Abstract
Protein design is a powerful tool for elucidating mechanisms of function and engineering new therapeutics and nanotechnologies. Although soluble protein design has advanced, membrane protein design remains challenging because of difficulties in modeling the lipid bilayer. In this work, we developed an implicit approach that captures the anisotropic structure, shape of water-filled pores, and nanoscale dimensions of membranes with different lipid compositions. The model improves performance in computational benchmarks against experimental targets, including prediction of protein orientations in the bilayer, ΔΔG calculations, native structure discrimination, and native sequence recovery. When applied to de novo protein design, this approach designs sequences with an amino acid distribution near the native amino acid distribution in membrane proteins, overcoming a critical flaw in previous membrane models that were prone to generating leucine-rich designs. Furthermore, the proteins designed in the new membrane model exhibit native-like features including interfacial aromatic side chains, hydrophobic lengths compatible with bilayer thickness, and polar pores. Our method advances high-resolution membrane protein structure prediction and design toward tackling key biological questions and engineering challenges.
Significance
Membrane proteins participate in many life processes, including transport, signaling, and catalysis. They constitute over 30% of all proteins and are targets for over 60% of pharmaceuticals. Computational design tools for membrane proteins will transform the interrogation of basic science questions such as membrane protein thermodynamics and the pipeline for engineering new therapeutics and nanotechnologies. Existing tools are either too expensive to compute or rely on manual design strategies. In this work, we developed a fast and accurate method for membrane protein design. The tool is available to the public and will accelerate the experimental design pipeline for membrane proteins.
Introduction
Membrane proteins partner with the surrounding lipid environment to perform essential life processes. They constitute 30% of all proteins (1) and are targets for over 60% of pharmaceuticals (2). However, experimental difficulties have limited our insights into their molecular mechanisms of function. Protein design tools are powerful for elucidating biological mechanisms and developing new therapeutics. Over the past 20 years, soluble protein design has advanced to atomic-level accuracy (3). A remaining challenge is to create robust tools for membrane proteins (4). There have been several achievements in membrane protein design, including a zinc-transporting tetramer Rocker (5), an ion-conducting protein based on the Escherichia coli Wza transporter (6), β-barrel pores with increased selectivity (7), receptors with new ligand-binding properties (8,9), and designed de novo α-helical bundles that insert into the membrane (10). A critical limitation is capturing the heterogeneous membrane environment: models are either too computationally expensive or severely approximate the bilayer. In fact, it has been common for membrane protein structure prediction and design to be carried out in a 30-Å hydrophobic slab. A slab is a poor proxy for the heterogeneous membranes found in biology with varying lipid composition across different organelles, cell types, and species. To apply membrane protein design to addressing biological questions, tools must sample a realistic distribution of amino acids tied to the diverse lipid composition.
The foundation of computational modeling and design tools is the energy function: a mathematical model of the physical rules that distinguish native from non-native membrane protein conformations and sequences. Currently, most computational studies of membrane proteins are molecular dynamics simulations with an all-atom lipid bilayer. In this conception, the lipid molecules are represented explicitly using force fields such as AMBER (11), CHARMM (12), or GROMOS (13), and the protein-lipid interactions are scored with a molecular mechanics energy function. All-atom models are attractive because they can feature hundreds of lipid types toward approximating the composition of biological membranes. With current technology, detailed all-atom models can be used to explore membrane dynamics for hundreds of nanoseconds (14): the timescale required to achieve equilibrated properties on a bilayer with ∼250 lipids (15). Coarse-grained representations such as MARTINI (16) and SIRAH (17) reduce computation time by mapping atoms onto representative beads. As a result, simulations have explored dynamics up to the millisecond timescale to access features of membrane organization and large protein domain motions (18).
Implicit solvent models enable simulations to reach longer timescales required to investigate biologically relevant conformational and sequence changes. Instead of using explicit molecules, implicit methods represent the solvent as a continuous medium (19,20), resulting in a 50- to 100-fold sampling speedup (21). The most detailed implicit model is the Poisson-Boltzmann (PB) equation, which relates electrostatic potential to dielectric properties of the solvent and solute through a second-order partial differential equation (22). Numerical solvers have enabled PB calculations on biomolecular systems (23); however, these calculations do not scale well. To reduce computational cost, the generalized Born (GB) approximation of the PB equation treats atoms as charged spheres (24). GB methods represent the low-dielectric membrane through various treatments ranging from a simple switching function (25) to heterogeneous dielectric approaches (26). However, evaluating the GB formalism is still computationally expensive.
A popular approach to overcoming the computational cost of solvent electrostatics models is the Lazaridis implicit membrane model (IMM1 (27)): a Gaussian solvent-exclusion model that uses experimentally measured transfer energies of side-chain analogs in organic solvents to emulate amino acid preferences in the bilayer (28). IMM1 has been applied to various biomolecular modeling problems, including studies of antimicrobial peptides (29), de novo folding (30), and de novo design of transmembrane helical bundles (10). However, organic solvent slabs differ from phospholipid bilayers because lipids are thermodynamically constrained to a bilayer configuration, resulting in a unique polarity gradient that influences side-chain preferences (31,32). An alternative is to directly calculate amino acid preferences by deriving statistical potentials from a database of known membrane protein structures (33, 34, 35, 36). Yet, statistical potentials do not capture varying physiochemical properties of the membrane.
In this work, we developed a biologically realistic implicit membrane model for protein structure prediction and design. We first developed the model from experimental and computational modeling of phospholipid bilayers to capture biologically important membrane features. Next, we tested the model on four benchmarks: 1) prediction of protein orientations in the membrane, 2) ΔΔG of mutation calculations, 3) native structure discrimination, and 4) native sequence recovery. We applied the model to protein design and investigated properties of the in silico designed membrane proteins including the amino acid composition. Finally, we share several design anecdotes that exhibit native-like membrane protein features, including interfacial aromatic side chains, hydrophobic lengths compatible with different lipid compositions, and polar pores.
Methods
Development of the implicit membrane model
Derivation of -values
The Moon and Fleming hydrophobicity scale provides a set of water-to-bilayer transfer energies for the 20 canonical amino acids (37) measured in the reversibly folding OmpLA scaffold. Note that the default ionization state for histidine in Rosetta is neutral and Glu and Asp are protonated because the Moon and Fleming scale was measured at pH 3.8. We used regression to derive energies that correspond to atom types (Table S5), called . Specifically, least-squares fitting was applied solve the equation Ax = b, where A is a matrix of atom type stoichiometric coefficients (Table S6), b is the vector of -values, and x is the desired vector of -values. Matrix rows for glycine, alanine, and proline were excluded to avoid overfitting. The resulting -values are in Table S7.
Molecular dynamics simulations of phospholipid bilayers
All-atom molecular dynamics simulations were performed to extract properties of membranes with different phospholipid compositions. We simulated phospholipid bilayers with hydrocarbon tails between 12 and 18 carbons long and either a phosphatidylethanolamine, phosphatidylcholine (PC), or phosphatidylglycerol headgroup (Table S1). The exceptions were 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dimyristoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (DMPG) because the liquid-to-gel phase transition temperatures are above physiological temperature (38,39). CHARMM-GUI (40) was used to configure each bilayer system with 75 lipids in each leaflet, 22.5 Å of water on each side, and 0.1 M NaCl. Simulations were performed using the NAMD molecular dynamics engine (41) at a constant pressure of 1 atm and a temperature of 37°C. We used the CHARMM36 (12) force field for lipid and the TIP3 model for water. The simulations were equilibrated with restraints according to the procedure outlined by Jo et al. (40). Then, each system was simulated for 50 ns.
Derivation of depth-dependent water-density profiles
MDAnalysis (42) was used to extract water-density information from each bilayer simulation. For each frame, the system was first recentered on the lipid center of mass. Then, we computed a normalized histogram of TIP3 z-coordinates with 1 Å bins to capture the distribution of water molecules. The histogram was recentered at z = 0 by fitting the histogram to a cosine function to estimate the midpoint. The time-averaged histogram was computed by averaging the histograms representing each frame (Fig. S9).
To generate analytic profiles, we used nonlinear regression to fit each histogram to the logistic function, fthk:
| (1) |
The function fthk depends on membrane depth (z) and has two adjustable parameters: steepness κ and width τ. We derived κ and τ for all simulated lipid compositions. The resulting parameters are in Table S2, and the analytic water-density profiles are in Fig. S10.
Calculation of water-filled pore shapes
For proteins with more than three transmembrane segments, we introduced a pore into the implicit membrane model. To determine the pore shape, we created a new method to transform discrete structural information into a smooth geometry described by differentiable functional forms. First, we used the convex-hull algorithm described in Koehler Leman et al. (43) to identify backbone and side-chain atoms that are in the transmembrane region (|z| ≤ T), face the protein interior, and are not buried. A side chain was defined as buried if it had 23 or more neighboring atoms within 12 Å of its Cα atom (44). Next, we computed a histogram of the z-coordinates of pore-facing atoms with a bin size of . For each bin, the (x, y) coordinates of the atoms were collected. Then, the Khachiyan algorithm (45) was used to compute the minimal-area ellipse that bounds these coordinates. Each ellipse is defined with the following parameters: major radius (a), minor radius (b), rotation angle (θ), and center (x0, y0). The radius of the ellipse, gradius, is calculated using rotation matrix M:
| (2) |
| (3) |
Cubic spline interpolation was used to fit polynomials to describe the depth dependence of each parameter. The result is five continuous and differentiable parametric functions: a(z), b(z), θ(z), x0(z), and y0(z). The transition between the water-filled pore and lipid phase is defined by gradius given the transition steepness n:
| (4) |
Validation of model parameters
-values
To verify -values, we first recalculated the side-chain transfer energies by solving Ax = b. The Pearson correlation coefficient between the calculated and experimentally measured side-chain transfer energies was R2 = 0.99 (Fig. S13). In addition, we used the procedure outlined in the Methods section to estimate the ΔΔGmut-values from Moon & Fleming (37). Specifically, we sought to verify that trends were preserved in context of the full energy function. The correlation between predicted and experimentally measured ΔΔGmut-values was R2 = 0.84 (Fig. S11) and the residuals are listed in Table S8. Note, the ΔΔGmut for proline was excluded from the correlation coefficients because steric clashes resulted in large energies.
Membrane thickness
We validated the water-density profiles computed from molecular dynamics by comparing the derived membrane thickness parameters with thickness measured at various temperatures via x-ray and neutron scattering experiments (46). First, we computed the membrane half thickness t from each logistic curve as the Gibbs dividing surface between the water and lipid phases (f(z) = 0.5). We then calculated a line of best fit through the measured thickness values at each temperature (Fig. S14).
RESULTS
Biologically realistic implicit membrane model
We developed a biologically realistic implicit membrane model inspired by IMM1 (27). Similar to IMM1, the membrane is modeled as a continuum of three phases: an isotropic phase representing bulk lipids, an isotropic phase representing bulk water, and an anisotropic phase representing the interfacial region. To accurately model the polarity gradient and dimensions of native membranes, we derived new equations and parameters from biophysical measurements. The result is a new energy term called ΔGmemb that computes protein stability given the water-to-bilayer transfer energy of atomic groups a and the fractional hydration fhyd:
| (5) |
The parameter captures the thermodynamics of protein-lipid interactions. We derived from the Moon and Fleming side-chain hydrophobicity scale (37) because the energies were measured in bilayers with phospholipids, a major component of biological membranes (47). Furthermore, we chose this scale because the measurements capture the stability of the final folded protein relative to the fully hydrated, unfolded state. Side-chain burial versus solvent exposure is accounted for through neighbor count calculations in the Rosetta energy functions. Then, following Lazaridis’ formalism (48), the function fhyd captures the three-dimensional shape of the implicit membrane as a dimensionless number that describes the phase given the position of an atomic group. When an atomic group is exposed to the lipid phase, fhyd = 0, whereas when an atomic group is exposed to the water phase, fhyd = 1.0. The transition between the two isotropic phases is modeled by a composition of two functions: fthk captures the membrane thickness, and fpore captures the geometry of a water-exposed pore:
| (6) |
The function fthk (Eq. 1; see Methods) models the transition between the water and lipid phase along the z axis and is thus an implicit representation of the hydrophobic thickness. We developed parameters for fthk by fitting to molecular dynamics simulations and scattering density profiles of phospholipid bilayers. The result is a logistic curve that depends on two parameters. We derived parameters for 13 phospholipid bilayer compositions (Table S1 and S2; see Supporting Materials and Methods). The membrane thickness can be derived by setting fthk = 0.5 (Fig. 1, A and B). Thus, the user can perform simulations with any of these 13 different phospholipid compositions or, in principle, with any mix of membrane components by using a molecular dynamics run and extracting the hydration profile parameters.
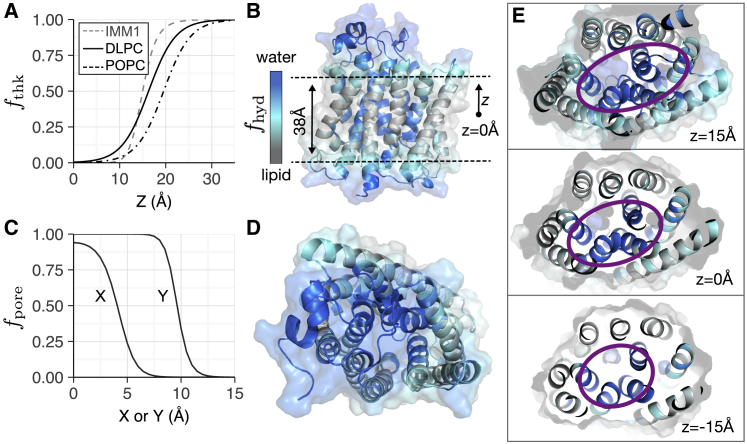
Figure 1.
Features of the biologically realistic implicit membrane model. The implicit membrane is modeled as three phases: two isotropic phases for water and lipid and a transition region that represents the interfacial headgroups. (A) The transition between phases in the z dimension is modeled by a logistic curve that can be parameterized for different lipid compositions. Example curves for DLPC (solid, black) and POPC (dot-dash, black) are shown in comparison to the sigmoid curve used in IMM1 (dashed, gray). (B) Implicit solvent phases are shown for the ammonium transporter Amt-1 (PDB: 2B2F) in the z dimension. The water phase is shown in blue, the interface is in teal, and the lipid is in gray. (C) The transition between phases due to an elliptical pore is modeled by a sigmoid curve. (D) Top view of implicit solvent phases due to a pore in Amt-1 is shown with the same coloring scheme as (B). The three panels of (E) demonstrate the variation in pore shape (purple) for different cross sections in the x, y plane along the z-axis. To see this figure in color, go online.
The function fpore defines the shape of a water-exposed pore (Fig. 1, C–E). Previously, Lazaridis developed a cylindrical model of pores for β-barrel proteins (48). This geometric assumption is straightforward for β-barrel proteins; however, α-helical protein pores require varied geometric descriptors such as cones, cylinders, and ellipses (49). To accommodate these geometries, we created a model that approximates pores as an elliptical tube with varying cross sections. This parameterization allows the model to describe cavities that do not penetrate through the membrane and pores that constrict, expand, or twist relative to z. The energy function accounts for the pore by first calculating a relative radius, gradius (Eqs. (2), (3); see Methods). The transition between the two phases is modeled by a sigmoid curve fpore (Eq. 4; see Methods) with two parameters: gradius and the transition steepness n (default n = 10). Additional examples for larger proteins with multiple pores and proteins with ellipsoidal architecture are shown in Fig. S12.
We integrated our model into the current all-atom energy function for modeling soluble proteins in Rosetta, called REF15 (50). REF15 computes macromolecular energies through a linear combination of terms for van der Waals, solvation, electrostatics, hydrogen bonding, backbone, and side-chain interactions. To account for the membrane environment, we added ΔGmemb with an empirically determined weight of 0.5. The resulting energy function, called franklin2019, is given by ΔEfranklin2019 = ΔEREF15 + ΔGmemb.
Computational benchmark performance of the biologically realistic implicit membrane
We evaluated the accuracy of franklin2019 using four computational benchmark tests against experimental targets. The tests were designed to evaluate an energy function’s ability to replicate measured membrane protein stabilities and perform accurate structure prediction and design. We compared the performance of franklin2019 to three existing models: 1) an implicit membrane parameterized from the behavior of side-chain analogs in organic solvents (M07 (30)), 2) a knowledge-based model that captures depth-dependent amino acid preferences (M12 (51)), and 3) the Rosetta all-atom energy function for soluble proteins (R15 (50,52)). For brevity, we will refer to franklin2019 as M19. We chose these models because the low computational cost enabled evaluation with structure prediction and design tests. Additional details describing the benchmark tests and command lines are provided in the Supporting Materials and Methods.
Test #1: prediction of membrane protein orientation and insertion energy
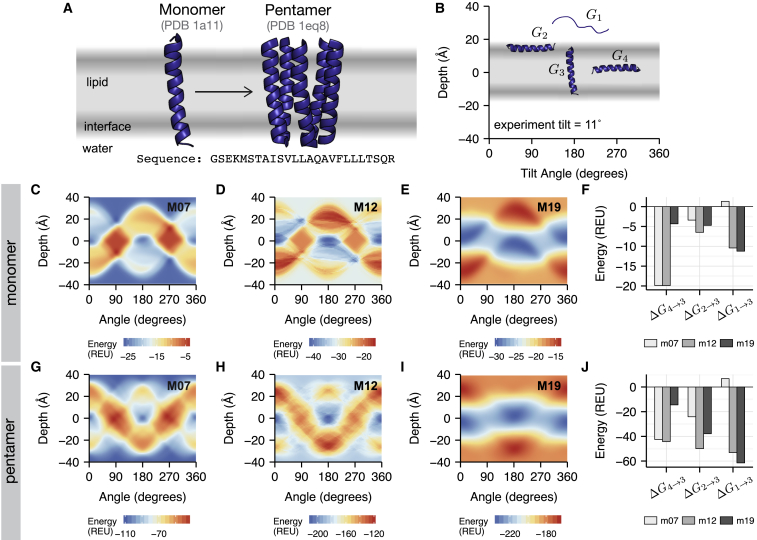
Membrane proteins are thermodynamically stable in the bilayer because of a favorable orientation and insertion energy. Therefore, implicit membrane energy functions must accurately estimate these quantities. First, we evaluated the partitioning properties of oligomeric proteins into the implicit membrane. Here, we chose to study oligomers because the single-transmembrane peptides may be marginally hydrophobic with insertion depending on the sequence context. We performed calculations for the acetylcholine receptor (pentamer, Fig. 2) and the influenza A M2 proton channel (tetramer, Fig. S2). Remarkably, M19 was the only model to predict a favorable insertion energy for both proteins. The mapping of peptide orientation to energies is shown for the acetylcholine receptor in Fig. 2. The M07 energy landscape (Fig. 2 C) has three small, low-energy wells, and they are isoenergetic with the water phase (because M07 was not parameterized for the water environment). This behavior is not physical. In contrast, the lipid phase is more thermodynamically favorable than the water phase for both M12 (Fig. 2 D) and M19 (Fig. 2 E). This result is quantified by a favorable transfer energy from the water phase (G1) to the lipid phase (G3; Fig. 2 F). Ultimately, M19 is the most native like because the model accurately captures the aqueous reference state relative to the bilayer phase.
Figure 2.
Prediction of membrane insertion and orientation for the acetylcholine receptor. (A) The sequence of the monomer and structures of both the monomer (PDB: 1A11) and pentamer (PDB: 1EQ8) are shown. (B) Important conformations are given as a function of peptide depth (z) and tilt angle (θ): G1 is the energy of the unfolded state in solution (z = 30 Å, θ = 90°); G2 is the energy of the folded state at the interface, parallel to the plane of the interface (z = 15 Å, θ = 90°); G3 is the energy of the peptide oriented vertically (z = 0 Å, θ = 0°); and G4 is the energy of a peptide buried in the membrane (z = 0 Å, θ = 90°). The mapping of protein orientations to energies calculated by the M07, M12, and M19 energy functions, respectively, is shown in (C)–(E) for the monomer and (G)–(I) for the pentamer. The partitioning energies between two lipid-buried conformations (ΔG4→3), from interface to lipid (ΔG2→3) and from water to lipid (ΔG1→3) are shown in (F) for the monomer and (J) for the pentamer. To see this figure in color, go online.
In addition, we predicted the tilt angle for five proteins with single transmembrane spans: influenza A M2 (Protein Data Bank, PDB: 1MP6), acetylcholine receptor segment 2 (PDB: 1A11), NR1 subunit of the NMDA receptor (PDB: 2NR1), VPU domain of HIV-1 (PDB: 1PJE), and WALP (WALP23). We chose the first four biological peptides from Ulmschneider et al. (53) because the sequences are less than 35% homologous and the tilt angles have been measured by solid-state NMR spectroscopy. We also included WALP because the sequence was rationally designed (54). The dependence of energy on orientation is shown in Fig. 2, C–E for PDB: 1A11 and Figs. S2 and S3 for the remaining targets. The dependence of energy on tilt angle is shown in Fig. S1, and the low-energy tilt angles are listed in Table S3. We found that M19 predicted tilt angles within ±10°of the experimentally measured value for four of the five peptides. Further, M19 predicted tilt angles closest to the measured value, in contrast to M07 and M12. Together, these results demonstrate that M19 is predictive for both insertion and orientation.
Test #2: predicting the ΔΔG of mutation
Predicting changes in protein stability upon single amino acid substitutions at lipid-exposed positions informs predictions of the effects of genetic mutations and de novo protein design. We evaluated the ability of M19 to capture the change in protein stability upon mutation, called ΔΔGmut, by comparing experimentally measured values with computational predictions. Here, we used a data set of mutations at position 111 on outer membrane palmitoyl transferase (PagP) (55). The data set contains mutations from the host amino acid (alanine) to all 19 other canonical amino acids. Therefore, the ΔΔG computed in this test represents side-chain stability relative to alanine. A summary of prediction accuracy relative to the experimentally measured values is given in Fig. 3. The raw predicted values are also listed in Table S4. Calculated energies are given in Rosetta energy units.
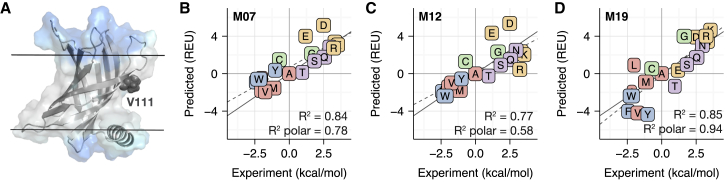
Figure 3.
Comparison between computationally predicted and experimentally measured ΔΔGmut for mutations in PagP. For all correlation plots (B–D), proline is not shown because of steric clashes resulting in a large ΔΔGmut-value. The dotted gray line is the line of best fit, and the solid gray line is y = x. In addition, amino acids are colored according to the following categories: charged (orange), nonpolar (red), aromatic (blue), polar (purple), and special case (green). (A) The structure of the PagP scaffold (PDB: 3GP6) with the mutation site V111 highlighted in dark gray is shown. The implicit solvent phases in (A) are colored in a similar manner as in Fig 1. The ΔΔGmut predictions for mutations in PagP by M07, M12, and M19 are shown in (B)–(D), respectively. To see this figure in color, go online.
The correlation between M19 predicted and experimentally measured ΔΔGmut-values was R2 = 0.85. Note that the ΔΔGmut for proline was excluded for all three energy functions because steric clashes resulted in large values. Although prediction accuracy was improved relative to M12 (R2 = 0.77), the accuracy was comparable to M07 (R2 = 0.84). We were surprised that M07 and M19 demonstrated similar predictive ability. This is because a second set of measurements in OmpLA (37) correlates well with PagP measurements, but not with M07 predictions (56). According to Marx et al. (55), the largest deviations were for side chains containing polar atoms. We therefore recalculated the correlation coefficient for polar and charged side chains. Here, the correlations were 0.78, 0.58, and 0.94 for M07, M12, and M19, respectively. Note that this is mainly due to Asp and Glu because the overall correlation coefficients without these side chains are 0.90, 0.87, and 0.85 for M07, M12, and M19, respectively. Nonetheless, we were encouraged by these results because they demonstrate the ability of our model to capture the behavior of polar side chains in the bilayer.
We examined ΔΔGmut predictions that deviate more than 1.5 Rosetta energy units from the measured value. For M19, this included predictions for G, T, V, Y, and L. To investigate, we analyzed contributions of the component energies to the overall ΔΔGmut (Figs. S4–S6). From the component energies, we found that glycine, threonine, and valine had errors arising from over- or underestimation of van der Waals energy. This suggests double counting between the physics-based terms and the water-to-bilayer energy that captures all of the enthalpic contributions to ΔΔGmut. On the other hand, tyrosine was predicted to be too favorable because of a large attractive van der Waals and water-to-bilayer energy, also suggesting double counting. We were most surprised by the prediction of leucine as less favorable relative to alanine because it is typically one of the most common side chains in the bilayer. This difference arises from a large positive contribution from the two-body solvation term (fa_sol), a term we have not yet refitted for the membrane because of insufficient experimental data.
Test #3: discrimination of native structures from decoys
Identification of native-like structures in an ensemble of candidate structures is a key function of biomolecular modeling energy functions. To evaluate native structure discrimination, we refined ensembles of candidate structures generated by molecular dynamics (57) and then computed the root mean-square deviation (RMSD) between the native crystal and the candidate models. We performed the analysis for five targets: bacteriorhodopsin (Brd7), fumarate reductase (Fmr5), lactose permease (LtpA), rhodopsin (RhoD), and V-ATPase (Vatp). To quantify decoy discrimination, we computed the Boltzmann-weighted average RMS value, called WRMS, for all targets (Table 1; see Supporting Materials and Methods for definition of WRMS). In addition, a mapping of energy versus RMSD for each target is shown in Fig. S7.
Table 1.
Weighted RMSD of Refined and Rescored Candidate Models by Each Energy Function
| Target | R15 (Å) | M07 (Å) | M12 (Å) | M19 (Å) |
|---|---|---|---|---|
| Brd7 | 1.95 | 3.21 | 5.89 | 2.59 |
| Fmr5 | 3.33 | 3.62 | 3.50 | 3.11 |
| LtpA | 2.25 | 1.65 | 1.69 | 2.20 |
| RhoD | 1.88 | 1.77 | 1.62 | 1.93 |
| Vatp | 1.38 | 1.52 | 1.36 | 1.55 |
| Average | 2.16 | 2.81 | 2.35 | 2.28 |
On average, all of the energy functions distinguished near-native from non-native conformations up to 2.1–2.3 Å from the native crystal structure, except M12, which distinguished conformations at 2.8 Å from the native crystal structure. In addition, for all targets except LtpA, all energy models score the native conformation as lower energy than the decoy structures. Upon examination of individual targets, we also found that no specific energy model was consistently better or worse.
We were surprised that the new implicit membrane model did not have an impact on native structure discrimination. Furthermore, R15, which does not consider the membrane, was able to distinguish near-native from non-native decoys at similar resolution. This result suggests although membrane environment energy terms are important, most of the high-resolution discrimination is driven by van der Waals and side-chain packing at high resolution. This finding complements recent work by Mravic et al. (58) that demonstrates side-chain packing is a key driver for stability.
Test #4: native sequence recovery
A fourth test evaluates sequence recovery: the fraction of amino acids recovered after performing complete redesign on naturally occurring proteins. High sequence recovery has long been correlated with strong energy function performance for soluble proteins (44). We therefore performed this test in the context of our membrane protein energy function. In this work, we used a test set of 133 α-helical and β-barrel membrane proteins. The test set is a subset of the 222-member data set from Koehler Leman et al. (43) and was chosen because it is the largest possible subset of high-resolution structures with diverse sequences, further filtered for proteins with known host lipid compositions.
To perform redesign, we used a Monte Carlo fixed-backbone design protocol which samples possible sequences using a full protein rotamer-and-sequence optimization and a multicool annealer-simulated annealing protocol (59). Each protein is initialized in the orientation computed from the Orientations of Proteins in Membranes database (60), and the orientation is kept fixed during sequence search. Then, we computed two metrics: 1) the fraction of all amino acids recovered and 2) the fraction of amino acid types with individual recovery rates greater than 0.05, the same probability of choosing an amino acid at random. Overall, 31.8% of the amino acids designed by M19 were identical to the native amino acid (Fig. 4 A). The soluble protein energy function R15 recovered the second highest percentage of amino acid positions at 29.9%. In contrast, the two existing implicit membrane models lagged behind, with M07 at 26.5% and M12 at 26.7%. The individual amino acid recovery rates were also revealing. Here, M19 and R15 recovered all 20 amino acids at rates above random, whereas M12 recovered 19 and M07 recovered 14.
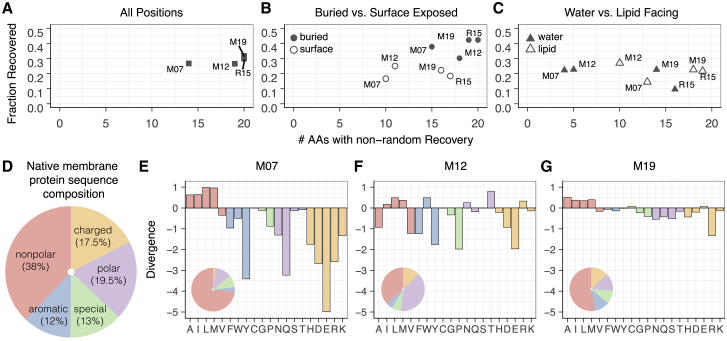
Figure 4.
Properties of designed membrane protein sequences relative to their native counterparts. (A)–(C) rank the performance of each energy function by two metrics: the fraction of all amino acids recovered on the y axis and the fraction of amino acid types with individual recovery rates greater than 0.05 on the x axis. An accurate energy function would have a high sequence-recovery rate both overall and for the individual amino acid types. The results are shown for all positions in (A), buried versus surface-exposed positions in (B), and water- versus lipid-exposed positions in (C). (D) shows the amino acid composition of the native sequences in the benchmark set. (E)–(G) show the KL divergence of the amino acid distribution of the designed proteins relative to the distribution in native membrane proteins. The designs by M07, M12, and M19 are shown in (E), (F), and (G), respectively. A positive value indicates that an amino acid is overenriched, whereas a negative value indicates that an amino acid is underenriched. Values are given on a logarithmic scale. An amino-acid-composition pie chart for sequence designed by each candidate energy function is also shown in the bottom left-hand corner of the divergence plots. To see this figure in color, go online.
To examine the influence of different solvent environments, we recomputed sequence recovery over subsets of residues. First, we compared buried versus solvent-exposed side chains (Fig. 4 B). For all energy functions, recovery was significantly higher for buried side chains than solvent-exposed side chains, as noted in previous studies, because of higher packing density (44). On the surface, M12 recovered 25% of acid positions, slightly higher than the 22% recovery rate by M19. However, M19 recovered 16 amino acids at rates above random, whereas M12 recovered only 12 amino acids. In essence, M19 gets the overall answer correct slightly less frequently; however, it is better at getting more amino acid types correct.
Next, we examined sequence-recovery differences between side chains facing the water and lipid phases (Fig. 4 C). In the lipid phase, all membrane energy functions recovered nearly the same fraction of amino acids. The main differentiating feature is the number of amino acids recovered with greater than random probability. Whereas M07 and M12 recovered four and five amino acids, respectively, M19 recovered 14 amino acids. We observed a similar trend in the water phase. Here, M12 has the highest overall sequence-recovery rate of 27%, next to M19 with a recovery rate of 23%. However, M12 only recovered 10 amino acid types, whereas M19 recovers 14. These results reveal that early energy functions used a rudimentary design strategy: prioritizing only some amino acid types. In contrast, M19 is capable of designing more chemically diverse sequences.
Looking ahead, there are many ways to expand this benchmark to provide more insight. Here, we used a fixed-backbone design algorithm to generate new sequences. An interesting future area would be to use flexible backbone design to enable a larger range of possible sequences. This is an easy extension because the pore shape calculation plus energy evaluation is efficient. In addition, we can compute sequence logos for each design relative to homologous sequences. This provides insight into recovered positions that are also conserved. Thus, our sequence-recovery test provides a foundation for learning more about energy function features in the future.
Comparison with the ref15_memb energy function
While this work was in revision, another membrane energy function was published by Weinstein et al. (61) (ref15_memb, R15M). This presented a good opportunity to compare the performance of franklin2019 with a more recent Rosetta model. We ran all four benchmark tests, and the results are reported in Fig. S8. Overall, M19 outperformed R15M on all tests. The largest discrepancy was performance on the ΔΔG of mutation test, with R15M incorrectly predicting ΔΔG-values for both OmpLA and PagP. The predicted tilt angles were correct for only one of five targets. The resolution of decoy discrimination was overall higher than for M19. Specifically, for R15M, the weighted RMS values were 6.00, 7.22, 2.47, 3.26, and 2.99 for Brd7, Fmr5, LtpA, RhoD, and Vatp, respectively. Further, although both methods predicted a more near-native distribution of amino acids during design, M19 outperformed in both Kullback-Leibler (KL) divergence and recovery metrics, especially for lipid-facing residues.
We were surprised about the discrepancy between M19 and R15M because both energy functions use the same foundation (R15), and the transfer energies in R15M from the dSTβL assay (62) have been shown to correlate with the Moon and Fleming scale. We hypothesize that the main challenge is consideration of side-chain exposure. R15M does not account for lipid composition or pores and cavities. Further, the method was predominantly benchmarked on docking and folding of single-span dimers, whereas the benchmarks in this work are larger and quantitatively more diverse. Therefore, these results suggest that although R15M may be specialized for single-transmembrane dimers, M19 is capable of handling more complex membrane protein topologies.
Designed membrane proteins exhibit native-like features
The sequence-recovery experiment enables us to study properties of in silico designed membrane proteins. These properties are crucial for demonstrating that the implicit model has native membrane properties and is capable of facilitating realistic design experiments. Below, we examine various sequence and structural features important for membrane protein stability and function.
Amino acid distribution in designed proteins mirrors the native distribution
We examined the distribution of amino acids in design protein sequences relative to their native counterparts. Specifically, we measured the KL divergence (DKL) (Eq. S2; see Supporting Materials and Methods) on our membrane protein data set. A negative DKL-value indicates that sequences are underenriched in specific amino acid types, whereas a positive DKL-value indicates that sequences are overenriched. An ideal KL-value is zero. Remarkably, sequences designed by M19 are near native with DKL = −2.7. This is in stark contrast to sequences designed by M07 and M12, which are strongly divergent from native membrane protein sequences, with DKL = −24.6 and DKL = −26.6, respectively.
To learn more about the design implications of each energy function, we computed the KL for each amino acid type (Fig. 4, D–G) and compared it to the composition of amino acids in the native set. The M07 sequences are overenriched in nonpolar amino acids and underenriched in all other categories. The deficits are large, with underenrichment values ranging from 10−2 to 10−4. The M12 sequences are less skewed, with the magnitude of underenrichment deficits ranging between 10−1 and 10−2. However, there is still a large overenrichment of nonpolar amino acids, including I, L, and M, as well as W and T. In contrast, the distribution of amino acids in M19 sequences is comparable to the native distribution, with the magnitude of under- and overenrichment values ranging between 101 and 10−1. Thus, M07 and M12 employ a rudimentary design strategy: only choosing nonpolar amino acids guaranteed to be compatible with the greasy membrane environment. The M19 model does not rely on this assumption and can design every amino acid type within each phase. As a result, M19 designs proteins with an amino acid distribution that is close to the native membrane protein sequence composition. We thus expect that M19 will more accurately evaluate the effects of genetic mutations on protein stability. Further, the diversified sequences will enable designed membrane proteins to achieve a broader range of architectures and functions.
Three-dimensional membrane geometry enables design of polar pores
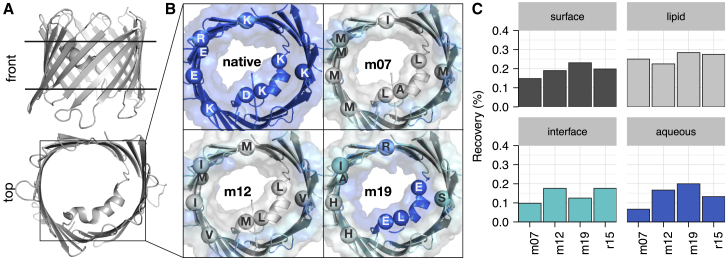
We were interested to see whether a three-dimensional implicit membrane shape facilitates accurate protein design. To do so, we investigated the native and designed sequence of the scaffold protein voltage-dependent anion channel 1 (VDAC1; PDB: 3EMN; Fig. 5). The native sequence of this β-barrel protein pore is rich in charged amino acids. In the two-dimensional membranes used by M07 and M12, the pore-facing residues are designed as if they are in the lipid phase, and as a result, the designed sequences are rich in nonpolar amino acids. In contrast, the three-dimensional implicit membrane geometry treats pore-facing residues as exposed to the water phase; thus, the designed sequence contains both polar and charged amino acids. These positive features are reflected in the sequence for this specific target. Here, M19 exhibits the highest recovery over all surface residues and lipid-facing and aqueous-facing residues when compared with other energy functions. This result suggests the potential of M19 to perform accurate design on both the lipid-facing and water-filled-pore-facing surfaces.
Figure 5.
An in silico redesigned β-barrel membrane protein with a polar aqueous pore. (A) Structure of the design scaffold protein voltage-dependent anion channel VDAC1 (PDB: 3EMN) is shown from a lateral and top view. The horizontal black lines denote the approximate position of the membrane. (B) Sequence composition and solvation properties of the pore redesigned by the M07 (top right), M12 (bottom left), and M19 (bottom right) energy functions are shown in contrast to the native sequence (top left). M07 and M12 treat the pore as lipid exposed, resulting in a nonpolar sequence. In contrast, the M19 energy function calculates a custom pore shape, resulting in a polar pore sequence. (C) Recovery of the native 3EMN sequence upon redesign is shown. In contrast to other energy functions, the M19 recovers the most native sequence for the total surface and lipid-exposed and aqueous-solvated residues. To see this figure in color, go online.
An unexpected result was that M19 outperformed R15 in the aqueous pore of VDAC1. In fact, we expected the performance of R15 to match M19 because in the pore region, fhyd = 0. We hypothesize that the pore size and transition steepness were underestimated, and thus, the calculation was influenced by M19. Although it is hard to draw a quantitative conclusion about the improved performance, we suggest a future step of investigating the amino acid composition of a larger set of β-barrel pores to understand the result.
Biologically relevant lipid composition parameters improve per-target sequence recovery
Finally, we were eager to explore whether implicit membrane parameters for different lipid compositions can improve design outcomes. This question is difficult to evaluate because the host membrane composition of proteins is not always known. At the same time, this question is crucial because of the long-standing criticism that implicit membrane models do not accurately capture the properties of different lipid membrane compositions. In this work, we investigated this question anecdotally by examining two examples from our membrane protein design data set.
First, we examined the β-barrel protein scaffold outer membrane transporter FecA from E. coli. The outer membranes of gram-negative bacteria are significantly thinner than eukaryotic plasma membranes. We therefore hypothesized that sequence recovery of lipid-facing residues in this protein would be higher in a thinner membrane. To test this hypothesis, we again searched for low-energy sequences in an M19 membrane with either 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) or 1-palmitoyl-2-oleoyl-glycerol-3-phosphocholine (POPC) parameters. Encouragingly, the recovery of lipid-facing residues in this protein was 33% in DLPC, in contrast to 28% in POPC. We also repeated this test on the α-helical protein scaffold VCX1 calcium-proton exchanger from Saccharomyces cerevisiae. Here, we expected the reverse trend: improved design in a POPC membrane over DLPC. Again, the design results followed: 22% sequence recovery in DLPC and 29% in POPC. These results demonstrate that lipid composition parameters facilitate more biologically realistic structure prediction and design.
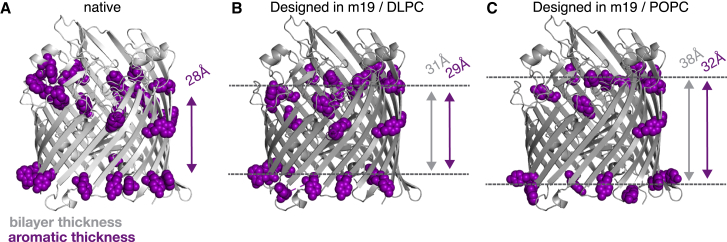
In addition, there was an inevitable question that we wanted to ask about our β-barrel protein scaffold. Experimental studies have long demonstrated that β-barrel membrane proteins have high concentrations of aromatic side chains near the interfacial headgroups (63). Although the thermodynamics of this phenomena are not completely understood, it has been suggested that stacking of the aromatics nearby polar headgroups stabilizes the protein (64). Thus, we asked the question: does M19 also design aromatics near the anisotropic phase representing interfacial headgroups? To answer, we calculated the apparent membrane thickness according to the average positions of aromatic side chains in native and designed FecA (Fig. 6). We found that M19 designed with a larger apparent thickness in POPC rather than DLPC membranes. Notably, the DLPC aromatic thickness is near the native aromatic thickness. Although still anecdotal, these results suggest that M19 designs proteins with native-like features.
Figure 6.
In silico redesigned β-barrel membrane proteins in native-like lipid compositions. (A) The structure of the design scaffold outer membrane transporter FecA (PDB: 1KMO) from E. coli is shown. The backbones redesigned by the M19 energy function with DLPC and POPC parameters are shown in (B) and (C), respectively. The native scaffold is colored in light gray, and the design scaffolds are colored in dark gray. Aromatic amino acids near the interface (>7 and <25 from the center) are colored in light pink. The gray arrow shows the bilayer thickness, and the pink arrow shows the thickness according to the average position of interfacial aromatic residues. The dotted lines denote the approximate position of the bilayer. The thickness from the DLPC design best matches the native and results in the highest recovery of lipid-facing residues in the transmembrane region. DLPC is closest to the thickness in E. coli. To see this figure in color, go online.
Discussion
In this work, we developed, implemented, and tested a new energy function for membrane protein structure prediction and design. The energy function, called franklin2019, uses an implicit approach to represent the anisotropic structure and nanoscale dimensions of membranes with varied phospholipid composition, a key component of biological membranes. Through computational benchmarking, we demonstrated that the model could replicate experimentally measured protein stabilities and orientations. With multiple diverse benchmark sets, we demonstrated that franklin2019 improves modeling and design of membrane proteins with complex topologies, pores, and juxtamembrane domains. Further, proteins designed by franklin2019 exhibit native-like features, including amino acid distribution, aromatic amino acids near interfacial headgroups, and hydrophobic match with specific lipid compositions. Together, these features demonstrate the potential of franklin2019 to advance high-resolution membrane protein structure prediction and design.
Through the goal of developing a new energy function, our study interrogated fundamental questions about the design rules for native membrane proteins. First, the implicit model is based on transfer energies from a thermodynamic hydrophobicity scale measured in a phospholipid bilayer. The high sequence-recovery rate demonstrates the importance of thermostability and bulk phospholipid chemistry in constraining membrane protein sequences. Furthermore, previous work relied on narrow membrane protein design rules such as enrichment of leucine side chains in the hydrophobic core. We demonstrated that native membrane protein sequences are diverse and not constrained to hydrophobic amino acids. Accordingly, our energy function uses the full palette of amino acid chemistries during design.
This work was enabled by the Moon and Fleming (37) hydrophobicity scale. Although there has been extensive work to quantify transfer energies (65), the Moon and Fleming scale captures the actual equilibrium change in free energy in the context of a membrane protein in a phospholipid bilayer (66). Thus, the implicit model captures more biologically realistic context relative to prior models that approximated the membrane as a slab of nonpolar organic solvent. One consideration of using the Moon and Fleming scale is that franklin2019 would not capture nonthermodynamic (kinetic) end states of ab initio folding in which chaperones are required. For example, in α-helical membrane protein folding, the intermediate states in the two-stage folding process may not be captured (67). However, because the goal of Rosetta calculations is to capture the free energy minima, this does not impose limitations. Second, the measurements were taken at pH 3.8. As a result, the energetics of Asp and Glu are undervalued because the side chains are protonated. This may affect estimates of transfer energies for soluble proteins and marginally hydrophobic proteins. Accurately assigning the protonation states of Asp and Glu is an ongoing challenge because of membrane-induced pKa shifts that alter the protonation equilibrium (68).
Next, we sought to develop a model that describes bilayers with different lipid compositions. We were inspired by prior studies that added more detail to implicit “slab” models, including anionic lipid parameters (69) and adjustable bilayer thickness (26). In this work, we focused on single-component phospholipid bilayers for two reasons: 1) there are significant small-angle x-ray scattering and neutron scattering data available for validation, and 2) many experiments are performed in single-component bilayers, enabling easy comparison. We coined our model “biologically realistic” to highlight the advance of using phospholipid models over prior organic slab models. Importantly, there are many future steps required to achieve a “biologically accurate” model. First, native membranes include hundreds of lipid types, distributed nonuniformly (47). Although all-atom models remain difficult, there has been excellent progress in coarse-grained modeling of native lipid bilayers (70). Thus, a possible step is to develop model parameters from these coarse-grained models. Another important step is to generate parameters for the asymmetric lipid composition to emulate the outer membrane of gram-negative bacteria (71). Additionally, the membrane bends and curves to accommodate the hydrophobic surface of proteins (72). A further challenge is accounting for local properties such as specific protein interactions with lipids and cholesterol, which may be captured by a hybrid implicit-explicit approach such as SPadES (73) or HMMM (74). Finally, an open question is how to account for mechanical properties such as lateral pressure and strain due to local curvature. In these scenarios, it is most likely that implicit membrane simulations will compliment information from emerging membrane protein modeling tools and molecular dynamics simulations to investigate structure, dynamics, and function.
Another important methodological step is modeling of membrane protein pores and cavities. Previously, implicit models approximated pores as cylinders (48) or segregated side chains using grid-based approaches (75). In contrast, franklin2019 uses continuous functions to model a wide range of pore geometries. We chose this approach over solvent-accessible surface area calculations to reduce computational cost, enabling scalability for more sophisticated molecular modeling applications such as flexible backbone design. For future work, we aim to capture membrane deformations (76) through the integration of continuum elastic models (77,78) or hybrid continuum-atomistic models (79). Additionally, more work is needed to account for fenestrations that alter the solvent exposure of lipid accessible residues (80). Ultimately, these features will advance franklin2019 from capturing static membrane features to incorporate dynamics important for protein function.
We evaluated our implicit membrane model using sparse, high-resolution experimental data. This approach contrasts soluble protein energy function evaluation, in which there is an abundance of thermodynamic and spectroscopic measurements of small molecules (81) and high-resolution protein structures (82). To overcome the possibility of overfitting, we limited the validation data to high-quality measurements. For instance, we did not use crystal structures with ≥3 Å resolution or ΔΔGmut-values that were not measured in a reversible system. Further, we benchmarked our energy function against both thermodynamic and structure prediction data. Previous studies have evaluated membrane energy functions on a single test such as tilt angles (53), native structure discrimination (57,83), predicting hydrophobic lengths (75), ΔΔG prediction (84), and sequence recovery (85). Simultaneously performing the benchmarks enables a well-rounded evaluation of the energy function for diverse biomolecular modeling tasks.
Looking ahead, a larger benchmark set will enable broader energy function development and optimization. This work focused on developing a single empirical term that captures water-to-bilayer transfer energetics that could be added to the existing Rosetta energy function. Naturally, this introduces double counting between the new term and existing physics-based terms such as solvation and electrostatics. Previous work on the soluble protein energy function used a Nelder-Mead optimization scheme (52) to remove double counting. Although there are currently insufficient data to apply this approach, we envision that as more data emerge, we will be able to apply more robust fitting techniques, including machine learning. Furthermore, additional benchmark data will enable adding membrane dependence to the solvation and electrostatic terms, which will improve the modeling of local side-chain environments. Important future requirements for a larger benchmark set include more diverse modeling tasks such as capturing multiple conformation states and diverse data sources such as models from x-ray crystallography, cryo-electron microscopy, and NMR spectroscopy.
An important remaining task is to compare the performance of franklin2019 with the latest methods in other molecular modeling packages. Currently, there are several technical hurdles: 1) alternate membrane representations are not implemented within the Rosetta package, and 2) other packages cannot generate all of the requisite data for each benchmark (e.g., design is computationally expensive for classical molecular dynamics packages). Notably, the latest energy functions for membrane protein modeling use a wide range of physical, empirical, and statistical models for energy calculations. Therefore, direct comparison will provide important information to the community about the best strategies for membrane protein structure prediction and design.
In summary, we developed a biologically realistic energy function for membrane protein structure prediction and design. The energy function is implemented within the Rosetta software and can be used for a wide range of macromolecular modeling tools. By pursuing a balance of efficiency and accuracy, we anticipate that the implicit membrane will enable high-throughput and high-resolution membrane protein structure prediction and design. Importantly, this model transforms once protein-centric tools to techniques that can predict and design structures tied to varied biologically relevant lipid compositions.
Data availability
The energy function and benchmark tests presented are available in the Rosetta software suite (https://www.rosettacommons.org). Rosetta is available to noncommercial users for free and to commercial users for a fee.
Author Contributions
Conceptualization and methodology, R.F.A, P.J.F., K.G.F., and J.J.G.; investigation, data curation, software, and analysis, R.F.A.; writing—original draft, R.F.A.; writing—review and editing, R.F.A., P.J.F., K.G.F., and J.J.G.; funding acquisition, R.F.A., K.G.F., and J.J.G.; resources and supervision, K.G.F. and J.J.G.
Acknowledgments
The authors thank Sergey Lyskov, Vikram Mulligan, and Julia Koehler for code reviews. We thank Sai Pooja Mahajan for critical reading of the manuscript. We also thank Michael Feig and Bercem Dutagaci for providing a high-resolution decoy set.
R.F.A. was supported by a Hertz Foundation Fellowship and a National Science Foundation Graduate Research Fellowship. This work was also supported by National Institute of Health grants GM-078221 (R.F.A. and J.J.G.) and GM-079440 (P.J.F. and K.G.F.). Computations were performed using the Maryland Advanced Research Computing Center and the National Science Foundation Extreme Science and Engineering Discovery Environment grant TG-MCB180056.
Editor: Michael Grabe.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.03.006.
Supporting Material
References
- 1.Tan S., Tan H.T., Chung M.C.M. Membrane proteins and membrane proteomics. Proteomics. 2008;8:3924–3932. doi: 10.1002/pmic.200800597. [DOI] [PubMed] [Google Scholar]
- 2.Overington J.P., Al-Lazikani B., Hopkins A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Huang P.-S., Boyken S.E., Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 4.Barth P., Senes A. Toward high-resolution computational design of the structure and function of helical membrane proteins. Nat. Struct. Mol. Biol. 2016;23:475–480. doi: 10.1038/nsmb.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joh N.H., Wang T., DeGrado W.F. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science. 2014;346:1520–1524. doi: 10.1126/science.1261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahendran K.R., Niitsu A., Bayley H. A monodisperse transmembrane α-helical peptide barrel. Nat. Chem. 2017;9:411–419. doi: 10.1038/nchem.2647. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury R., Ren T., Maranas C.D. PoreDesigner for tuning solute selectivity in a robust and highly permeable outer membrane pore. Nat. Commun. 2018;9:3661. doi: 10.1038/s41467-018-06097-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young M., Dahoun T., Barth P. Computational design of orthogonal membrane receptor-effector switches for rewiring signaling pathways. Proc. Natl. Acad. Sci. USA. 2018;115:7051–7056. doi: 10.1073/pnas.1718489115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng X., Ambia J., Barth P. Computational design of ligand-binding membrane receptors with high selectivity. Nat. Chem. Biol. 2017;13:715–723. doi: 10.1038/nchembio.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu P., Min D., Baker D. Accurate computational design of multipass transmembrane proteins. Science. 2018;359:1042–1046. doi: 10.1126/science.aaq1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson C.J., Rosso L., Gould I.R. GAFFlipid: a General Amber Force Field for the accurate molecular dynamics simulation of phospholipid. Soft Matter. 2012;8:9617. [Google Scholar]
- 12.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid N., Eichenberger A.P., van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 14.Phillips J.C., Sun Y., Kale L.V. Mapping to irregular torus topologies and other techniques for petascale biomolecular simulation. SC. Conf. Proc. 2014;2014:81–91. doi: 10.1109/SC.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marrink S.J., Corradi V., Sansom M.S.P. Computational modeling of realistic cell membranes. Chem. Rev. 2019;119:6184–6226. doi: 10.1021/acs.chemrev.8b00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 17.Barrera E.E., Frigini E.N., Pantano S. Modeling DMPC lipid membranes with SIRAH force-field. J. Mol. Model. 2017;23:259. doi: 10.1007/s00894-017-3426-5. [DOI] [PubMed] [Google Scholar]
- 18.Baoukina S., Rozmanov D., Tieleman D.P. Composition fluctuations in lipid bilayers. Biophys. J. 2017;113:2750–2761. doi: 10.1016/j.bpj.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warshel A., Russell S.T. Calculations of electrostatic interactions in biological systems and in solutions. Q. Rev. Biophys. 1984;17:283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- 20.Grossfield A. Chapter 5 implicit modeling of membranes. In: Feller S., Simon S., Benos D., editors. Computational Modeling of Membrane Bilayers: Current Topics in Membranes. Volume 60. Academic Press; 2008. pp. 131–157. [Google Scholar]
- 21.Ulmschneider J.P., Ulmschneider M.B. Sampling efficiency in explicit and implicit membrane environments studied by peptide folding simulations. Proteins. 2009;75:586–597. doi: 10.1002/prot.22270. [DOI] [PubMed] [Google Scholar]
- 22.Davis M.E., McCammon J.A. Electrostatics in biomolecular structure and dynamics. Chem. Rev. 1990;90:509–521. [Google Scholar]
- 23.Baker N.A., Sept D., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Im W., Lee M.S., Brooks C.L., III Generalized born model with a simple smoothing function. J. Comput. Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 25.Spassov V., Yan L., Sándor S. Introducing an implicit membrane in generalized born/solvent accessibility continuum solvent models. J. Phys. Chem. B. 2002;106:8726–8738. [Google Scholar]
- 26.Panahi A., Feig M. Dynamic Heterogeneous Dielectric Generalized Born (DHDGB): an implicit membrane model with a dynamically varying bilayer thickness. J. Chem. Theory Comput. 2013;9:1709–1719. doi: 10.1021/ct300975k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazaridis T. Effective energy function for proteins in lipid membranes. Proteins. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 28.Radzicka A., Wolfenden R. Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. Biochemistry. 1988;27:1664–1670. [Google Scholar]
- 29.Nepal B., Leveritt J., III, Lazaridis T. Membrane curvature sensing by amphipathic helices: insights from implicit membrane modeling. Biophys. J. 2018;114:2128–2141. doi: 10.1016/j.bpj.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barth P., Schonbrun J., Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. USA. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franks N.P., Abraham M.H., Lieb W.R. Molecular organization of liquid n-octanol: an X-ray diffraction analysis. J. Pharm. Sci. 1993;82:466–470. doi: 10.1002/jps.2600820507. [DOI] [PubMed] [Google Scholar]
- 32.MacCallum J.L., Bennett W.F.D., Tieleman D.P. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarov-Yarovoy V., Schonbrun J., Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senes A., Chadi D.C., Degrado W.F. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J. Mol. Biol. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 35.Schramm C.A., Hannigan B.T., Samish I. Knowledge-based potential for positioning membrane-associated structures and assessing residue-specific energetic contributions. Structure. 2012;20:924–935. doi: 10.1016/j.str.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh D., Davis A., Nanda V. A knowledge-based potential highlights unique features of membrane α-helical and β-barrel protein insertion and folding. Protein Sci. 2012;21:50–62. doi: 10.1002/pro.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon C.P., Fleming K.G. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc. Natl. Acad. Sci. USA. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamy-Freund M.T., Riske K.A. The peculiar thermo-structural behavior of the anionic lipid DMPG. Chem. Phys. Lipids. 2003;122:19–32. doi: 10.1016/s0009-3084(02)00175-5. [DOI] [PubMed] [Google Scholar]
- 39.Leonenko Z.V., Finot E., Cramb D.T. Investigation of temperature-induced phase transitions in DOPC and DPPC phospholipid bilayers using temperature-controlled scanning force microscopy. Biophys. J. 2004;86:3783–3793. doi: 10.1529/biophysj.103.036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jo S., Lim J.B., Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michaud-Agrawal N., Denning E.J., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koehler Leman J., Lyskov S., Bonneau R. Computing structure-based lipid accessibility of membrane proteins with mp_lipid_acc in RosettaMP. BMC Bioinformatics. 2017;18:115. doi: 10.1186/s12859-017-1541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhlman B., Baker D. Native protein sequences are close to optimal for their structures. Proc. Natl. Acad. Sci. USA. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khachiyan L.G., Todd M.J. On the complexity of approximating the maximal inscribed ellipsoid for a polytope. Math. Program. 1993;61:137–159. [Google Scholar]
- 46.Kučerka N., Nieh M.-P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 47.Harayama T., Riezman H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018;19:281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 48.Lazaridis T. Structural determinants of transmembrane β-Barrels. J. Chem. Theory Comput. 2005;1:716–722. doi: 10.1021/ct050055x. [DOI] [PubMed] [Google Scholar]
- 49.Pellegrini-Calace M., Maiwald T., Thornton J.M. PoreWalker: a novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PLoS Comput. Biol. 2009;5:e1000440. doi: 10.1371/journal.pcbi.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alford R.F., Leaver-Fay A., Gray J.J. The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yarov-Yarovoy V., DeCaen P.G., Catterall W.A. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc. Natl. Acad. Sci. USA. 2012;109:E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park H., Bradley P., DiMaio F. Simultaneous optimization of biomolecular energy functions on features from small molecules and macromolecules. J. Chem. Theory Comput. 2016;12:6201–6212. doi: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulmschneider M.B., Sansom M.S.P., Di Nola A. Evaluating tilt angles of membrane-associated helices: comparison of computational and NMR techniques. Biophys. J. 2006;90:1650–1660. doi: 10.1529/biophysj.105.065367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holt A., Koehorst R.B.M., Killian J.A. Tilt and rotation angles of a transmembrane model peptide as studied by fluorescence spectroscopy. Biophys. J. 2009;97:2258–2266. doi: 10.1016/j.bpj.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marx D.C., Fleming K.G. Influence of protein scaffold on side-chain transfer free energies. Biophys. J. 2017;113:597–604. doi: 10.1016/j.bpj.2017.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alford R.F., Koehler Leman J., Gray J.J. An integrated framework advancing membrane protein modeling and design. PLoS Comput. Biol. 2015;11:e1004398. doi: 10.1371/journal.pcbi.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dutagaci B., Wittayanarakul K., Feig M. Discrimination of native-like states of membrane proteins with implicit membrane-based scoring functions. J. Chem. Theory Comput. 2017;13:3049–3059. doi: 10.1021/acs.jctc.7b00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mravic M., Thomaston J.L., DeGrado W.F. Packing of apolar side chains enables accurate design of highly stable membrane proteins. Science. 2019;363:1418–1423. doi: 10.1126/science.aav7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leaver-Fay A., O’Meara M.J., Kuhlman B. Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 2013;523:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lomize M.A., Pogozheva I.D., Lomize A.L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinstein J.Y., Elazar A., Fleishman S.J. A lipophilicity-based energy function for membrane-protein modelling and design. PLoS Comput. Biol. 2019;15:e1007318. doi: 10.1371/journal.pcbi.1007318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elazar A., Weinstein J., Fleishman S.J. Mutational scanning reveals the determinants of protein insertion and association energetics in the plasma membrane. eLife. 2016;5:e12125. doi: 10.7554/eLife.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wallin E., Tsukihara T., Elofsson A. Architecture of helix bundle membrane proteins: an analysis of cytochrome c oxidase from bovine mitochondria. Protein Sci. 1997;6:808–815. doi: 10.1002/pro.5560060407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDonald S.K., Fleming K.G. Aromatic side chain water-to-lipid transfer free energies show a depth dependence across the membrane normal. J. Am. Chem. Soc. 2016;138:7946–7950. doi: 10.1021/jacs.6b03460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peters C., Elofsson A. Why is the biological hydrophobicity scale more accurate than earlier experimental hydrophobicity scales? Proteins. 2014;82:2190–2198. doi: 10.1002/prot.24582. [DOI] [PubMed] [Google Scholar]
- 66.Robertson J.L. We choose to go to the membrane. Proc. Natl. Acad. Sci. USA. 2011;108:10027–10028. doi: 10.1073/pnas.1107322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Popot J.L., Engelman D.M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 68.Teixeira V.H., Vila-Viçosa D., Machuqueiro M. pK(a) values of titrable amino acids at the water/membrane interface. J. Chem. Theory Comput. 2016;12:930–934. doi: 10.1021/acs.jctc.5b01114. [DOI] [PubMed] [Google Scholar]
- 69.Lazaridis T. Implicit solvent simulations of peptide interactions with anionic lipid membranes. Proteins. 2005;58:518–527. doi: 10.1002/prot.20358. [DOI] [PubMed] [Google Scholar]
- 70.Ingólfsson H.I., Melo M.N., Marrink S.J. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 71.Rothman J.E., Lenard J. Membrane asymmetry. Science. 1977;195:743–753. doi: 10.1126/science.402030. [DOI] [PubMed] [Google Scholar]
- 72.Jarsch I.K., Daste F., Gallop J.L. Membrane curvature in cell biology: an integration of molecular mechanisms. J. Cell Biol. 2016;214:375–387. doi: 10.1083/jcb.201604003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lai J.K., Ambia J., Barth P. Enhancing structure prediction and design of soluble and membrane proteins with explicit solvent-protein interactions. Structure. 2017;25:1758–1770.e8. doi: 10.1016/j.str.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vermaas J.V., Pogorelov T.V., Tajkhorshid E. Extension of the highly mobile membrane mimetic to transmembrane systems through customized in silico solvents. J. Phys. Chem. B. 2017;121:3764–3776. doi: 10.1021/acs.jpcb.6b11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dutagaci B., Feig M. Determination of hydrophobic lengths of membrane proteins with the HDGB implicit membrane model. J. Chem. Inf. Model. 2017;57:3032–3042. doi: 10.1021/acs.jcim.7b00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perozo E., Cortes D.M., Martinac B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 2002;418:942–948. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 77.Andersen O.S., Koeppe R.E. Bilayer thickness and membrane protein function: An energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 78.Choe S., Hecht K.A., Grabe M. A continuum method for determining membrane protein insertion energies and the problem of charged residues. J. Gen. Physiol. 2008;131:563–573. doi: 10.1085/jgp.200809959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Argudo D., Bethel N.P., Grabe M. New continuum approaches for determining protein-induced membrane deformations. Biophys. J. 2017;112:2159–2172. doi: 10.1016/j.bpj.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marcoline F.V., Bethel N., Grabe M. Membrane protein properties revealed through data-rich electrostatics calculations. Structure. 2015;23:1526–1537. doi: 10.1016/j.str.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 82.O’Meara M.J., Leaver-Fay A., Kuhlman B. Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J. Chem. Theory Comput. 2015;11:609–622. doi: 10.1021/ct500864r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuzlenko O., Lazaridis T. Membrane protein native state discrimination by implicit membrane models. J. Comput. Chem. 2013;34:731–738. doi: 10.1002/jcc.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kroncke B.M., Duran A.M., Sanders C.R. Documentation of an imperative to improve methods for predicting membrane protein stability. Biochemistry. 2016;55:5002–5009. doi: 10.1021/acs.biochem.6b00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duran A.M., Meiler J. Computational design of membrane proteins using RosettaMembrane. Protein Sci. 2018;27:341–355. doi: 10.1002/pro.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The energy function and benchmark tests presented are available in the Rosetta software suite (https://www.rosettacommons.org). Rosetta is available to noncommercial users for free and to commercial users for a fee.