Abstract
Prostate cancer (PC) incidence and mortality rates are higher in African-American (AA) than in European-American (EA) men. The main objective of this study was to elucidate the role of miR-130b as a contributor to PC health disparity in AA patients. We also determined whether miR-130b is a prognostic biomarker and a new therapeutic candidate for AA PC. A comprehensive approach of using cell lines, tissue samples and the TCGA database was employed. We performed a series of functional assays such as cell proliferation, migration, invasion, RT2-PCR-array, qRT-PCR, cell cycle, luciferase reporter, immunoblot and immunohistochemistry. Various statistical approaches such as Kaplan-Meier, Uni- and Multivariate analyses were utilized to determine the clinical significance of miR-130b. Our results showed that elevated levels of miR-130b correlated with race disparity and PSA levels/failure and acted as an independent prognostic biomarker for AA patients. Two tumor suppressor genes, CDKN1B and FHIT, were validated as direct functional targets of miR-130b. We also found race-specific cell cycle pathway activation in AA PC patients. Functionally, miR-130b inhibition reduced cell proliferation, colony formation, migration/invasion and induced cell cycle arrest. Inhibition of miR-130b modulated critical PC related biological pathways in AA compared to EA PC patients. In conclusion, attenuation of miR-130b expression has tumor suppressor effects in AA PC. miR-130b is a significant contributor to PC racial disparity as its overexpression is a risk factor for poor prognosis in AA PC patients. Thus, regulation of miR-130b may provide a novel therapeutic approach for the management of PC in AA patients.
Introduction
Prostate cancer (PC) is among the top estimated diagnosed cancers and second leading cause of cancer deaths in US American men (SEER Cancer Stat Facts 2019). African-American (AA) men have higher incidence and significantly higher PC mortality rates than European-American (EA) men (1). A number of groups have found that AA patients have malignant transformation and also greater prostate tumor volumes in radical prostatectomies as compared to similarly staged EA patients (2,3). While some of the differences in PC survival or mortality can be attributed to social-economic status (SES) (4,5), some studies have shown less or inconsistent associations between SES and PC mortality/ incidence (6,7). Another study found that Crude Cox models showed AA men had higher biochemical recurrence (BCR) risk that persisted after adjustment for covariates including SES though AA men were more likely to have lower SES than white men (8). A recent study also reported that there are no interactions between race and SES for BCR. AA men were found to have 10% to 11% increased BCR risk while SES was unrelated to BCR (9). These studies indicate that biological background accounts for a significant portion of PC disparity with regard to mortality, incidence and progression in AA men compared to EA men. However, further investigations are required to uncover the mechanisms underlying abnormal gene regulation and racial disparity. Also, there is an unmet need to develop prognostic biomarkers that contribute to AA PC health disparity.
miRNAs are a small non-coding RNAs that regulate gene expression either by regulating stability or by translational degradation of their cognate mRNAs in multiple cancers (10). miRNAs are potentially powerful diagnostic tools due to their stability in frozen biopsies, formalin fixed paraffin embedded (FFPE) tissue samples, blood and exosomes (11,12). Unlike studies of racial disparity in relation to protein-coding genes, the role of miRNAs has not been fully explored in prostate cancer.
Dysregulation of miR-130b is known in various human cancers. In hepatocellular carcinoma (HCC), PROM1(+) and PROM1(–) cells from human HCC clinical specimens and cell lines identified overexpression of miR-130b in CD133(+) tumor-initiating cells. Increased miR-130b also reduced TP53INP1 that is a known miR-130b target (13). In prostate cancer, it has been reported that castration resistant prostate cancers had c-Met activation that increases miR-130b levels, inhibits androgen receptor expression, promotes cancer spreading and resistance to hormone ablation therapy (14). Integrated public database analysis also supports that miR-130b is an oncogenic miRNA. Most published reports concerning miR-130b (27 out of 31) reveal up-regulation of the miR-130b cluster in PC that is correlated with adverse outcomes such as tumor malignancy and T-stage in PC (15). Despite increasing evidence for miR-130b in tumorigenesis, the clinical significance and functional role of miR-130b in PC racial disparity remains unexplored.
In this study, we sought to elucidate the role of miR-130b in the genetics of prostate cancer health disparity in AA patients. In addition, we examined the biomarker potential of miR-130b to predict prognosis of AA PC patients. We identified miR-130b as a significant contributor to PC racial disparity in AA men. Our results show that; i) miR-130b is significantly overexpressed in AA compared to EA tissue samples and cell lines; ii) miR-130b has prognostic biomarker potential in AA PC patients; iii) attenuation of miR-130b expression in AA cell lines results in suppression of tumorigenic characteristics as evidenced by repressed cell proliferation, migration, invasion and induction of cell cycle arrest; iv) downregulation of miR-130b modulates essential biological pathways in AA PC cell lines; v) FHIT and CDKN1B were confirmed to be direct functional targets of miR-130b. Thus, this study shows that miR-130b plays important role in PC racial disparity. Also, it may be a risk factor of poor prognosis in AA patients. Thus, regulation of miR-130b may offer a preventative and therapeutic approach for AA PC patients.
Materials & Methods
Research involving human subjects
Patient studies were conducted as per the “Declaration of Helsinki” ethical guidelines. Informed written consent was obtained from the patients involved in this study. The study was approved by the Institutional Review Board (IRB) on Human Research Protection Program (IRB approval #: 11–06755, Ref. #: 225293) at the San Francisco Veterans Affairs Medical Center and University of California San Francisco.
Cell lines
All cell lines were acquired from the American Type Culture Collection (ATCC) and were authenticated using DNA short- tandem repeat profiling by ATCC. EA derived cell lines DU-145, PC3, LNCaP were maintained in RPMI-1640 (Thermo Fisher Scientific) medium according to the manufacturer’s protocol. AA cell line, MDA-PCa-2B was maintained in BRFF-HPC1 (Athena ES, Fisher Scientific) medium with Poly-L-Lysine coated plates according to manufacturer’s protocol.
Clinical samples and Follow-up
Clinical Formalin Fixed Paraffin Embedded (FFPE) tissue samples from AA (n = 36) and EA (n = 57) were obtained from the Urology Tissue Bank of the San Francisco Veteran Affairs Medical Center (SFVAMC cohort). Clinical data is given in Supplementary Table 1 and 2 and the number of samples for which information was not available are indicated in each variable in same tables. After admission, all patients were followed until their deaths and recorded in the Computerized Patient Record System (CPRS) of the Department of Veterans Affairs. Follow-up was performed by phone or outpatient visits. Overall survival time endpoints were defined from the time of surgery until time of the death or last follow-up. RNA extraction, cDNA synthesis and TaqMan miRNA assays (Thermo Fisher Scientific) were performed as described previously (16).
Human prostate cancer mRNA and miRNA sequencing data
The expression levels of mRNAs and miRNAs in 455 (59 AA and 396 EA) PC patient samples (41 to 78 years, 61.0 ± 6.7 (mean±SD) years; Gleason score, 7.5 ± 0.9 (mean±SD)) were obtained from the Cancer Genome Atlas (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga/) and the Cancer Genetic Ancestry Atlas (TCGAA) (17).
Quantitative PCR
Real-time reverse transcription–polymerase chain reaction (RT–PCR) was carried out using a Quant Studio 7 PCR System, TaqMan Universal PCR Master Mix, TaqMan Reverse Transcription kit, TaqMan miRNA assays (Thermo Fisher Scientific; TaqMan MicroRNA Assays-hsa-miR-130b; Cat. # 000456) according to the manufacturer’s instructions. anti-miR-130b was measured by Custom TaqMan™ Small RNA Assay (Thermo Fisher Scientific; Cat. # 4440418; Designed from antisense of mature miR-130b; 3’-ATGCCCTTTCATCATTGCACTG-5’). The expression levels of miRNA were determined by normalizing with RNU48 (Thermo Fisher Scientific Cat. # 1006). A QuantiFast SYBR Green PCR Kit was also utilized for gene expression analysis of miR-130b targets. The primers used for SYBR Green-based qPCR analyses are listed in Supplementary Table 3.
Establishing stable miR-130b knock-down clones
7.5μg of psPAX2 and 2.5 μg of pMD2.g plasmids were used to pack 10μg of constructed pLenti-III-mir-Off viral vector (ABM, Canada). The filtered medium containing vector was used immediately for infection of prostate cancer cell lines for three days. Then, stable cells were selected with 5μg/mL puromycin for 1–4 weeks. Anti-sense miR-130b expression levels were confirmed by qPCR using a custom TaqMan small RNA assay kit (Thermo Fisher Scientific) according to manufacturer’s protocol.
Cell cycle analysis
Cell pellets were stained with PI/RNase Staining Buffer (BD Biosciences, San Jose, CA). Cells were analyzed with a BD FACSVerse (BD Biosciences). All analyses were performed in triplicate, and 10,000 gated events/sample were counted. Cell cycle related genes were referred from KEGG PATHWAY Database (https://www.kegg.jp/kegg/pathway.html).
Wound-healing assay
Wound-healing assays were performed with CytoSelect (Cell Biolabs. Inc., San Diego, CA) 24-well Wound Healing Assay kit following the manufacturer’s instructions. The wound regions calculated by ImageJ (MRI Wound Healing Tool).
Colony formation assay
For colony formation assays, anti-miR-130b/ control stable MDA-PCa-2b and DU-145 cells were plated onto 6-well plates (MDA; 2000 cells, DU; 200 cells). Following ten days for cell adherence, colonies of cells were fixed and stained with crystal violet reagent. ImageJ plug-in, “ColonyArea” was used to calculate the occupied areas of the cell colony as described in a published report (18).
RT2 profiler PCR array analysis
Expression profiling of 84 genes involved in cancer was performed utilizing human RT2 Profiler PCR Array (Qiagen, Cat. # PAHS-502Z). Using RNAs isolated from anti-miR-130b stable DU-145 and MDA-PCa-2b cells were applied to the qPCR-arrays. Fold- change calculation and Heatmap analysis were done using RT2 Profiler PCR Array Data Analysis (Qiagen) according to manufacturer’s protocol.
Over-Representative Analysis
Over-Representative Analysis was carried out as described in a published report (19) by using the Max Planck Institute, ConsensusPathDB (CPDB) (http://cpdb.molgen.mpg.de). Detailed methodology is described in the documentation of CPDB.
Western blot analysis
Western blot analyses were performed as described previously (16). The antibodies used were specific for FHIT (Millipore Sigma; 1:1000 Cat. # 71–9000), CDKN1B (p27 Kip1, Cell Signaling, Danvers, MA; 1:1000, Cat. # 2552), CDKN1A (p21 Waf1/Cip1 (12D1), Cell Signaling; 1:1000, Cat. # 2947), β-actin (Cell Signaling; 1;1000, Cat. # 3700) and GAPDH (SantaCruz; 1:1000, Cat. # sc-32233).
Measurement of cell viability
Cell viability assay was performed using CellTiter-Glo Luminescent Cell Viability Assay (Promega, Cat. # G7570) as described previously (16). Cell viability was measured every 24 hours for four days using established anti-miR-130b and control stable PC cells following the manufacturer’s instructions.
Invasion assay
Cell invasion was carried out in chambers coated with Matrigel (Corning, Corning, NY). Cells were seeded in serum-free medium in the upper chamber inserts and cells were allowed to invade into the lower chambers, which contained medium supplemented with 10% FBS. Inserts were then stained, and the invaded cells were photographed using a light microscope. Cell stain solution was then dissolved with methanol and absorbance was measured at 540nm using a SpectraMax plate reader according to manufacturer’s instruction.
Immunohistochemistry
Immunohistochemical (IHC) staining was performed with 15 (14 PC and 1 benign prostate hyperplasia (BPH) tissues) AA and 10 (9 PC and 1 BPH) EA specimens. The Vision™ UltraVision™ Detection System (Thermo Fisher Scientific) was used to stain FHIT proteins (Thermo Fisher Scientific, Cat. # 71–9000). The intensity of the staining was classified following Allred scoring (Harvey JM et al. 1999). Negative staning was “0”, low staining “1”, and high staining a “2–3” score. For correlation of FHIT staining with death of patients, we divided the staining intensity into negative group (0 staining) and positive group (1–3 staining score).
Dual-Luciferase reporter assay
Luciferase reporter vectors were constructed by ligation with annealed custom oligonucleotides containing the putative target binding sites of 3’-UTR into pmiR-GLO reporter vector (Promega, Cat. # E1330) (Supplementary Table 4). Anti-miR-130b stable MDA-PCa-2b cells were transfected with 0.1 ng of pmiR-GLO vector containing the 3’-UTR sequences and luciferase activity was measured 48 hr after transfection using a Dual-Luciferase Reporter Assay System (Promega) as per manufacturer’s instructions. Relative luciferase activity was calculated by normalizing to renilla luminescence.
Transient over-expression of miR-130b
miR-130b over-expression was carried out with mirVana miRNA mimic hsa-miR-130b-3p (Thermo Fisher Scientific; Cat. # 4464066, Product ID MC10777) and mirVana miRNA mimic, Negative Control #1 (Thermo Fisher Scientific; Cat. # 4464058). jetPRIME (Poplyplus, Cat. # 114–15) reagent was used for transient transfection of miR-130b mimic and Negative control #1.
Statistical analysis
Statistical analyses were conducted with GraphPad-Prism 8, and EZR (20). All quantified data represent the average of three or more independent experiments performed at different times or as indicated. Error bars show SEM of independent experiments. All tests were performed two-tailed and P values <0.05 were considered statistically significant. TCGA gene and miRNA sequencing data were normalized by quintile method in order to eliminate the bias of the differences between individual patients. miRNA sequencing data were analyzed using Mann–Whitney U-test. Youden’s index was calculated to generate a cut-off value to determine miR-130b high or low expression groups with the SFVAMC EA and AA samples (Supplementary Figure 1). The end points were defined from time of surgery until time of the death, or last follow-up. This cut-off value was used for Kaplan–Meier and uni-/ multivariate analyses. Kaplan–Meier analyses were used for overall survival curves and p-values were calculated with the Log-rank test. Uni- and Multivariate analyses were computed by the Cox proportional hazards model. The Pearson’s r correlation was computed to compare the correlation between miR-130b and cell cycle related genes in the TCGA cohort.
Results
miR-130b is up-regulated in AA tissue samples and cell lines
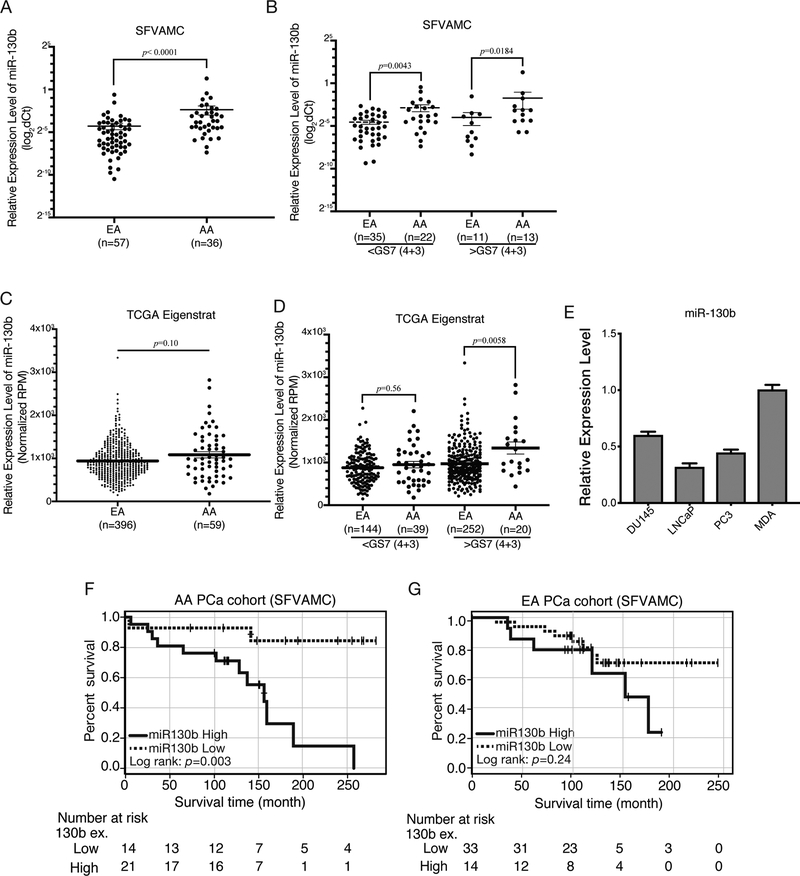
For the initial screening, we analyzed the expression of a panel of miRNAs in AA compared to EA PC samples. We observed significant differential expression profiles in our in-house cohort and in the published literature in AA compared to EA PC samples for miR-24 (16), miR-34b (21), miR-96 and miR-130b (22). In this study, we investigated the role of miR-130b in PC racial disparity. First, we confirmed over-expression of miR-130b in PC tissue samples from AA patient cohorts compared to EA patient samples. Higher expression levels of miR-130b were observed in AA (n=36) compared to EA patients (n=57) (Figure 1A; clinicopathological feature summary and its correlation with miR-130b expression in Supplementary Table 1 and 2, respectively). We further divided samples into low- (Gleason Score (GS)<7(4+3)) and high-grade (GS>7(4+3) groups and observed that miR-130b expression was consistently higher in AA compared to EA samples in both groups (Figure 1B). We validated the expression profiles in an independent sample cohort from TCGA PRAD and TCGAA (AA n=59; EA n=396) datasets (Figure 1C). A trend in miR-130b overexpression was observed in total (Figure 1C) and low-grade samples (Figure 1D, left panel) in AA compared to EA patients. However, significant miR-130b over-expression was observed in high grade AA compared to EA samples (Figure 1D, right panel). We also examined miR-130b expression in EA compared to AA cell lines. miR-130b was under-expressed in EA relative to AA cell lines (Figure 1E). These results confirm that miR-130b is overexpressed and can act as a contributive factor in aggressive AA PC.
Figure 1. Overview of the miRNA-130b expression pattern in AA and EA PC patient tissues.
(A) The comparison of miR-130b expression level between AA (n=36) and EA PC (n=57) patients in SFVAMC cohort (p<0.0001; Mann-Whitney U-test). (B) miR-130b expression level based on GS>7 (4+3) in SFVAMC cohort. p-value was calculated with Mann-Whitney U-test. (C) Dot plot of miR-130b expression levels in TCGA AA (n=59) compared to EA (n=396) PC tissues. (D) miR-130b expression level based on GS>7 (4+3) in TCGA patient. p-value was calculated with Mann-Whitney U-test. (E) miR-130b expression in PC cells (n=3) compared to MDA-PCa-2b cell. (F) Overall survival of AA and (G) EA patients from the SFVAMC cohort.
miR-130b expression is a risk factor for poor prognosis in AA PC patients
To determine the clinical significance of miR-130b in AA PC, we analyzed the correlation between miR-130b expression and overall survival time (OS) in the SFVAMC patient cohort. Kaplan-Meier survival curve analysis showed that higher miR-130b is expression significantly correlated (p=0.003) with poor OS in AA patients (Figure 1F). For EA patients, miR-130b expression was not associated with overall survival (Figure 1G). Univariate (p=0.0076) and multivariate (p=0.0078) analyses revealed miR-130b expression to be an independent risk factor linked to poor prognosis in the SFVAMC AA PC patient cohort (Table 1, upper panel). However, this association was not observed in EA patient cohort (Table 1, lower panel). These results confirm that miR-130b overexpression is a risk factor for poor prognosis in AA PC patients.
Table 1.
Univariate and multivariate analyses of factors for predicting overall survival in AA prostate cancer patients.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P value | HR | 95% CI | P value |
| miRNA expression (High vs Low) | 7.6 | 1.7–35.3 | 0.0076 | 22.4 | 2.3–222.4 | 0.0078 |
| Age (≥62 vs <62 years) | 2.5 | 0.8–7.5 | 0.1 | 0.2 | 0.03–1.6 | 0.1 |
| T classification (T3/T4 versus T2) | 1.6 | 0.3–4.2 | 0.81 | 8.9 | 0.9–85.6 | 0.057 |
| PSA (<10 vs ≥10) | 1.6 | 0.5–4.9 | 0.37 | 0.02 | 0.0006–1.0 | 0.048 |
| PSA failure (Positive vs Negative) | 1.4 | 0.4–4.1 | 0.59 | 0.7 | 0.10–4.7 | 0.68 |
| Gleason Score (8–10 vs 5–7) | 2.2 | 0.8–6.3 | 0.1 | 0.44 | 0.04–4.6 | 0.49 |
| Univariate and multivariate analyses of factors for predicting overall survival in EA prostate cancer patients. | ||||||
| Univariate analysis | Multivariate analysis | |||||
| Variables | HR | 95% CI | P value | HR | 95% CI | P value |
| miRNA expression (High vs Low) | 1.9 | 0.6–5.5 | 0.25 | 1.1 | 0.1–8.3 | 0.91 |
| Age (≥62 vs <62 years) | 0.8 | 0.3–2.5 | 0.59 | 0.5 | 0.1–2.2 | 0.34 |
| T classification (T3/T4 versus T2) | 4.2 | 1.1–16.0 | 0.036 | 2.0 | 0.2–17.0 | 0.53 |
| PSA (<10 vs ≥10) | 2.1 | 0.7–6.0 | 0.17 | 1.7 | 0.2–13.3 | 0.45 |
| PSA failure (Positive vs Negative) | 4.7 | 1.3–17.7 | 0.02 | 2.3 | 0.2–19.1 | 0.45 |
| Gleason Score (8–10 vs 5–7) | 2.1 | 0.6–8.0 | 0.25 | 0.8 | 0.1–4.0 | 0.74 |
Inhibition of miR-130b suppresses cell proliferation, colony formation, migration, invasion and induces cell cycle arrest in AA cells
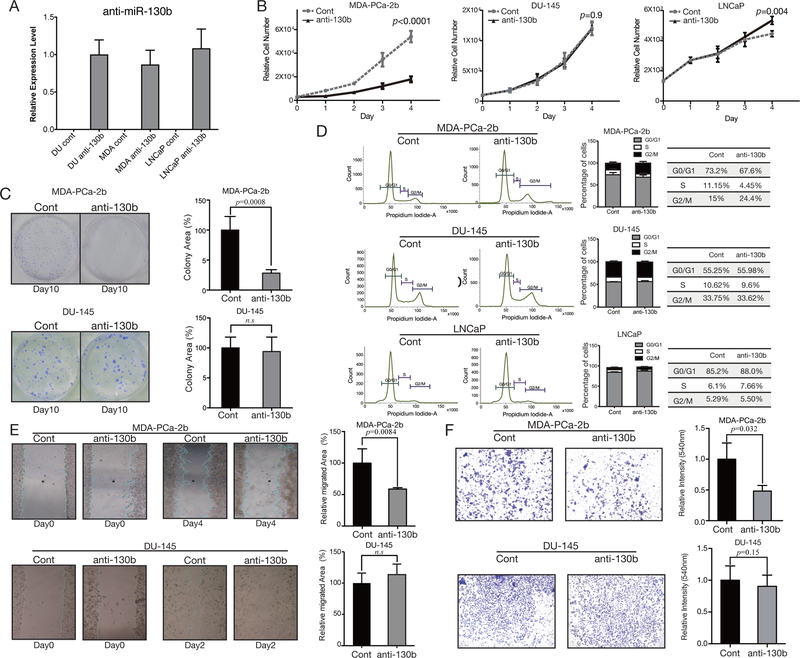
To determine the functional significance of inhibition of miR-130b expression, DU-145, LNCaP (EA cell lines) and MDA-PCa-2b (AA cell line) were transfected with either anti-miR-130b or control lentiviral vector. After establishing anti-miR-130b stable clones in all cell lines, we confirmed anti-miR-130b expression levels by Custom TaqMan qPCR analysis (Figure 2A). The efficiency of anti-miR-130b in all cell line stable clones was similar (Figure 2A). Next, these stable anti-miR-130b cells were used for a series of in vitro assays. miR-130b inhibition reduced cell proliferation exclusively in MDA-PCa-2b (Figure 2B) in comparison to controls, whereas no such effect was observed in the DU-145 and LNCaP cell lines (Figure 2B). Similarly, colony formation assay revealed that MDA-PCa-2b cells stably expressing anti-miR-130b resulted in significantly less colony area (p=0.0008) compared to control (Figure 2C, upper panel). This effect was not observed in DU-145 stable cells (Figure 2C, lower panel).
Figure 2. Functional analyses of anti-miR-130b stable PC cells.
(A) Anti-miR-130b expression level in established stable PC cells (n=3). (B) Proliferation of anti-miR-130b stable MDA-PCa-2b (left), DU-145 (middle) and LNCaP (right panel) cells. The p-values were calculated with the paired t-test (n=3). (C) Bar plots and images of colony formation assay using miR-130b stable DU-145 and MDA-PCa-2b. All plots show the mean of three independent assays (n=3). All p-values were calculated with the paired t-test. (D) Flow cytometry with anti-miR-130b stable MDA-PCa-2b, DU-145 and LNCaP cells. Each table beside the graph shows the percentage of cells in each cell cycle phase (n=3). (E) Wound-healing assay with chamber inserts using anti-miR-130b stable MDA-PCa-2b (n=6) and DU-145 (n=8) cells. All plots show the mean of three independent assays. All p-values were calculated with the paired t-test. (F) Invasion assay with anti-miR-130b stable MDA-PCa-2b and DU-145 cells. All plots show the mean of three independent assays and error indicates mean±SEM (n=6). All p-values were calculated with the paired t-test.
FACS cell cycle analysis showed no change in DU-145 and LNCaP cells between miR-130b knock-down and control cell (Figure 2D). However, G2/ M arrest was induced in stable MDA-PCa-2b anti-miR-130b cells (Controls 15% vs anti-miR-130b cells 24.4%, p = 0.001) (Figure 2D). Wound healing migration and chemotactic invasion assays revealed that miR-130b inhibition decreased migration (p=0.0084; Figure 2E, upper panel) and invasion (p=0.032; Figure 2F, upper panel) in stable anti-miR-130b MDA-PCa-2b cells These effects were not observed in DU-145 stable cells (Figure 2E, lower panel; Figure 2F, lower panel).
miR-130b expression is inversely correlated with cell cycle related gene expression in AA patients
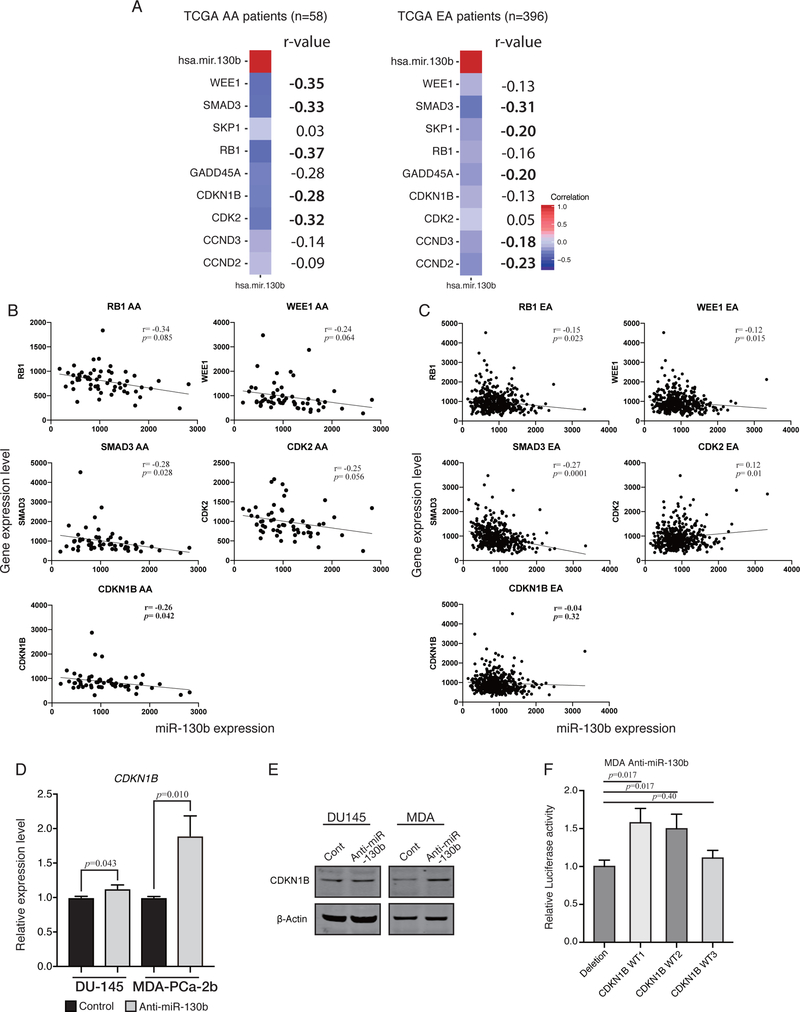
Since miR-130b inhibition increased G2/M arrest in AA PCa cells, we analyzed the correlation between miR-130b and cell cycle related gene expression by Pearson’s r correlation in TCGA PC samples. We used R package, “Hmisc” (23) to calculate the matrix of Pearson’s r correlation for all possible pairs of patient gene expression levels (Supplementary Figure 2). We arranged the r-value in decending order. Figure 3A lists the 9 genes that showed the highest negative correlation with miR-130b expression. These r-value are based on all possible pairs of patients’ gene expression. Next, we determined the correlation of the five topmost negatively correlated genes in AA patients individually (Figure 3B). The same genes were then correlated in EA patients individually (Figure 3C). Based on these computational analyses, we observed that CDKN1B showed the most inverse correlation (r=0.26, p=0.04) with miR-130b expression only in AA patients. However, other genes were more or less similar in both races and hence may not be race related. We also confirmed CDKN1B up-regulation by qPCR and Western blot analyses (Figure 3D and E). To confirm that CDKN1B is direct target of miR-130b, we performed luciferase reporter assays. Two out of three reporter vectors harboring the miR-130b site showed that luciferase activities were up-regulated in miR-130b stable knock-down AA cells compared to deletion vector transfected cells (Figure 3F).
Figure 3. Pearson’s r correlation analysis showing that miR-130b expression affects cell cycle related genes in TCGA PRAD dataset (AA, n=59; EA, n=396).
(A) Heatmap of Pearson’s r correlation for the five most negatively correlated genes in AA (RB1, WEE1, SMAD3, CDKN1B and GADD45A) and EA (SMAD3, CCND2, SKP1, GADD45A and CCND3). Pearson’s r correlation was computed using all possible pairs of patient’s gene expression level for each gene. The number on the right of each heatmap indicates r-value (five topmost gene are shown in bold). (B) Representative dot plot analysis of five most negatively correlated genes in AA. Pearson’s r correlation was computed using all individual patient’s gene expression level for each gene. Five plots show the results from TCGA AA patients and (C) the other five plots show TCGA EA patients. (D) qPCR analysis for CDKN1B expression level in anti-miR-130b stable EA derived DU-145 cells and AA cells, MDA-PCa-2b. (paired t-test; n=3) (E) Western blotting for CDKN1B in anti-miR-130b stable EA derived cell DU-145 and AA cell, MDA-PCa-2b. (F) Luciferase reporter assay with three different vectors harboring miR-130b binding site. Each CDKN1B WT1/ WT2/ WT3 was from a different miRNA target database, miRWalk, IntaRNA and RNAhybrid, respectively (paired t-test; n=3).
Inhibition of miR-130b expression modulates critical cancer related biological pathways
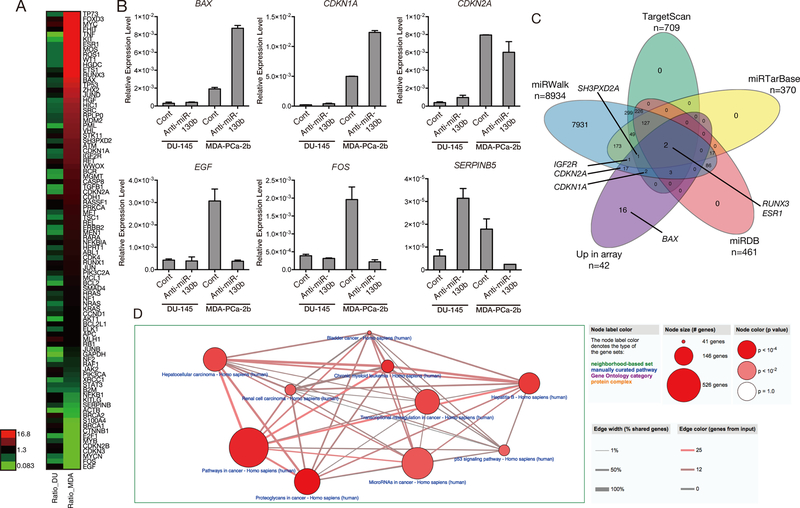
We conducted gene expression array analysis using a commercial assay containing oncogenic and tumor suppressive gene sets and compared gene expression patterns in anti-miR-130b stable cell lines to determine which biological pathways are involved in PC racial disparity (Figure 4A). These gene sets were curated by the manufacturer and represent oncogenic and tumor suppressor genes on the qPCR array plate. We randomly selected three up- and down-regulated genes from the array data to validate by qRT-PCR analysis. The qRT-PCR results were consistent with the array data and showed that the expression of these genes is modulated by miR-130b inhibition in MDA-PCa-2b cells (Figure 4B). Up-regulated genes were compared to predicted and validated miRNA targets by miRwalk, miRDB, TargetScan, and miRTarBase. Approximately 62% of up-regulated genes were predicted by at least one database (Figure 4C;26 out of 42). The genes that we validated by qRT-PCR analysis such as IGF2R, CDKN2A and CDKN1A were overlapped in miRWalk and TargetScan (Figure 4C). The other two, IGF2R and SH3PXD2A were validated by miRTarBase (Figure 4C). According to the Over-Representative Analysis with CPDB, 22 pathways were changed with miR-130b inhibition in MDA-PCa-2b cells compared to that of DU-145 and represent the top 10 pathways summarized in Figure 4D and Supplementary Table 5. In agreement with our functional assays, miR-130b significantly affected central cancer-related gene pathways, “Pathway in Cancer” (p= 0.0002), “p53 signaling pathway” (p= 0.002) and “miRNA in Cancer” (p= 0.002) (Supplementary Table 5). Focusing on single genes, we found that 16 genes were up-regulated more than four times and five genes were decreased by half in MDA-PCa-2b cells. However, these same genes were less affected in DU-145 compared to MDA-PCa-2b cells (Supplementary Table 6).
Figure 4. Potential miR-130b target genes by qPCR array.
(A) Heatmap analysis of the results of qPCR array with tumor suppressor and oncogenic genes. (B) Validation of array results by qPCR (n=3) (C) Venn diagram depicting the union, intersections and complements of predicted and/or validated genes in four different databases, miRWalk (blue), TargetScan (green), miRTarBase (yellow), miRDB (red) and up-regulated genes in qPCR array (purple). The representative genes in each group are given in the diagram. (D) The representative biological pathways from CPDB gene Over-Representative Analysis. These pathways were particularly affected by miR-130b inhibition in stable MDA-PCa-2b cell cells compared to DU-145 cells. The size of the node shows the total number of genes contained in the pathway. The width of the edge between nodes depicts the total number of overlapping genes between the pathways linked by nodes. The color density of the edge shows the number of genes contained in our input gene list.
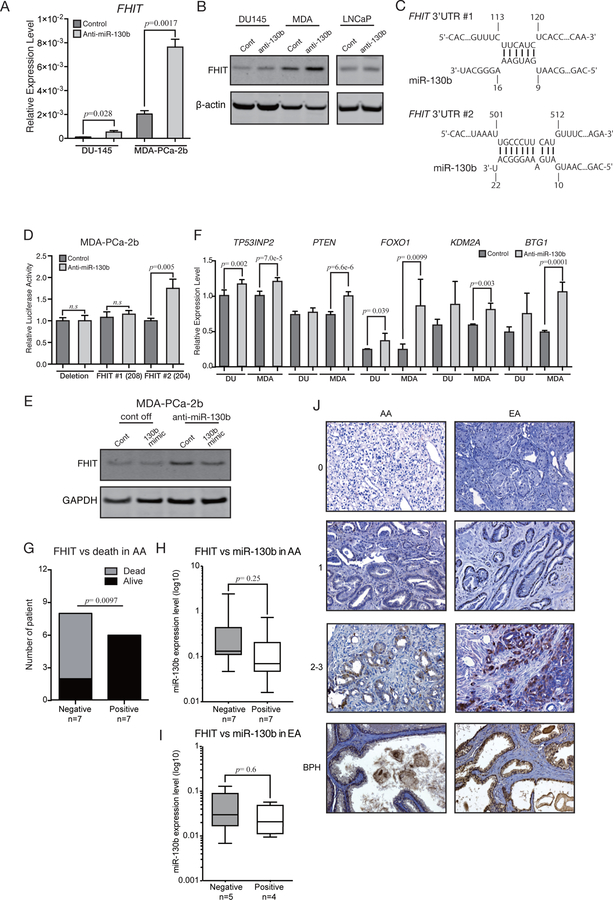
miR-130b directly binds to FHIT 3’-UTR and decreases its expression in AA PC cells
Based on the tumor-suppressive function of miR-130b inhibition, we focused our search for tumor-suppressor genes. Fragile Histidine Triad (FHIT) gene was the fourth-ranked significantly altered gene in our qPCR array results (Figure 4A). We confirmed the array results by qRT-PCR (Figure 5A). This gene is a tumor suppressor and is located at a fragile region in the human genome (24–26). We observed significant up-regulation of FHIT protein expression that was specific to anti-miR-130b stable MDA-PCa-2b cells as compared to DU-145 or LNCaP stable PC cells (Figure 5B). An in-silico algorithm (IntaRNA) showed two predicted miR-130b binding sites in the FHIT 3’UTR. We inserted both binding sites (Figure 5C) into pmiR-Glo (FHIT 3’UTR#1; FHIT 3’UTR#2) and transfected them into anti-miR-130b stable MDA-PCa-2b cells. The luciferase reporter activity confirmed the FHIT gene to be a direct target of miR-130b in MDA-PCa-2b cells (Figure 5D). Among the two binding sites, FHIT 3’UTR#2 was responsible for miR-130b binding as we observed that the luciferase activity of this binding site was high in anti-miR-130b stable MDA-PCa-2b cells. However, the FHIT 3’UTR#1 site did not show any changes (Figure 5D). Rescuing miR-130b expression using miR-130b mimic in anti-miR-130b stable MDA-PCa-2b cells resulted in FHIT down-regulation (Figure 5E). miR-130b expression levels after miR-130b mimic transfection are shown in Supplemental Figure 3A. We also randomly selected other miR-130b target genes from the validated or predicted miR-130b target gene list for qRT-PCR analysis and all these target genes were up-regulated in the anti-miR-130b stable MDA-PCa-2b cells compared to controls (Figure 5F). TP53INP2 (13) and PTEN, (27,28) reported miR-130b targets, were also up-regulated in our anti-miR-130b stable cells. FOXO1, KDM2A and BTG1 that are predicted miR-130b targets by in silico algorithm miRWalk were also confirmed to be up-regulated in our anti-miR-130b stable clones (Figure 5F). PCR array data showed another cell cycle regulator, CDKN1A as differentially expressed in anti-miR-130b stable MDA-PCa-2b cells compared to DU-145 cells. However, the results of protein expression by Western blots were inconsistent (Supplemental Figure 3B). Hence, we did not purse CDKN1A further. Similarly, RUNX3 and ESR were up-regulated in anti-miR-130b stable MDA cells in array data and these genes are predicted as miR-130b target in four prediction databases. Nevertheless, qPCR could not detect ESR expression in both cells. RUNX3 mRNA levels were increased in both DU-145 and MDA-PCa-2b anti-miR-130b stable cells (Supplemental Figure 3C) and hence appeared not to be race related. RUNX3 has been reported as a miR-130b target in gastric and ovarian cancers (29,30). These data reveal that among all the PCR array data predicted targets, only FHIT is a direct functional target of miR-130b relevant to AA race related PC.
Figure 5. miR-130b affects multiple pathways related to tumor metastasis in AA patients.
(A) qPCR validation for FHIT mRNA levels in stable anti-miR-130b cells (paired t-test; n=3). (B) FHIT Western blotting with stable anti-miR-130b PC cells. (C) FHIT binding sites for miR-130b predicted by IntaRNA. The numbers shown in the panel indicate the position from the 3’UTR start sites. (D) Luciferase reporter assay using vectors containing FHIT binding site or deletion site (paired t-test; n=3). (E) miR-130b rescue experiments to validate that FHIT up-regulation is due to knockdown of miR-130b (cont off, control vector stable MDA-PCa-2b cells; anti-miR-130b, anti-miR-130b stable MDA-PCa-2b cells; cont, cells transfected with negative control #1; 130b mimic, cells transfected with miR-130b mimic). (F) qPCR analysis for reported and predicted miR-130b target genes. RNA samples were isolated from two independent batches of control and anti-miR-130b stable DU-145/ MDA-PCa-2b cells. Then each triplicate well was amplified and measured by qPCR. Each of six expression values were compared by paired t-test (Error bar; mean±SD, n=3). (G) The results of Fisher’s exact test analyzing independency between FHIT expression and SFVAMC AA PC patient death (n=14). (H) The boxplot shows the miR-130b expression level in FHIT negative and positive SFVAMC AA and (I) EA patients. (J) Representative FHIT IHC results in SFVAMC AA, EA patients and BPH tissues. The numbers shown left of the images indicate staining intensity scores.
FHIT expression levels are related to AA PC patient death
We sought to determine the correlation of FHIT with AA and EA PC patient survival. We randomly selected a small sample number for IHC staining (AA=14, EA=10) from the same SFVAMC cohort that we used for expression analysis in Figure 1A and 1B. FHIT staining was observed mostly in the cytoplasm and inside nuclei of glandular epithelial cells. BPH samples from AA and EA tissues showed positive FHIT expression levels (2–3; Figure 5I, bottom panels). We classified FHIT expression levels into groups, 0 = negative, 1, 2–3= positive by Allred scoring and performed Fisher’s exact test to determine correlation of FHIT expression with patient death (Figure 5G). We observed a significant correlation between negative FHIT expression and AA patient death (Figure 5G) as there were more deceased patients with negative FHIT expression compared to AA patients still alive (Figure 5G). Further, we correlated FHIT protein expression in the same cohort in both AA and EA groups. IHC results indicated that miR-130b expression levels show an inverse trend with that of FHIT expression (Figure 5H). In EA, miR-130b expression was similar in negative and positive FHIT stained samples (Figure 5I). However, this needs to be confirmed in a large tissue cohort. Nevertheless, these data show that loss of FHIT is a risk factor in AA patients.
Discussion
In this study, for the first time, we identified miR-130b to be a significant contributor to PC racial disparity in AA men. miR-130b expression levels were higher in AA prostate cancer tissues in both TCGA and SFVAMC cohorts as well as in the AA-derived cell line, MDA-PCa-2b when compared to EA PC tissues and cells. miR-130b (chr22: 21653304–21653385) forms a cluster with miR-301b (chr22: 21652981–21653058; <10kb apart) and these two miRNAs harbor the same seed sequence. We compared the expression of miR-301b between AA and EA tissues in TCGA. There was no difference in expression of this miRNA between the two races. (Supplemental Figure 4A). This indicates that miR-130b may be the dominant miRNA than miR-301b that is related to racial disparity in AA patients. According to the UCSC genome browser, chromatin modification at the suspected miR-130b promoter region is more accessible to binding transcription factors in PC cells compared to normal prostate epithelial cells (PrEC) (15). Another study, not related to race, has reported that c-Met upregulated miR-130b expression and contributed to cancer progression and castration resistance through down-regulation of AR expression (14). However, we observed that the MET mRNA level is slightly lower in AA compared to EA PC patients according to the TCGA cohort (Supplemental Figure 4B). Hence it might not be the reason for miR-130b over-expression in AA patients. Meanwhile, the cell viability results in this report are consistent with ours. Taken together, miR-130b over-expression may be the result of open chromatin status in human PC leading to abnormal over-expression of miR-130b rather than MET driven regulation. A previously published study also showed that miR-130b tended to be up-regulated in a small sample of AA PC patients (22). However, our study is the first study to conclusively demonstrate the involvement of miR-130b in the AA PC health disparity. Clinically, we observed that elevated miR-130b expression positively correlated with poor overall survival of AA patients. Multivariate analysis also demonstrated that miR-130b independently serves as a prognostic marker for overall survival with increased significance (p=0.008) compared to PSA levels (p=0.05) for AA patients. Currently, clinicians are seeking non-invasive prognostic and diagnostic biomarkers for early PC detection. In nephroblastoma and hepatocellular cancer, high expression of miR-130b has been detected in patient serum and showed high diagnostic potential (31,32). The expression level of serum miR-130b was related to chemo-sensitivity and -resistance in colorectal cancer patients (33). In the future, we plan to analyze the potential for miR-130b to be used as a non-invasive biomarker in PC racial disparity.
Another analysis showed that inhibition of miR-130b caused a drastic change in 42 genes and differential gene expression patterns related to human cancer pathways in MDA-PCa-2b cells compared to DU-145 cells. Given that miR-130b caused different responses in a race-specific manner, there may be race-specific mutations within the miRNA binding site or differences in the basal expression level of miRNA target genes between the two races. As the qPCR array and its validation showed, this study supports the latter hypothesis, taking into account that most of the genes with altered expression in the array were in anti-miR-130b stable MDA-PCa-2b cells, whereas the majority of genes were not affected in anti-miR-130b stable DU-145 cells. Basal gene expression differences between DU-145 and MDA-PCa-2b control cells might generate distinct biological pathway activation by miR-130b inhibition in the two different cells.
We found that FHIT and CDKN1B, known as haplo-insufficient tumor suppressors (24,34,35), were significantly up-regulated in MDA-PCa-2b cells upon miR-130b inhibition and luciferase reporter assay showed that both genes are a direct miR-130b target. A previous study found that stimulation of CDKN1B caused G2 arrest by repressing cyclin D3 (36). In non–small cell lung cancer (NSCLC), SIRT1 silencing suppresses proliferation and induces senescence in a p27Kip1-dependent manner (37). In regard to the clinical relevance of CDKN1B, less CDKN1B prostate expression was an independent predictor of adverse outcomes for patients with local PC (38). CDKN1B expression levels also inversely correlated with BCR-free survival in those PCa patients who underwent radical prostatectomy with pathologic organ-confined disease (39). Furthermore, SKP2 gene negatively regulates CDKN1B. However, in AA patients, 46.3% showed lower expression of CDKN1B independent of SKP2 (40). Based on this report, we infer that CDKN1B might be sub-dominantly regulated by miR-130b in AA patients. Our analysis selected five cell cycle related genes that were correlated with PC disparity in AA patients. Among them, CDKN1B, a key cell cycle gene, was inversely correlated with miR-130b expression in TCGA AA patients while EA samples did not show any correlation. Consistent with this result, miR-130b inhibition in AA PC cells caused up-regulation of CDKN1B expression. These results suggest that the cell cycle pathway may be important prostate cancer racial disparity in AA PC patients. Further, FHIT is a tumor-suppressor gene which resides at a fragile chromosome site, FRA3B, and its genetic alterations have been observed in many types of human cancers (41). In human carcinoma, aberrant FHIT transcripts have been frequently observed in lung, esophageal and gastrointestinal carcinomas (42,43). Deficiency of FHIT contributed to tumorigenesis by N-nitrosomethylbenzylamine in murine gastric cancer (24,26). In prostate cancer, down-regulation and hyper-methylation of FHIT (44,45) along with its therapeutic intervention potential in human prostate cancer has been reported (44). We validated FHIT regulation via miR-130b by inhibition or rescuing of miR-130b expression. Our study suggested that miR-130b mediated down-regulation of FHIT induces malignant tumors in AA compared to EA patients. It was reported that FHIT up-regulated miR-30c and negatively controlled metastasis by modulating EMT genes in lung cancer (46). Similarly, FHIT down-regulation caused by miR-130b may mediate PC metastasis in AA PC patients. This is supported by the fact that suppression of miR-130b specifically inhibited tumorigenic characteristics such as cell migration and invasion in AA cells compared to EA cells. Thus, this is the first report indicating that FHIT may be involved in prostate cancer racial disparity.
This study documents the clinical significance of miR-130b in AA PC. Over-expression of miR-130b was observed to contribute to poor prognosis in AA patients. Understanding the relationships between miRNA expression and downstream regulation of mRNA targets in the AA population is important to comprehend the biological basis for racial health disparity in PC. Inhibition of miR-130b and rescue of FHIT and CDKN1B, may provide a novel therapeutic approach for the treatment of PC in AA patients. Furthermore, miR-130b might be a potential prognostic biomarker to predict mortality risk in AA PC patients.
Supplementary Material
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript.
Funding
This work was supported by the National Cancer Institute at the National Institutes of Health through grant numbers U01CA184966. Dr. Dahiya is the recipient of a Senior Research Career Scientist Award (# 1K6BX004473) from the Department of Veterans Affairs.
Footnotes
Conflicts of interest
There is no conflict of interest to disclose.
References
- 1.DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin 2019;69(3):211–33 doi 10.3322/caac.21555. [DOI] [PubMed] [Google Scholar]
- 2.Powell IJ, Bock CH, Ruterbusch JJ, Sakr W. Evidence supports a faster growth rate and/or earlier transformation to clinically significant prostate cancer in black than in white American men, and influences racial progression and mortality disparity. J Urol 2010;183(5):1792–6 doi 10.1016/j.juro.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundi D, Kryvenko ON, Carter HB, Ross AE, Epstein JI, Schaeffer EM. Pathological examination of radical prostatectomy specimens in men with very low risk disease at biopsy reveals distinct zonal distribution of cancer in black American men. J Urol 2014;191(1):60–7 doi 10.1016/j.juro.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byers TE, Wolf HJ, Bauer KR, Bolick-Aldrich S, Chen VW, Finch JL, et al. The impact of socioeconomic status on survival after cancer in the United States : findings from the National Program of Cancer Registries Patterns of Care Study. Cancer 2008;113(3):582–91 doi 10.1002/cncr.23567. [DOI] [PubMed] [Google Scholar]
- 5.DeRouen MC, Schupp CW, Koo J, Yang J, Hertz A, Shariff-Marco S, et al. Impact of individual and neighborhood factors on disparities in prostate cancer survival. Cancer Epidemiol 2018;53:1–11 doi 10.1016/j.canep.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbins AS, Whittemore AS, Thom DH. Differences in Socioeconomic Status and Survival among White and Black Men with Prostate Cancer. American Journal of Epidemiology 2000;151:409–16 doi 10.1093/oxfordjournals.aje.a010221. [DOI] [PubMed] [Google Scholar]
- 7.Krieger N, Quesenberry C Jr., Peng T, Horn-Ross P, Stewart S, Brown S, et al. Social class, race/ethnicity, and incidence of breast, cervix, colon, lung, and prostate cancer among Asian, Black, Hispanic, and White residents of the San Francisco Bay Area, 1988–92 (United States). Cancer Causes Control 1999;10(6):525–37. [DOI] [PubMed] [Google Scholar]
- 8.Chu DI, Moreira DM, Gerber L, Presti JC Jr., Aronson WJ, Terris MK, et al. Effect of race and socioeconomic status on surgical margins and biochemical outcomes in an equal-access health care setting: results from the Shared Equal Access Regional Cancer Hospital (SEARCH) database. Cancer 2012;118(20):4999–5007 doi 10.1002/cncr.27456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everist MM, Howard LE, Aronson WJ, Kane CJ, Amling CL, Cooperberg MR, et al. Socioeconomic status, race, and long-term outcomes after radical prostatectomy in an equal access health system: Results from the SEARCH database. Urol Oncol 2019;37(4):289 e11–e17 doi 10.1016/j.urolonc.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16(3):203–22 doi 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 11.Bhagirath D, Yang TL, Bucay N, Sekhon K, Majid S, Shahryari V, et al. microRNA-1246 Is an Exosomal Biomarker for Aggressive Prostate Cancer. Cancer Res 2018;78(7):1833–44 doi 10.1158/0008-5472.CAN-17-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng G Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev 2015;81:75–93 doi 10.1016/j.addr.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, et al. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 2010;7(6):694–707 doi 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Cannistraci A, Federici G, Addario A, Di Pace AL, Grassi L, Muto G, et al. C-Met/miR-130b axis as novel mechanism and biomarker for castration resistance state acquisition. Oncogene 2017;36(26):3718–28 doi 10.1038/onc.2016.505. [DOI] [PubMed] [Google Scholar]
- 15.Fort RS, Matho C, Oliveira-Rizzo C, Garat B, Sotelo-Silveira JR, Duhagon MA. An integrated view of the role of miR-130b/301b miRNA cluster in prostate cancer. Exp Hematol Oncol 2018;7:10 doi 10.1186/s40164-018-0102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto Y, Shiina M, Kato T, Yamamura S, Tanaka Y, Majid S, et al. The role of miR-24 as a race related genetic factor in prostate cancer. Oncotarget 2017;8(10):16581–93 doi 10.18632/oncotarget.15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, Pi J, et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell 2018;34(4):549–60e9 doi 10.1016/j.ccell.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One 2014;9(3):e92444 doi 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 2012;8(2):e1002375 doi 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanda Y Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant 2013;48(3):452–8 doi 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiina M, Hashimoto Y, Kato T, Yamamura S, Tanaka Y, Majid S, et al. Differential expression of miR-34b and androgen receptor pathway regulate prostate cancer aggressiveness between African-Americans and Caucasians. Oncotarget 2017;8(5):8356–68 doi 10.18632/oncotarget.14198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang BD, Ceniccola K, Yang Q, Andrawis R, Patel V, Ji Y, et al. Identification and Functional Validation of Reciprocal microRNA-mRNA Pairings in African American Prostate Cancer Disparities. Clin Cancer Res 2015;21(21):4970–84 doi 10.1158/1078-0432.CCR-14-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrell FE Jr. Package ‘Hmisc’: version 4.2–0. 2019. [Google Scholar]
- 24.Fong LYY, Fidanza V, Zanesi N, Lock LF, Siracusa LD, Mancini R, et al. Muir – Torre-like syndrome in Fhit-deficient mice. Proceedings of the National Academy of Sciences 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paisie CA, Schrock MS, Karras JR, Zhang J, Miuma S, Ouda IM, et al. Exome-wide single-base substitutions in tissues and derived cell lines of the constitutive Fhit knockout mouse. Cancer Sci 2016;107(4):528–35 doi 10.1111/cas.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanesi N, Fidanza V, Fong LY, Mancini R, Druck T, Valtieri M, et al. The tumor spectrum in FHIT-deficient mice. Proc Natl Acad Sci U S A 2001;98(18):10250–5 doi 10.1073/pnas.191345898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu T, Cao R, Li S, Fu M, Ren L, Chen W, et al. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer 2015;15:29 doi 10.1186/s12885-015-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Zhang B, Sun L, Yan Q, Zhang Y, Zhang Z, et al. MicroRNA-130b targets PTEN to induce resistance to cisplatin in lung cancer cells by activating Wnt/beta-catenin pathway. Cell Biochem Funct 2018;36(4):194–202 doi 10.1002/cbf.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paudel D, Zhou W, Ouyang Y, Dong S, Huang Q, Giri R, et al. MicroRNA-130b functions as a tumor suppressor by regulating RUNX3 in epithelial ovarian cancer. Gene 2016;586(1):48–55 doi 10.1016/j.gene.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Lai KW, Koh KX, Loh M, Tada K, Subramaniam MM, Lim XY, et al. MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer 2010;46(8):1456–63 doi 10.1016/j.ejca.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig N, Nourkami-Tutdibi N, Backes C, Lenhof HP, Graf N, Keller A, et al. Circulating serum miRNAs as potential biomarkers for nephroblastoma. Pediatr Blood Cancer 2015;62(8):1360–7 doi 10.1002/pbc.25481. [DOI] [PubMed] [Google Scholar]
- 32.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open 2012;2(2):e000825 doi 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Zhang K, Bi M, Jiao X, Zhang D, Dong Q. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer Drugs 2014;25(3):346–52 doi 10.1097/CAD.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 34.Payne SR, Zhang S, Tsuchiya K, Moser R, Gurley KE, Longton G, et al. p27kip1 deficiency impairs G2/M arrest in response to DNA damage, leading to an increase in genetic instability. Mol Cell Biol 2008;28(1):258–68 doi 10.1128/MCB.01536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fero ML, Randel E, Gurley KE, Roberts JM, Kemp CJ. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature 1998;396(6707):177–80 doi 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaharieva MM, Kirilov M, Chai M, Berger SM, Konstantinov S, Berger MR. Reduced expression of the retinoblastoma protein shows that the related signaling pathway is essential for mediating the antineoplastic activity of erufosine. PLoS One 2014;9(7):e100950 doi 10.1371/journal.pone.0100950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu L, Chiao CY, Enzer KG, Stankiewicz AJ, Faller DV, Dai Y. SIRT1 inactivation evokes antitumor activities in NSCLC through the tumor suppressor p27. Mol Cancer Res 2015;13(1):41–9 doi 10.1158/1541-7786.MCR-14-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang RM, Naitoh J, Murphy M, Wang HJ, Phillipson J, deKernion JB, et al. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol 1998;159(3):941–5. [PubMed] [Google Scholar]
- 39.Freedland SJ, de Gregorio F, Sacoolidge JC, Elshimali YI, Csathy GS, Elashoff DA, et al. Predicting biochemical recurrence after radical prostatectomy for patients with organ-confined disease using p27 expression. Urology 2003;61(6):1187–92 doi 10.1016/s0090-4295(03)00034-7. [DOI] [PubMed] [Google Scholar]
- 40.Drobnjak M, Melamed J, Taneja S, Melzer K, Wieczorek R, Levinson B, et al. Altered expression of p27 and Skp2 proteins in prostate cancer of African-American patients. Clin Cancer Res 2003;9(7):2613–9. [PubMed] [Google Scholar]
- 41.Croce CM, Sozzi G, Huebner K. Role of FHIT in human cancer. J Clin Oncol 1999;17(5):1618–24 doi 10.1200/JCO.1999.17.5.1618. [DOI] [PubMed] [Google Scholar]
- 42.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 1996;84(4):587–97. [DOI] [PubMed] [Google Scholar]
- 43.Sozzi G, Veronese ML, Negrini M, Baffa R, Cotticelli MG, Inoue H, et al. The FHIT gene at 3p14.2 is abnormal in lung cancer. Cell 1996;85:17–26 doi 10.1016/S0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- 44.Fouts RL, Sandusky GE, Zhang S, Eckert GJ, Koch MO, Ulbright TM, et al. Down-regulation of fragile histidine triad expression in prostate carcinoma. Cancer 2003;97(6):1447–52 doi 10.1002/cncr.11201. [DOI] [PubMed] [Google Scholar]
- 45.Maruyama R, Toyooka S, Toyooka KO, Virmani AK, Zochbauer-Muller S, Farinas AJ, et al. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin Cancer Res 2002;8(2):514–9. [PubMed] [Google Scholar]
- 46.Suh SS, Yoo JY, Cui R, Kaur B, Huebner K, Lee TK, et al. FHIT suppresses epithelial-mesenchymal transition (EMT) and metastasis in lung cancer through modulation of microRNAs. PLoS Genet 2014;10(10):e1004652 doi 10.1371/journal.pgen.1004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.