Abstract
We tested whether lifelong modification of vitamin D signaling can alter the progression of early prostate carcinogenesis in studies using mice that develop high-grade prostatic intraepithelial neoplasia (HGPIN) that is similar to humans. Two tissue-limited models showed that prostate vitamin D receptor (VDR) loss increased prostate carcinogenesis. In another study, we fed diets with three vitamin D3 levels (inadequate = 25 IU/kg diet, adequate for bone health = 150 IU/kg, or high = 1000 IU/kg) and two calcium levels (adequate for bone health = 0.5% and high = 1.5%). Dietary vitamin D caused a dose-dependent increase in serum 25-hydroxyvitamin D levels and a reduction in the percentage of mice with adenocarinoma but did not improve bone mass. In contrast, high calcium suppressed serum 1,25-dihydroxyvitamin D levels and improved bone mass but increased the incidence of adenocarcinoma. Analysis of the VDR cistrome in RWPE1 prostate epithelial cells revealed vitamin D-mediated regulation of multiple cancer-relevant pathways. Our data support the hypothesis that the loss of vitamin D signaling accelerates the early stages of prostate carcinogenesis and our results suggest that different dietary requirements may be needed to support prostate health or maximize bone mass.
Keywords: diet, calcium, vitamin D receptor, prevention, carcinogenesis
Introduction
Serum 25-hydroxyvitamin D (25OHD) concentrations beyond those needed for bone health may reduce cancer risk (1). However, the evidence relating vitamin D status to prostate cancer risk in humans is mixed. Early association studies with small sample size were supportive (2) but larger population-based studies have shown protective (3), conditional (4,5), and negative (6,7) effects of increased vitamin D status on prostate cancer. More recently, the VITAL trial showed that 2000 IU vitamin D3 per day over a median follow-up of 5.3 y reduced the hazard ratio for prostate cancer to 0.88 (8). However, the intervention was short relative to the process of human prostate carcinogenesis and since few cases were documented, the comparison was underpowered and not significant (8). There is also evidence that high dietary calcium (Ca) intake, a nutrient known to suppress renal production of the vitamin D hormone, 1,25-dihydroxyvitamin D (1,25(OH)2D), may increase prostate cancer risk (9). Thus, while the overall body of literature suggests a protective role for vitamin D on prostate cancer risk, the exact nature of the relationship, the protective serum vitamin D metabolite concentrations, and the mechanism of protective action have been elusive.
Animal and cell studies have provided essential proof for a direct causal relationship between vitamin D and prostate cancer development. In cultured prostate epithelial cells or prostate cancer cells, 1,25(OH)2D suppresses proliferation, induces apoptosis, and promotes differentiation (10). Consistent with these data, vitamin D deficiency (11) and VDR gene deletion (12) increase, while injections with 1,25(OH)2D or vitamin D analogs suppress (13), prostate tumor growth in various animal models. However, experiments linking prostate cancer development to human-relevant ranges of vitamin D status or Ca intake are limited (14–16).
Previously, we determined the vitamin D3 intake necessary to model human vitamin D status in mice (17) and found that intake as low as 100 IU vitamin D3/kg diet is sufficient to maintain the traditional vitamin D functions of bone growth and mineralization. Using this information, we showed that dietary vitamin D deficiency increased prostate epithelial cell (PEC) proliferation, reduced PEC apoptosis, and increased the incidence of HGPIN lesions in mice (18). Here, we report studies that extend our earlier work and directly address whether lifelong variation in the dietary levels of vitamin D and Ca can modify early stage prostate cancer. In addition, we examine the importance of signaling through the VDR during early prostate carcinogenesis and we identify potential candidate genes mediating the action of vitamin D on the prostate epithelial cell during carcinogenesis.
Materials and Methods
I. Animals:
In our studies we used TgAPT121 mice (TgAPT121+/−, C57BL/6 × DBA/2 background) (19) bred to male B6D2F1 mice (Jackson Laboratories, Bar Harbor, ME). This model has probasin promoter driven, prostate epithelial cell-specific expression of a truncated SV40 Large T antigen protein that inactivates pRb family proteins. It has strong histologic similarities to the earlier stages of human prostate cancer (i.e. demonstrating hyperplasia, PIN, and adenocarcinoma over 6 months). Other mouse models used include mice with a floxed exon 2 in the Vdr gene (Vdrfl/fl; C57BL/6) (18); mice with Cre recombinase expression driven by the rat probasin promoter (PB-Cre+/−; C57BL/6) (NCI Mouse Models of Human Cancer Consortium, # 01XF5; Frederick, MD); and Vdr knockout mice with intestine-specific transgenic expression of the human VDR gene (C57BL/6) (20). Mice were genotyped as previously described (18–21). Mice were housed with a 12 h light/12 h dark cycle, in shoebox cages with individual ventilation. Lights were covered with a UVB filter (Pegasus Associates Lighting, Beaver, PA). Diets and water were fed ad libitum. Except for Experiment 4, mice were fed AIN-76A diet (1000 IU vitamin D3/kg diet (22)). All experiments were approved by either the Purdue University or the Ohio State University Animal Care and Use Committees.
II. Experimental design
Experiment 1: Characterization of TgAPT121 mice.
For a microarray study, male TgAPT121+/− (n = 8) and TgAPT121−/− (n = 16) mice were used. After an overnight fast, 12 wk old mice were sacrificed and prostate lobes were dissected on ice, snap frozen in liquid nitrogen, and stored at −80 °C. The anterior prostates from these mice were used for microarray analysis while the dorsolateral prostate lobes were assessed for Vdr mRNA level by qPCR and VDR protein level by Western blot analysis. Prostates from a second 12 wk old cohort of 10 TgAPT121+/− and 4 TgAPT121−/− mice were used for histology and immunohistochemistry.
Experiment 2: Prostate epithelial cell-specific Vdr gene deletion on prostate cancer.
The TgAPT121+/−, Vdrfl/fl, and PB-Cre+/− lines were crossed to make mice with prostate epithelial cell-specific Vdr gene deletion (PEC-VDR KO: TgAPT121+/−,Vdrfl/fl,PB-Cre+/−; n = 32) and those with normal Vdr gene status (Cre negative littermate controls: TgAPT121+/−,Vdrfl/fl,Pb-Cre−/−; n = 33). At 28 wks of age, mice were sacrificed and the prostate was harvested and prepared for histology.
Experiment 3: Whole prostate Vdr gene deletion on prostate cancer.
TgAPT121 mice were crossed to Vdr knockout mice with intestine-specific transgenic expression of a hemaglutinin-tagged human VDR to generate mice lacking VDR in all cells in the prostate (HV2-VDR KO: TgAPT121+/−,HV2+/−,VDR−/−; n = 23) and littermate controls (TgAPT121+/−,HV2+/−,VDR+/−; n = 27). Transgenic expression of VDR in the intestine prevents abnormal Ca metabolism in Vdr knockout mice (20). At 26 wks of age, mice were sacrificed and the prostate was harvested and prepared for histology.
Experiment 4: Impact of diet on prostate tumorigenesis.
Male TgAPT121 transgenic mice were generated at Purdue University then shipped to Ohio State University at weaning. Mice were randomly assigned to one of 6 AIN-76A-based diets with varying levels of dietary vitamin D3 (25, 150, and 1000 IU /kg diet) and Ca (0.5, 1.5%) in a 2 × 3 factorial design (n = 34 mice per group). Diets and water were fed ad libitum. Mice were sacrificed at 28 wks of age when blood, the right femur, and the prostate was harvested. Serum was prepared from blood and a randomly selected subset of samples was analyzed for vitamin D metabolites (n=10 per diet group). Intact femora were prepared for DEXA analysis (n=25–27 per diet group). Prostates were processed for histology.
Experiment 5: VDR ChIP-seq analysis.
RWPE1 cells were cultured as we have described elsewhere (23). Cells were grown to 80% confluence and then treated with vehicle or 10 mM 1,25(OH)2D for 3 h. ChIP-seq and ChIP-qPCR analysis was conducted as described below.
III. Methods
A. Prostate preparation, histological grading, and scoring
The bladder, prostate, and seminal vesicles were removed en block and processed for histology as described previously (18). Histological examination of the anterior prostates was conducted using a modification of established guidelines (24) that better reflect the diversity of the early lesions in the TgAPT121 mouse prostate (see Table S1 and Fig. S1 for a more complete description of the grading criterion). Blinded assessment was conducted by one reviewer and was confirmed by a second reviewer.
Slides were digitized and the images were saved (ScanScope CS; Aperio, Vista, CA). For experiments 2 and 3, lesions were marked on the digital images using the ImageScope Viewer (Aperio). The area covered by each lesion and the numbers of independent foci were determined and the data were exported for further analysis. Total tissue area = the sum of the areas of all lesion foci plus the area covered by normal epithelium. Percent area = the proportion of total tissue area exhibiting the features of a lesion. Lesion Count = the number of independent lesion foci. Incidence = the percentage of mice in a group with a particular lesion. Average Lesion size = the ratio of the total area for a lesion type over the count of the lesion type.
For experiment 4, slides for the 204 mice were visually examined and scored for their highest histologic phenotype. Data were reported for the percentage of mice in a group with adenocarcinoma as their most severe lesion.
B. Immunohistochemical (IHC) analyses
a. p53:
Unstained sections were deparaffinized and rehydrated, followed by antigen retrieval for 30 min (Antigen Retrieval Citra Plus Solution, BioGenex, San Ramon, CA). Endogenous peroxidase activity was quenched (DAKO® Peroxidase Blocking Reagent, DAKO, Carpinteria, CA). Sections were incubated with rabbit polyclonal anti-mouse p53 antibody (NCL-p53-CM5p, 1:500; Leica Microsystems GmbH, Vienna, Austria) for 1 h at room temperature, followed by 30 min with DAKO EnVision+ Labeled Polymer-HRP anti-rabbit IgG secondary antibody. Antibody binding was visualized using the DAKO EnVision+ DAB+ Chromogen Solution and slides were counterstained with hematoxylin.
b. Apoptosis:
Unstained sections were evaluated by TUNEL staining with the ApopTag Plus Peroxidase In Situ Apoptosis Detection Kit following the manufacturer’s instructions (Millipore Corp., Billerica, MA). Slides were counterstained with methyl green.
c. Quantification:
3–5 representative, non-overlapping images without artifacts were captured by a high-resolution digital camera using bright field microscopy (Nikon ECLIPSE E800, Tokyo, Japan) and specific outcomes were analyzed using Image-Pro Plus 7.0 (Media Cybernetics, Inc., Bethesda, MD). A labeling index was calculated as the number of p53 or TUNEL positive nuclei divided by total number of nuclei in the fields examined: (%) = L / (L + C) × 100, where L = labeled nuclei and C = unlabeled nuclei.
C. Measurement of prostate VDR protein level:
Frozen dorsolateral prostates were homogenized on ice in lysis buffer (12 mM NaCl, 0.5% Nonidet P-40, 0.02 mM sodium orthovanadate, 5 mM Tris-HCl (pH 8.0), and one Protease Inhibitor Cocktail tablet per 5 ml (Roche)). Homogenates were centrifuged at 16,300 × g at 4 °C for 15 min and supernatants were harvested. Equal amounts of protein from 8 mice of each genotype were combined to generate two pooled samples. Samples (100 μg protein) were analyzed by Western blot analysis as previously described (23) and overnight incubation at 4 °C with mouse anti-human VDR antibody (1:500; D-6, sc-13133; Santa Cruz Biotechnology) or mouse anti-β-actin antibody (1:5000; AC-74; Sigma-Aldrich) followed by incubation with HRP-conjugated goat-anti-mouse IgG light chain antibody (1:5000; Jackson ImmunoResearch Laboratories; West Grove, PA) at room temperature for 1 h.
D. Prostate RNA preparation:
Frozen anterior prostates were crushed under liquid N2 and RNA was extracted with the SV Total RNA Isolation kit (Promega). Genomic DNA was removed by treating RNA with RQ1 RNase-Free DNase (Promega) prior to repurification of RNA with the RNeasy Mini Kit (Qiagen). RNA was quantified and RNA integrity was assessed visually from agarose gels.
E. Analysis of VDR mRNA levels:
RNA was reverse transcribed into cDNA and qPCR was conducted using primers and PCR conditions as we have previously reported (20).
F. Microarray analysis:
a. RNA isolation.
Equal amounts of RNA from two mice were pooled to yield 4 pooled TgAPT121+/− and 8 pooled TgAPT121−/− control samples. All samples used for microarray analysis had high RIN scores upon analysis on a Agilent Bioanalyzer. Gene expression profiling was conducted using the GeneChip® Mouse Gene 1.0 ST Array (ThermoFisher) and standard Affymetrix protocols at the Center for Medical Genomics at the Indiana University School of Medicine.
Microarray data were preprocessed using the Bioconductor package “xps” and data were normalized by the Robust Multichip Average method. The data for this experiment are available in GEO (GSE50662). All arrays passed quality control tests and were included in the analyses. The preprocessed data were filtered using the detection above background algorithm (p ≤ 0.05: score = 1; 0.05 < p ≤ 0.07: score = 0.6; p > 0.07: score = 0) and a probeset was called “present” if sum score was ≥ 4.8 (n=8, control) or 2.4 (n=4, TgAPT121+/−). Probesets “present” in at least one of the two genotype groups and those with a coefficient of variation < 50 were included in subsequent steps. We removed probesets with no gene annotation and only one probeset was used when multiple probesets were available. The final list contained 21,332 probesets. Differentially expressed transcripts were identified using the Significance Analysis of Microarray v3.09 (25) (log2-transformed data, two class, unpaired analysis, 900 permutations). Significant transcripts (false discovery rate (FDR) < 5%) with |fold change (FC)| > 1.5 were analyzed using MetaCore™ (Clarivate Analytics, Philadelphia, PA).
b. Reanalysis of data from Tomlins et al. (26).
Three normal prostate epithelial cells samples and 13 samples with HGPIN from GEO entry GSE6099 were examined for differential expression with GEO2R (15% FDR).
c. Reanalysis of Maund et al. (27) data.
CEL files from 1,25(OH)2D-treated (+/− 100 nM; 6 or 48 h) mouse prostate epithelial stem cells ((27) GEO entry GSE18993) were processed using RMAexpress (http://rmaexpress.bmbolstad.com/). One array was abnormal and was excluded from the reanalysis (GSM469894). Differential gene expression was determined with Significance Analysis for Microarray (10% FDR) within BRB ArrayTools v 4.5.1 (28) on data filtered to remove the transcripts whose variance was in the bottom 50th percentile.
G. Serum vitamin D metabolite analysis:
Serum 1,25(OH)2D and 25OHD were analyzed as previously described (20).
H. Bone analyses:
The right femur from each mouse was stripped of muscle and analyzed for bone mineral density (BMD; g/cm2) using DEXA (20).
I. ChIP-Seq analysis for VDR binding sites:
a. Sample preparation and ChIP-seq:
ChIP was conducted as previously described (29) using normal rabbit IgG polyclonal antibody (12–370, Millipore Sigma, Burlington, MA) or anti-VDR polyclonal antibody (C-20, sc-1008, Santa Cruz Biotechnology, Inc., Dallas, TX). ChIP-PCR (29) was used to identify three replicates with ligand-inducible VDR binding to the −252 to −51 bp region of the CYP24A1 gene promoter and no VDR binding to the GAPDH gene promoter. These were pooled by treatment to generate three samples (control treated/VDR IP, 1,25(OH)2D treated/VDR IP, and a pooled control + 1,25(OH)2D treated/IgG IP). ChIP was repeated on these samples and sent to the Michael Smith Genome Science Centre (Vancouver, Canada) for sequencing (library construction on 100–300 bp fragments, 36 bp single-end reads, Illumina sequencing (30)). The data for two lanes for each sample were pooled for analysis (GEO dataset GSE116843)
b. Bioinformatic analysis:
ChIP-seq data was analyzed as reported elsewhere (31). We also reanalyzed ChIP-seq data from THP-1 cells (+/− 100 nM 1,25(OH)2D, 2 h or 24 h), LPS-treated THP-1 cells (+/− 100 nM 1,25(OH)2D, 24 h), LS180 cells (100 nM 1,25(OH)2D, 3 h), and LX2 cells (100 nM 1,25(OH)2D, 16 h, (32) and GEO entry GSE53041) (Multi-cell analysis upon request). ChIP-seq peaks were annotated to the nearest neighbor gene and a search region of +/− 100 bp from the peak summit was examined for transcription factor binding motifs with HOMER (33).
RWPE VDR ChIP-seq peaks were compared to 1,25(OH)2D-regulated genes from: (a) RWPE1 cells (10% FDR (34)), (b) mouse prostate stem cells (10% FDR), and (c) 587 topologically associated domains (TADs) with vitamin D-regulated genes from THP-1 monocytes (35). Lists of genes with VDR binding sites were examined for function and pathway enrichment using the IPA pathway analysis tool (Qiagen).
c. ChIP-PCR validation:
80% confluent flasks of RWPE1 cells were treated with vehicle or 10 mM 1,25(OH)2D for 3 h (n=3/treatment). ChIP-qPCR was conducted as described above for peaks associated with MAPK6; CTSD/SYT8; DDIT4; KLF4; CA9; ETNK2/REN; and CYP24A1 (Fig. S2).
J. Statistical analysis of non-microarray data:
Statistical analyses were conducted using SPSS for Windows-v.19.0 (IBM Corp., Armonk, NY). In all experiments, differences were considered significant at p < 0.05, and a trend was defined as p < 0.10. Values are expressed as mean ± SEM of the non-transformed data. All sample size calculations were conducted for α=0.05 and β=0.8. In the tumor endpoint studies, 22 mice per group is necessary to detect 60% difference in the proportion of mice with adenocarcinoma using a Chi-Square test. For serum vitamin D metabolites, 10 mice per group is necessary to detect a significant main effect of diet or genotype of 100% based on a variance = 75% of the difference between means (17)). For bone, n = 17 mice is necessary to detect a significant main effect of diet or genotype of 4% based on a variance = difference between means (20).
The distribution of all continuous variables was examined by the Shapiro-Wilk test and Normal Q-Q plots and log- or square root transformations were used to achieve a normal distribution. If transformation did not normalize the distribution, the non-parametric Mann-Whitney U test was used. IHC and TUNEL data were analyzed by one-way ANOVA, followed by the Holm-Sidak step-down multiple comparison test to detect differences among lesion types. For the data from the mouse VDR deletion experiments and for Vdr mRNA levels, a Student’s t test was used to assess differences between genotype groups. For categorical variables in the cancer studies, the Chi-Square test or Fisher’s exact test were used to detect difference in frequencies. For the non-cancer endpoints in the diet study, data were analyzed by 2-way ANOVA to account for main effects and interactions between the dietary interventions.
Results
There were no observed adverse effects of the dietary interventions or genetic modifications on the growth, body weight, or appearance of the mice.
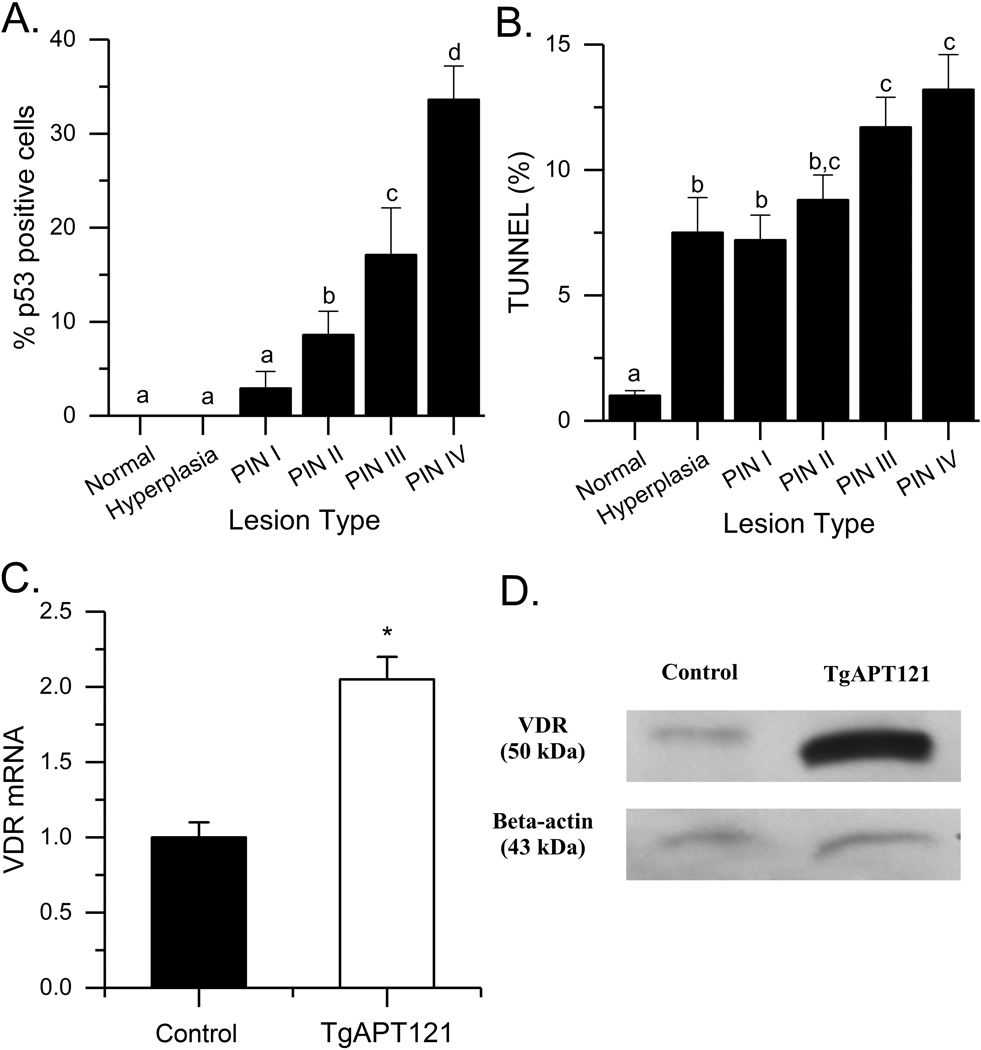
At 3 months of age the anterior prostate of TgAPT121 mice is filled with a mix of low grade (PIN I and II), and high grade (PIN III and IV) lesions (18). We used microarray analysis to determine whether the molecular phenotype of the anterior prostate in TgAPT121 mice adequately models the features of human HGPIN, the precursor lesion to human prostate cancer (36). TgAPT121 mouse prostates had 3440 upregulated and 2087-downregulated genes compared to normal prostate (data available upon request). There were similarities in the TgAPT121 mouse prostate transcriptome compared to published studies comparing normal prostate epithelium to HGPIN in humans, i.e. 17 of the 21 genes reported individually in the literature, 12 of 72 genes from (37), and 97 of 526 genes from (26) (Table S2). MetaCore™ analysis identified enriched pathways that link to prostate carcinogenesis within the TgAPT121 transcriptome including those regulating the DNA damage response, p53 signaling, apoptosis, TNF receptor signaling, TGFβ signaling, and signaling through the androgen receptor (Table S3). Bioinformatic analysis also found transcription factor networks centered on proto-oncogenic transcription factors (e.g. Myc, Ap-1, Creb1, Nfkb1, Ar, Egr1, Hnf4a) and Trp53 (encoding the protein p53). Increased p53 expression and apoptosis during the development of HGPIN lesions in TgAPT121 mouse prostate was confirmed by immunohistochemistry (Fig. 1A, 1B). Collectively, these analyses support the use of the anterior prostate from TgAPT121 as a model of human HGPIN.
Fig. 1.
Impact of prostate phenotype progression on (A) P53 levels, (B) TUNEL staining. Bars are mean ± SEM (n = 5–28 images/lesion type, n=4 Control, 10 TgAPT121 mice). Data were analyzed by ANOVA followed by the Holm-Sidak pairwise multiple comparisons. Bars without a common letter superscript differ significantly (p < 0.05). (C) Vdr mRNA level in the dosolateral prostates of TgAPT121 and control mice. Bars are mean ± SEM. (* p < 0.01, n = 8/genotype, Student’s t test). (D) VDR protein level in pooled dorsolateral prostates (n = 8/pool).
Vdr mRNA and protein levels were elevated in the dorsolateral lobe of 12 wk old TgAPT121+/− mice (Fig. 1C, D) and the microarray analysis shows that Vdr mRNA levels are also elevated in the anterior lobe (2.1 fold higher, Table S2). To test the importance of VDR signaling in the progression of early stages of prostate cancer in TgAPT121+/− mice, we conducted two VDR gene deletion studies.
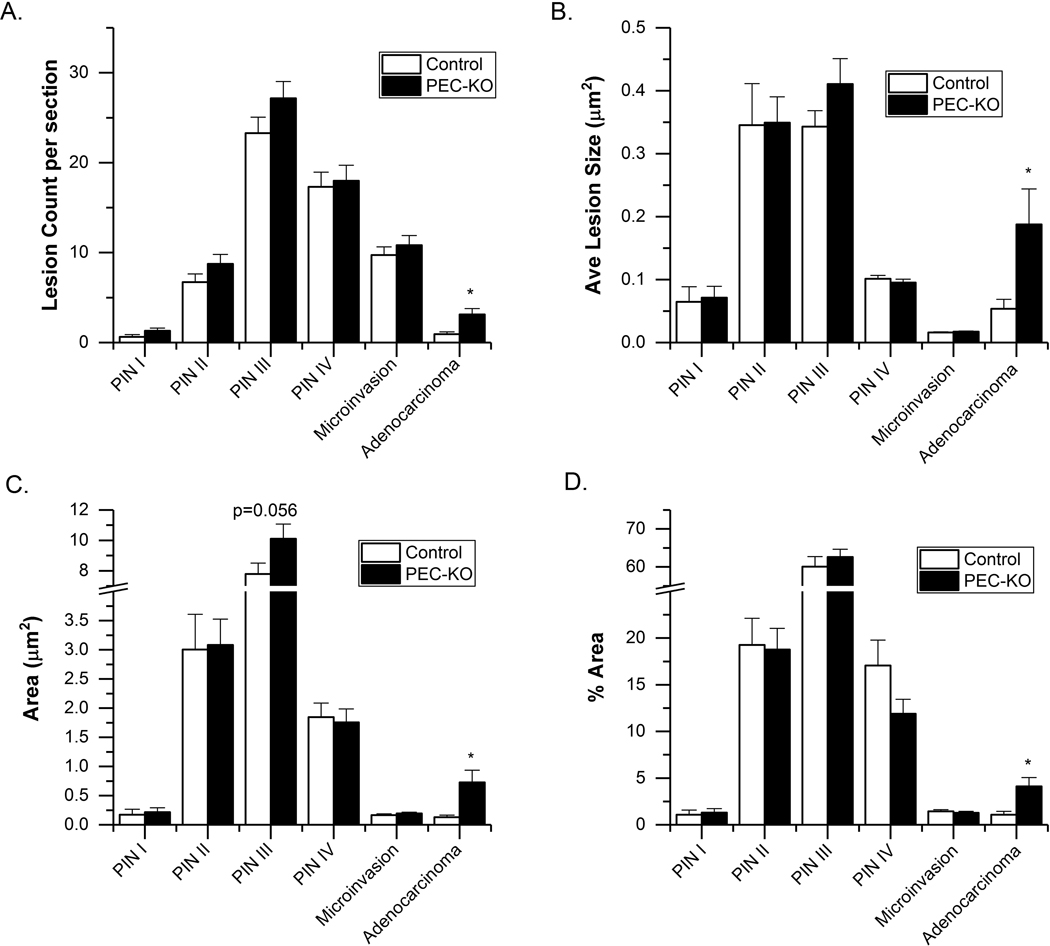
To test the role of VDR in the prostate epithelial cell we used PEC-VDR KO mice. At 28 wks of age, PIN III was the most prevalent lesion in the anterior prostate of TgAPT121 mice (60–70% of total prostate tissue area) and the next most common lesion types were PIN II and PIN IV (Fig. 2). Compared to controls, the PEC-VDR KO mice had more independent adenocarcinoma foci (p = 0.003, Fig. 2A), the average size of the adenocarcinoma foci was larger (p = 0.005, Fig. 2B), and the area covered by adenocarcinoma was larger (p = 0.004, Fig. 2C). In addition, a higher percentage of PEC-VDR KO mice developed focal adenocarcinoma than the control mice (73% vs. 38%; p = 0.004). Finally, while the % area of the total prostate tissue occupied by adenocarcinoma was low, it was 2.8-fold higher in the PEC-VDR KO mice than the control mice (p = 0.003, Fig. 2D).
Fig. 2.
Histological grading of the anterior prostates from 28-wk old PEC-VDR KO (n = 32) and control (PB-Cre-, VDRf/f; n = 33) mouse prostates. (A) Count of independent lesion foci per section; (B) average lesion size; (C) area of each lesion; (D) % area of each lesion type. Bars are mean ± SEM. (*p < 0.05 vs Control). Other comparisons where the difference approached significance are reported with the measured p-value.
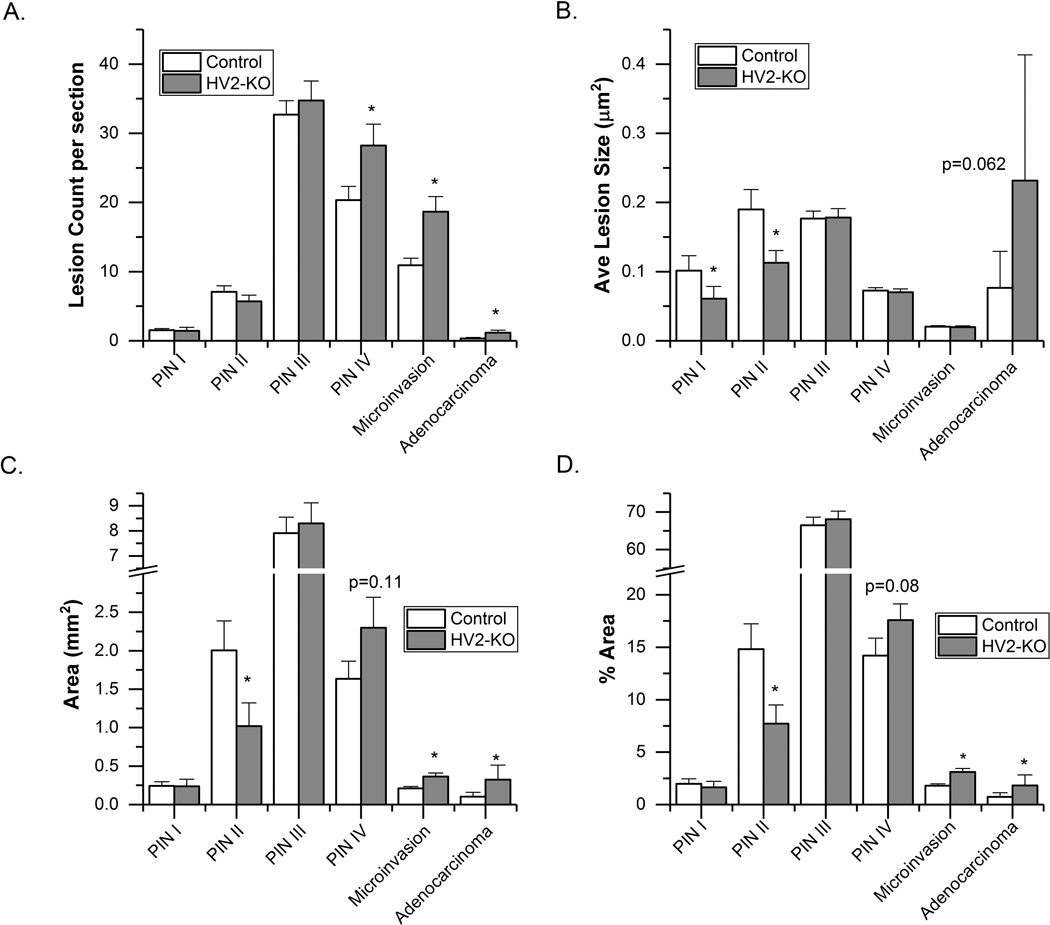
Mice with intestine specific expression of VDR in VDR KO mice (HV2-VDR KO) have normal calcium homeostasis but no VDR within the entire prostate micro-environment. In these mice, the lesion count was higher for all of the advanced lesions compared to controls: PIN IV (p = 0.032), microinvasion (p = 0.004) and adenocarcinoma (p = 0.026) (Fig. 3A). There was a trend towards larger adenocarcinoma lesion size (p = 0.062, Fig. 3B) and a greater area occupied by adenocarcinoma (p = 0.047, Fig. 3C) in the HV2-VDR KO mice. In addition, more HV2-VDR KO mice had adenocarcinoma than the control mice (48% vs. 19%; p = 0.027). Finally, HV2-VDR KO mice had higher % area of adenocarcinoma and microinvasion (p = 0.001), lower % area of the PIN II lesions (p = 0.024), and a trend towards higher % area for the PIN IV lesions (p = 0.08, Fig. 3D).
Fig. 3.
Histological grading of the anterior prostates from 26-wk old HV2-VDR KO (n = 23) and control mice (HV2+, VDR wild type, n = 27). (A) Count of independent lesion foci per section; (B) average lesion size; (C) area of each lesion; (D) % area of each lesion type. Bars are mean ± SEM. (*p < 0.05 vs Control). Other comparisons where the difference approached significance are reported with the measured p-value.
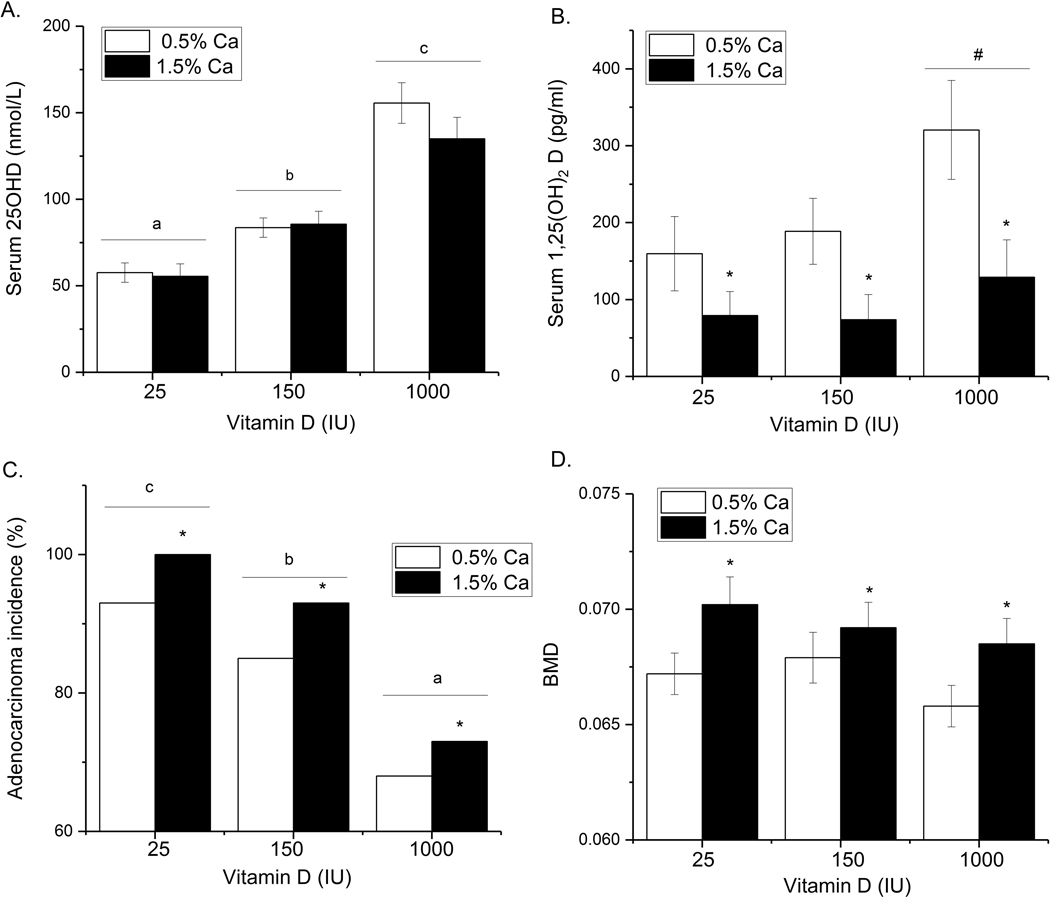
Serum 25OHD levels were increased by raising dietary vitamin D3 intake (p<0.001, Fig. 4A). The levels in the 25 IU/kg vitamin D3 group were slightly higher than we previously reported to due to the contribution of residual vitamin D3 from casein in the diet. Dietary Ca did not alter serum 25OHD levels but reduced serum 1,25(OH)2D levels by more than 57% (Fig. 4B). There was a trend towards higher serum 1,25(OH)2D levels in mice fed the 1000 IU/kg vitamin D3 diet (p=0.074).
Fig. 4:
The effect of dietary Ca and vitamin D on TgAPT121 mice. Diets were fed from weaning until 28 wks of age (n=34 per diet). (A) Serum 25OHD and (B) Serum 1,25(OH)2D are expressed as the mean + SEM (n=10/diet group). Values with different letter superscripts are significantly different (p < 0.05). (C) Incidence of prostate adenocarcinoma in the anterior prostate (n=34 per diet group). Values with different letter superscripts are significantly different between vitamin D groups (n=60/group) while * signifies a significant effect of Ca (n=90/diet Ca level, Chi Square test, p < 0.05). (D) BMD of the femur. * signifies a significant effect of Ca (p < 0.01). Bone data are expressed as mean + SEM (n=50–54/diet Ca level).
Low dietary vitamin D intake increased the percentage of mice with adenocarcinoma as their most advanced lesion, such that mice fed the 25 IU/kg vitamin D3 diet had the highest incidence of this phenotype (93% vs. 68% incidence in the 1000 IU/kg vitamin D3 group, p<0.001, Fig. 4C). Increasing dietary Ca to 3-times greater than the rodent requirement caused a uniform 8.5% increase in adenocarcinoma incidence across the vitamin D diet groups (p<0.05, Fig. 4C).
While higher dietary vitamin D suppressed the prostate cancer phenotype, higher vitamin D intake had no significant impact on bone mineral density (BMD) (Fig. 4D). However, increased dietary Ca increased BMD regardless of the dietary vitamin D intake level (p<0.01).
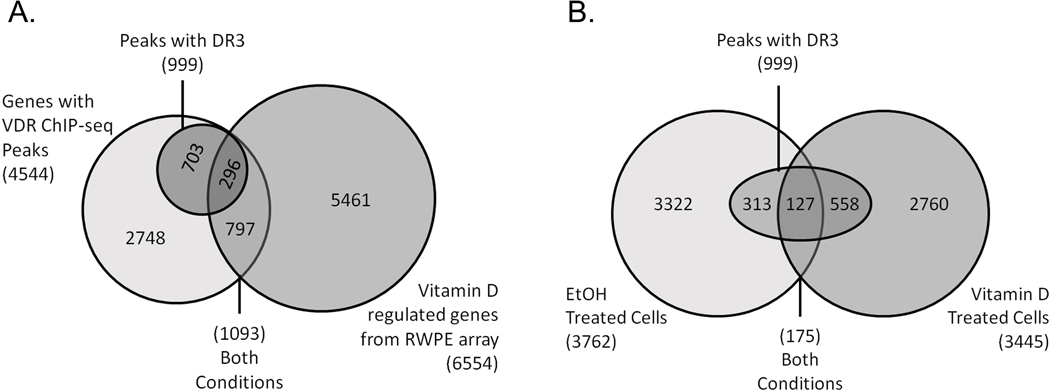
VDR ChIP-seq analysis in RWPE1 cells was used to identify vitamin D target genes in prostate that may contribute to the protection of prostate epithelial cells against prostate cancer. VDR ChIP-seq analysis revealed 3762 peaks in control cells and 3445 peaks in 1,25(OH)2D-treated cells with just 175 peaks common to both groups (Fig. 5A, Table S4). DR3-type VDR binding motifs that mediate VDR binding to DNA were found in 14.2% of the peaks, although more were found in the 1,25(OH)2 D group (20%) and the overlap group (72.6%). VDR ChIP-seq sites were annotated to 3390 protein coding genes, 682 long non-coding RNAs, and 472 micro RNAs. This includes VDR binding peaks in known vitamin D target genes like CYP24A1 (38) (5 peaks, 4 with DR3) and IGFBP3 (39) (2 peaks, 1 with DR3). The bulk of the peaks were intergenic (50.7%) or intronic (43.8%), reflecting long-distance regulation by enhancer elements. We confirmed the ChIP-seq results for 7 VDR binding peaks by ChIP-PCR in the genes for: CYP24A1, MAPK6, CTSD/SYT8, DDIT4, KLF4, CA9, and ETNK2 (Fig. S2). 492 of the VDR sites in RWPE1 cells (associated with 461 genes) were found in ChiP-seq data from more than one cell type. Most of these peaks were from 1,25(OH)2D-treated cells or were peaks found in both treatment groups (n=418, 73% with DR3 motifs). There was significant overlap between the VDR ChIP-seq peaks in RWPE1 cells and other datasets: (a) 1573 peaks in 1093 1,25(OH)2D-regulated genes from RWPE1 cells (34) (Fig. 5B), (b) 319 peaks in 193 1,25(OH)2D-regulated genes from mouse prostate epithelial stem cells (27), (c) 263 genes containing a VDR peak within a TAD for vitamin D regulated genes from human THP-1 cells (40).
Fig. 5.
Identification of VDR binding sites in RWPE1 cells. (A) A comparison of ethanol vehicle (EtOH) or 1,25(OH)2D (Vitamin D) treated cells (10 nM, 3 h). Traditional DR3-type VDR binding motifs were identified below the summits of VDR peaks. (B) Peaks were assigned to genes and then compared to transcriptome results from vitamin D treated RWPE1 cells. Overlap among the groups and the DR3 containing VDR peaks was assessed.
The potential functional impact of vitamin D signaling on the prostate epithelial cell was broad. Enriched canonical pathways included those with cancer relevant functions (Table 1, Table S5). Pathways regulating gene transcription included: one called “Transcriptional role for VDR in regulation of genes involved in osteoporosis” driven by genes like CYP24A1 and IGFBP3; four related to NF-κB signaling that included the genes NFKBIA and NFKBIZ; one for the oxidative stress response driven by the genes NFE2L2, TXNRD1, FOS and JUNB; and several that included transcription factors whose genes contain VDR binding sites, i.e. ESR1, CREB, HIF1A, and RBRJ. 109 transcription factors were vitamin D-regulated in RWPE1 cells and contained contain a VDR peak. In addition, we found 259 interaction networks in the VDR cistrome that were centered on transcription factors and 44 of these networks were centered on transcription factors whose genes had a DR3 motif below their VDR peak (Table 2).
Table 1.
Categories of Enriched Pathways for VDR ChIP-seq Peaks from RWPE1 Cells.
| Map Category | Representative Genes |
|---|---|
| Apoptosis | CALM2, FOXO1, p73, PTK2, ROCK1, SRF |
| Cancer | AREG, HGF, HGFR, ITGB1, MYC |
| Cell adhesion/Cytoskeleton | C3G, EFNA5, EPHB1, EPHA2, MLCK, PTK2, ROCK2 |
| Cell cycle | FOS, EGR1, MAPK8, MAD1, MDM2, NEK7 |
| Immune response | CCL16, CD40L, GNAI1, IL6, IL18RAP, IFNG, PTPRC, TNFSF5 |
| Signaling | ADCY1, 3, and 8, CAMKD, CAMKK2, MAP3K4, PIK3CA, PRKCE |
| Transcription | CBF1, FOS, MYC, CREB1, CREM, ESR1, HIF1A, NFKBIA, NFKBIZ, SMAD3 |
Table 2.
Enriched Transcription Factors with VDR ChIP-seq Peaks from RWPE1 Cells.
| TF Related Gene | TF Interaction Network | Peak signal1 | DR3 in Peak | VD effect in RWPE1 array | Peak in Multiple Cell Types |
|---|---|---|---|---|---|
| CREB1 | Y | VD | - | 48 h, − | N |
| EPAS1 | Y | VD | - | 24 h, + | N |
| FOS | Y | VD | + | all, + | Y |
| GLI3 | Y | VD | + | NS | Y |
| HIF1A | Y | VD | + | 6, 48 h, + | Y |
| KLF4 | Y | VD | + | 6 h, + | Y |
| KLF9 | Y | VD | + | NS | Y |
| NFKBIA | RELB | VD | + | 6, 24 h, + | Y |
| RARA | Y | VD | + | NS | Y |
| SMAD2 | Y | VD | + | NS | N |
| SRF | Y | VD | - | 48 h, + | N |
| TP73 | Y | VD | - | 46 h, − | N |
| DEC1 | Y | C | + | NS | N |
| ESR1 | Y | C | + | NS | N |
| ETS1 | Y | C | - | 6, 48 h, − | Y |
| GLI2 | Y | C | - | 48 h, − | N |
| MYC | Y | C | - | 6 h, − | N |
| SMAD3 | Y | C | - | 24 h, − | N |
| AHR | Y | B | + | 6 h, + | N |
| FOXO1 | Y | B | + | 6, 48 h, + | Y |
| RBPJ (CBF1) | Y | B | + | 48 h, − | Y |
| TBP | Y | B | + | 6 h, − | Y |
C = peak in ethanol treated control; B = peak in both groups; VD = peak in 1,25(OH)2D-treated group
Discussion
Vitamin D signaling through the VDR regulates cancer-relevant cellular events (10) and reduces prostate tumor growth (12,13,41) but definitive evidence for a beneficial effect of increasing vitamin D intake on prostate cancer risk has been lacking. Thus, specific recommendations for optimal vitamin D intake to reduce human prostate cancer risk are not yet possible. Rodent studies that manipulate dietary vitamin D intake have used either severe deficiency (11) or a large vitamin D excess (15,16) and thus have modest translational potential. Meanwhile, human vitamin D3 interventions for cancer prevention ((8,42,43) ClinicalTrials ID #NCT01463813) suffer from several limitations. First, they are typically conducted in older subjects with high baseline serum 25OHD levels (> 75 nM) and indolent cancer. Second, some studies use supplement levels that are too low for subjects who already have high vitamin D status (e.g. 800–2000 IU/d) (8,43) while other studies use pharmacologic, bolus doses (e.g. monthly doses at ≥ 10,000 IU) (42) that cause large swings in serum 25OHD and which may have adverse effects on other health outcomes (e.g. increased falls and fractures in older women (44)). Finally, these studies are typically short even though targeting prostate carcinogenesis may require decades of intervention to capture effects of vitamin D. This may explain why the VITAL study results were promising (i.e. reduced cancer mortality only when the first two years of follow-up were excluded, non-significant reduction in hazard ratio for prostate cancer to 0.88) but reported as negative for the entire 5.3 y median follow-up period (8).
With the challenges of conducting human intervention studies in mind, we designed mouse studies that test the lifelong effects of manipulating vitamin D signaling on early stages of prostate carcinogenesis. Many animal models for prostate cancer exist (45) but we chose the TgAPT121 model because it models the early steps of prostate carcinogenesis, its prostate cancer phenotype develops gradually over 6 months (19), and because our data show that the HGPIN in TgAPT121 mice has a molecular phenotype similar to human HGPIN (46). Our findings in TgAPT121 mice consistently show that disrupting vitamin D signaling increases the progression of HGPIN lesions to focal adenocarcinoma. These studies extend our earlier report (18) and our new findings are similar to the effect of whole body VDR gene deletion in the more aggressive LPB-Tag mouse (12). In addition, by using dietary vitamin D3 levels that allow us to model human relevant ranges of serum 25OHD (i.e. from adequate for bone health (50 nM) to high natural levels seen in humans (125–150 nM)), we can make two important conclusions. First, our data shows that extreme deficiency that disrupts Ca homeostasis is not necessary to increase cancer risk. Also, we found that raising serum 25OHD to levels that are 50–150% higher than those needed to protect bone can slow prostate cancer progression (Fig. 4). Still, before we can define the optimal vitamin D intake for prostate cancer prevention in humans, more research is needed to determine the life stages where increased vitamin D exposure is effective as well as to define the dose response curve for the anti-prostate cancer effects.
Although dietary vitamin D raises serum 25OHD levels, this metabolite is not a high affinity ligand for the VDR. As such, the benefit of higher serum 25OHD levels is likely not direct but may be due to local conversion to 1,25(OH)2 D. Consistent with this hypothesis, cell studies have shown that primary prostate epithelial cells and prostate cancer cells can synthesize 1,25(OH)2 D from 25OHD (47,48). In addition, Wagner et al. (49) found that supplementing preprostectomy patients with 40,000 IU vitamin D3 a day for ~30 days increased prostate 1,25(OH)2 D levels. They also showed that prostate 1,25(OH)2 D levels were inversely associated with proliferation, suggesting protection resulting from local 1,25(OH)2 D production. Our study using higher dietary Ca levels show that the severity of prostate lesions in mice is also increased by suppressing serum 1,25(OH)2 D levels. However, although high dietary Ca also increases prostate cancer two other mouse models with slow growing lesions (16), low serum 1,25(OH)2D levels caused by high Ca diets did not accelerate prostate cancer in the more rapidly progressing LPB-Tag model (12). This suggests that the negative impact of high dietary Ca on prostate carcinogenesis mediated through serum 1,25(OH)2D levels may be limited to specific stages of cancer development; this hypothesis has not been formally tested. Some data also suggests that the benefit of higher serum 25OHD on prostate cancer is modified by the level of dietary Ca (50) but we did not observe a significant Ca-by-vitamin D interaction in our study.
Our VDR ChIP-seq analysis demonstrates that activating vitamin D signaling affects multiple, complementary, cancer protective pathways in prostate epithelial cells and this is consistent with our previous microarray reports (27,34). The vitamin D-responsive gene signature in non-transformed prostate epithelial cells includes an anti-oxidant pathway signature that involves activation of NFE2L2, TXNRD1, FOS and JUNB. A similar vitamin D effect has been observed in other systems (e.g. diabetes (51)) and in the context of cancer, activation of this pathway may synergize with inhibition of genes controlling proliferation (e.g. TCF7L1 and KLF6 in our study), promoting apoptosis, and activating DNA protective pathways (e.g. GADD45A) to limit the accumulation of DNA mutations in prostate epithelial cells. Our VDR ChIP-seq data also suggest that vitamin D may amplify signals mediated through other transcription factors, e.g. by regulating genes for NFKBIA, an inhibitor of cancer promoting NF-κB signaling (52), or FOXO1, whose loss promotes prostate carcinogenesis when TMPRSS2-ERG is overexpressed (53). Finally, the ChIP-seq data revealed VDR binding peaks near genes that are involved in immune responses (e.g. IL1R2, Il20RB, IRAK2, and IRAK1BP1). These peaks may be responsible for the down-regulation of immune/inflammatory mRNAs we reported earlier (34) and make prostate epithelial cells less responsive to pro-inflammatory, mitogenic signals from immune cells.
A final interesting finding from our mouse studies is that they suggest that the benefits of vitamin D signaling in the prostate may extend beyond the prostate epithelial cell. While both prostate VDR deletion models had more advanced cancer, the impact of deletion was more dramatic in HV2-VDR KO mice that lack VDR in all prostate-associated cells than in the mice where VDR deletion was limited to prostate epithelial cells. Others have shown that prostate stromal cells are vitamin D target cells (54) and we previously showed that stromal cell proliferation is reduced in the prostates of PEC VDR KO mice (18). Collectively, these observations suggest that vitamin D signaling regulates multiple cells in the prostate and it may modulate complex interactions among those cells. This hypothesis requires further testing.
In summary, our data provide solid support for the proof-of-principle that vitamin D signaling modulates progression through the early stages of prostate carcinogenesis and for the idea that early, life-long dietary manipulation of serum vitamin D metabolites can modify the course of early stage prostate cancer. In addition, as Ames first suggested in his “triage theory” of nutrition and chronic disease (55), our data in mice demonstrate suggest that there can be different dietary requirements for vitamin D and Ca based on the health outcome examined (i.e. higher vitamin D and lower Ca for prostate protection; lower vitamin D and higher Ca for bone).
Supplementary Material
Statement of Significance:
This work shows that disrupting vitamin D signaling through diet or genetic deletion increases early prostate carcinogenesis through multiple pathways. Higher diet vitamin D levels are needed for cancer than bone.
Acknowledgements
We thank Ryan Schoch, Kimberly Carter, and Valerie DeGroff for technical support, and Sanjay Bhave, PhD and Hsueh-Li Tan, Ph.D. for assistance with necropsy and biosample collections.
Financial support for each author: This work was supported by National Institutes of Health / National Cancer Institute Award (NIH/NCI) R01 CA10113 (to JCF), American Institute for Cancer Research (AICR) Award #05A131 (to SKC), and a Showalter Trust Award (to JCF). Y. Li was supported in part through a Cancer Prevention Interdisciplinary Education Fellowship (NIH/NCI R25CA128770) and a Purdue Research Foundation Graduate Research Assistantship. The transgenic mouse studies were made possible by technical support from the Transgenic Mouse Core Facility, a core service of the Purdue University Center for Cancer Research (P30 CA023168). Additional support was provided by the Ohio State University Comprehensive Cancer Center (OSUCCC) (NIH/NCI P30 CA016058).
Footnotes
Conflict of interest statement: The authors declare no potential conflict of interest.
References
- 1.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer 2014;14(5):342–57. [DOI] [PubMed] [Google Scholar]
- 2.Corder EH, Guess HA, Hulka BS, Friedman GD, Sadler M, Vollmer RT, et al. Vitamin D and prostate cancer: a prediagnostic study with stored sera. Cancer Epidemiology Biomarkers and Prevention 1993;2(5):467–72. [PubMed] [Google Scholar]
- 3.Li H, Stampfer MJ, Hollis JB, Mucci LA, Gaziano JM, Hunter D, et al. A Prospective Study of Plasma Vitamin D Metabolites, Vitamin D Receptor Polymorphisms, and Prostate Cancer. PLoSMed 2007;4(3):e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kristal AR, Till C, Song X, Tangen CM, Goodman PJ, Neuhauser ML, et al. Plasma vitamin D and prostate cancer risk: results from the Selenium and Vitamin E Cancer Prevention Trial. Cancer Epidemiol Biomarkers Prev 2014;23(8):1494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schenk JM, Till CA, Tangen CM, Goodman PJ, Song X, Torkko KC, et al. Serum 25-hydroxyvitamin D concentrations and risk of prostate cancer: results from the Prostate Cancer Prevention Trial. Cancer Epidemiol Biomarkers Prev 2014;23(8):1484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandstedt J, Almquist M, Ulmert D, Manjer J, Malm J. Vitamin D, PTH, and calcium and tumor aggressiveness in prostate cancer: a prospective nested case-control study. Cancer Causes Control 2016;27(1):69–80. [DOI] [PubMed] [Google Scholar]
- 7.Travis RC, Perez-Cornago A, Appleby PN, Albanes D, Joshu CE, Lutsey PL, et al. A Collaborative Analysis of Individual Participant Data from 19 Prospective Studies Assesses Circulating Vitamin D and Prostate Cancer Risk. Cancer Res 2019;79(1):274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manson JE, Cook NR, Lee IM, Christen W, Bassuk SS, Mora S, et al. Vitamin D Supplements and Prevention of Cancer and Cardiovascular Disease. N Engl J Med 2019;380(1):33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannucci E, Rimm EB, Wolk A, Ascherio A, Stampfer MJ, Colditz GA, et al. Calcium and fructose intake in relation to risk of prostate cancer. Cancer Res 1998;58(3):442–47. [PubMed] [Google Scholar]
- 10.Fleet JC, DeSmet M, Johnson R, Li Y. Vitamin D and cancer: a review of molecular mechanisms. Biochem J 2012;441(1):61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ray R, Banks M, Abuzahra H, Eddy VJ, Persons KS, Lucia MS, et al. Effect of dietary vitamin D and calcium on the growth of androgen-insensitive human prostate tumor in a murine model. Anticancer Res 2012;32(3):727–31. [PMC free article] [PubMed] [Google Scholar]
- 12.Mordan-McCombs S, Brown T, Wang WL, Gaupel AC, Welsh J, Tenniswood M. Tumor progression in the LPB-Tag transgenic model of prostate cancer is altered by vitamin D receptor and serum testosterone status. J Steroid Biochem Mol Biol 2010;121(1–2):368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lokeshwar BL, Schwartz GG, Selzer MG, Burnstein KL, Zhuang SH, Block NL, et al. Inhibition of prostate cancer metastasis in vivo: a comparison of 1,23- dihydroxyvitamin D (calcitriol) and EB1089. Cancer Epidemiology Biomarkers and Prevention 1999;8(3):241–48. [PubMed] [Google Scholar]
- 14.Xue L, Yang K, Newmark H, Lipkin M. Induced hyperproliferation in epithelial cells of mouse prostate by a Western-style diet. Carcinogenesis 1997;18(5):995–99. [DOI] [PubMed] [Google Scholar]
- 15.Swami S, Krishnan AV, Wang JY, Jensen K, Horst R, Albertelli MA, et al. Dietary Vitamin D3 and 1,25-Dihydroxyvitamin D3 (Calcitriol) Exhibit Equivalent Anticancer Activity in Mouse Xenograft Models of Breast and Prostate Cancer. Endocrinology 2012;153(6):2576–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernichtein S, Pigat N, Barry Delongchamps N, Boutillon F, Verkarre V, Camparo P, et al. Vitamin D3 Prevents Calcium-Induced Progression of Early-Stage Prostate Tumors by Counteracting TRPC6 and Calcium Sensing Receptor Upregulation. Cancer Res 2017;77(2):355–65. [DOI] [PubMed] [Google Scholar]
- 17.Fleet JC, Gliniak C, Zhang Z, Xue Y, Smith KB, McCreedy R, et al. Serum metabolite profiles and target tissue gene expression define the effect of cholecalciferol intake on calcium metabolism in rats and mice. J Nutr 2008;138(6):1114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovalenko PL, Zhang Z, Yu JG, Li Y, Clinton SK, Fleet JC. Dietary vitamin D and vitamin D receptor level modulate epithelial cell proliferation and apoptosis in the prostate. Cancer Prev Res (Phila) 2011;4(10):1617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill R, Song Y, Cardiff RD, Van Dyke T. Heterogeneous tumor evolution initiated by loss of pRb function in a preclinical prostate cancer model. Cancer Res 2005;65(22):10243–54. [DOI] [PubMed] [Google Scholar]
- 20.Xue YB, Fleet JC. Intestinal Vitamin D Receptor Is Required for Normal Calcium and Bone Metabolism in Mice. Gastroenterology 2009;136(4):1317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y, Kato S, Fleet JC. Vitamin D receptor (VDR) knockout mice reveal VDR-independent regulation of intestinal calcium absorption and ECaC2 and calbindin D9k mRNA. J Nutr 2003;133(2):374–80. [DOI] [PubMed] [Google Scholar]
- 22.AIN. Report of the American Institute of Nutrition ad hoc committee on standards for nutritional studies. J Nutr 1977;107(7):1340–48. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Kovalenko P, Cui M, DeSmet M, Clinton SK, Fleet JC. Constitutive activation of the mitogen-activated protein kinase pathway impairs vitamin D signaling in human prostate epithelial cells. J Cell Physiol 2010;224(2):433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res 2004;64(6):2270–305. [DOI] [PubMed] [Google Scholar]
- 25.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences USA 2001;98(9):5116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 2007;39(1):41–51. [DOI] [PubMed] [Google Scholar]
- 27.Maund SL, Barclay WW, Hover LD, Axanova LS, Sui G, Hipp JD, et al. Interleukin-1α Mediates the Antiproliferative Effects of 1,25-Dihydroxyvitamin D3 in Prostate Progenitor/Stem Cells. Cancer Res 2011;71(15):5276–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 29.Cui M, Zhao Y, Hance KW, Shao A, Wood RJ, Fleet JC. Effects of MAPK signaling on 1,25-dihydroxyvitamin D-mediated CYP24 gene expression in the enterocyte-like cell line, Caco-2. J Cell Physiol 2009;219(1):132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 2007;4(8):651–57. [DOI] [PubMed] [Google Scholar]
- 31.Neme A, Seuter S, Carlberg C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim Biophys Acta 2017;1860(9):952–61. [DOI] [PubMed] [Google Scholar]
- 32.Tuoresmaki P, Vaisanen S, Neme A, Heikkinen S, Carlberg C. Patterns of genome-wide VDR locations. PLoS One 2014;9(4):e96105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38(4):576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovalenko PL, Zhang Z, Cui M, Clinton SK, Fleet JC. 1,25 dihydroxyvitamin D-mediated orchestration of anticancer, transcript-level effects in the immortalized, non-transformed prostate epithelial cell line, RWPE1. BMCGenomics 2010;11(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neme A, Seuter S, Carlberg C. Vitamin D-dependent chromatin association of CTCF in human monocytes. Biochim Biophys Acta 2016;1859(11):1380–88. [DOI] [PubMed] [Google Scholar]
- 36.Brawer MK. Prostatic intraepithelial neoplasia: an overview. Rev Urol 2005;7 Suppl 3:S11–8. [PMC free article] [PubMed] [Google Scholar]
- 37.Ashida S, Nakagawa H, Katagiri T, Furihata M, Iiizumi M, Anazawa Y, et al. Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res 2004;64(17):5963–72. [DOI] [PubMed] [Google Scholar]
- 38.Meyer MB, Goetsch PD, Pike JW. A Downstream Intergenic Cluster of Regulatory Enhancers Contributes to the Induction of CYP24A1 Expression by 1 alpha,25-Dihydroxyvitamin D-3. J Biol Chem 2010;285(20):15599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng LH, Malloy PJ, Feldman D. Identification of a functional vitamin D response element in the human insulin-like growth factor binding protein-3 promoter. Mol Endocrinol 2004;18(5):1109–19. [DOI] [PubMed] [Google Scholar]
- 40.Neme A, Nurminen V, Seuter S, Carlberg C. The vitamin D-dependent transcriptome of human monocytes. J Steroid Biochem Mol Biol 2016;164:180–87. [DOI] [PubMed] [Google Scholar]
- 41.Polek TC, Murthy S, Blutt SE, Boehm MF, Zou A, Weigel NL, et al. Novel nonsecosteroidal vitamin D receptor modulator inhibits the growth of LNCaP xenograft tumors in athymic mice without increased serum calcium. Prostate 2001;49(3):224–33. [DOI] [PubMed] [Google Scholar]
- 42.Scragg R, Khaw KT, Toop L, Sluyter J, Lawes CMM, Waayer D, et al. Monthly High-Dose Vitamin D Supplementation and Cancer Risk: A Post Hoc Analysis of the Vitamin D Assessment Randomized Clinical Trial. JAMA Oncol 2018;4(11):e182178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lappe J, Watson P, Travers-Gustafson D, Recker R, Garland C, Gorham E, et al. Effect of Vitamin D and Calcium Supplementation on Cancer Incidence in Older Women: A Randomized Clinical Trial. JAMA 2017;317(12):1234–43. [DOI] [PubMed] [Google Scholar]
- 44.Sanders KM, Stuart AL, Williamson EJ, Simpson JA, Kotowicz MA, Young D, et al. Annual high-dose oral vitamin D and falls and fractures in older women: a randomized controlled trial. JAMA 2010;303(18):1815–22. [DOI] [PubMed] [Google Scholar]
- 45.Grabowska MM, DeGraff DJ, Yu X, Jin RJ, Chen Z, Borowsky AD, et al. Mouse models of prostate cancer: picking the best model for the question. Cancer Metastasis Rev 2014;33(2–3):377–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bostwick DG, Liu L, Brawer MK, Qian J. High-grade prostatic intraepithelial neoplasia. Rev Urol 2004;6(4):171–9. [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz GG, Whitlatch LW, Chen TC, Lokeshwar BL, Holick MF. Human prostate cells synthesize 1,25-dihydroxyvitamin D3 from 25-hydroxyvitamin D3. Cancer EpidemiolBiomarkers Prev 1998;7(5):391–95. [PubMed] [Google Scholar]
- 48.Whitlatch LW, Young MV, Schwartz GG, Flanagan JN, Burnstein KL, Lokeshwar BL, et al. 25-Hydroxyvitamin D-1alpha-hydroxylase activity is diminished in human prostate cancer cells and is enhanced by gene transfer. J Steroid Biochem Mol Biol 2002;81(2):135–40. [DOI] [PubMed] [Google Scholar]
- 49.Wagner D, Trudel D, Van der Kwast T, Nonn L, Giangreco AA, Li D, et al. Randomized clinical trial of vitamin D3 doses on prostatic vitamin D metabolite levels and ki67 labeling in prostate cancer patients. J Clin Endocrinol Metab 2013;98(4):1498–507. [DOI] [PubMed] [Google Scholar]
- 50.Steck SE, Arab L, Zhang H, Bensen JT, Fontham ET, Johnson CS, et al. Association between Plasma 25-Hydroxyvitamin D, Ancestry and Aggressive Prostate Cancer among African Americans and European Americans in PCaP. PLoS One 2015;10(4):e0125151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakai K, Fujii H, Kono K, Goto S, Kitazawa R, Kitazawa S, et al. Vitamin D activates the Nrf2-Keap1 antioxidant pathway and ameliorates nephropathy in diabetic rats. Am J Hypertens 2014;27(4):586–95. [DOI] [PubMed] [Google Scholar]
- 52.Jin R, Yi Y, Yull FE, Blackwell TS, Clark PE, Koyama T, et al. NF-kappaB gene signature predicts prostate cancer progression. Cancer Res 2014;74(10):2763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Blee AM, Wang D, An J, Pan Y, Yan Y, et al. Loss of FOXO1 Cooperates with TMPRSS2-ERG Overexpression to Promote Prostate Tumorigenesis and Cell Invasion. Cancer Res 2017;77(23):6524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lou YR, Miettinen S, Kagechika H, Gronemeyer H, Tuohimaa P. Retinoic acid via RARalpha inhibits the expression of 24-hydroxylase in human prostate stromal cells. BiochemBiophys Res Commun 2005;338(4):1973–81. [DOI] [PubMed] [Google Scholar]
- 55.Ames BN. Low micronutrient intake may accelerate the degenerative diseases of aging through allocation of scarce micronutrients by triage. Proc Natl Acad Sci U S A 2006;103(47):17589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.