Abstract
The antibiotic and heavy metal resistome of a chronically polluted soil (3S) obtained from an automobile workshop in Ilorin, Kwara State, Nigeria was deciphered via functional annotation of putative ORFs (open reading frames). Functional annotation of antibiotic and heavy metal resistance genes in 3S metagenome was conducted using the Comprehensive Antibiotic Resistance Database (CARD), Antibiotic Resistance Gene-annotation (ARG-ANNOT) and Antibacterial Biocide and Metal Resistance Gene Database (BacMet). Annotation revealed detection of resistance genes for 15 antibiotic classes with the preponderance of beta lactamases, mobilized colistin resistance determinant (mcr), glycopepetide and tetracycline resistance genes, the OqxBgb and OqxA RND-type multidrug efflux pumps, among others. The dominance of resistance genes for antibiotics effective against members of the Enterobacteriaceae indicate possible contamination with faecal materials. Annotation of heavy metal resistance genes revealed diverse resistance genes responsible for the uptake, transport, detoxification, efflux and regulation of copper, zinc, cadmium, nickel, chromium, cobalt, mercury, arsenic, iron, molybdenum and several others. Majority of the antibiotic and heavy metal resistance genes detected in this study are borne on mobile genetic elements, which facilitate their spread and dissemination in the polluted soil. The presence of the heavy metal resistance genes is strongly believed to play a major role in the proliferation of antibiotic resistance genes. This study has established that soil is a huge repertoire of antibiotic and heavy metal resistome and due to the intricate link between human, animals and the soil environment, it may be a major contributor to the proliferation of multidrug-resistant clinical pathogens.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02219-z) contains supplementary material, which is available to authorized users.
Keywords: Heavy metals, Antibiotics, Shotgun metagenomics, Heavy metal resistome, Antibiotic resistome
Introduction
Soil, the naturally occurring unconsolidated mineral and organic material is home to a huge and diverse population of microorganisms due to its tremendous range of habitats determined by a complex interplay between several biotic and abiotic factors. Extensive inundation of the soil environment with a coterie of anthropogenic pollutants such as heavy metals and antibiotics imposes selective pressure on the microbial community and forces it to develop counteractive strategies for adaptation, survival and successful reproduction (Chandra and Kumar 2017). While intrinsic resistance against natural antibiotics produced by soil microorganisms could select for microorganisms with antibiotic resistance phenotypes, acquired resistance caused by the presence in the soil of potentially offensive compounds and conditions might select for specific and non-specific mechanisms of resistance (Martin and Liras 1989; Hopwood 2007; Tahlan et al. 2007; Allen et al. 2010).
It is widely believed that poor sanitary conditions and urban infrastructure, lack of regulatory and enforcement policies, and corruption, particularly in developing countries have allowed unrestricted environmental release of antibiotics from diverse point and non-point sources (Collignon et al. 2015). This has escalated the incidences of multidrug resistance particularly among clinical isolates due to the intricate link that exist between humans, animals and their environment (Allen et al. 2010). In soil, acquired resistance by microorganisms may be traced to various human activities such as excretion of antibiotics via open defecation and urination by medicated humans and pets; running urban sewage carrying antibiotics from houses and hospitals as well as wastewaters from fish ponds in open, exposed channel, and sometimes directly onto the streets (Amabile-Cuevas 2016). Others include leachates from antibiotic-containing wastes; surface runoff from solid waste municipal dumpsite; and the usage of antibiotic-containing wastewater and livestock manure in agriculture (Arun et al. 2017).
Microorganisms have deployed various strategies to counteract the toxic effects of antibiotics. These include active efflux of the antibiotic from the microbial cell; modification of antibiotic targets; enzymatic modification of the antibiotic; enzymatic degradation of the antibiotic; creating bypass or alternative metabolic pathways to those inhibited by the antibiotic; overproduction of the target enzyme; and access restriction of antibiotic to target sites via permeability changes in the bacterial cell wall (van Hoek et al. 2011; Panesyan et al. 2015).
Naturally, soil can have high concentrations of heavy metals due to weathering of parental material resulting in high concentrations of heavy metal minerals (Kamal et al. 2010). However, anthropogenic activities such as mining, long term application of sewage sludge, fertilizers, pesticides and herbicides, composted municipal solid wastes, improper waste disposal practices, smelting, wastewater irrigation, manufacturing and agrochemicals, and indiscriminate disposal of spent oils rich in heavy metals have exacerbated the heavy metals burden in the soil (Sandaa et al. 1999; Gans et al. 2005; Khan et al. 2017). Microorganisms have developed efficient detoxification strategies to counteract the toxic effects of heavy metal stress. These include intracellular sequestration, export, reduced permeability, extracellular sequestration, and extracellular detoxification (Rough et al. 2005). The resistance determinants are mostly encoded on mobile genetic elements such as plasmid and transposons, which facilitate promiscuous transmission and dissemination of the genes among members of the microbial community via horizontal gene transfer (Silver and Walderhaug 1992; Osborn et al. 1997).
Evidences abound suggesting that co-selection mechanisms such as co-resistance, cross-resistance and co-regulation contributed significantly to the maintenance and promotion of antibiotic resistance in the microbial population in the absence of antibiotics (Pal et al. 2015a, b, 2017). Cross-resistance mechanism, which occurs when a single mechanism provides resistance to different compounds, have been reported for efflux pumps with broad substrate specificity, which protect the cell from both antibiotics and heavy metals (Mata et al. 2000; Nies 2003a, b; Blanco et al. 2016). Co-selection of antibiotic and heavy metal resistance is also promoted by the co-resistance mechanism, which occurs when two or more different resistance genes are co-located on a plasmid or a transposon. This mechanism has been fingered in co-resistance to copper, erythromycin, vancomycin, and tetracycline (Hasman and Aarestrup 2002; Amachawadi 2011); copper, silver, β-lactam and fluoroquinolone (Fang et al. 2016); cadmium, zinc, and methicillin (Cavaco et al. 2011); copper, silver, mercury, colistin, ampicillin, sulphonamide, tetracycline, streptomycin, and chloramphenicol (Campos et al. 2016), and several others. Furthermore, there are reports that microorganisms use regulatory proteins, which control the expression of heavy metal resistance genes to confer resistance to antibiotics, a phenomenon termed co-regulatory mechanism (Perron et al. 2004; Pal et al. 2017). The concept of co-selection has heightened the rate of spread and dissemination of antibiotic resistance genes in the environments, which, in turn has increase the emergence of multidrug resistance clinical pathogens, constituting an alarming threat to public and environmental health.
The use of shotgun metagenomics to explore perturbed and pristine soil environments has gained ascendancy due to its ability to reveal intricate details about the structural and functional properties of the studied environment. In this study, an attempt was made to use shotgun metagenomics to unravel the antibiotic and heavy metal resistome of a chronically polluted soil and highlight the implications of their presence on public and environmental health.
Materials and methods
Sampling site description, microcosm set up, and determination of residual hydrocarbons
Hydrocarbon-polluted soil samples were collected from an automobile workshop at Taiwo, Ilorin, Nigeria. The coordinates of the sampling site were latitude 8°28′ 42.4ʺ N and longitude 4°32′15.6ʺ E. The site has a long history of hydrocarbon contamination spanning a period of more than 10 years. Prior to this time, small-scale agriculture is the predominant activity in this area. Until date, because the workshop is situated on an open expanse of land, traditional cattle rearing, as well as poor hygiene practices such as open defecation and urination is very common in this area. Samples were collected at a depth of 10–12 cm with a sterile hand trowel, sieved (4 mm) and thoroughly mixed in a large plastic bag to avoid variability among the results of replicate soil samples. Details on the soil microcosm (polluted soil, 3S) set up, incubation conditions, and residual hydrocarbons have been reported previously (Salam and Ishaq 2019). The physicochemical properties of the polluted soil indicate a pH of 6.76, organic matter content of 1.38%, total nitrogen of 0.13%, and phosphorus and potassium content of 6.38 and 0.15 mg/kg, respectively (Salam and Ishaq 2019).
DNA extraction, library construction, sequencing, and metagenome properties
Total DNA used for metagenomic analysis was extracted directly from 3S soil microcosm (0.25 g) using ZYMO soil DNA extraction Kit (Model D 6001, Zymo Research, USA) following the manufacturer’s instructions. DNA concentration and quality were ascertained using NanoDrop spectrophotometer and electrophoresed on a 0.9% (w/v) agarose gel, respectively. Shotgun metagenomic of 3S soil microcosm was prepared using the Illumina Nextera XT sample processing kit and sequenced on a MiSeq. The protocols for total DNA preparation for Illumina shotgun sequencing were as described previously (Salam 2018; Salam and Ishaq 2019). The sequence reads of 3S metagenome were deposited on the MG-RAST server with the ID 4704694.3 and can be accessed with the link https://www.mg-rast.org/linkin.cgi?project=mgp18598.
Sequence reads from the 3S microcosm set up were assembled individually by VelvetOptimiser v2.2.5, and the resulting contigs fed into the MG-RAST metagenomic analysis pipeline. The sequences were assembled into 1239 unique contigs with a total of 314,848 bp, an average sequence length of 254 ± 66 bp, and the mean GC content of 61 ± 6%, respectively. After dereplication and quality control by the MG-RAST, sequence reads in 3S metagenome reduced to 1,064 with 260,627 bp, and an average sequence length of 245 ± 55 bp (Salam and Ishaq 2019).
Accession number
The data, metadata and sequence reads of the metagenome used in this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB36986 (https://www.ebi.ac.uk/ena/data/view/PRJEB36986).
Functional analyses of 3S metagenome for antibiotic and heavy metal resistome
Gene calling was performed on the 3S contigs using FragGeneScan (Rho et al. 2010) to predict open reading frames (ORFs). The ORFs were functionally annotated for antibiotic and heavy metal resistance genes using the Comprehensive Antibiotic Resistance Database (CARD, McArthur et al. 2013), the Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT V6; Gupta et al. 2014), and BacMet (Pal et al. 2014), a function-specific bioinformatics resource for the detection of antibacterial biocide and metal-resistance genes. In CARD, the Resistance Gene Identifier (RGI 5.1.0, CARD 3.0.7) was used to predict antibiotic resistome from the protein sequences (ORFs) of the 3S metagenome. Strict significance was calculated based on CARD curated bitscore cut-offs for metagenomic reads, which allow prediction of partial genes. In ARG-ANNOT, 3S protein sequences (ORFs) were fed into the functional annotation resource, which uses NCBI blastp program to annotate the protein sequences and provide information on the existing and putative new antibiotic resistance genes (ARG) detected in the protein sequences based on coverage and similarity. In BacMet, the protein sequences (ORFs) were presented as a query to the BacMet database (version 2.0) using default parameters for identification of metal-resistance genes. A modified stand-alone version of the BLAST program (NCBI, version 2.2.2) implemented in the BacMet webserver was used for similarity searches against the BacMet sequence databases.
Results
Functional annotation of 3S metagenome ORFs using CARD, ARG-ANNOT and BacMet revealed the presence of antibiotic resistance genes for several antibiotic classes and resistance genes responsible for transport, efflux, and detoxification of heavy metals.
Antibiotic resistance genes (ARG)
β-lactamase genes
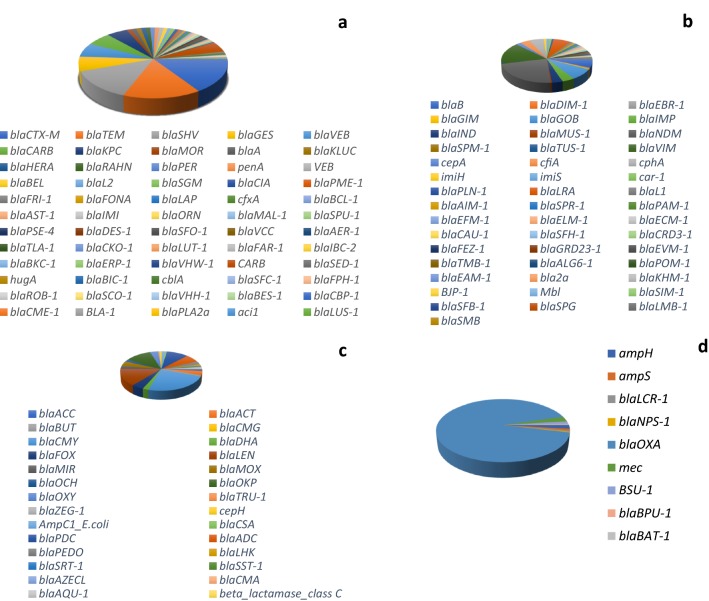
In 3S metagenome, 144 β-lactamase genes cutting across the four Ambler classes (A-D) were recovered (Fig. 1). In class A, 60 genes were detected. The predominant genes include blaCTX-M (146; 15.7%), blaTEM (142; 15.2%); blaSHV (126; 13.5%); blaGES (62; 6.7%), and blaVEB (59; 6.3%). Forty-six (46) β-lactamase genes belonging to class B were retrieved from 3S metagenome with the preponderance of blaNDM (151; 22.1%), blaVIM (106; 15.5%); blaGOB (55; 8.1%); blaLRA (46; 6.7%); and blaB (44; 6.5%), respectively. Twenty-eight (28) genes belonging to class C were recovered from 3S metagenome with blaCMY (273; 23.8%), blaLEN (161; 14.0%), blaOKP (135; 11.7%), blaPDC (110; 9.6%), and blaADC (74; 6.4%) preponderant. Only nine (9) genes belonging to class D were retrieved from 3S metagenome with the dominance of blaOXA gene (n = 507) constituting 91.7% of class D β-lactamase genes recovered (Fig. 1). An integral membrane β-lactamase regulatory protein blaR1, and 17 variants of penicillin-binding protein PBP were also recovered from the metagenome.
Fig. 1.
Distribution of resistance genes encoding class A (a), class B (b), class C (c), and class D β-lactamases in 3S metagenome (d). In class A, blaTEM, blaCTX-M, and blaSHV are the dominant resistance genes. In class B, blaNDM, blaVIM and blaGOB are the resistance genes with the highest representation. Class C β-lactamases are dominated by blaCMY, blaLEN, blaOKP and blaPDC while in class D, blaOXA is predominant. Majority of class A β-lactamases and blaOXA detected in this study belong to extended spectrum β-lactamases (ESBLs)
Aminoglycoside and MLS (macrolide-lincosamide-streptogramin) resistance genes
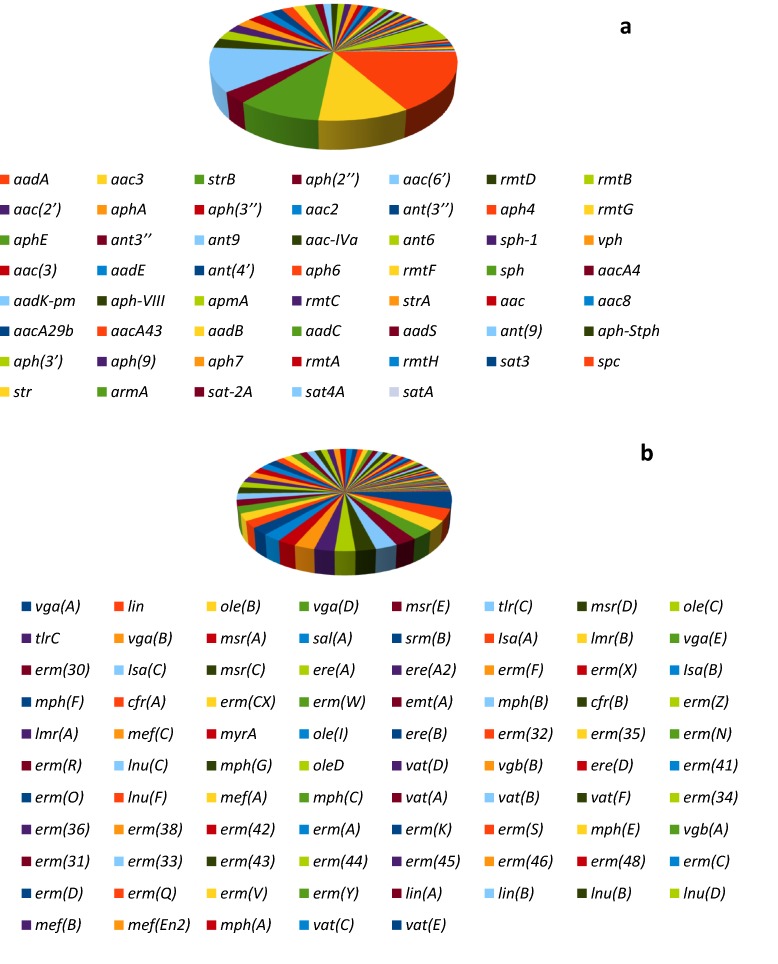
Fifty-six (56) aminoglycoside resistance genes were recovered from 3S metagenome (Fig. 2a). The recovered genes belong to the major classes of aminoglycoside resistance genes. This include acetyltransferase ACT (aac; sat; apmA), nucleotidyltransferase/adenyltransferase NUT (aad; sph; str; ant), methyltransferase MET (rmt; spc; armA), and phosphotransferase (aph; vph). Distribution of aminoglycoside resistance genes in 3S metagenome revealed the preponderance of aadA (80; 16.6%), aac(6′) (58; 12.1%), aac3 (48; 10.0%), strB (45; 9.4%), and aph(3′) (24; 5.0%), respectively.
Fig. 2.
Distribution of aminoglycoside (a) and MLS (b) resistance genes in 3S metagenome. The dominant aminoglycoside resistance genes are those belonging to the acetyltransferases ACT (aac) and the nucleotidyltransferase/adenyltransferase NUT (aadA, strB). Based on MLS resistance mechanisms, 37 genes (cfr; erm; tlr; myr; emt) is responsible for rRNA methylase, 23 genes (Isa; lmr; mef; msr; ole; srm; vga; sal(A)) is responsible for efflux, and 25 genes (esterase (ere); lyase (vgb); transferase (lnu, vat, lin); phosphorylase (mph)) is responsible for inactivating genes
Eighty-five (85) MLS resistance genes responsible for resistance mechanisms such as rRNA methylase, efflux and inactivating genes were retrieved from 3S metagenome (Fig. 2b). Of these, 37 genes (cfr; erm; tlr; myr; emt) is responsible for rRNA methylase, 23 genes (Isa; lmr; mef; msr; ole; srm; vga; sal(A)) is responsible for efflux, and 25 genes (esterase (ere); lyase (vgb); transferase (lnu, vat, lin); phosphorylase (mph)) is responsible for inactivating genes. Distribution of MLS resistance genes in the metagenome revealed the predominance of vga(A) (23; 5.1%), lin (16; 3.6%), ole(B) (15; 3.3%), vga(D) (14; 3.1%), and msr(E) (13; 2.9%), respectively. Other MLS resistance gene detected only in CARD is macB, a macrolide export ATP-binding cassette (ABC) efflux pump (Table 1).
Table 1.
Antibiotic resistome detected in 3S metagenome (using CARD) involved in resistance to different classes of antibiotics
| ORF ID | ARO term | SNP | Detection criteria | AMR gene family | Drug class | Resistance mechanism | % identity of matching region | % length of reference sequence |
|---|---|---|---|---|---|---|---|---|
| NODE_119_length_161_cov_1.937888.1 | arlR | Protein homolog model | Major facilitator superfamily (MFS) antibiotic efflux pump | Fluoroquinolone antibiotic, acridine dye | Antibiotic efflux | 44.9 | 34.25 | |
| NODE_206_length_233_cov_1.257511.1 | macB | Protein homolog model | ATP-binding cassette (ABC) antibiotic efflux pump | Macrolide antibiotic | Antibiotic efflux | 41.43 | 11.34 | |
| NODE_358_length_253_cov_1.557312.1 | Corynebacterium striatum tetA | Protein homolog model | Major facilitator superfamily (MFS) antibiotic efflux pump | Penam, tetracycline antibiotic | Antibiotic efflux | 61.22 | 20.47 | |
| NODE_453_length_289_cov_1.944637.1 | macB | Protein homolog model | ATP-binding cassette (ABC) antibiotic efflux pump | Macrolide antibiotic | Antibiotic efflux | 34.55 | 18.17 | |
| NODE_495_length_248_cov_1.419355.1 | TaeA | Protein homolog model | ATP-binding cassette (ABC) antibiotic efflux pump | Pleuromutilin antibiotic | Antibiotic efflux | 48.68 | 16.05 | |
| NODE_581_length_262_cov_1.881679.1 | MuxB | Protein homolog model | Resistance-nodulation-cell division (RND) antibiotic efflux pump | Macrolide antibiotic, monobactam, tetracycline antibiotic, aminocoumarin antibiotic | Antibiotic efflux | 59.8 | 10.45 | |
| NODE_694_length_211_cov_1.848341.1 | Bifidobacterium adolescentis rpoB mutants conferring resistance to rifampicin | Protein homolog model | Rifamycin-resistant beta-subunit of RNA polymerase (rpoB) | Rifamycin antibiotic | Antibiotic target alteration, antibiotic target replacement | 57.3 | 7.67 | |
| NODE_844_length_262_cov_2.870229.1 | MuxB | Protein homolog model | resistance-nodulation-cell division (RND) antibiotic efflux pump | Macrolide antibiotic, monobactam, tetracycline antibiotic, aminocoumarin antibiotic | antibiotic efflux | 64.08 | 10.45 | |
| NODE_893_length_142_cov_1.915493.1 | macB | Protein homolog model | ATP-binding cassette (ABC) antibiotic efflux pump | macrolide antibiotic | Antibiotic efflux | 38.78 | 10.71 | |
| NODE_1047_length_172_cov_1.453488.1 | msbA | Protein homolog model | ATP-binding cassette (ABC) antibiotic efflux pump | Nitroimidazole antibiotic | Antibiotic efflux | 32.73 | 13.4 | |
| NODE_1068_length_352_cov_1.073864.1 | vanSB | Protein homolog model | vanS, glycopeptide resistance gene cluster | Glycopeptide antibiotic | Antibiotic target alteration | 32.41 | 31.1 | |
| NODE_1294_length_172_cov_1.883721.1 | TRU-1 | Protein homolog model | TRU beta-lactamase | Cephalosporin, penam | Antibiotic inactivation | 36.84 | 20.68 | |
| NODE_1445_length_151_cov_2.655629.1 | golS | Protein homolog model | Resistance-nodulation-cell division (RND) antibiotic efflux pump | Monobactam, carbapenem, cephalosporin, cephamycin, penam, phenicol antibiotic, penem | Antibiotic efflux | 31.34 | 46.75 | |
| NODE_1538_length_228_cov_4.052631.1 | evgS | Protein homolog model | Major facilitator superfamily (MFS) antibiotic efflux pump, resistance-nodulation-cell division (RND) antibiotic efflux pump | Macrolide antibiotic, fluoroquinolone antibiotic, penam, tetracycline antibiotic | Antibiotic efflux | 35.14 | 8.1 | |
| NODE_1553_length_166_cov_2.765060.1 | mdtA | Protein homolog model | resistance-nodulation-cell division (RND) antibiotic efflux pump | Aminocoumarin antibiotic | Antibiotic efflux | 50 | 18.55 | |
| NODE_1650_length_140_cov_10.214286.1 | vanE | Protein homolog model | Glycopeptide resistance gene cluster, van ligase | Glycopeptide antibiotic | Antibiotic target alteration | 40 | 19.03 | |
| NODE_1661_length_323_cov_3.764706.1 | vanRF | Protein homolog model | Glycopeptide resistance gene cluster, vanr | Glycopeptide antibiotic | Antibiotic target alteration | 28.95 | 55.84 | |
| NODE_1668_length_361_cov_1.047091.1 | Bifidobacterium adolescentis rpoB mutants conferring resistance to rifampicin | Protein homolog model | Rifamycin-resistant beta-subunit of RNA polymerase (rpoB) | Rifamycin antibiotic | Antibiotic target alteration, antibiotic target replacement | 47.66 | 11.89 | |
| NODE_1676_length_210_cov_2.000000.1 | rpoB2 | Protein homolog model | Rifamycin-resistant beta-subunit of RNA polymerase (rpoB) | Rifamycin antibiotic | Antibiotic target alteration, antibiotic target replacement | 66.29 | 7.83 | |
| NODE_2311_length_373_cov_1.018767.1 | vanSB | Protein homolog model | vanS, glycopeptide resistance gene cluster | Glycopeptide antibiotic | Antibiotic target alteration | 27.34 | 32.66 | |
| NODE_2482_length_334_cov_1.107784.1 | sul2 | Protein homolog model | Sulfonamide resistant sul | Sulfonamide antibiotic | Antibiotic target replacement | 60.87 | 10.33 | |
| NODE_2884_length_294_cov_1.132653.1 | tetB(60) | Protein homolog model | ATP-binding cassette (ABC) antibiotic efflux pump | Tetracycline antibiotic | Antibiotic efflux | 36 | 19.69 | |
| NODE_2918_length_262_cov_4.240458.1 | mdtC | Protein homolog model | Resistance-nodulation-cell division (RND) antibiotic efflux pump | Aminocoumarin antibiotic | ANTIBIOTIC efflux | 76.47 | 10.63 | |
| NODE_3078_length_211_cov_3.127962.1 | Bifidobacterium adolescentis rpoB mutants conferring resistance to rifampicin | Protein homolog model | Rifamycin-resistant beta-subunit of RNA polymerase (rpoB) | Rifamycin antibiotic | Antibiotic target alteration, antibiotic target replacement | 56.82 | 7.67 | |
| NODE_3092_length_219_cov_2.251142.1 | hmrM | Protein homolog model | Multidrug and toxic compound extrusion (MATE) transporter | Fluoroquinolone antibiotic, acridine dye | Antibiotic efflux | 28.57 | 20.47 | |
| NODE_3146_length_148_cov_1.020270.1 | smeS | Protein homolog model | Resistance-nodulation-cell division (RND) antibiotic efflux pump | Aminoglycoside antibiotic, cephalosporin, cephamycin, penam | Antibiotic efflux | 43.48 | 15.2 | |
| NODE_3209_length_193_cov_1.471503.1 | macB | Protein homolog model | ATP-binding cassette (ABC) antibiotic efflux pump | Macrolide antibiotic | Antibiotic efflux | 29.41 | 13.35 | |
| NODE_3429_length_199_cov_1.839196.1 | Streptomyces rishiriensis parY mutant conferring resistance to aminocoumarin | Protein homolog model | Aminocoumarin resistant parY | Aminocoumarin antibiotic | Antibiotic target alteration | 44.71 | 12.52 | |
| NODE_3434_length_241_cov_1.609959.1 | otr(A) | Protein homolog model | Tetracycline-resistant ribosomal protection protein | Tetracycline antibiotic | Antibiotic target protection | 35.87 | 15.38 | |
| NODE_3458_length_256_cov_1.250000.1 | macB | Protein homolog model | ATP-binding cassette (ABC) antibiotic efflux pump | Macrolide antibiotic | Antibiotic efflux | 32.91 | 16.61 | |
| NODE_3466_length_199_cov_1.934673.1 | Streptomyces rishiriensis parY mutant conferring resistance to aminocoumarin | Protein homolog model | Aminocoumarin resistant parY | Aminocoumarin antibiotic | Antibiotic target alteration | 47.73 | 12.52 | |
| NODE_2500_length_249_cov_1.602410.1 | Escherichia coli fabI mutations conferring resistance to isoniazid and triclosan | F203L | Protein variant model |
Antibiotic resistant fabI |
Isoniazid, triclosan | Antibiotic target alteration | 30.77 | 37.4 |
Colistin, phenicol, rifampin, and fosfomycin resistance genes
Sixty-one (61) plasmid-borne mobilized colistin resistance determinant (mcr) comprising of mcr-1 to mcr-9 and their variants were recovered in 3S metagenome (Fig. 3a). The distribution of the mcr variants in the metagenome in decreasing order (abundance value in parentheses) indicate mcr-3 (27), mcr-1 (18), mcr-4 (6), mcr-5 (3), mcr-2 (2), mcr-8 (2), mcr-6 (1), mcr-7 (1), and mcr-9 (1), respectively.
Fig. 3.
Distribution of colistin (a), phenicol (b), rifampin (c), and fosfomycin (d) resistance genes in 3S metagenome. Colistin resistance genes are dominated by the mobilized colistin resistance (mcr) determinants, mcr-1 and mcr-3. Based on phenicol resistance mechanisms, 31 genes (cat, cmlV, cpt) are responsible for phenicol inactivation, while 18 genes (cmlA, cmlB, cmlR, cmr, cmrA, bcr1, dha1, fexA, floR, pexA) are responsible for phenicol efflux. Rifampin-inactivating phosphotransferase (rph, rphD) dominated rifampin resistance genes in 3S, however, all the resistance genes inactivate rifampin antibiotic. Fosfomycin resistance genes include fosfomycin modifying genes (fosA, fosB, fosC, fosD, fosE, fosF, fosG, fosH, fosX) and the fosfomycin kinases (fomA, fomB, fomC)
Forty-nine (49) phenicol resistance genes comprising cat (28), cml (10), cmr (4), cpt (2), dha1 (1), fexA (1), floR (1), and pexA (1) genes were recovered from 3S metagenome (Fig. 3b). Based on resistance mechanism, 31 genes (cat, cmlV, cpt) are responsible for phenicol inactivation, while the remaining 18 genes (cmlA, cmlB, cmlR, cmr, cmrA, bcr1, dha1, fexA, floR, pexA) are responsible for phenicol efflux.
Nine (9) rifampin resistance genes were retrieved from 3S metagenome (Fig. 3c). These include the rifampin-inactivating phosphotransferase (rph, rphD), rifampin resistance protein (rif), rifampin monooxygenase (iri), and integron (arr2, arr4, arr5, arr7) and plasmid (arr3) -encoded rifampin ADP ribosyltransferase. While the resistance mechanism of the 9 genes is antibiotic inactivation, annotation using CARD reveal the detection of rpoB gene (a β-subunit of RNA polymerase) (Table 1) whose mutations confers resistance to rifampin due to antibiotic target alteration and replacement.
Seventeen (17) fosfomycin resistance genes were retrieved from 3S metagenome (Fig. 3d). These include the fosfomycin modifying genes (fosA, fosB, fosC, fosD, fosE, fosF, fosG, fosH, fosX) and the fosfomycin kinases (fomA, fomB, fomC). The resistance mechanism of these genes is believed to be antibiotic inactivation.
Tetracycline, glycopeptide, fluoroquinolone, and oxazolidinone resistance genes
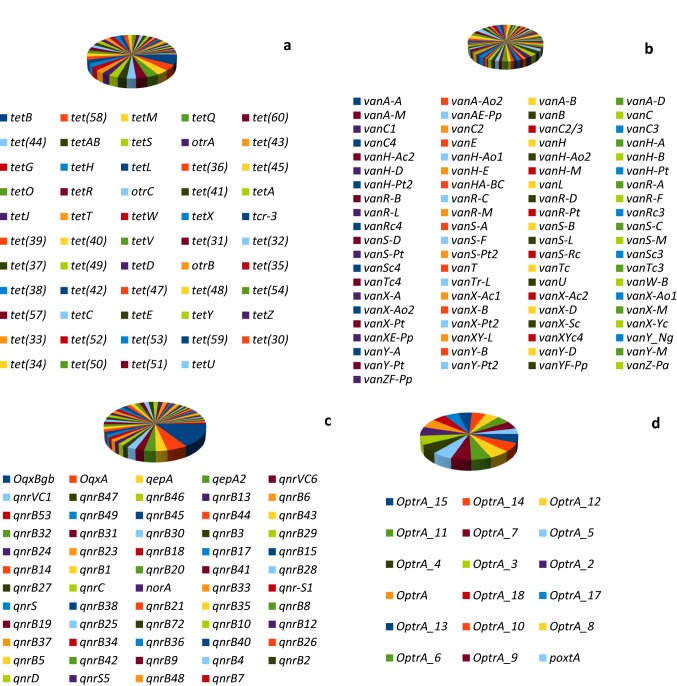
Fifty-four (54) tetracycline resistance genes were recovered from 3S metagenome (Fig. 4a). These are classified on the basis of resistance mechanism into resistance genes for ribosomal protection proteins (otrA, tetM, tetO, tetS, tetT, tetW, tet(32), tet(36), tet(44), tetQ), resistance genes for tetracycline-inactivating enzymes (tet(34), tet(37), tet(47), tet(48), tet(49), tet(50), tet(51), tet(52), tet(53), tet(54), tetX), and resistance genes for tetracycline efflux (otrB, otrC, tetA, tetB, tetAB, tetC, tetD, tetE, tetG, tetH, tetJ, tetL, tetR, tcr-3, tet(30), tet(31), tet(33), tet(35), tet(38), tet(39), tet(40), tet(41), tet(42), tet(43), tet(45), tet(57), tet(58), tet(59), tet(60), tetU, tetV, tetY, tetZ).
Fig. 4.
Distribution of tetracycline (a), glycopeptide (b), fluoroquinolone (c) and oxazolidinone (d) resistance genes in 3S metagenome. Tetracycline resistance genes are classified based on their resistance mechanisms into ribosomal protection proteins (otrA, tetM, tetO, tetS, tetT, tetW, tet(32), tet(36), tet(44), tetQ), tetracycline-inactivating enzymes (tet(34), tet(37), tet(47), tet(48), tet(49), tet(50), tet(51), tet(52), tet(53), tet(54), tetX), and tetracycline efflux (otrB, otrC, tetA, tetB, tetAB, tetC, tetD, tetE, tetG, tetH, tetJ, tetL, tetR, tcr-3, tet(30), tet(31), tet(33), tet(35), tet(38), tet(39), tet(40), tet(41), tet(42), tet(43), tet(45), tet(57), tet(58), tet(59), tet(60), tetU, tetV, tetY, tetZ). Glycopeptide resistance genes either modify peptidoglycan precursors (vanA, vanB, vanH, vanZ, vanC, van T, vanE, vanL), hydrolyze normal peptidoglycan precursors (vanX, vanY), regulate expression of vancomycin resistance (vanR, vanS) or serve as transcriptional activator of vancomycin resistance (vanU). Fluoroquinolone resistance genes are dominated by OqxBgb and OqxA RND-type multidrug efflux pump and genes responsible for fluoroquinolone target protection (qnrB, qnrC, qnrD, qnrS, qnrVC). Oxazolidinone resistance genes is dominated by OptrA (and its variant), a member of the ABC-F ATP-binding cassette ribosomal protection protein
Eighty-one glycopeptide resistance genes were retrieved from 3S metagenome (Fig. 4b). These are classified into resistance genes responsible for the production of modified peptidoglycan precursors ending in d-Ala-d-Lac (vanA, vanB, vanH, vanZ, and their variants) or d-Ala-d-Ser (vanC, van T, vanE, vanL and their variants), hydrolysis of normal peptidoglycan precursors d-Ala-d-Ala (vanX, vanY and their variants), regulation of expression of vancomycin resistance (vanR, vanS and their variants), and transcriptional activator of vancomycin resistance (vanU). The gene vanW is an accessory gene found on vancomycin resistance operons with unknown function.
Fifty-nine (59) fluoroquinolone resistance genes were recovered from 3S metagenome (Fig. 4c). In terms of abundance, both the integral membrane protein (OqxBgb) and membrane fusion protein (OqxA) RND-type multidrug efflux pumps are dominant among the fluoroquinolone resistance genes. Other fluoroquinolone efflux pump detected is qepA gene, a major facilitator superfamily (MFS) antibiotic efflux pump. Other resistance genes detected are those responsible for fluoroquinolone target protection (qnrB, qnrC, qnrD, qnrS, qnrVC and their variants) and those responsible for increased expression of efflux pump (norA).
Eighteen (18) oxazolidinone resistance genes were retrieved from 3S metagenome (Fig. 4d). The resistance genes were dominated by OptrA (and its variant), a member of the ABC-F ATP-binding cassette ribosomal protection protein. The other resistance gene detected is poxtA, also a member of the ABC-F ATP-binding cassette ribosomal protection protein that confers resistance to tetracycline, phenicol and oxazolidinone via modification of the bacterial ribosome.
Trimethoprim, Nitroimidazole, Fusidic acid, and Sulfonamide resistance genes.
Thirty-six (36) trimethoprim resistance genes dominated by the dfr genes responsible for the modification of trimethoprim target enzyme dihydrofolate reductase (dfr) were recovered from 3S metagenome (Fig. 5). Of these, twenty-three (23) belong to the dfrA group constituting 63.9% of the trimethoprim resistance genes in the metagenome.
Fig. 5.
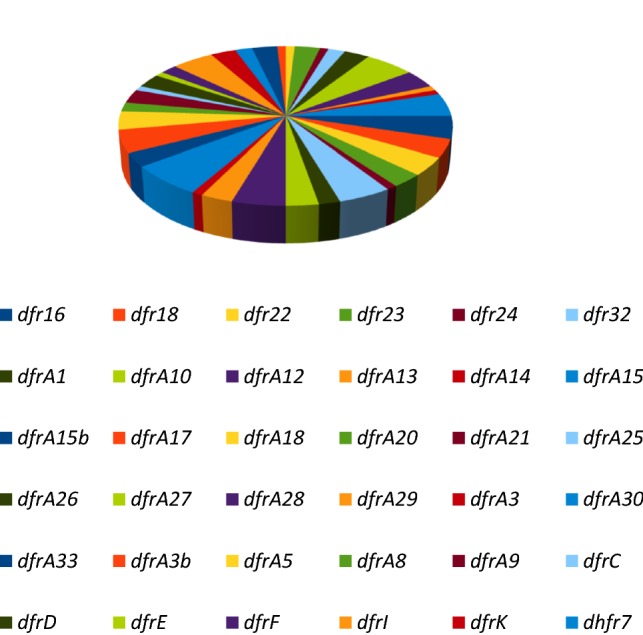
Distribution of trimethoprim resistance genes in 3S metagenome. The resistance genes are dominated by the dfr genes responsible for the modification of trimethoprim target enzyme dihydrofolate reductase (dfr)
Six (6) nitroimidazole (nim) genes were recovered from 3S metagenome. These are nimA (3), nimC (1), nimI (3), nimE (1), nimD (1), and nimJ (1). Functional annotation using CARD detected a nitroimidazole resistance gene msbA, an ATP-binding cassette (ABC) antibiotic efflux pump not detected by ARG-ANNOT (Table 1). Only four (4) fusidic acid resistance genes fusB (1), far1 (1), fusH (2), fusD (3) were detected in 3S metagenome. For sulfonamide resistance genes, four genes designated sul1 (1), sul2 (3), sul3 (5) and sul4 (4) were recovered in 3S metagenome.
Functional annotation of 3S metagenome using CARD revealed several resistance genes not detected by ARG-ANNOT. These include arlR, a major facilitator superfamily (MFS) antibiotic efflux pump that confer resistance to fluoroquinolones and acridine dye; TaeA, an ATP-binding cassette (ABC) antibiotic efflux pump that confer resistance to pleuromutilin antibiotic; and MuxB, a resistance-nodulation-cell division (RND) antibiotic efflux pump that confer resistance to macrolide, monobactam, tetracycline, and aminocoumarin antibiotics. Others include golS (monobactam, carbapenem, cephalosporin, cephamycin, penam, phenicol, penem); evgS (macrolide, fluoroquinolone, penam, and tetracycline antibiotics); mdtA, mdtC (aminocoumarin antibiotic) and several others (Table 1).
Heavy metal resistance genes
Copper (Cu) and silver (Ag) resistance genes
In 3S metagenome, copper resistance genes are represented by several systems (Table 2). These include the cop (copA, copB, copC, copD, copS, copR, copY, copZ, copJ, copP, copL), cut (cutA, cutC, cutE, cutO), cue (cueA, cueR, cueO), cus (cusA, cusB, cusS), cme (cmeB. CmeC) and the pco (pcoA, pcoS) systems as well as several transcriptional regulators (copR, cueR, copY, hmrR, crdR, baeR, yfmP) and transcriptional repressor (csoR). Also detected is a copper-exporting P-type ATPase ctpV gene and a gold/copper-translocating P-type ATPase golT. It is noteworthy that the genes cueA and cusS have dual resistance to copper and silver, respectively (Table 2).
Table 2.
Heavy metal resistome detected in 3S metagenome (using BacMet) involved in resistance to heavy metals
| Heavy metal | Enzymes/gene | Taxonomic affiliation |
|---|---|---|
|
Copper, zinc, silver Iron, zinc, cobalt, nickel, copper, cadmium, Manganese, gallium |
Copper resistance D; apolipoprotein N-acyltransferase actA (Cu/Zn); copper translocating P-type ATPase copB; cutE gene product; multicopper oxidase type 2 cutO; RND efflux system, inner membrane protein cmeB (Cu/Co); heavy metal translocating P-type ATPase copA; copper resistance protein copC; CopS protein; winged helix family two component heavy metal response transcriptional regulator copR; copper-sensing transcriptional repressor csoR; copper-translocating P-type ATPase cueA; MerR family transcriptional regulator cueR; metal-transporting P-type ATPase cueA (Cu/Ag); CutA1 divalent ion tolerance protein cutA; copper-binding protein cinA; RND efflux system, outer membrane protein cmeC (Cu/Co); multicopper oxidase type 3 cutO; molecular chaperone dnaK; transcriptional regulator copY; Cu(I)-responsive transcriptional regulator hmrR (Cu/HCl); Copper homeostasis protein cutC; Copper resistance protein L copL; laccase cueO; metal cation transporter P-type ATPase ctpV; heavy metal-translocating P-type ATPase actP; cation efflux system protein CusA; HAE1 family transporter acrD (Cu/Zn); sensor kinase cusS (Cu/Ag); thiol:disulphide interchange protein dsbC; integral membrane sensor signal transduction histidine kinase baeS (Cu/Zn/W); heavy metal translocating P-type ATPase golT (Cu/Au); response regulator crdR; type IIS restriction enzyme R protein crdS; copper resistance protein oxidase pcoA (plasmid); heavy metal sensor kinase pcoS (plasmid); CopJ protein (plasmid); heavy metal translocating P-type ATPase copZ; cation efflux system protein cusB (plasmid); winged helix family two component transcriptional regulator baeR (Cu/Zn/W); Heavy metal-translocating P-type ATPase Cd/Co/Hg/Pb/Zn-transporting ctpG (Cu); sensor histidine kinase corS; copper exporting ATPase copP; transcriptional regulator yfmP; crdB gene Zinc/iron ZIP family permease zupT (Zn/Fe/Co/Ni/Cu/Cd); iron-dependent repressor DxtR ideR; aconitase acn (Fe); transcriptional regulatory protein basR/pmrA (Fe); ferritin pfr (Fe/Cu/Mn); iron (III) ABC transporter, ATP binding protein fbpC (Fe/Ga); TonB-dependent siderophore receptor fpvA (Mn/Fe/Co/Zn/Ni/Cu/Cd/Ga); ABC transporter family protein ybtQ (Fe); cation efflux family protein fieF (Fe/Zn/Co/Cd/Ni); Fe2+/Zn2+/Mn2+ ABC transporter ATP-binding protein troB; iron chelate ABC transporter, ATP-binding protein yfeB (Fe/Mn); Fe3+ ABC superfamily ATP-binding cassette transporter, permease protein fbpB (Fe/Ga); manganese transport protein mntH (Mn/Fe/Cd/Co/Zn); permease and ATP-binding protein of yersiniabactin-iron ABC transporter ybtP (Fe); Mn2+/ Fe2+ transporter NRAMP family (mntH/yfeB); TonB-dependent siderophore receptor fptA (Fe/Co/Ni/Ga); ferric uptake regulator family protein furA (Fe); cation-activated repressor protein troR (Zn/Mn/Fe); Fe2+/Zn2+/Mn2+ ABC transporter membrane protein troD; ferritin Dps family protein dpsA (Fe/H2O2); chealated iron ABC transporter, permease yfeC (Fe/Mn); bacterioferritin bfrA (Fe); periplasmic iron-binding protein yfeA (Fe/Mn); putative ABC transport system permease fetB (Fe); iron transport inner membrane protein sitC (plasmid) (Fe/Mn/H2O2); fur regulated iron transporter sitB (plasmid) (Fe/Mn/H2O2); ferroxidase pfr (Fe/Cu/Mn); Chain A, Periplasmic Zinc Binding Protein troA (Zn/Mn/Fe); cation transport P-type ATPase nia (plasmid) (Fe/Ni) |
Rhodopseudomonas palustris BisB18; alphaproteobacterium BAL199; Methylobacterium sp. 4–46; Acidithiobacillus ferrooxidans ATCC 53993; Xenorhabdus bovienii SS-2004; Starkeya novella DSM 506; Campylobacter coli JV20; Paenibacillus dendritiformis C454; Thermobacillus composti KWC4; Ralstonia metallidurans CH34; Ralstonia pickettii 12J; Clostridium sp. M62/1; Pseudomonas syringae pv. Tomato NCPPB 1108; Pseudomonas mendocina ymp; Pseudomonas stutzeri A1501; Dickeya didantii Ech586; Marinobacter sp. Mn17-9; Campylobacter upsaliensis JV21; Streptomyces griseus XylebKG-1; Corynebacterium resistens DSM 45100; Oenococcus oeni ATCC BAA-1163; Pseudovibrio sp. JE062; Yersinia mollaretii ATCC 43969; Xanthomonas vesicatoria ATCC 35937; Aeromonas hydrophila; Mycobacterium marinum M; Beijerinckia indica subsp. indica ATCC 9039; Citrobacter youngae ATCC 29220; Pantoea stewartii subsp. stewartii DC283; Escherichia sp. TW09308; Enterococcus hirae; Photobacterium leiognathi subsp. mandapamensis svers.1.1; Glaciecola sp. 4H-3-7+YE-5; Agrobacterium tumefaciens F2; Helicobacter pylori 35A; Salmonella enterica ser.. Typhimurium; Helicobacter pylori 2017; Serratia marcescens; Ralstonia metallidurans CH34; Paenibacillus elgii B69; Escherichia coli STEC_DG131-3; Stenotrophomonas maltophilia R551-3; Saccharomonospora viridis DSM 43017; Myxococcus xanthus DK 1622; Streptococcus sp. C150; Bacillus pumilus SAFR-032 Methanosaeta harundinacea 6Ac; Corynebacterium accolens ATCC 49725; Corynebacterium nuruki S6-4; Pantoea vagans C9-1; Vibrio fischeri MJ11; Pseudomonas mendocina NK-1; Escherichia coli DEC10F; Shewanella benthica KT99; Treponema paraluiscuniculi Cuniculi A; Providencia rettgeri DSM 1131; Aggregatibacter segnis ATCC 33393; Pantoea sp. SL1_M5; Ralstonia solanacearum CFBP2957; Rhodobacter sp. SW2; Mycobacterium gilvum PYR-GCK; Treponema pallidum subsp. pallidum str. Nichols; Thioalkalimicrobium cyclium ALM1; Actinobacillus minor NM305; Rhodococcus pyridinivorans AK37; Pasteurella multocida 36950; Enterobacter cloacae subsp. cloacae ATCC 13047; Salmonella bongori NCTC 12419; Citrobacter rodentium ICC168; Cyanothece sp. PCC 7425; Treponema pallidum; Sinorhizobium meliloti 1021 |
| Zinc, cadmium, tellurium, tungsten, cobalt, chromium nickel, manganese | BaeS gene product (Zn/W); Zn/Cd/Hg/Pb-transporting ATPase zntA (Zn/Pb/Cd); DNA-binding response regulator irlR (Cd/Zn); winged helix two component transcriptional regulator baeR (Zn/W); inorganic phosphate transporter pitA (Zn/Te); integral membrane sensor signal transduction histidine kinase actS (Cd/Zn/HCl); acriflavin resistance protein mdtB (Zn); zinc transporter zitB; high-affinity zinc transporter ATPase znuC (Zn); Zn(II)-responsive transcriptional regulator zntR; multidrug resistance transporter mdrL (Zn/Co/Cr); probable RND efflux transporter mdtB (Zn); membrane fusion protein (MFP-RND) heavy metal cation tricomponent efflux zneB/hmxB (plasmid) (Zn); high-affinity zinc transporter periplasmic component znuA (Zn); sensor protein zraS (Zn/Pb); zinc/iron permease zipB (Cd/Zn); heavy metal-translocating P-type ATPase ctpC (Mn/Zn); zinc efflux system zitB (Zn); outer membrane efflux protein czcC (plasmid; Cd/Zn/Co); cation transporting ATPase, P-type ziaA (Zn); response regulator receiver protein actR (Cd/Zn/HCl); RND family efflux transporter MFP subunit 1 mdtA (Zn); transcriptional regulator zur (Zn); thiol:disulfide interchange protein dsbA (Cd/Zn/Hg); transcriptional regulatory protein zraR (Zn); AcrB/AcrD/AcrF family protein nczA (Ni/Co/Zn); RND divalent metal cation efflux transporter czrA (Zn/Cd); cadmium-zinc-nickel resistance protein cznA (Cd/Zn/Ni); cadmium-translocating P-type ATPase czcP (Cd/Zn/Co); heavy metal response regulator irlR (Cd/Zn); dsbA-like protein frnE (Cd/H2O2); twin-arginine translocation pathway signal mntA (Cd/Mn); MerR family transcriptional regulator cadR (plasmid, Cd) | Edwardsiella tarda EIB202; Pectobacterium carotovorum subsp. brasiliensis PBR1692; Burkholderia oklahomensis EO147; Geobacter sp. M18; Achromobacter piechaudii ATCC 43553; Sinorhizobium meliloti CCNWSX0020; Ralstonia pickettii 12J; Salmonella bongori NCTC 12419; Yokenella regensburgei ATCC 43003; Edwardsiella tarda ATCC 23685; Listeria monocytogenes J0161; Pseudomonas aeruginosa NCMG1179; Cupriavidus metallidurans CH34; Plautia stali symbiont; Escherichia coli IAI39; Pseudomonas sp. TJI-51; Kineococcus radiotolerans SRS30216; Pantoea stewartii subsp. stewartii DC283; Pyrococcus abyssi GE5; Rhizobium sp. PDO-076; Achromobacter arsenitoxydans SY8; Methylococcus capsulatus str. Bath; Aggregatibacter segnis ATCC 33393; Klebsiella pneumoniae 342; Caulobacter crescentus CB15; Pseudomonas aeruginosa PA7; Helicobacter mustelae 12198; Alicycliphilus denitrificans BC; Dermacoccus sp. Ellin185; Silicibacter sp. TrichCH4B; Pseudomonas putida S16 |
| Chromium, tellurium, selenium, molybdenum, vanadium, tungsten | ATP-dependent DNA helicase recG (Cr/Te/Se); Cr (VI) reductase (Cr, Fe, H2O2); NADPH-dependent FMN reductase chrR (Cr); putative NADH-dependent reductase yieF (Cr/V/Mo); malate dehydrogenase ruvB (Cr); chromate transporter chrA; nitroreductase A nfsA (Cr); NAD(P)H dehydrogenase (quinone) chrR (Cr); chromate resistance signal peptide protein chrB; Holliday junction DNA helicase ruvB (Cr/Te/Se); manganese/iron superoxide dismutase chrC (Cr); tellurite resistance protein tehB; tellurite resistance protein tehA; tellurite resistance protein trgB (Te/Cu); symporter actP (Te); KlaC protein klaC/telB (plasmid) (Te); molybdate ABC transporter inner membrane subunit modB (W/Mo); molybdate ABC transporter periplasmic protein wtpA (W/Mo); molybdenum ABC transporter ATP-binding protein modC (W/Mo); DNA-binding transcriptional regulator ModE (W/Mo); molybdenum ABC transporter periplasmic molybdate-binding protein modA (W/Mo); tungstate ABC transporter binding protein wtpA (W/Mo); ABC transporter, ATP-binding protein wtpC (W/Mo); molybdate/tungstate transport system permease protein wtpB; molybdenum-pterin binding domain protein/site-specific recombinase, phage integrase family tunR (W/Mo); sodA gene product (Se/H2O2); superoxide dismutase sodB (Se/H2O2) | Tolumonas auensis DSM 9187; Lysinibacillus fusiformis ZC1; Burkholderia multivorans ATCC 17616; Burkholderia cepacia; Shewanella frigidimarina NCIMB 400; Sinorhizobium meliloti BL225C; Vibrio vulnificus YJ016; Rhodobacter capsulatus SB 1003; Acinetobacter oleivorans DR1; Alkalilimnicola erhlichii MLHE-1; Alicycliphilus denitrificans K601; Haemophilus pittmaniae HK 85; Haemophilus haemolyticus M21621; Maritimibacter alkaliphilus HTCC2654; Magnetospirillum magneticum AMB-1; Ralstonia pickettii 12J; Desulfotomaculum kuznetsovii DSM 6115; Gillisia limnaea DSM 15749; Salmonella enterica subsp. enterica serovar Typhimurium str. T000240; Desulfotomaculum acetoxidans DSM 771; Methanolinea tarda NOBI-1; Thermococcus sibiricus MM 739; Thermococcus litoralis DSM 5473; Desulfovibrio vulgaris str. Hildenborough; Serratia symbiotica str. ‘Cinara cedri’; Burkholderia pseudomallei 1655 |
| Nickel, cobalt, manganese, magnesium | Major facilitator transporter nrsD/nreB (Ni/Co); P-ATPase superfamily, P-type ATPase cadmium transporter ctpD (Co/Ni); cation transporter dmeF (Co/Ni); magnesium and cobalt transport protein corA (Mg/Co/Ni); nickel ABC transporter substrate-binding protein nikA (Ni); iron-dicitrate transporter subunit, membrane component of ABC superfamily fecD (Ni/Co); nickel and cobalt resistance protein cnrA (Co/Ni); nickel transporter ATP-binding protein nikE (Ni); nickel ABC superfamily ATP binding cassette transporter, ABC protein nikD (Ni); integral membrane sensor signal transduction histidine kinase nrsS (Ni); high-affinity nickel transporter rcnA (Ni/Co/Fe); fecE gene product (Ni/Co); periplasmic nickel sensor cnrR/cnrX (plasmid, Ni); RNA polymerase sigma factor cnrH (plasmid, Co/Ni); nickel ABC transporter, permease subunit nikC (Ni); transcriptional regulator, copG family protein nikR (Ni); nickel ABC transporter permease nikB; czcA family heavy metal efflux pump nrsA (Ni); nickel/cobalt efflux system ncrC (Co/Ni); RNA polymerase sigma factor nccH (plasmid; Ni/Co/Cd); ArsR family transcriptional regulator cmtR (Co/Ni); conserved inner membrane protein mntP (Mn/Mg); magnesium-transporting ATPase P-type 1 mgtA; CO2+/Mg2+ efflux protein ApaG corD (Co/Mg); magnesium and cobalt efflux protein corB; Mg/Co transporter corC; magnesium/nickel/cobalt transporter corA | Roseiflexus sp. RS-1; Mycobacterium parascrofulaceum ATCC BAA-614; Pelobacter carbinolicus DSM 2380; Alkanivorax sp. DG881; Dickeya dadantii Ech703; Escherichia coli S88; Bradyrhizobium sp. STM 3843; Rhodospirillum rubrum ATCC 11170; Roseomonas cervicalis ATCC 49957; Cyanothece PCC 7424; Starkeya novella DSM 506; Xenorhabdus nematophila ATCC 19061; Cupriavidus metallidurans CH34; Brucella sp. NF 2653; Sagittula stellata E-37; Desulfotomaculum kuznetsovii DSM 6115; Klebsiella oxytoca 10-5245; Clavibacter michiganensis subsp. sepedonicus; Pantoea stewartii subsp. stewartii DC283; Klebsiella oxytoca 10-5246; Pseudomonas psychrotolerans L19; Vibrio sp. Ex25; Idiomarina loihensis L2TR; Actinobacillus succinogenes 130Z |
| Mercury, lead, arsenic, antimony | Mercuric reductase merA; periplasmic mercury transport protein merT; alkylmercury lyase merB2; MerR family transcriptional regulator merR1; heavy metal transport/detoxification protein merP; copper-transporting ATPase merT-P; putative mercury transport protein merC (plasmid); Pb-efflux ATPase pbrA; Pb-specific transcription regulator protein pbrR (plasmid); iron permease FTR1 family protein 2 pbrT (plasmid); phosphate import ATP binding protein pstB (As); arsenical resistance protein arsB (As); arsenical resistance protein acr3 (As); arsenite efflux pump ACR3 arsB (As/Sb); arsenate reductase arsC (As/Sb); molydopterin dinucleotide binding region (As); methyltransferase type 11 arsM; phosphate-binding protein pstS (As); phosphate ABC transporter permease protein pstC (As); MIP family channel protein glpF (As/Sb); phosphate transporter permease subunit A pstA (As); arsenical pump-driving ATPase arsA (plasmid, As/Sb); putative arsenic-efflux pump regulatory protein arsR (As/Sb/Bi); putative sensor histidine kinase aioS/aoxS (As); arsenite oxidase small subunit (As); arsenite oxidase large subunit | Micromonospora sp. L5; Thiobacillus sp.; Nocardioides sp. JS614; Xanthobacter autotrophicus Py2; Croceibacter atlanticus HTCC2559; Acidithiobacillus ferrooxidans ATCC 53993; Ralstonia metallidurans CH34; Herbaspirillum seropedicae SmR1; Achromobacter xylosoxidans A8; Sutterella wadsworthensis 3_1_45B; Moorella thermoacetica ATCC 39073; Campylobacter lari; Synechococcus sp. PCC 7002; Lactobacillus reuteri SD2112; Pyrobaculum calidifontis JCM 11548; Intrasporangium calvum DSM 43043; Cupriavidus basiliensis OR16; Nitrococcus mobilis Nb-231; Pseudomonas fulva 12-X; Leptothrix cholodnii SP-6; Klebsiella pneumoniae 342; Agrobacterium tumefaciens; Burkholderia multivorans ATCC 17616; Roseovarius sp. 217 |
Iron (Fe), manganese (Mn), and gallium (Ga) resistance genes
Majority of resistance genes detected for iron in 3S metagenome are genes that also confers resistance on other heavy metals such as zinc, cobalt, nickel, copper, cadmium, manganese and gallium (Table 2). These genes include zupT, pfr, fbpC, fpvA, fieF, yfeB, troB, troA, sitB, sitC, and several others. However, specific genes that confer resistance on iron alone such as ideR, ybtP, ybtQ, furA, bfrA, fetB, and several others were also detected. All the resistance genes detected for manganese and gallium confers resistance on other heavy metals. These include fbpC, fbpB, yfeB, mntH, mntA, troD, fptA, yfeA and several others (Table 2).
Zinc (Zn), cadmium (Cd), nickel (Ni), and cobalt (Co) resistance genes
Several zinc resistance genes were recovered from 3S metagenome (Table 2). These include zitB, znuC, mdtB, zntR, zneB/hmxB, znuA, ziaA, zur, zraR and several others. Resistance genes that confer resistance on zinc and other heavy metals such as zntA (Zn/Pb/Cd), pitA (Zn/Te), mdrL (Zn/Co/Cr), zipB (Zn/Cd) among others were also detected. Cadmium resistance genes such as czcC (Cd/Zn/Co), czrA (Cd/Zn), cznA (Cd/Zn/Ni), czcP (Cd/Zn/Co), cadR, among others, which confers resistance on cadmium and other heavy metals were also detected (Table 2).
In addition, representatives of nickel resistance genes (nikA, nikB, nikC, nikD, nikE, nrsA, nrsS, among others) and those that confer resistance to nickel and other heavy metals (nczA (Ni/Co/Zn), nia (Ni/Fe), nrsD/nreB (Ni/Co), dmeF (Co/Ni), fecD (Ni/Co), cnrA (Co/Ni), rcnA (Ni/Co/Fe)) and several others were also recovered from 3S metagenome (Table 2).
Chromium (Cr), tellurium (Te), selenium (Se), molybdenum (Mo), tungsten (W) and vanadium (V) resistance genes
In 3S metagenome, several chromium resistance genes were recovered. These include Cr (IV) reductase, chrA, chrB, chrC, chrR, nfsA, yieF (Cr/V/Mo), ruvB (Cr/Te/Se), recG (Cr/Te/Se) among others. Tellurium resistance genes detected include tehA, tehB, trgB (Te/Cu), and klaC/telB, respectively. Representatives of Molybdenum and tungsten resistance genes detected include modA (W/Mo), modB (mO/W), modC (W/Mo), modE (W/Mo), wtpA (W/Mo), wtpC (W/Mo) among others while sodA and sodB genes were recovered for selenium resistance (Table 2).
Magnesium (Mg), mercury (Hg), lead (Pb), arsenic (As), antimony (Sb) resistance genes
Functional annotation of 3S metagenome revealed the following genes responsible for magnesium, mercury, lead, aresenic and antimony resistance: Mg (mntP, mgtA, corA, corB, corC, corD); Hg (merA, merT, merB2, merR1, merP, merT-P, merC); Pb (pbrA, pbrR, pbrT); As/Sb (pstA, pstB, pstC, pstS, arsA, arsB, acr3, arsC, arsM, arsR, glpF, arsenite oxidase small/large subunits) (Table 2).
Discussion
Soil is a principal member of the ecosystem. Its inundation with diverse pollutants such as antibiotics and heavy metals from diverse sources has resulted in the proliferation of antibiotic and heavy metal resistance genes in the soil ecosystem with attendant consequences on public and environmental health. In this study, an attempt was made to unravel the antibiotic and heavy metal resistome in a chronically polluted soil using a shotgun metagenomic approach. This study is important for several reasons. First, the alarming rate and ease of dissemination and spread of antibiotic resistance genes between environmental compartments are disturbing, as several authors have indicated that the soil resistome may be a critical source for the emergence of novel antibiotic resistance genes in clinical pathogens (Willms et al. 2019; Zhu et al. 2019). Second, it is widely believed that pristine soil microbiomes comprise a wide array of novel and clinically characterized native antibiotic resistance genes, whose presence predates clinical use of antibiotics (Perry and Wright 2013; Durso et al. 2016). Third, co-selection of antibiotic and heavy metal resistance in microorganisms via cross-resistance, co-resistance and co-regulatory mechanisms has maintained and promote antibiotic resistance in microbial populations in the absence of antibiotics (Pal et al. 2015a, b).
β-lactamases, produced extracellularly by Gram-positive bacteria and in the periplasmic space by Gram-negative bacteria inactivate β-lactam antibiotics by binding covalently to their carbonyl moiety and hydrolyzing the β-lactam ring. Based on Ambler class A-D classification, Ambler classes A, C, and D are β-lactamases with serine at their active site while Ambler class B β-lactamases are metallo-enzymes that require zinc as a metal cofactor for their catalytic activities (van Hoek et al. 2011). In this study, 144 β-lactamase genes were recovered. Of these, 60 (41.7%) belong to class A β-lactamases. This is concerning as most of the extended-spectrum β-lactamases (ESBLs) belong to this class (Eiamphungporn et al. 2018). ESBLs, which are plasmid-borne are reputed for their ability to hydrolyze third and fourth generation cephalosporins and monobactams but not cephamycins (e.g. cefoxitin) and carbapenems (meropenem, imipenem, ertapenem, and doripenem) and are generally susceptible to β-lactamase inhibitors such as clavulanic acid, sulbactam, and tazobactam (Paterson and Bonomo 2005; Eiamphungporn et al. 2018; Rahman et al. 2018).
Expectedly, though with a narrow margin, the number of variants of blaCTX-M (cefotaximase) genes detected in this study trumped other Enterobacteriaceae ESBL enzymes blaTEM and blaSHV. This may be attributed to extraordinary dissemination of blaCTX-M genes in highly mobilizable plasmids and transposons and the phenomenon of co-resistance especially to aminoglycosides and fluoroquinolones synonymous with blaCTX-M gene-carrying microorganisms, which might facilitate co-selection processes (Canton and Ruiz-Garbajosa 2011; Rogers et al. 2011). blaCTX-M β-lactamases exhibit high hydrolytic activity against cefotaxime and a higher hydrolytic activity on cephalothin when compared to penicillins, cefaloxine and ceftazidime (Tzouvelekis et al. 2000). The second dominant class A β-lactamase in this study, blaTEM, the first plasmid-mediated β-lactamase is the most prevalent plasmid-encoded β-lactamase in Gram-negative bacteria responsible for about 90% of their resistance to ampicillin (van Hoek et al. 2011; Rahman et al. 2018). Its classic phenotype blaTEM-1 exhibits hydrolytic activity on penicillin and first-generation cephalosporin such as cephaloridine. Also detected are class A β-lactamases with carbapenem-hydrolyzing activity such as blaKPC, blaGES, and blaIMI. However, of great concern is the detection of blaKPC, as KPC β-lactamases are known to efficiently hydrolyze carbapenems, penicillins, cephalosporins and aztreonam. They also hydrolyze clinically available β-lactamase inhibitors (with the exception of azobactam, which hydrolyze KPC enzymes slowly) such as clavulanic acid, sulbactam, and tazobactam (Nguyen et al. 2016; Bonomo 2017).
The most prominent representatives of class B metalloenzymes β-lactamases detected in 3S metagenome are blaNDM and blaVIM constituting 22.1% and 15.5% of the class B β-lactamases. This is not surprising, as NDM (New Delhi MBL), which have been reported from diverse geographical locations are not inhibited by commercially available β-lactamases, genes are borne on transferable plasmids, chromosome and integrons and have tight hydrolytic activities on penicillins, cephalosporins and carbapenems (Bonomo 2017). In addition, the promiscuous spread of blaNDM genes among bacterial species, particularly members of Enterobacteriaceae (Darley et al. 2012; Jones et al. 2014), the possibility of its environmental spread (Walsh et al. 2011), and its concomitant carriage of loads of resistance determinants for β-lactamases, carbapenemases, and genes associated with resistance to fluoroquinolones and aminoglycosides (Bonomo 2017) makes blaNDM a public and environmental health menace.
Plasmid-mediated AmpC β-lactamases such as blaCMY, blaLEN, blaOKP dominated the class C β-lactamases recovered from 3S metagenome. These plasmid-mediated enzymes confer resistance to a broad-spectrum of β-lactam antibiotics including oxyimino-β-cephalosporins, penicillins, cephamycins and variably aztreonam (Jacoby 2009). Previous reports have also indicated co-selection of multiple resistance genes by plasmids carrying AmpC β-lactamases, which confer resistance to aminoglycosides, chloramphenicol, sulfonamides, quinolones, tetracycline and trimethoprim as well as genes for other β-lactamases (Jacoby 2009; Rahman et al. 2018).
In this study, the predominant class D β-lactamase genes are the blaOXA genes constituting 91.7% of the class members. The preponderance of the class D OXA-type enzymes may be attributed to mutation resulting in amino acid substitutions and the emergence of a high number of variants with an extended and narrow spectrum of activities against diverse β-lactam antibiotics such as penicillins, cephalosporins, extended-spectrum cephalosporins and carbapenems (van Hoek et al. 2011; Leonard et al. 2013; Bonomo 2017). The genes for OXA-type enzymes are also borne on plasmids and integrons, which allow promiscuous dissemination and spread of the resistance across not only members of Enterobacteriaceae (Castanheira et al. 2011; Patel and Bonomo 2013) but also Gram-positive bacteria belonging to the Bacillaceae, Clostridiaceae, and Eubacteriaceae families of the phylum Firmicutes (Toth et al. 2016).
Four mechanisms of resistance to aminoglycoside antibiotics have been reported. They are active efflux, decrease permeability, ribosome alteration, and antibiotic inactivation by aminoglycoside-modifying enzymes (van Hoek et al. 2011). In this study, majority of the aminoglycoside resistance genes detected code for aminoglycoside-modifying enzymes. Prominent among them is aadA gene, streptomycin 3′-adenyltransferase, a member of the ANT (3″) family, which confer resistance to streptomycin and spectinomycin and is encoded by plasmids, transposons and integrons in Enterobacteriaceae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Vibrio cholerae (Hollingshead and Vapnek 1985). Others include aac(6′) and aac(3) and their variants, which are plasmid-encoded aminoglycoside acetyltransferases that confers resistance to gentamicin, tobramycin, kanamycin (aac(3)), neomycin (aac(3)), amikacin (aac(6′)), and paromycin (aac(3)), respectively. These plasmid-encoded genes have been reported in Klebsiella pneumoniae, Enterobater cloacae, Actinobacillus pleuropneumoniae, Salmonella typhimurium, Citrobacter freundii, Acinetobacter spp., Stenotrophomonas spp., and Pseudomonas aeruginosa (Allmansberger et al. 1985; Shaw et al. 1993; Mugnier et al. 1996; Ramirez and Tolmansky 2010; van Hoek et al. 2011).
MLS antibiotics comprise three chemically distinct, but functionally similar antibiotic classes. However, due to similar mode of antibacterial action of macrolide and the two other antibiotic classes, lincosamide and streptogramin B and their comparable antibacterial spectra, they have been clustered together as macrolide-lincosamide-streptogramin (MLS) antibiotics (van Hoek et al. 2011; Marosevic et al. 2017). Three main resistance mechanisms have been reported for MLS antibiotics. These are rRNA methylation, active efflux and antibiotic inactivation. In this study, rRNA methylation mediated by rRNA methylases encoded by cfr; erm; tlr; myr; emt genes is the dominant MLS resistance mechanism in 3S metagenome. This is disheartening for two reasons. First, rRNA methylases, particularly those encoded by erm genes confer cross-resistance between macrolides, lincosamides and streptogramin B antibiotics. Second, there are several reports detailing co-selection of tetracycline and MLS resistance genes (especially erm) on large transposons such as Tn1545 (tet(M), macrolide-aminoglycoside-streptothricin element, erm(B)) (Cochetti et al. 2008), and CTnDOT family transposons (tet(Q), tet(X), erm(B), erm(F), erm(G)) (Shoemaker et al. 2001; Gupta et al. 2003). Moreover, some of these transposons belong to the Tn916 family, which has a broad host range and transfer promiscuously to a wide variety of Gram-positive and Gram-negative bacteria (Roberts and Mullany 2011).
Colistin, extensively used in agricultural production and treatment of clinical pathogens in spite of its potential nephrotoxicity and neurotoxicity, is the ultimate line of defence against critical infections caused by multidrug-resistant pathogens (Xu et al. 2018). However, the emergence of mobile colistin resistance (mcr) genes is compromising the efficacy of colistin as last-line antibiotic used to treat multidrug-resistant Gram-negative bacteria. In this study, mcr-3 and mcr-1 are the most prominent mcr genes recovered from 3S metagenome. This is not surprising as several reports have indicated the preponderance of mcr-1 and mcr-3 in clinical specimens, clinical isolates, MDR lineages of E. coli and in livestocks (pigs, poultries, cows, goats as a growth promoter and to treat infections) across several continents (Catry et al. 2015; Gao et al. 2016; Hu et al. 2016; Ma et al. 2016; Bi et al. 2017; Coates et al. 2017; Haenni et al. 2017; Litrup et al. 2017). In addition, most of the mcr-3 genes and its variants are located on IncHI2-type plasmids, IncP-like plasmid, and IncF plasmid, which facilitate their spread and dissemination (Haenni et al. 2017). Open defecation and urination by humans and unregulated rearing of livestock and its attendant defecation and urination along rearing paths, a regular activity in the geographical region where this study is conducted could be fingered as possible routes for spread and dissemination of the mcr genes in the soil environment.
Chloramphenicol acetyltransferases encoded by the cat genes dominated the phenicol resistance genes recovered from 3S metagenome. These enzymes inactivate chloramphenicol, thiamphenicol and azidamfenicol by attaching an acetyl group from acetyl-CoA to the antibiotics, which prevent them from binding to the ribosome (Murray and Shaw 1997; Schwarz et al. 2004; Wright 2005). Diverse Gram-positive and Gram-negative bacteria species have been reported to encode the cat genes. These include Acinetobacter calcoaceticus, Agrobacterium tumefaciens, Bacillus clausii, Bacillus subtilis, Campylobacter coli, Enterococcus faecalis, Enterococcus faecium, Lactococcus lactis, Listeria monocytogenes, Morganella morganii, Proteus mirabilis, Serratia marcescens, shigella flexneri, Staphylococcus aureus, S. haemolyticus, S. intermedius, Streptococcus agalactiae, S. suis, S. acrimycini (Projan et al. 1985; Schwarz et al. 1991; Tennigkeit and Matzura 1991; Cardoso and Schwarz 1992; Perreten et al. 1997; Parkhill et al. 2001; Grady and Hayes 2003; Takamatsu et al. 2003; Galopin et al. 2009).
Tetracycline antibiotics, well known for their broad spectrum of activity, encompasses a wide range of Gram-positive and Gram-negative bacteria, spirochaetes, atypical organisms such as mycoplasmas, chlamydiae and rickettsiae, as well as protozoan parasites (Kobashi et al. 2006). The drug has proven clinical safety, acceptable tolerability, and it is available in intravenous and oral formulations for most members of the class (Grossman 2016). Extensive use of tetracyclines in veterinary medicine and agricultural applications, including crop protection and intensive animal farming, has contributed to the widespread dissemination of tetracycline resistance (Surette and Wright 2017; Wu et al. 2020). In this study, resistance genes encoding ribosomal protection proteins, tetracycline-inactivating enzymes, and tetracycline efflux were detected. However, more disturbing is the detection in this study, of seven (tetX, tet(49), tet(50), tet(51), tet(52), tet(53), tet(54)) out of the nine tetracycline destructases that mediate enzymatic inactivation of third (tigecycline) and fourth (eravacycline and omadacycline) generation tetracyclines known to overcome resistance due to efflux and ribosome protection (Jenner et al. 2013; Tanaka et al. 2016; Zhanel et al. 2016). The destructases mediate covalent destruction of the antibiotic scaffold via enzymatic inactivation, which permanently eliminates the tetracycline antibiotic challenge by lowering intracellular and extracellular antibiotic concentrations (Markley and Wencewicz 2018). Moreover, the genes encoding the destructases, which are often borne on transferable plasmids, are normally present on operons that are co-transcribed (to ensure self-protection) with biosynthetic genes in the antibiotic-producing microorganism (Davies and Davies 2010; Mack et al. 2014; Li et al. 2018).
Majority of fluoroquinolone resistance genes recovered in this study such as OqxBgb, OqxA, qepA and qnr are plasmid encoded, which facilitate their spread and dissemination via horizontal gene transfer (Hooper and Jacoby 2016). OqxAB gene, first identified as a plasmid-mediated efflux pump conferring resistance to the olaquindox, also confer resistance to other quinolones ciprofloxacin and norfloxacin as well as other antibiotics such as chloramphenicol, nitrofurantoin, and trimethoprim (Hansen et al. 2007; Ho et al. 2016) and have been reported in multiple species of Enterobacteriaceae. Similarly, plasmid-borne qepA gene efflux pump, which has now been found worldwide (Habeeb et al. 2014; Zhao et al. 2015), have been reported in clinical E. coli isolates (often associated with aminoglycoside resistance) by researchers from Japan and France (Pe´richon et al. 2007; Yamane et al. 2007). In addition, qnr genes have been found regularly in multi-resistance plasmids that also encode genes for other resistance determinants such as β-lactamases, ESBLs, AmpC enzymes, and carbapenemases (Jacoby et al. 2014).
The detection of sulfonamide resistance genes sul1-4 in the 3S metagenome is interesting because sulfonamides are synthetic antibiotics, and naturally occurring enzymes that degrade or modify this antibiotic are not to be expected. Thus, its detection indicates active pollution of the polluted soil with the antibiotic, its residual or antibiotic resistance bacteria carrying the sul gene. Mobile genetic elements facilitate the spread and dissemination of the sul gene via horizontal gene transfer. This is evident in the location of the sul gene. While sul1 gene and other resistance determinants is located on class 1 integrons, sul2 gene is located on IncQ-type non-conjugative plasmid and large transmissible plasmids, and sul3 gene is reported to be encoded on E. coli conjugative plasmid pVP440 flanked by two copies of IS element IS15∆/26 (Razavi et al. 2017). sul4 gene is also believed to be mobile genetic element borne.
The preponderance of antibiotic resistance genes against β-lactam antibiotics as well as other antibiotics used to treat infection caused by members of the family Enterobacteriaceae at the sampling site point to possible direct/indirect contamination from faecal materials of medicated humans and animals from point and non-point sources.
While the presence of heavy metal resistance genes at the sampling site may be attributed to indiscriminate dumping of spent oils and other hydrocarbon products rich in heavy metals, which normally select for microorganisms with heavy metal resistance phenotype, various anthropogenic activities that predates this period (i.e. 15 years) may have also contributed to the soil heavy metal burden and the emergence of heavy metal resistance genes. In addition, protists are known to employ metal poisoning to inactivate their bacteria prey before killing them. To counteract this, bacteria have evolved diverse metal detoxification strategies such as copper/zinc resistance genes to avoid killing via metal poisoning (Hao et al. 2015, 2016, 2017). Thus, a non-anthropogenic scenario such as this also fuels the spread and dissemination of heavy metal resistance genes in bacteria.
Aside from being a constituent of spent oils and hydrocarbon products, copper have been used in aquaculture, horticulture and a pesticide or antimicrobial in livestock production. It is also added to animal feed as a growth promoter by influencing the gut microbiota (Pal et al. 2017). Bacterial cells have evolved systems for copper homeostasis to protect against excess copper-mediated toxicity while allowing the adequate supply of copper for cellular processes. In this study, four copper resistance systems cue, cop, cus, and pco used by members of the family Enterobacteriaceae were detected. In the cue system of E. coli used under both aerobic and anaerobic conditions (Bondarczuk and Piotrowska-Seget 2013), a MerR family copper (I)-responsive transcriptional activator, CueR, regulates expression of a copper efflux P1-type ATPase, CopA and of CueO, a periplasmic multi-copper oxidase that oxidize Cu(I) to Cu(II) (Hobman and Crossman 2014; Pal et al. 2017). Under anaerobic and extreme copper stress conditions, CueO is inactive due to oxygen limitation. As a result, E. coli use the cus system comprising a two-component regulator CusRS that activate expression of cusCFBA (in response to stress and elevated Cu(I) levels at the cell envelope); a tripartite RND family silver/copper efflux and CusF, a periplasmic metallochaperone (Munson et al. 2000; Outten et al. 2001). In the plasmid-borne copper resistance pco system, only PcoA, a multi-copper oxidase that oxidize Cu(I) to less toxic Cu(II) and PcoS, a member of the two-component regulator PcoRS, which regulate gene expression from the pco operon (Hobman and Crossman 2014) were detected in 3S metagenome. The detection of the Gram-positive copYZAB operon cop system (Pal et al. 2017); the Gram-negative copABCDRS operon cop system (Monchy et al. 2006; Zimmermann et al. 2012; Staehlin et al. 2016), cinA of Pseudomonas putida copper-induced cinAQ system; crdB, crdR, crdS of Helicobacter pylori CrdA-CrdB-CzcB-CzcA system; and csoR, ctpV of Mycobacterium tuberculosis (Waidner et al. 2005; Liu et al. 2007; Quaranta et al. 2009) and several others in 3S metagenome further highlight the diverse copper resistance systems employed by members of the microbial community for detoxification, transport and export of excess copper.
The detection of diverse resistance mechanisms for zinc, cadmium, cobalt, and nickel responsible for uptake, detoxification and efflux in 3S metagenome may be attributed to selective pressure imposed by the heavy metal burden on the microbial community, which naturally select for members of the microbial community with heavy metal resistance phenotypes. Resistance genes encoding for the transport of zinc and other divalent cations (zupT (Zn/Fe/Co/Ni/Cu/Cd), zipB (Zn/Cd) (Fox and Guerinot 1998), high-affinity zinc transporter znuA, znuC (Patzer and Hantke 1998), as well as nickel (nickel ABC transporter nikA, nikB, nikC, nikD, nikE and several others)) were detected. Also detected are genes responsible for efflux (ZntA (Zn/Cd/Pb), ZitB (Zn), ncrC (Co/Ni), czrA (Zn/Cd; Hassan et al. 1999), czcA, czcB, czcC (Cd/Zn/Co; Nies et al. 1989; Franke et al. 2003; Nies 2003a, b) and several others), and transcriptional regulation (zntR, zraR, zraS, cadR, cmtR) among others (Table 2).
Of great concern is the detection of mer resistance genes, which are usually borne on mer operon in 3S metagenome. In Hg2+ detoxification, periplasmic Hg2+-binding protein MerP bound to the Hg2+ and delivers it to the mercury transporter MerT for transport into the cytoplasm, where it is reduced in a NADPH-dependent manner to volatile, less reactive Hg0 by MerA, mercuric reductase (Nies 1999; Oregaard and Sorensen 2007). In this study all the genes encoding MerP, MerT, MerA (merP, merT, merA) were detected in 3S metagenome (Table 2). More so, resistance gene for alternative route of mercury uptake merC (Hamlett et al. 1992; Sahlman et al. 1997) was also detected in this study, thus, indicating that some members of the microbial community also use the alternative route for mercury uptake. In addition, as shown in Table 2, resistance to lead (Pb) in 3S metagenome is mediated a P-type ATPase, Pb-efflux ATPase pbrA, which facilitate effective efflux of Pb2+ from the cytoplasm.
Arsenic toxicity stem from its structural similarity to phosphate, which allows it to interfere in the metabolism of phosphorus, a major macronutrient required by microorganisms. Arsenate As(V) enters the cell via the phosphate uptake systems (PstA, PstB, PstC) where it is reduced to arsenite As(III) by arsenate reductase ArsC to facilitate its differentiation from phosphate. As(III) can also be taken up directly using GlpF, a MIP (Major Intrinsic Protein) family channel protein. The resultant As(III) is thereafter translocated across the cytoplasmic membrane via ArsB or Acr3 (Nies 1999; Ben Fekih et al. 2018). It is instructive to note that genes encoding PstA, PstB, PstC (pstA, pstB, pstC) as well as ArsC, ArsB, Acr3, GlpF (arsC, arsB, acr3, glpF) and several others were detected in the metagenome. ArsA, which catalyse the extrusion of arsenite (Wang et al. 2004); arsenite oxidase, which mediates the oxidation of toxic arsenite to less toxic arsenate (Ellis et al. 2001); and ArsM, which is involved in the conversion of As(III) to a number of methylated products (Wang et al. 2014) were also detected in this study (Table 2).
The mercury mer operon are usually borne on integrons, which of recent are found to be carried on transposons such as Tn21. The Tn21 transposons carries In2 integrons inserted between the mercury resistance genes and the transposition genes, which also carries the integrase gene intI1 that can acquire, express and reassort the antibiotic resistance cassettes carried by the integrons (Rosewarne et al. 2010; Mindlin and Petrova 2013). The consistent link between mercury resistance and antibiotic resistance in microorganisms from diverse habitats such as faecal and oral microbiota of primates (Wireman et al. 1997), fish gastrointestinal tracts (Akinbowale et al. 2007), mine sediments (Nemergut et al. 2004), and freshwater microcosms (Stepanauskas et al. 2006) is a cause for serious public health concerns and this has been largely fuelled by the presence of integron-associated integrases with antibiotics and heavy metal resistance determinants in mercury transposons (Pal et al. 2015a, b).
The co-resistance phenomenon is not limited to mercury as revealed in several reports. Co-existence of OqxAB, blaCTX-M, pco (copper) and silver (sil) resistance operons on the same plasmid have been reported (Mourao et al. 2015; Fang et al. 2016). The co-selection of cadmium and zinc for methicillin resistance in Staphylococcus aureus through HGT of plasmids encoding genes for methicillin (mec) and Cd/Zn resistance (czr) have also been reported (Cavaco et al. 2010). In addition, the mobile colistin resistance gene, mcr-1 has been found in copper-tolerant isolates (Campos et al. 2016). Furthermore, a Scottish study on randomly selected archived agricultural soils spanning a period of 30 years revealed the presence of 11 antibiotic resistance genes correlated to soil copper, chromium, lead and iron levels (Knapp et al. 2011).
Cross-resistance between antibiotics and heavy metals has also been documented. For instance, efflux systems have been reported to confer cross-resistance to metals and antibiotics (Cheng et al. 1996; Mata et al. 2000). In addition, co-regulation of antibiotic and metal resistance has been reported in Pseudomonas aeruginosa cadmium-zinc-cobalt regulatory system, CzcRS, which regulate not only the expression of the czcCBA efflux system that confers resistance to cadmium, zinc, and cobalt but also decrease expression of OprD porin, resulting in increased resistance to carbapenems (Perron et al. 2004).
From the foregoing, it is highly plausible that the spread and dissemination of antibiotic resistance genes in 3S metagenome are facilitated by the presence of heavy metal resistance genes, which are often borne on mobile genetic elements that also carries resistance determinant for various antibiotic classes. This assertion is further corroborated by the fact that most of the mobile genetic element-encoded heavy metal resistance genes detected in this study have been reported in the literature to also carry antibiotic resistance genes. Furthermore, exposure to multiple heavy metals, as obtained in this study, has been shown to be more effective for co-selection for antibiotic resistance (Guo et al. 2014). More so, it has been observed that the concentrations of heavy metals in natural environments, which at times can be lower than the minimum inhibitory concentration may be enough to drive the co-selection of antibiotic resistance genes (Seiler and Berendonk 2012), and this has also been demonstrated in in vitro studies (Gullberg et al. 2014).
Conclusions
In summary, this study has unravelled via next-generation shotgun metagenomics diverse antibiotic and heavy metal resistance genes constituting the resistome of 3S metagenome. Antibiotic resistance genes for 15 antibiotic classes and heavy metal resistance genes of various heavy metals were detected, their resistance mechanisms explained and the public and environmental health implications of their presence enunciated. The preponderance in this study, of resistance genes against antibiotics used to treat infections caused by members of the family Enterobacteriaceae points to possible contamination from faecal materials of medicated humans and animals via point and non-point sources. Attempt was made to establish the possible contributions of the heavy metal resistance genes to the spread and dissemination of antibiotic resistance determinants in the metagenome. The huge repository of antibiotic and heavy metal resistance genes in the soil environment poses a significant threat to public and environmental health, and due to the intricate link between humans and the soil, it may be a huge contributor to the proliferation of multidrug-resistant clinical pathogens.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
LBS designed, execute, analyse and wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The author declares that there is no conflict of interest.
References
- Akinbowale OL, Peng H, Grant P, Barton MD. Antibiotic and heavy metal resistance in motile aeromonads and pseudomonads from rainbow trout (Oncorhynchus mykiss) farms in Australia. Int J Antimicrob Agents. 2007;30(2):177–182. doi: 10.1016/j.ijantimicag.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- Allmansberger R, Bräu B, Piepersberg W (1985) Genes for gentamicin-(3)-N-acetyl-transferases III and IV. II. Nucleotide sequences of three AAC(3)-III genes and evolutionary aspects. Mol Gen Genet 198(3):514–520 [DOI] [PubMed]
- Amábile-Cuevas CF. Antibiotics and antibiotic resistance in the environment. Leiden: CRC Press/Balkema; 2016. p. 133. [Google Scholar]
- Amachawadi RG, Shelton NW, Shi X, et al. Selection of fecal enterococci exhibiting tcrB-mediated copper resistance in pigs fed diets supplemented with copper. Appl Environ Microbiol. 2011;77(16):5597–5603. doi: 10.1128/AEM.00364-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun S, Mukhopadhyay M, Chakraborty P (2017) A review on antibiotics consumption, physico-chemical properties and their sources in Asian soil. In: Hashmi MZ, Strezov V, Varma A (eds) Antibiotics and antibiotics resistance genes in soils. Soil biology, vol 51. 10.1007/978-3-319-66260-2_1. Springer, New York
- Ben Fekih I, Zhang C, Li YP, Zhao Y, Alwathnani HA, Saquib Q, Rensing C, Cervantes C. Distribution of arsenic resistance genes in prokaryotes. Front Microbiol. 2018;9:2473. doi: 10.3389/fmicb.2018.02473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Z, Berglund B, Sun Q, et al. Prevalence of the mcr-1 colistin resistance gene in extended-spectrum beta-lactamase-producing Escherichia coli from human faecal samples collected in 2012 in rural villages in Shandong Province, China. Int J Antimicrob Agents. 2017;49(4):493–497. doi: 10.1016/j.ijantimicag.2016.12.018. [DOI] [PubMed] [Google Scholar]
- Blanco P, Hernando-Amado S, Reales-Calderon JA, et al. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms. 2016;4(1):4. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarczuk K, Piotrowska-Seget Z. Molecular basis of active copper resistance mechanisms in Gram-negative bacteria. Cell Biol Toxicol. 2013;29(6):397–405. doi: 10.1007/s10565-013-9262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo RA. β-Lactamases: a focus on current challenges. Cold Spring Harb Perspect Med. 2017;7:a025239. doi: 10.1101/cshperspect.a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J, Cristino L, Peixe L, Antunes P (2016) MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Eurosurveillance 21(26) [DOI] [PubMed]
- Cantón R, Ruiz-Garbajosa P. Co-resistance: an opportunity for the bacteria and resistance genes. Curr Opin Pharmacol. 2011;11:477–485. doi: 10.1016/j.coph.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Cardoso M, Schwarz S. Chloramphenicol resistance plasmids in Staphylococcus aureus isolated from bovine subclinical mastitis. Vet Microbiol. 1992;30(2–3):223–232. doi: 10.1016/0378-1135(92)90116-b. [DOI] [PubMed] [Google Scholar]
- Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY antimicrobial surveillance program, 2006–2007. Antimicrob Agents Chemother. 2011;55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catry B, Cavaleri M, Baptiste K, et al. Use of colistin-containing products within the European Union and European economic area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int J Antimicrob Agents. 2015;46(3):297–306. doi: 10.1016/j.ijantimicag.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Cavaco LM, Hasman H, Aarestrup FM. Zinc resistance of Staphylococcus aureus of animal origin is strongly associated with methicillin resistance. Vet Microbiol. 2011;150(3–4):344–348. doi: 10.1016/j.vetmic.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Cavaco LM, Hasman H, Stegger M, et al. Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrob Agents Chemother. 2010;54(9):3605–3608. doi: 10.1128/AAC.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra N, Kumar S (2017) Antibiotics producing soil microorganisms. In: Hashmi MZ, Strezov V, Varma A (eds) Antibiotics and antibiotics resistance genes in soils. Soil biology, vol 51. 10.1007/978-3-319-66260-2_1. Springer, New York
- Cheng J, Hicks DB, Kruwlich TA. The purified Bacillus subtilis tetracycline efflux protein TetA(L) reconstitutes both tetracycline–cobalt/H+ and Na+(K+)/H+ exchange. Proc Natl Acad Sci USA. 1996;93(25):14446–14451. doi: 10.1073/pnas.93.25.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates K, Walsh TR, Spencer J, Hinchliffe P (2017) 1.12 A resolution crystal structure of the catalytic domain of the plasmid-mediated colistin resistance determinant MCR-2. Acta Crystallogr F Struct Biol Commun 73 (Pt 8):443–449 [DOI] [PMC free article] [PubMed]
- Cochetti I, Tili E, Mingoia M, Varaldo PE, Montanari MP. Erm(b)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob Agents Chemother. 2008;52:1285–1290. doi: 10.1128/AAC.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon P, Athukorala P, Senanayake S, Khan F. Antimicrobial resistance: the major contribution of poor governance and corruption to this growing problem. PLoS ONE. 2015;10:e0116746. doi: 10.1371/journal.pone.0116746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley E, Weeks J, Jones L, Daniels V, Wootton M, MacGowan A, Walsh T. NDM-1 polymicrobial infections including Vibrio cholerae. Lancet. 2012;380:1358. doi: 10.1016/S0140-6736(12)60911-8. [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso LM, Wedin DA, Gilley JE, Miller DN, Marx DB. Assessment of selected antibiotic resistances in ungrazed native Nebraska prairie soils. J Environ Qual. 2016;45(2):454–462. doi: 10.2134/jeq2015.06.0280. [DOI] [PubMed] [Google Scholar]
- Eiamphungporn W, Schaduangrat N, Malik AA, Nantasenamat C. Tackling the antibiotic resistance caused by class A β-lactamases through the use of β-lactamase inhibitory protein. Int J Mol Sci. 2018;19(8):2222. doi: 10.3390/ijms19082222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis PJ, Conrads T, Hille R, Kuhn P (2001) Crystal structure of the 100 kDa aresenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A. Structure 9(2):125–132 [DOI] [PubMed]
- Fang L, Li X, Li L et al (2016) Co-spread of metal and antibiotic resistance within ST3-IncH12 plasmids from E. coli isolates of food-producing animals. Sci Rep 6:25312 [DOI] [PMC free article] [PubMed]
- Fox TC, Guerinot ML. Molecular biology of cation transport in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:669–696. doi: 10.1146/annurev.arplant.49.1.669. [DOI] [PubMed] [Google Scholar]
- Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galopin S, Cattoir V, Leclercq R. A chromosomal chloramphenicol acetyltransferase determinant from a probiotic strain of Bacillus clausii. FEMS Microbiol Lett. 2009;296(2):185–189. doi: 10.1111/j.1574-6968.2009.01633.x. [DOI] [PubMed] [Google Scholar]
- Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- Gao R, Hu Y, Li Z, et al. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. 2016;12(11):e1005957. doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady R, Hayes F. Axe-Txe, a broad-spectrum proteic toxin–antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol Microbiol. 2003;47(5):1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- Grossman TH. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med. 2016;6(4):a025387. doi: 10.1101/cshperspect.a025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI (2014) Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 5(5):e01918.14 [DOI] [PMC free article] [PubMed]
- Guo X, Liu S, Wang Z, Zhang XX, Li M, Wu B. Metagenomic profiles and antibiotic resistance genes in gut microbiota of mice exposed to arsenic and iron. Chemosphere. 2014;112:1–8. doi: 10.1016/j.chemosphere.2014.03.068. [DOI] [PubMed] [Google Scholar]
- Gupta A, Vlamakis H, Shoemaker N, Salyers AA. A new bacteroides conjugative transposon that carries an erm(b) gene. Appl Environ Microbiol. 2003;69:6455–6463. doi: 10.1128/AEM.69.11.6455-6463.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58(1):212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeeb MA, Haque A, Iversen A, Giske CG. Occurrence of virulence genes, 16S rRNA methylases, and plasmid-mediated quinolone resistance genes in CTXM-producing Escherichia coli from Pakistan. Eur J Clin Microbiol Infect Dis. 2014;33:399–409. doi: 10.1007/s10096-013-1970-1. [DOI] [PubMed] [Google Scholar]
- Haenni M, Beyrouthy R, Lupo A, Chatre P, Madec JY, Bonnet R. Epidemic spread of Escherichia coli ST744 isolates carrying mcr-3 and blaCTX-M-55 in cattle in France. J Antimicrob Chemother. 2017;73(2):533–536. doi: 10.1093/jac/dkx418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlett NV, Landale EC, Davis BH, Summers AO. Roles of the Tn21 merT, merP, and merC gene products in mercury resistance and mercury binding. J Bacteriol. 1992;174:6377–6385. doi: 10.1128/jb.174.20.6377-6385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LH, Jensen LB, Sorensen HI, Sorensen SJ. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. 2007;60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- Hao X, Li X, Pal C, et al. Widespread resistance to arsenic in bacteria is influenced by its use in protists to kill bacterial prey. Biometals. 2017 doi: 10.1007/s10534-017-0003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X, Lűthje F, Rønn R, et al. A role for copper in protozoan grazing—two billion years selecting for bacterial copper resistance. Mol Microbiol. 2016;102(4):628–641. doi: 10.1111/mmi.13483. [DOI] [PubMed] [Google Scholar]
- Hao X, Lűthje FL, QinY et al. (2015) Survival in amoeba—a major selective pressure on the presence of bacterial copper and zinc resistance determinants? Identification of a “copper pathogenicity island”. Appl Microbiol Biotechnol 99(14):5817–5824 [DOI] [PubMed]
- Hasman H, Aarestrup FM. TcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob Agents Chemother. 2002;48(5):1410–1416. doi: 10.1128/AAC.46.5.1410-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MT, van der Lelie D, Springael D, Romling U, Ahmed N, Mergeay M. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene. 1999;238:417–425. doi: 10.1016/s0378-1119(99)00349-2. [DOI] [PubMed] [Google Scholar]
- Ho PL, Ng KY, LoWU LPY, Lai EL, Wang Y, Chow KH. Plasmid-mediated OqxAB is an important mechanism for nitrofurantoin resistance in Escherichia coli. Antimicrob Agents Chemother. 2016;60:537–543. doi: 10.1128/AAC.02156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman JL, Crossman LC. Bacterial antimicrobial metal ion resistance. J Med Microbiol. 2014;64:471–497. doi: 10.1099/jmm.0.023036-0. [DOI] [PubMed] [Google Scholar]
- Hollingshead S, Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. 1985;13(1):17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- Hooper GC, Jacoby GA. Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb Perspect Med. 2016;6:a025320. doi: 10.1101/cshperspect.a025320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol Microbiol. 2007;63:937–940. doi: 10.1111/j.1365-2958.2006.05584.x. [DOI] [PubMed] [Google Scholar]
- Hu M, Guo J, Cheng Q, et al. Crystal structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for colistin resistance. Sci Rep. 2016;6:38793. doi: 10.1038/srep38793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22(1):161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectr. 2014 doi: 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Starosta AL, Terry DS, et al. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci USA. 2013;110:3812–3816. doi: 10.1073/pnas.1216691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LS, Toleman MA, Weeks JL, Howe RA, Walsh TR, Kumarasamy KK. Plasmid carriage of bla NDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob Agents Chemother. 2014;58:4211–4213. doi: 10.1128/AAC.02500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal S, Prasad R, Varma A (2010) Soil microbial diversity in relation to heavy metals. In: Sherameti I, Varma A (eds) Soil heavy metals. Soil biology, vol 19. 10.1007/978-3-642-02436-8_3. Springer, Berlin
- Khan MA, Khan S, Khan A, Alam M. Soil contamination with cadmium, consequences and remediation using organic amendments. Sci Total Environ. 2017;601–602:1591–1605. doi: 10.1016/j.scitotenv.2017.06.030. [DOI] [PubMed] [Google Scholar]
- Knapp CW, McCluskey SM, Singh BK, Campbell CD, Hudson G, Graham DW. Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS ONE. 2011;6(11):e27300. doi: 10.1371/journal.pone.0027300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashi Y, Hasebe A, Nishio M, Uchiyama H. Diversity of tetracycline resistance genes in bacteria isolated from various agricultural environments. Microbes Environ. 2007;22(1):44–51. [Google Scholar]
- Leonard DA, Bonomo RA, Powers RA. Class D β-lactamases: a reappraisal after five decades. Acc Chem Res. 2013;46:2407–2415. doi: 10.1021/ar300327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-X, Zhong Z, Hou P, Zhang W-P, Qian P-Y. Resistance to nonribosomal peptide antibiotics mediated by D-stereospecific peptidases. Nat Chem Biol. 2018;14:381–387. doi: 10.1038/s41589-018-0009-4. [DOI] [PubMed] [Google Scholar]
- Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl M (2017) Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009–17. Euro Surveill 22(31) [DOI] [PMC free article] [PubMed]
- Liu T, Ramesh A, Ma Z, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- Ma G, Zhu Y, Yu Z, Ahmad A, Zhang H. High resolution crystal structure of the catalytic domain of MCR-1. Sci Rep. 2016;6:39540. doi: 10.1038/srep39540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack S, Xu Y, Nodwell JR. The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol Microbiol. 2014;93:391–402. doi: 10.1111/mmi.12689. [DOI] [PubMed] [Google Scholar]
- Markley JL, Wencewicz TA. Tetracycline-inactivating enzymes Front Microbiol. 2018;9:1058. doi: 10.3389/fmicb.2018.01058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marosevic D, Kaevska M, Jaglic Z. Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage- a review. Ann Agric Environ Med. 2017;24(2):338–344. doi: 10.26444/aaem/74718. [DOI] [PubMed] [Google Scholar]
- Martin MF, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- Mata MT, Baquero F, Perez-Dıaz JC. A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol Lett. 2000;187(2):185–188. doi: 10.1111/j.1574-6968.2000.tb09158.x. [DOI] [PubMed] [Google Scholar]
- McArthur AG, Waglechner N, Nizam F, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57(7):3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindlin S, Petrova M. Mercury resistance transposons. In: Roberts AP, Mullany P, editors. Bacterial integrative mobile genetic elements. Texas: Landes Bioscience; 2013. pp. 33–52. [Google Scholar]
- Monchy S, Benotmane MA, Wattiez R, et al. Transcriptomic and proteomic analyses of the pMOL30-encoded copper resistance in Cupriavidus metallidurans strain CH34. Microbiology. 2006;152(6):1765–1776. doi: 10.1099/mic.0.28593-0. [DOI] [PubMed] [Google Scholar]
- Mourao J, Novais C, Machado J, Peixe L, Antunes P. Metal tolerance in emerging clinically relevant multi-drug resistant Salmonella enterica serotype 4, [5], 12:i:- clones circulating in Europe. Int J Antimicrob Agents. 2015;45(6):610–616. doi: 10.1016/j.ijantimicag.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Mugnier P, Dubrous P, Casin I, Arlet G, Collatz E. A TEM-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40(11):2488–2493. doi: 10.1128/aac.40.11.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson GP, Lam DL, Outten FW, O’Halloran TV. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol. 2000;182(20):5864–5871. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Shaw WV. O-acetyltransferases for chloramphenicol and other natural products. Antimicrob Agents Chemother. 1997;41(1):1–6. doi: 10.1128/aac.41.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut DR, Martin AP, Schmidt SK. Integron diversity in heavy metal contaminated mine tailings and inferences about integron evolution. Appl Environ Microbiol. 2004;70(2):1160–1168. doi: 10.1128/AEM.70.2.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NQ, Krishnan NP, Rojas LJ, Prati F, Caselli E, Romagnoli C, Bonomo RA, van den Akker F. Crystal structures of KPC-2 and SHV-1 β-lactamases in complex with the boronic acid transition state analog S02030. Antimicrob Agents Chemother. 2016;60:1760–1766. doi: 10.1128/AAC.02643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- Nies DH, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oregaard G, Sorensen SJ. High diversity of bacterial mercuric reductase genes from surface and subsurface floodplain soil (Oak Ridge, USA) ISME J. 2007;1:453–467. doi: 10.1038/ismej.2007.56. [DOI] [PubMed] [Google Scholar]
- Osborn AM, Bruce KD, Strike P, Ritchie DA. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- Outten FW, Huffman DL, Hale JA, O’Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276(33):30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- Pal C, Asiani K, Arya S, Rensing C, Stekel DJ, Larsson DGJ, Hobman JL. Metal resistance and its association with antibiotic resistance. Adv Microb Physiol. 2017 doi: 10.1016/bs.ampbs.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics. 2015;16:964. doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics. 2015;6:964. doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DG (2014) BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res 42(database issue):D737–D743 [DOI] [PMC free article] [PubMed]
- Parkhill J, Dougan G, James KD et al (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413(6858):848–852 [DOI] [PubMed]
- Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:48. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28(6):1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- Pe´richon B, Courvalin P, Galimand M (2007) Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA mediated efflux in Escherichia coli. Antimicrob Agents Chemother 51:2464–2469 [DOI] [PMC free article] [PubMed]
- Penesyan A, Gillings M, Paulsen IT. Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules. 2015;20:5286–5298. doi: 10.3390/molecules20045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreten V, Schwarz F, Cresta L, Boeglin M, Dasen G, Teuber M. Antibiotic resistance spread in food. Nature. 1997;389:801–802. doi: 10.1038/39767. [DOI] [PubMed] [Google Scholar]
- Perron K, Caille O, Rossier C, Delden CV, Dumas J, Kohler T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J Biol Chem. 2004;279(10):8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- Perry JA, Wright GD. The antibiotic resistance “mobilome”: searching for the link between environment and clinic. Front Microbiol. 2013;4:138. doi: 10.3389/fmicb.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan SJ, Kornblum J, Moghazeh SL, Edelman I, Gennaro ML, Novick RP. Comparative sequence and functional analysis of pT181 and 6221, cognate plasmid replicons from Staphy1ococcus aureus. Mol Gen Genet. 1985;199:452–464. doi: 10.1007/BF00330758. [DOI] [PubMed] [Google Scholar]
- Quaranta D, McEvoy MM, Rensing C. Site-directed mutagenesis identifies a molecular switch involved in copper sensing by the histidine kinase CinS in Pseudomonas putida KT2440. J Bacteriol. 2009;191:5304–5311. doi: 10.1128/JB.00551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Ali T, Ali I, Khan NA, Han B, Gao J (2018) The growing genetic and functional diversity of extended spectrum beta-lactamases. BioMed Res Int 2018 (article ID 9519718) [DOI] [PMC free article] [PubMed]
- Ramirez MS, Tolmansky ME. Aminoglycoside modifying enzymes. Drug Resist Update. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi M, Marathe NP, Gillings MR, Flach C-F, Kristiansson E, Joakim Larsson DG. Discovery of the fourth mobile sulfonamide resistance gene. Microbiome. 2017;5:160. doi: 10.1186/s40168-017-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acid Res. 2010;38:20–191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev. 2011;35:856–871. doi: 10.1111/j.1574-6976.2011.00283.x. [DOI] [PubMed] [Google Scholar]
- Rogers BA, Sidiabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66(1):1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- Rosewarne CP, Pettigrove V, Stokes HW, Parsons YM. Class 1 integrons in benthic bacterial communities: Abundance, association with Tn402-like transposition modules and evidence for co-selection with heavy metal resistance. FEMS Microbiol Ecol. 2010;72(1):35–46. doi: 10.1111/j.1574-6941.2009.00823.x. [DOI] [PubMed] [Google Scholar]
- Rough DA, Lee BTO, Morby AP. Understanding cellular responses to toxic agents: a model for mechanism-choice in bacterial metal resistance. J Ind Microbiol. 1995;14:132–141. doi: 10.1007/BF01569895. [DOI] [PubMed] [Google Scholar]
- Sahlman L, Wong W, Powlowski J. A mercuric ion uptake role for the integral inner membrane protein, MerC, involved in bacterial mercuric ion resistance. J Biol Chem. 1997;272:29518–29526. doi: 10.1074/jbc.272.47.29518. [DOI] [PubMed] [Google Scholar]
- Salam LB. Detection of carbohydrate-active enzymes and genes in a spent engine oil-perturbed agricultural soil. Bull Natl Res Center. 2018;42:10. [Google Scholar]
- Salam LB, Ishaq A (2019) Biostimulation potentials of corn steep liquor in enhanced hydrocarbon degradation in chronically polluted soil. 3 Biotech 9:46 [DOI] [PMC free article] [PubMed]
- Sandaa RA, Torsvik V, Enger O, Daae LF, Castberg T, Hahn D. Analysis of bacterial communities in heavy metal-contaminated soils at different levels of resolution. FEMS Microbiol Ecol. 1999;30:237–251. doi: 10.1111/j.1574-6941.1999.tb00652.x. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. 2004;28(5):519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Spies U, Cardoso M. Cloning and sequence analysis of a plasmid-encoded chloramphenicol acetyltransferase gene from Staphylococcus intermedius. J Gen Microbiol. 1991;137(4):977–981. doi: 10.1099/00221287-137-4-977. [DOI] [PubMed] [Google Scholar]
- Seiler C, Berendonk TU. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol. 2012;3:339. doi: 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KJ, Rather PN, Hare RS, Miller GH. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57(1):138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol. 2001;67:561–568. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S, Walderhaug M. Gene-regulation of plasmid-determined and chromosome-determined inorganic ion transport in bacteria. Microbiol Mol Biol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehlin BM, Gibbons JG, Rokas A, O’Halloran TV, Slot JC. Evolution of a heavy metal homeostasis/resistance island reflects increasing copper stress in enterobacteria. Gen Biol Evol. 2016;8(3):811–826. doi: 10.1093/gbe/evw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas R, Glenn TC, Jagoe CH, et al. Co-selection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ Microbiol. 2006;8(9):1510–1514. doi: 10.1111/j.1462-2920.2006.01091.x. [DOI] [PubMed] [Google Scholar]
- Surette MD, Wright GD. Lessons from the environmental antibiotic resistome. Annu Rev Microbiol. 2017;71:309–329. doi: 10.1146/annurev-micro-090816-093420. [DOI] [PubMed] [Google Scholar]
- Tahlan K, Ahn SK, Sing A, Bodnaruk TD, Willems AR, Davidson AR, Nodwell JR. Initiation of actinorhodin export in Streptomyces coelicolor. Mol Microbiol. 2007;63:951–961. doi: 10.1111/j.1365-2958.2006.05559.x. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Osaki M, Sekizaki T. Chloramphenicol resistance transposable element TnSs1 of Streptococcus suis, a transposon flanked by IS6-family elements. Plasmid. 2003;49(2):143–151. doi: 10.1016/s0147-619x(02)00149-x. [DOI] [PubMed] [Google Scholar]
- Tanaka SK, Steenbergen J, Villano S. Discovery, pharmacology, and clinical profile of omadacycline, a novel aminomethylcycline antibiotic. Bioorg Med Chem. 2016;24:6409–6419. doi: 10.1016/j.bmc.2016.07.029. [DOI] [PubMed] [Google Scholar]
- Tennigkeit J, Matzura H. Nucleotide sequence analysis of a chloramphenicol-resistance determinant from Agrobacterium tumefaciens and identification of its gene product. Gene. 1991;98:113–116. doi: 10.1016/0378-1119(91)90112-o. [DOI] [PubMed] [Google Scholar]
- Toth M, Antunes NT, Stewart NK, Frase H, Bhattacharya M, Smith C, Vakulenko S. Class D β-lactamases do exist in Gram-positive bacteria. Nat Chem Biol. 2016;12(1):9–14. doi: 10.1038/nchembio.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouvelekis LS, Tzelepi E, Tassios PT, Legakis NJ. CTX-M-type beta-lactamases: an emerging group of extended-spectrum enzymes. Int J Antimicrob Agents. 2000;14(2):137–142. doi: 10.1016/s0924-8579(99)00165-x. [DOI] [PubMed] [Google Scholar]
- van Hoek AH, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJ. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011;2:203. doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidner B, Melchers K, Stähler FN, Kist M, Bereswill S. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J Bacteriol. 2005;187:4683–4688. doi: 10.1128/JB.187.13.4683-4688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- Wang G, Kennedy SP, Fasiludeen S, Rensing C, DasSarma S. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J Bacteriol. 2004;186:3187–3194. doi: 10.1128/JB.186.10.3187-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PP, Sun GX, Zhu YG. Identification and characterization of arsenite methyltransferase from an archaeon Methanosarcina acetivorans C2A. Environ Sci Technol. 2014;48:12706–12713. doi: 10.1021/es503869k. [DOI] [PubMed] [Google Scholar]
- Willms IM, Kamran A, Aßmann NF, Krone D, Bolz SH, Fiedler F, Nacke H. Discovery of novel antibiotic resistance determinants in forest and grassland soil metagenomes. Front Microbiol. 2019;10:460. doi: 10.3389/fmicb.2019.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wireman J, Liebert CA, Smith T, Summers AO. Association of mercury resistance with antibiotic resistance in the gram-negative fecal bacteria of primates. Appl Environ Microbiol. 1997;63(11):4494–4503. doi: 10.1128/aem.63.11.4494-4503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Zhang W, Xie S, Zheng M, Liu H, Yang J, Liu X, Yang F. Increasing prevalence of antibiotic resistance genes in manured agricultural soils in northern China. Front Environ Sci Eng. 2020;14:1. [Google Scholar]
- Xu Y, Zhong L-L, Srinivas S, et al. Spread of MCR-3 colistin resistance in China: an epidemiological, genomic and mechanistic study. EbioMedicine. 2018;34:139–157. doi: 10.1016/j.ebiom.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Wachino J, Suzuki S, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2007;51:3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel GG, Cheung D, Adam H, et al. Review of eravacycline, a novel fluorocycline antibacterial agent. Drugs. 2016;76:67–588. doi: 10.1007/s40265-016-0545-8. [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhang J, Zheng B, Wei Z, Shen P, Li S, Li L, Xiao Y. Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J Clin Microbiol. 2015;53:766–770. doi: 10.1128/JCM.02594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Udagedara SR, Sze CM et al. (2012) PcoE—a metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance. J Inorg Biochem 115(1):186–197 [DOI] [PubMed]
- Zhu Y-G, Zhao Y, Gillings M, Prenulas J, Ok YS, Capon A, Banwart S. Soil biota, antimicrobial resistance and planetary health. Environ Int. 2019;131:105059. doi: 10.1016/j.envint.2019.105059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.