Abstract
The genus Conidiobolus is an important group in entomophthoroid fungi and is considered to be polyphyletic in recent molecular phylogenies. To re-evaluate and delimit this genus, multi-locus phylogenetic analyses were performed using the large and small subunits of nuclear ribosomal DNA (nucLSU and nucSSU), the small subunit of the mitochondrial ribosomal DNA (mtSSU) and the translation elongation factor 1-alpha (EF-1α). The results indicated that the Conidiobolus is not monophyletic, being grouped into a paraphyletic grade with four clades. Consequently, the well-known Conidiobolus is revised and three new genera Capillidium, Microconidiobolus and Neoconidiobolus are proposed along with one new record and 22 new combinations. In addition, the genus Basidiobolus is found to be basal to the other entomophthoroid taxa and the genus Batkoa locates in the Entomophthoraceae clade.
Keywords: Zygomycetes , Entomophthorales , Morphology, Phylogeny, New taxa
Introduction
More than 250 species of entomophthoroid fungi were isolated from insects, soil and litter throughout the world (Gryganskyi et al. 2013). For a long time, this group has been considered to be polyphyletic (Nagahama et al. 1995; Jensen et al. 1998; James et al. 2006; Liu and Voigt 2010) and was classified into a subphylum Entomophthoromycotina and a pending taxon Basidiobolus (Hibbett et al. 2007). However, a recent phylogeny using the multi-gene dataset, 18S rDNA, 28S rDNA, mtSSU and RPB2, indicated that this group formed a monophyletic lineage including Basidiobolus and it was consequently reclassified as a new fungal phylum Entomophthoromycota. More recently, a phylogenomic analysis (192 clusters of orthologous proteins) has divided traditional zygomycotan into two phyla Mucoromycota and Zoopagomycota and the entomophthoroid fungi have been re-assigned into the subphylum Entomophthoromycotina under the latter phylum (Spatafora et al. 2016). This taxonomic scheme was supported by the phylogeny of mitochondrial genomes (Nie et al. 2019).
Together with other two genera Ancylistes and Macrobiotophthora, the genus Conidiobolus belongs to Ancylistaceae, Entomophthorales, Entomophthoromycetes, Entomophthoromycotina (Humber 2012). There are six and two accepted species within the Ancylistes and Macrobiotophthora, respectively, while Conidiobolus, one of the largest groups in the entomophthoroid fungi, contains 76 names (http://www.indexfungorum.org/). The genus Conidiobolus is typified by C. utriculosus Bref. 1884 and characterised morphologically by simple sporophores, globose to pyriform multinucleate primary conidia, various types of secondary conidia and resting spores (Brefeld 1884; Humber 1997). Up to the 1940s, for half a century, only three more species were reported, C. minor Bref., C. villosus Martin and C. brefeldianus Couch (Brefeld 1884; Martin 1925; Couch 1939). In the 1950s–1960s, 38 Conidiobolus species and a variety were isolated from the United States and India (Drechsler 1952, 1953a, b, 1954, 1955a, b, c, 1956, 1957a, b, c, 1960, 1961, 1962, 1965; Srinivasan and Thirumalachar 1961, 1962a, b, 1965, 1967, 1968a, b). Based on a numerical taxonomy, King (1976a, b, 1977) recognised 27 definitive species. Since then, along with some new combinations, 10 more species have been added to Conidiobolus (Bałazy et al. 1987; Waters and Callaghan 1989; Bałazy 1993; Huang et al. 2007; Waingankar et al. 2008; Nie et al. 2012, 2016, 2017, 2018). A total of 37 species are currently accepted in this genus (Nie et al. 2018).
Three subgenera – Capillidium, Conidiobolus and Delacroixia – were proposed within the Conidiobolus, based on shape of the secondary conidia and, amongst them, the subgenus Delacroixia was reduced from generic rank (Ben-Ze’ev and Kenneth 1982). This subgeneric criterion provided a valuable contribution for the taxonomy of the genus Conidiobolus (Humber 1989). Since the 1990s, molecular analysis has become an increasingly important tool for fungal taxonomy (Bruns et al. 1991; Taylor et al. 2000). The nucLSU rDNA and EF-1α regions proved to be distinguishable amongst Conidiobolus species (Nie et al. 2012), while nucSSU rDNA indicated the genus Conidiobolus might be a polyphyletic group (Jensen et al. 1998). The subgeneric circumscription was not defined because of instability to form a certain type of secondary conidia for each phylogenetic clade (Callaghan et al. 2000; Gryganskyi et al. 2013; Nie et al. 2018). Besides, the phylogenetic relationships amongst species of Conidiobolus have not been fully resolved due to the absence of types. The genus Batkoa, morphologically similar to Conidiobolus, was phylogenetically closely related to Entomophthoraceae rather than Ancylistaceae (Gryganskyi et al. 2012, 2013).
In the present study, a reclassification of the entomophthoroid fungi, including as many as available Conidiobolus types, was constructed based on four loci (nucSSU, nucLSU, EF-1α and mtSSU) to present the taxonomic delimitation of the genus Conidiobolus and to re-evaluate the phylogenetic relationship between Basidiobolus and Batkoa.
Materials and methods
Isolates and morphology
A total of 26 ex-types of Conidiobolus were purchased from the American Type Culture Collection, Manassas, USA (ATCC) and collected from the China General Microbiological Culture Collection Center, Beijing, China (CGMCC) and the Research Center for Entomogenous Fungi of Anhui Agricultural University, Anhui Province, China (RCEF). Dried cultures were deposited in the Herbarium Mycologicum Academiae Sinicae, Beijing, China (HMAS). Morphology was observed with an Olympus BX51 research microscope and photographed by an Olympus DP25 microscope-camera system. Growth diameter on PDA (potato 200 g, dextrose 20 g, agar 20 g, H2O 1 l), Mycelia, primary conidiophores, primary conidia, microconidia, capilliconidia and resting spores were measured and described with the method of King (1976a).
DNA extraction, PCR amplification and sequencing
Fungal strains were incubated on PDA for 7 d at 21 °C. Total genomic DNA was extracted from the fresh fungal mycelia by using modified CTAB method (Watanabe et al. 2010). Four gene portions from cell nuclei and mitochondria and one protein coding gene were used in this study: the large subunit of nuclear ribosomal RNA genes (nucLSU), the small subunit of nuclear ribosomal RNA genes (nucSSU), the small subunit of mitochondrial ribosomal RNA genes (mtSSU) and the translation elongation factor 1-alpha gene (EF-1α). The nucLSU region was amplified with the primers LR0R and LR5 (Vilgalys and Hester 1990), the nucSSU region with nucSSU-0021-5’ (Gargas and DePriest 1996) and nucSSU-1780-3’ (DePriest 1993) and EF-1α region with the primers EF983 and EF1aZ-1R (http://www.aftol.org/primers.php). These PCR reactions have been described by Liu et al. (2005), Jensen et al. (1998) and Nie et al. (2012). The primers used for the mtSSU region were mtSSU1 and mtSSU2R and the PCR reaction was performed using the following cycling parameters: denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 1 min, extension at 72 °C for 1 min and finalised with an extra extension at 72 °C for 7 min (Zoller et al. 1999). PCR products were purified and sequenced by Shanghai Genecore Biotechnologies Company (Shanghai, China) with the same primers as relative PCR. The nucleotide sequence data have been deposited in the GenBank (Table 1).
Table 1.
The species used in phylogenetic analyses.
| Species | Strains* | GenBank accession numbers | |||
|---|---|---|---|---|---|
| nucSSU | nucLSU | EF-1α | mtSSU | ||
| Allomyces arbusculus | AFTOL 300 | AY552524 | DQ273806 | DQ275334 | – |
| Basidiobolus haptosporus | ARSEF 261 | JX242606 | JX242586 | – | JX242626 |
| B. heterosporus | CBS 311.66 | JX242607 | JX242587 | – | JX242627 |
| B. magnus | CBS 205.64 | JX242608 | JX242588 | – | JX242628 |
| B. meristosporus | CBS 931.73 | JX242609 | JX242589 | – | JX242629 |
| B. microsporus | CBS 130.62 (T) | JX242610 | JX242590 | – | JX242630 |
| B. ranarum | NRRL 34594 | AY635841 | DQ273807 | DQ275340 | EF392490 |
| Batkoa apiculata | ARSEF 3130 | DQ177437 | EF392404 | – | EF392513 |
| B. gigantea | ARSEF 214 | JX242611 | JX242591 | – | JX242631 |
| B. major | ARSEF 2936 | EF392559 | EF392401 | – | EF392511 |
| B. obscurus** | CBS 182.60 | JX242614 | JX242595 | – | JX242635 |
| B. pseudapiculata** | ARSEF 395 | EF392557 | EF392378 | – | EF392508 |
| Coemansia reversa | AFTOL 140 | AY546685 | AY546689 | DQ282615 | – |
| Conidiobolus adiaeretus | ARSEF 451 (T) | – | KC461182 | – | – |
| C. adiaeretus | CGMCC 3.15888 | – | MN061284 | MN061481 | MN061287 |
| C. antarcticus | ARSEF 6913 (T) | – | DQ364207 | – | DQ364227 |
| C. bangalorensis | ARSEF 449 (T) | – | DQ364204 | – | DQ364225 |
| C. brefeldianus | ARSEF 452 (T) | AF368506 | EF392382 | – | EF392495 |
| C. chlamydosporus | ATCC 12242 (T) | – | JF816212 | JF816234 | MK301178 |
| C. coronatus | NRRL 28638 | AF113418 | AY546691 | DQ275337 | – |
| C. coronatus | RCEF 4518 | – | JN131537 | JN131543 | – |
| C. couchii | ATCC 18152 (T) | – | JN131538 | JN131544 | MK301179 |
| C. dabieshanensis | CGMCC 3.15763 (T) | – | KY398125 | KY402206 | MK301180 |
| C. denaeosporus | ATCC 12940 (T) | – | JF816215 | JF816228 | MK301181 |
| C. firmipilleus | ARSEF 6384 | JX242612 | JX242592 | – | JX242632 |
| C. gonimodes | ATCC 14445 (T) | – | JF816221 | JF816226 | MK301182 |
| C. heterosporus | RCEF 4430 | – | JF816225 | JF816239 | MK301183 |
| C. humicolus | ATCC 28849 (T) | – | JF816220 | JF816231 | MK301184 |
| C. incongruus | NRRL 28636 | AF113419 | AF113457 | – | – |
| C. iuxtagenitus | ARSEF 6378 (T) | – | KC788410 | – | – |
| C. iuxtagenitus | RCEF 4445 | – | JX946695 | JX946700 | MK333391 |
| C. khandalensis | ATCC 15162 (T) | – | KX686994 | KY402204 | MK301185 |
| C. lachnodes | ARSEF 700 | – | KC788408 | – | – |
| C. lamprauges | ARSEF 2338 | AF296754 | DQ364206 | – | DQ364226 |
| C. lichenicolus | ATCC 16200 (T) | – | JF816216 | JF816232 | MK301186 |
| C. lobatus | ATCC 18153 (T) | – | JF816218 | JF816233 | MK301187 |
| C. marcosporus | ATCC 16578 (T) | – | KY398124 | KY402209 | MK301188 |
| C. megalotocus | ATCC 28854 (T) | – | MF616383 | MF616385 | MK301189 |
| C. mirabilis | CGMCC 3.17763 (T) | – | MH282852 | MH282853 | MK333389 |
| C. mycophagus | ATCC 16201 (T) | – | JX946694 | JX946698 | MK301190 |
| C. mycophilus | ATCC 16199 (T) | – | KX686995 | KY402205 | MK301191 |
| C. nodosus | ATCC 16577 (T) | – | JF816217 | JF816235 | MK333388 |
| C. osmodes | ARSEF 79 | AF368510 | EF392371 | – | DQ364219 |
| C. osmodes | RCEF4447 | – | JN131539 | JN131545 | MK333392 |
| C. pachyzygosporus | CGMCC 3.17764 (T) | – | KP218521 | KP218524 | MK333390 |
| C. paulus | ARSEF 450 (T) | – | vv | – | – |
| C. polyspermus | ATCC 14444 (T) | – | MF616382 | MF616384 | MK301193 |
| C. polytocus | ATCC 12244 (T) | – | JF816213 | JF816227 | MK301194 |
| C. pumilus | ARSEF 453 (T) | JX242615 | EF392383 | – | EF392496 |
| C. rhysosporus | ATCC 12588 (T) | – | JN131540 | JN131546 | MK301195 |
| C. sinensis | RCEF 4952 (T) | – | JF816224 | JF816238 | MK301196 |
| C. stilbeus | RCEF 5584 (T) | – | KP218522 | KP218525 | MK301197 |
| C. stromoideus | ATCC 15430 (T) | – | JF816219 | JF816229 | MK301198 |
| C. terrestris | ATCC 16198 (T) | – | KX752050 | KY402208 | MK301199 |
| C. thromboides | ATCC 12587 (T) | – | JF816214 | JF816230 | MK301200 |
| C. thromboides | FSU 785 | JX242616 | JX242597 | – | JX242637 |
| C. thromboides | RCEF 4492 | – | JF816223 | JF816236 | MK333393 |
| C. undulatus | ATCC 12943 (T) | – | JX946693 | JX946699 | MK301201 |
| Dimargaris bacillispora | AFTOL 136 | AB016020 | DQ273791 | DQ282609 | – |
| Endogone pisiformis | AFTOL 539 | DQ322628 | DQ273811 | DQ282618 | – |
| Entomophaga aulicae | ARSEF 172 | EF392542 | EF392372 | – | EF392487 |
| E. conglomerata | ARSEF 2273 | AF368509 | – | – | – |
| E. maimaga | ARSEF 1400 | EF392556 | EF392395 | – | EF392505 |
| Eryniopsis caroloniana | ARSEF 640 | EF392552 | EF392387 | – | EF392500 |
| Entomophthora chromaphidis | ARSEF 1860 | AF353725 | – | – | – |
| E. culicis | ARSEF 387 | AF368516 | – | – | – |
| E. grandis | ARSEF 6701 | – | DQ481229 | – | – |
| E. scatophaga | ARSEF 6704 | – | DQ481226 | – | – |
| E. muscae | ARSEF 3074 | AY635820 | DQ273772 | DQ275343 | – |
| E. planconiana | ARSEF 6252 | AF353723 | GQ285878 | – | – |
| E. schizophorae | ARSEF 5348 | AF052402 | GQ285883 | – | – |
| E. syrphi | ARSEF 5595 | – | DQ481230 | – | – |
| E. tripidium | ARSEF 6518 | AF296755 | – | – | – |
| Erynia conica | ARSEF 1439 | AF368513 | EF392396 | – | EF392506 |
| E. ovispora | ARSEF 400 | JX242620 | JX242601 | – | JX242641 |
| E. rhizospora | ARSEF 1441 | AF368514 | EF392397 | – | EF392507 |
| E. sciarae | ARSEF 1870 | AF368515 | EF392399 | – | EF392509 |
| Furia americana | ARSEF 742 | EF392554 | EF392389 | – | – |
| F. gastropachae | ARSEF 5541 | EF392562 | EF392407 | – | EF392516 |
| F. ithacensis | ARSEF 663 | EF392553 | EF392388 | – | EF392501 |
| F. neopyralidarum | ARSEF 1145 | AF368518 | EF392394 | – | EF392504 |
| F. pieris | ARSEF 781 | AF368519 | EF392390 | – | EF392502 |
| F. virescens | ARSEF 1129 | EF392555 | EF392393 | – | EF392503 |
| Gaertneriomyces semiglobiferus | AFTOL 34 | AF164247 | DQ273778 | DQ275338 | – |
| Macrobiotophthora vermicola | ARSEF 650 | AF052400 | – | – | – |
| Massospora cicadina | ARSEF 374 | EF392548 | EF392377 | – | EF392492 |
| Mortierella verticillata | AFTOL 141 | AF157145 | DQ273794 | – | – |
| Pandora blunckii | ARSEF 217 (T) | JX242621 | JX242602 | – | – |
| P. delphacis | ARSEF 459 | AF368521 | EF392384 | – | EF392497 |
| P. dipterigena | ARSEF 397 | AF368522 | EF392380 | – | EF392565 |
| P. kondoiensis | CBS 642.92 | JX242622 | JX242603 | – | JX242642 |
| P. neoaphidis | ARSEF 3240 | EF392560 | EF392405 | – | EF392514 |
| Piptocephalis corymbifera | AFTOL 145 | AB016023 | AY546690 | DQ282619 | – |
| Rhizophagus intraradices | AFTOL 845 | DQ322630 | FJ461839 | DQ282611 | – |
| Rozella allomycis | AFTOL 297 | AY635838 | DQ273803 | DQ275342 | – |
| Schizangiella serpentis | ARSEF 2237 | AF368523 | EF392428 | – | EF392488 |
| Strongwellsea castrans | – | AF052406 | – | – | – |
| Zancudomyces culisetae | AFTOL 29 | AF277007 | DQ273773 | – | – |
| Zoophthora anglica | ARSEF 396 | – | EF392379 | – | EF392493 |
| Z. lanceolata | ARSEF 469 | EF392550 | EF392385 | – | EF392498 |
| Z. phalloides | ARSEF 2281 | EF392558 | EF392400 | – | EF392510 |
| Z. radicans | ARSEF 388 | JX242624 | JX242605 | – | JX242644 |
* AFTOL, Assembling the Fungal Tree of Life; ARSEF, ARS Entomopathogenic Fungus Collection (Ithaca, U.S.A.); ATCC, American Type Culture Collection (Manassas, U.S.A); CGMCC, China General Microbiological Culture Collection Center (Beijing, China); FSU, Jena Microbial Resource Collection (Friedrich-Schiller-University of Jena, Germany); NRRL, ARS Culture Collection (Peoria, U.S.A); RCEF, Research Center for Entomogenous Fungi (Hefei, China). T = ex-type. **Batkoa sp. CBS 182.60 was received as Conidiobolus obscurus, while B. pseudapiculataARSEF 395 was received as C. pseudapiculatus.
Phylogenetic analyses
More available nucLSU, nucSSU, mtSSU and EF-1α sequences of 14 Conidiobolus species and 47 other entomophthoroid fungi were obtained from GenBank. Ten species of Glomeromycotina, Mortierellomycotina, Mucoromycotina, Kickxellomycotina, Zoopagomycotina, Blastocladiomycota, Chytridiomycota and Cryptomycota, were chosen as outgroups. Alignments were constructed separately for each locus with MUSCLE 3.8.31 (Edgar 2004) and the concatenated matrices were assembled by SequenceMatrix 1.7.8 (Vaidya et al. 2011). The best model for the phylogenetic analysis was selected with Akaike Information Criterion (AIC) by using Modeltest 3.7 (Posada and Crandall 1998). Phylogenetic relationships were inferred using Maximum Likelihood (ML) and Bayesian Inference (BI). The best-scoring ML tree analysis was performed using raxmlGUI 1.5b1 with GTRGAMMA model and 1000 replicates (Silvestro and Michalak 2012). The BI analysis was performed using MrBayes 3.2.2 (Ronquist and Huelsenbeck 2003). Markov Chain Monte Carlo (MCMC) chains ran until the convergences met and the standard deviation fell below 0.01. The first 25% of trees were discarded as burn-in. The combined dataset was deposited at TreeBase (No. S25064). Phylogenetic trees were checked and modified in FigTree 1.4 (Rambaut 2012).
Results
Phylogenetic analyses
The combined dataset contained 4521 characters of nucLSU (1–1326), nucSSU (1327–3424), EF-1α (3425–4062) and mtSSU (4063–4521) after alignment. With the optimal model GTR+I+G and random starting trees, four Markov chains were run for 7 million generations and every 100th generation was sampled once. ML and BI analyses of the combined dataset resulted in phylogenetic reconstructions with almost similar topologies and the average standard deviation of split frequencies was 0.006721 (BI).
In the ML phylogenetic tree (Figure 1), the Basidiobolaceae lineage (88/0.94) is located at the base of the entomophthoroid fungi and is closely related to the Ancylistaceae group (56/0.91). The Batkoa lineage is grouped within the Entomophthoraceae Clade (60/0.89). All Conidiobolus lineages are clustered into a paraphyletic grade and therefore cannot be considered congeneric. Moreover, the Conidiobolus grade consists of four well supported clades. In detail, there are 7, 10, 16 and 3 species in Clade I (100/1.00), II (77/1.00), III (100/1.00) and IV (99/1.00), respectively.
Figure 1.
Phylogenetic tree constructed by maximum likelihood analyses of nucLSU, nucSSU, EF-1α and mtSSU sequences for Entomophthoromycotina, with some chytrid and mucoralean fungi as outgroups. Three new genera and one Chinese new record are shown in red. Maximum likelihood bootstrap values (≥ 50%) / Bayesian posterior probabilities (≥ 0.50) of main clades are indicated along branches. Scale bar indicates substitutions per site.
Taxonomy
In order to provide a more natural taxonomic classification, four genera (Capillidium, Conidiobolus, Microconidiobolus and Neoconidiobolus) and their type species (Ca. heterosporum, C. utriculosus, M. paulus and N. thromboides) are described here in this paper. Additionally, a new record Ca. adiaeretum, C. coronatus and C. iuxtagenitus with new isolates from China and C. khandalensis being first reported to produce microconidia are illustrated herein.
Capillidium
B. Huang & Y. Nie gen. nov.
D46F46A3-E0E1-5C9B-AD08-2A5C10757790
831596
Etymology.
Referring to unique ellipsoidal secondary conidia (capilliconidia).
Type species.
Capillidium heterosporum (Drechsler) B. Huang & Y. Nie.
Description.
Mycelia colourless. Primary conidiophores simple, bearing a single primary conidia. Primary conidia forcibly discharged multinucleate, colourless, globose, pyriform to obovoid. Two kinds of replicative conidia, the first one is similar and smaller than primary conidia, the second one (capilliconidia) arises from elongate and slender conidiophores. Zygospores present or absent, formed in axial alignment with conjugating segments, globose to subglobose, often smooth, sometimes rough, colourless or yellowish.
Notes.
Conidiobolus subgen. Capillidium Ben-Ze’ev & Kenneth was firstly established to include species with capilliconidia (Ben-Ze’ev and Kenneth 1982). In this phylogenetic analysis, all members of the subgenus Capillidium grouped with good support (100/1.00) and, therefore, it was raised from subgenus to genus status based on the monophyly, as well as the stability to form ellipsoidal secondary conidia (capilliconidia). In addition to capilliconidia, C. adiaeretus also produces microconidia.
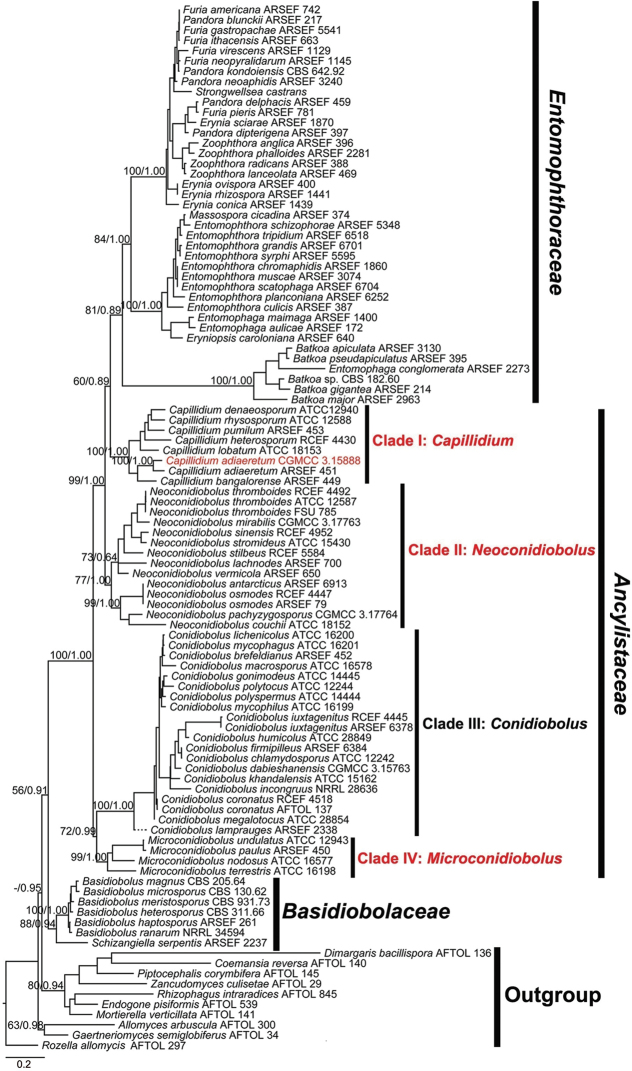
Capillidium heterosporum
(Drechsler) B. Huang & Y. Nie comb. nov.
033D89FA-FE07-5B76-ABF5-D805C655AA30
831601
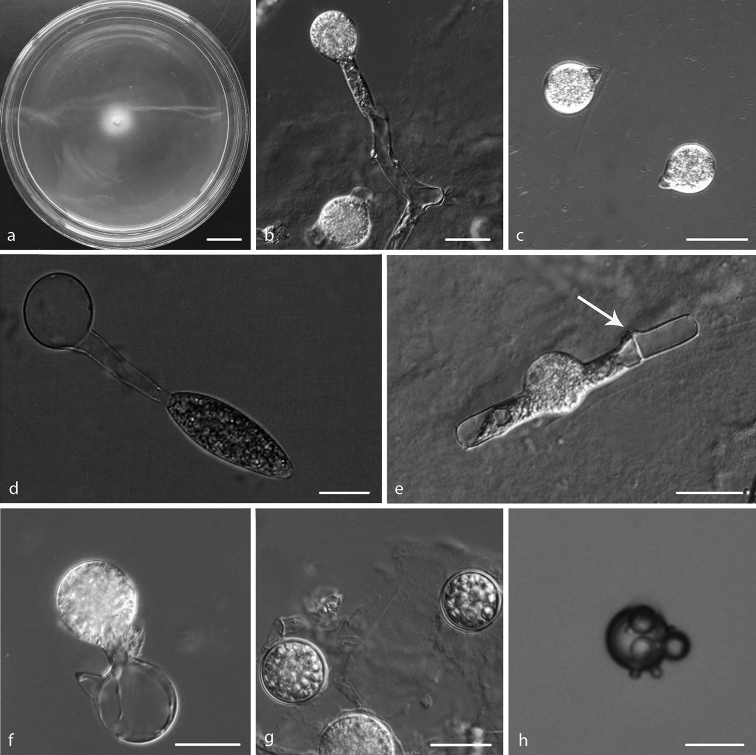
Figure 2.
Capillidium heterosporuma colony on PDA after 3 d at 25 °C b primary conidiophores bearing primary conidia c primary conidia d, e, f ellipsoidal secondary conidia arising from slender conidiophores g, h production of secondary conidia. Scale bars: 10 mm (a); 20 μm (b, c, d, g, h); 100 μm (e, f).
Conidiobolus heterosporus Drechsler, Am. J. Bot. 40: 107 (1953). Basionym.
=Conidiobolus rugosus Drechsler, Am. J. Bot. 42: 437 (1955).
Specimens examined.
China, Anhui Province, Plant detritus, 8 Nov 2008, C.F. Wang, RCEF 4430.
Description.
Colonies on PDA at 25 °C after 3 d, white, reaching ca. 21 mm in diameter. Mycelia colourless, 5–9 μm wide. Primary conidiophores, colourless, unbranched and producing a single globose conidium with widening upwards, extending to a length of 30–245 μm into the air, 8–17 μm wide. Primary conidia forcibly discharged, colourless, globose to subglobose, measuring 12–37 μm in greatest length and 11–31 μm in total width, including a basal papilla 1.5–5 μm high and 5–12 μm wide. After discharging on to 2% water-agar, similar and smaller secondary conidia arise from primary conidia, 1–6 ellipsoidal secondary conidia (capilliconidia, 10–20 × 12–38 μm) arise from slender conidiophores (50–250 × 2.5–4 μm). Resting spores not observed.
Notes.
The ex-type living culture is ATCC 12941 (United States, Maryland, 18 Mar 1952, Drechsler).
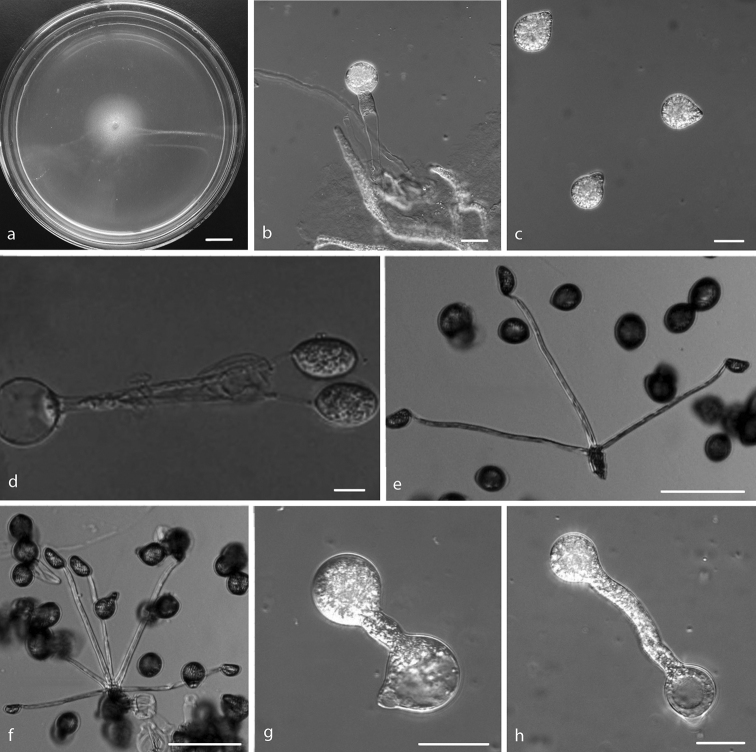
Capillidium adiaeretum
(Drechsler) B. Huang & Y. Nie comb. nov.
348681A3-0E38-5892-8481-DEEAF76FC24A
831602
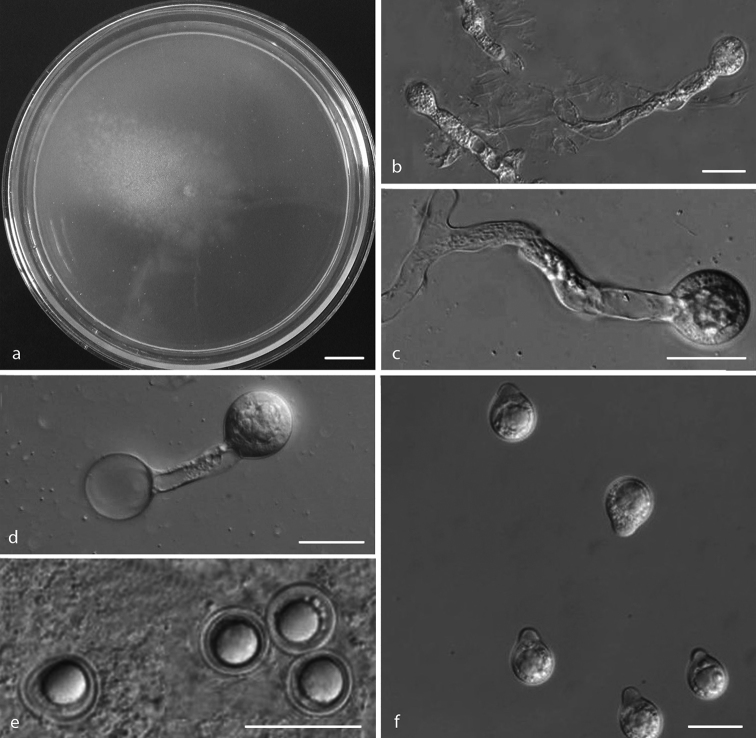
Figure 3.
Capillidium adiaeretuma colony on PDA after 3 d at 25 °C b mycelia c, d primary conidiophores bearing primary conidia e, f primary conidia g Production of secondary conidia h first stage of forming microconidia i second stage of forming microconidia j, k ellipsoidal secondary conidia arising from slender conidiophores l chlamydospores. Scale bars: 10 mm (a); 100 μm (b); 20 μm (c–l).
Conidiobolus adiaeretus Drechsler, J. Wash. Acad. Sci. 43: 42 (1953). Basionym.
Specimens examined.
China, Jiangsu Province, Nanjing City, Laoshan Forest Park, 32°5'58"N, 118°35'53"E, Plant detritus, 1 Dec 2018, Y. Nie and Y. Gao, HMAS 248358, culture CGMCC 3.15888 (=RCEF 6550).
Description.
Colonies on PDA at 25 °C after 3 d, white, reaching ca. 7–10 mm in diameter. Mycelia colourless, 3–4.5 μm wide. Primary conidiophores, colourless, unbranched and producing a single globose conidium with widening upwards; they offer a pronounced dimensional contrast with the mycelial filaments, extending to a length of 50–210 μm into the air, 3–25 μm wide. Primary conidia forcibly discharged, colourless, globose, measuring 15–45 μm in greatest length and 13–42 μm in total width, including a basal papilla 2–6 μm high and 5–17 μm wide. After discharging on to 2% water-agar, similar and smaller secondary conidia arise from primary conidia, two generations of multiple spherical units forming on the parent globose conidia Microconidia only formed from the second set, 5–12 × 9–10 μm. Capilliconidia formed readily from discharged microconidia, 16–24 × 5–6 μm. Chlamydospores formed within the substratum, colourless, globose to ellipsoidal, 13–40 × 15–45 μm.
Notes.
The species was firstly reported from America (Drechsler 1953a). The ex-type living culture is ATCC 12589 isolated by Drechsler (1953a). It is mainly characterised and differs from other Capillidium species by its ability to form both microconidia and capilliconidia (Callaghan et al. 2000). The Chinese specimen CGMCC 3.15888 clusters completely (100/1.00) with an isotype ARSEF 451 (98% sequence similarity in nucLSU) and fits well with its morphological descriptions. It is reported in China for the first time.
Conidiobolus
Bref., Mykol. Untersuch. 6(2): 35 (1884), emend.
A6638E2E-FF16-53E3-B738-84069BF5E7DF
20144
= Delacroixia Sacc. & P. Syd., Syll. fung. (Abellini) 14(1): 457 (1899).
Conidiobolus subgen. Delacroixia (Sacc. & P. Syd.) Tyrrell & Macleod, J. Invert. Pathol. 20: 12 (1972).
Type species.
Conidiobolus utriculosus Bref.
Description.
Mycelia colourless. Primary conidiophores simple or branched dichotomously, positively phototropic, bearing a single or 2–4 primary conidia. Primary conidia forcibly discharged, multinucleate, colourless, pyriform, obovoid, globose to subglobose. Secondary conidia usually with shape of primary conidia but smaller, formed singly on short secondary conidiophores. Microspores arising from primary or secondary conidia. Villose appendaged globose conidia and formed villose conidia. Chlamydospores formed intercalarily within assimilative hyphae. Zygospores formed in axial alignment with one or two (homothallic or heterothallic) conjugating segments.
Notes.
C. utriculosus, the type species of the genus Conidiobolus, has not been re-collected since Brefeld isolated it in 1884 and most taxonomists working on entomophthoroid fungi now universally recognised it as C. coronatus (Gryganskyi et al. 2013). However, the smaller pear-shaped conidia of C. utriculosus are different from the larger globose conidia of C. coronatus and villose spores in C. coronatus are not observed in C. utriculosus (Brefeld 1884; King 1977). Consequently, C. coronatus is not synonymised with C. utriculosus in this study. Instead, this study agrees with Srinivasan and Thirumalachar (1967) and King (1977) to place C. minor in synonymy with C. utriculosus because the small conidia of C. minor were probably replicative conidia of C. utriculosus. Nevertheless, neither C. utriculosus nor C. minor has available living cultures. Therefore, we have not yet designated an epitype and thus no DNA sequences for explaining this type. Fortunately, we are able to recognise clade III (Fig. 1) as Conidiobolus on the basis of its synapomorph, namely microspores.
Conidiobolus utriculosus
Bref., Mykol. Untersuch. 6(2): 35 (1884)
BAD66584-64D0-538B-90BE-223F06941DD7
144259 (MBT391377)
= Conidiobolus minor Bref., Mykol. Untersuch. 6(2): 35, 68 (1884).
Specimens examined.
No ex-type.
Description.
Refer to Brefeld (1884) and King (1977).
Notes.
Due to the lack of ex-type, plates 3, 4, and 5 in Brefeld, Mykol. Untersuch. 6(2): 35 (1884) are designated here as the lectotype for Conidiobolus utriculosus.
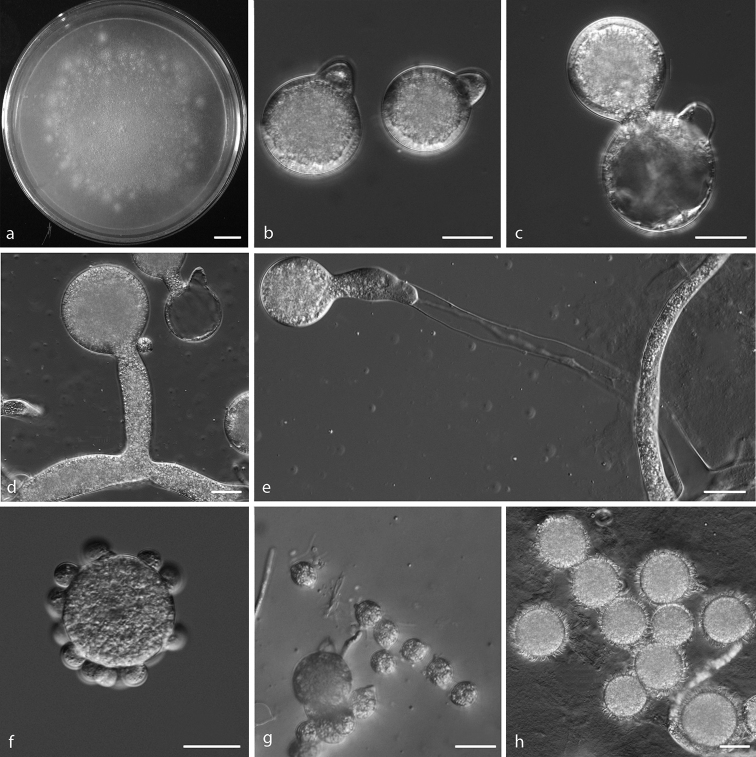
Conidiobolus coronatus
(Costantin) A. Batko, Entomophaga, Mémoires hors série 2: 129 (1964)
C501E77E-6761-514E-8E81-5DC66636ED0F
283037
Figure 4.
Conidiobolus coronatusa colony on PDA after 3 d at 21 °C b primary conidia c production of secondary conidia d, e primary conidiophores bearing primary conidia f, g microconidia h villose spores. Scale bars: 10 mm (a); 20 μm (b–h).
Boudierella coronata Costantin, Bull. Soc. mycol. Fr. 13: 40 (1897). Basionym.
Delacroixia coronata (Costantin) Sacc. & P. Syd., Syll. fung. (Abellini) 14(1): 457 (1899).
Entomophthora coronata (Costantin) Kevorkian, J. Agric. Univ. Puerto Rico 21(2): 191 (1937).
= Conidiobolus villosus G.W. Martin, Bot. Gaz. 80(3): 317 (1925).
Specimens examined.
China, Shandong Province, Plant detritus, 20 Mar 2009, C.F. Wang, RCEF 4518.
Description.
Colonies grown on PDA for 3 d at 21 °C, reaching ca. 65 mm in diameter. Mycelia colourless, 8–20 μm wide. Primary conidiophores, positively phototropic, colourless, unbranched and producing a single globose conidium, extending to a length of 53–287 μm into the air, 7.5–20.5 μm wide. Primary conidia forcibly discharged, colourless, globose, measuring 36–52 μm in greatest width and 42–65 μm in total length, including a basal papilla 12–18 μm high and 6.5–14 μm wide. After discharging on to 2% water-agar, similar and smaller secondary conidia arise from primary conidia. Microconidia produced readily from primary conidia, globose or almond-shaped, 13–19 × 11–15 μm. Villose spores formed after 4–5 d, globose, 20–42 μm.
Notes.
The ex-type living culture is ATCC 28691 (United States, Louisiana, Plant detritus, 3 January 1972). Due to the absence of molecular data of ex-type strain ATCC 28691, the molecular data of the authentic strain NRRL 28638, which has been applied in many other phylogenetic analysis (James et al. 2006; Liu and Voigt 2011; Gryganskyi et al. 2012; Tretter et al. 2014; Spatafora et al. 2016), was used in this study instead. The monotypic genus Delacroixia was typified by D. coronata which was transferred from an ascomycete Boudierella coronata Costantin (Costantin 1897; Saccardo and Sydow 1899). After that, it was reclassified as a subgenus of Conidiobolus, namely Conidiobolus sub. Delacroixia (Sacc. & P. Syd.) Tyrrell & MacLeod to define all those Conidiobolus species capable of forming microspores and, consequently, D. coronata was recombined as C. coronatus (Tyrrell and MacLeod 1972; Ben-Ze’ev and Kenneth 1982).
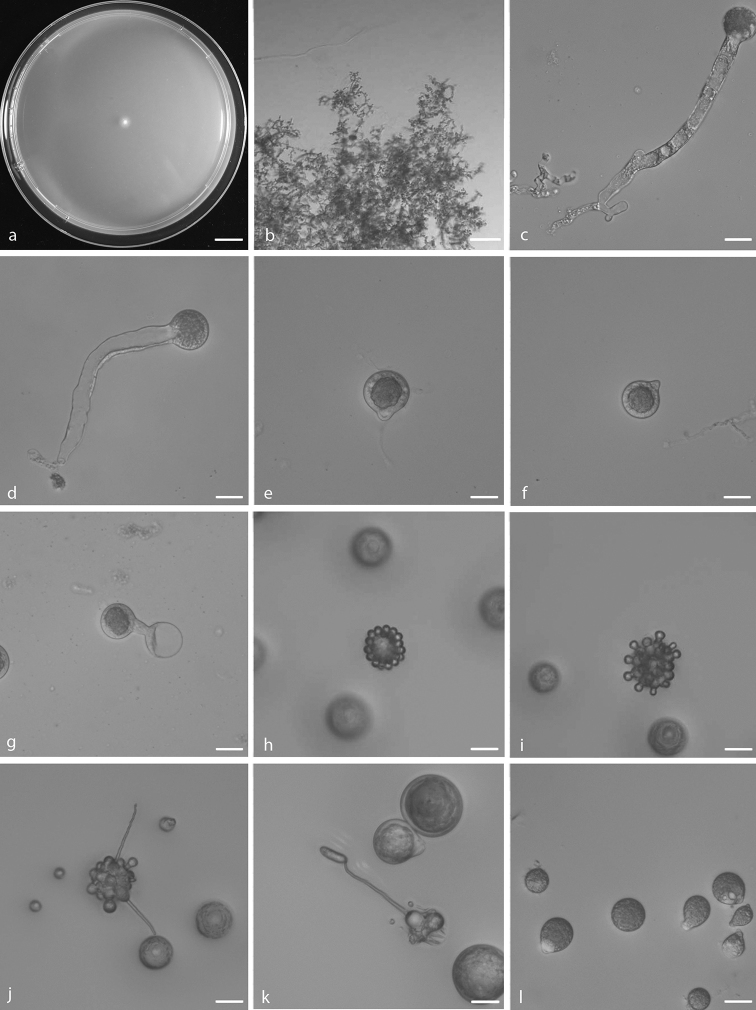
Conidiobolus iuxtagenitus
S.D. Waters & Callaghan, Mycol. Res. 93(2): 223 (1989)
2BC62ED2-2D2F-5A03-82F3-B6D6572047FA
135617
Figure 5.
a–gConidiobolus iuxtagenitushConidiobolus khandalensisa colony on PDA after 3 d at 21 °C b primary conidiophores bearing primary conidia c primary conidia d tertiary fusiform conidium from a globose spore e zygospore formation with the beak almost emptied of protoplasm f production of secondary conidia g zygospores h microconidia produced from global conidia. Scale bars: 10 mm (a); 20 μm (b–h).
Specimens examined.
China, Anhui Province, Plant detritus, 8 Nov 2008, C.F. Wang, RCEF 4445.
Description.
Colonies on PDA at 21 °C after 3 d white, flat, slow-growing, reaching ca. 13 mm in diameter. Mycelia colourless, 5.5–11 μm wide. Primary conidiophores, positively phototropic, arising from hyphal segments, colourless, 28–75 × 7.5–10 μm, unbranched and producing a single globose conidium. Primary conidia forcibly discharged, globose, 27–37 × 21–28 μm, with a basal papilla 6–10 μm wide. Secondary conidia arising from primary conidia, similar to, but smaller than the primary ones, forcibly discharged. Tertiary conidium fusiform arising from primary conidia, 30–45 × 16–22 μm. Zygospores in a position separated by a short beak near a lateral conjugation, globose to subglobose, smooth, 21–25 × 17–24 μm, with a 1–2 μm thick wall.
Notes.
The ex-type living culture is ARSEF 6378 (United Kingdom, Staffordshire, Plant detritus, 31 October 1983, M. F. Smith).
Conidiobolus khandalensis
Sriniv. & Thirum., Mycologia 54(6): 692 (1963) [1962]
E009E85B-7E6F-59CA-A1AA-BC7893AB6585
328754
Specimens examined.
India, Khandala, Dec. 1961, Srinivasan and Thirumalachar, ATCC 15162.
Description.
Refer to Srinivasan and Thirumalachar (1962b). Microconidia produced from global conidia on the 2% water-agar at 16 °C (Fig. 5h).
Notes.
According to the original morphological description (Srinivasan and Thirumalachar 1962b) and the re-examination by King (1977), microconidia have not been reported. However, we observed the microconidia produced from global conidia on 2% water-agar at 16 °C. Moreover, this specimen was located in the Conidiobolus lineage (Figure 1) which was supported by our morphological analyses.
Microconidiobolus
B. Huang & Y. Nie gen. nov.
744CDD7E-6E4B-56B4-BF10-59FE98E3F107
831597
Etymology.
Referring to small discharged primary conidia.
Type species.
Microconidiobolus paulus (Drechsler) B. Huang & Y. Nie.
Description.
Mycelia colourless. Primary conidiophores simple and short, bearing a single primary conidia. Primary conidia forcibly discharged, multinucleate, colourless, globose to obovoid, usually small, mostly less than 20 μm. Only globose replicative conidia produced, similar and smaller than primary conidia. Chlamydospores globose, formed terminally on hyphae or from globose cells by thickening of the wall. Zygospores formed in axial alignment with two conjugating segments, globose to ellipsoidal, smooth and yellowish.
Notes.
This genus includes three species producing smaller primary conidia (mostly less than 20 μm) without microspores or capilliconidia compared to other Conidiobolus spp. These three species are C. nodosus, C. paulus and C. terrestris. According to the taxonomic scheme of Conidiobolus by King (1977), C. undulatus is a synonym of C. paulus, which is supported herein by molecular evidence (Figure 1). However, the phylogeny does not support C. nodosus and C. terrestris as synonyms of C. lachnodes, since the former two were located in clade IV and the latter in clade II (Figure 1). Therefore, we accept the taxonomic status at species level for C. nodosus and C. terrestris, based on the morphological and phylogenetic analyses.
Microconidiobolus paulus
(Drechsler) B. Huang & Y. Nie comb. nov.
E2B29A52-D392-543D-A5BF-D98B97B87E44
831605
Conidiobolus paulus Drechsler, Bull. Torrey bot. Club. 84: 269 (1957). Basionym.
= Conidiobolus undulatus Drechsler, Bull. Torrey bot. Club. 84: 275 (1957).
= Conidiobolus parvus Drechsler, Bull. Torrey bot. Club. 89: 233 (1962).
Description.
Refer to Drechsler (1957a).
Notes.
The ex-type living culture is ATCC 12942 (United States, Wisconsin, 18 November 1954, Drechsler).
Neoconidiobolus
B. Huang & Y. Nie gen. nov.
8A004282-B8EF-516F-A99D-77EBD48BE926
831598
Etymology.
Referring to the subgenus Conidiobolus raised to generic rank.
Type species.
Neoconidiobolus thromboides (Drechsler) B. Huang & Y. Nie.
Description.
Mycelia colourless. Primary conidiophores simple, sometimes branched from hyphal knots or differentiated from aerial hyphae, positively phototropic, bearing a single primary conidium. Primary conidia forcibly discharged, multinucleate, colourless, globose, pyriform to obovoid. Replicative conidia similar and smaller than primary conidia. Chlamydospores globose, formed terminally on hyphae or from globose cells by thickening of the wall. Zygospores formed in axial alignment with two conjugating segments, globose to ellipsoidal, smooth, colourless, rarely pale yellowish.
Notes.
The genus Neoconidiobolus is strikingly similar to the subgenus Conidiobolus which produces neither microconidia nor capilliconidia. All members in the clade of Neoconidiobolus share the following characteristics: forcibly discharged, colourless, globose, pyriform to obovoid primary conidia. Two kinds of replicative conidia produced. One is discharged, similar and smaller than primary conidia and the other is elongate and forcibly discharged. Two types of resting spores produced: zygospores and chlamydospores.
Neoconidiobolus thromboides
(Drechsler) B. Huang & Y. Nie comb. nov.
672EE869-86FB-5516-BDC0-D34E25189680
831606
Figure 6.
Neoconidiobolus thromboidesa colony on PDA after 3 d at 25 °C b, c primary conidiophores bearing primary conidia d production of secondary conidia e zygospores f primary conidia. Scale bars: 10 mm (a); 20 μm (b–d, f); 40 μm (e).
Conidiobolus thromboides Drechsler, J. Wash. Acad. Sci. 43: 38 (1953). Basionym.
Specimens examined.
China, Anhui Province, Plant detritus, 21 Feb 2009, C.F. Wang, RCEF 4492.
Description.
Colonies grown on PDA for 3 d at 25 °C, white, reaching ca. 30 mm diameter. Mycelium colourless, filamentous, 5–7.5 µm wide. Primary conidiophores colourless, unbranched and producing a single conidium, 50–122.5 × 6–16.5 µm. Primary conidia forcibly discharged, colourless, globose to subglobose, 20–26.5 µm wide, 26.5–34 µm long, including a basal papilla 6–10 µm wide. Secondary conidia globose, forming from the primary conidia. Zygospores most often formed between segments of separate hyphae. Mature zygospores smooth, globose to subglobose, 25–30 μm in diameter with wall 2–3 μm thick.
Notes.
The ex-type living culture is ATCC 12587 (United States, New Hampshire, September 1957, Drechsler).
More new combinations
In addition to previously described taxa, more new combinations were proposed herein and their descriptions refer to relevant protologues.
Capillidium bangalorense
(Sriniv. & Thirum.) B. Huang & Y. Nie comb. nov.
49E94C4C-B2DA-5D26-98C6-CCA98B7576B6
831607
Conidiobolus bangalorensis Sriniv. & Thirum., Mycologia 59(4): 702 (1967). Basionym.
Capillidium denaeosporum
(Drechsler) B. Huang & Y. Nie comb. nov.
A9419A69-2648-5350-A992-B432913ECAD1
831608
Conidiobolus denaeosporus Drechsler, J. Wash. Acad. Sci. 47: 309 (1957). Basionym.
Capillidium lobatum
(Sriniv. & Thirum.) B. Huang & Y. Nie comb. nov.
63DB8CDB-AF6B-5281-8373-450ED3B2764B
831609
Conidiobolus lobatus Sriniv. & Thirum., J. Elisha Mitchell scient. Soc. 84: 212 (1968). Basionym.
Capillidium pumilum
(Drechsler) B. Huang & Y. Nie comb. nov.
098895D0-4C8A-590C-A70A-E436E6E987D9
831610
Conidiobolus pumilus Drechsler, J. Wash. Acad. Sci. 45: 115 (1955). Basionym.
= Conidiobolus globuliferus Drechsler, Am. J. Bot. 43: 783 (1957) [1956].
= Conidiobolus inordinatus Drechsler, J. Wash. Acad. Sci. 47: 312 (1957).
Capillidium rhysosporum
(Drechsler) B. Huang & Y. Nie comb. nov.
CDF80202-031E-5743-956C-E7B685CB859F
831611
Conidiobolus rhysosporus Drechsler, Am. J. Bot. 41: 567 (1954). Basionym.
Microconidiobolus nodosus
(Sriniv. & Thirum.) B. Huang & Y. Nie comb. nov.
0645FC92-B51F-5002-A8B6-0B825ACBE86D
831624
Conidiobolus nodosus Sriniv. & Thirum., Mycologia 59(4): 705 (1967). Basionym.
Microconidiobolus terrestris
(Sriniv. & Thirum.) B. Huang & Y. Nie comb. nov.
2F620080-2CCB-5A7C-897C-E4D1D3C2DCDE
831625
Conidiobolus terrestris Sriniv. & Thirum., Mycopathol. Mycol. appl. 36(3–4): 344 (1968). Basionym.
Neoconidiobolus couchii
(Sriniv. & Thirum.) B. Huang & Y. Nie comb. nov.
7C8ECD45-9291-5244-9F60-A808D89F7ECA
831626
Conidiobolus couchii Sriniv. & Thirum., J. Elisha Mitchell scient. Soc. 84: 211 (1968). Basionym.
Neoconidiobolus lachnodes
(Drechsler) B. Huang & Y. Nie comb. nov.
A8105E3E-8740-529B-AF8F-B699C93DBD05
831627
Conidiobolus lachnodes Drechsler, Am. J. Bot. 42: 442 (1955). Basionym.
Neoconidiobolus mirabilis
(Y. Nie & B. Huang) B. Huang & Y. Nie comb. nov.
6531CB5C-3134-596D-A5BB-EF4FF6757255
831628
Conidiobolus mirabilis Y. Nie & B. Huang, Mycol. Progr. 17(10): 1204 (2018). Basionym.
Neoconidiobolus osmodes
(Drechsler) B. Huang & Y. Nie comb. nov.
FEAA69F5-A749-5BB0-9095-AEF429D1BAFA
831629
Conidiobolus osmodes Drechsler, Am. J. Bot. 41: 571 (1954). Basionym.
= Conidiobolus antarcticus S. Tosi, Caretta & Humber, Mycotaxon 90(2): 344 (2004).
Neoconidiobolus pachyzygosporus
(Y. Nie & B. Huang) B. Huang & Y. Nie comb. nov.
50B684D1-0579-57E4-808E-4B1C906FDFEE
831630
Conidiobolus pachyzygosporus Y. Nie & B. Huang, Mycol. Progr. 17(10): 1206 (2018). Basionym.
Neoconidiobolus sinensis
(Y. Nie, X.Y. Liu & B. Huang) B. Huang & Y. Nie comb. nov.
278F1289-BCF8-5E30-B7FA-6090B1923C42
831631
Conidiobolus sinensis Y. Nie, X.Y. Liu & B. Huang, Mycotaxon 120: 432 (2012). Basionym.
Neoconidiobolus stilbeus
(Y. Nie & B. Huang) B. Huang & Y. Nie comb. nov.
A39E4A47-D4E5-532D-934E-D305571D2923
831632
Conidiobolus stilbeus Y. Nie & B. Huang, Mycosphere 7(6): 804 (2016). Basionym.
Neoconidiobolus stromoideus
(Sriniv. & Thirum.) B. Huang & Y. Nie comb. nov.
EBB4B068-7277-517B-B39B-9E1C0CA6D6FF
831633
Conidiobolus stromoideus Sriniv. & Thirum., Sydowia 16(1–6): 65 (1963) [1962]. Basionym.
Neoconidiobolus vermicola
(J.S. McCulloch) B. Huang & Y. Nie comb. nov.
0D7F7877-D73B-57D1-9B8E-5C0662E1290C
831634
Entomophthora vermicola J.S. McCulloch, Trans. Br. mycol. Soc. 68(2): 173 (1977). Basionym.
Macrobiotophthora vermicola (J.S. McCulloch) B.E. Tucker, Mycotaxon 13(3): 499 (1981).
Discussion
The phylogenetic position of Basidiobolus in the Kingdom Fungi has been problematic for a long time. Previous phylogenetic analyses of the rDNA (18S, 28S and 5.8S) sequences grouped Basidiobolus outside or basal in the Entomophthorales (Nagahama et al. 1995; Jensen et al. 1998; White et al. 2006). Combined with the study of other protein-coding molecular markers, Basidiobolus was located inside the Entomophthorales (James et al. 2006). Recently, according to the phylogeny of much more available molecular data of entomophthoroid fungi in three families, Basidiobolus was grouped basal to other entomophthoroid taxa (Gryganskyi et al. 2012) which was also supported by the phylogenomic analyses of zygomycete fungi (Spatafora et al. 2016) and by the multi-gene analyses in this study. Although the morphological characteristics of Batkoa were similar to Conidiobolus, the Batkoa lineage appeared to be most closely related to the other taxa in the Entomophthoraceae Clade and should be distinguished from Conidiobolus lineage by its obligate pathogenicity for invertebrates and by staining readily, while most members of Conidiobolus are saprobic and non-staining.
The phylogenetic relationship of the genus Conidiobolus has been unclear for a long time, because of its high heterology (Gryganskyi et al. 2013). This article used more available ex-type strains to revise this genus, based on phylogeny and morphology. According to Figure 1, four main clades were reconstructed and the results showed that Conidiobolus s.l. is not a monophyletic group but paraphyletic with Macrobiotophthora vermicola. The M. vermicola was originally placed in Entomophthora (Mcculloch 1977) and transferred to Macrobiotophthora, based on the morphological characters of primary spores, secondary spores and zygospores (Tucker 1981). The paraphyletic relationship between Macrobiotophthora vermicola and Conidiobolus s.l. was also revealed by Gryganskyi et al. (2012). In this paper, we treated it as a new combination and, therefore, proposed a monophyletic group of the new genus Neoconidiobolus.
In Clade I of the genus Capillidium, seven species grouped in a monophyletic clade with good support (100/1.00) and the synapomorph of producing capilliconidia: Conidiobolus adiaeretus (= Capillidium adiaeretum), Co. bangalorensis (= Ca. bangalorensis), Co. denaeosporus (= Ca. denaeosporum), Co. heterosporus (= Ca. heterosprum), Co. lobatus (= Ca. lobatum), Co. pumilus (= Ca. pumilum) and Co. rhysosporus (= Ca. rhysosporum). As a note, Co. denaeosporus was synonymised with Co. pumilus (King 1976b), but herein its taxonomic status of species level was accepted according to the phylogeny. Co. adiaeretus forms not only capilliconidia but also microspores (Callaghan et al. 2000).
In Clade II of the genus Neoconidiobolus, all 14 strains comprising 10 species produce neither microspores nor capilliconidia. Amongst these, C. antarcticus was identified as a synonym of C. osmodes (Chen and Huang 2018), which was confirmed here as they grouped into a robust clade.
Considering its long history and significant impact, we kept and emended the genus Conidiobolus and the original illustrations of the type species C. utriculosus (Brefeld 1884) were designated as its lectotype. Thus, we were able to recognise clade III under the genus name Conidiobolus on the basis of its synapomorph, namely microspores. In Clade III of the genus Conidiobolus, all species definitely produce microspores, except Conidiobolus dabieshanensis, C. iuxtagenitus, C. khandalensis and C. lichenicolus. Microspores have never been observed in C. dabieshanensis and C. iuxtagenitus (King 1977; Waters and Callaghan 1989; Nie et al. 2017), but cases for C. khandalensis and C. lichenicolus are somewhat different. For C. khandalensis, the protologue did not document any microspores (Srinivasan and Thirumalachar 1962b; King 1977), but they can be observed on 2% water-agar at 16 °C (Fig. 5h). Although the microspore of C. lichenicolus was not mentioned in the original description, the ability to produce microspores has been exhibited in accordance with original illustrations (Srinivasan and Thirumalachar 1968a). The phylogeny also resulted in the following taxonomic treatments. On the one hand, some previously synonymised taxa recover their specific status, for example, C. gonimodes, C. megalotocus and C. mycophagus should be separated from C. incongruus, C. macrosporus and C. mycophilus, respectively. On the other hand, C. chlamydosporus is synonymised with C. firmipilleus.
In Clade IV of the genus Microconidiobolus, Conidiobolus undulatus was identified as a synonym of C. paulus (= M. paulus) by King (1976b), which is supported by our molecular data. Otherwise, C. nodosus (= M. nodosus) and C. terrestris (= M. terristris) were classified as synonyms of C. lachnodes (= Neoconidiobolus lachnodes) in the study of King (1976b). Morphologically, C. lachnodes bears larger primary conidia (9–25 × 10–27 μm) than C. nodosus (13–16 × 17–22 μm) and C. terrestris (8–12 μm in width) (Drechsler 1955b; Srinivasan and Thirumalachar 1967, 1968a). Furthermore, C. lachnodes was located in Clade II and is distantly related to C. nodosus and C. terrestris. Therefore, C. nodosus and C. terrestris are accepted as two distinct species. This clade comprises four ex-type strains, all producing smaller primary conidia (mostly less than 20 μm) and can be morphologically easily distinguished from other Conidiobolus species.
Phylogenetically, Conidiobolus lamprauges does group with Clade III and received strong bootstrap support (100/1.00). Morphologically, this species produces small primary conidia (12.5–20 × 15–22 μm) without microconidia or capilliconidia and is similar to species within Clade IV. Its taxonomic status remains unclear in the present study.
Supplementary Material
Acknowledgements
We thank Dr. Z.F. Yu (Yunnan University) for improving the manuscript. We also thank Y. Gao (Jiangxi Agricultural University) for collecting some Conidiobolus strains. This project was supported by the National Natural Science Foundation of China (Nos. 31900008, 30770008 and 31670019).
Citation
Nie Y, Yu D-S, Wang C-F, Liu X-Y, Huang B (2020) A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): four clades including three new genera. MycoKeys 66: 55–81. https://doi.org/10.3897/mycokeys.66.46575
Contributor Information
Xiao-Yong Liu, Email: liuxiaoyong@im.ac.cn.
Bo Huang, Email: bhuang@ahau.edu.cn.
References
- Bałazy S, Wiśniewski J, Kaczmarek S. (1987) Some noteworthy fungi occurring on mites. Bulletin of the Polish Academy of Sciences, Biological Sciences 35: 199–224. [Google Scholar]
- Balazy S. (1993) Entomophthorales Flora of Poland (Flora Polska), Fungi (Mycota) 24: 1–356. Polish Academy of Sciences, W. Szafer Institute of Botany, Kraków, Poland.
- Ben-Ze’ev IS, Kenneth RG. (1982) Features-criteria of taxonomic value in the Entomophthorales: I. A revision of the Batkoan classification. Mycotaxon 14: 393–455. [Google Scholar]
- Brefeld O. (1884) Conidiobolus utriculosus und minor. Untersuchungen aus der Gesammtgebiete der Mykologie 6(2): 35–78. [Google Scholar]
- Bruns TD, White TJ, Taylor JW. (1991) Fungal molecular systematics. Annual Review of Ecology and Systematics 22: 525–564. 10.1146/annurev.es.22.110191.002521 [DOI] [Google Scholar]
- Callaghan AA, Waters SD, Manning RJ. (2000) Alternative repetitional conidia in Conidiobolus adiaeretus: development and germination. Mycological Research 104: 1270–1275. 10.1017/S0953756200003063 [DOI] [Google Scholar]
- Chen MJ, Huang B. (2018) Conidiobolus antarcticus, a synonym of C. osmodes. Mycotaxon 133(4): 635–641. 10.5248/133.635 [DOI] [Google Scholar]
- Costantin J. (1897) Sur une entomophthoree nouvelle. Bulletin de la Societe Mycologique de France 13: 38–43. [Google Scholar]
- Couch JN. (1939) A new Conidiobolus with sexual reproduction. American Journal of Botany 26: 119–130. 10.1002/j.1537-2197.1939.tb12878.x [DOI] [Google Scholar]
- DePriest PT. (1993) Small subunit rDNA variation in a population of lichen fungi due to optional group I introns. Gene 134: 67–74. 10.1016/0378-1119(93)90175-3 [DOI] [PubMed] [Google Scholar]
- Drechsler C. (1952) Widespread distribution of Delacroixia coronata and other saprophytic Entomophthoraceae in plant detritus. Science 115: 575–576. 10.1126/science.115.2995.575 [DOI] [PubMed] [Google Scholar]
- Drechsler C. (1953a) Three new species of Conidiobolus isolated from leaf mold. Journal of the Washington Academy of Science 43(2): 29–34. [Google Scholar]
- Drechsler C. (1953b) Two new species of Conidiobolus occurring in leaf mold. American Journal of Botany 40(3): 104–115. 10.1002/j.1537-2197.1953.tb06458.x [DOI] [Google Scholar]
- Drechsler C. (1954) Two species of Conidiobolus with minutely ridged zygospores. American Journal of Botany 41: 567–575. 10.1002/j.1537-2197.1954.tb14380.x [DOI] [Google Scholar]
- Drechsler C. (1955a) A small Conidiobolus with globose and with elongated secondary conidia. Journal of Washington Academy of Sciences 45(4): 114–117. [Google Scholar]
- Drechsler C. (1955b) Three new species of Conidiobolus isolated from decaying plant detritus. American Journal of Botany 42(5): 437–443. 10.1002/j.1537-2197.1955.tb11144.x [DOI] [Google Scholar]
- Drechsler C. (1955c) Two new species of Conidiobolus that produce microconidia. American Journal of Botany 42(9): 793–802. 10.1002/j.1537-2197.1955.tb10424.x [DOI] [Google Scholar]
- Drechsler C. (1956) Two new species of Conidiobolus. American Journal of Botany 43(10): 778–787. 10.1002/j.1537-2197.1956.tb11168.x [DOI] [Google Scholar]
- Drechsler C. (1957a) Two small species of Conidiobolus forming lateral zygospores. Bulletin of the Torrey Botanical Club 84(4): 268–280. 10.2307/2482673 [DOI] [Google Scholar]
- Drechsler C. (1957b) A new species of Conidiobolus with distended conidiophores. Sydowia Annales Mycologici 9: 189–192. [Google Scholar]
- Drechsler C. (1957c) Two medium-sized species of Conidiobolus occurring in Colorado. Journal of Washington Academy of Sciences 47: 309–315. [Google Scholar]
- Drechsler C. (1960) Two new species of Conidiobolus found in plant detritus. American Journal of Botany 47: 368–377. 10.1002/j.1537-2197.1960.tb07138.x [DOI] [Google Scholar]
- Drechsler C. (1961) Two species of Conidiobolus often forming zygospores adjacent to antheridium-like distentions. Mycologia 53(3): 278–303. 10.2307/3756275 [DOI] [Google Scholar]
- Drechsler C. (1962) A small Conidiobolus with resting spores that germinate like zygospores. Bulletin of the Torrey Botanical Club 89(4): 233–240. 10.2307/2483199 [DOI] [Google Scholar]
- Drechsler C. (1965) A robust Conidiobolus with zygospores containing granular parietal protoplasm. Mycologia 57(6): 913–926. 10.2307/3756891 [DOI] [Google Scholar]
- Edgar RC. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargas A, DePriest PT. (1996) A nomenclature for fungal PCR primers with examples from intron-containing SSU rDNA. Mycologia 88: 745–748. 10.2307/3760969 [DOI] [Google Scholar]
- Gryganskyi AP, Humber RA, Smith ME, Miadlikovska J, Wu S, Voigt K, Walther G, Anishchenko IM, Vilgalys R. (2012) Molecular phylogeny of the Entomophthoromycota. Molecular Phylogenetics and Evolution 65: 682–694. 10.1016/j.ympev.2012.07.026 [DOI] [PubMed] [Google Scholar]
- Gryganskyi AP, Humber RA, Smith ME, Hodge K, Huang B, Voigt K, Vilgalys R. (2013) Phylogenetic lineages in Entomophthoromycota. Persoonia 30: 94–105. 10.3767/003158513X666330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischo JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Lumbsch HT, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Grith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüßler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N. (2007) A higher-level phylogenetic classification of the Fungi. Mycological Research 111: 509–547. 10.1016/j.mycres.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Huang B, Humber RA, Hodge KT. (2007) A new species of Conidiobolus from Great Smoky Mountains National Park. Mycotaxon 100: 227–233. [Google Scholar]
- Humber RA. (1989) Synopsis of a revised classification for the Entomophthorales (Zygomycotina). Mycotaxon 34: 441–460. 10.1098/rspa.1937.0056 [DOI] [Google Scholar]
- Humber RA. (1997) Fungi: identification. In: LA Lacey (Ed.) Manual of Techniques in Insect Pathology. Academic Press, London. 10.1016/B978-012432555-5/50011-7 [DOI]
- James TY, Kau F, Schoch C, Matheny PB, Hofstetter V, Cox CJ, Celio G, Geuidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüßler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MW, Grith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman A, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443: 818–822. 10.1038/nature05110 [DOI] [PubMed] [Google Scholar]
- Jensen AB, Gargas A, Eilenberg J, Rosendahl S. (1998) Relationships of the insect-pathogenic order Entomophthorales (Zygomycota, Fungi) based on phylogenetic analyses of nucleus small subunit ribosomal DNA sequences (SSU rDNA). Fungal Genetics and Biology 24(3): 325–334. 10.1006/fgbi.1998.1063 [DOI] [PubMed] [Google Scholar]
- King DS. (1976a) Systematics of Conidiobolus (Entomophthorales) using numerical taxonomy I. Taxonomic considerations. Canadian Journal of Botany 54: 45–65. 10.1139/b76-008 [DOI] [Google Scholar]
- King DS. (1976b) Systematics of Conidiobolus (Entomophthorales) using numerical taxonomy II. Taxonomic considerations. Canadian Journal of Botany 54: 1285–1296. 10.1139/b76-141 [DOI] [Google Scholar]
- King DS. (1977) Systematics of Conidiobolus (Entomophthorales) using numerical taxonomy III. Descriptions of recognized species. Canadian Journal of Botany 55: 718–729. 10.1139/b77-086 [DOI] [Google Scholar]
- Liu M, Rombach MC, Humber RA, Hodge KT. (2005) What’s in a name? Aschersonia insperata: a new pleoanamorphic fungus with characteristics of Aschersonia and Hirsutella. Mycologia 97: 249–256. 10.3852/mycologia.97.1.246 [DOI] [PubMed] [Google Scholar]
- Liu XY, Voigt K. (2011) Molecular Characters of Zygomycetous Fungi. In: Gherbawy Y, Voigt K. (Eds) Molecular Identification of Fungi.Springer-Verlag, Heidelberg, Berlin, 461–488. 10.1007/978-3-642-05042-8_20 [DOI]
- Martin GW. (1925) Morphology of Conidiobolus villosus. Botanical Gazette 80: 311–318. 10.1086/333540 [DOI] [Google Scholar]
- Mcculloch JS. (1977) New species of nematophagous fungi from Queensland. Transactions of the British Mycological Society 68(2): 173–179. 10.1016/S0007-1536(77)80005-3 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, 8 pp 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Nagahama T, Sato H, Shimazu M, Sugiyama J. (1995) Phylogenetic divergence of the entomophthoralean fungi: evidence from nuclear 18S ribosomal RNA gene sequences. Mycologia 87: 203–209. 10.2307/3760906 [DOI] [Google Scholar]
- Nie Y, Yu CZ, Liu XY, Huang B. (2012) A new species of Conidiobolus (Ancylistaceae) from Anhui, China. Mycotaxon 120: 427–435. 10.5248/120.427 [DOI] [Google Scholar]
- Nie Y, Tang XX, Liu XY, Huang B. (2016) Conidiobolus stilbeus, a new species with mycelial strand and two types of primary conidiophores. Mycosphere 7(6): 801–809. 10.5943/mycosphere/7/6/11 [DOI] [Google Scholar]
- Nie Y, Tang XX, Liu XY, Huang B. (2017) A new species of Conidiobolus with chlamydospores from Dabie Mountains, eastern China. Mycosphere 8(7): 809–816. 10.5943/mycosphere/8/7/1 [DOI] [Google Scholar]
- Nie Y, Qin L, Yu DS, Liu XY, Huang B. (2018) Two new species of Conidiobolus occurring in Anhui, China. Mycological Progress 17(10): 1203–1211. 10.1007/s11557-018-1436-z [DOI] [Google Scholar]
- Nie Y, Wang L, Cai Y, Tao W, Zhang YJ, Huang B. (2019) Mitochondrial genome of the entomophthoroid fungus Conidiobolus heterosporus provides insights into evolution of basal fungi. Applied Microbiology and Biotechnology 103(3): 1379–1391. 10.1007/s00253-018-9549-5 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2012) FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/
- Ronquist F, Huelsenbeck JP. (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Saccardo PA, Sydow P. (1899) Delacroixia Sacc. et Syd. Sylloge Fungorum 14: 1–457. [Google Scholar]
- Silvestro D, Michalak I. (2012) RaxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, James TY, O’Donnell K, Roberson RW, Taylor TN, Uehling J, Vilgalys R, White MM, Stajich JE. (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108(5): 1028–1046. 10.3852/16-042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1961) Studies on species of Conidiobolus from India-I. Sydowia, Annales Mycologici 15: 237–241. [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1962a) Studies on species of Conidiobolus from India-II. Sydowia, Annales Mycologici 16: 60–66. [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1962b) Studies on species of Conidiobolus from India-III. Mycologia 54(6): 685–693. 10.2307/3756504 [DOI] [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1965) Studies on species of Conidiobolus from India-IV. Sydowia 19: 86–91. [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1967) Evaluation of taxonomic characters in the genus Conidiobolus with key to known species. Mycologia 59: 698–713. 10.2307/3757098 [DOI] [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1968a) Studies on species of Conidiobolus from India-V. Mycopathologica et Mycologia Applicata 36: 341–346. 10.1007/BF02050380 [DOI] [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1968b) Two new species of Conidiobolus from India. Journal of the Mitchell Society 84: 211–212. [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- Tretter ED, Johnson EM, Benny GL, Lichtwardt RW, Wang Y, Kandel P, Novak SJ, Smith JF, White MM. (2014) An eight-gene molecular phylogeny of the Kickxellomycotina, including the first phylogenetic placement of Asellariales. Mycologia 106(5): 912–935. 10.3852/13-253 [DOI] [PubMed] [Google Scholar]
- Tucker BE. (1981) A review of the nonentomogenous Entomophthorales. Mycotaxon 13: 481–505. [Google Scholar]
- Tyrrell D, MacLeod DM. (1972) A taxonomic proposal regarding Delacroixia coronata (Entomophthoraceae). Journal of Invertebrate Pathology 20(1): 11–13. 10.1016/0022-2011(72)90074-2 [DOI] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2): 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waingankar VM, Singh SK, Srinivasan MC. (2008) A new thermophilic species of Conidiobolus from India. Mycopathologia 165: 173–177. 10.1007/s11046-007-9088-6 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Lee K, Goto K, Kumagai S, Sugita-Konishi Y, Hara-Kudo Y. (2010) Rapid and effective DNA extraction method with bead grinding for a large amount of fungal DNA. Journal of Food Protection 73(6): 1077–1084. 10.4315/0362-028X-73.6.1077 [DOI] [PubMed] [Google Scholar]
- Waters SD, Callaghan AA. (1989) Conidiobolus iuxtagenitus, a new species with discharge delongate repetitional conidia and conjugation tubes. Mycological Research 93: 223–226. 10.1016/S0953-7562(89)80121-2 [DOI] [Google Scholar]
- White MM, James TY, O’Donnell K, Cafaro MJ, Tanabe Y, Sugiyama J. (2006) Phylogeny of the Zygomycota based on nuclear ribosomal sequence data. Mycologia 98: 872–884. 10.3852/mycologia.98.6.872 [DOI] [PubMed] [Google Scholar]
- Zollera S, Scheideggera C, Sperisena C. (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. The Lichenologist 31(5): 511–516. 10.1006/lich.1999.0220 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.