Significance
Photoenzymes use light as an energy source to catalyze chemical reactions and can be engineered as environmental-friendly biocatalysts. The LPOR, an enzyme late in the biosynthesis of chlorophyll, is one of the few naturally occurring photoenzymes. However, limited structural information prevents understanding of its working mechanism. In this study, we report two structures of cyanobacterial LPOR bound to NADPH, which reveals the molecular basis of a NADPH-binding pocket, a substrate cavity, and the proton-relay path. Our findings will help to elucidate and design the light-driven enzymes for the future development of photocatalysis.
Keywords: chlorophyll biosynthesis, photocatalysis, NADPH, proton relay, crystal structure
Abstract
The reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide) is the penultimate step of chlorophyll biosynthesis. In oxygenic photosynthetic bacteria, algae, and plants, this reaction can be catalyzed by the light-dependent Pchlide oxidoreductase (LPOR), a member of the short-chain dehydrogenase superfamily sharing a conserved Rossmann fold for NAD(P)H binding and the catalytic activity. Whereas modeling and simulation approaches have been used to study the catalytic mechanism of this light-driven reaction, key details of the LPOR structure remain unclear. We determined the crystal structures of LPOR from two cyanobacteria, Synechocystis sp. PCC 6803 and Thermosynechococcus elongatus. Structural analysis defines the LPOR core fold, outlines the LPOR–NADPH interaction network, identifies the residues forming the substrate cavity and the proton-relay path, and reveals the role of the LPOR-specific loop. These findings provide a basis for understanding the structure-function relationships of the light-driven Pchlide reduction.
POR catalyzes the stereoselective reduction of the C17 = C18 double bond of Pchlide to form Chlide, the penultimate step of chlorophyll biosynthesis (1–3). There are two structurally unrelated PORs in photosynthetic organisms: the dark-operative POR (DPOR; EC 1.3.7.7) and the LPOR (EC 1.3.1.33). DPOR is a three-subunit enzyme complex, powered by ATP hydrolysis and structurally homologous to nitrogenase (4, 5). LPOR is a single protein with a molecular weight of approximately 40 kDa, which requires light and NADPH for catalysis, and belongs to the short-chain dehydrogenase (SDR) superfamily (6, 7). Anoxygenic photosynthetic bacteria only have DPOR; cyanobacteria, algae, and nonflowering plants contain both DPOR and LPOR; the flowering higher plants only retain LPOR (8–11). Aside from its role in chlorophyll biosynthesis, LPOR also protects the etiolated and green plants by binding to the photosensitive Pchlide (12–14).
LPOR is one of the few naturally occurring photoenzymes (15). It has served as a model enzyme for studying enzyme catalysis and reaction dynamics (1, 16, 17). Extensive biochemical and biophysical studies have been performed on LPOR from two cyanobacteria, S. sp. PCC 6803 (18–26) and T. elongatus (27–35). The catalytic process has been demonstrated to be initiated by the absorption of light. The activated enzyme then enables hydride transfer from the pro-S face of the nicotinamide ring of NADPH to the C17 position of Pchlide, and a proton is transferred most likely from a conserved Tyr residue at the active site to the C18 position.
The overall fold and the active site of LPOR have been predicted based on sequence similarity between the LPOR and the SDR superfamily member alcohol dehydrogenase (6, 7). Their common features include the Rossmann fold with a conserved NAD(P)H binding site, a typical active site tetrad, and a variable C-terminal segment that defines substrate specificity (36, 37). LPORs in plants are classified as the SDR73C family, which are highly similar to cyanobacterial LPORs (38, 39). Three-dimensional models have been built for several LPORs based on their homology to the SDR superfamily members with known structure. These models include the Synechocystis LPOR (SyLPOR) (20), T. elongatus LPOR (TeLPOR) (34, 40), and LPOR from the higher plants Pisum sativum (41), Hordeum vulgare (14), and Arabidopsis thaliana (42, 43). They have helped in describing the catalytic mechanism of the light-driven Pchlide reduction, but they lack details, such as the boundary of secondary structure elements, the configuration of the key residues, and the conformation of the C-terminal segment. Hence, a well-defined LPOR structure is needed to delineate the mechanism of this light- and NADPH-dependent reaction. Here, we report the crystal structures of SyLPOR and TeLPOR, both in complex with NADPH at respective resolutions of 2.2 and 2.4 Å. This paper provides a clear insight into the structure-function relationships of this unusual enzyme. Very recently, Zhang et al. have reported the structures of SyLPOR and TeLPOR and, thus, proposed a ternary LPOR–NADPH–Pchlide complex model (44). Here, we present two structures of SyLPOR and TeLPOR, which suggest different scenarios than those of Zhang et al. (44).
Results
Overall Structure of SyLPOR and TeLPOR.
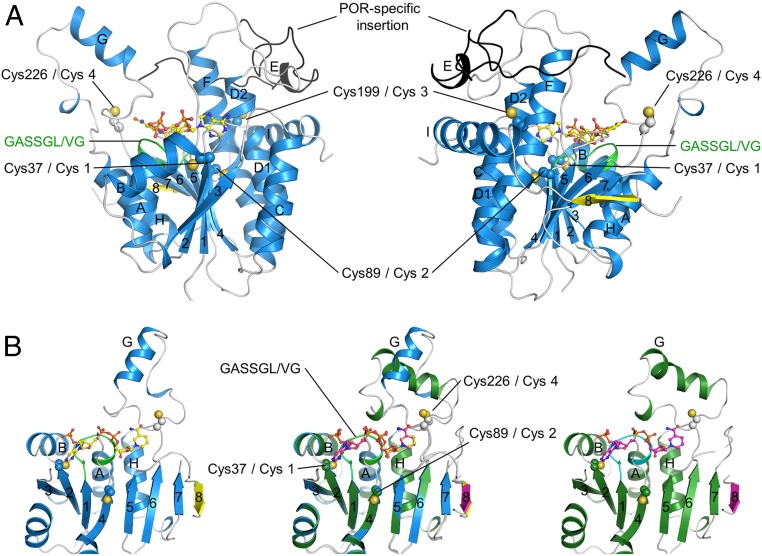
The recombinant SyLPOR and TeLPOR were generated (SI Appendix, Table S1) and purified (SI Appendix, Fig. S1). We obtained crystals of both SyLPOR and TeLPOR in their NADPH-bound form, but our attempt to crystallize the apo form was unsuccessful. The structures of NADPH-bound SyLPOR and TeLPOR were determined and refined to crystallographic Rwork/Rfree factors of 0.179/0.207 and 0.181/0.221 (SI Appendix, Table S2). Their secondary structure elements (SI Appendix, Fig. S2) were defined by the dictionary of the secondary structure of proteins algorithm (45). Here, we use the SyLPOR structure to present the characteristics of LPOR, whose central β-sheet consists of eight β-strands on the order of 3-2-1-4-5-6-7-8 with β8 antiparallel to the rest (Fig. 1A). Each side of the β-sheet is flanked by three α-helices, which are αA, αB, and αH on one side, and αC, αD, and αF on the other side. The slightly distorted αD has been named as αD1 and αD2 but is described as a continuous helix. The β-sheet and its six flanking α-helices constitute the αβα-core. The cofactor NADPH is located at the carboxy ends of β2-β1-β4-β5-β6. A 45-residue region (Gly144–Lys188) connects β5 and αF. Within this region, residues Glu169–Glu172 form a short four-residue helix (αE); residues Asn149–Ile157 in chain A and residues Ser150–Lys156 in chain B are not observed in the electron density. It is noteworthy that this region harbors a 33-residue insertion (Gly154–Lys186 in SyLPOR) that is unique for LPORs when compared with other SDR superfamily members and is suggested to participate in Pchlide binding (7, 20, 26, 34, 41, 43, 46). While a major portion of this insertion lies outside the αβα-core, the amino-terminal portion (although partially unobserved) is in the vicinity of the NADPH nicotinamide ring to where the substrate Pchlide is proposed to bind.
Fig. 1.
Ribbon representation of the overall structures of SyLPOR and TeLPOR. (A) Two side views of SyLPOR. The secondary structure elements are colored in blue except the antiparallel β8 in yellow. The loop region is in gray. The LPOR-specific insertion is colored in black. The NADPH-binding sequence is colored in green. Four cysteine residues are shown in sphere mode. The cofactor NADPH is shown in stick-and-ball mode. (B) Front view of SyLPOR (Left), TeLPOR (Right), and their superimposition (Middle). The secondary structure elements of TeLPOR are colored in deep green except β8 in magenta; the NADPH-binding sequence is colored in cyan. The α-helices are labeled alphabetically, and the β-strands are labeled numerically.
The amino acid sequence of TeLPOR shares 75.2% identity with that of SyLPOR, and these two structures are highly conserved (Fig. 1B and SI Appendix, Fig. S2). The αβα-core of TeLPOR is almost identical to that of the SyLPOR with a root mean square deviation of 0.58 Å over 269 aligned Cα atoms. The major structural difference of the two LPORs lies at αG, the helix that protrudes away from the core (Fig. 1B). The TeLPOR αG is formed by Pro238–Lys250 and is preceded by a 310 helix (Pro231–L232–Phe233); the SyLPOR αG is formed by Arg234–Ile243 and is followed by a 310 helix (Lys249–Asn250–Val251). This structural difference does not arise from the amino acid discrepancy as this region is highly conserved among LPORs (SI Appendix, Fig. S2).
LPORs contain four conserved cysteines (Cys-1–4). In SyLPOR and TeLPOR, they correspond to Cys38, Cys89, Cys199, and Cys226. Cys-1 and Cys-2 locate at the ends of β2 and β4, respectively, and Cys-3 locates within αF. Cys-4 is in the loop between β6 and αG (named as L6/G) and has been predicted to be involved in Pchlide binding (14, 19, 32).
NADPH-Binding Site.
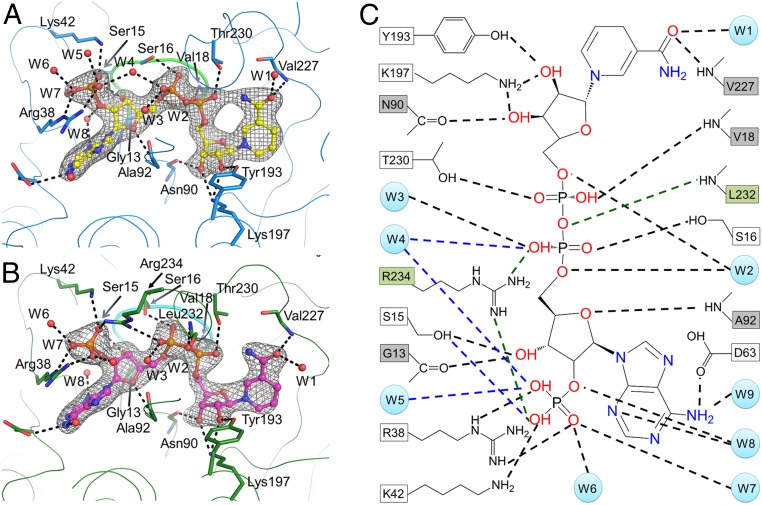
Both structures clearly define the LPOR–NADPH interactions (Fig. 2 A and B). Abundant intermolecular polar interactions among NADPH, the protein, and nine surrounding water molecules are observed that may stabilize the cofactor (Fig. 2 and SI Appendix, Table S3). The side chains of Tyr193 and Lys197, the backbone atoms of Val227, Asn90, and a water molecule (W1), interact with the nucleotide group at the nicotinamide end of NADPH. The backbone oxygen of Gly13, the hydroxyl group of Ser15, and the backbone NH of Ala92 interact with the ribose at the adenine end. Arg38 in L2/B, Lys42 in αB, and three water molecules (W5, W6, and W7) interact with the monophosphate group at the O2′ position of the adenine ribose. The hydroxyl group of Thr230 and Ser16, the backbone NH of Val18, together with three water molecules (W2, W3, and W4) interact with the pyrophosphate group. The side chain of Asp63 and two water molecules (W8 and W9) interact with the adenine group. The SDR active site motif YxxxK (YKDSK in LPOR), which is located on a long helix (αF), participates in the proper coordination of NADPH and possibly in the Pchlide binding. The Tyr193–Lys197 pair interacts with the ribose moiety at the nicotinamide end. Arg38 and Lys42 in both SyLPOR and TeLPOR participate in the coordination of the monophosphate group.
Fig. 2.
The NADPH-binding site. (A and B) NADPH interactions with (A) SyLPOR and (B) TeLPOR. The protein backbone is traced with thin lines. Residues directly interacting with NADPH are shown as sticks. The water molecules are shown as red spheres. The consensus sequence GASSGV/LG conserved for all LPORs is shown as thick lines. The color scheme is the same as in Fig. 1. The 2Fo − Fc maps are shown in gray mesh contoured at 1.0σ. The polar interactions are indicated by black dashed lines. (C) Detailed diagram showing NADPH interactions with SyLPOR/TeLPOR and water molecules. The residues that interact with NADPH by backbone atoms are shown in gray boxes, and the residues that interact with NADPH by their side-chain atoms are shown in white boxes. Water molecules are presented as light-blue circles. The black dashed lines indicate the interactions present in both SyLPOR and TeLPOR, the blue dashed lines indicate interactions only in SyLPOR, and the green dashed lines and green boxes indicate interactions only in TeLPOR.
The only noticeable difference between the NADPH-binding sites of SyLPOR and TeLPOR is the interaction with the phosphate groups. In SyLPOR, a water molecule (W4) interacts with both the pyrophosphate and the monophosphate groups; in TeLPOR, a conserved Arg234 occupies the W4 site. The W5 water is absent in TeLPOR. This difference can be attributed to the discrepancy of αG. In SyLPOR, Arg234 locates within αG (Arg234–Ile243) and is distant from NADPH; in TeLPOR, it locates within L6/G, the loop preceding αG (Pro238–Lys250) and directly interacts with NADPH. The structural difference suggests that αG and adjacent residues (including Cys-4) mediate the NADPH binding and catalysis process. The backbone NH of Leu232 interacts with the NADPH pyrophosphate group in TeLPOR. The side chain of Leu232 extends to the cavity near the nicotinamide end of NADPH, forming a hydrophobic surface together with its succeeding residues (Fig. 3).
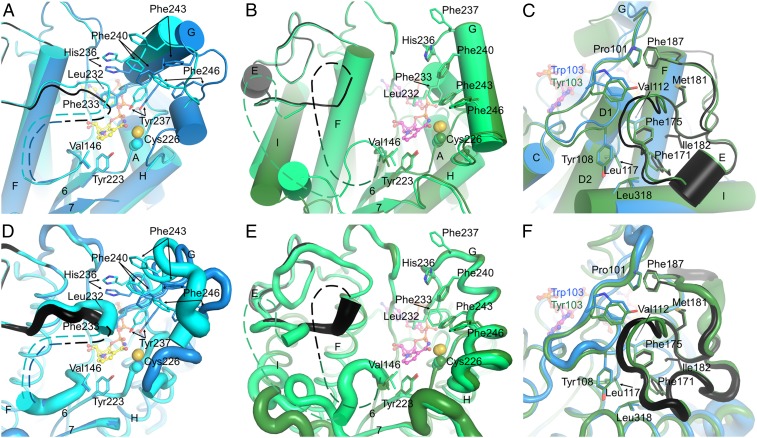
Fig. 3.
The substrate-binding site and the LPOR-specific insertion. (A) The substrate cavity of SyLPOR. The two polypeptide chains in an asymmetric unit are superimposed and colored blue and cyan. For clarity, only one NADPH molecule is shown (yellow stick). The side chains of residues possibly participating in Pchlide binding are shown as sticks. The LPOR-specific insertion of one chain is colored black. The missing fragment is depicted as dashed lines. (B) The substrate cavity of TeLPOR. The polypeptide chains are colored deep green and green, and only one NADPH is shown (magenta). Other representations are same as in A. (C) The majority of the LPOR-specific insertion is folded with the αβα-core. The core-insertion interacting residues are shown as sticks. SyLPOR and TeLPOR, respectively, are colored blue and deep green except that the insertion sequence in SyLPOR is colored black. (D–F) Representation of the flexibility of A, B, and C. The backbone is shown as tubes whose radius corresponds to the temperature factor of the Cα-atoms.
The LPOR-Specific Insertion Participates in Substrate Binding.
The formation of the LPOR–NADPH–Pchlide complex is the initiation step of the light-driven catalysis. Although we did not obtain the Pchlide-bound structures, the two NADPH-bound structures provide clues about the Pchlide-binding site (Fig. 3). Near the nicotinamide end, a clam-shaped cavity is formed by the residues conserved in LPORs (SI Appendix, Fig. S2). Seven residues, Leu232, Phe233, His236, Tyr237, Phe240, Phe243, and Phe246, constitute one shell of the clam (Fig. 3 A and B); Val146 in the L5/E loop together with the following nine to 13 missing residues (from 147 to 151 to 156 to 162) constitute the other shell; Tyr223 at the end of β6 forms the joint. The aforementioned residues are mostly hydrophobic or aromatic except His236 whose side-chain imidazole could interact with the Pchlide Mg2+ ion. This possible substrate-binding site is away from the αβα-core and shields the NADPH nicotinamide ring. The two sides of the clam-shaped cavity are quite dynamic, especially the side with missing residues (Fig. 3 D and E), and the clam joint Tyr223 is stably positioned.
The 33-residue insertion (Gly154–Lys186 in SyLPOR and TeLPOR) is unique in LPORs and was suggested to participate in the Pchlide binding (7, 20, 26, 34, 41, 43, 46). In the current structures, the amino end of this insertion overlaps with the missing fragment (Asn149–Ile157 in chain A, Ser150–Lys156 in chain B for SyLPOR, Thr147–Pro160 in chain A, and Lys151–Pro162 in chain B for TeLPOR). The missing fragment is adjacent to the nicotinamide ring and on one side of the clam-shaped pocket, implying that it is related to the Pchlide-binding/Chlide-releasing process. The majority of this LPOR-specific insertion, corresponding to residues around 163–186, is structurally stable. It folds with the αβα-core mainly by hydrophobic interaction (Fig. 3 C and F). The insertion residues Phe171, Phe175, Met181, Ile182, and an adjacent residue Phe187, fold inward and contact with the αβα-core residues Pro101and Trp103 (in SyLPOR), or Tyr103 (in TeLPOR), Tyr108, Val112, Leu117, and Leu318.
Dimerization and Structural Comparison.
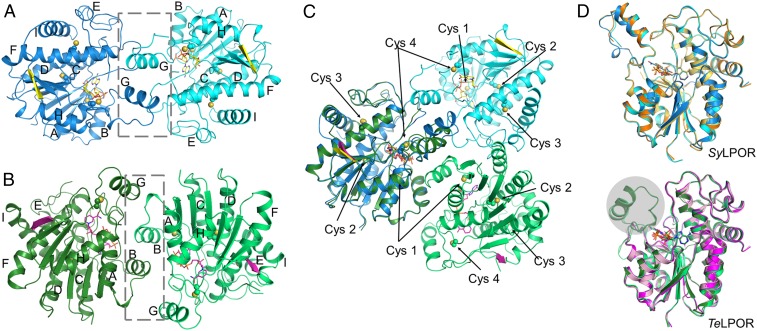
The size exclusion chromatography (SEC) profile revealed that SyLPOR and TeLPOR exist as monomers in solution (SI Appendix, Fig. S1). In crystal, they form homodimers, respectively (Fig. 4). The dimerization interfaces clearly differ from each other. For SyLPOR, the dimeric interface consists of L4/D, L5/E, and primarily αG (Fig. 4A). The predominant interactions are van der Waals contacts between two αGs and between αG and neighboring loops. For TeLPOR, the dimeric interface mainly consists of αB and LB/3 with a small contribution from αG (Fig. 4B). None of the four conserved cysteines are involved in dimerization.
Fig. 4.
Dimerization and structural comparison. (A) The SyLPOR dimer, (B) the TeLPOR dimer, and (C) their superimpositions with one monomer fixed in position. The gray box in the dashed lines indicates the dimeric interface. (D) Structural comparison of SyLPOR (Upper) and TeLPOR (Lower). SyLPOR chains A and B (Protein Data Bank [accession no. 6R48]) are in orange and light orange; the apo-TeLPOR (Protein Data Bank [accession no. 6RNV]) and NADPH-bound TeLPOR (Protein Data Bank [accession no. 6RNW]) structures are in pink and magenta. The gray sphere indicates the alternative conformation of αG not observed previously (44).
Recently, Zhang et al. has reported the structures of the NADPH-bound SyLPOR, apo-TeLPOR, and NADPH-bound TeLPOR (44). A comparison reveals that their overall structures are highly similar to those described here (Fig. 4D). The NADPH-bound SyLPOR structures are nearly the same, but the region between β6 and αH (and, hence, αG) was missing in the apo- and NADPH-bound TeLPOR structures reported previously (44). The TeLPOR αG displays a conformation alternative to that found in SyLPOR.
Discussion
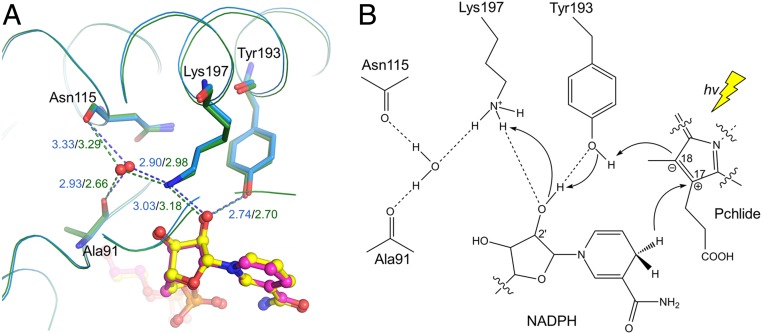
As a specialized SDR, LPOR catalyzes the light-dependent reduction of Pchlide. Its catalytic mechanism has been investigated on the micro- to picosecond timescales (1, 16, 24, 26, 33, 35, 47). The process can be briefly described as: The photoactivated Pchlide receives a hydride from the electron donor NADPH at C17 of the C17 = C18 double bond, then a proton is transferred through a conserved tyrosine (Tyr193 in SyLPOR and TeLPOR) to C18, resulting in two chiral centers at C17 and C18. However, the proton-relay path to this tyrosine is not clear. A key feature of the SDR superfamily is its catalytic tetrad Ser-Asn-Tyr-Lys by which a proton-relay path is formed with a water molecule bound to the tetrad Asn (37). The polar side chain of the tetrad Ser in classical SDRs usually stabilizes the hydroxy/keto group of the substrates, but this does not apply to the double-bond reduction of Pchlide catalyzed by LPOR. In the structures of SyLPOR and TeLPOR, there is a well-positioned water molecule that interacts with the ε-amino group of Lys197 and the backbone oxygens of Asn115 and Ala91 (Fig. 5A and SI Appendix, Fig. S3). Both Asn115 and Ala91 are conserved in LPORs (SI Appendix, Fig. S2), suggesting their side chains are required for their correct folding and, thus, to form the water-stable network by their backbone atoms. This water molecule, together with the Tyr193–Lys197 pair and the 2′-hydroxy group of NADPH, constitute a possible proton-relay path in which Lys197 extracts a proton from the solvent (Fig. 5B). The extracted proton is then transferred to Tyr193 and ultimately to C18 of Pchlide. Recently, systematic analyses of hydride and proton transfer dynamics in LPORs from cyanobacteria, algae, embryophytes, and angiosperms (47–49) have shown that hydride/proton transfer in eukaryotic LPORs is faster than those in prokaryotic LPORs, suggesting an optimized active site architecture in eukaryotic LPORs following endosymbiosis. Further structure-based phylogenetic analysis is expected to uncover the molecular basis for the high efficiency observed in eukaryotic LPORs.
Fig. 5.
Proposed proton-relay path. (A) The hydrogen bond network bridging the Tyr193 ηO and a solvent water molecule within the SyLPOR and TeLPOR structures. The well-positioned water, shown in the red sphere, is fixed by the backbone oxygens of Ala91 and Asn115, and the ε-amino group of Lys197. The hydrogen bonds are shown in dashed lines and the bond lengths (Å) are in blue for SyLPOR and dark green for TeLPOR. (B) A proposed proton-relay path following the hydride transfer from NADPH to C17. The photon energy (hv) is represented by a yellow thunderbolt.
The four LPOR-conserved cysteines locate around the active site (Figs. 1 and 4). Cys-1, Cys-2, and Cys-3 (corresponding to Cys38, Cys89, and Cys199 in SyLPOR and TeLPOR) are more distant than Cys-4 (Cys226 in SyLPOR and TeLPOR) from the NADPH nicotinamide ring. Cys-4 possibly participates in substrate binding and/or catalysis (Figs. 3 and 4C). An alanine mutation of Cys-4 severely impaired the enzymatic activity (14). Time-resolved absorption and emission spectroscopies indicate that a serine mutant of Cys-4 could bind Pchlide by a 180° turn, thus, the hydride from NADPH is transferred to C18 instead of C17 as in the wild type (32, 35).
The difference between the SyLPOR and the TeLPOR homodimers suggests that dimerization is not conserved for cyanobacteria LPORs (Fig. 4). In contrast, plant LPORs were found to form oligomers (14, 46, 50, 51). Two regions (residues 85–88 and 240–270 of A. thaliana PORA) were indicated to participate in oligomerization (50). The first region includes the amino end of SyLPOR and TeLPOR; the second region includes the LPOR-specific insertion (SI Appendix, Fig. S2). The insertion sequences are less conserved than the well-defined secondary structural elements. It is possible that plant LPORs have evolved species-specific motifs within this region that mediate oligomerization.
Zhang et al. proposed a ternary LPOR–NADPH–Pchlide complex model by docking and molecular-dynamic simulations (44). In the simulated binding process, αG remains relatively stable. The alternative conformation of αG in our structures (Figs. 1B, 3 B and E, and 4D) suggests different scenarios where large conformational changes could accompany Pchlide binding. In addition, the LPOR–NADPH–Pchlide model showed that the propionic acid group of Pchlide was stabilized by Lys197 (44). However, this essential lysine is covered by NADPH (Fig. 2). It is possible that the conserved basic residues Arg234, Arg241/Lys242, and Lys249 between β6 and αH (SI Appendix, Fig. S2) provide a candidate partner for the propionic acid group during Pchlide binding, and His236 interacts with the Pchlide Mg2+ ion. The involvement of the conserved histidine has been proposed previously (20, 43) but was suggested to be unlikely as the His236-to-Ala mutant of TeLPOR retained the Pchlide-binding ability (34). It should be pointed out that, in TeLPOR, the residue preceding His236 is also a histidine (SI Appendix, Fig. S2), which may substitute the conserved one in the His236-to-Ala mutant. The molecular details for formation of the ternary complex need further experimentation, especially structural characterization on the Pchlide-bound LPOR.
Methods
Protein Expression and Purification.
The slr0506 gene encoding SyLPOR and the tlr0575 gene encoding TeLPOR were condon optimized for Escherichia coli and commercially synthesized (Sango Biotech, Shanghai, China). The synthesized slr0506 and tlr0575 sequences delivered in pUC57 vector were PCR amplified and inserted into the Novagen expression vectors pET-28(+) and pET-22b(+), respectively (SI Appendix, Table S1). The expression product for SyLPOR contains an N-terminal His6-tag, and the product for TeLPOR contains a C-terminal His6-tag. The transformed E. coli BL21(DE3) cells harboring each vector were grown at 37 °C until optical density at 600 nm reached ∼0.6. Expression of the recombinant protein was induced with 0.4 mM isopropyl β-d-thiogalactopyranoside, and the cells were grown at 16 °C for 18 h before harvested by centrifugation.
The same procedure was used for purification of SyLPOR and TeLPOR. The cell pellets were resuspended in buffer A (0.5 M NaCl, 20 mM MES, pH 6.0), lysed by sonication, and cleared by centrifugation. The aqueous supernatant of cell lysate was incubated with buffer A-equilibrated nickel nitrilotriacetic acid resin (QIAGEN, Shanghai, China) for 1 h at 4 °C, and the recombinant protein was eluted with 200 mM imidazole. Further purification was performed by SEC on a HiLoad Superdex 75 pg column (GE Healthcare, Shanghai, China), which was preequilibrated and eluted with buffer A supplemented with 1 mM (ethylenedinitrilo)tetraacetic acid (EDTA). The purity of recombinant SyLPOR and TeLPOR was monitored by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Crystallization and Diffraction.
Crystallization was performed using the vapor diffusion method in a sitting drop consisting of 1 μL protein sample (in buffer A supplemented with 1 mM EDTA) and 1 μL reservoir solution. Purified SyLPOR was concentrated by ultrafiltration to 12 mg mL−1. The SyLPOR crystals grew within 1 wk at 16 °C using the reservoir solution of 3 mM NADPH, 0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), pH 7.5, and 1.5 M lithium sulfate. Purified TeLPOR was concentrated to 10 mg mL−1. The TeLPOR crystals grew within 1 wk at 4 °C using the reservoir solution of 6 mM NADPH, 0.1 M Hepes, pH 7.5, 20% (wt/vol) PEG 4,000, 10% (wt/vol) isopropanol, and 0.05% (vol/vol) n-octyl-β-d-glucoside. Attempts to crystalize the Pchlide-bound complex were unsuccessful. For data collection, the crystals were cryoprotected with 20% (vol/vol) glycerol and flash cooled in liquid nitrogen. The X-ray diffraction data sets were collected at the Shanghai Synchrotron Radiation Facility beamlines BL17U1 and BL18U1 and processed with the program package HKL-3000 (52). The X-ray data collection and refinement statistics are listed in SI Appendix, Table S2.
Structure Determination and Refinement.
The SyLPOR structure was solved by the automatic molecular replacement pipeline MoRDa (53) with the coordinates of a putative oxidoreductase from Mycobacterium paratuberculosis (27% sequence identity; Protein Data Bank [accession no. 3RD5]) (54) as the search model. The initial model was then automatically built by the PHENIX AutoBuild program (55). The iterative model adjustment and refinement were performed using Coot (56) and the phenix.refine program (57), respectively. The quality of the final model was checked with MolProbity (58). A similar procedure was used for the TeLPOR structure determination except that the initial model was built from the SyLPOR structure. All protein structure figures were prepared using the program PyMOL (Schrödinger, LLC).
Data Availability.
The coordinates and the structure factors of SyLPOR–NADPH and TeLPOR–NADPH have been deposited to the RCSB Protein Data Bank (accession nos. 6L1G and 6L1H).
Supplementary Material
Acknowledgments
We thank the beamline staff at the Shanghai Synchrotron Radiation Facility and Ming-Zhu Wang at Anhui University for their assistance during diffraction data collection. We also thank Shun Zhao at the Institute of Botany of the Chinese Academy of Sciences for his contribution at the early stage of this work. This work was supported by the National Key R&D Program of China (Grant 2017YFA0503703), the Ministry of Education Chang Jiang Scholars Program (Award Q2017241), and the Anhui Provincial Wanjiang Scholars Program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: RCSB Protein Data Bank crystallography, atomic coordinates, and structure factors can be found at https://www.rcsb.org/ (accession nos. 6L1G and 6L1H).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920244117/-/DCSupplemental.
References
- 1.Heyes D. J., Hunter C. N., Making light work of enzyme catalysis: Protochlorophyllide oxidoreductase. Trends Biochem. Sci. 30, 642–649 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Reinbothe C., et al. , Chlorophyll biosynthesis: Spotlight on protochlorophyllide reduction. Trends Plant Sci. 15, 614–624 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Gabruk M., Mysliwa-Kurdziel B., Light-dependent protochlorophyllide oxidoreductase: Phylogeny, regulation, and catalytic properties. Biochemistry 54, 5255–5262 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Muraki N., et al. , X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110–114 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Bröcker M. J., et al. , Crystal structure of the nitrogenase-like dark operative protochlorophyllide oxidoreductase catalytic complex (ChlN/ChlB)2. J. Biol. Chem. 285, 27336–27345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker M. E., Protochlorophyllide reductase is homologous to human carbonyl reductase and pig 20 β-hydroxysteroid dehydrogenase. Biochem. J. 300, 605–607 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilks H. M., Timko M. P., A light-dependent complementation system for analysis of NADPH:protochlorophyllide oxidoreductase: Identification and mutagenesis of two conserved residues that are essential for enzyme activity. Proc. Natl. Acad. Sci. U.S.A. 92, 724–728 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Von Wettstein D., Gough S., Kannangara C. G., Chlorophyll biosynthesis. Plant Cell 7, 1039–1057 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki J. Y., Bollivar D. W., Bauer C. E., Genetic analysis of chlorophyll biosynthesis. Annu. Rev. Genet. 31, 61–89 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Chew A. G., Bryant D. A., Chlorophyll biosynthesis in bacteria: The origins of structural and functional diversity. Annu. Rev. Microbiol. 61, 113–129 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Tanaka R., Tanaka A., Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 (2007). [DOI] [PubMed] [Google Scholar]
- 12.McCormac A. C., Terry M. J., Loss of nuclear gene expression during the phytochrome A-mediated far-red block of greening response. Plant Physiol. 130, 402–414 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda T., Takamiya K., Novel insights into the enzymology, regulation and physiological functions of light-dependent protochlorophyllide oxidoreductase in angiosperms. Photosynth. Res. 81, 1–29 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Buhr F., et al. , Photoprotective role of NADPH:protochlorophyllide oxidoreductase A. Proc. Natl. Acad. Sci. U.S.A. 105, 12629–12634 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmermund L., et al. , Photo-biocatalysis: Biotransformations in the presence of light. ACS Catal. 9, 4115–4144 (2019). [Google Scholar]
- 16.Scrutton N. S., Groot M. L., Heyes D. J., Excited state dynamics and catalytic mechanism of the light-driven enzyme protochlorophyllide oxidoreductase. Phys. Chem. Chem. Phys. 14, 8818–8824 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Belyaeva O. B., Litvin F. F., Mechanisms of phototransformation of protochlorophyllide into chlorophyllide. Biochemistry (Mosc.) 79, 337–348 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Suzuki J. Y., Bauer C. E., A prokaryotic origin for light-dependent chlorophyll biosynthesis of plants. Proc. Natl. Acad. Sci. U.S.A. 92, 3749–3753 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heyes D. J., Martin G. E., Reid R. J., Hunter C. N., Wilks H. M., NADPH:protochlorophyllide oxidoreductase from Synechocystis: Overexpression, purification and preliminary characterisation. FEBS Lett. 483, 47–51 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Townley H. E., Sessions R. B., Clarke A. R., Dafforn T. R., Griffiths W. T., Protochlorophyllide oxidoreductase: A homology model examined by site-directed mutagenesis. Proteins 44, 329–335 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Heyes D. J., Ruban A. V., Wilks H. M., Hunter C. N., Enzymology below 200 K: the kinetics and thermodynamics of the photochemistry catalyzed by protochlorophyllide oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 99, 11145–11150 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heyes D. J., Hunter C. N., Site-directed mutagenesis of Tyr-189 and Lys-193 in NADPH: Protochlorophyllide oxidoreductase from Synechocystis. Biochem. Soc. Trans. 30, 601–604 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Heyes D. J., Ruban A. V., Hunter C. N., Protochlorophyllide oxidoreductase: “dark” reactions of a light-driven enzyme. Biochemistry 42, 523–528 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Heyes D. J., Hunter C. N., van Stokkum I. H., van Grondelle R., Groot M. L., Ultrafast enzymatic reaction dynamics in protochlorophyllide oxidoreductase. Nat. Struct. Biol. 10, 491–492 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Heyes D. J., et al. , The first catalytic step of the light-driven enzyme protochlorophyllide oxidoreductase proceeds via a charge transfer complex. J. Biol. Chem. 281, 26847–26853 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Sytina O. A., et al. , Conformational changes in an ultrafast light-driven enzyme determine catalytic activity. Nature 456, 1001–1004 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Heyes D. J., Hunter C. N., Identification and characterization of the product release steps within the catalytic cycle of protochlorophyllide oxidoreductase. Biochemistry 43, 8265–8271 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Heyes D. J., Kruk J., Hunter C. N., Spectroscopic and kinetic characterization of the light-dependent enzyme protochlorophyllide oxidoreductase (POR) using monovinyl and divinyl substrates. Biochem. J. 394, 243–248 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heyes D. J., Menon B. R., Sakuma M., Scrutton N. S., Conformational events during ternary enzyme-substrate complex formation are rate limiting in the catalytic cycle of the light-driven enzyme protochlorophyllide oxidoreductase. Biochemistry 47, 10991–10998 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Heyes D. J., Sakuma M., de Visser S. P., Scrutton N. S., Nuclear quantum tunneling in the light-activated enzyme protochlorophyllide oxidoreductase. J. Biol. Chem. 284, 3762–3767 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Menon B. R., Waltho J. P., Scrutton N. S., Heyes D. J., Cryogenic and laser photoexcitation studies identify multiple roles for active site residues in the light-driven enzyme protochlorophyllide oxidoreductase. J. Biol. Chem. 284, 18160–18166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menon B. R., Davison P. A., Hunter C. N., Scrutton N. S., Heyes D. J., Mutagenesis alters the catalytic mechanism of the light-driven enzyme protochlorophyllide oxidoreductase. J. Biol. Chem. 285, 2113–2119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyes D. J., et al. , Excited-state charge separation in the photochemical mechanism of the light-driven enzyme protochlorophyllide oxidoreductase. Angew. Chem. Int. Ed. Engl. 54, 1512–1515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menon B. R., Hardman S. J., Scrutton N. S., Heyes D. J., Multiple active site residues are important for photochemical efficiency in the light-activated enzyme protochlorophyllide oxidoreductase (POR). J. Photochem. Photobiol. B 161, 236–243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archipowa N., Kutta R. J., Heyes D. J., Scrutton N. S., Stepwise hydride transfer in a biological system: Insights into the reaction mechanism of the light-dependent protochlorophyllide oxidoreductase. Angew. Chem. Int. Ed. Engl. 57, 2682–2686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jörnvall H., et al. , Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Kavanagh K. L., Jörnvall H., Persson B., Oppermann U., Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65, 3895–3906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moummou H., Kallberg Y., Tonfack L. B., Persson B., van der Rest B., The plant short-chain dehydrogenase (SDR) superfamily: Genome-wide inventory and diversification patterns. BMC Plant Biol. 12, 219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kramm A., Kisiela M., Schulz R., Maser E., Short-chain dehydrogenases/reductases in cyanobacteria. FEBS J. 279, 1030–1043 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Gholami S., et al. , Theoretical model of the protochlorophyllide oxidoreductase from a hierarchy of protocols. J. Phys. Chem. B 122, 7668–7681 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Dahlin C., et al. , The role of protein surface charge in catalytic activity and chloroplast membrane association of the pea NADPH: Protochlorophyllide oxidoreductase (POR) as revealed by alanine scanning mutagenesis. Plant Mol. Biol. 39, 309–323 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Gabruk M., Grzyb J., Kruk J., Mysliwa-Kurdziel B., Light-dependent and light-independent protochlorophyllide oxidoreductases share similar sequence motifs-in silico studies. Photosynthetica 50, 529–540 (2012). [Google Scholar]
- 43.Gabruk M., Mysliwa-Kurdziel B., Kruk J., MGDG, PG and SQDG regulate the activity of light-dependent protochlorophyllide oxidoreductase. Biochem. J. 474, 1307–1320 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Zhang S., et al. , Structural basis for enzymatic photocatalysis in chlorophyll biosynthesis. Nature 574, 722–725 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W., Sander C., Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- 46.Reinbothe C., Lepinat A., Deckers M., Beck E., Reinbothe S., The extra loop distinguishing POR from the structurally related short-chain alcohol dehydrogenases is dispensable for pigment binding but needed for the assembly of light-harvesting POR-protochlorophyllide complex. J. Biol. Chem. 278, 816–822 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Hoeven R., Hardman S. J., Heyes D. J., Scrutton N. S., Cross-species analysis of protein dynamics associated with hydride and proton transfer in the catalytic cycle of the light-driven enzyme protochlorophyllide oxidoreductase. Biochemistry 55, 903–913 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Heyes D. J., Levy C., Sakuma M., Robertson D. L., Scrutton N. S., A twin-track approach has optimized proton and hydride transfer by dynamically coupled tunneling during the evolution of protochlorophyllide oxidoreductase. J. Biol. Chem. 286, 11849–11854 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrone A., Archipowa N., Zipfel P. F., Hermann G., Dietzek B., Plant protochlorophyllide oxidoreductases A and B: Catalytic efficiency and initial reaction steps. J. Biol. Chem. 290, 28530–28539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gabruk M., et al. , Insight into the oligomeric structure of PORA from A. thaliana. Biochim. Biophys. Acta 1864, 1757–1764 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Reinbothe C., Lebedev N., Reinbothe S., A protochlorophyllide light-harvesting complex involved in de-etiolation of higher plants. Nature 397, 80–84 (1999). [Google Scholar]
- 52.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M., HKL-3000: The integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Vagin A., Lebedev A., MoRDa, an automatic molecualr replacement pipeline. Acta Crystallogr. A Found. Adv. 71, s19 (2015). [Google Scholar]
- 54.Baugh L., et al. , Increasing the structural coverage of tuberculosis drug targets. Tuberculosis (Edinb.) 95, 142–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger T. C., et al. , Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and the structure factors of SyLPOR–NADPH and TeLPOR–NADPH have been deposited to the RCSB Protein Data Bank (accession nos. 6L1G and 6L1H).