Abstract
Background.
We analyzed whole transcriptome sequencing in tumors from 23 patients with stage III or IV melanoma from a pilot trial of the anti-GD2 immunocytokine, hu14.18-IL2, to identify predictive immune and/or tumor biomarkers in melanoma patients at high risk for recurrence.
Methods.
Patients were randomized to receive the first of 3 monthly-courses of hu14.18-IL2 immunotherapy either before (Group A) or after (Group B) complete surgical resection of all known disease. Tumors were evaluated by histology and whole transcriptome sequencing.
Results.
Tumor infiltrating lymphocyte (TIL) levels directly associate with relapse-free survival (RFS) and overall survival (OS) in resected tumors from Group A, where early responses to the immunotherapy agent could be assessed. TIL levels directly associated with a previously reported immune signature, which associated with RFS and OS, particularly in Group A tumors. In Group A tumors there were decreased cell cycling gene RNA transcripts, but increased RNA transcripts for repair and growth genes. We found that outcome (RFS and OS) was directly associated with several immune signatures and immune-related RNA transcripts and inversely associated with several tumor growth-associated transcripts, particularly in Group A tumors. Most of these associations were not seen in Group B tumors.
Conclusion.
We interpret these data to signify that both immunologic and tumoral cell processes, as measured by RNAseq analyses detected shortly after initiation of hu14.18-IL2 therapy are associated with long term survival and could potentially be used as prognostic biomarkers in tumor resection specimens obtained after initiating neoadjuvant immunotherapy.
Statement of translational relevance:
This study further advances our understanding of the clinical and immunological activity of a novel immunocytokine and discusses plans for subsequent clinical testing. Our data suggest that both immune and tumor cell processes, as measured by RNAseq analyses, are associated with OS and RFS when evaluated in tumors resected approximately 2 weeks after starting the first of 3 scheduled monthly courses of hu14.18-IL2 immunotherapy. We identify specific immune- and tumor response-related molecular pathways associated with improved outcome after starting immunotherapy. We provide new insights into immunological processes involved in effective melanoma immunotherapy as well as prognostic information derived from the RNA-seq data that can facilitate patient management. These findings may be utilized for functionally similar forms of immunotherapy for other malignancies.
Introduction
Melanoma is considered an immunogenic and relatively immunoresponsive tumor with a relatively high tumor mutation burden (1, 2), often harboring many tumor infiltrating lymphocytes (TILs), which have prognostic value (3, 4). Improved biomarkers of outcome following melanoma immunotherapy are needed. Immune checkpoint inhibition is an effective therapy for some patients with melanoma (5–8). The mechanisms of melanoma response to checkpoint inhibition immunotherapy are complex and multifactorial (9). Previous transcriptomics analyses have found adaptive immune signatures in melanoma tumor biopsies soon after checkpoint blockade, which are predictive of response (10). Recently, whole transcriptomic interrogation of MYCN non-amplified pediatric neuroblastoma has shown immune signatures are associated with outcomes in children (11). In contrast melanoma-derived driver mutations (12–14) as well as anti-immunity signaling pathways (15, 16) also contribute to decreased long-term survival (17). The relative contribution of tumoral versus immunologic parameters to cancer survival is unclear; in colorectal carcinoma each side contributes approximately 50% (18).
The hu14.18-IL2 immunocytokine (IC) is a humanized monoclonal antibody (mAb) that binds to GD2 and is linked, as a fusion protein, to IL-2 at its Fc region (19, 20). GD2 is a cell membrane disialoganglioside found in neuroectodermal tumors (melanoma, neuroblastoma, sarcomas), but demonstrates only low expression in select normal tissues (cerebellum, peripheral nerves) (21, 22). Hu14.18-IL2 has been studied in vitro and in mouse models against melanoma and neuroblastoma (23–25). In mice, the antitumor effects of hu14.18-IL2 involve cytotoxic T cells and NK cells (26–28). In addition, mice with smaller tumors or with minimal residual disease (MRD) at treatment initiation elicit the most robust responses to single-agent IC therapy (29, 30). Hu14.18-IL2 has undergone Phase I and II testing in adults with metastatic melanoma and children with neuroblastoma (31–35), and shows reproducible antitumor activity, particularly in the MRD setting.
Here we present tumor analyses from a clinical trial (CO05601), wherein 21 of 23 patients with advanced melanoma received surgical resection of all evident disease to attain a complete response (CR) together with systemic IC administration (36). Patients were randomized to have their first of 3 monthly-courses of hu14.18-IL2 started either just before (Group A, n=13) or just after (Group B, n=8) their complete surgical resection. Our primary goal for this report is to identify differences in gene expression in these tumors prior to and after treatment with hu14.18-IL2; our secondary goal is to determine if gene expression patterns before treatment, or induced by the treatment, are associated with outcome. Published data from this trial reported prolonged tumor-free survival in some patients at high-risk for recurrence (36). In this report, we analyze whole transcriptome sequencing from these resected recurrent/refractory melanoma tumors and compare results for tumors obtained before vs. after 1 course of hu14.18-IL2. This is the first transcriptomic analysis of tumors from human patients receiving hu14.18-IL2. The literature is becoming rich in transcriptomic studies in the context of checkpoint inhibition, but it is relatively poor for experimental treatment outside this category of drugs. The data presented extend prior work evaluating the predictive ability of immune signatures for long term outcome associated with other forms of immunotherapy (particularly checkpoint blockade), and indicates the greater predictive power of transcriptomic analyses of tumor tissue obtained just after starting this form of immunotherapy over that obtained prior to initiating the therapy.
Methods
Clinical Trial Design
Twenty-three patients with advanced melanoma participated in this trial (UWCCC Protocol CO05601). The UW Human Subjects Committee and the FDA approved the study (ClinicalTrials.gov: NCT00590824; IND-12220). This study was conducted in accordance with the Declaration of Helsinki. Informed written consent was obtained from each subject. These human investigations were performed after approval by the UW institutional review board and in accordance with an assurance filed with and approved by the U.S. Department of Health and Human Services. Details of study design, conduct, and clinical results have been reported previously (36). All patients had recurrent stage III (recurrent regional metastasis), or stage IV (any distant metastasis) melanoma with biopsy-proven (current or previous) Stage III or Stage IV disease (36). Eligibility criteria required that at study entry, all patients have recurrent melanoma involving ≤3 sites, judged to be totally resectable, where resection would be clinically recommended. Patients signed informed consent-forms and were randomized into group A or B prior to treatment. In addition to hu14.18-IL2 treatment, three of these 23 patients also received cilengitide, an anti-angiogenic RGD-pentapeptide, as part of the original study (36). After enrolling those 3 patients the study was amended, removing cilengitide-treatment because of toxicity. The three cilengitide-treated patients were excluded from the previously reported clinical report (36). Two of ten patients randomized to Group B were not treated with hu14.18-IL2 as they didn’t achieve the required CR status following surgery. One of these 2 patients left the protocol due to extensive residual disease after surgery; the second was taken off protocol because melanoma was detected by PET/CT prior to the first hu14.18-IL2 course. The remaining eight patients were designated as Group B patients. All thirteen Group A patients received one cycle of hu14.18-IL2 prior to surgery and remained on study after resection.
Our previous report focused on clinical tolerance and treatment results for patients receiving only hu14.18-IL2 (36); this manuscript focuses on the biological analyses of the tumors and differences in the tumors for patients in Group A vs. Group B. To augment statistical power in this relatively small study, we’ve included tumors from the 3 cilengitide-treated patients. Therefore, this study includes histological and transcriptomic analyses for all tumors that were resected from all 23 patients. Associations with RFS and OS were performed for patients that received any amount (1 to 3 cycles) of hu14.18-IL2 and included the three patients (two patients from Group A and one patient from Group B) also given cilengitide.
Tumor Processing and Histological Analyses
Each patient’s surgical resection specimen (obtained following IC course-1 for Group A, and prior to IC course-1 for Group B) was processed by the UWHC Surgical Pathology Unit. Two representative 1 mm punch cores were taken from intra-tumoral regions within one representative formalin-fixed paraffin-embedded (FFPE) block of each patient’s resected melanoma. FFPE cores were coded and shipped to NCI on dry ice.
For histologic assessment, representative sections of each patient’s tumor were coded and analyzed by our board-certified anatomic pathologist (author EAR) for diagnosis confirmation and TIL assessment on H&E slides that were masked for treatment. Quantitation of TILs (%) was defined as (Cross-sectional area of TILs / Area of tumor) x 100 (see Supplemental Methods).
RNA Extraction and Transcriptome Sequencing
RNA was extracted from FFPE tumor cores using RNeasy FFPE kits according to the manufacturer’s protocol (Qiagen, Germantown, MD). RNA-seq libraries were generated using >50 ng of RNA and TruSeq RNA Access Library Prep Kits according to the manufacturer’s protocol (TruSeq RNA Exome kits; Illumina) and sequenced on NextSeq500 sequencers using 80bp paired-end sequencing method (Illumina, San Diego, CA). On average, >50 million aligned sequencing reads were generated from each RNA-seq library (>100X coverage of the coding regions in human genome). Quality control data are shown in Supplemental Table ST1. Gene expression level was performed as previously described (11). The expression and raw sequencing data have been deposited respectively in the NCBI’s Gene Expression Ominibus (GEO) under the accession GSE133713 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133713) and in the database of Genotypes and Phenotypes (dbGaP) under the accession phs001947. The processed RNA-seq data can be visualized and explored at https://clinomics.ccr.cancer.gov/clinomics/public/login using the database named “Melanoma_NCT00590824_CCR”.
Gene Signature Analysis
We determined the immune and stromal scores for each sample by using the Single-Sample Gene Set Enrichment Analysis (ssGSEA) and the ESTIMATE gene sets (37) (Supplemental Tables ST2–ST3) as described previously (11). We also utilized 22 CIBERSORT immune cell–specific gene sets (38) and a cytolytic score (average expression of GZMA, GZMB, GZMH, GZMK, GZMM, and PRF1) to assess the immune infiltrates in each sample. These signatures underwent validation in 3,809 TCGA transcriptional profiles (37), 3,061 human transcriptomes (38), and a cohort of 150 neuroblastoma patients (11), respectively.
Regarding tumor associated biomarkers, a more exploratory analysis interrogating the data on an individual gene level was undertaken based on the catalog of “cancer-related” genes under the list of protein classes on the Human Protein Atlas website (https://www.proteinatlas.org/). A total of 1,713 cancer-related genes were screened against OS and RFS with the top pathologically relevant (recognized as a neoplastic molecular biomarker aberration in molecular pathologic diagnosis) hits listed in Figure 4 and Supplemental Figure SF15. Molecular pathologic relevance was cross-referenced against the known genomic and epigenomic landscape of cutaneous melanoma (12, 14).
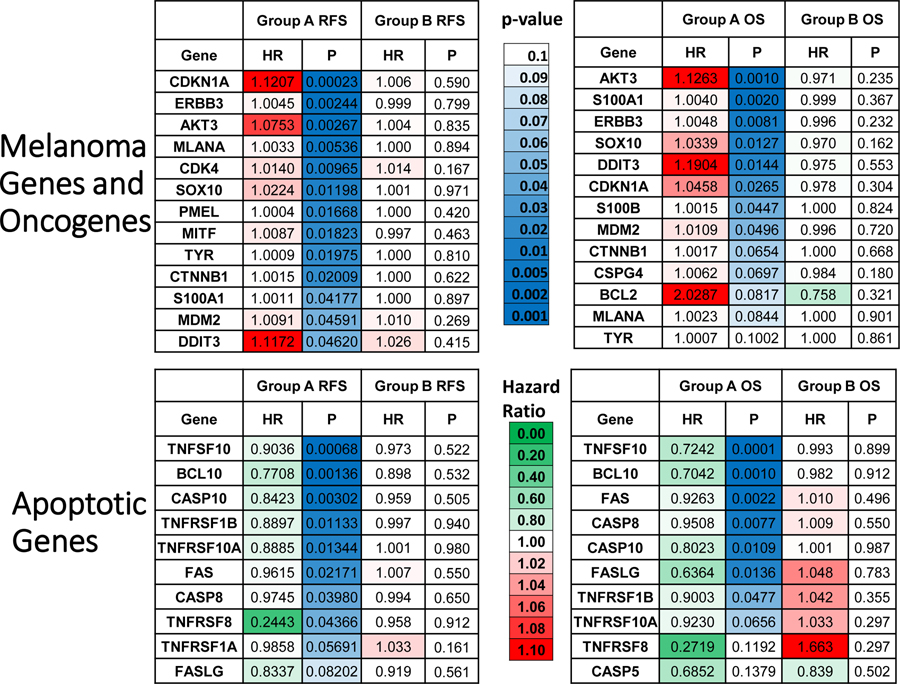
Figure 4. Expression of Melanoma Markers and Oncogenes Portends Unfavorable RFS and OS in Group A but not Groups B. Expression of Apoptotic Marker Genes Portends Favorable RFS and OS in Group A but not Group B.
Gene expression level relationships between melanoma related (top) and apoptosis related (bottom) expression and RFS and OS are shown in heat map form. Melanoma related genes were selected from molecular markers routinely used in diagnostic anatomic pathology. The green to red legend demonstrates either favorable prognosis (HR <1, green) or unfavorable prognosis (HR >1, red), respectively. The blue coloring represents level of significance starting with light blue at p=0.10 and becoming deeper blue with increasing significance. These melanoma and apoptotic parameters tend to prognosticate unfavorable or favorable survival, respectively, in Group A more than they do in Group B.
Statistics
RFS and OS were defined in the prior report (36), as detailed in Supplementary Methods. Gene- and gene signature-level survival analyses were performed using the univariate Cox proportional hazard regression with the likelihood ratio test (39). This study assumes the proportional hazards requirement holds. The Cox regression was implemented in R version 3.2.3 (https://www.R-project.org/). Estimated hazard ratios and p-values from the Cox regression model were used for screening in gene- and gene signature-level survival analyses. Survival data were also analyzed using the Kaplan-Meier methodology with the log rank test. Descriptive tumor statistics, TIL data analysis, Kaplan-Meier analysis, correlation analysis and significance tests were performed, and hazard ratio plots generated using Prism 8, version 8.01, software (GraphPad). Student’s t-tests, Fisher’s exact tests, Spearman rank and Pearson product-moment correlation coefficients, and corresponding p-values were calculated in Microsoft Excel and results confirmed with GraphPad Prism software. Due to the small sample sizes and exploratory nature of the statistical inference for the TIL data focused primarily on comparing groups A and B across the same set of genes/signatures, we report the nominal p-values, rather than p values corrected for multiple comparisons. To mitigate the issue of multiple comparisons, we use a p-value of < 0.01 to indicate significance in this study. Given small sample size for group B (n=8), power calculations are provided in Supplemental Methods.
Naïve tumors refer to an aggregate of resected tumors from eight Group B patients as well as from two never treated patients (these two were randomized to Group B, but non-evaluable for outcome as they did not achieve CR following resection).
Network- and Pathway-Level Analysis of Genes Whose Expression is Correlated with Survival
The ConsensusPathDB web-server (http://cpdb.molgen.mpg.de) (40) was used to perform two types of analyses: 1) Network-level analysis, and 2) Pathway-level enrichment analysis (41, 42) on genes whose expression are correlated with OS time. These are detailed in Supplementary Methods and data are presented in Supplemental Figure SF16.
Results
Description of Patient Treatment Timelines Tumor Characteristics
The study treatment schematic is found in Supplementary Figure SF1. Specific patient treatment timelines and tumor characteristics are found in Supplemental Table ST4. There was no significant difference in age, sex distribution, largest tumor diameter or tumor burden between groups A and B (Supplemental Table ST5). Group A patients had tumor resection 12.5 days (mean, range 9 to 17 days) after starting hu14.18-IL2 and Group B patients had their resection 28.6 days (mean, range 18 to 43 days) before starting hu14.18-IL2 (Supplemental Table ST6). The median diameter of largest tumor focus for Groups A and B were 0.7 cm and 1.1 cm, respectively (Supplemental Table ST7). The median estimated tumor burden for Groups A and B were 0.25 cm3 and 0.466 cm3, respectively (Supplemental Table ST8). None of these parameters were significantly different between Groups A and B.
Tumor Infiltrating Lymphocyte (TIL) Levels and Stromal Signature are Prognostic of RFS and OS in Group A, but not Group B Patients
We found a significant prognostic effect of TILs on survival endpoints (RFS p=0.0088, OS p=0.0307) when separated by median values, irrespective of treatment group, with low TILs associated with worse outcomes (Figure 1A). When stratified by treatment groups, however, these effects were only seen in Group A patients (RFS p=0.0030, OS p=0.0417), but not in Group B patients (Figure 1B, C). Additionally, the stromal signature (37, 11) demonstrated significant prognostic value for survival endpoints (RFS p=0.0072, OS p=0.0036) when separated by the median, irrespective of treatment group, with low stromal signature demonstrating worse outcomes (Figure 1D). When stratified by treatment group, however, these effects were seen in Group A patients (RFS p=0.0082, OS p=0.0145), but not in Group B patients (Figure 1E, F). In addition, these relationships are somewhat similar for the cytolytic and immune signature (Supplemental Figure SF2).
Figure 1. Morphologic Tumor Infiltrating Lymphocytes (TILs) Assessment and the Stromal Gene Signature Prognosticate Recurrence Free and Overall Survival after hu14.18-IL2 Immunocytokine (IC) Treatment, but not Before IC Treatment.
(A-C) Kaplan Meier curves of patients stratified by TIL assessment are shown. Panel A demonstrates stratification by median TIL assessment is prognostic for recurrence free survival (RFS) and overall survival (OS) within all patients. Patients with below the median TIL levels demonstrate worse outcomes. Panels B and C show this prognostic effect on RFS and OS, respectively, retained in treatment Group A, but not in treatment Group B. (D-F) Kaplan Meier curves of patients stratified by the stromal signature (37) are shown. Panel D demonstrates that the stromal signature is also prognostic of RFS and OS within all patients. Patients with below the median Stromal Signature demonstrate worse outcomes. Similarly, panels E and F show this prognostic effect on RFS and OS, respectively, retained in treatment Group A, but not in treatment Group B.
We applied Cox regression analyses to assess if the gene expression-based signatures provide adjunct prognostic value when combined with TILs. In this analysis, the immune and stromal signatures were handled as continuous variables, while TILs were handled as a categorical variable with ‘low’/’high’ categories (as in Figure 1). In the case of RFS time, the univariate Cox regression shows that in group A both TILs and the stromal signature (SS) are significantly associated with survival (TILs: HR=0.073 and p=0.018, SS: HR=0.097 and p=0.006). The immune signature in this univariate model shows a trend towards statistical significance (HR=0.38 and p=0.051). The bivariate Cox regression that combines TILs and the stromal signature shows that the overall model is significant (p-value of 0.01), with both covariates also being significant (TILs: HR=0.095 and p=0.038, SS: HR=0.067 and p=0.013). The bivariate Cox regression that combines TILs and the immune signature shows that the overall model is significant (p-value of 0.02), with TILs being significant (HR=0.095 and p=0.031) and the immune signature not significant (HR=0.405 and p=0.117). Thus, the results of this analysis indicate that, in the case of RFS time in group A, both TILs and the stromal signature are significantly associated with survival, even after controlling for the other variable (that is, the stromal signature provides an adjunct prognostic value). The results of a similar analysis performed for OS time in group A indicate that only the stromal signature is significantly associated with survival after controlling for the other variable (HR=0.027 and p=0.04). In group B, for both OS and RFS time, no significant associations are observed in either univariate or bivariate Cox regression models.
Positive Correlations Between Myeloid and Lymphoid Cells Gene Signatures in Group A (Pre-Treatment) Tumors
Spearman correlation matrices of the 25-whole transcriptomics-based immunologic gene signatures were created on treatment-naïve tumors (n=10, Supplemental Figure SF3A) as well as on Group A tumors previously treated with hu14.18-IL2 (n=13, Supplemental Figure SF3B). Significance levels in the form of Log10 (1/p-value) estimates are shown in Supplemental Figure SF3A–B. While naïve tumors demonstrated high levels of correlation between lymphocytic signatures (B, T, and NK cells) in Supplemental Figure S3A, these were less pronounced correlations than seen with treated tumors between lymphocytic and myeloid signatures (monocytes, macrophages, dendritic cells, eosinophils, and neutrophils) in Supplemental Figure SF3B. Subtraction of these Spearman correlation coefficients between hu14.18-IL2 treated and naïve tumors demonstrates a marked increase in correlation of myeloid signatures with lymphoid signatures (Supplemental Figure SF3C) in tumors treated with hu14.18-IL2; this implies that the hu14.18-IL2 treatment caused parallel changes in lymphoid and myeloid compartments. A comparison using alternative methods of determining correlation (Spearman and Pearson) confirmed these findings (Supplemental Figure SF4C–D).
Immunologic Gene Signatures Prognosticate Favorable Patient Survival in hu14.18-IL2 Treated (Group A) Tumors, but not in Untreated (Group B) Tumors
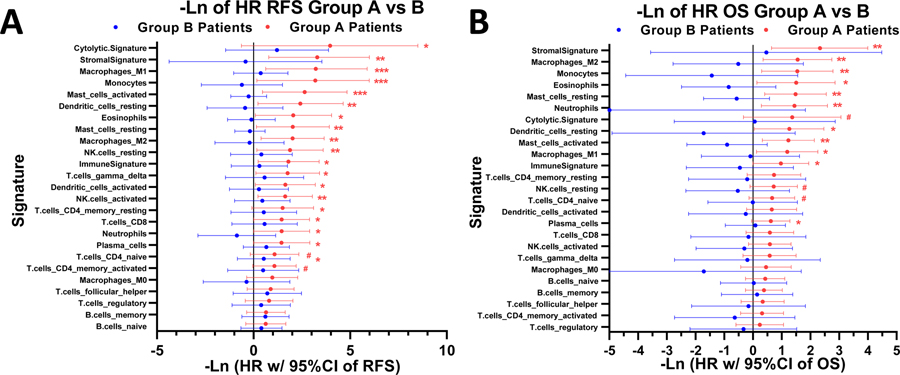
The 25 immunologic gene signatures consistently demonstrated favorable hazard ratios for RFS and OS (Figure 2A, B). For Group A patients, ten (40%) of these signatures showed statistically significant favorable hazard ratios (HRs) of RFS (and ten others showed a trend, 0.01< p ≤ 0.1); in addition 6 (24%) were statistically significant favorable HRs of OS (and eight others showed a trend, 0.01< p ≤ 0.1). In contrast, none of these relationships between gene signatures and survival demonstrated statistical significance (or showed a trend, 0.01< p ≤ 0.1) within Group B patients (RFS in Figure 2A and OS in Figure 2B). The sample size of Group B is smaller than Group A (n=8 versus n=13, respectively), but the difference in p-values between Groups A and B is large, and thus is unlikely to only be attributed to the smaller sample size. Another observation emphasizing the key difference between groups A and B independently of sample size is that in group A the HRs for all 25 immunologic signatures in both the OS and RFS analyses are favorable (less than 1, Figure 2), whereas in group B in the OS analysis the HRs for 20 of 25 signatures are unfavorable (greater than 1) and in the RFS analysis the HRs for 9 of 25 signatures are unfavorable. Moreover, the HRs for all 25 signatures are more favorable (to the R of in Figure 2) in Group A than in Group B, (in both the OS and RFS analyses). Thus, the immunologic signatures do not show significant associations with improved outcome in group B because the HRs for Group B are larger than those for group A, not because of a smaller sample size. A more granular representation of the data as a heatmap with patient level tumor gene signatures is found in Supplemental Figure SF5.
Figure 2. Gene Signatures Prognosticate Favorable Patient Survival in hu14.18-IL2 Treated Tumors, but Not in Group-B (Untreated) Tumors.
Tree plots of the 25 immunologic signatures consistently demonstrated favorable hazard ratios for RFS and OS. (A) For Group A patients, ten (40%) of these signatures showed statistically significant favorable hazard ratios (HRs) of RFS (and ten others showed a trend, 0.01< p ≤ 0.1) but none were significant (or showed a trend) for Group B patients. (B) For Group A patients six (24%) of these signatures showed statistically significantly favorable HRs of OS (and eight others showed a trend, 0.01< p ≤ 0.1), but none were significant (or showed a trend) for Group B patients. Hazard ratios are –Ln (natural log) transformed such that a point estimate of 1.0 denotes a HR of 0.368 and a point estimate of −1.0 denotes a HR of 2.72, thus points plotted to the right of 0.0 indicate favorable outcome and those to the left indicate unfavorable outcome. Legend: *** = p-value <0.001; ** = p-value <0.01, * = p-value <0.05, # = p-value <0.1. The p-values are from the likelihood ratio test (it performs better when sample size is small), whereas the 95% confidence intervals are based on the Wald test.
Immunologic Gene Expression Levels Demonstrate Favorable Survival and Increased Significance in Hu14.18-IL2 Treated (Group A) Tumors, but Not in Untreated (Group B) Tumors
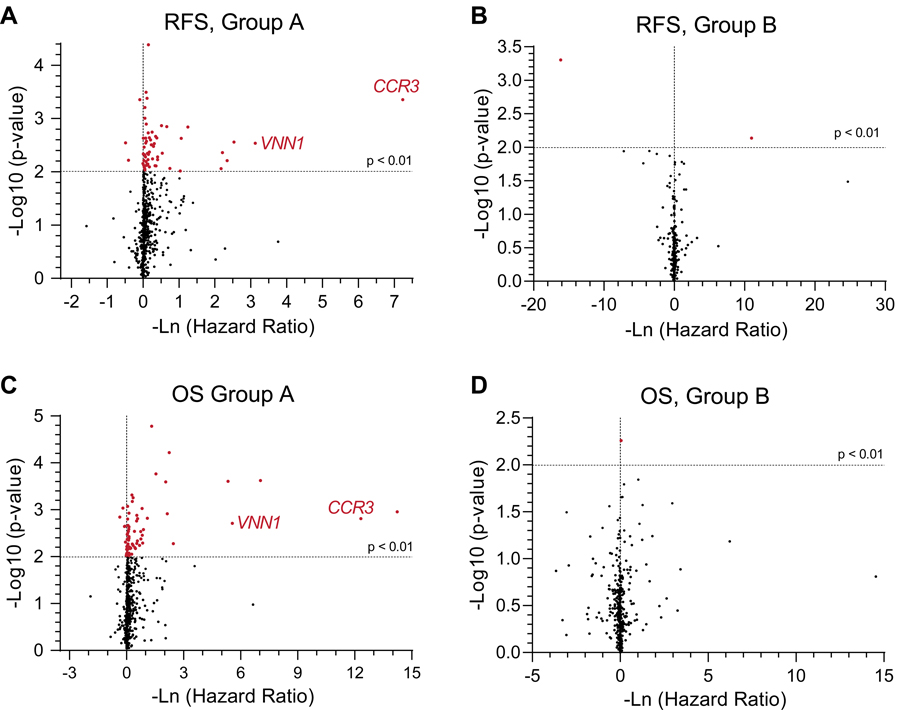
The transcriptomic data were interrogated on a more granular level by analyzing individual expression levels of 644 immune related genes (Supplemental Table ST9) within Groups A and B. These relationships are represented in volcano plots between Groups A and B for associations with RFS and for OS (Figure 3). We found several immune related genes that demonstrated significantly favorable prognostic values for both OS and RFS in Group A tumors (Figures 3A and 3C). In Figure 3A, there were 59 genes associated with RFS versus 4 genes associated with decreased RFS in Group A patients (Supplemental Tables ST10–ST11). In Figure 3C, there were 67 genes associated with OS versus 6 genes associated with decreased OS in Group A patients (Supplemental Tables ST12–ST13). Interestingly, expression levels of CCR3 (highlighted in red), a chemokine receptor on eosinophils and helper T cells, stood out as highly significant and associated with a very skewed HR, prognostic of favorable RFS and OS in Group A (Figure 3A and 3C). While not as significant or potently influencing HR, expression of VNN1 (a membrane protein influencing hematopoietic and T cell trafficking, highlighted in red) is also significantly associated with a favorable HR for both RFS and OS in Group A (Figure 3A and 3C). In contrast, we found very few immunologic genes that demonstrated statistically significant favorable prognostic values for OS or RFS in Group B tumors (Figure 3B and 3D). Associations between expression levels of individual immunologic genes and RFS and OS are presented (Supplemental Figures SF6). Again, these relationships were most robustly seen in Group A, but not in Group B, patients.
Figure 3. Granular Immunologic Gene Expression Demonstrates a Predominance of Parameters with Favorable Prognostic Value Rather than Unfavorable Prognostic Value.
Volcano plots of granular immunologic gene expression are plotted with hazard ratios against significance levels (A-D). Panels A and C represent the Group A patients’ (n=13) RFS and OS relationships, respectively, against these immunologic genes. Panels B and D represent the Group B patients’ (n=8) RFS and OS relationships, respectively, against these immunologic genes. Several genes show significantly favorable hazard ratios (red points) in Panels A and C. Among these CCR3 (a chemokine receptor on eosinophils and helper T cells) and VNN1 (a membrane protein felt to influence hematopoietic and T cell trafficking, both labelled in red) stood out as highly significant, and associated with a very skewed HR, prognostic of favorable RFS and OS in Group A. The horizontal dotted line represents a p-value of 0.01. The vertical dotted line represents a hazard ratio of 1.0.
We also examined the association of HR with the gene expression of class based immune molecules and RFS and OS times. Heatmaps of HR and p-values for RFS and OS times for TLRs (Toll-like Receptor), KIRs (Killer-like Ig Receptors), FcRs (Antibody Fragment Constant Region Receptors), TNFSFs (Tumor Necrosis Factor Superfamily proteins and receptors), ILs (Interleukins), and immune-related CD (Clusters of Differentiation) markers are shown in Supplemental Figures SF7–SF12, respectively. The above gene sets were taken from existing literature, showing these classes of genes having significant contribution towards anti-tumor immunity (43). Consistent with our observations described above, the gene expression of many of these were more significantly associated with improved HR (green) in Group A tumors, but not in Group B tumors.
Melanoma-Related Gene Expression Increases HR and Apoptosis Related Gene Expression Decreases HR of RFS and OS in Group A, But Not Group B Patients
Next, we interrogated transcriptomic data of 1,713 cancer related genes (found at https://www.proteinatlas.org/search/protein_class:Cancer-related+genes), which may have an influence on survival times. We found a number of clinically relevant melanoma related genes that demonstrated significantly increased (unfavorable) HRs for patients whose tumors express these genes (Figure 4, top panels) in Group A patients but not in Group B. These include MLANA (Melan A), SOX10, S100A, S100B, MITF, and PMEL (HMB-45). These markers are used regularly as melanoma markers for clinical pathologic identification and assessment by immunohistochemistry (IHC). In contrast, apoptosis related genes (including: BCL10, CASP8 [Caspase 8], CASP10 [Caspase 10], FAS, TNFSF10 [TRAIL], and others shown) revealed significantly decreased (favorable) HR in Group A patients, but not Group B (Figure 4, bottom panels). A more granular, individual patient level, breakdown of these melanoma-related and apoptosis-related genes may be found in Supplemental Figure SF13.
Hu14.18-IL2 Treated Tumors Demonstrate Increased Levels of Growth Factors and Repair, Overall Gene Expression, but Decreased Levels of Melanoma Related Genes
Next, we examined the global gene expression alterations resulting from one course of Hu14.18-IL2 treatment by comparing genes significantly different in Group A versus Group B and untreated (Naïve) tumors; such individual genes may shed light on hu14.18-IL2 effects on the tumor microenvironment (Figure 5). There are 52 genes (at p<0.01) and 610 genes (at p<0.05) that show significant fold difference between treated and hu14.18-IL2 naïve tumors (Supplemental Table ST14). In general, the data demonstrate a marked preponderance of genes that are significantly increased in expression in 13 Group A vs. 10 naïve (8 Group B and 2 never treated) tumors, indicating an overall increase in gene expression of many genes after treatment with hu14.18-IL2. A more granular assessment indicates that Hu14.18-IL2 treated tumors show increased levels of repair (growth factor and DNA damage repair) genes and immunologic genes, but decreased levels of melanoma, Warburg metabolism and cell-cycling related genes (Supplemental Figure SF14).
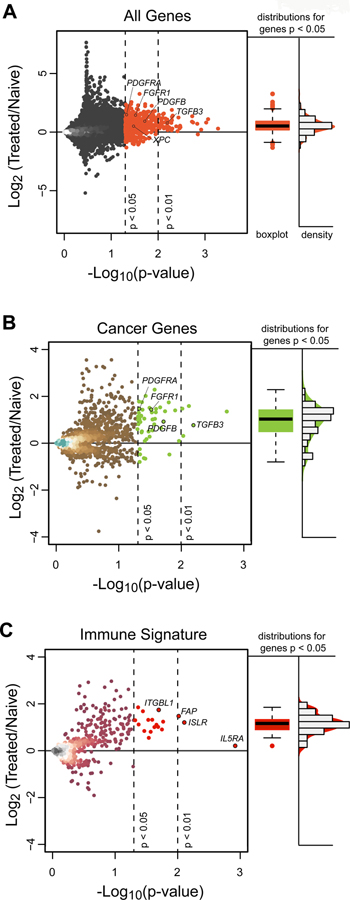
Figure 5. IC Treatment reveals a Predominance of Increased Rather than Deceased Gene Expression.
(A) Gene expression for individual genes are plotted with Log 2 fold changes from naïve [8 in Group B and 2 untreated (No IC)] patients to 13 Group A hu14.18-IL2 treated patients (y-axis) against significance levels (x-axis). There is a marked preponderance of genes that are significantly increased (upper right quadrant), indicating an overall increase in gene expression of transcripts after treatment with hu14.18-IL2. (B) Gene expression plot for individual cancer-related genes (annotated from the Human Protein Atlas) with Log 2 fold changes from 10 naïve patients to 13 Group A patients demonstrates a preponderance of significantly increased (upper right quadrant), but not decreased, expression of genes in Group A vs. naïve tumors. (C) Gene expression plot for individual ssGSEA CIBERSORT immune genes with Log 2 fold changes from 10 naïve patients to 13 Group-A patients demonstrates a preponderance of significantly increased, but not decreased, expression of genes in Group A vs. naïve tumors. There are 52 genes (at p<0.01) and 610 genes (at p<0.05) that show significant fold difference between treated and hu14.18-IL2 naïve tumors. The vast majority of these were upregulated rather than downregulated. Fold change is Log2 transformed such that a point estimate of 1.0 is denotes a fold change of 2.0 and a point estimate of −1.0 denotes a fold change of 0.5. In A, B, and C, the vertical dotted lines represent p-values of 0.05 and 0.01. The horizontal dotted line represents a ratio of 1.0 for expression of the individual gene in the tumors from 13 Group A patients vs. in the 10 naïve tumors. Immunologic genes were taken from all 645 immunologically related genes that are included in our ssGSEA analysis. For Fig. 5A, 5B and 5C, to the right of the dot plots for gene expression ratio vs. p value are box plots and density histograms of those points that are significant (p<0.01), together with those that have 0.01< p < 0.05.
Pathway-Level Analysis Reveals Over-representation of genes involved in Immunologic Pathways in Group A, but not in Group B.
A more integrative pathway-level enrichment analysis performed by using the ConsensusPathDB web-server indicates that the elevated expression of genes involved in innate immune response pathways is linked to favorable survival in the hu14.18-IL2 treated tumors (Group A), but not in the yet untreated tumors (Group B) (Figure 6, green left sided panels). A more extensive catalogue of 53 pathways associated with favorable OS (HR < 1) also demonstrated innate immunologic processes (Supplemental Figure SF15). Conversely, elevated expression of genes involved in glycolysis pathways was linked to unfavorable survival in the hu14.18-IL2 treated tumors (Group A), but not in the treatment naïve tumors (Group B) (Figure 6, pink right sided panels). Additionally, the elevated expression of genes involved in de-ubiquitination and processing of ubiquitinated proteins appears to be linked to unfavorable survival in Group B, (Figure 6, pink right sided panels) but not in Group A.
Figure 6. Pathway -Level Enrichment Analysis of Gene Sets Whose Expression Correlates with Survival.
The pathway-level enrichment analysis indicates that genes involved in innate immune response are significantly over-represented in patients with improved OS (HR < 1) in the IC treated tumors (n = 13, Group A, green top left panels), but not in the untreated tumors (n = 8, Group B). A complete list of 53 pathways significant associated with favorable OS in Group A patients may be found in Supplemental Figure S15. In contrast, only a few pathways including ectodermal differentiation and activation of SMO were found to associate with Group A patients with decreased OS (pink top right panel). Pathways associated with improved RFS (HR < 1) in Group A patients include chemokine and cytokine signaling (green middle left panels). In contrast, pathways associated with unfavorable RFS (HR > 1) in Group A patients include glycolysis and other glucose metabolism (pink middle right panels). In Group B patients, pathways associated with focal adhesion associated with favorable OS in Group B (green bottom left panel). In contrast, genes involved in de-ubiquitination and processing of ubiquitinated proteins are over-represented in patients with unfavorable OS (HR > 1) in Group B (pink bottom right panels). Pathways are ranked by their log (1/q-value) values and colored blue when the q-value <0.05 with darkening blue representing increasing statistical significance. The q-value is the false discovery rate (FDR) adjusted p-value and provides a correction for multiple tests (that is, takes into account the total number of pathways used in the analysis). The q-value is commonly used in genome-wide studies.
Network-level analysis of genes correlated with the overall survival (OS) time in Group A indicates that ELAVL1 is a hub protein that interacts with the products of many of these genes.
Intriguingly, the network-level analysis of genes whose expression is correlated with the OS time in Group A revealed a complex network of physical interactions (Supplemental Figure SF16A) in which ELAVL1 is a hub protein that interacts with the products of 25 of these genes. The majority of these genes (20 out of 25) were genes whose expression positively correlated with the OS time for Group A in this study. In contrast to Group A, the network-level analysis of genes whose expression was correlated with the OS time in Group B produced a very limited network of physical interactions that did not include ELAVL1 and did not contain any hub proteins (Supplemental Figure SF16B). This implies that hu14.18-IL2 treatment may result in a coordinated immune response centered with ELAVL1 which may impact on patient’s outcome.
Discussion
We analyzed histology and tumor mRNA whole transcriptomes from stage III and IV melanoma patients to screen for predictive biomarkers for patients participating in this hu14.18-IL2 immunotherapy trial. Patients were scheduled to receive 3 courses of hu14.18-IL2 and were randomized to have course 1 of the hu14.18-IL2 begin just before (Group A) or just after (Group B) surgical resection of all disease. No significant differences in RFS or OS between Groups A and B were seen in the overall trial (36). However, when evaluating tumor immune microenvironment using immune scores generated from ssGSEA (11), we found that TIL levels, as measured by histologic assessment, strongly and directly correlate with immune scores for all patients. Patients’ TIL levels in their resected tumors directly correlated with RFS and OS in the tumors from Group A patients who had been treated with hu14.18-IL2 prior to resection but no such correlation was seen in Group B patients who had not yet received hu14.18-IL2 prior to resection (Group B). Additionally, stromal signature is associated with improved HR of RFS and OS, in tumors that were resected after the first course of hu14.18-IL2 treatment (Group A), but not observed in tumors that were resected before initiating the hu14.18-IL2 (Group B).
We found a number of other immune signatures associated with decreased (favorable) HR of RFS and OS for Group A patients, but these findings were not found in Group B tumors. Specifically, several individual immune gene transcripts were associated with decreased HR of RFS and OS for Group A patients (but not for Group B patients). Amongst these transcripts associated with improved outcome for Group A patients were those related to cytotoxic T cells (Granzymes), NK cells (KIRs), and innate immune cells (TLRs). Additionally, there appeared to be a number of tumor-related genes where increased gene expression was significantly associated with increased HR of RFS/OS (melanoma marker transcripts) or decreased HR of survival (apoptosis transcripts) in Group A patients; no similar associations were seen for Group B. These findings suggest that the associations with outcome for these tumor transcripts were the result of changes in transcript expression induced by the hu14.18-IL2 therapy that were associated with RFS and OS. The inability of the immunotherapeutic intervention to induce these changes in a portion of the evaluable patients in Group A strongly predicted poorer clinical outcomes in these patients.
In addition, the data revealed that compared to naïve (untreated) tumors, hu14.18-IL2 treated tumors (Group A) demonstrated decreased levels of cell cycling transcripts (MKI67, CDK1), melanoma markers (MLANA [Melan A], MAGED1) and glycolysis transcripts (LDHA, HK1 [Supplemental Figure SF14]). Conversely, Group A tumors demonstrated increased levels of growth factor (PDGFRA, FGFR1, PDGFB, TGFB3 [Figure S14]) and repair transcripts (XPC) compared to naïve tumors. We take relationships between tumor-related transcripts as evidence of differences in tumor biology (or differences in tumor biology due to immune effects) that may indicate a change from a tumor replicative phenotype to an inflamed/healing-wound phenotype following hu14.18-IL2 therapy.
When evaluating Group A tumors, we found that expression of a number of individual genes were strongly and significantly associated with outcome (Figure 3 and Supplemental Figures SF7–SF12). Some of these may provide clues to mechanisms involved in improved outcome using hu14.18-IL2 like therapy and could potentially indicate additional molecules/pathways for targeting effective immunotherapy. We believe that the differences in transcript expression reflect changes in tumor-biology induced by the anti-tumor immune effects of hu14.18-IL2 treatment that was initiated only 12.5 days (mean) prior to the tumor resection for Group A. These changes in tumor biology are somewhat similar to that seen after a subacute (~2 weeks prior) injurious event (44).
Recently, an immuno-predictive score (IMPRES), a predictor of immune checkpoint blockade (ICB) response in melanoma which encompasses 15 pairwise transcriptomics parameters between immune checkpoint genes, has been described (45). The authors developed this gene expression predictor to indicate whether melanoma in a specific patient is likely to respond to treatment with ICB. We have done an analysis of IMPRES scores for these 13 Group A and 8 Group B patients; all patients have somewhat similar scores (ranging from 9 to 13). We see no statistical association of RFS or OS with IMPRES score within Group A or within Group B (data not shown). In contrast, we demonstrate the utility of the immune score as a predictor for response to hu14.18-IL2 when evaluated after treatment in this study of hu14.18-IL2. Checkpoint blockade is considered a mechanism to “release the brakes” on an immune response that is already underway. Thus, one would hypothesize that pre-treatment scores indicating pre-existing immune-activation (like the IMPRES) might be associated with clinical benefit from checkpoint blockade. In contrast, the induction of innate immune recognition by antibody-dependent cell-mediated cytotoxicity, together with IL2 induced activation-proliferation of immune cells, as shown for hu14.18-IL2 (46), might be considered an approach towards “starting the engine and providing some gas” to the immune response (47). From this perspective, one might expect that increased expression of immune genes seen following treatment initiation with this form of therapy, rather than gene expression seen before treatment, would be a better predictor of outcome, as observed in this report.
Finally, a network level analysis of genes associated with OS in this study revealed a complex interaction of many molecules associated with outcome for Group A (Figure S16A), with no similar complex network identified for Group B (Figure S16B). The network for Group A indicates a key position for ELAVL1, which is known to bind 3’-UTR regions of mRNAs with A-U rich elements and stabilize them, thus augmenting gene expression (48). ELAVL1 expression is known to be associated with better prognosis in stage-II melanoma, increased C-reactive protein and inflammation, increased TNF and TLR4, as well as regulation of GATA-3 and Th2 cytokine genes (49). These functions may put ELAVL1 in a key position for further analyses and possible targeting.
In summary, these data suggest that both immune and tumor cell processes, as measured by RNAseq analyses, are associated with OS and RFS when evaluated in tumors resected approximately 2 weeks after starting the first of 3 scheduled monthly courses of hu14.18-IL2 immunotherapy. These results may help point to specific immune-related and tumor response related molecular pathways associated with improved outcome after immunotherapy. Furthermore, if this approach is validated, it may enable analyses of RNAseq immune and tumor cell signatures obtained after initiating immune therapy to potentially predict which patients should continue their current regimen and which might need a change or addition to their therapy, due to the absence of a favorable histological or RNAseq response.
Supplementary Material
Acknowledgments:
We thank the UWCCC Melanoma clinical research and laboratory support team for the clinical research involved. We acknowledge contributions by the surgical oncologists participating in this study including Drs. Sharon Weber, Heather Neuman, Greg Hartig, Tracey Weigel, David Mahvi and also contributions by Mary Beth Henry, NP. This study was facilitated by the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://biowulf.nih.gov), and was enabled by funding from the Division of Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Funding. This work was supported by NIH Grants CA032685, CA166105, CA197078 (P. Sondel), GM067386, Clinical and Translational Science Award 1TL1RR025013–01 to the University of Wisconsin Institute for Clinical and Translational Research (E. Ranheim, Z. Morris, M. Albertini, K Kim and P Sondel), Cancer Center Support Grant, P30 CA014520, to the University of Wisconsin Carbone Cancer Center (E. Ranheim, Z. Morris, M. Albertini, J. Hank, K. Kim and P Sondel), The Midwest Athletes for Childhood Cancer Fund (J. Hank and P. Sondel), Ann’s Hope Foundation (M. Albertini), the Tim Eagle Memorial (M. Albertini), the Jay Van Sloan Memorial from the Steve Leuthold Family (M. Albertini), and The Stand Up To Cancer – St. Baldrick’s Pediatric Dream Team Translational Research Grant (SU2C-AACR-DT1113). Stand Up To Cancer is a division of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Footnotes
Conflict of interest statement: The authors report no financial or other conflicts of interest to disclose related to this publication.
Trial registration. ClinicalTrials.gov identifier: NCT00590824
Data and materials availability: The RNA sequencing data are uploaded on the NCBI’s Gene Expression Ominibus (GEO) website with Accession Number GSE133713 and with a released date set to be July 1, 2020. [NCBI tracking system #20118519] The raw sequencing data has been deposited in the database of Genotypes and Phenotypes (dbGaP) under the accession phs001947.
References and Notes:
- 1.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013. August 22; 500(7463):415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013. July 11; 499(7457):214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee N, Zakka LR, Mihm MC Jr, Schatton T. Tumour-infiltrating lymphocytes in melanoma prognosis and cancer immunotherapy. Pathology 2016. February; 48(2):177–87. [DOI] [PubMed] [Google Scholar]
- 4.Saldanha G, Flatman K, Teo KW, Bamford M. A Novel Numerical Scoring System for Melanoma Tumor-infiltrating Lymphocytes Has Better Prognostic Value Than Standard Scoring. Am J Surg Pathol 2017. July; 41(7):906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albertini MR. The age of enlightenment in melanoma immunotherapy. J Immunother Cancer. 2018. August 22; 6(1):80. doi: 10.1186/s40425-018-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med 2013. July 11; 369(2):122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larkin J, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 2015. July 2; 373(1):23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schadendorf D, et al. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J Clin Oncol 2015. June 10; 33(17):1889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews MC, Reuben A, Gopalakrishnan V, Wargo JA. Concepts Collide: Genomic, Immune, and Microbial Influences on the Tumor Microenvironment and Response to Cancer Therapy. Front Immunol 2018. May 4; 9:946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen PL, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov 2016. August; 6(8):827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei JS, et al. Clinically Relevant Cytotoxic Immune Cell Signatures and Clonal Expansion of T Cell Receptors in High-risk MYCN-not-amplified Human Neuroblastoma. Clin Cancer Res 2018. May 21 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahl C, Guldberg P. The genome and epigenome of malignant melanoma. APMIS 2007. October; 115(10):1161–76. Review. [DOI] [PubMed] [Google Scholar]
- 13.Ibrahim N, Haluska FG. Molecular Pathogenesis of Cutaneous Melanocytic Neoplasms. Annu. Rev. Pathol. Mech. Dis 20094:551–579. [DOI] [PubMed] [Google Scholar]
- 14.Zhang T, Dutton-Regester K, Brown KM, Hayward NK. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res 2016. May; 29(3):266–83. Review. [DOI] [PubMed] [Google Scholar]
- 15.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015. July 9; 523(7559):231–5. [DOI] [PubMed] [Google Scholar]
- 16.Hu-Lieskovan S, Homet Moreno B, Ribas A. Excluding T Cells: Is β-Catenin the Full Story? Cancer Cell 2015. June 8; 27(6):749–50. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Cao M, et al. ; Spanish Melanoma Group. Early evolution of BRAFV600 status in the blood of melanoma patients correlates with clinical outcome and identifies patients refractory to therapy. Melanoma Res 2018. June; 28(3):195–203. [DOI] [PubMed] [Google Scholar]
- 18.Pagès F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018. May 10. [DOI] [PubMed] [Google Scholar]
- 19.Gillies SD, Reilly EB, Lo KM, Reisfeld RA. Antibody-targeted interleukin 2 stimulates T-cell killing of autologous tumor cells. Proc Natl Acad Sci U S A 1992; 89:1428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reisfeld RA, Gillies SD. Antibody-interleukin 2 fusion proteins: a new approach to cancer therapy. J Clin Lab Anal 1996; 10:160–6. [DOI] [PubMed] [Google Scholar]
- 21.Mujoo K, Cheresh DA, Yang HM, Reisfeld RA. Disialoganglioside GD2 on human neuroblastoma cells: target antigen for monoclonal antibody-mediated cytolysis and suppression of tumor growth. Cancer Res 1987; 47:1098–104. [PubMed] [Google Scholar]
- 22.Saleh MN, Khazaeli MB, Wheeler RH, Dropcho E, Liu T, Urist M, Miller DM, Lawson S, Dixon P, Russell CH. Phase I trial of the murine monoclonal anti-GD2 antibody 14G2a in metastatic melanoma. Cancer Res 1992; 52:4342–7. [PubMed] [Google Scholar]
- 23.Hank JA, Surfus JE, Gan J, Jaeger P, Gillies SD, Reisfeld RA, Sondel PM. Activation of human effector cells by a tumor reactive recombinant anti-ganglioside GD2 interleukin-2 fusion protein (ch14.18-IL2). Clin Cancer Res 1996; 2:1951–9. [PubMed] [Google Scholar]
- 24.Lode HN, Xiang R, Varki NM, Dolman CS, Gillies SD, Reisfeld RA. Targeted interleukin-2 therapy for spontaneous neuroblastoma metastases to bone marrow. J Natl Cancer Inst 1997; 89:1586–94. [DOI] [PubMed] [Google Scholar]
- 25.Imboden M, Murphy KR, Rakhmilevich AL, Neal ZC, Xiang R, Reisfeld RA, et al. The level of MHC class I expression on murine adenocarcinoma can change the antitumor effector mechanism of immunocytokine therapy. Cancer Res 2001; 61:1500–7. [PubMed] [Google Scholar]
- 26.Becker JC, Pancook JD, Gillies SD, Furukawa K, Reisfeld RA. T cell-mediated eradication of murine metastatic melanoma induced by targeted interleukin 2 therapy. J Exp Med 1996; 183:2361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang RK, Kalogriopoulos NA, Rakhmilevich AL, Ranheim EA, Seo S, Kim K, Alderson KL, Gan J, Reisfeld RA, Gillies SD, Hank JA, Sondel PM. Intratumoral hu14.18-IL-2 (IC) induces local and systemic antitumor effects that involve both activated T and NK cells as well as enhanced IC retention. J Immunol 2012. September 1; 189(5):2656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris ZS, Guy EI, Francis DM, Gressett MM, Carmichael LL, Yang RK, Armstrong EA, Huang S, Navid F, Gillies SD, Korman A, Hank JA, Rakhmilevich AL, Harari PM, Sondel PM. In situ Tumor Vaccination by Combining Local Radiation and Tumor-specific Antibody or Immunocytokine Treatments. Cancer Research 2016; 76:3929–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang RK, Kalogriopoulos NA, Rakhmilevich AL, Ranheim EA, Seo S, Kim K, Alderson KL, Gan J, Reisfeld RA, Gillies SD, Hank JA, Sondel PM. Intratumoral treatment of smaller mouse neuroblastoma tumors with a recombinant protein consisting of IL-2 linked to the hu14.18 antibody increases intratumoral CD8+ T and NK cells and improves survival. Cancer Immunol Immunother 2013. August; 62(8):1303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neal ZC, Yang JC, Rakhmilevich AL, Buhtoiarov IN, Lum HE, Imboden M, Hank JA, Lode HN, Reisfeld RA, Gillies SD, Sondel PM. Enhanced activity of hu14.18-IL2 immunocytokine against murine NXS2 neuroblastoma when combined with interleukin 2 therapy. Clin Cancer Res 2004; 10:4839–47. [DOI] [PubMed] [Google Scholar]
- 31.King DM, et al. Phase I clinical trial of the immunocytokine EMD 273063 in melanoma patients. J Clin Oncol 2004; 22:4463–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osenga KL, et al. ; Children’s Oncology Group. A phase I clinical trial of the hu14.18-IL2 (EMD 273063) as a treatment for children with refractory or recurrent neuroblastoma and melanoma: a study of the Children’s Oncology Group. Clin Cancer Res 2006; 12:1750–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shusterman S, et al. Antitumor activity of hu14.18-IL2 in patients with relapsed/refractory neuroblastoma: a Children’s Oncology Group (COG) phase II study. J Clin Oncol 2010. November 20;28(33):4969–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang RK, Sondel PM. Anti-GD2 Strategy in the Treatment of Neuroblastoma. Drugs Future 2010;35(8):665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albertini MR, et al. Phase II trial of hu14.18-IL2 for patients with metastatic melanoma. Cancer Immunology, Immunotherapy 2012; 61:2261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albertini MR, Yang RK, Ranheim EA, Hank JA, Zuleger CL, Weber S, Neuman H, Hartig G, Weigel T, Mahvi D, Henry MB, Quale R, McFarland T, Gan J, Carmichael L, KyungMann K, Loibner H, Gillies SD, Sondel PM. Pilot trial of the hu14.18-IL2 immunocytokine in patients with completely resectable recurrent stage III or stage IV melanoma. Cancer Immunology, Immunotherapy 2018. August 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013; 4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453–7, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou M, et al. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J Exp Clin Cancer Res 2015, 34:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nature Protocols, 2016, 11(10):1889–1907. [DOI] [PubMed] [Google Scholar]
- 41.Fabregat A, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 2018, 46(D1):D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slenter DN, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res 2018, 46(D1):D661–D667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson M, Denardo DG, Coussens LM. Polarized immune responses differentially regulate cancer development. Immunol Rev 2008. April;222:145–54. doi: 10.1111/j.1600-065X.2008.00600.x. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008. May 15; 453(7193):314–21. [DOI] [PubMed] [Google Scholar]
- 45.Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T, Tian T, Wei Z, Madan S, Sullivan RJ, Boland G, Flaherty K, Herlyn M, Ruppin E. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med 2018. August 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sondel PM, Gillies SD. Current and Potential Uses of Immunocytokines as Cancer Immunotherapy. Antibodies (Basel). 2012. July 4; 1(2):149–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tirapu I, Mazzolini G, Rodriguez-Calvillo M, Arina A, Palencia B, Gabari I, Melero I. Effective tumor immunotherapy: start the engine, release the brakes, step on the gas pedal, and get ready to face autoimmunity. Arch Immunol Ther Exp (Warsz). 2002; 50(1):13–8. [PubMed] [Google Scholar]
- 48.Takeuchi O HuR keeps interferon-β mRNA stable. Eur J Immunol 2015. May; 45(5):1296–9. [DOI] [PubMed] [Google Scholar]
- 49.Stellato C, et al. Coordinate regulation of GATA-3 and Th2 cytokine gene expression by the RNA-binding protein HuR. J Immunol 2011. July 1; 187(1):441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.