Abstract
Germ-cell transcription factors control gene networks that regulate oocyte differentiation and primordial follicle formation during early, postnatal mouse oogenesis. Taking advantage of gene-edited mice lacking transcription factors expressed in female germ cells, we analyzed global gene expression profiles in perinatal ovaries from wildtype, FiglaNull, Lhx8Null and Sohlh1Null mice. Figla deficiency dysregulates expression of meiosis-related genes (e.g. Sycp3, Rad51, Ybx2) and a variety of genes (e.g. Nobox, Lhx8, Taf4b, Sohlh1, Sohlh2, Gdf9) associated with oocyte growth and differentiation. The absence of FIGLA significantly impedes meiotic progression, causes DNA damage and results in oocyte apoptosis. Moreover, we find that FIGLA and other transcriptional regulator proteins (e.g. NOBOX, LHX8, SOHLH1, SOHLH2) are co-expressed in the same subset of germ cells in perinatal ovaries and Figla ablation dramatically disrupts KIT, NOBOX, LHX8, SOHLH1 and SOHLH2 abundance. In addition, not only do FIGLA, LHX8 and SOHLH1 cross-regulate each other, they also cooperate by direct interaction with each during early oocyte development and share downstream gene targets. Thus, our findings substantiate a major role for FIGLA, LHX8 and SOHLH1 as multifunctional regulators of networks necessary for oocyte maintenance and differentiation during early folliculogenesis.
INTRODUCTION
In mice, primordial germ cells migrate to the gonad and undergo rapid mitosis with incomplete cytokinesis to form germline syncytia during embryonic day 10.5 (E10.5) to E12.5. Female germ cells enter meiosis at E13.5 but arrest at the diplotene stage of the first meiotic division. Shortly after birth, germ cell cysts break down and pre-granulosa cells invade oocyte syncytia to form primordial follicles (1–3). Postnatally, there are no germline stem cells and the reservoir of primordial follicles provides a steady source for folliculogenesis with subsequent ovulation of MII eggs essential for fertility during the reproductive life of females (4,5). Abnormalities in early folliculogenesis cause premature ovarian failure (POF) and sterility (6). Thus, investigations of fetal follicle formation provide insight into molecular mechanisms that support folliculogenesis and ensure female fertility in mammals, including humans.
FIGLA (Factor in the germline alpha) is a basic helix-loop-helix (bHLH) transcription factor that was initially defined as a regulator of zona pellucida gene expression (7). Female mice lacking FIGLA have normal embryonic gonad development, but are unable to form primordial follicles after birth which results in massive loss of oocytes and sterility (8). Although FIGLA is reported to up-regulate female-specific and down-regulate male-specific genes during early oogenesis (9,10), the molecular mechanisms underlying the control of FIGLA related networks remains incompletely understood. Previous studies have defined a group of transcriptional regulators that play crucial roles in oocyte differentiation and early folliculogenesis. These genes include spermatogenesis and oogenesis bHLH transcription factor 1 (Sohlh1) (11), Sohlh2 (12,13), LIM homeobox protein 8 (Lhx8) (14), NOBOX oogenesis homeobox (Nobox) (15), TATA-box binding protein associated factor 4b (Taf4b) (16–18) as well as Figla (8). Sohlh1, Sohlh2 and Taf4b are expressed in both male and female germlines and play important roles in male and female germ cell differentiation (19–24). Nobox, Lhx8 and Figla are primarily expressed in female germ cells and are essential for female germline development, without affecting male germline development. Sohlh1, Sohlh2, Lhx8 and Nobox regulate oocyte differentiation without affecting meiosis (25), while Taf4b regulates oocyte-specific genes essential for meiosis (17) and Figla deficiency appears to impair oocyte meiotic progression to the diplotene stage (8).
Ovaries lacking these transcription factors share characteristics in that gonads appear normal with similar histology and germ cell number in embryonic and newborn wildtype mice. However, within the first few days after birth, Figla- and Taf4b-deficient ovaries fail to establish primordial follicles and have massive germ cell loss. Deficiencies of other factors cause obvious defects in primordial follicle formation and development, with significant oocyte loss in the first few postnatal weeks. These females have premature ovarian failure and are sterile as adults. A recent study documents that SOHLH1 and SOHLH2 coordinate oocyte differentiation without affecting meiosis and regulate co-expression of oocyte-specific transcriptional regulators. Figla, Nobox, Kit and Lhx8 transcripts overlap with Sohlh1 expression in perinatal ovaries (25). However, because of the absence of a commercially available antibody for FIGLA, the regulatory network of FIGLA and the functional relationships among these transcriptional regulators remain unclear.
In this study, we show that FIGLA and other oocyte transcriptional regulators, including LHX8 and SOHLH1, are co-expressed in the same subset of germ cells in perinatal ovaries. RNA-seq analysis documents that Figla deficiency dysregulates expression of meiosis-related genes (e.g. Sycp3, Rad51, Cpeb1, Ybx2) and numerous oogenesis-associated genes (e.g. Nobox, Lhx8, Sohlh1, Sohlh2, Gdf9). Immunostaining analyses show that Figla deficiency impedes meiotic progression and disrupts KIT, NOBOX, LHX8, SOHLH1 and SOHLH2 expression. Additionally, not only do FIGLA, LHX8 and SOHLH1 cross-regulate each other as Figla expression is significantly diminished in Lhx8- and Sohlh1-deficient ovaries, but they also appear to function in a cooperative fashion as evidenced by the shared genetic downstream targets. Co-IP analyses in heterologous cells suggest that FIGLA, LHX8 and SOHLH1 physically interact with each other and form a nuclear complex in oocytes. Taken together, our findings document that FIGLA, LHX8 and SOHLH1 participate in a multifunctional network to regulate germ cell maintenance and differentiation in early mouse oogenesis.
MATERIALS AND METHODS
Animals
FiglaNull (8) and Lhx8Null (26) mouse lines were previously generated and primers for genotyping these mice are listed in Supplementary Table S1. All animal studies were performed in accordance with guidelines of the Animal Care and Use Committee of the National Institutes of Health under a Division of Intramural Research, NIDDK approved animal study protocol (protocol numbers K018LCDB18 and K044LCDB19).
Generation of FiglaFLAG mice by CRISPR/Cas9
A guide RNA (5′-TAGGAGTTACTTCACTCATT-3′) was designed to target the DNA sequence upstream of the Figla stop codon. Double-stranded synthetic DNA was cloned into pDR274 (Addgene, #42250) expressing a single guide RNA (sgRNA). After linearization by digestion with DraI, the DNA fragment was purified with a PCR Clean-up Kit (Thermo Fisher Scientific) and in vitro transcribed using AmpliScribe T7-Flash Transcription Kit (Lucigen). Cas9 mRNA (Addgene, #42251) was generated by linearization with PmeI, purified with the PCR clean-up kit, and in vitro transcribed with the mMESSAGE mMACHINE T7 Kit (Thermo Fisher Scientific). After transcription, the sgRNA and Cas9 mRNAs were purified with a MEGAclear Transcription Clean-Up Kit (Thermo Fisher Scientific). Single-strand oligo DNA donor containing a linker and encoding a FLAG tag was synthesized by Integrated DNA Technologies. Hormonally stimulated B6D2F1 (C57LB/6 × DBA2) female mice were mated to B6D2F1 male mice and zygotes were collected from oviducts at embryonic day 0.5 (E0.5). sgRNA (50 ng/μl), Cas9 mRNA (100 ng/μl) and DNA oligo donor (200 ng/μl) were mixed and injected into zygotes. Injected zygotes were cultured in KSOM (37°C, 5% CO2) to the two-cell embryo stage and transferred into the oviducts of pseudo-pregnant ICR female mice at E0.5. Genotyping primers (Supplementary Table S1) were designed to amplify a 181-bp wildtype band and a 232-bp band containing the FLAG tag sequence.
Histology and immunofluorescence
Mouse ovaries were fixed in 4% paraformaldehyde (PFA) overnight at 4°C, embedded in paraffin, sectioned at 5 μm and mounted on slides prior to staining with periodic acid-Schiff (PAS) and hematoxylin. Apoptosis was measured by terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate (TUNEL) assay using an In situ Apoptosis Detection Kit (Millipore) according to the manufacturer's instructions. After de-waxing, rehydration, and antigen retrieval with 0.01% sodium citrate buffer (pH 6.0) (Sigma Aldrich), tissue sections were blocked with blocking buffer containing 0.05% Tween-20 at RT for 1 h and incubated with primary antibodies (Supplementary Table S2) overnight at 4°C. Specific secondary antibodies (Supplementary Table S2) were used to detect the antigen and DNA was stained with Hoechst 33342.
For whole-mount staining, freshly isolated ovaries were fixed in 4% PFA overnight at 4°C, permeabilized in blocking buffer overnight at 4°C and then incubated with primary antibodies (Supplementary Table S2) for 1–3 days at 4°C. Secondary antibodies conjugated to Alexa Fluor were incubated for additional 1–3 days at 4°C. After washing in PBS, ovaries were mounted with PBS on the slides. Bright field images were obtained with an inverted microscope (AxioPlan 2; Carl Zeiss). Fluorescent images were captured by confocal microscopy (LSM 780; Carl Zeiss).
Immunoblot
Total protein was extracted in 1× LDS sample buffer with 1x NuPAGE sample reducing agent (Thermo Fisher Scientific). Proteins were separated on 4–12% Bis–Tris gels and electrophoretically transferred to PVDF membranes. The membranes were blocked with 5% non-fat milk in Tris-buffered saline containing 0.05% Tween-20 (TBST) at RT for 1 h and probed with primary antibodies (Supplementary Table S2) overnight at 4°C. The membranes were washed three times with TBST and incubated at RT for 1 h with secondary antibodies, washed with TBST and developed using SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific). Signals were detected with PXi Touch (Syngene) or Hyperfilm ECL (GE Healthcare) according to the manufacturer's instructions. Uncropped blots are presented in Supplementary Figure S1.
RNA isolation and quantitative real-time RT-PCR
Total RNA was isolated from mouse ovaries using a RNeasy Mini Kit (Qiagen) and cDNA was synthesized with SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). Quantitative RT-PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) and the QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). The primers used in the experiments are listed in Supplementary Table S3. The relative abundance of each transcript was calculated by 2−ΔΔCt normalized to endogenous β-actin expression (27).
Plasmid transfection
All plasmids in this study were purchased from GenScript except 3xMYC-SOHLH1 plasmid which was kindly provided by Dr Aleksandar Rajkovic. HEK-293T cells were cultured in DMEM (Gibco) supplement with 10% fetal bovine serum at 37°C with 5% CO2. Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific) was used for plasmid transfection based on the manufacturer's instructions. Vectors (∼14 μg total) were co-transfected into cells for each 10-cm wide dish. After 48 hr, the transfected cells were harvested for immunoprecipitation.
Immunoprecipitation and co-IP
Harvested cells were lysed in immunoprecipitation (IP) lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA) supplemented with 1× protease inhibitor cocktail (Thermo Fisher Scientific) on ice for 30 min. The cell lysates were clarified by centrifugation at 12 000 rpm for 30 min at 4°C. As appropriate, the supernatants were incubated overnight at 4°C with either anti-FLAG (Sigma Aldrich), anti-HA or anti-c-MYC (Thermo Fisher Scientific) magnetic beads. The protein-bead complexes were precipitated, washed three times with lysis buffer, resuspended in 1× LDS sample buffer and incubated at 70°C for 10 min. The eluates were then separated on 4–12% Bis–Tris gel as described above. Uncropped blots are presented in Supplementary Figure S1.
RNA-seq library preparation
Total RNA was isolated using a RNeasy Mini Kit and mRNA was purified with a Dynabeads mRNA Purification Kit (Thermo Fisher Scientific). First strand cDNA was synthesized with SuperScript III Reverse Transcriptase and the second strand was synthesized in 100 μl containing: 20 μl of the first strand cDNA synthesis mix, 10 μl of 10× second strand buffer (500 mM Tris–HCl, pH 7.5; 50 mM MgCl2; 10 mM DTT), 3 μl of dNTP mix (10 mM), 1 μl of RNase H (2 U μl−1), 5 μl of DNA Pol I (10 U μl−1) and 61 μl H2O. The mixture was incubated for 2 h at 16°C. The libraries were constructed with a Nextera DNA Sample Preparation Kit (Illumina) per the manufacturer's protocol. In summary, double-strand cDNA was fragmented, subjected to adapter ligation and amplified. The Lhx8 RNA-seq libraries were prepared using the QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen) according to the manufacturer's instructions. The final PCR amplified libraries were sequenced at the NIDDK Genomic Core Facility.
Common library analyses
All libraries were analyzed by Agilent 2100 Bioanalyzer system with the High Sensitivity DNA Kit (Agilent) for quality and PicoGreen (Thermo Fisher Scientific) was used for DNA quantification before sequencing on the Illumina HiSeq 2500 system. For RNA-seq, the sequencing length was single-end 50-bp reads for the Figla library and 100-bp reads for the Lhx8 library.
RNA-seq analysis
Raw sequence reads were trimmed with cutadapt v1.16 to remove any adapters while performing light quality trimming using parameters -q 20 -a AGATCGGAAGAGC -minimum-length 25. Trimmed reads were mapped to the mm10 reference genome using HISAT v2.1.0 with default parameters (28) and multimapping reads were filtered using SAMtools v1.7 (29). Uniquely aligned reads were then mapped to gene features using subread featureCounts v1.6.0 with default parameters (30). Differential expression between groups of samples was tested in R v3.4.1 with DESeq2 v1.18.1 (31). The raw RNA-seq data for SOHLH1 were obtained from NCBI’s Sequence Read Archive (accession no. PRJNA293873).
Total RNA was isolated from pools of six ovaries for each library. For Figla RNA-seq, four replicate libraries per genotype were prepared at E18.5 and three replicate libraries per genotype were prepared at P0. For Lhx8 RNA-seq, three replicate libraries per genotype were prepared at P0. The detailed information about the library quality in terms of mapped reads, reads mapping within exons, number of genes detected in each library and sample clustering by principal component analysis are provided in Supplementary Table S4.
Statistical analysis
Unless otherwise noted, data are presented as the mean ± S.D. The two-tailed Student's t-test was used to calculate P values. P < 0.05 was considered statistically significant.
RESULTS
Figla deficiency alters expression of genes involved in meiosis and oogenesis
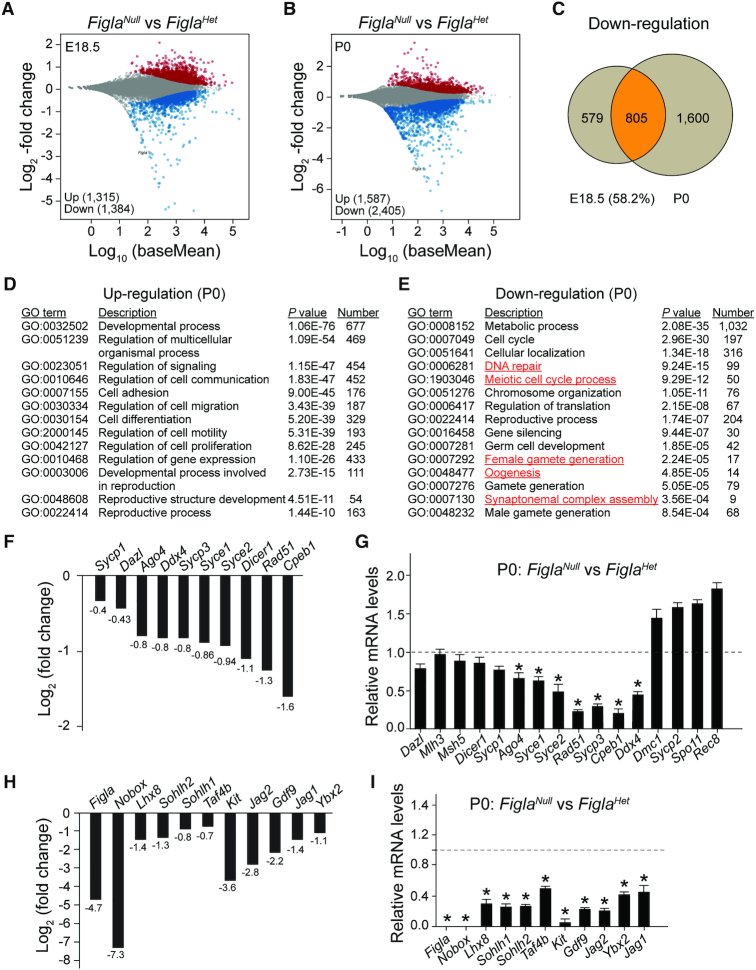
Our previous study documented that Figla-deficient ovaries at E18 and postnatal day 0 (P0) did not show gross differences in morphology, histology or the total number of germ cells compared to wildtype controls. However, the null phenotype began to evolve in the P1 ovary which developed massive germ cell loss (8). To investigate the molecular consequences of Figla deficiency in early oogenesis, we examined gene expression profiles of FiglaHet and FiglaNull ovaries and compared the transcriptome at E18.5 and P0 by high-throughput RNA sequencing (RNA-seq). We rationalize the use of whole ovaries because FIGLA is only expressed in germ cells, but there may be indirect effects on gene expression via the somatic compartment. We note that the total number of oocytes at each time point in FiglaHet and FiglaNull ovaries was similar and that we observed a substantial number of differentially expressed genes at the two time points in FiglaNull ovaries. Specifically, RNA-seq results documented that 1315 genes were significantly up-regulated, and 1384 genes were down-regulated at E18.5 whereas 1587 genes were up-regulated, and 2405 genes were down-regulated at P0 in FiglaNull ovaries (Figure 1A and B). Among down-regulated genes at the two time points in FiglaNull ovaries, most of the genes down-regulated at E18.5 (805/1384, 58.2%) were also down-regulated at P0 by which time the number of down-regulated genes had increased from 1384 (E18.5) to 2405 (Figure 1C). Gene Ontology (GO) enrichment analysis showed that up-regulated transcripts at P0 were primarily involved in developmental processes, cell communication, cell adhesion, cell migration, cell differentiation, cell motility, cell proliferation, reproductive structure development and reproduction processes (Figure 1D). Down-regulated transcripts were significantly enriched for those that function in metabolic processes, cellular localization, DNA repair, meiotic cell cycle processes, reproductive processes, gene silencing, germ cell development, female gamete generation, oogenesis, synaptonemal complex assembly and male gamete generation (Figure 1E). RNA-seq results indicated that some meiosis-related genes (e.g. Syce1, Syce2, Sycp3, Cpeb1, Rad51) were significantly reduced in P0 FiglaNull ovaries (Figure 1F), suggesting a defect in meiosis consistent with our previous findings (8). Additional germ cell-specific genes, well-known to be required for oocyte differentiation and early oogenesis also were down-regulated including Nobox (log2 fold change, −7.3), Lhx8 (log2 fold change, −1.4), Taf4b (log2 fold change, −0.7), Sohlh1 (log2 fold change, −0.8), Sohlh2 (log2 fold change, −1.3) and Gdf9 (log2 fold change, −2.2) (Figure 1H). The expression pattern of the genes involved in meiosis and oogenesis was validated by real-time quantitative PCR using RNA isolated from P0 ovaries. The qRT-PCR results confirmed the significant down-regulation of genes associated with meiosis (e.g. Syce1, Syce2, Rad51, Sycp3, Cpeb1, Ybx2) and genes essential for oogenesis (e.g. Nobox, Lhx8, Sohlh1, Sohlh2, Taf4b, Kit, Jag2, Jag1, Gdf9) in FiglaNull ovaries (Figure 1G and I). Taken together, these results suggest that Figla deficiency globally influences the expression of genes that control multiple processes in oogenesis including meiosis, differentiation and growth.
Figure 1.
Figla deficiency alters expression patterns of meiotic and oogenic genes. (A, B) MA plot (log ratio RNA abundance versus abundance) of RNA-seq data from FiglaHet and FiglaNull ovaries at E18.5 (A) and P0 (B), respectively, using adjusted P < 0.1 as the cut off. (C) Venn diagram depicting the overlap of down-regulated genes examined at two developmental time points, E18.5 and P0. (D, E) The enriched Gene Ontology terms of biological process categories of up- (D) and down-regulated (E) transcripts in P0 FiglaNull ovaries. (F) RNA-seq results of selected transcripts (log2-fold change) related to meiosis in P0 FiglaNull ovaries. (G) Quantitative RT-PCR validation of meiosis-related genes in FiglaNull ovaries at P0. The control genes relative to β-actin were set to 1. Data are presented as mean ± S.D. for n = 3 biologically independent experiments. *P <0.05 by two-tailed Student's t test. (H) Same as (F), but of oogenesis-related transcripts. (I) Same as (G), but of genes related to oogenesis.
Figla deficiency impedes meiotic progression, causes DNA damage and oocyte apoptosis
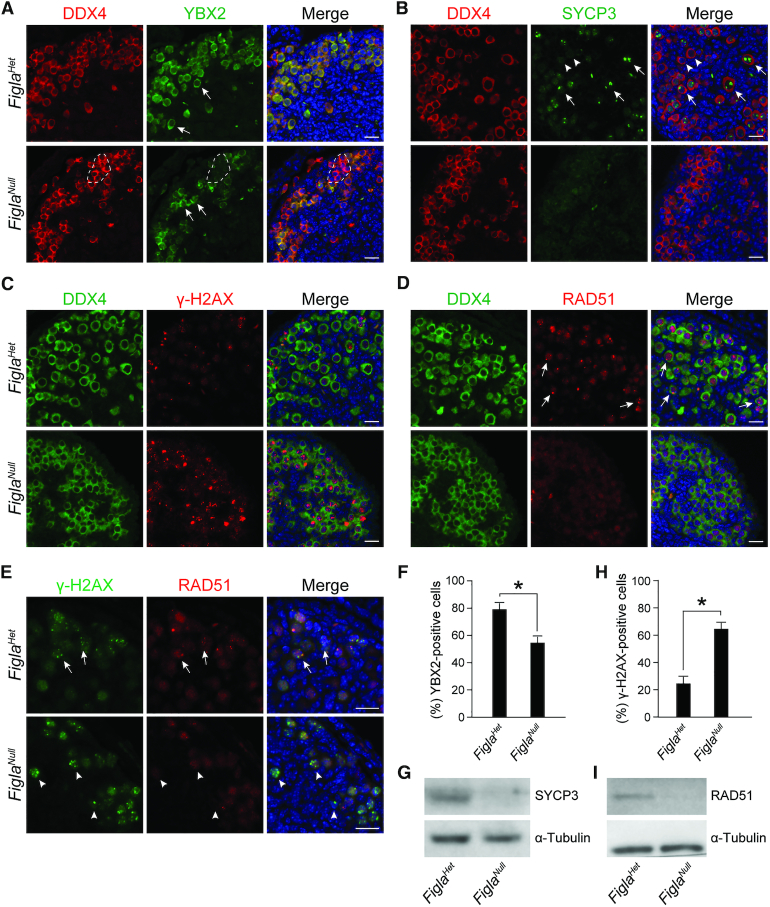
Based on RNA-seq results, genes involved in meiosis were defective in Figla-deficient oocytes. To assess meiotic progression in FiglaNull ovaries, we used immunofluorescence staining with antibodies to DEAD-box helicase 4 (DDX4, a germ cell-specific marker) and Y box protein 2 (YBX2, formerly MSY2) in P0 FiglaHet and FiglaNull ovaries. YBX2 expression is first detected in the cytoplasm of oocytes at diplotene stage of meiosis and persists in the arrested dictyate stage (32), making it a marker for the diplotene stage and later. In FiglaHet ovaries, most of the oocytes were positive for YBX2, suggesting normal entry into the diplotene stage of meiotic prophase I. However, there was an apparent decrease in the number of oocytes at the diplotene stage in FiglaNull ovaries (FiglaHet 78.9% ± 5.1% versus FiglaNull 56.8% ± 4.6%) (Figure 2A, F and Supplementary Figure S2A), indicating that Figla deficiency impairs meiotic progression to the diplotene stage in oocytes. The normal meiotic arrest at the dictyate stage is crucial for primordial follicle formation and can be identified by the presence of two to four visible nucleoli signals after staining with an antibody to synaptonemal complex protein 3 (SYCP3) (33). Therefore, immunostaining was performed with antibodies to DDX4 and SYCP3 to define whether oocytes reached to the dictyate stage. We found that a subset of oocytes had developed to the dictyate stage in P0 FiglaHet ovaries as indicated by the presence of visible nucleoli signals and virtually no oocytes reached this stage in FiglaNull ovaries (Figure 2B and Supplementary Figure S2B). Moreover, SYCP3 protein abundance was significantly reduced in P0 FiglaNull ovaries on immunoblots (Figure 2G). These results suggest that the few oocytes in FiglaNull ovaries that even reach the diplotene stage cannot develop further to the dictyate.
Figure 2.
Figla deficiency impairs meiotic progression and causes DNA damage. (A) Immunofluorescence staining with antibodies to DDX4 (red) and YBX2 (green) on ovarian sections from P0 FiglaHet and FiglaNull mice. DNA was stained with Hoechst 33342. Arrows indicate YBX2-positive oocytes. The dashed lines indicate DDX4-positive, but YBX2-negative oocytes. Scale bar, 20 μm. (B) Same as (A), but with antibodies to DDX4 (red) and SYCP3 (green). Some oocytes showed visible nucleoli (arrows) after staining with SYCP3 antibodies indicating that these oocytes had reached the dictyate stage. Oocytes that were negative for nucleoli (arrowheads) had not reached the dictyate. (C) Same as (A), but with antibodies to DDX4 (green) and γ-H2AX (red). (D) Same as (A), but with antibodies to DDX4 (green) and RAD51 (red). Arrows indicate oocytes expressing RAD51 foci in the nucleus. (E) Same as (A), but with antibodies to γ-H2AX (green) and RAD51 (red). Arrows indicate γ-H2AX-positive and RAD51-positive oocytes. Arrowheads indicate γ-H2AX-positive, but RAD51-negative oocytes. (F, H) Statistical analysis of the percentage of YBX2-positive cells (F) and γ-H2AX-positive cells (H) per section in FiglaHet and FiglaNull ovaries. Mean ± s.d, n = 3 biologically independent ovaries from three different animals. *P <0.05 by two-tailed Student's t test. (G, I) Immunoblot assay of SYCP3 (G) and RAD51 (I) in P0 FiglaHet and FiglaNull ovaries. α-Tubulin was used as an internal control. Representative of n = 3 (A–E, G and I) independent biological replicates with similar results per condition.
Because one of the categories highlighted in the gene ontology analysis was DNA repair, we examined DNA damage and repair in P0 FiglaHet and FiglaNull ovaries. Phosphorylated histone H2AX (γ-H2AX) is generated in response to DNA damage and marks DNA double stranded breaks (DSBs). Immunostaining documented distinct γ-H2AX signals in FiglaNull oocytes but few oocytes in FiglaHet ovaries had positive staining (Figure 2C and Supplementary Figure S2C). Statistical analysis confirmed the increase of the γ-H2AX-positive cells (FiglaHet 24.8 ± 3.8% versus FiglaNull 64.1 ± 4.1%) (Figure 2H) which suggested DNA damage in Figla-deficient oocytes. RAD51 recombinase, a RecA homolog, plays a pivotal role in homologous recombination of DNA during double strand break repair. Immunofluorescence results indicated that only a small portion of FiglaHet oocytes expressed distinct RAD51 foci in their nuclei (Figure 2D and Supplementary Figure S2D), suggesting a response to the DNA damage and subsequent DNA damage repair. In contrast, no obvious RAD51 foci signals were detected in FiglaNull oocytes (Figure 2D and Supplementary Figure S2D), indicating a failure to initiate DNA damage repair. Co-immunostaining also demonstrated that γ-H2AX-positive oocytes were RAD51 positive in FiglaHet ovaries. However, γ-H2AX-positive oocytes were RAD51 negative in FiglaNull ovaries (Figure 2E). These data suggest that Figla-deficient oocytes have impaired capacity to initiate DNA damage repair. Immunoblot analysis further documented a significant reduction of RAD51 protein in FiglaNull ovaries (Figure 2I). In addition, a TUNEL (TdT-mediated dUTP Nick-End Labeling) assay showed that P0 and P1 FiglaNull ovaries had significantly more TUNEL-positive cells than the FiglaHet controls (FiglaHet 16.1 ± 3.3 versus FiglaNull 63.7 ± 8.2 at P0 and FiglaHet 8.7 ± 2.5 versus FiglaNull 36.7 ± 5.1 at P1) (Supplementary Figure S3), implying excessive cell apoptosis in the absence of efficient DNA damage repair. These observations suggest that Figla deficiency in oocytes causes DNA damage, impairs DNA damage repair and leads to oocyte apoptosis. Taken together, these data indicate that FIGLA is required to ensure proper meiotic progression of prophase I and is indispensable for oocyte survival during early folliculogenesis.
Co-expression of transcription regulators in perinatal ovaries
A previous study reported that Figla, Nobox, Lhx8 and Sohlh1 gene expression increase in parallel, and NOBOX, LHX8 and SOHLH1 protein are present in the same subset of germ cells in embryonic ovaries (25). To define whether FIGLA co-localizes with them and how FIGLA affects these transcription regulators, we analyzed co-expression of FIGLA, NOBOX, LHX8, SOHLH1 and SOHLH2 proteins using immunofluorescent staining in perinatal ovaries. To localize the protein in the absence of a commercially available antibody to FIGLA, we established a FiglaFLAG knock in mouse line using CRISPR/Cas9 with a FLAG tag inserted at the C-terminus of Figla cDNA (Supplementary Figure S4A–C). Whole mount staining of P5 FiglaFLAG ovary with FLAG antibody exhibited correct nuclear staining of FIGLA in oocytes (Supplementary Figure S4D), suggesting successful expression of the FLAG tag. Our previous studies indicated that Figla transcripts were detected at low levels as early as E13.5. FIGLA protein, however, is not detected until E19 based on sensitive gel mobility shift assays (8,9,34). Co-immunostaining analyses documented that FIGLA was first detected in the nuclei of a very small subset of NOBOX- and LHX8-positive germ cells in E17.5 ovaries. This expression increased to include a substantial number of oocytes in E19 ovaries that co-expressed FIGLA, NOBOX and LHX8. Nearly all oocytes co-expressed FIGLA together with NOBOX and LHX8 in P0 and P4 ovaries (Figure 3 and Supplementary Figure S5). A similar expression pattern of FIGLA, SOHLH1 and SOHLH2 was observed in FiglaFLAG ovaries from E17.5 to P4 and co-immunostaining indicated that the three transcription factors were present in germ cells (Supplementary Figure S6). In summary, these results suggest that FIGLA, NOBOX, LHX8, SOHLH1 and SOHLH2 are co-expressed in the same subset of germ cells in perinatal mouse ovaries.
Figure 3.
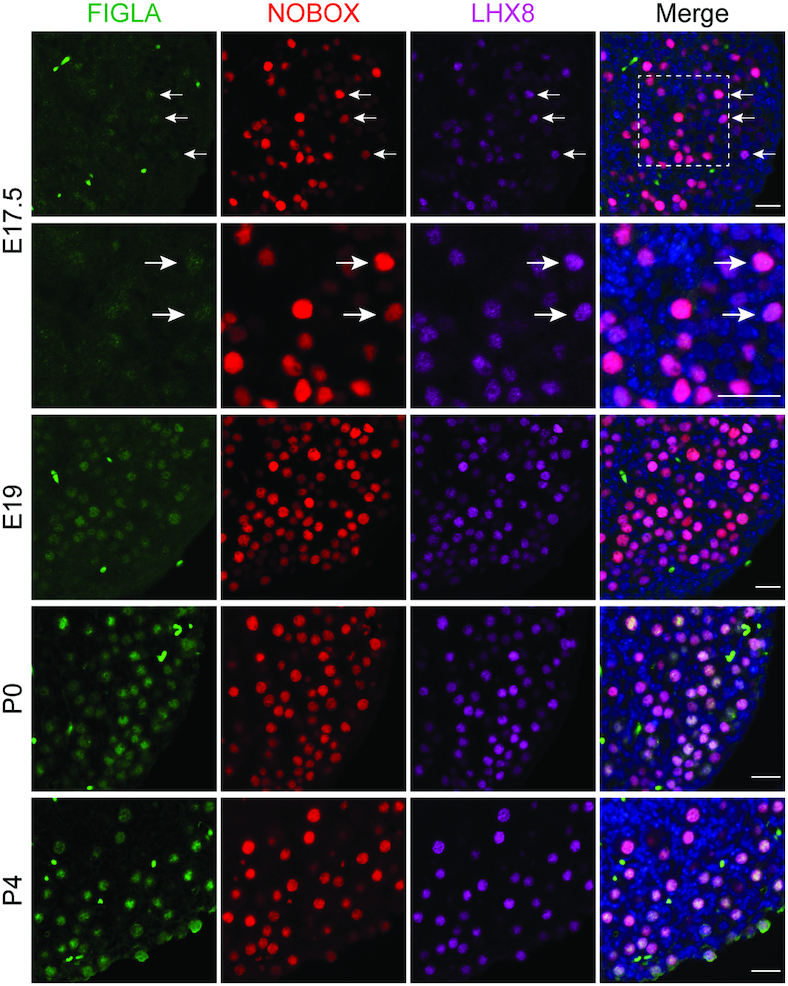
Co-expression of FIGLA, LHX8 and NOBOX in perinatal ovaries. Images showing immunofluorescence results from FiglaFLAG ovarian sections stained with antibodies against FLAG (FIGLA, green), NOBOX (red) and LHX8 (purple) at the indicated time points. DNA was stained with Hoechst 33342. Arrows indicate that FIGLA is expressed solely in a small subset of NOBOX- and LHX8-positive oocytes in E17.5 ovaries. FIGLA was co-expressed with NOBOX and LHX8 in the great majority of oocytes in E19, P0 and P4 ovaries. Scale bar, 20 μm. Data are representative of n = 3 independent ovaries from three different animals with similar results per condition.
Figla deficiency disrupts oocyte-specific expression of transcription regulators
KIT is a well-investigated germline receptor and NOBOX is an oocyte-specific homeobox protein. Both are essential for oocyte growth and differentiation (15,35) and their transcripts were significantly reduced in FiglaNull ovaries at P0. Immunofluorescence with antibodies to DDX4 and KIT or DDX4 and NOBOX demonstrated that KIT expression was completely abolished in FiglaNull oocytes, a result which was further confirmed by immunoblot (Figure 4A and C). We found NOBOX expression also was totally absent in FiglaNull ovaries as determined by immunostaining and immunoblot (Figure 4B and D). LHX8 is essential for oocyte maintenance and differentiation during early oogenesis (14). To determine if FIGLA affects LHX8 expression, we performed immunofluorescence staining with antibodies against DDX4 and LHX8 using ovarian sections from P0 FiglaHet and FiglaNull mice. LHX8 was expressed in the nuclei of most FiglaHet oocytes but was significantly decreased in FiglaNull oocytes where we observed only background staining in the cytoplasm and weak expression in the nuclei of only a few oocytes (Figure 4E and Supplementary Figure S7). In Lhx8Null ovaries at P0, only background levels of LHX8 were present in the cytoplasm of oocytes (Figure 4E and Supplementary Figure S7) consistent with a previous report (14). Over 90% of the oocytes displayed nuclear localization of LHX8 in FiglaHet ovaries, while only 20.6% oocytes exhibited weak nuclear localization in FiglaNull ovaries (FiglaHet 93.9 ± 1.9% versus FiglaNull 20.6 ± 3.9%) (Figure 4F). These results document that Figla deficiency abolishes KIT and NOBOX expression and severely disrupts LHX8 expression in oocytes.
Figure 4.
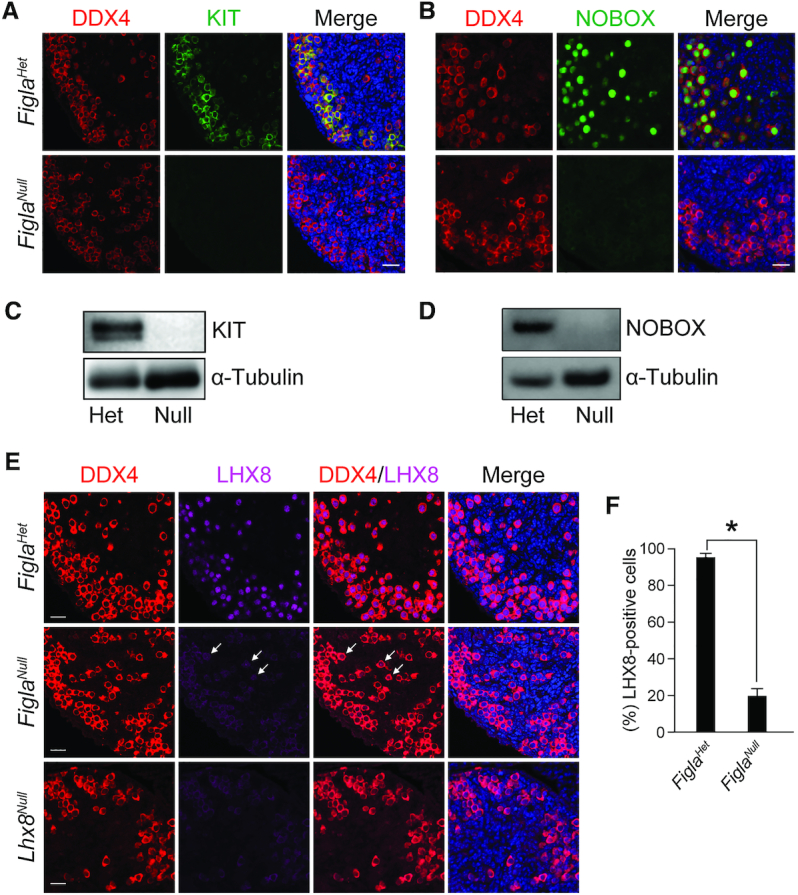
Figla deficiency disrupts KIT, NOBOX and LHX8 expression. (A) Immunofluorescence staining with DDX4 (red) and KIT (green) in FiglaHet and FiglaNull ovaries at P0. DNA was stained with Hoechst 33342. KIT expression was completely abolished in FiglaNull ovaries. Scale bar, 20 μm. (B) Same as (A), but with antibodies to DDX4 (red) and NOBOX (green). NOBOX expression was significantly diminished in Figla-deficient ovaries. (C) Immunoblot analysis of KIT protein expression in FiglaHet and FiglaNull ovaries at P0. α-Tubulin was used as a load control. (D) Same as (C), but with antibodies to NOBOX which was absent in Figla-deficient ovaries. (E) Immunostaining of DDX4 (red) and LHX8 (purple) in P0 FiglaHet, FiglaNull and Lhx8Null ovaries. DNA was stained with Hoechst 33342. Arrows indicate weak expression of LHX8 in the nucleus of Figla-deficient oocytes. Scale bar, 20 μm. (F) Statistical analysis of LHX8-positive cells per section in FiglaHet and FiglaNull ovaries at P0. Mean ± S.D., n = 3 biologically independent ovaries from three different animals. *P <0.05 by two-tailed Student's t test. Representative of n = 3 (A–E) independent biological replicates with similar results per condition.
A recent study reported that transcription factors SOHLH1 and SOHLH2 coordinate oocyte differentiation (25). Immunofluorescence was used to define whether Figla deficiency influences the expression of SOHLH1 and SOHLH2 in P0 ovaries from FiglaHet and FiglaNull mice. We found that SOHLH1 expression was detected in both the nucleus and cytoplasm in more than 70% of the germ cells (75.7% ± 3.9%) and 24.3% of the germ cells (24.3 ± 3.9%) showed only cytoplasmic localization in FiglaHet ovaries, while nearly all germ cells exhibited only cytoplasmic localization (96.2 ± 2.6%) without obvious nuclear localization (3.8 ± 2.6%) in FiglaNull ovaries (Supplementary Figure S8A and B). This observation suggests that FIGLA not only regulates expression of the Sohlh1 gene but also the cellular localization of the cognate protein. SOHLH2 localization was primarily confined to the nuclei of FiglaHet control oocytes, while only a few of the oocytes were positive for SOHLH2 in FiglaNull ovaries (Supplementary Figure S8C). Statistical analysis verified that most of the oocytes (87.2 ± 2.7%) were SOHLH2 positive in FiglaHet control ovaries, whereas significantly fewer oocytes (31.1 ± 3.1%) from FiglaNull ovaries were SOHLH2 positive (Supplementary Figure S8D). These data indicate that Figla deficiency significantly disrupts SOHLH1 and SOHLH2 expression in oocytes.
Lhx8 deficiency disrupts oogenesis
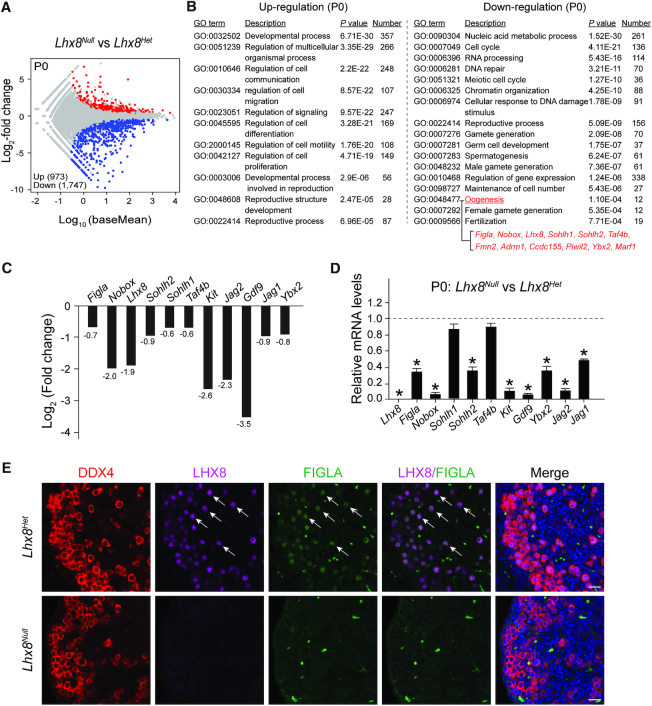
Previous studies reported that Lhx8Null ovaries appear normal at P0 but showed severe germ cell attrition by P7 (14,36) which resembles the phenotype of FiglaNull mice. RNA-seq and immunostaining data indicate that Lhx8 is a downstream target of FIGLA. Thus, we isolated mRNA from Lhx8Het and Lhx8Null ovaries and compared their transcriptomes at P0. RNA-seq analysis identified 973 up-regulated and 1,747 down-regulated genes using adjusted P < 0.1 in Lhx8Null mice (Figure 5A). Gene ontology analysis documented that up-regulated transcripts were primarily involved in developmental processes, cell communication, cell migration, cell differentiation, cell motility, cell proliferation, reproductive structure development and reproductive processes. Down-regulated transcripts were principally associated with nucleic acid metabolic process, cell cycle, RNA processing, DNA repair, reproductive processes, gamete generation, oogenesis and fertilization (Figure 5B). Multiple oogenesis-related genes were significantly reduced in Lhx8Null ovaries (Figure 5C) and qRT-PCR confirmed significant down-regulation of nine genes involved in early folliculogenesis (Figla, Nobox, Lhx8, Sohlh2, Kit, Ybx2, Gdf9, Jag1, Jag2) (Figure 5D). These genes were also down-regulated in FiglaNull ovaries suggesting that FIGLA and LHX8 co-regulate common downstream targets. Notably, we found that Figla transcripts were significantly less abundant in Lhx8Null ovaries, indicating that FIGLA and LHX8 can cross-regulate the expression of each other in oocytes. FiglaFLAG knock in mice were crossed into the Lhx8Null background and immunofluorescence staining was performed using antibodies to LHX8 and FLAG. At P0, FIGLA immunoreactivity was detected in the nuclei of oocytes in Lhx8Het ovaries. However, FIGLA protein is not detectable in Lhx8Nulloocytes (Figure 5E), indicating diminished abundance of FIGLA protein in Lhx8-deficient ovaries. Moreover, immunostaining for SOHLH2, NOBOX and KIT in Lhx8Null ovaries demonstrated that the abundance of all three proteins was dramatically reduced in Lhx8-deficient oocytes at P0 compared to controls (Supplementary Figures S8C and D, S9A and B). Taken together, these data indicate that Lhx8 deficiency disrupts the expression of genes essential for early oogenesis.
Figure 5.
Lhx8 deficiency disrupts oogenesis-associated genes. (A) MA plot of RNA-seq data from Lhx8Het and Lhx8Null ovaries at P0, using adjusted P < 0.1 as the cut off. (B) Enriched Gene Ontology terms of the biological process categories for up- and down-regulated transcripts in Lhx8Null ovaries. (C) RNA-seq results of selected transcripts (log2-fold change) related to oogenesis in Lhx8Null ovaries. (D) Quantitative RT-PCR validation of oogenesis-related genes in Lhx8Null ovaries at P0. Control genes relative to β-actin were set to 1. Data are presented as mean ± s.d for n = 3 biologically independent experiments. *P <0.05 by two-tailed Student's t test. (E) Immunofluorescence staining with antibodies to DDX4 (red), LHX8 (purple) and FIGLA (green) on ovarian sections from P0 Lhx8Het and Lhx8Null mice after crossing with FiglaFLAG mice. The DNA was stained with Hoechst 33342. Arrows indicate both LHX8- and FIGLA-positive germ cells. Scale bar, 20 μm. Representative of n = 3 independent biological replicates with similar results per condition.
FIGLA and LHX8 coordinate oocyte differentiation
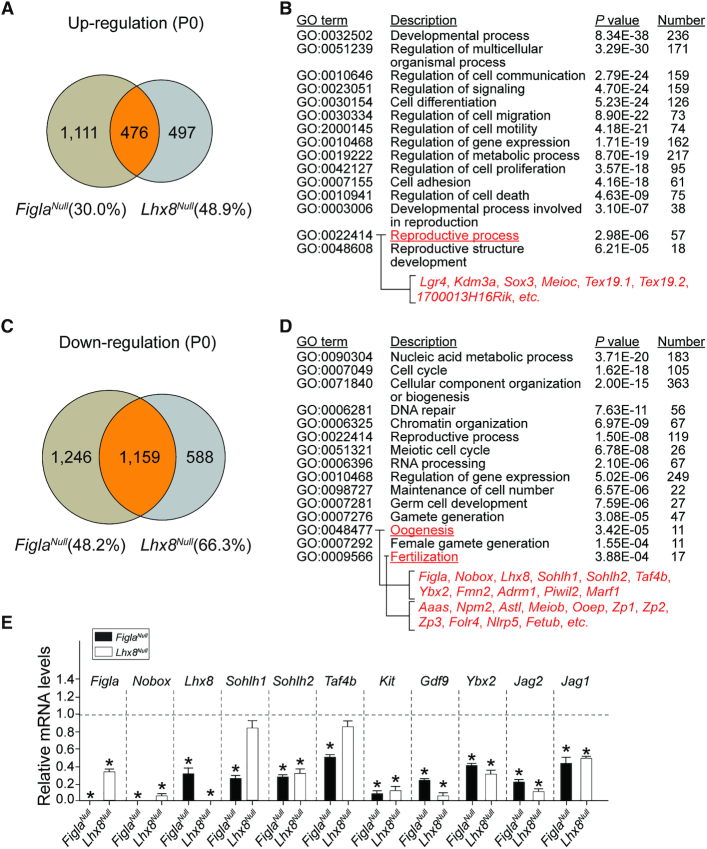
To further investigate the overlap of affected genes in FiglaNull and Lhx8Null mice, we compared the RNA-seq data at P0 from each mutant mouse line. Among up-regulated genes, Figla and Lhx8 mutants shared 476 of 1,587 genes among Figla mutant (30.0%) and 973 genes among Lhx8 mutant (48.9%) (Figure 6A). Gene ontology analysis documented that overlapping up-regulated transcripts were significantly enriched for developmental processes, cell differentiation, cell migration, cell motility, cell proliferation, cell adhesion, cell death and reproductive processes (Figure 6B). The up-regulated transcripts enriched in reproductive processes included testis-specific genes (e.g. Tex19.1, Tex19.2, 1700013H16Rik) and spermatogenesis-related genes (e.g. Lgr4, Kdm3a, Sox3, Meioc). The overexpression of these male germ cell-associated genes observed in both Figla and Lhx8 mutants has been previously observed in other oocyte-specific mutants of Nobox, Sohlh1 and Sohlh2 genes (25,37), which implies that FIGLA and LHX8 expression in oocytes plays an important role in repressing male-determining genes to ensure normal oogenesis.
Figure 6.
FIGLA and LHX8 coordinate oocyte differentiation. (A) Venn diagram showing overlap and difference in the number of up-regulated transcripts from FiglaNull and Lhx8Null ovaries when compared with control newborn ovary transcriptomes. (B) The enriched Gene Ontology terms of the biological process categories of up-regulated genes in both FiglaNull and Lhx8Null ovaries at P0. (C) Same as (A) but showing the down-regulated transcripts. (D) Same as (B), but of down-regulated transcripts. (E) Quantitative RT-PCR validation of oogenesis-related genes in both FiglaNull and Lhx8Null ovaries at P0. The control genes relative to β-actin were set to 1. Data are presented as mean ± S.D. for n = 3 biologically independent experiments. *P <0.05 by two-tailed Student's t test.
Among down-regulated transcripts, FiglaNull and Lhx8Null mice shared 1159 genes of a total of 2405 genes among Figla mutant (48.2%) and 1747 genes among Lhx8 mutant (66.3%) (Figure 6C). The overlapping down-regulated transcripts were significantly enriched for cell cycle, DNA repair, reproductive processes, meiotic cell cycle, RNA processing, gamete generation, oogenesis and fertilization (Figure 6D). We verified the down-regulation of oogenesis-related genes at P0 by real-time quantitative PCR in both FiglaNull and Lhx8Null mice. The qRT-PCR data confirmed the significant down-regulation of genes involved in early folliculogenesis (Figla, Nobox, Lhx8, Sohlh2, Kit, Ybx2, Gdf9, Jag1, Jag2) in both mutants, but down-regulation of Sohlh1 and Taf4b was restricted to FiglaNull mice (Figure 6E). The significant overlap in affected genes in Figla and Lhx8 mutants suggests that FIGLA and LHX8 coordinately regulate oocyte differentiation in early oogenesis.
FIGLA, LHX8 and SOHLH1 networks in early oogenesis
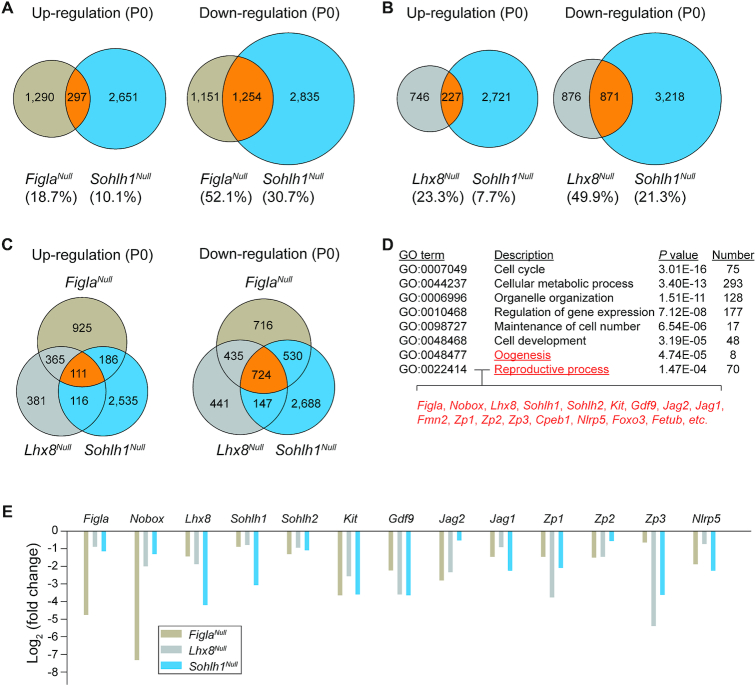
We have determined that some of the down-regulated transcripts in the Sohlh1 mutant (25) overlap with those in Figla and Lhx8 mutants (9,14) including Nobox, Gdf9, Kit, Jag1, Jag2, Zp1, Zp2, Zp3 and Nlrp5. To more systematically investigate the overlap of affected genes in Figla, Lhx8 and Sohlh1 mutant mice, we compared the published RNA-seq data of ovaries from newborn Sohlh1Null mice (25) with our RNA-seq data derived from FiglaNull and Lhx8Null mice. RNA-seq analysis identified 2948 up-regulated and 4089 down-regulated transcripts using adjusted P < 0.1 in Sohlh1Null mice (Supplementary Figure S10A). Gene ontology analysis revealed that up-regulated transcripts were primarily enriched in male reproductive processes including spermatogenesis and spermatid development, while down-regulated transcripts were involved in maintenance of cell number and female reproductive processes (Supplementary Figure S10B and C). The 297 up-regulated genes that were shared in Figla and Sohlh1 mutants represent 18.7% of FiglaNull (1587) and 10.1% of Sohlh1Null (2948) up-regulated ovarian genes (Figure 7A). The 1254 decreased transcripts that were shared represent 52.1% (2405) and 30.7% (4089), respectively, of the down-regulated genes in Figla and Sohlh1 null ovaries (Figure 7A). These data suggest that a significant overlap of mis-expressed genes was regulated by both FIGLA and SOHLH1. In addition, we compared the RNA-seq data between Lhx8 and Sohlh1 mutants. The data document that the two null mutants share 227 transcripts with increased abundance which represents 23.3% (973) and 7.7% (2,948), respectively, of up-regulated genes in Lhx8Null and Sohlh1Null ovaries. Likewise, the 871 transcripts that were decreased in both mutants, reflect 49.9% (1747) and 21.3% (4089) of down-regulated genes in Lhx8Null and Sohlh1Null ovaries, respectively, (Figure 7B), implying that SOHLH1 shared similar downstream genes and pathways with LHX8. Furthermore, we determined the overlap of affected genes among Figla, Lhx8 and Sohlh1 null mutants. Among the three mutant mouse lines, 111 and 724 genes were concomitantly up-regulated or down-regulated, respectively (Figure 7C). Gene ontology analysis showed that the 724 down-regulated transcripts were associated with oogenesis and reproductive processes (Figure 7D). The RNA-seq data verified that some of the oogenesis- and reproductive processes-associated genes were significantly reduced in Figla, Lhx8 and Sohlh1 mutants (Figure 7E). Even using an adjusted P value <0.05 as the cut off for the RNA-seq data, oogenesis- and reproductive processes-associated genes still appeared in the differentially expressed gene list and overlapped among the three mutant mouse lines (Supplementary Tables S5–S7). Taken together, analyses of the RNA-seq profiles indicate that FIGLA, LHX8 and SOHLH1 coordinately regulate similar downstream targets and participate in a multifunctional network necessary for germ cell maintenance and differentiation during early oogenesis.
Figure 7.
Transcriptome profiles and networks in P0 FiglaNull, Lhx8Null and Sohlh1Null ovaries. (A) Venn diagram of overlap and difference in the number of up-regulated and down-regulated transcripts in FiglaNull and Sohlh1Null ovaries. (B) Same as (A), but in Lhx8Null and Sohlh1Null ovaries. (C) Same as (A), but in FiglaNull, Lhx8Null and Sohlh1Null ovaries. (D) The enriched Gene Ontology terms of the biological process categories of down-regulated genes from FiglaNull, Lhx8Null and Sohlh1Null ovaries. (E) RNA-seq results of selected transcripts (log2-fold change) associated with oogenesis and reproductive processes from FiglaNull, Lhx8Null and Sohlh1Null ovaries.
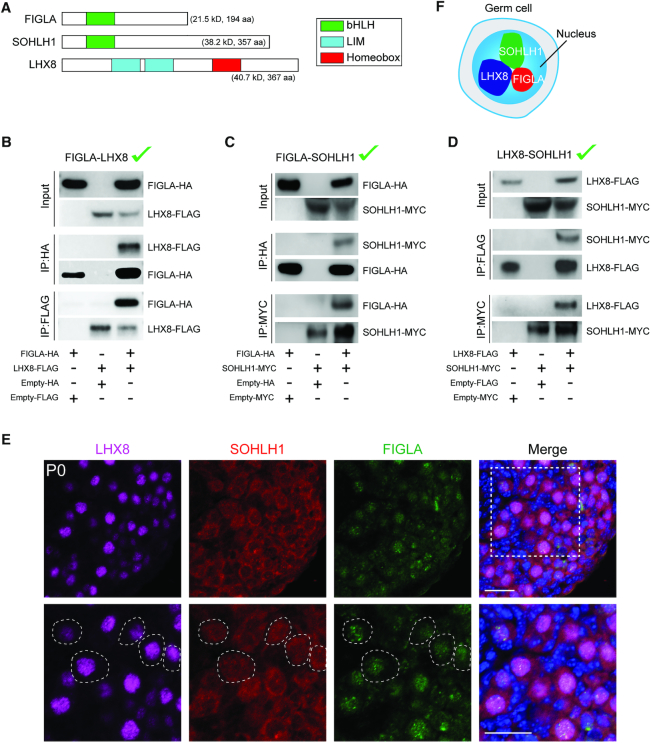
FIGLA, LHX8 and SOHLH1 physically interact
Because FIGLA, LHX8 and SOHLH1 have a significant overlap in downstream target genes, we investigated whether they could physically interact with one another. FIGLA and SOHLH1 proteins contain a well conserved bHLH domain and LHX8 has two tandem LIM domains and one homeobox domain (Figure 8A). To examine physical interactions of FIGLA, LHX8 and SOHLH1, we co-transfected pairs of the three recombinant vectors with different tags into HEK-293T cells and performed co-immunoprecipitation (co-IP) experiments. Co-transfections with FIGLAHA and LHX8FLAG vectors showed each could be co-precipitated by the other, indicating direct interaction between the two proteins (Figure 8B). Co-transfections with FIGLAHA and SOHLH1MYC (Figure 8C) or with LHX8FLAG and SOHLH1MYC (Figure 8D) also documented direct binding between the pairs of proteins. Co-expression of the three proteins in P0 FiglaFLAG ovaries indicated that FIGLA, LHX8 and SOHLH1 could co-localize in the nucleus of the same oocytes (Figure 8E). Taken together, these co-IP and immunostaining analyses indicate that FIGLA, LHX8 and SOHLH1 may directly interact with each other to form a nuclear complex in oocytes (Figure 8F) which could explain coordination regulation of similar downstream genes and pathways in early oocyte development.
Figure 8.
Interactions of FIGLA, LHX8 and SOHLH1. (A) Representative protein domains (bHLH, LIM, homeobox) of FIGLA, SOHLH1 and LHX8. (B) FIGLAHA and LHX8FLAG expression vectors were co-transfected into HEK-293T cells. Cell lysates were probed with HA and FLAG antibodies to detect input protein FIGLA and LHX8, respectively. After immunoprecipitation with HA and FLAG antibody, immunoblots were performed to detect FIGLA and associated LHX8 protein or LHX8 and associated FIGLA protein, respectively. (C) Same as (B) except that FIGLAHA and SOHLH1MYC were co-transfected into HEK-293T cells. Antibodies to HA and MYC were used to detect input proteins and immunoprecipitate FIGLA and SOHLH1, respectively, and their associated proteins. (D) Same as (B) except that LHX8FLAG and SOHLH1MYC were co-transfected into HEK-293T cells. Antibodies to FLAG and MYC were used to detect input proteins and immunoprecipitate LHX8 and SOHLH1, respectively, and their associated proteins. (E) Co-expression of FIGLA, LHX8 and SOHLH1 in P0 ovaries from FiglaFLAG mice. The dashed circles indicate co-expression of FIGLA, LHX8 and SOHLH1 in the same oocytes. Scale bar, 20 μm. (F) FIGLA, LHX8 and SOHLH1 appear to form a nuclear complex in oocytes. Representative of n = 3 (B–E) independent biological replicates with similar results per condition.
Model of FIGLA in driving oocyte differentiation
Our findings indicate that FIGLA is a multifunctional regulator of oocyte differentiation during early folliculogenesis (Figure 9). FIGLA regulates meiotic gene expression to ensure normal meiotic progression in oocytes. It represses male-specific genes and transcriptionally up-regulates female germline genes including Nobox, Lhx8, Sohlh1, Sohlh2, Taf4b and Gdf9 that stringently control downstream oocyte-specific gene expression. FIGLA, LHX8 and SOHLH1 proteins bind to each other to form a nuclear complex in oocytes that recruits co-activators or co-repressors to regulate transcriptional activity during oocyte growth and differentiation. FIGLA, LHX8 and SOHLH1 coordinate expression of genes during oocyte differentiation. It is also possible that FIGLA, LHX8 and SOHLH1 have additional and distinct target genes and pathways during early folliculogenesis.
Figure 9.
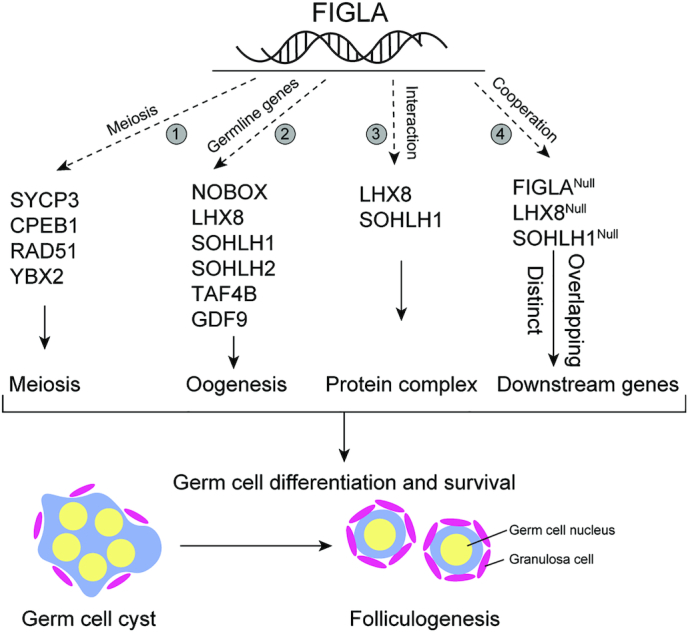
Hypothetical model of FIGLA function in early folliculogenesis. FIGLA concomitantly functions in multiple pathways during early oogenesis: 1, FIGLA regulates meiotic genes (e.g. Sycp3, Cpeb1, Rad51, Ybx2) to ensure proper meiotic progression; 2, FIGLA up-regulates a subset of germline genes (Nobox, Lhx8, Sohlh1, Sohlh2, Taf4b, Gdf9) and their downstream genes that are essential for early oocyte development; 3, FIGLA, LHX8 and SOHLH1 interact to form a nuclear complex in the oocytes which could recruit transcriptional co-activators or co-repressors during oocyte growth and differentiation and 4, FIGLA regulates the transcription of downstream targets in conjunction with LHX8 and SOHLH1, but each factor also has distinct, non-overlapping target genes.
DISCUSSION
To investigate networks that regulate early mouse oogenesis, we have integrated experimental results of downstream gene targets of FIGLA, LHX8 and SOHLH1 transcription factors. Each of the three proteins is present in the nucleus of oocytes in vivo and tagged proteins form a nuclear complex in heterologous cells. The absence of FIGLA sequentially affects meiosis and oocyte gene expression through nuclear interactions with LHX8 and SOHLH1 to coordinate appropriate gene expression during oogenesis. There is a substantial overlap of down-regulated genes in the absence of each factor which substantiates their cooperation in regulating genes essential for successful oogenesis.
Meiosis and oocyte growth
To elucidate functions specific to FIGLA, we performed RNA-seq and found that a series of genes involved in meiosis and oogenesis were down-regulated in FiglaNull ovaries. Cpeb1, essential for germ cell differentiation and synaptonemal complex formation (38) and Ybx2, required for oogenesis (39) are significantly decreased in FiglaNull ovaries. The disruption of meiotic genes suggests meiotic defects in Figla-deficient oocytes, a result which was observed in a previous study by immunostaining with YBX2 antibody (8). In the current study, we further investigated the role of FIGLA in meiosis. We found that diplotene stage oocytes (identified by YBX2 staining) are significantly reduced and dictyate stage oocytes (classified by the presence of two to four visible nucleoli by staining of SYCP3 antibody) are rarely detected in Figla-deficient oocytes, implying impairment of meiotic progression. Our previous findings document that comparable numbers of control and mutant oocytes reach the pachytene and diplotene stages of the prophase of MI in E19 ovaries (8) and our current study shows that FIGLA is only detectable in a subset of germ cells in E17.5 ovaries. The meiotic defects observed in Figla-deficient ovaries may be in part due to the diminished expression of Taf4b or to secondary effects of mis-expressed meiotic genes. Additionally, we observe increased DNA damage and less effective DNA damage repair based on the abundance of γ-H2AX and RAD51, respectively, in Figla-deficient oocytes. These defects in meiotic progression may reflect, at least in part, increased cell apoptosis that is observed by more TUNEL-positive cells in FiglaNull ovaries as γ-H2AX is also a hallmark of early cell death. Nevertheless, it is clear that FIGLA is indispensable for oocyte survival.
Gene networks in oogenesis
Figla ablation also disrupts the expression of other transcription factors (e.g. Nobox, Lhx8, Taf4b, Sohlh1, Sohlh2) that play crucial roles in postnatal oocyte differentiation. This suggests a role for FIGLA in regulating oogenesis genes by acting upstream of these transcription factors. However, a recent study proposed a model that TAF4B occupies the proximal promoters of Stra8, Dazl, Figla and Nobox and functions as direct upstream regulator of meiosis and oogenesis genes (17). Furthermore, SOHLH1 directly targets Lhx8, Zp1, Zp2 and Zp3 genes through the E-box elements in their promoters (7,11). LHX8 directly regulates the transcription of genes containing LHX8 DNA binding element (TGATTG) in oocytes and NOBOX containing a TAATTG sequence at its promoter region is confirmed to be a direct target of LHX8 (40). NOBOX has been reported to regulate transcription of Pou5f1 and Gdf9 in oocytes by binding their promoters directly (41). Thus, although they were down-regulated in Figla-deficient oocytes, whether the expression of these genes is regulated by FIGLA directly or indirectly remains to be determined.
A previous study has shown that oocyte-specific transcriptional regulators NOBOX, LHX8, SOHLH1 and SOHLH2 are co-expressed in the same subset of germ cells and are co-dependent on one another for their appropriate expression (25). However, because of the absence of commercially available FIGLA antibody, the expression pattern of FIGLA and these transcriptional regulators was unknown. In our study, we generated FiglaFLAG knock-in mice which allowed us to investigate the expression pattern of oocyte-specific transcriptional regulators during folliculogenesis. Our data, together with previous findings (13,25), indicated that FIGLA and other transcription factors (e.g. LHX8, NOBOX, SOHLH1, SOHLH2) are co-expressed in the same germ cell population and cross-regulate each other in perinatal ovaries.
FIGLA/LHX8/SOHLH1 protein complex
SOHLH1, SOHLH2 and other known oocyte-specific transcription factors (NOBOX and LHX8) have been shown to be co-dependent on one another for appropriate expression in the ovary (25). It is unknown whether the expression of these transcriptional regulators is dependent on Figla expression. Our findings document that Lhx8, Nobox, Taf4b, Sohlh1 and Sohlh2 expression is significantly decreased in FiglaNull ovaries, suggesting that FIGLA regulates numerous oocyte-specific genes in the ovaries. However, down-regulation of Figla mRNA is also observed in Lhx8-, Sohlh1-, Sohlh2- and Taf4b-deficient ovaries. Therefore, it is likely that these transcription factors cross-regulate each other, either directly or indirectly, in oocytes during early folliculogenesis.
Transcription factors-deficient ovaries share similar phenotypes and common genetic pathways, suggesting interactions. In this respect, we demonstrate in the present study that FIGLA, LHX8 and SOHLH1 can physically interact with each other to form a complex in heterologous cells. A previous study reported that FIGLA, but not SOHLH1, is a specific interacting partner of bovine LHX8 (42). Although interactions of SOHLH1 with LHX8 were not detected by yeast two-hybrid analysis in that study, our co-IP experiments clearly document physical interactions between SOHLH1 and LHX8. The different result of the interaction between SOHLH1 and LHX8 possibly results from different methodology and assay sensitivity. In the earlier study, the authors also documented that LHX8 interacts with FIGLA via LIM rather than homeobox domains (42), representing an example of protein-protein interactions mediated by the LIM and bHLH motifs. Thus, the bHLH domains of FIGLA and SOHLH1 and the LIM domains of LHX8 may mediate the interactions that form the nuclear complex. This complex may function through recruitment of transcriptional co-activators or co-repressors to regulate gene expression during early oocyte growth and differentiation. It is also possible that the common genetic pathways involved in Figla, Lhx8 and Sohlh1 mutants result from the formation of the complex in oocytes. However, we have no evidence to show that the overlapping downstream genes are direct targets of these transcription factors. Additional experiments using chromatin immunoprecipitation sequencing (ChIP-seq) will be needed to identify direct downstream targets of this protein complex. Furthermore, we cannot exclude the possibility that specific pathways are involved in each of these mutants and the formation of the complex needs to be further confirmed in vivo.
Similar downstream genes regulated by FIGLA, LHX8 and SOHLH1
Since Figla, Lhx8 and Sohlh1-deficient ovaries share many similar characteristics and interact as a complex in heterologous cells, we analyzed RNA-seq profiles to identify the downstream targets and pathways from each mutant mouse line. The down-regulated transcripts in Lhx8 and Sohlh1 mutants include similar oogenesis genes that are identified in Figla mutant, indicating an overlap of mis-expressed genes. Therefore, we systematically compared the up-regulated and down-regulated transcripts among Figla, Lhx8 and Sohlh1 mutants and found a significant overlap in affected genes. Among up-regulated transcripts, 30% of the genes in Figla mutant and 48.9% of the genes in Lhx8 mutant are shared. The overlapping up-regulated transcripts involve male-determining genes which have been previously observed in other mutants (25,37), signifying the importance of oocytes in repressing testis-specific genes that could otherwise disrupt normal oogenesis. Among the down-regulated transcripts, Figla and Lhx8 mutants showed a substantially higher percentage of overlapping genes, 48.2% of the transcripts in the Figla mutant and 66.3% in the Lhx8 mutant. These shared genes are involved in oogenesis and fertilization. Our study indicates that FIGLA regulates oocyte differentiation in conjunction with LHX8. Although analysis of the RNA-seq data from Sohlh1Null ovaries shows higher number of mis-expressed genes, 52.1% of FIGLA and 49.9% of LHX8 downstream genes are involved in SOHLH1 downstream targets. There is also a significant overlap in genes mis-expressed among all three Figla, Lhx8 and Sohlh1 mutants, indicating that these transcription factors coordinately regulate similar downstream targets and participate in a multifunctional network to regulate germ cell maintenance and differentiation during early oogenesis.
In humans, FIGLA, NOBOX, LHX8 and GDF9 mutations are associated with primary ovarian insufficiency and female infertility due to premature loss of germ cells in the ovary (43–46). Therefore, FIGLA, LHX8, NOBOX, SOHLH1 and the downstream genes that they regulate are all potential candidate genes for non-syndromic ovarian insufficiency. In this study, we investigated the functional relationship among these germ cell-specific transcription factors and document downstream target genes of FIGLA, LHX8 and SOHLH1. Thus, not only do these findings provide new insights to expand our understanding of physiological and pathological process in early folliculogenesis, they also contribute to opening new avenues of research to pursue the pathogenic mechanisms of primary ovarian insufficiency and female infertility in humans.
DATA AVAILABILITY
The accession number for the sequencing data reported in this study has been deposited in the Gene Expression Omnibus website with accession code GSE139966.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Jurrien Dean lab for helpful suggestions on the project and thank Quin Waterbury for editorial assistance. We thank Dr Aleksandar Rajkovic for providing NOBOX, LHX8, SOHLH1, SOHLH2 antibodies and the 3xMYC-SOHLH1 plasmid. We thank Dr Boris Baibakov for help with confocal microscopy and Dr Yuning Huang for animal husbandry of the Lhx8Null mice. We thank the NIDDK Genomic Core for sequencing and Dr Ryan Dale and Dr. Guanghui Yang for preliminary analyses of the resultant data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Disease (NIDDK). Funding for open access charge: NIDDK; NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pepling M.E., Spradling A.C.. Female mouse germ cells form synchronously dividing cysts. Development. 1998; 125:3323–3328. [DOI] [PubMed] [Google Scholar]
- 2. Pepling M.E., Spradling A.C.. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev. Biol. 2001; 234:339–351. [DOI] [PubMed] [Google Scholar]
- 3. Pepling M.E. From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis. 2006; 44:622–632. [DOI] [PubMed] [Google Scholar]
- 4. Zhang H., Liu L., Li X., Busayavalasa K., Shen Y., Hovatta O., Gustafsson J.A., Liu K.. Life-long in vivo cell-lineage tracing shows that no oogenesis originates from putative germline stem cells in adult mice. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:17983–17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lei L., Spradling A.C.. Female mice lack adult germ-line stem cells but sustain oogenesis using stable primordial follicles. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8585–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qin Y., Jiao X., Simpson J.L., Chen Z.J.. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum. Reprod. Update. 2015; 21:787–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang L.F., Soyal S.M., Dean J.. FIG alpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development. 1997; 124:4939–4947. [DOI] [PubMed] [Google Scholar]
- 8. Soyal S.M., Amleh A., Dean J.. FIG alpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development. 2000; 127:4645–4654. [DOI] [PubMed] [Google Scholar]
- 9. Joshi S., Davies H., Sims L.P., Levy S.E., Dean J.. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev. Biol. 2007; 7:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu W., Gauthier L., Baibakov B., Jimenez-Movilla M., Dean J.. FIGLA, a basic helix-loop-helix transcription factor, balances sexually dimorphic gene expression in postnatal oocytes. Mol. Cell Biol. 2010; 30:3661–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pangas S.A., Choi Y., Ballow D.J., Zhao Y., Westphal H., Matzuk M.M., Rajkovic A.. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:8090–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ballow D.J., Xin Y., Choi Y., Pangas S.A., Rajkovic A.. Sohlh2 is a germ cell-specific bHLH transcription factor. Gene Expr. Patterns. 2006; 6:1014–1018. [DOI] [PubMed] [Google Scholar]
- 13. Choi Y., Yuan D., Rajkovic A.. Germ cell-specific transcriptional regulator Sohlh2 is essential for early mouse folliculogenesis and oocyte-specific gene expression. Biol. Reprod. 2008; 79:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi Y., Ballow D.J., Xin Y., Rajkovic A.. Lim homeobox gene, Lhx8, is essential for mouse oocyte differentiation and survival. Biol. Reprod. 2008; 79:442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rajkovic A., Pangas S.A., Ballow D., Suzumori N., Matzuk M.M.. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004; 305:1157–1159. [DOI] [PubMed] [Google Scholar]
- 16. Grive K.J., Seymour K.A., Mehta R., Freiman R.N.. TAF4b promotes mouse primordial follicle assembly and oocyte survival. Dev. Biol. 2014; 392:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grive K.J., Gustafson E.A., Seymour K.A., Baddoo M., Schorl C., Golnoski K., Rajkovic A., Brodsky A.S., Freiman R.N.. TAF4b regulates oocyte-specific genes essential for meiosis. PLos Genet. 2016; 12:e1006128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Falender A.E., Shimada M., Lo Y.K., Richards J.S.. TAF4b, a TBP associated factor, is required for oocyte development and function. Dev. Biol. 2005; 288:405–419. [DOI] [PubMed] [Google Scholar]
- 19. Suzuki H., Ahn H.W., Chu T., Bowden W., Gassei K., Orwig K., Rajkovic A.. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 2012; 361:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Falender A.E., Freiman R.N., Geles K.G., Lo K.C., Hwang K., Lamb D.J., Morris P.L., Tjian R., Richards J.S.. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005; 19:794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hao J., Yamamoto M., Richardson T.E., Chapman K.M., Denard B.S., Hammer R.E., Zhao G.Q., Hamra F.K.. Sohlh2 knockout mice are male-sterile because of degeneration of differentiating type A spermatogonia. Stem Cells. 2008; 26:1587–1597. [DOI] [PubMed] [Google Scholar]
- 22. Toyoda S., Miyazaki T., Miyazaki S., Yoshimura T., Yamamoto M., Tashiro F., Yamato E., Miyazaki J.. Sohlh2 affects differentiation of KIT positive oocytes and spermatogonia. Dev. Biol. 2009; 325:238–248. [DOI] [PubMed] [Google Scholar]
- 23. Lovasco L.A., Gustafson E.A., Seymour K.A., de Rooij D.G., Freiman R.N.. TAF4b is required for mouse spermatogonial stem cell development. Stem Cells. 2015; 33:1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ballow D., Meistrich M.L., Matzuk M., Rajkovic A.. Sohlh1 is essential for spermatogonial differentiation. Dev. Biol. 2006; 294:161–167. [DOI] [PubMed] [Google Scholar]
- 25. Shin Y.H., Ren Y., Suzuki H., Golnoski K.J., Ahn H.W., Mico V., Rajkovic A.. Transcription factors SOHLH1 and SOHLH2 coordinate oocyte differentiation without affecting meiosis I. J. Clin. Invest. 2017; 127:2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao Y., Guo Y.J., Tomac A.C., Taylor N.R., Grinberg A., Lee E.J., Huang S., Westphal H.. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:15002–15006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 28. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao Y., Smyth G.K., Shi W.. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013; 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gu W., Tekur S., Reinbold R., Eppig J.J., Choi Y.C., Zheng J.Z., Murray M.T., Hecht N.B.. Mammalian male and female germ cells express a germ cell-specific Y-Box protein, MSY2. Biol. Reprod. 1998; 59:1266–1274. [DOI] [PubMed] [Google Scholar]
- 33. Wang Y., Teng Z., Li G., Mu X., Wang Z., Feng L., Niu W., Huang K., Xiang X., Wang C. et al.. Cyclic AMP in oocytes controls meiotic prophase I and primordial folliculogenesis in the perinatal mouse ovary. Development. 2015; 142:343–351. [DOI] [PubMed] [Google Scholar]
- 34. Millar S.E., Lader E.S., Dean J.. ZAP-1 DNA binding activity is first detected at the onset of zona pellucida gene expression in embryonic mouse oocytes. Dev. Biol. 1993; 158:410–413. [DOI] [PubMed] [Google Scholar]
- 35. Jones R.L., Pepling M.E.. KIT signaling regulates primordial follicle formation in the neonatal mouse ovary. Dev. Biol. 2013; 382:186–197. [DOI] [PubMed] [Google Scholar]
- 36. D’Ignazio L., Michel M., Beyer M., Thompson K., Forabosco A., Schlessinger D., Pelosi E.. Lhx8 ablation leads to massive autophagy of mouse oocytes associated with DNA damage. Biol. Reprod. 2018; 98:532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi Y., Qin Y., Berger M.F., Ballow D.J., Bulyk M.L., Rajkovic A.. Microarray analyses of newborn mouse ovaries lacking Nobox. Biol. Reprod. 2007; 77:312–319. [DOI] [PubMed] [Google Scholar]
- 38. Tay J., Richter J.D.. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell. 2001; 1:201–213. [DOI] [PubMed] [Google Scholar]
- 39. Yang J., Medvedev S., Yu J., Tang L.C., Agno J.E., Matzuk M.M., Schultz R.M., Hecht N.B.. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5755–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park M., Jeon S., Jeong J.H., Park M., Lee D.R., Yoon T.K., Choi D.H., Choi Y.. Identification and characterization of LHX8 DNA binding elements. Dev. Reprod. 2012; 16:379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi Y., Rajkovic A.. Characterization of NOBOX DNA binding specificity and its regulation of Gdf9 and Pou5f1 promoters. J. Biol. Chem. 2006; 281:35747–35756. [DOI] [PubMed] [Google Scholar]
- 42. Fu L., Zhang M., Mastrantoni K., Perfetto M., Wei S., Yao J.. Bovine Lhx8, a germ cell-specific nuclear factor, interacts with Figla. PLoS One. 2016; 11:e0164671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao H., Chen Z.J., Qin Y.Y., Shi Y.H., Wang S., Choi Y., Simpson J.L., Rajkovic A.. Transcription factor FIGLA is mutated in patients with Premature Ovarian Failure. Am. J. Hum. Genet. 2008; 82:1342–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qin Y., Choi Y., Zhao H., Simpson J.L., Chen Z.J., Rajkovic A.. NOBOX homeobox mutation causes premature ovarian failure. Am. J. Hum. Genet. 2007; 81:576–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qin Y., Zhao H., Kovanci E., Simpson J.L., Chen Z.J., Rajkovic A.. Analysis of LHX8 mutation in premature ovarian failure. Fertil. Steril. 2008; 89:1012–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao H., Qin Y., Kovanci E., Simpson J.L., Chen Z.J., Rajkovic A.. Analyses of GDF9 mutation in 100 Chinese women with premature ovarian failure. Fertil. Steril. 2007; 88:1474–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequencing data reported in this study has been deposited in the Gene Expression Omnibus website with accession code GSE139966.