Abstract
Background
Obesity and its associated diseases are major health problems characterized by extensive metabolic disturbances. Understanding the causal connections between these phenotypes and variation in metabolite levels can uncover relevant biology and inform novel intervention strategies. Recent studies have combined metabolite profiling with genetic instrumental variable (IV) analysis (Mendelian randomization) to infer the direction of causality between metabolites and obesity, but often omitted a large portion of untargeted profiling data consisting of unknown, unidentified metabolite signals.
Methods
We expanded upon previous research by identifying body mass index (BMI)-associated metabolites in multiple untargeted metabolomics datasets, and then performing bidirectional IV analysis to classify metabolites based on their inferred causal relationships with BMI. Meta-analysis and pathway analysis of both known and unknown metabolites across datasets were enabled by our recently developed bioinformatics suite, PAIRUP-MS.
Results
We identified 10 known metabolites that are more likely to be causes (e.g. alpha-hydroxybutyrate) or effects (e.g. valine) of BMI, or may have more complex bidirectional cause-effect relationships with BMI (e.g. glycine). Importantly, we also identified about 5 times more unknown than known metabolites in each of these three categories. Pathway analysis incorporating both known and unknown metabolites prioritized 40 enriched (p < 0.05) metabolite sets for the cause versus effect groups, providing further support that these two metabolite groups are linked to obesity via distinct biological mechanisms.
Conclusions
These findings demonstrate the potential utility of our approach to uncover causal connections with obesity from untargeted metabolomics datasets. Combining genetically informed causal inference with the ability to map unknown metabolites across datasets provides a path to jointly analyze many untargeted datasets with obesity or other phenotypes. This approach, applied to larger datasets with genotype and untargeted metabolite data, should generate sufficient power for robust discovery and replication of causal biological connections between metabolites and various human diseases.
INTRODUCTION
Abnormal blood metabolite levels are important, frequent, and quantifiable features of obesity and its associated phenotypes, which are major health problems globally1–5. Recently, systematic metabolite profiling (metabolomics) studies have described widespread alterations in the obesity metabolome and identified metabolite markers associated with risk of obesity-related diseases6–10. However, these studies broadly have two key analytic challenges limiting the biological interpretation and scope of their findings: these correlative studies have not generally been able to distinguish the cause and effect relationships between metabolites and phenotypes, and only a portion of the thousands of metabolite signals measured by untargeted profiling technology could be chemically identified and thereby routinely investigated.
Genetic instrumental variable (IV) analysis (for causal inference) and novel bioinformatics tools (for analysis of untargeted metabolite data) now provide the means to overcome these limitations and enhance our understanding of the metabolome of any phenotype. The genetic IV framework, also known as Mendelian randomization, uses genetic variants as instruments to infer causality from observational data in the presence of unmeasured confounding, provided certain methodological assumptions are met11,12. Bidirectional genetic IV analysis, using in turn genetic variants affecting metabolite levels and variants affecting a phenotype such as body mass index (BMI), offers a way to ascribe directionality of causal relationships and to prioritize potentially causal metabolite-phenotype associations. Previous genetic IV studies have utilized variants identified in genome-wide association studies (GWAS) to infer causality between obesity-related phenotypes and curated sets of metabolites (e.g. branched-chain amino acids [BCAAs] and aromatic amino acids)13–17. However, most studies did not perform comprehensive bidirectional IV analysis to assess causality and only focused on the metabolites that could be identified and curated from profiling data, thus likely capturing only a limited slice of obesity biology.
Previously, metabolites of unknown chemical identities – a large portion of untargeted profiling data – were mostly excluded from analyses (including GWAS) because inter-study comparison and biological interpretation were technically onerous or intractable18,19. To address these issues, we recently developed a bioinformatics suite, PAIRUP-MS19, to match up unknown metabolites across mass spectrometry-based untargeted profiling datasets, thereby enabling meta-analysis of multiple datasets and increasing statistical power for detecting biologically interesting unknowns. In addition, PAIRUP-MS provides a framework for annotating unknown metabolites using preexisting metabolic pathways and performing pathway analysis incorporating both known and unknown metabolites.
In this study, we demonstrate how the combination of bidirectional genetic IV framework and PAIRUP-MS can be used to analyze multiple untargeted metabolomics datasets and characterize causal connections between a phenotype and the metabolome. We identified both known and unknown BMI-associated metabolites, and then performed GWAS for each metabolite and for BMI, followed by bidirectional genetic IV analysis to identify metabolites likely to be causes or effects of obesity. In addition, we highlighted distinct biological pathways enriched for the cause versus effect metabolites, confirming that the bidirectional IV approach prioritized two distinct sets of BMI-associated metabolites. This initial work illustrates an approach that can now be generalized and scaled up to much larger datasets, which will enable well-powered studies to uncover novel metabolic causes and effects of obesity or any other phenotype of interest.
MATERIALS AND METHODS
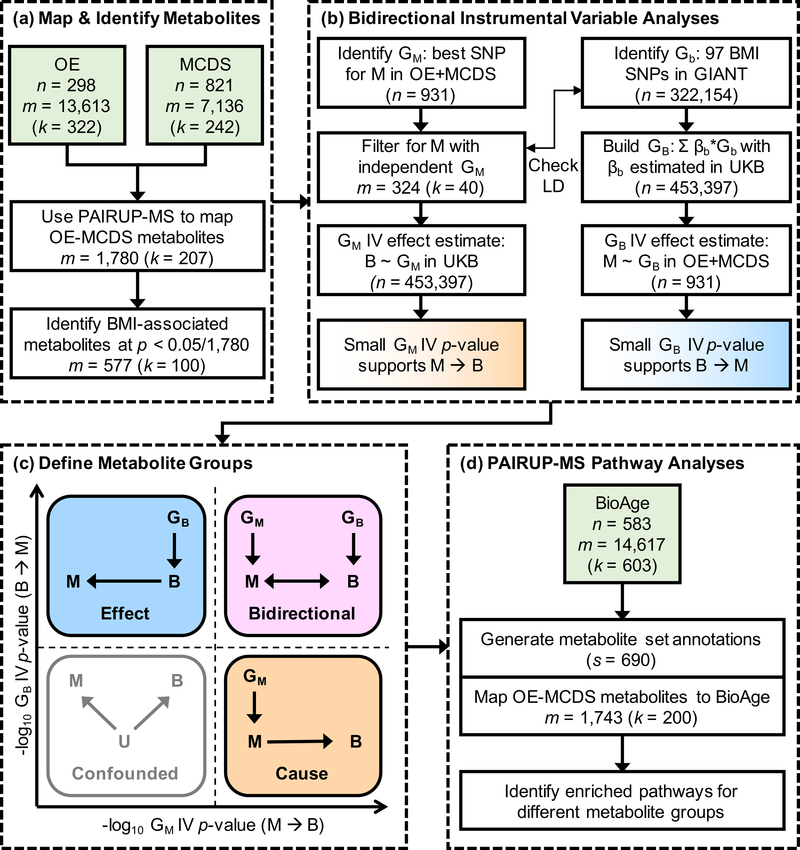
A schematic overview of our analysis plan is shown in Figure 1 and each step is described in more detail below. Supplementary Text 1 lists all supplementary materials referenced in this and all subsequent sections.
Figure 1. Overview for identifying and characterizing causal connections in the obesity metabolome.
(a) OE and MCDS metabolomics datasets, matched using PAIRUP-MS, were used to identify known and unknown metabolites associated with BMI. (b) Independent genetic instruments (GM) for the BMI-associated metabolites were selected using OE and MCDS data, and then used to test for a metabolite-to-BMI (M → B) causal effect in UKB; in parallel, BMI genetic instrument (GB), a polygenic risk score built using GIANT BMI-associated SNPs (Gb) and UKB effect estimate weights (βb), was used to test for a BMI-to-metabolite (B → M) causal effect in OE and MCDS. (c) A subset of metabolites was categorized into “cause”, “effect”, and “bidirectional” groups based on the magnitude of the evidence according to the GM and GB IV effect estimate p-values, reflecting different types of causal connections between the metabolites and BMI. (d) Pathway analyses of the three metabolite groups were performed using metabolite set annotations generated using PAIRUP-MS and an independent dataset (BioAge). Y ~ X, regression of Y on X; U, unmeasured confounder; n, number of samples; m, number of metabolites; k, number of known (or shared known) metabolites; s, number of metabolite sets.
Metabolomics datasets and data processing
Study populations
The study populations have been described previously19–21: (1) Obesity Extremes (OE): N = 300 sampled equally from lean, obese, and the general Estonian Biobank (EB) population, (2) Mexico City Diabetes Study (MCDS): N = 865 in a prospective study, and (3) BioAge Labs Mortality Study (BioAge): N = 583 in a retrospective mortality study nested in EB. All participants provided informed consent. Individual studies were approved by their respective local ethics committees. Boston Children’s Hospital Institutional Review Board approved this research.
Metabolite data processing
Untargeted liquid chromatography-mass spectrometry (LC-MS) profiling of plasma samples and subsequent log-transformation, normalization, quality control, and missing value imputation of the data have been described previously19. The processed OE dataset contains 298 samples and 13,613 metabolite signals (322 known); MCDS contains 821 samples and 7,136 signals (242 known); BioAge contains 583 samples and 14,617 signals (603 known). In order to derive easily interpretable metabolite abundance scores, within each dataset, we performed rank-based inverse normal transformation on each signal and used the resulting abundance z-scores in downstream analyses. For OE and MCDS data used in BMI and genetic association analyses, we performed covariate adjustment (age, sex, and fasting time for OE; age and sex for MCDS) before the transformation. In this paper, we refer to both known and unknown metabolite signals as “metabolites” for simplicity, recognizing that an unknown signal does not always represent an independent, functional circulating metabolite.
Mapping and identifying BMI-associated metabolites (Figure 1a)
Mapping metabolites across datasets
Using the imputation-based matching algorithm in PAIRUP-MS19, we identified 1,780 metabolite pairs (207 shared known metabolites measured in both datasets and 1,573 matched unknown or unshared known metabolites) that could be compared directly across OE and MCDS and restricted subsequent analyses to these metabolites. For pathway analyses requiring the BioAge-based metabolite set annotations (see below), we furthered mapped 1,743 (200 shared known and 1,543 matched) of these metabolite pairs to metabolites measured in BioAge.
Identifying BMI-associated metabolites
Within each cohort, we adjusted raw BMI (available for 298 OE and 818 MCDS samples, calculated from weight and height measured at the same study visit as sample collection) for age and sex, performed rank-based inverse normal transformation on the residuals, and used the resulting BMI z-scores in all further analyses. (Since the OE lean and obese samples were drawn from the BMI extremes of EB, all EB samples were used to calculate population-based z-scores.) To identify BMI-associated metabolites, we performed linear regression of BMI on each metabolite within each cohort, followed by inverse variance weighted meta-analysis across the two cohorts, and applied a Bonferroni significance threshold (p < 0.05/1,780) in meta-analysis.
Bidirectional IV analyses (Figure 1b)
Metabolite instrument (GM) selection
GWAS and meta-analysis of the BMI-associated metabolites using 294 OE and 637 MCDS samples (with available genetic data) were performed as described previously19. Briefly, linear mixed model GWAS were performed using imputed genotype dosages and EPACTS22 (v3.2.6) within each cohort, followed by inverse variance weighted meta-analysis using METAL23 (2011–03-25 version). To select GM, we first identified the SNP (single nucleotide polymorphism) with the best meta-analyzed p-value for each metabolite. We restricted to a single SNP per metabolite given the modest sample size for discovery and limited number of loci near genome-wide (p < 5 × 10−8) or sub-genome-wide (p < 1 × 10−5) significance. Next, to avoid using redundant GM, we used PLINK24 (v1.9) and 1000 Genomes phase 3 reference panel25 to “clump” the best SNPs for all metabolites, selecting independent SNPs that have r2 < 0.5 or are > 250 kb apart, and only kept the independent SNPs as GM in further analyses (along with their best-associated metabolites and respective effect estimates in meta-analysis). For known metabolites in our causality groups (see below), we performed an additional sensitivity analysis using (where available) genome-wide significant (p < 5 × 10−8) SNPs and their respective effect estimates from published metabolite GWAS26–31 as individual GM.
BMI instrument (GB) selection
We used 97 BMI-associated SNPs (Gb) previously identified in GIANT32 and their effect estimates (βb) in our UK Biobank (UKB) GWAS to calculate a weighted genetic risk score for use as GB (i.e. GB = Σ βb × Gb). We performed BMI GWAS in UKB using 453,397 European-ancestry samples and sex-combined BMI z-scores, using BOLT-LMM33 (v2.3.2) to account for relatedness and population structure (Supplementary Text 2). Analysis of UKB data was approved by its governing Research Ethics Committee and the Broad Institute Institutional Review Board. The GIANT, UKB, and metabolomics cohorts have no known sample overlap. In terms of unknown sample overlap, OE is contained within EB, a participating cohort in GIANT; OE therefore comprises, at maximum, less than 0.1% of GIANT. We confirmed that GB is associated with BMI in OE and MCDS and that none of the Gb are in linkage disequilibrium (r2 > 0.3) with the selected GM (using PLINK and 1000 Genomes reference panel described above).
Testing for metabolite-to-BMI causal effect using GM
The association between BMI and each GM was extracted from the UKB GWAS summary statistics and used to calculate the Wald ratio IV effect estimate of each metabolite on BMI. The p-value for the Wald estimate was calculated using an asymptotic standard error estimate described previously34. This p-value – a test of the null hypothesis of no causal effect of the metabolite – was used to rank metabolites as more or less likely to be causal for BMI.
Testing for BMI-to-metabolite causal effect using GB
We performed linear regression of each metabolite on GB in OE and MCDS separately, followed by inverse variance weighted meta-analysis. The Wald ratio IV effect estimate of BMI on each metabolite was calculated using the meta-analyzed statistics, and the corresponding p-value was used to rank metabolites as more or less likely to be effects of BMI. As a sensitivity analysis, we performed the MR-PRESSO global test35 (using 10,000 permutations for each metabolite) to assess overall horizontal pleiotropy among the individual SNPs (Gb) contained within GB, using metabolite-Gb association in the OE-MCDS meta-analysis and BMI-Gb association in UKB for 96 of 97 BMI SNPs (rs2033529 was excluded due to absence in our metabolite GWAS).
Defining cause, effect, and bidirectional metabolite groups (Figure 1c)
To rank BMI-associated metabolites as more or less likely to be the causes or effects of obesity, we used the -log10 p-value of the IV effect estimate for either the metabolite (GM) or BMI (GB) instrument, reasoning that the magnitude of these p-values informs the likelihood of the respective null hypotheses, provided the assumptions for IV analyses are met. The primary IV assumptions include the genetic variant is a valid instrument for the exposure, and the instrument is not associated with confounders or the outcome (except via the exposure-outcome effect)12. Metabolites in the top and bottom quartiles of these two p-value-based rankings were assigned to three distinct groups corresponding to different types of causal connections with BMI: (1) “cause”: metabolites that were ranked in the top quartile using GM and the bottom using GB, and thus are likely to be upstream causes for BMI; (2) “effect”: metabolites that were ranked in the bottom quartile using GM and the top using GB, and thus are likely to be downstream effects of BMI; (3) “bidirectional”: metabolites that were in the top quartiles of both rankings, suggesting complex bidirectional cause-effect relationships with BMI.
Pathway analyses of the defined metabolite groups (Figure 1d)
The PAIRUP-MS pathway annotation method and BioAge metabolite data were used to generate metabolite set annotations as described previously19 (Supplementary Figure 1a–b), resulting in 690 metabolite sets in which each metabolite was assigned a numeric membership score in each set. Next, we applied the PAIRUP-MS pathway analysis framework to identify enriched metabolite sets for the cause, effect, and bidirectional metabolite groups we defined. We compared each of the three groups individually versus all other BMI-associated metabolites and, in a fourth analysis, compared the cause versus effect groups. First, for each metabolite set in each comparison analysis, a two-tailed Wilcoxon rank-sum test was performed to compare the membership scores of the two groups of metabolites (Supplementary Figure 1c). Next, to account for correlation structure in our data, iterations of this procedure were performed using “null” metabolite groups to calculate a permutation-based enrichment p-value for each metabolite set (Supplementary Figures 1c and 2). All procedures described above can be performed using PAIRUP-MS source code, except for the generation of null metabolite groups, which is specific to the current study.
Performing m/z query for unknown metabolites
To assess if the unknown metabolites captured information redundant to the known metabolites in our dataset (and to look up potential identities of unknowns classified in the three causality groups), we performed m/z query as described previously19, using the “LC-MS Search” tool in the Human Metabolome Database (HMDB)36. The unknowns were annotated as an m/z-matched adduct of a known metabolite in our data, an m/z-matched adduct of an HMDB metabolite not identified in our data, or a metabolite without a match in HMDB.
Additional information on analysis software and data availability are provided in Supplementary Text 4 and 5, respectively. The study protocol and details were not pre-registered.
RESULTS
Identifying known and unknown metabolites associated with BMI
We used untargeted metabolomics data from OE and MCDS to identify metabolites associated with BMI. First, we identified 207 pairs of shared known metabolites measured in both cohorts, and used PAIRUP-MS to match 1,573 additional pairs of unknown or unshared known metabolites likely to represent identical or highly correlated metabolites. Then, by performing meta-analysis of both the shared known and matched pairs across the cohorts, we identified 577 BMI-associated metabolites at Bonferroni significance (p < 0.05/1,780), the majority of which are unknown metabolites: 418 (72.4%) consist of two paired unknown metabolites, 59 (10.2%) consist of a known metabolite matched to an unknown, and only 100 (17.3%) consist of shared known metabolites. When we clustered these metabolites based on their pairwise correlations, we observed clusters comprising mostly or entirely of matched metabolite pairs with unknown identities (Supplementary Figure 3). Therefore, including these unknowns in downstream analyses increased the number of candidate metabolites by nearly five-fold, and allowed us to investigate aspects of obesity biology not represented by the curated, known metabolites.
Identifying metabolites more likely to be causal for BMI
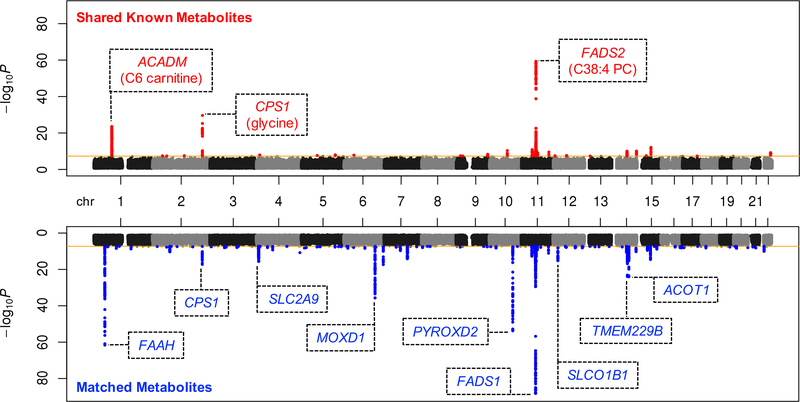
Before we could determine whether the BMI-associated metabolites are likely to be causal for BMI, we first needed to identify the SNP best-associated with each metabolite to use as genetic instrument (GM in Figure 1). We therefore performed GWAS of metabolite levels in both OE and MCDS, followed by meta-analysis. We identified genome-wide significant (p < 5 × 10−8) SNPs for 204 (35 shared known and 169 matched) of the BMI-associated metabolites (Figure 2); 66 (14 shared known and 52 matched) of these are also significant after correction for multiple hypothesis testing (p < 5 × 10−8/577). Overall, the matched, unknown metabolites show comparable degree of genetic associations as the shared known metabolites, even in loci not associated with any of the knowns. Analyzing the unknowns thus greatly improved our ability to obtain genome-wide significant and novel genetic instruments for metabolite signals, despite a relatively small GWAS sample size.
Figure 2. Joint Manhattan plots summarizing GWAS of BMI-associated metabolites in OE and MCDS.
Genetic associations for 100 shared known (top) or 477 matched (bottom) BMI-associated metabolites were consolidated to plot the best p-value for each SNP (i.e. only the p-value for the best associated metabolite was plotted for each SNP). Genome-wide significance threshold (p < 5 × 10−8) is marked by the orange lines. Genome-wide significant SNPs are plotted in red or blue, for shared known or matched metabolites, respectively. Lead SNPs of the most significant loci (p < 1 × 10−15) are annotated with nearest genes (within 5kb), along with the best associated known metabolites if applicable.
We observed that all 577 BMI-associated metabolites have best-associated SNPs with at least suggestive sub-genome-wide significance (maximum p = 2.5 × 10−6) and thus considered the best-associated SNP for each metabolite as a potential instrument. To avoid analyzing metabolites sharing the same instruments, we included only genetically independent GM (r2 < 0.5 or > 250kb apart) and the 324 (40 shared known and 284 matched) metabolites best-associated with these instruments in subsequent IV analyses (Supplementary Table 1). Supplementary Figure 4 summarizes the overall level of association between the 324 GM and the 324 metabolites, with 38.6% of GM being associated with more than one metabolite. Next, for each metabolite, we estimated the association between GM and BMI using a large independent cohort, UKB, in a two-sample design to calculate the metabolite-to-BMI IV effect estimate. We identified 50 (11 shared known and 39 matched) metabolites with metabolite-to-BMI IV p-values < 0.05, which indicates that they are more likely to be upstream causes for BMI (Supplementary Table 1).
Identifying metabolites more likely to be effects of BMI
To determine if the 324 BMI-associated metabolites are likely to be effects of BMI, we combined 97 BMI SNPs previously identified in GIANT into a weighted genetic risk score using UKB effect estimates as weights (Supplementary Table 2). As expected, the score is a valid genetic instrument for BMI (GB in Figure 1) in OE and MCDS (meta-analyzed BMI-GB association p = 5.9 × 10−7, Supplementary Table 3). For each metabolite, we estimated the association between GB and the metabolite using OE and MCDS data (Supplementary Table 3) to calculate the BMI-to-metabolite IV effect estimate. A total of 56 (8 shared known and 48 matched) metabolites have BMI-to-metabolite IV p-values < 0.05 and thus are more likely to be downstream effects of BMI (Supplementary Table 1).
Defining cause, effect, and bidirectional metabolite groups
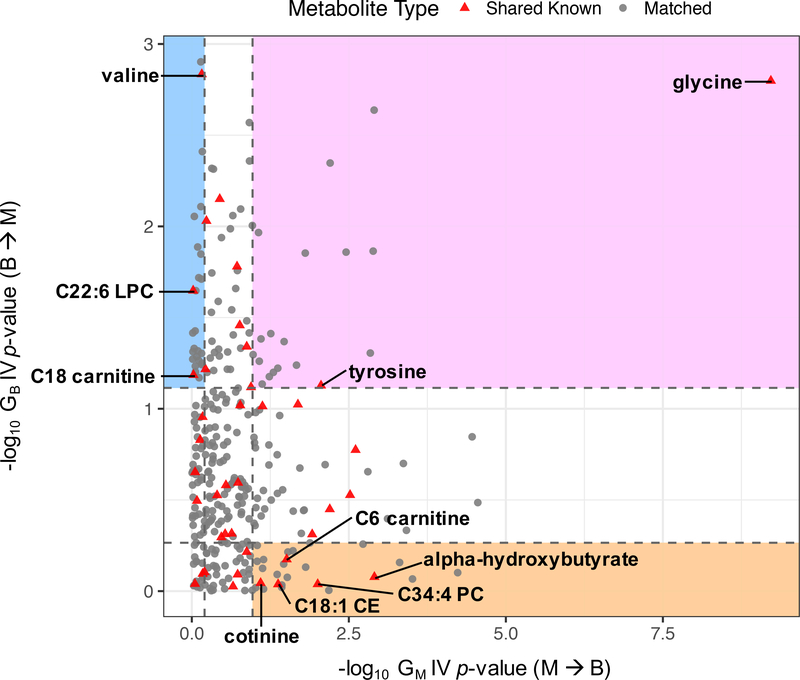
In order to further characterize the causal relationships between BMI and its associated metabolites, we ranked the metabolites based on the magnitude of the evidence according to their metabolite-to-BMI (GM) and BMI-to-metabolite (GB) IV p-values, and classified a subset of them into “cause”, “effect”, or “bidirectional” group using quartile cutoffs of the rankings (Figure 3). We defined 25 metabolites as more likely to be cause (5 shared known and 20 matched), 26 as more likely to be effect (3 shared known and 23 matched), and 19 as more likely to be bidirectional (2 shared known and 17 matched) with respect to BMI. The shared known metabolites in each group are listed in Table 1; the top cause, effect, and bidirectional known metabolites are alpha-hydroxybutyrate, valine, and glycine, respectively. Details for all metabolites in each group are in Supplementary Table 1. We also performed m/z query in HMDB to obtain potential identities for the unknowns in the matched metabolite pairs (Supplementary Table 4) and found only 6 out of the 60 matched pairs to be potentially redundant with the known metabolites curated in our data. Hence, we identified about 5 times more matched, unknown metabolites in the three causality categories compared to the known metabolites. In addition, we performed sensitivity analyses to assess how our genetic IV and classification scheme would be influenced by weak instrument or pleiotropy bias (Supplementary Text 3, Supplementary Tables 5 and 6); we obtained results that generally support the robustness of our approach.
Figure 3. Classifying BMI-associated metabolites using IV effect estimate p-values for GM (metabolite-to-BMI direction, x-axis) and GB (BMI-to-metabolite direction, y-axis).
Top and bottom quartile cutoffs along each axis are shown as dashed lines. Shared known metabolites in “cause” (orange), “effect” (blue), and “bidirectional” (pink) regions are labeled with their names.
Table 1. BMI-associated known metabolites classified into cause, effect, or bidirectional group based on their IV effect estimate p-values.
Y ~ X, regression of Y on X; B, BMI; M, metabolite; covariate adjustment for B and M described in Methods; β, effect size estimate; SNP, hg19 chromosome:position is shown; EA, effect allele (i.e. metabolite level-increasing allele). IV effect estimate p-values < 0.05 are in bold italic.
| Group | Metabolite | Observational Association | Metabolite Instrument | GM IV Estimate (M → B) | GB IV Estimate (B → M) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| β B ~ M | P B ~ M | SNP | EA | P M ~ GM | β | P | β | P | ||
| Cause | alpha-hydroxybutyrate | 0.235 | 7.09E-13 | 11:119745598 | T | 4.82E-07 | −0.040 | 1.24E-03 | −0.036 | 8.36E-01 |
| C34:4 PC | 0.153 | 3.45E-06 | 3:182171263 | A | 1.42E-07 | −0.027 | 9.90E-03 | 0.019 | 9.15E-01 | |
| C6 carnitine | 0.207 | 2.66E-10 | 1:76224010 | C | 2.88E-24 | −0.010 | 3.09E-02 | −0.076 | 6.69E-01 | |
| C18:1 CE | −0.215 | 5.54E-11 | 20:38984849 | T | 3.44E-07 | −0.016 | 4.23E-02 | −0.018 | 9.17E-01 | |
| cotinine | −0.169 | 3.10E-07 | 10:123918365 | C | 1.07E-06 | 0.012 | 8.03E-02 | −0.022 | 9.02E-01 | |
| Effect | valine | 0.445 | 1.20E-47 | 14:32724292 | A | 7.01E-08 | −0.003 | 6.94E-01 | 0.708 | 1.46E-03 |
| C22:6 LPC | −0.180 | 4.38E-08 | 18:71068347 | A | 3.26E-07 | −0.001 | 9.52E-01 | −0.450 | 2.25E-02 | |
| C18 carnitine | −0.193 | 4.65E-09 | 6:110760008 | A | 1.27E-07 | 0.001 | 9.41E-01 | −0.351 | 6.53E-02 | |
| Bidirectional | glycine | −0.308 | 7.55E-22 | 2:211540507 | A | 2.35E-30 | 0.030 | 6.03E-10 | −0.712 | 1.59E-03 |
| tyrosine | 0.376 | 6.22E-33 | 6:111477887 | C | 1.12E-07 | −0.022 | 8.77E-03 | 0.334 | 7.46E-02 | |
Prioritizing enriched pathways for cause, effect, and bidirectional metabolites
We identified many more matched, unknown metabolite pairs in the cause, effect, and bidirectional groups compared to the shared known metabolites, but it is difficult to hypothesize on their roles in obesity biology without knowing their chemical identities. Therefore, to extract useful information from the unknowns and to gain clues about the biology broadly captured by the three causality groups, we performed PAIRUP-MS pathway analyses encompassing both known and unknown metabolites, using metabolite set annotations generated from a separate cohort, BioAge. First, we carried out three separate analyses to identify pathways with enrichment p < 0.05 for metabolites in the cause, effect, or bidirectional group, respectively, when compared against all other BMI-associated metabolites (Supplementary Table 7). While the most enriched metabolite sets in each analysis are associated with different pathways, several metabolite sets are enriched in multiple analyses (e.g. “NAD de novo biosynthesis” is enriched for both cause and effect metabolites).
Hence, in order to identify pathways that are the most distinct between the defined metabolite groups, we next performed a pathway analysis directly comparing the cause versus effect metabolites, prioritizing 40 metabolite sets with enrichment p < 0.05 (Supplementary Table 7). The 13 cause metabolite sets (in which cause metabolites have higher membership scores than effect metabolites) are associated with various pathways, such as those connected to inflammation (e.g. nitric oxide signaling), redox metabolism (e.g. cysteine/methionine metabolism), and appetite regulation (e.g. endocannabinoid signaling). The 27 effect metabolite sets also contain varied pathways including those related to lysine catabolism, neurobiology (e.g. addiction and catecholamine biosynthesis), and stress response (e.g. FoxO signaling). While the known metabolites in our analysis have been linked to some of the enriched metabolite sets in literature, the unknowns contributed most of the data used to prioritize these sets.
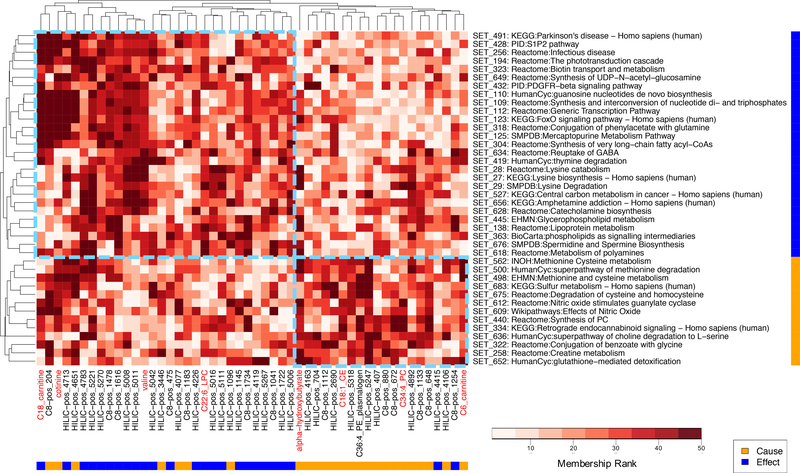
Finally, to better visualize the distinguishing features between the cause versus effect metabolites in terms of their roles in biological pathways, we constructed a clustered heat map of their membership scores in the enriched metabolite sets (Figure 4). The metabolites form two major clusters consisting of mostly cause or mostly effect metabolites, with a handful of metabolites clustering with the contrasting group (i.e. cause metabolite “misclassified” in the effect cluster or vice versa). In the other dimension, the cause and effect metabolite sets form two pure clusters consisting of all cause or all effect sets, agreeing with their pathway enrichment statistics. Overall, the pathway results suggest that the cause and effect metabolites we defined are involved in distinct biological processes and thus may be associated with BMI through different mechanisms.
Figure 4. Clustered heat map of cause and effect metabolites’ memberships in metabolite sets prioritized by pathway analysis.
Euclidean distance-based hierarchical clustering was performed using metabolite membership ranks in the BioAge-based metabolite set annotations. Each column is a shared known (red label) or matched (black label) metabolite from the cause (yellow bar) or effect (blue bar) metabolite group. Each row is an enriched (p < 0.05) metabolite set in pathway analysis in either the cause (yellow bar) or effect (blue bar) direction (with representative pathway name shown in label; see Supplementary Table 7 for full pathway list). Larger number in membership rank (darker red) indicates higher membership score. Dashed light blue boxes highlight the two major cause and effect clusters according to the clustering dendrograms.
DISCUSSION
The study of comprehensive metabolite profiles defines an exciting frontier in human pathophysiology. However, metabolite-phenotype associations discovered in metabolomics studies are often correlative in nature and additional causal inference approaches, such as genetic IV analysis, are required to help assess causality between metabolites and phenotypes. Furthermore, unknown metabolite signals are often filtered out prior to analysis of untargeted metabolomics data, greatly limiting investigation to a priori candidate metabolites, reducing the search space, and hindering downstream analyses such as pathway enrichment. Here we present a paradigm for combining untargeted metabolomics, genomics, and our recently described bioinformatics suite, PAIRUP-MS, to overcome these challenges. Using obesity as an exemplar state of metabolic dysregulation, we illustrate the potential utility of this approach to advance our understanding of causal connections in metabolic diseases.
In this study, we meta-analyzed hundreds of unknown metabolites from two cohorts using PAIRUP-MS, identifying novel associations between the unknowns, BMI, genetic variants, and biological pathways. Indeed, using bidirectional genetic IV analysis, we discovered about 5 times as many unknown than known metabolites with potential causal connections to BMI. While these unknowns are likely not all fully independent and functional circulating molecules, their associations with genetic variants and BMI, distinct from those with known metabolites, suggest that a sizable number of them reflects aspects of BMI biology not captured by known metabolites. Furthermore, the much larger number of candidate metabolites allowed us to perform PAIRUP-MS pathway analyses that account for potential redundancy, prioritizing biological pathways specific to the metabolites with cause or effect relationships to BMI. Despite the modest power of our metabolite GWAS, our study demonstrates a useful and generalizable analytic framework to probe the metabolome of obesity and other diseases.
We identified novel metabolites that may be causes of obesity, as well as replicating two known metabolites, valine and tyrosine, that may be effects of BMI15. The associations between BMI and known metabolites were broadly consistent with those observed in a recent comprehensive metabolomics study of BMI10, which used longitudinal data to suggest that BMI-associated metabolites were predictive of future cardiometabolic outcomes, but did not use the same bidirectional IV approach that we employed for causal inference between metabolites and obesity itself (see Supplementary Text 6 for additional discussion). The top cause metabolite we defined among the known metabolites is alpha-hydroxybutyrate, which has been linked to insulin resistance, oxidative stress, glutathione biosynthesis, and mitochondrial dysfunction6,37,38. The oxidative stress and glutathione links are especially intriguing since “glutathione-mediated detoxification” emerged as an enriched causal pathway when we compared the cause and effect metabolite groups in pathway analysis. It is also notable that the IV effect estimate of alpha-hydroxybutyrate on BMI is protective while the observational association suggests this metabolite is obesogenic. We postulate that a mitochondrial dysfunction/altered redox state linked to high alpha-hydroxybutyrate level could lead to decreased weight gain, while shared common causes, such as an obesogenic diet, may lead to increases in both alpha-hydroxybutyrate level and BMI. This example highlights the advantage of genetic IV analyses over observational studies alone to explore the potential impact of a theoretical intervention targeted to obesity-associated metabolites39,40.
When using genetic IV to classify metabolites into our three causality groups, weak instrument bias towards the null (for two-sample IV) and pleiotropy bias away from the null may lead to misclassification. To address weak instrument bias for classification of our known metabolites, we performed sensitivity analysis using stronger instruments from published metabolite GWAS, showing that our results are generally robust against weak instrument bias, although some misclassification is possible due to limited power of our internal instruments. Different instruments for the same metabolite sometimes show discordant results, indicating that there is heterogeneity in the underlying biology. This discordance is illustrated by the “effect” metabolite valine, whose top published GWAS instrument (rs9637599 on chr4 near PPM1K) yielded strong “cause” evidence (metabolite-to-BMI IV p = 2.8 × 10−7) and would reclassify valine into the “bidirectional” group. This locus has been linked with all three BCAAs, including valine, in previous studies31,41. In our GWAS, this SNP is only nominally associated with the three BCAAs (p = 0.011 to 0.038). On the other hand, our valine instrument (rs79674166 on chr14) is on a different chromosome, has no genes within 5 kb, and yet is also suggestively associated with all three BCAAs (p = 7.0 × 10−8 to 9.7 × 10−7). Thus, while the associations between this locus and BCAAs need to be replicated and validated in the future, it may underpin BCAA-related biology that is distinct from that captured by the published PPM1K locus. We could not conduct similar analyses for misclassification of the unknown metabolite instruments since there is not yet a straightforward way to obtain external instruments for comparison.
To address pleiotropy bias for our BMI instrument, we used a recently developed method, MR-PRESSO, to show that our BMI IV estimates are likely robust against extreme cases of pleiotropy bias. We could not examine pleiotropy in the metabolite instruments due to lack of multiple instruments for each metabolite (especially for the unknowns where additional instruments could not be obtained from published GWAS). More discussion on IV analysis limitations with respect to our study are provided in Supplementary Text 7.
Larger GWAS of both known and unknown metabolites, conducted across multiple datasets and populations with different ancestral backgrounds, will make it possible to generalize and extend our paradigm to understand causal biological mechanisms for various metabolic diseases and alleviate the limitations described above. With more candidate metabolites and genetic instruments emerging from better-powered studies, our approach can be expanded to leverage multiple IV per metabolite, mediation analyses42, pathway Mendelian randomization43, or metabolite IV subsetting according to predicted biological pathway memberships44. In conclusion, this study showcases the benefit of combining untargeted metabolomics with a bidirectional genetic IV approach to define the metabolome of a major human disease state, obesity. We therefore advocate for broader sharing of untargeted metabolomics and genetic datasets, similar to the approach taken by international efforts to optimize GWAS of many other phenotypes. Broader sharing would improve power and reliability of methodological frameworks such as the one presented here and would enable a fuller realization of the potential of metabolomics to generate important insights into human diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Broad Metabolomics Platform and SIGMA T2D Consortium for sharing data resources. This research has been conducted using the UK Biobank Resource under Application Number 11898. This work was supported by the National Heart, Lung, and Blood Institute grant F31HL126581 (Y.H.H.), National Institute of Diabetes and Digestive and Kidney Diseases grants T32DK110919 (Y.H.H.), K12DK094721 (C.M.A), and R01DK075787 (J.N.H.), Endocrine Scholars Award (C.M.A.), Doris Duke Charitable Foundation grant 215205 (J.N.H.), Estonian Research Council grants IUT20-60 (A.M.), PUT1665 (K.Fischer), and PUT1660 (T.E.), European Union through Horizon 2020 grant 692145 (A.M.), and European Union through the European Regional Development Fund Project No. 2014-2020.4.01.15-0012 (A.M.). The funding sources had no role in the design, analysis, and interpretation of this research.
Footnotes
COMPETING INTERESTS
K.Fortney and E.K.M. are affiliated with BioAge Labs, Inc.; J.N.H. serves on the Scientific Advisory Board of Camp4 Therapeutics.
REFERENCES
- 1.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014; 384: 766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006; 444: 840–846. [DOI] [PubMed] [Google Scholar]
- 3.Poirier P, Giles TD, Bray GA, Hong Y, Stern JS, Pi-Sunyer FX et al. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss. Arter Thromb Vasc Biol 2006; 26: 968–976. [DOI] [PubMed] [Google Scholar]
- 4.Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet 2008; 371: 569–578. [DOI] [PubMed] [Google Scholar]
- 5.Flegal KM, Kit BK, Orpana H, Graubard BI. Association of all-cause mortality with overweight and obesity using standard body mass index categories: a systematic review and meta-analysis. JAMA 2013; 309: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF et al. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab 2009; 9: 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho JE, Larson MG, Ghorbani A, Cheng S, Chen MH, Keyes M et al. Metabolomic Profiles of Body Mass Index in the Framingham Heart Study Reveal Distinct Cardiometabolic Phenotypes. PLoS One 2016; 11: e0148361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 2012; 125: 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E et al. Metabolite profiles and the risk of developing diabetes. Nat Med 2011; 17: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirulli ET, Guo L, Leon Swisher C, Shah N, Huang L, Napier LA et al. Profound Perturbation of the Metabolome in Obesity Is Associated with Health Risk. Cell Metab 2019. doi: 10.1016/j.cmet.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 2004; 33: 30–42. [DOI] [PubMed] [Google Scholar]
- 12.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014; 23: R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fall T, Hagg S, Magi R, Ploner A, Fischer K, Horikoshi M et al. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med 2013; 10: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes M V, Lange LA, Palmer T, Lanktree MB, North KE, Almoguera B et al. Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. Am J Hum Genet 2014; 94: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wurtz P, Wang Q, Kangas AJ, Richmond RC, Skarp J, Tiainen M et al. Metabolic Signatures of Adiposity in Young Adults: Mendelian Randomization Analysis and Effects of Weight Change. PLoS Med 2014; 11: e1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, van Klinken JB, Semiz S, van Dijk KW, Verhoeven A, Hankemeier T et al. A Mendelian Randomization Study of Metabolite Profiles, Fasting Glucose, and Type 2 Diabetes. Diabetes 2017; 66: 2915–2926. [DOI] [PubMed] [Google Scholar]
- 17.Haase CL, Tybjaerg-Hansen A, Qayyum AA, Schou J, Nordestgaard BG, Frikke-Schmidt R. LCAT, HDL cholesterol and ischemic cardiovascular disease: a Mendelian randomization study of HDL cholesterol in 54,500 individuals. J Clin Endocrinol Metab 2012; 97: E248–56. [DOI] [PubMed] [Google Scholar]
- 18.Patti GJ, Tautenhahn R, Siuzdak G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat Protoc 2012; 7: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu Y-HH, Churchhouse C, Pers TH, Mercader JM, Metspalu A, Fischer K et al. PAIRUP-MS: Pathway analysis and imputation to relate unknowns in profiles from mass spectrometry-based metabolite data. PLoS Comput Biol 2019; 15: 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leitsalu L, Haller T, Esko T, Tammesoo ML, Alavere H, Snieder H et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol 2015; 44: 1137–1147. [DOI] [PubMed] [Google Scholar]
- 21.Williams Amy AL, Jacobs Suzanne SBR, Moreno-Macías H, Huerta-Chagoya A, Churchhouse C, Márquez-Luna C et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 2014; 506: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang HM. EPACTS (Efficient and Parallelizable Association Container Toolbox). http://genome.sph.umich.edu/wiki/EPACTS.
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee EP, Ho JE, Chen MH, Shen D, Cheng S, Larson MG et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab 2013; 18: 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Draisma HHM, Pool R, Kobl M, Jansen R, Petersen AK, Vaarhorst AAM et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat Commun 2015; 6: 7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin S-YY, Fauman EB, Petersen A-KK, Krumsiek J, Santos R, Huang J et al. An atlas of genetic influences on human blood metabolites. Nat Genet 2014; 46: 543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long T, Hicks M, Yu HC, Biggs WH, Kirkness EF, Menni C et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet 2017; 49: 568–578. [DOI] [PubMed] [Google Scholar]
- 30.Burkhardt R, Kirsten H, Beutner F, Holdt LM, Gross A, Teren A et al. Integration of Genome-Wide SNP Data and Gene-Expression Profiles Reveals Six Novel Loci and Regulatory Mechanisms for Amino Acids and Acylcarnitines in Whole Blood. PLoS Genet 2015; 11: e1005510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kettunen J, Demirkan A, Wurtz P, Draisma HH, Haller T, Rawal R et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun 2016; 7: 11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015; 518: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh PR, Tucker G, Bulik-Sullivan BK, Vilhjalmsson BJ, Finucane HK, Salem RM et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 2015; 47: 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res 2017; 26: 2333–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018; 50: 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018; 46: D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gall WE, Beebe K, Lawton KA, Adam KP, Mitchell MW, Nakhle PJ et al. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One 2010; 5: e10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson Legault J, Strittmatter L, Tardif J, Sharma R, Tremblay-Vaillancourt V, Aubut C et al. A Metabolic Signature of Mitochondrial Dysfunction Revealed through a Monogenic Form of Leigh Syndrome. Cell Rep 2015; 13: 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgess S, Harshfield E. Mendelian randomization to assess causal effects of blood lipids on coronary heart disease: Lessons from the past and applications to the future. Curr. Opin. Endocrinol. Diabetes Obes. 2016. doi: 10.1097/MED.0000000000000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK et al. Plasma HDL cholesterol and risk of myocardial infarction: A mendelian randomisation study. Lancet 2012. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lotta LA, Scott RA, Sharp SJ, Burgess S, Luan J, Tillin T et al. Genetic Predisposition to an Impaired Metabolism of the Branched-Chain Amino Acids and Risk of Type 2 Diabetes: A Mendelian Randomisation Analysis. PLoS Med 2016. doi: 10.1371/journal.pmed.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Der weele T, Vansteelandt S. Mediation analysis with multiple mediators. Epidemiol Method 2013. doi: 10.1515/em-2012-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burgess S, Thompson SG. Multivariable Mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol 2015; 181: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wittemans LBL, Lotta LA, Oliver-Williams C, Stewart ID, Surendran P, Karthikeyan S et al. Assessing the causal association of glycine with risk of cardio-metabolic diseases. Nat Commun 2019; 10: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.