Abstract
Background. A unique genomic difference between human and civet severe acute respiratory syndrome coronaviruses (SARS-CoVs) is that the former has a deletion of 29 nucleotides from open reading frame (orf) 8d that results in the generation of orf8a and orf8b. The objectives of the present study were to analyze antibody reactivity to ORF8a in patients with SARS and to elucidate the function of ORF8a.
Methods. Western-blot and immunofluorescent antibody assays were used to detect anti-ORF8a antibody. SARS-CoV HKU39849 was used to infect stable clones expressing ORF8a and cells transfected with small interfering RNA (siRNA). The virus loads (VLs) and cytopathic effects (CPEs) were recorded. Confocal microscopy and several mitochondria-related tests were used to study the function of ORF8a.
Results. Two (5.4%) of 37 patients with SARS had anti-ORF8a antibodies. The VLs in the stable clones expressing ORF8a were significantly higher than those in control subjects 5 days after infection. siRNA against orf8a significantly reduced VLs and interrupted the CPE. ORF8a was found to be localized in mitochondria, and overexpression resulted in increases in mitochondrial transmembrane potential, reactive oxygen species production, caspase 3 activity, and cellular apoptosis.
Conclusions. ORF8a not only enhances viral replication but also induces apoptosis through a mitochondria-dependent pathway.
The severe acute respiratory syndrome coronavirus (SARS-CoV) has been identified as the causal agent of SARS [1-3]. The 29.7-kb RNA genome of SARS-CoV contains open reading frames (ORFs) for 5 structural proteins-replicase, spike, envelope, membrane, and nucleocapsid-as well as a number of accessory proteins that may participate in the viral life cycle [4, 5]. It is believed that human SARS-CoV may originate via zoonosis, given that similar viruses have been identified in the civet cat and bats [6, 7].
A feature of the human SARS-CoV that distinguishes it from its animal counterparts is a 29-nt deletion from orf8a′ (previously known as “orf10” ) resulting in orf8a (orf10), which has the potential to encode a protein with only 39 aa [7]. The 29-nt sequence in the animal versions of the virus results in the merging of orfs 8a and 8b into a new orf8a′ that encodes a putative protein 122 aa in length [7]. It is interesting to note that, among the >80 human SARS-CoVs whose full lengths have been sequenced, all of them contained orf8a, with 1 exception: GD01, a strain identified early in the SARS endemic in Guangdong Province that contains orf8a′ [8]. We hypothesized that orf8a may have important functions in human SARS-CoV and its pathogenesis in cells that enabled SARS-CoV containing orf8a to become the predominant strain in the 2003 pandemic.
In the present study, we used Western-blot (WB) assays with a recombinant glutathione S-transferase (GST)-ORF8a fusion protein to demonstrate that 2 patients with SARS had antiORF8a antibody reactivity. Subsequently, we used 2 stable clones expressing ORF8a and cells transfected with small interfering RNA (siRNA) to demonstrate that ORF8a enhances viral replication and the cytopathic effect (CPE). Finally, we showed that ORF8a was localized in mitochondria, where it could perturb the mitochondrial membrane potential and induce apoptosis via a caspase 3-dependent pathway.
Patients, Materials, and Methods
Patients. Serum samples from 37 patients infected with SARS-CoV were collected from Taipei Veterans General Hospital and Taipei Ho-Ping Hospital. Infection in all patients had been confirmed serologically using a WB assay with recombinant spike and nucleocapsid proteins [9]. In addition, serum samples collected from 31 healthy subjects were used as normal controls.
Construction of plasmids. Total RNA was extracted from SARS-CoV-infected cells using Trizol (Invitrogen). A cDNA fragment of orf8a containing Eco RI and Xho I restriction enzyme sites at the 5′ and 3′ ends, respectively, was generated by use of reverse-transcription polymerase chain reaction (RT-PCR). The RT-PCR product was digested by Eco RI and Xho I and inserted between the Eco RI and Xho I sites of a pcDNA3 vector containing a hemagglutinin (HA) tag sequence and pGEX-5X-1 containing a GST gene to generate pHA-ORF8a and pGEXORF8a plasmids. The plasmid called “pORF8a-EGFP” (expressing green fluorescent protein [GFP]-tagged ORF8a) was constructed by inserting the cDNA of orf8a that contained Sal I and Hin dIII flanking sequences at the 5′ and 3′ ends into the upstream region of the EGFP gene in pEGFP-1 vector. All plasmids were confirmed by DNA sequencing using an ABI PRISM 3700 DNA Analyzer (Applied Biosystems).
Generation of rabbit anti-ORF8a antiserum and WB assay. A synthetic peptide (SP) containing amino acid residues 25- 37 of ORF8a was generated using the solid-phase method (Kelowna). The SP was coupled with keyhole limpet hemocyanin in accordance with procedures published elsewhere [10] and was used to immunize rabbits. ORF8a recombinant protein (RP) expressed in Escherichia coli (BL21) as a GST fusion protein was purified by glutathione-sepharose affinity chromatography in accordance with the manufacturer’s recommendation. The GST-ORF8a RP (0.5 μg for each strip) was used to evaluate the antibody reactivity of serum from SARS-CoV-infected patients. In addition to patients’ serum, an anti-GST monoclonal antibody (MAb; Santa Cruz Biotechnology), rabbit preimmunized serum, and rabbit anti-ORF8a-SP antiserum (R26) were also used to examine recombinant GST-ORF8a protein. Patients’ serum samples were diluted at 1:100 and incubated with WB strips for 1 h at 37°C. Details of the WB have been described elsewhere [11].
Cell lines and stable clones expressing ORF8a. Three cell lines-VeroE6, HEK 293T, and HuH-7-were used in this study [12-14]. They were cultured in Dulbecco’s modified Eagle medium (DMEM; GIBCO) with 10% fetal calf serum (HyClone), penicillin, streptomycin, and l-glutamine. For transfection experiments, either VeroE6 or HEK 293T cells were seeded in plates and cultured overnight at 37°C with 5% CO2. HuH-7 cells were transfected with pHA-ORF8a plasmid DNA using lipofectamine 2000 (Invitrogen) in DMEM that contained 600 μgG418 (Sigma). Two clones, designated 18 and 24, were obtained and confirmed by both RT-PCR and WB. A negative control clone-ORF8a-neg, which was obtained through transfection of HuH-7 cells with pcDNA-HA plasmid DNA-was also used as a control.
Immunofluorescent antibody assay (IFA). For IFA, either VeroE6 or HEK 293T cells were seeded on cover slides and transfected with 0.5 μg of either pHA-ORF8a or pORF8a-EGFP plasmid. A commercial IFA test (Euroimmun) with SARSCoV-infected VeroE6 cells was also used in this study. All slides were incubated with patient serum (at 1:200 dilution) or rabbit R26 antiserum (1:500 dilution) for 1 h at 37°C. Detailed procedures have been published elsewhere [15].
Ex vivo infection of SARS-CoV HKU39849 strain. HKU39849 strain [16] was used to infect VeroE6 cells for 5 days, and the cultural supernatant was harvested. The TCID50 was determined using the method of Reed and Muench (described in [17]). To evaluate the effect of ORF8a on viral replication, stable clones 18 and 24 were seeded in a 6-well plate and infected with 100 TCID50 of HKU39849 virus. The CPEs were recorded, and viral titers from supernatant were determined at various time intervals. To determine the effect of siRNA of orf8a on viral replication, VeroE6 cells that had been transfected with siRNA for 36 h were infected with 100 TCID50 of HKU39849. The CPEs were recorded, and the cultural supernatant was harvested for viral load (VL) tests at various time points.
Preparation of siRNA against orf8a. The 21-nt dsRNA was chemically synthesized as 2′ bis(acetoxyethoxy)-methyl ether- protected oligonucleotides (Dharmacon) and stored in 0.1% diethylpyrocarbonate-treated water at 80°C. Sequences of siRNA were subjected to a BLAST search of the expressed sequence tag library of the National Center for Biotechnology Information (available at: http:\www.ncbi.nlm.nih.gov), to ensure that no endogenous genes of the genome were targeted. The sequences of 2 sets of siRNA were set 1 ds, 5CCUCAUGUGCUUGAAGAUCdTdT-3′, and set 2 ds, 5′-GAUCCUUGUAAGGUACAACdTdT-3′ (sense). The sequence of a control siRNA against GFP was 5′-AAGCAGCACGACUUCUUCAAGdTdT- 3′.
Viral RNA extraction and real-time PCR for the measurement of VLs. One hundred forty microliters of the cultural supernatant was used to extract viral RNA using a QIAamp Viral RNA mini kit (Qiagen). Real-time RT-PCR (Applied Biosystems) with primers for replicase Ib or nucleocapsid mRNAs was used to measure the VLs. The primers and probes used have been reported elsewhere [18].
Determination of mitochondrial membrane potential (ΔΨm). The lipophilic fluorochrome 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenz-imidazolylcarbocyanine iodide (JC-1; Molecular Probes) was used to measure ΔΨm in accordance with procedures described elsewhere [19]. The cells ( 1 × 106 cells) were incubated with 5 mgJC-1 for 20 min at 37°Cin the dark and were washed with PBS before they were analyzed using flow cytometry (FACS; Becton Dickinson).
Measurement of the reactive oxygen species (ROS), oxygen consumption, and caspase 3 activity. Production of hydrogen peroxide from cultured cells was measured using 2′,7′-dichlorofluorescin diacetate (DCFH-DA; Molecular Probes). Cells were incubated with 5 μmolDCFH-DA for 60 min at 37°C, washed with PBS twice, trypsinized, and resuspended in 0.5 mL of PBS before they were subjected to FACS analysis. Oxygen uptake was measured polarographically with a Clark-type oxygen electrode (model 110; Instech). The respiration rates were calculated with respect to calibration of the oxygen electrode with air-saturated medium that contained 155 μmolO2 at 37°C. Caspase 3 activity was measured using a method modified from that of DiPietrantonio et al. [20]. In brief, 50-μg cell lysates were added to 148 mL of reaction buffer (100 mmolHEPES [pH 7.5], 20% glycerol, 0.5 mmolEDTA, and 5 mmoldithiothreitol); then, 2 μL of the caspase 3 colorimetric substrate DEVD-pNA was added to the mixture and incubated for 4hat 37°C. Fluorescence was recorded using an MRX model ELISA reader (Dynatech Laboratories) at 405 nm wavelength.
Quantification of apoptotic cells. An annexin V-fluorescein isothiocyanate (FITC) kit (BD Biosciences) was used to quantify apoptotic cells in accordance with procedures described elsewhere [21]. In brief, cells were suspended in buffer (10 mmolHEPES[pH 7.4], 140 mmolNaCl, and 2.5 mmolCaCl2) that contained 50 μgpropidium iodide and 1:20 (vol) annexin V-FITC labeling solution and were incubated on ice for 15 min before they were mixed with 400 mL of binding buffer. Fluorescence was analyzed using FACS.
Statistical methods. Means and SDs are shown in the figures. Means among experimental cell lines and HuH-7 cell line were compared using the generalized linear model. All statistical procedures were performed using SAS software (version 9.1; SAS Institute), and statistical significance was set at P < .05.
Results
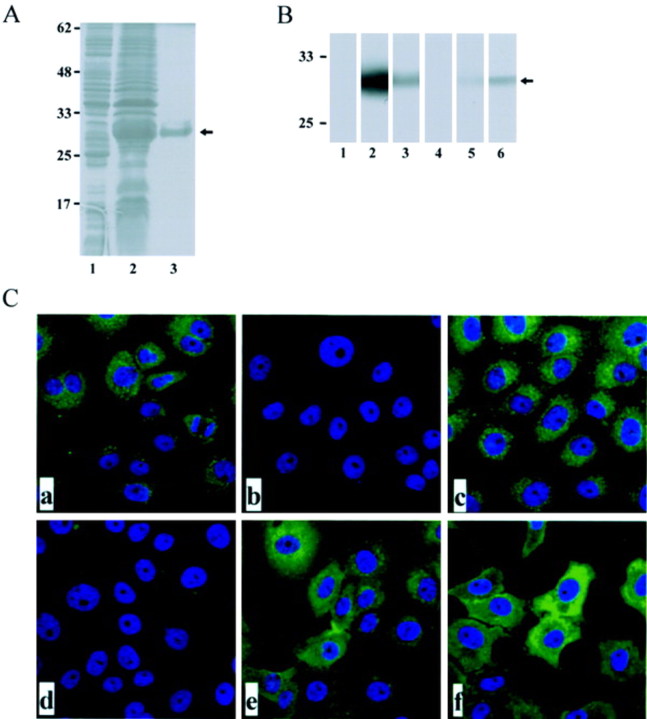
Induction and purification of ORF8a RP. To induce the GSTORF8a fusion protein, a colony of E. coli BL21 cells containing pGEX-ORF8a was grown at 37°C in Luria-Bertani broth that contained 100 μgampicillin and was treated with isopropyl-b-d-thiogalactopyranoside (IPTG). As shown in figure 1A, a protein ⩾30 kDa in size was found in the bacterial lysates induced by IPTG (lane 2) but was not found in the lysates without IPTG induction (lane 1). The ORF8a RP was further identified using an WB assay with rabbit antiserum (R26) against a synthetic peptide containing partial amino acid sequence of ORF8a (figure 1B, lane 2 ) and an anti-GST MAb (figure 1B, lane 3 ).
Figure 1.
Detection of anti-open reading frame (ORF)-8a antibody reactivity in patients infected with severe acute respiratory syndrome coronavirus. A, Induction and purification of the recombinant glutathione S-transferase (GST)-ORF8a protein. Lanes 1-3, Coomassie blue staining of an SDS-PAGE gel containing bacterial lysates (lanes 1 and 2) or GST-ORF8a fusion protein purified from glutathione-sepharose 4B bead column (lane 3). Lane 1, Before isopropyl-β-D-thiogalactopyranoside (IPTG) induction; lane 2, after IPTG induction. B, Western-blot assay with a recombinant GST-ORF8a protein. Lane 1, Preimmunized rabbit serum; lane 2, rabbit anti-ORF8a antiserum R26; lane 3, anti-GST monoclonal antibody (MAb); lane 4, normal human serum; lane 5, serum sample from patient HP631; and lane 6, serum sample from patient C596. Nos. along the left margin of panels A and B are molecular-weight markers in kilodaltons. C, Immunofluorescent antibody assay test using VeroE6 cells transfected with pHA-ORF8a. a, Anti-hemagglutinin antibody; b, preimmunized rabbit serum; c, rabbit anti-ORF8a antiserum R26 at 1:500 dilution; d, normal human serum; e, serum from patient HP631; f, serum from patient C596. The nucleus was stained with Hoechst H33258.
Antibody reactivity to ORF8a in SARS-CoV-infected patients. Purified GST-ORF8a RP was used as an antigen in a WB assay to detect anti-ORF8a antibody reactivity in patients infected with SARS-CoV. Two (5.4%; patients HP631 and C596) of 37 patients with SARS-CoV infection had anti-ORF8a antibody reactivity (figure 1B ). None of 31 control subjects had antibody reactivity (data not shown). The antibody reactivity of patients HP631 and C596 was studied using IFA. As shown in figure 1C, anti-HA antibody, R26 rabbit antiserum, and patients HP631 and C596 had positive reactivity in the cytoplasm of VeroE6 cells that had been transfected with pHA-ORF8a (figure 1C, a, c, e, and f ).
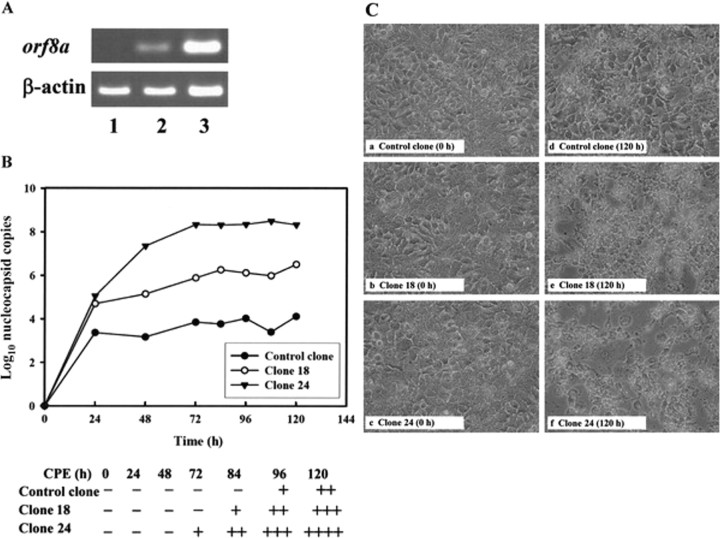
Overexpression of ORF8a-enhanced SARS-CoV replication and SARS-CoV-mediated CPE formation. Two stable cell clones (designated clones 18 and 24) expressing different levels of ORF8a were generated to study ORF8a function. As shown in figure 2A, orf8a expression in clone 24 was ⩾5 times greater than that expressed in clone 18 (lanes 2 and 3). After infection with SARS-CoV HKU39849, VLs and CPEs were monitored in both clones for 5 days. The results showed that, 24 h after infection, the VLs in both stable clones were significantly higher than those in controls ( P < .05; 1-way analysis of variance) (figure 2B ). At 48 h after infection, clone 24 showed significantly higher VLs than clone 18; this continued for at least 3 days ( P < .05) (figure 2B ). Five days after infection with SARS-CoV HKU39849, supernatant VLs in clones 18 and 24 were 60-and 13,400-fold higher, respectively, than those in controls.
Figure 2.
Enhancement viral replication and cytopathic effects (CPEs) of severe acute respiratory syndrome coronavirus (SARS-CoV) HKU39849 in 2 stable clones by open reading frame (ORF)-8a protein. A, Reverse-transcription polymerase chain reaction (PCR) analysis of orf8a mRNA levels in 3 stable clones from the HuH-7 cell line. Lane 1, control clone transfected with vector plasmid pcDNA3 DNA; lanes 2 and 3, stable clones expressing ORF8a (lane 2, clone 18; lane 3, clone 24). B, Measurements of viral loads in different stable clones infected with 100 TCID50 of SARS-CoV HKU39849. After infection, the cultural supernatants were collected at various time points for 5 days. The no. of copies of nucleocapsid mRNA was determined using real-time PCR. Plus symbols (+) refer to the percentage of cells showing a CPE in cell cultures: +, <25%; ++, 25%-50%; +++, 50%-75%; ++++, >75%. C, CPEs of SARS-CoV HKU39849 in 3 stable clones for 5 days after the viral infection.
The first appearance of CPE formation in clone 24 occurred 72 h after infection, but no CPE activity was noted in either clone 18 or the control (figure 2B, bottom ). Five days after infection, CPE formation was noted in the control, and CPE in clones 18 and 24 were measured as 3+ and 4+, respectively (figure 2C, d,e, and f ).
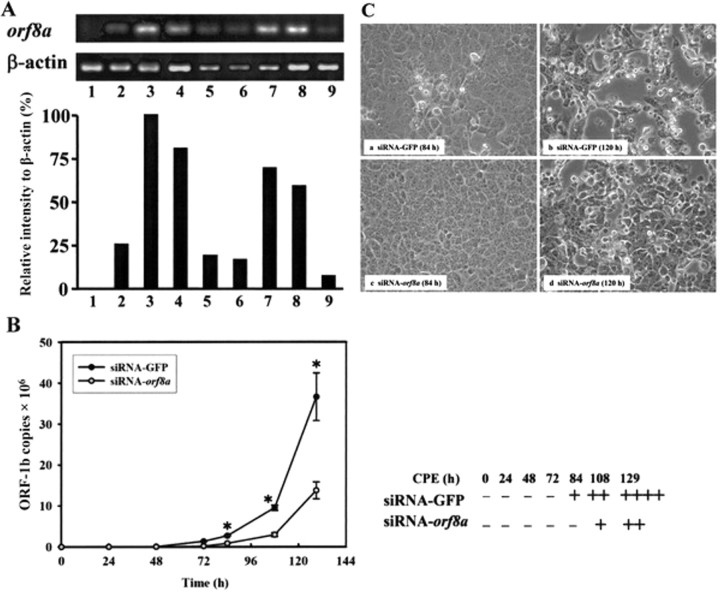
siRNA inhibition of SARS-CoV replication and CPE formation by orf8a. The effects of siRNA on orf8a mRNA expression were evaluated using VeroE6 cells cotransfected with pHA-ORF8a plasmid DNA and siRNAs targeted at orf8a. The results shown in figure 3A indicate RT-PCR detection of orf8a mRNA at 18 and 56 h after transfection (lanes 2 and 3). The suppressive effect of siRNA-orf8a set 1 was observed at 36 h, whereas the effect of siRNA-orf8a set 2 was not observed until 56 h (top and bottom, lanes 4–6 and 7–9). Compared with the control (siRNA-GFP), 80%-90% of the orf8a mRNA was suppressed by siRNAs in both sets of experiments; therefore, both sets were chosen for analysis. Next, siRNA-transfected VeroE6 cells were infected with SARS-CoV HKU39849, and their supernatant VLs were determined using real-time PCR. VLs were detected in VeroE6 cells transfected with the GFP-specific siRNA control 72 h after infection, but VLs in VeroE6 cells transfected with orf8a siRNA were not detected until 84 h after infection (figure 3B ). VL differences in the 2 cultures were more significant at 108 and 129 h after infection (figure 3B ).
Figure 3.
The effect of orf8a small interfering RNA (siRNA) on viral replication and cytopathic effects of severe acute respiratory syndrome coronavirus (SARS-CoV) HKU39849. A, Verification of the effects of 2 sets of orf8a siRNAs. Top, Results of reverse-transcription polymerase chain reaction (RTPCR) of orf8a mRNA using total RNA extracted from VeroE6 cells cotransfected with pHA-ORF8a and different siRNAs. Lane 1, pcDNA3-HA; lanes 2-9, pHA-ORF8a. The siRNAs used in different assays were as follows: lanes 2 and 3, siRNA-GFP; lanes 4-6, orf8a siRNA set 1; and lanes 7-9, orf8a siRNA set 2. The cells were harvested at different time points after transfection (18 h for lanes 2, 4, and 7; 36 h for lanes 5 and 8; and 56 h for lanes 1, 3, 6, and 9). The relative intensity (ratio) of the orf8a, compared with b-actin, in the top panel was calculated and normalized using the ratio of lane 3 (56 h of siRNA-GFP) as the 100% value. B, Measurements of the viral loads in the cultural supernatants from VeroE6 cells that had been transfected with orf8a siRNA for 36 h before they were inoculated with SARS-CoV HKU39849. After infection, the cultural supernatants were collected at various time points for 5 days. Nos. of replicase open reading frame (ORF)-1b mRNA copies were determined using real-time PCR. Error bars indicate SDs. * P < .05 (Student's t test). Plus symbols (+) refer to percentage of cells showing CPE in cell cultures: +, <25%; ++, 25%-50%; +++, 50%-75%; and ++++, 175%. C, Cytopathic effects of SARS-CoV HKU39849 in VeroE6 cells infected 120 h after transfection with different siRNAs for36h.
The results shown in figure 3B indicate CPEs in the siRNA-GFP control 84 h after infection and in siRNA-orf8a at 108 h after infection. At 129 h, cells transfected with siRNA-orf8a had lower CPEs than cells transfected with siRNA-GFP (figure 3C, d vs. b ).
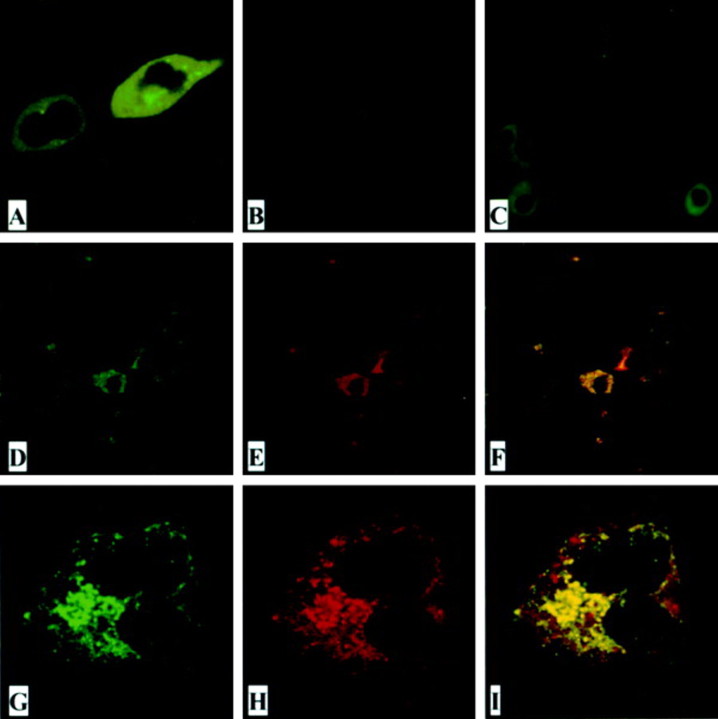
ORF8a protein expression in mitochondria. The subcellular localization of SARS-CoV ORF8a was analyzed by expressing an ORF8a-EGFP fusion protein in HEK 293T cells and examining the result using confocal microscopy. As shown in figure 4A, the fusion protein was distributed in the cytoplasm. A punctate pattern of ORF8a protein expression was observed in VeroE6 cells infected with SARS-CoV when IFA was used with anti-ORF8a synthetic peptide antiserum (R26) (figure 4C ). When we used confocal microscopic examination with a panel of fluorescence-tagged markers for endoplasmic reticulum, Golgi bodies, actin, mitochondria, and nucleus, we found that ORF8a was localized to mitochondria (figure 4D -4I ).
Figure 4.
The subcellular localization of open reading frame (ORF)-8a. A, HEK 293T cells transfected with plasmid pORF8a-EGFP. B, Severe acute respiratory syndrome coronavirus (SARS-CoV)-infected VeroE6 cells immunostained with preimmunized rabbit serum. C, SARS-CoV-infected VeroE6 cells immunostained with rabbit anti-ORF8a antiserum R26 (1:100 dilution). D-I, VeroE6 cells cotransfected with a mitochondrial expression plasmid (pEF-CKMt2-green fluorescent protein [GFP]) and pHA-ORF8a and immunostained with rabbit anti-ORF8a antiserum R26 (1:500 dilution). Rhodamine-conjugated goat anti-rabbit antibodies were used as a secondary antibody in the immunofluorescent antibody assay test. Green represents GFP (D and G); red represents ORF8a protein (E and H). Yellowish areas in merged panels (F and I) indicate ORF8a localized in mitochondria. D-F, Low magnification; G-I, high magnification.
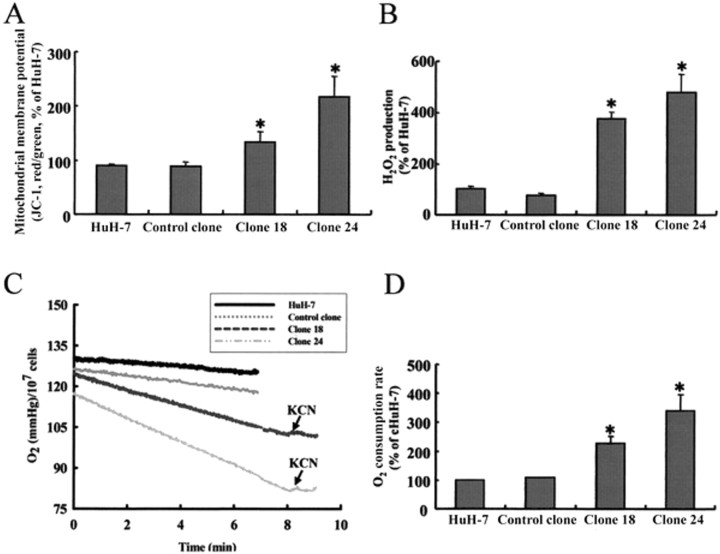
Effects of ORF8a on mitochondria. Mitochondria are essential for the survival and proliferation of eukaryotic cells. We therefore investigated the effect of ORF8a on ΔΨm. The results of a quantitative assay using JC-1 cationic dye indicated 50% and 140% increases in ΔΨm (red fluorescence) in stable clones 18 and 24, respectively, compared with a basal level in HuH7 cells (figure 5A ).
Figure 5.
The effects of open reading frame (ORF)-8a on mitochondria. A, Mitochondrial membrane potential. B, H2O2 production. C, Oxygen content. D, Oxygen consumption. Two stable ORF8a-expressing stable clones (18 and 24), a vector control clone, and the parental cell line HuH-7 were used. Data are averages of experiments performed in triplicate. Error bars indicate SDs. * P < .05. JC-1, 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenz-imidazolylcarbocyanine iodide; KCN, potassium cyanide.
Mitochondria are primary sources of ROS inside cells. To verify that the effect of ORF8a on membrane potential paralleled alterations in redox state in cells, we examined ROS production using a fluorescent dye (DCFH-DA). Results indicated significantly higher ROS production in clone 18 and 24 cells than in cells from control subjects (figure 5B ). Next, we used an oxygen microelectrode to monitor the effect of SARS-ORF8a on cellular oxygen consumption. Cells were cultured in air-saturated medium that contained 155 μmoloxygen before oxygen uptake was measured. Compared with HuH-7 cells and vector control clones, oxygen accumulation was repressed in ORF8a-expressing clones-especially clone 24 (figure 5C )- although both clones had significantly higher levels of oxygen consumption than control cells ( P < .05) (figure 5D ). We noted that this repression could be reversed by the addition of potassium cyanide, a potent mitochondrial respiratory chain inhibitor.
Induction of apoptosis by ORF8a through the induction of caspase 3 activity. Knowing that mitochondrial dysfunction can result in cellular apoptosis, we decided to use both transient transfection and stable clones to study the association between apoptosis and orf8a gene expression. As shown in figure 6A, the percentages of apoptotic cells in HuH-7 cells transfected with 3.75 and 7.5 mg of pHA-ORF8a plasmid DNA were 7.4% and 19.9%, respectively, when annexin V staining and FACS were used, whereas only 2.9% of the cells transfected with vector plasmid DNA underwent apoptosis. In this assay, the HIV-1 Tat protein served as a positive control, and ⩾10.7% cells underwent apoptosis after they were transfected with 7.5 μgpf pcDNA3-Tat plasmid DNA (figure 6A ). In terms of the stable clones, ⩾3.0% and ⩾4.2% of clones 18 and 24 were apoptotic. Clone 24 cells had significantly higher percentage of apoptotic cells than the control clone ( P < .05) (figure 6B ). Subsequently, we analyzed caspase 3 activity in the stable clones and found that both clones 18 and 24 had significantly higher activity than the vector control clone ( P < .05) (figure 6C ).
Figure 6.
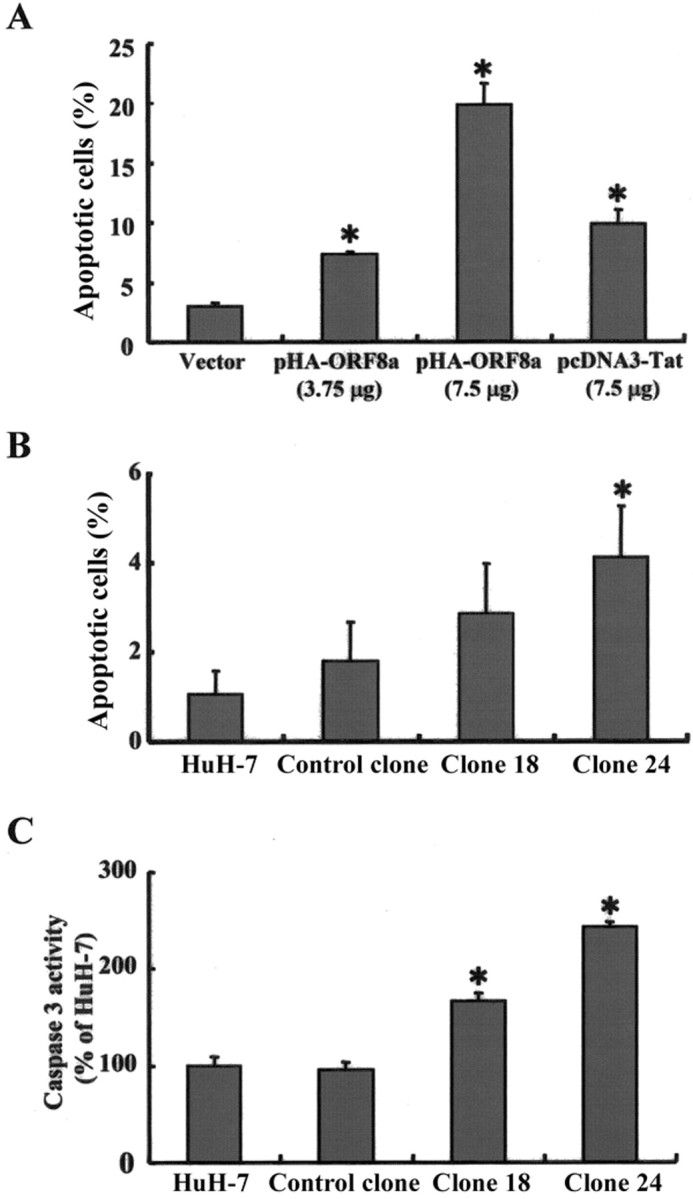
Open reading frame (ORF)-8a triggering apoptosis via a caspase 3-dependent pathway. A, Flow cytometry (FACS) with annexin V staining of HuH-7 cells that had been transfected with the following plasmid DNAs for 24 h: a vector control (pcDNA3-HA, 7.5 μg), pHAORF8a (3.75 μg), pHA-ORF8a (7.5 μg), and a positive control (pcDNA3Tat, 7.5 μg). B, FACS with annexin V staining of 2 stable clones expressing ORF8a (18 and 24), a control clone, and its parental cell line, HuH-7. C, Caspase 3 activity of the clones and cells mentioned above. Data are averages of experiments performed in triplicate. Error bars indicate SDs. *P < .05.
Discussion
In the present study, we induced and purified a GST-ORF8a fusion protein in E. coli for WB analysis. As shown in figure 1A, the size of the GST-ORF8a RP was ⩾30 kDa, which is very close to our estimation, given that the predicted sizes of GST and ORF8a were 26 and 4.4 kDa, respectively. Subsequently, we used WB with GST-ORF8a RP to detect anti-ORF8a antibody reactivity in 37 patients with SARS, and we found that 2 of them were seropositive (figure 1B ). The anti-ORF8a antibody reactivity was further confirmed by IFA with cells expressing HA-tagged ORF8a (figure 1C, e and f ). To our knowledge, this is the first study demonstrating anti-ORF8a antibody reactivity in patients with SARS. It has been reported that the region immediately upstream of orf8a gene contains a consensus transcription regulating sequence (TRS) [22, 23]. A 2-kb subgenomic RNA has been identified in a Northern-blot analysis as potentially responsible for ORF8a translation [24]. Therefore, orf8a encodes a protein that can induce antibody reactivity in some patients infected with SARS-CoV.
In general, viral structural proteins have higher immunogenicity than regulatory proteins [25, 26]. In addition, antibodies to virus regulatory proteins may be detectable at particular disease stages [27, 28]. According to clinical histories, none of the 7 patients in our study who died of SARS had antiORF8a antibody. Both patients with anti-ORF8a antibody (C596 and HP631) recovered from SARS-CoV infection, and neither of them developed acute respiratory distress syndrome (ARDS). Recently, we evaluated, using a WB assay, anti-spike and anti-nucleocapsid antibody reactivity among 108 medical staffpersons who had acquired SARS coronavirus infection in 2003, and we found 8 persons had reactivity to both antibodies. Among them, 1 person who had ARDS did not have antiORF8a antibody, whereas 2 of 7 individuals who did not have ARDS had anti-ORF8a antibody reactivity. It seems that patients with anti-ORF8a antibody reactivity had less-severe symptoms and a better prognosis. More research is needed to determine the association between the presence of anti-ORF8a antibody and the prognosis of SARS-CoV infection.
In this study, we found that ORF8a enhances SARS-CoV HK39849 replication very efficiently (figure 2B ). Furthermore, compared with the siRNA-GFP control, siRNA-orf8a inhibited >50% of the replication of SARS-CoV (figure 3B ). Previous studies have shown that both cellular and viral proteins can enhance the transcription of CoV via interaction with its leader RNA and TRS [29-31]. We performed a UV cross-linking assay to examine whether ORF8a has RNA-binding activity. Preliminary results showed that neither positive-strand nor negative-strand leader RNA of SARS-CoV interacted with GST-ORF8a fusion protein directly. It cannot be ruled out that the ORF8a protein interacts with leader RNA indirectly through other cellular proteins or that orf8a RNA other than the ORF8a protein is the basis of these effects. Further studies are needed to determine the mechanism of transactivation of ORF8a.
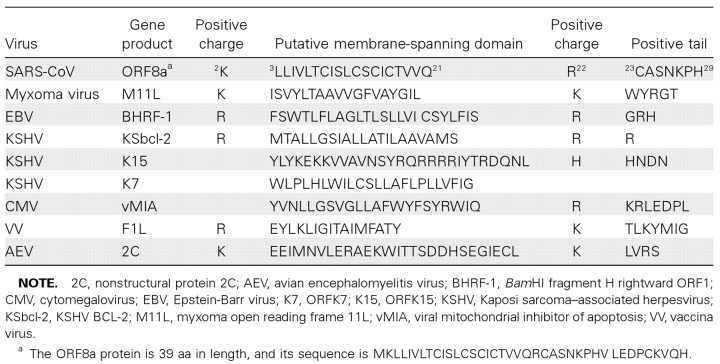
Results from an IFA and a series of functional assays show that the subcellular localization of ORF8a is in mitochondria. A consensus sequence motif containing a hydrophobic membrane-spanning domain 18–24 aa in length and flanked by positively charged residues for directing proteins to mitochondria has already been defined [32]. This motif exists in many viral proteins that target mitochondria, including the Bam HI fragment H rightward ORF1 in Epstein-Barr virus, F1L in vaccinia virus, and vBcl-2 in human herpesvirus type 8 [33-35]. According to the results of our sequence analysis, there is a hydrophobic membrane-spanning domain at the N-terminal region of ORF8a. This membrane-spanning domain is 19 aa in length and is surrounded by positively charged amino acids (table 1). Therefore, ORF8a may be located in mitochondria through a mitochondria-targeting signal at its N-terminal region.
Table 1.
Putative mitochondrial targeting signal found in the open reading frame (ORF)-8a protein of severe acute respiratory syndrome coronavirus (SARS-CoV) and gene products of other viruses.
In this study, we found that nearly 20% of cells underwent apoptosis after they were transfected with ORF8a-expressing plasmid DNA (figure 63A ). In addition, the annexin V intensity in ORF8a-expressing cells was greater than that of cells transfected with vector plasmid, which indicates that ORF8a induces severe apoptosis. Caspase 3, a potent enzyme in apoptotic signaling, was found to be activated by ORF8a. Further studies are needed to elucidate whether other pathways are also activated by ORF8a.
Many viral genomes encode proteins that can modulate apoptosis in their host cells [36-38]. It has been suggested that both apoptotic suppression and induction can be beneficial to viral replication, persistence, dissemination, and pathogenesis. Our results indicate that overexpression of ORF8a resulted in the hyperpolarization of mitochondria membrane potential, an increase in ROS production, and cell apoptosis. Mitochondrial hyperpolarization is known to be associated with yeast and lymphoblastoid cells committed to apoptosis [39, 40]. ORF8a therefore regulates the production of free radicals and causes hyperpolarization of membrane potential, which is a sensitive indicator of mitochondrial function, resulting in the occurrence of apoptosis [41, 42].
Previously, several reports have shown that patients with SARS frequently had lymphocytopenia, thrombocytopenia, hypocalcemia, and abnormal liver function [43-45]. Furthermore, apoptotic cells have been found in epithelial cells taken from the lungs of patients with SARS [46, 47]. Recently, several studies found that SARS-CoV ORF3a and ORF7a proteins induce apoptosis in VeroE6 cells, but whether ORF3a or ORF7a protein can transactivate viral replication is unclear [48-50]. Because ORF8a not only enhances viral replication but also induces apoptosis, it plays an important role in the pathogenesis of human SARS-CoV infection.
Acknowledgments
We thank Drs. Yu-Ching Lan and Shao-Yuan Chuang for their helpful discussions; Shu-Fen Lai, Chun-Chih Li, and Chi-Wei Wu for technical assistance; and Dr. Shin-Ru Shih (Chang Gung University) for providing the recombinant hnRNP A1 protein and PTB protein. We also acknowledge Dr. Tze-Tze Liu of the National Yang-Ming University Genome Research Center for help with sequencing.
Footnotes
Potential conflicts of interest: none reported.
Presented in part: Fourteenth Symposium on the Recent Advances in Cellular and Molecular Biology, Kenting, Taiwan, 18-20 January 2006 (poster 204).
Financial support: Veterans General Hospitals University System of Taiwan Joint Research Program, Tsou Foundation (grant VGHUST95-P7-22).
References
- 1.Fouchier RAM, Kuiken T, Schutten M, et al. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–9. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 5.Marra MA, Jones SJM, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Lau SK, Woo PC, Li KS, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–5. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–8. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 8.Lan YC, Liu HF, Shih YP, Yang JY, Chen HY, Chen YM. Phylogenetic analysis and sequence comparisons of structural and non-structural SARS coronavirus proteins in Taiwan. Infect Genet Evol. 2005;5:261–9. doi: 10.1016/j.meegid.2004.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YM, Liang SY, Shih YP, et al. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J Clin Microbiol. 2006;44:359–65. doi: 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YM, Zhang XQ, Dahl CE, et al. Delineation of type-specific regions on the envelope glycoproteins of human T cell leukemia viruses. J Immunol. 1991;147:2368–76. [PubMed] [Google Scholar]
- 11.Chen YM, Hu CP, Chen PH, et al. Nuclear antigens reacted with sera and ascites of hepatocellular carcinoma patients. Hepatology. 1988;8:547–52. doi: 10.1002/hep.1840080319. [DOI] [PubMed] [Google Scholar]
- 12.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–63. [PubMed] [Google Scholar]
- 13.Kitamura T, Morita C, Komatsu T, et al. Isolation of virus causing hemorrhagic fever with renal syndrome (HFRS) through a cell culture system. Jpn J Med Sci Biol. 1983;36:17–25. doi: 10.7883/yoken1952.36.17. [DOI] [PubMed] [Google Scholar]
- 14.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 15.Chen SY, Lin JR, Darbha R, Lin P, Liu TY, Chen YM. Glycine Nmethyltransferase tumor susceptibility gene in the benzo(a)pyrene-detoxification pathway. Cancer Res. 2004;64:3617–23. doi: 10.1158/0008-5472.CAN-03-3726. [DOI] [PubMed] [Google Scholar]
- 16.Ruan YJ, Wei CL, Ee AL, et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–85. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pizzi M. Sampling variation of the fifty percent end-point, determined by the Reed-Muench (Behrens) method. Hum Biol. 1950;22:151–90. [PubMed] [Google Scholar]
- 18.Emery SL, Erdman DD, Bowen MD, et al. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004;10:311–6. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smiley ST, Reers M, Mottola-Hartshorn C, et al. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci USA. 1991;88:3671–5. doi: 10.1073/pnas.88.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiPietrantonio AM, Hsieh T, Wu JM. Activation of caspase 3 in HL60 cells exposed to hydrogen peroxide. Biochem Biophys Res Commun. 1999;255:477–82. doi: 10.1006/bbrc.1999.0208. [DOI] [PubMed] [Google Scholar]
- 21.van EM, Ramaekers FC, Schutte B, Reutelingsperger CP. A novel assay to measure loss of plasma membrane asymmetry during apoptosis of adherent cells in culture. Cytometry. 1996;24:131–9. doi: 10.1002/(SICI)1097-0320(19960601)24:2<131::AID-CYTO5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Hu J, Wang J, et al. Genome organization of the SARS-CoV. Genomics Proteomics Bioinformatics. 2003;1:226–35. doi: 10.1016/S1672-0229(03)01028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snijder EJ, Bredenbeek PJ, Dobbe JC, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiel V, Ivanov KA, Putics A, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–15. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 25.Chen YM, Essex M. Immunogenicity of human retrovirus-specific proteins. In: Papas TS, ed. Gene regulation and AIDS. Houston: Gulf Publishing, 1990:239–45. [Google Scholar]
- 26.Chen YM, Chen SH, Fu CY, Chen JY, Osame M. Antibody reactivities to tumor-suppressor protein p53 and HTLV-I Tof, Rex and Tax in HTLV-I-infected people with differing clinical status. Int J Cancer. 1997;71:196–202. doi: 10.1002/(sici)1097-0215(19970410)71:2<196::aid-ijc12>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda Z, Chou MJ, atsuda M, et al. Human immunodeficiency virus type 1 has an additional coding sequence in the central region of the genome. Proc Natl Acad Sci USA. 1988;85:6968–72. doi: 10.1073/pnas.85.18.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YM, Lin RH, Lee CM, Fu CY, Chen SC, Syu WJ. Decreasing levels of anti-Nef antibody correlate with increasing HIV type 1 viral loads and AIDS disease progression. AIDS Res Hum Retroviruses. 1999;15:43–50. doi: 10.1089/088922299311691. [DOI] [PubMed] [Google Scholar]
- 29.Baric RS, Nelson GW, Fleming JO, et al. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–7. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li HP, Zhang X, Duncan R, Comai L, Lai MM. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiel V, Herold J, Schelle B, Siddell SG. Viral replicase gene products suffice for coronavirus discontinuous transcription. J Virol. 2001;75:6676–81. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everett H, Barry M, Lee SF, et al. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J Exp Med. 2000;191:1487–98. doi: 10.1084/jem.191.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boya P, Pauleau AL, Poncet D, Gonzalez-Polo RA, Zamzami N, Kroemer G. Viral proteins targeting mitochondria: controlling cell death. Biochim Biophys Acta. 2004;1659:178–89. doi: 10.1016/j.bbabio.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Cheng EH, Nicholas J, Bellows DS, et al. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–4. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khanim F, Dawson C, Meseda CA, Dawson J, ackett M, Young LS. BHRF1, a viral homologue of the Bcl-2 oncogene, is conserved at both the sequence and functional level in different Epstein-Barr virus isolates. J Gen Virol. 1997;78:2987–99. doi: 10.1099/0022-1317-78-11-2987. [DOI] [PubMed] [Google Scholar]
- 36.Park IW, Ullrich CK, Schoenberger E, Ganju RK, Groopman JE. HIV1 Tat induces microvascular endothelial apoptosis through caspase activation. J Immunol. 2001;167:2766–71. doi: 10.4049/jimmunol.167.5.2766. [DOI] [PubMed] [Google Scholar]
- 37.Perry SW, Norman JP, Litzburg A, Zhang D, Dewhurst S, Gelbard HA. HIV-1 transactivator of transcription protein induces mitochondrial hyperpolarization and synaptic stress leading to apoptosis. J Immunol. 2005;174:4333–44. doi: 10.4049/jimmunol.174.7.4333. [DOI] [PubMed] [Google Scholar]
- 38.Singh IN, Goody RJ, Dean C, et al. Apoptotic death of striatal neurons induced by human immunodeficiency virus-1 Tat and gp120: differential involvement of caspase-3 and endonuclease G. J Neurovirol. 2004;10:141–51. doi: 10.1080/13550280490441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matarrese P, Testa U, Cauda R, Vella S, Gambardella L, Malorni W. Expression of P-170 glycoprotein sensitizes lymphoblastoid CEM cells to mitochondria-mediated apoptosis. Biochem J. 2001;355:587–95. doi: 10.1042/bj3550587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross A, Pilcher K, Blachly-Dyson E, et al. Biochemical and genetic analysis of the mitochondrial response of yeast to BAX and BCL-X(L). Mol Cell Biol. 2000;20:3125–36. doi: 10.1128/mcb.20.9.3125-3136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narita M, himizu S, Ito T, et al. Bax interacts with the permeability transition pore to induce permeability transition and cytochrome c release in isolated mitochondria. Proc Natl Acad Sci USA. 1998;95:14681–6. doi: 10.1073/pnas.95.25.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starkov AA, Fiskum G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. J Neurochem. 2003;86:1101–7. doi: 10.1046/j.1471-4159.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- 43.Lang ZW, Zhang LJ, Zhang SJ, et al. A clinicopathological study of three cases of severe acute respiratory syndrome (SARS). Pathology. 2003;35:526–31. doi: 10.1080/00313020310001619118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong RS, Wu A, To KF, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326:1358–62. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Donnell R, Tasker RC, Roe MF. SARS: understanding the corona-virus: apoptosis may explain lymphopenia of SARS. BMJ. 2003;327:620. doi: 10.1136/bmj.327.7415.620-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lang Z, Zhang L, Zhang S, et al. Pathological study on severe acute respiratory syndrome. Chin Med J. 2003;116:976–80. [PubMed] [Google Scholar]
- 47.Zhang QL, Ding YQ, He L, et al. Detection of cell apoptosis in the pathological tissues of patients with SARS and its significance. Di Yi Jun Yi Da Xue Xue Bao. 2003;23:770–3. [PubMed] [Google Scholar]
- 48.Law PT, Wong CH, Au TC, et al. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J Gen Virol. 2005;86:1921–30. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- 49.Tan YJ, Fielding BC, Goh PY, et al. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol. 2004;78:14043–7. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kopecky-Bromberg SA, Martinez-Sobrido L, alese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J Virol. 2006;80:785–93. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]