This review provides a wide-ranging assessment of approaches to reduce the impact of heat stress on food crops.
Keywords: Breeding, climate change, cultivated plants, food crops, food security, global warming, heat stress, omics, phenomics
Abstract
To ensure the food security of future generations and to address the challenge of the ‘no hunger zone’ proposed by the FAO (Food and Agriculture Organization), crop production must be doubled by 2050, but environmental stresses are counteracting this goal. Heat stress in particular is affecting agricultural crops more frequently and more severely. Since the discovery of the physiological, molecular, and genetic bases of heat stress responses, cultivated plants have become the subject of intense research on how they may avoid or tolerate heat stress by either using natural genetic variation or creating new variation with DNA technologies, mutational breeding, or genome editing. This review reports current understanding of the genetic and molecular bases of heat stress in crops together with recent approaches to creating heat-tolerant varieties. Research is close to a breakthrough of global relevance, breeding plants fitter to face the biggest challenge of our time.
Introduction
Current analysis conducted by scientific communities including NASA’s Goddard Institute for Space Studies (GISS) indicates that the average global temperature on Earth has increased by ~0.8 °C since 1880. Two-thirds of the warming has occurred since 1975, at a rate of roughly 0.15–0.20 °C per decade (Lorenz et al., 2019). Moreover, the Intergovernmental Panel on Climate Change (IPCC) in its last report assesses that even limiting global warming to just 1.5 °C would require an unprecedented change in many aspects of society (https://www.ipcc.ch/sr15/).
For many years, scientists have studied the physiological mechanisms underlying heat stress response (HSR) and tolerance in plants (Fahad et al., 2017; Prasad et al., 2017; Govindaraj et al., 2018). However, after the discovery of the molecular responses and regulation of heat shock proteins (HSPs) and the activation of other genes, understanding of the HSR process became more mechanistic (Key et al., 1981; Altschuler and Mascarenhas, 1982). Major advances have been made in the molecular mechanisms of HSR (Ohama et al., 2017) and identification of genes or quantitative trait loci (QTLs) associated with heat tolerance in plants.
The production of plants with new alleles in HS-responsive genes and the possibility of testing these plants on innovative phenotyping platforms offer the opportunity fully to establish the role of these genes in the HSR and to develop crop plants with an increased tolerance to heat (Comastri et al., 2018).
Temperature stress, crop yield, and food security
Temperature is one of the major environmental factors which affect plant growth, development, and yield. Temperatures persistently above optimal for plant growth may induce HS and reduce yields. At some threshold, they will be lethal. Extreme heat events can be measured in different ways: the maximum temperatures reached (intensity), how often the events occur (frequency), and how long they last (duration). HS has negative effects on crop physiology (e.g. decreased photosynthesis, increased respiration) and plant growth and yield (Prasad et al., 2017). Another adverse effect of HS is the negative influence on the plant root system, which provides support, nutrient and water uptake, and transport to other plant organs (Valdés-López et al., 2016), resulting in disrupted pollination, flowering, root development, and root growth stages (Sehgal et al., 2017; Cho, 2018).
In many species, for both cool and warm seasons, yield reduction after HS is observed as a consequence of decreasing seed- or fruit-set percentage (Otero et al., 2011; Shah et al., 2011; Singh and Jwa, 2013; Jangid and Dwivedi, 2016; Fahad et al., 2017; Sehgal et al., 2017; Comastri et al., 2018; Devasirvatham and Tan, 2018; Govindaraj et al., 2018). In wheat, exposure to short episodes (2–5 d) of HS (>24 °C) at the reproductive stage (start of heading) resulted in substantial damage to floret fertility, while a mean daily temperature of 35 °C caused total failure. Increasing the duration of high temperature at this stage reduced the grain weight linearly (Maestri et al., 2002; Prasad and Djanaguiraman, 2014); similarly for pea (Bhattacharya, 2019), lentil (Barghi et al., 2012), and chickpea (Wang et al., 2006). An extensive review on the threshold temperatures for several crop species was reported by Kaushal et al., 2016. Table 1 shows threshold temperatures in vegetative and reproductive development for several crop species, together with literature indications about the losses (or gains) attributed to HS alone.
Table 1.
Yield losses due to heat stress in cool and warm season crops
| Species | Threshold temperatures for the speciesa | World production in 2017b (kg ha–1) | Average yield reduction (%) | Reference |
|---|---|---|---|---|
| Cool-season crops | ||||
| Barley (Hordeum vulgare L.) | Not reported | 3136 | 15 | Weichert et al. (2017) |
| Chick pea (Cicer arietinum L.) | 15–30 °C for growth, 25 °C for reproductive growth | 1015 | 19–50 | Devasirvatham and Tan (2018) |
| Citrus (Citrus spp.) | 35 °C for vegetative growth | 9600 | N/A | N/A |
| Lentils (Lens culinaris Medik.) | Not reported | 1153 | 38–58 | Sita et al. (2018) |
| Spinach (Spinacia oleracea L.) | Not reported | 29 993 | 50 | Yan et al. (2016) |
| Wheat (Triticum spp.) | 20–30 °C for vegetative growth, 15 °C for reproductive growth | 3531 | 6 | Lobell et al. (2011); Zampieri et al. (2017); Comastri et al. (2018) |
| Warm-season crops | ||||
| Grapes (Vitis vinifera L.) | Not reported | 10 716 | 35–50 | Greer and Weedon (2013) |
| Maize (Zea mays L.) | 33–38 °C for photosynthesis and pollen viability | 5755 | 7–40 | Valdés-López et al. (2016); Zhao et al. (2017); Meseka et al. (2018); Prasad et al. (2018) |
| Peanut (Arachis hypogaea L.) | 29–33 °C for vegetative growth, 39–40 °C for seed set and yield | 1686 | 6 | Prasad et al. (2001) |
| Potato (Solanum tuberosum L.) | Not reported | 20 111 | 18–23 | Hancock et al. (2014) |
| Rapeseed (Brassica napus L.) | About 30 °C for flowering | 2195 | Up to 85% | Koscielny et al. (2018); Sparks (2018) |
| Rice (Oryza sativa L.) | 33 °C for biomass, 35 °C limiting for grain formation and yield | 4602 | 3 | Zhao et al. (2017) |
| Sorghum [Sorghum bicolor (L.) Moench] | 26–34 °C for vegetative growth, 40 °C for reproductive growth and yield | 1416 | 17–44 | Tack et al. (2017) |
| Soybean [Glycine max (L.) Merr.] | 26–36 °C for reproductive development, 39 °C lethal | 2854 | 3–7 | Valdés-López et al., 2016; Zhao et al. (2017) |
| Sunflower (Helianthus annuus L.) | Not reported | 1804 | 10–70 | Debaeke et al. (2017) |
| Tomato (Solanum lycopersicum L.) | 37 °C for vegetative growth, 28–30 °C for reproductive development | 37 600 | 28 | Snider et al. (2012); Lamaoui et al. (2018) |
a Data from Luo (2011) and Kaushal et al. (2016).
b Data obtained by world production and world cultivated extension for each crop, from FAOSTAT 2017, http://www.fao.org/faostat/en/#data/QC/visualize.
HS can impair several physiological processes linked to seed size and quality. HS during grain filling markedly decreased starch accumulation in wheat (Hurkman et al., 2003), rice (Yamakawa and Hakata, 2010), and maize (Yang et al., 2018). The levels of sugars such as fructose, sugar nucleotides, and hexose phosphate also declined due to HS (Yang et al., 2018). The decrease in sugars may be related to assimilate utilization for purposes other than edible component production (Asthir et al., 2012). In maize, waxy grain starch content decreased, whereas protein content increased, resulting in a change of grain quality (Yang et al., 2018). Moreover, increasing temperature and CO2 reduced protein and micronutrient content in grain (Chakraborty and Newton, 2011) and soybean (Li et al., 2018). In soybean under HS, the nutritional value of total free amino acids was reduced together with total protein concentration, while the oil concentration was significantly increased. As a general evaluation, reductions in total yield are mainly due to an alteration in balance of the source and sink activities that take place under HS.
The capacity of crop plants to overcome temperature stress has been interpreted in terms of avoidance, escape, or tolerance (Osmond et al., 1987). Avoidance is any mechanism that permits the plant to be or to become non-susceptible to the stressor effect, annulling in this way most of its deleterious effects (Hasanuzzaman et al., 2013). In escape, plants can alter their growth cycle before the stress hits hard, such as anticipating reproduction or shedding vegetative structures. From the agronomic point of view, tolerance is certainly more interesting; being based on the endurance of the plant in stress conditions, it does not involve substantial modification of the growth habit with potential deleterious effects on yield. A heat-tolerant plant is therefore inclined to continue its growth cycle quite independently of the stress (Barnabás et al., 2008).
The contributions of ‘omics’
Conventional plant breeding strategies based mainly on phenotype selection and qualitative genetics have led breeders, after the green revolution, to achieve continuous increases in seed yield and improved yield stability, independently of the environmental cost of achieving them (Setia and Setia, 2008). However, the growing worldwide demand for enhancing yields of major crops is placing pressure on breeding programs to provide elite cultivars more adaptable to the ongoing changes. The complexity of the information needed to meet this challenge has prompted advances in understanding the biochemical and molecular processes that underlie important metabolic, physiological, and developmental traits which affect the ability of plants to cope with the upcoming climate change. This has provided new insights into how plants function, and promoted the development of new scientific disciplines, mainly based on high-throughput approaches.
The progress of ‘omics’ technologies, in particular genomics, transcriptomics, proteomics, and metabolomics, has enabled direct and unbiased monitoring of the factors affecting crop growth and yield in response to environmental threats. Overall, omics constitute powerful tools to reveal the complex molecular mechanisms underlying plant growth and development, and their interactions with the environment, which ultimately determine yield, nutritional value (Setia and Setia, 2008; Soda and Wallace, 2015), and the required level of agricultural inputs. The following sections provide some examples showing the successful application of omics in crop improvement.
Genomics and transcriptomics
Molecular responses to HS have been investigated in many plant species to identify the complex processes and pathways regulated in acclimation and protection against temperature stress (Qu et al., 2013; Driedonks et al., 2016). Regulation of gene expression through transcriptional and post-transcriptional mechanisms is certainly linked to both stress recognition and stress response; this aspect has been investigated at different stress levels and comparing different genetic resources. The conventional transcriptomic approach has been applied to study gene expression changes under stress conditions in many crop species, such as rice (Sarkar et al., 2014; González-Schain et al., 2016; Mangrauthia et al., 2017), tomato (Frank et al., 2009; Bita et al., 2011), barley (Mangelsen et al., 2011), maize (Frey et al., 2015; Shi et al., 2017b), wheat (Qin et al., 2008), soybean (Chung et al., 2013; Gillman et al., 2019), brassica (Dong et al., 2015), and grape (Liu et al., 2012a). More recently, application of advanced genome-wide transcriptome analyses has elucidated the roles of relevant genes from HS perception to signal transduction and stress response (Table 2). In general, most of the HS-responsive genes are involved in primary and secondary metabolism, translation, transcription, regulation, and responses to processes such as calcium, phytohormone, sugar, and lipid signaling, or protein modifications including phosphorylation (Jha et al., 2014; Takahashi and Shinozaki, 2019). Up-regulation under HS conditions is confined mainly to transcription factors and HSPs.
Table 2.
Review of the transcriptomic, proteomic and metabolomic analyses performed to dissect the mechanisms involved in the heat stress response
| Species | Primary stress | Secondary stresses | Tissue or development stage analyzed | Simulation conditions | Molecular techniques applied | References |
|---|---|---|---|---|---|---|
| Barley | Heat | N/A | Anthesis | 42 °C | Transcriptomics (Affymetrix Barley1 GeneChips) | Mangelsen et al. (2011) |
| Barley | Heat | Drought | Plant leaf | 26 °C | Metabolomics IC-MS/MS | Templer et al., (2017) |
| Barley | Heat | Mild drought and combination | Flag leaves | 30 °C+50% FC | Transcriptomics (RNA-seq) | Cantalapiedra et al. (2017) |
| Chickpea | Heat | N/A | Whole plant development | Up to 42 °C | Proteomics (LC-MS) | Parankusam et al. (2017) |
| Chickpea | Heat | General oxidative stress | Plant leaf | 37 °C | Targeted metabolomics | Salvi et al. (2018) |
| Citrus | Heat | Drought | Plant leaf | 40 °C | Metabolomics GC-MS LC-MS | Zandalinas et al. (2017) |
| Grape | Heat | Various temperatures | Leaves | 35, 40, and 45 °C | Proteomics iTRAQ LC-MS/MS | Jiang et al. (2017) |
| Grape | Heat | Followed by recovery | Plant leaves | 43 °C | Proteomics (iTRAQ LC-MS/MS) | Liu et al. (2014) |
| Grapes | Heat | N/A | Leaves | 45 °C | Transcriptomics (Affymetrix gene chip) | Liu et al. (2012a) |
| Lentils | Heat | N/A | Pods | Up to 33 °C | Enzyme activities | Sita et al. (2018) |
| Lentils | Heat | N/A | Leaves and pods | 30–50 °C | Enzyme activities | Chakraborty and Pradhan (2011) |
| Lentils | Heat | Drought, also combined | Leaves and pods | 30 °C | Enzyme activities | Sehgal et al. (2017) |
| Maize | Heat | Drought | Leaves | 42 °C | Proteomics (iTRAQ LC-MS/MS) | Hu et al. (2015); Zhao et al. (2016) |
| Maize | Heat | Drought | Chloroplasts | 42 °C | Proteomics (MALDI TOF/ TOF MS/MS) | Hu et al. (2015) |
| Maize | Heat | N/A | Seedlings | 32–38 °C | Transcriptomics (RNA-seq) | Frey et al. (2015) |
| Maize | Heat | N/A | Seedlings | 42 °C | Transcriptomics (RNA-seq) | Shi et al. (2017b) |
| Maize | Heat | High CO2 | Plant leaf | 32 °C | Metabolomics (GC-MS) | Qu et al. (2018) |
| Mung bean | Heat | Exogenous glutathione | Plant leaves | 42° | Targeted lipidomics | Nahar et al., 2015 |
| Peanut | Heat | N/A | Seedlings | 40 °C and 30 °C | Physiological measurements, metabolomics | Singh et al. (2016) |
| Peanut | Heat | N/A | Leaves | 45 °C | Transcriptomics (qRT–PCR), physiological measurements | Chakraborty et al. (2018) |
| Potato | Heat | N/A | Leaves, tubers | Up to 30 °C | Metabolomic (GC/MS), physiological measurements, transcriptomic (microarray) | Hancock et al. (2014) |
| Potato | Heat, drought | Leaves | 35 °C | Transcriptomics (RNAseq) | Tang et al. (2016) | |
| Potato | Heat | Potato virus Y (PVY) | Leaves | 28 °C | Transcriptomics (qRT–PCR) | Makarova et al. (2018) |
| Potato | Heat | Tubers, skin and phelloderm | 33–35 °C | Transcriptomics (qRT–PCR) | Ginzberg et al. (2009) | |
| Rapeseed | Heat | N/A | Flowering | Up to 35 °C | GWAs | Rahaman et al. (2018) |
| Rapeseed | Heat | N/A | Developing seed | Up to 35 °C | Transcriptomics (95k EST microarray) | Yu et al. (2014) |
| Rapeseed | Heat, drought | Combination of the two stresses | 29±0.5 °C from the 38th day after sowing, 30% field capacity | Metabolomics, physiological measurements | Elferjani and Soolanayakanahally (2018) | |
| Rice | Heat | N/A | Anthers | Up to 37 °C in | Proteomics (iTRAQ LC-MS/MS) | Mu et al. (2017) |
| Rice | Heat | N/A | Grain and Leaves | 30 °C | Transcriptomics (qRT–PCR) | Phan et al. (2013) |
| Rice | Heat | Open field | Leaves, early milky stage of rice grains, spikelets | 42/32, 38, 28 °C | Proteomics (MALDI TOF/TOF MS/MS) | Liao et al. (2014); Das et al. (2015) |
| Rice | Heat | Cold | Suspension cell | 44 °C | Proteomics (label-free in-gel digestion together with LC-MS/MS) | Gammulla et al. (2010) |
| Rice | Heat | Various temperatures | Seedlings, anthesis | 35 °C, 40 °C and 45 °C, 38 °C | Proteomics (MALDI TOF MS/MS) | Han et al. (2009); Jagadish et al. (2010) |
| Rice | Heat | N/A | Anthers | 38 °C | Proteomics (label-free in-gel digestion together with LC-MS/MS) | Kim et al. (2015) |
| Rice | Heat | N/A | Plants at flowering | 38 °C for 1 d | Transcriptomics (RNAseq, large-scale qRT–PCR) | González-Schain et al. (2016) |
| Rice | Heat | N/A | Seeds | Germination at 30 °C (control) and 42 °C (stress) | Transcriptomics (RNA-seq, qRT–PCR) | Mangrauthia et al. (2016) |
| Rice | Heat and drought | Sugar starvation | Floral organs | Metabolomic and Transcriptomics | Li et al. (2015) | |
| Rice | Heat | Oxidative stress | 10-day-old seedlings | 42±1 °C for 30 min with 10 mM H2O2 | Transcriptomics (60mer rice 44k array, qRT–PCR) | Mittal et al. (2012) |
| Rice | Heat | N/A | Pollen | 37 °C | Targeted metabolomics | Feng et al. (2018) |
| Rice | Heat | N/A | Leaves | 45 °C | Transcriptomics (RNA-seq) | Fang et al. (2018) |
| Rice | Heat | N/A | Grains | 38 °C | Transcriptomics (RNA-seq) | Liao et al. (2015) |
| Rice | Heat | Drought exogenous IAA | Plant leaf | 40 °C | Target lipidomics | Sharma et al. (2018) |
| Rice | Heat | Exogenous brassinosteroids | Plant leaf | 47 °C | Target lipidomics | Thussagunpanit et al. (2015) |
| Sorghum | Heat | Drought | Seedlings | 28 °C and 50 °C, water withholding. Combination of the stress | Transcriptomics (microarray) | Johnson et al. (2014) |
| Soybean | Heat | High humidity | Pre-harvest seed | 40 °C | Proteomics (MALDI TOF/TOF MS/MS) | Wang et al. (2012) |
| Soybean | Heat | N/A | 3- to 4-week-old plants | 45 °C for 3, 6, 24 h | Transcriptomics (qRT–PCR) | Chung et al. (2013) |
| Soybean | Heat | High CO2 | Plant leaf | 36 °C | Metabolomics (GC-MS) | Xu et al. (2016) |
| Soybean | Heat | N/A | Roots | 40 °C | Proteomics (LC-MS/MS); Transcriptomics (RNA-seq) | Valdés-López et al. (2016) |
| Soybean | Heat | N/A | Dry, imbibed, or germinated seeds from heat-tolerant and heat-sensitive cultivars | Field conditions with heat stress | Transcriptomics (RNA-seq) | Gillman et al. (2019) |
| Spinach | Heat | N/A | Leaves | 37 °C, 35 °C | Physiological, enzyme activity, proteomic (trypsin digestion and iTRAQ labeling), transcriptomics (NGS) | Yan et al. (2016); Zhao et al. (2018) |
| Sunflower | Heat | Light | Leaves and immature seed | 37 °C | Transcriptomics | Hewezi et al. (2008) |
| Tomato | Heat | Melatonin | Up to 42 °C | Metabolomics | Xu et al. (2016) | |
| Tomato | Heat | Oxidative stress | Flower buds | 36 °C\25 °C for 3 months | Proteomics (SDS–PAGE followed by LC-MS/MS) | Mazzeo et al. (2018) |
| Tomato | Heat | Lipid antioxidant and galactolipid remodeling | 5- to 6-week-old plants | 38 °C | Metabolomics (MS) | Spicher et al. (2016) |
| Tomato | Heat | N/A | Different developmental stages of tomato pollen | 38 °C | Proteomics (mass accuracy precursor alignment (MAPA) plus LC/MS) | Chaturvedi et al. (2015) |
| Tomato | Heat | Waterlogging in open field | Leaves | 39 °C | Proteomics (MALDI TOF/TOF MS/MS) | Lin et al. (2016) |
| Tomato | Heat | N/A | Anthers | 32 °C for 2, 6, 16 or 30 h | Transcriptomics (cDNA-AFLP; 90 K Custom TomatoArray 1.0) | Bita et al. (2011) |
| Tomato | Heat | N/A | Flower buds | 43–45 °C for 2 h | Transcriptomics (Affymetrix GeneChip® Tomato Genome Array) | Frank et al. (2009) |
| Tomato | Heat | N/A | week‐old tomato plants | 1 h at 39 °C | Transcriptomics (massive analysis of cDNA ends (MACE) | Fragkostefanakis et al. (2015) |
| Tomato | Heat | N/A | Pollen | 38 °C | Metabolomics (LC-QTOF-MS plus LTQ Orbitrap XL, mass spectrometer) | Paupière et al. (2017) |
| Tomato | Heat | N/A | Pollen tubes | 37 °C | Targeted metabolomics | Muhlemann et al. (2018) |
| Tomato | Heat | Cold | Plant leaves | 38, 20, 10 °C | Untargeted lipidomics LC/MS | Spicher et al. (2016) |
| Wheat | Heat | N/A | Seeds in filling stage | 37 °C for 4 h | Metabolomics, transcriptomics | Wang et al. (2015) |
| Wheat | Heat | Lipid alteration | Flowering | Up to 35 °C | Metabolomics | Narayanan et al. (2016) |
| Wheat | Heat | N/A | Seedlings | Up to 42 °C | Transcriptomics | Comastri et al. (2018) |
| Wheat | Heat | N/A | Seedlings | 35 °C | Proteomics (MALDI TOF/TOF MS/MS) | Gupta et al. (2015) |
| Wheat | Heat | N/A | Leaves, stems, and spikes, flag leaf | 38 °C, 37 °C | Proteomics (iTRAQ LC-MS/MS) | Lu et al. (2017); Kumar et al. (2019) |
| Wheat | Heat | Open field | Grain | 40 °C | Proteomics (MALDI TOF/TOF MS/MS) | Wang et al. (2018) |
| Wheat | Heat | Low nitrogen | Plant leaf | 32 °C | Proteomics (MALDI TOF/TOF MS/MS) | Yousuf et al. (2017) |
| Wheat | Heat | Drought | Post-anthesis | 35 °C | Proteomics (MALDI TOF/TOF MS/MS) | Zhang et al. (2018) |
| Wheat | Heat | N/A | Flag leaf | 35 °C | Proteomics | Wang et al. (2015) |
| Wheat | Heat | N/A | Leaf of heat-susceptible ‘Chinese Spring’ (CS) and heat-tolerant ‘TAM107’ (TAM) | 40 °C for 1 h with and without heat acclimation (34 °C, 3 h) | Transcriptomics (wheat genome array) | Qin et al. (2008) |
| Wheat | Heat | N/A | Flag leaf, seedlings | 37/17 °C for 3 days in anthesis stage | Proteomics | Lu et al. (2017) |
| Wheat | Heat | N/A | Spikelet post-anthesis | 35 °C | Metabolomics (LC/HRMS) | Thomason et al. (2018) |
| Wheat | Heat | Drought | Seedlings | 40 °C, 20 % (w/v) PEG-6000 | Transcriptomics (RNA-seq) | Liu et al. (2015) |
| Wheat | Heat | N/A | Plant leaf | 35 °C | Lipidomics ICP-MS | Narayanan et al. (2016) |
| Wheat | Heat | N/A | Pollen | 35 °C | Lipidomics ICP-MS | Narayanan et al. (2018) |
| Wheat | Heat | N/A | Plant leaf | 35 °C | Lipidomics ICP-MS/MS | Djanaguiraman et al. (2018) |
Abbreviations: cDNA-AFLP, cDNA-amplified fragment length polymorphism; GWAS, genome-wide association study; IAA, indole-3-acetic acid; IC-MS/MS, ion chromatography tandem MS; ICP-MS, inductively coupled plasma MS; iTRAQ, isobaric tags for relative and absolute quantitation; LC/HRMS, liquid chromatography-high resolution MS; MACE, massive analysis of cDNA ends; MALDI TOF/TOF MS/MS, matrix assisted laser desorption/ionization-time of flight tandem MS; MAPA, mass accuracy precursor alignment; N/A, not applicable; NGS, next-generation sequencing; qRT–PCR, quantitative reverse transcription–PCR; QTOF, quadrupole time of flight; RNA-seq, RNA sequencing,
The activation and production of heat shock factors (HSFs) and HSPs and the increase in reactive oxygen species (ROS)-scavenging activity play a key role in the responses and acclimation of plants to HS (Maestri et al., 2002; Driedonks et al., 2016; Comastri et al., 2018). ROS-related genes are within the main up-regulated category under HS in different plant species (Chao et al., 2008; Chou et al., 2012; Mittal et al., 2012; Suzuki et al., 2014) and have been associated with basal heat tolerance (Almeselmani et al., 2006, 2009; Kang et al., 2009; Bhattacharjee, 2012). Moreover, ROS play an important role as signal molecules involved in the transduction of intracellular and intercellular signals controlling gene expression and activity of anti-stress systems (Petrov and Van Breusegem, 2012), suggesting a role for ROS in the activation of HSFs during HS (Driedonks et al., 2016).
Both HSFs and HSPs are central to the HSR and in the acquisition of thermotolerance in plants (Scharf et al., 2012; Ohama et al., 2017). As a result of advances in whole-genome sequencing, additional information has been gathered for the Hsp gene family (Chen et al., 2018). HS transcription factors (TFs) involved in heat sensing and signaling are highly conserved both structurally and functionally throughout the eukaryotic kingdom and are key players for the expression of Hsp genes (Scharf et al., 2012). The HSF gene family has been identified and characterized in several crop species such as grape (Liu et al., 2012b), soybean (Scharf et al., 2012; Chung et al., 2013), maize (Lin et al., 2011), wheat (Xue et al., 2014), rice (Mittal et al., 2009; Jin et al., 2013; Lavania et al., 2018), and tomato (Yang et al., 2016). Progress in genome annotation has led to the development of the specialized HEATSTER database (http://www.cibiv.at/services/hsf/info#anfang) which can identify and correctly annotate new HSF genes in plant species (Berz et al., 2019).
The involvement of HSPs in the defense response following HS has been extensively reported in several plant species (Park and Seo, 2015; Comastri et al., 2018). Gene expression analysis was performed in tomato leaves applying MACE (massive analysis of cDNA ends), which showed that 2203 genes (9.6% of the total) had enhanced transcript abundance in response to HS (Fragkostefanakis et al., 2015). Those encoding small HSP (sHSP) family members showed the strongest induction, with nine out of 100 Hsp40 genes up-regulated in response to heat. HSF genes showed basal and stable expression in non-stressed conditions and, for some, a strong induction in response to HS. These studies also reported the importance of TFs in the regulation of HSR, since 92 of the genes up-regulated upon HS are TF genes (bHLH, bZIP, ERF, MYB, and WRKY families). These are well-known major elements in HSR and thermotolerance, highlighting the complex regulatory networks activated beyond those directly controlled by HSFs. The tomato HsfA1a TF seems to have a unique function as a master regulator for acquired thermotolerance and cannot be replaced by any other HSF (Scharf et al., 2012).
In wheat, 6560 probe sets displayed changes in expression after heat treatment at 40 °C, with or without acclimation at 34 °C; representing Hsp genes, HSF genes, and genes encoding proteins involved in phytohormone biosynthesis/signaling, calcium and sugar signal pathways, RNA metabolism and ribosomes, as well as primary and secondary metabolism (Kotak et al., 2007; Qin et al., 2008). In barley, the effects of a short-term HS on central developmental functions of the caryopsis were studied at the transcriptional level (Mangelsen et al., 2011). Over 2000 differentially expressed genes were identified, equally divided between up- and down-regulation. The core HSR includes some conserved processes such as abscisic acid- (ABA) responsive gene activation, HSP-mediated protein folding, calcium signaling, ROS scavenging, and the biosynthesis of compatible solutes, which are rapidly induced. Within the down-regulated genes, it is noticeable that those involved in carbon and nitrogen metabolism and cellular growth, which determine long-term negative effects, differ significantly depending on the plant developmental stage at the time of the stress.
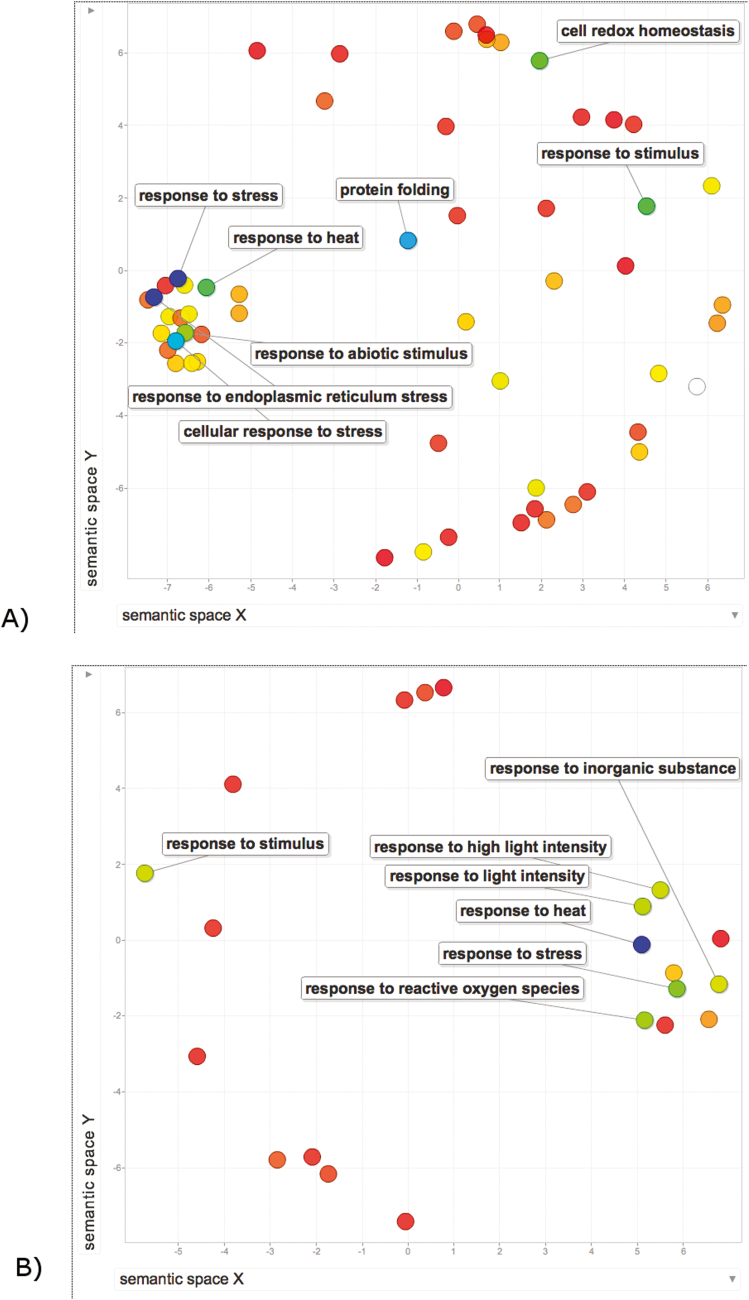
During grain development, the main negative effects of HS are related to the decreased accumulation of storage compounds, which can have a detrimental effect on both seed quality and final yield (Mangelsen et al., 2011; Hurkman et al., 2013). In wheat, HSPs can play a crucial role in gluten formation as a ‘glue’ between the reserve proteins (glutenins and gliadins) and starch (Maestri et al., 2002). During flowering, a significant reduction in the expression of non-essential photosynthetic genes may constitute an energy-saving strategy to facilitate other key stress responses with the aim of maintaining sugar metabolism and consequently protecting pollen viability (X. Li et al., 2015). Unraveling the complexity of high temperature tolerance may benefit from a systems biology approach (see Fig. 1).
Fig. 1.
Scatterplots obtained by multidimensional scaling of the matrix of biological process Gene Ontology (GO) terms across transcriptomic data obtained from heat-stressed tomato (A) and maize (B). The color of each spot indicates the level of enrichment from red (lowest p) to blue (highest p). The labels refer only to the most frequent GO terms. (A) Tomato transcripts identified by Fragkostefanakis et al. (2015) are analyzed through their Arabidopsis orthologs, 67 genes in total; (B) maize transcripts have been identified in Frey et al. (2015) as significantly induced, 454 genes in total. Enriched GO classes have been identified with AgriGO v.2 (http://systemsbiology.cau.edu.cn/agriGOv2/;Du et al., 2010) and the results visualized with ReviGO (http://revigo.irb.hr;Supek et al., 2011).
Proteomics
Protein profiling under HS conditions helps to identify stress-responsive proteins, and the detailed investigation of these proteins reveals their function in stress tolerance mechanisms (Priya et al., 2019). The synthesis and accumulation of HSPs is a prompt response after exposure to high temperature and it is considered one of the most important adaptive strategies to overcome the deleterious effects of HS (Wahid et al., 2007; Keller and Simm, 2018). The majority of HSPs are molecular chaperones involved in protein stabilization and signal transduction during HS (Sung et al., 2001; Arce et al., 2018). HSPs prevent accumulation of proteins with anomalous conformations and eliminate non-native aggregations formed during stress, with ubiquitin-mediated degradation of these proteins (Kotak et al., 2007).
Proteomics allows the study of the direct gene products, which often differ with the level of gene regulation. This technique has been applied to investigate heat responses in rice, wheat, tomato, maize, soybean, grape, and chickpea (for references, see Table 2). The types of technique utilized for these studies have been mostly 2D gel electrophoresis (SDS–PAGE) for protein separation, followed by MS:MALDI-TOF (matrix-assisted laser desorption/ionization-time of flight), MS, or TOF/TOF MS/MS for protein isolation, and iTRAQ (isobaric tags for relative and absolute quantitation) with LC-MS/MS for relative quantification (Zhang et al., 2006; Wiese et al., 2007). Other techniques have been deployed including label-free in-gel digestion together with LC-MS/MS or Orbitrap; SDS–PAGE followed by LC-MS/MS; and mass accuracy precursor alignment (MAPA) plus LC-MS (Chaturvedi et al., 2015). Many successful examples of proteomics applied to the study of HSPs and other HS-induced proteins have been reported (Malcevschi and Marmiroli, 2012; Table 2).
Different stages of plant development have been investigated using proteomics, from seedling, to flower (anthers and pollen especially in rice and tomato), to fruit, but also spikelet and grain maturation. In rice, where HS has been studied in isolation, all classes of HSPs (high and low molecular weight) have been found among the predominant protein categories, although other protein types and Gene Ontology (GO) classes varied extensively according to the part of the plant investigated. For instance, in pollen and anthers, late embryogenesis abundant (LEA) proteins were present in high numbers. Interestingly, proteins related to oxidative stress were identified in most of the studies on high HS, alone or in combination with other stresses. ROS are produced during HSR, which results in an oxidative imbalance within the cell. Furthermore, chloroplast function and photosynthesis are both strongly affected by ROS generated during HS, and it seems that HSPs can prevent ROS damage to the photosystems or the organelle structure. As an example, in maize, it was found that sHSP26 protects chloroplasts under HS (Hu et al., 2015). Other GO categories involved were sugar metabolism, the tricarboxylic acid (TCA) cycle, regulatory proteins, and proteins that function in energy metabolism.
More information on stress-associated active proteins (SAAPs) in wheat has recently been reported (Kumar et al., 2019). This study identified ~4272 SAAPs in wheat using absolute quantification methods. Some of the differentially abundant SAAPs identified were Rubisco, Rubisco activase (RCA), oxygen-evolving extrinsic protein (OEEP), HSP17, superoxide dismutase (SOD), catalase (CAT), and calcium-dependent protein kinase (CDPK). Pathway analysis showed the carbon assimilation pathway, followed by starch metabolism, to be most perturbed under HS in wheat. A positive correlation was established between the expression of SAAPs at transcript and protein levels in wheat under HS (Kumar et al., 2019).
Adaptive responses to HS also involve various post-translational modifications (PTMs) of proteins. There is a detailed literature on the effects of HS on PTMs such as protein phosphorylation in rice and wheat (Chen et al., 2011; Wu et al., 2014). Currently, >300 PTMs have been detected and partly characterized, and the numbers are increasing (Wu et al., 2016). HS has also been shown to enhance the small ubiquitin-like modification (SUMOylation) of particular proteins (Miller and Vierstra, 2011). SUMOylation of accessible lysines on target proteins is mediated in plants by a sequential three enzyme conjugation pathway: the E1 activating enzyme heterodimer (SAE1a or b combined with SAE2), a single E2 conjugating enzyme (SCE1), and at least two E3 ligases (SIZ1 and MMS21/HPY2). In Arabidopsis thaliana L. (Heynh), the activity of the HS TF HSFA2 was shown to be regulated by SUMOylation (Cohen-Peer et al., 2010). Moreover, in potato, rapid changes in protein SUMOylation and serine phosphorylation were observed in response to HS (Colignon et al., 2019). In addition, the overexpression of SlSIZ1 E3 ligase was shown to enhance heat tolerance in tomato (Zhang et al., 2018).
Metabolomics and lipidomics
Major environmental stresses cause metabolic reorganization towards homeostasis, maintaining essential metabolism and synthesizing metabolites with stress-protective and signaling characteristics. In plants such as tomato (Paupière et al., 2017), maize (Qu et al., 2018), barley (Templer et al., 2017), wheat (Thomason et al., 2018), soybean (Xu et al., 2016), and citrus (Zandalinas et al., 2017), metabolism reprogramming under HS has been studied by untargeted metabolomics. For example, in maize, under CO2 stress and a sudden HS, malate, valine, isoleucine, glucose, starch, sucrose, proline, glycine, and serine were successfully found as key players in the combined stress response (Qu et al., 2018). Under both heat and drought stress, amounts of amino acids, and antioxidants such as glutathione and α-tocopherol, were highly affected in two cultivars of barley (Templer et al., 2017); in particular, this combination of stresses led to accumulation of free amino acids in the leaf. In post-anthesis wheat, the amounts of some amino acids increased, while others decreased [l-arginine, l-histidine, l-tryptophan. l-threonine, 4-aminobutanoate (GABA), l-aspartate, and l-phenylalanine]; sugars, sugar alcohols, and other organic compounds were among the major groups of metabolites whose concentration decreased in tissue due to HS.
Thomason et al. (2018) identified the metabolites that exhibited the greatest decreases during HS in wheat: drummondol, anthranilate, dimethylmaleate, galactoglycerol, guanine, and also glycerone. Soybean subjected to both HS and excess CO2 had decreased starch, fructose, glucose, sucrose, and maltose concentrations, whereas pinitol increased with temperature (Xu et al., 2016). Zandalinas et al. (2017) found in citrus that water stress and HS, and their combination, altered metabolite levels related to glycolysis and the TCA cycle, the chloroplastic phase of the glyoxylate/dicarboxylate cycle, and the quantity of polar metabolites participating in the phenylpropanoid pathway arising from shikimic acid, depending on the level of stress. In tomato pollen, under HS, most of the putatively identified secondary metabolites belonged to three major groups: flavonoids, polyamines, and alkaloids (Paupière et al., 2017). Other examples come from untargeted metabolomics experiments where it was found that salicylic acid, ascorbic acid, sulfur-containing molecules (glutathione), phenolic secondary metabolites, and in general all antioxidant molecules can reduce HS symptoms in many plant species (Mobin et al., 2017; Feng et al., 2018; Muhlemann et al., 2018; Niu and Xiang, 2018; Salvi et al., 2018; Ihsan et al., 2019). Other major aspects of high temperature stress are cell signaling, molecular transport, and the changes in lipid metabolism as a basic mechanism to regulate membrane fluidity.
Lipids, which are major components of the membranes of cells and organelles, are among the first targets of ROS produced during HS and directly by high temperatures (Narayanan et al., 2016, 2018). Stress-induced lipid peroxidation of unsaturated fatty acids and other changes in membrane lipid profiles can lead to membrane damage, electrolyte leakage, and cell death (Liu and Huang, 2000). Under severe HS and long-term HS conditions, both chloroplastic and extra-plastidial glycerolipids are oxidized, yielding oxylipin-containing glycerolipids and other cytotoxic molecules such as acrolein and methyl vinyl ketone (MVK), derived from peroxidation of trienoic ω-3 fatty acids (Vu et al., 2012).
Accumulation of highly saturated fatty acids helps confer HS tolerance by reducing structural membrane fluidity, which is increased at high temperatures (Escandón et al., 2018). There is an interesting correlation between the type of metabolites involved and the need to protect specific cellular functions or cell compartments from the adverse effects of stress. In addition to acyl oxidation, plants regulate other aspects of membrane lipid composition in response to changes in temperature; in particular, the chloroplast membranes that host the photosynthetic apparatus are thought to be highly vulnerable to damage caused by HS (Welti et al., 2007; Zheng et al., 2011). Severe HS results in increasing frequency of membrane phase separation of non-bilayer-forming glycerolipids. Moderate HS (30–45 °C) results in thylakoid grana membrane destacking (Higashi and Saito, 2019). During HS, the endoplasmic reticulum (ER) is the major site for membrane phosphatidylcholine (PC) synthesis in plants (Jessen et al., 2015). Free fatty acids are exported from chloroplasts and re-esterified to CoA in the cytosol. PC synthesis in the ER utilizes a mixed pool of acyl-CoA substrates via the Kennedy pathway and the PC acyl editing pathway (Lands cycle) (Wang et al., 2012a). Fatty acid desaturase 2 (FAD2) converts PC-bound 18:1 to 18:2 in the ER and, during HS, FAD2 mediates polyunsaturation of ER glycerolipids and plays a role in the plant ER stress response to HS (Martinière et al., 2011). Chloroplastic galactolipid biosynthesis through the eukaryotic pathway needs a transfer of glycerolipids from the ER to chloroplasts by phospholipid flippase ALA10 which interacts with FAD2 affecting the degree of PC saturation. LACS4/9 proteins are also involved in this mechanism (Botella et al., 2016).
At the chloroplastic outer and inner membranes, five trigalactosyldiaglycerols (TGDs) and proteins TGD1, 2, 3, 4, and 5 form a transporter complex that mediates ER to plastid lipid trafficking (Xu et al., 2008). However, there is no concerted induction of these membrane proteins at the transcriptional level under HS in Arabidopsis (Higashi et al., 2015). In chloroplasts, long-term HS (>1 d) decreases the levels of glycerolipids containing 16:3 and/or 18:3, and reduces the activity of all FAD enzymes (Higashi and Saito, 2019). In general, during HS and other environmental stresses, conversion of monogalactosyldiacylglycerol (MGDG) into digalactosyldiacylglycerol (DGDG) increases through the action of DGDG synthases, encoded by DGD1 and DGD2. Terrestrial plants increase the ratio of DGDG to MGDG to improve thylakoid membrane stability under various abiotic stresses (Higashi et al., 2018). MGDGs are also decreased through the action of specific lipases (DAD1; PLIP1, 2, and 3; and HIL1). These recent studies suggest that lipases which are localized in chloroplasts have an important role in the lipid remodeling process under HS (Higashi and Saito, 2019).
Glycolipids and phospholipids are converted to triacylglycerol (TAG) in the form of oil bodies, as a transient storage depot for acyl groups prior to membrane lipid recycling/degradation: phospholipid:diacylglycerol acyltransferase 1 (PDAT1) transfers an acyl group from PC to diacylglycerol (DAG), and produces TAG. Mutant pdat1 seedlings are more sensitive to HS than the wild type; therefore, PDAT1 contributes to HS-induced TAG accumulation, and plays a role in HS response in Arabidopsis seedlings (Mueller et al., 2017). Other chloroplast-localized enzymes that maintain the structure of the thylakoid membranes during HS are FAX 1, 2, and 3, and VIPP1(Zhang et al., 2012; N. Li et al., 2015). HS also changes the lipid composition of the plasma membrane and of the ER; PC and PE (phosphatidylethanolamine) phospholipids are major components of extra-plastidial membranes. A decrease in the PE polyunsaturation level is likely to improve membrane stability under HS (Narayanan et al., 2016). Long-term HS increases phosphatidylserine (PS) content in the leaves of wheat (Narayanan et al., 2016).
Most of the lipidomics investigations of the response to HS have been performed on Arabidopsis, with only a few studies on crops such as wheat and rice. One example of lipidomics applied to HS in wheat found that the decrease in the photosynthetic rate under HS is due to lipid desaturation, oxidation, acylation, and consequent damage to organelles (Djanaguiraman et al., 2018). The membrane protein–lipid associations are of great importance for the maintenance of membrane fluidity; in this context, overexpression of the gene OsFBN1 (coding for a fibrillin) facilitates the import of lipids to the chloroplast during HS and the consequent grain filling (Djanaguiraman et al., 2018).
It has been suggested that diminishing the ROS pool directly has positive consequences on the state of the lipids. For example, modulating antioxidant defenses and the detoxification of methylglyoxal (MG) through the application of exogenous glutathione to mung bean [Vigna radiata (L.) R.Wilczek] increased the tolerance of the plants to HS in the short term (48 h) (Nahar et al., 2015). The increase in certain plant hormones can also be beneficial to the lipid status in rice during HS (Lim et al., 2017). For example, high concentrations of indole-3-acetic acid (IAA) or brassinosteroids can successfully initiate a mechanism for the decrease of primary ROS caused by HS that attack lipid membranes (MGDGs and sulfoquinovosyldiacylglycerols) and thylakoids (Thussagunpanit et al., 2015; Sharma et al., 2018). In a non-targeted lipidomics experiment in rice under both HS and water stress, 298 lipids responded to the changes in conditions; of these, 128 were identified. Interestingly, in the case of HS, a decrease in the unsaturation of lipids was predominant and can be linked to an increase in the cell membranes’ rigidity (Navarro-Reig et al., 2019). In a lipidome-wide study of tomato leaf under HS, changes in 791 molecules were detected between 20 °C and 38 °C. These results indicate that the most important changes at the lipidome-wide level occur in tocopherols, plastoquinone/plastoquinol (and their metabolites), and in the degree of fatty acid saturation of galactolipids (Spicher et al., 2016).
Epigenetic modifications
The epigenetic changes that occur at the DNA level through methylation of cytosine residues or at the level of chromatin by post-translational modifications of histones can result in altered gene transcription, and are an important mechanism in regulating gene expression during development and in response to environmental stimuli (Zhang and Hsieh, 2013). The persistence of a memory of temperature stress is dependent not only on the persistence of HSPs and their TFs but also on the expression of certain genes relevant for epigenetic signatures (Liu et al., 2015). A primary stress episode can sensitize plants for acclimation to subsequent stresses, resulting in faster or stronger changes in gene expression upon repeated exposure (Vriet et al., 2015), constituting a physiological priming mechanism. This primary event is fundamental in the acquisition of thermotolerance (Sanyal et al., 2018). A comprehensive RNA-Seq (RNA sequencing) analysis of gene expression and splicing events in HS-primed and non-primed plants has been carried out (Ling et al., 2018). This study showed alternative splicing as a novel and significant part of HS priming-induced memory and that this is critical for enhanced stress tolerance. However, most studies have been carried out in Arabidopsis, and in cultivated plants the understanding of the molecular mechanisms involved still represents a challenge (Friedrich et al., 2019).
Further epigenetic control is due to miRNAs, which are emerging as factors involved in transcriptional regulation and HS memory. miRNAs are involved directly in HS adaptation by acting as post-transcriptional regulators. Plant miRNAs bind their target transcripts and cleave and/or degrade the target mRNA molecule (Budak and Akpinar, 2015; Alptekin et al., 2017). The regulation of stress-responsive genes through activation of miRNAs is particularly important under abiotic stress conditions (Liu et al., 2017; Gahlaut et al., 2018).
Experimental validation of miRNA targets is still a challenge in polyploid crops including wheat. Recently, HS-responsive miRNAs have been reported from several susceptible and tolerant cultivars, with responses primarily being invoked within 24 h of treatment (Qin et al., 2008; Kumar et al., 2015). Furthermore, Ravichandran et al. (2019) performed a detailed miRNA and isomiR (miRNA isoforms) annotation and validated the HS-regulated miRNA target genes associated with thermotolerance. They confirmed a high degree of conservation between dicots and monocots for miR156 regulation of squamosa promoter-binding-like protein (Stief et al., 2014), miR159 regulation of MYB transcription factor (Wang et al., 2012b), miR166 regulation of homeobox leucine-zipper protein (Arikit et al., 2014), and miR398 regulation of SOD (Jagadeeswaran et al., 2009). New miRNA targets were also identified for organelle-specific transcripts such as pentatricopeptide repeat-containing proteins and mitochondrial transcription termination factor-like proteins.
Interestingly, several miRNAs showed altered expression patterns under heat and cold stresses in wheat, including miR164 (targeting HSP17) and miR319 (targeting a MYB transcription factor). These are up-regulated during a cold stress response but down-regulated in response to HS. On the contrary, miR160 and miR164 were down-regulated in response to heat, resulting in the induction of HSP expression, and up-regulated under cold stress. Regulating mechanisms triggered by high temperature can be reversed by cold stress, although the underlying mechanisms may be similar (Alptekin et al., 2017).
Breeding for heat tolerance
To ensure food security for future generations, agriculture must double the current crop production rate despite the predicted threats, which include climate change (Sedeek et al., 2019), decrease in arable land and desertification, salinization, and emerging diseases. Plant breeders need affordable and scalable solutions to achieve a more sustainable production. Approaches to reach the essential goal of producing more with less include (i) harnessing natural and artificial mutations; (ii) exploiting the available genetic resources to produce new genetic material more tolerant to HS and related secondary stresses; (iii) improving the ability to screen and identify the available sources of resilience; and (iv) developing new breeding techniques.
Conventional breeding approach
The effects of HS and prolonged heat waves on food production are heightened by a greater genetic uniformity in crop plants resulting from the narrowing of the varieties used in developed countries (Fu, 2015; Lopes et al., 2015). This has prompted increased efforts to identify new genetic resources and useful traits to mitigate or counteract the effects of climate change on crop productivity (Pignone et al., 2015; Janni et al., 2018) (Table 3).
Table 3.
Harnessing plant genetic resources for heat stress breeding
| Species | Countries of origin | Number of accessions | Source | Type of study | Reference |
|---|---|---|---|---|---|
| Barley | Spain and Germany | N/A | Saatzucht Breun GmbH | Gene expression studies | Cantalapiedra et al. (2017) |
| Chickpea | Several countries | 300 | N/A | GWAS | Thudi et al. (2014) |
| Chickpe0a | India | 280 | ICRISAT | Identification of heat tolerance superior lines | Krishnamurthy et al. (2011) |
| Maize | N/A | 100 inbred lines | N/A | Field trial | Naveed et al. (2016) |
| Rice | Several countries | 455 | N/A | QTL identification | Ye et al. (2015) |
| Rice | Several countries | 511 | IRRI | Identification of heat tolerance superior lines | Tenorio et al. (2013) |
| Tomato | Several countries | 81 | University of Naples (Italy) | GWAS | Ruggieri et al. (2019) |
| Tomato | N/A | 44 | TGRC, University of California, Davis | Identification of heat tolerance superior lines | Alsamir et al. (2017) |
| Wheat | Several countries | 1711 | CYMMIT | GWAS | Singh et al. (2018) |
| Wheat | Mexico | 2255 | CYMMIT | Identification of heat tolerance superior lines | Hede et al. (1999) |
CYMMIT, International Maize and Wheat Improvement Center; GWAS, genome-wide association study; ICRISAT, International Crops Research Institute for the Semi-Arid Tropics; IRRI, International Rice Research Institute; N/A not applicable; QTL, quantitative trait locus; TGRC, Tomato Genetics Resource Center
It is well established that responses in crop species to abiotic stress, including HS, form a complex quantitative trait whose inheritance is controlled by a synergy between genes identified as QTLs, distributed throughout the genome. Traditionally QTL mapping has been used to identify new genetic variability and new sources of tolerance for introduction to breeding programs. However, QTL regions can be quite large and may contain many genes to be investigated as potential candidate genes, and many QTL studies had limited value for breeding because of low marker density. More recently, genotyping-by-sequencing (GBS) has allowed an increase in the number of markers, in particular SNPs (single nuclear polymorphisms), evenly distributed throughout the genome (Spindel and Iwata, 2018). In this way, it is now possible to obtain genetic maps with high resolution and precise mapping of QTLs, and in some cases identifying candidate genes controlling associated quantitative traits (Bhat et al., 2016). The application of genome-wide association studies (GWAS) permits narrowing down the candidate regions to explore specific haplotypes in natural populations and even wild species (George and Cavanagh, 2015; Verdeprado et al., 2018).
In wheat, several genomic regions associated with heat tolerance have been mapped using QTL analysis, often combined with GWAS and GBS. Major QTL clusters associated with drought and heat tolerance have been mapped on several chromosomes (Vijayalakshmi et al., 2010; Paliwal et al., 2012; Talukder et al., 2014; Acuña-Galindo et al., 2015; Shirdelmoghanloo et al., 2016; Sharma et al., 2017; Maulana et al., 2018). GWAS of sorghum seedlings under HS has identified associated key genomic regions, and specific alleles from these regions can be used to develop heat-tolerant sorghum cultivars (Chopra et al., 2017). Through inheritance studies, Marfo and Hall (1992) reported two dominant genes controlling most of the heritable tolerance to heat at pod set in cowpea. Four QTLs were identified in cowpea as associated with pod set number per peduncle under HS, and markers were utilized in breeding applications (Lucas et al., 2013; Pottorff et al., 2014). Syntenic analysis of the closely related soybean genome identified HSPs and HSFs in these QTL regions (Liu et al., 2019). In rice, heat tolerance at the flowering stage is controlled by several QTLs that have been used in breeding programs based on gene pyramiding (Ye et al., 2012, 2015; Kilasi et al., 2018).
A major QTL for thermotolerance was successfully identified and cloned in African rice (Oryza glaberrima Steud.); thermo-tolerance 1 (TT1) encodes an α2 subunit of the 26S proteasome involved in the degradation of ubiquitinated proteins. The allele OgTT1 of the thermotolerant accession permitted increased thermotolerance through a more efficient elimination of cytotoxic denatured proteins and effective maintenance of heat response processes than the non-tolerant cultivar carrying the OsTT1 allele. Overexpression of OgTT1 was associated with enhanced thermotolerance in rice, Arabidopsis, and Festuca elata Keng f. ex E.B.Alexeev (X.M. Li et al., 2015).
In tomato, several QTLs involved in heat tolerance have been identified (Xu et al., 2017), while two QTL hot spots for heat tolerance with respect to grain yield were found in maize (Jha et al., 2014; Frey et al., 2016).
Screening for genotypic differences, and effective selection techniques, are crucial for identifying heat-tolerant parental sources and for inheritance studies, and also for crop improvement by combining other traits which are influenced by heat tolerance (Marfo and Hall, 1992; Hall, 2004). Since photosynthesis and reproductive development processes are directly adversely affected by HS (Prasad et al., 2008), desired characteristics of a heat-tolerant variety will be higher photosynthetic rates (e.g. stay-green leaves), enhanced membrane thermostability, and stable pod set or grain production under high temperature conditions (Bita and Gerats, 2013). Methods for breeding for heat tolerance in cowpea involved selection for abundant flower production and greater pod set under higher night temperatures and long-day growing conditions (Marfo and Hall, 1992). These efforts resulted in release of the HStolerant cowpea variety California Blackeye 27 (CB27) with better yield (Ehlers et al., 2000). This variety was crossed with Pima (a Nigerian heat-tolerant germplasm) giving Apagbaala, a cowpea variety intended for Ghana (Padi et al., 2004). However, this was not heat tolerant in Ghana even though it performed well under Californian growing conditions.
The common bean (Phaseolus vulgaris L.) has also been investigated. Initial screening at CIAT (Centro Internacional de Agricultura Tropical) of a germplasm core set for HS tolerance identified 30 resistant lines. Breeding programs utilizing these genetic resources helped to develop heat-tolerant bush (CIAT, 2006) and climbing bean lines (Blair et al., 2006). Moreover, introgression of Tepary bean (Phaseolus acutifolious A.Gray) genes into P. vulgaris gave an interspecific line that was used as a parent for breeding heat-tolerant lines (Polanía et al., 2017). Recent screening of chickpea genotypes indicated the existence of extensive genotypic variation for reproductive stage heat tolerance; further studies led to the release of heat-tolerant breeding line ICCV 92944 for late-sown conditions (Gaur et al., 2019).
Targeted breeding efforts will help to build heat tolerance in crops (Reynolds et al., 2011). A conceptual model to improve heat tolerance in wheat was proposed involving genetically determined physiological traits such as light interception, radiation use efficiency, and partitioning of total assimilates. The physiological breeding approach combines all these traits towards generating a cumulative genetic effect on yield (Cossani and Reynolds, 2012). Data sets from three different spring bread wheat nurseries at CIMMYT (International Maize and Wheat Improvement Center) with different breeding goals were extensively analyzed, and the results showed that spring wheat breeding targeted against abiotic stress delivers better genetic gains in warmer environments (Gourdji et al., 2013). The performance of yield traits under heat and non-stressed environments has been used to identify heat-tolerant genotypes for breeding programs (Ni et al., 2018; Gaur et al., 2019). The identification of critical genes controlling heat tolerance in common wheat led to the release of new cultivars of bread wheat and durum wheat capable of withstanding severe heat (Tadesse et al., 2019), for example the cultivar Faraj, which is able to maintain yield under heat and drought conditions El Hassouni et al. (2019).
It has been shown that compared with direct selection for grain yield, indirect selection through secondary traits with high heritability and significant association with grain yield under stress is a more effective approach in stress tolerance breeding (Bänziger and Lafitte, 1997; Bänziger et al., 2000; Bheemanahalli et al., 2017). In maize, traits associated with reproductive success under heat stress (anthesis–silking interval, pollen viability, stigma receptivity, tassel blast, tassel sterility, and seed set percentage under open pollinated conditions) and other morpho-physiological traits (leaf firing, senescence, and chlorophyll content) were studied along with grain yield for the selection of HS-tolerant germplasms (Alam et al., 2017). As a result, two maize genotypes, VL05728 and VL05799, with better seed setting due to reproductive success under stress were identified as tolerant lines for heat stress (Alam et al., 2017). Several maize breeding programs have successfully increased yields in HS conditions (Cairns and Prasanna, 2018). The identification of suitable donors, highly tolerant to HS and to the combination of HS and drought stress, was successfully achieved by evaluating 300 (Cairns et al., 2013) or 29 (CIMMYT) maize inbred lines (Dinesh et al., 2018). These studies are the best examples to show the significant molecular diversity among maize inbred lines selected for heat tolerance
Reproductive stage HS in rice causes substantial yield loss. Delayed flowering, reduced pollen dispersal, low pollen production, spikelet sterility due to poor anther dehiscence, and impaired starch synthesis during grain development are the major problem areas (Jagadish et al., 2016; Arshad et al., 2017). Natural genetic variation for HS tolerance exists in rice, and significant genetic components controlling these variations have been identified. Several QTLs and genes associated with HS tolerance have been reported (Ye et al., 2015;Shanmugavadivel et al., 2017; Kilasi et al., 2018), and some have been characterized (Krishnan et al., 2011), but more translational research is needed in this area. Guodao 6 and Xieyou 46 are heat-tolerant hybrids developed with stable high rates of grain setting and spikelet fertility under HS (Tao et al., 2008). Other varieties including Fusaotome (tolerant), Hanahikari, Koshijiwase, and Tentakaku (moderately tolerant) were used in breeding for grain quality under heat stress (Ishizaki, 2006). In rice, HS-tolerant traits from line N22 (Jagadish et al., 2008) have been used for introgression into other varieties for developing climate change-ready rice (Ye et al., 2015). QTLs for HS tolerance during the reproductive stage were identified using a recombinant inbred population between N22 and IR64 (Ye et al., 2012).
In tomatoes, reproduction is particularly sensitive to continuous mild heat, which mainly affects pollen viability. The Asian Vegetable Research and Development Center (AVRDC) has identified 39 tolerant lines after several years of natural genetic variation studies. Some of these were used in their tomato breeding programs which produced HS-tolerant lines, such as Equinox (Scott et al., 1995) and Sun Leaper (Gardner, 2000). Another example is the development of ‘Amelia’, a heat-tolerant tomato cultivar for tropical conditions (Gil et al., 2004). Identification of more QTLs associated with heat tolerance in tomatoes (Wen et al., 2019) could enhance breeding efforts for development of stress-tolerant tomato varieties.
The development of molecular tools to accelerate breeding is crucial for the identification of useful traits and their application in breeding programs. In particular, marker-assisted selection (MAS) to improve breeding efficiency has become commonplace. Many MAS strategies have been developed, including marker-assisted backcrossing with foreground and background selection, enrichment of favorable alleles in early generations, and selection for quantitative traits using markers at multiple loci and across multiple cycles of selection (Bassi et al., 2016). MAS requires the use of markers flanking the target locus, and is considered one of the most efficient methods when working on complex traits having a quantitative hereditary characteristic, such as HS tolerance (Tayade et al., 2018). MAS can be used in forward breeding to enrich the allelic frequency for a few desired traits with strong additive QTLs in early selection cycles, to introgress favorable alleles into an elite background, and for integration of (native) traits into a breeding pipeline. This approach has been used in maize to develop improved lines for stress-prone environments (Beyene et al., 2016).
The exploitation of GWAS has led to the approach called genomic selection (GS) a promising tool to design novel breeding programs and to develop new marker-based models for genetic evaluation. The most important factor for its successful and effective implementation in crop species is the availability of genome-wide high-throughput, cost-effective, and flexible molecular markers, having low ascertainment bias, suitable for large population sizes, as well as for both model and non-model crop species with or without the reference genome sequence (Bhat et al., 2016). As a pre-breeding tool, it can serve to identify genetic materials with beneficial variation for complex traits (Wang et al., 2018), predicting the breeding value of an individual within a breeding population. It also provides new opportunities to increase genetic gain of complex traits efficiently. GS has been applied to active breeding programs in several crop species including wheat (Song et al., 2017), rice (Spindel and Iwata, 2018), soybean (Duhnen et al., 2017; Matei et al., 2018), sunflower (Dimitrijevic and Horn, 2017), maize (Cerrudo et al., 2018), chickpea (Roorkiwal et al., 2016), and grapes (Fodor et al., 2014; Viana et al., 2016).
GS and marker-assisted recurrent selection (MARS), widely used in the private sector, are proving efficient for the development of novel cultivars in many crops (Tayade et al., 2018). The major advantage of GS with respect to MAS/MARS is that alleles with minor effects can be captured and used in selection (Cairns and Prasanna, 2018). For both approaches, success depends on excellent phenotypic characterization during the discovery or training phase, respectively.
New breeding techniques to increase heat tolerance
Genetic modification through biotechnology and other new breeding techniques (NBTs) is a powerful strategy that offers novel opportunities to improve crop adaptability. However, general public concerns and complex legislation are limiting the application of NBTs. Encouraging data collected from genetics can be exploited to significantly increase tolerance to both biotic stresses and abiotic stresses such as salinity, drought, heat, and cold (Zou et al., 2011; Raza et al., 2019). Several studies have used TF genes and other genes associated with abiotic stress tolerance as targets for development of new varieties (Lamaoui et al., 2018). Crops including wheat, maize, tomato, and rice have been genetically modified to enhance thermotolerance, targeting mainly HSPs and HSFs (Fu et al., 2008; Qi et al., 2011; Xue et al., 2015; Casaretto et al., 2016; Wang et al., 2016; Trapero-Mozos et al., 2018). Transgenic cotton plants developed to overexpress an HSP, AtHSP101, in pollen had improved pollen germination and pollen tube growth under high temperature. This significantly enhanced the overall heat tolerance of reproductive tissues and reduced yield losses due to high temperature, supported by both greenhouse and field evaluation of transgenic plants (Burke and Chen, 2015). A survey of the transgenic lines identified to enhance heat tolerance in several species is reported in Table 4.
Table 4.
List of selected heat stress- (HS) tolerant transgenic plants
| Crop | Target gene or protein/sources | Promoter | Stress or trait | Reference |
|---|---|---|---|---|
| Maize | OsMYB55/rice | Maize ubiquitin Ubi1 promoter/overexpression | Increased drought and HS tolerance | Casaretto et al. (2016) |
| Maize | ZmNF-YB2/maize | Rice actin 1 constitutive promoter/overexpression | Enhanced drought tolerance and photosynthetic capacity | Nelson et al. (2007) |
| Potato | HSc70 allelic variant/ potato | HSc70 native promoter | Greater tolerance to HS as determined by improved yield | Trapero-Mozos et al. (2018) |
| Rice | HSP70/rice | CaMV 35S | Overexpression manifested enhanced tolerance to HS | Qi et al. (2011) |
| Rice | Athsp101 /Arabidopsis | CaMV 35S promoter | Increased tolerance to high temperature | Katiyar-Agarwal et al. (2003) |
| Rice | OsRab7 | CaMV 35S | Greater tolerance to HS as determined by improved yield | El-Esawi and Alayafi (2019) |
| Rice | TaMBF1c/wheat | Maize ubiquitin 1 | Higher thermotolerance than control plants at both seedling and reproductive stages | Qin et al. (2015) |
| Soybean | P5CR/Arabidopsis | Enhanced HS tolerance | De Ronde et al. (2004) | |
| Tobacco (Nicotiana tabacum L.) | HSP70-1/tobacco | CaMV 35S | Transgenics possessed enhanced tolerance to HT stress | Montero-Barrientos et al. (2008) |
| Tobacco | HSP70-1/brassica | Enhanced tolerance to HT stress | Wang et al. (2016) | |
| Tomato | HSP21/tomato | CaMV 35S | Overexpression protected PSII from temperature-dependent oxidative stress; early accumulation of carotenoids noted | Neta-Sharir et al. (2005) |
| Wheat | Hsf6A /wheat | Barley HVA1s promoter/drought inducible, up-regulated | Improved thermotolerance | Xue et al. (2014) |
| Wheat | EF-Tu/maize | Maize ubiquitin 1 promoter/overexpression | Improved thermotolerance | Fu et al. (2008) |
| Wheat | Sucrose transporter gene HvSUT1/barley | Hordein B1 promoter | Increased enhanced in sucrose transport and shows a superior performance for many yield-related traits compared with control | Weichert et al. (2017) |
| Wheat | TaHsfA6f/wheat | HVA1s | Improved thermotolerance | Xue et al. (2015) |
| Wheat | TaFER-5B/wheat | Maize ubiquitin 1 | Enhance thermotolerance | Zang et al. (2017) |
| Wheat | TaGASR1/wheat | NA | Improved tolerance to HS and oxidative stress | Zhang et al. (2017) |
| Wheat | TaHsfC2a/wheat | NA | Improved thermotolerance | Hu et al. (2018) |
The availability of genomic sequences for several crops together with genome editing techniques has opened up new breeding possibilities for almost any given desirable trait (Jaganathan et al., 2018). The most common technique available for genome editing, clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), modifies a genome in a targeted manner, and has been used in nearly 20 crop species so far including rice, tomato, potato, cotton, soybean, maize, sorghum, and wheat (Table 5) (Ricroch et al., 2017; Jaganathan et al., 2018; Zaidi et al., 2018). There are at least 13 different patents on new CRISPR/Cas9 approaches, which makes editing even more simple and secure, and new perspectives in the production of novel genetic variability are opened up by its application as an allelic-drive tool, engineering and repairing pathways, and introducing specific point mutations or insertions (Guichard et al., 2019).
Table 5.
Genome editing approaches used for heat stress breeding
| Crop | Target gene | Stress or trait | Reference |
|---|---|---|---|
| Maize | ARGOS8 | Improved yield under drought stress condition | Shi et al. (2017a) |
| Rice | OsPDS, OsMPK2, OsBADH2 | Abiotic stress tolerance | Shan et al. (2013) |
| Rice | OsMPK2, OsDEP1 | Yield under stress | Shan et al. (2014) |
| Rice | GS3, Gn1a | Grain size and number increase | Shen et al. (2017) |
| Rice | GW2, GW5, TGW6 | Grain weight increase | Xu et al. (2016) |
| Rice | Gn1a, DEP1, GS3 | Grain size and number Increase in dense, erect panicles | Li et al. (2016) |
| Wheat | TaDREB2, TaERF3 | Abiotic stress tolerance | Kim et al. (2018) |
Currently, attempts exploiting genome editing to increase heat tolerance target several genes mainly involved in the ethylene response and TFs, with the final aim to increase yield under abiotic stresses including HS (Li et al., 2016; Shi et al., 2017a; Kim et al., 2018). However, despite the abundance of novel genetic material generated through CRISPR, few field experiments have been conducted associated with breeding programs.
Mutational breeding
Mutational breeding strategies have been applied since the late 1990s to generate new variability in plants (Gilliham et al., 2017; Uauy, 2017). An important breakthrough came with the development of the TILLING (targeting induced local lesions in genome) approach, which provides a relatively simple strategy to identify mutations (lesions) in a target sequence independently of their phenotypic effect (Uauy, 2017). TILLING approaches require a population of, typically, ethyl methanesulfonate- (EMS) induced mutants based on the capacity of the chemical agent to generate point mutations distributed randomly in the genome, and a screening method to identify individuals with mutations in the target gene (Wang et al., 2010). TILLING and its updated form, established for polyploidy species through the use of exome capture and the development of the in silico TILLING database (Krasileva et al., 2017), have been used extensively to investigate genetic variability in several species mainly targeting quality traits.
Although in TILLING approaches mutagenesis is untargeted and does not provide the versatility of genome editing, crops improved using chemical or radiation mutagenesis via TILLING are not regulated as transgenic organisms in most jurisdictions, increasing their commercial competitiveness with the more precise genome editing approaches (Kumar et al., 2017).
Recently, Comastri et al. (2018) identified a collection of four new small Hsp26 (sHsp26) alleles suitable for enhancing heat tolerance in durum wheat using both in silico and in vivo TILLING approaches. Following application of TILLING, the ability of a mutated HSP to enhance heat tolerance in tomato has also been demonstrated (Marko et al., 2019). In rice, a TILLING population has been screened for mutations in the HSP genes, and a number of lines showing preliminary enhanced tolerance to HS may be useful for future breeding programs (Yona, 2015; Table 6).
Table 6.
Mutational breeding in crops
| Species | Target trait /genes | Reference |
|---|---|---|
| Barley | Starch increases | Sparla et al. (2014) |
| Bread wheat (Triticum aestivum L.) | Starch branching enzyme | Botticella et al. (2011) |
| Chickpea | Salt tolerance | Kaashyap et al. (2017) |
| Durum wheat (Triticum durum Desf) | Heat stress | Comastri et al., 2018 |
| Durum wheat (Triticum durum Desf) | Amylose content | Sestili et al. (2015) |
| Durum wheat(Triticum durum Desf), bread wheat (Triticum aestivum L.) | High amylose | Slade et al. (2012) |
| Oat (Avena sativa L.) | Improved β-glucan, antioxidants and omega-3 fatty acid | Chawade et al. (2010) |
| Peanut | Allergen reduction | Knoll et al. (2011) |
| Peanut | LOX gene | Guo et al. (2015) |
| Potato | Waxy mutant | Muth et al. (2008) |
| Rapeseed | Erucic acid synthesis | Wang et al. (2008) |
| Rapeseed | Sinapine biosynthesis | Harloff et al. (2012) |
| Sorghum | Reduction of cyanogenic glucosides | Blomstedt et al. (2012) |
| Sunflower | Accumulation of fatty acids | Kumar et al. (2013) |
| Tobacco | Leaf yield | Reddy et al. (2012) |
| Tomato | Fruit biology and ripening | Okabe et al. (2011, 2013) |
| Tomato | Virus resistance | Piron et al. (2010) |
| Tomato | Increased pigment and nutrient content | Jones et al. (2012) |
| Tomato | Early flowering, a solitary flower | MacAlister et al. (2012) |
| Tomato | Reduced ethylene sensitivity, delayed fruit ripening, prolonged fruit shelf life | Okabe et al. (2011) |
| Tomato | Decreased ascorbate | Baldet et al. (2013) |
| Tomato | Decreased carotenoid content | Gady et al. (2012) |
| Tomato | Increased lycopene content | Silletti et al. (2013) |
| Tomato | Reduced phenolics content | Di Matteo et al. (2013) |
Conclusions
There are several examples of heat-tolerant varieties in major crops such as wheat, maize, rice, tomato, and legumes successfully developed through conventional breeding. These efforts can be fast-tracked using modern genomic and phenotyping tools. The first HS gene was identified and cloned in tobacco with the pioneering work of Barnett et al. (1979). This gene was shown to encode HSP70, which strongly accumulated in heat-stressed tissue.
Subsequent molecular studies revealed a more complex system with the discovery of HSFs (reviewed in Guo et al., 2016), organelle-specific HSPs (Waters and Vierling, 1999; Waters, 2013), and linkage between HS and the induction of an alternative splicing system (Ling et al., 2018) capable of directing de novo HSP synthesis in conditions where most proteins were either produced abnormally or were degraded (Vierling, 1991), remodeling the global protein machinery in cells. However, neither these discoveries nor the networking of all the known elements have produced a satisfying picture of the plant heat stress response. QTLs for heat tolerance offer a different, potentially complementary, interpretation with a stronger emphasis on physiology of reproduction in heat stress conditions.
Current data indicate that at least two different genetic systems can protect plants from the otherwise lethal effects of heat stress: (i) a series of Mendelian HS genes acting possibly in a dominant or semi-dominant way; and (ii) a small number of QTLs which allow plant growth and reproduction under different stress conditions. There has been little progress using transgenic approaches in studies of model plant to crop plant translational genetics. Extensive knowledge has become available on physiological, biochemical, and molecular regulation through advanced phenotyping and multi-omics tools developed over the last decade. All of this suggests that significant advances in crop improvement will be achieved, resulting in the development of new high-yielding varieties with greater heat tolerance and adaptation to climate change.
Author contributions
The authors contributed equally to the preparation of the review. MJ took on the responsibility of connecting all the different tasks distributed as follows: EM, gene ontology and system biology; MM, protein and metabolites; MG, gene expression and epigenetic modification; BV, gene expression and breeding; and MJ, induction of HS response; NM and HTN coordinated the planning and the writing of the review.
Acknowledgements
The authors wish to thank Consortium CINSA for the financial support for the preparation of this review. This review is dedicated to the pioneers of HS response research in plants: Professor Joseph P. Mascarenhas, Department of Biological Sciences, State University of New York at Albany, New York and Joe L. Key, Department of Botany, University of Georgia Athens (GA). The authors also recognize the dedication of Dr Natale Di Fonzo, Former Director of CREA Foggia (Italy), breeder, researcher, and friend. The authors wish to acknowledge the support of the project SUSTAINOLIVE, grant agreement no. 1811, PRIMA Partnership for Research and innovation in the Mediterranean Area and RGV FAO DM 10271.
References
- Acuña-Galindo MA, Mason RE, Subramanian NK, Hays DB. 2015. Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Science 55, 477–492. [Google Scholar]
- Alam MA, Seetharam K, Zaidi PH, Dinesh A, Vinayan MT, Nath UK. 2017. Dissecting heat stress tolerance in tropical maize (Zea mays L.). Field Crops Research 204, 110–119. [Google Scholar]
- Almeselmani M, Deshmukh P, Sairam R. 2009. High temperature stress tolerance in wheat genotypes: role of antioxidant defence enzymes. Acta Agronomica Hungarica 57, 1–14. [Google Scholar]
- Almeselmani M, Deshmukh PS, Sairam RK, Kushwaha SR, Singh TP. 2006. Protective role of antioxidant enzymes under high temperature stress. Plant Science 171, 382–388. [DOI] [PubMed] [Google Scholar]
- Alptekin B, Langridge P, Budak H. 2017. Abiotic stress miRNomes in the Triticeae. Functional & Integrative Genomics 17, 145–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsamir M, Ahmad NM, Keitel C, Mahmood T, Trethowan R. 2017. Identification of high-temperature tolerant and agronomically viable tomato (S. lycopersicum) genotypes from a diverse germplasm collection. Advances in Crop Science and Technology 5, 1000299. [Google Scholar]
- Altschuler M, Mascarenhas JP. 1982. Heat shock proteins and effects of heat shock in plants. Plant Molecular Biology 1, 103–115. [DOI] [PubMed] [Google Scholar]
- Arce D, Spetale F, Krsticevic F, Cacchiarelli P, Las Rivas J, Ponce S, Pratta G, Tapia E. 2018. Regulatory motifs found in the small heat shock protein (sHSP) gene family in tomato. BMC Genomics 19, 860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit S, Xia R, Kakrana A, et al. . 2014. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. The Plant Cell 26, 4584–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad MS, Farooq M, Asch F, Krishna JSV, Prasad PVV, Siddique KHM. 2017. Thermal stress impacts reproductive development and grain yield in rice. Plant Physiology and Biochemistry 115, 57–72. [DOI] [PubMed] [Google Scholar]
- Asthir B, Koundal A, Bains NS. 2012. Putrescine modulates antioxidant defense response in wheat under high temperature stress. Biologia Plantarum 56, 757–761. [Google Scholar]
- Baldet P, Bres C, Okabe Y, Mauxion J-P, Just D, Bournonville C, Ferrand C, Mori K, Ezura H, Rothan C. 2013. Investigating the role of vitamin C in tomato through TILLING identification of ascorbate-deficient tomato mutants. Plant Biotechnology 30, 309–314. [Google Scholar]
- Bänziger M, Edmeades GO, Beck DL, Bellon MR. 2000. Breeding for drought and nitrogen stress tolerance in maize: from theory to practice. Mexico: CIMMYT. [Google Scholar]
- Bänziger M, Lafitte HR. 1997. Efficiency of secondary traits for improving maize for low-nitrogen target environments. Crop Science 37, 1110–1117. [Google Scholar]
- Barghi SS, Mostafaii H, Peighami F, Zakaria RA. 2012. Path analysis of yield and its components in lentil under end season heat condition. International Journal of Agriculture: Research and Review 2, 969–974. [Google Scholar]
- Barnabás B, Jäger K, Fehér A. 2008. The effect of drought and heat stress on reproductive processes in cereals. Plant, Cell & Environment 31, 11–38. [DOI] [PubMed] [Google Scholar]
- Barnett T, Altschuler M, McDaniel CN, Mascarenhas JP. 1979. Heat shock induced proteins in plant cells. Genesis 1, 331–340. [Google Scholar]
- Bassi FM, Bentley AR, Charmet G, Ortiz R, Crossa J. 2016. Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Science 242, 23–36. [DOI] [PubMed] [Google Scholar]
- Berz J, Simm S, Schuster S, Scharf KD, Schleiff E, Ebersberger I. 2019. HEATSTER: a database and web server for identification and classification of heat stress transcription factors in plants. Bioinformatics and Biology Insights 13, 1177932218821365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene Y, Semagn K, Mugo S, et al. . 2016. Performance and grain yield stability of maize populations developed using marker-assisted recurrent selection and pedigree selection procedures. Euphytica 208, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat JA, Ali S, Salgotra RK, et al. . 2016. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Frontiers in Genetics 7, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S. 2012. An inductive pulse of hydrogen peroxide pretreatment restores redox-homeostasis and oxidative membrane damage under extremes of temperature in two rice cultivars. Plant Growth Regulation 68, 395–410. [Google Scholar]
- Bhattacharya A. 2019. Effect of high temperature on crop productivity and metabolism of macro molecules. London: Academic Press. [Google Scholar]
- Bheemanahalli R, Sathishraj R, Manoharan M, Sumanth HN, Muthurajan R, Ishimaru T, Krishna JS. 2017. Is early morning flowering an effective trait to minimize heat stress damage during flowering in rice? Field Crops Research 203, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bita CE, Gerats T. 2013. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Frontiers in Plant Science 4, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bita CE, Zenoni S, Vriezen WH, Mariani C, Pezzotti M, Gerats T. 2011. Temperature stress differentially modulates transcription in meiotic anthers of heat-tolerant and heat-sensitive tomato plants. BMC Genomics 12, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair MW, Iriarte G, Beebe S. 2006. QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean × wild common bean (Phaseolus vulgaris L.) cross. Theoretical and Applied Genetics 112, 1149–1163. [DOI] [PubMed] [Google Scholar]
- Blomstedt CK, Gleadow RM, O’Donnell N, et al. . 2012. A combined biochemical screen and TILLING approach identifies mutations in Sorghum bicolor L. Moench resulting in acyanogenic forage production. Plant Biotechnology Journal 10, 54–66. [DOI] [PubMed] [Google Scholar]
- Botella C, Sautron E, Boudiere L, Michaud M, Dubots E, Yamaryo-Botté Y, Albrieux C, Marechal E, Block MA, Jouhet J. 2016. ALA10, a phospholipid flippase, controls FAD2/FAD3 desaturation of phosphatidylcholine in the ER and affects chloroplast lipid composition in Arabidopsis thaliana. Plant Physiology 170, 1300–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botticella E, Sestili F, Hernandez-Lopez A, Phillips A, Lafiandra D. 2011. High resolution melting analysis for the detection of EMS induced mutations in wheat Sbella genes. BMC Plant Biology 11, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak H, Akpinar BA. 2015. Plant miRNAs: biogenesis, organization and origins. Functional & Integrative Genomics 15, 523–531. [DOI] [PubMed] [Google Scholar]
- Burke JJ, Chen J. 2015. Enhancement of reproductive heat tolerance in plants. PLoS One 10, e0122933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns JE, Crossa J, Zaidi PH, et al. . 2013. Identification of drought, heat, and combined drought and heat tolerant donors in maize. Crop Science 53, 1335. [Google Scholar]
- Cairns JE, Prasanna BM. 2018. Developing and deploying climate-resilient maize varieties in the developing world. Current Opinion in Plant Biology 45, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalapiedra CP, García-Pereira MJ, Gracia MP, Igartua E, Casas AM, Contreras-Moreira B. 2017. Large differences in gene expression responses to drought and heat stress between elite barley cultivar Scarlett and a Spanish landrace. Frontiers in Plant Science 8, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaretto JA, El-Kereamy A, Zeng B, Stiegelmeyer SM, Chen X, Bi YM, Rothstein SJ. 2016. Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 17, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo D, Cao S, Yuan Y, Martinez C, Suarez EA, Babu R, Zhang X, Trachsel S. 2018. Genomic selection outperforms marker assisted selection for grain yield and physiological traits in a maize doubled haploid population across water treatments. Frontiers in Plant Science 9, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K, Bishi SK, Singh AL, Zala PV, Mahatma MK, Kalariya KA, Jat RA. 2018. Rapid induction of small heat shock proteins improves physiological adaptation to high temperature stress in peanut. Journal of Agronomy and Crop Science 204, 285–297. [Google Scholar]
- Chakraborty S, Newton AC. 2011. Climate change, plant diseases and food security: an overview. Plant Pathology 60, 2–14. [Google Scholar]
- Chakraborty U, Pradhan D. 2011. High temperature-induced oxidative stress in Lens culinaris, role of antioxidants and amelioration of stress by chemical pre-treatments. Journal of Plant Interactions 6, 43–52. [Google Scholar]
- Chao Y-Y, Hsu YT, Kao CH. 2008. Involvement of glutathione in heat shock- and hydrogen peroxide-induced cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant and Soil 318, 37. [Google Scholar]
- Chaturvedi P, Doerfler H, Jegadeesan S, Ghatak A, Pressman E, Castillejo MA, Wienkoop S, Egelhofer V, Firon N, Weckwerth W. 2015. Heat-treatment-responsive proteins in different developmental stages of tomato pollen detected by targeted mass accuracy precursor alignment (tMAPA). Journal of Proteome Research 14, 4463–4471. [DOI] [PubMed] [Google Scholar]
- Chawade A, Sikora P, Bräutigam M, Larsson M, Vivekanand V, Nakash MA, Chen T, Olsson O. 2010. Development and characterization of an oat TILLING-population and identification of mutations in lignin and β-glucan biosynthesis genes. BMC Plant Biology 10, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Feder ME, Kang L. 2018. Evolution of heat-shock protein expression underlying adaptive responses to environmental stress. Molecular Ecology 27, 3040–3054. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang W, Zhang B, Zhou J, Wang Y, Yang Q, Ke Y, He H. 2011. Phosphoproteins regulated by heat stress in rice leaves. Proteome Science 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R. 2018. How climate change will alter our food. State of the Planet. New York: Columbia University Press. [Google Scholar]
- Chopra R, Burow G, Burke JJ, Gladman N, Xin Z. 2017. Genome-wide association analysis of seedling traits in diverse Sorghum germplasm under thermal stress. BMC Plant Biology 17, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TS, Chao YY, Kao CH. 2012. Involvement of hydrogen peroxide in heat shock- and cadmium-induced expression of ascorbate peroxidase and glutathione reductase in leaves of rice seedlings. Journal of Plant Physiology 169, 478–486. [DOI] [PubMed] [Google Scholar]
- Chung E, Kim KM, Lee JH. 2013. Genome-wide analysis and molecular characterization of heat shock transcription factor family in Glycine max. Journal of Genetics and Genomics 40, 127–135. [DOI] [PubMed] [Google Scholar]
- CIAT 2006. https://cgspace.cgiar.org/bitstream/handle/10568/73453/CIAT_Annual_Report_2015-2016_Synthesis.pdf?sequence=6 Annual Report of the International Center for Tropical Agriculture (CIAT)
- Cohen-Peer R, Schuster S, Meiri D, Breiman A, Avni A. 2010. Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Molecular Biology 74, 33–45. [DOI] [PubMed] [Google Scholar]
- Colignon B, Delaive E, Dieu M, Demazy C, Muhovski Y, Antoine A, Raes M, Mauro S. 2019. Dual coordination of the SUMOylation and phosphorylation pathways during the response to heat stress in Solanum tuberosum. Environmental and Experimental Botany 162, 192–200. [Google Scholar]
- Comastri A, Janni M, Simmonds J, Uauy C, Pignone D, Nguyen HT, Marmiroli N. 2018. Heat in wheat: exploit reverse genetic techniques to discover new alleles within the Triticum durum sHsp26 family. Frontiers in Plant Science 9, 1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossani CM, Reynolds MP. 2012. Physiological traits for improving heat tolerance in wheat. Plant Physiology 160, 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Krishnan P, Mishra V, Kumar R, Ramakrishnan B, Singh NK. 2015. Proteomic changes in rice leaves grown under open field high temperature stress conditions. Molecular Biology Reports 42, 1545–1558. [DOI] [PubMed] [Google Scholar]
- De Ronde JA, Cress WA, Krüger GHJ, Strasser RJ, Van Staden J. 2004. Photosynthetic response of transgenic soybean plants, containing an Arabidopsis P5CR gene, during heat and drought stress. Journal of Plant Physiology 161, 1211–1224. [DOI] [PubMed] [Google Scholar]
- Debaeke P, Casadebaig P, Flenet F, Langlade N. 2017. Sunflower crop and climate change: vulnerability, adaptation, and mitigation potential from case-studies in Europe. OCL; 24, D102. [Google Scholar]
- Devasirvatham V, Tan D. 2018. Impact of high temperature and drought stresses on chickpea production. Agronomy 8, 145. [Google Scholar]
- Di Matteo A, Ruggieri V, Sacco A, Rigano MM, Carriero F, Bolger A, Fernie AR, Frusciante L, Barone A. 2013. Identification of candidate genes for phenolics accumulation in tomato fruit. Plant Science 205–206, 87–96. [DOI] [PubMed] [Google Scholar]
- Dimitrijevic A, Horn R. 2017. Sunflower hybrid breeding: from markers to genomic selection. Frontiers in Plant Science 8, 2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh A, Patil A, Zaidi PH, Kuchanur PH, Vinayan MT, Seetharam K. 2018. Genetic diversity, linkage disequilibrium and population structure among CIMMYT maize inbred lines, selected for heat tolerance study. Maydica 61, 7. [Google Scholar]
- Djanaguiraman M, Boyle DL, Welti R, Jagadish SVK, Prasad PVV. 2018. Decreased photosynthetic rate under high temperature in wheat is due to lipid desaturation, oxidation, acylation, and damage of organelles. BMC Plant Biology 18, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Yi H, Lee J, Nou IS, Han CT, Hur Y. 2015. Global gene-expression analysis to identify differentially expressed genes critical for the heat stress response in Brassica rapa. PLoS One 10, e0130451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driedonks N, Rieu I, Vriezen WH. 2016. Breeding for plant heat tolerance at vegetative and reproductive stages. Plant Reproduction 29, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhnen A, Gras A, Teyssèdre S, Romestant M, Claustres B, Daydé J, Mangin B. 2017. Genomic selection for yield and seed protein content in soybean: a study of breeding program data and assessment of prediction accuracy. Crop Science 57, 1325. [Google Scholar]
- El-Esawi MA, Alayafi AA. 2019. Overexpression of rice Rab7 gene improves drought and heat tolerance and increases grain yield in rice (Oryza sativa L.). Genes 10, E56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferjani R, Soolanayakanahally R. 2018. Canola responses to drought, heat, and combined stress: shared and specific effects on carbon assimilation, seed yield, and oil composition. Frontiers in Plant Science 9, 1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers JD, Hall AE, Patel PN, Roberts PA, Matthews WC. 2000. Registration of ‘California Blackeye 27’ cowpea. Crop Science 40, 854–855. [Google Scholar]
- El Hassouni K, Belkadi B, Filali-Maltouf A, Tidiane-Sall A, Al-Abdallat A, Nachit M, Bassi FM. 2019. Loci controlling adaptation to heat stress occurring at the reproductive stage in durum wheat. Agromomy 9, 414. [Google Scholar]
- Escandón M, Meijón M, Valledor L, Pascual J, Pinto G, Cañal MJ. 2018. Metabolome integrated analysis of high-temperature response in Pinus radiata. Frontiers in Plant Science 9, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahad S, Bajwa AA, Nazir U, et al. . 2017. Crop production under drought and heat stress: plant responses and management options. Frontiers in Plant Science 8, 1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Dou L, Liu Y, Yu J, Tu J. 2018. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant rice by high-throughput sequencing. Ecological Genetics and Genomics 6, 33–40. [Google Scholar]
- Feng B, Zhang C, Chen T, Zhang X, Tao L, Fu G. 2018. Salicylic acid reverses pollen abortion of rice caused by heat stress. BMC Plant Biology 18, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor A, Segura V, Denis M, Neuenschwander S, Fournier-Level A, Chatelet P, Homa FA, Lacombe T, This P, Le Cunff L. 2014. Genome-wide prediction methods in highly diverse and heterozygous species: proof-of-concept through simulation in grapevine. PLoS One 9, e110436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S, Simm S, Paul P, Bublak D, Scharf KD, Schleiff E. 2015. Chaperone network composition in Solanum lycopersicum explored by transcriptome profiling and microarray meta-analysis. Plant, Cell & Environment 38, 693–709. [DOI] [PubMed] [Google Scholar]
- Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, Shen S, Firon N. 2009. Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. Journal of Experimental Botany 60, 3891–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey FP, Presterl T, Lecoq P, Orlik A, Stich B. 2016. First steps to understand heat tolerance of temperate maize at adult stage: identification of QTL across multiple environments with connected segregating populations. Theoretical and Applied Genetics 129, 945–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey FP, Urbany C, Hüttel B, Reinhardt R, Stich B. 2015. Genome-wide expression profiling and phenotypic evaluation of European maize inbreds at seedling stage in response to heat stress. BMC Genomics 16, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T, Faivre L, Bäurle I, Schubert D. 2019. Chromatin-based mechanisms of temperature memory in plants. Plant, Cell & Environment 42, 762–770. [DOI] [PubMed] [Google Scholar]
- Fu J, Momcilović I, Clemente TE, Nersesian N, Trick HN, Ristic Z. 2008. Heterologous expression of a plastid EF-Tu reduces protein thermal aggregation and enhances CO2 fixation in wheat (Triticum aestivum) following heat stress. Plant Molecular Biology 68, 277–288. [DOI] [PubMed] [Google Scholar]
- Fu YB. 2015. Understanding crop genetic diversity under modern plant breeding. Theoretical and Applied Genetics 128, 2131–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gady ALF, Vriezen WH, Van de Wal MHBJ, Huang P, Bovy AG, Visser RGF, Bachem CWB. 2012. Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Molecular Breeding 29, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlaut V, Baranwal VK, Khurana P. 2018. miRNomes involved in imparting thermotolerance to crop plants. 3 Biotech 8, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammulla CG, Pascovici D, Atwell BJ, Haynes PA. 2010. Differential metabolic response of cultured rice (Oryza sativa) cells exposed to high- and low-temperature stress. Proteomics 10, 3001–3019. [DOI] [PubMed] [Google Scholar]
- Gardner RG. 2000. ‘Sun Leaper’, a hybrid tomato, and its parent, NC HS-1. HortScience 35, 960–961. [Google Scholar]
- Gaur PM, Samineni S, Thudi M, et al. . 2019. Integrated breeding approaches for improving drought and heat adaptation in chickpea (Cicer arietinum L.). Plant Breeding 138, 389–400. [Google Scholar]
- George AW, Cavanagh C. 2015. Genome-wide association mapping in plants. Theoretical and Applied Genetics 128, 1163–1174. [DOI] [PubMed] [Google Scholar]
- Gil MA, López CM, Cuadra MED, Sánchez JA, Coca BM, Martínez SP, Zueco JC. 2004. ‘Amalia’: a medium-fruit-size, heat-tolerant tomato cultivar for tropical conditions. HortScience 39, 1503–1504. [Google Scholar]
- Gilliham M, Able JA, Roy SJ. 2017. Translating knowledge about abiotic stress tolerance to breeding programmes. The Plant Journal 90, 898–917. [DOI] [PubMed] [Google Scholar]
- Gillman JD, Biever JJ, Ye S, Spollen WG, Givan SA, Lyu Z, Joshi T, Smith JR, Fritschi FB. 2019. A seed germination transcriptomic study contrasting two soybean genotypes that differ in terms of their tolerance to the deleterious impacts of elevated temperatures during seed fill. BMC Research Notes 12, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzberg I, Barel G, Ophir R, Tzin E, Tanami Z, Muddarangappa T, de Jong W, Fogelman E. 2009. Transcriptomic profiling of heat-stress response in potato periderm. Journal of Experimental Botany 60, 4411–4421. [DOI] [PubMed] [Google Scholar]
- González-Schain N, Dreni L, Lawas LM, Galbiati M, Colombo L, Heuer S, Jagadish KS, Kater MM. 2016. Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant & Cell Physiology 57, 57–68. [DOI] [PubMed] [Google Scholar]
- Gourdji SM, Mathews KL, Reynolds M, Crossa J, Lobell DB. 2013. An assessment of wheat yield sensitivity and breeding gains in hot environments. Proceedings of the Royal Society B: Biological Sciences 280, 20122190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraj M, Pattanashetti SK, Patne N, Kanatti AA. 2018. Breeding cultivars for heat stress tolerance in staple food crops. In: Çiftçi YÖ, ed. Next generation plant breeding. InTechOpen. [Google Scholar]
- Greer DH, Weedon MM. 2013. The impact of high temperatures on Vitis vinifera cv. Semillon grapevine performance and berry ripening. Frontiers in Plant Science 4, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard A, Haque T, Bobik M, Xu XS, Klanseck C, Kushwah RBS, Berni M, Kaduskar B, Gantz VM, Bier E. 2019. Efficient allelic-drive in Drosophila. Nature Communications 10, 1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Abernathy B, Zeng Y, Ozias-Akins P. 2015. TILLING by sequencing to identify induced mutations in stress resistance genes of peanut (Arachis hypogaea). BMC Genomics 16, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Liu JH, Ma X, Luo DX, Gong ZH, Lu MH. 2016. The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Frontiers in Plant Science 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta OP, Mishra V, Singh NK, Tiwari R, Sharma P, Gupta RK, Sharma I. 2015. Deciphering the dynamics of changing proteins of tolerant and intolerant wheat seedlings subjected to heat stress. Molecular Biology Reports 42, 43–51. [DOI] [PubMed] [Google Scholar]
- Hall AE. 2004. Breeding for adaptation to drought and heat in cowpea. European Journal of Agronomy 21, 447–454. [Google Scholar]
- Han F, Chen H, Li X-J, Yang M-F, Liu G-S, Shen S-H. 2009. A comparative proteomic analysis of rice seedlings under various high-temperature stresses. Biochimica et Biophysica Acta 1794, 1625–1634. [DOI] [PubMed] [Google Scholar]
- Hancock RD, Morris WL, Ducreux LJM, et al. . 2014. Physiological, biochemical and molecular responses of the potato (Solanum tuberosum L.) plant to moderately elevated temperature: systems biology of heat stress in potato. Plant, Cell & Environment 37, 439–450. [DOI] [PubMed] [Google Scholar]
- Harloff H-J, Lemcke S, Mittasch J, Frolov A, Wu JG, Dreyer F, Leckband G, Jung C. 2012. A mutation screening platform for rapeseed (Brassica napus L.) and the detection of sinapine biosynthesis mutants. Theoretical and Applied Genetics 124, 957–969. [DOI] [PubMed] [Google Scholar]
- Hasanuzzaman M, Nahar K, Alam MM, Roychowdhury R, Fujita M. 2013. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. International Journal of Molecular Sciences 14, 9643–9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hede AR, Skovmand B, Reynolds MP, Crossa J, Vilhelmsen AL, Stølen O. 1999. Evaluating genetic diversity for heat tolerance traits in Mexican wheat landraces. Genetic Resources and Crop Evolution 46, 37–45. [Google Scholar]
- Hewezi T, Léger M, Gentzbittel L. 2008. A comprehensive analysis of the combined effects of high light and high temperature stresses on gene expression in sunflower. Annals of Botany 102, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Okazaki Y, Myouga F, Shinozaki K, Saito K. 2015. Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Scientific Reports 5, 10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Okazaki Y, Takano K, Myouga F, Shinozaki K, Knoch E, Fukushima A, Saito K. 2018. HEAT INDUCIBLE LIPASE1 remodels chloroplastic monogalactosyldiacylglycerol by liberating α-linolenic acid in Arabidopsis leaves under heat stress. The Plant Cell 30, 1887–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Saito K. 2019. Lipidomic studies of membrane glycerolipids in plant leaves under heat stress. Progress in Lipid Research 75, 100990. [DOI] [PubMed] [Google Scholar]
- Hu X-J, Chen D, Lynne Mclntyre C, Fernanda Dreccer M, Zhang Z-B, Drenth J, Kalaipandian S, Chang H, Xue G-P. 2018. Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant, Cell & Environment; 41, 79–98. [DOI] [PubMed] [Google Scholar]
- Hu X, Wu L, Zhao F, Zhang D, Li N, Zhu G, Li C, Wang W. 2015. Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Frontiers in Plant Science 6, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman WJ, McCue KF, Altenbach SB, et al. . 2003. Effect of temperature on expression of genes encoding enzymes for starch biosynthesis in developing wheat endosperm. Plant Science 164, 873–881. [Google Scholar]
- Hurkman WJ, Tanaka CK, Vensel WH, Thilmony R, Altenbach SB. 2013. Comparative proteomic analysis of the effect of temperature and fertilizer on gliadin and glutenin accumulation in the developing endosperm and flour from Triticum aestivum L. cv. Butte 86. Proteome Science 11, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihsan MZ, Daur I, Alghabari F, Alzamanan S, Rizwan S, Ahmad M, Waqas M, Shafqat W. 2019. Heat stress and plant development: role of sulphur metabolites and management strategies. Acta Agriculturae Scandinavica, Section B—Soil & Plant Science 69, 332–342. [Google Scholar]
- Ishizaki K. 2006. Evaluation of various screening systems for high grain quality in rice cultivars under high-temperature grain-filling conditions, and the selection of their standard cultivars. Japanese Journal of Crop Science 75, 502–506. [Google Scholar]
- Jagadeeswaran G, Saini A, Sunkar R. 2009. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229, 1009–1014. [DOI] [PubMed] [Google Scholar]
- Jagadish SV, Bahuguna RN, Djanaguiraman M, Gamuyao R, Prasad PV, Craufurd PQ. 2016. Implications of high temperature and elevated CO2 on flowering time in plants. Frontiers in Plant Science 7, 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadish SVK, Craufurd PQ, Wheeler TR. 2008. Phenotyping parents of mapping populations of rice for heat tolerance during anthesis. Crop Science 48, 1140–1146. [Google Scholar]
- Jagadish SVK, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ. 2010. Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). Journal of Experimental Botany 61, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaganathan D, Ramasamy K, Sellamuthu G, Jayabalan S, Venkataraman G. 2018. CRISPR for crop improvement: an update review. Frontiers in Plant Science 9, 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangid KK, Dwivedi P. 2016. Physiological responses of drought stress in tomato: a review. International Journal of Agriculture, Environment and Biotechnology 9, 53. [Google Scholar]
- Janni M, Cadonici S, Bonas U, Grasso A, Dahab AAD, Visioli G, Pignone D, Ceriotti A, Marmiroli N. 2018. Gene-ecology of durum wheat HMW glutenin reflects their diffusion from the center of origin. Scientific Reports 8, 16929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen D, Roth C, Wiermer M, Fulda M. 2015. Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiology 167, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha UC, Bohra A, Singh NP. 2014. Heat stress in crop plants: its nature, impacts and integrated breeding strategies to improve heat tolerance. Plant Breeding 133, 679–701. [Google Scholar]
- Jiang J, Liu X, Liu C, Liu G, Li S, Wang L. 2017. Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiology 173, 1502–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin GH, Gho HJ, Jung KH. 2013. A systematic view of rice heat shock transcription factor family using phylogenomic analysis. Journal of Plant Physiology 170, 321–329. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lim F-L, Finkler A, Fromm H, Slabas AR, Knight MR. 2014. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genomics 15, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MO, Piron-Prunier F, Marcel F, et al. . 2012. Characterisation of alleles of tomato light signalling genes generated by TILLING. Phytochemistry 79, 78–86. [DOI] [PubMed] [Google Scholar]
- Kaashyap M, Ford R, Bohra A, Kuvalekar A, Mantri N. 2017. Improving salt tolerance of chickpea using modern genomics tools and molecular breeding. Current Genomics 18, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang NJ, Kang YI, Kang KH, Jeong BR. 2009. Induction of thermotolerance and activation of antioxidant enzymes in H2O2 pre-applied leaves of cucumber and tomato seedlings. Journal of the Japanese Society for Horticultural Science 78, 320–329. [Google Scholar]
- Katiyar-Agarwal S, Agarwal M, Grover A. 2003. Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Molecular Biology 51, 677–686. [DOI] [PubMed] [Google Scholar]
- Kaushal N, Bhandari K, Siddique KHM, Nayyar H. 2016. Food crops face rising temperatures: an overview of responses, adaptive mechanisms, and approaches to improve heat tolerance. Cogent Food & Agriculture 2, 1134380. [Google Scholar]
- Keller M, Simm S; SPOT-ITN Consortium 2018. The coupling of transcriptome and proteome adaptation during development and heat stress response of tomato pollen. BMC Genomics 19, 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key JL, Lin CY, Chen YM. 1981. Heat shock proteins of higher plants. Proceedings of the National Academy of Sciences, USA 78, 3526–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilasi NL, Singh J, Vallejos CE, Ye C, Jagadish SVK, Kusolwa P, Rathinasabapathi B. 2018. Heat stress tolerance in rice (Oryza sativa L.): identification of quantitative trait loci and candidate genes for seedling growth under heat stress. Frontiers in Plant Science 9, 1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Alptekin B, Budak H. 2018. CRISPR/Cas9 genome editing in wheat. Functional & Integrative Genomics 18, 31–41. [DOI] [PubMed] [Google Scholar]
- Kim M, Kim H, Lee W, Lee Y, Kwon S-W, Lee J. 2015. Quantitative shotgun proteomics analysis of rice anther proteins after exposure to high temperature. International Journal of Genomics 2015, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll JE, Ramos ML, Zeng Y, Holbrook CC, Chow M, Chen S, Maleki S, Bhattacharya A, Ozias-Akins P. 2011. TILLING for allergen reduction and improvement of quality traits in peanut (Arachis hypogaea L.). BMC Plant Biology 11, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny CB, Gardner SW, Duncan RW. 2018. Impact of high temperature on heterosis and general combining ability in spring canola (Brassica napus L.). Field Crops Research 221, 61–70. [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. 2007. Complexity of the heat stress response in plants. Current Opinion in Plant Biology 10, 310–316. [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Vasquez-Gross HA, Howell T, et al. . 2017. Uncovering hidden variation in polyploid wheat. Proceedings of the National Academy of Sciences, USA 114, E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy L, Gaur PM, Basu PS, Chaturvedi SK, Tripathi S, Vadez V, Rathore A, Varshney RK, Gowda CLL. 2011. Large genetic variation for heat tolerance in the reference collection of chickpea (Cicer arietinum L.) germplasm. Plant Genetic Resources 9, 59–69. [Google Scholar]
- Krishnan A, Gupta V, Ritvik, Nongkynrih B, Thakur J. 2011. How to effectively monitor and evaluate NCD programmes in India. Indian Journal of Community Medicine 36, S57–S62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AP, Boualem A, Bhattacharya A, Parikh S, Desai N, Zambelli A, Leon A, Chatterjee M, Bendahmane A. 2013. SMART—Sunflower Mutant population And Reverse genetic Tool for crop improvement. BMC Plant Biology 13, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar APK, McKeown PC, Boualem A, Ryder P, Brychkova G, Bendahmane A, Sarkar A, Chatterjee M, Spillane C. 2017. TILLING by sequencing (TbyS) for targeted genome mutagenesis in crops. Molecular Breeding 37, 14. [Google Scholar]
- Kumar RR, Pathak H, Sharma SK, Kala YK, Nirjal MK, Singh GP, Goswami S, Rai RD. 2015. Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.). Functional & Integrative Genomics 15, 323–348. [DOI] [PubMed] [Google Scholar]
- Kumar RR, Singh K, Ahuja S, et al. . 2019. Quantitative proteomic analysis reveals novel stress-associated active proteins (SAAPs) and pathways involved in modulating tolerance of wheat under terminal heat. Functional & Integrative Genomics 19, 329–348. [DOI] [PubMed] [Google Scholar]
- Lamaoui M, Jemo M, Datla R, Bekkaoui F. 2018. Heat and drought stresses in crops and approaches for their mitigation. Frontiers in Chemistry 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavania D, Dhingra A, Grover A. 2018. Analysis of transactivation potential of rice (Oryza sativa L.) heat shock factors. Planta 247, 1267–1276. [DOI] [PubMed] [Google Scholar]
- Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H. 2016. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Frontiers in Plant Science 7, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Gügel IL, Giavalisco P, Zeisler V, Schreiber L, Soll J, Philippar K. 2015. FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biology 13, e1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lawas LM, Malo R, et al. . 2015. Metabolic and transcriptomic signatures of rice floral organs reveal sugar starvation as a factor in reproductive failure under heat and drought stress. Plant, Cell & Environment 38, 2171–2192. [DOI] [PubMed] [Google Scholar]
- Li XM, Chao DY, Wu Y, et al. . 2015. Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nature Genetics 47, 827–833. [DOI] [PubMed] [Google Scholar]
- Li Y, Yu Z, Jin J, Zhang Q, Wang G, Liu C, Wu J, Wang C, Liu X. 2018. Impact of elevated CO2 on seed quality of soybean at the fresh edible and mature stages. Frontiers in Plant Science 9, 1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J-L, Zhou H-W, Peng Q, Zhong P-A, Zhang H-Y, He C, Huang Y-J. 2015. Transcriptome changes in rice (Oryza sativa L.) in response to high night temperature stress at the early milky stage. BMC Genomics 16, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J-L, Zhou H-W, Zhang H-Y, Zhong P-A, Huang Y-J. 2014. Comparative proteomic analysis of differentially expressed proteins in the early milky stage of rice grains during high temperature stress. Journal of Experimental Botany 65, 655–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GH, Singhal R, Kachroo A, Kachroo P. 2017. Fatty acid- and lipid-mediated signaling in plant defense. Annual Review of Phytopathology 55, 505–536. [DOI] [PubMed] [Google Scholar]
- Lin H-H, Lin K-H, Syu J-Y, Tang S-Y, Lo H-F. 2016. Physiological and proteomic analysis in two wild tomato lines under waterlogging and high temperature stress. Journal of Plant Biochemistry and Biotechnology 25, 87–96. [Google Scholar]
- Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ. 2011. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Serrano N, Gao G, et al. . 2018. Thermopriming triggers splicing memory in Arabidopsis. Journal of Experimental Botany 69, 2659–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-W, Chang T-S, Hsu Y-K, Wang AZ, Yen H-C, Wu Y-P, Wang C-S, Lai C-C. 2014. Comparative proteomic analysis of early salt stress responsive proteins in roots and leaves of rice. Proteomics 14, 1759–1775. [DOI] [PubMed] [Google Scholar]
- Liu GT, Wang JF, Cramer G, Dai ZW, Duan W, Xu HG, Wu BH, Fan PG, Wang LJ, Li SH. 2012a Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biology 12, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GT, Wang JF, Cramer G, Dai ZW, Duan W, Xu HG, Wu BH, Fan PG, Wang LJ, Li SH. 2012b Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biology 12, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yan S, Yang T, Zhang S, Chen YQ, Liu B. 2017. Small RNAs in regulating temperature stress response in plants. Journal of Integrative Plant Biology 59, 774–791. [DOI] [PubMed] [Google Scholar]
- Liu X, Huang B. 2000. Heat stress injury in relation to membrane lipid peroxidation in creeping bentgrass. Crop Science 40, 503–510. [Google Scholar]
- Liu Y, Li J, Zhu Y, Jones A, Rose RJ, Song Y. 2019. Heat stress in legume seed setting: effects, causes, and future prospects. Frontiers in Plant Science 10, 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xin M, Qin J, Peng H, Ni Z, Yao Y, Sun Q. 2015. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biology 15, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell DB, Schlenker W, Costa-Roberts J. 2011. Climate trends and global crop production since 1980. Science 333, 616–620. [DOI] [PubMed] [Google Scholar]
- Lopes MS, El-Basyoni I, Baenziger PS, et al. . 2015. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. Journal of Experimental Botany 66, 3477–3486. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Stalhandske Z, Fischer EM. 2019. Detection of a climate change signal in extreme heat, heat stress, and cold in europe from observations. Geophysical Research Letters 46, 8363–8374. [Google Scholar]
- Lu Y, Li R, Wang R, Wang X, Zheng W, Sun Q, Tong S, Dai S, Xu S. 2017. Comparative proteomic analysis of flag leaves reveals new insight into wheat heat adaptation. Frontiers in Plant Science 8, 1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas MR, Ehlers JD, Huynh B-L, Diop N-N, Roberts PA, Close TJ. 2013. Markers for breeding heat-tolerant cowpea. Molecular Breeding 31, 529–536. [Google Scholar]
- Luo Q. 2011. Temperature thresholds and crop production: a review. Climate Change 109, 583–598. [Google Scholar]
- MacAlister CA, Park SJ, Jiang K, Marcel F, Bendahmane A, Izkovich Y, Eshed Y, Lippman ZB. 2012. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nature Genetics 44, 1393–1398. [DOI] [PubMed] [Google Scholar]
- Maestri E, Klueva N, Perrotta C, Gulli M, Nguyen HT, Marmiroli N. 2002. Molecular genetics of heat tolerance and heat shock proteins in cereals. Plant Molecular Biology 48, 667–681. [DOI] [PubMed] [Google Scholar]
- Makarova S, Makhotenko A, Spechenkova N, Love AJ, Kalinina NO, Taliansky M. 2018. Interactive responses of potato (Solanum tuberosum L.) plants to heat stress and infection with Potato virus Y. Frontiers in Microbiology 9, 2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcevschi A, Marmiroli N. 2012. Plant protein analysis. In: Hazelwood J, ed. Proteomic applications in biology. IntechOpen. [Google Scholar]
- Mangelsen E, Kilian J, Harter K, Jansson C, Wanke D, Sundberg E. 2011. Transcriptome analysis of high-temperature stress in developing barley caryopses: early stress responses and effects on storage compound biosynthesis. Molecular Plant 4, 97–115. [DOI] [PubMed] [Google Scholar]
- Mangrauthia SK, Agarwal S, Sailaja B, Sarla N, Voleti SR. 2016. Transcriptome analysis of Oryza sativa (rice) seed germination at high temperature shows dynamics of genome expression associated with hormones signalling and abiotic stress pathways. Tropical Plant Biology 9, 215–228. [Google Scholar]
- Mangrauthia SK, Bhogireddy S, Agarwal S, Prasanth VV, Voleti SR, Neelamraju S, Subrahmanyam D. 2017. Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. Journal of Experimental Botany 68, 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfo KO, Hall AE. 1992. Inheritance of heat tolerance during pod set in Cowpea. Crop Science 32, 912. [Google Scholar]
- Marko D, El-shershaby A, Carriero F, Summerer S, Petrozza A, Iannacone R, Schleiff E, Fragkostefanakis S. 2019. Identification and characterization of a thermotolerant TILLING allele of heat shock binding protein 1 in tomato. Genes 10, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinière A, Shvedunova M, Thomson AJ, Evans NH, Penfield S, Runions J, McWatters HG. 2011. Homeostasis of plasma membrane viscosity in fluctuating temperatures. New Phytologist 192, 328–337. [DOI] [PubMed] [Google Scholar]
- Matei G, Woyann LG, Milioli AS, de Bem Oliveira I, Zdziarski AD, Zanella R, Coelho ASG, Finatto T, Benin G. 2018. Genomic selection in soybean: accuracy and time gain in relation to phenotypic selection. Molecular Breeding 38, 117. [Google Scholar]
- Maulana F, Ayalew H, Anderson JD, Kumssa TT, Huang W, Ma XF. 2018. Genome-wide association mapping of seedling heat tolerance in winter wheat. Frontiers in Plant Science 9, 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzeo MF, Cacace G, Iovieno P, Massarelli I, Grillo S, Siciliano RA. 2018. Response mechanisms induced by exposure to high temperature in anthers from thermo-tolerant and thermo-sensitive tomato plants: a proteomic perspective. PLoS One 13, e0201027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseka S, Menkir A, Bossey B, Mengesha W. 2018. Performance assessment of drought tolerant maize hybrids under combined drought and heat stress. Agronomy 8, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Vierstra RD. 2011. Mass spectrometric identification of SUMO substrates provides insights into heat stress-induced SUMOylation in plants. Plant Signaling & Behavior 6, 130–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A. 2009. Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiology and Biochemistry 47, 785–795. [DOI] [PubMed] [Google Scholar]
- Mittal D, Madhyastha DA, Grover A. 2012. Gene expression analysis in response to low and high temperature and oxidative stresses in rice: combination of stresses evokes different transcriptional changes as against stresses applied individually. Plant Science 197, 102–113. [DOI] [PubMed] [Google Scholar]
- Mobin M, Khan MN, Abbas ZK, Ansari HR, Al-Mutairi KA. 2017. Significance of sulfur in heat stressed cluster bean (Cymopsis tetragonoloba L. Taub) genotypes: responses of growth, sugar and antioxidative metabolism. Archives of Agronomy and Soil Science 63, 288–295. [Google Scholar]
- Montero-Barrientos M, Hermosa R, Nicolás C, Cardoza RE, Gutiérrez S, Monte E. 2008. Overexpression of a Trichoderma HSP70 gene increases fungal resistance to heat and other abiotic stresses. Fungal Genetics and Biology 45, 1506–1513. [DOI] [PubMed] [Google Scholar]
- Mu Q, Zhang W, Zhang Y, Yan H, Liu K, Matsui T, Tian X, Yang P. 2017. iTRAQ-based quantitative proteomics analysis on rice anther responding to high temperature. International Journal of Molecular Sciences 18, 1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SP, Unger M, Guender L, Fekete A, Mueller MJ. 2017. Phospholipid:diacylglycerol acyltransferase-mediated triacylglyerol synthesis augments basal thermotolerance. Plant Physiology 175, 486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann JK, Younts TLB, Muday GK. 2018. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proceedings of the National Academy of Sciences, USA 115, E11188–E11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth J, Hartje S, Twyman RM, Hofferbert H-R, Tacke E, Prüfer D. 2008. Precision breeding for novel starch variants in potato. Plant Biotechnology Journal 6, 576–584. [DOI] [PubMed] [Google Scholar]
- Nahar K, Hasanuzzaman M, Alam MdM, Fujita M. 2015. Exogenous glutathione confers high temperature stress tolerance in mung bean (Vigna radiata L.) by modulating antioxidant defense and methylglyoxal detoxification system. Environmental and Experimental Botany 112, 44–54. [Google Scholar]
- Narayanan S, Prasad PV, Welti R. 2016. Wheat leaf lipids during heat stress: II. Lipids experiencing coordinated metabolism are detected by analysis of lipid co-occurrence. Plant, Cell & Environment 39, 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan S, Prasad PVV, Welti R. 2018. Alterations in wheat pollen lipidome during high day and night temperature stress. Plant, Cell & Environment 41, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Reig M, Tauler R, Iriondo-Frias G, Jaumot J. 2019. Untargeted lipidomic evaluation of hydric and heat stresses on rice growth. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences 1104, 148–156. [DOI] [PubMed] [Google Scholar]
- Naveed M, Ahsan M, Akram HM, Aslam M, Ahmed N. 2016. Genetic effects conferring heat tolerance in a cross of tolerant × susceptible maize (Zea mays L.) Genotypes. Frontiers in Plant Science 7, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, et al. . 2007. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proceedings of the National Academy of Sciences, USA 104, 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neta-Sharir I, Isaacson T, Lurie S, Weiss D. 2005. Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. The Plant Cell 17, 1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Li H, Zhao Y, Peng H, Hu Z, Xin M, Sun Q. 2018. Genetic improvement of heat tolerance in wheat: recent progress in understanding the underlying molecular mechanisms. The Crop Journal 6, 32–41. [Google Scholar]
- Niu Y, Xiang Y. 2018. An overview of biomembrane functions in plant responses to high-temperature stress. Frontiers in Plant Science 9, 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. 2017. Transcriptional regulatory network of plant heat stress response. Trends in Plant Science 22, 53–65. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Ariizumi T, Ezura H. 2013. Updating the Micro-Tom TILLING platform. Breeding Science 63, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Asamizu E, Saito T, Matsukura C, Ariizumi T, Brès C, Rothan C, Mizoguchi T, Ezura H. 2011. Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from micro-Tom mutant libraries. Plant & Cell Physiology 52, 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond CB, Austin MP, Berry JA, Billings WD, Boyer JS, Dacey JWH, Nobel PS, Smith SD, Winner WE. 1987. Stress physiology and the distribution of plants. BioScience 37, 38–48. [Google Scholar]
- Otero A, Goni C, Jifon JL, Syvertsen JP. 2011. High temperature effects on citrus orange leaf gas exchange, flowering, fruit quality and yield. Acta Horticulturae 1069–1075. [Google Scholar]
- Padi FK, Denwar NN, Kaleem FZ, Salifu AB, Clottey VA, Kombiok J, Haruna M, Hall AE, Marfo KO. 2004. Registration of ‘Apagbaala’ cowpea. Crop Science 44, 1486. [Google Scholar]
- Paliwal R, Röder MS, Kumar U, Srivastava JP, Joshi AK. 2012. QTL mapping of terminal heat tolerance in hexaploid wheat (T. aestivum L.). Theoretical and Applied Genetics 125, 561–575. [DOI] [PubMed] [Google Scholar]
- Parankusam S, Bhatnagar-Mathur P, Sharma KK. 2017. Heat responsive proteome changes reveal molecular mechanisms underlying heat tolerance in chickpea. Environmental and Experimental Botany 141, 132–144. [Google Scholar]
- Park CJ, Seo YS. 2015. Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathology Journal 31, 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupière MJ, van Haperen P, Rieu I, Visser RGF, Tikunov YM, Bovy AG. 2017. Screening for pollen tolerance to high temperatures in tomato. Euphytica 213, 130. [Google Scholar]
- Petrov VD, Van Breusegem F. 2012. Hydrogen peroxide—a central hub for information flow in plant cells. AoB Plants 2012, pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TTT, Ishibashi Y, Miyazaki M, Tran HT, Okamura K, Tanaka S, Nakamura J, Yuasa T, Iwaya-Inoue M. 2013. High temperature-induced repression of the rice sucrose transporter (OsSUT1) and starch synthesis-related genes in sink and source organs at milky ripening stage causes chalky grains. Journal of Agronomy and Crop Scence 199, 178–188. [Google Scholar]
- Pignone D, De Paola D, Rapanà N, Janni M. 2015. Single seed descent: a tool to exploit durum wheat (Triticum durum Desf.) genetic resources. Genetic Resources and Crop Evolution 62, 1029–1035. [Google Scholar]
- Piron F, Nicolaï M, Minoïa S, Piednoir E, Moretti A, Salgues A, Zamir D, Caranta C, Bendahmane A. 2010. An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS One 5, e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanía JA, Chaves N, Lobaton JD, Cajiao VCH, Rao IM, Raatz B, Beebe SE. 2017. Heat tolerance in common bean derived from interspecific crosses. International Center for Tropical Agriculture (CIAT) https://hdl.handle.net/10568/89450 [Google Scholar]
- Pottorff M, Roberts PA, Close TJ, Lonardi S, Wanamaker S, Ehlers JD. 2014. Identification of candidate genes and molecular markers for heat-induced brown discoloration of seed coats in cowpea [Vigna unguiculata (L.) Walp]. BMC Genomics 15, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad PVV, Bheemanahalli R, Jagadish SVK. 2017. Field crops and the fear of heat stress—opportunities, challenges and future directions. Field Crops Research 200, 114–121. [Google Scholar]
- Prasad PVV, Craufurd PQ, Kakani VG, Wheeler TR, Boote KJ. 2001. Influence of high temperature during pre- and post-anthesis stages of floral development on fruit-set and pollen germination in peanut. Functional Plant Biology 28, 233–240. [Google Scholar]
- Prasad PVV, Djanaguiraman M. 2014. Response of floret fertility and individual grain weight of wheat to high temperature stress: sensitive stages and thresholds for temperature and duration. Functional Plant Biology 41, 1261–1269. [DOI] [PubMed] [Google Scholar]
- Prasad PVV, Pisipati SR, Mutava RN, Tuinstra MR. 2008. Sensitivity of grain sorghum to high temperature stress during reproductive development. Crop Science 48, 1911. [Google Scholar]
- Priya M, Dhanker OP, Siddique KHM, HanumanthaRao B, Nair RM, Pandey S, Singh S, Varshney RK, Prasad PVV, Nayyar H. 2019. Drought and heat stress-related proteins: an update about their functional relevance in imparting stress tolerance in agricultural crops. Theoretical and Applied Genetics 132, 1607–1638. [DOI] [PubMed] [Google Scholar]
- Prasad R, Gunn SK, Rotz CA, Karsten H, Roth G, Buda A, Stoner AMK. 2018. Projected climate and agronomic implications for corn production in the Northeastern United States. PLoS One 13, e0198623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Wang H, Zou Y, Liu C, Liu Y, Wang Y, Zhang W. 2011. Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Letters 585, 231–239. [DOI] [PubMed] [Google Scholar]
- Qin D, Wang F, Geng X, Zhang L, Yao Y, Ni Z, Peng H, Sun Q. 2015. Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) Multiprotein Bridging Factor, confers heat tolerance in both yeast and rice. Plant Molecular Biology; 87, 31–45. [DOI] [PubMed] [Google Scholar]
- Qin D, Wu H, Peng H, Yao Y, Ni Z, Li Z, Zhou C, Sun Q. 2008. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using wheat genome array. BMC Genomics 9, 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu AL, Ding YF, Jiang Q, Zhu C. 2013. Molecular mechanisms of the plant heat stress response. Biochemical and Biophysical Research Communications 432, 203–207. [DOI] [PubMed] [Google Scholar]
- Qu M, Chen G, Bunce JA, Zhu X, Sicher RC. 2018. Systematic biology analysis on photosynthetic carbon metabolism of maize leaf following sudden heat shock under elevated CO2. Scientific Reports 8, 7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman M, Mamidi S, Rahman M. 2018. Genome-wide association study of heat stress-tolerance traits in spring-type Brassica napus L. under controlled conditions. The Crop Journal 6, 115–125. [Google Scholar]
- Ravichandran S, Ragupathy R, Edwards T, Domaratzki M, Cloutier S. 2019. MicroRNA-guided regulation of heat stress response in wheat. BMC Genomics 20, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A, Razzaq A, Mehmood S, Zou X, Zhang X, Lv Y, Xu J. 2019. Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants 8, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TV, Dwivedi S, Sharma NK. 2012. Development of TILLING by sequencing platform towards enhanced leaf yield in tobacco. Industrial Crops and Products 40, 324–335. [Google Scholar]
- Reynolds M, Bonnett D, Chapman SC, Furbank RT, Manès Y, Mather DE, Parry MA. 2011. Raising yield potential of wheat. I. Overview of a consortium approach and breeding strategies. Journal of Experimental Botany 62, 439–452. [DOI] [PubMed] [Google Scholar]
- Ricroch A, Clairand P, Harwood W. 2017. Use of CRISPR systems in plant genome editing: toward new opportunities in agriculture. Emerging Topics in Life Sciences 1, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roorkiwal M, Rathore A, Das RR, et al. . 2016. Genome-enabled prediction models for yield related traits in chickpea. Frontiers in Plant Science 7, 1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri V, Calafiore R, Schettini C, Rigano M, Olivieri F, Frusciante L, Barone A. 2019. Exploiting genetic and genomic resources to enhance heat-tolerance in tomatoes. Agronomy 9, 22. [Google Scholar]
- Salvi P, Kamble NU, Majee M. 2018. Stress-inducible galactinol synthase of chickpea (CaGolS) is implicated in heat and oxidative stress tolerance through reducing stress-induced excessive reactive oxygen species accumulation. Plant & Cell Physiology 59, 155–166. [DOI] [PubMed] [Google Scholar]
- Sanyal RP, Misra HS, Saini A. 2018. Heat-stress priming and alternative splicing-linked memory. Journal of Experimental Botany 69, 2431–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar NK, Kim YK, Grover A. 2014. Coexpression network analysis associated with call of rice seedlings for encountering heat stress. Plant Molecular Biology 84, 125–143. [DOI] [PubMed] [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L. 2012. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochimica et Biophysica Acta 1819, 104–119. [DOI] [PubMed] [Google Scholar]
- Scott JW, Olson SM, Howe TK, Stoffella PJ, Bartz JA, Bryan HH. 1995. ‘Equinox’ heat-tolerant hybrid tomato. HortScience 30, 647–648. [Google Scholar]
- Sedeek KEM, Mahas A, Mahfouz M. 2019. Plant genome engineering for targeted improvement of crop traits. Frontiers in Plant Science 10, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Sita K, Kumar J, Kumar S, Singh S, Siddique KHM, Nayyar H. 2017. Effects of drought, heat and their interaction on the growth, yield and photosynthetic function of lentil (Lens culinaris Medikus) genotypes varying in heat and drought sensitivity. Frontiers in Plant Science 8, 1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestili F, Palombieri S, Botticella E, Mantovani P, Bovina R, Lafiandra D. 2015. TILLING mutants of durum wheat result in a high amylose phenotype and provide information on alternative splicing mechanisms. Plant Science 233, 127–133. [DOI] [PubMed] [Google Scholar]
- Setia RC, Setia N. 2008. The ‘-OMICS’ technologies and crop improvement. In: Setia RC, Nayyar H, Setia N, eds. Crop improvement:strategies and applications . New Dehli: International Publishing House Pvt. Ltd, 1–18. [Google Scholar]
- Shah F, Huang J, Cui K, Nie L, Shah T, Chen C, Wang K. 2011. Impact of high-temperature stress on rice plant and its traits related to tolerance. Journal of Agricultural Science 149, 545–556. [Google Scholar]
- Shan Q, Wang Y, Li J, et al. . 2013. Targeted genome modification of crop plants using a CRISPR–Cas system. Nature Biotechnology 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C. 2014. Genome editing in rice and wheat using the CRISPR/Cas system. Nature Protocols 9, 2395–2410. [DOI] [PubMed] [Google Scholar]
- Shanmugavadivel PS, Amitha Mithra SV, Prakash C, MK R, Tiwari R, Mohapatra T, Singh NK. 2017. High resolution mapping of QTLs for heat tolerance in rice using a 5K SNP array. Rice 10, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DK, Torp AM, Rosenqvist E, Ottosen CO, Andersen SB. 2017. QTLs and potential candidate genes for heat stress tolerance identified from the mapping populations specifically segregating for Fv/Fm in wheat. Frontiers in Plant Science 8, 1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma L, Dalal M, Verma RK, Kumar SVV, Yadav SK, Pushkar S, Kushwaha SR, Bhowmik A, Chinnusamy V. 2018. Auxin protects spikelet fertility and grain yield under drought and heat stresses in rice. Environmental and Experimental Botany 150, 9–24. [Google Scholar]
- Shen L, Hua Y, Fu Y, et al. . 2017. Rapid generation of genetic diversity by multiplex CRISPR/Cas9 genome editing in rice. Science China Life Sciences 60, 506–515. [DOI] [PubMed] [Google Scholar]
- Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE. 2017a ARGOS8 variants generated by CRISPR–Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnology Journal 15, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Yan B, Lou X, Ma H, Ruan S. 2017b Comparative transcriptome analysis reveals the transcriptional alterations in heat-resistant and heat-sensitive sweet maize (Zea mays L.) varieties under heat stress. BMC Plant Biology 17, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirdelmoghanloo H, Taylor JD, Lohraseb I, Rabie H, Brien C, Timmins A, Martin P, Mather DE, Emebiri L, Collins NC. 2016. A QTL on the short arm of wheat (Triticum aestivum L.) chromosome 3B affects the stability of grain weight in plants exposed to a brief heat shock early in grain filling. BMC Plant Biology 16, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silletti MF, Petrozza A, Stigliani AL, Giorio G, Cellini F, D’Ambrosio C, Carriero F. 2013. An increase of lycopene content in tomato fruit is associated with a novel Cyc-B allele isolated through TILLING technology. Molecular Breeding 31, 665–674. [Google Scholar]
- Singh D, Balota M, Collakova E, Isleib TG, Welbaum GE, Tallury SP. 2016. Heat stress related physiological and metabolic traits in peanut seedlings. Peanut Science 43, 24–35. [Google Scholar]
- Singh S, Vikram P, Sehgal D, et al. . 2018. Harnessing genetic potential of wheat germplasm banks through impact-oriented-prebreeding for future food and nutritional security. Scientific Reports 8, 12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Jwa NS. 2013. Understanding the responses of rice to environmental stress using proteomics. Journal of Proteome Research 12, 4652–4669. [DOI] [PubMed] [Google Scholar]
- Sita K, Sehgal A, Bhandari K, Kumar J, Kumar S, Singh S, Siddique KH, Nayyar H. 2018. Impact of heat stress during seed filling on seed quality and seed yield in lentil (Lens culinaris Medikus) genotypes. Journal of the Science of Food and Agriculture 98, 5134–5141. [DOI] [PubMed] [Google Scholar]
- Slade AJ, McGuire C, Loeffler D, et al. . 2012. Development of high amylose wheat through TILLING. BMC Plant Biology 12, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider JL, Russo VM, Roberts W, Wann EV, Raper RL. 2012. Cultural and environmental factors governing tomato production: local production under elevated temperatures. HortScience 47, 1022–1028. [Google Scholar]
- Soda N, Wallace SA. 2015. Omics study for abiotic stress responses in plants. Advances in Plants & Agricultural Research 2, 00037. [Google Scholar]
- Song J, Carver BF, Powers C, Yan L, Klápště J, El-Kassaby YA, Chen C. 2017. Practical application of genomic selection in a doubled-haploid winter wheat breeding program. Molecular Breeding 37, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks DL. 2018. Advances in agronomy. New York: Academic Press. [Google Scholar]
- Sparla F, Falini G, Botticella E, Pirone C, Talamè V, Bovina R, Salvi S, Tuberosa R, Sestili F, Trost P. 2014. New starch phenotypes produced by TILLING in barley. PLoS One 9, e107779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicher L, Glauser G, Kessler F. 2016. Lipid antioxidant and galactolipid remodeling under temperature stress in tomato plants. Frontiers in Plant Science 7, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindel J, Iwata H. 2018. Genomic selection in rice breeding. In: Sasaki T, Ashikari M, eds. Rice genomics, genetics and breeding. Singapore: Springer Singapore, 473–496. [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I. 2014. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. The Plant Cell 26, 1792–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D-Y, Kaplan F, Guy CL. 2001. Plant Hsp70 molecular chaperones: protein structure, gene family, expression and function. Physiologia Plantarum 113, 443–451. [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS One 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R. 2014. Abiotic and biotic stress combinations. New Phytologist 203, 32–43. [DOI] [PubMed] [Google Scholar]
- Tack J, Lingenfelser J, Jagadish SVK. 2017. Disaggregating sorghum yield reductions under warming scenarios exposes narrow genetic diversity in US breeding programs. Proceedings of the National Academy of Sciences, USA 114, 9296–9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse W, Suleiman S, Tahir I, Sanchez-Garcia M, Jighly A, Hagras A, Thabet SH, Baum M. 2019. Heat-tolerant QTLs associated with grain yield and its components in spring bread wheat under heat-stressed environments of Sudan and Egypt. Crop Science 59, 199. [Google Scholar]
- Takahashi F, Shinozaki K. 2019. Long-distance signaling in plant stress response. Current Opinion in Plant Biology 47, 106–111. [DOI] [PubMed] [Google Scholar]
- Talukder SK, Babar MA, Vijayalakshmi K, Poland J, Prasad PV, Bowden R, Fritz A. 2014. Mapping QTL for the traits associated with heat tolerance in wheat (Triticum aestivum L.). BMC Genetics 15, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Zhu W, Song X, Lin X, Cai J, Wang M, Yang Q. 2016. Genome-wide identification and function analyses of heat shock transcription factors in potato. Frontiers in Plant Science 7, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L-X, Tan H-J, Wang X, Cao L-Y, Song J, Cheng S-H. 2008. Effects of high-temperature stress on flowering and grain-setting characteristics of guodao 6. Acta Agronomica Sinica 34, 609–674. [Google Scholar]
- Tayade R, Nguyen T, Oh SA, Hwang YS, Yoon IS, Deshmuk R, Jung K-H, Park SK. 2018. Effective strategies for enhancing tolerance to high-temperature stress in rice during the reproductive and ripening stages. Plant Breeding and Biotechnology 6, 1–18. [Google Scholar]
- Templer SE, Ammon A, Pscheidt D, et al. . 2017. Metabolite profiling of barley flag leaves under drought and combined heat and drought stress reveals metabolic QTLs for metabolites associated with antioxidant defense. Journal of Experimental Botany 68, 1697–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenorio FA, Ye C, Redoña E, Sierra S, Laza M, Argayoso MA. 2013. Screening rice genetic resources for heat tolerance. SABRAO Journal of Breeding and Genetics 45, 371–381. [Google Scholar]
- Thomason K, Babar MA, Erickson JE, Mulvaney M, Beecher C, MacDonald G. 2018. Comparative physiological and metabolomics analysis of wheat (Triticum aestivum L.) following post-anthesis heat stress. PLoS One 13, e0197919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thudi M, Upadhyaya HD, Rathore A, et al. . 2014. Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS One 9, e96758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thussagunpanit J, Jutamanee K, Kaveeta L, Chai-arree W, Pankean P, Homvisasevongsa S, Suksamrarn A. 2015. Comparative effects of brassinosteroid and brassinosteroid mimic on improving photosynthesis, lipid peroxidation, and rice seed set under heat stress. Journal of Plant Growth Regulation 34, 320–331. [Google Scholar]
- Trapero-Mozos A, Morris WL, Ducreux LJM, McLean K, Stephens J, Torrance L, Bryan GJ, Hancock RD, Taylor MA. 2018. Engineering heat tolerance in potato by temperature-dependent expression of a specific allele of HEAT-SHOCK COGNATE 70. Plant Biotechnology Journal 16, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C. 2017. Wheat genomics comes of age. Current Opinion in Plant Biology 36, 142–148. [DOI] [PubMed] [Google Scholar]
- Valdés-López O, Batek J, Gomez-Hernandez N, et al. . 2016. Soybean roots grown under heat stress show global changes in their transcriptional and proteomic profiles. Frontiers in Plant Science 7, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdeprado H, Kretzschmar T, Begum H, Raghavan C, Joyce P, Lakshmanan P, Cobb JN, Collard BCY. 2018. Association mapping in rice: basic concepts and perspectives for molecular breeding. Plant Production Science 21, 159–176. [Google Scholar]
- Viana AP, de Resende MDV, Riaz S, Walker MA, Viana AP, de Resende MDV, Riaz S, Walker MA. 2016. Genome selection in fruit breeding: application to table grapes. Scientia Agricola 73, 142–149. [Google Scholar]
- Vierling E. 1991. The roles of heat shock proteins in plants. Annual Review of Plant Physiology and Plant Molecular Biology 42, 579–620. [Google Scholar]
- Vijayalakshmi K, Fritz AK, Paulsen GM, Bai G, Pandravada S, Gill BS. 2010. Modeling and mapping QTL for senescence-related traits in winter wheat under high temperature. Molecular Breeding 26, 163–175. [Google Scholar]
- Vriet C, Hennig L, Laloi C. 2015. Stress-induced chromatin changes in plants: of memories, metabolites and crop improvement. Cellular and Molecular Life Sciences 72, 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu HS, Tamura P, Galeva NA, Chaturvedi R, Roth MR, Williams TD, Wang X, Shah J, Welti R. 2012. Direct infusion mass spectrometry of oxylipin-containing Arabidopsis membrane lipids reveals varied patterns in different stress responses. Plant Physiology 158, 324–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid A, Gelani S, Ashraf M, Foolad M. 2007. Heat tolerance in plants: an overview. Environmental and Experimental Botany 61, 199–223. [Google Scholar]
- Wang J, Gan YT, Clarke F, McDonald CL. 2006. Response of chickpea yield to high temperature stress during reproductive development. Crop Science 46, 2171–2178. [Google Scholar]
- Wang L, Ma H, Song L, Shu Y, Gu W. 2012a Comparative proteomics analysis reveals the mechanism of pre-harvest seed deterioration of soybean under high temperature and humidity stress. Journal of Proteomics 75, 2109–2127. [DOI] [PubMed] [Google Scholar]
- Wang L, Shen W, Kazachkov M, Chen G, Chen Q, Carlsson AS, Stymne S, Weselake RJ, Zou J. 2012b Metabolic interactions between the Lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. The Plant Cell 24, 4652–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Wang Y, Tian F, King GJ, Zhang C, Long Y, Shi L, Meng J. 2008. A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytologist 180, 751–765. [DOI] [PubMed] [Google Scholar]
- Wang T, Uauy C, Till B, Liu CM. 2010. TILLING and associated technologies. Journal of Integrative Plant Biology 52, 1027–1030. [DOI] [PubMed] [Google Scholar]
- Wang X, Dinler BS, Vignjevic M, Jacobsen S, Wollenweber B. 2015. Physiological and proteome studies of responses to heat stress during grain filling in contrasting wheat cultivars. Plant Science 230, 33–50. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu Y, Hu Z, Xu C. 2018. Genomic selection methods for crop improvement: current status and prospects. The Crop Journal 6, 330–340. [Google Scholar]
- Wang X, Yan B, Shi M, Zhou W, Zekria D, Wang H, Kai G. 2016. Overexpression of a Brassica campestris HSP70 in tobacco confers enhanced tolerance to heat stress. Protoplasma 253, 637–645. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sun F, Cao H, Peng H, Ni Z, Sun Q, Yao Y. 2012. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS One 7, e48445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER. 2013. The evolution, function, structure, and expression of the plant sHSPs. Journal of Experimental Botany 64, 391–403. [DOI] [PubMed] [Google Scholar]
- Waters ER, Vierling E. 1999. Chloroplast small heat shock proteins: evidence for atypical evolution of an organelle-localized protein. Proceedings of the National Academy of Sciences, USA 96, 14394–14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert H, Högy P, Mora-Ramirez I, Fuchs J, Eggert K, Koehler P, Weschke W, Fangmeier A, Weber H. 2017. Grain yield and quality responses of wheat expressing a barley sucrose transporter to combined climate change factors. Journal of Experimental Botany 68, 5511–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti R, Shah J, Li W, Li M, Chen J, Burke JJ, Fauconnier ML, Chapman K, Chye ML, Wang X. 2007. Plant lipidomics: discerning biological function by profiling plant complex lipids using mass spectrometry. Frontiers in Bioscience 12, 2494–2506. [DOI] [PubMed] [Google Scholar]
- Wen J, Jiang F, Weng Y, Sun M, Shi X, Zhou Y, Yu L, Wu Z. 2019. Identification of heat-tolerance QTLs and high-temperature stress-responsive genes through conventional QTL mapping, QTL-seq and RNA-seq in tomato. BMC Plant Biology 19, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese S, Reidegeld KA, Meyer HE, Warscheid B. 2007. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350. [DOI] [PubMed] [Google Scholar]
- Wu X, Gong F, Cao D, Hu X, Wang W. 2016. Advances in crop proteomics: PTMs of proteins under abiotic stress. Proteomics 16, 847–865. [DOI] [PubMed] [Google Scholar]
- Wu X, Gong F, Yang L, Hu X, Tai F, Wang W. 2014. Proteomic analysis reveals differential accumulation of small heat shock proteins and late embryogenesis abundant proteins between ABA-deficient mutant vp5 seeds and wild-type Vp5 seeds in maize. Frontiers in Plant Science 5, 801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Cornish AJ, Benning C. 2008. Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. The Plant Cell 20, 2190–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Driedonks N, Rutten MJM, Vriezen WH, de Boer GJ, Rieu I. 2017. Mapping quantitative trait loci for heat tolerance of reproductive traits in tomato (Solanum lycopersicum). Molecular Breeding 37, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Cai SY, Zhang Y, et al. . 2016. Melatonin enhances thermotolerance by promoting cellular protein protection in tomato plants. Journal of Pineal Research 61, 457–469. [DOI] [PubMed] [Google Scholar]
- Xue GP, Drenth J, McIntyre CL. 2015. TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. Journal of Experimental Botany 66, 1025–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP, Sadat S, Drenth J, McIntyre CL. 2014. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. Journal of Experimental Botany 65, 539–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Hakata M. 2010. Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant & Cell Physiology 51, 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Yu L, Xuan J, Lu Y, Lu S, Zhu W. 2016. De novo transcriptome sequencing and gene expression profiling of spinach (Spinacia oleracea L.) leaves under heat stress. Scientific Reports 6, 19473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gu X, Ding M, Lu W, Lu D. 2018. Heat stress during grain filling affects activities of enzymes involved in grain protein and starch synthesis in waxy maize. Scientific Reports 8, 15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhu W, Zhang H, Liu N, Tian S. 2016. Heat shock factors in tomatoes: genome-wide identification, phylogenetic analysis and expression profiling under development and heat stress. PeerJ 4, e1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Argayoso MA, Redoña ED, et al. . 2012. Mapping QTL for heat tolerance at flowering stage in rice using SNP markers. Plant Breeding 131, 33–41. [Google Scholar]
- Ye C, Tenorio FA, Argayoso MA, Laza MA, Koh HJ, Redoña ED, Jagadish KS, Gregorio GB. 2015. Identifying and confirming quantitative trait loci associated with heat tolerance at flowering stage in different rice populations. BMC Genetics 16, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona N. 2015. Genetic characterization of heat tolerant (HT) upland mutant rice (Oryza sativa L.) lines selected from rice genotypes. Master thesis, University of Agriculture Morogoro, Tanzania.
- Yousuf PY, Abd_Allah EF, Nauman M, Asif A, Hashem A, Alqarawi AA, Ahmad A. 2017. Responsive proteins in wheat cultivars with contrasting nitrogen efficiencies under the combined stress of high temperature and low nitrogen. Genes 8, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu E, Fan C, Yang Q, Li X, Wan B, Dong Y, Wang X, Zhou Y. 2014. Identification of heat responsive genes in Brassica napus siliques at the seed-filling stage through transcriptional profiling. PLoS One 9, e101914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SS, Mukhtar MS, Mansoor S. 2018. Genome editing: targeting susceptibility genes for plant disease resistance. Trends in Biotechnology 36, 898–906. [DOI] [PubMed] [Google Scholar]
- Zampieri M, Ceglar A, Dentener F, Toreti A. 2017. Wheat yield loss attributable to heat waves, drought and water excess at the global, national and subnational scales. Environmental Research Letters 12, 064008. [Google Scholar]
- Zang X, Geng X, Wang F, et al. . 2017. Overexpression of wheat ferritin gene TaFER-5B enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging. BMC Plant Biology 17, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandalinas SI, Balfagón D, Arbona V, Gómez-Cadenas A. 2017. Modulation of antioxidant defense system is associated with combined drought and heat stress tolerance in citrus. Frontiers in Plant Science 8, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. 2006. Detecting differential and correlated protein expression in label-free shotgun proteomics. Journal of Proteome Research 5, 2909–2918. [DOI] [PubMed] [Google Scholar]
- Zhang C, Hsieh T-F. 2013. Heritable epigenetic variation and its potential applications for crop improvement. Plant Breeding and Biotechnology 1, 307–319. [Google Scholar]
- Zhang L, Geng X, Zhang H, et al. . 2017. Isolation and characterization of heat-responsive gene TaGASR1 from wheat (Triticum aestivum L.). Journal of Plant Biology 60, 57–65. [Google Scholar]
- Zhang L, Kato Y, Otters S, Vothknecht UC, Sakamoto W. 2012. Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis. The Plant Cell 24, 3695–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Chen W, Bian J, et al. . 2018. Proteomics and phosphoproteomics of heat stress-responsive mechanisms in Spinach. Frontiers in Plant Science; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Liu B, Piao S, et al. . 2017. Temperature increase reduces global yields of major crops in four independent estimates. Proceedings of the National Academy of Sciences, USA 114, 9326–9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Zhang D, Zhao Y, Wang W, Yang H, Tai F, Li C, Hu X. 2016. The difference of physiological and proteomic changes in maize leaves adaptation to drought, heat, and combined both stresses. Frontiers in Plant Science 7, 1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Högy P, Wu X, Schmid I, Wang X, Schulze WX, Jiang D, Fangmeier A. 2018. Physiological and proteomic evidence for the interactive effects of post-anthesis heat stress and elevated CO2 on wheat. Proteomics 18, 1800262. [DOI] [PubMed] [Google Scholar]
- Zheng G, Tian B, Zhang F, Tao F, Li W. 2011. Plant adaptation to frequent alterations between high and low temperatures: remodelling of membrane lipids and maintenance of unsaturation levels. Plant, Cell & Environment 34, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Liu C, Chen X. 2011. Proteomics of rice in response to heat stress and advances in genetic engineering for heat tolerance in rice. Plant Cell Reports 30, 2155–2165. [DOI] [PubMed] [Google Scholar]