Class I BPCs repress FUSCA3 during reproductive and seed development to promote ovule development while coordinating embryo and endosperm growth.
Abstract
Spatiotemporal regulation of gene expression is critical for proper developmental timing in plants and animals. The transcription factor FUSCA3 (FUS3) regulates developmental phase transitions by acting as a link between hormonal pathways in Arabidopsis (Arabidopsis thaliana). However, the mechanisms governing its spatiotemporal expression pattern are poorly understood. Here, we show that FUS3 is repressed in the ovule integuments and seed endosperm. FUS3 repression requires class I BASIC PENTACYSTEINE (BPC) proteins, which directly bind GA/CT cis-elements in FUS3 and restrict its expression pattern. During vegetative and reproductive development, FUS3 derepression in bpc1-1 bpc2 (bpc1/2) double mutant or misexpression in ProML1:FUS3 lines causes dwarf plants carrying defective flowers and aborted ovules. After fertilization, ectopic FUS3 expression in bpc1/2 endosperm or ProML1:FUS3 endosperm and endothelium increases endosperm nuclei proliferation and seed size, causing delayed or arrested embryo development. These phenotypes are rescued in bpc1/2 fus3-3. Finally, class I BPCs interact with FIS-PRC2 (FERTILIZATION-INDEPENDENT SEED-Polycomb Repressive Complex2), which represses FUS3 in the endosperm during early seed development. We propose that BPC1 and 2 promote the transition from reproductive to seed development by repressing FUS3 in ovule integuments. After fertilization, BPC1 and 2 and FIS-PRC2 repress FUS3 in the endosperm to coordinate early endosperm and embryo growth.
INTRODUCTION
Because seeds provide ∼70% of the world’s human caloric intake in the form of food and animal feed, plant fertility is one of the most important traits for agricultural crop production (Sreenivasulu and Wobus, 2013). Seed abortion, which can be triggered by environmental stress, can cause large food and economic losses. Therefore, a thorough understanding of the molecular mechanisms regulating plant fertility is paramount. Sexual reproduction in seed plants starts with the production of female and male gametes. In the mature ovule, the maternal sporophytic integuments, which originate from the chalaza, enclose the female gametophyte (embryo sac), which contains two gametes: the haploid egg cell and the diploid central cell (Gasser and Skinner, 2019). After fertilization of the central cell, the triploid endosperm nuclei undergo multiple rounds of division, which are followed by cellularization. In most angiosperms, the function of the endosperm is to nourish the developing embryo.
Fertilization of the egg cell generates the diploid zygote, which divides asymmetrically producing two daughter cell lineages that form the apical embryo proper and basal suspensor, respectively (Lafon-Placette and Köhler, 2014; Dresselhaus et al., 2016; Gasser and Skinner, 2019). In angiosperms, the seed comprises three genetically distinct tissues: the diploid embryo, the triploid endosperm, and the diploid sporophytic seed coat. Auxin has been shown to play a major role in coordinating the development of these three tissues (Lau et al., 2012; de Vries and Weijers, 2017; Figueiredo and Köhler, 2018; Robert, 2019). In the absence of fertilization, seed development is repressed by the Polycomb-repressive complex2 (PRC2). In particular, the FERTILIZATION INDEPENDENT SEED (FIS)-PRC2 complex represses autonomous endosperm development, while EMBRYONIC FLOWER (EMF)-PRC2 and VERNALIZATION INDEPENDENT (VRN)-PRC2 prevent seed coat development in the absence of fertilization. Notably, mutations in FIS-PRC2 or impairment of auxin synthesis and signaling affect the development of these tissues leading to seed abortion (Roszak and Köhler, 2011; Figueiredo and Köhler, 2018; Robert, 2019). However, the molecular mechanisms that regulate the transition from reproductive to seed development downstream of auxin and PRC2 are largely unknown.
During seed maturation, the embryo accumulates storage compounds such as storage proteins and lipids, acquires dormancy, and establishes desiccation tolerance, whereas the endosperm undergoes programmed cell death and is progressively absorbed by the embryo (Sreenivasulu and Wobus, 2013). The transition from embryo-pattern formation to seed maturation is largely controlled by the LAFL proteins, which include the B3-domain transcription factors LEC2 (LEAFY COTYLEDON2), ABI3 (ABSCISIC ACID INSENSTIVE3), and FUS3 (FUSCA3), as well as the NF-YB subunits of the CCAAT binding complex LEC1 and LEC1-LIKE. LAFL genes are sequentially expressed during seed development, and loss-of-function mutations in these genes profoundly affects seed maturation, leading overall to reduced accumulation of seed storage proteins and lipids, reduced dormancy, and desiccation intolerance (Jia et al., 2014; Fatihi et al., 2016; Carbonero et al., 2017).
During germination, the maturation program is repressed by epigenetic mechanisms to allow the transition to vegetative growth, the next phase of development. These mechanisms include CHROMODOMAIN HELICASE DNA BINDING3 (CHD3)/PICKLE (PKL)-dependent chromatin remodeling and PRC2-mediated histone 3 Lys-27 trimethylation (H3K27me3); H2AK119ub1 deposition by the PRC1 components RING-finger homologs AtBMI1A and AtBMI1B; and VIP1/ABI3/LEC (VAL)-mediated recruitment of a histone deacetylase complex (Jia et al., 2014; Lepiniec et al., 2018). Derepression of LAFL genes in mutants affected in these processes results in the expression of seed-specific traits and arrested vegetative development. Similar phenotypes are found when LAFL genes are ectopically expressed postembryonically (Lotan et al., 1998; Stone et al., 2001; Gazzarrini et al., 2004; Braybrook et al., 2006). Clearly, multiple pathways are required to ensure stable repression of the embryonic program in order to allow vegetative growth. However, repression of LAFL genes by epigenetic mechanisms, as well as by posttranscriptional regulation, has also been shown during early embryonic development (Makarevich et al., 2006; Nodine and Bartel, 2010; Willmann et al., 2011; Tang et al., 2012; Vashisht and Nodine, 2014). This suggests that LAFL expression is tightly controlled not only during vegetative growth but also in specific seed tissues. However, the mechanisms regulating LAFL expression patterns during seed development are less clear.
Recently, we have shown that FUS3 plays a critical role also in reproductive development. The fus3-3 loss-of-function mutant displays ovule and seed abortion, which is enhanced in plants grown at elevated temperature and is dependent on phosphorylation of maternally derived FUS3 (Chan et al., 2017). Interestingly, ProML1:FUS3-GFP plants that misexpress FUS3 during reproductive development show aborted siliques, suggesting that spatiotemporal expression of FUS3 must be tightly regulated at this stage of development (Gazzarrini et al., 2004). Here, we show that FUS3 is indeed expressed during ovule development. Before fertilization, FUS3 is confined to the chalaza and funiculus of mature ovules, whereas following fertilization FUS3 is localized in the funiculus, seed coat, and chalaza. Class I BASIC PENTACYSTEINE1 (BPC1) binds to FUS3 in vivo during reproductive development and represses FUS3 in the integuments of mature ovules, as well as in the endosperm of developing seeds. FUS3 misexpression in the bpc1-1 bpc2 (bpc1/2) double mutant reduces plant height and impairs the development of flowers, ovules, and endosperm leading to ovule abortion and delayed or arrested embryogenesis, the latter causing seed abortion. Similar phenotypes are recapitulated in ProML1:FUS3-GFP misexpression plants. Furthermore, the strong vegetative phenotype (dwarf plants) and reproductive phenotype (ovule and seed abortion) of bpc1/2 can be partially rescued in the fus3-3 background, strongly indicating that they are partially caused by ectopic FUS3 expression. Last, we show that BPC1-3 interact with the FIS-PRC2 complex in planta. We propose that during reproductive development, BPC1- BPC2- and PRC2-mediated restriction of FUS3 expression is required for ovule development, whereas after fertilization FUS3 repression in the endosperm is necessary to coordinate endosperm and embryo growth. Hence, correct spatiotemporal expression of FUS3 plays a critical function during plant reproduction and early stages of seed development.
RESULTS
Spatiotemporal Regulation of FUS3 Expression during Reproductive Development Is Required for Proper Ovule Development
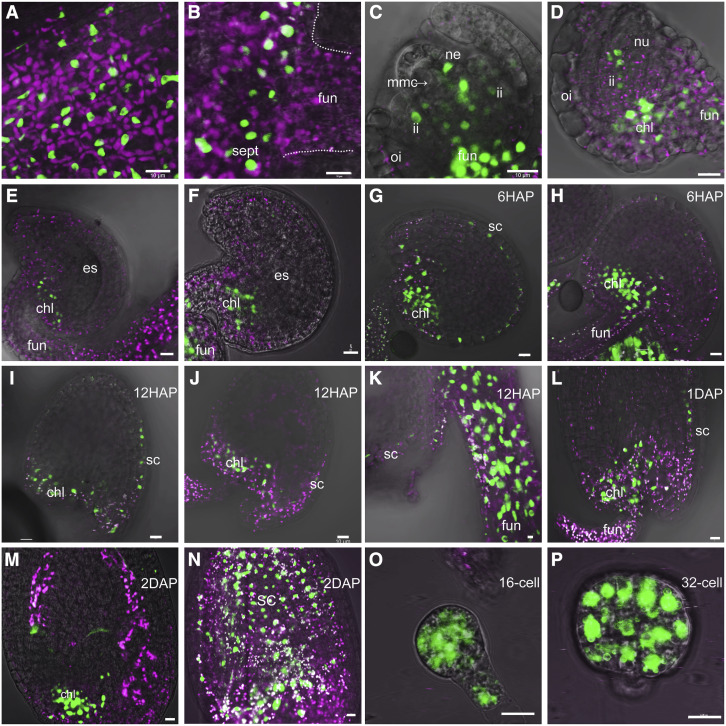
Both FUS3 transcript and FUS3 protein have been detected in embryos as early as the globular stage, but no expression was reported in reproductive organs (Kroj et al., 2003; Gazzarrini et al., 2004; Tsuchiya et al., 2004). To investigate the role of FUS3 in reproductive development, we first determined the FUS3 localization pattern in flower buds using a ProFUS3:FUS3-GFP translational reporter (Gazzarrini et al., 2004). However, no FUS3-GFP fluorescence was detected, likely due to the fast turnover rate of FUS3 (Lu et al., 2010). We then used a ProFUS3:FUS3ΔC-GFP reporter, which lacks the activation domain and the PEST instability motif (enriched in Pro, Glu, Ser, Thr) of FUS3 and allows detection of low FUS3 protein levels (Lu et al., 2010). This reporter is nonfunctional (it does not rescue fus3-3), but recapitulates FUS3 expression patterns determined by qRT-PCR, ProFUS3:GUS, and ProFUS3:GFP reporters (Lu et al., 2010). With use of the ProFUS3:FUS3ΔC-GFP reporter, the FUS3 protein was found to be localized to the pistil (septum, valves, and funiculus) and ovules, in agreement with microarray data (Figures 1A to 1F; Supplemental Figure 1A). In developing ovules, FUS3ΔC-GFP was localized to the epidermis of the nucellus, the chalaza, and funiculus, whereas in mature ovules (female gametophyte stage FG7; Yadegari and Drews, 2004) it was localized to the chalaza and funiculus (Figures 1C to 1F). After fertilization, (6 to 48 h) FUS3ΔC-GFP was present in the funiculus, the outer layer of the seed coat, the chalaza, and the micropyle; it was also localized to the embryo at early stages of embryogenesis (Figures 1G to 1P; Supplemental Figure 1B).
Figure 1.
FUS3 Localization in Developing Ovules and Early Stages of Seed Development.
(A) to (P) Confocal images showing ProFUS3:FUS3ΔC-GFP localization in Arabidopsis.
(A) and (B) FUS3ΔC-GFP in the epidermis of the valve (A) and septum (B) of the pistil. Dotted lines outline the septum and funiculus.
(C) to (F) Ovules during female megasporogenesis (C) and megagametogenesis at FG1-FG7 (D) to (F); FUS3ΔCGFP was localized to the nucellar epidermis (C), inner and outer integuments in (C) and (D), funiculus and chalaza in (C) to (F).
(G) to (N) Seeds at 6 h (6HAP) to 2 d (2DAP) after pollination. FUS3ΔC-GFP was localized to the outer seed coat, chalaza, and funiculus. (M) and (N) were taken at two different focal planes to show GFP in chalaza (M) or seed coat (N) of 2DAP seeds. 12HAP, 12 h after pollination; 1DAP, 1 d after pollination.
(O) and (P) FUS3ΔC-GFP was localized to the suspensor and to the 16-cell stage (16-cell; O) and 32-cell stage (32-cell; P) of the embryo proper. chl, chalaza; es, embryo sac; fun, funiculus; ii, inner integument; mmc, megaspore mother cell; ne, nucellar epidermis; nu, nucellus; oi, outer integument; sc, seed coat; sept, septum. Magenta, autofluorescence. Bars = 10 μm.
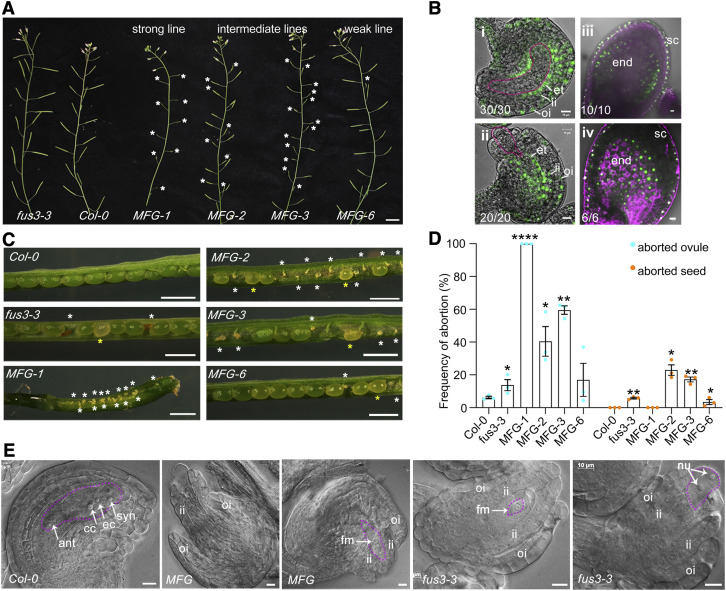
To further address the role of FUS3 in reproduction, we monitored ovule development in fus3-3 loss-of-function mutant and ProML1:FUS3-GFP misexpression lines (Gazzarrini et al., 2004). The MERISTEM LAYER1 (ML1) promoter drives expression in the L1 layer of the meristems (vegetative, inflorescence, and floral meristems), in the integuments of the ovule primordia, and in the endothelium of mature ovules, the protoderm of the embryo, as well as in the seed coat, endothelium, and endosperm of fertilized seeds (Supplemental Figures 2A and 2B; Lu et al., 1996; Huang et al., 2016). Although ProML1:FUS3-GFP was shown to rescue all fus3-3 seed maturation defects, including desiccation intolerance, misexpression in the meristems during postembryonic development caused additional phenotypes (Gazzarrini et al., 2004). Strong ProML1:FUS3-GFP lines show delayed vegetative growth and flowering, reduced plant height. and aborted siliques, as previously described (Figure 2A; Gazzarrini et al., 2004; Lu et al., 2010). We found that in ProML1:FUS3-GFP lines, FUS3-GFP was mislocalized to the endothelium, outer and inner integuments of FG5-FG7 ovules, whereas in aborted ovules FUS3-GFP surrounded the aborted embryo sac (Figures 2B-i and 2B-ii; Supplemental Figures 2C and 2D). During early seed development (1 to 3 d after fertilization, or DAF), FUS3-GFP was localized to the seed coat and mislocalized to the endothelium and endosperm (Figures 2B-iii and 2B-iv; Supplemental Figures 2E to H). Moreover, we found that siliques of intermediate-to-strong ProML1:FUS3-GFP lines contained (1) aborted ovules (appear as very small, white fists); (2) aborted seeds containing arrested embryos (very small and brown seeds); and (3) seeds with delayed embryo development (regular size, pale green seeds), as well as a variable number of regular green seeds (Figures 2C and 2D). Strong ProML1:FUS3-GFP lines, such as MFG-1, were sterile (Figures 2C and 2D).
Figure 2.
Loss and Misexpression of FUS3 Negatively Impacts Ovule and Seed Development.
(A) Aborted siliques (asterisks) of ProML1:FUS3-GFP (MFG) lines. Bars = 1 cm.
(B) FUS3-GFP localization to the integuments and endothelium of MFG ovules at the FG7 stage (i), the tissue surrounding an aborted embryo sac (ii), and the outer layer of the seed coat, endothelium, and endosperm of two (iii) and three (iv) d-after-pollination (DAP) seeds. Numbers refer to the number of ovules/seeds displaying the same GFP patterns shown in (B). Bars = 10 μM.
(C) Aborted ovules (small size, white) or aborted seeds (small size, brown) are marked by a white asterisk, and delayed-embryogenesis seeds (regular size, pale yellow) are marked by a yellow asterisk in MFG and fus3-3 siliques. Bars = 1 mm.
(D) Frequency of aborted ovules and seeds. Averages of three biological replicates ±sd (n = five siliques) are shown. *P < 0.05, **P < 0.01, ****P < 0.0001, two-sided t test (Supplemental File).
(E) DIC images of wild type (WT), MFG, and fus3-3 FG7 ovules. Pink dashed lines outline the embryo sac. ant, antipodal cell; cc, central cell; ec, egg cell; end, endosperm; et, endothelium; fm, functional megaspore; ii, inner integument; nu, nuclei; oi, outer integument; syn, synergid cell nuclei. Bars = 10 μm.
Next, we analyzed fus3-3 and ProML1:FUS3-GFP ovule development. The embryo sac of wild-type ovules at FG7 stage contained the egg cell nucleus, the central cell nucleus, and the synergids, and was surrounded by inner and outer integuments. However, at this stage the embryo sac of some fus3-3 and ProML1:FUS3-GFP lines was delayed at various stages, from FG1 to FG6, arrested or not fully wrapped by the integuments (Figure 2E). The arrest of female megagametogenesis resulted in ovule abortion in fus3-3 and more so in strong ProML1:FUS3-GFP lines (Figures 2C and 2D). Taken together, these results show that spatiotemporal localization of FUS3 is tightly regulated and that misexpression of FUS3 severely impairs embryo sac and integument development. Furthermore, fus3-3 also shows impaired ovule development, indicating that precise spatiotemporal regulation of FUS3 expression is required for proper ovule development.
Class I BPC Transcription Factors Bind to (GA/CT)n Motifs in FUS3
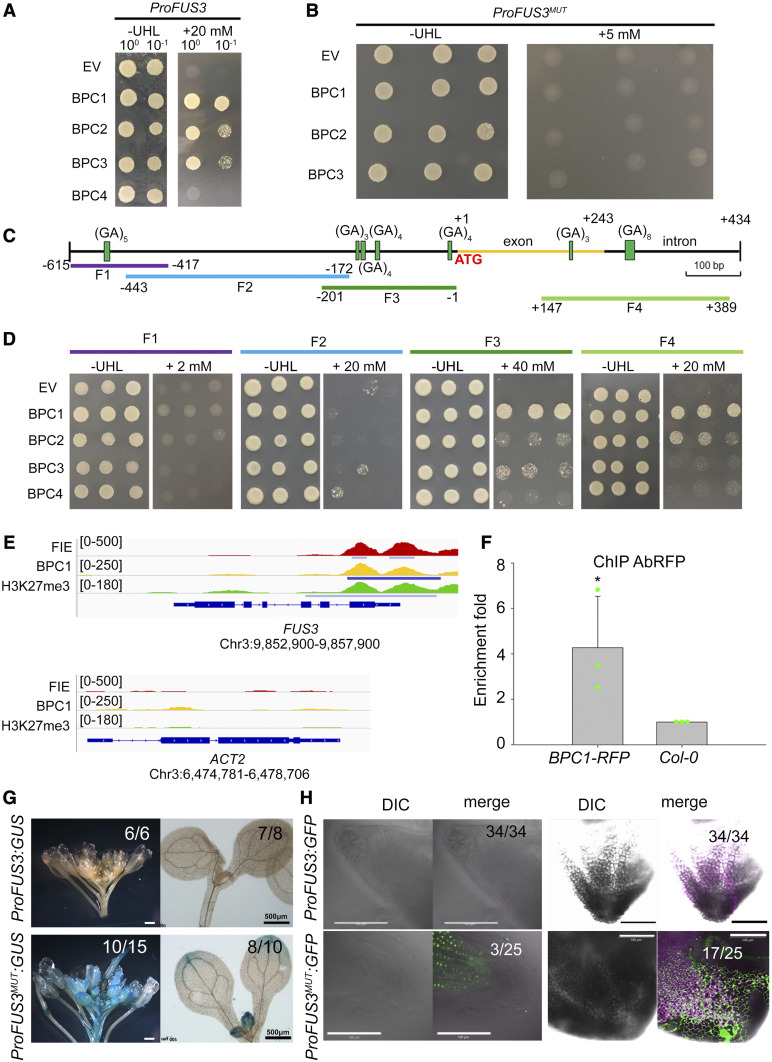
To understand the mechanisms responsible for the spatiotemporal patterns of FUS3 expression, we identified upstream regulators of FUS3 by yeast one-hybrid (Y1H). To increase screening specificity, a short genomic region 615 bp upstream of the FUS3 translation start codon (ProFUS3) was used to screen an Arabidopsis (Arabidopsis thaliana) transcription factor library (Figures 3A and 3C; Mitsuda et al., 2010). Sequencing of the resulting cDNA inserts revealed that all colonies contained BPC3. Also, a second Y1H screen identified BPC1. BPCs are a small family of plant specific transcription factors consisting of six genes and a pseudogene (BPC5) that are divided into 3 classes based on sequence similarity: class I (BPC1, 2, and 3), class II (BPC4, 5, and 6), and class III (BPC7; Meister et al., 2004). We individually retested all class I BPCs (BPC1-3) and also included class II BPC4, which is not present in the cDNA library but is highly expressed in embryos and flowers (Berger et al., 2011). All three class I BPCs bound to ProFUS3 based on yeast one-hybrid analysis, but not class II BPC4 (Figure 3A). This is in partial agreement with a recent study, which showed that BPC1-4 bound to FUS3 in Y1H (Roscoe et al., 2019).
Figure 3.
Class I BPCs Bind to the FUS3 Genomic Region Proximal to the Transcription Start Site.
(A) BPC1, 2, and 3 bound to a FUS3 genomic region [ProFUS3(0.6 kb); −615 to +1 base pairs] in Y1H. 10−1 is a 10-fold dilution of 10°.
(B) BPC1, 2, and 3 did not bind the FUS3 genomic sequence carrying mutations in all (GA/CT)n sequences [ProFUS3MUT(0.6 kb)]. Colonies in (A) and (B) were selected on -Ura-His-Leu medium (-UHL) with or without 5 or 20 mM 3-amino-1,2,4-triazole. EV, empty vector.
(C) Distribution of (GA/CT)n motifs in the FUS3 genomic sequence (−615 to +434).
(D) Binding of BPC1, 2, and 3 to regions of the FUS3 genomic sequence shown in (C; F1 to F4).
(E) Bowser view of chromatin occupancy of FIE, BPC1, and H3K27me3 at FUS3 and ACT2 (negative control) in 30-h-old seedlings using ChIP-seq data from Xiao et al. (2017). Significant peaks (Q < 10−10) according to MACS2 are marked by horizontal bars.
(F) Real-time quantitative PCR analysis of ChIP assay using chromatin from Pro35S:BPC1-RFP and Col-0 (negative control) inflorescences and primers for the F3 region of ProFUS3. Antibodies against the RFP tag were used in the IP. Error bars represent the propagated error value using three biological replicates (*p < 0.05; two-sided t test; Supplemental File).
(G) ProFUS3(1.5kb):GUS and ProFUS3MUT(1.5kb):GUS staining in 10-d-old seedlings and flower buds. White bars = 2 mm; black bars = 500 μm.
(H) ProFUS3(1.5kb):GFP and ProFUS3MUT(1.5kb):GFP fluorescence in the leaf and root tips of 15-d-old seedlings. Bars = 100 μm. Numbers in (G) and (H) refer to the number of transgenic lines displaying the same GUS/GFP pattern as the one shown.
BPCs were shown to bind to (GA/CT)n cis elements in several plant species, with a preference for different numbers of repeats (Berger and Dubreucq, 2012; Simonini and Kater, 2014). When all (GA/CT)n motifs of the ProFUS3 were mutated (ProFUS3MUT), none of the class I BPCs interacted with the FUS3 sequence, confirming binding specificity (Figure 3B; Supplemental Figure 3). To identify the binding location of BPCs on ProFUS3, we generated constructs containing ∼200-bp regions of ProFUS3 (fragments F1 to F3) for Y1H analysis. The first exon/intron region containing 2 (GA/CT)n repeats (F4) was also tested (Figure 3C). BPC1-3 bound to the FUS3 5′UTR (F3) and to the first exon/intron regions (F4) where (GA/CT)n motifs are enriched (Figure 3D). BPC1-4 did not bind the promoter region further upstream (F1 or F2), where there is only one (GA)5 or no (GA/CT)n motif, respectively (Figure 3D). To determine whether BPC1 also binds to the FUS3 locus in vivo during reproductive development, we generated BPC1 overexpression lines and performed chromatin immunoprecipitation (ChIP) in inflorescences. Our results showed that BPC1 directly binds to this region (Figure 3F). Altogether, this indicates that class I BPCs bind to the 5′UTR and first intron/exon regions of FUS3 in Y1H. Furthermore, BPC1 directly binds to FUS3 in vivo during reproductive development.
Class I BPCs Repress FUS3 during Vegetative Growth
In a genome-wide study, BPC1 interacted with and recruited the conserved PRC2-complex subunit FERTILIZATION INDEPENDENT ENDOSPERM (FIE) in vivo and triggered polycomb-mediated gene silencing in 30-h germinating seeds (Xiao et al., 2017). We first analyzed ChIP-seq data from Xiao et al. (2017) and found that the first exon/intron and 5′UTR of FUS3 was bound by BPC1 (Figure 3E). Furthermore, this same region was bound by FIE and associated with H3K27me3, a repressive mark (Figure 3E). Finally, BPC1 and -2 interacted with EMBRYONIC FLOWER2 (EMF2), which belongs to the EMF-PRC2 complex involved in repressing the vegetative-to-reproductive and embryo-to-seedling phase transitions (Mozgova et al., 2015; Xiao et al., 2017). This suggests that FUS3 may be repressed in germinating seeds by BPC1 recruitment of EMF-PRC2. To confirm this, we cloned 1.5 kb of the FUS3 promoter (+1/−1.5 kb), mutated all BPC binding sites (GA/CT)n shown in Figure 3C and Supplemental Figure 3, and fused it separately to GUS and GFP to generate ProFUS3MUT:GUS and ProFUS3MUT:GFP. Analysis of several independent transgenic lines showed that ProFUS3MUT is indeed derepressed postembryonically in the leaf and root tips, albeit derepression in root tips was observed only in a small number of lines (Figures 3G and 3H). Together with previous data showing that FUS3 was strongly upregulated in swinger curly leaf (swn clf) seedlings (Makarevich et al., 2006), these results strongly suggest that BPC1 binds to and represses FUS3 during vegetative development by recruiting the EMF-PRC2 complex.
Class I BPCs Repress FUS3 During Reproductive and Seed Development
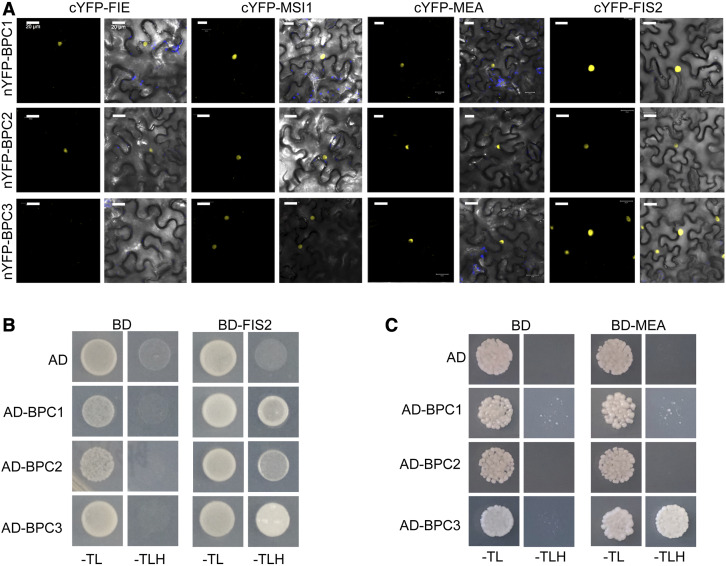
Single or higher order bpc mutants do not show phenotypes that are expected from ectopic expression of FUS3 or other embryonic regulators such as LEC1 and LEC2, including the development of embryonic tissues/organs in seedlings (Jia et al., 2014; Lepiniec et al., 2018). Instead, higher order bpc mutants show dramatic defects in ovule development leading to severe ovule abortion (Monfared et al., 2011). This suggests that BPCs have a more prominent role during reproductive development, but the mechanisms are unknown. Previous ChIP assays showed that, in closed flowers, the FUS3 locus associates also with the FIS-PRC2 complex component MEDEA (MEA) and H3K27me3 repressive marks and that FUS3 is upregulated in the endosperm of mea/MEA seeds at 3 DAF (Makarevich et al., 2006). Given that BPC1 binds to the FUS3 locus in closed flowers (Figure 3F), we reasoned that during reproductive development FUS3 may be repressed by BPCs through FIS-PRC2 recruitment and that this repression may be required for proper ovule and seed development. To test this hypothesis, we first monitored ProFUS3MUT:GUS staining and found that FUS3 is upregulated in flower buds (Figure 3G). Next, we determined whether class I BPCs interact in planta with the FIS-PRC2 complex, which acts during gametophyte and endosperm development (Supplemental Figure 4). We found that BPC1-3 interacts not only with the unique subunits of this complex, FIS2 and MEA, but also with the PRC2-shared component, MSI1, in bimolecular fluorescence complementation (BiFC) assays. All but BPC3 also interacted with FIE (Figure 4A).
Figure 4.
Class I BPC Family Members Interact with the FIS-PRC2 Complex.
(A) Interaction between class I BPC family members and the FIS-PRC2 complex assayed by BiFC. Lack of interaction between FUS3 and BPCs or FIS-PRC2 in BiFC assays is shown as the negative control in Supplemental Figure 6. Bars = 20 μm.
(B) and (C) All class I BPCs interacted with FIS2 (B), whereas only BPC3 interacted with MEA (C) in yeast two-hybrid assays. Yeast colonies were grown on -Trp-Leu (-TL) medium and interactions were tested on -Trp-Leu-His (-TLH) medium for three d. (B) AD, Gal4 activation domain (pDEST22); BD, Gal4 binding domain (pDEST32); yeast strain, AH109. (C) AD, Gal4 activation (pGADT7); BD, Gal4 Binding domain (pGBKT7); yeast strain, YRG-2.
To confirm these results, we tested the interactions between BPC1-3 and FIS-PRC2 specific components, MEA and FIS2, by yeast two-hybrid (Y2H) analysis, which allows the detection of binary interactions similarly to BiFC. BPC1-3 interacted with FIS2, but only BPC3 interacted with MEA (Figure 4B). Thus, our data confirmed the interaction between FIS2 and BPC1-3, but only MEA-BPC3 could be confirmed by Y2H and BiFC. In previous studies, interactions between FIE/MSI and BPC1 and -2 could not be detected by Y2H, although FIE and BPC1 could interact in BiFC (Figure 4; Mu et al., 2017; Xiao et al., 2017). Thus, it is possible that in planta assays are better suited to detect interactions between select BPCs and PRC2. This is in agreement with the literature showing that although the overlap between the two binary protein-protein interaction assays is small, Y2H and BiFC are highly complementary in detecting pairwise protein–protein interactions (Boruc et al., 2010; Braun et al., 2013). Alternatively, PRC2 components may directly interact with select BPCs. Thus, we conclude that BPC1-3 can recruit FIS-PRC2 through FIS2.
In agreement with previous Y2H results, we showed that class I BPCs interacted with each other also in planta and that BPC2 and -3 could also form homodimers (Supplemental Figure 5; Simonini et al., 2012). No class I BPC member or FIS-PRC2 subunit interacted with FUS3, suggesting that these BiFC interactions are specific (Supplemental Figure 6). Last, given that BPC6 recruits PRC2 by interacting with LIKE HETEROCHROMATIN PROTEIN1 (LHP1; Hecker et al., 2015), we also tested the interaction between class I BPCs and LHP1 in planta. However, no interaction was found, suggesting that class I and class II BPCs recruitment of the PRC2 complex may differ (Supplemental Figure 7). We conclude that class I BPCs can form homo- and heterodimers and recruit the FIS-PRC2 complex in planta.
Class I BPCs have been shown to be expressed in ovules (Monfared et al., 2011). To better understand their role in reproductive and seed development, we determined their expression patterns before fertilization (FG4 to FG7) and after fertilization (1 to 11 DAF). BPC1-3 had largely overlapping expression patterns before fertilization, and all were highly expressed in almost all tissues of developing ovules and embryos, as well as in the endosperm and seed coat (Supplemental Figure 8). BPC1 had a more restricted pattern before (chalaza and) and after (chalaza, micropyle, seed coat) fertilization. This suggests that class I BPCs may act redundantly during ovule and embryo development.
The FIS-PRC2 complex subunits FIS2 and MEA were only expressed in the central cell of developing ovules and in the endosperms at 2DAF, as previously shown (Supplemental Figure 8; Wang et al., 2006). Collectively, these data show that BPCs can interact with each other and with FIS-PRC2 to regulate gene expression. Given the specific localization of FIS2 and MEA to the central cell and endosperm, and FUS3 derepression in the endosperm of mea/MEA, we conclude that, aside from their role in silencing FUS3 during vegetative growth through EMF-PRC2, class I BPCs repress FUS3 during reproductive and seed development by recruiting FIS-PRC2 in the central cell and endosperm. Furthermore, BPCs may recruit sporophytic PRC2 (EMF/VRN PRC2) to repress FUS3 in the integuments and inner seed coat.
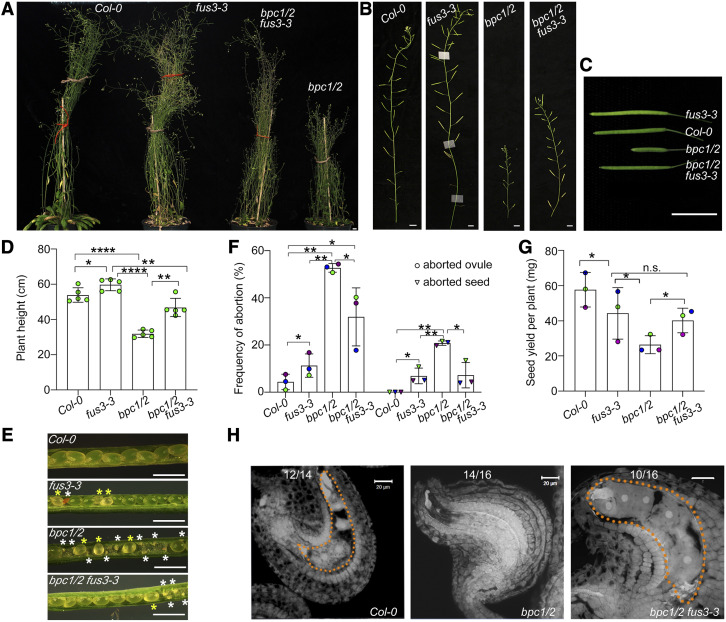
Reproductive Defects of bpc1/2 Are Partially Rescued by fus3-3
Previously, bpc mutants were shown to display pleiotropic phenotypes during vegetative and reproductive development (Monfared et al., 2011). Higher order double bpc1/2 and triple bpc1/2/3 mutants are dwarf, have shorter or aborted siliques, and display severe defects in embryo sac and seed development leading to ovule and seed abortion, whereas most single bpc mutants resemble wild type or display a low frequency of ovule/seed defects, suggesting functional redundancy (Figures 5A to 5F; Supplemental Figure 9; Monfared et al., 2011). Surprisingly, the bpc1/2 mutant phenotypes are partially rescued in the bpc1/2/3 triple mutant. Monfared et al. (2011) proposed that BPC3 may have opposite functions to those of BPC1 and -2. However, no mechanism has been shown so far.
Figure 5.
Partial Rescue of bpc1/2 Stunted Growth, Aborted Ovules and Seeds in bpc1/2 fus3-3.
(A) to (D) Stunted growth and silique elongation of bpc1/2 was partially rescued in bpc1/2 fus3- 3. Bars in (A) to (C) = 1 cm. (D) Quantification of plant height (n = five plants per genotype).
(D) to (G) bpc1/2 ovule/seed abortion and seed yield was partially rescued in bpc1/2 fus3-3. (E) Images of opened siliques showing aborted ovules and seeds (white asterisks), delayed embryogenesis seeds (yellow asterisks). Bars = 1 mm. (F) Frequencies of aborted ovules and seeds in bpc1/2 fus3-3 mutants (n = 10 peeled, half side siliques). (G) Seed yield (n = 5 plants per genotype). (D) to (G) Averages of at least three biological repeats ±sd are shown (*P < 0.05, **P < 0.01, ****P < 0.0001; (D) Tukey multiple comparison of means; (F) and (G) paired t test; n.s., no significant difference (Supplemental File). (H) bpc1/2 ovule and seed defects were partially rescued in bpc1/2 fus3-3. Images were taken at 1 DAP; Bars = 20 μm. Numbers refer to the number of samples displaying the phenotype shown. Dotted lines outline the endosperm.
The phenotypes displayed by bpc mutants are remarkably similar to those shown by ProML1:FUS3 misexpression lines (Figure 2; Gazzarrini et al., 2004), suggesting that they may be caused by ectopic expression of FUS3. To address the genetic relationship between class I BPCs and FUS3, we crossed bpc1/2 with fus3-3. The bpc1/2 fus3-3 indeed showed partial rescue of these phenotypes, including plant height (Figures 5A and 5D), silique abortion (Figures 5B and 5C), ovule and seed abortion (Figures 5E and 5F), supporting our hypothesis that FUS3 is misexpressed in bpc1/2.
At 1 to 2 d after fertilization, several bpc1/2 ovules lacked the embryo sac (Figure 5H; Supplemental Figure 9E), whereas most bpc1/2 fertilized seeds displayed delayed or arrested embryo development (Figures 5E and 5F; Supplemental Figures 9A, 9B, 9E to 9H). Overall, defects in ovule and seed development of higher order bpc mutants resulted in severe reduction of seed yield (Figure 5G; Supplemental Figure 9C). The bpc1/2 fus3-3 mutant partially rescued bpc1/2 ovule and seed development (Figures 5E, 5F, and 5H). These data strongly suggest that BPCs repress FUS3 during reproductive and seed development.
BPC1 and 2 Repress FUS3 to Promote Inflorescence Stem Elongation, Ovule, and Endosperm Development
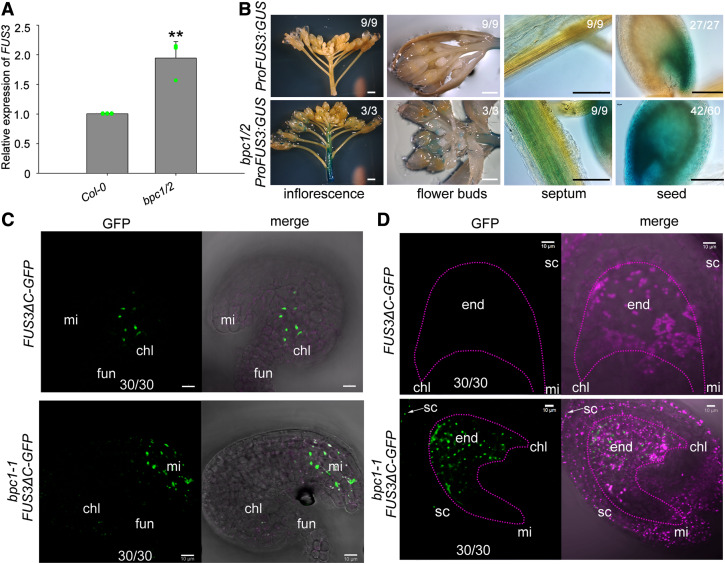
To confirm a repressive role of BPCs on FUS3 function, we analyzed the FUS3 expression level and patterns in bpc1/2 mutants. The FUS3 transcript level was indeed increased in bpc1/2 inflorescence stem (Figure 6A). Consistent with the transcript analysis, ProFUS3:GUS activity was also increased in bpc1/2 inflorescence stem and flower buds (Figure 6B). In wild type, low FUS3 expression in the inflorescence stem is shown by transcriptomic data and detected with the ProFUS3:FUS3ΔC-GFP sensitive reporter (Supplemental Figure 1). Together with previous findings showing that (1) plant height is reduced in ProML1:FUS3-GFP misexpression plants (Gazzarrini et al., 2004), but increased in the fus3-3 mutant (Figure 5D) and (2) bpc1/2 short stature is rescued in fus3-3 bpc1/2; these results indicate that BPC1 and 2 downregulate FUS3 in the stem to promote stem elongation.
Figure 6.
BPC1 and 2 Negatively Regulate FUS3 Expression in Reproductive Organs and Seeds.
(A) qRT-PCR showing increased FUS3 transcript level in bpc1/2 inflorescence stem. Error bars represent the SD of three biological replicates (**P < 0.01; two-sided t test; Supplemental File).
(B) GUS staining in the inflorescence stem, flower buds, septum and 2DAF of ProFUS3:GUS and bpc1/2 ProFUS3:GUS. GUS staining was enhanced in the inflorescence stem and septum, and ectopically expressed in the endosperm of bpc1/2. White bars = 2 mm; black bars = 100 μm.
(C) and (D) ProFUS3:FUS3ΔC-GFP and bpc1 ProFUS3:FUS3ΔC-GFP ovules were imaged before (C) and 2 d after (D) fertilization by confocal microscopy. FUS3ΔC-GFP was localized to the chalaza of developing wild type (WT) ovules before fertilization, and ectopically localized to the integuments at the micropylar region of bpc1-1 ovules (FG7 stage) and the endosperm of 2DAF bpc1-1 seeds. chl, chalaza; end, endosperm; fun, funiculus; mi, micropyle; sc, seed coat. See also Supplemental Figure 10 for bpc1/2 ProFUS3:FUS3ΔC-GFP. Bars = 10 μm.
We then crossed ProFUS3:FUS3ΔC-GFP into bpc1-1 and bpc1/2. During reproductive development, FUS3ΔC-GFP was mislocalized to the integuments at the micropylar region of developing bpc1-1 and bpc1/2 ovules, but after fertilization, ectopic ProFUS3:GUS activity and FUS3ΔC-GFP localization were detected in bpc1-1 and bpc1/2 endosperms (Figures 6B to 6D; Supplemental Figure 10). Combined with the above functional analysis, these results suggest that before fertilization BPCs restrict FUS3 expression to the funiculus and chalaza to promote ovule development, whereas after fertilization FUS3 is repressed by BPCs in most of the endosperm to coordinate embryo and endosperm growth.
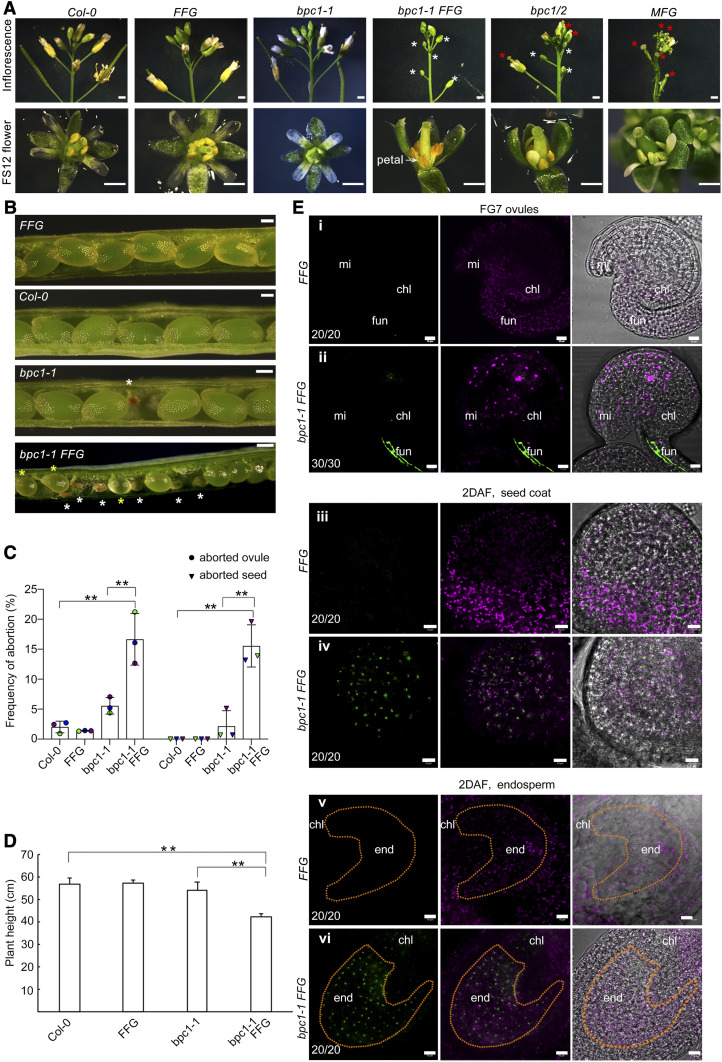
To analyze the repressive role of class I BPCs, we also crossed the ProFUS3:FUS3-GFP translational reporter, which rescues the fus3-3 mutant phenotypes (Gazzarrini et al., 2004; Chan et al., 2017), with the bpc1/2 mutant. However, we were only able to isolate bpc1-1 ProFUS3:FUS3-GFP lines. In agreement with previous research, bpc1-1 develops normal flowers and displays a very low frequency of ovule and seed phenotypes, although not statistically significant when compared to the wild type (Figures 7A to 7C; Supplemental Figure 9B; Monfared et al., 2011). However, in bpc1-1 ProFUS3:FUS3-GFP some flower buds were arrested and never opened, petals and anthers filaments did not elongate, and anthers were aborted, similar to bpc1/2 double mutant (Figure 7A). Ovule and seed abortion were increased, and delayed embryogenesis was evident in bpc1-1 ProFUS3:FUS3-GFP plants (Figures 7B, 7C, and E8E). The bpc1-1 ProFUS3:FUS3-GFP plants were shorter and resembled the bpc1/2 double mutant (Figures 5A, 5D, and D7D). Thus, our inability to isolate bpc1/2 ProFUS3:FUS3-GFP line may be due to the severe phenotype of such a mutant.
Figure 7.
Ectopic or Overexpression of FUS3 Negatively Impacts Flower, Ovule and Seed Development.
(A) Introduction of a ProFUS3:FUS3-GFP (FFG) transgene into bpc1-1 resulted in arrested flower buds that never opened (white asterisk), similar to bpc1/2. Arrested flower buds in bpc1-1 FFG had underdeveloped petals, nonelongated filaments, and aborted anthers, similar to bpc1/2. ProML1:FUS3-GFP (MFG) also showed shorter filaments and underdeveloped anthers, but flower buds opened prematurely. Bars = 1 mm for inflorescences and 5 mm for flowers.
(B) bpc1-1 FFG developed aborted seeds (white asterisk) and had delayed embryogenesis (yellow asterisk). Bars = 100 μm.
(C) Frequencies of aborted ovules and seeds. Averages of three biological replicates ±sd (n = 10 half side siliques); **P < 0.01, Tukey multiple comparison of means (Supplemental File).
(D) bpc1-1 FFG plants displayed stunted growth. Error bars represent SD of three biological replicates (n = 5 plants). **P < 0.01, Tukey multiple comparison of means (Supplemental File).
(E) FUS3-GFP was strongly expressed in the funiculus (ii) of bpc1-1 ovules (FG7 stage). At 2DAF, FUS3-GFP was strongly expressed in bpc1-1 seed coat (iv) and mis-expressed in bpc1-1 endosperm (vi). chl, chalaza; end, endosperm; fun, funiculus; mi, micropyle. Bars = 10 μm.
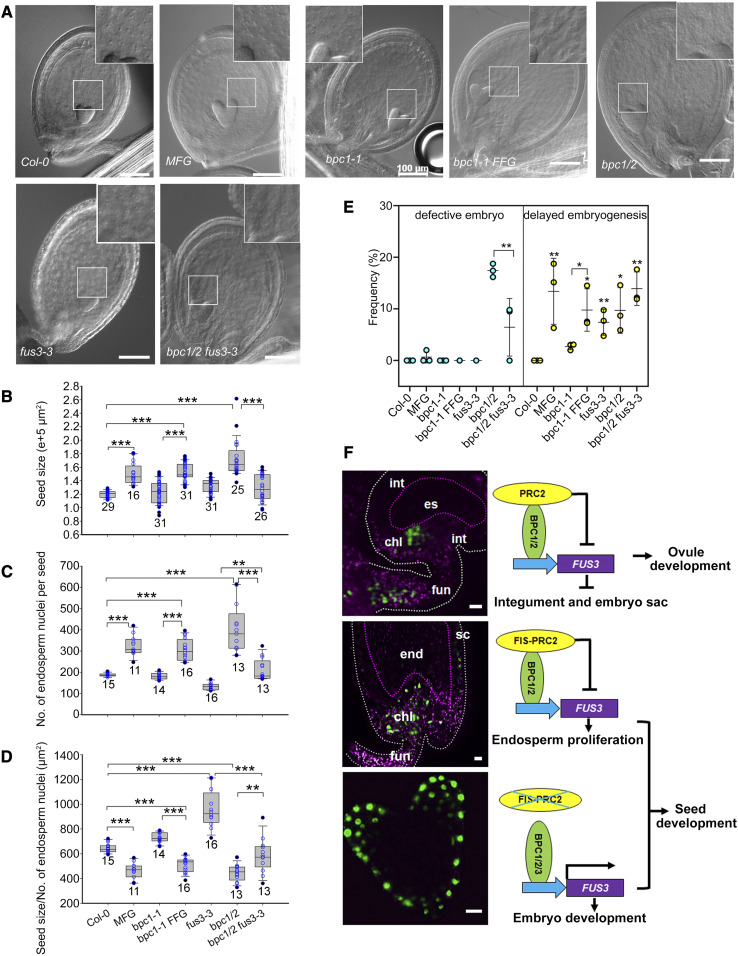
Figure 8.
Negative Regulation of Endosperm Nuclei Proliferation by BPC1 and 2-Mediated FUS3 Repression and Model of Spatiotemporal Regulation of FUS3 in Reproductive and Seed Development.
(A) Whole-mount clearing of seeds 3 d after fertilization (early heart-stage embryo). Bars = 100 μm.
(B) to (E) Ectopic expression of FUS3 in ProML1:FUS3-GFP (MFG), bpc1-1 ProFUS3:FUS3-GFP (FFG), and bpc1/2 led to increased seed size (B), increased endosperm nuclei proliferation (C) and density (D), and delayed embryogenesis (E), which were partially rescued in bpc1/2 fus3-3. (B) to (E) Averages of at least three biological replicates ±sd (n per genotype are listed in the graph). *P < 0.05, **P < 0.01, ****P < 0.0001, Tukey multiple comparison of means (Supplemental File).
(F) Model depicting spatiotemporal regulation of FUS3 by BPCs and PRC2 and its role in regulating reproductive and seed development. Before fertilization (top images), FUS3 becomes restricted to the funiculus and chalaza of mature ovules through BPC1 and 2-mediated FUS3 repression in the integuments. This is required to promote integument and embryo sac development. After fertilization (middle and bottom images), FUS3 is localized to the outer seed coat layer, chalaza, funiculus and embryo, but is repressed in the endosperm by BPC1 and 2. This is required to decrease endosperm nuclei proliferation and promote embryo development. In the integuments, BPC1 and -2–mediated FUS3 repression may be orchestrated by sporophytic EMF/VRN-PRC2 (Liu et al., 2016; Xiao et al., 2017). After fertilization, FIS-PRC2 represses FUS3 in the endosperm (Makarevich et al., 2006). chl, chalaza; es, embryo sac; fun, funiculus; int, integument; sc, seed coat. Bars = 10 μm.
The presence of the ProFUS3:FUS3-GFP transgene enhanced the bpc1-1 phenotype likely due to higher or ectopic FUS3 expression. Accordingly, we could detect strong GFP fluorescence in the integuments, seed coat, and funiculus of bpc1-1 ProFUS3:FUS3-GFP, whereas ProFUS3:FUS3-GFP showed no fluorescence in wild type (in contrast to the stable ProFUS3:FUS3ΔC-GFP). Furthermore, FUS3-GFP was mislocalized in bpc1-1 ProFUS3:FUS3-GFP endosperm after fertilization (Figure 7E), in agreement with FUS3ΔC-GFP mislocalization and ProFUS3:GUS misexpression in bpc1-1 and bpc1/2 endosperm (Figure 6; Supplemental Figure 10). These results further support a repressive role of BPCs on FUS3 expression in different tissues during reproductive and seed development.
Last, we show that bpc1/2, bpc1-1 ProFUS3:FUS3-GFP, and ProML1:FUS3-GFP ovules that were successfully fertilized had an increased number of endosperm nuclei, and this correlated with increased seed size (Figures 8A to 8D; Supplemental Figure 11), and some embryos were delayed or arrested at various stages of development (globular to early torpedo; Figure 8E; Supplemental Figures 9 and 11). bpc1/2 also showed aberrant cell division patterns in the embryo proper and suspensor, which resulted in defective embryos that were partially rescued by fus3-3 (Figure 8E; Supplemental Figure 11). Collectively, these data show that repression of FUS3 in the ovule and endosperm of developing seeds is required to coordinate ovule development, endosperm, and embryo growth.
DISCUSSION
PRC2 plays important roles in balancing cell proliferation with differentiation and regulating developmental phase transitions in plants and animals. Recently, genome-wide studies have shown that class I BPC transcription factors bind Polycomb response elements, recruit EMF-PRC2, and trigger gene silencing during germination to allow the embryonic-to-vegetative phase transition (Xiao et al., 2017). Similar to GAGA factors in Drosophila melanogaster, BPCs recognize (GA/CT)n cis elements, despite the lack of sequence similarity between these transcription factors, suggesting convergent evolution (Berger and Dubreucq, 2012). However, BPCs have been shown to play essential roles during vegetative and reproductive development, as shown by the dwarf stature and severe seed abortion displayed by higher order bpc mutants, but the molecular mechanisms were largely unknown (Kooiker et al., 2005; Monfared et al., 2011; Simonini et al., 2012; Simonini and Kater, 2014). Here we showed that class I BPCs interacted with FIS-PRC2 and bound to the FUS3 chromatin to restrict FUS3 expression to specific tissues during reproductive and seed development. We provide evidence that BPC-mediated spatiotemporal regulation of FUS3 expression is required to (1) promote stem elongation during vegetative-to-reproductive phase change; (2) promote ovule development; and (3) coordinate embryo and endosperm development after fertilization (Figure 8F).
Several lines of evidence support these conclusions. First, our Y1H analyses show that class I BPCs bind to (GA/CT)n around the FUS3 transcription start, and ChIP assays in flower buds show that BPC1 binds in vivo to the FUS3 chromatin. Mutations in these (GA/CT)n sites abolish BPCs binding and derepress FUS3. Furthermore, FUS3 is upregulated in the inflorescence stem of bpc1/2 dwarf plants, which is consistent with fus3-3 tall plant and ML1:FUS3-GFP dwarf plant phenotypes, as well as FUS3 role as repressor of vegetative-to-reproductive phase change (Gazzarrini et al., 2004; Lumba et al., 2012). Second, class I BPCs interact with FIS-PRC2 complex in planta, and the in vivo BPC1 binding region on FUS3 was shown to associate with MEA and H3K27me3 repressive marks (Makarevich et al., 2006), strongly suggesting BPC1 recruits FIS-PRC2 to repress FUS3 during reproductive/seed development. Third, FUS3 is transiently localized to the integuments during early ovule development and later restricted to the funiculus and chalaza of mature wild type ovules. Ectopic and persistent expression of FUS3 in the integuments of bpc1/2 and ML1:FUS3 misexpression lines impairs integument and embryo sac development leading to ovule abortion, which can be partially rescued in bpc1/2 fus3-3. Finally, after fertilization FUS3 is localized to the funiculus, chalaza and outer integument, aside from its known localization to the embryo (Gazzarrini et al., 2004). Ectopic expression of FUS3 in bpc1/2 and ML1:FUS3 endosperm leads to increased proliferation of the endosperm nuclei and delayed or arrested embryo development, which are rescued in bpc1/2 fus3-3. The latter phenotypes are also displayed by mutants in FIS-PRC2 subunits (Kiyosue et al., 1999; Köhler and Grossniklaus, 2002). We conclude that BPCs recruit PRC2 to restrict spatiotemporal FUS3 expression during reproductive and seed development. This is required to regulate tissue development locally and modulate developmental phase transitions in Arabidopsis. The genomic sequences of FUS3 orthologs in other species show conservation of (GA/CT)n repeats (Supplemental Figure 12), suggesting that similar mechanisms may regulate the expression of FUS3-like transcription factors in other species.
Inflorescence Stem Elongation and Flower Development Require Repression of FUS3 by Class I BPCs
During germination, BPC1 interacts with PRC2 and directly binds to the genomic region of FUS3 proximal to the transcription start, which is marked by H3K27me3 repressive marks and associates with FIE (Figure 3; Xiao et al., 2017). Furthermore, FUS3 is ectopically expressed in swn clf seedlings, which show embryonic traits (Makarevich et al., 2006). This suggests that, during germination, FUS3 is repressed through BPC1-recruitment of EMF/VRN-PRC2. Here we showed that mutations of all BPC binding sites on the FUS3 promoter derepressed FUS3 in vegetative and reproductive organs and that lack of BPCs resulted in ectopic FUS3 expression in leaves, inflorescence stem, and flower buds. Furthermore, ectopic FUS3 in bpc1/2, bpc1 ProFUS3:FUS3-GFP, or ProML1:FUS3-GFP led to similar phenotypes, including reduced internode elongation and defective flowers (arrested flower bud development, flowers with a protruding carpel and shorter floral organs), suggesting FUS3 inhibits the elongation of the stem and floral organs during flowering. Recently, deletion of a small region in the FUS3 promoter near the BPC binding sites and corresponding to the PRC2 recruitment region also caused ectopic FUS3 expression in vegetative and reproductive tissues (Roscoe et al., 2019). Thus, we propose that class I BPCs recruit sporophytic VRN/EMF-PRC2 to repress FUS3 postembryonically, allowing vegetative and reproductive phase transitions.
BPC-mediated Restriction of FUS3 Expression in Developing Ovules and Seeds Is Required to Promote Ovule Development and to Coordinate Endosperm and Embryo Growth
During ovule development, the funiculus supplies nutrients and signaling molecules from the mother plant to the chalaza, whereas the chalaza initiates the development of the integuments that grow around the nucellus and protect the developing female gametophyte (Schneitz et al., 1995). Our data show that, during megagametogenesis, FUS3 is initially localized to the nucellus epidermis and tissues surrounding the nucellus, including the integuments and chalaza. However, BPC1 and -2 later repress FUS3 in the integuments of mature ovules, and ectopic FUS3 expression inhibits integuments and embryo sac development, triggering ovule abortion. This is in agreement with previous findings showing that the integuments are required for female gametogenesis (Elliott et al., 1996; Klucher et al., 1996; Baker et al., 1997). Given that both fus3-3 and ectopic FUS3 trigger ovule abortion, we conclude that both low FUS3 expression in the funiculus and chalaza and FUS3 repression in the integuments of mature ovules are required for embryo sac and integument development (Figure 8F). BPCs ubiquitous expression in the ovule, however, also suggests that other mechanisms prevent FUS3 repression in the chalaza and funiculus. Thus, the identification of upstream and downstream regulators of FUS3 in the ovule will be required to further understand its role in reproductive development. Aside from auxin, sugars have also been implicated as signaling molecules coordinating seed and fruit development (Robert et al., 2018; Robert, 2019). Notably, FUS3 phosphorylation by the SnRK1 kinase, a conserved energy sensor in eukaryotes, is required to rescue fus3-3 seed abortion, suggesting that FUS3 may integrate endogenous signals (sugar) to coordinate ovule development (Tsai, Gazzarrini, 2012; Chan et al., 2017).
Following fertilization, the zygote together with the endosperm and the integuments develop in a coordinated manner to form the embryo and the seed coat of the mature seed. Here, we show that FUS3 localizes to the funiculus, chalaza, and outer seed coat of developing seeds, partially mirroring its expression pattern in the ovules, but is repressed in the endosperm. Ectopic FUS3 localization to the endosperm of bpc1/2 or endosperm and endothelium of ProML1:FUS3-GFP increases cell proliferation, resulting in enlarged endosperm and larger seed at the expense of embryo development (delayed or arrested). These phenotypes are reminiscent of some FIS-PRC2 mutant alleles of mea (Kiyosue et al., 1999). Given that FUS3 is derepressed in mea endosperm and that MEA associates with a repressive region of the FUS3 locus that has H3K27me3 repressive marks (Makarevich et al., 2006) and is also bound by BPC1, we propose that BPC1 and -2 recruit FIS-PRC2 to repress FUS3 in the endosperm and that this is required to reduce the rate of endosperm nuclei proliferation, promoting endosperm differentiation and embryo growth (Figure 8F).
The FIS-PRC2 specific subunits, MEA and FIS2, are targeted solely to the central cell in the ovule and to the endosperm in the seed, and thus are likely to participate in FUS3 repression in these tissues (Luo et al., 2000; Wang et al., 2006). The MEA homolog SWN, which belongs to the VRN-PRC2 and FIS-PRC2 complexes, has a broader localization pattern, but plays a partially redundant function with MEA in repressing central cell/endosperm nuclei proliferation in the absence of fertilization (Wang et al., 2006). Thus, SWN may also be involved in repressing FUS3 in the central cell/endosperm. In contrast, autonomous seed coat development in the ovule is repressed by the sporophytic complexes VRN-PRC2 and EMF-PRC2, which may be involved in repressing FUS3 in the integuments (Köhler and Grossniklaus, 2002; Roszak and Köhler, 2011). In accordance, FUS3 and other seed-specific genes were derepressed and showed reduced H3K27me3 repressive marks in siliques of a weak curly leaf (clf) allele, although the tissue specific expression was not investigated (Liu et al., 2016).
Although BPCs can recruit EMF- and FIS-PRC2 complexes for transcriptional silencing, BPCs were also shown to positively regulate a close FUS3 family member, LEC2 (Berger et al., 2011). This is in accordance with the role of GAGA binding proteins in animals, which have a dual function as both activators and repressors (Berger and Dubreucq, 2012). Interestingly, FUS3 is expressed in the embryo and in specific sporophytic tissues of the ovule and seed (chalaza, funiculus, seed coat) where all class I BPCs are expressed. Thus, it is possible that class I BPCs act as both positive and negative regulators of LAFL genes, depending on the interacting partners. The mechanisms of this dual function, however, has not been fully explored.
An important question is: How does FUS3 regulate reproductive tissue development and the transition from reproductive to seed development? FUS3 was shown to inhibit embryonic-to-vegetative and vegetative-to-reproductive phase changes by increasing ABA/GA and counteracting ethylene signaling, with ABA and GA stabilizing and destabilizing the FUS3 protein, respectively (Curaba et al., 2004; Gazzarrini et al., 2004; Lumba et al., 2012; Chiu et al., 2016a, 2016b). A positive feedback regulatory loop has also been established between auxin and FUS3 in the embryo, whereby auxin induces FUS3 expression and FUS3 promotes auxin synthesis. Auxin also positively regulates FUS3 (Gazzarrini et al., 2004; Wang and Perry, 2013). Given that auxin is required for the synchronized growth of the fruit, the different tissues within the seed (integuments, endosperm, and embryo) and that FUS3 localization patterns in ovules, seeds, and embryos largely mirror those of auxin, we propose that FUS3 may regulate auxin level/localization and that auxin may in turn regulate FUS3 expression/activity (Gazzarrini et al., 2004; Figueiredo et al., 2015; Figueiredo et al., 2016; Larsson et al., 2017; Robert et al., 2018). Reduced auxin accumulation in the chalaza and funiculus of fus3-3 or increased auxin levels in the integuments and endosperm of ProML1:FUS3 or bpc1/2 would impair ovule and seed development resulting in seed abortion and delayed embryo development, respectively, as shown by delayed endosperm cellularization and embryo growth arrest triggered by auxin overproduction in the endosperm (Figueiredo and Köhler, 2018; Batista et al., 2019; Robert, 2019). Other LAFLs have also been shown to regulate the expression of auxin biosynthesis or signaling genes, suggesting that the LAFL network may be involved in coordinating not only seed maturation processes but also early phases of seed development.
In conclusion, mutations affecting FIS-PRC2 or Polycomb response elements binding transcription factors such as BPCs cause severe seed abortion; however, the molecular mechanisms are not fully understood (Monfared et al., 2011; Wang and Köhler, 2017; Figueiredo and Köhler, 2018). Here we show that BPC1- and BPC2-mediated spatiotemporal restriction of FUS3, a target of the PRC2 complex, is required for the transition from reproductive to seed development, as well as from early embryogenesis to seed maturation in Arabidopsis.
METHODS
Plant Material
T-DNA insertion lines bpc1-1 (SALK_072966C), bpc2 (SALK_090810), bpc1-1 bpc2 (bpc1/2; CS68700), and bpc1-1 bpc2 bpc3-1 (CS68699), and an ethyl methanesulfonate (EMS) mutant bpc3-1 (CS68805) were previously described (Monfared et al., 2011). The fus3-3 loss-of-function mutant was previously described (Keith et al., 1994). T-DNA insertion lines bpc1_salk (SALK_101466C), bpc2_salk (SALK_110830C), and bpc3_sail (SAILseq_553_B09.0) were obtained from ABRC, and homozygous lines were confirmed by PCR with primers listed in Supplemental Table. The ProFIE:FIE-GFP, ProMSI1:MSI1-GFP, ProMEA:MEA-YFP, and ProFIS2:GUS reporter lines were previously described (de Lucas et al., 2016). The ProML1:FUS3-GFP construct (containing 3 kb of promoter sequence upstream of the translational start codon) was previously described and shown to be nuclear localized and to rescue fus3-3 (Gazzarrini et al., 2004); twenty independent lines were isolated, and four homozygous lines were selected for further analysis. The ProFUS3:FUS3ΔC-GFP construct (containing 1.5 kb of promoter sequence upstream of the start codon) previously described and shown to be nuclear localized was transformed into Col-0 (Lu et al., 2010); seven independent lines were isolated, and two homozygous lines were selected for further analysis. The ProFUS3:FUS3ΔC-GFP does not rescue fus3-3 and is therefore not functional (Lu et al., 2010). The ProFUS3:FUS3-GFP construct (containing 1.5 kb of promoter sequence upstream of the start codon) was previously described and shown rescues fus3-3 (Gazzarrini et al., 2004; Lu et al., 2010; Chan et al., 2017). ProFUS3 (1.5 kb):GUS/GFP and ProFUS3MUT (1.5 kb):GUS/GFP were generated as described in the Supplemental Methods. Six to 15 ProFUS3 (1.5 kb):GUS and ProFUS3MUT (1.5 kb):GUS transgenic lines per constructs were generated and analyzed for GUS staining. Then 25 to 34 transgenic lines of ProFUS3(1.5kb):GFP and ProFUS3MUT(1.5kb):GFP were generated per constructs and analyzed for GFP fluorescence. ProFUS3(1.5kb):GUS/GFP and ProFUS3MUT(1.5 kb):GUS/GFP lines contain 1.5 kb of sequence upstream the ATG, including the 5′UTR, which has GA/CT repeats. GA/CT repeats were mutated in ProFUS3MUT, as shown Supplemental Figure 3. Transformed lines were selected on kanamycin or hygromycin plates. Sterilized Arabidopsis (Arabidopsis thaliana) seeds were germinated on half-strength Murashige and Skoog medium, transferred to soil, and grown under 16-h/8-h light/darkness 22°C/18°C. Frequencies of aborted ovules and seeds, as well as other seed phenotypes displayed by various genotypes, were calculated using half dissected siliques (n = 5-10 siliques), which had a variable number of seeds depending on the genotype. Experiments were repeated three times, and averages of three biological replicates ±sd are shown. Total seed yield per plant was calculated with five plants per pot. Experiments were repeated three times, and averages of three biological replicates ±sd are shown.
Yeast One-Hybrid Screening
Yeast one-hybrid (Y1H) library screening and one-on-one retests were performed as described by Deplancke et al. (2006) with some modifications. See the Supplemental Methods for bait construction and Y1H screen.
Yeast Two-Hybrid Assay
To test interactions between FIS2 and BPCs, FIS2 was cloned in pDEST32 by Gateway (Invitrogen) and fused to the GAL4 binding domain (BD), and BPCs were cloned in pDEST22 and fused to the GAL4 activation domain (AD). The yeast stain AH109 was cotransformed with AD and BD vectors by PEG/LiAC (Gietz and Schiestl, 2007). Because MEA cloned in the pDEST32 vectors autoactivated, to test the interaction between MEA and BPCs, MEA was cloned in pGBKT7 and fused to the GAL4 BD, and BPC1-3 were cloned in pGAD424 and fused to the GAL4 AD. The yeast stain YRG-2 was transformed with AD and BD vectors. Despite their similarity, the pDEST32/22 and pGBKT7/pGAD424 vector pairs result in different expression levels of bait/pray and show different sensitivity when tested in large scale screens, thus yielding a higher number of positive and reproducible interactions when both vectors are used in parallel (Rajagopala et al., 2009). All colonies grew on -Trp -Leu (-TL) medium and interactions were tested on -Trp -Leu -His (-TLH) medium for 3 to 4 d. Interaction assays were repeated three times with very similar results, and one is shown.
BiFC
The CDS of BPCs, FUSCA3, FIE, FIS2, LHP1 MSI1, and MEA were cloned into BiFC vectors pB7WGYN2 (YNE) or pB7WGYC2 (YCE; Tsuda et al., 2017) by Gateway (Invitrogen), transformed into Agrobacterium tumefaciens strain GV2260, and infiltrated into Nicotiana benthamiana leaves as previously described by Duong et al. (2017). At least three biological replicates were performed with similar results and one is shown. For each replicate, a minimum of three discs were randomly selected from three infiltrated leaves and imaged. Different areas of the disk were imaged, and interactions were observed in several cells of each disk (10 or more interactions out of 50 to 200 cells in each area screened, depending on the interaction pairs).
Differential Interference Contrast Microscopy
Pistils or siliques were dissected and immersed in fixing solution (9:1, ethanol:acetic acid [v/v]) for 2 h before washing them twice with 90% (v/v) ethanol. The siliques were then cleared with clearing solution (2.5 g/mL chloral hydrate and 30% [v/v] glycerol) overnight. Images were taken with a Zeiss Axioplant 2 microscope equipped with differential interference contrast (DIC) optics. The quantifications of seed sizes and numbers of endosperm nuclei were performed by ImageJ (National Institutes of Health).
Confocal Microcopy
To observe GFP fluorescence in transgenic Arabidopsis, fresh tissue was dissected and mounted on the slides in 10% (v/v) glycerol. Visualization was done with a LSM510 confocal microscope (488 nm excitation and a 515- to 535-nm band-pass filter; Zeiss).
GUS Staining
GUS staining assays were performed as previously described by Wu et al. (2019) with some modifications. The concentration of ferri/ferrocyanide used for ProBPC3:GUS was 2 mM, and 5 mM was used for ProBPC1:GUS and ProBPC2:GUS. To detect low expression of FUS3 in inflorescences, leaves or flowers of ProFUS3 (1.5 kb):GUS and ProFUS3MUT(1.5 kb):GUS lines, ferri/ferrocyanide was not included in the buffer. Cleared tissues were imaged by DIC microscopy using a Axioplant 2 microscope (Zeiss).
Glutaraldehyde Staining
To visualize ovule/seed structures, whole pistils at FS12, and 1DAF or 2DAF siliques were fixed in 3% (w/v) paraformaldehyde in PBS (0.145 M NaCl, 0.0027 M KCl, 0.0081 M Na2HPO4, 0.0015 M KH2PO4, pH 7.4) for 15 min at room temperature and rinsed twice with PBS. The treated tissues were stained in 5% (v/v) glutaraldehyde in PBS at 4°C overnight in the dark. Tissues were washed three times with PBS and cleared for ∼1 to 2 weeks with ClearSee buffer (Kurihara et al., 2015). The images were photographed with a Zeiss LSM510 confocal microscope (530-nm excitation and a 560-nm long-pass filter).
Gene Expression Assay
RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). About 1 μg of RNA was used for reverse transcription. Quantitative real-time PCR was performed using Step One Plus real-time PCR system (Applied Biosystems) with SYBR premix. PP2AA3 was chosen as the internal reference gene to normalize the gene expression data using the relative quantification method (2–ΔΔCT). Primers used are listed in the Supplemental Table. Three biological replicates were performed.
ChIP Assay
To generate Pro35S:BPC1-red fluorescent protein (RFP), the BPC1 coding sequence was first cloned into pDONR221 (Life Technologies) and subsequently transferred to pB7RWG2 (Flanders Interuniversity Institute for Biotechnology). Arabidopsis plants were transformed with the Pro35S:BPC1-RFP using the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998). Transformed plants were sown on soil and selected with a 50 μg/mL BASTA solution (Bayer); the presence of the construct was assessed by genotyping and analysis of RFP expression. Arabidopsis plants were directly sown on soil and kept under short-day conditions for 2 weeks (22°C, 8-h light/16-h dark) and then moved to long-day conditions (22°C, 16-h light/8-h dark). ChIP assays were performed in inflorescences (from the inflorescence meristem to open flowers) and young siliques, as described by Gregis et al. (2009) using for BPC1-RFP, an anti-RFP VHH coupled to magnetic agarose beads RFP-trap_Magnetic Agarose (Chromotek). Real-time PCR assays were performed to determine the enrichment of the fragments. The detection was performed in triplicate using the iQ SYBR Green Supermix (Bio-Rad) and the Bio-Rad iCycler iQ Optical System (software version 3.0a), with the primers listed in Supplemental Table. ChIP-qPCR experiments and relative enrichments were calculated as reported by Gregis et al. (2009).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BPC1 (AT2G01930), BPC2 (AT1G14685), BPC3 (AT1G68120), BPC4 (AT2G21240), FIE (AT3G20740), FIS2 (AT2G35670), FUSCA3 (At3g26790), LHP1 (AT5G17690), MEA (AT1G02580), ML1 (At4G21750), MSI1 (AT5G58230), and PP2AA3 (At1G13320). Germplasm used included T-DNA insertion lines bpc1-1 (SALK_072966C), bpc2 (SALK_090810), bpc1-1 bpc2 (bpc1/2; CS68700), bpc1-1 bpc2 bpc3-1 (CS68699), bpc1_salk (SALK_101466C), bpc2_salk (SALK_110830C), and bpc3_sail (SAILseq_553_B09.0), as well as fus3-3 EMS mutant (CS68731) and bpc3-1 EMS mutant (CS68805).
SUPPLEMENTAL DATA
Supplemental Figure 1. FUS3 expression profile and FUS localization in reproductive tissues and stem epidermis.
Supplemental Figure 2. ML1 expression profile and FUS3-GFP localization in ovules and seeds.
Supplemental Figure 3. Location of (GA/CT)n mutated in the FUS3 genomic region used in Y1H.
Supplemental Figure 4. The VRN-, EMF- and FIS-PRC2 complexes.
Supplemental Figure 5. Homo- and heterodimerization between class I BPCs in planta.
Supplemental Figure 6. Negative controls for bimolecular fluorescence complementation assays.
Supplemental Figure 7. Class I BPC proteins do not interact with LHP1 in planta.
Supplemental Figure 8. Expression/localization patterns of class I BPCs and FIS-PRC2 in ovules and seeds.
Supplemental Figure 9. Class I bpc mutants show delayed megagametogenesis, ovule and seed abortion, and delayed embryogenesis.
Supplemental Figure 10. Ectopic expression of FUS3ΔC-GFP in bpc1/2 ovules and seeds.
Supplemental Figure 11. Overexpression of FUS3 results in embryo defects and overproliferation of the endosperm nuclei.
Supplemental Figure 12. Conserved (GA/CT)n motifs in orthologous FUS3 genes.
Supplemental Table. Primers used in this study.
Supplemental Methods. Cloning and Y1H screen.
Supplemental References. References to Supplemental Figures and Supplemental Methods.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Charles S. Gasser (University of California, Davis) for the ProBPC3:GUS reporter construct; François Parcy (Centre National de la Recherche Scientifique) for ProFUS3:GUS; Siobhan M. Brady (University of California, Davis) and Miguel De Lucas (Durham University) for ProFIE:FIE:GFP, ProMSI1:MSI1:GFP, ProMEA:MEA:YFP, and ProFIS2:GUS reporter lines as well as the FIE and MSI1 vectors; Claudia Koehler (Swedish University of Agricultural Sciences) and Ramin Yadegari (University of Arizona) for the MEA/pBluescript II KS and FIS2/pGBKT7 vectors; Daniel Riggs (University of Toronto Scarborough) for the YRG-2 yeast strain; and Jumi A. Shin (University of Toronto Mississauga) for the AH109 yeast strain. This work was supported by the National Natural Science Foundation (31701952 to J.W.); the China Postdoctoral Council (scholarships to J.W.); the Ministero dell’Istruzione, Università e Ricerca (MIUR; SIR2014 MADSMEC [RBSI14BTZR]; to V.G.); the Doctorate School in Molecular and Cellular Biology, Università degli Studi di Milano (Fellowship to R.P.); the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (grant CEFF-PXM2019_014207_000032 to J.W. and J.L.); and a Natural Science and Engineering Research Council (NSERC) of Canada (Discovery Grant 480529 to S.G.).
AUTHOR CONTRIBUTIONS
J.W. conducted most of the experiments; J.W. and D.M. performed MFG confocal localization; J.W. and S.G. conceived the study and wrote the article; S.D. contributed to the identification of higher order mutants; R.P. and V.G. conducted ChIP assays; J.L. and L.W. performed Y2H assays; all authors read and approved the article.
References
- Baker S.C., Robinson-Beers K., Villanueva J.M., Gaiser J.C., Gasser C.S.(1997). Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145: 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista R.A., Figueiredo D.D., Santos-González J., Köhler C.(2019). Auxin regulates endosperm cellularization in Arabidopsis. Genes Dev. 33: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger N., Dubreucq B.(2012). Evolution goes GAGA: GAGA binding proteins across kingdoms. Biochim. Biophys. Acta 1819: 863–868. [DOI] [PubMed] [Google Scholar]
- Berger N., Dubreucq B., Roudier F., Dubos C., Lepiniec L.(2011). Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23: 4065–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J., Van den Daele H., Hollunder J., Rombauts S., Mylle E., Hilson P., Inzé D., De Veylder L., Russinova E.(2010). Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P., Aubourg S., Van Leene J., De Jaeger G., Lurin C.(2013). Plant protein interactomes. Annu. Rev. Plant Biol. 64: 161–187. [DOI] [PubMed] [Google Scholar]
- Braybrook S.A., Stone S.L., Park S., Bui A.Q., Le B.H., Fischer R.L., Goldberg R.B., Harada J.J.(2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA 103: 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonero P., Iglesias-Fernández R., Vicente-Carbajosa J.(2017). The AFL subfamily of B3 transcription factors: Evolution and function in angiosperm seeds. J. Exp. Bot. 68: 871–880. [DOI] [PubMed] [Google Scholar]
- Chan A., Carianopol C., Tsai A.Y.L., Varatharajah K., Chiu R.S., Gazzarrini S.(2017). Corrigendum: SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J. Exp. Bot. 68: 5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R.S., Pan S., Zhao R., Gazzarrini S.(2016a). ABA-dependent inhibition of the ubiquitin proteasome system during germination at high temperature in Arabidopsis. Plant J. 88: 749–761. [DOI] [PubMed] [Google Scholar]
- Chiu R. Shun, Saleh Y., Gazzarrini S.(2016b). Inhibition of FUSCA3 degradation at high temperature is dependent on ABA signaling and is regulated by the ABA/GA ratio. Plant Signal Behav 11: e1247137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curaba J., Moritz T., Blervaque R., Parcy F., Raz V., Herzog M., Vachon G.(2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 136: 3660–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Pu L., Turco G., Gaudinier A., Morao A.K., Harashima H., Kim D., Ron M., Sugimoto K., Roudier F., Brady S.M.(2016). Transcriptional regulation of arabidopsis polycomb repressive complex 2 coordinates cell-type proliferation and differentiation. Plant Cell 28: 2616–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S.C., Weijers D.(2017). Plant embryogenesis. Curr. Biol. 27: R870–R873. [DOI] [PubMed] [Google Scholar]
- Deplancke B., Vermeirssen V., Arda H.E., Martinez N.J., Walhout A.J.(2006). Gateway-compatible yeast one-hybrid screens. CSH Protoc 2006: pdb.prot4590. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T., Sprunck S., Wessel G.M.(2016). Fertilization mechanisms in flowering plants. Curr. Biol. 26: R125–R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong S., Vonapartis E., Li C.Y., Patel S., Gazzarrini S.(2017). The E3 ligase ABI3-INTERACTING PROTEIN2 negatively regulates FUSCA3 and plays a role in cotyledon development in Arabidopsis thaliana. J. Exp. Bot. 68: 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R.C., Betzner A.S., Huttner E., Oakes M.P., Tucker W.Q., Gerentes D., Perez P., Smyth D.R.(1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatihi A., Boulard C., Bouyer D., Baud S., Dubreucq B., Lepiniec L.(2016). Deciphering and modifying LAFL transcriptional regulatory network in seed for improving yield and quality of storage compounds. Plant Sci. 250: 198–204. [DOI] [PubMed] [Google Scholar]
- Figueiredo D.D., Batista R.A., Roszak P.J., Hennig L., Köhler C.(2016). Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 5: e20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo D.D., Batista R.A., Roszak P.J., Köhler C.(2015). Auxin production couples endosperm development to fertilization. Nat. Plants 1: 15184. [DOI] [PubMed] [Google Scholar]
- Figueiredo D.D., Köhler C.(2018). Auxin: A molecular trigger of seed development. Genes Dev. 32: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser C.S., Skinner D.J.(2019). Development and evolution of the unique ovules of flowering plants. Curr. Top. Dev. Biol. 131: 373–399. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P.(2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7: 373–385. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H.(2007). Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 38–41. [DOI] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Dorca-Fornell C., Kater M.M.(2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60: 626–637. [DOI] [PubMed] [Google Scholar]
- Hecker A., Brand L.H., Peter S., Simoncello N., Kilian J., Harter K., Gaudin V., Wanke D.(2015). The Arabidopsis GAGA-binding factor BASIC PENTACYSTEINE6 recruits the POLYCOMB-REPRESSIVE COMPLEX1 component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA motifs. Plant Physiol. 168: 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wijeratne A.J., Tang C., Zhang T., Fenelon R.E., Owen H.A., Zhao D.(2016). Ectopic expression of TAPETUM DETERMINANT1 affects ovule development in Arabidopsis. J. Exp. Bot. 67: 1311–1326. [DOI] [PubMed] [Google Scholar]
- Jia H., Suzuki M., McCarty D.R.(2014). Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip. Rev. Dev. Biol. 3: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K., Kraml M., Dengler N.G., McCourt P.(1994). fusca3: A heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T., Ohad N., Yadegari R., Hannon M., Dinneny J., Wells D., Katz A., Margossian L., Harada J.J., Goldberg R.B., Fischer R.L.(1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher K.M., Chow H., Reiser L., Fischer R.L.(1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Grossniklaus U.(2002). Epigenetic inheritance of expression states in plant development: The role of Polycomb group proteins. Curr. Opin. Cell Biol. 14: 773–779. [DOI] [PubMed] [Google Scholar]
- Kooiker M., Airoldi C.A., Losa A., Manzotti P.S., Finzi L., Kater M.M., Colombo L.(2005). BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17: 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj T., Savino G., Valon C., Giraudat J., Parcy F.(2003). Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073. [DOI] [PubMed] [Google Scholar]
- Kurihara D., Mizuta Y., Sato Y., Higashiyama T.(2015). ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142: 4168–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C., Köhler C.(2014). Embryo and endosperm, partners in seed development. Curr. Opin. Plant Biol. 17: 64–69. [DOI] [PubMed] [Google Scholar]
- Larsson E., Vivian-Smith A., Offringa R., Sundberg E.(2017). Auxin homeostasis in Arabidopsis ovules is anther-dependent at maturation and changes dynamically upon fertilization. Front Plant Sci 8: 1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S., Slane D., Herud O., Kong J., Jürgens G.(2012). Early embryogenesis in flowering plants: Setting up the basic body pattern. Annu. Rev. Plant Biol. 63: 483–506. [DOI] [PubMed] [Google Scholar]
- Lepiniec L., Devic M., Roscoe T.J., Bouyer D., Zhou D.X., Boulard C., Baud S., Dubreucq B.(2018). Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 31: 291–307. [DOI] [PubMed] [Google Scholar]
- Liu J., Deng S., Wang H., Ye J., Wu H.W., Sun H.X., Chua N.H.(2016). CURLY LEAF Regulates Gene Sets Coordinating Seed Size and Lipid Biosynthesis. Plant Physiol. 171: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A.L., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J.(1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lu P., Porat R., Nadeau J.A., O’Neill S.D.(1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8: 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q.S., Paz J.D., Pathmanathan A., Chiu R.S., Tsai A.Y.L., Gazzarrini S.(2010). The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J. 64: 100–113. [DOI] [PubMed] [Google Scholar]
- Lumba S., Tsuchiya Y., Delmas F., Hezky J., Provart N.J., Shi Lu Q., McCourt P., Gazzarrini S.(2012). The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis. BMC Biol. 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E.S., Peacock W.J., Chaudhury A.(2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G., Leroy O., Akinci U., Schubert D., Clarenz O., Goodrich J., Grossniklaus U., Köhler C.(2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7: 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister R.J., Williams L.A., Monfared M.M., Gallagher T.L., Kraft E.A., Nelson C.G., Gasser C.S.(2004). Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 37: 426–438. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Ikeda M., Takada S., Takiguchi Y., Kondou Y., Yoshizumi T., Fujita M., Shinozaki K., Matsui M., Ohme-Takagi M.(2010). Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 51: 2145–2151. [DOI] [PubMed] [Google Scholar]
- Monfared M.M., Simon M.K., Meister R.J., Roig-Villanova I., Kooiker M., Colombo L., Fletcher J.C., Gasser C.S.(2011). Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis. Plant J. 66: 1020–1031. [DOI] [PubMed] [Google Scholar]
- Mozgova I., Köhler C., Hennig L.(2015). Keeping the gate closed: Functions of the polycomb repressive complex PRC2 in development. Plant J. 83: 121–132. [DOI] [PubMed] [Google Scholar]
- Mu Y., Zou M., Sun X., He B., Xu X., Liu Y., Zhang L., Chi W.(2017). BASIC PENTACYSTEINE proteins repress ABSCISIC ACID INSENSITIVE4 expression via direct recruitment of the polycomb -repressive complex 2 in Arabidopsis root development. Plant Cell Physiol. 58: 607–621. [DOI] [PubMed] [Google Scholar]
- Nodine M.D., Bartel D.P.(2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 24: 2678–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopala S.V., Hughes K.T., Uetz P.(2009). Benchmarking yeast two-hybrid systems using the interactions of bacterial motility proteins. Proteomics 9: 5296–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S.(2019). Molecular communication for coordinated seed and fruit development: What can we learn from auxin and sugars? Int. J. Mol. Sci. 20: E936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S., Park C., Gutièrrez C.L., Wójcikowska B., Pěnčík A., Novák O., Chen J., Grunewald W., Dresselhaus T., Friml J., Laux T.(2018). Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 4: 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe T.J., Vaissayre V., Paszkiewicz G., Clavijo F., Kelemen Z., Michaud C., Lepiniec L.C., Dubreucq B., Zhou D.X., Devic M.(2019). Regulation of FUSCA3 expression during seed development in Arabidopsis. Plant Cell Physiol. 60: 476–487. [DOI] [PubMed] [Google Scholar]
- Roszak P., Köhler C.(2011). Polycomb group proteins are required to couple seed coat initiation to fertilization. Proc. Natl. Acad. Sci. USA 108: 20826–20831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K., Hülskamp M., Pruitt R.E.(1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7: 731–749. [Google Scholar]
- Simonini S., Kater M.M.(2014). Class I BASIC PENTACYSTEINE factors regulate HOMEOBOX genes involved in meristem size maintenance. J. Exp. Bot. 65: 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini S., Roig-Villanova I., Gregis V., Colombo B., Colombo L., Kater M.M.(2012). Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant Cell 24: 4163–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N., Wobus U.(2013). Seed-development programs: A systems biology-based comparison between dicots and monocots. Annu. Rev. Plant Biol. 64: 189–217. [DOI] [PubMed] [Google Scholar]
- Stone S.L., Kwong L.W., Yee K.M., Pelletier J., Lepiniec L., Fischer R.L., Goldberg R.B., Harada J.J.(2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98: 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., et al. (2012). MicroRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet. 8: e1003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A.Y., Gazzarrini S.(2012). AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 69: 809–821. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Nambara E., Naito S., McCourt P.(2004). The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37: 73–81. [DOI] [PubMed] [Google Scholar]
- Tsuda K., Abraham-Juarez M.J., Maeno A., Dong Z., Aromdee D., Meeley R., Shiroishi T., Nonomura K.I., Hake S.(2017). KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. Plant Cell 29: 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht D., Nodine M.D.(2014). MicroRNA functions in plant embryos. Biochem. Soc. Trans. 42: 352–357. [DOI] [PubMed] [Google Scholar]
- Wang D., Tyson M.D., Jackson S.S., Yadegari R.(2006). Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13244–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Köhler C.(2017). Epigenetic processes in flowering plant reproduction. J. Exp. Bot. 68: 797–807. [DOI] [PubMed] [Google Scholar]
- Wang F., Perry S.E.(2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161: 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann M.R., Mehalick A.J., Packer R.L., Jenik P.D.(2011). MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 155: 1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wu W., Liang J., Jin Y., Gazzarrini S., He J., Yi M.(2019). GhTCP19 transcription factor regulates corm dormancy release by repressing GhNCED expression in gladiolus. Plant Cell Physiol. 60: 52–62. [DOI] [PubMed] [Google Scholar]
- Xiao J., et al. (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49: 1546–1552. [DOI] [PubMed] [Google Scholar]
- Yadegari R., Drews G.N.(2004). Female gametophyte development. Plant Cell 16 (Suppl): S133–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]