The associations between roots and bacteria are important for rhizosheath formation in rice plants under moderate soil drying and involve the ethylene response.
Abstract
The rhizosheath is a layer of soil around the root that provides a favorable environment for soil microbe enrichment and root growth. Rice (Oryza sativa) roots form rhizosheaths under moderate soil drying (MSD) conditions, but how the rhizosheath forms associations with microbes is unclear. To investigate rice rhizosheath formation under MSD, we employed a multiphasic approach, integrating data from high-throughput sequencing and root-bacteria interactions. Rice roots formed a pronounced rhizosheath under MSD, but not under continuous flooding regimens. Plant growth-promoting rhizobacteria of the Enterobacteriaceae were enriched in rhizosheaths of two different rice varieties, ‘Gaoshan 1’ (drought tolerant) and ‘Nipponbare’ (drought sensitive). RNA-sequencing analysis revealed that the ethylene pathway was induced in the rhizosheath-root system under MSD. Enterobacter aerogenes, a bacterium isolated from the rhizosheath, degrades the ethylene precursor 1-aminocyclopropane-1-carboxylate, thereby increasing rhizosheath formation. Furthermore, a 1-aminocyclopropane-1-carboxylate deaminase-deficient mutant of E. aerogenes failed to enhance rice rhizosheath formation. Our results suggest that root-bacteria associations substantially contribute to rhizosheath formation in rice under MSD conditions by mechanisms that involve the ethylene response. These data inform strategies to reduce water consumption in rice production, one of the most water-intensive human activities.
The rhizosheath is a layer of soil around the root, and it adheres to the root upon excavation of the root system, as a consequence of adherence of soil particles to root hairs and mucilage secreted by roots or microbes (Liu et al., 2019; Zhang et al., 2020). The rhizosheath was first described in desert grasses as cylindrical sand grain sheaths encasing the roots, and then was found in some major cereals (including barley [Hordeum vulgare], wheat [Triticum aestivum], maize [Zea mays], and sorghum [Sorghum bicolor]; Duell and Peacock, 1985). Recently, Brown et al. (2017) investigated 58 species in 19 orders of angiosperms and found that 81% of the 58 species and 89% of the 19 orders formed rhizosheaths, which suggests that rhizosheath formation occurs in orders throughout the angiosperms.
Rhizosheaths preferentially form in drier soil, in which adhesiveness of rhizosheath mucilage is enhanced and root hair growth is stimulated, both increasing the stability of rhizosheaths (Watt et al., 1994). Therefore, rhizosheaths are thought to improve drought resistance, which has been validated in several studies (Brown et al., 2017; Rabbi et al., 2018). Moreover, rhizosheaths increase resistance to other abiotic stresses, including the stresses associated with soil strength (Haling et al., 2013; Haling et al., 2014), soil acidity (Haling et al., 2010; Delhaize et al., 2012), and nutrient deficiencies such as reduced phosphorus (James et al., 2016), nitrogen (Wullstein et al., 1979; Bergmann et al., 2009), and zinc (Nambiar, 1976). Because rhizosheaths play such important roles in many abiotic stresses, rhizosheath formation has been identified as a potential trait for improvement of agricultural sustainability if optimized in major crops (Brown et al., 2017).
The rhizosphere is commonly used to describe the narrow zone affected by root activity, while the rhizosheath describes a casing of soil or sand around the roots. Therefore, in plants with rhizosheaths, the rhizosheath is a part of the rhizosphere. However, sometimes roots have no rhizosheath, because rhizosheath formation is related to root hairs, mucilage secreted by roots, or microbes and soil structure. For example, mutant barley plants lacking root hairs form little or no rhizosheath (George et al., 2014). We also found that rice roots can form a rhizosheath under conditions of moderate soil drying (MSD) but not in continuous flooding (CF; Zhang et al., 2020). In comparison with the normal rhizosphere, the rhizosheath is related to root hair cylinder volume and within a 0–0.5 mm radius of the root in rice, while the rhizosphere (a few millimeters wide) is less related to root hairs and typically extends significantly beyond the boundaries of the rhizosheath (Philippot et al., 2013; Pang et al., 2017). Put another way, the rhizosheath is soil closely attached to roots, while the rhizosphere is soil affected by root activity, whether the soil attached or did not attach to roots (Brown et al., 2017).
Plants drive colonization by rhizo-associated microbiota by producing and secreting a variety of metabolites, including many primary metabolites and a diverse set of secondary metabolites (Hu et al., 2018). In addition to providing food for microbial growth, root exudates act as signaling molecules that can attract, stimulate, but also inhibit or repel specific microbes (Baetz and Martinoia, 2014; Coskun et al., 2017). There is now a significant body of research showing that the rhizosphere represents a highly dynamic environment for initiation of associations between roots and soil microbes (Venturi and Keel, 2016).
Many bacteria isolated from the plant rhizosphere and endosphere exhibit plant growth-promoting (PGP) activity (Marasco et al., 2012; Rolli et al., 2015). First, bacteria can alter the plant’s physiology and metabolic processes to enhance plant-induced systemic tolerance to drought (Cho et al., 2013). Second, some bacteria produce 1-aminocyclopropane-1-carboxylate deaminase (ACCd) to degrade the ethylene (ET) biosynthetic precursor ACC into α-ketobutyrate and ammonia within roots or in exudates, leading to a decrease in the ET level in plant roots, and promoting plant growth and development (Nascimento et al., 2018). Third, some bacteria produce plant-growth-promoting auxins and contribute to plant growth and hormone homeostasis (Remans et al., 2008). Moreover, some bacteria solubilize poorly available nutrients, such as iron and phosphorus, thereby indirectly increasing plant growth (Marschner et al., 2011). Even though the rhizosheath typically contains a mixture of exudates, mucilage, and exopolymers, which increase the wettability and water-use efficiency of the root system and produce a favorable environment for microbial enrichment (Marasco et al., 2018a), few studies have focused on the roles of microbes within the rhizosheath, and the roles of microbial communities and microbe-root associations in rhizosheath formation under soil drying are unclear.
Rice (Oryza sativa) is one of the major food crops, and billions of the world’s people rely on it. However, rice is vulnerable to drought, and its cultivation requires very large quantities of irrigation water (Zhang et al., 2009). Indeed, the production of 1 kg of rice at maturity in Asia typically requires the input of 1,000–5,000 L of water in the field over the growing season, and the majority of irrigation water use in Asia is committed to rice cultivation (Bouman and Tuong, 2001; Wu et al., 2017; Britto and Kronzucker, 2018). Our previous study showed that the drought-tolerant rice ssp. japonica ‘Gaoshan 1’ (Up1) can induce root growth under moderate soil drying compared with a drought-sensitive rice ssp. japonica ‘Nipponbare’ (Nip; Shi et al., 2015).
Moreover, root microbiota composition is also important in rice (Zhang et al., 2019). Some bacterial communities may play more important roles in crop rhizosheath formation compared to other microbial groups (Hu et al., 2018; Marasco et al., 2018a). Therefore, in the current study, bacterial communities and predicted function of endosphere root tissues, the rhizosheath under MSD, and the rhizosphere under CF were investigated in drought-tolerant Up1 and drought-sensitive Nip cultivars, coupled with bulk examination of the soil. Sequence analysis for bacterial communities and roots bounded by rhizosheaths (rhizosheath roots) under MSD and without rhizosheath (bald-roots) under CF were analyzed, and effects of isolated bacteria on rhizosheath formation were evaluated, with the goal of illuminating the associations of root and bacteria in the rice rhizosheath, and the effects of water stress and microbial communities upon rhizosheath formation.
RESULTS
Rhizosheath Formation in Rice Plants during MSD and CF Irrigation
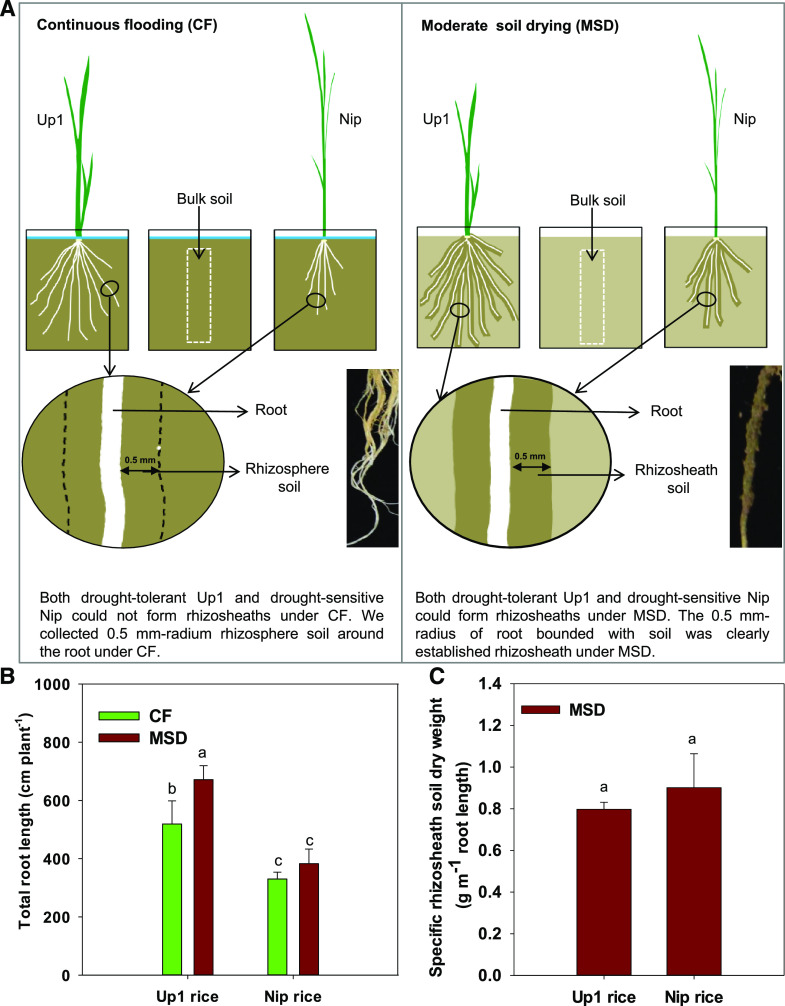
Root growth was enhanced in Up1 under MSD, and Up1 had greater root length under MSD compared to CF, while Nip maintained the same root length under MSD as under CF (Fig. 1). Both Up1 and Nip could form rhizosheaths under MSD, but not under CF. Under MSD, there was no significant difference in the specific rhizosheath between the two rice varieties (Fig. 1).
Figure 1.
Experimental design for determination of bacterial community composition and root traits of rice used in this study. A, Experimental design for determining bacterial community composition in samples from different root-system compartments (endosphere root tissues and rhizosheath or rhizosphere) of two rice varieties (Up1 and Nip) under MSD (with rhizosheath) and CF (without rhizosheath) conditions. Both Up1 and Nip rice varieties could form rhizosheaths under MSD, but not under CF. Since the rhizosheath was clearly established as a 0.5-mm radius of root bounded with soil under MSD in rice, soil was also collected from a 0.5-mm-radius rhizosphere around the root under CF for comparison with the rhizosheath under MSD. B, Total root length of Up1 and Nip rice varieties under the MSD and CF conditions. One-way ANOVA was performed and Duncan’s test was used for post hoc multiple comparisons. The bars with different letter notations were significantly different at P < 0.05. C, Specific rhizosheath soil dry weight under the MSD condition. Student’s t test was performed. Lowercase letters indicate a significant difference at P < 0.05. Data in B and C are the means ± se of five replicates (one replicate per plant).
Diversity of Bacterial Communities Associated with Rice Root-Zone Compartments
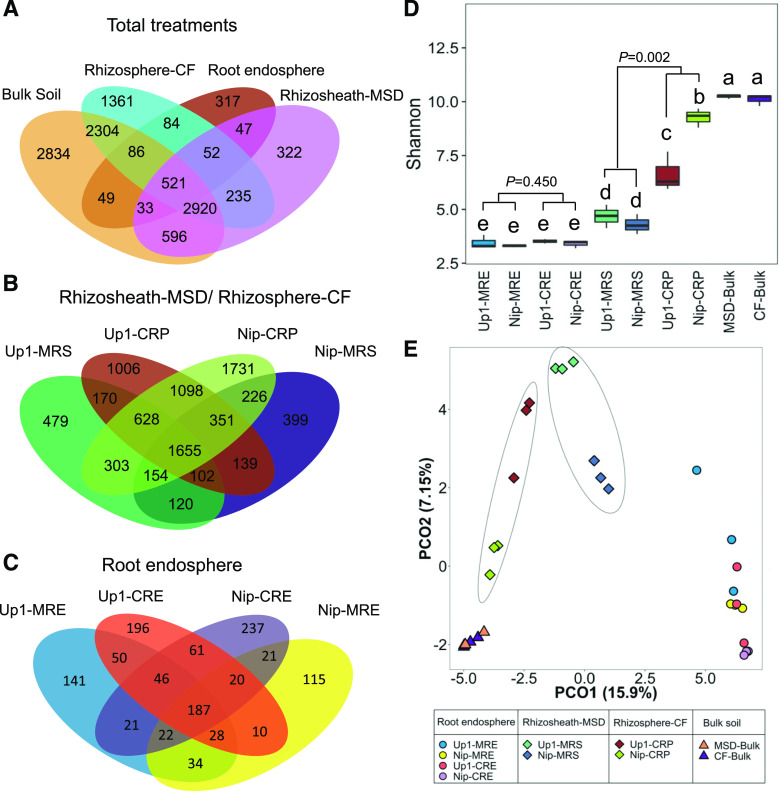
A total of 11,761 prokaryotic operational taxonomic units (OTUs) with 97% sequence similarity were identified (470 ± 111, 3,579 ± 298, 5,432 ± 248, and 6,918 ± 402 OTUs corresponding to 130,399 ± 11,257, 125,693 ± 13,678, 133,789 ± 10,382, and 128,553 ± 8,192 paired-end reads in the root endosphere, rhizosheath [under the MSD condition], rhizosphere [under the CF condition], and bulk soil, respectively). Among the rhizosheath OTUs (4,726), 76% were only from bulk soil, 2% were only from the root, 12% were shared between bulk soil and root, and 79% were shared between the rhizosheath and the rhizosphere (Fig. 2A). The Up1 rhizosheath shared 56% of its 3,611 OTUs with the Nip rhizosheath (Fig. 2B). MSD not only affected the OTUs in the rhizosheath, but also impacted the OTUs in the root endosphere compared with CF (Fig. 2, B and C).
Figure 2.
Comparison of bacterial OTUs in samples from different root-system compartments (endosphere root tissues and rhizosheath or rhizosphere) of two rice varieties (Up1 and Nip) under MSD (with rhizosheath) and CF (without rhizosheath). A to C, Venn diagrams of shared OTUs across sample types or groups. D, Estimated Shannon index across sample groups, shown as means ± sd. Lowercase letters indicate a significant difference at P < 0.05. E, PCoA for samples. Both Up1 and Nip rice varieties could form rhizosheaths under MSD, but not under CF. There were three biological replicates for each treatment and each replicate used two plants. MRE, Root endosphere under MSD; CRE, root endosphere under CF; MRS, rhizosheath under MSD; CRP, rhizosphere under CF; MSD-Bulk, bulk soil under MSD; CF-Bulk, bulk soil under CF.
The level of alpha diversity, measured by the Shannon diversity index, decreased in value from bulk soil to rhizosphere to rhizosheath to root endosphere (Fig. 2D). Differences in Shannon diversity between the MSD and CF conditions were not significant among communities in the root endosphere (P = 0.450, Student’s t test). However, differences in Shannon diversity between the rhizosheath under MSD and the rhizosphere under CF were significant (P = 0.002, Student’s t test). Principal coordinate analysis (PCoA; Fig. 2E) revealed a strong clustering of bacteria at the OTU level (97% identity) according to the different root zone compartments (root, rhizosheath, rhizosphere, and bulk soil). A global segregation between the bacterial communities associated with the rhizosheath (under MSD) and the rhizosphere (under CF) was observed using permanent multivariate ANOVA (PERMANOVA; F = 5.02, P = 0.003). Furthermore, a global segregation was observed between the bacterial communities associated with the rhizo-soil (including the rhizosheath under MSD and the rhizosphere under CF) and the root endosphere or bulk soil (F = 20.86, P = 0.001 compared with the root endosphere, and F = 7.33, P = 0.001 compared with bulk soil, based on PERMANOVA).
The Enterobacteriaceae Family Was Enriched in Both the Rhizosheath and the Root Endosphere under MSD
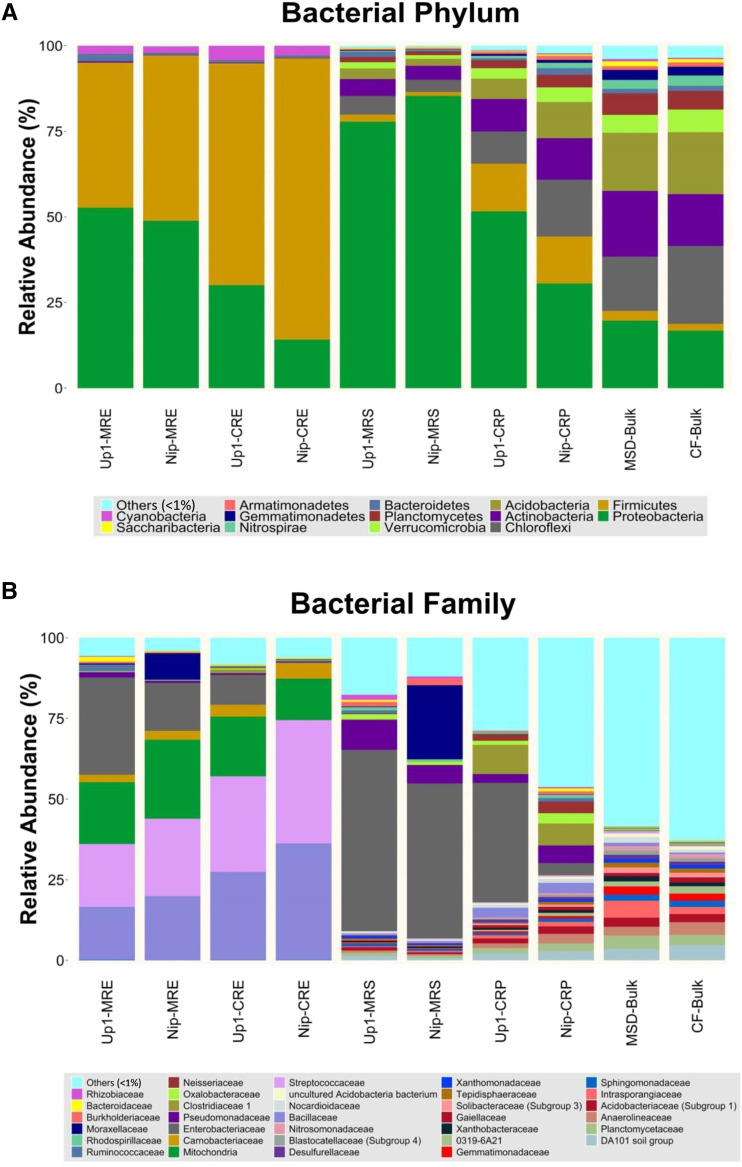
The bacterial phylum Proteobacteria in the rhizosheath under the MSD condition significantly increased in average relative abundance (82% [Gammaproteobacteria, 72%]) compared with that in the rhizosphere under the CF condition (41% [Gammaproteobacteria, 25%]; P = 0.001, Student’s t test; Fig. 3A; Supplemental Fig. S1A; Supplemental Table S1). By contrast, the bacterial phylum Firmicutes in the rhizosheath under the MSD condition significantly decreased in average relative abundance (1.6% [Clostridia, 1%; Bacilli, 0.6%]) compared with that in the rhizosphere under the CF condition (14% [Clostridia, 9.6%; Bacilli, 4%]; P = 0.006, Student’s t test; Fig. 3A; Supplemental Fig. S1A; Supplemental Table S1). Although the relative abundance of the phyla Proteobacteria and Firmicutes in the rhizosphere appeared similar between Up1 and Nip (P = 0.06 and P = 0.973, respectively, Student’s t test; Fig. 3A, Supplemental Fig. S1A; Supplemental Table S1), the relative abundances of the classes Gammaproteobacteria and Betaproteobacteria in the rhizosphere were significantly different (P = 0.025 and P = 0.038, respectively, by Student’s t test; Fig. 3A, Supplemental Fig. S1A; Supplemental Table S1). However, in the rhizosheath, relative abundances of these phyla and classes were similar in Up1 and Nip (P > 0.05, Student’s t test; Fig. 3A; Supplemental Fig. S1A; Supplemental Table S1). We found that the rhizosheath can enrich bacteria belonging to the Gammaproteobacteria class, and the most abundant family representation was from members of the Enterobacteriaceae (56% in Up1 and 48% in Nip; P = 0.146, Student’s t test; Fig. 3B) in the Enterobacteriales order. Moreover, the abundance of Enterobacteriaceae in the rhizosphere of Up1 (37%) was much higher than that in the rhizosphere of Nip (4%; P = 0.012, Student’s t test; Fig. 3B). These results show that the Enterobacteriaceae family was enriched in the rhizosheath of both rice varieties.
Figure 3.
The Enterobacteriaceae family of the Proteobacteria phylum was enriched in the rhizosheath of two rice varieties (Up1 and Nip) under MSD conditions (with rhizosheath). A and B, Phylum-level (A) and family-level (B) relative abundance showed >1% relative abundance of all reads in samples from different root-system compartments (endosphere root tissues and rhizosheath or rhizosphere) of two rice varieties (Up1 and Nip) along with bulk soil under MSD (with rhizosheath) and CF (without rhizosheath) conditions. Both Up1 and Nip rice varieties could form rhizosheaths under MSD, but not under CF. MRE, Root endosphere under MSD; CRE, root endosphere under CF; MRS, rhizosheath under MSD; CRP, rhizosphere under CF; MSD-Bulk, bulk soil under MSD; CF-Bulk, bulk soil under CF. Each treatment had three biological replicates and each replicate had two plants.
Proteobacteria and Firmicutes were the dominant phyla in the root endosphere, comprising on average >96% of each sample. The relative abundance of Firmicutes in the root endosphere was 73% (Bacilli, 72%; Clostridia, 2%) under the CF condition, while it was decreased to 45% (Bacilli, 44%; Clostridia, 2%) under the MSD condition (P = 0.006, Student’s t test; Fig. 3A, Supplemental Fig. S1A; Supplemental Table S1). By contrast, the relative abundance of Proteobacteria in the root endosphere was elevated from 22% (Gammaproteobacteria, 5%; Alphaproteobacteria, 16%) under the CF condition to 51% (Gammaproteobacteria, 28%; Alphaproteobacteria, 22%) under the MSD condition (P = 0.002, Student’s t test; Fig. 3A, Supplemental Fig. S1; Supplemental Table S1). Further, the abundance of Enterobacteriaceae in the root endosphere of Up1 (30% under MSD and 9% under CF) was much higher than that in Nip (15% under MSD and 0.3% under CF; Fig. 3B). The relative abundance of members of the Gammaproteobacteria and the Enterobacteriaceae family (order Enterobacteriales) was similar in the rhizosheath/rhizosphere and the root endosphere (Supplemental Fig. S2).
In the bulk soil, six bacterial phyla, Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, Verrucomicrobia, and Planctomycetes, comprised >80% of each sample under both CF and MSD conditions. The relative abundance of these phyla was not significantly different under the CF and MSD conditions, except for that of Verrucomicrobia (5% under MSD, 6% under CF; P = 0.037, Student’s t test; Fig. 3A; Supplemental Table S1).
For prediction of bacterial functions, the samples were first clustered according to sample type, and there was a clear distinction among root endosphere, rhizosheath/rhizosphere, and bulk soil (Supplemental Fig. S3A). In rhizosheath and rhizosphere samples, replicate samples clustered closely together, and bacterial functional clusters varied between the two rice varieties and the different irrigation conditions (Supplemental Fig. S3B). However, bacterial functions in the root endosphere could not be clustered based on the variety or irrigation conditions (Supplemental Fig. S3C). Regarding the functional profile, secretion system was abundant in the rhizosheath of both rice varieties under the MSD condition (Supplemental Fig. S3, D and E).
Transcriptomic Responses of the Root with or without a Rhizosheath
The analysis found 2,164 and 1,684 significantly upregulated genes and 1,361 and 1,478 significantly downregulated genes in the rhizosheath-root continuum under the MSD condition compared with roots without a rhizosheath under the CF condition, respectively (Supplemental Datasets S1 and S2). Moreover, 13 genes were subjected to reverse-transcription quantitative PCR (RT-qPCR) to validate the RNA-sequencing (RNA-seq) data (Supplemental Table S2). These genes showed similar expression trends in RT-qPCR and RNA-seq analyses (Supplemental Table S3).
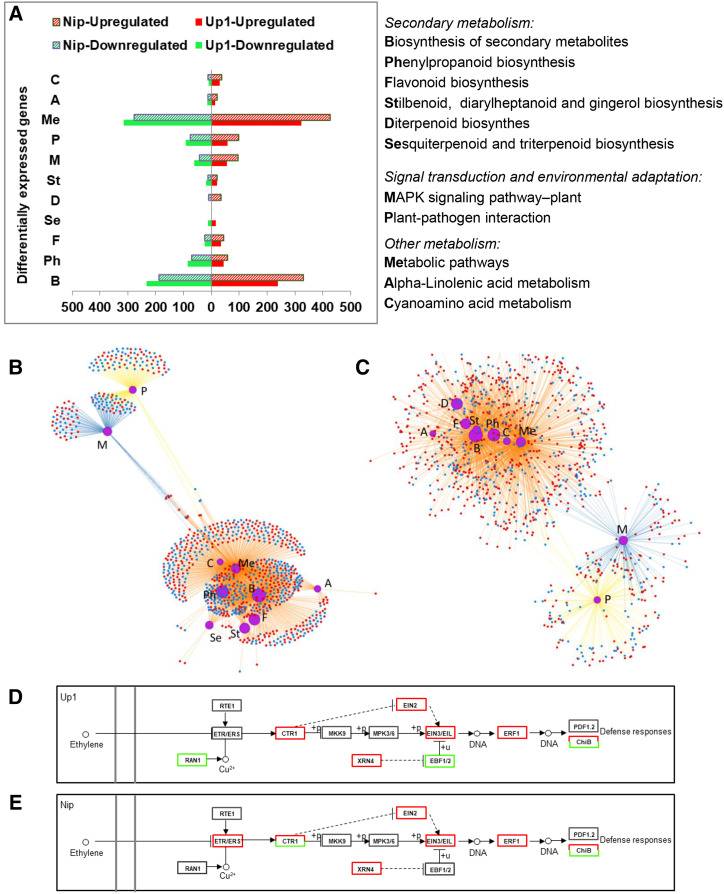
According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, the biosynthesis pathway of secondary metabolites (240/232 and 330/187 genes were upregulated/downregulated in Up1 and Nip, respectively) was the top enriched pathway in both Up1 and Nip under the MSD condition (Fig. 4). More upregulated genes involved in the biosynthesis of secondary metabolites were found in Nip than in Up1. Meanwhile, the biosynthesis subsystems of secondary metabolite pathways, such as phenylpropanoid biosynthesis (45/84 and 58/71 genes upregulated/downregulated in Up1 and Nip, respectively), flavonoid biosynthesis (35/24 and 44/24 genes), stilbenoid, diarylheptanoid and gingerol biosynthesis (21/19 and 22/12 genes), and terpenoid (diterpenoid, triterpenoid, sesquiterpenoid) biosynthesis, were also enriched under the MSD condition in the rice root (Fig. 4). In addition, among the top 10 enriched pathways, sesquiterpenoid and triterpenoid biosynthesis was enriched only in Up1 (15/12 genes upregulated/downregulated), while diterpenoid biosynthesis was enriched only in Nip (34/9 genes were upregulated/ downregulated).
Figure 4.
Enrichment of functional categories in root transcriptomes of Up1 and Nip under MSD (with rhizosheath) and CF (without rhizosheath) conditions. A, Number of DEGs in the top 10 KEGG pathways of Up1 and Nip. B and C, Network view of the top 10 KEGG pathways of Up1 (B) and Nip (C). Uppercase letters in A to C represent the functional categories as described in A. In B and C, red dots represent upregulated genes and blue dots represent downregulated genes. Purple circles represent pathways of enrichment. The larger the size of the purple circle, the higher the enrichment. Orange lines represent the pathway of metabolism, blue lines the pathway of environmental information processing, and yellow lines the pathway of organismal systems. D and E, Profiles of the ET pathway (belonging to the MAPK-regulated pathway-plant category) in Up1 (D) and Nip (E). The double gray lines on the left side represent the cell membrane. Boxes represent the gene products, with red boxes indicating all upregulated genes, green boxes indicating all downregulated genes, and red and green boxes indicating that some genes were upregulated and others were downregulated. Solid lines represent direct effects and dashed lines represent indirect effects. Each treatment had three biological replicates and each replicate used two plants. RAN1, P-type Cu+ transporter; RTE1, transmembrane protein 222; ETR/ERS, ET receptor; CTR1, Ser/Thr-protein kinase CTR1; MKK9, MEK 9; MPK3/6, MAPK 3; XRN4, 5′-3′ exoribonuclease4; EBF1/2, EIN3-binding F-box protein; PDF1.2, defensin-like protein 16; CHIB, basic endochitinase B.
Pathways related to plant defense, including plant-pathogen interaction (58/91 and 99/75 genes upregulated/downregulated in Up1 and Nip, respectively) and mitogen-activated protein kinase (MAPK) signaling pathway-plant interaction (55/62 and 96/42 genes) were ranked in the top 10 enriched pathways (Fig. 4). For example, 81 and 98 genes related to pathogen- and microbe-associated molecular pattern-triggered immunity (PTI) were regulated in Up1 (30/51 upregulated/downregulated genes) and Nip (67/31 upregulated/downregulated genes), respectively (Supplemental Tables S4 and S5), and 98 genes and 115 genes related to effector-triggered immunity (ETI) were regulated in Up1 and Nip, respectively (35/63 and 52/63 upregulated/downregulated genes, respectively; Supplemental Tables S4 and S5).
Moreover, genes involved in the jasmonic acid (JA) and ET pathways were commonly upregulated in both Up1 (24 upregulated and 7 downregulated genes) and Nip (30 upregulated and 5 downregulated genes; Supplemental Tables S4 and S5). In this study, many genes, including ETHYLENE INSENSITIVE2 (EIN2), EIN3 and its homolog EIN3-LIKE1 (EIL1), and genes encoding ETHYLENE RESPONSE FACTORS (ERFs), were upregulated under MSD conditions compared to CF conditions in both Up1 (from 2.1- to 8.6-fold) and Nip (from 2- to 10.6-fold; Fig. 4, D and E; Supplemental Tables S4 and S5). Five of these genes upregulated in both Up1 and Nip were validated by RT-qPCR (Supplemental Table S3). These results show that when accumulated in the rhizosheath, rice roots initiated a complex defense response including PTI, ETI, and hormone-mediated defense.
Isolated and Functionally Assessed PGP Rhizobacteria Belong to the Enterobacteriaceae Family
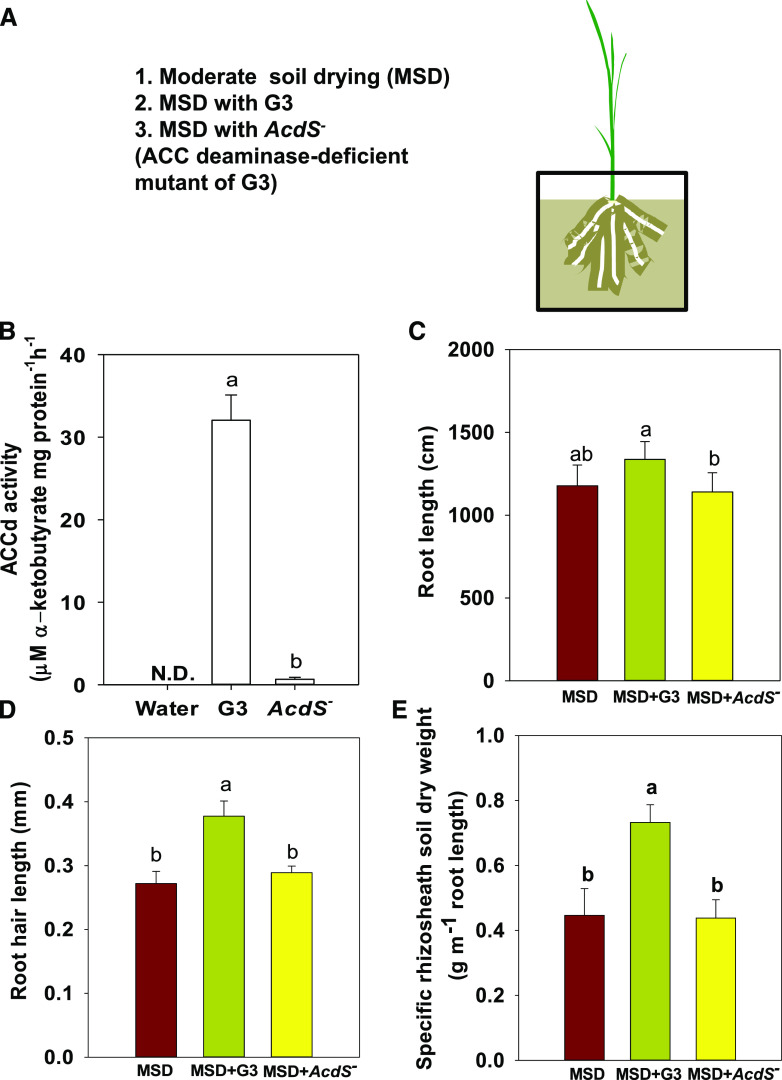
Twenty-one cultivable bacteria were isolated from the rhizosheaths of the two rice varieties and were identified using the 16S ribosomal RNA (rRNA) gene. The Enterobacteriaceae dominated the entire collection (19 of the total isolates, with 9 and 10 isolates in the rhizosheaths of Up1 and Nip, respectively; Supplemental Table S6). Among the isolates, ACCd production ranged from 0.88 ± 0.07 to 31.79 ± 4.64 μm·α-ketobutyrate mg protein−1 h−1, with production exceeding 10 μm·α-ketobutyrate mg protein−1 h−1 in four strains (Enterobacter aerogenes [G3], 31.79 ± 4.64 μm·α-ketobutyrate mg protein−1 h−1; Enterobacter sp. [N11], 11.42 ± 2.07 μm·α-ketobutyrate mg protein−1 h−1; Enterobacteriaceae bacterium [G6], 10.98 ± 0.06 μm·α-ketobutyrate mg protein−1 h−1; and Klebsiella sp. [G5], 10.84 ± 0.2 μm·α-ketobutyrate mg protein−1 h−1). Expression of ET pathway genes in the root, including EIL5 (LOC107278750), ERF15 (LOC9266984), and ERF094 (LOC107278456), was increased following exposure to E. aerogenes (G3) compared with No-inoculation under MSD (Supplemental Fig.. S4). These results are consistent with sequencing results showing that bacteria in the rhizosheath can induce the ET pathway under MSD (Fig. 4, D and E).
E. aerogenes (G3), isolated from the rhizosheath of Up1, showed the highest ACCd production out of 21 strains. The ACCd-deficient mutant (AcdS−) of E. aerogenes (G3), obtained by gene replacement, showed low ACCd production at 0.66 ± 0.23 μm·α-ketobutyrate mg protein−1 h−1 (Fig. 5B, Supplemental Figs. S5 and S6). G3 and AcdS− were used to evaluate the role of ET in rice rhizosheath formation. Compared to MSD, MSD with G3 maintained root length and promoted root hair length and rhizosheath formation in rice under MSD, while MSD with AcdS− yielded no significant changes in root length, root hair length, and rhizosheath formation (Fig. 5). These results show that ACCd-contained G3, a bacterium that can potently degrade the principal ET precursor, facilitated rhizosheath formation by mediating the ET response.
Figure 5.
E. aerogenes (G3) isolated from rice rhizosheath increased rhizosheath formation in Nip under MSD. A, Experimental design. B, ACCd activity of G3 and the ACCd-deficient mutant (AcdS−) of G3. C to E, Dry weight of root length (C), root hair length (D), and dry weight of specific rhizosheath soil (E) in rice under MSD, MSD with G3, and MSD with AcdS−. Data are the means ± se of five replicates (one replicate per plant). Duncan’s test was used for post hoc multiple comparisons. Lowercase letters indicate significant difference at P < 0.05.
DISCUSSION
Rice cultivation is water intensive and, in most cultivation systems, involves CF. Recently, alternate wetting and drying (AWD) has been employed increasingly in an effort to effectuate water-saving irrigation of rice. During AWD, irrigation is interrupted and water is allowed to subside until the soil reaches a certain moisture level, after which the field is reflooded (Wang et al., 2016). Our previous research has shown that moderate AWD, similar to MSD, not only decreases water usage, but also maintains or even increases grain yield of rice (Song et al., 2018). In this study, we found that both Up1 and Nip can form rhizosheaths under MSD irrigation, but not under CF irrigation (Fig. 1). This is very different from dryland-farmed crops, such as maize, wheat, or barley, which are planted in dryer soil and can form rhizosheaths continuously. By exploiting this unique characteristic of rice rhizosheath formation during MSD, we aimed to address the associations of root and bacteria in the rice rhizosheath and identify the factors governing rice rhizosheath formation under soil drying.
Rhizosheath-Associated Microbes Induce ET-Related Pathways in Rice
The relative abundance of members of the Enterobacteriaceae was increased in the rhizosheath under MSD compared with the rhizosphere under CF, in turn also reflecting the relative abundance of Enterobacteriaceae in the root endosphere (Fig. 3; Supplemental Figs. S1 and S2). The Enterobacteriaceae family has been shown to preferentially inhabit the rhizosphere of many plants and is known to harbor many PGP rhizobacteria (PGPRs) that could promote root development and/or protection against pathogens (Vessey, 2003; Haichar et al., 2008; Mendes et al., 2014; Niu et al., 2017; Marasco et al., 2018b). The enrichment by Enterobacteriaceae in the rhizosheath and the rhizosheath-root continuum is supportive of the notion of rhizosheath-resident bacteria acting as PGPRs and being beneficial for plant growth under the MSD condition.
The composition of the rhizosphere community is often driven by root exudation, which can be changed by plants in response to biotic and abiotic stresses (Stringlis et al., 2018). Here, transcriptional results indicate that the biosynthesis of secondary metabolites, including phenylpropanoid, flavonoid, terpenoid, stilbenoid, diarylheptanoid, and gingerol biosynthesis, was significantly affected in the rhizosheath-root continuum under the MSD condition compared with the CF condition in both Up1 and Nip (Fig. 4), suggesting a significant change in root exudation and in turn impacting the composition of the rhizosheath microbial community. The enhancement of terpenoid-synthesis pathways was significantly different between Up1 and Nip, with sesquiterpenoid and triterpenoid biosynthesis enriched only in Up1 and diterpenoid biosynthesis enriched only in Nip (Fig. 4). Studies have shown that sesquiterpenes, such as (E)‐β‐caryophyllene, can aid in the defense against bacterial pathogens (Huang et al., 2012). Moreover, phenylpropanoids are known to possess strong external antibacterial properties and can function effectively in attracting soil-borne microorganisms (Baetz and Martinoia, 2014). Previous research has shown that coumarins, which are synthesized via the phenylpropanoid pathway, can shape root-associated microbial communities (Stringlis et al., 2018). These findings suggest that the enrichment in PGPRs (Enterobacteriaceae) in the rhizosheath relates to a change in root exudates under the MSD condition and that the differential expression of secondary metabolite biosynthetic pathways between Up1 and Nip may also influence the bacterial communities in the rhizosheath.
Numerous studies show that PGPRs are initially recognized by the plant as potential pathogens, which induces the immune system response by the plant (De Coninck et al., 2015). The immune system response often triggers induced systemic resistance against pathogens and herbivores (Pieterse et al., 2014; Shi et al., 2015; Venturi and Keel, 2016). Previous studies have suggested that induced systemic resistance acts via the JA and ET signaling pathways, which are commonly induced by beneficial soil microbes, necrotrophic pathogens, and herbivorous insects (Venturi and Keel, 2016). Meanwhile, we also found that soil bacteria belonging to the Enterobacteriaceae family, many of whose members have been shown to act as PGPRs, was enriched in the rhizosheath under MSD (Fig. 3).
In our study, compared to the JA pathway, many genes involved in the ET pathway were regulated in the rhizosheath-root continuum in the two rice varieties under MSD (Fig. 4, D and E; Supplemental Tables S4 and S5). Rice has five ET receptors (ETHYLENE RESPONSE SENSOR1 [ERS1], ERS2, ETHYLENE RESPONSE2 [ETR2], ETR3, and ETR4), and they are negative regulators of ET responses (Yang et al., 2015). The ET response pathway also includes the downstream CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), EIN2, EIN3 and its homolog EIL1, and ERFs. Activated CTR1 and the receptors cooperatively inhibit EIN2 through physical interaction and protein phosphorylation. Upon ET binding to its receptors, CTR1 kinase activity is deactivated. EIN2 is a positive regulator of the rice ET response, and the active form of EIN2 protects the transcription factors of the EIN3/EIL family stabilization by repressing the function of two F-box proteins, EIN3-BINDING F-BOX1 (EBF1) and EBF2 (De Vleesschauwer et al., 2010; Mao et al., 2016). The EIN3/EIL proteins subsequently bind to the promoters of the ERF genes and then activate their transcription (Mao et al., 2016). In this study, genes encoding EIN2, EIN3/EIL (EIL5), and ERFs (ERF15 and ERF094) were upregulated under MSD in both Up1 and Nip (Fig. 4, D and E; Supplemental Tables S3–S5). When ET binds to receptors, ET receptors deactivate CTR1, which leads to positive regulation of EIN2. Activated EIN2 protects EIN3, and its functionally redundant homolog EIL1, from proteasomal degradation. EIN3 acts as a master transcription factor that activates a transcriptional cascade by binding as homodimers to the promoters of ERF genes (Ju and Chang, 2015). Thus, ET-related pathways can be induced by beneficial bacteria enriched in the rice rhizosheath under MSD.
In Arabidopsis, EIN3 regulates the vast majority of ET-directed gene expression, including events leading to ET-inhibited root growth and ET-promoted root hair growth (Klay et al., 2018). In rice, there are six family members of EIN3-like homologs (EIL1–EIL6). However, the functions of most EIL proteins in rice, including those of EIL5, are still unclear. In this study, expression of the gene encoding EIL5 was upregulated in the rhizosheath-root continuum under MSD in both Up1 and Nip (Supplemental Tables S3–S5), which implies that EIL5 is involved in ET-responsive pathways that regulate root growth in rice. Our results showed that some positively regulated genes of the ET pathway were upregulated in the rhizosheath-root continuum under MSD in both Up1 and Nip, Thus, these results suggest that rhizosheath-associated microbes induce ET-related pathways in rice plants under soil drying.
E. aerogenes, a Bacterium That Can Degrade the ET Precursor ACC, Promotes Rice Rhizosheath Formation under Soil Drying
Previously research has shown that bacteria in the rhizosphere can contribute to plant growth (Huang et al., 2019). However, how beneficial bacteria influence rhizosheath formation is still unclear. In this study, we found that bacteria belonging to the Enterobacteriaceae family were highly enriched in the rhizosheath under MSD (Fig. 3). Further, 21 bacteria were randomly isolated from the rhizosheath of the two rice varieties examined, and most of these isolates had high ACCd activity (Supplemental Table S6). The enzyme ACCd cleaves ACC, the immediate precursor to ET in plants, thereby lowering the ET concentration in the plant (Glick et al., 2007). Bacteria that produce the enzyme ACCd play an active role in modulating ET levels in plants when they are present either on the root surface (rhizosphere or rhizosheath) or within plant tissues (endosphere; Gamalero and Glick, 2015). Previous studies have shown that many bacteria facilitate plant growth especially in the presence of environmental stresses and that many of these are ACCd-containing bacteria (Marasco et al., 2012).
E. aerogenes (G3), isolated from the rhizosheath of Up1, had the highest ACCd activity of 21 isolates (Supplemental Table S6). Hence, Nip was exposed to E. aerogenes (G3) from the Up1 rhizosheath, and the consequences to root growth and rhizosheath formation under MSD were evaluated. Comparison of the results with those from exposure of Nip to the mutant AcdS− of E. aerogenes (G3) showed that E. aerogenes (G3) can promote root hair length and maintain root length under MSD, which further promotes rhizosheath formation of rice under MSD. However, when the ACCd gene was deleted in G3, the mutant AcdS− could not increase rhizosheath formation (Fig. 5). Thus, E. aerogenes, a bacterium that can greatly degrade the ET precursor ACC, increases rice rhizosheath formation under MSD.
In conclusion, ET-related pathways were induced in roots of both drought-tolerant and drought-sensitive rice under MSD. E. aerogenes increases rice rhizosheath formation under soil drying. These results suggest that root-bacteria associations are critical for rice rhizosheath formation under moderate soil drying. This enhanced understanding of rice root performance under water shortage is especially important with a view to designing new strategies that help maintain and promote rice root growth under water-saving rice production regimes, which will become progressively critical as global water shortages increase under a changing climate.
MATERIALS AND METHODS
Plant Materials and Treatments
Two rice (Oryza sativa) varieties, ssp. japonica ‘Gaoshan 1’ (Up1, drought-tolerant) and ssp. japonica ‘Nipponbare’ (Nip, drought-sensitive) were used in this study (Shi et al., 2015). Seeds were surface-sterilized using 1.5% (v/v) sodium hypochlorite for 10 min and rinsed with double-distilled water five times. They were then placed on moistened filter paper in the dark at 30°C for 3 d. Uniform seedlings were transplanted into pots (12-cm diameter, 14-cm height) with 1.8 kg dry soil. Soil was collected from a paddy rice field in the town of Huayang, Jiangxi Province, China (115°09′32′′E, 28°32′29′′N), from a depth of 0–20 cm, air-dried, mixed, and passed through a 4-mm sieve to remove coarse material and vegetative matter. Soil contained 20.5 g kg−1 soil organic carbon, 1.75 g kg−1 total nitrogen, 0.65 g kg−1 total P, 27.7 g kg−1 total potassium, 42.6 mg kg−1 Olsen-P, and 92.0 mg kg−1 exchangeable potassium.
After sowing a single seed in each pot, a 3- to 5-cm water layer above the soil surface was maintained for 3 d to allow for full seed imbibitions, after which pots were drained and no water was supplied for 4 d. Thereafter, half of the pots were kept in CF conditions (a water layer of 3–5 cm above the soil surface was maintained in pots), while the other half were subjected to a MSD regime. For the MSD treatment, rice was watered every 3 d to 80% (w/w) field capacity, equivalent to 38% (w/w) soil water content. Both the CF and MSD regimes were maintained for 9 d. The greenhouse environment was set to a 14 h/10 h light/dark cycle, a 26°C/22°C day/night temperature cycle, 40% (w/w) relative humidity, and a photosynthetic photon flux density of 300 μmol photons m−2 s−1.
Analysis of Root Length and Rhizosheath Weight
Under the MSD regime, roots of rice at the four-leaf stage were carefully collected and shaken after pots were disassembled, and the soil tightly adhering to roots upon excavation was further analyzed as rhizosheath soil. Roots and rhizosheath soil were washed with water, and both soil and washing water were collected in a tray and dried to determine the dry weight of the rhizosheaths. Root length was measured using winRHIZO software (Regent Instruments). Specific rhizosheath weight was determined as the total rhizosheath soil dry weight per plant divided by total root length. Root hair length was measured as described previously (George et al., 2014). Images were taken using a SMZ18 stereomicroscope and a DS-U3 camera (Nikon). Ten fully elongated root hairs were measured in each image using Image J software (National Institutes of Health). One-way ANOVA or Student’s t test was used to determine the level of significance (P < 0.05) in SPSS (version 17.0; IBM).
Root and Soil Sampling for Microbial Community Structure Analysis
Up1 and Nip were chosen for analysis of the microbial communities in the root endosphere and rhizosheath under MSD, the rhizosphere under CF, and bulk soil under the MSD and CF conditions (Fig. 1). Testing occurred using a Vernier caliper; the 0.5-mm radius of root bounded with soil was clearly established as rhizosheath under MSD, and samples of rhizosphere under the CF condition were therefore also collected from the soil around the root within a 0.5-mm radius. After 9 d of irrigation treatment, three biological replicates were collected for each treatment condition. For the MSD treatment, root plus rhizosheath soil were collected. For the CF treatment, the root was taken from the pot after water was removed, and the soil around the root within a 0.5-mm radius was kept, while other soil attached to the root was removed with sterile tweezers. Each root with the soil from the MSD or CF condition was washed with 30 mL sterilized PBS-S buffer (130 mm NaCl, 7 mm Na2HPO4, 3 mm NaH2PO4 [pH 7.0], and 0.02% [v/v] Silwet l-77) in 50-mL Falcon tubes (Bulgarelli et al., 2012). The washing buffer was subjected to centrifugation (1,500g for 20 min) and the resulting pellet was defined as rhizosheath soil or rhizosphere soil. Soil from control pots (no plants in pots) was considered as bulk soil. Soil samples were frozen in liquid nitrogen and stored at −80°C. After the roots were removed from the PBS-S buffer, they were surface-sterilized with 1.5% (v/v) sodium hypochlorite for 15 min while mixing in a table-top shaker, then washed with sterilized double-distilled water four times. The efficacy of the sterilization was verified by plating the final wash water volume onto Luria-Bertani medium (Marasco et al., 2012). Following surface sterilization, root tissues were placed in 50-mL sterile tubes and stored at −80°C.
DNA Extraction
The root tissues were preground with a sterile mortar and pestle in liquid nitrogen prior to DNA extraction. Total DNA extraction of root tissues and soil samples was conducted using the Mag-Bind Soil DNA Kit (Omega Bio-Tek). DNA quality and quantity were measured by 1% (w/v) agarose gel electrophoresis and NanoDrop ONE spectrophotometry (Thermo Scientific). All DNA extracts were stored at −80°C for further analysis.
Illumina Sequencing of 16S rRNA Gene Amplicons and Data Processing
The V4 region of 16S rRNA genes in all DNA samples was amplified with the following primer set: F515 (GTGCCAGCMGCCGCGGTAA) and R806 (GGACTACHVGGGTWTCTAAT) with a six-base barcode and attached Illumina flow cell adapter sequences (Carini et al., 2016). Sample-specific paired-end six-base barcodes (Supplemental Table S8) were incorporated into the TrueSeq adaptor for multiplex sequencing. Fifty-microliter PCRs of single DNA extracts were performed as follows: 25 μL Phusion High-Fidelity PCR Master Mix with HF buffer (New England Biolabs), 3 μL of each primer (10 μm), 10 μL template DNA, 6 μL double-distilled water and 3 μL dimethyl sulfoxide. Thermocycling conditions were as follows: an initial denaturation of 98°C for 30 s; 25 cycles of 98°C for 15 s, 58°C for 15 s, 72°C for 15 s; and a final elongation step of 72°C for 60 s. PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter) to remove unspecific products and quantified using a PicoGreen dsDNA assay kit (Invitrogen). Samples were then pooled in equimolar concentrations and paired-end 2 × 150-bp sequencing was performed using an Illumina HiSeq4000 platform.
Quantitative Insights into Microbial Ecology (QIIME, v1.9.1) was used for sequencing-data analysis (Caporaso et al., 2010). Paired-end reads were merged using Vsearch (v2.4.4, –fastq_mergepairs–fastq_minovlen5). According to barcode sequences, the sequencing data of each sample were demultiplexing from the original data. The low-quality reads (length of <150 bp, average Phred scores of <20, containing ambiguous bases or mononucleotide repeats of >8 bp) were filtered (Gill et al., 2006; Chen and Jiang, 2014). OTUs were clustered using Vsearch (v2.4.4), including dereplication (–derep_fulllength), cluster (–cluster_fast,–id0.97), detection of chimeras (–uchime_ref; Rognes et al., 2016). Sequences were clustered into OTUs at 97% sequence similarity. OTU classification was conducted by VSEARCH, searching the representative sequence set against the SILVA132 database (Quast et al., 2013). An OTU table was further generated to record the abundance of each OTU in each sample and the taxonomy of these OTUs. OTUs containing <0.001% of total reads across all samples were discarded. A total of 3,908,135 reads (maximum 144,131, minimum 100,106) were obtained from the 30 samples. To minimize the difference in sequencing depth across samples, an averaged, rounded rarefied OTU table was resampled to identify OTU subsets with <90% of the minimum sequencing depth for further analysis.
OTU-level α-diversity (the Shannon index) was applied for analyzing the complexity of species diversity among samples, and PCoA (Bray) and permutational multivariate ANOVA (PERMANOVA) were applied to display the differences of OTUs in different samples. Venn diagrams were developed to display the number of common or unique OTUs in multi- groups using R package Venn Diagram setting. Taxonomic composition was summarized in profiling tables and histograms as Phylum, Order, Class, Family, and Genus levels. To predict bacterial functional profiles, the Statistical Analysis of Metagenomic Profiles (STAMP) software package was used as previously described (Mendes et al., 2014; Parks et al., 2014). A table showing the frequency of hits of taxa and the functional subsystem (SEED database) for each amplicon base was generated from MetaGenomics Rapid Annotation using Subsystem Technology and used as input. SEED subsystem level 3 was used to predict bacterial functions. P-values in STAMP were calculated using the two-sided Fischer’s exact test, while confidence intervals were calculated using the Newcombe-Wilson method and correction was made using Welch’s t test (P < 0.05).
Root Sampling for RNA Isolation and Sequencing
For RNA-seq and RT-qPCR analysis, roots with rhizosheath (under MSD) and without rhizosheath (under CF) of Up1 and Nip rice were rapidly harvested, cleaned, immediately frozen in liquid nitrogen, and stored at −80°C for subsequent RNA extraction and sequencing. Each treatment had three replicates. Total RNA of roots was extracted according to Liu et al. (2016). RNA was qualified and quantified using a Nano Drop (Thermo Fisher Scientific) and Agilent 2100 bioanalyzer (Thermo Fisher Scientific). Sequencing libraries were generated according to the methods of Zhang et al., (2016).
RNA-Seq Data Analysis and Validation
Raw reads were preprocessed using SOAPnuk (v1.5.2). Adapter sequences, each read containing ≥10% poly-N, and low-quality reads (Q-score ≤15 for ≥50% of nucleotides per read) were filtered. The resulting clean data consisting of only high-quality reads were mapped to the reference genome (Oryza_sativa_Japonica_Group, IRGSP-1.0) using HISAT (v2.0.4). RSEM (version 1.2.12) was used to calculate fragments per kilobase of transcript per million mapped fragment. Differentially expressed genes (DEGs) were identified using DEGseq (fold change ≥2; adjusted P-value ≤0.001). Pathway enrichment was analyzed using the KEGG database (http://www.genome.jp/kegg) . To validate the RNA-seq results, 13 genes were chosen for RT-qPCR, of which five (LOC107278000, LOC107278750, LOC107278456, LOC9266984, and LOC4343006) were related to the ET pathway and the other eight were selected randomly from among the KEGG classifications of interest.
Isolation and in Vitro Characterization of Rhizosheath Bacteria
To investigate the role of ET in rice rhizosheath formation, 1 g of rhizosheath soil was used as inoculum to isolate the ACCd-containing bacteria using an ACCd enrichment method described previously (Penrose and Glick, 2003). A total of 21 strains were purified and identified by the 16S rRNA gene using the universal primers 27F and 1492R. ACCd activity of bacteria was determined as previously described (Ali et al., 2014). The knockout AcdS− mutant of strain G3 was constructed using the suicide T-vector pLP12 (http://www.biovector.net/product/1717948.html) as described previously (Li et al., 2000). Briefly, fragment A was generated by PCR using primer pair ACCD-MF1 and ACCD-MR1 with the bacteria DNA as template. Fragment B was generated using the same PCR system as for fragment A, but with primers ACCD-MF2 and ACCD-MR2. Then, the recovered fragments A and B were mixed together as a template for PCR to generate target fragment AB (Supplemental Fig. S6) using ACCD-MF1 and ACCD-MR2 as primers. Finally, the target fragment AB was inserted into plasmid plP12 between restriction sites Xba1 and Bhe1. The final vector was identified by Sanger sequencing. The plasmids, bacterial strains, and primers used in this study are listed in Supplemental Table S8.
Evaluation of Bacterial Effects on Root Growth of Rice under MSD
After seedlings (Nip) were established in pots as described above, half of the seedlings were inoculated with a bacterial suspension at a concentration of 108 cells g−1 of soil, while the other half, the uninoculated seedlings, were watered with tap water (Rolli et al., 2015). After 7 d of inoculation, roots of some seedlings were collected for RNA isolation and RT-qPCR, while others were subjected to MSD treatment for 9 d, as described above. Then, root length, root hair length, and rhizosheath weight were determined as described above.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SAMN14239335, SAMN14239336, SAMN14239337, SAMN14239338, SAMN14239339, SAMN14239340, SAMN14239341, SAMN14239342, SAMN14239343, SAMN14239344, SAMN14239345, SAMN14239346, SAMN14253255, SAMN14253256, SAMN14253257, SAMN14253258, SAMN14253259, SAMN14253260, SAMN14253261, SAMN14253262, SAMN14253263, SAMN14253264, SAMN14253265, SAMN14253266, SAMN14253267, SAMN14253268, SAMN14253269, SAMN14253270, SAMN14253271, SAMN14253272, SAMN14253273, SAMN14253274, SAMN14253275, SAMN14253276, SAMN14253277, SAMN14253278, SAMN14253279, SAMN14253280, SAMN14253281, SAMN14253282, SAMN14253283, and SAMN14253284.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Different levels of relative abundance of bacterial communities in samples from different root-system compartments (endosphere root tissues and rhizosheath or rhizosphere) of Up1 and Nip under MSD (with rhizosheath) and CF (without rhizosheath) conditions.
Supplemental Figure S2. Relative abundances of members of the Gammaproteobacteria class and the Enterobacteriaceae family (order Enterobacteriales) were similar in the rhizosheath/rhizosphere and root endosphere.
Supplemental Figure S3. Predicting bacterial functions in samples from different root-system compartments (endosphere root tissues and rhizosheath or rhizosphere) of two rice varieties (Up1 and Nip) under MSD (with rhizosheath) and CF (without rhizosheath) conditions.
Supplemental Figure S4. Expression of ET pathway genes in Nip roots after exposure to E. aerogenes (G3) by RT-qPCR.
Supplemental Figure S5. Verification of AcdS deletion strain using PCR.
Supplemental Figure S6. Verification of the AcdS deletion strain using DNA sequencing.
Supplemental Table S1. Mean estimated bacterial taxa (>1% relative abundance) at phylum and class levels with ses.
Supplemental Table S2. Primers for RT-qPCR used in this study.
Supplemental Table S3. Fold changes in gene expression in roots bounded by rhizosheaths under MSD compared with no rhizosheath under CF in two rice varieties (Up1 and Nip), determined by RNA-seq and RT-qPCR.
Supplemental Table S4. Enrichment of DEGs related to plant immunity and resistance in Up1 rice under MSD (with rhizosheath) compared with CF (without rhizosheath) conditions.
Supplemental Table S5. Enrichment of DEGs related to plant immunity and resistance in Nip rice under MSD (with rhizosheath) compared with CF (without rhizosheath) conditions.
Supplemental Table S6. ACCd activity of rhizosheath bacteria from rice.
Supplemental Table S7. Barcodes used in Illumina sequencing of 16S rRNA gene amplicons.
Supplemental Table S8. Primers, plasmids, and bacterial strains used in construction of the AcdS− mutant.
Supplemental Dataset 1. DEGs in Up1 rice under MSD (with rhizosheath) compared with CF (without rhizosheath) conditions.
Supplemental Dataset 2. DEGs in Nip rice under MSD (with rhizosheath) compared with CF (without rhizosheath) conditions.
Acknowledgments
We thank Ian Dodd (Lancaster University, UK) for the useful discussion and Herbert J. Kronzucker (University of Melbourne, Australia) for correcting the English.
Footnotes
This work was supported by the National Key R&D Program of China (grant nos. 2017YFE0118100 and 2018YFD02003025), the National Natural Science Foundation of China (grant nos. 31761130073, 31872169, and 31901428), a Newton Advanced Fellowship (NSFC-RS: grant no. NA160430), Fujian Province Education Department Funding (JK2017015), and Fujian Agriculture and Forestry University (research grant no. KXGH17005).
References
- Ali SZ, Sandhya V, Venkateswar Rao L(2014) Isolation and characterization of drought-tolerant ACC deaminase and exopolysaccharide-producing fluorescent Pseudomonas sp. Ann Microbiol 64: 493–502 [Google Scholar]
- Baetz U, Martinoia E(2014) Root exudates: The hidden part of plant defense. Trends Plant Sci 19: 90–98 [DOI] [PubMed] [Google Scholar]
- Bergmann D, Zehfus M, Zierer L, Smith B, Gabel M(2009) Grass rhizosheaths: Associated bacterial communities and potential for nitrogen fixation. West N Am Nat 69: 105–114 [Google Scholar]
- Bouman BAM, Tuong TP(2001) Field water management to save water and increase its productivity in irrigated lowland rice. Agric Water Manag 49: 11–30 [Google Scholar]
- Britto DT, Kronzucker HJ(2018) From aquaporin to ecosystem: Plants in the water cycle. J Plant Physiol 227: 1–2 [DOI] [PubMed] [Google Scholar]
- Brown LK, George TS, Neugebauer K, White PJ(2017) The rhizosheath—a potential trait for future agricultural sustainability occurs in orders throughout the angiosperms. Plant Soil 418: 115–128 [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95 [DOI] [PubMed] [Google Scholar]
- Carini P, Marsden PJ, Leff JW, Morgan EE, Strickland MS, Fierer N(2016) Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol 2: 16242. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Jiang W(2014) Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol 5: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S-M, Kang BR, Kim YC(2013) Transcriptome analysis of induced systemic drought tolerance elicited by Pseudomonas chlororaphis O6 in Arabidopsis thaliana. Plant Pathol J 29: 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun D, Britto DT, Shi W, Kronzucker HJ(2017) Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat Plants 3: 17074. [DOI] [PubMed] [Google Scholar]
- De Coninck B, Timmermans P, Vos C, Cammue BPA, Kazan K(2015) What lies beneath: Belowground defense strategies in plants. Trends Plant Sci 20: 91–101 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D, Yang Y, Cruz CV, Höfte M(2010) Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol 152: 2036–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, James RA, Ryan PR(2012) Aluminium tolerance of root hairs underlies genotypic differences in rhizosheath size of wheat (Triticum aestivum) grown on acid soil. New Phytol 195: 609–619 [DOI] [PubMed] [Google Scholar]
- Duell RW, Peacock GR(1985) Rhizosheaths on mesophytic grasses. Crop Sci 25: 880–883 [Google Scholar]
- Gamalero E, Glick BR(2015) Bacterial modulation of plant ethylene levels. Plant Physiol 169: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George TS, Brown LK, Ramsay L, White PJ, Newton AC, Bengough AG, Russell J, Thomas WT(2014) Understanding the genetic control and physiological traits associated with rhizosheath production by barley (Hordeum vulgare). New Phytol 203: 195–205 [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE(2006) Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B(2007) Promotion of plant growth by bacterial ACC deaminase. CRC Crit Rev Plant Sci 26: 227–242 [Google Scholar]
- Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W(2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Haling RE, Brown LK, Bengough AG, Valentine TA, White PJ, Young IM, George TS(2014) Root hair length and rhizosheath mass depend on soil porosity, strength and water content in barley genotypes. Planta 239: 643–651 [DOI] [PubMed] [Google Scholar]
- Haling RE, Brown LK, Bengough AG, Young IM, Hallett PD, White PJ, George TS(2013) Root hairs improve root penetration, root-soil contact, and phosphorus acquisition in soils of different strength. J Exp Bot 64: 3711–3721 [DOI] [PubMed] [Google Scholar]
- Haling RE, Richardson AE, Culvenor RA, Lambers H, Simpson RJ(2010) Root morphology, root-hair development and rhizosheath formation on perennial grass seedlings is influenced by soil acidity. Plant Soil 335: 457–468 [Google Scholar]
- Hu L, Robert CAM, Cadot S, Zhang X, Ye M, Li B, Manzo D, Chervet N, Steinger T, van der Heijden MGA, et al. (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9: 2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Jiang T, Liu Y-X, Bai Y-C, Reed J, Qu B, Goossens A, Nützmann H-W, Bai Y, Osbourn A(2019) A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364: eaau6389. [DOI] [PubMed] [Google Scholar]
- Huang M, Sanchez-Moreiras AM, Abel C, Sohrabi R, Lee S, Gershenzon J, Tholl D(2012) The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol 193: 997–1008 [DOI] [PubMed] [Google Scholar]
- James RA, Weligama C, Verbyla K, Ryan PR, Rebetzke GJ, Rattey A, Richardson AE, Delhaize E(2016) Rhizosheaths on wheat grown in acid soils: Phosphorus acquisition efficiency and genetic control. J Exp Bot 67: 3709–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Chang C(2015) Mechanistic insights in ethylene perception and signal transduction. Plant Physiol 169: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klay I, Gouia S, Liu M, Mila I, Khoudi H, Bernadac A, Bouzayen M, Pirrello J(2018) Ethylene Response Factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci 274: 137–145 [DOI] [PubMed] [Google Scholar]
- Li J, Ovakim DH, Charles TC, Glick BR(2000) An ACC deaminase minus mutant of Enterobacter cloacae UW4 no longer promotes root elongation. Curr Microbiol 41: 101–105 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang C, Wei C, Liu X, Wang M, Yu F, Xie Q, Tu J(2016) The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol 170: 429–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-Y, Ye N, Song T, Cao Y, Gao B, Zhang D, Zhu F, Chen M, Zhang Y, Xu W, et al(2019) Rhizosheath formation and involvement in foxtail millet (Setaria italica) root growth under drought stress. J Integr Plant Biol 61: 449–462 [DOI] [PubMed] [Google Scholar]
- Mao J-L, Miao Z-Q, Wang Z, Yu L-H, Cai X-T, Xiang C-B(2016) Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet 12: e1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco R, Mosqueira MJ, Fusi M, Ramond J-B, Merlino G, Booth JM, Maggs-Kölling G, Cowan DA, Daffonchio D(2018a) Rhizosheath microbial community assembly of sympatric desert speargrasses is independent of the plant host. Microbiome 6: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, Borin S, Abou-Hadid AF, El-Behairy UA, Sorlini C, Cherif A, et al. (2012) A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One 7: e48479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco R, Rolli E, Fusi M, Michoud G, Daffonchio D(2018b) Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P, Crowley D, Rengel Z(2011) Rhizosphere interactions between microorganisms and plants govern iron and phosphorus acquisition along the root axis—model and research methods. Soil Biol Biochem 43: 883–894 [Google Scholar]
- Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM(2014) Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar E.(1976) The uptake of zinc-65 by oats in relation to soil water content and root growth. Soil Res 14: 67–74 [Google Scholar]
- Nascimento FX, Rossi MJ, Glick BR(2018) Ethylene and 1-aminocyclopropane-1-carboxylate (ACC) in plant-bacterial interactions. Front Plant Sci 9: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Paulson JN, Zheng X, Kolter R(2017) Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci USA 114: E2450–E2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J, Ryan MH, Siddique KHM, Simpson RJ(2017) Unwrapping the rhizosheath. Plant Soil 418: 129–139 [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG(2014) STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30: 3123–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrose DM, Glick BR(2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118: 10–15 [DOI] [PubMed] [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH(2013) Going back to the roots: The microbial ecology of the rhizosphere. Nat Rev Microbiol 11: 789–799 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PAHM(2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347–375 [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO(2013) The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbi SMF, Tighe MK, Flavel RJ, Kaiser BN, Guppy CN, Zhang X, Young IM(2018) Plant roots redesign the rhizosphere to alter the three-dimensional physical architecture and water dynamics. New Phytol 219: 542–550 [DOI] [PubMed] [Google Scholar]
- Remans R, Beebe S, Blair M, Manrique G, Tovar E, Rao I, Croonenborghs A, Torres-Gutierrez R, El-Howeity M, Michiels J, et al. (2008) Physiological and genetic analysis of root responsiveness to auxin-producing plant growth-promoting bacteria in common bean (Phaseolus vulgaris L.). Plant Soil 302: 149–161 [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F(2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ 4: e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R, et al. (2015) Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol 17: 316–331 [DOI] [PubMed] [Google Scholar]
- Shi L, Guo M, Ye N, Liu Y, Liu R, Xia Y, Cui S, Zhang J(2015) Reduced ABA accumulation in the root system is caused by ABA exudation in upland rice (Oryza sativa L. var. Gaoshan1) and this enhanced drought adaptation. Plant Cell Physiol 56: 951–964 [DOI] [PubMed] [Google Scholar]
- Song T, Xu F, Yuan W, Zhang Y, Liu T, Chen M, Hu Q, Tian Y, Xu W, Zhang J(2018) Comparison on physiological adaptation and phosphorus use efficiency of upland rice and lowland rice under alternate wetting and drying irrigation. Plant Growth Regul 86: 195–210 [Google Scholar]
- Stringlis IA, Yu K, Feussner K, de Jonge R, Van Bentum S, Van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, Pieterse CMJ(2018) MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci USA 115: E5213–E5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Keel C(2016) Signaling in the rhizosphere. Trends Plant Sci 21: 187–198 [DOI] [PubMed] [Google Scholar]
- Vessey JK.(2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255: 571–586 [Google Scholar]
- Wang ZQ, Zhang WY, Beebout SS, Zhang H, Liu LJ, Yang JC, Zhang JH(2016) Grain yield, water and nitrogen use efficiencies of rice as influenced by irrigation regimes and their interaction with nitrogen rates. Field Crops Res 193: 54–69 [Google Scholar]
- Watt M, McCully ME, Canny MJ(1994) Formation and stabilization of rhizosheaths of Zea mays L. (effect of soil water content). Plant Physiol 106: 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XH, Wang W, Yin CM, Hou HJ, Xie KJ, Xie XL(2017) Water consumption, grain yield, and water productivity in response to field water management in double rice systems in China. PLoS One 12: e0189280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullstein LH, Bruening ML, Bollen WB(1979) Nitrogen fixation associated with sand grain root sheaths (rhizosheaths) of certain xeric grasses. Physiol Plant 46: 1–4 [Google Scholar]
- Yang C, Lu X, Ma B, Chen S-Y, Zhang J-S(2015) Ethylene signaling in rice and Arabidopsis: Conserved and diverged aspects. Mol Plant 8: 495–505 [DOI] [PubMed] [Google Scholar]
- Zhang H, Xue Y, Wang Z, Yang J, Zhang J(2009) An alternate wetting and moderate soil drying regime improves root and shoot growth in rice. Crop Sci 49: 2246–2260 [Google Scholar]
- Zhang J, Liu Y-X, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X, et al. (2019) NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol 37: 676–684 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen S, Hao X, Su JQ, Xue X, Yan Y, Zhu YG, Ye J(2016) Transcriptomic analysis reveals adaptive responses of an Enterobacteriaceae strain LSJC7 to arsenic exposure. Front Microbiol 7: 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Du H, Gui Y, Xu F, Liu J, Zhang J, Xu W (2020) Moderate water stress in rice induces rhizosheath formation associated with abscisic acid and auxin responses. J Exp Bot 71: 2740–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]