SUMMARY
CodY is a global transcriptional regulator that controls, directly or indirectly, expression of dozens of genes and operons in Listeria monocytogenes. We used in vitro DNA affinity purification combined with massively parallel sequencing (IDAP-Seq) to identify genome-wide L. monocytogenes chromosomal DNA regions that CodY binds in vitro. The total number of CodY-binding regions exceeded 2,000, but they varied significantly in their strengths of binding at different CodY concentrations. The 388 strongest CodY-binding regions were chosen for further analysis. A strand-specific analysis of the data allowed pinpointing CodY-binding sites at close to single-nucleotide resolution. Gel shift and DNase I footprinting assays confirmed the presence and locations of several CodY-binding sites. Surprisingly, most of the sites were located within genes’ coding regions. The binding site within the beginning of the coding sequence of the prfA gene, which encodes the master regulator of virulence genes, has been previously implicated in regulation of prfA, but this site was weaker in vitro than hundreds of other sites. The L. monocytogenes CodY protein was functionally similar to Bacillus subtilis CodY when expressed in B. subtilis cells. Based on the sequences of the CodY-binding sites, a model of CodY interaction with DNA is proposed.
Keywords: Listeria monocytogenes, CodY-binding sites, gene expression, IDAP-Seq
Graphical Abstract
The CodY protein is a global transcriptional regulator of metabolic and virulence genes in low G+C Gram-positive bacteria, including pathogenic Listeria monocytogenes. Hundreds of CodY-binding sites were found in the L. monocytogenes genome at near single-nucleotide resolution, most of them within genes’ coding sequences. In many cases, CodY appears to bind to two overlapping 15-nt motifs.
INTRODUCTION
Listeria monocytogenes is a low G+C Gram-positive foodborne pathogen that causes listeriosis, an infection with a 30% mortality rate in susceptible humans. It mostly affects pregnant women, newborns, the elderly and the immunocompromised population (Schlech, 2019). L. monocytogenes infection has been an important model system for the study of host-pathogen interactions and mechanisms of intracellular parasitism (Radoshevich and Cossart, 2018; D’Orazio, 2019; Johansson and Freitag, 2019). L. monocytogenes is able to grow intracellularly in a variety of mammalian cells, but it can also adapt to saprophytic growth on decaying soil vegetation (Weis and Seeliger, 1975). It is therefore interesting to examine how this bacterium senses the environment to regulate expression of its genes.
CodY, first identified in Bacillus subtilis (Slack et al., 1995), is a DNA-binding protein and a global regulator of metabolism and virulence genes in nearly all low G+C Gram-positive pathogens, including Bacillus, Clostridium, Staphylococcus, Streptococcus, Enterococcus, and Listeria spp. (Geiger and Wolz, 2014; Richardson et al., 2015; Brinsmade, 2017; Li et al., 2017; Mlynek et al., 2018; Colomer-Winter et al., 2019; Daou et al., 2019). The DNA-binding affinity of CodY from B. subtilis and most other species is increased by interaction with two types of ligands, the branched-chain amino acids (isoleucine, leucine, and valine) and GTP (Sonenshein, 2007; Richardson et al., 2015; Han et al., 2016). The response of CodY to these effectors ties its activity to the ability of the cell to make RNA, protein, and branched-chain fatty acids, the primary membrane fatty acids in CodY-encoding species. CodY is most active when cells are in a relatively rich environment; its inactivation occurs under conditions of nutritional limitation and frequently correlates with the onset of stationary phase of growth.
In L. monocytogenes, CodY has been shown to regulate expression of virulence genes and affect virulence in animal models. In one study, CodY-mediated regulation was found to contribute to the virulence defect of a relA mutant of L. monocytogenes strain EGD-e (Bennett et al., 2007). That is, a relA codY double mutant was much more virulent than a relA single mutant, but the mechanism of this negative CodY-mediated effect was not elucidated (Bennett et al., 2007). In strain 10403S, however, CodY was shown to be a positive regulator of the PrfA regulon, which includes the most important virulence genes of L. monocytogenes, and a codY null mutant strain exhibited a virulence defect (Lobel et al., 2015). The exact mechanism of CodY action in the latter case is also uncertain since the positive regulation appears to occur through binding within the beginning of the prfA coding sequence and only under conditions in which CodY is generally found to be poorly active, i.e., in a medium containing a low concentration of branched-chain amino acids (Lobel et al., 2015).
Genome-wide targets of L. monocytogenes CodY have been detected previously in DNA microarray, RNA-Seq, and ChIP-Seq experiments (Bennett et al., 2007; Lobel and Herskovits, 2016). However, expression analyses do not distinguish between direct and indirect targets of regulation. Although ChIP-Seq experiments can reveal extended regions of protein binding, they do not pinpoint binding sites, i.e., exact sequences that directly contribute to protein binding, especially when several binding sites are located in close vicinity of each other. In addition, regulation by CodY and binding of CodY to DNA can be masked by the activities of other gene-specific or global regulators, some of which are themselves under CodY control (Belitsky et al., 2015; Barbieri et al., 2016). Regulators that compete with CodY for binding may be active under laboratory conditions but less active during infection allowing CodY-mediated regulation to dominate.
To discover the full range of genes that can be regulated directly by CodY, it is important to identify the entire set of genome-wide CodY-binding sites in vitro. We have previously developed a novel method, in vitro DNA affinity purification coupled with massively parallel sequencing (IDAP-Seq), that allows genome-wide identification of protein-binding regions in vitro using purified DNA-binding proteins in the absence of potential interference by other proteins. This method was successfully applied to analyze CodY binding to Staphylococcus aureus and Clostridiodes (formerly Clostridium) difficile genomes (Dineen et al., 2010; Majerczyk et al., 2010). More recently, a modification of this method was used to identify B. subtilis and Bacillus anthracis CodY-binding sites at near single-nucleotide resolution (Belitsky and Sonenshein, 2013; Chateau et al., 2013). This approach has been recently used to analyze the binding sites of several other proteins (Smith and Grossman, 2015; Tran et al., 2018). The resulting data can be used to compare the sequences and locations of sites associated with strong and weak regions of binding, to identify common motifs and possible mechanisms of regulation, and to predict the conditions under which a particular gene would be regulated. Although detection of a binding site in vitro does not constitute evidence for regulation, the presence of a site identifies candidate targets for direct regulation and its absence makes direct regulation very unlikely.
In the present work, we have identified, at near single-nucleotide resolution, virtually all sites within the L. monocytogenes chromosome to which CodY is able to bind in vitro. By varying the concentration of CodY we were also able to rank the CodY-binding regions with respect to their relative strengths of binding. Using gel shift and DNase I footprinting assays, we confirmed the presence and locations of binding sites for a number of L. monocytogenes genes. We also compared the properties of the L. monocytogenes and B. subtilis CodY proteins in vitro and in vivo and proposed a model of CodY interaction with DNA.
RESULTS
Genome-wide identification of L. monocytogenes CodY-binding regions in vitro
We have used the IDAP-Seq approach (Belitsky and Sonenshein, 2013)(see Experimental Procedures) to identify regions of the L. monocytogenes strain 10403S chromosome that interact in vitro with L. monocytogenes CodY. Purified His-tagged CodY was incubated with randomly fragmented (~150–250 bp), adaptor-ligated chromosomal DNA in the presence of a 10 mM mixture of isoleucine, leucine, and valine (ILV), known positive effector molecules of CodY. The CodY-DNA complexes were selected using immobilized metal-ion affinity purification; the CodY-binding fragments were released from the complexes after treatment with proteinase K, amplified by PCR using adaptor-specific primers, and subjected to sequencing en masse.
In order to identify all binding regions and improve the resolution power of our experiments to determine the relative CodY-binding strengths of different DNA regions, six different CodY concentrations ranging from 0.32 nM to 1 μM were used and four successive rounds of IDAP were performed at each CodY concentration using the PCR-amplified output of the previous round as the input for the next round, resulting in the total of 24 different assays. To be considered as a binding region, a sequence needed to have at least 3-fold higher-than-average coverage by sequencing reads. Then, the total number of reads was determined for each binding region, normalized per the number of reads in each sample, and an enrichment factor was determined by dividing the number of normalized reads in each region under any particular condition by the number of reads for the same region in the samples that were not incubated with CodY. We assumed that the relative binding strength of a region correlates with its enrichment factor.
The list of 2,311 detected CodY-binding regions with their enrichment factors achieved under each of the conditions used in our IDAP experiments is presented in Dataset S1A (Dataset S1B tabulates the relative numbers of counted reads for each region under every condition used). The CodY-binding regions formed a continuum of regions of various strengths. Similar results reflecting promiscuous, although specific, binding by CodY were previously obtained during analysis of CodY-binding regions in the B. subtilis chromosome (Belitsky and Sonenshein, 2013). The lengths of the L. monocytogenes CodY-binding regions ranged from 95 to 856 bp, but only 31 regions were longer than 400 bp and only 6 regions were longer than 500 bp (the expected length of the strongest binding regions was between ~300 and ~500 bp, i.e., twice the length of the fragments used for binding because each binding site was surrounded by 150–250 bp sequences from the (+) strand on one side and the (−) strand on the other side; weaker regions were shorter due to lower coverage, especially at the ends of binding regions).
Only a limited number of high-affinity binding regions should be recovered if very low protein concentrations are used in an IDAP assay. Indeed, only 16 CodY-binding regions enriched by ≥10-fold were recovered at 0.32 nM CodY after 4 IDAP cycles; the highest enrichment was 2,750-fold (Dataset S1A). By contrast, more than 200 binding regions with the same enrichment threshold were identified at 200 nM or 1 μM CodY, i.e., the concentrations that are likely higher than the apparent dissociation constant (KD) for physiologically relevant binding sites (Dataset S1A).
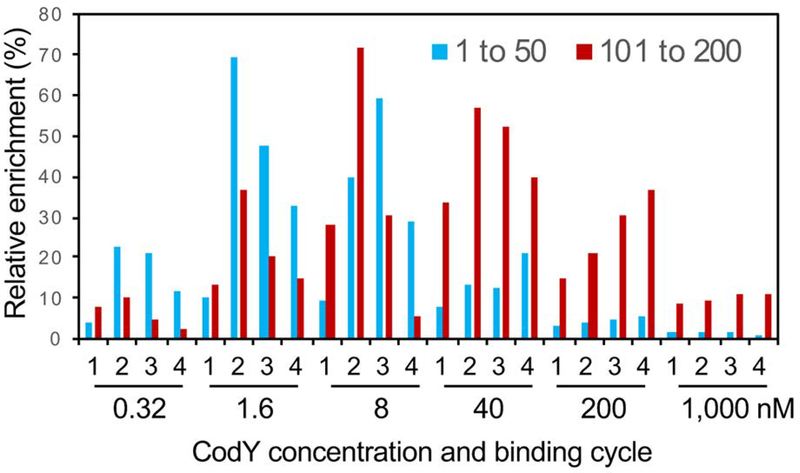
Since many of the weaker regions detected in vitro may not have physiological significance, we arbitrarily selected for further analysis the 388 strongest CodY-binding regions, as defined by the maximal enrichment factor of ≥10 in IDAP-Seq experiments performed at CodY concentrations of 0.32, 1.6, 8, and 40 nM, which we believe to be closer to physiological conditions of binding. Furthermore, results from only the first two rounds of IDAP at each protein concentration, i.e., from 8 different assays, were used for the calculation of the maximal strength to reduce a bias towards recovering only the strongest sites after multiple cycles of binding. The relative strengths and the corresponding rankings of many regions varied significantly in experiments performed at different CodY/DNA ratios and depended on the reiteration cycle used (Dataset S2) similar to the situation described previously for B. subtilis (Belitsky and Sonenshein, 2013). This is likely to be partly due to the known cooperativity of CodY binding, the extent of which varies for individual genes and is dependent on the protein/DNA ratio. As expected, the highest enrichment of stronger binding regions was in general observed at lower CodY concentrations, with a sharp decrease at higher concentrations of CodY when these sites had to compete with many weaker sites (Fig. 1). In contrast, the highest enrichment of weaker binding regions was observed at higher CodY concentrations, and the differences in enrichment at different CodY concentrations were less pronounced (Fig. 1). Importantly, the changes in relative strengths of CodY-binding regions observed in IDAP-Seq experiments at different CodY concentrations are likely to correlate with changes in relative expression levels of the target genes under conditions of varying CodY activities in vivo.
Fig. 1.
Relative enrichments of CodY-binding regions under different binding conditions. The relative enrichments were compared for two groups of CodY-binding regions comprising the 50 strongest regions (enrichment rankings 1–50) or regions with enrichment rankings from 101 to 200. The maximum enrichment for each region under any of the 24 binding conditions was defined as 100%, and the enrichments under other 23 conditions were normalized to this value. The obtained relative enrichments for individual regions of each group were averaged for each binding condition. The 24 binding assays were performed at CodY concentrations of 0.32, 1.6, 8, 40, 200, and 1,000 nM with four successive cycles of binding (labeled as 1–4) at each concentration.
Identification of CodY-binding sites
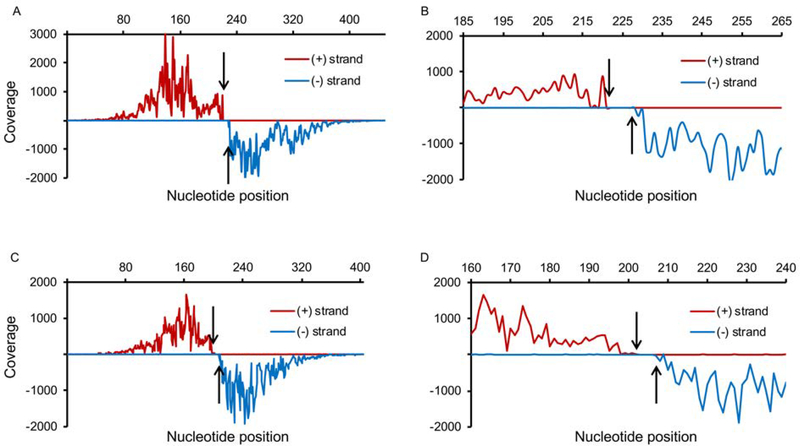
To identify the specific nucleotides required for CodY binding we used an approach described previously (Belitsky and Sonenshein, 2013) to create strand-specific chromosomal coverage maps by counting only the 5’ nucleotide of each sequenced DNA fragment after having defined where within the chromosome the total sequence lies. As shown in Fig. 2 and Fig. S1 and in the previous publication (Belitsky and Sonenshein, 2013), each simple CodY-binding site, reading left-to-right, is surrounded by a region with a high number of sequencing reads at each position on the (+) DNA strand and a non-overlapping sequence with a high number of reads on the (−) strand. The zone of few or no reads on either strand separates the two zones of high reads and corresponds to the sequence that is essential for binding, since fragments lacking even one of these nucleotides would not be able to interact with CodY (Belitsky and Sonenshein, 2013). The sequence corresponding to the gap in coverage on both DNA strands together with its boundary nucleotides on each side is referred to here as the core binding site. In most cases, several additional nucleotides on either side of the core are likely required to form a site maximally competent for binding.
Fig. 2.
Coverage maps of CodY-binding regions using strand-specific counting of 5’ nucleotides of sequencing reads. The 453-bp mfd (A, B) and 405-bp lmaA (C, D) CodY-binding regions were identified after two rounds of purification using 1.6 or 8 nM CodY, respectively. The coverage of the (+) and (−) strands is shown above and below the x-axis, respectively. Arrows indicate a gap in coverage. Non-uniform coverage is likely due to varying position-specific efficiency of DNA shearing during sonication and to non-uniform size of DNA fragments. (A, C) Coverage of the entire binding region. (B, D) Coverage of the 80 bp surrounding the core CodY-binding site.
Manual inspection of CodY-binding regions from the 388-region dataset revealed that 111 regions contained two-to-five CodY-binding sites resulting in a total number of 518 sites in the entire Dataset S2. This is a much more frequent occurrence of closely spaced binding sites than we detected for a similar dataset in B. subtilis (only 30 out of 323 B. subtilis CodY-binding regions contained more than one binding site) and explains the extended length of some binding regions. The ability of IDAP-Seq to fully resolve locations of nearby sites depends on the distance between the sites and the size of the DNA fragments used in the experiment (Belitsky and Sonenshein, 2013). Only one boundary (upstream or downstream) could be determined for most closely spaced sites. The region between the two boundaries detected for such sites is characterized by overlapping coverage on both strands instead of a gap in coverage (Fig. S1)(Belitsky and Sonenshein, 2013). Both boundaries could be identified for 359 out of 518 sites (69%). The length of the corresponding core sequences varied from 3 to 105 nt, although 92% of the core sequences were between 3 and 25 nt in length and only 5 core sites were longer than 60 bp (Dataset S2). Given the short length of an average core binding site, the availability of coordinates for just a single boundary does not compromise significantly the ability to locate the binding site. The larger length of a small number of binding sites is not likely to reflect the actual length of these sites but rather a low coverage of the corresponding binding regions that was insufficient to determine confidently the boundaries of the site. Table 1 lists core binding sites of the 15 highest enriched CodY-binding regions in the L. monocytogenes genome.
Table 1.
CodY-binding sites within the 15 highest enriched CodY-binding regions of the L. monocytogenes genomea.
| Region ranking and putative target gene(s) | Alternative gene name(s) | Distanceb (nt) from: |
Site length (nt) | Site locationc with respect to the target gene(s) | Maximal enrichment factor of the binding region | Site detection by ChIP-Seqd | |
|---|---|---|---|---|---|---|---|
| first gene | second gene | ||||||
| 1. LMRG_RS01035 | mfd | −24 | 8 | UPSTREAM | 554.0 | yes | |
| 2. LMRG_RS00580 | lmaA | 282 | 7 | INTERNAL | 335.1 | yes | |
| 3. LMRG_RS07545,LMRG_RS07550 | amtB,glnK | 1110 | −104 | 6 | INTERNAL,UPSTREAM | 302.1 | no |
| 4. LMRG_RS04970,LMRG_RS04975 | 797 | −99 | 9 | INTERNAL,UPSTREAM | 288.0 | yes | |
| 5. LMRG_RS01845,LMRG_RS01850 | 377 | −63 | NDe | INTERNAL,UPSTREAM | 274.6 | yes | |
| 5. LMRG_RS01845,LMRG_RS01850 | 399 | −41 | ND | INTERNAL,UPSTREAM | 274.6 | yes | |
| 6. LMRG_RS13490 | 136 | 6 | INTERNAL | 257.1 | yes | ||
| 7. LMRG_RS11140/LMRG_RS11145 | oppA | −11 | −593 | ND | UPSTREAM/UPSTREAM | 250.1 | no |
| 7. LMRG_RS11140/LMRG_RS11145 | oppA | −149 | −460 | 16 | UPSTREAM/UPSTREAM | 250.1 | no |
| 7. LMRG_RS11140/LMRG_RS11145 | oppA | −179 | −425 | ND | UPSTREAM/UPSTREAM | 250.1 | no |
| 8. LMRG_RS00110,LMRG_RS00115 | 179 | −323 | 10 | INTERNAL,UPSTREAM | 217.5 | no | |
| 8. LMRG_RS00110,LMRG_RS00115 | 390 | −114 | 8 | INTERNAL,UPSTREAM | 217.5 | yes | |
| 9. LMRG_RS10840,LMRG_RS10835 | 1331 | −138 | 7 | INTERNAL,UPSTREAM | 211.0 | yes | |
| 10. LMRG_RS09960 | 796 | ND | INTERNAL | 168.2 | no | ||
| 10. LMRG_RS09960 | 751 | ND | INTERNAL | 168.2 | yes | ||
| 11. LMRG_RS00240 | agrB | −124 | 3 | UPSTREAM | 156.7 | yes | |
| 12. LMRG_RS08950,LMRG_RS08945 | purC,purS | 647 | −78 | 8 | INTERNAL,UPSTREAM | 140.0 | yes |
| 13. LMRG_RS00135 | 110 | 6 | INTERNAL | 116.8 | yes | ||
| 14. LMRG_RS02565/LMRG_RS02570 | −176 | −186 | ND | UPSTREAM/UPSTREAM | 115.1 | no | |
| 14. LMRG_RS02565/LMRG_RS02570 | −276 | −91 | 16 | UPSTREAM/UPSTREAM | 115.1 | yes | |
| 14. LMRG_RS02570 | 86 | 17 | INTERNAL | 115.1 | no | ||
| 15. LMRG_RS02730 | 462 | 3 | INTERNAL | 111.2 | yes | ||
See Dataset S2 for additional information.
Distances from the translation start of putative target genes.
“upstream” indicates the location of a CodY-binding site in the regulatory region of a gene; “upstream/upstream” indicates the location of a CodY-binding site in the regulatory region between two divergent genes; “internal” indicates the location of a CodY-binding site within the coding region of a gene; for some internal sites, nearby adjacent genes are shown as candidate targets for regulation (the gene names are separated by a comma).
Only the strongest IDAP-Seq site was detected for some regions.
ND (not determinable) indicates that the site boundary and the site length could not be determined because of the presence of a second, nearby CodY-binding site.
CodY-binding motifs
The rules of CodY binding to DNA are not fully understood. The 15-nt CodY-binding consensus motif, AATTTTCWGAAAATT (den Hengst et al., 2005; Guedon et al., 2005; Belitsky and Sonenshein, 2008), is relatively well defined, but is rarely found in its entirety. Although the Staphylococcus aureus chromosome has one “perfect” site, the L. monocytogenes chromosome, like that of B. subtilis, does not contain even one sequence with perfect adherence to the canonical 15-nt CodY-binding consensus; instead, it contains 2, 28, and 431 sequences with 1, 2 or 3 mismatches to the consensus motif, respectively; 3,465 sequences contain 4 mismatches.
To analyze further the sequences of individual CodY-binding sites, we created a new dataset consisting of the 518 IDAP-Seq core sequences that were extended by 21 nt at each end to allow for adjacent sequences to be included in the analysis (for the purpose of this analysis, we assumed all sites with one undefined boundary to be 21 nt long before extension). The extended sequences overlap with only 1, 5, and 47 of in silico identified motifs with 1, 2 or 3 mismatches, respectively (53 sites total; 10%; some sites overlap with multiple motifs). An additional 149 overlapping motifs have 4 mismatches to the consensus. Altogether, only 181 of the 518 binding sites (35%) overlap with motifs with ≤5 mismatches with respect to the conventional consensus sequence.
An unbiased search for a common DNA motif on both strands of the dataset of 518 extended CodY-binding sites using the MEME motif-searching algorithm (Bailey and Elkan, 1994) yielded a 14-nt motif (Fig. 3A). The first 13 nt of the MEME motif corresponded well to nucleotides 3 to 15 of the canonical 15-nt CodY-binding motif, although the central nucleotide is not conserved. If we analyzed only one, “given” strand of each of our sequences, a motif with 24 prominent positions (2 to 25) was found (Fig. 3B). These 24 nucleotides perfectly corresponded to two canonical 15-nt motifs overlapping by 6 nt (Fig. 4A). It was previously suggested that such overlapping arrangement is a common feature of CodY binding to DNA (Wray and Fisher, 2011); our genome-wide analysis of S. aureus, Clostridioides difficile, and B. subtilis CodY-binding sites (Dineen et al., 2010; Majerczyk et al., 2010; Belitsky and Sonenshein, 2013) supported this suggestion. We believe that the “given strand only” analysis reveals the overlapping arrangement of 15-nt canonical CodY-binding motifs because it prevents extra flexibility in a number of ways by which the MEME algorithm can align multiple copies of the symmetrical 24 nt motif and disfavors alignment of the strongest 15-nt motifs of the overlapping pair that occurs during analysis of both DNA strands. Though “given strand only” alignment of multiple 24-bp motifs, which contains directly repeated nucleotide sequences, can potentially generate a 42-bp sequence, the conservation of nucleotides at the ends of this sequence is apparently too weak to be detected (Fig. 4B).
Fig. 3.
Motif logos for the L. monocytogenes CodY-binding site. The logos were generated by the MEME function of the MEME suite (Bailey and Elkan, 1994). All 518 CodY core binding sequences of Dataset S2 extended by 21 nt on each end were used for the analysis of the motifs on both DNA strands (A) or only one, given strand (B). The sequence of the canonical 15-nt CodY-binding motif(s) is shown below the logos.
Fig. 4.
24-nt CodY-binding motif. (A) Generation of a 24-bp motif from two overlapping 15-nt motifs. (B) Three possible ways to align 24-nt motifs and the resulting potential consensus 42-nt sequence. W stands for A or T.
The stronger conservation of A and T nucleotides at positions 11–16 in the motif presented in Fig. 3B partly reflects that they correspond to the overlapping sequences of two 15-nt motif, but also suggests an important role for some of these nucleotides, especially those at positions 11 and 16, in CodY binding. High conservation of the same nucleotides can be seen in the CodY-binding motifs from other bacteria (Fig. S2) (Majerczyk et al., 2010; Belitsky and Sonenshein, 2013; Chateau et al., 2013). Interestingly, the two most conserved of these nucleotides are significantly less conserved in chromosomal sequences that resemble CodY-binding motifs but are unable to bind CodY compared to motifs that correspond to actual CodY-binding sites (Fig. S3, see positions 10 and 15).
Validation of IDAP-Seq results
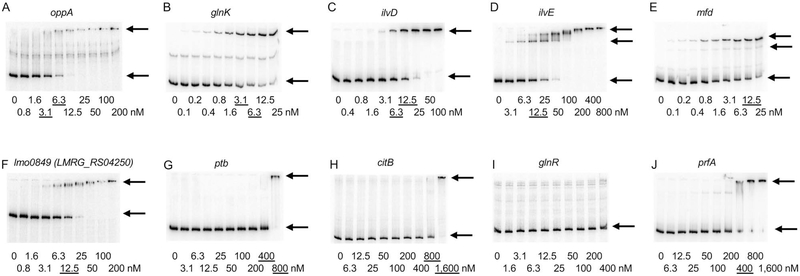
Previously, we have demonstrated a very good correlation between the results of IDAP-Seq and data obtained by conventional DNA-binding assays (Belitsky and Sonenshein, 2013). To verify the value of the IDAP-Seq results obtained in this work, we determined the strength and location of several CodY-binding sites using gel shift and DNase I footprinting experiments performed in the presence of 10 mM ILV. In gel shift assays, we detected CodY binding to the sequences upstream of the oppA (lmo2196, LMRG_RS11140, LMRG_01636) (KD≈5 nM), glnK (lmo1517, LMRG_RS07550, LMRG_01453) (KD≈5 nM), ilvD (lmo1983, LMRG_RS10020, LMRG_01131) (KD≈10 nM), ilvE (lmo0978, LMRG_RS04930, LMRG_02078) (KD≈12.5 nM), mfd (lmo0214, LMRG_RS01035, LMRG_02636) (KD≈12.5 nM), and lmo0849 (LMRG_RS04250, LMRG_02272) (KD≈12.5 nM) genes (Fig. 5A–F). All but one of these genes were found in the IDAP-Seq experiments to have CodY-binding sites in the intergenic regions upstream of their coding sequences; only the glnK site was located at the end of the coding sequence of the upstream gene, amtB (Dataset S2). No binding at CodY concentrations less than 800 nM was detected for the regulatory regions of the ptb (lmo1369, LMRG_RS06810, LMRG_00819), citB (lmo1641, LMRG_RS08180, LMRG_01325), or glnR (lmo1298, LMRG_RS06455, LMRG_00748) genes (Fig. 5G–I). None of these three regions were found in our 2,311-region dataset (Dataset S1); the probed sequences did not contain the internal citB and glnR sites detected by IDAP-Seq (Dataset S2).
Fig. 5.
Binding of CodY to the regulatory regions of L. monocytogenes genes as detected by a gel shift assay. Radioactively labeled DNA fragments were incubated with increasing amounts of purified L. monocytogenes CodY in the presence of 10 mM ILV. CodY monomer concentrations used (nM) are indicated below each lane, and the concentrations needed to shift ~50% of DNA fragments, are underlined. The arrows indicate the bands corresponding to unbound DNA fragments (lower bands) and the complexes of CodY with DNA (upper bands). Each experiment was performed at least twice.
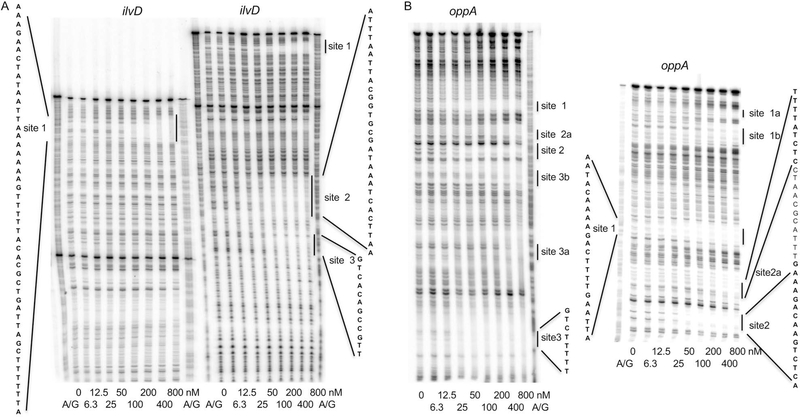
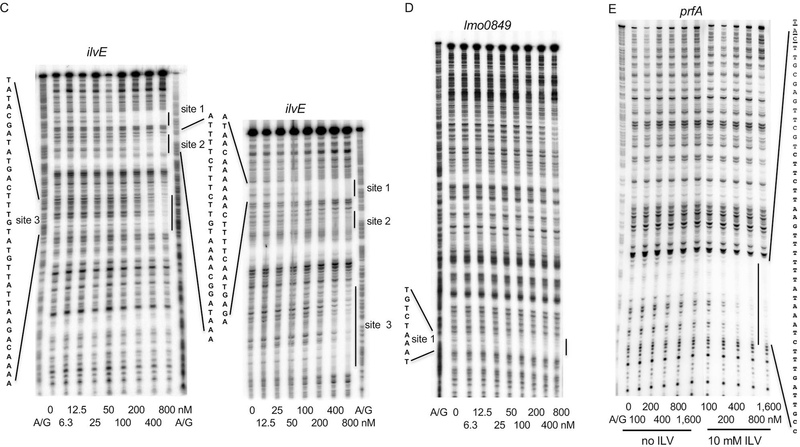
DNase I footprinting experiments were also in very good accord with the IDAP-Seq results. The regulatory region of the ilvD gene was found to have 5 sites in IDAP-Seq experiments. Using DNase I footprinting, we found 3 sites and all of them overlapped with sites determined by IDAP-Seq; the other two sites were located too far from the promoter to be resolved in our footprinting experiment (Fig. 6A and Table 2). The regulatory region of the oppA gene was found to have 3 sites in IDAP-Seq experiments. DNase I footprinting experiments revealed 3 strong sites, one weaker site (site 2a) and several very weak sites; three of the stronger sites overlapped with sites determined by IDAP-Seq; the remaining site, site 1, was very close to a weak IDAP-Seq site that was not included in Dataset S2 (Fig. 6B and Table 2). Similar results were obtained for the regulatory regions of ilvE (two out of three sites detected by footprinting overlapped with the IDAP-Seq sites) and lmo0849 (LMRG_RS04250, LMRG_02272) (a very similar site determined by both methods) (Fig. 6C and D and Table 2). No footprint was detected for the ptb regulatory region, as expected from its absence from Dataset S2 (data not shown). Importantly, IDAP-Seq is a genome-wide competition experiment. Even if a given sequence is able to bind CodY in vitro, as measured in a standard gel shift or DNase I footprinting experiment, the presence of hundreds of competing CodY-binding sites might reduce the binding significantly. Thus, it is possible that some sites detected by footprinting were not detected by IDAP-Seq because of the competition with other genomic CodY-binding sites.
Fig. 6.
DNase I footprinting analysis of CodY binding to the regulatory regions of L. monocytogenes genes. DNA fragments, radioactively labeled on the template strand, were incubated with increasing concentrations of purified CodY in the presence (A-E) or absence (E) of 10 mM ILV. Short and long gel runs are presented in panels A-C. CodY monomer concentrations used (nM) are indicated below each lane. The corresponding A + G sequencing ladders are shown in the left or right lane. The protected areas are shown by vertical lines. Fig. S5 shows the sequence of the beginning of the prfA coding sequence and the location of the CodY-binding site.
Table 2.
Correlation between coordinates of L. monocytogenes CodY-binding sites determined by IDAP-Seq and DNase I footprinting.
| Gene and sites | Site coordinates as determined by IDAP-Seq | Site coordinates as determined by DNase I footprinting | ||
|---|---|---|---|---|
| ilvD | ||||
| site | NDa | 2010412 | NAb | |
| site | 2010555 | ND | NA | |
| site 1 | 2010658 | ND | 2010630 | 2010677 |
| site 2 | ND | 2010951 | 2010921 | 2010949 |
| site 3 | 2010952 | ND | 2010959 | 2010970 |
| oppA | ||||
| site 1c | 2245658 | ND | 2245679 | 2245700 |
| site 2a | 2245619 | ND | 2245627 | 2245635 |
| site 2 | 2245589 | 2245604 | 2245600 | 2245614 |
| site 3 | ND | 2245471 | 2245462 | 2245469 |
| ilvE | ||||
| site 1 | ND | 990735 | 990717 | 990740 |
| site 2 | 990761 | ND | 990760 | 990785 |
| site 3 | not found | 990837 | 990871 | |
| lmo0849 (LMRG_RS04250, LMRG_02272) | ||||
| site 1 | 870581 | 870596 | 870567 | 870575 |
| prfA | ||||
| site 1 | 198774 | 198825 | 198766 | 198797 |
ND - not determinable; one boundary of the CodY-bonding site could not be determined due to the presence of another site in close proximity.
NA - information is not available, because this part of the regulatory region was not footprinted.
The oppA site 1 had very low coverage in the IDAP-Seq experiment and was not included in Dataset 2.
CodY-binding site within the prfA gene
Positive CodY-mediated regulation of the prfA gene (LMRG_RS00965, LMRG_02622, lmo0200), encoding a master regulator of virulence genes in L. monocytogenes, has been described (Lobel et al., 2012; Lobel et al., 2015; Lobel and Herskovits, 2016). Surprisingly, this regulation was observed only under conditions of low ILV concentrations (Lobel et al., 2012), i.e., when CodY activity is expected to be low (Petranovic et al., 2004; Shivers and Sonenshein, 2004; Richardson et al., 2015). A relatively weak binding site was identified within the 5’ end of the coding region of the prfA gene (Lobel et al., 2015). Using IDAP-Seq, we were also able to identify a weak CodY-binding region within the beginning of the prfA coding region (enrichment factor 6.84; ranking number 647; Dataset S1) that contained a CodY-binding site at coordinates (198774 to 198794) that corresponded to the position of the binding site determined previously by the deletion and mutational analyses (Lobel et al., 2015). Using gel shift and DNase I footprinting assays, we confirmed the presence and location of this site and its weak binding strength in vitro (Fig. 5J and 6E). Binding of CodY to the prfA site was increased in the presence of ILV (Fig. 6E), as is the case so far for all other CodY-regulated genes in various bacterial species, including L. monocytogenes (Shivers and Sonenshein, 2004; den Hengst et al., 2005; Dineen et al., 2007; Hendriksen et al., 2008; Lobel et al., 2015). Even in the presence of ILV, CodY binding was very weak (KD about 400 nM in a gel shift assay), a strength level that raises questions about its physiological significance and suggests that regulation of prfA by CodY is primarily indirect (Fig. 5J). Another region further within the prfA coding sequence contains three CodY-binding sites, but their combined binding strength was also low (enrichment factor 9.98; ranking number 389; Dataset S1).
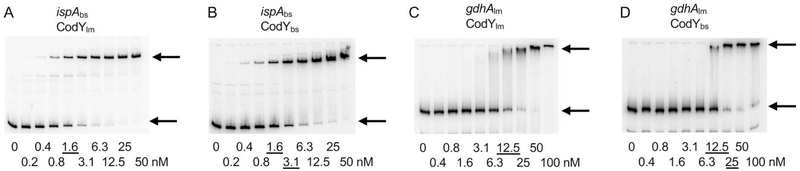
Functional comparison of L. monocytogenes and B. subtilis CodY proteins in vitro and in vivo
In gel shift assays, purified L. monocytogenes CodY in the presence of ILV bound to the regulatory region of the CodY-regulated B. subtilis ispA gene (Belitsky and Sonenshein, 2013) with affinity that was very similar to that of B. subtilis CodY (Fig. 7A and B). Similarly, both B. subtilis CodY and L. monocytogenes CodY bound to the regulatory region of the L. monocytogenes gdhA (lmo0560, LMRG_RS02800, LMRG_00242) gene with very similar affinities (Fig. 7C and D).
Fig. 7.
Comparison of binding by CodY proteins from L. monocytogenes and B. subtilis by gel shift assays. Radioactively labeled DNA fragments of the B. subtilis ispA gene (A, B) or the L. monocytogenes gdhA gene (C, D) were incubated with increasing amounts of purified L. monocytogenes (A, C) or B. subtilis (B, D) CodY in the presence of 10 mM ILV. CodY monomer concentrations used (nM) are indicated below each lane, and the concentrations needed to shift ~50% of DNA fragments, are underlined. The arrows indicate the bands corresponding to unbound DNA fragments (lower bands) and the complexes of CodY with DNA (upper bands).
To compare the regulatory properties of the two proteins in vivo, we replaced the open reading frame of the B. subtilis codY gene within the B. subtilis chromosome by the open reading frame of the L. monocytogenes codY gene. Expression from two B. subtilis promoters, bcaP and ybgE, and one L. monocytogenes promoter, gdhA, that are regulated by their cognate CodY proteins (Bennett et al., 2007; Belitsky and Sonenshein, 2011; Belitsky and Sonenshein, 2011; Lobel and Herskovits, 2016) was monitored using corresponding lacZ fusions in B. subtilis cells under conditions of low, moderate, and high activity of B. subtilis CodY (i.e., in TSS glucose-ammonium medium or the same medium that was enriched with ILV, or a mixture of other 13 amino acids, or both). We were unable to detect any significant difference between the regulatory effects of B. subtilis and L. monocytogenes CodY proteins indicating that they have very similar activities and respond in a very similar way to the same effectors (Table 3). This result is fully consistent with the high degree of similarity of the two proteins (79.5% identity); moreover, their helix-turn-helix DNA-binding motifs are completely identical.
Table 3.
Expression of lacZ fusions in B. subtilis cells
| Strain | Fusion type | Genotype (codY allele) | Additions to the medium | β-galactosidase activity (Miller units) |
|---|---|---|---|---|
| BB2505 | bcaPbs | codY+bs | none | 53.9 |
| ILV | 7.44 | |||
| 13 aa | 13.6 | |||
| 13 aa + ILV | 0.14 | |||
| BB3781 | codY+lm | none | 33.7 | |
| ILV | 6.90 | |||
| 13 aa | 6.03 | |||
| 13 aa + ILV | 0.13 | |||
| BB2548 | codY::spc | none | 119.0 | |
| 13 aa + ILV | 137.0 | |||
| BB2770 | ybgEbs | codY+bs | none | 19.2 |
| ILV | 3.49 | |||
| 13 aa | 11.7 | |||
| 13 aa + ILV | 1.12 | |||
| BB3782 | codY+lm | none | 14.4 | |
| ILV | 5.92 | |||
| 13 aa | 9.48 | |||
| 13 aa + ILV | 1.00 | |||
| BB2771 | codY::spc | none | 268.0 | |
| 13 aa + ILV | 428.0 | |||
| BB3776 | gdhAlm | codY+bs | none | 676.1 |
| ILV | 660.9 | |||
| 13 aa | 490.1 | |||
| 13 aa + ILV | 236.6 | |||
| BB3784 | codY+lm | none | 655.2 | |
| ILV | 635.1 | |||
| 13 aa | 516.8 | |||
| 13 aa + ILV | 221.8 | |||
| BB3778 | codY::spc | none | 689.3 | |
| 13 aa + ILV | 636.1 |
Cells were grown in TSS glucose-ammonium medium with or without mixtures of ILV or 13 aa or both (see Experimental procedures). β-Galactosidase specific activity was assayed and expressed in Miller units. All values are averages of at least two experiments, and the relative standard errors of the mean did not exceed 20%. Some data for the bcaP283-lacZ and ybgE292-lacZ fusions in a wild-type and codY::spc mutant strains were published previously (Belitsky and Sonenshein, 2011).
DISCUSSION
We report here genome-wide identification of virtually all L. monocytogenes CodY-binding sites in vitro at near single-nucleotide resolution. As far as we know, for global transcription regulators this has been previously achieved only for B. subtilis CodY (Belitsky and Sonenshein, 2013). Although direct interaction with CodY can also be determined by in vitro DNA-binding tests of individual genes, such experiments are very labor-intensive and therefore are restricted only to a small subset of the hundreds of CodY-regulated genes. Moreover, binding of CodY to individual sites in vitro occurs in the absence of competition with other binding sites and may overestimate or underestimate the in vivo binding strength of these sites. Varying the protein concentration during the purification step and performing reiterative purification at a given protein concentration allowed us to distinguish among regions of different binding strength under conditions that mimic the genome-wide competition occurring in vivo. A similar procedure, genomic SELEX, was used to analyze binding sites of numerous transcription regulators but the results were not analyzed at this level of precision (Ishihama et al., 2016; Shimada et al., 2018). Other procedures have been developed to detect protein-binding sites with high resolution in vivo (Perreault and Venters, 2016; Chumsakul et al., 2017; Chumsakul et al., 2018; Rossi et al., 2018).
The large number of binding regions found by IDAP-Seq reflects the AT richness and the degenerate nature of the CodY-binding motif, which allows a very large number of potential binding sites of different strengths to exist in the low G+C bacterial genomes. Some parameters of subsets of CodY-binding regions that differ in their binding strengths are summarized in Table 4.
Table 4.
Comparison of subsets of CodY-binding regions with different strengths
| Enrichment (fold) | Number of: |
|||||
|---|---|---|---|---|---|---|
| CodY-binding regions | CodY-binding sites | upstream sites | overlapping CodY-binding motifs | ChIP-Seq regionsa | RNA-Seq regulated genesa | |
| Analysis I | ||||||
| >10 | 388 | 518 | 98 (19%) | 202 (39%) | 46 (12%) | 84 (22%) |
| >15 | 224 | 313 | 72 (23%) | 132 (42%) | 41 (18%) | 49 (22%) |
| >20 | 146 | 203 | 54 (27%) | 94 (46%) | 37 (25%) | 34 (23%) |
| >30 | 78 | 107 | 40 (37%) | 56 (52%) | 27 (35%) | 20 (25%) |
| >40 | 58 | 81 | 32 (40%) | 47 (58%) | 26 (45%) | 14 (24%) |
| >60 | 31 | 46 | 17 (37%) | 30 (65%) | 19 (61%) | 9 (29%) |
| >100 | 17 | 26 | 9 (35%) | 13 (50%) | 14 (82%) | 5 (29%) |
| Analysis II | ||||||
| 10–15 | 164 | 205 | 26 (13%) | 70 (34%) | 6 (3%) | 35 (21%) |
| 15–20 | 78 | 110 | 18 (16%) | 38 (35%) | 4 (5%) | 15 (19%) |
| 20–30 | 68 | 96 | 14 (15%) | 38 (40%) | 10 (15%) | 14 (21%) |
| 30–60 | 47 | 61 | 23 (38%) | 26 (43%) | 8 (17%) | 11 (23%) |
| >60 | 31 | 46 | 17 (37%) | 30 (65%) | 19 (61%) | 9 (29%) |
| ChIP-Seqb | 58 | 59 | 7 (12%) | 25 (42%) | 58 (100%) | 13 (22%) |
Various subsets of 388 CodY-binding regions and associated 518 CodY-binding sites identified by IDAP-Seq were analyzed and compared to the CodY-binding regions and sites identified by ChIP-Seq.
Data taken from our analysis of the previously obtained results (Lobel and Herskovits, 2016)
All regions with CodY-binding sites identified by ChIP-Seq in cells grown in BHI (Lobel and Herskovits, 2016)
Location of CodY-binding sites
Of the 518 CodY-binding sites identified by IDAP-Seq in the 388-region dataset, only 19% were located in putative regulatory regions upstream of the coding sequence of a gene (59 sites) or between the coding sequences of two divergent genes (38 sites) (Dataset S2A). Four sites were located between two convergent genes (Dataset S2A). A surprisingly large number of CodY-binding sites, 417 or 81%, were located within genes’ coding sequences. This is significantly more than the fraction (47%) of such internal sites that was found in B. subtilis (Belitsky and Sonenshein, 2013). Carefully executed ChIP-Seq experiments are required to find out how many of these sites are bound by CodY in vivo (see below). The roles of these sites remain rather mysterious. Previously we found that at least three of the internal B. subtilis sites cause efficient repression of gene expression, apparently by a roadblock mechanism (Belitsky and Sonenshein, 2011). Thus, binding of CodY to at least some of the internal sites in the L. monocytogenes chromosome is also likely to cause premature termination of transcription. Still, the ability of such internal sites to confer strong regulation is likely to be smaller than that of sites that are located within or very near to the promoter regions. Indeed, the frequency of upstream binding sites was increased about two-fold (to 37%) within stronger CodY-binding regions with an enrichment factor >60 compared to the entire group of 518 sites (Table 4).
It should be noted that the stated locations of some CodY-binding sites may be misleading. For instance, two sites located between the convergent genes LMRG_RS10855 (LMRG_02799, lmo2141) and LMRG_RS10860 (LMRG_02798, lmo2142) are internal to the sequence of an sRNA, rli47 (Toledo-Arana et al., 2009). Some sites within coding sequences may be located upstream of currently unidentified promoters. The assumption that a site found within an intergenic region is more likely to have regulatory significance than internal sites may be also incorrect. Some binding sites located in genes’ regulatory regions may be positioned in such a way (e.g., too far upstream or downstream of the promoter) that binding of CodY to such a site has little or no impact on gene regulation. For example, using the RACE assay (Frohman, 1994) and analyzing our unpublished RNA-Seq data, we failed to detect a promoter in the intergenic region upstream of the mfd gene indicating that mfd is transcribed from the promoter of an upstream gene. Therefore, the functional role of one of the strongest L. monocytogenes CodY-binding sites located upstream of the mfd coding sequence (Table 1) may be to block read-through transcription from the upstream gene.
CodY-binding motif and organization of CodY-binding sites
The rules of CodY interaction with DNA have not been fully determined, but it is assumed that all or most of CodY-binding sites contain one or two degenerate versions of a consensus 15-nt motif, AATTTTCWGAAAATT (den Hengst et al., 2005; Guedon et al., 2005; Belitsky and Sonenshein, 2008; Wray and Fisher, 2011). Only 53 of 518 sites (10%) found within the subset of 388 strongest regions overlap with CodY-binding motifs containing ≤3 mismatches with respect to the consensus sequence. This is much less than the 27.8% of such sites found in the B. subtilis 354-site dataset (Belitsky and Sonenshein, 2013), indicating that L. monocytogenes CodY-binding sites on average are weaker than B. subtilis sites. In accord with this observation, the average compliance of L. monocytogenes CodY-binding sites determined in this work to a canonical CodY-binding motif appears to be weaker than those in B. subtilis (Fig. S2). This difference becomes even more apparent if we consider that the smaller L. monocytogenes genome contains more CodY motif-like sequences than the larger B. subtilis genome.
A group of 129 L. monocytogenes CodY-binding sites from the 518-site dataset (25%) overlap with sequences with four mismatches to the canonical 15-nt motif, suggesting that a more degenerate version of the canonical motif can be an important component of CodY-binding sites. Indeed, the regulatory region of the ilvD gene subject to strong, 16-fold, CodY-mediated repression (Lobel and Herskovits, 2016) does not contain a single sequence with 1, 2 or 3 mismatches to the CodY-binding consensus and contains just two motifs with 4 mismatches (at coordinates 2010643 to 2010657 and 2010771 to 2010785). Only one of these motifs overlaps with one of the 5 ilvD binding sites detected by the IDAP-Seq or footprinting experiments (Table 2) (this is the highest number of sites detectable in any region). Three other binding sites overlap with sequences that have 5 mismatches to the consensus, and the remaining site overlaps with a sequence that has 6 mismatches to the consensus. We conclude, as we did previously for B. subtilis (Belitsky and Sonenshein, 2008), that even sites with multiple mismatches to the consensus may be functional (it is possible that clustering of binding sites may contribute to CodY-mediated regulation).
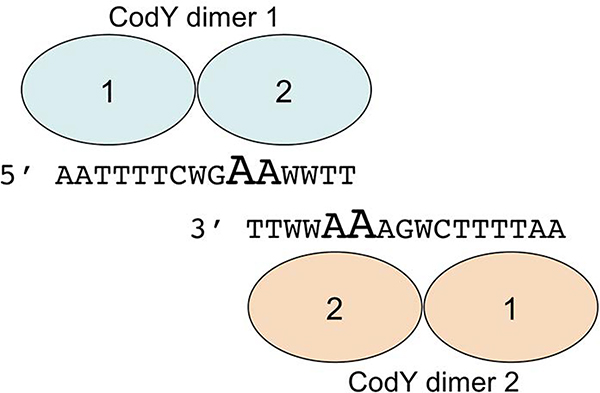
Based on the current results, other analyses of CodY-binding motifs (den Hengst et al., 2005; Guedon et al., 2005; Belitsky and Sonenshein, 2008; Belitsky and Sonenshein, 2011; Levdikov et al., 2017), and the overlapping motifs model created by Wray and Fisher (Wray and Fisher, 2011), we suggest that two dimers of CodY bind in the opposite orientations to two different strands of DNA containing two overlapping sequences resembling the 15-nt consensus CodY-binding motif, forming a 24-nt site (Fig. 8). Each of these 15-nt motifs may contain multiple mismatches and only the adenine at position 10 (and to a lesser degree at position 11) appears to be the most critical for binding; these nucleotides create the central conserved region of the 24-nt motif (Fig. 3B). Poor conservation of these nucleotides and/or the absence of a second overlapping motif may explain why many genomic sequences resembling a consensus CodY-binding motif do not serve as actual CodY-binding sites. However, we do not know whether CodY binding always requires the presence of two (or more) 15-nt motifs as, at least in vitro, CodY is able to bind to a short, 19-bp long fragment of DNA that could accommodate only a single, albeit very strong, CodY-binding motif (Levdikov et al., 2017).
Fig. 8.
A model of interaction between two CodY dimers and overlapping CodY-binding 15-nt sequences located on opposite strands of DNA. The most highly conserved A10 and A11 nucleotides of each motif are shown in fonts of larger size.
The detection by IDAP-Seq of many CodY-binding sites that are longer than 15 bp is consistent with the model of overlapping motifs. If each individual motif of an overlapping pair is essential for CodY binding, the gap in coverage observed in IDAP-Seq experiments will be longer (Fig. S4A). However, if each individual motif of an overlapping pair is able to bind CodY independently of its partner, the gap in coverage observed in IDAP-Seq experiments should be no more than the length of the overlap and potentially IDAP-Seq can detect two overlapping binding sites (Fig. S4B). In fact, 72 sites that are <8 bp long and even some longer sites are likely to represent two overlapping sites in which each site can bind CodY individually though at a reduced efficiency. Also, the coordinates of 25 pairs of closely spaced CodY-binding sites were separated by less than 28 bp, indicating that the sites of each pair may overlap (Dataset S2).
Interestingly, the pattern of coverage by sequencing reads revealed that CodY binding to individual fragments that extend ≤20–30 bp beyond each side of the core site was progressively decreased (Fig. 2A and C). This may correspond to the presence of only one site (or part of a site) of an overlapping pair of sites within this particular fragment and the resulting decrease in binding. It is also possible that additional CodY dimers bind outside a simple or overlapping site either via protein-protein interaction or interaction with adjacent highly degenerate CodY-binding motifs or both.
Comparison of the IDAP-Seq results to ChIP-Seq data
Previous ChIP-Seq experiments revealed 302 extended regions of CodY binding in vivo (131 in BHI and 270 in LBMM, a defined medium containing low concentrations of ILV) (Lobel and Herskovits, 2016). The coordinates of the binding regions and their peaks were not reported; therefore, we reanalyzed the raw sequencing data of this experiment using the procedure that was developed for the analysis of IDAP-Seq. In BHI, we identified 102 regions that were enriched more than 3.5-fold (Dataset S3). Of these regions, 21 corresponded to tRNA and rRNA genes, which are present in the genome in multiple copies, and the LMRG_RS09100 (LMRG_02823, lmo1799) gene containing numerous repeated sequences and may be false-positive signals for CodY binding. An additional 13 regions were found to be parts of other regions present in the same dataset. Thus, only 68 regions were selected as candidates for CodY-binding regions.
Using our analysis of single-stranded coverage by 5’ nucleotides of sequencing reads, CodY-binding sites were found in 58 of these regions (one region contained two sites) (Dataset S3). All these regions were also found by IDAP-Seq in vitro. Moreover, the coordinates of all but one binding site identified in vivo corresponded very well, within 10–20 bp or less, to coordinates of binding sites identified in vitro, a remarkable correlation, especially considering very low counts and missing coverage at many positions in the case of the ChIP-Seq experiment (Dataset S3). All regions identified by ChIP-seq in which we were not able to identify CodY-binding sites, were enriched only weakly; only one of these regions was enriched more than 5-fold (our more detailed analysis showed that this region contains only half of an actual CodY-binding region).
Of the 15 strongest IDAP-Seq sites, 13 were also observed in the ChIP-Seq experiment (Table 1, Dataset S3). We hypothesize that the other two strong sites, for the amtB and oppA genes, were not accessible to CodY in vivo due to binding of other transcriptional regulators, e.g, GlnR in the case of amtB (Kaspar et al., 2014). However, the general correlation between the apparent strengths of binding regions between the two experiments was not perfect. For example, only 46 of 59 ChIP-Seq sites were found within our 518-site IDAP-Seq dataset subject to a detailed analysis in this work. Other sites identified by ChIP-Seq were found among weaker, sometimes very weak, binding regions identified by IDAP-Seq (Dataset S3). Currently, we do not know whether the discrepancy between the strength of binding sites in vivo and in vitro is due to technical imperfection of one or both approaches or due to significant increase of strength of several CodY-binding sites occurring in vivo. It is not surprising that the number of binding regions identified by ChIP-Seq is much smaller than that in IDAP-Seq experiments as it was expected that IDAP-Seq should be a more sensitive approach that identifies even very weak binding regions.
Probably, the most interesting observation from the analysis of CodY-binding sites identified by ChIP-Seq is that only 7 of them (12%) are located in the intergenic regions upstream of the putative target genes. This is even less frequent than for sites identified in vitro (19%) and proves that widespread CodY binding within coding sequences is not an artefact of in vitro binding conditions. The physiological significance of such binding remains to be established. Frequent intragenic binding has been demonstrated for other transcriptional regulators (Chumsakul et al., 2011; Picossi et al., 2014; Knapp et al., 2015; Prestel et al., 2015).
Using the ChIP-Seq data from cells grown in the low ILV medium, LBMM (Lobel and Herskovits, 2016), we identified 13 regions that were enriched more than 3.0-fold and considered as candidates for CodY-binding regions (Dataset S3). CodY-binding sites were found in all of these regions; 12 of these regions (and the corresponding sites) were also identified in BHI, with a higher enrichment factor; 11 of these 12 regions were among the 12 strongest in BHI. Two of the LBMM regions contained additional, weaker CodY-binding sites that were not seen in BHI but were found in vitro. The site of the only binding region that was not found in BHI was located in the intergenic region between two divergent genes, LMRG_RS00730 (LMRG_02397, lmo0152) and LMRG_RS00735 (LMRG_02398, lmo0153); neither of these genes was regulated by CodY in LBMM (or BHI) (Lobel and Herskovits, 2016). Interestingly, this region did not contain an in vitro CodY-binding site, and the mechanism of CodY binding to this site in vivo remains unknown.
ChIP-Seq sites were not associated with CodY-regulated genes or with sequences containing CodY-binding motifs more frequently than IDAP-seq sites (Table 4). Importantly, most of the CodY-binding sites detected by ChIP-Seq appeared to contain two closely spaced or overlapping sites (Fig. S1B and S4B), i.e., the site coordinates on the (+) strand, as detected by our analysis, were downstream of the coordinates on the (−) strand (Dataset S3). Alternatively, the sites appeared to be closely spaced due to the fact that in ChIP-Seq experiments, in contrast to IDAP-Seq, DNA-CodY complexes are formed before DNA fragmentation and CodY may remain bound to DNA even if some nucleotides that are required for initial binding are removed during fragmentation. Therefore, DNA fragments that initiate from nucleotides comprising the central core of a CodY-binding site would contribute to counting of sequencing reads on both strands of DNA. This would lead to a short overlap in coverage of the two strands of chromosomal DNA on the combined coverage map, which coincides with the location of the binding site but is characteristic for closely spaced sites in IDAP-Seq (Fig. S1B).
Comparison of the IDAP-Seq results to other published data
Using gel shift assays, CodY was previously found to bind regulatory regions upstream of the ilvD and codV genes (Lobel et al., 2015). We have detected binding sites within both of these regions (Dataset S2). Eleven additional regulatory regions were subsequently identified as direct CodY-binding fragments (hisZ, rsbV, glpF, argG, gadC, feoA, actA, gdhA, poxB, glnR, fliN) (Lobel and Herskovits, 2016). Our IDAP-Seq assays confirmed the presence of CodY-binding sites in only two of these regions, gdhA and poxB (Dataset S2). Similarly, we could not confirm binding of CodY to the promoter region of the argC gene (Bennett et al., 2007). The discrepancy may be due to the ability of CodY to bind to most DNA fragments if nonspecific, competitor DNA is not present in sufficient concentration. The lack of CodY binding to the regulatory region of the glnR gene under our experimental conditions was confirmed in a gel shift assay (Fig. 5I).
The L. monocytogenes CodY regulon has been determined both by microarray and RNA-Seq experiments. However, growth of cells for the microarray experiment was performed in a defined medium in which CodY is not expected to be fully active; indeed, the authors observed only small effects of a codY null mutation on gene expression (Bennett et al., 2007). Using the published RNA-Seq data and the transcriptome map of the listerial genome (Wurtzel et al., 2012; Lobel and Herskovits, 2016), we found that 50 of about 142 transcriptional units negatively regulated ≥1.8-fold in the complex BHI medium and 27 of about 66 transcriptional units positively regulated ≥1.8-fold in the BHI medium are associated with CodY-binding sites from the 518-site IDAP-Seq dataset. A likely interpretation is that these transcriptional units are regulated by CodY directly.
Despite the uncertainty about the exact number of regulated genes, it is obvious that the number of CodY-binding regions in the L. monocytogenes genome is much larger than the number of CodY-regulated transcriptional units identified by in vivo transcription assays. There are several possible explanations for this result. First, CodY binding to some regions detected by IDAP-Seq may have little or no effect on regulation in vivo. Second, a substantial number of CodY-binding sites detected in vitro are within coding regions. As a result, at least parts of those genes will be transcribed even when CodY is actively blocking the completion of transcription. Third, binding of CodY to some regions in vivo and the resulting CodY-mediated regulation of the corresponding genes may be prevented under certain conditions by the presence or absence of other regulators (Belitsky et al., 2015; Barbieri et al., 2016).
Many genes and operons are regulated by CodY indirectly and therefore should not be expected to contain a CodY-binding site. Although the size of the L. monocytogenes CodY regulon in vivo is comparable to that of B. subtilis, L. monocytogenes contains a smaller number of highly regulated genes or operons under the various conditions tested that may be explained by weaker conservation of the CodY-binding motif (Molle et al., 2003; Bennett et al., 2007; Brinsmade et al., 2014; Lobel and Herskovits, 2016).
EXPERIMENTAL PROCEDURES
CodY overexpression and purification
The L. monocytogenes CodY protein containing six additional histidine residues at its C terminus was purified from Escherichia coli strain BL21(DE3) carrying plasmid pCodY (Bennett et al., 2007) in which the codY gene is transcribed from the bacteriophage T7 promoter. The cells were induced in L-broth cultures (at an optical density of A600=0.35) by addition of 0.1 mM isopropyl-β-D-thiogalactoside and incubated for 4 more hours. L. monocytogenes CodY was purified to virtual homogeneity as described previously for B. subtilis CodY (Belitsky and Sonenshein, 2008).
Construction of a fragmented chromosomal DNA library for IDAP-Seq
Ten μg of total chromosomal DNA of L. monocytogenes strain 10403S were fragmented using the Covaris M220 Focused-Ultrasonicator with a setting for generating 150-bp fragments. The DNA fragments were blunted and phosphorylated at the 5’ end using the Quick Blunting Kit (NEB), followed by addition of dA to the fragments’ 3’ ends using Klenow Fragment (exo-) (NEB). The resulting fragments were ligated (Quick Ligation Kit, NEB) to a universal non-barcoded, partially double-stranded adapter with a 5’ dT overhang formed by annealing oligonucleotides olj543 and olj331. The ligated products were fractionated using a 2% agarose gel to isolate fragments of 250- to 300-bp in length (including the length of the ligated oligonucleotides) and purified using the Gel Extraction Kit (Qiagen). The size-fractionated fragments were amplified by PCR using oligonucleotides olj139 and olj331 as primers.
One μg samples of the amplified DNA fragments (~30 nM) were incubated with varying concentrations of purified L. monocytogenes CodY-His6 (0, 0.32, 1.6, 8, 40, 200, and 1,000 nM) in 200 μl of binding buffer (20 mM Tris-Cl (pH 8.0)-50 mM KCl-2 mM MgCl2–5% glycerol-0.05% Nonidet P-40) in the presence of the CodY effectors isoleucine, leucine, and valine (ILV, 10 mM each) and GTP (2 mM). After 20 min at room temperature, 20 μl of Ni2+-charged His·Bind-resin (Novagen) was added and the CodY-DNA complexes were allowed to absorb to the resin with slight agitation for an additional 20 min at room temperature. The resin was collected by centrifugation at 600 rpm for 60 sec and washed 4 times with the binding buffer containing 10 mM ILV. The final pellet was resuspended in 100 μl of 20 mM Tris-Cl (pH 8.0)-1 mM CaCl2 with 1 μl of proteinase K (Sigma P4850) and incubated at 37oC for 2 h with occasional mixing. The resin was removed by centrifugation, and DNA was purified using the Qiagen PCR Purification Kit. For each individual IDAP reaction, the purified fragments were amplified by PCR using one of the barcoded oligonucleotides of the BC33 to BC56 series, or olj533, or olj534 as a forward primer and olj139 as a common reverse primer. When reiterative rounds of IDAP were performed, 0.1 μg of the amplified DNA fragments from the preceding round (~3 nM) and the same concentration of CodY were used.
The IDAP buffer was identical to the conventional binding buffer (Belitsky and Sonenshein, 2008), except for the absence of EDTA and DTT, which would interfere with the downstream steps of IDAP, and salmon sperm DNA. Salmon sperm DNA, used to prevent nonspecific low affinity binding, was omitted so that its presence would not affect competition for CodY binding among various DNA fragments. The enrichment of fragments containing known CodY-binding sites during IDAP was followed by real-time PCR using gene-specific oligonucleotides as primers, and the amount of PCR products was normalized to the amount of total recovered DNA determined with olj139 and olj331 as primers. PCR products that contained no CodY-binding sites served as negative controls for enrichment tests.
Samples (1 to 9 nM in a total volume of 20 μl) of amplified DNA from each of the IDAP reactions, as well as a sample of the original size-fractionated and amplified DNA library (input), were subjected to massively parallel sequencing using the Illumina HiSeq2500 system under conditions of 26-fold multiplexing. A total of 160 million 50-nt reads was obtained.
Analysis of IDAP-Seq and ChIP-Seq results
The Galaxy suite (Goecks et al., 2010) was used for the analysis of sequencing results. The Illumina reads were aligned to the reference genome of L. monocytogenes strain 10403S using the Bowtie program (Langmead et al., 2009). Coverage maps (numbers of sequencing reads for each genome position) were generated using a custom Galaxy-integrated script. Importantly, only 5’ nucleotides of each read were counted for coverage determination. The CodY-binding regions ≥25 nt in length and having coverage at each position at least 3-fold higher than average were determined using the custom Peak Finder script, which was set for 70-nt (35 nt each way) shift and 6-nt smoothing. For further analysis, the regions were extended by 35 nt on each side to ensure that all reads associated with the binding sites(s) are included in the analysis. Groups of overlapping extended regions identified under different conditions were consolidated into single regions that were used for further analysis.
The relative number of reads for each consolidated region under each binding condition was presented as the fraction of the double-stranded coverage of each region in the coverage of total recovered DNA multiplied by 10,000. The background, determined in the same way from the input sample, was subtracted for each region. The total number of reads in each region of the control sample that was purified in the absence of CodY was almost identical to that in the input DNA. The enrichment factor for each region was determined by dividing the total number of reads in this region under any particular condition of CodY binding by the average of the total numbers of counts in this region in the input and no-CodY samples.
CodY-binding sites were identified as described previously (Belitsky and Sonenshein, 2013) by manual visual inspection of the original (no shifting and smoothing) strand-specific coverage of each individual CodY-binding region as very short gaps in the combined coverage maps of the two strands of chromosomal DNA. That is, if either end of a DNA fragment is lacking even just one bp required for CodY binding, that fragment will not be represented in the group of molecules that co-purify with CodY in vitro resulting in the sharp drop in coverage by the counted reads on each of the two DNA strands of the chromosome (Fig. S1). For closely spaced binding sites, including some overlapping sites, the drop in coverage can be detected only on one DNA strand of the chromosome (Fig. S1 and S4).
The analysis of ChIP-Seq data was performed essentially as described for IDAP-Seq. The CodY-binding regions ≥25 nt in length and having coverage at each position at least 3-fold higher than average were determined using the custom Peak Finder script, which was set for 100-nt (50 nt each way) shift and 12-nt smoothing; the last two parameters were changed in an attempt to compensate for the low or missing coverage at many genome positions.
Bacterial strains and culture media
L. monocytogenes strains 10403S and EGD-e (Becavin et al., 2014) were used in this study. All B. subtilis strains created in this work were derivatives of strain SMY (Zeigler et al., 2008) and are described in Table 5. L. monocytogenes cells were grown in Brain Heart Infusion (BHI) complex medium. E. coli strain JM107 (Yanisch-Perron et al., 1985) was used for isolation of plasmids and grown in L broth (Miller, 1972). B. subtilis cells were grown in DS nutrient broth or TSS 0.5% (w/v) glucose-0.2% (w/v) NH4Cl minimal medium (Belitsky and Sonenshein, 2011). The TSS medium was supplemented as indicated with amino acids (Atkinson et al., 1990). The 16-amino acid mixture contained all amino acids commonly found in proteins except for glutamine, asparagine, histidine, and tyrosine. Concentrations in μg/ml were: glutamate-Na, 800; aspartate-K, 665; serine, 525; alanine, 445; arginine-HCl, 400; glycine, 375; isoleucine, leucine, and valine, 200 each; methionine, 160; tryptophan, 150; proline, threonine, phenylalanine, and lysine, 100 each; cysteine, 40. In some experiments, ILV were omitted from the amino acid mixture, creating a 13 amino acid mixture, or added without other amino acids.
Table 5.
B. subtilis strains used
| Strain | Genotype | Source or reference |
|---|---|---|
| PS251 | codY::(erm::spc) trpC2 | P. Serror |
| BB1043 | codY::(erm::spc) | (Barbieri et al., 2015) |
| BB1888 | lacA::tet | (Belitsky and Sonenshein, 2008) |
| BB2505 | ΔamyE::[Φ(bcaP283-lacZ) erm] lacA::tet | (Belitsky and Sonenshein, 2011) |
| BB2511 | ΔamyE::spc lacA::tet | (Belitsky and Sonenshein, 2008) |
| BB2548 | ΔamyE::[Φ(bcaP283-lacZ) erm] codY::(erm::spc) lacA::tet | (Belitsky and Sonenshein, 2011) |
| BB2770 | ΔamyE::[Φ(ybgE292-lacZ) erm] lacA::tet | (Belitsky and Sonenshein, 2011) |
| BB2771 | ΔamyE::[Φ(ybgE292-lacZ) erm] codY::(erm::spc) lacA::tet | (Belitsky and Sonenshein, 2011) |
| BB3764 | codY::(erm::spc) lacA::tet | BB1043 xDNA(BB1888) |
| BB3766 | codY::codY+lm lacA::tet | BB3764 xpBB1789 |
| BB3776 | ΔamyE::[Φ(gdhAlm489-lacZ erm] lacA::tet | BB2511 xpBB1796 |
| BB3778 | ΔamyE::[Φ(gdhAlm489-lacZ erm] codY::(erm::spc) lacA::tet | BB3776 xDNA(BB1043) |
| BB3781 | ΔamyE::[Φ(yhdG-lacZ erm] codY+lm lacA::tet | BB3766 xDNA(BB2548) |
| BB3782 | ΔamyE::[Φ(ybgE-lacZ erm] codY+lm lacA::tet | BB3766 xDNA(BB2771) |
| BB3784 | ΔamyE::[Φ(gdhAlm489-lacZ erm] codY+lm lacA::tet | BB3766 xDNA(BB3778) |
For growth of bacterial cells on plates, the same media with addition of agar were used. The following antibiotics were used when appropriate: for B. subtilis strains, tetracycline, 15 μg/ml; spectinomycin, 50 μg/ml; or the combination of erythromycin, 0.5 μg/ml, and lincomycin, 12.5 μg/ml; for E. coli strains, ampicillin, 50–100 μg/ml, and kanamycin, 40 μg/ml.
General molecular genetic methods
Methods for common DNA manipulations, E. coli electroporation and B. subtilis chromosomal DNA isolation and transformation were as previously described (Belitsky and Sonenshein, 1998). Chromosomal DNA of L. monocytogenes was isolated using phenol:chloroform extraction after the cells were disrupted using 0.1 mm silica beads and a Mini-BeadBeater (Biospec Products) for two 30-sec cycles at the maximal setting. All oligonucleotides used in this work are described in Tables S1 and S2. All cloned PCR-generated fragments were verified by sequencing.
Replacement of the B. subtilis codY gene by its L. monocytogenes counterpart
An integrative plasmid containing the coding sequence of the L. monocytogenes codY gene was created in several steps. First, the 0.52-kb downstream flanking fragment containing the B. subtilis codY-flgB intergenic region and the 5’ part of the flgB gene was generated by PCR using oligonucleotides oKK34 and oKK35 as primers, digested with EcoRI and XhoI, and cloned in the integrative plasmid pBB544 (carrying a neomycin-resistance gene) (Belitsky et al., 1997) to create pBB1788. Second, two fragments were synthesized: i) the 0.53-kb sequence lying upstream of B. subtilis codY and containing the 3’ part of the clpY gene, the clpY-codY intergenic region, and the codY ribosome-binding site was generated by PCR using oligonucleotides oKK32 and oBB635 as primers and ii) a fragment containing the 0.81-kb coding sequence of the L. monocytogenes codY gene was generated by PCR using oligonucleotides oBB636 and oBB637 as primers. These two PCR products were used in a second, splicing step of PCR mutagenesis as overlapping templates to generate a fragment containing the entire codYlm coding region fused at the 5’ end to the B. subtilis clpY-codY intergenic region; oligonucleotides oKK32 and oBB637 served as PCR primers. Finally, the resulting spliced fragment was digested with SacI and EcoRI and cloned in pBB1788 to create pBB1789 (‘clpYbs-codYlm-flgB’bs).
Plasmid pBB1789 was integrated by a single-crossover, homologous recombination event into the codY locus of strain BB3764 codY::(erm::spc). Neos Spcs colonies indicating spontaneous excision of pBB1789 from the chromosome and replacement of the codY::(erm::spc) allele by the L. monocytogenes codY coding sequence were searched for. The desired replacement was confirmed in strain BB3766 by sequencing the PCR product corresponding to the chromosomal codY locus.
Construction of a transcriptional gdhAlm-lacZ fusion
The 0.5-kb gdhA PCR product, containing the entire L. monocytogenes gdhA regulatory region, was synthesized with oKZ8 and oKZ9 as primers. Plasmid pBB1796 (gdhAlm489-lacZ) was created by cloning the EcoRI- and BglII-treated PCR product in an integrative plasmid pHK23 (erm) (Belitsky and Sonenshein, 2008).
B. subtilis strain carrying the gdhAlm489-lacZ fusion at the amyE locus was isolated after transforming strain BB2511 (amyE::spc lacA::tet) with pBB1796, by selecting for resistance to erythromycin conferred by the plasmid, and screening for loss of the spectinomycin-resistance marker, which indicated a double crossover, homologous recombination event. Strain BB2511 and all its derivatives have very low endogenous β-galactosidase activity due to a null mutation in the lacA gene (Daniel et al., 1997).
Labeling of DNA fragments
The PCR products containing the entire intergenic regions upstream of various L. monocytogenes genes were synthesized using chromosomal DNA of strain 10403S [ilvD (492 bp), mfd (116 bp), glnK (168 bp), prfA (a 325-bp fragment was cloned in pHK23 and amplified as a 466-bp fragment using vector primers)] or EGD-e [oppA (692 bp), ilvE (361 bp), gdhA (510 bp), ptb (352 bp), citB (261 bp), glnR (297 bp), lmo0849 (593 bp)] and primers specified in Table S2. The EGD-e fragments used had from 0 to 4 mismatches with respect to corresponding 10403S fragments (oppA - 2; ilvE - 4; gdhA - 1; ptb - 2; citB - 0; glnR - 1; lmo0849 – 2); these mismatches did not affect CodY-binding sites determined in this work. The 197-bp B. subtilis ispA regulatory region was synthesized by PCR using oBB607 and vector-specific oligonucleotide oBB102 as primers and pBB1755 (Belitsky and Sonenshein, 2013) as template. One of the primers for each PCR reaction was labeled using T4 polynucleotide kinase and [γ−32P]-ATP. The labeled PCR products were purified on an 8% non-denaturing polyacrylamide gel or used without purification.
Gel shift assays and DNase I protection experiments
Incubation of CodY with the 32P-labeled promoter fragments was performed in a binding buffer containing 20 mM Tris-Cl (pH 8.0), 50 mM KCl, 2 mM MgCl2, 5% glycerol, 0.5 mM EDTA, 1 mM DTT, 0.05% Nonidet P-40, 25 μg/ml sonicated salmon sperm DNA and 10 mM ILV. Samples (10–11 μl) containing varying amounts of CodY and less than 1 fmole of DNA were incubated for 16 min at room temperature and separated on 8% non-denaturing 50 mM Tris, 384 mM glycine, 1 mM EDTA polyacrylamide gels in 35 mM HEPES, 43 mM imidazole buffer.
For DNase I protection experiments, samples containing 20–40 fmoles of labeled DNA were incubated with CodY as described above. In some experiments, ILV was omitted from the incubation mixture, as indicated. One μl of the binding buffer containing 0.1–0.2 U RQ1 DNase I (Promega), 10 mM MgCl2 and 20 mM CaCl2 was then added, followed by addition, after 1 min, of 4 μl of 20 mM EDTA-95% formamide dye solution and subsequent heating of the samples at 80°C for 5 min. The samples were loaded without further purification on 7 M urea - 6% polyacrylamide DNA sequencing gels. The G+A sequencing ladder, generated according to a published procedure by boiling the appropriate samples of labeled DNA for 20 min (Liu and Hong, 1998), served to locate precisely the protected region.
The gels were dried, and the radioactive bands were detected and quantified using storage screens, a Storm PhosphorImager, and ImageQuant software (GE Healthcare).
Enzyme assays
β-Galactosidase specific activity was determined as described previously (Belitsky and Sonenshein, 1998).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to David Lazinski for help with the design of DNA library construction, sharing the oligonucleotides, and multiple discussions. This work was supported by a research grant from the US National Institute of Allergy and Infectious Diseases (R01AI109048) to A. L. Sonenshein, A. Herskovits and M. O’Riordan. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. No conflict of interest is declared. Data available in article supplementary material.
REFERENCES
- Atkinson MR, Wray LV Jr. and Fisher SH (1990) Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol 172, 4758–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL and Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol 2, 28–36. [PubMed] [Google Scholar]
- Barbieri G, Albertini AM, Ferrari E, Sonenshein AL and Belitsky BR (2016) Interplay of CodY and ScoC in the regulation of major extracellular protease genes of Bacillus subtilis. J. Bacteriol 198, 907–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri G, Voigt B, Albrecht D, Hecker M, Albertini AM, Sonenshein AL, et al. (2015) CodY regulates expression of the Bacillus subtilis extracellular proteases Vpr and Mpr. J. Bacteriol 197, 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, et al. (2014) Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio 5, e00969–00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Barbieri G, Albertini AM, Ferrari E, Strauch MA and Sonenshein AL (2015) Interactive regulation by the Bacillus subtilis global regulators CodY and ScoC. Mol. Microbiol 97, 698–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Gustafsson MC, Sonenshein AL and Von Wachenfeldt C (1997) An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol 179, 5448–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR and Sonenshein AL (1998) Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol 180, 6298–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR and Sonenshein AL (2008) Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol 190, 1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR and Sonenshein AL (2011) Contributions of multiple binding sites and effector-independent binding to CodY-mediated regulation in Bacillus subtilis. J. Bacteriol 193, 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR and Sonenshein AL (2011) Roadblock repression of transcription by Bacillus subtilis CodY. J. Mol. Biol 411, 729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR and Sonenshein AL (2013) Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc. Natl. Acad. Sci. USA 110, 7026–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, et al. (2007) Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol 63, 1453–1467. [DOI] [PubMed] [Google Scholar]
- Brinsmade SR (2017) CodY, a master integrator of metabolism and virulence in Gram-positive bacteria. Curr. Genet 63, 417–425. [DOI] [PubMed] [Google Scholar]
- Brinsmade SR, Alexander EL, Livny J, Stettner AI, Segre D, Rhee KY and Sonenshein AL (2014) Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc. Natl. Acad. Sci. USA 111, 8227–8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateau A, van Schaik W, Joseph P, Handke LD, McBride SM, Smeets FM, et al. (2013) Identification of CodY-targets in Bacillus anthracis by genome-wide in vitro binding analysis. J. Bacteriol 195, 1204–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumsakul O, Anantsri DP, Quirke T, Oshima T, Nakamura K, Ishikawa S and Nakano MM (2017) Genome-Wide Analysis of ResD, NsrR, and Fur Binding in Bacillus subtilis during Anaerobic Fermentative Growth by In Vivo Footprinting. J. Bacteriol 199, e00086–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumsakul O, Nakamura K, Ishikawa S and Oshima T (2018) GeF-seq: A Simple Procedure for Base Pair Resolution ChIP-seq. Methods Mol. Biol 1837, 33–47. [DOI] [PubMed] [Google Scholar]
- Chumsakul O, Takahashi H, Oshima T, Hishimoto T, Kanaya S, Ogasawara N and Ishikawa S (2011) Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 39, 414–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Winter C, Flores-Mireles AL, Kundra S, Hultgren SJ and Lemos JA (2019) (p)ppGpp and CodY Promote Enterococcus faecalis Virulence in a Murine Model of Catheter-Associated Urinary Tract Infection. mSphere 4, e00392–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio SEF (2019) Innate and Adaptive Immune Responses during Listeria monocytogenes Infection. Microbiol. Spectr 7, 10.1128/microbiolspec.GPP1123-0065-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Haiech J, Denizot F and Errington J (1997) Isolation and characterization of the lacA gene encoding beta-galactosidase in Bacillus subtilis and a regulator gene, lacR. J. Bacteriol 179, 5636–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou N, Wang Y, Levdikov VM, Nandakumar M, Livny J, Bouillaut L, et al. (2019) Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027. PLoS One 14, e0206896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hengst CD, Curley P, Larsen R, Buist G, Nauta A, van Sinderen D, et al. (2005) Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol 187, 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hengst CD, van Hijum SA, Geurts JM, Nauta A, Kok J and Kuipers OP (2005) The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem 280, 34332–34342. [DOI] [PubMed] [Google Scholar]
- Dineen SS, McBride SM and Sonenshein AL (2010) Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol 192, 5350–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineen SS, McBride SM and Sonenshein AL (2010) Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol 192, 5350–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineen SS, Villapakkam AC, Nordman JT and Sonenshein AL (2007) Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol 66, 206–219. [DOI] [PubMed] [Google Scholar]
- Frohman MA (1994) On beyond classic RACE (rapid amplification of cDNA ends). PCR Methods Appl. 4, S40–58. [DOI] [PubMed] [Google Scholar]
- Geiger T and Wolz C (2014) Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int. J. Med. Microbiol 304, 150–155. [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J and Team G (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology 11, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedon E, Sperandio B, Pons N, Ehrlich SD and Renault P (2005) Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151, 3895–3909. [DOI] [PubMed] [Google Scholar]
- Han AR, Kang HR, Son J, Kwon DH, Kim S, Lee WC, et al. (2016) The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism. Nucleic Acids Res. 44, 9483–9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen WT, Bootsma HJ, Estevao S, Hoogenboezem T, de Jong A, de Groot R, et al. (2008) CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol 190, 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A, Shimada T and Yamazaki Y (2016) Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors. Nucleic Acids Res. 44, 2058–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson J and Freitag NE (2019) Regulation of Listeria monocytogenes virulence. Microbiol. Spectr 7, 10.1128/microbiolspec.GPP1123-0064-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar D, Auer F, Schardt J, Schindele F, Ospina A, Held C, et al. (2014) Temperature- and nitrogen source-dependent regulation of GlnR target genes in Listeria monocytogenes. FEMS Microbiol. Lett. 355, 131–141. [DOI] [PubMed] [Google Scholar]
- Knapp GS, Lyubetskaya A, Peterson MW, Gomes AL, Ma Z, Galagan JE and McDonough KA (2015) Role of intragenic binding of cAMP responsive protein (CRP) in regulation of the succinate dehydrogenase genes Rv0249c-Rv0247c in TB complex mycobacteria. Nucleic Acids Res. 43, 5377–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M and Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdikov VM, Blagova E, Young VL, Belitsky BR, Lebedev A, Sonenshein AL and Wilkinson AJ (2017) Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis. J. Biol. Chem 292, 2714–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Freedman JC, Evans DR and McClane BA (2017) CodY Promotes Sporulation and Enterotoxin Production by Clostridium perfringens Type A Strain SM101. Infect. Immun 85, pii: e00855–00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ST and Hong GF (1998) Three-minute G + A specific reaction for DNA sequencing. Anal. Biochem 255, 158–159. [DOI] [PubMed] [Google Scholar]
- Lobel L and Herskovits AA (2016) Systems Level Analyses Reveal Multiple Regulatory Activities of CodY Controlling Metabolism, Motility and Virulence in Listeria monocytogenes. PLoS Genet. 12, e1005870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L, Sigal N, Borovok I, Belitsky BR, Sonenshein AL and Herskovits AA (2015) The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol. Microbiol 95, 624–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L, Sigal N, Borovok I, Ruppin E and Herskovits AA (2012) Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 8, e1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, et al. (2010) Direct targets of CodY in Staphylococcus aureus. J. Bacteriol 192, 2861–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH (1972). Experiments in molecular genetics Cold Spring Harbor, N.Y., Cold Spring Harbor Laboratory. [Google Scholar]
- Mlynek KD, Sause WE, Moormeier DE, Sadykov MR, Hill KR, Torres VJ, et al. (2018) Nutritional Regulation of the Sae Two-Component System by CodY in Staphylococcus aureus. J. Bacteriol 200, pii: e00012–00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y and Sonenshein AL (2003) Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol 185, 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault AA and Venters BJ (2016) The ChIP-exo Method: Identifying Protein-DNA Interactions with Near Base Pair Precision. J. Vis. Exp doi: 10.3791/55016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranovic D, Guedon E, Sperandio B, Delorme C, Ehrlich D and Renault P (2004) Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol 53, 613–621. [DOI] [PubMed] [Google Scholar]
- Picossi S, Flores E and Herrero A (2014) ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genomics 15, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestel E, Noirot P and Auger S (2015) Genome-wide identification of Bacillus subtilis Zur-binding sites associated with a Zur box expands its known regulatory network. BMC Microbiol. 15, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshevich L and Cossart P (2018) Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol 16, 32–46. [DOI] [PubMed] [Google Scholar]
- Richardson AR, Somerville GA and Sonenshein AL (2015) Regulating the Intersection of Metabolism and Pathogenesis in Gram-positive Bacteria. Microbiol. Spectr 3, doi: 10.1128/microbiolspec.MBP-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi MJ, Lai WKM and Pugh BF (2018) Simplified ChIP-exo assays. Nat. Commun 9, 2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlech WF (2019) Epidemiology and clinical manifestations of Listeria monocytogenes infection. Microbiol. Spectr 7. doi: 10.1128/microbiolspec.GPP1123-0014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Ogasawara H and Ishihama A (2018) Genomic SELEX Screening of Regulatory Targets of Escherichia coli Transcription Factors. Methods Mol. Biol 1837, 49–69. [DOI] [PubMed] [Google Scholar]
- Shivers RP and Sonenshein AL (2004) Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol 53, 599–611. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Serror P, Joyce E and Sonenshein AL (1995) A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol 15, 689–702. [DOI] [PubMed] [Google Scholar]
- Smith JL and Grossman AD (2015) In Vitro Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA. PLoS Genet. 11, e1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein AL (2007) Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol 5, 917–927. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, et al. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. [DOI] [PubMed] [Google Scholar]
- Tran NT, Stevenson CE, Som NF, Thanapipatsiri A, Jalal ASB and Le TBK (2018) Permissive zones for the centromere-binding protein ParB on the Caulobacter crescentus chromosome. Nucleic Acids Res. 46, 1196–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis J and Seeliger HP (1975) Incidence of Listeria monocytogenes in nature. Appl. Microbiol 30, 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray LV Jr. and Fisher SH (2011) Bacillus subtilis CodY operators contain overlapping CodY binding sites. J. Bacteriol 193, 4841–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, et al. (2012) Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol 8, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J and Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33, 103–119. [DOI] [PubMed] [Google Scholar]
- Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, et al. (2008) The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol 190, 6983–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.