Clostridioides difficile infection is the most common health care-associated infection in the United States with more than 20% patients experiencing symptomatic recurrence. The complex nature of host-bacterium interactions makes it difficult to predict the course of the disease based solely on clinical parameters. In the present study, we built a robust prediction model using representative plasma biomarkers and clinical parameters for 90-day all-cause mortality. Risk prediction based on immune biomarkers and clinical variables may contribute to treatment selection for patients as well as provide insight into the role of immune system in C. difficile pathogenesis.
KEYWORDS: Clostridium difficile, Clostridioides, inflammation, mortality, predictive modeling
ABSTRACT
There is a pressing need for biomarker-based models to predict mortality from and recurrence of Clostridioides difficile infection (CDI). Risk stratification would enable targeted interventions such as fecal microbiota transplant, antitoxin antibodies, and colectomy for those at highest risk. Because severity of CDI is associated with the immune response, we immune profiled patients at the time of diagnosis. The levels of 17 cytokines in plasma were measured in 341 CDI inpatients. The primary outcome of interest was 90-day mortality. Increased tumor necrosis factor alpha (TNF-α), interleukin 6 (IL-6), C-C motif chemokine ligand 5 (CCL-5), suppression of tumorigenicity 2 receptor (sST-2), IL-8, and IL-15 predicted mortality by univariate analysis. After adjusting for demographics and clinical characteristics, the mortality risk (as indicated by the hazard ratio [HR]) was higher for patients in the top 25th percentile for TNF-α (HR = 8.35, P = 0.005) and IL-8 (HR = 4.45, P = 0.01) and lower for CCL-5 (HR = 0.18, P ≤ 0.008). A logistic regression risk prediction model was developed and had an area under the receiver operating characteristic curve (AUC) of 0.91 for 90-day mortality and 0.77 for 90-day recurrence. While limited by being single site and retrospective, our work resulted in a model with a substantially greater predictive ability than white blood cell count. In conclusion, immune profiling demonstrated differences between patients in their response to CDI, offering the promise for precision medicine individualized treatment.
INTRODUCTION
Clostridioides difficile infection (CDI) has emerged as a leading cause of nosocomial diarrhea and an important public health threat. CDI causes an estimated half million infections and at least 13,000 deaths annually (1, 2). Health care costs associated with CDI management have been estimated to be around $40 billion per year in the United States (3, 4). Thus, the development of novel approaches to treat and prevent CDI is essential.
Although a major risk factor for CDI is antibiotic-associated dysbiosis, other factors, including the use of gastric acid-suppressing agents, nonsteroidal anti-inflammatory drugs, chemotherapy, inflammatory bowel disease, and prolonged hospital stay, are shown to play a role (5). Clinical manifestations associated with CDI range from asymptomatic colonization and mild diarrhea to toxic megacolon and life-threatening fulminant colitis (6). Toxins A and B are major virulence factors of C. difficile that disrupt the cytoskeletal structure and tight junctions of target cells, leading to cell rounding and death. The emergence of a hypervirulent strain, BI/NAP1/027, has altered traditional epidemiology. This strain is capable of producing a binary toxin (C. difficile toxin [CDT]) in addition to toxins A and B and has been implicated in C. difficile outbreaks associated with increased morbidity and mortality since the early 2000s (7–10).
Recent mouse and human studies have shown that C. difficile toxins as well as pathogen-associated molecular patterns (PAMPs) elicit a multifaceted immune response that can impact the disease outcome (11–14). For example, early recruitment of neutrophils and eosinophils (15–17), gamma interferon (IFN-γ)-producing type 1 innate lymphoid cells (18), leptin (19), and microbiota-dependent interleukin 33 (IL-33) (20) were associated with protection. On the other hand, IL-23 (21, 22), IL-17A (23, 24), Toll-like receptor 2-mediated signaling (16), chemokine (C-X-C motif) ligand 5 (CXCL-5) (25), IL-6 (23), IL-8 and C-C motif chemokine ligand 5 (CCL5) (26) were associated with unfavorable outcome during CDI.
A picture is therefore emerging that the immune response to C. difficile is a predominant factor determining clinical outcome. While a limited number of studies have evaluated systemic biomarkers in humans with CDI (24, 26–28), this is the first study to apply multiple immune biomarkers in a model to predict mortality. Since CDI is a result of an exaggerated host immune response, we hypothesized that systemic cytokine signature during CDI can be modeled to predict survival as well as recurrence.
RESULTS
Population characteristics.
Plasma samples from a total of 341 CDI patients were included in this study. Baseline patient characteristics are described in Table 1. The median age was 63 years, 50.7% patients were female, and the majority of the patients were of European descent. Severe CDI was present in 34% patients based on a peak white blood cell (WBC) count of >15,000/μl (29) within 48 h of CDI diagnosis. Some patients (13%) had received immunosuppressive therapy at some point within a 90-day period preceding CDI diagnosis.
TABLE 1.
Demographics and clinical characteristics of CDI inpatients at the University of Virginia hospitala
| Demographic or clinical characteristic | Value for patients |
||
|---|---|---|---|
| All | Moderate CDI | Severe CDI | |
| No. of patients | 341 | 218 | 123 |
| % Patients | 65.8 | 34.2 | |
| Females (%) | 50.7 | 54.5 | 46.8 |
| Median age (yr) (25th to 75th percentile) |
63 (51.2−72) | 63 (51−72) | 65 (52−73) |
| Race (%) | |||
| Whites | 77.8 | 76 | 79.5 |
| Blacks | 21 | 23 | 18 |
| Others | <1 | <1 | <1 |
| Mean BMI (SD) | 28 (±7.7) | 28 (±8) | 27.7 (±7.4) |
| Median Charlson score (25th to 75th percentile) |
3 (1−7) | 3 (1−7) | 3 (1−7) |
| Mean WBCC (SD) | 13.6 (±8.3) | 8.7 (±3.6) | 22.9 (±6.8) |
| 90-day all-cause death (%) | 13 | 9.6 | 17.2 |
| 30-day all-cause death (%) | 8 | 3.7 | 14.7 |
| % of ICU patients | 30 | 22.3 | 42.3 |
| % receiving immunosuppressive therapy* | 12.8 | 12.8 | 13 |
Abbreviations: CDI, Clostridioides difficile infection; BMI, body mass index; WBCC, white blood cell count; SD, standard deviation; ICU, intensive care unit; *, medical record searched from 90 days prior to 30 days post detection.
Association between plasma cytokine levels and CDI severity.
A total of 341 plasma samples were analyzed (actual sample size numbers for each cytokine varied due to missing values). As shown in Table 2, plasma levels of seven analytes were higher in the severe CDI group (WBC count > 15,000/μl). These analytes included five proinflammatory cytokines: MIF (macrophage migration inhibitory factor) (P < 0.0001), IL-6 (P < 0.0001), IL-1β (P = 0.004), IL-16 (P = 0.01) and IL-15 (P = 0.04). HGF (hepatocyte growth factor) (P < 0.0001) and type 2 cytokine IL-4 (P = 0.03) were also significantly elevated. Ninety-day mortality was associated with elevated IL-6, IL-15, sST-2 (suppression of tumorigenicity 2 receptor), IL-8 and TNF-α (tumor necrosis factor alpha) and decreased CCL-5, CCL-4 and EGF (epidermal growth factor) (see Table S1 in the supplemental material).
TABLE 2.
Plasma cytokine levels in moderate versus severe CDI patientsa
| Biomarker | Moderate CDI (WBCC ≤ 15 × 109/liter) |
Severe CDI (WBCC > 15 × 109/liter) |
P value |
|---|---|---|---|
| HGF | 252.6 (3,155.7−513.9) | 584.1 (286.8−1,166) | <0.0001 |
| MIF | 12,055 (5,363−25,898) | 23,935 (9,642−42,371) | <0.0001 |
| IL-6 | 7.73 (2.72−22.67) | 16.60 (5.94−35.96) | <0.0001 |
| IL-1β | 1.43 (1.07−2.78) | 2.46 (1.07−5.58) | 0.004 |
| IL-16 | 674.5 (394.8−1,177) | 941.1 (516.5−1,315) | 0.01 |
| IL-4 | 16.5 (14.6−62.7) | 29.21 (14.6−58.05) | 0.03 |
| IL-15 | 2.21 (1.41−3.39) | 2.6 (1.67−3.85) | 0.04 |
| EGF | 171.7 (94.5−272) | 190.3 (110.3−329) | 0.06 |
| sST-2 | 169,458 (41,260−488,414) | 207,281 (71,536−636,327) | 0.057 |
| IL-23 | 4.88 (4.88−14.7) | 4.88 (4.88−21.78) | 0.20 |
| CCL-4 | 2,223 (1,686−2,698) | 2,036 (1,729−2,677) | 0.54 |
| IL-8 | 69.36 (44.21−117.5) | 82.66 (47.22−140.8) | 0.23 |
| TNF-α | 7.36 (4.61−11.84) | 7.28 (4.71−13.7) | 0.81 |
| IL-17A | 0.32 (0.12−0.79) | 0.35 (0.08−1.17) | 0.87 |
| IL-10 | 4.8 (3−5.4) | 4.68 (2.99−7.67) | 0.56 |
| Eotaxin | 529.2 (324.3−819.5) | 500.5 (326.8−720.3) | 0.30 |
| CCL-5 | 35,807 (23,050−50,219) | 33,487 (18,726−55,342) | 0.68 |
Patients were stratified into moderate or severe (WBC count of >15,000 per ml) CDI groups based on the Infectious Diseases Society of America (IDSA) recommendations. Plasma biomarker levels are expressed as medians (25th to 75th percentiles). Values for undetectable samples were set to the lowest standard value for the respective target. Statistical significance was calculated by the Mann-Whitney test. The sample numbers (n) for biomarkers were as follows: n = 345 for HGF, MIF, IL-4, EGF, IL-16, IL-10 and eotaxin; n = 326 for IL-6, IL-15 and TNF-α; n = 333 for IL-1β; n = 326 for sST-2; n = 302 for CCL-4, IL-8 and CCL-5; n = 283 for IL-17A and IL-23.
Descriptive statistics of biomarkers by mortality at 90 days. Plasma cytokine levels (in picograms per milliliter) were compared and are shown as medians (25th to 75th percentiles). Values for undetectable samples were set to the lowest standard value for the respective target. Statistical significance was calculated using Mann-Whitney test. Download Table S1, PDF file, 0.2 MB (195.8KB, pdf) .
Copyright © 2020 Abhyankar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predictive biomarkers of survival.
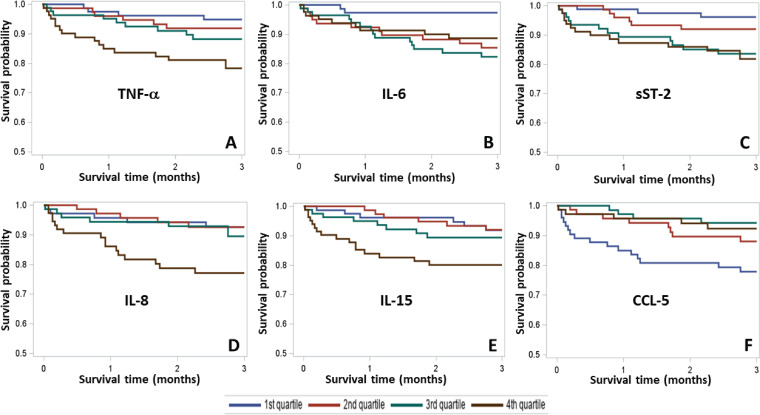
Higher levels of TNF-α (fourth quartile, P = 0.006), IL-6 (second, third, and fourth quartiles, P = 0.03), sST-2 (third and fourth quartiles, P = 0.01), IL-8 (fourth quartile, P = 0.009) and IL-15 (fourth quartile, P = 0.03) were indicative of an increased risk for mortality (Fig. 1A to E). In contrast, higher levels of chemokine CCL-5 (second, third, and fourth quartiles, P = 0.008) were associated with better survival (Fig. 1F). Multivariable Cox regression analysis indicated clinical parameters, including WBC count of >15,000/μl (hazard ratio [HR] = 2.13, P = 0.04) and age (HR = 1.03, P = 0.008) to be associated with 90-day all-cause mortality. After adjustment for age and WBC count, patients in the highest quartile for TNF-α were 8.3 times more likely to die (P = 0.005), and patients in the highest quartile for IL-8 were 4.4 times more likely to die (P = 0.01) (Table 3). In contrast, patients in the highest quartile for CCL-5 were 5.5 times more likely to survive (P = 0.001) (Table 3).
FIG 1.
Kaplan-Meier survival curves for biomarker quartiles. Patients were divided into lower quartile (blue), second quartile (red), third quartile (green), and top quartile (brown) for comparison based on the levels of biomarkers in plasma. The relationship of biomarker quartiles with 90-day survival time is shown. (A) TNF-α, (B) IL-6, (C) sST-2, (D) IL-8, (E) IL-15, and (F) CCL-5. The y axes represent survival probability. Abbreviations: TNF-α, tumor necrosis factor alpha; IL-6, interleukin 6; CCL-5, C-C motif chemokine ligand 5; sST-2, suppression of tumorigenicity 2 receptor.
TABLE 3.
TNF-α, IL-8 and CCL-5 as independent predictors of 90-day survival in a Cox regression modela
| Biomarker | Quartile | Hazard ratio (95% CI)b |
P value |
|---|---|---|---|
| TNF-α | 1st (reference) | ||
| 2nd | 3.06 (0.57−16.28) | 0.18 | |
| 3rd | 4.52 (0.93−21.87) | 0.06 | |
| 4th | 8.35 (1.86−37.5) | 0.005 | |
| IL-8 | 1st (reference) | ||
| 2nd | 1.29 (0.30−5.43) | 0.72 | |
| 3rd | 1.55 (0.41−5.80) | 0.51 | |
| 4th | 4.45 (1.38−14.34) | 0.01 | |
| CCL-5 | 1st (reference) | ||
| 2nd | 0.52 (0.21−1.28) | 0.15 | |
| 3rd | 0.18 (0.05−0.64) | 0.008 | |
| 4th | 0.18 (0.06−0.52) | 0.001 | |
CDI patients were divided into quartiles based on the plasma levels of biomarkers. A Cox proportional hazard model was used to adjust for clinical variables, including age and WBC count at the time of diagnosis.
The hazard ratio represents the factor by which the hazard changes for each one-unit increase of the cytokine expression. 95% CI, 95% confidence interval, or the upper and lower limits of the confidence interval with a significance level of 0.05.
Prediction performance of biomarker-based survival and recurrence models.
We employed receiver operating characteristic (ROC) curve analysis to assess prediction performance of biomarkers for the mortality and recurrence at 3 months. A basic model without biomarkers comprising age and WBC count showed modest ability to predict 90-day survival (area under the receiver operating characteristic curve [AUC] = 0.69) (Fig. 2A). Inclusion of three independent predictors of survival TNF-α, IL-8 and CCL-5 significantly improved predictive capacity over the basic model (AUC increased to 0.83). Inclusion of all six biomarkers identified through univariate and multivariable analysis (TNF-α, IL-8, CCL-5, IL-6, IL-15 and sST-2) further improved the AUC value to 0.86 (Fig. 2A). However, such improvement was not statistically significant. Further, inclusion of immunosuppression as an additional predictor did not have any influence on the AUC value. PCR cycle threshold (Ct) value was not an independent predictor of survival in the present cohort but improved performance of the model when included (AUC = 0.91) (Fig. 2B). Additionally, although none of these biomarkers independently predicted 90-day recurrence, collectively, they gave an AUC value of 0.77 for 90-day recurrence upon integration with the basic model (see Fig. S1 in the supplemental material).
FIG 2.
ROC models to predict 3-month survival. Receiver operating characteristic curve analysis was performed using clinical variables and biomarkers most associated with 3-month survival. The basic model comprised of age +plus WBC count. (A) ROC curves of biomarkers associated with survival (n = 265). The final model consisted of the basic model + TNF-α + IL-8 + CCL5 + IL-6 + sST2 + IL-15. (B) ROC curves of biomarkers in combination with PCR cycle threshold (Ct) data (n = 207). The final model consisted of the basic model + Ct + TNF-α + IL-8 + CCL5 + IL-6 + sST2 + IL-15.
ROC models to predict 3-month recurrence. Receiver operating characteristic curve analysis was performed (n = 245). The basic model comprises of age + gender + WBC count. The final model consists of the basic model + Charlson index + TNF-α + IL-8 + CCL5 + IL-6 + sST2 + IL-15. Download FIG S1, PDF file, 0.1 MB (81.3KB, pdf) .
Copyright © 2020 Abhyankar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
We investigated association of systemic biomarkers and readily available clinical parameters with survival during CDI. The most important finding from this work was identification of an immune signature that, in combination with basic clinical variables, substantially improved survival prediction. Unadjusted analysis identified several biomarkers (HGF, MIF, IL-4, IL-1β, IL-6, IL-15, and IL-16) that were elevated in severe CDI. Unadjusted analysis of biomarkers between dead and surviving patients identified IL-15, sST-2, IL-8, TNF-α, and IL-6 to be associated with 90-day mortality, whereas CCL-5, EGF, and CCL-4 to be associated with survival. Kaplan-Meier survival analysis showed an association of increased TNF-α, IL-8, IL-6, IL-15, and sST-2 and decreased CCL-5 with all-cause mortality. Cox regression further confirmed these associations for TNF-α, IL-8, and CCL-5. Finally, the biomarker-based model showed excellent predictability for 90-day survival.
Severe CDI was present in 34% patients based on a WBC count of >15,000/μl. A diverse set of circulating biomarkers showed upregulation during severe CDI. These biomarkers included HGF, which is primarily secreted by fibroblasts and plays a major role in wound healing and tissue regeneration (30). Increased levels of IL-4 might also be due to its anti-inflammatory effects. IL-1β was also increased in patients with severe CDI. We had previously reported an increase in IL-1β levels for CDI patients compared to outpatients (16). In a recent study, IL-1β was seen upregulated by toxins A and B in vitro, and it was elevated in CDI patients compared to healthy controls (24). Increased serum levels of HGF and IL-1β have also been previously reported in CDI patients compared to outpatients (26). Perhaps most importantly, we have shown in the mouse model of CDI that the IL-1β/Th17 axis plays a major role in promoting inflammatory damage (23).
MIF, IL-15, and IL-16 are pleiotropic cytokines and were seen elevated in severe CDI in this study. These cytokines predominantly support inflammatory immune response. MIF promotes secretion of inflammatory mediators leading to severe pathology (31). Recently, it was shown that MIF levels were increased after CDI both in humans and mice and that neutralization of MIF could protect mice (32). It was suggested that MIF induced a type 17 response to exaggerate CDI, and our results are consistent with this hypothesis. IL-15 is mainly produced by macrophages as well as nonlymphoid cells (33). Monocytes, macrophages, and dendritic cells are the primary targets which in response to IL-15 secrete proinflammatory cytokines, including IL-6, IL-8, and TNF-α (32). IL-15 has been reported to be upregulated in the inflamed tissue from patients with inflammatory bowel disease (IBD) and celiac disease (34). Studies from other laboratories have shown higher levels of IL-15 in CDI patients than in outpatient controls (26) and in patients with severe CDI (24). IL-16 is produced by lymphocytes as well as some epithelial cells and is a chemoattractant for CD4-expressing cells, including T cells, monocytes, dendritic cells, and eosinophils (35). IL-16 was elevated in CDI patients than in healthy donors (24) and was also elevated in patients with Crohn’s disease and ulcerative colitis (36). Thus, MIF, IL-15, and IL-16 may contribute to an exaggerated type 17 response.
In line with our previous studies showing association of IL-6 with mortality (23), higher IL-6 levels correlated with disease severity in this study. Additionally, when patients were divided into quartiles based on IL-6 levels, patients with higher IL-6 (second, third, and fourth quartiles) showed significantly reduced survival probability. Previous reports have shown that serum IL-6 levels were 40-fold higher in CDI patients than in healthy controls (24), and IL-6 was also seen to be associated with severity, although the number of patients analyzed was modest (n = 8) (26). We hypothesize that IL-6 might worsen prognosis of CDI because of its ability to promote Th17 cell differentiation.
Overall, there was a substantial increase in proinflammatory cytokines during severe CDI, and many of these, including MIF, IL-1β, and IL-6 were type 17 promoting cytokines. We and others have previously shown that the Th17 axis plays a central role during CDI pathogenesis. The present study confirms these observations and reiterates the Th17 axis as a target for intervention.
In addition to IL-6, five other biomarkers, sST-2, IL-8, IL-15, TNF-α, and CCL-5, showed association with mortality following Kaplan-Meier analysis. Similar to IL-6, higher levels of either sST-2 or TNF-α showed association with an increased mortality. We have previously shown that increased sST-2 levels predicted CDI-associated mortality likely via dysregulation of protective IL-33 signaling (20). In a recent study, systemic TNF-α levels were higher in CDI patients than in healthy donors (24), but TNF-α has not been studied extensively in the context of CDI. In the present study, higher levels of this cytokine (fourth quartile) were indicative of decreased survival by Kaplan-Meier as well as Cox regression analysis. TNF-α is a well-known player in inflammation. C. difficile toxins have been shown to induce TNF-α in vitro (37).
IL-8 is a CXC family inflammatory chemokine and the principal human chemoattractant for neutrophils. IL-8 was shown to be secreted by monocytes in vitro upon exposure to toxin A (24, 38). A common single nucleotide polymorphism (SNP) in the IL-8 gene promoter was an independent predictor of recurrent CDI (39). IL-8 was also one of the most upregulated biomarkers in CDI patients compared to healthy controls (24), and fecal IL-8 was seen upregulated in severe CDI patients (40). Neutrophils are the primary cells that respond to C. difficile invasion, and neutrophilic inflammation is the hallmark of C. difficile-associated disease. Thus, IL-8 and TNF-α seem to be the robust biomarkers of mortality as identified through univariate and multivariate analyses.
In the present study, EGF, CCL-4, and CCL-5 showed association with protection in univariate analysis. EGF has been reported to exhibit a cytoprotective effect on gastrointestinal epithelia via a receptor-mediated mechanism (41) and also showed inverse association with CDI (26). Similar to EGF, CCL-4 (macrophage inflammatory protein 1β [MIP1β]) showed an inverse correlation with CDI severity and is a chemoattractant for NK cells, monocytes, and other immune cells (26).
CCL-5 (also known as RANTES [42, 43]) was the only biomarker associated with protection in adjusted analysis. Higher levels (second, third, and fourth quartiles) were prosurvival according to Kaplan-Meier analysis. Similarly, patients with higher levels (third and fourth quartiles) were seen protected according to Cox regression analysis. Rao et al. identified CCL-5 as the only biomarker that differentiated between CDI cases and inpatient diarrheal controls and suggested that it was an important mediator of acute intestinal inflammation (26). CCL-5 is expressed by many immune as well as nonimmune cells and actively recruits leukocytes to the inflammatory sites (36, 37). The mechanism of CCL-5-mediated disease modulation remains unknown, and this chemokine warrants further study.
Kaplan-Meier survival analysis did not show association of absolute eosinophil counts (n = 338), body mass index (BMI) (n = 338), or inflammatory bowel disease status (n = 19) with 90-day survival (data not shown). However, patients in the top quartile for absolute neutrophil counts showed decreased probability of survival (P = 0.002; data not shown). Eosinophil count at admission was shown to be an independent predictor of mortality (44), and this association was not seen in the present study. Compared to the Kulaylat et al. study (44) which included two centers (n = 2,065), the present single-center study had a smaller sample size and looked at survival for the first 90 days after diagnosis, and not in-hospital mortality.
A basic prediction model comprised of age and white blood cell count showed poor prediction ability for survival (AUC = 0.69). Integration of all six biomarkers with the basic model substantially improved the survival prediction capacity of the model (AUC = 0.86). The Charlson comorbidity index did not have any significant impact on survival prediction (data not shown). Inclusion of toxin B PCR Ct values in the survival model comprising biomarkers and clinical parameters further improved the prediction potential (AUC = 0.91).
Currently, there is no biomarker to predict CDI recurrence. Interestingly, when data from all the patients irrespective of their prior CDI history were included in the analysis, CCL-5 was the sole biomarker that showed a strong trend (P = 0.07) toward association with 90-day recurrence.
Models based solely on detection of stool toxins A/B (10) or microbiome composition (45) were not able to predict severe CDI or recurrence. Peripheral eosinopenia (44) was identified as an independent predictor of mortality, as was infection with ribotype 027 strain (46). Therefore, an advance from this work is the demonstration that immune profiling adds value to the prediction of mortality in patients with CDI.
This study has several limitations that should be addressed, most importantly that it is retrospective and single site and requires prospective validation before implementation. The study was limited by a lack of information regarding recent history of antibiotic usage, serum albumin levels, toxin status of the infecting strain, microbiome composition, and other chronic inflammatory or major fatal health conditions that could affect systemic cytokine levels. We believe that this information would all be important to include in a prospective evaluation of our model. The study also has significant strengths, most notably being the first to our knowledge to use immune biomarkers to risk stratify patients with CDI for death.
Cytokine-targeted therapies have transformed the treatment of chronic inflammatory diseases, including IBD, providing control of symptoms and longer relapse-free periods (47). We have shown protection from death in murine models by depletion of Th17 cells and by administration of IL-33 (20). The development of predictive models of CDI mortality is an important and necessary first step toward immune therapy of CDI.
MATERIALS AND METHODS
Patient population and clinical samples.
CDI patients were retrospectively identified from the University of Virginia (UVA) Medical Center’s electronic database between 2013 to 2016. UVA Medical Center at the time used the Xpert C. difficile PCR (Cepheid, CA, USA) alone for CDI diagnosis. Clinical data were matched to discarded plasma and stool samples that were banked within 48 h of C. difficile testing by the microbiology laboratory. This study evaluated patients with newly diagnosed CDI; patients with a history of CDI within 90 days were excluded. Demographics (age, gender, and race) and clinical characteristics, such as white blood cell count and Charlson comorbidity index, were extracted from the electronic health record database. An immunosuppressant pharmacy grouper was used to identify receipt of immunosuppressants or systemic corticosteroids. Archived and available PCR cycle threshold (Ct) values were collected from the Xpert PCR machine. After initial storage at 4°C for a maximum of 24 h, samples were stored at −80°C until testing. The collection of patient data was approved by the institutional review board (protocol IRB-HSR 16926). This project met the criteria of research involving coded private information or biological specimens.
Measurement of cytokines in plasma samples.
The concentrations of various target analytes in patient plasma samples were measured on a Bio-Plex 200 suspension array system (Bio-Rad, Hercules, CA, USA) using R&D Systems custom Luminex assays (R & D Systems, Minneapolis, MN). Some analytes, including sST-2 (catalog no. DST 200; R&D Systems), IL-23 (catalog no. D2300B; R&D Systems), and IL-17A (catalog no. HS170; R&D Systems), were measured by using an enzyme-linked immunosorbent assay (ELISA). The analytes that did not give satisfactory signal included IFN-γ, IL-22, IL-13, IL-5, IL-2, and IL-33. All analytes are expressed as picograms per milliliter.
Outcomes.
The outcomes of interest in this study were 90-day survival and recurrence. Patients were analyzed from the initial CDI diagnosis to death for the outcomes of interest for 90 days or the last time of follow-up, whichever occurred earlier. Due to the small number of events, no further analyses were performed for these outcomes at 30 days.
Statistical analysis.
Our primary objective was to evaluate effects and predictability of elevated biomarkers on mortality and CDI recurrence. Cytokines in moderate versus severe CDI patient groups were assessed with the Mann-Whitney U test. In survival analysis, patients were categorized into quartiles based on the plasma cytokine levels due to skewed distributions of these biomarkers. Univariate biomarker effect was analyzed using the Kaplan-Meier method, where survival probabilities were estimated and survival differences in the quartiles of each biomarker were evaluated by the log rank test. Those biomarkers with a log rank P value of <0.1 were further analyzed using Cox regression, and the final model was determined with stepwise selection for joint effects of these biomarkers on the outcomes. Cox regression analyses were adjusted for demographics and clinical characteristics.
The predictability of biomarkers for the outcomes were estimated using the area under the receiver operating characteristic (ROC) curve. A receiver operating characteristic curve plots the true positive rate (sensitivity) against the false-positive rate (1 – specificity) for all possible cutoff values to predict a dichotomous outcome. In this study, ROC curves were constructed at possible values of demographics, comorbidities, and biomarkers to predict the 90-day mortality and recurrence using logistic regression, and the areas under the ROC curves (AUC) were used to measure predictabilities. Further immunosuppression was also included in evaluating the predictability. A perfect model would have an AUC of 1. The higher the AUC, the better the model is at distinguishing between patients who had the event and those who did not at 90 days. Usually, the optimal cutoff points would be the values of predictors that correspond to the highest sensitivity and lowest specificity. Differences in ROC curves under different models were tested using the empirical method available in SAS (Proc Logistic). Values of P < 0.05 were considered statistically significant. The descriptive statistics were analyzed using GraphPad Prism, and survival analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01 AI124214-04) to W.A.P.
W. A. Petri is a consultant for TechLab Inc., a company that makes diagnostic tests for C. difficile toxins. All of the other authors declare no conflict of interest.
Footnotes
This article is a direct contribution from William A. Petri, Jr., a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Wes Vanvoorhis, University of Washington, and David Aronoff, Vanderbilt.
Citation Abhyankar MM, Ma JZ, Scully KW, Nafziger AJ, Frisbee AL, Saleh MM, Madden GR, Hays AR, Poulter M, Petri WA, Jr. 2020. Immune profiling to predict outcome of Clostridioides difficile infection. mBio 11:e00905-20. https://doi.org/10.1128/mBio.00905-20.
REFERENCES
- 1.Lessa FC, Gould CV, McDonald LC. 2012. Current status of Clostridium difficile infection epidemiology. Clin Infect Dis 55(Suppl 2):S65–S70. doi: 10.1093/cid/cis319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2020. Clostridioides difficile fact sheet. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/drugresistance/pdf/threats-report/clostridioides-difficile-508.pdf. Accessed 13 March 2020.
- 3.Kassam Z, Cribb Fabersunne C, Smith MB, Alm EJ, Kaplan GG, Nguyen GC, Ananthakrishnan AN. 2016. Clostridium difficile associated risk of death score (CARDS): a novel severity score to predict mortality among hospitalized patients with Clostridium difficile infection. Aliment Pharmacol Ther 43:725–733. doi: 10.1111/apt.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopkins RJ, Wilson RB. 2018. Treatment of recurrent Clostridium difficile colitis: a narrative review. Gastroenterol Rep (Oxf) 6:21–28. doi: 10.1093/gastro/gox041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hookman P, Barkin JS. 2009. Clostridium difficile associated infection, diarrhea and colitis. World J Gastroenterol 15:1554–1580. doi: 10.3748/wjg.15.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leffler DA, Lamont JT. 2015. Clostridium difficile infection. N Engl J Med 373:287–288. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 7.Gerding DN, Johnson S, Rupnik M, Aktories K. 2014. Clostridium difficile binary toxin CDT. Gut Microbes 5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.See I, Mu Y, Cohen J, Beldavs ZG, Winston LG, Dumyati G, Holzbauer S, Dunn J, Farley MM, Lyons C, Johnston H, Phipps E, Perlmutter R, Anderson L, Gerding DN, Lessa FC. 2014. NAP1 strain type predicts outcomes from Clostridium difficile infection. Clin Infect Dis 58:1394–1400. doi: 10.1093/cid/ciu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao K, Micic D, Natarajan M, Winters S, Kiel MJ, Walk ST, Santhosh K, Mogle JA, Galecki AT, LeBar W, Higgins PDR, Young VB, Aronoff DM. 2015. Clostridium difficile ribotype 027: relationship to age, detectability of toxins A or B in stool with rapid testing, severe infection, and mortality. Clin Infect Dis 61:233–241. doi: 10.1093/cid/civ254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JY, Kim H, Cha MY, Park HG, Kim Y-J, Kim IY, Kim JM. 2009. Clostridium difficile toxin A promotes dendritic cell maturation and chemokine CXCL2 expression through p38, IKK, and the NF-kappaB signaling pathway. J Mol Med (Berl) 87:169–180. doi: 10.1007/s00109-008-0415-2. [DOI] [PubMed] [Google Scholar]
- 12.Papatheodorou P, Carette JE, Bell GW, Schwan C, Guttenberg G, Brummelkamp TR, Aktories K. 2011. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT). Proc Natl Acad Sci U S A 108:16422–16427. doi: 10.1073/pnas.1109772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jefferson KK, Smith MF, Bobak DA. 1999. Roles of intracellular calcium and NF-kappa B in the Clostridium difficile toxin A-induced up-regulation and secretion of IL-8 from human monocytes. J Immunol 163:5183–5191. [PubMed] [Google Scholar]
- 14.Hemmasi S, Czulkies BA, Schorch B, Veit A, Aktories K, Papatheodorou P. 2015. Interaction of the Clostridium difficile binary toxin CDT and its host cell receptor, lipolysis-stimulated lipoprotein receptor (LSR). J Biol Chem 290:14031–14044. doi: 10.1074/jbc.M115.650523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarchum I, Liu M, Shi C, Equinda M, Pamer EG. 2012. Critical role for MyD88-mediated neutrophil recruitment during Clostridium difficile colitis. Infect Immun 80:2989–2996. doi: 10.1128/IAI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, Kuehne SA, Schwan C, Eichhoff AM, Koch-Nolte F, Lyras D, Aktories K, Minton NP, Petri WA. 2016. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol 1:16108. doi: 10.1038/nmicrobiol.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonomo EL, Cowardin CA, Wilson MG, Saleh MM, Pramoonjago P, Petri WA. 2016. Microbiota-regulated IL-25 increases eosinophil number to provide protection during Clostridium difficile infection. Cell Rep 16:432–443. doi: 10.1016/j.celrep.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Sušac B, Ling L, Leiner I, Pamer EG. 2015. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madan R, Guo X, Naylor C, Buonomo EL, Mackay D, Noor Z, Concannon P, Scully KW, Pramoonjago P, Kolling GL, Warren CA, Duggal P, Petri WA Jr.. 2014. Role of leptin-mediated colonic inflammation in defense against Clostridium difficile colitis. Infect Immun 82:341–349. doi: 10.1128/IAI.00972-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frisbee AL, Saleh MM, Young MK, Leslie JL, Simpson ME, Abhyankar MM, Cowardin CA, Ma JZ, Pramoonjago P, Turner SD, Liou AP, Buonomo EL, Petri WA Jr.. 2019. IL-33 drives group 2 innate lymphoid cell-mediated protection during Clostridium difficile infection. Nat Commun 10:2712. doi: 10.1038/s41467-019-10733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buonomo EL, Madan R, Pramoonjago P, Li L, Okusa MD, Petri WA. 2013. Role of interleukin 23 signaling in Clostridium difficile colitis. J Infect Dis 208:917–920. doi: 10.1093/infdis/jit277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDermott AJ, Falkowski NR, McDonald RA, Pandit CR, Young VB, Huffnagle GB. 2016. Interleukin-23 (IL-23), independent of IL-17 and IL-22, drives neutrophil recruitment and innate inflammation during Clostridium difficile colitis in mice. Immunology 147:114–124. doi: 10.1111/imm.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saleh MM, Frisbee AL, Leslie JL, Buonomo EL, Cowardin CA, Ma JZ, Simpson ME, Scully KW, Abhyankar MM, Petri WA. 2019. Colitis-induced Th17 cells increase the risk for severe subsequent Clostridium difficile infection. Cell Host Microbe 25:756–765.e5. doi: 10.1016/j.chom.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Chen K, Sun Y, Carter M, Garey KW, Savidge TC, Devaraj S, Tessier ME, von Rosenvinge EC, Kelly CP, Pasetti MF, Feng H. 2017. Cytokines are markers of the Clostridium difficile-induced inflammatory response and predict disease severity. Clin Vaccine Immunol 24:e00037-17. doi: 10.1128/CVI.00037-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Feghaly RE, Stauber JL, Tarr PI, Haslam DB. 2013. Intestinal inflammatory biomarkers and outcome in pediatric Clostridium difficile infections. J Pediatr 163:1697–1704.e2. doi: 10.1016/j.jpeds.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao K, Erb-Downward JR, Walk ST, Micic D, Falkowski N, Santhosh K, Mogle JA, Ring C, Young VB, Huffnagle GB, Aronoff DM. 2014. The systemic inflammatory response to Clostridium difficile infection. PLoS One 9:e92578. doi: 10.1371/journal.pone.0092578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limsrivilai J, Rao K, Stidham RW, Govani SM, Waljee AK, Reinink A, Johnson L, Briggs E, Higgins PDR. 2018. Systemic inflammatory responses in ulcerative colitis patients and Clostridium difficile infection. Dig Dis Sci 63:1801–1810. doi: 10.1007/s10620-018-5044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czepiel J, Biesiada G, Brzozowski T, Ptak-Belowska A, Perucki W, Birczynska M, Jurczyszyn A, Strzalka M, Targosz A, Garlicki A. 2014. The role of local and systemic cytokines in patients infected with Clostridium difficile. J Physiol Pharmacol 65:695–703. [PubMed] [Google Scholar]
- 29.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH, Society for Healthcare Epidemiology of America, Infectious Diseases Society of America. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto K, Nakamura T. 1996. Emerging multipotent aspects of hepatocyte growth factor. J Biochem 119:591–600. doi: 10.1093/oxfordjournals.jbchem.a021283. [DOI] [PubMed] [Google Scholar]
- 31.Calandra T, Roger T. 2003. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol 3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jose S, Mukherjee A, Abhyankar MM, Leng L, Bucala R, Sharma D, Madan R. 2018. Neutralization of macrophage migration inhibitory factor improves host survival after Clostridium difficile infection. Anaerobe 53:56–63. doi: 10.1016/j.anaerobe.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budagian V, Bulanova E, Paus R, Bulfone-Paus S. 2006. IL-15/IL-15 receptor biology: a guided tour through an expanding universe. Cytokine Growth Factor Rev 17:259–280. doi: 10.1016/j.cytogfr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Pagliari D, Cianci R, Frosali S, Landolfi R, Cammarota G, Newton EE, Pandolfi F. 2013. The role of IL-15 in gastrointestinal diseases: a bridge between innate and adaptive immune response. Cytokine Growth Factor Rev 24:455–466. doi: 10.1016/j.cytogfr.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Cruikshank WW, Center DM, Nisar N, Wu M, Natke B, Theodore AC, Kornfeld H. 1994. Molecular and functional analysis of a lymphocyte chemoattractant factor: association of biologic function with CD4 expression. Proc Natl Acad Sci U S A 91:5109–5113. doi: 10.1073/pnas.91.11.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seegert D, Rosenstiel P, Pfahler H, Pfefferkorn P, Nikolaus S, Schreiber S. 2001. Increased expression of IL-16 in inflammatory bowel disease. Gut 48:326–332. doi: 10.1136/gut.48.3.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun X, He X, Tzipori S, Gerhard R, Feng H. 2009. Essential role of the glucosyltransferase activity in Clostridium difficile toxin-induced secretion of TNF-alpha by macrophages. Microb Pathog 46:298–305. doi: 10.1016/j.micpath.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linevsky JK, Pothoulakis C, Keates S, Warny M, Keates AC, Lamont JT, Kelly CP. 1997. IL-8 release and neutrophil activation by Clostridium difficile toxin-exposed human monocytes. Am J Physiol 273:G1333–G1340. doi: 10.1152/ajpgi.1997.273.6.G1333. [DOI] [PubMed] [Google Scholar]
- 39.Garey KW, Jiang Z-D, Ghantoji S, Tam VH, Arora V, DuPont HL. 2010. A common polymorphism in the interleukin-8 gene promoter is associated with an increased risk for recurrent Clostridium difficile infection. Clin Infect Dis 51:1406–1410. doi: 10.1086/657398. [DOI] [PubMed] [Google Scholar]
- 40.Steiner TS, Flores CA, Pizarro TT, Guerrant RL. 1997. Fecal lactoferrin, interleukin-1beta, and interleukin-8 are elevated in patients with severe Clostridium difficile colitis. Clin Diagn Lab Immunol 4:719–722. doi: 10.1128/CDLI.4.6.719-722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bodnar RJ. 2013. Epidermal growth factor and epidermal growth factor receptor: the yin and yang in the treatment of cutaneous wounds and cancer. Adv Wound Care (New Rochelle) 2:24–29. doi: 10.1089/wound.2011.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Appay V, Rowland-Jones SL. 2001. RANTES: a versatile and controversial chemokine. Trends Immunol 22:83–87. doi: 10.1016/S1471-4906(00)01812-3. [DOI] [PubMed] [Google Scholar]
- 43.Nelson PJ, Pattison JM, Krensky AM. 1997. Gene expression of RANTES. Methods Enzymol 287:148–162. doi: 10.1016/S0076-6879(97)87012-7. [DOI] [PubMed] [Google Scholar]
- 44.Kulaylat AS, Buonomo EL, Scully KW, Hollenbeak CS, Cook H, Petri WA, Stewart DB. 2018. Development and validation of a prediction model for mortality and adverse outcomes among patients with peripheral eosinopenia on admission for Clostridium difficile infection. JAMA Surg 153:1127–1133. doi: 10.1001/jamasurg.2018.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pakpour S, Bhanvadia A, Zhu R, Amarnani A, Gibbons SM, Gurry T, Alm EJ, Martello LA. 2017. Identifying predictive features of Clostridium difficile infection recurrence before, during, and after primary antibiotic treatment. Microbiome 5:148. doi: 10.1186/s40168-017-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao K, Higgins PDR, Young VB. 2018. An observational cohort study of Clostridium difficile ribotype 027 and recurrent infection. mSphere 3:e00033-18. doi: 10.1128/mSphere.00033-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedrich M, Pohin M, Powrie F. 2019. Cytokine networks in the pathophysiology of inflammatory bowel disease. Immunity 50:992–1006. doi: 10.1016/j.immuni.2019.03.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Descriptive statistics of biomarkers by mortality at 90 days. Plasma cytokine levels (in picograms per milliliter) were compared and are shown as medians (25th to 75th percentiles). Values for undetectable samples were set to the lowest standard value for the respective target. Statistical significance was calculated using Mann-Whitney test. Download Table S1, PDF file, 0.2 MB (195.8KB, pdf) .
Copyright © 2020 Abhyankar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ROC models to predict 3-month recurrence. Receiver operating characteristic curve analysis was performed (n = 245). The basic model comprises of age + gender + WBC count. The final model consists of the basic model + Charlson index + TNF-α + IL-8 + CCL5 + IL-6 + sST2 + IL-15. Download FIG S1, PDF file, 0.1 MB (81.3KB, pdf) .
Copyright © 2020 Abhyankar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.