Abstract
Circadian clock proteins are endogenous timing mechanisms that control the transcription of hundreds of genes. Their integral role in coordinating metabolism has led to their scrutiny in a number of diseases, including NAFLD. Discoordination between central and peripheral circadian rhythms is a core feature of nearly every genetic, dietary, or environmental model of metabolic syndrome and NAFLD. Restricting feeding to a defined daily interval (time-restricted feeding) can synchronize the central and peripheral circadian rhythms, which in turn can prevent or even treat the metabolic syndrome and hepatic steatosis. Importantly, a number of proteins currently under study as drug targets in NAFLD (SREBP, ACC, PPARs, and incretins) are modulated by circadian proteins. Thus, the clock can be used to maximize the benefits and minimize the side effects of pharmaceutical agents for NAFLD. The circadian clock itself has the potential for use as a target for the treatment of NAFLD.
Keywords: Dyssynchrony, gut microbiome, steatohepatitis, lipogenesis, bile acids
The 2017 Nobel Prize in Medicine signaled a recognition of the fundamental role that circadian clock proteins play in physiology and disease. It was awarded to Jeffrey C. Hall, Michael Rosbash, and Michael W. Young for their discovery of molecular mechanisms controlling circadian rhythms in Drosophila. Clock proteins are ubiquitous endogenous timing mechanisms that help maintain energy homeostasis by coordinating cellular processes within and between organ systems. They do so by regulating the transcription of hundreds of genes, including themselves. Their integral role in coordinating metabolism has led to closer scrutiny of their contribution to a number of diseases including non-alcoholic fatty liver disease (NAFLD).
Though the term “circadian rhythms” usually conjures thoughts of jet lag and sleep deprivation, physiologically they are core cellular processes that affect every organ system. The pervasiveness of clock proteins, which are found in diverse organisms from cyanobacteria to mammals, shows their evolutionary importance to life. Development of a molecular program to anticipate and prepare for periodic, predictable changes gives a tremendous adaptative advantage. Organisms have developed specialized sensory cells that allow them to adjust their own rhythmicity to external cues (called zeitgebers – German for “time-giver”). The daily light-dark cycle on earth governs rhythmic changes in the behavior and physiology of most species, including humans. Almost all organisms consolidate their biological processes (e.g. sleeping, feeding) to particular times of day. To acknowledge that the periodicity of these oscillations stays approximately 24 hours, Franz Halberg, a pioneer in the field of biological rhythms, introduced the term “circadian rhythms” (from Latin, circa – “about”, diem – “a day”) in 1959.
In mammals, the circadian pacemaker resides in hypothalamic structures called the suprachiasmatic nuclei (SCN), and it is entrained by specialized retinal ganglion cells that measure ambient light (Figure 1). However, circadian clock proteins are not restricted to the SCN. Explants from lung, liver, and other organs exhibit sustained oscillations in vitro.1, 2 Thus, endogenous functional clocks are also active in peripheral tissues.3 Temporal signals such as timing of food intake can reset the hepatic clock without affecting the SCN central clock rhythms. Consequently, different entrainment signals can activate peripheral and central circadian clocks separately.4, 5 However, this does not undermine the importance of the SCN in coordinating physiological processes across multiple tissues.
Figure 1: Central and Peripheral Circadian Rhythms.
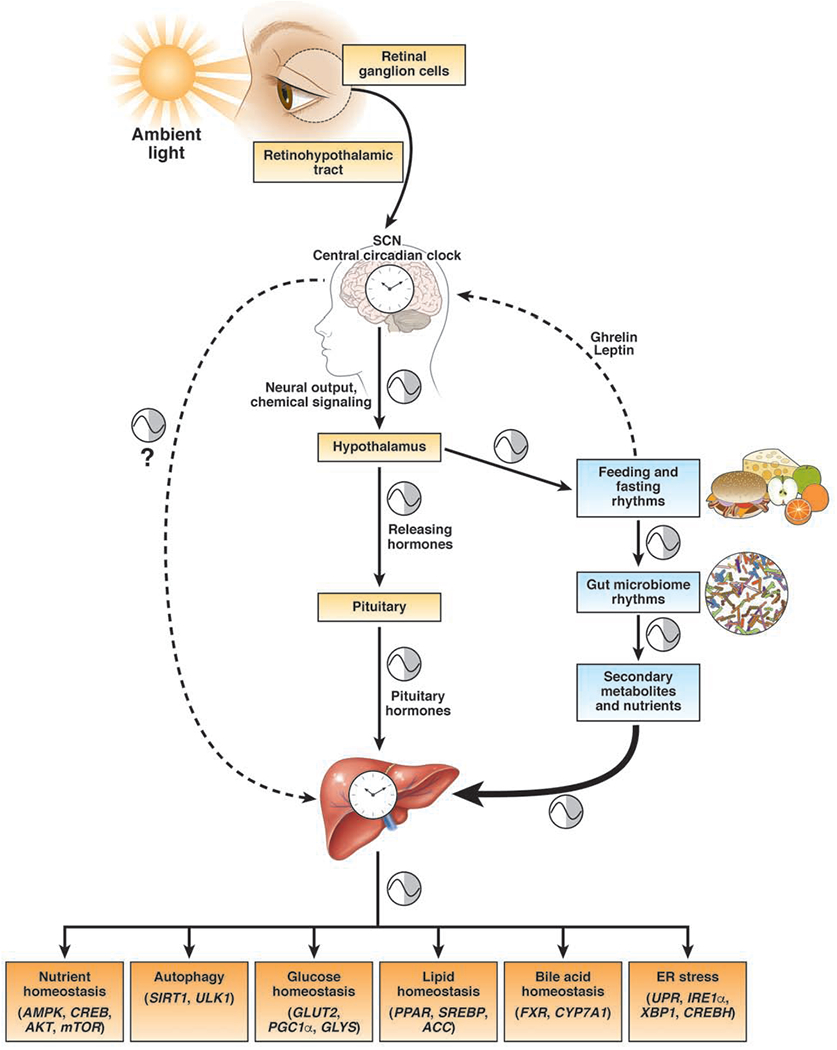
Retinal ganglion cells detect ambient light and entrain the suprachiasmatic nucleus (SCN). Their projects travel through the retinohypothalamic tract, though indirect signals are also received from the thalamus. The SCN contains the central circadian clock which can modulate other oscillatory networks including sleep/wake, temperature, and feeding/fasting homeostasis through the hypothalamus. In addition, the SCN regulates the circadian release of hypothalamic releasing hormones (e.g. CRH), and thus, pituitary hormones (e.g. ACTH). Hormones released from pituitary gland are one way that the central clock can regulate the peripheral organs. The SCN also regulates peripheral clock through poorly understood neurohumoral mechanisms. SCN can drive feeding/rhythms through the hypothalamus. Fasting induces the release of ghrelin which promotes feeding while feeding induces the release of leptin which promotes satiety, and thus affecting feeding/fasting rhythms. Diet and feeding/fasting rhythms modulate cyclical fluctuations in the gut microbiome, which then creates cyclical changes in secondary metabolite production, such as SCFAs and bile acids. Secondary metabolites and/or bacterial products are necessary for the maintenance of peripheral circadian rhythms. Feeding/fasting rhythms are powerful drivers of hepatic circadian clock. The rhythmic expression of genes in the liver generates rhythmicity in the transcripts and activity of genes involved in critical metabolic processes such as glucose, lipid, and bile acid metabolism, nutrient homeostasis, autophagy and ER stress. Metabolic syndrome occurs in the setting of central and peripheral circadian rhythm dyssynchrony.
In this review, we will describe the molecular organization of the circadian clock proteins, the relationship between circadian clock and metabolic master-regulators, and clock regulation of metabolic processes important to the pathophysiology of NAFLD. Environmental cues that entrain peripheral circadian rhythms will be also discussed in detail. The effects of circadian dyssynchrony on host metabolism will be summarized next along with recent preclinical and clinical studies which show that maintaining peripheral circadian rhythms by consolidating feeding within a narrow time frame (i.e. time-restricted feeding [TRF]) can restore metabolic health. Over the last five years, there have been many excellent reviews of pre-clinical data regarding of the role of circadian clock in hepatic metabolic processes specifically,6–11 and metabolic syndrome in general.12–16 Ours will build on this strong body of work by focusing on key regulatory pathways that play an important role in the treatment of steatohepatitis and fibrosis, thus demonstrating the vital role of circadian machinery in helping our understanding of NAFLD.
MOLECULAR ORGANIZATION OF THE CLOCK
The circadian clock in mammals, expressed in nearly every cell, is comprised of a series of transcription-translation feedback loops (Figure 2). These include circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1) as transcriptional activators. These are opposed by period (PER) and cryptochrome (CRY) as transcriptional repressors, (Figure 3A). CLOCK and BMAL1 are two central components that make up the positive (or activation) limb of the molecular clock. The CLOCK:BMAL1 heterodimer then translocates into the nucleus to initiate transcription at the E-box (5′-CACGTG-3′), a specific DNA binding sequence (DBS) on the promoters of its target genes.17 The main downstream targets of CLOCK:BMAL1 include their own repressors, period (PER1, PER2 and PER3) and cryptochrome (CRY1, CRY2), in addition to over 300 other genes.18 Over time, PERs and CRYs accumulate in the cytoplasm, where they are further regulated by casein kinase 1 (CK1) ε, CK1δ, and F-box/LRR-repeat protein 3 (FBXL3). CK1ε and CK1δ phosphorylate PER for degradation; FBXL3 promotes the degradation of CRY.19, 20 However, if Ck^ phosphorylates PER after it is bound to CRY, the three-protein complex migrates into the nucleus and inhibits the CLOCK:BMAL1 heterodimer.21 By inhibiting their own activators, PERs and CRYs repress the transcription of their own genes. Post-translational modification of PERs and CRYs leads to their degradation. This then initiates a new circadian cycle with increased binding of CLOCK:BMAL1 to the now open E-Box binding sites.
Figure 2: Overview of Circadian Clock Machinery.
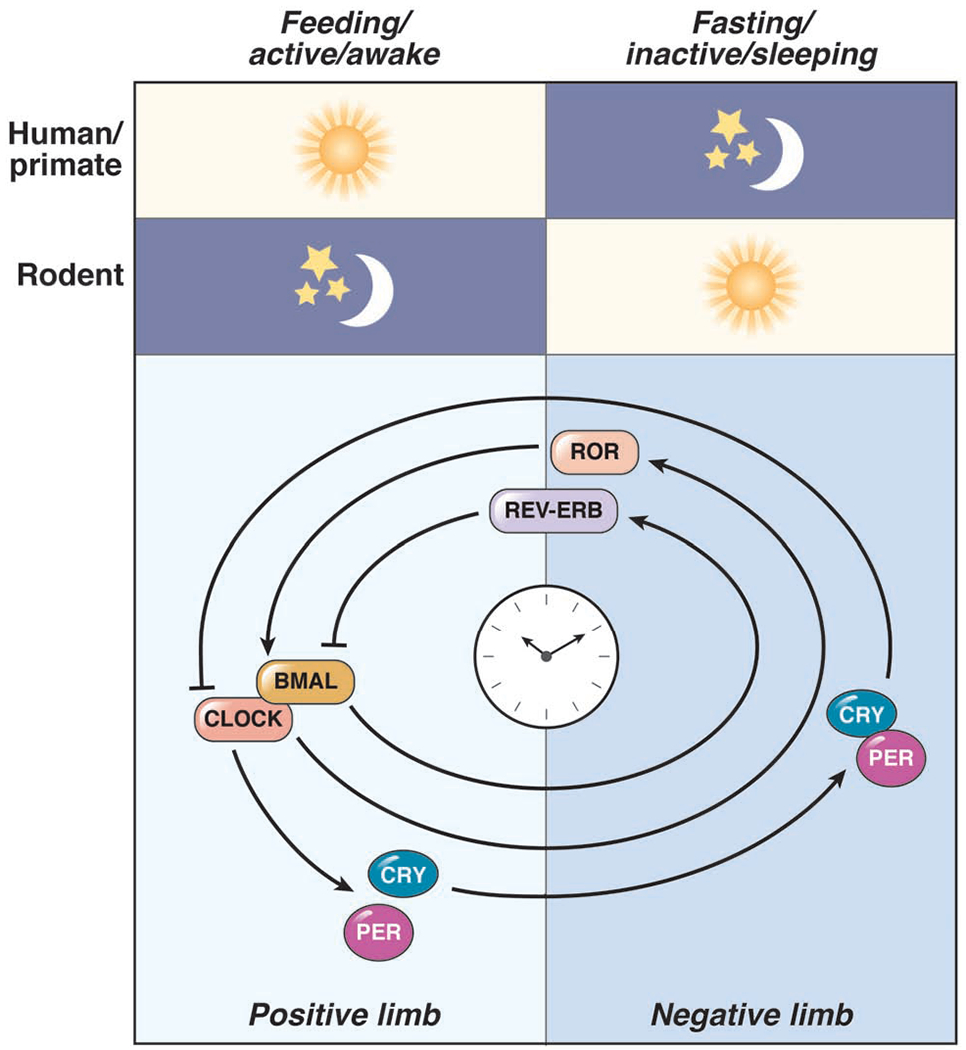
Humans/primates are diurnal animals with the feeding/active period occurs during the light hours. However, mice (and most rodents) are nocturnal, with the feeding/active period occurring in the dark hours. The cellular molecular clock is a series of transcription-translation feedback loops that include the transcriptional acti vators (CLOCK and BMAL1; the positive limb) and repressors (PER and CRY; the negative limb). The PER:CRY complex repress their own activators CLOCK:BMAL1. CLOCK:BMAL1 also promote REV-ERBs and RORs which add another layer of regulation over the circadian clock and regulate hundreds of additional genes.
Figure 3: Circadian Clock Machinery.
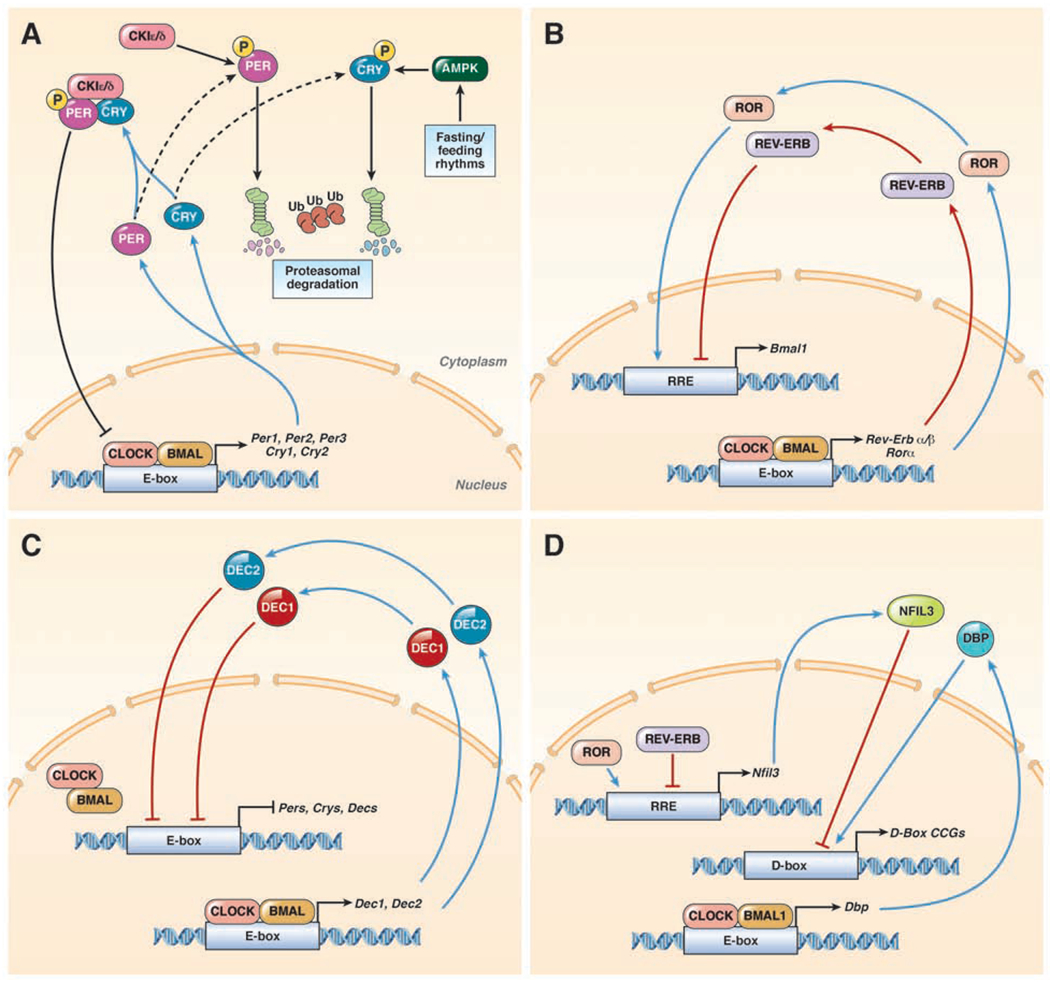
(A) In mammals, the core molecular clock is comprised of a series of transcription-translation feedback loops that include the transcriptional activators (CLOCK and BMAL1). The CLOCK:BMAL1 heterodimer regulate genes at a specific DNA binding sequence, the E-box to regulate expression of hundreds of genes, including their own repressors (Per1, Per2, Per3, Cry1, and Cry2). Once translated, PERs and CRYs accumulate in the cytoplasm, where they are regulated by CK1ε/δ and AMPK, respectively. If these PER or CRY are individually phosphorylated, they will undergo ubiquitylation (Ub) and proteasomal degradation. However, if PER and CRY form a heterodimer prior to phosphorylation by CK1ε/δ, the three protein complex is transported to the nucleus, where it directly inhibits the CLOCK:BMAL1 heterodimer. (B) CLOCK:BMAL1 also regulates the expression of Rev-Erbs and Rors. Once translated REV-ERBs and RORs bind to the RRE DNA binding sequence but with opposite effects. RORs promote, while REV-ERBs suppress at this DNA binding sequence. Together these clock proteins control the regulation of hundreds of genes including the expression of Bmal1. (C) A third loop of the circadian clock involves DEC1 and DEC2 that are transcriptionally activated by CLOCK:BMAL1. DEC1 and DEC2 proteins migrate back into the nucleus and inhibit Per1 transactivation by competing for E-Box binding. (D) CLOCK:BMAL1 promote the transcription of Dbp, and REV-ERB/ROR regulate the expression of Nfil3. Once translated, DBP and NFIL3 promote or suppress, respectively, gene expression at the D-Box DNA binding sequence. The D-box DNA binding sequence controls expression of hundreds of clock-controlled genes (CCGs).
In a second regulatory loop, CLOCK:BMAL1 drives the transcription of the nuclear hormone receptors retinoic acid receptor-related orphan receptor (Ror) α and Rory and Rev-Erbα and Rev-Erbβ by binding E-box elements on their promoters (Figure 3B).22–24 Both of these receptors can bind the ROR-responsive element (RORE) promoter DNA binding sequence (5’-AGGTCA-3’), but with opposite effects; REV-ERBα/β suppresses expression at the site, where as RORα/γ promotes expression. 25, 26 Together, REV-ERBs and RORs generate large scale cyclical fluctuations in transcription of hundreds of clock-controlled genes, including regulation of the transcription of Bmal1.27
Recent studies have revealed an additional autonomous feedback loop involving basic helix-loop-helix family member e40 (BHLHE40, more commonly DEC1) and BHLHE41 (more commonly DEC2) which also suppress CLOCK:BMAL1 activity (Figure 3C).28, 29 CLOCK:BMAL1 induces transcription of Dec1 and Dec2 by binding E-box elements on their promoters. Unlike PERs and CRYs which act directly on the CLOCK:BMAL1 heterodimer, DECs compete with CLOCK:BMAL1 for the E-box, thus suppressing all E-box genes, including transcription of their own genes as well as that of PERs and CRYs.
Other circadian clock-controlled genes bring additional refinement to this complex interplay of feedback loops. CLOCK:BMAL1 modulates the expression of hundreds of other genes by promoting the transcription of D-box binding protein (DBP) by binding E-box elements on its promoter (Figure 3D).11, 30 DBP rhythmically activates transcription of genes that have D-box elements in their promoter regions. Nuclear factor interleukin 3 (NFIL3, also E4BP4), which is regulated by a RORE promoter region (and thus susceptible to activation by RORs and suppression by REV-ERBs), represses DBP-dependent transactivation by competing for the D-box. Computational genomic analysis of D-box elements has identified nearly 1500 regions that can be recognized by DBP and NFIL3.31
The transcription-translation feedback loops generate large scale rhythms in the activities and levels of downstream clock-controlled genes.32 Since these processes are temporally controlled, it is important to regulate the abundance and activity of each component to ensure it is in the right place at the right time. This is achieved by post-translational modifications of clock components such as phosphorylation, acetylation, SUMOylation, O-GlcNAcylation, and ubiquitylation. Gene expression studies show that approximately 10-40% of the rodent genome exhibits a 24-hour rhythm in a highly tissue-specific manner.33 However, humans are diurnal animals that feed during the day whereas most rodents are nocturnal animals that feed primarily at night. The recent publication of a transcriptome atlas of a baboon, a diurnal primate, shows that the aforementioned percentages from rodent studies do not convey the full picture.3 Of the approximately 11,000 genes that are commonly expressed in all tissues in the baboon, 96.6% have 24-hour rhythmicity in at least one tissue. Greater than 80% of the 18,000 protein-coding baboon genes were circadian. More importantly, more than 80% of genes encoding proteins targeted by pharmaceuticals are transcribed cyclically. This implies that most physiological processes are under circadian control, and thus the disruption of circadian rhythms can result in widespread downstream pathology.
THE CIRCADIAN CLOCK REGULATES METABOLIC PROCESSES
Hepatic metabolic processes, including glucose, lipid, and cholesterol/bile acid metabolism are highly dynamic, influenced by feeding/fasting and circadian rhythms. Data supporting these relationships come from several lines of research, including metabolic phenotyping data from circadian clock transgenic mouse lines, molecular biology studies showing direct interactions between regulators of nutrient homeostasis and circadian clock proteins, and the relationship of feeding/fasting cycles and metabolic gene expression. In this section, we will discuss the relationship between circadian clock machinery with various aspects of metabolism that are dysfunctional in patients with NAFLD.
Circadian Clock and Regulators of Nutrient Homeostasis and Autophagy
The liver contains several master metabolic regulators that sense feeding/fasting states and control the expression of hundreds of downstream metabolic targets. These include AMP-activated protein kinase (AMPK), cAMP response element-binding protein (CREB), v-Akt murine thymoma viral oncogene homolog 1 (AKT/PKB), and sirtuin 1 (SIRT1). All have cyclical activity mainly driven by feeding behavior and can regulate, and are regulated by, circadian clock proteins.
AMPK, a fasting-sensitive protein kinase that regulates cellular energy homeostasis, can modulate, and is modulated by, the circadian clock machinery. AMPK can affect the length of circadian cycling by phosphorylating CRY and targeting it for degradation.34 Moreover, RORα activation can modulate lipid metabolism and prevent hepatic steatosis by activating AMPK which controls key lipid regulators (discussed below).35 Feeding, on the other hand, activates CREB, which upregulates Per1 transcription.36 This relationship connects G protein coupled receptors (GPCRs) that use cAMP as a second messenger to transcription of circadian clock genes.37 Several metabolic hormones including glucagon and epinephrine, both of which activate hepatic gluconeogenesis, can influence circadian clock machinery through CREB activation.38–40 In addition, CRYs can independently suppress CREB thereby not only affecting hepatic gluconeogenesis but also modulating Per transcription.39 Finally, AKT, a protein kinase that regulates glucose metabolism among other cellular processes (e.g. cell proliferation, apoptosis), and is also activated by feeding, can modulate the circadian clock machinery by phosphorylating BMAL1 and CLOCK in the cytosol and preventing their translocation across the nuclear membrane.41
Sensing the nutrient status is a critical regulation step in the maintenance of homeostasis. Metabolic stressors such as starvation, induce autophagy to utilize glycogen and lipid components for generating fuel.42 Circadian induction of autophagy during low nutrient states (e.g. inactive state) allows efficient coordination of these cellular processes. AMPK, under glucose starvation conditions, phosphorylates and activates the autophagy-initiating kinase ULK1 (Unc-51 like autophagy activating kinase 1), thus promoting autophagy. However, under nutrient sufficiency, mammalian target of rapamycin (mTOR), another metabolic master-regulator, phosphorylates ULK1 and disrupts its interaction with AMPK, thus inhibiting autophagy.43 Interestingly, liver specific Bmal1-null mice display dampened rhythms of autophagy gene expression, along with increased mTOR activation.44 Thus, autophagy has multiple layers of regulation from metabolic regulators and the circadian clock.
SIRT1, an anti-aging and nutrient sensing protein, can upregulate the expression of a number of genes involved in autophagy and energy metabolism by binding to the E-box elements in a complex with the CLOCK:BMAL1 heterodimer.45, 46 This complex in particular creates daily oscillations in nicotinamide phosphoribosyltransferase (NAMPT), which is the rate-limiting enzyme in NAD+ (nicotinamide adenine dinucleotide) salvage pathway, thus inducing circadian oscillation of intracellular NAD+.47, 48 SIRT1 can further act on circadian clock proteins by deacetylating BMAL1 and PER2, as well as other key metabolic regulators, through its regulation of NAD+.48 SIRT1 histone deacetylation leads to a repressive chromatin configuration, essentially down regulating their downstream targets.49 As a result of SIRT1 interaction with CLOCK:BMAL1 and deacetylation of key regulator proteins, these genes have day-night rhythmicity to their transcriptional activity and temporal regulation of their target genes. Thus, the role of autophagy in the pathogenesis of NAFLD is well described and compounds that target autophagy pathways have been proposed to have therapeutic potential.50–52
Circadian Clock and Glucose Homeostasis
Glucose metabolism is regulated by the circadian clock machinery. The relationship between circadian changes in metabolically important hormones (i.e. glucocorticoids, insulin, ghrelin, incretins, adiponectin, and leptin) and ambient light provided the initial clues that the central clock plays an important role in glucose and nutrient homeostasis.53, 54 Moreover, these hormones can directly affect circadian clock in their target cells. Insulin is a regulator of Per2 and Rev-erbα, and both glucose and insulin can affect the expression of Dec1 and Dec2.55, 56 Low glucose/fasting induction of AMPK can repress the cryptochromes as described in the previous section. Thus, the relationship between circadian clock and glucose homeostasis is bidirectional.
Of particular interest, incretins, which are already the target of FDA approved pharmaceuticals aimed to treat type 2 diabetes (T2D) and obesity, are being investigated as therapeutic agents in NAFLD.57 Glucagon-like peptide 1 (GLP-1), an incretin secreted by the intestinal L-cells located primarily in the ileum, is an insulin sensitizer.58 GLP-1 release correlates with the pattern of insulin secretion, demonstrating an increase immediately preceding the feeding period. Thus, GLP-1 contributes to the decrease in blood sugar levels by enhancing the glucose-dependent insulin secretion.57, 59 Patients with NAFLD display diminished concentrations of active incretins.60–62 GLP-1’s release is circadian in rodents59 and humans,63 and this rhythmicity is lost with the consumption of a high fat/palmitate diet64, 65 and sleep deprivation.66 Knockdown of Bmal1 or Clock results in impairment of insulin release from β-cells in response to exendin-4.67, 68 Thus, circadian clock control of glucose-modifying hormones is not limited to the hypothalamic-pituitary axis.
Circadian clock machinery regulates glucose at a cellular level as well. Glucose homeostasis is altered in nearly all transgenic lines where the circadian clock is perturbed (Table 1).12 The liver specific Bmal1 knockout (KO) mice helped elucidate the relationship between circadian clock and GLUT2, the main hepatic glucose transporter.69–71 The expression of Glut2 in mice is characterized by the peak and trough of expression corresponding to the inactive/daytime and active/nighttime states, respectively. This homeostatic stage is disrupted with loss of Bmal1, which results in constitutively low expression of both transcript and protein levels of Glut2. Hepatocyte-specific deletion of Bmal1 in mice led to fasting hypoglycemia, hypoinsulinemia, and loss of rhythmicity in the expression of hepatic glucose metabolic genes.69–71
TABLE 1:
Circadian Clock Transgenic Lines and their Effects on NAFLD
| Mouse Strain | Diet/Intervention | Liver Phenotype | Δ Glucose | Δ Insulin | Δ serum TGs | Δ Weight | Δ Adiposity | Refs |
|---|---|---|---|---|---|---|---|---|
| Rev-erbα KO + Hepatic Reverb β KO | NCD | Marked hepatic steatosis with deficiency of both Reverbs, but only subtle changes with loss of either subtype alone | ↓ | 177 | ||||
| Rev-erbα KO | NCD | Higher hepatic TG (trend) | ↑ | ↑ Normal ITT | ↔ | ↑ | 178 | |
| HFD (53%) | Higher hepatic TG (trend) | ↑ | ↑ | ↑ | ↑ | |||
| Per1/2 KO | NCD | Lower hepatic TG depending on what time of day they were eating (oscillating) | 179 | |||||
| Cry1/2 double KO | NCD | ↑ | ↔ | ↓ | 180 | |||
| HFD (45%) | Hepatic steatosis | Impaired GTT | ↑ Impaired ITT | ↑ | ↑ | |||
| Bmal1 KO | NCD | ↑ | ↓ | 181 | ||||
| HFD (60%) | Hepatic steatosis in global (not tissue-specific) Bmal1 KO mice, but not in control mice | ↑ | ↓ | ↓ | ||||
| ClockΔ19 KO | NCD | ↑ | ↔ | ↑ | ↑ | ↑ | 73 | |
| HFD (45%) | Lipid engorgement of hepatocytes in Clock KO mice compared to WT | ↑ | ↑ | |||||
| ClockΔ19 KO | NCD | Normal | ↔ | 182 | ||||
| HFD | Lower hepatic TG | ↔ | ||||||
| mPer2−/− KO | NCD & CCl4 | Increased hepatic fibrosis in KO mice | 183 | |||||
| Rev-erbα KO (and HDCA3 KO) | NCD | Increased hepatic TGs/steatosis in KO mice | 184 |
ALAN: artificial light at night; CCl4: carbon tetrachloride; DIO: diet-induced obesity; GTT: glucose tolerance test; HFD: high fat diet (with percentage of fat in diet); ITT: insulin tolerance test; NCD: normal chow diet; TGs: triglycerides; WT: wild type; KO: knockout;
The influence of the circadian clock is not limited to glucose transport. BMAL1 also regulates the expression of Pgc1α (peroxisome proliferator-activated receptor-gamma coactivator 1α), a co-activator of gluconeogenesis.72 Hepatic glycogenesis is controlled by CLOCK through transcriptional activation of glycogen synthase 2 (Gys2), the rate limiting enzyme in glycogen synthesis.70 The increased expression of Gys2 is temporally timed with surge of post-prandial glucose. Clock mutant mice display severely disturbed feeding rhythms and glucose dysregulation (i.e hyperglycemia, hypoinsulinemia).71, 73 In addition, negative mutations in Clock and Bmal1 lead to impaired gluconeogenesis.74 Per1/Per2 double KO mice have severe hypoglycemia in the fasting/inactive period and impaired glucose tolerance.69 Cry1/Cry2 double KO mice also show glucose intolerance with elevated circulating corticosterone levels.75 CRYs are transcriptional repressors that regulate gluconeogenic enzymes through the repression of glucocorticoid receptors75 as well as CREB.39, 76
Not all circadian clock transgenic mice have unfavorable glucose homeostatic responses. Liver-specific KO of Rory are protected against insulin resistance and hyperglycemia.77 Incidentally, RorY KO are also protected against diet-induced hepatic steatosis.35 Likewise, mice with hepatic overexpression of Cry1 had normoglycemia and improved insulin sensitivity in response to dietary challenge.39 These findings have bolstered the argument for therapeutic agents targeting circadian clock components as potential therapeutic target for T2D and
Circadian Clock and Lipid and Bile Acid Homeostasis
In addition to glucose homeostasis, the circadian clock regulates lipid and bile acid metabolism. This includes regulatory transcription factors such as peroxisome proliferator-activated receptor (PPARs) and sterol regulatory element-binding proteins (SREBP) 1c (lipid metabolism) or rate limiting enzymes such as acetyl-CoA carboxylase (ACC; fatty acid biosynthesis) and cholesterol 7 alpha-hydroxylase (CYP7A1; cholesterol and bile acid metabolism). Dysfunction in these regulatory pathways are part of the pathophysiology of NAFLD.
One of the main reasons for lipid accumulation in hepatocytes of patients with NAFLD is dysregulation of de novo lipogenesis. SREBP1c is one of several transcription factors that can drive the induction of lipogenic genes. Overexpression of SREBP1c in the liver is associated with a two-fold increase in ALT, AST, hepatic triglyceride levels, and serum free fatty acids.78 Transgenic mice that overexpress SREBP1c displayed a fatty liver phenotype along with hyperglycemia and hyperinsulinemia.78 The activity of this regulator is dependent on the fastingfeeding cycle and nutritional status.79 However, more recent studies demonstrate that SREBP1c, and its targets, are also regulated by the circadian clock machinery. SREBP1c transcripts, as well as protein levels, display daily rhythmicity.80 This rhythmicity is also reflected in the binding pattern of SREBP1c to some of its canonical target genes.81 SREBP1c is regulated by BMAL1, REV-ERBα, RORα, RORγ, and DEC1.80, 82, 83 For example, REV-ERBα can control hepatic cholesterol and lipid metabolism through the controlled expression of Insig2 (insulin induced gene 2) which is an inhibitor of SREBP1c.83
Cyp7a1, the gene for the rate limiting enzyme that converts cholesterol to bile acids, is also regulated by the circadian clock and expressed cyclically. REV-ERBα 84 and DBP 85 regulate its transcription. Per1/Per2 double KO mice have abnormal expression of Cyp7a1 and other key bile acid enzymes, suggesting that they have at least indirect regulation of bile acid synthesis.86 Moreover, Cyp7a1 transcription is regulated by hepatic farnesoid X receptor (FXR) and small heterodimer partner (SHP), as well as fibroblast growth factor (FGF) 15/19 which is released as a result of FXR activation in the ileum. Fxr and Shp transcription (and thus, also transcription of Fgf15/19) are also clock-gated.87, 88 FXR agonists, such as obeticholic acid are currently being investigated as a potential therapeutic agent for NAFLD.89 Finally, it should be noted that BMAL1 can further influence triglyceride metabolism through PGC1α regulation of Fxr further demonstrating circadian clock’s multiple layers of regulatory control.90 Altogether, cyclical activity of CYP7A1 and its regulating enzymes generate circadian fluctuation of serum and fecal bile acids which has implications on lipid absorption and microbiome composition.91, 92
Another important regulator of lipogenesis that participates in clock controlled metabolic processes is ACC. Phosphorylation and inactivation of ACC is driven by feeding and follows a 24-hour rhythm.11, 93 Circadian clock regulation of AMPK (e.g. with RORα) represses ACC and provides temporal control of lipogenesis.34 Mice with liver-specific deletion of ACC1 exhibit lowered accumulation of hepatic triglycerides and decreased de novo fatty acid synthesis.94 Similarly, ACC2 KO mice fed a high fat high carbohydrate diet display lower body weight and less epididymal fat compared to wild type mice.95 Recently an allosteric inhibitor of ACC1/2 showed promising liver specific effects, including decreased lipogenesis, hepatic steatosis, and improved insulin sensitivity.96
Genes involved in lipid metabolism and lipogenic processes are regulated by PPARs, a class of ligand-activated transcription factors.97 PPARα KO mice fed a high fat diet (HFD) accumulate more hepatic triglycerides and show a much higher NAFLD activity score.98, 99 Conversely, activation of PPARs with fenofibrate (PPARα) and rosiglitazone (PPARγ) leads to amelioration of certain parameters of NAFLD, including decreased ALT, AST, steatosis, and inflammation in both mouse and clinical studies. Several randomized controlled trials have demonstrated improvement in liver inflammation in both diabetic and non-diabetic patients with pioglitazone, another PPARγ agonist.100–102 BMAL1 and CLOCK transcriptionally regulate the circadian rhythmicity of Pparα. PPARα in turn directly binds to PPAR response elements (PPRE) on the Bmal1 promoter and promotes its expression in a positive feedback loop.103, 104 In addition, PPARα directly interacts with PER2 in hepatic tissue to modulate transcription of their target genes.105 Since PPARs are highly circadian proteins, it is unclear whether dosing at different times of day can alter the bioavailability and effectiveness of the drug in treating NAFLD.
Circadian Clock and Endoplasmic Reticulum Stress
One of the major functions of the endoplasmic reticulum (ER) is to facilitate the correct folding of proteins. In addition, ER is the site of lipid synthesis in hepatocytes.106 Stressors such as changes in redox states or high-protein demands lead to an accumulation of unfolded or mis-folded proteins, ultimately leading to ER stress. The risk of ER stress is higher at certain times of the day; thus, many of its regulatory proteins are under circadian control. As an adaptation response, the unfolded protein response (UPR), along with its component proteins, such as inositol-requiring enzyme 1α (IRE1α), is triggered to restore cellular homeostasis.107, 108 The UPR plays a critical role in the pathogenesis of NAFLD.109–111 In mice, UPR-regulated genes exhibit a biphasic 12-hour periodicity which is driven by circadian clock control of IRE1α as well XBP1 (X-box binding protein 1), a UPR master regulator.112 In arrhythmic Cry1/Cry2 double KO mice, hepatic Ire1α is constitutively expressed, suggesting that this pathway is under clock control. These mice also had perturbed levels of hepatic and serum triglycerides.112
Finally, the ER-associated transcription factor CREBH (hepatocyte specific CREB) regulates stress-related lipogenesis and fatty acid oxidation in mice.113–115 CREBH is an important mediator of the circadian oscillator in the liver and exhibits rhythmicity in its proteolytic cleavage but not mRNA level, indicating that circadian clock regulates it through posttranslational modification.115, 116 CREBH is regulated indirectly by BMAL1 and AKT, and its activity is modified by DBP and NFIL3.116, 117 Mice with CREBH deficiency leads to reduced levels of genes involved in fatty acid metabolism. These mice display hypertriglyceridemia, increased fatty acids, and are more susceptible to diet-induce hepatitis and hepatic fibrosis, whereas CREBH activation protects against steatosis.118–120
LUMINAL CONTENT ENTRAINS HEPATIC CLOCK
The SCN functions as the master pacemaker and is sensitive to light signals but largely unresponsive to feeding patterns. On the other hand, the peripheral clock in the liver is dependent on feeding pattern for the amplitude and phase of oscillation of its transcripts.4, 121 Restricting feeding in mice to only the daytime (when they normally sleep) results in a phase shift between the central and peripheral clocks.81 Recent studies show that feeding is a stronger driver of rhythmic gene expression than circadian clock proteins with 70% of hepatic transcripts losing rhythmicity under arrhythmic feeding.122 Thus, feeding is an important cue (i.e. zeitgeber) for hepatic circadian clock function. It should be noted that the impetus for feeding comes from the hypothalamus, where the SCN is located. Therefore, dyssynchrony in feeding and light cycles can be considered root disorders of the SCN/hypothalamus.12
The gut microbiome is widely recognized for its importance in regulating metabolic physiology, and as playing an important role in the pathophysiology of metabolic syndrome and NAFLD.123–125 Recent studies show that the gut microbiome can contribute to dysmetabolism by disruption of epithelial and hepatic circadian rhythms. Though the gut microbiome is not exposed to light, it exhibits cyclical variations that are closely intertwined to host gene expression, feeding pattern, and gender.126–130 Indeed, circadian disruption induced by jet-lag,126 genetic mutation of host circadian genes,126–128 or by diet126, 129, 130 are associated with disruptions in microbe-host circadian dynamics that can result in increased adiposity, insulin resistance, and inflammation.12 Luminal secondary (bacterially-produced) metabolites demonstrate cyclical rhythms over a 24-hour period.126, 127, 129 The cyclical variations of the bacterial taxa is one of the most robustly reproducible characteristics of the gut microbiome in murine models.131 However, the cyclical dynamics of the human gut microbiome have been difficult to study. Nevertheless, salivary samples collected over a 24-hour period132, 133 and analysis of stool microbiome collected at different times134, 135 suggest that the human microbiome is also highly dynamic.
Gut microbes can drive host circadian rhythms.12, 125 Germ-free mice, antibiotic-induced microbiome depleted mice, or mice who lose cyclical oscillation of their gut microbiome through dietary interventions lack hepatic and intestinal circadian rhythms.129, 136 Circadian variation of the gut microbiome can cyclically activate luminal toll-like receptors (TLRs).136 During the rest phase, intestinal TLRs are inactive and there is an increase in the transcription factor PPARα, which promotes the transcription of Rev-Erbα. During the active phase, TLRs respond to bacterial proteins and suppress Pparα transcription. With microbiome depletion, TLRs are less activated and both PPARα and REV-ERBα are elevated regardless of time. Though others have reported an increase in TLR expression137 and no changes in Pparα expression,137, 138 likely due to differences in antibiotic depletion protocols, the observation that REV-ERBα does not cycle in microbiome-depleted mice has been reproduced in multiple studies.138, 139 The circadian relationship between the gut microbiome and REV-ERBα was recently reproduced in another study investigating the role of microbiome on circadian clock.139 Cyclical expression of REV-ERBα leads to cyclical transcription of Nfil3 (See Figure 3D). Nfil3 can regulate body composition through circadian expression of the intestinal lipid metabolic program, regulating lipid absorption and its export from intestinal epithelial cells.139 Germ-free and antibiotic treated mice have low expression of Nfil3, and, thus, much lower body lipid absorption.
Though these studies show a relationship between cyclical fluctuations of the gut microbiome and the circadian machinery of the host intestinal epithelial cells, it is not clear which microbially-derived mediators are responsible for changes in hepatic oscillations and, thus, metabolic functions. Two potential mediators are bile acids and short-chain fatty acids (SCFAs) (Supplemental Figure 1).125 As discussed above, bile acids and key enzymes in their production and uptake are cyclically expressed and regulated by circadian clock genes.84, 91, 140 Though incompletely characterized, secondary bile acids can affect intestinal and hepatic circadian gene expression in vivo.141 Increased expression of bile salt hydrolase, a bacterial enzyme that deconjugates bile acids and allows for the production of secondary bile acids, induces a shift in ileal and hepatic circadian gene expression in antibiotic treated mice.142 Moreover, sleep disruption, altered feeding pattern, and circadian gene KOs alter the expression of key bile acid regulatory genes.85, 91 Cecal SCFAs have cyclical fluctuations that are perturbed with a HFD.129 Though the mechanism is incompletely understood, supplementation with SCFAs can cause changes in Bmal1 and Per2 gene expression. Interestingly, Bmal1 KO mice lose cyclical fluctuation of their fecal SCFAs.143 Overall, these studies suggest a reciprocal interaction between the circadian clock and luminal bile acids and SCFAs.
DYSSYNCHRONY AND METABOLIC SYNDROME
Synchrony between the SCN and hepatic circadian clock temporally organizes the expression of a large number of metabolic regulatory genes to the daily pattern of food availability. In the absence of a robust hepatic circadian clock, the organism becomes susceptible to various metabolic disorders, including increased adiposity, ectopic steatosis, and insulin resistance.12 Transgenic mouse lines affecting various clock genes demonstrate the interconnectedness between circadian rhythms and metabolic regulators. Circadian gene transgenic lines nearly universally have perturbed metabolic phenotypes (e.g. insulin resistance, increased adiposity) (Table 1).12 Moreover, nearly all dietary-induced dysmetabolic mouse models have perturbed feeding patterns, often characterized by the loss of discrete meals and the spread of caloric intake throughout the day and night.12 Thus, circadian dyssynchrony, characterized by phase shifts and dampening of circadian gene oscillations that lead to misalignment of peripheral and central circadian rhythms, is common to nearly all models of metabolic syndrome, including NAFLD.
Murine Models
Efforts to decipher the mechanistic relationship between circadian clock and host metabolism have been quite fruitful, especially over the last two decades. In general, decreased expression of the clock genes in the positive limb result in worse metabolic outcomes. Conversely, decreased expression of the negative limb (see Figure 2) clock genes leads to improved metabolic outcomes and protection from hepatic steatosis. A summary of selected studies of clock transgenic mice and their metabolic characteristics are presented in Table 1.
Environmental stressors, such as changes in diet, sleeping pattern, or lighting conditions can also cause circadian dyssynchrony. For example, in the diet-induced obesity (DIO) model – where mice are given a HFD and develop increased adiposity, hyperlipidemia, steatosis, and insulin resistance – mice spread their caloric intake throughout the day and night, thereby doubling the percentage of food eaten during the day/inactive period.144 This altered feeding pattern causes a disruption of the hepatic circadian clock. Ob/Ob mice, a genetic model of obesity characterized by hyperphagia due to leptin deficiency, also have dampened oscillation of their circadian clock prior to weight gain.145, 146 In addition to feeding, changes to light (e.g. light at night) or disruption of sleep in mice also leads to dysmetabolic states characterized by increased features of metabolic syndrome including steatosis.16
Human Studies
As in mice, human studies reveal a strong relationship between the circadian dyssynchrony and dysmetabolism. Genetic studies have been instrumental in showing this relationship. Haplotype analysis of patients with NAFLD revealed an association between steatosis and CLOCK, where gene variants have a potential role in the susceptibility, pathogenesis, and disease progression.147 A similar study reported the putative role of CLOCK polymorphisms which was associated with a 1.8-fold increase in susceptibility to obesity.148 SNPs in CLOCK have also been associated with metabolic syndrome, hypoglycemia, obesity, and T2D.149 Genome wide association studies provide strong evidence for the role of circadian gene variants in susceptibility to metabolic syndrome. They have demonstrated a link between PER3 and BMAL1 to T2D, BMAL1 to hypertension, and CRY2 with insulin resistance.150
Environmental and behavioral effects on the circadian clock also have metabolic consequences, which are summarized in Table 2. Though human studies show significant association between sleep disturbances, shift-work, and genetic predispositions to metabolic syndrome, it is not clear whether the mechanisms driving these metabolic phenotypes are similar to what has been observed in mice. Provocatively, one of the studies that investigated the circadian dynamics of the gut microbiome suggests that the microbiota could be mediating the dysmetabolic phenotype of circadian dyssynchrony in humans.126 Gnotobiotic mice that received a fecal transplant from jet-lagged individuals had increased adiposity and insulin resistance compared to gnotobiotic mice who received pre-jet lag and post-jet lag stool from the same individuals.126 However, only two patients were used in this small cohort and more vigorous investigation of the role of the gut microbiome in circadian dyssynchrony is necessary.
Table 2:
Selected studies investigating associations between sleep or food behaviors and metabolic syndrome or NAFLD
| Article | Study Type | Population | Control | Comparator | Key findings |
|---|---|---|---|---|---|
| Selected studies investigating associations between sleep disturbance or time-restricted feeding and metabolic syndrome | |||||
| Spiegel 1999 185 | Observation study | Healthy volunteers (n=11) | n/a | Sleep deprivation | • Sleep-deprivation reduced glucose tolerance and evening cortisol concentrations |
| Wang 2014 186 | Meta-analysis of 13 observational studies | Workers exposed to night shift (n=2,286) | Day shift work | Night shift workers | • Higher risk of metabolic syndrome in workers exposed to night shift (OR 1.5-1.6) |
| Vyas 2012 187 | Meta-analysis of 9 case control studies, 11 prospective cohort studies, and 6 retrospective cohort studies | Workers exposed to shift work (n=2,000,000) | Shit workers | Day workers | • Shift work is associated with an increased risk of myocardial infarction and coronary events (OR 1.23) |
| Karlsson 2001 188 | Observational study | Working adults (n=27,485) | Day workers | Shift workers | • OR of obesity ~1.3 higher in shift workers compared to day workers • Dyslipidemia clustered with shit workers compared to day workers (p<0.05) |
| Wilkinson 2019 189 | Paired-sample trial | Patients with metabolic syndrome (n=19) | 2-week baseline of eating interval of ≥14 h per day | 12-weeks of time-restricted eating (TRE) to a 10-hour interval | • After 12-weeks of TRF, there was a 3% reduction in weight (p=0.0003), 11% reduction in LDL-C (=0.016), and 8% reduction in estimated calorie intake (p=0.007) , but no difference in insulin resistance (p=0.107) |
| Selected studies investigating associations between sleep disturbances and/or shiftwork and NAFLD | |||||
| Hsieh 2011 190 | Observational | All-male Tokyo office workers (n=8157) | Sleep >7hours | Sleep <7 hours | • Rates of NAFLD were higher in patients who slept <7 hours (OR 1.23) |
| Kim 2013 191 | Observational | Workers and spouses at a Korean conglomerate (n= 69,463) | Sleep >5 hours | Sleep <5 hours | • Rates of NAFLD were higher among women (but not men) who slept 5 hours (OR 1.59 in women; 1.03 in men) |
| Imaizumi 2015 192 | Observational studies | General patients (n=3986) | Sleep duration 6 to ≤7 h | Sleep duration ≤ 6 hours | • Rates of NAFLD were higher among women (but not men) who slept ≤ 6hours (OR 1.44 in women, 0.98 in men) |
| Bernsmeier 2015 193 | Case-control study | Cohort in Basel (n= 68) | Healthy volunteers (n=22) | Biopsy-proven NAFLD group (n=46) | • NAFLD group had shorter sleep duration and poorer sleep quality compared to controls (p<0.01). In NAFLD patients, but not in controls, daytime sleepiness was correlated with higher liver enzymes, and insulin resistance. • NAFLD patients with fibrosis had stronger correlation to daytime sleepiness (p=0.02) |
| Marin-Alejandre 2019 194 | Cross-sectional study | Fatty Liver in Obesity (FLiO) participants in Spain (n=134) | Normal weight non-NAFLD patients (n=40) | Overweight patients with NAFLD (n=94) | • Adjusted odds of sleep disturbance was higher in NAFLD group (OR 1.59) • Regression models predicted ~20% of variability in liver stiffness could be attributed to sleep disturbance or sleep quality |
| Liu 2016 195 | Cohort study | Patients who developed NAFLD after 5 years of follow-up (n=2197) | Sleep duration 7-8 hours | Sleep duration 8-9 hours; sleep duration >9 hours | • Adjusted odds of NAFLD was higher with longer sleep durations (OR 1.21 for 8-9 hours group 1.31 in >9 hours group) |
| Balakrishnan 2017 196 | Observational study | General workers (n=8159) | Non-shift workers | Shift workers | • Adjusted odds of NAFLD did not differ between shift-workers and non-shift workers (OR 1.11) |
| Katsagoni 2018 197 | Randomized controlled trial | Patients with NAFLD (n=63) | Controls with NAFLD without intervention | Mediterranean lifestyle (adequate sleep and activity) | • Mediterranean lifestyle was associated with a decrease in liver stiffness (adjusted OR 0.78) |
| Katsagoni 2017198 | Case Control Study | Patients with NAFLD and controls (n=155) | Controls without NAFLD (n=55) | Patients with NAFLD (n=100) | • Optimal sleep duration was inversely associated with NAFLD presence (OR = 0.38, p=.0.05) |
| Selected studies investigating link between OSA and NAFLD | |||||
| Musso 2013 199 | Meta-analysis of 18 cross-sectional studies | 18 cross-sectional studies (n=2, 183) of patients with OSA who were assessed for NAFLD | OSA patients without NAFLD | OSA patients with NAFLD | • OSA was associated with NASH (OR 2.37), and advanced fibrosis (OR 2.3) in biopsy proven NAFLD patients |
| Pamidi 2012200 | Observational | Healthy and young OSA patients | Control subjects without OSA (n=20) | Patients with OSA (n=12) | • OSA was associated with a 27% lower insulin sensitivity compared to controls |
CORRECTION OF DYSSYNCHRONY WITH TIME RESTRICTED FEEDING
It was not clear until recently whether correcting circadian dyssynchrony is sufficient to reverse the dysmetabolic effects of these various insults. Time-restricted feeding (TRF), a behavioral paradigm where feeding is consolidated to the active period, aligns peripheral and central circadian rhythms.151 Correction of circadian dyssynchrony with TRF prevents and treats the metabolic consequences of a large variety of insults.12, 152–155 It should be noted that TRF is not synonymous with intermittent fasting. The former is focused on aligning peripheral and central circadian rhythms. When applied to human populations, the number of calories consumed is irrelevant in the time-restricted diet, which is part of its appeal. Intermittent fasting, on the other hand, focuses on creating periods of fasting, usually in alternate days, to induce changes to metabolism. Caloric intake is strictly regulated on fasting days.
Mice in the TRF condition only have access to food at the nocturnal (active) phase, which promotes natural feeding rhythms and restores the amplitude and phase of oscillations of peripheral circadian clock genes, which are obliterated in DIO mice.151 TRF also restores cycling of metabolic regulators (e.g. AMPK, mTOR, CREB) as well as their downstream targets. The metabolic effects of restoring synchrony can be profound. Though TRF mice consume the same number of calories and have similar activity levels, they do not have the dysmetabolic phenotype of DIO mice. TRF prevents and treats the increased adiposity, insulin and leptin resistance, steatohepatitis, and dyslipidemia that occurs with the DIO condition.130, 151, 156, 157 Subsequent studies show that TRF protects against the metabolic effects of a variety of dysmetabolic diets, including the Western diet, high fructose, and high sucrose diets.156
TRF also protects against the dysmetabolism induced by circadian gene KOs. Per1, hepatic Bmal1, hepatic Rev-Erbα/β, Cry1/Cry2 KOs all have dysmetabolic phenotypes when given free access to HFD. In all cases, TRF prevented the dysmetabolic phenotype.82 TRF reduces hepatic steatosis and enhances cellular response to metabolic stress. Hence, the benefits from TRF likely arise from restoring daily feeding/fasting rhythms and balancing nutrient absorptions with cellular stress response. TRF also protected against the dysmetabolic phenotypes due to aging, obesity, and central circadian rhythm dysfunction in Drosophila.158, 159
Contrary to popular belief, instead of consuming three distinct meals, humans consume calories sporadically and over a large range of time within a 24-hour period.160 Thus, TRF could potentially be an easily adoptable behavioral intervention that improves outcomes in patients with metabolic syndrome. Several small-scale human TRF studies have demonstrated interesting, though sometimes contradictory results. In one randomized crossover trial, healthy volunteers were first either assigned to a standard three meals per day diet or one meal in the evening, both without caloric restriction.161 The two groups did not have a difference in the number of calories consumed. The one-meal a day group had higher morning fasting plasma glucose levels and a delayed insulin response in an oral glucose tolerance test (GTT). A different study compared healthy males consuming a standard three meals per day diet with TRF in the afternoon-to-evening group. In this case, the TRF group had decreased fat mass, without any difference in triglycerides, total cholesterol, or LDL.162 A subsequent randomized, supervised controlled feeding trial in humans demonstrated that participants in the early TRF group had improved insulin sensitivity, appetite, and blood but had higher levels of triglycerides and total cholesterol.163 A more recent study aimed to compare early vs. late TRF among men at risk for T2D (monitored with continuous glucose monitors) found that the early group had lower mean fasting glucose whereas both TRF groups had an improvement in glucose response to meals.164 However, it is not clear whether the investigators controlled for the length of time the subjects were fasting during their sample collection, necessitating additional larger studies. A recent study investigated the effects of TRF on patients with metabolic syndrome undergoing pharmacotherapy. In this single-arm, paired-sample trial of 19 patients who had an initial eating window of >14 hours and were undergoing statin or antihypertensive therapy, imposing a 10-hour feeding window (monitored with continuous glucose monitors) led to improved cardiometabolic health. Though these studies are promising, a study investigating the effects of TRF on NAFLD endpoints have not yet been investigated.
CONCLUSION
NAFLD is a complex disease that is associated with a multitude of metabolic perturbations.7, 165 Since many circadian clock controlled genes are vital participants in metabolic processes of the body, it is not surprising that some of these rhythmic genes can be potential targets for therapy. Behavioral interventions, such as TRF, may have benefits in NAFLD that is independent of its weight loss effects. TRF may be easier for patients to adopt since it does not restrict calories nor require a specific macronutrient dietary profile.160
Chemical modulators of clock-regulated processes such as bile acid metabolism (FXR agonist obeticholic acid, FGF15/19 analogue NGM282), incretin response (GLP-1 receptor agonist liraglutide), PPAR-controlled lipogenic mechanisms (PPARα activator Elafibranor) are under various stages of the clinical trial process.11, 166, 167 However, since some components of the core clock directly participate in metabolic processes and the oscillator itself is responsive to resetting stimuli, small molecules directed towards clock components might provide alternative targets for therapeutic intervention.47, 168 In this light, administration of compound KL001, an activator of CRY proteins showed improved glucose tolerance in DIO mice.169, 170 This is consistent with the known role of CRYs in the inhibition of gluconeogenesis.75 Recent studies show that CRY1 is targeted for degradation by autophagy, thus depressing hepatic gluconeogenesis in a temporal manner.76 Further research into these sites on CRY1 might direct researchers towards novel drug targets for the treatment of hyperglycemia and insulin resistance. Nobiletin, a drug that enhances the amplitude of Ror expression, prevents weight gain and improves metabolic parameters in DIO and Ob/Ob mice.171, 172 Additionally, REV-ERB agonists SR9009 and SR9011 treatment led to improved metabolic endpoints in DIO mice.173, 174 These drugs could potentially be beneficial in NAFLD.
Furthermore, the circadian clock extends its functional network to the control of global physiological processes including the targets of nearly all drugs affecting metabolic syndrome spectrum disorders.175 The rhythmicity imparted by the circadian clock in drug metabolic and detoxification processes can be a potential target for the treatment of NAFLD.10, 11 By better accounting for the circadian cycling of their targets, new drugs can be administered in a way that maximizes benefits, and minimizes side effects, potentially arriving at a therapeutic dose with fewer administrations and lower dosages.176 Further study of the chronopharmacology and circadian xenobiotics of small molecule therapeutics might provide better insight into the proper temporal profile and contribute to improved efficacy.
Supplementary Material
Acknowledgments
Grant Support: AZ is supported by K08 DK102902, R03 DK114536, R21 MH117780, R01 HL148801. All authors receive institutional support from P30 DK120515, P30 DK063491, and UL1 TR001442.
Abbreviations:
- ACC
acetyl-CoA carboxylase
- AKT (or PKB)
v-Akt murine thymoma viral oncogene homolog 1 or (protein kinase B)
- ALAN
artificial light at night
- AMPK
AMP-activated protein kinase
- BMAL1
brain and muscle ARNTL-like protein 1
- CCGs
clock-controlled genes
- CCl4
carbon tetrachloride
- CK1
casein kinase 1
- CLOCK
circadian locomotor output cycles kaput
- CREB
cAMP response element-binding protein
- CREBH
cAMP response element binding protein hepatocyte specific
- CRY
cryptochrome
- CYP7A1
cholesterol 7 alpha-hydroxylase
- DBP
D-box binding protein
- DEC (or BHLH)
basic helix-loop-helix family member
- DIO
diet-induced obesity
- dTRF
delayed time-restricted feeding
- eTRF
early time-restricted feeding
- FBXL3
F-box/LRR-repeat protein 3
- FGF
fibroblast growth factor 15
- FXR
farnesoid X receptor
- GLP-1
glucagon-like peptide
- GLUT2
glucose transporter 2
- GPCRs
G-protein coupled receptors
- GTT
glucose tolerance test
- GYS2
glycogen synthase 2
- HFD
high fat diet
- INSIG2
insulin induced gene 2
- IRE1α
inositol-requiring 1α
- ITT
insulin tolerance test
- KO
knockout
- LDLR
low-density lipoprotein receptor
- mTOR
mammalian target of rapamycin
- NAD+
nicotinamide adenine dinucleotide
- NAFLD
non-alcoholic fatty liver disease
- NAMPT
nicotinamide phosphoribosyltransferase
- NASH
non-alcoholic steatohepatitis
- NCD
normal chow diet
- NFIL3 (or E4BP4)
nuclear factor, interleukin 3 regulated
- Ob/Ob
leptin knockout mouse
- OR
Odds ratio
- OSA
obstructive sleep apnea
- PER
period
- PGC1α
PPAR-gamma coactivator 1α
- PPAR
peroxisome proliferator-activated receptor
- PPRE
PPAR response elements
- ROR
retinoic acid receptor-related orphan receptors
- RORE
ROR-responsive element
- SCFA
short-chain fatty acid
- SCN
suprachiasmatic nucleus
- SHP
small heterodimer partner
- SIRT1
sirtuin 1
- SREBP
sterol regulatory element-binding proteins
- T2D
type 2 diabetes
- TGs
triglycerides
- TLR
toll-like receptor
- TRF
time-restricted feeding
- Ub
ubiquitylation
- ULK1
unc-51 like autophagy activating kinase 1
- UPR
unfolded protein response
- WT
wild type
- XBP1
X-box binding protein 1
Biographies



Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: All authors have no relevant disclosures. Dr. Zarrinpar is a co-founder and “equity holder” of Tortuga Biosciences, a company that engineers native bacteria to functionally manipulate the gut microbiome, and wholly unrelated to the circadian research described in this review.
REFERENCES
- 1.Gachon F, Nagoshi E, Brown SA, et al. The mammalian circadian timing system: from gene expression to physiology. Chromosoma 2004;113:103–12. [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki S, Numano R, Abe M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000;288:682–5. [DOI] [PubMed] [Google Scholar]
- 3.Mure LS, Le HD, Benegiamo G, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018;359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damiola F, Le Minh N, Preitner N, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 2000;14:2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buijs R, Salgado R, Sabath E, et al. Peripheral circadian oscillators: time and food. Prog Mol Biol Transl Sci 2013;119:83–103. [DOI] [PubMed] [Google Scholar]
- 6.Shetty A, Hsu JW, Manka PP, et al. Role of the Circadian Clock in the Metabolic Syndrome and Nonalcoholic Fatty Liver Disease. Dig Dis Sci 2018;63:3187–3206. [DOI] [PubMed] [Google Scholar]
- 7.Shi D, Chen J, Wang J, et al. Circadian Clock Genes in the Metabolism of Non-alcoholic Fatty Liver Disease. Front Physiol 2019;10:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnocchi D, Custodero C, Sabba C, et al. Circadian rhythms: a possible new player in non-alcoholic fatty liver disease pathophysiology. J Mol Med (Berl) 2019;97:741–759. [DOI] [PubMed] [Google Scholar]
- 9.Reinke H, Asher G. Circadian Clock Control of Liver Metabolic Functions. Gastroenterology 2016;150:574–80. [DOI] [PubMed] [Google Scholar]
- 10.Tahara Y, Shibata S. Circadian rhythms of liver physiology and disease: experimental and clinical evidence. Nat Rev Gastroenterol Hepatol 2016;13:217–26. [DOI] [PubMed] [Google Scholar]
- 11.Mukherji A, Bailey SM, Staels B, et al. The circadian clock and liver function in health and disease. J Hepatol 2019;71:200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarrinpar A, Chaix A, Panda S. Daily Eating Patterns and Their Impact on Health and Disease. Trends Endocrinol Metab 2016;27:69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cederroth CR, Albrecht U, Bass J, et al. Medicine in the Fourth Dimension. Cell Metab 2019;30:238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challet E The circadian regulation of food intake. Nat Rev Endocrinol 2019;15:393–405. [DOI] [PubMed] [Google Scholar]
- 15.Panda S Circadian physiology of metabolism. Science 2016;354:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenvers DJ, Scheer F, Schrauwen P, et al. Circadian clocks and insulin resistance. Nat Rev Endocrinol 2019;15:75–89. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Wu Y, Li L, et al. Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix-loop-helix domains with E-box DNA. Cell Res 2013;23:213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan J, Wang H, Liu Y, et al. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol 2008;4:e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knippschild U, Gocht A, Wolff S, et al. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal 2005;17:675–89. [DOI] [PubMed] [Google Scholar]
- 20.Busino L, Bassermann F, Maiolica A, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 2007;316:900–4. [DOI] [PubMed] [Google Scholar]
- 21.Vielhaber E, Eide E, Rivers A, et al. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol 2000;20:4888–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Fang B, Emmett MJ, et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science 2015;348:1488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato TK, Panda S, Miraglia LJ, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004;43:527–37. [DOI] [PubMed] [Google Scholar]
- 24.Preitner N, Damiola F, Lopez-Molina L, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002;110:251–60. [DOI] [PubMed] [Google Scholar]
- 25.Everett LJ, Lazar MA. Nuclear receptor Rev-erbalpha: up, down, and all around. Trends Endocrinol Metab 2014;25:586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol 2005;12:441–8. [DOI] [PubMed] [Google Scholar]
- 27.Guillaumond F, Dardente H, Giguère V, et al. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms 2005;20:391–403. [DOI] [PubMed] [Google Scholar]
- 28.Kato Y, Kawamoto T, Fujimoto K, et al. DEC1/STRA13/SHARP2 and DEC2/SHARP1 coordinate physiological processes, including circadian rhythms in response to environmental stimuli. Curr Top Dev Biol 2014;110:339–72. [DOI] [PubMed] [Google Scholar]
- 29.Honma S, Kawamoto T, Takagi Y, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 2002;419:841–4. [DOI] [PubMed] [Google Scholar]
- 30.Yamajuku D, Shibata Y, Kitazawa M, et al. Cellular DBP and E4BP4 proteins are critical for determining the period length of the circadian oscillator. FEBS Lett 2011;585:2217–22. [DOI] [PubMed] [Google Scholar]
- 31.Yoshitane H, Asano Y, Sagami A, et al. Functional D-box sequences reset the circadian clock and drive mRNA rhythms. Commun Biol 2019;2:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duffield GE. DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocrinol 2003;15:991–1002. [DOI] [PubMed] [Google Scholar]
- 33.Zhang R, Lahens NF, Ballance HI, et al. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 2014; 111:16219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamia KA, Sachdeva UM, DiTacchio L, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 2009;326:437–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim EJ, Yoon YS, Hong S, et al. Retinoic acid receptor-related orphan receptor alpha-induced activation of adenosine monophosphate-activated protein kinase results in attenuation of hepatic steatosis. Hepatology 2012;55:1379–88. [DOI] [PubMed] [Google Scholar]
- 36.Travnickova-Bendova Z, Cermakian N, Reppert SM, et al. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A 2002;99:7728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neill JS, Maywood ES, Chesham JE, et al. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 2008;320:949–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun X, Dang F, Zhang D, et al. Glucagon-CREB/CRTC2 signaling cascade regulates hepatic BMAL1 protein. J Biol Chem 2015;290:2189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang EE, Liu Y, Dentin R, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 2010;16:1152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalsbeek A, la Fleur S, Fliers E. Circadian control of glucose metabolism. Mol Metab 2014;3:372–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luciano AK, Zhou W, Santana JM, et al. CLOCK phosphorylation by AKT regulates its nuclear accumulation and circadian gene expression in peripheral tissues. J Biol Chem 2018;293:9126–9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabinowitz JD, White E. Autophagy and metabolism. Science 2010;330:1344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khapre RV, Kondratova AA, Patel S, et al. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany NY) 2014;6:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakahata Y, Sahar S, Astarita G, et al. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009;324:654–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eckel-Mahan K, Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol Rev 2013;93:107–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 2011;13:125–37. [DOI] [PubMed] [Google Scholar]
- 48.Zhou B, Zhang Y, Zhang F, et al. CLOCK/BMAL1 regulates circadian change of mouse hepatic insulin sensitivity by SIRT1. Hepatology 2014;59:2196–206. [DOI] [PubMed] [Google Scholar]
- 49.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008;134:329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka S, Hikita H, Tatsumi T, et al. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology 2016;64:1994–2014. [DOI] [PubMed] [Google Scholar]
- 51.Lin CW, Zhang H, Li M, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol 2013;58:993–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fucho R, Martinez L, Baulies A, et al. ASMase regulates autophagy and lysosomal membrane permeabilization and its inhibition prevents early stage non-alcoholic steatohepatitis. J Hepatol 2014;61:1126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albrecht U Timing to perfection: the biology of central and peripheral circadian clocks. Neuron 2012;74:246–60. [DOI] [PubMed] [Google Scholar]
- 54.Van Cauter E Sleep disturbances and insulin resistance. Diabet Med 2011;28:1455–62. [DOI] [PubMed] [Google Scholar]
- 55.Tahara Y, Otsuka M, Fuse Y, et al. Refeeding after fasting elicits insulin-dependent regulation of Per2 and Rev-erbalpha with shifts in the liver clock. J Biol Rhythms 2011;26:230–40. [DOI] [PubMed] [Google Scholar]
- 56.Sato F, Muragaki Y, Kawamoto T, et al. Rhythmic expression of DEC2 protein in vitro and in vivo. Biomed Rep 2016;4:704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zarrinpar A, Loomba R. Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and nonalcoholic fatty liver disease. Aliment Pharmacol Ther 2012;36:909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev 2007;87:1409–39. [DOI] [PubMed] [Google Scholar]
- 59.Gil-Lozano M, Mingomataj EL, Wu WK, et al. Circadian secretion of the intestinal hormone GLP-1 by the rodent L cell. Diabetes 2014;63:3674–85. [DOI] [PubMed] [Google Scholar]
- 60.Seghieri M, Christensen AS, Andersen A, et al. Future Perspectives on GLP-1 Receptor Agonists and GLP-1/glucagon Receptor Co-agonists in the Treatment of NAFLD. Front Endocrinol (Lausanne) 2018;9:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernsmeier C, Meyer-Gerspach AC, Blaser LS, et al. Glucose-induced glucagon-like Peptide 1 secretion is deficient in patients with non-alcoholic fatty liver disease. PLoS One 2014;9:e87488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Firneisz G, Varga T, Lengyel G, et al. Serum dipeptidyl peptidase-4 activity in insulin resistant patients with non-alcoholic fatty liver disease: a novel liver disease biomarker. PLoS One 2010;5:e12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galindo Munoz JS, Jimenez Rodriguez D, Hernandez Morante JJ. Diurnal rhythms of plasma GLP-1 levels in normal and overweight/obese subjects: lack of effect of weight loss. J Physiol Biochem 2015;71:17–28. [DOI] [PubMed] [Google Scholar]
- 64.Martchenko A, Oh RH, Wheeler SE, et al. Suppression of circadian secretion of glucagon-like peptide-1 by the saturated fatty acid, palmitate. Acta Physiol (Oxf) 2018;222:e13007. [DOI] [PubMed] [Google Scholar]
- 65.Gil-Lozano M, Wu WK, Martchenko A, et al. High-Fat Diet and Palmitate Alter the Rhythmic Secretion of Glucagon-Like Peptide-1 by the Rodent L-cell. Endocrinology 2016;157:586–99. [DOI] [PubMed] [Google Scholar]
- 66.Gil-Lozano M, Hunter PM, Behan LA, et al. Short-term sleep deprivation with nocturnal light exposure alters time-dependent glucagon-like peptide-1 and insulin secretion in male volunteers. Am J Physiol Endocrinol Metab 2016;310:E41–50. [DOI] [PubMed] [Google Scholar]
- 67.Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 2010;466:627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brubaker PL, Gil-Lozano M. Glucagon-like peptide-1: The missing link in the metabolic clock? J Diabetes Investig 2016;7 Suppl 1:70–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A 2008;105:15172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frese T, Bazwinsky I, Muhlbauer E, et al. Circadian and age-dependent expression patterns of GLUT2 and glucokinase in the pancreatic beta-cell of diabetic and nondiabetic rats. Horm Metab Res 2007;39:567–74. [DOI] [PubMed] [Google Scholar]
- 71.Doi R, Oishi K, Ishida N. CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J Biol Chem 2010;285:22114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu C, Li S, Liu T, et al. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature 2007;447:477–81. [DOI] [PubMed] [Google Scholar]
- 73.Turek FW, Joshu C, Kohsaka A, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005;308:1043–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rudic RD, McNamara P, Curtis AM, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2004;2:e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamia KA, Papp SJ, Yu RT, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 2011;480:552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toledo M, Batista-Gonzalez A, Merheb E, et al. Autophagy Regulates the Liver Clock and Glucose Metabolism by Degrading CRY1. Cell Metab 2018;28:268–281 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeda Y, Kang HS, Freudenberg J, et al. Retinoic acid-related orphan receptor gamma (RORgamma): a novel participant in the diurnal regulation of hepatic gluconeogenesis and insulin sensitivity. PLoS Genet 2014;10:e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Knebel B, Haas J, Hartwig S, et al. Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass. PLoS One 2012;7:e31812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y, Xu S, Mihaylova MM, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab 2011;13:376–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Papazyan R, Damle M, et al. The hepatic circadian clock fine-tunes the lipogenic response to feeding through RORalpha/gamma. Genes Dev 2017;31:1202–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vollmers C, Gill S, DiTacchio L, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 2009;106:21453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaix A, Lin T, Le HD, et al. Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Le Martelot G, Claudel T, Gatfield D, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol 2009;7:e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duez H, van der Veen JN, Duhem C, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology 2008;135:689–98. [DOI] [PubMed] [Google Scholar]
- 85.Ferrell JM, Chiang JY. Short-term circadian disruption impairs bile acid and lipid homeostasis in mice. Cell Mol Gastroenterol Hepatol 2015;1:664–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma K, Xiao R, Tseng HT, et al. Circadian dysregulation disrupts bile acid homeostasis. PLoS One 2009;4:e6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang X, Downes M, Yu RT, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell 2006;126:801–10. [DOI] [PubMed] [Google Scholar]
- 88.Stroeve JH, Brufau G, Stellaard F, et al. Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab Invest 2010;90:1457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet 2015;385:956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y, Castellani LW, Sinal CJ, et al. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev 2004;18:157–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang YK, Guo GL, Klaassen CD. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS One 2011;6:e16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Galman C, Angelin B, Rudling M. Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology 2005;129:1445–53. [DOI] [PubMed] [Google Scholar]
- 93.Davies SP, Carling D, Munday MR, et al. Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP-activated protein kinase, demonstrated using freeze-clamping. Effects of high fat diets. Eur J Biochem 1992;203:615–23. [DOI] [PubMed] [Google Scholar]
- 94.Mao J, DeMayo FJ, Li H, et al. Liver-specific deletion of acetyl-CoA carboxylase 1 reduces hepatic triglyceride accumulation without affecting glucose homeostasis. Proc Natl Acad Sci U S A 2006;103:8552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abu-Elheiga L, Wu H, Gu Z, et al. Acetyl-CoA carboxylase 2−/− mutant mice are protected against fatty liver under high-fat, high-carbohydrate dietary and de novo lipogenic conditions. J Biol Chem 2012;287:12578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goedeke L, Bates J, Vatner DF, et al. Acetyl-CoA Carboxylase Inhibition Reverses NAFLD and Hepatic Insulin Resistance but Promotes Hypertriglyceridemia in Rodents. Hepatology 2018;68:2197–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol 2015;62:720–33. [DOI] [PubMed] [Google Scholar]
- 98.Stienstra R, Mandard S, Patsouris D, et al. Peroxisome proliferator-activated receptor alpha protects against obesity-induced hepatic inflammation. Endocrinology 2007;148:2753–63. [DOI] [PubMed] [Google Scholar]
- 99.Patsouris D, Reddy JK, Muller M, et al. Peroxisome proliferator-activated receptor alpha mediates the effects of high-fat diet on hepatic gene expression. Endocrinology 2006;147:1508–16. [DOI] [PubMed] [Google Scholar]
- 100.Aithal GP, Thomas JA, Kaye PV, et al. Randomized, placebo-controlled trial of pioglitazone in nondiabetic subjects with nonalcoholic steatohepatitis. Gastroenterology 2008;135:1176–84. [DOI] [PubMed] [Google Scholar]
- 101.Cusi K, Orsak B, Bril F, et al. Long-Term Pioglitazone Treatment for Patients With Nonalcoholic Steatohepatitis and Prediabetes or Type 2 Diabetes Mellitus: A Randomized Trial. Ann Intern Med 2016;165:305–15. [DOI] [PubMed] [Google Scholar]
- 102.Sanyal AJ, Chalasani N, Kowdley KV, et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med 2010;362:1675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kawai M, Rosen CJ. PPARγ: a circadian transcription factor in adipogenesis and osteogenesis. Nature reviews. Endocrinology 2010;6:629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Inoue I, Shinoda Y, Ikeda M, et al. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb 2005;12:169–74. [DOI] [PubMed] [Google Scholar]
- 105.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, et al. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 2010;24:345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang Z, Kim H, Ali A, et al. Interaction between stress responses and circadian metabolism in metabolic disease. Liver Res 2017;1:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chaix A, Zarrinpar A, Panda S. The circadian coordination of cell biology. J Cell Biol 2016;215:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007;8:519–29. [DOI] [PubMed] [Google Scholar]
- 109.Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab 2012;15:623–34. [DOI] [PubMed] [Google Scholar]
- 110.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov 2013;12:703–19. [DOI] [PubMed] [Google Scholar]
- 111.Dara L, Ji C, Kaplowitz N. The contribution of endoplasmic reticulum stress to liver diseases. Hepatology 2011. ;53:1752–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab 2010;11:47–57. [DOI] [PubMed] [Google Scholar]
- 113.Zhang K, Shen X, Wu J, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 2006;124:587–99. [DOI] [PubMed] [Google Scholar]
- 114.Zhang C, Wang G, Zheng Z, et al. Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology 2012;55:1070–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nakagawa Y, Shimano H. CREBH Regulates Systemic Glucose and Lipid Metabolism. Int J Mol Sci 2018; 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim H, Zheng Z, Walker PD, et al. CREBH Maintains Circadian Glucose Homeostasis by Regulating Hepatic Glycogenolysis and Gluconeogenesis. Mol Cell Biol 2017;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng Z, Kim H, Qiu Y, et al. CREBH Couples Circadian Clock With Hepatic Lipid Metabolism. Diabetes 2016;65:3369–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Park JG, Xu X, Cho S, et al. CREBH-FGF21 axis improves hepatic steatosis by suppressing adipose tissue lipolysis. Sci Rep 2016;6:27938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakagawa Y, Oikawa F, Mizuno S, et al. Hyperlipidemia and hepatitis in liver-specific CREB3L3 knockout mice generated using a one-step CRISPR/Cas9 system. Sci Rep 2016;6:27857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakagawa Y, Satoh A, Tezuka H, et al. CREB3L3 controls fatty acid oxidation and ketogenesis in synergy with PPARalpha. Sci Rep 2016;6:39182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stokkan KA, Yamazaki S, Tei H, et al. Entrainment of the circadian clock in the liver by feeding. Science 2001;291:490–3. [DOI] [PubMed] [Google Scholar]
- 122.Greenwell BJ, Trott AJ, Beytebiere JR, et al. Rhythmic Food Intake Drives Rhythmic Gene Expression More Potently than the Hepatic Circadian Clock in Mice. Cell Rep 2019;27:649–657 e5. [DOI] [PubMed] [Google Scholar]
- 123.Tripathi A, Debelius J, Brenner DA, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 2018;15:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leung C, Rivera L, Furness JB, et al. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol 2016;13:412–25. [DOI] [PubMed] [Google Scholar]
- 125.Frazier K, Chang EB. Intersection of the Gut Microbiome and Circadian Rhythms in Metabolism. Trends Endocrinol Metab 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thaiss CA, Zeevi D, Levy M, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014;159:514–29. [DOI] [PubMed] [Google Scholar]
- 127.Thaiss CA, Levy M, Korem T, et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 2016;167:1495–1510 e12. [DOI] [PubMed] [Google Scholar]
- 128.Liang X, Bushman FD, FitzGerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci U S A 2015;112:10479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leone V, Gibbons SM, Martinez K, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 2015;17:681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zarrinpar A, Chaix A, Yooseph S, et al. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 2014;20:1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Uhr GT, Dohnalova L, Thaiss CA. The Dimension of Time in Host-Microbiome Interactions. mSystems 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takayasu L, Suda W, Takanashi K, et al. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res 2017;24:261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Collado MC, Engen PA, Bandin C, et al. Timing of food intake impacts daily rhythms of human salivary microbiota: a randomized, crossover study. FASEB J 2018;32:2060–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaczmarek JL, Musaad SM, Holscher HD. Time of day and eating behaviors are associated with the composition and function of the human gastrointestinal microbiota. Am J Clin Nutr 2017;106:1220–1231. [DOI] [PubMed] [Google Scholar]
- 135.Skarke C, Lahens NF, Rhoades SD, et al. A Pilot Characterization of the Human Chronobiome. Sci Rep 2017;7:17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mukherji A, Kobiita A, Ye T, et al. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013;153:812–27. [DOI] [PubMed] [Google Scholar]
- 137.Zarrinpar A, Chaix A, Xu ZZ, et al. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat Commun 2018;9:2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oh HYP, Ellero-Simatos S, Manickam R, et al. Depletion of Gram-Positive Bacteria Impacts Hepatic Biological Functions During the Light Phase. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Y, Kuang Z, Yu X, et al. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 2017;357:912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lavery DJ, Schibler U. Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev 1993;7:1871–84. [DOI] [PubMed] [Google Scholar]
- 141.Govindarajan K, MacSharry J, Casey PG, et al. Unconjugated Bile Acids Influence Expression of Circadian Genes: A Potential Mechanism for Microbe-Host Crosstalk. PLoS One 2016;11:e0167319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Joyce SA, MacSharry J, Casey PG, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A 2014;111:7421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Segers A, Desmet L, Thijs T, et al. The circadian clock regulates the diurnal levels of microbial short-chain fatty acids and their rhythmic effects on colon contractility in mice. Acta Physiol (Oxf) 2019;225:e13193. [DOI] [PubMed] [Google Scholar]
- 144.Kohsaka A, Laposky AD, Ramsey KM, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 2007;6:414–21. [DOI] [PubMed] [Google Scholar]
- 145.Ando H, Kumazaki M, Motosugi Y, et al. Impairment of peripheral circadian clocks precedes metabolic abnormalities in ob/ob mice. Endocrinology 2011;152:1347–54. [DOI] [PubMed] [Google Scholar]
- 146.Ho A, Chin A. Circadian feeding and drinking patterns of genetically obese mice fed solid chow diet. Physiol Behav 1988;43:651–6. [DOI] [PubMed] [Google Scholar]
- 147.Sookoian S, Castano G, Gemma C, et al. Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J Gastroenterol 2007;13:4242–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sookoian S, Gemma C, Gianotti TF, et al. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr 2008;87:1606–15. [DOI] [PubMed] [Google Scholar]
- 149.Willer CJ, Bonnycastle LL, Conneely KN, et al. Screening of 134 single nucleotide polymorphisms (SNPs) previously associated with type 2 diabetes replicates association with 12 SNPs in nine genes. Diabetes 2007;56:256–64. [DOI] [PubMed] [Google Scholar]
- 150.Potter GD, Skene DJ, Arendt J, et al. Circadian Rhythm and Sleep Disruption: Causes, Metabolic Consequences, and Countermeasures. Endocr Rev 2016;37:584–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hatori M, Vollmers C, Zarrinpar A, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 2012;15:848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Longo VD, Panda S. Fasting, Circadian Rhythms, and Time-Restricted Feeding in Healthy Lifespan. Cell Metab 2016;23:1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Manoogian ENC, Panda S. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res Rev 2017;39:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Melkani GC, Panda S. Time-restricted feeding for prevention and treatment of cardiometabolic disorders. J Physiol 2017;595:3691–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chaix A, Manoogian ENC, Melkani GC, et al. Time-Restricted Eating to Prevent and Manage Chronic Metabolic Diseases. Annu Rev Nutr 2019;39:291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chaix A, Zarrinpar A, Miu P, et al. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab 2014;20:991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sherman H, Genzer Y, Cohen R, et al. Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB J 2012;26:3493–502. [DOI] [PubMed] [Google Scholar]
- 158.Gill S, Le HD, Melkani GC, et al. Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science 2015;347:1265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Villanueva JE, Livelo C, Trujillo AS, et al. Time-restricted feeding restores muscle function in Drosophila models of obesity and circadian-rhythm disruption. Nat Commun 2019;10:2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gill S, Panda S. A Smartphone App Reveals Erratic Diurnal Eating Patterns in Humans that Can Be Modulated for Health Benefits. Cell Metab 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Carlson O, Martin B, Stote KS, et al. Impact of reduced meal frequency without caloric restriction on glucose regulation in healthy, normal-weight middle-aged men and women. Metabolism 2007;56:1729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Moro T, Tinsley G, Bianco A, et al. Effects of eight weeks of time-restricted feeding (16/8) on basal metabolism, maximal strength, body composition, inflammation, and cardiovascular risk factors in resistance-trained males. J Transl Med 2016;14:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sutton EF, Beyl R, Early KS, et al. Early Time-Restricted Feeding Improves Insulin Sensitivity, Blood Pressure, and Oxidative Stress Even without Weight Loss in Men with Prediabetes. Cell Metab 2018;27:1212–1221 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hutchison AT, Regmi P, Manoogian ENC, et al. Time-Restricted Feeding Improves Glucose Tolerance in Men at Risk for Type 2 Diabetes: A Randomized Crossover Trial. Obesity (Silver Spring) 2019;27:724–732. [DOI] [PubMed] [Google Scholar]
- 165.Haas JT, Francque S, Staels B. Pathophysiology and Mechanisms of Nonalcoholic Fatty Liver Disease. Annu Rev Physiol 2016;78:181–205. [DOI] [PubMed] [Google Scholar]
- 166.Harrison SA, Rinella ME, Abdelmalek MF, et al. NGM282 for treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2018;391:1174–1185. [DOI] [PubMed] [Google Scholar]
- 167.Hartmann P, Hochrath K, Horvath A, et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology 2018;67:2150–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell 2012;47:158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hirota T, Lee JW, St John PC, et al. Identification of small molecule activators of cryptochrome. Science 2012;337:1094–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen Z, Yoo SH, Takahashi JS. Development and Therapeutic Potential of Small-Molecule Modulators of Circadian Systems. Annu Rev Pharmacol Toxicol 2018;58:231–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lee YS, Cha BY, Saito K, et al. Nobiletin improves hyperglycemia and insulin resistance in obese diabetic ob/ob mice. Biochem Pharmacol 2010;79:1674–83. [DOI] [PubMed] [Google Scholar]
- 172.He B, Nohara K, Park N, et al. The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab 2016;23:610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Solt LA, Wang Y, Banerjee S, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012;485:62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen Z, Yoo SH, Takahashi JS. Small molecule modifiers of circadian clocks. Cell Mol Life Sci 2013;70:2985–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Erkekoglu P, Baydar T. Chronopharmacokinetics of drugs in toxicological aspects: A short review for pharmacy practitioners. J Res Pharm Pract 2012;1:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dallmann R, Brown SA, Gachon F. Chronopharmacology: new insights and therapeutic implications. Annu Rev Pharmacol Toxicol 2014;54:339–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bugge A, Feng D, Everett LJ, et al. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev 2012;26:657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Delezie J, Dumont S, Dardente H, et al. The nuclear receptor REV-ERBalpha is required for the daily balance of carbohydrate and lipid metabolism. FASEB J 2012;26:3321–35. [DOI] [PubMed] [Google Scholar]
- 179.Adamovich Y, Rousso-Noori L, Zwighaft Z, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab 2014;19:319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Barclay JL, Shostak A, Leliavski A, et al. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am J Physiol Endocrinol Metab 2013;304:E1053–63. [DOI] [PubMed] [Google Scholar]
- 181.Shimba S, Ogawa T, Hitosugi S, et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One 2011;6:e25231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kudo T, Tamagawa T, Kawashima M, et al. Attenuating effect of clock mutation on triglyceride contents in the ICR mouse liver under a high-fat diet. J Biol Rhythms 2007;22:312–23. [DOI] [PubMed] [Google Scholar]
- 183.Chen P, Han Z, Yang P, et al. Loss of clock gene mPer2 promotes liver fibrosis induced by carbon tetrachloride. Hepatol Res 2010;40:1117–27. [DOI] [PubMed] [Google Scholar]
- 184.Feng D, Liu T, Sun Z, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 2011. ;331:1315–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Spiegel K, Leproult R, Van Cauter E. Impact of sleep debt on metabolic and endocrine function. Lancet 1999;354:1435–9. [DOI] [PubMed] [Google Scholar]
- 186.Wang F, Zhang L, Zhang Y, et al. Meta-analysis on night shift work and risk of metabolic syndrome. Obes Rev 2014;15:709–20. [DOI] [PubMed] [Google Scholar]
- 187.Vyas MV, Garg AX, Iansavichus AV, et al. Shift work and vascular events: systematic review and meta-analysis. BMJ 2012;345:e4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med 2001;58:747–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wilkinson MJ, Manoogian ENC, Zadourian A, et al. Ten-hour time-restricted eating reduces weight, blood pressure, and atherogenic lipids in patients with metabolic syndrome. Cell Metab 2020;31:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hsieh SD, Muto T, Murase T, et al. Association of short sleep duration with obesity, diabetes, fatty liver and behavioral factors in Japanese men. Intern Med 2011;50:2499–502. [DOI] [PubMed] [Google Scholar]
- 191.Kim CW, Yun KE, Jung HS, et al. Sleep duration and quality in relation to non-alcoholic fatty liver disease in middle-aged workers and their spouses. J Hepatol 2013;59:351–7. [DOI] [PubMed] [Google Scholar]
- 192.Imaizumi H, Takahashi A, Tanji N, et al. The Association between Sleep Duration and Non-Alcoholic Fatty Liver Disease among Japanese Men and Women. Obesity facts 2015;8:234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bernsmeier C, Weisskopf DM, Pflueger MO, et al. Sleep Disruption and Daytime Sleepiness Correlating with Disease Severity and Insulin Resistance in Non-Alcoholic Fatty Liver Disease: A Comparison with Healthy Controls. PLoS One 2015;10:e0143293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Marin-Alejandre BA, Abete I, Cantero I, et al. Association between Sleep Disturbances and Liver Status in Obese Subjects with Nonalcoholic Fatty Liver Disease: A Comparison with Healthy Controls. Nutrients 2019; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Liu C, Zhong R, Lou J, et al. Nighttime sleep duration and risk of nonalcoholic fatty liver disease: the Dongfeng-Tongji prospective study. Ann Med 2016;48:468–476. [DOI] [PubMed] [Google Scholar]
- 196.Balakrishnan M, El-Serag HB, Kanwal F, et al. Shiftwork Is Not Associated with Increased Risk of NAFLD: Findings from the National Health and Nutrition Examination Survey. Dig Dis Sci 2017;62:526–533. [DOI] [PubMed] [Google Scholar]
- 197.Katsagoni CN, Papatheodoridis GV, Ioannidou P, et al. Improvements in clinical characteristics of patients with non-alcoholic fatty liver disease, after an intervention based on the Mediterranean lifestyle: a randomised controlled clinical trial. Br J Nutr 2018;120:164–175. [DOI] [PubMed] [Google Scholar]
- 198.Katsagoni CN, Georgoulis M, Papatheodoridis GV, et al. Associations Between Lifestyle Characteristics and the Presence of Nonalcoholic Fatty Liver Disease: A Case-Control Study. Metab Syndr Relat Disord 2017;15:72–79. [DOI] [PubMed] [Google Scholar]
- 199.Musso G, Cassader M, Olivetti C, et al. Association of obstructive sleep apnoea with the presence and severity of non-alcoholic fatty liver disease. A systematic review and meta-analysis. Obes Rev 2013;14:417–31. [DOI] [PubMed] [Google Scholar]
- 200.Pamidi S, Tasali E. Obstructive sleep apnea and type 2 diabetes: is there a link? Front Neurol 2012;3:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


