Abstract
Vitamin A is an essential nutrient necessary for numerous basic physiological functions, including reproduction and development, immune cell differentiation and communication, as well as the perception of light. To evade the dire consequences of vitamin A deficiency, vertebrates have evolved specialized metabolic pathways that enable the absorption, transport, and storage of vitamin A acquired from dietary sources as preformed retinoids or provitamin A carotenoids. This evolutionary advantage requires a complex interplay between numerous specialized retinoid-transport proteins, receptors, and enzymes. Recent advances in molecular and structural biology resulted in a rapid expansion of our understanding of these processes at the molecular level. This progress opened new avenues for the therapeutic manipulation of retinoid homeostasis. In this review, we summarize current research related to the biochemistry of carotenoid and retinoid-processing proteins with special emphasis on the structural aspects of their physiological actions.
Keywords: carotenoids, vitamin A, retinol, retinoid metabolism
1. INTRODUCTION
Human health depends on numerous micronutrients, which serve as, or are precursors to essential cofactors, chromophores, and signaling molecules. Among them, fat-soluble vitamin A (alL-trans-retinol, ROL) plays a pivotal role throughout the vertebrate life cycle. It is a precursor for at least two bioactive molecules, all-trans-retinoic acid (RA) and 11-cis-retinaldehyde [1, 2]. RA regulates a large number of physiological processes by acting as a highly potent agonist of nuclear retinoic acid receptors (RARs) [3]. RARs, in conjunction with their heterodimeric partners, retinoid X receptors, regulate the activity of diverse target genes, controlling the differentiation and development of fetal and adult tissues, reproductive functions, immune responses, and the regulation of energy balance [4-8]. 11-cis-retinaldehyde serves as a visual chromophore by covalently binding to rod and cone opsin protein scaffolds [9, 10]. Its geometric isomerization upon excitation by a photon triggers the activation of G protein-coupled opsins, and thus constitutes the chemical basis for the perception of light [11, 12].
To avoid the dire consequences of vitamin A deficiency, vertebrates have evolved specialized metabolic pathways that enable the efficient absorption and storage of preformed dietary retinoids (Fig. 1). They have also retained the metabolic ability to convert selected C40 isoprenoid precursor molecules (carotenoids) into ROL [13-15]. From over 1100 naturally occurring carotenoids around 50 can be found in the human diet [16]. However, only α-carotene, β-carotene, and β-cryptoxanthin that contain at least one unsubstituted β-ionone ring can be metabolized to ROL, whereas many abundant xanthophylls such as lutein and zeaxanthin, or acyclic lycopene are classified as nonprovitamin A carotenoids [17, 18].
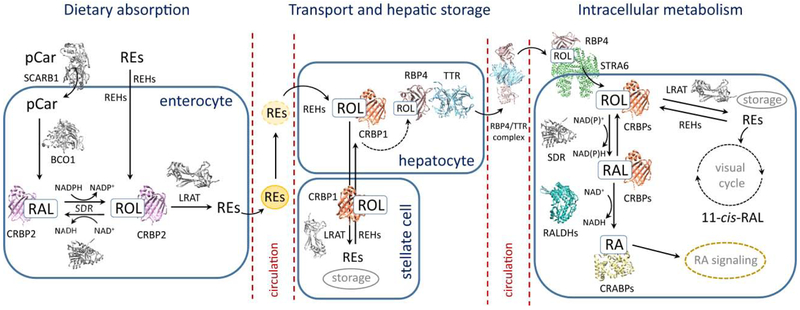
Figure 1– Schematic representation of vitamin A uptake, transport, and metabolism.
Dietary retinyl esters (REs) and provitamin A carotenoids (pCars) are absorbed by enterocytes and converted into all-trans-retinol (ROL). The subsequent esterification of ROL enables the effective packaging and transport of REs by chylomicrons (represented by yellow spheres). Circulating chylomicrons deliver REs to the peripheral tissues, including the liver, where upon hydrolysis of REs resulting ROL is transported to hepatic stellate cells, re-esterified and stored. The mobilization of the hepatic pool of retinoids requires the hydrolysis of REs, transport of ROL back to the hepatocytes, and binding to RBP4. The complex of RBP4 and TTR is subsequently secreted to the blood stream. The effective uptake of ROL bound to RBP4 is achieved by the interaction of holo-RBP4 with its cell surface receptor, STRA6. Inside the peripheral tissues, ROL can be enzymatically oxidized to all-trans-retinaldehyde (RAL) and all-trans-retinoic acid (RA). In specialized RPE and photoreceptor cells, ROL can be utilized for the production of visual chromophore (11-cis-RAL) via the visual cycle metabolic pathway. Each step of the vitamin A metabolic pathways is accompanied by a set of specialized receptors, enzymes, and binding proteins. Those with known 3D structures are represented by colored cartoon structures. Proteins for which detailed molecular architectures remain unknown are depicted as grey colored models. Abbreviations used are the following: BCDO1, β,β-carotene-15,15-dioxygenase; CRABPs, cellular retinoic acid-binding proteins; CRBP1, cellular retinol-binding protein 1; CRBP2, cellular retinol-binding protein 2; CRBPs, cellular retinol-binding proteins; LRAT, lecithin:retinol acyltransferase; RALDHs: retinaldehyde dehydrogenases; REHs, retinyl ester hydrolases; RBP4, serum retinol-binding protein; SCARB1, scavenger receptor class B type 1; SDRs, short-chain dehydrogenases/reductases; STRA6, stimulated by retinoic acid 6; TTR, transthyretin.
The complexity of the processes involved in the absorption and metabolism of carotenoids and retinoids has been acknowledged by the pioneers of this field. As summarized by Thomas Moor in 1957 “There are many complicating factors, both chemical and physiological, which will make it difficult to give an account of the absorption of vitamin A, and its provitamins, which is both clear and reasonably comprehensive” [19]. This notion was reiterated by John Erdman and his colleagues in the early nineties, indicating the major experimental challenges associated with the study of fat-soluble vitamins [20]. However, the subsequent era of molecular cloning, the phenotypic characterization of genetically modified animal models, improvements in analytical methods, and the contribution of structural biology fueled rapid progress in understanding the diverse physiology of vitamin A at the cellular and molecular levels. Consequently, many intracellular and extracellular proteins have been identified that specifically interact with retinoids or carotenoids facilitating their uptake, protecting from chemical modification or catabolism, converting into desired metabolites, and transporting to the site of biological activity (Fig. 1). Unveiling the secrets harbored by this biological machinery is essential to identify the precipitating causes of pathological conditions related to impairments in retinoid homeostasis, as well as uncovering new avenues for therapeutic intervention. Thus, in this review, we aim to summarize current research related to the biochemistry of carotenoids and retinoids in vertebrates. We focus predominantly on the tremendous progress that has been made in delineating the molecular basis for absorption, transport, and metabolism of these compounds through advances in structural biology.
2. INTESTINAL ABSORPTION OF CAROTENOIDS
2.1. Significance of a receptor-mediated uptake
It is assumed that upon digestion, ingested carotenoids share the fate of other lipophilic nutrients. They are liberated from the food matrix, emulsified, and incorporated into the mixed micelles composed of phospholipids, cholesterol, free fatty acids, acylglycerols, and bile salts [21]. A fraction of carotenoids also partitions into phospholipid vesicles, liposome-like structures composed of either single or multiple layers of phospholipids. The processes of extraction and solubilization are critical for the efficient absorption of dietary carotenoids and often determine the overall bioavailability of these nutrients [22].
It was long believed that, similarly to preformed retinoids, the intestinal absorption of carotenoids occurs via passive diffusion and is not facilitated by a receptor present at the brush border of the enterocytes. However, this supposition failed to explain a profound interindividual variability in the efficiency of dietary carotenoid absorption in both animals and humans [23]. Also, the passive diffusion model does not account for the isomer selectivity of the absorption and the competition for cellular uptake between carotenoids and other lipids, such as phytosterols, fatty acids, or vitamin E [24-26]. It was not until the discovery of the role of the Drosophila melanogaster ninaD gene that the dogma shifted in favor of a protein-facilitated transport model of carotenoid absorption [27]. In vertebrates, the orthologue of this insect gene encodes a class B scavenger receptor type 1 (SCARB1). This multi-ligand membrane receptor was first characterized in the context of its role in the bidirectional transport of cholesterol or cholesterol esters between circulating high-density lipoproteins (HDL) and target cells [28]. SCARB1 is expressed in many mammalian tissues and cell types, including the small intestine. The immunoblot analysis of mouse intestinal cell extracts demonstrated a gradation of SCARB1 expression along the gastrocolic axis of the intestine [29]. The highest level of expression was observed in the proximal intestine, whereas minimal expression levels were detected in the distal intestine. Thus, the spatial distribution of SCARB1 corresponds well with the areas of the highest dietary absorption of carotenoids. Indeed, in addition to the impaired cholesterol homeostasis exemplified by reduced cholesterol in secreted bile and accelerated atherosclerosis [30, 31], studies on SCARB1-deficient mice (Scarb1−/−) confirmed that this receptor is required for the efficient uptake of β-carotene and zeaxantin [32, 33]. SCARB1 is also involved in facilitating systemic tissue-specific distribution of circulating carotenoids. A knockdown of SCARB1 in retinal pigmented epithelium (RPE) cells leads to a dramatic decrease in the cellular uptake of β-carotene and zeaxantin [34, 35]. Conversely, the overexpression of this receptor causes an increase in carotenoid uptake [33]. Consistent with these observations, the physiological significance of SCARB1 was confirmed in numerous other species, including silkworms, salmon, canaries, and most significantly in humans, where a mutation in this receptor causes not only an increase in high-density lipoproteins (HDL) levels, but also a decrease in the serum and ocular concentration of specific carotenoids [36-42]. Importantly, the function of SCARB1 is linked to the vitamin A status of an organism via intestinal-specific homeobox transcriptional factor (ISX) [43, 44]. The expression of ISX is under the control of the retinoic acid-response element identified in the Isx promoter region [44, 45]. The induction of ISX directly suppresses the transcription of Scarb1, and thus limits the intestinal uptake of carotenoids. This elegant negative feedback mechanism explains the molecular basis for the absence of β-carotene toxicity [33, 46].
2.2. Structures of class B scavenger receptors – a glance at the SCARB1 activity
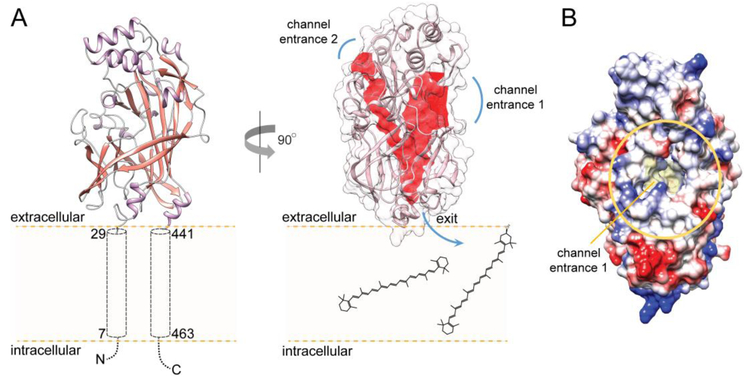
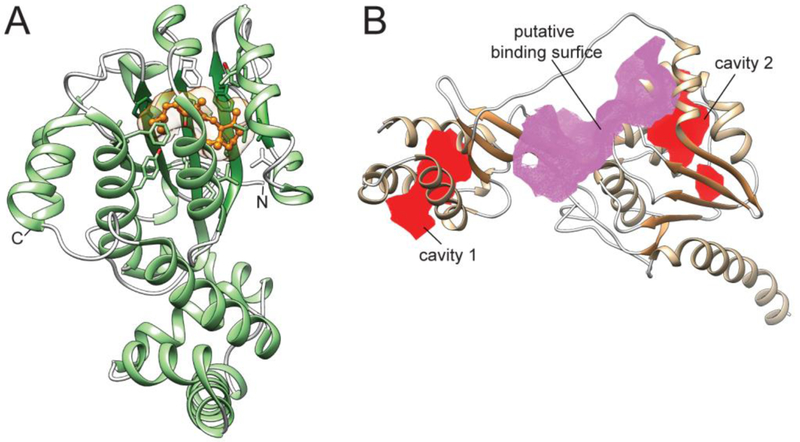
Our knowledge related to the structure/function relationship of SCARB1 remains incomplete. Despite substantial progress in studies on the closely related members of the class B scavenger receptor family, such as cluster determinant 36 receptor (CD36) and lysosomal integral membrane protein-2 (LIMP-2), a structure of SCARB1 has not yet been determined. Nevertheless, crystal structures of CD36 and LIMP-2 and a wealth of biochemical data provide valuable insight into the putative mechanism of carotenoid uptake by SCARB1 [47, 48]. Among several isoforms of SCARB1, the most abundant corresponds to the 509 amino acid variant of the receptor [49, 50]. It is also the most efficient in the selective uptake of cholesterol, lutein, and meso-zeaxanthin in tissue culture [35]. Analysis of the primary sequence of human SCARB1 reveals a large extracellular domain flanked by two single transmembrane α-helices (amino acids 7-29 and 441-463, respectively) (Fig. 2A). Thus, both N- and C-termini are located at the cytoplasmic side of the cell membrane. The extracellular domains of human LIMP-2 and CD36 shares considerable sequence homology with SCARB1 (33.5% and 34.4% identity, respectively), allowing for building a three-dimensional (3D) model of this receptor with relatively high confidence. By analogy to LIMP-2, the polypeptide chain of SCARB1 fold into an antiparallel β-barrel core with several short α-helices that compose extended loops between β-strands (Fig. 2A). Three of these α-helices form a bundle at the top of the receptor. Two others link the soluble portion of the receptor with the transmembrane α-helices. Thus, the ectodomain of SCARB1 is most likely docked at the exofacial leaflet of the cell membrane. Significant differences in the length of the β-strands contribute to the irregularity of the barrel, in which one of the sides is noticeably elongated. This structural asymmetry forms a wide entrance to a hydrophobic lipid transport tunnel that traverses the entire length of the receptor and exits at the lipid membrane (Fig. 2A, channel entrance 1). It is believed that this structural component facilitates transport of the lipid molecules from mixed micelles into the phospholipid membrane. Remarkably, the opening of the channel is imbedded within an extended amphiphilic patch with predominantly positive changes that surround the entrance (Fig. 2B). Because lipids in the intestine are solubilized by bile acids (predominantly cholic and deoxycholic acids), one can argue that the molecular properties of the channel entrance favor the interaction of SCARB1 with the negatively charged mixed micelles, and thus facilitate the transfer of carotenoids into the receptor. Interestingly, the model revealed an additional narrower groove that is connected with the main tunnel, but opens on the opposite side of the ectodomain.
Figure 2– Molecular model of SCARB1 receptor.
(A) A putative membrane topology of SCARB1. The model of the human receptor was built based on the crystal structure of CD36 (PDB #5LGD) using SWISS-MODEL on the ExPASy web server and further optimized in CHIMERA [56, 57]. The central core channel present in SCARB1. The cavity spanning across the receptor involved in lipid transport is marked in red. Channel entrance 1 represents a site where a transported lipid molecule is acquired from mixed micelles, whereas channel entrance 2 may correspond to an interaction site with small molecule modulators of the receptor’s activity. The CavityPlus server was used for the identification of the intermolecular surface of the channel and the entry and exit sites [58]. (B) The vicinity of the entrance to the channel. The channel opens into a partially positively charged amphiphilic patch (marked by a yellow oval) that may facilitate interaction with intestinal mixed micelles. The color scheme represents charge distribution on the surface of the soluble part of the receptor. Negative charges are shown in red and positive charges in blue.
Although the function of this cavity is unknown, it may represent a binding site for small molecule modulators of lipid transport (Fig. 2A, channel entrance 2). One of the prospective ligands postulated for CD36 is oleic acid [51]. However, whether fatty acids can regulate SCARB1 activity remains an open question. Interestingly, several cysteine residues are conserved between LIMP-2, CD36, and SCARB1 [52, 53]. At least two intramolecular disulfide bonds stabilize the fold of the CD36 and SCARB1 receptors, whereas other conserved cysteine residues are required for the HDL binding activity or trafficking of the receptor to the plasma membrane [53, 54]. Moreover, the mode of action of the synthetic inhibitor of SCARB1, BLT-1, involves covalent modification of the Cys384 thiol group by thiosemicarbazone moiety of the inhibitor [55].
Despite the structural and biochemical data, the mechanism of the selective uptake of lipids by SCARB1 and its regulation remains elusive. This process requires the sequential coupling of the transient interaction of lipid-rich vesicles or micelles with the receptor, the selective extraction of lipid molecules, and their efficient channeling into the cell membrane. Studies on the uptake of cholesterol esters indicate the critical role of apolipoprotein A1 in the recognition and binding of HDLs [59, 60]. However, the absence of well-defined HDLs in the intestinal lumen does not affect the SCARB1-mediated absorption of carotenoids. Moreover, the interaction with lipid vesicles is essential, but not sufficient for effective lipid transfer, indicating that additional factors influence the receptor’s activity. They may include the effectiveness of the intracellular metabolism of carotenoids that contributes to the chemical gradient across the cell membrane or the direct regulation of the receptor’s activity through dynamic posttranslational modifications. Increase of the lipid transport efficiency due to the phosphorylation of the C-terminal domain of SCARB1 by salt-inducible kinase 1 provides evidence for such regulation [61]. Moreover, it has been proposed that the regulation of the receptor’s activity by posttranslational modifications is cell specific. Thus, this phenomenon represents one of the potential mechanisms by which SCARB1 function can be modified in a tissue-specific manner.
3. CAROTENOIDS CLEAVAGE ENZYMES
3.1. A functional link between ‘yellow pigment’ and the bioactivity of vitamin A
Early studies on the physiological function of vitamin A in vertebrates performed over 100 years ago revealed a surprising functional relationship between “plant lipochrome pigments” (exemplified by carotene) and vitamin A [62-64]. Experiments in which dietary vitamin A deficiency could be remedied by carotene supplementation raised a fundamental question of “how molecules, differing so obviously as do carotene and vitamin A can be interchangeable in function?” [65]. Despite numerous nutritional studies, the enzymatic activity allowing for the production of vitamin A from β,β-carotene was reported nearly 40 years later in an extract from rat’s intestine and liver [66, 67]. However, it was not until 2000 that the genetic analysis of blind Drosophila mutants led to the identification of β-carotene-15,15’-dioxygenase (BCDO1), an enzyme responsible for the oxidative symmetric cleavage of β-carotene. This process yields two molecules of all-trans-retinal per β-carotene molecule [68, 69]. Subsequently, the vertebrate orthologous genes and their protein products possessing equivalent enzymatic activity were cloned and characterized [70-74]. The analysis of BCDO1’s substrate specificity revealed that it utilizes provitamin A carotenoids with at least one nonoxidized β-ionone ring, such as β,β-carotene, α-carotene, and β-cryptoxanthin. However, this enzyme is unable to process non-provitamin A carotenoids, such as lycopene or zeaxanthin [71, 75]. Consequently, Bcdo1 knockout mice (Bcdo1−/−) become susceptible to vitamin A deficiency when kept on a diet with β-carotene as the main source of retinoids [76]. These mice also accumulate carotenoids in peripheral tissues, such as white adipocytes, the liver, and lungs as well as exhibit elevated levels of β-carotene in the blood. Importantly, the latter has also been reported in humans affected by a missense mutation in the BCDO1 gene [77].
Along with the symmetric cleavage of carotenoids, early biochemical studies indicated the presence of an alternative enzymatic reaction that yielded apo-carotenoids (8’, 10’, or 12’), fueling a decade long scientific discussion about the metabolic pathway of vitamin A biosynthesis [78, 79]. Cloning a second representative of mammalian carotenoid cleavage enzymes, β-carotene-9’,10’-dioxygenase (BCDO2) provided evidence for two independent proteins responsible for the symmetric and eccentric cleavage of carotenoids [80, 81]. Importantly, the difference between BCDO1 and BCDO2 is not limited to the position at which the polyene chain is split. Unlike BCDO1, the substrate specificity of BCDO2 is very broad. In addition to β-carotene, this enzyme preferably accepts carotenoids with assorted ionone rings, including xanthophylls and 4-oxo-carotenoids as well as various apocarotenoids [82-86]. Although BCDO2 may contribute to vitamin A production from specific carotenoid substrates, its biological role is more diverse. BCDO2 localizes predominantly in the mitochondria, where it plays an important role in preventing an accumulation of carotenoids in these organelles [75]. As evidenced in Bcdo2−/− mice kept on lutein rich diet, the absence of BCDO2 enzymatic activity adversely affected the mitochondrial electron transport chain, which resulted in carotenoid-induced oxidative stress [75, 87]. Spontaneous mutations and genetic variations in the BCDO2 gene have been reported to cause elevated levels of carotenoids in mammalian tissues [88-90]. In humans, polymorphisms in the BCDO2 gene have been linked to elevated levels of proinflammatory interleukin 18 associated with diabetes type 2 and cardiovascular diseases [91]. Although the correlation between diet, genetic variations in BCDO2, and a disease state requires further investigation, it is plausible that BCDO2 activity is part of a protection mechanism against the undesirable biological effects of dietary carotenoids that cannot be utilized for the biogenesis of vitamin A.
3.2. Oxygen at work – structural and mechanistic determinants of BCDOs’ activity
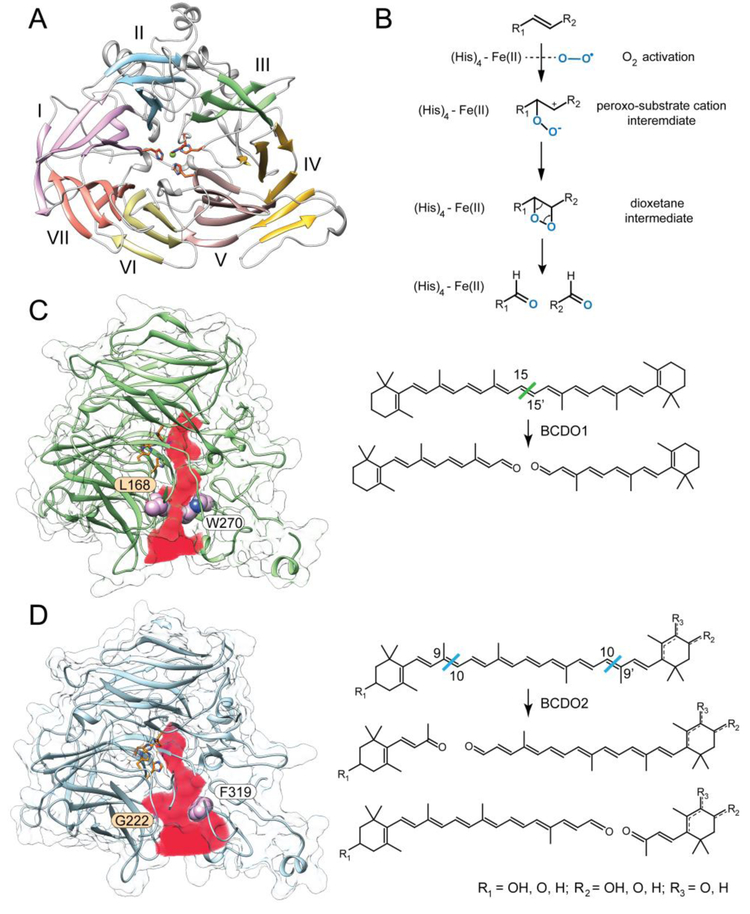
Mammalian BCDO1 and BCDO2 belong to a large and diverse protein family of carotenoid cleavage oxygenases (CCOs). Representatives of CCOs can be found in all kingdoms of life, except Archeobacteria, contributing not only to the metabolism of carotenoids but also to that of lignin-related phenylpropanoids [92-94]. Although direct structural information about BCDOs is not currently available, valuable insight into the mechanistic aspects of catalysis can be inferred from the structures of related proteins. Four different CCOs representing cyanobacterial apocarotenoid oxygenase (ACO), bacterial lignostilbene-α,β-dioxygenase A, plant VP14enzyme, and mammalian retinoid isomerase (RPE65) have been crystalized so far [95-98]. Despite the lack of significant sequence homology, they reveal a strikingly conserved 3D architecture of the protein molecules. The overall fold is composed of a well-defined seven-bladed β-propeller (Fig. 3A) [99]. The β-strands are unequally distributed among the propeller. Blades I, II, IV, and V consist of four antiparallel β-strands, whereas VI and VII are enlarged by an additional strand. The main difference in the architecture of the bacterial and plant CCOs and RPE65 is seen within blade III, which contains a two-strand extension in the mammalian enzyme. The α-helical components of the structures form a dome on the top of the β-propeller that covers the iron cofactor-binding site in the catalytic center of these enzymes (Fig. 3A). This Fe2+ ion is coordinated by Nε atoms donated by four histidine residues. The second coordination sphere is formed by three glutamic acid side chains that hydrogen bond to three out of the four histidines involved in the iron binding [97]. All of these residues are absolutely conserved among CCOs [99]. Thus, in contrast to the heme-dependent systems, CCOs belong to the class of non-heme iron enzymes that use Fe2+ to activate dioxygen (O2) in the active site [100]. The exception in regards to O2 utilization is RPE65. It acts on an ester bond of retinyl esters through the Lewis acid property of the iron ion, and thus does not require O2 for enzymatic catalysis [101, 102].
Figure 3– Structure and enzymatic function of mammalian BCDOs.
(A) The overall molecular organization of BCDOs represented by a homology model of human BCDO1. The model was built based on the crystal structure of RPE65 (PDB #4F2Z). The side chains of conserved histidine residues involved in the coordination of an iron ion are shown in the center. (B) A proposed mechanism for the enzymatic cleavage of carotenoids by BCDOs. The dioxygenase mechanism is supported by the incorporation of both oxygen atoms derived from O2 into the products of the reaction. Panels C and D represent structural models and reactions catalyzed by BCDO1 and BCDO2, respectively. The channels leading to the active sites of the enzymes are marked in red. Marked residues are responsible for the difference in the aperture of the binding cavities and thus contribute to the enzymes’ substrate selectivity. An alternative position of a double bond in the substrates for BCDO2 are marked with a dashed line.
Much attention has been dedicated to the elucidation of the catalytic mechanisms of CCOs. The heated debate has been fueled by conflicting data indicating that both monooxygenase and dioxygenase reaction patterns may exist within the CCO protein family [103-107]. The major difficulty in the interpretation of the isotopic labeling experiments in the presence of H218O and 18O2 is the use of crude cell extract, the low enzymatic activity of the enzymes, and the poorly controlled solvent back-exchange of the aldehyde cleavage products. Addressing these problems in more recent studies has shifted the experimental evidence in favor of the dioxygenase mechanism for the Synechocystis ACO, Novosphingobium NOV2, and Galleria mellonella NinaB enzymes [101, 108]. Importantly, the analogous labeling experiments performed on human BCDO1 also revealed the exclusive incorporation of O2-derived oxygen atoms into the retinaldehyde (RAL) products of the reaction (Fig. 3B) [109]. Thus, the cleavage of carotenoids by BCDOs also occurs via the dioxygenase mechanism. Despite this unquestionable progress, our knowledge about the key steps of the catalysis remains incomplete. Particularly, experimental scrutiny requires the initial step of the catalysis that includes activation of O2. It may involve a single electron transfer from Fe2+ followed by the generation of a stabilized substrate radical intermediate [110]. Subsequently, electron transfer to Fe3+ would then generate a stabilized substrate carbocation intermediate. Alternatively, upon the formation of the enzyme-O2-substrate complex, π electron density from the scissile double bond can be redistributed to the iron-oxy complex to directly form a Fe2+-peroxo-substrate carbocation intermediate [106, 111]. Also unclear is the role of the ferrous iron in the decomposition of the dioxetane intermediate. Theoretical calculations indicate that the Fe2+-facilitated cleavage of the dioxetane O-O bond is thermodynamically preferable over a spontaneous collapse of this intermediate [106].
The fundamental differences in substrate specificity observed for BCDO1 and BCDO2 define their physiological functions. Structural homology models of BCDOs as well as mutagenesis studies provide evidence that these differences arise from a subtle alteration in the architecture of the substrate-binding sites [82]. Carotenoids are relatively large lipophilic and nearly planar molecules that need to be channeled through a long hydrophobic cavity that extends beyond the catalytic center to properly align the polyene chain with respect to the iron cofactor (Fig. 3C, D). This substrate-binding tunnel is lined up predominantly with hydrophobic residues (Phe, Leu, Ile, and Met) along with the side chains of aromatic amino acids (Tyr and Trp). These general commonalities are less obvious towards the far end of the substrate-binding cavity. In comparison to BCDO1, the terminus of the tunnel in BCDO2 is assorted with a cluster of polar side residues represented by Gln375, His400, Ser406, and Glu460 (in the human sequence). Also this part of the binding side is slightly wider due to the substitution of a bulky Tyr332 with a Leu residue. This increase in polarity may facilitate the protein’s interaction with carotenoids containing an oxygen substituted ionone ring. In addition to the altered physiochemical properties of the binding site, the entrance to the cavity in BCDO2 is much wider (~9 Å in diameter) as compared to BCDO1’s (~5 A) (Figs. 3C, D). The narrowing of the channel is attributed to the presence of the two bulky side chains, Leu168 and Trp270 in BCDO1, which are substituted with the smaller residues of glycine (Gly222) and phenylalanine (Phe319) respectively in BCDO2. Importantly, the functional significance of these differences was examined experimentally by generating a mutated BCDO1 in which Trp270 was exchanged for phenylalanine and Leu168 for glycine [82]. Remarkably, this chimeric enzyme exhibited robust activity towards zeaxanthin, converting this substrate to all-trans-3-hydroxy-retinal, a reaction not detectable for the wild-type BCDO1. Despite a wealth of information provided by the comparative biochemistry approach, the elucidation of the high-resolution structures of mammalian BCDOs will represent a significant step towards a comprehensive understanding of the evolution and functional determinants of CCO enzymatic activity.
4. REDOX REACTIONS OF RETINOIDS – AT THE CROSSROAD OF METABOLIC FATES
4.1. The interconversion of retinaldehyde and retinol
The enzymatic cleavage of provitamin A carotenoids and apocarotenoids by BCDOs yields RAL (Fig. 1). Similar to other aldehydes present in biological systems, RAL acts as a reactive electrophile, which readily conjugates with thiols and primary amines, forming adducts with proteins and phospholipids [112-115]. The high reactivity of RAL is responsible for the primary cytotoxicity of this compound [116-119]. Additionally, RAL is a direct precursor for RA, a potent signaling molecule and activating ligand of nuclear retinoid acid receptors [120-122]. Thus, the concentration of RAL is tightly controlled by the metabolic activity of numerous retinoid-processing enzymes collectively classified as oxidoreductases. Based on sequence homology, they are representatives of three major protein families: microsomal retinol dehydrogenases (RDHs) that represent the short-chain dehydrogenase/reductases (SDRs), cytosolic alcohol dehydrogenases (ADHs) that belong to the medium-chain dehydrogenase/reductase family, and selected members of aldo–keto reductases (AKRs) [123-127]. Each of these classes of enzymes is comprised of several distinct members [122, 124, 128]. Interestingly, none of these enzymes reveal exclusive selectivity towards retinoids. Their broad substrate promiscuity includes ketosteroids, ketoprostaglandins, and a variety of xenobiotic compounds [129-131]. The main evidence for the contribution of particular proteins to retinoid metabolism is predominantly derived from the phenotypic characterization of genetically modified mice. Consequently, the role of ADHs has diminished over the last year as they turned out to be essential only in specific conditions of vitamin A excess (ADH1), or deficiency (ADH4), but dispensable for maintaining retinoid homeostasis under normal physiological conditions [132]. Contrary to ADHs, the in vivo characterization of RDHs revealed their critical role in tissue-specific RA signaling and visual chromophore regeneration [122, 133, 134].
The universal redox carriers for oxidoreductases are the oxidized or reduced forms of dinucleotides NAD(H) or NADP(H). Because the redox reactions of ROL to RAL conversion are fully reversible, their net direction is determined by the substrates’ availability and the enzymes’ specificity for binding either NAD(H) or NADP(H). In eukaryotic cells, the concentration of NAD+ is a thousand times higher as compared to its reduced form (NADH), whereas the NADP+/NADPH ratio is 1:100 [135, 136]. Thus, the oxidoreductases that bind NADP(H) can contribute significantly only to RAL reduction. Conversely, the enzymes that prefer NAD(H) catalyze the oxidation of ROL.
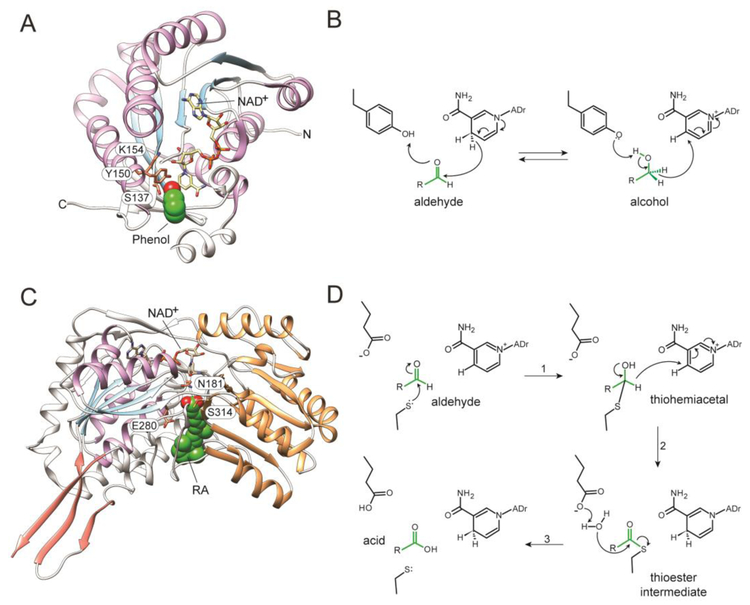
In ADHs and RDHs the nucleotide cofactors are bound by a classic Rossmann fold [137]. This is one of the most common structural elements composed of 6 to 7 twisted central parallel β-strands flanked by 3–4 α-helices from each side (Fig. 4A) [138, 139]. This structural motif contains a highly variable Gly-rich sequence (GXXXGXG) responsible for its structural integrity and binding of the diphosphate portion of the nucleotide cofactors. An acidic residue interacting with the 2′ and 3′ hydroxyls of the adenine ribose and located downstream of the Gly-rich motif is responsible for NAD(H) specificity, whereas NADP(H) binding is dictated by the presence of a basic residue within the Gly-rich segment [139, 140]. Interestingly, the common mode of cofactor binding does not result in identical mechanisms of catalysis. The active site of RDHs contains a catalytic Tyr residue, whose hydroxyl group donates or accepts a proton from the substrate (Fig. 4B). Lowering the pKa of this hydroxyl by nearly 3 units is facilitated by the interaction of the hydroxyl with the ε-amino group of an adjacent Lys residue. Additional hydrogen bonding between the Lys residue and the 2′- and the 3′-hydroxyl groups of the cofactor’s ribose defines the orientation of the nicotinamide with respect to the catalytic residues, allowing exclusively for a 4-pro-S hydride transfer [141, 142]. For the oxidation reaction, the deprotonated Tyr residue acts as a catalytic base. The hydride ion, extracted from the substrate hydroxyl group, is then transferred to position 4 of the nicotinamide ring. In addition to the interaction with the nicotinamide ring of the cofactor, the reaction intermediate is stabilized by the hydroxyl group of an adjacent serine residue. Thus, the active site of RDHs comprises a catalytic triad of conserved Ser-Tyr-Lys residues [143].
Figure 4– Structure and catalytic mechanisms of RDHs and ALDHs.
(A) A cartoon representation of RDH from Drosophila melanogaster (PDB # 1B2L). The labeled amino acids correspond to key residues involved in the catalysis. A putative orientation of the retinoid substrate is marked by a phenol molecule present in the structure (green and red). The secondary structures that constitute the Rossmann fold are colored blue (β-strains) and purple (α-helices). (B) The reversible transfer of hydride from the S4-face of the nucleotide cofactor to RAL to produce pro-R-ROL. (C) The structure of human ALDH1A3 in complex with RA (PDB #5FHZ). Residues essential for the catalysis are contoured and labeled. RA molecule is shown in green and red. The secondary structures corresponding to the Rossmann-like fold are indicated as in panel A. The catalytic domain is colored orange, whereas the dimerization domain is shown in salmon. (D) The catalytic mechanism of RAL oxidation to RA involves the transient covalent addition of the retinoid moiety to the catalytic cysteine residues in the form of a thioester.
Despite the presence of the common nucleotide-binding motif, ADHs depend on a catalytic Zn2+ ion. In the absence of a substrate, it is coordinated by a water molecule and the side chains of two Cys and a His residues. Displacing water by the substrate’s oxygen atom in the zinc coordination shell enables the transfer of a proton to the solvent, resulting in alkoxide reaction intermediate. The transfer of a hydride to the oxidized cofactor finalizes the catalysis, yielding an aldehyde product [144, 145].
AKRs represent an interesting example of the convergent evolution of oxidoreductase enzymatic activities. Their single-domain structure does not comprise the canonical Rossmann fold. Instead, it adopts a TIM barrel motif with (α/β)8 topology [146, 147]. This difference determines the stereospecificity of the hydride transfer, making AKRs 4-pro-R enzymes. Nevertheless, the overall catalytic mechanism of AKRs is very similar to that found in RDHs, with an active site Tyr and assisting Lys residue that enable the general acid-base catalytic mechanism [148].
It is important to note that the majority of the mechanistic aspects of RDH function are derived from structural studies on SDRs that do not utilize retinoids as substrates. In fact, RDHs turned out to be difficult targets for structural studies, thus none of the vertebrate representatives of RDHs have been crystalized so far. One of the major obstacles to the successful purification and crystallization of these enzymes is their amphiphilic character needed for their interaction with lipid membranes. However, since there is no well-defined and conserved membrane domain in these enzymes, the mode of membrane binding and topology seems to vary between specific RDHs [12, 125]. For example, according to biochemical studies, RDH1 is anchored in endoplasmic reticulum (ER) membranes with the catalytic domain facing the cytoplasm [149]. The N-terminal residues are essential for the membrane localization and topology of this enzyme, whereas the C-terminus was postulated to be involved in the stabilization of the protein’s membrane orientation. A similar role for the N-terminal segment was postulated for RDH11 [150, 151]. Contrary to these models, the presence of endoglycosidase H-sensitive N-linked sugar oligosaccharides on RDH12 suggest a luminal orientation for this enzyme [152]. An alternative mode of RDHs-membrane interaction involves the posttranslational lipidation of the enzymes as exemplified by RDH8. Studies on the localization of this protein in transgenic Xenopus implied that its membrane association was mainly mediated by the fatty acid acylation of the C-terminal Cys residues [153]. Yet other models indicate that the hydrophobic stretch of the catalytic domain in RDH11 and RDH4 can contribute to the membrane binding [151, 154]. This fact is supported by the structure of photoreceptor RDH from Drosophila melanogaster, the only crystalized representative of retinoid-processing SDRs (Fig. 4A) [155]. In the structure of this enzyme, the entry to the putative retinoid binding site is enclosed by a hydrophobic patch that most likely associates with the lipid membrane interface, whereas the hydrophilic entrance for the cofactors is localized at the opposite site of the catalytic domain [155]. This arrangement allows a membrane-residing substrate, such as RAL, to have access to the enzyme’s catalytically active site independently from the water soluble NAD(H).
4.2. Unidirectional production of retinoic acid
The enzymatic reduction of RAL into ROL is necessary for the efficient transport, storage, and tissue-specific distribution of vitamin A (Fig. 1). However, RAL also serves as a direct substrate for the biosynthesis of RA. It is impossible to overestimate the significance of this regulatory molecule for the physiology and pathophysiology of vertebrates as it is involved in the transcriptional regulation of over 500 different genes [5, 156-159]. Thus, it is intriguing that the production of RA is accomplished by representatives of a promiscuous class of enzymes belonging to the aldehyde dehydrogenase (ALDH) protein family. There are 19 functional members of ALDHs found in the human genome classified into 11 families and 4 subfamilies [160]. They are predominantly involved in the clearance of all kinds of potentially cytotoxic endobiotic and xenobiotic aldehydes [161-163]. However, three members of the ALDH1A family have been shown to be critical for the production of RA. ALDH1A2 (also known as RALDH2) is the primary enzyme responsible for the synthesis of RA during embryogenesis [164, 165]. Consequently, the genetic ablation of this enzyme leads to prenatal death due to defects in heart development. The role of closely related ALDH1A3 (RALDH3) in RA production has also been documented in vivo. Although Aldh1a3 null mice are born alive, they succumb to fatal defects in their nasal cavities shortly after [166]. Less dramatic is a deficiency in ALDH1A1 (RALDH1). Although, this enzyme is not essential for embryogenesis, it contributes to retinoid homeostasis as evidenced by diminished retinoic acid synthesis in the livers of Aldh1a3−/− mice [167, 168]. Importantly, deregulation of RA signaling is one of the hallmarks of carcinogenesis and autoimmune diseases [169, 170]. In this context, ALDHs activity may be considered not only as a prognostic marker but also as a potential therapeutic target [171-174]. In fact, compelling evidence indicates the ALDHs involvement in regulation of cell proliferation, survival, and chemoresistance of cancer stem cells [175, 176]. Moreover. RA inhibits differentiation of naive T cells but induces regulatory T cells and gut-specific homing of activated T cells. Imbalance in RA biosynthesis has been associated with autoimmune pathologies, such as inflammatory bowel disease, rheumatoid arthritis, systemic lupus erythematosus, and multiple sclerosis [177, 178]. Therefore, restoring the retinoid homeostasis in autoimmune diseases has been of increasing interest in the development of therapeutics against these conditions.
The high sequence homology between RALDHs (reaching ~70% of identity) is reflected in the structural similarities between these enzymes. All RALDHs form functional homotetramers in both solution and crystal forms. Each monomer acts independently as an autonomous catalytic unit. There are three distinct domains in each protomer: the N-terminal NAD+-binding domain, the catalytic domain, and the oligomerization domain (Fig. 4C). The architecture of the cofactor binding domain and the catalytic domain resembles a βαβ polypeptide topology, similar to a canonical Rossmann fold. The recent determination of the structure of human RALDH3 in complex with NAD+ and RA significantly contributed to our understanding of the substrate-enzyme interactions as well as dynamic changes within the protein domains that are associated with the catalysis [179]. The tunnel leading to the active site is located in-between the NAD+-binding and catalytic domains. The side chains of the residues from both domains contribute to the hydrophobic environment found inside this binding pocket. Similarly to BCDOs, the substrate specificity of RALDHs is most likely determined by subtle changes in the amino acid composition of the binding site. However, rigorous mutagenesis studies verifying this stipulation have not yet been conducted. The β-ionone ring of the substrate is shelled from the solvent by making van der Waals contacts with conserved hydrophobic residues at the entrance to the tunnel. In the closed, catalytic competent conformation, the carboxylic carbon of RA (equivalent of the carbonyl carbon in RAL) is 3.1 Å away from the thiol group of the catalytic Cys314 residue. The presence of Cys at the active site defines the mechanism of catalysis that is distinct from RDHs. The NAD+-dependent oxidation of RAL is initiated by the Glu280-mediated abstraction of a proton from the sulfhydryl group of Cys, which acts as a nucleophile that attacks the carbon on the carbonyl group of the substrate (Fig. 4D) [180, 181]. The resulting thiohemiacetal with a negatively charged oxygen atom is stabilized by the side chain of an Asn residue. The spontaneous transfer of the hydride ion from the thiohemiacetal to the cofactor is accompanied by formation of a thioester bond between the aldehyde and the catalytic Cys residue. The activation of a water molecule by the Glu residue enables the hydrolysis of the covalent thioacyl-enzyme intermediate and release of the RA product. Interestingly, the trapping of multiple conformations of NAD+ and RA in the active site of RALDH3 led to the discovery that both the cofactor and substrate molecules can move in a concerted manner, either towards the active site Cys residue, facilitating the catalysis, or outwards promoting the post-reaction release of the products [179].
5. NOT ONLY TRANSPORT AND STORAGE – THE ROLE OF RETINYL ESTERS
5.1. Physiological significance of retinol esterification
Molecules of ROL derived from carotenoids or preformed retinoids share a common metabolic fate (Fig. 1). They are efficiently esterified in the enterocytes by the action of two enzymes: lecithin:retinol acyltransferase (LRAT) and diacylglycerol O-acyltransferase 1 (DGAT1) [182]. The formation of highly hydrophobic and chemically inert long-chain fatty acid esters of ROL serves two main functions: the creation of a mass-action that drives the absorption of retinoids, and the facilitation of retinyl ester packaging into vesicles that are secreted into the lymphatic system as chylomicrons. Approximately 90% of acyltransferase activity towards ROL is attributed to LRAT, whereas the remaining 10% is contributed by DGAT1 [183]. These two acyltransferases belong to unrelated classes of enzymes. DGAT1 depends on a pre-activated form of fatty acids carried by acyl-CoAs and reveals broad substrate specificity as it catalyzes the esterification of variety of abundant mono- and diacylglycerols in addition to ROL [184]. On the contrary, LRAT’s substrate specificity is limited to ROL and its derivatives such as endogenous apo-carotenoids or xenobiotic retinylamine [185-187]. Also, LRAT does not require acyl-CoA. Instead, it utilizes phosphatidylcholine as a direct acyl donor [185, 188-190]. Importantly, only the acyl chain at the sn-1 position of the phospholipid substrate can be transferred, thus retinyl palmitate is the predominant form of esterified ROL found in vivo [185, 191].
In addition to the enterocytes, LRAT is robustly expressed in hepatic stellate cells. This subset of liver cells plays a central role in the storage and mobilization of hepatic vitamin A as they contain over 90% of total liver retinoids [192-194]. To maintain this reservoir of retinoids, the majority of the circulating chylomicrons are uptaken by the liver in an apolipoprotein E cell surface receptor-dependent manner [195]. Upon being internalized to hepatocytes, retinyl esters undergo rapid hydrolysis and the resulting ROL molecules are subsequently transported to adjacent stellate cells [196, 197]. Although this process is not fully understood on the molecular level, studies on Lrat−/− mice clearly indicate that the accumulation of retinyl esters in stellate cells depends exclusively on LRAT’s enzymatic activity [183, 194]. Notably, the size of the retinyl ester droplets strongly correlates with the dietary supply of vitamin A and carotenoids, indicating the regulatory role of the stellate cells in maintaining retinoid homeostasis [194].
The esterification of ROL is an efficient mechanism that supplies a readily available pool of retinoids that can be mobilized during dietary deficiencies. In fact, the ability of vitamin A to be stored in hepatic cells makes it unique among the fat-soluble vitamins [14, 197, 198]. Importantly, retinyl esters also play a direct role in the biosynthesis of key vitamin A metabolites. In the RPE cells, LRAT provides a direct substrate for the production of 11-cis-retinol, a precursor of visual chromophore (11-cis-retinylaldehyde) [12]. The geometric isomerization of the polyene chain is catalyzed by RPE65 [102, 199, 200]. This enzyme combines the O-alkyl cleavage of all-trans-retinyl esters (mostly palmitate) with a trans to cis geometric conversion of the polyene chain [97, 201]. Such an unusual mode of ester cleavage gives rise to the formation of intermediate retinyl cation and provides energy for the thermodynamically unfavorable trans to cis isomerization [202, 203]. Consequently, LRAT-deficient mice as well as humans affected by LRAT mutations are blind and undergo progressive retinal degeneration due to the inability to provide photoreceptor cells with visual chromophore [183, 204]. Importantly, the analysis of the phenotype of Lrat−/− mice revealed not only the lack of cis isomers, but also the general absence of retinoids within the retina and RPE cells [205]. This observation led to yet another important conclusion that ROL esterification is needed for the efficient cellular uptake of circulating vitamin A in peripheral tissues, a phenomenon described in detail in the following chapters.
Acyl-CoA-dependent ROL esterification has been proposed to be a part of the cone-specific visual chromophore regeneration pathway [206-208]. In contrast to mice or rats, the retinas and RPE cells of cone-dominant species accumulate predominantly 11-cis-retinyl esters [209, 210]. Moreover, chicken and ground squirrel retina homogenates exhibit robust retinoid isomerase activity that, unlike RPE65, converts ROL directly into its 11-cis isomer [211, 212]. This reaction is driven by the subsequent esterification of newly produced 11-cis-retinol. Notably, DGAT1 activity does not contribute to the formation of 11-cis-retinyl esters [188, 213]. Based on biochemical analysis, the specific esterification of 11-cis-retinol was attributed to acyl-CoA wax alcohol acyltransferase 2 (AWAT2), also known as multifunctional O-acyltransferase (MFAT), a distinct member of the DGAT2 protein family [214]. Although the substrate preference of AWAT2 toward cis-retinols has been independently verified, the role of this enzyme in the regeneration of cone light sensitivity awaits verification in vivo [215].
5.2. Catalysis at the lipid membrane/water interface
Membrane associated enzymes constitute a challenging target for structural studies. This is particularly true for the members of DGAT1 and DGAT2 protein families. Currently, even the most fundamental information about the architecture of these proteins is not available. The main obstacle in structural studies on these enzymes is their amphiphilic character. The hydrophobic domains seem not to localize within the defined transmembrane segment, but spread throughout the protein molecule. Consequently, these enzymes are anchored to the lipid membrane by one or two transmembrane α-helices and multiple secondary protein-lipid interactions facilitated by hydrophobic patches on the catalytic domains [216, 217]. Thus, the stabilization of the tertiary structure of DGATs outside of a phospholipid environment becomes problematic.
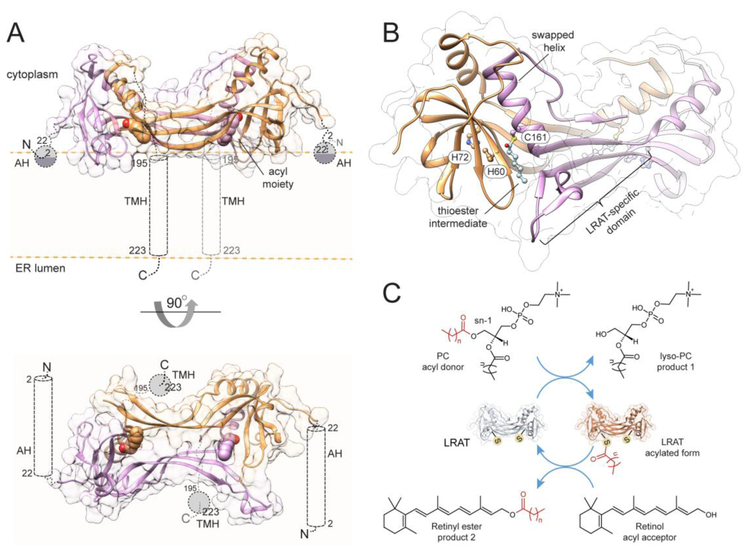
Substantially more is known about the molecular organization of LRAT. Located in the endoplasmic reticulum, this enzyme adopts a bitopic membrane topology with a transmembrane helix at the C-terminus and an additional amphiphilic domain present at the N-terminus (Fig. 5A) [218, 219]. LRAT belongs to an ancient NlpC/P60 thiol peptidase protein superfamily that includes seven representatives encoded in the human genome: two neurological sensory proteins (NSE1-2), and five H-ras-like tumor suppressors (HRASLS1-5) [220, 221]. They predominantly act as phospholipases, cleaving the ester bonds at the sn-1 or sn-2 positions. However, in the presence of a proper substrate, the esterase activity is accompanied by a transfer of the acyl moiety onto enzyme-specific substrates, including phosphatidylethanolamine and short chain alcohols, but not ROL [222-225]. The functional difference between HRASLSs and LRAT is surprising as they share a high degree of sequence similarity and use the same catalytic machinery, derived from a common cysteine peptidase ancestor [225, 226]. The reconciliation of these functional differences led to the designing of chimeric enzymes, where an LRAT-specific 30 amino acid sequence was inserted into the catalytic domains of HRASLS2, 3, and 4. Remarkably, this short insertion was sufficient for these artificial enzymes to acquire reaction specificity similar to that of authentic LRAT [191]. The mechanistic explanation of this phenomenon was inferred from the crystal structure of the HRASLS3/LRAT chimeric enzyme, which revealed dramatic structural changes as compared to the native HRASLS3 [191, 225]. The LRAT-specific domain folds into a β-hairpin structure (two subsequent antiparallel strands) that provides a primary dimerization interface, and when paired with a β-strand of the catalytic domain causes the swapping of an adjacent α-helix (Fig. 5B). This arrangement resulted in a mixed catalytic triad whereby the two His residues are contributed from one protomer and the catalytic Cys from the other. Thus, unlike the native HRASL3, the chimeric enzyme and presumably LRAT act as functional homodimers [191, 227]. Additionally, the LRAT-specific domain forms a central, highly lipophilic patch that is probably important for attracting membrane-dissolved ROL to the active site. The structure of the HRASLS3/LRAT chimera revealed the thioester intermediate of the enzymatic reaction, providing ultimate proof for the previously postulated mechanism of catalysis by LRAT [228-230]. The catalytic triad is composed of two His residues that assist in the deprotonation of the sulfhydryl group of the catalytic Cys side chain (Fig. 5C). Thus, Cys serves as a nucleophile attacking the carbonyl carbon of an sn-1 ester bond of phosphatidylcholine, leading to the formation of a tetrahedral intermediate [12]. The collapse of this intermediate results in the transient acylation of the protein by the formation of a thioester bond at the Cys sulfhydryl group and the liberation of the first product of the reaction, lyso-phosphatidylcholine. Limited water access to the catalytic thiol nucleophile is the key structural adaptation that enables efficient acyl transfer by blocking competing hydrolysis. The thioester is surprisingly stable in the absence of ROL [191]. The lack of measurable hydrolytic activity suggests that LRAT persists in the cells in an acylated form. In the second stage of the reaction, the His-mediated deprotonation of the hydroxyl group on the ROL molecule permits nucleophilic attack of the activated hydroxyl group followed by the decomposition of the thioester intermediate and transfer of the acyl group from the enzyme onto its retinoid acceptor to yield retinyl ester [134].
Figure 5– Structural model of human LRAT.
(A) Ribbon representations of the enzyme in a dimeric form positioned at the lipid membrane. Each protomer contains an N-terminal amphiphilic α-helix (AH) as well as a transmembrane one (TMH) at the C-terminus. The acyl moieties forming thioester bonds with the catalytic cysteine on each monomer are represented with atomic spheres. The model was built using the SWISS-MODEL server based on PDB # 4Q95. (B) Zoomed in view of the architecture of the catalytic domain of LRAT. Each active site is formed by residues donated by both protomers. The labeled histidine and serine residues constitute the catalytic triad. (C) A schematic representation of the acyl transfer catalyzed by the enzyme indicating the transient thioester intermediate of the reaction.
The overall topology of LRAT and the orientation of its active site, facing the lipid/water interface, suggest that the retinoid substrate is acquired directly from the membranes [198]. Thus, it remains unclear what factors dictate the narrow substrate specificity of this enzyme [187]. Also, the kinetic data indicate that the enhanced esterification of ROL delivered by cellular retinol-binding proteins (CRBPs) cannot be explained by the chimeric structure [231-233]. However, one cannot exclude the possibility that the N-terminal α-helix or the catalytic domain of LRAT absent in the chimeric enzyme may assist in the release of ROL from holo-CRBPs near the esterification site.
6. CARRIER PROTEINS INTEGRATE RETINOID METABOLIC PATHWAYS
Retinoids and carotenoids are lipophilic compounds that contain a system of conjugated double bonds. These physiochemical properties limit their ability to diffuse in an aqueous environment and make them susceptible to various catabolic reactions. To alleviate the barriers in the transport and biological utilization of retinoids, several specialized carrier proteins have evolved. Their main function is to sequester and protect labile retinoids as well as facilitate their systemic and intracellular transport. Therefore, they play a critical role in interconnecting compartmentalized metabolic and signaling pathways of retinoids.
6.1. RBP4 facilitates the systemic transport of vitamin A
Absorbed retinoids are stored in large quantities in form of hepatic retinyl esters that can be mobilized depending on physiological needs. It is astonishing that despite its physiological significance, a mechanism responsible for sensing the vitamin A level in the organism and mobilizing hepatic retinyl esters has not yet been discovered. Nevertheless, it is clear that this process is not simply a reversal of the hepatic retinoid absorption pathway. The recruitment of ROL requires the hydrolysis of stellate cell retinyl esters. Despite substantial research efforts to identify the specific enzyme responsible, none of the examined hydrolases, esterases, or lipases exhibited a limiting or regulatory role in this process [197]. At the same time, the example of hormone-sensitive lipase in adipose tissues suggests that it is unlikely that numerous redundant enzymes carry out the hydrolysis of retinyl esters. Thus, the discovery of cell and function specific retinyl ester hydrolases remains one of the key challenges in the field.
After the cleavage of retinyl esters, the liberated ROL needs to become available to extrahepatic tissues. For this purpose, ROL is transported to the hepatocytes where it forms a complex with a specific 21-kDa protein called retinol-binding protein 4 (RBP4) [234-236]. The holo-RBP4 is subsequently secreted into systemic circulation (Fig. 1). Notably, over 95% of the total ROL present in the blood serum during fasting is bound to RBP4. Thus, the carrier protein-dependent transport of vitamin A is exclusively responsible for delivering ROL from liver storages to extrahepatic tissues. RBP4-deficient mice are viable and fertile, but develop progressive retinal dysfunction due to inadequate vitamin A supplementation of the ocular tissues [234]. In humans, mutations in the RBP4 gene have been identified as a precipitating cause of RPE and retinal atrophy [237, 238]. They may also lead to developmental eye defects as exemplified by a dominant-negative mutant, which increased the affinity of RBP4 to its cellular receptor (as discussed in the next section of this review) [239].
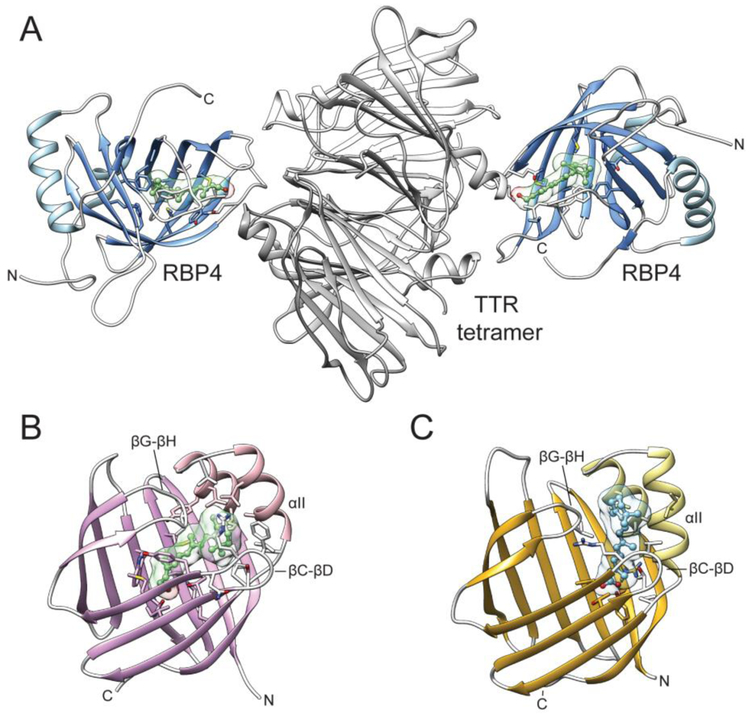
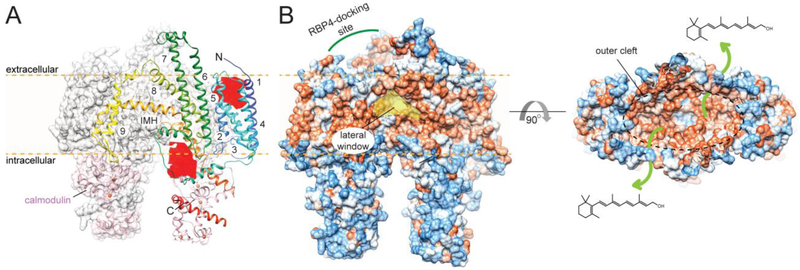
RBP4 is the earliest discovered and the most studied protein involved in retinoid metabolism. It belongs to a widespread and diverse group of transport proteins that are collectively referred to as lipocalins [240]. This class of carrier proteins evolved to facilitate the transport of small hydrophobic molecules, including steroids, fatty acids, and retinoids. Although members of the lipocalin protein superfamily share very little primary sequence homology, they all contain similar 3D architectures known as lipocalin folds. The main structural component of these carrier proteins is an antiparallel β-barrel composed of 8 to 10 β-strands (Fig. 6A). It forms a single binding pocket the size and hydrophobicity of which is defined by the side chains of the amino acids lining the interior of the β-barrel. Thus, the lipocalin fold serves as a universal and highly modifiable platform for numerous carrier proteins. Along with RBP4, the representative members of this class of proteins in the human genome are fatty acid-binding proteins, CRBPs, and cellular retinoic acid-binding proteins (CRABPs) [241]. RBP4 accommodates one molecule of ROL with the β-ionone ring positioned in the center of the β-barrel with the isoprene tail extending along the barrel axis toward the entrance to the binding pocket [242]. The contacts between the protein and retinoid moiety involve predominantly hydrophobic alkyl and van der Waals interactions with a single hydrogen bond between the hydroxyl group of the ligand and the main chain Gln98 (in the human sequence) [243]. The comparison of the apo- and holo-forms of RBP4 did not reveal major conformational changes upon ligand binding, with the exception of a loop that includes amino acids 33-36 [244]. This portion of the protein exerts a significant functional role, acting as a molecular sensor of the ligand binding. To avert rapid clearance by glomerular filtration, holo-RBP4 binds to a tetramer (dimer of dimers) of transthyretin (TTR) [236, 245]. The maximum stoichiometry of the complex revealed by in vitro studies is 4:2. However, due to the limited concentration of RBP4 in the plasma, the complex isolated from the serum contains only one holo-RBP4 per four molecules of TTR [246]. The binding interface involves the open end of the RBP barrel and includes amino acids 33-36 [246, 247]. Importantly, the conformation of these residues in the apo-protein does not favorably interact with TTR, explaining the observed four-fold diminished affinity of apo-RBP for this protein [248].
Figure 6– Structures of retinoid-binding proteins involved in extra- and intracellular transport.
(A) A complex of holo-RBP4 with a tetramer of TTR (PDB # 3BSZ). ROL molecules bound to RBP4 are shown in green. (B) The molecular architecture of human holo-CRBP1 (PDB # 5H8T). Labels indicate the parts of the protein that form the portal region. (C) A crystal structure of CRABP2 in complex with RA. The bound ligand is colored blue. Analogously to panel B, parts of the protein corresponding to the portal region are marked.
Although ROL is the major physiological ligand for RBP4, the orientation of the retinoid moiety in the binding site and the lack of specific interaction with the hydroxyl group prohibits a high selectivity towards just one class of retinoid ligands. Consequently, RBP4 has been shown to bind RAL, RA, and retinyl acetate with affinities comparable to ROL [249-252]. However, these ligands have never been found bound to RBP4 purified from human or bovine serum. On the contrary, more recent studies unambiguously identified long chain fatty acids as endogenous and physiologically relevant binding partners for RBP4 [253]. The protein isolated from human urine or amniotic fluid carried predominantly palmitic acid, but also palmitoleic, oleic, and stearic acids. As evidenced by X-ray crystallography, these new ligands compete with ROL for the same binding site. Similarly to ROL, the polar group of the fatty acids is exposed to the surface of the protein molecule, whereas their hydrophobic tails are buried inside the β-barrel. Importantly, the interaction with fatty acids does not trigger the conformational changes at the 33-36 loop characteristic for binding of ROL, leading to a diminished affinity for TTR. Thus, it is reasonable to assume that the ROL-free fraction of RBP4 is in fact saturated with free fatty acids. However, the physiological significance of this finding awaits further investigation. Nevertheless, RBP4 should not be considered a specific transporter of ROL, but rather as a lipid binding protein with specificity that has yet to be completely evaluated. As argued in the next section, this notion might also be true for other representatives of retinol-binding proteins.
The manipulation of the vitamin A metabolism by acting on RBP4 appears to be an attractive method for the treatment of diseases that involve the deregulation of RA signaling or an imbalance in retinoid homeostasis. They include certain types of breast cancers and lymphomas as well as retinal degenerative disorders such as Stargardt disease [254]. Although somewhat conflicting, recent years have provided data for an association between elevated RBP4 blood levels and insulin resistance, obesity, and metabolic syndrome, making RBP4 a potentially important therapeutic target [255-260]. Several small molecule antagonists of RBP4 have been developed, including the oldest fenretinide (N-(4-hydroxyphenyl)retinamide) as well as highly potent non-retinoid compounds such as A1120 (2-(4-(2-(trifluoromethyl)phenyl)piperidine-1-carboxamido)benzoic acid), and BPN-14136 2-((3aR,5r,6aS)-5-(2-(trifluoromethyl)phenyl)octahydrocyclopenta[c]pyrrole-2-carboxamido)benzoic acid [258, 261, 262]. They all exert their biological activity by disrupting the retinol-induced RBP4-TTR interactions through a combination of steric hindrance and changes in the polypeptide chain conformation at the TTR binding interface. The net result is the enhanced clearance of RBP4, lower concentrations of circulating ROL and its accessibility to peripheral tissues. Although this mechanism of action has been well documented in animals, the efficacy of the RBP4 antagonist in the treatment of human conditions still remains to be rigorously validated.
6.2. Diverse functions of homologous cellular retinoid-binding proteins
A subclass of intracellular lipocalins exhibits high specificity for ROL and RAL. They have been classified as cellular retinoid-binding proteins (CRBPs). Four representatives of CRBPs are encoded in the human genome (CRBP1-4) [263-266]. The tissue distribution and relative expression levels differ among these carrier proteins. The most widely expressed is CRBP1 with the highest abundance in the liver, kidneys, lungs, and RPE cells. In contrast, CRBP2 is present exclusively in the enterocytes of the small intestine, where it constitutes ~1% of total soluble proteins [267-270]. The concentrations of CRBP3 and CRBP4 are much lower. Their expression is apparent in several organs, including the kidneys, liver, heart, white adipocytes, and mammary glands [263, 264, 266]. In addition to diverse tissue distribution, CRBPs reveal significant differences in their affinity for retinoid ligands. The highest affinity was reported for CRBP1. The dissociation constant (Kd) was estimated to be between 0.1 and 20 nM depending on the measurement methodology [271-273]. A similar affinity (~11 nM) was calculated for CRBP2, whereas the interaction of ROL with CRBP3 and CRBP4 is significantly weaker (Kd of ~60 and 200 nM, respectively) [263, 264, 274].
These differences in expression and binding affinities are reflected in the diverse physiological functions of CRBPs. In addition to serving as chaperones for labile retinoids, CRBP1 plays an important role in the regulation of vitamin A uptake and metabolism. CRBP1-deficient (Rbp1−/−) mice revealed a lower rate of retinyl ester synthesis associated with 6-fold increase in retinoid turnover [275]. Consequently, retinyl ester storage in RPE and hepatic stellate cells was diminished by 50% as compared to wild-type mice. This aberration in retinoid homeostasis makes Rbp1−/− mice more susceptible to vitamin A deficiency and causes a 2-fold reduction in the rate of visual chromophore regeneration [275, 276]. This phenotype is in agreement with the postulated role of CRBP1 in the intracellular transport of ROL, enabling its efficient esterification by LRAT [233, 277, 278]. However, the involvement of CRBP1 in controlling the concentration of RAL and the biosynthesis of RA remains more controversial. Unfortunately, biochemical studies often yielded contradictory results that did not allow for a final conclusion [122, 279-281]. It is possible that the function of CRBP1 in this matter may strongly depend on the type of tissue and available enzymatic partners. This notion is supported by in vivo studies. Although Rbp1−/− mice do not reveal the developmental abnormalities characteristic of an imbalance in RA signaling, the absence of CRBP1 disrupts RA homeostasis in the mammary tissue of adult animals, resulting in microenvironmental defects similar to those observed in the early stages of tumorigenesis [282].
Consistent with its localization, CRBP2 is critical for the uptake and channeling of newly absorbed ROL to LRAT [283, 284]. However, the abundance of CRBP2 may also play an important role in protecting enterocytes against RA toxicity by sequestering RAL produced by the cleavage of dietary pro-vitamin A carotenoids and facilitating its reduction to ROL [281, 285]. The biological functions of CRBP3 and CRBP4 are less understood. The deletion of the CRBP3 gene in mice does not cause adverse health problems. These animals are vital and healthy, but reveal significantly reduced levels of retinyl esters in breast milk [286]. This relatively mild phenotype is most likely a consequence of the compensatory overexpression of CRBP1 in selected tissues that allows for the maintenance of retinoid-dependent functions [286]. Interestingly, when fed a high-fat diet CRBP3-deficient mice were less susceptible to developing hepatic steatosis, had lower concentrations of serum fatty acids and decreased adiposity, suggesting the involvement of CRBP3 in the regulation of lipid homeostasis [287]. However, it is currently unclear whether this function is related to retinoid signaling or reflects the ability of CRBP3 to interact with other classes of endogenous lipids.
Although CRBPs and RBP4 belong to the lipocalin protein family and bind to the same ligand, the topological organization of the secondary structures of these proteins is largely dissimilar [288]. The β-barrel that constitutes the binding cavity in CRBPs is composed of two additional β-strands (ten total) as compared to RBP4 (Fig. 6B). Additionally, the entrance to the binding site in CRBPs is guarded by two short α-helices and two extended hairpin loops between β-strands. These α-helices and turns constitute a so-called ‘portal region’ [289-291]. This part of CRBPs exhibits significant conformational flexibility, providing a passageway for ROL to enter the binding site. The interaction with the β-ionone ring of the ligand greatly affects the conformation of the portal region, prompting a change in the orientation of several side chains that otherwise protrude into the binding cavity in the apo-protein [273]. This mode of interaction with the retinoid moiety implies an inverted orientation of ROL as compared to RBP4 (Fig. 6B). The polyene chain reaches deep into the binding pocket where the hydroxyl group of ROL is imbedded into a polar patch at the base of the cavity. It forms hydrogen bonds with the side chain of Lys and Glu residues in CRBP1 and CRBP2 or Lys and His in CRBP3 and CRBP4. Consequently, the β-ionone ring of the ligand is located at the entrance to the pocket. It is protected from the aqueous environment by the side chain of the portal region. These binding characteristics ensure a high specificity for ROL and RAL, limiting the affinity of CRBPs for other retinoids.
The discovery of the interaction of CRBP2 with the canonical endocannabinoid, 2-arachidonoylglycerol and the endocannabinoid-like 2-lineoylglycerol, 2-oleoylglycerol, as well as other monoacylglycerols opened a new chapter in studies on CRBPs. As evidenced by the X-ray crystallography, these endogenous lipids compete with ROL for the same binding site (PDB # 6BTH and 6BTI). Moreover, their affinities for CRBP2 are comparable to that observed for vitamin A. Interestingly, the same phenomenon is not true for CRBP1. The extensive screening for alternative endogenous binding partners for this protein has proved its high selectivity for ROL [292]. It is therefore possible that each of the CRBPs may selectively bind a different subset of lipids. Notably, the electron density for a retinoid moiety has never been observed in the structures of CRBP3 or CRBP4. This fact, in conjunction with the metabolic phenotype of CRBP3-deficient mice suggests the existence of alternative endogenous ligands for these carrier proteins. Although more studies are needed to clarify the ligand specificity, the classification of CRBPs as strictly retinoid-binding proteins is deceptive and may conceal alternative physiological functions of some these proteins.
6.3. CRABPs in the signaling and catabolism of retinoic acid
Two highly homologous intracellular proteins are specialized for sequestering RA: CRABP1 and CRABP2 [293, 294]. Similar to CRBPs, they differ only in their expression profiles and cellular functions. CRABP1 is expressed ubiquitously, whereas CRABP2 is found only in tissues that synthesize high levels of RA, such as the ovaries, uterus, skin, and choroid plexus [295-299]. Based on enzyme kinetic studies, it has been postulated that both CRABPs facilitate the oxidative catabolism of RA by delivering RAL substrate to CYP enzymes, particularly CYP2B1 [300, 301]. Despite the uncertainty over whether the carrier proteins interact directly with the CYP enzymes, the Km values for holo-CRBPs were ~3-fold lower than for “free” RA. The evidence supporting the channeling of RA for catabolism comes from studies on tumorigenic cell lines. The overexpression of CRABP1 in F9, AMC-HN-7 or A-498 carcinoma lines alleviated the anti-proliferative effect of RA [302-304]. Elevated levels of CRABPs have also been associated with a poor prognosis in several other types of cancer [305, 306]. The precise effect of these proteins on cancer cell proliferation and migration is likely tumor specific and involves the roles CRABPs in the redistribution of RA into nuclei. Both proteins were found to translocate to the nucleus [298, 307]. However, only CRABP2 increased the transcription rates of genes dependent on RA response elements [308, 309]. Thus, the function of CRABP1 might be to sequester excess RA, whereas CRABP2 acts as a specific delivery carrier for apo-RA receptors. Surprisingly, the disruption of either gene in mice did not affect development, fertility, life span or general behavior [310]. This lack of apparent phenotype was not due to the compensatory effect of CRABPs since Crabp1−/−/Crabp2−/− double-knockout mice also appeared to be essentially normal [310]. The possibility that other lipid-binding proteins may compensate for the absence of CRABPs has not yet been verified experimentally.
The tertiary structures of CRBPs and CRABPs are completely superimposable with Rmsd = 1.5 Å [288]. Despite the high overall structural similarity, the organization of binding sites in CRABPs differs dramatically. In fact, none of the ligand-interacting residues are conserved between these classes of proteins. Consequently, the spatial position of the RA molecule in CRABPs does not overlap with ROL bound to CRBPs (Fig. 6C). RA binds much shallower, persisting near the entrance of the barrel. The portal region does not completely fold over the β-ionone ring, which is partially exposed to the solvent [311]. This phenomenon explains the ability of CRABPs to accommodate 4-oxo- and 2-hydroxo retinoic acids [312, 313]. Because the retinoid moiety is shifted upwards, the polar patch inside the binding site that accommodates the carboxyl group of RA is arranged differently than in CRBPs. The carboxyl oxygen atoms form an extended network of hydrogen bonds. These interactions directly involve the side chains of Arg and Tyr residues, as well as the ordered water molecules present in the binding site. The striking differences in the modes of interaction between retinoid ligands within the same structural scaffold prove the high universality and plasticity of the common lipocalin fold.
6.4. Specialized ocular retinol-binding proteins
The primary function of vitamin A metabolism in the eye is to support the perception of light by providing visual chromophore, 11-cis-retinaldehyde [11, 12]. 11 -cis retinoids are produced enzymatically via the canonical visual cycle or by light-induced trcms to cis isomerization [12, 314, 315]. However, regardless of the mechanism of biosynthesis, the 11-cis isomers require specialized binding proteins to facilitate their intra- and intercellular transport. Two retinoid-binding proteins are specifically expressed in the eye: cellular retinaldehyde-binding protein (CRALBP) and interphotoreceptor retinol-binding protein (IRBP) [316].
CRALBP is found predominantly in the RPE and Muller cells of the retina [317, 318]. This protein binds specifically 9- or 11-cis-retinol and RALs with low nanomolar affinities in vitro. However, only 11-cis isomers are associated with CRALBP isolated from natural sources [319-321] . In RPE cells, CRALBP sequesters 11-cis-retinaldehyde produced by the consecutive action of LRAT, RPE65, and RDH5. The bound ligand is protected from thermal re-isomerization or side reactions, and thus can be efficiently delivered to the plasma membrane for transport to the photoreceptors [322, 323]. This role of CRALBP was supported by in vivo studies. Although, Rlbp1−/− mice show no spontaneous retinal degeneration, the rate of visual chromophore regeneration after exposure to light was 10-fold slower than in the wild-type [324]. The effect of CRALBP-deficiency on cone function seems to be more complex, mostly due to our insufficient understanding of the cone regeneration process. The knockout mice revealed premature M-cone cell death, aberrant localization of M-opsin, and changes in M-cone cell dark adaptation rate [325]. This phenotype partially recapitulates rod/cone dystrophies observed in humans affected by mutations in the RLBP1 gene [326-328]. Importantly, M-cone sensitivity was restored upon viral-driven expression of CRALBP in Müller glia, but not in RPE cells, indicating the functional significance of Müller cells in the ocular retinoid metabolism [325].
CRALBP is not structurally related to lipocalins. Instead, it is a founding member of a family of carrier proteins containing the CRAL–TRIO domain, which is also present in proteins that bind non-retinoid lipids such as tocopherol, squalene, and phosphatidylinositol [329, 330]. There are two distinct domains within the CRALBP structure, the α-helical N-terminal and the binding site containing αβα sandwich domain at the C-terminus (Fig. 7A) [331]. The cis retinoids-binding cavity is defined by the convex side of the β-sheet and the four adjacent helices. It adapts a curved shape, which is complementary to the 11-cis- and 9-cis-configurations. When bound to CRALBP, the retinoid moiety is completely isolated from the bulk solvent. The β-ionone ring and the polyene chain is immobilized by multiple van der Waals interactions with the surrounding hydrophobic side chains. The carbonyl oxygen of 11-cis-retinaldehyde is positioned within hydrogen bonding distance from the side chains of Tyr and Glu residues (residues 180 and 220 in the human protein, respectively). The hydroxyl group of Tyr serves as a hydrogen donor for the carbonyl oxygen of the ligand, whereas the acidic oxygen of the Glu becomes a hydrogen acceptor for the hydroxyl of 11-cis-retinol.
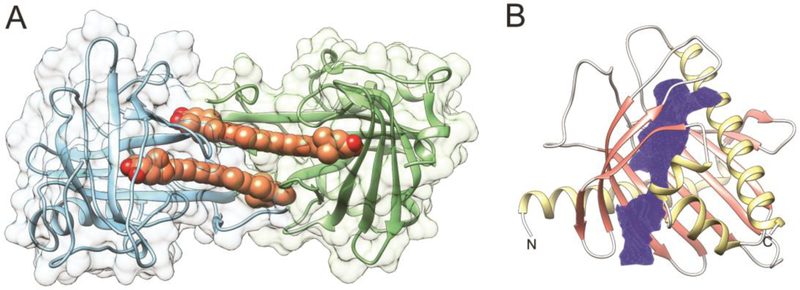
Figure 7– Structures of eye-specific retinoid-binding proteins.
(A) CRALBP in complex with 11-cis-retinaldehyde (PDB # 3HY5). The location of the retinoid molecule within the binding pocket is shown in orange. (B) The structure of module 2 from Xenopus laevis IRBP (PDB #1J7X). Two cavities that represent binding sites are marked in red, whereas a lipophilic hinge region is colored purple. The CavityPlus server was used to identify the intramolecular cavities and binding sites [58].
The second eye-specific retinoid-binding protein is IRBP. This relatively large (143 kDa) glycoprotein is secreted by photoreceptor cells into the intercellular space, where it constitutes the majority of soluble proteins [332]. IRBP is thought to be involved in the transport and buffering of visual cycle retinoids, and thus facilitates metabolic interactions between the neural retina and the RPE. In this model, IRBP promotes the release of 11-cis-retinaldehyde from the RPE and the re-uptake of ROL from rod outer segments [333, 334]. Surprisingly, although Irbp−/− mice show a progressive loss of photoreceptor nuclei and changes in the structure and organization of the receptor’s outer segments, they do not exhibit acute problems with vision or dark adaptation [335, 336]. These findings suggest that IRBP plays a subtler role, buffering the potentially cytotoxic effects of retinoids released from photoreceptors. It is also possible that IRBP is not explicitly involved in trafficking of retinoids. The lack of geometric specificity for the ligands, a relatively low affinity for retinoids (>100 nM), as well as the ability to interact with numerous other lipids, including fatty acids, α-tocopherol, and cholesterol indicates the more complex involvement of this protein in lipid transport [337-340].
A complete high resolution structure of IRBP has yet to be reported. The early biochemical and electron microscopy data indicate that the protein folds into four distinct modules of significant homology [340-342]. The polypeptide chain adopts a flexible, elongated shape, which undergoes conformational rearrangement upon saturation with ROL [341]. Each of the modules represents a functional unit as 3 to 4 independent binding sites have been postulated within IRBP [343, 344]. Partial information about the molecular organization of this protein can be inferred from the crystal structure of an individual module of Xenopus laevis IRBP [345]. This functional unit is composed of two domains (A and B) and a lipophilic hinge region (Fig. 7B). Interestingly, the two domain architecture as well as α-β-α sandwich motif present in IRBP resembles the structure of CRALBP (Figs. 7A, B). Two putative binding sites were identified within this structure, one in the hydrophobic cleft at the hinge region, the second within the α-β-α-fold of domain B. An additional smaller cavity can be seen in the β-β-α-spiral of domain A. Unfortunately, the lack of clear electron density for a retinoid ligand does not allow for the precise identification of the site of interaction. However, the crystal structure of a single module of related IRBP from zebrafish clearly indicated the presence of a single molecule of oleic acid that occupied the cavity within domain A [346]. The fluorescence titration of IRBP with ROL in the presence of fatty acids indicated only partial competition for the binding. Thus, retinoids and fatty acids interact with the protein simultaneously utilizing distinct binding sites. This finding only adds to the complexity of IRBP’s role in ocular lipid transport and requires further investigation.
6.5. A quest for carotenoid-binding proteins in vertebrates
Carotenoid-binding proteins (CBPs) have been identified and extensively studied in plants and cyanobacteria, where they are involved in the fundamental processes of light harvesting and protection against photo- and free radical-driven cell damage [347-349]. Representatives of CBPs have also been characterized in invertebrates. Crustacyanin isolated from the exoskeleton of a lobster, Homarus gammarus, is a carotenoprotein biological pigment (Fig. 8A) [350, 351]. The interaction of astaxanthin with the protein scaffold causes a bathochromic shift in the absorbance spectrum of this carotenoid that gives a deep blue color to the lobster’s shell [352]. A lutein-specific binding protein was identified in the silkworm, Bombyx mori [353]. This StART domain-containing protein has been genetically and functionally identified as an intracellular carrier involved in the facilitated intestinal uptake of carotenoids [354]. Despite these well-studied examples, relatively little information is available about specific CBPs in vertebrates. Yet, similarly to retinoid-binding proteins, they are likely to play a pivotal role in carotenoid absorption, transport between cellular organelles, and tissue-specific deposition. A great example of the latest is the selective accumulation of lutein and zeaxanthin in the macula of human and primate eyes. The concentration of these carotenoids in the foveal region of the macula exceeds 1 mM and is 100-fold higher than in the peripheral retina [355, 356]. Notably, the amount of lutein and zeaxanthin present in the macula is dynamic and depends on the dietary intake of these carotenoids [357-359]. This phenomenon suggests a specific mechanism for the deposition of selected non-provitamin A xanthophylls that might be driven by CBPs.
Figure 8– Carotenoid-binding proteins.
(A) The mode of astaxanthin binding by β-crustacyanin from Homarus gammarus (PDB # 1GKA). The accommodation of the relatively large ant stiff carotenoid ligand is achieved by the homodimerization of the protein. Notably, the protein environment at each end of the carotenoid molecule is dissimilar. (B) A crystal structure of human StARD3 (PDB # 5I9J). The large internal cavity representing the lipid-interaction site is shown in blue.
So far, the quest for vertebrate CBPs has not been decisive. Attempts to purify CBPs from livers isolated from ferrets or quails failed to report the amino acid sequences of the candidate proteins [360, 361]. Other studies indicated that ubiquitous albumin, β-lactoglobulin, or α-actinin can serve as non-specific carrier proteins for carotenoids [362-364]. Because of the high selectivity and enhanced deposition of carotenoids in the retina, this tissue becomes a source of putative CBPs. The fractionation of proteins associated with xanthophylls from human macula led to identification of glutathione-S-transferase P1 isoform (GSTP1) as a putative zeaxanthin-binding protein [365]. However, it seems controversial that this ubiquitous enzyme of established physiological significance can act as a specific zeaxanthine binding partner in the ocular tissue. GSTP1 is a small (~24 kDa) globular protein with a single glutathione-binding site located within the thioredoxin-like domain [366]. Thus, the accommodation of large planar ligands such as zeaxanthins might be problematic without exposing a large portion of the hydrophobic ligand to the bulk solvent. Even more so at the proposed carotenoid/protein stoichiometry of 2:1 [365]. Also, human GSTP1 is readily crystallizable as evidenced by dozens of PDB depositions. Thus, the determination of the specificity of this protein’s interaction with zeaxanthins at the atomic level is achievable and awaits ultimate structural verification.
Interestingly, the isolation of carotenoid-rich fractions from human retina revealed an additional protein associated with lutein that immunologically cross-reacted with antibodies against the silkworm lutein-binding protein [367, 368]. This finding led to the identification of human steroidogenic acute regulatory domain protein 3 (StARD3) as a lutein-binding protein in the retina. Unlike GSTP1, StARD3 contains a canonical StART domain that serves as a versatile binding interface for lipids common in plants and animals [369, 370]. The main structural feature of StARD3 is the helix-grip fold with a nine-stranded curved β-sheet and three α-helices enclosing a large, predominantly hydrophobic tunnel that extends through nearly the entire length of the protein (~24 Å) (Fig. 8B) [371, 372]. The most unusual feature of StARD3 as compared to others StART domain proteins is that the binding cavity is accessible by two openings at each end of the molecule. Unfortunately, attempts to co-crystallize StARD3 with lutein did not reveal the electron density for this ligand [372]. Docking experiments exposed several limitations that might prevent the efficient interaction of carotenoids with this carrier protein. The binding site is significantly shorter than zeaxanthin, which stretches over 30 Å long. Thus, it would be impossible to completely fit the entire molecule into the cavity, exposing ionone rings to the solvent. However, this mode of binding has its precedence in recently discovered small helical carotenoid proteins from cyanobacteria [373]. Nevertheless, the preferred orientation of the zeaxanthin moiety in StARD3 incurs at least 14 steric clashes with atoms of the protein scaffold, which is unprecedented in other carotenoid binding proteins [372, 374, 375]. Also, the binding pocket features a large polar side chain of Arg351 that potentially restricts a ligand from penetrating the binding pocket. StARD3 has been extensively studied in the context of cholesterol binding and intracellular trafficking [376, 377]. Notably, Arg351 has been proposed to serve as the main hydrogen bonding contact for cholesterol’s 3-hydroxyl group [371]. However, like zeaxanthin, StARD3 has never been co-crystalized with a sterol ligand. In the light of inconclusive biochemical data, the future phenotypic examination of Stard3−/− mice in the context of carotenoid uptake and accumulation seems to be a rational approach for verifying the function of this protein in vivo [378].
7. RECEPTOR-FACILITATED CELLULAR UPTAKE OF CIRCULATING VITAMIN A
7.1. STRA6 represents a unique class of cell surface receptors
The main benefit of a complex system of vitamin A absorption and liver storage is the ability to maintain retinoid homeostasis independently from a varying amount of dietary vitamin A intake. The concentration of circulating holo-RBP4 is highly regulated and remains constant (2-3 μM), except in cases of extreme vitamin A deficiency or certain physiological abnormalities [379-381]. However, the accessibility of the vitamin A released by the liver does not explain the mechanism or specificity of ROL uptake by selected cell types. Although earlier studies indicated the existence of an RBP4-specific receptor on the surface of epithelial cells, its role has remained controversial for over three decades [382-384]. Finally, a biochemical approach using the photo-cross-linking method resulted in the identification of a candidate protein that constituted an integral membrane protein encoded by the STRA6 (stimulated by retinoic acid 6) gene [385]. The subsequent analysis of STRA6 activity revealed that this receptor facilitated the binding of holo-RBP4 to the cell surface, catalyzed the release of ROL from the binding protein, and promoted ROL partition into the plasma membrane [386]. Interestingly, the mechanism by which STRA6 exerts its biological function escapes the traditional classification of cellular receptors. STRA6’s action does not depend on endocytosis, does not require energy, and it is not driven by cotransport or an electrochemical gradient [387]. Instead, this receptor functions in association with two other proteins, CRBP1 and LRAT, the biological activities of which facilitate vitamin A uptake into a cell [387, 388]. Apo-CRBP1 stimulates STRA6’s activity by increasing retinol-binding capacity in the cytoplasm. However, the main factor is the esterification of ROL by LRAT, which creates a chemical gradient preventing the premature saturation of uptake [205, 385, 387, 388]. Remarkably, the action of STRA6 is bidirectional. In the presence of apo-RBP4, STRA6 promotes the efflux of ROL through loading of this carrier protein with the ligand as shown in vitro and in tissue culture assays [386, 388]. Thus, STRA6 can be considered as a founding member of a new class of cell-surface receptors [389].
Concomitantly with the identification of STRA6 as a RBP4 receptor, independent studies show that mutations in STRA6 cause Matthew–Wood syndrome [390, 391]. This rare inherited disease is associated with the combination of developmental abnormalities that affect the eyes (microphthalmia), heart and lungs. Additionally, brain anomalies, mental retardation, and malformations of the kidneys have been reported in affected individuals [392, 393]. The clinical manifestations of Matthew–Wood syndrome strongly suggests a link between the STRA6 gene and retinoid homeostasis. The spectrum of developmental abnormalities may reflect not only the severity of particular mutations, but also differences in maternal vitamin A status [394]. The effects of a loss of Stra6 function in Strct6−/− mice are less severe, and limited predominantly to the ocular tissues. When kept on a vitamin A sufficient diet, these mice show a slightly smaller eye diameter and shortened outer segments of photoreceptor cells [395-397]. Retinoid levels in the RPE and retina were dramatically reduced, impairing phototransduction; however, this phenotype can be partially reversed by giving the effected mice pharmacological doses of vitamin A [396]. Notably, the concentration of retinoids in other tissues was no different than in wild-type mice. The physiological role of STRA6 was best illustrated during dietary vitamin A restriction. In Stra6−/− mice, ocular vitamin A content dropped below detection level after 4 months, whereas the same dietary restrictions resulted in an increase of retinoid amount in wild-type mice [396]. This finding indicates that STRA6 is mandatory for ocular vitamin A uptake from circulating holo-RBP4.
Although the role of STRA6 in extrahepatic retinoid hemostasis is established, much less is known about the re-uptake of circulating retinol from holo-RBP4 by the liver. Despite the lack of expression of STAR6 in liver cells, hepatocytes reveal a high affinity for RBP4 and are capable of the efficient uptake of retinol bound to RBP4 [398, 399]. These initial observations led to the hypothesis that there is a liver-specific receptor for RBP4 [400]. It has been identified 25 years later as a product of the 1300002K09Rik gene and named RBP4 receptor-2 (Rbpr2) [401]. Consistently with its proposed function, RBPR2 is highly expressed in the liver and to a lesser degree in the small intestine, spleen, and the adipocytes of mice. While RBPR2 shares overall low sequence homology with STRA6, it has a similar molecular mass (70.1 kDa) and a predicted membrane topology that closely resembles that of STRA6 [401, 402]. Interestingly, RBPR2 is a single polypeptide chain protein in vertebrates with exception of human and other primates, where the protein is encoded by two separate genes located on chromosome 9. This unusual arrangement implies a need for the posttranscriptional assembly of the two unequal parts of the receptor into a functional unit. So far, the RBPR2-dependent uptake of retinol was confirmed in tissue culture experiments and in zebrafish models. The deletion of rbpr2 in zebrafish impaired the transport and distribution of retinol from the yolk of developing larvae, leading to developmental defects associated with vitamin A deficiency such as microphthalmia, hydrocephaly, and cardiac edema [403]. Because of the specificity of the zebrafish model, these results cannot be directly translated into the physiological function of RBPR2 in mammals. Therefore, the characterization of the phenotypic consequences of RBPR2 deficiency in mice is necessary to definitely elucidate the physiological role of this receptor.
7.2. A functional dimer of STRA6 is a gatekeeper for vitamin A entering the cells
In the absence of structural information, the intriguing mechanism by which STRA6 acquires vitamin A bound to RBP4 remained controversial. Initial attempts to crystalize the receptor failed despite highly purified, functional, and monodisperse protein preparations [402, 404]. More recent screening for stable orthologs of mammalian STRA6, however, has led to the selection of the zebra fish form of this receptor for further research. Cryo-electron microscopy images of the zebrafish receptor allowed for determination of the molecular architecture of this receptor [402]. STRA6 assembles as an intricate, symmetric dimer with a large surface area (4,811 Å2) buried at the promoter interface (Fig. 9A). The dimer is anchored in the phospholipid membrane by 18 transmembrane (nine per monomer) and two horizontal intramembrane α-helices. The first five N-terminal α-helices of STRA6 fold into a distinct bundle that is separated from the rest of the structure by an intracellular domain. Long α-helices 6 and 7 extend above the plane of the lipid membrane at the extracellular side. Interacting with the same structural motif of the neighboring protomer they form a “dome” towering above an extensive hydrophobic cleft (Fig. 9A). Mutagenesis studies identified the apex of the dome-like structure as a putative site for holo-RBP4 binding [405]. Thus, it is reasonable to assume that holo-RBP4 interacts with this portion of the dome, which in turn weakens its affinity for ROL, leading to the deposition of ROL into the STRA6 outer cleft [402]. Interestingly, the cleft does not extend across the lipid bilayer, but rather ends about halfway through with a lateral exit window (Fig. 9B). This finding confirmed the previously observed phenomenon that ROL diffuses from holo-RBP4 via STRA6 into the phospholipid membrane [387]. However, it remains to be determined whether ROL release is achieved through conformational changes in RBP4 or by the enforced proximity of ROL to the highly hydrophobic interface of STRA6. The structure of a complex between holo-RBP4 and its receptor would definitely be informative, bringing new insights into this aspect of STRA6 function.
Figure 9– Molecular architecture of STRA6 in complex with calmodulin.
(A) The cryo-EM dimeric structure and lipid membrane topology of the receptor. An individual protomer is shown in color. The numbers indicate transmembrane helices present within the protein. Calmodulin interacting at the intracellular site of STRA6 is shown in pink. Two cavities that represent putative small molecule-binding sites are colored in red. IMH – intramembrane helix. (B) A surface representation of STRA6. Colors from blue to red indicate increasing hydrophobicity. The location of the binding site for holo-RBP4 and a hydrophobic outer cleft as well as the presence of a well-defined lateral window suggest a putative ROL transfer pathway from the binding-protein to the plasma membrane (indicated with green arrows).
It is established that STRA6 is regulated transcriptionally in a retinoic acid-dependent manner [396, 406, 407]. Unexpectedly, the structure of STRA6 suggests alternative modes of regulation that can directly affect STRA6 activity. The receptor was found to co-purify with calmodulin, which binds tightly to the C-terminal cytoplasmic site of STRA6 (Fig. 9A) [402]. This Ca2+-dependent interaction occurs in a noncanonical manner. Importantly, the residues forming the STRA6/calmodulin interface are conserved across all species. In fact, the co-purification of calmodulin with STRA6 was also observed in the bovine proteins (communication by Dr. Hui Sun at the FASEB meeting 2013). Moreover, the peptide derived from the human receptor interacts with Ca2+-loaded calmodulin, forming a stable complex that readily crystalized [402]. Despite the structural data, the role of calmodulin in STRA6 function remains elusive. Possible scenarios of regulation may include calmodulin-induced conformational changes in the receptor that in turn affect ROL diffusion or block interaction with other cytoplasmic partners. Although two polymorphisms in human STRA6 (Arg655/Cys and Thr644/Met) located at the calmodulin binding interface are associated with Matthew-Wood syndrome, there is no experimental evidence showing a functional relationship between Ca2+ signaling and ROL uptake [390]. Another intriguing feature of STRA6 are two well-defined cavities within the N-terminal domain that constitute potential allosteric ligand binding sites (Fig. 9A). The larger pocket is located inside the bundle of α-helices and can be accessed from the extracellular side of the receptor, whereas a much smaller cavity is found on the cytoplasmic side. A potential ligand interacting at these binding sites has not yet been identified. Although, it is unclear whether the activity STRA6 can be modulated by small molecule compounds, studies in this direction could lead to establishing STRA6 as a potential pharmacological target for treating diseases related to an imbalance in retinoid homeostasis.
8. CONCLUSIONS AND FUTURE PROSPECTS
The past two decades have brought unprecedented progress in our understanding of carotenoid and retinoid metabolism at the molecular level. The advances in structural biology reveal the details of the atomic organizations of key receptors, enzymes and transport proteins, providing a mechanistic explanation for their physiological functions. High-resolution structures in combination with modern experimental approaches have also verified old assumptions and became the basis for new data-driven hypotheses. Although a lot has already been accomplished, it is important to keep this momentum, as there are still many fundamental aspects of vitamin A biochemistry that require further investigation. We list a few subjective areas of interest, in which progress will be impactful on the field.
While the facilitated uptake of carotenoids by SCARB1 is well documented, the principles of the activity and selectivity of this receptor cannot be fully understood without structural data. The recent successful crystallization of the related CD36 and LIMP-2 receptors suggests that the determination of a high-resolution structure for SCARB1 is not out of reach [47, 48]. Such a structure in complex with a lipid molecule would be particularly interesting. Observing the electron density of a carotenoid within the receptor’s cavity would provide insightful knowledge about the pathway channeling hydrophobic molecules from the food matrix into enterocytes and potential methods of controlling the bioavailability of carotenoids.
The cloning of BCDO1 and BCDO2 and the establishment of their role in retinoid production provided a molecular explanation for the vitamin A-like biological activity of certain carotenoids first reported nearly 100 years ago [17, 69]. Although the activity of these enzymes has been characterized in detail, the molecular principles for their substrate specificity and Fe2+-dependent catalysis await further investigation. Also, BCDOs have tremendous significance for carotenoid and retinoid homeostasis. Ongoing and future studies on the effects of polymorphisms in human BCDO1 and BCDO2 genes may have important consequences for understanding the complex role of carotenoids in the health status of a population [15].
The molecular identification and structural determination of the architecture of STRA6 greatly improved our knowledge of how vitamin A is distributed between tissues [385, 402, 408]. However, the interaction of holo-RBP4 with the receptor, as well as the details of ROL channeling through STRA6 remain to be clarified. In this context, the trapping and cryo-EM imaging of a complex between RBP4 and STAR6 would be of particular interest. Such a structure may also shed new light on the role of TTR in docking of holo-RBP4 at the receptor surface. It remains to be determined whether the regulation of STRA6 activity is due to interaction with calmodulin or some other small molecule modulators since the transcriptional regulation of STRA6 expression by RA seems not to be the only mechanism that governs the cellular uptake of vitamin A.
Although CRBPs are one of the most studied classes of proteins involved in retinoid trafficking, it remains unknown how they acquire ROL and which factors allow for the liberation and effective delivery of this ligand to retinol-processing enzymes. It is clear that this cannot be a stochastic event, but rather a controlled process facilitated by specific interactions with phospholipids or protein binding partners. Unfortunately, meaningful clues about the functional dynamics of CRBPs cannot be inferred directly from their structures. This important aspect of vitamin A biochemistry needs to be revisited using a combination of biophysical techniques and more advanced methods for detecting and stabilizing transient protein-protein or protein-lipid membrane interactions. Another emerging aspect of retinoid-binding protein activity is their ability to bind endogenous non-retinoid lipids, as this has been recently shown for RBP4 and CRBP2 [253]. This previously unanticipated competition between lipids may have important consequences for the absorption and trafficking of retinoids.
Finally, the components of a feedback loop that functionally links the concentration of ROL in the blood with the mobilization of retinoids stored in the liver are currently unidentified. Any progress in the matter would open new research avenues for a better understanding of retinoid biology and, in the long term, could provide key insight into the effects of an imbalance in retinoid homeostasis on the health of humans.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Loredana Quadro for the invitation to contribute this article to the special issue of Biochimica et Biophysica Acta. Also, we would like to acknowledge the contributions of Josie Silvaroli and Wiktor Golczak in the editing and revision of this manuscript. This work was supported by the National Institute of Health grant EY023948 (to M.G.).
Abbreviations:
- 3D
three-dimensional
- ALDH
aldehyde dehydrogenase
- AKRs
aldo–keto reductases
- AWAT2
acyl-CoA wax alcohol acyltransferase 2
- BCDO1
β-carotene-15,15’-dioxygenase; BCDO2 β-carotene-9’,10’-dioxygenase
- CBPs
carotenoid-binding proteins
- CCOs
carotenoids cleavage oxygenases
- CD36
cluster determinant 36 receptor
- CRABPs
cellular retinoic acid-binding proteins; CRALBP, cellular retinaldehyde-binding protein
- CRBPs
cellular retinol-binding proteins
- DGAT1
diacylglycerol O-acyltransferase 1
- GSTP1
glutathione-S-transferase P1 isoform
- HDL
high-density lipoproteins
- HRASLS
H-ras-like tumor suppressor
- IRBP
interphotoreceptor retinol-binding protein
- LIMP-2
lysosomal integral membrane protein-2
- LRAT
lecithin:retinol acyltransferase
- MFAT
multifunctional O-acyltransferase
- NSE
neurological sensory protein
- RA
all-trans-retinoic acid
- RAL
all-trans-retinaldehyde
- RARs
retinoic acid nuclear receptors
- RBP4
retinol-binding protein 4
- RBPR2
retinol-binding protein receptor-2
- RDHs
retinol dehydrogenases
- ROL
all-trans-retinol
- SDRs
short-chain dehydrogenase/reductases
- SCARB1
class B scavenger receptor type I
- StARD3
steroidogenic acute regulatory domain protein 3
- STRA6
stimulated by retinoic acid 6
- TTR
transthyretin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Wolf G, The discovery of the visual function of vitamin A, J Nutr, 131 (2001) 1647–1650. [DOI] [PubMed] [Google Scholar]
- [2].Duester G, Retinoic acid synthesis and signaling during early organogenesis, Cell, 134 (2008) 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mark M, Ghyselinck NB, Chambon P, Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis, Annu Rev Pharmacol Toxicol, 46 (2006) 451–480. [DOI] [PubMed] [Google Scholar]
- [4].Means AL, Gudas LJ, The roles of retinoids in vertebrate development, Annu Rev Biochem, 64 (1995) 201–233. [DOI] [PubMed] [Google Scholar]
- [5].Maden M, Retinoic acid in the development, regeneration and maintenance of the nervous system, Nat Rev Neurosci, 8 (2007) 755–765. [DOI] [PubMed] [Google Scholar]
- [6].Zhao Z, Murasko DM, Ross AC, The role of vitamin A in natural killer cell cytotoxicity, number and activation in the rat, Nat Immun, 13 (1994) 29–41. [PubMed] [Google Scholar]
- [7].Ross AC, Vitamin A supplementation and retinoic acid treatment in the regulation of antibody responses in vivo, Vitam Horm, 75 (2007) 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Herman MA, Kahn BB, Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony, J Clin Invest, 116 (2006) 1767–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wald G, The molecular basis of visual excitation, Nature, 219 (1968) 800–807. [DOI] [PubMed] [Google Scholar]
- [10].Palczewski K, G protein-coupled receptor rhodopsin, Annu Rev Biochem, 75 (2006) 743–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hubbard R, Wald G, Cis-trans isomers of vitamin A and retinene in the rhodopsin system, J Gen Physiol, 36 (1952) 269–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kiser PD, Golczak M, Palczewski K, Chemistry of the retinoid (visual) cycle, Chem Rev, 114 (2014) 194–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].D'Ambrosio DN, Clugston RD, Blaner WS, Vitamin A metabolism: an update, Nutrients, 3 (2011) 63–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Blaner WS, Li Y, Brun PJ, Yuen JJ, Lee SA, Clugston RD, Vitamin A Absorption, Storage and Mobilization, Subcell Biochem, 81 (2016) 95–125. [DOI] [PubMed] [Google Scholar]
- [15].von Lintig J, Provitamin A metabolism and functions in mammalian biology, Am J Clin Nutr, 96 (2012) 1234S–1244S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yabuzaki J, Carotenoids Database: structures, chemical fingerprints and distribution among organisms, Database (Oxford), 2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Moore T, Vitamin A and carotene: The absence of the liver oil vitamin A from carotene. VI. The conversion of carotene to vitamin A in vivo, Biochem J, 24 (1930) 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].von Lintig J, Hessel S, Isken A, Kiefer C, Lampert JM, Voolstra O, Vogt K, Towards a better understanding of carotenoid metabolism in animals, Biochim Biophys Acta, 1740 (2005) 122–131. [DOI] [PubMed] [Google Scholar]
- [19].Moore T, Vitamin A, Amsterdam (London, New York & Prince-ton): Elsevier Publishing Company, 1957. [Google Scholar]
- [20].Erdman JW Jr., Bierer TL, Gugger ET, Absorption and transport of carotenoids, Ann N Y Acad Sci, 691 (1993) 76–85. [DOI] [PubMed] [Google Scholar]
- [21].Borel P, Factors affecting intestinal absorption of highly lipophilic food microconstituents (fat-soluble vitamins, carotenoids and phytosterols), Clin Chem Lab Med, 41 (2003) 979–994. [DOI] [PubMed] [Google Scholar]
- [22].Cervantes-Paz B, Victoria-Campos CI, Ornelas-Paz JD, Absorption of Carotenoids and Mechanisms Involved in Their Health-Related Properties, Carotenoids in Nature: Biosynthesis, Regulation and Function, 79 (2016) 415–454. [DOI] [PubMed] [Google Scholar]
- [23].Bohn T, Desmarchelier C, Dragsted LO, Nielsen CS, Stahl W, Ruhl R, Keijer J, Borel P, Host-related factors explaining interindividual variability of carotenoid bioavailability and tissue concentrations in humans, Mol Nutr Food Res, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].During A, Hussain MM, Morel DW, Harrison EH, Carotenoid uptake and secretion by CaCo-2 cells: beta-carotene isomer selectivity and carotenoid interactions, J Lipid Res, 43 (2002) 1086–1095. [DOI] [PubMed] [Google Scholar]
- [25].Hageman SH, She L, Furr HC, Clark RM, Excess vitamin E decreases canthaxanthin absorption in the rat, Lipids, 34 (1999) 627–631. [DOI] [PubMed] [Google Scholar]
- [26].Fahy DM, O'Callaghan YC, O'Brien NM, Phytosterols: lack of cytotoxicity but interference with beta-carotene uptake in Caco-2 cells in culture, Food Addit Contam, 21 (2004) 42–51. [DOI] [PubMed] [Google Scholar]
- [27].Kiefer C, Sumser E, Wernet MF, Von Lintig J, A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila, Proc Natl Acad Sci U S A, 99 (2002) 10581–10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M, Identification of scavenger receptor SR-BI as a high density lipoprotein receptor, Science, 271 (1996) 518–520. [DOI] [PubMed] [Google Scholar]
- [29].Cai SF, Kirby RJ, Howles PN, Hui DY, Differentiation-dependent expression and localization of the class B type I scavenger receptor in intestine, J Lipid Res, 42 (2001) 902–909. [PubMed] [Google Scholar]
- [30].Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M, A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism, Proc Natl Acad Sci U S A, 94 (1997) 12610–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Van Eck M, Twisk J, Hoekstra M, Van Rij BT, Van der Lans CA, Bos IS, Kruijt JK, Kuipers F, Van Berkel TJ, Differential effects of scavenger receptor BI deficiency on lipid metabolism in cells of the arterial wall and in the liver, J Biol Chem, 278 (2003) 23699–23705. [DOI] [PubMed] [Google Scholar]
- [32].van Bennekum A, Werder M, Thuahnai ST, Han CH, Duong P, Williams DL, Wettstein P, Schulthess G, Phillips MC, Hauser H, Class B scavenger receptor-mediated intestinal absorption of dietary beta-carotene and cholesterol, Biochemistry, 44 (2005) 4517–4525. [DOI] [PubMed] [Google Scholar]
- [33].Widjaja-Adhi MA, Lobo GP, Golczak M, Von Lintig J, A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption, Hum Mol Genet, 24 (2015) 3206–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].During A, Doraiswamy S, Harrison EH, Xanthophylls are preferentially taken up compared with beta-carotene by retinal cells via a SRBI-dependent mechanism, J Lipid Res, 49 (2008) 1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shyam R, Vachali P, Gorusupudi A, Nelson K, Bernstein PS, All three human scavenger receptor class B proteins can bind and transport all three macular xanthophyll carotenoids, Arch Biochem Biophys, 634 (2017) 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tsuchida K, Sakudoh T, Recent progress in molecular genetic studies on the carotenoid transport system using cocoon-color mutants of the silkworm, Arch Biochem Biophys, 572 (2015) 151–157. [DOI] [PubMed] [Google Scholar]
- [37].Sundvold H, Helgeland H, Baranski M, Omholt SW, Vage DI, Characterisation of a novel paralog of scavenger receptor class B member I (SCARB1) in Atlantic salmon (Salmo salar), BMC Genet, 12 (2011) 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Toomey MB, Lopes RJ, Araujo PM, Johnson JD, Gazda MA, Afonso S, Mota PG, Koch RE, Hill GE, Corbo JC, Carneiro M, High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds, Proc Natl Acad Sci U S A, 114 (2017) 5219–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ljunggren SA, Levels JH, Hovingh K, Holleboom AG, Vergeer M, Argyri L, Gkolfinopoulou C, Chroni A, Sierts JA, Kastelein JJ, Kuivenhoven JA, Lindahl M, Karlsson H, Lipoprotein profiles in human heterozygote carriers of a functional mutation P297S in scavenger receptor class B1, Biochim Biophys Acta, 1851 (2015) 1587–1595. [DOI] [PubMed] [Google Scholar]
- [40].Borel P, Moussa M, Reboul E, Lyan B, Defoort C, Vincent-Baudry S, Maillot M, Gastaldi M, Darmon M, Portugal H, Planells R, Lairon D, Human plasma levels of vitamin E and carotenoids are associated with genetic polymorphisms in genes involved in lipid metabolism, J Nutr, 137 (2007) 2653–2659. [DOI] [PubMed] [Google Scholar]
- [41].McKay GJ, Loane E, Nolan JM, Patterson CC, Meyers KJ, Mares JA, Yonova-Doing E, Hammond CJ, Beatty S, Silvestri G, Investigation of genetic variation in scavenger receptor class B, member 1 (SCARB1) and association with serum carotenoids, Ophthalmology, 120 (2013) 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zanoni P, Khetarpal SA, Larach DB, Hancock-Cerutti WF, Millar JS, Cuchel M, DerOhannessian S, Kontush A, Surendran P, Saleheen D, Trompet S, Jukema JW, De Craen A, Deloukas P, Sattar N, Ford I, Packard C, Majumder A, Alam DS, Di Angelantonio E, Abecasis G, Chowdhury R, Erdmann J, Nordestgaard BG, Nielsen SF, Tybjaerg-Hansen A, Schmidt RF, Kuulasmaa K, Liu DJ, Perola M, Blankenberg S, Salomaa V, Mannisto S, Amouyel P, Arveiler D, Ferrieres J, Muller-Nurasyid M, Ferrario M, Kee F, Willer CJ, Samani N, Schunkert H, Butterworth AS, Howson JM, Peloso GM, Stitziel NO, Danesh J, Kathiresan S, Rader DJ, Consortium CHDE, Consortium CAE, Global Lipids Genetics C, Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease, Science, 351 (2016) 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bachmann H, Desbarats A, Pattison P, Sedgewick M, Riss G, Wyss A, Cardinault N, Duszka C, Goralczyk R, Grolier P, Feedback regulation of beta,beta-carotene 15,15'-monooxygenase by retinoic acid in rats and chickens, J Nutr, 132 (2002) 3616–3622. [DOI] [PubMed] [Google Scholar]
- [44].Lobo GP, Hessel S, Eichinger A, Noy N, Moise AR, Wyss A, Palczewski K, von Lintig J, ISX is a retinoic acid-sensitive gatekeeper that controls intestinal beta,beta-carotene absorption and vitamin A production, FASEB J, 24 (2010) 1656–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lobo GP, Amengual J, Baus D, Shivdasani RA, Taylor D, von Lintig J, Genetics and diet regulate vitamin A production via the homeobox transcription factor ISX, J Biol Chem, 288 (2013) 9017–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Widjaja-Adhi MAK, Palczewski G, Dale K, Knauss EA, Kelly ME, Golczak M, Levine AD, von Lintig J, Transcription factor ISX mediates the cross talk between diet and immunity, Proc Natl Acad Sci U S A, 114 (2017) 11530–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hsieh FL, Turner L, Bolla JR, Robinson CV, Lavstsen T, Higgins MK, The structural basis for CD36 binding by the malaria parasite, Nat Commun, 7 (2016) 12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Neculai D, Schwake M, Ravichandran M, Zunke F, Collins RF, Peters J, Neculai M, Plumb J, Loppnau P, Pizarro JC, Seitova A, Trimble WS, Saftig P, Grinstein S, Dhe-Paganon S, Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36, Nature, 504 (2013) 172–176. [DOI] [PubMed] [Google Scholar]
- [49].Webb NR, de Villiers WJ, Connell PM, de Beer FC, van der Westhuyzen DR, Alternative forms of the scavenger receptor BI (SR-BI), J Lipid Res, 38 (1997) 1490–1495. [PubMed] [Google Scholar]
- [50].Webb NR, Connell PM, Graf GA, Smart EJ, de Villiers WJ, de Beer FC, van der Westhuyzen DR, SR-BII, an isoform of the scavenger receptor BI containing an alternate cytoplasmic tail, mediates lipid transfer between high density lipoprotein and cells, J Biol Chem, 273 (1998) 15241–15248. [DOI] [PubMed] [Google Scholar]
- [51].Pepino MY, Kuda O, Samovski D, Abumrad NA, Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism, Annu Rev Nutr, 34 (2014) 281–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rasmussen JT, Berglund L, Rasmussen MS, Petersen TE, Assignment of disulfide bridges in bovine CD36, Eur J Biochem, 257 (1998) 488–494. [DOI] [PubMed] [Google Scholar]
- [53].Yu M, Lau TY, Carr SA, Krieger M, Contributions of a disulfide bond and a reduced cysteine side chain to the intrinsic activity of the high-density lipoprotein receptor SR-BI, Biochemistry, 51 (2012) 10044–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hu J, Zhang Z, Shen WJ, Nomoto A, Azhar S, Differential roles of cysteine residues in the cellular trafficking, dimerization, and function of the high-density lipoprotein receptor, SR-BI, Biochemistry, 50 (2011) 10860–10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yu M, Romer KA, Nieland TJ, Xu S, Saenz-Vash V, Penman M, Yesilaltay A, Carr SA, Krieger M, Exoplasmic cysteine Cys384 of the HDL receptor SR-BI is critical for its sensitivity to a small-molecule inhibitor and normal lipid transport activity, Proc Natl Acad Sci U S A, 108 (2011) 12243–12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T, SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res, 46 (2018) W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem, 25 (2004) 1605–1612. [DOI] [PubMed] [Google Scholar]
- [58].Xu Y, Wang S, Hu Q, Gao S, Ma X, Zhang W, Shen Y, Chen F, Lai L, Pei J, CavityPlus: a web server for protein cavity detection with pharmacophore modelling, allosteric site identification and covalent ligand binding ability prediction, Nucleic Acids Res, 46 (2018) W374–W379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Thuahnai ST, Lund-Katz S, Anantharamaiah GM, Williams DL, Phillips MC, A quantitative analysis of apolipoprotein binding to SR-BI: multiple binding sites for lipid-free and lipid-associated apolipoproteins, J Lipid Res, 44 (2003) 1132–1142. [DOI] [PubMed] [Google Scholar]
- [60].Connelly MA, Klein SM, Azhar S, Abumrad NA, Williams DL, Comparison of class B scavenger receptors, CD36 and scavenger receptor BI (SR-BI), shows that both receptors mediate high density lipoprotein-cholesteryl ester selective uptake but SR-BI exhibits a unique enhancement of cholesteryl ester uptake, J Biol Chem, 274 (1999) 41–47. [DOI] [PubMed] [Google Scholar]
- [61].Hu Z, Hu J, Shen WJ, Kraemer FB, Azhar S, A Novel Role of Salt-Inducible Kinase 1 (SIK1) in the Post-Translational Regulation of Scavenger Receptor Class B Type 1 Activity, Biochemistry, 54 (2015) 6917–6930. [DOI] [PubMed] [Google Scholar]
- [62].Moore T, The relation of carotin to vitamin A, Lancet, 2 (1929) 380–381. [Google Scholar]
- [63].Moore T, Vitamin A and carotene. The absence of the liver oil vitamin A from carotene. VI. The conversion of carotene to vitamin A in vivo., Biochemical Journal, 24 (1930) 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Steenbock H, White corn vs yellow corn and a probable relation between the fat-soluble vitamine and yellow plant pigments, Science, 50 (1919) 352–353. [DOI] [PubMed] [Google Scholar]
- [65].Moore T, Vitamin A and carotene. I. The association of vitamin A activity with carotene in the carrot root., Biochemical Journal, 23 (1929) 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Goodman DS, Huang HS, Biosynthesis of Vitamin a with Rat Intestinal Enzymes, Science, 149 (1965)879-&. [DOI] [PubMed] [Google Scholar]
- [67].Olson JA, Hayaishi O, The enzymatic cleavage of beta-carotene into vitamin A by soluble enzymes of rat liver and intestine, Proc Natl Acad Sci U S A, 54 (1965) 1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].von Lintig J, Dreher A, Kiefer C, Wernet MF, Vogt K, Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation invivo, Proc Natl Acad Sci U S A, 98 (2001) 1130–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].von Lintig J, Vogt K, Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal, J Biol Chem, 275 (2000) 11915–11920. [DOI] [PubMed] [Google Scholar]
- [70].Wyss A, Wirtz G, Woggon W, Brugger R, Wyss M, Friedlein A, Bachmann H, Hunziker W, Cloning and expression of beta,beta-carotene 15,15'-dioxygenase, Biochem Biophys Res Commun, 271 (2000) 334–336. [DOI] [PubMed] [Google Scholar]
- [71].Lindqvist A, Andersson S, Biochemical properties of purified recombinant human beta-carotene 15,15'-monooxygenase, J Biol Chem, 277 (2002) 23942–23948. [DOI] [PubMed] [Google Scholar]
- [72].Paik J, During A, Harrison EH, Mendelsohn CL, Lai K, Blaner WS, Expression and characterization of a murine enzyme able to cleave beta-carotene. The formation of retinoids, J Biol Chem, 276 (2001) 32160–32168. [DOI] [PubMed] [Google Scholar]
- [73].Takitani K, Zhu CL, Inoue A, Tamai H, Molecular cloning of the rat beta-carotene 15,15'-monooxygenase gene and its regulation by retinoic acid, Eur J Nutr, 45 (2006) 320–326. [DOI] [PubMed] [Google Scholar]
- [74].Redmond TM, Gentleman S, Duncan T, Yu S, Wiggert B, Gantt E, Cunningham FX Jr., Identification, expression, and substrate specificity of a mammalian beta-carotene 15,15'-dioxygenase, J Biol Chem, 276 (2001) 6560–6565. [DOI] [PubMed] [Google Scholar]
- [75].Amengual J, Lobo GP, Golczak M, Li HN, Klimova T, Hoppel CL, Wyss A, Palczewski K, von Lintig J, A mitochondrial enzyme degrades carotenoids and protects against oxidative stress, FASEB J, 25 (2011) 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hessel S, Eichinger A, Isken A, Amengual J, Hunzelmann S, Hoeller U, Elste V, Hunziker W, Goralczyk R, Oberhauser V, von Lintig J, Wyss A, CMO1 deficiency abolishes vitamin A production from beta-carotene and alters lipid metabolism in mice, J Biol Chem, 282 (2007) 33553–33561. [DOI] [PubMed] [Google Scholar]
- [77].Lindqvist A, Sharvill J, Sharvill DE, Andersson S, Loss-of-function mutation in carotenoid 15,15'-monooxygenase identified in a patient with hypercarotenemia and hypovitaminosis A, J Nutr, 137 (2007) 2346–2350. [DOI] [PubMed] [Google Scholar]
- [78].Glover J, The conversion of beta-carotene into vitamin A, Vitam Horm, 18 (1960) 371–386. [DOI] [PubMed] [Google Scholar]
- [79].Wolf G, The enzymatic cleavage of beta-carotene: still controversial, Nutr Rev, 53 (1995) 134–137. [DOI] [PubMed] [Google Scholar]
- [80].Kiefer C, Hessel S, Lampert JM, Vogt K, Lederer MO, Breithaupt DE, von Lintig J, Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A, J Biol Chem, 276 (2001) 14110–14116. [DOI] [PubMed] [Google Scholar]
- [81].Hu KQ, Liu C, Ernst H, Krinsky NI, Russell RM, Wang XD, The biochemical characterization of ferret carotene-9',10'-monooxygenase catalyzing cleavage of carotenoids in vitro and in vivo, J Biol Chem, 281 (2006) 19327–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kelly ME, Ramkumar S, Sun W, Colon Ortiz C, Kiser PD, Golczak M, von Lintig J, The Biochemical Basis of Vitamin A Production from the Asymmetric Carotenoid beta-Cryptoxanthin, ACS Chem Biol, 13 (2018) 2121–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Amengual J, Widjaja-Adhi MA, Rodriguez-Santiago S, Hessel S, Golczak M, Palczewski K, von Lintig J, Two carotenoid oxygenases contribute to mammalian provitamin A metabolism, J Biol Chem, 288 (2013) 34081–34096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Babino D, Palczewski G, Widjaja-Adhi MA, Kiser PD, Golczak M, von Lintig J, Characterization of the Role of beta-Carotene 9,10-Dioxygenase in Macular Pigment Metabolism, J Biol Chem, 290 (2015) 24844–24857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Dela Sena C, Sun J, Narayanasamy S, Riedl KM, Yuan Y, Curley RW Jr., Schwartz SJ, Harrison EH, Substrate Specificity of Purified Recombinant Chicken beta-Carotene 9',10'-Oxygenase (BCO2), J Biol Chem, 291 (2016) 14609–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lobo GP, Isken A, Hoff S, Babino D, von Lintig J, BCDO2 acts as a carotenoid scavenger and gatekeeper for the mitochondrial apoptotic pathway, Development, 139 (2012) 2966–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wu L, Guo X, Hartson SD, Davis MA, He H, Medeiros DM, Wang W, Clarke SL, Lucas EA, Smith BJ, von Lintig J, Lin D, Lack of beta, beta-carotene-9', 10'-oxygenase 2 leads to hepatic mitochondrial dysfunction and cellular oxidative stress in mice, Mol Nutr Food Res, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Strychalski J, Brym P, Czarnik U, Gugolek A, A novel AAT-deletion mutation in the coding sequence of the BCO2 gene in yellow-fat rabbits, J Appl Genet, 56 (2015) 535–537. [DOI] [PubMed] [Google Scholar]
- [89].Tian R, Pitchford WS, Morris CA, Cullen NG, Bottema CD, Genetic variation in the beta, beta-carotene-9', 10'-dioxygenase gene and association with fat colour in bovine adipose tissue and milk, Anim Genet, 41 (2010) 253–259. [DOI] [PubMed] [Google Scholar]
- [90].Vage DI, Boman IA, A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries), BMC Genet, 11 (2010) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].He M, Cornelis MC, Kraft P, van Dam RM, Sun Q, Laurie CC, Mirel DB, Chasman DI, Ridker PM, Hunter DJ, Hu FB, Qi L, Genome-wide association study identifies variants at the IL18-BCO2 locus associated with interleukin-18 levels, Arterioscler Thromb Vasc Biol, 30 (2010) 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kloer DP, Schulz GE, Structural and biological aspects of carotenoid cleavage, Cell Mol Life Sci, 63 (2006) 2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ, Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family, Plant J, 45 (2006) 982–993. [DOI] [PubMed] [Google Scholar]
- [94].Kamoda S, Saburi Y, Cloning, expression, and sequence analysis of a lignostilbene-alpha,beta-dioxygenase gene from Pseudomonas paucimobilis TMY1009, Biosci Biotechnol Biochem, 57 (1993) 926–930. [DOI] [PubMed] [Google Scholar]
- [95].Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE, The structure of a retinal-forming carotenoid oxygenase, Science, 308 (2005) 267–269. [DOI] [PubMed] [Google Scholar]
- [96].Messing SA, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM, Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid, Plant Cell, 22 (2010) 2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K, Crystal structure of native RPE65, the retinoid isomerase of the visual cycle, Proc Natl Acad Sci U S A, 106 (2009) 17325–17330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD, Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase, J Biol Chem, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Sui X, Kiser PD, Lintig J, Palczewski K, Structural basis of carotenoid cleavage: from bacteria to mammals, Arch Biochem Biophys, 539 (2013) 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Solomon EI, Decker A, Lehnert N, Non-heme iron enzymes: contrasts to heme catalysis, Proc Natl Acad Sci U S A, 100 (2003) 3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sui X, Golczak M, Zhang J, Kleinberg KA, von Lintig J, Palczewski K, Kiser PD, Utilization of Dioxygen by Carotenoid Cleavage Oxygenases, J Biol Chem, 290 (2015) 30212–30223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kiser PD, Zhang J, Badiee M, Li Q, Shi W, Sui X, Golczak M, Tochtrop GP, Palczewski K, Catalytic mechanism of a retinoid isomerase essential for vertebrate vision, Nat Chem Biol, 11 (2015) 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Leuenberger MG, Engeloch-Jarret C, Woggon WD, The Reaction Mechanism of the Enzyme-Catalyzed Central Cleavage of beta-Carotene to Retinal, Angew Chem Int Ed Engl, 40 (2001) 2613–2617. [DOI] [PubMed] [Google Scholar]
- [104].Marasco EK, Schmidt-Dannert C, Identification of bacterial carotenoid cleavage dioxygenase homologues that cleave the interphenyl alpha,beta double bond of stilbene derivatives via a monooxygenase reaction, Chembiochem, 9 (2008) 1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Schmidt H, Kurtzer R, Eisenreich W, Schwab W, The carotenase AtCCD1 from Arabidopsis thaliana is a dioxygenase, J Biol Chem, 281 (2006) 9845–9851. [DOI] [PubMed] [Google Scholar]
- [106].Borowski T, Blomberg MR, Siegbahn PE, Reaction mechanism of apocarotenoid oxygenase (ACO): a DFT study, Chemistry, 14 (2008) 2264–2276. [DOI] [PubMed] [Google Scholar]
- [107].Ryu JY, Seo J, Park S, Ahn JH, Chong Y, Sadowsky MJ, Hur HG, Characterization of an isoeugenol monooxygenase (Iem) from Pseudomonas nitroreducens Jin1 that transforms isoeugenol to vanillin, Biosci Biotechnol Biochem, 77 (2013) 289–294. [DOI] [PubMed] [Google Scholar]
- [108].Babino D, Golczak M, Kiser PD, Wyss A, Palczewski K, von Lintig J, The Biochemical Basis of Vitamin A3 Production in Arthropod Vision, ACS Chem Biol, 11 (2016) 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].dela Sena C, Riedl KM, Narayanasamy S, Curley RW Jr., Schwartz SJ, Harrison EH, The human enzyme that converts dietary provitamin A carotenoids to vitamin A is a dioxygenase, J Biol Chem, 289 (2014) 13661–13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Harrison PJ, Bugg TD, Enzymology of the carotenoid cleavage dioxygenases: reaction mechanisms, inhibition and biochemical roles, Arch Biochem Biophys, 544 (2014) 105–111. [DOI] [PubMed] [Google Scholar]
- [111].Sui X, Weitz AC, Farquhar ER, Badiee M, Banerjee S, von Lintig J, Tochtrop GP, Palczewski K, Hendrich MP, Kiser PD, Structure and Spectroscopy of Alkene-Cleaving Dioxygenases Containing an Atypically Coordinated Non-Heme Iron Center, Biochemistry, 56 (2017) 2836–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Katz ML, Gao CL, Rice LM, Formation of lipofuscin-like fluorophores by reaction of retinal with photoreceptor outer segments and liposomes, Mech Ageing Dev, 92 (1996) 159–174. [DOI] [PubMed] [Google Scholar]
- [113].Sparrow JR, Vitamin A-aldehyde adducts: AMD risk and targeted therapeutics, Proc Natl Acad Sci U S A, 113 (2016) 4564–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Niemela O, Aldehyde-protein adducts in the liver as a result of ethanol-induced oxidative stress, Front Biosci, 4 (1999) D506–513. [DOI] [PubMed] [Google Scholar]
- [115].Uchida K, Role of reactive aldehyde in cardiovascular diseases, Free Radic Biol Med, 28 (2000) 1685–1696. [DOI] [PubMed] [Google Scholar]
- [116].Park JW, Jung KH, Lee JH, Moon SH, Cho YS, Lee KH, Inhibition of aldehyde dehydrogenase 1 enhances the cytotoxic effect of retinaldehyde on A549 cancer cells, Oncotarget, 8 (2017) 99382–99393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Chen Y, Okano K, Maeda T, Chauhan V, Golczak M, Maeda A, Palczewski K, Mechanism of all-trans-retinal toxicity with implications for stargardt disease and age-related macular degeneration, J Biol Chem, 287 (2012) 5059–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Sawada O, Perusek L, Kohno H, Howell SJ, Maeda A, Matsuyama S, Maeda T, All-trans-retinal induces Bax activation via DNA damage to mediate retinal cell apoptosis, Exp Eye Res, 123 (2014) 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Maeda T, Golczak M, Maeda A, Retinal photodamage mediated by all-trans-retinal, Photochem Photobiol, 88 (2012) 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Chambon P, A decade of molecular biology of retinoic acid receptors, FASEB J, 10 (1996) 940–954. [PubMed] [Google Scholar]
- [121].Napoli JL, Retinoic acid biosynthesis and metabolism, FASEB J, 10 (1996) 993–1001. [DOI] [PubMed] [Google Scholar]
- [122].Kedishvili NY, Enzymology of retinoic acid biosynthesis and degradation, J Lipid Res, 54 (2013) 1744–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Porte S, Xavier Ruiz F, Gimenez J, Molist I, Alvarez S, Dominguez M, Alvarez R, de Lera AR, Pares X, Farres J, Aldo-keto reductases in retinoid metabolism: search for substrate specificity and inhibitor selectivity, Chem Biol Interact, 202 (2013) 186–194. [DOI] [PubMed] [Google Scholar]
- [124].Ruiz FX, Porte S, Pares X, Farres J, Biological role of aldo-keto reductases in retinoic Acid biosynthesis and signaling, Front Pharmacol, 3 (2012) 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Liden M, Eriksson U, Understanding retinol metabolism: structure and function of retinol dehydrogenases, J Biol Chem, 281 (2006) 13001–13004. [DOI] [PubMed] [Google Scholar]
- [126].Gallego O, Belyaeva OV, Porte S, Ruiz FX, Stetsenko AV, Shabrova EV, Kostereva NV, Farres J, Pares X, Kedishvili NY, Comparative functional analysis of human medium-chain dehydrogenases, short-chain dehydrogenases/reductases and aldo-keto reductases with retinoids, Biochem J, 399 (2006) 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Sahu B, Maeda A, Retinol Dehydrogenases Regulate Vitamin A Metabolism for Visual Function, Nutrients, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Duester G, Genetic dissection of retinoid dehydrogenases, Chem Biol Interact, 130-132 (2001) 469–480. [DOI] [PubMed] [Google Scholar]
- [129].Su J, Chai X, Kahn B, Napoli JL, cDNA cloning, tissue distribution, and substrate characteristics of a cis-Retinol/3alpha-hydroxysterol short-chain dehydrogenase isozyme, J Biol Chem, 273 (1998) 17910–17916. [DOI] [PubMed] [Google Scholar]
- [130].Gough WH, VanOoteghem S, Sint T, Kedishvili NY, cDNA cloning and characterization of a new human microsomal NAD+-dependent dehydrogenase that oxidizes all-trans-retinol and 3alpha-hydroxysteroids, J Biol Chem, 273 (1998) 19778–19785. [DOI] [PubMed] [Google Scholar]
- [131].Baker ME, Unusual evolution of 11beta- and 17beta-hydroxysteroid and retinol dehydrogenases, Bioessays, 18 (1996) 63–70. [DOI] [PubMed] [Google Scholar]
- [132].Pares X, Farres J, Kedishvili N, Duester G, Medium- and short-chain dehydrogenase/reductase gene and protein families : Medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism, Cell Mol Life Sci, 65 (2008) 3936–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Rhinn M, Dolle P, Retinoic acid signalling during development, Development, 139 (2012) 843–858. [DOI] [PubMed] [Google Scholar]
- [134].Kiser PD, Golczak M, Maeda A, Palczewski K, Key enzymes of the retinoid (visual) cycle in vertebrate retina, Biochim Biophys Acta, 1821 (2012) 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Veech RL, Eggleston LV, Krebs HA, The redox state of free nicotinamide-adenine dinucleotide phosphate in the cytoplasm of rat liver, Biochem J, 115 (1969) 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Merrill DK, Guynn RW, The calculation of the cytoplasmic free [NADP+]/[NADPH] ratio in brain: effect of electroconvulsive seizure, Brain Res, 221 (1981) 307–318. [DOI] [PubMed] [Google Scholar]
- [137].Rossmann MG, Argos P, Protein folding, Annu Rev Biochem, 50 (1981) 497–532. [DOI] [PubMed] [Google Scholar]
- [138].Hanukoglu I, Proteopedia: Rossmann fold: A beta-alpha-beta fold at dinucleotide binding sites, Biochem Mol Biol Educ, 43 (2015) 206–209. [DOI] [PubMed] [Google Scholar]
- [139].Lesk AM, NAD-binding domains of dehydrogenases, Curr Opin Struct Biol, 5 (1995) 775–783. [DOI] [PubMed] [Google Scholar]
- [140].Persson B, Kallberg Y, Bray JE, Bruford E, Dellaporta SL, Favia AD, Duarte RG, Jornvall H, Kavanagh KL, Kedishvili N, Kisiela M, Maser E, Mindnich R, Orchard S, Penning TM, Thornton JM, Adamski J, Oppermann U, The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative, Chem Biol Interact, 178 (2009) 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Yao Y, Han WW, Zhou YH, Luo Q, Li ZS, Catalytic Reaction Mechanism of Human Photoreceptor Retinol Dehydrogenase: A Theoretical Study, J Theor Comput Chem, 7 (2008) 565–578. [Google Scholar]
- [142].Tanaka N, Nonaka T, Nakamura KT, Hara A, SDR: Structure, mechanism of action, and substrate recognition, Curr Org Chem, 5 (2001) 89–111. [Google Scholar]
- [143].Selles Vidal L, Kelly CL, Mordaka PM, Heap JT, Review of NAD(P)H-dependent oxidoreductases: Properties, engineering and application, Biochim Biophys Acta Proteins Proteom, 1866 (2018) 327–347. [DOI] [PubMed] [Google Scholar]
- [144].Britton K, Langridge S, Baker PJ, Weeradechapon K, Sedelnikova SE, De Lucas JR, Rice DW, Turner G, The crystal structure and active site location of isocitrate lyase from the fungus Aspergillus nidulans, Structure, 8 (2000) 349–362. [DOI] [PubMed] [Google Scholar]
- [145].Lee C, Bedgar DL, Davin LB, Lewis NG, Assessment of a putative proton relay in Arabidopsis cinnamyl alcohol dehydrogenase catalysis, Org Biomol Chem, 11 (2013) 1127–1134. [DOI] [PubMed] [Google Scholar]
- [146].Wilson DK, Nakano T, Petrash JM, Quiocho FA, 1.7 A structure of FR-1, a fibroblast growth factor-induced member of the aldo-keto reductase family, complexed with coenzyme and inhibitor, Biochemistry, 34 (1995) 14323–14330. [DOI] [PubMed] [Google Scholar]
- [147].Oppermann UC, Maser E, Molecular and structural aspects of xenobiotic carbonyl metabolizing enzymes. Role of reductases and dehydrogenases in xenobiotic phase I reactions, Toxicology, 144 (2000) 71–81. [DOI] [PubMed] [Google Scholar]
- [148].Penning TM, The aldo-keto reductases (AKRs): Overview, Chem Biol Interact, 234 (2015) 236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zhang M, Hu P, Napoli JL, Elements in the N-terminal signaling sequence that determine cytosolic topology of short-chain dehydrogenases/reductases. Studies with retinol dehydrogenase type 1 and cis-retinol/androgen dehydrogenase type 1, J Biol Chem, 279 (2004) 51482–51489. [DOI] [PubMed] [Google Scholar]
- [150].Lhor M, Methot M, Horchani H, Salesse C, Structure of the N-terminal segment of human retinol dehydrogenase 11 and its preferential lipid binding using model membranes, Biochim Biophys Acta, 1848 (2015) 878–885. [DOI] [PubMed] [Google Scholar]
- [151].Belyaeva OV, Stetsenko AV, Nelson P, Kedishvili NY, Properties of short-chain dehydrogenase/reductase RalR1: characterization of purified enzyme, its orientation in the microsomal membrane, and distribution in human tissues and cell lines, Biochemistry, 42 (2003) 14838–14845. [DOI] [PubMed] [Google Scholar]
- [152].Janecke AR, Thompson DA, Utermann G, Becker C, Hubner CA, Schmid E, McHenry CL, Nair AR, Ruschendorf F, Heckenlively J, Wissinger B, Nurnberg P, Gal A, Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy, Nat Genet, 36 (2004) 850–854. [DOI] [PubMed] [Google Scholar]
- [153].Luo W, Marsh-Armstrong N, Rattner A, Nathans J, An outer segment localization signal at the C terminus of the photoreceptor-specific retinol dehydrogenase, J Neurosci, 24 (2004) 2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lapshina EA, Belyaeva OV, Chumakova OV, Kedishvili NY, Differential recognition of the free versus bound retinol by human microsomal retinol/sterol dehydrogenases: characterization of the holo-CRBP dehydrogenase activity of RoDH-4, Biochemistry, 42 (2003) 776–784. [DOI] [PubMed] [Google Scholar]
- [155].Hofmann L, Tsybovsky Y, Alexander NS, Babino D, Leung NY, Montell C, Banerjee S, von Lintig J, Palczewski K, Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family, Biochemistry, 55 (2016) 6545–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Clagett-Dame M, Knutson D, Vitamin A in Reproduction and Development, Nutrients, 3 (2011) 385–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Soprano DR, Teets BW, Soprano KJ, Role of retinoic acid in the differentiation of embryonal carcinoma and embryonic stem cells, Vitam Horm, 75 (2007) 69–95. [DOI] [PubMed] [Google Scholar]
- [158].Noy N, Between death and survival: retinoic acid in regulation of apoptosis, Annu Rev Nutr, 30 (2010) 201–217. [DOI] [PubMed] [Google Scholar]
- [159].Ross AC, Gardner EM, The function of vitamin A in cellular growth and differentiation, and its roles during pregnancy and lactation, Adv Exp Med Biol, 352 (1994) 187–200. [DOI] [PubMed] [Google Scholar]
- [160].Vasiliou V, Nebert DW, Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family, Hum Genomics, 2 (2005) 138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Sladek NE, Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact, J Biochem Mol Toxicol, 17 (2003) 7–23. [DOI] [PubMed] [Google Scholar]
- [162].Vasiliou V, Pappa A, Petersen DR, Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism, Chem Biol Interact, 129 (2000) 1–19. [DOI] [PubMed] [Google Scholar]
- [163].Bchini R, Vasiliou V, Branlant G, Talfournier F, Rahuel-Clermont S, Retinoic acid biosynthesis catalyzed by retinal dehydrogenases relies on a rate-limiting conformational transition associated with substrate recognition, Chem Biol Interact, 202 (2013) 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Niederreither K, Subbarayan V, Dolle P, Chambon P, Embryonic retinoic acid synthesis is essential for early mouse post-implantation development, Nat Genet, 21 (1999) 444–448. [DOI] [PubMed] [Google Scholar]
- [165].Napoli JL, Physiological insights into all-trans-retinoic acid biosynthesis, Biochim Biophys Acta, 1821 (2012) 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB, A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment, Proc Natl Acad Sci U S A, 100 (2003) 14036–14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G , Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina, Mol Cell Biol, 23 (2003) 4637–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Molotkov A, Duester G, Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid, J Biol Chem, 278 (2003) 36085–36090. [DOI] [PubMed] [Google Scholar]
- [169].Tang XH, Gudas LJ, Retinoids, retinoic acid receptors, and cancer, Annu Rev Pathol, 6 (2011) 345–364. [DOI] [PubMed] [Google Scholar]
- [170].Bazewicz CG, Dinavahi SS, Schell TD, Robertson GP, Aldehyde dehydrogenase in regulatory T-cell development, immunity and cancer, Immunology, 156 (2019) 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Vasiliou V, Thompson DC, Smith C, Fujita M, Chen Y, Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells, Chem Biol Interact, 202 (2013) 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Yang X, Yao R, Wang H, Update of ALDH as a Potential Biomarker and Therapeutic Target for AML, Biomed Res Int, 2018 (2018) 9192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V, Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application, Pharmacol Rev, 64 (2012) 520–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Erkelens MN, Mebius RE, Retinoic Acid and Immune Homeostasis: A Balancing Act, Trends Immunol, 38 (2017) 168–180. [DOI] [PubMed] [Google Scholar]
- [175].Marcato P, Dean CA, Giacomantonio CA, Lee PW, Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform, Cell Cycle, 10 (2011) 1378–1384. [DOI] [PubMed] [Google Scholar]
- [176].Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, Giacomantonio CA, Lee PW, Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis, Stem Cells, 29 (2011) 32–45. [DOI] [PubMed] [Google Scholar]
- [177].Sanders TJ, McCarthy NE, Giles EM, Davidson KL, Haltalli ML, Hazell S, Lindsay JO, Stagg AJ, Increased production of retinoic acid by intestinal macrophages contributes to their inflammatory phenotype in patients with Crohn's disease, Gastroenterology, 146 (2014) 1278–1288 e1271-1272. [DOI] [PubMed] [Google Scholar]
- [178].Bhattacharya N, Yuan R, Prestwood TR, Penny HL, DiMaio MA, Reticker-Flynn NE, Krois CR, Kenkel JA, Pham TD, Carmi Y, Tolentino L, Choi O, Hulett R, Wang J, Winer DA, Napoli JL, Engleman EG, Normalizing Microbiota-Induced Retinoic Acid Deficiency Stimulates Protective CD8(+) T Cell-Mediated Immunity in Colorectal Cancer, Immunity, 45 (2016) 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Moretti A, Li J, Donini S, Sobol RW, Rizzi M, Garavaglia S, Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD(+) and retinoic acid, Sci Rep, 6 (2016) 35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Hempel J, Perozich J, Chapman T, Rose J, Boesch JS, Liu ZJ, Lindahl R, Wang BC, Aldehyde dehydrogenase catalytic mechanism. A proposal, Adv Exp Med Biol, 463 (1999) 53–59. [DOI] [PubMed] [Google Scholar]
- [181].Hempel J, Kuo I, Perozich J, Wang BC, Lindahl R, Nicholas H, Aldehyde dehydrogenase. Maintaining critical active site geometry at motif 8 in the class 3 enzyme, Eur J Biochem, 268 (2001) 722–726. [DOI] [PubMed] [Google Scholar]
- [182].Wongsiriroj N, Piantedosi R, Palczewski K, Goldberg IJ, Johnston TP, Li E, Blaner WS, The molecular basis of retinoid absorption: a genetic dissection, J Biol Chem, 283 (2008) 13510–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K, Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver, J Biol Chem, 279 (2004) 10422–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Yen CL, Monetti M, Burri BJ, Farese RV Jr., The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters, J Lipid Res, 46 (2005) 1502–1511. [DOI] [PubMed] [Google Scholar]
- [185].Golczak M, Imanishi Y, Kuksa V, Maeda T, Kubota R, Palczewski K, Lecithin:retinol acyltransferase is responsible for amidation of retinylamine, a potent inhibitor of the retinoid cycle, J Biol Chem, 280 (2005) 42263–42273. [DOI] [PubMed] [Google Scholar]
- [186].Golczak M, Kuksa V, Maeda T, Moise AR, Palczewski K, Positively charged retinoids are potent and selective inhibitors of the trans-cis isomerization in the retinoid (visual) cycle, Proc Natl Acad Sci U S A, 102 (2005) 8162–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Zhang J, Dong Z, Mundla SR, Hu XE, Seibel W, Papoian R, Palczewski K, Golczak M, Expansion of first-in-class drug candidates that sequester toxic all-trans-retinal and prevent light-induced retinal degeneration, Mol Pharmacol, 87 (2015) 477–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Orland MD, Anwar K, Cromley D, Chu CH, Chen L, Billheimer JT, Hussain MM, Cheng D, Acyl coenzyme A dependent retinol esterification by acyl coenzyme A: diacylglycerol acyltransferase 1, Biochim Biophys Acta, 1737 (2005) 76–82. [DOI] [PubMed] [Google Scholar]
- [189].MacDonald PN, Ong DE, A lecithin:retinol acyltransferase activity in human and rat liver, Biochem Biophys Res Commun, 156 (1988) 157–163. [DOI] [PubMed] [Google Scholar]
- [190].Ruiz A, Winston A, Lim YH, Gilbert BA, Rando RR, Bok D, Molecular and biochemical characterization of lecithin retinol acyltransferase, J Biol Chem, 274 (1999) 3834–3841. [DOI] [PubMed] [Google Scholar]
- [191].Golczak M, Sears AE, Kiser PD, Palczewski K, LRAT-specific domain facilitates vitamin A metabolism by domain swapping in HRASLS3, Nat Chem Biol, 11 (2015) 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Batres RO, Olson JA, Distribution of Vitamin-a among Parenchymal and Stellate Cells of Livers of Rats with Low Vitamin-a Stores, Federation Proceedings, 46 (1987) 1191–1191. [Google Scholar]
- [193].Moriwaki H, Blaner WS, Piantedosi R, Goodman DS, Effects of dietary retinoid and triglyceride on the lipid composition of rat liver stellate cells and stellate cell lipid droplets, J Lipid Res, 29 (1988) 1523–1534. [PubMed] [Google Scholar]
- [194].Blaner WS, O'Byrne SM, Wongsiriroj N, Kluwe J, D'Ambrosio DM, Jiang H, Schwabe RF, Hillman EM, Piantedosi R, Libien J, Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage, Biochim Biophys Acta, 1791 (2009) 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Redgrave TG, Chylomicron metabolism, Biochem Soc Trans, 32 (2004) 79–82. [DOI] [PubMed] [Google Scholar]
- [196].Dixon JL, Goodman DS, Studies on the metabolism of retinol-binding protein by primary hepatocytes from retinol-deficient rats, J Cell Physiol, 130 (1987) 14–20. [DOI] [PubMed] [Google Scholar]
- [197].Grumet L, Taschler U, Lass A, Hepatic Retinyl Ester Hydrolases and the Mobilization of Retinyl Ester Stores, Nutrients, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Chelstowska S, Widjaja-Adhi MA, Silvaroli JA, Golczak M, Molecular Basis for Vitamin A Uptake and Storage in Vertebrates, Nutrients, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Gollapalli DR, Rando RR, All-trans-retinyl esters are the substrates for isomerization in the vertebrate visual cycle, Biochemistry, 42 (2003) 5809–5818. [DOI] [PubMed] [Google Scholar]
- [200].Sheridan C, Boyer NP, Crouch RK, Koutalos Y, RPE65 and the Accumulation of Retinyl Esters in Mouse Retinal Pigment Epithelium, Photochem Photobiol, 93 (2017) 844–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].McBee JK, Kuksa V, Alvarez R, de Lera AR, Prezhdo O, Haeseleer F, Sokal I, Palczewski K, Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: dependence on the specificity of retinoid-binding proteins, Biochemistry, 39 (2000) 11370–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Redmond TM, Poliakov E, Kuo S, Chander P, Gentleman S, RPE65, visual cycle retinol isomerase, is not inherently 11-cis-specific: support for a carbocation mechanism of retinol isomerization, J Biol Chem, 285 (2010) 1919–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Deigner PS, Law WC, Canada FJ, Rando RR, Membranes as the energy source in the endergonic transformation of vitamin A to 11-cis-retinol, Science, 244 (1989) 968–971. [DOI] [PubMed] [Google Scholar]
- [204].Thompson DA, Li Y, McHenry CL, Carlson TJ, Ding X, Sieving PA, Apfelstedt-Sylla E, Gal A, Mutations in the gene encoding lecithin retinol acyltransferase are associated with early-onset severe retinal dystrophy, Nat Genet, 28 (2001) 123–124. [DOI] [PubMed] [Google Scholar]
- [205].Amengual J, Golczak M, Palczewski K, von Lintig J, Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein, J Biol Chem, 287 (2012) 24216–24227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Mata NL, Radu RA, Clemmons RC, Travis GH, Isomerization and oxidation of vitamin a in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight, Neuron, 36 (2002) 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Muniz A, Betts BS, Trevino AR, Buddavarapu K, Roman R, Ma JX, Tsin AT, Evidence for two retinoid cycles in the cone-dominated chicken eye, Biochemistry, 48 (2009) 6854–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Wang JS, Kefalov VJ, The cone-specific visual cycle, Prog Retin Eye Res, 30 (2011) 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Rodriguez KA, Tsin AT, Retinyl esters in the vertebrate neuroretina, Am J Physiol, 256 (1989) R255–258. [DOI] [PubMed] [Google Scholar]
- [210].Babino D, Perkins BD, Kindermann A, Oberhauser V, von Lintig J, The role of 11-cis-retinyl esters in vertebrate cone vision, FASEB J, 29 (2015) 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Mata NL, Ruiz A, Radu RA, Bui TV, Travis GH, Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of all-trans-retinol to 11-cis-retinol, Biochemistry, 44 (2005) 11715–11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Muniz A, Villazana-Espinoza ET, Hatch AL, Trevino SG, Allen DM, Tsin AT, A novel cone visual cycle in the cone-dominated retina, Exp Eye Res, 85 (2007) 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Ables GP, Yang KJ, Vogel S, Hernandez-Ono A, Yu S, Yuen JJ, Birtles S, Buckett LK, Turnbull AV, Goldberg IJ, Blaner WS, Huang LS, Ginsberg HN, Intestinal DGAT1 deficiency reduces postprandial triglyceride and retinyl ester excursions by inhibiting chylomicron secretion and delaying gastric emptying, J Lipid Res, 53 (2012) 2364–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Kaylor JJ, Yuan Q, Cook J, Sarfare S, Makshanoff J, Miu A, Kim A, Kim P, Habib S, Roybal CN, Xu T, Nusinowitz S, Travis GH, Identification of DES1 as a vitamin A isomerase in Muller glial cells of the retina, Nat Chem Biol, 9 (2013) 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Arne JM, Widjaja-Adhi MA, Hughes T, Huynh KW, Silvaroli JA, Chelstowska S, Moiseenkova-Bell VY, Golczak M, Allosteric modulation of the substrate specificity of acyl-CoA wax alcohol acyltransferase 2, J Lipid Res, 58 (2017) 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].McFie PJ, Stone SL, Banman SL, Stone SJ, Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the n terminus in dimer/tetramer formation, J Biol Chem, 285 (2010) 37377–37387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Stone SJ, Levin MC, Farese RV Jr., Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2, J Biol Chem, 281 (2006) 40273–40282. [DOI] [PubMed] [Google Scholar]
- [218].Moise AR, Golczak M, Imanishi Y, Palczewski K, Topology and membrane association of lecithin: retinol acyltransferase, J Biol Chem, 282 (2007) 2081–2090. [DOI] [PubMed] [Google Scholar]
- [219].Chelstowska S, Widjaja-Adhi MAK, Silvaroli JA, Golczak M, Impact of LCA-Associated E14L LRAT Mutation on Protein Stability and Retinoid Homeostasis, Biochemistry, 56 (2017) 4489–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Anantharaman V, Aravind L, Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes, Genome Biol, 4 (2003) R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Albalat R, Evolution of the genetic machinery of the visual cycle: a novelty of the vertebrate eye?, Mol Biol Evol, 29 (2012) 1461–1469. [DOI] [PubMed] [Google Scholar]
- [222].Uyama T, Morishita J, Jin XH, Okamoto Y, Tsuboi K, Ueda N, The tumor suppressor gene H-Rev107 functions as a novel Ca2+-independent cytosolic phospholipase A1/2 of the thiol hydrolase type, J Lipid Res, 50 (2009) 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Jin XH, Uyama T, Wang J, Okamoto Y, Tonai T, Ueda N, cDNA cloning and characterization of human and mouse Ca(2+)-independent phosphatidylethanolamine N-acyltransferases, Biochim Biophys Acta, 1791 (2009) 32–38. [DOI] [PubMed] [Google Scholar]
- [224].Han BG, Cho JW, Cho YD, Kim SY, Yoon HJ, Song HK, Cheong HK, Jeon YH, Lee DK, Lee S, Lee BI, Expression, purification and biochemical characterization of the N-terminal regions of human TIG3 and HRASLS3 proteins, Protein Expr Purif, 71 (2010) 103–107. [DOI] [PubMed] [Google Scholar]
- [225].Golczak M, Kiser PD, Sears AE, Lodowski DT, Blaner WS, Palczewski K, Structural basis for the acyltransferase activity of lecithin:retinol acyltransferase-like proteins, J Biol Chem, 287 (2012) 23790–23807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Pang XY, Cao J, Addington L, Lovell S, Battaile KP, Zhang N, Rao JL, Dennis EA, Moise AR, Structure/function relationships of adipose phospholipase A2 containing a cys-his-his catalytic triad, J Biol Chem, 287 (2012) 35260–35274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Jahng WJ, Cheung E, Rando RR, Lecithin retinol acyltransferase forms functional homodimers, Biochemistry, 41 (2002) 6311–6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Shi YQ, Hubacek I, Rando RR, Kinetic mechanism of lecithin retinol acyl transferase, Biochemistry, 32 (1993) 1257–1263. [DOI] [PubMed] [Google Scholar]
- [229].Xue L, Rando RR, Roles of cysteine 161 and tyrosine 154 in the lecithin-retinol acyltransferase mechanism, Biochemistry, 43 (2004) 6120–6126. [DOI] [PubMed] [Google Scholar]
- [230].Golczak M, Palczewski K, An acyl-covalent enzyme intermediate of lecithin:retinol acyltransferase, J Biol Chem, 285 (2010) 29217–29222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Ong DE, Cellular transport and metabolism of vitamin A: roles of the cellular retinoid-binding proteins, Nutr Rev, 52 (1994) S24–31. [DOI] [PubMed] [Google Scholar]
- [232].Herr FM, Ong DE, Differential interaction of lecithin-retinol acyltransferase with cellular retinol binding proteins, Biochemistry, 31 (1992) 6748–6755. [DOI] [PubMed] [Google Scholar]
- [233].Yost RW, Harrison EH, Ross AC, Esterification by rat liver microsomes of retinol bound to cellular retinol-binding protein, J Biol Chem, 263 (1988) 18693–18701. [PubMed] [Google Scholar]
- [234].Quadro L, Blaner WS, Salchow DJ, Vogel S, Piantedosi R, Gouras P, Freeman S, Cosma MP, Colantuoni V, Gottesman ME, Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein, EMBO J, 18 (1999) 4633–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Biesalski HK, Frank J, Beck SC, Heinrich F, Illek B, Reifen R, Gollnick H, Seeliger MW, Wissinger B, Zrenner E, Biochemical but not clinical vitamin A deficiency results from mutations in the gene for retinol binding protein, Am J Clin Nutr, 69 (1999) 931–936. [DOI] [PubMed] [Google Scholar]
- [236].Kanai M, Raz A, Goodman DS, Retinol-binding protein: the transport protein for vitamin A in human plasma, J Clin Invest, 47 (1968) 2025–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Seeliger MW, Biesalski HK, Wissinger B, Gollnick H, Gielen S, Frank J, Beck S, Zrenner E, Phenotype in retinol deficiency due to a hereditary defect in retinol binding protein synthesis, Invest Ophthalmol Vis Sci, 40 (1999) 3–11. [PubMed] [Google Scholar]
- [238].Cukras C, Gaasterland T, Lee P, Gudiseva HV, Chavali VR, Pullakhandam R, Maranhao B, Edsall L, Soares S, Reddy GB, Sieving PA, Ayyagari R, Exome analysis identified a novel mutation in the RBP4 gene in a consanguineous pedigree with retinal dystrophy and developmental abnormalities, PLoS One, 7 (2012) e50205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Chou CM, Nelson C, Tarle SA, Pribila JT, Bardakjian T, Woods S, Schneider A, Glaser T, Biochemical Basis for Dominant Inheritance, Variable Penetrance, and Maternal Effects in RBP4 Congenital Eye Disease, Cell, 161 (2015) 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Asimakopoulou A, Weiskirchen R, Lipocalin 2 in the pathogenesis of fatty liver disease and nonalcoholic steatohepatitis, Clin Lipidol, 10 (2015) 47–67. [Google Scholar]
- [241].Du ZP, Wu BL, Wu X, Lin XH, Qiu XY, Zhan XF, Wang SH, Shen JH, Zheng CP, Wu ZY, Xu LY, Wang D, Li EM, A systematic analysis of human lipocalin family and its expression in esophageal carcinoma, Sci Rep, 5 (2015) 12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Cowan SW, Newcomer ME, Jones TA, Crystallographic refinement of human serum retinol binding protein at 2A resolution, Proteins, 8 (1990) 44–61. [DOI] [PubMed] [Google Scholar]
- [243].Zanotti G, Ottonello S, Berni R, Monaco HL, Crystal structure of the trigonal form of human plasma retinol-binding protein at 2.5 A resolution, J Mol Biol, 230 (1993) 613–624. [DOI] [PubMed] [Google Scholar]
- [244].Zanotti G, Berni R, Monaco HL, Crystal structure of liganded and unliganded forms of bovine plasma retinol-binding protein, J Biol Chem, 268 (1993) 10728–10738. [PubMed] [Google Scholar]
- [245].Vogel S, Piantedosi R, O'Byrne SM, Kako Y, Quadro L, Gottesman ME, Goldberg IJ, Blaner WS, Retinol-binding protein-deficient mice: biochemical basis for impaired vision, Biochemistry, 41 (2002) 15360–15368. [DOI] [PubMed] [Google Scholar]
- [246].Naylor HM, Newcomer ME, The structure of human retinol-binding protein (RBP) with its carrier protein transthyretin reveals an interaction with the carboxy terminus of RBP, Biochemistry, 38 (1999) 2647–2653. [DOI] [PubMed] [Google Scholar]
- [247].Monaco HL, Three-dimensional structure of the transthyretin-retinol-binding protein complex, Clin Chem Lab Med, 40 (2002) 1229–1236. [DOI] [PubMed] [Google Scholar]
- [248].Monaco HL, Rizzi M, Coda A, Structure of a complex of two plasma proteins: transthyretin and retinol-binding protein, Science, 268 (1995) 1039–1041. [DOI] [PubMed] [Google Scholar]
- [249].Zanotti G, Marcello M, Malpeli G, Folli C, Sartori G, Berni R, Crystallographic studies on complexes between retinoids and plasma retinol-binding protein, J Biol Chem, 269 (1994) 29613–29620. [PubMed] [Google Scholar]
- [250].Berni R, Formelli F, In vitro interaction of fenretinide with plasma retinol-binding protein and its functional consequences, FEBS Lett, 308 (1992) 43–45. [DOI] [PubMed] [Google Scholar]
- [251].Berni R, Clerici M, Malpeli G, Cleris L, Formelli F, Retinoids: in vitro interaction with retinol-binding protein and influence on plasma retinol, FASEB J, 7 (1993) 1179–1184. [DOI] [PubMed] [Google Scholar]
- [252].Cogan U, Kopelman M, Mokady S, Shinitzky M, Binding affinities of retinol and related compounds to retinol binding proteins, Eur J Biochem, 65 (1976) 71–78. [DOI] [PubMed] [Google Scholar]
- [253].Perduca M, Nicolis S, Mannucci B, Galliano M, Monaco HL, Human plasma retinol-binding protein (RBP4) is also a fatty acid-binding protein, Biochim Biophys Acta Mol Cell Biol Lipids, 1863 (2018) 458–466. [DOI] [PubMed] [Google Scholar]
- [254].De Laey JJ, Verougstraete C, Hyperlipofuscinosis and subretinal fibrosis in Stargardt's disease, Retina, 15 (1995) 399–406. [DOI] [PubMed] [Google Scholar]
- [255].Graham TE, Yang Q, Bluher M, Hammarstedt A, Ciaraldi TP, Henry RR, Wason CJ, Oberbach A, Jansson PA, Smith U, Kahn BB, Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects, N Engl J Med, 354 (2006) 2552–2563. [DOI] [PubMed] [Google Scholar]
- [256].Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB, Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes, Nature, 436 (2005) 356–362. [DOI] [PubMed] [Google Scholar]
- [257].Codoner-Franch P, Carrasco-Luna J, Allepuz P, Codoner-Alejos A, Guillem V, Association of RBP4 genetic variants with childhood obesity and cardiovascular risk factors, Pediatr Diabetes, 17 (2016) 576–583. [DOI] [PubMed] [Google Scholar]
- [258].Motani A, Wang Z, Conn M, Siegler K, Zhang Y, Liu Q, Johnstone S, Xu H, Thibault S, Wang Y, Fan P, Connors R, Le H, Xu G, Walker N, Shan B, Coward P, Identification and characterization of a non-retinoid ligand for retinol-binding protein 4 which lowers serum retinol-binding protein 4 levels in vivo, J Biol Chem, 284 (2009) 7673–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Kotnik P, Fischer-Posovszky P, Wabitsch M, RBP4: a controversial adipokine, Eur J Endocrinol, 165 (2011) 703–711. [DOI] [PubMed] [Google Scholar]
- [260].Henze A, Frey SK, Raila J, Tepel M, Scholze A, Pfeiffer AF, Weickert MO, Spranger J, Schweigert FJ, Evidence that kidney function but not type 2 diabetes determines retinol-binding protein 4 serum levels, Diabetes, 57 (2008) 3323–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Sporn MB, Dunlop NM, Newton DL, Smith JM, Prevention of chemical carcinogenesis by vitamin A and its synthetic analogs (retinoids), Fed Proc, 35 (1976) 1332–1338. [PubMed] [Google Scholar]
- [262].Racz B, Varadi A, Kong J, Allikmets R, Pearson PG, Johnson G, Cioffi CL, Petrukhin K, A non-retinoid antagonist of retinol-binding protein 4 rescues phenotype in a model of Stargardt disease without inhibiting the visual cycle, J Biol Chem, 293 (2018) 11574–11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Folli C, Calderone V, Ottonello S, Bolchi A, Zanotti G, Stoppini M, Berni R, Identification, retinoid binding, and x-ray analysis of a human retinol-binding protein, Proc Natl Acad Sci U S A, 98 (2001) 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Folli C, Calderone V, Ramazzina I, Zanotti G, Berni R, Ligand binding and structural analysis of a human putative cellular retinol-binding protein, J Biol Chem, 277 (2002) 41970–41977. [DOI] [PubMed] [Google Scholar]
- [265].Bass NM, Cellular binding proteins for fatty acids and retinoids: similar or specialized functions?, Mol Cell Biochem, 123 (1993) 191–202. [DOI] [PubMed] [Google Scholar]
- [266].Vogel S, Mendelsohn CL, Mertz JR, Piantedosi R, Waldburger C, Gottesman ME, Blaner WS, Characterization of a new member of the fatty acid-binding protein family that binds all-trans-retinol, J Biol Chem, 276 (2001) 1353–1360. [DOI] [PubMed] [Google Scholar]
- [267].Eriksson U, Das K, Busch C, Nordlinder H, Rask L, Sundelin J, Sallstrom J, Peterson PA, Cellular retinol-binding protein. Quantitation and distribution, J Biol Chem, 259 (1984) 13464–13470. [PubMed] [Google Scholar]
- [268].Kato M, Kato K, Goodman DS, Immunocytochemical studies on the localization of plasma and of cellular retinol-binding proteins and of transthyretin (prealbumin) in rat liver and kidney, J Cell Biol, 98 (1984)1696–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Saari JC, Bunt-Milam AH, Bredberg DL, Garwin GG, Properties and immunocytochemical localization of three retinoid-binding proteins from bovine retina, Vision Res, 24 (1984) 1595–1603. [DOI] [PubMed] [Google Scholar]
- [270].Li E, Demmer LA, Sweetser DA, Ong DE, Gordon JI, Rat cellular retinol-binding protein II: use of a cloned cDNA to define its primary structure, tissue-specific expression, and developmental regulation, Proc Natl Acad Sci U S A, 83 (1986) 5779–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Li E, Qian SJ, Winter NS, d'Avignon A, Levin MS, Gordon JI, Fluorine nuclear magnetic resonance analysis of the ligand binding properties of two homologous rat cellular retinol-binding proteins expressed in Escherichia coli, J Biol Chem, 266 (1991) 3622–3629. [PubMed] [Google Scholar]
- [272].Malpeli G, Stoppini M, Zapponi MC, Folli C, Berni R, Interactions with retinol and retinoids of bovine cellular retinol-binding protein, Eur J Biochem, 229 (1995) 486–493. [PubMed] [Google Scholar]
- [273].Silvaroli JA, Arne JM, Chelstowska S, Kiser PD, Banerjee S, Golczak M, Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures, J Biol Chem, 291 (2016) 8528–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Kane MA, Bright FV, Napoli JL, Binding affinities of CRBPI and CRBPII for 9-cis-retinoids, Biochim Biophys Acta, 1810 (2011) 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Ghyselinck NB, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson CB, Hakansson H, Sauvant P, Azais-Braesco V, Frasson M, Picaud S, Chambon P, Cellular retinol-binding protein I is essential for vitamin A homeostasis, EMBO J, 18 (1999) 4903–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Saari JC, Nawrot M, Garwin GG, Kennedy MJ, Hurley JB, Ghyselinck NB, Chambon P, Analysis of the visual cycle in cellular retinol-binding protein type I (CRBPI) knockout mice, Invest Ophthalmol Vis Sci, 43 (2002) 1730–1735. [PubMed] [Google Scholar]
- [277].Napoli JL, A gene knockout corroborates the integral function of cellular retinol-binding protein in retinoid metabolism, Nutr Rev, 58 (2000) 230–236. [DOI] [PubMed] [Google Scholar]
- [278].Rigtrup KM, Ong DE, A retinyl ester hydrolase activity intrinsic to the brush border membrane of rat small intestine, Biochemistry, 31 (1992) 2920–2926. [DOI] [PubMed] [Google Scholar]
- [279].Farjo KM, Moiseyev G, Nikolaeva O, Sandell LL, Trainor PA, Ma JX, RDH10 is the primary enzyme responsible for the first step of embryonic Vitamin A metabolism and retinoic acid synthesis, Dev Biol, 357 (2011) 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Boerman MH, Napoli JL, Characterization of a microsomal retinol dehydrogenase: a short-chain alcohol dehydrogenase with integral and peripheral membrane forms that interacts with holo-CRBP (type I), Biochemistry, 34 (1995) 7027–7037. [DOI] [PubMed] [Google Scholar]
- [281].Napoli JL, Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: Effects on retinoid metabolism, function and related diseases, Pharmacol Ther, 173 (2017) 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Pierzchalski K, Yu J, Norman V, Kane MA, CrbpI regulates mammary retinoic acid homeostasis and the mammary microenvironment, FASEB J, 27 (2013) 1904–1916. [DOI] [PubMed] [Google Scholar]
- [283].E X, Zhang L, Lu J, Tso P, Blaner WS, Levin MS, Li E, Increased neonatal mortality in mice lacking cellular retinol-binding protein II, J Biol Chem, 277 (2002) 36617–36623. [DOI] [PubMed] [Google Scholar]
- [284].Ong DE, Kakkad B, MacDonald PN, Acyl-CoA-independent esterification of retinol bound to cellular retinol-binding protein (type II) by microsomes from rat small intestine, J Biol Chem, 262 (1987) 2729–2736. [PubMed] [Google Scholar]
- [285].Kakkad BP, Ong DE, Reduction of retinaldehyde bound to cellular retinol-binding protein (type II) by microsomes from rat small intestine, J Biol Chem, 263 (1988) 12916–12919. [PubMed] [Google Scholar]
- [286].Piantedosi R, Ghyselinck N, Blaner WS, Vogel S, Cellular retinol-binding protein type III is needed for retinoid incorporation into milk, J Biol Chem, 280 (2005) 24286–24292. [DOI] [PubMed] [Google Scholar]
- [287].Zizola CF, Schwartz GJ, Vogel S, Cellular retinol-binding protein type III is a PPARgamma target gene and plays a role in lipid metabolism, Am J Physiol Endocrinol Metab, 295 (2008) E1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Zhang YR, Zhao YQ, Huang JF, Retinoid-binding proteins: similar protein architectures bind similar ligands via completely different ways, PLoS One, 7 (2012) e36772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Hodsdon ME, Cistola DP, Discrete backbone disorder in the nuclear magnetic resonance structure of apo intestinal fatty acid-binding protein: implications for the mechanism of ligand entry, Biochemistry, 36 (1997) 1450–1460. [DOI] [PubMed] [Google Scholar]
- [290].Franzoni L, Lucke C, Perez C, Cavazzini D, Rademacher M, Ludwig C, Spisni A, Rossi GL, Ruterjans H, Structure and backbone dynamics of Apo- and holo-cellular retinol-binding protein in solution, J Biol Chem, 277 (2002) 21983–21997. [DOI] [PubMed] [Google Scholar]
- [291].Xiao H, Kaltashov IA, Transient structural disorder as a facilitator of protein-ligand binding: native H/D exchange-mass spectrometry study of cellular retinoic acid binding protein I, J Am Soc Mass Spectrom, 16 (2005) 869–879. [DOI] [PubMed] [Google Scholar]
- [292].Silvaroli JA, Widjaja-Adhi MAK, Trischman T, Chelstowska S, Horwitz S, Banerjee S, Kiser PD, Blaner WS, Golczak M, Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1, ACS Chem Biol, 14 (2019) 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Ong DE, Chytil F, Cellular retinoic acid-binding protein from rat testis. Purification and characterization, J Biol Chem, 253 (1978) 4551–4554. [PubMed] [Google Scholar]
- [294].Giguere V, Lyn S, Yip P, Siu CH, Amin S, Molecular cloning of cDNA encoding a second cellular retinoic acid-binding protein, Proc Natl Acad Sci U S A, 87 (1990) 6233–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Zheng WL, Ong DE, Spatial and temporal patterns of expression of cellular retinol-binding protein and cellular retinoic acid-binding proteins in rat uterus during early pregnancy, Biol Reprod, 58 (1998)963–970. [DOI] [PubMed] [Google Scholar]
- [296].Wardlaw SA, Bucco RA, Zheng WL, Ong DE, Variable expression of cellular retinol- and cellular retinoic acid-binding proteins in the rat uterus and ovary during the estrous cycle, Biol Reprod, 56 (1997) 125–132. [DOI] [PubMed] [Google Scholar]
- [297].Yamamoto M, Drager UC, Ong DE, McCaffery P, Retinoid-binding proteins in the cerebellum and choroid plexus and their relationship to regionalized retinoic acid synthesis and degradation, Eur J Biochem, 257 (1998) 344–350. [DOI] [PubMed] [Google Scholar]
- [298].Gaub MP, Lutz Y, Ghyselinck NB, Scheuer I, Pfister V, Chambon P, Rochette-Egly C, Nuclear detection of cellular retinoic acid binding proteins I and II with new antibodies, J Histochem Cytochem, 46 (1998) 1103–1111. [DOI] [PubMed] [Google Scholar]
- [299].Astrom A, Tavakkol A, Pettersson U, Cromie M, Elder JT, Voorhees JJ, Molecular cloning of two human cellular retinoic acid-binding proteins (CRABP). Retinoic acid-induced expression of CRABP-II but not CRABP-I in adult human skin in vivo and in skin fibroblasts in vitro, J Biol Chem, 266 (1991) 17662–17666. [PubMed] [Google Scholar]
- [300].Fiorella PD, Napoli JL, Expression of cellular retinoic acid binding protein (CRABP) in Escherichia coli. Characterization and evidence that holo-CRABP is a substrate in retinoic acid metabolism, J Biol Chem, 266 (1991) 16572–16579. [PubMed] [Google Scholar]
- [301].Nelson CH, Peng CC, Lutz JD, Yeung CK, Zelter A, Isoherranen N, Direct protein-protein interactions and substrate channeling between cellular retinoic acid binding proteins and CYP26B1, FEBS Lett, 590 (2016) 2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Boylan JF, Gudas LJ, Overexpression of the cellular retinoic acid binding protein-I (CRABP-I) results in a reduction in differentiation-specific gene expression in F9 teratocarcinoma cells, J Cell Biol, 112 (1991) 965–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Won JY, Nam EC, Yoo SJ, Kwon HJ, Um SJ, Han HS, Kim SH, Byun Y, Kim SY, The effect of cellular retinoic acid binding protein-I expression on the CYP26-mediated catabolism of all-trans retinoic acid and cell proliferation in head and neck squamous cell carcinoma, Metabolism, 53 (2004) 1007–1012. [DOI] [PubMed] [Google Scholar]
- [304].Pfoertner S, Goelden U, Hansen W, Toepfer T, Geffers R, Ukena SN, von Knobloch R, Hofmann R, Buer J, Schrader AJ, Cellular retinoic acid binding protein I: expression and functional influence in renal cell carcinoma, Tumour Biol, 26 (2005) 313–323. [DOI] [PubMed] [Google Scholar]
- [305].Liu RZ, Garcia E, Glubrecht DD, Poon HY, Mackey JR, Godbout R, CRABP1 is associated with a poor prognosis in breast cancer: adding to the complexity of breast cancer cell response to retinoic acid, Mol Cancer, 14 (2015) 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Wu JI, Lin YP, Tseng CW, Chen HJ, Wang LH, Crabp2 Promotes Metastasis of Lung Cancer Cells via HuR and Integrin beta1/FAK/ERK Signaling, Sci Rep, 9 (2019) 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Takase S, Ong DE, Chytil F, Transfer of retinoic acid from its complex with cellular retinoic acid-binding protein to the nucleus, Arch Biochem Biophys, 247 (1986) 328–334. [DOI] [PubMed] [Google Scholar]
- [308].Dong D, Ruuska SE, Levinthal DJ, Noy N, Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid, J Biol Chem, 274 (1999) 23695–23698. [DOI] [PubMed] [Google Scholar]
- [309].Delva L, Bastie JN, Rochette-Egly C, Kraiba R, Balitrand N, Despouy G, Chambon P, Chomienne C, Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex, Mol Cell Biol, 19 (1999) 7158–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Lampron C, Rochette-Egly C, Gorry P, Dolle P, Mark M, Lufkin T, LeMeur M, Chambon P, Mice deficient in cellular retinoic acid binding protein II (CRABPII) or in both CRABPI and CRABPII are essentially normal, Development, 121 (1995) 539–548. [DOI] [PubMed] [Google Scholar]
- [311].Kleywegt GJ, Bergfors T, Senn H, Le Motte P, Gsell B, Shudo K, Jones TA, Crystal structures of cellular retinoic acid binding proteins I and II in complex with all-trans-retinoic acid and a synthetic retinoid, Structure, 2 (1994) 1241–1258. [DOI] [PubMed] [Google Scholar]
- [312].Fiorella PD, Giguere V, Napoli JL, Expression of cellular retinoic acid-binding protein (type II) in Escherichia coli. Characterization and comparison to cellular retinoic acid-binding protein (type I), J Biol Chem, 268 (1993) 21545–21552. [PubMed] [Google Scholar]
- [313].Fogh K, Voorhees JJ, Astrom A, Expression, purification, and binding properties of human cellular retinoic acid-binding protein type I and type II, Arch Biochem Biophys, 300 (1993) 751–755. [DOI] [PubMed] [Google Scholar]
- [314].Kaylor JJ, Xu T, Ingram NT, Tsan A, Hakobyan H, Fain GL, Travis GH, Blue light regenerates functional visual pigments in mammals through a retinyl-phospholipid intermediate, Nat Commun, 8 (2017) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Morshedian A, Kaylor JJ, Ng SY, Tsan A, Frederiksen R, Xu T, Yuan L, Sampath AP, Radu RA, Fain GL, Travis GH, Light-Driven Regeneration of Cone Visual Pigments through a Mechanism Involving RGR Opsin in Muller Glial Cells, Neuron, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Noy N, Retinoid-binding proteins: mediators of retinoid action, Biochem J, 348 Pt 3 (2000) 481–495. [PMC free article] [PubMed] [Google Scholar]
- [317].Futterman S, Saari JC, Blair S, Occurrence of a binding protein for 11-cis-retinal in retina, J Biol Chem, 252 (1977) 3267–3271. [PubMed] [Google Scholar]
- [318].Bunt-Milam AH, Saari JC, Immunocytochemical localization of two retinoid-binding proteins in vertebrate retina, J Cell Biol, 97 (1983) 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Saari JC, Crabb JW, Focus on molecules: cellular retinaldehyde-binding protein (CRALBP), Exp Eye Res, 81 (2005) 245–246. [DOI] [PubMed] [Google Scholar]
- [320].Saari JC, Bredberg L, Garwin GG, Identification of the endogenous retinoids associated with three cellular retinoid-binding proteins from bovine retina and retinal pigment epithelium, J Biol Chem, 257 (1982) 13329–13333. [PubMed] [Google Scholar]
- [321].Saari JC, Bredberg DL, Photochemistry and stereoselectivity of cellular retinaldehyde-binding protein from bovine retina, J Biol Chem, 262 (1987) 7618–7622. [PubMed] [Google Scholar]
- [322].McBee JK, Van Hooser JP, Jang GF, Palczewski K, Isomerization of 11-cis-retinoids to all-trans-retinoids in vitro and in vivo, J Biol Chem, 276 (2001) 48483–48493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Saari JC, Nawrot M, Stenkamp RE, Teller DC, Garwin GG, Release of 11-cis-retinal from cellular retinaldehyde-binding protein by acidic lipids, Mol Vis, 15 (2009) 844–854. [PMC free article] [PubMed] [Google Scholar]
- [324].Saari JC, Nawrot M, Kennedy BN, Garwin GG, Hurley JB, Huang J, Possin DE, Crabb JW, Visual cycle impairment in cellular retinaldehyde binding protein (CRALBP) knockout mice results in delayed dark adaptation, Neuron, 29 (2001) 739–748. [DOI] [PubMed] [Google Scholar]
- [325].Xue Y, Shen SQ, Jui J, Rupp AC, Byrne LC, Hattar S, Flannery JG, Corbo JC, Kefalov VJ, CRALBP supports the mammalian retinal visual cycle and cone vision, J Clin Invest, 125 (2015) 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Maw MA, Kennedy B, Knight A, Bridges R, Roth KE, Mani EJ, Mukkadan JK, Nancarrow D, Crabb JW, Denton MJ, Mutation of the gene encoding cellular retinaldehyde-binding protein in autosomal recessive retinitis pigmentosa, Nat Genet, 17 (1997) 198–200. [DOI] [PubMed] [Google Scholar]
- [327].Burstedt MS, Sandgren O, Holmgren G, Forsman-Semb K, Bothnia dystrophy caused by mutations in the cellular retinaldehyde-binding protein gene (RLBP1) on chromosome 15q26, Invest Ophthalmol Vis Sci, 40 (1999) 995–1000. [PubMed] [Google Scholar]
- [328].Morimura H, Berson EL, Dryja TP, Recessive mutations in the RLBP1 gene encoding cellular retinaldehyde-binding protein in a form of retinitis punctata albescens, Invest Ophthalmol Vis Sci, 40 (1999) 1000–1004. [PubMed] [Google Scholar]
- [329].Sha B, Phillips SE, Bankaitis VA, Luo M, Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein, Nature, 391 (1998) 506–510. [DOI] [PubMed] [Google Scholar]
- [330].Zimmer S, Stocker A, Sarbolouki MN, Spycher SE, Sassoon J, Azzi A, A novel human tocopherol-associated protein: cloning, in vitro expression, and characterization, J Biol Chem, 275 (2000) 25672–25680. [DOI] [PubMed] [Google Scholar]
- [331].He X, Lobsiger J, Stocker A, Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W, Proc Natl Acad Sci U S A, 106 (2009) 18545–18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Pepperberg DR, Okajima TL, Wiggert B, Ripps H, Crouch RK, Chader GJ, Interphotoreceptor retinoid-binding protein (IRBP). Molecular biology and physiological role in the visual cycle of rhodopsin, Mol Neurobiol, 7 (1993) 61–85. [DOI] [PubMed] [Google Scholar]
- [333].Qtaishat NM, Wiggert B, Pepperberg DR, Interphotoreceptor retinoid-binding protein (IRBP) promotes the release of all-trans retinol from the isolated retina following rhodopsin bleaching illumination, Exp Eye Res, 81 (2005) 455–463. [DOI] [PubMed] [Google Scholar]
- [334].Gonzalez-Fernandez F, Interphotoreceptor retinoid-binding protein--an old gene for new eyes, Vision Res, 43 (2003) 3021–3036. [DOI] [PubMed] [Google Scholar]
- [335].Ripps H, Peachey NS, Xu X, Nozell SE, Smith SB, Liou GI, The rhodopsin cycle is preserved in IRBP "knockout" mice despite abnormalities in retinal structure and function, Vis Neurosci, 17 (2000) 97–105. [DOI] [PubMed] [Google Scholar]
- [336].Palczewski K, Van Hooser JP, Garwin GG, Chen J, Liou GI, Saari JC, Kinetics of visual pigment regeneration in excised mouse eyes and in mice with a targeted disruption of the gene encoding interphotoreceptor retinoid-binding protein or arrestin, Biochemistry, 38 (1999) 12012–12019. [DOI] [PubMed] [Google Scholar]
- [337].Chen Y, Noy N, Retinoid specificity of interphotoreceptor retinoid-binding protein, Biochemistry, 33(1994) 10658–10665. [DOI] [PubMed] [Google Scholar]
- [338].Chen Y, Houghton LA, Brenna JT, Noy N, Docosahexaenoic acid modulates the interactions of the interphotoreceptor retinoid-binding protein with 11-cis-retinal, J Biol Chem, 271 (1996) 20507–20515. [DOI] [PubMed] [Google Scholar]
- [339].Lin ZY, Li GR, Takizawa N, Si JS, Gross EA, Richardson K, Nickerson JM, Structure-function relationships in interphotoreceptor retinoid-binding protein (IRBP), Mol Vis, 3 (1997) 17. [PubMed] [Google Scholar]
- [340].Gonzalez-Fernandez F, Baer CA, Ghosh D, Module structure of interphotoreceptor retinoid-binding protein (IRBP) may provide bases for its complex role in the visual cycle - structure/function study of Xenopus IRBP, BMC Biochem, 8 (2007) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Adler AJ, Stafford WF 3rd, Slayter HS, Size and shape of bovine interphotoreceptor retinoid-binding protein by electron microscopy and hydrodynamic analysis, J Biol Chem, 262 (1987) 13198–13203. [PubMed] [Google Scholar]
- [342].Gonzalez-Fernandez F, Ghosh D, Focus on Molecules: interphotoreceptor retinoid-binding protein (IRBP), Exp Eye Res, 86 (2008) 169–170. [DOI] [PubMed] [Google Scholar]
- [343].Gonzalez-Fernandez F, Baer CA, Baker E, Okajima TI, Wiggert B, Braiman MS, Pepperberg DR, Fourth module of Xenopus interphotoreceptor retinoid-binding protein: activity in retinoid transfer between the retinal pigment epithelium and rod photoreceptors, Curr Eye Res, 17 (1998) 1150–1157. [DOI] [PubMed] [Google Scholar]
- [344].Baer CA, Retief JD, Van Niel E, Braiman MS, Gonzalez-Fernandez F, Soluble expression in E. coli of a functional interphotoreceptor retinoid-binding protein module fused to thioredoxin: correlation of vitamin A binding regions with conserved domains of C-terminal processing proteases, Exp Eye Res, 66 (1998) 249–262. [DOI] [PubMed] [Google Scholar]
- [345].Loew A, Gonzalez-Fernandez F, Crystal structure of the functional unit of interphotoreceptor retinoid binding protein, Structure, 10 (2002) 43–49. [DOI] [PubMed] [Google Scholar]
- [346].Ghosh D, Haswell KM, Sprada M, Gonzalez-Fernandez F, Structure of zebrafish IRBP reveals fatty acid binding, Exp Eye Res, 140 (2015) 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Kirilovsky D, Kerfeld CA, Cyanobacterial photoprotection by the orange carotenoid protein, Nat Plants, 2 (2016) 16180. [DOI] [PubMed] [Google Scholar]
- [348].Llorente B, Martinez-Garcia JF, Stange C, Rodriguez-Concepcion M, Illuminating colors: regulation of carotenoid biosynthesis and accumulation by light, Curr Opin Plant Biol, 37 (2017) 49–55. [DOI] [PubMed] [Google Scholar]
- [349].Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK, A pigment-binding protein essential for regulation of photosynthetic light harvesting, Nature, 403 (2000) 391–395. [DOI] [PubMed] [Google Scholar]
- [350].Quarmby B, Norden DA, Zagalsky PF, Ceccaldi HJ, Daumas R, Studies on the quaternary structure of the lobster exoskeleton carotenoprotein, crustacyanin, Comp Biochem Physiol B, 56 (1977) 55–61. [DOI] [PubMed] [Google Scholar]
- [351].Wald G, Nathanson N, et al. , Crustacyanin, the blue carotenoid-protein of the lobster shell, Biol Bull, 95 (1948) 249. [PubMed] [Google Scholar]
- [352].Gamiz-Hernandez AP, Angelova IN, Send R, Sundholm D, Kaila VR, Protein-Induced Color Shift of Carotenoids in beta-Crustacyanin, Angew Chem Int Ed Engl, 54 (2015) 11564–11566. [DOI] [PubMed] [Google Scholar]
- [353].Tabunoki H, Sugiyama H, Tanaka Y, Fujii H, Banno Y, Jouni ZE, Kobayashi M, Sato R, Maekawa H, Tsuchida K, Isolation, characterization, and cDNA sequence of a carotenoid binding protein from the silk gland of Bombyx mori larvae, J Biol Chem, 277 (2002) 32133–32140. [DOI] [PubMed] [Google Scholar]
- [354].Sakudoh T, Sezutsu H, Nakashima T, Kobayashi I, Fujimoto H, Uchino K, Banno Y, Iwano H, Maekawa H, Tamura T, Kataoka H, Tsuchida K, Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the Yellow blood gene, Proc Natl Acad Sci U S A, 104 (2007) 8941–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [355].Bone RA, Landrum JT, Friedes LM, Gomez CM, Kilburn MD, Menendez E, Vidal I, Wang W, Distribution of lutein and zeaxanthin stereoisomers in the human retina, Exp Eye Res, 64 (1997) 211–218. [DOI] [PubMed] [Google Scholar]
- [356].Landrum JT, Bone RA, Lutein, zeaxanthin, and the macular pigment, Arch Biochem Biophys, 385 (2001) 28–40. [DOI] [PubMed] [Google Scholar]
- [357].Hammond BR Jr., Johnson EJ, Russell RM, Krinsky NI, Yeum KJ, Edwards RB, Snodderly DM, Dietary modification of human macular pigment density, Invest Ophthalmol Vis Sci, 38 (1997) 1795–1801. [PubMed] [Google Scholar]
- [358].Bone RA, Landrum JT, Dixon Z, Chen Y, Llerena CM, Lutein and zeaxanthin in the eyes, serum and diet of human subjects, Exp Eye Res, 71 (2000) 239–245. [DOI] [PubMed] [Google Scholar]
- [359].Bone RA, Landrum JT, Guerra LH, Ruiz CA, Lutein and zeaxanthin dietary supplements raise macular pigment density and serum concentrations of these carotenoids in humans, J Nutr, 133 (2003) 992–998. [DOI] [PubMed] [Google Scholar]
- [360].Rao MN, Ghosh P, Lakshman MR, Purification and partial characterization of a cellular carotenoid-binding protein from ferret liver, J Biol Chem, 272 (1997) 24455–24460. [DOI] [PubMed] [Google Scholar]
- [361].Bhosale P, Bernstein PS, Vertebrate and invertebrate carotenoid-binding proteins, Arch Biochem Biophys, 458 (2007) 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [362].Clevidence BA, Bieri JG, Association of carotenoids with human plasma lipoproteins, Methods Enzymol, 214 (1993) 33–46. [DOI] [PubMed] [Google Scholar]
- [363].Yemelyanov AY, Katz NB, Bernstein PS, Ligand-binding characterization of xanthophyll carotenoids to solubilized membrane proteins derived from human retina, Exp Eye Res, 72 (2001) 381–392. [DOI] [PubMed] [Google Scholar]
- [364].Matthews SJ, Ross NW, Lall SP, Gill TA, Astaxanthin binding protein in Atlantic salmon, Comp Biochem Physiol B Biochem Mol Biol, 144 (2006) 206–214. [DOI] [PubMed] [Google Scholar]
- [365].Bhosale P, Larson AJ, Frederick JM, Southwick K, Thulin CD, Bernstein PS, Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye, J Biol Chem, 279 (2004) 49447–49454. [DOI] [PubMed] [Google Scholar]
- [366].Oakley AJ, Lo Bello M, Battistoni A, Ricci G, Rossjohn J, Villar HO, Parker MW, The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution, J Mol Biol, 274 (1997) 84–100. [DOI] [PubMed] [Google Scholar]
- [367].Li B, Vachali P, Bernstein PS, Human ocular carotenoid-binding proteins, Photochem Photobiol Sci, 9 (2010) 1418–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [368].Bhosale P, Li B, Sharifzadeh M, Gellermann W, Frederick JM, Tsuchida K, Bernstein PS, Purification and partial characterization of a lutein-binding protein from human retina, Biochemistry, 48 (2009) 4798–4807. [DOI] [PubMed] [Google Scholar]
- [369].Soccio RE, Breslow JL, StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism, J Biol Chem, 278 (2003) 22183–22186. [DOI] [PubMed] [Google Scholar]
- [370].Silvaroli JA, Pleshinger MJ, Banerjee S, Kiser PD, Golczak M, Enzyme That Makes You Cry-Crystal Structure of Lachrymatory Factor Synthase from Allium cepa, ACS Chem Biol, 12 (2017) 2296–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [371].Tsujishita Y, Hurley JH, Structure and lipid transport mechanism of a StAR-related domain, Nat Struct Biol, 7 (2000) 408–414. [DOI] [PubMed] [Google Scholar]
- [372].Horvath MP, George EW, Tran QT, Baumgardner K, Zharov G, Lee S, Sharifzadeh H, Shihab S, Mattinson T, Li B, Bernstein PS, Structure of the lutein-binding domain of human StARD3 at 1.74 A resolution and model of a complex with lutein, Acta Crystallogr F Struct Biol Commun, 72 (2016) 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [373].Melnicki MR, Leverenz RL, Sutter M, Lopez-Igual R, Wilson A, Pawlowski EG, Perreau F, Kirilovsky D, Kerfeld CA, Structure, Diversity, and Evolution of a New Family of Soluble Carotenoid-Binding Proteins in Cyanobacteria, Mol Plant, 9 (2016) 1379–1394. [DOI] [PubMed] [Google Scholar]
- [374].Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W, Crystal structure of spinach major light-harvesting complex at 2.72 A resolution, Nature, 428 (2004) 287–292. [DOI] [PubMed] [Google Scholar]
- [375].Wan T, Li M, Zhao X, Zhang J, Liu Z, Chang W, Crystal structure of a multilayer packed major light-harvesting complex: implications for grana stacking in higher plants, Mol Plant, 7 (2014) 916–919. [DOI] [PubMed] [Google Scholar]
- [376].Wilhelm LP, Wendling C, Vedie B, Kobayashi T, Chenard MP, Tomasetto C, Drin G, Alpy F, STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites, EMBO J, 36 (2017) 1412–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [377].Reitz J, Gehrig-Burger K, Strauss JF 3rd, Gimpl G, Cholesterol interaction with the related steroidogenic acute regulatory lipid-transfer (START) domains of StAR (STARD1) and MLN64 (STARD3), FEBS J, 275 (2008) 1790–1802. [DOI] [PubMed] [Google Scholar]
- [378].Kishida T, Kostetskii I, Zhang Z, Martinez F, Liu P, Walkley SU, Dwyer NK, Blanchette-Mackie EJ, Radice GL, Strauss JF 3rd, Targeted mutation of the MLN64 START domain causes only modest alterations in cellular sterol metabolism, J Biol Chem, 279 (2004) 19276–19285. [DOI] [PubMed] [Google Scholar]
- [379].Quadro L, Gamble MV, Vogel S, Lima AA, Piantedosi R, Moore SR, Colantuoni V, Gottesman ME, Guerrant RL, Blaner WS, Retinol and retinol-binding protein: gut integrity and circulating immunoglobulins, J Infect Dis, 182 Suppl 1 (2000) S97–S102. [DOI] [PubMed] [Google Scholar]
- [380].Tacke F, Weiskirchen R, Trautwein C, Liver function critically determines serum retinol-binding protein 4 (RBP4) levels in patients with chronic liver disease and cirrhosis, Hepatology, 48 (2008) 1724–1725; author reply 1725-1726. [DOI] [PubMed] [Google Scholar]
- [381].Cho YM, Youn BS, Lee H, Lee N, Min SS, Kwak SH, Lee HK, Park KS, Plasma retinol-binding protein-4 concentrations are elevated in human subjects with impaired glucose tolerance and type 2 diabetes, Diabetes Care, 29 (2006) 2457–2461. [DOI] [PubMed] [Google Scholar]
- [382].Heller J, Interactions of plasma retinol-binding protein with its receptor. Specific binding of bovine and human retinol-binding protein to pigment epithelium cells from bovine eyes, J Biol Chem, 250 (1975) 3613–3619. [PubMed] [Google Scholar]
- [383].Heller M, Bok D, A specific receptor for retinol binding protein as detected by the binding of human and bovine retinol binding protein to pigment epithelial cells, Am J Ophthalmol, 81 (1976) 93–97. [DOI] [PubMed] [Google Scholar]
- [384].Sivaprasadarao A, Findlay JB, The interaction of retinol-binding protein with its plasma-membrane receptor, Biochem J, 255 (1988) 561–569. [PMC free article] [PubMed] [Google Scholar]
- [385].Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H, A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A, Science, 315 (2007) 820–825. [DOI] [PubMed] [Google Scholar]
- [386].Kawaguchi R, Zhong M, Kassai M, Ter-Stepanian M, Sun H, STRA6-catalyzed vitamin A influx, efflux, and exchange, J Membr Biol, 245 (2012) 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [387].Kawaguchi R, Yu J, Ter-Stepanian M, Zhong M, Cheng G, Yuan Q, Jin M, Travis GH, Ong D, Sun H, Receptor-mediated cellular uptake mechanism that couples to intracellular storage, ACS Chem Biol, 6 (2011) 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [388].Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, Imanishi Y, Palczewski K, von Lintig J, RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome, Cell Metab, 7 (2008) 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [389].Kawaguchi R, Zhong M, Kassai M, Ter-Stepanian M, Sun H, Vitamin A Transport Mechanism of the Multitransmembrane Cell-Surface Receptor STRA6, Membranes (Basel), 5 (2015) 425–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [390].Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernandez-Martinez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nurnberg P, Reis A, Rauch A, Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation, Am J Hum Genet, 80 (2007) 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [391].Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, Bonniere M, Delahaye S, Munnich A, Encha-Razavi F, Lyonnet S, Vekemans M, Attie-Bitach T, Etchevers HC, Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6, Am J Hum Genet, 80 (2007) 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [392].Chassaing N, Golzio C, Odent S, Lequeux L, Vigouroux A, Martinovic-Bouriel J, Tiziano FD, Masini L, Piro F, Maragliano G, Delezoide AL, Attie-Bitach T, Manouvrier-Hanu S, Etchevers HC, Calvas P, Phenotypic spectrum of STRA6 mutations: from Matthew-Wood syndrome to non-lethal anophthalmia, Hum Mutat, 30 (2009) E673–681. [DOI] [PubMed] [Google Scholar]
- [393].Chassaing N, Ragge N, Kariminejad A, Buffet A, Ghaderi-Sohi S, Martinovic J, Calvas P, Mutation analysis of the STRA6 gene in isolated and non-isolated anophthalmia/microphthalmia, Clin Genet, 83 (2013) 244–250. [DOI] [PubMed] [Google Scholar]
- [394].Casey J, Kawaguchi R, Morrissey M, Sun H, McGettigan P, Nielsen JE, Conroy J, Regan R, Kenny E, Cormican P, Morris DW, Tormey P, Chroinin MN, Kennedy BN, Lynch S, Green A, Ennis S, First implication of STRA6 mutations in isolated anophthalmia, microphthalmia, and coloboma: a new dimension to the STRA6 phenotype, Hum Mutat, 32 (2011) 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [395].Ruiz A, Mark M, Jacobs H, Klopfenstein M, Hu J, Lloyd M, Habib S, Tosha C, Radu RA, Ghyselinck NB, Nusinowitz S, Bok D, Retinoid content, visual responses, and ocular morphology are compromised in the retinas of mice lacking the retinol-binding protein receptor, STRA6, Invest Ophthalmol Vis Sci, 53 (2012) 3027–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [396].Amengual J, Zhang N, Kemerer M, Maeda T, Palczewski K, Von Lintig J, STRA6 is critical for cellular vitamin A uptake and homeostasis, Hum Mol Genet, 23 (2014) 5402–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [397].Berry DC, Jacobs H, Marwarha G, Gely-Pernot A, O'Byrne SM, DeSantis D, Klopfenstein M, Feret B, Dennefeld C, Blaner WS, Croniger CM, Mark M, Noy N, Ghyselinck NB, The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye, J Biol Chem, 288 (2013) 24528–24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [398].Blomhoff R, Norum KR, Berg T, Hepatic uptake of [3H]retinol bound to the serum retinol binding protein involves both parenchymal and perisinusoidal stellate cells, J Biol Chem, 260 (1985) 13571–13575. [PubMed] [Google Scholar]
- [399].Yamamoto Y, Yoshizawa T, Kamio S, Aoki O, Kawamata Y, Masushige S, Kato S, Interactions of transthyretin (TTR) and retinol-binding protein (RBP) in the uptake of retinol by primary rat hepatocytes, Exp Cell Res, 234 (1997) 373–378. [DOI] [PubMed] [Google Scholar]
- [400].Gjoen T, Bjerkelund T, Blomhoff HK, Norum KR, Berg T, Blomhoff R, Liver takes up retinol-binding protein from plasma, J Biol Chem, 262 (1987) 10926–10930. [PubMed] [Google Scholar]
- [401].Alapatt P, Guo F, Komanetsky SM, Wang S, Cai J, Sargsyan A, Rodriguez Diaz E, Bacon BT, Aryal P, Graham TE, Liver retinol transporter and receptor for serum retinol-binding protein (RBP4), J Biol Chem, 288 (2013) 1250–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [402].Chen Y, Clarke OB, Kim J, Stowe S, Kim YK, Assur Z, Cavalier M, Godoy-Ruiz R, von Alpen DC, Manzini C, Blaner WS, Frank J, Quadro L, Weber DJ, Shapiro L, Hendrickson WA, Mancia F, Structure of the STRA6 receptor for retinol uptake, Science, 353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [403].Shi Y, Obert E, Rahman B, Rohrer B, Lobo GP, The Retinol Binding Protein Receptor 2 (Rbpr2) is required for Photoreceptor Outer Segment Morphogenesis and Visual Function in Zebrafish, Sci Rep, 7 (2017) 16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [404].Breen CJ, Martin DS, Ma H, McQuaid K, O'Kennedy R, Findlay JB, Production of functional human vitamin A transporter/RBP receptor (STRA6) for structure determination, PLoS One, 10 (2015) e0122293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [405].Kawaguchi R, Yu J, Wiita P, Honda J, Sun H, An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism, J Biol Chem, 283 (2008) 15160–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [406].Laursen KB, Kashyap V, Scandura J, Gudas LJ, An alternative retinoic acid-responsive Stra6 promoter regulated in response to retinol deficiency, J Biol Chem, 290 (2015) 4356–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [407].Kelly M, Widjaja-Adhi MA, Palczewski G, von Lintig J, Transport of vitamin A across blood-tissue barriers is facilitated by STRA6, FASEB J, 30 (2016) 2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [408].Blaner WS, STRA6, a cell-surface receptor for retinol-binding protein: the plot thickens, Cell Metab, 5 (2007) 164–166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.