Abstract
Noncoding RNAs (ncRNAs) direct a remarkable number of diverse functions in development and disease through their regulation of transcription, RNA processing and translation. Leading the charge in the RNA revolution is a class of ncRNAs that are synthesized at active enhancers, called enhancer RNAs (eRNAs). Here, we review recent insights into the biogenesis of eRNAs and the mechanisms underlying their multifaceted functions and consider how these findings could inform future investigations into enhancer transcription and eRNA function.
The explosion of high-throughput sequencing data has revealed the complexity and diversity of the transcriptome. These data have also unexpectedly revealed that only 1–2% of the transcriptome provides instructions for the synthesis of functional proteins, while the remaining 98–99% gives rise to a plethora of ncRNAs, including transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), intronic RNAs, small nuclear (sn)RNAs, small nucleolar (sno)RNAs, microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). A recent addition to the expanding list of regulatory ncRNAs is the emerging class of enhancer RNAs (eRNAs), which are transcribed from enhancers in a tissue-specific manner. Increasing evidence that ncRNAs regulate gene expression has fundamentally altered how the scientific community views RNA-mediated gene regulation. These new advances in our understanding of ncRNAs have also piqued an interest in pursuing investigations of them, as the functions of the vast majority of ncRNAs remain to be determined.
Enhancers are classically defined as DNA sequences that regulate the gene expression networks underlying distinct cellular identities and cellular responses to environmental cues1–7. The ENCODE Consortium estimates that there are >400,000 putative enhancers encoded by the human genome8. Taken together, the evidence that enhancers account for a significant proportion of the genome and the identification of disease-associated genetic variants within enhancers underscores the importance of understanding how these elements are regulated and how they function in gene expression control8,9. While sequencing technologies have advanced the ability to predict enhancer location and activity, their functional dissection remains challenging due, in part, to their ability to act over long and variable distances with respect to their target genes and to the propensity of individual enhancers to regulate multiple genes (reviewed in ref.10). Additional challenges are that enhancer activity is dynamic and restricted to particular cell types or tissues and environmental signals3–7. Moreover, the prediction of specific sequences that contribute to enhancer function is problematic because of their modest sequence conservation across species11–15. It may be that a common function underlies enhancer evolution, given that enhancer function has been shown to be conserved without discernible sequence conservation16–18. Together, these studies indicate the need to continue improving methodologies that aid in both the functional dissection of enhancers and in uncovering the mechanisms directing their evolution.
Many different models for how enhancers function in gene control have been proposed since their initial discovery nearly four decades ago19–21. Specifically, there is considerable evidence demonstrating that looping of distal enhancers to their target promoters is required for enhancer function (reviewed in ref.22). For example, a key study revealed that experimental induction of chromatin looping between the mouse β-globin (Hbb) promoter and its associated enhancer region results in transcriptional activation of the Hbb gene23. Additional analyses of the forced looping of the Hbb enhancer and promoter regions revealed that enhancer-promoter contacts affect transcription by supporting an increase in the transcriptional burst fraction (number of transcribing alleles), although the burst size (number of transcripts produced) was unaltered24. The role of looping in the activation or increased rate of transcription may be due to the binding of transcription factors, cofactors and the general transcription machinery to enhancers, raising the local abundance of the transcription machinery within the vicinity of specific target genes. Enhancer looping has also been shown to play a role in RNA polymerase II (RNAPII)-mediated transcriptional elongation. Specifically, the LIM domain-binding protein 1 (LDB1), which establishes promoter-enhancer looping, plays a key role in regulating the pause release of RNAPII within the β-globin gene25,26. Enhancers may also regulate target genes via transcripts produced from the enhancer regions themselves. eRNA production is a widespread phenomenon that has been implicated in the regulation of gene expression in multiple cell types in response to various stimuli7,27–32. The emerging prevalence of noncoding eRNAs makes them well positioned to dynamically remodel cellular transcriptomes and adds a new layer of complexity to gene regulation. However, an issue that remains to be resolved is whether eRNAs have direct roles in gene control, as current efforts to determine this face the challenge of uncoupling eRNA function from the act of enhancer transcription. Thus, it remains necessary to develop tools to experimentally manipulate and model the direct functions of eRNAs.
Enhancers as functional noncoding RNA transcription units
Nearly a decade ago, two studies reported the intriguing finding that enhancer regions support transcription and give rise to noncoding eRNAs33,34. Since the discovery of eRNAs, there have been numerous reports that eRNA is synthesized in a cell-type and signal-dependent manner3–7. Because eRNAs are not readily detectable in steady-state RNA-sequencing data, their annotation depends on sequencing nascent RNA using approaches that include global run-on sequencing (GRO-seq)3,5–7,27,35–40, precision run-on nuclear sequencing (PRO-seq)41 and cap analysis gene expression (CAGE)42,43. These nascent transcription assays have been instrumental in uncovering a wide array of ncRNAs, including long noncoding RNAs, enhancer RNAs, promoter upstream transcripts and upstream antisense RNAs. Indeed, global annotation analyses have revealed that eRNA transcripts account for a large proportion of initiation events in the transcriptome, with approximately 40,000–65,000 eRNAs expressed in human cells4,44. The annotation of such transcripts in Drosophila melanogaster45,46 and Caenorhabditis elegans47 reinforces the finding that eRNAs are a common feature of active enhancers in metazoans.
eRNAs are produced from active enhancers that share several features: (i) an open chromatin state, reflected by the presence of DNase hypersensitive sites (DHSs); (ii) binding of transcription factors and cofactors, including the histone acetyltransferase p300 and cAMP response element-binding protein (CBP); and (iii) the co-occurrence of histone H3 lysine 4 monomethylation (H3K4me1) and histone H3 lysine 27 acetylation (H3K27ac)48–52. While these features have guided enhancer identification, not all active enhancers that share these features direct eRNA production53. Also, not all active enhancers support comparable levels of transcriptional activity. Higher levels of eRNA synthesis have been shown to correlate with an increased ratio of histone H3 lysine 4 trimethylation (H3K4me3) to H3K4me1 and increased levels of RNApII occupancy37,54,55. These findings suggest that measuring H3K4me1 accumulation alone may underestimate the number of functional enhancers and that H3K4me3 may be a superior predictor of the level of enhancer activity. Thus, identifying additional features that can be used to predict enhancers will improve the identification of functionally active enhancers and bona fide eRNAs. In turn, eRNAs themselves may prove to be the most reliable predictor of enhancer activity.
Regulation of enhancer transcription
Enhancers and promoter regions of protein-encoding genes share similar properties and rules for transcription initiation. Core promoter sequences, such as the TATA box, nucleosome spacing and the assembly of general transcription factors (TFIID/RNAPII) and cofactors (Mediator, p300) are observed at both enhancers and promoters55,56. Consistent with this, several studies have revealed that, similar to promoter regions, transcription at enhancers predominantly occurs in a bidirectional manner4,37,46,55,57–59. Enhancers and promoters have also been shown to be functionally interchangeable in supporting RNAPII initiation: intragenic enhancers behave as alternative promoters and can functionally substitute for promoters to drive mRNA and long noncoding RNA transcrip tion60. Studies in Drosophila have also revealed that bidirectionally transcribed enhancers behave as weak promoters and, conversely, that bidirectionally transcribed promoters can function as strong enhancers53. These findings blur the classical definitions of promoters and enhancers and raise the possibility that noncoding transcripts generated at both regulatory regions may be functional (reviewed in refs.61,62).
By comparison, there are discernible differences between the transcriptional elongation phases of eRNA and mRNA transcript production. These differences primarily reflect the well-defined cycles of RNAPII activity, which correlate with phosphorylation and dephosphorylation of the carboxyl terminal domain (CTD) of mammalian RNAPII63–65. The CTD domain consists of a heptameric sequence (YSPTSPS) repeated 52 times that is phosphorylated at S2, S5, S7, T4 and Y166–68. The general model of CTD phosphorylation during transcription is that the CTD becomes enriched with S5P at the 5’ ends of coding regions, and, as RNAPII elongates a transcript, S2P levels increase and S5P levels decrease69–71. On the other hand, the elongation of enhancer transcripts is distinguished by low levels of the S2P form of RNAPII and minimal levels of the elongation-specific histone mark H3K36me3, both of which are enriched within gene bodies of lncRNAs and mRNAs55,72. Low levels of S2P are consistent with the enrichment of the Tyr1P form of RNAPII that is specifically found at active enhancers and not at the sense strand of gene promoters73,74. The prevalence of Tyr1P coincides with the production of eRNAs and promoter-directed upstream antisense transcripts (PROMPTs), both of which are relatively unstable due to exosome-mediated degradation73. Notably, Tyr1P is required for transcription termination control in yeast68,75 and, more recently, has been shown to prevent transcription readthrough at mammalian gene ends76. However, no apparent contribution of Tyr1P to the regulation of transcription readthrough is observed at enhancers76. Thus, Tyr1P accumulation may contribute to different functional consequences at promoters and enhancers. Further demonstrating a role for elongation in enhancer regulation, the RNAPII-associated transcription elongation factor SPT6 contributes to the recruitment of Integrator to lncRNA genes. The Integrator subunit INTS3 is enriched at enhancer regions that generate bidirectional eRNAs, and SPT6 depletion substantially decreases INTS3 recruitment and eRNA production77,78. The elongation factor ELL3 has also been shown to regulate occupation of enhancers by RNAPII79. Additional studies are needed to determine whether distinct mechanisms regulate mRNA and eRNA elongation and to fully understand the factors underlying the regulation of transcription and termination at enhancers.
Classification of noncoding enhancer RNAs
A comprehensive definition of eRNAs has not yet been formulated, as annotated eRNAs are defined as both (i) short, bidirectional, non-polyadenylated, non-spliced and unstable and (ii) unidirectionally transcribed, spliced, polyadenylated and stable33,80,81 (Fig. 1). While the majority of eRNAs are not polyadenylated or spliced, they tend to be capped, as shown by nuclear run-on assays followed by 5’ cap sequencing (5’GRO-seq) and CAGE4,7,37. Consistent with their low abundance and instability, eRNAs are predominantly localized in the nucleus and chromatin-bound fractions4,33,34,36,82,83.
Fig. 1 |. Molecular features that define enhancer RNAs.
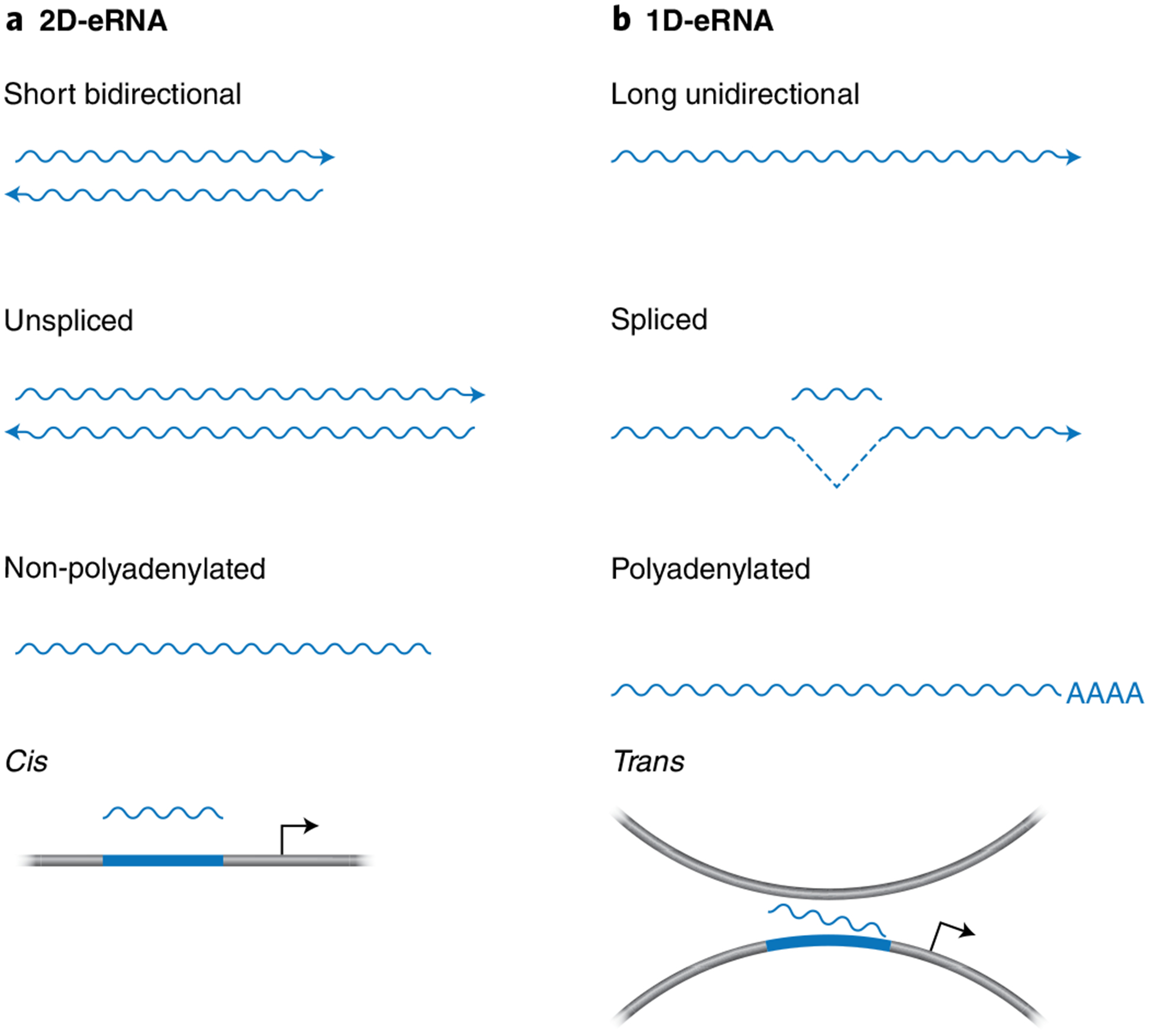
Schematic diagram to depict the differences between annotated eRNAs56. Distinct transcripts synthesized at enhancers are frequently classified as bidirectional (2D-eRNAs) or unidirectional (1D-eRNAs). a, The majority of enhancer transcripts are 2D-eRNAs that are comparatively short, non-polyadenylated and non-spliced and function in cis4,33,34,36,82,83. b, 1D-eRNAs are longer, unidirectional, polyadenylated and spliced33,80,81 and can function in trans29.
The early transcription termination of eRNAs is regulated by the Integrator complex in a manner that is probably dependent on termination-triggering polyadenylation (pA)-like signals6,37,84. In addition, the high turnover rate of eRNAs is mediated by the nuclear RNA exosome complex4,84–86. The length of eRNA transcripts is predicted to be less than 150 nucleotides, based on the identification of transcription termination sites within ~150 nucleotides of the transcription start site of enhancer loci46,87. Notably, current predictions of eRNA length that stem from PRO-seq and Start-seq datasets do not exclude the possibility of longer eRNA transcripts46,87.
The current predictions regarding eRNA length have led to their classification as a distinct family of lncRNAs. Nonetheless, lncRNAs are distinguished from eRNAs not only by their length, which is >200 nucleotides, but also by their processing, which is consistent with their stability88–90. Importantly, however, the definitions of lncRNAs and eRNAs are not mutually exclusive, and it is thus likely that some annotated eRNAs are actually lncRNAs and vice versa. This inherent complexity in classifying eRNAs is due, in part, to the difficulty in uncoupling specific lncRNAs from their associated enhancer elements33,81. For example, a recent study found that genomic regions defined by bidirectional transcription and enhancer features (termed eRNA-producing centers, or EPCs) are located in proximity to the transcription start sites of lncRNAs80. The ability of the EPCs to drive lncRNA production was shown to be correlated with higher enhancer activity and is in part linked to lncRNA maturation and the presence of evolutionarily conserved U1 splicing motifs80. As is the case for bidirectional promoters91, the enrichment of U1 splicing sites at EPCs may prevent premature termination and permit productive elongation by RNAPII to generate lncRNAs. It will therefore be of interest to determine whether this same mechanism also applies at enhancers and whether lncRNAs and eRNAs establish a positive transcriptional feedback loop that regulates eRNA and lncRNA synthesis. It should be noted, however, that only a minority (3–5%) of the total EPCs are in proximity to lncRNAs80. Similarly, the annotated lncRNAs linc-p21 (upstream of Cdknla) and Lockd (downstream of Cdknlb) regulate gene expression in a manner that is dependent on enhancer elements that are mapped to lncRNA loci rather than to the lncRNAs themselves92,93. In addition, reports that enhancer-associated lncRNAs function in a manner similar to eRNAs and, conversely, that eRNAs derived from super enhancers function as lncRNAs, reinforce the idea that these two classes of ncRNAs are not mutually exclusive94,95. These studies are also consistent with the notion that lncRNAs may have evolved from eRNAs. This suggestion is based on the presence of splicing and 3’ processing sequences in the vicinity of specific enhancers, which could promote eRNA stabilization and the evolutionary selection of new trans-acting functions96. Further analyses are needed to unravel the respective functions associated with particular DNA regions and the RNA transcripts they encode. Moreover, updates to our systems are needed to distinguish between eRNAs and lncRNAs and to resolve current investigations into the functional specialization of these classes of ncRNAs.
Functional roles of enhancer transcription and eRNAs
While eRNAs have become a hallmark of active enhancers, it remains to be resolved whether enhancer transcription, eRNAs themselves, or both, are important for enhancer activity. Studies have demonstrated that enhancer transcription is important for maintaining an open chromatin state that is readily accessible to transcription factors and cofactors83. Specifically, investigation of the formation of de novo enhancers during macrophage activation revealed that inhibition of transcriptional elongation at enhancers affects H3K4me1 and H3K4me2 deposition and is independent of eRNAs83. Also, transcription at intragenic enhancers, and not the eRNA itself, was found to interfere with and attenuate host gene expression during embryonic stem cell differentiation97. Similarly, a prior report revealed a requirement for the lncRNA Lockd in regulating gene expression in a manner that is dependent on its associated enhancer elements but not the Lockd transcripts92. Moreover, not all active enhancers support eRNA production, suggesting that eRNAs are not functionally important at all enhancers53. However, this remains a matter of continued debate, as it is presently unclear whether these enhancers produce low levels of eRNAs that are not readily detectable with current methods53. Nevertheless, a growing number of studies have shown that a subset of eRNAs are required to support the expression of cognate target genes, and we discuss these various eRNA functions7,27–32 in detail below.
A variety of experimental methods continue to advance our understanding of eRNA function. Perturbing nuclear decay pathways by knocking down the core components Exosc3 and nuclear RNase Exosc10 of the RNA exosome results in stabilization of eRNAs and permits their functional assessment4,85. It is important to note that, upon RNA exosome ablation, eRNA-expressing regions show an accumulation of R-loops, which is consistent with the involvement of the RNA exosome in resolving R-loops at active enhancers85. Another methodology to interrogate the functions of eRNAs includes CRISPR-Display, in which eRNAs are tethered to catalytically dead Cas9 (dCas9) for targeting to specific genomic loci98. Also, loss-of-function studies, such as eRNA knockdown analyses, could be used to target the enhancer region (deletion or insertion of genetic elements) or the enhancer-directed transcript (RNAi-mediated knockdown or incorporation of antisense oligonucleotides (ASO)). However, two recent studies have revealed that ASO-mediated knockdown is limited by its inability to discriminate between RNA function and the act of transcription because incorporation of ASOs results in premature transcription termination99,100. Several studies have also explored eRNA functions by employing inhibitors of transcription elongation, such as actinomycin D and flavopiridol28,36,83,101. These methods used to probe the functional consequences of eRNAs are often paired with approaches that can assess where in the genome the eRNAs are functioning. Specifically, chromatin isolation by RNA purification (ChIRP)-seq is a powerful method to determine the genomic sites bound by eRNAs27,102. In addition, single-molecule fluorescence in situ hybridization (smFISH), which provides a quantitative assessment of RNAs that are localized at distinct transcriptionally active regions, can be employed to investigate the cellular localization of eRNAs29,103.
While evidence for eRNA function is rapidly emerging, it is yet to be determined whether eRNAs act in cis versus in trans. Given that the majority of eRNAs are relatively unstable, it is not surprising that they are expected to act in cis4,33,34,36,82,83. Several recent studies have shown that eRNAs function in cis by demonstrating eRNA-dependent transcriptional regulation of mRNAs produced from loci adjacent to the corresponding eRNA-producing enhancer regions7,27–29,85. Specific eRNAs have been shown to function by interacting with CBP and BRD4 at the enhancers where these eRNAs are produced and transcriptional regulators are localized28,104. However, eRNAs have also been found to relocate to chromosomal regions distinct from those they are produced from to perform functions in trans29. A distal regulatory MyoD enhancer (distal regulatory region, or DRR) is transcribed into the DRR eRNA that mediates cohesin recruitment and promotes Myogenin gene expression in trans to control myogenic differentiation29. Similarly, an eRNA synthesized adjacent to the kallikrein related peptidase 3 (KLK3) gene functions in trans to enhance androgen receptor-dependent gene expression in human prostate cancer105. In both cases, the DRR and KLK3 eRNAs are polyadenylated, which raises the possibility that post-transcriptional regulation of eRNAs contributes to both increased eRNA stability and eRNA function in trans. Alternatively, the aforementioned eRNAs may be lncRNAs operating in trans. Future studies focused on establishing direct links between eRNAs and gene loci will aid in determining features that dictate whether eRNAs function in cis or in trans.
eRNAs contribute to gene control by altering the chromatin environment.
Several functional studies have revealed that eRNAs play an important role in regulating gene expression by modulating chromatin structure and function. Specifically, eRNAs direct chromatin accessibility at protein-coding promoters106. In addition, eRNAs function in cis to contribute to the dynamic stabilization of enhancer-promoter looping27,36,105 and in trans to regulate chromatin-remodeling events that modulate transcription factor complex assembly and gene regulation during myogenic differentiation29 (Fig. 2a). eRNAs also regulate the chromatin landscape through their interactions with epigenetic modifying enzymes that deposit (‘write’) post-translational modifications on the histone tails. Specifically, eRNAs have been shown to promote gene expression through their ability to augment histone acetylation at enhancers through interactions with the histone acetyltransferase CBP104 (Fig. 2b). Future analyses are required to extend the understanding of eRNAs in the regulation of chromatin structure and function.
Fig. 2 |. eRNA regulation of enhancer-promoter interactions and the epigenetic state of chromatin.
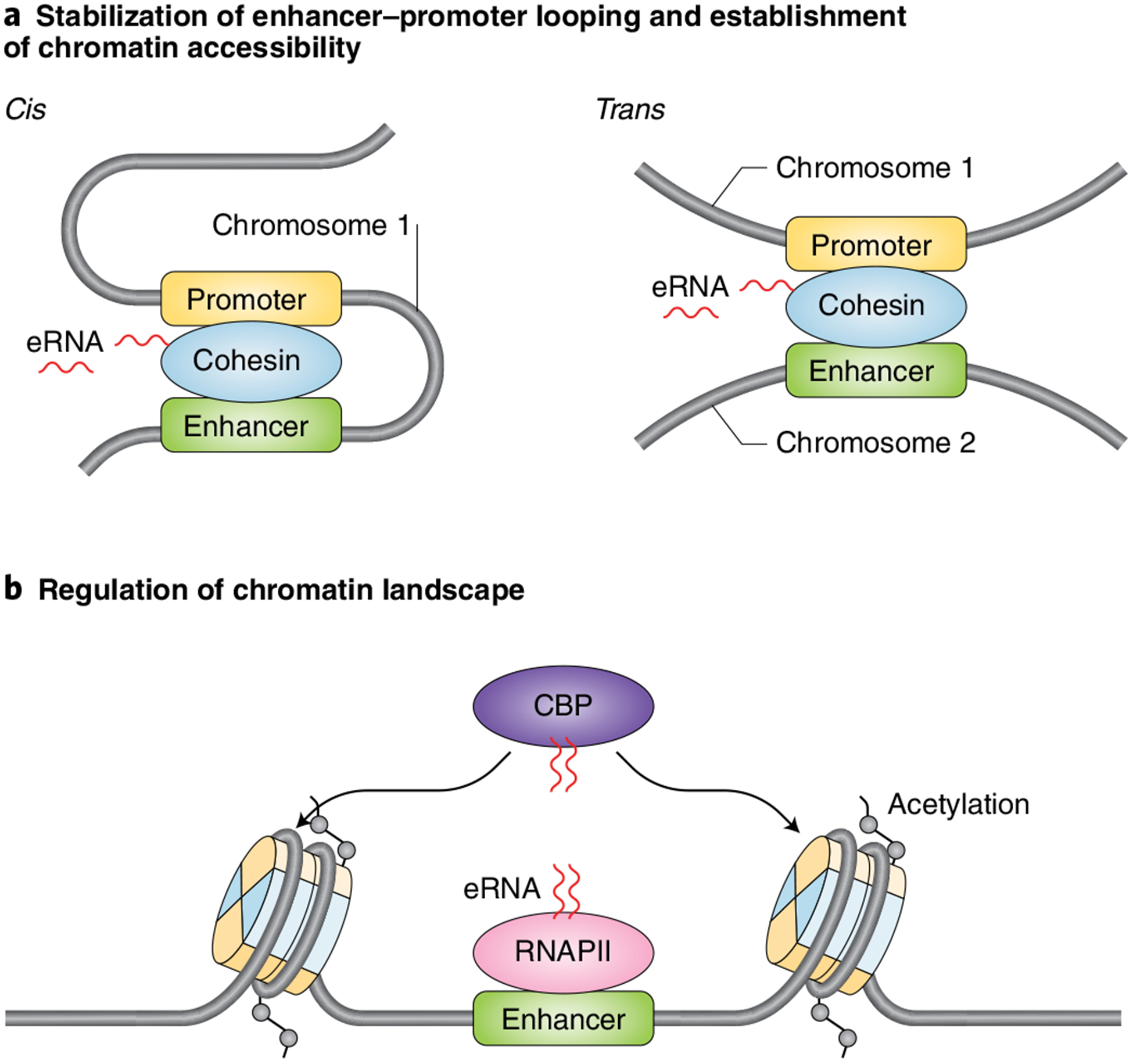
a, Schematic diagram depicting the roles of eRNAs acting in cis to regulate specific enhancer-promoter looping by supporting cohesin binding at enhancers27,106 (left) and influencing chromatin accessibility by modulating cohesin complex recruitment in trans to regulate gene expression (right). b, Schematic showing that eRNAs stimulate the catalytic activity of the histone acetyltransferase CBP to regulate increased deposition of histone acetylation, which in turn contributes to greater enhancer activation104.
eRNAs interact with transcriptional regulators to control gene expression.
To advance our understanding of how eRNAs control gene regulation, numerous studies have focused on the identification and classification of the eRNA-interacting proteome. The methods most frequently employed to identify direct binding partners and physiological associations of eRNAs include RNA electrophoretic mobility shift assays and RNA immunoprecipitation (UV-RIP), respectively. Purification of target eRNAs complexed with proteins is often performed by using biotinylated antisense oligonucleotide to capture the RNAs, coupled with mass spectrometry (RAP-MS). These assays have identified eRNAs that regulate gene expression via interactions with RNAPII, various transcription factors and cofactors. eRNAs have been shown to interact with Yin Yang 1 (YY1) to increase recruitment of this transcriptional regulator at enhancers107 (Fig. 3a, left). Similarly, eRNAs increase RNAPII occupancy at protein-coding loci28,106. It remains to be determined whether transcription factor and cofactor trapping at enhancers and gene loci is a widespread mechanism underlying eRNA function. In addition to regulating RNAPII binding, eRNAs also regulate RNA Pol II pause release by acting as a decoy for the negative elongation factor (NELF) complex30 and through activation of the positive transcription elongation factor b (P-TEFb) complex108 (Fig. 3b). Several recent studies have also linked eRNA functions to their interactions with several general cofactors, including cohesin27,36, Mediator31, CBP104 and BRD4 (ref.28). eRNA interactions with cohesin and CBP augment enhancer activation through the regulation of chromatin looping and increased deposition of histone acetylation, respectively27,36,104. eRNAs directly interact with BRD4 through its bromodomains (BDs) to promote greater binding of BRD4 to acetylated histones, which, in turn, contributes to the maintaining enhancers in an active state28 (Fig. 3a, right). Lastly, eRNA interactions with Mediator are required to support transcriptional activation by promoting chromatin looping31. Importantly, all of these studies are consistent with eRNAs exhibiting their functional roles in collaboration with different binding partners involved in transcriptional regulation.
Fig. 3 |. eRNAs modulate the chromatin interactions of transcriptional regulators.
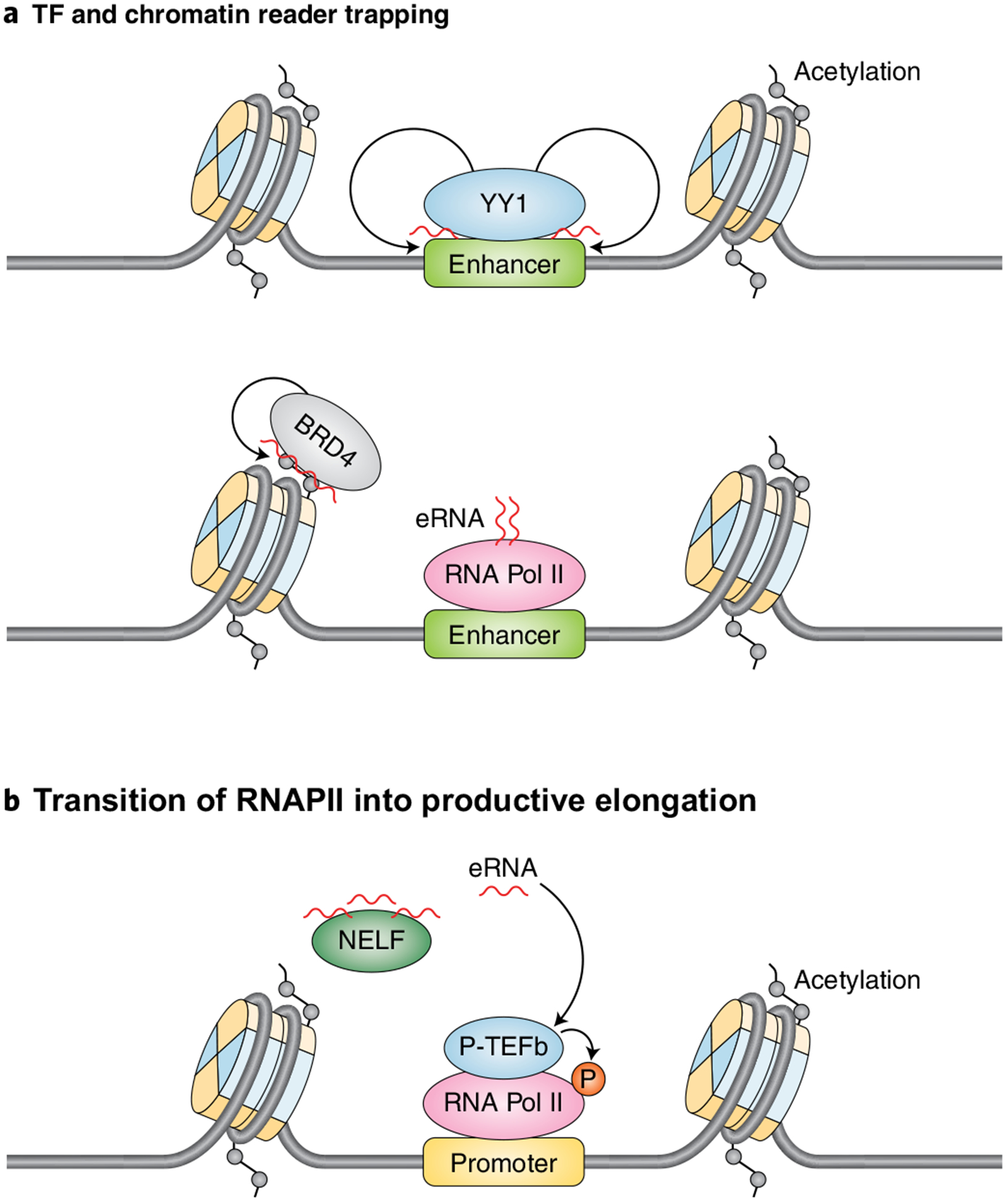
Schematic illustrating the role of eRNAs in directly regulating transcription. a, eRNAs regulate the enhancer occupancy of the transcription factor YY1107 and the transcriptional coactivator BRD4 (ref.28). b, eRNAs promote RNAPII chromatin engagement and the transition from RNAPII pause release to productive elongation by acting as a decoy for NELF30 and by contributing to the activation of P-TEFb108.
eRNAs in tumor-promoting gene regulation and genomic instability in cancer.
Since enhancers are known to control the selection and maintenance of hundreds of cell types, it is not surprising that enhancer misregulation has emerged as a driving force behind many types of human cancers. eRNAs are expressed across human cancer tissues3,5,21,27,31, supporting their potential relevance as therapeutic targets and biomarkers. The biggest barrier to moving these molecules to the clinic as a new generation of actionable targets is the lack of knowledge of their biological functions. However, recent studies have begun to uncover roles for eRNAs in the regulation of tumor-promoting gene expression programs. The ARIEL eRNA has recently been shown to promote activation of an oncogenic gene-expression program in T-cell acute lymphoblastic leukemia109. eRNAs have also been shown to regulate the chromatin interactions and transcriptional activities of BRD4, a potent cancer disseminator, that are required to regulate a subset of tumor-promoting genes in response to chronic TNF-a signaling in colon cancer28 (Fig. 3).
Beyond their roles in the regulation of gene expression in cancer, eRNAs are also linked to the maintenance of genome stability. Increased levels of eRNAs in human cancers have been shown to promote the formation of three-stranded nucleic acid DNA:RNA hybrids (R-loops) that interfere with DNA replication and, in turn, induce chromosome rearrangements and genome instability (reviewed in ref.110). Consistent with the possibility that aberrant eRNA synthesis contributes to tumorigenesis is the finding that enhancer transcription leads to activation-induced cytidine deaminase (AID) mistargeting, which promotes genome instability and malignancy111. Similarly, oncogenes, including mutant p53, have been shown to drive potent levels of eRNA synthesis in human colon cancer cell lines3, which may lead to aberrant R-loop formation and the loss of genome stability. Future studies that examine the mechanisms underlying the accumulation of eRNAs, R-loop formation and gene expression changes, and the connections between these events in normal versus cancer cells, will help to address remaining questions in the enhancer biology field. One unresolved issue relates to how enhancer regions are able to support high levels of eRNA synthesis while remaining uninhibited by increased formation of R-loops. Another possibility that remains to be explored is whether single nucleotide polymorphisms (SNPs) that are localized within enhancers alter or disrupt eRNA functions; a recent high-throughput analysis that identified SNPs that alter the regulatory functions of enhancers and promoters is consistent with this possibility9. Genes that exhibit transcriptional burst frequency differences were found to have higher densities of SNPs in their enhancer but not promoter regions. Identification of enhancer transcription ‘signatures’ and elucidation of the mechanisms underlying the precise functions of eRNAs in cancers will pave the way for the potential use of eRNAs as diagnostic markers and therapeutic targets.
Mechanisms underlying the functions of eRNAs
Recent studies have demonstrated that eRNA functions are largely mediated through interactions with eRNA binding partners27,28,31,36,104,105. Several studies suggest that RNA sequence is probably not involved in contributing to the specificity of eRNA-protein interactions. For example, both CBP104 and BRD428 were found to bind a broad spectrum of RNAs, which is consistent with these factors binding RNA in a non-sequence-specific manner. However, both of these studies suggest that the specificity of these eRNA interactions stems from eRNAs interacting with CBP and BRD4 in an enhancer-specific manner28,104. This property may be exploited by the pervasive binding of transcriptional cofactors throughout the genome, such that RNAs could stimulate activity locally, independently of RNA sequence. Nonetheless, only a few specific eRNAs were examined for direct interactions with CBP104 and BRD428, suggesting that additional analyses are needed to investigate the specificity of interactions between eRNAs and their respective binding partners. Consistent with the possibility that eRNAs may function through specific sequence motifs is the finding that a similar motif is present in a subset of androgen-receptor-regulated eRNAs that promotes transcription by activating P-TEFb108. In addition, current analyses cannot rule out the contributions of eRNA length, structure, and conformational flexibility. A further possibility that remains to be explored is whether post-translational modification of eRNAs contributes to the regulation and function of eRNA binding partners. To this end, deposition of 5-methylcytosine on eRNAs supports the coactivator functions of PGC1-a during metabolic stress112. Chemical modification of eRNAs may thus provide an additional layer of regulation in the interactions between eRNAs and binding partners and may thereby have important implications for enhancer and gene control.
Interestingly, eRNAs were shown to interact with binding partners through noncanonical RNA binding regions (RBRs), which are distinct from the well-characterized RNA-interacting domains, including the RNA-recognition motif (RRM)113, the hnRNPK-homology domain (KH)114 and the double-stranded RNA binding domain (dsRBD)115. For example, eRNAs have recently been shown to interact with the highly conserved acetyl-lysine-binding BDs of the BET family members and the single BDs of the non-BET proteins BRG1 and BRD728. This study raises the interesting questions of how prevalent eRNA interactions with chromatin reader domains are and what the significance of such interactions is in the regulation of chromatin and in gene control. Several RBRs in CBP/p300 were predicted to interact with eRNAs104; however, this was attributed to a single CBP RBR that resides within the catalytic histone acetyltransferase domain. eRNA interactions with this domain were shown to contribute to the displacement of the CBP activation loop that blocks substrate binding to the active site to allow for enhanced histone acetyltransferase activity104. Moreover, further analyses will be needed to investigate the significance of eRNA interactions with (i) both canonical and noncanonical RNA binding regions; (ii) multiple domains within a single protein, as was predicted for CBP104; and (iii) multiple subunits within a multiprotein complex. Taken together, these studies allude to the exciting possibilities for advancing our mechanistic understanding of the biological and biochemical roles of eRNAs and their corresponding binding partners. Yet additional studies are required to obtain a comprehensive list of eRNA binding partners and to uncover the molecular mechanisms underlying these interactions. Such analyses are likely to have broad implications, based on widespread eRNA synthesis in different cell types and because a large-scale analysis of eRNA binding partners has not yet been performed.
Future perspectives
The widespread involvement of ncRNAs in the regulation of gene expression suggests that a great deal remains to be discovered about the functional significance of these important molecules. On the basis of current studies, the causal roles of eRNAs are likely to be important for the regulation of a subset of enhancers and target loci. Nonetheless, additional evidence is required to definitively identify eRNAs and to elucidate their biological roles. Notably, these advances will require improvements to methodologies that cannot currently distinguish between the roles of enhancer transcription and eRNAs. As such, approaches that evaluate enhancer-mediated transcription dynamics and the direct roles of eRNAs are needed to advance our understanding of enhancer regulation and function. This includes cell-free systems, which could be used to manipulate and model the roles of eRNAs and eRNA-associating binding proteins and thus close an important gap in our knowledge of enhancer and gene regulation.
Future directions will also involve cataloging additional eRNAs, determining their cellular localization and expression patterns and exploring their mechanisms of action. Toward this goal, it will be necessary to define a more comprehensive list of eRNA binding partners and to determine the regulatory parameters underlying their interactions with eRNAs. Categorical data analyses and sorting of eRNA binding partners into functional and structural classes will provide new insights into how eRNAs interact with eRNA binding partners. These analyses could also provide new insights into whether eRNA binding partners display specificity for different RNA sequences and structural features and, if so, whether either or both of these possibilities impart the eRNA-protein complexes with varying functions. It will also be important to address the question of whether the activity of eRNAs and their corresponding eRNA binding partners is tailored to specific enhancers to control distinct gene expression programs and cellular processes. It will also be important to consider the recently identified role of eRNA-containing protein complexes in the regulation and function of the RNA-dependent architecture that directs the formation and integrity of phase-separated condensates116 (Fig. 4). Several questions that remain to be addressed include the following: how do eRNAs contribute to changes in the physical properties of condensates, and can eRNAs specifically regulate the sorting of proteins and regulatory DNA elements into specific condensates? Which eRNA motifs are sufficient for the formation of phase separated particles? Finally, it remains to be determined whether eRNA sequences that show increased affinity for binding partners exist. High-affinity binding sites and the potential for cooperative spreading of binding partners on eRNAs, which has been shown for mRNAs, could play an important role in facilitating multivalent interactions, which contribute to phase separation117. In addition, eRNAs provide the potential for multivalency in RNA-mediated chromatin associations28, as demonstrated by the recent finding that eRNAs interact with BDs to facilitate enhanced chromatin associations and the downstream transcription-associated functions of BRD4. Histone reader proteins tend to form weak interactions with chromatin through their recognition of and direct interactions with particular histone marks. This model would be consistent with previous findings that the overall affinity and specificity of chromatin binding is enhanced when multiple chromatin-interacting domains in a single protein or within a multiprotein complex are able to engage with chromatin118.
Fig. 4 |. The role of eRNAs in regulating condensate assembly on enhancers.
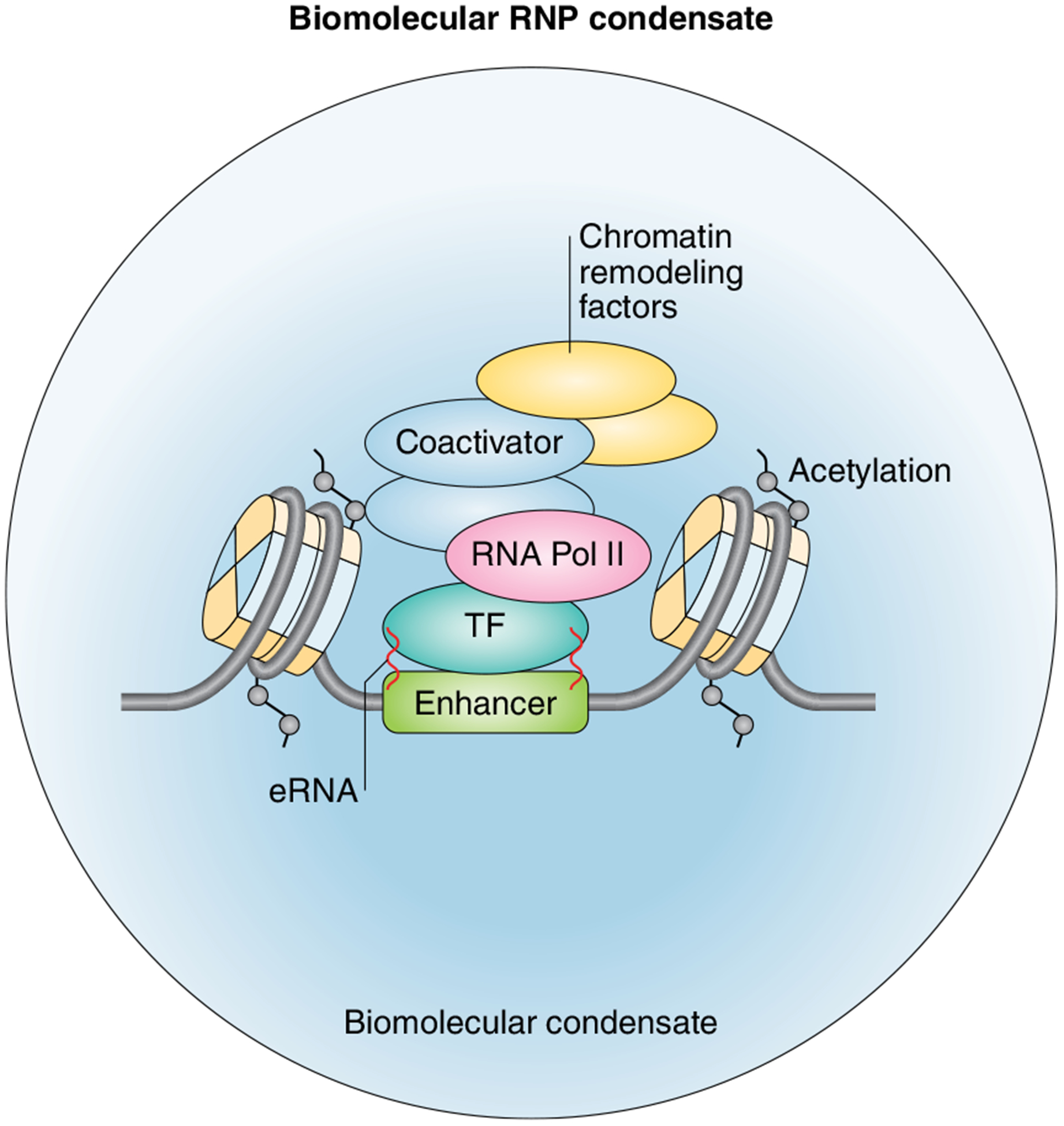
Schematic representation of an enhancer-RNA-dependent ribonucleoprotein (eRNP) complex exhibiting properties of a phase-separated condensate. A growing number of studies suggest that the formation of liquid-like droplets or phase condensates may be a general mechanism to compartmentalize biochemical reactions that support cellular activities (reviewed in refs.119,120). In support of the phase-separation model, recent studies have revealed that the recruitment of multimolecular assemblies to super-enhancers (SEs) imparts the ability to form phase condensates to SEs119. It has also been demonstrated that phase separation at SEs plays a key role in transcriptional control through the compartmentalization of transcriptional regulators119,121–123. Emerging evidence also shows that eRNAs produced from ligand-activated enhancers regulate enhancer activation by controlling the diffusion properties of phase-separated components116. Specifically, eRNAs and eRNA binding partners with intrinsically disordered regions were found to form ribonucleoprotein (eRNP) complexes at enhancers, which exhibited properties of dynamic phase-separated condensates116. This same study revealed that eRNAs play a pivotal role in regulating the fluid properties of the condensates by promoting a more dynamic and liquid-like state rather than a gel-like state, which is required for transcriptional activity116. Conversely, phase separation also regulates enhancer activity and eRNA production. Phase separation has recently been shown to regulate the spatial interactions between enhancers that are required for both cooperative ligand-mediated enhancer activation and the production of eRNAs116. Together, these recent findings reveal the importance of further dissecting the relationship between eRNAs and phase separation to understand key elements of their functional mechanisms.
In summary, an explosion of studies in the last decade has identified noncoding enhancer transcripts that are produced from a wide array of active enhancers in multiple cell types and tissues. Building on this foundation, the next decade will be an exciting time for uncovering the mechanisms that control enhancer transcription and eRNA functions in gene regulation and biological processes. Progress toward this goal will be the next fundamental step in advancing our understanding of gene regulation.
Acknowledgements
We thank Jim Kadonaga for reading the manuscript and providing helpful suggestions. Research in the Lauberth laboratory is supported by a grant from the NIH/National Institute of General Medical Sciences (R35 GM128900) to S.L., and research in the Sartorelli laboratory is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Intramural Research Program of the NIH (grants AR041126 and AR041164).
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Plank JL & Dean A Enhancer function: mechanistic and genome-wide insights come together. Mol. Cell 55, 5–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long HK, Prescott SL & Wysocka J Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahnamoun H et al. Mutant p53 shapes the enhancer landscape of cancer cells in response to chronic immune signaling. Nat. Commun 8, 754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson R et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franco HL, Nagari A & Kraus WL TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol. Cell 58, 21–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai F, Gardini A, Zhang A & Shiekhattar R Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam MT et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498, 511–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Arensbergen J et al. High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet 51, 1160–1169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenfelder S & Fraser P Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet 20, 437–455 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Villar D et al. Enhancer evolution across 20 mammalian species. Cell 160, 554–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold CD et al. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat. Genet 46, 685–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt D et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036–1040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blow MJ et al. ChIP-seq identification of weakly conserved heart enhancers. Nat. Genet 42, 806–810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May D et al. Large-scale discovery of enhancers from human heart tissue. Nat. Genet 44, 89–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang S et al. Functionally conserved enhancers with divergent sequences in distant vertebrates. BMC Genomics 16, 882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee S, Bourque G & Lufkin T Conserved and non-conserved enhancers direct tissue specific transcription in ancient germ layer specific developmental control genes. BMC Dev. Biol 11, 63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hare EE, Peterson BK, Iyer VN, Meier R & Eisen MB Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet 4, e1000106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beagrie RA & Pombo A Gene activation by metazoan enhancers: diverse mechanisms stimulate distinct steps of transcription. Bioessays 38, 881–893 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Furlong EEM & Levine M Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catarino RR & Stark A Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev 32, 202–223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krivega I & Dean A Enhancer and promoter interactions—long distance calls. Curr. Opin. Genet. Dev 22, 79–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng W et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartman CR, Hsu SC, Hsiung CC-S, Raj A & Blobel GA Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol. Cell 62, 237–247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawado T, Halow J, Bender MA & Groudine M The β-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev 17, 1009–1018 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song S-H et al. Multiple functions of Ldb1 required for β-globin activation during erythroid differentiation. Blood 116, 2356–2364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahnamoun H et al. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat. Struct. Mol. Biol 25, 687–697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai P-F et al. A muscle-specific enhancer RNA mediates cohesin recruitment and regulates transcription in trans. Mol. Cell 71, 129–141.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaukowitch K et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56, 29–42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai F et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivaldi MS et al. Fetal γ-globin genes are regulated by the BGLT3 long noncoding RNA locus. Blood 132, 1963–1973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Santa F et al. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol 8, e1000384 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim T-K et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franco HL et al. Enhancer transcription reveals subtype-specific gene expression programs controlling breast cancer pathogenesis. Genome Res 28, 159–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hah N, Murakami S, Nagari A, Danko CG & Kraus WL Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 23, 1210–1223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Core LJ et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet 46, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hah N et al. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc. Natl Acad. Sci. USA 112, E297–E302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hah N et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwak H, Fuda NJ, Core LJ & Lis JT Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahat DB et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc 11, 1455–1476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carninci P et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet 38, 626–635 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Shiraki T et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA 100, 15776–15781 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arner E et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347, 1010–1014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meers MP et al. Transcription start site profiling uncovers divergent transcription and enhancer-associated RNAs in Drosophila melanogaster. BMC Genomics 19, 157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henriques T et al. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev 32, 26–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen RA-J et al. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res 23, 1339–1347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chepelev I, Wei G, Wangsa D, Tang Q & Zhao K Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res 22, 490–503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heintzman ND et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Creyghton MP et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rada-Iglesias A et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikhaylichenko O et al. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev 32, 42–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pekowska A et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 30, 4198–4210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koch F et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol 18, 956–963 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Natoli G & Andrau JC Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet 46, 1–19 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Flynn RA, Almada AE, Zamudio JR & Sharp PA Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc. Natl Acad. Sci. USA 108, 10460–10465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scruggs BS et al. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol. Cell 58, 1101–1112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Preker P et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res 39, 7179–7193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kowalczyk MS et al. Intragenic enhancers act as alternative promoters. Mol. Cell 45, 447–458 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Andersson R & Sandelin A Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet 21, 71–87 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Kim TK & Shiekhattar R Architectural and functional commonalities between enhancers and promoters. Cell 162, 948–959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirose Y & Manley JL RNA polymerase II and the integration of nuclear events. Genes Dev 14, 1415–1429 (2000). [PubMed] [Google Scholar]
- 64.Proudfoot NJ, Furger A & Dye MJ Integrating mRNA processing with transcription. Cell 108, 501–512 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Shatkin AJ & Manley JL The ends of the affair: capping and polyadenylation. Nat. Struct. Biol 7, 838–842 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Hintermair C et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J 31, 2784–2797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsin JP & Manley JL The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26, 2119–2137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayer A et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336, 1723–1725 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Corden JL Tails of RNA polymerase II. Trends Biochem. Sci 15, 383–387 (1990). [DOI] [PubMed] [Google Scholar]
- 70.West ML & Corden JL Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140, 1223–1233 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eick D & Geyer M The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev 113, 8456–8490 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Bonn S et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet 44, 148–156 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Descostes N et al. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. Elife 3, e02105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsin JP, Li W, Hoque M, Tian B & Manley JL RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. Elife 3, e02112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bentley DL Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet 15, 163–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah N et al. Tyrosine-1 of RNA polymerase II CTD controls global termination of gene transcription in mammals. Mol. Cell 69, 48–61.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Nojima T et al. Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-loop formation, replication stress, and cellular senescence. Mol. Cell 72, 970–984.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang AH et al. The elongation factor Spt6 maintains ESC pluripotency by controlling super-enhancers and counteracting Polycomb proteins. Mol. Cell 68, 398–413.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin C, Garruss AS, Luo Z, Guo F & Shilatifard A The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 152, 144–156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gil N & Ulitsky I Production of spliced long noncoding RNAs specifies regions with increased enhancer activity. Cell Syst 7, 537–547.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vučićević D, Corradin O, Ntini E, Scacheri PC & Ørom UA Long ncRNA expression associates with tissue-specific enhancers. Cell Cycle 14, 253–260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Core LJ et al. Defining the status of RNA polymerase at promoters. Cell Rep 2, 1025–1035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaikkonen MU et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andersson R et al. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat. Commun 5, 5336 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Pefanis E et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lubas M et al. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep 10, 178–192 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Austenaa LM et al. Transcription of mammalian cis-regulatory elements is restrained by actively enforced early termination. Mol. Cell 60, 460–474 (2015). [DOI] [PubMed] [Google Scholar]
- 88.Ulitsky I & Bartel DP lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guttman M & Rinn JL Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cabili MN et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Almada AE, Wu X, Kriz AJ, Burge CB & Sharp PA Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paralkar VR et al. Unlinking an lncRNA from its associated cis element. Mol. Cell 62, 104–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Groff AF et al. In vivo characterization of Linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep 16, 2178–2186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kopp F & Mendell JT Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alvarez-Dominguez JR, Knoll M, Gromatzky AA & Lodish HF The super-enhancer-derived alncRNA-EC7/Bloodlinc potentiates red blood cell development in trans. Cell Rep 19, 2503–2514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Espinosa JM On the origin of lncRNAs: missing link found. Trends Genet 33, 660–662 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cinghu S et al. Intragenic enhancers attenuate host gene expression. Mol. Cell 68, 104–117.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shechner DM, Hacisuleyman E, Younger ST & Rinn JL Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods 12, 664–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee JS & Mendell JT Antisense-mediated transcript knockdown triggers premature transcription termination. Mol. Cell 77, 1044–1054.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lai F, Damle SS, Ling KK & Rigo F Directed RNase H cleavage of nascent transcripts causes transcription termination. Mol. Cell 77, 1032–1043.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 101.Yang Y et al. Enhancer RNA-driven looping enhances the transcription of the long noncoding RNA DHRS4-AS1, a controller of the DHRS4 gene cluster. Sci. Rep 6, 20961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chu C, Qu K, Zhong FL, Artandi SE & Chang HY Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Femino AM, Fay FS, Fogarty K & Singer RH Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). [DOI] [PubMed] [Google Scholar]
- 104.Bose DA et al. RNA binding to CBP stimulates histone acetylation and transcription. Cell 168, 135–149.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hsieh C-L et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl Acad. Sci. USA 111, 7319–7324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mousavi K et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 51, 606–617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sigova AA et al. Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao Y et al. Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep 15, 599–610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan SH et al. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood 134, 239–251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aguilera A & García-Muse T R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012). [DOI] [PubMed] [Google Scholar]
- 111.Meng FL et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell 159, 1538–1548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aguilo F et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep 14, 479–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maris C, Domínguez C & Allain FH The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272, 2118–2131 (2005). [DOI] [PubMed] [Google Scholar]
- 114.Grishin NV KH domain: one motif, two folds. Nucleic Acids Res 29, 638–643 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chang KY & Ramos A The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J 272, 2109–2117 (2005). [DOI] [PubMed] [Google Scholar]
- 116.Nair SJ et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol 26, 193–203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ruthenburg AJ, Li H, Patel DJ & Allis CD Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol 8, 983–994 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Polymenidou M The RNA face of phase separation. Science 360, 859–860 (2018). [DOI] [PubMed] [Google Scholar]
- 121.Boija A et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sabari BR et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boeynaems S et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


