Abstract
Protein complexes at the cell surface facilitate the detection of danger signals from diverse pathogens and initiate a series of complex intracellular signaling events that result in various immune responses.
Metazoans and plants employ innate immune systems that combat microbial infection (Wan et al., 2019a). Pattern-triggered immunity (PTI) relates to the ability of multicellular eukaryotic organisms to employ pattern recognition receptors (PRRs) for sensing microbial structures, termed pathogen- or microbe-associated molecular patterns (PAMP/MAMPs), and execute antimicrobial defenses. Recently, plants have also been shown to employ PRRs that detect multicellular pathogens including nematodes, herbivorous insects, and parasitic plants. Host-derived damage-associated molecular patterns (DAMPs) and immunomodulatory phytocytokines are recognized by the same types of cellular surface receptors. In plants, microbial effector-mediated suppression of PTI has driven the evolution of another set of immune receptors that either recognize microbial effectors directly or detect effector-mediated perturbations of host target proteins. This type of immunity is referred to as effector-triggered immunity. In many cases, the boundaries between PTI and effector-triggered immunity cannot be clearly defined (Thomma et al., 2011). Therefore, it has been suggested that the plant immune system should be viewed more uniformly as a surveillance system that detects invasion or danger (Cook et al., 2015; Gust et al., 2017).
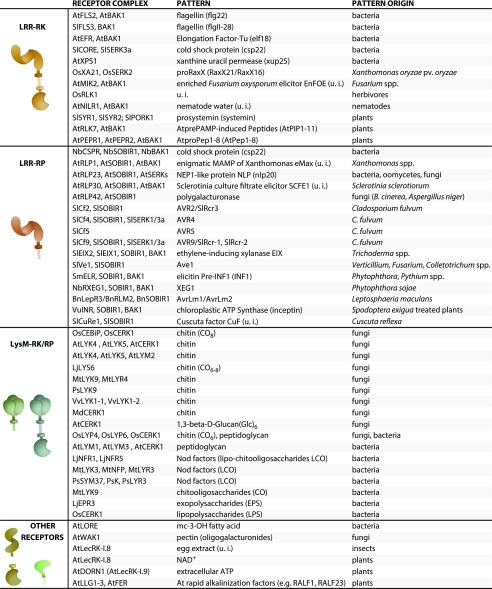
Plants recognize patterns of danger at the cell surface via membrane-localized PRRs (Fig. 1). Depending on the presence or absence of an intracellular kinase domain, PRRs are categorized as receptor kinases (RKs) or receptor proteins (RPs), respectively. Many macromolecules harboring patterns are degraded by plant enzymes to make the immunogenic epitopes accessible for detection. Immune signaling upon pattern perception relies not only on the PRRs themselves, but on immune signaling complexes consisting of membrane-bound and intracellular proteins that are implicated in scaffolding or signal transduction.
Figure 1.
Plant pattern recognition receptors, coreceptors, and corresponding ligands. Receptor complexes are grouped according to the structural class of the (putative) ligand binding receptors. Receptor complex components listed have biochemically or genetically been shown to be involved in perception of cognate patterns. Species prefixes for receptors and coreceptors: At, Arabidopsis; Bn, B. napus; Lj, L. japonicus; Md, Malus domestica; Mt, M. truncatula; Nb, N. benthamiana; Os, O. sativa; Ps, P. sativum; Sl, S. lycopersicum; Sm, Solanum microdontum; Vu, V. unguiculata; and Vv, Vitis vinifera. Species prefixes are indicated for coreceptors if studied in the same species as the receptor, but are excluded if studied in (multiple) heterologous organisms. Identified minimal epitopes of patterns are given in parentheses. Dependent on the range of analysis, pattern origins are stated as whole kingdoms or as species names, which does not exclude a broader, yet unknown, distribution.
LEUCINE-RICH REPEAT-RKS
The molecular variety of PAMP/DAMPs necessitates different ectodomain properties of the corresponding receptors for recognition (Saijo et al., 2018). Leucine-rich repeat (LRR)-RKs and -RPs are similar to PRRs in animals, most notably Toll-like receptors (Ronald and Beutler, 2010). Plant LRR-RKs typically recognize proteinaceous ligands and are involved in developmental and immunogenic processes. In plants, LRR-RKs comprise a prolific family of several hundred proteins grouped into different clades. Many LRR-RKs form signaling complexes with somatic embryogenesis receptor kinases (SERKs), LRR-RK coreceptors with relatively short ectodomains (five LRRs; Chinchilla et al., 2009).
Among the first reported PRRs in plants, flagellin-sensing2 (AtFLS2) recognizes fragments of bacterial flagellin (Gómez-Gómez and Boller, 2000). Immunogenic fragments are made accessible through the action of plant enzymes. In Arabidopsis (Arabidopsis thaliana), the β-galactosidase AtBGAL1 deglycosylates flagellin, making it susceptible to proteolytic degradation (Buscaill et al., 2019). Immunogenic flagellin fragments, including the 22-amino acid minimal epitope flg22, directly bind to AtFLS2 LRRs, leading to recruitment of the coreceptor AtBAK1/AtSERK3 (Felix et al., 1999; Gómez-Gómez et al., 1999; Chinchilla et al., 2006). The C terminus of the flg22 fragment is bound by both AtFLS2 and AtBAK1, thereby facilitating complex formation and induction of downstream signaling (Chinchilla et al., 2006; Sun et al., 2013). Flg22 perception through FLS2 homologs is widespread in land plants (Albert et al., 2010). Some solanaceous plants have evolved a second perception system for flagellin. The tomato (Solanum lycopersicum) LRR-RK SlFLS3 binds flgII-28, an epitope distinct from flg22 (Hind et al., 2016). Rice (Oryza sativa) evolved yet another perception system for Acidovorax avenae flagellin, although the cognate receptor remains to be identified (Katsuragi et al., 2015).
Another well-studied LRR-RK is the Arabidopsis elongation factor Tu (EF-Tu)-receptor (AtEFR), which perceives bacterial EF-Tu via the n-acetylated 18-amino acid peptide elf18 (Zipfel et al., 2006). An alternative EF-Tu epitope, the central fragment EFa50, is detected in rice indicating that multiple receptors have independently evolved for the detection of EF-Tu (Furukawa et al., 2014), as is the case for flagellin. Recognition of different epitopes within one protein may therefore be a widespread attribute of basal immunity in higher organisms and point to the selection of well-suited target molecules for pathogen perception. Additional LRR-RK/PAMP pairs have been identified in various plant species. These include OsXA21/RaxX16 (conserved Xanthomonas protein required for activation of XA21-mediated immunity, RaxX; Pruitt et al., 2015, 2017), bacterial cold shock protein (SlCORE/csp22; Wang et al., 2016), and xanthine/uracil permease family protein (AtXPS1/xup25; Mott et al., 2016). A common feature of all these PRRs is their plant genus-specific distribution. Intergeneric transfer of PRRs to crop plants lacking that particular perception system has been reported to enhance resistance to pathogens and could thus be a useful tool for disease management (Brutus et al., 2010; Lacombe et al., 2010; Rodriguez-Moreno et al., 2017; Pfeilmeier et al., 2019).
In addition to detecting microbial molecules, plants use LRR-RKs to monitor the presence of endogenous phytocytokines that regulate development and immunity (Luo, 2012; Gust et al., 2017). Tomato systemin, the first identified plant peptide hormone, is an 18-amino acid peptide that contributes to herbivorous insect resistance. The LRR-RK SlSYR1 was recently identified as the systemin receptor (Wang et al., 2018b). The closely related SlPORK1 associates with SlSYR1 and SlSYR2 and is critical for systemin response (Xu et al., 2018). AtPEPR1 and 2 bind endogenous AtPep1-8 peptides derived from the C-termini of their AtPROPEP1-8 precursors (Yamaguchi et al., 2006, 2010; Krol et al., 2010). Mature peptides are generated by Ca2+-dependent type-II metacaspases (Hander et al., 2019; Shen et al., 2019). The AtPep1 C terminus binds to the LRR domain of AtPEPR1 and induces AtPEPR1-AtBAK1 heterodimerization (Tang et al., 2015). A similar signaling mechanism is hypothesized for the 11-member AtPIP (PAMP‐induced secreted peptide) family. AtPIPs are cleaved by unknown proteases from AtPrePIPs. AtPIP1 has been shown to bind to and signal via AtRLK7. AtPep and AtPIP signaling work cooperatively to amplify immune responses (Hou et al., 2014).
Other LRR-RKs are implicated in immunity, but the ligands remain to be elucidated. Notable among these are AtNILR1, the first identified plant receptor involved in immunity to nematodes (Mendy et al., 2017); OsRLK1, which initiates defense against chewing herbivores (Hu et al., 2018); and AtMIK2, which has been described as a key component in sensing Fusarium (Coleman et al., 2019).
LRR-RPS
Multiple LRR-RPs from Arabidopsis and solanaceous plants have been shown to have a role in immunity. Unlike the widespread distribution of FLS2, thus far all known LRR-RPs display genus- or subgenus-specific distribution. Many characterized LRR-RPs constitutively associate with the LRR-RK SOBIR1/EVR (Liebrand et al., 2014). LRR-RP/SOBIR1 complexes have been described as “bimolecular RKs” with the LRR-RP providing the ligand specificity and SOBIR1 providing the kinase domain for intracellular signaling (Gust and Felix, 2014).
The first LRR-RPs involved in plant defense were classified as resistance genes. These include tomato SlCf2, SlCf4, SlCf5, and SlCf9, which confer resistance to Cladosporium fulvum (Cf) isolates carrying the corresponding avirulence (Avr) genes encoding the Cys-rich proteins Avr2, Avr4, Avr5, and Avr9, respectively (Jones et al., 1994; Joosten et al., 1994; Dixon et al., 1996, 1998). Rather than binding Avrs directly, Cfs are reported to detect the inhibitory effect of Avrs on specific plant proteases (Hammond-Kosack et al., 1994; Rooney et al., 2005). Since the identification of Cfs, many LRR-RPs have been shown to have a role in defense in solanaceous plants. SlEIX1 and SlEIX2 bind ethylene-inducing xylanase (EIX) from Trichoderma spp. (Dean et al., 1989; Ron and Avni, 2004). Whereas SlEIX2 is capable of initiating defense responses upon EIX treatment, SlEIX1 is involved in BAK1-dependent attenuation of EIX-mediated defense (Bar et al., 2010, 2011). Different surface-exposed regions within EIX and the related Botrytis xylanase BcXyn11A have been reported to be crucial for SlEIX2-dependent recognition, but the activity of these regions could not be verified using peptide epitopes. Recently, the 25-amino acid peptide xyn25 from BcXyn11A was reported to trigger defense responses in tomato and tobacco (Nicotiana tabacum; Frías et al., 2019). Whether this activity is EIX1/2-dependent remains to be shown. SlVe1 is implicated in resistance to Verticillium dahliae harboring the effector Ave1, a homolog of plant natriuretic peptides presumably acquired by horizontal gene transfer (Kawchuk et al., 2001; de Jonge et al., 2012; Postma et al., 2016). The wild potato LRR-RP SmELR mediates response to the Phytophthora-derived elicitin INF1 in a SOBIR1-dependent manner (Du et al., 2015; Domazakis et al., 2018). Nicotiana benthamiana harbors the receptor NbRXEG1 that is responsible for recognition of the glycoside hydrolase family 12 member XEG1 from Phytophthora sojae (Ma et al., 2015; Wang et al., 2018c). While csp22 is perceived by an LRR-RK in tomato (Wang et al., 2016), the csp22 response in N. benthamiana is reported to require the LRR-RP NbCSPR (Saur et al., 2016).
LRR-RPs are also known to play multiple roles in microbial resistance in Brassicaceae. Resistance of Brassica napus to Leptosphaeria maculans is determined by BnLepR3/BnRLM2-mediated recognition of AvrLm1/AvrLm2 (Long et al., 2011; Larkan et al., 2013, 2014, 2015). The Arabidopsis LRR-RPs AtRLP1/ReMAX, AtRLP23, AtRLP30, and AtRLP42/RBPG1 have been described to perceive an enigmatic MAMP from Xanthomonas spp. (Jehle et al., 2013); Nep1-like proteins (NLPs) from various bacteria, oomycetes, and fungi (Böhm et al., 2014; Albert et al., 2015); Sclerotinia sclerotiorum filtrate elicitor1 (SCFE1; Zhang et al., 2013) and fungal polygalacturonases (Zhang et al., 2014), respectively. Nlp20, the 20-amino acid minimal immunogenic motif from NLPs, binds directly to AtRLP23 and induces complex formation with AtSERKs (Böhm et al., 2014; Albert et al., 2015, 2019). SCFE1 also induces complex formation between AtRLP30 and AtSERKs (Albert et al., 2019). A similar mechanism is anticipated for the activation of other RPs.
In addition to microbial pathogens, LRR-RPs are also involved in defense against higher organisms. SlCuRe1 detects a proteinaceous pattern from Cuscuta reflexa and represents the first receptor implicated in defense against parasitic plants (Hegenauer et al., 2016). Cowpea (Vigna unguiculata) inceptin receptor binds the 11-amino acid inceptin (Vu-In) that is proteolytically cleaved from the chloroplastic ATP synthase during caterpillar herbivory (Steinbrenner et al., 2019).
LYSIN MOTIF-RKS AND RPS
Plants sense polysaccharide patterns using lysin motif (LysM) receptors. LysM-RKs contain three extracellular LysM domains, a transmembrane domain, and an intracellular kinase domain (Buendia et al., 2018; Schlöffel et al., 2019). LysM-RPs lack kinase and transmembrane domains but are instead attached to the plasma membrane by GPI-anchors. Compared to the LRR-receptor family, the LysM-receptor family is rather small, comprising five to 22 LysM-RKs and two to five LysM-RPs (Buendia et al., 2018). LysM-RKs and -RPs contribute to both immunity and symbiosis by sensing n-acetylglucosamine-containing patterns such as lipochitooligosaccharides, chitooligosaccharides (CO), and peptidoglycan (PGN). Interplay between receptors and coreceptors seems to be complex, particularly in respect to distinguishing between defense and symbiosis.
The LysM-RK chitin elicitor receptor kinase1 (CERK1) seems to be a key player in n-acetylglucosamine perception for most defense pathways (Shinya et al., 2015). Rice perceives octameric CO fragments by OsCERK1 and the LysM-RP chitin elicitor-binding protein (OsCEBiP; Kaku et al., 2006; Shimizu et al., 2010). Structural and biochemical studies revealed that CO binding within the second LysM domain induces homodimerization of OsCEBiP, which is likely followed by the recruitment of OsCERK1 (Shimizu et al., 2010; Hayafune et al., 2014; Liu et al., 2016). OsCERK1 is also required for perception of symbiotic short chain CO, a process that is independent of OsCEBiP (Carotenuto et al., 2017). Intriguingly, OsCERK1 has been reported to play a role in lipopolysaccharide (LPS) signaling as well (Desaki et al., 2018).
In Arabidopsis, chitin is perceived by a complex of LysM-RKs comprising AtLYK4, AtLYK5, and AtCERK1 (Miya et al., 2007; Cao et al., 2014; Xue et al., 2019). AtLYK4 and AtLYK5 interact constitutively (Xue et al., 2019), whereas AtLYK5 and AtCERK1 interaction is ligand-dependent (Cao et al., 2014). Downstream signaling is dependent on AtCERK1 homodimerization (Liu et al., 2012b; Cao et al., 2014). A second chitin-sensing mechanism in Arabidopsis has been proposed for regulation of fluxes across plasmodesmata. This pathway is dependent on AtLYK4 and AtLYK5 along with AtLYM2/AtCEBiP (Faulkner et al., 2013), which has been shown to bind chitin (Shinya et al., 2012).
Putative orthologs of OsCEBiP in other plant species have also been shown to be involved in CO response. Lotus (Lotus japonicus) LjLYS6/LjCERK6 binds CO6-8. The structure of LjLYS6 is highly similar to AtCERK1 with a highly conserved chitin binding site (Bozsoki et al., 2017). MtLYK9 (a putative ortholog of LjLYS6) and MtLYR4 are indispensable for chitin signaling in Medicago truncatula (Bozsoki et al., 2017). PsLYK9 is proposed as a CERK1-like receptor for chitin in Pisum sativum (Leppyanen et al., 2017), although chitin binding has not yet been demonstrated. LysM-RKs are also involved in CO-triggered immunity in grapevine (Vitis vinifera; VvLYK1-1 and VvLYK1-2) and apple (Malus domestica; MdCERK1; Zhou et al., 2018; Brulé et al., 2019).
PGN signaling in Arabidopsis and rice is mediated by tripartite complexes of LysM-RPs and -RKs. In Arabidopsis, AtLYM1 and AtLYM3 form a PGN receptor complex with AtCERK1 (Willmann et al., 2011). As with AtBGAL1-mediated breakdown of flagellin, the hydrolase AtLYS1 promotes elicitor accessibility by releasing PGN fragments from insoluble bacterial cell walls (Liu et al., 2014). In addition to CO and PGN perception, AtCERK1 is involved in the perception of 1,3-β-d-(Glc)6 from fungal cell walls (Mélida et al., 2018). In rice, two LysM-RPs, OsLYP4 and OsLYP6, together with OsCERK1, contribute to PGN and CO6 recognition using the same binding site (Liu et al., 2012a). Structure–function analysis of PGN perception is lacking.
Legumes use LysM-receptors not only for danger surveillance but also to regulate symbiosis. In lotus, LjNFR1 (LYK) and LjNFR5 (LYR) bind lipochitooligosaccharide Nod factors and are essential for symbiosis (Broghammer et al., 2012). Homologs of LjNFR1 and LjNFR5 have been identified in M. truncatula (MtLYK3; MtNFP, MtLYR3) and P. sativum (PsSym37, PsK; PsSym10, PsLYR3). In Medicago, MtLYK9 is involved in symbiosis with arbuscular mycorrhizal fungi (Gibelin-Viala et al., 2019). The legume LysM-RK LjEPR3 binds bacterial exopolysaccharides involved in symbiosis (Kawaharada et al., 2015).
OTHER TYPES OF RECEPTORS
The malectin-like-domain RK FERONIA (AtFER) was initially found to be critical for female fertility in Arabidopsis (Huck et al., 2003; Rotman et al., 2003) and later identified as a receptor for the peptide rapid alkalinization factor1 (AtRALF1) involved in regulating primary root growth (Haruta et al., 2014). In the past few years, AtFER has emerged as an important component of immune signaling complexes with a proposed role as a RALF-regulated scaffold. Regulation of immunity by AtFER depends on the GPI-anchored protein AtLLG1, which constitutively associates with AtFER, AtFLS2, and AtEFR (Shen et al., 2017). AtFER and AtLLG1 enhance the association of AtFLS2/AtEFR with AtBAK1 upon flg22/elf18 perception, but this association is hampered by AtRALF23 (Stegmann et al., 2017). A recent study shows that AtRALF23 directly interacts with both AtFER and AtLLG1 and promotes assembly of an AtRALF23-AtLLG1-AtFER heterocomplex (Xiao et al., 2019). How AtRALF23 negatively regulates AtFLS2/AtEFR-mediated response through the AtLLG1-AtFER complex remains unknown. Other roles have also been assigned to AtFER. During salt-stress, AtFER is necessary to maintain cell-wall integrity by sensing the damage of pectin (Feng et al., 2018). Cell-wall LRR extensin 3 (LRX3),, LRX4, and LXR5 associate with AtRALF23 and AtFER, and function together as a module to transduce cell-wall signals that regulate growth and salt tolerance (Zhao et al., 2018). Whether AtLRX3/4/5, pectin, or other cell-wall components are also part of the scaffolds that regulate immune receptor complexes is an interesting topic for further investigation.
Several nonproteinaceous elicitors are detected by other lectin-type RKs. The bulb-type lectin s-domain-1 RK AtLORE was initially reported to sense the conserved lipid A moiety of LPS (Ranf et al., 2015). Recently, however, medium-chain 3-hydroxy fatty acids (mc-3-OH-FAs) that copurify with LPS, but not LPS itself, were shown to elicit AtLORE-dependent immunity (Kutschera et al., 2019). How the mc-3-OH-FA metabolites are derived from bacteria remains unknown. A legume-type lectin RK, AtLecRK-I.9/AtDORN1, was identified as a high affinity receptor for extracellular ATP (eATP; Choi et al., 2014). eATP triggers AtLecRK-I.9/AtDORN1-mediated AtRBOHD phosphorylation to regulate stomatal aperture and bacterial resistance (Chen et al., 2017). Recently, it was reported that the root endophytic fungus Serendipita indica secretes an ecto-5′-nucleotidase to modulate eATP levels and thereby prevent detection (Nizam et al., 2019). Another legume-type lectin RK, LecRK-I.8, is also involved in basal resistance against bacterial pathogens. LecRK-I.8 is reported to directly sense extracellular NAD+ as a DAMP (Wang et al., 2017). Surprisingly, AtLecRK-I.8 was also recently reported to mediate immune response to lipid extracts from the eggs of the insect Pieris brassicae (Gouhier-Darimont et al., 2019). The explanation for these paradoxical findings are not yet clear. The lipidic extracts are unlikely to contain NAD+, but it is possible that they could trigger NAD+ release to the cytoplasm (Gouhier-Darimont et al., 2019).
The EGF-like–domain RK AtWAK1 perceives oligogalacturonide (OG), which is released from pectin during fungal infection (Brutus et al., 2010). Hyperaccumulation of OGs causes growth inhibition and eventually cell death. Oxidization of OGs by OG oxidases 1–4 dampens OG-triggered immune responses (Benedetti et al., 2018). Oxidization of cellodextrin, another DAMP released from cellulose, by AtCELLOX also attenuates immune responses (Locci et al., 2019), suggesting that oxidation of cell-wall–derived oligosaccharides is a common mechanism to modulate DAMP-triggered immune signaling.
PRR COMPLEXES AT THE PLASMA MEMBRANE
As mentioned previously, SERKs serve as coreceptors for various LRR-RK and -RP receptors involved in developmental and immunity signaling pathways (Yasuda et al., 2017; He et al., 2018). Structural analyses have revealed a common activation mechanism for LRR-RKs, in which the LRR domains of receptors and shape-complementary SERKs heterodimerize in the presence of ligand. Ligands promote dimerization either by binding both proteins directly as “molecular glue” or allosterically through stabilization of an RK island domain (Sun et al., 2013; Wang et al., 2015; Hohmann et al., 2017). LRR-RPs likely employ a similar activation mechanism, but the structural role of SOBIR1 in formation of the tripartite complex is unknown.
In the absence of ligand, receptor–SERK complex assembly is negatively regulated by AtBIR2 and AtBIR3, two closely related LRR-RKs with structures similar to SERKs but lacking kinase activity (Halter et al., 2014; Imkampe et al., 2017). AtBIR2/3 constitutively associate with AtBAK1 in the absence of ligand and AtBAK1 heterodimerization with AtFLS2 and other LRR-RKs (Halter et al., 2014; Imkampe et al., 2017). In the presence of flg22, AtBIR2 and AtBIR3 dissociate from AtBAK1, which then becomes accessible for the interaction with AtFLS2 (Halter et al., 2014; Imkampe et al., 2017). Recent structural analyses revealed that AtBIR2/3 have a conserved SERK binding mechanism (Hohmann et al., 2018). Additional quantitative biochemical assays revealed that AtBRI1 (an LRR-RK involved in brassinolide-mediated growth) and AtBIR2/3 compete for binding of SERKs. AtBRI1 is able to out-compete AtBIR2/3 in the presence of ligand (Hohmann et al., 2018). It is plausible that a similar competition system exists for AtFLS2 and AtBIR2/3. It is unknown whether such a regulatory system exists for LysM-RKs or other types of receptors.
AtNIK1, a LRR-RK, has previously been shown to be a positive regulator of plant immunity to viral infections (Zorzatto et al., 2015). Recently, AtNIK1 has been reported to negatively regulate flg22-induced immune responses by physical interaction with both FLS2 and BAK1 (Li et al., 2019). Whereas BIR2/BIR3 function to prevent FLS2–BAK1 complex formation in the absence of flg22, NIK1 suppression of FLS2–BAK1 complex formation is enhanced upon flg22 perception (Li et al., 2019). Thus, NIK1 may be involved in attenuating FLS2 signaling or controlling optimum levels of FLS2–BAK1 complexes.
Like AtFER described previously, several other RKs also appear to function as scaffolds to regulate immune receptor complex formation. AtIOS1, a malectin-like LRR-RK, associates with multiple PRRs and coreceptors and promotes AtFLS2–AtBAK1 complex formation upon flg22 perception (Yeh et al., 2016). Two other malectin-like-domain RKs, AtANX1 and AtANX2, also associate with AtFLS2 and AtBAK1, but negatively regulate AtFLS2–AtBAK1 complex formation (Mang et al., 2017). It is not known if AtANX1 and AtANX2 function as scaffolds for other PRRs. A comprehensive interaction network analysis of receptor ectodomains identified the LRR-RK AtFIR as an additional AtFLS2 interactor that promotes AtFLS2–AtBAK1 complex formation. AtFIR is required for flg22 response and microbial resistance (Smakowska-Luzan et al., 2018). This study further revealed that the LRR-RK AtAPEX associates with AtPEPR1 and AtPEPR2 in a ligand-independent manner and negatively regulates AtPEPR1/AtPEPR2-mediated and AtFLS2-mediated immune responses. In contrast, AtAPEX acts as a positive regulator in AtBRI1 signaling (Smakowska-Luzan et al., 2018).
Single-particle tracking live-cell imaging revealed that AtFLS2 and AtBRI1 localize within distinct plasma membrane nanodomains of different sizes, which suggests that spatiotemporal separation of receptor complexes may play a role in maintenance of signal specificity (Bücherl et al., 2017; McKenna et al., 2019). Nanodomain size and diffusion of AtFLS2 is regulated by both the cytoskeleton and the cell wall (McKenna et al., 2019). How the dynamics of AtFLS2-containing nanodomains regulate signaling remains unknown.
POST-TRANSLATIONAL MODIFICATIONS IN EARLY IMMUNE SIGNALING
Perception of danger triggers numerous defense responses including stomatal closure, callose deposition, reactive oxygen species (ROS) burst, and production of defensive secondary metabolites. Generation and regulation of these responses involves a complex signaling network that employs several types of post-translational modifications (Bhattacharjee et al., 2015; Withers and Dong, 2017; de Vega et al., 2018). Here we highlight early events in PRR signaling (i.e. those occurring in the proximity of the PRR complexes). Presumably, the first intracellular signaling event upon PAMP perception is transphosphorylation between the kinase domains of coreceptors and PRRs (or adaptor kinases like SOBIR1). Phosphorylation patterns are not the same in all cases. In vitro studies show that AtBAK1 is differentially phosphorylated by the AtBRI1, AtBIK1, AtFLS2, and AtEFR kinase domains (Wang et al., 2014). Perraki et al. (2018) identified AtBAK1 phosphosites that are required for immunity but not brassinolide signaling. These results imply that kinases may not be simply on/off switches in certain pathways, but could have multiple modes of activation depending on the pattern of phosphorylation or phosphocode.
Upon activation, AtBAK1 phosphorylates positive regulators of immunity such as the receptor-like cytoplasmic kinase (RLCK) AtBIK1 and negative regulators of immunity such as AtBIR2/3. Phosphorylation of AtBIR2/3 promotes their dissociation from the receptor complex, facilitating formation of an active complex (Halter et al., 2014; Imkampe et al., 2017). AtBIK1 also dissociates from the receptor complex and phosphorylates multiple downstream targets including AtRGS1, which regulates heterotrimeric G proteins, and AtRBOHD, the NADPH oxidase responsible for PRR-mediated ROS burst (Liang et al., 2018; Liang and Zhou, 2018; Mithoe and Menke, 2018). AtBIK1 has also been shown to partially localize to the nucleus, where it can phosphorylate several WRKY transcription factors involved in transcriptional reprogramming (Lal et al., 2018). Recently, AtBIK1 was further shown to activate a calmodulin-gated calcium channel composed of AtCNGC2 and AtCNGC4 (Tian et al., 2019). Phosphorylation of AtCNGC2/4 by AtBIK1 is critical for calcium spiking in response to immune elicitors (Tian et al., 2019). The related channel proteins AtCNGC19 and 20 have recently been implicated in regulation of AtBAK1-dependent cell death containment in plant immunity (Yu et al., 2019), and AtCNGC19 has further been shown to have a role in resistance against insect herbivory (Meena et al., 2019). It is not yet known if BIK1 also has a role in activating AtCNGC19 and AtCNGC20.
AtBIK1 is part of the 46-member Arabidopsis RLCK-VII family (Rao et al., 2018), which contains many members thought to play roles in development and immunity. Rao et al. (2018) performed an extensive analysis in which higher order mutants of nine RLCK-VII subfamilies were tested for their roles in immunity and development. This study revealed that multiple RLCK-VIIs play roles in immunity including subfamilies 4, 5, 7, and 8. AtPBL27, from the RLCK-VII-1 subfamily, was recently shown to promote chitin-induced stomatal closure by phosphorylating the s-type anion channel AtSLAH3 (Liu et al., 2019). AtPBL27 is critical for this activity, as a pbl27 mutant is defective in chitin-induced stomatal closure and more susceptible to Botrytis infection (Liu et al., 2019). AtPBL27 was also reported to phosphorylate AtMAPKKK5, initiating a MAP kinase signaling cascade including AtMPK3/6 (Yamada et al., 2016). However, a subsequent report claims that RLCK-VII-4 subfamily members, but not AtPBL27, mediate this process (Bi et al., 2018).
In most cases, RLCK-VII single mutants do not show significant defects in immunity, likely due to overlapping functions. Nevertheless, RLCK-VIIs play selective roles in triggering different outputs. AtBIK1, for example, is critical for ROS and calcium signaling (Tian et al., 2019) but not MAPK activation (Feng et al., 2012). RLCK-VIIs also play differential roles in responses to different elicitors. For example, the RLCK-VII-4 subfamily is important for ROS responses to chitin but not flg22 or elf18 (Rao et al., 2018). In Arabidopsis, nlp20 triggers notably different outputs than flg22 or elf18 (Wan et al., 2019b). Intriguingly, although AtBIK1 is a positive regulator of flg22- and elf18-triggered immunity, bik1 mutants show stronger responses to nlp20, suggesting an opposing regulatory role of BIK1 in LRR-RP- and LRR-RK signaling (Wan et al., 2019b).
The 12-member RLCK-XII family also has important roles in immune and developmental signaling. AtBSK1 associates with AtFLS2 and is critical for several immune responses including flg22-induced ROS (Shi et al., 2013). Like PBL27 and RLCK-VII-4s mentioned previously, AtBSK1 is also reported to phosphorylate AtMAPKKK5 and thereby initiate MAPK signaling (Yan et al., 2018). The related AtBSK5 was recently shown to be involved in AtFLS2-, AtEFR-, and AtPEPR-mediated responses (Majhi et al., 2019). In addition to RLCK-VII and XII RLCKs, other important kinases in early signaling include calcium-dependent kinases (CPK) such as AtCPK5 (Yip Delormel and Boudsocq, 2019), the MAP4Ks AtSIK1 and AtMAP4K4 (Zhang et al., 2018; Jiang et al., 2019), and Cys-rich receptor-like kinases such as AtCRK2 (Kimura et al., 2019). All of these kinases act cooperatively with AtBIK1 to trigger ROS burst via AtRBOHD. Some kinases, such as AtCPK28 and AtPBL13, negatively regulate immune signaling (Monaghan et al., 2014; Lin et al., 2015).
The phosphorylation status of immune signaling components must be tightly regulated. Arabidopsis encodes more than 100 phosphatase catalytic subunits (Wang et al., 2007). AtPP2A negatively regulates immunity by modulating AtBAK1 phosphorylation (Segonzac et al., 2014), while AtPP2C38 has a similar function on AtBIK1 (Couto et al., 2016). In tomato, the PP2C phosphatase SlPic1 was recently shown to negatively regulate flg22- and csp22-induced immunity through its action on the RLCK SlPti1b (Giska and Martin, 2019). The small BSU1 family of phosphatases, on the other hand, positively regulates immune and brassinolide signaling (Park et al., 2019). AtBSU1 is phosphorylated by AtBIK1 on Ser-251 in a flg22-dependent manner, an event that appears to be necessary for AtMPK3/6 activation (Park et al., 2019). Notably, Ser-251 phosphorylation is required for immune signaling but not brassinolide signaling indicating that, like AtBAK1, AtBSU1 activity is also phosphocode-dependent. The target(s) of AtBSU1 are not yet known.
The above examples highlight only a small number of kinases and phosphatases that play roles in early defense signaling. Given the number and diversity of kinases and phosphatases involved, the partial redundancies, and the unknown complexities of the phosphocode for each kinase or phosphatase, elucidation of the full role of phosphorylation in immune signaling remains a formidable challenge.
In addition to phosphorylation, ubiquitination plays an established role in plant immunity (Duplan and Rivas, 2014; Zhou and Zeng, 2017; Mithoe and Menke, 2018). The ubiquitin E3 ligases PUB12/13, for example, are important for attenuation of AtFLS2 signaling (Lu et al., 2011). Upon AtFLS2 activation, AtBAK1 phosphorylates AtPUB12/13. The activated E3 ligases, in turn, polyubiquitinate AtFLS2, targeting it for degradation (Lu et al., 2011). The ubiquitin E3 ligases AtPUB25/26 work together with AtCPK28 and heterotrimeric G-protein to tightly control AtBIK1 levels (Monaghan et al., 2014; Liang et al., 2016; Wang et al., 2018a). Although ubiquitination is mostly used as a means to attenuate immune signaling, it can also play a positive role. AtPUB4, for example, is a positive regulator of chitin-induced immune responses, but the target(s) of AtPUB4 is unknown (Desaki et al., 2019).
SUMOylation, or incorporation of a small ubiquitin-like modifier (SUMO), is a reversible process that can affect protein structure, localization, and interaction with partners. A recent study showed that AtFLS2 is SUMOylated on Lys-1120 rapidly upon flg22 treatment (Orosa et al., 2018). Mutation of the SUMOylation site alters its interaction with AtBIK1 and blocks downstream signaling (Orosa et al., 2018). SUMOylation is regulated by the deSUMOylating enzyme AtDesi3a, which is rapidly degraded upon flg22 perception (Orosa et al., 2018).
Proteolytic processing also plays important roles in immune modulation, notably in the production of phytocytokines such as Peps, PIPs, and RALFs. Several studies show that proteolytic processing of RKs may also play a role in early signaling events. Zhou et al. (2019) found that AtBAK1 is processed by a conserved protease and that its cleavage is PAMP-induced. Mutation of Asp-287 prevents AtBAK1 processing, and impairs flg22-induced responses, but this may be in part due to mislocalization of the receptor. Proteolytic processing has also been reported for two different PRRs (Park and Ronald, 2012; Petutschnig et al., 2014), but the role of RK processing in immune signaling remains unknown.
A number of posttranslational modifications are important for the appropriate folding or localization of RKs, RPs, and RLCKs. These include glycosylation, myristoylation, and s-acylation (Saijo, 2010; Boyle and Martin, 2015; Withers and Dong, 2017; Majeran et al., 2018). s-acylation is notable among these because it is a reversible modification, and a recent study reports that PAMP-induced de-acylation regulates the activity of AtFLS2, AtDORN1, and AtCERK1 (Chen et al., 2019). The study suggests that acylation regulates the activity of the receptors by controlling their nanodomain localization (Chen et al., 2019). However, these results conflict with another recent study that shows that s-acylation is not required for AtFLS2 signaling (Hurst et al., 2019). It will be interesting to learn to what extent SUMOylation, proteolytic processing of RKs, and s-acylation play roles in various immune signaling pathways.
PATHOGEN EFFECTORS TARGETING PRR COMPLEXES
Plant-adapted pathogens suppress or evade plant immunity through the use of secreted protein effectors, many of which directly target the plant surface receptor complex. Bacterial pathogens utilize a type-III secretion system to transport effectors that suppress the catalytic activity of RKs (Göhre et al., 2008; Shan et al., 2008; Macho et al., 2014), degrade PRRs (Göhre et al., 2008; Gimenez-Ibanez et al., 2009; Li et al., 2016), or inhibit phosphorylation of RLCKs (Zhang et al., 2010; Feng et al., 2012; Yamaguchi et al., 2013). Prototypical examples include Pseudomonas syringae AvrPto, which can target multiple LRR-RKs (e.g. AtFLS2, AtEFR, and AtBAK1; Shan et al., 2008; Xiang et al., 2008); AvrPtoB, which can target both LRR-RKs (e.g. AtFLS2 and AtBAK1) and LysM-RK CERK1 (Göhre et al., 2008; Gimenez-Ibanez et al., 2009); the tyrosine phosphatase HopAO1, which can target AtFLS2 and AtEFR (Macho et al., 2014); and the cysteine protease AvrPphB, which degrades RLCKs, and the 5′-monophosphate transferase AvrAC that uridylates RLCKs (Zhang et al., 2010; Feng et al., 2012). More detailed information and additional examples have been well-reviewed (Macho and Zipfel, 2015; Melotto et al., 2017). No bacterial effectors have been found to target the extracellular domain of PRRs.
Two important functions were found recently for the P. syringae effectors AvrPto and HopB1 (Li et al., 2016; Wu et al., 2018). For over 10 years, AvrPto has been known to act on multiple LRR-RKs such as AtFLS2 and AtEFR, as well as the coreceptor AtBAK1 (Shan et al., 2008; Xiang et al., 2008). Recently, the coreceptor SOBIR1 was identified as an additional target of AvrPto. AvrPto suppresses Avr4/SlCf4-triggered cell death in N. benthamiana and forms a complex with SlSOBIR1 in planta. AvrPto binding does not require SOBIR1 kinase activity and does not affect SlCf-4/SlSOBIR1/SlSERK3a complex formation (Wu et al., 2018). The detailed mechanism of cell death suppression by AvrPto remains unclear. The previously uncharacterized HopB1 was recently identified as a Ser protease that cleaves AtBAK1 (Li et al., 2016). HopB1 constitutively associates with the AtFLS2 kinase domain but does not bind AtBAK1 or AtBIK1. Upon flg22 treatment, HopB1 cleaves the kinase-activated form of AtBAK1. With this novel strategy to cleave only activated AtBAK1, HopB1 can suppress immunity while causing minimal cellular disturbances that might be perceived by host surveillance systems (Li et al., 2016).
The vascular fungal pathogen Fusarium oxysporum produces small secreted peptides homologous to plant RALFs (F-RALFs). F-RALFs induce host alkalinization, which is required for the infection of some hemibiotrophic fungi (Masachis et al., 2016). Loss of F-RALF in F. oxysporum results in increased ROS accumulation, callose deposition, and PR gene expression during fungal infection. F-RALF-mediated immune suppression is not observed in fer mutant plants, suggesting that F-RALFs mimic endogenous FER-targeting RALF peptides (Masachis et al., 2016). Many other phytopathogenic fungi produce RALF-like peptides with functions that remain to be elucidated (Thynne et al., 2017). Thus, RALF mimicry could represent a common mode of action for fungal apoplastic effectors. Recently, another conserved fungal effector named necrosis-inducing secreted protein1 (NIS1) from Colletotrichum spp. and Magnaporthe oryzae was found to inhibit autophosphorylation of AtBAK1 and AtBIK1. NIS1 also disrupts AtBIK1 association with AtRBOHD upon flg22-treatment. Interestingly, NIS1 not only suppresses immune responses triggered by the extracellular patterns flg22, INF1, and chitin, but also responses triggered upon recognition of the Phytophthora infestans effector Avr3a (Irieda et al., 2019).
Thus far, no oomycete effectors have been definitively shown to target PRRs. However, some P. infestans RxLR effectors suppressing plant early immune responses are worth noting (Zheng et al., 2014). The RxLR effector SFI5 suppresses ROS accumulation and MAPK activation triggered by flg22 in a calcium/calmodulin-dependent manner in both Arabidopsis and tomato. Although the host targets of SFI5 have not been found, its subcellular localization highly resembles that of AvrPto (Zheng et al., 2018). The eATP receptor AtLecRK-I.9/AtDORN1 is implicated as a possible target of the P. infestans RxLR effector IPI-O (Bouwmeester et al., 2011). There are no reports of insect or nematode effectors that directly target PRRs. However, these pests produce numerous effectors that suppress immunity through as-yet unknown mechanisms (Hogenhout and Bos, 2011; Mejias et al., 2019), and nematodes are known to manipulate RKs in other signaling pathways through ligand mimicry (Hu and Hewezi, 2018). The identification of additional effectors and their activities is important both to better understand pathogen strategies and to further elucidate the proteins involved in immune signaling and their functions.
CONCLUSION
Tremendous efforts from laboratories all over the world have led to the discovery of dozens of plant PRRs in numerous plant species. Comparative analyses of PRR structures, ligand specificity, and genetic distribution have led to unexpected insights. For example, only a minor fraction of microbe-derived patterns (bacterial flagellin-derived flg22, bacteria-derived PGN, fungus-derived chitin) are recognized by PRRs that are widespread among plants and can be found in both monocot and eudicot plant species. Sensor systems for such patterns should thus be considered an ancient set of PRRs. In contrast, the majority of PRRs known to date exhibit genus-specific distribution patterns, a feature that is particularly inherent to LRR-RP-type PRRs. Lack (or loss) of PRR activities in closely related plant species or among accessions of the same species must further be interpreted as evidence for substantial evolutionary dynamics within PRR protein families. Likewise, the discovery of nonhomologous PRRs recognizing different structural epitopes within one and the same microbial molecule (e.g. SlFLS2 and SlFLS3 recognition of flg22 and flgII-28, respectively) is an important finding.
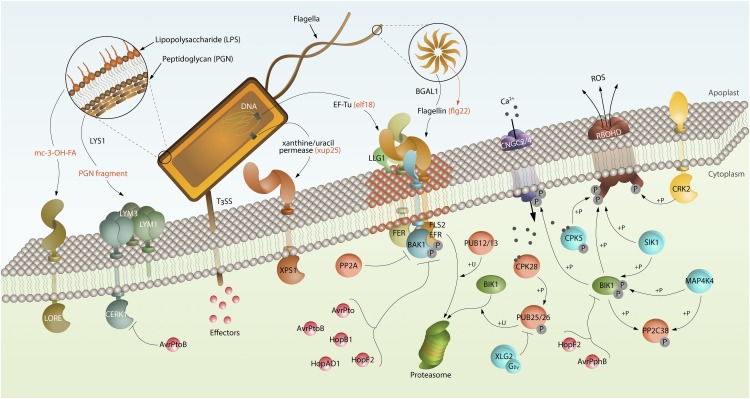
In our view, one of the most astonishing lessons learned from plant PRR research is the sheer complexity of patterns and PRRs that underlie PTI activation within a given plant-microbe interaction. Figure 2 exemplifies this complexity for the interaction of the bacterial pathogen P. syringae with its host, Arabidopsis. P. syringae can be detected by at least five different PRRs, which recognize three proteinaceous bacterial patterns, one lipophilic compound, and PGN. This list is expected to increase, and it is reasonable to assume that similarly complex scenarios may also be found for less-studied plant–microbe interactions (see Outstanding Questions). Past and current research into PRR identification and characterization also hold implications for biotechnological applications. For example, single plant-genus–specific PRRs have been used successfully to engineer crop plants with enhanced immunity to microbial infection (Lacombe et al., 2010; Albert et al., 2015; Boutrot and Zipfel, 2017). Broad spectrum disease resistance in crops may further be boosted by the use of combinations of PRR stacking (PRRs; Wan et al., 2019a).
Figure 2.
Interactions between the bacterial pathogen P. syringae and Arabidopsis surface sensor systems. Arabidopsis employs several plasma membrane immune receptors that detect multiple patterns derived from P. syringae. These include LORE, which perceives mc-3-OH-FA; the CERK1–LYM1–LYM3 complex, which perceives PGN fragments released by LYS1; XPS1, which perceives xanthine/uracil permease (xup25); EFR, which perceives EF-Tu (elf18); and FLS2, which perceives flagellin (flg22) and is deglycosylated by BGAL1. FER and LLG1 associate with FLS2/EFR and regulate immune complexes, which may reside in specific plasma membrane nanodomains (rust-colored). BIK1 plays a central role in early responses after PAMP recognition. Upon flg22 perception, BAK1 associates with and transphosphorylates FLS2, leading to BIK1 activation and disassociation from FLS2–BAK1 complex. Activated BIK1 phosphorylates CNGC2/4 and RBOHD to regulate Ca2+ influx and ROS production, respectively. CPK5 and SIK1 also phosphorylate RBOHD, and SIK1 and MAP4K4 phosphorylate and stabilize BIK1 to ensure robust ROS production. BIK1 function is negatively regulated by PP2C38 in the resting state, whereas PP2C38 is phosphorylated by BIK1 and MAP4K4 and is released from BIK1 upon flg22 perception. To tightly control the immune responses, BAK1 is negatively regulated by PP2A. FLS2 and nonphosphorylated BIK1 are polyubiquitinated by E3 ligases PUB12/13 and PUB25/26, respectively, targeting them for degradation. Furthermore, heterotrimeric G-protein (XLG2/Gβ/Gγ) stabilizes BIK1 by inhibiting PUB25/26 activity in the resting state. Upon flg22 perception, the heterotrimeric G-protein disassociates from FLS2–BIK1 complex. CPK28 is further activated to phosphorylate PUB25/26 to accelerate nonphosphorylated BIK1 degradation, which in turn allows the phosphorylated BIK1 to activate downstream signaling. To suppress plant immune responses, P. syringae transports effectors (eg. AvrPto, AvrPtoB, HopB1, HopAO1, HopF2) through a type-III secretion system (T3SS) to inhibit the functions of receptor complex (e.g. FLS2, EFR, BAK1, and CERK1) and other key signaling components (e.g. BIK1).
Footnotes
Articles can be viewed without a subscription.
References
- Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang H, et al. (2015) An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat Plants 1: 15140. [DOI] [PubMed] [Google Scholar]
- Albert I, Zhang L, Bemm H, Nürnberger T (2019) Structure-function analysis of immune receptor AtRLP23 with its ligand nlp20 and coreceptors AtSOBIR1 and AtBAK1. Mol Plant Microbe Interact 32: 1038–1046 [DOI] [PubMed] [Google Scholar]
- Albert M, Jehle AK, Lipschis M, Mueller K, Zeng Y, Felix G (2010) Regulation of cell behaviour by plant receptor kinases: Pattern recognition receptors as prototypical models. Eur J Cell Biol 89: 200–207 [DOI] [PubMed] [Google Scholar]
- Bar M, Sharfman M, Avni A (2011) LeEix1 functions as a decoy receptor to attenuate LeEix2 signaling. Plant Signal Behav 6: 455–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Sharfman M, Ron M, Avni A (2010) BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J 63: 791–800 [DOI] [PubMed] [Google Scholar]
- Benedetti M, Verrascina I, Pontiggia D, Locci F, Mattei B, De Lorenzo G, Cervone F (2018) Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J 94: 260–273 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Noor JJ, Gohain B, Gulabani H, Dnyaneshwar IK, Singla A (2015) Post-translational modifications in regulation of pathogen surveillance and signaling in plants: The inside- (and perturbations from) outside story. IUBMB Life 67: 524–532 [DOI] [PubMed] [Google Scholar]
- Bi G, Zhou Z, Wang W, Li L, Rao S, Wu Y, Zhang X, Menke FLH, Chen S, Zhou JM (2018) Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30: 1543–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H, Albert I, Oome S, Raaymakers TM, van den Ackerveken G, Nürnberger T (2014) A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog 10: e1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55: 257–286 [DOI] [PubMed] [Google Scholar]
- Bouwmeester K, de Sain M, Weide R, Gouget A, Klamer S, Canut H, Govers F (2011) The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog 7: e1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle PC, Martin GB (2015) Greasy tactics in the plant-pathogen molecular arms race. J Exp Bot 66: 1607–1616 [DOI] [PubMed] [Google Scholar]
- Bozsoki Z, Cheng J, Feng F, Gysel K, Vinther M, Andersen KR, Oldroyd G, Blaise M, Radutoiu S, Stougaard J (2017) Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc Natl Acad Sci USA 114: E8118–E8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109: 13859–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulé D, Villano C, Davies LJ, Trdá L, Claverie J, Héloir MC, Chiltz A, Adrian M, Darblade B, Tornero P, et al. (2019) The grapevine (Vitis vinifera) LysM receptor kinases VvLYK1-1 and VvLYK1-2 mediate chitooligosaccharide-triggered immunity. Plant Biotechnol J 17: 812–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6: e25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia L, Girardin A, Wang T, Cottret L, Lefebvre B (2018) LysM receptor-like kinase and LysM receptor-like protein families: An update on phylogeny and functional characterization. Front Plant Sci 9: 1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill P, Chandrasekar B, Sanguankiattichai N, Kourelis J, Kaschani F, Thomas EL, Morimoto K, Kaiser M, Preston GM, Ichinose Y, et al. (2019) Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364: eaav0748. [DOI] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotenuto G, Chabaud M, Miyata K, Capozzi M, Takeda N, Kaku H, Shibuya N, Nakagawa T, Barker DG, Genre A (2017) The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol 214: 1440–1446 [DOI] [PubMed] [Google Scholar]
- Chen D, Ahsan N, Thelen JJ, Stacey G (2019) s-Acylation of plant immune receptors mediates immune signaling in plasma membrane nanodomains. bioRxiv 720482, doi:10.1101/720482 [Google Scholar]
- Chen D, Cao Y, Li H, Kim D, Ahsan N, Thelen J, Stacey G (2017) Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat Commun 8: 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Shan L, He P, de Vries S, Kemmerling B (2009) One for all: The receptor-associated kinase BAK1. Trends Plant Sci 14: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- Coleman AD, Raasch L, Maroschek J, Ranf S, Hückelhoven R (2019) The Arabidopsis leucine-rich repeat receptor kinase MIK2 is a crucial component of pattern-triggered immunity responses to Fusarium fungi. bioRxiv 720037, doi:10.1101/720037 [DOI] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BP (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol 53: 541–563 [DOI] [PubMed] [Google Scholar]
- Couto D, Niebergall R, Liang X, Bücherl CA, Sklenar J, Macho AP, Ntoukakis V, Derbyshire P, Altenbach D, Maclean D, et al. (2016) The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog 12: e1005811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, Zhang Z, Usami T, Lievens B, Subbarao KV, et al. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA 109: 5110–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vega D, Newton AC, Sadanandom A (2018) Post-translational modifications in priming the plant immune system: Ripe for exploitation? FEBS Lett 592: 1929–1936 [DOI] [PubMed] [Google Scholar]
- Dean J, Gamble R, Anderson JD (1989) The ethylene biosynthesis-inducing xylanase: Its induction in Trichoderma viride and certain plant pathogens. Phytopathology 79: 1071–1078 [Google Scholar]
- Desaki Y, Kouzai Y, Ninomiya Y, Iwase R, Shimizu Y, Seko K, Molinaro A, Minami E, Shibuya N, Kaku H, et al. (2018) OsCERK1 plays a crucial role in the lipopolysaccharide-induced immune response of rice. New Phytol 217: 1042–1049 [DOI] [PubMed] [Google Scholar]
- Desaki Y, Takahashi S, Sato K, Maeda K, Matsui S, Yoshimi I, Miura T, Jumonji JI, Takeda J, Yashima K, et al. (2019) PUB4, a CERK1-interacting ubiquitin ligase, positively regulates MAMP-triggered immunity in Arabidopsis. Plant Cell Physiol 60: 2573–2583 [DOI] [PubMed] [Google Scholar]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JD (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10: 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84: 451–459 [DOI] [PubMed] [Google Scholar]
- Domazakis E, Wouters D, Visser RGF, Kamoun S, Joosten MHAJ, Vleeshouwers VGAA (2018) The ELR-SOBIR1 complex functions as a two-component receptor-like kinase to mount defense against Phytophthora infestans. Mol Plant Microbe Interact 31: 795–802 [DOI] [PubMed] [Google Scholar]
- Du J, Verzaux E, Chaparro-Garcia A, Bijsterbosch G, Keizer LC, Zhou J, Liebrand TW, Xie C, Govers F, Robatzek S, et al. (2015) Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat Plants 1: 15034. [DOI] [PubMed] [Google Scholar]
- Duplan V, Rivas S (2014) E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ (2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou JM (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118 [DOI] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, et al. (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 28: 666–675 e665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frías M, González M, González C, Brito N (2019) A 25-residue peptide from Botrytis cinerea xylanase BcXyn11A elicits plant defenses. Front Plant Sci 10: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Inagaki H, Takai R, Hirai H, Che FS (2014) Two distinct EF-Tu epitopes induce immune responses in rice and Arabidopsis. Mol Plant Microbe Interact 27: 113–124 [DOI] [PubMed] [Google Scholar]
- Gibelin-Viala C, Amblard E, Puech-Pages V, Bonhomme M, Garcia M, Bascaules-Bedin A, Fliegmann J, Wen J, Mysore KS, le Signor C, et al. (2019) The Medicago truncatula LysM receptor-like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol 223: 1516–1529 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Giska F, Martin GB (2019) PP2C phosphatase Pic1 negatively regulates the phosphorylation status of Pti1b kinase, a regulator of flagellin-triggered immunity in tomato. Biochem J 476: 1621–1635 [DOI] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Gouhier-Darimont C, Stahl E, Glauser G, Reymond P (2019) The Arabidopsis lectin receptor kinase LecRK-I.8 is involved in insect egg perception. Front Plant Sci 10: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Felix G (2014) Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr Opin Plant Biol 21: 104–111 [DOI] [PubMed] [Google Scholar]
- Gust AA, Pruitt R, Nürnberger T (2017) Sensing danger: Key to activating plant immunity. Trends Plant Sci 22: 779–791 [DOI] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bücherl C, Kiefer C, Stahl M, Chinchilla D, Wang X, et al. (2014) The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol 24: 134–143 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones DA, Jones J (1994) Identification of two genes required in tomato for full Cf-9-dependent resistance to Cladosporium fulvum. Plant Cell 6: 361–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hander T, Fernandez-Fernandez AD, Kumpf RP, Willems P, Schatowitz H, Rombaut D, Staes A, Nolf J, Pottie R, Yao P, et al. (2019) Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides. Science 363: eaar7486. [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayafune M, Berisio R, Marchetti R, Silipo A, Kayama M, Desaki Y, Arima S, Squeglia F, Ruggiero A, Tokuyasu K, et al. (2014) Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc Natl Acad Sci USA 111: E404–E413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhou J, Shan L, Meng X (2018) Plant cell surface receptor-mediated signaling: A common theme amid diversity. J Cell Sci 131: jcs209353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegenauer V, Fürst U, Kaiser B, Smoker M, Zipfel C, Felix G, Stahl M, Albert M (2016) Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science 353: 478–481 [DOI] [PubMed] [Google Scholar]
- Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O’Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR, et al. (2016) Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants 2: 16128. [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Bos JI (2011) Effector proteins that modulate plant–insect interactions. Curr Opin Plant Biol 14: 422–428 [DOI] [PubMed] [Google Scholar]
- Hohmann U, Lau K, Hothorn M (2017) The structural basis of ligand perception and signal activation by receptor kinases. Annu Rev Plant Biol 68: 109–137 [DOI] [PubMed] [Google Scholar]
- Hohmann U, Nicolet J, Moretti A, Hothorn LA, Hothorn M (2018) The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling. Nat Plants 4: 345–351 [DOI] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Di Pietro A, Zhang W (2014) The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog 10: e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Ye M, Kuai P, Ye M, Erb M, Lou Y (2018) OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol 219: 1097–1111 [DOI] [PubMed] [Google Scholar]
- Hu Y, Hewezi T (2018) Nematode-secreted peptides and host factor mimicry. J Exp Bot 69: 2866–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U (2003) The Arabidopsis mutant Feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159 [DOI] [PubMed] [Google Scholar]
- Hurst CH, Wright KM, Turnbull D, Leslie K, Jones S, Hemsley PA (2019) Juxta-membrane s-acylation of plant receptor-like kinases is likely fortuitous and does not necessarily impact upon function. Sci Rep 9: 12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkampe J, Halter T, Huang S, Schulze S, Mazzotta S, Schmidt N, Manstretta R, Postel S, Wierzba M, Yang Y, et al. (2017) The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29: 2285–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irieda H, Inoue Y, Mori M, Yamada K, Oshikawa Y, Saitoh H, Uemura A, Terauchi R, Kitakura S, Kosaka A, et al. (2019) Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc Natl Acad Sci USA 116: 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle AK, Lipschis M, Albert M, Fallahzadeh-Mamaghani V, Fürst U, Mueller K, Felix G (2013) The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 25: 2330–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Han B, Zhang H, Mariappan KG, Bigeard J, Colcombet J, Hirt H (2019) MAP4K4 associates with BIK1 to regulate plant innate immunity. EMBO Rep 20: e47965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JD (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793 [DOI] [PubMed] [Google Scholar]
- Joosten MH, Cozijnsen TJ, De Wit PJ (1994) Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367: 384–386 [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y, Takai R, Furukawa T, Hirai H, Morimoto T, Katayama T, Murakami T, Che FS (2015) CD2-1, the C-terminal region of flagellin, modulates the induction of immune responses in rice. Mol Plant Microbe Interact 28: 648–658 [DOI] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312 [DOI] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, et al. (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98: 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Hunter K, Vaahtera L, Tran C, Vaattovaara A, Rokka A, Stolze SC, Harzen A, Meißner L, Wilkens M, Hamann T, et al. (2019) CRK2-mediated control of ROS production by phosphorylation of the RBOHD C-terminus in Arabidopsis. bioRxiv 618819, doi:10.1101/618819 [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera A, Dawid C, Gisch N, Schmid C, Raasch L, Gerster T, Schäffer M, Smakowska-Luzan E, Belkhadir Y, Vlot AC, et al. (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364: 178–181 [DOI] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B, et al. (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28: 365–369 [DOI] [PubMed] [Google Scholar]
- Lal NK, Nagalakshmi U, Hurlburt NK, Flores R, Bak A, Sone P, Ma X, Song G, Walley J, Shan L, et al. (2018) The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23: 485–497 e485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan NJ, Lydiate DJ, Parkin IA, Nelson MN, Epp DJ, Cowling WA, Rimmer SR, Borhan MH (2013) The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol 197: 595–605 [DOI] [PubMed] [Google Scholar]
- Larkan NJ, Lydiate DJ, Yu F, Rimmer SR, Borhan MH (2014) Co-localisation of the blackleg resistance genes Rlm2 and LepR3 on Brassica napus chromosome A10. BMC Plant Biol 14: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan NJ, Ma L, Borhan MH (2015) The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol J 13: 983–992 [DOI] [PubMed] [Google Scholar]
- Leppyanen IV, Shakhnazarova VY, Shtark OY, Vishnevskaya NA, Tikhonovich IA, Dolgikh EA (2017) Receptor-like kinase LYK9 in Pisum sativum L. is the CERK1-like receptor that controls both plant immunity and AM symbiosis development. Int J Mol Sci 19: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Ferreira MA, Huang M, Camargos LF, Yu X, Teixeira RM, Carpinetti PA, Mendes GC, Gouveia-Mageste BC, Liu C, et al. (2019) The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat Commun 10: 4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kim P, Yu L, Cai G, Chen S, Alfano JR, Zhou JM (2016) Activation-dependent destruction of a co-receptor by a Pseudomonas syringae effector dampens plant immunity. Cell Host Microbe 20: 504–514 [DOI] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, et al. (2016) Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5: e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ma M, Zhou Z, Wang J, Yang X, Rao S, Bi G, Li L, Zhang X, Chai J, et al. (2018) Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res 28: 529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhou JM (2018) Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol 69: 267–299 [DOI] [PubMed] [Google Scholar]
- Liebrand TW, van den Burg HA, Joosten MH (2014) Two for all: Receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci 19: 123–132 [DOI] [PubMed] [Google Scholar]
- Lin ZJ, Liebrand TW, Yadeta KA, Coaker G (2015) PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol 169: 2950–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li JF, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J, Feng D, Qi K, et al. (2012a) Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24: 3406–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang J, Han Z, Gong X, Zhang H, Chai J (2016) Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure 24: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, et al. (2012b) Chitin-induced dimerization activates a plant immune receptor. Science 336: 1160–1164 [DOI] [PubMed] [Google Scholar]
- Liu X, Grabherr HM, Willmann R, Kolb D, Brunner F, Bertsche U, Kühner D, Franz-Wachtel M, Amin B, Felix G, et al. (2014) Host-induced bacterial cell wall decomposition mediates pattern-triggered immunity in Arabidopsis. eLife 3: e01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Maierhofer T, Rybak K, Sklenar J, Breakspear A, Johnston MG, Fliegmann J, Huang S, Roelfsema MRG, Felix G, et al. (2019) Anion channel SLAH3 is a regulatory target of chitin receptor-associated kinase PBL27 in microbial stomatal closure. eLife 8: e44474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci F, Benedetti M, Pontiggia D, Citterico M, Caprari C, Mattei B, Cervone F, De Lorenzo G (2019) An Arabidopsis berberine bridge enzyme-like protein specifically oxidizes cellulose oligomers and plays a role in immunity. Plant J 98: 540–554 [DOI] [PubMed] [Google Scholar]
- Long Y, Wang Z, Sun Z, Fernando DW, McVetty PB, Li G (2011) Identification of two blackleg resistance genes and fine mapping of one of these two genes in a Brassica napus canola cultivar ‘Surpass 400’. Theor Appl Genet 122: 1223–1231 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. (2012) Plant cytokine or phytocytokine. Plant Signal Behav 7: 1513–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Song T, Zhu L, Ye W, Wang Y, Shao Y, Dong S, Zhang Z, Dou D, Zheng X, et al. (2015) A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27: 2057–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Schwessinger B, Ntoukakis V, Brutus A, Segonzac C, Roy S, Kadota Y, Oh MH, Sklenar J, Derbyshire P, et al. (2014) A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2015) Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol 23: 14–22 [DOI] [PubMed] [Google Scholar]
- Majeran W, Le Caer JP, Ponnala L, Meinnel T, Giglione C (2018) Targeted profiling of Arabidopsis thaliana subproteomes illuminates co- and posttranslationally N-terminal myristoylated proteins. Plant Cell 30: 543–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majhi BB, Sreeramulu S, Sessa G (2019) BRASSINOSTEROID-SIGNALING KINASE5 associates with immune receptors and is required for immune responses. Plant Physiol 180: 1166–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang H, Feng B, Hu Z, Boisson-Dernier A, Franck CM, Meng X, Huang Y, Zhou J, Xu G, Wang T, et al. (2017) Differential regulation of two-tiered plant immunity and sexual reproduction by ANXUR receptor-like kinases. Plant Cell 29: 3140–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, Leon-Ruiz M, Fürst U, El Ghalid M, Leonard G, López-Berges MS, Richards TA, Felix G, et al. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol 1: 16043. [DOI] [PubMed] [Google Scholar]
- McKenna JF, Rolfe DJ, Webb SED, Tolmie AF, Botchway SW, Martin-Fernandez ML, Hawes C, Runions J (2019) The cell wall regulates dynamics and size of plasma-membrane nanodomains in Arabidopsis. Proc Natl Acad Sci USA 116: 12857–12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena MK, Prajapati R, Krishna D, Divakaran K, Pandey Y, Reichelt M, Mathew MK, Boland W, Mithöfer A, Vadassery J (2019) The Ca2+ channel CNGC19 regulates Arabidopsis defense against Spodoptera herbivory. Plant Cell 31: 1539–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejias J, Truong NM, Abad P, Favery B, Quentin M (2019) Plant proteins and processes targeted by parasitic nematode effectors. Front Plant Sci 10: 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélida H, Sopeña-Torres S, Bacete L, Garrido-Arandia M, Jordá L, López G, Muñoz-Barrios A, Pacios LF, Molina A (2018) Non-branched β-1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J 93: 34–49 [DOI] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, He SY (2017) Stomatal defense a decade later. Plant Physiol 174: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendy B, Wang’ombe MW, Radakovic ZS, Holbein J, Ilyas M, Chopra D, Holton N, Zipfel C, Grundler FM, Siddique S (2017) Arabidopsis leucine-rich repeat receptor-like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog 13: e1006284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithoe SC, Menke FL (2018) Regulation of pattern recognition receptor signalling by phosphorylation and ubiquitination. Curr Opin Plant Biol 45(Pt A): 162–170 [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, et al. (2014) The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615 [DOI] [PubMed] [Google Scholar]
- Mott GA, Thakur S, Smakowska E, Wang PW, Belkhadir Y, Desveaux D, Guttman DS (2016) Genomic screens identify a new phytobacterial microbe-associated molecular pattern and the cognate Arabidopsis receptor-like kinase that mediates its immune elicitation. Genome Biol 17: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizam S, Qiang X, Wawra S, Nostadt R, Getzke F, Schwanke F, Dreyer I, Langen G, Zuccaro A (2019) Serendipita indica E5’NT modulates extracellular nucleotide levels in the plant apoplast and affects fungal colonization. EMBO Rep 20: e47430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosa B, Yates G, Verma V, Srivastava AK, Srivastava M, Campanaro A, De Vega D, Fernandes A, Zhang C, Lee J, et al. (2018) SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat Commun 9: 5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Youn J-H, Xu S-L, Kim J-G, Bi Y, Xu N, Mudgett MB, Kim S-K, Kim T-W, Wang Z-Y (2019) BSU1 family phosphatases mediate flagellin-FLS2 signaling through a specific phosphocode. bioRxiv 685610, doi:10.1101/685610 [Google Scholar]
- Park CJ, Ronald PC (2012) Cleavage and nuclear localization of the rice XA21 immune receptor. Nat Commun 3: 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, DeFalco TA, Derbyshire P, Avila J, Séré D, Sklenar J, Qi X, Stransfeld L, Schwessinger B, Kadota Y, et al. (2018) Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561: 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petutschnig EK, Stolze M, Lipka U, Kopischke M, Horlacher J, Valerius O, Rozhon W, Gust AA, Kemmerling B, et al. (2014) A novel Arabidopsis CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) mutant with enhanced pathogen-induced cell death and altered receptor processing. New Phytol 204: 955–967 [DOI] [PubMed] [Google Scholar]
- Pfeilmeier S, George J, Morel A, Roy S, Smoker M, Stransfeld L, Downie JA, Peeters N, Malone JG, Zipfel C (2019) Expression of the Arabidopsis thaliana immune receptor EFR in Medicago truncatula reduces infection by a root pathogenic bacterium, but not nitrogen-fixing rhizobial symbiosis. Plant Biotechnol J 17: 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma J, Liebrand TW, Bi G, Evrard A, Bye RR, Mbengue M, Kuhn H, Joosten MH, Robatzek S (2016) Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol 210: 627–642 [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Joe A, Zhang W, Feng W, Stewart V, Schwessinger B, Dinneny JR, Ronald PC (2017) A microbially derived tyrosine-sulfated peptide mimics a plant peptide hormone. New Phytol 215: 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJ, Luu DD, Chen H, et al. (2015) The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv 1: e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Gisch N, Schäffer M, Illig T, Westphal L, Knirel YA, Sánchez-Carballo PM, Zähringer U, Hückelhoven R, Lee J, et al. (2015) A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol 16: 426–433 [DOI] [PubMed] [Google Scholar]
- Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, Zhou JM (2018) Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol 177: 1679–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Moreno L, Song Y, Thomma BP (2017) Transfer and engineering of immune receptors to improve recognition capacities in crops. Curr Opin Plant Biol 38: 42–49 [DOI] [PubMed] [Google Scholar]
- Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald PC, Beutler B (2010) Plant and animal sensors of conserved microbial signatures. Science 330: 1061–1064 [DOI] [PubMed] [Google Scholar]
- Rooney HC, van’t Klooster JW, van der Hoorn RA, Joosten MH, Jones JD, de Wit PJ (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308: 1783–1786 [DOI] [PubMed] [Google Scholar]
- Rotman N, Rozier F, Boavida L, Dumas C, Berger F, Faure JE (2003) Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr Biol 13: 432–436 [DOI] [PubMed] [Google Scholar]
- Saijo Y. (2010) ER quality control of immune receptors and regulators in plants. Cell Microbiol 12: 716–724 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Loo EP, Yasuda S (2018) Pattern recognition receptors and signaling in plant-microbe interactions. Plant J 93: 592–613 [DOI] [PubMed] [Google Scholar]
- Saur IM, Kadota Y, Sklenar J, Holton NJ, Smakowska E, Belkhadir Y, Zipfel C, Rathjen JP (2016) NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc Natl Acad Sci USA 113: 3389–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlöffel MA, Käsbauer C, Gust AA (2019) Interplay of plant glycan hydrolases and LysM proteins in plant–bacteria interactions. Int J Med Microbiol 309: 252–257 [DOI] [PubMed] [Google Scholar]
- Segonzac C, Macho AP, Sanmartín M, Ntoukakis V, Sánchez-Serrano JJ, Zipfel C (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J 33: 2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Bourdais G, Pan H, Robatzek S, Tang D (2017) Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc Natl Acad Sci USA 114: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Liu J, Li JF (2019) Type-II metacaspases mediate the processing of plant elicitor peptides in Arabidopsis. Mol Plant 12: 1524–1533 [DOI] [PubMed] [Google Scholar]
- Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D (2013) BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T, Motoyama N, Ikeda A, Wada M, Kamiya K, Hayafune M, Kaku H, Shibuya N (2012) Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol 53: 1696–1706 [DOI] [PubMed] [Google Scholar]
- Shinya T, Nakagawa T, Kaku H, Shibuya N (2015) Chitin-mediated plant–fungal interactions: Catching, hiding and handshaking. Curr Opin Plant Biol 26: 64–71 [DOI] [PubMed] [Google Scholar]
- Smakowska-Luzan E, Mott GA, Parys K, Stegmann M, Howton TC, Layeghifard M, Neuhold J, Lehner A, Kong J, Grünwald K, et al. (2018) An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rövenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Steinbrenner AD, Muñoz-Amatriaín M, Venegas JMA, Lo S, Shi D, Holton N, Zipfel C, Abagyan R, Huffaker A, Close TJ, et al. (2019) A receptor for herbivore-associated molecular patterns mediates plant immunity. bioRxiv 10.1101/679803 [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2–BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Tang J, Han Z, Sun Y, Zhang H, Gong X, Chai J (2015) Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res 25: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH (2011) Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thynne E, Saur IML, Simbaqueba J, Ogilvie HA, Gonzalez-Cendales Y, Mead O, Taranto A, Catanzariti AM, McDonald MC, Schwessinger B, et al. (2017) Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol Plant Pathol 18: 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L, et al. (2019) A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572: 131–135 [DOI] [PubMed] [Google Scholar]
- Wan WL, Fröhlich K, Pruitt RN, Nürnberger T, Zhang L (2019a) Plant cell surface immune receptor complex signaling. Curr Opin Plant Biol 50: 18–28 [DOI] [PubMed] [Google Scholar]
- Wan WL, Zhang L, Pruitt R, Zaidem M, Brugman R, Ma X, Krol E, Perraki A, Kilian J, Grossmann G, et al. (2019b) Comparing Arabidopsis receptor kinase and receptor protein-mediated immune signaling reveals BIK1-dependent differences. New Phytol 221: 2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou M, Zhang X, Yao J, Zhang Y, Mou Z (2017) A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. eLife 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chevalier D, Larue C, Ki Cho S, Walker JC (2007) The protein phosphatases and protein kinases of Arabidopsis thaliana. Arabidopsis Book 5: e0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L, et al. (2018a) A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell 69: 493–504 [DOI] [PubMed] [Google Scholar]
- Wang J, Li H, Han Z, Zhang H, Wang T, Lin G, Chang J, Yang W, Chai J (2015) Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525: 265–268 [DOI] [PubMed] [Google Scholar]
- Wang L, Albert M, Einig E, Fürst U, Krust D, Felix G (2016) The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat Plants 2: 16185. [DOI] [PubMed] [Google Scholar]
- Wang L, Einig E, Almeida-Trapp M, Albert M, Fliegmann J, Mithöfer A, Kalbacher H, Felix G (2018b) The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat Plants 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Z, Liu D, Xu J, Wei X, Yan L, Yang C, Lou Z, Shui W (2014) Assessment of BAK1 activity in different plant receptor-like kinase complexes by quantitative profiling of phosphorylation patterns. J Proteomics 108: 484–493 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu Y, Sun Y, Wang H, Qi J, Wan B, Ye W, Lin Y, Shao Y, Dong S, et al. (2018c) Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat Commun 9: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono JJ, Cullimore JV, et al. (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA 108: 19824–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers J, Dong X (2017) Post-translational regulation of plant immunity. Curr Opin Plant Biol 38: 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, van der Burgh AM, Bi G, Zhang L, Alfano JR, Martin GB, Joosten MHAJ (2018) The bacterial effector AvrPto targets the regulatory coreceptor SOBIR1 and suppresses defense signaling mediated by the receptor-like protein Cf-4. Mol Plant Microbe Interact 31: 75–85 [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J (2019) Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572: 270–274 [DOI] [PubMed] [Google Scholar]
- Xu S, Liao CJ, Jaiswal N, Lee S, Yun DJ, Lee SY, Garvey M, Kaplan I, Mengiste T (2018) Tomato PEPR1 ORTHOLOG RECEPTOR-LIKE KINASE1 regulates responses to systemin, necrotrophic fungi, and insect herbivory. Plant Cell 30: 2214–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue DX, Li CL, Xie ZP, Staehelin C (2019) LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. J Exp Bot 70: 5507–5516 [DOI] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T, Nakagami H, Mine A, Ishikawa K, Fujiwara M, Narusaka M, Narusaka Y, Ichimura K, et al. (2016) The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J 35: 2468–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, Yoshimura S, Hayashi N, Uchihashi K, Ishihama N, Kishi-Kaboshi M, Takahashi A, Tsuge S, et al. (2013) A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13: 347–357 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA (2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhao Y, Shi H, Li J, Wang Y, Tang D (2018) BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol 176: 2991–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Okada K, Saijo Y (2017) A look at plant immunity through the window of the multitasking coreceptor BAK1. Curr Opin Plant Biol 38: 10–18 [DOI] [PubMed] [Google Scholar]
- Yeh YH, Panzeri D, Kadota Y, Huang YC, Huang PY, Tao CN, Roux M, Chien HC, Chin TC, Chu PW, et al. (2016) The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28: 1701–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip Delormel T, Boudsocq M (2019) Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol 224: 585–604 [DOI] [PubMed] [Google Scholar]
- Yu X, Xu G, Li B, de Souza Vespoli L, Liu H, Moeder W, Chen S, de Oliveira MVV, Ariadina de Souza S, Shao W, et al. (2019) The receptor kinases BAK1/SERK4 regulate Ca2+ channel-mediated cellular homeostasis for cell death containment. Curr Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhang L, Kars I, Essenstam B, Liebrand TW, Wagemakers L, Elberse J, Tagkalaki P, Tjoitang D, van den Ackerveken G, van Kan JAL (2014) Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol 164: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chiang YH, Toruno TY, Lee D, Ma M, Liang X, Lal NK, Lemos M, Lu YJ, Ma S, et al. (2018) The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe 24: 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fraiture M, Kolb D, Löffelhardt B, Desaki Y, Boutrot FF, Tör M, Zipfel C, Gust AA, Brunner F (2013) Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25: 4227–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zayed O, Yu Z, Jiang W, Zhu P, Hsu CC, Zhang L, Tao WA, Lozano-Durán R, Zhu JK (2018) Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc Natl Acad Sci USA 115: 13123–13128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, McLellan H, Fraiture M, Liu X, Boevink PC, Gilroy EM, Chen Y, Kandel K, Sessa G, Birch PR, et al. (2014) Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathog 10: e1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wagener N, McLellan H, Boevink PC, Hua C, Birch PRJ, Brunner F (2018) Phytophthora infestans RXLR effector SFI5 requires association with calmodulin for PTI/MTI suppressing activity. New Phytol 219: 1433–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zeng L (2017) Conventional and unconventional ubiquitination in plant immunity. Mol Plant Pathol 18: 1313–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang P, Claus LAN, Savatin DV, Xu G, Wu S, Meng X, Russinova E, He P, Shan L (2019) Proteolytic processing of SERK3/BAK1 regulates plant immunity, development, and cell death. Plant Physiol 180: 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Tian Y, Cong P, Zhu Y (2018) Functional characterization of an apple (Malus x domestica) LysM domain receptor encoding gene for its role in defense response. Plant Sci 269: 56–65 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zorzatto C, Machado JP, Lopes KV, Nascimento KJ, Pereira WA, Brustolini OJ, Reis PA, Calil IP, Deguchi M, Sachetto-Martins G, et al. (2015) NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 520: 679–682 [DOI] [PMC free article] [PubMed] [Google Scholar]