Cicadas establish an intimate symbiosis with microorganisms to obtain essential nutrients that are extremely deficient in host plant sap. Previous studies on bacterial communities of cicadas mainly focused on a few widely distributed species, but knowledge about mountain-habitat species is quite poor. We initially revealed the physical distribution of the primary symbionts “Ca. Sulcia” and “Ca. Hodgkinia” and the dominant secondary symbionts Rickettsia and Arsenophonus in the mountain-habitat specialist Pycna repanda and then clarified the transovarial transmission process of bacteriome-associated symbionts in this species. Our observations suggest that “Ca. Hodgkinia” may have split into cytologically distinct lineages within this cicada species, and related cicadas might have developed complex mechanisms for the vertical transmission of the bacteriome-associated symbionts. We also revealed that Arsenophonus can be transovarially transmitted in auchenorrhynchan insects when it is not harbored in the cytoplasm of other endosymbionts. Our results highlight transovarial transmission mechanisms of bacteriome-associated symbionts in sap-feeding insects.
KEYWORDS: symbiosis, sap-feeding insects, bacteriocytes, transovarial transmission, cellular mechanism
ABSTRACT
Although transovarial transmission of bacteriome-associated symbionts in hemipteran insects is extremely important for maintaining intimate host-symbiont associations, our knowledge of cellular mechanisms underlying the transmission process is quite limited. We investigated bacterial communities of salivary glands, bacteriomes, and digestive and reproductive organs and clarified the transovarial transmission of bacteriome-associated symbionts of the mountain-habitat specialist Pycna repanda using integrated methods. The bacterial communities among different gut tissues and those of bacteriomes of males and females both show similarity, whereas differences are exhibited among bacterial communities in testes and ovaries. The primary symbionts “Candidatus Sulcia muelleri” (hereafter “Ca. Sulcia”) and “Candidatus Hodgkinia cicadicola” (hereafter “Ca. Hodgkinia”) were not only restricted to but also dominant in the bacteriomes and ovaries. “Ca. Hodgkinia” cells in the bacteriomes of both sexes exhibited different colors by histological and electron microscopy. Also considering the results of a restriction fragment length polymorphism (RFLP)-based cloning approach, we hypothesize that “Ca. Hodgkinia” may have split into cytologically different cellular lineages within this cicada species. Regarding the dominant secondary symbionts, Rickettsia was detected in the salivary glands, digestive organs, and testes, whereas Arsenophonus was detected in the bacteriomes and ovaries. Our results show that Arsenophonus can coexist with “Ca. Sulcia” and “Ca. Hodgkinia” within bacteriomes and can be transovarially transmitted with these obligate symbionts together from mother to offspring in cicadas, but it is not harbored in the cytoplasm of “Ca. Sulcia.” The change in the shape of “Ca. Sulcia” and “Ca. Hodgkinia” during the transovarial transmission process is hypothesized to be related to the limited space and novel microenvironment.
IMPORTANCE Cicadas establish an intimate symbiosis with microorganisms to obtain essential nutrients that are extremely deficient in host plant sap. Previous studies on bacterial communities of cicadas mainly focused on a few widely distributed species, but knowledge about mountain-habitat species is quite poor. We initially revealed the physical distribution of the primary symbionts “Ca. Sulcia” and “Ca. Hodgkinia” and the dominant secondary symbionts Rickettsia and Arsenophonus in the mountain-habitat specialist Pycna repanda and then clarified the transovarial transmission process of bacteriome-associated symbionts in this species. Our observations suggest that “Ca. Hodgkinia” may have split into cytologically distinct lineages within this cicada species, and related cicadas might have developed complex mechanisms for the vertical transmission of the bacteriome-associated symbionts. We also revealed that Arsenophonus can be transovarially transmitted in auchenorrhynchan insects when it is not harbored in the cytoplasm of other endosymbionts. Our results highlight transovarial transmission mechanisms of bacteriome-associated symbionts in sap-feeding insects.
INTRODUCTION
Plant-sucking insects of the order Hemiptera, including aphids, planthoppers, leafhoppers, treehoppers, cicadas, spittlebugs, and whiteflies, etc., exclusively feed on plant xylem or phloem sap (1, 2). Plant sap is relatively abundant in carbohydrates but nutritionally deficient in nitrogenous nutrients (1, 3, 4). Most sap-feeding insects are dependent on symbionts for the provisioning of vitamins and essential amino acids that are extremely unbalanced or limited in the diet (1, 5–7).
Mutually beneficial interactions between symbionts and host insects are especially ubiquitous in nature, which have been extensively studied among a great diversity of phytophagous hemipteran insects, particularly those of the suborder Auchenorrhyncha, including planthoppers, leafhoppers, treehoppers, spittlebugs, and cicadas (1, 6). The symbionts can be classified into two categories: primary symbionts (obligate) and secondary symbionts (facultative). The primary symbionts are strictly maternally inherited, residing in highly specialized and host-derived organs termed bacteriomes, which comprise numerous bacteriocytes (1, 5, 6, 8–10). Primary symbionts supply the host insects with essential nutrients that are necessary for the survival and reproduction of the hosts (6, 7, 11). In contrast, secondary symbionts are not required for the survival and development of the host insects but may be involved in manipulating reproduction or providing the hosts with benefits under certain environmental conditions (12–15). Secondary symbionts are reported to reside in the bacteriomes (16, 17), reproductive organs (18–20), Malpighian tubules (21), and salivary glands (19, 22). These localization patterns vary considerably within the body among different insects (12, 23).
The common ancestor of the auchenorrhynchan insects developed an intimate symbiosis with “Candidatus Sulcia muelleri” (hereafter “Ca. Sulcia”) dating back to approximately 260 million years ago (2). During the diversification of auchenorrhynchan insects, many lineages acquired an additional symbiotic microorganism belonging to the phylum Proteobacteria, e.g., “Candidatus Hodgkinia cicadicola” (hereafter “Ca. Hodgkinia”), in some cicada species (2, 24). Complicated interdependence, especially the metabolic complementarity of symbiont genomes, has been described for host-symbiont associations over the past few years. The “Ca. Sulcia” genome can synthesize 8 of the 10 essential amino acids, whereas the “Ca. Hodgkinia” genome is complementarily retained for the biosynthesis of the remaining 2 essential amino acids, cobalamin, and vitamins to maintain the survival and development of host cicadas relying on nutritionally unbalanced plant sap (24, 25). Although such mutually beneficial associations between hosts and symbionts can certainly maintain continuity and stability, host-symbiont associations may experience collapse and instability over evolutionary time (26). Recent studies have shown that the bacteriome-associated symbiont “Ca. Sulcia,” whose genome is highly conserved, seems to be present in all auchenorrhynchan insects, including cicadas (2), while the genome of the coresident symbiont “Ca. Hodgkinia” can split into two or more cytologically distinct but metabolically interdependent cellular lineages in some related cicadas (e.g., species of the genus Tettigades) (27, 28) or is even absent from some cicada species (29). This is a surprising finding, as the “Ca. Hodgkinia” genome carries biosynthetic pathway genes for the biosynthesis of essential nutrients that are nutritionally important for the host cicadas (24, 25, 29). More recent investigations revealed that “Ca. Hodgkinia” has been replaced in some cicada species by a yeast-like fungal symbiont, which may be functionally similar to “Ca. Hodgkinia,” thereby compensating for the absence of “Ca. Hodgkinia” (29).
Transovarial transmission is extremely important for maintaining host-symbiont associations, but our knowledge about the cellular mechanism of such a process is quite poor. Recently, a few microscopic studies of the vertical transmission of bacteriome-associated symbionts in auchenorrhynchan insects were conducted. For example, it has been described that both “Ca. Sulcia” and betaproteobacteria were harbored in the bacteriomes of the leafhopper Evacanthus interruptus, gathered around the posterior pole of the oocytes after leaving the bacteriocytes, passed through the cytoplasm of follicular cells, entered the perivitelline space, and finally formed a “symbiont ball” at the posterior end of the oocytes (20). It was confirmed using confocal imaging that “Ca. Sulcia” together with the coinfected symbiont “Ca. Hodgkinia” or the yeast-like fungal symbiont also formed a symbiont ball at the posterior pole of the developing oocytes during transovarial transmission in related cicada species (29). We provide a schematic diagram (Fig. 1) to summarize the transovarial transmission process of the bacteriome-associated endosymbionts in auchenorrhynchan insects. Secondary symbionts, transmitted vertically or horizontally, can also be of vital importance in host fitness and reproductive manipulation (14). Recently, it was revealed that the symbiotic microorganism Arsenophonus in the leafhopper Macrosteles laevis can be transovarially transmitted with “Ca. Sulcia” and “Candidatus Nasuia deltocephalinicola” between host generations, whereas Arsenophonus is harbored in the cytoplasm of “Ca. Sulcia” in the transmission process (30). However, our understanding of the transmission patterns of secondary symbionts in auchenorrhynchan insects is very limited. This is particularly the case for the lineage of Cicadoidea.
FIG 1.
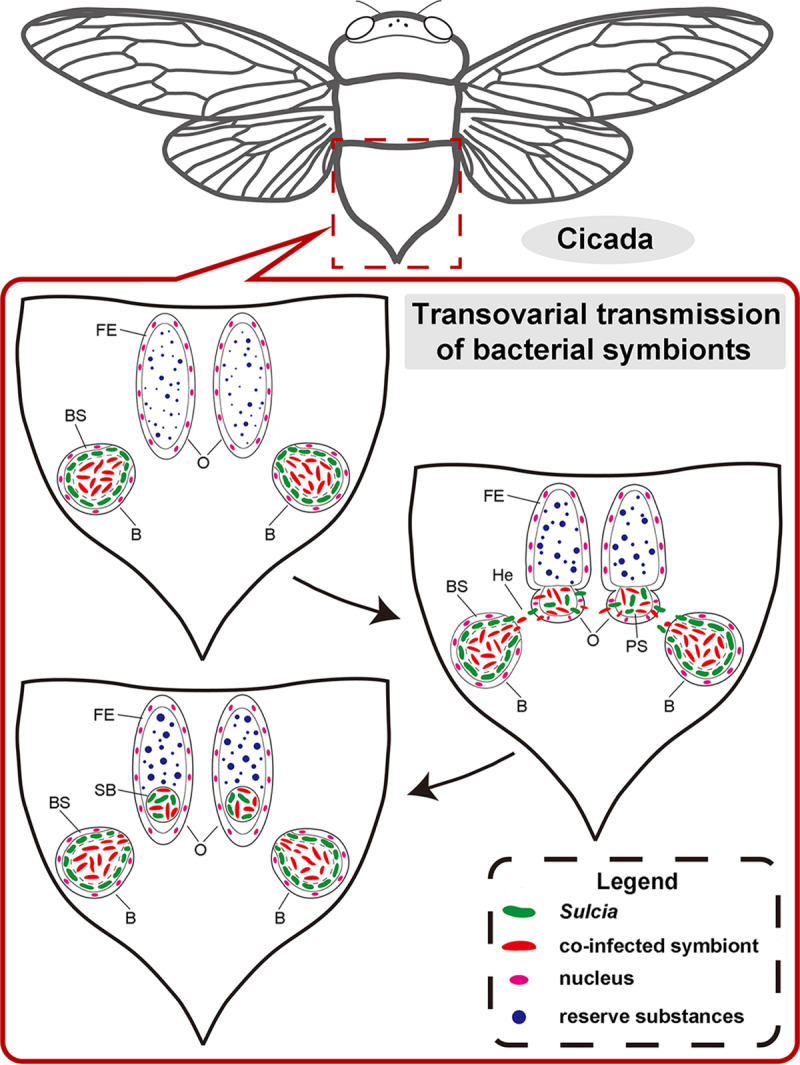
Schematic representation showing the transovarial transmission process for the bacterial symbionts of representative auchenorrhynchan insects. In mature females, “Ca. Sulcia” and the coinfected symbiont leave the bacteriome (B) for the hemolymph (He), invade the posterior pole of the oocytes (O), migrate through the follicular epithelium (FE) into the perivitelline space (PS), and finally form a characteristic symbiont ball (SB) at the posterior pole of the oocytes. BS, bacteriome sheath.
Previous studies of host-symbiont associations in Auchenorrhyncha mainly focused on the presence/absence and the replacement of bacteriome-associated symbionts (i.e., “Ca. Sulcia” and “Ca. Hodgkinia”) and their genomes (27, 29, 31) as well as changes in related symbiont complexity (32, 33). The nutritional function and metabolic versatility of bacterial symbionts were also featured in some auchenorrhynchan insects (5, 24, 25). However, knowledge about the bacterial communities and distribution of obligate and facultative symbionts residing in cicadas is quite limited, although bacterial communities of the alimentary canal and some other organs of a few cicada species have been investigated (34–37). To date, nothing is known about the transmission patterns of secondary symbionts in cicadas. The mountain-habitat specialist cicada Pycna repanda is mainly distributed in natural forests at elevations of over 1,500 m and primarily feeds on xylem sap of the Chinese red birch Betula albosinensis Burk. The natural habitat of Py. repanda is quite distinct from that of the majority of other cicadas. Here, we investigated the bacterial communities and physical distribution of dominant symbionts in the bacteriomes, reproductive organs, salivary glands, and digestive organs of Py. repanda using integrated culture-independent methods. We further clarified the morphology and transovarial transmission of the bacteriome-associated symbionts in this cicada species using histological and ultrastructural microscopy combined with confocal imaging. The study hypothesis was that a combination of these methods would provide accurate characterization and clarify the transovarial transmission process of bacteriome-associated endosymbionts within an auchenorrhynchan species.
RESULTS
Illumina amplicon analyses.
In total, 2,975,564 effective tags were obtained from 36 samples after the removal of low-quality sequences. Subsequently, the high-quality sequences were clustered into operational taxonomic units (OTUs) using Uparse software at a 97% similarity threshold (38). A total of 9,562 OTUs were obtained from the samples, and the numbers of OTUs were different among different tissues (for details, see Table S1 in the supplemental material). A total of 110 bacterial OTUs were shared among the salivary glands, filter chamber, conical segment, midgut, and hindgut of Py. repanda males (Fig. S1A). Thirty-three bacterial OTUs were shared among the reproductive organs and the bacteriomes of both sexes (Fig. S1B).
Bacterial compositions of bacteriomes, salivary glands, and reproductive and digestive organs based on Illumina amplicon analyses.
We categorized the assigned phyla or genera with low abundances as “others” and the unassigned tags as “unclassified.” The others were present at an abundance of <2% across all samples. As a result, the identified sequences mainly belonged to 10 assigned bacterial phyla or genera. The compositions of bacterial communities varied considerably across different samples.
At the genus level, “Ca. Sulcia” was dominant in the ovaries (11.91 to 25.70%) and bacteriomes (males, 64.14 to 90.80%; females, 56.80 to 89.07%). However, Rickettsia exhibited extremely high abundances in the salivary glands (72.81 to 89.88%), filter chamber (78.51 to 91.56%), conical segment (79.0 to 95.80%), midgut (79.39 to 97.96%), hindgut (59.93 to 85.71%), and testes (50.64 to 79.17%). Arsenophonus accounted for relatively high proportions in the ovaries (36.31 to 45.79%) and bacteriomes (6.81 to 36.76%) of females, but it exhibited a relatively low abundance in the bacteriomes (0.59 to 3.00%) of males (Fig. 2). A phylum-level analysis revealed similar overall differences and shifts between the major groups, the Proteobacteria and the Bacteroidetes (for details, see Fig. S2 in the supplemental material).
FIG 2.
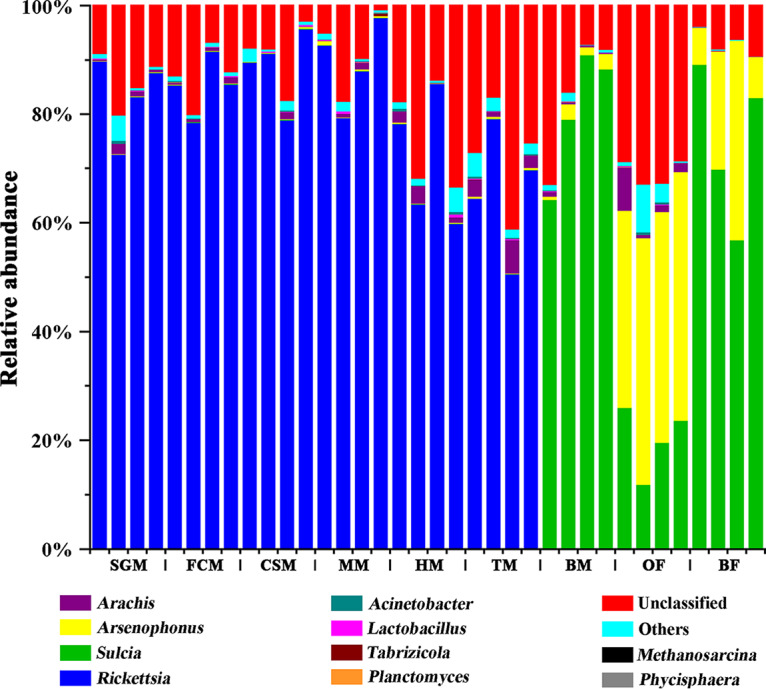
Bacterial compositions of the salivary glands, bacteriomes, and reproductive and digestive organs of Pycna repanda at the genus level. Abbreviations: SGM, salivary glands of males; FCM, filter chamber of males; CSM, conical segment of males; MM, midgut of males; HM, hindgut of males; TM, testes of males; BM, bacteriomes of males; OF, ovaries of females; BF, bacteriomes of females.
Similarity of bacterial communities among different tissues.
Principal-component analysis (PCA) was conducted to reveal the similarity of bacterial communities among different tissues. The PCA yielded two main axes, which accounted for 96.0% of the total variation in bacterial communities. In the PCA plot, the salivary glands and gut tissues clustered closely together, while the bacteriomes and reproductive organs were extremely dispersed across all samples (Fig. S3). Different gut tissues exhibited high similarity in the bacterial communities based on permutation multivariate analysis of variance (PERMANOVA) (P > 0.05). In addition, the bacteriomes of both sexes also exhibited no statistical differences in the bacterial communities according to PERMANOVA results (P > 0.05). However, there were significant differences in the bacterial communities between testes and ovaries (0.01 < P < 0.05).
Bacterial composition of bacteriomes analyzed by RFLP analysis.
The 16S rRNA gene sequences obtained by restriction fragment length polymorphism (RFLP) analysis were used as queries in BLAST searches of the NCBI GenBank nucleotide database. The bacteria harbored in the bacteriomes of males exhibit the highest similarity to “Ca. Sulcia” (99% similarity) and “Ca. Hodgkinia” (∼94% similarity) (Tables S2 and S3). “Ca. Sulcia” harbored in Py. repanda shows 99% similarity to that of platypleurine relatives, e.g., Platypleura kaempferi, Pl. yaeyamana, and Pl. kuroiwae (Table S3). However, “Ca. Hodgkinia” harbored in Py. repanda shows only <94% similarity to that of platypleurine relatives (Table S3). We further identified unassigned OTUs in the bacteriomes and ovaries obtained via high-throughput sequencing. The results reveal that the majority of unassigned OTUs exhibit a similarity level of 97 to 99% to “Ca. Hodgkinia” sequences obtained by RFLP analysis, indicating that they belong to “Ca. Hodgkinia” (data not shown).
Reconstruction of phylogenetic relationships of “Ca. Sulcia” and “Ca. Hodgkinia” in cicadas using 16S rRNA gene sequences.
The phylogenetic trees of “Ca. Sulcia” and “Ca. Hodgkinia” obtained from Py. repanda and other representative cicadas were reconstructed based on 16S rRNA gene sequences using Bayesian inference (BI) and maximum likelihood (ML). The Bayesian topology is completely congruent with the maximum likelihood topology in the trees for both “Ca. Sulcia” and “Ca. Hodgkinia” (data not shown). The phylogenetic analysis of “Ca. Sulcia” clearly demonstrates that all the “Ca. Sulcia” symbionts form a well-defined monophyletic group, with strong support in both the BI and ML trees (Fig. S4). In contrast, “Ca. Hodgkinia” of Py. repanda displays relatively low similarity (<94%) to those of other representative cicadas, but all the “Ca. Hodgkinia” symbionts form a well-defined monophyletic group, with strong support in both the BI and ML trees (Fig. S5).
Physical distribution of dominant symbionts in different tissues determined using diagnostic PCR.
Diagnostic PCR amplification was performed to monitor the physical distribution of dominant symbionts, including “Ca. Sulcia,” “Ca. Hodgkinia,” Rickettsia, and Arsenophonus. Arsenophonus together with “Ca. Sulcia” and “Ca. Hodgkinia” were detected in the ovaries and bacteriomes, whereas Rickettsia was detected in the salivary glands, digestive organs, and testes (Table S4). The amplified 16S rRNA gene sequences of Rickettsia and Arsenophonus are listed in Table S2.
Ultrastructure and localization of symbionts in the bacteriomes.
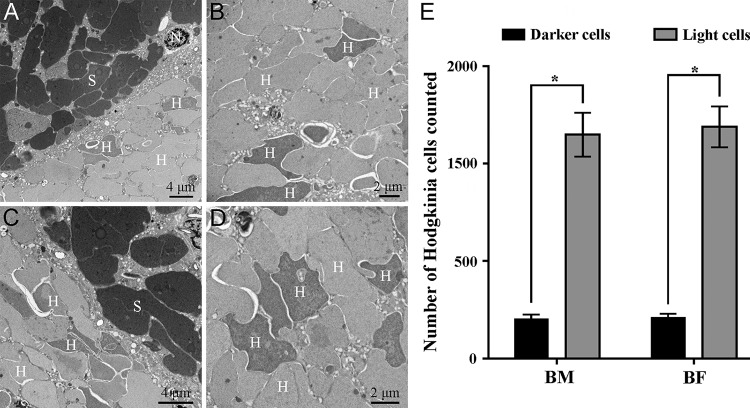
Qualitatively, the bacteriome units (viz., sphere-like clusters composed of numerous bacteriocytes) of males were smaller than those of females (Fig. S6A and B). Histological microscopy revealed that two morphologically distinct obligate symbionts were harbored in the bacteriocytes (Fig. S6C and D). Based on ultrastructural observations, electron-dense bacteria were localized in the peripheral bacteriocytes, whereas the electron-translucent symbiont occupied the central bacteriocytes (Fig. S6C and D). Interestingly, the electron-translucent symbiont exhibited dark and light cells via both histological and electron microscopy in the bacteriomes of both sexes (Fig. 3A to D and Fig. S6E and F). The numbers of dark and light cells in the bacteriomes were significantly different for both males and females (Fig. 3E), and the ratio of dark cells to light cells was estimated to be nearly 1:5.609 in both sexes (Table S5).
FIG 3.
Localization of primary symbionts and differences in numbers of dark and light “Ca. Hodgkinia” cells in the bacteriomes of Pycna repanda males and females by ultrastructural microscopy. (A) Fragment of the bacteriocytes of males containing “Ca. Sulcia” (S) and “Ca. Hodgkinia” (H). (B) Central part of the bacteriocytes of males containing “Ca. Hodgkinia” cells of different colors. (C) Fragment of the bacteriocytes of females containing “Ca. Sulcia” and “Ca. Hodgkinia.” (D) Central part of the bacteriocytes of females containing “Ca. Hodgkinia” cells of different colors. (E) Numbers of dark and light “Ca. Hodgkinia” cells counted in the bacteriomes of both sexes. We randomly selected multiple sections for statistical analysis, aiming to test the differences in numbers for dark and light “Ca. Hodgkinia” cells in the bacteriomes. Asterisks indicate a statistically significant difference (P < 0.01 by a Mann-Whitney test). BM, bacteriomes of males; BF, bacteriomes of females; N, nucleus.
Fluorescence microscopy showed that “Ca. Hodgkinia” and “Ca. Sulcia” were cytologically distinct and isolated from each other in the bacteriome units (Fig. S6G to I). Fluorescence in situ hybridization (FISH) analysis, histological studies, and ultrastructural observations clarified that “Ca. Sulcia” was harbored in the peripheral bacteriocytes, whereas “Ca. Hodgkinia” occupied the central bacteriocytes. The third type of symbiont was revealed from the peripheral bacteriocytes, which has been identified as the secondary symbiont Arsenophonus according to 16S rRNA community surveys (Fig. 2), diagnostic PCR (Tables S2 and S4), and FISH analysis (Fig. 4A).
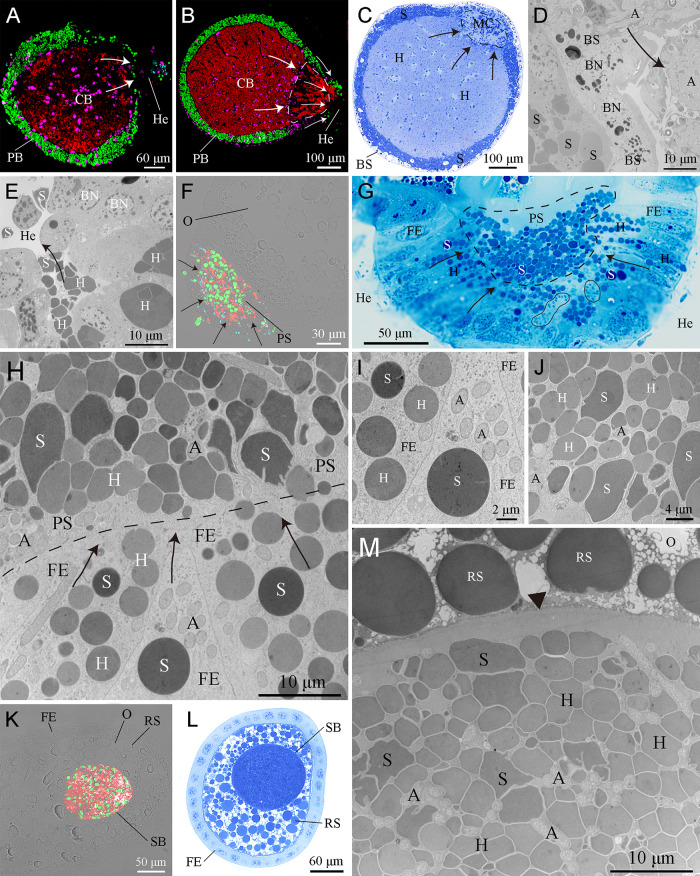
FIG 4.
Transovarial transmission of bacteriome-associated symbionts. (A to E) FISH analysis combined with histological and ultrastructural observations showing Arsenophonus (A), “Ca. Sulcia” (S), and “Ca. Hodgkinia” (H) released from the bacteriocytes into the hemolymph (He). In mature females, Arsenophonus and “Ca. Sulcia” were directly released from the peripheral bacteriocytes (PB) into the hemolymph, but “Ca. Hodgkinia” emigrated from central bacteriocytes (CB) through the multinuclear compartment (MC) (encircled with black/white dotted lines) into the hemolymph. (F to H) Arsenophonus (encircled with a black closed line in panel G), “Ca. Sulcia,” and “Ca. Hodgkinia” toward the posterior pole of the ovarioles, migrating through the cytoplasm of the follicular epithelium (FE) into the perivitelline space (PS) (encircled with a black dotted line in panel G). (I) “Ca. Sulcia” and “Ca. Hodgkinia” remained spherical in the cytoplasm of the follicular epithelium. (J) “Ca. Sulcia” and “Ca. Hodgkinia” seem irregular in the perivitelline space. (K to M) Intermixed Arsenophonus, “Ca. Sulcia,” and “Ca. Hodgkinia” cells forming a characteristic symbiont ball (SB) in the posterior pole of the oocytes (O). Magenta, green, cyan, and red represent the bacteriocyte nucleus (BN), “Ca. Sulcia,” Arsenophonus, and “Ca. Hodgkinia,” respectively. Black and white arrows represent the emigration of the symbionts. The black arrowhead represents the oolemma. BS, bacteriome sheath; RS, reserve substances.
Transovarial transmission of bacteriome-associated symbionts Arsenophonus, “Ca. Sulcia,” and “Ca. Hodgkinia” in Py. repanda.
The paired ovaries of Py. repanda consist of numerous telotrophic ovarioles (Fig. S7A). Each ovariole contains several linearly arranged oocytes, which are surrounded by a single layer of follicular epithelial cells (Fig. S7B). Based on detailed FISH analysis and histological and ultrastructural observations, we revealed the transovarial transmission process of the bacteriome-associated symbionts in Py. repanda (Table S6). In mature females, Arsenophonus and “Ca. Sulcia” were directly released from the peripheral bacteriocytes into the hemolymph, but “Ca. Hodgkinia” emigrated from central bacteriocytes through the multinuclear compartment into the hemolymph (Fig. 4A to E). Notably, only the darker “Ca. Hodgkinia” cells could be observed to emigrate from central bacteriocytes through the multinuclear compartment into the hemolymph (Fig. S8 and Table S7). “Ca. Sulcia” and “Ca. Hodgkinia” could be observed to change their shape from irregular to roughly spherical when leaving the bacteriocytes (Table S8). Arsenophonus, “Ca. Sulcia,” and “Ca. Hodgkinia” were then transported to the posterior pole of the oocytes, migrating through the cytoplasm of the single-layered follicular epithelium into the perivitelline space (Fig. 4F to J). Qualitatively, the volume of the follicular epithelium seems to increase as the number of symbionts entering the cytoplasm of the follicular epithelium increases (Fig. S9). Before entering the perivitelline space from the follicular epithelium, “Ca. Sulcia” and “Ca. Hodgkinia” rechanged their shape from roughly spherical to irregular. In contrast, Arsenophonus did not exhibit any obvious change in shape during transovarial transmission (Table S8). Finally, the intermixed symbionts formed a characteristic symbiont ball at the posterior pole of the oocytes (Fig. 4K to M). Statistical analysis revealed that the ratio of the numbers of “Ca. Sulcia”/“Ca. Hodgkinia”/Arsenophonus cells within symbiont balls was estimated to be nearly 1:9:3.7 (Table S9). The transmission process distinctly shows that Arsenophonus can be transovarially transmitted together with “Ca. Sulcia” and “Ca. Hodgkinia” from mother to offspring in Py. repanda.
DISCUSSION
In the present study, we investigated the bacterial communities of salivary glands, bacteriomes, and digestive and reproductive organs of Py. repanda using integrated approaches. The primary symbionts “Ca. Sulcia” and “Ca. Hodgkinia” were not only restricted to but also dominant in the ovaries and bacteriomes. The secondary symbiont Arsenophonus was predominant in the bacteriomes and ovaries, whereas Rickettsia was prominent in the salivary glands, digestive organs, and testes. Arsenophonus together with “Ca. Sulcia” and “Ca. Hodgkinia” was released from the bacteriocytes, entered the cytoplasm of the follicular epithelium, migrated into the perivitelline space, and finally formed a characteristic symbiont ball at the posterior pole of the oocytes. This study confirms that Arsenophonus can coexist with “Ca. Sulcia” and “Ca. Hodgkinia” in the bacteriomes and can be transovarially transmitted together with these two obligate symbionts from mother to offspring in Py. repanda, but it is not harbored in the cytoplasm of “Ca. Sulcia.” We clarified the transovarial transmission of bacteriome-associated symbionts in this mountain-habitat specialist. These results are helpful for a better understanding of the bacterial communities of different tissues and transovarial transmission mechanisms of bacteriome-associated symbionts in sap-sucking insects.
Comparison of high-throughput 16S rRNA amplicon sequencing and RFLP-based cloning approaches.
We used both high-throughput 16S rRNA amplicon sequencing and RFLP-based cloning approaches to investigate the bacterial communities of Py. repanda, and the results highlight a clear difference between the two methods. In comparison with the RFLP-based cloning approach, more microbial species were detected using high-throughput 16S rRNA amplicon sequencing, which greatly extends our knowledge of the composition and diversity of microbial communities. The failure to detect the secondary symbiont Arsenophonus using RFLP-based cloning is possibly due to either the relatively low abundance or absence of Arsenophonus in the bacteriomes of males or the insensitivity of this method for microbial identification. However, the majority of the unassigned OTUs could not be assigned to “Ca. Hodgkinia” using 16S rRNA amplicon sequencing through a BLAST search of the NCBI database because the similarities between these unassigned OTUs and “Ca. Hodgkinia” sequences are <97%. Amplicon sequencing in this study may have impacted the resolution of OTU identification and taxonomic assignment because it analyzed only the V3-V4 hypervariable regions of the 16S rRNA gene. Although the RFLP-based cloning approach may reveal relatively limited microbial species, it provided nearly complete 16S rRNA gene sequences; thus, the accuracy of OTU identification and taxonomic assignment could be greatly improved. This method may facilitate further studies of microorganisms using other methods, such as gene-specific PCR, fluorescence microscopy, and phylogenetic analysis.
Transovarial transmission of primary symbionts “Ca. Sulcia” and “Ca. Hodgkinia” in Py. repanda.
Our results revealed that the primary symbionts “Ca. Sulcia” and “Ca. Hodgkinia” were not only restricted to but also dominant in the bacteriomes of both sexes and the ovaries of Py. repanda. They were not detected in any other sampled tissues, including the testes. It has been estimated that a shared ancestor of the majority of auchenorrhynchan insects established symbiosis with “Ca. Sulcia” dating back to ∼260 million years ago (2). The “Ca. Sulcia” symbiont provides the phytophagous host insects with 8 of the 10 essential amino acids, including lysine, arginine, phenylalanine, tryptophan, leucine, threonine, isoleucine, and valine (24). In contrast to our study, previous studies indicated that “Ca. Sulcia” is not confined to the bacteriomes and reproductive organs but can also be found in some other tissues in auchenorrhynchan insects, e.g., the gut of the planthopper Hyalesthes obsoletus (39) and the “filter chamber plus conical segment” and testes of the cicadas Pl. kaempferi and Meimuna mongolica (36). “Ca. Sulcia” was found in the bacteriomes and ovaries but was not detected in the salivary glands, digestive organs, and testes of Py. repanda. The presence or absence of “Ca. Sulcia” in related organs/tissues may be related to the microenvironment, developmental stages, as well as competition between symbionts within the host cicadas.
In comparison with the ubiquitous presence of “Ca. Sulcia” in Cicadidae and allies, the coresident symbiont “Ca. Hodgkinia” is present in only some cicada species. Notably, “Ca. Hodgkinia” can evolve into complexes of cytologically distinct lineages with more reduced but complementary genomes in some cicadas (e.g., species of the genus Tettigades) (27, 28) or has been replaced by a yeast-like fungal symbiont that may be functionally similar to “Ca. Hodgkinia” in some other cicada species (29). Our present study and a previous study (29) showed that “Ca. Hodgkinia” is present in all the sampled platypleurine cicadas to date, including Py. repanda, Pl. kaempferi, Pl. kuroiwae, and Pl. yaeyamana. We hypothesize that “Ca. Hodgkinia” is present in all members of the Platypleurini.
Distinct “Ca. Hodgkinia” variants have been revealed to coexist with but not overlap the “Ca. Hodgkinia”-occupied syncytium in the bacteriomes of cicadas of the genus Tettigades (27, 28). A previous study revealed that the ratio of the numbers of transmitted “Ca. Hodgkinia”/“Ca. Sulcia” cells was nearly 1:1 in cicadas having a single “Ca. Hodgkinia” lineage, whereas it was 11.2:1 in the species harboring the most complex “Ca. Hodgkinia” lineages (33). In our present study, the ratio of the numbers of transmitted “Ca. Hodgkinia”/“Ca. Sulcia” cells was estimated to be nearly 9:1 in Py. repanda (see Table S9 in the supplemental material). We are not aware of a prior description of “Ca. Hodgkinia” in the bacteriomes of Py. repanda exhibiting dark and light cells (Fig. 3). Furthermore, RFLP profiles were observed for “Ca. Hodgkinia” cells in the bacteriomes of Py. repanda, and 16S rRNA gene sequences of “Ca. Hodgkinia” were dissimilar (Table S3). In addition, only the darker “Ca. Hodgkinia” cells could be observed after emigrating from the central bacteriocytes through the multinuclear compartments in our study (Fig. S8 and Table S7). Therefore, we suggest that “Ca. Hodgkinia” may have split into cytologically distinct lineages within this cicada species and that related cicadas might have developed quite complex mechanisms for the vertical transmission of the bacteriome-associated symbionts. Further investigations are required to confirm whether “Ca. Hodgkinia” variants are a particularly common evolutionary phenomenon within cicadas, and the results may provide a better understanding of vertical transmission mechanisms of bacteriome-associated symbionts in hemipteran insects.
Although previous studies have revealed that “Ca. Sulcia” and its coresident symbionts have a route of vertical transmission between cicada generations (29), our knowledge of the detailed transmission routes of these symbionts is very limited. Histological, ultrastructural, and fluorescence microscopy analyses in our study revealed that “Ca. Sulcia” is present in the peripheral bacteriocytes and that “Ca. Hodgkinia” is harbored in the central bacteriocytes of Py. repanda. The localization of “Ca. Sulcia” and “Ca. Hodgkinia” within the bacteriocytes of Py. repanda is generally similar to previous observations of a few other cicada species (27–29). In comparison with the majority of auchenorrhynchan insects, cicadas develop a similar mechanism of transovarial transmission for the primary symbionts from one generation to the next. We clarified that Arsenophonus together with these two primary symbionts were released from the bacteriocytes into the hemolymph of cicadas, gathered around the posterior pole of the oocytes, emigrated through the cytoplasm of the follicular epithelium into the perivitelline space, and finally formed a characteristic symbiont ball in each egg (Fig. 5). Interestingly, “Ca. Sulcia” and “Ca. Hodgkinia” changed their shape from irregular to roughly spherical when leaving the bacteriocytes for the hemolymph, and both of them rechanged their shape from roughly spherical to irregular before entering the perivitelline space from the follicular epithelium of the posterior pole of the oocytes (Fig. 5 and Table S8). The reason for this remains unknown. Some hypotheses are that the transformation of “Ca. Sulcia” and “Ca. Hodgkinia” may be related to the extremely limited space and/or the novel microenvironments in the ovaries and bacteriomes. We presumed that the spherical shape of symbionts could significantly reduce energy consumption during the transovarial transmission process, which may be beneficial for them to successfully adapt to certain novel microenvironments. In contrast, primary symbionts with irregular shapes in the bacteriomes, perivitelline space, and oocyte space may increase their extracellular surface, which probably enhances nutrient absorption and transport, energetic metabolism, and biosynthetic pathways. However, an alternative explanation for this phenomenon could be that transformation is an inherent characteristic of “Ca. Sulcia” and “Ca. Hodgkinia,” which merits further investigation.
FIG 5.
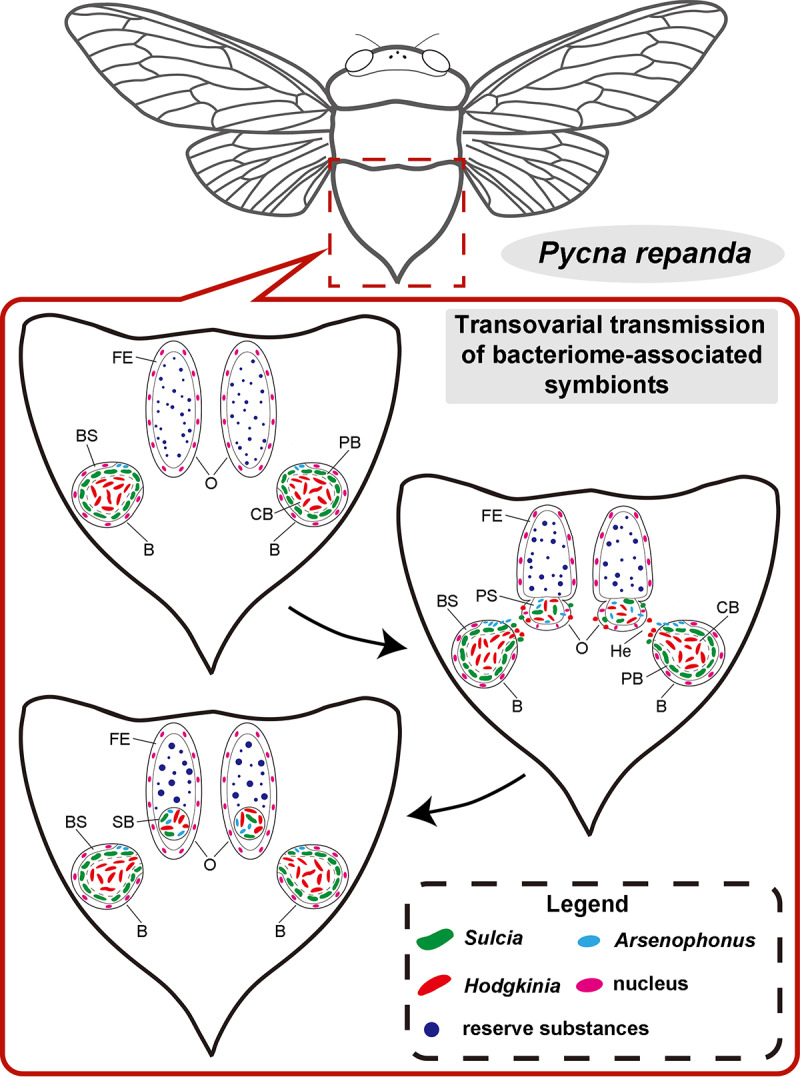
Schematic representation showing the transovarial transmission process for the bacteriome-associated symbionts of Pycna repanda. In mature females, Arsenophonus together with “Ca. Hodgkinia” and “Ca. Sulcia” leave the bacteriome (B) for the hemolymph (He), gather toward the posterior pole of the oocytes (O), migrate through the single-layered follicular epithelium (FE) into the perivitelline space (PS), and finally form a characteristic symbiont ball (SB) at the posterior pole of the oocytes. BS, bacteriome sheath; CB, central bacteriocyte; PB, peripheral bacteriocyte.
The prominent secondary symbionts Rickettsia and Arsenophonus are associated with cicadas.
The facultative symbionts Rickettsia and Arsenophonus are also prominent in Py. repanda, among which Rickettsia was detected in the salivary glands, digestive organs, and testes. Rickettsia was reported to reside in the salivary glands and some digestive and reproductive organs in hemipteran insects, e.g., most organs in the body cavity except the bacteriomes in the whitefly Bemisia tabaci (19); the salivary glands, Malpighian tubules, midgut, and testes of the cicada Pl. kaempferi; and the midgut of the cicada M. mongolica (36). Rickettsia was hypothesized to take part in making the gelling saliva needed for stylet penetration in host plant leaves as well as facilitating food digestion (19) and was reported to be horizontally transmitted through host plant sap (40). This symbiont might help Py. repanda digest food, detoxify plant chemicals, and/or manipulate reproduction. However, future studies are needed to clarify the potential functions and transmission mechanism of this secondary symbiont in Cicadidae.
Previous studies have revealed that Arsenophonus may be horizontally and vertically transmitted (41, 42). It was revealed that Arsenophonus is harbored in the cytoplasm of “Ca. Sulcia” in the leafhopper Macrosteles laevis, thereby ensuring its transovarial transmission between host generations (30). Arsenophonus was restricted to the bacteriocytes at all developmental stages of B. tabaci, which may be involved in conferring functional benefits to the host rather than manipulating reproduction (16). However, it has been demonstrated that Arsenophonus nasoniae killed males of the wasp Nasonia vitripennis by inhibiting maternal centrosome formation (43). Additionally, Arsenophonus together with Hamiltonella contributed to the fitness of Aphis gossypii by enhancing its performance but not through parasitoid resistance (44). It has been shown that Arsenophonus infection increased A. gossypii requirements for the amino acid phenylalanine but decreased requirements for leucine (45). Our results revealed that Arsenophonus can coexist with “Ca. Sulcia” and “Ca. Hodgkinia” in bacteriocytes and can be transovarially transmitted together with these two obligate symbionts from mother to offspring in Py. repanda, but it is not harbored in the cytoplasm of “Ca. Sulcia” (Fig. 5). Therefore, we hypothesize that Arsenophonus may rely on bacteriomes for essential nutrients and protect itself from host immunity and that it can benefit from coexisting with the primary symbionts inside the bacteriomes to ensure its transovarial transmission from mother to offspring. We describe here that Arsenophonus is not harbored in the cytoplasm of other endosymbionts, but it can be transovarially transmitted from mother to offspring in cicadas. Arsenophonus is probably not essential for host survival, but it might be involved in conferring functional benefits to the host. The exact function of this symbiont and whether it is commonly harbored in cicadas need to be investigated in the future. It is also unclear whether it can be horizontally transmitted in cicadas, which merits further investigation.
MATERIALS AND METHODS
Sample collection and dissection.
During the adult emergence period, adults of Py. repanda were captured in the Tongtian River National Forest Park (34°11′N, 106°35′E; Fengxian County, Shaanxi Province, China) from late July to the middle of August each year from 2017 to 2019. Live specimens were immediately transferred to the laboratory for subsequent dissection.
Before dissection, cicadas were anesthetized at −20°C for 5 min, externally sterilized with 75% ethanol, and then rinsed with sterile water five times. Dissection was performed under a stereoscopic zoom microscope (Motic SMZ168; Xiamen, China) under sterile conditions with sterile forceps to obtain intact salivary glands, digestive organs (i.e., filter chamber, conical segment, midgut, and hindgut), bacteriomes, and reproductive organs. To prevent cross-contamination, all tools were strictly flame sterilized when reused. The dissected samples were fixed for ultrastructural and fluorescence microscopy or transferred to 1.5-ml centrifuge tubes and then stored in a −80°C freezer for subsequent DNA extraction. Additionally, the dissected bacteriomes of both sexes were photographed under the stereoscopic zoom microscope.
High-throughput sequencing analysis of bacterial communities.
For high-throughput sequencing, each tissue sample contained four biological replicates. The bacteriomes and reproductive organs of both sexes and the digestive organs (i.e., filter chamber, conical segment, midgut, and hindgut) as well as the salivary glands of Py. repanda males were used for DNA extraction. Genomic DNA of each sample was extracted using a DNeasy blood and tissue kit (Tiangen Inc.) according to the manufacturer’s instructions. PCR amplification was performed using template-specific primers with barcodes 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′), targeting the V3-V4 hypervariable regions of the 16S rRNA gene sequences (46, 47). The amplified region is considered suitable for analyzing the bacterial community (48). PCR cycle conditions were previously described (47). The PCR products were examined by 1% agarose gel electrophoresis and then purified with a universal DNA purification kit (Tiangen Inc.) according to the manufacturer’s standard protocol. After determining the concentration and quality by using QuantiFluor (Promega, USA), the purified PCR products were sent for sequencing on the HiSeq 2500 PE250 platform (Gene Denovo Biotechnology Co., Ltd., Guangzhou, China).
QIIME software was used to remove low-quality reads in raw Illumina fastq data sets (49). Paired reads were merged into raw tags by using Flash (version 1.2.11; http://ccb.jhu.edu/software/FLASH/), and the raw tags were filtered in QIIME (version 1.9.1; http://qiime.org/) to obtain high-quality tags for further analysis. Chimeras were checked and removed using the mothur package (version 1.39.1; https://www.mothur.org/) (50). The cleaned tags were aligned into operational taxonomic units (OTUs) with Uparse at a 97% similarity level (49). The Ribosome Database Project (RDP) classifier was used to assign clustered bacterial OTUs against the SILVA database for taxonomic classifications. We performed a nonparametric test (Kruskal-Wallis test) to test the differences of Proteobacteria and Bacteroidetes among different tissues. To reveal the similarities of bacterial communities, we performed principal-component analysis (PCA) using R software. In addition, permutational multivariate analysis of variance (PERMANOVA) was conducted to determine the differences in bacterial communities among different tissues.
Bacterial composition of bacteriomes in males analyzed by RFLP-based cloning.
For RFLP-based cloning to analyze the bacterial composition of the bacteriomes in males, samples contained three biological replicates. PCR amplification of the 16S rRNA gene was performed using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (51). The PCR mixture contained 12.5 μl Premix Taq DNA polymerase, 8.5 μl double-distilled water (ddH2O), 2 μl template DNA, and 1 μl each of the forward and reverse primers (10 μΜ each). The following PCR cycle conditions were used. There was an initial denaturation step at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 2 min and a final elongation step at 72°C for 5 min. The PCR products were checked by 1% agarose gel electrophoresis, purified with a universal DNA purification kit (Tiangen Inc.), cloned into the pMD 19-T vector (TaKaRa Inc.), and subsequently transformed into Escherichia coli DH5α competent cells (TaKaRa Inc.).
For each biological replicate, ∼200 positive clones were selected randomly from a white-blue selection system containing ampicillin and X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and then grown overnight in 900 μl liquid lysogeny broth (LB) medium at 37°C. Extracted plasmid DNA was used as the template for subsequent PCR amplification with M13 vector primers to check positive clones. Positive clones were determined as previously described (52). If the PCR product was the expected size of nearly 1.5 kb, it was digested with the restriction endonucleases HhaI and XspI at 37°C for 4 h according to the manufacturer’s protocol (TaKaRa Inc.). The restriction fragments were separated by 1% agarose gel electrophoresis and analyzed under UV light. Three representative clones for each RFLP profile were randomly selected for sequencing. In total, 15 representative clones were subjected to sequencing at Sangon Biotech Co., Ltd. (Shanghai, China).
Reconstruction of phylogenetic relationships of “Ca. Sulcia” and “Ca. Hodgkinia” in cicadas using 16S rRNA gene sequences.
Chromas Pro software (Technelysium Pty., Ltd., Australia) was used to check the sequence chromatograms. Multiple alignments of the 16S rRNA gene sequences of the “Ca. Sulcia” and “Ca. Hodgkinia” symbionts were performed using the program Clustal X (53, 54), and the reliability of the aligned sequences was determined using the program GUIDANCE2 (55). Subsequently, we removed the gappy columns at the beginning and end of the aligned sequences with BioEdit software (56). The phylogenetic trees were constructed using Bayesian inference (BI) with the program MrBayes version 3.1.2 (57) and using maximum likelihood (ML) with the program RAxML version 8.1.5 (58).
Diagnostic PCR analyses of dominant symbionts in different tissues.
Diagnostic PCR amplification was performed for 30 males and 25 females of Py. repanda to confirm the physical distribution of dominant symbionts (i.e., “Ca. Sulcia,” “Ca. Hodgkinia,” Rickettsia, and Arsenophonus) in different tissues (i.e., salivary glands, filter chamber, conical segment, midgut, hindgut, Malpighian tubules, testes, ovaries, and bacteriomes). Target-specific primers were designed using Primer-BLAST, which can place primers according to single-nucleotide polymorphism (SNP) locations and exon/intron boundaries (59). Although the Primer-BLAST program can check the specificity of the generated PCR primers, the amplified PCR products were also sent for sequencing to further test specific PCR amplification. PCR primers used for the amplification of 16S rRNA gene sequences of dominant symbionts are summarized in Table 1. PCR amplification was carried out under the following cycling conditions: an initial denaturation step at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 2 min and a final extension step at 72°C for 5 min. The PCR products were determined by 1% agarose gel electrophoresis and then purified with a universal DNA purification kit (Tiangen Inc.). Representative PCR products were sent for sequencing at Sangon Biotech Co., Ltd. (Shanghai, China).
TABLE 1.
Primers used for amplification of dominant symbionts in this study
| Target | Primer name | Primer sequence (5′–3′) | Reference |
|---|---|---|---|
| “Ca. Hodgkinia” | Hod-F | TCTTACGACTTCACCTCGGTC | This study |
| Hod-R | CACATGCAAGTCAAGCGAAC | ||
| “Ca. Sulcia” | 10_CFB_FF | AGAGTTTGATCATGGCTCAGGATG | 2 |
| 1515_R | GTACGGCTACCTTGTTACGACTTAG | ||
| Arsenophonus | Ars-F | CAATGGGCGAAAGCCTGATG | This study |
| Ars-R | ACCCCAGTCATGAACCACAAA | ||
| Rickettsia | Rick-F | GTGGGAATCTGCCCATCAGT | This study |
| Rick-R | GCAGTGTGTACAAGRCCCGA | ||
Histological and ultrastructural microscopy.
The bacteriomes (18 males and 18 females) of both sexes and ovaries (18 females) were dissected rapidly in 0.1 M phosphate-buffered saline (PBS) (pH 7.2) and fixed in 2.5% glutaraldehyde in 0.1 M PBS (pH 7.2) at 4°C overnight. After rinsing with PBS five times, the samples were postfixed with 1% osmium tetroxide (OsO4) in 0.1 M PBS for 1.5 h at 4°C. After washing with PBS six times, the samples were dehydrated in a graded ethanol series (30%, 50%, 70%, 80%, and 90% for 10 min twice; 95% for 15 min twice; and 100% for 30 min twice). Next, the samples were infiltrated with a graded mixture of ethanol and White London resin (LR) (Sigma-Aldrich, USA), subsequently infiltrated with LR for 24 h twice, and eventually embedded in pure LR, polymerized at 60°C for 48 h.
Semithin sections (1 μm) were cut with a glass knife, stained with 1% methylene blue, and photographed under a DM6 B light microscope (Leica, Germany). Ultrathin sections (70 nm) for this study were cut with a diamond knife on the ultramicrotome, doubly stained with uranyl acetate and lead citrate, and finally examined under a Tecnai G2 Spirit Bio Twin microscope (FEI, Czech Republic).
Fluorescence in situ hybridization.
The bacteriomes and ovaries of female adults (25 individuals) were fixed in 4% paraformaldehyde, dehydrated in a graded ethanol series, cleared four times in xylene for 2 h, and finally embedded in melted paraffin. Paraffin blocks were sectioned to 4 μm. Thin sections were used for histological or fluorescence microscopy. Sections for histological microscopy were stained with hematoxylin-eosin and photographed under a DM6 B light microscope (Leica, Germany).
Fluorescence in situ hybridization (FISH) was conducted in this study to further reveal the distribution and vertical transmission of Arsenophonus, “Ca. Hodgkinia,” and “Ca. Sulcia” in the ovaries and bacteriomes of Py. repanda. The FISH assay was performed as previously described (27, 28). The probe sequences were Cy5-CCAATGTGGCTGACCGT or Cy5-TTGCGACTTTCTGTCTCCCA for “Ca. Hodgkinia” (28, 29), Cy3-CCAATGTGGGGGWACGC or Cy3-CCACACATTCCAGTTACTCC for “Ca. Sulcia” (27, 28), and Alexa Fluor 488-TCATGACCACAACCTCCAAA for Arsenophonus (16). Briefly, analysis of each slide was carried out using a final volume of 25 μl of hybridization buffer, which contained 0.25% bovine serum albumin (BSA), 2.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 12.5% dextran sulfate, and fluorescently labeled probes at 200 nM. Hybridization was performed overnight at 37°C in a humidified chamber. Also, a negative control was done to check the specificity of hybridization using only one symbiont-targeted probe and no probe staining. Slides were eventually observed and imaged under an FV1000 IX confocal microscope (Olympus, Japan).
Accession number(s).
The Illumina data obtained in this study have been deposited in the GenBank database under accession no. SRP189619.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhenpeng Wang, Songshan Wei (Northwest A&F University, China), and Nan Zhang (Tongtian River National Forest Park, China) for collecting samples and Jing Chen, Guoliang Pei, and Mi Huo (State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, China) and Aihua Liang, Zhen Wang, and Kerang Huang (Life Science Research Core Services, Northwest A&F University, China) for the assistance in ultrastructural observations. We also thank J. Billen (Zoological Institute, KU Leuven, Belgium) for revising the manuscript.
This study was supported by the National Natural Science Foundation of China (grant no. 31772505 and 31572302). We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 2.Moran NA, Tran P, Gerardo NM. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol 71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redak RA, Purcell AH, Lopes JR, Blua MJ, Mizell RF, Andersen PC. 2004. The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu Rev Entomol 49:243–270. doi: 10.1146/annurev.ento.49.061802.123403. [DOI] [PubMed] [Google Scholar]
- 4.Ishii Y, Matsuura Y, Kakizawa S, Nikoh N, Fukatsu T. 2013. Diversity of bacterial endosymbionts associated with Macrosteles leafhoppers vectoring phytopathogenic phytoplasmas. Appl Environ Microbiol 79:5013–5022. doi: 10.1128/AEM.01527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moran NA, Plague GR, Sandström JP, Wilcox JL. 2003. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc Natl Acad Sci U S A 100:14543–14548. doi: 10.1073/pnas.2135345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 7.Douglas AE. 2009. The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47. doi: 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- 8.Bright M, Bulgheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koga R, Meng XY, Tsuchida T, Fukatsu T. 2012. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci U S A 109:E1230–E1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuura Y, Kikuchi Y, Miura T, Fukatsu T. 2015. Ultrabithorax is essential for bacteriocyte development. Proc Natl Acad Sci U S A 112:9376–9381. doi: 10.1073/pnas.1503371112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dale C, Moran NA. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Fukatsu T, Tsuchida T, Nikoh N, Koga R. 2001. Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol 67:1284–1291. doi: 10.1128/AEM.67.3.1284-1291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montllor CB, Maxmen A, Purcell AH. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195. doi: 10.1046/j.1365-2311.2002.00393.x. [DOI] [Google Scholar]
- 14.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brumin M, Kontsedalov S, Ghanim M. 2011. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci 18:57–66. doi: 10.1111/j.1744-7917.2010.01396.x. [DOI] [Google Scholar]
- 16.Gottlieb Y, Ghanim M, Gueguen G, Kontsedalov S, Vavre F, Fleury F, Zchori-Fein E. 2008. Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J 22:2591–2599. doi: 10.1096/fj.07-101162. [DOI] [PubMed] [Google Scholar]
- 17.Skaljac M, Zanic K, Ban SG, Kontsedalov S, Ghanim M. 2010. Co-infection and localization of secondary symbionts in two whitefly species. BMC Microbiol 10:142. doi: 10.1186/1471-2180-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zchori-Fein E, Gottlieb Y, Kelly SE, Brown JK, Wilson JM, Karr TL, Hunter MS. 2001. A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. Proc Natl Acad Sci U S A 98:12555–12560. doi: 10.1073/pnas.221467498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brumin M, Levy M, Ghanim M. 2012. Transovarial transmission of Rickettsia spp. and organ-specific infection of the whitefly Bemisia tabaci. Appl Environ Microbiol 78:5565–5574. doi: 10.1128/AEM.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarzewicz T, Grzywacz B, Szwedo J, Michalik A. 2016. Bacterial symbionts of the leafhopper Evacanthus interruptus (Linnaeus, 1758) (Insecta, Hemiptera, Cicadellidae: Evacanthinae). Protoplasma 253:379–391. doi: 10.1007/s00709-015-0817-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bution ML, Caetano FH, Zara FJ. 2008. Contribution of the Malpighian tubules for the maintenance of symbiotic microorganisms in cephalotes ants. Micron 39:1179–1183. doi: 10.1016/j.micron.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Gonella E, Crotti E, Rizzi A, Mandrioli M, Favia G, Daffonchio D, Alma A. 2012. Horizontal transmission of the symbiotic bacterium Asaia sp. in the leafhopper Scaphoideus titanus Ball (Hemiptera: Cicadellidae). BMC Microbiol 12(Suppl 1):S4. doi: 10.1186/1471-2180-12-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukatsu T, Nikoh N, Kawai R, Koga R. 2000. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol 66:2748–2758. doi: 10.1128/aem.66.7.2748-2758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCutcheon JP, McDonald BR, Moran NA. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A 106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol 2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett GM, Moran NA. 2015. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci U S A 112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Leuven JT, Meister RC, Simon C, McCutcheon JP. 2014. Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one. Cell 158:1270–1280. doi: 10.1016/j.cell.2014.07.047. [DOI] [PubMed] [Google Scholar]
- 28.Łukasik P, Nazario K, Van Leuven JT, Campbell MA, Meyer M, Michalik A, Pessacq P, Simon C, Veloso C, McCutcheon JP. 2018. Multiple origins of interdependent endosymbiotic complexes in a genus of cicadas. Proc Natl Acad Sci U S A 115:E226–E235. doi: 10.1073/pnas.1712321115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuura Y, Moriyama M, Łukasik P, Vanderpool D, Tanahashi M, Meng XY, McCutcheon JP, Fukatsu T. 2018. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc Natl Acad Sci U S A 115:E5970–E5979. doi: 10.1073/pnas.1803245115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobiałka M, Michalik A, Walczak M, Junkiert Ł, Szklarzewicz T. 2016. Sulcia symbiont of the leafhopper Macrosteles laevis (Ribaut, 1927) (Insecta, Hemiptera, Cicadellidae: Deltocephalinae) harbors Arsenophonus bacteria. Protoplasma 253:903–912. doi: 10.1007/s00709-015-0854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koga R, Bennett GM, Cryan JR, Moran NA. 2013. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 32.Campbell MA, Van Leuven JT, Meister RC, Carey KM, Simon C, McCutcheon JP. 2015. Genome expansion via lineage splitting and genome reduction in the cicada endosymbiont Hodgkinia. Proc Natl Acad Sci U S A 112:10192–10199. doi: 10.1073/pnas.1421386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell MA, Łukasik P, Meyer MC, Buckner M, Simon C, Veloso C, Michalik A, McCutcheon JP. 2018. Changes in endosymbiont complexity drive host-level compensatory adaptations in cicadas. mBio 9:e02104-18. doi: 10.1128/mBio.02104-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W, Nan X, Zheng Z, Wei C, He H. 2015. Analysis of inter-individual bacterial variation in gut of cicada Meimuna mongolica (Hemiptera: Cicadidae). J Insect Sci 15:131. doi: 10.1093/jisesa/iev113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Huang Z, He H, Wei C. 2018. Comparative analysis of microbial communities associated with bacteriomes, reproductive organs and eggs of the cicada Subpsaltria yangi. Arch Microbiol 200:227–235. doi: 10.1007/s00203-017-1432-8. [DOI] [PubMed] [Google Scholar]
- 36.Zheng Z, Wang D, He H, Wei C. 2017. Bacterial diversity of bacteriomes and organs of reproductive, digestive and excretory systems in two cicada species (Hemiptera: Cicadidae). PLoS One 12:e0175903. doi: 10.1371/journal.pone.0175903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Wei C. 2020. Bacterial communities in digestive and excretory organs of cicadas. Arch Microbiol 202:539–553. doi: 10.1007/s00203-019-01763-4. [DOI] [PubMed] [Google Scholar]
- 38.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 39.Gonella E, Negri I, Marzorati M, Mandrioli M, Sacchi L, Pajoro M, Crotti E, Rizzi A, Clementi E, Tedeschi R, Bandi C, Alma A, Daffonchio D. 2011. Bacterial endosymbiont localization in Hyalesthes obsoletus, the insect vector of bois noir in Vitis vinifera. Appl Environ Microbiol 77:1423–1435. doi: 10.1128/AEM.02121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caspi-Fluger A, Inbar M, Mozes-Daube N, Katzir N, Portnoy V, Belausov E, Hunter MS, Zchori-Fein E. 2012. Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc Biol Sci 279:1791–1796. doi: 10.1098/rspb.2011.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran NA, Baumann P. 2000. Bacterial endosymbionts in animals. Curr Opin Microbiol 3:270–275. doi: 10.1016/s1369-5274(00)00088-6. [DOI] [PubMed] [Google Scholar]
- 42.Jousselin E, Cœur d’Acier A, Vanlerberghe-Masutti F, Duron O. 2013. Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol Ecol 22:260–270. doi: 10.1111/mec.12092. [DOI] [PubMed] [Google Scholar]
- 43.Ferree PM, Avery A, Azpurua J, Wilkes T, Werren JH. 2008. A bacterium targets maternally inherited centrosomes to kill males in Nasonia. Curr Biol 18:1409–1414. doi: 10.1016/j.cub.2008.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayoubi A, Talebi AA, Fathipour Y, Mehrabadi M. 2020. Coinfection of the secondary symbionts, Hamiltonella defensa and Arsenophonus sp. contribute to the performance of the major aphid pest, Aphis gossypii (Hemiptera: Aphididae). Insect Sci 27:86–98. doi: 10.1111/1744-7917.12603. [DOI] [PubMed] [Google Scholar]
- 45.Tian P, Chang C, Miao N, Li M, Liu X. 2019. Infections with Arsenophonus facultative endosymbionts alter performance of aphids (Aphis gossypii) on an amino-acid-deficient diet. Appl Environ Microbiol 85:e01407-19. doi: 10.1128/AEM.01407-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeibich L, Schmidt O, Drake HL. 2018. Protein- and RNA-enhanced fermentation by gut microbiota of the earthworm Lumbricus terrestris. Appl Environ Microbiol 84:e00657-18. doi: 10.1128/AEM.00657-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinclair L, Osman OA, Bertilsson S, Eiler A. 2015. Microbial community composition and diversity via 16S rRNA gene amplicons: evaluating the Illumina platform. PLoS One 10:e0116955. doi: 10.1371/journal.pone.0116955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunger S, Gößner AS, Drake HL. 2015. Anaerobic trophic interactions of contrasting methane-emitting mire soils: processes versus taxa. FEMS Microbiol Ecol 91:fiv045. doi: 10.1093/femsec/fiv045. [DOI] [PubMed] [Google Scholar]
- 53.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. 1998. Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 55.Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res 43:W7–W14. doi: 10.1093/nar/gkv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 41:95–98. [Google Scholar]
- 57.Ronquist F, Huelsenbeck JP. 2003. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 58.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.