Abstract
Biogeography is an implicit and fundamental component of almost every dimension of modern biology, from natural selection and speciation to invasive species and biodiversity management. However, biogeography has rarely been integrated into human or veterinary medicine nor routinely leveraged for global health management. Here we review the theory and application of biogeography to the research and management of human infectious diseases, an integration we refer to as ‘pathogeography’. Pathogeography represents a promising framework for understanding and decomposing the spatial distributions, diversity patterns and emergence risks of human infectious diseases into interpretable components of dynamic socio‐ecological systems. Analytical tools from biogeography are already helping to improve our understanding of individual infectious disease distributions and the processes that shape them in space and time. At higher levels of organization, biogeographical studies of diseases are rarer but increasing, improving our ability to describe and explain patterns that emerge at the level of disease communities (e.g. co‐occurrence, diversity patterns, biogeographic regionalisation). Even in a highly globalized world most human infectious diseases remain constrained in their geographic distributions by ecological barriers to the dispersal or establishment of their causal pathogens, reservoir hosts and/or vectors. These same processes underpin the spatial arrangement of other taxa, such as mammalian biodiversity, providing a strong empirical ‘prior’ with which to assess the potential distributions of infectious diseases when data on their occurrence is unavailable or limited. In the absence of quality data, generalized biogeographic patterns could provide the earliest (and in some cases the only) insights into the potential distributions of many poorly known or emerging, or as‐yet‐unknown, infectious disease risks. Encouraging more community ecologists and biogeographers to collaborate with health professionals (and vice versa) has the potential to improve our understanding of infectious disease systems and identify novel management strategies to improve local, global and planetary health.
Keywords: diversity, mapping, biodiversity
Introduction
In a globalized world where the spread of infectious diseases appears to ignore all boundaries and the risk of emerging pathogens is on the rise (Jones et al. 2008, Fisher et al. 2012), there has been a resurgence in interest by academics, the general public and both national and international government authorities in the geography of human infectious diseases at all spatial scales. Since most endemic and emerging human pathogens utilise non‐human animal species at some stage in their transmission cycles (e.g. reservoir and intermediate hosts and vector species) (Taylor et al. 2001, Woolhouse and Gowtage‐Sequeria 2005), biogeographers and community ecologists are increasingly involved in this pursuit. Their contributions have yielded a range of novel and complementary insights on the spatial and temporal patterns of infectious disease occurrence, emergence and burden, their underlying ecological processes, and their surveillance and management (Guernier et al. 2004, Smith et al. 2007, Peterson 2008, 2014, Reperant 2010, Johnson et al. 2015, Murray et al. 2015, Stephens et al. 2016b).
Medical geography has a long and rich history (Barrett 2000, Cliff et al. 2000, Rogers et al. 2002, Cliff and Haggett 2004) and its methods and objectives have numerous parallels to those of modern biogeography, with its broad aim of determining how multiple processes (e.g. speciation, adaptation, extinction, ecology, geology, climate) interact with one another to produce distributional patterns in the world's biota (Myers and Giller 1988). For infectious diseases, this history stretches as far back as to the time of the debate between contagionists and anticontagionists, when experts disagreed about the very existence of infectious disease‐causing agents; for example, several ‘spot maps’ in the context of yellow fever in the US were developed in the late 18th and early 19th centuries to identify patterns and attempt to infer environmental causes for the disease, well before Pasteur and Koch's eventual and definitive development of the ‘germ theory of disease’ in the late 1800s (Howe 1989, Jones 1990, Lederberg 2000).
However, the largely correlative methods and findings of medical geography seemed to lose ground as modern medicine developed in favour of a relatively narrow focus on molecules, individuals, individual diseases or sub‐components thereof, and small and homogenous populations and areas (e.g. cohort studies, randomized‐ and case‐control trials, small area health statistics), in which causality is presumed easier to stalk (Schwartz 1994, McLaren and Hawe 2005). As a consequence, and despite the availability of theory and methods in other disciplines to overcome, negate or manage key issues related to correlation and scale complexity (Chesson 2012), modern epidemiology has been arguably caught off‐guard in a rapidly changing world.
So called ‘prisoners of the proximate’, in reference to a limiting preoccupation with direct individual‐level disease risk factors, McMichael (1999) suggested that modern epidemiologists and public health managers have been sluggish and ill‐equipped to recognise, prepare for and proactively respond to some of the most pressing and emerging health challenges of the 21st century, such as climate change, habitat alteration and degradation, biodiversity loss, invasive species including vectors, demographic shifts, migration and epidemiological transitions. Although many global health metrics, such as life expectancy and childhood mortality, have nevertheless continued to improve, researchers from a range of disciplines are increasingly forecasting a collision between ongoing improvements in human health and a range of large‐scale and accelerating global change processes, particularly those relating to environmental factors and declining environmental quality (Foley et al. 2005, MEA 2005, Raudsepp‐Hearne et al. 2010, Suk and Semenza 2011, Costanza et al. 2014, Watts et al. 2015, Whitmee et al. 2015).
Health funders are also beginning to recognise these complex risks to human health (e.g. Wellcome Trust < https://wellcome.ac.uk/what-we-do/our-work/our-planet-our-health >, Rockefeller Foundation < http://www.rockefellerfoundation.org/our-work/initiatives/planetary-health/ >). Given that this type of multi‐scale, multi‐disciplinary complexity is commonplace in biogeography, and much precedence exists from the study of parasitism, plant and animal diseases and global change ecology, there has never been a better time for biogeographers and ecologists to contribute their knowledge, theory and methods to public and global health. Critically, where strong links between human health and the environment are identified and quantified, such collaborations could stimulate novel streams of funding and yield cost‐effective co‐benefits for health and the environment (Myers et al. 2013, Waldron et al. 2017).
This is particularly salient for research on human infectious diseases, most of which involve animal hosts and vectors in shaping their distributions and are therefore influenced by many of the same ecological processes that govern biodiversity patterns more generally (Guernier et al. 2004, Murray et al. 2015). Analogous to its utility for understanding wildlife distributions, biodiversity patterns and improving conservation management, biogeography has the potential to improve our understanding of infectious disease distributions, describe and explain patterns and processes underlying pathogen diversity (‘pathodiversity’) and contribute to infectious disease forecasting, risk management and threat abatement. Through the analysis of historical disease, host and/or vector occurrence and co‐occurrence patterns, biogeographic approaches could even yield some of the earliest, and in some cases the only, insights into poorly known, burgeoning and future infectious disease risks (Murray et al. 2015).
Here, we review the building blocks of biogeography and illustrate how it has and could continue to provide novel insights for the study and management of human infectious diseases, an integration we refer to as ‘pathogeography’ (resurrecting a term coined by plant pathologist Israel Reichert; Reichert and Palti 1967). Better understanding of pathogeography among ecologists and improved biogeographic knowledge among veterinary and medical scientists and public health managers should help improve disease surveillance, combat the global burden of human infectious diseases, and improve environmental management. It may even help re‐bridge a divide that has opened between the medical and ecological sciences, two powerful, explanatory and potentially predictive disciplines that ultimately share common roots in basic scientific inquiry.
Pathogeography – a framework for the biogeographic study of infectious diseases
The overarching objectives of pathogeography, following biogeography (Myers and Giller 1988), are to determine how the interactions among varying biotic (e.g. speciation, adaptation, extinction) and abiotic (e.g. topography, geology, climate) factors and processes combine to produce distributional patterns in infectious diseases through time. Johnson et al. (2015) and Stephens et al. (2016b) recently reviewed the community ecology and macroecology of infectious diseases, respectively, and a natural intersection between these disciplines and the biogeography of infectious diseases occurs where these fields become spatially explicit. Macroecology deals with ecological questions that demand large‐scale analysis (Brown 1995, Brown and Lomolino 1998, Cox et al. 2016). The limits between biogeography and macroecology are fuzzy. They converge when biogeography is focused on the study of how population‐ or community‐level parameters vary along geographic dimensions (Lomolino et al. 2006), or when macroecology deals with spatial patterns (e.g. the geographic distribution of a certain pathogen species or diversity patterns). The divergence occurs when spatially explicit components are not crucial to answering questions being addressed by macroecology (Blackburn and Gaston 2006), and when biogeography does not invoke structural and functional patterns of ecological systems (e.g. areography and some evolutionary biogeography approaches) (Morrone 2009). By contrast, community ecology offers further insights on the mechanisms and processes that bridge fine scale processes of individuals and populations with the ecology and evolutionary processes of species and disease distributions at larger spatial scales (Johnson et al. 2015).
Whereas biogeography is concerned with the analysis of spatial patterns of biological diversity, pathogeography (i.e. the biogeography of pathogens) is concerned with the analysis of spatial patterns of pathogen (or disease) diversity (‘pathodiversity’ being the obvious analogue, but also referred to as the ‘pathogen pool’, ‘pathogen community’, ‘pathosphere’ or ‘pathobiome’). Taylor et al. (2001) constructed the first list of distinct species at a global scale known to be infectious to and capable of causing disease in humans under natural conditions, tallying 1415 species (217 viruses and prions, 538 bacteria and rickettsia, 307 fungi, 66 protozoa and 287 helminths). Discovery and recognition of new human pathogens occurs regularly (Woolhouse et al. 2008), with the most recent comprehensive inventory that we are aware of listing 2107 pathogens in humans (274 viruses, 1003 bacteria, 447 fungi, 82 protozoa and 301 helminths) (Wardeh et al. 2015).
A comprehensive global database of clinically relevant human infectious diseases, GIDEON (< http://www.gideononline.com/ >), tracks more than 350 of these pathogens (Berger 2005, Victor and Edberg 2005). Surprisingly, the distributions of most of these are very poorly known. As recently as 2013, only 7 infectious diseases were considered ‘comprehensively mapped’ (including Old World coltiviruses, Plasmodium falciparum and P. vivax, monkey pox, dengue, Lassa fever and Mayaro) (Hay et al. 2013), fundamentally limiting the ability of public and global health resources to be systematically and efficiently directed towards precise geographic areas and populations at highest risk from other impactful diseases. Indeed, distributional patterns of human infectious diseases are generally far more poorly compiled and characterized (e.g. often at only country or regional level and as coarse presence vs absence data) than many plant and animal species, for which numerous global stock takes, status assessments, occurrence databases and detailed distribution maps exist following a long tradition of biogeographic study (Wallace 1876, Murray et al. 2015) (see also Supplementary material Appendix 1 Table A1). However, with the development of big data approaches and curated databases, resources are slowly improving for human infectious diseases (Wardeh et al. 2015, Stensgaard et al. 2017). Data may even on occasion be far richer or more geographically precise than for many plant and animal species (especially invasive species) owing, for example, to notifiable disease reporting requirements, such as those in place in EU member states for reporting human cases of certain diseases to ECDC and zoonotic and food‐borne diseases to other EU registries (e.g. haemorrhagic cases of dengue and Rift Valley fever, Crimean Congo haemorrhagic fever, West Nile fever, cholera, campylobacteriosis) (Lindgren et al. 2012). In addition, databases for specific groups of diseases (e.g. helminths, neglected tropical diseases) and host‐pathogen (including human) associations are increasing (e.g. < http://www.thiswormyworld.org/ >).
In global studies that have classified pathogens according to their epidemiological traits, the majority of known human pathogens (58–61%) are classed as zoonotic, defined as diseases and/or pathogens that are naturally transmissible from vertebrate animals to humans, and 14% are arthropod vector‐borne (WHO 1959, Palmer et al. 1998, Taylor et al. 2001, Woolhouse and Gowtage‐Sequeria 2005). The close association between animals and human pathogens means that the diversity of potentially pathogenic microorganisms that occur in animals including wildlife is also of major interest for human health (Morse 1995, Murray and Daszak 2013). However, at present, the full dimensions of this broader ‘pathogen pool’ are almost entirely unknown. For example, estimates of the total number of viruses within just nine target viral families from the first intensively sampled wildlife species (the natural host of Nipah virus, fruit bat Pteropus giganteus) suggest that the number of known human pathogens is just a tiny fraction of the total viral diversity that occurs in wildlife (Anthony et al. 2013).
Units of analysis
The ‘operational taxonomic unit’ (OTU) of pathogeography may differ somewhat from conventional OTUs in biogeography (e.g. genes, species). While infectious diseases are all caused by specific pathogenic microbial species (which could, or perhaps should, themselves be the relevant OTU), it is their infection/presence in human and in some cases livestock or wild animal hosts or vectors that is of primary interest to health stakeholders and the usual target of surveillance programs.
‘Occurrence’ is the presence of a disease or its causative agent in a particular place at a particular time. This can in turn be represented as a presence (i.e. in a human host or a specific geographic unit) or some measure of relative abundance either within single hosts (e.g. the infection load, particularly for macroparasites, such as helminths), within a defined geographic area (e.g. density), time period (incidence) or within the host population (e.g. prevalence). It might also be represented to reflect the process of transmission itself (e.g. ‘force of infection’). With some information on the average impact of infections in humans, public health practitioners often also describe spatial and temporal patterns of disease in terms of ‘burden’, with various available measures that broadly seek to capture the loss of healthy life (e.g. death and disability) attributable to the presence of certain diseases within a population (e.g. disability adjusted life years, DALYs) (Murray et al. 2013). Conversely, the absence of disease is of equal interest for pattern and process analysis, but harder to obtain given the sampling difficulties of asserting freedom from disease (Cameron 2012). Furthermore, occurrence records (and its derivatives) will generally be a subset of actual occurrences, because in most cases they will be heavily influenced by observation effort. Given the diversity of health studies, all of these epidemiological metrics could be potentially relevant for pathogeographic analysis.
The same units of measure (occurrence, abundance, density) for known or potential hosts and vector species are also relevant and will already be familiar to ecologists. Some estimates of disease ‘risk’ (or, perhaps more accurately, ‘hazards’) are based on these (e.g. tick nymphal abundance as a measure of Lyme disease risk) and their ecologies may contribute directly or indirectly to disease patterns (Civitello et al. 2015), such that data on their potential occurrence can improve biogeographically‐informed risk mapping of disease outcomes in humans (Messina et al. 2015, Pigott et al. 2016, Olivero et al. 2017a). One major challenge for biogeographic analyses of human infectious diseases, however, is the availability and utility of appropriate data, which we discuss further in Box 1. Supplementary material Appendix 1 Table A1 provides some example data types, and some useful databases and sources relevant to biogeographic analyses of human infectious diseases.
Box 1. Box 1. Data requirements for pathogeographic studies.
The use of biogeographical methods for examining the distributions of human infectious diseases will be dependent on the degree of existing knowledge (e.g. about the epidemiology of a particular pathogen or pathogen assemblage in a particular geographic context), data availability (e.g. from publicly accessible databases, surveillance data) and the potential for new data collection (e.g. targeted field collections). These elements could range from no knowledge, no data and large barriers to the collection of new data (e.g. for many emerging infections in developing country contexts, such as at the beginning of the 2014 Ebola outbreak in west Africa), to high degree of knowledge from existing scientific studies, well developed and accessible databases on relevant geographic/environmental, reservoir host, vector, pathogen, human and disease data, and existing structures to streamline the collection of new data (e.g. high impact diseases in developed country contexts, such as malaria in the EU).
Data on human infectious diseases are normally collected for the needs of a particular discipline or research focus, which may typically limit the extent to which it can be used laterally or opportunistically for answering non‐target questions. This may be particularly the case for macroecological and biogeographical studies, which are often data intensive and conducted at scales that may be difficult or impractical to undertake new data collection. There is thus a clear need to expand the scope of research programmes on infectious diseases to encompass the geographic, biological and temporal scales relevant to biogeographical analysis. This involves informing monitoring and surveillance activities on what types of data would be most useful and urgent. Improving data capture and quality with standardized sampling methodologies could help lead to analyses and discoveries that transcend the specific epidemiological details of a single site, geographical context or disease. To this end, the following information would be helpful to allow coherent data collection and analysis of infectious disease occurrence and transmission in space and time:
Operational taxonomic units: potential complications for biogeographic pattern and process analysis may arise when the ‘presence’ of a specific disease type is in fact confounded. This could occur, for example, if multiple causal agents result in disease complexes (e.g. Leishmaniasis) or if the causal agent is unknown and diseases are instead classified by their symptoms (e.g. syndromes). Distributional data and databases on infectious diseases should thus strive for the highest ‘taxonomic resolution’ possible (Murray et al. 2015, Stensgaard et al. 2017).
Observation effort: a persistent issue in robustly quantifying occurrence, in any form, is its relationship to observation effort (Allen et al. 2017). Observation effort could vary spatially, temporally or taxonomically. Presence, prevalence, burden and diversity patterns of infectious diseases, hosts and vectors may all increase proportionally to observation effort, and confidence surrounding reported absences also increases with observation effort. Failing to account for this has the potential to introduce biases in biogeographic studies. Numerous studies have taken steps to incorporate measures of observation effort to limit the potential effects of observation bias in biogeographic studies of infectious diseases, typically by including factors hypothesised to be related to observation effort, such as sampling intensity, GDP, health expenditure or publication output, in statistical models (Jones et al. 2008, Hopkins and Nunn 2010, Yang et al. 2012, Murray et al. 2015) (see also O in Box 2).
Sampling protocols: a lack of communication between biostatisticians and field workers in both ecological and health fields before collecting samples can lead to a breakdown in robustness of subsequent analyses. For example, sampling too many host individuals of one species could be as problematic as not sampling enough from a target host (e.g. humans), particularly when research resources are constrained. Best practices involving probability sampling should be pursued where possible (Nusser et al. 2008).
Geographic coverage and resolution: sampling should cover a sufficiently large area(s) to reproduce in space what really exists in the field; for example, metapopulation or metacommunity geographic distributions with sources and sinks of pathogen transmission. The effects of uneven sampling across space and one‐shot sampling should be avoided (Peterson 2014). At the other extreme, improving precision on available data points is a high priority for developing robust geo‐referenced databases of disease (or pathogen, host, vector) occurrence. All data should be collected and stored for subsequent access at the highest spatial resolution possible (i.e. GPS coordinate locations).
Temporal coverage: infected hosts including humans may travel long distances during disease incubation periods, introducing uncertainty in the attribution of the geographic location of infection (Peterson 2014). Surveillance and sampling strategies should thus allow the capture of appropriate temporal scales (i.e. matching scale of disease‐relevant processes) so that data can be better aligned with covariate information such as environmental variables and human activities.
Phenology: animal and plant phenology should be monitored; the behavior and biology of most species, including humans, are influenced by often relatively predictable annual changes in climate that determine when they start or finish natural events, such as flowering and fruiting, breeding and mass gatherings. Many of these activities and departures from norms due to, for example, unusual weather events have implications for disease risks and spatio‐temporal distributions, and could have large implications when considering the influences of large scale change (e.g. climate change).
Humans as hosts and dispersers: humans will often serve not only as hosts but also as effective dispersal mechanisms for infectious agents. Although this can lead to unpredictability and extreme long distance invasion events (Peterson 2008), data on human populations and their movement are being increasingly well resolved at both fine and coarse spatial scales and this is proving invaluable in decomposing biogeographical components of many infectious disease systems including emergence risks (e.g. use of flight or road traffic data, mobile phone use data, social media) (Colizza et al. 2007, Balcan et al. 2009, Wesolowski et al. 2012, Jurdak et al. 2015).
Concurrent covariate sampling and time‐lagging: ideally, assessment of environmental and social variables should be conducted at the same time as human or wildlife disease sampling (if it is not available at the appropriate scale retrospectively i.e. from remote sensing data). Peterson (2014) discusses a range of issues relevant to modeling the distribution of infectious diseases, including quality control of input occurrence data, sampling design with the reduction of oversampling in certain areas, and design of analysis (see also Hosseini et al. 2017).
Single diseases
The study of infectious disease spatial distributions is not new (Barrett 2000), nor are integrated approaches to studying infectious disease distributions, whether they are labeled biogeographic (Peterson 2008) or not (Lambin et al. 2010). Indeed, the emergence of informatics, geographic information systems (GIS) and satellite technology has increased the availability of tools and high quality spatial datasets relevant to both ecologists and medical geographers, driving a recent convergence in the data and in some cases the methods used to examine the distributions of wild species and infectious diseases alike and for developing or evaluating causal hypotheses underpinning them (Hay et al. 2006, 2013, Kraemer et al. 2016).
Such developments have facilitated the study of the spatial structure of some infectious diseases in unprecedented detail (Fig. 1). In some cases, there has been innovation in the integration of these approaches with mechanistic models traditionally used to explore the population dynamics of infectious diseases (Redding et al. 2016), and some analyses have been developed and updated in close to real time (e.g. during the west African Ebola outbreak) (Pigott et al. 2014, 2016). These advances complement an already strong suite of tools already used in epidemiological studies, which might equally flow the other way into the toolboxes of ecologists (Magalhães et al. 2011, Caprarelli and Fletcher 2014, Bhatt et al. 2017).
Figure 1.
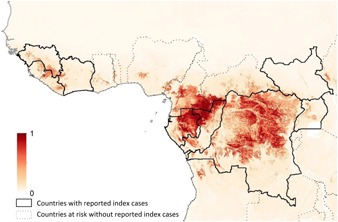
Ebola risk map (Pigott et al. 2016). Illustrating the detail of modern cartographic projections of disease risk based on models of environmental suitability for the zoonotic transmission of Ebola virus (shaded colours) and the spatial variation in disease risk that would be masked from country‐level chloropleths (thick black lines). Dotted lines indicate regions that have reported no index cases to date but are predicted to be partially at risk based on environmental suitability models that utilize a thresholding approach on the model output that captures 95% of the occurrence records used for model fitting (see also Fig. 6). Boosted regression trees were used to develop the model based on georeferenced records of Ebola virus disease outbreaks in humans and infection records in bat reservoirs and a range of environmental covariates. The scale reflects the relative probability that zoonotic transmission of Ebola virus could occur at these locations; areas closer to 1 (dark red) are more environmentally similar to locations reporting Ebola virus occurrences; areas closer to 0 (light yellow) are least similar.
Peterson (2008) defined the first explicit ‘biogeographic framework’ for human infectious diseases. Drawing on the work of Soberón (2007) on the Grinnellian niche and geographic distributions of species, the framework is characterized by considerations of a pathogen's dispersal ability combined with the abiotic and biotic factors that interact to determine whether a disease is able to fulfill its full geographic potential. Although not explicitly demonstrated, Peterson (2008) emphasizes that a key difference between pathogens and diseases in comparison to free ranging wild species (with the exception of invasive species) is the relative unpredictability of dispersal events (e.g. rapid, long distance introductions), and the relatively lower importance of abiotic and relatively higher importance of biotic factors. This applies particularly to the high degree of inter‐specific interactions, from the infection of a pathogen in a host, to the numerous other species that may be involved in pathogen transmission cycles. The distribution of an infectious disease is thus defined by the joint distributions of all species involved in its transmission cycle as dictated by the suitable ecological conditions and dispersal limitations for each.
In contrast, Lambin et al. (2010) present an alternative framework for understanding generalized disease risks across landscapes, drawing on principals from ‘spatial epidemiology’ (Ostfeld et al. 2005) and ‘landscape epidemiology’ (Pavlovsky 1966). Pavlovsky (1966) proposed that infectious disease occurrence is determined by ‘a continuous interaction’ of five landscape factors: 1) animal ‘donors’ (e.g. reservoir hosts), 2) vectors, 3) animal ‘recipients’ (including humans), 4) an infective pathogen, and 5) environmental factors that are conducive to transmission. Lambin et al. (2010) emphasise the dynamic nature of the spatial and temporal interactions between these ‘prerequisites’ at multiple scales when assessing the impact of landscape changes on vector‐borne and zoonotic diseases. Although they did not identify their study explicitly as ‘biogeographic’, there are clear parallels with Peterson's (2008) framework (as well as many others, such as Plowright et al.'s (2017) recent treatment of spillover ecology).
We develop these ideas further in Box 2, focussing on the interactions between a number of inter‐dependent elements, including: physical geography (G), environment (E), reservoir host(s) (R), vector(s) (V), pathogen(s) (P), human factors (H), and the management (M) and observation (O) landscapes, the latter serving as the lens through which all other elements and disease distributions (D) are ultimately observed. From this framework, we can envisage how these elements may act and interact to influence both the real and observed distributions of specific diseases in space and through time, as well as multi‐disease patterns that may emerge at higher levels of organization (see ‘Emergent patterns and multiple diseases’ below).
Box 2. Box 2. Pathogeography: decomposing the key elements and interactions shaping the distributions and diversity patterns of human infectious diseases.
Figure 2.
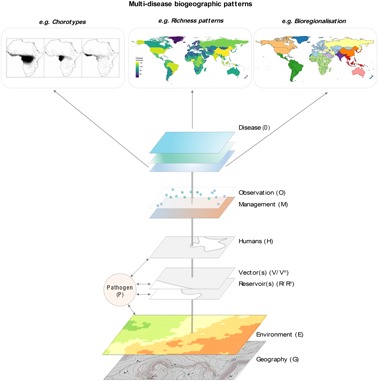
We may represent the challenge of simultaneously understanding patterns and processes of infectious disease systems with respect to a series of interacting elements; including G, the physical geography context (e.g. topography) and E, the abiotic (e.g. climate) and biotic (e.g. habitat) environment; R n and V n, the single or multiple (denoted by superscript n) species of reservoir hosts or vectors; P, the pathogen being transmitted; H, the human population itself; O, the observation effort that may apply to each of the other elements (e.g. surveillance and data collation from existing sources); and M, the management landscape (e.g. interventions). The combinations of these elements ultimately give rise to D, the observed disease distributions. Where these elements have consistent effects across multiple diseases, ‘higher order’ biogeographic patterns emerge; for example, we can observe biogeographic regionalization as a consequence of the more pervasive effects of components of G (e.g. ocean or mountain barriers to dispersal) or E (e.g. unsuitable climates), while the effects of components of H (e.g. population growth, global movement, socio‐economic status, immunity heterogeneity) and M (e.g. health infrastructure, vaccination) have fundamentally reshaped the global diversity and burden patterns of infectious diseases. In addition, each layer potentially has additional modifying elements that could further shape disease distributions and diversity patterns, such as temporal fluxes (see main text), or life‐history and epidemiological traits of hosts, vectors and pathogens (Smith et al. 2007, Woolhouse et al. 2016).
For pathogeographic analyses and as a starting point for risk assessments, a series of ‘profiling’ steps could help integrate existing information at a scale relevant to a particular research question. This could include geographic and/or environmental profiling (e.g. detailed assessments of G and E for diseases/hosts/vectors in regions of interest relevant to single diseases or disease assemblages) (Eisenberg et al. 2007), disease trait profiling (classifying epidemiological features of diseases, such as the presence or absence of the E, V, R and H elements as sources of pathogen transmission (see Box 3), and the species known to be implicated in each), and human profiling (assessments of the human population distribution, density and movement patterns and the existing management and observation landscapes). Intersection of these geographic, disease trait or human profiles will ultimately yield yet deeper understanding or management relevant insights (Semenza et al. 2016).
Depending on epidemiological characteristics, not all of the elements illustrated in Box 2 will be relevant for all human infectious diseases, resulting in a range of ‘biogeographic complexity’ among human infectious diseases. Whereas G and E will almost always have some kind of modifying influences, single‐element transmission source systems (i.e. diseases only involving human‐human, environmental, single reservoir species or single vector species transmission) represent the least biogeographically complex examples in this framework. Examples include tetanus (E), measles (H), and Lassa fever (R). The more biogeographically complex diseases involve multiple elements; for example, multiple reservoir host or vector species (R n and V n, respectively) in addition to human‐human or environmental transmission (e.g. dengue (HV n R n); infection with Mycobacterium ulcerans (EV n R n)). We describe the potential utility and demonstrate an application of this general framework further in Box 3, by undertaking a ‘disease trait profiling’ exercise whereby we classify a large number of clinically relevant human infectious diseases according to their transmission sources (i.e. the E, H, V and R elements described in Box 2). The trends that emerge illustrate how certain disease characteristics are far more common than others (e.g. transmission from animal reservoirs vs arthropod vectors) and the extent to which mapping efforts are already underway. However, the analysis also highlights some important gaps. For instance, 31.9% of diseases with a strong rationale for mapping (as rated by Hay et al. 2013) have not been mapped in any study (Box 3, Fig. panel F), and declining overall quality of mapping efforts correlates with increasing biogeographic complexity (Box 3, Fig. panel G), highlighting a need to expand the breadth of ecological interactions considered within disease systems to improve mapping quality in future studies.
Box 3. Box 3. Disease trait profiling: disease diversity, biogeographic complexity and mapping.
Figure 3.
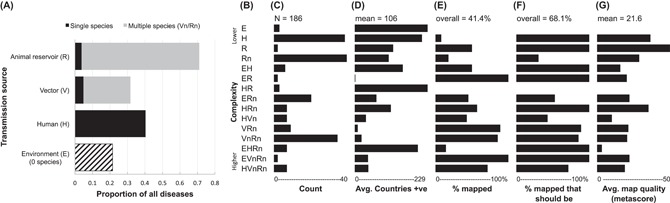
Disease trait profiling could help synthesize and summarize the range of potential ecological interactions of diseases and highlight important priorities or gaps for biogeographic analyses. To illustrate, we classified all clinically relevant diseases within the GIDEON database with single causative pathogens (n = 186 diseases) into each of the combinations possible considering whether disease transmission sources included the E, H, V and R elements outlined in Box 2 (note, although H is by definition always involved for human infectious diseases, here it is treated more specifically as a transmission source i.e. human–human transmission). We then examined how variations in the complexity of diseases, as indicated by the number of elements involved in their transmission (complexity score, CS), was related to geographic range size (as broadly indicated by the number of countries in which the disease is present), the rationale for mapping, whether mapping efforts had already taken place, and the quality of existing mapping as rated by Hay et al. (2013).
Consistent with other studies, 71.0% of diseases in our dataset involved (but do not necessarily require) transmission to humans from animal reservoirs (zoonotic) (Fig. A). Human–human transmission was also common (40.3%), while diseases including potential transmission from vectors (31.7%) or environmental sources (21.5%) were less common. For diseases involving vectors or reservoirs, the great majority involved multiple vector (95%) or reservoir (85%) species rather than single species (Fig. A). Diseases ranged considerably in their degree of ‘biogeographic complexity’, but only 15 of 35 possible combinations were represented in our dataset (Fig. B). The least complex examples involved transmission from the environment (E) only, a single reservoir (R) only, or human–human (H) only (CS = 1), while the most biogeographically complex diseases involved multiple vectors, reservoirs and could include either environmental (EVnRn) or human–human (HVnRn) transmission as well (CS = 8). On average, diseases including human–human transmission were the least biogeographically complex (‘H’ mean CS = 3.0), followed by diseases including transmission from reservoirs (‘R’ CS = 4.0), while diseases involving transmission from the environment (‘E’ mean CS = 4.3) and vectors (‘V’ mean CS = 5.2) were more complex. On average, complexity was not obviously related to commonness (Fig. C), but more complex diseases tended to be more geographically restricted (Fig. D), and had both a stronger rationale for mapping (Fig. E) as well as a higher proportion of diseases that had already been mapped (Fig. F), particularly when human–human or environmental transmission were never involved (traits that can radically increase their distributions to the point of making them essentially ubiquitous). However, the quality ratings of existing mapping efforts for more complex diseases were considerably lower on average than for simpler diseases (Fig. G). These trends illustrate the extent to which mapping efforts are already underway for clinically relevant infectious diseases but also highlight some important gaps. For instance, as of Hay et al.'s (2013) study, 31.9% of diseases with a strong rationale for mapping had not been mapped in any study (Fig. F), and declining overall quality of mapping efforts for more restricted and biogeographically complex diseases highlights a clear need to address the breadth of ecological interactions within disease systems in future studies.
Temporal effects
An additional consideration not explicitly included in Box 2 is that changes in interactions through time can influence the occurrence, transmission and emergence of many diseases. For example, short or longer term changes in land‐use and climate (both components of E) can influence D directly or through their influences on V, R and/or H (Epstein 2001, Patz et al. 2004, Nakazawa et al. 2007, Jones et al. 2013, Hoberg and Brooks 2015, Mackenstedt et al. 2015). Ebola virus disease (EVD) outbreaks, for example, have been closely associated with inter‐annual anomalies in meteorological seasonality, whereby sharply drier conditions at the end of the rainy season seem to favour the occurrence of outbreaks (Pinzon et al. 2004). More recently, Ebola outbreaks have been linked to fragmentation (Rulli et al. 2017) and recent (< 2 yr) deforestation (Olivero et al. 2017b).
Species distribution modeling (SDM) for infectious diseases
A promising approach emerging from ecology warranting additional attention here is the increasingly widespread use of species distribution modelling (SDM) (also called ecological niche modelling (ENM)). When applied to pathogens, hosts and/or reservoirs and vectors, SDM is useful for understanding risk factors conditioning the distributions, emergence or the accumulation of new outbreaks of disease. In SDMs, the available information on the presence or the incidence of disease is linked to a diverse set of environmental variables and allows the evaluation of the degree to which certain environments are favourable for the occurrence of disease, even in areas where it has not been detected before. In the absence of, or in combination with, local‐scale data suitable for mechanistic models, SDMs can take advantage of the large geographic scale to explore macroecological processes that are able to explain and predict the occurrence of disease. This can reveal emergent patterns and processes not perceptible at the local scale (Brown 1995), and can help lay a foundation for hypothesis testing or provide a geographic context for further studies on the ground (Allen et al. 2017).
The outputs provided by SDMs can be summarized in three main categories: probability (e.g. logistic regression, generalized additive models, random forests, boosted regression trees), suitability (e.g. MaxEnt, GARP) and favourability (e.g. favourability function). Suitability is an idiosyncratic way of ranking local sites according to their capacity to hold the species or pathogen that is not directly related to the probability of occurrence (Guisan and Zimmermann 2000, Royle et al. 2012). In contrast, favourability values can be obtained from probability and prevalence (here defined as the proportion of presences in the set of observations) (Real et al. 2006). Probability and suitability are biased in their outputs when working with samples differing in prevalence, which is not the case with favourability (Acevedo and Real 2012). This quality of favourability enables direct comparison and combination when several species are involved in the analytical design; for example, when using models for deriving indices based on multiple species (Estrada et al. 2008), and for the study of biogeographical relationships between species (Real et al. 2009, Acevedo et al. 2010), including relationships between pathogen and host complexes (Olivero et al. 2017a).
SDM approaches represent one of the major recent advances in infectious disease cartography (Hay et al. 2013, Peterson 2014, Kraemer et al. 2016), producing a diverse range of predictions on the presence or risk of human infectious diseases or their causative organisms (Peterson et al. 2004, Peterson 2006, Neerinckx et al. 2008, Reed et al. 2008, Samy et al. 2014, Zhu and Peterson 2014), and their animal hosts and vectors (Sweeney et al. 2006, de Oliveira et al. 2013, Giles et al. 2014). Modelling the distribution of biotic interactions is a relatively recent advance in macroecology and biogeography (Kissling et al. 2012, Wisz et al. 2013, Ovaskainen et al. 2016) and could similarly provide a further way forward for the analysis of pathogens with zoonotic cycles based on joint distributions and multispecies assemblages. Other methods of representing potential interactions at a community level (e.g. network modeling) or inferring potential hosts/vectors and host/vector ranges from geographic co‐occurrence are similarly being developed alongside (or in some cases integrated with) niche modeling approaches to yield novel insights on the spatial distribution of some infectious diseases (Stephens et al. 2009, 2016a).
Emergent patterns and multiple diseases
Returning to the framework in Box 2, G, E together with time (t) could each have more pervasive effects through their simultaneous influences on the other elements, giving rise to higher‐order biogeographic patterns that are defined by multiple diseases, such as co‐occurrences (including co‐infection patterns), chorotypes, diversity gradients, or biogeographic regionalisation. While biogeography has already made significant contributions to providing a generalized framework for disease mapping (Peterson 2014, Escobar and Craft 2016), we emphasise that it is the comparative power of biogeography that could help take pathogeography a step further, diverging more radically from medical geography, to address the factors that govern the structure, assembly, dynamics and spatial patterns of multiple or entire assemblages of human infectious diseases over a more diverse range of spatial and temporal scales. Below we provide some illustrative examples relevant to what has or potentially could be applied to human infectious diseases.
Chorotypes
A chorotype is a type of distribution pattern that is followed by one or several species (Baroni Urbani et al. 1978, Real et al. 2008, Passalacqua 2015). As chorotypes represent the shared geographical, ecological and evolutionary context of several species (Real et al. 2008, Fattorini 2015), these patterns could be useful for generating hypotheses about the causes and origins of host, reservoir and pathogen distributions. Although not yet widely explored for infectious diseases, chorotype analysis could significantly contribute to the study of the complex interactions characteristic of human infectious disease systems (Peterson 2008, Roche et al. 2013).
In the analysis of disease distributions, the relative importance of these interactions, compared to the relevance of other environmental factors, is variable. Some studies have raised this issue through the comparative analysis of pathogen models and host models based on their respective responses to environmental conditions (Maher et al. 2010, Costa and Peterson 2012). However, with the exception of using host distributions as a simple proxy for the potential distributions of pathogens (Daszak et al. 2012), the distribution of reservoir species has only recently been used as an explanatory variable, together with other environmental descriptors, to define a pathogen distribution model (e.g. compare Peterson et al. Walsh and Haseeb, and Pigott et al.'s models for Ebola virus) (Peterson et al. 2004, Walsh and Haseeb 2015, Pigott et al. 2016).
Although we are aware of no studies examining chorotypes of human infectious diseases, mammalian chorotypes have been recently incorporated into an infectious disease distribution model (Olivero et al. 2017a). When the ecology of a pathogen is complex and unresolved (e.g. Ebola virus, Leroy et al. 2004, Groseth et al. 2007, Olival and Hayman 2014), imposing restrictions to the selection of host or vector species considered in a model might under‐represent the zoological substrate conditioning a pathogen's transmission and distribution (Roche et al. 2012). Olivero et al. (2017a) thus addressed the mapping of favourable areas for the Ebola virus in the wild by combining two biogeographical approaches: SDM and chorotype analysis. Mammalian chorotypes in Africa were employed as surrogates of the types of distributions shown by reservoirs and any wildlife species implicated in the virus spillover cycle. Olivero et al. (2017a) found that a model based on a number of diversity patterns, each one associated with a different mammalian chorotype, defined favourable areas for the presence of Ebola virus with higher accuracy than did a model based on environmental variables alone (i.e. climate, forest type), concluding that mammalian biogeography contributes significantly to explaining the distribution of Ebola virus in Africa. In addition, vegetation was identified as a factor placing clear limits to the presence of the virus. Favourable areas for Ebola virus were thus determined from information provided by both models (Fig. 2).
Figure 4.
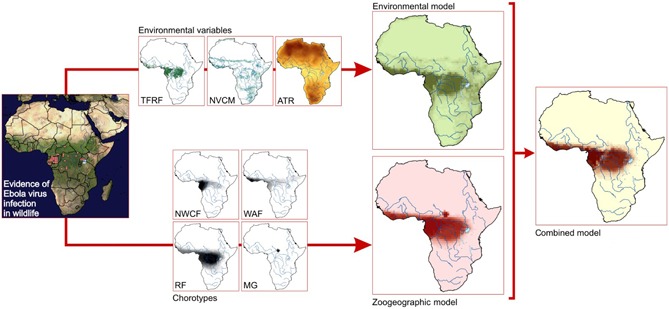
Modelling of the environmental/zoogeographic favourability for the presence of Ebola virus in wildlife. The model is based on serological evidence and observations of increased wildlife mortality attributed to Ebola virus disease. The environmental model is based on terra‐firme rain forests (TFRF), natural vegetation/cropland mosaics (NVCM) and annual temperature range (ATR, with increasing values from yellow to red). The zoogeographic model is derived from four types of mammalian distributions or chorotypes (see main text) (following Olivero et al. 2011): North‐Western Congolian Forest (NWCF), West‐African Forest (WAF), Rain Forest (RF) and the distribution of Mus goundae (MG). The two models are then combined according to fuzzy logic, requiring both environmentally and zoogeographically favourable conditions (from Olivero et al. 2017a, b).
Diversity patterns
It is now widely recognized that multiple pathogens may act independently or interact through a variety of different mechanisms to influence disease outcomes in human populations (Pederson and Fenton 2006, Jolles et al. 2008, Scholthof 2011), and yet studies of human infectious disease, host and vector diversity or community assembly patterns are rare in comparison to distributional studies of single infectious diseases (see ‘Single diseases’ above). Characterizing diversity patterns can thus provide a range of insights on the distributions and processes underlying multiple species of pathogens or diseases.
Based on components first proposed by Whittaker (1960), inventory diversity quantifies diversity within an environment, where alpha (α) diversity is used to refer to diversity at the local scale (i.e. smallest scale being measured). α‐diversity of human infectious diseases has been analysed in a number of studies, typically by comparing the number of diseases occurring in different countries at a global or continental scale due to limited comparative data availability at higher spatial resolutions. Although strongly heterogeneous (Fig. 3) (see also Stensgaard et al. 2017), some striking patterns suggest that human infectious disease communities are shaped by the same ecological processes that shape the diversity of life more generally (Guernier et al. 2004).
Figure 5.
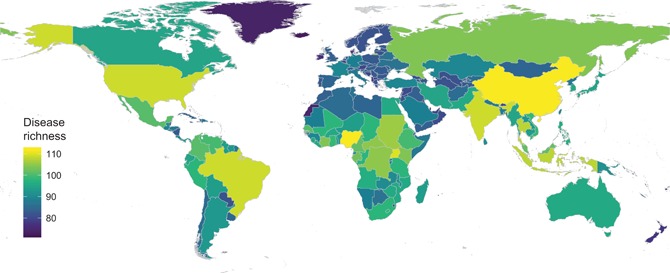
Alpha diversity (richness, or total number of different diseases) of clinically relevant human infectious diseases. Data are plotted at country level as derived from the GIDEON global infectious disease database (< http://www.gideononline.com/ >). Depiction is restricted to human infectious diseases for which a single causative agent is known (n = 187 diseases) (following Murray et al. 2015).
This is evident, for example, from observations of a clear latitudinal gradient in disease diversity and disease range sizes, whereby disease richness decreases and range size increases towards the poles (Fig. 4) (Guernier et al. 2004, Guernier and Guégan 2009). Other patterns are consistent with island biogeography theory, such as a positive relationship between land surface area and disease richness (Smith and Guégan 2010), and reduced richness on smaller islands and with distance to the nearest mainland (Jean et al. 2016). Together, these findings likely explain why the strongest predictor of human infectious disease richness known to date is wildlife richness (Dunn et al. 2010), while some vector groups show similar patterns (Foley et al. 2007). These patterns also illustrate that a correlation between human disease diversity and wildlife/vector diversity may not necessarily imply direct causation.
Nevertheless, other evidence points to the importance of animals, particularly mammalian and bird wildlife, as a key source of endemic and emerging human pathogens (Taylor et al. 2001, Woolhouse and Gowtage‐Sequeria 2005, Jones et al. 2008, Allen et al. 2017). In addition to the causal links, such parallels in patterns of human disease and other taxa reinforce the importance and potential utility of considering wildlife and vector biogeography alongside or as a central component of studies of infectious diseases, including distributional and diversity studies of known or potential zoonotic hosts and vectors (Foley et al. 2007, Cooper and Nunn 2013, Han et al. 2016, Olivero et al. 2017a).
In contrast to inventory diversity, proportional diversity measures the difference in diversity between environments or across gradients of habitats, commonly expressed as beta (β) diversity (Whittaker 1960, Jost 2007). β‐diversity patterns can be expressed in a number of ways, such as biogeographic regionalisations, which define biotic boundaries according to between‐area gaps in species composition, and biotic regions based on biotic similarities (Olivero et al. 2013). Such approaches, however, are yet to be widely applied to human infectious diseases.
In one study, β‐diversity patterns of human infectious diseases appear to parallel patterns in other taxa, consistent with patterns identified to date for α‐diversity (richness); Murray et al. (2015) show that human infectious disease assemblages exhibit biogeographic regionalisation reminiscent of zoogeographic patterns, particularly for zoonotic (Fig. 7A, B), vector‐borne and parasitic diseases, and that mammalian assemblage similarity is consistently among the strongest predictors of human infectious disease assemblage similarity among countries (Fig. 7C). Such an effect is very likely predictive of as‐yet undescribed patterns of microbial diversity, such as the geographic structure recently demonstrated among novel coronaviruses detected in wild bat hosts (Anthony et al. 2017). In addition to this dominant explanatory effect of biodiversity, other factors, including environmental (climate, land area, population size), social (human connectivity, health expenditure, observation effort) and epidemiological characteristics (e.g. pathogen type, transmission mode) also affect infectious disease α‐ and β‐diversity patterns (Fig. 7C). The strongest β‐diversity patterns, for example, can be observed in zoonotic, vector‐borne and parasitic infectious diseases, likely due to a more dominant role of environmental factors and persistence of historical dispersal barriers limiting their geographic distributions, while patterns of human‐specific diseases are far more homogenous at the global scale (Smith et al. 2007, Dunn et al. 2010, Just et al. 2014, Murray et al. 2015, Jean et al. 2016) (see also Box 3 Fig. panel D).
Figure 7.
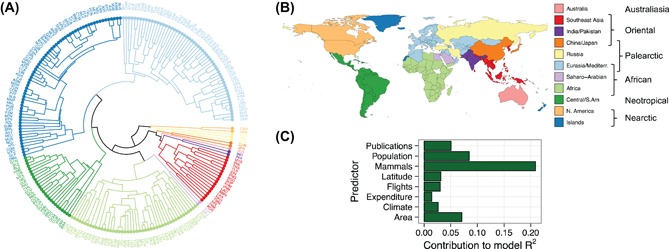
Global β‐diversity patterns of zoonotic diseases. (A) Hierarchical cluster analysis (UPGMA method) of a global disease‐by‐country presence–absence matrix represented as a circular dendrogram showing how countries group (regionalization) on the basis of the similarity (as measured by Sørensen β‐diversity) of their zoonotic infectious disease assemblages (following Kreft and Jetz 2010, Murray et al. 2015). Colours represent statistically supported groups (n = 11 groups) of countries that share similar diseases, as derived by evaluating results from the Silhouette, Elbow, CH index and Gap statistic tests; (B) global pathogeographic realms for zoonotic diseases derived from (A) (colours for mapping match country clusters identified in (A)). Legend labels indicate the statistically supported regions (first column) and how these align with ‘classic’ zoogeographic realms (second column) (note although the realm label for Nearctic includes Greenland for illustration purposes, the ‘Islands’ group (dark blue) actually includes a large number of small islands plus a few other countries scattered globally (A) that may cluster on the basis of being a depauperate or data deficient group); (C) the relative explanatory value of a range of social and environmental covariates for explaining these global patterns in disease beta diversity, illustrating that mammalian biodiversity is the best predictor of zoonotic disease diversity at a global scale (as derived from a relative importance analysis following multiple regression on distance matrices controlling for the effects of spatial autocorrelation (following Murray et al. (2015)).
β‐diversity can be further decomposed into two separable components, nestedness and turnover, which may further help characterize the processes driving differences in the composition of assemblages between sites (Harrison et al. 1992, Baselga 2010). Nestedness occurs where species assemblages are subsets of the biotas at more diverse sites (Wright and Reeves 1992, Ulrich and Gotelli 2007), and indicates a non‐random process arising from any factor that promotes the orderly disaggregation of assemblages (Gaston et al. 2000). The latitudinal gradient of human infectious diseases exhibits such a pattern, with disease assemblages occurring at higher latitudes being subsets of those occurring closer to the equator (Fig. 6C) (Guernier et al. 2004). In contrast, turnover indicates the replacement of some species by others as a consequence of environmental sorting or spatial and historical constraints (Qian et al. 2005, Baselga 2010). In the only assessment of nestedness vs turnover undertaken so far for human infectious diseases that we are aware of, both nestedness and turnover appear to contribute to overall differences in infectious disease assemblages among countries at a global scale, with the relative contribution varying between major epidemiological classes (Murray et al. 2015). For example, nestedness dominates differences in human‐specific diseases, whereas turnover dominates differences in zoonotic and vector‐borne diseases.
Figure 6.
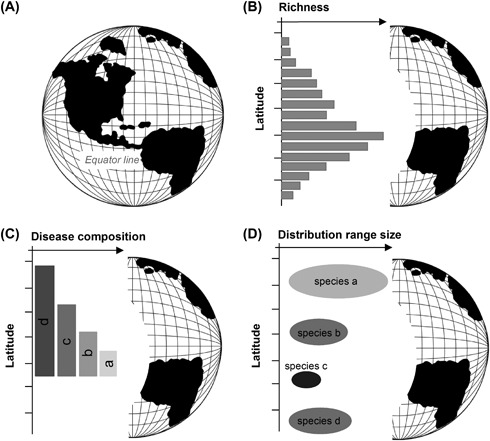
Schematic illustrating the latitudinal variation and diversity patterns of human infectious diseases. (A) A global view of Earth showing latitudinal bands; (B) disease richness (grey bars): the total number of different human infectious diseases present per latitudinal unit (e.g. 40–45°) increases towards the tropics (adapted from Guernier et al. (2004)); (C) nestedness: a hierarchical pattern of human pathogen composition with the pathogen species found at higher latitudes (darker bars) constituting nested subsets of those in progressively richer communities at lower latitudes (lighter bars) (adapted from Guernier et al. (2004)); (D) disease range size: narrower distributional ranges occur in the tropics (darker circles) for human pathogens compared to higher latitudes (lighter bars) (adapted from Guernier and Guégan (2009)).
Concluding remarks – leveraging pathogeography for health research and management
Biogeographic methods and outputs have already contributed and continue to show great promise for a number of health management or research applications on infectious diseases, which could help direct the allocation of scarce public and global health resources more efficiently and effectively. As our abilities to assemble ecological datasets and conduct infectious disease surveillance and analyses are steadily improving, multidimensional ecological data can be mapped and relationships can be identified as data accumulate in close to real‐time, providing decision‐relevant information for health managers and researchers to respond to.
This has already lead to rapid advances in improving disease mapping for single infectious diseases and a closing of the gap between the data types and methods used to characterise disease and species distributions by medical geographers and ecologists, respectively. Many other applications are conceivable albeit so far poorly explored.
For example, taking inspiration from conservation and ecological applications, diversity analyses and biogeographic regionalisation could be used to test and propose hypotheses about ecological factors and historical events that could underlie the current organization of disease assemblages or combined disease risks (e.g. identifying processes of disease dispersal, establishment, and extinction and the ‘upstream’ risk factors or drivers of novel health threats) (following Carmona et al. 2000, Báez et al. 2005); to define contexts for representativeness (e.g. improved disease surveillance design) (following Austin and Margules 1986, Carey et al. 1995, Mackey 2008); to provide consistent units for environmental management and for sampling stratification (e.g. for the optimal discovery of novel pathogens) (following Bunce et al. 1996, Wright et al. 1998); and as geographic contexts for imputation/extrapolation or forecasting when data from a unit within a region is missing or unavailable (e.g. where disease surveillance coverage is low or patchy) (Cooper and Nunn 2013).
Biogeographic approaches may also be useful for examining the risks associated with emerging infectious diseases (EIDs), since data are often extremely limited on EIDs and yet the priorities for management revolve around anticipating (through forecasting) when, where and why emergence of pathogens in human populations occurs (Peterson 2008, Morse et al. 2012). Large‐scale demographic and environmental factors and changes in these factors are increasingly being recognized as key drivers underlying disease emergence, with shifts in the distributions of disease hosts and vectors being central to this process (Jones et al. 2008, Semenza et al. 2016).
Given that most pathogen distributions are very poorly characterized or completely unknown (Hay et al. 2013), and that most of the microbial diversity from which novel and potentially pathogenic agents could originate are as yet undiscovered (Anthony et al. 2013), biogeographic pattern definition and process identification based on historical patterns of disease occurrence, recent emergence events, or proxy taxa (e.g. mammalian or arthropod vector biodiversity) may provide some of the earliest and in some cases the only insights into such burgeoning or future disease risks (Fig. 8A, B) (Murray et al. 2015), which may give way to more refined models as data quality or availability increases (Fig. 8C, Fig. 1). Increasing pathogeographic awareness, participation and collaboration among ecologists, biogeographers and veterinary and medical practitioners could thus contribute to closing the gap between environmental and health management, increased inter‐disciplinary research and management efficiency, and reductions in the global burden of disease.
Figure 8.
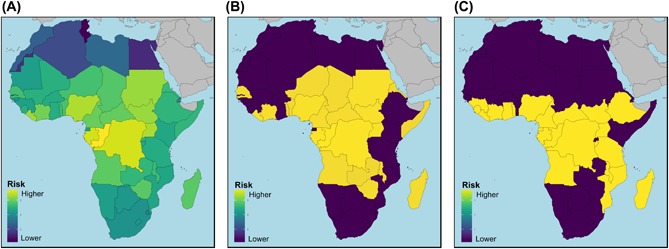
Comparing approaches to country‐level Ebola risk assessment for Africa. (A) Co‐occurrence (β‐diversity) analysis of historical zoonotic disease occurrence among countries in which Ebola outbreaks have occurred in humans. ‘Higher risk’ countries are defined as those with more similar zoonotic disease diversity to Ebola‐positive countries (human index cases only) (following Murray et al. 2015). Countries with index cases are shown in Fig. 1 (thick black line). (B) Top 22 ranked countries from (A), for comparison with the 22 ‘at risk’ countries as determined by Pigott et al. (2014) (C). (C) ‘At risk’ countries (n = 22, yellow colour) as determined by high resolution spatial modeling aggregated to country level. The underlying model is based on Ebola outbreaks (index cases) in humans and infection in wildlife and analysis of spatial covariates (data from Pigott et al. 2014) (see Fig. 1 for raw model output and description). Yellow indicates countries that contain some environmentally suitable areas for Ebola. Darker colour indicates countries that do not contain areas predicted to be suitable for Ebola. The overlap in top 22 priority countries between (B) and (C) is ~70%. The approach taken in (A/B) requires no specific information about the target disease and could provide a relevant biogeographic ‘prior’ for planning (e.g. having an emergency response plan in place), reacting to novel appearance of diseases (e.g. for prioritizing surveillance), in data poor settings, and for conservative risk assessment. The approach taken in (C) is more refined and specific but also more data intensive (see Fig. 1 for more detail).
Data deposition
Data available from the Dryad Digital Repository: < http://dx.doi.org/10.5061/dryad.p1n10dv > (Murray et al. 2018).
Funding – KM is partially supported by UK Medical Research Council grant MR/P024513/1. BR and JFG are funded by an ‘Investissement d'Avenir’ Laboratoire d'Excellence Centre d'Etude de la Biodiversité Amazonienne Grant (ANR‐10‐LABX‐2501). JO is partially supported by the project CGL2016‐76747‐R of the Spanish Ministry of Economy and Competitiveness and FEDER Funds.
Supplementary material (Appendix ECOG‐03625 at < http://www.ecography.org/appendix/ecog-03625 >). Appendix 1.
References
- Acevedo P. Real R. 2012. Favourability: concept, distinctive characteristics and potential usefulness. – Naturwissenschaften 99: 515–522. [DOI] [PubMed] [Google Scholar]
- Acevedo P. et al. 2010. Assessing biogeographical relationships of ecologically related species using favourability functions: a case study on British deer. – Divers. Distrib. 16: 515–528. [Google Scholar]
- Allen T. et al. 2017. Global hotspots and correlates of emerging zoonotic diseases. – Nat. Commun. 8: 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S. J. et al. 2013. A strategy to estimate unknown viral diversity in mammals. – mBio 4: e00598‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S. J. 2017. Global patterns in coronavirus diversity. – Virus Evol. 3: vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M. P. Margules C. R. 1986. Assessing representativeness. – In: Usher M. (ed.), Wildlife conservation evaluation. Chapman and Hall, pp. 45–67. [Google Scholar]
- Báez J. C. et al. 2005. A biogeographical analysis of the genera Audoinella (Rhodophyta), Cystoseira (Phaeophyceae) and Cladophora (Chlorophyta) in the western Mediterranean Sea and Adriatic Sea. – Phycol. Res. 53: 255–265. [Google Scholar]
- Balcan D. et al. 2009. Multiscale mobility networks and the spatial spreading of infectious diseases. – Proc. Natl Acad. Sci. USA 106: 21484–21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni Urbani C. et al. 1978. Materiali per una biogeografia italiana fondata su alcuni generi di coleotteri Cicindelidi, Carabidi e Crisomelidi. – Memoire‐Societa Entomol. Italiana 56: 35–92. [Google Scholar]
- Barrett F. A. 2000. Disease & geography: the history of an idea. – Atkinson College, Dept of Geography. [Google Scholar]
- Baselga A. 2010. Partitioning the turnover and nestedness components of beta diversity. – Global Ecol. Biogeogr. 19: 134–143. [Google Scholar]
- Berger S. A. 2005. GIDEON: a comprehensive web‐based resource for geographic medicine. – Int. J. Health Geogr. 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S. et al. 2017. Improved prediction accuracy for disease risk mapping using Gaussian process stacked generalization. – J. R. Soc. Interface 14, doi:10.1098/rsif.2017.0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn T. M. Gaston K. J. 2006. There's more to macroecology than meets the eye. – Global Ecol. Biogeogr. 15: 537–540. [Google Scholar]
- Brown J. H. Lomolino M. V. 1998. Biogeography. – Sinauer Associates. [Google Scholar]
- Brown J. H. S. 1995. Macroecology. – Univ. of Chicago Press. [Google Scholar]
- Bunce R. G. H. et al. 1996. Land classification for strategic ecological survey. – J. Environ. Manage. 47: 37–60. [Google Scholar]
- Cameron A. 2012. The consequences of risk‐based surveillance: developing output‐based standards for surveillance to demonstrate freedom from disease. – Prev. Vet. Med. 105: 280–286. [DOI] [PubMed] [Google Scholar]
- Caprarelli G. Fletcher S. 2014. A brief review of spatial analysis concepts and tools used for mapping, containment and risk modelling of infectious diseases and other illnesses. – Parasitology 141: 581–601. [DOI] [PubMed] [Google Scholar]
- Carey P. D. et al. 1995. An environmentally defined biogeographical zonation of Scotland designed to reflect species distributions. – J. Ecol. 83: 833–845. [Google Scholar]
- Carmona J. A. et al. 2000. Testing for inter‐drainage connections on the basis of the distribution pattern of endemic freshwater. – Arch. Hydrobiol. 150: 101–116. [Google Scholar]
- Chesson P. 2012. Scale transition theory: its aims, motivations and predictions. – Ecol. Complex. 10: 52–68. [Google Scholar]
- Civitello D. J. et al. 2015. Biodiversity inhibits parasites: broad evidence for the dilution effect. – Proc. Natl Acad. Sci. USA 112: 8667–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff A. Haggett P. 2004. Time, travel and infection. – Br. Med. Bull. 69: 87–99. [DOI] [PubMed] [Google Scholar]
- Cliff A. et al. 2000. Island epidemics. – Oxford Univ. Press. [Google Scholar]
- Colizza V. et al. 2007. Modeling the worldwide spread of pandemic influenza: baseline case and containment interventions. – PLoS Med. 4: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N. Nunn C. L. 2013. Identifying future zoonotic disease threats: where are the gaps in our understanding of primate infectious diseases? – Evol. Med. Public Health 2013: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J. Peterson A. T. 2012. Ecological niche modeling as a tool for understanding distributions and interactions of vectors, hosts, and etiologic agents of Chagas disease. – Adv. Exp. Med. Biol. 710: 59–70. [DOI] [PubMed] [Google Scholar]
- Costanza R. et al. 2014. Changes in the global value of ecosystem services. – Global Environ. Change 26: 152–158. [Google Scholar]
- Cox C. B. 2016. Biogeography: an ecological and evolutionary approach. – John Wiley and Sons. [Google Scholar]
- Daszak P. et al. 2012. Interdisciplinary approaches to understanding disease emergence: the past, present, and future drivers of Nipah virus emergence. – Proc. Natl Acad. Sci. USA (Suppl. 1) 110: 3681–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira S. V. et al. 2013. Potential geographic distribution of hantavirus reservoirs in Brazil. – PLoS One 8: e85137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R. R. et al. 2010. Global drivers of human pathogen richness and prevalence. – Proc. R. Soc. B 277: 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg J. N. S. et al. 2007. Environmental determinants of infectious disease: a framework for tracking causal links and guiding public health research. – Environ. Health Perspect. 115: 1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein P. R. 2001. Climate change and emerging infectious diseases. – Microbes Infect. 3: 747–54. [DOI] [PubMed] [Google Scholar]
- Escobar L. E. Craft M. E. 2016. Advances and limitations of disease biogeography using ecological niche modeling. – Front. Microbiol. 7: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada A. et al. 2008. Using crisp and fuzzy modelling to identify favourability hotspots useful to perform gap analysis. – Biodivers. Conserv. 17: 857–871. [Google Scholar]
- Fattorini S. 2015. On the concept of chorotype. – J. Biogeogr. 42: 2246–2251. [Google Scholar]
- Fisher M. C. et al. 2012. Emerging fungal threats to animal, plant and ecosystem health. – Nature 484: 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley D. H. et al. 2007. Insight into global mosquito biogeography from country species records. – J. Med. Entomol. 44: 554–567. [DOI] [PubMed] [Google Scholar]
- Foley J. A. et al. 2005. Global consequences of land use. – Science 309: 570–574. [DOI] [PubMed] [Google Scholar]
- Gaston K. J. et al. 2000. Abundance–occupancy relationships. – J. Appl. Ecol. 37: 39–59. [Google Scholar]
- Giles J. R. et al. 2014. Invasive potential of cattle fever ticks in the southern United States. – Parasites Vectors 7: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A. et al. 2007. The ecology of Ebola virus. – Trends Microbiol. 15: 408–416. [DOI] [PubMed] [Google Scholar]
- Guernier V. Guégan J.‐F. 2009. May Rapoport's rule apply to human associated pathogens? – EcoHealth 6: 509–521. [DOI] [PubMed] [Google Scholar]
- Guernier V. et al. 2004. Ecology drives the worldwide distribution of human diseases. – PLoS Biol. 2: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisan A. Zimmermann N. E. 2000. Predictive habitat distribution models in ecology. – Ecol. Model. 135: 147–186. [Google Scholar]
- Han B. A. et al. 2016. Global patterns of zoonotic disease in mammals. – Trends Parasitol. 32: 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. et al. 1992. Beta diversity on geographic gradients in Britain. – J. Anim. Ecol. 151–158. [Google Scholar]
- Hay S. et al. 2006. Global environmental data for mapping infectious disease distribution. – Adv. Parasitol. 62: 37–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay S. I. et al. 2013. Global mapping of infectious disease. – Phil. Trans. R. Soc. B 368: 20120250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoberg E. P. Brooks D. R. 2015. Evolution in action: climate change, biodiversity dynamics and emerging infectious disease. – Phil. Trans. R. Soc. B 370: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M. E. Nunn C. L. 2010. Gap analysis and the geographical distribution of parasites. – In: Krasnov S. M. A. B. (ed.), The biogeography of host–parasite interactions. Oxford Univ. Press, pp. 129–142. [Google Scholar]
- Hosseini P. R. et al. 2017. Does the impact of biodiversity differ between emerging and endemic pathogens? The need to separate the concepts of hazard and risk. – Phil. Trans. R. Soc. B 372: 20160129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G. M. 1989. Historical evolution of disease mapping in general and specifically of cancer mapping. – In: Boyle P. et al. (eds), Cancer mapping. Springer, pp. 1–21. [DOI] [PubMed] [Google Scholar]
- Jean K. et al. 2016. An equilibrium theory signature in the island biogeography of human parasites and pathogens. – Global Ecol. Biogeogr. 25: 107–116. [Google Scholar]
- Johnson P. T. et al. 2015. Why infectious disease research needs community ecology. – Science 349: 1259504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolles A. E. et al. 2008. Interactions between macroparasites and microparasites drive infection patterns in free‐ranging African buffalo. – Ecology 89: 2239–2250. [DOI] [PubMed] [Google Scholar]
- Jones B. A. et al. 2013. Zoonosis emergence linked to agricultural intensification and environmental change. – Proc. Natl Acad. Sci. USA 110: 8399–8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H. 1990. Population geography. – Paul Chapman Publishing. [Google Scholar]
- Jones K. E. et al. 2008. Global trends in emerging infectious diseases. – Nature 451: 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. 2007. Partitioning diversity into independent alpha and beta components. – Ecology 88: 2427–2439. [DOI] [PubMed] [Google Scholar]
- Jurdak R. et al. 2015. Understanding human mobility from Twitter. – PLoS One 10: e0131469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just M. G. 2014. Global biogeographic regions in a human‐dominated world: the case of human diseases. – Ecosphere 5: art143. [Google Scholar]
- Kissling W. D. et al. 2012. Towards novel approaches to modelling biotic interactions in multispecies assemblages at large spatial extents. – J. Biogeogr. 39: 2163–2178. [Google Scholar]
- Kraemer M. U. et al. 2016. Progress and challenges in infectious disease cartography. – Trends Parasitol. 32: 19–29. [DOI] [PubMed] [Google Scholar]
- Kreft H. Jetz W. 2010. A framework for delineating biogeographical regions based on species distributions. – J. Biogeogr. 37: 2029–2053. [Google Scholar]
- Lambin E. F. et al. 2010. Pathogenic landscapes: interactions between land, people, disease vectors, and their animal hosts. – Int. J. Health Geogr. 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J. 2000. Infectious history. – Science 288: 287–293. [DOI] [PubMed] [Google Scholar]
- Leroy E. M. et al. 2004. Multiple Ebola virus transmission events and rapid decline of central African wildlife. – Science 303: 387–390. [DOI] [PubMed] [Google Scholar]
- Lindgren E. et al. 2012. Monitoring EU emerging infectious disease risk due to climate change. – Science 336: 418–419. [DOI] [PubMed] [Google Scholar]
- Lomolino M. V. et al. 2006. Biogeography. – Sinauer Associates. [Google Scholar]
- Mackenstedt U. et al. 2015. The role of wildlife in the transmission of parasitic zoonoses in peri‐urban and urban areas. – Int. J. Parasitol. 4: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey B. G. 2008. Boundaries, data and conservation. – J. Biogeogr. 35: 392–393. [Google Scholar]
- Magalhães R. J. S. et al. 2011. The applications of model‐based geostatistics in helminth epidemiology and control. – Adv. Parasitol. 74: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher S. P. et al. 2010. Range‐wide determinants of plague distribution in North America. – Am. J. Trop. Med. Hyg. 83: 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren L. Hawe P. 2005. Ecological perspectives in health research. – J. Epidemiol. Commun. Health 59: 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael A. J. 1999. Prisoners of the proximate: loosening the constraints on epidemiology in an age of change. – Am. J. Epidemiol. 149: 887–897. [DOI] [PubMed] [Google Scholar]
- MEA 2005. Millennium ecosystem assessment: ecosystems and human well‐being. – Island Press. [Google Scholar]
- Messina J. P. et al. 2015. The many projected futures of dengue. – Nat. Rev. Microbiol. 13: 230–239. [DOI] [PubMed] [Google Scholar]
- Morrone J. J. 2009. Evolutionary biogeography: an integrative approach. – Columbia Univ. Press. [Google Scholar]
- Morse S. S. 1995. Factors in the emergence of infectious diseases. – Emerg. Infect. Dis. 1: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S. S. et al. 2012. Prediction and prevention of the next pandemic zoonosis. – Lancet 380: 1956–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. J. et al. 2013. Disability‐adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. – Lancet 380: 2197–2223. [DOI] [PubMed] [Google Scholar]
- Murray K. A. Daszak P. 2013. Human ecology in pathogenic landscapes: two hypotheses on how land use change drives viral emergence. – Curr. Opinion Virol. 3: 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K. A. et al. 2015. Global biogeography of human infectious diseases. – Proc. Natl Acad. Sci. USA 112: 12746–12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K. A. et al. 2018. Data from: Pathogeography: leveraging the biogeography of human infectious diseases for global health management. – Dryad Digital Repository, < 10.5061/dryad.p1n10dv >. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. A. Giller P. S. 1988. Process, pattern and scale in biogeography. – In: Myers A. A. and Giller P. S. (eds), Analytical biogeography. Springer, pp. 3–12. [Google Scholar]
- Myers S. S. et al. 2013. Human health impacts of ecosystem alteration. – Proc. Natl Acad. Sci. USA 110: 18753–18760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y. et al. 2007. Climate change effects on plague and tularemia in the United States. – Vector‐Borne Zoonotic Dis. 7: 529–540. [DOI] [PubMed] [Google Scholar]
- Neerinckx S. B. et al. 2008. Geographic distribution and ecological niche of plague in sub‐Saharan Africa. – Int. J. Health Geogr. 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusser S. M. et al. 2008. Sampling considerations for disease surveillance in wildlife populations. – J. Wildl. Manage. 72: 52–60. [Google Scholar]
- Olival K. J. Hayman D. T. 2014. Filoviruses in bats: current knowledge and future directions. – Viruses 6: 1759–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivero J. et al. 2011. Fuzzy chorotypes as a conceptual tool to improve insight into biogeographic patterns. – Syst. Biol. 60: 645–660. [DOI] [PubMed] [Google Scholar]
- Olivero J. et al. 2013. Integrating fuzzy logic and statistics to improve the reliable delimitation of biogeographic regions and transition zones. – Syst. Biol. 62: 1–21. [DOI] [PubMed] [Google Scholar]
- Olivero J. et al. 2017a. Mammalian biogeography and the Ebola virus in Africa. – Mammal. Rev. 47: 24–37. [Google Scholar]
- Olivero J. et al. 2017b. Recent loss of closed forests is associated with Ebola virus disease outbreaks. – Sci. Rep. 7: 14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostfeld R. S. et al. 2005. Spatial epidemiology: an emerging (or re‐emerging) discipline. – Trends Ecol. Evol. 20: 328–336. [DOI] [PubMed] [Google Scholar]
- Ovaskainen O. et al. 2016. Uncovering hidden spatial structure in species communities with spatially explicit joint species distribution models. – Methods Ecol. Evol. 7: 428–436. [Google Scholar]
- Palmer S. R. et al. 1998. Zoonoses: biology, clinical practice and public health control. – Oxford Univ. Press. [Google Scholar]
- Passalacqua N. G. 2015. On the definition of element, chorotype and component in biogeography. – J. Biogeogr. 42: 611–618. [Google Scholar]
- Patz J. A. et al. 2004. Unhealthy landscapes: policy recommendations on land use change and infectious disease emergence. – Environ. Health Perspect. 112: 1092–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovsky E. N. 1966. Natural nidality of transmissible diseases with special reference to the landscape epidemiology of zooanthroponoses. – Univ. Illinois Press. [Google Scholar]
- Pederson A. B. Fenton A. 2006. Emphasizing the ecology in parasite community ecology. – Trends Ecol. Evol. 22: 133–139. [DOI] [PubMed] [Google Scholar]
- Peterson A. T. 2006. Ecological niche modeling and spatial patterns of disease transmission. – Emerg. Infect. Dis. 12: 1822–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson A. T. 2008. Biogeography of diseases: a framework for analysis. – Naturwissenschaften 95: 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson A. T. 2014. Mapping disease transmission risk: enriching models using biogeography and ecology. – JHU Press. [Google Scholar]
- Peterson A. T. et al. 2004. Ecologic and geographic distribution of filovirus disease. – Emerg. Infect. Dis. 10: 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigott D. M. et al. 2014. Mapping the zoonotic niche of Ebola virus disease in Africa. – Elife 3: e04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigott D. M. et al. 2016. Updates to the zoonotic niche map of Ebola virus disease in Africa. – Elife 5: e16412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzon J. E. et al. 2004. Trigger events: enviroclimatic coupling of Ebola hemorrhagic fever outbreaks. – Am. J. Trop. Med. Hyg. 71: 664–674. [PubMed] [Google Scholar]
- Plowright R. K. et al. 2017. Pathways to zoonotic spillover. – Nat. Rev. Microbiol. 15: 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H. et al. 2005. Beta diversity of angiosperms in temperate floras of eastern Asia and eastern North America. – Ecol. Lett. 8: 15–22. [Google Scholar]
- Raudsepp‐Hearne C. et al. 2010. Untangling the environmentalist's paradox: why is human well‐being increasing as ecosystem services degrade? – Bioscience 60: 576–589. [Google Scholar]
- Real R. et al. 2006. Obtaining environmental favourability functions from logistic regression. – Environ. Ecol. Stat. 13: 237–245. [Google Scholar]
- Real R. et al. 2008. Using chorotypes to deconstruct biogeographical and biodiversity patterns: the case of breeding waterbirds in Europe. – Global Ecol. Biogeogr. 17: 735–746. [Google Scholar]
- Real R. et al. 2009. Conservation biogeography of ecologically interacting species: the case of the Iberian lynx and the European rabbit. – Divers. Distrib. 15: 390–400. [Google Scholar]
- Redding D. W. et al. 2016. Environmental–mechanistic modelling of the impact of global change on human zoonotic disease emergence: a case study of Lassa fever. – Methods Ecol. Evol. 7: 646–655. [Google Scholar]
- Reed K. D. et al. 2008. Ecologic niche modeling of Blastomyces dermatitidis in Wisconsin. – PLoS One 3: e2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert I. Palti J. 1967. Prediction of plant disease occurrence a patho‐geographical approach. – Mycopathol. Mycol. Appl. 32: 337–355. [Google Scholar]
- Reperant L. A. 2010. Applying the theory of island biogeography to emerging pathogens: toward predicting the sources of future emerging zoonotic and vector‐borne diseases. – Vector‐Borne Zoonotic Dis. 10: 105–110. [DOI] [PubMed] [Google Scholar]
- Roche B. et al. 2012. Linking community and disease ecology: the impact of biodiversity on pathogen transmission. – Phil. Trans. R. Soc. B 367: 2807–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B. et al. 2013. The impact of community organization on vector‐borne pathogens. – Am. Nat. 181: 1–11. [DOI] [PubMed] [Google Scholar]
- Rogers D. J. et al. 2002. Satellite imagery in the study and forecast of malaria. – Nature 415: 710–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle J. A. et al. 2012. Likelihood analysis of species occurrence probability from presence‐only data for modelling species distributions. – Methods Ecol. Evol. 3: 545–554. [Google Scholar]
- Rulli M. C. et al. 2017. The nexus between forest fragmentation in Africa and Ebola virus disease outbreaks. – Sci. Rep. 7: 41613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samy A. M. et al. 2014. Mapping the potential risk of mycetoma infection in Sudan and South Sudan using ecological niche modeling. – PLoS Negl. Trop. Dis. 8: e3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof K. B. 2011. Taking some of the mystery out of host–virus interactions. – PLoS Pathogen. 7: e1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. 1994. The fallacy of the ecological fallacy: the potential misuse of a concept and the consequences. – Am. J. Public Health 84: 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza J. C. et al. 2016. Determinants and drivers of infectious disease threat events in Europe. – Emerg. Infect. Dis. 22: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. F. Guégan J.‐F. 2010. Changing geographic distributions of human pathogens. – Annu. Rev. Ecol. Evol. Syst. 41: 231–250. [Google Scholar]
- Smith K. F. et al. 2007. Globalization of human infectious disease. – Ecology 88: 1903–1910. [DOI] [PubMed] [Google Scholar]
- Soberón J. 2007. Grinnellian and Eltonian niches and geographic distributions of species. – Ecol. Lett. 10: 1115–1123. [DOI] [PubMed] [Google Scholar]
- Stensgaard A.‐S. 2017. The neglected geography of human pathogens and diseases. – Nat. Ecol. Evol. 1: s41559‐017‐0190. [DOI] [PubMed] [Google Scholar]
- Stephens C. R. et al. 2009. Using biotic interaction networks for prediction in biodiversity and emerging diseases. – PLoS One 4: e5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C. R. et al. 2016a. Can you judge a disease host by the company it keeps? Predicting disease hosts and their relative importance: a case study for Leishmaniasis. – PLoS Negl. Trop. Dis. 10: e0005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P. R. et al. 2016b. The macroecology of infectious diseases: a new perspective on global‐scale drivers of pathogen distributions and impacts. – Ecol. Lett. 19: 1159–1171. [DOI] [PubMed] [Google Scholar]
- Suk J. E. Semenza J. C. 2011. Future infectious disease threats to Europe. – Am. J. Public Health 101: 2068–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney A. W. et al. 2006. Environmental factors associated with distribution and range limits of malaria vector Anopheles farauti in Australia. – J. Med. Entomol. 43: 1068–1075. [DOI] [PubMed] [Google Scholar]
- Taylor L. H. et al. 2001. Risk factors for human disease emergence. – Phil. Trans. R. Soc. B 356: 983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich W. Gotelli N. J. 2007. Null model analysis of species nestedness patterns. – Ecology 88: 1824–1831. [DOI] [PubMed] [Google Scholar]
- Victor L. Y. Edberg S. C. 2005. Global infectious diseases and epidemiology network (GIDEON): a world wide web‐based program for diagnosis and informatics in infectious diseases. – Clin. Infect. Dis. 40: 123–126. [DOI] [PubMed] [Google Scholar]
- Waldron A. et al. 2017. Reductions in global biodiversity loss predicted from conservation spending. – Nature 551: 364. [DOI] [PubMed] [Google Scholar]
- Wallace A. R. 1876. The geographical distribution of animals, with a study of the relations of living and extinct faunas as elucidating the past changes of the earth's surface. – Macmillan. [Google Scholar]
- Walsh M. G. Haseeb M. 2015. The landscape configuration of zoonotic transmission of Ebola virus disease in west and central Africa: interaction between population density and vegetation cover. – PeerJ 3: e735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardeh M. et al. 2015. Database of host‐pathogen and related species interactions, and their global distribution. – Sci. Data 2: 150049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts N. et al. 2015. Health and climate change: policy responses to protect public health. – Lancet 386: 1861–1914. [DOI] [PubMed] [Google Scholar]
- Wesolowski A. et al. 2012. Quantifying the impact of human mobility on malaria. – Science 338: 267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmee S. et al. 2015. Safeguarding human health in the Anthropocene epoch: report of The Rockefeller Foundation–Lancet Commission on planetary health. – Lancet 386: 1973–2028. [DOI] [PubMed] [Google Scholar]
- Whittaker R. H. 1960. Vegetation of the Siskiyou mountains, Oregon and California. – Ecol. Monogr. 30: 279–338. [Google Scholar]
- WHO 1959. Zoonoses: second report of the joint WHO/FAO expert committee. – World Health Organization. [Google Scholar]
- Wisz M. S. et al. 2013. The role of biotic interactions in shaping distributions and realised assemblages of species: implications for species distribution modelling. – Biol. Rev. 88: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M. E. et al. 2008. Temporal trends in the discovery of human viruses. – Proc. R. Soc. B 275: 2111–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M. E. et al. 2016. Assessing the epidemic potential of RNA and DNA viruses. – Emerg. Infect. Dis. 22: 2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M. E. J. Gowtage‐Sequeria S. 2005. Host range and emerging and reemerging pathogens. – Emerg. Infect. Dis. 11: 1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. H. Reeves J. H. 1992. On the meaning and measurement of nestedness of species assemblages. – Oecologia 92: 416–428. [DOI] [PubMed] [Google Scholar]
- Wright R. G. et al. 1998. Ecoregions as a level of ecological analysis. – Biol. Conserv. 86: 207–213. [Google Scholar]
- Yang K. et al. 2012. Global distribution of outbreaks of water‐associated infectious diseases. – PLoS Negl. Trop. Dis. 6: e1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G. Peterson A. T. 2014. Potential geographic distribution of the novel avian‐origin influenza A (H7N9) virus. – PLoS One 9: e93390. [DOI] [PMC free article] [PubMed] [Google Scholar]


