The implementation of multiple hurdles in the beef production chain has resulted in substantial improvement in the microbial safety of beef in Canada. In this study, we characterized a large number of Escherichia coli isolates (n = 1,450) from various sources/stages of beef processing to determine whether the commonly used antimicrobial interventions would give rise to heat-resistant E. coli on meat, which in turn may require alternatives to the current control of pathogens and/or modifications to the current cooking recommendations for meat. The findings show that the degree and rate of heat resistance in E. coli did not increase along the production chain or with time. This furthers our understanding of man-made ecological niches that are required for the development of heat resistance in E. coli.
KEYWORDS: D value, Escherichia coli, antimicrobial interventions, beef packing plant, cattle, heat resistance, locus of heat resistance
ABSTRACT
Decontamination practices, which often involve thermal treatments, are routinely performed in beef packing plants and have generally improved the safety of meat in North America. We investigated whether Escherichia coli in the beef production chain is becoming more heat resistant due to those treatments. Cattle isolates (n = 750) included seven serogroups (O157, O103, O111, O121, O145, O26, and O45) which were collected between 2002 and 2017. Beef plant isolates (n = 700) from carcasses, fabrication equipment, and beef products were included. Heat resistance was determined in Luria-Bertani broth at 60°C and by PCR screening for the locus of heat resistance (LHR). The decimal reduction for E. coli at 60°C (D60ºC values) ranged from 0 to 7.54 min, with 97.2% of the values being <2 min. The prevalence of E. coli with D60ºC values of >2 min was not significantly different (P > 0.05) among cattle and meat plant isolates. E. coli from equipment before sanitation (median, 1.03 min) was more heat resistant than that after sanitation (median, 0.9 min). No significant difference in D60ºC values was observed among E. coli isolates from different years, from carcasses before and after antimicrobial interventions, or from before and during carcass chilling. Of all isolates, 1.97% harbored LHR, and the LHR-positive isolates had greater median D60ºC values than the LHR-negative isolates (3.25 versus 0.96 min). No increase in heat resistance in E. coli was observed along the beef production chain or with time.
IMPORTANCE The implementation of multiple hurdles in the beef production chain has resulted in substantial improvement in the microbial safety of beef in Canada. In this study, we characterized a large number of Escherichia coli isolates (n = 1,450) from various sources/stages of beef processing to determine whether the commonly used antimicrobial interventions would give rise to heat-resistant E. coli on meat, which in turn may require alternatives to the current control of pathogens and/or modifications to the current cooking recommendations for meat. The findings show that the degree and rate of heat resistance in E. coli did not increase along the production chain or with time. This furthers our understanding of man-made ecological niches that are required for the development of heat resistance in E. coli.
INTRODUCTION
Escherichia coli is a Gram-negative, facultative anaerobe that inhabits the gut of warm-blooded animals. Most strains of E. coli are nonpathogenic, but some strains are pathogenic to humans and animals (1). Clinical signs of intestinal diseases caused by pathogenic E. coli range from self-limiting diarrhea to life-threatening hemolytic uremic syndrome (HUS), the latter of which is most often caused by Shiga toxin-producing E. coli (STEC) strains. Cattle are natural reservoirs of STEC, but they lack vascular receptors for Shiga toxins (2). The STEC which has caused the most cases of human disease is E. coli O157:H7, which can result in up to 10% of infected people developing severe and potentially fatal HUS (3). In North America, more than 50% of STEC infections are caused by E. coli O157:H7/NM, and the rest, primarily by serogroups O103, O111, O121, O145, O26, and O45 (4, 5). Together, they are commonly referred to as the “Top 7” serogroups.
Hides of cattle are often heavily contaminated with E. coli, which may be transferred to previously bacterium-free carcasses or the environment of beef processing plants (6, 7). To control the contamination of beef carcasses with E. coli, hazard analysis critical control point (HACCP) programs have been required in federally inspected meat plants in North America since the 1990s, and antimicrobial interventions are often incorporated at various stages of meat processing (6–10). Commonly used antimicrobial interventions may include washing hides on carcasses with a solution of 1.5% sodium hydroxide at 55°C (11, 12), washing skinned carcasses with organic acids such as lactic, acetic, and peroxyacetic acids, and pasteurization of carcass sides with hot water (85°C) or steam (>90°C) (10). To prevent bacterial growth, dressed carcasses are chilled to a low temperature before further processing (7, 8). Beef processing equipment surfaces are cleaned and sanitized daily, if not more often (13, 14). These decontamination practices can reduce the number of bacteria, including E. coli, on carcasses and beef products to a very low number, as evidenced by recent studies carried out in commercial beef packing plants (11, 15–18), and in turn, led to the decline of human disease incidence associated with E. coli O157:H7/NM in Canada (18). However, the use of these physical and chemical treatments may also lead to development of bacterial resistance.
E. coli is considered to be a species sensitive to heat, with the decimal reduction at 60°C (D60ºC values; i.e., the time required to inactivate 90% of a bacterial population at 60°C) being mostly less than 2 min (19, 20). However, in recent years, a remarkably heat-resistant E. coli strain (AW1.7), originally isolated from beef carcasses at a Canadian beef packing plant, has been reported (21), followed by isolation of heat-resistant E. coli strains from other sources with D60ºC values of >10 min (22–24). Thus, it would be important to look into whether the benefits of antimicrobial interventions, which often include thermal treatments at various temperatures, currently implemented at beef packing plants are compromised by potentially giving rise to heat-resistant E. coli. The extreme heat resistance in E. coli strains is attributable to a genomic island, the locus of heat resistance (LHR), which contains genes encoding heat shock proteins and genes related to membrane and envelope integrity and oxidative stress (25–27).
Therefore, the present study characterized the relative heat resistance in E. coli isolates recovered from cattle over time and from various sources in meat plants. All isolates were screened for the presence of LHR, with the objectives to (i) better understand the flow of potentially heat-resistant E. coli in the beef production chain and (ii) determine whether antimicrobial interventions currently in use in beef processing plants select for heat-resistant E. coli. Considering the large strain variability in the bacterial response to stress, a population-based approach involving 1,450 isolates was taken in this study.
RESULTS
Overall heat resistance.
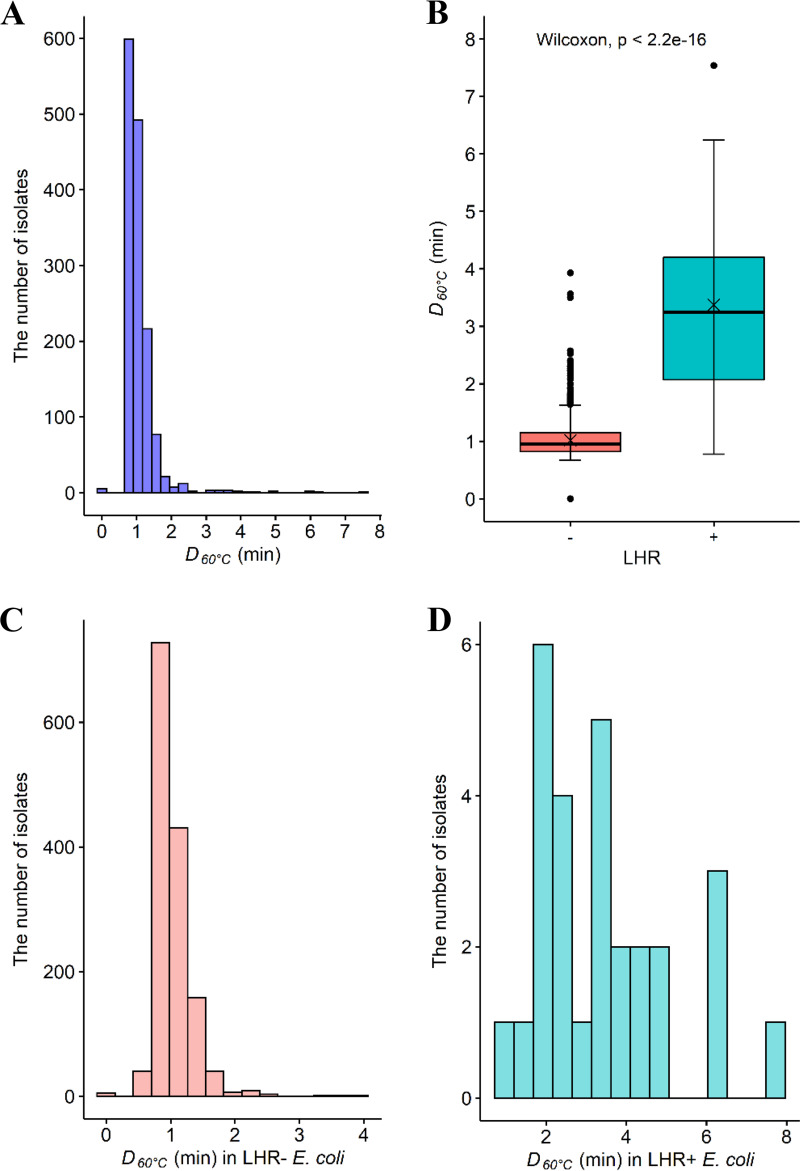
Heat resistance (D60°C [min]) was determined for a total of 1,450 isolates comprising both generic (n = 700) and the Top 7 (n = 750) E. coli (Tables 1 and 2). The D60°C values did not follow normal distribution (P < 0.05; Fig. 1A); hence, nonparametric analysis was performed for pairwise comparison, and median values instead of means were reported. The D60°C values ranged from 0 to 7.54 min, with the median value being 0.96 min. The great majority of isolates (97.2%, 1,410) had D60°C values of <2 min. For the E. coli isolates with D60°C values of >2 min, there was no significant difference (P > 0.05) in their distribution among cattle (24/750) and meat plant (16/700) isolates (Tables 1 and 2).
TABLE 1.
Escherichia coli isolates from cattleb
| Serogroupa | No. of strains isolated in yr: |
Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2011 | 2013 | 2014 | 2015 | 2017 | ||
| O103 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 86 | 25 | 6 (2) | 157 (2) |
| O111 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 7 | 0 | 12 |
| O121 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 41 (3) | 33 (1) | 28 | 1 | 103 (4) |
| O145 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 16 (1) | 18 | 0 | 35 (1) |
| O157 | 14 | 28 (1) | 10 | 9 | 6 | 57 | 67 | 11 (2) | 19 (1) | 12 (1) | 6 (1) | 239 (6) |
| O26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 42 (1) | 37 (6) | 21 (3) | 0 | 100 (10) |
| O45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 34 | 40 (1) | 28 | 2 | 104 (1) |
| Total | 14 | 28 (1) | 10 | 9 | 6 | 57 | 67 | 172 (6) | 233 (10) | 139 (4) | 15 (3) | 750 (24) |
TABLE 2.
Escherichia coli isolates from beef packing plants
| Beef processing plant | Isolation source | Stage of process | No. of isolatesb | Reference(s) |
|---|---|---|---|---|
| B | Hide-on carcasses | Dressing/before hide-on wash | 100 (1) | 11 |
| Hide-on/dressed carcasses | Dressing/after hide-on wash | 100 (3) | ||
| A | Dressed carcasses | Before chilling | 100 (6) | 54, 55 |
| During chilling | 100 | |||
| A | Fabrication equipment surface | Before sanitation | 100 (4) | 9 |
| After sanitation | 100 | |||
| A | Beef productsa | Fabrication | 100 (2) | 16 |
Isolates were recovered from samples collected from beef cuts and trimmings during routine production.
Numbers in parentheses indicate E. coli isolates with D60°C values of >2 min.
FIG 1.
D60°C values (min) for Escherichia coli isolates (n = 1,450) from various sources. (A, C, and D) The distribution of D60°C in all E. coli isolates (A), in LHR-negative (–, n = 1,422) isolates (C), and in LHR-positive (+, n = 28) isolates (D). (B) The comparison of D60°C between the LHR-positive (+, green shading) and -negative (–, orange shading) E. coli. Central horizontal lines and “x” marks indicate medians and means, respectively; whiskers indicate the lowest data point within 1.5 interquartile range (IQR) of the first quartile and the highest data point within 1.5 IQR of the third quartile. Filled circles indicate data points below or above the 1.5 IQR of the first or third quartile, respectively.
The LHR was found in 28 (1.97%) isolates (see Table S1 in the supplemental material). No significant difference (P > 0.05) in LHR prevalence was found between cattle (Top 7; 17/750) and meat plant (generic; 11/700) E. coli. Among Top 7 E. coli, the LHR was found in O157 (8 isolates), O26 (6 isolates), O103 (1 isolate), O145 (1 isolate), and O45 (1 isolate). LHR-positive generic E. coli isolates were from carcasses after hide-on wash (3 isolates), carcasses before chilling (6 isolates), and beef products (2 isolates).
D60°C values for the LHR-positive E. coli isolates (median, 3.25 min; range, 0.78 to 7.54 min) were significantly larger (P < 0.05) than those for the LHR-negative isolates (median, 0.96 min; range, 0 to 3.93 min) (Fig. 1B). The D60°C values within the respective LHR-positive and -negative E. coli groups varied widely (Fig. 1C and D). Among the LHR-positive E. coli group (Fig. 1D; Table S1), four generic E. coli isolates, which were recovered from carcasses before chilling, showed D60°C values of ≥6 min, comparable to the positive-control E. coli AW1.7 (6.16 ± 2 min), while all the other LHR-positive E. coli isolates had D60°C values of less than 6 min, with the lower limit being 0.78 min. Among the LHR-negative E. coli group, five of the Top 7 isolates, comprising O26 (1 isolate), O103 (1 isolate), and O157 (3 isolates), were all extremely sensitive to heat and were killed during the come-up time; the D60°C values for these isolates were arbitrarily assigned as 0 min. In contrast, two O103 isolates and one O26 isolate that were LHR negative had D60°C values of 3.93, 3.50, and 3.57 min (Table S2), and these values were numerically higher than the median D60°C value for the LHR-positive isolates, 3.25 min.
Effect of antimicrobial interventions on heat resistance in E. coli.
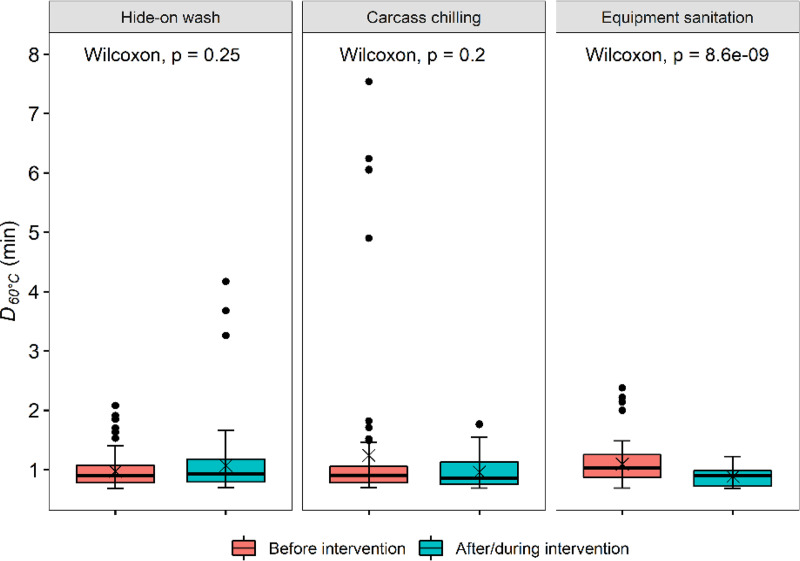
Various antimicrobial interventions are generally in use in meat processing plants in North America (7, 8). This study compared the heat resistance in E. coli recovered from hide-on carcasses before any antimicrobial treatment was applied (before hide-on wash) and from carcasses along the dressing line where multiple antimicrobial interventions were applied (after hide-on wash), from dressed carcasses before and during carcass chilling from a beef plant where no antimicrobial interventions were used during the dressing process, and from beef processing equipment surfaces before and after sanitation to investigate whether these interventions select for heat-resistant E. coli (Table 2). The median D60°C value for the isolates recovered from beef processing equipment before sanitation (1.03 min) was larger (P < 0.05) than that for isolates recovered after sanitation (0.9 min; Fig. 2). No significant difference (P > 0.05) in heat resistance was found between E. coli isolates from carcasses before and after hide-on wash or from carcasses before and during chilling (Fig. 2).
FIG 2.
D60°C values (min) for Escherichia coli isolates (n = 600) recovered from beef packing plants. From left to right are E. coli from carcasses before and after the hide-on wash intervention, E. coli from dressed carcasses before and during carcass chilling, and E. coli from fabrication equipment before and after the daily routine sanitation. Central horizontal lines and “x” marks indicate medians and means, respectively; whiskers indicate the lowest data point within 1.5 IQR of the first quartile and the highest data point within 1.5 IQR of the third quartile. Filled circles indicate data points below or above the 1.5 IQR of the first or third quartile, respectively. Each group included 100 E. coli isolates.
Year of isolation and heat resistance in E. coli.
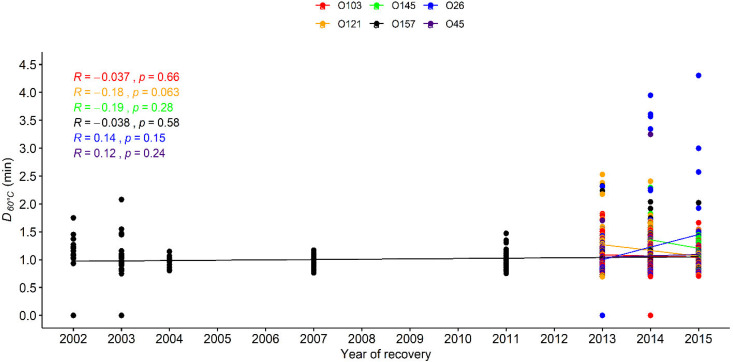
The Top 7 E. coli isolates in this study were recovered over 15 years, from 2002 to 2017 (Table 1). To investigate whether the heat resistance in E. coli changed over time, the correlation between D60°C values and years of recovery was determined using Spearman’s method. The serogroup O111 had fewer than 10 isolates from individual years and, as such, was excluded from the comparison to avoid bias in the statistical analysis. No significant correlation (P > 0.05) between D60°C values for each of the other six serogroups by year was noted (Fig. 3).
FIG 3.
D60°C values (min) for six Escherichia coli serogroups (O157, O26, O45, O103, O121, and O145) recovered from different years. The data points show the D60°C values for E. coli of the six serogroups, and each line shows the trend of the D value change for each group along the years of isolation. As the D value of each serogroup did not follow normal distribution, the correlation between heat resistance in each serogroup and recovery year was examined using Spearman’s method. The Spearman’s rank correlation coefficients (R) and P values are shown in the figure.
Heat resistance in generic versus the Top 7 E. coli.
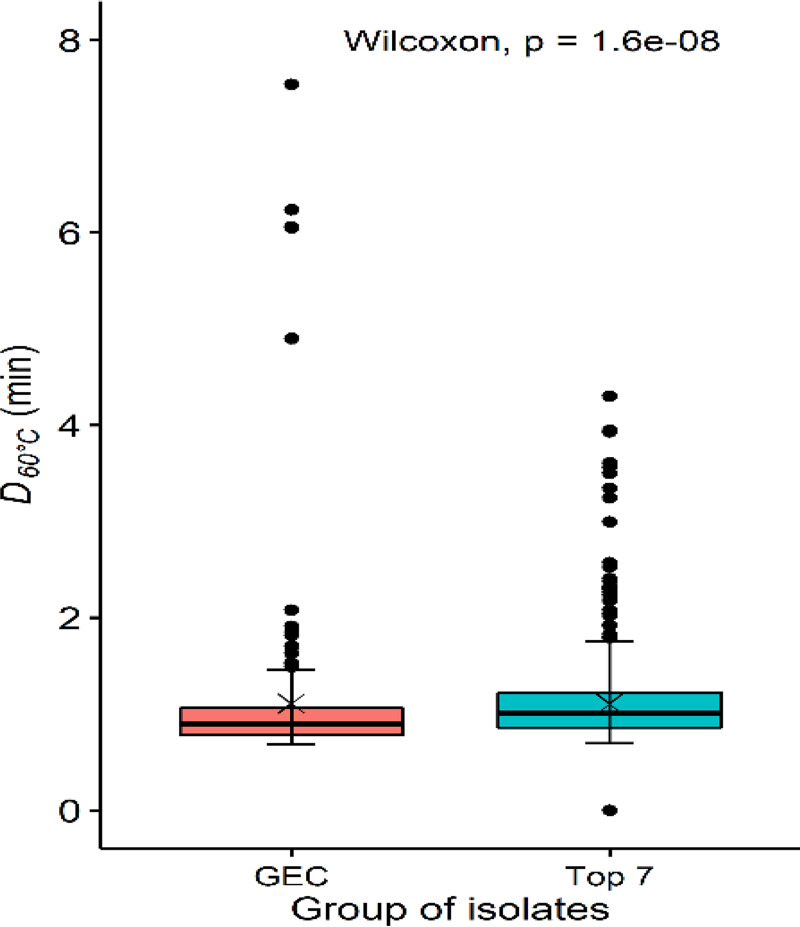
To investigate whether the heat resistance was different in the Top 7 and generic E. coli, the two groups of isolates from the common source, cattle, were involved. The Top 7 group comprised 750 STEC isolates from cattle, and the generic E. coli group comprised 200 isolates from carcasses that did not experience any antimicrobial treatment, 100 from hide-on carcasses before hide-on wash from plant B, and 100 from dressed carcasses before carcass chilling from plant A. The Top 7 isolates had a slightly higher (P < 0.05) median D60°C value than the generic E. coli isolates (1.0 versus 0.9 min; Fig. 4).
FIG 4.
D60°C values (min) for generic (GEC, n = 200) and the Top 7 (n = 750) E. coli isolates. Central horizontal lines and “x” marks indicate medians and means, respectively; whiskers indicate the lowest data point within 1.5 IQR of the first quartile and the highest data point within 1.5 IQR of the third quartile. Filled circles indicate data points below or above the 1.5 IQR of the first or third quartile, respectively.
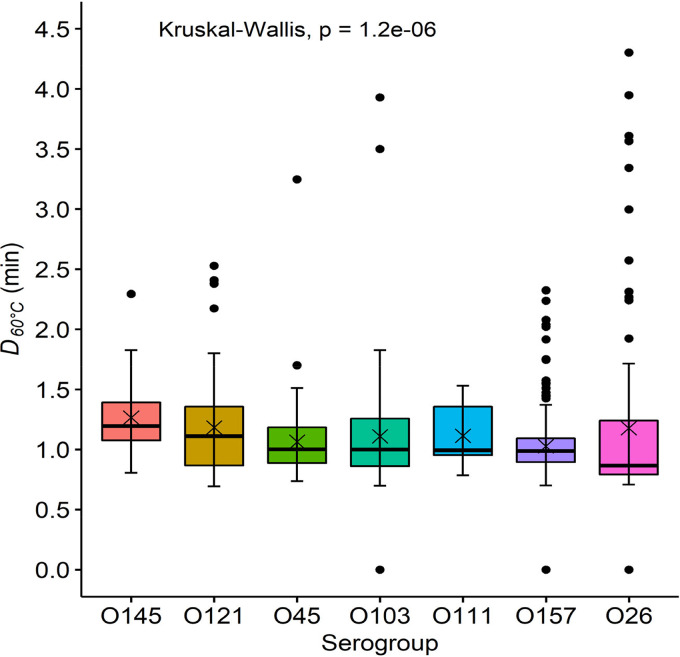
Among the Top 7 E. coli, O145 isolates were the most heat resistant, with a median D60°C value of 1.2 min, followed by O121, O111, O103, O45, O157, and O26 with D60°C values of 1.11, 1.0, 1.0, 1.0, 0.99, and 0.87 min, respectively (Fig. 5).
FIG 5.
D60°C values (min) for the Top 7 Escherichia coli isolates. Central horizontal lines and “x” marks indicate medians and means, respectively; whiskers indicate the lowest data point within 1.5 IQR of the first quartile and the highest data point within 1.5 IQR of the third quartile. Filled circles indicate data points below or above the 1.5 IQR of the first or third quartile, respectively. The Top 7 E. coli included serogroups O103 (n = 157), O111 (n = 12), O121 (n = 103), O145 (n = 35), O157 (n = 239), O26 (n = 100), and O45 (n = 104). The serogroups in the figure are ordered according to the level of the median D60°C value in each group.
Heat resistance in E. coli recovered from different seasons and sample types.
This study also explored the potential difference in heat resistance in generic E. coli from different sample sources and different sampling seasons. No significant difference (P > 0.05) was noted between E. coli from hide-on carcasses, dressed carcasses, equipment surfaces, and beef products or between different seasons in which they were recovered (Fig. S1 and S2).
DISCUSSION
This study compared the relative heat resistance of 1,450 E. coli isolates from live cattle and meat plant environments to better understand whether antimicrobial interventions commonly employed at commercial beef packing plants in North America would enrich for heat-resistant E. coli. Muscle tissues of healthy animals are largely free of bacteria. Most bacteria found on beef carcasses have been transferred to the meat from the hide during hide removal operations (6, 28, 29). Therefore, STEC from cattle and generic E. coli on carcasses can all be regarded as being of cattle origin. The finding that the median D60ºC values for STEC isolates from cattle and generic E. coli from carcasses prior to antimicrobial treatments only differed slightly (1.0 versus 0.9 min; Fig. 4) further confirms the similarity of these populations of E. coli. Due to the large number of isolates used in the study, a fixed incubation time, i.e., 6 min, was used for determination of D values for most of the isolates. This may have led to overestimation of D values for some isolates that were completely inactivated by this incubation time.
Overall, LHR-positive E. coli showed higher D60ºC values than LHR-negative E. coli, which is in agreement with previous reports that LHR increases heat resistance of E. coli (25, 30). However, of the 28 LHR-positive E. coli isolates, 5 had D60ºC values of <2 min, with one being as low as 0.78 min. LHR-positive isolates with relatively low heat resistance were also reported in three recent studies (23, 31, 32). The LHR is also present in other Enterobacteriaceae species, including Salmonella enterica, Enterobacter cloacae, and Klebsiella pneumoniae, and it provides genus-specific protection from heat treatment (25, 30). In addition, two copies of LHR have been found in a few strains of E. coli from raw milk cheese, with one in the chromosome and the other in a plasmid, and the strains with the extra copy had higher heat resistance (33, 34). Interestingly, a number (1.2%, 17/1,422) of LHR-negative isolates had D60ºC values of >2 min, with 3.93 min being the largest value, which is comparable to the median value (3.25 min) for LHR-positive E. coli isolates. Taken together, these findings suggest that factors in addition to the LHR are required to confer extreme heat resistance to E. coli and that more than the simple presence or absence of the LHR dictates the extent of heat resistance in E. coli.
The majority (97.2%) of the 1,450 E. coli isolates had D60ºC values of <2 min, which is consistent with published accounts (19, 20, 35–39). The D60ºC values ranged from 0 min (5 isolates from cattle were killed during the come-up time) to 7.54 min (an isolate from a carcass that did not encounter any antimicrobial treatments before chilling), suggesting large strain variation in heat resistance for naturally occurring E. coli. Isolates that had D60ºC values of >2 min were similarly distributed among cattle and meat plant E. coli sources. In addition, there was no difference observed in D60ºC values among E. coli from before or after antimicrobial interventions (before hide-on wash versus after hide-on wash and during the dressing process, before hide-on wash and beef products), suggesting that the proportion of heat-resistant E. coli found on carcasses and meat largely reflects the naturally occurring heat resistance in E. coli of cattle origin. The prevalence of LHR in E. coli recovered from the beef production chain (1.97%) is similar to that in the published genomes (2%) of E. coli (25). In contrast, a much larger fraction of LHR-positive E. coli was found in raw milk cheese (36.3%, 93/256) and treated municipal wastewater (59%, 41/70) (24, 34, 40). The former was attributed to the commercial practice of thermization of milk at 57 to 68°C for at least 15 s. However, information on the prevalence level of heat-resistant E. coli in raw milk cheese before heat treatment was unavailable. The much-increased prevalence of heat-resistant E. coli in treated wastewater, compared to 5% in untreated wastewater, was likely from coselection of chlorine and heat resistance by the wastewater treatment practice (24, 26, 27). Bacteria in general can develop resistance responses when exposed to stress conditions, and in the case of heat stress, “heat shock response” will be elicited when exposed to sublethal temperatures (41). Several studies have demonstrated that a higher temperature, especially with a step-wise increase (up to 50°C), and extended incubation time (up to 2 to 3 h) in rich medium that can support the development of a heat response can aid thermotolerance acquisition (42–44). The carcass E. coli populations before and after hide-on wash were collected from a plant where the following multiple antimicrobial interventions were used: washing hide-on carcasses with 1.5% NaOH at 55°C, spraying skinned carcasses with 5% lactic acid, and pasteurization of carcass sides for 12 s with steam at a temperature of >90°C (11, 15, 45). Considering the short duration of heat exposure of carcasses (a few seconds) and environments with limited nutrients (equipment surfaces), the lack of increased heat resistance level in the E. coli populations exposed to multiple antimicrobial interventions is not surprising. It is also possible that the after hide-on wash E. coli population survived the treatments not owing to their higher tolerance of the treatments but owing to evading the treatments through the topostructural properties of carcass surfaces. Interestingly, the range of D60ºC values and the prevalence of LHR-positive isolates were much more reduced after than before air chilling even though the median values did not differ significantly. This corroborates the finding that the E. coli population after sanitation was more heat sensitive than the population before sanitation in that the latter population had a large fraction of E. coli isolates that were adapted to the low-temperature environment of the fabrication floor of a beef processing plant through persistence (9, 46). Whether chilling would help combat heat-resistant E. coli warrants further investigation.
The levels of heat resistance of the Top 7 E. coli isolates collected in different years were comparable, suggesting that they are not naturally evolving to be more heat resistant. Of the seven serogroups, O26 (0.87 min) and O145 (1.20 min) had the smallest and largest median D60ºC values, respectively. A similar observation was made in a study conducted by Liu et al. (35), where O26 (n = 10) had smaller D60ºC values in phosphate-buffered saline (PBS) than serogroups O111 (n = 10), O121 (n = 10), and O145 (n = 10), though the latter groups did not differ. A study conducted by Enache et al. (37) also showed that O26 (n = 3) and O157 (n = 3) were the least and the most heat resistant, respectively, among the Top 7 in apple juice at 60°C. Median log reductions of O157 (n = 50) and non-O157 STEC (n = 63) at 60°C did not differ, but the median reduction of O145 (n = 10) was less than that of O111 (n = 10) by 1.4 log units (47). These findings seem to suggest that the O group may play a role in heat resistance. On the other hand, low variability in heat resistance between closely related E. coli O157 strains has been reported (48). It is possible that the disparities between studies could also be attributed to the narrow phylogenetic lineage for some serogroups or the limited number of strains being compared. However, the Top 7 E. coli in the present study had diverse origins, and 100 or more isolates were evaluated for all serogroups except for O111 and O145.
In conclusion, most E. coli strains from cattle and beef packing plant environments are heat sensitive, and the commonly used antimicrobial interventions in meat packing plants do not seem to select for heat-resistant E. coli. The likelihood of a cold environment such as the fabrication facility for meat to be a reservoir for heat-resistant E. coli seems low.
MATERIALS AND METHODS
Bacterial isolates and culture conditions.
Escherichia coli isolates (n = 1,450), comprising 750 Top 7 and 700 generic E. coli isolates, were used in this study (Tables 1 and 2). These isolates were from the culture collections of Alberta Agriculture and Forestry and of the Lacombe Research and Development Centre of Agriculture and Agri-Food Canada, respectively. The Top 7 E. coli isolates were recovered between 2002 and 2017 from feces or hides of cattle in feedlots or transportation trucks going to slaughter plants in Alberta (Table 1) (49–53; Tim Reuter, Shaun Cook, and Lisa Tymensen, unpublished data). Studies including non-O157 E. coli used methods described by Stanford et al. (49) for isolation and confirmation as STEC. Earlier, O157:H7-only studies used methods described by Stephens et al. (52) for isolation and STEC confirmation. The cattle E. coli did not experience antimicrobial interventions, and all were classified as STEC at initial isolation. Generic E. coli isolates were obtained between 2013 and 2015 from two federally inspected beef processing plants (A and B) in Alberta (Table 2) (9, 11, 16, 54, 55). At plant A, no decontaminating treatment was used other than carcass trimming and cold-water wash, and dressed carcasses were air chilled (16, 54). Sanitation of fabrication equipment occurred at the end of each fabrication day, which primarily included physical removal of soil, prerinsing with pressurized water at 40 to 50°C, spraying with a chlorine-based alkaline foaming agent, and sanitization with a quaternary ammonium compound sanitizer (9). During the period of strain isolation, plant B employed the following decontamination interventions: hide-on carcass wash with 1.5% sodium hydroxide at 55°C, spraying skinned carcasses with 5% lactic acid, and pasteurization of carcass sides at the end of the dressing process with steam at >90°C (11, 15). Isolates obtained from plant A after air chilling and after routine sanitation and from plant B after hide-on wash were regarded as isolates after antimicrobial interventions.
All bacterial stock and working cultures were maintained at –80°C and 4°C, respectively, as described previously (56). The species identity of all E. coli isolates was verified via PCR using the primers URL-301, 5′-TGTTACGTCCTGTAGAAAGCCC-3′, and URL-432, 5′-AAAACTGCCTGGCACAGCAATT-3′, as described previously (57). Each E. coli isolate was streaked on MacConkey agar (Oxoid) and incubated at 35°C for 24 ± 2 h and was then subcultured in Luria-Bertani (LB; BD Difco, Fisher Scientific, Canada) at 35°C with shaking at 80 to 100 rpm for 16 h. The LB overnight cultures were used for heat resistance determination.
Phenotypic characterization of heat resistance.
Heat inactivation experiments were carried out in a water bath (Isotemp 228; Fisher Scientific, Nepean, ON, Canada) with an SC100 heated immersion circulator (Fisher Scientific) measuring 13.2 by 5.4 by 7.8 in. (height [H] by width [W] by diameter [D]) to ensure even water temperature distribution. The come-up time, i.e., the time required to reach the target temperature, was determined by measuring the time it took for 1.5 ml of LB (n = 40) to reach 60°C using a type K thermocouple probe connected to a portable EasyLog USB temperature logger (Thermoworks, Lindon, UT). The come-up time was also verified twice on the day of heat treatment. A portion of 1.5 ml (∼9 log CFU/ml) of each overnight culture was added to a 2-ml microcentrifuge tube (Eppendorf, Canada), with 4 tubes included for each culture. Samples were heated in a 60°C water bath for the duration of the come-up time (2.51 ± 0.004 min), at which point two tubes for each culture were immediately removed from the water bath (T0), and then the remaining two tubes were removed after another 6 min (T6). Upon removal from the water bath, the tubes were immediately placed in an ice water bath for rapid cooling. The cultures were then serially diluted in 0.1% (wt/vol) peptone water, and 1 ml of appropriate dilutions or the original undiluted bacterial suspensions was plated onto Petrifilm aerobic count plates (AC; 3M Corp., St. Paul, MN, USA) following the manufacturer’s instructions. The plates were incubated at 35°C for 18 to 24 h, after which the CFU were enumerated, with a preference for plates bearing 20 to 200 CFU. E. coli AW1.7, the benchmark heat-resistant E. coli, was included as a positive control.
Bacterial counts were transformed to log values. Mean log transformed values (log CFU/ml) were determined from duplicate technical replicates, and log reduction was calculated (log CFU/mlT0 to log CFU/mlT6). For heat-treated samples from which no bacteria were recovered at the detection limit of 1 CFU/ml, a value of –0.5 was arbitrarily assigned for calculation. For the isolates with a log reduction of >2, the D value at 60°C (D60°C, min) was calculated by dividing the incubation time (6 min) by the log reduction. To obtain more accurate D60°C values for the isolates with relatively smaller log reductions (≤2), a longer incubation time was used. Each culture was incubated in a 60°C water bath as before, but for up to 30 min, with duplicate tubes withdrawn at 5-min intervals. Surviving populations were determined as described before. For those isolates, log counts were plotted against incubation time, and the regression of the plot was used to calculate D60°C for each isolate (58).
Screening for LHR.
DNA of each overnight culture was extracted using a water boiling method described by Yang et al. (16). All isolates were screened for LHR using a PCR method (25, 59) with modifications (31). DNA of E. coli AW1.7 was used as a positive control, and sterile distilled water was used as the negative control. An E. coli isolate was considered to be LHR positive if all three open reading frames (ORFs)/fragments were amplified and negative otherwise.
Data analysis.
All statistical analyses in this study were performed in R version 3.6.0. The D60°C values were grouped into sets according to isolation source, LHR characteristics, and serogroups and year of isolation for the Top 7. Most data sets did not follow normal distribution, as determined by the Shapiro-Wilk test. As such, nonparametric analyses, namely, Wilcoxon rank sum test and Kruskal-Wallis test, were performed to compare two and more sets of data, respectively. Dunn’s test was used for pairwise comparison following the Kruskal-Wallis test. To determine whether the heat resistance of cattle isolates changed over time, potential correlation between the heat resistance of each serogroup and the year of recovery was examined using Spearman’s method. A contingency table containing the composition of E. coli with D60°C values of >2 min was analyzed using Fisher’s exact test. The difference/effect in each analysis was regarded as significant if the P value was <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the funding provided by the Beef Cattle Research Council in Canada for this study.
Technical assistance from Yidong Graham and undergraduate students Katie Petrella, Simon Zhou, and Angela Ippolito is greatly appreciated.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Yang X, Wang H. 2014. Pathogenic E. coli (introduction), p 695–701. In Tortorello ML. (ed), Encyclopedia of food microbiology, 2nd ed Academic Press, Oxford, UK. [Google Scholar]
- 2.Pruimboom-Brees IM, Morgan TW, Ackermann MR, Nystrom ED, Samuel JE, Cornick NA, Moon HW. 2000. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc Natl Acad Sci U S A 97:10325–10329. doi: 10.1073/pnas.190329997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorpe CM. 2004. Shiga toxin-producing Escherichia coli infection. Clin Infect Dis 38:1298–1303. doi: 10.1086/383473. [DOI] [PubMed] [Google Scholar]
- 4.Heiman KE, Mody RK, Johnson SD, Griffin PM, Gould LH. 2015. Escherichia coli O157 outbreaks in the United States, 2003–2012. Emerg Infect Dis 21:1293–1301. doi: 10.3201/eid2108.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill A, Gill CO. 2010. Non-O157 verotoxigenic Escherichia coli and beef: a Canadian perspective. Can J Vet Res 74:161–169. [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur TM, Bosilevac JM, Nou X, Shackelford SD, Wheeler TL, Kent MP, Jaroni D, Pauling B, Allen DM, Koohmaraie M. 2004. Escherichia coli O157 prevalence and enumeration of aerobic bacteria, Enterobacteriaceae, and Escherichia coli O157 at various steps in commercial beef processing plants. J Food Prot 67:658–665. doi: 10.4315/0362-028x-67.4.658. [DOI] [PubMed] [Google Scholar]
- 7.Yang X. 2017. Microbial ecology of beef carcasses and beef products, p 442–462. In de Souza Sant’Ana A. (ed), Quantitative microbiology in food processing. John Wiley and Sons, Chichester, UK. doi: 10.1002/9781118823071.ch22. [DOI] [Google Scholar]
- 8.Gill CO. 2009. Effects on the microbiological condition of product of decontaminating treatments routinely applied to carcasses at beef packing plants. J Food Prot 72:1790–1801. doi: 10.4315/0362-028X-72.8.1790. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Wang H, He A, Tran F. 2017. Microbial efficacy and impact on the population of Escherichia coli of a routine sanitation process for the fabrication facility of a beef packing plant. Food Control 71:353–357. doi: 10.1016/j.foodcont.2016.07.016. [DOI] [Google Scholar]
- 10.Gill CO, Bryant J. 1997. Decontamination of carcasses by vacuum-hot water cleaning and steam pasteurizing during routine operations at a beef packing plant. Meat Sci 47:267–276. doi: 10.1016/S0309-1740(97)00058-2. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Badoni M, Tran F, Gill CO. 2015. Microbiological effects of a routine treatment for decontaminating hide-on carcasses at a large beef packing plant. J Food Prot 78:256–263. doi: 10.4315/0362-028X.JFP-14-226. [DOI] [PubMed] [Google Scholar]
- 12.Bosilevac JM, Nou X, Osborn MS, Allen DM, Koohmaraie M. 2005. Development and evaluation of an on-line hide decontamination procedure for use in a commercial beef processing plant. J Food Prot 68:265–272. doi: 10.4315/0362-028x-68.2.265. [DOI] [PubMed] [Google Scholar]
- 13.Quintavalla S. 2010. Plant cleaning and sanitation, p 287–297. In Toldrá F. (ed), Handbook of meat processing. Wiley-Blackwell, Ames, IA. [Google Scholar]
- 14.Wang H, He A, Yang X. 2018. Dynamics of microflora on conveyor belts in a beef fabrication facility during sanitation. Food Control 85:42–47. doi: 10.1016/j.foodcont.2017.09.017. [DOI] [Google Scholar]
- 15.Yang X, Badoni M, Youssef MK, Gill CO. 2012. Enhanced control of microbiological contamination of product at a large beef packing plant. J Food Prot 75:144–149. doi: 10.4315/0362-028X.JFP-11-291. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Tran F, Youssef MK, Gill CO. 2015. Determination of sources of Escherichia coli on beef by multiple-locus variable-number tandem repeat analysis. J Food Prot 78:1296–1302. doi: 10.4315/0362-028X.JFP-15-014. [DOI] [PubMed] [Google Scholar]
- 17.Youssef MK, Badoni M, Yang X, Gill CO. 2013. Sources of Escherichia coli deposited on beef during breaking of carcasses carrying few E. coli at two packing plants. Food Control 31:166–171. doi: 10.1016/j.foodcont.2012.09.045. [DOI] [Google Scholar]
- 18.Pollari F, Christidis T, Pintar KDM, Nesbitt A, Farber J, Lavoie M-C, Gill A, Kirsch P, Johnson RP. 2017. Evidence for the benefits of food chain interventions on E. coli O157:H7/NM prevalence in retail ground beef and human disease incidence: a success story. Can J Public Health 108:e71–e78. doi: 10.17269/cjph.108.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stringer SC, George SM, Peck MW. 2000. Thermal inactivation of Escherichia coli O157:H7. J Appl Microbiol 88(Suppl):79S–89S. doi: 10.1111/j.1365-2672.2000.tb05335.x. [DOI] [PubMed] [Google Scholar]
- 20.Sörqvist S. 2003. Heat resistance in liquids of Enterococcus spp., Listeria spp., Escherichia coli, Yersinia enterocolitica, Salmonella spp. and Campylobacter spp. Acta Vet Scand 44:1–19. doi: 10.1186/1751-0147-44-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dlusskaya EA, McMullen LM, Ganzle MG. 2011. Characterization of an extremely heat-resistant Escherichia coli obtained from a beef processing facility. J Appl Microbiol 110:840–849. doi: 10.1111/j.1365-2672.2011.04943.x. [DOI] [PubMed] [Google Scholar]
- 22.Ma A, Glassman H, Chui L. 2020. Characterization of Escherichia coli possessing the locus of heat resistance isolated from human cases of acute gastroenteritis. Food Microbiol 88:103400. doi: 10.1016/j.fm.2019.103400. [DOI] [PubMed] [Google Scholar]
- 23.Stanford K, Reuter T, Bach SJ, Chui L, Ma A, Conrad CC, Tostes R, McAllister TA. 2017. Effect of severe weather events on the shedding of Shiga toxigenic Escherichia coli in slaughter cattle and phenotype of serogroup O157 isolates. FEMS Microbiol Ecol 93:fix098. doi: 10.1093/femsec/fix098. [DOI] [PubMed] [Google Scholar]
- 24.Zhi S, Banting G, Li Q, Edge TA, Topp E, Sokurenko M, Scott C, Braithwaite S, Ruecker NJ, Yasui Y, McAllister T, Chui L, Neumann NF. 2016. Evidence of naturalized stress-tolerant strains of Escherichia coli in municipal wastewater treatment plants. Appl Environ Microbiol 82:5505–5518. doi: 10.1128/AEM.00143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercer RG, Zheng J, Garcia-Hernandez R, Ruan L, Gänzle MG, McMullen LM. 2015. Genetic determinants of heat resistance in Escherichia coli. Front Microbiol 6:932. doi: 10.3389/fmicb.2015.00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercer R, Nguyen O, Ou Q, McMullen L, Ganzle MG. 2017. Functional analysis of genes comprising the locus of heat resistance in Escherichia coli. Appl Environ Microbiol 83:e01400-17. doi: 10.1128/AEM.01400-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Fang Y, Zhi S, Simpson DJ, Gill A, McMullen LM, Neumann NF, Ganzle MG. 2019. The locus of heat resistance confers resistance to chlorine and other oxidizing chemicals in Escherichia coli. Appl Environ Microbiol 86:e02123-19. doi: 10.1128/AEM.02123-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bacon RT, Belk KE, Sofos JN, Clayton RP, Reagan JO, Smith GC. 2000. Microbial populations on animal hides and beef carcasses at different stages of slaughter in plants employing multiple-sequential interventions for decontamination. J Food Prot 63:1080–1086. doi: 10.4315/0362-028x-63.8.1080. [DOI] [PubMed] [Google Scholar]
- 29.Bell RG. 1997. Distribution and sources of microbial contamination on beef carcasses. J Appl Microbiol 82:292–300. doi: 10.1046/j.1365-2672.1997.00356.x. [DOI] [PubMed] [Google Scholar]
- 30.Mercer RG, Walker BD, Yang X, McMullen LM, Ganzle MG. 2017. The locus of heat resistance (LHR) mediates heat resistance in Salmonella enterica, Escherichia coli and Enterobacter cloacae. Food Microbiol 64:96–103. doi: 10.1016/j.fm.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Tran F, Klassen MD. 2020. Heat resistance in Escherichia coli and its implications on ground beef cooking recommendations in Canada. J Food Safety 40:e12769. doi: 10.1111/jfs.12769. [DOI] [Google Scholar]
- 32.de Souza Figueiredo EE, Yang X, Zhang P, Reuter T, Stanford K. 2019. Comparison of heating block and water bath methods to determine heat resistance in Shiga-toxin producing Escherichia coli with and without the locus of heat resistance. J Microbiol Methods 164:105679. doi: 10.1016/j.mimet.2019.105679. [DOI] [PubMed] [Google Scholar]
- 33.Boll EJ, Frimodt-Moller J, Olesen B, Krogfelt KA, Struve C. 2016. Heat resistance in extended-spectrum beta-lactamase-producing Escherichia coli may favor environmental survival in a hospital setting. Res Microbiol 167:345–349. doi: 10.1016/j.resmic.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Boll EJ, Marti R, Hasman H, Overballe-Petersen S, Stegger M, Ng K, Knochel S, Krogfelt KA, Hummerjohann J, Struve C. 2017. Turn up the heat: food and clinical Escherichia coli isolates feature two transferrable loci of heat resistance. Front Microbiol 8:579. doi: 10.3389/fmicb.2017.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Gill A, McMullen L, Ganzle MG. 2015. Variation in heat and pressure resistance of verotoxigenic and nontoxigenic Escherichia coli. J Food Prot 78:111–120. doi: 10.4315/0362-028X.JFP-14-267. [DOI] [PubMed] [Google Scholar]
- 36.Brar JS, Waddell JN, Bailey M, Corkran S, Velasquez C, Juneja VK, Singh M. 2018. Thermal inactivation of Shiga toxin-producing Escherichia coli in ground beef with varying fat content. J Food Prot 81:986–992. doi: 10.4315/0362-028X.JFP-17-455. [DOI] [PubMed] [Google Scholar]
- 37.Enache E, Mathusa EC, Elliott PH, Black DG, Chen Y, Scott VN, Schaffner DW. 2011. Thermal resistance parameters for Shiga toxin-producing Escherichia coli in apple juice. J Food Prot 74:1231–1237. doi: 10.4315/0362-028X.JFP-10-488. [DOI] [PubMed] [Google Scholar]
- 38.Jin T, Zhang H, Boyd G, Tang J. 2008. Thermal resistance of Salmonella enteritidis and Escherichia coli K12 in liquid egg determined by thermal-death-time disks. J Food Eng 84:608–614. doi: 10.1016/j.jfoodeng.2007.06.026. [DOI] [Google Scholar]
- 39.Juneja VK, Marmer BS. 1999. Lethality of heat to Escherichia coli O157:H7: D- and z-value determinations in turkey, lamb and pork. Food Res Int 32:23–28. doi: 10.1016/S0963-9969(99)00060-5. [DOI] [Google Scholar]
- 40.Marti R, Muniesa M, Schmid M, Ahrens CH, Naskova J, Hummerjohann J. 2016. Short communication: heat-resistant Escherichia coli as potential persistent reservoir of extended-spectrum beta-lactamases and Shiga toxin-encoding phages in dairy. J Dairy Sci 99:8622–8632. doi: 10.3168/jds.2016-11076. [DOI] [PubMed] [Google Scholar]
- 41.Cotto JJ, Morimoto RI. 1999. Stress-induced activation of the heat-shock response: cell and molecular biology of heat-shock factors. Biochem Soc Symp 64:105–118. [PubMed] [Google Scholar]
- 42.Mackey BM, Derrick C. 1990. Heat shock protein synthesis and thermotolerance in Salmonella Typhimurium. J Appl Bacteriol 69:373–383. doi: 10.1111/j.1365-2672.1990.tb01527.x. [DOI] [PubMed] [Google Scholar]
- 43.Mackey BM, Derrick CM. 1987. Changes in the heat resistance of Salmonella typhimurium during heating at rising temperatures. Lett Appl Microbiol 4:13–16. doi: 10.1111/j.1472-765X.1987.tb01571.x. [DOI] [Google Scholar]
- 44.Cebrián G, Sagarzazu N, Pagán R, Condón S, Mañas P. 2010. Development of stress resistance in Staphylococcus aureus after exposure to sublethal environmental conditions. Int J Food Microbiol 140:26–33. doi: 10.1016/j.ijfoodmicro.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 45.Gill CO, Landers C. 2003. Microbiological effects of carcass decontaminating treatments at four beef packing plants. Meat Sci 65:1005–1011. doi: 10.1016/S0309-1740(02)00319-4. [DOI] [PubMed] [Google Scholar]
- 46.Yang X, He A, Badoni M, Tran F, Wang H. 2017. Mapping sources of contamination of Escherichia coli on beef in the fabrication facility of a commercial beef packing plant. Food Control 75:153–159. doi: 10.1016/j.foodcont.2016.12.004. [DOI] [Google Scholar]
- 47.Gill A, Tamber S, Yang X. 2019. Relative response of populations of Escherichia coli and Salmonella enterica to exposure to thermal, alkaline and acidic treatments. Int J Food Microbiol 293:94–101. doi: 10.1016/j.ijfoodmicro.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 48.Lee K-i, French NP, Jones G, Hara-Kudo Y, Iyoda S, Kobayashi H, Sugita-Konishi Y, Tsubone H, Kumagai S. 2012. Variation in stress resistance patterns among stx genotypes and genetic lineages of Shiga toxin-producing Escherichia coli O157. Appl Environ Microbiol 78:3361–3368. doi: 10.1128/AEM.06646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanford K, Johnson RP, Alexander TW, McAllister TA, Reuter T. 2016. Influence of season and feedlot location on prevalence and virulence factors of seven serogroups of Escherichia coli in feces of western-Canadian slaughter cattle. PLoS One 11:e0159866. doi: 10.1371/journal.pone.0159866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanford K, Bach SJ, Marx TH, Jones S, Hansen JR, Wallins GL, Zahiroddini H, Mcallister TA. 2005. Monitoring Escherichia coli O157:H7 in inoculated and naturally colonized feedlot cattle and their environment. J Food Prot 68:26–33. doi: 10.4315/0362-028X-68.1.26. [DOI] [PubMed] [Google Scholar]
- 51.Stanford K, Gibb D, McAllister TA. 2013. Evaluation of a shelf-stable direct-fed microbial for control of Escherichia coli O157 in commercial feedlot cattle. Can J Anim Sci 93:535–542. doi: 10.4141/cjas2013-100. [DOI] [Google Scholar]
- 52.Stephens TP, McAllister TA, Stanford K. 2009. Perineal swabs reveal effect of super shedders on the transmission of Escherichia coli O157:H7 in commercial feedlots. J Anim Sci 87:4151–4160. doi: 10.2527/jas.2009-1967. [DOI] [PubMed] [Google Scholar]
- 53.Stanford K, Hannon S, Booker CW, Jim GK. 2014. Variable efficacy of a vaccine and direct-fed microbial for controlling Escherichia coli O157:H7 in feces and on hides of feedlot cattle. Foodborne Pathog Dis 11:379–387. doi: 10.1089/fpd.2013.1693. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, Youssef MK, Yang X. 2016. Effects of dry chilling on the microflora on beef carcasses at a Canadian beef packing plant. J Food Prot 79:538–543. doi: 10.4315/0362-028X.JFP-15-476. [DOI] [PubMed] [Google Scholar]
- 55.Visvalingam J, Liu Y, Yang X. 2017. Impact of dry chilling on the genetic diversity of Escherichia coli on beef carcasses and on the survival of E. coli and E. coli O157. Int J Food Microbiol 244:62–66. doi: 10.1016/j.ijfoodmicro.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 56.Visvalingam J, Ells TC, Yang X. 2017. Impact of persistent and nonpersistent generic Escherichia coli and Salmonella sp. recovered from a beef packing plant on biofilm formation by E. coli O157. J Appl Microbiol 123:1512–1521. doi: 10.1111/jam.13591. [DOI] [PubMed] [Google Scholar]
- 57.Bej AK, DiCesare JL, Haff L, Atlas RM. 1991. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl Environ Microbiol 57:1013–1017. doi: 10.1128/AEM.57.4.1013-1017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Asselt ED, Zwietering MH. 2006. A systematic approach to determine global thermal inactivation parameters for various food pathogens. Int J Food Microbiol 107:73–82. doi: 10.1016/j.ijfoodmicro.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 59.Ma A, Chui L. 2017. Identification of heat resistant Escherichia coli by qPCR for the locus of heat resistance. J Microbiol Methods 133:87–89. doi: 10.1016/j.mimet.2016.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.