Abstract
It is now widely appreciated that the spatial organization of the genome is non-random, and its complex 3D folding has important consequences for many genome processes. Recent developments in multiplexed, super-resolution microscopy have enabled an unprecedented view of the polymeric structure of chromatin, from the loose folds of whole chromosomes, to the detailed loops of cis-regulatory elements that regulate gene expression. Facilitated by the use of robotics, microfluidics, and improved approaches to super-resolution, thousands to hundreds of thousands of individual cells can be analyzed in an individual experiment. This has led to new insights into the nature of genomic structural features identified by sequencing, such as topologically associated domains (TADs), and the nature of enhancer-promoter interactions underlying transcriptional regulation. Here, we review these recent improvements.
Keywords: super-resolution microscopy, chromatin, TADs
Nuclear microscopy goes molecular
Work in recent decades has shown that the spatial organization of the genome is non-random, and its complex 3D folding has important consequences for all genome processes, including replication, segregation, repair and transcriptional regulation [1–12]. Microscopy has long been a mainstay technique used by researchers to visualize nuclear organization, though the resolution of conventional approaches did not reach the the important molecular scale of many of these processes. This is now changing, as recent developments in super-resolution microscopy (see Glossary) approaches provide access to an unprecedented view of the polymeric structure of chromatin, from the length scales of whole chromosomes, to the few kilobase interactions among nearby cis-regulatory elements. In place of pairwise or three-way interaction measurements, new highly-multiplexed methods allow thousands of potential interactions to be probed simultaneously in each cell, for unbiased image-based discovery. Facilitated by robotics and microfluidics, along with tricks to accelerate super-resolution imaging, thousands to hundreds of thousands of individual cells can be analyzed in an individual experiment, providing a robust statistical power to these methods. These advances have given us new insights into the relationships between chromatin structure and epigenetics [13–17], the nature of topological associated domains (TADs) [16,18,19], the nature of enhancer-promoter interactions underlying transcriptional regulation [19], and opened a multitude of new opportunities for future investigations.
Here, we review the technologies that have driven these improvements. We first highlight significant technical advances in labeling and imaging respectively, which have marked a new chapter in microscopy studies of chromatin structure. In the process, we will also explain a labeling paradox - why a small volume protocol outperforms a larger volume alternative, and an imaging paradox - why some super-resolution techniques like photoblinking fluors can worsen resolution in certain high-resolution chromatin imaging experiments. With this appreciation of recent technical developments, we will describe new insights into nuclear architecture revealed by these imaging techniques, highlighting some of the ways these newly revealed structures differ from the textbook picture and earlier reviews based on bulk approaches. Finally, we touch on the exciting emerging frontiers and new challenges for the field.
Technical Advances
Super-resolution imaging of precisely defined genomic coordinates
Two recent innovations have transformed sequence specific imaging of chromatin organization: the ability to produce synthetic DNA at high complexity and large volumes, most notably through the Oligopaints technology [20–24], and the advent of super-resolution fluorescence microscopy techniques which allow resolution of 50 nanometers or better [25–27] (Fig. 1). To appreciate the contribution of these innovations and their foundations in earlier work, it will be helpful to consider a brief summary of earlier established approaches.
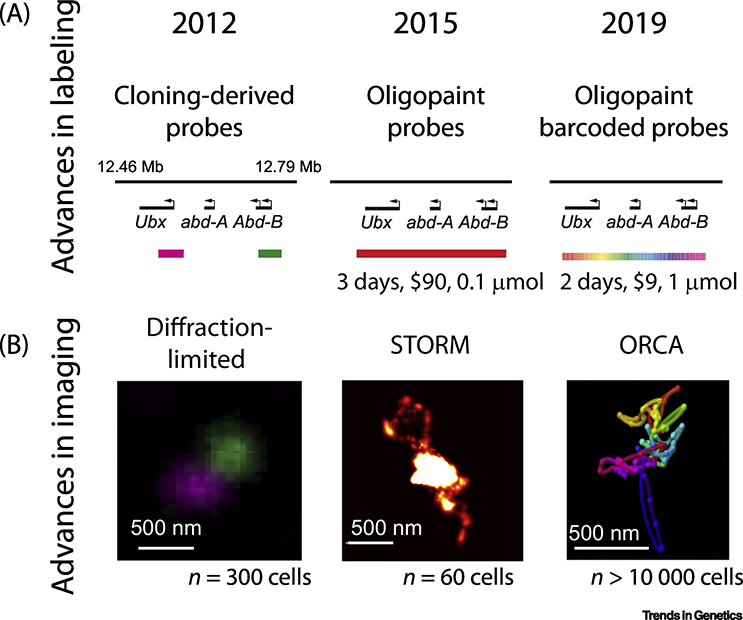
Figure 1. Improvements in labeling and imaging chromatin.
A. Schematic of evolution in probes targeting the BX-C from Drosophila melanogaster over the last decade, taken from our own experiments. Changes in probe cost and production time are noted. B. Example images of the structure of the BX-C using the probe sets in A and the techniques indicated. Typical number of cells analyzed per experiment are noted. The STORM image is adapted from [21] with permission. In the ORCA image, data are represented as colored spheres; the polymer interpolates these data in 3D to guide the eye in the correct order.
Oligonucleotide probes for imaging the 3D organization of genomic sequences have been produced through approaches dependent on molecular cloning, such as PCR and nick translation [28–32]. These classical methods enabled numerous insights into the spatial organization of nucleic acids in cells and nuclei over the last 50 years [33–47]. Despite notable innovations [32,48,49] these methods provide limited control over the probe properties; producing a pool of dsDNA carrying randomly interspersed modified bases, typically fragmented in variable lengths of 50–500 basepairs. Variability in probe quality among in this approach arises due to uncompensated variation in sequence composition across the genome in GC content, repeat content, and enzyme interaction bias. Many conventional experiments mitigate this variation by using larger genomic target windows. This has typically limited the genomic resolution of conventional FISH probes to 10s of kilobases.
Oligopaints: precision, control, and affordability
The advent of low cost, high complexity oligonucleotide-synthesis, enabled by the proliferation of microarrays, allowed synthetic, rationally and computationally designed oligonucleotides (oligos) to circumvent many of the limitations of cloning dependent approaches [20,32,50–52]. Control of the nucleic acid sequence at the base pair level allows for computational design of an optimal array of short homologous sequence (typically 20–50mers). All oligos can be made to the same length (for more uniform diffusion), and selected for similar GC content, melting point, and degree of secondary structure. Computational tools identify the best way to tile the oligos across the target to maximize their number and uniformity of hybridization properties, while avoiding potential for off-target binding. Thus, though genomic targets vary substantially in features which affect hybridization, the computationally designed probes reduce this variation, enabling increasingly high-throughput and multi-target applications. The use of array-based oligo probes was introduced by several groups [20,51,52], though the variation called Oligopaints [20] has been most widely adopted, due in part to its clear and improving pipelines for computational design and probe synthesis [22,24,53] and the integration of synthetic handles for downstream applications [17,21,24,54].
Approaches for making Oligopaints have become steadily easier and more cost efficient in recent years. An original emulsion-PCR-based approach costing ~$900/umol probe was replaced with a traditional PCR amplification and labeling step, requiring only 3 days at one tenth the cost in reagents [20]. Further improvements followed by removing gel-based purification [22] and minimizing the use of milliliter-scale PCR [24]. The evolved production protocols reduced reaction volumes needed for umol-scale probe production by two orders of magnitude, cutting cost by at least an order of magnitude [24]. This was achieved by relying on in vitro transcription (IVT) as a key amplification step followed by reverse transcription (RT) to convert probes back to more stable DNA [24,55,56].
How does a smaller volume reaction help increase yield and how can it be more cost and time efficient to convert from DNA to RNA and back, rather than clean up a labeled PCR product? The key insight is that IVT and RT reactions can be carried out at a very high concentration of substrate and product, unlike PCR. Amplification by IVT reduces the loss of diversity inherent in competitive exponential growth of PCR. RT also produces single-stranded DNA, removing the need for enzymatic or gel-based separation. Small volume (high concentration) protocols dramatically reduce enzyme and purification costs, and enable 96-well plate parallel processing. These advances facilitated the production of the enormous amounts of probe required for hybridization based spatial transcriptomic methods such as Multiplex Error Robust FISH (MERFISH)[24] and subsequent massively parallel RNA imaging [53,57–63]. It also enabled targeted whole-chromosome scale imaging [54,64,65] and subsequent super-resolution experiments probing genomic structure [14,17–19,21,64,66].
Another advantage of Oligopaints has been the ability to multiplex experiments using sequential rounds of imaging to expand the number of targets beyond the chromatic limits set by spectral overlap. This is achieved by appending distinct synthetic barcode sequences to the probes. These barcodes can then be labeled through hybridization with fluorescently labeled secondary oligos, without requiring a distinct fluorescent dye for each target. Use of adapter/bridge oligos to link the unique barcode to a common fluorescently-labeled oligo can further increase the flexibility and decrease the cost of labeling [17–19,54]. Aided by advances in robotic fluid-handling, the number of sequential labeling rounds used in published experiments has increased from 16 [24] to around 100 [19], with individual rounds requiring only 10–30 minutes [19,24,53]. Methods of extinguishing fluorescent signal after imaging have also evolved, from photobleaching [24,64] (which requires significant time and is not compatible with re-labeling controls) to DNase [67] (not compatible with DNA targets) to fluor-cleavage [53] (fast, but not compatible with re-labeling) to strand displacement [18,19]. The latter method is not only the fastest, as it can be performed in the same reaction in which new labels are added and thus adds no time, but is also cheaper than fluor-cleavage (disulfide linked fluors and TCEP for cleavage are among the most costly reagents in the experiment) and allows repeated labeling of the same target.
Super-resolution chromatin imaging
The ability to densely label chromatin with oligo-probes that specifically match genomic features like chromatin boundaries, without restrictions based on the existing borders of BACs or other templates, enabled new investigations into the links between epigenetic state and 3D chromatin structure. Single molecule localization microscopy (SMLM) based super-resolution methods, such as STORM/PALM provided the resolution boost needed to exploit this dense, precise labeling. These methods circumvent the diffraction limit by separating in time fluorescent signals that are too close to resolve in space. To achieve this, all the fluorophores are initially converted into a non-fluorescent dark state, and a subset are stochastically converted back into the bright state. This subset must be sparse enough that no two molecules within a diffraction limited distance are bright (Fig. 2a). The position of these molecules can be found with high precision by fitting the center of their fluorescent signal, with a precision limited only by the number of photons (brightness) and the noise of the detector (camera). Other super-resolution methods such as structured illumination microscopy (SIM) have also been applied to chromatin imaging and can acquire data faster, though the more modest 2x increase in resolution of linear SIM is notably less than that afforded by SMLM [68]. STORM-based methods have been especially effective to study chromatin structural properties that do not depend on knowing the precise polymer trajectory of the chromosome, such as compaction, asphericity, variable or smooth density, and degree of intermixing of select regions (Fig 2d). The first major insights from super-resolution imaging explored how these structural properties varied with distinct epigenetic states (see below).
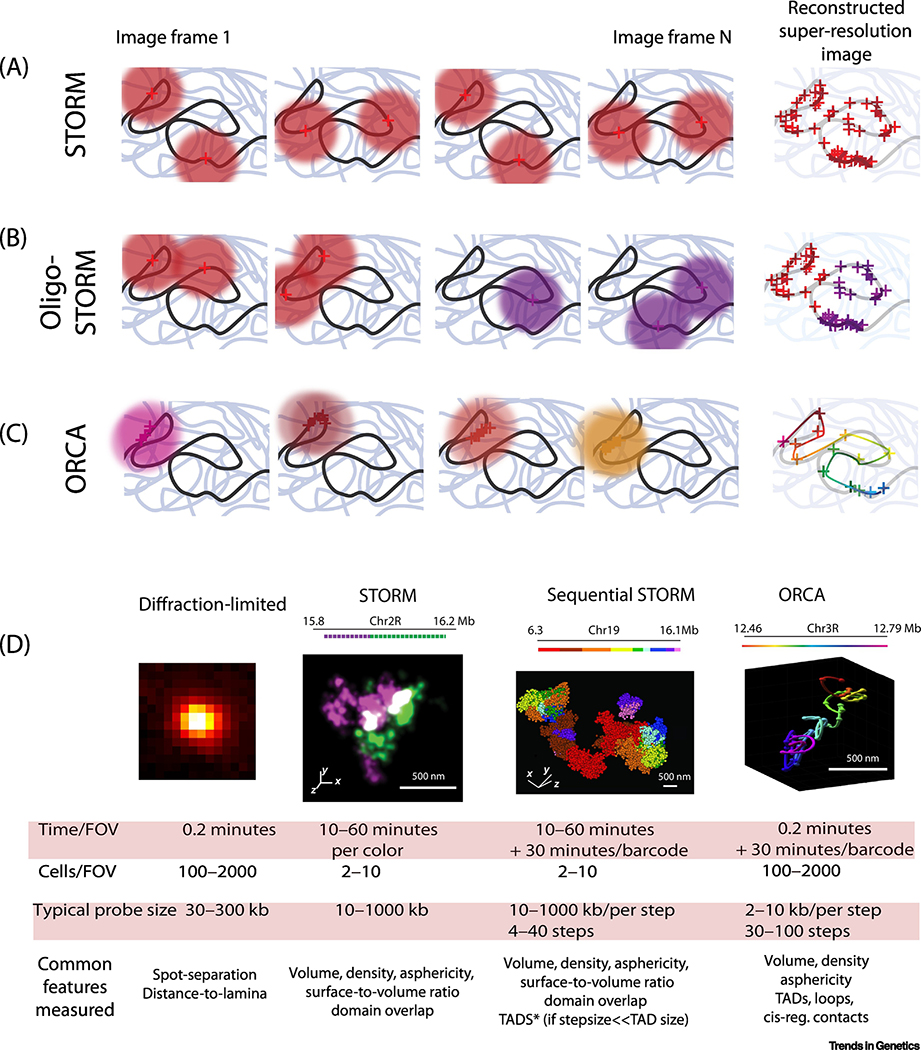
Figure 2. Similarities and differences explained: Diffraction-limited, STORM imaging, OligoSTORM, and ORCA.
A. in STORM (e.g. [14]) a sparse subset of fluors are activated at a time, so that their detected diffraction patterns (a.k.a. point-spread functions) don’t overlap. The centroids can be fit computationally (red crosses). After many frames of photoswitching, the position of all individual fluorophore centroids are combined to form a super-resolution image. Spectrally distinct fluorophores have been used for multi-color imaging, though different color channels have quantitatively different performance, affecting resolution [14,166]. B. In OligoSTORM (Nir 2018, Bintu 2018) different portions of the chromatin are labeled with different barcodes, enabling reliable STORM in arbitrary number of pseudocolors by sequential rounds of STORM and then hybridization. C. In ORCA (Mateo 2019), every short segment (1–10 kb) of chromatin is given its own barcode. No stochastic switching is used, allowing lower laser powers, wider field of view and faster imaging. A larger number of sequential hybridization rounds is necessary to complete the domain. A smooth interpolation of the recorded centroids approximates the chromatin path. In all the methods, temporally separated images circumvent the diffraction limit for spatial separation. Fiducial labels are imaged in parallel with data collection to correct nanoscopic drift during imaging. D. Table comparing the different features which can be measured with each approach, with example images adapted from [14,17,19], with permission. By measuring TADs, we mean de novo mapping of TAD boundaries independent of 3C-methods or related sequencing dependent technologies.
Rather than exploiting the precision labeling of Oligopaints, an innovative approach from Wang and colleagues exploited the use of orthogonal barcode sequences of Oligopaints to “walk” along whole chromosomes in megabase-scale steps, providing a megabase-resolution view of the trajectory of individual chromosomes. The average interactions across the chromosomes from a mere ~100 cells exhibited remarkable agreement with the checkerboard compartment pattern seen in Hi-C experiments which average millions of cells - suggesting these interactions are surprisingly stereotyped [64]. Merging these approaches, investigators have recently combined the higher degree of multiplexing enabled by barcoded Oligopaints with the resolution of densely-labeled chromatin imaged by STORM [17,18] to study the finer, sub-megabase scale structure of chromosomes (Fig. 2b). Nir and colleagues used sequential STORM imaging with larger 100–1,000 kb step sizes, each matched to boundaries previously identified by Hi-C, and spanning up to 8 Mb of the chromosome [17]. Concurrently, Bintu, Mateo and colleagues used uniform 30 kb steps. The degree of overlap between the 3D point clouds for any pair of steps showed a box-like organization that corresponded perfectly to the TADs identified by Hi-C, the first demonstration of mapping architectural features of TADs and loops de novo from microscopy approaches [18]. In all these experiments, the qualitative and quantitative agreement between indirect, sequencing based Hi-C and the microscopy results have provided encouraging cross-validation of both methods. Where Hi-C remains much superior in coverage, the microscopy has opened new biological insights of higher-order physical 3D structures and cell-cell variation (discussed more in the next section).
An imaging paradox: understanding resolution
As the step size for each barcode in OligoSTORM becomes smaller, the physical size of the domain approaches the resolution limit of STORM (Fig. 3). At this point, using STORM - that is, localizing the fluors attached to the domain one at a time by stochastic activation, can actually reduce the resolution of the data. This leads to our second paradox: How can a super-resolution approach make the resolution worse? If the physical separation of the fluors is smaller than the photon-limited resolution of an individual fluor, a more accurate measurement is achieved just by combining the photons of all fluors and ignoring the minute physical separation (Fig. 3). Dispensing with photo-switching has further advantages for resolution as well. Since molecules blink stochastically and briefly, STORM techniques must sacrifice x-y resolution to approximate z-position from a 2D image. Popular z-estimation methods for STORM have limited dynamic range (typically <1 um), making it necessary to sample several focal planes (typically >4 um), during which time out-of-focus fluors are bleaching, reducing resolution. In contrast, the centroid of each unresolved cluster of fluors yields many more photons to enhance localization precision and can be scanned axially with no tradeoff in lateral resolution. Moreover, it allows the use of optimally bright non-photoswitching fluors and buffers. Imaging is much faster, as there is no need to record long movies of stochastic blinking events, in which the region of interest is primarily blank. The high laser power density required for stochastic photoswitching can also be avoided, allowing the user to spread limited power over a larger field of view (laser power density is typically the major limitation to field of view size). Finally, the collected data occupies ten- to hundredfold less disk space. With typical STORM experiments requiring 5–50 Tb of storage, data volume is a major limitation in storage and data-processing costs. Collectively, these differences contribute substantially to the difference in number of cells analyzed by recently by STORM (e.g. <100–7000 cells) [14,17,18] and non-stochastic blinking super-resolution approaches that we will describe next (>100,000 cells) [18,19,69].
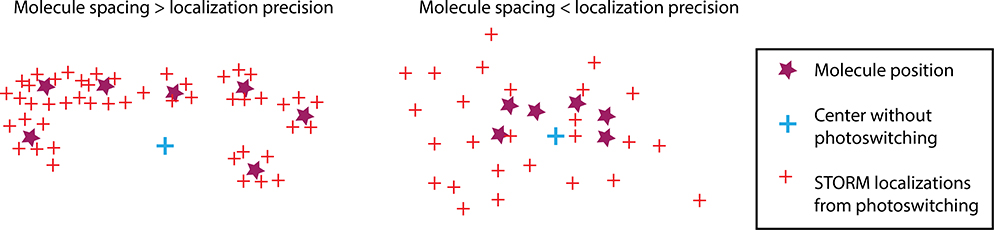
Figure 3. Limits of STORM.
For STORM (stochastic localization of individual fluors) to be advantageous, the absolute spread of the fluorescent molecules positions (purple stars) should be greater than the precision at which individual fluors can be localized by photoswitching (red crosses). In this regime, the localizations from photoswitching (red crosses) provide a better description of the true location and physical extent of the fluors than the centroid (blue cross). If the distance between molecules is smaller than the localization precision, the centroid from the combined emission of all fluors provides a more accurate estimate of the position than individual photoswitching events.
By reducing the step size of sequentially imaged regions to 2–10 kb, Mateo and colleagues were therefore able to scale up the throughput of data collection to easily acquire more cells, while still tracing domains ranging from 100 to 700 kilobases [19]. As discussed above, this smaller step size allows for sub-diffraction spatial resolution without the need for STORM and provides enhanced genomic resolution, allowing for better measure of the path of the DNA polymer. As the approach is still a single-point-source-based optical reconstruction technique, like STORM, but no longer stochastic, and unlike STORM is specific to DNA, it was named ‘optical reconstruction of chromatin architecture’, (ORCA) - (Fig 2c). This method builds on previous sequential imaging at megabase resolution [64] and the insights in labeling consecutive 30 kb domains with STORM compared to centroid fitting only [18]. Mateo and colleagues optimized the approach for use in tissue sections, simultaneously mapping the cell’s spatial context (its connectome in the embryo) with its genome structure. The authors combined it with sequential labeling of dozens of mRNA to sample the transcriptome and provide information on the different cell types. Additionally, nascent transcripts were labeled with intronic probes, enabling new studies of enhancer promoter interactions on nascent transcription state. A conceptually similar approach appeared at the same time called ‘Hi-M’, (high-throughput, high-resolution, high-coverage microscopy). It differed in its use of a non-uniform distribution of barcode probes, resulting in an average resolution of 17 kb, and in its application in much thicker whole-mount Drosophila embryos in place of cryosections [69].
Technical advances to watch for
We expect to see considerable technical advances in imaging of fixed cells in coming years, especially in terms of enhanced throughput, more analyses in primary tissues, new multi-modal super-resolution imaging of DNA with protein, and the development of less perturbative methods. While we anticipate some improvement in resolution, dramatic advances may need to await the development of different labeling approaches. Currently, labeling of sequence, rather than localization accuracy, is the major challenge to super-resolution of DNA, and cannot be readily addressed through advances in light microscopy such as signal amplification [70–76] or electron microscopy with immuno-gold [77–80]. Moreover, it is uncertain to what extent chromatin structure has been preserved at length scales finer than current super-resolution imaging described here.
Technological innovations that improve the throughput of these methods will enable their use in screens to identify how chromatin structure responds to a wide range of cis mutations, trans perturbations, and environmental perturbations, all with high temporal resolution relative to time of perturbation. Given the large number of cells required by sequencing based methods like Hi-C, the stagnating costs of sequencing, and the poor scaling of resolution with sequencing depth, it will be expensive to conduct such screens across an array of timepoints with these methods. By contrast, ORCA is already able to image many different cell types within a single experiment and identify unique and shared structural features of these populations. In place of using mRNA expression profiles to separate cell types [19], new methods using spatial position or barcodes will allow users to distinguish cells from different treatments, genetic backgrounds, and timepoints. With as few as 100 cells providing decent mapping of TADs, subTADs and loops, increasing the number of cells per experiment to millions of cells will allow potentially hundreds of distinct conditions to be screened in a single experiment, rapidly and at low cost. While it is unlikely that ORCA will provide a whole-genome view of response to perturbation, such whole-genome views are not essential to understanding the effects or dynamics of many perturbations. These same properties of spatial resolution and throughput will also greatly facilitate exploration of chromatin structure in tissue samples, which to date has largely been concentrated in cell lines. While live-cell imaging methods will be needed to resolve the stochastic dynamics in these experiments, live approaches are not poised to be able to map TADs or process large numbers of cells (Box 1).
Box 1: When is a TAD not a TAD?
Methods with progressively finer resolution of the polymer trajectory of the chromatin have now started to clarify some of the most studied and debated questions of chromatin structure, such as the nature of the “TAD”. TADs were originally reported from Hi-C data [116–118] and since also found by related techniques that infer 3D structures from a sequencing readout of a large population of cells [119,120]. They were described as domains of the chromatin which exhibit more frequent intra-domain than interdomain contacts and appear as boxes on the diagonal of a chromatin-contact-map. From flies to humans, the genome partition into TADs of variable size, leading some to call them the ‘building blocks of the genome’ [121,122]. The correlation of TAD boundaries with other important biological features such as the boundaries of epigenetic domains, replication domains, and of clusters of commonly co-regulated genes suggests these structures may be functionally important [1,2,10]. Consistent with this, mutations that disrupt TAD organization in many cases lead to aberrant gene expression [19,123–126]. Despite this interest, there has been much controversy over defining a TAD. Do these patterns in population average data reflect the existence of a globular domain, isolated from neighboring globular domains by loops pinched by CTCF and cohesin, as TADs are frequently depicted in reviews and summary figures? That is to say are TADs a structural feature the way the nucleus is a structural feature - or are TADs only a property of an ensemble, much like the temperature of the room is an emergent property of the motion of many molecules. Polymer simulations can reproduce the population average distribution of TADs without globular domains or even detectably separated domains at the single cell level [127].
The single-cell contact maps of Bintu et al and Mateo et al frequently exhibit TAD-like structures - i.e. local domains of enhanced contact well separated from one another, as to create boxes on the diagonal of the single-cell contact map (Fig 4b) [18,19]. In 3D, these structures look like separated globules, as TADs are commonly illustrated in schematics and review articles. But are they TADs? No. Where TAD boundaries are defined by particular genomic coordinates, these globular domain transitions can be found at different frequencies throughout the region. The transitions have a statistical preference for certain positions, and when all the transitions are averaged together in population data the TADs emerge from the microscopy data in the precise pattern measured with bulk Hi-C [18,19,69]. These data indicate TADs are not structural building blocks the way the chromosome territory is a structure. Despite numerous reviews drawing the parallel of chromosome territories and TADs as different scales of genome folding, the microscopy data now show one is a physical structure that can be seen in single cells and the other is a statistical feature which only exists in a population. The distinction is potentially confusing as single cells do exhibit globular domains which in some cells align with TADs. Still, imaging in perturbed cell-lines makes the difference clear: The globular, TAD-like features of single cells persist in the absence of cohesin, while TADs within a common epigenetic state (e.g. A-compartment) disappear [128]. The microscopy data show exactly how this pattern emerges: the statistical positioning of the boundaries becomes uniform instead of biased to CTCF sites, such that the population average is free of boundaries even though individual cells retain globules [18]. These differences observed in the distribution of conformations at the single cell level in both wildtype and perturbed conditions also provide valuable new data with which to distinguish competing mechanistic models of chromatin folding [127,129–133].
Imaging the nanoscale distribution of chromatin proteins in combination with ORCA is another developing area in which we expect progress. Significant advances have been made in imaging nuclear proteins in both live and fixed cells using super resolution, too numerous to summarize here [81–96], though most of protein-labeling data to date lack information on sequences associated with different features. A major challenge to multimodal (protein + DNA) imaging will be achieving sufficiently high labeling efficiency of the protein targets [97,98]. Recent advances in protein labeling [99–107] aided by new benchmarking approaches to measure efficiency [97,98] augur for exciting advances.
An essential area of research will be the development of less perturbative labeling and imaging strategies. Key to such work will be robust multi-scale metrics of chromatin structure. For example, a common objection to hybridization-based approaches is that they denature the DNA doublèhelix, and generally unfold proteins. However, if proteins and DNA denature in situ (in place, to nanoscale accuracy) these molecular scale perturbations would be unresolved and of no consequence to the nanoscale super-resolution methods discussed here. We look forward to the development of methods which chemically freeze DNA and protein in place with a physical constraint equal or better than the resolution. Some molecular-scale flexibility will still be desired in these approaches, to facilitate entry/diffusion and binding of labeled probes. The method will also need to be rapid -- without time for degradation or activation of cell stress response pathways during the chemical freezing process. Existing fixative approaches such as aldehyde crosslinking, high-pressure freezing, and heat-fixation may be part of the way there. The use of heat-denaturation-free methods such as exo-nuclease digestion down to single-strand DNA as in RASER-FISH [108], or Cas9-mediated denaturation and binding, as in CASFISH [109], provide promising new directions. In any case, imaging of relevant distance scale markers (kilobase to megabase features separated by 10s of nanometers, not angstroms), should be used to assess perturbation. Considerable recent advances in labeling regions of chromatin for live imaging [110–114] will make these challenging measurements increasingly feasible.
Biological insights
Links between epigenetic state and chromatin folding
STORM-based chromatin studies that resolve compaction, spreading, and mixing of chromatin below the diffraction limit without resolving the polymer trajectory have provided exciting insights into the links between epigenetic state and chromatin folding. For example, a survey of three major epigenetic states of chromatin in Drosophila, active, inactive, and Polycomb-repressed, revealed that each chromatin state followed distinct power-law scaling of domain size as a function of length, suggesting common physical principles that track with epigenetic state may dominate nuclear chromatin folding [14]. The three epigenetic classes also differed in their packaging density, the degree of self-interaction within a domain, and the degree to which they intermix or separate from one-another (Fig 4a).
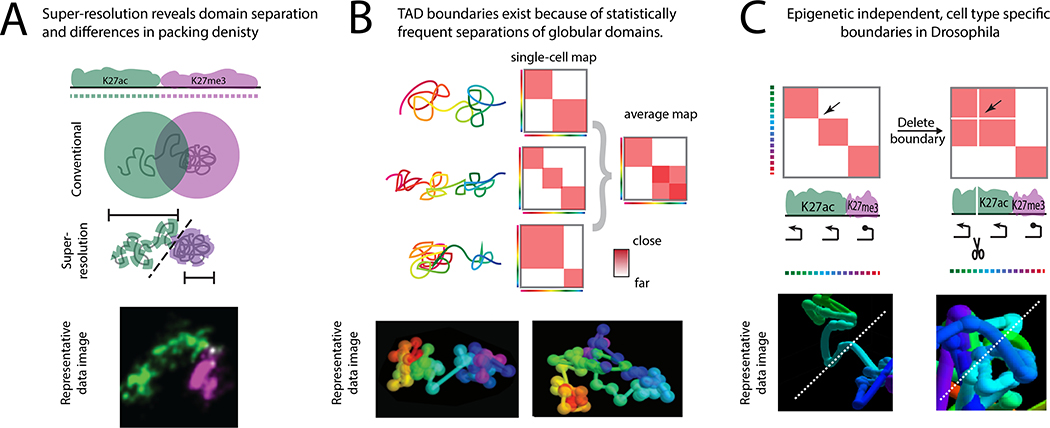
Figure 4. Key Figure. Recent biological insights from advances in super-resolution imaging of chromatin.
A. Epigenetic domains are generally not resolved using conventional microscopy but reveal structurally distinct organization such as compaction, asphericity, and lack-of-mixing when resolved in super-resolution [13–16,66]. An example image from [14], with permission. B. Heterogeneous TAD-like domains are observed in single cells when using with multiplexed super-resolution imaging. There is not a 1-to-1 correspondence between the TAD-like domains of single cells and the TADs in the population data, but TAD-like domain boundaries occur more frequently at population TAD boundary positions [18]. Images adapted from [18] with permission. C. In certain cell types of Drosophila embryos, neighboring active hox genes are physically separated from one another into distinct TADs. This separation depends on boundary elements at the border of these TADs [19]. Image adapted from [19], with permission.
Super-resolution imaging has been particularly informative in revealing the organization of Polycomb (Pc) bound chromatin – an epigenetic state associated with facultatively repressed genes. In both Drosophila and mouse, Pc domains were found to be largely compact, globular structures, contrasting the extended, often multi-lobed organization of active chromatin [13–16]. Pc-repressed domains also exhibit greater intradomain mixing and greater separation from neighboring active chromatin than other forms of transcriptionally silent chromatin [14]. The compacted, globular structure is lost upon removal of the Pc-group protein, Polyhomeotic (Ph) [14,15,115], which suggests that PRC1 activity (of which Ph is a core component), rather than the K27me3 deposited by PRC2, is they key determinant of these structural features. As Pc-chromatin plays an important role in epigenetic memory, maintaining transcriptional silencing across cell divisions and for months to decades in post-mitotic cells, it has been postulated that its unique structural features contribute to this ability [14,15], though further investigation will be required to test this hypothesis and dissect the key features.
Epigenetic independent boundaries
While recent super-resolution chromatin microscopy experiments have supported the conclusion that epigenetic states (e.g. active, Pc-repressed, HP1-repressed) contribute to the 3D partitioning of the genome through homotypic association, the roles of epigenetic-independent features have been less clear - especially outside of in vitro mammalian cell culture. Taking advantage of ORCA’s ability to map even very small TADs (<20 kb) with high resolution in small populations of cells, Mateo and colleagues identified a strong, cell-type-specific physical partition of the posterior hox complex separating the transcriptionally active Ubx and abd-A genes (Fig. 4c). Unlike most structural boundaries studied in cultured Drosophila cells in vitro, this separation does not correspond to a difference in epigenetic state and cannot be explained a homotypic association or heterotypic avoidance. Intriguingly, deleting a small (4 kb) element at the boundary of these two larger (~60 kb) physically separated domains abrogated this partitioning and led to mis-expression of both genes [19]. This showed that epigenetic state is not the only structural organization mechanism functioning in Drosophila [134]. It also shows that border elements can contribute to separation of much larger flanking regions of chromatin in vivo, consistent with interpretations from systems where much more abundant cell-populations have made 3C analysis feasible [123,126,135–139]. How such relatively small border elements (a.k.a. insulators) influence physical interactions across such substantial distances of a soft polymer remains a mystery. Yet certainly single cell images of these separated domains should help figure this out by allowing a direct test of some hypothesized models.
New biological frontiers
These recent advances leave the community ready to investigate a range of exciting long-standing questions pertaining to chromatin structure. Lately, there has been considerable interest in the potential of multiway interactions to achieve novel forms of regulation. For example, it has been hypothesized that separate enhancers (or elements of a super-enhancer) may interact with one another to seed a high local concentration of certain transcription factors (TFs) and trigger condensate formation to regulate gene expression [90,140–142]. It has also been hypothesized that the formation of TF condensates 100s of nanometers across could allow for enhancer-promoter communication without proximity [110,141,143]. ORCA provides the ability to resolve clusters of enhancers and their target promoters and test if multiway hubs form and if hub-formation correlates with enhanced gene expression. Similarly, it provides a chance to test the action-at-a-distance proposal by relating enhancer-promoter positions to RNA expression, especially if combined with imaging of TF condensates.
Advances in imaging also promise to provide new insights into the longstanding mystery of insulation. Originally identified by transgenic approaches, insulators are genomic elements which can negate the effect of an enhancer specifically when positioned between the enhancer and its cognate promoter [144–153]. Several hypotheses have been posited over the years to explain this phenomenon, including functioning as a competitive promoter bait, a directed tether, an anchor of an isolating loop domain, and a stiff or greatly decondensed chromatin region [144–153]. With the ability to visualize the chromatin path on the length scale of these enhancer-promoter-insulator interactions, many of these structural models may soon be tested directly for the first time.
Concluding Remarks and Future Perspectives
From pairs of spots separated by hundreds of kilobases of unseen turns of DNA to nanoscale 3D trajectories with sequence features resolved at a couple kilobases in thousands of cells, the last decade has witnessed a significant revolution in chromatin imaging of fixed nuclei.
While for the sake of brevity we have focused on advances in imaging fixed cells at high spatial resolution, recent advances in live imaging have confirmed the dynamic and frequently transient nature of these interactions suggested by heterogeneity of structural properties measured by fixed approaches [18,19,47]. The development of the CARGO system has allowed live imaging of non-repetitive regulatory sequences using chimeric arrays of guide RNAs and dCAS9 [154]. This approach has been combined with imaging of chromatin associated proteins such as mediator [89]. Tandem arrays of heterologous TF binding sites (like TetR) have provided a first glimpse of the dynamics of chromatin interactions of sites proximal to interacting enhancers and promoters [110,113,114]. More widespread application of the MS2 system to label RNAs in live cells is providing new insight about transcriptional dynamics, including regulation of bursting kinetics [89,110,113,155–161], new evidence of rapidly encoded transcriptional memory [162], and coordinated activation of two genes by the same promoter [157,161]. We look forward to seeing more minimally perturbative imaging approaches with smaller arrays, approaches that allow fluorescent labels to be positioned closer to enhancer/promoter elements for more accurate tracking, brighter live-compatible labels to improve resolution, and more efficient protocols for engineering labels into cells and embryos.
Recent improvements in super-resolution microscopy for chromatin organization have the promise to address many gaps in the field, but to achieve the impact of sequencing-based approaches, innovations are needed to make the technologies more widely accessible. Many features make microscopy appealing, like the perseveration of spatial organization in tissues, the small number of cells required to detect features like TADs, the ability to process many thousands of cells per experiment easily and cheaply, the integration with RNA detection and the low cost per experiment once the equipment and expertise is in place. Sequencing-based approaches like Hi-C have set a remarkable standard for portability of both experimental procedures and computational analysis pipelines. The advances in super-resolution and high-throughput microscopy have relied largely on home-built setups integrating optics and automated fluid handling [17–19,69]. Some commercial setups are emerging [17], though significant further advances are necessary to match the scalability of home-built methods and to integrate with established optical setups. A variety of resources have appeared to aid probe design [20,53,163–165], but tools that combine both flexibility and ease of use are sorely needed. The large datasets produced highlight a need for improved tools for data processing, data sharing, and data browsing. With a robust computational community interested in genome structure we hope to see advances in this analysis as well in the coming years.
Outstanding Questions.
How should we store, analyze, and annotate large imaging datasets measuring genome structure?
At what length scale is the structure of chromatin preserved by current imaging and labeling methods and at what length scales is it perturbed? What is the best way to measure this?
How can we image protein and DNA structure simultaneously to better test models of how chromatin proteins shape chromatin structure?
What proteins play important roles in shaping chromatin structure in animals, and can we develop image-based screens to identify them? By what mechanisms do they work -- as motors, as self-associating liquid droplets, as sticky tethers on a polymer, as polymerizing agents? How can we image these factors in single cells with their DNA substrates to distinguish these mechanisms?
How do multiple enhancers regulate a single gene?
How do insulators prevent aberrant interaction between enhancers and promoters?
Highlights.
Microscopy now allows measurement of the polymeric structure of chromatin in intact, fixed nuclei, with a resolution of a few kilobases.
Multiway contact interactions and physical separation of domains can be probed directly.
Structural information can be combined with measurements of nascent transcription, nuclear position, cellular position, and cellular morphology.
Single cell imaging validates many of the features previously identified by bulk, proximity-based approaches and challenges existing models of chromatin structure.
In situ super-resolution imaging offers new potential for multi-omic single cell analyses.
Acknowledgements
We thank the members of the Boettiger lab and Chao-ting Wu for helpful comments and suggestions on the manuscript. This work is supported by an NIH New Investigator’s grant to ANB (DGM132935A).
Glossary
- Cis regulatory element
A portion of the DNA sequence that influences the expression of genes on the same chromosome (in cis) and not its homolog (in trans).
- FISH
fluorescence in situ hybridization, a method for labeling RNA or DNA by hybridization (base-pairing) of fluorescently tagged nucleic acid probes.
- Hi-C
a genome-wide variant of chromosome conformation capture that uses restriction digestion, followed by ligation and deep sequencing to detect spatial proximity among DNA sequences.
- Oligopaints
a method for producing renewable complex pools of synthetic oligonucleotides using array-based synthesis and PCR.
- Spectral overlap
The portion of light spectra shared by two fluorescent probes.
- STORM
a super-resolution approach which uses stochastic blinking of fluorescent molecules to separate detection events in time (in different image frames), allowing resolution of emitters. Also known as PALM, especially when the emitters are genetically encoded fluors instead of small molecule dyes.
- Strand displacement
a process by which one a longer strand of DNA displaces a shorter strand from its longer complementary partner.
- Super-resolution microscopy
Fluorescence microscopy with resolution better than the Abbe-diffraction limit of fluorescent label: Wavelength/ 2 NA
- TADs
topologically associated domains -- domains of the genome that exhibit higher intra-domain contact than inter-domain contact.
- Tile
(verb) to cover uniformly with separate non-overlapping units.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Szabo Q et al. (2019) Principles of genome folding into topologically associating domains. Sci Adv 5, eaaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu M and Ren B (2017) The Three-Dimensional Organization of Mammalian Genomes. Annu. Rev. Cell Dev. Biol. 33, 265–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowley MJ and Corces VG (2018) Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spielmann M et al. (2018) Structural variation in the 3D genome. Nat. Rev. Genet. 7, 85–97 [DOI] [PubMed] [Google Scholar]
- 5.Spitz F (2016) Gene regulation at a distance: From remote enhancers to 3D regulatory ensembles. Semin. Cell Dev. Biol. 57, 57–67 [DOI] [PubMed] [Google Scholar]
- 6.van Steensel B and Furlong EEM (2019) The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. DOI: 10.1038/s41580-019-0114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson MI et al. (2019) Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D. Mol. Cell 74, 1110–1122 [DOI] [PubMed] [Google Scholar]
- 8.Mirny LA et al. (2019) Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol. 58, 142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furlong EEM and Levine M (2018) Developmental enhancers and chromosome topology. Science 361, 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long HK et al. (2016) Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 167, 1170–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekker J and Mirny LA (2016) The 3D Genome as Moderator of Chromosomal Communication. Cell 164, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bickmore W and Pombo A (2019) Editorial overview: Genome architecture and expression. Curr. Opin. Genet. Dev. 55, iii–iv [DOI] [PubMed] [Google Scholar]
- 13.Fabre PJ et al. (2015) Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states. Proc. Natl. Acad. Sci. U. S. A. 112, 13964–13969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boettiger AN et al. (2016) Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kundu S et al. (2017) Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol. Cell 65, 432–446.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szabo Q et al. (2018) TADs are 3D structural units of higher-order chromosome organization in Drosophila. Science advances 4, eaar8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nir G et al. (2018) Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. 14, e1007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bintu B et al. (2018) Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateo LJ et al. (2019) Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beliveau BJ et al. (2012) Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proceedings of the National Academy of Sciences 109, 21301–21306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beliveau BJ et al. (2015) Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt TL et al. (2015) Scalable amplification of strand subsets from chip-synthesized oligonucleotide libraries. Nat. Commun. 6, 8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beliveau BJ et al. (2017) In Situ Super-Resolution Imaging of Genomic DNA with OligoSTORM and OligoDNA-PAINT. 1663 (Erfle H, ed), pp. 231–252, Springer New York; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen KH et al. (2015) Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betzig E et al. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 [DOI] [PubMed] [Google Scholar]
- 26.Rust MJ et al. (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang B et al. (2008) Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardue ML and Gall JG (1969) Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc. Natl. Acad. Sci. U. S. A. 64, 600–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauman JG et al. (1980) A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridization of fluorochromelabelled RNA. Exp. Cell Res. 128, 485–490 [DOI] [PubMed] [Google Scholar]
- 30.Itzkovitz S and van Oudenaarden A (2011) Validating transcripts with probes and imaging technology. Nat. Methods 8, S12–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levsky JM and Singer RH (2003) Fluorescence in situ hybridization: past, present and future. J. Cell Sci. 116, 2833–2838 [DOI] [PubMed] [Google Scholar]
- 32.Huber D et al. (2018) Fluorescence in situ hybridization (FISH): History, limitations and what to expect from micro-scale FISH? Nanoscale Microscale Thermophys. Eng. 1, 15–24 [Google Scholar]
- 33.McGinnis W et al. (1984) A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308, 428–433 [DOI] [PubMed] [Google Scholar]
- 34.Harding K et al. (1985) Spatially regulated expression of homeotic genes in Drosophila. Science 229, 1236–1242 [DOI] [PubMed] [Google Scholar]
- 35.Chambeyron S and Bickmore WA (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18, 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eskeland R et al. (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Molecular cell 38, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bantignies F et al. (2011) Polycomb-Dependent Regulatory Contacts between Distant Hox Loci in Drosophila. Cell 144, 214–226 [DOI] [PubMed] [Google Scholar]
- 38.Cheutin T and Cavalli G (2012) Progressive Polycomb Assembly on H3K27me3 Compartments Generates Polycomb Bodies with Developmentally Regulated Motion. PLoS Genet. 8, e1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheutin T and Cavalli G (2018) Loss of PRC1 induces higher-order opening of Hox loci independently of transcription during Drosophila embryogenesis. Nat. Commun. 9, 3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahy NL et al. (2002) Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159, 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabin M et al. (1985) Two homoeo box loci mapped in evolutionarily related mouse and human chromosomes. Nature 314, 175–178 [DOI] [PubMed] [Google Scholar]
- 42.Williamson I et al. (2016) Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development 143, 2994–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joyce EF et al. (2012) Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8, e1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolzer A et al. (2005) Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3, e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shachar S et al. (2015) Identification of Gene Positioning Factors Using High-Throughput Imaging Mapping Resource Identification of Gene Positioning Factors Using High-Throughput Imaging Mapping. Cell 162, 911–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giorgetti L et al. (2015) High-Resolution 3D DNA FISH Using Plasmid Probes and Computational Correction of Optical Aberrations to Study Chromatin Structure at the Sub-megabase Scale. 1262 pp. 37–53 [DOI] [PubMed] [Google Scholar]
- 47.Finn EH et al. (2019) Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell 176, 1502–1515.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bienko M et al. (2012) A versatile genome-scale PCR-based pipeline for high-definition DNA FISH. Nat. Methods [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cremer M et al. (2008) Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol. Biol. 463, 205–239 [DOI] [PubMed] [Google Scholar]
- 50.Müller P et al. (2010) COMBO-FISH enables high precision localization microscopy as a prerequisite for nanostructure analysis of genome loci. Int. J. Mol. Sci. 11, 4094–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyle S et al. (2011) Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 19, 901–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada N. a. et al. (2011) Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet. Genome Res. 132, 248–254 [DOI] [PubMed] [Google Scholar]
- 53.Moffitt JR et al. (2016) High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proceedings of the National Academy of Sciences 113, 11046–11051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fields BD et al. (2019) A multiplexed DNA FISH strategy for assessing genome architecture in Caenorhabditis elegans. Elife 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murgha YE et al. (2014) Methods for the preparation of large quantities of complex single-stranded oligonucleotide libraries. PLoS One 9, e94752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murgha Y et al. (2015) Combined in vitro transcription and reverse transcription to amplify and label complex synthetic oligonucleotide probe libraries. Biotechniques 58, 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eng C-HL et al. (2017) Profiling the transcriptome with RNA SPOTs. Nat. Methods 14, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eng C-HL et al. (2019) Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature DOI: 10.1038/s41586-019-1049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moffitt JR et al. (2016) High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proceedings of the National Academy of Sciences 113, 14456–14461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moffitt JR et al. (2016) Spatial organization shapes the turnover of a bacterial transcriptome. Elife 5, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah S et al. (2016) In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron 92, 342–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shah S et al. (2017) seqFISH Accurately Detects Transcripts in Single Cells and Reveals Robust Spatial Organization in the Hippocampus. Neuron 94, 752–758.e1 [DOI] [PubMed] [Google Scholar]
- 63.Shah S et al. (2018) Dynamics and Spatial Genomics of the Nascent Transcriptome by Intron seqFISH. Cell 174, 363–376.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang S et al. (2016) Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosin LF et al. (2018) Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLoS Genet. 14, e1007393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cattoni DI et al. (2017) Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat. Commun. 8, 1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lubeck E et al. Single-cell in situ RNA profiling by sequential hybridization., Nature methods, 11 April-(2014), 360–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sigal YM et al. (2018) Visualizing and discovering cellular structures with super-resolution microscopy. Science 361, 880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cardozo Gizzi AM et al. (2019) Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Mol. Cell 74, 212–222.e5 [DOI] [PubMed] [Google Scholar]
- 70.Kishi JY et al. (2019) SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods 16, 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Battich N et al. (2013) Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods 10, 1127–1133 [DOI] [PubMed] [Google Scholar]
- 72.Wang F et al. (2012) RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ke R et al. (2013) In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 [DOI] [PubMed] [Google Scholar]
- 74.Lee JH et al. (2014) Highly Multiplexed Subcellular RNA Sequencing in Situ. Science 343, 1360–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rouhanifard SH et al. (2018) ClampFISH detects individual nucleic acid molecules using click chemistry-based amplification. Nat. Biotechnol. DOI: 10.1038/nbt.4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi HMT et al. (2018) Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Albiez H et al. (2006) Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 14, 707–733 [DOI] [PubMed] [Google Scholar]
- 78.Solovei I et al. (2002) Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp. Cell Res. 276, 10–23 [DOI] [PubMed] [Google Scholar]
- 79.Iwano M et al. (2003) Three-dimensional architecture of ribosomal DNA within barley nucleoli revealed with electron microscopy. Scanning 25, 257–263 [DOI] [PubMed] [Google Scholar]
- 80.Ou HD et al. (2017) ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ricci MA et al. (2015) Chromatin Fibers Are Formed by Heterogeneous Groups of Nucleosomes In Vivo. Cell 160, 1145–1158 [DOI] [PubMed] [Google Scholar]
- 82.Kieffer-Kwon K-R et al. (2017) Myc Regulates Chromatin Decompaction and Nuclear Architecture during B Cell Activation. Mol. Cell 67, 566–578.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bystricky K (2015) Chromosome dynamics and folding in eukaryotes: insights from live cell microscopy. FEBS Lett. 589, 3014–3022 [DOI] [PubMed] [Google Scholar]
- 84.Chen J et al. (2014) Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hansen AS et al. (2017) CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 6, 1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chong S et al. (2018) Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2555, eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cisse II et al. (2013) Real-Time Dynamics of RNA Polymerase II Clustering in Live Human Cells. Science 664, [DOI] [PubMed] [Google Scholar]
- 88.Guo YE et al. (2019) Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature DOI: 10.1038/s41586-019-1464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cho W-K et al. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 4199, eaar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 3958, eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holzmann J et al. (2019) Absolute quantification of cohesin, CTCF and their regulators in human cells. Elife 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shaban HA et al. (2018) Formation of correlated chromatin domains at nanoscale dynamic resolution during transcription. Nucleic Acids Res. 46, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Otterstrom J et al. (2019) Super-resolution microscopy reveals how histone tail acetylation affects DNA compaction within nucleosomes in vivo. Nucleic Acids Res. 47, 8470–8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu J et al. (2018) Super-Resolution Imaging of Higher-Order Chromatin Structures at Different Epigenomic States in Single Mammalian Cells. Cell Rep. 24, 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fang K et al. (2018) Super-resolution Imaging of Individual Human Subchromosomal Regions in Situ Reveals Nanoscopic Building Blocks of Higher-Order Structure. ACS Nano 12, 4909–4918 [DOI] [PubMed] [Google Scholar]
- 96.Nozaki T et al. (2017) Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Mol. Cell 67, 282–293.e7 [DOI] [PubMed] [Google Scholar]
- 97.Wang S et al. (2014) Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proceedings of the National Academy of Sciences 111, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thevathasan JV et al. (2019) Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat. Methods 16, 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Los GV et al. (2008) HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 [DOI] [PubMed] [Google Scholar]
- 100.Duc HN and Ren X (2017) Labelling HaloTag Fusion Proteins with HaloTag Ligand in Living Cells. Bio Protoc 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Keppler A et al. (2004) Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. U. S. A. 101, 9955–9959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grimm JB et al. (2017) A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods 14, 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grimm JB et al. (2017) General Synthetic Method for Si-Fluoresceins and Si-Rhodamines. ACS Cent Sci 3, 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grimm JB et al. (2016) Bright photoactivatable fluorophores for single-molecule imaging. Nat. Methods 13, 985–988 [DOI] [PubMed] [Google Scholar]
- 105.Grimm JB et al. (2017) Synthesis of Janelia Fluor HaloTag and SNAP-Tag Ligands and Their Use in Cellular Imaging Experiments. Methods Mol. Biol. 1663, 179–188 [DOI] [PubMed] [Google Scholar]
- 106.Zheng Q et al. (2019) Rational Design of Fluorogenic and Spontaneously Blinking Labels for Super-Resolution Imaging. ACS Cent Sci 5, 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Legant WR et al. (2016) High-density three-dimensional localization microscopy across large volumes. Nat. Methods [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brown JM et al. (2018) A tissue-specific self-interacting chromatin domain forms independently of enhancer-promoter interactions. Nat. Commun. 9, 3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Deng W et al. (2015) CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proceedings of the National Academy of Sciences [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alexander JM et al. (2019) Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. Elife 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mariamé B et al. (2018) Real-Time Visualization and Quantification of Human Cytomegalovirus Replication in Living Cells Using the ANCHOR DNA Labeling Technology. J. Virol. 92, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Germier T et al. (2017) Real-Time Imaging of a Single Gene Reveals Transcription-Initiated Local Confinement. Biophys. J. 113, 1383–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen H et al. (2018) Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 50, 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lucas JS et al. (2014) 3D trajectories adopted by coding and regulatory DNA elements: First-passage times for genomic interactions. Cell 158, 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wani AH et al. (2016) Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat. Commun. 7, 10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sexton T et al. (2012) Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 148, 458–472 [DOI] [PubMed] [Google Scholar]
- 117.Nora EP et al. (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dixon JR et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Quinodoz SA et al. (2018) Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Beagrie RA et al. (2017) Complex multi-enhancer contacts captured by genome architecture mapping. Nature DOI: 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ciabrelli F and Cavalli G (2015) Chromatin-driven behavior of topologically associating domains. J. Mol. Biol. 427, 608–625 [DOI] [PubMed] [Google Scholar]
- 122.Krefting J et al. (2018) Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 16, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lupiáñez DG et al. (2015) Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 161, 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo Y et al. (2015) CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162, 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flavahan WA et al. (2015) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Symmons O et al. (2016) The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Dev. Cell 39, 529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fudenberg G et al. (2016) Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 15, 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rao SSP et al. (2017) Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barbieri M et al. (2012) Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. U. S. A. 109, 16173–16178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brackley CA et al. (2018) Extrusion without a motor: a new take on the loop extrusion model of genome organization. Nucleus 1034, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jost D et al. (2014) Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sanborn AL et al. (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U. S. A. 112, E6456–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Giorgetti L et al. (2014) Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157, 950–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang H-LV and Corces VG (2019) Seeing Is Believing: ORCA Allows Visualization of Three-Dimensional Genome Organization at Single-Cell Resolution. Biochemistry DOI: 10.1021/acs.biochem.9b00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Franke M et al. (2016) Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature [DOI] [PubMed] [Google Scholar]
- 136.Williamson I et al. (2019) Developmentally regulated Shh expression is robust to TAD perturbations. Development DOI: 10.1242/dev.179523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Paliou C et al. (2019) Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc. Natl. Acad. Sci. U. S. A. 116, 12390–12399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Despang A et al. (2019) Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 51, 1263–1271 [DOI] [PubMed] [Google Scholar]
- 139.Rodríguez-Carballo E et al. (2017) The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes and Development 31, 2264–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hnisz D et al. (2017) A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boija A et al. (2018) Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell DOI: 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shrinivas K et al. (2019) Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 75, 549–561.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Heist T et al. (2019) Large distances separate coregulated genes in living Drosophila embryos. Proc. Natl. Acad. Sci. U. S. A. 116, 15062–15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Özdemir I and Gambetta MC (2019) The Role of Insulation in Patterning Gene Expression. Genes 10, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stadler MR et al. (2017) Convergence of topological domain boundaries, insulators, and polytene interbands revealed by high-resolution mapping of chromatin contacts in the early Drosophila melanogaster embryo. Elife 6, 1–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li H-B et al. (2010) Insulators, not Polycomb Response Elements, are required for long-range interactions between Polycomb targets in Drosophila. Mol. Cell. Biol. 31, 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang J and Corces VG (2012) Insulators, long-range interactions, and genome function. Curr. Opin. Genet. Dev. 22, 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Core LJ and Lis JT (2009) Paused Pol II captures enhancer activity and acts as a potent insulator. Genes Dev. 23, 1606–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kyrchanova O et al. (2015) The boundary paradox in the Bithorax complex. Mech. Dev. 138, 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bonchuk A et al. (2015) Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 13, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kyrchanova O et al. (2008) Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 36, 7019–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chetverina D et al. (2014) Making connections: Insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays 36, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fujioka M et al. (2016) Determinants of Chromosome Architecture: Insulator Pairing in cis and in trans. PLoS Genet. 12, e1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gu B et al. (2018) Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359, 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bothma JP et al. (2014) Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. U. S. A. 7, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bothma JP et al. (2015) Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife 4, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fukaya T et al. (2016) Enhancer Control of Transcriptional Bursting. Cell 166, 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lim B et al. (2017) Transcriptional Pre-patterning of Drosophila Gastrulation. Curr. Biol. [DOI] [PubMed] [Google Scholar]
- 159.Fukaya T et al. (2017) Rapid Rates of Pol II Elongation in the Drosophila Embryo. Curr. Biol. 27, 1387–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Garcia HG and Gregor T (2018) Live Imaging of mRNA Synthesis in Drosophila. Methods Mol. Biol. 1649, 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lim B et al. (2018) Temporal dynamics of pair-rule stripes in living Drosophila embryos. Proc. Natl. Acad. Sci. U. S. A. 115, 8376–8381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ferraro T et al. (2015) Transcriptional Memory in the Drosophila Embryo. Curr. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rouillard J-M (2003) OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31, 3057–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gelali E et al. (2019) iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture. Nat. Commun. 10, 1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Beliveau BJ et al. (2018) OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl. Acad. Sci. U. S. A. 115, E2183–E2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dempsey GT et al. (2011) Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]