Abstract
As sessile organisms, plants must properly coordinate their growth and developmental programs with changes in the environment. The integration of exogenous environmental cues with endogenous plant hormone responses often occurs through physical protein–protein interactions (PPIs). However, a comprehensive PPI network that mediates environmental and hormonal responses has not been established. In this study, we initially cloned 113 phytohormone‐related genes and 29 light signaling components of Arabidopsis and then individually tested their mutual interactions (in total 2,655 tests) using a yeast‐two‐hybrid approach to ultimately identify 141 interactions. Based on these interaction results, we next revealed the signaling cross talk between jasmonate and abscisic acid by characterizing the JAZ1‐PYL4 and JAZ1‐ABI1 interactions. Thus, we generated a useful resource for the community to explore the molecular mechanisms underlying signaling interactions between plant hormones and/or with light.
Keywords: abscisic acid, jasmonate, JAZ1, plant hormone, protein–protein interaction
1. INTRODUCTION
Phytohormone and environmental cues coordinately regulate plant growth and development partially though protein–protein interactions (PPIs) among regulatory proteins in various signaling pathways (Chaiwanon, Wang, Zhu, Oh, & Wang, 2016). PPIs in hormone and environmental signaling pathways must first be tested to elucidate the interactions among signaling pathways. Brassinosteroids (BR), auxins, and light have recently been shown to coordinate seedling morphogenesis through PPIs. Three transcription factors BRASSINAZOLE‐RESISTANT 1 (BZR1, a transcription factor involved in BR signaling), PHYTOCHROME‐INTERACTING FACTOR 4 (PIF4, a transcription factor involved in light signaling) and AUXIN RESPONSE FACTOR 6 (ARF6, a transcription factor involved in auxin signaling) physically interact and enhance the transcriptional activities of the other transcription factors to promote cell elongation (Oh et al., 2014; Oh, Zhu, & Wang, 2012). On the other hand, DELLAs, repressor proteins of gibberellin (GA) signaling, also interact with these transcription factors and inhibit their transcriptional activities to antagonize the stimulatory effects of BR or auxin (Chaiwanon et al., 2016; Wang, Bai, & Wang, 2014). In addition, photoreceptors (Cryptochrome [CRY] or Phytochrome B [phyB]) interact with AUX/IAA proteins (Auxin/INDOLE‐3‐ACETIC ACID proteins, negative regulators of auxin signaling) to mediate the coherent regulation of light and auxin signaling (Mao et al., 2019; Xu, He, & Zhang, 2018).
Nonetheless, numerous gaps exist in our current understanding of hormone signal cross talk. For example, the biotic stress hormone jasmonate (JA) has been reported to interact with the abiotic stress hormone abscisic acid (ABA; Aleman et al., 2016; Howe, Major, & Koo, 2018; Lackman et al., 2011; Pauwels et al., 2015). The ABA receptor PYL4 is implicated in the co‐regulatory effects of ABA and JA on plant metabolism and growth (Lackman et al., 2011). Another ABA receptor, PYL6, directly interacts with and alters the transcriptional activity of MYC2 (a key transcription factor involved in JA signaling; Aleman et al., 2016). The E3 ubiquitin ligase KEEP ON GOING (KEG), a known repressor of ABA INSENSITIVE 5 (ABI5) in the ABA signaling pathway, directly interacts with JASMONATE‐ZIM‐DOMAIN PROTEIN 12 (JAZ12) and modulates its stability (Pauwels et al., 2015). Furthermore, JAZ proteins interact with and regulate MYC/MYB TFs, which mediated partially ABA signaling pathway (Abe et al., 2003; Adie et al., 2007). Recently, ABA has been shown to modulate the expressions of JA‐responsive genes (Bodenhausen & Reymond, 2007). In addition, exogenous JA has been reported to increase the ABA concentration fourfold and sevenfold (de Ollas, Arbona, & Gomez‐Cadenas, 2015).
In the past decade, researchers have generated protein interactome networks through large‐scale yeast‐two‐hybrid (Y2H) screens (Arabidopsis Interactome Mapping Consortium, 2011). Although these researchers have generated Arabidopsis PPI networks on the proteome scale, these screening methods are not specifically designed for testing PPIs in the hormone or light signaling field. Although 795 proteins involved in plant hormone signaling and 47 proteins involved in light signaling have been selected as bait proteins in previous screens (Arabidopsis Interactome Mapping Consortium, 2011), the studies screened these baits with a collection of open reading frames, but not with specific signaling proteins. Thus, a point‐by‐point protein interactome is need to identify the PPIs among plant hormone and light signaling pathways. In this study, we initially chose 113 phytohormone‐related and 29 light signaling components of Arabidopsis, and then cloned them into pGADT7 and pGBKT7 through EXIN reactions, respectively. After 2,655 individual yeast‐two‐hybrid assays, 141 PPIs were successfully identified. We then focused on JAZ1‐PYL4 and JAZ1‐ABI1 interactions to address the crosstalk between JA and ABA signaling. Taken together, we have generated a useful resource to identify novel signaling interactions between endogenous and exogenous signaling pathways.
2. MATERIALS AND METHODS
2.1. Plant material and growth conditions
The Arabidopsis thaliana 35Spro:JAZ1‐GUS transgenic lines (Columbia ecotype) were surface‐sterilized and sown on 1× Murashige and Skoog (MS) medium supplemented with 1% agar (Sigma). The seeds were stratified in the darkness for three days at 4°C and cultured in light chambers for seven days at 22°C.
The ABI1‐FLAG constructs were transformed into Agrobacterium tumefaciens GV3101 using the freeze‐thaw method and then transformed into 35Spro:JAZ1‐GUS plants using the floral dip method. Transgenic plants were selected on MS medium containing 50 mg/L hygromycin.
2.2. Library construction and PPI identification
The regulators (or transcription factors) involved in light or circadian rhythm signaling pathways were amplified with specific primers (Table S1) and individually cloned into the expression vector pGBKT7 though EXIN reactions (Biogle), according to the manufacturer's instructions. The cDNAs of hormone‐related genes were cloned into the pGADT7 vector. The pGADT7 and pGBKT7 vectors were transformed into the Y187 and AH109 yeast strains, respectively, using the LiAc‐mediated method. Mating was performed by simply mixing 20 μL of the donor and host strains. Then, the mixture was transferred to a new sterile 96‐well plate containing 100 μL of YPDA medium. The mixture was cultured at 30°C (200 rpm) for 16 hr. The mating products (10 μL/well) were pipetted onto SD/‐Leu/‐Trp and SD/‐His/‐Leu/‐Trp plates and incubated for 4–7 days at 30°C.
2.3. In vitro pull‐down assay
The cDNA fragments encoding PYL4 and JAZ1 (full length and N‐terminus) were cloned into pGEX5x and pMAL‐c5X, respectively. The recombinant proteins were expressed in the Escherichia coli BL21 strain. PYL4‐GST and JAZ1‐GST proteins were purified using reagents from NEB, according to the manufacturer's instructions. In the pull‐down assay, proteins were incubated with glutathione Sepharose 4B at 4°C for 2 hrs in binding buffer (150 mmol/l NaCl, 100 mmol/l Tris, pH 7.5, 1 mmol/l EDTA, 0.1% TritonX‐100, and 1 mmol/l PMSF). The beads were washed five times with washing buffer (50 mmol/l NaCl, 100 mmol/l Tris pH 8.0, 1 mmol/l EDTA, and 0.1% TritonX‐100) and boiled with 4× loading buffer and 1 mol/l DTT. Pull‐down products were separated on SDS‐PAGE gels and analyzed by performing immunoblot analyses.
2.4. Co‐IP
The four‐day‐old ABI1‐FLAG/JAZ1‐GUS Arabidopsis seedlings were ground in liquid nitrogen. For anti‐FLAG immunoprecipitation, proteins were extracted with buffer (50 mmol/l Tris‐HCl (pH 7.4), 100 mmol/l NaCl, 10% glycerol, 0.1% Tween‐20, 1 mmol/l DTT, 1×Protease Inhibitor Mixture (Roche), and 50 μmol/l MG132). After centrifugation, the supernatant was incubated with anti‐FLAG M2 Affinity Gel (F1804; Sigma‐Aldrich) for 1 hr at 4°C. The immunoprecipitation was washed with the extraction buffer 3 times. The immunoprecipitation product was boiled with 4× loading buffer and 1 mol/L DTT. The eluate was subjected to Western blot analyses with anti‐FLAG (F1804; Sigma) and anti‐GUS (A5790; Thermo) antibody.
2.5. Immunoblot analysis and quantification
For JAZ1‐GUS immunoblots, nine‐day‐old 35Spro:JAZ1‐GUS transgenic seedlings were transferred to liquid MS medium containing 100 μmol/l MeJA or 100 μmol/l MeJA plus 60 μmol/l ABA. Seedlings were collected at different time points for protein extraction. JAZ1‐GUS fusion proteins were extracted and visualized by performing immunoblots using the GUS antibody (Thermo; A5790), and a nonspecific HSP90 protein served as a loading control. The relative levels of the JAZ1‐GUS protein were measured after normalization to the loading control using the Image J software.
2.6. RT‐qPCR
After treated with MeJA in the presence or absence of ABA, seven‐day‐old Col‐0 seedlings were collected at different time points. Total RNA was isolated with TRIzol reagent (Invitrogen; 03877) and then reverse‐transcribed into cDNAs using a reverse transcription system (TaKaRa RR820) according to the manufacturer's instructions. The transcript levels of individual genes were measured using gene‐specific primers. RT‐qPCR was performed on a LightCycler 96 PCR instrument (Roche) with SYBR Green Master Mix (TaKaRa RR047A). The relative expression levels of the genes were quantified via the 2−ΔΔCT Ct method, and ACTIN2 served as the internal control.
3. RESULTS
3.1. Construction of a regulatory factor library
We initially selected receptors, important regulatory factors, and key transcriptional factors involved in phytohormone and light signaling pathways, including 113 phytohormone signaling‐related genes and 29 light signaling components of Arabidopsis, from literature to prepare constructs and identify the PPIs that participate in the hormone or light signaling pathways (Figure 1 and Table S2). The cDNAs of phytohormone‐related genes and light signaling components were cloned into pGADT7 and pGBKT7 vector, respectively, through EXIN reactions. The EXIN reaction system enables us to easily transfer the constructs from the pGADT7 or pGBKT7 vector into the compatible binary vectors through a one‐step reaction, which will save time and labor in the downstream plant transformation process (Figure 2).
FIGURE 1.
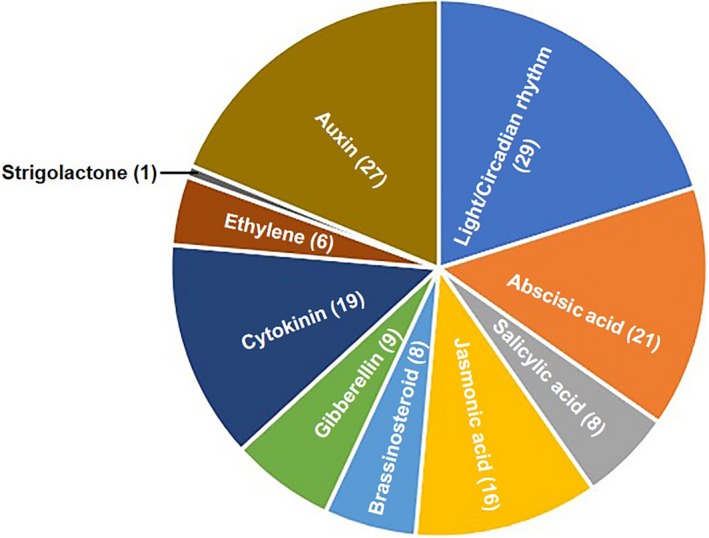
Overview of proteins included in the library. One hundred forty‐two proteins involved diverse signaling pathways are included in the library. The number of proteins in each signaling pathway is listed in brackets after the name of hormones and environmental cues
FIGURE 2.
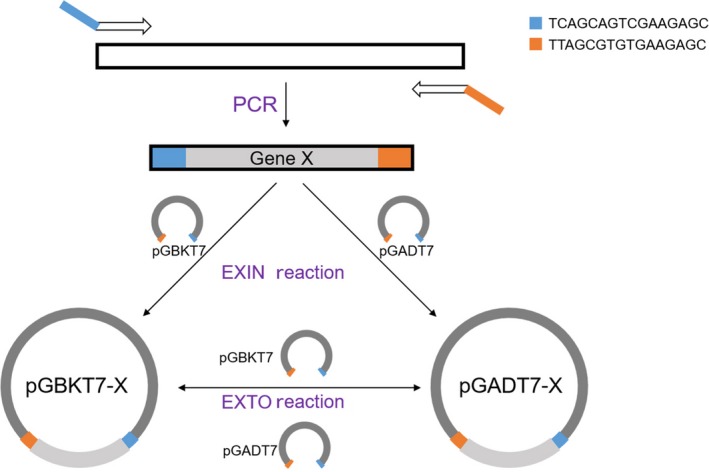
Schematic of the library construction procedure. All cDNAs were amplified with specific primers and cloned into pGBKT7 or pGADT7 (Biologe) though EXIN reactions. EXTO recombination reactions were performed to transfer cDNAs between pGBKT7 and pGADT7
3.2. Identification of interacting proteins that involved in cross talk between the light and hormone signaling pathways
We next performed a point‐by‐point analysis the mutual interactions between light and hormone signaling pathways using a high‐throughput screening system to identify PPIs involved in signaling crosstalk. After screening 2,655 individual yeast clones, we finally identified 141 putative PPIs (Table S3). As shown in Table 1 and Table S3, 129 PPIs were identified in this screening. These interactions might mediate the cross talk between light and hormone signaling. Among these interactions, many Aux/IAA proteins were identified to interact with core components (CRY2, CCA1 (CCA1 (CIRCADIAN CLOCK ASSOCIATED 1)), and CDF1) of the light or circadian signaling pathways (Table 1 and Table S3). Aux/IAA proteins are considered co‐receptors of TIR1/AFB for auxin and interact with the auxin receptors TIR1/AFB and transcriptional factors (auxin response factors ARFs) that link auxin perception to auxin response (Leyser, 2018). Based on these results, auxin, light, and circadian signaling synergistically regulate plant growth and development. CRY1, the homologue of CRY2, interacted with IAA7, IAA12, IAA17, ARF6, and ARF8 and mediated the light‐auxin signaling‐induced antagonistic regulation of hypocotyl elongation in recent studies (Mao et al., 2020; Xu et al., 2018). These PPIs confirm that auxin and light co‐regulate plant development.
TABLE 1.
Overview of interactions involved in phytohormone and light signaling pathways
| AD BD | Abscisic acid | Salicylic acid | Jasmonate | Brassinosteroid | Gibberellin | Cytokinin | Ethylene | Auxin |
|---|---|---|---|---|---|---|---|---|
| CRY2 | MYC2 | GID1A | ARR5, CKH1 | EIL1 | ARF16, IAA3, IAA12, IAA16 | |||
| ELF3 | NINJA | AHP4, AHP5, ARR6, CRF1 | CTR1, EIL1, ERF1, EBF1 | AFB2, ARF16, AXR4, IAA3, IAA16 | ||||
| ELF4 | SnRK2.6 | TGA3 | EIL1 | IAA3, IAA12, IAA16 | ||||
| PIF4 | EIN3, EBF1 | TPR1 | ||||||
| CCA1 | SLY1 | AHP2 | EIL1 | ARF16, IAA3, IAA12, IAA16 | ||||
| PHOT1 | MYC2 | ARR15 | AFB2, AFB3 | |||||
| CDF3 | CPK32 | ARR4 | EIL1 | ARF16, IAA3, IAA12 | ||||
| CKB2 | AHP2 | |||||||
| CIB1 | PYL4, PYL7, HAI1 | BEN1 | RGL1, RGL2 | ARR4, CRF6 | EIL1 | AFB4, AXR4 | ||
| CDF2 | IAA3 | |||||||
| TOC1 | CPK32 | MYC2 | CTR1, EIL1 | ARF16, IAA3, IAA16 | ||||
| CDF1 | HAB2, AHG3 | GID1B, RGL2 | ARR4 | EIL1 | IAA3, IAA12, IAA16 | |||
| CKA1 | TGA3 | CKH1 | EIL1 | ARF16, IAA3, IAA12 | ||||
| PPK1 | SnRK2.2 | BAK1 | GID1A, GID1B, RGL1, SLY1 | CKH1 | ARF8, TPR2 | |||
| PPK2 | ARR8 | EIL1 | ARF16, IAA3, IAA12, IAA16 | |||||
| PPK3 | ABF3, CPK4 | WAK2 | BAK1 | RGL3, SLY1 | ARF16, IAA3, IAA12, IAA16 | |||
| PPK4 | PYL1, ABF3, CPK32 | BAK1 | ARR6, ARR16, CRF6 | CTR1, EIL1 | ARF16, IAA3, IAA7, IAA12, IAA17 | |||
| BIC1 | GID1A, RGL3 | AHP2 | ARF10, IAA3, IAA16 |
PPK proteins, such as casein kinase, interact with, and phosphorylate RYR/PYLs, CRY2, phyB, and PIF3, and they are involved in light signaling and the ABA response (Chen, Qu, Xu, Zhu, & Xue, 2018; Liu, Wang, & Deng, 2017; Ni, Xu, & González‐Gr&ío, E., Chalkley, R. J. Huhmer, A. F. R., Burlingame, A. L., Wang, Z. Y., & Quail, P. H., 2017). We identified 39 interactions between PPK proteins and potential target proteins. Among these interactions, 14 interactions between PPK proteins and Aux/IAAs or ARFs suggest that the PPKs‐mediated IAA/ARF phosphorylation regulates auxin signaling. PPK proteins may be new kinases involved in phosphorylating IAAs/ARFs (Table 1 and Table S3). Additionally, PPK proteins interacted with kinase, such as SnRK2.2, CPK4, CPK32, BAK1, and CTR1 (Table 1 and Table S3), suggesting that the protein kinase cascade plays a pivotal role in the integration of various cues to regulate plant growth and development.
3.3. Identification of JAZ interactors
We transferred coding sequences of JAZs from pGADT7 into pGBKT7 though EXTO reactions and screened PPIs between JAZs and key components of ABA signaling pathways to assess the convenience and effectiveness of transferring clone vectors between pGADT7 and pGBKT7 We identified seven proteins that interacted with JAZ1, including the ABA receptors PYL4 and PYL7, the clade‐A PP2C proteins HAB1, ABI1, and AHG3, and the kinases SnRK2.2 and SnRK2.3 (Figure 3a, Figure S1 and Table S3). In addition, JAZ9 interacted with PYL4, AHG3, SnRK2.2, and SnRK2.3 (Figure 3a and Table S3).
FIGURE 3.
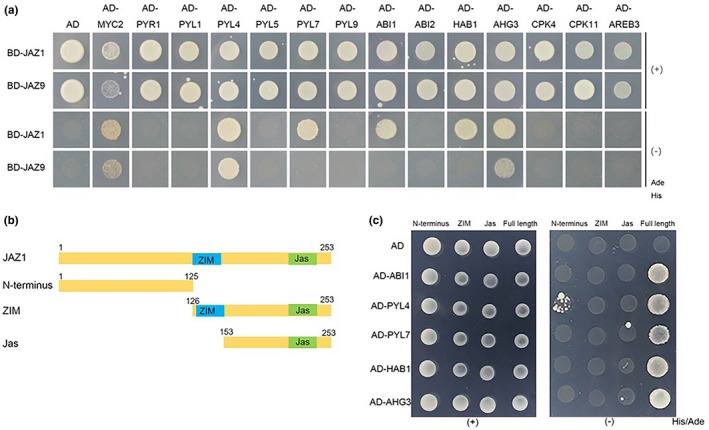
JAZ1/JAZ9 interacted with key components of the ABA signaling pathway. (a) Y2H assays designed to test the interactions between JAZs and key components of the ABA signaling pathway. (b) Schematic showing full‐length JAZ1 and its deletion proteins. The ZIM domain and Jas domain are shown in blue and green, respectively. The numbers indicate the positions of the first and last amino acid of the domain constructs. (c) Y2H assays designed to test the interactions between truncated versions of JAZ1 and key components of the ABA signaling pathways
3.4. Mapping the binding domains of JAZ1
We analyzed the interactions in yeast to confirm the interactions identified using the screening system described above. As shown in Figure 3a, JAZ1 indeed interacted with PYL4, PYL7, HAB1, AHG3, and ABI1 in yeast. JAZ9 interacted with PYL4 and AHG3 (Figure 3a). We generated constructs expressing the N‐terminus (amino acids 1–120), ZIM domain and Jas domain‐comprising C‐terminus (amino acid 126–253), and Jas domain‐comprising C‐terminus (amino acid 153–253) of JAZ1 to map the domains of JAZ1 that mediate the interactions with PYL4, PYL7, HAB1, ABI1, and AHG3 (Figure 3b). The yeast‐two‐hybrid results showed that the N‐terminus, rather than the C‐terminus of JAZ1, interacted with PYL4 (Figure 3c), indicating that the N‐terminus of JAZ1 is essential for the interaction between PYL4 and JAZ1. Additionally, the N‐terminal region may participate in the cross talk between JA signaling and ABA signaling. The N‐terminus was also shown to interact with DELLA proteins to mediate the interactions between JA and GA signaling (Pauwels & Goossens, 2011). However, none of the truncated versions of JAZ1 interacted with PYL7, HAB1, and AHG3 (Figure 3c), suggesting that different motifs of JAZ1 or proper folding are required for these interactions.
3.5. Confirmation of the interactions of two proteins with JAZ1
We performed pull‐down assays with maltose‐binding protein (MBP)‐tagged JAZ1, GST‐tagged PYL4, and the N‐terminus of JAZ1 fusion proteins expressed in E. coli to verify the interaction between JAZ1 and PYL4. As expected, PYL4 was pulled down by both the full‐length protein and N‐terminus of JAZ1 (Figure 4a), consistent with the results from the yeast‐two‐hybrid screen.
FIGURE 4.
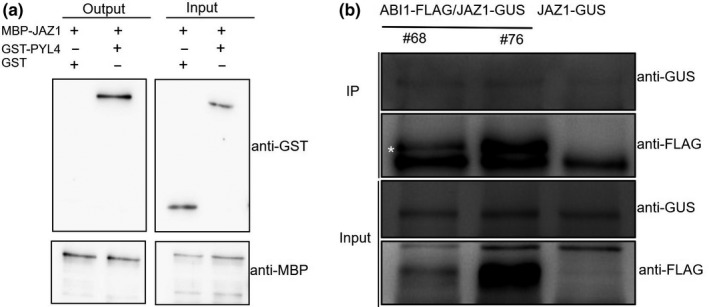
Interactions between JAZ1, PYL4, and ABI1. (a) Pull‐down assay showing interactions between PYL4 and JAZ1 (full length or the N‐terminus). (b) Co‐IP assay showing that JAZ1 interacts with ABI1 in vivo. The asterisk indicates the target protein
We performed Co‐IP experiments with proteins extracts prepared from transgenic lines expressing both ABI1‐FLAG and JAZ1‐GUS to confirm whether JAZ1 might interact with ABI1 in vivo. The coimmunoprecipitation assays revealed the coimmunoprecipitation of ABI1‐FLAG with JAZ1‐GUS (Figure 4b). The results further confirmed the interaction between JAZ1 and ABI1.
Based on our findings, JAZ1 interacts with PYL4 and ABI1, and the N‐terminal region is required for the interaction between JAZ1 and PYL4. A reasonable hypothesis is that ABA and JA co‐regulate plant growth and development and that JAZ1 is involved in this process. In addition, these interactions may explain the JA‐hypersensitive phenotype of pyl4 mutant and the ABA‐hypersensitive phenotype of osjaz1 mutant (Fu et al., 2017; Lackman et al., 2011).
3.6. Exogenous ABA enhanced the JA response
Protein interactions suggested that JAZ1 is a node for cross talk between the JA and ABA signaling pathways. JA‐induced JAZ degradation provides an efficient mechanism to dissociate corepressor modules and relieve the repression of downstream transcription factors. We initially analyzed the effect of ABA on JAZ1 degradation to investigate the potential effects of ABA on JA signaling. Nine‐day‐old 35S:JAZ1‐GUS seedlings were treated with MeJA or MeJA and ABA for 0, 5, 10, 15, 20, 30, and 40 min, and then, seedlings were harvested for protein extraction. JAZ1 was detected by immunoblotting with an anti‐GUS antibody. After treatment with MeJA treatment alone, the JAZ1‐GUS protein was degraded (Figure 5a). Notably, more rapid degradation of JAZ1 was observed after the simultaneous treatment MeJA and ABA (Figure 5a). A significant decrease in the levels of the JAZ1‐GUS protein abundance was observed at 10 min after treatment with MeJA and ABA, and by 20 and 30 min, more rapid degradation was observed (Figure 5b), suggesting that ABA accelerates JA‐induced JAZ1 degradation.
FIGURE 5.
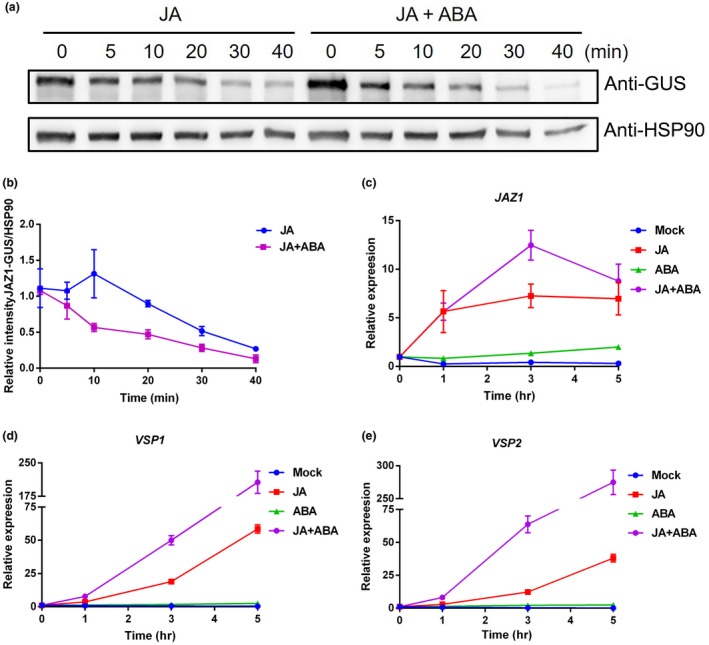
ABA treatment potentiates the JA signaling output. (a) and (b) Immunoblot analysis (a) and quantitative analysis (b) of the JAZ1 protein in 35S:JAZ1‐GUS plants cultivated under the indicated conditions. The level of JAZ1 was analyzed by immunoblotting using an anti‐GUS antibody. HSP90 was used as the loading control. (c)‐(e) RT‐qPCR analysis of the expression of the JAZ1 (c), VSP1 (d), and VSP2 (e) transcripts in plants cultivated under the indicated conditions. Error bars represent the SE of three replicates
The expression of the JAZ1 gene is rapidly induced by JA‐Ile accumulation (Chini et al., 2007; Thines et al., 2007). We then detected the expression of JAZ1 under different conditions. Seven‐day‐old Col‐0 seedlings were treated with MeJA (10 μmol/l) or MeJA (10 μmol/l) and ABA (60 μmol/l) for different time. JAZ1 expression was induced by MeJA (10 μmol/l), consistent with previous reports (Chini et al., 2007). After treatment with ABA (60 μmol/l) alone, no detectable change in the expression level of JAZ1 expression was observed (Figure 5c). Notably, a significant increase in expression was observed the treatment with the combination of MeJA and ABA (Figure 5c), suggesting that ABA may enhance JA‐induced JAZ1 expression.
Because JAZ1 interacts with core components of the ABA signaling pathway and ABA accelerates JA‐induced JAZ1 degradation, we sought to explore whether ABA positively regulates JA signaling. Therefore, we examined the expression of the JA‐responsive genes VEGETATIVE STORAGE PROTEIN 1 (VSP1) and VSP2 after treatment with MeJA in the presence or absence of ABA. The expression of VSP1 and VSP2 was induced by MeJA, but not by ABA (Figure 5d,e). However, markedly higher levels of the VSP1 and VSP2 transcripts were detected after treatment with the combination of MeJA and ABA than treatment with MeJA alone (Figure 5d,e), suggesting that ABA enhances the function of JAZ1‐MYC2‐VSP in plant defense mechanisms.
4. DISCUSSION
4.1. Identification of PPIs involved in the cross talk among endogenous and environmental signaling pathways
Signal transduction pathways are partly based on PPIs that transduce information to produce specific cellular outputs. The identification of PPIs among key components in different signaling pathways, which provide an excellent initial step toward revealing the biological networks, is essential for understanding the cross talk between different signaling pathways and the signaling interactions among plant hormones and environmental cues. Recently, some methods have been developed to reveal signaling interaction networks in Arabidopsis, such as mass spectrometry, yeast‐two‐hybrid, and Co‐IP. To date, cross talk between diverse signaling pathways has been studied using genetic and molecular methods. Given the high degree of integration between diverse signaling pathways and the complexity of the information processing system, a high‐throughput analysis of PPIs is required to obtain a better understanding of the regulatory system in Arabidopsis. Although the Arabidopsis interactome map has been reported (Arabidopsis Interactome Mapping Consortium, 2011), we constructed a library with specific signaling proteins. The library contains 113 phytohormone‐related genes and 29 light signaling components, which function as receptors, important regulatory factors and key transcriptional factors in phytohormone and light signaling pathways (Figure 1). Half of the 113 phytohormone‐related genes overlap with the 795 proteins in Arabidopsis interactome map (Arabidopsis Interactome Mapping Consortium, 2011). In addition, the exchange of cloning vectors between pGADT7 and pGBKT7 in one‐step reaction is simple and convenient (Figure 2). Thus, the library can be used to screen PPIs between any two signaling pathways. The library is now available for other researchers to use in their studies.
According to recent studies, the light, auxin, ethylene, GA, and BR pathways converge in plant growth and development (Chaiwanon et al., 2016). The basic helix‐loop‐helix transcription factors, phytochrome‐interacting factors (PIFs), not only function in photomorphogenesis, but also modulate the expression levels of essential regulators in phytohormone signaling pathways (Duek & Fankhauser, 2005; Leivar et al., 2008; Quail, 2011; Shin et al., 2009; Verma, Ravindran, & Kumar, 2016). For example, PIF1 directly induces the expression of RGA and GAI, which encode two key DELLA proteins that function as repressors of GA signaling (Oh et al., 2007). In addition, PIF proteins are also regulated by phytohormones and play important roles in the crosstalk between light and hormone signaling pathways (Feng et al., 2008; Zhong et al., 2012). DELLA proteins directly interact with PIF3 and prevent it from binding to its target gene promoters and regulating gene expression (Feng et al., 2008). The expression level of PIF3 is modulated by ethylene‐insensitive 3 (EIN3, Zhong et al., 2012). In the present study, EIN3 interacted with PIF4, and EIL1 interacted with most of components of the light signaling pathway (Table 1), suggesting that ethylene and light co‐regulate plant growth and development, and EIN/EIL proteins play important roles in this process.
Auxin plays important roles in the spatial regulation of plant growth and development. Aux/IAA proteins are considered co‐receptors of TIR1/AFB for auxin, interact with transcription factors (auxin response factors ARFs), and inhibit their transcriptional activities. The transcriptional activities of ARFs are also regulated by kinase‐mediated phosphorylation. Many kinase proteins (BIN2, phytochrome, MAPK, and PINOID) are involved in the phosphorylation of IAAs/ARFs (Cho et al., 2014; Colón‐Carmona, Chen, Yeh, & Abel, 2000; Dharmasiri & Estelle, 2004). However, PPKs have not been reported to regulate IAAs/ARFs. We identified 14 interactions between PPK proteins and IAAs/ARFs in the yeast‐two‐hybrid screen, suggesting that PPK may be a novel type of kinase regulating IAAs/ARFs. The validity and biological implications of those interactions must be explored in further studies.
Auxin helps plants properly respond to light (Mao et al., 2020; Xu et al., 2018). In our screen, many IAA proteins were identified to interact with CRY2, a core component of the light signaling pathway (Table 1). CRY1, a homologue of CRY2, interacted with IAA7, IAA12, IAA17, ARF6, and ARF8 and mediated light‐auxin signaling‐induced antagonistic regulation of hypocotyl elongation in recent studies (Mao et al., 2020; Xu et al., 2018). These PPIs confirm that auxin and light co‐regulate plant development. On the other hand, auxin signal transduction is regulated by the circadian clock (Covington & Harmer, 2007). The circadian clock controls the sensitivity to auxin at both transcriptional level and stem growth level (Covington & Harmer, 2007). CCA1, an MYB domain transcription factor, is postulated to be an important component of the circadian clock. In the present study, we identified three Aux/IAA family members that interacted with CCA1 (Table 1). CCA1 may compete with ARFs for binding to IAAs, disturb the interactions between IAAs and ARFs, and relieve Aux/IAA‐mediated transcriptional repression. Additional physiological and biochemical evidence is needed to confirm this hypothesis.
Casein kinase PPK proteins have been reported to interact with and phosphorylate RYR/PYLs, CRY2, phyB, and PIF3, and to participate in light signaling and the ABA response (Chen et al., 2018; Liu et al., 2017; Ni et al., 2017). PPK proteins were identified as interactors of the kinase, SnRK2.2, CPK4, CPK32, BAK1, and CTR1 (Table 1). These PPIs suggest that phosphorylation signaling cascades may play pivotal roles in integrating various cues to regulate plant growth and development. In this screen, PPK4 was identified as a PYL1 interactor, consistent with a previous report (Ni et al., 2017), validating the effectiveness of the screening system.
Because one protein interacts with many proteins, one possible explanation would be that this protein has multiple functions in integrating multiple informational cues. The system may also produce false‐positive results. Hence, all the PPIs obtained from the yeast‐two‐hybrid must be verified in a further study. Many members of the Aux/IAA family interacted with the same protein, suggesting that these proteins might exhibit functional redundancy. Therefore, multiple mutants are required to explore the significance of these interactions.
4.2. JAZ1‐PYL4 and JAZ1‐ABI1 interactions mediate cross talk between JA and ABA signaling
Previous studies have reported both synergistic and antagonistic interactions between the JA and ABA signaling pathways, which depend on the developmental stage, tissue type, and response (Adie et al., 2007; Munemasa et al., 2007; Nahar, Kyndt, Nzogela, & Gheysen, 2012). JAZ proteins integrate multiple cues and mediate cross talk between the JA and ABA signaling pathways. MYC2 mediates the synergistic regulation of JA and ABA signaling to inhibit primary root growth. The transcriptional activity of MYC2 is not only repressed by JAZ proteins but also modulated by the ABA coreceptor PYL6 (Aleman et al., 2016). JAZ12 stability is modulated by the E3 ubiquitin ligase KEG (Pauwels et al., 2015). In addition, OsJAZ1, whose mutant plants are hypersensitive to MeJA and ABA, plays a role in the drought resistance of rice partially via the ABA and JA signaling pathways (Fu et al., 2017). The pyl4 mutant plants are hypersensitive to a JA treatment, and the expression levels of this subgroup of ABA receptors change upon JA treatment (Lackman et al., 2011). The phenotypes of osjaz1 and pyl4 mutants suggest that JAZ1 and PYL4 are involved in the cross talk between the JA and ABA signaling pathways. Recently, we reported direct interactions between JAZs (JAZ1 and JAZ9), ABA receptors (PYL4 and PYL7), and protein phosphatase 2C proteins (ABI1, HAB1, and AHG3) in yeast‐two‐hybrid experiments (Figure 3a and S1), consistent with the JA‐hypersensitive phenotype of the pyl4 mutant. As homologous proteins, PYL4 and PYL7 interact with JAZ1, suggesting that PYL4 and PYL7 might be functionally redundant. JAZ proteins contain a JA‐associated (Jas) domain in the C‐terminus, ZIM domain, and N‐terminal region. The N‐terminal region of JAZ1 interacts with DELLAs (RGA, GAI, and RGL1), and links JA signaling and GA signaling (Pauwels & Goossens, 2011). Furthermore, using an array of truncated versions of JAZ1, we determined that only the mutated JAZ1 version (N‐terminal region) interacted with PYL4 (Figure 3c). Thus, the N‐terminal region may also participate in the cross talk between JA signaling and ABA signaling. Our study improved our understanding of the function of the N‐terminal region. Interestingly, the interactions between JAZ1 and PYL7, ABI1, HAB1, or AHG1 require the full‐length JAZ1 protein, suggesting that different motifs of JAZ1 or a proper folding are required for these interactions.
The interaction between JAZ1 and key components of the ABA signaling pathway imply that JAZ1 links JA signaling to the ABA signaling pathway. MeJA and ABA treatments were administered to examine the JA‐ and ABA‐ responsiveness of JAZ1. The changes in JAZ1 expression revealed that JAZ1 was responsive to JA, but not ABA, whereas ABA enhanced JA‐induced JAZ1 expression (Figure 5c). The result is consistent with the data showing that JAZ1 is involved in the crosstalk between the JA and ABA signaling pathways though JAZ1‐PYL interactions. The expression of OsJAZ1 not only responds to JA but also induced by ABA (Fu et al., 2017). The different ABA responses of JAZ1 and OsJAZ1 may be derived from their species.
Based on these results, we provide evidence to support the hypothesis that ABA promotes JA signaling outputs by accelerating the degradation of the JAZ1 protein and increasing the expression of JA‐responsive genes. However, confirmation of the synergistic regulation of the plant immune system by the JAZ1 and PYL proteins modules requires more physiological and biochemical evidence.
AUTHOR CONTRIBUTIONS
HL conceptualized the study; KS and XX validated and investigated the study; KS, XX, and NL formally analyzed the study; KS, ZZ, and HL wrote the manuscript; HL supervised the study; ZZ and HL acquired funding.
Supporting information
Figure S1
Table S1
Table S2
Table S3
ACKNOWLEDGMENTS
We thank Hongwei Guo (Southern University of Science and Technology) for providing us with 35S:JAZ1‐GUS seeds. This work is supported by the National Natural Science Foundation of China (31800198), the Fok Ying Tung Education Foundation (161023), the Natural Science Foundation of Jiangsu Higher Education Institutions (18KJB180010), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Sun K, Xue X, Liu N, Zhu Z, Li H. A point‐to‐point protein–protein interaction assay reveals the signaling interplays among plant hormones and environmental cues. Plant Direct. 2020;4:1–10. 10.1002/pld3.228
REFERENCES
- Abe, H. , Urao, T. , Ito, T. , Seki, M. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell, 15, 63–78. 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie, B. A. , Perez‐Perez, J. , Perez‐Perez, M. M. , Godoy, M. , Sanchez‐Serrano, J. J. , Schmelz, E. A. , & Solano, R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis . The Plant Cell, 19, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman, F. , Yazaki, J. , Lee, M. , Takahashi, Y. , Kim, A. Y. , Li, Z. , … Schroeder, J. I. (2016). An ABA‐increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: A putative link of ABA and JA signaling. Scientific Reports, 6, 28941–28950. 10.1038/srep28941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for Network Evolution in an Arabidopsis Interactome Map. Science, 333, 601–607. 10.1126/science.1203877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen, N. , & Reymond, P. (2007). Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Molecular Plant‐Microbe Interactions, 20, 1406–1420. [DOI] [PubMed] [Google Scholar]
- Chaiwanon, J. , Wang, W. , Zhu, J. Y. , Oh, E. , & Wang, Z. Y. (2016). Information integration and communication in plant growth regulation. Cell, 164, 1257–1268. 10.1016/j.cell.2016.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. H. , Qu, L. , Xu, Z. H. , Zhu, J. K. , & Xue, H. W. (2018). EL1‐like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis . Molecular Plant, 11, 706–719. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Fernández, G. , Adie, B. , Chico, J. M. , Lorenzo, O. , … Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature, 448, 666–671. 10.1038/nature06006 [DOI] [PubMed] [Google Scholar]
- Cho, H. , Ryu, H. , Rho, S. , Hill, K. , Smith, S. , Audenaert, D. , … Hwang, I. (2014). A secreted peptide acts on BIN2‐mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nature Cell Biology, 16, 66–76. 10.1038/ncb2893 [DOI] [PubMed] [Google Scholar]
- Colón‐Carmona, A. , Chen, D. L. , Yeh, K. C. , & Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiology, 124, 1728–1738. 10.1104/pp.124.4.1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington, M. F. , & Harmer, S. L. (2007). The circadian clock regulates auxin signaling and responses in Arabidopsis . PLoS Biology, 5, e222 10.1371/journal.pbio.0050222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ollas, C. , Arbona, V. , & Gomez‐Cadenas, A. (2015). Jasmonoyl isoleucine accumulation is needed for abscisic acid build‐up in roots of Arabidopsis under water stress conditions. Plant, Cell & Environment, 38, 2157–2170. 10.1111/pce.12536 [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N. , & Estelle, M. (2004). Auxin signaling and regulated protein degradation. Trends in Plant Science, 9, 302–308. 10.1016/j.tplants.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Duek, P. D. , & Fankhauser, C. (2005). bHLH class transcription factors take center stage in phytochrome signaling. Trends in Plant Science, 10, 51–54. [DOI] [PubMed] [Google Scholar]
- Feng, S. , Martinez, C. , Gusmaroli, G. , Wang, Y. U. , Zhou, J. , Wang, F. , … Deng, X. W. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature, 451, 475–479. 10.1038/nature06448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J. , Wu, H. , Ma, S. , Xiang, D. , Liu, R. , & Xiong, L. (2017). OsJAZ1 Attenuates drought resistance by regulating JA and ABA signaling in rice. Frontiers in Plant, 8, 2018 10.3389/fpls.2017.02108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G. A. , Major, I. T. , & Koo, A. J. (2018). Modularity in jasmonate signaling for multistress resilience. Annual Review of Plant Biology, 69, 387–415. 10.1146/annurev-arplant-042817-040047 [DOI] [PubMed] [Google Scholar]
- Lackman, P. , Gonzalez‐Guzman, M. , Tilleman, S. , Carqueijeiro, I. , Perez, A. C. , Moses, T. , … Goossens, A. (2011). Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proceedings of the National Academy of Sciences of the United States of America, 108, 5891–5896. 10.1073/pnas.1103010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P. , Monte, E. , Oka, Y. , Liu, T. , Carle, C. , Castillon, A. , … Quail, P. H. (2008). Multiple phytochrome‐interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Current Biology, 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, O. (2018). Auxin signaling. Plant Physiology, 176, 465–479. 10.1104/pp.17.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Wang, Q. , Deng, W. , Wang, X. , Piao, M. , Cai, D. , … Liu, B. (2017). Molecular basis for blue light‐dependent phosphorylation of Arabidopsis cryptochrome 2. Nature Communications, 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Z. , He, S. , Xu, F. , Wei, X. , Jiang, L. , Liu, Y. , … Li, L. (2020). Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA‐binding activity and auxin‐induced hypocotyl elongation in Arabidopsis . New Phytologist, 225(2), 848–865. [DOI] [PubMed] [Google Scholar]
- Munemasa, S. , Oda, K. , Watanabe‐Suqimoto, M. , Nakamura, Y. , Shimoishi, Y. , & Murata, Y. (2007). The coronatine‐insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiology, 143, 1398–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar, K. , Kyndt, T. , Nzogela, Y. B. , & Gheysen, G. (2012). Abscisic acid interacts antagonistically with classical defense pathways in rice–migratory nematode interaction. New Phytologist, 196, 901–913. 10.1111/j.1469-8137.2012.04310.x [DOI] [PubMed] [Google Scholar]
- Ni, W. , Xu, S. L. , González‐Gr&ío, E. , Chalkley, R. J. , Huhmer, A. F. R. , Burlingame, A. L. , … Quail, P. H. (2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nature Communications, 8(1), 1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E. , Yamaguchi, S. , Hu, J. , Yusuke, J. , Jung, B. , Paik, I. , … Choi, G. (2007). PIL5, a phyto‐chrome‐interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. The Plant Cell, 19, 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E. , Zhu, J. Y. , Bai, M. Y. , Arenhart, R. A. , Sun, Y. , & Wang, Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the arabidopsis hypocotyl. Elife, 3, e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E. , Zhu, J. Y. , & Wang, Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology, 14, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, L. , & Goossens, A. (2011). The JAZ proteins: A crucial interface in the jasmonate signaling cascade. The Plant Cell, 23, 3089–3100. 10.1105/tpc.111.089300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, L. , Ritter, A. , Goossens, J. , Durand, A. N. , Liu, H. , Gu, Y. , … Goossens, A. (2015). The RING E3 ligase KEEP ON GOING modulates JASMONATE ZIM‐DOMAIN12 stability. Plant Physiology, 169, 1405–1417. 10.1104/pp.15.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P. H. (2011). Phytochromes. Current Biology, 20, R504–R507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J. , Kim, K. , Kang, H. , Zulfugarov, I. S. , Bae, G. , Lee, C. H. , … Choi, G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively‐acting phyto‐chrome‐interacting factors. Proceedings of the National Academy of Sciences of the United States of America, 106, 7660–7665. 10.1073/pnas.0812219106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G. , … Browse, J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature, 2007, 18. [DOI] [PubMed] [Google Scholar]
- Verma, V. , Ravindran, P. , & Kumar, P. P. (2016). Plant hormone‐mediated regulation of stress responses. BMC Plant Biology, 16, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Bai, M. Y. , & Wang, Z. Y. (2014). The brassinosteroid signaling network‐a paradigm of signal integration. Current Opinion in Plant Biology, 21, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , He, S. , Zhang, J. , Mao, Z. , Wang, W. , Li, T. , … Lian, H. (2018). Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis . Molecular Plant, 11, 523–541. [DOI] [PubMed] [Google Scholar]
- Zhong, S. , Shi, H. , Xue, C. , Wang, L. , Xi, Y. , Li, J. , … Guo, H. (2012). A molecular framework of light‐controlled phytohormone action in Arabidopsis . Current Biolology, 22, 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Table S2
Table S3


