Abstract
The NAC gene family is one of the important plant-specific transcription factor families involved in variety of physiological processes. It has been found in several plant species; however, little is known about the gene family in ginseng, Panax ginseng C.A. Meyer. Here we report identification and systematic analysis of this gene family in ginseng. A total of 89 NAC genes, designated PgNAC01 to PgNAC89, are identified. These genes are alternatively spliced into 251 transcripts at fruiting stage of a four-year-old ginseng plant. The genes of this gene family have five conserved motifs and are clustered into 11 subfamilies, all of which are shared with the genes of the NAC gene families identified in the dicot and monocot model plant species, Arabidopsis and rice. This result indicates that the PgNAC gene family is an ancient and evolutionarily inactive gene family. Gene ontology (GO) analysis shows that the functions of the PgNAC gene family have been substantially differentiated; nevertheless, over 86% the PgNAC transcripts remain functionally correlated. Finally, five of the PgNAC genes, PgNAC05-2, PgNAC41-2, PgNAC48, PgNAC56-1, and PgNAC59, are identified to be involved in plant response to cold stress, suggesting that this gene family plays roles in response to cold stress in ginseng. These results, therefore, provide new insights into functional differentiation and evolution of a gene family in plants and gene resources necessary to comprehensively determine the functions of the PgNAC gene family in response to cold and other abiotic stresses in ginseng.
Introduction
Plants often suffer from a variety of adverse environments, such as salt, drought, cold, pathogens, and insect pests, due to their sessile lifestyle [1]. These biotic and abiotic stresses directly or indirectly affect plant growth and development, thus influencing plant production. To cope with these various environmental stresses, plants have evolved a series of defense mechanisms to resile from these stresses. As the molecular switch that can activate or inactivate gene expression, transcription factors play important roles in plant response to these stresses. The NAC transcription factor gene family is one of the large plant-specific gene families that plays vital roles in response to adverse environmental stresses [1,2].
The NAC gene family is designated, after the initials of three genes, NAM (NO APICAL MERISTEM, petunia), ATAF1/2 (ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR, Arabidopsis) and CUC2 (CUP-SHAPE COTYLEDON, Arabidopsis). The NAC domain at the N-terminus of the NAC protein is highly conserved across all the NAC proteins identified to date in plants. This domain contains five converted motifs, designated from A to E. Motif A may be involved in the formation of dimers; Motifs C and D are highly conserved and involved in DNA binding [3]; Motifs B and E are relatively variable and may be involved in the functional diversification of the NAC gene family. The C-terminus of the NAC protein is a highly diverged transcriptional regulatory region with transcriptional activator or repressor activity. In addition, some NAC transcription factors contain an α-helical transmembrane motif at their C-termini that is responsible for anchoring to plasma membrane or endoplasmic reticulum, known as the NAC membrane-bound transcription factor [4]. The NAC gene family widely exists in plant species [5–7]. After the first NAC transcription factor-encoding gene, NAM, was identified from petunia, numerous genes coding NAC proteins have been identified from 57 plant species, including Arabidopsis thaliana, Oryza sativa, Zea mays, Nicotiana tabacum, and so on. Nevertheless, little is known about this gene family in ginseng, Panax ginseng C.A. Meyer.
The NAC transcription factors have been shown to participate in plant response to several abiotic stresses [8–10]. For instance, ANAC096 is a positive response element to dehydration and osmotic stress in the ABA-dependent signal transduction pathway in Arabidopsis. The mutant of this gene, anac096, is less sensitive to exogenous ABA, Consequently, ABA-induced stomatal closure of the mutant was impaired and water loss was increased under drought stress conditions. On the other hand, the ANAC096-overexpressing (ANACO96OX) plants showed higher sensitive to exogenous ABA and enhanced tolerance to dehydration stress [11]. NTL4 (NAC with transmembrane motif 1-like) is another stress-related NAC transcription factor identified in Arabidopsis. Leaf senescence triggered by ROS (reactive oxygen species) can help plant survival under drought condition. By stimulating ROS production, NTL4 induced leaf senescence and sustained plant survival [12]. The rice OsNAP gene was significantly induced by ABA and abiotic stresses. The overexpression of OsNAP enhanced tolerance to drought and salinity, and increased grain yields [13]. The overexpression of tomato SlNAM1 increased the content of osmotic adjustment substance and decreased the content of H2O2 and O2•- under low temperature stress [14]. The overexpression of MlNAC9 gene increased the germination rate and root growth of seedlings in Miscanthus, and enhanced tolerance to drought and cold stresses in Arabidopsis transgenic plants [15]. Therefore, as one of the gene families that are responsible for plant response to different abiotic stresses, identification of new members of the PgNAC gene family that are involved in abiotic stress responses will facilitate research in the functions of the gene family in response to abiotic stresses in ginseng.
Ginseng is a perennial overwintering herb in the Araliaceae family. It has been historically widely used for human health and medicine for thousands of years, and has been a model species for medicinal plant research in medicinal chemistry, genetics and genomics [16]. Ginseng is vulnerable to environmental stresses, and its production and quality are significantly affected by adverse environments. Therefore, it has become the urgent issue for ginseng production and industry to improve its tolerance to these adverse environmental stresses. It has been shown that ginseng responded to environmental stresses by enhancing the expressions of defense-related genes [17]. The present study took advantages of several transcriptomes recently developed in ginseng, identified the gene members of the NAC gene family in ginseng, designated hereafter PgNAC gene family, and characterized them in several aspects. Then, we identified five genes of the family that are responsive to cold stress. The findings of this study, therefore, provide new insights into functional differentiation and evolution of the NAC gene family in plants and NAC gene resources necessary to comprehensively elucidate the functions of the NAC gene family in ginseng, especially in plant growth, development, and response to abiotic stresses.
Materials and methods
Database resources
A transcriptome database consisting of 248,993 transcripts [17], herein designated Database A, was used for this study. This database was previously generated from 14 tissues of a four-year-old plant of Jilin ginseng cv. Damaya sampled at the fruiting stage, including fiber root, leg root, main root epiderm, main root cortex, rhizome, arm root, stem, leaf peduncle, leaflet pedicel, leaf blade, fruit peduncle, fruit pedicel, fruit flesh, and seed. Moreover, two other transcriptome databases, Database B and Database C, were also used for this study. Database B was developed from the roots of 5-, 12-, 18- and 25-year-old Jilin ginseng plants and Database C developed from the four-year-old plant roots of 42 genotypes (coded from S1 to S42) of Jilin ginseng [18].
Identification of PgNAC genes
Two methods were used to identify the PgNAC genes in ginseng from Jilin ginseng Database A [17]. The first one used the NAC domain of the NAC proteins (PF02365) (http://pfam.xfam.org) as queries to tblastn search Database A for PgNAC genes at E-value ≤ le-05. The second method used the NAC domain of these NAC proteins (PF02365) to establish the HMM (Hidden Markov Model) profile using the Hummer software. The protein sequences obtained by HMM were used as queries to also search Database A for PgNAC genes. Then, the NAC protein sequences obtained from the above two searches were combined and verified by existence of the NAC domain using NCBI CDD database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The genes confirmed to contain the NAC domain were designated PgNAC genes.
Protein conserved motifs and phylogeny of the PgNAC genes
Open-reading frame (ORF) search of the PgNAC genes was performed using the ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The conserved domains of the PgNAC genes that had complete ORFs were predicted using the NCBI CDD database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Then, these PgNAC genes were subjected to motif prediction using the MEME software [19]. A neighbor joining (NJ) phylogenetic tree was constructed using MEGA 5.0 based on multiple alignment [20]. The phylogenetic tree was constructed with 10,000 bootstrap replications and visualized using the iTOL software (https://itol.embl.de/itol.cgi).
Functional categorization and enrichment analysis of the PgNAC genes
The PgNAC genes were functionally categorized with gene ontology (GO) terms using the Blast2GO version 4.1.9 software [21]. The GO functional categorization of the transcripts of the entire Database A [17] was used as the background control or theoretical number of transcripts categorized into each subcategory (Level 2) for enrichment analysis of the PgNAC transcripts. Enrichment of the number of PgNAC transcripts categorized into each subcategory was determined by Chi-square test.
Construction of co-expression network of the PgNAC genes
All 251 PgNAC transcripts were subjected to co-expression network analysis. The co-expression network was constructed by BioLayout Express3D Version 3.2 software [22].
Roles of the PgNAC gene family in ginseng response to cold stress
Ginseng is a perennial winter herb to which cold tolerance is crucial. Therefore, we further examined the roles of the PgNAC gene family in response to cold stress. The hairy roots of ginseng were cultured on 1/2 MS medium at 22°C in dark for 28 days and then subjected to experimental treatment. For cold stress treatment, the hairy roots were stressed at 4°C with three biological replicates. For non-cold stress control, the hairy roots were continuously cultured at 22°C, also with three biological replicates. The hairy root samples cultured at 4°C (cold treated) and 22°C (non-cold treated) were collected at 6 h, 12 h, 24 h, 48 h, and 72 h, respectively, and stored at -80°C for further analysis.
Malondialdehyde assay
Malondialdehyde (MDA), a product of peroxidation of polyunsaturated fatty acids in phospholipids, has been widely used as an indicator for cell membrane damage resulting from abiotic stresses, including cold stress [23–25]. Therefore, we employed the content variations of MDA in cold-treated ginseng hairy roots relative to that of non-cold-treated ginseng hairy roots to confirm efficiency of the cold stress treatment. The content of MDA was measured according to Zhu et al. [26].
Expressions of PgNAC genes
The collected samples of hairy roots were subjected to RT-qPCR, with three biological and three technical replicates, to estimate the expressions of the PgNAC genes that were likely involved in ginseng response to abiotic stresses, including cold stress (for the PgNAC genes, see Results), in the cold-treated hairy roots relative to in the non-cold-treated hairy roots, Total RNA was extracted using the TRIzol reagent (Invitrogen) according to manufacturer's instructions (BioTeke Corporation). Reverse transcription was performed using a commercial kit (Beijing ComWin Biotech). GADPH was used as a reference gene. The RT-qPCR reactions were carried out with a pre-denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation (95°C for 15 s), annealing (60°C for 1 min), and extension (68°C for 10 s), and a final stage of 60°C - 95°C to determine the melting curves of the amplified products. The relative expression of a target gene was determined by the 2-ΔΔCt method [27].
Results
Identification and number variation of PgNAC genes
A total of 251 transcripts containing partial or complete NAC domains were obtained. These transcripts were alternatively spliced from 89 PgNAC genes. These 89 PgNAC genes were designated PgNAC01 to PgNAC89, with different transcripts derived from a PgNAC gene suffixed with “-01, -02…” (S1 Table). The nucleotide sequences of these 251 transcripts had a length ranging from 202 bp to 7,965 bp, with an average length of 1,627 bp.
When the 251 transcripts were aligned to three ginseng databases, with criteria of alignment length ≥ 200 nucleotides or amino acids, identity ≥ 90%, and e-value ≤ 1.0E-06, 105 (42%) of them were aligned to the ginseng cv. ChP genome [28], 116 (46%) aligned to the ginseng cv. ChP NAC gene database [29], and 111 (44%) aligned to the Strain IR826 protein database [30] (S2 Table). One hundred seventeen (47%) of them that were spliced from 55 (61.8%) of the 89 PgNAC genes identified in this study were aligned to none of the three ginseng databases, suggesting that they were highly likely specific for the genotype, Jilin ginseng cv. Damaya, analyzed in this study. This result also suggested that the number of genes in the PgNAC gene family varied among genotypes within P. ginseng. This discrepancy could be attributed to the variation in number of the PgNAC genes among genotypes, because it has been documented that the number of genes in a gene family or a genome varied dramatically among genotypes within a species [26, 31, 32].
Sequence analysis and motif identification
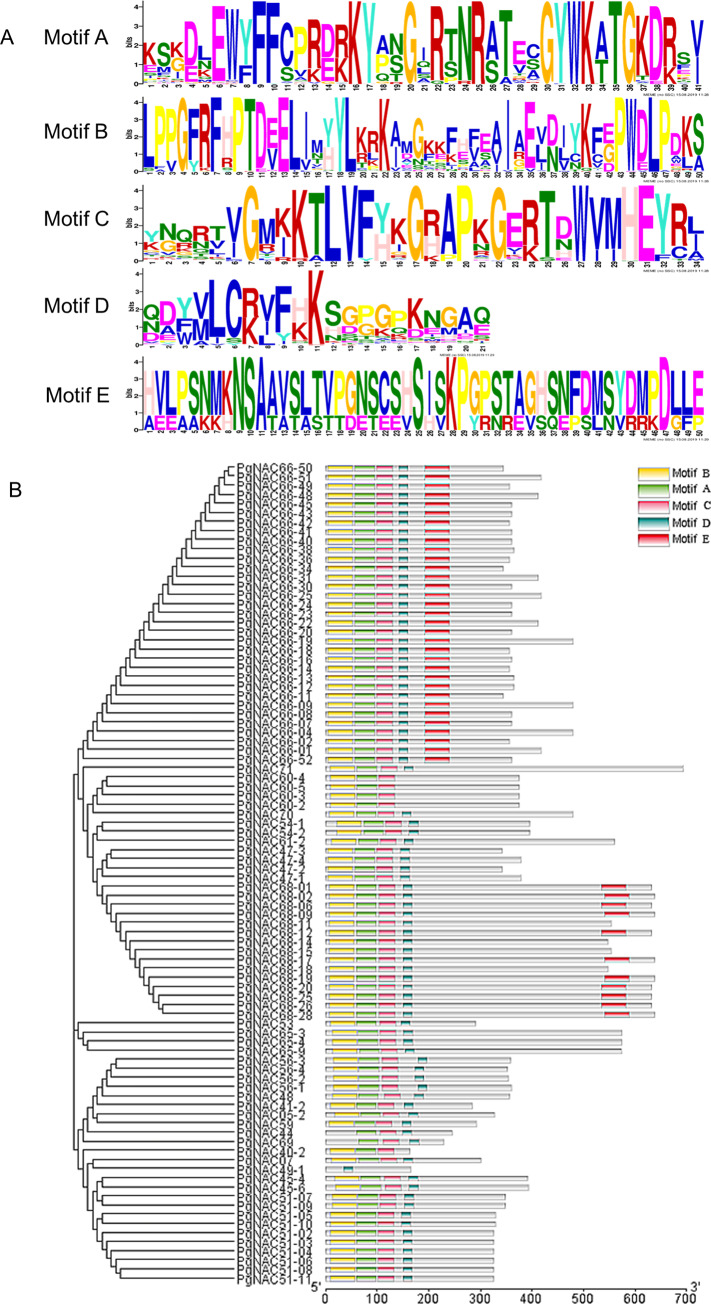
ORF analysis showed that 90 (35.9%) of the 251 transcripts that derived from 22 of the 89 PgNAC genes had complete NAC domains within ORFs. Therefore, these 90 PgNAC transcripts were used for motif identification. Five conserved motifs, A, B, C, D and E, were found (Fig 1A). The N-termini of the PgNAC putative proteins were highly conserved, suggesting that the conservation may be essential for the function of the NAC proteins. Of the 90 PgNAC putative proteins, 44 (48.9%) contained all five motifs; 33 (36.7%) contained four of the five motifs, A, B, C, and D; 6 (6.7%) contained three motifs, A, B, and C; 3 (3.3%) contained three motifs, B, C, and D; 3 (3.3%) contained two motifs, B and C; and 1 (1.1%) contained one motif, D (Fig 1B). Except for PgNAC49-1 whose putative protein only contained Motif D, the putative proteins of all remaining 89 PgNAC transcripts contained both Motifs B and C. Compared with Motifs B and C, Motif E was more variable. The results showed that Motifs B and C of the PgNAC putative proteins were highly conserved and shared with the NAC proteins of other species [33–35].
Fig 1.
The conserved motifs (A) and their distributions (B) in PgNAC putative proteins.
Phylogenetic analysis of the PgNAC gene family
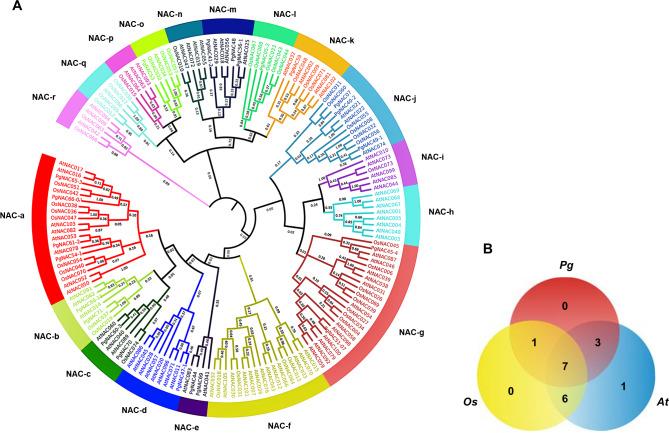
To determine the phylogeny of the PgNAC gene family, one PgNAC transcript that had the longest sequence and/or the largest number of conserved motifs were selected for each PgNAC gene. As a result, 22 PgNAC transcripts that were representative for 22 PgNAC genes were selected and used to construct the phylogenetic tree of the PgNAC gene family. Eighty-six AtNAC genes from Arabidopsis thaliana (At) and 50 OsNAC genes from Oryza sativa (Os) were used as evolutionary controls (S3 Table). The selected transcripts of the 22 PgNAC genes were translated into putative proteins, aligned and constructed into the phylogenetic tree of the PgNAC gene family (Fig 2A). The PgNAC genes were classified into 11 subfamilies, while the AtNAC genes and OsNAC genes were classified into 17 and 14 subfamilies, respectively. Of the 11 PgNAC gene subfamilies, 7 (63.6%) were classified with both the AtNAC genes and the OsNAC genes, 10 (90.9%) with the AtNAC genes, and 8 (72.7%) with the OsNAC genes (Fig 2B). No PgNAC gene was classified into a new subfamily. This result indicated that the PgNAC gene family is an ancient gene family originated before the divergence between the monocotyledon (rice) and dicotyledon (Arabidopsis and ginseng) plants. Nevertheless, the evolution of the PgNAC gene family had actively occurred after the separation of the dicotyledon (Arabidopsis and ginseng) plants from the monocotyledon (rice) plants because the PgNAC genes shared more subfamilies with the AtNAC genes than with the OsNAC genes. No significant evolution of the PgNAC gene family occurred afterward.
Fig 2. Phylogenetic tree of the PgNAC gene family.
(A) The neighbor-joining (NJ) phylogenetic tree of the PgNAC genes with the AtNAC genes in Arabidopsis thaliana (At) and OsNAC genes in rice (Oryza sativa, Os). The subfamilies of the NAC gene families are defined from NAC-a through NAC-r. The tree was constructed with 10,000 bootstrap replications. (B) Comparison of the subfamilies among the PgNAC, AtNAC and OsNAC gene families. The numbers in the Venn diagram indicate the numbers of subfamilies shared among the three NAC gene families and the number of subfamilies specific for each species.
The phylogenetic analysis also showed that some of PgNAC gene putative proteins were highly homologous with those of AtNAC and OsNAC genes, such as PgNAC61-2 with AtNAC053, PgNAC47-1 with AtNAC011, PgNAC44 with AtNAC083, PgNAC51-07 with AtNAC098, PgNAC07 with OsNAC060, PgNAC49-1 with AtNAC074, PgNAC41-2 with AtNAC029, PgNAC56-1 with AtNAC025, PgNAC53 with AtNAC036, and PgNAC60-3 with AtNAC060 (Fig 2A). These results indicated that these PgNAC genes may have the same or similar functions in ginseng to those in Arabidopsis and/or rice.
Functional differentiation of the PgNAC gene family
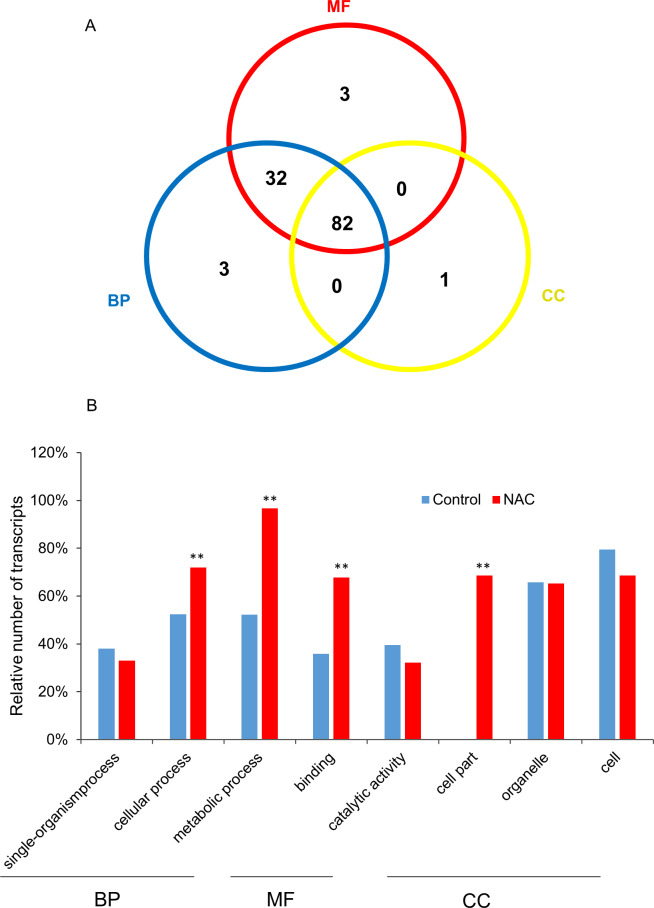
The gene members in a gene family, because they share high sequence identity and/or the same conserved domain(s), have been often considered to have the same or similar functions. Nevertheless, recent studies showed that the functions of the genes in a gene family have been dramatically differentiated, indicating that they may not have same or similar functions [26, 36, 37]. To understand the functional differentiation of the PgNAC gene family, the 251 PgNAC transcripts were annotated and functionally categorized. Only 121 (48%) of the 251 PgNAC transcripts were annotated, while the remaining 130 could not be annotated, suggesting the uniqueness of the PgNAC genes in ginseng. The annotated PgNAC transcripts were categorized into all three primary functional categories: biological process (BP), molecular function (MF) and cellular component (CC). Of the 121 transcripts annotated, 82 were categorized into all three primary categories (67.8%), MF, BP and CC; 32 (26.5%) into the BP and CC categories; and 7 (5.8%) into one of the three primary categories (Fig 3A). Of these seven transcripts, PgNAC67-2, PgNAC67-3, and PgNAC81 were categorized into BP; PgNAC63-78, PgNAC63-16, and PgNAC63-6 categorized into MF; and PgNAC10 categorized into CC (Fig 3A; S4 Table).
Fig 3. Functional categorization of the PgNAC transcripts.
(A) Venn diagram of the primary functional categorization of the PgNAC transcripts. BP, biological process; MF, molecular function; and CC, cellular component. (B) functional categorization of the PgNAC transcripts at Level 2 and their enrichments. The GO terms of the transcripts expressed in the 14 tissues of a four-year-old plant were used as the background control for the enrichment analysis. **, significant at P ≤ 0.01.
At Level 2, the 121 PgNAC transcripts were categorized into eight subcategories, including single-organism process (33.1%), cellular process (71.9%), metabolic process (96.7%), binding (67.8%), catalytic activity (32.2%), cell part (68.6%), organelle (65.3%), and cell (68.6%) (Fig 3B; S4 Table). Of the eight subcategories, the number of the PgNAC transcripts were significantly up-enriched, relative to the background control, in the cell process, metabolic process, binding, and cell part subcategories. These results indicated that the functions of PgNAC genes have substantially differentiated, ever though they have high sequence identity and all contain the NAC domain, further supporting the findings that the different genes in a gene family may not have same or similar functions [26, 36, 37].
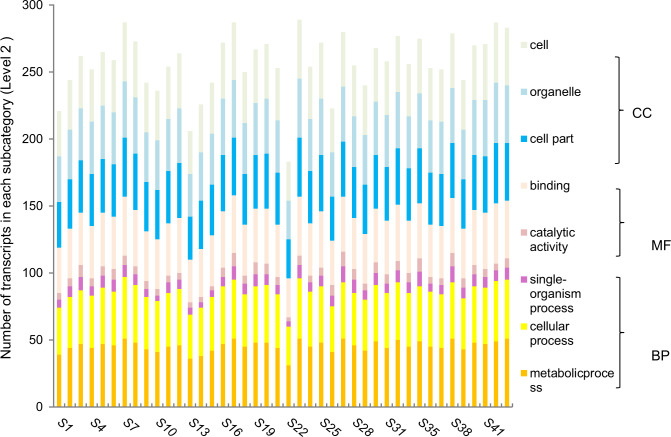
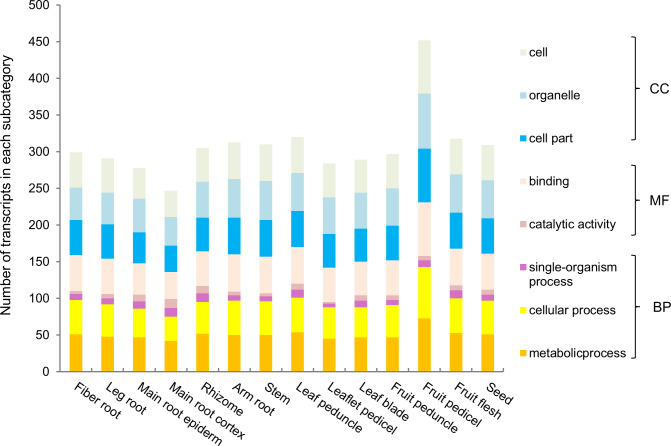
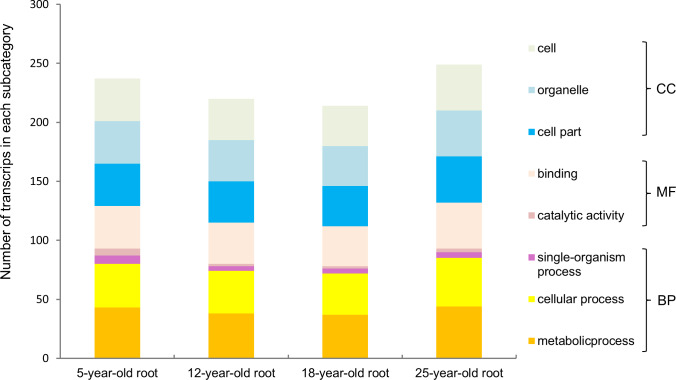
Furthermore, we functionally categorized the 121 annotated PgNAC transcripts in the four-year-old plant roots of 42 genotypes (Fig 4), in the 14 tissues of a four-year-old plant (Fig 5), and the roots of 5-, 12-, 18- and 25-year-old plants (Fig 6). Although the number of the PgNAC transcripts categorized into each subcategory (Level 2) substantially varied among tissues, across genotypes, and at different developmental stages, the PgNAC transcripts expressed in the four-year-old plant roots of 42 genotypes, the 14 tissues of a four-year-old plant, and the roots of 5-, 12-, 18- and 25-year-old plants were all categorized into the eight subcategories. These results further confirmed the functional differentiation of the PgNAC gene family and also demonstrated the functional consistency of the PgNAC gene family at different developmental stages, in different tissues and among genotypes.
Fig 4. Variation of the functional categorization of the PgNAC transcripts among the four-year-old plant roots of 42 genotypes sampled from Jilin, China.
Fig 5. Variation of the functional categorization of the PgNAC transcripts among 14 tissues of a four-year-old plant.
Fig 6. Variation of the functional categorization of the PgNAC transcripts among the roots of 5-, 12-, 18- and 25-year-old plants.
Temporal-spatial expressions of the PgNAC transcripts and their variation across genotypes
To characterize the expression patterns of the PgNAC transcripts, the expression profiles of all 251 PgNAC transcripts in the roots of 5-, 12-, 18- and 25-year-old plants, the 14 tissues of a four-year-old plant, and the four-year-old plant roots of 42 genotypes were quantified. The result showed that the expressions of the PgNAC transcripts varied from 0.00 TPM to 400 TPM temporally, spatially and across genotypes. Of the 251 PgNAC transcripts, 160 (63.7%) expressed in all four different year-old plant roots, 226 (90%) expressed in all 14 tissues of a four-year-old plant, and 218 (86.9%) expressed in the four-year-old roots of all 42 genotypes (S5 Table). Eight of the 251 PgNAC transcripts, PgNAC63-33, PgNAC63-51, PgNAC63-56, PgNAC63-80, PgNAC66-13, PgNAC66-18, PgNAC66-36, and PgNAC79, were silent at all of the developmental stages, tissues and genotypes studied (S5 Table).
Functional relationship among the gene members of the PgNAC gene family
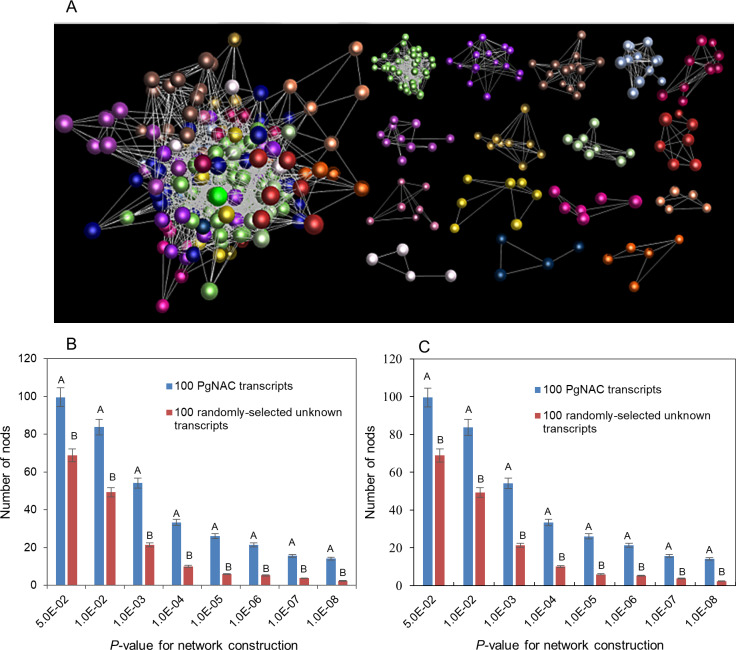
To find out whether the gene members of the PgNAC gene family are functionally related, we further analyzed their co-expression networks in the four-year-old plant roots of 42 ginseng genotypes and in the 14 tissues of a four-year-old plant, respectively. As a result, 218 of these 251 PgNAC transcripts formed a co-expression network in the four-year-old plant roots of 42 genotypes (Fig 7A). The network consisted of 16 clusters, 218 transcript nodes and 2,245 transcript-transcript co-expression edges. In comparison, the PgNAC transcripts were much more likely to form a co-expression network than the transcripts randomly selected from Database A in both number of nodes (Fig 7B) and number of edges (Fig 7C).
Fig 7. Co-expression network of the PgNAC transcripts in the four-year-old plant roots of 42 genotypes.
These 42 ginseng genotypes were collected from the origin and diversity center of ginseng, Jilin, China. (A) The co-expression network and its clusters constructed from 218 of the 251 PgNAC transcripts at P ≤ 5.0E-02. The network consisted of 218 PgNAC transcript nodes and 2,245 co-expression edges. (B) Statistics of the tendency of the PgNAC transcripts forming a co-expression network: number of nodes. (C) Statistics of the tendency of the PgNAC transcripts forming a co-expression network: number of edges. The 100 PgNAC transcripts were randomly selected from the 251 PgNAC transcripts identified in this study by bootstrap sampling, with 10 replications, while the 100 randomly-selected unknown transcripts were randomly selected from Database A [17]. Different capital letters indicate that the difference is significant at a two-tailed significance of P ≤ 0.01.
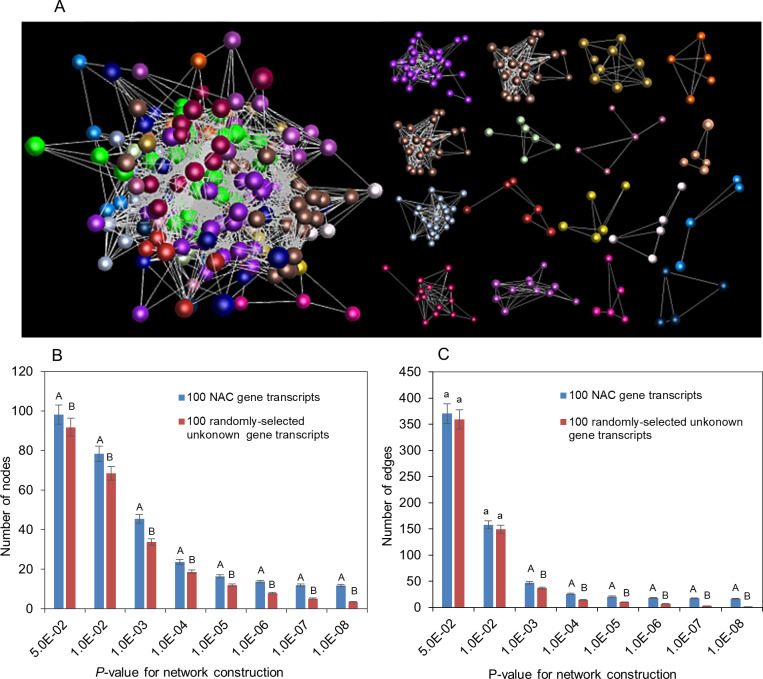
Similarly, 226 of the 251 PgNAC transcripts formed a co-expression network in the 14 tissues of a four-year-old plant. The network consisted of 17 clusters, 226 transcript nodes and 1,935 transcript-transcript co-expression edges (Fig 8A). These PgNAC transcripts were also much more likely to form a co-expression network than the transcripts randomly selected from Database A in both number of nodes (Fig 8B) and number of edges (Fig 8C). These results indicated that the gene members of the PgNAC gene family functionally differentiated, but they were still functionally correlated in ginseng.
Fig 8. Co-expression network of the PgNAC transcripts in 14 tissues of a four-year-old plant.
These 14 tissues were collected at the fruiting stage of the plant. (A) The co-expression network and its clusters constructed from 226 of the 251 PgNAC transcripts at P ≤ 5.0E-02. The network consisted of 226 PgNAC transcript nodes and 1,935 transcript co-expression edges. (B) Statistics of the tendency of the PgNAC transcripts forming a co-expression network: number of nodes. (C) Statistics of the tendency of the PgNAC transcripts forming a co-expression network: number of edges. The 100 PgNAC transcripts were randomly selected from the 251 PgNAC transcripts identified in this study by bootstrap sampling, with 10 replications, while the 100 randomly-selected unknown transcripts were randomly selected from Database A [17]. Different capital letters indicate that the difference is significant at a two-tailed significance of P ≤ 0.01. The same small letters indicate that the difference is not significant at a two-tailed significance of P ≤ 0.05.
Roles of the PgNAC gene family in plant response to cold stress
Studies showed that the NAC gene family responded to a variety of abiotic and biotic stresses in plants, including cold stress [38, 39]. Nakashima et al. [39] grouped the NAC genes of Arabidopsis (A. thaliana), rice (O. sativa), lycophyte (Selaginella moellendorffii), and moss (Physcomitrella patens) into six ancient groups: AM/CUC3, SND, TIP, SNAC, ANAC034, and ONAC4. Zhu et al. [4] and Nakashima et al. [39] showed that many of the NAC genes grouped into the SNAC group played roles in response to abiotic stresses. Of the 22 PgNAC genes identified in this study, five were grouped into the SNAC group (S1 Fig). Nuruzzaman et al. [9] found that the SNAC group had a highly conserved motif (WVLCR) in the region outside the NAC domain (S2 Fig). Therefore, we further investigated in this study these five PgNAC genes, PgNAC05-2, PgNAC41-2, PgNAC48, PgNAC56-1, and PgNAC59, to explore whether the PgNAC gene family has roles in plant response to abiotic stresses.
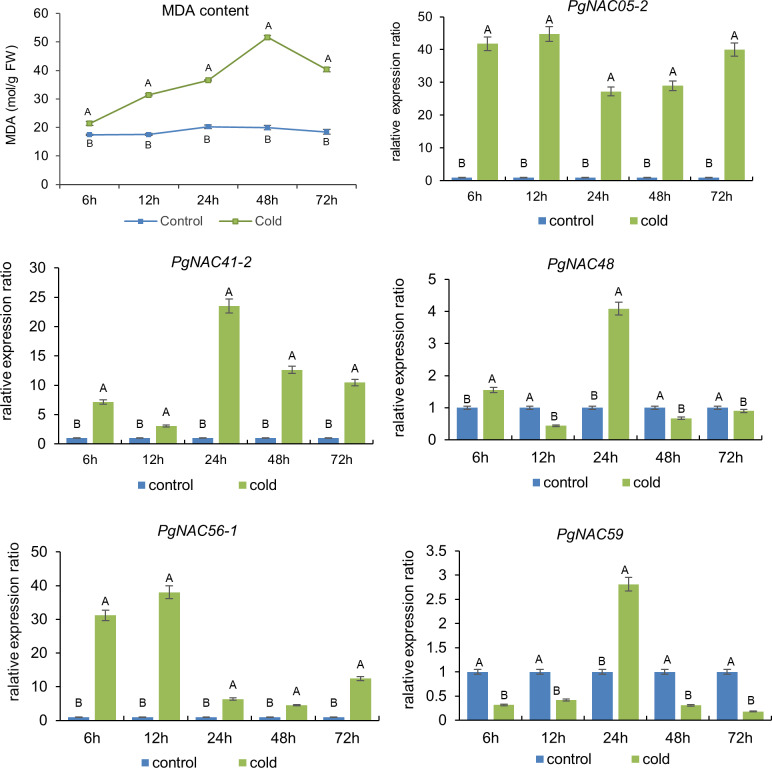
Cold is one of the abiotic stresses crucial to plants, especially to winter plants, such as ginseng and winter wheat. Therefore, we investigated in this study the roles of the five PgNAC genes, PgNAC05-2, PgNAC41-2, PgNAC48, PgNAC56-1, and PgNAC59, in plant response to cold stress. Because previous studies showed that cold stress induced oxidative damage on cell membrane and MDA is an indicator of cell membrane damage, such as membrane lipid peroxidation and cell membrane permeability, due to cold stress [40], we measured the MDA contents of ginseng hairy roots, after exposed to 4°C for 6 h, 12 h, 24 h, 48 h, and 72 h, with three biological replicates. Two-tailed t-test showed that the contents of MDA in the cold-treated hairy roots were significantly higher than those in the non-cold treated controls (Fig 9) for each of the five time points, suggesting that the cold stress significantly increased the contents of MDA in the cold-treated ginseng hairy roots. The contents of MDA in the cold-treated roots increased along with the treatment time increase (Fig 9). The MDA content of the cold-treated hairy roots reached its peak at 48 hours after cold stress, which was nearly 3 times higher than that of the non-cold treated roots, and then decreased. These results indicated that the cold treatments were proper for assaying expression response of the PgNAC genes to cold stress.
Fig 9. Increase of the MDA content and expression regulation of PgNAC transcripts induced by cold treatment in ginseng hairy roots.
The experiments for both cold treatment and non-cold-treated control were carried out with three biological replicates. MDA, malondialdehyde, has been widely used as an indicator of plant response to cold stress [23–25].
Accordingly, the expression levels of all five PgNAC transcripts (PgNAC05-2, PgNAC41-2, PgNAC48, PgNAC56-1, and PgNAC59) analyzed were significantly up- or down-regulated in the cold-treated roots, relative to the non-cold-treated roots. The patterns of root response to the cold treatment varied dramatically among the five PgNAC transcripts studied. For instance, PgNAC05-2 and PgNAC56-1 reached their highest expression levels at 12 h, while PgNAC41-2, PgNAC48, and PgNAC59 reached their expression peaks at 48 h (Fig 9). These results demonstrated that the PgNAC gene family was involved in plant response to cold stress, even though the expression patterns of the PgNAC genes varied, indicating that the PgNAC gene family plays roles in plant response to cold stress.
Discussion
The NAC gene family has been shown to play important roles in plant response to varying environments. This study has systematically analyzed the NAC gene family in ginseng, designated the PgNAC gene family. The results reveal that the PgNAC gene family consists of at least 89 gene members, because they are identified from the transcriptome of 14 tissues of a four-year-old ginseng plant, and the number of the genes in the family varies dramatically among genotypes within the ginseng species. This number of the genes in the PgNAC gene family is largely consistent with those identified in other species, such as Arabidopsis (117), rice (151), Chinese cabbage (188), sugarcane (88), soybean (152), durum wheat (168), and strawberry (112) [41–46]. The variation in number of genes in the PgNAC gene family supports the discovery of Zhang et al. [31] that the number of genes in a gene family may vary by multiple fold. The gene number variation of the PgNAC gene family, as demonstrated by Zhang et al. [31], may be regulated by several factors, the environmental variation of which, i.e., natural selection, may play a more important role in ginseng.
The PgNAC gene family is an ancient and evolutionarily inactive gene family. As the NAC genes identified in Arabidopsis (AtNAC) and rice (OsNAC), the PgNAC gene family is shown to also contain five conserved motifs (A though E), in addition to the NAC domain. Moreover, phylogenetic analysis clusters the PgNAC gene family into 11 subfamilies, but none of them is the P. ginseng linkage-specific or diverged after P. ginseng split from Arabidopsis. These results consistently indicate the ancient and inactive evolution nature of the PgNAC gene family.
Furthermore, this study, for the first time, reveals that the functions of the PgNAC gene family has been substantially differentiated. This finding is supported by both the lower annotation rate (48%) and functional categorization of the PgNAC transcripts into multiple GO categories. However, the degree of functional differentiation of the PgNAC gene family is much smaller than those of the PgNBS gene family [18], the PgRLK gene family [36], the PgCYP gene family [37], and the AP2/ERF gene family [47] in ginseng. Co-expression analysis, which is an indicator of functional correlation between genes, suggests that a majority of the PgNAC genes remain functionally correlated, in spite to their sequence divergence. Zhang et al. [48] reported that the genes controlling a polygenic trait or involved a a common biological process were several times more likely to form a co-expression network, regardless of their sequence identities and biochemical functions.
Finally, this study provides a first line of evidence that the PgNAC gene family also plays roles in plant response to cold stress. This result is consistent with those of the NAC genes obtained in other plant species, such as rice [39]. This study examined five of the 89 PgNAC genes that were grouped into the same group (the SNAC group) as those NAC genes shown to respond to abiotic stresses [39], but they may not be all genes that have such a role in the gene family. It is highly likely that more genes in the PgNAC gene family also play the roles in plant response to cold stress. Given that ginseng is a winter plant species to which plant response to cold stress is crucial and Oh et al. [49] revealed that the content of total saponins, the major bioactive components of ginseng, increased significantly after its plant root was treated with low temperature (4°C), additional research is deserved to identify all the gene members of the PgNAC gene family involved in and comprehensively understand the molecular mechanisms of the family in response to cold stress. Such research will allow not only understanding of the molecular mechanism underlying plant response to cold stress, but also provides molecular tools useful for enhanced genetic improvement in ginseng and other winter plant species.
Supporting information
(PPTX)
(PPTX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by an award from the China 863 Project (2013AA102604-3), the Bureau of Science and Technology of Jilin Province (20170101010JC, 20180414077GH, 20180101027JC, 20190201264JC, and 20190201171JC), the Education Department of Jilin Province (JJKH20200318KJ and JJKH20200320KJ), and the Development and Reform Commission of Jilin Province (2016C064 and 2018C047-3).
References
- 1.Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Frontiers in Plant Science. 2015; 6:902 10.3389/fpls.2015.00902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu Y, Duan M, Zhang Z, Dong J, Wang T. Overexpression of the Medicago falcata NAC transcription factor MfNAC3 enhances cold tolerance in Medicago truncatula. Environmental Experimental Botany. 2016; 129:67–76. [Google Scholar]
- 3.Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends in Plant Science. 2012; 17(6):369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 4.Zhu G, Chen G, Zhu J, Zhu Y, Lu X, Li X, et al. Molecular characterization and expression profiling of NAC transcription factors in Brachypodium distachyon L. PLoS One. 2015; 10(10):e0139794 10.1371/journal.pone.0139794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Zhou Y, Li H, Wang T, Zhang J, Ouyang B, et al. Molecular and functional characterization of ShNAC1, an NAC transcription factor from Solanum habrochaites. Plant Science. 2018; 271:9–19. 10.1016/j.plantsci.2018.03.005 [DOI] [PubMed] [Google Scholar]
- 6.Souer E, Van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996; 85(2):159–70. 10.1016/s0092-8674(00)81093-4 [DOI] [PubMed] [Google Scholar]
- 7.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant cell. 1997; 9(6):841–857. 10.1105/tpc.9.6.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold and heat. Frontiers in Plant Science. 2014; 5:170 10.3389/fpls.2014.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuruzzaman M, Sharoni M, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Frontiers Microbiology. 2013; 4:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends in Plant Science. 2005; 10(2):79–87. 10.1016/j.tplants.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 11.Xu ZY, Kim SY, Hyeon DY, Kim DH, Dong T, Park Y, et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-Type transcription factors in dehydration and osmotic stress responses. The Plant Cell. 2013; 25(11):4708–4724. 10.1105/tpc.113.119099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Seo PJ, Lee HJ, Park CM. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. The Plant Journal. 2012; 70(5):831–844. 10.1111/j.1365-313X.2012.04932.x [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Wang Y, Lv B, Li J, Luo L, Lu S, et al. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiology. 2014; 55(3):604–619. 10.1093/pcp/pct204 [DOI] [PubMed] [Google Scholar]
- 14.Li XD, Zhuang KY, Liu ZM, Yang DY, Ma NN, Meng QW. Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic tobacco. Journal of Plant Physiology. 2016; 204:54–65. 10.1016/j.jplph.2016.06.024 [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Yang X, Pei S, He G, Wang X, Tang Q. The Miscanthus NAC transcription factor MlNAC9 enhances abiotic stress tolerance in transgenic Arabidopsis. Gene. 2016; 586(1):158–169. 10.1016/j.gene.2016.04.028 [DOI] [PubMed] [Google Scholar]
- 16.Parvin S, Pulla RK, Kim YJ, Sathiyaraj G, Jung SK, Khorolragchaa A, et al. Isolation and characterization of glycolate oxidase gene from Panax ginseng C.A. Meyer. Journal of Ginseng Research. 2009; 33:249–255. [Google Scholar]
- 17.Wang K, Jiang S, Sun C, Lin Y, Yin R, Wang Y, et al. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C.A. Meyer. Scientific Reports. 2015; 5:18283 10.1038/srep18283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin R, Zhao M, Wang K, Lin Y, Wang Y, Sun C, et al. Functional differentiation and spatial-temporal coexpression networks of the NBS-encoding gene family in Jilin ginseng, Panax ginseng C.A. Meyer. PLoS One. 2017; 12:e0181596 10.1371/journal.pone.0181596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009; 37:W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony Methods. Molecular Biology Evolution. 2011; 28(10):2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genetics. 2000; 25(1):25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theocharidis A, Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout Express3D. Nature Protocols. 2009; 4(10):1535–1550. 10.1038/nprot.2009.177 [DOI] [PubMed] [Google Scholar]
- 23.Tian Y, Zhang H, Pan X, Chen X, Zhang Z, Lu X, et al. Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings. Transgenic Research. 2011; 20:857–866. 10.1007/s11248-010-9463-9 [DOI] [PubMed] [Google Scholar]
- 24.Feng HL, Ma NN, Meng X, Zhang S, Wang JR, Chai S, et al. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiology and Biochemistry. 2013; 73:309–320. 10.1016/j.plaphy.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Yu K, He T, Li F, Zhang D, Liu J. The low temperature induced physiological responses of Avena nuda L., a cold-tolerant plant species. Scientific World Journal. 2013; 2013:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L, Zhao M, Chen M, Li L, Jiang Y, Liu S, et al. The bHLH gene family and its response to saline stress in Jilin ginseng, Panax ginseng C.A. Meyer. Molecular Genetics and Gnomics. 2020; 10.1007/s00438-020-01658-w [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001; 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 28.Kim NH, Jayakodi M, Lee SC, Choi BS, Jang W, Lee J, et al. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnology Journal. 2018; 16:1904–1917. 10.1111/pbi.12926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayakodi M, Choi BS, Lee SC, Kim NH, Park JY, Jang W, et al. Ginseng Genome Database: an open-access platform for genomics of Panax ginseng. BMC Plant Biology. 2018; 18(1):62 10.1186/s12870-018-1282-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Chu Y, Liao B, Xiao S, Yin Q, Bai R, et al. Panax ginseng genome examination for ginsenoside biosynthesis. GigaScience. 2017; 6:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang MP, Wu Y-H, Lee M-K, Liu Y-H, Rong Y, Santos FS, et al. Numbers of genes in the NBS and RLK families vary by more than four-fold within a plant species and are regulated by multiple factors. Nucleic Acids Research. 2010; 38:6513–6525. 10.1093/nar/gkq524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao L, Gonda I, Sun H, Ma Q, Bao K, Tieman DM, et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nature Genetics. 2019; 51:1044–1051. 10.1038/s41588-019-0410-2 [DOI] [PubMed] [Google Scholar]
- 33.Singh AK, Sharma V, Pal AK, Acharya V, Ahuja PS. Genome-wide organization and expression profiling of the NAC transcription factor family in Potato (Solanum tuberosum L.). DNA Research. 2013; 20(4):403–423. 10.1093/dnares/dst019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Li D, Wang Y, Zhou R, Wang L, Zhang Y, et al. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum. PLoS One. 2018; 13(6):e0199262 10.1371/journal.pone.0199262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Wang L, Wang S. Comprehensive analysis and discovery of drought-related NAC transcription factors in common bean. BMC Plant Biology. 2016; 16(1):193 10.1186/s12870-016-0882-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y, Wang K, Li X, Sun C, Yin R, Wang Y, et al. Evolution, functional differentiation, and co-expression of the RLK gene family revealed in Jilin ginseng, Panax ginseng C.A. Meyer. Molecular Genetics Genomics. 2018; 293:845–849. 10.1007/s00438-018-1425-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Li X, Lin Y, Wang Y, Wang K, Sun C, et al. Structural variation, functional differentiation, and activity correlation of the cytochrome P450 gene superfamily revealed in ginseng. Plant Genome. 2018; 11:1701–1706. [DOI] [PubMed] [Google Scholar]
- 38.Nuruzzaman M, Manimekalai R, Sharoni AM. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010; 465:30–40. 10.1016/j.gene.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 39.Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-shinozaki K. NAC transcription factors in plant abiotic stress responses. BBA—Gene Regulatory Mechanisms. 2012; 1819(2):97–103. 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed] [Google Scholar]
- 40.Kuk YI, Shin JS, Burgos NR, Hwang TE, Han O, Cho BH, et al. Antioxidative enzymes offer protection from chilling damage in rice plants. Crop Science. 2003; 43(6):2109–2117. [Google Scholar]
- 41.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research. 2003; 10(6):239–247. 10.1093/dnares/10.6.239 [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Wang F, Li MY, Jiang Q, Tan GF, Xiong AS. Genome wide analysis of the NAC transcription factor family in Chinese cabbage to elucidate responses to temperature stress. Scientia Horticulturae. 2014; 165:82–90. [Google Scholar]
- 43.Ramaswamy M, Narayanan J, Manickavachagam G, Athiappan S, Arun M, R G, et al. Genome wide analysis of NAC gene family sequences in sugarcane and its comparative phylogenetic relationship with rice, sorghum, maize and Arabidopsis for prediction of stress associated NAC genes. Agri Gene. 2017; 3:1–11. [Google Scholar]
- 44.Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-chinozaki K, Chinozaki K, et al. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dhydration stress. DNA Research. 2011; 18(4):263–276. 10.1093/dnares/dsr015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saidi MN, Mergby D, Brini F. Identification and expression analysis of the NAC transcription factor family in durum wheat (Triticum turgidum L. ssp. durum). Plant Physiology and Biochemistry. 2017; 112:117–128. 10.1016/j.plaphy.2016.12.028 [DOI] [PubMed] [Google Scholar]
- 46.Enriqueta M, Martínez-Rivas Félix J, Rosario BP, Francisco Javier MH, Pablo RV, Matas-Arroyo Antonio J, et al. Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria × ananassa fruits. PLoS One. 2018; 13(5):e0196953 10.1371/journal.pone.0196953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Zhou Y, Zhang Q, Liu Q, Li L, Sun C, et al. Structural variation, functional differentiation and expression characteristics of the AP2/ERF gene family and its response to cold stress and methyl jasmonate in Panax ginseng C.A. Meyer. PLoS One. 2020; 15(3):e0226055 10.1371/journal.pone.0226055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang MP, Liu Y-H, Xu W, Smith CW, Murray SC, Zhang H-B. Analysis of the genes controlling three quantitative traits in three diverse plant species reveals the molecular basis of quantitative traits. Scientific Reports. 2020; (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh JY, Kim YJ, Jang MG, Joo SC, Kwon WS, Kim SY, et al. Investigation of ginsenosides in different tissues after elicitor treatment in Panax ginseng. Journal of Ginseng Research. 2014; 38(4):270–277. 10.1016/j.jgr.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX)
(PPTX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.