Abstract
Selection of the translational initiation site in most eukaryotic mRNAs appears to occur via a scanning mechanism which predicts that proximity to the 5′ end plays a dominant role in identifying the start codon. This ‘position effect’ is seen in cases where a mutation creates an AUG codon upstream from the normal start site and translation shifts to the upstream site. The position effect is evident also in cases where a silent internal AUG codon is activated upon being relocated closer to the 5′ end. Two mechanisms for escaping the first-AUG rule – reinitiation and context-dependent leaky scanning – enable downstream AUG codons to be accessed in some mRNAs. Although these mechanisms are not new, many new examples of their use have emerged. Via these escape pathways, the scanning mechanism operates even in extreme cases, such as a plant virus mRNA in which translation initiates from three start sites over a distance of 900 nt. This depends on careful structural arrangements, however, which are rarely present in cellular mRNAs. Understanding the rules for initiation of translation enables understanding of human diseases in which the expression of a critical gene is reduced by mutations that add upstream AUG codons or change the context around the AUGSTART codon. The opposite problem occurs in the case of hereditary thrombocythemia: translational efficiency is increased by mutations that remove or restructure a small upstream open reading frame in thrombopoietin mRNA, and the resulting overproduction of the cytokine causes the disease. This and other examples support the idea that 5′ leader sequences are sometimes structured deliberately in a way that constrains scanning in order to prevent harmful overproduction of potent regulatory proteins. The accumulated evidence reveals how the scanning mechanism dictates the pattern of transcription – forcing production of monocistronic mRNAs – and the pattern of translation of eukaryotic cellular and viral genes.
Keywords: Translational control, AUG context, 5′ untranslated region, Reinitiation, Leaky scanning, Dicistronic mRNA, Internal ribosome entry site
Abbreviations: AdoMetDC, S-adenosylmethionine decarboxylase; A2AR, A2A adenosine receptor; C/EBP, CCAAT/enhancer binding protein; CTL, cytotoxic T-lymphocyte; EGFP, enhanced green fluorescent protein; eIF, eukaryotic initiation factor; HIV-1, human immunodeficiency virus 1; IRES, internal ribosome entry site; LEF1, lymphoid enhancer factor-1; OGP, osteogenic growth peptide; ORF, open reading frame; R, purine; TPO, thrombopoietin; upORF, upstream open reading frame; UTR, untranslated region
1. Introduction
The scanning mechanism for initiation of translation postulates that the small (40S) ribosomal subunit enters at the 5′ end of the mRNA and migrates linearly, stopping when the first AUG codon is reached. Consistent with the postulated 5′ end-dependent entry of ribosomes, translation in vivo is strongly augmented by the m7G cap (Furuichi and Shatkin, 2000, Horikami et al., 1984, Lo et al., 1998, Neeleman et al., 2001) and ribosome binding in vitro is prevented by circularization of the mRNA (Kozak, 1979a, Konarska et al., 1981).
Perhaps because the scanning mechanism has been around for a while, the evidence for some basic points has been forgotten. One recent commentary even questions whether the 40S ribosomal subunit has anything to do with it (Mathews, 2002). The easiest answer is that the stop-scanning step is clearly mediated by pairing of the initiation codon with the anticodon in Met-tRNAi (Cigan et al., 1988a), and the 40S ribosomal subunit is the carrier of Met-tRNAi eIF2. But the 40S subunit was already implicated by experiments done earlier.
The experiments that gave rise to the scanning model concerned unusual polysome-like complexes formed in the presence of edeine, an antibiotic which blocks recognition of the AUG codon (Kozak and Shatkin, 1978). Analysis of the rapidly sedimenting complexes revealed 40S ribosomal subunits distributed throughout the body of the mRNA. Because control experiments showed that, even in the presence of edeine, ribosomes can enter only from the 5′ end, the simplest explanation was that 40S subunits enter at the 5′ end and then migrate into the interior of the mRNA; in the absence of edeine, the migration would stop when an AUG codon is reached. Independent experiments confirmed that edeine is targeted to the ribosome (Herrera et al., 1986), and use of a fractionated translation system confirmed that the edeine-induced complexes are formed by 40S but not 60S ribosomal subunits (Kozak and Shatkin, 1978, Kozak, 1979b). Subsequent experiments, with edeine omitted, showed that scanning can be interrupted by inserting a base-paired structure between the cap and the AUG codon; the resulting abortive complexes sediment around 40S (Kozak, 1989, Kozak, 1998, Paraskeva et al., 1999).
We are not yet sure which initiation factors are associated with the 40S ribosomal subunit during the scanning phase. The only factors whose role in scanning has been defined clearly are the GTP-binding protein eIF2, which escorts Met-tRNAi onto the 40S subunit, and eIF5, which activates GTP hydrolysis by eIF2 (Asano et al., 2001, Das et al., 2001). By controlling the rate of GTP hydrolysis, eIF5 controls the fidelity of initiation, i.e. the fidelity of the stop-scanning step (Huang et al., 1997). Other protein factors have not yet been fitted in. (The voluminous literature on factors focuses on modifications – phosphorylation, cleavages – rather than on defining the initiation pathway. Basic questions, such as when each factor enters and leaves, have not yet been answered.) One untested possibility is that the large initiation factor eIF3, bound to the 40S ribosomal subunit, might form a clamp around the mRNA that is opened and closed by cycles of ATP hydrolysis. Scanning appears to be dependent on ATP hydrolysis (Kozak, 1980), thereby implicating eIF4A, an RNA-dependent ATPase which might control the hypothetical clamp. Some ideas about the function of other initiation factors are reviewed elsewhere (Dever, 2002, McCarthy, 1998, Pestova et al., 2001).
The strongest evidence that the scanning 40S ribosome/factor complex advances linearly is the position effect on selection of the start codon: initiation at the first potential start codon has been demonstrated in rigorous experimental tests (Cigan et al., 1988a, Kozak, 1983, Kozak, 1995) and confirmed in many ‘natural tests’ wherein addition or removal of an AUG codon produces the expected shift in the site of initiation (see below). The aforementioned blockade caused by inserting a base-paired structure between the 5′ cap and the AUGSTART codon is further evidence that 40S ribosomal subunits traverse the leader sequence linearly, rather than hopping (discontinuous scanning) or entering directly at the AUG codon.
Although the scanning mechanism predicts that translation should initiate at the AUG codon nearest the 5′ end of the mRNA, two ancillary mechanisms – reinitiation and context-dependent leaky scanning – enable additional initiation events at downstream AUG codons in some mRNAs. These well-defined mechanisms for escaping the first-AUG rule are discussed below. An additional escape mechanism might involve direct entry of ribosomes at an internal site in the mRNA. While there is evidence suggestive of direct internal initiation with picornavirus mRNAs, the evidence for internal ribosome entry sites (IRES) in cellular mRNAs is problematic (Kozak, 2001a). The absence of shared structural features among candidate cellular IRES elements makes it impossible to predict which mRNAs, if any, might use such a mechanism. Rather than attempting to summarize the extensive literature on internal initiation, I refer the reader to other detailed reviews on that subject (Dever, 2002, Hellen and Sarnow, 2001, Pestova et al., 2001).
The next section provides a terse summary of points that are easily explained by the scanning model. The bulk of the review then focuses on complicated examples and issues.
2. Constraints imposed by the scanning mechanism explain many common aspects of gene expression in higher eukaryotes
2.1. Silent downstream cistrons
Many plant and animal viruses produce dicistronic or polycistronic mRNAs from which only the 5′ cistron can be translated (Table 1). All these viruses solve the problem of ‘silent 3′ cistrons’ by producing – via splicing or discontinuous transcription or an internal promoter – additional forms of mRNA in which the downstream cistron is repositioned closer to the 5′ end. The reason for the complicated pattern of splicing seen with human immunodeficiency virus type 1 (HIV-1), for example, is simply to produce mRNAs that allow downstream open reading frames (ORFs) to be translated. The broad range of viruses represented in Table 1 merits attention.
Table 1.
Partial list of structurally polycistronic viral mRNAs which are functionally monocistronic, i.e. only the first cistron is translateda
| Virus | Expressed 5′ cistronb | Silent 3′ cistron(s) | Source of short transcriptc | References |
|---|---|---|---|---|
| Polyoma virus | Capsid protein VP2 | Capsid protein VP1 | Splicing | Siddell and Smith, 1978 |
| Bovine papillomavirus | Numerous examples | Numerous examples | Promoter switch and splicing | Lambert et al., 1988 |
| Cytomegalovirus | UL98d | UL99 (pp28) | Promoter switch | Wing and Huang, 1995 |
| Adenovirus | Numerous examples | Numerous examples | Splicing | Wold et al., 1995, Ziff, 1985 |
| Parvovirus: adeno-associated | Capsid protein A | Capsid proteins B/C | Splicing | Muralidhar et al., 1994 |
| Hepatitis B virus | Core protein | S proteins (envelope) | Promoter switch | Schaller and Fischer, 1991 |
| Retrovirus: avian, murine | Gag (capsid) protein | Env protein | Splicinge | Pawson et al., 1977, Van Zaane et al., 1977 |
| Retrovirus: human foamy | Gag (capsid) protein | Pol precursor | Splicing | Jordan et al., 1996 |
| Lentivirus: HIV-1 | Tat | Rev and Neff | Splicing | Schwartz et al., 1992 |
| Alphavirus: Semliki Forest | Nonstructural proteins | Capsid protein | Internal promoter | Glanville et al., 1976, Strauss and Strauss, 1994 |
| Calicivirus: felineg | Nonstructural proteins | Capsid protein | Independent replication | Carter, 1990, Neill et al., 1991 |
| Coronavirus: mouse hepatitis | Membrane protein | Nucleocapsid protein | Discontinuous transcription | Lai and Cavanagh, 1997 |
| Equine arteritis virus | Replicase polyprotein | Gs glycoprotein | Discontinuous transcription | Pasternak et al., 2000 |
| Brome mosaic virus | RNA polymerase | Coat protein | Internal promoter | Miller et al., 1985, Shih and Kaesberg, 1976 |
| Tobacco mosaic virus | Replicase | Coat and movement proteins | Internal promoters | Grdzelishvili et al., 2000, Hunter et al., 1976 |
| Potato virus X | 25 kDa movement protein | 12 and 8 kDa movement proteinsf | ?? | Verchot et al., 1998 |
| Carmovirus: turnip crinkleg | Replicase (p28/p88) | p8 and p9 movement proteins | Internal promoters | Li et al., 1998, Wang and Simon, 1997 |
| Tombusvirus: tobacco necrosisg | RNA polymerase | Coat protein (ORF5)h | Internal promoters? | Meulewaeter et al., 1992 |
| Southern bean mosaic virusg | Movement protein and polymerasef | Coat protein | Internal promoter | Hacker and Sivakumaran, 1997 |
| Luteovirus: barley yellow dwarfg | Protease/polymerase | Coat protein and p17f | Internal promoters | Koev and Miller, 2000, Mayo and Ziegler-Graff, 1996 |
| Turnip yellow mosaic tymovirus | p69 and p206 replicasef | Coat protein | Internal promoter | Schirawski et al., 2000, Szybiak et al., 1978 |
| Closterovirus: citrus tristeza; beet yellows | Polymerase precursor | Eight to ten downstream ORFs | Internal transcription elements | Gowda et al., 2001, Peremyslov and Dolja, 2002 |
| Geminivirus: tomato leaf curl | C1 replication proteind | C2 transcription factor | Internal promoter | Mullineaux et al., 1993 |
| Pararetrovirus: rice tungro bacilliform | ORFs 1, 2, 3f | ORF4 | Splicing | Fütterer et al., 1994 |
The silent downstream cistron is expressed only upon being moved closer to the 5′ end via production of a second, shorter mRNA.
Translation of most genes derived from these viruses follows straightforward predictions of the scanning mechanism, although occasional deviations have been reported. In rare instances where a 3′ cistron appears to be translated from a dicistronic mRNA (Grundhoff and Ganem, 2001, Kirshner et al., 1999, Nador et al., 2001, Stacey et al., 2000), the virus in question employs a complicated pattern of splicing and therefore the existence of an undetected monocistronic mRNA is not beyond the realm of reason. In some other cases only a small amount of the protein encoded by the 3′ cistron was produced, and the published RNA analyses were not sufficiently sensitive to rule out the presence of an additional subgenomic mRNA (Herbert et al., 1996).
In some cases the listed example is arbitrary, i.e. with retroviruses, coronaviruses, closteroviruses, etc., there are additional polycistronic mRNAs wherein translation is restricted to the 5′ cistron.
Whereas DNA viruses and retroviruses use conventional promoter-switching or splicing mechanisms to generate alternative forms of mRNA that allow translation of the downstream cistron, more complicated mechanisms underlie the production of subgenomic mRNAs by some RNA viruses (Miller and Koev, 2000).
The presence of internal promoters that produce a shorter transcript for each downstream ORF is suggestive, but testing of translation is still needed for the mRNAs produced by cytomegalovirus and geminivirus.
Whereas all retroviruses employ splicing to produce the subgenomic mRNA from which envelope protein (Env) is translated, some retroviruses also employ an internal promoter which is postulated to mediate expression of novel ORFs, such as the superantigen of mouse mammary tumor virus (Reuss and Coffin, 1998) and orf-x of the virus that causes lung cancer in sheep (Palmarini et al., 2002).
In place of the usual m7G cap, the 5′ end of these viral RNAs carries a covalently linked protein (VPg) or is unblocked. The need for a subgenomic mRNA even in these cases emphasizes that translation is 5′ end-dependent even when it is not cap-dependent.
The full-length genomic mRNA supports translation of the 3′ cistron in vitro but the 3′ cistron is silent in vivo. The latter result is considered more reliable (Meulewaeter et al., 1992).
The same problem and same solution – post-transcriptional processing of polycistronic mRNAs – underlie the expression of many genes in Caenorhabditis elegans (Blumenthal et al., 2002, Hough et al., 1999). In mammalian cells, mRNAs that contain two full-length nonoverlapping cistrons are extremely rare and, as with the aforementioned viruses, actual translation of the 3′ cistron probably occurs from a second, monocistronic mRNA (Pardigol et al., 1998, Westerman et al., 2001) or from a second mRNA in which the two cistrons are fused into a single translation unit (Gray and Nicholls, 2000, Hänzelmann et al., 2002). A dicistronic transcript derived from the mouse Snurf-Snrpn locus (Gray et al., 1999) barely supports translation of the second cistron, as discussed below in the section on reinitiation (Section 4). Recently discovered dicistronic transcripts produced from the mouse Hyal locus support translation of only the 5′ cistron (Shuttleworth et al., 2002). A few other reported dicistronic mRNAs await testing.
Notwithstanding the documented inability to translate the 3′ cistron in natural dicistronic mRNAs, synthetic dicistronic mRNAs – constructed by inserting a putative IRES element between two reporter genes – appear sometimes to allow translation of the downstream cistron. The interpretation that this occurs via direct internal initiation of translation has been questioned (Kozak, 2001a) and defended (Hellen and Sarnow, 2001) in other reviews.
2.2. First-AUG rule
The position effect, indicative of scanning, is seen when a mutation creates an AUG codon upstream from the normal start codon and translation shifts to the upstream site (Bergenhem et al., 1992, Cai et al., 1992, Gross et al., 1998, Harington et al., 1994, Liu et al., 1999, Lock et al., 1991, Mével-Ninio et al., 1996, Muralidhar et al., 1994, Wada et al., 1995). In the most stringent test of the rule, the first AUG codon was shown to be the exclusive site of initiation even when the second AUG was positioned just a few bases downstream from, and in the same optimal context as, the first (Kozak, 1995).
The position effect is seen also when removal of the first start codon activates initiation from the next AUG downstream. Some genes require production of two versions of the encoded protein, wherein the shorter version, initiated from an internal AUG codon, lacks the N-terminal domain of the longer isoform. The problem of how ribosomes can gain access to an internal start codon is solved by producing, via splicing or a downstream promoter, a second form of mRNA from which the first AUGSTART codon has been removed. Table 2 lists some examples. The N-terminally truncated isoform thus produced may reside in a different cellular compartment, or may function as an antagonist to the full-length protein (as seen with various transcription factors listed in Table 2), or may function in a surprising way. One such surprise was the discovery that a truncated form of tryptophanyl-tRNA-synthetase (‘miniTrpRS’) has angiostatic activity (Wakasugi et al., 2002).
Table 2.
Partial list of vertebrate genes that produce a second, shorter version of the encoded protein via a second form of mRNA in which an internal AUG codon becomes a functional start site upon elimination of the upstream AUGSTART codona
| Gene | Source | References |
|---|---|---|
| Tryptophanyl-tRNA synthetase | Human | Tolstrup et al., 1995, Wakasugi et al., 2002 |
| Stromelysin 3 | Human | Luo et al., 2002 |
| Procaspase-8 | Human | Breckenridge et al., 2002 |
| ATBF1 transcription factorb | Human | Berry et al., 2001 |
| Hepatocyte-nuclear factor 3βb | Mouse | Philippe, 1995 |
| ZAC transcription factor | Human | Bilanges et al., 2001 |
| HOF transcription factor | Mouse | Mitchelmore et al., 2002 |
| Lymphoid enhancer factor-1 (LEF1)b | Human | Hovanes et al., 2001 |
| RIZ transcription factor | Human | Jiang et al., 1999, Liu et al., 1997 |
| Estrogen receptor-αb | Human | Denger et al., 2001, Flouriot et al., 2000 |
| Thyroid hormone receptor-β | Rat | Williams, 2000 |
| Progesterone receptorb | Human | Kastner et al., 1990a |
| CCAAT enhancer binding protein (C/EBP) ε | Human | Yamanaka et al., 1997 |
| Smoothelin | Human | Rensen et al., 2002 |
| Protein kinase Ntk | Mouse | Chow et al., 1994 |
| Protein kinase Chkb | Rat | Shann and Hsu, 2001 |
| MDM2 oncogeneb | Mouse | Perry et al., 2000, Saucedo et al., 1999 |
| MXI1 tumor suppressor gene | Human, mouse | Schreiber-Agus et al., 1995, Wechsler et al., 1996 |
| Dopamine-regulated phosphoprotein | Human | El-Rifai et al., 2002 |
| Adenosine deaminase | Human | Kawakubo and Samuel, 2000 |
| Caveolin-1 | Murine | Kogo and Fujimoto, 2000 |
| Nitric-oxide synthase | Human | Wang et al., 1997 |
| Gelsolinc | Human | Kwiatkowski et al., 1988 |
| Serine:pyruvate aminotransferasec | Rat | Oda et al., 1990 |
| Alanine:glyoxylate aminotransferasec | Rat, frog | Holbrook and Danpure, 2002 |
| Phospholipid-hydroperoxide GTH peroxidasec | Rodent | Knopp et al., 1999, Pushpa-Rekha et al., 1995 |
| Folylpoly-γ-glutamate synthetasec | Human | Freemantle et al., 1995, Turner et al., 2000 |
| Porphobilinogen deaminased | Human, mouse | Chretien et al., 1988, Porcher et al., 1991 |
| Erythroid membrane protein 4.1d | Human, mouse | Conboy et al., 1991, Huang et al., 1993 |
| p120ctn cateninc, d | Human, mouse | Aho et al., 2002, Keirsebilck et al., 1998, Mo and Reynolds, 1996 |
| Carbonic anhydrase VI | Mouse | Sok et al., 1999 |
| Water channel aquaporin 4 | Human, mouse | Lu et al., 1996, Zelenin et al., 2000 |
| Sterol carrier protein 2 | Rat liver | Seedorf and Assmann, 1991 |
| β1,4-galactosyltransferasee | Mouse | Harduin-Lepers et al., 1993 |
| Calmodulin-kinase IV/calspermin | Rodent | Sun et al., 1995 |
Production of long and short protein isoforms via this mechanism is seen also with genes from insects (Mével-Ninio et al., 1996), plants (Cunillera et al., 1997, Wimmer et al., 1997), yeast (Beltzer et al., 1988, Carlson et al., 1983, Chatton et al., 1988, Ellis et al., 1989, Gammie et al., 1999, Natsoulis et al., 1986, Wolfe et al., 1996) and viruses (Barbosa and Wettstein, 1988, Lambert et al., 1987, Liu and Roizman, 1991, Liu and Biegalke, 2002, Weimer et al., 1987, Welch et al., 1991, Wu et al., 1993b, Zheng et al., 1994).
In these cases, the long and short protein isoforms have different functional effects. Other genes that resemble this pattern, producing long and short isoforms with contrasting functions, are not listed in the table because the AUGSTART codon for the shorter protein is carried on an alternative exon present only in the shorter mRNA (e.g. Koski et al., 1999, Molina et al., 1993). That arrangement does not illustrate the main point of the table, which is that a silent internal AUG codon in the longer mRNA can be activated simply by truncating the transcript.
The long and short isoforms are targeted to different cellular compartments.
The long and short isoforms are expressed in different tissues.
The long and short forms of β1,4-galactosyltransferase appear to function identically. The main significance of the promoter switch, which eliminates the first AUGSTART codon, is that the shorter 5′ UTR supports translation more efficiently (Charron et al., 1998).
The entries in Table 2 and some other examples (Aichem and Mutzel, 2001, Beuret et al., 1999, Falvey et al., 1995, Nagpal et al., 1992) are what I call natural tests of the position rule. Additional evidence comes from experimental manipulations wherein removal of the first AUG was shown to activate initiation from a downstream site (Cahana et al., 2001, Chenik et al., 1995, Tailor et al., 2001, Thoma et al., 2001).
2.3. Inhibition by upstream AUG codons
The scanning mechanism predicts that the 5′ untranslated region (5′ UTR, an unfortunate misnomer) is actually traversed by ribosomes. This explains why translation of the major coding domain is reduced when adventitious out-of-frame AUG codons occur upstream. The upstream AUG codons often create small ORFs (upORFs) which are indeed translated, as shown by detecting the encoded peptide (Hackett et al., 1986, Raney et al., 2000, Wang and Wessler, 2001) or by fusing a reporter gene to the upORF (Abastado et al., 1991, Donzé et al., 1995, Liu et al., 1999, Steel et al., 1996, Tanaka et al., 2001, Xu et al., 2001). The fusion test is the more reliable, as small peptides are usually degraded rapidly.
Even if upstream AUG codons are arranged in a way that allows reinitiation, there is a penalty because reinitiation is usually inefficient. This topic will be discussed at length in Section 4.
2.4. Inhibition by secondary structure
The hypothesis that the 5′ UTR is traversed by ribosomes explains why a highly structured 5′ leader sequence is so detrimental to translation. Vertebrate mRNAs characteristically have long, GC-rich – hence highly structured – leader sequences (Kozak, 1991a, Macleod et al., 1998), and the resulting difficulty in translation has been discovered over and over in the course of cloning. Even a short GC-rich 5′ UTR can inhibit profoundly, as illustrated in cases where a gene produces a mixture of mRNAs with different leader sequences, and the worst-translated mRNA was found to be the form with the shortest 5′ UTR (Jiang and Lucy, 2001, Yang et al., 1998). A stem-and-loop structure, stabilized in some cases by a repressor protein, is most inhibitory when its proximity to the 5′ end blocks ribosome binding (Goossen and Hentze, 1992, Kozak, 1989, Wang and Wessler, 2001). If the structure is far enough from the 5′ end to allow ribosome entry, the advancing 40S ribosome/factor complex apparently has some ability to disrupt base pairing, but this ability is notably less than that of 80S elongating ribosomes (Kozak, 1986a, Kozak, 2001b, Lingelbach and Dobberstein, 1988, Paraskeva et al., 1999) and is curtailed in yeast (Koloteva et al., 1997).
While there are mechanisms for reducing the inhibitory effects of upstream AUG codons, as discussed below, no mechanism has yet been defined for modulating the inhibitory effects of secondary structure. Some studies suggest that secondary structure might be less inhibitory to translation in vivo than in vitro (Charron et al., 1998, Curnow et al., 1995, Hensold et al., 1997, Hoover et al., 1997, Morrish and Rumsby, 2001, Van der Velden et al., 2002). This could be due to production of an alternative transcript that simply eliminates the secondary structure – a reasonable possibility given that GC-rich domains often harbor promoter elements – or to modification of the translation machinery. Interpretation of in vivo tests of translation could also be complicated by effects of secondary structure on mRNA stability (Stefanovic et al., 1999). Whether and how translation of GC-rich leader sequences might improve in exponentially growing cells (Nielsen et al., 1995) remains an important open question.
2.5. Non-AUG start codons
The scanning mechanism rationalizes the occurrence of initiation at upstream ACG or CUG codons in some mRNAs. These alternative codons are usually too weak to actually substitute for the AUGSTART codon (reviewed by Kozak, 1999; for some exceptions see Falvey et al., 1995, Kiefer et al., 1994, Riechmann et al., 1999, Sadler et al., 1999). It is not uncommon, however, for initiation to occur at an upstream non-AUG codon in addition to the first AUG (see leaky scanning in Section 3). This is observed frequently with cellular genes that have highly structured, GC-rich leader sequences (Kozak, 1991b), perhaps because secondary structure slows scanning and thus allows more time for the mismatched codon to pair with Met-tRNAi.
With some viruses, the extra protein isoform initiated from an upstream non-AUG codon serves an essential function (Muralidhar et al., 1994, Portis et al., 1994, Portis et al., 1996). While the N-terminally-extended isoforms derived from some cellular genes also display distinct functions (Arnaud et al., 1999, Calkhoven et al., 2000, Spotts et al., 1997) or patterns of localization (Acland et al., 1990, Lock et al., 1991, Packham et al., 1997), it would not be surprising if some other upstream-initiated proteins turn out to be inadvertent byproducts generated in the course of slowly traversing a GC-rich leader sequence.
2.6. Introduction to harder cases
Because the ability of the scanning mechanism to explain the big picture is generally accepted, the remainder of this review directs attention, not to examples that can be seen readily to support the model, but to mRNAs that seem to be poorly designed for a scanning mode of initiation. The main point is that the scanning mechanism applies even in these difficult cases.
Understandably, such mRNAs are translated inefficiently and this brings out a second important point: some critical regulatory genes require protein synthesis to be inefficient. An earlier review raised awareness that genes that encode potent regulatory proteins – cytokines, growth factors, kinases, transcription factors – often produce mRNAs in which the 5′ leader sequence is GC-rich or burdened by upstream AUG codons (Kozak, 1991a). Some examples described herein validate the prediction that these encumbered 5′ sequences are nature's way of limiting the synthesis of potent proteins that would be harmful if overproduced.
I also suggested in earlier reviews that, when a cDNA sequence has so many upstream AUG codons as to challenge the applicability of the scanning mechanism, it is wise to ask whether the cDNA correctly reflects the structure of the mRNA. That advice is not changed by what is written here. Very often, cDNA sequences that appear incompatible with scanning have been found to derive from incompletely spliced transcripts or to have been misinterpreted in other ways (Kozak, 1996, Kozak, 2000). In other cases, although an encumbered cDNA sequence is correct, it derives from a transcript that does not support translation (Hake and Hecht, 1993, Han et al., 2002, Larsen et al., 2002, Lee et al., 2000). Only after one is certain of the mRNA structure should the mechanisms below be considered.
3. Context-dependent leaky scanning
3.1. Definition, conventional examples, exceptions
In mammals, the optimal context for recognition of the AUGSTART codon is GCCRCCaugG. Within this motif, the purine (R) in position −3 is the most highly conserved (see Section 6.1) and functionally the most important position. The importance of A or G (A is somewhat better than G) in position −3 was proved by mutagenesis experiments on a wide variety of genes (Kozak, 1986b; and see entries marked ‘tested’ in Table 3). The G in position +4 is also highly conserved and, especially in the absence of A in position −3, contributes strongly (Kozak, 1997). Adherence to the rest of the GCCRCCaugG motif varies, without major consequences as long as positions −3 and +4 conform; the upstream GCC motif can be seen to contribute, however, in the absence of other elements (Kozak, 1987b).
Table 3.
Partial list of cellular and viral mRNAs that produce two separately-initiated proteins by context-dependent leaky scanninga
| Source of mRNA | Identifying information | Sequence flanking first AUG codonb | Protein productsc | References |
|---|---|---|---|---|
| Glucocorticoid receptor gene | Human | CUGaugG; tested | Long and short transactivators | Yudt and Cidlowski, 2001 |
| NFAT transcription factor gene | Human | CGGaugC | 90 and 86 kDa isoforms | Lyakh et al., 1997 |
| C/EBPα gened | Mouse | CCCaugG; tested | 42 and 30 kDa isoforms | Lin et al., 1993, Ossipow et al., 1993 |
| Rx/rax homeobox gene | Mouse | UCCaugC; tested | Long and short isoforms | Tucker et al., 2001 |
| GATA-1 gene | Human, mouse | CCCaugG (mouse); UCCaugG (human) | 50 and 40 kDa isoforms | Calligaris et al., 1995, Wechsler et al., 2002 |
| Peripherin gene | Rat | UGAaugC; tested | Long and short isoforms | Ho et al., 1995 |
| MxB protein gene | Human | CACaugU; tested | Nuclear and cytoplasmic isoforms | Melén et al., 1996 |
| Ubiquitin-activating enzyme E1 gene | Human | UUGaugU | Nuclear and cytoplasmic isoforms | Handley-Gearhart et al., 1994, Shang et al., 2001 |
| Microtubule-associated protein gene | Human | CCAaugC | Long and short isoforms | Su and Qi, 2001 |
| Von Hippel-Lindau genee | Human | GGAaugC | 24 and 18 kDa isoforms | Iliopoulos et al., 1998, Schoenfeld et al., 1998 |
| S6 kinase gene | Human, rat | CCCaugA | Long and short isoforms | Grove et al., 1991, Reinhard et al., 1992 |
| Rlk/Txk tyrosine kinase gene | Mouse | GCCaugA | Long and short isoforms | Debnath et al., 1999 |
| Vitamin D receptor gene | Chicken | UCCaugU; tested | Long and short isoforms | Lu et al., 1997 |
| Val-tRNA synthetase gene | Arabidopsis | UCUaugU; tested | Mitochondrial and cytoplasmic isoforms | Souciet et al., 1999 |
| Simian virus 40 | Late 19S mRNA | UCCaugG; tested | Capsid proteins VP2 and VP3 | Sedman and Mertz, 1988 |
| Cytomegalovirus | UL4 mRNA | GUGaugC; tested | Inhibitory peptide and gp48 | Cao and Geballe, 1995 |
| Adenovirus type 5 | Region E3 | UAUaugA | 6.7 kDa protein and gp19K | Wilson-Rawls et al., 1990, Wold et al., 1986 |
| Adenovirus type 5 | Region E1B | UCCaugG | 21 kDa and 55kDa proteins | Bos et al., 1981 |
| Hepatitis B virus | 2.1 kb mRNA | GCCaugC; tested | Middle (pre-S2) and small (p24) surface proteins | Sheu and Lo, 1992 |
| Feline leukemia retrovirusf | Genomic mRNA | CUGaugU | gp80gag and pr65gag | Laprevotte et al., 1984 |
| HIV-1 | Spliced mRNA | GUAaugC; tested | Vpu and Env | Schwartz et al., 1992 |
| HIV-1 | Spliced mRNA | CCUaugG; tested | Rev and Nef | Schwartz et al., 1992 |
| Reovirus (mammalian) | RNA segment S1 | CGGaugG; tested | σ1 and 14kDa proteins | Fajardo and Shatkin, 1990, Kozak, 1982 |
| Reovirus (baboon)g | RNA segment S4 | UACaugG | p15 and p16 proteins | Dawe and Duncan, 2002 |
| Bunyavirus | RNA segment S | UCAaugA | Nucleocapsid (N) and NSs | Bridgen et al., 2001, Elliott and McGregor, 1989 |
| Influenza A virusg | RNA segment 2 | UGAaugG | Polymerase subunit PB1 and mitochondrialprotein | Chen et al., 2001 |
| Barley yellow dwarf luteovirus | Subgenomic mRNA | UGAaugA; tested | Coat protein and p17 | Dinesh-Kumar and Miller, 1993 |
| Turnip yellow mosaic viruse | Genomic mRNA | CAAaugA | p69 and p206replicase | Weiland and Dreher, 1989 |
| Cucumber necrosis virus | 0.9 kb subgenomic mRNA | UUCaugG; tested | p21 and p20 | Johnston and Rochon, 1996 |
| Peanut clump furovirus | RNA segment 2 | CUUaugU; tested | p23 (coat) and p39 | Herzog et al., 1995 |
| Potato virus X potexvirus | Subgenomic mRNA | CAUaugU; tested | 12 and 8kDa movement proteins | Verchot et al., 1998 |
| Southern bean mosaic virusg | Genomic mRNA | UUUaugA; tested | p21 movement protein and p105polymerase | Sivakumaran and Hacker, 1998 |
| Baculovirusd | IE0 mRNA | GACaugA | Long and short forms of transactivator (IE0, IE1) | Theilmann et al., 2001 |
In all mRNAs here listed, the sequence flanking the first start codon deviates from the consensus sequence in position −3 and/or position +4, highlighted by underlining. When the postulated link between context and leaky scanning was tested (so marked in this column), mutations that improved the context at the first start site diminished access to the downstream start site. This test failed only with cucumber necrosis virus, where the short distance between the m7G cap and AUG#1 allowed some leaky scanning even when the context was optimized.
In some cases the first and second AUG codons are in the same reading frame, generating long and short versions of the encoded protein which may function differently. In cases where the first and second start codons are in different reading frames, indicated by italicizing the second product, the extent of overlap between the two ORFs ranges from a few codons (peanut clump virus, southern bean mosaic virus) to 626 codons (turnip yellow mosaic virus).
Access to the downstream initiation site via leaky scanning is augmented by a reinitiation shunt, as explained in the text (Section 4.3) and diagrammed in Fig. 1 for C/EBPβ mRNA.
Mutations that eliminate AUG#1 usually increase production of the second, downstream protein. In rare cases where the expected increase was not seen (e.g. von Hippel-Lindau, turnip yellow mosaic virus), it might be because translation of the second protein was restricted at the level of elongation. For a similar reason, improving the context around AUG#1 occasionally fails to elevate production of the protein there initiated (Fajardo and Shatkin, 1990). These entries nevertheless satisfy the main prediction of the leaky scanning mechanism, which is that improving the context around AUG#1 prevents initiation from the second, downstream site (Fajardo and Shatkin, 1990, Iliopoulos et al., 1998).
Whereas feline leukemia virus produces an N-terminally-extended, glycosylated form of Gag (gp80gag) from the indicated weak AUG codon, the corresponding upstream start site in murine leukemia virus is ACCCUGG (Portis et al., 1994). When that site was experimentally ablated, however, revertants expressed the extended protein from a weak upstream AUG codon (UUUaugG) created by a point mutation. Those revertants were selected because the extra glycosylated form of Gag contributes to viral spread (Portis et al., 1996).
In the mRNAs from baboon reovirus, influenza A virus, and southern bean mosaic virus, the indicated proteins derive from the first (weak) and fourth AUG codons. AUG#2 and AUG#3 initiate small ORFs that terminate before AUG#4. Thus, a combination of leaky scanning and reinitiation probably mediates access to the downstream start site.
The aforementioned mutagenesis experiments define two extremes: (i) when the first AUG codon occurs in a strong context – ANNaugN or GNNaugG – all or almost all ribosomes stop and initiate at that point; (ii) when the first AUG resides in a very weak context, lacking both R in position −3 and G in position +4, some ribosomes initiate at that point but most continue scanning and initiate farther downstream. This leaky scanning enables the production of two separately initiated proteins from one mRNA, as documented below. It is harder to predict what happens at start sites that fall between the extremes, i.e. mRNAs in which the first potential start codon has the sequence YNNaugG, GNNaugY or GNNaugA. Leaky scanning is seen in some but not all such cases. A possible explanation suggested by studies with test transcripts (Kozak, 1990a) is that initiation might be restricted to the first AUG codon, despite a suboptimal context, when downstream secondary structure slows scanning and thus provides more time for codon/anticodon pairing. Suppression of leaky scanning via this mechanism requires a critical distance (13–15 nt, which corresponds to half the diameter of the ribosome) between the AUG codon and the downstream structured element.
Table 3 lists some examples in which two proteins are produced from one mRNA via leaky scanning. The postulated link between context and leaky scanning has been tested in many of these cases by showing that, upon improving the context at the upstream site, initiation from the second site is reduced or abolished. (Whether the second AUG codon resides in a strong or weak context is not relevant; the ribosome reads the mRNA linearly and thus the decision to stop or to bypass the first AUG is not influenced by whether there is a better initiation site downstream.) The large number of genes that employ leaky scanning precludes discussion of the biological significance of the proteins thereby produced, but it merits noting that, for many of the viruses in Table 3, replication requires production of both listed proteins. For some other viruses, the second protein is a virulence factor that weakens host defenses (Bridgen et al., 2001, Chen et al., 2001, Weber et al., 2002). The biological importance of these downstream-initiated proteins shows that leaky scanning is a deliberately employed tool; it does not simply reflect sloppiness on the part of the translational machinery.
The long list of examples in Table 3 conforms to expectations in that the first start codon resides in a suboptimal context. There are, however, rare instances of leaky scanning despite a good context (R−3 and G+4) around the first AUG. This can happen when the first AUG codon is too close to the 5′ end to be recognized efficiently (Kaneda et al., 2000, Kozak, 1991c, Ruan et al., 1994, Sedman et al., 1990, Slusher et al., 1991, Spiropoulou and Nichol, 1993, Werten et al., 1999) or when the facilitating effect of G in position +4 is canceled by U in position +5 (Kozak, 1997, Sloan et al., 1999, Stallmeyer et al., 1999). Other occasional claims of leaky scanning despite a strong context at the first AUG codon were simply mistaken (Scherer et al., 1995); the shorter protein turned out to be translated from a second form of mRNA (Kogo and Fujimoto, 2000).
At the opposite extreme, there are rare mammalian mRNAs in which, despite a very unfavorable context flanking the first AUG codon, translation appears to initiate exclusively at that site (Arai et al., 1991, Hickey and Roth, 1993, Leslie et al., 1992, McNeil et al., 1992, Plowman et al., 1990, Wu et al., 1993a). Leaky scanning might be suppressed in these few cases because of downstream secondary structure, or because the wider context (C in positions −1, −2, −4, −5; G in position −6) compensates to some extent for the absence of R−3 and G+4, or for other unknown reasons.
The same principle that allows initiation from the first and second AUG codons when the first AUG is in a suboptimal context (Table 3) applies in cases where translation initiates at an upstream non-AUG codon in addition to the first AUG (Acland et al., 1990, Arnaud et al., 1999, Carroll and Derse, 1993, Fajardo et al., 1993, Florkiewicz and Sommer, 1989, Fütterer et al., 1996, Fuxe et al., 2000, Lock et al., 1991, Muralidhar et al., 1994, Packham et al., 1997, Saris et al., 1991, Spotts et al., 1997). Recognition of an upstream ACG, CUG or GUG start codon requires a strong context (Portis et al., 1994), despite which scanning is usually leaky because the initiator codon itself is weak.
Production of long and short protein isoforms via leaky scanning is harder to regulate – e.g. to achieve tissue specific expression of one or the other form – than when a unique mRNA encodes each isoform, as in Table 2. There are hints, however, that dual initiation via leaky scanning might be regulable (Probst-Kepper et al., 2001, Spotts et al., 1997). This could conceivably be accomplished via proteins that stabilize downstream secondary structure or, perhaps, via a combination of leaky scanning and regulated reinitiation, if the mRNA also has small upORFs.
Among the examples in Table 3 are many plant viruses, indicating that the basic context rules extend to plant systems. Mutagenesis experiments confirm the functional importance of R in position −3 and G in position +4 in plant mRNAs (Jones et al., 1988, Lukaszewicz et al., 2000) and surveys of plant cDNA sequences confirm the conservation of those key positions (Pesole et al., 2000, Rogozin et al., 2001). Unlike mammalian mRNAs, however, plant mRNAs do not show a predominance of C in positions −1, −2, −4 and −5.
The foregoing discussion pertains to mRNAs from plants and vertebrate animals. There is some evidence for context-dependent leaky scanning in fungi (Arst and Sheerins, 1996), but context effects on initiation have not yet been studied carefully in protozoa, insects, and various other systems. The observation that trans-splicing of mRNAs in C. elegans sometimes brings a purine into position −3 is interesting (Hough et al., 1999) but the significance awaits testing. With a number of yeast genes, there is a hint of leaky scanning when the usual A in position −3 is replaced by a pyrimidine (Gaba et al., 2001, Slusher et al., 1991, Vilela et al., 1998, Welch and Jacobson, 1999, Wolfe et al., 1994). Context effects were not evident, however, in other studies of translation in yeast (Cigan et al., 1988b). For whatever reason, leaky scanning is rare in yeast, apart from a few cases attributable to the first AUG codon residing too close to the 5′ end.
3.2. Pushing the limits: maximally leaky mRNAs
mRNAs that initiate translation from three sites provide a striking illustration of how far leaky scanning, alone or in combination with reinitiation, can be pushed. Fig. 1 shows some examples.
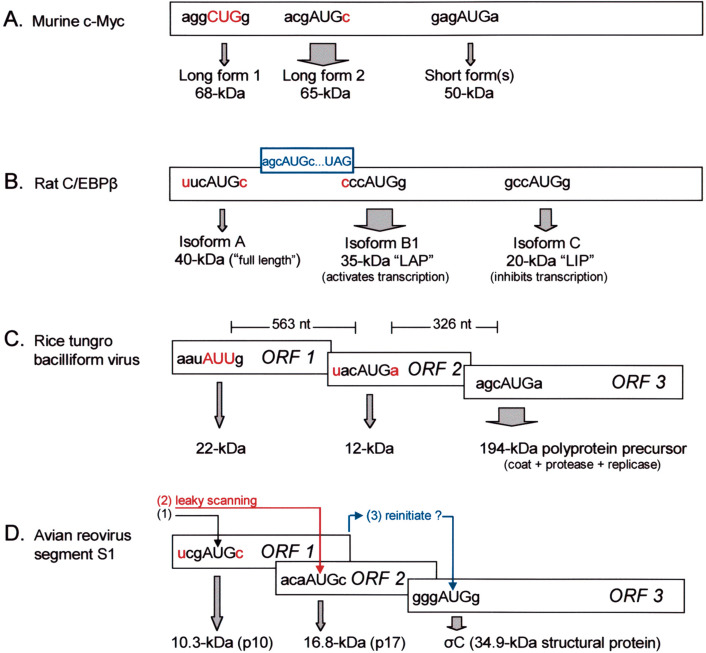
Fig. 1.
Examples of ‘maximally leaky’ scanning wherein one mRNA produces three independently initiated proteins. Major (thick arrow) and minor (thin arrow) translation products are identified below their respective start codons. Sequences that cause the initiation site to be weak, and thus promote leaky scanning, are highlighted in red. Offset rectangles represent ORFs in different reading frames. (A) With c-myc mRNA, a leaky scanning mechanism was inferred from experiments in which optimizing the context around the first AUG codon suppressed production of the 50 kDa isoform, while changing the upstream CUG codon to AUG suppressed production of both the 65 and 50 kDa isoforms (Spotts et al., 1997). Access to the downstream start site might be more complicated than here depicted, as there is a small out-of-frame ORF between the 65 and 50 kDa start sites. (B) With C/EBPβ mRNA, a mutation that strengthens the first start codon (UUCaugC→ACCaugG) blocked production of all shorter isoforms, implicating a leaky scanning mechanism (Calkhoven et al., 2000). A small upORF (blue) superimposes another level of control, causing more ribosomes to bypass the start site for isoform B1 than would be expected from leaky scanning alone. Presumably because the AUGSTART codon for isoform B1 is positioned close to the termination site of the upORF, reinitiation at site B1 is inefficient and some ribosomes thus reach the far downstream start site for the 20 kDa isoform (LIP). As evidence for this reinitiation shunt, Calkhoven et al. (2000) showed that eliminating the AUG codon of the upORF abolished production of LIP and that strengthening or weakening the context around the upORF start codon caused corresponding changes in the yield of LIP. Although the smallest form of C/EBPβ can be generated in some situations by proteolysis (Dearth et al., 2001), the effects of the aforementioned mutations clearly implicate a translational mechanism. The LAP/LIP ratio shows tissue and stage specific variation (Dearth et al., 2001, Descombes and Schibler, 1991). (C) Whereas leaky scanning allows initiation at multiple sites within a single ORF in C/EBPβ and c-myc mRNAs, leaky scanning allows translation of three separate ORFs in the pregenomic mRNA of rice tungro bacilliform virus. These ORFs (not drawn to scale) have overlapping start and stop codons of the form AUGA. Translation via leaky scanning was inferred from the strong reduction (>13-fold) in translation of ORF2 and ORF3 when the start codon of ORF1 was changed from AUU to AUG (Fütterer et al., 1997) and from the inhibitory effect on expression of ORF3 when an adventitious AUG codon was inserted into ORF2. The 5′ leader sequence that precedes ORF1 has ten small upORFs which are not depicted here because that peculiar leader sequence, postulated to be translated by ribosome hopping (Fütterer et al., 1996), is not required for the leaky scanning mechanism that underlies translation of ORFs 1, 2 and 3. (D) The avian reovirus S1 mRNA supports translation of one structural and two nonstructural proteins (Bodelón et al., 2001). The depicted mechanism postulates that ORF1 has a dual function, encoding its own polypeptide (p10) and facilitating translation of ORF3 by shunting some ribosomes past the strong AUGSTART codon for ORF2. The absence of extraneous AUG codons in the 310 nt region between the end of ORF1 and the start of ORF3 is consistent with the idea that ORF3 might be translated by reinitiation. Some ribosomes would be expected to translate p17 (ORF2) by leaky scanning, engendered by the poor context at the start of ORF1. Improving the context at the start of ORF1 indeed increased production of p10 (Shmulevitz et al., 2002); unfortunately, the yield of p17, which would be expected to decrease, was not monitored. The observation that strengthening the context at the start of ORF1 had no effect on the yield of σC is not surprising because the reinitiation mechanism postulated to underlie translation of ORF3 would probably be limited by other features, such as the relatively large size of ORF1.
The predominant translation product obtained from c-myc mRNA is a 65 kDa ‘long form 2’ which initiates at the first AUG codon (Fig. 1A). A small amount of a longer isoform (68 kDa) derives from an upstream CUG codon which is a weak start site (i.e. very leaky) because the codon is not AUG. Although the first AUG codon has the required A in position −3, a small percentage of ribosomes bypass that site and initiate at the next AUG, producing a third (50 kDa) form of c-myc. This happens apparently because the context flanking the first AUG codon is good but not perfect. Thus, production of the 50 kDa isoform was eliminated when the upstream site was changed from ACGaugC to ACCaugG (Spotts et al., 1997).
Fig. 1B depicts another example in which ribosomes initiate from three in-frame AUG codons. With the mRNA that encodes C/EBPβ, access to the far downstream site via leaky scanning is augmented by a reinitiation shunt, as explained in the legend to Fig. 1 and discussed further in Section 4. Translation of C/EBPα mRNA occurs by a mechanism similar to that depicted for C/EBPβ except that the first start site in C/EBPα is a CUG codon (Calkhoven et al., 2000), which generates a smaller amount of the longest protein (isoform A) than does the AUG codon in C/EBPβ. With c-myc, C/EBPβ and C/EBPα mRNAs, leaky scanning is biologically important because the long and short versions of the protein have opposing effects as regulators of transcription.
It is striking that leaky scanning can operate even when the second initiation site resides far downstream from the first. With synthetic transcripts designed to test the processivity of scanning, there was no reduction in initiation from the downstream site when the inter-AUG distance was expanded stepwise from 11 to 251 nt (Kozak, 1998). In some remarkable viral mRNAs, the second functional initiation site is more than 500 nt downstream from the first AUG (Herzog et al., 1995, Sivakumaran and Hacker, 1998).
The pregenomic mRNA of rice tungro bacilliform virus (Fig. 1C) provides the most dramatic illustration of these points. Use of a weak (non-AUG) codon to initiate ORF1 and an unfavorable context at the start of ORF2 (UACaugA) enables the majority of ribosomes to reach and initiate at the start of ORF3. The ORF3 polyprotein is thought to be a precursor from which coat protein, protease and reverse transcriptase are derived by proteolysis. The remarkable absence of AUG codons from the long (563 nt) coding domain of ORF1 and the presence of but one weak AUG codon within ORF2 underscore how carefully this mRNA is constructed to support translation via scanning. The careful construction includes minimizing the overlap between adjacent cistrons. Without that precaution, elongational occlusion might work against utilization of a far downstream start codon, as documented in other cases (Kozak, 1995).
The avian reovirus RNA diagrammed in Fig. 1D offers another example of initiation from three sites in one mRNA. Additional experiments are needed to validate the postulated mechanism.
3.3. Pushing the limits: minimally leaky mRNAs
In contrast with the ‘maximally leaky’ mRNAs in Fig. 1, the mRNAs in Fig. 2 are minimally leaky: only a small fraction of ribosomes bypass the first AUG codon and initiate downstream. Here the leaky scanning mechanism has been pushed to the limits in the sense that there is (low-level) initiation from a second site despite the presence of a strong context around the first AUG codon. The explanation is that the context flanking the first AUGSTART codon is good but not perfect. The resulting low-level leaky scanning enables the viruses depicted in Fig. 2A,B to produce two proteins – one abundant, the second in small amounts – from a single mRNA. Experimental manipulations that support this interpretation are summarized in the legend to Fig. 2. A few other viral genes that might fit this category have been described (Chenik et al., 1995, Jayakar and Whitt, 2002). The hepatitis B virus example is noteworthy because, via the Rube Goldberg mechanism diagrammed in Fig. 2B, reverse transcriptase encoded by the P gene is initiated independently from a far downstream site, unlike most other reverse transcriptase genes which lack an independent start codon and therefore require frameshifting during translation of the preceding core gene.
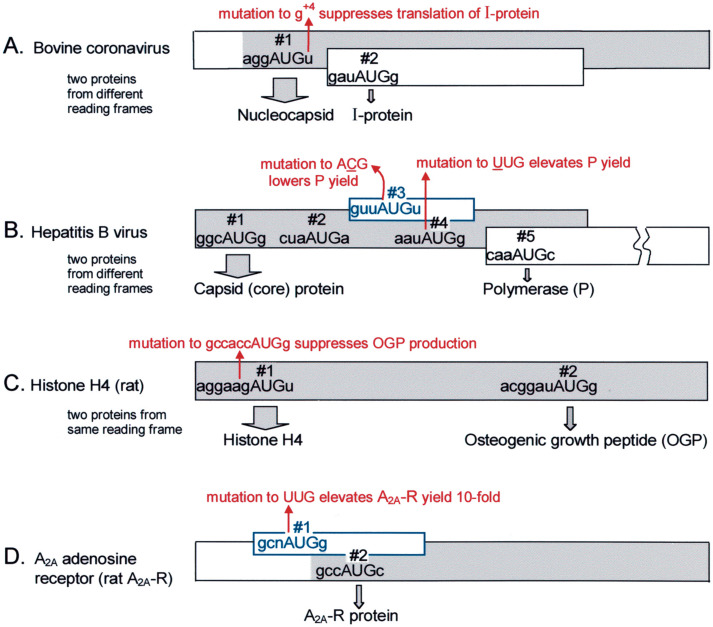
Fig. 2.
Examples of minimally leaky scanning in which a strong, but not quite perfect, context at AUG#1 causes most ribosomes to initiate there while allowing a low level of initiation downstream. With the depicted viral mRNAs (A,B), the predominant product of translation is the capsid protein initiated from AUG#1. Low-level leaky scanning generates a small but adequate amount of the indicated second protein. With bovine coronavirus, a mutation in position +4 (U→G, indicated in red) flanking AUG#1 strongly reduced translation from the downstream site (Senanayake and Brian, 1997), supporting the interpretation that the natural mRNA is slightly leaky because the context flanking AUG#1 is not a perfect match to the consensus sequence. With hepatitis B virus, ribosomes en route to the P start site (AUG#5) apparently bypass the weak AUG#2 by leaky scanning, while translation of the small ORF initiated at AUG#3 enables some ribosomes to miss the inhibitory AUG#4 (inhibitory because it resides in a strong context and overlaps the P ORF) and thus to reach AUG#5. Whereas the core protein start codon (AUG#1) here depicted resides in a context which allows a low level of leaky scanning, a slightly longer mRNA which encodes the pre-core protein has a stronger start codon (A in position −3) and polymerase cannot be translated from that form of mRNA (Fouillot and Rossignol, 1996). The publications on which the scheme shown here is based (Fouillot et al., 1993, Hwang and Su, 1998) also discuss some alternative possibilities vis-à-vis translation of polymerase. (C) The first AUG codon in rat histone H4 mRNA initiates translation of the full-length protein. The second AUG, 85 codons downstream and in the same reading frame, initiates production of a peptide which has growth-regulatory properties (Bab et al., 1999). (D) With rat A2AR adenosine receptor mRNA, an overlapping upORF that initiates at an AUG codon in a strong context is used to minimize production of A2AR protein. The overlapping arrangement precludes reinitiation but the not-quite-perfect context at the upstream start site allows low-level leaky scanning. This interpretation is supported by the observed ten-fold increase in translation of A2AR in vivo when the start codon of the upORF was eliminated (Lee et al., 1999). Via a second promoter, the rat A2A-R gene produces some transcripts with additional upORFs, but no transcript has yet been found that lacks the inhibitory upORF discussed here. Here and in Fig. 3, the major coding domain is shaded gray. Small regulatory ORFs (blue rectangles) are not drawn to scale.
Production of a second protein isoform via low-level leaky scanning is seen also with some cellular mRNAs. An interesting example is the production in rats of an osteogenic growth peptide (OGP) initiated from codon 85 in the histone H4 gene (Fig. 2C). The leaky scanning explanation was tested by showing that production of OGP increased upon deleting the upstream H4 start codon, and that production of OGP was suppressed upon changing the H4 start codon from a good (AGGAAGaugU) to a perfect (GCCACCaugG) context. A similar mechanism might operate with a few other cellular genes that produce a trace amount of a second protein isoform (Liu et al., 2000, Short and Pfarr, 2002; it is not clear whether leaky scanning or a change in splicing underlies the translational switch described by Land and Rouault, 1998).
The fourth example in Fig. 2 differs from the others in that a good-but-not-perfect context at the first start site serves, not to enable production of two proteins, but simply to modulate the yield of A2A-R from the second AUG. Examination of A2A-R genes from various organisms shows conservation of the overlapping ORF, with the upstream AUG codon always in a context that allows only low-level leaky scanning (Lee et al., 1999). Conservation of the structure supports the interpretation that this is a device contrived to limit the production of A2A-R protein.
Low-level leaky scanning caused by a not-quite-perfect context around the first AUG codon might occur with other mRNAs where it normally goes unnoticed because the downstream start site(s) are out-of-frame. Antigenic peptides recognized by cytotoxic T-lymphocytes (CTLs) might be produced in this way, as discussed in Section 5.4. A small degree of leaky scanning that normally goes unnoticed could become significant if a mutation that shifts the normal start codon out of frame moves a downstream AUG codon into the main reading frame. In some cases where low-level internal initiation was observed with such a mutated gene (e.g. Maser et al., 2001), the possibility that the downstream site is reached via a combination of leaky scanning and reinitiation – a mechanism such as that proposed for hepatitis B virus (Fig. 2B) – merits consideration.
4. Reinitiation
4.1. Definition and general rules
Reinitiation occurs with mRNAs, such as those depicted in Fig. 3 , that have small ORFs near the 5′ end. Our rudimentary understanding of what happens following translation of the first upORF may be summarized as follows.
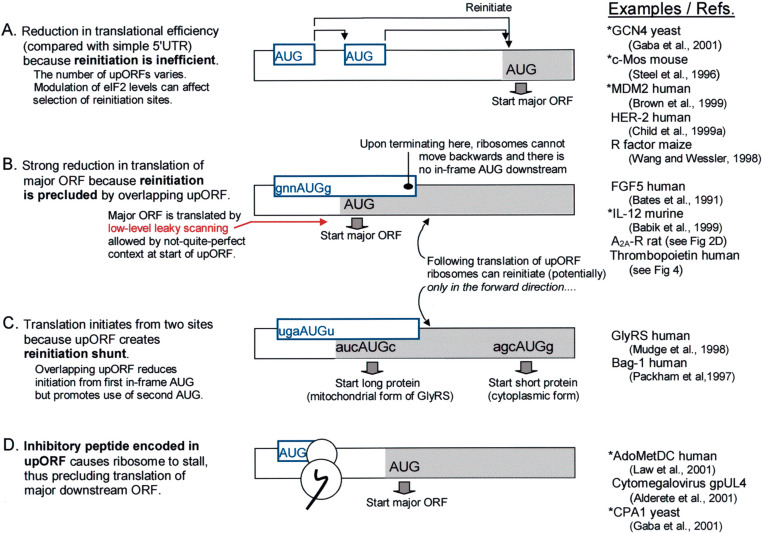
Fig. 3.
Small upstream ORFs in eukaryotic mRNAs function in various ways to modulate translation. Only the 5′ end of each mRNA is depicted. (A) The presence of upORFs forces translation of the major ORF to occur by a reinitiation mechanism, which is usually inefficient. The extent of inhibition depends on the number and arrangement of upORFs and whether the context flanking the upstream start codon(s) allows some escape via leaky scanning. (B) Because reinitiation can occur only in the forward direction, an overlapping upORF strongly impairs translation of the major ORF. (C) Whereas type B mRNAs have a single in-frame start codon which is bypassed due to the overlapping upORF, type C mRNAs initiate from two in-frame start codons; the upORF serves to divert some ribosomes to the downstream start site. The depicted sequence is a simplified representation of GlyRS mRNA (Mudge et al., 1998). Translation of Bag-1 mRNA can also be fitted to this pattern: the first start site is an in-frame CUG codon which produces the 50 kDa form of Bag-1; the next start site (AUG#1, out-of-frame) initiates a small upORF within which the first in-frame AUG codon (AUG#2) resides, and that AUG is thereby skipped; the 36 kDa form of Bag-1 is produced from AUG#3 which is accessed by reinitiation following translation of the small upORF (Packham et al., 1997). Some other mRNAs that use an upORF to dodge one AUG codon in favor of another are described elsewhere (Mittag et al., 1997, Sarrazin et al., 2000). Note that the reinitiation shunt as here defined adheres to the linear scanning mechanism, unlike a shunt postulated to operate with cauliflower mosaic virus mRNA (Ryabova et al., 2000). (D) The common feature of mRNAs that use mechanism D is inhibition of translation in cis by a peptide encoded within the upORF. The amino acid sequence of the inhibitory peptide is different in each case (Morris and Geballe, 2000). In the column at the far right, asterisks indicate examples in which the translational control mechanism is regulated, e.g. via a change in concentration of eIF2 (GCN4) or arginine (CPA1) or polyamines (AdoMetDC) or, more commonly, via an alternative promoter that generates a simpler form of mRNA devoid of upORFs (c-mos, MDM2, IL-12; see text for other examples).
When the 80S ribosome reaches the termination site of the upORF, the 60S ribosomal subunit is thought to be released (this has not actually been shown) while the 40S subunit remains bound to the mRNA, resumes scanning, and may initiate another round of translation at a downstream AUG codon. For the downstream reinitiation event to occur, the 40S subunit must reacquire Met-tRNAi and this appears to be an important point of control. Reacquisition of Met-tRNAi is promoted by lengthening the intercistronic domain (Abastado et al., 1991, Kozak, 1987c), which provides more time for Met-tRNAi to bind, or by increasing the concentration of eIF2 (Abastado et al., 1991, Hinnebusch, 1997). Genetic experiments also implicate eIF3 in the Met-tRNAi rebinding step (Garcia-Barrio et al., 1995). Another potential point of control is at the termination site of the upORF, where certain features – perhaps nearby secondary structure (Grant and Hinnebusch, 1994, Vilela et al., 1998) – might prevent the resumption of scanning or, in some other way, prevent reinitiation. This brief summary is based on studies carried out in yeast and mammals. Some studies of reinitiation in plants suggest that the intercistronic sequence may have effects beyond simply providing time for ribosomes to reacquire Met-tRNA (Wang and Wessler, 1998).
Some results obtained in early experiments with mammalian vectors were interpreted as evidence that ribosomes can scan backwards and thus reinitiate at an AUG codon positioned upstream from the termination site (Peabody et al., 1986), but recent experiments contradict that view (Kozak, 2001b). Indeed many studies have shown that the strongest inhibition is caused by an upORF that overlaps the start of the downstream cistron (Babik et al., 1999, Bates et al., 1991, Byrne et al., 1995, Cao and Geballe, 1995, Ghilardi et al., 1998, Hansen et al., 2002, Kos et al., 2002, Lee et al., 1999, Liu et al., 1999), which would not be the case if ribosomes could move backwards to reinitiate.
The size of the first ORF is a major limitation on reinitiation in eukaryotes: reinitiation can occur following the translation of a ‘minicistron’ (a small first ORF) but not following the translation of a full-length 5′ cistron. The long list of mRNAs that contain silent 3′ cistrons (Table 1) underscores the point. The only apparent exception occurs with cauliflower mosaic virus, where a protein encoded by the virus appears to promote reinitiation following the translation of a full-length first cistron (Park et al., 2001). The reason why reinitiation is usually restricted by the size of the first ORF is not known, but a possible explanation is that certain initiation factors dissociate from the ribosome only gradually during the course of elongation. If the elongation phase is brief – i.e. if the first ORF is a minicistron – the factors required for reinitiation would still be present when the 40S subunit resumes scanning. Although the postulated factors have not been identified, there is evidence for the idea that the duration of the elongation phase matters: when a short upORF which normally permits reinitiation was reconfigured to contain a pseudoknot that is known to slow elongation, reinitiation failed (Kozak, 2001b). That result makes it difficult to specify a cutoff size, i.e. one cannot say ‘an upORF this long will allow reinitiation’ while a longer ORF will not. The permissible size is likely to vary depending on features, such as secondary structure or codon usage, that affect the rate of elongation. As a rough guide, however, one may note that reinitiation often has been observed following translation of a ten to 12 codon upORF, and that reinitiation was substantially reduced, but not abolished, when a 13 codon upORF was lengthened to 33 codons (Kozak, 2001b). In a different study, reinitiation occurred following a 24 codon upORF but not when the ORF was lengthened to 40 codons (Luukkonen et al., 1995).
Some naturally occurring upORFs that strongly inhibit translation, perhaps because their size precludes reinitiation, include a 36 codon upORF in mitochondrial uncoupling protein 2 mRNA (Pecqueur et al., 2001), a 71 codon upORF in polyoma virus JC mRNA (Shishido-Hara et al., 2000), and a 53 codon upORF in plant S-adenosylmethionine decarboxylase (AdoMetDC) mRNA (Hanfrey et al., in press). That the size of the upORF might be what limits translation of AdoMetDC is suggested from the five-fold increase in translation observed when the upORF was shortened from 53 to 25 codons, but that result could also be explained in other ways. (The suggested interpretation is not contradicted by the fact that an alternative upORF in AdoMetDC mRNA caused little inhibition even when lengthened to 66 codons; the alternative upORF initiates from an AUG codon in a weak context which would allow it to be bypassed to some extent by leaky scanning. The 53 codon upORF, in contrast, has a strong start codon.)
With the mouse Snurf-Snrpn transcript, where the first cistron is 71 codons long (Gray et al., 1999), a very low level of reinitiation might account for translation of the downstream SNRPN cistron. A naturally occurring ATG-to-AGG mutation in the start codon of the upstream SNURF cistron was found to elevate translation of SNRPN >15-fold (Tsai et al., 2002), which implicates a scanning/reinitiation mechanism and rules out direct internal initiation.
4.2. Identifying candidate genes that might be regulated by reinitiation
From cDNA sequencing data, it is clear that many vertebrate mRNAs have small ORFs upstream from the start of the major coding domain, but an accurate count of genes in this class is difficult. The tallies that have been attempted (e.g. Pesole et al., 2001, Suzuki et al., 2000) are invariably flawed by inclusion of misinterpreted cDNA sequences, such as cDNAs in which a putative 5′ UTR with ‘upstream’ AUG codons turned out to be part of the coding domain or part of an intron that gets removed from the functional mRNA (Di Fruscio et al., 1998, Kozak, 1996, Kozak, 2000, Kubu et al., 2000, Nishitani et al., 2001, Santamarina-Fojo et al., 2000, Wagner et al., 1998). Some transcripts with long, AUG-burdened leader sequences are not associated with polysomes (Hake and Hecht, 1993, Sanchez-Góngora et al., 2000) or not able to support protein synthesis (Foo et al., 1994, Larsen et al., 2002, Lee et al., 2000), emphasizing that not all cDNAs correspond to functional mRNAs.
A more fundamental complication vis-à-vis which genes to count is the propensity for a single gene to produce transcripts with different 5′ leader sequences, only some of which have upstream AUG codons (Anant et al., 2002, Aplan et al., 1991, Eerola et al., 2001, Huo and Scarpulla, 1999, Kawakubo and Samuel, 2000, Laurin et al., 2000, Perälä et al., 1994, Perrais et al., 2001, Sanchez-Góngora et al., 2000, Suva et al., 1989, Tanaka et al., 2001, Tsuda et al., 2000, Zimmermann et al., 2000). The significance of a particular form of RNA cannot always be deduced from its abundance, inasmuch as a minor transcript is sometimes the major functional mRNA (Andrea and Walsh, 1995, Babik et al., 1999, Ghilardi et al., 1998, Mitsuhashi and Nikodem, 1989, Nielsen et al., 1990) and incompletely processed transcripts are sometimes more abundant than the fully-spliced, translatable mRNA (Boularand et al., 1995, Frost et al., 2000, Xie et al., 1991, Zachar et al., 1987). Translational regulation mediated by small upORFs is important, as discussed below, but equally important are non-translational mechanisms – use of alternative promoters or splice sites – that simplify the 5′ UTR by eliminating upORFs in certain tissues or at certain times when elevated synthesis of the protein is required (Aizencang et al., 2000, Anusaksathien et al., 2001, Arrick et al., 1994, Babik et al., 1999, Brown et al., 1999, Horiuchi et al., 1990, Landers et al., 1997, Lee et al., 2000, Nonaka et al., 1989, Phelps et al., 1998, Ren and Stiles, 1994, Steel et al., 1996, Teruya et al., 1990).
Because vertebrate mRNA leader sequences are often GC-rich (Section 2.4), secondary structure near the 5′ end might impair translation even more than the presence of upstream AUG codons. Thus, it is not surprising that eliminating upstream AUG codons does not improve translation in every case (Rao et al., 1988, Wood et al., 1996). In many cases, however, mutations targeted to the upstream AUG codons confirmed their role in restricting translation from downstream (Anant et al., 2002, Babik et al., 1999, Bates et al., 1991, Brown et al., 1999, Child et al., 1999a, Gereben et al., 2002, Ghilardi et al., 1998, Griffin et al., 2001, Harigai et al., 1996, Kos et al., 2002, Lee et al., 1999, Marth et al., 1988, Meijer et al., 2000, Pecqueur et al., 2001, Ren and Stiles, 1994, Steel et al., 1996, Tanaka et al., 2001, Tsai et al., 2002, Wang and Wessler, 1998, Wang and Rothnagel, 2001, Wera et al., 1995, Wu et al., 2002). This occurs by a variety of mechanisms, as summarized in Fig. 3 and discussed next.
4.3. Yeast GCN4 as an example of regulation via small upstream ORFs
While the efficiency of reinitiation varies, there is almost always a penalty – demonstrable by showing an increase in translation when the upORF is deleted – and the penalty can be severe. Thus, the simplest function of small upORFs is to limit production of the protein encoded in the full-length ORF by making downstream translation dependent on an inefficient reinitiation mechanism (Fig. 3A).
The best studied example is yeast GCN4, which initiates from the fifth AUG codon in the mRNA; the long leader sequence contains four small upORFs. In a series of classic experiments, Hinnebusch (1997) was able to reconstruct GCN4 regulation using only the first and fourth upORFs, and I will explain what happens in that simplified case. UpORF1 is always translated efficiently (it is the first AUG in the mRNA), after which ribosomes resume scanning and reinitiate, usually, at upORF4. UpORF4 is unusual in that its translation precludes further reinitiation events: thus, when upORF4 is translated, GCN4 is not. That is the situation in yeast cultures that have adequate nutrients. Starvation for amino acids, however, causes some ribosomes to bypass the inhibitory upORF4 and reinitiate instead farther downstream. This happens because starvation creates a pool of uncharged tRNAs which activate a protein kinase that phosphorylates, and thus partially inactivates, eIF2. When eIF2 levels fall, it takes longer for ribosomes to reacquire Met-tRNAi and thus become competent to reinitiate. The slower acquisition of competence means that some ribosomes, scanning in the reinitiation mode, will bypass the nearby upORF4 and can thus reach the downstream GCN4 start site.
Three general lessons from the GCN4 story appear to carry over to mammals. (i) Fig. 3A lists some examples of mammalian mRNAs that are translated inefficiently due to small upORFs; many other examples were cited in Section 4.2. (ii) Experimental manipulations with C/EBPβ mRNA (Fig. 1B) support the interpretation that an AUG codon which follows the upORF too closely is skipped (presumably because ribosomes have not yet reacquired Met-tRNAi), allowing reinitiation to occur farther downstream. The same mechanism might be invoked to explain how an internal start codon is accessed in miniTrpRS mRNA (Wakasugi et al., 2002) and baculovirus IE0 mRNA (Theilmann et al., 2001), and how c-myb gets translated from a rearranged transcript generated by retrovirus insertion (Jiang et al., 1997). In each of these mRNAs, the first AUG codon that follows a small upORF must be bypassed to reach the functional start codon downstream. (iii) The third lesson from GCN4 pertains to regulation of reinitiation by manipulation of eIF2 levels. Although hints of this have been described with mammalian genes that encode C/EBP transcription factors (Calkhoven et al., 2000), macrophage receptor protein CD36 (Griffin et al., 2001) and activating transcription factor 4 (Harding et al., 2000), the point requires much more careful study.
4.4. Human thrombopoietin – an example of severely restricted translation
The mRNAs discussed in connection with Fig. 3A have upORFs that terminate before the start of the major coding domain, thus allowing (inefficient) translation of the main ORF by reinitiation. In Fig. 3B, however, the upORF overlaps the start of the major coding domain. This precludes reinitiation and profoundly reduces the translational yield. Limited access to the main ORF in some of these mRNAs might be achieved by leaky scanning, as was discussed for A2A-R (Fig. 2D).
mRNAs derived from the human thrombopoietin (TPO) gene have structures similar to that depicted in Fig. 3B and much can be learned from the TPO story, as outlined in Fig. 4 . The normal gene produces a mixture of transcripts with different leader sequences, all of which translate TPO poorly because of an overlapping upORF (upORF7 in Fig. 4). Targeted mutagenesis (Ghilardi et al., 1998) confirmed that upstream AUG#7 is primarily responsible for blocking translation of TPO. This is because its near-optimal context (GCCGCCUCCaugG) prevents leaky scanning and the overlapping arrangement precludes reinitiation.
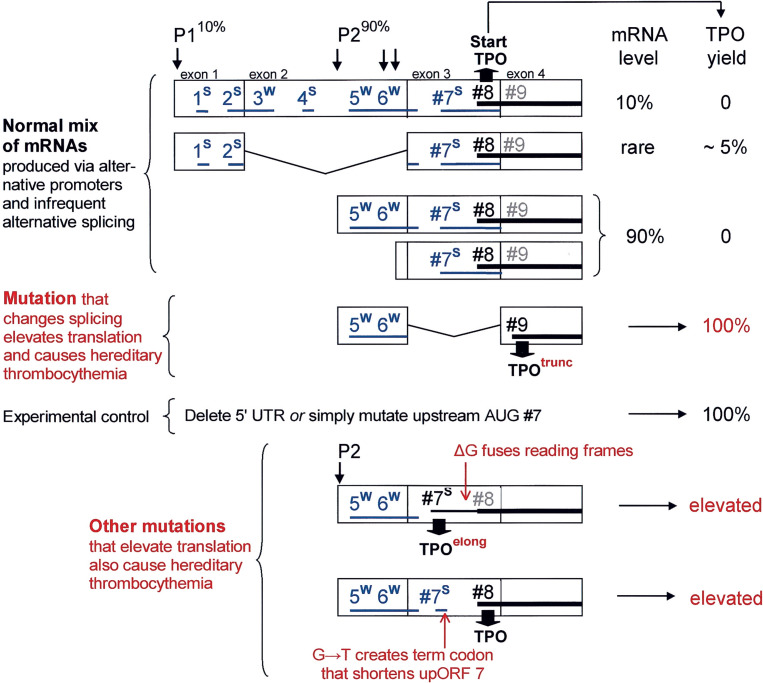
Fig. 4.
A low-level reinitiation mechanism normally prevents overproduction of TPO. Translational yields from various forms of TPO mRNA in transfected COS cells (far right column) are expressed relative to a control transcript that has a short, unencumbered 5′ UTR. P1 and P2 are alternative promoters; a cluster of arrows indicates that P2 produces staggered start sites. The TPO coding domain (horizontal black bar) begins at an AUG codon which is labeled #8 because, in the longest form of mRNA (line 1), it is preceded by seven AUG codons that initiate small upORFs. Superscript letters indicate whether each upstream AUG resides in a strong (S) or weak (W) context and horizontal blue lines depict the approximate length and arrangement of the upORFs. Vertical lines demarcate the boundaries of exons; carets depict the introns in alternatively spliced transcripts. Only the beginning of the TPO coding domain (exons 3–7) is shown. The key point is that the normal set of transcripts supports translation poorly because upORF7 overlaps the TPO start site. Various mutations (shown in red) that relieve this constraint elevate the translation of TPO, and this overproduction causes hereditary thrombocythemia. Among the normal set of mRNAs, the ‘rare’ transcript from promoter P1 (line 2) supports translation slightly better than the others, perhaps because the short distance between upORF2 and AUG#7 enables some reinitiating ribosomes to bypass AUG#7 and thus reach AUG#8. Because of the strong context at AUGs #1 and #2, upORFs 1 and 2 would be more effective than upORFs 5 and 6 in setting up this reinitiation shunt. The depicted scheme is based on experiments described by Ghilardi et al. (1998) and Wiestner et al. (1998). Additional mutations diagrammed near the bottom of the figure were described by Ghilardi and Skoda (1999), Ghilardi et al. (1999), and Kondo et al. (1998).
Various mutations that restructure the 5′ UTR in ways that increase production of TPO cause hereditary thrombocythemia. Translation of TPO normally initiates at AUG#8 in exon 3, but a splice-site mutation that causes deletion of exon 3 causes initiation to shift to a previously silent in-frame codon (AUG#9) in exon 4; this is diagrammed in the center of Fig. 4. The resulting truncated form of TPO lacks only the first four amino acids and appears to function normally (Wiestner et al., 1998). The problem – the cause of the pathology – is that the mutation greatly increases translation of TPO by removing the inhibitory upORF7. In two other families affected with hereditary thrombocythemia, production of TPO is elevated by mutations that restructure upORF7. In one case, deletion of a G residue shifts upORF7 into the same reading frame as TPO, thereby causing overproduction of an elongated form of TPO initiated from AUG#7 (Ghilardi and Skoda, 1999, Kondo et al., 1998). In the other case, a G→T mutation creates a terminator codon within upORF7 and this shortening of the ORF, which now terminates 31 nt before AUG#8, enables efficient reinitiation at AUG#8 (Ghilardi et al., 1999).
These insightful studies of TPO expression make two important points: (i) the bulk of the transcripts produced by the wild type gene are virtually untranslatable; and (ii) it is necessary for this potent cytokine to be translated poorly; overproduction results in disease. With TPO as precedent, one suspects that in other cases where – despite the production of alternative leader sequences – it is hard to find even one form of mRNA devoid of upstream AUG codons (e.g. Larsen et al., 2002, Lee et al., 1999, Pecci et al., 2001, Peterson and Morris, 2000, Wang et al., 1999), the goal is to ensure that translation is very, very inefficient. The wig-1 growth-regulatory gene might be another example: an overlapping upORF initiates from a strong AUG codon, while the wig-1 start codon itself is weak, and these distinctive features are conserved between the human and mouse genes (Hellborg et al., 2001).
4.5. Other uses of upstream ORFs
Whereas an overlapping upORF functions simply to down-modulate translation in the examples depicted in Fig. 3B, with the mRNAs in Fig. 3C the overlapping upORF qualitatively affects the protein output. Ribosomes that translate the upORF thereby miss the first in-frame AUG codon but proceed to reinitiate at another start codon downstream. If the upORF itself has a suboptimal initiation site (U in position −3 in the depicted example), leaky scanning will allow some production of the long protein isoform from the first in-frame AUG codon while the reinitiation shunt promotes production of the shorter protein isoform. The operation of a reinitiation shunt is most obvious when the upORF overlaps a potential start codon, as shown in Fig. 3C, but the same principle applies in cases (discussed in Section 4.3) where, although the upORF terminates prior to a potential downstream start codon, the intervening distance is too short to allow reinitiation.
The fourth regulatory mechanism diagrammed in Fig. 3 is used only rarely. Mammalian AdoMetDC mRNA is the best studied example in which a small upORF encodes a peptide which functions in cis to inhibit downstream translation. The nascent peptide (MAGDIS) produced during translation of the upORF is thought to interact with ribosomes in a way that prevents completion of the termination process and thus prevents reinitiation (Law et al., 2001). The stalled ribosome, held at the termination site of the upORF, would also block other ribosomes from reaching the downstream start site via leaky scanning. Biologically, this mechanism is important because AdoMetDC is a key enzyme in polyamine biosynthesis and, at least in vitro, elevated polyamine levels stabilize the ribosome complex stalled at the end of the upORF (Law et al., 2001). In other words, elevated polyamine levels down-regulate translation of AdoMetDC. It is interesting to note parenthetically that antizyme, a protein that down-regulates polyamine levels, is also translated via a polyamine-sensitive mechanism. Elevated polyamine levels up-regulate production of antizyme by promoting a ribosomal frameshift needed to translate the full-length protein (Ivanov et al., 2000).
The foregoing examples illustrate how reinitiation operates as part of the normal translation mechanism in cases where upORFs are constitutively present in mRNAs. There are other cases in which a reinitiation mechanism kicks in only when a nonsense mutation is introduced in a way that truncates the coding domain. In effect, the normal AUG initiator codon becomes the start of an upORF, following which reinitiation occurs at a normally silent internal AUG codon (Chang and Gould, 1998, Ledley et al., 1990, Zoppi et al., 1993). The N-terminally truncated protein thus produced sometimes retains enough function to mitigate the pathological effects of the nonsense mutation (Chang and Gould, 1998). This potential rescue device often fails, however, because many mRNAs are rapidly degraded when a nonsense codon is introduced (Frischmeyer and Dietz, 1999, He and Jacobson, 2001). The mRNA decay pathway that targets these abnormal mRNAs is activated in part by cis elements located in the coding domain (Gudikote and Wilkinson, 2002), which might explain why normal upORF-containing mRNAs (e.g. those discussed in Fig. 3, Fig. 4) are not rapidly degraded.
5. Changes in mRNA structure and translation linked to human diseases
Initiation factor eIF2 plays a key role in translational control (Clemens, 2001, Dever, 2002) and mutations that perturb regulation of eIF2 have profound pathological consequences (Delépine et al., 2000, Han et al., 2001, Harding et al., 2001, Van der Knaap et al., 2002). Human genetic disorders have been traced also to disruption of regulatory mechanisms mediated by mRNA binding proteins (Brown et al., 2001, Cazzola and Skoda, 2000, Cazzola et al., 2002, Kaytor and Orr, 2001, Mikulits et al., 1999). Here, however, I focus on pathologies resulting from increases or decreases in translation caused directly by changes in mRNA structure. The preceding paragraph mentioned some cases in which an N-terminally truncated protein is produced, apparently by reinitiation, when a mutation introduces a premature nonsense codon. The effects of some other types of mutations can also be understood in light of the scanning mechanism, as outlined next and discussed elsewhere in more detail (Kozak, 2002).
Recent investigations have identified diseases that result from failure to produce one of the two protein isoforms derived from genes that encode certain transcription factors (Table 4). Because the second isoform often functions as a modulator, the transcriptional imbalance caused by these changes in translation can have serious consequences.
Table 4.
Pathologies resulting from a change in mRNA structure which selectively abolishes production of the long or short form of a transcription factor
| Gene | Translational mechanism that normally generates two protein isoforms | Disease-associated change in mRNA structure and translation | References |
|---|---|---|---|
| C/EBPα (human) | Two proteins from one mRNA via leaky scanning+reinitiation shunt | In acutemyeloidleukemia, mutations near amino terminus eliminate production of longer isoform. | Pabst et al., 2001 |
| GATA1 (human) | Two proteins from one mRNA via leaky scanning | In Downsyndrome-relatedleukemia, premature stop codon eliminates production of longer isoform. | Wechsler et al., 2002 |
| LEF1 (human) | Two proteins from two mRNAs (via two promoters) | In coloncancer, failure to activate downstream promoter prevents production of shorter (inhibitory) isoform. | Hovanes et al., 2001 |
| Rx/rax (mouse) | Two proteins from one mRNA via leaky scanninga | In eyeless mice, mutation of second AUG, leaving only the weak upstream start codon, results in inadequate yield. | Tucker et al., 2001 |
Here the long and short isoforms appear to function identically; the significance of the second AUGSTART codon pertains to boosting the overall protein yield. The eyeless mouse serves as a spontaneous model for human anophthalmia.
5.1. AUG start codon and context mutations
Hereditary diseases have been traced occasionally to point mutations that alter the context flanking the AUGSTART codon. The list includes α-thalassaemia caused by an A→C change in position −3 of the α-globin gene (Morlé et al., 1985), androgen insensitivity syndrome caused by a G→A mutation in position +4 of the androgen receptor gene (Choong et al., 1996), and vitamin E deficiency associated with a C→T mutation in position −1 of the α-tocopherol transfer protein gene (Usuki and Maruyama, 2000). There is an interesting report of a somatic point mutation (G→C in position −3) in the BRCA1 gene in a highly aggressive case of sporadic breast cancer (Signori et al., 2001). In mice, a screen for mutations that cause defects in eye development uncovered an A→T change in position −3 of the Pax6 gene (Favor et al., 2001). Each of these mutations was shown to cause a decrease (generally two- to four-fold) in translation.
Not every mutation or polymorphism within the consensus motif can be explained simply, however. Other considerations, such as codon usage, might prevent an increase in translation even when the context is improved (i.e. translation might be limited at the level of elongation rather than initiation), and some mutations near the AUG codon might affect mRNA processing or stability rather than translation per se. A clinically relevant polymorphism in position −1 of annexin V appears to have an effect on translation which is inconsistent with the context rules (González-Conejero et al., 2002), but the effect was small and documented only by assaying translation in vitro, which is not always reliable (Section 6.2). A natural polymorphism in position −5 of the glycoprotein Ibα gene displayed a small effect on translation in vitro that was consistent with the rules (C worked better than T; Afshar-Kharghan et al., 1999) but, in the same study, mutations that changed position +4 from C to G did not augment translation. Testing mutations in position +4 is tricky, however, because the change in identity of the penultimate amino acid might affect protein stability in ways that obscure the effects on translation. The solution is to use an assay that directly monitors the initiation step of translation (Kozak, 1997).
The scanning mechanism predicts that a mutation which weakens or destroys the normal start codon should activate initiation from the next AUG downstream. In some hereditary diseases in which the AUGSTART codon is ablated, a truncated protein is indeed produced in this way but it does not function well enough to prevent the disease (Cahana et al., 2001, Huang et al., 1999, O'Neill et al., 2001). In the case of a mutated vasopressin gene in which the G of the AUGSTART codon is deleted, the shorter signal peptide initiated from the second AUG codon is not recognized by signal peptidase (Beuret et al., 1999). The resulting uncleaved vasopressin-precursor protein folds incorrectly, causing subsequent processing steps to fail, and therefore vasopressin never gets released from the endoplasmic reticulum. The second AUG is only four codons downstream from the first, but the processing defect caused by this slight shift in the site of initiation causes diabetes insipidus.
5.2. Restructuring cellular mRNAs by addition or removal of upstream AUG codons
The scanning mechanism predicts that, when an out-of-frame AUG codon is introduced into the 5′ UTR, the adventitious upstream start codon should supplant the normal start site. A number of pathologies result from this kind of translational block. Sometimes the upstream AUG codon is created by a rare mutation (Cai et al., 1992, Liu et al., 1999). Other times it derives from a common polymorphism (Bergenhem et al., 1992, Endler et al., 2001, Kanaji et al., 1998, Kraft et al., 1998, Zysow et al., 1995). The reduction in translation is more or less severe depending on the context of the upstream AUG codon and whether reinitiation is possible.
I have already explained how hereditary thrombocythemia is caused by mutations that elevate translation of TPO by restructuring or eliminating an inhibitory upORF (Fig. 4). Translation of proto-oncogenes is also elevated in some cases by eliminating small upORFs from the mRNA. The MDM2 oncogene is one example: whereas the normal mRNA has a long 5′ UTR that includes two upstream AUG codons, in tumor cells the use of a different promoter eliminates the upstream AUGs and thus increases translational efficiency 20-fold (Brown et al., 1999, Landers et al., 1997). In the case of oncogene GLI1, the upstream AUG codons that restrict translation in normal cells reside in an intron which is eliminated by more efficient splicing in basal cell carcinomas (Wang and Rothnagel, 2001). Translation of many other human or rodent oncogenes is restricted in normal cells by an encumbered (AUG-burdened or GC-rich) leader sequence (Arrick et al., 1991, Bates et al., 1991, Child et al., 1999a, Harigai et al., 1996, Hoover et al., 1997, Horvath et al., 1995, Manzella and Blackshear, 1990, Sarrazin et al., 2000); in some of these cases, a shorter 5′ UTR that better supports translation is produced in transformed cells (Arrick et al., 1994, Marth et al., 1988). For other oncogenes, although there are alternative leader sequences that might regulate expression in normal tissues (Link et al., 1992, Sasahara et al., 1998), there is no evidence that switching leader sequences contributes to tumorigenesis.
Whereas removal of small upORFs elevates the translation of the aforementioned MDM2 and GLI1 oncogenes in tumor cells, addition of small upORFs shuts off the translation of some tumor suppressor genes. In the case of HYAL1, retention of an intron which contains eight upstream AUG codons renders the mRNA untranslatable in squamous cell carcinomas (Frost et al., 2000). A striking example of translational inactivation of a tumor suppressor gene is seen in some individuals with a predisposition to melanoma. In certain families, a point mutation (G→T) creates an upstream, out-of-frame AUG codon in the CDKN2 gene (Liu et al., 1999). The small upORF initiated from this new AUG codon overlaps the CDKN2 start codon, and the resulting inhibition of translation is profound.
5.3. Modulation of viral infectivity by restructuring mRNAs
Structural changes that attenuate the translation of viral mRNAs can contribute to the development of persistent infections. The 5′ leader sequence on bovine coronavirus mRNAs, for example, was found to evolve – by acquiring a small upORF – during the course of establishing a persistent infection (Hofmann et al., 1993). Shishido-Hara et al. (2000) speculate that human polyomavirus JC might cause persistent rather than acute infection because all capsid-protein encoding transcripts produced by the JC virus have a small upORF. With the related simian virus 40, in contrast, the upORF is sometimes eliminated by splicing, generating transcripts that better support translation of the major capsid protein. Attenuating effects caused by introducing an upstream AUG codon have been described also with other viruses (Petty et al., 1990, Slobodskaya et al., 1996).
A more drastic restructuring of mRNAs sometimes occurs during the establishment of persistent infections by the measles virus. Instead of the normal monocistronic mRNA for the fusion protein, the predominant transcript in some persistently infected cells was a dicistronic mRNA from which the F cistron, located at the 3′ end, could not be translated (Hummel et al., 1994). A similar problem encountered in studies with recombinant rhabdoviruses provides insight into the transcriptional defects that can generate untranslatable dicistronic mRNAs (Quiñones-Kochs et al., 2001).
In the case of a human parvovirus, productive infection is restricted to a subset of erythroid cells in which splicing generates a monocistronic mRNA for each of the major capsid proteins. In nonpermissive cells, a slight shift in the position of a splice site imposes an upstream ORF which is postulated to restrict translation of the capsid proteins (Brunstein et al., 2000).
5.4. Production of CTL antigens
Translational twists sometimes generate antigens which, by stimulating the CTL response, are important in the host defense against tumor cells and viruses (Shastri et al., 2002). Leaky scanning is a likely explanation in several cases where the major ORF starts with an AUG codon in a suboptimal context and the CTL antigen derives from initiation at the next (out-of-frame) AUG (Aarnoudse et al., 1999, Bullock et al., 1997, Probst-Kepper et al., 2001, Rimoldi et al., 2000). In one notable case, translation shifts upstream to an in-frame AUG codon created during insertion of a provirus, and the resulting novel N-terminal amino acid extension functions as a tumor rejection antigen (Wada et al., 1995).
The scanning mechanism cannot explain the translation of CTL antigens for which the start codon resides far in the interior of the mRNA (Ronsin et al., 1999, Wang et al., 1996). In these cases the antigenic peptide might be produced from an undetected alternative form of mRNA. Sensitive new assay techniques employed with some genes indeed reveal an array of alternative transcripts from which novel tumor antigens can be translated (Behrends et al., 2002). In another study, a potent tumor rejection peptide, which maps to an internal AUG codon in the full-length cDNA, was expressed experimentally from a truncated cDNA wherein the start codon for the antigenic peptide was made the first AUG (Rosenberg et al., 2002). Additional analyses are needed to determine whether, in the melanoma cells wherein this antigen is expressed naturally, a transcript similar to the experimentally truncated cDNA is produced via a downstream promoter or splice site.
6. Surveys and assays and problems therein
6.1. cDNA surveys
Surveys of mRNA/cDNA sequences differ in other details, but every survey confirms the presence of a purine in position −3 in most (≥90%) vertebrate mRNAs (Kozak, 1987a, Pesole et al., 2000, Rogozin et al., 2001, Sakai et al., 2001). The occasional survey that purports to challenge the context rules involves distortions, such as emphasizing the low percentage of cDNAs that have the full consensus sequence while ignoring the high percentage of cDNAs that have the critical purine in position −3 (Peri and Pandey, 2001). A major uncertainty pertaining to all cDNA surveys concerns the validity of the database. When I re-examined the entries in one study (Suzuki et al., 2000), I found numerous instances in which the AUGSTART codon had been misidentified; the corrected start sites adhered more closely to the consensus motif (Kozak, 2000). Some authors pre-emptively defend their conclusions on the grounds that the (unidentified) cDNA sequences used for their analysis derive from RefSeq, which is a curated database (Pruitt et al., 2000). But the entries in RefSeq are not without errors, some of which – e.g. misidentified start codons, mistaken claims of upstream AUG codons – can be traced by comparing curated GenBank entries NM_005493, NM_005502, NM_000282 and NM_003605 with results published elsewhere (Campeau et al., 2001, Nishitani et al., 2001, Nolte and Müller, 2002, Santamarina-Fojo et al., 2000).
Some cDNA surveys use misleading terminology, e.g. referring to upstream AUG codons as ‘unused’ (Peri and Pandey, 2001). Given that upstream AUG codons are used, as proved by detecting the encoded peptide or by fusing the upORF to a reporter gene, it is not anomalous to find a good context around some upstream AUG codons. All surveys tend to overestimate the incidence of upstream AUGs by scoring only the longest cDNA isoform, ignoring the existence of alternative transcripts that have shorter, unencumbered 5′ UTRs (Section 4.2). The significance of upstream AUG codons also tends to be misstated: the presence of small upORFs in vertebrate mRNAs which are thereby translated inefficiently (see the foregoing discussion of TPO, oncogenes, etc.) constitutes evidence for, rather than against, the scanning model. The first-AUG rule, which I cite as evidence for the scanning mechanism, derives not from statistical analysis of cDNA sequences but from the experimentally observed fact (Section 2.2) that translation shifts predictably upstream or downstream when an AUG codon is added or removed. In short, it makes more sense to use the scanning/context rules to evaluate cDNA sequences (Hatzigeorgiou, 2002) than to attempt the reverse.
6.2. Assays used to study translation
While conclusions about translation derived from experimental studies are arguably more meaningful than those derived from statistics, the interpretation of experimental results can be complicated. In vivo assays avoid the problem of reaction-conditions-dictating-the-outcome (see next paragraph), but there are other potential traps. The usually-valid assumption that polysomal association identifies actively translated mRNAs is called into question by the recent discovery of mRNAs trapped on large polysomes from which there is no polypeptide production (Rüegsegger et al., 2001). The major problem when translation is studied in vivo is uncertainty about the structure of the mRNA. For example, a claim that IRES-mediated translation is developmentally regulated (Créancier et al., 2000, Créancier et al., 2001) is premature, inasmuch as those studies monitored the amount but not the form of mRNA. Other studies wherein translation of an encumbered leader sequence appears to improve under certain conditions or in certain cell types require better analyses to rule out a possible change in structure of the 5′ UTR (Bernstein et al., 1995, Child et al., 1999b, Li et al., 2001, Zimmer et al., 1994). Some useful hints may be found in reports that describe the belated discovery of alternative forms of mRNA that were missed the first time around (Chen et al., 1999, Cortner and Farnham, 1990, Déjardin et al., 2000, Deng et al., 2002, Frost et al., 2000, Grundhoff and Ganem, 2001, Jordan et al., 1996, Kastner et al., 1990b, Kiss-László et al., 1995, Laurin et al., 2000, Peremyslov and Dolja, 2002, Zhang and Liu, 2000, Zheng et al., 1994).
In vitro translation assays pose a different set of problems. The commercial availability of in vitro translation kits is both a blessing – the systems are easy to use – and a curse. The latter because insufficient attention is paid to reaction conditions that can affect the selection of AUG start codons. When the magnesium concentration is too low, the first AUG codon may be bypassed despite an adequate context; when the magnesium concentration is too high, initiation may occur at upstream non-AUG codons that are not naturally used. One solution is to include control transcripts for which start-codon selection was determined in vivo, and to adjust the in vitro reaction conditions to give the same result (Kozak, 1990b). Some suppliers of translation kits make it possible to adjust the magnesium concentration, but there is little awareness of the need to do so and the use of coupled transcription/translation systems makes it difficult.
For whatever reason, in vitro translation results sometimes deviate significantly from what is seen in vivo vis-à-vis access to internal AUG codons (Grove et al., 1991, Land and Rouault, 1998, Meijer et al., 2000, Meulewaeter et al., 1992, Mitchelmore et al., 2002, Saucedo et al., 1999) and the degree of inhibition caused by small upORFs (Ghilardi et al., 1998, Harigai et al., 1996, Pecqueur et al., 1999, Pecqueur et al., 2001, Tanaka et al., 2001, Wang and Wessler, 1998). The fidelity of initiation in vitro is clearly impaired, possibly due to degradation of the mRNA, in cases where extraneous, low molecule weight polypeptides are produced (Herbert et al., 1996, Liu and Biegalke, 2002, Lekven et al., 2001, Maser et al., 2001, Packham et al., 1997).
The possibility that the input mRNA might undergo cleavage during incubation in vitro complicates attempts to study the expression of dicistronic mRNAs, as discussed in the next section. This type of artifact is not ruled out by finding that only certain downstream ORFs are translated (O'Connor and Brian, 2000). Extrapolating from what is seen when mRNAs are deliberately cleaved in vivo (Thoma et al., 2001), activation of internal start codons in vitro would depend on where the accidental cleavage occurs and whether the endolytic cleavage product persists long enough for a ribosome to engage the newly created 5′ end before exonucleases take over. This line of reasoning could explain the claim that an ‘artificial IRES’, consisting of a multiple cloning region and a portion of the Escherichia coli lacI gene, supports internal initiation of translation in starved yeast cells (Paz et al., 1999): starvation is likely to promote mRNA degradation, and the ‘IRES’ might fortuitously stabilize certain intermediates in degradation. The discovery that IRES elements are actually targeted by some ribonucleases (Elgadi and Smiley, 1999, Nadal et al., 2002) should be remembered.
Recent studies that use a primer-extension inhibition (toeprinting) assay to monitor the binding of ribosomes to mRNAs have the advantage of focusing directly on the initiation step, but care is needed to distinguish authentic initiation complexes from artifactual pauses in primer extension caused by base-paired structures or extraneous proteins bound to the mRNA. The complicating effects of mRNA secondary structure, which are prominent when avian reverse transcriptase is used for toeprinting, can be minimized by using a form of the enzyme derived from murine leukemia virus (Kozak, 1998).
6.3. Studies of eIF4G translation illustrate some common problems
Attempts to explain the origin of multiple isoforms of eIF4G (Bradley et al., 2002) illustrate how challenging it can be to interpret translation assays. In vitro experiments presented in support of the idea that translation can initiate from five AUG codons, in a single form of eIF4G mRNA, might have been compromised by mRNA breakage; this would explain the production of an array of extraneous smaller polypeptides (Byrd et al., 2002, Fig. 3C, lanes 1, 3 and 5). Translation of some eIF4G isoforms from broken mRNAs could also explain the variability in yields noted throughout that study. When the endogenous eIF4G gene is expressed in vivo, access to certain downstream AUG codons might occur via alternative splicing or internal promoters; both mechanisms have been documented in studies of eIF4G by other investigators (Han and Zhang, in press).
Thus, even though one could rationalize the production of at least three isoforms of eIF4G from one mRNA via established translational mechanisms – an overlapping upORF could shunt some 40S ribosomal subunits past the first in-frame AUG codon (position 275), and the unfavorable context at AUG 395 might allow some ribosomes to reach AUG 536 by leaky scanning – it would be premature to propose that solution. The in vitro experiments need to be repeated with careful attention to magnesium levels and with efforts to minimize mRNA breakage. The latter might be accomplished by lowering the temperature to 25 °C and limiting the window for initiation to 5 or 10 min. (Addition of edeine after the first 5 or 10 min, followed by another period of incubation, would allow polypeptides to be elongated without further initiation events.) The possible production of some eIF4G isoforms by proteolysis also needs to be ruled out, as this protein is notoriously susceptible to cleavage.
The study by Byrd et al. (2002) included experiments carried out with dicistronic transcripts, predicated on the belief that eIF4G mRNA contains IRES elements which allow direct internal initiation of translation. An eIF4G/EGFP (enhanced green fluorescent protein) fusion gene positioned at the 3′ end of a dicistronic transcript was translatable in vitro, but the aforementioned possibility of mRNA breakage complicates the interpretation. Indeed, the unexpected translation of EGFP from the 3′ position even without fusion to eIF4G (Byrd et al., 2002, Fig. 7A, lane 3) is most easily explained by mRNA cleavage. (The authors invoke reinitiation as the explanation, but reinitiation cannot occur following translation of a large 5′ cistron.) Fragmentation of the mRNA could explain why translation of the 3′ eIF4G/EGFP cistron persisted when translation of the 5′ cistron was blocked by a hairpin structure (Byrd et al., 2002, Fig. 7B). The hairpin test, widely used to test for internal initiation, is meaningless without evidence that the dicistronic input mRNA remains intact.
The in vivo tests of eIF4G translation (Byrd et al., 2002, Fig. 8) also require careful RNA analyses to document that the vector produces only the intended dicistronic mRNA; the quality of the Northern blot in that figure falls far short of what is required. To rule out the possibility that the 3′ cistron might be translated from an unintended monocistronic mRNA, a promoter-deletion control is needed – a control which shows that, upon deleting the promoter that precedes the 5′ cistron, expression of the 3′ cistron is also abolished. This test failed in studies with some other sequences, revealing that the candidate IRES actually harbors a cryptic promoter (Han and Zhang, 2002, Han et al., 2002, Larsen et al., 2002).
The foregoing discussion of eIF4G translation alludes to only some of the problems associated with dicistronic vectors; a more detailed critique may be found elsewhere (Kozak, 2001a, Kozak, 2001b). Use of a certain popular vector which harbors an intron near the 5′ end (Jopling and Willis, 2001) increases the likelihood of producing an unintended monocistronic mRNA via splicing; the candidate IRES need contribute only a cryptic 3′ splice site. This indeed happens in some cases (Grundhoff and Ganem, 2001, Pinkstaff et al., 2001). Claims of IRES activity are problematic when supported by in vitro assays in which translation of the 3′ ORF is very, very weak (e.g. Deffaud and Darlix, 2000, Lekven et al., 2001, Maser et al., 2001). The simple idea that an IRES can be identified based on the ability to support translation of a 3′ cistron runs into trouble when, for example, the β-globin mRNA leader sequence, intended to serve as a negative control, turns out somehow to allow translation of a downstream cistron (Van der Velden et al., 2002). In another study, merely lengthening the intercistronic domain enabled substantial translation of the 3′ cistron (Gallie et al., 2000), perhaps by providing room for RNases to cleave and thus release a translatable 3′ fragment. Even with the paradigmatic IRES elements derived from picornaviruses, the ability to support internal initiation was found to depend inexplicably on the choice and arrangement of 5′ and 3′ reporter genes (Hennecke et al., 2001). These oddities – along with the notable inability to translate the 3′ cistron in natural dicistronic mRNAs (Table 1) – are reason to worry about the validity of experiments that employ synthetic dicistronic constructs.
The proffered rationale for a cap-independent internal initiation mechanism is that it would enable certain mRNAs to be translated when eIF4E levels decline, but recent experiments presented in support of that idea (Li et al., 2002) used the 5′ UTR from poliovirus rather than 5′ UTRs from cellular mRNAs, such as eIF4G, that are supposedly regulated via ‘a dynamic interplay between cap-dependent and cap-independent processes’. Even if the proffered rationale is valid, convincing evidence for direct internal initiation with particular mRNAs is needed. The widely used dicistronic assay has flaws, as outlined above. An alternative assay which involves circularization of the mRNA has been attempted with only one viral IRES element (Chen and Sarnow, 1995); the results await independent verification and extension to other sequences.
7. Conclusions and closing notes
The scanning model provides a framework for understanding basic patterns of eukaryotic gene expression, such as the reliance on monocistronic mRNAs, and for understanding how translation is perturbed by mutations that restructure the 5′ UTR. A growing number of human diseases have been traced to such mutations. The scanning mechanism has been shown to operate not only with simple mRNAs that have a short 5′ UTR and initiate at the first AUG codon, but also with mRNAs that have complicated leader sequences and multiple start codons.
One often hears the suggestion that an alternative, IRES-mediated mechanism of initiation is required when a long leader sequence is encumbered by secondary structure or upstream AUG codons (Dever, 2002, Pestova et al., 2001). That view is not well taken. Scanning can occur over long distances, as evidenced by some bifunctional viral mRNAs in which the second start site is more than 500 nt downstream from the first (e.g. peanut clump virus, southern bean mosaic virus, rice tungro bacilliform virus; Table 3 and Fig. 1C). The structure-prone, GC-rich leader sequences on mammalian mRNAs strongly reduce translational efficiency but do not preclude operation of the scanning mechanism (Van der Velden et al., 2002). Upstream AUG codons also reduce translational efficiency and that is why they are there. To postulate the need for an alternative mechanism is to miss the point that an encumbered leader sequence ensures that translation via scanning will be inefficient, and thus ensures against harmful overproduction of cytokines (Fig. 4) and other potent proteins.
The high frequency of intron-containing cDNA sequences (Kozak, 1991a, Kozak, 1996) might reflect another type of regulation. Inefficient or regulated removal of the first intron has been documented in some cases (Boularand et al., 1995, Frost et al., 2000, Van der Leij et al., 2002, Wang and Rothnagel, 2001, Xie et al., 1991, Zachar et al., 1987) and I suspect that additional examples might be found – miscategorized – among the aforementioned cDNAs that are postulated to require an alternative mechanism of initiation. Removal of the intron, or use of a cryptic promoter therein, would eliminate the upstream AUG codons that are barriers to scanning. Examples in which translation is prevented deliberately by splicing-out the exon that contains the AUGSTART codon (Lin et al., 1998) or by other regulated splicing events (Rueter et al., 1999) underscore the point that not every transcript – hence not every cDNA – corresponds to a functional mRNA. Before postulating the need for a new mechanism to explain how a funny looking cDNA gets translated, one must be certain that it is translated.
The mechanisms discussed herein for escaping the first-AUG rule, within the constraints imposed by the scanning model, obviously cannot explain every report of initiation from an internal position. More information is needed to understand how N-terminally truncated versions of some proteins are produced apparently without truncation of the mRNA (Goss et al., 2002, Maser et al., 2001, Santagata et al., 2000, Scharnhorst et al., 1999, Vanhoutte et al., 2001). An IRES element was postulated in some of those cases, based on the dicistronic test, but in one study there were no accompanying analyses of RNA structure in vivo (Goss et al., 2002), and in another study the use of an in vitro translation system produced too little of the truncated protein to be convincing (Maser et al., 2001). Speculation about how some other interesting genes are translated (Klemke et al., 2001) also must be postponed pending a search for possible additional forms of mRNA. Although I listed the von Hippel-Lindau tumor suppressor gene as a possible example of leaky scanning (Table 3), definitive tests are needed to distinguish between that and other mechanisms for producing the short isoform (Iliopoulos et al., 1998).
Those of us with an interest in translation have a tendency to interpret every change in mRNA structure as a means to control translation, but transcriptional requirements – the need to turn on a gene in various tissues via whatever promoter works in each tissue – underlie most switches in 5′ leader sequences. In some cases the actual sequence of the 5′ UTR is dictated by the presence therein of transcriptional control elements (Akiri et al., 1998, Minami et al., 2001, Solecki et al., 1997, Yin and Blanchard, 2000, Yu et al., 2001, Zimmermann et al., 2000). Regulation of transcription is the major reason for the GC-rich domains near the 5′ end of many mammalian genes; the accompanying down-modulation of translation is an inevitable consequence – arguably a useful consequence because, given the long half-life of most mammalian mRNAs, inefficient translation might be a necessity.
It merits repeating that, although the m7G cap strongly promotes ribosome binding, the scanning mechanism is not dependent on the presence of the cap. The essence of the scanning model is 5′ entry of ribosomes and position-dependent selection of the AUGSTART codon. Those key points hold with naturally uncapped mRNAs produced by some viruses (footnote g in Table 1) and with synthetic uncapped mRNAs used to study translation in vitro (Kozak, 1979a, Kozak, 1998). The inclination to invoke internal initiation based on indirect criteria – absence of a cap, or the ability to be translated in extracts from poliovirus-infected cells – should be resisted. It is a mistake to think that, because archaeal mRNAs lack a 5′ cap, translation in that system cannot occur via scanning. The discovery in archaea of proteins similar to certain eukaryotic initiation factors (Kyrpides and Woese, 1998) is intriguing for other reasons but has no direct bearing on whether the start codon in archaeal mRNAs might be recognized via a prokaryotic- or eukaryotic-type mechanism. That interesting question, which bears on the evolutionary origin of scanning, awaits answering.
Fundamental questions about the molecular workings of the scanning mechanism also await answering. What drives migration of the 40S subunit during the scanning phase? How does the 40S subunit hold on at a terminator codon, in order to reinitiate? What prevents reinitiation when the size of the first ORF exceeds a certain length? We know nothing about how recognition of the start codon is aided by a purine in position −3 and G in position +4. There is no evidence for base pairing between the GCCRCC motif and rRNA (or for binding of rRNA to any other sequence in eukaryotic mRNAs). There is as yet no convincing evidence for recognition of GCCRCC by a trans-acting protein factor. It would be easy, and meaningless, simply to find proteins that bind an RNA fragment which contains the motif. Credible experiments would require controls based on what we know about the consensus sequence: that the purine (A>G) in position −3 plays a dominant role, and the full effect requires that the GCCRCC motif abut the AUG codon (Kozak, 1999, Fig. 1).
With so much effort being directed to searching for possible exceptions to the scanning mechanism, one can only wish that some enterprising soul would tackle these important questions.
8. Uncited References
Acknowledgements
Research in my laboratory is supported by grant GM33915 from the National Institutes of Health.
Received by A. Bernardi
References
- Aarnoudse C.A., van den Doel P.B., Heemskerk B., Schrier P.I. Interleukin-2-induced, melanoma-specific T cells recognize CAMEL, an unexpected translation product of LAGE-1. Int. J. Cancer. 1999;82:442–448. doi: 10.1002/(sici)1097-0215(19990730)82:3<442::aid-ijc19>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Abastado J.-P., Miller P.F., Jackson B.M., Hinnebusch A.G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 1991;11:486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acland P., Dixon M., Peters G., Dickson C. Subcellular fate of the Int-2 oncoprotein is determined by choice of initiation codon. Nature. 1990;343:662–665. doi: 10.1038/343662a0. [DOI] [PubMed] [Google Scholar]
- Afshar-Kharghan V., Li C.Q., Khoshnevis-Asi M., López J.A. Kozak sequence polymorphism of the glycoprotein (GP) Ibα gene is a major determinant of the plasma membrane levels of the platelet GP Ib-IX-V complex. Blood. 1999;94:186–191. [PubMed] [Google Scholar]
- Aho S., Levänsuo L., Montonen O., Kari C., Rodeck U., Uitto J. Specific sequences in p120ctn determine subcellular distribution of its multiple isoforms involved in cellular adhesion of normal and malignant epithelial cells. J. Cell Sci. 2002;115:1391–1402. doi: 10.1242/jcs.115.7.1391. [DOI] [PubMed] [Google Scholar]
- Aichem A., Mutzel R. Unconventional mRNA processing in the expression of two calcineurin B isoforms in Dictyostelium. J. Mol. Biol. 2001;308:873–882. doi: 10.1006/jmbi.2001.4645. [DOI] [PubMed] [Google Scholar]
- Aizencang G.I., Bishop D.F., Forrest D., Astrin K.H., Desnick R.J. Uroporphyrinogen III synthase. An alternative promoter controls erythroid-specific expression in the murine gene. J. Biol. Chem. 2000;275:2295–2304. doi: 10.1074/jbc.275.4.2295. [DOI] [PubMed] [Google Scholar]
- Akiri G., Nahari D., Finkelstein Y., Le S.-Y., Elroy-Stein O., Levi B.-Z. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- Alderete J.P., Child S.J., Geballe A.P. Abundant early expression of gpUL4 from a human cytomegalovirus mutant lacking a repressive upstream open reading frame. J. Virol. 2001;75:7188–7192. doi: 10.1128/JVI.75.15.7188-7192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anant S., Mukhopadhyay D., Hirano K.-I., Brasitus T.A., Davidson N.O. Apobec-1 transcription in rat colon cancer: decreased apobec-1 protein production through alterations in polysome distribution and mRNA translation associated with upstream AUGs. Biochim. Biophys. Acta. 2002;1575:54–62. doi: 10.1016/s0167-4781(02)00250-6. [DOI] [PubMed] [Google Scholar]
- Andrea J.E., Walsh M.P. Identification of a brain-specific protein kinase Cξ pseudogene (ΨPKCξ) transcript. Biochem. J. 1995;310:835–843. doi: 10.1042/bj3100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anusaksathien O., Laplace C., Li X., Ren Y., Peng L., Goldring S.R., Galson D.L. Tissue-specific and ubiquitous promoters direct the expression of alternatively spliced transcripts from the calcitonin receptor gene. J. Biol. Chem. 2001;276:22663–22674. doi: 10.1074/jbc.M007104200. [DOI] [PubMed] [Google Scholar]
- Aplan P.D., Lombardi D.P., Kirsch I.R. Structural characterization of SIL, a gene frequently disrupted in T-cell acute lymphoblastic leukemia. Mol. Cell. Biol. 1991;11:5462–5469. doi: 10.1128/mcb.11.11.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M., Alpert N.R., Periasamy M. Cloning and characterization of the gene encoding rabbit cardiac calsequestrin. Gene. 1991;109:275–279. doi: 10.1016/0378-1119(91)90621-h. [DOI] [PubMed] [Google Scholar]
- Arnaud E., Touriol C., Boutonnet C., Gensac M.-C., Vagner S., Prats H., Prats A.-C. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol. Cell. Biol. 1999;19:505–514. doi: 10.1128/mcb.19.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrick B.A., Lee A.L., Grendell R.L., Derynck R. Inhibition of translation of transforming growth factor-β3 mRNA by its 5′ untranslated region. Mol. Cell. Biol. 1991;11:4306–4313. doi: 10.1128/mcb.11.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrick B.A., Grendell R.L., Griffin L.A. Enhanced translational efficiency of a novel transforming growth factor β3 mRNA in human breast cancer cells. Mol. Cell. Biol. 1994;14:619–628. doi: 10.1128/mcb.14.1.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arst H.N., Jr., Sheerins A. Translational initiation competence, ‘leaky scanning’ and translational reinitiation in areA mRNA of Aspergillusnidulans. Mol. Microbiol. 1996;19:1019–1024. doi: 10.1046/j.1365-2958.1996.470976.x. [DOI] [PubMed] [Google Scholar]
- Asano K., Shalev A., Phan L., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bab I., Smith E., Gavish H., Attar-Namdar M., Chorev M., Chen Y.-C., Muhlrad A., Birnbaum M.J., Stein G., Frenkel B. Biosynthesis of osteogenic growth peptide via alternative translational initiation at AUG85 of histone H4 mRNA. J. Biol. Chem. 1999;274:14474–14481. doi: 10.1074/jbc.274.20.14474. [DOI] [PubMed] [Google Scholar]
- Babik J.M., Adams E., Tone Y., Fairchild P.J., Tone M., Waldmann H. Expression of murine IL-12 is regulated by translational control of the p35 subunit. J. Immunol. 1999;162:4069–4078. [PubMed] [Google Scholar]
- Barbosa M.S., Wettstein F.O. The two proteins encoded by the cottontail rabbit papillomavirus E6 open reading frame differ with respect to localization and phosphorylation. J. Virol. 1988;62:1088–1092. doi: 10.1128/jvi.62.3.1088-1092.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates B., Hardin J., Zhan X., Drickamer K., Goldfarb M. Biosynthesis of human fibroblast growth factor-5. Mol. Cell. Biol. 1991;11:1840–1845. doi: 10.1128/mcb.11.4.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends U., Jandl T., Golbeck A., Lechner B., Müller-Weihrich S., Schmid I., Till H., Berthold F., Voltz R., Mautner J.M. Novel products of the HUD, HUC, NNP-1 and α-INTERNEXIN genes identified by autologous antibody screening of a pediatric neuroblastoma library. Int. J. Cancer. 2002;100:669–677. doi: 10.1002/ijc.10550. [DOI] [PubMed] [Google Scholar]
- Beltzer J.P., Morris S.R., Kohlhaw G.B. Yeast LEU4 encodes mitochondrial and non-mitochondrial forms of α-isopropylmalate synthase. J. Biol. Chem. 1988;263:368–374. [PubMed] [Google Scholar]
- Bergenhem N.C.H., Venta P.J., Hopkins P.J., Kim H.J., Tashian R.E. Mutation creates an open reading frame within the 5′ untranslated region of macaque erythrocyte carbonic anhydrase (CA) I mRNA that suppresses CA I expression and supports the scanning model for translation. Proc. Natl. Acad. Sci. USA. 1992;89:8798–8802. doi: 10.1073/pnas.89.18.8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J., Shefler I., Elroy-Stein O. The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA leader is relieved during megakaryocytic differentiation. J. Biol. Chem. 1995;270:10559–10565. doi: 10.1074/jbc.270.18.10559. [DOI] [PubMed] [Google Scholar]
- Berry F.B., Miura Y., Mihara K., Kaspar P., Sakata N., Hashimoto-Tamaoki T., Tamaoki T. Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J. Biol. Chem. 2001;276:25057–25065. doi: 10.1074/jbc.M010378200. [DOI] [PubMed] [Google Scholar]
- Beuret N., Rutishauser J., Bider M.D., Spiess M. Mechanism of endoplasmic reticulum retention of mutant vasopressin precursor caused by a signal peptide truncation associated with diabetes insipidus. J. Biol. Chem. 1999;274:18965–18972. doi: 10.1074/jbc.274.27.18965. [DOI] [PubMed] [Google Scholar]
- Bilanges B., Varrault A., Mazumdar A., Pantaloni C., Hoffmann A., Bockaert J., Spengler D., Journot L. Alternative splicing of the imprinted candidate tumor suppressor gene ZAC regulates its antiproliferative and DNA binding activities. Oncogene. 2001;20:1246–1253. doi: 10.1038/sj.onc.1204237. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Evans D., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M., Kim S.K. A global analysis of Caenorhabditiselegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- Bodelón G., Labrada L., Martínez-Costas J., Benavente J. The avian reovirus genome segment S1 is a functionally tricistronic gene that expresses one structural and two nonstructural proteins in infected cells. Virology. 2001;290:181–191. doi: 10.1006/viro.2001.1159. [DOI] [PubMed] [Google Scholar]
- Bos J.L., Polder L.J., Bernards R., Schrier P.I., van den Elsen P.J., van der Eb A.J., van Ormondt H. The 2.2 kb E1b mRNA of human Ad12 and Ad5 codes for two tumor antigens starting at different AUG triplets. Cell. 1981;27:121–131. doi: 10.1016/0092-8674(81)90366-4. [DOI] [PubMed] [Google Scholar]
- Boularand S., Darmon M.C., Mallet J. The human tryptophan hydroxylase gene. An unusual splicing complexity in the 5′-untranslated region. J. Biol. Chem. 1995;270:3748–3756. doi: 10.1074/jbc.270.8.3748. [DOI] [PubMed] [Google Scholar]
- Bradley C.A., Padovan J.C., Thompson T.L., Benoit C.A., Chait B.T., Rhoads R.E. Mass spectrometric analysis of the N terminus of translational initiation factor eIF4G-1 reveals novel isoforms. J. Biol. Chem. 2002;277:12559–12571. doi: 10.1074/jbc.M111134200. [DOI] [PubMed] [Google Scholar]
- Breckenridge D.G., Nguyen M., Kuppig S., Reth M., Shore G.C. The procaspase-8 isoform, procaspase-8L, recruited to the BAP31 complex at the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2002;99:4331–4336. doi: 10.1073/pnas.072088099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgen A., Weber F., Fazakerley J.K., Elliott R.M. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA. 2001;98:664–669. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.Y., Mize G.J., Pineda M., George D.L., Morris D.R. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene. 1999;18:5631–5637. doi: 10.1038/sj.onc.1202949. [DOI] [PubMed] [Google Scholar]
- Brown V., Jin P., Ceman S., Darnell J.C., O'Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., Darnell R.B., Warren S.T. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Brunstein J., Söderlund-Venermo M., Hedman K. Identification of a novel RNA splicing pattern as a basis of restricted cell tropism of erythrovirus B19. Virology. 2000;274:284–291. doi: 10.1006/viro.2000.0460. [DOI] [PubMed] [Google Scholar]
- Bullock T.N.J., Patterson A.E., Franlin L.L., Notidis E., Eisenlohr L.C. Initiation codon scanthrough versus termination codon readthrough demonstrates strong potential for major histocompatibility complex class I-restricted cryptic epitope expression. J. Exp. Med. 1997;186:1051–1058. doi: 10.1084/jem.186.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd M.P., Zamora M., Lloyd R.E. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 2002;22:4499–4511. doi: 10.1128/MCB.22.13.4499-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne P.C., Sanders P.G., Snell K. Translational control of mammalian serine hydroxymethyl-transferase expression. Biochem. Biophys. Res. Commun. 1995;214:496–502. doi: 10.1006/bbrc.1995.2314. [DOI] [PubMed] [Google Scholar]
- Cahana A., Escamez T., Nowakowski R.S., Hayes N.L., Giacobini M., von Holst A., Shmueli O., Sapir T., McConnell S.K., Wurst W., Martinez S., Reiner O. Targeted mutagenesis of Lis1 disrupts cortical development and LIS1 homodimerization. Proc. Natl. Acad. Sci. USA. 2001;98:6429–6434. doi: 10.1073/pnas.101122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S.-P., Eng B., Francombe W.H., Olivieri N.F., Kendall A.G., Waye J.S., Chui D.H.K. Two novel β-thalassemia mutations in the 5′ and 3′ noncoding regions of the β-globin gene. Blood. 1992;79:1342–1346. [PubMed] [Google Scholar]
- Calkhoven C.F., Müller C., Leutz A. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- Calligaris R., Bottardi S., Cogoi S., Apezteguia I., Santoro C. Alternative translation initiation site usage results in two functionally distinct forms of the GATA-1 transcription factor. Proc. Natl. Acad. Sci. USA. 1995;92:11598–11602. doi: 10.1073/pnas.92.25.11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau E., Desviat L.R., Leclerc D., Wu X., Pérez B., Ugarte M., Gravel R.A. Structure of the PCCA gene and distribution of mutations causing propionic acidemia. Mol. Genet. Metab. 2001;74:238–247. doi: 10.1006/mgme.2001.3210. [DOI] [PubMed] [Google Scholar]
- Cao J., Geballe A.P. Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J. Virol. 1995;69:1030–1036. doi: 10.1128/jvi.69.2.1030-1036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Taussig R., Kustu S., Botstein D. The secreted form of invertase in Saccharomycescerevisiae is synthesized from mRNA encoding a signal sequence. Mol. Cell. Biol. 1983;3:439–447. doi: 10.1128/mcb.3.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll R., Derse D. Translation of equine infectious anemia virus bicistronic tat-rev mRNA requires leaky ribosome scanning of the tat CTG initiation codon. J. Virol. 1993;67:1433–1440. doi: 10.1128/jvi.67.3.1433-1440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.J. Transcription of feline calicivirus RNA. Arch. Virol. 1990;114:143–152. doi: 10.1007/BF01310744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzola M., Skoda R.C. Translational pathophysiology: a novel molecular mechanism of human disease. Blood. 2000;95:3280–3288. [PubMed] [Google Scholar]
- Cazzola M., Foglieni B., Bergamaschi G., Levi S., Lazzarino M., Arosio P. A novel deletion of the L-ferritin iron-responsive element responsible for severe hereditary hyperferritinaemia-cataract syndrome. Br. J. Haematol. 2002;116:667–670. doi: 10.1046/j.0007-1048.2001.03310.x. [DOI] [PubMed] [Google Scholar]
- Chang C.-C., Gould S.J. Phenotype-genotype relationships in complementation group 3 of the peroxisome-biogenesis disorders. Am. J. Hum. Genet. 1998;63:1294–1306. doi: 10.1086/302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron M., Shaper J.H., Shaper N.L. The increased level of β1,4-galactosyltransferase required for lactose biosynthesis is achieved in part by translational control. Proc. Natl. Acad. Sci. USA. 1998;95:14805–14810. doi: 10.1073/pnas.95.25.14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton B., Walter P., Ebel J.-P., Lacroute F., Fasiolo F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J. Biol. Chem. 1988;263:52–57. [PubMed] [Google Scholar]
- Chen B., Rigat B., Curry C., Mahuran D.J. Structure of the GM2A gene: identification of an exon 2 nonsense mutation and a naturally occurring transcript with an in-frame deletion of exon 2. Am. J. Hum. Genet. 1999;65:77–87. doi: 10.1086/302463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- Chen W., Calvo P.A., Malide D., Gibbs J., Schubert U., Bacik I., Basta S., O'Neill R., Schickli J., Palese P., Henklein P., Bennink J.R., Yewdell J.W. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- Chenik M., Chebli K., Blondel D. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J. Virol. 1995;69:707–712. doi: 10.1128/jvi.69.2.707-712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child S.J., Miller M.K., Geballe A.P. Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem. 1999;274:24335–24341. doi: 10.1074/jbc.274.34.24335. [DOI] [PubMed] [Google Scholar]
- Child S.J., Miller M.K., Geballe A.P. Cell type-dependent and -independent control of HER-2/neu translation. Int. J. Biochem. Cell Biol. 1999;31:201–213. doi: 10.1016/s1357-2725(98)00068-5. [DOI] [PubMed] [Google Scholar]
- Choong C.S., Quigley C.A., French F.S., Wilson E.M. A novel missense mutation in the amino-terminal domain of the human androgen receptor gene in a family with partial androgen insensitivity syndrome causes reduced efficiency of protein translation. J. Clin. Invest. 1996;98:1423–1431. doi: 10.1172/JCI118930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L.M.L., Davidson D., Fournel M., Gosselin P., Lemieux S., Lyu M.S., Kozak C.A., Matix L.A., Veillette A. Two distinct protein isoforms are encoded by ntk, a csk-related tyrosine protein kinase gene. Oncogene. 1994;9:3437–3448. [PubMed] [Google Scholar]
- Chretien S., Dubart A., Beaupain D., Raich N., Grandchamp B., Rosa J., Goossens M., Romeo P.-H. Alternative transcription and splicing of the human porphobilinogen deaminase gene result either in tissue-specific or in housekeeping expression. Proc. Natl. Acad. Sci. USA. 1988;85:6–10. doi: 10.1073/pnas.85.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A.M., Feng L., Donahue T.F. tRNAmet functions in directing the scanning ribosome to the start site of translation. Science. 1988;242:93–97. doi: 10.1126/science.3051379. [DOI] [PubMed] [Google Scholar]
- Cigan A.M., Pabich E.K., Donahue T.F. Mutational analysis of the HIS4 translational initiator region in Saccharomycescerevisiae. Mol. Cell. Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J. Initiation factor eIF2α phosphorylation in stress responses and apoptosis. Prog. Mol. Subcell. Biol. 2001;27:57–89. doi: 10.1007/978-3-662-09889-9_3. [DOI] [PubMed] [Google Scholar]
- Conboy J.G., Chan J.Y., Chasis J.A., Kan Y.W., Mohandas N. Tissue- and development-specific alternative RNA splicing regulates expression of multiple isoforms of erythroid membrane protein 4.1. J. Biol. Chem. 1991;266:8273–8280. [PubMed] [Google Scholar]
- Cortner J., Farnham P.J. Identification of the serum-responsive transcription initiation site of the zinc finger gene Krox-20. Mol. Cell. Biol. 1990;10:3788–3791. doi: 10.1128/mcb.10.7.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Créancier L., Morello D., Mercier P., Prats A.-C. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell Biol. 2000;150:275–281. doi: 10.1083/jcb.150.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Créancier L., Mercier P., Prats A.-C., Morello D. c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol. Cell. Biol. 2001;21:1833–1840. doi: 10.1128/MCB.21.5.1833-1840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunillera N., Boronat A., Ferrer A. The ArabidopsisthalianaFPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J. Biol. Chem. 1997;272:15381–15388. doi: 10.1074/jbc.272.24.15381. [DOI] [PubMed] [Google Scholar]
- Curnow K.M., Pascoe L., Davies E., White P.C., Corvol P., Clauser E. Alternatively spliced human type 1 angiotensin II receptor mRNAs are translated at different efficiencies and encode two receptor isoforms. Mol. Endocrinol. 1995;9:1250–1262. doi: 10.1210/mend.9.9.7491117. [DOI] [PubMed] [Google Scholar]
- Das S., Ghosh R., Maitra U. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J. Biol. Chem. 2001;276:6720–6726. doi: 10.1074/jbc.M008863200. [DOI] [PubMed] [Google Scholar]
- Dawe S., Duncan R. The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein. J. Virol. 2002;76:2131–2140. doi: 10.1128/jvi.76.5.2131-2140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearth L.R., Hutt J., Sattler A., Gigliotti A., DeWille J. Expression and function of CCAAT/enhancer binding protein β (C/EBPβ) LAP and LIP isoforms in mouse mammary gland, tumors and cultured mammary epithelial cells. J. Cell. Biochem. 2001;82:357–370. doi: 10.1002/jcb.1167. [DOI] [PubMed] [Google Scholar]
- Debnath J., Chamorro M., Czar M.J., Schaeffer E.M., Lenardo M.J., Varmus H.E., Schwartzberg P.L. rlk/TXK encodes two forms of a novel cysteine string tyrosine kinase activated by Src family kinases. Mol. Cell. Biol. 1999;19:1498–1507. doi: 10.1128/mcb.19.2.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffaud C., Darlix J.-L. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J. Virol. 2000;74:846–850. doi: 10.1128/jvi.74.2.846-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjardin J., Bompard-Maréchal G., Audit M., Hope T.J., Sitbon M., Mougel M. A novel subgenomic murine leukemia virus RNA transcript results from alternative splicing. J. Virol. 2000;74:3709–3714. doi: 10.1128/jvi.74.8.3709-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delépine M., Nicolino M., Barrett T., Golamaully M., Lathrop G.M., Julier C. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- Deng H., Song M.J., Chu J.T., Sun R. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) J. Virol. 2002;76:8252–8264. doi: 10.1128/JVI.76.16.8252-8264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denger S., Reid G., Koš M., Flouriot G., Parsch D., Brand H., Korach K.S., Sonntag-Buck V., Gannon F. ERα gene expression in human primary osteoblasts: evidence for the expression of two receptor proteins. Mol. Endocrinol. 2001;15:2064–2077. doi: 10.1210/mend.15.12.0741. [DOI] [PubMed] [Google Scholar]
- Descombes P., Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Dever T.E. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Di Fruscio M., Gilchrist C.A., Baker R.T., Gray D.A. Genomic structure of Unp, a murine gene encoding a ubiquitin-specific protease. Biochim. Biophys. Acta. 1998;398:9–17. doi: 10.1016/s0167-4781(98)00035-9. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Miller W.A. Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell. 1993;5:679–692. doi: 10.1105/tpc.5.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzé O., Damay P., Spahr P.-F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eerola I., McIntyre B., Vikkula M. Identification of eight novel 5′-exons in cerebral capillary malformation gene-1 (CCM1) encoding KRIT1. Biochim. Biophys. Acta. 2001;1517:464–467. doi: 10.1016/s0167-4781(00)00303-1. [DOI] [PubMed] [Google Scholar]
- Elgadi M.M., Smiley J.R. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host-shutoff protein. J. Virol. 1999;73:9222–9231. doi: 10.1128/jvi.73.11.9222-9231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R.M., McGregor A. Nucleotide sequence and expression of the small (S) RNA segment of Maguari bunyavirus. Virology. 1989;171:516–524. doi: 10.1016/0042-6822(89)90621-1. [DOI] [PubMed] [Google Scholar]
- Ellis S.R., Hopper A.K., Martin N.C. Amino-terminal extension generated from an upstream AUG codon increases the efficiency of mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferases. Mol. Cell. Biol. 1989;9:1611–1620. doi: 10.1128/mcb.9.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Rifai W., Smith M.F., Jr., Li G., Beckler A., Carl V.S., Montgomery E., Knuutila S., Moskaluk C.A., Frierson H.F., Jr., Powell S.M. Gastric cancers overexpress DARPP-32 and a novel isoform, t-DARPP. Cancer Res. 2002;62:4061–4064. [PubMed] [Google Scholar]
- Endler G., Exner M., Mannhalter C., Meier S., Ruzicka K., Handler S., Panzer S., Wagner O., Quehenberger P. A common C→T polymorphism at nt 46 in the promoter region of coagulation factor XII is associated with decreased factor XII activity. Thrombosis Res. 2001;101:255–260. doi: 10.1016/s0049-3848(00)00404-7. [DOI] [PubMed] [Google Scholar]
- Fajardo J.E., Shatkin A.J. Translation of bicistronic viral mRNA in transfected cells: regulation at the level of elongation. Proc. Natl. Acad. Sci. USA. 1990;87:328–332. doi: 10.1073/pnas.87.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo J.E., Birge R.B., Hanafusa H. A 31-amino acid N-terminal extension regulates c-Crk binding to tyrosine-phosphorylated proteins. Mol. Cell. Biol. 1993;13:7295–7302. doi: 10.1128/mcb.13.12.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvey E., Fleury-Olela F., Schibler U. The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favor J., Peters H., Hermann T., Schmahl W., Chatterjee B., Neuhäuser-Klaus A., Sandulache R. Molecular characterization of Pax62Neu through Pax610Neu: an extension of the Pax6 allelic series and the identification of two possible hypomorph alleles in the mouse Musmusculus. Genetics. 2001;159:1689–1700. doi: 10.1093/genetics/159.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florkiewicz R.Z., Sommer A. Human basic fibroblast growth factor gene encodes four polypeptides: three initiate translation from non-AUG codons. Proc. Natl. Acad. Sci. USA. 1989;86:3978–3981. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouriot G., Brand H., Denger S., Metivier R., Kos M., Reid G., Sonntag-Buck V., Gannon F. Identification of a new isoform of the human estrogen receptor-alpha (hER-α) that is encoded by distinct transcripts and that is able to repress hER-α activation function 1. EMBO J. 2000;19:4688–4700. doi: 10.1093/emboj/19.17.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo N.-C., Funkhouser J.M., Carter D.A., Murphy D. A testis-specific promoter in the rat vasopressin gene. J. Biol. Chem. 1994;269:659–667. [PubMed] [Google Scholar]
- Fouillot N., Rossignol J.-M. Translational stop codons in the precore sequence of hepatitis B virus pre-C RNA allow translation reinitiation at downstream AUGs. J. Gen. Virol. 1996;77:1123–1127. doi: 10.1099/0022-1317-77-6-1123. [DOI] [PubMed] [Google Scholar]
- Fouillot N., Tlouzeau S., Rossignol J.-M., Jean-Jean O. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 1993;67:4886–4895. doi: 10.1128/jvi.67.8.4886-4895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemantle S.J., Taylor S.M., Krystal G., Moran R.G. Upstream organization of and multiple transcripts from the human folylpoly-γ-glutamate synthetase gene. J. Biol. Chem. 1995;270:9579–9584. doi: 10.1074/jbc.270.16.9579. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P.A., Dietz H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Frost G.I., Mohapatra G., Wong T.M., Csóka A.B., Gray J.W., Stern R. HYAL1LUCA-1, a candidate tumor suppressor gene on chromosome 3p21.3, is inactivated in head and neck squamous cell carcinomas by aberrant splicing of pre-mRNA. Oncogene. 2000;19:870–877. doi: 10.1038/sj.onc.1203317. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Shatkin A.J. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Potrykus I., Brau M.P.V., Dasgupta I., Hull R., Hohn T. Splicing in a plant pararetrovirus. Virology. 1994;198:663–670. doi: 10.1006/viro.1994.1078. [DOI] [PubMed] [Google Scholar]
- Fütterer J., Potrykus I., Bao Y., Li L., Burns T.M., Hull R., Hohn T. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J. Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Rothnie H.M., Hohn T., Potrykus I. Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J. Virol. 1997;71:7984–7989. doi: 10.1128/jvi.71.10.7984-7989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxe J., Raschperger E., Pettersson R.F. Translation of p15.5INK4B, an N-terminally extended and fully active form of p15INK4B, is initiated from an upstream GUG codon. Oncogene. 2000;19:1724–1728. doi: 10.1038/sj.onc.1203496. [DOI] [PubMed] [Google Scholar]
- Gaba A., Wang Z., Krishnamoorthy T., Hinnebusch A.G., Sachs M.S. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 2001;20:6453–6463. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D.R., Ling J., Niepel M., Morley S.J., Pain V.M. The role of 5′-leader length, secondary structure and PABP concentration on cap and poly(A) tail function during translation in Xenopus oocytes. Nucleic Acids Res. 2000;28:2943–2953. doi: 10.1093/nar/28.15.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie A.E., Stewart B.G., Scott C.F., Rose M.D. The two forms of karyogamy transcription factor Kar4p are regulated by differential initiation of transcription, translation, and protein turnover. Mol. Cell. Biol. 1999;19:817–825. doi: 10.1128/mcb.19.1.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio M.T., Naranda T., Vazquez de Aldana C.R., Cuesta R., Hinnebusch A.G., Hershey J.W.B., Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- Gereben B., Kollár A., Harney J.W., Larsen P.R. The mRNA structure has potent regulatory effects on type 2 iodothyronine deiodinase expression. Mol. Endocrinol. 2002;16:1667–1679. doi: 10.1210/mend.16.7.0879. [DOI] [PubMed] [Google Scholar]
- Ghilardi N., Skoda R.C. A single-base deletion in the thrombopoietin (TPO) gene causes familial essential thrombocythemia through a mechanism of more efficient translation of TPO mRNA. Blood. 1999;94:1480–1482. [PubMed] [Google Scholar]
- Ghilardi N., Wiestner A., Skoda R.C. Thrombopoietin production is inhibited by a translational mechanism. Blood. 1998;92:4023–4030. [PubMed] [Google Scholar]
- Ghilardi N., Wiestner A., Kikuchi M., Ohsaka A., Skoda R.C. Hereditary thrombocythaemia in a Japanese family is caused by a novel point mutation in the thrombopoietin gene. Br. J. Haematol. 1999;107:310–316. doi: 10.1046/j.1365-2141.1999.01710.x. [DOI] [PubMed] [Google Scholar]
- Glanville N., Ranki M., Morser J., Kääriäinen L., Smith A.E. Initiation of translation directed by 42S and 26S RNAs from Semliki Forest virus in vitro. Proc. Natl. Acad. Sci. USA. 1976;73:3059–3063. doi: 10.1073/pnas.73.9.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Conejero R., Corral J., Roldán V., Martinez C., Marin F., Rivera J., Iniesta J.A., Lozano M.L., Marco P., Vicente V. A common polymorphism in the annexin V Kozak sequence (−1 C>T) increases translation efficiency and plasma levels of annexin V, and decreases the risk of myocardial infarction in young patients. Blood. 2002;100:2081–2086. [PubMed] [Google Scholar]
- Goossen B., Hentze M.W. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol. Cell. Biol. 1992;12:1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss K.H., Trzepacz C., Tuohy T.M.F., Groden J. Attenuated APC alleles produce functional protein from internal translation initiation. Proc. Natl. Acad. Sci. USA. 2002;99:8161–8166. doi: 10.1073/pnas.112072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S., Satyanarayana T., Ayllón M.A., Albiach-Marti M.R., Mawassi M., Rabindran S., Garnsey S.M., Dawson W.O. Characterization of the cis-acting elements controlling subgenomic mRNAs of Citrustristezavirus: production of positive- and negative-stranded 3′-terminal and positive-stranded 5′-terminal RNAs. Virology. 2001;286:134–151. doi: 10.1006/viro.2001.0987. [DOI] [PubMed] [Google Scholar]
- Grant C.M., Hinnebusch A.G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T.A., Nicholls R.D. Diverse splicing mechanisms fuse the evolutionarily conserved bicistronic MOCS1A and MOCS1B open reading frames. RNA. 2000;6:928–936. doi: 10.1017/s1355838200000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T.A., Saitoh S., Nicholls R.D. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc. Natl. Acad. Sci. USA. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grdzelishvili V.Z., Chapman S.N., Dawson W.O., Lewandowski D.J. Mapping of the tobacco mosaic virus movement protein and coat protein subgenomic RNA promoters. Virology. 2000;275:177–192. doi: 10.1006/viro.2000.0511. [DOI] [PubMed] [Google Scholar]
- Griffin E., Re A., Hamel N., Fu C., Bush H., McCaffrey T., Asch A.S. A link between diabetes and atherosclerosis: glucose regulates expression of CD36 at the level of translation. Nat. Med. 2001;7:840–846. doi: 10.1038/89969. [DOI] [PubMed] [Google Scholar]
- Gross C., Krishnan A.V., Malloy P.J., Eccleshall T.R., Zhao X.-Y., Feldman D. The vitamin D receptor gene start codon polymorphism: a functional analysis of FokI variants. J. Bone Miner. Res. 1998;13:1691–1699. doi: 10.1359/jbmr.1998.13.11.1691. [DOI] [PubMed] [Google Scholar]
- Grove J.R., Banerjee P., Balasubramanyam A., Coffer P.J., Price D.J., Avruch J., Woodgett J.R. Cloning and expression of two human p70 S6 kinase polypeptides differing only at their amino termini. Mol. Cell. Biol. 1991;11:5541–5550. doi: 10.1128/mcb.11.11.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A., Ganem D. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2001;75:1857–1863. doi: 10.1128/JVI.75.4.1857-1863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudikote J.P., Wilkinson M.F. T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J. 2002;21:125–134. doi: 10.1093/emboj/21.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker D.L., Sivakumaran K. Mapping and expression of southern bean mosaic virus genomic and subgenomic RNAs. Virology. 1997;234:317–327. doi: 10.1006/viro.1997.8667. [DOI] [PubMed] [Google Scholar]
- Hackett P.B., Petersen R.B., Hensel C.H., Albericio F., Gunderson S.I., Palmenberg A.C., Barany G. Synthesis invitro of a seven amino acid peptide encoded in the leader RNA of Rous sarcoma virus. J. Mol. Biol. 1986;190:45–57. doi: 10.1016/0022-2836(86)90074-4. [DOI] [PubMed] [Google Scholar]
- Hake L.E., Hecht N.B. Utilization of an alternative transcription initiation site of somatic cytochrome c in the mouse produces a testis-specific cytochrome c mRNA. J. Biol. Chem. 1993;268:4788–4797. [PubMed] [Google Scholar]
- Han A.-P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G., Fleming M., Leboulch P., Orkin S.H., Chen J.-J. Heme-regulated eIF2α kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 2001;20:6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Zhang J.-T. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol. Cell. Biol. 2002;22 doi: 10.1128/MCB.22.21.7372-7384.2002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Liu Y., Chen Q., Dong Z., Hashimoto K., Zhang J.-T. Dual role of the 5′ untranslated region of mouse PTEN in translational and transcriptional regulation of PTEN expression. J. Biol. Chem. 2002 in press. [Google Scholar]
- Handley-Gearhart P.M., Stephen A.G., Trausch-Azar J.S., Ciechanover A., Schwartz A.L. Human ubiquitin-activating enzyme, E1. Indication of potential nuclear and cytoplasmic subpopulations using epitope-tagged cDNA constructs. J. Biol. Chem. 1994;269:33171–33178. [PubMed] [Google Scholar]
- Hanfrey C., Franceschetti M., Mayer M.J., Illingworth C., Michael A.J. Abrogation of upstream open reading frame-mediated translational control of a plant S-adenosylmethionine decarboxylase results in polyamine disruption and growth perturbations. J. Biol. Chem. 2002;277 doi: 10.1074/jbc.M206161200. in press. [DOI] [PubMed] [Google Scholar]
- Hansen M.B., Mitchelmore C., Kjaerulff K.M., Rasmussen T.E., Pedersen K.M., Jensen N.A. Mouse Atf5: molecular cloning of two novel mRNAs, genomic organization, and odorant sensory neuron localization. Genomics. 2002;80:344–350. doi: 10.1006/geno.2002.6838. [DOI] [PubMed] [Google Scholar]
- Hänzelmann P., Schwarz G., Mendel R.R. Functionality of alternative splice forms of the first enzymes involved in human molybdenum cofactor biosynthesis. J. Biol. Chem. 2002;277:18303–18312. doi: 10.1074/jbc.M200947200. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H., Sabatini D.D., Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in Perk −/− mice reveals a role for translational control in secretory cell survival. Mol. Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A., Shaper J.H., Shaper N.L. Characterization of two cis-regulatory regions in the murine β1,4-galactosyltransferase gene. J. Biol. Chem. 1993;268:14348–14359. [PubMed] [Google Scholar]
- Harigai M., Miyashita T., Hanada M., Reed J.C. A cis-acting element in the BCL-2 gene controls expression through translational mechanisms. Oncogene. 1996;12:1369–1374. [PubMed] [Google Scholar]
- Harington A., Schwarz E., Slonimski P.P., Herbert C.J. Subcellular relocalization of a long-chain fatty acid CoA ligase by a suppressor mutation alleviates a respiration deficiency in Saccharomycescerevisiae. EMBO J. 1994;13:5531–5538. doi: 10.1002/j.1460-2075.1994.tb06890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzigeorgiou A.G. Translation initiation start prediction in human cDNAs with high accuracy. Bioinformatics. 2002;18:343–350. doi: 10.1093/bioinformatics/18.2.343. [DOI] [PubMed] [Google Scholar]
- He F., Jacobson A. Upflp, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellborg F., Qian W., Mendez-Vidal C., Asker C., Kost-Alimova M., Wilhelm M., Imreh S., Wiman K.G. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene. 2001;20:5466–5474. doi: 10.1038/sj.onc.1204722. [DOI] [PubMed] [Google Scholar]
- Hellen C.U.T., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Hennecke M., Kwissa M., Metzger K., Oumard A., Kröger A., Schirmbeck R., Reimann J., Hauser H. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29:3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensold J.O., Stratton C.A., Barth D. The conserved 5′-untranslated leader of Spi-1 (PU.1) mRNA is highly structured and potently inhibits translation invitro but not invivo. Nucleic Acids Res. 1997;25:2869–2876. doi: 10.1093/nar/25.14.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert T.P., Brierley I., Brown T.D.K. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic subgenomic mRNA. J. Gen. Virol. 1996;77:123–127. doi: 10.1099/0022-1317-77-1-123. [DOI] [PubMed] [Google Scholar]
- Herrera F., Franceschi F., Zambrano R., Alvarado M. An altered ribosomal protein in an edeine-resistant mutant of Saccharomycescerevisiae. Mol. Gen. Genet. 1986;202:120–124. doi: 10.1007/BF00330527. [DOI] [PubMed] [Google Scholar]
- Herzog E., Guilley H., Fritsch C. Translation of the second gene of peanut clump virus RNA 2 occurs by leaky scanning invitro. Virology. 1995;208:215–225. doi: 10.1006/viro.1995.1145. [DOI] [PubMed] [Google Scholar]
- Hickey M.J., Roth G.J. Characterization of the gene encoding human platelet glycoprotein IX. J. Biol. Chem. 1993;268:3438–3443. [PubMed] [Google Scholar]
- Hinnebusch A.G. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Ho C.-L., Chin S.S.M., Carnevale K., Liem R.K.H. Translation initiation and assembly of peripherin in cultured cells. Eur. J. Cell Biol. 1995;68:103–112. [PubMed] [Google Scholar]
- Hofmann M.A., Senanayake S.D., Brian D.A. A translation-attenuating intraleader open reading frame is selected on coronavirus mRNAs during persistent infection. Proc. Natl. Acad. Sci. USA. 1993;90:11733–11737. doi: 10.1073/pnas.90.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J.D., Danpure C.J. Molecular basis for the dual mitochondrial and cytosolic localization of alanine:glyoxylate aminotransferase in amphibian liver cells. J. Biol. Chem. 2002;277:2336–2344. doi: 10.1074/jbc.M107047200. [DOI] [PubMed] [Google Scholar]
- Hoover D.S., Wingett D.G., Zheng J., Reeves R., Magnuson N.S. Pim-1 protein expression is regulated by its 5′-untranslated region and translation initiation factor eIF-4E. Cell Growth Differ. 1997;8:1371–1380. [PubMed] [Google Scholar]
- Horikami S.M., De Ferra F., Moyer S.A. Characterization of the infections of permissive and nonpermissive cells by host range mutants of vesicular stomatitis virus defective in RNA methylation. Virology. 1984;138:1–15. doi: 10.1016/0042-6822(84)90142-9. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Macon K.J., Kidd V.J., Volanakis J.E. Translational regulation of complement protein C2 expression by differential utilization of the 5′-untranslated region of mRNA. J. Biol. Chem. 1990;265:6521–6524. [PubMed] [Google Scholar]
- Horvath P., Suganuma A., Inaba M., Pan Y.-B., Gupta K.C. Multiple elements in the 5′ untranslated region down-regulate c-sis messenger RNA translation. Cell Growth Differ. 1995;6:1103–1110. [PubMed] [Google Scholar]
- Hough R.F., Lingam A.T., Bass B.L. Caenorhabditiselegans mRNAs that encode a protein similar to ADARs derive from an operon containing six genes. Nucleic Acids Res. 1999;27:3424–3432. doi: 10.1093/nar/27.17.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanes K., Li T.W.H., Munguia J.E., Truong T., Milovanovic T., Marsh J.L., Holcombe R.F., Waterman M.L. β-Catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat. Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- Huang C.-H., Cheng G.-J., Reid M.E., Chen Y. Rhmod syndrome: a family study of the translation-initiator mutation in the Rh50 glycoprotein gene. Am. J. Hum. Genet. 1999;64:107–117. doi: 10.1086/302215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Yoon H., Hannig E.M., Donahue T.F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomycescerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-P., Tang C.-J.C., Kou G.-H., Marchesi V.T., Benz E.J., Jr., Tang T.K. Genomic structure of the locus encoding protein 4.1. Structural basis for complex combinatorial patterns of tissue-specific alternative RNA splicing. J. Biol. Chem. 1993;268:3758–3766. [PubMed] [Google Scholar]
- Hummel K.B., Vanchiere J.A., Bellini W.J. Restriction of fusion protein mRNA as a mechanism of measles virus persistence. Virology. 1994;202:665–672. doi: 10.1006/viro.1994.1388. [DOI] [PubMed] [Google Scholar]
- Hunter T.R., Hunt T., Knowland J., Zimmern D. Messenger RNA for the coat protein of tobacco mosaic virus. Nature. 1976;260:759–764. doi: 10.1038/260759a0. [DOI] [PubMed] [Google Scholar]
- Huo L., Scarpulla R.C. Multiple 5′-untranslated exons in the nuclear respiratory factor 1 gene span 47 kb and contribute to transcript heterogeneity and translational efficiency. Gene. 1999;233:213–224. doi: 10.1016/s0378-1119(99)00135-3. [DOI] [PubMed] [Google Scholar]
- Hwang W.-L., Su T.-S. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J. Gen. Virol. 1998;79:2181–2189. doi: 10.1099/0022-1317-79-9-2181. [DOI] [PubMed] [Google Scholar]
- Iliopoulos O., Ohh M., Kaelin W.G., Jr. pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc. Natl. Acad. Sci. USA. 1998;95:11661–11666. doi: 10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Matsufuji S., Murakami Y., Gesteland R.F., Atkins J.F. Conservation of polyamine regulation by translational frameshifting from yeast to mammals. EMBO J. 2000;19:1907–1917. doi: 10.1093/emboj/19.8.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakar H.R., Whitt M.A. Identification of two additional translation products from the matrix (M) gene that contribute to vesicular stomatitis virus cytopathology. J. Virol. 2002;76:8011–8018. doi: 10.1128/JVI.76.16.8011-8018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Liu L., Buyse I.M., Simon D., Huang S. Decreased RIZ1 expression but not RIX2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int. J. Cancer. 1999;83:541–546. doi: 10.1002/(sici)1097-0215(19991112)83:4<541::aid-ijc17>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Jiang H., Lucy M.C. Variants of the 5′-untranslated region of the bovine growth hormone receptor mRNA: isolation, expression and effects on translational efficiency. Gene. 2001;265:45–53. doi: 10.1016/s0378-1119(01)00356-0. [DOI] [PubMed] [Google Scholar]
- Jiang W., Kanter M.R., Dunkel I., Ramsay R.G., Beemon K.L., Hayward W.S. Minimal truncation of the c-myb gene product in rapid-onset B-cell lymphoma. J. Virol. 1997;71:6526–6533. doi: 10.1128/jvi.71.9.6526-6533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.C., Rochon D.M. Both codon context and leader length contribute to efficient expression of two overlapping open reading frames of a cucumber necrosis virus bifunctional subgenomic mRNA. Virology. 1996;221:232–239. doi: 10.1006/viro.1996.0370. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dean C., Gidoni D., Gilbert D., Bond-Nutter D., Lee R., Bedbrook J., Dunsmuir P. Expression of bacterial chitinase protein in tobacco leaves using two photosynthetic gene promoters. Mol. Gen. Genet. 1988;212:536–542. [Google Scholar]
- Jopling C.L., Willis A.E. N-myc translation is initiated via an internal ribosome entry segment that displays enhanced activity in neuronal cells. Oncogene. 2001;20:2664–2670. doi: 10.1038/sj.onc.1204404. [DOI] [PubMed] [Google Scholar]
- Jordan I., Enssle J., Güttler E., Mauer B., Rethwilm A. Expression of human foamy virus reverse transcriptase involves a spliced pol mRNA. Virology. 1996;224:314–319. doi: 10.1006/viro.1996.0534. [DOI] [PubMed] [Google Scholar]
- Kanaji T., Okamura T., Osaki K., Kuroiwa M., Shimoda K., Hamasaki N., Niho Y. A common genetic polymorphism (46 C to T substitution) in the 5′-untranslated region of the coagulation factor XII gene is associated with low translation efficiency and decrease in plasma factor XII level. Blood. 1998;91:2010–2014. [PubMed] [Google Scholar]
- Kaneda S., Yura T., Yanagi H. Production of three distinct mRNAs of 150 kDa oxygen-regulated protein (ORP150) by alternative promoters: preferential induction of one species under stress conditions. J. Biochem. 2000;128:529–538. doi: 10.1093/oxfordjournals.jbchem.a022783. [DOI] [PubMed] [Google Scholar]
- Kastner P., Krust A., Turcotte B., Stropp U., Tora L., Gronemeyer H., Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P., Bocquel M.-T., Turcotte B., Garnier J.-M., Horwitz K.B., Chambon P., Gronemeyer H. Transient expression of human and chicken progesterone receptors does not support alternative translational initiation from a single mRNA as the mechanism generating two receptor isoforms. J. Biol. Chem. 1990;265:12163–12167. [PubMed] [Google Scholar]
- Kawakubo K., Samuel C.E. Human RNA-specific adenosine deaminase (ADAR1) gene specifies transcripts that initiate from a constitutively active alternative promoter. Gene. 2000;258:165–172. doi: 10.1016/s0378-1119(00)00368-1. [DOI] [PubMed] [Google Scholar]
- Kaytor M.D., Orr H.T. RNA targets of the fragile X protein. Cell. 2001;107:555–557. doi: 10.1016/s0092-8674(01)00590-6. [DOI] [PubMed] [Google Scholar]
- Keirsebilck A., Bonné S., Staes K., van Hengel J., Nollet F., Reynolds A., van Roy F. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics. 1998;50:129–146. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- Kiefer P., Acland P., Pappin D., Peters G., Dickson C. Competition between nuclear localization and secretory signals determines the subcellular fate of a single CUG-initiated form of FGF3. EMBO J. 1994;13:4126–4136. doi: 10.1002/j.1460-2075.1994.tb06730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner J.R., Staskus K., Haase A., Lagunoff M., Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J. Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-László Z., Blanc S., Hohn T. Splicing of cauliflower mosaic virus 35S RNA is essential for viral infectivity. EMBO J. 1995;14:3552–3562. doi: 10.1002/j.1460-2075.1995.tb07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemke M., Kehlenbach R.H., Huttner W.B. Two overlapping reading frames in a single exon encode interacting proteins – a novel way of gene usage. EMBO J. 2001;20:3849–3860. doi: 10.1093/emboj/20.14.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopp E.A., Arndt T.L., Eng K.L., Caldwell M., LeBoeuf R.C., Deeb S.S., O'Brien K.D. Murine phospholipid hydroperoxide glutathione peroxidase: cDNA sequence, tissue expression, and mapping. Mamm. Genome. 1999;10:601–605. doi: 10.1007/s003359901053. [DOI] [PubMed] [Google Scholar]
- Koev G., Miller W.A. A positive-strand RNA virus with three very different subgenomic RNA promoters. J. Virol. 2000;74:5988–5996. doi: 10.1128/jvi.74.13.5988-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo H., Fujimoto T. Caveolin-1 isoforms are encoded by distinct mRNAs. FEBS Lett. 2000;465:119–123. doi: 10.1016/s0014-5793(99)01730-5. [DOI] [PubMed] [Google Scholar]
- Koloteva N., Müller P.P., McCarthy J.E.G. The position dependence of translational regulation via RNA-RNA and RNA-protein interactions in the 5′-untranslated region of eukaryotic mRNA is a function of the thermodynamic competence of 40S ribosomes in translational initiation. J. Biol. Chem. 1997;272:16531–16539. doi: 10.1074/jbc.272.26.16531. [DOI] [PubMed] [Google Scholar]
- Konarska M., Filipowicz W., Domdey H., Gross H.J. Binding of ribosomes to linear and circular forms of the 5′-terminal leader fragment of tobacco mosaic virus RNA. Eur. J. Biochem. 1981;114:221–227. doi: 10.1111/j.1432-1033.1981.tb05139.x. [DOI] [PubMed] [Google Scholar]
- Kondo T., Okabe M., Sanada M., Kurosawa M., Suzuki S., Kobayashi M., Hosokawa M., Asaka M. Familial essential thrombocythemia associated with one-base deletion in the 5′-untranslated region of the thrombopoietin gene. Blood. 1998;92:1091–1096. [PubMed] [Google Scholar]
- Kos M., Denger S., Reid G., Gannon F. Upstream open reading frames regulate the translation of multiple mRNA variants of the estrogen receptor alpha. J. Biol. Chem. 2002 doi: 10.1074/jbc.M206325200. in press. [DOI] [PubMed] [Google Scholar]
- Koski C., Saharinen J., Keski-Oja J. Independent promoters regulate the expression of two amino terminally distinct forms of latent transforming growth factor-β binding protein-1 (LTBP-1) in a cell type-specific manner. J. Biol. Chem. 1999;274:32619–32630. doi: 10.1074/jbc.274.46.32619. [DOI] [PubMed] [Google Scholar]
- Kozak M. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 1979;280:82–85. doi: 10.1038/280082a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Migration of 40S ribosomal subunits on messenger RNA when initiation is perturbed by lowering magnesium or adding drugs. J. Biol. Chem. 1979;254:4731–4738. [PubMed] [Google Scholar]
- Kozak M. Role of ATP in binding and migration of 40S ribosomal subunits. Cell. 1980;22:459–467. doi: 10.1016/0092-8674(80)90356-6. [DOI] [PubMed] [Google Scholar]
- Kozak M. Analysis of ribosome binding sites from the s1 message of reovirus. Initiation at the first and second AUG codons. J. Mol. Biol. 1982;156:807–820. doi: 10.1016/0022-2836(82)90143-7. [DOI] [PubMed] [Google Scholar]
- Kozak M. Translation of insulin-related polypeptides from messenger RNAs with tandemly reiterated copies of the ribosome binding site. Cell. 1983;34:971–978. doi: 10.1016/0092-8674(83)90554-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res. 1990;18:2828. doi: 10.1093/nar/18.9.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc. Natl. Acad. Sci. USA. 1995;92:2662–2666. doi: 10.1073/pnas.92.7.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm. Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Primer-extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res. 1998;26:4853–4859. doi: 10.1093/nar/26.21.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Kozak M. Do the 5′ untranslated domains of human cDNAs challenge the rules for initiation of translation (or is it vice versa)? Genomics. 2000;70:396–406. doi: 10.1006/geno.2000.6412. [DOI] [PubMed] [Google Scholar]
- Kozak M. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Emerging links between initiation of translation and human diseases. Mamm. Genome. 2002;13:401–410. doi: 10.1007/s00335-002-4002-5. [DOI] [PubMed] [Google Scholar]
- Kozak M., Shatkin A.J. Migration of 40S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- Kraft H.-G., Windegger M., Menzel H.J., Utermann G. Significant impact of the +93 C/T polymorphism in the apolipoprotein(a) gene on Lp(a) concentrations in Africans but not in Caucasians: confounding effect of linkage disequilibrium. Hum. Mol. Genet. 1998;7:257–264. doi: 10.1093/hmg/7.2.257. [DOI] [PubMed] [Google Scholar]
- Kubu C.J., Goldhawk D.G., Barker P.A., Verdi J.M. Identification of the translational initiation codon in human MAGED1. Genomics. 2000;70:150–152. doi: 10.1006/geno.2000.6356. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski D.J., Mehl R., Yin H.L. Genomic organization and biosynthesis of secreted and cytoplasmic forms of gelsolin. J. Cell Biol. 1988;106:375–384. doi: 10.1083/jcb.106.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides N.C., Woese C.R. Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B α-β-δ subunit families. Proc. Natl. Acad. Sci. USA. 1998;95:3726–3730. doi: 10.1073/pnas.95.7.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert P.F., Spalholz B.A., Howley P.M. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell. 1987;50:69–78. doi: 10.1016/0092-8674(87)90663-5. [DOI] [PubMed] [Google Scholar]
- Lambert P.F., Baker C.C., Howley P.M. The genetics of bovine papillomavirus type 1. Annu. Rev. Genet. 1988;22:235–258. doi: 10.1146/annurev.ge.22.120188.001315. [DOI] [PubMed] [Google Scholar]
- Land T., Rouault T.A. Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell. 1998;2:807–815. doi: 10.1016/s1097-2765(00)80295-6. [DOI] [PubMed] [Google Scholar]
- Landers J.E., Cassel S.L., George D.L. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 1997;57:3562–3568. [PubMed] [Google Scholar]
- Laprevotte I., Hampe A., Sherr C.J., Galibert F. Nucleotide sequence of the gag gene and gag-pol junction of feline leukemia virus. J. Virol. 1984;50:884–894. doi: 10.1128/jvi.50.3.884-894.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen L.K., Amri E.-Z., Mandrup S., Pacot C., Kristiansen K. Genomic organization of the mouse peroxisome proliferator activated receptor β/γ gene. Alternative promoter usage and splicing yield transcripts exhibiting differential translational efficiency. Biochem. J. 2002 doi: 10.1042/BJ20011821. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin N.N., Wang S.P., Mitchell G.A. The hormone-sensitive lipase gene is transcribed from at least five alternative first exons in mouse adipose tissue. Mamm. Genome. 2000;11:972–978. doi: 10.1007/s003350010185. [DOI] [PubMed] [Google Scholar]
- Law G.L., Raney A., Heusner C., Morris D.R. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 2001;276:38036–38043. doi: 10.1074/jbc.M105944200. [DOI] [PubMed] [Google Scholar]
- Ledley F.D., Jansen R., Nham S.-U., Fenton W.A., Rosenberg L.E. Mutation eliminating mitochondrial leader sequence of methylmalonyl-CoA mutase causes muto methylmalonic acidemia. Proc. Natl. Acad. Sci. USA. 1990;87:3147–3150. doi: 10.1073/pnas.87.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.-H., Kim S.J., Kang S.W., Lee K.-K., Rhee S.G., Yu D.-Y. Molecular cloning and characterization of the mouse peroxiredoxinV gene. Biochem. Biophys. Res. Commun. 2000;270:356–362. doi: 10.1006/bbrc.2000.2430. [DOI] [PubMed] [Google Scholar]
- Lee Y.C., Chang C.-W., Su C.-W., Lin T.-N., Sun S.H., Lai H.-L., Chern Y. The 5′ untranslated regions of the rat A2A adenosine receptor gene function as negative translational regulators. J. Neurochem. 1999;73:1790–1798. doi: 10.1046/j.1471-4159.1999.0731790.x. [DOI] [PubMed] [Google Scholar]
- Lekven A.C., Thorpe C.J., Waxman J.S., Moon R.T. Zebrafish wnt8 encodes two Wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev. Cell. 2001;1:103–114. doi: 10.1016/s1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- Leslie N.D., Immerman E.B., Flach J.E., Florez M., Fridovich-Keil J.L., Elsas L.J. The human galactose-1-phosphate uridyltransferase gene. Genomics. 1992;14:474–480. doi: 10.1016/s0888-7543(05)80244-7. [DOI] [PubMed] [Google Scholar]
- Li S., Sonenberg N., Gingras A.-C., Peterson M., Avdulov S., Polunovsky V.A., Bitterman P.B. Translational control of cell fate: availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Mol. Cell. Biol. 2002;22:2853–2861. doi: 10.1128/MCB.22.8.2853-2861.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Qu F., Morris T.J. Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function intrans. Virology. 1998;244:405–416. doi: 10.1006/viro.1998.9125. [DOI] [PubMed] [Google Scholar]
- Li Y., DeFatta R., Anthony C., Sunavala G., De Benedetti A. A translationally regulated Tousled kinase phosphorylates histone H3 and confers radioresistance when overexpressed. Oncogene. 2001;20:726–738. doi: 10.1038/sj.onc.1204147. [DOI] [PubMed] [Google Scholar]
- Lin F.-T., MacDougald O.A., Diehl A.M., Lane M.D. A 30-kDa alternative translation product of the CCAAT/enhancer protein α message: transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. USA. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.C., Naujokas M., Zhu H., Nolet S., Park M. Intron-exon structure of the MET gene and cloning of an alternatively-spliced Met isoform reveals frequent exon-skipping of a single large internal exon. Oncogene. 1998;16:833–842. doi: 10.1038/sj.onc.1201599. [DOI] [PubMed] [Google Scholar]
- Lingelbach K., Dobberstein B. An extended RNA/RNA duplex structure within the coding region of mRNA does not block translational elongation. Nucleic Acids Res. 1988;16:3405–3414. doi: 10.1093/nar/16.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link D.C., Gutkind S.J., Robbins K.C., Ley T.J. Characterization of the 5′ untranslated region of the human c-fgr gene and identification of the major myelomonocytic c-fgr promoter. Oncogene. 1992;7:877–884. [PubMed] [Google Scholar]
- Liu F., Roizman B. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J. Virol. 1991;65:206–212. doi: 10.1128/jvi.65.1.206-212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Prolla G., Rostagno A., Chiarle R., Feiner H., Inghirami G. Initiation of translation from a downstream in-frame AUG codon on BRCA1 can generate the novel isoform protein ΔBRCA1(17aa) Oncogene. 2000;19:2767–2773. doi: 10.1038/sj.onc.1203599. [DOI] [PubMed] [Google Scholar]
- Liu L., Shao G., Steele-Perkins G., Huang S. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain-lacking product through an internal promoter. J. Biol. Chem. 1997;272:2984–2991. doi: 10.1074/jbc.272.5.2984. [DOI] [PubMed] [Google Scholar]
- Liu L., Dilworth D., Gao L., Monzon J., Summers A., Lassam N., Hogg D. Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat. Genet. 1999;21:128–132. doi: 10.1038/5082. [DOI] [PubMed] [Google Scholar]
- Liu Y., Biegalke B.J. The human cytomegalovirus UL35 gene encodes two proteins with different functions. J. Virol. 2002;76:2460–2468. doi: 10.1128/jvi.76.5.2460-2468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.-J., Huang H.-K., Donahue T.F. RNA polymerase I-promoted HIS4 expression yields uncapped, polyadenylated mRNA that is unstable and inefficiently translated in Saccharomycescerevisiae. Mol. Cell. Biol. 1998;18:665–675. doi: 10.1128/mcb.18.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock P., Ralph S., Stanley E., Boulet I., Ramsay R., Dunn A.R. Two isoforms of murine hck, generated by utilization of alternative translational initiation codons, exhibit different patterns of subcellular localization. Mol. Cell. Biol. 1991;11:4363–4370. doi: 10.1128/mcb.11.9.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Lee M.D., Smith B.L., Jung J.S., Agre P., Verdijk M.A.J., Merkx G., Rijss J.P.L., Deen P.M.T. The human AQP4 gene: definition of the locus encoding two water channel polypeptides in brain. Proc. Natl. Acad. Sci. USA. 1996;93:10908–10912. doi: 10.1073/pnas.93.20.10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Hanson K., DeLuca H.F. Cloning and origin of the two forms of chicken vitamin D receptor. Arch. Biochem. Biophys. 1997;339:99–106. doi: 10.1006/abbi.1996.9864. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz M., Feuermannl M., Jerouville B., Stas A., Boutry M. In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 2000;154:89–98. doi: 10.1016/s0168-9452(00)00195-3. [DOI] [PubMed] [Google Scholar]
- Luo D., Mari B., Stoll I., Anglard P. Alternative splicing and promoter usage generates an intracellular stromelysin 3 isoform directly translated as an active matrix metalloproteinase. J. Biol. Chem. 2002;277:25527–25536. doi: 10.1074/jbc.M202494200. [DOI] [PubMed] [Google Scholar]
- Luukkonen B.G.M., Tan W., Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyakh L., Ghosh P., Rice N.R. Expression of NFAT-family proteins in normal human T cells. Mol. Cell. Biol. 1997;17:2475–2484. doi: 10.1128/mcb.17.5.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod D., Ali R.R., Bird A. An alternative promoter in the mouse major histocompatibility complex class II I-Aβ gene: implications for the origin of CpG islands. Mol. Cell. Biol. 1998;18:4433–4443. doi: 10.1128/mcb.18.8.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzella J.M., Blackshear P.J. Regulation of rat ornithine decarboxylase mRNA translation by its 5′-untranslated region. J. Biol. Chem. 1990;265:11817–11822. [PubMed] [Google Scholar]
- Marth J.D., Overell R.W., Meier K.E., Krebs E.G., Perlmutter R.M. Translational activation of the lck proto-oncogene. Nature. 1988;332:171–173. doi: 10.1038/332171a0. [DOI] [PubMed] [Google Scholar]
- Maser R.S., Zinkel R., Petrini J.H.J. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- Mathews M.B. Lost in translation. Trends Biochem. Sci. 2002;27:267–269. doi: 10.1016/s0968-0004(02)02087-x. [DOI] [PubMed] [Google Scholar]
- Mayo M.A., Ziegler-Graff V. Molecular biology of luteoviruses. Adv. Virus Res. 1996;46:413–460. doi: 10.1016/s0065-3527(08)60077-9. [DOI] [PubMed] [Google Scholar]
- McCarthy J.E.G. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil H.P., Reynolds D.S., Schiller V., Ghildyal N., Gurley D.S., Austen K.F., Stevens R.L. Isolation, characterization, and transcription of the gene encoding mouse mast cell protease 7. Proc. Natl. Acad. Sci. USA. 1992;89:11174–11178. doi: 10.1073/pnas.89.23.11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer H.A., Dictus W.J.A.G., Keuning E.D., Thomas A.A.M. Translational control of the Xenopuslaevis connexin-41 5′-untranslated region by three upstream open reading frames. J. Biol. Chem. 2000;275:30787–30793. doi: 10.1074/jbc.M005531200. [DOI] [PubMed] [Google Scholar]
- Melén K., Keskinen P., Ronni T., Sareneva T., Lounatmaa K., Julkunen I. Human MxB protein, an interferon-α-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem. 1996;271:23478–23486. doi: 10.1074/jbc.271.38.23478. [DOI] [PubMed] [Google Scholar]
- Meulewaeter F., Cornelissen M., Van Emmelo J. Subgenomic RNAs mediate expression of cistrons located internally on the genomic RNA of tobacco necrosis virus strain A. J. Virol. 1992;66:6419–6428. doi: 10.1128/jvi.66.11.6419-6428.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mével-Ninio M., Fouilloux E., Guénal I., Vincent A. The three dominant female-sterile mutations of the Drosophilaovo gene are point mutations that create new translation-initiator AUG codons. Development. 1996;122:4131–4138. doi: 10.1242/dev.122.12.4131. [DOI] [PubMed] [Google Scholar]
- Mikulits W., Schranzhofer M., Beug H., Mullner E.W. Post-transcriptional control via iron-responsive elements: the impact of aberrations in hereditary disease. Mutat. Res. 1999;437:219–230. doi: 10.1016/s1383-5742(99)00085-x. [DOI] [PubMed] [Google Scholar]
- Miller W.A., Koev G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- Miller W.A., Dreher T.W., Hall T.C. Synthesis of brome mosaic virus subgenomic RNA invitro by internal initiation on (−)-sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- Minami T., Rosenberg R.D., Aird W.C. Transforming growth factor-β1-mediated inhibition of the flk-1/KDR gene is mediated by a 5′-untranslated region palindromic GATA site. J. Biol. Chem. 2001;276:5395–5402. doi: 10.1074/jbc.M008798200. [DOI] [PubMed] [Google Scholar]
- Mitchelmore C., Kjaerulff K.M., Pedersen H.C., Nielsen J.V., Rasmussen T.E., Fisker M.F., Finsen B., Pedersen K.M., Jensen N.A. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J. Biol. Chem. 2002;277:7598–7609. doi: 10.1074/jbc.M110023200. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi T., Nikodem V.M. Regulation of expression of the alternative mRNAs of the rat α-thyroid hormone receptor gene. J. Biol. Chem. 1989;264:8900–8904. [PubMed] [Google Scholar]
- Mittag M., Eckerskorn C., Strupat K., Hastings J.W. Differential translational initiation of lbp mRNA is caused by a 5′ upstream open reading frame. FEBS Lett. 1997;411:245–250. doi: 10.1016/s0014-5793(97)00701-1. [DOI] [PubMed] [Google Scholar]
- Mo Y.-Y., Reynolds A.B. Identification of murine p120cas isoforms and heterogeneous expression of p120cas isoforms in human tumor cell lines. Cancer Res. 1996;56:2633–2640. [PubMed] [Google Scholar]
- Molina C.A., Foulkes N.S., Lalli E., Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- Morlé F., Lopez B., Henni T., Godet J. α-Thalassaemia associated with the deletion of two nucleotides at position −2 and −3 preceding the AUG codon. EMBO J. 1985;4:1245–1250. doi: 10.1002/j.1460-2075.1985.tb03767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish B.C., Rumsby M.G. The 5′ UTR of protein kinase C ε confers translational regulation in vitro and in vivo. Biochem. Biophys. Res. Commun. 2001;283:1091–1098. doi: 10.1006/bbrc.2001.4909. [DOI] [PubMed] [Google Scholar]
- Mudge S.J., Williams J.H., Eyre H.J., Sutherland G.R., Cowan P.J., Power D.A. Complex organisation of the 5′-end of the human glycine tRNA synthetase gene. Gene. 1998;209:45–50. doi: 10.1016/s0378-1119(98)00007-9. [DOI] [PubMed] [Google Scholar]
- Mullineaux P.M., Rigden J.E., Dry I.B., Krake L.R., Rezaian M.A. Mapping of the polycistronic RNAs of tomato leaf curl geminivirus. Virology. 1993;193:414–423. doi: 10.1006/viro.1993.1138. [DOI] [PubMed] [Google Scholar]
- Muralidhar S., Becerra S.P., Rose J.A. Site-directed mutagenesis of adeno-associated virus type 2 structural protein initiation codons: effects on regulation of synthesis and biological activity. J. Virol. 1994;68:170–176. doi: 10.1128/jvi.68.1.170-176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal A., Martell M., Lytle J.R., Lyons A.J., Robertson H.D., Cabot B., Esteban J.I., Esteban R., Guardia J., Gómez J. Specific cleavage of hepatitis C virus RNA genome by human RNase P. J. Biol. Chem. 2002;277:30606–30613. doi: 10.1074/jbc.M203595200. [DOI] [PubMed] [Google Scholar]
- Nador R.G., Milligan L.L., Flore O., Wang X., Arvanitakis L., Knowles D.M., Cesarman E. Expression of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor monocistronic and bicistronic transcripts in primary effusion lymphocytes. Virology. 2001;287:62–70. doi: 10.1006/viro.2001.1016. [DOI] [PubMed] [Google Scholar]
- Nagpal S., Zelent A., Chambon P. RAR-β4, a retinoic acid receptor isoform is generated from RAR-β2 by alternative splicing and usage of a CUG initiator codon. Proc. Natl. Acad. Sci. USA. 1992;89:2718–2722. doi: 10.1073/pnas.89.7.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis G., Hilger F., Fink G.R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986;46:235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- Neeleman L., Olsthoorn R.C.L., Linthorst H.J.M., Bol J.F. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc. Natl. Acad. Sci. USA. 2001;98:14286–14291. doi: 10.1073/pnas.251542798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill J.D., Reardon I.M., Heinrikson R.L. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J. Virol. 1991;65:5440–5447. doi: 10.1128/jvi.65.10.5440-5447.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen F.C., Gammeltoft S., Christiansen J. Translational discrimination of mRNAs coding for human insulin-like growth factor II. J. Biol. Chem. 1990;265:13431–13434. [PubMed] [Google Scholar]
- Nielsen F.C., Østergaard L., Nielsen J., Christiansen J. Growth-dependent translation of IGF-II mRNA by a rapamycin-sensitive pathway. Nature. 1995;377:358–362. doi: 10.1038/377358a0. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Hirose E., Uchimura Y., Nakamura M., Umeda M., Nishii K., Mori N., Nishimoto T. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 2001;272:25–33. doi: 10.1016/s0378-1119(01)00553-4. [DOI] [PubMed] [Google Scholar]
- Nolte D., Müller U. Human O-GlcNAc transferase (OGT): genomic structure, analysis of splice variants, fine mapping in Xq13.1. Mamm. Genome. 2002;13:62–64. doi: 10.1007/s00335-001-2108-9. [DOI] [PubMed] [Google Scholar]
- Nonaka M., Ishikawa N., Passwell J., Natsuume-Sakai S., Colten H.R. Tissue-specific initiation of murine complement factor B mRNA transcription. J. Immunol. 1989;142:1377–1382. [PubMed] [Google Scholar]
- O'Connor J.B., Brian D.A. Downstream ribosomal entry for translation of coronavirus TGEV gene 3b. Virology. 2000;269:172–182. doi: 10.1006/viro.2000.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T., Funai T., Ichiyama A. Generation from a single gene of two mRNAs that encode the mitochondrial and peroxisomal serine:pyruvate aminotransferase of rat liver. J. Biol. Chem. 1990;265:7513–7519. [PubMed] [Google Scholar]
- O'Neill G., Aoki M., Brown R.H., Jr. ABCD1 translation-initiator mutation demonstrates genotype-phenotype correlation for AMN. Neurology. 2001;57:1956–1962. doi: 10.1212/wnl.57.11.1956. [DOI] [PubMed] [Google Scholar]
- Ossipow V., Descombes P., Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc. Natl. Acad. Sci. USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst T., Mueller B.U., Zhang P., Radomska H.S., Narravula S., Schnittger S., Behre G., Hiddemann W., Tenen D.G. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat. Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- Packham G., Brimmell M., Cleveland J.L. Mammalian cells express two differently localized Bag-1 isoforms generated by alternative translation initiation. Biochem. J. 1997;328:807–813. doi: 10.1042/bj3280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmarini M., Murgia C., Fan H. Spliced and prematurely polyadenylated Jaagsiekte sheep retrovirus-specific RNAs from infected or transfected cells. Virology. 2002;294:180–188. doi: 10.1006/viro.2001.1323. [DOI] [PubMed] [Google Scholar]
- Paraskeva E., Gray N.K., Schläger B., Wehr K., Hentze M.W. Ribosomal pausing and scanning arrest as mechanisms of translational regulation from cap-distal iron-responsive elements. Mol. Cell. Biol. 1999;19:807–816. doi: 10.1128/mcb.19.1.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardigol A., Forssmann U., Zucht H.-D., Loetscher P., Schulz-Knappe P., Baggiolini M., Forssmann W.-G., Mägert H.-J. HCC-2, a human chemokine: gene structure, expression pattern, and biological activity. Proc. Natl. Acad. Sci. USA. 1998;95:6308–6313. doi: 10.1073/pnas.95.11.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-S., Himmelbach A., Browning K.S., Hohn T., Ryabova L.A. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001;106:723–733. doi: 10.1016/s0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., Gultyaev A.P., Spaan W.J.M., Snijder E.J. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 2000;74:11642–11653. doi: 10.1128/jvi.74.24.11642-11653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T., Harvey R., Smith A.E. The size of Rous sarcoma virus mRNAs active in cell-free translation. Nature. 1977;268:416–420. doi: 10.1038/268416a0. [DOI] [PubMed] [Google Scholar]
- Paz I., Abramovitz L., Choder M. Starved Saccharomycescerevisiae cells have the capacity to support internal initiation of translation. J. Biol. Chem. 1999;274:21741–21745. doi: 10.1074/jbc.274.31.21741. [DOI] [PubMed] [Google Scholar]
- Peabody D.S., Subramani S., Berg P. Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol. Cell. Biol. 1986;6:2704–2711. doi: 10.1128/mcb.6.7.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecci A., Viegas L.R., Barañao J.L., Beato M. Promoter choice influences alternative splicing and determines the balance of isoforms expressed from the mouse bcl-X gene. J. Biol. Chem. 2001;276:21062–21069. doi: 10.1074/jbc.M008665200. [DOI] [PubMed] [Google Scholar]
- Pecqueur C., Cassard-Doulcier A.-M., Raimbault S., Miroux B., Fleury C., Gelly C., Bouillaud F., Ricquier D. Functional organization of the human uncoupling protein-2 gene, and juxtaposition to the uncoupling protein-3 gene. Biochem. Biophys. Res. Commun. 1999;255:40–46. doi: 10.1006/bbrc.1998.0146. [DOI] [PubMed] [Google Scholar]
- Pecqueur C., Alves-Guerra M.-C., Gelly C., Lévi-Meyrueis C., Couplan E., Collins S., Ricquier D., Bouillaud F., Miroux B. Uncoupling protein 2, in vivo distribution, induction upon oxidative stress, and evidence for translational regulation. J. Biol. Chem. 2001;276:8705–8712. doi: 10.1074/jbc.M006938200. [DOI] [PubMed] [Google Scholar]
- Perälä M., Elima K., Metsäranta M., Rosati R., de Crombrugghe B., Vuorio E. The exon structure of the mouse α2(IX) collagen gene shows unexpected divergence from the chick gene. J. Biol. Chem. 1994;269:5064–5071. [PubMed] [Google Scholar]
- Peremyslov V.V., Dolja V.V. Identification of the subgenomic mRNAs that encode 6-kDa movement protein and Hsp70 homolog of Beetyellowsvirus. Virology. 2002;295:299–306. doi: 10.1006/viro.2002.1396. [DOI] [PubMed] [Google Scholar]
- Peri S., Pandey A. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- Perrais M., Pigny P., Ducourouble M.-P., Petitprez D. Characterization of human mucin gene MUC4 promoter. J. Biol. Chem. 2001;276:30923–30933. doi: 10.1074/jbc.M104204200. [DOI] [PubMed] [Google Scholar]
- Perry M.E., Mendrysa S.M., Saucedo L.J., Tannous P., Holubar M. p76MDM2 inhibits the ability of p90MDM2 to destabilize p53. J. Biol. Chem. 2000;275:5733–5738. doi: 10.1074/jbc.275.8.5733. [DOI] [PubMed] [Google Scholar]
- Pesole G., Gissi C., Grillo G., Licciulli F., Liuni S., Saccone C. Analysis of oligonucleotide AUG start codon context in eukariotic mRNAs. Gene. 2000;261:85–91. doi: 10.1016/s0378-1119(00)00471-6. [DOI] [PubMed] [Google Scholar]
- Pesole G., Mignone F., Gissi C., Grillo G., Licciulli F., Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U.T. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.J., Morris J.F. Human myeloid zinc finger gene MZF produces multiple transcripts and encodes a SCAN box protein. Gene. 2000;254:105–118. doi: 10.1016/s0378-1119(00)00281-x. [DOI] [PubMed] [Google Scholar]
- Petty I.T.D., Edwards M.C., Jackson A.O. Systematic movement of an RNA plant virus determined by a point substitution in a 5′ leader sequence. Proc. Natl. Acad. Sci. USA. 1990;87:8894–8897. doi: 10.1073/pnas.87.22.8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps D.E., Hsiao K.-M., Li Y., Hu N., Franklin D.S., Westphal E., Lee E.Y.-H.P., Xiong Y. Coupled transcriptional and translational control of cyclin-dependent kinase inhibitor p18INK4c expression during myogenesis. Mol. Cell. Biol. 1998;18:2334–2343. doi: 10.1128/mcb.18.4.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe J. Hepatocyte-nuclear factor 3β gene transcripts generate protein isoforms with different transactivation properties on the glucagon gene. Mol. Endocrinol. 1995;9:368–374. doi: 10.1210/mend.9.3.7776982. [DOI] [PubMed] [Google Scholar]
- Pinkstaff J.K., Chappell S.A., Mauro V.P., Edelman G.M., Krushel L.A. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc. Natl. Acad. Sci. USA. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman G.D., Green J.M., McDonald V.L., Neubauer M.G., Disteche C.M., Todaro G.J., Shoyab M. The amphiregulin gene encodes a novel epidermal growth factor-related protein with tumor-inhibitory activity. Mol. Cell. Biol. 1990;10:1969–1981. doi: 10.1128/mcb.10.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C., Pitiot G., Plumb M., Lowe S., de Verneuil H., Grandchamp B. Characterization of hypersensitive sites, protein-binding motifs, and regulatory elements in both promoters of the mouse porphobilinogen deaminase gene. J. Biol. Chem. 1991;266:10562–10569. [PubMed] [Google Scholar]
- Portis J.L., Spangrude G.J., McAtee F.J. Identification of a sequence in the unique 5′ open reading frame of the gene encoding glycosylated gag which influences the incubation period of neurodegenerative disease induced by a murine retrovirus. J. Virol. 1994;68:3879–3887. doi: 10.1128/jvi.68.6.3879-3887.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis J.L., Fujisawa R., McAtee F.J. The glycosylated gag protein of MuLV is a determinant of neuroinvasiveness: analysis of second site revertants of a mutant MuLV virus lacking expression of this protein. Virology. 1996;226:384–392. doi: 10.1006/viro.1996.0666. [DOI] [PubMed] [Google Scholar]
- Probst-Kepper M., Stroobant V., Kridel R., Gaugler B., Landry C., Brasseur F., Cosyns J.-P., Weynand B., Boon T., Van den Eynde B.J. An alternative open reading frame of the human macrophage colony-stimulating factor gene is independently translated and codes for an antigenic peptide of 14 amino acids recognized by tumor-infiltrating CD8 T lymphocytes. J. Exp. Med. 2001;193:1189–1198. doi: 10.1084/jem.193.10.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Katz K.S., Sicotte H., Magiott D.R. Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. doi: 10.1016/s0168-9525(99)01882-x. [DOI] [PubMed] [Google Scholar]
- Pushpa-Rekha T.R., Burdsall A.L., Oleksa L.M., Chisolm G.M., Driscoll D.M. Rat phospholipid-hydroperoxide glutathione peroxidase. cDNA cloning and identification of multiple transcription and translation start sites. J. Biol. Chem. 1995;270:26993–26999. doi: 10.1074/jbc.270.45.26993. [DOI] [PubMed] [Google Scholar]
- Quiñones-Kochs M.I., Schnell M.J., Buonocore L., Rose J.K. Mechanisms of loss of foreign gene expression in recombinant vesicular stomatitis viruses. Virology. 2001;287:427–435. doi: 10.1006/viro.2001.1058. [DOI] [PubMed] [Google Scholar]
- Raney A., Baron A.C., Mize G.J., Law G.L., Morris D.R. In vitro translation of the upstream open reading frame in the mammalian mRNA encoding S-adenosylmethionine decarboxylase. J. Biol. Chem. 2000;275:24444–24450. doi: 10.1074/jbc.M003364200. [DOI] [PubMed] [Google Scholar]
- Rao C.D., Pech M., Robbins K.C., Aaronson S.A. The 5′ untranslated sequence of the c-sis platelet-derived growth factor 2 transcript is a potent translational inhibitor. Mol. Cell. Biol. 1988;8:284–292. doi: 10.1128/mcb.8.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard C., Thomas G., Kozma S.C. A single gene encodes two isoforms of the p70 S6 kinase: activation upon mitogenic stimulation. Proc. Natl. Acad. Sci. USA. 1992;89:4052–4056. doi: 10.1073/pnas.89.9.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Stiles G.L. Posttranscriptional mRNA processing as a mechanism for regulation of human A1 adenosine receptor expression. Proc. Natl. Acad. Sci. USA. 1994;91:4864–4866. doi: 10.1073/pnas.91.11.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensen S.S.M., Thijssen V.L.J.L., De Vries C.J., Doevendans P.A., Detera-Wadleigh S.D., Van Eys G.J.J.M. Expression of the smoothelin gene is mediated by alternative promoters. Cardiovasc. Res. 2002;55:850–863. doi: 10.1016/s0008-6363(02)00491-1. [DOI] [PubMed] [Google Scholar]
- Reuss F.U., Coffin J.M. Mouse mammary tumor virus superantigen expression in B cells is regulated by a central enhancer within the pol gene. J. Virol. 1998;72:6073–6082. doi: 10.1128/jvi.72.7.6073-6082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Ito T., Meyerowitz E.M. Non-AUG initiation of AGAMOUS mRNA translation in Arabidopsisthaliana. Mol. Cell. Biol. 1999;19:8505–8512. doi: 10.1128/mcb.19.12.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi D., Rubio-Godoy V., Dutoit V., Lienard D., Salvi S., Guillaume P., Speiser D., Stockert E., Spagnoli G., Servis C., Cerottini J.-C., Lejeune F., Romero P., Valmori D. Efficient simultaneous presentation of NY-ESO-1/LAGE-1 primary and nonprimary open reading frame-derived CTL epitopes in melanoma. J. Immunol. 2000;165:7253–7261. doi: 10.4049/jimmunol.165.12.7253. [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Kochetov A.V., Kondrashov F.A., Koonin E.V., Milanesi L. Presence of ATG triplets in 5′ untranslated regions of eukaryotic cDNAs correlates with a ‘weak’ context of the start codon. Bioinformatics. 2001;17:890–900. doi: 10.1093/bioinformatics/17.10.890. [DOI] [PubMed] [Google Scholar]
- Ronsin C., Chung-Scott V., Poullion I., Aknouche N., Gaudin C., Triebel F. A non-AUG-defined alternative open reading frame of the intestinal carboxyl esterase mRNA generates an epitope recognized by renal cell carcinoma-reactive tumor-infiltrating lymphocytes in situ. J. Immunol. 1999;163:483–490. [PubMed] [Google Scholar]
- Rosenberg S.A., Tong-On P., Li Y., Riley J.P., El-Gamil M., Parkhurst M.R., Robbins P.F. Identification of BING-4 cancer antigen translated from an alternative open reading frame of a gene in the extended MHC class II region using lymphocytes from a patient with a durable complete regression following immunotherapy. J. Immunol. 2002;168:2402–2407. doi: 10.4049/jimmunol.168.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H., Hill J.R., Fatemie-Nainie S., Morris D.R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Influence of the structure of the 5′ transcript leader on regulation by the upstream open reading frame. J. Biol. Chem. 1994;269:17905–17910. [PubMed] [Google Scholar]
- Rüegsegger U., Leber J.H., Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- Rueter S.M., Dawson T.R., Emeson R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–79. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Ryabova L.A., Pooggin M.M., Dominguez D.I., Hohn T. Continuous and discontinuous ribosome scanning on the cauliflower mosaic virus 35S RNA leader is controlled by short open reading frames. J. Biol. Chem. 2000;275:37278–37284. doi: 10.1074/jbc.M004909200. [DOI] [PubMed] [Google Scholar]
- Sadler R., Wu L., Forghani B., Renne R., Zhong W., Herndier B., Ganem D. A complex translational program generates multiple novel proteins from the latently expressed Kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 1999;73:5722–5730. doi: 10.1128/jvi.73.7.5722-5730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Washio T., Saito R., Shinagawa A., Itoh M., Shibata K., Carninci P., Konno H., Kawai J., Hayashizaki Y., Tomita M. Correlation between sequence conservation of the 5′ untranslated region and codon usage bias in Musmusculus genes. Gene. 2001;276:101–105. doi: 10.1016/s0378-1119(01)00671-0. [DOI] [PubMed] [Google Scholar]
- Sanchez-Góngora E., Lisbona C., de Gregorio R., Ballester A., Calvo V., Pérez-Jurado L., Alemany S. COT kinase proto-oncogene expression in T cells. J. Biol. Chem. 2000;275:31379–31386. doi: 10.1074/jbc.M000382200. [DOI] [PubMed] [Google Scholar]
- Santagata S., Gomez C.A., Sobacchi C., Bozzi F., Abinum M., Pasic S., Cortes P., Vezzoni P., Villa A. N-terminal RAG1 frameshift mutations in Omenn's syndrome: internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role invivo for the N-terminal domains. Proc. Natl. Acad. Sci. USA. 2000;97:14572–14577. doi: 10.1073/pnas.97.26.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamarina-Fojo S., Peterson K., Knapper C., Qui Y., Freeman L., Cheng J.-F., Osorio J., Remaley A., Yang X.-P., Haudenschild C., Prades C., Chimini G., Blackmon E., Francois T., Duverger N., Rubin E.M., Rosier M., Denèfle P., Fredrickson D.S., Brewer H.B., Jr. Complete genomic sequence of the human ABCA1 gene: analysis of the human and mouse ATP-binding cassette A promoter. Proc. Natl. Acad. Sci. USA. 2000;97:7987–7992. doi: 10.1073/pnas.97.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saris C.J.M., Domen J., Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10:655–664. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S., Starck J., Gonnet C., Doubeikovski A., Melet F., Morlé F. Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5′ upstream open reading frames in Fli-1 mRNA. Mol. Cell. Biol. 2000;20:2959–2969. doi: 10.1128/mcb.20.9.2959-2969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahara M., Amano S., Sato H., Yang J.G., Hayase Y., Kaneko M., Sato I., Suzuki M., Hazama F. Normal developing rat brain expresses a platelet-derived growth factor B chain (c-sis) mRNA truncated at the 5′ end. Oncogene. 1998;16:1571–1578. doi: 10.1038/sj.onc.1201679. [DOI] [PubMed] [Google Scholar]
- Saucedo L.J., Myers C.D., Perry M.E. Multiple murine double minute gene 2 (MDM2) proteins are induced by ultraviolet light. J. Biol. Chem. 1999;274:8161–8168. doi: 10.1074/jbc.274.12.8161. [DOI] [PubMed] [Google Scholar]
- Schaller H., Fischer M. Transcriptional control of hepadnavirus gene expression. Curr. Top. Microbiol. Immunol. 1991;168:21–39. doi: 10.1007/978-3-642-76015-0_2. [DOI] [PubMed] [Google Scholar]
- Scharnhorst V., Dekker P., van der Eb A.J., Jochemsen A.G. Internal translation initiation generates novel WT1 protein isoforms with distinct biological properties. J. Biol. Chem. 1999;274:23456–23462. doi: 10.1074/jbc.274.33.23456. [DOI] [PubMed] [Google Scholar]
- Scherer P.E., Tang Z., Chun M., Sargiacomo M., Lodish H.F., Lisanti M.P. Caveolin isoforms differ in their N-terminal protein sequence and subcellular distribution. J. Biol. Chem. 1995;270:16395–16401. doi: 10.1074/jbc.270.27.16395. [DOI] [PubMed] [Google Scholar]
- Schirawski J., Voyatzakis A., Zaccomer B., Bernardi F., Haenni A.-L. Identification and functional analysis of the turnip yellow mosaic tymovirus subgenomic promoter. J. Virol. 2000;74:11073–11080. doi: 10.1128/jvi.74.23.11073-11080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld A., Davidowitz E.J., Burk R.D. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc. Natl. Acad. Sci. USA. 1998;95:8817–8822. doi: 10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N., Chin L., Chen K., Torres R., Rao G., Guida P., Skoultchi A.I., DePinho R.A. An amino-terminal domain of Mxi1 mediates anti-myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- Schwartz S., Felber B.K., Pavlakis G.N. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol. Cell. Biol. 1992;12:207–219. doi: 10.1128/mcb.12.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman S.A., Mertz J.E. Mechanisms of synthesis of virion proteins from the functionally bigenic late mRNAs of simian virus 40. J. Virol. 1988;62:954–961. doi: 10.1128/jvi.62.3.954-961.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman S.A., Gelembiuk G.W., Mertz J.E. Translation initiation at a downstream AUG occurs with increased efficiency when the upstream AUG is located very close to the 5′ cap. J. Virol. 1990;64:453–457. doi: 10.1128/jvi.64.1.453-457.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf U., Assmann G. Cloning, expression, and nucleotide sequence of rat liver sterol carrier protein 2 cDNAs. J. Biol. Chem. 1991;266:630–636. [PubMed] [Google Scholar]
- Senanayake S.D., Brian D.A. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 1997;48:101–105. doi: 10.1016/S0168-1702(96)01423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang F., Deng G., Obin M., Wu C.C.N., Gong X., Smith D., Laursen R.A., Andley U.P., Reddan J.R., Taylor A. Ubiquitin-activating enzyme (E1) isoforms in lens epithelial cells: origin of translation, E2 specificity and cellular localization determined with novel site-specific antibodies. Exp. Eye Res. 2001;73:827–836. doi: 10.1006/exer.2001.1091. [DOI] [PubMed] [Google Scholar]
- Shann Y.-J., Hsu M.-T. Cloning and characterization of liver-specific isoform of Chk1 gene from rat. J. Biol. Chem. 2001;276:48863–48870. doi: 10.1074/jbc.M108253200. [DOI] [PubMed] [Google Scholar]
- Shastri N., Schwab S., Serwold T. Producing nature's gene-chips: the generation of peptides for display by MHC class I molecules. Annu. Rev. Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- Sheu S.Y., Lo S.J. Preferential ribosomal scanning is involved in the differential synthesis of the hepatitis B viral surface antigens from subgenomic transcripts. Virology. 1992;188:353–357. doi: 10.1016/0042-6822(92)90764-g. [DOI] [PubMed] [Google Scholar]
- Shih D.S., Kaesberg P. Translation of the RNAs of brome mosaic virus: the monocistronic nature of RNA1 and RNA2. J. Mol. Biol. 1976;103:77–88. doi: 10.1016/0022-2836(76)90053-x. [DOI] [PubMed] [Google Scholar]
- Shishido-Hara Y., Hara Y., Larson T., Yasui K., Nagashima K., Stoner G.L. Analysis of capsid formation of human polyomavirus JC (Tokyo-1 strain) by a eukaryotic expression system: splicing of late RNAs, translation and nuclear transport of major capsid protein VP1, and capsid assembly. J. Virol. 2000;74:1840–1853. doi: 10.1128/jvi.74.4.1840-1853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmulevitz M., Yameen Z., Dawe S., Shou J., O'Hara D., Holmes I., Duncan R. Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation. J. Virol. 2002;76:609–618. doi: 10.1128/JVI.76.2.609-618.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J.D., Pfarr C.M. Translational regulation of the JunD messenger RNA. J. Biol. Chem. 2002;277:32697–32705. doi: 10.1074/jbc.M204553200. [DOI] [PubMed] [Google Scholar]
- Shuttleworth T.L., Wilson M.D., Wicklow B.A., Wilkins J.A., Triggs-Raine B.L. Characterization of the murine hyaluronidase gene region reveals complex organization and cotranscription of Hyal1 with downstream genes, Fus2 and Hyal3. J. Biol. Chem. 2002;277:23008–23018. doi: 10.1074/jbc.M108991200. [DOI] [PubMed] [Google Scholar]
- Siddell S.G., Smith A.E. Polyoma virus has three late mRNAs: one for each virion protein. J. Virol. 1978;27:427–431. doi: 10.1128/jvi.27.2.427-431.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signori E., Bagni C., Papa S., Primerano B., Rinaldi M., Amaldi F., Fazio V.M. A somatic mutation in the 5′ UTR of BRCA1 gene in sporadic breast cancer causes down-modulation of translation efficiency. Oncogene. 2001;20:4596–4600. doi: 10.1038/sj.onc.1204620. [DOI] [PubMed] [Google Scholar]
- Sivakumaran K., Hacker D.L. The 105-kDa polyprotein of southern bean mosaic virus is translated by scanning ribosomes. Virology. 1998;246:34–44. doi: 10.1006/viro.1998.9183. [DOI] [PubMed] [Google Scholar]
- Sloan J., Kinghorn J.R., Unkles S.E. The two subunits of human molybdopterin synthase: evidence for a bicistronic messenger RNA with overlapping reading frames. Nucleic Acids Res. 1999;27:854–858. doi: 10.1093/nar/27.3.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodskaya O.R., Gmyl A.P., Maslova S.V., Tolskaya E.A., Viktorova E.G., Agol V.I. Poliovirus neurovirulence correlates with the presence of a cryptic AUG upstream of the initiator codon. Virology. 1996;221:141–150. doi: 10.1006/viro.1996.0360. [DOI] [PubMed] [Google Scholar]
- Slusher L.B., Gillman E.C., Martin N.C., Hopper A.K. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc. Natl. Acad. Sci. USA. 1991;88:9789–9793. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok J., Wang X.-Z., Batchvarova N., Kuroda M., Harding H., Ron D. CHOP-dependent stress-inducible expression of a novel form of carbonic anhydrase VI. Mol. Cell. Biol. 1999;19:495–504. doi: 10.1128/mcb.19.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki D., Schwarz S., Wimmer E., Lipp M., Bernhardt G. The promoters for human and monkey poliovirus receptors. J. Biol. Chem. 1997;272:5579–5586. doi: 10.1074/jbc.272.9.5579. [DOI] [PubMed] [Google Scholar]
- Souciet G., Menand B., Ovesna J., Cosset A., Dietrich A., Wintz H. Characterization of two bifunctional Arabidopsisthaliana genes coding for mitochondrial and cytosolic forms of valyl-tRNA synthetase and threonyl-tRNA synthetase by alternative use of two in-frame AUGs. Eur. J. Biochem. 1999;266:848–854. doi: 10.1046/j.1432-1327.1999.00922.x. [DOI] [PubMed] [Google Scholar]
- Spiropoulou C.F., Nichol S.T. A small highly basic protein is encoded in overlapping frame within the P gene of vesicular stomatitis virus. J. Virol. 1993;67:3103–3110. doi: 10.1128/jvi.67.6.3103-3110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spotts G.D., Patel S.V., Xiao Q., Hann S.R. Identification of downstream-initiated c-myc proteins which are dominant-negative inhibitors of transactivation by full-length c-myc proteins. Mol. Cell. Biol. 1997;17:1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey S.N., Jordan D., Williamson A.J.K., Brown M., Coote J.H., Arrand J.R. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J. Virol. 2000;74:7284–7297. doi: 10.1128/jvi.74.16.7284-7297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallmeyer B., Drugeon G., Reiss J., Haenni A.L., Mendel R.R. Human molybdopterin synthase gene: identification of a bicistronic transcript with overlapping reading frames. Am. J. Hum. Genet. 1999;64:698–705. doi: 10.1086/302295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel L.F., Telly D.L., Leonard J., Rice B.A., Monks B., Sawicki J.A. Elements in the murine c-mos messenger RNA 5′-untranslated region repress translation of downstream coding sequences. Cell Growth Differ. 1996;7:1415–1424. [PubMed] [Google Scholar]
- Stefanovic B., Hellerbrand C., Brenner D.A. Regulatory role of the conserved stem-loop structure at the 5′ end of collagen α1(I) mRNA. Mol. Cell. Biol. 1999;19:4334–4342. doi: 10.1128/mcb.19.6.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L.-K., Qi Y. Characterization of human MAPRE genes and their proteins. Genomics. 2001;71:142–149. doi: 10.1006/geno.2000.6428. [DOI] [PubMed] [Google Scholar]
- Sun Z., Sassone-Corsi P., Means A.R. Calspermin gene transcription is regulated by two cyclic AMP response elements contained in an alternative promoter in the calmodulin kinase IV gene. Mol. Cell. Biol. 1995;15:561–571. doi: 10.1128/mcb.15.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva L.J., Mather K.A., Gillespie M.T., Webb G.C., Ng K.W., Winslow G.A., Wood W.I., Martin T.J., Hudson P.J. Structure of the 5′ flanking region of the gene encoding human parathyroid-hormone-related protein (PTHrP) Gene. 1989;77:95–105. doi: 10.1016/0378-1119(89)90363-6. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Ishihara D., Sasaki M., Nakagawa H., Hata H., Tsunoda T., Watanabe M., Komatsu T., Ota T., Isogai T., Suyama A., Sugano S. Statistical analysis of the 5′ untranslated region of human mRNA using “oligo-capped” cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- Szybiak U., Bouley J.P., Fritsch C. Evidence for the existence of a coat protein messenger RNA associated with the top component of each of three tymoviruses. Nucleic Acids Res. 1978;5:1821–1831. doi: 10.1093/nar/5.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor C.S., Marin M., Nouri A., Kavanaugh M.P., Kabat D. Truncated forms of the dual function human ASCT2 neutral amino acid transporter/retroviral receptor are translationally initiated at multiple alternative CUG and GUG codons. J. Biol. Chem. 2001;276:27221–27230. doi: 10.1074/jbc.M100737200. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Ito S., Kiuchi K. The 5′-untranslated region of the mouse glial cell line-derived neurotrophic factor gene regulates expression at both the transcriptional and translational levels. Mol. Brain Res. 2001;91:81–95. doi: 10.1016/s0169-328x(01)00125-5. [DOI] [PubMed] [Google Scholar]
- Teruya J.H., Kutsunai S.Y., Spear D.H., Edwards P.A., Clarke C.F. Testis-specific transcription initiation sites of rat farnesyl pyrophosphate synthetase mRNA. Mol. Cell. Biol. 1990;10:2315–2326. doi: 10.1128/mcb.10.5.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilmann D.A., Willis L.G., Bosch B.-J., Forsythe I.J., Li Q. The baculovirus transcriptional transactivator ieO produces multiple products by internal initiation of translation. Virology. 2001;290:211–223. doi: 10.1006/viro.2001.1165. [DOI] [PubMed] [Google Scholar]
- Thoma C., Hasselblatt P., Köck J., Chang S.-F., Hockenjos B., Will H., Hentze M.W., Blum H.E., von Weizsäcker F., Offensperger W.-B. Generation of stable mRNA fragments and translation of N-truncated proteins induced by antisense oligonucleotides. Mol. Cell. 2001;8:865–872. doi: 10.1016/s1097-2765(01)00364-1. [DOI] [PubMed] [Google Scholar]
- Tolstrup A.B., Bejder A., Fleckner J., Justesen J. Transcriptional regulation of the interferon-γ-inducible tryptophanyl-tRNA synthetase includes alternative splicing. J. Biol. Chem. 1995;270:397–403. doi: 10.1074/jbc.270.1.397. [DOI] [PubMed] [Google Scholar]
- Tsai T.F., Chen K.S., Weber J.S., Justice M.J., Beaudet A.L. Evidence for translational regulation of the imprinted Snurf-Snrpn locus in mice. Hum. Mol. Genet. 2002;11:1659–1668. doi: 10.1093/hmg/11.14.1659. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Egashira M., Niikawa N., Wada Y., Honke K. Cancer-associated alternative usage of multiple promoters of human GalCer sulfotransferase gene. Eur. J. Biochem. 2000;267:2672–2679. doi: 10.1046/j.1432-1327.2000.01281.x. [DOI] [PubMed] [Google Scholar]
- Tucker P., Laemie L., Munson A., Kanekar S., Oliver E.R., Brown N., Schlecht H., Vetter M., Glaser T. The eyeless mouse mutation (ey1) removes an alternative start codon from the Rx/rax homeobox gene. Genesis. 2001;31:43–53. doi: 10.1002/gene.10003. [DOI] [PubMed] [Google Scholar]
- Turner F.B., Taylor S.M., Moran R.G. Expression patterns of the multiple transcripts from the folylpolyglutamate synthetase gene in human leukemias and normal differentiated tissues. J. Biol. Chem. 2000;275:35960–35968. doi: 10.1074/jbc.M005228200. [DOI] [PubMed] [Google Scholar]
- Usuki F., Maruyama K. Ataxia caused by mutations in the α-tocopherol transfer protein gene. J. Neurol. Neurosurg. Psychiatry. 2000;69:254–256. doi: 10.1136/jnnp.69.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Knaap M.S., Leegwater P.A.J., Könst A.A.M., Visser A., Naidu S., Oudejans C.B.M., Schutgens R.B.H., Pronk J.C. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann. Neurol. 2002;51:264–270. doi: 10.1002/ana.10112. [DOI] [PubMed] [Google Scholar]
- Van der Leij F.R., Cox K.B., Jackson V.N., Huijkman N.C.A., Bartelds B., Kuipers J.R.G., Dijkhuizen T., Terpstra P., Wood P.A., Zammit V.A., Price N.T. Structural and functional genomics of the CPT1B gene for muscle-type carnitine palmitoyltransferase I in mammals. J. Biol. Chem. 2002;277:26994–27005. doi: 10.1074/jbc.M203189200. [DOI] [PubMed] [Google Scholar]
- Van der Velden A.W., van Nierop K., Voorma H.O., Thomas A.A.M. Ribosomal scanning on the highly structured insulin-like growth factor II-leader 1. Int. J. Biochem. Cell Biol. 2002;34:286–297. doi: 10.1016/s1357-2725(01)00116-9. [DOI] [PubMed] [Google Scholar]
- Vanhoutte P., Nissen J.L., Brugg B., Gaspera B.D., Besson M.-J., Hipskind R.A., Caboche J. Opposing roles of Elk-1 and its brain-specific isoform, short elk-1, in nerve growth factor-induced PC12 differentiation. J. Biol. Chem. 2001;276:5189–5196. doi: 10.1074/jbc.M006678200. [DOI] [PubMed] [Google Scholar]
- Van Zaane D., Gielkens A.L.J., Hesselink W.G., Bloemers H.P.J. Identification of Rauscher murine leukemia virus-specific mRNAs for the synthesis of gag- and env-gene products. Proc. Natl. Acad. Sci. USA. 1977;74:1855–1859. doi: 10.1073/pnas.74.5.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J., Angell S.M., Baulcombe D.C. In vivo translation of the triple gene block of potato virus X requires two subgenomic mRNAs. J. Virol. 1998;72:8316–8320. doi: 10.1128/jvi.72.10.8316-8320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C., Linz B., Rodrigues-Pousada C., McCarthy J.E.G. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 1998;26:1150–1159. doi: 10.1093/nar/26.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H., Matsuo M., Uenaka A., Shimbara N., Shimizu K., Nakayama E. Rejection antigen peptides on BALB/c RL♂1 leukemia recognized by cytotoxic T lymphocytes: derivation from the normally untranslated 5′ region of the c-Akt proto-oncogene activated by long terminal repeat. Cancer Res. 1995;55:4780–4783. [PubMed] [Google Scholar]
- Wagner K.-U., Dierisseau P., Rucker E.B., III, Robinson G.W., Hennighausen L. Genomic architecture and transcriptional activation of the mouse and human tumor susceptibility gene TSG101: common types of shorter transcripts are true alternative splice variants. Oncogene. 1998;17:2761–2770. doi: 10.1038/sj.onc.1202529. [DOI] [PubMed] [Google Scholar]
- Wakasugi K., Slike B.M., Hood J., Otani A., Ewalt K.L., Friedlander M., Cheresh D.A., Schimmel P. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Simon A.E. Analysis of the two subgenomic RNA promoters for turnip crinkle virus invivo and invitro. Virology. 1997;232:174–186. doi: 10.1006/viro.1997.8550. [DOI] [PubMed] [Google Scholar]
- Wang L., Wessler S.R. Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell. 1998;10:1733–1745. doi: 10.1105/tpc.10.10.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wessler S.R. Role of mRNA secondary structure in translational repression of the maize transcriptional activator Lc. Plant Physiol. 2001;125:1380–1387. doi: 10.1104/pp.125.3.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.-F., Parkhurst M.R., Kawakami Y., Robbins P.F., Rosenberg S.A. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J. Exp. Med. 1996;183:1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-Q., Rothnagel J.A. Post-transcriptional regulation of the GLI1 oncogene by the expression of alternative 5′ untranslated regions. J. Biol. Chem. 2001;276:1311–1316. doi: 10.1074/jbc.M005191200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Goligorsky M.S., Lin M., Wilcox J.N., Marsden P.A. A novel, testis-specific mRNA transcript encoding an NH2-terminal truncated nitric-oxide synthase. J. Biol. Chem. 1997;272:11392–11401. [PubMed] [Google Scholar]
- Wang Y., Newton D.C., Robb G.B., Kau C.-L., Miller T.L., Cheung A.H., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A. RNA diversity has profound effects on the translation of neuronal nitric oxide synthase. Proc. Natl. Acad. Sci. USA. 1999;96:12150–12155. doi: 10.1073/pnas.96.21.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Bridgen A., Fazakerley J.K., Streitenfeld H., Kessler N., Randall R.E., Elliott R.M. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 2002;76:7949–7955. doi: 10.1128/JVI.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D.S., Shelly C.A., Dang C.V. Genomic organization of human MXI1, a putative tumor suppressor gene. Genomics. 1996;32:466–470. doi: 10.1006/geno.1996.0144. [DOI] [PubMed] [Google Scholar]
- Wechsler J., Greene M., McDevitt M.A., Anastasi J., Karp J.E., LeBeau M.M., Crispino J.D. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- Weiland J.J., Dreher T.W. Infectious TYMV RNA from cloned cDNA: effects invitro and invivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res. 1989;17:4675–4687. doi: 10.1093/nar/17.12.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer T., Salfeld J., Will H. Expression of the hepatitis B virus core gene in vitro and in vivo. J. Virol. 1987;61:3109–3113. doi: 10.1128/jvi.61.10.3109-3113.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch A.R., McNally L.M., Gibson W. Cytomegalovirus assembly protein nested gene family: four 3′-coterminal transcripts encode four in-frame overlapping proteins. J. Virol. 1991;65:4091–4100. doi: 10.1128/jvi.65.8.4091-4100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch E.M., Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wera S., Fernandez A., Lamb N.J.C., Turowski P., Hemmings-Mieszczak M., Mayer-Jaekel R.E., Hemmings B.A. Deregulation of translational control of the 65-kDa regulatory subunit (PR65α) of protein phosphatase 2A leads to multinucleated cells. J. Biol. Chem. 1995;270:21374–21381. doi: 10.1074/jbc.270.36.21374. [DOI] [PubMed] [Google Scholar]
- Werten P.J., Stege G.J., de Jong W.W. The short 5′ untranslated region of the betaA3/A1-crystallin mRNA is responsible for leaky ribosomal scanning. Mol. Biol. Rep. 1999;26:201–205. doi: 10.1023/a:1007046926233. [DOI] [PubMed] [Google Scholar]
- Westerman B.A., Poutsma A., Looijenga L.H.J., Wouters D., van Wijk I.J., Oudejans C.B.M. The human achaete scute homolog 2 gene contains two promoters, generating overlapping transcripts and encoding two proteins with different nuclear localization. Placenta. 2001;22:511–518. doi: 10.1053/plac.2001.0695. [DOI] [PubMed] [Google Scholar]
- Wiestner A., Schlemper R.J., van der Maas A.P.C., Skoda R.C. An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nat. Genet. 1998;18:49–52. doi: 10.1038/ng0198-49. [DOI] [PubMed] [Google Scholar]
- Williams G.R. Cloning and characterization of two novel thyroid hormone receptor β isoforms. Mol. Cell. Biol. 2000;20:8329–8342. doi: 10.1128/mcb.20.22.8329-8342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Rawls J., Saha S.K., Krajcsi P., Tollefson A.E., Gooding L.R., Wold W.S.M. A 6700 MW membrane protein is encoded by region E3 of adenovirus type 2. Virology. 1990;178:204–212. doi: 10.1016/0042-6822(90)90395-8. [DOI] [PubMed] [Google Scholar]
- Wimmer B., Lottspeich F., van der Klei I., Veenhuis M., Gietl C. The glyoxysomal and plastid molecular chaperones (70-kDa heat shock protein) of watermelon cotyledons are encoded by a single gene. Proc. Natl. Acad. Sci. USA. 1997;94:13624–13629. doi: 10.1073/pnas.94.25.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing B.A., Huang E.-S. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences of human cytomegalovirus open reading frames UL93 through UL99. J. Virol. 1995;69:1521–1531. doi: 10.1128/jvi.69.3.1521-1531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W.S.M., Deutscher S.L., Takemori N., Bhat B.M., Magie S.C. Evidence that AGUAUAUGA and CCAAGAUGA initiate translation in the same mRNA in region E3 of adenovirus. Virology. 1986;148:168–180. doi: 10.1016/0042-6822(86)90412-5. [DOI] [PubMed] [Google Scholar]
- Wold W.S.M., Tollefson A.E., Hermiston T.W. E3 transcription unit of adenovirus. Curr. Top. Microbiol. Immunol. 1995;199:237–274. doi: 10.1007/978-3-642-79496-4_13. [DOI] [PubMed] [Google Scholar]
- Wolfe C.L., Lou Y., Hopper A.K., Martin N.C. Interplay of heterogeneous transcriptional start sites and translational selection of AUGs dictate the production of mitochondrial and cytosolic/nuclear tRNA nucleotidyltransferase form the same gene in yeast. J. Biol. Chem. 1994;269:13361–13366. [PubMed] [Google Scholar]
- Wolfe C.L., Hopper A.K., Martin N.C. Mechanisms leading to and the consequences of altering the normal distribution of ATP(CTP):tRNA nucleotidyltransferase in yeast. J. Biol. Chem. 1996;271:4679–4686. doi: 10.1074/jbc.271.9.4679. [DOI] [PubMed] [Google Scholar]
- Wood M.W., VanDongen H.M.A., VanDongen A.M.J. The 5′-untranslated region of the N-methyl-D-aspartate receptor NR2A subunit controls efficiency of translation. J. Biol. Chem. 1996;271:8115–8120. doi: 10.1074/jbc.271.14.8115. [DOI] [PubMed] [Google Scholar]
- Wu H., Ross C.R., Blecha F. Characterization of an upstream open reading frame in the 5′ untranslated region of PR-39, a cathelicidin antimicrobial peptide. Mol. Immunol. 2002;39:9–18. doi: 10.1016/s0161-5890(02)00093-7. [DOI] [PubMed] [Google Scholar]
- Wu J., Harrison J.K., Dent P., Lynch K.R., Weber M.J., Sturgill T.W. Identification and characterization of a new mammalian mitogen-activated protein kinase kinase, MKK2. Mol. Cell. Biol. 1993;13:4539–4548. doi: 10.1128/mcb.13.8.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Stewart S., Theilmann D.A. Alternative transcriptional initiation as a novel mechanism for regulating expression of a baculovirus trans activator. J. Virol. 1993;67:5833–5842. doi: 10.1128/jvi.67.10.5833-5842.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Chipman J.G., Robertson D.L., Erikson R.L., Simmons D.L. Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing. Proc. Natl. Acad. Sci. USA. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Rabadan-Diehl C., Nikodemova M., Wynn P., Spiess J., Aguilera G. Inhibition of corticotropin releasing hormone type-1 receptor translation by an upstream AUG triplet in the 5′ untranslated region. Mol. Pharmacol. 2001;59:485–492. doi: 10.1124/mol.59.3.485. [DOI] [PubMed] [Google Scholar]
- Yamanaka R., Kim G.-D., Radomska H.S., Lekstrom-Himes J., Smith L.T., Antonson P., Tenen D.G., Xanthopoulos K.G. CCAAT/enhancer binding protein ε is preferentially up-regulated during granulocyte differentiation and its functional versatility is determined by alternative use of promoters and differential splicing. Proc. Natl. Acad. Sci. USA. 1997;94:6462–6467. doi: 10.1073/pnas.94.12.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Mumy M., Romeo D., Wakefield L.M. Identification of the start sites for the 1.9- and 1.4-kb rat transforming growth factor-β1 transcripts and their effect on translational efficiency. Gene. 1998;219:81–89. doi: 10.1016/s0378-1119(98)00402-8. [DOI] [PubMed] [Google Scholar]
- Yin H., Blanchard K.L. DNA methylation represses the expression of the human erythropoietin gene by two different mechanisms. Blood. 2000;95:111–119. [PubMed] [Google Scholar]
- Yu J.J., Thornton K., Guo Y., Kotz H., Reed E. An ERCC1 splicing variant involving the 5′-UTR of the mRNA may have a transcriptional modulatory function. Oncogene. 2001;20:7694–7698. doi: 10.1038/sj.onc.1204977. [DOI] [PubMed] [Google Scholar]
- Yudt M.R., Cidlowski J.A. Molecular identification and characterization of A and B forms of the glucocorticoid receptor. Mol. Endocrinol. 2001;15:1093–1103. doi: 10.1210/mend.15.7.0667. [DOI] [PubMed] [Google Scholar]
- Zachar Z., Chou T.-B., Bingham P.M. Evidence that a regulatory gene autoregulates splicing of its transcript. EMBO J. 1987;6:4105–4111. doi: 10.1002/j.1460-2075.1987.tb02756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenin S., Gunnarson E., Alikina T., Bondar A., Aperia A. Identification of a new form of AQP4 mRNA that is developmentally expressed in mouse brain. Pediatric Res. 2000;48:335–339. doi: 10.1203/00006450-200009000-00012. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu R. Identification of a noncanonical signal for transcription of a novel subgenomic mRNA of mouse hepatitis virus: implication for the mechanism of coronavirus RNA transcription. Virology. 2000;278:75–85. doi: 10.1006/viro.2000.0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.-W., Riegler J., Wu J., Yen T.S.B. Novel short transcripts of hepatitis B virus X gene derived from intragenic promoter. J. Biol. Chem. 1994;269:22593–22598. [PubMed] [Google Scholar]
- Ziff E.B. Splicing in adenovirus and other animal viruses. Int. Rev. Cytol. 1985;93:327–358. doi: 10.1016/s0074-7696(08)61377-7. [DOI] [PubMed] [Google Scholar]
- Zimmer A., Zimmer A.M., Reynolds K. Tissue specific expression of the retinoic acid receptor-β2: regulation by short open reading frames in the 5′-noncoding region. J. Cell Biol. 1994;127:1111–1119. doi: 10.1083/jcb.127.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann N., Daugherty B.L., Kavanaugh J.L., El-Awar F.Y., Moulton E.A., Rothenberg M.E. Analysis of the CC chemokine receptor 3 gene reveals a complex 5′ exon organization, a functional role for untranslated exon 1, and a broadly active promoter with eosinophil-selective elements. Blood. 2000;96:2346–2354. [PubMed] [Google Scholar]
- Zoppi S., Wilson C.M., Harbison M.D., Griffin J.E., Wilson J.D., McPhaul M.J., Marcelli M. Complete testicular feminization caused by an amino-terminal truncation of the androgen receptor with downstream initiation. J. Clin. Invest. 1993;91:1105–1112. doi: 10.1172/JCI116269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zysow B.R., Lindahl G.E., Wade D.P., Knight B.L., Lawn R.M. C/T polymorphism in the 5′ untranslated region of the apolipoprotein(a) gene introduces an upstream AUG and reduces in vitro translation. Arterioscler. Thromb. Vasc. Biol. 1995;15:58–64. doi: 10.1161/01.atv.15.1.58. [DOI] [PubMed] [Google Scholar]