Abstract
Objectives
Treatment of non-small cell lung cancer (NSCLC) with immune checkpoint blockade (ICB) has resulted in striking clinical responses, but only in a subset of patients. The goal of this study was to evaluate transcriptional signatures previously reported in the literature in an independent cohort of NSCLC patients receiving ICB.
Materials and Methods
This retrospective study analyzed transcriptional profiles from pre-treatment tumor samples of 52 chemotherapy-refractory advanced NSCLC patients treated with anti-PD1/PD-L1 therapy. Gene signatures based on published reports were created and examined for their association with response to therapy and progression-free and overall survival (PFS, OS).
Results
Two signatures predicting response and outcomes were identified. One reflected the degree of immune infiltration and upregulation of interferon-gamma-induced genes. A second reflected the EMT status. Compared to those not responding to therapy, patients whose tumors responded to ICB had higher scores in an inflammatory gene signature (6.0±2.9 vs −5.5±3.4, p=0.014) or a more epithelial phenotype (−1.7±1.0 vs 2.1±1.2, p=0.016). Both signatures demonstrated a satisfactory predictive accuracy for response: AUC of 0.69 (95% CI: 0.54, 0.84) for the inflammatory and 0.70 (95% CI: 0.55, 0.85) for EMT signatures, respectively. A weighted score combining EMT and inflammatory signatures showed increased predictive value with AUC of 0.92 (95% CI: 0.85, 0.99). Kaplan-Meier curves for patients above and below the median combined score showed a significant separation for PFS and OS (all p<0.01, log rank test).
Conclusions
The EMT/Inflammation signature score may be useful in directing checkpoint inhibitor therapy in lung cancer and suggests that reversal of EMT might augment efficacy of ICB.
Keywords: Non-small cell lung cancer, Immunotherapy, Gene Signatures, Cancer Biomarkers
1. INTRODUCTION
Immune checkpoint blockade (ICB) using anti-PD11,2 or anti-PD-L1 antibodies3,4 has demonstrated enhanced response rates, progression free survival (PFS), and overall survival (OS) in non-small cell lung cancer (NSCLC) patients. However, only a subset of patients achieve durable clinical benefit. Since these therapies are associated with immune-related adverse events and have significant financial implications,5,6 a need exists for robust predictive biomarkers.
Measurement of PD-L1 protein expression on tumor cells by immunohistochemistry (IHC) has been the most commonly used predictive marker, however its utility has been limited by poor uniformity in the anti-PDL1 IHC antibodies, different thresholds for PD-L1 positivity across different trials, and inherent fluidity in PD-L1 expression.7 In addition, several trials have shown no correlation between PD-L1 expression and response rates.1,8 Assessments of the tumor-immune cell infiltrate within the tumor microenvironment suggest that “hot” or more inflammatory tumors have better responses.3,9–11
Other non-IHC-based biomarker strategies have shown promise. For example, in some studies, tumor somatic mutation burden (TMB) seems to predict response and PFS, presumably because these tumors have a higher neoantigen burden which makes them more sensitive to T cell activation therapy.12,13
Another strategy is to use tumor gene-expression profiling to develop genomic signatures of response.3,14–17 The most well-developed of these signatures reflects the inflammatory state of the tumor and includes genes that identify T cells, T cell activation, interferon-responsive genes, chemokine expression and adaptive immune resistance. This has been used to predict response to pembrolizumab in melanoma, gastric, and head and neck squamous cell carcinomas.14 Recently, the inflammation scores were combined with TMB scores and showed higher predictive value than either score alone in a similar range of cancers.16,18
Other tumor factors may also regulate response to immunotherapy. Hugo et al. found a transcriptional signature referred to as IPRES (innate anti-PD1 resistance) that included genes involved in cell adhesion, extracellular matrix remodeling, angiogenesis, wound healing, and mesenchymal transition that predicted response to anti-PD1 antibody therapy in melanoma.15 Genes reflecting TGF-β expression and signaling of EMT/stroma-related genes have been reported to associate with resistance to PD-1 blockade in urothelial cancers.19,20
There is also increasing evidence from preclinical studies to suggest that the level of epithelial-to-mesenchymal transition (EMT) of tumor cells contributes to the level of immunosuppression, with more mesenchymal tumors being more resistant to immunotherapy.21,22 To our knowledge, the EMT hypothesis has not been rigorously examined in human cancers.
The goal of this study was to evaluate whether genomic data could predict response to ICB in NSCLC. To do so, we analyzed transcriptomic data from pre-treatment tumor specimens from 52 metastatic NSCLC patients treated with anti-PD1/PD-L1 monotherapy. Specifically, we assessed the predictive power of expression levels of macrophage, T cells, and TGF-β related genes, as well as a series of established genomic signatures reflecting inflammation, chronic interferon stimulation, the IPRES genes, and the EMT status of the tumors.
2. MATERIALS AND METHODS
2.1. Study Design and Patients
This single-center, retrospective, observational study was conducted at the Hospital of the University of Pennsylvania from September 2013 to August 2018 and was approved by the University’s Institutional Review Board. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement was followed to ensure the quality of data reported in this study.23 Patients with metastatic NSCLC treated with anti-PD1 or anti-PD-L1 antibodies in the second-line setting that had sufficient residual formalin-fixed paraffin-embedded (FFPE) tumor material for detailed RNA analysis were included. A pathologist confirmed the presence of sufficient tumor material in FFPE slides. Initially, a total of 67 subjects were identified; 52 had RNA of sufficient quality for analysis. Baseline demographics and clinical variables were obtained from the electronic medical record.
2.2. Tissue Collection and RNA Sequencing
RNA was analyzed using the AmpliSeq™ Transcriptome Human Gene Expression Kit (ThermoFisher Scientific). RNA was extracted using the RNAstorm, RNA Isolation kit for FFPE (CELLDATA) from 3 FFPE slides per patient as per the manufacturer’s instructions. The quality of FFPE RNA was measured using the Agilent 2100 Bioanalyzer system (RNA 6000 Pico Kit). Sequencing libraries were prepared according to the AmpliSeq Library prep kit protocol as previously described.24 Pooled libraries were amplified using emulsion PCR on an Ion Torrent OneTouch2 instrument and enriched following the manufacturer’s instructions. Libraries were then loaded onto an Ion P1 chip V2 and sequenced on the Ion Torrent Proton™ sequencing system, using the Ion PI sequencing 200 Kit v3 chemistry.
The data are raw read counts per gene. The primers used for each gene target regions of approximately the same length in all genes so do not need to be corrected for gene length. The data were normalized as “counts per million”, where the raw counts for each gene were divided by the total number of counts, multiplied by a constant (106), and log2 transformed.
2.3. Gene Signature Generation
The goal of this study was to validate existing signatures from the literature in our independent patient population. By summing the log2 Z scores of genes, we calculated: (1) the IPRES score for innate anti-PD1 resistance using 16 of 17 (expression of CCL8 was too low) reported genes (see Supplemental Table 1);15 and (2) a chronic IFN activation score using STAT1, IFI44, IFIT1, IFIT3, OAS1,OAS2, MX1, IRF7, and ISG15 as described.25,26 The TGF-β genes analyzed were those described by Mariathasan19 as TGFB1, TGFBR2, ACTA2, COL4A2, and TAGLN.
Our inflammation signature was based on a previously published gene score14 and created by summing (unweighted) log2 Z scores for 27 inflammation related genes (Supplemental Table 1).
There is not yet a validated lung cancer EMT signature, so we prospectively generated a gene list based on previous publications describing “classic” EMT genes in cancer23,24 with the addition of some genes specifically mentioned in studies evaluating EMT in NSCLC.27–29 Selected genes had levels of expression that were clearly above baseline (using a cutoff of 10 reads in our data set). Supplemental Table 2 shows the list of genes included along with their average expression level. Although the genes SNAI1, TWIST1, TWIST2, CDH2, and ZEB1 are classic mesenchymal markers, their expression levels were very low in our dataset and thus not included. Based on these criteria, we generated the EMT signature by adding the sum of the log2 Z scores of 6 established mesenchymal genes (AGER, FN1, MMP2, SNAI2, VIM, ZEB2) and subtracting the sum of the log2 Z scores of 6 established epithelial genes (CDH1, CDH3, CLDN4, EPCAM, MAL2, and ST14) (Supplemental Table 2). In this signature, the most mesenchymal tumors have the most positive EMT scores and the most epithelial tumors have the most negative scores.
2.4. Statistical Analyses
Comparisons among response groups was done using Pearson’s chi-squared test for categorical characteristics or Fisher’s exact test. Response variables were grouped into the binary categories of responders (including patients with partial responses and stable disease) versus non-responders. The responder group was defined by a radiologist reporting a decrease in size of overall disease burden or stability of disease ≥ 6 months. The association between a gene signature score and response was examined using a t-test after the normality of the gene scores were established. Descriptive statistics are displayed as mean±sem. The area under a receiving operating characteristics (ROC) curve (AUC) was obtained by fitting a logistic regression model with response status as the dependent variable and the signature score as an independent variable. The significance level of AUC against the null value of 0.5 was tested.
We created a combined score for inflammation and EMT using a simple unweighted score by taking the difference between the inflammation and EMT scores (higher scores are more inflammatory and more epithelial). We also generated a weighted score with the estimated regression weights from the multivariable logistic regression model that included both the inflammation and EMT scores. Based on the maximum likelihood estimation, the weighted score was computed as −0.60*EMT+0.19*inflammation.
The significance level for the change in AUC from the single signature model to 2-signature model was tested.30 Kaplan-Meier curves for PFS and OS were plotted for patients above or below the median weighted combined score and compared by Cox proportional hazards models. PFS was defined as months since the start of ICB therapy until the date of progression or death, or censored at the cut-off date of August 31, 2018. All statistical analyses were two sided and performed using Graphpad Prism version 7.0 (La Jolla, CA) or Stata version 15.1 (College Station, TX).
3. RESULTS
3.1. Patient Characteristics
52 stage IV NSCLC patients were ultimately analyzed (Table 1). 63% had an adenocarcinoma histology and 85% were current or former smokers. 36 (69%) patients were treated with nivolumab, 14 (27%) with pembroluzimab, and 2 (4%) with atezoluzimab. 25 patients (48%) had a clinical response (partial response or stable disease) with 27 (52%) patients with progressive disease (non-responders). Response rates were not statistically different by histology, gender, age at diagnosis, age at treatment, pack years, or types of ICB treatment (all p>0.05, Table 1). The median PFS was 1.7 months in non-responders and 25 months in responders. The median OS was 6.0 months in non-responders and 25 months in responders.
Table 1.
Clinical Data Table
| Total | Non-responder | Responder | P-value | |
|---|---|---|---|---|
| 52 | 27 (52%) | 25(48%) | ||
| Immunotherapy Treatment | ||||
| Nivolumab | 36 (69%) | 19 (70%) | 17 (68%) | 0.98 |
| Pembrolizumab | 14 (27%) | 7 (26%) | 7 (28%) | |
| Atezolizumab | 2 (4%) | 1 (4%) | 1 (4%) | |
| Histology | ||||
| Adenocarcinoma | 33 (63%) | 17 (63%) | 16 (64%) | 0.94 |
| Squamous | 19 (37%) | 10 (37%) | 9 (36%) | |
| Driver Mutation* | ||||
| Yes | 9 (17%) | 6 (22%) | 3 (12%) | 0.47 |
| Gender | ||||
| Male | 23 (44%) | 12 (44%) | 11 (44%) | 0.97 |
| Female | 29 (56%) | 15 (56%) | 14 (56%) | |
| Age at Diagnosis | ||||
| Median | 62 | 62 | 63 | 0.59 |
| Age at ICB Initiation | ||||
| Median | 64 | 64 | 65 | 0.79 |
| Smoking History | ||||
| Former/Current | 44 (85%) | 23 (85%) | 21 (84%) | 0.91 |
| Never | 8 (15% | 4 (15%) | 4 (16%) | |
| Smoking (Pack Years) | ||||
| Median | 20 | 25 | 20 | 0.77 |
| PFS (mo) | ||||
| Events | 42 | 27 | 15 | <0.001 |
| Median | 3.3 | 1.7 | 25 | |
| OS (mo) | ||||
| Events | 41 | 25 | 16 | <0.001 |
| Median | 9.6 | 6.0 | 25 | |
Age at ICB: Age at time of starting immunotherapy; PFS (mo): Progression-free Survival in months; OS (mo): Overall survival in months
Driver mutations were defined as actionable EGFR mutations or EML4-ALK fusions
Footnote: Fisher’s exact test for categorical variables, t-test or median test for continuous variables, log-rank test for PFS and OS.
3.2. Gene Signatures and Response to ICB
PD-L1 gene expression levels were not significantly different between responder and non-responders (Supplemental Figure 1).
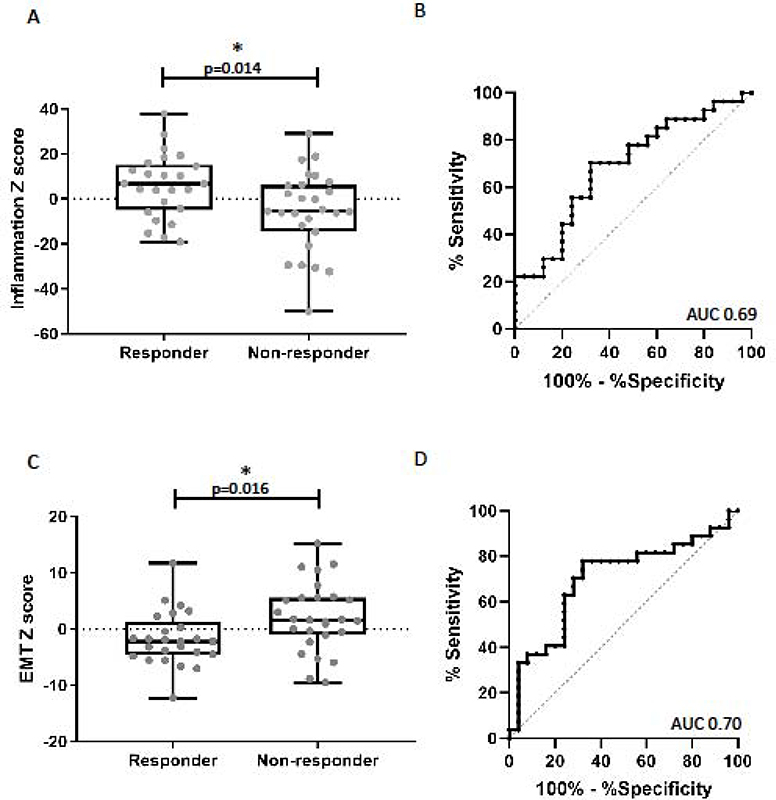
The inflammation signature scores were significantly higher in responders compared to non-responders. (Figure 1A, +6.0±2.9 vs −5.5±3.4, t test p=0.014). A ROC curve evaluating the inflammatory score had an AUC of 0.69 (Figure 1B, p =0.011). To assess the effects of tumor infiltration with macrophages and T cells, we examined the expression levels of a set of established macrophage genes (CD68, CD14, CD163, and CSF1R) and T cell genes (CD4, CD8A, and CD8B) and found no significant differences when comparing responder and non-responder groups (Supplemental Figure 2).
Figure 1. Analysis of the inflammatory and EMT signatures and response to checkpoint blockade.
(A) Comparison of the log2 z-scores of the 27-gene inflammatory gene signature between responders and non-responders. The mean inflammatory score was significantly higher in the responders compared to the non-responder group (6.0 vs. −5.5, p=0.014). (B) Receiver Operator Characteristics curve (ROC curve) utilizing the inflammatory signature to predict response to checkpoint blockade, AUC 0.69 (p=0.011). (C) Comparison of the log2 z-scores of the EMT signature between responders and non-responders. The mean EMT score was significantly lower (more epithelial) in the responders compared to non-responders (−1.7 vs. 2.1, p=0.016). (B) ROC curve utilizing the EMT signature to predict response to checkpoint blockade, AUC 0.70 (p=0.01).
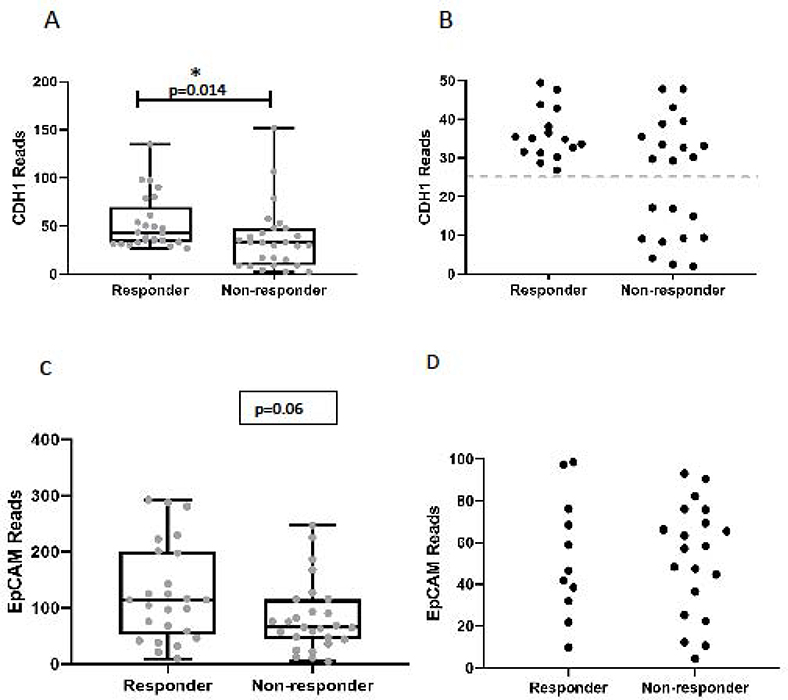
The EMT signature scores were significantly lower (more epithelial) in the responder cohort (average score= −1.7±1.0) compared to non-responders (average score = +2.1±1.2) (Figure 1C; t test p=0.016). The area under the ROC curve was 0.70 (p=0.01) (Figure 1D). Similar to previous reports in melanoma,15,31 (the epithelial marker E-cadherin (CDH1 was significantly higher in the responder group compared to the non-responders (Figure 2A; average reads: 53.0 ±5.6 vs 36.4 ±6.4, p=0.014). Interestingly, the non-responder group included a number of patients with very low levels of E-cadherin expression that had no overlap with the responding patients (Figure 2B). Analysis of EpCAM expression between responders and non-responders showed a near statistically significant difference (p=0.06) (Figure 2C), however, we did not observe a difference in the number of patients with very low expression as we did with E-cadherin (Figure 2D).
Figure 2. Analysis of E-cadherin expression levels and response to checkpoint blockade.
(A) Comparison of CDH1 gene expression levels in responders and non-responders. The mean CDH1 expression levels were significantly higher in the responder group compared to non-responders (53.0 ±5.6 vs 36.4 ±6.4, p=0.014). (B) Closer view of low-level CDH1 expression in responders and non-responders demonstrating enrichment of very low levels of CDH1 expression in the non-responder group. (C) Comparison of EpCAM gene expression levels in responders and non-responders. The mean EpCAM expression levels in responders compared to non-responders (125.8 vs 82.8, p=0.06). (D) Closer view of low-level EpCAM expression showing similar values between responder and non-responder groups.
We also assessed previously published gene signatures that have been associated with primary resistance to ICB. We did not observe a significant difference in the IPRES scores (p=0.1) (Supplemental Figure 3A), the chronic IFN scores (p=0.63) (Supplemental 3B), or the TGF-β signature scores (Supplemental Figure 4) between responders and non-responders.
3.3. Relationship of Inflammation and EMT Gene Scores
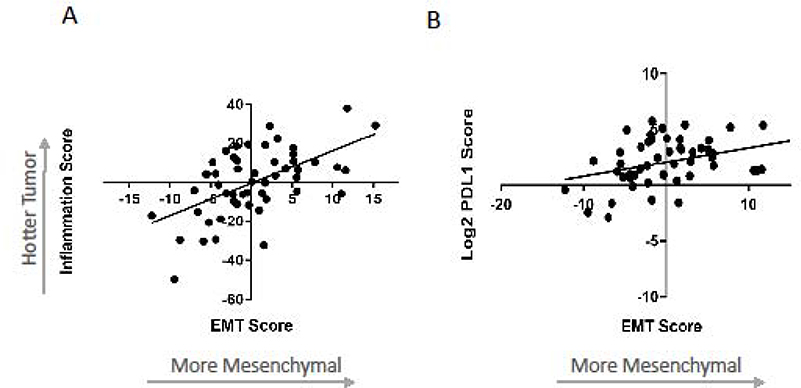
Because both the inflammatory and EMT gene signatures were significantly associated with response rates, we evaluated their relationship and noted a positive correlation between the two signatures (r2=0.32, p<0.0001) (Figure 3A). This relationship signifies that the more mesenchymal tumors (most positive EMT scores) tended to be the “hottest” (most inflammatory). Of interest, we noted (as has been previously reported),32 a significant positive correlation (r2=0.13, p=0.009) between PD-L1 expression levels and EMT scores, with the more mesenchymal tumors showing higher PD-L1 gene expression levels (Figure 3B).
Figure 3. Relationship between the EMT signature, Inflammatory signature and PD-L1 gene expression levels.
(A) Correlation between the log2 z-scores of the Inflammatory and EMT gene signatures (r2=0.32, p=0.0001). (B) Correlation between the log2 z-scores of the EMT gene signature and PD-L1 gene expression levels (r2=0.13, p=0.009).
3.4. Combined EMT and Inflammation Scores
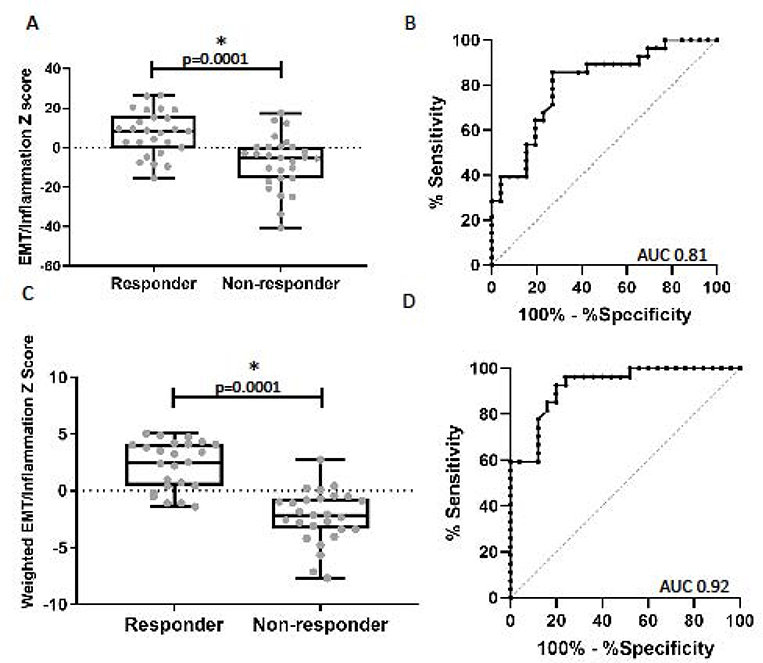
Because the Inflammation and EMT signatures appear to assess different tumor programs, we generated a combined EMT/Inflammation score. The unweighted combination score was significantly higher in responders compared to non-responders (+7.7±2.3 vs −7.7±2.6) (p<0.0001) (Figure 4A). The weighted combination score was also significantly higher in responders compared to non-responders (2.2±0.4 vs −2.3±0.4) (p<0.0001) Figure 4C). A ROC analysis evaluating the ability of the combined scores to predict response had an AUC of 0.81 (p<0.001) for the unweighted score (Figure 4B) and AUC of 0.92 (p<0.001) for the weighted scores (Figure 4D). This value was higher than the AUCs using either the Inflammatory or EMT score alone (p=0.0007 and p=0.0051, respectively). Using the median value −0.4 for the weighted score, we could correctly classify 83% of patients’ response status with sensitivity of 84% and specificity of 81%.
Figure 4. Relationship between the combined EMT and Inflammatory gene signatures with response.
(A) Comparison of the log2 z-scores of the combined unweighted EMT/Inflammatory gene signature between responders and non-responders. The mean EMT/Inflammatory score was significantly higher in the responders compared to the non-responder group (7.7 vs. −7.7, p=0.0001). (B) ROC curve utilizing the unweighted EMT/Inflammatory signature to predict response to checkpoint blockade, AUC 0.81 (p=0.001). (C) Comparison of the log2 z-scores of the combined weighted EMT/Inflammatory gene signature between responders and non-responders. The mean EMT/Inflammatory score was significantly higher in the responders compared to the non-responders (2.2 vs. −2.3, p=0.0001). (B) ROC curve utilizing the weighted EMT/Inflammatory signature to predict response to checkpoint blockade, AUC 0.92 (p<0.001).
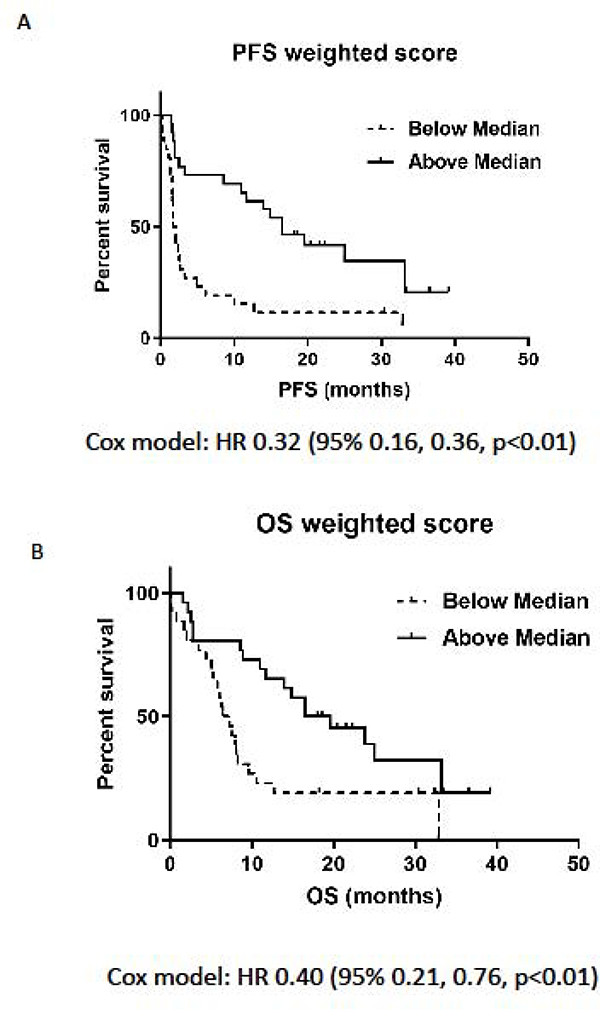
The weighted combined score was also highly predictive of PFS and OS. Kaplan-Meier curves for patients above and below the median showed a good separation for PFS and OS (Figure 5A and 5B, log rank test, all p<0.01) indicating a strong ability to discriminate patients with longer and shorter survival. The hazard ratio (HR) comparing high and low weighted combined scores was 0.32 (95% CI: 0.16, 0.60) for PFS and 0.40 (95% CI: 0.21, 0.76) for OS.
Figure 5. Association between the combined weighted EMT/Inflammatory score and outcome.
Patients were categorized as having an EMT/Inflammatory score above or below the median value for the full cohort and Progression-free Survival (PFS) and overall Survival (OS) were calculated. (A) Kaplan-Meier curves for PFS for patients above or below the median weighted combined score: Cox HR 0.32 (95% 0.16, 0.36, p<0.01). (B) OS for patients above or below the median weighted combined score: Cox HR 0.40 (95% 0.21, 0.76, p<0.01).
4. Discussion
We found that two distinct genomic signatures in lung cancer tumor samples obtained before the initiation of ICB can predict response to therapy. Higher scores of a 27 gene inflammation signature were associated with better responses in lung cancer, as has been shown in other tumors.14 Although the calculated IPRES scores, chronic interferon-gamma stimulation scores, and expression of TGF-β related genes failed to predict ICB responsiveness, the signature that specifically reflected the epithelial vs mesenchymal transition characteristics of the tumor was significantly associated with response to ICB. As predicted in preclinical models,21,22 patients whose tumors were more mesenchymal had poorer responses and shorter PFS and OS than those with more epithelial tumors. Importantly, the EMT score was significantly positively correlated with the inflammation score, and a combined score predicted response with higher accuracy than either score alone suggesting these features drive independent programs within the tumor microenvironment. To our knowledge, the weighted EMT/Inflammation score is more predictive of response (an AUC of 0.92) and survival than any previously published score.16,18
Our findings associating the mesenchymal phenotype with a poor response to ICB in lung cancer are novel, but consistent with previous studies in humans, as well as previous preclinical work, showing that forced mesenchymal transformation can lead to immunosuppression.21,33 A proteomic analysis in a small cohort of melanoma patients (n=8) identified that mesenchymal protein features were associated with lack of response to ICB.31 Wang et al. found a higher gene score associated with stromal-related gene expression in a subgroup of patients with metastatic urothelial cancers who responded to nivolumab.20 Chakravarthy et al. showed that a set of TGF-β associated extracellular matrix genes (many of which would qualify as “mesenchymal” ) was linked to a worse prognosis in two out of three published data sets of PD-1 blocked patients (two melanoma and one bladder).34
Disagreement exists in the literature about the relationship of inflammatory genes to the mesenchymal phenotype. When examining TCGA data, Chae et al. found that a more mesenchymal signature was associated with lower T cell gene expression in NSCLC.35 In contrast, our data (Figure 3A) showing that tumors with higher inflammation scores had higher (more mesenchymal) EMT scores is similar to reports from Lou et al.29 and Chen et al. 32
An interesting question raised by our findings is why mesenchymal differentiation was associated with decreased response to checkpoint blockade? One possibility is that EMT might simply reflect high levels of the immunosuppressive cytokine TGF-β in the tumor; however, we saw no genomic evidence of increased TGF-β related genes. We feel a more likely possibility is that the biological process of mesenchymal transition (which results in changes such as loss of HLA Class 1 antigen, upregulation of PD-L1, 32,36 downregulation of tumor antigens, decreased expression of immunoproteosome subunits,36 stimulation of autophagy,37 and a well- recognized general resistance to any type of cell killing28 makes cancer cells more resistant to T cell killing38, as has been shown in a number of in vitro studies.37,39,40 Our data also suggest that expression levels of E-cadherin might play a role. Although loss of both EPCAM and E-cadherin (classic epithelial to epithelial adhesion molecules) are hallmarks of mesenchymal transformation,41 only E-cadherin (in contrast to EPCAM) serves as the ligand for the adhesion molecule CD103 which is highly expressed on tissue resident memory CD8 T cells. A higher density of tissue resident memory cells in the tumor microenvironment has been associated with improved outcomes in lung cancer42 and other tumor types.43 Mechanistically, the binding of CD103 to E-cadherin on target cells provides costimulatory signals and promotes effector T-cell function.44 These findings suggest that the downregulation of E-cadherin may dampen the CD103-mediated immune attack thus serving as a potential mechanism of resistance to ICB. Intriguingly, we observed significantly higher levels of E-cadherin in the responder group compared to the non-responders. In addition, we observed that very low levels of E-cadherin expression were only found in the non-responders (Figure 2), suggesting that loss of E-cadherin might be a good predictor of non-response to checkpoint blockade.
An interesting implication of our study is that if EMT is a cause of resistance to anti-PD1 therapy, a possible therapeutic approach might be to first use an EMT-reversing agent and then follow this with checkpoint inhibition.45 New drugs are being developed to reverse EMT46 including inhibitors of the tyrosine kinase AXL.47,48 It might also be possible to use drugs developed for other pathways that also affect EMT, such as ERK1–2 inhibitors49 or the multi-kinase inhibitor drug nintedanib.50
The strengths and weaknesses of this study should be considered. As strengths, we used samples from actual clinical practice rather than a formalized clinical trial. Our findings appear to be robust, in that we were able to obtain good data from small biopsy specimens (transbronchial biopsies and needle biopsies) that were not macrodissected and thus contained variable amounts of non-tumor tissue. We were also able to obtain highly congruent results using different analytic approaches (Nanostring®, HTGseq®, data not shown). This study also has limitations. Our data set was only moderate in size (52 patients) and derived from only one center. Thus, replication of these findings in other patient populations is needed. This study exclusively analyzed relapsed, second-line NSCLC patients who primarily received two different anti-PD1 inhibitor antibodies. Given the changing clinical standards of care, a goal for future studies will be to examine the value of these signatures in front-line patients receiving checkpoint antibodies alone or in combination with chemotherapy. The other major limitations to our study are that we did not consistently have PD-L1 immunohistochemical staining or measurements of TMB; our patients were treated in the second-line setting a number of years ago when neither of these tests were routinely performed. Although mRNA and protein levels do not always correlate perfectly, we did analyze the expression of the PD-L1 gene and found it did not correlate with response.
There are a number of possible extensions to our signatures that could make our findings more clinically applicable. This includes further optimization of weighting specific genes within each signature. As an alternative to genomic analysis, it may also be possible to obtain prognostic information about the “EMT status” of tumors by adding additional immunohistochemical stains to standard PD-L1 staining. Finally, one long-range goal would be to develop blood tests that might reflect the EMT status of tumors.
5. CONCLUSIONS
In summary, we have found that two genomic signatures, reflecting aspects of tumor inflammation and the EMT status of the tumor, when combined, predict response to immune checkpoint inhibitors in the second-line treatment of advanced NSCLC patients with very high accuracy. A higher combined EMT/Inflammatory score was independently associated with improved progression-free and overall survival. Application of these signatures to this population could provide a needed tool to direct checkpoint therapy to those patients most likely to benefit. Success in predicting responses in front-line therapy needs to be tested, but could extend this approach to a larger number of patients.
Supplementary Material
Highlights.
Evaluation of gene signatures to predict response to immunotherapy in NSCLC.
An EMT/Inflammation score was highly predictive of response and survival to ICB.
Identifies potential role of EMT in primary resistance to immunotherapy in NSCLC.
Acknowledgments
FUNDING/SUPPORT
This work was supported in part by the Abramson Cancer Center Lung Cancer Translational Center of Excellence (WTH, CD, CD, SJ, CJL, SMA), by K08 CA234335-01 from the National Cancer Institute (NIH), LUNGevity Foundation (JCT) and by Janssen Research and Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ROLE OF THE FUNDER/SPONSOR
Janssen Research and Development provided support for RNA sequencing and assisted in the bioinformatics analysis, as well as preparation and approval of the final manuscript. Janssen had no role in the design and conduct of the study.
CONFLICT OF INTEREST DISCLOSURES
Dr. Albelda has received research funding from Janssen Research and Development.
Dr. Thompson reported a consulting or advisory role for Oncocyte Corporation, Guardant Health, AstraZeneca, and Honeywell.
Dr. Langer reported honoraria from Bristol-Myers Squibb, Genentech/Roche, and Lilly/ImClone; a consulting or advisory role with Genentech/Roche, Lilly/ImClone, Merck Sharp & Dohme, Abbott Biotherapeutics, Inc, Bayer/Onyx, Clariant, Clovis Oncology, Celgene Corporation, Cancer Support Community, Bristol-Myers Squibb, Ariad Pharmaceuticals, Inc, Takeda PharmaceuticalCompany Ltd, and AstraZeneca; institutional research funding from Merck Sharp & Dohme, Advantagene, Inc, Clovis Oncology, Celgene Corporation, Inovio Pharmaceuticals, Ariad Pharmaceuticals, Inc, GlaxoSmithKline, Genentech/Roche, and Stemcentrx, Inc; and other relationships with Eli Lilly and Company, Amgen, Inc, Peregrine Pharmaceuticals, Inc, and Synta Pharmaceuticals, Inc.
Rajpurohit Yashoda, Vinod Krishna, Denis Smirnov, Raluca Verona, Mathew V. Lorenzi work for Janssen Research and Development.
No other disclosures were reported.
Footnotes
Appendix A. Supplementary data
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brahmer J, Reckamp KL, Baas P, et al. : Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 373:123–35, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbst RS, Baas P, Kim DW, et al. : Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387:1540–50, 2016 [DOI] [PubMed] [Google Scholar]
- 3.Fehrenbacher L, Spira A, Ballinger M, et al. : Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387:1837–46, 2016 [DOI] [PubMed] [Google Scholar]
- 4.Rittmeyer A, Barlesi F, Waterkamp D, et al. : Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 389:255–265, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmer JR, Lacchetti C, Schneider BJ, et al. : Management of Immune-Related Adverse Events in Patients Treated With Immune Checkpoint Inhibitor Therapy: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 36:1714–1768, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matter-Walstra K, Schwenkglenks M, Aebi S, et al. : A Cost-Effectiveness Analysis of Nivolumab versus Docetaxel for Advanced Nonsquamous NSCLC Including PD-L1 Testing. J Thorac Oncol 11:1846–1855, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Hansen AR, Siu LL: PD-L1 Testing in Cancer: Challenges in Companion Diagnostic Development. JAMA Oncol 2:15–6, 2016 [DOI] [PubMed] [Google Scholar]
- 8.Langer CJ, Gadgeel SM, Borghaei H, et al. : Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol 17:1497–1508, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tumeh PC, Harview CL, Yearley JH, et al. : PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515:568–71, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taube JM, Klein A, Brahmer JR, et al. : Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 20:5064–74, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thommen DS, Koelzer VH, Herzig P, et al. : A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 24:994–1004, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellmann MD, Nathanson T, Rizvi H, et al. : Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 33:843–852 e4, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. : Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 378:2093–2104, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayers M, Lunceford J, Nebozhyn M, et al. : IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127:2930–2940, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugo W, Zaretsky JM, Sun L, et al. : Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 165:35–44, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristescu R, Mogg R, Ayers M, et al. : Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prat A, Navarro A, Pare L, et al. : Immune-Related Gene Expression Profiling After PD-1 Blockade in Non-Small Cell Lung Carcinoma, Head and Neck Squamous Cell Carcinoma, and Melanoma. Cancer Res 77:3540–3550, 2017 [DOI] [PubMed] [Google Scholar]
- 18.Ott PA, Bang YJ, Piha-Paul SA, et al. : T-Cell-Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated With Pembrolizumab Across 20 Cancers: KEYNOTE-028. J Clin Oncol:JCO2018782276, 2018 [DOI] [PubMed] [Google Scholar]
- 19.Mariathasan S, Turley SJ, Nickles D, et al. : TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554:544–548, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Saci A, Szabo PM, et al. : EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun 9:3503, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dongre A, Weinberg RA: New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol, 2018 [DOI] [PubMed] [Google Scholar]
- 22.Terry S, Savagner P, Ortiz-Cuaran S, et al. : New insights into the role of EMT in tumor immune escape. Mol Oncol 11:824–846, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Elm E, Altman DG, Egger M, et al. : The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Epidemiology 18:800–4, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Li W, Turner A, Aggarwal P, et al. : Comprehensive evaluation of AmpliSeq transcriptome, a novel targeted whole transcriptome RNA sequencing methodology for global gene expression analysis. BMC Genomics 16:1069, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weichselbaum RR, Ishwaran H, Yoon T, et al. : An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A 105:18490–5, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benci JL, Xu B, Qiu Y, et al. : Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 167:1540–1554 e12, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mak MP, Tong P, Diao L, et al. : A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res 22:609–20, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byers LA, Diao LX, Wang J, et al. : An Epithelial-Mesenchymal Transition Gene Signature Predicts Resistance to EGFR and PI3K Inhibitors and Identifies Axl as a Therapeutic Target for Overcoming EGFR Inhibitor Resistance. Clinical Cancer Research 19:279–290, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lou Y, Diao L, Cuentas ER, et al. : Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin Cancer Res 22:3630–42, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLong ER, DeLong DM, Clarke-Pearson DL: Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–45, 1988 [PubMed] [Google Scholar]
- 31.Shields BD, Mahmoud F, Taylor EM, et al. : Indicators of responsiveness to immune checkpoint inhibitors. Sci Rep 7:807, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Gibbons DL, Goswami S, et al. : Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 5:5241, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudo-Saito C, Shirako H, Takeuchi T, et al. : Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 15:195–206, 2009 [DOI] [PubMed] [Google Scholar]
- 34.Chakravarthy A, Khan L, Bensler NP, et al. : TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat Commun 9:4692, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chae YK, Chang S, Ko T, et al. : Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC). Sci Rep 8:2918, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tripathi SC, Peters HL, Taguchi A, et al. : Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc Natl Acad Sci U S A 113:E1555–64, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akalay I, Janji B, Hasmim M, et al. : EMT impairs breast carcinoma cell susceptibility to CTL-mediated lysis through autophagy induction. Autophagy 9:1104–6, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song KA, Niederst MJ, Lochmann TL, et al. : Epithelial-to-Mesenchymal Transition Antagonizes Response to Targeted Therapies in Lung Cancer by Suppressing BIM. Clin Cancer Res 24:197–208, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woods K, Pasam A, Jayachandran A, et al. : Effects of epithelial to mesenchymal transition on T cell targeting of melanoma cells. Front Oncol 4:367, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricciardi M, Zanotto M, Malpeli G, et al. : Epithelial-to-mesenchymal transition (EMT) induced by inflammatory priming elicits mesenchymal stromal cell-like immune-modulatory properties in cancer cells. Br J Cancer 112:1067–75, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Roy F: Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer 14:121–34, 2014 [DOI] [PubMed] [Google Scholar]
- 42.Ganesan AP, Clarke J, Wood O, et al. : Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol 18:940–950, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duhen T, Duhen R, Montler R, et al. : Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat Commun 9:2724, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Floc’h A, Jalil A, Franciszkiewicz K, et al. : Minimal engagement of CD103 on cytotoxic T lymphocytes with an E-cadherin-Fc molecule triggers lytic granule polarization via a phospholipase Cgamma-dependent pathway. Cancer Res 71:328–38, 2011 [DOI] [PubMed] [Google Scholar]
- 45.Hamilton DH, Huang B, Fernando RI, et al. : WEE1 inhibition alleviates resistance to immune attack of tumor cells undergoing epithelial-mesenchymal transition. Cancer Res 74:2510–9, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis FM, Stewart TA, Thompson EW, et al. : Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharmacol Sci 35:479–88, 2014 [DOI] [PubMed] [Google Scholar]
- 47.Ying X, Chen J, Huang X, et al. : Effect of AXL on the epithelial-to-mesenchymal transition in non-small cell lung cancer. Exp Ther Med 14:785–790, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Z, Li Y, Zhang D, et al. : Axl inhibition induces the antitumor immune response which can be further potentiated by PD-1 blockade in the mouse cancer models. Oncotarget 8:89761–89774, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buonato JM, Lazzara MJ: ERK1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res 74:309–19, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang RY, Kuay KT, Tan TZ, et al. : Functional relevance of a six mesenchymal gene signature in epithelial-mesenchymal transition (EMT) reversal by the triple angiokinase inhibitor, nintedanib (BIBF1120). Oncotarget 6:22098–113, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.