β-Lactam resistance in Staphylococcus aureus limits treatment options. Stp1 and Stk1, a serine-threonine phosphatase and kinase, respectively, mediate serine-threonine kinase (STK) signaling. Loss-of-function point mutations in stp1 were detected among laboratory-passaged β-lactam-resistant S. aureus strains lacking mecA and blaZ, the major determinants of β-lactam resistance in the bacteria.
KEYWORDS: Stp1, β-lactam resistance, Staphylococcus aureus
ABSTRACT
β-Lactam resistance in Staphylococcus aureus limits treatment options. Stp1 and Stk1, a serine-threonine phosphatase and kinase, respectively, mediate serine-threonine kinase (STK) signaling. Loss-of-function point mutations in stp1 were detected among laboratory-passaged β-lactam-resistant S. aureus strains lacking mecA and blaZ, the major determinants of β-lactam resistance in the bacteria. Loss of Stp1 function facilitates β-lactam resistance of the bacteria.
INTRODUCTION
Over several decades, Staphylococcus aureus has successfully bypassed the action of antimicrobials through sophisticated resistance mechanisms (1). Its ability to develop resistance together with its vast array of virulence genes has made it a serious threat to global health (2). β-Lactam drugs have been a benchmark for treatment of S. aureus due to their superior efficacy and safety over other drugs. However, rampant β-lactam resistance among clinical strains of S. aureus makes it difficult to treat infections caused by the bacteria.
The two major factors that traditionally drive β-lactam resistance in S. aureus are penicillin binding protein 2a (PBP2a) and β-lactamase (3), encoded by mecA and blaZ, respectively. We previously reported that S. aureus strains (COLnex and SF8300ex; derivatives of COL and SF8300 strains, respectively, lacking both mecA and blaZ) can also produce high-level β-lactam resistance when passaged in the laboratory (4, 5). These results indicated that unknown factors that are independent of mecA and blaZ were responsible for resistance in these strains. Whole-genome sequence analysis of the resistant passaged strains (i.e., COLnex and SF8300ex strains passaged in ceftobiprole, ceftaroline, and nafcillin; i.e., six strains total) revealed mutations in several interesting genes, which suggested that they might play a role in this uncanonical mode of β-lactam resistance (5). One of these candidate genes, which codes for a serine-threonine phosphatase (stp1), had point mutations G169S and Q31X in the SF8300ex strain that was passaged in ceftobiprole and ceftaroline, respectively. Interestingly, none of the COLnex background strains showed any mutations in stp1 (5).
Although both stk1 and stp1 are conserved in COLnex and SF8300ex strains, the former possesses a truncated Stk1 due to a natural point mutation that has been reported in COL (the parent of COLnex) (6). Thus, an absence of stp1 point mutation may be due to an inherent defect in STK signaling among COLnex passaged strains.
STK signaling is predominantly present in eukaryotes, and it was recently shown to be present in bacteria. Since the discovery of the first serine-threonine kinase in Myxococcus xanthus isolates, several studies have elucidated the roles of STK signaling in bacteria (7). In S. aureus, STK signaling is mediated by a serine-threonine kinase and phosphatase, encoded by stk1 and stp1, respectively (8). Stk1 and Stp1, through their opposing functions, maintain the balance of STK signaling.
Recent studies have shown that STK signaling plays important roles in S. aureus pathogenesis (9). An stp1 deletion mutant strain displayed attenuated hemolysis due to low-level production of alpha-toxin compared with its isogenic wild-type (wt) strain (10). Stp1 has also been shown to play a role in resistance of cell wall-reactive antibiotics in S. aureus isolates (11, 12). On the other hand, an stk1 deletion mutant was found to be susceptible to many cell wall-active antibiotics (6, 9, 13). Although much of the mechanistic detail on how STK signaling mediates these processes is currently lacking, it was recently shown that Stk1 controls the activity of β-lactamase in S. aureus through phosphorylation of its sensory inducer, BlaR1 (14). These results indicated that STK signaling mediates β-lactam resistance in S. aureus, at least in part, via the known mediators of resistance. Our results, i.e., detection of Stp1 mutations among resistant passaged strains lacking mecA and blaZ, indicated that STK signaling may also mediate β-lactam resistance independent of the known mediators of resistance. Thus, in this study, we sought to determine the role of STK signaling in β-lactam resistance in strains that lacked mecA and blaZ and to evaluate the role of the Stp1 mutations (G169S and Q31X) detected among our resistant passaged strains.
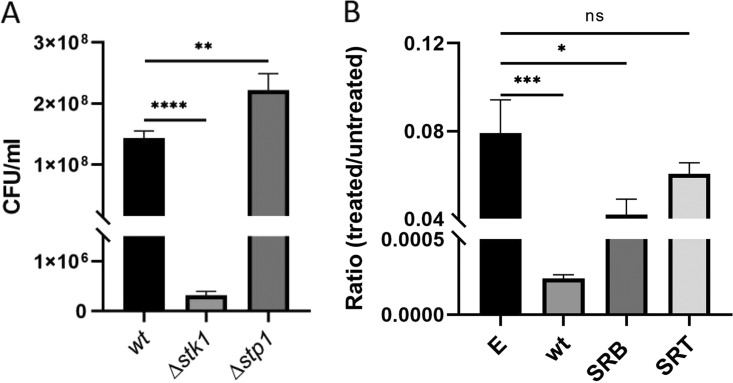
We created stk1 and stp1 deletion mutants (Δstk1 and Δstp1, respectively) in the SF8300ex strain and compared their susceptibilities to nafcillin, a β-lactam antibiotic commonly used for staphylococcal infection (Fig. 1A) (15). Creation of isogenic strains in this manner allowed us to determine the roles of stk1 and stp1 in β-lactam resistance and to compare them with their isogenic parental strain (SF8300ex). For this assay, bacteria were grown in the presence of nafcillin 0.25 μg/ml (half the MIC of SF8300ex) for 4 h and then plated to determine bacterial CFU counts (15). Our results indicated that the Δstk1 mutant strain was highly susceptible to nafcillin treatment, whereas Δstp1 survived the nafcillin challenge better than the SF8300ex strain. Comparison of the growth profiles of these strains in the absence of nafcillin ensured that the difference in survival of these strains on nafcillin challenge was not due to any inherent growth defect (see Fig. S1 in the supplemental material). The resistance phenotype was confirmed with another β-lactam drug, cefoxitin, at a dose that represented half its MIC (15) (see Fig. S3 in the supplemental material). Thus, STK signaling appeared to be important in mediating survival against a β-lactam challenge in strains that lacked both mecA and blaZ. Furthermore, our results suggested that deletion of stk1 and stp1 had opposite effects on β-lactam resistance in the SF8300ex strain.
Fig 1.
Deletion of stp1 facilitates resistance in SF8300ex. (A) CFU/ml of knockout strains (Δstk1 and Δstp1) and wild-type SF8300ex after 4 h of treatment with 0.25 μg/ml nafcillin. (B) Ratio of survival of Δstp1-complemented strains after 4 h of treatment with 0.25 μg/ml nafcillin. Δstp1 complemented with empty-vector pTXΔ (E), wild-type stp1 (wt), mutant G169S stp1 (SRB), and mutant Q31X stp1 (SRT). *, P < 0.01; **, P < 0.009; ***, P < 0.0008; ****, P < 0.0001. Student's t test was used to analyze the P values using GraphPad Prism software. All experiments were repeated at least twice. ns, not significant.
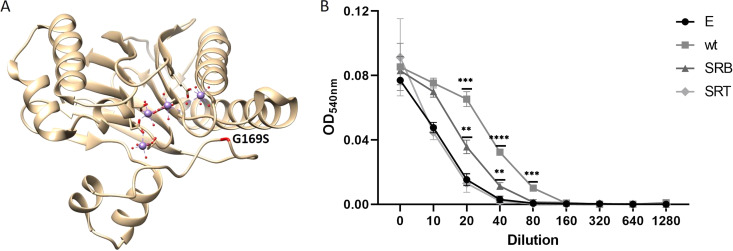
To determine the role of stp1 point mutations G169S and Q31X detected among our resistant passaged strains SRB and SRT, respectively, we cloned stp1 genes from the wt SF8300ex strain and the mutants in a constitutively expressing vector (pTXΔ) (15). The resultant vectors along with the empty-vector control were transformed to the Δstp1 strain to generate Δstp1-complemented strains. Growth assay of the resultant strains in rich medium showed attenuated growth of the strains that were complemented with the wt and SRB stp1 compared with those complemented with the empty-vector or the SRT stp1 (see Fig. S2 in the supplemental material). Because the SRT mutation (Q31X) produced a truncated Stp1 after 31 amino acids, we hypothesized that the resultant growth defects of the above-mentioned strains were likely due to the overexpression of full-length Stp1 (wt and SRB). Because of this growth difference, we challenged the complemented strains with or without nafcillin, and bacterial survival was presented as the ratio of bacterial CFUs between these treatment conditions. Complementation of wt stp1 reduced bacterial survival compared with the empty-vector control (Fig. 1B), supporting an important role for Stp1 in modulating a β-lactam challenge. The survival of the strain with SRB stp1 was detected to be significantly less than that of the empty-vector complemented strain, whereas the strain with SRT stp1 showed bacterial survival that was comparable to that of the strain that received the empty vector. These results suggested that G169S stp1 likely had a loss of Stp1 function mutation, and the Q31X stp1 mutant did not have any Stp1 function, as expected due to its premature truncation caused by a nonsense mutation. Stp1 belongs to the protein phosphatase 2C (PP2C) family, which is known to use metal ions for activity. On mapping the G169S mutation onto the stp1 crystal structure, we found that the point mutation is near the metal binding site of the protein, which may affect its catalytic activity (Fig. 2A) (16, 17).
Fig 2.
Loss of function of Stp1 reduces hemolytic activity of SF8300ex. (A) G169S mutation annotated in the crystal structure of Stp1 bound to metals (purple spheres). The figure was generated using UCSF Chimera and PDB entry 5F1M. (B) Decrease in hemolysis in Δstp1 strains expressing mutant Stp1s. Δstp1 complemented with empty-vector pTXΔ (E), wild-type stp1 (wt), and mutant stp1 strains SRB (G169S) and SRT (Q31X). *, P < 0.01; **, P < 0.009; ***, P < 0.0008; ****, P < 0.0001. Student's t test used to analyze the P values using GraphPad Prism software. Experiments were repeated at least twice.
Stp1 is known to modulate S. aureus hemolysis. As shown previously, a Δstp1 strain causes attenuated hemolysis of sheep blood compared with its wt strain (10). To examine whether the Stp1 mutants had a loss of function mutation through an alternative way, we checked the hemolytic activity of the Δstp1-complemented strains. Culture filtrates from 12-h-grown bacterial cultures were used in various dilutions to check their hemolytic potential. The 0.5% (vol/vol) washed sheep erythrocytes were incubated with culture filtrates at 37°C for 1 h, as described previously (6). Both SRB and SRT complements had attenuated hemolytic activity compared with wt Stp1 (Fig. 2B). These results further indicated that the mutated Stp1s (G169S and Q31X) among our resistant passaged strains were loss-of-function mutations.
Our results indicate that loss of Stp1 mutation facilitates β-lactam resistance in S. aureus isolates that lack mecA and blaZ. Previously, we showed that pbp4 played a pivotal role in β-lactam resistance of SRB and SRT (15). To determine whether the Stk1/Stp1 pathway modulates pbp4 expression, we performed reporter assays that suggested that deletion of stk1 or stp1 did not play any role in pbp4 regulation (see Fig. S4 in the supplemental material). Thus, the mechanistic basis of how the Stk1/Stp1 pathway mediates β-lactam resistance that lacks the classical mediators (mecA and blaZ) remains to be elucidated. In a recent study, Stp1 was shown to regulate the SarA/MgrA family of transcription regulators, which in turn affect staphylococcal virulence and resistance (18–21). Thus, it is possible that Stp1-mediated β-lactam resistance is carried out via the SarA/MgrA family of transcriptional factors. The proteins that mediate this mode of β-lactam resistance are currently unknown.
Supplementary Material
ACKNOWLEDGMENTS
We thank Henry F. Chambers for carefully reading the manuscript. S.C. thanks Dr. Chambers for outstanding mentorship and support over the past several years.
This work was funded by NIH grants 2R01AI100291 and R21AI142501.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foster TJ. 2017. Antibiotic resistance in Staphylococcus aureus: current status and future prospects. FEMS Microbiol Rev 41:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 3.Fuda CC, Fisher JF, Mobashery S. 2005. Beta-lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cell Mol Life Sci 62:2617–2633. doi: 10.1007/s00018-005-5148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. 2010. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level β-lactam resistance contains mutations in three genes. Antimicrob Agents Chemother 54:4900–4902. doi: 10.1128/AAC.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee SS, Chen L, Gilbert A, da Costa TM, Nair V, Datta SK, Kreiswirth BN, Chambers HF. 2017. PBP4 mediates β-lactam resistance by altered function. Antimicrob Agents Chemother 61:e00932-17. doi: 10.1128/AAC.00932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamber S, Schwartzman J, Cheung AL. 2010. Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect Immun 78:3637–3646. doi: 10.1128/IAI.00296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manuse S, Fleurie A, Zucchini L, Lesterlin C, Grangeasse C. 2016. Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol Rev 40:41–56. doi: 10.1093/femsre/fuv041. [DOI] [PubMed] [Google Scholar]
- 8.Debarbouille M, Dramsi S, Dussurget O, Nahori MA, Vaganay E, Jouvion G, Cozzone A, Msadek T, Duclos B. 2009. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J Bacteriol 191:4070–4081. doi: 10.1128/JB.01813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlsen K, Donat S. 2010. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int J Med Microbiol 300:137–141. doi: 10.1016/j.ijmm.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Burnside K, Lembo A, de Los Reyes M, Iliuk A, Binhtran NT, Connelly JE, Lin WJ, Schmidt BZ, Richardson AR, Fang FC, Tao WA, Rajagopal L. 2010. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One 5:e11071. doi: 10.1371/journal.pone.0011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron DR, Ward DV, Kostoulias X, Howden BP, Moellering RC Jr, Eliopoulos GM, Peleg AY. 2012. Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J Infect Dis 205:1677–1687. doi: 10.1093/infdis/jis252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Q, Peng H, Rao X. 2016. Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front Microbiol 7:1601. doi: 10.3389/fmicb.2016.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beltramini AM, Mukhopadhyay CD, Pancholi V. 2009. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun 77:1406–1416. doi: 10.1128/IAI.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boudreau MA, Fishovitz J, Llarrull LI, Xiao Q, Mobashery S. 2015. Phosphorylation of BlaR1 in manifestation of antibiotic resistance in methicillin-resistant Staphylococcus aureus and its abrogation by small molecules. ACS Infect Dis 1:454–459. doi: 10.1021/acsinfecdis.5b00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan LC, Gilbert A, Basuino L, da Costa TM, Hamilton SM, Dos Santos KR, Chambers HF, Chatterjee SS. 2016. PBP 4 mediates high-level resistance to new-generation cephalosporins in Staphylococcus aureus. Antimicrob Agents Chemother 60:3934–3941. doi: 10.1128/AAC.00358-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng W, Cai X, Xie M, Liang Y, Wang T, Li Z. 2016. Structure-based identification of a potent inhibitor targeting Stp1-mediated virulence regulation in Staphylococcus aureus. Cell Chem Biol 23:1002–1013. doi: 10.1016/j.chembiol.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 18.Cheung AL, Nishina KA, Trotonda MP, Tamber S. 2008. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol 40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun F, Ding Y, Ji Q, Liang Z, Deng X, Wong CC, Yi C, Zhang L, Xie S, Alvarez S, Hicks LM, Luo C, Jiang H, Lan L, He C. 2012. Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc Natl Acad Sci U S A 109:15461–15466. doi: 10.1073/pnas.1205952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun F, Zhou L, Zhao BC, Deng X, Cho H, Yi C, Jian X, Song CX, Luan CH, Bae T, Li Z, He C. 2011. Targeting MgrA-mediated virulence regulation in Staphylococcus aureus. Chem Biol 18:1032–1041. doi: 10.1016/j.chembiol.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingavale S, van Wamel W, Luong TT, Lee CY, Cheung AL. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect Immun 73:1423–1431. doi: 10.1128/IAI.73.3.1423-1431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.