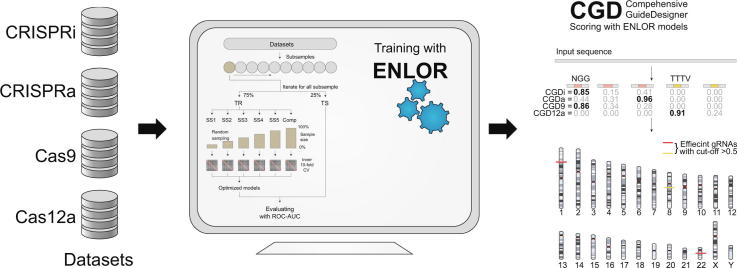
Graphical abstract
Keywords: CRISPR system, Cas9, Cas12a, dCas9, gRNA design, Machine learning, Logistic regression
Abstract
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas systems, including dead Cas9 (dCas9), Cas9, and Cas12a, have revolutionized genome engineering in mammalian somatic cells. Although computational tools that assess the target sites of CRISPR-Cas systems are inevitably important for designing efficient guide RNAs (gRNAs), they exhibit generalization issues in selecting features and do not provide optimal results in a comprehensive manner. Here, we introduce a Comprehensive Guide Designer (CGD) for four different CRISPR systems, which utilizes the machine learning algorithm, Elastic Net Logistic Regression (ENLOR), to autonomously generalize the models. CGD contains specific models trained with public datasets generated by CRISPRi, CRISPRa, CRISPR-Cas9, and CRISPR-Cas12a (designated as CGDi, CGDa, CGD9, and CGD12a, respectively) in an unbiased manner. The trained CGD models were benchmarked to other regression-based machine learning models, such as ElasticNet Linear Regression (ENLR), Random Forest and Boruta (RFB), and Extreme Gradient Boosting (Xgboost) with inbuilt feature selection. Evaluation with independent test datasets showed that CGD models outperformed the pre-existing methods in predicting the efficacy of gRNAs. All CGD source codes and datasets are available at GitHub (https://github.com/vipinmenon1989/CGD), and the CGD webserver can be accessed at http://big.hanyang.ac.kr:2195/CGD.
1. Introduction
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) system is an adaptive defense mechanism employed by bacteria and archaea against invading viruses or foreign plasmids [1]. Reprogramming of these defense mechanisms to generate gene knockouts [2] has revolutionized the use of the CRISPR system in various applications, from medicine to crop seed enhancement [3]. In human genetics, in particular, CRISPR-based functional screening approaches have become popular in genome-wide studies [4], [5], [6], [7]. CRISPR-based functional screening systems can be divided into three categories: dead Cas9(dCas9)-mediated knockdown (CRISPRi) and activation (CRISPRa) [5], Cas9-mediated knockout [4], [6], and Cas12a-mediated knockout [7].
Previous studies evaluated thousands of guide RNAs (gRNAs), assigning them efficiency scores that were utilized to develop computational models such as CRISPRko v1 [8], SSC [9], and CRISPRko v2 [10] of effective target sites. However, these gRNA datasets appear to display a skewed distribution, according to the selection methods and the essentiality of the target genes, causing biased training and overfitting. To design efficient gRNAs for generating DNA double strand breaks, a large-scale dataset of gRNAs and their associated indel ratios are required. Recently, CINDEL [7], CRISPR-DT [11], DeepCpf1 [12], and DeepCas9 [13] studies have produced high-throughput datasets of gRNA sequences with efficiencies to train their own models. In the process of model development, diverse features, including sequences as well as the epigenetic status of target sites, were incorporated to improve these models. However, the algorithms utilized to build these models were selected without proper benchmarking or generalization in selecting features. Due to these limitations, the predictive ability of a given model is less effective across all CRISPR systems. Thus, an optimal algorithm suitable for the all of the datasets is necessary.
In this study, we developed the Comprehensive Guide Designer (CGD), a consortium of regression-based machine learning models for CRISPRi, CRISPRa, Cas9, and Cas12a. Our Elastic Net Logistic Regression (ENLOR) models, suitable for tackling the generalization problem over datasets generated by the respective CRISPR-Cas systems, outperformed other previous models. Therefore, CGD could provide useful guidelines for selecting effective target sites and assist users in designing more efficient gRNAs.
2. Materials and methods
2.1. Workflow of CGD
CGD integrates our four predictive models for gRNAs, designated as CGDi, CGDa, CGD9, and CGD12a, built based on ENLOR. The step-by-step process for designing gRNAs with high target efficiency is shown in Fig. 1.
Fig. 1.
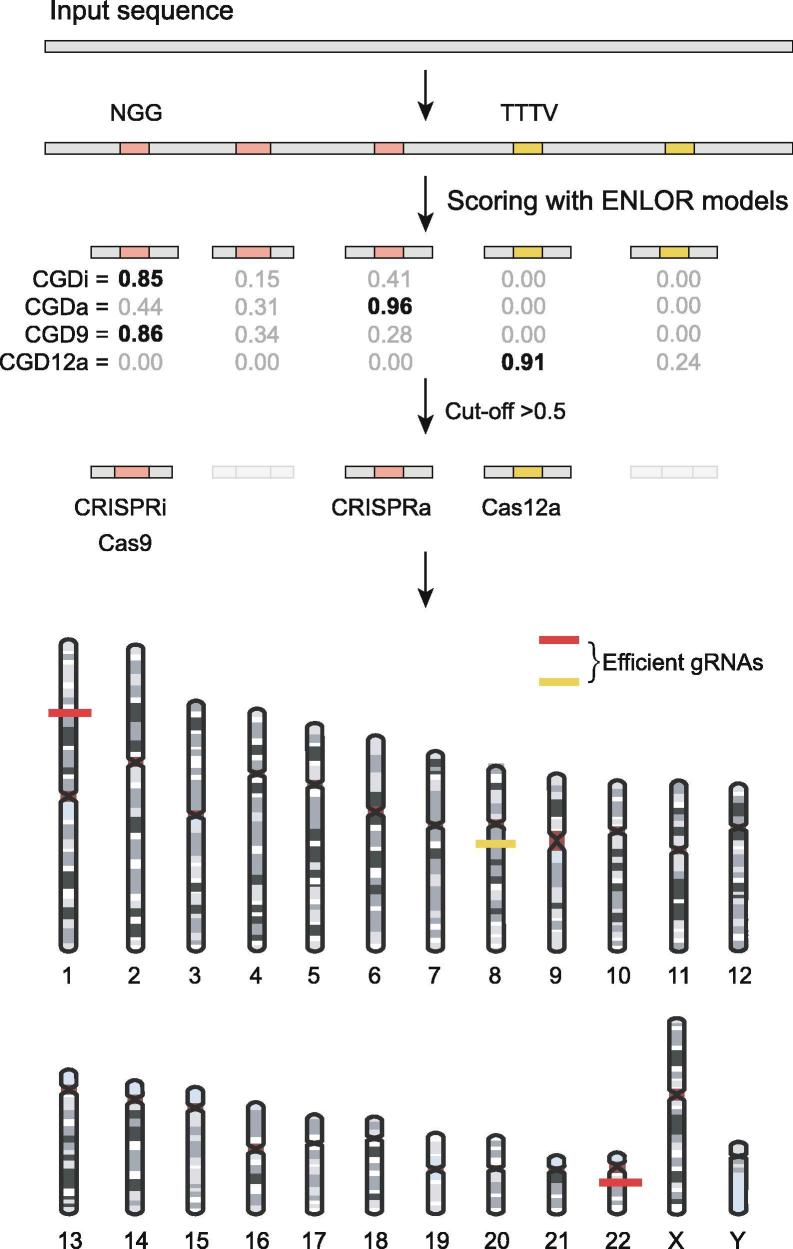
Schematic CGD workflow. CGD processes the input DNA sequence (genome/contig/chromosome) in FASTA format, calculates scores for all possible target sites and corresponding gRNAs, and displays all efficient gRNAs and their target sites with their scores.
2.1.1. Input sequence
An input DNA sequence(s) of a genome, - contig or chromosome is required. The minimum and maximum lengths of the input DNA sequence are 100 nt and 10,000 nt, respectively. Multiple input sequences can be submitted in a FASTA format.
2.1.2. PAM identification and segregation of gRNA sequences
CGD predicts gRNA sequences based on the PAM motifs of the respective nuclease (NGG for Cas9, CRISPRi and CRISPRa, and TTTV for Cas12a).
2.1.3. Scoring of gRNA sequences
The score of each gRNA sequence is calculated based on weights estimated by the ENLOR algorithm for each CRISPR system. The weights of features vary depending on the CRISPR system (Supplementary Table 1). The final score is generated by summing the of the features. The value of each feature was standardized with the Z score.
2.1.4. Potential target sites
The score range is [0, 1]; gRNAs with scores > 0.5 were selected as efficient gRNAs along with their target sequences and position.
2.2. Datasets
The datasets used in this study were either downloaded from our previous study [7] or other previous studies [4], [5], [6], [12], [14], [15], [16], [17], [18], [19], [20]. Because different activity scores, such as depletion, enrichment, or indel ratio, were applied to each dataset (which correspond to different CRISPR systems), the activity scores were standardized by Z scoring. For CRISPRi, CRISPRa, and Cas9 systems, genome-wide screening datasets [4], [5], [6], [21], [22] were used for training for several reasons, such as 1) a higher number of gRNAs targeted to each gene (6–20 gRNAs per gene) as well as a high number of targeted genes; 2) gRNAs targeted to different sites within genes, giving a comprehensive view of targeting efficiency; and 3) the incorporation of control gRNAs, which reduces false positives. For Cas12a, the high-throughput data, with 15,000 gRNAs sequences, generated by Kim et al. were used for training [12].
The datasets generated by different CRISPR-Cas systems (CRISPRi, CRISPRa, Cas9, and Cas12a) were categorized based on the essentiality and functions of the target genes (Supplementary Fig. 1 and Supplementary Table 2). If a dataset contained >100 gRNAs and associated activities, we categorized efficient and inefficient gRNAs using the method adopted by Xu et al. [9] (Supplementary Fig. 2). If a dataset contained fewer than 100, we categorized the top 20% gRNAs as efficient and considered the rest as inefficient. The efficient and inefficient gRNAs were designated as class 1 and class 0, respectively.
For training and testing of the models, the datasets for each CRISPR system were randomly divided into training (75%) and test (25%) data (Supplementary Table 3; Supplementary Data). To evaluate the trained models, independent test datasets were compiled from small-scale datasets (Supplementary Table 4). Here, Evers’ and Radzisheuskaya’s datasets [16], [17] for CRISPRi, Beottcher’s and Simeonov’s datasets [23], [24] for CRISPRa, Indel and Shalem’s datasets [6], [9], [14], [15], [18], [19], [20] for Cas9, and Kleinstiver-Chari’s [25], [26] and Kim’s datasets [7] for Cas12a were applied in the evaluation.
2.3. Features
The features utilized in this study were categorized into two parts: sequence and structure. In total, 609 features (604 for sequence and 5 for structure) were selected for the CRISPRi, CRISPRa, and Cas9 models, and 689 (684 for sequence and 5 for structure) were selected for the Cas12a model.
2.3.1. Sequence features
1) Position-independent nucleotides: the identity of each nucleotide (A/T/G/C) in a gRNA sequence, irrespective of the position; 2) Position-independent dinucleotides: the composition of dinucleotides in a guide RNA sequence, irrespective of the position; 3) Position-specific nucleotides: a “one-hot” encoding of nucleotide sequences, in which the presence or absence of a nucleotide (A/T/G/C) at a specific position of the target sequence is designated by 1 or 0, respectively (4 30 = 120 features for Cas9, CRISPRi, and CRISPRa; 4 34 = 136 features for Cas12a); 4) Position-specific dinucleotides: a “one-hot” encoding of dinucleotide sequences, in which the presence or absence of a dinucleotide at a specific position of a target sequence is designated by 1 or 0, repsectively (16 29 = 464 features for Cas9, CRISPRi, and CRISPRa; 16 33 = 528 features for Cas12a).
2.3.2. Structural features
1) GC count: the number of G or C in a given gRNA sequence. Because the GC count for a specific gRNA has a non-linear relationship with its activity, the GC counts were converted into a binary value: GC high (1 for GC count > 9 else 0 for CRISPRi, CRISPRa, and Cas9; 1 for GC count > 10 else 0 for Cas12a) and GC low (1 for GC count ≤ 9 else 0 for CRISPRi, CRISPRa, and Cas9; 1 for GC count ≤ 10 else 0 for Cas12a); 2) Melting temperature: the thermodynamic feature that indicates the stability of gRNA-target pairs, computed using Le Novere’s equation [10], [27]; 3) Self-folding energy: the secondary structure of the gRNA was computed using Vienna RNA package [28], [29]; 4) Shannon entropy: the information content of gRNA sequences [30], which was calculated using the Shannon equation [31].
2.4. Benchmarking machine learning algorithms
We have trained and benchmarked four regression-based machine learning algorithms—ENLOR, ENLR, RFB, and Xgboost—with inbuilt feature selection methods (Supplementary Note). ENLOR and ENLR are regularization techniques, incorporating logistic or linear equations. They implement LASSO [32] and Ridge [33] regularization modules for feature selection and bias reduction [34]. These algorithms were implemented using glmnet package in R. RFB is an ensemble learning algorithm that uses bagging to develop a prediction model. This algorithm was implemented using the Random Forest and Boruta package in R. Xgboost is another ensemble learning method that uses extreme gradient boosting framework. It employs optimal regularization and penalization to boosted trees [35].
2.5. Training and validation of CGD models
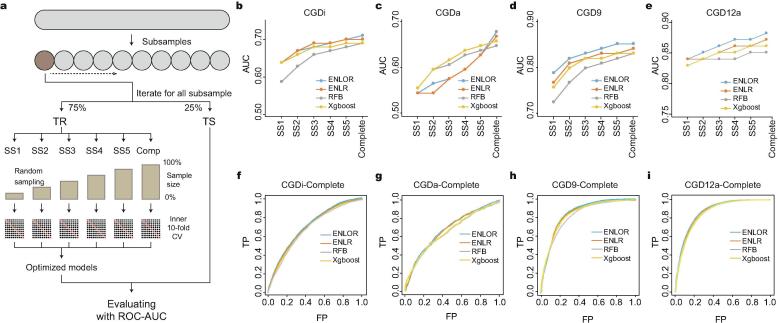
To optimally train and benchmark machine learning algorithms, we applied a nested cross-validation (CV) approach that reduces overfitting and stringent bias and minimizes a loss function in algorithms by selecting features and optimizing parameters. The workflow of nested CV is shown in Fig. 2a. The data were randomly subsampled to construct ten cohorts. Each cohort included 75% training and 25% test data. Subsets and the complete set of each cohort were randomly sampled across CRISPR systems (Supplementary Table 5). All subsets and the complete training dataset across the CRISPR system underwent inner ten-fold CV to optimize the parameters. Then, the subsets and the complete training dataset were tested using the aforementioned algorithms with fixed test data for each CRISPR system. This procedure was repeated for ten cohorts, and the results were averaged.
Fig. 2.
Schematic of the nested CV procedure and performance comparison. a The nested CV procedure used for CRISPRi, CRISPRa, Cas9, and Cas12a. TR: Training; TS: Test. b–e The plots depict AUC values for each algorithm across CRISPR-Cas systems. AUC values for subsets as well as complete training data are shown. The x-axis of each plot denotes subset and complete training data, and the y-axis denotes AUC values. f–i ROC curves depict the algorithms’ performance on the complete training data. In the ROC plots, the x-axis denotes the average False Positive rate (FP) and the y-axis denotes the average True Positive rate (TP).
2.6. Performance metrics
Receiver operating characteristics (ROC) and area under the curve (AUC) values were used to evaluate the metrics by varying the threshold. To further evaluate the classification power of our models and existing methods with independent test data, Kolmogorov–Smirnov (KS) test was performed for predicted efficient and inefficient gRNAs.
3. Results
3.1. Performance analyses of benchmarking machine learning algorithms
Performance analyses using ROC and AUC showed that ENLOR robustly outperformed other algorithms across all CRISPR-Cas systems (Fig. 2b–i and Supplementary Fig. 3). The performance analyses with subsets of data showed that none of the models were fully converged yet, indicating that more data would help improve the models. With these results, we implemented ENLOR in our CGD models.
3.2. Evaluation of our CGD models with existing tools using independent test data
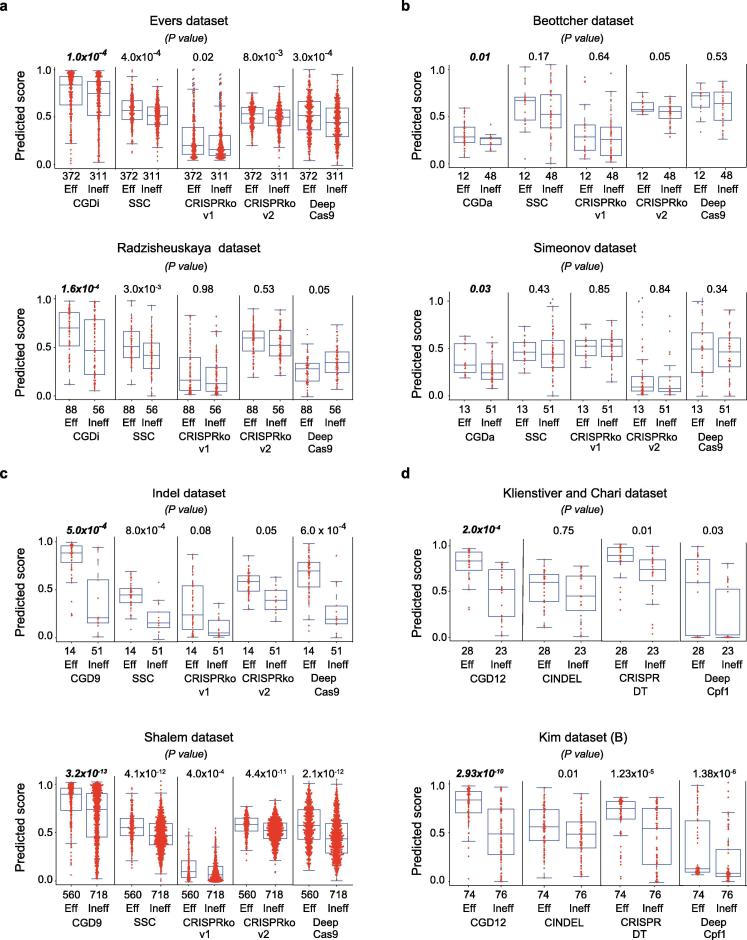
Our CGD models were compared with other existing tools, such as CRISPRko v1 [8], CRISPRko v2 [10], SSC [9], CINDEL [7], CRISPR-DT [11], DeepCas9 [13], and DeepCpf1 [12], using independent datasets (Supplementary Table 4). Our CGD models outperformed the other models in predicting efficient gRNAs (Fig. 3). The CGD models stratified efficient gRNAs in a better manner than other methods for all independent datasets generated by the respective CRISPR systems. For some datasets, previous methods failed to classify efficient gRNAs (Fig. 3).
Fig. 3.
Evaluation of CGD using independent test data. Comparison of existing machine learning algorithms, including deep learning algorithms, and our CGD models on independent test data using the KS test. The numbers of efficient (Eff) and inefficient (Ineff) gRNAs are indicated on the x-axis for machine learning (CGD9, CGDi/a, CGD12a, CINDEL 1.0, SSC, and CRISPRko v1/2) or deep learning (DeepCas9/Cpf1) algorithms. a–d Box plots representing the classification efficiency of different algorithms with P values as an indicator of competence for the respective datasets for CRISPRi (a), CRISPRa (b), Cas9 (c), and Cas12a (d).
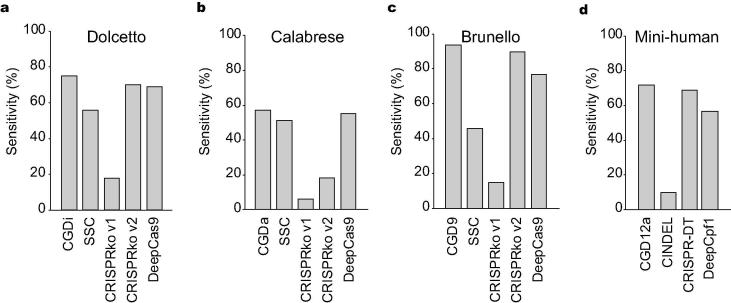
We also compared the sensitivity of the models identifying efficient gRNAs in CRISPR libraries developed for humans [36], [37]. Across CRISPR systems, our models outperformed previous models (Fig. 4).
Fig. 4.
Sensitivity of models for identifying efficient gRNAs in human gRNA libraries. The bar charts represent the percentage of efficient gRNAs identified by different models for each CRISPR library, namely Dolcetto (n = 55,521) for CRISPRi (a), Calabrese (n = 16,254) for CRISPRa (b), Brunello (n = 76,442) for Cas9 (c), and Mini human (n = 1963) for Cas12a (d). In each plot, the models are shown along the x-axis, and the sensitivity (the percentage of efficient gRNAs identified) is shown on the y-axis.
3.3. Common features of ENLOR models
The ENLOR algorithms select features relevant to on-target gRNA activity along with their weights, which indicate their contribution to the model. Thus, the features for calculating gRNA activity can vary according to the model. In the final models, 83 features for CGDi, 82 for CGDa, 84 for CGD9, and 215 for CGD12a were selected with positive or negative weights (Supplementary Table 1). When the feature sets of each model were compared, the majority of features appeared to be distinct to individual models; however, some were found in more than one model. In particular, four features were fully shared, two of which were related to thermodynamic measures (GC content and self-folding energy); the other two were sequential features related to the seed region of the respective CRISPR system (Supplementary Fig. 4). In fact, thermodynamic features and seed sequences have been shown to be common factors that determine gRNA efficiency [38], [39]. Although the four features were selected across all CRISPR systems, their weights varied, probably due to differences in the acting mechanisms of gRNAs [7], [10], [40]. CGD12a and CGDi consider GC content to be a more important contributor, indicating that Cas12a and CRISPRi activities are more dependent on the thermodynamic status of the target sites and gRNAs.
3.4. CGD web system
We built a web-based CGD model (http://big.hanyang.ac.kr:2195/CGD). The web program runs with a Python script; it finds all candidate gRNAs with CGD scores for each CRISPR system in the input DNA sequence. The candidate gRNAs are then mapped to the human reference genome (hg19) to extract their genomic coordinates with scores.
4. Discussion and conclusion
Here, we introduced CGD computational models that predict efficient gRNAs and their target sites for CRISPRi, CRISPRa, Cas9, and Cas12a systems. By benchmarking regression-based machine learning algorithms with a generalization algorithm and optimization, we selected ENLOR as a machine learning algorithm for CGD models. CGD models outperformed previous models that implement heuristic and machine learning methods.
Our study developed computational models for canonical PAMs (NGG for Cas9 and TTTV for Cas12a). However, recent CRISPR studies have also employed non-canonical PAMs recognized by Cas9 and Cas12a to improve the specificity of gene editing [41], [42], [43], [44], [45]. We thus applied our machine learning framework to non-canonical PAMs (NGH) using a dataset published by Kim et al. [45], resulting in performances comparable with that of the previous deep learning model (Supplementary Fig. 5). The CGD model for non-canonical PAMs has also been integrated into the CGD webserver.
The performance of our computational models varies over the different datasets generated by each CRISPR system because the experiments that produced the data were intentionally designed by different groups with distinct purposes, such as targeting essential or all genes; targeting coding sequences, introns, or untranslated regions; genome-wide screening approaches with positive or negative selection; and targeting synthetically designed target sequences (Fig. 3). These heterogeneous designs could lead to biases in gRNA sequence composition, changing the training data space. In fact, gRNA activities appeared to vary greatly depending on whether positive or negative selection was used for functional screening and whether essential genes were targeted (Supplementary Fig. 2). Hence, to train a robust model for each CRISPR system, the biggest datasets with the same type of selection and with gRNAs targeting essential genes were integrated to form a large-scale dataset. We used approximately 6000 gRNAs and their activities for training and validation of CRISPRi, 1000 for CRISPRa, approximately 3000 for Cas9, and approximately 15,000 for Cas12a (Supplementary Table 3).
Integration of all possible datasets is hindered by the different distributions of gRNA activity that result from the different types of selection and experiments (Supplementary Fig. 2). In fact, some independent test datasets (the Radzisheuskaya, Boettcher, and Simenov datasets) display relatively worse results than others (Fig. 3a and b), partly due to the data-specific distribution of gRNA activity. The performance of the CGD models in terms of the prediction of efficient gRNAs can be diminished just by integrating such heterogeneous datasets. This issue could be mitigated by incorporating proper normalization and regularization methods to penalize and reduce data-associated noise and biases.
Conflicts of interest
The authors declare no conflicts of interest.
CRediT authorship contribution statement
A Vipin Menon: Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing - original draft. Jang-il Sohn: Writing - review & editing. Jin-Wu Nam: Supervision, Conceptualization, Writing - review & editing, Funding acquisition.
Acknowledgements
We thank all of Jin-Wu Nam’s BIG Lab members, and Professor Suresh Ramakrishna of Hanyang University for critical reading and comments. This work was supported by the Bio and Medical Technology Development Program and the Basic Science Research Program through the National Research Foundation (NRF), funded by the Ministry of Science and ICT, South Korea (grant numbers 2014M3C9A3063541 and 2018R1A2B2003782).
Biographies
A Vipin Menon, a Ph.D. candidate in Department of Life Science, College of Natural Sciences at Hanyang University, has studied and developed an algorithm (CINDEL) for analysis of CRISPR-Cas12a.
Jang-il Sohn received a Ph.D. in Physics from Korea University in 2017, and has worked in Nam’s laboratory as a researcher (2015-17) and a research assistant professor (since 2017) at Hanyang University.
Jin-Wu Nam has been an associate professor at Hanyang University since 2012. He received a Ph.D. in Bioinformatics from Seoul National University and studied RNA computational biology at the Whitehead Institute for Biomedical Research, affiliated with MIT, as a Postdoctoral associate.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.03.020.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Jinek M., Chylinski K., Fonfora I., Hauer M. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora L., Narula A. Gene editing and crop improvement using CRISPR-Cas9 system. Front Plant Sci. 2017;8:1932. doi: 10.3389/fpls.2017.01932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert L.A., Horlbeck M.A., Adamson B., Villata J.E., Chen Y. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shalem O., Sanjana N.E., Hartenian E., Xi Shi, Scott D.A. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H.K., Song M., Lee J., Menon A.V., Jung S. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods. 2017;14(2):153–159. doi: 10.1038/nmeth.4104. [DOI] [PubMed] [Google Scholar]
- 8.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32(12):1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H., Xiao T., Chen C.H., Li W., Meyer C.A. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25(8):1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H., Liang C. CRISPR-DT: designing gRNAs for the CRISPR-Cpf1 system with improved target efficiency and specificity. Bioinformatics. 2019;35(16):2783–2789. doi: 10.1093/bioinformatics/bty1061. [DOI] [PubMed] [Google Scholar]
- 12.Kim H.K., Min S., Song M., Jung S., Choi J.W. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat Biotechnol. 2018;36(3):239–241. doi: 10.1038/nbt.4061. [DOI] [PubMed] [Google Scholar]
- 13.Xue L., Tang B., Wei Chen, Jiesi Luo. Prediction of CRISPR sgRNA Activity Using a Deep Convolutional Neural Network. J Chem Inf Model. 2019;59(1):615–624. doi: 10.1021/acs.jcim.8b00368. [DOI] [PubMed] [Google Scholar]
- 14.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 15.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radzisheuskaya A., Shlyueva D., Muller I., Kristian H. Optimizing sgRNA position markedly improves the efficiency of CRISPR/dCas9-mediated transcriptional repression. Nucl Acids Res. 2016;44(18):e141. doi: 10.1093/nar/gkw583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evers B., Jastrzebski K., Heijmans J.P., Grernrum W., Beijersbergen R.L. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34(6):631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 18.Ramakrishna S., Cho S.W., Kim S., Song M., Gopalappa R. Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat Commun. 2014;5:3378. doi: 10.1038/ncomms4378. [DOI] [PubMed] [Google Scholar]
- 19.Ramakrishna S., Cho S.W., Kim S., Song M., Gopalappa R. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24(6):1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15(5):321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 21.Konermann S., Brigham M.D., Trevino A.E., Joung J. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koike-Yusa H., Li Y., Tan E.P., Velasco-Herrera Mdel C., Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 23.Boettcher M., Tian M., Blau J.A., Markegard E., Wagner R.T. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat Biotechnol. 2018;36:170–178. doi: 10.1038/nbt.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simeonov D.R., Gowen B.G., Bootanrart M., Roth T.L., Gagnon J.D. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature. 2017;549:111–115. doi: 10.1038/nature23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chari R., Yeo N.C., Chavez A., Church G.M. sgRNA scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth Biol. 2017;6(5):902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Novere N. MELTING, computing the melting temperature of nucleic acid duplex. Bioinformatics. 2001;17(12):1226–1227. doi: 10.1093/bioinformatics/17.12.1226. [DOI] [PubMed] [Google Scholar]
- 28.Wong N., Liu W., Wang X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015;16:218. doi: 10.1186/s13059-015-0784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz R., Bernhart S.H., Honer Zu., Siederdissen C., Tafer H. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6(1):26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo C.Y., Long J.D., Campo-Fernandez B., de Oliveira S., Cooper A.R. Site-specific gene editing of human hematopoietic stem cells for X-linked hyper-IgM syndrome. Cell Rep. 2018;23(9):2606–2616. doi: 10.1016/j.celrep.2018.04.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon C.E. A mathematical theory of communication. Bell Syst Tech J. 1948;27(3):379–423. [Google Scholar]
- 32.Tibshirani R. Regression shrinkage and selection via the lasso. J Roy Stat Soc: Ser B (Methodol) 1996;58(1):267–288. [Google Scholar]
- 33.Hoerl A.E., Kennard R.W. Ridge regression: biased estimation for nonorthogonal problems. Technometrics. 1970;12(1):55–67. [Google Scholar]
- 34.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 35.Chen T, Guestrin C. XGBoost, in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining – KDD '16: 2016. 785–794.
- 36.Sanson K.R., Hanna R.E., Hegde M., Donovan K.F., Strand C. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun. 2018;9(1):5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J., Srinivasan S., Li C.Y., Ho I.L., Rose J. Pooled library screening with multiplexed Cpf1 library. Nat Commun. 2019;10(1):3144. doi: 10.1038/s41467-019-10963-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim Y., Bak S.Y., Sung K., Jeong E., Lee S.H. Structural roles of guide RNAs in the nuclease activity of Cas9 endonuclease. Nat Commun. 2016;7:13350. doi: 10.1038/ncomms13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swarts D.C., van der Oost J., Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol Cell. 2017;66(2):221–233.e4. doi: 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horlbeck M.A., Gilbert L.A., Villalta J.E., Adamson B., Pak R.A. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 2016;5 doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H.K., Kim Y., Lee S., Min S., Bae J.Y. SpCas9 activity prediction by DeepSpCas9, a deep learning–based model with high generalization performance. Sci Adv. 2019;5(11) doi: 10.1126/sciadv.aax9249. eaax9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D., Zhang C., Wang B., Li B., Wang Q. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat Commun. 2019;10(1):4284. doi: 10.1038/s41467-019-12281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zetsche B., Strecker J., Abudayyeh O.O., Gootenberg J.S., Scott D.A. A survey of genome editing activity for 16 Cas12a orthologs. Keio J Med. 2019 doi: 10.2302/kjm.2019-0009-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeong Y.K., Yu J., Bae S. Construction of non-canonical PAM-targeting adenosine base editors by restriction enzyme-free DNA cloning using CRISPR-Cas9. Sci Rep. 2019;9(1):4939. doi: 10.1038/s41598-019-41356-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H.K., Lee S., Kim Y., Park J., Min S. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells. Nat Biomed Eng. 2020;4(1):111–124. doi: 10.1038/s41551-019-0505-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.