Abstract
Biological nanopores are emerging as powerful tools for single-molecule analysis and sequencing. Here, we engineered the two-component pleurotolysin (PlyAB) toxin to assemble into 7.2 × 10.5 nm cylindrical nanopores with a low level of electrical noise in lipid bilayers, and we addressed the nanofluidic properties of the nanopore by continuum simulations. Surprisingly, proteins such as human albumin (66.5 kDa) and human transferrin (76–81 kDa) did not enter the nanopore. We found that the precise engineering of the inner surface charge of the PlyAB induced electro-osmotic vortices that allowed the electrophoretic capture of the proteins. Once inside the nanopore, two human plasma proteins could be distinguished by the characteristics of their current blockades. This fundamental understanding of the nanofluidic properties of nanopores provides a practical method to promote the capture and analysis of folded proteins by nanopores.
Keywords: biological nanopore, single-molecule sensing, electro-osmosis, plasma proteins, nanofluidics
Introduction
Nanopore currents allow identifying and studying unlabeled analytes lodged inside the nanopore at the single-molecule level in real time and under physiological aqueous conditions.1−5 Many initial efforts focused on using biological nanopores to detect unfolded polymers such as PEG,6−9 DNA,10−16 unfolded proteins,17,18 peptides19−30 and small analytes.31 Despite ingenious solutions where sensing elements were attached to nanopores,32−35 folded proteins have long resisted direct nanopore analysis, mainly because the biological nanopores available for analysis (e.g., α-hemolysin) were too small to allow the entry of large molecules. Among the few biological nanopores with known crystal structure that are large enough to accommodate folded proteins, only the α-helical cytolysin A (ClyA) has been used thus far.36−43 It has been shown that proteins up to ∼40 kDa remain trapped between the roughly cylindrical 5.5 × 10 nm (diameter × height) vestibule region and the narrower 3.3 nm trans entrance.44 In turn, this allowed the real-time observation of protein conformation changes and function dynamics such as enzyme catalysis and binding with small metabolite molecules.39,43,45 However, globular proteins larger than the ClyA lumen cannot be studied using this method.
Nanopores with a variety of sizes can now be fabricated in synthetic materials and have been employed extensively for folded protein detection and analysis.46 Notably, it was shown that the shape47,48 and the conformational flexibility49,50 of proteins can be deduced from correlation analysis of their current blockades and that small proteins such as ubiquitin (8.5 kDa) can be detected using 3 nm nanopores.51 However, the analysis of folded proteins using solid-state nanopores is challenging. Proteins often clog the pore,52 most likely because of unspecific absorption to the inorganic nanopore surface.53−56 In addition, proteins might stall at various locations inside the pore,57 and they often translocate too quickly to allow accurate analysis.58 Finally, the surface charge of solid-state nanopores, which plays a major role in the nanofluidic properties of the pore,59−61 cannot be easily engineered with atomic precision. This contrasts with protein nanopores, whose proteinaceous nature allows introducing, removing, or reversing individual charges at specific locations within the pore.26,62−64
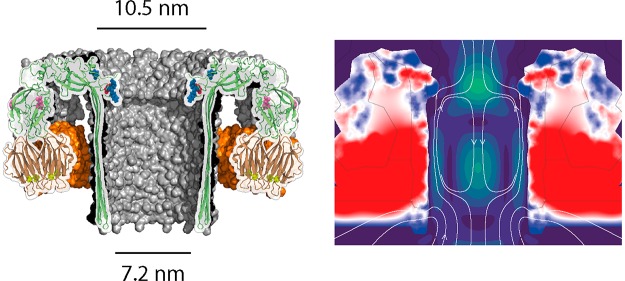
In this study, we engineered pleurotolysin (PlyAB, Figure 1a) oligomers to form nanopores into lipid bilayers with low-noise electrical properties. PlyAB consists of two distinct components.65,66 Pleurotolysin A (PlyA, 16 kDa) acts as a scaffold to recruit the second component pleurotolysin B (PlyB, 54 kDa), which spans the lipid bilayer. Cryogenic electron microscopy revealed a nanopore with a cis entry of ∼10.5 nm, a trans entry of ∼7.2 nm, and a constriction with a diameter of ∼5.5 nm.66 Proteins come with a variety of charges, sizes, and shapes, and one of the main challenges in nanopore analysis is to promote the capture of proteins.26 Here, we describe the engineering of PlyAB nanopores to enable the capture of large folded proteins that otherwise would not enter the nanopore. Using continuum simulations, we were able to unravel the differences of the nanofluidic properties of these engineered pores, most notably the electro-osmotic flow, that allow folded protein capture.
Figure 1.
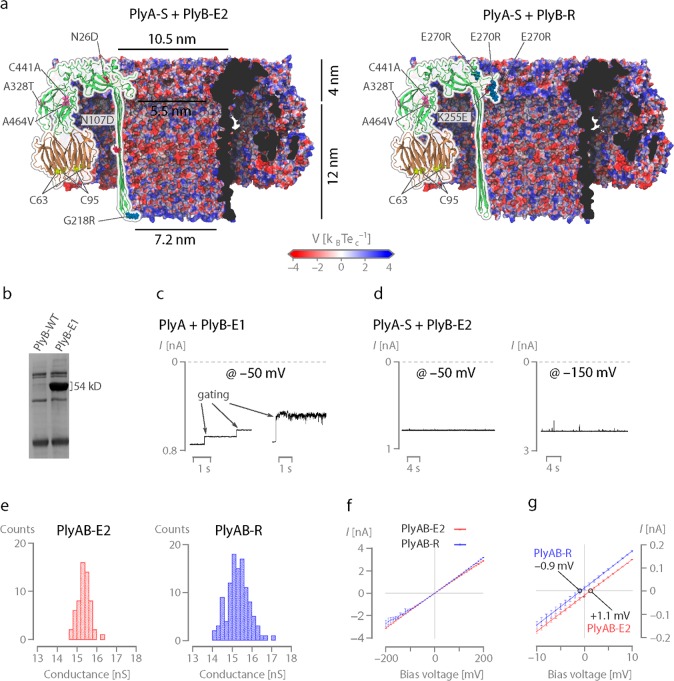
Engineering of PlyAB nanopores. (a) Cut through of the surfaces of PlyAB-E2 (left) and PlyA-R (right) nanopores with the mutations relative to the wild type shown as spheres on top of the overlaying cartoon representation. The surface is colored according to the electrostatic potential at 1 M salt, as computed by the adaptive Poisson–Boltzmann solver (APBS). (b) 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis of PlyB-WT and PlyB-E1 monomers. (c) Typical gating events for PlyAB-E1 nanopores under −50 mV applied bias. (d) 30 s open pore traces of PlyAB-E2 nanopores at −50 and −150 mV bias potentials. (e) Single channel distributions of PlyAB-E2 and PlyAB-R in 1 M NaCl at pH 7.5. (f) I–V curves of PlyAB-E2 and PlyAB-R collected in 1 M NaCl at pH 7.5. (g) Reversal potentials (Vr) measured for the PlyAB-E2 and PlyAB-R at pH 7.5, which correspond with the ion selectivities of 1.07 ± 0.02 and −0.94 ± 0.04, respectively (eq S1, Table S4). The ionic concentration was 500 mM NaCl in trans and 2 M NaCl in cis. Solutions were buffered with 15 mM Tris–HCl (pH 7.5). Error bars represent the standard deviations calculated from a minimum of three repeats.
Results and Discussion
Engineering the Electrical Properties of PlyAB Nanopores
The overexpression of PlyB in E. coli leads to inclusion bodies. Hence, we used directed evolution to improve soluble expression (Figure S1) and obtained PlyB-E1 (Figure 1b, Table S1). After oligomerization with PlyA using sphingomyelin–cholesterol (1:1 mass ratio) liposomes (Figure S2),67 PlyAB-E1 nanopores were reconstituted into an artificial lipid bilayer. We found that the PlyAB in proteoliposomes inserted efficiently into planar lipid bilayers in 1 M NaCl solutions but much less so in 300 mM NaCl. The majority of nanopores showed spontaneous opening and closing (gating, Figure 1c, Figure S3), which could not be suppressed by two additional rounds of directed evolution (Figure S4, Table S1). Simultaneously, we replaced the cysteine residues in both PlyA (PlyA-C62S-C94S, or PlyA-S) and PlyB (PlyB-E1-N107D-G218R-C441A, or PlyB-E2, Figure 1a). Interestingly, PlyA-S readily oligomerized with cysteine-free PlyB-E2, producing nanopores that routinely remained open at −150 mV for tens of seconds (Figure 1d). Oligomers formed with PlyA-S and PlyB-E1, the latter containing a cysteine residue at position 441, were also stable (Figure S4a), while oligomers formed using PlyA (with cysteine residues) and PlyB-E2 (cysteine-less) often gated (Figure S4b). Unexpectedly, therefore, the electrical stability in PlyAB is most likely inferred by removing the cysteine residues in PlyA, which are located at the interface with the lipid membrane and are known to be involved in the lipid binding.68,69
In planar lipid bilayers, oligomers formed by PlyA-S and PlyB-E2 (PlyAB-E2 nanopores) showed an average conductance of 15.4 ± 0.3 nS in 1 M NaCl buffered with 15 mM Tris–HCl (pH 7.5, −50 mV, Figure 1e, Table S2) and slightly asymmetric I–V curves (Figure 1f, Table S3), with higher currents recorded under a negatively applied bias. The reversal potential, which was measured using asymmetric salt concentration in cis and trans (trans/cis: 0.5 M NaCl/2 M NaCl), allowed calculating the ion selectivity of the nanopore (PlyAB-E2: PNa+/PCl– = 1.07 ± 0.02, Figure 1g; PlyAB-E1: PNa+/PCl– = 1.08 ± 0.02, Figure S5, Tables S4 and S5). The reversal potential (1.24 ± 0.2 and 1.10 ± 0.28 for PlyAB-E1 and PlyAB-E2, respectively, Table S4) suggests that pleurotolysin nanopores are slightly cation selective, as expected from the negatively charged lumen of these nanopores (Figure 1a). In nanopores, the degree of ion selectivity and the direction of the electro-osmotic flow are often correlated, as they both result from the interaction of the electrolyte ions with the fixed charges on the nanopore walls. Hence, inspired by previous work with FraC70 and ClyA,40,64 while exploring the stability of PlyAB-E1 nanopores, we also exchanged the negative charges at the constriction of PlyB-E1 with positively charged residues by site-directed mutagenesis (E260R, E261R, and E270R) with the aim of creating an altered electro-osmotic flow (EOF). Unfortunately, these new PlyB proteins were not soluble. Hence, three additional rounds of directed evolution were performed to obtain a mutant with a sufficiently high solubility to justify further electrophysiological characterization. A selected PlyB variant (PlyB-R, Figure 1a, Table S1) displayed the desired positively charged constriction (E260R, E261R, E270R), together with an additional compensating mutation (K255E), the original PlyB-E1 mutations (N26D, A328T, A464V), and the removal of a cysteine (C441A). Nanopores formed by PlyA-S and PlyB-R (PlyAB-R) were stable and displayed a slightly asymmetric conductance, with higher currents at positive applied bias (Table S3). The current asymmetry was more accentuated at lower ionic strengths (Figure S6). Rewardingly, the pore was weakly anion selective (PlyAB-R, PNa+/PCl– = 0.94 ± 0.04, Figure 1g). PlyAB-R and PlyAB-E2 showed a similar single channel distribution (Figure 1e) and power spectra (Figure S7).
Nanofluidic Properties of PlyAB Nanopores
The opposite ion selectivity of PlyAB-E2 and PlyAB-R suggests that the direction of their electro-osmotic flows might also be reversed. To investigate their nanofluidic behavior, we set up a computational model of both pores, in which we solved the extended Poisson–Nernst–Planck and Navier–Stokes (ePNP-NS) equations,71 a continuum simulation framework geared toward simulating biological nanopores (eqs S2–S15), with the finite element method (COMSOL Multiphysics v5.4). These simulations enabled us to obtain detailed distribution profiles of the ion concentrations inside the pore (Figure 2a–c), the electro-osmotic flow velocity (Figure 2d and e), and the electric field (Figure 2f and g). To increase the computational efficiency of our simulations, we reduced the geometry and charge distribution of the full atomic structures of both pores from a 3D- to a 2D-axisymmetric representation—a reasonable approximation given PlyAB’s high degree of rotational symmetry (13 identical subunits). We defined the 2D-axisymmetric geometry as the 25% density line of the radially averaged atomic density map (eq S16 and Figure S8a) and the charge density as the summation of all atomic partial charges, represented by 2D-Gaussian functions, on a single plane (eq S17 and Figure S8b,c). As validation, we found the experimental and simulated I–V curves to be in excellent agreement (Figure S9). Furthermore, the electrostatic potential profiles obtained from the ePNP-NS equations closely matched those computed directly from the full-atom model using the adaptive Poisson–Boltzmann solver (APBS, Figure S10), justifying our 2D-axisymmetric approximation a posteriori.
Figure 2.
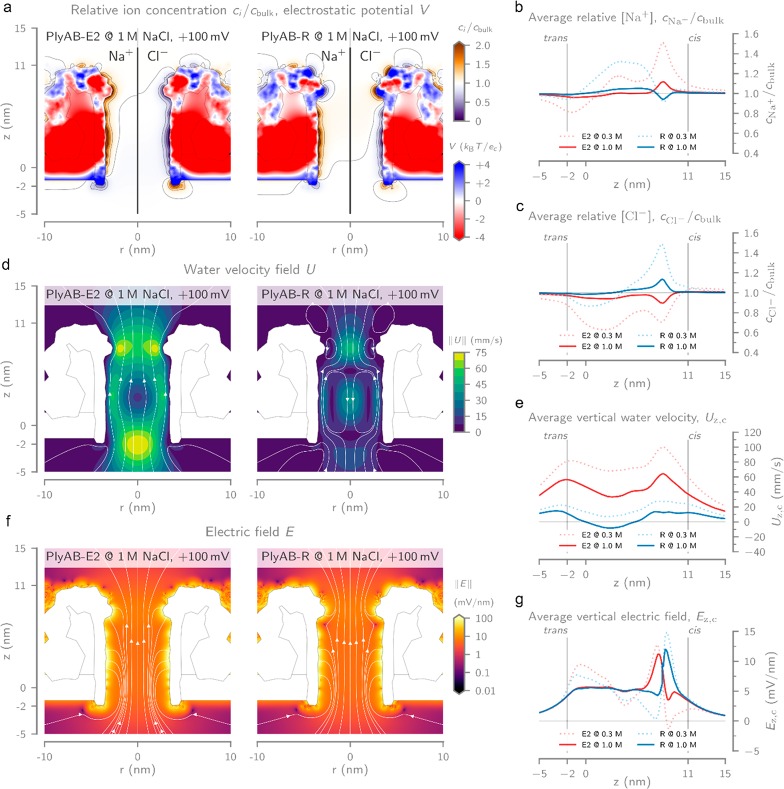
Computational modeling of PlyAB at +100 mV bias voltage and in 1 M NaCl. (a) Heatmaps of the relative Na+ and Cl– ion concentrations (ci/cbulk) inside PlyAB-E2 (left) and PlyAB-R (right). The coloring inside the pore represents the computed electric potential (V), expressed as units of thermal voltage. The geometry of the pore and bilayer is shown as a gray outline. Radially averaged ion concentrations for (b) Na+ and (c) Cl– inside PlyAB for both the E2 and R variants at reservoir ionic strengths of 0.3 and 1.0 M NaCl. Values were computed by averaging the concentration within 2.5 nm distance from the longitudinal axis or the pore. (d) Contour plots of the electro-osmotic flow velocity field magnitude (U) of PlyAB-E2 (left) and PlyAB-R (right). The white field lines indicate the direction of the flow and reveal the existence of vortices in the cis and trans chambers of PlyAB-R. (e) Radially averaged vertical water velocity (Uz,c, computed as in part b) at reservoir ionic strengths of 0.3 and 1.0 M NaCl. (f) Heatmap with streamlines showing the electric field vector magnitude (E) and its directionality inside the electrolyte for PlyAB-E2 (left) and PlyAB-R (right). The electric field inside the pore and bilayer is not shown for clarity. (g) Average vertical electrical field (Ez,c) inside both PlyAB variants at reservoir ionic strengths of 0.3 and 1.0 M NaCl. All values were obtained with 2D-axisymmetric models of PlyAB-E2 and PlyAB-R by solving the extended Poisson–Nernst–Planck Navier–Stokes (ePNP-NS) equations, implemented in the software package COMSOL Multiphysics, using the finite element method. Detailed information can be found in the Supporting Information.
Plotting the ion concentrations inside the pore at 1 M NaCl bulk ionic strength (Figure 2a) revealed an excess of cations (Na+) and a depletion of anions (Cl–) along the walls of the trans lumen for both PlyAB-E2 and PlyAB-R. For PlyAB-E2, this results in a positively charged electrical double layer (EDL) that continues inside the cis constriction. The additional arginine residues introduced at the constriction of PlyAB-R, however, promoted the accumulation of anions and induced a negatively charged EDL. These ion distributions correspond with the fixed charges of the pore and the resulting electrostatic potential (Figure 2a). Our simulation results show that, at positive applied bias voltages, PlyAB-E2 exhibits a strong unidirectional water flow (≈12.8 nm3·ns–1·V–1, Figure S11) from trans to cis (Figure 2d, left). Opposite electro-osmotic flow rates are observed at negative applied potentials (Figure S12). The electro-osmotic flow originates from the predominantly negatively charged interior lining of the pore and is consistent with the observations of other negatively charged pores such as ClyA.40,64 This can be explained by the excess of cations in the EDL of both the cis constriction and the trans lumen (Figure 2a–c), which results in a net force exerted on the fluid from trans to cis at positive potentials. In the case of PlyAB-R, the charge reversal of the constriction results in a more complex flow profile (Figure 2d, right). Notably, the net volumetric flow rate is reduced ≈7-fold (≈1.8 nm3·ns–1·V–1, Figure S11) compared to PlyAB-E2 and the unidirectional flow inside the lumen of the pore is replaced by a vortex. The positively charged cis constriction now attracts a negatively charged EDL, which in turn exerts on the liquid an opposing force compared to the force on the water in the lumen of the pore. These two opposing flows mix right below the constriction, and the conservation of the momentum of the fluid gives rise to the observed vortices. Such nanoscale vortices have also been observed recently in truncated pyramidal nanopores and can significantly impact the capture frequency of proteins.72
The reduction of the flow rate is also reflected by the average vertical water velocity (Uz,c, i.e., the average value within 2.5 nm distance from the center of the pore, Figure 2e). For PlyAB-E2, Uz,c is positive (Figure 2e) along the entire length of the pore, with peak values of ≈60 mm·s–1 at the cis and trans entries and a velocity of ≈40 mm·s–1 inside the trans chamber. The water velocities observed in PlyAB-R are significantly lower, with values of ≈10 mm·s–1 at the entries of the pore. Moreover, the vortex in the trans chamber results in a negative (i.e., Figure 2e) velocity of ≈ –5 mm·s–1. In contrast to the electro-osmotic flow, the electric fields (E) of PlyAB-E2 and PlyAB-R at 1 M NaCl and +100 mV bias are primarily determined by the geometry of the pore (Figure 2f). Our simulations reveal a relatively constant value inside the trans chamber of ≈5 mV·nm–1 and a larger value inside the constriction of ≈10 mV·nm–1 (Figure 2g). The location of the maximum electric field magnitude is modulated by the charge at the constriction and is shifted 0.7 nm upward (i.e., closer to cis) for PlyAB-R relative to PlyAB-E2 (Figure 2g).
At lower ionic strengths (e.g., 0.3 M NaCl bulk ionic strength), the ionic enhancements and depletions are significantly more prominent, as shown by the radially averaged (within a 2.5 nm radius from the center) concentration profiles along the longitudinal axis of the pore (Figure 2b and c). Higher accumulation and depletion, coupled with a reduced ionic screening at lower salt concentrations, in turn results in faster water velocities (Figure 2e) and stronger fluctuations of the electric field inside the nanopore for both PyA-E2 and PlyA-R nanopores (Figure 2g).
An overview of the distribution of ion concentrations, electrostatic potential, water velocity, and electric field for PlyAB-E2 and PlyAB-R at 0.15, 0.3, and 1.0 M NaCl and for +100, 0, and −100 mV bias voltages can be found in the Supporting Information (Figures S13–S16).
Protein Capture with PlyAB Nanopores
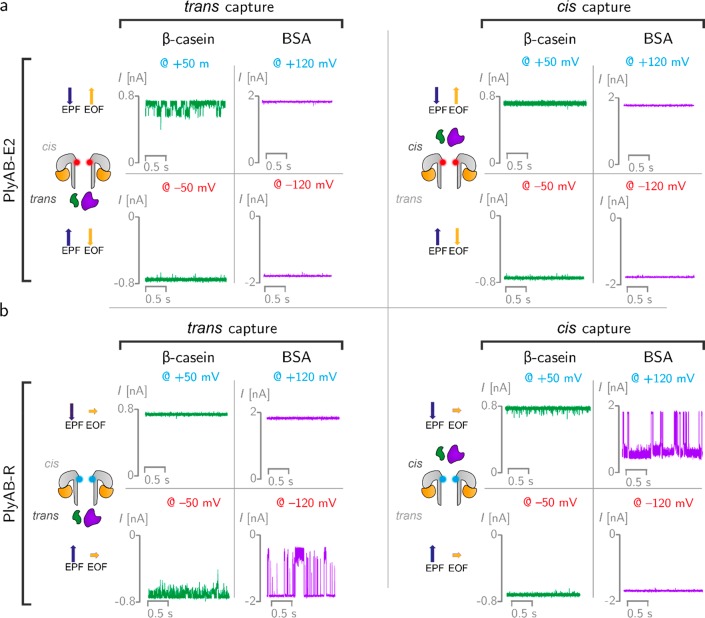
The ability of PlyAB nanopores to capture and analyze proteins was tested in 1 M NaCl and at pH 7.5 solutions using two proteins with different sizes: β-casein (24 kDa, pI = 5.1, net charge −5.8) and bovine serum albumin (BSA, 66.5 kDa, pI = 4.7, net charge −18.5). Protein blockades to PlyAB-E2 nanopores were only observed with β-casein and only when the protein was added to the trans side under positive applied potentials (at trans) (Figure 3a). Although we cannot exclude that under certain conditions proteins translocate across the nanopore too quickly to be observed, it is likely that the entry of proteins inside PlyAB is governed by a fine balance between electrophoretic and electro-osmotic forces. Under “capturing” conditions, the EOF promotes the entry of the protein, while the electrophoretic force (EPF) acts in the opposite direction. It is likely that for β-casein, which is smaller than the nanopore constriction, the competition between the electrophoretic and electro-osmotic forces allows the trapping of the protein within the lumen of the nanopore. By contrast, when using PlyAB-R, both β-casein and BSA blockades were observed, and proteins could be captured from either side of the nanopore according to the direction of the EPF. During trans capture, β-casein blockades in PlyAB-R (dwell time: 1.6 ± 0.1 ms, −50 mV) were shorter than those in PlyAB-E2 (25.0 ± 6.3 ms, +50 mV, Table S6) or PlyA-E1 (8.1 ± 1.3 ms, +50 mV, Table S6), suggesting that the competition between electrophoretic and electro-osmotic forces is important for obtaining long residence times inside the nanopore.
Figure 3.
Protein capture with PlyAB nanopores in 1 M NaCl at pH 7.5. (a) β-casein (24 kDa, pI 5.1) and bovine serum albumin (BSA, 66.5 kDa, pI 4.7) were measured with PlyAB-E2 nanopores. The PlyAB-E2 constriction is negatively charged and shown in red in the cartoon. β-Casein (green) and BSA (purple) were added to the trans and cis sides separately and tested by applying both positive and negative biases to the trans side. The direction of electrophoretic force (EPF) and electro-osmotic flow (EOF) are shown with blue and yellow arrows, respectively. (b) β-Casein and BSA were also measured with PlyAB-R nanopores (cyan constriction) from both sides. Recordings were collected with a 50 kHz sampling rate and a 10 kHz low-pass Bessel filter.
BSA, which could not be captured by PlyAB-E2 nanopores, entered PyAB-R nanopores, although only at relatively high applied potentials (e.g., > +100 mV, Figure 3b). Most likely, the large electrophoretic and entropic barriers of BSA (net charge −18.5 at pH 7.5, 66.5 kDa) compared to β-casein (net charge −5.8 at pH 7.5, 24 kDa) prevented BSA entry into PlyAB-E2 nanopores. Once the electro-osmotic flow is reduced, BSA can be electrophoretically captured by the nanopore, although the electro-osmotic vortices observed in PlyAB-R (Figure 2d) are also likely to play a role in trapping BSA inside the nanopore. Notably, we found that cis capture was more efficient than trans capture (Table S6), possibly reflecting the larger capture radius of the cis side. Increasing the applied potentials, BSA blockades became longer and multiple levels were observed within individual blockades (Figure S17), suggesting that the protein does not translocate the nanopore and that multiple residence sites might be occupied by BSA inside PlyAB-R. The latter was observed for the residence of thrombin inside ClyA nanopores.73 Finally, the duration of the BSA blockade was different depending on the direction of entry, suggesting that the interaction between the constriction and the protein during translocation plays a role.
Discrimination of Human Plasma Proteins with PlyAB-R Nanopores
PlyAB-R nanopores were used to identify two plasma proteins: human albumin serum (HSA, 66.5 kDa, pI = 4.7), which accounts for 55% of blood protein and behaves as an important transporter for many substrates like lipids, steroid hormones, and drugs, and human transferrin (HTr, 76–81 kDa, pI = 5.8), which is a glycoprotein that controls the level of iron in biological fluids. Because the electro-osmotic flow influences the capture and the residence of proteins inside the nanopore, we used 300 mM NaCl solutions, which are expected to increase the relative force of the electro-osmotic flow (Figure 2e) and improve the detection of the plasma proteins. Blockades were characterized by measuring the Ires%, which is defined as the ionic current associated with a protein-blocked pore IB divided by the open pore current IO percent. Homogeneous and well-defined single current blockades were observed with PyAB-R nanopores for both HSA and HTr (Figure 4a and b, Tables S7 and S8) from the cis side. Higher applied potentials were required to observe blockades when HTr and HSA were added to the trans side (Figure S18), reflecting the higher entropic barrier for trans entry compared to that for cis entry. Under +50 mV, HSA and HTr added to the cis side showed distinct Ires% (46.3 ± 0.9 and 33.5 ± 1.1%, respectively), which reflected the different volumes excluded by the two proteins. HSA showed longer dwell time blockades (118.5 ± 43.0 ms) than HTr (30.3 ± 5.4 ms), possibly reflecting the different electrostatic interaction between the two negatively charged proteins with the positively charged constriction of PyAB-R, although entropic contributions might also play an important role. Importantly, a HSA and HTr mixture could be identified on the bases of individual blockades (Figure 4c). Notably, protein blockades in solid-state nanopores with similar dimensions to PlyAB typically induce microsecond long blockades,58 which complicates protein identification.74−77 The millisecond long blockades recorded here for HSA and HTr (118.5 ± 43.0 and 30.3 ± 5.4 ms, respectively, at +50 mV) indicate that the precise engineering of the nanopore surface is important to allow protein analysis.
Figure 4.
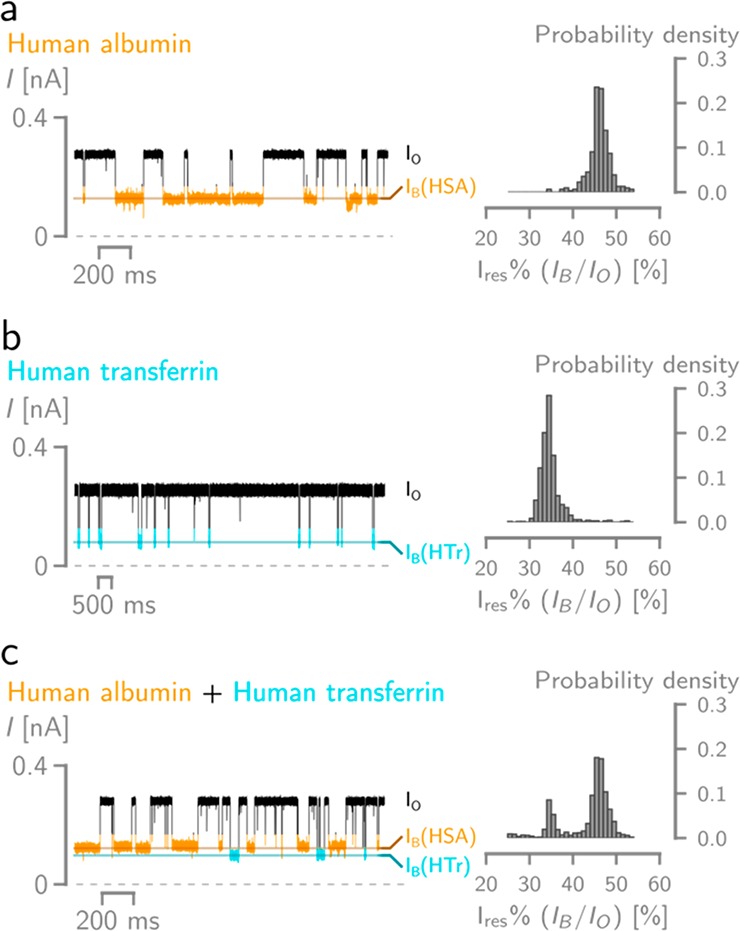
Electrical recordings of human albumin (HSA) and transferrin (HTr) using PlyAB-R nanopores. On the left are typical traces under +50 mV applied potential, on the right are the probability density histogram distribution of the Ires%. (a) HSA blockades, (b) HTr blockades, (c) mixture of HTr and HSA blockades (0.15 μM HSA and 0.375 μM HTr). Recordings were conducted in 300 mM NaCl at pH 7.5, with a 50 kHz sampling and a 10 kHz Bessel filter.
Conclusions
It has been shown that biological nanopores can be used as sensors for the identification of proteins or studying single enzymes.4,39,44,45 This approach is advantageous because it does not require the labeling of proteins and allows monitoring conformational changes for extended periods of time. One of the main challenges in nanopore analysis is to obtain biological nanopores with a wide range of sizes and shapes that allow accommodating different proteins. In this work, we engineered the pore forming toxin PlyAB for single-molecule analysis. This β-barrel nanopore comprises two communicating reaction chambers: a cylindrical trans chamber (7.2 nm × 12 nm, diameter × height) and a truncated cone cis chamber (10.5 nm × 5.5 nm × 4 nm, large diameter × small diameter × height), describing the largest biological nanopore available for protein analysis.
Surprisingly, large proteins such as BSA, HSA, and HTr could not overcome the entropic and electrostatic energy barrier to enter the nanopore. This is because for negatively charged proteins the electro-osmotic flow across the nanopore opposes the electrophoretic force. We found, however, that the precise engineering of the nanopore inner surface charge allowed overcoming this limitation. Our simulations showed that the introduction of positive charges into the inner constriction of PlyAB created an electro-osmotic vortex that greatly reduced the electro-osmotic flow across the nanopore. This enabled the electrophoretic capture and the unambiguous identification of human serum proteins by analysis of their blockade currents.
Acknowledgments
We thank the University of Groningen and ERC consolidator grant (726151, DeE-Nano) for funding (G.H. and G.M.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.0c00877.
Extensive description of the materials and methods including the preparation of nanopores, a description of the model used in the simulation, and the modified PNP-NS simulations and supporting figures describing the characterization of the PyAB nanopores and simulations (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Restrepo-Pérez L.; Joo C.; Dekker C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 2018, 13 (9), 786–796. 10.1038/s41565-018-0236-6. [DOI] [PubMed] [Google Scholar]
- Varongchayakul N.; Song J.; Meller A.; Grinstaff M. W. Single-molecule protein sensing in a nanopore: a tutorial. Chem. Soc. Rev. 2018, 47, 8512–8524. 10.1039/C8CS00106E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. W. F.; Reiner J. E. The utility of nanopore technology for protein and peptide sensing. Proteomics 2018, 18, 1800026. 10.1002/pmic.201800026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems K.; Van Meervelt V.; Wloka C.; Maglia G. Single-molecule nanopore enzymology. Philos. Trans. R. Soc., B 2017, 372 (1726), 20160230. 10.1098/rstb.2016.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastoriza-Gallego M.; Betton J.-M.; Pelta J.; Bacri L.; Oukhaled A. Sensing Proteins through Nanopores: Fundamental to Applications. ACS Chem. Biol. 2012, 7 (12), 1935–1949. 10.1021/cb300449t. [DOI] [PubMed] [Google Scholar]
- Baaken G.; Halimeh I.; Bacri L.; Pelta J.; Oukhaled A.; Behrends J. C. High-Resolution Size-Discrimination of Single Nonionic Synthetic Polymers with a Highly Charged Biological Nanopore. ACS Nano 2015, 9 (6), 6443–6449. 10.1021/acsnano.5b02096. [DOI] [PubMed] [Google Scholar]
- Robertson J. W.; Rodrigues C. G.; Stanford V. M.; Rubinson K. A.; Krasilnikov O. V.; Kasianowicz J. J. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (20), 8207–8211. 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov O. V.; Rodrigues C. G.; Bezrukov S. M. Single polymer molecules in a protein nanopore in the limit of a strong polymer-pore attraction. Phys. Rev. Lett. 2006, 97 (1), 018301. 10.1103/PhysRevLett.97.018301. [DOI] [PubMed] [Google Scholar]
- Kasianowicz J. J.; Brutyan R. A.; Vodyanoy I.; Bezrukov S. M. Noise-Induced by Differently Sized Polyethylene-Glycols Shows Interaction of Polymer with the Alpha-Toxin Ion-Channel. Biophys. J. 1994, 66 (2), A214–A214. [Google Scholar]
- Maglia G.; Restrepo M. R.; Mikhailova E.; Bayley H. Enhanced translocation of single DNA molecules through α-hemolysin nanopores by manipulation of internal charge. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (50), 19720–19725. 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrington I. M.; Butler T. Z.; Collins M. D.; et al. Nanopore DNA sequencing with MspA. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (37), 16060–16065. 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasianowicz J. J.; Brandin E.; Branton D.; Deamer D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U. S. A. 1996, 93 (24), 13770–13773. 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner S.; Chen R. J. A.; Wilson N. A.; et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat. Nanotechnol. 2007, 2 (11), 718–724. 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J.; Wu H. C.; Jayasinghe L.; Patel A.; Reid S.; Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009, 4 (4), 265–270. 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Patil K. M.; Yan S.; et al. Nanopore Sequencing Accurately Identifies the Mutagenic DNA Lesion O6-Carboxymethyl Guanine and Reveals Its Behavior in Replication. Angew. Chem., Int. Ed. 2019, 58 (25), 8432–8436. 10.1002/anie.201902521. [DOI] [PubMed] [Google Scholar]
- Ying Y. L.; Li Z. Y.; Hu Z. L.; et al. A Time-Resolved Single-Molecular Train Based on Aerolysin Nanopore. Chem. 2018, 4 (8), 1893–1901. 10.1016/j.chempr.2018.05.004. [DOI] [Google Scholar]
- Pastoriza-Gallego M.; Rabah L.; Gibrat G.; et al. Dynamics of unfolded protein transport through an aerolysin pore. J. Am. Chem. Soc. 2011, 133 (9), 2923–2931. 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- Pastoriza-Gallego M.; Breton M. F.; Discala F.; Auvray L.; Betton J. M.; Pelta J. Evidence of unfolded protein translocation through a protein nanopore. ACS Nano 2014, 8 (11), 11350–11360. 10.1021/nn5042398. [DOI] [PubMed] [Google Scholar]
- Huang G.; Voet A.; Maglia G. FraC nanopores with adjustable diameter identify the mass of opposite-charge peptides with 44 Da resolution. Nat. Commun. 2019, 10 (1), 835. 10.1038/s41467-019-08761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Pérez L.; Huang G.; Bohländer P. R.; et al. Resolving Chemical Modifications to a Single Amino Acid within a Peptide Using a Biological Nanopore. ACS Nano 2019, 13 (12), 13668–13676. 10.1021/acsnano.9b05156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland T. C.; Long Y.-T.; Stefureac R.-I.; Bediako-Amoa I.; Kraatz H.-B.; Lee J. S. Structure of Peptides Investigated by Nanopore Analysis. Nano Lett. 2004, 4 (7), 1273–1277. 10.1021/nl049413e. [DOI] [Google Scholar]
- Stefureac R.; Long Y. T.; Kraatz H. B.; Howard P.; Lee J. S. Transport of alpha-helical peptides through alpha-hemolysin and aerolysin pores. Biochemistry 2006, 45 (30), 9172–9179. 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- Restrepo-Pérez L.; Wong C. H.; Maglia G.; Dekker C.; Joo C. Label-Free Detection of Post-translational Modifications with a Nanopore. Nano Lett. 2019, 19, 7957–7964. 10.1021/acs.nanolett.9b03134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z.; Guo P. Channel from bacterial virus T7 DNA packaging motor for the differentiation of peptides composed of a mixture of acidic and basic amino acids. Biomaterials 2019, 214 (5), 119222. 10.1016/j.biomaterials.2019.119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet F.; Ouldali H.; Pastoriza-Gallego M.; Manivet P.; Pelta J.; Oukhaled A. Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat. Commun. 2018, 9 (1), 966. 10.1038/s41467-018-03418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; Willems K.; Soskine M.; Wloka C.; Maglia G. Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores. Nat. Commun. 2017, 8 (1), 935. 10.1038/s41467-017-01006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavis A. E.; Brady K. T.; Hatmaker G. A.; et al. Single Molecule Nanopore Spectrometry for Peptide Detection. ACS Sensors 2017, 2 (9), 1319–1328. 10.1021/acssensors.7b00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asandei A.; Schiopu I.; Chinappi M.; Seo C. H.; Park Y.; Luchian T. Electroosmotic Trap Against the Electrophoretic Force Near a Protein Nanopore Reveals Peptide Dynamics During Capture and Translocation. ACS Appl. Mater. Interfaces 2016, 8 (20), 13166–13179. 10.1021/acsami.6b03697. [DOI] [PubMed] [Google Scholar]
- Li S.; Cao C.; Yang J.; Long Y. Detection of peptides with different charge and length by aerolysin nanopore. ChemElectroChem 2019, 6, 126–129. 10.1002/celc.201800288. [DOI] [Google Scholar]
- Movileanu L.; Schmittschmitt J. P.; Scholtz J. M.; Bayley H. Interactions of peptides with a protein pore. Biophys. J. 2005, 89 (2), 1030–1045. 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma A. J.; Bayley H. Continuous Stochastic Detection of Amino Acid Enantiomers with a Protein Nanopore. Angew. Chem., Int. Ed. 2012, 51 (38), 9606–9609. 10.1002/anie.201205687. [DOI] [PubMed] [Google Scholar]
- Movileanu L.; Howorka S.; Braha O.; Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 2000, 18 (10), 1091–1095. 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- Fahie M. A.; Yang B.; Mullis M.; Holden M. A.; Chen M. Selective Detection of Protein Homologues in Serum Using an OmpG Nanopore. Anal. Chem. 2015, 87 (21), 11143–11149. 10.1021/acs.analchem.5b03350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A. K.; Movileanu L. Real-time measurement of protein–protein interactions at single-molecule resolution using a biological nanopore. Nat. Biotechnol. 2019, 37, 96. 10.1038/nbt.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.-W.; Van Meervelt V.; Tsai K.-C.; De Temmerman P.-J.; Mast J.; Maglia G. Engineering a nanopore with co-chaperonin function. Sci. Adv. 2015, 1 (11), e1500905. 10.1126/sciadv.1500905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M.; Biesemans A.; Moeyaert B.; Cheley S.; Bayley H.; Maglia G. An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Lett. 2012, 12 (9), 4895–4900. 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M.; Biesemans A.; De Maeyer M.; Maglia G. Tuning the size and properties of ClyA nanopores assisted by directed evolution. J. Am. Chem. Soc. 2013, 135 (36), 13456–13463. 10.1021/ja4053398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C.; Van Meervelt V.; van Gelder D.; et al. Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore. ACS Nano 2017, 11 (5), 4387–4394. 10.1021/acsnano.6b07760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galenkamp N. S.; Soskine M.; Hermans J.; Wloka C.; Maglia G. Direct electrical quantification of glucose and asparagine from bodily fluids using nanopores. Nat. Commun. 2018, 9, 4085. 10.1038/s41467-018-06534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems K.; Ruić D.; Biesemans A.; Galenkamp N. S.; Van Dorpe P.; Maglia G. Engineering and Modeling the Electrophoretic Trapping of a Single Protein Inside a Nanopore. ACS Nano 2019, 13 (9), 9980–9992. 10.1021/acsnano.8b09137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meervelt V.; Soskine M.; Maglia G. Detection of Two Isomeric Binding Con fi gurations in a Protein À Aptamer Complex with a Biological Nanopore. ACS Nano 2014, 8 (12), 12826–12835. 10.1021/nn506077e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems K.; Van Meervelt V.; Wloka C.; Maglia G. Single-molecule nanopore enzymology. Philos. Trans. R. Soc., B 2017, 372 (1726), 20160230. 10.1098/rstb.2016.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernia S.; van der Heide N. J.; Galenkamp N. S.; Gouridis G.; Maglia G. Current Blockades of Proteins inside Nanopores for Real-Time Metabolome Analysis. ACS Nano 2020, 14 (2), 2296–2307. 10.1021/acsnano.9b09434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M.; Biesemans A.; Maglia G. Single-Molecule Analyte Recognition with ClyA Nanopores Equipped with Internal Protein Adaptors. J. Am. Chem. Soc. 2015, 137 (17), 5793–5797. 10.1021/jacs.5b01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meervelt V.; Soskine M.; Singh S.; et al. Real-Time Conformational Changes and Controlled Orientation of Native Proteins Inside a Protein Nanoreactor. J. Am. Chem. Soc. 2017, 139 (51), 18640–18646. 10.1021/jacs.7b10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.; Gatterdam V.; Wieneke R.; Tampé R.; Rant U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 2012, 7 (4), 257–263. 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]
- Houghtaling J.; Ying C.; Eggenberger O. M.; et al. Estimation of Shape, Volume, and Dipole Moment of Individual Proteins Freely Transiting a Synthetic Nanopore. ACS Nano 2019, 13 (5), 5231–5242. 10.1021/acsnano.8b09555. [DOI] [PubMed] [Google Scholar]
- Yusko E. C.; Bruhn B. R.; Eggenberger O. M.; et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2017, 12 (4), 360–367. 10.1038/nnano.2016.267. [DOI] [PubMed] [Google Scholar]
- Hu R.; Rodrigues J. V.; Waduge P.; et al. Differential Enzyme Flexibility Probed Using Solid-State Nanopores. ACS Nano 2018, 12 (5), 4494–4502. 10.1021/acsnano.8b00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waduge P.; Hu R.; Bandarkar P.; et al. Nanopore-Based Measurements of Protein Size, Fluctuations, and Conformational Changes. ACS Nano 2017, 11 (6), 5706–5716. 10.1021/acsnano.7b01212. [DOI] [PubMed] [Google Scholar]
- Nir I.; Huttner D.; Meller A. Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores. Biophys. J. 2015, 108 (9), 2340–2349. 10.1016/j.bpj.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko E. C.; Johnson J. M.; Majd S.; et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 2011, 6 (4), 253–260. 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oukhaled A.; Cressiot B.; Bacri L.; et al. Dynamics of Completely Unfolded and Native Proteins through Solid-State Nanopores as a Function of Electric Driving Force. ACS Nano 2011, 5 (5), 3628–3638. 10.1021/nn1034795. [DOI] [PubMed] [Google Scholar]
- Sexton L. T.; Horne L. P.; Sherrill S. A.; Bishop G. W.; Baker L. A.; Martin C. R. Resistive-pulse studies of proteins and protein/antibody complexes using a conical nanotube sensor. J. Am. Chem. Soc. 2007, 129 (43), 13144–13152. 10.1021/ja0739943. [DOI] [PubMed] [Google Scholar]
- Niedzwiecki D. J.; Grazul J.; Movileanu L. Single-molecule observation of protein adsorption onto an inorganic surface. J. Am. Chem. Soc. 2010, 132 (31), 10816–10822. 10.1021/ja1026858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. R.; Sexton L. T.; Mukaibo H.; et al. An Adsorption-Based Model for Pulse Duration in Resistive-Pulse Protein Sensing. J. Am. Chem. Soc. 2010, 132 (24), 6755–6763. 10.1021/ja100693x. [DOI] [PubMed] [Google Scholar]
- Talaga D. S.; Li J. Single-molecule protein unfolding in solid state nanopores. J. Am. Chem. Soc. 2009, 131 (26), 9287–9297. 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesa C.; Kowalczyk S. W.; Zinsmeester R.; Grosberg A. Y.; Rabin Y.; Dekker C. Fast translocation of proteins through solid state nanopores. Nano Lett. 2013, 13 (2), 658–663. 10.1021/nl3042678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.; Lu B.; Zhao Q.; Wang J.; Luo K.; Yu D. Surface modification of solid-state nanopores for sticky-free translocation of single-stranded DNA. Small 2014, 10 (21), 4332–4339. 10.1002/smll.201401091. [DOI] [PubMed] [Google Scholar]
- Kapinos L.; Lim R. Y. H.; Kowalczyk S. W.; et al. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat. Nanotechnol. 2011, 6 (7), 433–438. 10.1038/nnano.2011.88. [DOI] [PubMed] [Google Scholar]
- Wanunu M.; Meller A. Chemically modified solid-state nanopores. Nano Lett. 2007, 7 (6), 1580–1585. 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
- Maglia G.; Rincon Restrepo M.; Mikhailova E.; Bayley H. Enhanced translocation of single DNA molecules through α-hemolysin nanopores by manipulation of internal charge. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 19720–19725. 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler T. Z.; Pavlenok M.; Derrington I. M.; Niederweis M.; Gundlach J. H. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (52), 20647–20652. 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini L.; Brouns T.; Willems K.; Carlon E.; Maglia G. DNA Translocation through Nanopores at Physiological Ionic Strengths Requires Precise Nanoscale Engineering. ACS Nano 2016, 10 (9), 8394–8402. 10.1021/acsnano.6b03159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul C. F.; Whisstock J. C.; Dunstone M. A. Giant MACPF/CDC pore forming toxins: A class of their own. Biochim. Biophys. Acta, Biomembr. 2016, 1858 (3), 475–486. 10.1016/j.bbamem.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Lukoyanova N.; Kondos S. C.; Farabella I.; et al. Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin. PLoS Biol. 2015, 13 (2), e1002049. 10.1371/journal.pbio.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T.; Noguchi K.; Mimuro H.; et al. Pleurotolysin, a novel sphingomyelin-specific two-component cytolysin from the edible mushroom Pleurotus ostreatus, assembles into a transmembrane pore complex. J. Biol. Chem. 2004, 279 (26), 26975–26982. 10.1074/jbc.M402676200. [DOI] [PubMed] [Google Scholar]
- Berne S.; Križaj I.; Pohleven F.; Turk T.; Maček P.; Sepčić K. Pleurotus and Agrocybe hemolysins, new proteins hypothetically involved in fungal fruiting. Biochim. Biophys. Acta, Gen. Subj. 2002, 1570 (3), 153–159. 10.1016/S0304-4165(02)00190-3. [DOI] [PubMed] [Google Scholar]
- Berne S.; Sepčić K.; Anderluh G.; Turk T.; Maček P.; Ulrih N. P. Effect of pH on the pore forming activity and conformational stability of ostreolysin, a lipid raft-binding protein from the edible mushroom Pleurotus ostreatus. Biochemistry 2005, 44 (33), 11137–11147. 10.1021/bi051013y. [DOI] [PubMed] [Google Scholar]
- Wloka C.; Mutter N. L.; Soskine M.; Maglia G. Alpha-Helical Fragaceatoxin C Nanopore Engineered for Double-Stranded and Single-Stranded Nucleic Acid Analysis. Angew. Chem., Int. Ed. 2016, 55 (40), 12494–12498. 10.1002/anie.201606742. [DOI] [PubMed] [Google Scholar]
- Willems K.; Ruić D.; Lucas F.; Barman U.; Hofkens J.; Maglia G.; Van Dorpe P.. Modeling of Ion and Water Transport in the Biological Nanopore ClyA. 2020, bioRxiv 2020.01.08.897819. bioRxiv.org e-Print archive. https://www.biorxiv.org/content/10.1101/2020.01.08.897819v1.
- Zeng S.; Wen C.; Solomon P.; Zhang S. L.; Zhang Z. Rectification of protein translocation in truncated pyramidal nanopores. Nat. Nanotechnol. 2019, 14 (11), 1056–1062. 10.1038/s41565-019-0549-0. [DOI] [PubMed] [Google Scholar]
- Soskine M.; Biesemans A.; De Maeyer M.; Maglia G. Tuning the size and properties of ClyA nanopores assisted by directed evolution. J. Am. Chem. Soc. 2013, 135 (36), 13456–13463. 10.1021/ja4053398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A.; Schürmann G.; Mondin G.; et al. Sensing protein molecules using nanofabricated pores. Appl. Phys. Lett. 2006, 88 (9), 093901. 10.1063/1.2180868. [DOI] [Google Scholar]
- Fologea D.; Ledden B.; McNabb D. S.; Li J. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 2007, 91 (5), 053901. 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Bell N. A. W.; Hernández-Ainsa S.; et al. Single protein molecule detection by glass nanopores. ACS Nano 2013, 7 (5), 4129–4134. 10.1021/nn4004567. [DOI] [PubMed] [Google Scholar]
- Steinbock L. J.; Krishnan S.; Bulushev R. D.; et al. Probing the size of proteins with glass nanopores. Nanoscale 2014, 6 (23), 14380–14387. 10.1039/C4NR05001K. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.