Abstract
The A-type lamins (lamin A/C), encoded by the LMNA gene, are important structural components of the nuclear lamina. LMNA mutations lead to degenerative disorders known as laminopathies, including the premature aging disease Hutchinson-Gilford progeria syndrome. In addition, altered lamin A/C expression is found in various cancers. Reports indicate that lamin A/C plays a role in DNA double strand break repair, but a role in DNA base excision repair (BER) has not been described. We provide evidence for reduced BER efficiency in lamin A/C-depleted cells (Lmna null MEFs and lamin A/C-knockdown U2OS). The mechanism involves impairment of the APE1 and POLβ BER activities, partly effectuated by associated reduction in poly-ADP-ribose chain formation. Also, Lmna null MEFs displayed reduced expression of several core BER enzymes (PARP1, LIG3 and POLβ). Absence of Lmna led to accumulation of 8-oxoguanine (8-oxoG) lesions, and to an increased frequency of substitution mutations induced by chronic oxidative stress including GC>TA transversions (a fingerprint of 8-oxoG:A mismatches). Collectively, our results provide novel insights into the functional interplay between the nuclear lamina and cellular defenses against oxidative DNA damage, with implications for cancer and aging.
INTRODUCTION
The nuclear lamina is a dense network of fibers physically associated with the inner nuclear membrane and peripheral chromatin, and consists mostly of architectural proteins called lamins, along with associated proteins (1). Lamins are type V intermediate filament proteins that form coiled-coil parallel dimers that assemble into higher order filamentous structures that carry out important scaffolding roles in the nucleus (2). In somatic cells of most vertebrates, the major forms of lamins are expressed from three genes: the LMNA gene codes for both lamins A and C (A-type lamins) via alternative splicing; the LMNB1 and LMNB2 genes code for lamin B1 and lamin B2, respectively (B-type lamins). B-type lamins are constitutively expressed in all cell types. However, A-type lamins are only expressed after the onset of cell differentiation; thus, A-type lamins are not expressed in stem cells or early embryonic cells. A-type and B-type lamins form distinct, yet overlapping, lattices in the nuclear lamina (2); unlike B-type lamins, A-type lamins are also found in a mobile and dynamic pool throughout the nucleoplasm. In addition to providing the structural support of the nucleus, lamins regulate a variety of nuclear processes, such as protein transport, gene expression, DNA replication and DNA repair (1). Thus, depletion or mutations in lamins can impact cellular phenotypes such as cell proliferation, differentiation, apoptosis and senescence (2). Regulation of these nuclear activities occurs in part from interaction of lamins with nuclear envelope proteins, transcription factors and chromatin (directly or through interaction with histones or other lamin-associated proteins). Moreover, nearly 40% of the human genome is organized into lamina-associated domains (LADs) that help organize the genome and have been associated with gene repression (3). Changes in chromatin organization can modulate gene expression by altering accessibility of gene promoters to transcription factors.
Mutations in the LMNA gene have been linked to degenerative disorders, broadly termed laminopathies, including Emery-Dreifuss muscular dystrophy, neuropathies, lipodystrophies and the premature aging syndrome Hutchinson-Gilford progeria (HGPS) (4,5). In addition to the impact of LMNA mutations, altered cellular levels of wild-type lamin A/C can be deleterious. Forms of dilated cardiomyopathy and muscular dystrophy are generally associated with reduced A-type lamin function and can be phenocopied by knockout of the Lmna locus in the mouse (6). Accumulating evidence indicates that altered expression levels of lamins can influence cell proliferation, tumor formation and cancer progression (7). Several studies indicate that the levels of A-type lamins are either absent, lower or higher in various human cancers (7–10), and that these changes have a significant impact on the cancer severity (11).
The specific molecular mechanisms that lead to disease from defective A-type lamins are poorly understood. Certainly, LMNA mutations and lamin A/C depletion affect the structural integrity of the nucleus and the nuclear–cytoskeleton interactions, resulting in abnormal mechanoregulation and ultimately disease progression (12,13). Studies have demonstrated that lamins play an important role in physically connecting the nucleus to the cytoskeleton (via LINC complex) to enable mechanosignaling and cell migration, which play important roles in laminopathies and cancer metastasis (2,14,15). Recent studies have implicated a role for lamin A/C in promoting DNA double strand break (DSB) repair (12), thus implying a role in carcinogenesis (16). For example, mouse embryo fibroblast (MEF) null for Lmna has a marked decrease in the accumulation of 53BP1 at IR-induced foci; moreover, these Lmna−/− MEFs displayed transcriptional repression of the BRCA1 and RAD51 genes, which encode proteins essential for homologous recombination repair (17). The structural defects in the nucleus of lamin A/C-defective cells contribute to the impaired DSB repair (12,18). Similarly to DSBs, single strand breaks and oxidative DNA lesions, repaired by base excision repair (BER), are important in carcinogenesis (19). However, despite the links between lamin A/C and cancer (7) and between BER and cancer (20), there has been no reported investigation into the role of lamin A/C in BER; thus, that is the aim of this study.
To overcome genotoxic stress, several DNA repair pathways have evolved, classified on the basis of the target lesion in the DNA (21). BER is the major pathway for removal of small, non-helix-distorting, base lesions from the genome, predominantly oxidized, alkylated and deaminated bases (19). In this pathway, a damaged base is recognized and removed by a DNA glycosylase, leaving an apurinic/apyrimidinic site (AP site). The AP site is then cleaved (i.e. DNA incision) by AP endonuclease 1 (APE1) to form a 3′-OH end and a 5′-deoxyribose phosphate (5′-dRP) end. DNA polymerase β (POL β) catalyzes both excision of the 5′-dRP moiety and DNA synthesis at the 3′-hydroxyl terminus prior to completion of BER (DNA ligation) by DNA ligase 3 (LIG3) (22,23). If the DNA glycosylase has both glycosylase activity and additional 3′AP lyase activity (bifunctional), it can also incise the DNA at the AP site. 8-Oxoguanine (8-oxoG) is one of the most common DNA lesions resulting from reactive oxygen species. This lesion is recognized and removed by 8-oxoguanine DNA glycosylase (OGG1) via its glycosylase activity. OGG1 is a bifunctional DNA glycosylase; however, its lyase activity is weak and thus, in the presence of APE1, is not a dominant activity (24). There is evidence that APE1 coordinates with upstream OGG1 and downstream POLβ in BER and that these activities can be important in facilitating an optimal rate of nuclear BER of oxidative lesions (19,24,25). The repair of acute oxidative DNA damage requires many rounds of the BER pathway, depending on the amount of available BER enzymes and the extent of DNA damage, and thus the repair of oxidative DNA lesions can take many hours (26).
There are many accessory proteins involved in BER, such as PARP1 that helps mediate BER by catalyzing the poly-ADP-ribosylation (PARylation) of several acceptor proteins (including PARP1 itself) involved in chromatin architecture (19). PARylation is a widespread post-translational modification that occurs in response to DNA damage, appearing rapidly at DNA damage sites, catalyzed by PARPs, using donor nicotinamide adenine dinucleotide (NAD+) molecules. This modification regulates a number of biological processes, including chromatin structure, the DNA damage response (DDR), protein turnover, transcription, apoptosis (27), as well as aging and metabolic regulation (28). Interestingly, a recent report found that the NAD+ salvage pathway was altered in the heart of mice and humans carrying a LMNA mutation, leading to altered PARP1 PARylation (29). Another report has shown that lamin A promotes SIRT6-mediated PARP1 mono-ADP ribosylation, in response to DNA damage (30).
Relative mRNA expression data by DNA microarray analysis of Lmna null (Lmna−/−) MEFs, compared to wild-type (Lmna+/+) MEFs, have recently been reported (31), indicating associations of lamin A/C with diseases, such as cancer, and significant alterations of several gene classes that include the DDR. Moreover, antibody microarrays and bioinformatics analysis on HeLa clones with reduced expression of lamin A/C revealed an association of lamin A/C depletion with significant alterations in expression of proteins involved in the cellular stress response (32). Based on the above studies and our own DNA microarray data from Lmna−/− and Lmna+/+ MEFs, reported here, we examined the effect of lamin A/C depletion (Lmna knockout in MEFs and lamin A/C siRNA knockdown in U2OS cells) on BER rate and associated cellular and molecular activities. We report here an unprecedented role for lamin A/C in promoting BER. We find that a contributing mechanism to this lamin A/C function entails PARylation-augmented APE1 DNA incision and POLβ nucleotide incorporation activities. We also demonstrate that the lamin A/C knockout leads to mutations that are fingerprints of oxidative lesion accumulation, thus linking the lamin A/C-dependent defect in BER to increased likelihood of initiation of multistage carcinogenesis and as a potential contributing factor in aging (33,34).
MATERIALS AND METHODS
Cell culture and treatments
Spontaneously immortalized MEFs [wild-type (Lmna+/+) and Lmna knockout (Lmna−/−)], and U2OS cells, were grown in Dulbecco’s modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum (FBS) (Gibco BRL), 50 μg/ml streptomycin and 50 units/ml penicillin, at 37°C in 20% oxygen and 5% CO2. The Lmna+/+ and Lmna−/− MEFs were generated in the laboratory of Colin L. Stewart (6). Retroviral vectors for expression of lamin A and lamin C were a gift from Brian Kennedy (National University of Singapore). Retroviral transductions were performed as described (35). Briefly, HEK-293T cells were transfected with viral packaging (pUMVC3-gag-pol) and envelope (p-CMV-VSV-G) plasmids along with the vectors containing lamin A or lamin C, or with empty vector (EV) as a control. After 48 h, virus-containing media were harvested to infect target cells. Retroviral transductions were performed as two 4−6 h infections on sequential days. Cells were allowed to recover for 48 h and selected with the appropriate drugs.
To inhibit PARylation, the PARP inhibitor 3-aminobenzamide (3-AB; 10 mM) or PARP inhibitor olaparib (2 μM) was added to the medium (DMEM containing 10% FBS) of cultured cells, and incubation carried out for 2 h. To activate PARylation, 2 mM NAD+ was added to the medium and incubation carried out for 2 h.
Microarray
DNA microarray experiments and analysis were carried out at the microarray facility of the National Institute on Aging, Intramural Research Program, National Institute of Health (NIH). A total of 2 × 106 cells were seeded in 100-mm dishes and grown overnight. The next day, cells were washed in cold PBS and RNA was extracted using trizol (Invitrogen). Total RNA quantity and quality were tested using the Agilent Bioanalyzer RNA 6000 Chip (Agilent, Santa Clara, CA). Five hundred nanograms of total RNA was labeled according to the manufacturer’s instructions using the Illumina™ TotalPrep™ RNA amplification kit (Illumina, San Diego, CA). A total of 750 ng biotinylated amplified RNA were hybridized to each lane of an Illumina Mouse Ref-8v2 Expression BeadChip overnight. Following posthybridization rinses, arrays were incubated with streptavidin-conjugated Cy3, and scanned at a resolution of 0.53 μm using an Illumina iScan scanner. Hybridization intensity data were extracted from the scanned images using Illumina BeadStudio GenomeStudio software, V2011.1. Raw hybridization intensity data were log-transformed and normalized to yield z-scores. The z-ratio was calculated as the difference between the observed gene z-scores for the experimental and the control comparisons, and dividing by the standard deviation associated with the distribution of these differences. Z-ratio values = +2.0 or = –2.0 were chosen as cut-off values, defining increased and decreased expression, respectively. A complete set of 522 cellular pathways was obtained from the Molecular Signatures Database (MSigDB, Broad Institute, USA). The complete set was tested for Geneset enrichment using Parametric Analysis of Gene set Enrichment (PAGE). For each pathway z-score, a P-value was computed using JMP 6.0 software to test for the significance of the z-score obtained. Principal component analysis was done using JMP 6.0. These tools were part of DIANE 1.0 (see http://www.grc.nia.nih.gov/branches/rrb/dna/diane_software.pdf for information).
Ingenuity Pathway Analysis (IPA) was performed according to vendor’s specifications (Qiagen). The IPA ‘Top Canonical Pathways’ analysis identified the pathways, from the IPA library of canonical pathways, which were most significant to the input data set. The significance of the association between the data set and the canonical pathway was determined based on two parameters: (i) A ratio of the number of genes from the data set that maps to the pathway divided by the total number of genes that maps to the canonical pathway and (ii) a P-value calculated using Fischer’s exact test determining the probability that the association between the genes in the data set and the canonical pathway is due to chance alone. The IPA ‘Top Upstream Regulators’ analysis is based on prior knowledge of expected effects between transcriptional regulators and their target genes stored in the Ingenuity Knowledge Base. The analysis examines how many known targets of each transcription regulator are present in the user’s data set, and also compares their direction of change (i.e. expression in the experimental sample(s) relative to control) to what is expected from the literature in order to predict likely relevant transcriptional regulators. If the observed direction of change is mostly consistent with a particular activation state of the transcriptional regulator (‘activated’ or ‘inhibited’), then a prediction is made about that activation state. The ‘P-value of overlap’ calls likely upstream regulators based on significant overlap between data set genes and known targets regulated by a transcriptional regulator.
Transfection of APE1 expression plasmids
Lmna +/+ and Lmna−/− MEFs were transiently transfected with the following APE1 constructs: wild-type (wt) APE1-pAcGFP, D210N-pAcGFP (D210N is a mutant of APE1 that lacks endonuclease activity) and empty vector (EV; pAcGFP). These constructs were provided by David Wilson III (National Institute on Aging). For 60-mm dishes, transfection was carried out as follows: 5 μg of DNA was added to 500 μl of Opti-MEM and mixed. Then, this DNA/Opti-MEM was added to 500 μl of OptiMEM containing 10 μl of Lipofectamine 2000 (Thermo Fisher Scientific) and incubated for 20 min at room temperature to allow the formation of DNA–Lipofectamine 2000 complexes. The 1000 μl of DNA–Lipofectamine solution was then added to each dish of cells containing 4 ml of DMEM/10% FBS medium. The dishes were incubated for 24 h to enable significant GFP expression.
RNA interference
U2OS cells, growing in 60-mm dishes in DMEM medium supplemented with 10% FBS, were transfected with 25 nM siRNA oligo using DharmaFECT 1 transfection reagent (Dharmacon, T-2001–02), according to the manufacturer’s protocol. The siRNAs (ON-TARGETplus) were chemically synthesized by Dharmacon. The target sequence for human lamin A/C (Dharmacon, J-004978–05) is 5′-GAAGGAGGGUGACCUGAUA-3′. The negative control (scrambled siRNA, designed to target no known genes in human, mouse or rat) was ON-TARGETplus non-targeting number #1 (Dharmacon, D-001810–010). After 24 h incubation, the transfection medium was replaced with fresh culture medium. The cells were harvested, or used for further experiments, 72 h after transfection. Also, Lmna+/+ MEFs were transfected with mouse lamin A/C. The target sequence for mouse siLamin A/C (Dharmacon ON TARGETplus, J-040758–05-0005) is 5′-GGAAGCAGCGAGAGUUUGA-3′.
Cell proliferation and viability assays
For cell proliferation assays, cells were seeded in 6-well plates at 25 000 cells per well and counted by hemocytometer every day for 4 days (MEFs) or 5 days (U2OS cells). The cells were grown in DMEM containing 10% FBS, at 20% oxygen and 5% CO2; MEFs were also grown in 200 μM H2O2 or 20 μM methyl methane sulfonate (MMS). Each graph data point represents the average of three wells. For viability assays, 20 000 cells were plated in triplicate in 96-well plates in DMEM containing 10% FBS. After 24 h incubation, the cells were washed in PBS and exposed to the indicated stressors (in DMEM without FBS) over a range of doses as indicated. Oxidative DNA damage was induced and sustained by treatment for 4 h with hydrogen peroxide (H2O2) or 2 h with menadione. Alkylation DNA damage was induced and sustained by MMS treatment for 1 h. At the end of treatments, the cells were washed with PBS and then 100 μl of fresh medium was added and then immediately 10 μl of WST-1 (Roche). After 3 h of incubation in WST-1, the absorbance (450 nm minus 630 nm) was read to estimate the number of viable metabolically active cells. Each graph data point represents the average of six wells.
Western blotting analysis
Proteins were separated on 12% Tris-glycine gels (Bio-Rad) in Novex tris-glycine SDS running buffer (Life Technologies). Whole cell extracts were prepared in lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% triton X-100, with 1× mini EDTA-free protease inhibitor cocktail (Sigma), and 1× PhosSTOP phosphatase inhibitor (Sigma). Tris-glycine SDS sample buffer was then added for loading. Transfer to PVDF membranes (Life Technologies) was carried out by electroblotting in Novex tris-glycine transfer buffer (Life Technologies) containing 15% methanol for 1 h at 100 V. Membranes were blocked 1 h at room temperature in 5% nonfat dry milk in TBST (20 mM Tris-HCl, pH 7.2, 137 mM NaCl, 0.1% Tween-20). All antibodies were diluted in 3% milk in TBST. Antibodies: anti-β actin (AMB-7229; Nordic Biosite), anti-lamin A/C (sc-20681; Santa Cruz), anti-αOGG1 (ab124741: Abcam), anti-APE1 (ab97296; Abcam), anti-LIG3 (sc-135883; Santa Cruz), anti-PARP1 (614302; Nordic Biosite), anti-PAR (ab14460; Abcam), anti-POLβ (ab26343; Abcam), anti-NRF2 (ab62352; Abcam), anti-p53 (sc-126; Santa Cruz) and anti-phospho(Ser15)-p53 (9284; Cell Signaling). Secondary HRP-conjugated antibodies (Jackson ImmunoResearch) and Pierce ECL Plus (Thermo Scientific) were used to visualize the protein bands on high-performance chemiluminescence film (GE Healthcare). The films were scanned and saved as 16-bit grayscale tiff files.
Cyclohexamide chase assay
Cyclohexamide (Sigma-Aldrich) was added to MEFs (100-mm dishes, ∼70% confluent) at a concentration of 50 μg/ml and cells collected at the various time points of incubation (untreated, 4, 8, 12 and 24 h). Lysates were prepared from the cells as described for western blotting.
Quantitative real-time PCR
Total RNA samples were isolated using RNeasy Mini Kit (Qiagen Sciences) according to the manufacturer’s protocol. First strand cDNA was synthesized from 2 μg of total RNA with random hexamer primers using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). qRT-PCR was performed using the ABI Prism 7300 system (Applied Biosystems) and SYBR Select Master Mix containing SYBR Green dye (Applied Biosystems). The relative quantity of cDNA was estimated by the Delta-Delta-Ct (ddCt) algorithm and data were normalized to β-actin. The primers were purchased from Eurofins Genomics (the sequences are shown in Supplementary Table S1).
Comet assay
The comet assay (single cell gel electrophoresis assay) was performed as described previously (36). Cells were grown in 60-mm dishes and treated with 100 μM H2O2 for 30 min at 37°C, and then the medium was changed to complete medium (DMEM with 10% FBS) and allowed to repair for 5 min and 8 h; an untreated (UT) condition was also included. Harvested cells were collected and suspended in 200 μl PBS, of which 5–10 μl was mixed with 75 μl 0.5% low-melting-point agarose in PBS, and spread on a microscope slide (precoated with 1% agarose in PBS) and allowed to cool for 5 min. The slides were then incubated overnight in lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 1% Triton X-100, pH 10). Cells were rinsed three times, 5 min each, in neutralization buffer (0.4 M Tris-HCl, pH 7.4). In order to specifically determine the extent of oxidative DNA damage, the slides were then treated with 5 U of formamidopyrimidine DNA glycosylase (FPG) (4040–500-01; Trevigen, 5 U/μl) per slide [1 μl FPG in 99 μl FLARE buffer I (Trevigen) containing 0.1 mg/ml BSA] and incubated for 1 h at 37°C; corresponding slides with buffer/BSA-only (‘no-FPG’) were included as controls. Slides were then washed in PBS three times before incubation in cold unwinding solution (300 mM NaOH, 1 mM EDTA, pH > 13) at 4°C for 1 h. Electrophoresis was carried out in the unwinding solution at 25 V, 300 mA, for 30 min. Slides washed for 5 min in neutralization buffer, then in 95% ethanol for 5 min. The slides were stained with SYBR Gold nucleic acid dye (Life Technologies) at 1/10 000 dilution in Tris-EDTA buffer, pH 7.5. Assay results were visualized using fluorescence microscopy (Axiovert 200M, Carl Zeiss) and analyzed using Comet Assay IV v4.2 software (Perceptive Instruments). A larger and more intense comet tail intensity indicates more DNA damage. The intensity of each comet was represented as ‘Tail Intensity’ (i.e. % DNA in tail) = total intensity of tail/total intensity of comet × 100. For estimation of oxidative damage (FPG-sensitive sites), the tail intensity for buffer treated cells (‘no FPG’) was subtracted from the tail intensity for FPG-treated cells. This was done for all time points (including untreated cells) in order to calculate percent DNA repair: the calculated FPG-sensitive sites for the untreated condition were subtracted from the calculated FPG-sensitive sites for the 5 min and 8 h time points and then the resultant value for 8 h was compared to that for 5 min as a percent.
APE1 DNA incision activity assay
Preparation of cell extracts and performance of the assay for APE1 incision activity were performed based on our previously described method (37). Specifically, the cultured cells (100-mm dishes) were washed with PBS, scraped into 1 ml of PBS and pelleted by centrifugation. Cells pellets were then stored at −80°C until extraction. Cells were extracted by resuspending in 100 μl of extraction buffer [100 mM KCl, 2 mM EDTA, 40% (v/v) glycerol, 0.2% (v/v) Nonidet P-40, 2 mM dithiothreitol (DTT). 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease inhibitor mixture (Roche Applied Science; pH 7.8)]. The lysate was briefly sonicated to disrupt cell and nuclear membranes. A 16 000 × g centrifugation at 4°C for 10 min was performed to remove cellular debris and DNA. The cell extract were dialyzed overnight in dialysis buffer (25 mM HEPES-KOH, 100 mM KCl, 12 mM MgCl2, 1 mM EDTA, 17% glycerol, 1 mM DTT, pH 8.0) at 4°C. The amount of total protein used for incision activity assays was 2.5 ng. 5′-End labeling was carried out using T4 polynucleotides kinase (New England BioLabs) and [γ-32P] ATP. Unincorporated [32P] ATP was removed with a Sephadex G25 spin column (GE Healthcare). The incision reaction was performed in 10 μl of reaction buffer (25 mM HEPES–KOH (pH 7.4), 25 mM KCl, 0.1 mg/ml BSA, 5 mM MgCl2, 10% glycerol and 0.05% Triton X-100) containing 95 fmol of radiolabeled 32P THF28/OG 28-comp oligo (substrate) (Supplementary Table S2A). The THF substrate is an abasic site (AP site) analog. The reaction was incubated for 10 min at 37°C and terminated by the addition of 3× loading dye (90% formamide, 10 mmol/l EDTA, 1 mg/ml xylene cyanol FF and 1 mg/ml bromophenol blue) and heat inactivated at 80°C for 10 min. The reaction products were resolved in 20% acrylamide containing 7 M urea, visualized by phosphorimaging (Typhoon 9410, Amersham Biosciences) and analyzed using ImageQuant 5.2 (Molecular Dynamics). Incision activity was determined as the intensity of product band relative to the combined intensities of substrate and product bands. ‘Activity’ determined from this assay is a measure of effectiveness of the extract at producing an incision product from the substrate and may represent enzymatic activity on the substrate and/or ability of the enzyme to get to the DNA damage site.
POLβ nucleotide incorporation assay
The assay for POLβ activity (nucleotide incorporation assay) was performed as described below and verified in extracts of MEFs subjected to POLβ siRNA knockdown (Supplementary Figure S11). Whole cell extracts (WCEs) were prepared in 20 mM HEPES-KOH, pH 7.5, 200 mM KCl, 10% glycerol, 1% Triton X-100, 1% IGEPAL, 1 mM EDTA, 1 mM DTT, 1× complete-ULTRA protease inhibitor (Sigma) and 1× PhosSTOP phosphatase inhibitor (Sigma) and left on ice for 60 min. The supernatant from the WCEs was separated by centrifugation at 15 000 × g for 10 min and used in the experiments. The substrate oligonucleotides (TAG Copenhagen A/S) (Supplementary Table S2B) were annealed by heating at 90°C for 5 min and slowly cooling to room temperature in 100 mM NaCl and 20 mM HEPES-NaOH, pH 8. Oligonucleotide labeling was by way of the fluorescent dye carboxy tetramethylrhodamine (TAMRA). The extracts were diluted in reaction buffer (50 mM HEPES-KOH, pH 7.5, 5 mM MgCl2, 70 mM KCl, 1 mM DTT, 50 μM of each dNTPs, 0.36 mg/ml BSA and 0.6 mM N-ethylmaleimide) to inhibit DNA ligases and non-β polymerases (38)) to 1 mg/ml to reduce sample-to-sample variation. The extracts (5 μg) were incubated on ice for 5 min in the reaction buffer before adding DNA substrate (1 pmol per reaction) and initiation of repair in final 25 μl reaction volume. The repair was carried out at 37°C for 10 min. The repair reaction was stopped by adding EDTA (10 mM), then SDS (0.5%) and proteinase K (0.2 mg/ml) to the samples and incubating at 55°C for 30 min. About 20 μl of loading buffer (10 mM EDTA, 95% formamide, 0.01% bromophenol blue, 0.01% xylene cyanol) was added to each sample, which was then heated at 90°C for 5 min and DNA was separated in 15% denaturing acrylamide gel (19:1), 300 V, 2 h. The reaction products were visualized by Typhoon 9410 (Amersham Biosciences) and analyzed using ImageJ. Incorporation activity was determined as the combined intensity of the three incorporation bands (one, two and three nucleotide incorporation) relative to the combined intensity of the three incorporation bands plus the intensity of the substrate (i.e. ‘unrepaired’) band. The property of POLβ to enable detection of one, two and three nucleotide incorporations in the nucleotide incorporation assay has been described previously (39,40). ‘Activity’ determined from this assay is a measure of effectiveness of the extract at producing an elongated product from the substrate and may represent enzymatic activity on the substrate and/or ability of the enzyme to get to the DNA damage site.
Immunofluorescence
For 8-oxoG detection, Lmna+/+ and Lmna−/− MEFs were seeded into 4-chamber covered slides at 200 000 cells/well and incubated for 24 h. Exponentially growing cells were treated with 100 μM H2O2 in DMEM for 30 min and allowed to repair in complete media (DMEM with 10% FBS) for 5 min and 8 h; an untreated (UT) condition was also included. Preparation of stained slides was then carried out as described previously (41), including fixation with ice-cold 1:1 methanol–acetone for 20 min, and using anti-8-oxoG antibody (clone 2E2) (4354-MC; Trevigen). This technique involved denaturing with HCl that results in good nuclear staining but not staining mitochondrial DNA (42). The negative control was normal mouse IgG in place of the primary antibody (which is also mouse IgG). The slides were mounted with DAPI-containing vectashield (Vector laboratories) and analyzed using a Zeiss Axiovert 200M microscope. Signal intensity was determined using ImageJ software and displayed as the average intensity from 25 cells.
Genome-wide analysis of point mutation rates
Whole genomic DNA sequencing was performed by BGI (Copenhagen, Denmark). The strategy entailed short insert size library preparation, PE150 sequencing with 30X coverage (96 Gb clean data) per sample on BGISEQ (DNBseq tech). We isolated substitution mutations by selecting loci homozygous for reference A, C, G or T alleles (mm10, Genome Reference Consortium Mouse Build 38 (GCA_000001635.2)) in untreated Lmna−/− MEF and Lmna+/+ MEFs. To enhance sensitivity, we included loci with a sequencing depth of at least 40 reads in both the H2O2 and untreated samples. We also discarded base reads and alignments with Phred quality scores <30 (99.9% accuracy). We calculated the allele frequency of each base at each homozygous A, C, G or T locus in the Lmna−/− MEF and Lmna+/+ MEFs after a 3-week treatment with 50 μM H2O2 (relative to the allele frequencies in the untreated condition). To account for sequencing and alignment errors, we weighted base reads according to their associated base and alignment Phred quality scores. All source code used in the analysis is available at github.com/scheibye-knudsen-lab/genome-wide-point-mutation-rates.
Statistics
Graphs: statistical comparisons and graphing were performed using GraphPad Prism 5.04 software (La Jolla, CA, USA). Statistical significance was determined by two-tailed Student’s t-test, or, in the case of proliferation and survival assays, two-way ANOVA Sidak’s multiple comparisons test. ***P < 0.0001, **P <0.005, *P < 0.05 were considered as statistically significant. Genome-wide analysis of point mutation rates: unpaired t-test was used to compare the genome-wide allele frequencies of the H2O2-treated Lmna+/+ MEFs and the H2O2-treated Lmna−/− MEFs (relative to the mutation frequencies in the untreated condition).
RESULTS
Microarray analysis
We performed microarray gene expression analysis on Lmna−/− and wild-type (Lmna+/+) MEFs (Supplementary Figure S1). We found gene ontology (GO) terms that supported the premise that lamin A/C has effects on DNA repair processes, and that lamin A/C activities likely span a wider array of DNA repair processes than the already characterized role in double strand break repair (Table 1). The GO term ‘Metabolic Processes’ (the chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances) was highly downregulated. These include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation. GO terms ‘DNA Repair’ and ‘Mitochondrial Genome Maintenance’ (notably, BER is a robust DNA repair pathway in mitochondria due to constant oxidative stress on mitochondrial DNA) were upregulated. This potentially indicates higher DNA damage accumulation and consequent DDR. Also, the GO term ‘Aldehyde Dehydrogenase (ALDH) NAD(P)+ activity’ was downregulated; ALDH activity consumes NAD+ and thus other NAD+-dependent enzymes may be affected by or affect ALDH activity, such as PARylation by PARP1. PARP1 activation (PAR chain formation, i.e. PARylation) requires and depletes intracellular NAD+. A recent study has shown that PARP1 activation plays a role the ROS-induced inhibition of mitochondrial ALDH2 dehydrogenase (43).
Table 1.
Gene ontology (GO) terms that suggest enhanced DNA damage and DNA damage responses in Lmna null MEFs, including responses to DNA damage repaired by BER
| GO accession number | GO term | Z-score | P-value |
|---|---|---|---|
| GO0008152 | Metabolic process | -4.215669497 | 0.00052508 |
| GO0004030 | Aldehyde dehydrogenase NAD(P)+ activity | -3.387727434 | 0.0046726 |
| GO0006281 | DNA repair | 2.362503986 | 0.0011468 |
| GO0000002 | Mitochondrial Genome Maintenance* | 2.054294833 | 0.0024042 |
Microarray analyses were performed on control and Lmna knockout MEFs, followed by Gene Ontology (GO) analysis of the downregulated and upregulated genes. For each pathway z-score, a P-value was computed using JMP 6.0 software to test for the significance of the z-score obtained.
*This GO term points to BER since mitochondria are exposed to persistent oxidative stress, and BER has a key role in counteracting mitochondrial oxidative DNA damage.
The Ingenuity Pathway Analysis (IPA) identified Top Canonical Pathways (Table 2A), which is expanded in Supplementary Table S3 to show the IPA ‘molecules’ of each pathway. One of the pathways identified was ‘NRF2-mediated Oxidative Stress Response’. NRF2 is an important transcription factor that plays a significant role in protecting cells from endogenous and exogenous stresses and helps control ROS levels. Supplementary Table S4 shows that 17 out of 28 genes of the NRF2 IPA pathway are downregulated in the Lmna−/− MEFs. ‘Molecular Mechanisms of Cancer’ was also identified as a top canonical pathway, thus indicating that Lmna knockout has a strong relevance in cancer. IPA analysis also identified Top Upstream Regulators (Table 2B). The top five regulators are shown, all of which are known to be associated with tumor cells. TP53 (i.e. p53) was the highest on the list and predicted to be inhibited, thus suggesting that Lmna knockout negatively impacts expression of many genes downstream of this tumor suppressor protein.
Table 2A.
Ingenuity Top Canonical Pathways
| Name | P-value | Ratio |
|---|---|---|
| Pancreatic adenocarcinoma signaling | 1.01E-08 | 23/106 ( = 0.217) |
| Hepatic fibrosis / Hepatic stellate cell activation | 2.79E-08 | 32/197 ( = 0.162) |
| Aryl hydrocarbon receptor signaling | 1.40E-07 | 25/140 ( = 0.179) |
| Molecular mechanisms of cancer | 1.68E-06 | 43/365 ( = 0.118) |
| NRF2-mediated oxidative stress response* | 5.26E-06 | 26/180 ( = 0.144) |
IPA to determine top canonical pathways associated with Lmna knockout in MEFs. Statistical significance and the ratio of genes included in the pathway are shown. Target gene prediction and pathway enrichment analysis were performed using IPA; NRF2, nuclear factor erythroid 2-related factor 2.
*This pathway points to altered oxidative DNA damage (repaired by BER).
Table 2B.
Ingenuity top upstream regulators
| Upstream regulator | P-value of overlap | Predicted activation state |
|---|---|---|
| TP53* | 7.05E-35 | Inhibited |
| TGFB1 | 8.04E-33 | Inhibited |
| TNF | 2.44E-26 | |
| ERBB2 | 1.70E-25 | |
| HRAS | 5.35E-25 |
IPA to determine top upstream transcriptional regulators associated with Lmna knockout in MEFs. The overlap P-value measures whether there is a statistically significant overlap between the data set genes and the genes that are regulated by an upstream transcriptional regulator. It was calculated using Fisher’s Exact Test, and significance is attributed to P-values < 0.01. If the observed direction of change is mostly consistent with a particular activation state of the transcriptional regulator (‘activated’ or ‘inhibited’), then a prediction is made about that activation state, as shown; TP53, tumor protein p53; TGFB1, transforming growth factor β1; TNF, tumor necrosis factor; ERBB2, erb-b2 receptor tyrosine kinase 2; HRAS, HRas proto-oncogene, GTPase.
*Important tumor suppressor and upstream regulator of several DNA repair pathways, including BER.
Lamin A/C promotes base excision repair of oxidative lesions
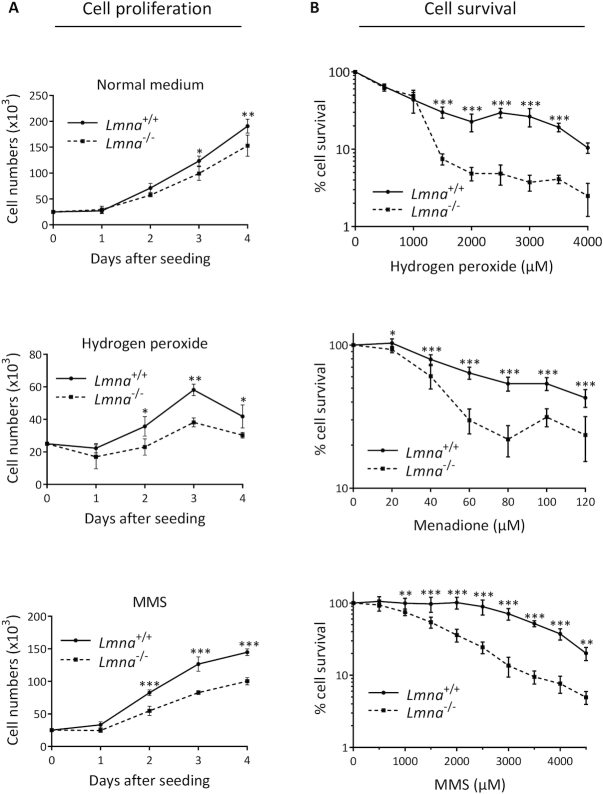
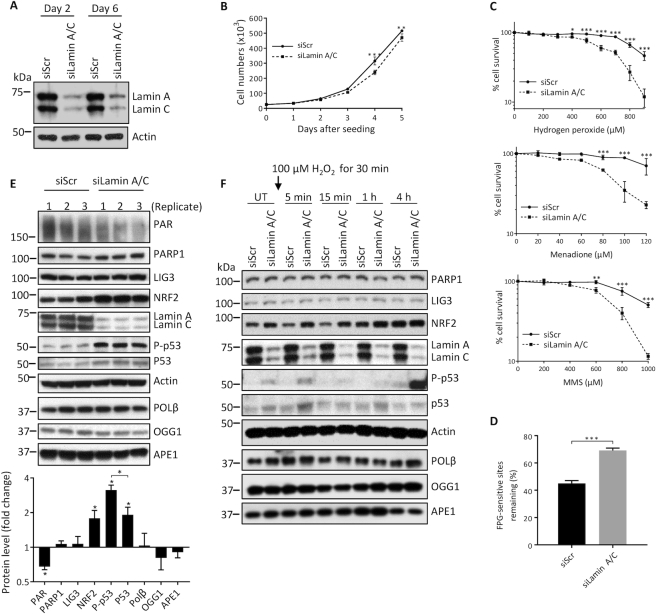
Studies suggest that lamins have a role in regulating cell proliferation (44), including as a component of the oxidative DDR (45). Thus, our early investigations into the role of lamin A/C in genome maintenance involved comparing cellular proliferation of Lmna−/− MEFs with Lmna+/+ MEFs. The Lmna−/− MEFs displayed reduced cellular proliferation, and this effect was enhanced when the cells were grown in low dose genotoxic stressors known to produce DNA damage that is repaired by BER (oxidative damage by H2O2 and alkylation damage by methyl methane sulfonate, (MMS)) (Figure 1A). It has been shown that MEFs accumulate more DNA damage when grown in 20% oxygen compared to the much lower oxygen tension of 3%, contributing to slower proliferation and earlier senescence (46,47). Thus, to verify our premise that Lmna−/− MEFs are more sensitive to genotoxic stress than Lmna+/+ MEFs and that this contributes to slower proliferation, we performed the growth curve in a 3% oxygen incubator, and found that the negative effect of lamin A/C removal on proliferation no longer occurs (Supplementary Figure S2). Since BER deficiency is also often reflected by poor cell survival after oxidative or alkylation DNA damage (41,48), we performed cell survival assays (colorimetric, using WST-1 reagent), and found that Lmna−/− MEFs were more sensitive to both oxidative and alkylation DNA damage, compared to Lmna+/+ MEFs (Figure 1B). Using an alternative approach for the oxidative (H2O2) damage survival analysis, based on propidium iodide staining to monitor cell viability, we obtained the same conclusion (Supplementary Figure S3).
Figure 1.
Loss of lamin A/C in MEFs results in slower cell proliferation and reduced cell survival. (A) Cell proliferation assays were performed by seeding 25 000 cells per well in 6-well plates, with our without a stressor that causes DNA damage known to be repaired by BER (hydrogen peroxide or MMS). The cells were incubated at standard 37°C/20% oxygen/5% CO2, and counted each day on a hemocytometer (n = 3, mean ± SD). (B) Cell survival was assessed by the WST-1 assay to estimate the relative number of viable cells at the indicated stressor concentrations. All data points are the mean WST-1 absorbance (as percent of the value from untreated cells) from six wells (in 96-well plates) ± SD. Cells were seeded at 20 000 cells per well, 24 h prior to treatment. Treatment durations for the stressors were as follows: H2O2, 4 h; Menadione, 2 h; MMS; 1 h. P values for were determined by two-way ANOVA Sidak’s multiple comparisons test; ***P < 0.0001; **P < 0.005; *P < 0.05
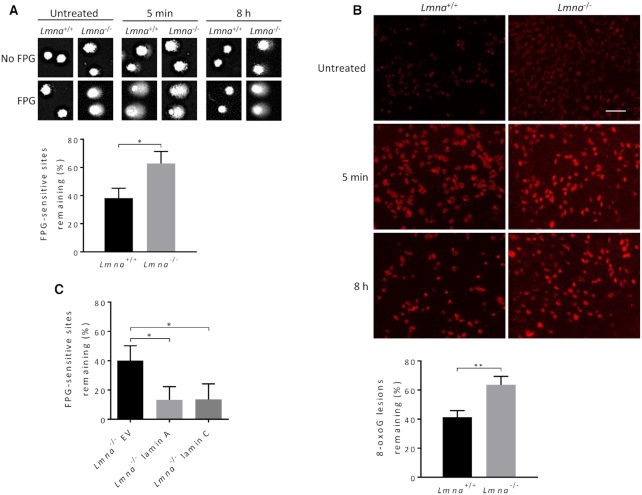
To examine directly the effect of Lmna knockout on DNA repair of oxidative DNA lesions, we performed the FPG-comet assay. This is a variant of the comet assay that incorporates the FPG enzyme to convert oxidative lesions (primarily 8-oxoG and the less abundant FapyG; i.e. FPG-sensitive sites) to DNA breaks (36). We measured the comet tail intensities (i.e. broken DNA) before and after (5 min, 8 h) exposure to H2O2. To determine comet tail intensities that estimate the amount of oxidative lesions, the mean comet tail DNA intensity generated from the non-FPG treated cells is subtracted from the mean comet tail intensity of the FPG-treated cells. The Lmna−/− MEFs had significantly less efficient DNA repair of FPG-sensitive sites, relative to Lmna+/+ MEFs, during the 8 h repair time (Figure 2A). Notably, the FPG-sensitive sites accumulate in the Lmna−/− MEFs even before the H2O2-treatment (Supplementary Figure S4A), presumably due to the oxygen stress that exists at standard 20% oxygen incubation. The mean comet tail intensities at each time point are shown in Supplementary Figure S4B. To support the comet assay data, we carried out immunofluorescence to monitor the level of the 8-oxoG lesions in nuclear DNA of Lmna−/− and Lmna+/+ MEFs, under the same conditions (using an antibody against the 8-oxoG DNA lesion). The Lmna−/− MEFs had significantly less efficient repair of 8-oxoG lesions, relative to Lmna+/+ MEFs (Figure 2B). The average 8-oxoG intensities at each time point are shown in Supplementary Figure S5. The staining from DAPI and negative control (normal mouse IgG) are shown in Supplementary Figure S6A. High magnification images, including brightfield (to show that the cytoplasm is not stained) are shown in Supplementary Figure S6B. In order to determine if both lamin A and C are promoting BER, we performed the FPG-comet assay (as usual comparing 5 min to 8 h repair after exposure to H2O2), this time using Lmna−/− MEFs stably expressing (via retroviral vector transfection) either lamin A or lamin C or empty vector. Both lamin A and lamin C significantly corrected the BER defect of the empty vector (EV)-Lmna−/− MEFs (Figure 2C; Supplementary Figure S7A and B). In conclusion, lamin A/C loss hinders the repair of oxidative DNA lesions, increasing cellular sensitivity to oxidative stress.
Figure 2.
Loss of lamin A/C in MEFs significantly impairs BER. (A) FPG-comet assay. The MEFs were treated with oxidative stress (100 μM H2O2 for 30 min) and then allowed to repair in fresh medium for 8 h. DNA repair efficiency was expressed as percent of FPG-sensitive sites remaining, after 8 h of repair, relative to 5 min of repair, after correction for the FPG-sensitive sites in untreated cells (n = 100 comet tails, mean ± SEM). (B) Immunofluorescence, using the 8-oxoG antibody, was used to monitor the removal of 8-oxoG lesions after 8 h of repair from H2O2 treatment (100 μM H2O2 for 30 min), relative to 5 min repair, after correction for 8-oxoG accumulation in untreated cells; images from staining with DAPI (to reveal nuclei) and with normal mouse IgG (negative control) are shown in Supplementary Figure S6A. Bar represents 10 μm. Data are expressed as mean fluorescence of 25 cells ± SEM. (C) FPG-comet assays on Lmna−/− MEFs with forced expression of empty vector (Lmna−/− EV), lamin A (Lmna−/− lamin A), or lamin C (Lmna−/− lamin C); n = 100 comet tails, mean ± SEM. P values were determined by Student’s t-test; **P < 0.005; *P < 0.05.
Several BER genes display reduced protein and mRNA levels in Lmna−/− MEFs
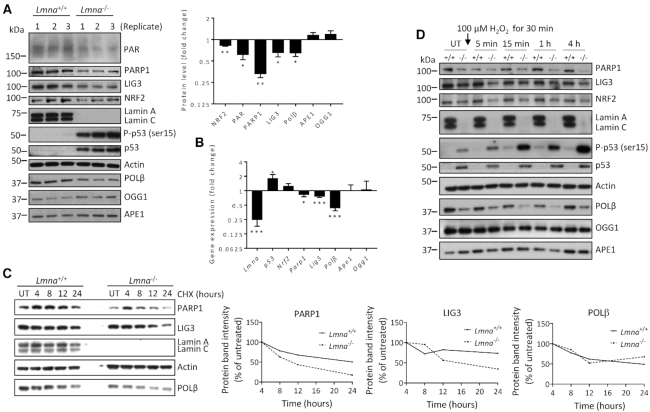
We next compared the levels of several core BER proteins in Lmna−/− MEFs relative to Lmna+/+ MEFs, via western blotting analysis of triplicate lysates (Figure 3A). Given the above data from IPA, we also examined the levels of the p53 and serine 15 phosphorylated (activated) p53, herein referred to as p-p53 (ser15), as well as transcription factor NRF2 (a master regulator of antioxidant response). Also, studies suggest that the accumulation of PAR chains (PARylation), especially on PARP1 itself, may promote BER (27) and therefore we also examined PAR levels. We found that the Lmna−/− MEFs displayed lower levels of PAR, PARP1, LIG3, POLβ and NRF2 proteins, and enhanced p53 and p-p53 (ser15) levels. Since lamins are major factors in the structural organization and function of the chromatin (49), it is likely that the altered protein levels are due to modified gene expression. Indeed, qPCR analysis (Figure 3B) revealed that Parp1, Lig3 and Polβ mRNA expression are reduced, and p53 mRNA expression is enhanced, in Lmna−/− MEFs, thus showing that regulation of the levels of these proteins by lamin A/C occurs at the level of transcription. Unlike its protein level, the Nrf2 mRNA expression was not significantly altered. This is likely because NRF2 level is largely regulated by proteasomal degradation (50). Protein and mRNA levels from the Ogg1 and Ape1 genes were not significantly affected by Lmna knockout.
Figure 3.
The expression and stability of specific BER-associated proteins are reduced in Lmna−/− MEFs, relative to Lmna+/+ MEFs. (A) Western blotting was performed on MEF lysates prepared in three separate experiments. Band intensities were quantitated using ImageJ. Bars represent the mean intensity of the proteins bands displayed as fold change in Lmna−/− MEFs (relative to Lmna+/+ MEFs) (n = 3, mean ± SEM). Band intensities of p53 and p-p53 (ser15) from Lmna+/+ MEF lysates were too low for quantification. (B) Real-time qPCR was performed on the indicated genes, in three separate experiments, displayed as fold change in Lmna−/− MEFs (relative to Lmna+/+ MEFs) ± SEM. (C) Cycloheximide chase assay. Cycloheximide (CHX) was added to the MEFs at a concentration of 50 μg/ml and cells collected for lysate preparation (and western blotting) at the indicated time points. (D) Western blotting was performed on lysates prepared from the Lmna+/+ and Lmna−/− MEFs that were either untreated (UT) or exposed to 100 μM H2O2 for 30 min, and left to repair for the indicated times in fresh medium. P values were determined by Student’s t-test; ***P < 0.0001; **P < 0.005; *P < 0.05; +/+, Lmna+/+; -/-, Lmna−/−.
Since there was only a small decrease in Parp1 and Lig3 mRNA expression (compared to Polβ) due to the Lmna knockout, we tested for reduced protein stability as a potential contributing factor in the observed reduction of protein levels, using the protein synthesis inhibitor cycloheximide (CHX) (Figure 3C). We found that PARP1 and LIG3 have reduced protein stability in the Lmna−/− MEFs; the Polβ protein stability was not changed. Interestingly, one can clearly see on the western blot that the PARP1 protein level was markedly elevated at the first time point (4-h CHX treatment; relative to the untreated condition). This may be a cellular stress response to the CHX, perhaps via transient blocking of ubiquitin/proteasomal degradation, early in CHX treatment (51). To circumvent this issue, the plots all began at the 4-h time point. This experiment suggests that altered protein stability of PARP1 and LIG3 may contribute to the reduced BER efficiency in Lmna−/− MEFs.
We then examined the effect of H2O2 treatment on the levels of the BER-associated proteins in Lmna−/− relative to Lmna+/+ MEFs (Figure 3D and Supplementary Figure S8). Consistent with above data, PARP1, LIG3, POLβ and NRF2 showed lower levels in Lmna−/− MEFs in all conditions tested; the relative levels of each of these proteins, as well as OGG1 and p53, did not change in any consistent manner during the repair period. There was a robust increase in p-p53 (ser15) levels in the Lmna−/− MEFs during the DNA repair period that became most intense at 4 h, thus suggesting a strong genotoxic stress response, consistent with the DDR function of p-p53 (ser15) (52). There was a slight increase in APE1 in the Lmna+/+ MEFs (such that the level was now higher than in Lmna−/− MEFs) starting immediately after treatment that was maintained but not enhanced further during the repair period; this would be predicted to benefit BER in Lmna+/+ MEFs.
Lamin A/C promotes BER by enhancing APE1 DNA incision and POLβ nucleotide incorporation in a PARylation-associated manner
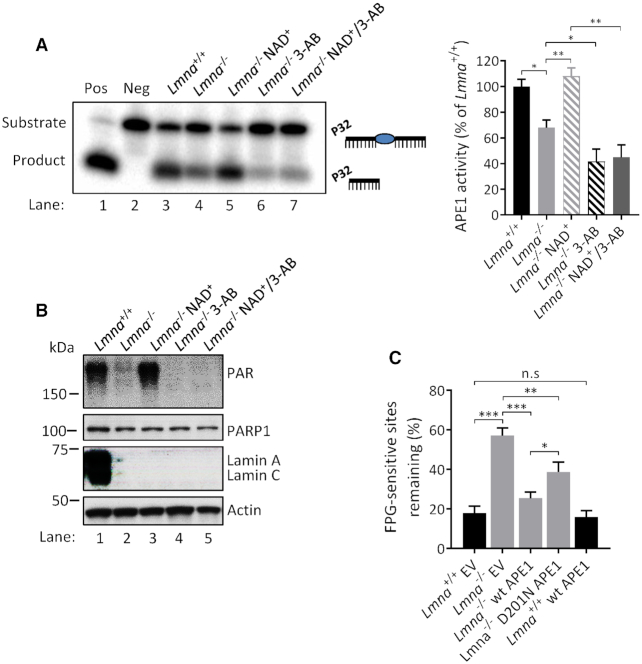
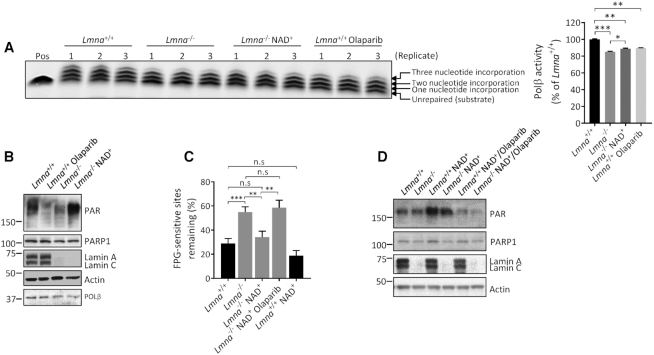
To understand in depth how lamin A/C regulates the response to oxidative DNA damage, we investigated the effect of Lmna knockout on the DNA AP-site incision activity of APE1 and the nucleotide incorporation (DNA polymerization) activity of POLβ. ‘Activity’ determined from these assay is a measure of the effectiveness of the extracts at producing an incision product (by APE1) or an elongated product (by POLβ) on the substrate, and may represent enzymatic activity (quality or activation state of the enzyme) on the substrate and/or the ability of the enzymes to get to the DNA damage site. Previous research suggests that APE1 DNA incision activity coordinates with POLβ nucleotide incorporation activity in sequential reactions in BER to facilitate an optimal rate of DNA repair (24,53,54). Moreover, examining the repair of AP sites is particularly important due to their potential cytotoxicity and mutagenicity (55). We also looked for potential regulation of these two BER activities by PARylation, since previous data suggest that this post-translational process may promote BER at the level of the APE1-mediated DNA incision step (27,56) and the POLβ-mediated DNA polymerization step (27,54,56,57). We first verified that media supplementation with the PARylation substrate NAD+ enhances PAR formation, as previous studies have shown (58,59). We did this by showing that the NAD+-induced PAR band on a western blot is reduced by inclusion of a PARP inhibitor [either 3-aminobenzamide (3-AB) or olaparib] together with the NAD+ (Supplementary Figure S9A and B). Our data (Figure 4A and B) showed that Lmna−/− MEFs have reduced APE1 incision activity and that this defect is corrected by addition of NAD+ into the culture medium (i.e. by boosting PAR formation). We also found that addition of 3-AB to the Lmna−/− MEFs (i.e. reducing PAR formation) reduced the APE1 DNA incision activity. To verify that the positive effects of NAD+ on DNA incision was at least in part due to its enhancement of PARylation (and not solely due to PARylation-independent effects of NAD+ on BER), we incubated the NAD+-supplemented Lmna−/− MEFs together with 3-AB and found that the 3-AB significantly reduced the APE1 incision activity (to the same level as the Lmna−/− 3-AB condition) despite the presence of NAD+ (compare bars 3, 4 and 5, right panel).
Figure 4.
DNA incision activity of APE1 is reduced by Lmna knockout in MEFs in a PARylation-dependent manner. (A) APE1 DNA incision product formation from extracts of Lmna+/+ and Lmna−/− MEFs and the effects of NAD+ (2 mM, 2 h) and/or PARP1 inhibitor 3-AB (10 mM, 2 h) media supplementation on product formation. A representative gel image is shown, as well as the APE1 activity quantitation based on three separate experiments (mean ± SD). APE1 DNA incision activity was determined as the intensity of product band relative to the combined intensities of substrate and product bands. (B) A representative western blot showing the PAR bands (PARylation) from lysates that were made at the same time as the extracts used for the APE1 DNA incision assay. (C) FPG-comet assay on MEFs overexpressing wild-type APE1, empty vector (EV) or DNA incision mutant D201N. The MEFs were treated with oxidative stress (100 μM H2O2) for 30 min and then allowed to repair for 8 h in fresh medium. DNA repair efficiency was expressed as percent of FPG-sensitive sites remaining, after 8 h of repair, relative to 5 min of repair, after correction for the FPG-sensitive sites in untreated cells (n = 100 comet tails, mean ± SEM); Pos, positive control (purified APE1 instead of lysate); Neg, negative control (no lysate); n.s, not significant. P values were determined by Student’s t-test; ***P < 0.0001; **P < 0.005; *P < 0.05.
We then tested if overexpression of wild-type APE1 in Lmna−/− MEFs has a better effect at correcting the BER defect than does overexpression of a DNA incision defective APE1 mutant (D210N), using the FPG-comet assay (Figure 4C and Supplementary Figure S10). The wild-type APE1 reversed the defect to near normal levels of DNA repair efficiency; the D201N APE1 DNA incision mutant caused some reversal of the BER defect (perhaps due to the redox activity of APE1 (60)), but to a significantly lesser extent than wild-type APE1. This demonstrates that the DNA-incising enzymatic activity, which was defective in the APE1 mutant, is an important component in BER rate. Notably, overexpression of wild-type APE1 in the Lmna+/+ MEFs did not enhance the DNA repair efficiency.
We also observed that POLβ BER activity (nucleotide incorporation effectiveness) was impaired in Lmna−/− MEFs and that PAR activation, by NAD+ supplementation, significantly reversed this defect (Figure 5A and B); as positive controls, olaparib-mediated inhibition of PARP1 and siRNA depletion of POLβ (Supplementary Figure S11) led to reduced POLβ nucleotide incorporation activity. We then performed the FPG-comet assay to test for the effect of NAD+ media supplementation (i.e. PARylation enhancement) on BER at the cell culture level. We found that NAD+ supplementation significantly corrected the impaired BER rate in the Lmna−/− MEFs and that inhibiting PAR formation (using PARP inhibitor olaparib), while maintaining NAD+ addition, significantly reversed the correction effect of NAD+ on BER efficiency (Figure 5C and D) thus confirming that PAR formation had a significant role in the correction of the BER defect in Lmna−/− MEFs. This comet assay experiment was repeated a second time, but using a different PARP inhibitor (3-AB); the same conclusion can be reached from that data (Supplementary Figure S12). In sum, these data suggest lamin A/C promotes BER efficiency by enhancing the DNA incision and nucleotide incorporation steps of BER, assisted by lamin A/C-boosted PARylation.
Figure 5.
DNA nucleotide incorporation activity of POLβ is reduced by Lmna knockout in MEFs, in a PARylation-dependent manner. (A) POLβ nucleotide incorporation product formation from extracts of Lmna+/+ and Lmna−/− MEFs and the effects of NAD+ (2 mM, 2 h) or PARP1 inhibitor olaparib (2 μM, 2 h) media supplementation on product formation. The gel image is shown, as well as quantitation of POLβ nucleotide incorporation activity based on three separate experiments (mean ± SD). Incorporation activity was determined as the combined intensity of the three incorporation bands (one, two and three nucleotide incorporation) relative to the combined intensity of the three incorporation bands plus the intensity of the substrate (i.e. ‘unrepaired’) band (n = 3, mean ± SD). (B) A representative western blot showing the PAR bands (PARylation) from lysates that were made at the same time as the extracts used for the POLβ nucleotide incorporation assay. (C) FPG-comet assay in which the MEFs were treated with oxidative stress (100 μM H2O2) for 30 min and then allowed to repair for 8 h in either normal medium or medium supplemented with NAD+ (2 mM) or NAD+ plus PARP1 inhibitor olaparib (2 μM); treatment duration for NAD+ and olaparib was 2 h and continued for the duration of the repair period. DNA repair efficiency was expressed as percent of FPG-sensitive sites remaining, after 8 h of repair, relative to 5 min of repair, after correction for the FPG-sensitive sites in untreated cells (n = 100 comet tails, mean ± SEM). (D) Western blotting shows the PAR levels under each condition in the FPG-comet assay, at 1 h into the repair period. Pos, positive control (purified POLβ instead of lysate); n.s, not significant. P values were determined by Student’s t-test; ***P < 0.0001; **P < 0.005; *P < 0.05.
Lmna −/− MEFs accumulate enhanced levels of transition mutations from chronic oxidative stress
Defective DNA repair of lesions can lead to mutations in the genome that can be inherited to daughter cells. When defective DNA repair leads to mutations or chromosomal aberrations affecting oncogenes and tumor suppressor genes, cells undergo malignant transformation resulting in cancerous growth. The GC>TA transversion mutation (i.e. C>A) is a marker of defective BER since it arises from 8-oxoG:A mismatches (61). Also, the GC>AT transition mutation (i.e. C>T) is an abundant genetic change induced as a consequence of chronic oxidative damage; this substitution commonly results from spontaneously hydrolytic deamination of oxidized cytosines (62–64). Both of these substitutions are known signatures of oxidative damage persistence (65,66). Thus, to support our findings that BER is defective in lamin A/C-depleted cells leading to enhanced oxidative damage, we isolated genomic DNA from the Lmna+/+ and Lmna−/− MEFs for whole genome deep sequencing and examined the frequency of these substitution markers. Our analysis of the sequenced data showed the enhanced accumulation of C>A transversions and C>T transitions under chronic oxidative stress in Lmna−/− MEFs, relative to Lmna+/+ MEFs treated in the same way (Table 3). These data support or finding in this report that lamin A/C promotes BER. The data also implicate lamin A/C in helping to suppress potentially carcinogenic mutations that arise from chronic oxidative stress.
Table 3.
Effect of Lmna deletion on known base substitution fingerprints of persistent oxidative damage
| Base substitution | Frequency (per 104 bases) | Fold change | |
|---|---|---|---|
| MEF Lmna+/+ | MEF Lmna−/− | ||
| C>A* | 0.58 | 0.63 | 1.07 |
| C>T** | 0.78 | 0.91 | 1.16 |
Mutational analysis of genomic DNA isolated from Lmna+/+ and Lmna−/− MEFs that were treated with 50 μM H2O2 for 3 weeks. Frequencies represent the extent of substitution mutations at each locus in MEFs treated with H2O2, relative to the untreated condition. Fold change represents the effect of Lmna deletion on mutation frequency in the treated cells. The substitution mutation frequencies in H2O2-treated Lmna−/− were significantly higher than the frequencies in H2O2-treated Lmna+/+ MEFs (P< 0.001, unpaired t-test).
*Base substitution that reflects the GC>TA transversion mutations that result from 8-oxoG:A mismatches.
**Base substitution that reflects GC>AT transition mutations that often result from oxidative deamination of cytosines.
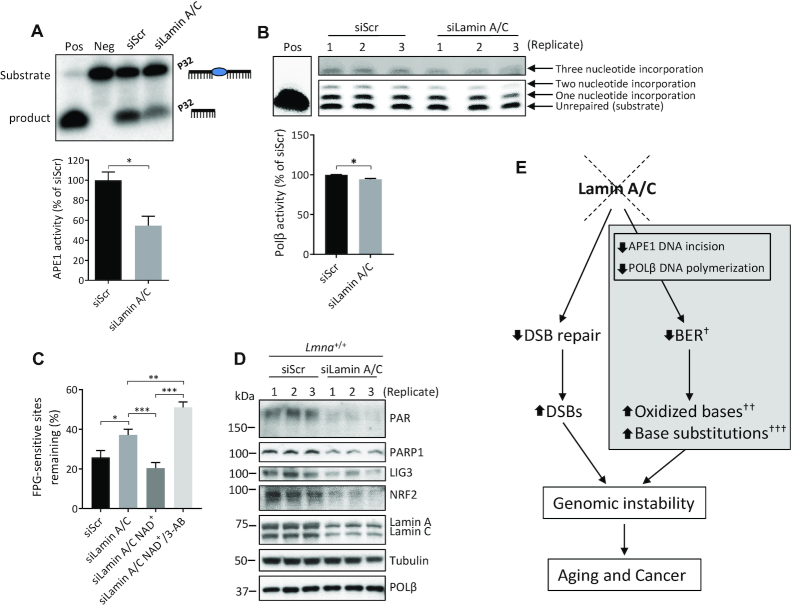
Lamin A/C depletion leads to impaired BER in U2OS cells
To assess if the BER defects we observed in Lmna−/− MEFs can be recapitulated in human cells, we repeated the above assays on lamin A/C siRNA-depleted (i.e. siLamin A/C) U2OS cells (Figures 6 and 7). The extent of knockdown in siLamin A/C cells compared to control cells (U2OS transfected with scrambled siRNA; i.e. siScr) at day 2 and day 6 post-transfection is shown in a representative western blot (Figure 6A). Like the MEFs, we found that, relative to siScr, the siLamin A/C U2OS cells displayed reduced cell proliferation (Figure 6B), enhanced sensitivity to oxidative and alkylation stress (Figure 6C), impaired BER efficiency (Figure 6D), lower PAR formation (Figure 6E), higher p53 and p-p53 (ser15) protein levels before and after H2O2 addition (Figure 6E and F), high activation of p-p53 (ser15) in siLamin A/C at 4h of repair (Figure 6F), reduced APE1 activity (Figure 7A), reduced POLβ activity (Figure 7B) and less efficient BER that is significantly corrected by PARylation (Figure 7C and Supplementary Figure S13). However, unlike the Lmna knockout in MEFs, the lamin A/C depletion in U2OS cells resulted in no change (instead of a reduction) in PARP1, LIG3 or POLβ protein levels, and a higher (instead of lower) level of NRF2 (Figure 6E). We have also demonstrated that lamin A/C depletion in another human cell line (HeLa) leads to less efficient BER (Supplementary Figure S14).
Figure 6.
BER is also impaired in U2OS cells that are siRNA-depleted of lamin A/C. (A) A representative western blot showing effective siRNA knockdown of lamin A/C in U2OS cells at the indicated days after transfection. (B) Cell proliferation assays were performed by seeding 25 000 cells per well in 6-well plates and counting the cells each day on a hemocytometer (n = 3, mean ± SD). (C) Cell survival was assessed by the WST-1 assay at the indicated stressor concentrations. All data points are the mean WST-1 absorbance (as percent of the value from untreated cells) from six wells (in 96-well plates) ± SD. Cells were seeded at 20 000 cells per well, 24 h prior to treatment. Treatment durations for the stressors were as follows: H2O2, 4 h; Menadione, 2 h; MMS; 1 h. (D) FPG-comet assay. DNA repair efficiency was expressed as percent of FPG-sensitive sites remaining after 8 h of repair, relative to 5 min of repair, after correction for the FPG-sensitive sites in untreated cells (n = 100 comet tails, mean ± SEM). (E) Western blotting was performed on lysates prepared from untreated siScr and siLamin A/C U2OS cells in triplicate. Fold change in protein levels from siLamin A/C cells relative to siScr cells was quantitated using ImageJ (n = 3, mean ± SD). (F) Western blotting was also performed at the indicated time points, pre- and post- treatment (100 μM H2O2 for 30 min). P values from the growth curves and survival assays were determined by two-way ANOVA Sidak’s multiple comparisons test. The other P values were determined by Student’s t-test. ***P < 0.0001; **P < 0.005; *P < 0.05.
Figure 7.
The APE1 DNA incision and POLβ nucleotide incorporation activities are also defective in the lamin A/C siRNA-depleted U2OS cells; moreover, the BER defect in these cells is PARylation dependent. (A) APE1 DNA incision product generation from extracts of siScr and siLamin A/C U2OS cells (representative image); incision activity was determined as the intensity of product band relative to the combined intensities of substrate and product bands (n = 3, mean ± SD). (B) POLβ nucleotide incorporation product formation from extracts of siScr and siLamin A/C U2OS cells (representative image); incorporation activity was determined as the combined intensity of the three incorporation bands (one, two and three nucleotide incorporation) relative to the combined intensity of the three incorporation bands plus the intensity of the substrate (i.e. ‘unrepaired’) band (n = 3, mean ± SD). (C) FPG-comet assay. DNA repair efficiency was expressed as percent of FPG-sensitive sites remaining after 8 h of repair, relative to 5 min of repair, after correction for the FPG-sensitive sites in untreated cells (n = 100 comet tails, mean ± SEM). Cell culture medium was supplemented with NAD+ (2 mM) or NAD+ together with PARP1 inhibitor 3-AB (10 mM), as indicated in the figure, for 2 h prior to H2O2 treatment and sustained for the duration of the repair period. (D) Western blot of triplicate lysates from Lmna+/+ MEFs transfected with either siScr or siLamin A/C siRNA. (E) Model for the role of lamin A/C in BER. New pathway insight from this study is shown in the gray shaded box. Loss of lamin A/C leads to reduction in APE1 DNA incision and POLβ nucleotide incorporation activities that negatively affect the rate of BER. This leads to accumulation of oxidized bases. Chronic exposure to oxidative stress leads to enhanced DNA base substitution mutations in lamin A/C-deficient cells that are signatures of oxidative damage persistence. As previously characterized, lamin A/C depletion also leads to defective double strand break (DSB) repair. Like the BER defect, this can lead to genomic instability and contribute to aging and cancer. †In Lmna−/− MEFs (but not in siLamin A/C U2OS cells) there was reduced expression of PARP1, LIG3 and POLβ; ††The other main two DNA lesion types that accumulate when BER rate is impaired are alkylated and deaminated bases; †††Tested in MEFs (but not in U2OS cells); Pos, positive control (purified APE1 or POLβ instead of lysate), Neg, negative control (no lysate). P values were determined by Student's t-test; ***P < 0.0001; **P < 0.005; *P < 0.05.
To assess if the reduced levels of PARP1, LIG3, POLβ and NRF2 that we see in Lmna−/− MEFs, but do not see in siLamin A/C U2OS cells, is due to the complete removal of lamin A/C (i.e. Lmna deletion), we carried out western blotting on triplicate lysates made from Lmna+/+-siLamin A/C MEFs (Figure 7D). We found that the levels of PARP1, LIG3 and NRF2 were reduced (as we saw in Lmna−/− MEFs); however, POLβ was unchanged. Thus, the reduced levels of PARP1, LIG3 and NRF2 that we see in Lmna−/− MEFs are not because of the complete removal of lamin A/C. A lamin A/C-dependent reduction of POLβ appears to require a more extensive reduction, or complete removal, of lamin A/C. Note that PAR was also reduced in the Lmna+/+-siLamin A/C MEFs, thus demonstrating that, like in the case of U2OS cells, complete removal of lamin A/C is not necessary to have a noticeable reduction in PAR formation.
DISCUSSION
Here we present evidence that lamin A/C depletion leads to deficient BER in MEFs and U2OS cells, effectuated, in part, by impaired APE1 and POLβ BER activities and reduced PARylation. Moreover, in the Lmna−/− MEFs (but not in the siLamin A/C U2OS cells) there was a reduction in the expression (mRNA and protein levels) of several core BER proteins (PARP1, LIG3 and POLβ) and the protein level of the redox-sensitive transcription factor NRF2. Notably, our IPA data identified ‘NRF-2 mediated oxidative stress response’ as a top canonical pathway in Lmna−/− MEFs. Our data also revealed that the lamin A/C-depleted MEFs and U2OS cells attempt to compensate for their DNA repair defects (i.e. respond to excessive DNA damage) via activation of p53 (in the form of phospho-p53 (ser15)), before, and to a larger degree after, H2O2 insult. In contrast to Lmna−/− MEFs, the NRF2 level in siLamin A/C U2OS cells is higher than in control U2OS cells, even before the addition of H2O2. Since the cells are growing at standard 20% oxygen, which is considered high oxygen tension, the higher levels of NRF2 in the siLamin A/C U2OS cells may be an oxidative stress response that entails enhanced NRF2 expression (and/or decreased NRF2 protein degradation) (67). In fact, a recent study used 20% oxygen as the high oxygen tension condition (relative to more biologically relevant 5% oxygen) to show that cultured bovine embryo survival under oxidative-stress conditions is associated with activation of the NRF2-mediated oxidative stress response pathway; specifically the expression levels of NRF2 and its downstream antioxidant genes were higher, whereas the expression of KEAP1 (which promotes proteasomal degradation of NRF2) was down-regulated, under 20% oxygen tension (68). In sum, there are differences in the levels of some DNA repair-associated proteins in our two lamin-depletion models; however, the common defects are impaired enzyme activity (of APE1 and POLβ) and reduced PARylation, which exist prior to the H2O2 treatment, thus rendering these cells not properly prepared for optimal BER. Future studies dealing with the impact of lamin A/C alterations on DNA repair could involve measuring another important BER activity of POLβ, namely its lyase activity for the removal of the 5′-dRP moiety (23).
Our data illustrate that the measured BER enzymatic activities can be impaired even without reduced levels of the respective protein (recall that APE1 in MEFs, and both APE1 and POLβ in U2OS cells, showed no reduction in protein level from lamin A/C depletion, despite having reduced BER enzymatic activities). An obvious potential mechanism by which lamin A/C can impact the APE1 and POLβ BER activities (under the in vitro assay conditions used in this study) is post-translational modification via interaction with lamin A/C or with other proteins whose expression or activity are modified by lamin A/C. Studies seeking to identify lamin A interactors in a human ORF library (69), and in a ‘One-STrEP-tag’ precipitation in MEFs (70), revealed no evidence for binding of lamin A to any of the core BER proteins. We performed co-immunoprecipitation analysis to test for physical interaction of lamin A/C with APE1, OGG1 or PARP1, in MEFs and U2OS cells; no such interactions were found (Supplementary Figure S15).
PARylation has been shown to occur on several DNA repair proteins and alter their functions (71,72). A recent report identifying PARylated proteins did not identify APE1 as a target of PAR. However, POLβ was identified as a PAR target (only under unstressed conditions: there was no significant increase in PAR binding after oxidative or alkylation stress) (72). Thus, it is possible that direct PAR covalent interaction with POLβ may contribute to the regulation of POLβ in vitro nucleotide incorporation activity. Other repair proteins associated with BER that were identified to be PARylated (and the PARylation was significantly increased after both oxidative and alkylation stress) were PARP1 (as expected, being the primary PAR target), XRCC1 and LIG3. Further research will be needed to determine to what extent and what direction (positive or negative), the PARylation of these BER-associated enzymes modulate their function, and if the identified PAR-BER protein regulations are effected by lamin A/C level.
Our comet assay using the D201N APE1 DNA incision mutant clearly showed that a contributing factor to the reduced BER rate in the Lmna−/− MEFs is impaired intrinsic enzymatic activity of the APE1 protein to incise the DNA. There are likely modifications (as discussed above) to APE1 and POLβ that affect their intrinsic enzymatic activity in lamin A/C-depleted cells that we have not characterized here. However, aside from impaired intrinsic enzymatic activity, the hindrance of these enzymes to properly reach and interact with their target sites may also contribute to the observed impairment of the in vitro activity (product formation) and BER rate. It is known that non-PARylated PARP1 physically hinders the ability of BER enzymes to their target sites and that PARP1-PARylation reduces this physical hindrance: specifically, the consensus from several studies is that PARP1 effectuates recruitment of BER proteins via physical association (including binding to XRCC1, DNA ligase III, DNA POLβ and OGG1) and that PARylation of PARP1 at DNA repair sites enhances BER by allowing PARP1 to dissociate from the repair site, to allow access of BER proteins (54,56,73–76). For example, Parsons et al. (56) demonstrated that inhibition of PARylation (by 3-AB) blocks dissociation of PARP1 from damaged DNA (specifically at AP sites) and prevents further DNA repair. We found here that PARylation promotes the in vitro BER activities of APE1 and POLβ (and the BER rate in cell culture) and that lamin A/C-depleted cells have reduced PARylation; this suggests a model in which the higher ratio of non-PARylated/PARylated PARP1 in lamin A/C-depleted cells impairs the recruitment and proper access of APE1 and POLβ BER enzymes to their repair sites.
Nuclear lamins play an important role in the control of gene expression, chromatin access to regulatory proteins (such as DNA repair proteins) and other nuclear functions through effects on the organization and regulation of chromatin (49,77). These regulations may occur via direct interaction of lamin A/C with chromatin or by interacting with factors that epigenetically modify the chromatin or that directly regulate replication or transcription (2,77). Lamin A/C regulation of chromatin may impact cell differentiation, cellular response to mechanical and genotoxic stresses, cell cycle progression and DNA repair (77–80). There is accumulating evidence that lamin A promotes chromatin decompaction (chromatin accessibility) (77), which can allow better access to repair proteins or transcription factors. PARylation of nucleosomes also causes relaxation of chromatin structure (81). The synthesis of PAR can be considered a marker of chromatin accessibility (82). Thus, the reduced levels of PAR that we observed in both Lmna−/− MEFs and siLamin A/C U2OS cells suggests a more compact chromatin in these cells compared to their control cells.
Our approach for activating PARylation by NAD+ supplementation into the medium of the lamin A/C-depleted cells is not a common approach, but we found it be very effective. Typically, researchers use a DNA damage agent, such as alkylating agent MNNG (83), to induce PARylation via DDR; however, that approach can activate multiple potentially confounding DDR pathways including DNA repair. Cell culture supplementation with NAD+ (84) or NAD+ precursor nicotinamide riboside (NR) (85), or feeding an Alzheimer’s mouse model with NR (86), has been used to show benefits of NAD+ in neurons. Previous work has shown that addition of the NAD+ precursor nicotinic acid into the medium of human peripheral blood mononuclear cells (87), and NAD+ supplementation to hepatocytes (59), can drive PARylation. Moreover, it has been demonstrated that NAD+ can penetrate into mouse fibroblasts to increase intracellular NAD+ levels (58). It is likely that some of the effects of NAD+ supplementation are unrelated to PARylation. For example, Wang et al. showed that NAD+ supplementation restores BER in oxygen-glucose deprived rat primary neurons by inhibiting serine-specific phosphorylation of APE1 and POLβ (84). However, our strategy of reversing the NAD+-mediated BER enhancement by including PARP1 inhibitor with the NAD+ showed that the stimulating effect of NAD+ on APE1 activity in vitro and overall BER (using comet assay) was at least in part due to PARylation.
Studies indicate that alterations in the levels of A-type lamins impact cancer progression (88), such as by associated alterations in mechanosensitivity (89–91) and nuclear rigidity (90,92) that affects cell migration and tumor growth. Lamin A/C had been implicated in DDR pathways (93,94); optimal DDR guards against genomic instability, activated oncogenes, and tumor progression (95). Cancer cells often have abnormalities in the DDR, including defects in cell cycle checkpoints and/or DNA repair, rendering them particularly sensitive to the induction of DSBs (96). Moreover, it is well established that the genotoxic DNA lesions that are repaired by BER contribute to the etiology of cancer (97,98). Studies have revealed that reduced BER rate is associated with cancer risk (20). Interestingly, BER enzymes are being pursued as potential biomarkers of cancer (99), and as targets (via pharmacological inhibition) to enhance chemosensitivity in cancer cells (97). Studies indicate that low APE1 level or enzymatic activity, and germline mutations in the APE1 gene, are associated with increased cancer risk, in accordance with a role in preventing genomic instability and mutations (100–102). Recent small-scale studies have shown that about one third of all human tumors express POLβ variant proteins; in fact, the POLβ colon cancer-associated mutants, K289M, has been shown to have impaired DNA synthesis (103). The above studies, combined with our findings in this study that lamin A/C depletion impairs APE1 DNA incision and POLβ DNA synthesis, points to lamin A/C as an upstream modulator of cancer via regulation of these APE1 and POLβ BER activities.
We also reported the novel finding that Lmna−/− MEFs accumulate transversions (GC>TA) and transitions (GC>AT) under chronic oxidative stress that are signatures of accumulated oxidative DNA damage that would result from defective BER. The studies that have shown these substitutions to be oxidative damage signature include Viel et al. (66), who used whole exome sequencing to identify a strongly conserved signature (C>A and C>T base substation) associated with 8-oxoG persistence in colorectal cancer tumor samples (CRCs) from several patients with MUTYH-Associated Polyposis (MAP) syndrome. Another study showed that accumulated 8-oxoG lesions lead to mainly GC>TA transversion (and to a lesser degree GC>AT transitions) in the inactive transgenic gpt gene in the kidney of Ogg1+/+ and Ogg1−/− mice that were administered a known oxidative agent (KBrO3) for 12 weeks (65). These reports, and our current mutational data, demonstrate the importance of mutational signatures as guides to determine the presence of altered DNA repair processes that can lead to genomic instability and eventually cancer. Several studies have used mutational signatures in this way to identify potential alterations in specific DNA repair pathway (104–106). Further research on the roles of lamins in cancer and DNA damage may lead to new therapies or identification of cancer biomarkers.
Previous studies have established a role for BER in neurodegenerative disease (107). BER dysfunction is a general feature of Alzheimer’s disease brains (108). However, lamin A levels are low in brain, and yet BER is quite proficient in normal brains. This might be explained by recent data showing that lamin C transcripts are present at relatively high levels in the brain (109,110). This suggests that lamin C is compensating for low lamin A in BER in normal brain. It has been demonstrated that gene targeted mice containing only lamin C, but no lamin A or pre-lamin A, are phenotypically indistinguishable from wild-type mice; the mice are healthy and the cells from the mice exhibited only minimal alterations in nuclear shape and deformability (111). This is consistent with our data in this report, in which we demonstrate that either lamin A or C can correct BER defect in the Lmna−/− MEFs; this finding is potentially important in better understanding brain BER, and the associated diseases that occur when brain BER is defective. The down-regulation of pre-lamin A expression in the brain could explain why children with Hutchinson Gilford Progeria syndrome (a premature aging disease caused by the toxic effects of a mutant form of lamin A) have a lack of neurological deficit, in contrast to the premature aging features that appear in many other tissues and organs.
As mentioned, other studies have shown that lamin A depletion lead to impaired DSB repair; defects in DSB repair have also been linked to cancer and aging (112) (Figure 7E). There is no reported defect in nucleotide excision repair (NER) or mismatch repair by lamin A depletion; however, a report has demonstrated that both global-and transcription coupled–NER are impaired in knockdown of lamin B1 in U2OS cells. The mechanism appears to involve down-regulation of several NER proteins including DDB1, CSB, and PCNA (113).
As schematically summarized in Figure 7E, data are presented showing that lamin A/C depletion leads to impaired BER, mediated in part by defective APE1 and POLβ BER activities, leading to the accumulation of oxidized bases and eventually to an increased frequency of base substitutions that are fingerprints of accumulated oxidative damage. These data add to previous data showing that lamin A/C depletion leads to defective DSB repair and genomic instability (114,115). Genomic instability can lead to cancer and aging (33,34). In this report, we uncovered additional influences that A-type lamins have on the preservation of genomic integrity, with implications for associated diseases. As the roles of lamins become increasingly characterized, including roles in DNA instability, opportunities develop to harness this knowledge for cancer therapeutics and biomarker development.
DATA AVAILABILITY
The microarray data sets used in this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE120389.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the scientists at the Gene Expression and Genomics Core of the Laboratory of Genetics and Genomics Unit within the National Institute on Aging Intramural Research Program for provided expert assistance in performing microarray experiments and analysis, specifically Dr Elin Lehrmann (for performing the microarray experiments, and for preliminary data analysis) and Dr Yongqing Zhang (for processing the microarray data. We thank Deborah Croteau and Apoliner Maya-Mendoza for critically reading the manuscript. We thank Thuan-Son T. Dinh and Ray Kreienkamp for their participation in the early stages of this project, producing preliminary data that helped direct the project design.
Author contributions: S.M., J.B. and V.A.B. designed the study. S.M., G.K., M.A., A.H., M.M. and M.S-K. performed experiments. M.B.E performed the mutation frequency analysis. S.M. wrote the manuscript. J.B., S.G., and V.A.B. helped develop the manuscript. All authors helped to interpret the data and commented on the manuscript.
Notes
Present address: Marya Morevati, University of Copenhagen, Nephrological Department P, Rigshospitalet, DK-2100 Copenhagen, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nordea-fonden; Danish Council for Independent Research; Intramural Program of the National Institute on Aging; National Institute of Health; Lundbeck Foundation; Danish Cancer Society. Funding for open access charge: NIH Intramural Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Burke B., Stewart C.L.. The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 2013; 14:13–24. [DOI] [PubMed] [Google Scholar]
- 2. Dechat T., Adam S.A., Taimen P., Shimi T., Goldman R.D.. Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2010; 2:a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bickmore W.A., van Steensel B.. Genome architecture: domain organization of interphase chromosomes. Cell. 2013; 152:1270–1284. [DOI] [PubMed] [Google Scholar]
- 4. Bonne G., Di Barletta M.R., Varnous S., Becane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A. et al.. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999; 21:285–288. [DOI] [PubMed] [Google Scholar]
- 5. Rankin J., Ellard S.. The laminopathies: a clinical review. Clin. Genet. 2006; 70:261–274. [DOI] [PubMed] [Google Scholar]
- 6. Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B.. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999; 147:913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Broers J.L., Ramaekers F.C.. The role of the nuclear lamina in cancer and apoptosis. Adv. Exp. Med. Biol. 2014; 773:27–48. [DOI] [PubMed] [Google Scholar]
- 8. Kong L., Schafer G., Bu H., Zhang Y., Zhang Y., Klocker H.. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 2012; 33:751–759. [DOI] [PubMed] [Google Scholar]
- 9. Stadelmann B., Khandjian E., Hirt A., Luthy A., Weil R., Wagner H.P.. Repression of nuclear lamin A and C gene expression in human acute lymphoblastic leukemia and non-Hodgkin's lymphoma cells. Leuk. Res. 1990; 14:815–821. [DOI] [PubMed] [Google Scholar]
- 10. Broers J.L., Raymond Y., Rot M.K., Kuijpers H., Wagenaar S.S., Ramaekers F.C.. Nuclear A-type lamins are differentially expressed in human lung cancer subtypes. Am. J. Pathol. 1993; 143:211–220. [PMC free article] [PubMed] [Google Scholar]
- 11. Sakthivel K.M., Sehgal P.. A novel role of lamins from genetic disease to cancer biomarkers. Oncol. Rev. 2016; 10:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gonzalo S. DNA damage and lamins. Adv. Exp. Med. Biol. 2014; 773:377–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hah J., Kim D.H.. Deciphering nuclear mechanobiology in laminopathy. Cells. 2019; 8:E231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chambliss A.B., Khatau S.B., Erdenberger N., Robinson D.K., Hodzic D., Longmore G.D., Wirtz D.. The LINC-anchored actin cap connects the extracellular milieu to the nucleus for ultrafast mechanotransduction. Sci. Rep. 2013; 3:1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu L., Luo Q., Sun J., Song G.. Nucleus and nucleus-cytoskeleton connections in 3D cell migration. Exp. Cell Res. 2016; 348:56–65. [DOI] [PubMed] [Google Scholar]
- 16. Aparicio T., Baer R., Gautier J.. DNA double-strand break repair pathway choice and cancer. DNA Repair. 2014; 19:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Redwood A.B., Perkins S.M., Vanderwaal R.P., Feng Z., Biehl K.J., Gonzalez-Suarez I., Morgado-Palacin L., Shi W., Sage J., Roti-Roti J.L. et al.. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011; 10:2549–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parnaik V.K., Manju K.. Laminopathies: multiple disorders arising from defects in nuclear architecture. J. Biosci. 2006; 31:405–421. [DOI] [PubMed] [Google Scholar]
- 19. Maynard S., Schurman S.H., Harboe C., de Souza-Pinto N.C., Bohr V.A.. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009; 30:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tudek B. Base excision repair modulation as a risk factor for human cancers. Mol. Aspects Med. 2007; 28:258–275. [DOI] [PubMed] [Google Scholar]
- 21. Keijzers G., Bakula D., Scheibye-Knudsen M.. Monogenic Diseases of DNA Repair. N. Engl. J. Med. 2017; 377:1868–1876. [DOI] [PubMed] [Google Scholar]
- 22. Sobol R.W., Prasad R., Evenski A., Baker A., Yang X.P., Horton J.K., Wilson S.H.. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000; 405:807–810. [DOI] [PubMed] [Google Scholar]
- 23. Srivastava D.K., Berg B.J., Prasad R., Molina J.T., Beard W.A., Tomkinson A.E., Wilson S.H.. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998; 273:21203–21209. [DOI] [PubMed] [Google Scholar]
- 24. Hill J.W., Hazra T.K., Izumi T., Mitra S.. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001; 29:430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennett R.A., Wilson D.M. III, Wong D., Demple B.. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dianov G.L., Hubscher U.. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013; 41:3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wei H., Yu X.. Functions of PARylation in DNA Damage Repair Pathways. Genomics Proteomics Bioinformatics. 2016; 14:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vida A., Marton J., Miko E., Bai P.. Metabolic roles of poly(ADP-ribose) polymerases. Semin. Cell Dev. Biol. 2017; 63:135–143. [DOI] [PubMed] [Google Scholar]
- 29. Vignier N., Chatzifrangkeskou M., Morales Rodriguez B., Mericskay M., Mougenot N., Wahbi K., Bonne G., Muchir A.. Rescue of biosynthesis of nicotinamide adenine dinucleotide protects the heart in cardiomyopathy caused by lamin A/C gene mutation. Hum. Mol. Genet. 2018; 27:3870–3880. [DOI] [PubMed] [Google Scholar]
- 30. Ghosh S., Liu B., Wang Y., Hao Q., Zhou Z.. Lamin A Is an Endogenous SIRT6 Activator and Promotes SIRT6-Mediated DNA Repair. Cell Rep. 2015; 13:1396–1406. [DOI] [PubMed] [Google Scholar]
- 31. Singh M., Hunt C.R., Pandita R.K., Kumar R., Yang C.R., Horikoshi N., Bachoo R., Serag S., Story M.D., Shay J.W. et al.. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol. Cell Biol. 2013; 33:1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen S., Martin C., Maya-Mendoza A., Tang C.W., Lovric J., Sims P.F., Jackson D.A.. Reduced expression of lamin A/C results in modified cell signaling and metabolism coupled with changes in expression of structural proteins. J. Proteome Res. 2009; 8:5196–5211. [DOI] [PubMed] [Google Scholar]
- 33. De Luca G., Ventura I., Sanghez V., Russo M.T., Ajmone-Cat M.A., Cacci E., Martire A., Popoli P., Falcone G., Michelini F. et al.. Prolonged lifespan with enhanced exploratory behavior in mice overexpressing the oxidized nucleoside triphosphatase hMTH1. Aging Cell. 2013; 12:695–705. [DOI] [PubMed] [Google Scholar]
- 34. Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D.E.. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gonzalez-Suarez I., Redwood A.B., Perkins S.M., Vermolen B., Lichtensztejin D., Grotsky D.A., Morgado-Palacin L., Gapud E.J., Sleckman B.P., Sullivan T. et al.. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009; 28:2414–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maynard S., Swistowska A.M., Lee J.W., Liu Y., Liu S.T., Da Cruz A.B., Rao M., de Souza-Pinto N.C., Zeng X., Bohr V.A.. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells. 2008; 26:2266–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J.L., Tadokoro T., Keijzers G., Mattson M.P., Bohr V.A.. Neurons efficiently repair glutamate-induced oxidative DNA damage by a process involving CREB-mediated up-regulation of apurinic endonuclease 1. J. Biol. Chem. 2010; 285:28191–28199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Akbari M., Visnes T., Krokan H.E., Otterlei M.. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair. 2008; 7:605–616. [DOI] [PubMed] [Google Scholar]
- 39. Asagoshi K., Liu Y., Masaoka A., Lan L., Prasad R., Horton J.K., Brown A.R., Wang X.H., Bdour H.M., Sobol R.W. et al.. DNA polymerase beta-dependent long patch base excision repair in living cells. DNA Repair. 2010; 9:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Akbari M., Pena-Diaz J., Andersen S., Liabakk N.B., Otterlei M., Krokan H.E.. Extracts of proliferating and non-proliferating human cells display different base excision pathways and repair fidelity. DNA Repair. 2009; 8:834–843. [DOI] [PubMed] [Google Scholar]
- 41. de Souza-Pinto N.C., Maynard S., Hashiguchi K., Hu J., Muftuoglu M., Bohr V.A.. The recombination protein RAD52 cooperates with the excision repair protein OGG1 for the repair of oxidative lesions in mammalian cells. Mol. Cell Biol. 2009; 29:4441–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohno M., Oka S., Nakabeppu Y.. Quantitative analysis of oxidized guanine, 8-oxoguanine, in mitochondrial DNA by immunofluorescence method. Methods Mol. Biol. 2009; 554:199–212. [DOI] [PubMed] [Google Scholar]
- 43. Wei S.J., Xing J.H., Wang B.L., Xue L., Wang J.L., Li R., Qin W.D., Wang J., Wang X.P., Zhang M.X. et al.. Poly(ADP-ribose) polymerase inhibition prevents reactive oxygen species induced inhibition of aldehyde dehydrogenase2 activity. Biochim. Biophys. Acta. 2013; 1833:479–486. [DOI] [PubMed] [Google Scholar]
- 44. Kennedy B.K., Pennypacker J.K.. RB and lamins in cell cycle regulation and aging. Adv. Exp. Med. Biol. 2014; 773:127–142. [DOI] [PubMed] [Google Scholar]
- 45. Shimi T., Goldman R.D.. Nuclear lamins and oxidative stress in cell proliferation and longevity. Adv. Exp. Med. Biol. 2014; 773:415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maynard S.P., Miller R.A.. Fibroblasts from long-lived Snell dwarf mice are resistant to oxygen-induced in vitro growth arrest. Aging Cell. 2006; 5:89–96. [DOI] [PubMed] [Google Scholar]
- 47. Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., Campisi J.. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 2003; 5:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim Y.J., Wilson D.M. 3rd.. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012; 5:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D.K., Solimando L., Goldman R.D.. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008; 22:832–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kensler T.W., Wakabayashi N., Biswal S.. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007; 47:89–116. [DOI] [PubMed] [Google Scholar]
- 51. Shenkman M., Tolchinsky S., Kondratyev M., Lederkremer G.Z.. Transient arrest in proteasomal degradation during inhibition of translation in the unfolded protein response. Biochem. J. 2007; 404:509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Loughery J., Cox M., Smith L.M., Meek D.W.. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 2014; 42:7666–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bennett R.A., Wilson D.M. 3rd, Wong D., Demple B.. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sukhanova M.V., Khodyreva S.N., Lebedeva N.A., Prasad R., Wilson S.H., Lavrik O.I.. Human base excision repair enzymes apurinic/apyrimidinic endonuclease1 (APE1), DNA polymerase beta and poly(ADP-ribose) polymerase 1: interplay between strand-displacement DNA synthesis and proofreading exonuclease activity. Nucleic Acids Res. 2005; 33:1222–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Demple B., Harrison L.. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994; 63:915–948. [DOI] [PubMed] [Google Scholar]
- 56. Parsons J.L., Dianova I.I., Allinson S.L., Dianov G.L.. Poly(ADP-ribose) polymerase-1 protects excessive DNA strand breaks from deterioration during repair in human cell extracts. FEBS J. 2005; 272:2012–2021. [DOI] [PubMed] [Google Scholar]
- 57. Horton J.K., Stefanick D.F., Prasad R., Gassman N.R., Kedar P.S., Wilson S.H.. Base excision repair defects invoke hypersensitivity to PARP inhibition. Mol. Cancer Res. 2014; 12:1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bruzzone S., Guida L., Zocchi E., Franco L., De Flora A.. Connexin 43 hemi channels mediate Ca2+-regulated transmembrane NAD+ fluxes in intact cells. FASEB J. 2001; 15:10–12. [DOI] [PubMed] [Google Scholar]
- 59. Pang J., Cui J., Gong H., Xi C., Zhang T.M.. Effect of NAD on PARP-mediated insulin sensitivity in oleic acid treated hepatocytes. J. Cell Physiol. 2015; 230:1607–1613. [DOI] [PubMed] [Google Scholar]
- 60. Rajapakse A., Suraweera A., Boucher D., Naqi A., O’Byrne K., Richard D.J., Croft L.V.. Redox regulation in the base excision repair pathway: old and new players as cancer therapeutic targets. Curr. Med. Chem. 2019; doi:10.2174/0929867326666190430092732. [DOI] [PubMed] [Google Scholar]
- 61. Shibutani S., Takeshita M., Grollman A.P.. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991; 349:431–434. [DOI] [PubMed] [Google Scholar]
- 62. An Q., Robins P., Lindahl T., Barnes D.E.. C → T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005; 24:2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Millar C.B., Guy J., Sansom O.J., Selfridge J., MacDougall E., Hendrich B., Keightley P.D., Bishop S.M., Clarke A.R., Bird A.. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002; 297:403–405. [DOI] [PubMed] [Google Scholar]
- 64. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 65. Arai T., Kelly V.P., Minowa O., Noda T., Nishimura S.. High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis. 2002; 23:2005–2010. [DOI] [PubMed] [Google Scholar]
- 66. Viel A., Bruselles A., Meccia E., Fornasarig M., Quaia M., Canzonieri V., Policicchio E., Urso E.D., Agostini M., Genuardi M. et al.. A Specific Mutational Signature Associated with DNA 8-Oxoguanine Persistence in MUTYH-defective Colorectal Cancer. EBioMedicine. 2017; 20:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Davies K.J. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life. 2000; 50:279–289. [DOI] [PubMed] [Google Scholar]
- 68. Amin A., Gad A., Salilew-Wondim D., Prastowo S., Held E., Hoelker M., Rings F., Tholen E., Neuhoff C., Looft C. et al.. Bovine embryo survival under oxidative-stress conditions is associated with activity of the NRF2-mediated oxidative-stress-response pathway. Mol. Reprod. Dev. 2014; 81:497–513. [DOI] [PubMed] [Google Scholar]
- 69. Dittmer T.A., Sahni N., Kubben N., Hill D.E., Vidal M., Burgess R.C., Roukos V., Misteli T.. Systematic identification of pathological lamin A interactors. Mol. Biol. Cell. 2014; 25:1493–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kubben N., Voncken J.W., Demmers J., Calis C., van Almen G., Pinto Y., Misteli T.. Identification of differential protein interactors of lamin A and progerin. Nucleus. 2010; 1:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Teloni F., Altmeyer M.. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016; 44:993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M.O., Nielsen M.L.. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 2013; 52:272–285. [DOI] [PubMed] [Google Scholar]
- 73. Leppard J.B., Dong Z., Mackey Z.B., Tomkinson A.E.. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell Biol. 2003; 23:5919–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Masson M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., de Murcia G.. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998; 18:3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. El-Khamisy S.F., Masutani M., Suzuki H., Caldecott K.W.. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003; 31:5526–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Noren Hooten N., Kompaniez K., Barnes J., Lohani A., Evans M.K.. Poly(ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1). J. Biol. Chem. 2011; 286:44679–44690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Naetar N., Ferraioli S., Foisner R.. Lamins in the nuclear interior - life outside the lamina. J. Cell Sci. 2017; 130:2087–2096. [DOI] [PubMed] [Google Scholar]
- 78. Oldenburg A., Briand N., Sorensen A.L., Cahyani I., Shah A., Moskaug J.O., Collas P.. A lipodystrophy-causing lamin A mutant alters conformation and epigenetic regulation of the anti-adipogenic MIR335 locus. J. Cell Biol. 2017; 216:2731–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Redwood A.B., Gonzalez-Suarez I., Gonzalo S.. Regulating the levels of key factors in cell cycle and DNA repair: new pathways revealed by lamins. Cell Cycle. 2011; 10:3652–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zuela N., Bar D.Z., Gruenbaum Y.. Lamins in development, tissue maintenance and stress. EMBO Rep. 2012; 13:1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Poirier G.G., de Murcia G., Jongstra-Bilen J., Niedergang C., Mandel P.. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. U.S.A. 1982; 79:3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Khoronenkova S.V., Dianova I.I., Parsons J.L., Dianov G.L.. USP7/HAUSP stimulates repair of oxidative DNA lesions. Nucleic Acids Res. 2011; 39:2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Keil C., Grobe T., Oei S.L.. MNNG-induced cell death is controlled by interactions between PARP-1, poly(ADP-ribose) glycohydrolase, and XRCC1. J. Biol. Chem. 2006; 281:34394–34405. [DOI] [PubMed] [Google Scholar]
- 84. Wang S., Xing Z., Vosler P.S., Yin H., Li W., Zhang F., Signore A.P., Stetler R.A., Gao Y., Chen J.. Cellular NAD replenishment confers marked neuroprotection against ischemic cell death: role of enhanced DNA repair. Stroke. 2008; 39:2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Brown K.D., Maqsood S., Huang J.Y., Pan Y., Harkcom W., Li W., Sauve A., Verdin E., Jaffrey S.R.. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014; 20:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hou Y., Lautrup S., Cordonnier S., Wang Y., Croteau D.L., Zavala E., Zhang Y., Moritoh K., O’Connell J.F., Baptiste B.A. et al.. NAD(+) supplementation normalizes key Alzheimer's features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E1876–E1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Weidele K., Kunzmann A., Schmitz M., Beneke S., Burkle A.. Ex vivo supplementation with nicotinic acid enhances cellular poly(ADP-ribosyl)ation and improves cell viability in human peripheral blood mononuclear cells. Biochem. Pharmacol. 2010; 80:1103–1112. [DOI] [PubMed] [Google Scholar]
- 88. Irianto J., Pfeifer C.R., Ivanovska I.L., Swift J., Discher D.E.. Nuclear lamins in cancer. Cell Mol. Bioeng. 2016; 9:258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lammerding J., Schulze P.C., Takahashi T., Kozlov S., Sullivan T., Kamm R.D., Stewart C.L., Lee R.T.. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004; 113:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.W., Tewari M. et al.. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013; 341:1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Broders-Bondon F., Nguyen Ho-Bouldoires T.H., Fernandez-Sanchez M.E., Farge E.. Mechanotransduction in tumor progression: The dark side of the force. J. Cell Biol. 2018; 217:1571–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Harada T., Swift J., Irianto J., Shin J.W., Spinler K.R., Athirasala A., Diegmiller R., Dingal P.C., Ivanovska I.L., Discher D.E.. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 2014; 204:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gibbs-Seymour I., Markiewicz E., Bekker-Jensen S., Mailand N., Hutchison C.J.. Lamin A/C-dependent interaction with 53BP1 promotes cellular responses to DNA damage. Aging Cell. 2015; 14:162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gonzalez-Suarez I., Gonzalo S.. Nurturing the genome: A-type lamins preserve genomic stability. Nucleus. 2010; 1:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bartek J., Bartkova J., Lukas J.. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007; 26:7773–7779. [DOI] [PubMed] [Google Scholar]
- 96. Huhn D., Bolck H.A., Sartori A.A.. Targeting DNA double-strand break signalling and repair: recent advances in cancer therapy. Swiss Med. Wkly. 2013; 143:w13837. [DOI] [PubMed] [Google Scholar]
- 97. Visnes T., Grube M., Hanna B.M.F., Benitez-Buelga C., Cazares-Korner A., Helleday T.. Targeting BER enzymes in cancer therapy. DNA Repair. 2018; 71:118–126. [DOI] [PubMed] [Google Scholar]
- 98. Scott T.L., Rangaswamy S., Wicker C.A., Izumi T.. Repair of oxidative DNA damage and cancer: recent progress in DNA base excision repair. Antioxid. Redox Signal. 2014; 20:708–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gossage L., Perry C., Abbotts R., Madhusudan S.. Base excision repair factors are promising prognostic and predictive markers in cancer. Curr. Mol. Pharmacol. 2012; 5:115–124. [DOI] [PubMed] [Google Scholar]
- 100. Park J.S., Kim H.L., Kim Y.J., Weon J.I., Sung M.K., Chung H.W., Seo Y.R.. Human AP endonuclease 1: a potential marker for the prediction of environmental carcinogenesis risk. Oxid. Med. Cell Longev. 2014; 2014:730301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Sevilya Z., Leitner-Dagan Y., Pinchev M., Kremer R., Elinger D., Lejbkowicz F., Rennert H.S., Freedman L.S., Rennert G., Paz-Elizur T. et al.. Development of APE1 enzymatic DNA repair assays: low APE1 activity is associated with increase lung cancer risk. Carcinogenesis. 2015; 36:982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sweasy J.B., Lang T., DiMaio D.. Is base excision repair a tumor suppressor mechanism. Cell Cycle. 2006; 5:250–259. [DOI] [PubMed] [Google Scholar]
- 103. Starcevic D., Dalal S., Sweasy J.B.. Is there a link between DNA polymerase beta and cancer. Cell Cycle. 2004; 3:998–1001. [PubMed] [Google Scholar]
- 104. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L. et al.. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Galanos P., Pappas G., Polyzos A., Kotsinas A., Svolaki I., Giakoumakis N.N., Glytsou C., Pateras I.S., Swain U., Souliotis V.L. et al.. Mutational signatures reveal the role of RAD52 in p53-independent p21-driven genomic instability. Genome Biol. 2018; 19:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ma J., Setton J., Lee N.Y., Riaz N., Powell S.N.. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun. 2018; 9:3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Akbari M., Morevati M., Croteau D., Bohr V.A.. The role of DNA base excision repair in brain homeostasis and disease. DNA Repair. 2015; 32:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Weissman L., Jo D.G., Sorensen M.M., de Souza-Pinto N.C., Markesbery W.R., Mattson M.P., Bohr V.A.. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007; 35:5545–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jung H.J., Tu Y., Yang S.H., Tatar A., Nobumori C., Wu D., Young S.G., Fong L.G.. New Lmna knock-in mice provide a molecular mechanism for the ‘segmental aging’ in Hutchinson-Gilford progeria syndrome. Hum. Mol. Genet. 2014; 23:1506–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jung H.J., Coffinier C., Choe Y., Beigneux A.P., Davies B.S., Yang S.H., Barnes R.H. 2nd, Hong J., Sun T., Pleasure S.J. et al.. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E423–E431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fong L.G., Ng J.K., Lammerding J., Vickers T.A., Meta M., Cote N., Gavino B., Qiao X., Chang S.Y., Young S.R. et al.. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J. Clin. Invest. 2006; 116:743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Aguilera A., Garcia-Muse T.. Causes of genome instability. Annu. Rev. Genet. 2013; 47:1–32. [DOI] [PubMed] [Google Scholar]
- 113. Butin-Israeli V., Adam S.A., Goldman R.D.. Regulation of nucleotide excision repair by nuclear lamin b1. PLoS One. 2013; 8:e69169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gonzalez-Suarez I., Redwood A.B., Gonzalo S.. Loss of A-type lamins and genomic instability. Cell Cycle. 2009; 8:3860–3865. [DOI] [PubMed] [Google Scholar]
- 115. Redwood A.B., Perkins S.M., Vanderwaal R.P., Feng Z., Biehl K.J., Gonzalez-Suarez I., Morgado-Palacin L., Shi W., Sage J., Roti-Roti J.L. et al.. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011; 10:2549–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data sets used in this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE120389.