The taxonomic and functional diversity inherent to the soil microbiome complicate assessments of the metabolic potential carried out by the community members. An alternative approach is to break down the soil microbiome into reduced-complexity subsets based on metabolic capacities (functional modules) prior to sequencing and analysis. Here, we demonstrate that this approach successfully identified specific phylogenetic and biochemical traits of the soil microbiome that otherwise remained hidden from a more top-down analysis.
KEYWORDS: functional modules, microbiome, reduced complexity, soil metatranscriptome, soil microbiome
ABSTRACT
The soil microbiome represents one of the most complex microbial communities on the planet, encompassing thousands of taxa and metabolic pathways, rendering holistic analyses computationally intensive and difficult. Here, we developed an alternative approach in which the complex soil microbiome was broken into components (“functional modules”), based on metabolic capacities, for individual characterization. We hypothesized that reproducible, low-complexity communities that represent functional modules could be obtained through targeted enrichments and that, in combination, they would encompass a large extent of the soil microbiome diversity. Enrichments were performed on a starting soil inoculum with defined media based on specific carbon substrates, antibiotics, alternative electron acceptors under anaerobic conditions, or alternative growing conditions reflective of common field stresses. The resultant communities were evaluated through 16S rRNA amplicon sequencing. Less permissive modules (anaerobic conditions, complex polysaccharides, and certain stresses) resulted in more distinct community profiles with higher richness and more variability between replicates, whereas modules with simple substrates were dominated by fewer species and were more reproducible. Collectively, approximately 27% of unique taxa present in the liquid soil extract control were found across functional modules. Taxa that were underrepresented or undetected in the source soil were also enriched across the modules. Metatranscriptomic analyses were carried out on a subset of the modules to investigate differences in functional gene expression. These results demonstrate that by dissecting the soil microbiome into discrete components it is possible to obtain a more comprehensive view of the soil microbiome and its biochemical potential than would be possible using more holistic analyses.
INTRODUCTION
The soil microbiome carries out key ecosystem services, including biogeochemical cycling, soil-atmosphere gas exchange, and plant growth promotion. Better understanding of the soil microbiome will aid predictions of how these beneficial services may be affected by environmental perturbations such as climate change. However, the high taxonomic and functional diversity of the soil microbiome is currently a bottleneck to understanding and prediction of the biochemical reactions that they carry out (1, 2). Soil is a heterogeneous mixture of distinct microenvironments (1), containing numerous microbial guilds of disparate metabolic capacities (2, 3). In addition, the soil microbiome harbors a significant fraction of rare and/or quiescent members (4), significantly contributing to process rates (5, 6) but existing below the threshold of detection of current technologies. Finally, the vast amounts of information generated through holistic analyses, such as bulk metagenomics, represents an intense computational burden (7, 8) hindering assessment of the total diversity contained within the soil microbiome.
Here, we aimed to dissect the biochemical capacity of the soil microbiome into discrete “functional modules” through targeted enrichments. The functional module concept circumvents numerous limitations in soil microbiome analysis. First, by selective enrichment, the soil microbiome diversity (richness) can be reduced into a tractable number of species. These reduced-complexity consortia can then be studied in detail to determine how individual species interact to carry out a specific biochemical process, such as soil organic matter decomposition. Reduced-complexity communities are also less computationally challenging for bioinformatics applications (9). Second, individual species within a functional module can be isolated and studied in different combinations to define specific interspecies interactions that underlie a metabolic process of interest (10). Third, enrichment can allow for growth of rare or otherwise-undetectable microbes from the source soil (9). Finally, this approach is aligned with the paradigm of considering microbiome-associated functions and phenotypes rather than strictly phylogenetic structures (11).
We hypothesized that, through targeted enrichments of a starting soil inoculum, we could obtain functional module communities with distinct characteristics. These functional modules would have lower diversity relative to the soil extract controls and would be reproducible and predictable. In addition, we hypothesized that the functional modules would encapsulate a significant extent of soil phylogenetic diversity and would enrich for underrepresented soil taxa. In total, the results presented here illustrate how this approach can illuminate both the known and the hidden diversity of a complex microbiome.
RESULTS
Designing functional modules.
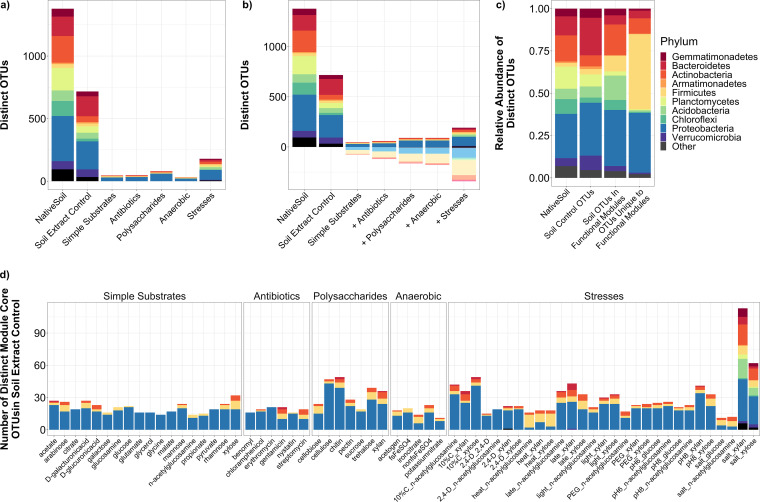
Functional modules were designed to dissect the complex soil microbiome into discrete functional units and, in doing so, to maximize capture of the soil phylogenetic and functional diversity. Native field soil was incubated under a wide range of incubation conditions that represented potential resources and/or environmental conditions that would be experienced by soil microorganisms. Module categories included: “simple substrates” (e.g., sugars and organic acids), “antibiotics,” “polysaccharides,” “anaerobic” (i.e., modules under anoxic conditions with alternative electron acceptors), and “stresses,” where three types of carbon modules were exposed to common field stresses, including heat, low pH, and high salt. Control “soil extract” modules were cultured in liquid soil extract media, and “native soil” modules were obtained by sequencing the source soil.
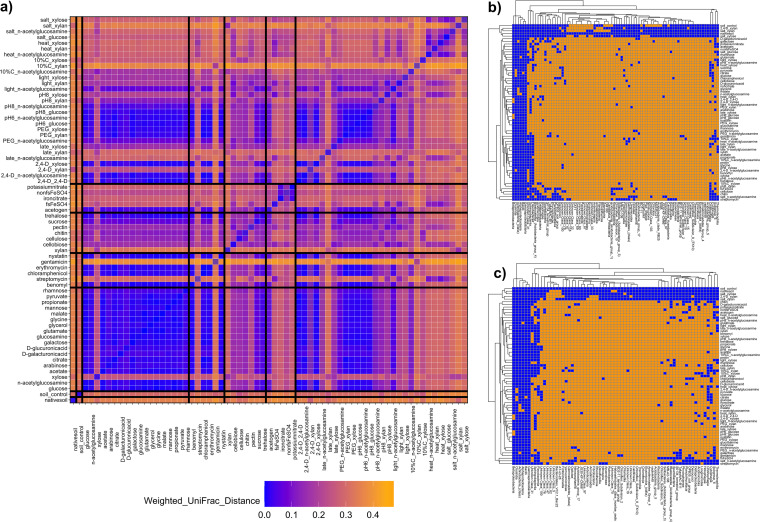
In total, 324 communities were obtained with up to 5 replicates each for 66 distinct functional modules. Community composition was determined by 16S rRNA amplicon analysis. Using weighted UniFrac as a distance metric, the factors “category” (which of the 6 module categories) and “modulename” (which of the 66 distinct modules) were both significant (P < 0.001) for permutational multivariate analysis of variance (PERMANOVA) tests in explaining variation across the full data set (see Table S1 in the supplemental material). Comparable results were found for unweighted UniFrac (Table S1); for this reason, analyses here will be generally restricted to weighted UniFrac (statistics on unweighted and weighted UniFrac data are available in Tables S1 to S3). Similarities in community structure between modules largely depended on the module category; e.g., simple substrate modules were similar to one another and to certain polysaccharide and stress modules (Fig. 1a). Meanwhile, anaerobic modules and other stress modules were distinct from most other modules (Fig. 1a). When using an unweighted approach, the majority of taxonomic classes detected in either the liquid soil extract control or the native soil control were not detected in the modules, with the exception of some of the “stress” modules (Fig. 1b and c), where more classes present in the native soil or liquid soil extract control were detected.
FIG 1.
Distance between functional module communities and presence/absence heatmaps. (a) Heatmap of the mean pairwise weighted UniFrac distance between all sample replicates for one functional module and all replicates for another module. Each box represents the average across all replicates for the module. Shades of orange and blue represent high and low average weighted UniFrac distances, respectively. Black lines between samples represent the demarcation between functional module categories (i.e., “soil,” “simple substrates,” “antibiotics,” “polysaccharides,” “anaerobic,” and “stresses”). (b and c) Heatmaps representing how functional modules captured soil taxa at the class level. The x axes represent all classes that had nonzero counts in either liquid soil extract control (b) or native soil control (c); the y axes represent all functional modules, grouped by hierarchical clustering. Orange squares are for modules by class combinations that have no counts for the corresponding class in any module replicate; blue squares represent module by class combinations that have at least one count in at least one module replicate.
PERMANOVA of weighted and unweighted UniFrac distances for full dataset and stress dataset. In full dataset (excluding native soil controls), PERMANOVA analyses were run to determine the significance of factors “category” and “replicate” and of “modulename” and “replicate” (note that these analyses were run separately due to the interdependence of “category” and “modulename”), in explaining the variation in beta-diversity (weighted and unweighted UniFrac distances). In the stress dataset, analyses were run to determine the significance of “stress,” “substrate,” “replicate,” and the interaction term “stress:substrate” or of “modulename” and “replicate” (again, analyses were run separately due to interdependence). Analyses were run using the “adonis” function within the vegan package in R. Download Table S1, PDF file, 0.04 MB (46.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Functional modules are reduced in diversity and richness.
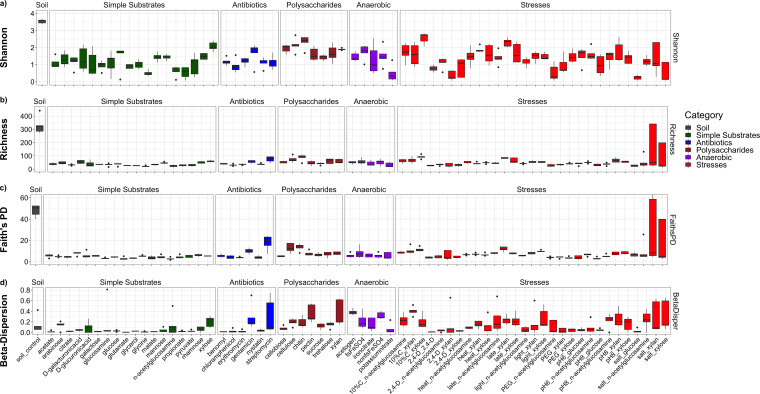
In the full data set, alpha-diversity metrics (Shannon’s diversity, richness, and Faith’s phylogenetic diversity [PD]) (Fig. 2a to c) were significantly (P < 0.001) explained by “category” and “modulename” (Table S2) in ANOVA tests. However, the significances of these effects were often driven by the presence of the liquid soil extract controls. Upon their omission, richness was no longer significantly explained by “category” or “modulename,” although Shannon’s diversity was (P < 0.001) (Table S2). Tukey’s post hoc tests confirmed that no functional module categories differed regarding richness. For Shannon’s diversity, the “polysaccharides” modules were higher than all other modules and for Faith’s PD, “stresses” were significantly different from “simple substrates” (Table S2).
FIG 2.
Full dataset of alpha-diversity and beta-diversity trends. (a to d) Boxplots for the alpha-diversity metrics: Shannon’s diversity (a), OTU richness (b), Faith’s PD (c), and beta-dispersion (d). Samples are separated by individual module (x axis). Samples are further segregated and colored by module category (soil, gray; simple substrates, green; antibiotics, blue; polysaccharides, brown; anaerobic, purple; and stresses, red). Beta-dispersion is represented by the distance for all replicates within a module from their respective median values for weighted UniFrac. All boxplots were generated using default parameters, where lower and upper hinges represent first and third quartiles (25th and 75th percentiles). Upper whiskers represent the largest value at or below the upper hinge + 1.5× the interquartile range; lower whiskers represent the smallest value at or above the lower hinge – 1.5× the interquartile range. All outliers fall outside these ranges.
ANOVA of alpha-diversity metrics (Shannon’s diversity and richness) and beta-dispersion across the full dataset with Tukey’s post hoc tests. ANOVA analyses for Shannon’s diversity, richness, and beta-dispersion across full dataset for significance of the “factors” category (soil control, simple substrates, antibiotics, polysaccharides, anaerobic, or stress modules) and “replicate,” and by individual “modulename” (the specific module) and “replicate.” These factors have to be run in separate ANOVA analyses due to the interdependence of factors “category” and “modulename”; furthermore, separate analyses were run for the full dataset where soils were either included or excluded (as indicated below) to examine the influence of the control soils on alpha-diversity metrics. (Control soils refer to liquid soil extract controls and not native soils). ANOVA calculations used the “aov” function in the stats package in R. The “betadisper” function within the vegan package was used for calculation of beta-dispersion values. Tukey’s for alpha-diversity and beta-dispersion on full dataset by “category” and “modulename.” Tukey’s results for Shannon’s diversity/richness/beta-dispersion were done for the full dataset (or full dataset excluding soil controls) by “category” or by “modulename.” Generation of Tukey’s values was accomplished using the “HSD.test” function as part of the agricolae package in R. Native soils are not included in this set of analyses. Download Table S2, PDF file, 0.1 MB (89.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Functional modules vary in reproducibility.
Reproducibility (consistent community composition among replicates) was quantified by “beta-dispersion” (average weighted UniFrac distance between module replicates and the median), where higher values indicate lower reproducibility. ANOVA confirmed “category” and “modulename” were significant (P < 0.001) in explaining variation in beta-dispersion (Table S2). Beta-dispersion was highest for the “anaerobic” and “polysaccharides” modules (Fig. 2d, Table S3), and “simple substrates” had a lower beta-dispersion than other categories according to Tukey’s post hoc tests (though only statistically different from “anaerobic” and “polysaccharides”) (Table S2). These results suggest that the simple substrate modules were more reproducible. Modules with a high beta-dispersion generally had a higher alpha-diversity (Fig. 2), suggesting a link between diverse community structures and variation in community assembly. Within the stress modules subset, “stress” and “substrate” (though not the interaction term “stress:substrate”) significantly (P < 0.007) influenced beta-dispersion (Table S3), the least reproducible stresses being salt and dilute carbon.
ANOVA of alpha-diversity metrics and beta-dispersion for stress subset with Tukey’s post hoc tests. ANOVA analyses for Shannon’s diversity, richness, and beta-dispersion across stress subset including the factors “stress” (which of the eight stresses or control were considered), “substrate” (which of the three substrates in this dataset), “replicate,” and the interaction term “stress:substrate,” or by individual “modulename” (the specific stress × substrate module) and “replicate.” These factors have to be run in separate ANOVA analyses due to the interdependence of factors “stress/substrate” and “modulename.” ANOVA calculations used the “aov”’ function in the stats package in R. Tukey’s calculations used the “HSD.test”’ function as part of the agricolae package in R. Note that this subset does not include control soils, so as to focus on specific comparisons between stresses and the substrates used within this subset. Download Table S3, PDF file, 0.1 MB (71.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Functional module community profiles.
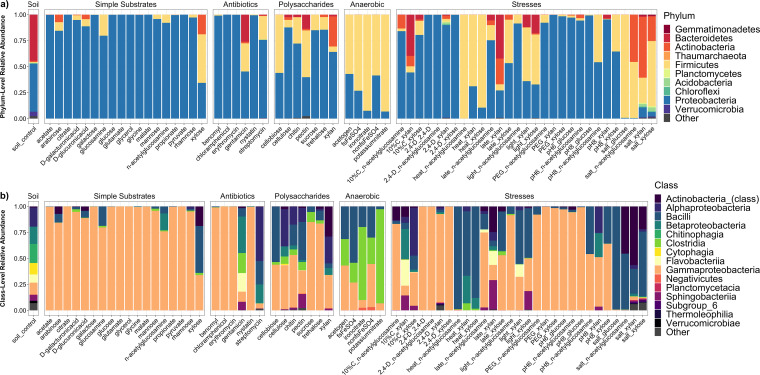
Dominant phyla in liquid soil extract controls included Proteobacteria (mean relative abundance, 46.2%) and Bacteroidetes (44.2%), distantly followed by Verrucomicrobia (4.3%) and Actinobacteria (1.3%) (Fig. 3a). Major classes included Chitinophagia (18.3%), Alphaproteobacteria (18.2%), Betaproteobacteria (16.4%), and Gammaproteobacteria (11.5%) (Fig. 3b). Taxonomic trends for each module category are examined in detail below.
FIG 3.
Community trends for full data set, phylum level and class level. (a and b) Relative abundance plots for all modules encompassing either the top 10 most abundant phyla (a) or the top 15 most abundant classes (b). Modules are grouped by the overlying module category (soil, simple substrates, antibiotics, anaerobic, or stresses). Plots represent the average of all replicates for each given module. “Soil” refers to liquid soil extract control.
(i) Simple substrate modules. The intent of the “simple substrates” modules was to represent a diverse range of metabolic capabilities from a variety of taxa present in soils, by including common metabolic intermediates (simple sugars, amino sugars, organic acids, and small carbon compounds). Taxonomic profiles for these modules were largely dominated by Proteobacteria (Fig. 3a), specifically the genus Pseudomonas, whose relative abundance increased from an average of 10.8% in soil controls to 90.7% in “simple substrates,” reflecting their known ability to rapidly grow on these substrates in liquid medium (12). Some modules also were enriched for Actinobacteria and/or Firmicutes (Fig. 3a). For example, with xylose, Actinobacteria increased from <1% (the average across all simple substrates) to 18.9%, and Firmicutes increased from <10%, to 46.8%.
(ii) Antibiotic modules. Antibiotics are relevant to soil systems, since they are produced by some soil microorganisms, with roles in microbe-microbe competition or signaling (13). Here, we designed modules based on antibiotic supplementation to enrich for antibiotic-resistant populations. The antibiotics included those with modes of action against Gram-negative bacteria (e.g., gentamicin) and against Gram-positive bacteria (erythromycin), broad-spectrum activity (chloramphenicol and streptomycin), and antifungal activity (benomyl and nystatin). While growth rates were negatively impacted in all cases, Proteobacteria (largely Pseudomonas) were still predominant in modules for chloramphenicol, erythromycin, and fungicides (Fig. 3a). However, gentamicin and streptomycin enriched for Bacteroidetes, and the Clostridia and Flavobacteriia classes (Fig. 3a and b).
(iii) Polysaccharide modules. “Polysaccharides” modules included plant or fungal polysaccharides or disaccharides, ones that would be commonly found in soils containing degraded lignocellulosic biomass. Shannon’s diversity was significantly higher for “polysaccharides” than for “simple substrates” according to Tukey’s post hoc tests (Table S2), an effect more pronounced for the more complex polysaccharides (chitin, cellulose, and xylan) (Fig. 2a). Apart from Proteobacteria, “polysaccharides” modules were largely enriched for Actinobacteria, Firmicutes, and Bacteroidetes (Fig. 3a). Bacteroidetes in particular represents a major soil phylum only previously substantially found in our gentamicin module.
(iv) Anaerobic modules. For the “anaerobic” modules we took a basic glucose module and supplied alternative electron acceptors to oxygen, aiming to enrich for the significant extent of soil taxa that are anaerobes. Specifically, the modules included the following: no alternative e– acceptor and N2 gas (referred to as “acetogen” media), potassium nitrate under 9:1 N2:CO2, iron (III) citrate under 9:1 N2:H2, and iron sulfate under 9:1 N2:H2. Trends for these modules included a decrease in representation of Proteobacteria and increases in Firmicutes (Fig. 3a); in particular, increases in the Negativicutes and Clostridia classes.
(v) Stress modules. Certain major soil phyla (e.g., Chloroflexi, Planctomycetes, and Verrucomicrobia) were not substantially captured in any modules up to this point. As opposed to testing more substrates, we incubated a set of modules under alternative growth conditions, namely, stresses that represented extremes of normal field conditions, to vary the community composition and capture missing groups. Three carbon substrate modules (N-acetylglucosamine, xylose, and xylan) were subjected to treatments including: exposure to the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D), osmotic stress (addition of polyethylene glycol 8000), increasing pH from 7 to 8, constant light, diluting carbon concentrations 10-fold, imposing constant heat stress (40°C), imposing salt stress (0.8 M NaCl), and “late” modules (harvested after 1 full week). We note that certain conditions in this analysis represent permissive conditions for certain taxa and thus the term “stresses” may not be completely accurate. However, the goal was to induce growing conditions indicative of environmental/geochemical fluctuations in the native soil, not solely obtaining stress-tolerant microbes.
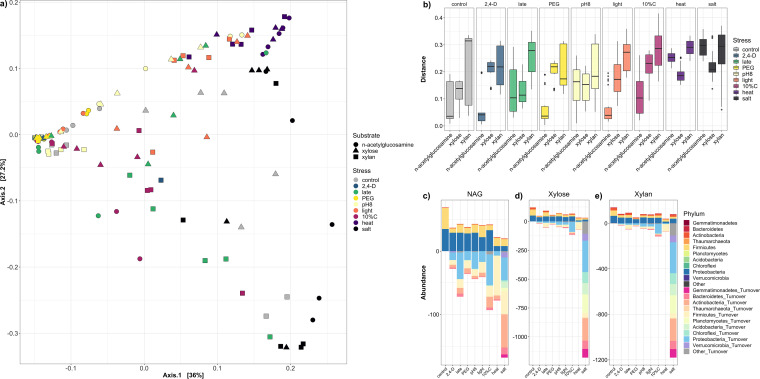
The factors “stress” and “substrate” and their interaction term (“stress:substrate”’) were all significant (P < 0.001) in explaining community dissimilarity according to PERMANOVA (Table S1). Salt and heat stress modules clustered separately from most other modules on an ordination plot (Fig. 4a), with the greatest distances from their respective unstressed controls (Fig. 4b). Effects varied by substrate, with N-acetylglucosamine stress modules showing a clearer segregation for salt and heat stress modules compared to xylose or xylan, although these effects were somewhat attenuated if unweighted UniFrac was used in lieu of weighted UniFrac (Fig. S1). Furthermore, N-acetylglucosamine stress modules had higher turnover for novel taxa relative to unstressed control, compared to xylan or xylose stress modules (Fig. 4c to e). There were some common taxonomic trends across stresses (e.g., heat enriching for class Bacilli, salt enriching for Actinobacteria), although in others (dilute carbon and “late” stresses) profiles differed by substrate (Fig. 3a and b). Notably, in xylose and xylan salt stress modules there was emergence of soil phyla (Gemmatimonadetes, Acidobacteria, and Verrucomicrobia) not seen elsewhere across the modules (Fig. 3a).
FIG 4.
Stress dataset of weighted UniFrac distance and nestedness/turnover trends. (a) Principal coordinate analysis of weighted UniFrac Distance for samples within the “stress” subset. Substrate is delineated by shape: ●, N-acetylglucosamine; ▲, xylose; and ■, xylan. Colors indicate the specific stress imposed (gray, control; blue, 2,4-D; green, late harvesting; yellow, PEG [osmotic stress]; light yellow, pH 8; orange, constant light; magenta, 10%C [dilute carbon]; purple, heat stress; and black, salt stress). (b) Boxplots representing the weighted UniFrac distances for all replicates for a stress module from its unstressed control of the same substrate, segregated into stress × substrate combinations. Control boxplots (gray) indicate distances for control replicates from one another. (c, d, and e) Nestedness and turnover plots for stress modules, segregated by substrate (for N-acetylglucosamine or NAG [c], for xylose [d], and for xylan [e]). Above the x axis at 0 represents nestedness, or the distinct OTUs present in the control unstressed module that were also found in the stress modules for the same substrate. Below the x axis at 0 represents turnover, or the distinct OTUs uniquely found in stress modules not present in their respective unstressed control. Bars are colored by which parent phylum to which the OTUs belong.
Ordination plots for stress samples for weighted and unweighted UniFrac distances, segregated by substrate. Principal coordinate analysis of weighted UniFrac distance (a, c, and e) or unweighted UniFrac distance (b, d, and f) for samples within the “stress” subset. Substrate is delineated by shape: ●, N-acetylglucosamine; ▲, indicates xylose; and ■, xylan. Substrates are further separated into individual plots for N-acetylglucosamine (a and b), xylose (c and d), and xylan (e and f). Colors indicate the specific stress imposed (gray, control; blue, 2,4-D; green, late harvesting; yellow, PEG [osmotic stress]; light yellow, pH 8; orange, constant light; magenta, 10%C [dilute carbon]; purple, heat stress; and black, salt stress). Download FIG S1, PDF file, 0.04 MB (41.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(vi) Indicator genera. We ran indicator species analysis (at the genus level) to determine which genera were significantly (P < 0.05) associated with a given module (Table S4). Gentamicin and streptomycin modules were enriched for genera known to harbor genes for gentamicin resistance (Stenotrophomonas [14]), streptomycin resistance (Lactobacillus [15]), or streptomycin synthesis (Leifsonia, Burkholderia-Paraburkholderia, Rhodanobacteria, and Dyella), whereas indicators for polysaccharide modules included known cellulose degraders (Kaistia, Devosia, and Bradyrhizobium [16]) (Table S4).
Indicator species analysis (set to “genus” level) for full dataset. Analysis was conducted using the “indval” function within the labdsv package in R using default parameters. Data generated include “indval” (indicator value; this value is a product of relative abundance and frequency, and serves as a measure of exclusivity for a given taxon within a group), “P value” (significance of the aforementioned indicator value, and “freq” (relative frequency of the taxon across all replicates for a module). Only indicator genera with a P value of <0.05 were retained. Download Table S4, PDF file, 0.1 MB (88.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Soil diversity represented in functional modules.
We compared functional modules to native soil control communities to determine how well we captured soil taxonomic diversity. A core microbiome was calculated for each functional module (operational taxonomic units [OTUs] present in ≥40% of replicates at ≥0.01% of cumulative relative abundance, similar to definitions in previous studies [17, 18]) to obtain a sense of the microbes that were consistently enriched and thus representative of a given module. The (full, noncore) control liquid soil extract microbiome contained 717 unique OTUs across 25 phyla, where 192 (∼26.7%) were present in >1 functional module core (Fig. 5b). The most inclusive module categories were “stresses” (179 soil OTUs) and “polysaccharides” (80 soil OTUs) (Fig. 5a), and the most inclusive individual modules were xylan and xylose under salt stress (Fig. 5d). Nevertheless, there were still phyla in the liquid soil extract controls that were not found (Planctomycetes) or were rare (Verrucomicrobia) in our modules (Fig. 5c) and likely require alternative strategies for their enrichment. We repeated this analysis using native soil (solid) to provide a comparison to the original source soil. There were 1,378 unique OTUs in the native soil, of which 416 were found in the liquid soil extract controls, 185 in “stresses,” 59 in “polysaccharides,” with all other categories having below 30 (data not shown). We obtained 341 unique OTUs in module cores that were absent in the control liquid soil extract microbiome (Fig. 5b), a number which only decreased to 331 when comparing module cores to native soil (data not shown).
FIG 5.
Microbiome comparisons between soil controls and functional modules. Core microbiomes were generated for each module through calculation of distinct OTUs represented in at least 40% of replicates and comprising at least 0.01% of cumulative read counts for that module. (a) Absolute abundance plot of distinct OTUs, with a comparison of the distinct OTUs present in native soils, distinct OTUs present in liquid soil extract controls, and the OTUs in liquid soil extract controls represented in at least one module core for each of the functional module categories. (b) Stepwise absolute abundance plot of distinct core OTUs in module categories, where each successive module category represents the distinct module core OTUs present in the category, along with those in all previous categories. Above the x axis represents OTUs also found in liquid soil extract control; below the x axis represents OTUs uniquely found in functional module cores. (c) Relative abundance plot of the distinct OTUs—either those present in native soil, those present in liquid soil extract controls, those present in both liquid soil extract control and functional module cores, or those uniquely present in functional module cores. (d) Absolute abundance plot of distinct liquid soil extract OTUs found in each individual functional module. Bars in plots are colored according to the phylum to which each core OTU belongs.
Modules are functionally distinct.
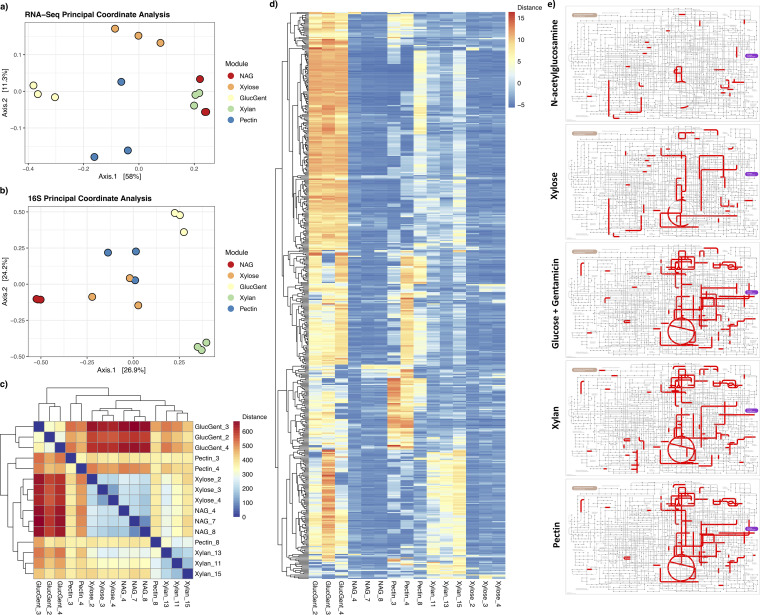
A subset of taxonomically diverse modules was selected to investigate their phylogenetic and functional gene expression profiles. Metatranscriptomes were obtained from 15 samples: 3 replicates each from the modules “N-acetylglucosamine,” “xylose,” “gentamicin” (note that glucose was added together with gentamicin and is designated “GlucGent” on figures), “xylan,” and “pectin.” Transcripts were annotated against the UniRef90 database to get taxonomic information and against the EggNOG database to obtain KEGG functional information.
Bray-Curtis dissimilarity objects were constructed for the normalized transcript abundance table and corresponding 16S rRNA gene amplicon data. In both data sets, “modulename” significantly (P < 0.001) explained variation in Bray-Curtis dissimilarity (Table S5), and replicates within modules clustered together (apart from one outlier for pectin) (Fig. 6a to c), demonstrating consistency in module enrichment patterns for both functional and taxonomic profiles. In addition, a Mantel’s test confirmed that a significant correlation existed between 16S rRNA gene amplicon and transcript intersample Bray-Curtis distances (Mantel’s statistic = 0.42, P = 0.0001), indicating that the more dissimilar samples are taxonomically, the more dissimilar they are functionally. Functional reproducibility also varied between the modules. Relatively consistent expression patterns were seen for N-acetylglucosamine, xylose, and gentamicin, while pectin and xylan were more variable (Fig. 6d).
FIG 6.
RNA-Seq functional trends. Bray-Curtis distance objects were generated for the RNA-Seq data set as well as the corresponding samples within the 16S amplicon data set, generating principal coordinate analysis plots for both the RNA-Seq (a) and 16S (b) data sets. Points are colored by module (red, NAG/N-acetylglucosamine; orange, xylose; yellow, GlucGent/glucose + gentamicin; green, xylan; blue, pectin). (c) Heatmap of Euclidean distances between individual samples within the RNA-Seq data set, where red indicates high distance/dissimilarity between samples, and blue indicates low distance/similarity between samples. (d) Heatmap of expression values for the 15 samples in the RNA-Seq data set. To subset the data set to the transcripts that were most informative with regard to sample-to-sample variation, in this plot the transcripts were first reduced to the 5,000 with the highest average counts, then further to the 500 of those with the highest coefficient of variation (the ratio of standard deviation to the mean) for counts. The data in panels a, c, and d are based on applying variance-stabilizing transformation to the original DESeq-normalized data object so as to more easily visualize differences between samples. (e) Significantly upregulated KEGG orthologs for each of the five modules used in RNA-Seq. Lists of enriched KOs were generated by performing comparisons between all pairs of modules, where KOs that were significantly upregulated in a module relative to at least two other modules were retained (as well as having a shrunken log-fold change of >1.0 and an adjusted P value of <0.05). Subsequent visualization of KOs on a microbial metabolism map was accomplished using iPath 3.0.
PERMANOVA of Bray-Curtis distances for 16S and RNA-Seq datasets. PERMANOVA is performed specifically for the 15 samples used for RNA-Seq and the corresponding 16S data. Significance of experimental factors “modulename” and “replicate” were calculated using the “adonis” function within the R vegan package. Download Table S5, PDF file, 0.05 MB (50.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To determine functional enrichment patterns, we made pairwise comparisons between all combinations of modules. Transcripts significantly (adjusted P value of <0.05, shrunken log-fold change of >1.0) elevated in one module relative to at least two of the remaining four modules were retained (Table S6). Subsequently, KEGG categories or subcategories disproportionately represented in elevated transcripts, relative to their percentage in the full annotated data set, were considered “enriched” in a module. Distribution of categories and subcategories differed by module (Fig. S2a and b); for example, the KEGG categories “transport and binding proteins” and “glycan biosynthesis and metabolism” were enriched in gentamicin, and pectin and xylan modules, respectively (Fig. S2a), and the subcategory “amino acids, peptides, and amines” was enriched in gentamicin (Fig. S2b). Furthermore, the enriched KEGG orthologs (KOs) in xylan and pectin modules encompassed more of the microbial metabolic map than did enriched KOs in xylose or N-acetylglucosamine (Fig. 6e), suggesting either that individual microbes have a greater range of expression in these modules or that increased community richness means a greater diversity of functional capacity were encompassed.
Taxonomic and functional abundance trends for samples used in RNA-Seq analysis. (a and b) Functional trends for RNA-Seq dataset, KEGG category and subcategory level. DESeq-normalized counts for the RNA-Seq database were annotated with KEGG categories and subcategories through the use of the EggNOG database. Transcripts that were successfully annotated were retained, and the relative abundances for transcripts were plotted for the top 20 most abundant KEGG categories (a) and top 30 subcategories (b). In the event that a transcript matched to multiple KEGG categories or subcategories, transcript counts were evenly divided among them. Download FIG S2, PDF file, 0.02 MB (16.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Enriched or depleted KEGG categories or subcategories in a given module relative to at least two of four remaining modules. The Fisher exact test was used to confirm significance of KEGG categories/subcategories in enriched transcripts relative to their proportion in the overall annotated dataset. Download Table S6, PDF file, 0.1 MB (54.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The module incubated with the gentamicin antibiotic plus glucose had the most significantly differing enriched transcripts (n = 1,233) relative to all other modules (Table S7 and Fig. 6d). Of the transcripts with the highest average log2-fold expression changes, the majority were for housekeeping functions (e.g., oxidative phosphorylation and ribosomal proteins), although a select few had putative roles in aminoglycoside antibiotic resistance, such as gentamicin efflux or modification (Table S7) (19, 20). For example, two of the most highly enriched transcripts in gentamicin relative to all other modules were for “transport and binding proteins” (Table S7), one of which was for a “porin” protein, which belongs to a family with noted roles in antibiotic resistance through efflux (19, 20). Similarly, another transcript was for “pyridoxine 5′-phosphate synthase” (EC 2.6.99.2), the gene responsible for pyridoxal-5-phosphate, which is known to combat aminoglycoside toxicity (21), while another was for an adenylyltransferase (EC 2.7.7.42), which has known activity in aminoglycoside resistance (22). However, the majority of enriched transcripts had no explicit connection to gentamicin resistance (Fig. 6d). Instead, these findings were likely a consequence of gentamicin exerting selective pressure to alter community composition and thus community-wide gene expression. Indeed, 16S amplicon sequencing data confirmed different community compositions for the “gentamicin” module relative to its control (i.e., basic glucose with no gentamicin), such as a higher relative abundance of class Betaproteobacteria at the expense of Gammaproteobacteria (Fig. 3b). Approximately 47.3% of all significantly enriched transcripts in the gentamicin module belonged to Betaproteobacteria, an increase from 16.5% in the full data set (data not shown). Aminoglycoside-resistant soil bacteria belonging to class Betaproteobacteria (e.g., order Burkholderiales, members of which have demonstrated resistance to gentamicin [23]) are likely responsible for these metabolic shifts.
List of significantly enriched transcripts in gentamicin modules (“GlucGent”) relative to all four other modules (renamed “OtherSubstrates”). The function “results” from the DESeq2 package was used on the DESeq2 data object for transcriptomic data to determine enrichment patterns for “GlucGent” versus “OtherSubstrate” modules. Transcripts that had a significant (adjusted P value < 0.05) log2FoldChange value of greater than 1 or less than −1 were retained. KEGG and UniRef annotations (including taxonomic information) were then added for transcripts that had a match to one or both databases. The results are sorted by the Log2FoldChange to determine which transcripts were most consistently highly enriched (log2FoldChange > 1) or depleted (log2FoldChange < −1) in gentamicin. Note that transcripts with more than one KEGG hit have only the first hit listed. Download Table S7, PDF file, 0.6 MB (664.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Another striking trend was enrichment of more KEGG subcategories in the polysaccharide modules (14 and 12 subcategories for xylan and pectin, respectively) compared to those for simple substrates (6 and 5 for xylose and N-acetylglucosamine, respectively) (Table S6). This was also reflected at the individual transcript level, with significantly more enriched transcripts in polysaccharide modules relative to those for simple substrates (1,291 versus 213 transcripts). Polysaccharide-enriched transcripts included many with functions relevant to polysaccharide metabolism (endo-1,4-β-xylanase [EC 3.2.1.37] or an SBP domain-containing protein, which is associated with binding plant polysaccharides [24]), while many transcripts enriched with simple substrates were associated with amino acid metabolism (Table S8). There were also a number of enriched transcripts responding to polysaccharides that were uncharacterized and may serve as attractive targets for elucidating mechanisms behind microbial polysaccharide metabolism. In particular, a larger proportion of enriched transcripts in polysaccharide modules were attributable to Bacteroidetes compared to those in simple substrate modules (13.6% versus 0.8%, respectively) (Table S8), which may be attributable to a high presence of CAZymes for glycan breakdown in this phylum (25). Because only a small number of polysaccharide-enriched transcripts were specifically implicated in polysaccharide breakdown (Table S8), the differences in gene expression patterns are more likely due to shifts in active members of the community rather than changes of specific transcripts being upregulated. This is supported by the amplicon sequencing data, which showed a higher abundance of Bacteroidetes in the polysaccharide modules (chitin, cellulose, pectin, and xylan) compared to the simple substrate modules (Fig. 3a), as well as indicator analysis confirming presence of polysaccharide degraders in polysaccharide modules (Table S4). In addition, there was a higher community diversity and a corresponding greater metabolic diversity in the polysaccharide modules as a whole, compared to that of the simple substrates (Fig. 2a to c and Fig. 6).
List of significantly enriched transcripts in polysaccharide modules (“xylan” and “pectin”) relative to simple substrate (“xylose” and “NAG”) modules. The “GlucGent” module was excluded from this analysis. The function “results” from the DESeq2 package was used on the DESeq2 data object for transcriptomic data to determine enrichment patterns for “polysaccharides” versus “SimpleSubstrate” modules, where the six replicates for each (e.g., 3 xylan + 3 pectin, 3 xylose + 3 NAG) were pooled for said comparison. Transcripts that had a significant (adjusted P value < 0.05) log2FoldChange value greater than 1 or less than −1 were retained. KEGG and UniRef annotations (including taxonomic information) were then added for transcripts that had a match to one or both databases. The results are sorted by the average Log2FoldChange to determine which transcripts were most consistently highly enriched (log2FoldChange > 1) or depleted (log2FoldChange < −1) in polysaccharides. Note that transcripts with more than one KEGG hit have only the first hit listed. Download Table S8, PDF file, 0.8 MB (857.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Network analysis of functional modules.
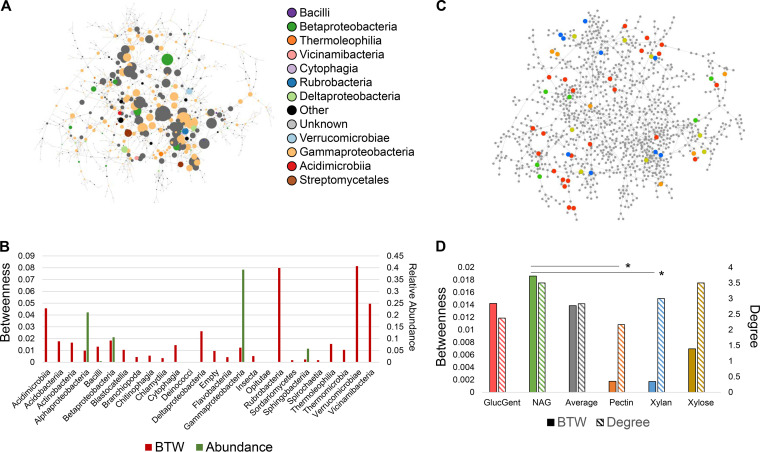
We integrated the metatranscriptomic data to identify genes/taxa occupying central and potentially metabolically important positions in the soil microbiome. The context likelihood of relatedness (CLR) algorithm was used to infer a coexpression network linking genes based on mutual information; then, taxonomic and centrality information were integrated to identify highly central genes and taxa. For taxa, in contrast to findings from 16S rRNA gene amplicon analysis (Fig. 3b), the centrality of genes from Gammaproteobacteria and Betaproteobacteria was much lower compared to genes from other taxa (Fig. 7a and b). Betaproteobacteria had a betweenness centrality value roughly in line with its abundance level. In contrast, other classes (Rubrobacteria, Verrucomicrobia, Vicinamibacteria, and Acidimicrobiia) were low in relative abundance based on 16S amplicon sequencing (Fig. 3b) but expressed genes of high centrality, including an extracellular solute binding protein (Rubrobacteria), a catalase peroxidase (Verrucomicrobia; EC 1.11.1.21), NADH-quinone oxidoreductase subunit C (Vicinamibacteria; EC 1.6.5.3), and an ABC1 domain-containing protein (Acidimicrobiia). These genes occupied bottleneck positions of high betweenness and acted to link a large number of other genes throughout the network.
FIG 7.
Network analysis of metatranscriptomic data. (a) Gene coexpression network of metatranscriptomic data. Circles represent genes, and lines instances of high mutual information (Z-score above 13.35). Genes are sized by betweenness centrality, with a larger size indicating higher betweenness. Genes are colored by the class of the species expressing them (see inset). (b) Bar chart showing average betweenness (red bars, left y axis) and abundance (green bars, right y axis) for each taxonomic class in the network. Certain species could not be detected by 16S and have no abundance bar. (c) Network with genes colored by which functional module perturbation they are uniquely associated with (red, glucose with gentamicin; green, NAG; orange, pectin; blue, xylan; yellow, xylose). (d) Bar chart showing average betweenness (filled bars, left y axis) and degree (hashed bars, right y axis) for each set of genes uniquely associated with each perturbation. Gray bars represent the average betweenness or degree of all genes in the network. Asterisks indicate statistically significantly higher betweenness values determined by a Student t test (P > 0.03).
To investigate centrality of functions, transcripts significantly associated with each of the five modules were located in the network (Fig. 7c). Genes responding to incubation with N-acetylglucosamine and xylose had higher centrality compared both to those belonging to other modules and to the average centrality of all genes in the network (Fig. 7d). N-acetylglucosamine-enriched genes had statistically higher betweenness centrality compared to pectin- and xylan-enriched genes (P < 0.03). Genes responding to xylan, pectin, and xylose had much lower betweenness centrality values (7.9, 8.0, and 1.99-fold, respectively) compared to average betweenness, although these shifts were not statistically significant.
DISCUSSION
In this work, we deconstructed the soil microbiome into discrete modules based on functional capacities. The resulting module communities were reduced in complexity, varied in reproducibility, and represented a significant extent of diversity present in either liquid soil extract controls or native soils, while also enriching for microbes that were rare or undetectable in either soil. We also demonstrated that low-abundance taxa, despite their rarity, substantially contributed to module functional profiles through expression of a few highly central genes. Targeted enrichments for distinct consortia from a complex parent microbiome have previously been conducted using substrate enrichments (12, 26-28) or stress perturbations (9) to investigate substrate uptake and decomposition trends (26), to determine taxonomic diversity and/or growth on carbon sources (12, 28), or to obtain reduced complexity consortia for optimized whole genome binning in metagenomes (9). Here, we provide new knowledge about the breadth of soil phylogenetic and functional capacity through high-resolution analyses of the soil microbiome using a functional module approach.
Diversity and phylogenetic trends for functional module consortia.
The functional modules had lower diversity metrics compared to liquid soil extract controls. Opportunistic growth by Pseudomonas was the cause of this trend in numerous modules enriched with simple substrates. As an r-selected organism, Pseudomonas grows rapidly in liquid cultures containing labile carbon substrates (12, 29), outcompeting less opportunistic microbes. Thus, the results observed here are consistent with its life strategy (30). However, even the comparatively complex polysaccharide modules were less diverse than liquid soil extract control.
We also hypothesized that functional module enrichments would be highly reproducible given the tight restrictions on medium composition, especially relative to the undefined soil extract media used as controls. However, more than 20 modules had higher values for beta dispersion than liquid soil extract controls (although not statistically significant [Table S2]). These modules primarily belonged to the “stress” and “polysaccharides” categories which tended toward more diverse communities with a prevalence of Firmicutes and Actinobacteria. Previous reports have associated Actinobacteria-rich communities with higher dispersion (31), possibly due to their lower growth rates preventing any one taxon from crowding out all others. Indeed, under “late” and dilute carbon stresses (both conditions that would encourage slow-growing microbes), Actinobacteria were present in all module replicates (Fig. 3a).
Polysaccharide breakdown generates a variety of carbon substrates, enriching for a larger repertoire of metabolic functions (32). Higher functional diversity means more chances for the “lottery hypothesis” to take effect, in which whoever is able to seize opportunity first exerts priority effects (33, 34), increasing community dispersion. One potential activity is metabolic cross-feeding: secondary consumers utilize polysaccharide breakdown products but possess no intrinsic breakdown activity. Consequently, caution must be taken when inferring which community members actively participate in polysaccharide breakdown, although it is unlikely secondary consumers outnumber primary consumers (35). Although outside the scope of the present study, the influence of multiple habitat niches on community succession could be tested by incorporating a solid matrix together with complex substrates.
We also addressed to what extent functional modules typified the parent soil microbiome. Approximately 27% of OTUs found in the liquid soil extract controls were also found in module cores. Therefore, 1/4 of the control soil microbiome was connected to at least one metabolic function tested in our study. While most major soil phyla had a substantial presence across module cores, other phyla (e.g., Bacteroidetes and Verrucomicrobia) were underrepresented. Bacteroidetes are predominantly copiotrophic and associated with C mineralization (30) but were likely outcompeted by rapid Pseudomonas growth in simple substrate modules. However, Bacteroidetes were enriched (though not significantly so) in polysaccharide modules compared to simple substrate modules (mean relative abundances of 2.86% and 0.03%, respectively), consistent with their putative role in polysaccharide breakdown (25), and growth in xylan-containing medium (36). For this reason, inclusion of more complex polysaccharide modules (or altering the growing conditions of existing ones) could serve to better enrich for this phylum. Depletion of Verrucomicrobia was more striking, as they were a substantial proportion of liquid soil extract controls but only present in four module cores. Verrucomicrobia are characteristically slow growing and difficult to cultivate, although highly diluted nutrient broth has been shown to be an effective enrichment strategy (37) that could be harnessed in future functional module approaches to capture this phylum. Interestingly, despite the low abundance of the Verrucomicrobia, this phylum was highly central in the gene coexpression network (Fig. 7b), suggesting that it carried out key functions for the community.
Including various stress perturbations also extended the range of soil diversity we could reconstitute. Salt stress was particularly inclusive in enriching for soil taxa, which is line with previous reports implicating salinity as a strong determinant on soil community structures (38, 39). However, our finding that salt stress increased diversity contrasts with previous findings showing the opposite (39). These results highlight the importance of considering microbial functional capacity in aspects other than substrate utilization, since stress imposition often provoked substantially different communities than the unstressed control (Fig. 4b).
Our approach also demonstrated the potential of capturing rare soil microbes through targeted enrichments. In total, 341 unique OTUs across a wide range of taxa were found in functional module cores but not in liquid soil extract controls. Functional modules augmented the prevalence of members of Firmicutes (mainly class Bacilli) and Actinobacteria (mainly class Actinobacteria), as well as the classes Gammaproteobacteria, Negativicutes, and Clostridia. While the majority of Gammaproteobacteria enrichment was due to Pseudomonas OTUs not found in the soil controls, the other trends are less easily explained. Firmicutes and Actinobacteria are associated with carbon cycling processes in fertilized prairie soils (40), which could explain their prevalence in polysaccharide modules. In addition, Firmicutes were prevalent in anaerobic and heat stress modules. Given Firmicutes’ involvement in anaerobic processes (sulfate/iron reduction, fermentation [41], and endospore formation under high heat [42]), these modules may be suitably tailored to Firmicutes’ metabolic capacities. Similarly, Clostridia were a small minority (0.0125%) of liquid soil extract controls but were enriched in most polysaccharide and anaerobic modules (Fig. 3b). Taken together, these results suggest that complex microbiomes such as soil have substantial “hidden” diversity that require alternative approaches (such as functional modules) to be unearthed.
Mechanistic insights and functional trends across functional modules.
Previous studies have attempted to relate microbial guilds to the functional capacity for nutrient usage or carbohydrate metabolism (10, 43), to explain the functional contributions of predetermined groups of taxa (44), or to relate functional shifts under a certain treatment to the major contributing taxa (45). However, to our knowledge this is the first study demonstrating the deconstruction of a complex microbiome’s collective functional capacity (i.e., the “metaphenome” [11]) into component parts with distinct expression profiles. In our study, through analyzing gene expression data on a subset of modules, we confirmed that different modules were characteristically enriched or depleted in different functions or functional categories. For example, xylan was uniquely enriched for ascorbate metabolism, pectin for nitrogen metabolism, and gentamicin for pyrimidine metabolism (Table S6). Patterns for KEGG category abundances could often be related to the nature of the module itself. Xylan and pectin modules had high levels of “glycan biosynthesis and metabolism” (Fig. S2a), which is likely attributable to these complex carbohydrates (i.e., glycans) requiring a greater variety of relevant functions for their catabolism. Similarly, gentamicin modules had high levels of “transport and binding proteins” (Fig. S2a), possibly for transporters involved in antibiotic efflux or secretion (46), which is line with the types of genera significantly enriched in this module by indicator analysis (Table S4). For KEGG subcategories, gentamicin modules were elevated in “amino acids, peptides, and amines” (Fig. S2b), potentially explained by gentamicin interacting with chaperone proteins’ peptide-binding domains (47). Both xylan and pectin were less reproducible at the KEGG subcategory level, with functions such as “polysaccharides and lipopolysaccharide metabolism” and “DNA replication, recombination, and repair” showing variable relative abundances between replicates.
There were 1,233 transcripts significantly elevated in gentamicin compared to the other four modules (Table S7), many with putative functions in antibiotic resistance (efflux pumps and antibiotic modification). Interestingly, a higher proportion of transcripts matched to Betaproteobacteria in the gentamicin-enriched subset relative to the full metatranscriptome. We also observed strong community shifts in the gentamicin module relative to the antibiotic-free glucose module, in particular an increase in Betaproteobacteria at the expense of Gammaproteobacteria. These results point toward changing gene expression patterns being a consequence of gentamicin selecting against susceptible Gammaproteobacteria, while enriching for Betaproteobacteria. For polysaccharides, there were far more significantly enriched transcripts in polysaccharide modules over simple substrate modules than for the reverse comparison (Table S8). These shifts in gene expression are likely reflective of the higher community diversity found among polysaccharide modules (Fig. 2a to c) and the diverse sets of pathways needed for polysaccharide metabolism, resulting in more opportunity for variability in functional profiles and metabolic cross-feeding.
Replicates within modules were generally more similar to one another than they were to other module replicates (Fig. 6a), confirming this approach as viable for obtaining functionally distinct consortia. However, complex polysaccharide modules had less consistency in gene expression and also community dispersion based on 16S rRNA gene sequencing, implicating microbial relative abundance shifts, rather than alternative expression patterns, as the root cause of disparate transcriptomic profiles. Such findings may be attributable to the diverse nature of the soil microbiome: within its extensive array of functional guilds, those with the inherent capacity to survive in a given module will grow rapidly and contribute more to transcriptomic profiles over persistent, metabolically plastic species that simply alter their expression patterns.
Our finding that genes associated with N-acetylglucosamine and xylose occupy highly central network positions suggests that processes that respond to these carbon and nitrogen sources are central, and these substrates are important to the overall soil microbiome. Indeed, xylose is one of the most highly abundant saccharides found in plant material, being a major substituent of the plant cell wall hemicelluloses xylan and xyloglucan (48). Chitin, of which N-acetylglucosamine is a monomer, is abundant in many soils, as a component of some fungal cell walls and insect exoskeletons (49). As such, xylose and N-acetylglucosamine represent important sources of carbon and energy that soil microbial communities must be prepared to metabolize.
We acknowledge some potential drawbacks in our approach. First, in the interests of controllability and throughput, all enrichments were conducted in liquid (rather than soil) media. Due to the relative homogeneity of this media type, liquid cultures are predisposed to enrich for one or a few OTUs (10, 35), are potentially biased against rare taxa due to the use of soil dilutions (5), and will evoke different community profiles than would be obtained in soil (29, 50). We observed domination by fast-growing microbes under certain substrates in liquid cultures, but these findings do not equate to these microbes being the only ones capable of metabolizing the substrates. In contrast, in soils, various factors (e.g., inaccessibility of substrates due to irregular spatial distribution or physical protection mechanisms [51]) will invoke competitive or synergistic interactions and, by extension, a more diverse community, even with the same substrate (29). However, where applicable we included native bulk soil as a point of comparison to partially address this issue. Second, the carbon substrates supplied were largely labile substrates, while in native soils, microbial diversity is limited by low C availability (52) and (aside from root exudates) available carbon sources are mixtures of nonlabile biomacromolecules (51). For that reason, future functional module approaches based on a soil system could incorporate additional complex substrates and lower nutrient availabilities to more accurately mirror native soil conditions. Third, amplicon sequencing cannot be used to extrapolate functional capacity of a community without complementary approaches. Methods such as stable isotope probing represent an advantageous approach in that the microbes directly metabolizing a given labeled substrate can be accurately identified but are comparatively low throughput and require labeled versions of the desired substrates to be commercially available (12). However, due to the limitations in relying on amplicon sequencing alone, we chose to supplement this data set with metatranscriptomics on a subset of samples. The advantage of metatranscriptomics is that this data type provides a more comprehensive approach in determining functional profiles than does stable isotope probing, which targets active taxa (18O) or transformation and assimilation of substrates (13C). In addition, generating and sequencing the high number of modules (48) used in this analysis represented a significant effort. Our approach could be streamlined through inclusion of only a subset (e.g., 5 to 10) of modules that together encompass the largest extent of soil diversity of any module subset, such as focusing on a combination of stress and polysaccharide modules. In this way, the process can be optimized to provide the greatest amount of information with the least amount of work. Ultimately, by deep exploration of functional modules representative of specific types of metabolism that are found in native soil, we should be able to further expand our knowledge about the potential organismal interactions and the types of metabolic routes carried out by different taxa in soil.
MATERIALS AND METHODS
Functional module design.
Distinct media types were created for each functional module. All functional module media were based on 1× minimal M9 media (in 1 liter of double-distilled water [ddH2O], addition of 6 g of sodium phosphate dibasic, 3 g of potassium phosphate monobasic, 1 g of ammonium chloride, 0.5 g of sodium chloride, 0.0111 g of calcium chloride, with 0.24 g of magnesium sulfate added after autoclaving), with subsequent modifications that varied by the specific module.
For all modules based on utilization of simple substrates, M9 medium was supplemented with 10 mM carbon substrate. These substrates included sodium acetate, arabinose, sodium citrate, d-galacturonic acid, d-glucuronic acid, galactose, glucosamine, glucose, glutamate, glycerol, glycine, sodium malate, mannose, N-acetylglucosamine, sodium propionate, sodium pyruvate, rhamnose, and xylose. All substrates were added prior to autoclaving.
All antibiotic modules were based on supplementation of a M9-plus-glucose (M9+glucose) module (as prepared above) with antibiotics, added after autoclaving to avoid denaturation then filter sterilizing the new media. Multiple concentrations were tested for a level that would impose a selective force while still allowing for growth, as measured by increases in optical density at 600 nm (OD600) over time relative to nonsupplemented glucose control. Concentrations were finalized for benomyl at 15 μg/ml culture media, chloramphenicol at 128 μg/ml, erythromycin at 100 μg/ml, nystatin at 50 μg/ml, and streptomycin sulfate at 100 μg/ml. Additional antibiotics (ampicillin sodium salt, tetracycline hydrochloride, and vancomycin) were tested and discarded due to producing no or very limited microbial growth.
Polysaccharide modules were based on adding 10 mM a carbon source to minimal M9 media. Substrates included: carboxymethyl cellulose (to represent cellulose), xylan (specifically, purified beechwood xylan), pectin, chitin (purified from crab shells), cellobiose, trehalose, and sucrose. Apart from pectin, all polysaccharide/disaccharide substrates were added prior to autoclaving. Pectin was added to autoclaved minimal M9 media by dissolving at 70°C with stirring, followed by filter sterilizing (pore diameter, 0.22 μm) to remove possible contaminants.
Anaerobic modules were based on supplementation of M9+glucose with an alternative redox acceptor and flushing with a specific gas mixture for 30 min to remove residual O2 (both culture media and the starting soil inoculum were flushed for 15 min separately; then, after the addition of the inoculum to the media, the new solution was flushed for an additional 15 min). A basic anaerobic module, referred to as the “acetogen” module, was constructed with M9+glucose and flushing with pure N2 gas. Iron sulfate medium was constructed by either filter sterilizing (“fs”) or autoclaving (“nonfs”) M9+glucose with 0.5 g/liter iron sulfate heptahydrate and then flushing with 9:1 N2:H2. Potassium nitrate medium was constructed by adding 0.5 g/liter potassium nitrate to M9+glucose, autoclaving, and then flushing with 4:1 N2:CO2. Iron citrate medium was constructed by adding 13 g of iron(III) citrate to M9+glucose, autoclaving, and then flushing with 9:1 N2:H2.
Several functional modules were designed to impose further stress perturbations on the soil microbiome. Preliminary stress tests were performed by modifying glucose and N-acetylglucosamine modules with respect to pH or salt concentration. In these tests, pH of the starting minimal M9 medium was changed from 7 to either 6 or 8, altering the ratios of sodium phosphate monobasic to potassium phosphate dibasic accordingly (while maintaining a constant phosphate concentration), prior to the addition of glucose or N-acetylglucosamine. Salt modules were created through supplementation of 0.8 M sodium chloride to M9+glucose or M9+N-acetylglucosamine media. Certain preliminary samples (i.e., glucose at pH 6, pH 8, glucose+salt, and NAG at pH 6) were excluded from further downstream analyses if either the stress condition or substrate was not being used for other stress modules. After it was determined by 16S rRNA gene sequencing that the resultant communities were affected by stress imposition relative to unstressed glucose or N-acetylglucosamine modules, additional stress modules were designed. These modules used a combination of one of three carbon substrates (xylose, N-acetylglucosamine, or xylan) and one of eight alternative growing conditions meant to mimic stresses that could be experienced by soil microbes. These included: (i) “10%C,” diluting the carbon substrate 10-fold (1 mM rather than 10 mM); (ii) “2,4-D,” supplementation with the herbicide 2,4-dichlorophenoxyacetic acid at a 10 mM final concentration); (iii) “heat,” constant incubation temperature of 40°C; (iv) “late,” harvesting after 1 week; (v) “light,” growth under a constant light source; (bi) “PEG,” supplementing media with polyethylene glycol 8000 at an amount sufficient to impose a final osmotic pressure of −1.0 MPa (28.66 g of PEG-8000 per 100 ml of media, followed by autoclaving); (vii) “pH 8,” altering the initial M9 medium from pH 7 to pH 8; and (viii) “salt,” adding sodium chloride to a final concentration of 0.8 M to impose a salt stress.
Soil control modules consisted of liquid soil extract that was used to represent nutrient sources present in the native soil, but in a liquid environment comparable to that of the functional modules. Briefly, the soil extract was generated by suspending 0.5 kg of the native field soil (see field information below) in 2 liters of ddH2O with shaking at 4°C for 48 h, followed by centrifugation for 15 min at 8,000 × g to pellet soil particles. The resultant supernatant was filter sterilized (0.22 μm) to remove residual particulate matter and autoclaved.
Native soil controls were also included for comparison (although any instance where the terminology “soil control” is used refers to liquid soil extract). The soil was collected from the field as described below. DNA was extracted from two 0.25-g samples using a DNEasy PowerSoil kit (Qiagen, Carlsbad, CA), and 16S rRNA gene sequences were generated as described below.
Functional module sample generation.
Soil used in these experiments was the same as described in a previous study (29). Briefly, the soil was collected in October 2017 from an experimental field site (Washington State University Irrigated Agriculture Research and Extension Center located in Prosser, WA), and then 4 mm sieved to remove soil aggregates, rocks, and extraneous plant matter before long-term storage (0.5 to 1.5 years) at 4°C. For inoculation into the different functional module media, 0.5 g of soil was first added to 50 ml of phosphate-buffered saline to achieve a 10−2 dilution and mixed to resuspend by vortexing. Subsequently, 1 ml of the soil inoculum was added to 9 ml of the respective functional module media in Balch tubes to achieve a final soil dilution of 10−3, and the tubes were capped and incubated in a benchtop shaker.
All functional modules samples were incubated at room temperature apart from the heat stress modules, which were incubated at 40°C. Similarly, no functional modules were exposed to light apart from “light stress” modules, which were illuminated at all times by 23-W fluorescent lights. Anaerobic modules were incubated with shaking at 50 rpm, while all nonanaerobic modules were incubated with shaking at 170 rpm to maintain aeration. The OD600 values were recorded every 1 to 2 days as a proxy for microbial growth using a Spectronic 20D+ spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Samples were harvested when the OD600 readings were roughly 0.2 to 0.8, i.e., when a pellet could be obtained with sufficient biomass to extract nucleic acids. The duration of incubation necessary to reach this OD600 measurement varied from days to weeks, depending on the module in question (simple substrates were typically between 3 and 7 days, whereas complex substrates and anaerobic modules could take as long as 3 to 4 weeks). To harvest samples, two replicates using 3 ml for each respective module were centrifuged for 10 min at 8,000 × g, generating one pellet for DNA extraction and (optionally) one for RNA extraction. The supernatant was removed, and pellets were quick-frozen in liquid N2 before long-term storage at –80°C.
16S rRNA gene amplicon library preparation and sequencing.
Total DNA was extracted from cell pellets by using a DNeasy blood and tissue DNA isolation kit (Qiagen, Carlsbad, CA) with the specific modification of pretreatment for Gram-positive bacteria as described in the kit protocol. DNA was then quantified using Qubit dsDNA HS assay kit (Thermo Fisher). 16S rRNA gene amplicon library preparation, sequencing, and downstream processing was performed as described previously (29); briefly, sequencing was performed on a MiSeq instrument (Illumina, San Diego, CA) using 16S primers (515F and 806R) that targeted the V4 hypervariable region. The resultant sequence data were processed using Hundo (53), an in-house protocol for amplicon quality control and annotation that wraps multiple programs (BBDuk, VSEARCH, FastTree2, etc.) in a streamlined pipeline.
16S rRNA gene community analysis.
Statistical analyses on 16S data sets were performed using the program R (54), incorporating the R packages phyloseq (55) and vegan (56) packages. The original data object consisted of 13,316,501 reads across 324 samples; after rarefying to an even depth of 8,000 reads per sample, this was reduced to 2,592,000 reads. This data set represented at least five biological replicates for each of the 66 unique functional modules, except for those that only had three replicates (glycerol) or four replicates (galactose, glycine, malate, and propionate) that passed the rarefaction threshold. For the remaining modules, the five replicates with the highest values for Shannon’s diversity were retained for downstream statistical analyses, to maximize the taxa represented. While this may represent a potential source of bias, the goal of this study was to capture as much of the phylogenetic and functional representation of the native soil as possible. A separate phyloseq data object incorporating the two native soil replicates, rarefied to 6,000 reads per sample, was used for tests incorporating explicit comparisons to native soils.
Alpha-diversity metrics (Shannon’s diversity and richness) were calculated using the function “estimate_richness” within phyloseq or, for Faith’s PD, using the function “pd” within the picante package (57). Beta-dispersion was calculated using the “betadisper” function within vegan, which estimates multivariate dispersion for a group of samples from their median and then tests to determine whether dispersions are distinct between groups through ANOVA. ANOVA for significances of experimental factors in explaining alpha-diversity or beta-dispersion metrics was conducted using the “aov” function within the stats package (54). Tukey’s post hoc tests for significant differences between groups for the above metrics were performed using the “HSD.test” function within the agricolae package (58), which calculates group means and uses ANOVA to determine which pairwise combinations of group means are significantly different from one another. Beta-diversity calculations were based on the weighted UniFrac distance metric so as to account for phylogenetic relationships between OTUs, and these distances were generated using the “UniFrac” function within phyloseq. Principal coordinate analyses were performed using the “ordinate” function on weighted UniFrac distance objects in phyloseq. Statistical tests were also conducted using unweighted UniFrac for comparison. To determine significances of experimental factors in explaining beta-diversity, PERMANOVA was conducted using the adonis package within vegan. Relative abundance plots for phylum- and class-level profiles were graphed using ggplot2 (59). Indicator species analysis was conducted using the “indval” function in the package labdsv (60), which uses the product of the relative frequency and the average relative abundance across groups for a taxon and then calculates the indicator value for this taxon.
RNA library preparation and sequencing.
Total RNA was extracted from cell pellets using TRIzol reagent (Invitrogen, Carlsbad, CA), incorporating the kit protocol specific to “cells grown in suspension.” Eluted RNA was quantified using a Qubit RNA HS assay kit (Thermo Fisher). Residual DNA was removed with Ambion Turbo DNase according to the manufacturer’s protocol (Thermo Fisher). Enrichment of mRNA (through rRNA depletion) was performed using the MicrobExpress kit (Thermo Fisher) according to the manufacturer’s protocol, including the optional step of initial ethanol precipitation of RNA. Prior to mRNA enrichment RNA quality was assessed on an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA), and samples with an RNA integrity number between 7 and 10 were retained for library preparation. Samples were also examined on a bioanalyzer after mRNA enrichment to confirm depletion of rRNA species. The resultant mRNA samples were sequenced by GENEWIZ (GENEWIZ, South Plainfield, NJ). Raw reads from all samples were aligned to a previously obtained soil metagenome from the native field site using the Burrows-Wheeler aligner (BWA) (61), retaining reads that successfully mapped only once to the soil metagenome. The resulting SAM files were converted to raw counts using HTSeq (62). To focus on taxa with at least moderate contributions to the functional response of the modules, we only included transcripts with 75 cumulative counts across the 15 samples, with no more than three absences (zero counts).
Downstream statistical analyses were conducted using R. The subsequent data set was normalized with “DESeq” in the DESeq2 package (63), resulting in a data set consisting of 6,947 unique transcripts across the 15 samples and 185,920,068 reads. For visualization purposes, the “VarianceStabilizing Transformation” function within DESeq2 was applied to the data set. Gene annotations were obtained using the EggNOG (for functional annotations [64], namely, KEGG categories and subcategories) or UniRef90 databases (for taxonomic annotations [65]). Of the 6,947 transcripts, 4,352 had taxonomic annotations through the UniRef90 database to the superkingdom level, while only 1,728 were annotated to the species level. Likewise, 4,975 transcripts had at least one functional role assigned to them from the EggNOG database.
RNA-Seq analysis.
Heatmaps were generated using the “pheatmap” function within the package of the same name (66) on the 500 genes with the highest coefficient of variation (within the 5,000 genes with the highest read counts). To generate lists of up- or downregulated genes in each of the five modules used in metatranscriptome sequencing (RNA-Seq) analysis, pairwise comparisons were conducted for all pairs of modules. Genes determined as significantly upregulated had a shrunken log-fold change (“lfcShrink” function in DESeq2) of more than 1.0 and an adjusted P value of less than 0.05; significantly downregulated genes had a shrunken log-fold change of less than −1.0 and an adjusted P value of less than 0.05. Only transcripts that were significantly enriched in a given module relative to two or more remaining modules were retained. Polysaccharide-enriched transcripts (Table S7) were determined by comparing pectin and xylan (as one pool) to xylose and N-acetylglucosamine (as a second pool), while gentamicin-enriched transcripts (Table S8) were determined by comparing gentamicin to all other modules at once. KEGG ortholog (KO) annotations obtained from the EggNOG database were mapped to these lists of enriched or depleted genes. The lists of upregulated KOs were used to generate Fig. 6e, using the online program iPath 3.0 (67) to visually represent where they landed on the microbial metabolic map.
To determine which functional enrichment or depletion ratios for KEGG categories or subcategories in a given module, the lists of up- or downregulated genes generated above were first annotated with KEGG categories or subcategories obtained from the EggNOG database. Then, the Fisher exact test was used to see whether a given KEGG category or subcategory was disproportionately represented among the enriched/depleted transcripts relative to its representation in the transcripts of the full annotated data set of the soil metagenome, as well as the corresponding P value.
Comparisons between RNA-Seq data and 16S rRNA gene amplicon data.
To compare RNA-Seq and 16S rRNA gene data for the 15 samples selected for RNA-Seq, the 16S rRNA data were imported and rarefied to 5,000 reads per sample. Bray-Curtis dissimilarity objects were generated for both data sets using the “vegdist” function within vegan. Bray-Curtis was used since the weighted UniFrac is incompatible with data sets lacking a phylogenetic structure (in this case, gene expression data sets), and we wished to retain a consistent distance metric for more accurate comparisons. PERMANOVA and Mantel analyses were conducted using the functions “adonis” and “mantel,” also within the vegan package.
Network analysis.
From the gene expression data set generated as described above, a network was inferred using the context likelihood of relatedness (CLR) program (68), with genes as nodes and edges as instances of high coexpression between nodes. The CLR program was run using default settings with the output a matrix of Z-scores of mutual information values between all gene pairs. A cutoff of 13.35 was used to define edges in the network, indicating that the mutual information for a linked gene pair in the final network was at least 13.35 standard deviations above the mean of all gene pairs (68). The weighted Z-score matrix was converted to an unweighted matrix that replaced all Z-scores with either a “0” (if below 13.35) or a “1” (if above 13.35). The resulting unweighted networks were viewed in Cytoscape (69), and centrality values of betweenness and degree were also calculated using Cytoscape. To identify genes uniquely associated with each perturbation, we first calculated which KOs responded to perturbations as described above under “RNA-Seq analysis.” From this list, we then selected only those genes that responded to a particular perturbation and to no other.
Data availability.
Raw 16S amplicon sequence data are deposited in the NCBI Short Read Archive (SRA) under BioProject PRJNA594403 and BioSample numbers SAMN13560171 to SAMN13560497. For RNA-Seq data, the soil metagenome used for transcript alignment is publicly available at BioProject PRJNA597444, BioSample SAMN13675441, accession number WUUC00000000. RNA-Seq data have been deposited in NCBI’s Gene Expression Omnibus (70) and are available through GEO Series accession number GSE143587. All codes used to generate statistical inferences within the manuscript are publicly available at https://github.com/dtnaylor124/FunctionalModules.
ACKNOWLEDGMENTS
This research was supported by the US Department of Energy Office of Biological and Environmental Research (BER) and is a contribution to the Scientific Focus Area ‘Phenotypic response of the soil microbiome to environmental perturbations.’ Pacific Northwest National Laboratory is operated for the Department of Energy by Battelle Memorial Institute under Contract DE-AC05-76RLO1830.
Footnotes
This article is a direct contribution from Janet K. Jansson, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Ashley Shade, Michigan State University, and Étienne Yergeau, Institut National de la Recherche Scientifique.
Citation Naylor D, Fansler S, Brislawn C, Nelson WC, Hofmockel KS, Jansson JK, McClure R. 2020. Deconstructing the soil microbiome into reduced-complexity functional modules. mBio 11:e01349-20. https://doi.org/10.1128/mBio.01349-20.
REFERENCES
- 1.Bach EM, Williams RJ, Hargreaves SK, Yang F, Hofmockel KS. 2018. Greatest soil microbial diversity found in micro-habitats. Soil Biol Biochem 118:217–226. doi: 10.1016/j.soilbio.2017.12.018. [DOI] [Google Scholar]
- 2.Perez-Garcia O, Lear G, Singhal N. 2016. Metabolic network modeling of microbial interactions in natural and engineered environmental systems. Front Microbiol 7:673. doi: 10.3389/fmicb.2016.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song H-S, Cannon W, Beliaev A, Konopka A. 2014. Mathematical modeling of microbial community dynamics: a methodological review. Processes 2:711–752. doi: 10.3390/pr2040711. [DOI] [Google Scholar]
- 4.Blagodatskaya E, Kuzyakov Y. 2013. Active microorganisms in soil: critical review of estimation criteria and approaches. Soil Biol Biochem 67:192–211. doi: 10.1016/j.soilbio.2013.08.024. [DOI] [Google Scholar]
- 5.Shade A, Gilbert JA. 2015. Temporal patterns of rarity provide a more complete view of microbial diversity. Trends Microbiol 23:335–340. doi: 10.1016/j.tim.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Dawson W, Hör J, Egert M, van Kleunen M, Pester M. 2017. A small number of low-abundance bacteria dominate plant species-specific responses during rhizosphere colonization. Front Microbiol 8:975. doi: 10.3389/fmicb.2017.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misra BB, Langefeld C, Olivier M, Cox LA. 2019. Integrated omics: tools, advances and future approaches. J Mol Endocrinol 2018:R21–R45. doi: 10.1530/JME-18-0055. [DOI] [PubMed] [Google Scholar]
- 8.Prosser JI. 2015. Dispersing misconceptions and identifying opportunities for the use of “omics” in soil microbial ecology. Nat Rev Microbiol 13:439–446. doi: 10.1038/nrmicro3468. [DOI] [PubMed] [Google Scholar]
- 9.Delmont TO, Eren AM, Maccario L, Prestat E, Esen ÖC, Pelletier E, Le Paslier D, Simonet P, Vogel TM. 2015. Reconstructing rare soil microbial genomes using in situ enrichments and metagenomics. Front Microbiol 6:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn TM, Koval JC, Greenwald SM, Owens SM, Kemner KM, Antonopoulos DA. 2017. Parallelized, aerobic, single carbon-source enrichments from different natural environments contain divergent microbial communities. Front Microbiol 8:2321. doi: 10.3389/fmicb.2017.02321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansson JK, Hofmockel KS. 2018. The soil microbiome: from metagenomics to metaphenomics. Curr Opin Microbiol 43:162–168. doi: 10.1016/j.mib.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Goldfarb KC, Karaoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK, Wallenstein MD, Brodie EL. 2011. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front Microbiol 2:94. doi: 10.3389/fmicb.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouskill NJ, Wood TE, Baran R, Hao Z, Ye Z, Bowen BP, Lim HC, Nico PS, Holman H-Y, Gilbert B, Silver WL, Northen TR, Brodie EL. 2016. Belowground response to drought in a tropical forest soil. II. Change in microbial function impacts carbon composition. Front Microbiol 7:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klare I, Konstabel C, Werner G, Huys G, Vankerckhoven V, Kahlmeter G, Hildebrandt B, Müller-Bertling S, Witte W, Goossens H. 2007. Antimicrobial susceptibilities of Lactobacillus, Pediococcus, and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J Antimicrob Chemother 59:900–912. doi: 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, Sheng P, Zhang H. 2012. Isolation and identification of cellulolytic bacteria from the gut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). Int J Mol Sci 13:2563–2577. doi: 10.3390/ijms13032563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martinez LP, Tringe SG. 2016. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209:798–811. doi: 10.1111/nph.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. 2017. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J 11:2691–2704. doi: 10.1038/ismej.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi U, Lee C-R. 2019. Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front Microbiol 10:953. doi: 10.3389/fmicb.2019.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Mah T-F. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol 190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asif AH, Rasool ST. 2012. Pyridoxal phosphate: a possible intervention to prevent aminoglycoside induced electrolyte imbalance. J Clin Res Bioethics 3. doi: 10.4172/2155-9627.1000124. [DOI] [Google Scholar]
- 22.Papadovasilaki M, Oberthür D, Gessmann R, Sarrou I, Betzel C, Scoulica E, Petratos K. 2015. Biophysical and enzymatic properties of aminoglycoside adenylyltransferase AadA6 from Pseudomonas aeruginosa. Biochem Biophys Rep 4:152–157. doi: 10.1016/j.bbrep.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh F, Duffy B. 2013. The culturable soil antibiotic resistome: a community of multi-drug resistant bacteria. PLoS One 8:e65567. doi: 10.1371/journal.pone.0065567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokoyama H, Yamashita T, Morioka R, Ohmori H. 2014. Extracellular secretion of noncatalytic plant cell wall-binding proteins by the cellulolytic thermophile Caldicellulosiruptor bescii. J Bacteriol 196:3784–3792. doi: 10.1128/JB.01897-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapébie P, Lombard V, Drula E, Terrapon N, Henrissat B. 2019. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat Commun 10:2043. doi: 10.1038/s41467-019-10068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinnan R, Baath E. 2009. Differential utilization of carbon substrates by bacteria and fungi in tundra soil. Appl Environ Microbiol 75:3611–3620. doi: 10.1128/AEM.02865-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monard C, Binet F, Vandenkoornhuyse P. 2008. Short-term response of soil bacteria to carbon enrichment in different soil microsites. Appl Environ Microbiol 74:5589–5592. doi: 10.1128/AEM.00333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wawrik B, Kerkhof L, Kukor J, Zylstra G. 2005. Effect of different carbon sources on community composition of bacterial enrichments from soil. Appl Environ Microbiol 71:6776–6783. doi: 10.1128/AEM.71.11.6776-6783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zegeye EK, Brislawn CJ, Farris Y, Fansler SJ, Hofmockel KS, Jansson JK, Wright AT, Graham EB, Naylor D, McClure RS, Bernstein HC. 2019. Selection, succession, and stabilization of soil microbial consortia. mSystems 4:e00055-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 31.Hartmann M, Frey B, Mayer J, Mäder P, Widmer F. 2015. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J 9:1177–1194. doi: 10.1038/ismej.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupatini M, Korthals GW, de Hollander M, Janssens TKS, Kuramae EE. 2016. Soil microbiome is more heterogeneous in organic than in conventional farming system. Front Microbiol 7:2064. doi: 10.3389/fmicb.2016.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. 2011. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci U S A 108:14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukami T. 2015. Historical contingency in community assembly: integrating niches. Annu Rev Ecol Evol Syst 46:1–23. doi: 10.1146/annurev-ecolsys-110411-160340. [DOI] [Google Scholar]
- 35.Wang X, Sharp CE, Jones GM, Grasby SE, Brady AL, Dunfield PF. 2015. Stable-isotope probing identifies uncultured planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl Environ Microbiol 81:4607–4615. doi: 10.1128/AEM.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlos C, Fan H, Currie CR. 2018. Substrate shift reveals roles for members of bacterial consortia in degradation of plant cell wall polymers. Front Microbiol 9:364. doi: 10.3389/fmicb.2018.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396. doi: 10.1128/aem.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rath KM, Fierer N, Murphy DV, Rousk J. 2019. Linking bacterial community composition to soil salinity along environmental gradients. ISME J 13:836–846. doi: 10.1038/s41396-018-0313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K, Shi Y, Cui X, Yue P, Li K, Liu X, Tripathi BM, Chu H. 2019. Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems 4:e00225-18. doi: 10.1128/mSystems.00225-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howe A, Yang F, Williams RJ, Meyer F, Hofmockel KS. 2016. Identification of the core set of carbon-associated genes in a bioenergy grassland soil. PLoS One 11:e0166578. doi: 10.1371/journal.pone.0166578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta A, Dutta A, Sarkar J, Panigrahi MK, Sar P. 2018. Low-abundance members of the Firmicutes facilitate bioremediation of soil impacted by highly acidic mine drainage from the Malanjkhand Copper Project, India. Front Microbiol 9:2882. doi: 10.3389/fmicb.2018.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galperin MY. 2013. Genome diversity of spore-forming Firmicutes. Microbiol Spectr 1. doi: 10.1128/microbiolspectrum.TBS-0015-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartman WH, Ye R, Horwath WR, Tringe SG. 2017. A genomic perspective on stoichiometric regulation of soil carbon cycling. ISME J 11:2652–2665. doi: 10.1038/ismej.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cernava T, Erlacher A, Aschenbrenner IA, Krug L, Lassek C, Riedel K, Grube M, Berg G. 2017. Deciphering functional diversification within the lichen microbiota by meta-omics. Microbiome 5:82. doi: 10.1186/s40168-017-0303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manor O, Borenstein E. 2017. Systematic characterization and analysis of the taxonomic drivers of functional shifts in the human microbiome. Cell Host Microbe 21:254–267. doi: 10.1016/j.chom.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Méndez C, Salas JA. 2001. The role of ABC transporters in antibiotic-producing organisms: drug secretion and resistance mechanisms. Res Microbiol 152:341–350. doi: 10.1016/s0923-2508(01)01205-0. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto S, Nakano S, Owari K, Fuziwara K, Ogawa N, Otaka M, Tamaki K, Watanabe S, Komatsuda A, Wakui H, Sawada K, Kubota H, Itoh H. 2010. Gentamicin inhibits HSP70-assisted protein folding by interfering with substrate recognition. FEBS Lett 584:645–651. doi: 10.1016/j.febslet.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 48.Matsuzawa T, Kimura N, Suenaga H, Yaoi K. 2016. Screening, identification, and characterization of α-xylosidase from a soil metagenome. J Biosci Bioeng 122:393–399. doi: 10.1016/j.jbiosc.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 49.Jacquiod S, Franqueville L, Cécillon S, M Vogel T, Simonet P. 2013. Soil bacterial community shifts after chitin enrichment: an integrative metagenomic approach. PLoS One 8:e79699. doi: 10.1371/journal.pone.0079699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Van Nostrand JD, Yang Y, He Z, Wu L, Stahl DA, Hazen TC, Tiedje JM, Arkin AP. 2014. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci U S A 111:E836–E845. doi: 10.1073/pnas.1324044111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lueders T, Kindler R, Miltner A, Friedrich MW, Kaestner M. 2006. Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl Environ Microbiol 72:5342–5348. doi: 10.1128/AEM.00400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Xia B, Treves DS, Wu L-Y, Marsh TL, O’Neill RV, Palumbo AV, Tiedje JM. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol 68:326–334. doi: 10.1128/aem.68.1.326-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown J, Zavoshy N, Brislawn CJ, McCue LA. 2020. Hundo: a Snakemake workflow for microbial community sequence data. PeerJ Prepr 6:e27272v1. [Google Scholar]
- 54.R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 55.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oksanen JF, Blanchet Fg, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymas P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: Community Ecology Package. R Development Core Team, Vienna, Austria. [Google Scholar]
- 57.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 58.de Mendiburu F. 2019. agricolae: statistical procedures for agricultural research. https://CRAN.R-project.org/package=agricolae. [Google Scholar]
- 59.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 60.Roberts DW. 2019. labdsv: ordination and multivariate analysis for ecology. https://CRAN.R-project.org/package=labdsv. [Google Scholar]
- 61.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH, UniProt Consortium. 2015. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolde R. 2019. pheatmap: pretty heatmaps. R package version 1.0.12 https://CRAN.R-project.org/package=pheatmap. [Google Scholar]
- 67.Darzi Y, Letunic I, Bork P, Yamada T. 2018. iPath3.0: interactive pathways explorer v3. Nucleic Acids Res 46:W510–W513. doi: 10.1093/nar/gky299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. 2007. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol 5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PERMANOVA of weighted and unweighted UniFrac distances for full dataset and stress dataset. In full dataset (excluding native soil controls), PERMANOVA analyses were run to determine the significance of factors “category” and “replicate” and of “modulename” and “replicate” (note that these analyses were run separately due to the interdependence of “category” and “modulename”), in explaining the variation in beta-diversity (weighted and unweighted UniFrac distances). In the stress dataset, analyses were run to determine the significance of “stress,” “substrate,” “replicate,” and the interaction term “stress:substrate” or of “modulename” and “replicate” (again, analyses were run separately due to interdependence). Analyses were run using the “adonis” function within the vegan package in R. Download Table S1, PDF file, 0.04 MB (46.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ANOVA of alpha-diversity metrics (Shannon’s diversity and richness) and beta-dispersion across the full dataset with Tukey’s post hoc tests. ANOVA analyses for Shannon’s diversity, richness, and beta-dispersion across full dataset for significance of the “factors” category (soil control, simple substrates, antibiotics, polysaccharides, anaerobic, or stress modules) and “replicate,” and by individual “modulename” (the specific module) and “replicate.” These factors have to be run in separate ANOVA analyses due to the interdependence of factors “category” and “modulename”; furthermore, separate analyses were run for the full dataset where soils were either included or excluded (as indicated below) to examine the influence of the control soils on alpha-diversity metrics. (Control soils refer to liquid soil extract controls and not native soils). ANOVA calculations used the “aov” function in the stats package in R. The “betadisper” function within the vegan package was used for calculation of beta-dispersion values. Tukey’s for alpha-diversity and beta-dispersion on full dataset by “category” and “modulename.” Tukey’s results for Shannon’s diversity/richness/beta-dispersion were done for the full dataset (or full dataset excluding soil controls) by “category” or by “modulename.” Generation of Tukey’s values was accomplished using the “HSD.test” function as part of the agricolae package in R. Native soils are not included in this set of analyses. Download Table S2, PDF file, 0.1 MB (89.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ANOVA of alpha-diversity metrics and beta-dispersion for stress subset with Tukey’s post hoc tests. ANOVA analyses for Shannon’s diversity, richness, and beta-dispersion across stress subset including the factors “stress” (which of the eight stresses or control were considered), “substrate” (which of the three substrates in this dataset), “replicate,” and the interaction term “stress:substrate,” or by individual “modulename” (the specific stress × substrate module) and “replicate.” These factors have to be run in separate ANOVA analyses due to the interdependence of factors “stress/substrate” and “modulename.” ANOVA calculations used the “aov”’ function in the stats package in R. Tukey’s calculations used the “HSD.test”’ function as part of the agricolae package in R. Note that this subset does not include control soils, so as to focus on specific comparisons between stresses and the substrates used within this subset. Download Table S3, PDF file, 0.1 MB (71.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Ordination plots for stress samples for weighted and unweighted UniFrac distances, segregated by substrate. Principal coordinate analysis of weighted UniFrac distance (a, c, and e) or unweighted UniFrac distance (b, d, and f) for samples within the “stress” subset. Substrate is delineated by shape: ●, N-acetylglucosamine; ▲, indicates xylose; and ■, xylan. Substrates are further separated into individual plots for N-acetylglucosamine (a and b), xylose (c and d), and xylan (e and f). Colors indicate the specific stress imposed (gray, control; blue, 2,4-D; green, late harvesting; yellow, PEG [osmotic stress]; light yellow, pH 8; orange, constant light; magenta, 10%C [dilute carbon]; purple, heat stress; and black, salt stress). Download FIG S1, PDF file, 0.04 MB (41.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Indicator species analysis (set to “genus” level) for full dataset. Analysis was conducted using the “indval” function within the labdsv package in R using default parameters. Data generated include “indval” (indicator value; this value is a product of relative abundance and frequency, and serves as a measure of exclusivity for a given taxon within a group), “P value” (significance of the aforementioned indicator value, and “freq” (relative frequency of the taxon across all replicates for a module). Only indicator genera with a P value of <0.05 were retained. Download Table S4, PDF file, 0.1 MB (88.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
PERMANOVA of Bray-Curtis distances for 16S and RNA-Seq datasets. PERMANOVA is performed specifically for the 15 samples used for RNA-Seq and the corresponding 16S data. Significance of experimental factors “modulename” and “replicate” were calculated using the “adonis” function within the R vegan package. Download Table S5, PDF file, 0.05 MB (50.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Taxonomic and functional abundance trends for samples used in RNA-Seq analysis. (a and b) Functional trends for RNA-Seq dataset, KEGG category and subcategory level. DESeq-normalized counts for the RNA-Seq database were annotated with KEGG categories and subcategories through the use of the EggNOG database. Transcripts that were successfully annotated were retained, and the relative abundances for transcripts were plotted for the top 20 most abundant KEGG categories (a) and top 30 subcategories (b). In the event that a transcript matched to multiple KEGG categories or subcategories, transcript counts were evenly divided among them. Download FIG S2, PDF file, 0.02 MB (16.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Enriched or depleted KEGG categories or subcategories in a given module relative to at least two of four remaining modules. The Fisher exact test was used to confirm significance of KEGG categories/subcategories in enriched transcripts relative to their proportion in the overall annotated dataset. Download Table S6, PDF file, 0.1 MB (54.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of significantly enriched transcripts in gentamicin modules (“GlucGent”) relative to all four other modules (renamed “OtherSubstrates”). The function “results” from the DESeq2 package was used on the DESeq2 data object for transcriptomic data to determine enrichment patterns for “GlucGent” versus “OtherSubstrate” modules. Transcripts that had a significant (adjusted P value < 0.05) log2FoldChange value of greater than 1 or less than −1 were retained. KEGG and UniRef annotations (including taxonomic information) were then added for transcripts that had a match to one or both databases. The results are sorted by the Log2FoldChange to determine which transcripts were most consistently highly enriched (log2FoldChange > 1) or depleted (log2FoldChange < −1) in gentamicin. Note that transcripts with more than one KEGG hit have only the first hit listed. Download Table S7, PDF file, 0.6 MB (664.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of significantly enriched transcripts in polysaccharide modules (“xylan” and “pectin”) relative to simple substrate (“xylose” and “NAG”) modules. The “GlucGent” module was excluded from this analysis. The function “results” from the DESeq2 package was used on the DESeq2 data object for transcriptomic data to determine enrichment patterns for “polysaccharides” versus “SimpleSubstrate” modules, where the six replicates for each (e.g., 3 xylan + 3 pectin, 3 xylose + 3 NAG) were pooled for said comparison. Transcripts that had a significant (adjusted P value < 0.05) log2FoldChange value greater than 1 or less than −1 were retained. KEGG and UniRef annotations (including taxonomic information) were then added for transcripts that had a match to one or both databases. The results are sorted by the average Log2FoldChange to determine which transcripts were most consistently highly enriched (log2FoldChange > 1) or depleted (log2FoldChange < −1) in polysaccharides. Note that transcripts with more than one KEGG hit have only the first hit listed. Download Table S8, PDF file, 0.8 MB (857.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Raw 16S amplicon sequence data are deposited in the NCBI Short Read Archive (SRA) under BioProject PRJNA594403 and BioSample numbers SAMN13560171 to SAMN13560497. For RNA-Seq data, the soil metagenome used for transcript alignment is publicly available at BioProject PRJNA597444, BioSample SAMN13675441, accession number WUUC00000000. RNA-Seq data have been deposited in NCBI’s Gene Expression Omnibus (70) and are available through GEO Series accession number GSE143587. All codes used to generate statistical inferences within the manuscript are publicly available at https://github.com/dtnaylor124/FunctionalModules.