Uropathogenic Escherichia coli strains cause most uncomplicated urinary tract infections (UTI), one of the most common infectious diseases worldwide. Random transposon mutagenesis techniques have been utilized to identify essential bacterial genes during infection; however, this has been met with limitations when applied to the murine UTI model. Conventional high-throughput transposon mutagenesis screens are not feasible because of inoculum size restrictions due to a bottleneck during infection. Our study utilizes a condensed ordered transposon library, limiting the number of mutants while maintaining the largest possible genome coverage. Screening of this library in vivo, and in human urine in vitro, identified numerous candidate fitness factors. Additionally, we have developed a novel technique using qPCR to quantify bacterial outputs following infection with small subgroups of transposon mutants. Molecular approaches developed in this study will serve as useful tools to probe in vivo models that are restricted by anatomical, physiological, or genetic bottleneck limitations.
KEYWORDS: Escherichia coli, urinary tract infection
ABSTRACT
Urinary tract infections (UTI), the second most diagnosed infectious disease worldwide, are caused primarily by uropathogenic Escherichia coli (UPEC), placing a significant financial burden on the health care system. High-throughput transposon mutagenesis combined with genome-targeted sequencing is a powerful technique to interrogate genomes for fitness genes. Genome-wide analysis of E. coli requires random libraries of at least 50,000 mutants to achieve 99.99% saturation; however, the traditional murine model of ascending UTI does not permit testing of large mutant pools due to a bottleneck during infection. To address this, an E. coli CFT073 transposon mutant ordered library of 9,216 mutants was created and insertion sites were identified. A single transposon mutant was selected for each gene to assemble a condensed library consisting of 2,913 unique nonessential mutants. Using a modified UTI model in BALB/c mice, we identified 36 genes important for colonizing the bladder, including purB, yihE, and carB. Screening of the condensed library in vitro identified yigP and ubiG to be essential for growth in human urine. Additionally, we developed a novel quantitative PCR (qPCR) technique to identify genes with fitness defects within defined subgroups of related genes (e.g., genes encoding fimbriae, toxins, etc.) following UTI. The number of mutants within these subgroups circumvents bottleneck restriction and facilitates validation of multiple mutants to generate individual competitive indices. Collectively, this study investigates the bottleneck effects during UTI, provides two techniques for evading those effects that can be applied to other disease models, and contributes a genetic tool in prototype strain CFT073 to the field.
IMPORTANCE Uropathogenic Escherichia coli strains cause most uncomplicated urinary tract infections (UTI), one of the most common infectious diseases worldwide. Random transposon mutagenesis techniques have been utilized to identify essential bacterial genes during infection; however, this has been met with limitations when applied to the murine UTI model. Conventional high-throughput transposon mutagenesis screens are not feasible because of inoculum size restrictions due to a bottleneck during infection. Our study utilizes a condensed ordered transposon library, limiting the number of mutants while maintaining the largest possible genome coverage. Screening of this library in vivo, and in human urine in vitro, identified numerous candidate fitness factors. Additionally, we have developed a novel technique using qPCR to quantify bacterial outputs following infection with small subgroups of transposon mutants. Molecular approaches developed in this study will serve as useful tools to probe in vivo models that are restricted by anatomical, physiological, or genetic bottleneck limitations.
INTRODUCTION
Urinary tract infections (UTI) are one of the most common human infections and generate a significant health burden worldwide (1, 2). An estimated 80% of uncomplicated UTI in otherwise healthy women are caused by uropathogenic Escherichia coli (UPEC) (3, 4). Multidrug resistance in E. coli is becoming increasingly common and has amplified the difficulty of treating these infections (1). This fact underlines the urgent need for defining fitness and essential genes required for the infection so that new targets for antimicrobial therapy may be exploited. The ability of UPEC to successfully colonize and survive within the urinary tract is facilitated by genes that reside in the accessory genome, which helps classify these strains as extraintestinal pathogenic E. coli (ExPEC). The accessory genome includes genes encoding specialized adhesins, iron acquisition systems, autotransporters, and toxins, all of which promote infection within the host urinary tract (5–7). Virulence properties that promote UPEC colonization and infection of the bladder have been well studied, though unbiased and systematic approaches to screening the entire genome simultaneously for infection-specific and host-specific fitness factor genes have been met with many limitations.
Historically, random transposon mutagenesis has been utilized in numerous bacterial genera to generate a large pool of mutants for use in various animal models of infection (8–11). This technique, involving a pool of hundreds of thousands of mutants, was used by us to determine UPEC fitness factors during systemic infection (12). However, in a murine UTI model, proposed factors including micturition, exfoliation of bladder epithelial cells, and the innate immune response stochastically reduce the diversity of the inoculum and have prevented the use of in vivo transposon screens of bladder colonization using large-format mutant libraries (13, 14). A notable example of limitations associated with bottlenecks was quantified by Walters et al. (13) in a study that examined the population dynamics during infection. The results demonstrated the concept of a bottleneck during the traditional ascending mouse model of UTI, which was validated by the randomness of population dynamics that occurred within 48 h of initial infection in CBA/J mice as bacteria ascended the ureters to the kidneys. These results prompted a more thorough investigation of the bottleneck effect in order to provide the optimal conditions for screening UPEC fitness factors during UTI in an unbiased and comprehensive manner.
Random mutant libraries are the most common genetic approach due to the simplicity of their construction; however, limitations include a large gene bias and loss of information due to intergenic transposon insertions. Construction of a defined mutant library allows for optimization within the composition of a given inoculum, avoiding transposon insertions in intergenic regions and reducing gene size bias (15, 16). For our studies, we chose to generate a defined mutant library for a more thorough and optimized characterization of genes involved in UTI fitness. The construction and in vivo screening of an ordered mutant library ease the burden of limitations in the ascending UTI model and have many advantages. By consolidating and categorizing each individual transposon mutant, a minimal number of mutants can be tested to represent the largest possible genome coverage (17). Additionally, if there are multiple transposon insertion sites within the gene, the insertion site with the most potential for disrupting the resulting protein product and the least possibility for polar effects can be chosen to represent the gene in the library. Ordered libraries have been made and utilized in numerous model bacterial organisms, such as Enterococcus and Staphylococcus spp. (18, 19). Here, we employed this strategy in E. coli CFT073 to overcome the bottleneck effect in the murine model of ascending UTI. Creation of the ordered library required the generation of 9,216 transposon mutants, which disrupted 54% of the 5,369 annotated genes in E. coli CFT073.
The previously described bottleneck during ascending UTI was further assessed in the present study and adjusted to allow for the testing of larger mutant input pools without stochastic loss. An E. coli CFT073 transposon ordered library of 9,216 mutants was created, and insertion sites were identified by multiplexed Cartesian pooling-coordinate sequencing. Single transposon mutants were selected to assemble defined low-complex inoculums, and 2,913 genes were tested in vivo in the optimized UTI model. By transposon sequencing, 36 genes were identified as candidate fitness factors during bladder colonization. We also utilized this ordered library to screen for essential genes during in vitro growth in human urine. This assessment allowed us to tease apart which bacterial genes are important for growth in the urinary tract milieu (infection-specific fitness factors) versus the genes that are exclusively essential in the host environment (host-specific fitness factors). Additionally, we developed a novel quantitative PCR (qPCR) strategy, which does not require deep sequencing, for testing co-challenge of smaller pools of transposon mutants as a means to generate hierarchies of fitness factors for UTI. These fitness hierarchies will be useful to design mechanistic studies to better understand the biology of uropathogenic E. coli infection.
RESULTS
Bottleneck assessment.
The anatomy of the urinary tract leads to stochastic loss of mutants within a pool due to urination after inoculation. Also, an early clonal expansion of a subpopulation in a confined space, such as the bladder, may reduce the population diversity during prolonged infections (13). Such bottlenecks may result in false-positive identification of fitness defects during transposon sequencing (Tn-seq) analysis. To overcome these obstacles and optimize the UTI model for Tn-seq, a bottleneck assessment was performed examining murine bladders. To define the capacity for clonal diversity over time, mice were infected with 1:1, 1:100, 1:500, and 1:1,000 ratios of wild-type (WT) strain CFT073 and a nondeleterious kanamycin-resistant CFT073 mutant, the ΔaraF::Kanr mutant, which exhibits no growth defect in vivo (20). In separate pools of mice, bladder samples were harvested at 16 and 24 hours postinoculation (hpi) to determine the appropriate duration of infection. In this simulated transposon mutant library, the mutant strain CFT073 ΔaraF::Kanr represents any given mutant in a mixed population, and a log10 competitive index (CI) for CFT073 ΔaraF::Kanr of <−1 would indicate that colonization was limited by bottleneck effects (8, 13).
Various inoculum doses of CFT073 were tested in numerous mouse strains to examine optimal colonization of the urinary tract (data not shown). Ultimately, BALB/c mice inoculated with 108 CFU of statically cultured bacteria were the selected conditions for the optimized UTI model. In BALB/c mice at 24 hpi, the bladder was well colonized; however, bacterial input ratios of 1:500 and 1:1,000 (ΔaraF::Kanr mutant to wild type) showed spontaneous loss of the resistant mutant strain from the colonizing population as indicated by a log10 CI of <−1 or >1 (see Fig. S1A and B in the supplemental material). The severity of this bottleneck effect was reduced by limiting the duration of the infection by 4 h. Following a 16-h infection, input ratios of 1:500 (ΔaraF::Kanr mutant to wild type) showed a median log10 CI of −0.29 with CFU bladder counts remaining similar to 24 hpi (Fig. S1C and D). Based on these findings, a 16-h model of infection was determined to accommodate pools consisting of up to 500 unique mutants, which requires an ordered library to achieve a robust genome-wide screen.
Construction of the CFT073 ordered transposon library.
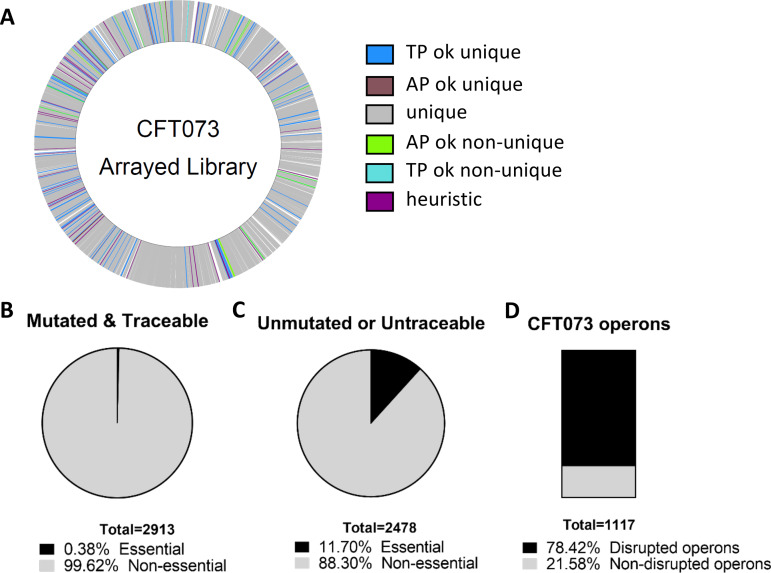
To construct the E. coli CFT073 ordered library, we used the CP-CSeq protocol previously described by Vandewalle et al. (17). The CP-CSeq protocol simultaneously determines transposon insertion coordinates within the genome and the physical location of the corresponding mutant archived in a 96-by-96-well plate library (see Data Set S1 in the supplemental material). This approach resulted in 12,573 transposon insertions within the genome with an additional 542 sites where reads could not be uniquely mapped (Fig. 1A). Over 64% (8,105/12,573) of uniquely mapped insertion events had sufficient sequencing coverage for customized software-based automated identification of the mutants’ positions in the archived clone library (see Table S1 in the supplemental material). Specifically, 2,913 genes had associated insertions with traceable reads and 99.6% aligned to nonessential genes (21) (Fig. 1B). About 88% (7,123) of the total insertions map within the open reading frames (ORFs) of known genes, targeting 54% of the total number of genes in the CFT073 genome (GenBank annotation [22]). A total of 2,478 genes either were not mutated or had untraceable reads, and 11.7% of these aligned to essential genes (Fig. 1C). Of the total 1,117 annotated operons in CFT073, 78.42% (876) had at least one transposon insertion (Fig. 1D).
FIG 1.
Construction and identification of transposon mutants within the library. A total of 9,216 E. coli CFT073 mutants were generated and sequenced to identify the locations of transposon insertions. (A) The CFT073 genome is marked with each identified transposon insertion site. Unique, unique identification of the plate and well coordinates within the library; TPok_unique, unique identification of the plate coordinate and the most highly probable unique location for the well coordinate; TPok_nonunique, unique identification of the plate coordinate and multiple locations for the well coordinate; APok_unique, unique identification of the well coordinate and the most highly probable unique location for the plate coordinate; APok_nonunique, unique identification of the well coordinate and multiple locations for the plate coordinate; heuristic, the most highly probable unique location for the plate and well coordinates. (B) A total of 2,913 genes had a traceable transposon insertion; 99.9% of these were in nonessential genes. (C) Of the 2,478 nonmutated genes in the CFT073 genome, 290 are annotated as essential. (D) There are 1,117 annotated operons in the CFT073 genome. Sequencing of the ordered library confidently identified transposon insertions in 876 unique operons.
To ensure the accuracy of mapping the transposon insertion sites, 24 mutants with traceable reads were selected at random for screening. The presence of all 24 mutants that were assigned by CP-CSeq to particular wells across the library were verified as accurate by PCR analysis. The PCR products of all specific CP-CSeq assignments resulted in the expected fragment size (see Fig. S2 and Table S2 in the supplemental material). Having high confidence of the determined transposon insertion sites based on sequence analysis, a condensed library was created by selecting one unique mutant per disrupted gene and, when possible, picking those with an insertion in the first 80% of the gene sequence.
Identification of UPEC fitness factors during UTI.
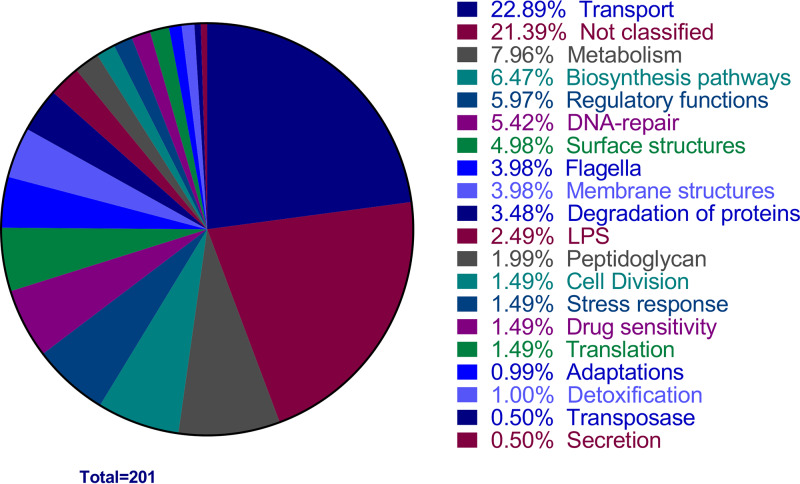
The condensed library consisting of 2,913 unique disrupted genes (ultimately represented by 3,373 mutants [see Materials and Methods]) was transurethrally inoculated via seven pools, each containing <500 mutants, into groups of 5 BALB/c mice (see Fig. S3A in the supplemental material). The CFU per organ was determined for individual mice (Fig. S3B). The bladder samples were harvested at 16 hpi to determine the ratio of input to output for each individual mutant, and fitness was assessed using fold change (FC) (see Data Set S2 in the supplemental material). Due to the observed variability between biological replicates, the adjusted P value was greater than 0.05 for all mutants tested. Therefore, a standard P value of <0.05 was chosen to detect the most promising fitness defect candidates. A total of 201 mutants displayed a P value of <0.05 and FC of >2. These preliminary fitness factors were separated into functional groups based on annotated categories listed in the KEGG database, with over 21% being associated with unknown function and therefore providing opportunity for novel discoveries (Fig. 2). To narrow down the list to the most likely fitness factors and facilitate further analysis, a stricter threshold was applied. Only mutants displaying a P value of <0.05 and an FC greater than the median FC plus standard deviation (i.e., fold change of >513) were selected as CFT073 fitness factors (Table 1). This decreased the number of candidate fitness factors to a total of 36 genes (Table 1).
FIG 2.
Functional classification of preliminary fitness factors for murine UTI. Tn-seq preliminary fitness factors are shown by functional categories. Primary analysis of the Tn-seq from mouse bladders identified 201 preliminary fitness factors. The percentage of the 201 preliminary CFT073 fitness factors belonging to each annotated functional category is documented in the pie chart. Categories of transport, unknown, and metabolism represented the largest proportion of hits with 22.9%, 21.4%, and 8%, respectively.
TABLE 1.
E. coli CFT073 genes tested in the UTI model showing significant fitness defects (≥513-fold change; P < 0.05) for colonization of the murine bladder
| Locus |
Protein | Fold change | P value | Adjusted P value | |
|---|---|---|---|---|---|
| RefSeq | GenBank | ||||
| C_RS19445 | c4105_pshM | Putative general secretion pathway protein M (Transport protein PshM) | 2,737.2 | 0.0211 | 0.5358 |
| C_RS25485 | c5399_fimG | FimG protein precursor | 2,032.6 | 0.0269 | 0.5711 |
| C_RS24715 | c5232_efp | Elongation factor P | 1,623.2 | 0.0296 | 0.5711 |
| C_RS22205 | c4704_rfe | Undecaprenyl-phosphate alpha-N-acetylglucosaminyltransferase | 1,583.6 | 0.0005 | 0.2269 |
| C_RS00190 | c0041_carB | Carbamoyl-phosphate synthase large chain | 1,456.1 | 0.0159 | 0.5042 |
| C_RS21030 | c4456_rfaQ | Lipopolysaccharide core biosynthesis glycosyl transferase RfaQ | 1,380.3 | 0.0167 | 0.5042 |
| C_RS23585 | c4979_yjbB | Hypothetical protein YjbB | 1,346.3 | 0.0287 | 0.5711 |
| C_RS25490 | c5400_fimH | FimH protein precursor | 1,340.6 | 0.0384 | 0.6299 |
| C_RS10865 | c2294_flhB | Flagellar biosynthetic protein FlhB | 1,340.3 | 0.0172 | 0.5042 |
| C_RS24925 | c5277_yjfO | Hypothetical lipoprotein YjfO precursor | 1,185.5 | 0.0193 | 0.5291 |
| C_RS07675 | c1641_ldcA | Muramoyltetrapeptide carboxypeptidase | 1,174.8 | 0.0011 | 0.2269 |
| C_RS10925 | c2308_flhD | Flagellar transcriptional activator FlhD | 1,126.5 | 0.0203 | 0.5304 |
| C_RS22705 | c4803_yihE | Hypothetical protein YihE | 1,110.5 | 0.0164 | 0.5042 |
| C_RS11185 | c2361_fliL | Flagellar FliL protein | 1,061.3 | 0.0012 | 0.2269 |
| C_RS14420 | c3035_yfgM | Hypothetical protein YfgM | 1,052.3 | 0.0038 | 0.3253 |
| C_RS06310 | c1345_flgE | Flagellar hook protein FlgE | 1,023.6 | 0.0012 | 0.2269 |
| C_RS27145 | c2497 | Transposase | 947.5 | 0.0240 | 0.5711 |
| C_RS17605 | c3712 | Putative saframycin Mx1 synthetase B | 938.1 | 0.0013 | 0.2269 |
| C_RS06175 | c1314_mdoH | Periplasmic glucans biosynthesis protein MdoH | 937.4 | 0.0024 | 0.3082 |
| C_RS21025 | c4455_rfaG | Lipopolysaccharide core biosynthesis protein RfaG (glucosyltransferase I) | 923.7 | 0.0017 | 0.2625 |
| C_RS08200 | c1750_pyrF | Orotidine 5′-phosphate decarboxylase | 903.9 | 0.0250 | 0.5711 |
| C_RS22120 | c4687_yifB | Hypothetical protein YifB (ORF III) | 751.3 | 0.0057 | 0.3493 |
| C_RS19985 | c4222 | Putative DNA-processing protein | 746.8 | 0.0012 | 0.2269 |
| C_RS07045 | c1510_purB | Adenylosuccinate lyase | 738.2 | 0.0304 | 0.5772 |
| C_RS18670 | c3936_ftsJ | Ribosomal RNA large subunit methyltransferase J | 717.2 | 0.0067 | 0.3584 |
| C_RS03055 | c0660_cusB | Putative copper efflux system protein CusB precursor | 707.7 | 0.0011 | 0.2269 |
| C_RS23265 | c4922_oxyR | Hydrogen peroxide-inducible genes activator (morphology and autoaggregation control protein) | 707.3 | 0.0271 | 0.5711 |
| C_RS24085 | c5093_yjcW | d-Allose transport ATP-binding protein AlsA | 671.0 | 0.0365 | 0.6258 |
| C_RS06240 | c1329_pyrC | Dihydroorotase | 652.1 | 0.0026 | 0.3118 |
| C_RS11080 | c2338_fliC | Flagellin | 551.4 | 0.0026 | 0.3118 |
| C_RS22910 | c4847_fdhD | FdhD protein | 541.0 | 0.0356 | 0.6233 |
| C_RS14270 | c3004_purC | Phosphoribosylaminoimidazole-succinocarboxamide synthase | 531.0 | 0.0031 | 0.3253 |
| C_RS02790 | c0606_ybaT | Hypothetical transport protein YbaT | 530.6 | 0.0067 | 0.3584 |
| C_RS08560 | c1822_ompN | Outer membrane protein N precursor (porin OmpN) | 522.5 | 0.0248 | 0.5711 |
| C_RS16250 | c3425_ygdP | (Di)nucleoside polyphosphate hydrolase | 518.6 | 0.0094 | 0.4128 |
| C_RS11255 | c2378_yedA | Hypothetical transport protein YedA | 513.9 | 0.0230 | 0.5520 |
To validate the finalized list of 36 hits in Table 1, we first scrutinized the literature for previously established fitness or virulence factors. Type 1 fimbriae are the preeminent virulence factor during experimental UTI (23), and our work also found them to be a crucial fitness factor; fimG and fimH mutants are known to be attenuated (24). Similarly, rfaQ and rfaG were shown to contribute significantly to the fitness of UPEC strain NU14 in the mouse bladder (25). These lipopolysaccharide (LPS) core biosynthesis-encoding genes were determined to have 1-log and 3-log decreases in colonization, respectively (25). Identification of already-known fitness factors within this data set provide confidence that the screen will provide valuable information. After eliminating previously identified fitness factors, a total of 32 newly identified gene candidates remained.
Selection of genes for validation by Tn-seq.
To validate our candidate fitness factor genes, we conducted twelve cochallenge experiments in which the wild type was competed against each mutant, individually. A total of 5 to 10 BALB/c mice were inoculated via the transurethral route with an inoculum of 108 CFU containing of 1:1 ratio of wild-type and transposon mutant strains selected from the ordered library. After 16 h, the urine, bladder, and kidneys were aseptically collected and processed to enumerate the CFU for the wild-type and mutant strains to calculate a competitive index (CI) for defining fitness factors. The following mutant strains were selected for validation experiments: yihE, purB, carB, yfgM, rfe, efp, flhB, cusB, mdoH, yjbB, pshM, and yfjO.
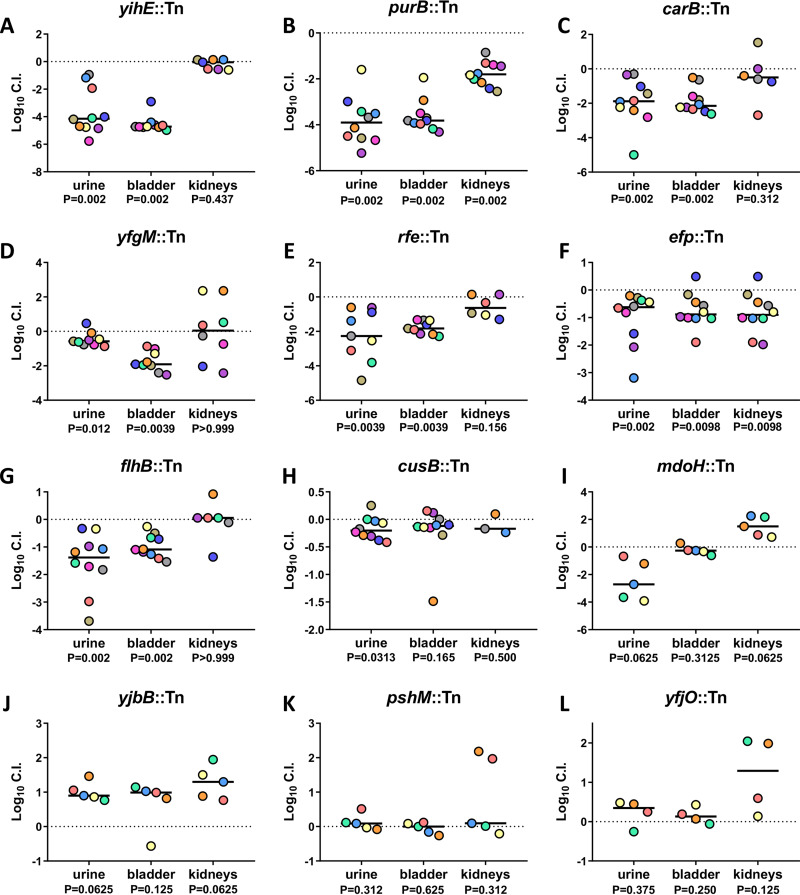
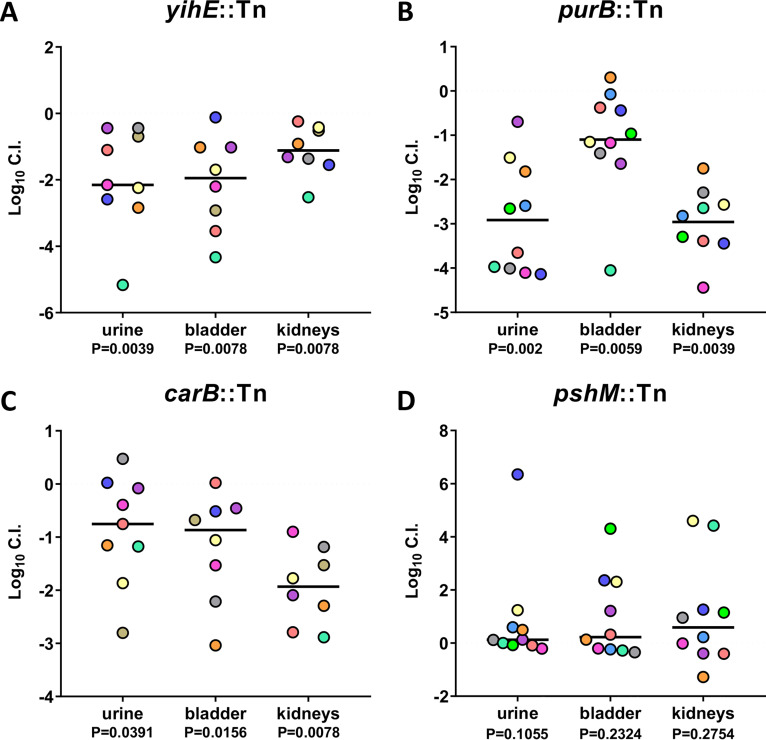
The urinary tract has potent antimicrobial defenses and is nutrient limited, making it difficult for bacteria to cause infection; therefore, genes that facilitate survival under stressful conditions could be fitness factors for UPEC during cystitis. Indeed, yihE encodes a protein kinase important for handling induced stress from reactive oxygen species and is a central regulator of cell death in response to antimicrobial and environmental stressors (26). The yihE::Tn mutant had one of the most dramatic and consistent reductions in colonization ability in both the urine and the bladder samples, with a 4-log deficit in each following co-challenge with the wild type (Fig. 3A; see Fig. S4A in the supplemental material). Purine biosynthesis is important for the intracellular survival of UPEC and biofilm formation (27, 28). purB encodes the adenylosuccinate lyase crucial to this de novo process. The purB::Tn mutant was outcompeted 10,000-fold by wild-type CFT073 in both the urine and bladder. In the kidneys, the purB::Tn strain had a 2-log defect, which is a greater fitness defect than any other validated ordered library mutant tested in the kidneys (Fig. 3B and S4B). Amino acids and peptides have been demonstrated as the primary carbon sources for UPEC during UTI (20). Strains deficient in peptide import (ΔoppB and ΔdppB) had reduced fitness in vivo (20). carB encodes an important synthase for amino acid biosynthesis and pyrimidine metabolism. A transposon mutant of carB displayed a significant 10-fold defect during co-challenge in both the urine and bladder (Fig. 3C and S4C). Also identified was yfgM, which encodes a periplasmic chaperone protein required for the proper folding of several inner membrane and periplasmic proteins (29, 30). In co-challenge, the yfgM::Tn mutant had a significant fitness defect in the urine and bladder but not in the kidneys (Fig. 3D and S4D). The rfe::Tn ordered library mutant had a 2-log decrease in both the urine and bladder, with a 1-log decrease in the kidneys (Fig. 3E and S4E). Without functional rfe, O antigen does not localize to the bacterial surface (31). The efp::Tn elongation factor protein mutant had a significant defect in colonization of all three organ sites. A 10-fold fitness defect was observed in the urine, bladder, and kidneys (Fig. 3F and S4F). The importance of efp is consistent with recent work from our laboratory demonstrating that the growth rate of CFT073 is higher in vivo than in any tested growth medium (32). Four of the top 36 hits were genes related to flagellum structure and function. We chose one of these genes, flhB, to represent this functional group. flhB regulates the switching and systematic export of two main flagellar structural protein functional groups, and thus the flhB mutant does not assemble a functional flagellum (33, 34). In the co-challenge model, the flhB::Tn mutant had an ∼10-fold defect in colonization of both the urine and the bladder and no defect in the kidneys (Fig. 3G and S4G).
FIG 3.
Validation of in vivo Tn-seq hits as bladder fitness factors for murine UTI. BALB/c mice were transurethrally inoculated with 108 CFU of UPEC, which consisted of a 1:1 mixture of wild-type CFT073 and an isogenic transposon mutant. Urine, bladder, and kidneys were collected at 16 hpi, and resulting homogenates were plated on LB agar with and without kanamycin to determine the respective bacterial burdens of the mutant and wild type. Five mice were tested per mutant. If initial results indicated successful validations, then 5 additional mice were added to the experiment. Each data point represents the log10 competitive index calculated for each individual mouse organ; each mouse is represented by a single color. Black lines indicate the median. A log10 CI of 0 indicates that the wild type and mutant have equal fitness. P values determined by the Wilcoxon signed-rank test are listed below each organ tested.
Another mutant that our Tn-seq study identified has defects in response to metal toxicity. The cus operon was found to be upregulated in UPEC collected from the urine of women with significant E. coli bacteriuria (35). This operon encodes proteins that are formed in response to metal stress and coordinate an efflux system to efficiently export toxic metals to promote resistance to copper toxicity (36). The cusB::Tn mutant obtained significant colonization defects in the urine during co-challenge infection (Fig. 3H and S4H), and a similar trend was observed in bladder and kidneys. mdoH encodes a membrane-bound glucosyltransferase involved in the translocation of unfolded polyglucose chains to the periplasm (37). Although the mdoH::Tn mutant exhibited a 3-log-decreased colonization in the urine, it had no defect in either the bladder or kidneys (Fig. 3I and S4I). yjbB encodes a hypothetical sodium-dependent phosphate exporter (38). In co-challenge with wild-type CFT073, the yjbB::Tn mutant outcompeted CFT073 by 1 log in the urine, bladder, and kidneys; therefore, it was not validated as a fitness candidate (Fig. 3J and S4J). pshM, which encodes a putative type II secretion protein, was the highest-ranked fitness factor as determined by fold change. When the pshM::Tn mutant was simultaneously inoculated into mice with wild-type CFT073, no significant difference in CI was observed in the urine, bladder, or kidneys (Fig. 3K and S4K); therefore, pshM was unable to be validated as a fitness factor. yjfO has been renamed bsmA for biofilm stress and motility, as it was determined to be crucial for withstanding biofilm stressors and enhancing motility in soft agar (39). In our study, the yjfO::Tn mutant had no fitness defect in vivo (Fig. 3L and S4L).
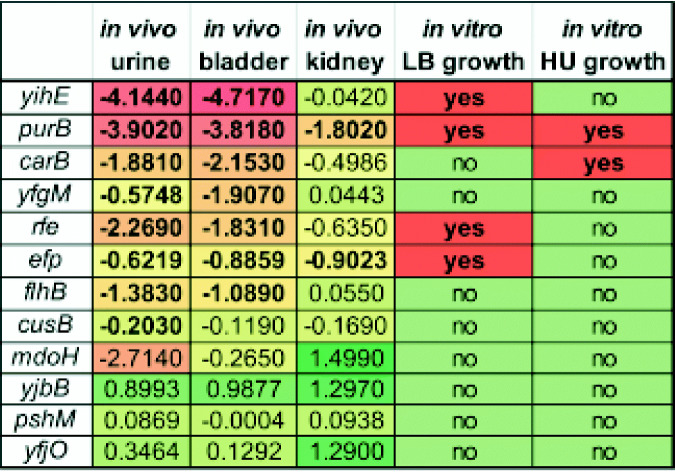
In summary, 7 of our 12 tested transposon mutants were validated during co-challenge experiments in the mouse bladder (Table 2). Therefore, our high-throughput results yielded a 58% success rate, although we selected mutants by avoiding those which had been previously studied. Considering the four genes that can be validated by literature and previous studies, our validation rate improves to 69%. Similarly, since 4 flagellar genes were on the fitness factor list but only one was selected for testing and all prevent the formation of functional organelles, we infer that all would be important (Table 1). Considering these two factors, the validation rate was increased to 75%, a rate typical for Tn-seq studies of Gram-negative pathogens in murine models of infection such as bacteremia and pneumonia (9, 11).
TABLE 2.
Classification of validation hitsa
All 12 mutants that were chosen for 1:1 co-challenge are listed here. The median log10 CI value in each of the organ sites is recorded, with a color scheme for red having the most severe mutant defect and green the least. The presence of growth defects in either human urine (HU) or LB medium is also indicated. Bold text indicates that values were statistically significant.
Growth defects of mutants selected for validation experiments.
Urine serves as the growth medium during UTI, providing a dilute mixture of amino acids as the main carbon source (40, 41). The ability of UPEC to successfully grow in this nutrient-limited medium is an adaptation considered to be an infection-specific fitness trait. As an initial test to begin defining a mechanism for potential in vivo fitness defects, we examined the growth of the selected mutants in human urine and LB medium. Overnight cultures were back diluted into fresh medium, and the optical density at 600 nm (OD600) was taken every 15 min for up to 24 h.
The poor growth of a mutant strain in human urine does not indicate a fitness defect, but faulty growth can be a contributing factor for reduced colonization in vivo. The purB::Tn mutant had growth defects in both human urine and LB medium; however, the growth in LB medium was deficient only during the exponential growth phase, and the final OD reached wild-type strain levels (see Fig. S5A in the supplemental material). There were defects for carB::Tn in both the exponential growth phase and saturation of OD when grown in human urine, but there were none in LB (Fig. S5A and B). In both LB medium and human urine, the efp::Tn mutant was found to have a lag during the exponential growth phase (Fig. S5A and B); however, only the growth in LB medium was statistically significantly impacted. The rfe::Tn mutant displayed a decreased growth rate in LB medium and had slightly lower final cell density than the wild type, which was statistically significant as determined by area under the curve (AUC) analysis (Fig. S5A). The yihE::Tn mutant also had statistically significant growth defects in LB as determined by AUC analysis (Fig. S5A). All other mutants tested in vivo exhibited no significant growth defect in either LB medium or human urine in vitro.
Two mutants (purB::Tn and carB::Tn) had a statistically significant in vitro growth defect in human urine and also had an in vivo fitness defect in the mouse bladder (Table 2). A total of five additional mutants displayed an in vivo defect in the bladder but did not have a statistically significant in vitro growth defect in human urine. Eight mutants displayed a fitness defect in the urine during co-challenge, but only two of them had deficient in vitro growth in human urine (Table 2). All four mutants that had statistically significant growth defects in LB medium also presented an in vivo fitness defect in the urine and bladder. However, four other mutants also had statistically significantly reduced colonization in the urine and bladder but with no observable decrease in growth in LB medium (Table 2). Taken together, growth in LB medium or human urine was unable to explain mutants with defects during UTI.
Validation of Tn-seq hits as fitness factors in the standard ascending UTI model.
As previously described, the bottleneck during murine UTI dictated that the Tn-seq experiments be performed for a duration of 16 h in a BALB/c model. To test if findings from this modified infection model were comparable to those from the traditional model of ascending murine UTI, four ordered library transposon mutants (yihE::Tn, purB::Tn, carB::Tn, and pshM::Tn) were tested in a 48-h infection in CBA/J mice (Fig. 4). Compared to infections in BALB/c mice, the bacterial burden in the bladders of CBA/J mice was lower and with greater deviation among mice; however, the kidneys contained more total bacteria. Ten CBA/J mice were infected via the transurethral route with a 1:1 co-challenge of mutant and wild-type E. coli CFT073. After 48 h, the urine, bladder, and kidneys were harvested, and the corresponding CFU of each strain was determined via differential plating.
FIG 4.
Validation of Tn-seq hits as fitness factors in the traditional ascending UTI model. Ten CBA/J mice were transurethrally inoculated with 1 × 108 CFU of UPEC, which consisted of a 1:1 mixture of wild-type CFT073 and an isogenic transposon mutant. Urine, bladder, and kidneys were collected at 48 hpi, and resulting homogenates were plated on LB agar with and without kanamycin to determine each contributing bacterial burden of the mutant and wild type, respectively. Each data point represents the log10 competitive index calculated for individual mouse organ; each mouse is represented by a single color. Black lines indicate the median. A competitive index of 0 concludes that the wild-type and mutant have equal fitness. P values determined by the Wilcoxon signed-rank test are listed below.
During a 16 h infection, the yihE::Tn kinase mutant displayed a consistent and dramatic loss of fitness in the urine and bladders of BALB/c mice (Fig. 3A). Similarly, the yihE::Tn mutant displayed a 2-log deficiency in both the urine and bladder at 48 h and reduced fitness in the kidneys (Fig. 4A; see S6B in the supplemental material). The purB::Tn mutant demonstrated a statistically significant fitness defect in all three organs during infection of BALB/c mice following a 16-h infection (Fig. 3B). Likewise, the purB::Tn mutant exhibited a fitness defect in the urine, bladder, and kidneys at 48 hpi, resulting in 3-log, 1-log, and 3-log losses, respectively (Fig. 4B and S6B). The carB::Tn mutant was observed to have a 10-fold fitness defect in the urine and bladder (Fig. 3C), and these findings were validated in the 48-h infection model (Fig. 4C and S6C). The defect of the carB::Tn mutant in the kidneys was even more severe than that in the 16-h BALB/c model (Fig. 3C), with a mean 2-log CI deficiency, which was statistically significant (Fig. 5C and S6C). Even though pshM::Tn was not validated in the 16-h BALB/c experiment (Fig. 3K), which mimicked the methodology of Tn-seq, we wanted to test whether this candidate was perhaps relevant in an infection model with a longer time point. Consistent with the 16-hpi data, the pshM::Tn mutant did not have a fitness defect in the 48-h infection model in the CBA/J mice (Fig. 4D and S6D). Mutant CFU were slightly higher in all three organs than wild-type CFU, although the difference was not statistically significant. Overall, we found that our modified 16-h BALB/c UTI model was also predictive of fitness during the traditional 48-h infection in CBA/J mice.
FIG 5.
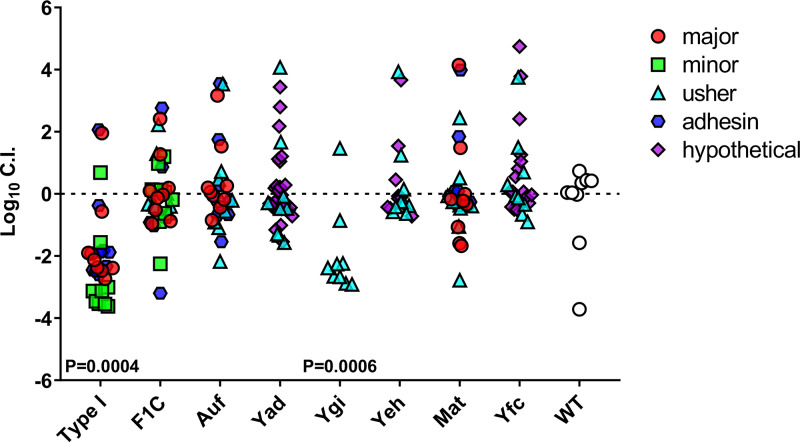
Utilizing qPCR to determine fimbria mutant fitness during UTI. Ten BALB/c mice were transurethrally inoculated with 108 CFU of mixed CFT073 mutants. Twenty-three unique gene transposon mutants with disruptions in known fimbria genes were selected to compete with the marked wild-type ΔaraF::Kan strain in co-challenge. qPCR on the gDNA harvested from the mouse bladders was used to determine log10 CI compared to the total bacterial gDNA. Ordered library mutants have been categorized by the domain containing the insertion: major, minor, usher, adhesion, and hypothetical components. The type 1 fimbrial gene cluster has 3 represented mutants (30 total symbols), F1C has 4, Auf has 3, Yad has 4, Ygi is represented by a single transposon mutant, Yeh has 2, Mat has 3, and Yfc has 3. P values determined by the Wilcoxon signed-rank test are listed if significant.
Circumventing the bottleneck during UTI by utilizing subgroup co-challenge with output determined by qPCR.
We also developed an alternate method to quantify bacteria that utilizes qPCR to quantify bacteria recovered following in vivo UTI. This method allows custom groups of mutants, assigned by gene function, to be pooled for direct competition, and it results in a mutant hierarchy based on fitness defect. This novel technique also eliminates the expensive and time-consuming costs of deep-sequencing methods for each in vivo Tn-seq competition experiment. This better allowed us to study multiple fitness factor candidates in the UTI murine model, reduced bottleneck limitations, and eliminated the need for multiple antibiotic resistance cassettes to distinguish mutants from each other.
To test this proof of concept, a well-known virulence factor, type 1 fimbriae, and seven other fimbrial types (F1C, Auf, Yad, Ygi, Yeh, Mat, and Yfc) were selected for our first experiment. From the available fimbrial gene mutants in the transposon library, a collection of 23 mutants, showing insertions in the major pilin, minor pilin, usher, adhesin, or a hypothetical domain, were utilized. Mutants were competed against wild-type CFT073 containing a mutation in araF, the same strain used in bottleneck studies (Fig. S1). A total of 14 BALB/c mice were inoculated with 108 CFU consisting of an equal ratio of transposon mutants as well as the marked wild-type ΔaraF::Kanr strain. Mouse organs were harvested 16 hpi, and homogenates were spread onto LB agar plates to generate lawns. Bacterial lawns were collected into sterile phosphate-buffered saline (PBS), and genomic extraction was performed for qPCR. Primers were designed to amplify each specific transposon mutation and ΔaraF::Kanr, as well as a control primer to amplify total bacterial genomic DNA (gDNA). The absolute qPCR quantification was used to calculate the copy number for each strain in the mixed population. Log10 competitive indices were generated by calculating the input and output of each mutant over the total bacterial input and output for each mouse bladder. This method displays the variation within the wild-type control and shows the dynamics of the group as a whole, rather than generating a competitive index in relation to the wild type. Type 1 fimbriae were confirmed as a fitness factor with a median log10 CI of −2.5 (Fig. 5). This proof-of-concept experiment demonstrated that known virulence factors with fitness defects in the traditional co-challenge model could also be detected with this method.
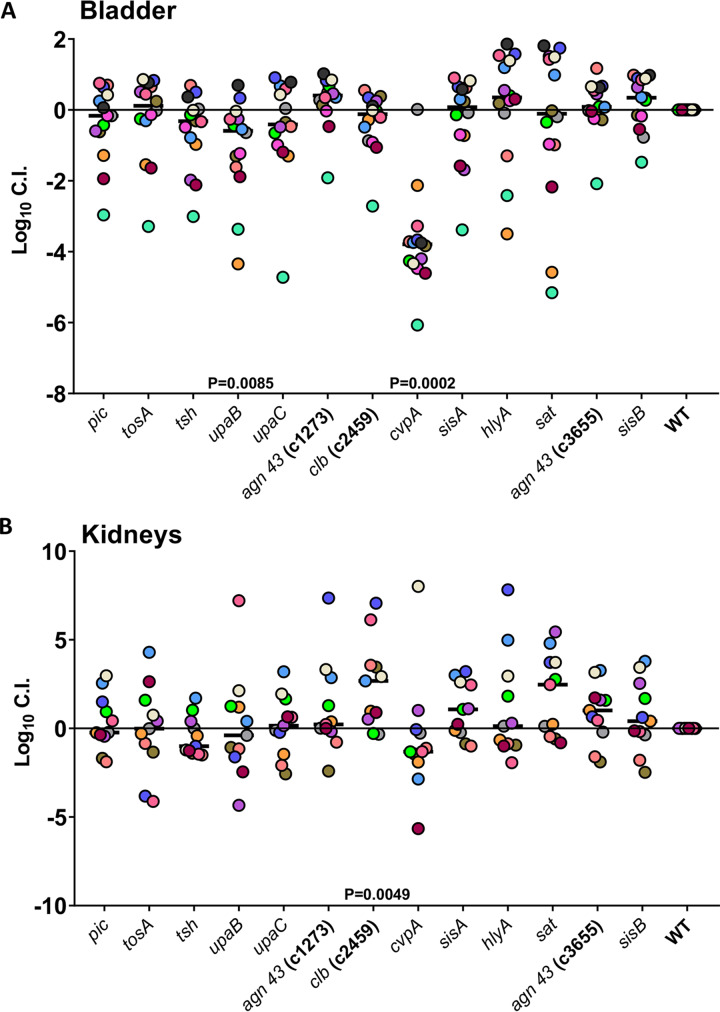
For the second experiment, toxins and virulence factors, such as hemolysin, autotransporters, colibactin, and other elements known for interacting with the host during infection, were also examined to determine if their fitness defects could be recapitulated by this qPCR method. Three predicted fitness factors (pic, upaB, and cvpA) identified in the original Tn-seq data set of this study were tested. Additionally, tosA, tsh, upaB, upaC, agn43, clb, sisA, hlyA, sat, and sisB were included in this experiment. We utilized qPCR to determine the copy number of each mutant and the marked wild type in each mouse bladder and kidney sample. The copy number determined from input and output samples of each mutant was normalized to the marked wild type for calculating log10 CI. Although displaying a trend toward reduced fitness, the pic::Tn mutant did not confirm a statistically significant decrease in fitness in the bladders of BALB/c mice (Fig. 6A). However, the upaB::Tn and cvpA::Tn mutants had statistically significantly reduced log10 CIs in the bladder compared to the marked wild-type CFT073 strain, with 1-log and 3-log decreases, respectively (Fig. 6A). Both the ΔupaB and ΔupaC mutants were previously tested for colonization in murine UTI models. Loss of upaB, but not upaC, was found to cause a statistically significant fitness defect during co-challenge (42). Previous studies utilized C57BL/6 mice, and samples collected at 18 hpi resulted in a 2-log fitness defect (42). CvpA has also been suggested to play a role in bladder colonization. A ΔcvpA purF double mutant was found to have a 2-log reduction in fitness of the C3H/HeN mouse bladder at 16 hpi (27). Several other genes encoding known virulence proteins were previously reported not to have a fitness defect in vivo: ΔsisA, ΔsisB, Δc1273, Δc3655, and ΔhlyA (43, 44). These findings were consistent with our qPCR co-challenge infection model; these mutants did not have a reduction in fitness. The fitness impacts of these mutations during kidney infection are less clear. Our qPCR data suggest that colibactin perhaps plays a negative role during kidney infection, because the mutant outcompeted the wild type by 1 log in the kidneys (Fig. 7B). Our overall findings, especially from bladder tissue, give us confidence that qPCR performed on co-challenge subgroups can provide an accurate and more convenient way to simultaneously assess the fitness of small groups of mutants in the UTI model.
FIG 6.
Applying the qPCR co-challenge technique to groups of toxin mutants from bladder and kidneys. Fourteen BALB/c mice were transurethrally inoculated with 108 CFU of mixed CFT073 mutants. We combined 13 transposon mutants, pulled from the ordered library, with mutations in known toxins and virulence genes and ranked these in relation to wild-type CFT073. Each mouse is represented by a single color; the black line indicates the median. qPCR was performed on the gDNA harvested from mouse bladders (A) or kidneys (B) to determine Log10 CI compared to WT. If significant, P values determined by the Wilcoxon signed-rank test are listed.
FIG 7.
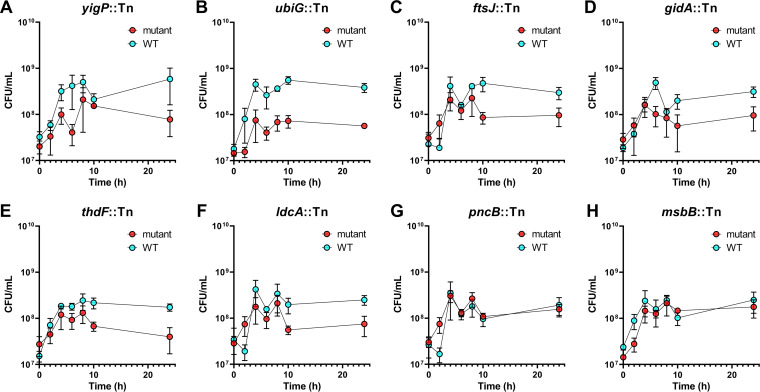
Co-challenge validation of select genes discovered using human urine in vitro Tn-seq. Cocultures of 1:100 back diluted wild-type CFT073 and a single isogenic transposon mutant were incubated statically at 37°C in human urine for 24 h. Differential plating for CFU was done at 0, 2, 4, 6, 8, 10, and 24 h. The mean from three independent trials is plotted by a single dot at every time point, displayed with standard error of the mean (SEM). Each mutant is individually plotted with wild-type CFT073 for comparison.
Identification of UPEC fitness factors during growth in human urine.
The entire condensed library of CFT073 was subjected to growth in human urine. Similarly to the Tn-seq experiments, the input versus output of each mutant was compared (see Data Set S3 in the supplemental material). Fitness genes required for in vitro growth in human urine are listed in Table 3. A total of 87 potential fitness genes were identified in our screen, showing a fold change (FC) from 2 to 12.2. Interestingly, eight of these genes (oxyR, fdhD, purB, yjfO, carB, yfgM, ldcA, and ftsJ) were also detected as fitness genes in the UTI model (Table 1). To validate the observed fitness defect for growth in human urine, eight mutants showing different FC values (yigP [FC = 8.0], ubiG [FC = 3.6], ftsJ [FC = 2.2], gidA [FC = 4.1], thdF [FC = 3.7], ldcA [FC = 2.3], msbB [FC = 4.0], and pncB [FC = 12.2]) (Table 3) were selected from the ordered library and cultured independently in human urine and LB medium (see Fig. S7 and S8 in the supplemental material). All mutants except the msbB::Tn and pncB::Tn mutants (75%) displayed a visible growth defect in human urine compared to wild-type strain CFT073, although only three were calculated to be significant (Fig. S7). Three of the 8 mutants tested also have significant growth defects in LB medium (Fig. S8). To better mimic the original Tn-seq experimental design, we also conducted co-challenge growth curves of each individual transposon mutant mixed with the wild type (Fig. 7). The mixture of strains was back diluted into human urine and incubated statically, and CFU were enumerated at several time points. In total, 6 out of 8 mutants tested were found to be at least 5-fold outcompeted by the wild type after 24 h of incubation (Fig. 7).
TABLE 3.
E. coli CFT073 genes with significant fitness defects (≥2.0-fold change; adjusted P < 0.05) for growth in human urine
| Locus |
Protein | Fold change | P value | Adjusted P value | |
|---|---|---|---|---|---|
| RefSeq | GenBank | ||||
| C_RS00815 | c0175_folK | 2-Amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase | 20.9 | 0.0000 | 0.0000 |
| C_RS00820 | c0176_pcnB | Poly(A) polymerase | 12.2 | 0.0000 | 0.0000 |
| C_RS23240 | c4917_argC | N-Acetyl-gamma-glutamyl-phosphate reductase | 9.1 | 0.0000 | 0.0000 |
| C_RS19495 | c4118_yheM | Hypothetical protein YheM | 8.1 | 0.0000 | 0.0000 |
| C_RS23250 | c4919_argH | Argininosuccinate lyase | 8.1 | 0.0000 | 0.0000 |
| C_RS22590 | c4783_yigP | Hypothetical protein YigP | 8.0 | 0.0000 | 0.0000 |
| C_RS16210 | c3416_recC | Exodeoxyribonuclease V gamma chain | 7.1 | 0.0000 | 0.0000 |
| C_RS07955 | c1700_galU | UTP-glucose-1-phosphate uridylyltransferase | 6.9 | 0.0000 | 0.0000 |
| C_RS23265 | c4922_oxyR | Hydrogen peroxide-inducible gene activator (morphology and autoaggregation control protein) | 6.7 | 0.0000 | 0.0000 |
| C_RS22910 | c4847_fdhD | FdhD protein | 5.9 | 0.0000 | 0.0000 |
| C_RS20980 | c4446_rfaF | ADP-heptose-LPS heptosyltransferase II | 5.5 | 0.0000 | 0.0000 |
| C_RS23155 | c4899_metF | 5,10-Methylenetetrahydrofolate reductase | 5.2 | 0.0000 | 0.0000 |
| C_RS25275 | c5353_argI | Ornithine carbamoyltransferase chain I | 5.1 | 0.0000 | 0.0000 |
| C_RS18635 | c3929_argG | Argininosuccinate synthase | 4.8 | 0.0000 | 0.0000 |
| C_RS23545 | c4970_metA | Homoserine O-succinyltransferase | 4.7 | 0.0000 | 0.0000 |
| C_RS07045 | c1510_purB | Adenylosuccinate lyase | 4.7 | 0.0000 | 0.0000 |
| C_RS15475 | c3253_recA | RecA protein (recombinase A) | 4.6 | 0.0000 | 0.0000 |
| C_RS22450 | c4751_metE | 5-Methyltetrahydropteroyltriglutamate–homocysteine methyltransferase | 4.3 | 0.0000 | 0.0000 |
| C_RS22445 | c4750_metR | Transcriptional activator protein MetR | 4.2 | 0.0000 | 0.0000 |
| C_RS00670 | c0142_aceE | Pyruvate dehydrogenase E1 component | 4.2 | 0.0000 | 0.0000 |
| C_RS22005 | c4669_gidA | Glucose inhibited division protein A | 4.1 | 0.0000 | 0.0000 |
| C_RS10745 | c2269_msbB | Lipid A biosynthesis (KDO)2-(lauroyl)-lipid IVA acyltransferase | 4.0 | 0.0000 | 0.0000 |
| C_RS24865 | c5261_purA | Adenylosuccinate synthetase | 3.8 | 0.0000 | 0.0000 |
| C_RS24925 | c5277_yjfO | Hypothetical lipoprotein YjfO precursor | 3.8 | 0.0000 | 0.0000 |
| C_RS21835 | c4630_thdF | Probable tRNA modification GTPase TrmE | 3.7 | 0.0000 | 0.0000 |
| C_RS13160 | c2774_ubiG | 3-Demethylubiquinone-9,3-methyltransferase | 3.6 | 0.0000 | 0.0000 |
| C_RS16190 | c3412_argA | Amino acid acetyltransferase | 3.4 | 0.0000 | 0.0000 |
| C_RS18695 | c3941_yhbE | Hypothetical transport protein YhbE | 3.3 | 0.0010 | 0.0036 |
| C_RS03320 | c0715_crcB | Protein CrcB | 3.2 | 0.0000 | 0.0000 |
| C_RS16485 | c3469_lysS | Lysyl-tRNA synthetase | 3.2 | 0.0000 | 0.0000 |
| C_RS20870 | c4417_mtlD | Mannitol-1-phosphate 5-dehydrogenase | 3.2 | 0.0000 | 0.0000 |
| C_RS22140 | c4692_ilvE | Branched-chain amino acid aminotransferase | 3.1 | 0.0000 | 0.0000 |
| C_RS19690 | c4157_dam | DNA adenine methylase | 3.0 | 0.0000 | 0.0000 |
| C_RS19695 | c4158_damX | DamX protein | 2.9 | 0.0000 | 0.0000 |
| C_RS22355 | c4732_xerC | Integrase/recombinase XerC | 2.9 | 0.0000 | 0.0000 |
| C_RS21050 | c4460_rpmG | 50S ribosomal protein L33 | 2.8 | 0.0000 | 0.0000 |
| C_RS17965 | c3781_tolC | Outer membrane protein TolC precursor | 2.8 | 0.0000 | 0.0000 |
| C_RS13375 | c2819_nuoL | NADH dehydrogenase I chain L | 2.7 | 0.0000 | 0.0000 |
| C_RS03535 | c0753_nagB | Glucosamine-6-phosphate isomerase | 2.7 | 0.0000 | 0.0000 |
| C_RS16565 | c3489_ubiH | 2-Octaprenyl-6-methoxyphenol hydroxylase | 2.7 | 0.0000 | 0.0000 |
| C_RS22585 | c4782_ubiE | Ubiquinone/menaquinone biosynthesis methyltransferase UbiE | 2.6 | 0.0000 | 0.0000 |
| C_RS13430 | c2830_lrhA | Probable transcriptional regulator LrhA | 2.6 | 0.0000 | 0.0000 |
| C_RS19570 | c4132_crp | Catabolite gene activator (cAMP receptor protein) | 2.4 | 0.0000 | 0.0001 |
| C_RS00420 | c0091_leuA | 2-Isopropylmalate synthase | 2.4 | 0.0000 | 0.0000 |
| C_RS04715 | c0996_artI | Arginine-binding periplasmic protein 1 precursor | 2.4 | 0.0000 | 0.0000 |
| C_RS18280 | c3850_uxaC | Uronate isomerase | 2.4 | 0.0000 | 0.0000 |
| C_RS00190 | c0041_carB | Carbamoyl-phosphate synthase large chain | 2.4 | 0.0000 | 0.0000 |
| C_RS00830 | c0178_dksA | DnaK suppressor protein | 2.4 | 0.0000 | 0.0000 |
| C_RS19475 | c4112_fusA | Elongation factor G | 2.4 | 0.0000 | 0.0000 |
| C_RS04860 | c1027_ftsK | Cell division protein FtsK | 2.4 | 0.0000 | 0.0000 |
| C_RS21670 | c4596_ilvB | Acetolactate synthase isozyme I large subunit | 2.4 | 0.0000 | 0.0000 |
| C_RS23235 | c4916_argE | Acetylornithine deacetylase | 2.3 | 0.0000 | 0.0000 |
| C_RS00665 | c0140_pdhR | Pyruvate dehydrogenase complex repressor | 2.3 | 0.0000 | 0.0000 |
| C_RS14420 | c3035_yfgM | Hypothetical protein YfgM | 2.3 | 0.0000 | 0.0000 |
| C_RS07675 | c1641_ldcA | Muramoyltetrapeptide carboxypeptidase | 2.3 | 0.0000 | 0.0000 |
| C_RS15895 | c3347_relA | GTP pyrophosphokinase | 2.3 | 0.0000 | 0.0000 |
| C_RS00415 | c0090_leuB | 3-Isopropylmalate dehydrogenase | 2.3 | 0.0000 | 0.0000 |
| C_RS00900 | c0195_pfs | MTA/SAH nucleosidase (P46) | 2.2 | 0.0000 | 0.0000 |
| C_RS13580 | c2864_usg | USG-1 protein | 2.2 | 0.0000 | 0.0000 |
| C_RS18670 | c3936_ftsJ | Ribosomal RNA large subunit methyltransferase J | 2.2 | 0.0000 | 0.0001 |
| C_RS02240 | c0483_yaiW | Hypothetical protein YaiW | 2.2 | 0.0000 | 0.0000 |
| C_RS02560 | c0557_ybaU | Peptidyl-prolyl cis-trans isomerase D | 2.2 | 0.0000 | 0.0000 |
| C_RS16560 | c3488_visC | Protein VisC | 2.2 | 0.0000 | 0.0000 |
| C_RS14930 | c3141_yfjG | Hypothetical protein YfjG | 2.2 | 0.0000 | 0.0001 |
| C_RS15990 | c3371_fucI | l-Fucose isomerase | 2.1 | 0.0005 | 0.0020 |
| C_RS20385 | c4316_chuY | ORF; hypothetical protein | 2.1 | 0.0000 | 0.0000 |
| C_RS11625 | c2450 | Hypothetical protein | 2.1 | 0.0000 | 0.0000 |
| C_RS21130 | c4477_recG | ATP-dependent DNA helicase RecG | 2.1 | 0.0000 | 0.0000 |
| C_RS08235 | c1757_rnb | Exoribonuclease II | 2.1 | 0.0000 | 0.0000 |
| C_RS09455 | c2005_manA | Mannose-6-phosphate isomerase | 2.1 | 0.0000 | 0.0000 |
| C_RS12710 | c2682_mglC | Galactoside transport system permease protein MglC | 2.1 | 0.0000 | 0.0000 |
| C_RS18405 | c3879_yhaD | Glycerate kinase 2 | 2.1 | 0.0000 | 0.0000 |
| C_RS05060 | c1075_pepN | Aminopeptidase N | 2.1 | 0.0000 | 0.0000 |
| C_RS22255 | c4715_wecF | Putative enterobacterial common antigen polymerase | 2.0 | 0.0000 | 0.0000 |
| C_RS00410 | c0089_leuC | 3-Isopropylmalate dehydratase large subunit | 2.0 | 0.0000 | 0.0000 |
| C_RS21965 | c4659_atpG | ATP synthase gamma chain | 2.0 | 0.0000 | 0.0001 |
| C_RS04380 | c0926_ybjG | Hypothetical protein YbjG | 2.0 | 0.0000 | 0.0000 |
| C_RS18355 | c3867 | Conserved hypothetical protein | 2.0 | 0.0000 | 0.0000 |
| C_RS22820 | c4829 | Hypothetical protein | 2.0 | 0.0000 | 0.0000 |
| C_RS18790 | c3963_yhbH | Probable sigma 54 modulation protein (ORF3) | 2.0 | 0.0000 | 0.0000 |
| C_RS01720 | c0374_yafK | Hypothetical protein YafK precursor | 2.0 | 0.0000 | 0.0002 |
| C_RS16205 | c3415_ptr | Protease III precursor | 2.0 | 0.0000 | 0.0000 |
| C_RS21360 | c4524 | Putative propanol dehydrogenase | 2.0 | 0.0000 | 0.0000 |
| C_RS20580 | c4354_yhjV | Hypothetical transport protein YhjV | 2.0 | 0.0000 | 0.0000 |
| C_RS20325 | c4299_gor | Glutathione reductase | 2.0 | 0.0000 | 0.0000 |
| C_RS04850 | c1025_trxB | Thioredoxin reductase | 2.0 | 0.0000 | 0.0000 |
| C_RS22645 | c4794_pepQ | Xaa-Pro dipeptidase | 2.0 | 0.0000 | 0.0000 |
Of the identified fitness genes, 17.2% represented amino acid biosynthesis operons, including arginine (argACEGHI), leucine (leuABC), methionine (metAER), and isoleucine/valine (ilvBE) genes. UPEC mutants that were auxotrophic for arginine were previously reported to demonstrate a severe growth defect in human urine, while leucine and methionine autotrophs displayed a moderate growth rate (45). According to previously published findings, argCEG and metA biosynthesis genes were also described to be upregulated in the UTI89 strain during growth in human urine (46). Thus, our results are consistent with those findings (45) in showing that arginine and methionine biosyntheses are important for UPEC fitness in urine and that mutations affecting these processes can affect the virulence of UPEC.
Genes involved in degradation of biomolecules were also well represented in our screening (11.5%). This functional group included genes involved in degradation of protein and peptides (pepQN and protease III [ptr]), carbon compounds (galU, manA, fucI, uxaC, and mtlD), and nucleic acids (recC and rnb). Six genes involved in cell division were detected as less competitive in growth in urine: gydA, which regulates the initiation of replication (47); ftsK, required for normal septation during cell division (48); ftsJ, encoding a methyltransferase; the cell division gene damX, mediator of reversible filamentation during UTI in strain UTI89 (49); xerC, encoding integrase/recombinase which are homologous to those in Staphylococcus aureus and contribute to biofilm-associated infections and acute bacteremia (50); and tolC, an outer membrane protein-coding gene. Ubiquinone is a lipid-soluble electron transporter in the plasma membrane of prokaryotes. Although there are no studies reporting the importance of this molecule during E. coli growth in urine, our screening detected that the disruption of four ubiquinone biosynthesis genes mutations (ubiEGH and yigP) resulted in a less competitive phenotype.
We chose four of the studied mutants with known growth defects in vitro (the yigP::Tn, ubiG::Tn, efp::Tn, and purB::Tn mutants) to complement using the pGEN vector (see Fig. S9 in the supplemental material). We compared the growth of wild-type CFT073 containing the empty vector to that of each respective mutant containing either empty pGEN or pGEN carrying the gene of interest. The successful complementation can be best seen in Fig. S9B, where all four empty-vector transposon mutants display visible growth defects in human urine compared to the wild type. The growth rates of all 4 mutants were improved when genetically complemented in trans compared to empty vector (Fig. S9). These data demonstrate the ability of the detected fitness factor mutants from this study to be complemented by the single gene that was originally mutated.
DISCUSSION
Uropathogenic E. coli (UPEC) is the primary causative organism of uncomplicated urinary tract infections (UTI) and has been thoroughly studied in murine ascending models of UTI. Specifically, E. coli CFT073 has served as the model strain for nearly 300 studies and has been extensively annotated, with the genome sequence published in 2002, representing only the third E. coli genome completed and annotated (22). This strain has been cited in the literature over 10,326 times (Web of Science). Many large screens using transposon mutants have been completed using this model organism; however, they were met with two major limitations. Previous random transposon studies utilized uncatalogued mutants, which decreased the genome coverage and increased the number of mutant strains required to obtain that coverage. The second limitation occurs specifically in the urinary tract model, where we have identified a severe bottleneck effect that causes loss of random mutants, not as a result of fitness defects. This observation was first documented in signature-tagged mutagenesis studies and has been expanded upon in this study (13). Despite numerous attempts to identify fitness factors in the CFT073 genome during UTI, there has yet to be a comprehensive, large, unbiased study.
This work utilized an ordered library of catalogued CFT073 mutants generated using previously defined protocols in other model organisms (17). A total of 9,216 mutants were generated and sequenced to identify the specific site of transposon insertion within the CFT073 genome. A total of 7,123 unique transposon mutations were identified, occurring in 2,913 genes. A transposon mutant was chosen to represent each of the 2,913 interrupted genes; this collection created the condensed library that was tested in the ascending murine model of UTI. The advantage of this approach is the limited number of mutants required to obtain maximum nonessential genome coverage, which is especially important when considering the bottleneck effect. Although using ordered libraries helps alleviate some aspects related to bottleneck during infection, other intrinsic limitations exist that cannot be completely circumvented. For example, although 54% of genes were successfully mutated, only mutations in coding sequences were included in this study. Regulatory sequences such as small RNAs (sRNAs) are not considered in our approach. Additionally, the idea of potential transcomplementation between strains within a given library can mask deficient phenotypes, particularly with secreted molecules such as toxins and siderophores.
After carefully calculating the bottleneck effect and adjusting the model of ascending infection to utilize BALB/c mice with a 16-h infection time point, we determined that up to 500 mutants could be tested in a single pool. After testing the 3,373 mutants divided into seven pools for infection, 201 genes were identified as preliminary fitness factors during bladder infection. Applying a stricter statistical threshold (P value of <0.05 and >513-fold change) to focus on the strongest results, 36 genes were ultimately presented as fitness factors from this study. Within this list of genes are several fitness factors that have previously been identified using the ascending model of UTI. These findings provide confidence that conclusions made from this modified murine model are relevant. However, nearly 60% of the genes identified have yet to be thoroughly studied in the context of fitness factors during UTI. Another strong feature of this data set is the redundancy of multiple genes within operons or similar systems being identified. For example, five flagellum genes, two lipopolysaccharide genes, two type 1 fimbria genes, and two purine biosynthesis genes were classified as fitness factors. This work is the first use of an ordered library in a uropathogen for screening in vivo during the ascending model of UTI and therefore serves as guidance for future studies investigating important virulence properties of uropathogens during UTI.
Using a traditional co-challenge infection, we were able to confirm 7 out of 12 selected transposon mutations as fitness factors in the mouse bladder, as defined by a significantly reduced competitive index. Some of these genes, such as yihE, yfgM, rfe, and efp, have not been studied in UPEC. Thus, this large nonbiased genomic screen has produced valuable results and candidate genes which can be pursued for mechanistic and follow-up studies. Importantly, when four mutants tested in the modified UTI model were competed in the traditional murine model of ascending UTI, they displayed the same colonization trends during infection. We therefore have assurance that the candidate genes presented in this study are highly likely to be important bacterial factors during UTI in multiple murine models.
The murine model bottleneck causes a considerable limitation in the deconvolution of large data sets and elimination of the false-positive hits. To circumvent these limitations in our model, in addition to the creation of an ordered library, we sought to develop an alternate method for testing multiple mutants during infection. We demonstrate a novel approach to co-challenge experiments in which multiple mutants can be competed against the wild type to assign competitive indices. The bioinformatic pipelines used for the analysis of high-throughput sequencing experiments do not generate competitive indices for individual mutants. Additionally, deep-sequencing techniques have a much greater associated cost and are more time-consuming than the qPCR method established here. This method provides a relative ranking of fitness for small groups of transposon mutants (up to 23) compared to a marked wild-type strain. This method was validated first with a group of fimbria mutants, in which the well-known type 1 fimbria served as an internal control (23). Indeed, the fim mutants were outcompeted when mixed within a group co-challenge during ascending UTI. This confirms that mutants of interest can be competed against one another in order to identify and rank the greatest fitness defects worth pursuing. This method was tested further by generating a hierarchy of toxin mutants. Several of the included mutants had been previously assessed in infection; for example, hemolysin, sisA, sisB, agn43, sat, and upaC mutants are all reported to have no fitness defects in a traditional 1:1 co-challenge infection (42–44). Likewise, cvpA and upaB are known to have fitness defects, and these were detected in our qPCR co-challenge experiment (27, 42). We have confidence that this novel approach will serve as a valuable alternative to deep-sequencing techniques when appropriate. This method also significantly reduces the number of animals required to test multiple mutants compared to the traditional 1:1 co-challenge approach.
This work provides insight into the bottleneck associated with UTI and offers methods to circumvent this effect to decrease the randomness of data. We provide several candidate fitness genes in UPEC, many of which have yet to be investigated. Our method for utilizing qPCR will reduce the number of animals compared to traditional co-challenge experiments and provide an alternate method for testing multiple transposon mutants in pools. Overall, we have provided invaluable insight into the kinetics of the UTI model and have provided methodological tools to help overcome the limitations detected in this model in order to promote the discovery of novel fitness genes. This ordered library in prototype UPEC strain E. coli CFT073 will serve as a tool for many future screens and studies, as it provides a readily available source of annotated transposon mutants.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Strains and plasmids used in this study are listed in Table S4 in the supplemental material. Bacteria were cultured statically in lysogeny broth (LB) or filter-sterilized pooled human urine at 37°C. Antibiotics and reagents were added as required at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; and diaminopimelic acid (DAP), 50 μg/ml. Human urine was collected from anonymized healthy female adult volunteers at the University of Michigan who had not had UTI or undergone any antibiotic treatment in the prior 6 weeks. After filter sterilization (0.22 μm), pooled urine specific gravity was measured at the University of Michigan Hospital via refractometer and adjusted to 1.006, and the urine was stored at –20°C. Urine collection was performed as approved by the University of Michigan Institutional Review Board (HUM00004949).
Animal protocols.
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Michigan Medical School (PRO00009173), in accordance with the Office of Laboratory Animal Welfare (OLAW) and the U.S. Department of Agriculture (USDA), as well as guidelines specified by the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC, Intl.). Mice were anesthetized with a weight-appropriate dose (0.1 ml for a mouse weighing 20 g) of ketamine-xylazine (80 to 120 mg/kg ketamine and 5 to 10 mg/kg xylazine) by intraperitoneal injection. Mice were euthanized by inhalant anesthetic overdose, which was followed by organ removal.
Mouse model of UTI.
Bacteria were cultured statically in ∼30 ml LB overnight at 37°C. The next day, bacteria were pelleted by centrifugation at 2,754 × g for 30 min at room temperature. The bacterial pellet was resuspended in 6 ml phosphate-buffered saline (PBS) (pH 7.4) and adjusted to an optical density at 600 nm (OD600) of ∼4.0 (3.2 × 109 CFU/ml). BALB/c or CBA/J mice were inoculated transurethrally with 50 μl of inoculum (1.6 × 108 CFU/mouse). Mice were euthanized at 16, 24, or 48 h postinoculation (hpi), and urine, bladders, or kidneys were harvested into PBS (pH 7.4) and homogenized using an Omni TH homogenizer (Omni International). A 100-μl sample was removed and plated onto LB agar plates (50 μl/plate) with and without kanamycin using an Autoplate 4000 spiral plater (Spiral Biotech) for enumeration of colonies. When required, the remaining bladder homogenates were spread in their entirety onto LB agar plates supplemented with kanamycin, and colonies were collected, pelleted, and frozen for sequencing or qPCR.
Bottleneck assessment.
Colonization bottlenecks were assessed by competition infections as described previously (11) using the wild-type strain CFT073 and the nonattenuated isogenic mutant CFT073 ΔaraF::Kanr. Both strains were cultivated as described above and mixed in 1:1, 1:100, and 1:500 ratios (mutant to WT) prior to inoculation of BALB/c mice. Animals were sacrificed at 16 or 24 hpi, and the bladders were harvested and homogenized. The proportion of each strain in the input and output samples was determined by plating on LB agar plates with or without kanamycin. The competitive index (CI) was calculated as follows: CI = (mutant output CFU/WT output CFU)/(mutant input CFU/WT input CFU).
Generation of transposon mutants.
A mariner-based transposon carried by suicide plasmid pSAM_AraC containing an arabinose-inducible mariner himar1C9 transposase (8) was used for generation of the E. coli CFT073 transposon mutant library.
A library of random transposon mutants was created by mating a mid-log-phase culture of DAP-auxotrophic E. coli MGN-617-λpir carrying pSAM_AraC (donor strain) with a pre-heat-shocked (30 min at 50°C) mid-log-phase culture of E. coli CFT073 (recipient strain) at a 1:1 ratio of donor to recipient. Ten mating mixes were pelleted, incubated at room temperature for 5 min, and gently resuspended in 150 μl LB-DAP. Mating mixtures were then spread onto 0.22-μm filter disks (Millipore) on the surface of LB-DAP agar plates and incubated at 37°C for 4 h. The filters then were transferred to LB agar plates supplemented with 0.02% arabinose (Sigma) and incubated at 37°C for 1 h to induce transposase expression via the pBAD promoter. Filter disks were transferred to tubes, and cells were washed from filters with 2 ml of LB. Suspensions diluted 10−2 were plated on LB agar supplemented with 25 μg/ml kanamycin and incubated at 37°C overnight to isolate E. coli mutants harboring the transposon and remove the MGN-617 DAP-auxotrophic bacteria and were plated on LB agar supplemented with ampicillin (100 μg/ml) to determine the frequency of pSAM_AraC retention. Fewer than 0.00001% of mutants retained the pSAM_AraC vector.
The occurrence of one transposon insertion event per genome and randomness of insertions were verified by extracting genomic DNA from individual mutants, digesting with HindIII, and performing a Southern blot with a digoxigenin (Roche)-labeled probe targeting the Kanr cassette within the transposon.
CFT073 ordered mutant library construction.
Upon mutagenesis, 9,216 transposon mutants from ten mating reactions were picked from LB agar plates containing kanamycin (25 μg/ml) using a Qpix2 colony picker (Molecular Devices, LLC) and cultured in LB containing kanamycin (25 μg/ml) in 96- by 96-well plates at 37°C overnight. After overnight culture, a replicate of the mutant library was made to determine the chromosomal position of each transposon insertion and the position of each mutant in the archived library by CP-Cseq. Thirty percent glycerol was added to the original 96- by 96-well plate mutant library to be stored at −80°C.
The position of each mutant in the archived library was determined by CP-Cseq (17) with some modifications (see Data Set S1 in the supplemental material). Briefly, the 9,216 mutants of the replicated mutant library were combined following the specific CP-Cseq pooling strategy, which captures the positional information in 40 master pools. Genomic DNA of each of the 40 pools was sheared to yield fragments of ∼500 bp (Covaris) and ligated to customized adaptors containing different unique 6-bp barcodes for multiplex sequencing. Transposon-gDNA junctions were enriched by PCR amplification with specific primers hybridizing to the transposon ends and to the adaptor. The 40 PCR products were then combined and size selected before sequencing on a single lane of an Illumina HiSeq2500 chip using the standard Illumina sequencing primers (single-read 100-bp Cycle Rapid) with barcode sequencing. The lane was spiked with 15% bacteriophage ϕX DNA to overcome low-diversity sequences. Sequencing was performed at the University of Michigan DNA core facility. Primers and adaptors used in this CP-Cseq experiment are described in see Data Set S4 in the supplemental material. The Illumina sequencing data were processed with CLC Genomics Workbench and the open source Galaxy platform (http://galaxyproject.org/) with the Galaxy workflows described by Vandewalle et al. (17).
The mutant deconvolution was carried out using the CP-CSeq algorithm (17). To determine the number of disrupted operons, the E. coli CFT073 operon list from the DOOR2.0 operon database was used (51). An operon was considered disrupted when it harbored at least one transposon insertion. To determine the percentage of disrupted essential/nonessential genes, the Keio collection list of essential genes (21) was used and compared with the ordered library. In CP-Cseq, transposon mutants are archived in 96- by 96-well plates in a manner by which each mutant position is defined by three Cartesian coordinates. Each mutant’s x and y coordinates correspond to its row and column, and each mutant’s z coordinate corresponds to its plate within the stack. In CP-CSeq, mutants from a stack of plates are subpooled in two master plates. The first master plate (xy) combines a small culture volume of one specific well position (for example, A1) in all of the 96-well plates in the same specific well (for example, A1) of the master plate. The second master plate is constructed by collapsing the stack into a single master plate (z coordinate). Next, each row and each column of both master plates are pooled in columns (n = 12) and rows (n = 8), allowing for pool to capture the positional information of the 9,216 mutants in just 40 samples. These master plates are then profiled by sequencing, and the coordinates of each mutant are deconvoluted using the specific CP-CSeq algorithm (17).
Verification of mutant deconvolution.
Twenty-four mutants with unique transposon positions were randomly picked from the library, inoculated in 2 ml of LB, and cultured at 37°C overnight. Bacteria were washed with PBS and pelleted, and the genomic DNA was isolated by hexadecyltrimethyl ammonium bromide (CTAB) precipitation. DNA preparations were used as templates in a PCR (Phusion polymerase) with a primer hybridizing to the transposon P7 sequence and gene-specific primers hybridizing 200 to 400 bp upstream (see Table S2 in the supplemental material). PCR products were run on a 1.2% agarose gel, stained with ethidium bromide, and visualized under UV light. Only PCR products showing one single band with the expected fragment size were considered verified.
Identification of CFT073 fitness factors for bladder colonization and growth in human urine.
In order to reduce the mutant library size and fit it to the detected bottleneck, a condensed ordered library was created. One mutant per disrupted gene was selected to be picked out and represent the 2,913 disrupted genes in the transposon library. The criterion to choose among different mutants with insertions in the same gene was the proximity of the insertion to the initial codon ATG, avoiding, when possible, insertions in the last 20% of the gene sequence. These 2,913 were grouped in six pools to be tested in six mouse groups, respecting the 500-mutant size rule (named pools 1 to 6). An extra pool (named pool 9) was added to amend the selection of 460 mutants with insertions in the last 20% of the gene sequence when better candidates were available. Together, 3,373 transposon mutants were picked from the CFT073 mutant library to represent the 2,913 disrupted genes. Selected mutants were then cultured independently in 200 μl of fresh LB onto 96-well plates at 37°C for 18 h.
For the mouse model of UTI, cultures were combined in seven exclusive pools, named input pools 1, 2, 3, 4, 5, 6 and 9, grouping fewer than 500 mutants per pool, and then adjusted to an OD600 of ∼4.0 (∼3 × 109 CFU/ml) (input samples). For each transposon pool input, five BALB/c mice (Jackson Laboratory) were inoculated transurethrally with 108 CFU (average mutant coverage = 4.4 × 105). Mice were euthanized 16 h postinoculation, and bladders were harvested into 1 ml of PBS (pH 7.4). Tissues were homogenized using an Omni TH homogenizer (Omni International), and a 100-μl aliquot sample was removed and plated for enumeration of colonies. The remaining bladder homogenates were spread plated in their entirety, and colonies were collected, pelleted, and frozen for sequencing (output samples).
For growth in human urine, cultures were pooled and adjusted to an OD600 of ∼4.0 (3 × 109 CFU/ml). Twenty microliters of bacterial suspension (6 × 107 CFU; average mutant coverage = 1.77 × 104) was inoculated in 20 ml of pooled human urine (four replicates) and cultured statically for 16 h at 37°C. Cultures were pelleted and frozen for sequencing (output samples).
Illumina sequencing and mapping of transposon insertion sites.
Genomic DNA was isolated from each input inoculum (two technical replicates) and from output samples by hexadecyltrimethyl ammonium bromide (CTAB) precipitation. Samples were enriched for transposon insertion junctions as outlined by Goodman et al. (52). TapeStation analysis was used to confirm the concentration and purity of the transposon insertion junctions, which were then multiplexed and subjected to V4 single-end 50 HiSeq-2500 high-output sequencing. Sequencing was performed at the University of Michigan DNA core facility. The Goodman IN-seq pipeline (52) was applied on the raw reads to perform read filtration, transposon nucleotide removal, debarcoding, alignment, and insertion calling. A script was written to map the insertions onto the CFT073 genome. For each gene, this script used 100% of the gene body as the effective gene region. Sequence data were deposited in the SRA database under accession number PRJNA596215.
Input pool quality analysis.
To evaluate whether all the transposon mutants that were selected from the ordered library are certainly present in the output pool samples, mapped insertions at TA sites showing a read coverage over 3 were compared to the list of selected insertions (see Fig. S10 in the supplemental material). Table S3 in the supplemental material summarizes the number of expected and unexpected insertions found in each input pool sample and the insertion read count median. Overall, the number of expected insertions found in the input pool samples was high, ranging from a minimum of 91.24% (input pool 6) up to a maximum of 95.04% (input pool 5). In the other hand, a high number of unexpected insertions at TA sites that passed over read coverage filter (>3) was also found, ranging from 299 reads (input pool 6, replicate 2) up to 2,507 reads (input pool 9, replicate 2). However, the median of the insertion read count of those putative unexpected insertions was very low in all the samples (from 6 up to 47 reads), indicating that those insertions could be artifacts or noise. A graphic representation of the insertion read coverage of each mapped insertion at TA sites (read coverage > 3) is shown in Fig. S10 in the supplemental material. Expected and unexpected insertions grouped in two different populations separated by a gap of read count. Expected insertions showed the highest read coverages, and unexpected insertions showed the lowest coverages. To reduce false insertions or artifacts, only expected insertions were analyzed by the TnseqDiff package.
Identification of CFT073 fitness factors.
Only expected insertions from the ordered transposon mutant library identified in input samples were considered for estimation of conditional fitness genes. When indicated, potential read count bias caused by the DNA replication process was first corrected. Specifically, a mean count in each genomic region was estimated from the LOESS function to obtain a bias factor for each region. The observed insertion counts at any particular location were then divided by the bias factor for their genomic region. The nonzero mean (NZmean) method in the TRANSIT software (53) was used to normalize data sets, and then TnseqDiff (54) was used to estimate the fitness contribution of each tested gene. TnseqDiff takes into account the unique features of Tn-seq data and identifies conditionally essential genes using insertion-level data. First, it collects evidence of conditional essentiality for each insertion by comparing read counts of that insertion between conditions, with the counts at each insertion site modeled as a linear function of the condition (input/output). To account for the overdispersion of the count data produced from the sequencing technique, a precision weight was estimated for each observation from the mean-variance relationship of the log counts and incorporated into the linear modeling. The CD function was constructed using the slope (log fold change) estimates (mean, standard error, and degrees of freedom) from the linear model. Finally, insertion-level CD functions were combined to obtain a single CD function for the corresponding gene, and a P value was derived from this combined CD function to infer the fitness effect of that gene. These P values were further adjusted to control for the false-discovery rate (55). Except when indicated, significant genes for further analysis were selected based on a P value of <0.05 and a >2-fold ratio of output to input (see Data Set S2 in the supplemental material).
Co-challenge model of UTI (1:1 infection).
Transposon mutants and wild-type CFT073 were cultured as described above. Both strains were adjusted to an OD600 of ∼4.0 (3.2 × 109 CFU/ml) in PBS and mixed in equal volume to create the inoculum. The input inoculum was plated for enumeration. BALB/c or CBA/J mice were inoculated transurethrally with 50 μl of inoculum (1.6 × 108 CFU/mouse). For each co-challenge, 5 to 10 mice were infected. Urine was collected into a sterile microcentrifuge tube, and the total volume was measured and adjusted to 150 μl using 1× PBS. Mice were sacrificed 16 h (BALB/c) or 48 h (CBA/J) postinfection. Bladders and kidneys were harvested, weighed, and homogenized into 3 ml of 1× PBS. Tissues were homogenized using an Omni TH homogenizer (Omni International), and a 1-ml aliquot was put into a microcentrifuge tube for further dilution and plating. Urine samples were plated undiluted and at a 10−2 dilution; bladders and kidneys were plated undiluted and at a 10−1 dilution on both plain and kanamycin-containing LB agar plates. Fifty microliters was spiral plated using an Autoplate 4000 (Spiral Biotech), plates were incubated at 37°C overnight, and counting was done using a QCount automated plate counter (Spiral Biotech). A log10 competitive index (CI) was calculated using the formula cited above. If the mutant came out in the same ratio relative to the wild type, indicating that neither strain had a fitness advantage, the log10 CI was 0; the log10 CI was >0 if the mutant had a fitness advantage over the wild-type strain and was <0 if the wild type had a competitive advantage over the isogenic mutant.
Co-challenge model of UTI using qPCR to quantify bacterial burden of individual mutants.
Transposon mutants and CFT073 ΔaraF were cultured statically in ∼15 ml LB overnight at 37°C. The next day, bacteria were pelleted by spinning at 2,754 × g for 30 min at room temperature. The bacterial pellet was resuspended in 3 ml 1× PBS. All cultures were adjusted to an OD600 of ∼4.0 (3.2 × 109 CFU/ml) and mixed in equal volumes to create the inoculum. One-milliliter samples of input inoculum were collected for the gDNA sample. Fifteen BALB/c mice were inoculated transurethrally with 50 μl of inoculum (1.6 × 108 CFU/mouse). Mice were sacrificed 16 h postinfection (hpi). Bladders and kidneys were harvested, weighed, and homogenized into 1 ml of PBS. CFU were determined as described above. After spiral plating, remaining tissue homogenate was spread-plated onto 10 LB agar plates containing kanamycin (25 μg/ml) and incubated overnight at 37°C. The next day, bacterial lawns were collected into 10 ml PBS using sterile cotton swabs. Each lawn sample (one per organ for each mouse) was adjusted to an OD600 of ∼4.0 and aliquoted into 1-ml volumes. The aliquots (in addition to the input gDNA sample) were spun at 16,770 × g for 5 min, the supernatant was aspirated, and pellets were stored at −20°C. Genomic DNA was isolated from the pellet using the Wizard genomic purification kit (Promega). The DNA concentration for each sample was determined using the Nanodrop instrument. All samples were normalized to 10 ng/μl for qPCR.
qPCR to determine CI during co-challenge.
Primer pairs were designed for each mutant using one common primer embedded within the transposon sequence (CP7) and one unique primer inside the CFT073 genome within 250 bp of the insertion site. Primers for the wild-type strain (CFT073 ΔaraF) were designed within the kanamycin cassette of pKD4. Control primers were designed to target the adenylate kinase gene (adk). All primer efficiencies and concentrations were determined prior to running experimental samples, using previously described methods (56). Primers are listed in Table S4 in the supplemental material. Experimental qPCR was performed using Power SYBR green master mix (Invitrogen) with 10 μl of 10-ng/μl gDNA in a total reaction volume of 25 μl. For each mutant primer pair, a control template gDNA sample was added at a concentration of 1 ng/μl. Values for adk detection were used for normalization of samples and to calculate the threshold cycle (2−ΔΔCT) values. Each mouse organ output sample, and the input inoculum sample, was compared to the template control to determine the gDNA concentration for that respective mutant within the total sample. The concentration of gDNA (total ng in the sample) was divided by 9.974 × 10−6 to obtain the copy number. The competitive index (CI) for each individual transposon mutant strain relative to the wild-type was then determined.
Growth curves.
LB cultures were inoculated from a single colony and incubated overnight with shaking at 37°C, with kanamycin (25 μg/ml) for transposon mutants. Overnight cultures were washed one time in PBS and diluted 1:100 into fresh LB or filter-sterilized pooled human urine from healthy donors (no antibiotics). A Bioscreen-C automated growth curve analysis system (Growth Curves USA) took OD600 readings every 15 min for 24 h. Cultures were incubated with continuous shaking at 37°C.
Statistical analysis.
The Wilcoxon signed-rank test was used for all co-challenge experiments to conclude which transposon mutant strains were significantly outcompeted by the wild type. The area under the curve (AUC) was calculated for growth curves. AUC values were compared to that for the wild type by Kruskal-Wallis one-way analysis of variance (ANOVA) to determine statistical power. GraphPad Prism version 7 (GraphPad Software, San Diego, CA) was used to perform the analysis.
Data availability.
Sequence data were deposited in the SRA database under accession number PRJNA596215.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Weisheng Wu for bioinformatic expertise, Charles M. Dozois for providing E. coli MGN-617, Sébastien Crépin and Valerie Forsyth for their contributions to experiments, and Mark Anderson, Melanie Pearson, and Chris Alteri for their edits and input to the text of the manuscript.
J.M. acknowledges the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia, for his postdoctoral grant (ED481B 2014/018-0). This work was funded in part by Public Health Service grants R01 AI059722 and R01 DK094777 from the National Institutes of Health.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Foxman B. 2014. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 28:1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jancel T, Dudas V. 2002. Management of uncomplicated urinary tract infections. West J Med 176:51–55. doi: 10.1136/ewjm.176.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooton TM. 2012. Clinical practice. Uncomplicated urinary tract infection. N Engl J Med 366:1028–1037. doi: 10.1056/NEJMcp1104429. [DOI] [PubMed] [Google Scholar]
- 5.Subashchandrabose S, Mobley H. 2015. Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol Spectr 3:UTI-0015-2012. doi: 10.1128/microbiolspec.UTI-0015-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielubowicz GR, Mobley HL. 2010. Host-pathogen interactions in urinary tract infection. Nat Rev Urol 7:430–441. doi: 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 7.Johnson JR. 2003. Microbial virulence determinants and the pathogenesis of urinary tract infection. Infect Dis Clin North Am 17:261–278. doi: 10.1016/S0891-5520(03)00027-8. [DOI] [PubMed] [Google Scholar]
- 8.Armbruster CE, Forsyth-DeOrnellas V, Johnson AO, Smith SN, Zhao L, Wu W, Mobley H. 2017. Genome-wide transposon mutagenesis of Proteus mirabilis: essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog 13:e1006434. doi: 10.1371/journal.ppat.1006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachman MA, Breen P, Deornellas V, Mu Q, Zhao L, Wu W, Cavalcoli JD, Mobley HL. 2015. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6:e00775. doi: 10.1128/mBio.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armbruster CE, Forsyth VS, Johnson AO, Smith SN, White AN, Brauer AL, Learman BS, Zhao L, Wu W, Anderson MT, Bachman MA, Mobley H. 2019. Twin arginine translocation, ammonia incorporation, and polyamine biosynthesis are crucial for Proteus mirabilis fitness during bloodstream infection. PLoS Pathog 15:e1007653. doi: 10.1371/journal.ppat.1007653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson MT, Mitchell LA, Zhao L, Mobley H. 2018. Citrobacter freundii fitness during bloodstream infection. Sci Rep 8:11792. doi: 10.1038/s41598-018-30196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HL. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9:e1003788. doi: 10.1371/journal.ppat.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walters MS, Lane MC, Vigil PD, Smith SN, Walk ST, Mobley HL. 2012. Kinetics of uropathogenic Escherichia coli metapopulation movement during urinary tract infection. mBio 3:e00303-11. doi: 10.1128/mBio.00303-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz DJ, Chen SL, Hultgren SJ, Seed PC. 2011. Population dynamics and niche distribution of uropathogenic Escherichia coli during acute and chronic urinary tract infection. Infect Immun 79:4250–4259. doi: 10.1128/IAI.05339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drenkard E, Hibbler RM, Gutu DA, Eaton AD, Silverio AL, Ausubel FM, Hurley BP, Yonker LM. 2018. Replication of the ordered, nonredundant Library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. J Vis Exp 2018:e57298. doi: 10.3791/F57298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol 14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandewalle K, Festjens N, Plets E, Vuylsteke M, Saeys Y, Callewaert N. 2015. Characterization of genome-wide ordered sequence-tagged Mycobacterium mutant libraries by Cartesian pooling-coordinate sequencing. Nat Commun 6:7106. doi: 10.1038/ncomms8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dale JL, Beckman KB, Willett JLE, Nilson JL, Palani NP, Baller JA, Hauge A, Gohl DM, Erickson R, Manias DA, Sadowsky MJ, Dunny GM. 2018. Comprehensive functional analysis of the Enterococcus faecalis core genome using an ordered, sequence-defined collection of insertional mutations in strain OG1RF. mSystems 3:e00062-18. doi: 10.1128/mSystems.00062-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connolly J, Boldock E, Prince LR, Renshaw SA, Whyte MK, Foster SJ. 2017. Identification of Staphylococcus aureus factors required for pathogenicity and growth in human blood. Infect Immun 85:e00337-17. doi: 10.1128/IAI.00337-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alteri CJ, Smith SN, Mobley HL. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog 5:e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch RA, Burland V, Plunkett G III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahrani-Mougeot FK, Buckles EL, Lockatell CV, Hebel JR, Johnson DE, Tang CM, Donnenberg MS. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol Microbiol 45:1079–1093. doi: 10.1046/j.1365-2958.2002.03078.x. [DOI] [PubMed] [Google Scholar]
- 24.Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A 93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguiniga LM, Yaggie RE, Schaeffer AJ, Klumpp DJ. 2016. Lipopolysaccharide domains modulate urovirulence. Infect Immun 84:3131–3140. doi: 10.1128/IAI.00315-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorsey-Oresto A, Lu T, Mosel M, Wang X, Salz T, Drlica K, Zhao X. 2013. YihE kinase is a central regulator of programmed cell death in bacteria. Cell Rep 3:528–537. doi: 10.1016/j.celrep.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaffer CL, Zhang EW, Dudley AG, Dixon B, Guckes KR, Breland EJ, Floyd KA, Casella DP, Algood HMS, Clayton DB, Hadjifrangiskou M. 2017. Purine biosynthesis metabolically constrains intracellular survival of uropathogenic Escherichia coli. Infect Immun 85:e00471-16. doi: 10.1128/IAI.00471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadjifrangiskou M, Gu AP, Pinkner JS, Kostakioti M, Zhang EW, Greene SE, Hultgren SJ. 2012. Transposon mutagenesis identifies uropathogenic Escherichia coli biofilm factors. J Bacteriol 194:6195–6205. doi: 10.1128/JB.01012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotzke H, Muheim C, Altelaar AF, Heck AJ, Maddalo G, Daley DO. 2015. Identification of putative substrates for the periplasmic chaperone YfgM in Escherichia coli using quantitative proteomics. Mol Cell Proteomics 14:216–226. doi: 10.1074/mcp.M114.043216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gotzke H, Palombo I, Muheim C, Perrody E, Genevaux P, Kudva R, Muller M, Daley DO. 2014. YfgM is an ancillary subunit of the SecYEG translocon in Escherichia coli. J Biol Chem 289:19089–19097. doi: 10.1074/jbc.M113.541672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander DC, Valvano MA. 1994. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J Bacteriol 176:7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forsyth VS, Armbruster CE, Smith SN, Pirani A, Springman AC, Walters MS, Nielubowicz GR, Himpsl SD, Snitkin ES, Mobley H. 2018. Rapid growth of uropathogenic Escherichia coli during human urinary tract infection. mBio 9:e00186-18. doi: 10.1128/mBio.00186-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol 48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- 34.Williams AW, Yamaguchi S, Togashi F, Aizawa SI, Kawagishi I, Macnab RM. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol 178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subashchandrabose S, Hazen TH, Brumbaugh AR, Himpsl SD, Smith SN, Ernst RD, Rasko DA, Mobley HL. 2014. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci U S A 111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Outten FW, Huffman DL, Hale JA, O'Halloran TV. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem 276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 37.Debarbieux L, Bohin A, Bohin JP. 1997. Topological analysis of the membrane-bound glucosyltransferase, MdoH, required for osmoregulated periplasmic glucan synthesis in Escherichia coli. J Bacteriol 179:6692–6698. doi: 10.1128/jb.179.21.6692-6698.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Motomura K, Hirota R, Ohnaka N, Okada M, Ikeda T, Morohoshi T, Ohtake H, Kuroda A. 2011. Overproduction of YjbB reduces the level of polyphosphate in Escherichia coli: a hypothetical role of YjbB in phosphate export and polyphosphate accumulation. FEMS Microbiol Lett 320:25–32. doi: 10.1111/j.1574-6968.2011.02285.x. [DOI] [PubMed] [Google Scholar]
- 39.Weber MM, French CL, Barnes MB, Siegele DA, McLean RJ. 2010. A previously uncharacterized gene, yjfO (bsmA), influences Escherichia coli biofilm formation and stress response. Microbiology 156:139–147. doi: 10.1099/mic.0.031468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruckner H, Haasmann S, Friedrich A. 1994. Quantification of d-amino acids in human urine using GC-MS and HPLC. Amino Acids 6:205–211. doi: 10.1007/BF00805848. [DOI] [PubMed] [Google Scholar]
- 41.Patzold R, Schieber A, Bruckner H. 2005. Gas chromatographic quantification of free d-amino acids in higher vertebrates. Biomed Chromatogr 19:466–473. doi: 10.1002/bmc.515. [DOI] [PubMed] [Google Scholar]
- 42.Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo JM, Schembri MA. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect Immun 80:321–332. doi: 10.1128/IAI.05322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lloyd AL, Smith SN, Eaton KA, Mobley HL. 2009. Uropathogenic Escherichia coli Suppresses the host inflammatory response via pathogenicity island genes sisA and sisB. Infect Immun 77:5322–5333. doi: 10.1128/IAI.00779-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haugen BJ, Pellett S, Redford P, Hamilton HL, Roesch PL, Welch RA. 2007. In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain CFT073 dsdA. Infect Immun 75:278–289. doi: 10.1128/IAI.01319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hull RA, Hull SI. 1997. Nutritional requirements for growth of uropathogenic Escherichia coli in human urine. Infect Immun 65:1960–1961. doi: 10.1128/IAI.65.5.1960-1961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madelung M, Kronborg T, Doktor TK, Struve C, Krogfelt KA, Moller-Jensen J. 2017. DFI-seq identification of environment-specific gene expression in uropathogenic Escherichia coli. BMC Microbiol 17:99. doi: 10.1186/s12866-017-1008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lies M, Visser BJ, Joshi MC, Magnan D, Bates D. 2015. MioC and GidA proteins promote cell division in E. coli. Front Microbiol 6:516. doi: 10.3389/fmicb.2015.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubarry N, Possoz C, Barre FX. 2010. Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol Microbiol 78:1088–1100. doi: 10.1111/j.1365-2958.2010.07412.x. [DOI] [PubMed] [Google Scholar]
- 49.Khandige S, Asferg CA, Rasmussen KJ, Larsen MJ, Overgaard M, Andersen TE, Moller-Jensen J. 2016. DamX controls reversible cell morphology switching in uropathogenic Escherichia coli. mBio 7:e00642-16. doi: 10.1128/mBio.00642-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atwood DN, Beenken KE, Loughran AJ, Meeker DG, Lantz TL, Graham JW, Spencer HJ, Smeltzer MS. 2016. XerC contributes to diverse forms of Staphylococcus aureus infection via agr-dependent and agr-independent pathways. Infect Immun 84:1214–1225. doi: 10.1128/IAI.01462-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao F, Dam P, Chou J, Olman V, Xu Y. 2009. DOOR: a database for prokaryotic operons. Nucleic Acids Res 37:D459–D463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodman AL, Wu M, Gordon JI. 2011. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc 6:1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT—a software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11:e1004401. doi: 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao L, Anderson MT, Wu W, Mobley HLT, Bachman MA. 2017. TnseqDiff: identification of conditionally essential genes in transposon sequencing studies. BMC Bioinformatics 18:326. doi: 10.1186/s12859-017-1745-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data were deposited in the SRA database under accession number PRJNA596215.