Abstract
Mitochondrial dysfunction underlies many heritable diseases, acquired pathologies, and aging-related declines in health. Szeto–Schiller (SS) peptides comprise a class of amphipathic tetrapeptides that are efficacious toward a wide array of mitochondrial disorders and are believed to target mitochondrial membranes because they are enriched in the anionic phospholipid cardiolipin (CL). However, little is known regarding how SS peptides interact with or alter the physical properties of lipid bilayers. In this study, using biophysical and computational approaches, we have analyzed the interactions of the lead compound SS-31 (elamipretide) with model and mitochondrial membranes. Our results show that this polybasic peptide partitions into the membrane interfacial region with an affinity and a lipid binding density that are directly related to surface charge. We found that SS-31 binding does not destabilize lamellar bilayers even at the highest binding concentrations; however, it did cause saturable alterations in lipid packing. Most notably, SS-31 modulated the surface electrostatics of both model and mitochondrial membranes. We propose nonexclusive mechanisms by which the tuning of surface charge could underpin the mitoprotective properties of SS-31, including alteration of the distribution of ions and basic proteins at the interface, and/or modulation of bilayer physical properties. As a proof of concept, we show that SS-31 alters divalent cation (calcium) distribution within the interfacial region and reduces the energetic burden of calcium stress in mitochondria. The mechanistic details of SS-31 revealed in this study will help inform the development of future compound variants with enhanced efficacy and bioavailability.
Keywords: membrane biophysics, mitochondria, drug action, lipid structure, peptides, cardiolipin, elamipretide, electrostatics, peptide therapeutic, SS-31, Szeto-Schiller peptide, inner membrane, bioenergetics
Introduction
Mitochondria are eukaryotic organelles that orchestrate the majority of cellular energy metabolism as well as a range of other processes, including lipid biosynthesis, ion homeostasis, and programmed cell death (apoptosis). Mitochondrial dysfunction encompasses a wide array of both genetically encoded and acquired pathologies. Several mitochondrial diseases originate from heritable mutations in genes encoding mitochondrial proteins, either in nuclear or mitochondrial DNA (1). Moreover, reduced mitochondrial function accompanies the general decline in cellular bioenergetic capacity that occurs with aging as well as with many complex pathologies, including frailty, cardiomyopathy, cancer, and neurodegeneration (2). Features of mitochondrial dysfunction include gross alterations of the inner membrane cristae morphology, destabilization of respiratory complexes of the oxidative phosphorylation (OXPHOS)5 system, and overproduction of reactive oxygen species (ROS) and reactive nitrogen species. Yet despite recent progress aimed at finding pharmacological approaches for mitochondrial disorders (3), there are presently no Food and Drug Administration–approved therapeutic compounds to treat them.
Szeto–Schiller (SS) peptides are among the most promising compounds currently under investigation for the treatment of mitochondrial dysfunction (4, 5). These synthetic tetrapeptides have a characteristic motif of alternating cationic and aromatic side chains (Fig. 1). Early in their development, SS peptides were shown to be cell-permeable in a wide range of cell types and to specifically target mitochondria. Despite their strong positive charge density (formal charge of +3 at neutral pH), exogenously added SS peptides traverse the plasma membrane in an energy-independent and nonsaturable manner and accumulate strongly (1000–5000-fold) at the mitochondrial inner membrane (6, 7). The intrinsic therapeutic activity of the lead compound, SS-31 (Fig. 1A), was first demonstrated in cell culture studies, wherein the peptides were shown to protect cells against induced oxidative stress by curtailing oxidative cell death, reducing intracellular ROS, maintaining membrane potential (ΔΨm), and preventing lipid peroxidation, all in a dose-dependent manner (6, 8). SS peptides have since been shown by many independent groups using cell culture and animal disease models to have significant efficacy in restoring mitochondrial function with a wide range of pathologies, including cardiomyopathy and heart failure, skeletal muscle injury/atrophy, ischemia and ischemia-reperfusion injury, kidney injury and disease, neurodegenerative diseases, cancer, and the heritable disease Friedreich's ataxia (summarized in Ref. 5). These studies have all underscored the safety and favorable pharmacokinetic profile of SS peptides. Currently, Stealth BioTherapeutics is conducting early to advanced-phase clinical trials with SS-31 (elamipretide) for a range of primary mitochondrial and aging-related diseases.
Figure 1.
Chemical structures of SS peptides. A, SS-31 (d-Arg-2′6′-dimethyl-Tyr-Lys-Phe-NH2); B, SS-20 (Phe-d-Arg-Phe-Lys-NH2); C, [ald]SS-31, SS-31 with an aladan moiety (red) in place of Phe.
Despite abundant evidence for the broad therapeutic potential of SS peptides, the molecular mechanism of action (MoA) of these compounds is poorly understood. It was originally proposed that SS peptides act as mitochondria-targeted antioxidants. Peptide analogs, such as SS-31, which contain a free radical-scavenging tyrosine (or dimethyltyrosine) moiety, could in principle serve to reduce ROS burden (9). However, antioxidant chemistry is not likely to be the primary mechanism of SS peptides, given (i) that such scavenging would occur stoichiometrically, not catalytically, and (ii) that other peptide variants, such as SS-20 (Fig. 1B), which do not contain scavenging side chains, have also proven effective in preclinical studies.
Perhaps the most critical insight into the MoA of SS peptides is that they are believed to target the mitochondrial inner membrane (MIM) by virtue of its enrichment in the anionic phospholipid cardiolipin (CL) (10–13). CL has an unusual dimeric structure with a two-phosphate headgroup and four acyl chains (Fig. S1A) and comprises roughly 10–20 mol % of the total lipids within the MIM. CL plays a central role in many mitochondrial processes, including apoptosis, mitophagy, signaling, fission/fusion dynamics, and energy metabolism (14, 15). Within the protein-rich MIM, CL mediates many interactions with peripheral and integral membrane proteins (16, 17), underpinning its role in the assembly of OXPHOS complexes into supercomplexes (18, 19). Moreover, with its unique physicochemical properties and propensity for conical molecular geometry and structural polymorphism, CL has a complex phase behavior in lipid bilayers (20, 21). Thus, local enrichment of CL can promote negative curvature, which likely helps to stabilize cristae architecture (22, 23). Nascent CL is synthesized de novo within the MIM and subsequently undergoes a remodeling process to render mature species with a highly unsaturated complement of acyl chains that is species- and tissue-specific (24) (Fig. S1B). Alterations in CL distribution and biogenesis are associated with a number of diseases (25), including Barth syndrome (26, 27), which is caused by defects in the transacylase tafazzin with concurrent aberrant remodeling of CL and buildup of its lysolipid form, monolysocardiolipin (MLCL). Hence, being critical for mitochondrial structure and function, the anionic phospholipids of mitochondrial membranes are promising targets for therapeutic agents.
A primary mode of interaction between SS peptides and CL-rich lipid bilayers would be a truly unparalleled type of MoA. This is because the vast majority of drug compounds are designed to target and act upon specific proteins, not the lipid bilayer (with rare exceptions, such as general anesthetics, whose mechanisms may involve alteration of bilayer properties (28)). In the present study, we analyzed the nature of the SS-31 interaction with biomimetic model membranes and intact mitochondria to understand the forces that drive the peptide-membrane interaction and how SS-31 binding affects physical properties of model and natural lipid bilayers. Our results support a molecular MoA that involves alteration of bulk lipid bilayer properties, most notably the membrane surface electrostatic profile. We discuss multiple nonexclusive mechanisms by which the regulated tuning of surface electrostatics could underpin the broad therapeutic efficacy of SS peptides.
Results
Binding of SS-31 to model membranes
The interaction of basic amphipathic peptides with negatively charged membranes depends on the electric field originating from the bilayer surface as well as specific electrostatic and hydrophobic interactions between the peptide and lipids (Fig. S2). The first objective of this study was to quantitatively evaluate the binding of SS-31 to model membranes with specific lipid composition. For spectroscopic analysis of SS-31 binding, we made use of the endogenous spectral properties of the 2′,6′-dimethyltyrosine (2′,6′-Dmt) side chain at the second position in the peptide. Our initial spectral characterization of SS-31 confirmed the feasibility of using endogenous peptide fluorescence to quantify peptide-membrane interactions (supporting Results A and B and Figs. S3 and S4). We therefore proceeded to measure the binding of SS-31 to large unilamellar vesicles (LUVs) containing anionic lipids in a host background of the zwitterionic lipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, 16:0/18:1 PC). The anionic lipids included 1′,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol (tetraoleoylcardiolipin (TOCL), all acyl chains 18:1), monolysocardiolipin (MLCL, all acyl chains 18:1), or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG, 16:0/18:1 PG). Binding measurements were performed using two complementary approaches: (i) titration of fixed concentrations of LUVs with peptide and (ii) titration of fixed concentrations of peptide with LUVs (supporting Results C and Fig. S5). Note that in our analyses, we assume that peptide does not cross the bilayer of our model systems and can only access the outer leaflet; for LUVs of this size, the “effective lipid concentration” of the outer leaflet (herein referenced as [L]eff, [Lip]eff, or [lipid]eff) is taken as half of the total lipid concentration in the system (29).
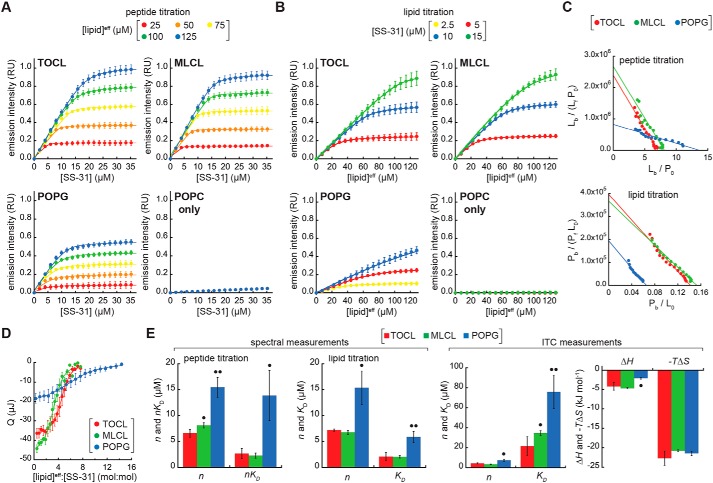
Binding isotherms based on SS-31 emission intensity are shown in Fig. 2. First, peptide titrations were conducted by the progressive addition of SS-31 to LUVs of different composition at lipid concentrations ranging from 25 to 125 μm (Fig. 2A; see also supporting Results D and Figs. S6 and S7). Second, lipid titrations were conducted by the progressive addition of LUVs to SS-31 at concentrations up to 15 μm (Fig. 2B). Taken together, these binding curves reveal the following: (i) in the absence of anionic lipid (LUVs with POPC only), SS-31 binding is negligible even at the highest lipid concentrations used, consistent with the established requirement of anionic lipids for binding (10–13); (ii) in the presence of anionic membranes, SS-31 binding displays saturation binding behavior; and (iii) membranes containing dianionic lipid (TOCL and MLCL) have a higher SS-31 binding capacity than membranes containing monoanionic POPG, reflected in the roughly 2-fold higher [SS-31] required to saturate binding in peptide titrations and in the roughly 2-fold lower [lipid] required to saturate binding in lipid titrations.
Figure 2.
SS-31 binding isotherms. A–C, fluorescence-based measurements. A, peptide titrations. Liposomes containing anionic lipid (20 mol % TOCL, MLCL, or POPG in a POPC background) or POPC only at the lipid concentrations indicated were titrated with SS-31 (4-nmol increments). Values shown are means ± S.D. (error bars) (n = 3), and lines show data fits based on Equation S2. B, lipid titrations. Peptides at the concentrations indicated were titrated with liposomes (30-nmol total lipid increments). Values shown are means ± S.D. (error bars) (n = 3), and lines show data fits based on Equation S3. C, Scatchard analyses. Titration data with liposomes containing 20 mol % TOCL, MLCL, and POPG are shown as Scatchard plots. Top, peptide titrations ([lipid]eff = 125 μm) fit according to Equation S6; bottom, lipid titrations (10 μm SS-31, TOCL, and MLCL; 5 μm SS-31, POPG) fit according to Equation S8. D, ITC analyses. A Wiseman plot shows blank-subtracted average integrated heats (means ± S.D. (error bars); n = 3) as a function of [lipid]eff/[SS-31] molar ratio for LUVs containing TOCL, MLCL, and POPG. E, summary of binding parameter data. Statistical differences within each data set were determined relative to TOCL-containing membranes: •, p < 0.05; ••, p < 0.01.
To obtain equilibrium binding parameters n (the number of lipid molecules per peptide bound) and KD (the dissociation constant), we analyzed binding isotherm data by Scatchard analysis (Fig. 2C) and by fits of the data to Langmuir adsorption models (Equations S2 and S3 for peptide and lipid titrations, respectively). The equilibrium binding parameters from this work (Fig. 2E, left panels) reveal two key points. First, for membranes containing cardiolipin variants, the average lipid/peptide stoichiometry (n = 6.9 and 7.4 for TOCL and MLCL, respectively) is roughly half the value for membranes with POPG (n = 15.4). Hence, if we assume ideal lipid mixing and approximate lipid cross-sectional areas (70 Å2 for POPC and POPG, 130 Å2 for TOCL, and 110 Å2 for MLCL (30, 31)), then for membranes composed of 20% TOCL or MLCL, the SS-31 “binding site” comprises an area of ∼560–580 Å2 in which each peptide associates, on average, with 1.4–1.5 dianionic lipids. By comparison, for membranes containing 20% POPG, SS-31 binds an area of roughly 1080 Å2 and associates on average with three monoanionic lipids. Based on these stoichiometric relationships, then, there is an approximate charge balance between each peptide and its corresponding lipid binding site. Second, the affinity of SS-31 for membranes containing TOCL and MLCL is significantly higher than it is for membranes containing POPG. Specifically, the average dissociation constants for lipid monomers (reflected in KD values for lipid titrations and in nKD values for peptide titrations) are 2.0–2.9 μm for membranes composed of 20% TOCL and MLCL, whereas they range from 6.0 to 13 μm for membranes composed of 20% POPG. These values of n and KD are consistent with parameters measured for other amphipathic membrane-active peptides (32, 33). They are also consistent with previous studies showing a strong correlation between binding energies of basic peptides and the negative surface charge of lipid bilayers (e.g. see Ref. 34). As an independent measure of peptide binding, the molar partition coefficients (KP, Equation S9) for membranes containing 20% TOCL (KP = 5.07 ± 0.76 × 104) and 20% MLCL (KP = 5.90 ± 0.44 × 104) are significantly higher than that of 20% POPG-containing membranes (KP = 1.28 ± 0.63 × 104).
As a complementary approach for measuring SS-31 membrane interactions, we performed isothermal titration calorimetry (ITC) measurements by the progressive injection of LUVs into solutions of peptide in the sample cell (35). The interaction of SS-31 with membranes containing anionic lipid was shown to be exothermic in all cases (Fig. S8), and fits of complete binding isotherms (Fig. 2D) yielded the thermodynamic binding parameters shown (Fig. 2E, right panels). The values of n and KD from fits to the ITC data showed consistent differences between TOCL/MLCL- and POPG-containing membranes when compared with our spectral-based measurements (Fig. 2). However, compared with our spectral analyses, there were differences in the absolute values from ITC-derived parameters (mean n values slightly lower and KD values ∼2–3 times higher). This difference could be related to different phenomena being measured: our spectral analyses may only reflect the binding event per se, whereas ITC measurements could be reflecting binding as well as peptide-dependent alterations of bilayer properties. Apropos of this point, we did observe a slight asymmetry in our exothermic peaks, particularly for TOCL- and MLCL-containing membranes, which could be related to thermotropic alterations in the bilayers following peptide interaction.
Comparison of the binding-associated changes in enthalpy (ΔH) and entropy (ΔS) provides insights into the membrane-binding mechanism. Namely, exothermic (ΔH < 0) values of peptide-lipid interaction, as we observe for SS-31, originate mainly from the establishment of polar contacts; by contrast, increases in the system entropy (ΔS > 0), as we also observe for SS-31, originate mainly from the burial of hydrophobic side chains in the acyl chain region with concurrent release of water and ions from nonpolar surfaces of the peptide and bilayer (36). For membranes containing TOCL, MLCL, and POPG, the interaction of SS-31 was associated with a favorable enthalpic change (ΔH = −4.2, −4.7, and −2.1 kJ mol−1, respectively); the 2-fold difference in ΔH between TOCL/MLCL- and POPG-containing membranes is consistent with the difference in formal charges of these anionic lipids. The peptide-bilayer association, however, was predominantly entropy-driven (−TΔS = −22.7, −20.8, and −21.5 kJ mol−1, respectively), showing that peptide association is stabilized mostly by burial of the nonpolar residues. Indeed, earlier studies on the interaction of small basic peptides with anionic lipid bilayers indicated a dominant role of entropy over enthalpy in stabilizing the interaction (e.g. see Ref. 37). Given that the hydrophobic contribution to peptide binding energy is proportional to the exposed side-chain area (∼80 J mol−1 Å−2) (38), the burial of Tyr (similar to 2′,6′-Dmt) and Phe side chains (accessible surface areas 187 and 175 Å2, respectively) would theoretically contribute a combined ∼28 kJ mol−1, in reasonable agreement with our measurements. Moreover, because SS peptides are short and probably do not form any appreciable secondary structure upon membrane binding, their membrane binding does not likely incur an entropic penalty that would be associated with stabilizing a secondary structure.
Taken together, our spectral and calorimetric binding data show that the bilayer binding density and affinity of SS-31 are directly related to the anionic lipid composition, that the lipid-dependent differences in interaction energy are mostly on the net charge of anionic lipids, and that membrane interactions are enthalpically and entropically favorable.
Dependence of SS-31 binding on model membrane surface electrostatics
The electrostatic profile of biomembranes consists of multiple potential energy functions (Fig. S2). Among them, the surface potential (ψs) originates from charge distribution at the membrane interface based on ionizable functional groups (e.g. lipid headgroups) and adsorbed ions, creating a strong electrostatic driving force for the binding of charged moieties to the bilayer surface (39). Electrolytes partitioned at the bilayer interface can have a complex effect on membrane interactions of amphipathic peptides. By attenuating the ψs, solution cations can reduce the electrostatic attraction of basic peptides to the anionic lipid surface; however, increasing solution ionic strength may also favor the burial of hydrophobic side chains in the nonpolar core of the bilayer. We therefore explored the relationship between SS-31 binding and the ψs of model membranes and the influence of ionic strength (Fig. 3).
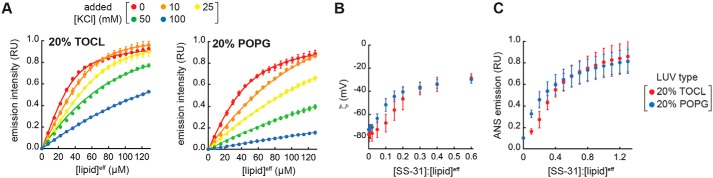
Figure 3.
SS-31 binding and surface electrostatics. A, binding isotherms. Peptide was titrated with LUVs of the indicated composition (20% TOCL, [SS-31] = 7.5 μm and 20% POPG, [SS-31] = 3.8 μm) at 30-nmol total lipid increments in the presence of different concentrations of added KCl. Values shown are means ± S.D. (error bars) (n = 3), and lines show data fits based on Equation S3. Because isotherms at higher [KCl] did not reach saturation, curves within each membrane set were fit globally, assuming a common saturation point. B and C, effect of SS-31 binding on surface charge. Measurements of ζ (B) and ANS fluorescence (C) are shown as a function of [SS-31]/[lipid]eff for membranes containing 20% TOCL and 20% POPG as indicated. Values shown are means ± S.D. (error bars) (n = 3).
SS-31 binding curves measured by titration of SS-31 with LUVs containing 20% TOCL or POPG revealed a strong effect of monovalent electrolyte (Fig. 3A). We confirmed that SS-31 binding to model bilayers was reversible in a manner that depends upon solution ionic strength used for these binding isotherms (supporting Results E and Fig. S9 (A and B)). When binding parameters n and KD calculated from the curves of Fig. 3A are considered as a function of salt concentration, there is a large disparity between membranes containing divalent TOCL and monovalent POPG (Fig. S9C, top panels). However, recasting these parameters as a function of ψs, calculated from Gouy-Chapman-Stern (GCS) formalism (40, 41) (Equations S10–S12), reveals a much smaller difference between membranes containing the two anionic lipids (Fig. S9C, bottom panels). This GCS analysis, which accounts for the roughly 2-fold larger intrinsic surface charge (σmax) of TOCL- versus POPG-containing bilayers, indicates that SS-31 binding is more related to the prevailing surface charge (σ) per se, rather than to any specific features of CL.
To continue this analysis, we determined the relationship between SS-31 binding and surface electrostatics by two complementary approaches. We first performed zeta potential (ζ) measurements of LUVs containing 20 mol % TOCL or POPG with increasing [SS-31] (Fig. 3B). The ζ represents the electrostatic potential at the hydrodynamic shear plane of membranes and is related to the ψs by the Debye constant (κ), which describes the position-dependent decay of electrostatic potential from the membrane surface to the bulk solution. As expected for the binding of a polybasic peptide to a negatively charged surface, SS-31 caused a saturable reduction in the magnitude of ζ for both anionic bilayers. As an independent technique, we measured the effect of SS-31 on surface electrostatics using 1-anilinonaphthalene-8-sulfonic acid (ANS), a fluorescent reporter whose binding to anionic bilayers increases as the magnitude of the ψs is reduced, measured as an increase in probe emission intensity (42) (Fig. 3C and Fig. S9D). Consistent with our ζ data, SS-31 caused a saturable ANS-detected reduction in the ψs of anionic model membranes. We observed the same response with SS-20 (Fig. S9E), supporting that this effect on membrane surface electrostatics is a general feature of SS peptides. Beyond the observation that SS peptides attenuate membrane surface potential, two features are notable from these experiments. First, for both ζ and ANS measurements, SS-31 elicited a hyperbolic decay of ψs for PG-containing bilayers, whereas the response was more sigmoidal for CL-containing bilayers (Fig. 3 (B and C) and Fig. S9 (D and E), blue versus red). This suggests that at low peptide concentration, there is not a linear correspondence between peptide binding and charge attenuation for CL-containing membranes. Second, the ζ responses, which provide an absolute measure of ψs, consistently saturated near −30 mV with increasing peptide. This indicates that when the model bilayers are maximally bound with peptide, there remains an appreciable negative surface charge density. This observation is in contrast to some other basic peptides (43) and multivalent cations (44) that cause charge overcompensation (lead to positive surface charge density) upon binding anionic bilayers at high concentration. Two nonexclusive explanations may account for this: (i) at binding saturation, bound SS-31 may render some ionized phosphate groups unavailable for further interaction, and/or (ii) the formal charge on SS-31 may become reduced upon binding. Regardless of the mechanism, the fact that SS-31 attenuates the ψs, but does not completely reverse it, is a fundamental feature of its effect on the membrane surface electrostatic profile.
SS-31 maintains the lamellarity of model membranes but alters lipid interactions at the interface
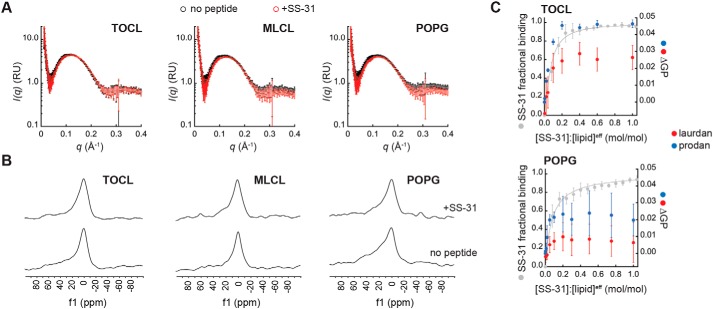
Amphipathic molecules such as antimicrobial peptides can induce large structural changes in membranes, including micellization, pore formation, and induction of inverted topologies (45). We therefore addressed whether SS-31 induced structural polymorphism within model bilayers using a range of approaches (Fig. 4). Based on synchrotron small-angle X-ray scattering (SAXS) measurements, LUVs composed of POPC alone or with 20% TOCL, MLCL, or POPG yielded scattering profiles typical of lipid vesicles (46), and the presence of SS-31 did not cause structural perturbation (Fig. 4A, supporting Results F, and Fig. S10A). Similarly, based on 31P solid-state NMR (ssNMR) measurements, these model membranes yielded line shapes consistent with lamellar bilayers (47), and we observed no effect of SS-31 on bilayer structure (Fig. 4B, supporting Results F, and Fig. S10B). We conclude that even at the highest peptide concentration used in this study ([P]/[L]eff = 1:5), SS-31 did not cause major topological alterations of our model membranes. Thus, these membranes are expected to exist stably in the liquid crystalline lamellar mesophase over all peptide concentrations.
Figure 4.
Effects of SS-31 on structural properties of model membranes. A and B, SS-31 does not cause major structural perturbation of model membranes. LUVs composed of 20% anionic lipid (TOCL, MLCL, or POPG) in POPC background were measured in the absence or presence of peptide ([SS-31]/[lipid]eff = 1:5) as indicated. A, synchrotron SAXS measurements showing background-subtracted scattering density profiles for LUVs in the absence (black) or presence (red) of SS-31. B, 31P solid-state NMR spectra of LUVs in the absence (bottom spectra) or presence (top spectra) of SS-31. C, SS-31 alters lipid packing interactions. Shown are changes in laurdan and prodan GP values (ΔGP) with increasing [SS-31] for LUVs containing 20% TOCL or POPG in POPC background as indicated. Data and fits in gray show SS-31 fractional saturation (calculated from Fig. 2A, [lipid]eff = 25 μm). Values shown are means ± S.D. (error bars) (n = 3).
We therefore addressed whether SS-31 altered lipid interactions within these model bilayers. To this end, we used membrane-bound fluorescent reporters of lipid dynamics and packing that partition into the bilayer at different depths (Fig. S10C). Using 1,6-diphenyl-1,3,5-hexatriene (DPH) anisotropy (〈r〉DPH) as a readout of the fluid dynamics of hydrocarbon tails, we observed no effect of SS-31 in TOCL- or POPG-containing membranes (Fig. S10, D and E). We then used the solvatochromic probes 6-dodecanoyl-2-dimethylaminonaphthalene (laurdan) and 6-propinoyl-2-dimethylaminonaphthalene (prodan), whose spectral properties are quantified as the generalized polarization (GPLAU and GPPRO, respectively) that increases with enhanced lipid packing (reduced interfacial hydration) (48) (see supporting Results F and Fig. S10F). We observed modest, yet repeatable and saturable, increases in GPLAU and GPPRO that corresponded to the fractional occupancy of peptide on the membrane (Fig. 4C). Notably, the response of GP values in TOCL-containing bilayers was much higher in magnitude compared with the response in those with POPG. Based on these results, SS-31 binding causes a change in the hydration/polarity of the bilayer interface that likely correlates with an increase in lipid headgroup packing density.
Molecular dynamics analysis of the interaction between SS-31 and lipid bilayers
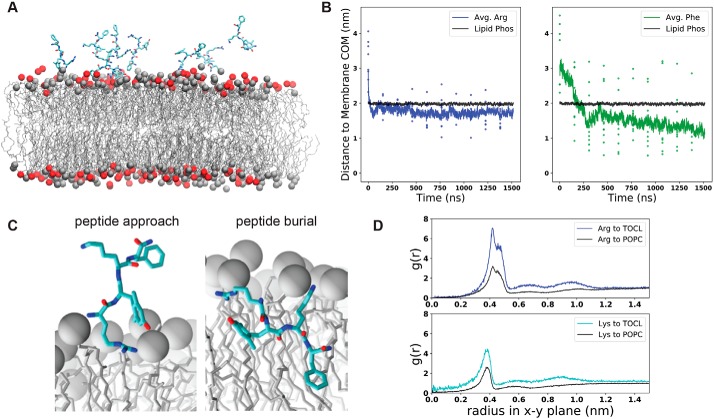
We next used all-atom molecular dynamics (MD) simulations to analyze the interaction between SS-31 and lipid bilayers (Fig. 5, supporting Results G, and Figs. S11–S14). These simulation systems contained solvated bilayers composed of 20 mol % TOCL, MLCL, or POPG in a host background of POPC and were conducted in the presence or absence of SS-31 to evaluate SS-31–bilayer interactions and how the presence of SS-31 may affect bilayer properties (Fig. 5A). In our initial investigations, we performed spontaneous membrane binding simulations by placing 10 SS-31 peptides in the aqueous phase in random orientations at distances of 1–3 nm from the bilayer surface and conducting simulations for up to 1.6 μs. Throughout each trajectory, we quantified the distance between each side chain and the bilayer center of mass along an axis perpendicular to the membrane (the z-coordinate, Fig. S11 and Fig. 5B). We observed rapid association (<200 ns) of all peptides to the bilayer surface, in which all side chains had docked to the membrane near the interfacial region, presumably driven by the electrostatic attraction between the polybasic SS-31 and the negative surface charge of the lipid bilayer. However, the membrane association time differed among the side chains. For example, the N-terminal Arg adsorbed to the bilayer rapidly (within 50 ns), whereas the C-terminal Phe took significantly longer to localize to the bilayer (within 180 ns) (Fig. 5B).
Figure 5.
Molecular dynamics simulations of SS-31 bilayer interactions. A, snapshot of a typical MD simulation with SS-31 peptides in the aqueous phase relative to the upper leaflet of a 20% TOCL bilayer. Peptides are shown in licorice representation (cyan, carbon; blue, nitrogen; red, oxygen), lipid acyl chains are in wireframe, and lipid phosphates are shown as van der Waals spheres for POPC (gray) and TOCL (red). B, temporal evolution of SS-31 Arg (blue) and Phe (green) side-chain positions relative to the membrane center of mass (COM). Black lines show the average position of all phosphates in the upper leaflet, colored dots show z-coordinates (zPOS) of individual side chains, and colored lines represent average zPOS values. C, orientation of SS-31 bound to a lipid bilayer consisting of 20 mol % TOCL in a POPC background in the peptide approach (left) and buried (right) states. Peptide representation is as in A, with all lipid phosphates shown as gray van der Waals spheres. D, lipid distributions around basic SS-31 residues. Shown are lateral (x-y plane) radial distribution profiles from Arg and Lys side chains to lipid headgroup phosphate atoms of TOCL or POPC as indicated.
From these simulations, we observed two main types of SS-31 poses on the membrane (Fig. 5C): a “peptide approach” state, in which the primary points of contact with the membrane are the N terminus and Arg side chain anchored to phosphate headgroups, and a “peptide burial” state, in which the aromatic side chains have partitioned into the nonpolar core while the basic side chains remain electrostatically tethered to headgroup phosphates. Notably, the burial of the aromatic side chains of SS-31 in the acyl chain region is consistent with previous NMR studies (10). Once the peptides reached the burial state, each side chain assumed a distinct, stable membrane insertion depth, quantified as the average z-coordinate between the side chain and the membrane center of mass (zpos) (Fig. S11, B and C). In the 20:80 TOCL/POPC system, the basic side chains occupied the interfacial region, with Arg (zpos Arg = 1.75 nm) residing just below the average position of lipid phosphates (zpos Phos = 1.98 nm) and Lys residing at a slightly more distal position near the headgroups (zpos Lys = 1.99 nm). By comparison, the aromatic residues assumed stable positions within the nonpolar core with the Phe side chain burying deeper (zpos Phe = 1.12 nm) than the 2′,6′-Dmt side chain (zpos Dmt = 1.37 nm). These residue-specific membrane insertion depths were consistent across the three different anionic lipid compositions tested (20% TOCL, 20% MLCL, and 20% POPG) (Fig. S11C).
To evaluate the association of SS-31 side chains with lipid headgroups, we analyzed radial distribution profiles from our MD trajectories (Fig. 5D and Fig. S12A). This analysis revealed that basic side chains of SS-31 preferentially interacted with anionic lipids. For example, based on g(r) peak height, in lipid systems with 20% TOCL, Arg and Lys showed 2.25- and 1.72-fold relative increases in local concentration of phosphate groups from TOCL compared with those from POPC (Fig. 5D). These results are consistent with peptide-lipid co-diffusion, suggesting that the SS-31 Arg can concurrently associate with multiple lipid phosphates with a preference for TOCL over POPC (Fig. S12B). As a complement to these analyses, we evaluated how the presence of SS-31 may influence lipid bilayer properties. First, lipid-to-lipid radial distribution profiles showed a slight peptide-dependent local clustering of anionic lipids (Fig. S12C). Second, based on the lateral mean square displacement of lipid phosphates, we observed marked peptide-dependent decreases in the lateral diffusivity (Dxy) of all tested lipids (Fig. S13). Finally, to study how SS-31 binding could affect membrane packing defects, we measured the solvent-accessible surface area of acyl chains in systems with and without SS-31 (Fig. S14). This analysis showed peptide-dependent decreases in acyl chain solvent accessibility of all tested lipids, consistent with our observed effects of SS-31 on interfacial hydration and packing density (Fig. 4C). Hence, taken together, our MD simulations elucidate the nature of polar and nonpolar interactions that mediate the SS-31-lipid bilayer interaction and how peptide binding may affect lipid distribution, reduce lateral lipid mobility, and decrease the accessibility of solvent (and solvated ions) to acyl chains that may otherwise be exposed to the interface by packing defects.
SS peptide interactions with mitochondrial membranes
Having evaluated the binding of SS-31 with model membranes, the next objective was to quantitatively assess how this peptide interacts with mitochondria. We first evaluated peptide binding interactions. The spectral complexity of mitochondria precludes analysis of SS-31 binding by endogenous peptide fluorescence, so we used instead the variant [ald]SS-31, which contains the environment-sensitive fluorophore aladan (49) in place of the C-terminal Phe residue (Fig. 1C). As shown previously, [ald]SS-31 displays a blue-shifted emission spectrum and an increase in fluorescence intensity when bound to bilayers, making it an excellent reporter for membrane interaction. This peptide variant has also been shown to target mitochondria in a manner similar to SS-31 (11, 12). We first thoroughly evaluated the binding of [ald]SS-31 to model membranes composed of POPC with TOCL, MLCL, and POPG in different molar ratios and under different ionic conditions (Fig. S15, A and B). Equilibrium binding parameters for [ald]SS-31 with model membranes compared favorably with those of SS-31 (Fig. S15C). Hence, this variant served as a suitable model for SS-31 binding to mitochondria.
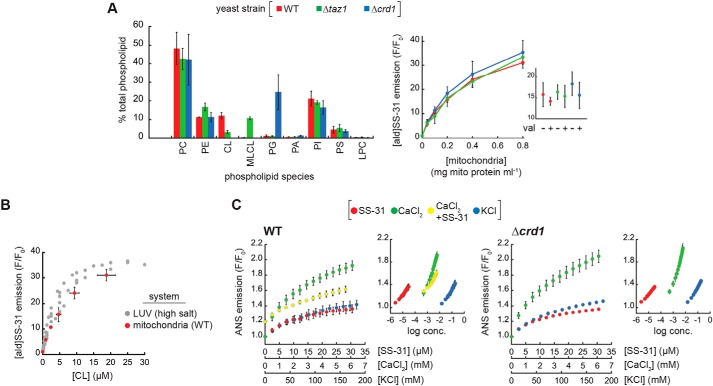
In our evaluation of SS-31 mitochondrial interactions, our first goal was to assess the CL dependence of peptide binding. Because of the relative simplicity of the CL biosynthesis and remodeling pathway in yeast compared with higher eukaryotes (Fig. S1B), Saccharomyces cerevisiae is an excellent model organism to directly compare effects of altered CL metabolism among otherwise isogenic systems. We therefore used mitochondria isolated from S. cerevisiae strains with normal CL metabolism or with knockouts in cardiolipin synthase (Δcrd1) or the transacylase tafazzin (Δtaz1) (50). Shotgun lipidomics data confirmed the expected phospholipidome of mitochondria from these three strains (Fig. 6A, left), namely that Δcrd1 mitochondria lack CL and have a buildup of PG, the immediate biosynthetic precursor of CL and that Δtaz1 mitochondria show an accumulation of MLCL and reduced CL. Further, the acyl chain distribution of anionic lipids among these strains shows the expected patterns of fatty acid saturation (50) (Fig. S16A). Emission scans of [ald]SS-31 revealed spectral changes with increasing mitochondria consistent with membrane interaction (Fig. S16B and Fig. 6A, right), showing two important features. First, as shown previously (8), collapse of the transmembrane potential (ΔΨm) by the ionophore valinomycin had no effect on peptide interaction (Fig. 6A, inset). Second, the relative binding of [ald]SS-31 was nearly identical among the WT, Δcrd1, and Δtaz1 strains. Similar peptide binding between WT and Δtaz1 strains is expected, given that tetraacyl TOCL and triacyl MLCL equally support SS-31 binding in our reductionist systems (Figs. 2 and 3). Uninhibited peptide binding by the Δcrd1 strain is perhaps surprising given that POPG supports significantly less peptide binding than CL (Figs. 2 and 3). However, our lipidomics data indicate that PG levels are dramatically increased when CL synthesis is blocked. This increase in PG likely creates a negative surface charge density of the MIM large enough to promote peptide binding at WT levels. Again, this supports a model in which SS-31 interaction is related more to surface charge than it is to specific lipid headgroup identity.
Figure 6.
Interaction of SS-31 with isolated mitochondria. A, [ald]SS-31 binding with differing phospholipid composition. Left, percentages of total phospholipid of each species (phosphatidylcholine (PC), phosphatidylethanolamine (PE), cardiolipin (CL), monolyso-CL (MLCL), phosphatidylglycerol (PG), phosphatidic acid (PA), phosphatidylinositol (PI), phosphatidylserine (PS), and lysophosphatidylcholine (LPC)) in mitochondria from WT, Δtaz1, and Δcrd1 strains measured by shotgun lipidomics. Right, binding of 1 μm [ald]SS-31 with mitochondria isolated from WT, Δtaz1, and Δcrd1 yeast strains quantified as the mean fractional increase (F/F0) in aladan emission with increasing mitochondria concentration for fully energized mitochondria. Inset, relative binding of [ald]SS-31 to mitochondria (0.2 mg ml−1) in the presence or absence of 1 μm valinomycin (val) as indicated. Values shown are means ± S.D. (error bars) (n = 3). B, comparison of [ald]SS-31 binding to mitochondria and model membranes. [ald]SS-31 binding is shown as a function of CL concentration for (i) LUVs of defined TOCL composition under high-salt conditions (gray; Fig. S15) and (ii) mitochondria isolated from yeast WT strain (red; Fig. S16 and Fig. 6A). Calculation of [CL] for LUVs accounts for outer leaflet (accessible) CL only from known lipid concentrations; calculation of [CL] for mitochondrial samples was based on lipidomics analyses of Fig. 6A, accounting for total CL content. C, effects of SS-31 and salt cations on mitochondrial surface potential. Fractional change in ANS emission is shown for titration of mitoplasts from WT (left) and Δcrd1 (right) yeast with SS-31, CaCl2, or KCl at the indicated concentrations. For WT samples, CaCl2 titrations were also conducted following the addition of 20 μm SS-31 (CaCl2 + SS-31). Insets, ANS emission data plotted as a function of log concentration for all three titrants. Values shown are means ± S.D. (error bars) (n = 3).
The second goal of these binding experiments was to directly compare peptide binding between model membranes and mitochondria. We reasoned that the best comparison would come from quantifying [ald]SS-31 binding as a function of a common independent variable (CL composition), made possible using the known mitochondrial CL concentration from our lipidomics data. When evaluated in this way, the binding curves of [ald]SS-31 to LUVs and WT mitochondria are strikingly similar (Fig. 6B). This comparison, although indirect, is consistent with a model in which binding of SS-31 to mitochondria is governed by the same interactions in our model systems (i.e. through lipid bilayer interactions).
The final goal of these experiments was to evaluate the effect of SS peptides on the surface electrostatic properties of mitochondrial membranes. Based on our observation that SS peptides attenuated the ψs of model membranes (Fig. 3 and Fig. S9), we again used the ANS fluorescent reporter to measure the ψs of mitoplasts (mitochondria with osmotically ruptured outer membrane to allow probe access to the MIM). We therefore compared the titration of mitoplasts with SS-31 and mono- and divalent cations (K+ and Ca2+, respectively) to evaluate the relative effects of each species on ANS-detected ψs (Fig. 6C). All three cationic species caused an increase in ANS fluorescence, reflecting a decrease in the ψs of mitochondrial membranes from WT (left) and Δcrd1 (right) yeast. Consistent with the exponential dependence of formal charge on accumulation of ionized species in the electric field of membranes (Equation S11), the effective concentration ranges of SS-31, Ca2+, and K+ progressively differed by about 2 orders of magnitude (Fig. 6C, insets). The first notable feature of this analysis is the relative effect of each species on surface potential. Compared with K+, Ca2+ ions caused a much stronger impact on the relative ψs, as expected. Interestingly, however, within its effective concentration range, SS-31 had a reduced effect on the relative ψs that was closer to that of the monovalent cation, an observation consistent with the fact that SS-31 does not completely reverse negative surface potential of model membranes even upon binding saturation (Fig. 3C). The second notable feature of this analysis is the effect of SS-31 on Ca2+-mediated decreases in the relative ψs. SS-31 addition to WT mitoplasts prior to CaCl2 titration strongly blunted the effects of Ca2+ on surface potential (Fig. 6C, left, compare green and yellow symbols), supporting a binding model in which SS-31 preferentially binds to mitochondrial membranes over Ca2+ ions. Taken together, we conclude from the results of Fig. 6 that the binding of SS peptides to mitochondrial membranes is strongly dependent on surface charge of the lipid bilayer, that SS peptide binding causes a “controlled down-tuning” of the electric field originating from the membrane surface, and that SS peptides at the membrane interface can mitigate the effects of polyvalent metal cations on membrane surface electrostatics.
The effects of SS peptides on interfacial cation accumulation
The final analyses of this study were focused on testing the functional implications of our observed effects of SS peptides on membrane surface electrostatics. We began with a theoretical consideration of how SS peptides may affect the decay of the electric field and ion distribution at the membrane interface based on GCS formalism (Fig. 7A and supporting Results H). By this analysis, SS peptides accumulate at the membrane surface with the effect of (i) reducing the magnitude of the surface charge and (ii) decreasing concentrations of other cationic species at the interface. This effect is particularly relevant for Ca2+, because mitochondria serve as high-capacity calcium stores in mediating cellular ion homeostasis and Ca2+ overload can cause severe damage to anionic lipid bilayers, particularly within mitochondria (see “Discussion”). This analysis shows that SS peptides can theoretically cause a strong reduction in surface accumulation of divalent cations, by over an order of magnitude. Hence, this model, albeit a simplified one, makes testable predictions about how SS peptides may mitigate divalent cation stress by reducing Ca2+ accumulation at the membrane interface. Our model was tested as follows.
Figure 7.
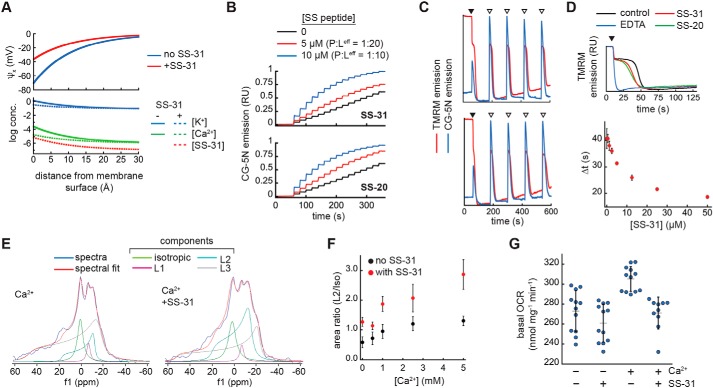
SS peptide effects on Ca2+-membrane interactions and mitochondrial cation stress. A, profiles of surface potential decay (top) and ion distribution (bottom) based on GCS theory in the presence or absence of SS-31 (see supporting Results H for model description). B, effect of SS peptides on Ca2+ accumulation at model membrane interfaces. Time course measurements of CG-5N emission were performed in solutions with LUVs (20 mol % TOCL in host POPC background, [lipid]eff = 100 μm) prebound with SS peptide at the indicated concentration followed by titration of CaCl2 in increments of 1 nmol at 20-s intervals starting at t = 60 s. C and D, effect of SS peptides on mitochondrial Ca2+ uptake. C, time courses of TMRM or CG-5N emission in mitochondrial suspensions with ETH-129 in the absence (top) or presence (bottom) of SS-31 following the addition of NADH (black arrowheads) and CaCl2 (open arrowheads). See Fig. S18A for experimental details. D, top, TMRM traces of ΔΨm generation following the addition of 1 mm NADH (black arrowhead) in samples supplemented with 12.5 μm CaCl2 and vehicle only (control), 25 μm EDTA, SS-20, or SS-31 as indicated. Bottom, dose response of SS-31 on ΔΨm generation with 25 μm CaCl2 (defined in Fig. S18D); values shown are means ± S.D. (error bars) (n = 3). E–G, effect of SS-31 on external Ca2+ stress. E and F, 31P ssNMR analysis. Mitochondria (70 mg ml−1) were given acute treatment of SS-31 (0.6 mm) or vehicle only followed by calcium treatment (up to 5 mm CaCl2). E, static wide-line 31P NMR spectra of mitochondria subject to calcium stress (5 mm CaCl2) following treatment without (left) or with (right) SS-31. Spectral deconvolution yielded the indicated components (see Fig. S19, A and B), which were used to quantify the area ratios of L2/iso in F (values shown are means ± S.D. (error bars) (n = 3). G, respirometry analysis. Mitochondria (1 mg ml−1) were treated in the absence or presence of SS-31 (54.4 μm) or CaCl2 (1 mm) as indicated and measured for state 2 respiration (n = 10–12 each).
First, we measured the effects of SS peptides on the accumulation of Ca2+ in the interfacial region of model membranes. We used a fluorescence-based approach with Calcium Green-5N (CG-5N), a membrane-impermeant probe that exhibits an emission intensity increase upon binding Ca2+ (characterized for our system in supporting Results H and Fig. S17). We reasoned that the binding of Ca2+ to the headgroup region of anionic lipid bilayers would reduce the bulk Ca2+ detected by the CG-5N probe, providing a direct readout of Ca2+ accumulation at the membrane interface. As a proof of principle, we titrated Ca2+ into solutions of CG-5N containing LUVs with monoanionic (POPG) or dianionic (TOCL) lipids at different concentrations (Fig. S17B). In the absence of LUVs, there was a saturating increase in CG-5N emission as the probe became maximally bound with Ca2+. However, the presence of POPG- and TOCL-containing LUVs caused a reduction in the CG-5N response to Ca2+ that was directly related to the amount of bilayer surface charge in the system. Hence, this assay provided a direct measure of Ca2+ sequestration within the headgroup region of anionic bilayers. To test the effects of SS peptides on Ca2+ binding to membrane surfaces, we first titrated SS-20 and SS-31 into solutions containing LUVs with 20% TOCL prebound with Ca2+ (Fig. S17C). SS-20 and SS-31 identically released Ca2+ from the membrane interface in a dose-dependent manner. Importantly, the CG-5N response began to saturate near P/Leff of ∼1:10 (Fig. S17C, green traces), corresponding to the peptide binding saturation point, suggesting that Ca2+ release into the bulk corresponded to peptide-bilayer binding. As an independent assay, we performed Ca2+ titrations with solutions of LUVs that had been preincubated without peptide or with SS peptides at molar P/Leff of 1:20 and 1:10 (Fig. 7B), again showing that SS peptides reduced the equilibrium binding of Ca2+ to anionic bilayer surfaces in a dose-dependent manner. Taken together, these results support a model in which SS peptide binding to bilayer surfaces reduces the accumulation of Ca2+ ions within the interfacial region of model membranes.
Second, we measured the effects of SS peptides on mitochondrial Ca2+ flux. External Ca2+ is taken up into the mitochondrial matrix electrophoretically by the ΔΨm (matrix negative). We reasoned that if SS peptides altered the accumulation of cations at the bilayer surface (Fig. 7B and Fig. S17C), then they may have a measurable effect on mitochondrial Ca2+ uptake. We therefore performed time course measurements of isolated mitochondria supplemented with respiratory substrate NADH followed by additions of calcium. In these experiments, we monitored the relative ΔΨm using the potentiometric probe tetramethylrhodamine methyl ester (TMRM) (the emission of which is inversely related to membrane potential; Fig. S17E) and, in parallel, monitored the external [Ca2+] using CG-5N (Fig. 7C and Fig. S18A). Note that because S. cerevisiae mitochondria do not contain a Ca2+ uniporter, Ca2+ uptake does not occur spontaneously but can occur in the presence of a calcium ionophore, for which we used ETH-129. These time courses show two key processes: (i) following NADH addition, there is a TMRM-detected establishment of the ΔΨm, and (ii) after each calcium addition, there is a CG-5N-detected spike in external [Ca2+] followed by a decrease in CG-5N emission as Ca2+ is taken into the matrix, which is accompanied by a transient TMRM-detected partial depolarization and re-establishment of the ΔΨm (see Fig. S18B for expanded views of each of these transient processes). These experiments revealed two notable effects of SS peptides. The first was related to the establishment of the ΔΨm in samples containing ETH-129. Upon reaching a threshold potential, there was a temporal lag in potential generation, after which the mitochondria resumed establishing the maximal ΔΨm. This is attributable to the energetic demands coupled to the uptake of external calcium. Interestingly, both SS-20 and SS-31 reduced the extent of this temporal delay in mitochondrial energization observed in the presence of increasing [Ca2+] (Fig. S18, B (ΔΨm establishment) and C). Quantitative analysis of Ca2+-dependent ΔΨm generation (described in Fig. S18D) revealed that SS-31 reduced this temporal delay in a dose-dependent and saturable manner (Fig. 7D). The second related effect was associated with subsequent calcium spikes. Following calcium additions, mitochondria transiently depolarized to near the same ΔΨm associated with the temporal lag. The kinetics of repolarization during the first calcium additions were faster in the presence of SS peptides; however, with increasing calcium load, the peptides no longer enhanced repolarization (see Fig. S18A, Ca2+ transient 1–4). By contrast, external calcium flux, judging by the extent of the CG-5N signal and by the kinetics of Ca2+ uptake, was not affected at all by SS peptides. Hence, under these conditions, SS peptides do not measurably alter the calcium storage capacity of mitochondria, but do improve the ability to respond energetically to calcium load. These results may provide a mechanistic underpinning to previous observations that acute treatment of isolated mitochondria with SS peptides protected against Ca2+-induced onset of the mitochondrial permeability transition (MPT) (8).
Finally, we measured the effects of SS peptides on mitochondrial calcium stress that may occur independently of Ca2+ uptake into the matrix. To this end, we used a 31P ssNMR–based strategy to monitor mitochondrial membrane structural integrity, based on previous studies using NMR analysis of membranes from prokaryotic cells (51) or isolated mitochondria (52–54). We first characterized 31P ssNMR spectra of our isolated mitochondria compared with model membranes (supporting Results H and Fig. S19), showing that mitochondria spectra contained a complex superposition of line shapes that could be deconvoluted for the direct analysis of phospholipid bilayers. Based on previous work using NMR analysis to measure the effects of calcium (up to 10 mm) on mitochondrial membranes (54), we then tested the ability of SS-31 to mitigate damage to mitochondrial membranes by calcium stress. We found that acute treatment of mitochondria with SS-31 (8.5 nmol of peptide/mg of mitochondrial protein) reduced NMR-detected membrane degradation caused by high calcium load (up to 5 mm CaCl2) (Fig. 7E). By spectral deconvolution, we observed that SS-31 treatment resulted in a significant preservation of the L2 peak, which corresponds to lamellar phospholipid bilayers (Figs. 7F and Fig. S19A). To corroborate these findings, we measured mitochondrial oxygen consumption rates under similar conditions of SS peptide treatment and calcium stress. Under these conditions, we observed a significant increase in basal respiration caused by calcium addition that was prevented by SS-31 pretreatment (Fig. 7G). Hence, acute SS-31 treatment rendered protective effects under these harsh conditions of calcium stress manifested as the preservation of membrane integrity and OXPHOS activity.
Discussion
It is well-established that SS-31 specifically targets mitochondria and ameliorates the decrease in mitochondrial function associated with aging, cellular stress, and heritable diseases. Further, SS-31 is known to have affinity for aqueous dispersions of anionic lipids (12), particularly membranes containing CL (10). However, the nature of the interaction between SS-31 and mitochondrial membranes has remained elusive. Toward the goal of understanding the molecular MoA of SS-31, the present study addressed how this peptide interacts with lipid bilayers and the effect that it has on membrane properties. Our results support a mechanism in which the modulation of mitochondrial membrane surface electrostatic properties plays a critical role.
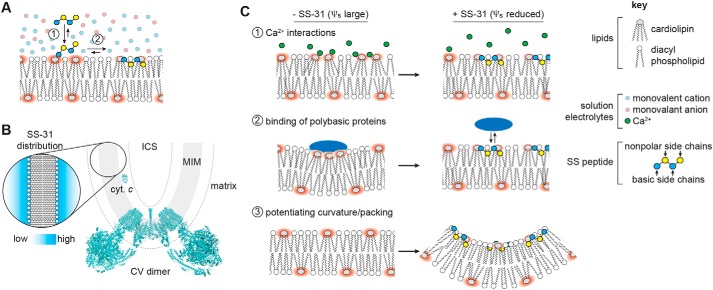
The first key finding of this study was on the lipid determinants of SS-31 membrane interactions, showing that (i) peptide binding depends on surface charge density, not the identity of particular component lipids; (ii) a key role of anionic headgroups is the establishment of a strong electric field that increases local concentration of SS peptides, enabling binding that is energetically dominated by hydrophobic contacts; and (iii) CL, although not strictly required for membrane interaction, is likely critical for SS peptide activity because it is the dominant anionic lipid of mitochondria and its structural polymorphism may be important for post-binding events in the SS peptide MoA (see supporting Discussion A). The second key finding was on the membrane binding equilibrium parameters of SS-31, including lipid binding densities (n) and affinities (KD) under different conditions. The relatively low affinity (KD in the micromolar range) determined for SS-31 is consistent with the interaction between a peptide and thermally disordered lipid bilayer and with the pharmacokinetic profile and cellular clearance reported previously (see supporting Discussion B). The third key finding was on the effects of SS peptides on membrane physical properties. Even at the highest concentrations used, SS peptides did not disrupt lamellar bilayer structural integrity; however, they did affect properties such as lipid packing and diffusivity and, most notably, membrane surface charge (and hence the ψs) (see supporting Discussion C). We propose a binding model (Fig. 8A) in which the primary outcome of SS peptide binding is partial neutralization of charge on anionic lipids, which will thereby reduce the hydration, cross-sectional area, and Coulombic repulsion among headgroups and facilitate local increases in lipid packing. A critical corollary of this model is that this effect on surface electrostatics should be shared by all SS peptides due to their common positive charge density, and not depend on specific side-chain features such as the ROS-scavenging potential of the 2′,6′-Dmt moiety of SS-31. Indeed, we found that SS-20 and SS-31 are indistinguishable in terms of their effects on tuning membrane surface potential. A key feature of our binding model is that mitochondria-bound SS peptides will reside within the MIM diffuse double layer in a distribution governed by Boltzmann statistics, likely varying among mitochondrial subcompartments with different surface charge characteristics (Fig. 8B and supporting Discussion D).
Figure 8.
SS peptide distribution and modulation of membrane surface electrostatics. A, equilibrium binding of SS-31. 1, polybasic peptide is drawn to the bilayer surface under the force of the electric field originating from negative surface charge density. 2, peptide interaction stabilized by polar contacts (basic side chains and headgroup phosphates) and hydrophobic interactions (burial of nonpolar side chains in acyl chain region). B, depiction of SS peptide equilibrium distribution at mitochondrial membranes showing mitochondrial cristae region, including the MIM, which delineates aqueous matrix and intracristal space (ICS). Topological positioning of the amphitropic protein cytochrome c (cyt. c, Protein Data Bank entry 1YCC) and FOF1-ATP synthase (CV dimer, Protein Data Bank entry 6B8H) are shown for comparison. The dashed line represents a region of ∼30 Å (2–3 Debye lengths) from the membrane surface. Inset, representation of SS peptide distribution within the diffuse double layer. C, working models for how regulation of mitochondrial surface charge may underpin the efficacy of SS peptides. Divalent cations (1) and basic protein binding (e.g. cytochrome c) (2) cause lipid demixing and sequestration of CL with implications for membrane integrity, lipid-protein interactions, and lipid peroxidation. The binding of SS peptides to CL-containing bilayers shifts the equilibrium binding of cations and basic proteins away from the membrane surface (3). SS peptide binding alters physical properties of CL-containing bilayers. The ionized CL headgroup creates a strong local electric field (red) that alters lateral lipid interactions and creates a strong ψs. Interfacial binding of SS-31 reduces ψs, thereby enhancing lipid packing and promoting local curvature.
How might the down-regulation of the ψs of anionic mitochondrial membranes underpin the broad therapeutic efficacy of SS peptides? We propose several nonexclusive possibilities (Fig. 8C). First, reducing the magnitude of the membrane surface charge will alter cation distribution at the interfacial region (Fig. 8C, 1). This is particularly relevant for Ca2+ because mitochondria must accumulate large concentrations of this divalent cation in their role as cellular calcium stores. Ca2+ binding to the lipid headgroup region severely alters properties of anionic bilayers (53). More relevant to mitochondrial membranes, Ca2+ interacts strongly with CL, altering bilayer properties (e.g. demixing, phase behavior, and headgroup conformation) (21); Ca2+ can disrupt protein-lipid interactions, causing effects such as respiratory complex disintegration (55); and mitochondrial Ca2+ dyshomeostasis underpins a wide variety of mitochondrial diseases (56). In this study, we have directly shown that the binding of SS peptides reduces the effects of Ca2+ on mitochondrial surface electrostatics (Fig. 6C) and reduces the equilibrium binding of Ca2+ to the interface of anionic synthetic membranes, which act as a “sink” for divalent cations (Figs. 7B and Fig. S17 (B and C)). We also demonstrated that SS peptides appear to reduce the energetic burden associated with calcium uptake (Fig. 7, C and D) and preserve mitochondrial function and membrane integrity with calcium stress (Fig. 7, E–G). Although we do not directly show that the latter two physiological effects originate specifically from the reduction of Ca2+ accumulation at the bilayer surface, evidence that the site of action of SS peptides is at the membrane interface supports this as a likely mechanism. This proposed effect of SS peptides on mitigating bilayer Ca2+ accumulation is consistent with their demonstrated efficacy toward processes coupled to pathogenic mitochondrial Ca2+ overload, including ischemia-reperfusion, induction of the MPT, and apoptosis/necrosis (5). A second related mechanism is that reducing the magnitude of the ψs could reduce the mitotoxic interaction of basic proteins with CL-rich mitochondrial membranes (Fig. 8C, 2). These include cytochrome c (cyt. c), whose binding to the MIM can alter lipid phase properties and cause lipid demixing (57) as well as lead to lipid peroxidation and initiate the intrinsic apoptotic pathway (58), and neurotoxic amyloid peptides, known to aggregate upon and damage mitochondrial membranes (59, 60). To this point, it is noteworthy that SS-31 has been shown to alter cytochrome c interactions with CL-containing bilayers (10, 11) and to reduce mitochondrial damage in models of Alzheimer's and Parkinson's disease (61, 62). Third, by down-regulating the surface charge density of mitochondrial membranes, SS peptides may act in part by altering the physical properties of CL-containing bilayers (Fig. 8C, 3). Full ionization of CL phosphate groups creates strong electrostatic repulsion in the lipid headgroup region and increases the effective headgroup area, imparting a more cylindrical lipid geometry. Reduction of CL headgroup charge promotes more conical molecular geometry, thereby altering lipid packing interactions and making the establishment of membrane curvature more energetically accessible (22, 63–65). SS peptides may therefore help stabilize local regions of high curvature in the morphologically complex MIM. A final potential mechanism could relate to recent findings that dysfunctional mitochondrial protein complex assembly is coupled to the release of CL into the bulk lipid phase (66); under such pathological conditions, SS peptides could help regulate the resulting increase in membrane surface charge.
With these proposed mechanisms in mind, there are two ways to consider the broad efficacy of SS peptides in treating mitochondrial dysfunction. First, the controlled down-regulation of the mitochondrial ψs could have direct and specific impacts on different molecular pathomechanisms: for instance, by altering the molecular geometry of MLCL to impart more conical-like character, SS peptides could help stabilize cristae curvature in patients with Barth syndrome. But from a more general standpoint, SS peptides could also serve to reduce the constant stresses that mitochondria face under normal physiological conditions, including spikes in calcium levels and mitotoxic interactions between CL and polybasic proteins. Such stresses could present a manageable burden on mitochondria of healthy individuals but an excessively taxing one on mitochondria whose functional integrity is compromised, as occurs with aging and heritable mitochondrial disease. Hence, the remarkably broad effects of SS peptides in treating mitochondrial dysfunction could also be based partly on their ability to mitigate general molecular stressors at the membrane.
In summary, the results of this study support a model in which SS-31 binding to mitochondrial membranes is largely driven by, and subsequently down-regulates, the ψs as a core component of its molecular MoA. Considering SS peptides as mitochondria-targeted hydrophobic polyvalent cations, this model explains how these compounds act upon the general property of bilayer surface electrostatics to serve as broad-based therapeutic compounds for a wide array of mitochondrial disorders. Finally, it is noteworthy that a recent chemical cross-linking/MS-based study has identified several potential mitochondrial proteins with which SS-31 may interact (67); therefore, it remains possible that the SS peptide MoA may involve a combination of interactions with CL-enriched lipid bilayers and CL-associated proteins.
Experimental procedures
Reagents
Peptides SS-31 and SS-20 were prepared by solid-phase synthesis by Phoenix Pharmaceuticals (Burlingame, CA), and [ald]SS-31 was synthesized as described (12). All peptides were prepared as aqueous stocks. Synthetic phospholipids were purchased as chloroform stocks from Avanti Polar Lipids (Alabaster, AL), including POPC, POPG, and variants of cardiolipin with 18:1 acyl chains, including TOCL and trioleoyl MLCL. All lipid stocks were stored at −20 °C in clear vials with Teflon-lined cap closures until use. Fluorescent probes TMRM, laurdan, prodan, DPH, ANS, and CG-5N were purchased from Molecular Probes (Thermo Fisher Scientific). Solutions were prepared with deionized ultrapure water, and all buffers, salts and other reagents were obtained from Sigma–Aldrich.
Liposome preparation
Liposomes of a specific lipid composition were prepared by mixing chloroform stocks at the appropriate molar ratios in glass vials and drying under N2 flow, followed by incubation in a vacuum desiccator for a minimum of 2 h to remove all organic solvent. Lipid films were hydrated in aqueous solutions to produce dispersions of multilamellar vesicles. To prepare LUVs, lipid suspensions were extruded through polycarbonate membranes with 0.1-μm pores (Whatman, Maidstone, UK) (68). Multilamellar vesicles and LUVs were prepared in aqueous solutions of buffer (20 mm HEPES-KOH, pH 7.5) in the presence or absence of different concentrations of salt (KCl), depending on the application. Hydration and extrusion steps were all performed at a temperature above the main (gel to liquid phase) temperature of the highest-melting lipid. The phospholipid concentration of each preparation was determined by a colorimetric ammonium ferrothiocyanate assay (69) against standards based on the mole fraction of lipids of a given preparation. Liposome suspensions were routinely monitored for size and monodispersity by dynamic light scattering.
Yeast strains and isolation of mitochondria
The yeast strains used in this study are isogenic to GA74-1A, including genetic knockouts Δcrd1 and Δtaz1, which were described previously (50). Yeast cells were grown at 30 °C in lactate medium containing 0.1% glucose. For the preparation of active mitochondria, yeast cells were cultivated to mid-log phase, and mitochondria were isolated by Dounce homogenization of spheroplasts and differential centrifugation as described (70). Isolated mitochondria were resuspended in buffer (600 mm sorbitol, 20 mm HEPES-KOH, 1 mm EDTA, pH 7.5), and aliquots (1–2 mg of mitochondrial protein each) were snap-frozen in liquid nitrogen and stored at −80 °C. Mitoplasts were prepared by osmotically rupturing the outer membrane as described (70) by incubating intact mitochondria in buffer (60 mm sorbitol, 20 mm HEPES, pH 7.5) on ice for 30 min.
Analytical fluorescence spectroscopy
Steady-state fluorescence measurements were performed with a Fluorolog 3-22 spectrofluorometer (HORIBA Jobin-Yvon) equipped with photon-counting electronics, double-grating excitation and emission monochromators, automated Glan–Thompson polarizers, and a 450-watt xenon lamp. Measurements were made either in 4 × 4-mm quartz microcells or in 1 × 1-cm quartz cuvettes with a stir disc seated in a thermostated cell holder. For all spectral measurements, samples containing the fluorescent moiety (peptide or extrinsic fluorophore) were recorded in the L-configuration. Endogenous fluorescence measurements were made in parallel with blanks (lacking fluorophore but otherwise identical), which were used for background subtraction to obtain the signal originating solely from the fluorescent molecule. The following steady-state measurements were made. (a) Endogenous fluorescence from the 2′,6′-Dmt moiety of the native SS-31 peptide was measured by emission (λex = 283 nm; λem = 290–400 nm) or excitation (λex = 260–300 nm; λem = 308 nm) scans using 1-nm increments and 2-s integration times. To suppress scattered light from lipid-containing samples and eliminate spectral distortions, in-line polarizers were used in the cross-oriented configuration (Expol = 90°, Empol = 0°) as described (71). (b) Endogenous steady-state anisotropy (〈r〉) was determined by measuring emission intensities at four different polarizer orientations (excitation with vertical polarization, IVV and IVH, and excitation with horizontal polarization, IHV and IHH). Each sample was background-subtracted with the cognate blank to measure the G-factor (G = IHV/IHH) and the steady-state anisotropy 〈r〉 = (IVV − GIVH)/(IVV + 2GIVH). The steady-state anisotropy of 2′,6′-Dmt was determined using λex = 283 nm; λem = 308 nm, and the steady state anisotropy of DPH was determined using λex = 360 nm; λem = 430 nm. (c) Dynamic (collisional) quenching of endogenous fluorescence was performed by titration of quenching agent (Q) and measurement of steady-state fluorescence (F) relative to unquenched fluorescence (F0) with linear fits of (F0/F) − 1 versus [Q] based on Stern–Volmer formalism, (F0/F) − 1 = KSV[Q], to yield the Stern–Volmer constant, KSV. (d) Fluorescence measurements of extrinsic fluorophores were performed by emission scans (1-nm increments with 1–2-s integration times) or by kinetic measurements (0.5–1-s integration times) as follows. Aladan fluorescence from the [ald]SS-31 peptide was measured by emission scans (λex = 360 nm; λem = 400–600 nm) with excitation and emission bandpass at 4 nm. The fluorescence of laurdan and prodan-containing samples was measured by emission scans (λex = 340 nm; λem = 370–600 nm) with excitation and emission bandpass at 2 nm. Emission intensities at 440 nm and 490 nm (I440 and I490, representing emission maxima of pure Lβ and Lα phases, respectively) were used to calculate the generalized polarization (GP) of laurdan as GPLAU = (I440 − I490)/(I440 + I490). The GP of prodan was calculated by the three-wavelength excitation GP (3wGP) (72) as GPPRO = (R12 − 1)/(R12 + 1), where R12 = I420k/(I480k − I530); I420, I480, and I530 are emission intensities at 420, 480, and 530 nm; and k is a constant independently verified to be 2.8. The fluorescence of ANS was measured by time course (λex = 380 nm; λem = 460 nm) or emission scan (λex = 380 nm; λem = 400–500 nm) measurements with excitation and emission bandpass at 4 nm. The fluorescence of CG-5N was measured by time course (λex = 506 nm; λem = 532 nm) or emission scan (λex = 506 nm; λem = 512–562 nm) measurements with excitation and emission bandpass at 2 nm. And the fluorescence of TMRM was measured by time course measurements (λex = 546 nm; λem = 573 nm) with excitation and emission bandpass at 4 nm.
Time-resolved fluorescence lifetime measurements were performed with a Fluorolog 3-22 instrument equipped with a TCSPC module and pulsed nano-LED light source (peak wavelength 280 nm ± 10 nm; pulse width <1.2 ns).
Microcalorimetry
ITC measurements were performed as described (35) with a low-volume nano-ITC microcalorimeter (TA Instruments, New Castle, DE). Solutions of SS-31 (titrate) and LUVs (titrant) were prepared in degassed buffer, equivalent to that used for liposome preparation. Lipid-into-peptide titrations were performed with 87.5–175 μm SS-31 in the calorimeter cell (volume 170 μl), and LUVs (8 mm total lipid) were injected in aliquots of 2.5 μl (20 total injections) at time intervals of 300 s at 25 °C. To account for heats of dilution, experiments were performed by the addition of titrant into solutions of buffer only, which were used for baseline subtraction. Data from dilution-corrected and integrated heat flow time courses were fit as Wiseman plots, from which equilibrium binding and thermodynamic parameters (KA, n, ΔH, and ΔS) were determined by nonlinear regression fits (NanoAnalyze software version 3.10.0, TA Instruments).
NMR spectroscopy
NMR data were recorded at 162 MHz on a Bruker AVANCE III 400-MHz WB spectrometer equipped with an HXY probe tuned for 1H and 31P. High-power decoupled spectra were acquired with a recycle delay of 2 s, a 90° pulse of 5 μs, and a 50-kHz decoupling field. All 31P NMR spectra were externally referenced by assigning the 85% H3PO4 peak to 0 ppm. Model membranes (LUVs) were measured at 25 °C (total of 10,240 scans/sample) with a total lipid concentration of 2 mm lipid in the presence of different concentrations of SS-31 (0, 1:500, 1:50, and 1:5 [P]/[L]eff). Isolated mitochondria were measured in minimal buffer (600 mm sorbitol, 20 mm HEPES-KOH, pH 7.5) at 4 °C (total of 512 scans/sample) at a total concentration of 70 mg of mitochondrial protein ml−1 with different concentrations of SS-31 (up to 3 mm) and/or [CaCl2] (up to 5 mm). Peak fitting and deconvolution were performed using the software dmFit (73).
SAXS
SAXS scattering curves (scattered intensity, I(q), as a function of moment transfer, q) were recorded at the LiX beamline of the National Synchrotron Light Source II (NSLS-II), Brookhaven National Laboratory, operating at 13 keV. LUV samples (2 mm total lipid) were measured in 1.5-mm diameter quartz flow-through capillaries using a 1-s exposure, and the q-range covered was 0.005–3.2 Å−1.
Electrokinetic measurements
Measurements of zeta potential (ζ) of LUVs were performed as we have described (74), using a Zetasizer Nano ZS (Malvern). LUVs (100 μm total lipid) were added to the sample cell (final volume, 1 ml) with different concentrations of SS-31. Values of ζ represent the electrostatic potential ψx at the hydrodynamic shear plane of lipid vesicles based on the measured electrophoretic mobility (μ) by the Helmholtz–Smoluchowski relationship,
| (Eq. 1) |
where η is the viscosity of the solution, ϵr is the dielectric constant of the aqueous phase, and ϵ0 is the permittivity of free space. Calculation of surface potential (ψ0) was based on the distance-dependent decay of electrostatic potential from the membrane surface,
| (Eq. 2) |
where R is the molar gas constant, T is the absolute temperature, Z is ionic charge of solution electrolyte, x is the distance from the membrane surface, and κ (the Debye constant) is as follows.
| (Eq. 3) |
The electrostatic shear plane was assumed to be at x = 2 Å.
Mitochondrial activity assays
Mitochondrial calcium uptake was measured by monitoring the bulk [Ca2+] using the cell-impermeant calcium-indicating fluorophore CG-5N (prepared as an aqueous stock), adapted from a protocol described for measuring Ca2+ uptake into S. cerevisiae mitochondria (75). Reactions containing mitochondria (100 μg) in calcium flux buffer (300 mm sucrose, 20 mm HEPES-KOH, 2 mm KH2PO4, 0.5 mg ml−1 BSA, pH 7.5) were prepared with the probe (1 μm CG-5N) in the presence or absence of calcium ionophore (5 μm ETH-129) and variable concentrations of CaCl2. Measurements of mitochondrial membrane potential were made using the potentiometric fluorophore TMRM under identical conditions, but in the presence of 50 nm TMRM (prepared as a methanol stock). CG-5N and TMRM measurements were performed as time course measurements in stirred cuvettes with the progressive addition of respiratory substrate (1 mm NADH) and CaCl2 at specified time intervals in the presence or absence of the indicated concentrations of SS peptides.
Oxygen consumption rates of isolated mitochondria were measured using a closed, thermostated chamber equipped with a Clark-type oxygen electrode (Oxygraph Plus System, Hansatech Instruments, UK). Reactions were set up with 100 μg of mitochondria in one of two respiration measurement buffers. Standard respiration buffer (300 mm sucrose, 20 mm HEPES-KOH, 10 mm KH2PO4, 10 mm KCl, 5 mm MgCl2, 1 mg ml−1 BSA, 0.5 mm EDTA, pH 7.5) was used for routine testing of mitochondrial activity and coupling. Minimal respiration buffer (300 mm sucrose, 20 mm HEPES-KOH, 1 mg ml−1 BSA, 10 mm KH2PO4, pH 7.5) was used for measurements under NMR conditions. To test the effects of calcium stress and/or SS peptide on mitochondrial respiration, a concentrated stock of mitochondria (1 mg ml−1) was preincubated with CaCl2 (1 mm, equivalent to 1 μmol of Ca2+/mg of mitochondria) and/or SS-31 (54.4 μm, equivalent to 54.4 nmol of SS-31/mg of mitochondria) on ice for 20 min prior to polarographic measurements. The effect of calcium stress on mitochondrial respiration was measured by monitoring state 2 respiration for 3 min following the addition of respiratory substrate (19 mm succinate and 1 mm NADH) and by monitoring respiration without noncatalytic H+ flux through the FO of ATP synthase for 3 min following the addition of 14.4 μm oligomycin.
Lipidomics
Multidimensional MS-based shotgun lipidomic analysis of mitochondrial lipids was performed as described (50, 76). In brief, a mixture of internal standards was added to isolated mitochondria for quantitation of phospholipid species based on the content of mitochondrial protein, lipids were extracted using a modified Bligh and Dyer procedure (77), and each lipid extract was reconstituted in 1:1 (v/v) chloroform/methanol at a volume of 200 μl mg−1 mitochondrial protein. For shotgun lipidomics, lipid extracts were diluted to a final concentration of ∼500 fmol of total lipids μl−1. Mass spectrometric analysis was performed on a triple quadrupole mass spectrometer (TSQ Altis, Thermo Fisher Scientific) and a Q Exactive mass spectrometer (Thermo Scientific), both of which were equipped with an automated nanospray device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, NY) as described (78). Identification and quantification of phospholipid species were performed using an automated software program (79, 80). Data processing (ion peak selection, baseline correction, data transfer, peak intensity comparison, and quantitation) was performed as described (80).
MD simulations
The SS-31 peptide structure was generated by modifying an extended tetrapeptide with sequence Arg-Tyr-Lys-Phe. Coordinates were modified using the VMD Molefacture plugin (81) to invert the stereochemistry of the N-terminal Arg residue (from l to d) and to replace the 2′ and 6′ hydrogen atoms of the Tyr side chain with methyl groups. Parameters for the 2′,6′-Dmt were modeled after the output parameters of 3′,5′-dimethylphenol after running its structure through ParamChem's CGenFF server (82). The fully parameterized SS-31 peptide was then run through the CGenFF server again to check for energy penalties.
MD simulations were then used to characterize the binding mechanism of SS-31 and to investigate its effects on membrane dynamics. All-atom systems with explicit membrane and solvent were prepared using CHARMM-GUI with the CHARMM-36m forcefield and the TIP3 water model (83–89). Bilayers were generated with molar ratios of TOCL/POPC (20:80), MLCL/POPC (20:80), and POPG/POPC (20:80). Each system contained a total of 300 lipids (150 per leaflet). As done in a previous study (30), the MLCL system was created from the TOCL system by replacing an sn-2 acyl chain with a hydroxyl, appending the topology, and using a GROMACS-compatible topology file (.itp) for MLCL. Following the CHARMM-GUI standard protocol for membranes, the bilayer systems were energy-minimized using the steepest-descent algorithm for 5000 steps, followed by canonical ensemble equilibration for 50 ps with 1-fs timestep, and 325 ps of NPT equilibration with a 2-fs timestep accomplished using the semiisotropic pressure-coupling scheme and the Berendsen barostat. Position and dihedral restraints were used during equilibration on the lipids to maintain lipid geometry and bilayer morphology. Production simulations of the bilayers alone were each run for 1 μs in an NVT ensemble with a timestep of 2 fs. In our simulations, we define the “upper leaflet” as the side of the membrane exposed to peptides and the “lower leaflet” as the opposing side, which was not exposed to peptides.
Peptide-bilayer systems were constructed by removing solvent from the equilibrated bilayers, placing 10 peptides 1–3 nm away from the upper leaflet headgroup region, and then resolvating and adding neutralizing sodium ions. The sizes of the systems were ∼88,000 atoms for the TOCL systems, ∼85,000 atoms for the MLCL systems, and ∼73,000 for the POPG systems. The systems were then energy-minimized for 5000 steepest-descent steps, followed by canonical ensemble equilibration for 50 ps with a 1-fs timestep and 425 ps of NPT equilibration with a 2-fs timestep accomplished using the semiisotropic pressure-coupling scheme and the Berendsen barostat. Position and dihedral restraints were used during equilibration on the lipids and peptides to maintain lipid geometry and bilayer morphology and to prevent the peptides from interacting with the bilayers during equilibration. Production simulations were run for 1.6 μs and saved every 50 ps for the GROMACS mean squared displacement and radial distribution function analyses, every 100 ps for GROMACS solvent-accessible surface area analyses, and every 250 ps for the MDTraj analyses. The first 50 ns of the production simulations were omitted from all analyses except the binding time dependence (Fig. 5A and Fig. S11A). To enforce peptide binding to only the upper leaflet (i.e. to impose an energy penalty on peptides crossing the z-dimension periodic boundary and binding to lower leaflet), two inverted flat-bottom restraints in the z-direction were placed ∼1 nm away from the headgroup region of the lower leaflet. A restraint was placed on the peptides with a force constant of 1000 kJ mol−1 nm−1 and a radius of 2 nm, which served to prevent peptides from binding to the lower leaflet. A second restraint was placed on the lower leaflet phosphates of POPC with a force constant of 200 kJ mol−1 nm−1 and a radius of 0.5 nm to prevent the lower leaflet from drifting in the negative z-direction while still allowing for natural membrane deformations.
All simulations were performed using GROMACS 2016 (90, 91). Electrostatic and Lennard–Jones interactions were cut off at 1.2 nm, with electrostatics shifted from 0 nm to the cutoff and Lennard–Jones interactions shifted from 1.0 nm to the cutoff. All production runs were simulated in the NPT ensemble using the Parrinello–Rahman coupling scheme, with the temperature maintained at 303.15 K and pressure kept at 1.0 bar with semiisotropic coupling. The time constants for pressure and temperature couplings were 5.0 and 1.0 ps, respectively, and the compressibility value was set to 4.5 × 10−5 bar−1. Simulations were performed using periodic boundary conditions in all dimensions, and the simulation time step was 2 fs.
Statistical analysis
All reported means are the average values of a minimum of three measurements from independent experimental samples. Statistical comparisons were performed by t test (not significant (ns), p > 0.05; •, p < 0.05; ••, p < 0.01; •••, p < 0.001; ••••, p < 0.0001).
Data availability
All data are contained within the article.
Author contributions
W. M., E. A. N., J. D. T., K. J. B., M. S., A. C., E. R. M., and N. N. A. conceptualization; W. M., E. A. N., J. D. T., K. J. B., M. S., A. C., M. P., X. H., N. A. E., E. R. M., and N. N. A. data curation; W. M., E. A. N., J. D. T., K. J. B., M. S., A. C., M. P., X. H., N. A. E., E. R. M., H. H. S., and N. N. A. formal analysis; W. M., E. A. N., J. D. T., K. J. B., E. R. M., and N. N. A. validation; W. M., E. A. N., J. D. T., K. J. B., M. S., A. C., M. P., X. H., N. A. E., E. R. M., and N. N. A. investigation; W. M., E. A. N., J. D. T., K. J. B., M. S., A. C., M. P., X. H., N. A. E., E. R. M., and N. N. A. methodology; W. M., E. A. N., J. D. T., K. J. B., E. R. M., H. H. S., and N. N. A. writing-original draft; W. M., E. A. N., J. D. T., K. J. B., E. R. M., H. H. S., and N. N. A. writing-review and editing; E. R. M. and N. N. A. supervision; E. R. M. and N. N. A. resources; E. R. M. and N. N. A. funding acquisition; N. N. A. visualization; N. N. A. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Steven Claypool (Johns Hopkins University School of Medicine) for the synthesis of yeast mutants, Drs. Mu-Ping Nieh and Kuo-Chih Shih (University of Connecticut) for technical assistance with SAXS measurements, Dr. Georg Pabst (University of Graz) for providing the GAP program for analysis of SAXS data, Drs. Peter Rabinovitch and David Marcinek for useful discussions, and the UT Health-San Antonio Mass Spectrometry Core Facility.
This work was supported by National Institutes of Health Grants R01-GM113092 (to N. N. A.), R35-GM119762 (to E. R. M.), and RF1-AG061872 (to X. H.); by National Science Foundation GRFP Award 1247393 (to K. J. B.); and by a charitable contribution from the Social Profit Network (to N. N. A.). H. H. S. is the inventor of the mitochondria-targeted peptides described in this article, and the Founder of Stealth Biotherapeutics, a clinical stage biopharmaceutical company that licensed this peptide technology from the Cornell Research Foundation for research and development. H. H. S. does not currently hold any position in Stealth Biotherapeutics but has financial interests in the company. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supporting Analytic Approaches, Results, and Discussion and Figs. S1–S19.
- OXPHOS
- oxidative phosphorylation
- ROS
- reactive oxygen species
- 2′,6′-Dmt
- 2′,6′-dimethyltyrosine
- ANS
- 1-anilinonaphthalene-8-sulfonic acid
- CL
- cardiolipin
- DPH
- 1,6-diphenyl-1,3,5-hexatriene
- GCS
- Gouy-Chapman-Stern
- GP
- generalized polarization
- ITC
- isothermal titration calorimetry
- LUV
- large unilamellar vesicle
- MD
- molecular dynamics
- MIM
- mitochondrial inner membrane
- MLCL
- trioleoyl monolysocardiolipin
- MoA
- mechanism of action
- MPT
- mitochondrial permeability transition
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- SAXS
- small-angle X-ray scattering
- SS peptides
- Szeto–Schiller peptides
- TMRM
- tetramethylrhodamine methyl ester
- TOCL
- 1′,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol (tetraoleoyl cardiolipin)
- ssNMR
- solid-state NMR
- CG-5N
- Calcium Green-5N
- laurdan
- 6-dodecanoyl-2-dimethylaminonaphthalene
- prodan
- 6-propinoyl-2-dimethylaminonaphthalene.
References
- 1. Gorman G. S., Chinnery P. F., DiMauro S., Hirano M., Koga Y., McFarland R., Suomalainen A., Thorburn D. R., Zeviani M., and Turnbull D. M.. 2016) Mitochondrial diseases. Nat. Rev. Dis. Primers 2, 16080 10.1038/nrdp.2016.80 [DOI] [PubMed] [Google Scholar]
- 2. Nunnari J., and Suomalainen A.. 2012) Mitochondria: in sickness and in health. Cell 148, 1145–1159 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sorrentino V., Menzies K. J., and Auwerx J.. 2018) Repairing mitochondrial dysfunction in disease. Annu. Rev. Pharmacol. Toxicol. 58, 353–389 10.1146/annurev-pharmtox-010716-104908 [DOI] [PubMed] [Google Scholar]
- 4. Szeto H. H., and Liu S.. 2018) Cardiolipin-targeted peptides rejuvenate mitochondrial function, remodel mitochondria, and promote tissue regeneration during aging. Arch. Biochem. Biophys. 660, 137–148 10.1016/j.abb.2018.10.013 [DOI] [PubMed] [Google Scholar]
- 5. Szeto H. H. 2018) Stealth peptides target cellular powerhouses to fight rare and common age-related diseases. Protein Pept. Lett. 25, 1108–1123 10.2174/0929866525666181101105209 [DOI] [PubMed] [Google Scholar]
- 6. Zhao K., Luo G., Giannelli S., and Szeto H. H.. 2005) Mitochondria-targeted peptide prevents mitochondrial depolarization and apoptosis induced by tert-butyl hydroperoxide in neuronal cell lines. Biochem. Pharmacol. 70, 1796–1806 10.1016/j.bcp.2005.08.022 [DOI] [PubMed] [Google Scholar]
- 7. Zhao K., Luo G., Zhao G. M., Schiller P. W., and Szeto H. H.. 2003) Transcellular transport of a highly polar 3+ net charge opioid tetrapeptide. J. Pharmacol. Exp. Ther. 304, 425–432 10.1124/jpet.102.040147 [DOI] [PubMed] [Google Scholar]
- 8. Zhao K., Zhao G. M., Wu D., Soong Y., Birk A. V., Schiller P. W., and Szeto H. H.. 2004) Cell-permeable peptide antioxidants targeted to inner mitochondrial membrane inhibit mitochondrial swelling, oxidative cell death, and reperfusion injury. J. Biol. Chem. 279, 34682–34690 10.1074/jbc.M402999200 [DOI] [PubMed] [Google Scholar]
- 9. Winterbourn C. C., Parsons-Mair H. N., Gebicki S., Gebicki J. M., and Davies M. J.. 2004) Requirements for superoxide-dependent tyrosine hydroperoxide formation in peptides. Biochem. J. 381, 241–248 10.1042/BJ20040259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Birk A. V., Chao W. M., Bracken C., Warren J. D., and Szeto H. H.. 2014) Targeting mitochondrial cardiolipin and the cytochrome c/cardiolipin complex to promote electron transport and optimize mitochondrial ATP synthesis. Br. J. Pharmacol. 171, 2017–2028 10.1111/bph.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Birk A. V., Chao W. M., Liu S., Soong Y., and Szeto H. H.. 2015) Disruption of cytochrome c heme coordination is responsible for mitochondrial injury during ischemia. Biochim. Biophys. Acta 1847, 1075–1084 10.1016/j.bbabio.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Birk A. V., Liu S., Soong Y., Mills W., Singh P., Warren J. D., Seshan S. V., Pardee J. D., and Szeto H. H.. 2013) The mitochondrial-targeted compound SS-31 re-energizes ischemic mitochondria by interacting with cardiolipin. J. Am. Soc. Nephrol. 24, 1250–1261 10.1681/ASN.2012121216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Szeto H. H. 2014) First-in-class cardiolipin-protective compound as a therapeutic agent to restore mitochondrial bioenergetics. Br. J. Pharmacol. 171, 2029–2050 10.1111/bph.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paradies G., Paradies V., De Benedictis V., Ruggiero F. M., and Petrosillo G.. 2014) Functional role of cardiolipin in mitochondrial bioenergetics. Biochim. Biophys. Acta 1837, 408–417 10.1016/j.bbabio.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 15. Agrawal A., and Ramachandran R.. 2019) Exploring the links between lipid geometry and mitochondrial fission: emerging concepts. Mitochondrion 49, 305–313 10.1016/j.mito.2019.07.010 [DOI] [PubMed] [Google Scholar]
- 16. Musatov A., and Sedlák E.. 2017) Role of cardiolipin in stability of integral membrane proteins. Biochimie 142, 102–111 10.1016/j.biochi.2017.08.013 [DOI] [PubMed] [Google Scholar]
- 17. Planas-Iglesias J., Dwarakanath H., Mohammadyani D., Yanamala N., Kagan V. E., and Klein-Seetharaman J.. 2015) Cardiolipin interactions with proteins. Biophys. J. 109, 1282–1294 10.1016/j.bpj.2015.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mileykovskaya E., and Dowhan W.. 2014) Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem. Phys. Lipids 179, 42–48 10.1016/j.chemphyslip.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu Y., Anjaneyulu M., Donelian A., Yu W., Greenberg M. L., Ren M., Owusu-Ansah E., and Schlame M.. 2019) Assembly of the complexes of oxidative phosphorylation triggers the remodeling of cardiolipin. Proc. Natl. Acad. Sci. U.S.A. 116, 11235–11240 10.1073/pnas.1900890116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alessandrini A., and Muscatello U.. 2009) AFM and FTIR spectroscopy investigation of the inverted hexagonal phase of cardiolipin. J. Phys. Chem. B 113, 3437–3444 10.1021/jp809705d [DOI] [PubMed] [Google Scholar]
- 21. Lewis R. N., and McElhaney R. N.. 2009) The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochim. Biophys. Acta 1788, 2069–2079 10.1016/j.bbamem.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 22. Boyd K. J., Alder N. N., and May E. R.. 2017) Buckling under pressure: curvature-based lipid segregation and stability modulation in cardiolipin-containing bilayers. Langmuir 33, 6937–6946 10.1021/acs.langmuir.7b01185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ikon N., and Ryan R. O.. 2017) Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 1859, 1156–1163 10.1016/j.bbamem.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schlame M., and Greenberg M. L.. 2017) Biosynthesis, remodeling and turnover of mitochondrial cardiolipin. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 3–7 10.1016/j.bbalip.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu Y. W., and Claypool S. M.. 2015) Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front Genet. 6, 3 10.3389/fgene.2015.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ikon N., and Ryan R. O.. 2017) Barth syndrome: connecting cardiolipin to cardiomyopathy. Lipids 52, 99–108 10.1007/s11745-016-4229-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ren M., Miller P. C., Schlame M., and Phoon C. K. L.. 2019) A critical appraisal of the tafazzin knockdown mouse model of Barth syndrome: what have we learned about pathogenesis and potential treatments? Am. J. Physiol. Heart Circ. Physiol. 317, H1183–H1193 10.1152/ajpheart.00504.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kopp Lugli A., Yost C. S., and Kindler C. H.. 2009) Anaesthetic mechanisms: update on the challenge of unravelling the mystery of anaesthesia. Eur. J. Anaesthesiol. 26, 807–820 10.1097/EJA.0b013e32832d6b0f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seelig J. 1997) Titration calorimetry of lipid-peptide interactions. Biochim. Biophys. Acta 1331, 103–116 10.1016/S0304-4157(97)00002-6 [DOI] [PubMed] [Google Scholar]
- 30. Boyd K. J., Alder N. N., and May E. R.. 2018) Molecular dynamics analysis of cardiolipin and monolysocardiolipin on bilayer properties. Biophys. J. 114, 2116–2127 10.1016/j.bpj.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan J., Cheng X., Sharp M., Ho C. S., Khadka N., and Katsaras J.. 2015) Structural and mechanical properties of cardiolipin lipid bilayers determined using neutron spin echo, small angle neutron and X-ray scattering, and molecular dynamics simulations. Soft Matter 11, 130–138 10.1039/C4SM02227K [DOI] [PubMed] [Google Scholar]
- 32. Walrant A., Vogel A., Correia I., Lequin O., Olausson B. E., Desbat B., Sagan S., and Alves I. D.. 2012) Membrane interactions of two arginine-rich peptides with different cell internalization capacities. Biochim. Biophys. Acta 1818, 1755–1763 10.1016/j.bbamem.2012.02.024 [DOI] [PubMed] [Google Scholar]
- 33. Witte K., Olausson B. E., Walrant A., Alves I. D., and Vogel A.. 2013) Structure and dynamics of the two amphipathic arginine-rich peptides RW9 and RL9 in a lipid environment investigated by solid-state NMR and MD simulations. Biochim. Biophys. Acta 1828, 824–833 10.1016/j.bbamem.2012.11.014 [DOI] [PubMed] [Google Scholar]
- 34. Ben-Tal N., Honig B., Peitzsch R. M., Denisov G., and McLaughlin S.. 1996) Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys. J. 71, 561–575 10.1016/S0006-3495(96)79280-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Swamy M. J., and Sankhala R. S.. 2013) Probing the thermodynamics of protein-lipid interactions by isothermal titration calorimetry. Methods Mol. Biol. 974, 37–53 10.1007/978-1-62703-275-9_3 [DOI] [PubMed] [Google Scholar]
- 36. Kessel A., and Ben-Tal N.. 2002) Free energy determinants of peptide association with lipid bilayers. Curr. Top. Membr. 52, 205–253 10.1016/S1063-5823(02)52010-X [DOI] [Google Scholar]
- 37. Montich G., Scarlata S., McLaughlin S., Lehrmann R., and Seelig J.. 1993) Thermodynamic characterization of the association of small basic peptides with membranes containing acidic lipids. Biochim. Biophys. Acta 1146, 17–24 10.1016/0005-2736(93)90333-U [DOI] [PubMed] [Google Scholar]
- 38. Thorgeirsson T. E., Russell C. J., King D. S., and Shin Y. K.. 1996) Direct determination of the membrane affinities of individual amino acids. Biochemistry 35, 1803–1809 10.1021/bi952300c [DOI] [PubMed] [Google Scholar]
- 39. Seelig J. 2004) Thermodynamics of lipid-peptide interactions. Biochim. Biophys. Acta 1666, 40–50 10.1016/j.bbamem.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 40. McLaughlin S. 1977) Electrostatic potentials at membrane-solution interfaces. Curr. Top. Membr. Transp. 9, 71–144 10.1016/S0070-2161(08)60677-2 [DOI] [Google Scholar]
- 41. Oshima H. 2010) Biophysical Chemistry of Interfaces, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 42. Slavík J. 1982) Anilinonaphthalene sulfonate as a probe of membrane composition and function. Biochim. Biophys. Acta 694, 1–25 10.1016/0304-4157(82)90012-0 [DOI] [PubMed] [Google Scholar]
- 43. Willumeit R., Kumpugdee M., Funari S. S., Lohner K., Navas B. P., Brandenburg K., Linser S., and Andrä J.. 2005) Structural rearrangement of model membranes by the peptide antibiotic NK-2. Biochim. Biophys. Acta 1669, 125–134 10.1016/j.bbamem.2005.01.011 [DOI] [PubMed] [Google Scholar]
- 44. McLaughlin S., Mulrine N., Gresalfi T., Vaio G., and McLaughlin A.. 1981) Adsorption of divalent cations to bilayer membranes containing phosphatidylserine. J. Gen. Physiol. 77, 445–473 10.1085/jgp.77.4.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumar P., Kizhakkedathu J. N., and Straus S. K.. 2018) Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 8, E4 10.3390/biom8010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tyler A. I., Law R. V., and Seddon J. M.. 2015) X-ray diffraction of lipid model membranes. Methods Mol. Biol. 1232, 199–225 10.1007/978-1-4939-1752-5_16 [DOI] [PubMed] [Google Scholar]
- 47. Seelig J. 1978) 31P nuclear magnetic resonance and the head group structure of phospholipids in membranes. Biochim. Biophys. Acta 515, 105–140 10.1016/0304-4157(78)90001-1 [DOI] [PubMed] [Google Scholar]
- 48. Demchenko A. P., Mély Y., Duportail G., and Klymchenko A. S.. 2009) Monitoring biophysical properties of lipid membranes by environment-sensitive fluorescent probes. Biophys. J. 96, 3461–3470 10.1016/j.bpj.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cohen B. E., McAnaney T. B., Park E. S., Jan Y. N., Boxer S. G., and Jan L. Y.. 2002) Probing protein electrostatics with a synthetic fluorescent amino acid. Science 296, 1700–1703 10.1126/science.1069346 [DOI] [PubMed] [Google Scholar]
- 50. Baile M. G., Sathappa M., Lu Y. W., Pryce E., Whited K., McCaffery J. M., Han X., Alder N. N., and Claypool S. M.. 2014) Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J. Biol. Chem. 289, 1768–1778 10.1074/jbc.M113.525733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Overall S. A., Zhu S., Hanssen E., Separovic F., and Sani M. A.. 2019) In situ monitoring of bacteria under antimicrobial stress using 31P solid-state NMR. Int. J. Mol. Sci. 20, E181 10.3390/ijms20010181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gasanov S. E., Shrivastava I. H., Israilov F. S., Kim A. A., Rylova K. A., Zhang B., and Dagda R. K.. 2015) Naja naja oxiana cobra venom cytotoxins CTI and CTII disrupt mitochondrial membrane integrity: implications for basic three-fingered cytotoxins. PLoS ONE 10, e0129248 10.1371/journal.pone.0129248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Melcrová A., Pokorna S., Pullanchery S., Kohagen M., Jurkiewicz P., Hof M., Jungwirth P., Cremer P. S., and Cwiklik L.. 2016) The complex nature of calcium cation interactions with phospholipid bilayers. Sci. Rep. 6, 38035 10.1038/srep38035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sani M. A., Keech O., Gardeström P., Dufourc E. J., and Gröbner G.. 2009) Magic-angle phosphorus NMR of functional mitochondria: in situ monitoring of lipid response under apoptotic-like stress. FASEB J. 23, 2872–2878 10.1096/fj.09-134114 [DOI] [PubMed] [Google Scholar]
- 55. Hwang M. S., Schwall C. T., Pazarentzos E., Datler C., Alder N. N., and Grimm S.. 2014) Mitochondrial Ca2+ influx targets cardiolipin to disintegrate respiratory chain complex II for cell death induction. Cell Death Differ. 21, 1733–1745 10.1038/cdd.2014.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Granatiero V., De Stefani D., and Rizzuto R.. 2017) Mitochondrial calcium handling in physiology and disease. Adv. Exp. Med. Biol. 982, 25–47 10.1007/978-3-319-55330-6_2 [DOI] [PubMed] [Google Scholar]
- 57. Trusova V. M., Gorbenko G. P., Molotkovsky J. G., and Kinnunen P. K.. 2010) Cytochrome c-lipid interactions: new insights from resonance energy transfer. Biophys. J. 99, 1754–1763 10.1016/j.bpj.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kagan V. E., Bayir H. A., Belikova N. A., Kapralov O., Tyurina Y. Y., Tyurin V. A., Jiang J., Stoyanovsky D. A., Wipf P., Kochanek P. M., Greenberger J. S., Pitt B., Shvedova A. A., and Borisenko G.. 2009) Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic. Biol. Med. 46, 1439–1453 10.1016/j.freeradbiomed.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Camilleri A., Zarb C., Caruana M., Ostermeier U., Ghio S., Högen T., Schmidt F., Giese A., and Vassallo N.. 2013) Mitochondrial membrane permeabilisation by amyloid aggregates and protection by polyphenols. Biochim. Biophys. Acta 1828, 2532–2543 10.1016/j.bbamem.2013.06.026 [DOI] [PubMed] [Google Scholar]
- 60. Sciacca M. F. M., Tempra C., Scollo F., Milardi D., and La Rosa C.. 2018) Amyloid growth and membrane damage: current themes and emerging perspectives from theory and experiments on Aβ and hIAPP. Biochim. Biophys. Acta Biomembr. 10.1016/j.bbamem.2018.02.022 [DOI] [PubMed] [Google Scholar]
- 61. Manczak M., Mao P., Calkins M. J., Cornea A., Reddy A. P., Murphy M. P., Szeto H. H., Park B., and Reddy P. H.. 2010) Mitochondria-targeted antioxidants protect against amyloid-β toxicity in Alzheimer's disease neurons. J. Alzheimers Dis. 20, S609–S631 10.3233/JAD-2010-100564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reddy P. H., Manczak M., and Kandimalla R.. 2017) Mitochondria-targeted small molecule SS31: a potential candidate for the treatment of Alzheimer's disease. Hum. Mol. Genet. 26, 1483–1496 10.1093/hmg/ddx052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dahlberg M. 2007) Polymorphic phase behavior of cardiolipin derivatives studied by coarse-grained molecular dynamics. J. Phys. Chem. B 111, 7194–7200 10.1021/jp071954f [DOI] [PubMed] [Google Scholar]
- 64. Khalifat N., Fournier J. B., Angelova M. I., and Puff N.. 2011) Lipid packing variations induced by pH in cardiolipin-containing bilayers: the driving force for the cristae-like shape instability. Biochim. Biophys. Acta 1808, 2724–2733 10.1016/j.bbamem.2011.07.013 [DOI] [PubMed] [Google Scholar]
- 65. Lewis R. N., and McElhaney R. N.. 2000) Surface charge markedly attenuates the nonlamellar phase-forming propensities of lipid bilayer membranes: calorimetric and 31P-nuclear magnetic resonance studies of mixtures of cationic, anionic, and zwitterionic lipids. Biophys. J. 79, 1455–1464 10.1016/S0006-3495(00)76397-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu Y., Phoon C. K., Berno B., D'Souza K., Hoedt E., Zhang G., Neubert T. A., Epand R. M., Ren M., and Schlame M.. 2016) Loss of protein association causes cardiolipin degradation in Barth syndrome. Nat. Chem. Biol. 12, 641–647 10.1038/nchembio.2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chavez J. D., Tang X., Campbell M. D., Reyes G., Kramer P. A., Stuppard R., Keller A., Marcinek D. J., and Bruce J. E.. 2020) Mitochondrial protein interaction landscape of SS-31. bioRxiv 10.1101/739128 [DOI] [PMC free article] [PubMed]
- 68. Mayer L. D., Hope M. J., and Cullis P. R.. 1986) Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta 858, 161–168 10.1016/0005-2736(86)90302-0 [DOI] [PubMed] [Google Scholar]
- 69. Stewart J. C. 1980) Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 104, 10–14 10.1016/0003-2697(80)90269-9 [DOI] [PubMed] [Google Scholar]
- 70. Alder N. N., Sutherland J., Buhring A. I., Jensen R. E., and Johnson A. E.. 2008) Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol. Biol. Cell 19, 159–170 10.1091/mbc.e07-07-0669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ladokhin A. S., Jayasinghe S., and White S. H.. 2000) How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal. Biochem. 285, 235–245 10.1006/abio.2000.4773 [DOI] [PubMed] [Google Scholar]
- 72. Krasnowska E. K., Gratton E., and Parasassi T.. 1998) Prodan as a membrane surface fluorescence probe: partitioning between water and phospholipid phases. Biophys. J. 74, 1984–1993 10.1016/S0006-3495(98)77905-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Massiot D., Fayon F., Capron M., King I., Le Calve S., Alonso B., Durand J.-O., Bujoli B., Gan Z., and Hoatson G.. 2002) Modelling one- and two-dimensional solid-state NMR spectra. Magn. Reson. Chem. 40, 70–76 10.1002/mrc.984 [DOI] [Google Scholar]
- 74. Sathappa M., and Alder N. N.. 2016) The ionization properties of cardiolipin and its variants in model bilayers. Biochim. Biophys. Acta 1858, 1362–1372 10.1016/j.bbamem.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Carraro M., Checchetto V., Sartori G., Kucharczyk R., di Rago J. P., Minervini G., Franchin C., Arrigoni G., Giorgio V., Petronilli V., Tosatto S. C. E., Lippe G., Szabó I., and Bernardi P.. 2018) High-conductance channel formation in yeast mitochondria is mediated by F-ATP synthase e and g subunits. Cell Physiol. Biochem. 50, 1840–1855 10.1159/000494864 [DOI] [PubMed] [Google Scholar]
- 76. Calzada E., Avery E., Sam P. N., Modak A., Wang C., McCaffery J. M., Han X., Alder N. N., and Claypool S. M.. 2019) Phosphatidylethanolamine made in the inner mitochondrial membrane is essential for yeast cytochrome bc1 complex function. Nat. Commun. 10, 1432 10.1038/s41467-019-09425-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang M., and Han X.. 2014) Multidimensional mass spectrometry-based shotgun lipidomics. Methods Mol. Biol. 1198, 203–220 10.1007/978-1-4939-1258-2_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Han X., Yang K., and Gross R. W.. 2008) Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom. 22, 2115–2124 10.1002/rcm.3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang M., Wang C., Han R. H., and Han X.. 2016) Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 61, 83–108 10.1016/j.plipres.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang K., Cheng H., Gross R. W., and Han X.. 2009) Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 81, 4356–4368 10.1021/ac900241u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Humphrey W., Dalke A., and Schulten K.. 1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 82. Vanommeslaeghe K., Raman E. P., and MacKerell A. D. Jr.. 2012) Automation of the CHARMM general force field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 10.1021/ci3003649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Brooks B. R., Brooks C. L. 3rd, Mackerell A. D. Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., et al. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wu E. L., Cheng X., Jo S., Rui H., Song K. C., Dávila-Contreras E. M., Qi Y., Lee J., Monje-Galvan V., Venable R. M., Klauda J. B., and Im W.. 2014) CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 10.1002/jcc.23702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B. L., Grubmüller H., and MacKerell A. D. Jr.. 2017) CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 10.1038/nmeth.4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lee J., Cheng X., Swails J. M., Yeom M. S., Eastman P. K., Lemkul J. A., Wei S., Buckner J., Jeong J. C., Qi Y., Jo S., Pande V. S., Case D. A., Brooks C. L. 3rd, MacKerell A. D. Jr., et al. (2016) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 10.1021/acs.jctc.5b00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jo S., Lim J. B., Klauda J. B., and Im W.. 2009) CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 10.1016/j.bpj.2009.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jo S., Kim T., Iyer V. G., and Im W.. 2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- 89. Jo S., Kim T., and Im W.. 2007) Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2, e880 10.1371/journal.pone.0000880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Abraham M. J., Murtola T., Schulz R., Pail S., Smith J. C., Hess B., and Lindahl E.. 2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- 91. Hess B., Kutzner C., van der Spoel D., and Lindahl E.. 2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.