Abstract
Lipid metabolic reprogramming is one hallmark of cancer. Lipid metabolism is regulated by numerous enzymes, many of which are targeted by several drugs on the market. We aimed to characterize the lipid alterations in oral squamous cell carcinoma (OSCC) as a basis for understanding its lipid metabolism, thus identifying potential therapeutic targets. We compared lipid species, classes, and glycerophospholipid (GPL) fatty acid species between paired tumor tissue and healthy oral tongue mucosa samples from 10 OSCC patients using a QExactive mass spectrometer. After filtering the 1370 lipid species identified, we analyzed 349 species: 71 were significantly increased in OSCC. The GPL metabolism pathway was most represented by the lipids differing in OSCC (P = .005). Cholesterol and the GPLs phosphatidylcholines, phosphatidylethanolamines, and phosphatidylinositols were most significantly increased in OSCC tissue (FC 1.8, 2.0, 2.1, and 2.3 and, P = .003, P = .005, P = .002, P = .007). In conclusion, we have demonstrated a shift in the lipid metabolism in these OSCC samples by characterizing the detailed landscape. Predominantly, cholesterol and GPL metabolism were altered, suggesting that interactions with sterol regulatory binding proteins may be involved. The FA composition changes of the GPLs suggest increased de novo lipogenesis.
Introduction
With a worldwide annual incidence of around 275 000, oral squamous cell carcinoma (OSCC) is the most common type of head and neck cancer (HNC) [1]. Significant risk factors include smoking and alcohol. Despite advances in treatment strategies, the overall survival has only improved by 5% over the last two decades, with 5-year survival rates of around 60% [2].
Metabolic reprograming, of which lipid metabolism is a part, is a hallmark of many cancers; the ability of cancers to perform de novo lipid synthesis was first recognized over 60 years ago [3]. Increased de novo synthesis and uptake of lipids are early events in cancer development [4]. Lipids are involved in various malignant processes: glycerophospholipids (GPLs) have been shown to regulate various functions such as adhesion, migration, apoptosis, and signal transduction [5]; cholesterol is an integral part of lipid rafts, and also plays a key role in cellular signal transduction, in pathways governing carcinogenesis, drug resistance, and metastasis [6].
Due to the structural complexity of lipids, the ability to analyze them on a large scale has been problematic. With the advent of soft electrospray ionization techniques pioneered by Fenn in 1989 [7], the ability to identify and quantify many structurally different lipid classes has been made possible [8].
Lipidomics is a rapidly growing field that aims to quantify and comprehensively identify lipid classes and individual lipid species [9]. Its ultimate goal is to uncover the changes in lipid metabolism in pathological conditions, which should allow for discovery of prognostic or predictive biomarkers and drug targets, to facilitate more personalized treatment approaches. Drug targets would likely be the enzymes involved in lipid metabolism; drugs exist already for many of these [10]. The first step in being able to target lipid entities and pathways for theranostic purposes is to characterize the lipidome. Thus far, the lipid species’ composition has been investigated in a variety of cancers using this approach [[11], [12], [13], [14], [15], [16], [17], [18]], providing insights into candidate biomarkers as well as increasing knowledge of the function of these lipids.
In OSCC, the detailed composition of plasma lipid species has been described [19]. At the tissue level, Raman spectroscopy has been used to identify the topological distribution of molecules including lipids, carbohydrates, and proteins [20]; however, the use of a high-throughput, highly sensitive mass spectrometry (MS) approach gives more information about the lipid species involved. To the best of our knowledge, this more detailed quantitative assessment of lipidomic aberrations at the tissue level is yet to be done. This will help improve understanding of the role of lipid metabolism in OSCC pathophysiology, identifying the key lipid metabolic pathways. This may in turn help to provide targets for prognostication and treatment. Therefore, we aimed to use an MS-based lipidomic approach to characterize how the lipidome differs in OSCC compared to healthy tissue.
Methods
Patient Samples
Samples were collected from 10 patients treated for primary OSCC of the oral tongue at the Department of Otorhinolaryngology–Head and Neck Surgery and Department of Oral and Maxillofacial Surgery at the Helsinki University Hospital (Helsinki, Finland) between 2015 and 2017. They had had no preoperative radio- or chemotherapy. Fresh tumor samples and healthy epithelial tissue samples from the opposite side of the tongue were collected from each patient in the operating room, immediately snap-frozen using dry ice and ethanol, and then stored at −72°C.
Ethical approval was granted by the institutional Research Ethics Committee of Medical Sciences (Dnro: 64/13/03/02/2014). All patients provided informed written consent to participate in the study.
Lipid Extraction for MS Lipidomics
Frozen samples were homogenized in LC-MS–grade water (LicChrosolv) in homogenization bead tubes for six runs at 45 seconds at 6.5 m/s with 5-minute incubation on ice in between runs, diluted to a concentration of 5 mg/ml (wet tissue weight per volume), and then stored at −72°C.
MS-based lipid analysis was performed by Lipotype GmbH (Dresden, Germany), as described [21]. Lipids were extracted using a two-step chloroform/methanol procedure [22]. Samples were spiked with internal lipid standard mixture containing cardiolipin 16:1/15:0/15:0/15:0 (CL), ceramide 18:1;2/17:0 (Cer), diacylglycerol 17:0/17:0 (DAG), hexosylceramide 18:1;2/12:0 (HexCer), lysophosphatidate 17:0 (LPA), lysophosphatidylcholine 12:0 (LPC), lysophosphatidylethanolamine 17:1 (LPE), lysophosphatidylglycerol 17:1 (LPG), lysophosphatidylinositol 17:1 (LPI), lysophosphatidylserine 17:1 (LPS), phosphatidate 17:0/17:0 (PA), phosphatidylcholine 17:0/17:0 (PC), phosphatidylethanolamine 17:0/17:0 (PE), phosphatidylglycerol 17:0/17:0 (PG), phosphatidylinositol 16:0/16:0 (PI), phosphatidylserine 17:0/17:0 (PS), cholesterol ester 20:0 (CE), sphingomyelin 18:1;2/12:0;0 (SM), triacylglycerol 17:0/17:0/17:0 (TAG), and cholesterol D6. After extraction, the organic phase was transferred to an infusion plate and dried in a speed vacuum concentrator. The first-step dry extract was resuspended in 7.5 mM ammonium acetate in chloroform/methanol/propanol (1:2:4, v:v), and the second-step dry extract was resuspended in 33% ethanol solution of methylamine in chloroform/methanol (0.003:5:1; v:v). All liquid handling steps were performed using Hamilton Robotics STARlet robotic platform with the antidroplet control feature for organic solvents pipetting.
MS Data Acquisition
Samples were analyzed by direct infusion on a QExactive mass spectrometer (Thermo Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences). Samples were analyzed in both positive and negative ion modes with a resolution of Rm/z=200=280,000 for MS and Rm/z=200=17,500 for MS/MS experiments in a single acquisition. MS/MS was triggered by an inclusion list encompassing corresponding MS mass ranges scanned in 1-Da increments [23]. Both MS and MS/MS data were combined to monitor CE, DAG, and TAG ions as ammonium adducts; PC and PC O- as acetate adducts; and CL, PA, PE, PE O-, PG, PI, and PS as deprotonated anions. MS only was used to monitor LPA, LPE, LPE O-, LPI, and LPS as deprotonated anions; Cer, HexCer, SM, LPC, and LPC O- as acetate adducts; and cholesterol as ammonium adduct of an acetylated derivative [24].
Data Analysis and Postprocessing
Data were analyzed with in-house–developed lipid identification software based on LipidXplorer [25,26]. Data postprocessing and normalization were performed using an in-house–developed data management system. Only lipid identifications with a signal-to-noise ratio >5 and a signal intensity five-fold higher than in corresponding blank samples were considered for further data analysis.
As filtering steps, firstly, abundances of <1 pmol per sample per lipid species identified were removed. Secondly, if no specific lipid or fatty acid species was identified in >20% of samples in both groups, then that species was excluded from comparative analysis between OSCC and healthy tissue, although all values are included in the GPL fatty acid figures. For abundance of each lipid class, the abundances of all lipid species within that class were summed. To calculate the abundance of each fatty acid chain in the GPL classes, which have two fatty acid binding sites (see Figure S1), the abundance of each species containing each fatty acid was summed [i.e., to calculate the abundance of palmitate (C16:0) in PC, we calculated Σ [1 ∗ PC(16:0/X:X) + 2∗PC (16:0/16:0)]. Abundances of lysophospholipids and ether lipids were incorporated into one group; for example, LPC, PC and PC ether (PC O-) were combined. Table S1 provides an example calculation.
OSCC was compared with healthy tissue: lipid classes, species, and fatty acid chains within each GPL class.
Statistical Analysis
To identify differentially expressed lipids, the paired t test was performed using the t.test function in R. The significance level was set to P < .05. Multiple testing [false discovery rate (FDR)] was done using the Benjamini-Hochberg method, with the FDR set to <0.05. Power for each lipid species and class was calculated using R.
The volcano plot was created using Microsoft Excel for Mac v16.16.2 (Microsoft, Redmond, WA), and elements were highlighted using Sketch v25.2 (Sketch, the Netherlands).
Hierarchically clustered heatmaps, PCA, and box plots were created using the statistical analysis module on metaboanalyst.ca. Data normalization for the hierarchically clustered heatmap is as follows: sample normalization by median, cube-root data transformation, and data scaling by autoscaling. Results of normalization are in Figure S2. Euclidean distance measure and Ward D clustering algorithm were used.
Pathway Analysis
Pathway analysis was performed on the differentially expressed individual lipids (P < .05) by using Lipid Pathway Enrichment Analysis, a web tool for overrepresentation analysis of lipid signatures (https://lipea.biotec.tu-dresden.de/home).
Results
Patient and Tumor Characteristics
Patient demographics are shown in Table S2. The male:female ratio was 2:3, and average patient age was 55 years (median 65.5, range 24-78). Seven tumors were stage IVa, two were stage II, and one was stage I. All tumors were M0 at the time of tissue collection. One patient had had a kidney cancer 8 years prior to OSCC diagnosis. Four patients had disease recurrence or progression after the initial treatment.
Lipid Classes and Species
Using an MS-based, data-independent acquisition approach to lipids, 1370 separate lipid species were quantified (Table S3). Of 23 lipid classes (Figure S1 shows their structures), four were significantly different (Table 1, Table S4): three GPLs and cholesterol. The most abundant lipids were cholesterol, phosphatidylcholine, and triacylglycerol. After filtering, 349 species were included in the t test (Table S4). Figure 1 shows a volcano plot of the different species and their abundances, and Table 2 shows the 71 significantly different species. Boxplots for each lipid species and class are shown in Figure S3, Figure S4, respectively. The majority of significant lipid species were GPLs, the next largest group being sphingolipids.
Table 1.
Lipid Class Differences Between OSCC and Healthy Tissue
| Names | Healthy Mean Abundance | OSCC Mean Abundance | FC OSCC/Healthy | P Value | FDR | |
|---|---|---|---|---|---|---|
| Phosphatidylethanolamine | PE | 519.6 | 1194.9 | 2.3 | .002 | 0.038 |
| Phosphatidylcholine | PC | 2680.5 | 5517.4 | 2.1 | .005 | 0.039 |
| Phosphatidylinositol | PI | 461.2 | 903.9 | 2 | .007 | 0.042 |
| Cholesterol | Chol | 2336.6 | 4198.3 | 1.8 | .003 | 0.038 |
| Ceramide | Cer | 44.4 | 232.2 | 5.2 | .1 | 0.192 |
| Phosphatidylcholine ether | PC O- | 147.4 | 314.5 | 2.1 | .09 | 0.192 |
| Lysophosphatidylinositol | LPI | 2.3 | 4.7 | 2 | .066 | 0.192 |
| Phosphatidylglycerol | PG | 11.1 | 22.4 | 2 | .075 | 0.192 |
| Lysophosphatidylethanolamine ether | LPE O- | 5.1 | 9.1 | 1.8 | .228 | 0.327 |
| Cholesterol ester | CE | 391.6 | 676.4 | 1.7 | .076 | 0.192 |
| Lysophosphatidylethanolamine | LPE | 11.3 | 18.2 | 1.6 | .243 | 0.328 |
| Phosphatidylethanolamine ether | PE O- | 775.8 | 1269.8 | 1.6 | .092 | 0.192 |
| Lysophosphatidylcholine | LPC | 44.4 | 71.8 | 1.6 | .094 | 0.192 |
| Sphingomyelin | SM | 469 | 725.8 | 1.5 | .026 | 0.121 |
| Phosphatidylserine | PS | 703.9 | 959.2 | 1.4 | .189 | 0.29 |
| Diacylglycerol | DAG | 220.7 | 299.2 | 1.4 | .335 | 0.391 |
| Hexosylceramide | HexCer | 33.2 | 34.7 | 1 | .902 | 0.902 |
| Lysophosphatidylserine | LPS | 3.7 | 2.9 | 0.8 | .656 | 0.686 |
| Cardiolipin | CL | 124.1 | 100.6 | 0.8 | .329 | 0.391 |
| Phosphatidic acid | PA | 67.5 | 50.3 | 0.7 | .614 | 0.673 |
| Lysophosphatidylcholine ether | LPC O- | 6.9 | 3.6 | 0.5 | .114 | 0.202 |
| Lysophosphatidic acid | LPA | 5.6 | 2 | 0.4 | .34 | 0.391 |
| Triacylglycerol | TAG | 5036 | 836.3 | 0.2 | .174 | 0.286 |
Significantly different lipid classes are shown in bold. The “Healthy Mean Abundance” and “OSCC Mean Abundance” columns show the average absolute abundance of each lipid class in the healthy and OSCC samples, respectively. Abundance in pmol/mg tissue.
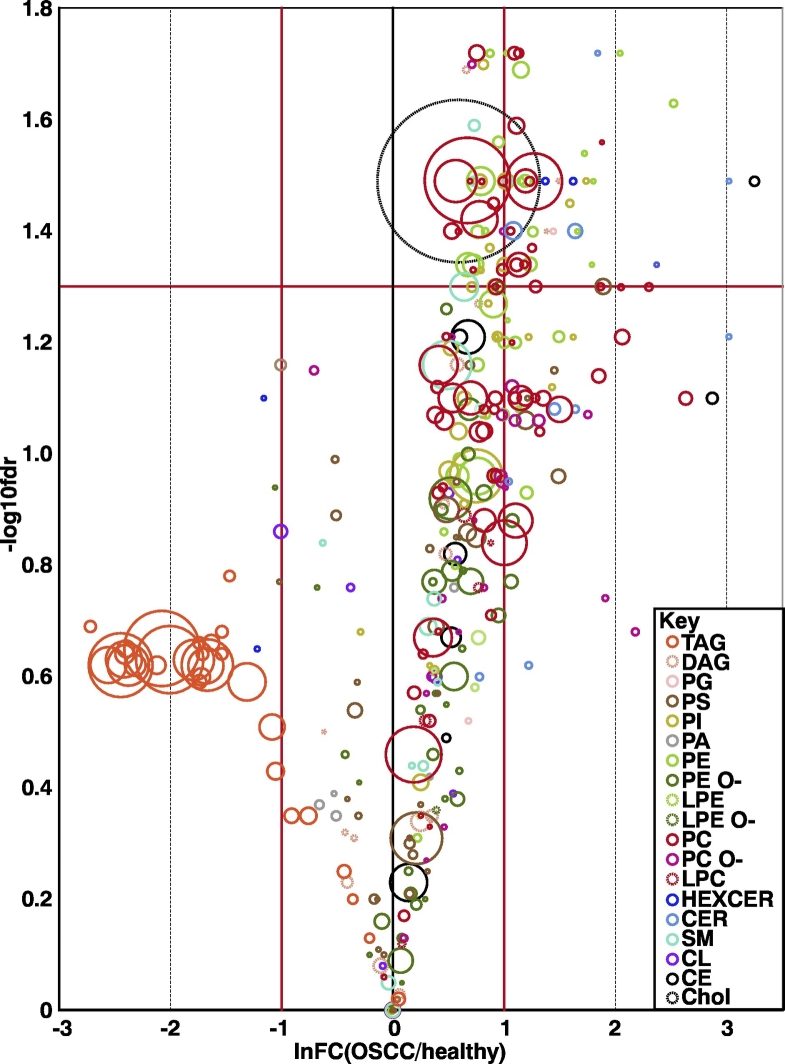
Figure 1.
Volcano plot of lipid species. The red horizontal line indicates the significance limit (FDR of 0.05). Lipid species above this line are statistically significant. The red vertical lines represent a fold change of >2 and <0.5 (OSCC/healthy), i.e., those to the right of the rightmost vertical red line are twice as abundant in OSCC, and those to the left of the leftmost vertical red line are twice as abundant in the healthy tissue. The size of the circles indicates the average abundance of the species in the most abundant group, and the colors of each circle show which class the species belong to (see the Key).
Table 2.
Significantly Different Lipid Species
| Average Abundance |
Fold Change | ||
|---|---|---|---|
| Species | Healthy | OSCC | |
| PC 16:0;0_22:2;0 | 3.2 | 20.8 | 6.58 |
| PC 16:0;0_16:0;0 | 1411.4 | 5020.7 | 3.56 |
| PC 18:1;0_20:5;0 | 27.7 | 96.9 | 3.5 |
| PC 18:2;0_20:2;0 | 35.7 | 121.7 | 3.41 |
| PC 14:0;0_16:0;0 | 253.8 | 833 | 3.28 |
| PC 18:0;0_20:5;0 | 27.8 | 90.2 | 3.25 |
| PC 16:1;0_16:1;0 | 283.9 | 877.3 | 3.09 |
| PC 18:1;0_20:2;0 | 42.2 | 130.3 | 3.08 |
| PC 15:0;0_16:0;0 | 127.1 | 387.4 | 3.05 |
| PC 16:0;0_20:2;0 | 83.5 | 254.4 | 3.05 |
| PC 16:0;0_20:1;0 | 83.1 | 246.4 | 2.96 |
| PC 18:2;0_20:3;0 | 29.1 | 83.7 | 2.87 |
| PC 18:2;0_20:1;0 | 42.7 | 115.1 | 2.69 |
| PC 14:0;0_18:0;0 | 77.5 | 207.8 | 2.68 |
| PC 18:1;0_20:1;0 | 75.7 | 186.6 | 2.47 |
| PC 16:0;0_16:1;0 | 947.1 | 2074.8 | 2.19 |
| PC 16:1;0_20:3;0 | 20.2 | 43.1 | 2.13 |
| PC 14:0;0_18:1;0 | 176 | 371.2 | 2.11 |
| PC 18:1;0_19:1;0 | 21.1 | 43.1 | 2.05 |
| PC 14:0;0_17:0;0 | 16.3 | 32.4 | 1.99 |
| PC 16:0;0_18:1;0 | 5928.1 | 11577.4 | 1.95 |
| PC 16:0;0_17:1;0 | 180.7 | 326.5 | 1.81 |
| PC 16:0;0_19:1;0 | 27.2 | 49 | 1.8 |
| PC 18:1;0_18:1;0 | 1607.2 | 2815.9 | 1.75 |
| PC 15:0;0_18:1;0 | 221.1 | 374.4 | 1.69 |
| PC O-17:0;0/15:0;0 | 19.6 | 52.8 | 2.69 |
| PC O-16:0;0/16:1;0 | 38.5 | 78.1 | 2.03 |
| PG 16:0;0_18:1;0 | 10.3 | 43.6 | 4.22 |
| PI 18:1;0_22:4;0 | 7.6 | 43.5 | 5.71 |
| PI 16:0;0_22:4;0 | 16.6 | 81.2 | 4.89 |
| PI 18:1;0_20:4;0 | 137.4 | 379.8 | 2.76 |
| PI 16:0;0_20:4;0 | 101.9 | 275.7 | 2.7 |
| PI 18:0;0_22:6;0 | 37.7 | 90 | 2.39 |
| PI 18:1;0_20:3;0 | 62.5 | 140.4 | 2.25 |
| PI 16:0;0_20:3;0 | 21.8 | 48.2 | 2.21 |
| PI 18:1;0_18:1;0 | 135.7 | 295.4 | 2.18 |
| Cer 32:1;2 | 1.4 | 29.7 | 20.57 |
| Cer 34:2;2 | 5.2 | 32.9 | 6.27 |
| Cer 42:2;2 | 60.4 | 310.4 | 5.14 |
| Cer 34:1;2 | 155.6 | 457.9 | 2.94 |
| CE 20:3;0 | 5.3 | 137.3 | 25.83 |
| Chol | 23365.7 | 41983.4 | 1.8 |
| DAG 16:0;0_20:3;0 | 5.6 | 24.6 | 4.42 |
| DAG 18:1;0_20:3;0 | 5 | 19.9 | 3.97 |
| DAG 16:0;0_20:4;0 | 27.9 | 76.6 | 2.75 |
| DAG 18:0;0_20:3;0 | 34.9 | 67.6 | 1.94 |
| HexCer 40:1;2 | 2.6 | 27.9 | 10.74 |
| HexCer 42:2;2 | 12.8 | 64.7 | 5.07 |
| HexCer 34:1;2 | 15.6 | 61.2 | 3.92 |
| PE 16:0;0_16:0;0 | 6.3 | 35.1 | 5.6 |
| PE 18:1;0_22:3;0 | 6.5 | 81.3 | 12.49 |
| PE 18:1;0_20:2;0 | 4.5 | 34.8 | 7.7 |
| PE 16:0;0_20:1;0 | 4.5 | 27.2 | 6.07 |
| PE 20:1;0_20:4;0 | 4.2 | 25 | 6.01 |
| PE 16:0;0_20:2;0 | 5.5 | 28.7 | 5.25 |
| PE 18:1;0_22:4;0 | 40.3 | 141.8 | 3.52 |
| PE 16:0;0_20:4;0 | 85.1 | 291.7 | 3.43 |
| PE 16:0;0_22:4;0 | 68.2 | 223.4 | 3.27 |
| PE 16:0;0_22:5;0 | 122.2 | 386.4 | 3.16 |
| PE 18:1;0_20:1;0 | 21.9 | 69.3 | 3.16 |
| PE 16:1;0_20:4;0 | 12.3 | 34.1 | 2.77 |
| PE 18:1;0_20:3;0 | 72.2 | 187.3 | 2.6 |
| PE 16:1;0_22:5;0 | 34.2 | 82 | 2.4 |
| PE 16:0;0_20:3;0 | 27.7 | 63.5 | 2.29 |
| PE 18:1;0_18:1;0 | 553.8 | 1221.9 | 2.21 |
| PE 18:1;0_22:5;0 | 112.8 | 242.1 | 2.15 |
| PE 18:1;0_18:2;0 | 313.4 | 645.9 | 2.06 |
| PE 16:0;0_18:1;0 | 430.9 | 841.8 | 1.95 |
| SM 44:2;2 | 41.6 | 117.4 | 2.82 |
| SM 32:1;2 | 89.5 | 186.5 | 2.08 |
| SM 40:2;2 | 64 | 133 | 2.08 |
Abundance is in pmol/mg. Fold change is OSCC/healthy.
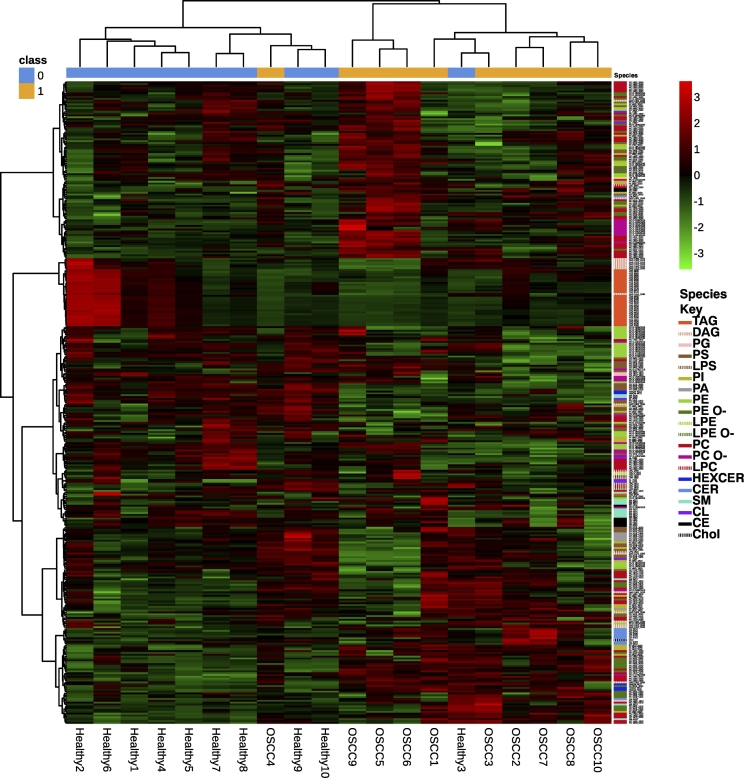
A hierarchically clustered heatmap did not separate healthy samples from OSCC using lipid classes (Figure S5). However, at a species level, healthy and OSCC tissue separated, except in patient 3 whose healthy sample was intermixed with the OSCC samples and in patient 4 whose OSCC sample was intermixed with the healthy samples (Figure 2). Results of the PCA for lipid species and classes can be seen in Figure S6, Figure S7. As can be expected, they are very similar to each other. There is some variation in the healthy samples, and healthy sample 2 is separated from the other healthy samples due to its high TAG content. Taken together, principle component 1 is influenced by the TAG levels, with all of the OSCC samples having a similar low level and TAG-level variation occurring in the healthy samples. Principle component 2 is most influenced by the classes PC and then by cholesterol, with increasing levels in most of the cancers. At the species level, certain TAGs most influence principle component 1 (especially TAG 52:2;0, TAG 53:2;0 and 50:2;0), and principle component 2 is most influenced by cholesterol and then by PC16:0;0_18:1;0 and PC PC16:0;0_ 16:0;0. It is worth noting that there is pairwise separation between all of the pairs at a class level, although OSCC 1 and healthy 1 are relatively similar at the class level using two principle components and OSCC 10 and healthy 10 are not separated well in the species-level PCA with two components. These further separate when including principle component 3.
Figure 2.
Hierarchically clustered heatmap containing the lipid species. Red indicates a relative high abundance of a lipid; green indicates a relative low abundance of a lipid. The species in each sample are shown on the right hand panel, with their classes labeled in the key.
Pathways
We identified pathways enriched with the altered lipid species (Table S5) using all lipid species with a paired t test P value < .05. The pathway containing the greatest number of altered lipid species was “GPL metabolism” followed by the sphingolipid signaling pathway and retrograde endocannabinoid signaling (Figure S8).
Fatty Acids
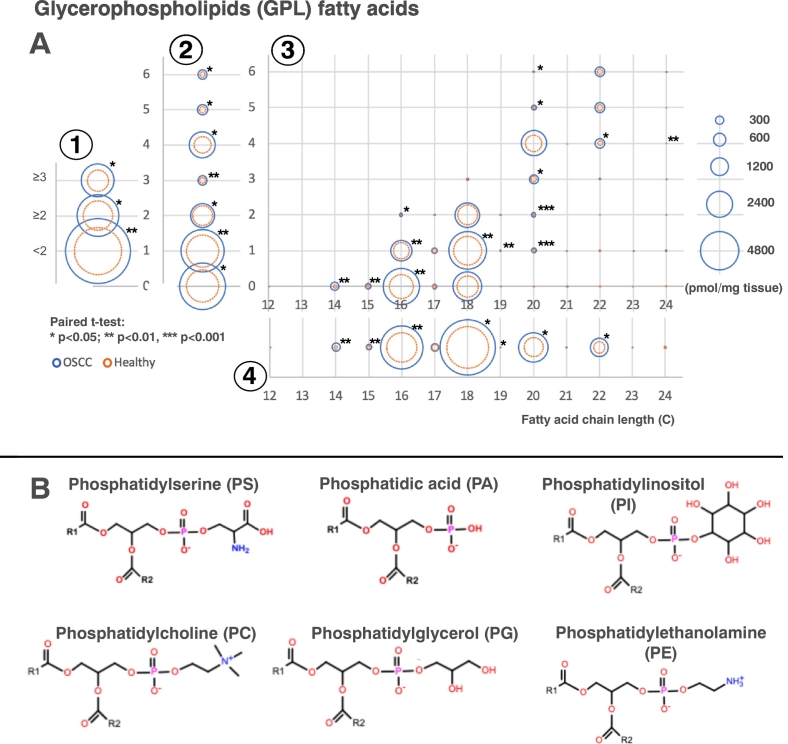
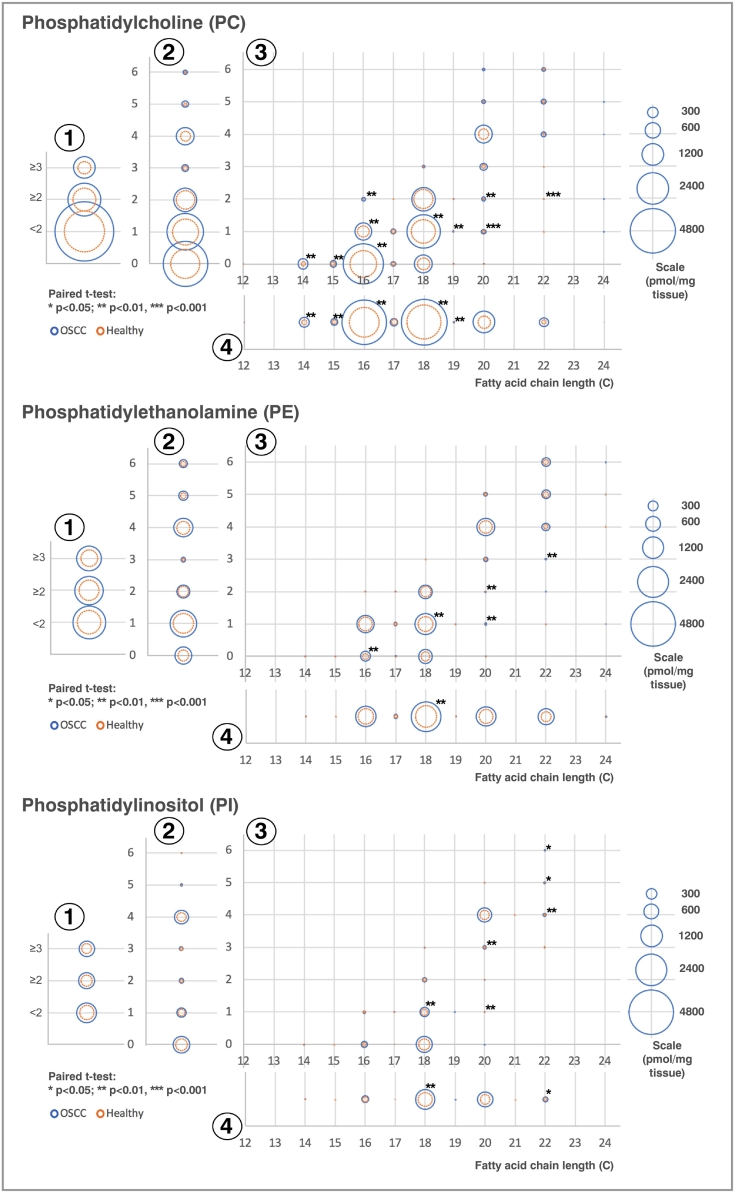
As GPLs were the majority of the significantly different species and the most represented pathway, we looked at the fatty acids connected to the GPLs to identify how their fatty acids changed with cancer. Figure 3 shows the composition of the fatty acids in all of the GPLs combined except cardiolipin. The majority of the fatty acids were increased in the OSCC compared with healthy tissue; those with the biggest increases were 16:0 (palmitate) followed by 18:1. Using this methodology, we were unable to determine the exact location of any double bonds within the fatty acids, so we cannot be certain which isomer the fatty acid 18:1 is. Figure 4 shows the composition of fatty acids in the significantly different GPL classes: PC, PE, and PI. Other GPL classes’ fatty acid compositions are available in Figure S9 and Table S6.
Figure 3.
GPLs’ fatty acid compositions in OSCC (blue) and healthy (orange) tissue. (A) Composition of fatty acid species within the GPLs. In the graph, fatty acids have been shown separated by 1) those with <2 double bonds and ≥2 double bonds, 2) the number of double bonds, 3) chain length and number of double bonds, and 4) chain length. The area of each circle is proportional to the abundance of each fatty acid moiety in pmol/mg tissue. Only lipids that are significant after using the Benjamini-Hochberg multiple testing correction are shown with an asterisk (*). (B) Structures of the different GPLs. Additionally, the compositions of the phosphatidylcholine ether and phosphatidylethanolamine ether were included (structures not shown). R1 and R2: variable fatty acid chains. (C) An overview of the Kennedy pathway.
Figure 4.
(A) Composition of fatty acid species within PC, PE, and PI in OSCC (blue) and healthy (orange) tissue. Fatty acids for each GPL subspecies have been separated by 1) those with <2 double bonds, ≥2 double bonds, etc.; 2) the number of double bonds; 3) chain length and number of double bonds; and 4) chain length. The area of each circle is proportional to the abundance of each fatty acid moiety pmol/mg tissue. Only lipids that are significant after using the Benjamini-Hochberg multiple testing correction are shown with an asterisk.
Discussion
Metabolic reprogramming including lipid metabolism alterations is considered one of the hallmarks of cancer and has been implicated in metastatic behavior as well as, for instance, resistance to cetuximab in HNC models [27,28]. Given this crucial role lipids play in different cancer behaviors, it is important to understand the overall tissue lipid alterations to begin identifying theranostic targets: either the lipids themselves or enzymes involved in their regulation.
We compared the lipidome of OSCC with healthy tissue from the same patient, reducing the chances of interindividual lipidomic alterations within the tissue that may result from other factors such as diet [27]. There was almost complete separation in the hierarchically clustered heatmap of lipid species (Figure 2): one healthy sample clustered together with the OSCC samples, although it was most similar to its OSCC counterpart. One potential explanation for this could be the phenomenon of field cancerization [29], especially since this patient later developed a fast growing neck metastasis after radiotherapy had finished. Additionally, one OSCC sample clustered with the healthy samples. This was a stage IVa tumor that was successfully treated so could perhaps reflect a tumor with less aggressive features.
From this unbiased approach of quantifying the lipidomic aberrations of OSCC, we found that lipid levels were greater in the cancer tissue than healthy tissue. We identified that the main differences lie in cholesterol and the GPLs (Table 1).
Cholesterol levels were higher in the cancer tissue than healthy tissue. Cholesterol accumulation in mitochondria can suppress apoptosis via inhibition of apoptotic proteins’ release from the mitochondria [30]. Cellular cholesterol levels are maintained in healthy cells by a tightly synchronized balance between uptake, efflux, and de novo synthesis, which is deregulated in many cancers [31]. Sterol regulatory element binding proteins (SREBPs) and liver X receptors (LXRs) are important transcription factor classes in cholesterol homeostasis [6,31]: when cholesterol levels fall, SREBPs are activated, leading to the transcription of genes involved in cholesterol uptake and biosynthesis. When cholesterol levels increase, LXRs are indirectly activated, leading to upregulation of the efflux-related genes ABCA1 and ABCG1 [6]. Interestingly, a small study showed upregulation of LXRs in OSCC along with increased cholesterol efflux via ABCA1 receptors [32]. If the levels of LXRs would be similarly raised in our cohort, then there could not be an increase of efflux as would be expected. Possible explanations could be increased uptake or synthesis of cholesterol that surpasses the efflux capacity, or due to molecular intermediaries: various miRNAs have been shown to target and inhibit ABCA1 and ABCG1 [6].
SREBPs, which upregulate cholesterol uptake and biosynthesis-related proteins, can be activated in many ways: Ras-oncogene activation, mutated p53, and PI3K/Akt/mTORC signaling [30,31]. Akt signaling is well known to be disturbed in OSCC as a downstream product of Ras-oncogene activation [33]. Recently, it has been shown that mTORC levels are also increased in OSCC tissues [34]. Traditional serum lipid profiling has shown that hypocholesterolemia occurs in OSCC patients, suggesting that increased uptake may occur in OSCC tissue [35]. Furthermore, many cancers have increased cholesterol synthesis [31]; however, this has not been studied in OSCC to our knowledge. Further research into the alterations of cholesterol metabolism in OSCC will be important to identify potential therapeutic targets.
Additionally, three GPLs classes’ abundances were significantly increased (PE, PC, and PI) in the cancer, and “GPL metabolism” was the lipid pathway including the greatest proportion of lipid alterations (Table S5, Figure S8A).
PI is an important cellular component that is synthesized in the endoplasmic reticulum. It is part of a complex network of eight phosphatidylinositides that can be interconverted by different enzymes [36] and which have important roles in cell signaling, transcription, RNA editing, and regulation of membrane proteins, among others. They signal through many axes, including via the PI3K/Akt/mTOR pathway, which influences among other things cell survival, metabolism, and growth and is well established as an aberrant pathway in HNC [37].
We found that the PI levels were twice as high in the cancer than healthy epithelium, which is the case also in prostate cancer. In prostate cancer, the acylation patterns were also different in different stages of disease, with changes in acyl saturation [38]. P53 is the most common mutation in HNCs, and this has also been shown to affect acylation patterns in PI, especially leading to an increase in PIs with 36 carbons in both acyl chains together [39]. The same pattern exists in our study, and several such 36-carbon PIs were significantly upregulated in the cancer, for instance, PI 16:0_20:4 and PI 18:1_18:1 (Table S3).
The two most abundant GPLs that we identified were PC and PE. These are mainly found in the cell membrane, with PE being also found in the mitochondrial membrane. Interestingly, all cancers tested hitherto display abnormal PC and PE metabolism [40]. They are formed, among other ways, via the Kennedy pathway [41] (Figure 3), which has potential for pharmacological targeting [4]. One enzyme in the Kennedy required for PC and PE synthesis, choline kinase, has increased levels in various cancers and correlate with poor prognosis [42]. Its inhibition has also promising results in terms of antimetastasis, induction of cell senescence, and tumor growth reduction in animal models; thus, it would be useful to see if this is also the case with OSCC [43]. Uptake from the serum could alternatively be a mechanism here, as a previous serum lipidomics study by Wang et al. [19] found that GPLs, especially PC and PE, were decreased in the serum of OSCC patients compared to controls. The SR-B1 cholesterol transporter has been shown to also uptake phospholipids PC, SM, and PE from HDL and LDL [44]; this could be one mechanism for uptake. SM also trended towards being upregulated in the OSCC tissue (FC=1.5) but was insignificant when multiple testing correction was applied.
One other interesting finding was the precipitous drop in TAG abundance in the tumor tissue (Table 2, Figure 1). It can be seen in the volcano plot (Figure 1) that there were several high-abundance TAG species, almost all of which were more abundant in the healthy tissue. However, not all healthy samples had such high levels, as can be seen in the heatmap in Figure 2; hence, this difference did not reach significance at the group level. TAGs are used as lipid storage within cells at a time of high nutrient availability. The large drop in TAGs suggests that they are either extensively used for energy requirements in the OSCC tissue or that the DAGs from which they are formed are instead being used as a substrate for PC and PE synthesis [45]. Based on our pilot results, larger studies should investigate the TAG alterations in OSCC.
GPL Fatty Acid Composition
The majority of cells acquire fatty acids via uptake from the blood stream. De novo synthesis of fatty acids occurs primarily in the liver, adipose tissue, and lactating breast. Many enzymes are involved, including fatty acid synthase (FASN), which creates palmitate, the 16-carbon saturated fatty acid, which can then be elongated and desaturated to form other fatty acids [42].
We examined the compositions of fatty acids within the GPLs except for CL, for which the fatty acid composition was undetermined (Figures 3A, Figure 4, Figure S9; Table S6). Alas, we were unable to assess the fatty acid composition of other lipid classes such as TAG and DAGs. In general, where there was a difference in abundance, most GPL fatty acid species were upregulated in the cancer tissue. The chain lengths and saturations in different GPLs varied even in the healthy tissue; however, the significance of this is unclear. In PC, the most abundant GPL, almost all of the fatty acids that were identified were increased in the cancer tissue. Palmitate (16:0) was the fatty acid most increased in cancer in the GPLs, namely, in PC and also in PE, which suggests that there is an increase in FASN expression, as has previously been found in OSCC [46]. Interestingly however, some classes had no significant changes in the fatty acid composition, examples being PA and PG. All GPL species that are newly synthesized via the Kennedy pathway undergo fatty acid chain remodeling in a process known as the Lands cycle. This complex cycle comprising many different enzymes involves the remodeling of GPL species into other GPL species, as well as the fatty acid composition of the GPLs [45], which will affect the properties of the cell membrane [47]. It would be interesting to further investigate what effect on membrane properties is caused by these altered GPLs. Furthermore, many of the enzymes involved in de novo fatty acid synthesis are also regulated by SREBPs [48].
C18:1 was the most abundant monounsaturated fatty acid and probably represents oleic acid, although other isomers such as vaccenic acid and elaidic acid are also known. This was also increased in PC and PE in the OSCC tissue. Overall, the biggest changes were seen in the monounsaturated and saturated fatty acids. The stearoyl CoA desaturases catalyze the rate-limiting step of desaturating fatty acids. This is overexpressed in some cancers, and in breast cancer, this overexpression correlates with reduced survival [49]. Moreover, the inhibition of stearoyl CoA desaturase in cancer cells has been shown to induce the endoplasmic reticulum stress pathway leading to cell death [50] and thus may be an interesting area to pursue in OSCC.
Limitations
Although these are exciting findings, there are several constraints at present with data interpretation. Though there are databases available for lipidomic profile interpretation, detailed knowledge of the functions of lipid species is still being unraveled. Additionally, although we know how many double bonds were present in each lipid species/fatty acid chain within a species, we were unable to identify where each double bond was within the chain, thus limiting our interpretation of which enzymes could have participated in their desaturation. As we have not detected the lipidomic changes in specific organelles, we are unable to describe with certainty the topographical alterations within the cells. Another limitation of this work is the small patient number. Due to this, we were unable to correlate the results with smoking, alcohol use, or other clinicopathological features and had no information on dietary habits of the participants, which may have influenced the results. Nevertheless, we had the benefit of being able to compare cancer and tissue from the same patients in a pairwise manner, which should mitigate for the effect of diet.
Conclusions
We found that cholesterol levels are upregulated in OSCC tissue, suggesting deregulation of the usually tightly controlled cholesterol homeostasis.
We identified that GPL metabolism is dysregulated, and specifically PC and PE are the lipid classes most increased in OSCC tissue, suggesting that the Kennedy pathway is upregulated. We ascertained the fatty acid alterations in these GPLs, whereby mono- and unsaturated fatty acids were most increased in cancer, especially in PCs. Palmitate was especially increased, which suggests increased de novo fatty acid synthesis via FASN.
SREBPs are involved in both cholesterol metabolism as well as de novo fatty acid synthesis and so may be a good starting place for future OSCC lipid research.
The following are the supplementary data related to this article.
Example of how glycerophospholipid fatty acid chains were calculated.
Patient Demographics/Tumor data. Explanation of the column headings and abbreviations: No. — sample number; Sex — of the patient (m, male; f, female); TNM — tumor, node, metastasis classification. All patients were M0; stage — tumor stage according to UICC eighth edition guidelines; Recurrence — whether the disease was recurrent posttreatment and whether this was locoregional recurrence or distant metastasis; Previous cancer — any previous tumor and its location; Status — the current situation of the patient; Smoking — history of smoking; Alcohol — alcohol use.
Raw data.
All lipid species and classes, along with p-values and FDR.
Pathways enriched with the altered lipid species.
Glycerophospholipid fatty acid chains, chain saturations and chain lengths.
Structures of the identified lipid classes.
Results of data normalisation from metaboanalyst.ca.
Lipid species boxplots. Statistically significant values are identified using an asterisk (*).
Lipid classes boxplots. Statistically significant values are identified using an asterisk (*).
Hierarchically clustered heatmap of lipid classes.
Principal component analyses of lipid species. Page 1 - the top panel shows principal components 1 and 2 and 95% confidence intervals are shown with coloured rings; the lower panel shows principal components 1 and 2 without 95% confidence intervals. Page 2 - loadings for principal components 1 and 2. Page 3 - the top panel shows principal components 1, 2 and 3; the bottom panel shows the loadings for principal components 1, 2, and 3.
Principal component analyses of lipid classes. Page 1 - the top panel shows principal components 1 and 2 and 95% confidence intervals are shown with coloured rings; the lower panel shows principal components 1 and 2 without 95% confidence intervals. Page 2 - loadings for principal components 1 and 2. Page 3 - the top panel shows principal components 1, 2 and 3; the bottom panel shows the loadings for principal components 1, 2, and 3.
Top 3 altered KEGG pathways.
Visualisations of fatty acid chains, chain lengths and saturation in individual glycerophospholipids.
Acknowledgments
Acknowledgements
Immense gratitude goes out to the participants in the study who donated their tissue samples for this research.
Thanks to Katja Tuomainen for collecting samples and to Toni Saarela for his statistical advice.
The processing of the homogenized tissue samples was performed by Lipotype GmbH on a commercial basis.
Funding
This work was supported by the University of Helsinki, Korvatautien Tutkimussäätiö, Jane ja Aatos Erkon Säätiö, Emil Aaltonen Research Foundation, and the Finnish-Norwegian Medical Foundation.
Data Statement
Research data supporting these conclusions are in Table S3.
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- 1.Annertz K., Anderson H., Palmer K., Wennerberg J. The increase in incidence of cancer of the tongue in the Nordic countries continues into the twenty-first century. Acta Otolaryngol. 2012;132(5):552–557. doi: 10.3109/00016489.2011.649146. [DOI] [PubMed] [Google Scholar]
- 2.Chinn S.B., Myers J.N. Oral cavity carcinoma: current management, controversies, and future directions. J Clin Oncol. 2015;33(29):3269–3276. doi: 10.1200/JCO.2015.61.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medes G., Thomas A., Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953;13(1):27–29. [PubMed] [Google Scholar]
- 4.Cheng C., Geng F., Cheng X., Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun (Lond). 2018;38(1):27. doi: 10.1186/s40880-018-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrotti F., Rosa C., Cicalini I., Sacchetta P., Del Boccio P., Genovesi D. Advances in lipidomics for cancer biomarkers discovery. Int J Mol Sci. 2016;17(12) doi: 10.3390/ijms17121992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma B., Agnihotri N. Role of cholesterol homeostasis and its efflux pathways in cancer progression. J Steroid Biochem Mol Biol. 2019;191:105377. doi: 10.1016/j.jsbmb.2019.105377. [DOI] [PubMed] [Google Scholar]
- 7.Fenn J.B., Mann M., Meng C.K., Wong S.F., Whitehouse C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 8.Gross R.W., Han X. Lipidomics at the interface of structure and function in systems biology. Chem Biol. 2011;18(3):284–291. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephenson D.J., Hoeferlin L.A., Chalfant C.E. Lipidomics in translational research and the clinical significance of lipid-based biomarkers. Transl Res. 2017;189:13–29. doi: 10.1016/j.trsl.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5 doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa S., Tateya I., Hayasaka T., Masaki N., Takizawa Y., Ohno S., Kojima T., Kitani Y., Kitamura M., Hirano S. Increased expression of phosphatidylcholine (16:0/18:1) and (16:0/18:2) in thyroid papillary cancer. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0048873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wojakowska A., Cole L.M., Chekan M., Bednarczyk K., Maksymiak M., Oczko-Wojciechowska M., Jarząb B., Clench M.R., Polańska J., Pietrowska M. Discrimination of papillary thyroid cancer from non-cancerous thyroid tissue based on lipid profiling by mass spectrometry imaging. Endokrynol Pol. 2018;69(1):2–8. doi: 10.5603/EP.a2018.0003. [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Guan M., Lin Y., Cui X., Zhang Y., Zhao Z., Zhu J. Aberrant lipid metabolism in hepatocellular carcinoma revealed by liver lipidomics. Int J Mol Sci. 2017;18(12) doi: 10.3390/ijms18122550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu C., Zhou D., Luo Y., Guo S., Wang T., Liu J., Liu Y., Li Z. Tissue and serum lipidome shows altered lipid composition with diagnostic potential in mycosis fungoides. Oncotarget. 2017;8(29):48041–48050. doi: 10.18632/oncotarget.18228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Ren S., Piao H.L., Wang F., Yin P., Xu C., Lu X., Ye G., Shao Y., Yan M. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Sci Rep. 2016;6:20984. doi: 10.1038/srep20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marien E., Meister M., Muley T., Gomez Del Pulgar T., Derua R., Spraggins J.M., Van de Plas R., Vanderhoydonc F., Machiels J., Mercedes Binda M. Phospholipid profiling identifies acyl chain elongation as a ubiquitous trait and potential target for the treatment of lung squamous cell carcinoma. Oncotarget. 2016;7(11):12582–12597. doi: 10.18632/oncotarget.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cifkova E., Holcapek M., Lisa M., Vrana D., Melichar B., Student V. Lipidomic differentiation between human kidney tumors and surrounding normal tissues using HILIC-HPLC/ESI-MS and multivariate data analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2015;1000:14–21. doi: 10.1016/j.jchromb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Hilvo M., Denkert C., Lehtinen L., Müller B., Brockmöller S., Seppänen-Laakso T., Budczies J., Bucher E., Yetukuri L., Castillo S. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71(9):3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 19.Wang L., Wang X., Li Y., Hou Y., Sun F., Zhou S., Li C., Zhang B. Plasma lipid profiling and diagnostic biomarkers for oral squamous cell carcinoma. Oncotarget. 2017;8(54):92324–92332. doi: 10.18632/oncotarget.21289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cals F.L.J., Bakker Schut T.C., Caspers P.J. Baatenburg de Jong RJ, Koljenovic S, Puppels GJ. Raman spectroscopic analysis of the molecular composition of oral cavity squamous cell carcinoma and healthy tongue tissue. Analyst. 2018;143(17):4090–4102. doi: 10.1039/c7an02106b. [DOI] [PubMed] [Google Scholar]
- 21.Sampaio J.L., Gerl M.J., Klose C., Ejsing C.S., Beug H., Simons K., Shevchenko A. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci U S A. 2011;108(5):1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ejsing C.S., Sampaio J.L., Surendranath V., Duchoslav E., Ekroos K., Klemm R.W., Simons K., Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106(7):2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Surma M.A., Herzog R., Vasilj A., Klose C., Christinat N., Morin-Rivron D., Simons K., Masoodi M., Sampaio J.L. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur J Lipid Sci Technol. 2015;117(10):1540–1549. doi: 10.1002/ejlt.201500145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liebisch G., Binder M., Schifferer R., Langmann T., Schulz B., Schmitz G. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS) Biochim Biophys Acta. 2006;1761(1):121–128. doi: 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Herzog R., Schuhmann K., Schwudke D., Sampaio J.L., Bornstein S.R., Schroeder M., Shevchenko A. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0029851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herzog R., Schwudke D., Schuhmann K., Sampaio J.L., Bornstein S.R., Schroeder M., Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12(1):R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascual G., Avgustinova A., Mejetta S., Martin M., Castellanos A., Attolini C.S., Berenguer A., Prats N., Toll A., Hueto J.A. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541(7635):41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 28.Luo J., Hong Y., Lu Y., Qiu S., Chaganty B.K., Zhang L., Wang X., Li Q., Fan Z. Acetyl-CoA carboxylase rewires cancer metabolism to allow cancer cells to survive inhibition of the Warburg effect by cetuximab. Cancer Lett. 2017;384:39–49. doi: 10.1016/j.canlet.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohan M., Jagannathan N. Oral field cancerization: an update on current concepts. Oncol Rev. 2014;8(1):244. doi: 10.4081/oncol.2014.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuzu OF, Noory M.A., Robertson G.P. The role of cholesterol in cancer. Cancer Res. 2016;76(8):2063–2070. doi: 10.1158/0008-5472.CAN-15-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding X., Zhang W., Li S., Yang H. The role of cholesterol metabolism in cancer. Am J Cancer Res. 2019;9(2):219–227. [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko T., Kanno C., Ichikawa-Tomikawa N., Kashiwagi K., Yaginuma N., Ohkoshi C., Tanaka M., Sugino T., Imura T., Hasegawa H. Liver X receptor reduces proliferation of human oral cancer cells by promoting cholesterol efflux via up-regulation of ABCA1 expression. Oncotarget. 2015;6(32):33345–33357. doi: 10.18632/oncotarget.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murugan A.K., Munirajan A.K., Tsuchida N. Ras oncogenes in oral cancer: the past 20 years. Oral Oncol. 2012;48(5):383–392. doi: 10.1016/j.oraloncology.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Kawasaki G., Naruse T., Furukawa K., Umeda M. mTORC1 and mTORC2 expression levels in oral squamous cell carcinoma: an immunohistochemical and clinicopathological study. Anticancer Res. 2018;38(3):1623–1628. doi: 10.21873/anticanres.12393. [DOI] [PubMed] [Google Scholar]
- 35.Acharya S., Rai P., Hallikeri K., Anehosur V., Kale J. Serum lipid profile in oral squamous cell carcinoma: alterations and association with some clinicopathological parameters and tobacco use. Int J Oral Maxillofac Surg. 2016;45(6):713–720. doi: 10.1016/j.ijom.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 37.Simpson D.R., Mell L.K., Cohen E.E. Targeting the PI3K/AKT/mTOR pathway in squamous cell carcinoma of the head and neck. Oral Oncol. 2015;51(4):291–298. doi: 10.1016/j.oraloncology.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Koizumi A., Narita S., Nakanishi H., Ishikawa M., Eguchi S., Kimura H., Takasuga S., Huang M., Inoue T., Sasaki J. Increased fatty acyl saturation of phosphatidylinositol phosphates in prostate cancer progression. Sci Rep. 2019;9(1):13257. doi: 10.1038/s41598-019-49744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naguib A., Bencze G., Engle D.D., Chio I.I., Herzka T., Watrud K., Bencze S., Tuveson D.A., Pappin D.J., Trotman L.C. p53 mutations change phosphatidylinositol acyl chain composition. Cell Rep. 2015;10(1):8–19. doi: 10.1016/j.celrep.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng M., Bhujwalla Z.M., Glunde K. Targeting phospholipid metabolism in cancer. Front Oncol. 2016;6:266. doi: 10.3389/fonc.2016.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibellini F., Smith T.K. The Kennedy pathway—de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62(6):414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 42.Santos C.R., Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 43.Mariotto E., Viola G., Ronca R., Persano L., Aveic S., Bhujwalla Z.M., Mori N., Accordi B., Serafin V., Carlota López-Cara L. Choline kinase alpha inhibition by EB-3D triggers cellular senescence, reduces tumor growth and metastatic dissemination in breast cancer. Cancers (Basel) 2018;10(10) doi: 10.3390/cancers10100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelmann B., Wiedmann M.K. Cellular phospholipid uptake: flexible paths to coregulate the functions of intracellular lipids. Biochim Biophys Acta. 2010;1801(6):609–616. doi: 10.1016/j.bbalip.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Shindou H., Shimizu T. Acyl-CoA:lysophospholipid acyltransferases. J Biol Chem. 2009;284(1):1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- 46.Silva S.D., Perez D.E., Nishimoto I.N., Alves F.A., Pinto C.A., Kowalski L.P., Graner E. Fatty acid synthase expression in squamous cell carcinoma of the tongue: clinicopathological findings. Oral Dis. 2008;14(4):376–382. doi: 10.1111/j.1601-0825.2007.01395.x. [DOI] [PubMed] [Google Scholar]
- 47.McMahon H.T., Boucrot E. Membrane curvature at a glance. J Cell Sci. 2015;128(6):1065–1070. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohrig F., Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16(11):732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 49.Holder A.M., Gonzalez-Angulo A.M., Chen H., Akcakanat A., Do K.A., Fraser Symmans W., Pusztai L., Hortobagyi G.N, Mills G.B., Meric-Bernstam F. High stearoyl-CoA desaturase 1 expression is associated with shorter survival in breast cancer patients. Breast Cancer Res Treat. 2013;137(1):319–327. doi: 10.1007/s10549-012-2354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minville-Walz M., Pierre A.S., Pichon L., Bellenger S., Fevre C., Bellenger J., Tessier C., Narce M., Rialland M. Inhibition of stearoyl-CoA desaturase 1 expression induces CHOP-dependent cell death in human cancer cells. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0014363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example of how glycerophospholipid fatty acid chains were calculated.
Patient Demographics/Tumor data. Explanation of the column headings and abbreviations: No. — sample number; Sex — of the patient (m, male; f, female); TNM — tumor, node, metastasis classification. All patients were M0; stage — tumor stage according to UICC eighth edition guidelines; Recurrence — whether the disease was recurrent posttreatment and whether this was locoregional recurrence or distant metastasis; Previous cancer — any previous tumor and its location; Status — the current situation of the patient; Smoking — history of smoking; Alcohol — alcohol use.
Raw data.
All lipid species and classes, along with p-values and FDR.
Pathways enriched with the altered lipid species.
Glycerophospholipid fatty acid chains, chain saturations and chain lengths.
Structures of the identified lipid classes.
Results of data normalisation from metaboanalyst.ca.
Lipid species boxplots. Statistically significant values are identified using an asterisk (*).
Lipid classes boxplots. Statistically significant values are identified using an asterisk (*).
Hierarchically clustered heatmap of lipid classes.
Principal component analyses of lipid species. Page 1 - the top panel shows principal components 1 and 2 and 95% confidence intervals are shown with coloured rings; the lower panel shows principal components 1 and 2 without 95% confidence intervals. Page 2 - loadings for principal components 1 and 2. Page 3 - the top panel shows principal components 1, 2 and 3; the bottom panel shows the loadings for principal components 1, 2, and 3.
Principal component analyses of lipid classes. Page 1 - the top panel shows principal components 1 and 2 and 95% confidence intervals are shown with coloured rings; the lower panel shows principal components 1 and 2 without 95% confidence intervals. Page 2 - loadings for principal components 1 and 2. Page 3 - the top panel shows principal components 1, 2 and 3; the bottom panel shows the loadings for principal components 1, 2, and 3.
Top 3 altered KEGG pathways.
Visualisations of fatty acid chains, chain lengths and saturation in individual glycerophospholipids.