Abstract
For several decades, researchers are working to develop improved major crops with better adaptability and tolerance to environmental stresses. Forage legumes have been widely spread in the world due to their great ecological and economic values. Abiotic and biotic stresses are main factors limiting legume production, however, alfalfa (Medicago sativa L.) shows relatively high level of tolerance to drought and salt stress. Efforts focused on alfalfa improvements have led to the release of cultivars with new traits of agronomic importance such as high yield, better stress tolerance or forage quality. Alfalfa has very high nutritional value due to its efficient symbiotic association with nitrogen-fixing bacteria, while deep root system can help to prevent soil water loss in dry lands. The use of modern biotechnology tools is challenging in alfalfa since full genome, unlike to its close relative barrel medic (Medicago truncatula Gaertn.), was not released yet. Identification, isolation, and improvement of genes involved in abiotic or biotic stress response significantly contributed to the progress of our understanding how crop plants cope with these environmental challenges. In this review, we provide an overview of the progress that has been made in high-throughput sequencing, characterization of genes for abiotic or biotic stress tolerance, gene editing, as well as proteomic and metabolomics techniques bearing biotechnological potential for alfalfa improvement.
Keywords: alfalfa, Medicago sativa, genomics, metabolomics, proteomics, stress resistance genes
Introduction
Legumes are important food crops for the exponentially growing population, owing to their micronutrient, macronutrient, and secondary metabolite content (Le et al., 2007). Some of these organic compounds (e.g., phytoalexins and chitinases) play roles in plant defense against pathogens and pests (He and Dixon, 2000). Moreover, Fabaceae is one of the most studied plant families, and it has gained high agricultural importance, especially owing to its ability to fix nitrogen in symbiosis with rhizobia (Doyle and Luckow, 2003).
Medicago sativa L., commonly known as alfalfa or “lucerne,” belongs to Fabaceae, and its first cultivated form most likely originates from western Persia. It then spread to many regions in Asia, Europe, and America. In addition, Rashmi et al. (1997) and Samac and Temple (2004) reported that alfalfa ranks fourth in terms of acreage and economic value, following corn, soybean, and wheat.
The genus Medicago includes both perennial and annual species. Alfalfa is a highly valuable perennial deep-rooted forage legume, especially because of its widespread production, soil protection, and ability to improve nitrogen-limited soils (Radović et al., 2009). It is also widely cultivated for livestock feed (Flajoulot et al., 2005), and is used as a biofuel feedstock for ethanol production, either as hay or silage (McCoy and Bingham, 1988). The biological and agronomical potential of alfalfa, like all other members of the whole legume family, is extraordinary because it requires little to no nitrogen fertilizer for optimal growth (Ebert, 2007). In addition, alfalfa plays an important role as a free fertilizer providing nitrogen to subsequent crops (Triboi and Triboi-Blondel, 2014).
Alfalfa shows a high content of proteins, enzymes (amylase, coagulase, peroxidase, erepsin, lipase, invertase, and pectinase), antioxidants, minerals, and vitamins A, C, K, and E, as well as valuable phytopharmaceutical components (Bora and Sharma, 2011 and references therein). Moreover, alfalfa and some other species of Fabaceae family possess two different thiol redox compounds, namely glutathione (GSH) and the homoglutathione (hGSH), with higher content of hGSH (Klapheck, 1988; Baldacci-Cresp et al., 2012). More specifically, alfalfa shows different ratios of hGSH/GSH in diverse organs such as leaves, stems, and roots (Pasternak et al., 2014). Thus, alfalfa represents one of the most valuable and important forage crops, and can also be used in grasslands as a cover crop for improved weed control. Finally, alfalfa is also suitable for use in the production of recombinant pharmaceutical proteins (Fu et al., 2015) and in phytoremediation (Nirola et al., 2016).
The tetraploid genome of alfalfa and outbreeding mating systems have made selective breeding harder (Zhou et al., 2011; Annicchiarico et al., 2015). Advanced methods such as genomic, proteomic, and metabolomic approaches, as well as gene editing, could lead to the practical applications of genes that have biotechnological value for alfalfa improvement, especially if applied in an integrated and targeted manner. As a result, single or multiple genes might show desirable effects on several agronomically important alfalfa traits, which can significantly accelerate research in comparison to conventional breeding (Singer et al., 2018). Alfalfa is a major source of proteins in the livestock and dairy industries. In the last years, alfalfa production has been displaced to saline environments by major cereals. Therefore, the incorporation of transgenic traits into alfalfa with varying degrees of tolerance to salinity has been developed and this robust approach can improve the productivity and quality of nitrogen-fixing crops (Kang et al., 2016; Stritzler et al., 2018). Genetically engineered glyphosate-resistant alfalfa was commercialized in the United States in 2010. Another alfalfa variety with reduced lignin content stacked to glyphosate resistance trait has been available since 2015. Reduced lignin content in forage legumes can improve their digestibility by animals, thus it is an important forage quality trait (Li et al., 2016; Barros et al., 2019).
The purpose of this review is to provide a perspective on the current state of alfalfa biotechnology research. It focuses mainly on the biotechnological potential of genomic and transcriptomic approaches, biotechnologically valuable genes, gene editing, proteomics, and metabolomics. When appropriate it is compared to barrel medic.
Genomic Approaches
The identification of genes that affect legume crop production represents an important aim of current genomic studies (Bevan et al., 2017), and this requires knowledge of their full genomic sequences. Technologies for sequencing DNA and RNA have undergone revolutionary improvements (Ari and Arikan, 2016). It is known that after the evolutionary split between monocots and eudicots, several whole genome duplications and triplications had occurred in legumes (Severin et al., 2011; Masonbrink et al., 2017), which might delay whole genome sequencing efforts. The major strength of next-generation sequencing (NGS) is its ability to detect abnormalities across the entire genome. NGS is less costly and has a faster turnaround time compared to classical sequencing methods. New NGS platforms, such as the Roche/454 system (Margulies et al., 2005), Illumina platform (Wang et al., 2012), real-time DNA sequencing by Pacific Biosciences (Eid et al., 2009), Oxford Nanopore system (Lu et al., 2016), and Ion Torrent system (Rothberg et al., 2011), were used for sequencing crop and legume genomes. They have had a major impact on plant research, since they enable the understanding of genomic complexity as well as the identification of genomic variations, such as single nucleotide polymorphisms (SNPs) or insertions/deletions (INDELs; Valliyodan et al., 2017; Abdelrahman et al., 2018). NGS and bioinformatics approaches for high-throughput data analysis are major tools in modern plant breeding programs (Abdelrahman et al., 2015, 2017a,b; Pavlovich, 2017). These modern technologies are also used in legume research, and several recent studies have been devoted to alfalfa genomics using high-throughput genome sequencing (reviewed by Hawkins and Yu, 2018).
High-Throughput NGS in Genomics and Transcriptomics
Genome sequencing and assembly have been applied to many plant species, including crops. Such genome assemblies serve as common references for alignment with re-sequenced plants (Huang et al., 2012; Schreiber et al., 2018). Large-scale systematic genome sequencing has been carried out in leguminous plants such as Lotus japonicus (Sato et al., 2008), M. truncatula (release 3.0)1, and Glycine max (Schmutz et al., 2010). The genome sequence of alfalfa has not yet been published, and current transcriptomic studies and SNP discoveries rely on the barrel medic genome sequence alignment (genome version2 Mt4.0v1; Young et al., 2011; Tang et al., 2014). Currently, the most advanced genome sequencing method is NGS. It has become the major tool for the development of new molecular markers and for gene identification (Edwards and Batley, 2010). Together with the rapid development of NGS, the number of plants with completely sequenced genomes has dramatically increased (Van et al., 2013; Le Nguyen et al., 2018; Kersey, 2019). Advantages of NGS include lower costs and shorter time requirements. The development of NGS technology contributed to the identification of new genes that had evolved by whole-genome duplication and structural variations in chromosomes (Barabaschi et al., 2012; Van et al., 2013). Reference genome sequences of several legume and crop species are now available, and candidate genes of important SNPs can be rapidly and easily identified (Gao et al., 2012; Van et al., 2013; Le Nguyen et al., 2018; Scheben et al., 2019). Alfalfa is an outbred, tetrasomic tetraploid (2n = 4x = 32) with eight basic chromosomes and a genome size of 800–1000 Mbp (Blondon et al., 1994). Genetic and genomic resources have been widely explored and developed, but in the absence of a fully sequenced and assembled reference genome for alfalfa, genome of closely related barrel medic is used as a model organism (Zhou et al., 2011). Barrel medic is a diploid species (2n = 2x = 16) with smaller genome (about 550 Mbp; Piano and Pecetti, 2010). NGS technologies could speed up the discovery of quantitative trait loci (QTLs) and candidate SNPs, which represent common sequence variations among plants and are functionally important. Numerous molecular markers are used in high-throughput genotyping by sequencing (GBS) platforms associated with alfalfa mapping (Hawkins and Yu, 2018), population diversity studies (Herrmann et al., 2018), and genomic selection (Annicchiarico et al., 2016). In the past years, low density linkage maps were constructed on diploid alfalfa (Brummer et al., 1993; Kiss et al., 1993; Echt et al., 1994; Julier et al., 2003). Although several genetic linkage maps have been constructed for tetraploid alfalfa, most of them were framework maps with only few markers (Brouwer and Osborn, 1999; Julier et al., 2003; Musial et al., 2007; Robins et al., 2007; Khu et al., 2013). Li X. et al. (2014) have constructed a saturated genetic linkage map of autotetraploid alfalfa by using GBS. They have shown high synteny between linkage groups of alfalfa and barrel medic, and clearly identified translocations between chromosomes 4 and 8, and small inversion on chromosome 1. The high-density linkage maps contained 3,591 SNP markers on 64 linkage groups across both maternal and paternal genomes of an autotetraploid alfalfa F1 population (Li X. et al., 2014).
Genome-wide associated studies (GWAS) are a modern and powerful strategy that can be used to overcome the limitations of conventional QTL mapping. GWAS map genetic loci in a breeding population, relying on linkage disequilibrium (LD; Liu X. P. et al., 2019). Recently, GWAS have been used in the identification of genetic loci in crop species such as soybean (Hwang et al., 2014), maize (Olukolu et al., 2016), barrel medic (Kang et al., 2015), and alfalfa. Zhang T. et al. (2015) evaluated two important features associated with drought resistance, namely drought resistance index (DRI) and relative leaf water content (RWC) under greenhouse conditions in 198 alfalfa cultivars and landraces. These results were then correlated with genomic data obtained through GBS. Subsequent to the QTL mapping approach, GWAS provided identification of 15 loci associated with DRI and RWC. Markers associated with DRI are located at all chromosomes, whereas markers associated with RWC are located at chromosomes 1, 2, 3, 4, 5, 6, and 7. Co-localization of markers for DRI and WRC were found on chromosomes 3, 5, and 7 (Zhang T. et al., 2015). A GWAS approach using more than 15,000 genome-wide SNPs obtained through GBS was applied to examine forage yield and nutritive value-related traits. Five genes, containing known SNPs aligned to the barrel medic genome, were found as candidates in determining fall dry matter yield (TUBBY-LIKE PROTEIN), summer dry matter yield (E3 SUMO-PROTEIN LIGASE SIZ1, RNA-DEPENDENT RNA POLYMERASE FAMILY PROTEIN), fall stem weight (UBIQUITIN-LIKE-SPECIFIC PROTEASE ESD4-LIKE PROTEIN), and cell wall biogenesis (NUCLEOTIDE-DIPHOSPHO-SUGAR TRANSFERASE FAMILY PROTEIN; Sakiroglu and Brummer, 2017). Aiming to find markers for alfalfa forage quality, 154 plants originating from the second generation prepared by the outcrossing of three alfalfa cultivars were subjected to GBS, while their half-sib progenies were phenotyped for forage quality parameters under three different growing conditions. Subsequently, GWAS of SNPs was carried out using barrel medic as a reference genome, confirming a polygenic control of quality traits and indicating a substantially different genetic control of a given trait in stems and leaves (Biazzi et al., 2017). Important alfalfa loci for salt tolerance during germination were identified by similar marker-trait association using a GWAS approach (Yu et al., 2016). Remarkably, they used 198 different accessions with potential drought tolerance, whereas DNA libraries were sequenced in two lanes of an Illumina Hi-Seq2000 instrument. Identified SNP markers were located on all chromosomes, with the exception of chromosome 3. Several alfalfa loci showed similar genetic locations to the reported QTLs associated with salt tolerance in barrel medic. The results suggest the similarity of mechanisms controlling salt stress responses in these two species. This study resulted in the identification of 14 genes connected to 23 markers associated with salt tolerance during germination. These include PEROXYGENASE, B3 DNA-BINDING PROTEIN, and CPR5 PROTEIN, which are linked to cuticle wax biosynthesis and ABA signaling (Yu et al., 2016).
Over the last two decades, several methods have been developed that allowed the examination of global transcriptional changes. The most used ones are the hybridization of cDNAs (DNA microarrays) and the deep sequencing of cDNA (RNA-Seq; Schena et al., 1995; Wang et al., 2009; Lardi and Pessi, 2018). RNA-Seq, a massive parallel sequencing method for transcriptome analysis, was developed 10 years ago (Wang et al., 2009). Transcriptomic studies analyze only the transcribed portion of the genome and provides in-depth sequencing coverage and additional qualitative information such as isoform-specific expression (Abdelrahman et al., 2018). In contrast to microarrays, ribosomal RNA (rRNA) does not hybridize to the chip, as homologous probes are not present. In RNA-Seq, the abundant rRNA is removed (Lardi and Pessi, 2018). Originally, transcriptomic studies were based on Sanger sequencing of expressed sequence tags (ESTs) or microarrays, which was used in alfalfa and barrel medic (Aziz et al., 2005; Cheung et al., 2006; Yang et al., 2010). It has also been applied for other legumes such as G. max (Le et al., 2012; Ha et al., 2015; Tripathi et al., 2016), L. japonicus (Asamizu et al., 2004), and Cicer arietinum (Deokar et al., 2011).
Several studies contributed to the transcriptome sequencing of alfalfa with various coverage. These studies relied on NGS technologies such as 454 technology (Han et al., 2011) or RNA-Seq (Yang et al., 2011; Li and Brummer, 2012; Liu et al., 2013; O’Rourke et al., 2015). Liu et al. (2013) performed de novo transcriptome sequencing of M. sativa L. subsp. sativa using Illumina paired-end sequencing. Plant material included 15 tissue types, and the transcriptome coverage was 5.64 Gbp of clean nucleotides. About 40,433 unigenes were obtained, and 1649 potential expressed sequence tags simple sequence repeat markers (EST-SSRs) were annotated by alignment with the following databases: the National Center for Biotechnology Information (NCBI) nonredundant protein (Nr) database, the NCBI non-redundant nucleotide sequence (Nt) database, Swiss-Prot, The Kyoto Encyclopedia of Genes and Genomes (KEGG), the Clusters of Orthologous Group (COG), Translated EMBL (TrEMBL), and the InterPro (Ipr) database (Liu et al., 2013). RNA-Seq analysis of two alfalfa subspecies, namely M. sativa ssp. sativa (B47) and M. sativa ssp. falcata (F56) using roots, nitrogen-fixing root nodules, leaves, flowers, elongating stem internodes, and post-elongation stem internodes resulted in 112,626 unique transcript sequences, which were assembled into the alfalfa Gene Index 1.2 (MSGI 1.2; O’Rourke et al., 2015). Chao et al. (2019) used PacBio SMRT technology and identified 72,606 open reading frames (ORFs) including 46,616 full-length ORFs, 1670 transcription factors and 44,040 SSRs. A total of 7568 alternative splicing events and 17,740 long non-coding RNAs supported the feasibility of deep sequencing full length RNA from alfalfa transcriptome on a single-molecule level (Chao et al., 2019). Another approach developed to provide long-read sequencing of transcripts is Oxford Nanopore Technologies®. The MinION device, which was developed by Oxford Nanopore, is a portable apparatus compatible with a PC or laptop (Jain et al., 2016; Lu et al., 2016). Fleming et al. (2018) evaluated changes in mRNA in dry soybean seeds with use of MinION-based pipeline technology. Li et al. (2019) used MinION-based technology for high-throughput mapping of transgenic alleles in soybean. They rapidly mapped the transgene insertion positions in 51 transgenic soybean plants in a single 1D sequencing run. This method was optimized using a population of soybean lines, but it can be adapted to map the transgenes in any other crops.
Transcriptomic Approaches and Gene Expression Modifications
Resistance to Abiotic Stress
Salinity stress interferes with plant growth because it causes two main stresses on plants: hyperosmotic pressure and ion toxicity, especially due to Na+ (Volkov et al., 2004). High salinity often triggers an increase in cytosolic Ca2+, reactive oxygen species (ROS), abscisic acid (ABA), and mitogen activated protein kinase (MAPK) signaling (Ovečka et al., 2014; Mittler and Blumwald, 2015). These activated signal molecules affect plant transcriptomes by regulating transcription factors (Xiong et al., 2002; Zhu, 2002). One of the basic strategies in plant stress responses is the accumulation of water-soluble compounds of low molecular weight, such as betaines, polyols, sugars, and amino acids (Chen and Murata, 2002). These compounds accumulate to high concentrations under water or salt stress and protect plants via ROS detoxification and membrane integrity maintenance (Bohnert and Jensen, 1996). For example, glycinebetaine (GB) is a particularly effective protectant against abiotic stress (Chen and Murata, 2008), and accumulates rapidly in plants exposed to salt, drought, and low temperature stresses (Rhodes and Hanson, 1993).
Previous studies have shown that overexpression of stress-related genes caused enhanced tolerance of alfalfa to the salinity stress (Luo et al., 2019b). Li H. et al. (2014) successfully targeted CHOLINE OXIDASE A (CODA) cDNA derived from Agrobacterium globiformis to alfalfa chloroplasts under the control of the strong stress inducible SWEETPOTATO PEROXIDASE ANIONIC 2 (SWPA2) promoter (Kim et al., 2003). Such transgenic alfalfa plants exhibited increased tolerance to oxidative, drought, and salt stress. Because salinity also causes cellular ionic imbalances, the Na+/H+ antiporter in the plasma membrane (SOS1 – SALT OVERLAY SENSITIVE 1) and tonoplast (NHX2 – SODIUM/HYDROGEN EXCHANGER 2) can maintain higher K+/Na+ ratios in the cytoplasm as a protection against sodium toxicity (Fukuda et al., 1999; Xia et al., 2002; Zhang L. Q. et al., 2014). Moreover, the expression of foreign genes, such as TaNHX2 (Triticum aestivum NHX2), AhBADH (Atriplex hortensis BETAINE ALDEHYDE DEHYDROGENASE), SsNHX1 (Suaeda salsa NHX1), and GmDREB1 (G. max DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 1), can increase salt tolerance in transgenic alfalfa plants (Zhang et al., 2012). As such, Zhang L. Q. et al. (2014) transformed the exogenous gene SeNHX1 (Salicornia europaea NHX1) into alfalfa using Agrobacterium-mediated transformation; this enhanced tolerance to salt stress was manifested by improved photosynthesis and membrane stability. Another attempt to improve salt tolerance in alfalfa was reported by Jin et al. (2010) using transformation with the soybean DREB ortholog, GmDREB1, under the control of Arabidopsis stress-inducible RD29A (RD – RESPONSIVE TO DESICCATION) promoter. Ion leakage, chlorophyll fluorescence, total soluble sugars, transcript level of Δ1-PYRROLINE-5-CARBOXYLATE SYNTHASE (P5CS), and free proline contents were correlated with the higher salt tolerance of transgenic lines (Jin et al., 2010). Wang et al. (2014) generated and characterized transgenic alfalfa plants with heterologous expression of AtNDPK2 (NUCLEOSIDE DIPHOSPHATE KINASE 2) under the control of oxidative stress inducible SWPA2 promoter. These transgenic plants showed increased tolerance to oxidative, high temperature, salt and drought stresses. Such enhanced tolerance was mediated by activation of ROS scavenging, enhanced activity of NDPK2 enzyme, improved protection of membrane integrity, and increased proline accumulation (Wang et al., 2014).
First studies on drought responses of alfalfa started in the 1990s (Luo et al., 1991, 1992; Laberge et al., 1993). Metabolite profiling and proteomic approaches identified soluble sugars, amino acids, and proteins that respond to drought in leaves and nodules of alfalfa variety Magali (Aranjuelo et al., 2011). Simultaneously, Kang et al. (2011) have shown systematic analysis of two alfalfa varieties, Wisfal and Chilean, with different tolerance/sensitivity to the drought stress. They have identified many genes involved in adaptation to the drought stress, including genes encoding transcription and regulatory factors, or genes involved in the biosynthesis of osmolytes and antioxidants. Knowledge of such genes can help in breeding programs. A number of microRNAs have been used to improve various crop species via genetic engineering (Macovei et al., 2012; Zhou and Luo, 2013; Aung et al., 2015). Researchers also characterized microRNAs and their target genes that respond to hypoxia, wounding, heat or oxidative stress (Zhao et al., 2007; Budak et al., 2015). Recent study by Arshad et al. (2017) suggested that overexpression of microRNA156 (miR156OE) is an emerging tool to improve drought tolerance of alfalfa since it silenced SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 13 (SPL13i) leading to reduced water loss and enhanced stomatal conductance and photosynthetic assimilation. Another study proposed a role of miR156OE and SPL13i in heat stress tolerance since plants carrying these constructs showed increased antioxidant levels (Matthews et al., 2019). As found by NGS, plants possessing miR156OE exhibited broad changes in gene expression, including genes involved in nodulation, root development and phytohormone biosynthesis (Aung et al., 2017). Taking together, miR156 can improve drought or heat stress tolerance in alfalfa, at least partially by silencing SPL13 (Feyissa et al., 2019; Matthews et al., 2019).
RNA-Seq analysis was utilized in the transcriptome profiling of alfalfa in order to study the molecular mechanisms underlying frost (Song et al., 2016), salinity (Postnikova et al., 2013; An et al., 2016; Lei et al., 2018), drought (Arshad et al., 2018), resistance to aluminum (Liu W. et al., 2017), lead (Xu et al., 2017) and waterlogging (Zeng et al., 2019), or fall dormancy (Zhang S. et al., 2015). For example, genes encoding membrane proteins, and proteins of hormonal signal transduction, and ubiquitin-mediated proteolysis pathways contribute to the freezing adaptation mechanisms in alfalfa (Song et al., 2016). Using high-throughput sequencing technology, Postnikova et al. (2013) have demonstrated that salinity stress affects a variety of alfalfa genes. Among the most affected ones were genes of known function, such as DIHYDROFLAVONOL REDUCTASE (DFR), transcription factor MYB59, SUGAR TRANSPORTER ERD6-like 16 (ERD – EARLY RESPONSE TO DEHYDRATION), and INOSITOL-145-TRISPHOSPHATE 5-PHOSPHATASE (IP5P2). This study revealed that 86 transcription factors responded to salinity stress; among them are those belonging to GRAS, ARR, JUMONJI, and MYB families that were preferentially upregulated in the tolerant alfalfa cultivar (Postnikova et al., 2013). Alfalfa fall dormancy is determined by genes involved in auxin (e.g., AUXIN-INDUCED PROTEIN 5NG4) and ethylene signaling (ethylene responsive TF RAP2-11) and carbohydrate transport (ERD6-LIKE PROTEIN; Zhang S. et al., 2015). Genes encoding BETA-AMYLASE, ETHYLENE RESPONSE FACTOR (ERF), CALCINEURIN B-LIKE (CBL) INTERACTING PROTEIN KINASES (CIPKs), GLUTATHIONE PEROXIDASE (GPX), and GLUTATHIONE S-TRANSFERASE (GST) are among those important for waterlogging stress resistance in alfalfa (Zeng et al., 2019).
Plant damage caused by saline stress is usually divided into three categories: high pH damage, osmotic shock, and toxic cation stress. Nutrient solution pH variation significantly affected growth of alfalfa seedlings with the optimal pH values in the range between 5.0 and 6.0, as estimated by length and fresh weight of roots, hypocotyls, epicotyls, first leaf petioles, and leaf blades (Köpp et al., 2011). Alfalfa is a saline-alkaline stress-tolerant species (Zhu, 2001; Wong et al., 2006; Gong et al., 2014; An et al., 2016). An et al. (2016) performed transcriptomic analysis of whole alfalfa seedlings treated with saline-alkaline solutions using ion torrent sequencing technology to study changes in the gene expression pattern. This method detects hydrogen ions that are released during DNA polymerization. DEG profiles were obtained and annotated using two methods. Firstly, generated reads were mapped to barrel medic, which has a sequence that is highly homologous to alfalfa. Secondly, functional annotations of assembled unigenes were performed using BLASTX search against the Swiss-Prot databases of barrel medic, thale cress, and soybean. Gene ontology analysis revealed 14 highly enriched pathways. Specific responses of peroxidases, the expression level of RUBISCO, and flavonoids indicated antioxidant capacity as one of the main mechanisms behind the saline-alkaline stress tolerance in alfalfa (An et al., 2016). Another study provided a comprehensive transcriptome analysis of alfalfa roots under prolonged ABA treatment (Luo et al., 2019a). Sequences were assembled for many isoforms and were analyzed for their potential role. Differentially expressed isoforms (DEIs) regulated by ABA were mainly involved in transcriptional regulation, plant immunity, plant hormone signal transduction, and anti-oxidative defense.
Nevertheless, these studies were mainly focused on genotype-specific stress mechanisms. Functional and structural genomics studies are fundamental for the understanding of plant biology. Access to high-quality genome and transcriptome sequences is important to perform studies of this kind. Recently, the third-generation sequencing technology PacBio RSII has emerged as a unique method for constructing full-length transcripts (Dong et al., 2015; Nakano et al., 2017). PacBio RSII is an ideal tool for whole genome sequencing, targeted sequencing, RNA-Seq, and epigenetic characterization. This technique allows the sequencing of single DNA molecules in real-time (SMRT) without amplification by PCR (Dong et al., 2015). Using PacBio RSII, Luo et al. (2019b) studied salt stress as a major environmental factor that impacts alfalfa development and production (Zhang S. et al., 2015). They have constructed the first full-length transcriptome database of alfalfa root tips treated with mannitol (a non-ionic osmotic stress) and NaCl (an ionic osmotic stress), which provided evidence that the response to salinity stress includes both osmotic and ionic components. They have found 8,016 mannitol-regulated DEGs and 8,861 NaCl-regulated DEGs. These DEGs are involved in signal transduction, transcriptional regulation, anti-oxidative defense, and signal perceptions (Luo et al., 2019b).
Resistance to Biotic Stress
Biotic stress also considerably affects alfalfa growth and yield. Current methods of plant protection focus mostly on the elimination of pathogenic organisms using pesticides (Shafique et al., 2014). However, the improvement of plant resistance against such pathogens seems like a more beneficial alternative, since it might be more effective and more environmentally friendly (Kudapa et al., 2013; Varshney and Kudapa, 2013). It is expected that climatic changes are linked to the spread of diseases and emergence of new ones and can raise the threat of parasites and pests (Kudapa et al., 2013; Shafique et al., 2014). Therefore, disease-resilient plants could provide higher production and yield, reflecting the importance of genetically engineering specific genes (de Zélicourt et al., 2011).
Disease resistance mechanisms in plants after encountering a pathogen have been well-described (Roumen, 1994; Zipfel, 2014; Rubiales et al., 2015). Plant infection is facilitated by effector molecules produced by pathogens, which can overcome the first line of plant defense, which is the pathogen−associated molecular pattern (PAMP) triggered immunity; subsequently, plant resistance is suppressed. On the other hand, specific plant resistance (R) proteins have been evolutionarily developed and can provide protection against specific pathogen effectors (Jones and Dangl, 2006; Singer et al., 2018). Nowadays, genes encoding R proteins are widely manipulated for introducing plant resistance to a specific pathogen (Rubiales et al., 2015).
Generally, the most frequently occurring pathogens are bacteria and fungi belonging to Ascomycetes and Basidiomycetes; these obtain nutrients by attacking various parts of the plant body (Shafique et al., 2014). Considerable declines in alfalfa production have been observed mostly due to root infections leading to wilting caused by the bacterium Clavibacter michiganensis, fungi Fusarium oxysporum and Verticillium alfalfae, and microscopic fungus Phytophthora medicaginis, or due to leaves infected by Colletotrichum trifolii (Nutter et al., 2002; Singer et al., 2018). Alfalfa varieties resistant to these diseases have been obtained by common breeding methods over decades (Toth and Bakheit, 1983; Elgin et al., 1988; Pratt and Rowe, 2002). However, it may not be enough to cover the world demand for crop yields, considering the influence of a retrogressive living environment. Because of alfalfa autopolyploidy and its out-crossing nature (Zhang T. et al., 2015; Yu et al., 2017), the comprehension of molecular and genetic mechanisms during pathogenesis leading to the introduction of specific resistance can be a demanding task. For this reason, barrel medic is widely used for such purposes. Different transcriptomic methods (Gao et al., 2012; Van et al., 2013; Le Nguyen et al., 2018; Scheben et al., 2019) were used to identify barrel medic loci correlated with QTLs, providing resistance to diseases caused by fungi such as Uromyces striatus and Erysiphe pisi (Bustos-Sanmamed et al., 2013).
C. trifolii is an agent of a highly destructive and prevalent foliar disease, anthracnose (Annicchiarico et al., 2015), which can cause up to 30% decrease in alfalfa yield (Yang et al., 2008). Recognition of this pathogen and induction of response in alfalfa are understudied and need further characterization by cloning techniques. Nevertheless, Yang et al. (2008) found out that overexpression of the gene for intracellular R protein, RCT1 encoding TIR-NBS-LRR (TOLL/INTERLEUKIN-1 RECEPTOR NUCLEOTIDE BINDING SITE LEUCINE-RICH REPEAT) from barrel medic, ensured anthracnose resistance in alfalfa. Mackie et al. (2007) and Tesfaye et al. (2007) identified tetrasomic dominant ANTHOCYANIN genes AN1 and AN2 regulating resistance against C. trifolii (Elgin and Ostazeski, 1985). Mackie et al. (2007) mapped locations of QTLs for C. trifolii traits 1, 2, and 4 in autotetraploid alfalfa clone W126, which is resistant to this pest. Interactions between particular QTLs and phenotypic variations for three C. trifolii traits have been described. Obtained markers may be usable in alfalfa breeding for introducing multiple sources of resistance. Although genes for a specific resistance have been identified, new pathotypes of C. trifolii are still being discovered; therefore, the generation of new, long-lasting resistant plants is more difficult (Shafique et al., 2014).
Using the suppression subtractive hybridization library, Bustos-Sanmamed et al. (2013) proved the importance of pathogenesis-related (PR) proteins of group 10, as well as proteins engaged in ABA signaling for resistance against harmful fungi, e.g., Aphanomyces enteiches. Bahramnejad et al. (2010) designated and isolated the MsPR10.1A gene in alfalfa based on its homology to PR10 genes from other Fabaceae plants, e.g., Lupinus luteus (Zhang, 2004). Expression levels of MsPR10.1A under different conditions such as ABA treatment, heat shock, wounding, and pathogen attack, were compared with the expression levels of a previously described gene, PPRG2 (termed as MsPR10.1B; Borsics and Lados, 2002). Bahramnejad et al. (2010) observed faster induction of MsPR10.1A gene expression than that of MsPR10.1B gene after ABA and ethylene treatment, and after application of the pathogenic bacterium Xanthomonas campestris. However, inoculation of alfalfa leaves with compatible X. campestris led to markedly higher expression of both genes. On the other hand, gene AAB41557 from the alfalfa PR10 group did not respond to X. campestris inoculation (Esnault et al., 1993). Generally, most examples regarding PR10 induction due to bacterial inoculation involve incompatible bacteria, such as activation of alfalfa genes AAB41557 (Esnault et al., 1993) and MsPR10.1B (Borsics and Lados, 2002) after Pseudomonas syringae pv. pisi inoculation. The promoter of YPR-10 (of the RIBONUCLEASE-LIKE PR PROTEIN-10 gene) from G. max fused with GUS showed activity in the vasculature of Nicotiana benthamiana leaves after transient transformation (Walter et al., 1996). Moreover, Bahramnejad et al. (2010) suggested the importance of MsPR10.1A promoter expression in the leaf vasculature, resulting in resistance against diseases. MsPR10.1A and MsPR10.1B promoters have many similar functions in stress responses, but notable differences were found in their reactions to wounding. Thus, promoters of PR10 genes may be potentially used in biotechnological applications for directing transgene expression in proper tissues.
Plant defense peptides are composed of five main groups: proteases, α-amylase inhibitors, lectins, chitinases, and polyphenol oxidases (Fürstenberg-Hägg et al., 2013). Singer et al. (2018) summarized several genes for the biosynthesis of substances with anti-pathogen effect, such as AGLUL encoding β-1,3-glucanase (Masoud et al., 1996), IOMT – Isoflavone-O-methyltransferase (He and Dixon, 2000), LF – encoding lactoferrin (Stefanova et al., 2013) and RS – encoding resveratrol synthase (Hipskind and Paiva, 2000). Highly effective protectants, such as protease inhibitors, naturally occur in plants, and they can inhibit proteolytic enzymes in the digestive system of insects or nematodes. Consequently, plant material is not digestible, leading to pathogen starvation and removal from the plant. Inhibitors of cysteine proteases called phytocystatins were identified in many plants, showing potential in conferring resistance against pathogens. Rice ORYZACYSTATIN-I (OC-I) and ORYZACYSTATIN II (OC-II) genes driven by a potato wound-inducible promoter (Protease inhibitor II, PinII) were transferred to alfalfa attacked by root lesion nematode and leaf beetle. Such transgenic plants revealed a reduction in the Pratylenchus penetrans population and enhanced mortality of Phytodecta fornicata larvae (Ninković et al., 1995; Samac and Smigocki, 2003).
Tesfaye et al. (2005) generated alfalfa plants that secreted a fungal endochitinase (ECH42). These transgenic plants showed up to 25.7 times increased chitinase activity in vegetative organs and root exudates. Such secreted endochitinases not only retained the lytic activity against glycol chitin, but also showed antifungal activity by the inhibition of spore germination of two fungal pathogens, namely, Phoma medicaginis and C. trifolii (Tesfaye et al., 2005).
Based on the expression distribution of SNAKIN gene StSN1 in Solanum tuberosum, Segura et al. (1999) hypothesized SN1 as a component of constitutive defense barriers in reproductive and storage plant organs. StSN2 is induced locally after wounding and pathogen attack; accordingly, it could play an important role in constitutive and inducible defense barriers (Kovalskaya and Hammond, 2009; Guzman-Rodriguez et al., 2013). Next, García et al. (2014) proposed SNAKIN proteins as antimicrobial compounds in plant innate immunity. Indeed, alfalfa transgenic plants carrying SNAKIN-1 (MsSN1) under the control of a constitutive promoter showed improved tolerance against pathogenic fungi. Three independent transgenic lines carrying the CaMV35S:MsSN1 construct showed significantly lower amounts of infected leaves than wild type plants when treated by C. trifolii and with the oomycete P. medicaginis (García et al., 2014).
Finally, it is worth mentioning that the genetic transformation of alfalfa with Bacillus thuringiensis gene Cry1C coding for δ-endotoxin has also been shown to be an effective protective strategy. After transformation, alfalfa was more resistant to Nemapogon granellus and Spodoptera exigua (Strizhov et al., 1996).
Transcriptomic studies contributed to the knowledge of alfalfa resistance to aphids, strips, and nematodes. Aphids are major insect pests causing a significant decrease of alfalfa yield. Tu et al. (2018b) performed a transcriptomic analysis of two alfalfa cultivars differing in aphid resistance. Genes involved in salicylic acid biosynthesis represented an important defense mechanism in both cultivars. The alfalfa resistance against aphids was mainly determined by induction of genes involved in linoleic acid synthesis important for jasmonic acid and flavonoid biosynthesis (Tu et al., 2018b). Genes participating in jasmonic acid biosynthesis, such as LIPOXYGENASE, SERINE PROTEINASE INHIBITOR, and SEED LINOLEATE 9S-LIPOXYGENASE were also important for alfalfa resistance to strips infestation. Moreover, genes involved in fatty acid degradation, chloroalkane and chloroalkene degradation, beta-alanine and phenylalanine metabolism and flavonoid biosynthesis also contributed to this resistance (Tu et al., 2018a). Another comparative transcriptomic analysis aimed to screen for genes determining alfalfa resistance to root-knot nematode Meloidogyne incognita (Postnikova et al., 2015). LRR AND NB-ARC DOMAIN DISEASE RESISTANCE PROTEIN (Medtr3g023030.1), RECEPTOR-LIKE PROTEIN (Medtr5g087320.1) and DISEASE RESISTANCE PROTEIN (TIR-NBS-LRR class, Medtr0277s0020.3) were up-regulated in the resistant cultivar, while susceptible one showed their down-regulation (Postnikova et al., 2015).
From the biotechnological point of view, ideal alfalfa cultivars should have better nutritional quality, enhanced biomass production and yield, and better resistance to biotic and abiotic stress. All such traits mentioned should be sustainable over a long period of time. Several experimental studies have been conducted to improve alfalfa, but detailed characterization and relationships between desired traits need further genetic and molecular research.
Proteomics and Metabolomics
Owing to its beneficial agronomical traits, alfalfa has been attracting substantial interest in the fields of proteomics and metabolomics during the past two decades. A strong effort was invested in the discovery of new proteins and metabolites involved in alfalfa development and abiotic stress response. In this section, we attempt to summarize the recent achievements of current alfalfa proteomic and metabolomic research. We also aim to highlight the relevance of these investigations for putative biotechnological applications.
Nitrogen and Carbon Metabolism in Alfalfa From a Proteomic Perspective
Proteomics and metabolomics have a remarkable capability to examine the balance between carbon and nitrogen metabolism under stress conditions in alfalfa during interactions with nitrogen-fixing bacteria (Aranjuelo et al., 2011, 2013). Water stress limits nitrogen fixation in nodules by the reduction of nitrogenase activity (Carter and Sheaffer, 1983; Aranjuelo et al., 2011) and Rubisco availability in leaves (Aranjuelo et al., 2005, 2011). The latter likely occurs due to Rubisco-enhanced proteolysis and lower abundance of RUBISCO ACTIVASE. Water stress also affected ammonia assimilation into amino acids, as evidenced by the upregulation of glutamine synthetase and decreased levels of glutamic acid and asparagine in leaves. The effects of water stress were followed by elevated photorespiration (exemplified by increased abundances of photorespiratory enzymes), lower demand for carbohydrates, and accumulation of soluble sugars. In nodules, water deprivation caused the attenuation of respiration, leading to CO2 recycling by PHOSPHOENOLPYRUVATE CARBOXYLASE. This likely occurred in order to support carbon skeletons for amino acid biosynthesis. The reduced respiration may also be a consequence of increased demand for compounds with osmoregulation capacity such as glycerol (Aranjuelo et al., 2013). The dynamic behavior of ammonia assimilation seems to be important for abiotic stress tolerance. It is likely that nitrogen is relocated from glutamic acid and asparagine, which are the main nitrogen sources in control conditions, to proline under stress conditions. Thus, proline might be an alternative nitrogen source under osmotic stress, and it seems that alfalfa may easily switch between proline biosynthesis and degradation (Zhang and Shi, 2018). Abiotic stresses caused accumulation of enzymes of nitrogen assimilation, such as GLUTAMINE SYNTHETASE and FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE (Rahman et al., 2016) as well as GLUTAMATE DEHYDROGENASE (Dai et al., 2017). Remarkably, heat stress positively affected the abundance of ASPARTATE AMINOTRANSFERASE and GLUTAMINE SYNTHETASE, indicating an enhancement of nitrogen metabolism (Li W. et al., 2013).
Clearly, Rubisco availability and homeostasis between carbon and nitrogen metabolism is crucial for plant performance under unfavorable environmental conditions. For this reason, the proteins regulating C and N metabolism, as well as stress related proteins (Table 1), appear to be prospective candidates for the biotechnological improvement of alfalfa.
TABLE 1.
Overview of proteins and metabolites important for biotechnological improvement of alfalfa as revealed by proteomic and metabolomic studies.
| Treatment, stress, condition | Sample | Methodological approach | Proteins and metabolites of biotechnological importance | References |
| Seed germination and osmopriming | Seeds | 2-D gel electrophoresis (nano-LC MS/MS) | Carbohydrate metabolism: UDP glucose pyrophosphorylase Protein destination and storage: HSP70 and HSP20, GroEL-like chaperone, ATPase, vicilin, protein disulfide-isomerase precursor Stress response: annexin, peroxiredoxins, manganese superoxide dismutase, glyoxalase, lipoxygenase, glutathione S-transferase, thioredoxin | Yacoubi et al. (2011) |
| Proteolysis: peptidase T1A, proteasome beta subunit, peptidase A1 pepsin | ||||
| Osmoprimed seeds germinating under salt stress | Seeds | 2-D gel electrophoresis (nano-LC MS/MS) | Small HSPs: 18.2 kDa class I HSP Methionine synthesis: methionine synthase, cysteine synthase Dehydration defense: LEA proteins, PM22 Others: annexin, RNA-binding protein, heme oxygenase, glutathione S-transferase 9 | Yacoubi et al. (2013) |
| PEG-induced osmotic stress | Roots of varieties contrasting in drought tolerance | iTRAQ (strong cation exchange fractionation and LC MS/MS) | Stress and defense: glutathione S-transferases, disease resistance response protein, epoxide hydrolase, chitinase, reticuline oxidase-like protein, low-temperature-induced 65 kDa protein, aldo/keto reductase, pirin-like plant protein, glucan endo-1,3-beta-glucosidase Protein metabolism: HSPs, lysine-ketoglutarate reductase/saccharopine dehydrogenase, phosphatidylethanolamine-binding protein, homoglutathione synthetase | Zhang and Shi (2018) |
| Signal transduction: monooxygenases, cysteine-rich RLK (receptor-like kinase) protein, 12-oxophytodienoate reductase | ||||
| Cell wall: beta xylosidase, xyloglucan-specific endoglucanase inhibitor protein, expansin-B1-like protein | ||||
| Salt stress | Roots of two cultivars contrasting in salt resistance | 2-D gel electrophoresis (MALDI TOF/TOF) | Oxidative stress: peroxidase, peroxiredoxin Protein folding: protein disulfide isomerase Metabolism: NAD synthetase, UTP-glucose 1 phosphate uridylyltransferase Fatty acid metabolism: biotin carboxylase 3 | Rahman et al. (2015) |
| Membrane transport: V-ATPase | ||||
| Salt and drought stress | Seedlings | 2-D gel electrophoresis (MALDI TOF-MS/MS) | Salt stress: caffeoyl-CoA 3-O-methyltransferase, peroxiredoxin, ubiquitin-conjugating enzyme, UV excision repair protein rad23, glutathione peroxidase | Ma et al. (2017) |
| Drought stress: ubiquitin-conjugating enzyme, putative alcohol dehydrogenase, chaperonin 10 | ||||
| Drought stress | Leaves of plants inoculated by S. meliloti | Proteomics: 2-D gel electrophoresis (LCMS/MS analysis) Metabolomics: GC TOF-MS | Rubisco availability and regeneration: rubisco activase, sedoheptulose-1,7-bisphosphatase, ribulose-phosphate 3-epimerase and phosphoribulokinase Nitrogen metabolism: glutamine synthetase | Aranjuelo et al. (2011) |
| Stress and defense response: superoxide dismutase, dehydroascorbate reductase, 2-cys peroxiredoxin-like protein, 14-3-3-like protein | ||||
| Osmoprotectant metabolites: proline, pinitol | ||||
| Drought stress | Nodules, roots, leaves | Proteomics: 2-D gel electrophoresis (LCMS/MS analysis) Metabolomics: GC TOF-MS | Nodule proteome: alpha 1,4-glucan protein synthase, lipoxygenase, PEP-carboxylase Nodule N containing metabolites: glutamine, asparagine Nodule osmoprotectant metabolites: glycerol, galactinol, myo-inositol, proline, sucrose, raffinose, fumaric acid and malate Nodule metabolites with antioxidant capacity: ascorbate, threonate | Aranjuelo et al. (2013) |
| Water deficit stress | Roots | 2-D gel electrophoresis (MALDI TOF) | Nitrogen metabolism: glutamine synthetase, ferredoxin-dependent glutamate synthase | Rahman et al. (2016) |
| ABA biosynthesis: 9-cis-epoxycarotenoid dioxygenase | ||||
| Stress response and oxidative stress: ascorbate peroxidase, peroxiredoxin, calreticulin, stress-induced phosphoprotein, annexin | ||||
| Transcription: basic helix-loop-helix (bHLH) transcription factor, agamous-like 65 Other functions: inward-rectifying potassium channel, auxin-independent growth promoter | ||||
| Heat stress | Leaves | 2-D gel electrophoresis (MALDI TOF/TOF) | Rubisco availability: Rubisco activase isoforms Nitrogen metabolism: aspartate aminotransferase and glutamine synthetase | Li W. et al. (2013) |
| Protein synthesis and processing: peptidyl-prolyl cis–trans isomerases, protein disulfide isomerase-like protein precursor, porin, proteasome subunit β type, eukaryotic translation initiation factor 3 subunit I, BiP isoform A/glycine max, cysteine proteinase, outer plastidial membrane protein porin | ||||
| Intracellular traffic, cell structure: protein TOC75, translocon Tic40, profilin | ||||
| Defense response: 17 kDa HSP, 18.2 kDa class I HSP, 20 kDa chaperonin, HSP23, HSP70, thaumatin-like protein, ubiquitin, ascorbate peroxidases, glucan endo-1,3-beta-glucosidase | ||||
| Cold acclimation | Leaves of cultivars tolerant or sensitive to freezing | 2-D gel electrophoresis (MALDI TOF/TOF) | Oxidative stress: monodehydroascorbate reductase, glutathione peroxidase, peptide methionine sulfoxide reductases A3, thioredoxin-like protein CDSP32, 2-cys peroxiredoxin BAS1-like | Chen et al. (2015) |
| Methionine biosynthesis: 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | ||||
| Lignin and terpenoid biosynthesis: cinnamoyl-CoA reductase, 1-deoxy-D-xylulose 5-phosphate reductoisomerase | ||||
| Photosynthesis and Rubisco availability: Rubisco large subunit-binding protein subunit beta, Rubisco activase B, chlorophyll A/B binding protein, oxygen-evolving enhancer protein 1, cytochrome b6-f complex iron-sulfur subunit | ||||
| Protein folding and disassembling: chaperone protein ClpC, GTPase, peptidyl-prolyl cis–trans isomerase CYP20-3 | ||||
| Cadmium stress | Cell walls and soluble proteins from stems | 2-D DIGE (MALDI TOF/TOF) | Cell wall modification: sucrose synthase, pectinesterase/pectinesterase inhibitor, polygalacturonase non-catalytic protein, polygalacturonase-inhibiting protein 1, b-xylosidase/alpha-L-arabinofuranosidase, trichome birefringence-like protein, xyloglucan endotransglucosylase/hydrolase family protein, dirigent protein 21-like | Gutsch et al. (2018a) |
| Defense: chitinase (Class Ib)/hevein, chitinase, class I chitinase, disease resistance response protein, pathogenesis-related protein 1, pathogenesis-related thaumatin family protein, plant basic secretory protein (BSP) family protein, pre-hevein-like protein, stromal 70 kDa heat shock-related protein, CAP, cysteine-rich secretory protein, antigen 5 | ||||
| Oxidation-reduction process: anionic peroxidase swpb3 protein, class III peroxidase, peroxidase family protein, peroxidase1b, peroxidase2 | ||||
| Cadmium stress | Stems (soluble and cell wall enriched proteins) | 2-D DIGE (MALDI TOF/TOF) | Cell wall modification: pectinesterase/pectinesterase inhibitor, polygalacturonase non-catalytic protein, polygalacturonase-inhibiting protein 1 Chloroplast protein degradation: chloroplastic aspartyl protease isoforms Cell wall: class III peroxidase, lignin biosynthetic peroxidase, chitinases | Gutsch et al. (2018b) |
| Stem growth | Different regions of stems (apical, intermediate, and basal) | 2-D gel electrophoresis (MALDI TOF/TOF) | Chloroplast protein synthesis: CSP41-b, EF-Tu, EF-G, Cpn 60, HSP70 Lignin biosynthesis: transketolase, enolase Cytoplasmic protein synthesis: eIF-5a, endoplasmic protein disulfide isomerase, HSP90, ribosomal protein P3-like | Printz et al. (2015) |
| Vesicular trafficking: clathrin light chain | ||||
| Stress response: peroxisomal membrane protein, monodehydroascorbate reductase, flavoprotein wrbA-like, Pprg2 | ||||
| Sieve element development: sieve element occlusion by forisomes 3 | ||||
| Cadmium stress and hydrogen- rich water | Roots | iTRAQ (nano-LC MS/MS) | Defense response: mitogen-activated protein kinase, pathogenesis-related thaumatin family protein, pathogenesis-related protein bet V I family protein, disease-resistance response protein Nitrogen metabolism: glutamate dehydrogenase Sulfur compound metabolic process: cysteine synthase, ATP sulfurylase | Dai et al. (2017) |
| Secondary metabolism: chalcone-flavonone isomerase family protein | ||||
| Waterlogging | Leaves of two cultivars contrasting in tolerance to waterlogging | iTRAQ (reverse-phase HPLC fractionation and LC-MS/MS) | Cell wall and defense response: acidic endochitinase, expansin-like B1, early nodulin-like protein 2, thaumatin-like protein, 1,4 alpha-glucan-branching enzyme 1, pathogenesis-related protein Stress response: glutathione S-transferase, protein C2-DOMAIN ABA-RELATED 9, aldo-keto reductase family 4 member C9, Fe superoxide dismutase 2, 1 aminocyclopropane-1-carboxylate oxidase homolog 5, | Zeng et al. (2019) |
| Proteolysis: vacuolar-processing enzyme | ||||
| Different developmental stages (budding and mid-flowering) | Leaves | TMT labeling (nano-LC MS/MS) | Metabolites: D-mannose hemicellulose precursor (upregulated in mid flowering), L-phenylalanine, L-tyrosine, L-phenylalanine Metabolism: alpha glucosidase, alpha amylase Cell wall modification: UDP-glucuronic acid decarboxylase (xylan production), cinnamyl alcohol dehydrogenase (lignin biosynthesis) | Fan et al. (2018) |
| Fall dormancy | Terminal buds of fall dormant and non-fall dormant cultivars | iTRAQ (SCX fractionation, LC MS/MS) | Nitrogen metabolism: L-asparaginase Auxin polar transport: stilbene synthase family protein, monothiol glutaredoxin-S17 protein Lignin biosynthesis: cinnamyl alcohol dehydrogenase Pyruvate metabolism and transport: pyruvate carrier protein | Du et al. (2018) |
| Vitamin B1 metabolism: thiazole biosynthetic enzyme |
Proteins and Pathways Found by Proteomics as Promising Candidates for Alfalfa Abiotic Stress Resistance Improvement
Seed priming involves a complex array of physiological as well as molecular processes leading to an improved ability of plants to withstand adverse environment (Paparella et al., 2015). A gel-based proteomic approach was employed to investigate proteome remodeling during osmoprimed alfalfa seed germination. This process was accompanied by intense accumulation of storage proteins (such as vicilins), proteins involved in protein folding, UDP glucose and methionine biosynthesis, annexins, and antioxidant enzymes, compared to seeds that were not osmoprimed. Osmopriming was also followed by remarkable induction of stress-related proteins and proteasome components (Table 1) (Yacoubi et al., 2011). A follow-up article highlighted that osmopriming has remarkable consequences on the proteome of seeds germinating under saline conditions. An increased seed vigor associated with osmopriming was related to the accumulation of storage proteins, annexins and RNA-BINDING PROTEIN. The last one indicated the possible importance of posttranscriptional regulation in the seedlings exposed to salt stress. On the other hand, seeds without osmopriming accumulated HEAT SHOCK PROTEINS (HSP), LATE EMBRYOGENESIS ABUNDANT (LEA) PROTEINS, SEED MATURATION PROTEINS, GLUTATHIONE S-TRANSFERASE 9, and HEME OXIDASE (Table 1) (Yacoubi et al., 2013). These data indicate that the transient genetic modification of genes encoding the above-mentioned stress-related proteins (for instance, by expression under an inducible tissue-specific promoter), might be of biotechnological importance.
Tolerance of alfalfa to the polyethylene glycol (PEG)-induced osmotic stress was accompanied by enhanced carbohydrate metabolism and energy production. Stress-related proteins such as glutathione S-transferases and LEA proteins are also correlated with osmotic stress tolerance (Table 1) (Zhang and Shi, 2018), and both represent promising candidates for biotechnological applications. A similar study revealed that proteins involved in protein folding (DISULFIDE ISOMERASE), NAD production (NAD SYNTHASE), methylation (ADENOSINE KINASE, S-ADENOSYL-METHIONINE) and antioxidant defense (represented mainly by peroxidases), are candidates to determine alfalfa salt tolerance (Rahman et al., 2015). Overabundance of proteins involved in the enzymatic antioxidant defense was commonly associated with an increased tolerance of alfalfa not only to the salt, but also to the drought and osmotic stresses (Table 1) (Rahman et al., 2015; Long et al., 2018; Zhang and Shi, 2018). According to a proteomic study, water stress increased the abundance of AGAMOUS-LIKE 65 and bHLH TRANSCRIPTION FACTORS, while it reduced the abundance of JADE-1 and JADE-3, transcriptional regulators belonging to a PHD (plant homeodomain)-type zinc fingers family (Table 1) (Rahman et al., 2016). These intriguing findings of transcriptional factors involved in water stress deserve further biotechnological investigations. Genetic modifications of hormone biosynthesis belong also to promising biotechnological approaches, since water stress elevated the abundances of ABA (9-CIS-EPOXYCAROTENOID DIOXYGENASE) and auxin (AUXIN-INDEPENDENT GROWTH PROMOTER) biosynthetic proteins in alfalfa (Rahman et al., 2016). In this regard, local stress-induced changes in the turnover of auxin regulatory proteins could modify plant developmental processes, such as cell elongation, lateral roots emergence, transition from cell division to cell differentiation, enabling plants to rapidly adapt to adverse environmental conditions (Korver et al., 2018). On the other hand, drought stress caused some common but also distinct responses when compared to salt stress at the level of the alfalfa proteome. Interestingly, both stresses targeted proteasome complex and translation. Nevertheless, the proteasome complex exhibits different sensitivity to these stressors, since the abundance of 26S PROTEASOME REGULATORY SUBUNIT 6 was increased by drought but subsequently reduced by salt stress (Ma et al., 2017).
Comparative proteomic studies point out to obvious similarities between alfalfa and barrel medic in their response to environmental stimuli. Proteome-wide comparison of salt-tolerant alfalfa and salt-sensitive barrel medic indicated that both species are capable of keeping photosynthetic activity during salt stress. Only heat shock protein (gi357476131) was differentially regulated under salt stress in these two Medicago species. It was upregulated in alfalfa but downregulated in barrel medic (Long et al., 2016), indicating its potential biotechnological significance for salt tolerance. A proteomic analysis of these two species at the early post-germination stage showed an important role of antioxidant defense, cell wall metabolism, and jasmonic acid biosynthesis during response to salt (Long et al., 2018). Enhanced salt tolerance of alfalfa, compared to salt sensitive barrel medic, was reflected by higher numbers of differentially regulated proteins, also suggesting higher proteome plasticity (Long et al., 2016, 2018).
Differences in the composition of differentially abundant proteins between two alfalfa cultivars with contrasting freezing tolerance were reported after cold stress treatment (Chen et al., 2015). Freezing-tolerant cultivar exhibited higher abundances of Rubisco subunits as compared to the freezing susceptible one, but showed downregulation of proteins involved in methionine, lignin and terpenoid biosynthesis, and energy metabolism under cold stress (Chen et al., 2015). Heat stress caused an upregulation of proteins involved in energy production, signaling, and intracellular transport and defense, including chaperones, antioxidant enzymes and PR proteins (Li W. et al., 2013). Interestingly, only prolonged heat stress caused downregulation of Rubisco and photosynthetic enzyme activities. Lower abundance of photosynthetic proteins was associated with altered abundance of proteins involved in plastid protein import.
It is known that the external application of bioactive molecules such as hydrogen (H2; Jin et al., 2013) may remarkably increase plant survival rate under adverse environmental conditions. Proteomic elucidation of the beneficial effects of H2 on the alfalfa response to cadmium revealed that this is mainly determined by the modification of proteins involved in the cellular redox homeostasis. Among these proteins, enzymes involved in cysteine biosynthesis and CYSTEINE DESULFURYLASE are elevated by external H2. Cysteine is a precursor for GSH and hGSH, an important redox buffering compounds (Baldacci-Cresp et al., 2012; Diaz-Vivancos et al., 2015), hGSH is specifically produced in species of Fabaceae family including alfalfa, in higher rate compared to GSH, having important role in nodulation (Klapheck, 1988; Matamoros et al., 1999; Frendo et al., 2005; Baldacci-Cresp et al., 2012; Pasternak et al., 2014). Similarly, the abundance of CuZn SUPEROXIDE DISMUTASE (SOD) increased along with a positive effect of external H2 treatment on alfalfa Cd tolerance. Gaseous H2 also enhances the abundance of defense related proteins such as PATHOGENESIS-RELATED PROTEIN BET V I FAMILY PROTEIN and PATHOGENESIS-RELATED THAUMATIN FAMILY PROTEIN (Dai et al., 2017). Such induction of defense related proteins, including chitinases and enzymes involved in cell wall modification, was also observed in alfalfa stems and leaves exposed to long-term Cd stress (Gutsch et al., 2018a, b). Remarkably, chitinases are also employed in the alfalfa response to osmotic stress and waterlogging (Table 1) (Zhang and Shi, 2018; Zeng et al., 2019). This implies that genetic modification of cell walls might improve alfalfa tolerance to multiple stresses.
Proteins Implicated in Development-Associated Agronomical Traits
Proteomics has also been proven as valuable for the evaluation of metabolic activities during alfalfa stem development. The apical region characterized by fiber development showed an overabundance of proteins involved in chloroplast protein synthesis and carbon fixation. The mature stem part possessed a pool of proteins involved in redox homeostasis (Printz et al., 2015). Moreover, the stem is an organ highly sensitive to perturbations of mineral nutrition. This was highlighted by recent proteomic studies reporting that copper availability greatly influenced the abundance of proteins involved in cell wall biogenesis, and in pectin and lignin biosynthesis (Printz et al., 2016). Thus, mineral homeostasis seems to be a crucial factor affecting alfalfa stem growth and rigidity, and also eventually affecting drought tolerance and pathogen resistance.
Flowering represents a critical developmental stage in alfalfa, mainly in terms of seed yield and quality. Pollination and post-pollination processes in alfalfa are linked to altered homeostasis of stress-related proteins such as DUAL SPECIFICITY KINASE SPLA-LIKE PROTEIN, NADPH: QUINONE OXIDOREDUCTASE-LIKE PROTEIN, and CARBONIC ANHYDRASE (Chen et al., 2016). Moreover, PROTEIN DISULFIDE ISOMERASE-LIKE PROTEIN, ASCORBATE PEROXIDASE, GLUTAREDOXIN, and PEROXIREDOXINS also showed fluctuations in their abundances. In addition, metabolic activity was enhanced during pollination and declined afterward.
Fall dormancy is a crucial phenomenon influencing alfalfa performance in autumn, but also during the following season. Based on a comparative proteomic study of terminal buds isolated from two alfalfa cultivars with contrasting fall dormancy, several new proteins were discovered as important for this physiological process (Du et al., 2018). It was suggested that lower abundance of L-ASPARAGINASE and CINNAMYL ALCOHOL DEHYDROGENASES may contribute to fall dormancy. In addition, CHALCONE AND STILBENE SYNTHASE FAMILY PROTEIN (a protein involved in flavonoid biosynthesis) and GLUTAREDOXIN S17 seemed to be important for shoot apical meristem maintenance. Both proteins also have a role in polar auxin transport (Table 1) (Du et al., 2018).
Finally, the nutritional value of alfalfa depends on the developmental stage. Cutting of alfalfa in later developmental stages, such as in full flowering, leads to increased fiber and decreased protein content in the biomass (Fan et al., 2018). Combined proteomic and metabolomic analyses underpinned this finding and showed changes in amino acid composition. These unfeasible nutritional changes are accompanied by increased hemicellulose content, due to the accumulation of D-mannose and higher abundance of ALPHA GLUCOSIDASE, ALPHA AMYLASE, and UDP-GLUCURONIC ACID DECARBOXYLASE, as well as lignin, due to the higher levels of lignin precursors and proteins involved in lignin biosynthesis (Table 1) (Fan et al., 2018).
Gene Editing Using Talen and CRISPR/Cas Technologies
The process of gene editing is based on sequence-specific nucleases (SSNs) creating in vivo loci-specific DNA double-stranded breaks (DSBs) that are subsequently repaired. There are two main DNA repair systems: homology-directed repair (HDR), and the more efficient but less precise non-homologous end joining (NHEJ). NHEJ can result in the insertion or deletion (indel) of nucleotides and a frameshift mutation, which can consequently create a premature stop codon, thus rendering the gene non-functional and creating a genetic knockout. Gene targeting technologies include meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TAL effector nucleases or TALENs), and clustered regularly interspaced short palindromic repeat/CRISPR–associated protein 9 (CRISPR/Cas9). Among these, TALENs and CRISPR/Cas9 are the preferred SSNs for research purposes (Kanaar et al., 1998; Pastwa and Blasiak, 2003; Smith et al., 2006; Pâques and Duchateau, 2007; Hartlerode and Scully, 2009; Sander et al., 2011; Qi, 2015; Steinert et al., 2016; Malzahn et al., 2017; Shan et al., 2020).
The history of gene targeting technologies started in 1988 when the first gene-targeting experiment was performed on tobacco (Nicotiana tabacum) protoplasts (Paszkowski et al., 1988). Later, Puchta et al. (1993) discovered that gene-targeting efficiency can be improved by DSBs in plant cells. More than a decade later, ZFNs were adapted in tobacco and were used in a few plant species for trait improvement (Wright et al., 2005). Subsequently, TALENs were introduced into the group of plant genome editing technologies (Christian et al., 2010). Finally, CRISPR/Cas9 technology has been used in plants such as Arabidopsis thaliana, N. benthamiana, Oryza sativa, and T. aestivum (Li J. F. et al., 2013; Nekrasov et al., 2013; Shan et al., 2013, 2020).
TALENs
TALENs are created by the fusion of DNA binding TALE repeats to the Fok1 nuclease domain. TALENs are less toxic and are easier to engineer than ZFNs. Each of these two platforms has unique limitations, and they are not routinely used in plants. The main advantages of TALENs over CRISPR are that they have less off-target effects due to their ∼30 bp target requirement, as well as their lack of PAM requirement, as unlike CRISPR, TALENs are able to target any sequence. On the other hand, TALENs have more disadvantages: an increased time and financial investment due to the difficulty in protein engineering, a highly variable efficiency for each construct, an inability to target methylated DNA, and the difficulties in engineering nickase (Christian et al., 2010; Li et al., 2011; Mahfouz et al., 2011; Miller et al., 2011; Malzahn et al., 2017; Chen et al., 2019). So far, a successful application of TALEN technology has not been published for either alfalfa or barrel medic. Nevertheless, TALENs have been used for the targeted mutagenesis of another legume, namely soybean (Haun et al., 2014; Demorest et al., 2016; Du et al., 2016; Curtin et al., 2018). The use of TALENs for the mutagenesis of higher plants was recently reviewed by Malzahn et al. (2017) and Khan et al. (2017).
CRISPR/Cas9
In bacteria and archaea, CRISPR and Cas9 function together against invading phages, plasmids, and viruses in adaptive immune system by cleaving the invader’s nucleic acids. The first component is single guide RNA (sgRNA) that associates with a Cas9 protein a Cas9/sgRNA complex. The second component Cas9 belongs to the single-protein effectors of Class 2 CRISPR-Cas systems and is composed of two endonuclease domains, namely, the RuvC-like domain and the HNH, each cutting one strand of DNA. The CRISPR/Cas9 constituents can be transformed into plant cells by different strategies, including Agrobacterium-mediated delivery, gene gun (biolistic delivery), or using virus-based guide RNA (gRNA). Out of the primary SSN classes, CRISPR/Cas9 technology has been the most used and adopted in recent years (Barrangou et al., 2007; Marraffini and Sontheimer, 2008; Wiedenheft et al., 2012; Graham and Root, 2015; Schiml and Puchta, 2016; Makarova et al., 2017; Malzahn et al., 2017; Chen et al., 2019). The CRISPR/Cas system has the potential for numerous applications, such as fusing dCas9 (deactivated Cas9) with other proteins, which can be used for DNA imaging, epigenome editing, gene regulation, and genomic labeling (Chen et al., 2019). One of the main limitations of CRISPR/Cas9 technology might be the generation of undesired off-target effects. Nevertheless, whole-genome sequencing revealed very limited off-target effect mutations in Arabidopsis (Feng et al., 2013), rice (Zhang H. et al., 2014; Tang et al., 2018), and tomato (Nekrasov et al., 2017). Using software tools such as CRISPR-P (Liu H. et al., 2017) and CRISPRGE (Xie et al., 2017) can further decrease any potential off-target occurrence by designing highly specific guide RNAs. Finally, breeding processes may remove any off-target mutations that have negative effects and may keep positive or neutral off-target mutations (Mao et al., 2019).
CRISPR/Cas9 in Alfalfa
CRISPR/Cas9 technology was very recently used for targeted mutagenesis in alfalfa. Selected SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (SPL9) gene was successfully mutagenized and transgenic lines were pre-selected by using droplet digital PCR (ddPCR) for high-throughput screening of large populations. It was further confirmed by restriction enzyme digestion after PCR amplification and sequencing of sub-clones. Comparison of editing efficiency with available data on barrel medic showed lower efficiency in alfalfa, which might be related to its tetraploid genome possessing highly repeated clusters (Meng et al., 2017, 2019; Gao et al., 2018). Gao et al. (2018) concluded that CRISPR/Cas9-mediated modifications of tetraploid alfalfa genome have been successfully performed, but there is still a need to improve editing efficiency. Alfalfa plants with silenced SPL9 had no visible phenotype so ddPCR-based estimation of concentration of the event per μl was a direct indicator of the genome editing rate. Sequencing analysis showed no off-target effects in the alfalfa genome and proved that the sgRNAs of SPL9 were highly specific to the recognition site. In other legumes such as barrel medic, CRISPR/Cas9 technology has been used as well (Michno et al., 2015; Meng et al., 2017, 2019; Curtin et al., 2018; Wen et al., 2019; Yin et al., 2020). Recently, Meng et al. (2019) developed an optimized Agrobacterium-dependent CRISPR/Cas9 system and successfully edited an endogenous PHYTOENE DESATURASE (MtPDS) gene. CRISPR/Cas9 technology for the mutagenesis was also used in L. japonicus (Wang et al., 2016, 2019), and G. max (Cai et al., 2015; Jacobs et al., 2015; Li et al., 2015; Sun et al., 2015; Du et al., 2016; Tang et al., 2016; Curtin et al., 2018; Bao et al., 2019; Wang et al., 2020). Utilization of CRISPR/Cas9-based mutagenesis in several non-leguminous plant species, including data on delivery method, integration into the genome, and editing efficiency, has been reviewed recently (Belhaj et al., 2013; Jaganathan et al., 2018; Liu X. et al., 2019; Kuluev et al., 2019; Mao et al., 2019; Moradpour and Abdulah, 2020; Shan et al., 2020). Approaches such as transgene integration and gene stacking developed for diploid crop species (e.g., corn, cotton, soybean) might be less suitable for alfalfa due to its auto-tetraploid character (Kumar et al., 2018), but the CRISPR/Cas9 technology seems to work well.
Phosphorylation-Dependent Post-Translational Modification by MAPKs
Multiple abiotic stress stimuli, such as wounding, cold, salinity, or drought, are perceived by plants through the activation of MAPKs (Šamajová et al., 2013b). Activated MAPKs phosphorylate, and thereby regulate, several intracellular targets including other protein kinases, cytoskeletal components, nuclear transcription factors, and proteins involved in vesicular trafficking (Komis et al., 2011; Šamajová et al., 2013a). In alfalfa, STRESS-INDUCED MAPK (SIMK), was identified as a salt- and elicitor- stress induced MAPK (Cardinale et al., 2002). SIMK in response to salt stress is specifically activated by upstream STRESS-INDUCED MAPKK (SIMKK; Kiegerl et al., 2000; Bekešová et al., 2015). SIMK is localized to nuclei and cytoplasm of root cells, while in developing root hairs it relocated from the nucleus to the growing tip (Šamaj et al., 2002). Moreover, stimulus-dependent activation and the subsequent subcellular relocation of both SIMK and its upstream SIMKK were induced by salt stress (Ovečka et al., 2014). Such activity-dependent and coordinated relocation of SIMK-SIMKK module from the nucleus to cytoplasm under salt stress were observed in alfalfa and thale cress. Transgenic thale cress plants stably producing SIMKK-YFP exhibited enhanced MITOGEN-ACTIVATED PROTEIN KINASE 3 (MPK3) and MITOGEN-ACTIVATED PROTEIN KINASE 6 (MPK6) activation and conferred altered sensitivity to salt stress. These data suggested that SIMKK may serve as a negative regulator of the salt stress response in alfalfa (Ovečka et al., 2014).
Conclusion and Perspectives
Alfalfa is a perennial, cross-pollinated, autotetraploid (2n = 4x = 32) plant with genome size of 800–900 Mbp. It is often mentioned as the “queen of forages” due to the very high production potential as hay, silage or as a biofuel feedstock for ethanol production (Blondon et al., 1994). However, tetraploid nature made understanding and improving of alfalfa by traditional breeding methods rather challenging. Therefore, the use of modern biotechnological, omics and genetic engineering approaches for alfalfa improvement is highly actual and desirable task for crop researchers.
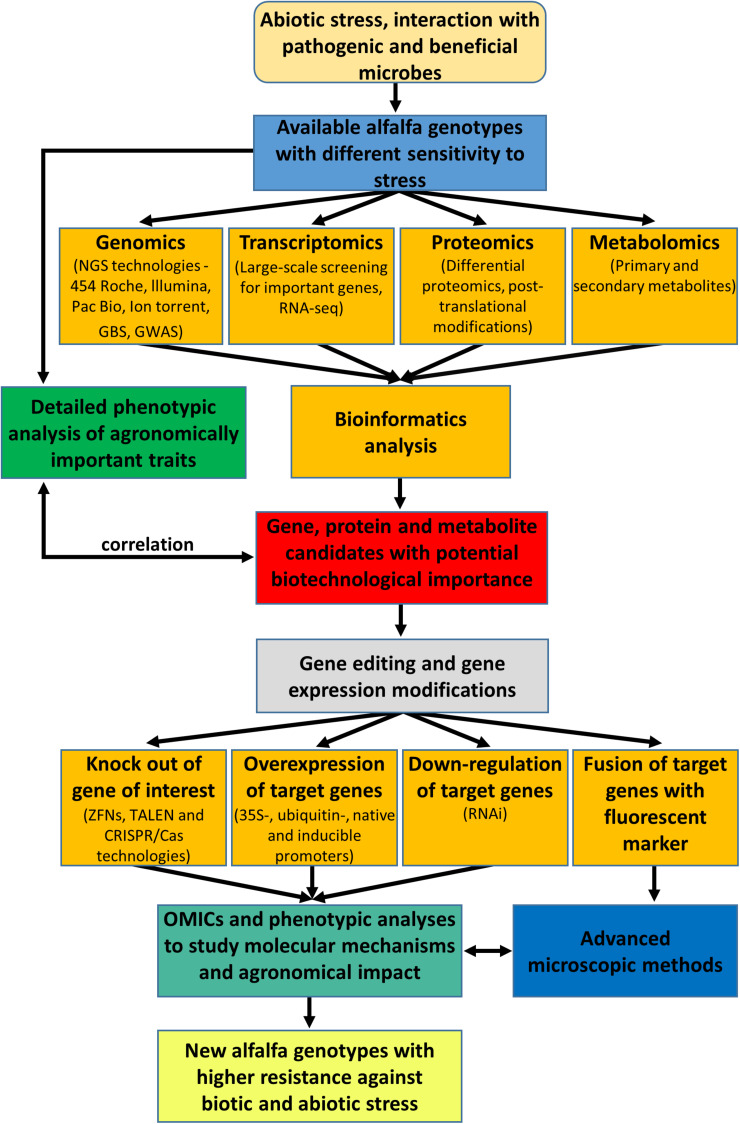
This review provides an overview of the biotechnological potential of alfalfa based on the integration of various omics and molecular tools as depicted in the Figure 1. Recent advances in high-throughput sequencing technology have opened another scientific boundary, and many species, including economically important crops, have been subjected to whole-genome sequencing by de novo assembly and resequencing. Several novel genes have been identified owing to whole-genome duplications and structural variations in chromosomes (Van et al., 2013). Since plant responses to stresses are often very specific, proteomic and transcriptomic approaches should be targeted to individual cell types and tissues at different developmental stages. Such approach was already reported for root hairs and root border cells of barrel medic (Breakspear et al., 2014; Watson et al., 2015). In this respect, the integration of fast-developing omics methods and bioinformatics into systems biology at the single cell level might bring new opportunities to improve plant stress tolerance (Libault et al., 2017).
FIGURE 1.
Overview and integration of omics and molecular genetics approaches aiming to improve agronomic traits and performance of alfalfa.
Biotechnological approaches provide a great potential to increase crop production for the constantly growing global population. Introducing tolerance to environmental abiotic and biotic stresses is crucial for improving the productivity of crop legumes (Farooq et al., 2017). Extensive research conducted on alfalfa stress tolerance suggests that it is able to cope with abiotic stresses using general mechanisms such as antioxidant defense, protein folding, and cell wall remodeling. Research in the field of alfalfa biotechnology also aimed to identify genes involved in the energy production pathway or in enhancing environmental tolerance (Pennycooke et al., 2008; Aranjuelo et al., 2011; Mo et al., 2011). Scientists grew alfalfa plants under different conditions in order to analyze gene expression profiles and to identify crucial genes and proteins, as well as to understand global correlations between genes, proteins, and metabolites using omics approaches.
The potentials of these methods have only partially been exploited in alfalfa research. Continued research toward the development of alfalfa proteome studies (Komatsu and Ahsan, 2009) should permit the rapid comparison of alfalfa cultivars, mutants, and transgenic lines.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. MH, PD, TT, MT, and IL drafted the review which was coordinated by OŠ, MO, and finally edited by JŠ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
Funding. This work was funded by the European Regional Development Fund, European Union (ERDF) project “Plants as a tool for sustainable global development” (CZ.02.1.01/0.0/0.0/16_019/0000827).
References
- Abdelrahman M., El-Sayed M., Sato S., Hirakawa H., Ito S. I., Tanaka K., et al. (2017a). RNA-sequencing-based transcriptome and biochemical analyses of steroidal saponin pathway in a complete set of Allium fistulosum-A. cepa monosomic addition lines. PLoS One 12:e0181784 10.1371/journal.pone.0181784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelrahman M., Suzumura N., Mitoma M., Matsuo S., Ikeuchi T., Mori M., et al. (2017b). Comparative de novo transcriptome profiles in Asparagus officinalis and A. kiusianus during the early stage of Phomopsis asparagi infection. Sci. Rep. 7:2608 10.1038/s41598-017-02566-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelrahman M., Jogaiah S., Burritt D. J., Tran L. S. P. (2018). Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ. 41 1972–1983. 10.1111/pce.13123 [DOI] [PubMed] [Google Scholar]
- Abdelrahman M., Sawada Y., Nakabayashi R., Sato S., Hirakawa H., El-Sayed M., et al. (2015). Integrating transcriptome and target metabolome variability in doubled haploids of Allium cepa for abiotic stress protection. Mol. Breed. 35:195 10.1007/s11032-015-0378-2 [DOI] [Google Scholar]
- An Y. M., Song L. L., Liu Y. R., Shu Y. J., Guo C. H. (2016). De novo transcriptional analysis of alfalfa in response to saline-alkaline stress. Front. Plant Sci. 7:931 10.3389/fpls.2016.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annicchiarico P., Barrett B., Brummer E. C., Julier B., Marshall A. H. (2015). Achievements and challenges in improving temperate perennial forage legumes. Crit. Rev. Plant Sci. 34 327–380. 10.1080/07352689.2014.898462 [DOI] [Google Scholar]
- Annicchiarico P., Nazzicari N., Brummer E. C. (2016). ““Alfalfa genomic selection: challenges, strategies, transnational cooperation“,” in Breeding in a World of Scarcity, eds Roldán-Ruiz I., Baert J., Reheul D. (Cham: Springer; ), 145–149. 10.1007/978-3-319-28932-8_22 [DOI] [Google Scholar]
- Aranjuelo I., Molero G., Erice G., Avice J. C., Nogués S. (2011). Plant physiology and proteomics reveals the leaf response to drought in alfalfa (Medicago sativa L.). J. Exp. Bot. 62 111–123. 10.1093/jxb/erq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranjuelo I., Perez P., Hernandez L., Irigoyen J. J., Zita G., Martinez-Carrasco R., et al. (2005). The response of nodulated alfalfa to water supply, temperature and elevated CO2: photosynthetic downregulation. Physiol. Plant. 123 348–358. 10.1111/j.1399-3054.2005.00459.x [DOI] [Google Scholar]
- Aranjuelo I., Tcherkez G., Molero G., Gilard F., Avice J.-C., Nogués S. (2013). Concerted changes in N and C primary metabolism in alfalfa (Medicago sativa) under water restriction. J. Exp. Bot. 64 885–897. 10.1093/jxb/ers367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ari S̨, Arikan M. (2016). “Next-generation sequencing: advantages, disadvantages, and future,” in Plant Omics: Trends and Applications, eds Hakeem K., Tombuloglu H., Tombuloglu G. (Cham: Springer; ), 109–135. 10.1007/978-3-319-31703-8_5 [DOI] [Google Scholar]
- Arshad M., Feyissa B. A., Amyot L., Aung B., Hannoufa A. (2017). MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 258 122–136. 10.1016/j.plantsci.2017.01.018 [DOI] [PubMed] [Google Scholar]
- Arshad M., Gruber M. Y., Hannoufa A. (2018). Transcriptome analysis of microRNA156 overexpression alfalfa roots under drought stress. Sci. Rep. 8:9363 10.1038/s41598-018-27088-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E., Nakamura Y., Sato S., Tabata S. (2004). Characteristics of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis. Plant Mol. Biol. 54 405–414. 10.1023/B:PLAN.0000036372.46942.b8 [DOI] [PubMed] [Google Scholar]
- Aung B., Gao R., Gruber M. Y., Yuan Z. C., Sumarah M., Hannoufa A. (2017). MsmiR156 affects global gene expression and promotes root regenerative capacity and nitrogen fixation activity in alfalfa. Transgenic Res. 26 541–557. 10.1007/s11248-017-0024-3 [DOI] [PubMed] [Google Scholar]
- Aung B., Gruber M. Y., Amyot L., Omari K., Bertrand A., Hannoufa A. (2015). Micro RNA 156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13 779–790. 10.1111/pbi.12308 [DOI] [PubMed] [Google Scholar]
- Aziz N., Paiva N. L., May G. D., Dixon R. A. (2005). Transcriptome analysis of alfalfa glandular trichomes. Planta 221 28–38. 10.1007/s00425-004-1424-1 [DOI] [PubMed] [Google Scholar]
- Bahramnejad B., Goodwin P. H., Zhang J., Atnaseo C., Erickson L. R. (2010). A comparison of two class 10 pathogenesis-related genes from alfalfa and their activation by multiple stresses and stress-related signaling molecules. Plant Cell Rep. 29 1235–1250. 10.1007/s00299-010-0909-6 [DOI] [PubMed] [Google Scholar]
- Baldacci-Cresp F., Chang C., Maucourt M., Deborde C., Hopkins J., Lecomte P., et al. (2012). Homoglutathione deficiency impairs root-knot nematode development in Medicago truncatula. PLoS Pathog. 8:2471 10.1371/journal.ppat.1002471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao A., Chen H., Chen L., Chen S., Hao Q., Guo W., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 19:131 10.1186/s12870-019-1746-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabaschi D., Guerra D., Lacrima K., Laino P., Michelotti V., Urso S., et al. (2012). Emerging knowledge from genome sequencing of crop species. Mol. Biotech. 50 250–266. 10.1007/s12033-011-9443-1 [DOI] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Barros J., Temple S., Dixon R. A. (2019). Development and commercialization of reduced lignin alfalfa. Curr. Opin. Biotech. 56 48–54. 10.1016/j.copbio.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Bekešová S., Komis G., Křenek P., Vyplelová P., Ovečka M., Luptovčiak I., et al. (2015). Monitoring protein phosphorylation by acrylamide pendant Phos-TagTM in various plants. Front. Plant Sci. 6:336 10.3389/fpls.2015.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj K., Chaparro-Garcia A., Kamoun S., Nekrasov V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39 10.1186/1746-4811-9-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. W., Uauy C., Wulff B. B., Zhou J., Krasileva K., Clark M. D. (2017). Genomic innovation for crop improvement. Nature 543 346–354. 10.1038/nature22011 [DOI] [PubMed] [Google Scholar]
- Biazzi E., Nazzicari N., Pecetti L., Brummer E. C., Palmonari A., Tava A., et al. (2017). Genome-wide association mapping and genomic selection for alfalfa (Medicago sativa) forage quality traits. PLoS One 12:e0169234 10.1371/journal.pone.0169234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondon F., Marie D., Brown S., Kondorosi A. (1994). Genome size and base composition in Medicago sativa and M. truncatula species. Genome 37 264–270. 10.1139/g94-037 [DOI] [PubMed] [Google Scholar]
- Bohnert H. J., Jensen R. G. (1996). Strategies for engineering water-stress tolerance in plants. Trends Biotechnol. 14 89–97. 10.1016/0167-7799(96)80929-2 [DOI] [Google Scholar]
- Bora K. S., Sharma A. (2011). Phytochemical and pharmacological potential of Medicago sativa: a review. Pharm. Biol. 49 211–220. 10.3109/13880209.2010.504732 [DOI] [PubMed] [Google Scholar]
- Borsics T., Lados M. (2002). Dodder infection induces the expression of a pathogenesis-related gene of the family PR-10 in alfalfa. J. Exp. Bot. 53 1831–1832. 10.1093/jxb/erf039 [DOI] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26 4680–4701. 10.1105/tpc.114.133496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer D. J., Osborn T. C. (1999). A molecular marker linkage map of tetraploid alfalfa (Medicago sativa L.). Theor. Appl. Gen. 99 1194–1200. 10.1007/s001220051324 [DOI] [Google Scholar]
- Brummer E. C., Bouton J. H., Kochert G. (1993). Development of an RFLP map in diploid alfalfa. Theor. Appl. Gen. 86 329–332. 10.1007/BF00222097 [DOI] [PubMed] [Google Scholar]
- Budak H., Kantar M., Bulut R., Akpinar B. A. (2015). Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 235 1–13. 10.1016/j.plantsci.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Bustos-Sanmamed P., Mao G., Deng Y., Elouet M., Khan G. A., Bazin J., et al. (2013). Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct. Plant Biol. 40 1208–1220. 10.1071/FP13123 [DOI] [PubMed] [Google Scholar]
- Cai Y., Chen L., Liu X., Sun S., Wu C., Jiang B., et al. (2015). CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS One 10:e0136064 10.1371/journal.pone.0136064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale F., Meskiene I., Ouaked F., Hirt H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14 703–711. 10.1105/tpc.010256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P. R., Sheaffer C. C. (1983). Alfalfa response to soil water deficits. III. Nodulation and N2 fixation. Crop Sci. 23 985–990. 10.2135/cropsci1983.0011183X002300050041x [DOI] [Google Scholar]
- Chao Y., Yuan J., Guo T., Xu L., Mu Z., Han L. (2019). Analysis of transcripts and splice isoforms in Medicago sativa L. by single-molecule long-read sequencing. Plant Mol. Biol. 99 219–235. 10.1007/s11103-018-0813-y [DOI] [PubMed] [Google Scholar]
- Chen J., Han G., Shang C., Li J., Zhang H., Liu F., et al. (2015). Proteomic analyses reveal differences in cold acclimation mechanisms in freezing-tolerant and freezing-sensitive cultivars of alfalfa. Front. Plant Sci. 6:105 10.3389/fpls.2015.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wang Y., Zhang R., Zhang H., Gao C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70 667–697. 10.1146/annurev-arplant-050718-100049 [DOI] [PubMed] [Google Scholar]
- Chen L., Chen Q., Zhu Y., Hou L., Mao P. (2016). Proteomic identification of differentially expressed proteins during alfalfa (Medicago sativa L.) flower development. Front. Plant Sci. 7:1502 10.3389/fpls.2016.01502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. H., Murata N. (2002). Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr. Opin. Plant Biol. 5 250–257. 10.1016/s1369-5266(02)00255-8 [DOI] [PubMed] [Google Scholar]
- Chen T. H., Murata N. (2008). Glycinebetaine: an effective protectant against abiotic stress in plants. Trends Plant Sci. 13 499–505. 10.1016/j.tplants.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Cheung F., Haas B. J., Goldberg S. M. D., May G. D., Xiao Y., Town C. D. (2006). Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genom. 7:272 10.1186/1471-2164-7-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., Hummel A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186 757–761. 10.1534/genetics.110.120717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S. J., Xiong Y., Michno J. M., Campbell B. W., Stec A. O., Čermák T., et al. (2018). CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotech. J. 16 1125–1137. 10.1111/pbi.12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Cui W., Pan J., Xie Y., Wang J., Shen W. (2017). Proteomic analysis provides insights into the molecular bases of hydrogen gas-induced cadmium resistance in Medicago sativa. J. Proteom. 152 109–120. 10.1016/j.jprot.2016.10.013 [DOI] [PubMed] [Google Scholar]
- de Zélicourt A., Diet A., Marion J., Laffont C., Ariel F., Moison M., et al. (2011). Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J. 70 220–230. 10.1111/j.1365-313X.2011.04859.x [DOI] [PubMed] [Google Scholar]
- Demorest Z. L., Coffman A., Baltes N. J., Stoddard T. J., Clasen B. M., Luo S., et al. (2016). Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol. 16:225 10.1186/s12870-016-0906-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deokar A. A., Kondawar V., Jain P. K., Karuppayil S. M., Raju N. L., Vadez V., et al. (2011). Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and -susceptible genotypes of chickpea under terminal drought stress. BMC Plant Biol. 11:70 10.1186/1471-2229-11-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Vivancos P., de Simone A., Kiddle G., Foyer C. H. (2015). Glutathione–linking cell proliferation to oxidative stress. Free Radical Biol. Med. 89 1154–1164. 10.1016/j.freeradbiomed.2015.09.023 [DOI] [PubMed] [Google Scholar]
- Dong L., Liu H., Zhang J., Yang S., Kong G., Chu J. S., et al. (2015). Single-molecule real-time transcript sequencing facilitates common wheat genome annotation and grain transcriptome research. BMC Genom. 16:1039 10.1186/s12864-015-2257-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Luckow M. A. (2003). The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 131 900–910. 10.1104/pp.102.018150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Shi Y., Li D., Fan W., Wang Y., Wang G., et al. (2018). Proteomics reveals key proteins participating in growth difference between fall dormant and non-dormant alfalfa in terminal buds. J. Proteom. 173 126–138. 10.1016/j.jprot.2017.11.029 [DOI] [PubMed] [Google Scholar]
- Du H., Zeng X., Zhao M., Cui X., Wang Q., Yang H., et al. (2016). Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 217 90–97. 10.1016/j.jbiotec.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Ebert J. (2007). Alfalfa’s bioenergy appeal. Ethanol Prod. Mag. 88–94. [Google Scholar]
- Echt C. S., Kidwell K. K., Knapp S. J., Osborn T. C., McCoy T. J. (1994). Linkage mapping in diploid alfalfa (Medicago sativa). Genome 37 61–71. 10.1139/g94-008 [DOI] [PubMed] [Google Scholar]
- Edwards D., Batley J. (2010). Plant genome sequencing: applications for crop improvement. Plant Biotechnol. J. 8 2–9. 10.1111/j.1467-7652.2009.00459.x [DOI] [PubMed] [Google Scholar]
- Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science 323 133–138. 10.1126/science [DOI] [PubMed] [Google Scholar]
- Elgin J. H., Jr., Ostazeski S. A. (1985). Inheritance of resistance to race 1 and race 2 anthracnose in Arc and Saranac AR alfalfa. Crop Sci. 25 861–865. 10.2135/cropsci1985.0011183X002500050032x [DOI] [Google Scholar]
- Elgin J. H., Jr., Welty R. E., Gilchrist D. B. (1988). Breeding for disease and nematode resistance. Alfalfa Alfalfa Impr. 29 827–858. 10.2134/agronmonogr29.c27 [DOI] [Google Scholar]
- Esnault R., Buffard D., Breda C., Sallaud C., Turk J., Kondorosi A. (1993). Pathological and molecular characterizations of alfalfa interactions with compatible and incompatible bacteria, Xanthomonas campestris pv. alfalfae and Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 6 655–664. 10.1094/MPMI-6-655 [DOI] [PubMed] [Google Scholar]
- Fan W., Ge G., Liu Y., Wang W., Liu L., Jia Y. (2018). Proteomics integrated with metabolomics: analysis of the internal causes of nutrient changes in alfalfa at different growth stages. BMC Plant Biol. 18:78 10.1186/s12870-018-1291-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M., Gogoi N., Hussain M., Barthakur S., Paul S., Bharadwaj N., et al. (2017). Effects, tolerance mechanisms and management of salt stress in grain legumes. Plant Physiol. Biochem. 118 199–217. 10.1105/10.1016/j.plaphy.2017.06.020 [DOI] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D. L., Wei P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23 1229–1232. 10.1038/cr.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyissa B. A., Arshad M., Gruber M. Y., Kohalmi S. E., Hannoufa A. (2019). The interplay between miR156/SPL13 and DFR/WD40–1 regulate drought tolerance in alfalfa. BMC plant Biol. 19:2059 10.1186/s12870-019-2059-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajoulot S., Ronfort J., Baudouin P., Barre P., Huguet T., Huyghe C., et al. (2005). Genetic diversity among alfalfa (Medicago sativa) cultivars coming from a breeding program, using SSR markers. Theor. Appl. Genet. 111 1420–1429. 10.1007/s00122-005-0074-4 [DOI] [PubMed] [Google Scholar]
- Fleming M. B., Patterson E. L., Reeves P. A., Richards C. M., Gaines T. A., Walters C. (2018). Exploring the fate of mRNA in aging seeds: protection, destruction, or slow decay? J. Exp. Bot. 69 4309–4321. 10.1093/jxb/ery215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo P., Harrison J., Norman C., Jiménez M. J. H. (2005). Glutathione and homoglutathione play a critical role in the nodulation process of Medicago truncatula. Mol. Plant-Mic. Int. 18 254–259. 10.1094/MPMI-18-0254 [DOI] [PubMed] [Google Scholar]
- Fu G., Grbic V., Ma S., Tian L. (2015). Evaluation of somatic embryos of alfalfa for recombinant protein expression. Plant Cell Rep. 34 211–221. 10.1007/s00299-014-1700-x [DOI] [PubMed] [Google Scholar]
- Fukuda A., Nakamura A., Tanaka Y. (1999). Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochem. Biophys. Acta 1446 149–155. 10.1016/s0167-4781(99)00065-2 [DOI] [PubMed] [Google Scholar]
- Fürstenberg-Hägg J., Zagrobelny M., Bak S. (2013). Plant defense against insect herbivores. Int. J. Mol. Sci. 14 10242–10297. 10.3390/ijms140510242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Feyissa B. A., Croft M., Hannoufa A. (2018). Gene editing by CRISPR/Cas9 in the obligatory outcrossing Medicago sativa. Planta 247 1043–1050. 10.1007/s00425-018-2866-1 [DOI] [PubMed] [Google Scholar]
- Gao Z., Luo W., Liu H., Zeng C., Liu X., Yi S., et al. (2012). Transcriptome analysis and SSR/SNP markers information of the blunt snout bream (Megalobrama amblycephala). PLoS One 7:42637 10.1371/journal.pone.0042637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A. N., Ayub N. D., Fox A. R., Gómez M. C., Diéguez M. J., Pagano E. M., et al. (2014). Alfalfa snakin-1 prevents fungal colonization and probably coevolved with rhizobia. BMC Plant Biol. 14:248 10.1186/s12870-014-0248-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B., Li X., Bloszies S., Wen D., Sun S., Wei M. (2014). Sodic alkaline stress mitigation by interaction of nitric oxide and polyamines involves antioxidants and physiological strategies in Solanum lycopersicum. Free Radic. Biol. Med. 71 36–48. 10.1016/j.freeradbiomed.2014.02.018 [DOI] [PubMed] [Google Scholar]
- Graham D. B., Root D. E. (2015). Resources for the design of CRISPR gene editing experiments. Genome Biol. 16:26 10.1186/s13059-015-0823-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsch A., Keunen E., Guerriero G., Renaut J., Cuypers A., Hausman J. F., et al. (2018b). Long-term cadmium exposure influences the abundance of proteins that impact the cell wall structure in Medicago sativa stems. Plant Biol. 20 1023–1035. 10.1111/plb.12865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsch A., Zouaghi S., Renaut J., Cuypers A., Hausman J. F., Sergeant K. (2018a). Changes in the proteome of Medicago sativa leaves in response to long-term cadmium exposure using a cell-wall targeted approach. Int. J. Mol. Sci. 19:2498 10.3390/ijms19092498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Rodriguez J. J., Ibarra-Laclette E., Herrera-Estrella L., Ochoa-Zarzosa A., Suarez-Rodriguez L. M., Rodriguez-Zapata L. C., et al. (2013). Analysis of expressed sequence tags (ESTs) from avocado seed (Persea americana var. drymifolia) reveals abundant expression of the gene encoding the antimicrobial peptide snakin. Plant Physiol. Biochem. 70 318–324. 10.1016/j.plaphy.2013.05.045 [DOI] [PubMed] [Google Scholar]
- Ha C. V., Watanabe Y., Tran U. T., Le D. T., Tanaka M., Nguyen K. H., et al. (2015). Comparative analysis of root transcriptomes from two contrasting drought-responsive Williams 82 and DT2008 soybean cultivars under normal and dehydration conditions. Front. Plant Sci. 6:551 10.3389/fpls.2015.00551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Kang Y., Torres-Jerez I., Cheung F., Town C. D., Zhao P. X., et al. (2011). Genome-wide SNP discovery in tetraploid alfalfa using 454 sequencing and high resolution melting analysis. BMC Genom. 12:350 10.1186/1471-2164-12-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlerode A. J., Scully R. (2009). Mechanisms of double-strand break repair in somatic mammalian cells. Biochem. J. 423 157–168. 10.1042/BJ20090942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun W., Coffman A., Clasen B. M., Demorest Z. L., Lowy A., Ray E., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12 934–940. 10.1111/pbi.12201 [DOI] [PubMed] [Google Scholar]
- Hawkins C., Yu L. X. (2018). Recent progress in alfalfa (Medicago sativa L.) genomics and genomic selection. Crop J. 6 565–575. 10.1016/j.cj.2018.01.006 [DOI] [Google Scholar]
- He X. Z., Dixon R. A. (2000). Genetic manipulation of isoflavone 7-O-methyltransferase enhances biosynthesis of 4’-O-methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell 12 1689–1702. 10.1105/tpc.12.9.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann D., Flajoulot S., Barre P., Huyghe C., Ronfort J., Julier B. (2018). Comparison of morphological traits and molecular markers to analyse diversity and structure of alfalfa (Medicago sativa L.) cultivars. Gen. Res. Crop Evol. 65 527–540. 10.1007/s10722-017-0551-z [DOI] [Google Scholar]
- Hipskind J. D., Paiva N. L. (2000). Constitutive accumulation of a resveratrol-glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol. Plant Microbe Int. 13 551–556. 10.1094/MPMI.2000.13.5.551 [DOI] [PubMed] [Google Scholar]
- Huang X., Kurata N., Wang Z. X., Wang A., Zhao Q., Zhao Y., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490 497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E. Y., Song Q., Jia G., Specht J. E., Hyten D. L., Costa J., et al. (2014). A genome-wide association study of seed protein and oil content in soybean. BMC Genom. 15:2164 10.1186/1471-2164-15-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T. B., LaFayette P. R., Schmitz R. J., Parrott W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16 10.1186/s12896-015-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaganathan D., Ramasamy K., Sellamuthu G., Jayabalan S., Venkataraman G. (2018). CRISPR for crop improvement: an update review. Front. Plant Sci. 9:985 10.3389/fpls.2018.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Olsen H. E., Paten B., Akeson M. (2016). The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Gen. Biol. 17:239 10.1186/s13059-016-1103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Zhu K., Cui W., Xie Y., Han B., Shen W. (2013). Hydrogen gas acts as a novel bioactive molecule in enhancing plant tolerance to paraquat-induced oxidative stress via the modulation of heme oxygenase-1 signalling system. Plant Cell Environ. 36 956–969. 10.1111/pce.12029 [DOI] [PubMed] [Google Scholar]
- Jin T., Chang Q., Li W., Yin D., Li Z., Wang D., et al. (2010). Stress-inducible expression of GmDREB1 conferred salt tolerance in transgenic alfalfa. Plant Cell Trans. Organ Cult. 100 219–227. 10.1007/s11240-009-9628-5 [DOI] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Julier B., Flajoulot S., Barre P., Cardinet G., Santoni S., Huguet T., et al. (2003). Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant B. 3:9 10.1186/1471-2229-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R., Hoeijmakers J. H., van Gent D. C. (1998). Molecular mechanisms of DNA double strand break repair. Trends Cell. Biol. 8 483–489. 10.1016/S0962-8924(98)01383-X [DOI] [PubMed] [Google Scholar]
- Kang P., Bao A. K., Kumar T., Pan Y. Q., Bao Z., Wang F., et al. (2016). Assessment of stress tolerance, productivity, and forage quality in T1 transgenic alfalfa co-overexpressing ZxNHX and ZxVP1-1 from Zygophyllum xanthoxylum. Front. Plant Sci. 7:1598 10.3389/fpls.2016.01598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Han Y., Torres-Jerez I., Wang M., Tang Y., Monteros M., et al. (2011). System responses to long-term drought and re-watering of two contrasting alfalfa varieties. Plant J. 68 871–889. 10.1111/j.1365-313X.2011.04738.x [DOI] [PubMed] [Google Scholar]
- Kang Y., Sakiroglu M., Krom N., Stanton-Geddes J., Wang M., Lee Y. C., et al. (2015). Genome-wide association of drought-related and biomass traits with HapMap SNPs in Medicago truncatula. Plant Cell Environ. 38 1997–2011. 10.1111/pce.12520 [DOI] [PubMed] [Google Scholar]
- Kersey P. J. (2019). Plant genome sequences: past, present, future. Curr. Opin. Plant Biol. 48 1–8. 10.1016/j.pbi.2018.11.001 [DOI] [PubMed] [Google Scholar]
- Khan Z., Khan S. H., Mubarik M. S., Sadia B., Ahmad A. (2017). Use of TALEs and TALEN technology for genetic improvement of plants. Plant Mol. Biol. Rep. 35 1–19. 10.1007/s11105-016-0997-8 [DOI] [Google Scholar]
- Khu D. M., Reyno R., Han Y., Zhao P. X., Bouton J. H., Brummer E. C., et al. (2013). Identification of aluminum tolerance quantitative trait loci in tetraploid alfalfa. Crop Sci. 53 148–163. 10.2135/cropsci2012.03.0181 [DOI] [Google Scholar]
- Kiegerl S., Cardinale F., Siligan C., Gross A., Baudouin E., Liwosz A., et al. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress–induced MAPK, SIMK. Plant Cell 12 2247–2258. 10.1105/tpc.12.11.2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Y., Kwon S. Y., Lee H. S., Hur Y., Bang J. W., Kwak S. S. (2003). A novel oxidative stress-inducible peroxidase promoter from sweetpotato: molecular cloning and characterization in transgenic tobacco plants and cultured cells. Plant Mol. Biol. 51 831–838. 10.1023/a:1023045218815 [DOI] [PubMed] [Google Scholar]
- Kiss G. B., Csanádi G., Kálmán K., Kaló P., Ökrész L. (1993). Construction of a basic genetic map for alfalfa using RFLP, RAPD, isozyme and morphological markers. Mol. Gen. Gen. 238 129–137. 10.1007/BF00279539 [DOI] [PubMed] [Google Scholar]
- Klapheck S. (1988). Homoglutathione: isolation, quantification and occurrence in legumes. Physiol. Plant. 74 727–732. 10.1111/j.1399-3054.1988.tb02044.x [DOI] [Google Scholar]
- Komatsu S., Ahsan N. (2009). Soybean proteomics and its application to functional analysis. J. Proteomics 72 325–336. 10.1016/j.jprot.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Komis G., Illés P., Beck M., Šamaj J. (2011). Microtubules and mitogen-activated protein kinase signalling. Curr. Oppin. Plant Biol. 14 650–657. 10.1016/j.pbi.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Köpp M., Passos L., Verneue R., Lédo F. J., Coimbra J. L., de Oliveira A. (2011). Effects of nutrient solution pH on growth parameters of alfalfa (Medicago sativa L.) genotypes. Comun. Sci.e 2 135–141. 10.14295/cs.v2i3.39 [DOI] [Google Scholar]
- Korver R. A., Koevoets I. T., Testerink C. (2018). Out of shape during stress: a key role for auxin. Trends Plant Sci. 23 783–793. 10.1016/j.tplants.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalskaya N., Hammond R. W. (2009). Expression and functional characterization of the plant antimicrobial snakin-1 and defensin recombinant proteins. Protein Expr. Purif. 63 12–17. 10.1016/j.pep.2008.08.013 [DOI] [PubMed] [Google Scholar]
- Kudapa H., Ramalingam A., Nayakoti S., Chen W., Zhuang W., Liang X., et al. (2013). Functional genomics to study stress responses in crop legumes: progress and prospects. Funct. Plant Biol. 40 1221–1233. 10.1071/FP13191 [DOI] [PubMed] [Google Scholar]
- Kuluev B. R., Gumerova G. R., Mikhaylova E. V., Gerashchenkov G. A., Rozhnova N. A., Vershinina Z. R., et al. (2019). Delivery of CRISPR/Cas components into higher plant cells for genome editing. Russ. J. Plant. Physl. 66 694–706. 10.1134/S102144371905011X [DOI] [Google Scholar]
- Kumar T., Bao A. K., Bao Z., Wang F., Gao L., Wang S. M. (2018). The progress of genetic improvement in alfalfa (Medicago sativa L.). Czech. J. Genet. Plant Breed. 54 41–51. 10.17221/46/2017-CJGPB [DOI] [Google Scholar]
- Laberge S., Castonguay Y., Vézina L. P. (1993). New cold-and drought-regulated gene from Medicago sativa. Plant Physiol. 101 1411–1411. 10.1104/pp.101.4.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardi M., Pessi G. (2018). Functional genomics approaches to studying symbioses between legumes and nitrogen-fixing rhizobia. High Throughput. 7:15 10.3390/ht7020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le B. H., Wagmaister J. A., Kawashima T., Bui A. Q., Harada J. J., Goldberg R. B. (2007). Using genomics to study legume seed development. Plant Physiol. 144 562–574. 10.1104/pp.107.100362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D. T., Nishiyama R., Watanabe Y., Tanaka M., Seki M., Ham L. H., et al. (2012). Differential gene expression in soybean leaf tissues at late developmental stages under drought stress revealed by genome-wide transcriptome analysis. PLoS One 7:e49522 10.1371/journal.pone.0049522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Nguyen K., Grondin A., Courtois B., Gantet P. (2018). Next-generation sequencing accelerates crop gene discovery. Trends Plant Sci. 24 8 10.1016/j.tplants.2018.11.008 [DOI] [PubMed] [Google Scholar]
- Lei Y., Xu Y., Hettenhausen C., Lu C., Shen G., Zhang C., et al. (2018). Comparative analysis of alfalfa (Medicago sativa L.) leaf transcriptomes reveals genotype-specific salt tolerance mechanisms. BMC Plant Biol. 18:35 10.1186/s12870-018-1250-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang Z., Ke Q., Ji C. Y., Jeong J. C., Lee H. S., et al. (2014). Overexpression of codA gene confers enhanced tolerance to abiotic stresses in alfalfa. Plant Physiol. Biochem. 85 31–40. 10.1016/j.plaphy.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Li J. F., Norville J. E., Aach J., McCormack M., Zhang D., Bush J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31 688–691. 10.1038/nbt.2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Jia S., Hou L., Nguyen H., Sato S., Holding D., et al. (2019). Mapping of transgenic alleles in soybean using a nanopore-based sequencing strategy. J. Exp. Bot. 70 3825–3833. 10.1093/jxb/erz202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Jiang W. Z., Wright D., Spalding M. H., Weeks D. P., et al. (2011). TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 39 359–372. 10.1093/nar/gkq704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wei Z., Qiao Z., Wu Z., Cheng L., Wang Y. (2013). Proteomics analysis of alfalfa response to heat stress. PLoS One 8:e82725 10.1371/journal.pone.0082725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Brummer E. C. (2012). Applied genetics and genomics in alfalfa breeding. Agronomy 2 40–61. 10.3390/agronomy2010040 [DOI] [Google Scholar]
- Li X., Hannoufa A., Zhang Y., Yu P. (2016). Gene-silencing-induced changes in carbohydrate conformation in relation to bioenergy value and carbohydrate subfractions in modeled plant (Medicago sativa) with down-regulation of HB12 and TT8 transcription factors. Int. J. Mol. Sci. 17:720 10.3390/ijms17050720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wei Y., Acharya A., Jiang Q., Kang J., Brummer E. C. (2014). A saturated genetic linkage map of autotetraploid alfalfa (Medicago sativa L.) developed using genotyping-by-sequencing is highly syntenous with the Medicago truncatula genome. G3 4 1971–1979. 10.1534/g3.114.012245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu Z. B., Xing A., Moon B. P., Koellhoffer J. P., Huang L., et al. (2015). Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 169 960–970. 10.1104/pp.15.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Pingault L., Zogli P., Schiefelbein J. (2017). Plant systems biology at the single-cell level. T. Plant Sci. 22 949–960. 10.1016/j.tplants.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Liu H., Ding Y., Zhou Y., Jin W., Xie K., Chen L. L. (2017). CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10 530–532. 10.1016/j.molp.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Liu W., Xiong C., Yan L., Zhang Z., Ma L., Wang Y., et al. (2017). Transcriptome analyses reveal candidate genes potentially involved in al stress response in alfalfa. Front. Plant Sci. 8:26 10.3389/fpls.2017.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wu S., Xu J., Sui C., Wei J. (2019). Application of CRISPR/Cas9 in plant biology. Acta Pharm. Sin. B. 7 292–302. 10.1016/j.apsb.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. P., Hawkins C., Peel M. D., Yu L. X. (2019). Genetic loci associated with salt tolerance in advanced breeding populations of tetraploid alfalfa using genome-wide association studies. Plant Genome 12:26 10.3835/plantgenome2018.05.0026 [DOI] [PubMed] [Google Scholar]
- Liu Z., Chen T., Ma L., Zhao Z., Zhao P. X., Nan Z., et al. (2013). Global transcriptome sequencing using the Illumina platform and the development of EST-SSR markers in autotetraploid alfalfa. PLoS One 8:e83549 10.1371/journal.pone.0083549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R., Gao Y., Sun H., Zhang T., Li X., Li M., et al. (2018). Quantitative proteomic analysis using iTRAQ to identify salt-responsive proteins during the germination stage of two Medicago species. Sci. Rep. 8:9553 10.1038/s41598-018-27935-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R., Li M., Zhang T., Kang J., Sun Y., Cong L., et al. (2016). Comparative proteomic analysis reveals differential root proteins in Medicago sativa and Medicago truncatula in response to salt stress. Front. Plant Sci. 7:424 10.3389/fpls.2016.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Giordano F., Ning Z. (2016). Oxford Nanopore MinION sequencing and genome assembly. Genom. Proteom. Bioinf. 14 265–279. 10.1016/j.gpb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Wu Y., Liu J., Zhou Q., Liu W., Wang Y., et al. (2019a). Comparative transcriptomic and physiological analyses of Medicago sativa L. indicates that multiple regulatory networks are activated during continuous aba treatment. Int. J. Mol. Sci. 20:E47 10.3390/ijms20010047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Zhou Q., Wu Y., Chai X., Liu W., Wang Y., et al. (2019b). Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago sativa L.). BMC Plant Biol. 19:32 10.1186/s12870-019-1630-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Lin L., Hill R. D., Mohapatra S. S. (1991). Primary structure of an environmental stress and abscisic acid-inducible alfalfa protein. Plant Mol. Biol. 17 1267–1269. 10.1007/bf00028745 [DOI] [PubMed] [Google Scholar]
- Luo M., Liu J. H., Mohapatra S., Hill R. D., Mohapatra S. S. (1992). Characterization of a gene family encoding abscisic acid-and environmental stress-inducible proteins of alfalfa. J. Biol. Chem. 267 15367–15374. [PubMed] [Google Scholar]
- Ma Q., Kang J., Long R., Zhang T., Xiong J., Zhang K., et al. (2017). Comparative proteomic analysis of alfalfa revealed new salt and drought stress-related factors involved in seed germination. Mol. Biol. Rep. 44 261–272. 10.1007/s11033-017-4104-5 [DOI] [PubMed] [Google Scholar]
- Mackie J. M., Musial J. M., Armour D. J., Phan H. T. T., Ellwood S. E. (2007). Identification of QTL for reaction to three races of Colletotrichum trifolii and further analysis of inheritance of resistance in autotetraploid lucerne. Theor. Appl. Genet. 114 1417–1426. 10.1007/s00122-007-0527-z [DOI] [PubMed] [Google Scholar]
- Macovei A., Gill S. S., Tuteja N. (2012). microRNAs as promising tools for improving stress tolerance in rice. Plant Sig. Beh. 7 1296–1301. 10.4161/psb.21586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz M. M., Li L., Shamimuzzaman M., Wibowo A., Fang X., Zhu J. K. (2011). De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 108 2623–2628. 10.1073/pnas.1019533108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Zhang F., Koonin E. V. (2017). SnapShot: Class 2 CRISPR-Cas systems. Cell 168 328–328. 10.1016/j.cell.2016.12.038 [DOI] [PubMed] [Google Scholar]
- Malzahn A., Lowder L., Qil Y. (2017). Plant genome editing with TALEN and CRISPR. Cell Biosci. 7:21 10.1186/s13578-017-0148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y. F., Botella J. R., Liu Y. G., Zhu J. K. (2019). Gene editing in plants: progress and challenges. Natl. Sci. Rev. 6 421–437. 10.1093/nsr/nwz005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437 376–380. 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322 1843–1845. 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masonbrink R. E., Severin A. J., Seetharam A. S. (2017). “Comparative genomics of soybean and other legumes,” in The Soybean Genome, eds Nguyen H., Bhattacharyya M. (Cham: Springer; ), 83–93. 10.1007/978-3-319-64198-0_6 [DOI] [Google Scholar]
- Masoud S. A., Zhu Q., Lamb C., Dixon R. A. (1996). Constitutive expression of an inducible β-1,3-glucanase in alfalfa reduces disease severity caused by the oomycete pathogen Phytophthora megasperma f. sp. medicaginis, but does not reduce disease severity of chitin-containing fungi. Transgenic Res. 5 313–323. 10.1007/BF01968941 [DOI] [Google Scholar]
- Matamoros M. A., Moran J. F., Iturbe-Ormaetxe I., Rubio M. C., Becana M. (1999). Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol. 121 879–888. 10.1104/pp.121.3.879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews C., Arshad M., Hannoufa A. (2019). Alfalfa response to heat stress is modulated by microRNA156. Physiol. Plant. 165 830–842. 10.1111/ppl.12787 [DOI] [PubMed] [Google Scholar]
- McCoy T. J., Bingham E. T. (1988). “Cytology and cytogenetics of alfalfa,” in Alfalfa and Alfalfa Improvement, ed. Hanson A. A. (Madison, WI: ASA; ), 737–776. [Google Scholar]
- Meng Y., Wang C., Yin P., Zhu B., Zhang P., Niu L., et al. (2019). “Targeted mutagenesis by an optimized agrobacterium-delivered CRISPR/Cas 9 system in the model legume Medicago truncatula,” in The Model Legume Medicago truncatula, ed. Bruijn F. D. (Hoboken, NJ: Wiley; ), 1015–1018. 10.1002/9781119409144.ch130 [DOI] [Google Scholar]
- Meng Y. Y., Hou Y. L., Wang H., Ji R. H., Liu B., Wen J. Q., et al. (2017). Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula. Plant Cell Rep. 36 371–374. 10.1007/s00299-016-2069-9 [DOI] [PubMed] [Google Scholar]
- Michno J. M., Wang X., Liu J., Curtin S. J., Kono T. J., Stupar R. M. (2015). CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6 243–252. 10.1080/21645698.2015.1106063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., Xia D. F., et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29 143–148. 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- Mittler R., Blumwald E. (2015). The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27 64–70. 10.1105/tpc.114.133090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y., Liang G., Shi W., Xie J. (2011). Metabolic responses of alfalfa (Medicago Sativa L.) leaves to low and high temperature induced stresses. Afr. J. Biotechnol. 10 1117–1124. 10.5897/AJB10.1433 [DOI] [Google Scholar]
- Moradpour M., Abdulah S. N. A. (2020). CRISPR/dCas9 platforms in plants: strategies and applications beyond genome editing. Plant Biotechnol. J. 18 32–44. 10.1111/pbi.13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musial J. M., Mackie J. M., Armour D. J., Phan H. T. T., Ellwood S. E., Aitken K. S., et al. (2007). Identification of QTL for resistance and susceptibility to Stagonospora meliloti in autotetraploid lucerne. Theor. Appl. Gen. 114 1427–1435. 10.1007/s00122-007-0528-y [DOI] [PubMed] [Google Scholar]
- Nakano K., Shiroma A., Shimoji M., Tamotsu H., Ashimine N., Ohki S., et al. (2017). Advantages of genome sequencing by long-read sequencer using SMRT technology in medical area. Hum. Cell 30 149–161. 10.1007/s13577-017-0168-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Staskawicz B., Weigel D., Jones J. D., Kamoun S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31 691–693. 10.1038/nbt.2655 [DOI] [PubMed] [Google Scholar]
- Nekrasov V., Wang C. M., Win J. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7:482 10.1038/s41598-017-00578-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninković S., Miljuš-Ðukić J., Nešković M. (1995). Genetic transformation of alfalfa somatic embryos and their clonal propagation through repetitive somatic embryogenesis. Plant Cell T. Organ Cult. 42 255–260. 10.1007/BF00029996 [DOI] [Google Scholar]
- Nirola R., Megharaj M., Beecham S., Aryal R., Thavamani P., Vankateswarlu K., et al. (2016). Remediation of metalliferous mines, revegetation challenges and emerging prospects in semi-arid and arid conditions. Env. Sci. Poll. Res. 23 20131–20150. 10.1007/s11356-016-7372-z [DOI] [PubMed] [Google Scholar]
- Nutter F. W., Guan J., Gotlieb A. R., Rhodes L. H., Grau C. R., Sulc R. M. (2002). Quantifying alfalfa yield losses caused by foliar diseases in Iowa, Ohio, Wisconsin, and Vermont. Plant Dis. 863 269–277. 10.1094/PDIS.2002.86.3.269 [DOI] [PubMed] [Google Scholar]
- Olukolu B. A., Tracy W. F., Wisser R., De Vries B., Balint-Kurti P. J. (2016). A genome-wide association study for partial resistance to maize common rust. Phytopath. 106 745–751. 10.1094/PHYTO-11-15-0305-R [DOI] [PubMed] [Google Scholar]
- O’Rourke J. A., Fu F., Bucciarelli B., Yang S. S., Samac D. A., Lamb J. F. S., et al. (2015). The Medicago sativa gene index 1.2: a web-accessible gene expression atlas for investigating expression differences between Medicago sativa subspecies. BMC Genom. 16:502 10.1186/s12864-015-1718-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovečka M., Takáč T., Komis G., Vadovič P., Bekešová S., Doskočilová A., et al. (2014). Salt-induced subcellular kinase relocation and seedling susceptibility caused by overexpression of Medicago SIMKK in Arabidopsis. J. Exp. Bot. 65 2335–2350. 10.1093/jxb/eru115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparella S., Araújo S. S., Rossi G., Wijayasinghe M., Carbonera D., Balestrazzi A. (2015). Seed priming: state of the art and new perspectives. Plant Cell Rep. 34 1281–1293. 10.1007/s00299-015-1784-y [DOI] [PubMed] [Google Scholar]
- Pâques F., Duchateau P. (2007). Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 7 49–66. 10.2174/156652307779940216 [DOI] [PubMed] [Google Scholar]
- Pasternak T., Asard H., Potters G., Jansen M. A. (2014). The thiol compounds glutathione and homoglutathione differentially affect cell development in alfalfa (Medicago sativa L.). Plant Phys. Biochem. 74 16–23. 10.1016/j.plaphy.2013.10.028 [DOI] [PubMed] [Google Scholar]
- Pastwa E., Blasiak J. (2003). Non-homologous DNA end joining. Acta. Biochim. Pol. 50 891–908. 10.18388/abp.2003_3622 [DOI] [PubMed] [Google Scholar]
- Paszkowski J., Baur M., Bogucki A., Potrykus I. (1988). Gene targeting in plants. EMBO J. 7 4021–4026. 10.1002/j.1460-2075.1988.tb03295.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovich M. (2017). Computing in biotechnology: omics and beyond. Trends Biotechnol. 35 450–497. 10.1016/j.tibtech.2017.03.011 [DOI] [PubMed] [Google Scholar]
- Pennycooke J. C., Cheng H., Stockinger E. J. (2008). Comparative genomic sequence and expression analyses of Medicago truncatula and alfalfa subspecies falcata COLD-ACCLIMATION-SPECIFIC genes. Plant Physiol. 146 1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano E., Pecetti L. (2010). “Minor legume species,” in Fodder Crops and Amenity Grassesm Handbook of Plant Breeding, Vol. 5 eds Boller B., Posselt U. K., Veronesi F. (NewYork, NY: Springer; ), 477–500. 10.1007/978-1-4419-0760-8_20 [DOI] [Google Scholar]
- Postnikova O. A., Hult M., Shao J., Skantar A., Nemchinov L. G. (2015). Transcriptome analysis of resistant and susceptible alfalfa cultivars infected with root-knot nematode Meloidogyne incognita. PLoS One 10:e0123157 10.1371/journal.pone.0118269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikova O. A., Shao J., Nemchinov L. G. (2013). Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol. 54 1041–1055. 10.1093/pcp/pct056 [DOI] [PubMed] [Google Scholar]
- Pratt R. G., Rowe D. E. (2002). Enhanced resistance to Sclerotium rolfsii in populations of alfalfa selected for quantitative resistance to Sclerotinia trifoliorum. Phytopathology 92 204–209. 10.1094/PHYTO.2002.92.2.204 [DOI] [PubMed] [Google Scholar]
- Printz B., Guerriero G., Sergeant K., Audinot J. N., Guignard C., Renaut J., et al. (2016). Combining-omics to unravel the impact of copper nutrition on alfalfa (Medicago sativa) stem metabolism. Plant Cell Physiol. 57 407–422. 10.1093/pcp/pcw001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printz B., Guerriero G., Sergeant K., Renaut J., Lutts S., Hausman J. F. (2015). Ups and downs in alfalfa: proteomic and metabolic changes occurring in the growing stem. Plant Sci. 238 13–25. 10.1016/j.plantsci.2015.05.014 [DOI] [PubMed] [Google Scholar]
- Puchta H., Dujon B., Hohn B. (1993). Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21 5034–5040. 10.1093/nar/21.22.5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y. (2015). “High efficient genome modification by designed zinc finger nuclease,” in Advances in New Technology for Targeted Modification of Plant Genomes, eds Zhang F., Puchta H., Thomson J. G. (New York, NY: Springer; ), 39–53. 10.1007/978-1-4939-2556-8_3 [DOI] [Google Scholar]
- Radović J., Sokolović D., Marković J. (2009). Alfalfa-most important perennial forage legume in animal husbandry. Biotechnol. Anim. Husb. 25 465–475. 10.2298/BAH0906465R [DOI] [Google Scholar]
- Rahman M. A., Alam I., Kim Y. G., Ahn N. Y., Heo S. H., Lee D. G., et al. (2015). Screening for salt-responsive proteins in two contrasting alfalfa cultivars using a comparative proteome approach. Plant Physiol. Biochem. 89 112–122. 10.1016/j.plaphy.2015.02.015 [DOI] [PubMed] [Google Scholar]
- Rahman M. A., Yong-Goo K., Iftekhar A., Liu G., Hyoshin L., Joo L. J., et al. (2016). Proteome analysis of alfalfa roots in response to water deficit stress. J. Integr. Agric. 15 1275–1285. 10.1016/S2095-3119(15)61255-2 [DOI] [Google Scholar]
- Rashmi R., Sarkar M., Vikramaditya T. (1997). Cultivaton of alfalfa (Medicago sativa L.). Anc. Sci. Life 17 117–119. [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Hanson A. D. (1993). Quaternary ammonium and tertiary sulfonium compounds in higher plants. Ann. Rev. Plant Biol. 44 357–384. 10.1146/annurev.pp.44.060193.002041 [DOI] [Google Scholar]
- Robins J. G., Luth D., Campbell T. A., Bauchan G. R., He C., Viands D. R., et al. (2007). Genetic mapping of biomass production in tetraploid alfalfa. Crop Sci. 47 1–10. 10.2135/cropsci2005.11.0401 [DOI] [Google Scholar]
- Rothberg J. M., Hinz W., Rearick T. M., Schultz J., Mileski W., Davey M., et al. (2011). An integrated semiconductor device enabling non-optical genome sequencing. Nature 475 348–352. 10.1038/nature10242 [DOI] [PubMed] [Google Scholar]
- Roumen E. C. (1994). “A strategy for accumulating genes for partial resistance to blast disease in rice within a conventional breeding program,” in Rice Blast Disease, eds Zeigler R. S., Leong S. A., Teng P. S. (Cambridge: CAB International; ), 245–265. [Google Scholar]
- Rubiales D., Fondevilla S., Chen W., Gentzbittel L., Higgins T. J. V., Castillejo M. A., et al. (2015). Achievements and challenges in legume breeding for pest and disease resistance. CRC Crit. Rev. Plant Sci. 34 195–236. 10.1080/07352689.2014.898445 [DOI] [Google Scholar]
- Sakiroglu M., Brummer E. C. (2017). Identification of loci controlling forage yield and nutritive value in diploid alfalfa using GBS-GWAS. Theor. Appl. Gen. 130 261–268. 10.1007/s00122-016-2782-3 [DOI] [PubMed] [Google Scholar]
- Samac D., Smigocki A. (2003). Expression of oryzacystatin I and II in alfalfa increases resistance to the root-lesion nematode. Phytopathology 93 799–804. 10.1094/PHYTO.2003.93.7.799 [DOI] [PubMed] [Google Scholar]
- Samac D. A., Temple S. J. (2004). “Development and utilization of transformation in Medicago species,” in Genetically Modified Crops, Their Development, Uses and Risks, eds Liang G. H., Skinner D. Z. (New York, NY: The Haworth Press; ), 165–202. [Google Scholar]
- Šamaj J., Ovečka M., Hlavačka A., Lecourieux F., Meskiene I., Lichtscheidl I., et al. (2002). Involvement of the mitogen-activated protein kinase SIMK in regulation of root hair tip growth. EMBO J. 21 3296–3306. 10.1093/emboj/cdf349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šamajová O., Komis G., Šamaj J. (2013a). Emerging topics in the cell biology of mitogen-activated protein kinases. Trans. Plant Sci. 18 140–148. 10.1016/j.tplants.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Šamajová O., Plíhal O., Al-Yousif M., Hirt H., Šamaj J. (2013b). Improvement of stress tolerance in plants by genetic manipulation of mitogen-activated protein kinases. Biotech. A. 31 118–128. 10.1016/j.biotechadv.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Sander J. D., Dahlborg E. J., Goodwin M. J., Cade L., Zhang F., Cifuentes D., et al. (2011). Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods 8 67–69. 10.1038/nmeth.1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Nakamura Y., Kaneko T., Asamizu E., Kato T., Nakao M., et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15 227–239. 10.1093/dnares/dsn008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheben A., Verpaalen B., Lawley C. T., Chan C. K. K., Bayer P. E., Batley J., et al. (2019). CropSNPdb: a database of SNP array data for Brassica crops and hexaploid bread wheat. Plant J. 98 142–152. 10.1111/tpj.14194 [DOI] [PubMed] [Google Scholar]
- Schena M., Shalon D., Davis R. W., Brown P. O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270 467–470. 10.1126/science.270.5235.467 [DOI] [PubMed] [Google Scholar]
- Schiml S., Puchta H. (2016). Revolutionizing plant biology: multiple ways of genome engineering by CRISPR/Cas. Plant Methods 12:8 10.1186/s13007-016-0103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Schreiber M., Stein N., Mascher M. (2018). Genomic approaches for studying crop evolution. Genome Biol. 19:140 10.1186/s13059-018-1528-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura A., Moreno M., Madueno F., Molina A., Garcia-Olmedo F. (1999). Snakin-1, a peptide from potato that is active against plant pathogens. Mol. Plant Microbe Interact. 12 16–23. 10.1094/MPMI.1999.12.1.16 [DOI] [PubMed] [Google Scholar]
- Severin A. J., Cannon S. B., Graham M. M., Grant D., Shoemaker R. C. (2011). Changes in twelve homoeologous genomic regions in soybean following three rounds of polyploidy. Plant Cell 23 3129–3136. 10.1105/tpc.111.089573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafique A., Rehman A., Khan A., Kazi A. G. (2014). “Chapter 1 - Improvement of legume crop production under environmental stresses through biotechnological intervention,” in Emerging Technologies and Management of Crop Stress Tolerance: Volume II - A Sustainable Approach, eds Ahmad P., Rehman S. (San Diego: Accademic Press; ), 1–22. 10.1016/B978-0-12-800875-1.00001-6 [DOI] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31 686–688. 10.1038/nbt.2650 [DOI] [PubMed] [Google Scholar]
- Shan S., Soltis P. S., Soltis D. E., Yang B. (2020). Considerations in adapting CRISPR/Cas9 in nongenetic model plant systems. Appl. Plant Sci. 8 e11314 10.1002/aps3.11314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S. D., Hannoufa A., Acharya S. (2018). Molecular improvement of alfalfa for enhanced productivity and adaptability in a changing environment. Plant Cell Environ. 41 1955–1971. 10.1111/pce.13090 [DOI] [PubMed] [Google Scholar]
- Smith J., Grizot S., Arnould S., Duclert A., Epinat J. C., Chames P., et al. (2006). A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 34 e149 10.1093/nar/gkl720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Jiang L., Chen Y., Shu Y., Bai Y., Guo C. (2016). Deep-sequencing transcriptome analysis of field-grown Medicago sativa L. crown buds acclimated to freezing stress. Func. Integr. Genomics 16 495–511. 10.1007/s10142-016-0500-5 [DOI] [PubMed] [Google Scholar]
- Stefanova G., Slavov S., Gecheff K., Vlahova M., Atanassov A. (2013). Expression of recombinant human lactoferrin in transgenic alfalfa plants. Biol. Plant. 57 457–464. 10.1007/s10535-013-0305-5 [DOI] [Google Scholar]
- Steinert J., Schiml S., Puchta H. (2016). Homology-based double-strand break-induced genome engineering in plants. Plant Cell Rep. 35 1429–1438. 10.1007/s00299-016-1981-3 [DOI] [PubMed] [Google Scholar]
- Strizhov N., Keller M., Mathur J., Koncz-Kálmán Z., Bosch D., Prudovsky E., et al. (1996). A synthetic cryIC gene, encoding a Bacillus thuringiensis δ-endotoxin, confers Spodoptera resistance in alfalfa and tobacco. Proc. Natl. Acad. Sci. U.S.A. 93 15012–15017. 10.1073/pnas.93.26.15012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritzler M., Elba P., Berini C., Gomez C., Ayub N., Soto G. (2018). High-quality forage production under salinity by using a salt-tolerant AtNXH1-expressing transgenic alfalfa combined with a natural stress-resistant nitrogen-fixing bacterium. J. Biotechnol. 276 42–45. 10.1016/j.jbiotec.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Sun X., Hu Z., Chen R., Jiang Q., Song G., Zhang H., et al. (2015). Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5:10342 10.1038/srep10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Yang S., Liu J., Zhu H. (2016). Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol. 170 26–32. 10.1104/pp.15.01661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Krishnakumar V., Bidwell S., Rosen B., Chan A., Zhou S., et al. (2014). An improved genome release (version Mt4. 0) for the model legume Medicago truncatula. BMC genom. 15:312 10.1186/1471-2164-15-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Liu G. Q., Zhou J. P., Ren Q., You Q., Tian L., et al. (2018). A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 19:84 10.1186/s13059-018-1458-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye M., Denton M. D., Samac D. A., Vance C. P. (2005). Transgenic alfalfa secretes a fungal endochitinase protein to the rhizosphere. Plant Soil 269 233–243. 10.1007/s11104-004-0520-0 [DOI] [Google Scholar]
- Tesfaye M., Liu J., Vance C. P. (2007). Genomic and genetic control of phosphate stress in legumes. Plant Physiol. 144 594–603. 10.1104/pp.107.097386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth E., Bakheit B. R. (1983). Results of resistance breeding in alfalfa. II. Resistance to Verticillium wilt. Acta Biol. Hung. 32 78–85. [Google Scholar]
- Triboi E., Triboi-Blondel A. M. (2014). “Towards sustainable, self-supporting agriculture: biological nitrogen factories as a key for future cropping systems,” in Soil as World Heritage, ed. Dent D. (Dordrecht: Springer; ), 329–342. 10.1007/978-94-007-6187-2_32 [DOI] [Google Scholar]
- Tripathi P., Rabara R. C., Reese R. N., Miller M. A., Rohila J. S., Subramanian S., et al. (2016). A toolbox of genes, proteins, metabolites and promoters for improving drought tolerance in soybean includes the metabolite coumestrol and stomatal development genes. BMC Genom. 17:102 10.1186/s12864-016-2420-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X., Liu Z., Zhang Z. (2018a). Comparative transcriptomic analysis of resistant and susceptible alfalfa cultivars (Medicago sativa L.) after thrips infestation. BMC Genom. 19:116 10.1186/s12864-018-4495-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X., Zhao H., Zhang Z. (2018b). Transcriptome approach to understand the potential mechanisms of resistant and susceptible alfalfa (Medicago sativa L.) cultivars in response to aphid feeding. J. Integr. Agric. 17 2518–2527. 10.1016/S2095-3119(17)61843-4 [DOI] [Google Scholar]
- Valliyodan B., Ye H., Song L., Murphy M., Shannon J. G., Nguyen H. T. (2017). Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J. Exp. Bot. 68 1835–1849. 10.1093/jxb/erw433 [DOI] [PubMed] [Google Scholar]
- Van K., Rastogi K., Kim K. H., Lee S. H. (2013). Next-generation sequencing technology for crop improvement. SABRAO J. Breed. Genet. 45 84–99. 10.3389/fpls.2014.00367 [DOI] [Google Scholar]
- Varshney R. K., Kudapa H. (2013). Legume biology: the basis for crop improvement. Funct. Plant Biol. 40 5–8. 10.1071/FPv40n12_FO [DOI] [PubMed] [Google Scholar]
- Volkov V., Wang B., Dominy P. J., Fricke W., Amtmann A. (2004). Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Environ. 27 1–14. 10.1046/j.0016-8025.2003.01116.x [DOI] [Google Scholar]
- Walter M. H., Liu J. W., Wünn J., Hess D. (1996). Bean ribonuclease-like pathogenesis-related protein genes Ypr10 display complex patterns of developmental, dark-induced and exogenous-stimulus-dependent expression. Eur. J. Biochem. 239 281–293. 10.1111/j.1432-1033.1996.0281u.x [DOI] [PubMed] [Google Scholar]
- Wang K., Wang Z., Li F., Ye W., Wang J. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nature Gen. 44 1098–1103. 10.1038/ng.2371 [DOI] [PubMed] [Google Scholar]
- Wang L., Rubio M. C., Xin X., Zhang B., Fan Q., Wang Q., et al. (2019). CRISPR/Cas9 knockout of leghemoglobin genes in Lotus japonicus uncovers their synergistic roles in symbiotic nitrogen fixation. New Phytol. 224 818–832. 10.1111/nph.16077 [DOI] [PubMed] [Google Scholar]
- Wang L., Sun S., Wu T., Liu L., Sun X., Cai Y., et al. (2020). Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 1–13. 10.1111/pbi.13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang L., Tan Q., Fan Q., Zhu H., Hong Z., et al. (2016). Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicus using CRISPR-Cas9. Front. Plant Sci. 7:1333 10.3389/fpls.2016.01333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li H., Ke Q., Jeong J. C., Lee H. S., Xu B., et al. (2014). Transgenic alfalfa plants expressing AtNDPK2 exhibit increased growth and tolerance to abiotic stresses. Plant Physiol. Biochem. 84 67–77. 10.1016/j.plaphy.2014.08.025 [DOI] [PubMed] [Google Scholar]
- Watson B. S., Bedair M. F., Urbanczyk-Wochniak E., Huhman D. V., Yang D. S., Allen S. N., et al. (2015). Integrated metabolomics and transcriptomics reveal enhanced specialized metabolism in Medicago truncatula root border cells. Plant Physiol. 167 1699–1716. 10.1093/jxb/erx308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Chen Y., Schnabel E., Crook A., Frugoli J. (2019). Comparison of efficiency and time to regeneration of Agrobacterium-mediated transformation methods in Medicago truncatula. Plant Met. 15:20 10.1186/s13007-019-0404-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482 331–338. 10.1038/nature10886 [DOI] [PubMed] [Google Scholar]
- Wong C. E., Li Y., Moffatt B. A. (2006). Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 140 1437–1450. 10.1104/pp.105.070508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. A., Townsend J. A., Winfrey R. J., Jr., Irwin P. A., Rajagopal J. (2005). High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 44 693–705. 10.1111/j.1365-313X.2005.02551.x [DOI] [PubMed] [Google Scholar]
- Xia T., Apse M. P., Aharon G. S., Blumwald E. (2002). Identification and characterization of a NaCl-inducible vacuolar Na+/H+ antiporter in Beta vulgaris. Physiol. Plant. 116 206–212. 10.1034/j.1399-3054.2002.1160210.x [DOI] [PubMed] [Google Scholar]
- Xie X., Ma X., Zhu Q., Zeng D., Li G., Liu Y. G. (2017). CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol. Plant. 10 1246–1249. 10.1016/j.molp.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Xiong L., Lee H., Ishitani M., Zhu J. K. (2002). Regulation of osmotic stress-responsive gene expression by theLOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277 8588–8596. 10.1074/jbc.M109275200 [DOI] [PubMed] [Google Scholar]
- Xu B., Wang Y., Zhang S., Guo Q., Jin Y., Chen J., et al. (2017). Transcriptomic and physiological analyses of Medicago sativa L. roots in response to lead stress. PLoS One 12:e0175307 10.1371/journal.pone.0175307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoubi R., Job C., Belghazi M., Chaibi W., Job D. (2011). Toward characterizing seed vigor in alfalfa through proteomic analysis of germination and priming. J. Proteome Res. 10 3891–3903. 10.1021/pr101274f [DOI] [PubMed] [Google Scholar]
- Yacoubi R., Job C., Belghazi M., Chaibi W., Job D. (2013). Proteomic analysis of the enhancement of seed vigour in osmoprimed alfalfa seeds germinated under salinity stress. Seed Sci. Res. 23 99–110. 10.1017/S0960258513000093 [DOI] [Google Scholar]
- Yang S., Gao M., Xu C., Gao J., Deshpande S., Lin S., et al. (2008). Alfalfa benefits from Medicago truncatula: the RCT1 gene from M. truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proc. Natl. Acad. Sci. U.S.A. 105 12164–12169. 10.1073/pnas.0802518105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. S., Tu Z. J., Cheung F., Xu W. W., Lamb J. F., Jung H. J. G., et al. (2011). Using RNA-Seq for gene identification, polymorphism detection and transcript profiling in two alfalfa genotypes with divergent cell wall composition in stems. BMC genom. 12:199 10.1186/1471-2164-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. S., Xu W. W., Tesfaye M., Lamb J. F., Jung H. J. G., VandenBosch K. A., et al. (2010). Transcript profiling of two alfalfa genotypes with contrasting cell wall composition in stems using a cross-species platform: optimizing analysis by masking biased probes. BMC genom. 11:323 10.1186/1471-2164-11-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Ma Q., Wang H., Feng D., Wang X., Pei Y., et al. (2020). SMALL Leaf and BUSHY1 controls organ size and lateral branching by modulating the stability of BIG SEEDS1 in Medicago truncatula. New Phytol. [Epub ahead of print] 10.1111/nph.16449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. D., Debellé F., Oldroyd G. E., Geurts R., Cannon S. B., Udvardi M. K., et al. (2011). The medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480 520–524. 10.1038/nature10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. X., Liu X., Boge W., Liu X. P. (2016). Genome-wide association study identifies loci for salt tolerance during germination in autotetraploid alfalfa (Medicago sativa L.) using genotyping-by-sequencing. Front. Plant Sci. 7:956 10.3389/fpls.2016.00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. X., Zheng P., Zhang T., Rodringuez J., Main D. (2017). Genotyping-by-sequencing-based genome-wide association studies on Verticillium wilt resistance in autotetraploid alfalfa (Medicago sativa L.). Mol. Plant Pathol. 18 187–194. 10.1111/mpp.12389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng N., Yang Z., Zhang Z., Hu L., Chen L. (2019). Comparative transcriptome combined with proteome analyses revealed key factors involved in alfalfa (Medicago sativa) response to waterlogging stress. Int. J. Mol. Sci. 20:1359 10.3390/ijms20061359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Shi S. (2018). Physiological and proteomic responses of contrasting alfalfa (Medicago sativa L.) varieties to PEG-induced osmotic stress. Front Plant Sci. 9:242 10.3389/fpls.2018.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12 797–807. 10.1111/pbi.12200 [DOI] [PubMed] [Google Scholar]
- Zhang J. (2004). Harvesting Inducible Gene And Promoters In Alfalfa. Dissertation thesis, University of Guelph, Guelph, ON. [Google Scholar]
- Zhang L. Q., Niu Y. D., Huridu H., Hao J. F., Qi Z., Hasi A. (2014). Salicornia europaea L. Na+/H+ antiporter gene improves salt tolerance in transgenic alfalfa (Medicago sativa L.). Genet. Mol. Res. 13 5350–5360. 10.4238/2014.July.24.14 [DOI] [PubMed] [Google Scholar]
- Zhang S., Shi Y., Cheng N., Du H., Fan W., Wang C. (2015). De novo characterization of fall dormant and nondormant alfalfa (Medicago sativa L.) leaf transcriptome and identification of candidate genes related to fall dormancy. PloS One 10:e0122170 10.1371/journal.pone.0122170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Yu L. X., Zheng P., Li Y., Rivera M., Main D., et al. (2015). Identification of loci associated with drought resistance traits in heterozygous autotetraploid alfalfa (Medicago sativa L.) using genome-wide association studies with genotyping by sequencing. PLoS One 10:e0138931 10.1371/journal.pone.0138931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. M., Liu Z. H., Wen Z. Y., Zhang H. M., Yang F., Guo X. L. (2012). The vacuolar Na+- H+ antiport gene TaNHX2 confers salt tolerance on transgenic alfalfa (Medicago sativa). Funct. Plant Biol. 39 708–716. 10.1071/FP12095 [DOI] [PubMed] [Google Scholar]
- Zhao B., Liang R., Ge L., Li W., Xiao H., Lin H., et al. (2007). Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Comm. 354 585–590. 10.1016/j.bbrc.2007.01.022 [DOI] [PubMed] [Google Scholar]
- Zhou C., Han L., Pislariu C., Nakashima J., Fu C., Jiang Q., et al. (2011). From model to crop: functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 157 1483–1496. 10.1104/pp.111.185140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Luo H. (2013). MicroRNA-mediated gene regulation: potential applications for plant genetic engineering. Plant Mol. Biol. 83 59–75. 10.1007/s11103-013-0089-1 [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6 66–71. 10.1016/S1360-1385(00)01838-0 [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Ann. Rev. Plant Biol. 53 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35 345–351. 10.1016/j.it.2014.05.004 [DOI] [PubMed] [Google Scholar]