Etravirine (ETR) is a nonnucleoside reverse transcriptase inhibitor (NNRTI) used in treatment-experienced individuals. Genotypic resistance test-interpretation systems can predict ETR resistance; however, genotype-based algorithms are derived primarily from HIV-1 subtype B and may not accurately predict resistance in non-B subtypes. The frequency of ETR resistance among recombinant subtype C HIV-1 and the accuracy of genotypic interpretation systems were investigated. HIV-1LAI containing full-length RT from HIV-1 subtype C-positive individuals experiencing virologic failure (>10,000 copies/ml and >1 NNRTI resistance-associated mutation) were phenotyped for ETR susceptibility.
KEYWORDS: ETR, HIVdb, NNRTI, etravirine, genotyping, phenotyping, subtype C, third line
ABSTRACT
Etravirine (ETR) is a nonnucleoside reverse transcriptase inhibitor (NNRTI) used in treatment-experienced individuals. Genotypic resistance test-interpretation systems can predict ETR resistance; however, genotype-based algorithms are derived primarily from HIV-1 subtype B and may not accurately predict resistance in non-B subtypes. The frequency of ETR resistance among recombinant subtype C HIV-1 and the accuracy of genotypic interpretation systems were investigated. HIV-1LAI containing full-length RT from HIV-1 subtype C-positive individuals experiencing virologic failure (>10,000 copies/ml and >1 NNRTI resistance-associated mutation) were phenotyped for ETR susceptibility. Fold change (FC) was calculated against a composite 50% effective concentration (EC50) from treatment-naive individuals and three classifications were assigned: (i) <2.9-FC, susceptible; (ii) ≥2.9- to 10-FC, partially resistant; and (iii) >10-FC, fully resistant. The Stanford HIVdb-v8.4 was used for genotype predictions merging the susceptible/potential low-level and low-level/intermediate groups for 3 × 3 comparison. Fifty-four of a hundred samples had reduced ETR susceptibility (≥2.9-FC). The FC correlated with HIVdb-v8.4 (Spearman’s rho = 0.62; P < 0.0001); however, 44% of samples were partially (1 resistance classification difference) and 4% completely discordant (2 resistance classification differences). Of the 34 samples with an FC of >10, 26 were HIVdb-v8.4 classified as low-intermediate resistant. Mutations L100I, Y181C, or M230L were present in 27/34 (79%) of samples with an FC of >10 but only in 2/46 (4%) of samples with an FC of <2.9. No other mutations were associated with ETR resistance. Viruses containing the mutation K65R were associated with reduced ETR susceptibility, but 65R reversions did not increase ETR susceptibility. Therefore, genotypic interpretation systems were found to misclassify ETR susceptibility in HIV-1 subtype C samples. Modifications to genotypic algorithms are needed to improve the prediction of ETR resistance for the HIV-1 subtype C.
INTRODUCTION
Over the last fifteen years, treatment programs in low and middle-income countries (LMIC) have rapidly expanded such that there are now over 21.7 million people accessing care (1). One of the obstacles to successful treatment programs is the development of drug resistance, which has resulted in the need to modify current first-line regimens and develop third-line regimens (2). The World Health Organization (WHO) has recommended the nonnucleoside reverse transcriptase inhibitor (NNRTI) etravirine (ETR) as an option for third-line antiretroviral therapy as it has been shown to safely and effectively suppress HIV-1 replication in treatment-experienced individuals in several studies (3–6). This efficacy is a result of ETR being effective against some HIV drug resistance mutations associated with reduced susceptibility to nevirapine (NVP) and efavirenz (EFV) (7–10).
Though ETR has a higher genetic barrier to resistance than first-generation NNRTIs, several NNRTI resistance-associated mutations (RAMs), including L100I, K101H/P, Y181C/I, G190E/S/A, and M230L, reduce viral susceptibility to ETR and may compromise ETR efficacy in treatment-experienced individuals (11, 12). Algorithms based on phenotypic (mainly from subtype B) and clinical outcome data have been developed to interpret genotypic drug resistance data (13). These algorithms are regularly updated to include new antiretrovirals (ARV), such as ETR (14–17). The algorithm on the Stanford HIV Drug Resistance Database (HIVdb) uses a weighted scoring system to determine ETR susceptibility, based predominantly on phenotypic data from subtype B samples (11, 18).
The Stanford drug resistance algorithm has been used to predict ETR susceptibility of HIV-1 subtype C from first-line antiretroviral therapy (ART) failures and these studies have shown ETR cross-resistance from previous NNRTI regimens (19–21). A recent report from South Africa on subtype C HIV-1 resistance after NNRTI-based first-line and protease inhibitor (PI)-based second-line treatment showed that 74% and 77% of 140 applicants for third-line ART had genotype evidence of resistance to EFV and NVP, respectively, while 37% (52/140) were estimated to have ETR resistance using the Stanford drug resistance algorithm (21). These studies have focused on the prevalence of first-line NNRTI drug resistance mutations as a means to predict the efficacy of incorporating ETR into future regimens (20). However, because drug resistance algorithms are based predominately on data from subtype B isolates, these estimates may not accurately reflect ETR resistance in non-subtype B HIV-1 (22, 23). For example, genotype and phenotype testing from forty-four HIV-1 subtype C samples collected as part of the AIDS Clinical Trial Group (ACTG) study A5230 found that, although genotype-based algorithms and phenotypic fold change (FC) were largely concordant for EFV and NVP, the Stanford drug resistance algorithm overestimated cross-resistance to ETR in 28% of subtype C samples (23).
Modeling has shown that the inclusion of genotypic resistance testing prior to third-line regimen initiation may be more cost-effective than switching individuals from second- to third-line based on virologic failure alone (24, 25). However, the accuracy of genotypic drug resistance algorithms in predicting ETR resistance in subtype C HIV-1 is uncertain. The current study investigated genotype-phenotype correlations for ETR in subtype C samples having failed an NNRTI-based first-line regimen.
(This work was presented in part as a poster at the Conference on Retroviruses and Opportunistic Infections, Seattle, WA, 4 to 7 March 2019 [26].)
RESULTS
Cross-resistance to etravirine of plasma-derived virus from individuals experiencing virologic failure on first-line ART.
Wild-type plasma-derived recombinant viruses from twelve treatment-naive individuals showed mean ETR 50% effective concentration (EC50) of 0.99 ± 0.28 nM. This EC50 was similar to previously reported EC50 values for ETR of 1.2 nM (0.4 to 1.6 nM) in subtype B/C chimeric viruses, using a similar TZM-bl assay, and 1.4 nM (0.3 to 1.6 nM) in xLAI virus, and was thus used as the wild-type, ETR-susceptible control value to calculate fold change (FC) for other viruses (27, 28).
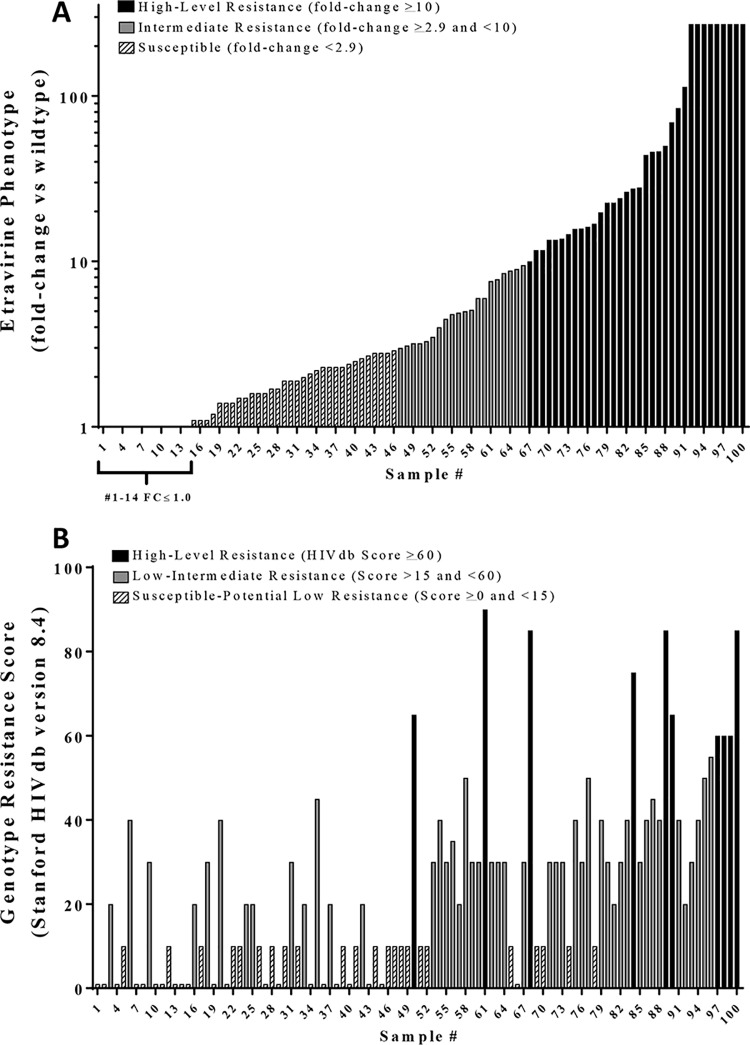
Fifty-four of the hundred (54%) recombinant plasma-derived viruses containing full-length RT from individuals accessing a failing first-line ART exceeded the clinical cutoff of 2.9 FC for ETR (Fig. 1A). Of those 54, nine exceeded the upper limit of the assay (EC50 >272 nM, 272 FC). The median EC50 and FC for the remaining viruses in the resistant category were 15 nM (Q1 to Q3: 6.0, 46.0) and 15-FC (Q1 to Q3: 6.0, 46.0), respectively.
FIG 1.
ETR resistance of 100 first-line antiretroviral treatment failures. (A) ETR fold change (FC) values were determined by dividing the EC50 generated for each plasma-derived virus by a composite EC50 from 12 treatment-naive plasma-derived viruses collected form the same geographical region. The bar color and pattern indicate the ETR phenotypic clinical cutoffs of <2.9 FC as susceptible (diagonal stripes), ≥2.9 FC (gray) as intermediate resistance, and >10 FC as high-level resistance (black). The EC50 values of samples 91 through 100 exceeded the highest concentration of ETR that could be tested in TZM-bl cells without cytotoxicity and are reported as >272 nM. (B) The GRT-IS scores were determined using the HIVdb resistance interpretation algorithm version 8.4 (13). The five HIVdb classifications were collapsed into three by merging susceptible and/potential low-level into “susceptible” and low-level and/intermediate into a “low-intermediate” for comparison.
Drug susceptibility scores were calculated using the HIVdb v8.4 for all 100 plasma-derived recombinant test viruses. Fifty-six of the hundred samples had drug susceptibility scores that exceeded the low-level resistance score cutoff of 15 and, of the 56 that exceeded the cutoff, 10 samples had resistance scores of 60 or greater (Fig. 1B).
Comparative analysis of ETR phenotype and genotype.
The phenotype FC and drug susceptibility classifications disagreed in several samples (Fig. 1A and B), therefore the Spearman’s rho correlation coefficient between the FC and the HIVdb v8.4 score was tested. Drug susceptibility classifications correlated (ρ = 0.62; P < 0.0001) with the phenotype FC; however, 48 of the 100 drug susceptibility scores were discrepant with the phenotype FC classification (Fig. 2A). There were 11 samples with drug susceptibility classification of “susceptible/potential low-level” that had reduced ETR phenotypic susceptibility (FC of ≥2.9; Fig. 2A). Twenty-six samples with a drug susceptibility classification of “susceptible/potential low-level” and “low-intermediate” resistance had an ETR resistance of >10-FC (summarized in the error matrices shown in Fig. 2B).
FIG 2.
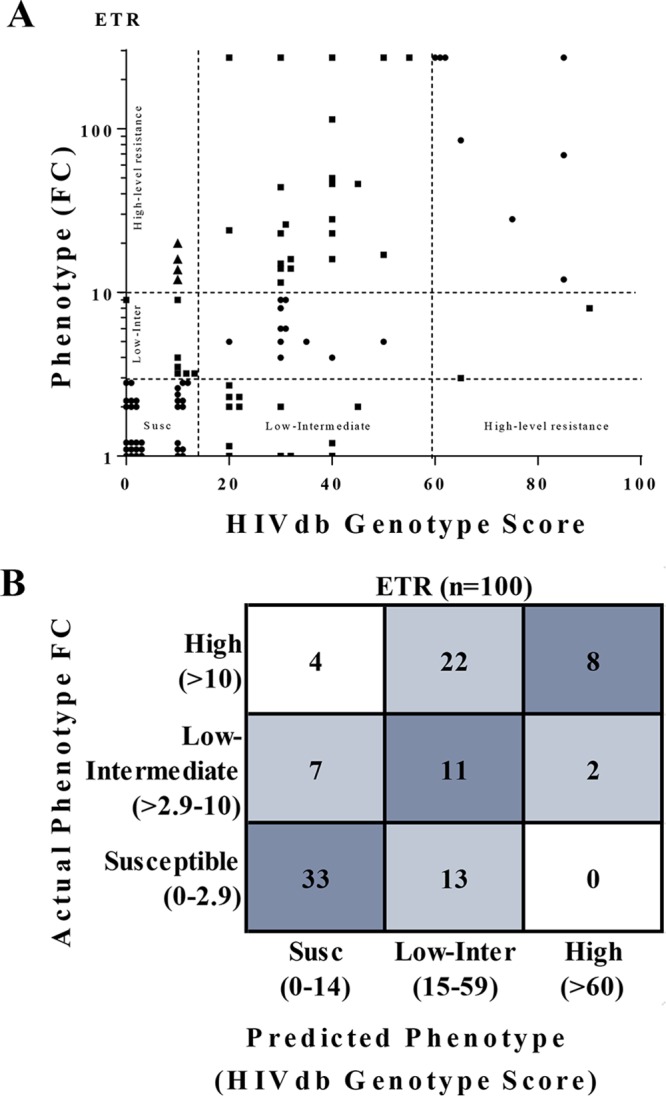
Comparison of ETR phenotype to genotype-based predicted phenotype. (A) ETR phenotype (fold change in EC50) does not strongly correlate with HIVdb score (r = 0.47) for HIV-1 subtype C isolates. Results show 52% of genotype scores were concordant (●, classifications matching), 44% were partially discordant (■, HIVdb predicted 1 classification different), and 4% were completely discordant (▲, HIVdb predicted 2 classifications different) relative to the phenotype clinical cutoffs. (B) Error matrixes of actual fold phenotypic resistance versus predicted resistance for ETR. More samples (26/100) with high phenotypic ETR resistance (FC >10) were misclassified as having low or intermediate resistance. GTR-IS scores were determined using the HIVdb v8.4.
To assess if the discordance was specific to ETR, we tested the correlation of phenotypic FC and HIVdb v8.4 classification on the same 100 recombinant viruses to the NNRTI rilpivirine (RPV) and found fewer (38 of 100) samples with discordance between drug susceptibility and phenotype FC (Fig. S1A and S1B in the supplemental materials). This difference was likely the result of the disproportionate number of samples that had an underestimated ETR resistance score with HIVdb v8.4 (n = 33; Fig. 2B) compared to the underestimation of the RPV resistance score with HIVdb v8.4 (n = 13; Fig. S1B). To assess if the discordances observed for ETR were specific to the HIVdb v8.4, the data set was reanalyzed with the Tibotec resistance algorithm for ETR and a significant correlation (ρ = 0.92; P < 0.0001) between the HIVdb v8.4 and Tibotec ETR scores (Fig. S2) (29) was observed. Interestingly, Tibotec scoring also did not accurately predict the ETR phenotype resistance in 42% of the subtype C isolates, which was only slightly improved from 48% HIVdb discrepancies (Fig. S3A and B). Thus, both ETR drug resistance algorithms tended to underestimate ETR resistance.
HIV-1 drug resistance mutations associated with ETR cross-resistance.
NNRTI and NRTI drug resistance mutations from the 100 donor-derived recombinant viruses were analyzed to determine if specific mutations were associated with resistance to ETR. After adjustment for multiple comparisons, L100I (q < 0.049), Y181C (q < 0.001), and M230L (q < 0.174) were the only NNRTI mutations significantly associated with ETR resistance (Table 1). The NRTI mutation K65R was found to be more prevalent in samples with ETR resistance.
TABLE 1.
Association of HIV-1 drug resistance mutations with ETR resistancef
| Mutation | No. ETR resistant (%)a | No. ETR susceptible (%)b | Oddsc | P value | q valued |
|---|---|---|---|---|---|
| NNRTI-associated resistance mutationse | |||||
| V90I | 5 (9) | 0 (0) | Inf | 0.024 | 0.567 |
| A98G | 10 (19) | 4 (7) | 3.068 | 0.087 | 0.775 |
| L100I | 12 (23) | 1 (2) | 16.286 | 0.001 | 0.049 |
| K101H | 0 (0) | 3 (5) | 0.000 | 0.244 | 0.952 |
| K101E | 5 (9) | 5 (8) | 1.082 | 1.000 | 1.000 |
| K103N | 31 (58) | 24 (41) | 1.909 | 0.130 | 0.812 |
| K103S | 3 (6) | 3 (5) | 1.078 | 1.000 | 1.000 |
| V106M | 18 (34) | 26 (44) | 0.615 | 0.248 | 0.952 |
| V108I | 8 (15) | 5 (8) | 1.843 | 0.382 | 1.000 |
| E138A | 4 (8) | 5 (8) | 0.848 | 1.000 | 1.000 |
| E138K | 2 (4) | 1 (2) | 2.192 | 0.608 | 1.000 |
| V179D | 11 (21) | 2 (3) | 7.163 | 0.007 | 0.233 |
| Y181C | 16 (30) | 0 (0) | Inf | <0.001 | 0.001 |
| Y188L | 5 (9) | 1 (2) | 5.816 | 0.104 | 0.775 |
| G190A | 9 (17) | 17 (29) | 0.597 | 0.273 | 0.970 |
| H221Y | 6 (11) | 2 (3) | 3.500 | 0.152 | 0.901 |
| P225H | 7 (13) | 6 (10) | 1.344 | 0.769 | 1.000 |
| M230L | 10 (19) | 1 (2) | 12.955 | 0.003 | 0.174 |
| NRTI-associated resistance mutations | |||||
| M41L | 7 (13) | 10 (17) | 0.746 | 0.610 | 1.000 |
| K65R | 27 (51) | 8 (14) | 6.620 | <0.001 | 0.006 |
| D67N | 6 (11) | 11 (19) | 0.557 | 0.306 | 0.957 |
| K70R | 7 (13) | 6 (10) | 1.344 | 0.769 | 1.000 |
| Y115F | 7 (13) | 7 (12) | 1.130 | 1.000 | 1.000 |
| M184V | 41 (77) | 41 (69) | 1.500 | 0.397 | 1.000 |
| M184I | 5 (9) | 0 (0) | Inf | 0.021 | 0.605 |
Resistance defined as ≥2.9-fold change; n = 54.
Susceptibility defined as <2.9-fold change; n = 58.
Inf, infinite odds.
q value, false-discovery rate (FDR)-adjusted P value; a q value of <0.2 was considered significant.
NNRTI mutations K101P, V106I, E138G, V179F, V179L, V179T, Y181I, Y181V, G190E, G190S and F227G were not found in this data set.
Boldface type indicates resistance mutations that were significantly associated with ETR resistance.
HIV-1 subtype C donor-derived viruses with V90I (P < 0.024) and V179D (P < 0.007) also showed a trend toward increased ETR resistance, but when adjusted for multiple comparisons (q < 0.567 and q < 0.233, respectively), these were no longer significant. No other NNRTI mutations in this sample set, including A98G, K101E/H/P, V106I, E138A/G/K/Q, V179F/T, Y181V or G190S/A were associated with ETR resistance (Table 1). Interestingly, three out of nine isolates with the RNase H domain mutations Q480H and Q483L (q < 0.18 and q < 0.18, respectively) had phenotypic FC values that exceeded the upper limit of the assay (>272 FC category; data not shown).
Interestingly, L100I, Y181C, and M230L were the only NNRTI RAMs associated with ETR resistance; however, these mutations are scored 15 (Y181C) and 30 (L100I and M230L) points lower in the HIVdb v8.4 predicted phenotype for ETR versus RPV. We recalculated the predicted phenotype using the HIVdb v8.4 scores for ETR with the additional HIVdb v8.4 points that would be allocated for RPV. The recalculation in samples containing these mutations reduced the HIVdb underestimation of ETR predicted phenotype (33/100 underestimated before and 18/100 after recalculation) and improved the overall concordance between ETR predicted and actual resistance (52/100 concordant before and 60/100 after recalculation).
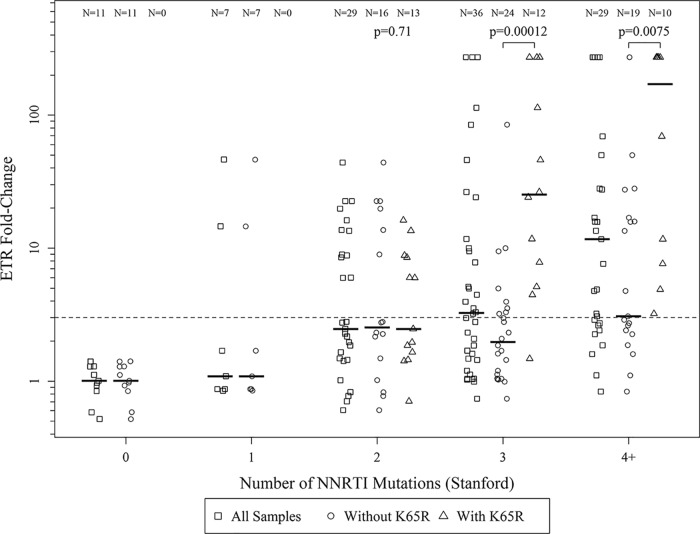
The K65R mutation is associated with ETR resistance but does not contribute directly to the resistance phenotype.
Fifty-one percent of samples with an FC of ≥2.9 had the K65R NRTI resistance mutation, whereas only 14% of samples that were susceptible to ETR had HIV-1 with K65R (Table 1). The mean phenotypic FC of K65R-containing samples was significantly higher (73.83 ± 18.86, n = 35) compared to the mean FC of samples without K65R (10.95 ± 3.78, n = 77; Fig. 3). In contrast, the NRTI mutation M184V was not associated with ETR resistance in these samples and there was no association between the K65R mutation and RPV or dapivirine (DPV) resistance (Fig. S4B), where these two NNRTIs share structural similarity to ETR (30, 31). To determine if the association of the K65R mutation with reduced ETR susceptibility was unique to our data set, phenotypic data collected from an independent study through the Stanford RT Phenotype Query was accessed (32). Using data from Melikian et al. 2012 (33) (the largest data set comparing codon 65 sequence with Phenosense score), a significant association of K65R with ETR resistance (Fig. S4A) was found. As above, the M184V mutation was not associated with ETR resistance in data from the Stanford RT Phenotype Query.
FIG 3.
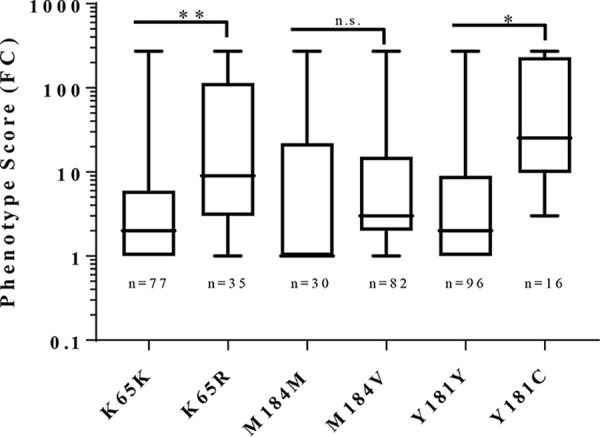
K65R is associated with higher ETR phenotypic resistance than samples with wild-type 65K. Fold change in ETR phenotype resistance was evaluated based on the presence or absence of K65R, M184V, or Y181C in this data set with differences evaluated by Fisher’s exact test: *, P < 0.05; **, P < 0.01; n.s., no significant difference.
To further investigate the impact of K65R and susceptibility to ETR, site-directed mutagenesis was used to make 65R reversions in 2 of the 35 K65R-containing recombinant virus clones. No change in virus susceptibility to ETR was observed in these K65R reverted recombinant virus clones (Table S1). Because 77% (27/35) of the HIV-1 genotypes with K65R had a reduced phenotypic susceptibility to ETR, we explored the phenotypic effects of K65R combined with individual NNRTI resistance mutations on ETR susceptibility. We found that the association between K65R and ETR phenotypic resistance was limited to viruses that had three or greater NNRTI-associated resistance mutations (Fig. 4). Moreover, K65R was found to be associated with reduced susceptibility to ETR in genotypes that contained the NNRTI mutations V179D/F/T, Y181C/I/V, and/or M230L, whereas, no association was found with A98G, L100I, K101E/H/P, V106I, E138A/G/K/Q, or G190S/A (Table S2).
FIG 4.
The correlation between K65R and greater ETR phenotypic resistance is limited to viral genomes containing 3 or more ETR NNRTI resistance-associated mutations. Fold change resistance was evaluated based on the contribution of the number of NNRTI-associated mutations A98G, L100I/V, K101E/H/P, K103H/N/S/T, V106A/I/M, V108I, E138A/G/K/Q/R, V179D/E/F/L, Y181C/F/G/I/V/S, Y188C/F/H/L, G190A/C/E/Q/S/T/V, H221Y, P225H, F227C/L, M230I/L, K238N/T, Y318F, or N348I (i.e., those with resistance scores ≥10 for one or more NNRTI as reported by the Stanford HIV Drug Resistance Database v8.4) per sample with and without K65R. Samples were grouped based on having zero, one, two, three, or four or more NNRTI-associated mutations. Samples with zero NNRTI-associated mutations include the 12 treatment-naive individuals (i.e., control samples derived from the ARV treatment-naive individuals from South Africa) that were used as wild-type comparators in this study. Each group displays individual FC values for all samples in the category (boxes), samples without K65R (circles), and samples containing K65R (triangles). The dotted line indicates the ETR phenotypic resistance clinical cutoff of 2.9-FC. ETR FC between samples with and without K65R were compared with the Mann-Whitney U-Test.
DISCUSSION
ETR is a potentially active drug for treatment of individuals who have experienced failure of first-line NVP- or EFV-based ART, and has been suggested by the WHO for use in third-line treatment in LMIC (6). However, there is considerable overlap in the resistance mutations associated with ETR, NVP, and EFV, increasing the likelihood that NNRTI treatment-experienced individuals will not respond to ETR-containing ART regimens. Genotypic HIV drug resistance testing is faster, less complex to perform, and more affordable than phenotypic assays, making it a more accessible diagnostic tool for treatment optimization in clinical practice. In LMIC, the use of genotypic HIV drug resistance testing and the accompanying algorithms to triage individuals to the most optimal third-line treatments has shown to be successful (34). It is therefore imperative that the genotypic algorithms accurately predict the level of drug resistance across all subtypes. Evidence suggests that current algorithms fail to accurately predict HIV-1 resistance to ETR in individuals with subtype C infections and with previous NVP and EFV exposure (22, 23). To add to this body of evidence, we investigated the correlation between genotypic and phenotypic susceptibility to ETR using recombinant viruses derived from persons living with HIV-1 subtype C who experienced virologic failure on an EFV- or NVP-based first-line treatment.
Over half (54%) of recombinant viruses were found to be phenotypically resistant to ETR (Fig. 1A). This finding supports previous studies showing that 42% and 49% of HIV-1 subtype C isolates with at least one NNRTI mutation or from individuals failing first-line NNRTI, respectively, were phenotypically resistant to ETR (23, 35). Furthermore, we found that, although results from the HIVdb v8.4 correlated with ETR phenotype (r = 0.47), there was disagreement between phenotypic and genotypic prediction to ETR in 48% of samples. More specifically, the HIVdb v8.4 algorithm under- and overestimated ETR resistance in 33% and 13% of subtype C samples, respectively (Fig. 2A and B). This underestimation of ETR resistance with genotypic interpretation algorithms is of concern because of the potential impact on clinical response to ETR-containing ART regimens. We found that stratifying our data according to alternate cutoffs used in the field did not alter the overall interpretation of our study; therefore, it is unlikely that the phenotyping cutoffs used for this analysis had an impact on the observed discordance (data not shown). We also compared our phenotyping data to another drug resistance algorithm, Tibotec (36), and we found that this algorithm also failed to accurately predict resistance in 42% of our HIV-1 subtype C samples (Fig. S2).
Several factors could explain the discrepancy between genotypic interpretation algorithms and phenotypic assessment of resistance. One possible explanation is that resistant variants could be present at frequencies that could not be detected by population-based sequencing but could be contributing to the phenotypic outcome. However, Agneskog and colleagues showed that even when detecting minority variants using a 0.5% cutoff with next generation sequencing (NGS), genotypic interpretation algorithms such as Monogram, Tibotec, and HIVdb all underestimated the level of phenotypic resistance in ETR (22).
Another explanation for the predicted phenotype discrepancy is that there are NNRTI mutations or combinations of mutations that contribute to the resistance phenotype that are not incorporated into the genotypic interpretation algorithms. For example, there have been a number of studies suggesting a role for RNase H domain mutations in antiretroviral resistance and such mutations may not be included in genotypic interpretation algorithms (37). Interestingly, 33% (3/9) of the samples that had >272 FC in ETR phenotype resistance had two RNase H mutations at Q480H and H483L (q < 0.18); however, we did not attempt genotypic reversions with these recombinant viruses because their phenotypes exceeded the maximum threshold for our phenotyping assay.
A third scenario is that the contributions of specific NNRTI mutations and combinations of mutations are not accurately accounted for in genotypic scoring. For example, we found that the current scoring for L100I, Y181C, and M230L may underestimate ETR phenotypic resistance while current scoring for A98G, K101H, V179D, Y188L, G190A, H221Y, and P225H may overestimate ETR phenotypic resistance. These findings are supported by a previous study on HIV-1 subtype C samples that concluded that EFV/NVP resistance mutations K103N, V106M, and G190A had no effect on ETR susceptibility and that the mutations L100I (n = 12/100) and M230L (n = 10/100) increased ETR resistance (35). Mutations at E138 have recently been shown to reduce ETR susceptibility; however, the mutation E138A was not associated with resistance in our data set (Table 1) and E138K/Q/R mutations were too few to reach significance (35, 38).
Interestingly, the NRTI mutation K65R was present in the majority of ETR-resistant viruses, and K65R-containing viruses had significantly higher ETR resistance than viruses without K65R (Fig. 3). However, K65R reversions in two recombinant viruses had no effect on ETR susceptibility and, consequently, we were unable to link a mechanism of K65R contribution to phenotypic resistance in these two samples. Site-directed mutations in wild-type virus may have elucidated the role of K65R, but we were unable to produce viable recombinant virions that contained K65R. Without an observable change in phenotype, the findings on K65R in these data remain unclear; however, because the association was limited to genomes of ≥3 NNRTI RAM, we hypothesize that the correlation between K65R and ETR phenotypic resistance is a consequence of prolonged ART failure and the accumulation of NNRTI mutations that conferred ETR resistance.
In summary, this study confirms that phenotypic cross-resistance to ETR is common in individuals that have experienced virologic failure on an NNRTI-based first-line regimen in South Africa. Both the HIVdb and Tibotec genotypic interpretation algorithms were found to misclassify the ETR susceptibility in HIV-1 subtype C samples. Modifications to current genotypic algorithms for ETR are needed to improve prediction of phenotypic resistance for HIV-1 subtype C.
MATERIALS AND METHODS
Clinical samples.
The donor-derived sample set used in this analysis has been described previously (30, 31). Briefly, plasma samples from 100 persons living with HIV-1 subtype C and experiencing virologic failure (>10,000 HIV-1 RNA copies/ml after 6 months of nevirapine (NVP)- and/or efavirenz (EFV)-containing ART) with at least one major NNRTI resistance mutation in HIV-1 reverse transcriptase (RT) (as defined by HIVdb v8.4) were used in this study (18, 39). These samples contained a median of three NNRTI-associated drug resistance mutations and the majority (94/100) of these samples also contained at least one NRTI resistance mutation. Control samples from 12 HIV-1 subtype C-infected treatment-naive individuals having no NRTI or NNRTI resistance mutations (as defined by HIVdb v8.4) were obtained from the same geographical location. All donor samples were anonymized, and testing was approved by the South African Medical Association Research Ethics Committee (SAMAREC) and the Institutional Review Board of the University of Pittsburgh.
Generation of full-length donor-derived HIV-1LAI.
The 100 recombinant HIV-1LAI-containing bulk-cloned, donor-derived, full-length RT have been previously reported (30, 31). Briefly, viral RNA was extracted using m2000sp (Abbott Molecular) and full-length HIV-1 RT was amplified using SuperScript III one-step RT-PCR System with Platinum Taq DNA polymerase (Invitrogen). The full-length RT amplicon was then bulk cloned into xxLAIxhoI (40) using the In-Fusion HD cloning system (Clonetech), followed by purification of plasmid DNA using the PureYield plasmid midiprep system (Promega). Lipofectamine2000 (Life Technologies) transfection into 293T cells was performed to generate infectious viral clones as reported (30, 31).
Clonal isolation and reversion to wild type at codon 65 in donor-derived HIV-1LAI.
Out of the 35 K65R-containing HIV-1LAI recombinants, two were selected to make 65R reversions based on the following criteria: (i) ≤2 NNRTI mutations, including V179D/F/T, Y181C/I/V, or M230L; (ii) moderate resistance phenotype (greater than 10, but less than 270); and (iii) limited (≤2) mixed viral populations identified by Sanger sequencing. QuikChange XL site-directed mutagenesis kit (Agilent Technologies) was used to make the point mutations according to the manufacturer’s instructions. QuikChange primers were designed using Agilent’s primer design website (https://www.agilent.com/store/primerDesignProgram.jsp) and then ordered from Integrated DNA Technologies, Inc. Single plasmid clones were isolated by picking single colonies from limiting dilutions of bacteria plated on agar plates. The HIV RT region from these colony-isolated xxLAIxhoI recombinants was amplified and sequenced to ensure that the plasmids were single clones and that they contained the 65R reversion.
HIV-1 phenotyping.
A relative light unit (RLU)-normalized input was used to infect untreated or ETR-treated TZM-bl cells in a luciferase-based single cycle drug susceptibility assay as previously reported (30, 31, 41). Briefly, TZM‐bl cells were plated at 10,000 cells per well overnight. The cells were treated with serial dilutions of ETR, and infected with a standardized input of infectious HIV‐1 wild-type or resistant virus. After a 48‐h incubation at 37°C, the cells were lysed, and luminescence was measured in RLU (Britelite Plus; Perkin-Elmer). Endpoints were measured as RLU generated by luciferase expression and, therefore, viruses used for the ETR drug susceptibility were normalized to 300 RLU for optimal signal within the dynamic range for the Thermo Scientific Luminoskan Ascent instrument (42). The 50% effective concentration (EC50) in cloned wild-type virus remained constant regardless of the virus input tested. GraphPad Prism 6 software (GraphPad Software, Inc.) four parameter, nonlinear regression for curve fitting was used to generate EC50 values. All ETR EC50s were evaluated as fold change values (FC) determined by dividing the EC50 generated for each recombinant plasma-derived virus by the mean EC50 from duplicate determinations of the twelve treatment-naive plasma-derived subtype C viruses collected from the same geographical region. The lower cutoff of <2.9-FC was used to define virus susceptibility based on the cutoff established in the PhenoSenseGT HIV assay and the upper cutoff >10-FC as fully resistant was used to be consistent with phenotyping assay resistance cutoffs established in the field (35, 43, 44). However, it is important to note that, although the lower clinical cutoffs from PhenoSenseGT HIV assay were used, the cutoff values are not linked to clinical correlates or outcomes by the phenotyping assay used in this study. ETR was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: etravirine (cat. number 11609) from Janssen Pharmaceutical Companies.
HIV-1 population-based genotype.
An in-house genotype assay was used to sequence HIV-1 from donor plasma at Lancet Laboratories, SA, as previously described (45). Full-length HIV-1 RT from plasma-derived recombinant xxLAIxhoI viral stocks were also sequenced using six bidirectional primers spanning the entire length of RT. Drug resistance mutations and virus subtypes were identified using the HIVdb resistance interpretation algorithm v8.4 (HIVdb v8.4) and REGA HIV-1 subtyping tool, respectively (13, 46). Phylogenetic analysis (Stanford Calibrated Population Resistance Tool v6.0) (47) was used to ensure that plasma-derived cloned virus sequences clustered with the parental virus in the starting plasma sample and the sequences had a median hamming distance of 99% (686/691). An ambiguity index was calculated as the percentage of ambiguous base calls (R, Y, K, M, S, W, B, D, H, V, or N) in each sequence over the number of bases in the sequence (48). The median ambiguity index for plasma-derived virus (0.0072) was not significantly different from the median ambiguity index for recombinant virus (0.0087) (P > 0.05).
HIVdb v8.4 genotypic interpretation algorithm for ETR drug resistance.
The drug susceptibility scores were determined using the HIVdb v8.4 by uploading HIV-1 population genotyping FASTA files into the HIVdb website (https://hivdb.stanford.edu/hivdb/by-sequences/) (13). HIVdb v8.4 computes a final weight factor for each drug resistance mutation or for pairs of drug resistance mutations as follows: K101E+Y181C, K101E+Y188L, K101E+G190A, K101E+G190S, and A98G+Y181C each have a factor of five; A98G, L100V, K101H, E138A/G/K/Q/R, V179D/E/L, Y188L, G190A/C/S/T/V, H221Y, V179T+Y181C, and Y181C+G190A/C/S/T/V each have a score of 10; K101E, V179F, Y181F/G/S, M230I, and V179F+Y181C each have a score of 15; L100I, Y181C, F227C, and M230L have a score of 30; G190E/Q has a score of 45; and K101P and Y181I/V both have a score of 60 for ETR. Weighted scores are tallied and the HIV-1 phenotype relative to ETR is predicted to be: susceptible at <10; potential low-level resistance at 10 to 14; low-level resistance at 15 to 29; intermediate resistance at 30 to 59; and high-level resistance at >60. For comparison between genotype-predicted resistance and phenotypic actual resistance, the HIVdb v8.4 weighted scores groups were collapsed to create three categories as follows: susceptible (HIVdb score 0 to 14), low-intermediate resistance (HIVdb score 15 to 59), and high resistance (HIVdb score >60).
Statistical analysis.
Statistical analysis was conducted as previously reported (30, 31). Fisher’s exact test (FET) was used to assess differences in the prevalence of amino acids at all RT codons between samples with ≥2.9-FC ETR resistance and samples with lower resistance, including those from 12 treatment-naive individuals (FC of <2.9). The individual effect of NNRTI resistance mutations (as defined by HIVdb v8.4) on ETR resistance was assessed using FET to evaluate significance (39). A mutation was included in the analysis if it was present at an estimated frequency of >25% within a sample, as determined by population sequencing, and if there were at least three observations of the mutation in the entire data set. Correction for multiple comparisons was performed by controlling for the false discovery rate (FDR) using the method of Benjamini and Hochberg (49). An FDR-adjusted P value (or “q value”) of <0.2 was considered significant. Statistical analyses were performed in R (v3.1.2) with the glmnet library.
Data availability.
All full-length RT sequences that were derived from the 112 subtype C HIV-1 isolates have been deposited in GenBank under the accession numbers MT109380 to MT109491.
Supplementary Material
ACKNOWLEDGMENTS
K.D.M., K.P., C.J.B., R.V.V., and U.M.P. declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. P.R.H. has received grants from, served as an ad hoc advisor to, or spoke at various events sponsored by Pfizer, Glaxo-SmithKline, Abbott, Merck, Tobira Therapeutics, Virco, and Quest Diagnostics, and has served as a consultant for ViiV Health Care, Tobira Therapeutics, Selah Genomics, Inc., and Quest Diagnostics. P.R.H. is supported by the CIHR/GSK Research Chair in Clinical Virology. J.W.M. is a consultant to Gilead Sciences, Merck, Accelevir Diagnostics, and Yufan Biotechnologies, and owns share options in Co-Crystal Pharma, Inc. C.L.W. has served on the expert panel for the Stanford genotypic interpretation algorithm, received honoraria from MSD, Janssen, Abbott, and AbbVie, and serves as a consultant for IPM.
Funding for this study was from the Bill and Melinda Gates Foundation, grant number OPP1019228. P.R.H. was supported by a Glen Hillson Professorship in Clinical Virology and the CIHR/GSK Research Chair in HIV/AIDS at the University of British Columbia. C.J.B. was supported by a Postgraduate Fellowship from the Canadian Institutes of Health Research (MFE-146750). This work was also made possible by the generous support of the American people through the United States Agency for International Development (USAID) and the U.S. President’s Emergency Plan for AIDS Relief (PEPFAR). The contents are the responsibility of the coauthors and do not necessarily reflect the views of USAID, PEPFAR, or the United States Government. Cooperative agreement AID-OAA-A-15-00031.0.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.World Health Organization. 2018. Antiretroviral therapy coverage data and estimates by WHO region. Data HIV/AIDS response. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.World Health Organization. 2017. HIV drug resistance report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Allavena C, Dat’AIDS Study group, Katlama C, Cotte L, Roger PM, Delobel P, Cheret A, Duvivier C, Poizot-Martin I, Hoen B, Cabie A, Cheret A, Lahoulou R, Raffi F, Pugliese P. 2016. Long-term efficacy and safety of etravirine-containing regimens in a real-life cohort of treatment-experienced HIV-1-infected patients. Infect Dis (Lond) 48:392–398. doi: 10.3109/23744235.2015.1133927. [DOI] [PubMed] [Google Scholar]
- 4.Nuttall J, Pillay V. 2018. Characteristics and early outcomes of children and adolescents treated with darunavir/ritonavir-, raltegravir- or etravirine-containing antiretroviral therapy in the Western Cape Province of South Africa. S Afr Med J 108:105–110. doi: 10.7196/SAMJ.2017.v108i2.12573. [DOI] [PubMed] [Google Scholar]
- 5.Potard V, Goujard C, Valantin MA, Lacombe JM, Lahoulou R, Chéret A, Girard PM, Costagliola D, French Hospital Database on HIV (FHDH-ANRS CO4). 2018. Impact of etravirine on hospitalization rate between 2005 and 2011 among heavily treated HIV-1-infected individuals on failing regimens. BMC Infect Dis 18:326. doi: 10.1186/s12879-018-3231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. 2016. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 7.Lazzarin A, Campbell T, Clotet B, Johnson M, Katlama C, Moll A, Towner W, Trottier B, Peeters M, Vingerhoets J, de Smedt G, Baeten B, Beets G, Sinha R, Woodfall B, DUET-2 study group. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet (London, England) 370:39–48. doi: 10.1016/S0140-6736(07)61048-4. [DOI] [PubMed] [Google Scholar]
- 8.Madruga JV, Cahn P, Grinsztejn B, Haubrich R, Lalezari J, Mills A, Pialoux G, Wilkin T, Peeters M, Vingerhoets J, de Smedt G, Leopold L, Trefiglio R, Woodfall B, DUET-1 study group. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet (London, England) 370:29–38. doi: 10.1016/S0140-6736(07)61047-2. [DOI] [PubMed] [Google Scholar]
- 9.Schrijvers R. 2013. Etravirine for the treatment of HIV/AIDS. Expert Opin Pharmacother 14:1087–1096. doi: 10.1517/14656566.2013.787411. [DOI] [PubMed] [Google Scholar]
- 10.Vingerhoets J, Azijn H, Fransen E, De Baere I, Smeulders L, Jochmans D, Andries K, Pauwels R, de Béthune M-P. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J Virol 79:12773–12782. doi: 10.1128/JVI.79.20.12773-12782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melikian GL, Rhee S-Y, Varghese V, Porter D, White K, Taylor J, Towner W, Troia P, Burack J, Dejesus E, Robbins GK, Razzeca K, Kagan R, Liu TF, Fessel WJ, Israelski D, Shafer RW. 2014. Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. J Antimicrob Chemother 69:12–20. doi: 10.1093/jac/dkt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramalingam VV, Demosthenes JP, Ghale BC, Rupali P, Varghese GM, Abraham OC, Kannangai R. 2018. Frequency of cross-resistance to rilpivirine and etravirine among HIV-1 subtype C infected individuals failing nevirapine/efavirenz based ART regimen. Infect Dis (Lond) 51:71–74. doi: 10.1080/23744235.2018.1510182. [DOI] [PubMed] [Google Scholar]
- 13.Liu TF, Shafer RW. 2006. Web resources for HIV Type 1 genotypic-resistance test interpretation. Clin Infect Dis 42:1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saravanan S, Kausalya B, Gomathi S, Sivamalar S, Pachamuthu B, Selvamuthu P, Pradeep A, Sunil S, Mothi SN, Smith DM, Kantor R. 2017. Etravirine and rilpivirine drug resistance among HIV-1 subtype C infected children failing non-nucleoside reverse transcriptase inhibitor-based regimens in South India. AIDS Res Hum Retroviruses 33:567–574. doi: 10.1089/AID.2016.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teeranaipong P, Sirivichayakul S, Mekprasan S, Ohata PJ, Avihingsanon A, Ruxrungtham K, Putcharoen O. 2016. Role of rilpivirine and etravirine in efavirenz and nevirapine-based regimens failure in a resource-limited country: a cross-sectional study. PLoS One 11:e0154221. doi: 10.1371/journal.pone.0154221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vingerhoets J, Nijs S, Tambuyzer L, Hoogstoel A, Anderson D, Picchio G. 2012. Similar predictions of etravirine sensitivity regardless of genotypic testing method used: comparison of available scoring systems. Antivir Ther 17:1571–1579. doi: 10.3851/IMP2275. [DOI] [PubMed] [Google Scholar]
- 17.Snoeck J, Kantor R, Shafer RW, Van Laethem K, Deforche K, Carvalho AP, Wynhoven B, Soares MA, Cane P, Clarke J, Pillay C, Sirivichayakul S, Ariyoshi K, Holguin A, Rudich H, Rodrigues R, Bouzas MB, Brun-Vezinet F, Reid C, Cahn P, Brigido LF, Grossman Z, Soriano V, Sugiura W, Phanuphak P, Morris L, Weber J, Pillay D, Tanuri A, Harrigan RP, Camacho R, Schapiro JM, Katzenstein D, Vandamme A-M. 2006. Discordances between interpretation algorithms for genotypic resistance to protease and reverse transcriptase inhibitors of human immunodeficiency virus are subtype dependent. Antimicrob Agents Chemother 50:694–701. doi: 10.1128/AAC.50.2.694-701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paredes R, Tzou PL, van Zyl G, Barrow G, Camacho R, Carmona S, Grant PM, Gupta RK, Hamers RL, Harrigan PR, Jordan MR, Kantor R, Katzenstein DA, Kuritzkes DR, Maldarelli F, Otelea D, Wallis CL, Schapiro JM, Shafer RW. 2017. Collaborative update of a rule-based expert system for HIV-1 genotypic resistance test interpretation. PLoS One 12:e0181357. doi: 10.1371/journal.pone.0181357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corrigan B, Mukui I, Mulenga L, Mthethwa N, Letsie M, Bruno S, Rakhmanina N. 2017. Characteristics of treatment-experienced HIV-infected African children and adolescents initiating darunavir and/or etravirine-based antiretroviral treatment. Pediatr Infect Dis J 37:669–672. doi: 10.1097/INF.0000000000001843. [DOI] [PubMed] [Google Scholar]
- 20.Diphoko T, Gaseitsiwe S, Kasvosve I, Moyo S, Okatch H, Musonda R, Wainberg M, Makhema J, Marlink R, Novitsky V, Essex M. 2018. Prevalence of rilpivirine and etravirine resistance mutations in HIV-1 subtype C-infected patients failing nevirapine or efavirenz-based combination antiretroviral therapy in Botswana. AIDS Res Hum Retroviruses 34:667–671. doi: 10.1089/AID.2017.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moorhouse M, Maartens G, Francois Venter WD, Moosa M-Y, Steegen K, Jamaloodien K, Fox MP, Conradie F. 2019. Third-line antiretroviral therapy program in the South African public sector: cohort description and virological outcomes. J Acquir Immune Defic Syndr 80:73–78. doi: 10.1097/QAI.0000000000001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agneskog E, Nowak P, Maijgren Steffensson C, Casadellà M, Noguera-Julian M, Paredes R, Källander CFR, Sönnerborg A. 2014. Decreased phenotypic susceptibility to etravirine in patients with predicted genotypic sensitivity. PLoS One 9:e101508. doi: 10.1371/journal.pone.0101508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derache A, Wallis CL, Vardhanabhuti S, Bartlett J, Kumarasamy N, Katzenstein D. 2016. Phenotype, genotype, and drug resistance in subtype C HIV-1 infection. J Infect Dis 213:250–256. doi: 10.1093/infdis/jiv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chimbetete C, Katzenstein D, Shamu T, Spoerri A, Estill J, Egger M, Keiser O. 2018. HIV-1 Drug Resistance and Third-Line Therapy Outcomes in Patients Failing Second-Line Therapy in Zimbabwe. Open Forum Infect Dis 5:ofy005. doi: 10.1093/ofid/ofy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzana SB, Hughes MD, Grinsztejn B, Collier AC, Luz PM, Freedberg KA, Wood R, Levison JH, Mugyenyi PN, Salata R, Wallis CL, Weinstein MC, Schooley RT, Walensky RP. 2012. Genotype assays and third-line ART in resource-limited settings: a simulation and cost-effectiveness analysis of a planned clinical trial. AIDS 26:1083–1093. doi: 10.1097/QAD.0b013e32835221eb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick KD, Penrose KJ, Brumme CJ, Harrigan PR, Viana RV, Mellors JW, Parikh UM, Wallis CL. 2019. Frequent discordance between etravirine phenotype & genotype in subtype C ART failure, poster 532. 2019 Conf Retrovir Opportun Infect, Seattle, WA. [Google Scholar]
- 27.Andries K, Azijn H, Thielemans T, Ludovici D, Kukla M, Heeres J, Janssen P, De Corte B, Vingerhoets J, Pauwels R, de Béthune M-P. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob Agents Chemother 48:4680–4686. doi: 10.1128/AAC.48.12.4680-4686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Njenda DT, Aralaguppe SG, Singh K, Rao R, Sönnerborg A, Sarafianos SG, Neogi U. 2018. Antiretroviral potency of 4’-ethnyl-2’-fluoro-2’-deoxyadenosine, tenofovir alafenamide and second-generation NNRTIs across diverse HIV-1 subtypes. J Antimicrob Chemother 73:2721–2728. doi: 10.1093/jac/dky256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penrose KJ, Wallis CL, Sluis-Cremer N, Scoulos-Hanson M, Viana RV, Harrigan PR, Mellors JP. 2014. Frequent etravirine cross-resistance in subtype C HIV-1 isolates among first-line ART failures in South Africa. International Workshop on Antiretroviral Drug Resistance, Berlin, Germany.
- 30.Penrose KJ, Wallis CL, Brumme CJ, Hamanishi KA, Gordon KC, Viana RV, Harrigan PR, Mellors JW, Parikh UM. 2016. Frequent cross-resistance to dapivirine in HIV-1 subtype C-infected individuals on failing first-line antiretroviral therapy in South Africa. Antimicrob Agents Chemother 61:e1805. doi: 10.1128/AAC.01805-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penrose KJ, Brumme CJ, Scoulos-Hanson M, Hamanishi K, Gordon K, Viana RV, Wallis CL, Harrigan PR, Mellors JW, Parikh UM. 2018. Frequent cross-resistance to rilpivirine among subtype C HIV-1 from first-line antiretroviral therapy failures in South Africa. Antivir Chem Chemother 26:2040206618762985. doi: 10.1177/2040206618762985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela JS, Shafer RW. 2003. Detailed RT phenotype query. https://hivdb.stanford.edu/cgi-bin/RT_Phenotype.cgi. Accessed 18 April 2018.
- 33.Melikian GL, Rhee S-Y, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia-Cancio PV, Zolopa A, Robbins GK, Kagan R, Israelski D, Shafer RW. 2012. Standardized comparison of the relative impacts of HIV-1 reverse transcriptase (RT) mutations on nucleoside RT inhibitor susceptibility. Antimicrob Agents Chemother 56:2305–2313. doi: 10.1128/AAC.05487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grinsztejn B, Hughes MD, Ritz J, Salata R, Mugyenyi P, Hogg E, Wieclaw L, Gross R, Godfrey C, Cardoso SW, Bukuru A, Makanga M, Faesen S, Mave V, Wangari Ndege B, Nerette Fontain S, Samaneka W, Secours R, van Schalkwyk M, Mngqibisa R, Mohapi L, Valencia J, Sugandhavesa P, Montalban E, Avihingsanon A, Santos BR, Kumarasamy N, Kanyama C, Schooley RT, Mellors JW, Wallis CL, Collier AC, A5288 Team. 2019. Third-line antiretroviral therapy in low-income and middle-income countries (ACTG A5288): a prospective strategy study. Lancet HIV 6:e588–e600. doi: 10.1016/S2352-3018(19)30146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basson AE, Rhee S-Y, Parry CM, El-Khatib Z, Charalambous S, De Oliveira T, Pillay D, Hoffmann C, Katzenstein D, Shafer RW, Morris L. 2015. Impact of drug resistance-associated amino acid changes in HIV-1 subtype C on susceptibility to newer nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 59:960–971. doi: 10.1128/AAC.04215-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vingerhoets J, Monika P, Hilde A, Tambuyzer L, Hoogstoel A, Nijs S, de Béthune M-P, Picchio G. 2008. Tibotec etravirine weighted genotype score to predict response. XVII International HIV Drug Resistance Workshop, Sitges, Spain.
- 37.Delviks-Frankenberry KA, Nikolenko GN, Pathak VK. 2010. The “connection” between HIV drug resistance and RNase H. Viruses 2:1476–1503. doi: 10.3390/v2071476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H-T, Colby-Germinario SP, Asahchop EL, Oliveira M, McCallum M, Schader SM, Han Y, Quan Y, Sarafianos SG, Wainberg MA. 2013. Effect of mutations at position E138 in HIV-1 reverse transcriptase and their interactions with the M184I mutation on defining patterns of resistance to nonnucleoside reverse transcriptase inhibitors rilpivirine and etravirine. Antimicrob Agents Chemother 57:3100–3109. doi: 10.1128/AAC.00348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee S-Y, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi C, Mellors JW. 1997. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob Agents Chemother 41:2781–2785. doi: 10.1128/AAC.41.12.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melody K, McBeth S, Kline C, Kashuba ADM, Mellors JW, Ambrose Z. 2015. Low frequency of drug-resistant variants selected by long-acting rilpivirine in macaques infected with simian immunodeficiency virus containing HIV-1 reverse transcriptase. Antimicrob Agents Chemother 59:7762–7770. doi: 10.1128/AAC.01937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thermo Fisher Scientific. 2018. Ultrafast flash kinetic assays with Luminoskan and Fluoroskan FL microplate readers. Thermo Fisher Scientific, Waltham, MA. [Google Scholar]
- 43.Coakley E, Chappey C, Benhamida J, Picchio G, Tambuyzer L, Vingerhoets J, deBethune M-P. 2008. Biological and clinical cut-off analyses for etravirine in the PhenoSense HIV assay. XVII International HIV Drug Resistance Workshop, Sitges, Spain.
- 44.Picchio G, Vingerhoets J, Tambuyzer L, Coakley E, Haddad M, Witek J. 2011. Short communication prevalence of susceptibility to etravirine by genotype and phenotype in samples received for routine HIV type 1 resistance testing in the United States. AIDS Res Hum Retroviruses 27:1271–1275. doi: 10.1089/AID.2011.0049. [DOI] [PubMed] [Google Scholar]
- 45.Wallis CL, Papathanasopoulos MA, Lakhi S, Karita E, Kamali A, Kaleebu P, Sanders E, Anzala O, Bekker L-G, Stevens G, de Wit TFR, Stevens W. 2010. Affordable in-house antiretroviral drug resistance assay with good performance in non-subtype B HIV-1. J Virol Methods 163:505–508. doi: 10.1016/j.jviromet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, Seebregts C, Snoeck J, van Rensburg EJ, Wensing AMJ, van de Vijver DA, Boucher CA, Camacho R, Vandamme A-M. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 47.Gifford RJ, Liu TF, Rhee S-Y, Kiuchi M, Hue S, Pillay D, Shafer RW. 2009. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics 25:1197–1198. doi: 10.1093/bioinformatics/btp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersson E, Shao W, Bontell I, Cham F, Cuong DD, Wondwossen A, Morris L, Hunt G, Sönnerborg A, Bertagnolio S, Maldarelli F, Jordan MR. 2013. Evaluation of sequence ambiguities of the HIV-1 pol gene as a method to identify recent HIV-1 infection in transmitted drug resistance surveys. Infect Genet Evol 18:125–131. doi: 10.1016/j.meegid.2013.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All full-length RT sequences that were derived from the 112 subtype C HIV-1 isolates have been deposited in GenBank under the accession numbers MT109380 to MT109491.