Abstract
It has been hypothesised that the 2‐year oscillations in abundance of Xestia moths are mediated by interactions with 1‐year Ophion parasitoid wasps. We tested this hypothesis by modelling a 35‐year time series of Xestia and Ophion from Northern Finland. Additionally, we used DNA barcoding to ascertain the species diversity of Ophion and targeted amplicon sequencing of their gut contents to confirm their larval hosts. Modelling of the time‐series data strongly supported the hypothesised host–parasitoid dynamics and that periodic occurrence of Xestia moths is mediated by Ophion. DNA barcodes revealed that Ophion included five species rather than just one while targeted amplicon sequencing verified that Ophion does parasitise Xestia. At least one Ophion species employs 1‐year Syngrapha interrogationis as an alternate host, but it did not detectably affect Xestia–Ophion dynamics. We also demonstrate the previously unrecognised complexity of this system due to cryptic parasitoid diversity.
Keywords: DNA barcoding, MAPL, Ophion, periodic occurrence, population dynamics, Xestia
We provide evidence that the periodic fluctuations in abundance of Xestia moths are mediated by Ophion parasitoids. By sequencing the gut contents of adult parasitoids, we provide genetic evidence of their larval hosts. We also demonstrate cryptic diversity in the studied parasitoids.

Introduction
Determining what drives the dynamics of natural populations is difficult. A group of about 20 species of Xestia moths that are very abundant in the circumboreal region show an unusual demographic pattern. Their populations show alternate year oscillations in abundance with numbers being roughly two orders of magnitude greater in years when moths are abundant than when they are rare (Mikkola 1976; Mikkola & Kononenko 1989; Várkonyi et al. 2002; Kankare et al. 2002). As a particularly remarkable feature, multiple species share similar patterns of abundance and rarity. Rearing studies show that the development time in these species is invariably two years so there are two more or less independent cohorts at each locality, one hatching in odd years and the other in even years (Várkonyi et al. 2002). This raises a key question – what mechanism sustains the systematic, persistent abundance difference between the two coexisting but temporally isolated cohorts? Two main hypotheses have been proposed: competition between the cohorts (e.g., Bulmer 1977; Heliövaara & Väisänen 1986; Heliövaara et al. 1994) and interaction between the cohorts via a natural enemy with a 1‐year life cycle, such as a parasitoid wasp, which by switching between the two host cohorts keeps the rare cohort rare (e.g., Mikkola 1976; Bulmer 1977; Heliövaara et al. 1994) (Figure S1). Empirical time series data for Xestia and a parasitoid wasp in the genus Ophion have provided strong indirect support for the host–parasitoid hypothesis (Rost et al. 2001; Várkonyi et al. 2002). A further feature of the dynamics is large‐scale spatial variation in the identity of the common cohort. For instance, in Western Finnish Lapland the even year cohort is abundant, whereas in Eastern Lapland the odd year cohort is abundant, with a narrow transition zone (Mikkola 1976; Rost et al. 2001; Várkonyi 2003). Rost et al. (2001) showed that a spatial host–parasitoid model that incorporates environmental heterogeneity predicts spatio‐temporal dynamics that are consistent with these empirical observations.
The Ophion species in the time series analysed by Várkonyi et al. (2002) was identified as O. luteus by GV and confirmed by J.P. Brock, whose revision of the genus (Brock 1982) was then the best resource for separating species in this genus which presents unusual taxonomic difficulty. Broad et al. (2015) elucidated the natural history of O. luteus in Britain and Germany. They found that it attacks late‐instar larvae of moths in another noctuid genus, Agrotis, a result contrary to the assumption of the Xestia–Ophion hypothesis which requires both host specialization and the attack of early‐instar host. However, it remains possible that the dominant autumnal Ophion species in Finnish Lapland is a separate cryptic species, morphologically close to O. luteus. If so, references to O. luteus in Rost et al. (2001), Kankare et al. (2002), Várkonyi et al. (2002) and Várkonyi (2003) would be incorrect, and the taxonomy of Ophion needs to be revisited. In fact, the taxonomy of Swedish Ophion have been recently revised (Johansson & Cederberg 2019), potentially providing new insights into the Ophion diversity in Finnish Lapland.
Unfortunately, extensive rearing studies have failed to prove that Ophion parasitise Xestia (Várkonyi et al. 2002). However, it has been shown, under controlled conditions, that host DNA can be retrieved from parasitoid wasps after metamorphosis using an approach termed Molecular Analysis of Parasitoid Linkages (MAPL) method (Rougerie et al. 2011), making it possible to directly ascertain the identity of their host species. Wirta et al. (2014) elucidated host–parasitoid interactions in an Arctic community by sequencing short, diagnostic segments of the DNA barcode region of the COI gene from the gut contents of adult parasitoids and compared these sequences with those of potential hosts.
Here, we aimed to provide new insights on Xestia–Ophion relationships and to rigorously test the underlying host–parasitoid hypothesis based on long time series collected for both hosts and parasitoids. First, we used MAPL to genetically ascertain the host species for Ophion. Second, we sequenced the barcode region of the Ophion wasps to ascertain if there was just a single or several cryptic species participating in the system, similar to the situation noted in other parasitoids wasps (Smith et al. 2008, 2013) and flies (Smith et al. 2006, 2007). Third, we modelled a 35‐year long time series of Ophion and Xestia to test if these empirical data support the host–parasitoid hypothesis. Finally, we evaluated the impacts of potential alternative hosts for Ophion apart from Xestia. Várkonyi et al. (2002) analysed a 22‐year time series (until 1999) from the same time series and found strong indirect support for the host–parasitoid hypothesis. By extending the time series by 13 years, we now test the generality of the previous result, and compare a set of alternative models to test for evidence of some other moths than Xestia acting as alternate hosts for Ophion.
We provide strong statistical support for the host–parasitoid hypothesis based on unusually long time series, demonstrate that the Ophion parasitoid community is a species complex, and provide, for the first time, direct genetic evidence of host use of Ophion.
Material and methods
Long‐term insect monitoring programme
Initiated in 1978, the Värriö insect monitoring programme was designed as a long‐term assessment of the moth fauna at this subarctic site based on specimens collected by deploying 11 light traps. The trap transect is about 1300 m long, and ranges in altitude from 340 to 470 m a.s.l. (Pulliainen & Itämies 1988). Three traps are in an old‐growth Scots pine (Pinus sylvestris) forest, three traps in a ravine of spruce‐dominated (Picea abies) mixed forest, three traps in a mountain birch (Betula pubescens ssp. czerepanovii) forest on the northern slope of the Värriötunturi fell, and two traps on the treeless summit of the fell. Traps are similar to those described by Jalas (1975) and use 500‐W blended light lamps which are illuminated from 20:00 to 08:00 from mid‐May until mid‐October. Samples were collected every morning, stored in a freezer, and moths were identified by JI. The voucher specimens are deposited in the Zoological Museum of the University of Oulu. Conveniently, the same traps also collected adult Ophion as they fly during the night and are strongly attracted to light.
Sampling of Ophion for DNA analyses
Ophion wasps have been separated from the rest of the trap samples since 1978 with dry specimens preserved in plastic jars. Material collected in 2012 was moved to 100% ethanol to slow DNA degradation. To clarify Ophion species diversity and their host species in the study area, we selected 190 specimens for barcode analysis. To analyse possible temporal variation in species composition, specimens from several years were examined with a focus on recent material to ensure high quality DNA. As a result, the specimens derived from 4 years: 2008 (50), 2009 (19), 2011 (31), and 2012 (90). One morphologically distinct specimen (later found to be O. costatus) was included for comparison.
The entire metasoma was used as a source of tissue for DNA extraction while the rest of the specimen was preserved as a voucher. To avoid contamination, each metasoma was separately washed in alcohol three times and subsequently cleaned of all visible adherent material, such as moth wing scales, with a brush under microscope. Tissue samples were placed in 96‐well microplates for subsequent DNA extraction.
DNA‐based characterisation of Ophion species diversity and their hosts
DNA was extracted from each metasoma by incubating it in lysis buffer (700 mM guanidine thiocyanate (Sigma), 30 mM EDTA pH 8.0 (Fisher Scientific), 30 mM Tris‐HCl pH 8.0 (Sigma), 0.5% Triton X‐100 (Sigma), 5% Tween‐20 (Fluka Analytical), 2 mg/mL Proteinase K (Promega)) at 56 °C for 18 h. DNA was purified using the silica membrane‐based method of Ivanova et al. (2006).
A 658 bp segment of the mitochondrial COI 5’ terminus, i.e. the standard DNA barcode (Hebert et al. 2003), was sequenced at the Centre for Biodiversity Genomics in Guelph to assess species diversity in the O. luteus complex. The COI barcode region was amplified using primers LepF1 + LepR1, and, in cases where they did not yield an amplicon, two shorter, overlapping fragments were amplified using the primer pairs LepF1 + C_ANTMR1D and RonMWASPdeg_t1 + LepR1. All amplification reactions consisted of 2 µL of Hyclone ultra‐pure water (Thermo Scientific), 6.25 µL of 10% D‐(+)‐trehalose dihydrate (Fluka Analytical), 0.0625 µL of 10 mM dNTP (KAPA Biosystems), 1.25 µL of 10X PlatinumTaq buffer (Invitrogen), 0.625 µL of 50 mM MgCl2 (Invitrogen), 0.125 µL of each primer, 0.060 µL of 5 U/µL PlatinumTaq DNA Polymerase (Invitrogen), and 2 µL of DNA for a total reaction volume of 12.5 µL. Sanger sequencing, each 96‐plate with a negative control, was performed on an ABI 3730Xl sequencer using a modified BigDye protocol (Hajibabaei et al. 2005).
Since host DNA had been in the gut for several weeks, exposed to unfavorable conditions for DNA preservation, we sought to amplify a short (148bp) fragment of the host COI gene. Fragments of this length are adequate for species identification, especially when a comprehensive reference library of potential target species is available (Hajibabaei et al. 2006; Mutanen et al. 2015). In the present case, all potential hosts of Ophion are Lepidoptera and there is a reference library for every lepidopteran species from the study region. Lepidoptera‐specific primers, MAPL_LepF1_t1 and MAPL_LepR1_t1 (Wirta et al. 2014), were used which selectively amplify a 148 bp region of the COI barcode sequence which was then sequenced on an ABI 3730XL sequencer following Hajibabaei et al. (2005). Trace files were edited in CodonCode (CodonCode Corporation, Dedham, Massachusetts) and uploaded to BOLD as stated below.
A recent revision of the Swedish fauna of Ophion (Johansson & Cederberg 2019) was used to assign each specimen to a species.
All sequence and voucher data, including specimen collection data, high‐resolution images of specimens, DNA sequences, GenBank accession numbers and raw trace files were deposited in BOLD (http://www.boldsystems.org) and all data are publicly available in the data set DS‐OPGUT at dx.doi.org/10.5883/DS‐OPGUT. The COI sequences were compared to those of Johansson & Cederberg (2019) for taxonomic identifications.
Host–parasitoid models
The Xestia–Ophion model
We use as our starting point the model described by Rost et al. (2001) and Várkonyi et al. (2002), given by
| (1) |
| (2) |
where and denote population sizes of the host (Xestia) and parasitoid (Ophion), respectively. The function describes the population growth rate of the host while the function models the fraction of hosts that are not parasitised. These equations assume the host has a 2‐year life cycle while the parasitoid has a 1‐year life cycle. There are thus two independent host populations, one enclosing in even and the other in odd years. The model assumes no direct interaction between the two host cohorts, but they are coupled through the parasitoid.
We assume the Ricker model for host population dynamics and the model of Rost et al. (2001) for the parasitoid numerical response
| (3) |
| (4) |
This deterministic model has three parameters, which are assumed to be the same for the two Xestia cohorts: is the search efficiency of the parasitoid, and and are, respectively, the growth rate and the carrying capacity of the host in the absence of the parasitoid. Rost et al. (2001) used the scaling , and , in which case eqn (3) simplifies to , and eqn (4) to , and thus the model has only two parameters ( and ). However, we proceed with the unscaled version of the model.
Model with an alternative host
Result from the MAPL analysis (see Results) suggested that Ophion might also parasitise Syngrapha interrogationis, a moth with a 1‐year life cycle. In the absence of empirical data, we assume Syngrapha follows the same model as Xestia, but with a 1‐year life cycle, and with different parameters. The model with the two host species (denoted by superscripts X and S for Xestia and Syngrapha, respectively) thus reads
| (5) |
| (6) |
| (7) |
and it has six parameters: and .
Fitting the model to empirical data
Data are available for the years 1978–2012, except that data for Ophion are missing for 2006 and 2010. As the abundance observations are based on 11 traps, they capture only a small proportion of the local populations. We log(x + 1) transformed the count data and assumed that observation error is normally distributed at this scale. We denote the error variance related to the observation process by , and assume it to be the same for the two hosts and the parasitoid.
We fitted the Xestia–Ophion model (eqns (1), (2)) and the Xestia–Ophion–Syngrapha model (eqns (5), (6), (7)) to the data using Bayesian inference. We computed the likelihood of the data as the product of the likelihoods of the yearly transitions for the hosts and the parasitoid, all conditional on the observed values in previous years. In the Xestia–Ophion model, the estimated parameters are the process model parameters , and , and the observation model parameter . The Xestia–Ophion–Syngrapha model additionally has the parameters , and for Syngrapha. We assumed for all log‐transformed parameters the essentially uninformative normal prior with mean 0 and standard deviation 10. The exception was , for which we set a normal prior with mean and standard deviation selected so that belonged to the interval from 0.5 to 1.0 with 95% prior credibility. These values were selected by examining the consistency among the counts in different traps which were pooled for the present analysis (see Appendix for details). We sampled the posterior with a random walk Metropolis–Hastings algorithm (programmed with Mathematica) with adaptive adjustment of the normal proposals during the initial 1,000 iterations. We ran the model for 20,000 iterations and discarded the initial half as a burn‐in.
Testing the relative strength of interaction between Syngrapha and Ophion
We examined this question in two ways, by asking (1) whether accounting for Ophion helps to predict the dynamics of Syngrapha, and (2) whether accounting for Syngrapha helps to predict the dynamics of Ophion. In this context, we consider the model given by eqns (5), (6), (7) as the full model, and the model
| (8) |
| (9) |
| (10) |
as the reduced model that lacks the interaction between Ophion and Syngrapha. Note that the full model has one more parameter () than the reduced model.
To address question (1), we compared the full and reduced models by computing difference in the log‐transformed maximum likelihood (ML) value related to the response variable . To address question (2), we compared the full and reduced models by computing their difference for the log‐transformed ML value related to the response variable .
We computed the null distribution of the ML likelihood values in two ways. Null Distribution A was obtained by repeating the model fitting after replacing the Syngrapha time series by one of nine other control moth species which are common in the study area but are unlikely to be hosts for Ophion. Null Distribution B was obtained by repeating the model fitting for both Syngrapha and the other nine moth species, but now randomly permuting the time series, twice for each of the ten species (Syngrapha and the nine control moth species).
There are two reasons why we did not apply DIC‐based model selection but derived the distribution of null values for the ML likelihood by approaches A and B. The first reason is that DIC (or other standard model selection tools) may deliver incorrect outcomes with multispecies time‐series data that contain dependencies generated by the dynamics. The second reason is that comparison between Null Distributions A and B makes it possible to disentangle cases of complete noise (data vs. Null Distribution B) and an apparent correlation between Syngrapha and Ophion that could be generated, for example, by covariation with some environmental feature that influences all moth species (data vs. Null Distribution A).
Results
Xestia and Ophion dynamics
Over the 35 years, the light traps sampled 15 682 individuals of eight species of Xestia. Two were dominant; X. tecta (9396 individuals) and X. alpicola (4600) accounted for 89% of all individuals. Table S1 in the Supplementary Material reports the number of each species collected in each year. In the same period, 8,932 individuals of Ophion were caught excluding 2006 and 2010 where the samples were lost.
Figure 1 shows the time series of log‐transformed total abundances for the eight species of Xestia (Figure 1a), Ophion (Figure 1b) and Syngrapha interrogationis (Figure 1c). The two‐year periodicity is striking in Xestia with the odd‐year cohort being far more abundant without exception than the even‐year cohort for all 35 years (Figure 1a). Similar periodicity is not seen in S. interrogationis which has a 1‐year life cycle. The dynamics of Ophion show a similar pattern to Xestia, though now we have a single population with a true 2‐year cycle, with population size greater in the even‐ than odd‐years (Figure 1b). The dynamics of Ophion are somewhat less regular than those of Xestia, and we describe a plausible biological explanation for this difference (see Discussion).
Figure 1.
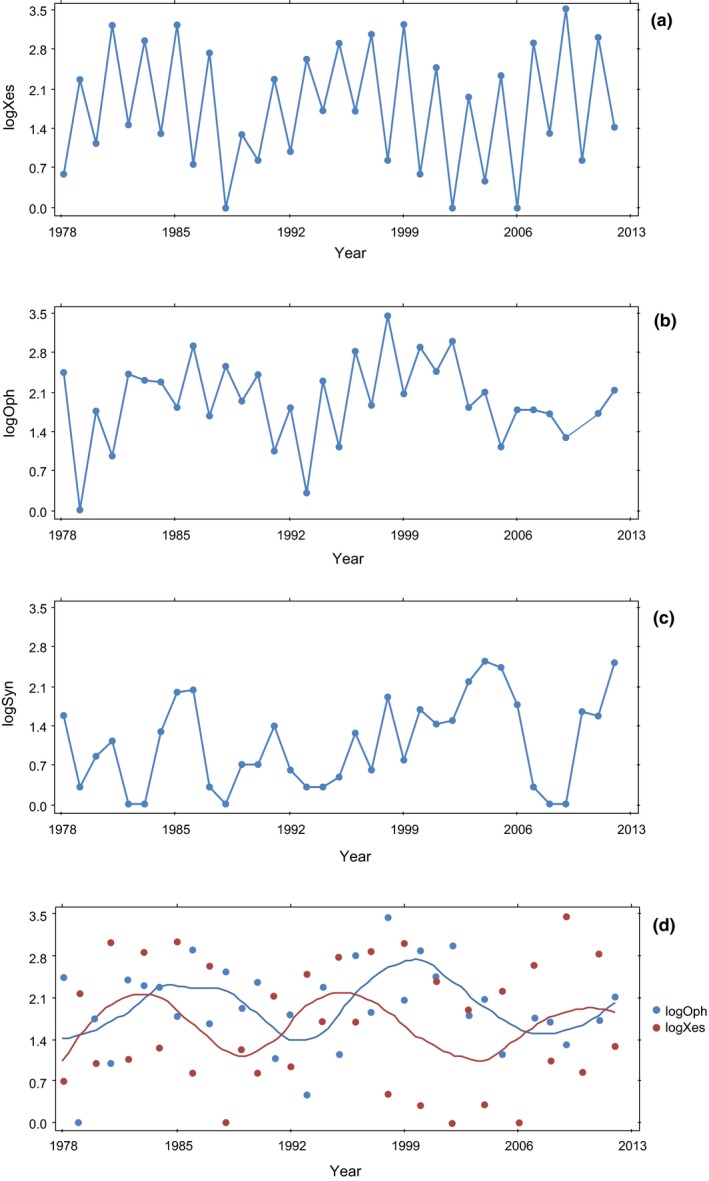
Time series of Xestia (a), Ophion (b), Syngrapha interrogationis (c) and the smoothed time series for Xestia + Ophion (d).
The time series suggests a longer cyclic component with a roughly 15‐year cycle period (Figure 1d), a pattern already noted by Várkonyi et al. (2002) and now strengthened by the current longer time series.
Species diversity of Ophion
Previous morphological studies (Rost et al. 2001; Várkonyi et al. 2002) assumed the presence of just a single species of Ophion (O. luteus) in the study area. We tested this assumption based on the COI sequences obtained from 180 of the 190 individuals that were analysed. Maximum p‐distance divergence within the 180 Ophion was 12.58%. Sequences for the O. luteus complex formed five distinct clusters with the morphologically distinct O. costatus comprising a sixth group (Figure 2). Maximum p‐distance divergences within each cluster ranged from 0% to 0.46%, while the minimum divergences between the clusters varied from 1.96% to 12.19% (Figure 2). Based on Johansson & Cederberg (2019), most (69%) of the specimens of Ophion actually represent O. kevoensis Jussila, 1965 (cluster #1), while 26% represent O. inclinans Johansson, 2019 (cluster #3). Ophion broadi Johansson, 2019 (cluster #2), Ophion sp. (cluster #4) and O. tenuicornis Johansson, 2019 (cluster #5) were uncommon, being represented by two, one, and five individuals, respectively (Figure S2).
Figure 2.
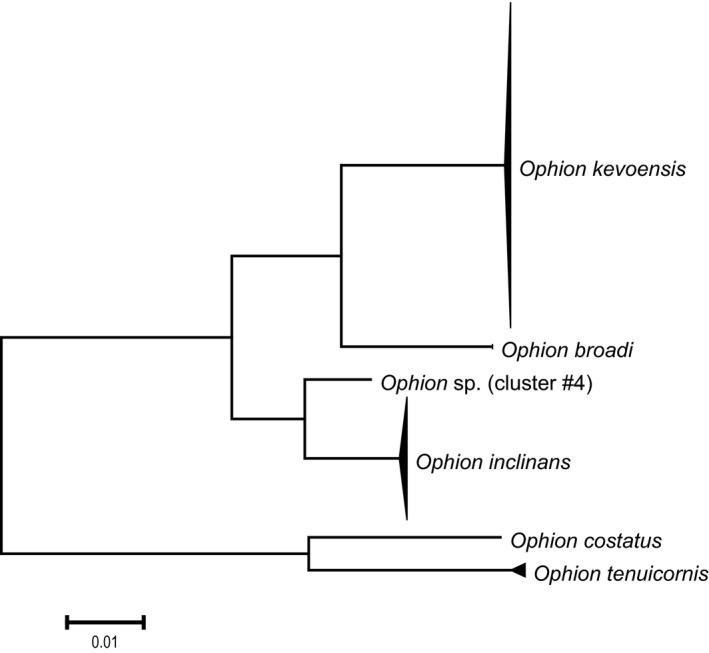
Neighbour‐Joining (p‐distance) tree for the Ophion. Height of the triangle is proportional to the sampling intensity, and its depth to the genetic variability within the species.
The relative abundance of the two dominant Ophion species (O. kevoensis and O. inclinans) was not random with respect to odd and even years. Just four individuals of O. kevoensis were collected in odd years versus 120 in even years. By contrast, 34 individuals of O. inclinans were collected in odd years versus 13 in even years. Considering all five species, the difference between the years is highly significant for O. kevoensis (χ2 = 69.5, P < 0.0001), whereas in O. inclinans the difference is only marginally so (χ2 = 4.6, P = 0.03).
Host species revealed by MAPL
Gut contents of 50 Ophion individuals produced a lepidopteran sequence that derived from 11 different species. Many (35) of these cases involved taxa that are very improbable hosts, suggesting contamination. Fifteen cases involved species that are too small to be hosts (Tortricidae: 4 species, 12 observations; Gelechiidae: 1 species, 1 observation, Geometridae: 2 species, 2 observations). Further 20 cases involved the noctuid Lithomoia solidaginis that is an unlikely host due to its life history. Ophion lay their eggs in the larvae of Lepidoptera (e.g., Quicke 2015), while Lithomoia overwinters as an egg (Mikkola & Jalas 1977) so its larvae are unavailable for the autumnal Ophion at Värriö.
The recovery of these sequences might reflect Ophion contaminated by other insects simultaneously attracted by the light trap. To test this possibility, we examined if the spurious hosts occurred in the same trap night as the Ophion specimen. For L. solidaginis, this was true in all 20 cases. Among the 170 cases where the Ophion gut content did not produce a L. solidaginis sequence, L. solidaginis was recorded in the same sample with an Ophion specimen in 101 cases. The 121 cases with co‐occurrence could be distributed among the 190 Ophion samples in ways. Out of these, are such for which the 20 co‐occurrences would have been found for those cases where the Ophion gut content produced L. solidaginis. Because the probability of obtaining this result by chance is , we consider it likely that the L. solidaginis host records reflect contamination of Ophion adults by other specimens in the trap.
The remaining 15 records represented three species, Syngrapha interrogationis (5), Xestia alpicola (5) and X. speciosa (5), which are all potential hosts based on their size and life history – although S. interrogationis, unlike the two Xestia species – has a 1‐yr life cycle so it is extremely unlikely to contribute to the alternate‐year dynamics of Ophion. All of these records derived from 2012. Among the 15 likely host records, 14 derived from Ophion kevoensis and one from Ophion inclinans (See Figure S2 for Ophion specimens with host confirmed). In the case of X. alpicola and X. speciosa, the host and Ophion never occurred together in a trap catch. In fact, no specimens of X. alpicola and only a single X. speciosa were captured during 2012. In the case of S. interrogationis, the host and Ophion occurred in the same trap in the same night in three of five cases. However, the first host records (13 August 2012) were made before the first moth was caught (16 August 2012) suggesting that S. interrogationis is likely a host.
Modelling results
Table 1 provides the parameter estimates for the Xestia–Ophion model. Parameters and are positively correlated (r = 0.42) in the posterior distribution, whereas other parameter pairs show no strong posterior correlations (maximum absolute correlation 0.06). The dynamics of the two‐species model are determined by and the parameter combination (Rost et al. 2001), of which the latter could not be estimated accurately (Table 1). Between these parameters, there is more uncertainty in the estimate of the host carrying capacity than for parasitoid searching efficiency .
Table 1.
Parameter estimates of the Xestia–Ophion model (X–O; eqns (1), (2)), the full Xestia‐Ophion–Syngrapha model (X–O–S full; eqns (5), (6), (7)), and the reduced Xestia–Ophion– Syngrapha model (X–O–S reduced; eqns (8), (9), (10)). All values show posterior median estimate (95% credibility interval)
| Model/parameter | X–O | X–O–S full | X–O–S reduced | |
|---|---|---|---|---|
|
|
2.5 (2.1…4.9) | 2.5 (2.1…4.6) | 2.5 (2.0…4.9) | |
|
|
0.12 (0.08…1.40) | 0.11 (0.07…0.93) | 0.12 (0.08…1.21) | |
|
|
2800 (2000…1.6 · 106) | 2500 (1900…6.7 · 105) | 2700 (2000…2.1 · 107) | |
|
|
410 (190…2.7 · 105) | 310 (170…1.1 · 105) | 400 (190…3.4 · 106) | |
|
|
1.41 (1.35…1.66) | 1.29 (1.24…1.45) | 1.44 (1.39…1.62) | |
|
|
NA | 0.004 (3 · 10‐5…0.91) | 2·10‐4(4 · 10‐6…0.26) | |
|
|
NA | 0.003 (0.002…0.009) | NA | |
|
|
NA | 210 (17…2.8 · 109) | 32 (1…1.6 · 109) |
Figure 3 shows the bifurcation diagram for Xestia and Ophion dynamics in the Xestia–Ophion model. We assume here the estimated values = 2.5 and = 2800 (Table 1), and vary the value of the parasitoid searching efficiency (on the horizontal axis). The diagram shows six distinct phases (Roman numerals I–VI in Figure 3). The dynamics representative of each phase are illustrated in Figure S3. For very small values of (Phase I), the parasitoid is at a stable equilibrium at a very low level and the host shows a two‐point cycle, generated by its intrinsic dynamics (note that is greater than 2), which are apparently synchronised by the interaction with the parasitoid. Somewhat greater values of (Phase II) lead to stable dynamics, with equally large odd‐year and even‐year cohorts. For still larger values of (Phase III), the dynamics become cyclic, but without a 2‐year component. For still larger values of (Phase IV), the dynamics turn into a 4‐point cycle, with 2 peak‐year and 2 low‐year densities, hence essentially representing a clear abundance difference between the two cohorts. Still larger values (Phase V), such as the posterior median estimate of , lead to more complex dynamics, which involve two main components, the alteration between the peak and low years, and a longer cyclic component, roughly 15 years in length. Finally, in Phase VI the dynamics are similar to Phase I, with the host showing a 2‐point cycle and the parasitoid stable dynamics at very low density. Figure 4 shows the predicted dynamics for the posterior median parameter values. The dynamics are qualitatively similar to the observed ones (Figure 1). Interestingly, the model replicates both the 2‐year cyclic component (Figure 4ac), and the longer‐term (ca. 15 years) cyclic component (Figure 4bd).
Figure 3.
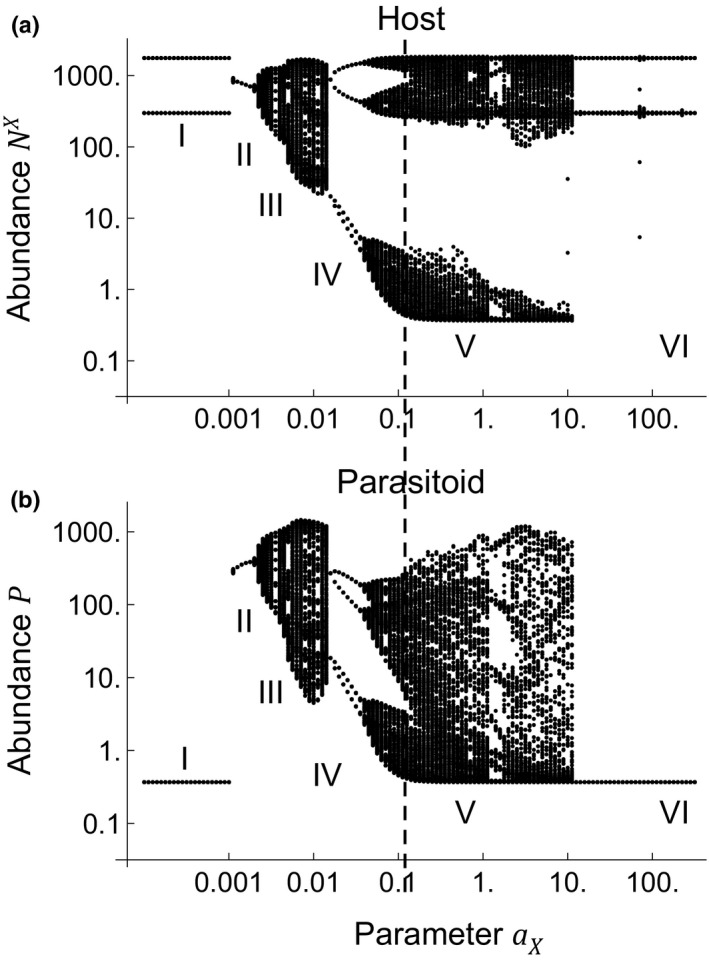
A bifurcation diagram of the Xestia–Ophion model. Panel a shows the host Xestia (eqn 1) while panel b shows the parasitoid Ophion (eqn. 2). We assumed the posterior median parameter values for the carrying capacity and growth rate , and varied search efficiency parameter . For each parameter value, we simulated the dynamics for 500 years, out of which the first 200 were ignored as a transient. The dashed vertical line shows the posterior median estimate of the parameter αx. The Roman numerals I‐VI refer to different phases of the bifurcation diagram. Note the logarithmic scales in both axes.
Figure 4.
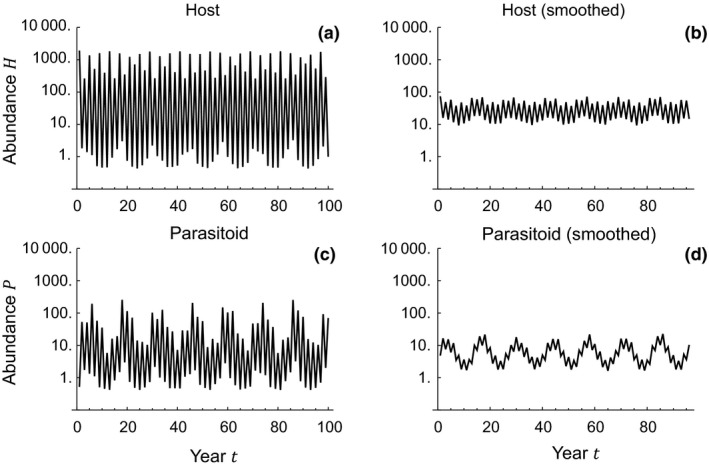
The predicted dynamics of the Xestia–Ophion model for posterior median parameter values (, , ). The left‐hand panels (a and c) show the yearly dynamics of the host and parasitoid, while the right‐hand panels (b and d) show the same data smoothed by a 5‐year moving average.
The estimates of the shared parameters are similar for the Xestia–Ophion model and the Xestia–Ophion–Syngrapha full model, suggesting that the presence of Syngrapha does not greatly affect the dynamics of Ophion and Xestia. Both the growth rate and the carrying capacity remained poorly estimated. The full model yielded higher log‐likelihood values than the reduced model, the difference being ca. 5 units for the response variable , ca. 10 units for the response variable , and ca 2 units for the response variable (Figure 5). However, the observed values are not distinct from null distributions, A and B, which moreover did not differ substantially from each other. Thus, the time‐series data do not provide evidence for Syngrapha being an ecologically important alternative host for Ophion in the sense that their dynamics would be strongly coupled.
Figure 5.
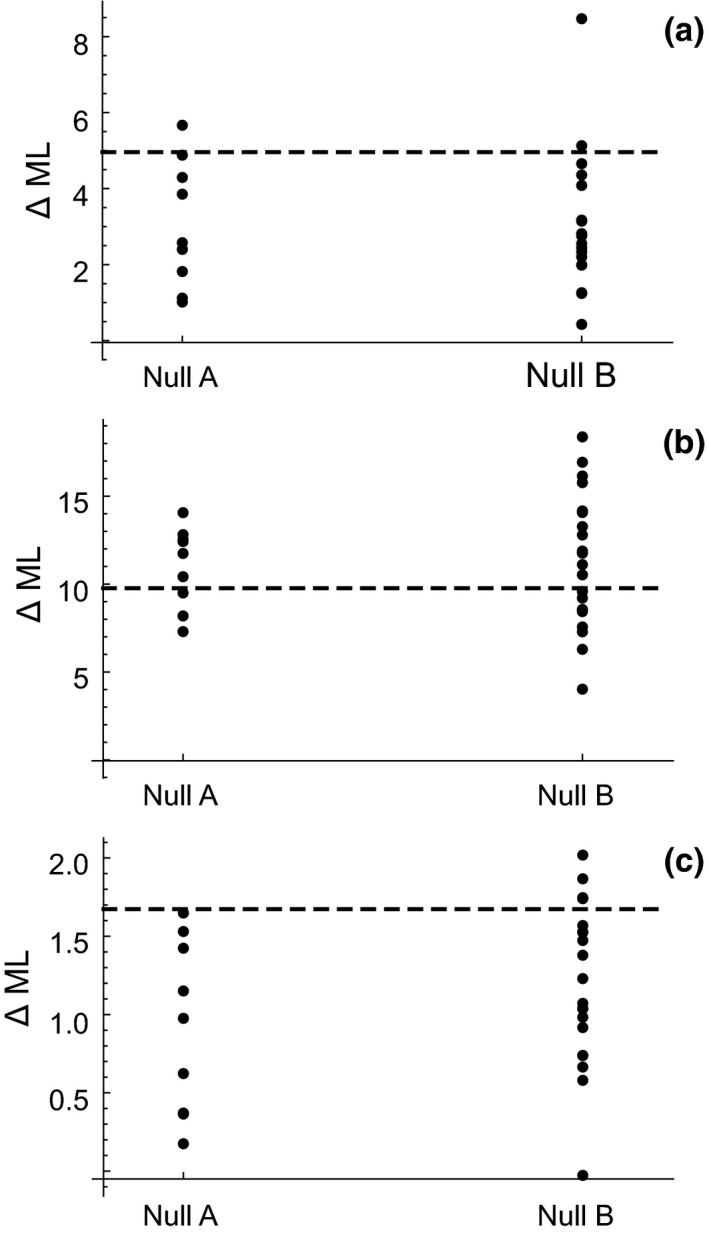
Statistical tests examining whether the time‐series data support the alternative host hypothesis for Syngrapha. The difference in log‐transformed maximal likelihood between the full and reduced models (ML). The panels correspond to the likelihoods computed for the response variables (a), (b), and (c). The dashed line shows the value for the data, and the dots show the Null Distributions A and B (see text).
Discussion
The statistical modelling of the time‐series data provided very strong evidence for a host–parasitoid interaction between Xestia and Ophion. The host–parasitoid model, when fitted to the data, replicated the very regular 2‐year periodic behaviour of the host (Figure 1a vs. Figure 4a), and the somewhat less regular 2‐year cyclic behaviour of the parasitoid (Figure 1b vs. Figure 4c), as well as the longer cyclic component apparent in the empirical data (Figure 1d vs. Figure 4b,d). By contrast, modelling did not give additional support for Syngrapha being an alternative host of Ophion, as including Syngrapha in the model did not improve the likelihood of observing the data more than expected by chance. One novelty of our statistical methodology is that we quantified the level of likelihood increase expected by chance under two ecologically meaningful null hypotheses, in which we either permuted Syngrapha data over the years, or replaced Syngrapha data with data from other species for which there was no molecular or other evidence for their role as potential hosts. As both types of null distributions generate the same result, our conclusion of the data not supporting Syngrapha as a dynamically important alternative host can be considered robust.
Our study demonstrates that DNA barcode reference libraries coupled with molecular techniques enable the dissection of species interactions in an unprecedented way. The present study benefitted from the fact that reference barcodes were available for all species of Lepidoptera known from Finland. Our study is among the first to apply the MAPL approach (Rougerie et al. 2011) to clarify host–parasitoid relationships, and we did so to resolve an enigma that had eluded resolution despite years of fieldwork and many rearing and laboratory experiments. The reasons for earlier failures remain unclear, but the possibilities include the (1) low probability of finding larvae of the rare cohort where the percent parasitism is presumed to be high and conversely the presumed very low parasitism rate in the common cohort, (2) changes in the behaviour of parasitised larvae which reduce their chances of discovery, or (3) high winter mortality of parasitised larvae under laboratory conditions. Since high‐throughput sequencing permits the simultaneous acquisition of large amounts of data from many specimens, such approaches will soon be widely used to probe species interactions. A huge advantage of molecular methods is their capacity to simultaneously determine both host and parasitoid species from the same specimen. Since parasitoids ordinarily cause the death of their host, such data were previously difficult to obtain.
Our DNA barcode analysis of 180 Ophion sp. cf. O. luteus revealed five lineages showing deep sequence divergences. Although deep intraspecific divergences in mtDNA may result from other mechanisms, such as incomplete lineage sorting or Wolbachia‐mediated spread of a distinct mtDNA sequence in a population through introgression (Funk & Omland 2003; Hurst & Jiggins 2005), we initially expected these lineages to represent five distinct but cryptic species. Three observations supported this conclusion. First, variation between the clusters is very high (14%) while intracluster is less than 0.5% in each case. Clusters are separated from each other by > 2% divergence with the mean divergence between closest clusters being 4.6%. Second, the five clusters were widely scattered among the many known species of the Ophion in a Neighbour‐Joining tree with all data accessible in BOLD. Such phylogenetically deep polyphyly in mtDNA is rare in DNA barcode data; most reported cases likely represent cryptic diversity (Mutanen et al. 2016). Thirdly, the Ophion clusters showed non‐random occurrence patterns in odd and even years. Furthermore, our data suggest that these taxa parasitise different hosts. During the review of this manuscript, a taxonomic revision of Swedish Ophion was published (Johansson & Cederberg 2019), enabling us to assign four of our five clusters to a named species. Both the host observations and host–parasitoid dynamics suggest that O. kevoensis is the main, or perhaps the only, parasitoid of periodic Xestia moths in the region. Perhaps the other four Ophion species occasionally parasitise Xestia, but O. inclinans must use a different species of Lepidoptera as its host. The presence of multiple species means that our modelling of Xestia–Ophion dynamics is based on empirical time series that contain noise because some of the Ophion specimens likely belong to species that do not parasitize Xestia. However, this weakness was not strong enough to blur resolution of the 2‐year dynamics in the Xestia–Ophion system.
In conclusion, by coupling DNA‐based approaches with long time series of both hosts and parasitoids, we obtained strong evidence that host–parasitoid interactions underpin the striking periodic occurrence of Xestia. We demonstrated that Ophion parasitoids have a crucial role in maintaining this system and showed that seemingly simple biological systems may be unexpectedly complex once cryptic diversity is evaluated. Furthermore, we showcased how DNA‐based methods can reveal food webs in unprecedented detail. For future directions and further elucidating the Xestia–Ophion system, we propose (1) reducing incidence of false positives in the dietary analyses by collecting parasitoids in a way that minimises contact with other species and using disinfectant techniques to remove external DNA and/or dissecting the gut tract; (2) examining a large number of Ophion specimens with high‐throughput technologies to advance understanding of their diversity and distribution; and (3) extending the investigations by Várkonyi et al. (2002) and Várkonyi (2003) whether other natural enemies of Xestia participate in the striking periodic dynamics of Xestia and Ophion.
Authorship
MM and IH designed the study. MM and GV designed and conducted the specimen sampling. JI identified moths from the Värriö monitoring programme. JI and GV separated Ophion wasps. OO and IH formulated the statistical time‐series models and OO fitted them to the data and performed the analyses. SP and PH conducted DNA sequencing while MM and SP performed the sequence analyses. MM, IH and OO drafted the manuscript and all other authors aided in its revision.
Supporting information
Fig S1
Fig S2
Fig S3
Table S1
Appendix S1
Supplementary Material
Acknowledgements
This study was enabled by support from the Academy of Finland (Grant 284601 to IH, Grant 309581 to OO and Grant 277984 to MM), the Jane and Aatos Erkko Foundation (Grant to OO), and the Research Council of Norway through its Centres of Excellence Funding Scheme (223257) to OO via the Centre for Biodiversity Dynamics. Sequence analyses were largely covered by a Discovery grant to PDNH from NSERC. We are grateful to Niklas Johansson (Habo, Sweden) for taxonomic help with Ophion and Bess Hardwick (Helsinki, Finland) and Pirjo Appelgren (Kuhmo, Finland) for their assistance with the figures. Similarly, we are grateful to the staff of Värriö research station as well as Arja Itämies and Päivi Tanner and for taking care of insect collection and pre‐sampling of specimens. Finally, we want to express our gratitude to three anonymous reviewers for useful comments that significantly improved the quality of our paper.
DATA AVAILABILITY STATEMENT
All specimen and sequence data are publicly available through the BOLD dataset DS‐OPGUT at dx.doi.org/10.5883/DS‐OPGUT and from the Dryad Digital Repository: https://doi.org/10.5061/dryad.nzs7h44nh.
References
- Broad, G.R. , Schnee, H. & Shaw, M.R. (2015). The hosts of Ophion luteus (Linnaeus) (Hymenoptera, Ichneumonidae, Ophioninae) in Europe. J. Hymenopt. Res., 46, 115–125. [Google Scholar]
- Brock, J.P. (1982). A systematic study of the genus Ophion in Britain (Hymenoptera, Ichneumonidae). Tijdschr. Entomol., 125, 57–97. [Google Scholar]
- Bulmer, M.G. (1977). Periodical insects. American Naturalist, 111, 1099–1117. [Google Scholar]
- Funk, D.J. & Omland, K.E. (2003). Species‐level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst., 34, 397–423. [Google Scholar]
- Hajibabaei, M. , deWaard, J.R. , Ivanova, N.V. , Ratnasingham, R. , Dooh, R.T. , Kirk, S.L. et al (2005). Critical factors for assembling a high volume of DNA barcodes. Philosophical Transactions of the Royal Society B‐Biological Sciences, 360, 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajibabaei, M. , Smith, M.A. , Janzen, D.H. , Rodriguez, J.J. , Whitfield, J.B. & Hebert, P.D.N. (2006). A minimalist barcode can identify a specimen whose DNA is degraded. Mol. Ecol. Notes, 6, 959–964. [Google Scholar]
- Hebert, P.D.N. , Cywinska, A. , Ball, S.L. & deWaard, J.R. (2003). Biological identifications through DNA barcodes. Proc R. Soc. Lond. B., 270, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliövaara, K. & Väisänen, R. (1986). Bugs in bags: intraspecific competition affects the biogeography of the alternate‐year populations of Aradus cinnamomius (Heteroptera). Oikos, 47, 327–334. [Google Scholar]
- Heliövaara, K. , Väisänen, R. & Simon, C. (1994). Evolutionary ecology of periodical insects. Trends Ecol. Evol., 9, 475–480. [DOI] [PubMed] [Google Scholar]
- Hurst, G.D.D. & Jiggins, F.M. (2005). Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc. R. Soc. B., 272, 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova, N.V. , deWaard, J.R. & Hebert, P.D.N. (2006). An inexpensive, automation‐friendly protocol for recovering high‐quality DNA. Mol. Ecol. Notes, 6, 998–1002. [Google Scholar]
- Jalas, I. (1975). Perhostenkeräilijän opas. Keuruu, p. 256. [Google Scholar]
- Johansson, N. & Cederberg, B. (2019). Review of the Swedish species of Ophion (Hymenoptera: Ichneumonidae: Ophioninae) with the description of 17 new species and an illustrated key to Swedish species. European Journal of Taxonomy, 550, 1–136. [Google Scholar]
- Kankare, M. , Várkonyi, G. & Saccheri, I. (2002). Genetic differentiation between alternate‐year cohorts of Xestia tecta (Lepidoptera, Noctuidae) in Finnish Lapland. Hereditas, 136, 169–176. [DOI] [PubMed] [Google Scholar]
- Mikkola, K. (1976). Alternate‐year flight of northern Xestia species (Lep., Noctuidae) and its adaptive significance. Ann. Entomol. Fenn., 42, 191–199. [Google Scholar]
- Mikkola, K. & Jalas, I. (1977). Suomen Perhoset. Yökköset, Vol. 1 Helsinki, p. 256. [Google Scholar]
- Mikkola, K. & Kononenko, V.S. (1989). Flight year of the alternate‐year Xestia moths (Lepidoptera, Noctuidae) in north‐eastern Siberia – A character from the Ice Ages? Nota lepidop., 12, 144–152. [Google Scholar]
- Mutanen, M. , Kekkonen, M. , Prosser, S.W.J. , Hebert, P.D.N. & Kaila, L. (2015). One species in eight: DNA barcodes from type specimens resolve a taxonomic quagmire. Mol. Ecol. Resour., 15, 967–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutanen, M. , Kivelä, S.M. , Vos, R.A. , Doorenweerd, C. , Ratnasingham, S. , Hausmann, A. et al (2016). Species‐level para‐ and polyphyly in DNA barcode gene trees: Strong operational bias in European Lepidoptera. Syst. Biol., 65, 1024–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliainen, E. & Itämies, J. (1988). Xestia communities (Lepidoptera, Noctuidae) in eastern Finnish Forest Lapland as indicated by light trapping sampling. Holarctic Ecol., 11, 235–240. [Google Scholar]
- Quicke, D.L.J. (2015). The braconid and ichneumonid parasitoid wasps. Biology, systematics, evolution and ecology. John Wiley & Sons, Chichester, UK, p. 681. [Google Scholar]
- Rost, M. , Várkonyi, G. & Hanski, I. (2001). Patterns of 2‐year population cycles in spatially extended host–parasitoid systems. Theor. Popul. Biol., 59, 223–233. [DOI] [PubMed] [Google Scholar]
- Rougerie, R. , Smith, M.A. , Fernandez‐Triana, J.L. , Lopez‐Vaamonde, C. , Ratnasingham, S. & Hebert, P.D.N. (2011). Molecular analysis of parasitoid linkages (MAPL): gut contents of adult parasitoid wasps reveal larval host. Mol. Ecol., 20, 179–186. [DOI] [PubMed] [Google Scholar]
- Smith, M.A. , Woodley, N. , Hallwachs, W. , Janzen, D.H. & Hebert, P.D.N. (2006). DNA barcodes reveal cryptic host‐specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc. Natl Acad. Sci. USA, 103, 3657–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.A. , Wood, D.M. , Janzen, D. , Hallwachs, W. & Hebert, P.D.N. (2007). DNA barcodes affirm that 16 species of apparently generalist tropical parasitoid flies (Diptera, Tachinidae) are not all generalists. Proc. Natl Acad. Sci. USA, 104, 4967–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.A. , Rodriguez, J.J. , Whitfield, J.B. , Deans, A.R. , Janzen, D.H. , Hallwachs, W. et al (2008). Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proc. Natl Acad. Sci. USA, 105, 12359–10.1073/pnas.0805319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.A. , Fernández‐Triana, J.L. , Eveleigh, E. , Gómez, J. , Guclu, C. , Hallwachs, W. et al (2013). DNA barcoding and the taxonomy of Microgastrinae wasps (Hymenoptera, Braconidae): impacts after eight years and nearly 20,000 sequences. Mol. Ecol. Resour., 13, 168–76. [DOI] [PubMed] [Google Scholar]
- Várkonyi, G. (2003). Population biology of periodic Xestia moths. Academic dissertation. University of Helsinki., 100, pp. http://urn.fi/URN:ISBN:952‐10‐1319‐2. [Google Scholar]
- Várkonyi, G. , Hanski, I. , Rost, M. , & Itämies, J., (2002). Host‐parasitoid dynamics in periodic boreal moths. Oikos, 98, 421–430. [Google Scholar]
- Wirta, H. , Hebert, P.D.N. , Kaartinen, R. , Prosser, S.W. , Várkonyi, G. & Roslin, T. (2014). Complementary molecular information changes our perception of food web structure. Proc. Natl Acad. Sci. USA, 111, 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Table S1
Appendix S1
Supplementary Material
Data Availability Statement
All specimen and sequence data are publicly available through the BOLD dataset DS‐OPGUT at dx.doi.org/10.5883/DS‐OPGUT and from the Dryad Digital Repository: https://doi.org/10.5061/dryad.nzs7h44nh.


