HapMap samples (from Coriell cell lines) on the MVP 1.0 array. 210,630 markers are present in both the MVP 1.0 array and HapMap release 27, and among these markers, 205,647 (97.20%) are classified as recommended (see Supplemental Materials and Methods, Standard Marker Quality Control). When these 205,647 markers were analyzed over the 96 HapMap samples, and when HapMap and Axiom uncalled genotypes were removed from the numerator and denominator, the sample concordance across all population groups is 99.70% (Table 2). The Axiom sample call rate for recommended markers is 99.85%.
Table 2.
Concordance Rates across 96 HapMap Samples Genotyped on the MVP 1.0 Array
| − | − | Metrics over RecommendedaMarkers | Metrics over All Markers | ||
|---|---|---|---|---|---|
| Population | Number of Samples | Average Sample Concordance (%) | Average Sample Call Rate (%) | Average Sample Concordance (%) | Average Sample Call Rate (%) |
| ALL | 96 | 99.70 | 99.85 | 99.35 | 99.49 |
| CEU | 28 | 99.70 | 99.85 | 99.34 | 99.47 |
| CHB | 20 | 99.70 | 99.86 | 99.37 | 99.51 |
| JPT | 20 | 99.68 | 99.84 | 99.35 | 99.51 |
| YRI | 28 | 99.71 | 99.86 | 99.34 | 99.49 |
Recommended markers are those that were classified into one of the recommended SNP classes after execution of the Axiom Best Practices Genotyping workflow for the 96 co-clustered samples.
Assessing Rare-Allele Genotyping Quality
Given the importance of rare markers in clinically related studies, we evaluated the analytical validity of MVP 1.0 rare markers by observing the concordance of MAFs for rare markers with overlap between MVP 1.0 and either the gnomAD or the UK Biobank (Figures 2D and 2E). These databases are large enough for detection of very low MAFs, and agreement of MVP 1.0 marker MAFs with MAFs from these databases provides evidence for the accuracy of MVP 1.0 calls. MAFs were considered to agree when the lower bound of the regression slope’s 95% confidence interval was ≥0.9. This value leaves some margin of error for expected differences between the databases in population structure (non-Finnish Europeans versus European Americans [EA]), technology (genotype arrays versus exome sequencing), technical processes (batch, user, etc.), and sample size. We used the MVP EA subgroup to benchmark performance because it has a larger sample size, which provides better confidence in assessing frequency of rare markers, and it has large complementary subgroups in gnomAD and the UK Biobank. We classified markers into three subgroups by MAF: rare variants (< 1%), low-frequency variants (1%–5%), and common variants (> 5%). The EA subgroup yielded 321,290 (48.1%) rare markers, 46,626 (6.97%) low-frequency markers, and 300,375 (44.9%) common markers.
From the gnomAD, we compared the allele frequencies derived from the non-Finnish European subgroup (n = 55,860) of the exome call set. This subgroup provided the largest cohort that was comparable in population structure. 69% (221,374 of 321,290 markers) of the rare variants in MVP were also found in gnomAD. Additionally, both MVP and gnomAD showed similar MAFs for these concordant rare variants (slope 0.9290, 95% CI: 0.9002, 0.9578).
From the UK Biobank, we compared allele frequencies derived from the self-reported white British ancestry group (N > 330,000). We found MAF agreement, as supported by the strong coefficient of determination (R2) of 0.9864 and a slope of 0.9536 (95% CI: 0.9841, 0.9887) between 46,872 overlapping markers.
Although comparison against both sources met the ≥0.9 agreement threshold, we observed a small set of about 6,000 extremely discrepant markers (defined as having MAF > 0.001 in one database but MAF < 0.001 in the other) between MVP and gnomAD. About 53% of these markers were also present in the UK Biobank. For these discrepant markers, MAFs in the UK Biobank were much closer to MVP MAFs than those in gnomAD, and only one-quarter of the overlapping UK Biobank markers retained the “extremely discrepant” label. This is expected and consistent with previous observations that MAFs of MVP and the UK Biobank are in close agreement. The extremely discrepant markers between MVP and gnomAD might be attributed to smaller sample size of the gnomAD-exome database in comparison to the UK Biobank. The lowest MAF limit for MVP’s EA subgroup is 1.6 × 10−6 (1 of 622,730 total alleles), 8.9 × 10−6 (1 of 111,720) for gnomAD’s non-Finnish subgroup, and 1.4 × 10−6 (1 of 674,398) for UK Biobank. At very low frequencies, the absolute difference between rare variants, but not necessarily the relative difference, will be small. A given marker with a MAF of 0.001 in MVP and 0.01 in gnomAD will have an absolute difference of 0.009, but a relative difference of 10-fold. This is a common situation in our pairwise marker comparisons because overlapping marker MAFs are heavily clustered near zero (Figures 2D and 2E). This could also explain the relatively higher variance observed in the lower extremes when MVP is compared to gnomAD versus the UK Biobank. Overall, our results nonetheless show that our rare variant calls are highly consistent and within a reasonable range of agreement with overlapping markers in gnomAD and the UK Biobank. However, it is important to note that the precision of calling very rare variants assayed with SNP chips has been reported to show variable quality.20 Thus, visual inspection of calls underlying initial association results are always required.
Population Analysis of MVP Samples and a Test GWAS on Height
The MVP Cohort
In addition to quality assessment of MVP 1.0 genotyping results, we also performed exploratory analysis of the current population represented in the MVP samples. On the basis of data from the VistA EHR, the genotyped participants in the MVP cohort had a median age of 65 years at time of enrollment, and 8.33% are female. Although the percentage of female participants is low, reflecting the demographics of the Veteran population, this percentage corresponds to 46,924 female participants in the current release.
In light of the samples that have already been genotyped, the MVP cohort is relatively more diverse than other large biobanks on which data are available. For example, more than 94% of UK Biobank participants self-report as British, Irish, or “any other white background”4,12, and 81% of individuals whose data are included in the Kaiser RPGEH biobank report as “white, non-Hispanic.” On the other hand, 70.9% of MVP participants self-report as “white” and “non-Hispanic or Latino,” in agreement with United States 2010 census information indicating that 63.7% of respondents self-report as “white alone” and “not Hispanic or Latino”21.
Analysis of Relatedness
We examined the degree to which samples in the MVP population are related. Of the approximately 105.70 billion possible MVP sample pairings, 15,384 pairs appeared to be third-degree relatives or closer. The number of pairs for each type of relative pair, including trios, is shown in Table S8. Compared with the UK Biobank, this installment of MVP samples has a reduced fraction of related pairs.
Analysis of Genetic Ancestry
Assessing genetic ancestry for genotyped samples is an important tool for many applications, such as correcting for biases caused by population structure, constructing tests for natural selection, and determining disease risk by genetic ancestry, among other tasks.22 To assess genetic ancestry in our sample, we visualized and then quantitatively assessed the genetic ancestry of MVP samples relative to external reference populations.
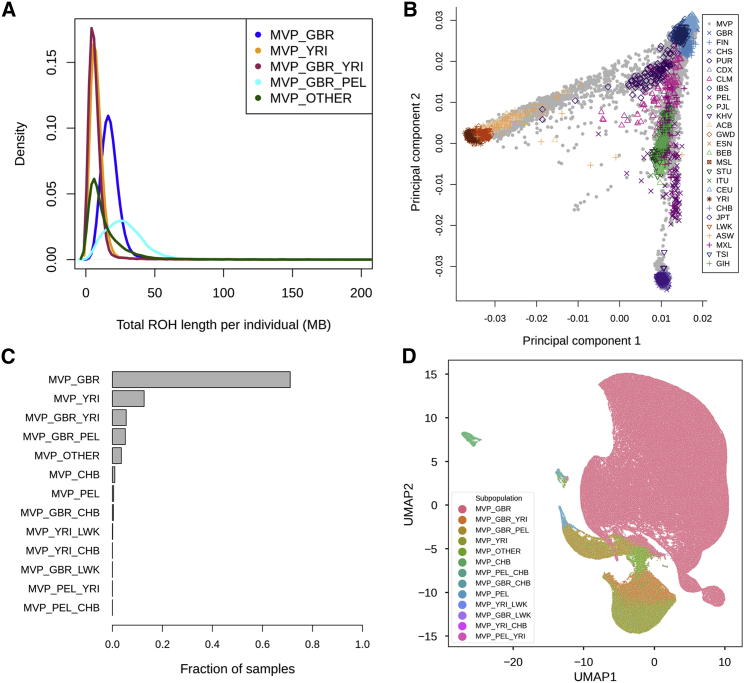
Runs of homozygosity (ROH) were measured via PLINK with a minimum ROH length of 1,000 Kb. The median total length of ROH is approximately 15.65 Mb, and the median number of blocks per sample is 10. In Figure 3A, we plotted the total length of ROH per individual by genetic ancestry subgroup for the five most common subgroups as defined in the Materials and Methods. MVP_GBR_PEL samples have a wide distribution of total ROH length but also some of the longest total lengths of all samples. Samples that had African ancestry or that were admixed between three or more reference populations (MVP_OTHER) have the shortest total length of ROH per sample. Samples of mainly European ancestry have intermediate total ROH length. The total length of ROH per sample varies depending on the genetic-ancestry subgroup.
Figure 3.
Analysis of Genetic Ancestry in the MVP Dataset
(A) Density plots of the total length of runs of homozygosity (ROH) per individual in each genetic-ancestry subgroup. Only the top five most common subgroups are shown.
(B) Principal-component analysis of the 1000 Genomes Project phase 3 dataset with MVP samples projected onto principal components 1 and 2.
(C) The number of MVP samples in each genetic-ancestry subgroup as inferred by ADMIXTURE percentages and our thresholds. For a single ancestry subgroup, such as MVP_GBR, the threshold is at least 80% inferred for that ancestry (e.g., MVP_GBR is GBR > 80%). For a pair of identified subgroups, the two ancestries must be at least 90% combined (e.g., MBP_GBR_YRI is GBR + YRI > 90%). MVP_OTHER includes all samples that had less than 80% ancestry aligned to any reference population and less than 90% combining any two populations.
(D) Visualization of ancestry subgroups via Uniform Manifold Approximation Projection (UMAP).
We also compared MVP samples to those in the 1000 Genomes Project. We first ran a PCA on the 1000 Genomes Project phase 3 samples and then projected the MVP samples onto these PCs. We found that most MVP samples lie close to reference populations of European origin. In addition, when we performed PCA on MVP samples alone, we found that genetic ancestry subgroups contain more complex intercontinental population structure, and a sizeable fraction of MVP samples exhibit admixture with respect to African and Asian references samples (Figures 3B and S9).
To assess ancestry proportion for each sample in MVP, we ran the program ADMIXTURE in supervised mode by using five 1000 Genomes Project phase 3 reference populations: Han Chinese in Beijing, China (CHB); British in England and Scotland (GBR); Luhya in Webuye, Kenya (LWK); Peruvians from Lima, Peru (PEL); and Yoruba in Ibadan, Nigeria (YRI).16 For most participants, the largest percentage of their genome aligns with the GBR population (Figure S8C). However, a substantial fraction of samples contains a moderate amount of genetic ancestry similar to the YRI reference population. Examples were also found of participants who have almost 100% of their genetic ancestry aligning to each of the five reference populations except for LWK. By using ADMIXTURE analysis results, we grouped the MVP samples into 16 subgroups and determined the proportion of MVP samples belonging to each (Figure 3C). For example, 326,777 samples have over 80% of their genome aligning with the GBR reference population (MVP_GBR), whereas 58,267 samples have 80% or more of their genome aligning with YRI (MVP_YRI). Excluding samples with more than 80% of their genome aligning to one reference population, 25,295 of the samples have 90% or more of their genome aligning with a combination of GBR and YRI reference populations (MVP_GBR_YRI). Approximately 16,351 samples (MVP_OTHER) have neither 80% of their genome aligning with one reference population nor 90% aligning with a combined pair, indicating substantial admixture between three or more reference populations.
Finally, we visualized the diverse ancestry composition of MVP by using a non-parametric dimensionality reduction method called uniform manifold approximation projection (UMAP) (Figure 3D). As shown through PCA and ADMIXTURE, the largest cluster corresponds to samples with largely European ancestry. In this visualization, the distance between samples and clusters is not to be directly interpreted as genetic distance. Although there are distinct clusters (such as the tight cluster of individuals with Asian ancestry on the top left corner, and another small cluster of probable Polynesians in the middle of the plot), most MVP samples of different ancestries form a large single cluster rather than individual ancestry clusters with distinct breaks. This large cluster shows a continuum of ancestry proportion that transitions from GBR on the top right to different levels of admixture with YRI and PEL proportions. This is in line with a previous report based on 32,000 US individuals in the National Geographic Genographic Project cohort.23
GWAS of Height
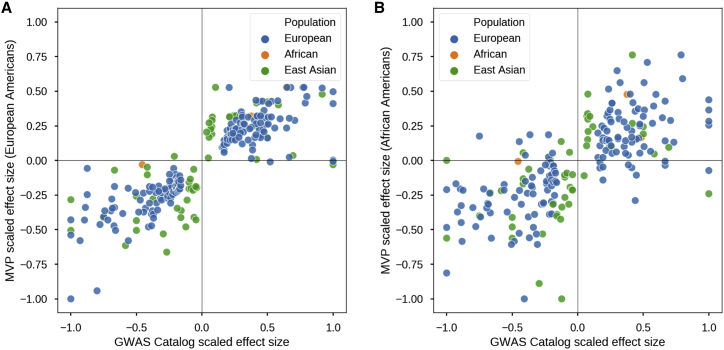
To further validate the quality of our genotype data and the utility of MVP 1.0 array, we conducted a GWAS of height in both the EA and African American (AA) MVP subpopulations. EAs were defined as individuals with a GBR proportion greater than 90%, and AAs were defined as individuals with a YRI proportion greater than 60% and less than 40% GBR on the basis of ADMIXTURE results (Figures S8A and S8B). Our GWAS of height within EA and AA cohorts showed moderate inflation of λGC = 1.12 and λGC = 1.13 and pseudo-heritability of 0.396 and 0.378, respectively,19,24,25 levels comparable to those found in previous height association studies without genotype imputation.26
Of the 822 reported height associations listed in the GWAS catalog,27 230 were present in the MVP EA GWAS, and 209 were present in the MVP AA GWAS. We assessed whether we could replicate effect sizes and direction of effects for markers present in MVP EA and AA GWASs by plotting these against the GWAS-catalog effect sizes and direction of effects (Figure 4). For the two subpopulations, the MVP associations perfectly replicated the directions of effect in most markers (two SNPs had an effect size near zero in EA). However, because most associations in the GWAS catalog are derived from Europeans, the overall correlation across all markers was lower for the AA cohort (r = 0.69) than for the EA cohort (r = 0.85).
Figure 4.
GWAS of Height with MVP Cohort
(A) Replication of the direction of effect for markers previously associated with height as annotated in the NHGRI-EBI GWAS Catalog in the MVP cohort of non-related European Americans (n = 291,609). Color coding denotes the genetic ancestry of the original cohort in which the markers were associated with height.
(B) Same as (A) except with the MVP cohort of non-related African Americans (N = 73,190).
Overall, we show that the performance of MVP 1.0 and the quality of its genotyping across 459,777 individuals of diverse ethnic backgrounds is very consistent and accurate by a variety of metrics.
Discussion
In this report, we provide an overview of the design of the MVP 1.0 genotyping array, the development of accompanying quality-control analyses, and our initial data exploration of an interim MVP genotyping dataset that consists of nearly 460,000 veterans. Our results demonstrate that the MVP 1.0 chip and the subsequent QC procedures have addressed notable challenges characteristic of large projects with individuals of diverse genetic backgrounds and that the resulting genotype calls are of high quality akin to that of other projects similar in scope. By using a single chip and unified quality control across the diverse cohort, we aimed to minimize batch effects between different ancestries and provide an initial genome-wide scan before whole-genome-sequenced samples become available.
Addressing the Challenges of MVP
MVP’s large, diverse, and still-growing cohort poses numerous challenges for designing genotyping procedures and their subsequent quality-assessment and quality-control protocols. Genotyping large and ethnically diverse cohorts along with clinically relevant markers is even more challenging because of the finite number of probesets that can fit on a single array. However, using different arrays for different ethnic groups can also exacerbate the differences between these groups and lead to batch effects.
To address the limitations of array-based genotyping in diverse cohorts, we carefully selected array content to maximize clinical utility while at the same time ensuring both broad coverage of variants and robust imputation capabilities across different ethnic groups. We also developed comprehensive quality controls for markers and samples both before and after genotyping. These controls included intentional duplication of ~5% randomly selected samples over time, blinded-to-assay technicians so that batch variation could be detected and mitigated; assessment of genotyping concordance with positive control samples and HapMap samples (Figure S7A, Table 2); comparison of MVP 1.0 MAFs to those in gnomAD and the UK Biobank (Figure 2); and a height GWAS intended to replicate previously reported results (Figure 4). Overall, we retained and released 459,777 samples and 668,418 markers after QC for the initial release of data. Although QC metrics vary slightly over time and genotyping vendors, the final genotyped sample set shows consistently high call rates (98.5%) and genotype concordance over intentional duplicates (99.8%) both within and between vendors and over time. Furthermore, marker concordance is also high even for rare markers. Additionally, genotype concordance, MAF, and GWAS association results are generally in strong agreement with external or previously reported results. These results indicate that the design of the MVP 1.0 array and the associated quality-control and assessment procedures provide a robust, reliable method for genotyping common, low-frequency, and rare variants in a large, ethnically diverse cohorts.
Challenges remain however, and the MVP 1.0 array has several limitations. Notably, although concordance rates were high, our results demonstrate that low-frequency and rare variants are still more difficult to genotype accurately with the MVP 1.0 array than are common variants. Additionally, while although added markers to MVP 1.0 to increase coverage for AAs, we lack boosters for other ethnic groups, such as Asian and Native American populations, which currently comprise smaller but growing proportions of the MVP population. In addition, although a standard imputation to the 1000 Genomes reference has been completed, we have yet to quantify the effect of imputation across different ancestries within MVP to devise an optimized imputation strategy. The strategy implemented by the UK Biobank was to use the Haplotype Reference Consortium (HRC) as the main imputation reference panel and to supplement with variants from 1000 Genomes if those variants were missing in HRC. However, this is not viable for MVP, which has a large proportion of non-European individuals. Further understanding and analyses of imputation strategies in a multi-ethnic and admixed cohort will be required if we are to obtain an optimized strategy for MVP.
The MVP Dataset Is Ethnically and Genetically Diverse
Our exploratory analysis indicates that the MVP dataset and samples offer unique value for disease research. One particularly valuable aspect of the MVP dataset is the ethnic diversity it encompasses. Genetic ancestry analysis suggests that the MVP dataset contains sub-populations with both homogeneous and admixed genetic ancestry from multiple global populations. The largest sub-population corresponds to 71% of samples of mostly European descent, and the remaining samples show substantial African, East Asian, and Native American ancestry.
Because MVP recruits participants from United States veterans who receive care at VA hospitals, the demographics of the MVP dataset diverge from those of the United States population. Approximately 8.5% of MVP samples are female, which is similar to the fraction of women in the Veteran population.28 With a median age of 68 years as opposed to 37.9 years, MVP participants are also substantially older than the United States population.29 However, the demographics of MVP might change with increasing use of the VA by more recent veterans. The proportion of female veterans is projected to continuously grow and nearly double, to 16.5%, by 2043.28 Meanwhile, the proportion of veterans from minority populations is expected to increase by approximately 50% over the same time period.28 Thus, the VA and MVP is in a unique position for further inclusion of participants from diverse backgrounds.
The MVP Dataset Is an Invaluable Disease Research Resource
MVP has several unique features that make it an invaluable resource for researching human disease. As evidence of the general utility of this resource, initial reports using an earlier tranche of ~300,000 genotyped participants have reported substantial new findings regarding the genetics of blood lipids, a major cardiovascular risk factor.30 Not only is MVP ideal for studying the burden of chronic disease, which increases with age, many of the clinical records in its EHR span several decades, allowing for robust longitudinal analysis. This is possible because patients using the VA health services do not lose coverage even after changing employers or residence. Additionally, MVP provides an opportunity to study diseases, such as PTSD and31 alcohol- and substance-abuse disorders,32 that disproportionately affect US veterans, as well as to study other deployment-related conditions and their impact on human health. The MVP phenotypes are collected in the VA EHR as part of the routine clinical care at VA national hospitals and clinics across the country, and clinical data spanning the past three decades are available to be integrated with genomic data on demand. A list of prevalent disease phenotypes surveyed from raw International Classification of Diseases (ICD) diagnostic codes in the VA EHR are provided in Table S9. Although ethnically diverse, the MVP cohort is not a true representation of the US general population, given the underrepresentation of women and requirements for military service. For instance, early-onset disease phenotypes (e.g., Mendelian diseases with large effect sizes) that might limit military service are underrepresented.
In conclusion, the high-quality genotype data generated with the MVP 1.0 array provides a valuable resource for researchers investigating the effect of both rare and common genetic variants within MVP. These quality-controlled genotype data as well as the results from genetic ancestry and relatedness analyses are made available to all approved researchers. MVP intends to make coded data available in a secure data and computing environment after periodic requests for proposals, where the data use intent and provenance will be clearly verified in accordance with the participant consent and MVP policies. The genotype data can be linked to participants’ full EHR, often covering decades of care provided by the VA. MVP is a continuously expanding research cohort made available by participants with diverse backgrounds and altruistic intentions to support research that will benefit their fellow veterans and others.
Consortia
Million Veteran Program (MVP) Consortium
Michael Gaziano, MD, MPH, Rachel Ramoni, DMD, ScD, Jim Breeling, MD, Kyong-Mi Chang, MD, Grant Huang, PhD, Sumitra Muralidhar, PhD, Christopher J. O’Donnell, MD, MPH, Philip S. Tsao, PhD, Sumitra Muralidhar, PhD, Jennifer Moser, PhD, Stacey B. Whitbourne, PhD, John Concato, MD, MPH, Stuart Warren, JD, Pharm D, Dean P. Argyres, MS, Brady Stephens, MS, Mary T. Brophy MD, MPH, Donald E. Humphries, PhD, Xuan-Mai T. Nguyen, PhD, Saiju Pyarajan, PhD, Kelly Cho, MPH, PhD, Peter Wilson, MD, Rachel McArdle, PhD, Louis Dellitalia, MD, John Harley, MD, Jeffrey Whittle, MD, Jean Beckham, PhD, John Wells, PhD, Salvador Gutierrez, MD, Gretchen Gibson, DDS, Laurence Kaminsky, PhD, Gerardo Villareal, MD, Scott Kinlay, PhD, Junzhe Xu, MD, Mark Hamner, MD, Kathlyn Sue Haddock, PhD, Sujata Bhushan, MD, Pran Iruvanti, PhD, Michael Godschalk, MD, Zuhair Ballas, MD, Malcolm Buford, MD, Stephen Mastorides, MD, Jon Klein, MD, Nora Ratcliffe, MD, Hermes Florez, MD, Alan Swann, MD, Maureen Murdoch, MD, Peruvemba Sriram, MD, Shing Shing Yeh, MD, Ronald Washburn, MD, Darshana Jhala, MD, Samuel Aguayo, MD, David Cohen, MD, Satish Sharma, MD, John Callaghan, MD, Kris Ann Oursler, MD, Mary Whooley, MD, Sunil Ahuja, MD, Amparo Gutierrez, MD, Ronald Schifman, MD, Jennifer Greco, MD, Michael Rauchman, MD, Richard Servatius, PhD, Mary Oehlert, PhD, Agnes Wallbom, MD, Ronald Fernando, MD, Timothy Morgan, MD, Todd Stapley, DO, Scott Sherman, MD, Gwenevere Anderson, RN, Philip Tsao, PhD, Elif Sonel, MD, Edward Boyko, MD, Laurence Meyer, MD, Samir Gupta, MD, Joseph Fayad, MD, Adriana Hung, MD, Jack Lichy, MD, PhD, Robin Hurley, MD, Brooks Robey, MD, Robert Striker, MD.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was funded by the United States Veterans Administration (VA) Office of Research and Development (ORD), Million Veteran Program Grant number MVP000 to S.P. and the VA Special Fellowship in Medical Informatics to H.H.-Z. We would like to thank all Veteran participants in the MVP for donating their samples, information, and time to this project. We additionally thank Anoop Sendamarai and Frederick Dong for critical discussions and providing editorial assistance and review. The content of this manuscript reflects the views of the authors and does not represent the views of the Department of Veterans Affairs, FDA, or the United States Government.
Published: April 2, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.03.004.
Web Resources
Supplemental Data
References
- 1.Gaziano J.M., Concato J., Brophy M., Fiore L., Pyarajan S., Breeling J., Whitbourne S., Deen J., Shannon C., Humphries D. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Banda Y., Kvale M.N., Hoffmann T.J., Hesselson S.E., Ranatunga D., Tang H., Sabatti C., Croen L.A., Dispensa B.P., Henderson M. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics. 2015;200:1285–1295. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kvale M.N., Hesselson S., Hoffmann T.J., Cao Y., Chan D., Connell S., Croen L.A., Dispensa B.P., Eshragh J., Finn A. Genotyping informatics and quality control for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics. 2015;200:1051–1060. doi: 10.1534/genetics.115.178905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manrai A.K., Funke B.H., Rehm H.L., Olesen M.S., Maron B.A., Szolovits P., Margulies D.M., Loscalzo J., Kohane I.S. Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrovski S., Goldstein D.B. Unequal representation of genetic variation across ancestry groups creates healthcare inequality in the application of precision medicine. Genome Biol. 2016;17:157. doi: 10.1186/s13059-016-1016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popejoy A.B., Fullerton S.M. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Affimetrix . 2016. Axiom genotyping solution data analysis guide. [Google Scholar]
- 10.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C.C., Chow C.C., Tellier L.C.A.M., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh P.R., Kichaev G., Gazal S., Schoech A.P., Price A.L. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018;50:906–908. doi: 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galinsky K.J., Loh P.R., Mallick S., Patterson N.J., Price A.L. Population Structure of UK Biobank and Ancient Eurasians Reveals Adaptation at Genes Influencing Blood Pressure. Am. J. Hum. Genet. 2016;99:1130–1139. doi: 10.1016/j.ajhg.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz-Papkovich A., Anderson-Trocme L., Gravel S. Revealing multi-scale population structure in large cohorts. bioRxiv. 2019 [Google Scholar]
- 18.Noël P.H., Copeland L.A., Pugh M.J., Kahwati L., Tsevat J., Nelson K., Wang C.P., Bollinger M.J., Hazuda H.P. Obesity diagnosis and care practices in the Veterans Health Administration. J. Gen. Intern. Med. 2010;25:510–516. doi: 10.1007/s11606-010-1279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Corvin A., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weedon M.N., Jackson L., Harrison J.W., Ruth K.S., Tyrrell J., Hattersley A.T., Wright C.F. Very rare pathogenic genetic variants detected by SNP-chips are usually false positives: implications for direct-to-consumer genetic testing. bioRxiv. 2019 [Google Scholar]
- 21.United States Census Bureau . 2010 Census Briefs. 2011. The White Population: 2010–c2010; pp. 1–20. [Google Scholar]
- 22.Padhukasahasram B. Inferring ancestry from population genomic data and its applications. Front. Genet. 2014;5:204. doi: 10.3389/fgene.2014.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai C.L., Vazifeh M.M., Yeang C.-H., Tachet R., Wells R.S., Vilar M.G., Daly M.J., Ratti C., Martin A.R. Population histories of the United States revealed through fine-scale migration and haplotype analysis. bioRxiv. 2019 doi: 10.1016/j.ajhg.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J., Manolio T.A., Pasquale L.R., Boerwinkle E., Caporaso N., Cunningham J.M., de Andrade M., Feenstra B., Feingold E., Hayes M.G. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Center for Veterans Analysis and Statistics . 2014. Table 3L: Living veterans by race/ethnicity, gender; pp. 2013–2043. [Google Scholar]
- 29.World Factbook C.I.A. The World Factbook; 2017. Central Intelligence Agency. [Google Scholar]
- 30.Klarin D., Damrauer S.M., Cho K., Sun Y.V., Teslovich T.M., Honerlaw J., Gagnon D.R., DuVall S.L., Li J., Peloso G.M. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelernter J., Sun N., Polimanti R., Pietrzak R., Levey D.F., Bryois J., Lu Q., Hu Y., Li B., Radhakrishnan K. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat. Neurosci. 2019;22:1394–1401. doi: 10.1038/s41593-019-0447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kranzler H.R., Zhou H., Kember R.L., Vickers Smith R., Justice A.C., Damrauer S., Tsao P.S., Klarin D., Baras A., Reid J. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat. Commun. 2019;10:1499. doi: 10.1038/s41467-019-09480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.