Abstract
RNA editing plays a key role in organelle gene expression. Little is known about how RNA editing factors influence soybean plant development. Here, we report the isolation and characterization of a soybean yl (yellow leaf) mutant. The yl plants showed decreased chlorophyll accumulation, lower PS II activity, an impaired net photosynthesis rate, and an altered chloroplast ultrastructure. Fine mapping of YL uncovered a point mutation in Glyma.20G187000, which encodes a chloroplast-localized protein homologous to Arabidopsis thaliana (Arabidopsis) ORRM1. YL is mainly expressed in trifoliate leaves, and its deficiency affects the editing of multiple chloroplast RNA sites, leading to inferior photosynthesis in soybean. Taken together, these results demonstrate the importance of the soybean YL protein in chloroplast RNA editing and photosynthesis.
Keywords: soybean, yellow leaf mutant, photosynthesis, chloroplast RNA editing
1. Introduction
Photosynthesis is a vital process in which plants convert light energy into chemical energy [1,2]. In higher plants, chlorophyll a and b are the two major pigments located in the thylakoid membrane of photosynthetic organisms [2]. These chlorophyll molecules play essential roles in harvesting light energy and transferring that energy to reaction centers of the photosystem [2,3]. In the past few decades, tremendous research has been conducted on chlorophyll-deficient mutants in model plants, but the molecular genetic mechanisms underlying these soybean chlorophyll-deficiency mutations are not well understood.
For soybean, 15 chlorophyll-deficient mutant genes were reported to be mapped to chromosomes [4]. Of these genes, six lethal yellow mutant genes, including Y11, Y18/Y18_1, Y18_2, YL_PR350, PsbP, and CD-5, were mapped to chromosomes 13, 14, 17, 15, 3 and 15, respectively [5,6,7,8,9]. The other nine viable yellow mutant genes, including Y9, Y10, Y12, Y13, Y17, Y20, Y23, Tic110, and Cd1, were mapped to chromosomes 15, 3, 6, 13, 15, 12, 13, 2, and 10, respectively [8,10,11,12,13,14,15,16]. However, only the function of Y11 and Y9 genes in yellow foliage was validated by complementary analysis [6,9].
RNA editing is a post-transcriptional modification process that changes the sequence of RNA molecules so that the information in the mature RNA differs from that defined in the genome [17]. In land plants, RNA editing occurs in transcripts of chloroplasts and mitochondria. There are 20–60 chloroplasts and over 300 RNA mitochondrial editing sites in most flowering plants [17,18]. RNA editing is performed by an editosome that is assembled via protein-protein/RNA interactions [19]. Several organelle RNA recognition motif-containing (ORRM) proteins are known to be essential RNA editing factors. ORRM1 controls 62% of chloroplast editing sites in Arabidopsis and 81% of editing sites in maize, with the Zm-orrm1 mutant exhibiting a pale green phenotype [20]. ORRM6 is primarily required for editing psbF-C77 and accD-C794 sites in Arabidopsis chloroplasts [21]. In addition, ORRM2, ORRM3, ORRM4 and ORRM5 are mitochondrial RNA editing factors [22,23,24]. Moreover, pentatricopeptide repeat (PPR) proteins, multiple organellar RNA editing factor (MORF)/RNA editing factor interacting proteins (RIPs), organelle zinc finger 1 (OZ1), protoporphyrinogen oxidase 1 (PPO1), and genomes uncoupled 1 (GUN1) have been identified as components of the plant RNA editosome [19,25,26,27,28,29,30].
Here, we report the characterization of a soybean yellow leaf (yl) mutant with chlorophyll deficiency and impaired photosynthesis. Fine mapping and DNA sequencing showed that YL encodes a GmORRM1. We showed that multiple chloroplast RNA editing sites were changed in yl mutants. Our findings provide new insight into the function of YL in soybean photosynthesis.
2. Results
2.1. yl Plants Show Yellow Leaves and Abnormal Photosynthesis at the Seedling Stage
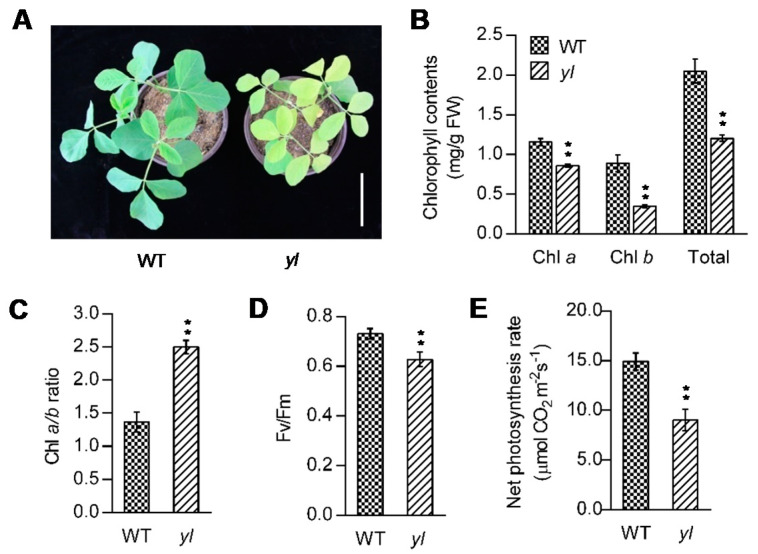
The yl mutant was identified from the soybean cultivar Jindou 23 mutagenized with ethyl methane sulfonate (EMS). The yl mutant plants exhibited conspicuous yellow leaves (Figure 1A). Chlorophyll (Chl) analysis revealed that the Chl a, b and total Chl contents of wild type (Jindou 23) leaves were 1.15 mg/g, 0.89 mg/g and 2.04 mg/g, respectively (Figure 1B). In contrast, the Chl a, b and total Chl contents of yl leaves were 0.86 mg/g, 0.34 mg/g and 1.20 mg/g, respectively (Figure 1B). In addition, the Chl a/b ratio of yl leaves was significantly increased compared to that of the wild type (Figure 1C). To evaluate whether the photosynthesis of yl leaves was affected, we measured the maximum photochemical efficiency of photosystem II (PSII), defined as the Fv/Fm and net photosynthesis rate. The yl leaves displayed obviously decreased Fv/Fm values (wild type, 0.73; yl, 0.62) (Figure 1D). Moreover, the net photosynthesis rate of yl leaves was 9.03 μmol CO2 m−2s−1, which was only 60% that of wild type leaves (14.93 μmol CO2 m−2s−1) (Figure 1E). Therefore, the mutation in yl causes not only a reduction in chlorophyll contents but also abnormalities in photosynthesis.
Figure 1.
Phenotypic characterization of the wild type and yl mutant plants. (A) Two-week-old wild type (left) and yl (right) seedlings. (B) Chlorophyll contents of the third leaf of the wild type (left) and yl (right) plants. (C) The Chl a/b ratio of the wild type (left) and yl (right) leaves. (D) The maximum photochemical efficiency of PSII measured by the Fv/Fm chlorophyll fluorescence ratio in the wild type (left) and yl (right) leaves. (E) The net photosynthesis rate of the wild type (left) and yl (right) plants. Error bars (B–E) represent the mean ± SD (n ≥ 3); ** indicates a significant difference at the 0.01 level. The p values were calculated by Student’s t-test. Bar = 5 cm in (A).
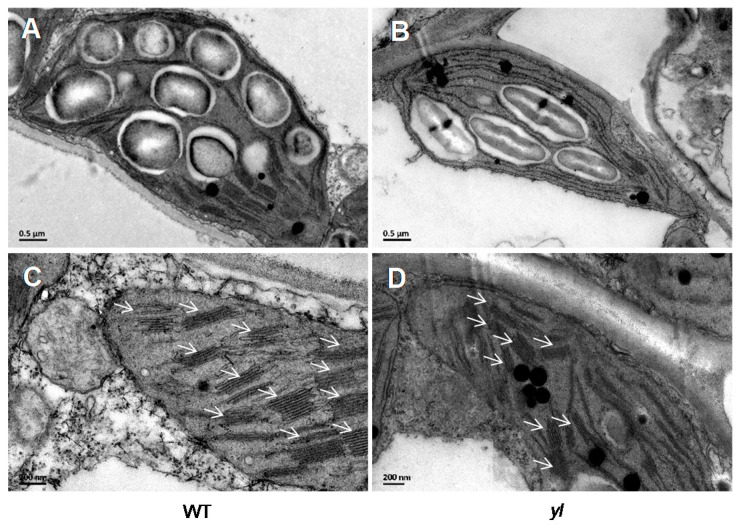
We further performed transmission electron microscopy (TEM) to compare the chloroplast ultrastructure between wild type and yl leaves. No differences in the number of chloroplasts per cell or chloroplast size were detected between the wild type and yl. However, the number of grana (stacked thylakoids) were slightly reduced in the yl chloroplasts (Figure 2A–D), indicating that yl chloroplasts were probably less functional than wild type chloroplasts.
Figure 2.
Chloroplast ultrastructure in the wild type and yl leaves. (A,B) Transmission electron micrographs (TEM) of chloroplasts in the wild type (A) and yl (B) leaves. (C,D) Magnified images of a chloroplast in the wild type (C) and yl (D) leaves. Arrows show grana. Bar = 0.5 μm in (A,B) and 200 nm in (C,D).
2.2. Fine Mapping of the YL Gene
To unravel the molecular mechanism of the yl phenotype, we performed genetic mapping to isolate the YL gene. The yl mutant was crossed to two soybean cultivars with normal green leaves, Williams 82 and Zhonghuang 13, and three F2 mapping populations were produced by reciprocal crosses, yl × Williams 82, yl × Zhonghuang 13 and Zhonghuang 13 × yl. The leaf color of all F1 plants was normal green. The segregation ratio of green to yellow leaves appeared to be 3:1 in F2 populations, indicating that the yl phenotype is controlled by a single recessive nuclear gene (Table 1).
Table 1.
Segregation pattern and chi-square tests for green/yellow leaves of F2 progeny from the crosses between yl and Williams 82 or Zhonghuang 13.
| Crosses | F1 | Number of F2 Plants | χ2 (3:1) | P | ||
|---|---|---|---|---|---|---|
| Wild Type | Mutant | Total | ||||
| yl × Williams 82 | Normal | 679 | 227 | 906 | 0 | >0.9 |
| yl × Zhonghuang13 | Normal | 440 | 163 | 603 | 1.221 | 0.50~0.25 |
| Zhonghuang13 × yl | Normal | 448 | 132 | 580 | 1.437 | 0.25~0.10 |
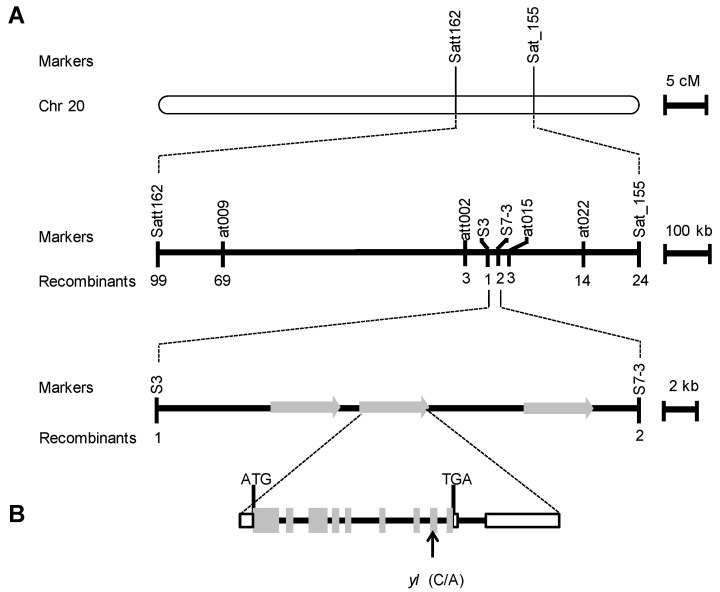
The yl mutation was primarily mapped on chromosome 20 between microsatellite markers Satt162 and Sat_155, which were 9.9 centimorgans (cM) and 1.1 cM from yl, respectively (Figure 3A). The YL locus was ultimately fine mapped into a 28-kb interval between single nucleotide polymorphism (SNP) markers S3 and S7-3 (Figure 3A). According to the soybean gene annotation database (www.phytozome.net) [31], there were three putative open reading frames (ORFs) within this 28-kb region (Figure 3A). We sequenced the 28-kb sequences between the wild type and yl mutant and found a C to A transition at the eighth exon of Glyma.20G187000 (Figure 3B).
Figure 3.
Fine mapping of the YL gene. (A) The YL locus was initially mapped to a region between markers Satt162 and Sat_155 on soybean chromosome 20. The gene was finally delimited to a 28 kb region between markers S3 and S7-3. Three predicted open reading frames (ORFs) were within this region. (B) YL (Glyma.20G187000) structure indicating nine exons (gray boxes), eight introns (line segments between the exons), and 5′ and 3′ untranslated regions (white boxes with black frame). Start (ATG) and stop (TGA) codons are marked. The yl mutation in the YL gene is shown.
2.3. YL Encodes an Organelle RNA Recognition Motif-Containing Protein 1 (GmORRM1)
The YL gene (Glyma.20G187000) encodes a protein of 390 amino acids with a C-terminal RNA recognition motif (Figure 4A). The yl mutation occurred in the RNA recognition motif and resulted in the substitution of an alanine with a glutamic acid (Figure 4A). The alignment of amino acid sequences homologous to YL from several plant species showed that this alanine residue in the RNA recognition motif was highly conserved (Figure 4B). In addition, the YL sequence shared 62.3% similarity with Arabidopsis ORRM1 (AtORRM1, At3G20930) and 58.2% similarity with maize ORRM1 (ZmORRM1, GRMZM5G899787), suggesting that YL (GmORRM1) and its homologs might have conserved functions.
Figure 4.
The YL protein contains an RNA recognition motif. (A) Amino acid sequence of YL. Position of the yl (asterisk) mutation is exhibited. The RNA recognition motif is underlined. (B) Partial alignment of the amino acid sequences of YL and its homologous proteins. The aligned sequences are from Glycine max (YL, Glyma.20G187000), Arabidopsis thaliana (AtORRM1, At3G20930), Zea mays (ZmORRM1, GRMZM5G899787), Oryza sativa (Os02g49820), Medicago truncatula (Medtr1g092860) and Phaseolus vulgaris (Phvul.007G103400). The red rectangle represents the conserved alanine residue in the RNA recognition motif.
2.4. Expression Pattern and Protein Subcellular Localization of YL
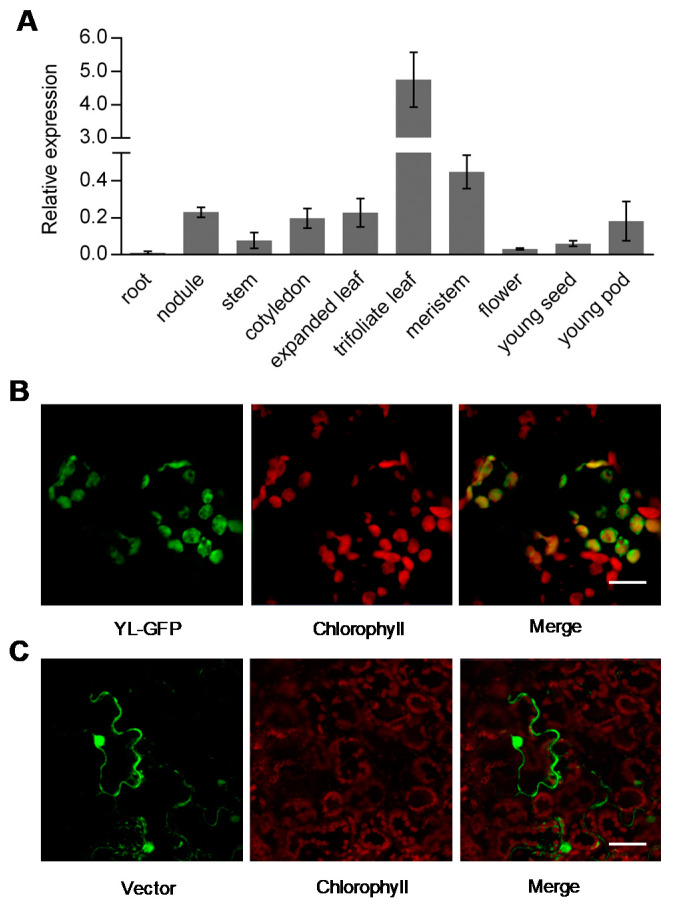
We performed quantitative real-time RT-PCR (qRT-PCR) to examine the expression of YL among various tissues. YL was expressed in all tissues tested, with the highest expression in trifoliate leaves, intermediate expression in meristem, nodules, expanded leaves, cotyledons, and young pods and weak expression in roots, stems, flowers, and young seeds (Figure 5A). The expression pattern of YL was consistent with the function of YL in affecting leaf color.
Figure 5.
Expression pattern and subcellular localization of YL. (A) Relative expression levels of the YL gene in various soybean tissues. The real-time RT-PCR assays were performed in three biological replicates. qRT-PCR values for YL are normalized with the actin and ATP synthase genes. Error bars represent the SD. (B,C) Subcellular localization of the YL-GFP fusion protein (B) and GFP (C) in epidermal cells of Nicotiana benthamiana leaves. Bar = 20 µm in (B) and 50 µm in (C).
Using the Predotar and TargetP programs, the YL protein was predicted to localize in chloroplasts [32,33]. To obtain experimental verification of this localization, we expressed a YL-GFP fusion protein under the 35S promoter in Nicotiana benthamiana leaves. The green fluorescence signals of the YL-GFP fusion protein were observed in the chloroplasts of epidermal cells (Figure 5B,C).
2.5. Dramatic Defects of Chloroplast RNA Editing in yl
To further understand the function of YL, we performed DNA resequencing and RNA sequencing (RNA-seq) for the wild type to screen out soybean chloroplast editing sites, obtaining 44 predicted sites from 22 chloroplast transcripts (Table 2, Tables S1 and S2). In addition, the rpl23-89 site was identified through comparative analyses with Arabidopsis chloroplast editing sites (Table 2). Most of these sites were verified by direct sequencing of PCR products of transcripts or the corresponding genomic DNA carrying them (Table S2). Among the 45 editing sites, 44 sites were C-to-U conversions, 43 sites were in the coding regions of genes, and most of them caused alteration of the encoded amino acids (Table 2).
Table 2.
RNA editing sites in soybean chloroplast transcripts.
| Gene | Position a | Codon Site b | Conversion |
|---|---|---|---|
| accD | 617 | 2 | S(uCg)→L(uUg) |
| atpF | 92 | 2 | P(cCa)→L(cUa) |
| clpP | 559 | 1 | H(Cau)→Y(Uau) |
| ndhA | 341 | 2 | S(uCa)→L(uUa) |
| 1073 | 2 | S(uCu)→F(uUu) | |
| ndhB | 9 | 3 | W(ugG)→stop codon (ugA) |
| 149 | 2 | S(uCa)→L(uUa) | |
| 542 | 2 | T(aCg)→M(aUg) | |
| 586 | 1 | H(Cau)→Y(Uau) | |
| 737 | 2 | P(cCa)→L(cUa) | |
| 746 | 2 | S(uCu)→F(uUu) | |
| 830 | 2 | S(uCa)→L(uUa) | |
| 836 | 2 | S(uCa)→L(uUa) | |
| 1112 | 2 | S(uCa)→L(uUa) | |
| 1255 | 1 | H(Cau)→Y(Uau) | |
| 1481 | 2 | P(cCa)→L(cUa) | |
| ndhC | 323 | 2 | S(uCa)→L(uUa) |
| ndhD | 2 | 2 | T(aCg)→M(aUg) |
| 383 | 2 | T(aCa)→I(aUa) | |
| 674 | 2 | S(uCa)→L(uUa) | |
| 878 | 2 | S(uCa)→L(uUa) | |
| 1298 | 2 | S(uCa)→L(uUa) | |
| ndhE | 233 | 2 | P(cCg)→L(cUg) |
| ndhF | 290 | 2 | S(uCa)→L(uUa) |
| petB | 611 | 2 | S(uCa)→L(uUa) |
| psaI | 79 | 1 | H(Cau)→Y(Uau) |
| psbE | 214 | 2 | S(uCc)→F(uUc) |
| psbF | 6 | 3 | T(acC)→T(acU) |
| 77 | 2 | S(uCu)→F(uUu) | |
| psbL | 2 | 2 | T(aCg)→M(aUg) |
| rpl23 | 89 | 2 | S(uCa)→L(uUa) |
| rpoA | 200 | 2 | S(uCa)→L(uUa) |
| rpoB | 338 | 2 | S(uCu)→F(uUu) |
| 551 | 2 | S(uCa)→L(uUa) | |
| 566 | 2 | S(uCg)→L(uUg) | |
| 2000 | 2 | S(uCu)→F(uUu) | |
| rpoC1 | 41 | 2 | S(uCa)→L(uUa) |
| 488 | 2 | S(uCa)→L(uUa) | |
| rps2 | 134 | 2 | T(aCa)→I(aUa) |
| 248 | 2 | S(uCa)→L(uUa) | |
| rps12 | 554 c | ||
| rps14 | 80 | 2 | S(uCa)→L(uUa) |
| rps16 | 499 c | ||
| 212 | 2 | S(uCa)→L(uUa) | |
| rps18 | 221 | 2 | S(uCg)→L(uUg) |
a Position is given with respect to the initiation codon of each chloroplast transcript. b Codon site is the order in the amino acid codon. c RNA editing sites are in introns of the chloroplast gene. The position here is given with respect to the initiation codon of each gene.
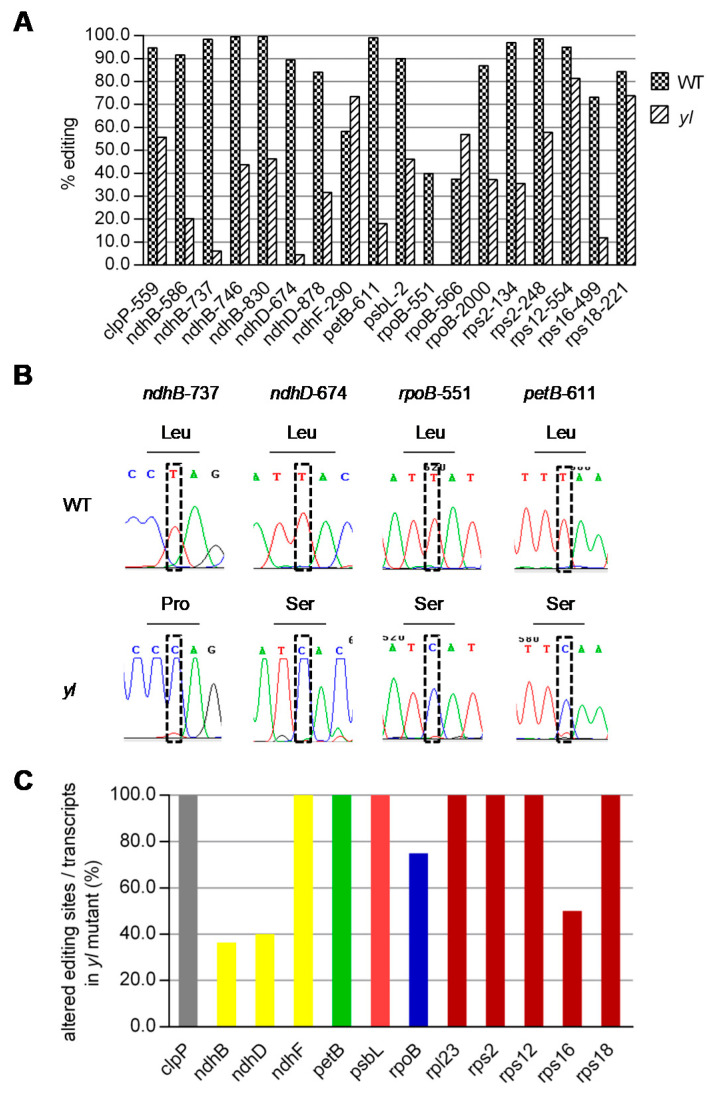
The RNA-seq method was combined with direct sequencing of PCR products of transcripts carrying chloroplast RNA editing sites to compare the RNA editing between wild type and yl leaves. The results showed that the editing was completely abolished for ndhB-737, ndhD-674 and rpoB-551 in the yl mutant (Figure 6A,B, Tables S1 and S2). These deficiencies caused changes in the encoded amino acid residues from Leu, Leu and Leu in the wild type to Pro, Ser and Ser in the yl mutant, respectively. In addition, the editing of 14 sites was decreased by 10% to 90% in the yl mutant (Figure 6A, Tables S1 and S2). However, the ndhF-290 and rpoB-566 sites exhibited higher editing levels in the yl mutant compared with the wild type (Figure 6A, Tables S1 and S2).
Figure 6.
RNA editing at multiple chloroplast sites is impaired in the yl mutant. (A) Eighteen sites exhibit a significant alteration in editing of more than 10% in the yl mutant through RNA-seq analysis. (B) The three abolished editing sites and petB-611 in the yl mutant. Rectangular frames indicate defective editing sites. The corresponding amino acids are underlined. (C) Percentage of altered editing sites/transcripts in the yl mutant. Each bar shows a transcript color-coded in accordance with the complex to which it belongs.
We then investigated the distribution of the affected editing sites in the yl mutant. The 19 affected editing sites were distributed in 12 chloroplast transcripts encoding components of the Clp protease proteolytic subunit, NDH complex, cytochrome b6f complex, PSII complex, RNA polymerase or ribosomal proteins. As shown in Figure 6C, the percentage of altered editing sites per transcript varied from 36.4% to 100%, suggesting that the effect of the YL mutation on editing was site specific but not transcript specific.
3. Discussion
In plants, characterization of yellow foliar mutants will present an interesting opportunity to understand the complex photosynthesis process. The yl mutant reported here showed reduced Chl a, b and total Chl contents (Figure 1B). Our study identified a point mutation in the YL gene responsible for the mutant phenotype (Figure 3B). In addition, we discovered decreased PSII activity and a decreased net photosynthesis rate of yl leaves (Figure 1D,E). These results reveal that YL plays an important role in the photosynthetic process. This conclusion is supported by two pieces of evidence: (1) Strong expression of YL was detected in the trifoliate leaves; (2) The yl mutant lacked stacked thylakoids in the chloroplast, where photosynthesis takes place. The YL protein was homologous to maize ORRM1. In addition, the Mu transposon insertional mutant in ZmORRM1 showed defects in the major photosynthetic enzyme complexes [20]. However, it is not known whether YL could influence similar photosynthetic proteins.
Based on bioinformatic analysis and protein subcellular localization experiments, we conclude that YL encodes a 390-amino-acid chloroplast-localized protein (Figure 4A and Figure 5B,C). Identification of the C-terminal RNA recognition motif of YL and the high level of homology with AtORRM1 and ZmORRM1 strongly suggest that YL functions in RNA editing. ORRM1 controlled the extent of editing in 62% of the chloroplast sites in Arabidopsis and 81% of sites in maize [20]. In our study, YL was required for the editing of 42% of chloroplast sites in 12 chloroplast transcripts (Figure 6A, Tables S1 and S2). Moreover, we compared RNA editing alterations in soybean, Arabidopsis and maize orrm1 mutants. Notably, the three completely lost editing sites, ndhB-737, ndhD-674 and rpoB-551, in the yl mutant also exhibited a pronounced reduction in editing in Arabidopsis or maize orrm1 mutants (Figure 6A,B, Tables S1 and S2). In contrast, the editing of ndhF-290 exhibited no change in the Arabidopsis orrm1 mutant, whereas we observed a slight increase in the editing of ndhF-290 in the yl mutant (Figure 6A, Table S1). Consequently, it seems that ORRM1 showed species-specific functions in plants. These results suggest that YL may be involved in chloroplast RNA editing.
Twelve chloroplast transcripts, which encode components of the Clp protease proteolytic subunit, NDH complex, cytochrome b6f complex, PSII complex, RNA polymerase or ribosomal proteins, could not be edited in the yl mutant (Figure 6C). The RNA editing defect in transcripts encoding the components of the Clp protease proteolytic subunit, NDH complex, and RNA polymerase or ribosomal proteins may not be the main reason for the yl mutant phenotype. Although the Arabidopsis orrm1 mutant exhibited severe editing defects in transcripts encoding these proteins, it did not show any phenotypic deficiencies [20]. In addition, partial knockout of clpP in tobacco resulted in an asymmetric, slender leaf shape but normal leaf color [34]. Moreover, no obvious phenotype was observed in tobacco mutants with disrupted NDH complexes [35].
The reduction in petB-611 and psbL-2 editing may have large contributions to the yl phenotype. The petB gene encodes cytochrome b6, which is one of the major subunits of the cytochrome b6f complex mediating electron transfer between PSII and I [36,37,38]. In the yl mutant, defective editing at petB-611 resulted in a change from the wild type residue at position 204 (Leu) to Ser (Figure 6B). The Leu204 residue belongs to the D helix span, which is involved in heme binding [37,39]. Leu204 to Ser204 alteration likely disrupts the assembly of cytochrome b6f complexes, as suggested by the behavior of petB mutants in Chlamydomonas reinhardtii [37]. The yl mutant is phenotypically similar to a tobacco petB mutant in which the portion of the petB coding region was replaced with an aadA cassette [38]. Both yl and petB mutants display yellow (pale green) leaf color and reduced chlorophyll contents and grana stacks. The psbL gene encodes a conserved low molecular weight protein of PS II [40]. In wild type soybean, the initiator codon (AUG) of psbL is formed by a C to U editing of the ACG codon. In the present yl mutant, defective editing at psbL-2 may partially influence the formation of the initiator codon (AUG). The lack of PsbL in tobacco has been implicated as impairing the assembly of PSII [41,42]. The reduced Fv/Fm in the yl mutant is similar to the behavior of Arabidopsis mutants deficient in RNA editing sites in plastid transcripts encoding PSII proteins, including orrm6 mutants [21].
The cytochrome b6f and PSII complexes belong to the main components of the photosynthetic electron transfer chain [2]. The yl mutant, exhibiting RNA editing deficiency at petB-611 and psbL-2, may be unable to assemble functional cytochrome b6f and PSII complexes. Since the localization of cytochrome b6f and PSII complexes is predominantly in the grana regions of the thylakoid membrane system, this inference may account for lacked grana in yl chloroplast. In soybean, some mutants with defects in photosynthetic electron carrier proteins display a yellow phenotype [4,8]. For instance, the mutation in the PSII extrinsic protein GmpsbP leads to a lethal-yellow phenotype, extremely low Fv/Fm, and failure of proplastid differentiation into normal chloroplasts with grana [8]. The yl mutant showed reduced Fv/Fm, indicating that the yl mutant cannot utilize the absorbed light in photochemistry as effectively as the wild type. When more light energy is absorbed than is converted in plants, the photosynthetic organism is subjected to photooxidative stress, known as photoinhibition, leading to pigment bleaching, inactivation of electron transport and damage of the reaction center [43,44]. A recent study has shown that a leaf yellowing mutant phenotype in soybean may be largely due to the abnormal light absorption in the photosynthesis process [45]. Thus, photoinhibition may also be the cause of decreased Chl contents and net photosynthesis rate in the yl mutant.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Seeds of soybean cultivar Jindou 23 (wildtype) were mutagenized with ethyl methane sulfonate (EMS). The phenotypes of the second mutant generation (M2 progeny) plants were observed and the yl mutant with yellow foliage was identified from the M2 progeny. The yl mutant has been continuously self-pollinated and selected based on the yellow foliar phenotype. For phenotype characterization of the yl mutant, the simultaneous field trial consisted of three replicates per genotype, planted in 0.50 m row spacing and 0.10 m spacing in the rows. To analyze the inheritance pattern of the yl mutant and fine-mapping of the YL gene, three F2 populations were generated from reciprocal crosses between the yl mutant and two soybean cultivars, Williams 82 and Zhonghuang 13 (yl × Williams 82, yl × Zhonghuang 13 and Zhonghuang 13 × yl). All soybean materials were planted in the experimental field at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (Beijing, China), during the natural growing seasons (May to October). The common meteorological conditions during soybean growing seasons in Beijing were as follows: (1) mean daily temperatures: about 24 °C. (2) overall precipitation: 400 ~ 600 mm.
4.2. Chl Contents, Net Photosynthetic Rate and Photochemical Efficiency Analysis
The third true leaves from the top of five-week-old soybean plants were used to estimate Chl contents, net photosynthetic rate and photochemical efficiency. The total Chl, Chl a and Chl b contents were determined as previously described [46]. Fresh leaves (200 mg) were immersed into 10 mL of 95% ethanol for pigments extraction under dark conditions. Then the absorbance of supernatant was quantified spectrophotometrically at 665 and 649 nm. Chlorophyll contents were calculated using the following formulas [46]: Chl a (mg/g) = [(13.95OD665 − 6.88OD649) × 10]/(tissue fresh weight (g) × 1000); Chl b (mg/g) = [(24.96OD649 − 7.32OD665) × 10]/(tissue fresh weight (g) × 1000); Total Chl (mg/g) = Chl a (mg/g) + Chl b (mg/g). Net photosynthetic rate was measured using an Li-6400 instrument (LI-COR, Lincoln, NE, USA) under a CO2 concentration of 250 μmol mol−1 and 1000 μmol m−2s−1 photosynthetically active radiation (PAR) between 9:00 and 11:00 in the morning. The maximum photochemical efficiency of photosystem II was measured as the Fv/Fm using an IMAGING-PAM Chlorophyll Fluorometer (Heinz Walz, Effeltrich, Germany).
Three to eight individual plants were examined for each material and the examination were repeated at least three times. The experimental data for each material are reported as mean ± standard deviation (SD). Statistical significance of differences between yl mutant and wildtype plants were tested using independent sample Student’s t test algorithm. All statistical analysis were performed in SPSS Statistics 17.0.
4.3. Transmission Electron Microscopy
The third leaves of 30-day-old wild type and yl plants were collected and fixed for at least 2 h at 4 °C in 0.1 M phosphate buffer, pH 7.4, with 2.5% glutaraldehyde and washed with the same buffer. The samples were fixed in 1% osmium tetroxide for 2 h and then dehydrated through an ethanol gradient, infiltrated and embedded in epoxy resin. Ultra-thin sections were obtained using a diamond knife and mounted on copper grids. Then the grids were stained with uranyl acetate and lead citrate, and examined using a JEM-1400 (JEOL, Tokyo, Japan) transmission electron microscope.
4.4. Fine Mapping of the YL Gene
Three F2 populations generated from reciprocal crosses between the yl mutant and two soybean cultivars, Williams 82 and Zhonghuang 13 (yl × Williams 82, yl × Zhonghuang 13 and Zhonghuang 13 × yl), were used for genetic analysis. In the F2 populations, genomic DNA was isolated from selected etiolated seedlings exhibiting mutant phenotype for gene mapping. The cetyl trimethyl ammonium bromide (CTAB) method was used for genomic DNA extraction from the fresh young trifoliate leaf tissue and for all subsequent DNA extractions using this method unless otherwise stated. Briefly, frozen tissue was powdered and dispersed in 2 × CTAB extraction buffer, and incubated at 65 °C for about 60 min. Chloroform/Tris-phenol, 1:1 (vol/vol), was added and mixed to form an emulsion that was centrifuged for 10 min. The upper aqueous phase was transferred to a new tube, and 2/3 vol of isopropanol was added for DNA precipitation. The extracted DNA was treated with RNase to remove RNA contamination.
For primary mapping, 92 mutant individuals were selected from two F2 populations between yl and Williams 82 or Zhonghuang 13. The markers used for primary mapping were 71 published SSR markers (http://soybase.org/) [47]. A total of 770 mutant individuals selected from the F2 population between the yl mutant and Williams 82 were used for fine mapping. New molecular markers were developed for fine mapping (Table S3). To identify the candidate gene, the corresponding DNA fragments within the fine mapping region were amplified from the wild type and yl mutant using special primers (Table S3) and sequenced.
4.5. RNA Extraction and Quantitative Real-Time RT-PCR
For analysis of the expression of YL in various soybean tissues, total RNA was isolated from root, nodule, stem, cotyledon, expanded leaf, trifoliate leaf, meristem, flower, young seed and pod collected from three individuals. For the analysis of chloroplast RNA editing in the yl mutant and wild type, total RNA was isolated from trifoliate leaves of five-week-old soybean plants. Fresh tissue was frozen immediately in liquid nitrogen and ground to powder using a mortar and pestle. Total RNA was extracted using 1 mL TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. After ethanol precipitation, the RNA was dissolved in RNase-free water.
Approximately 2 μg of total RNA were reverse transcribed using the Prime Script RT reagent Kit with gDNA Eraser (TaKaRa, Beijing, China), according to the manufacturer’s instructions. Quantitative real-time RT-PCR was performed in a 20 μL reaction mix containing 50 ng of cDNA, 1 × Light Cycler 480 SYBR Green I Master (Roche, Mannheim, Germany), 0.5 μM of each primer on a Lightcycler 480 (Roche, Mannheim, Germany) machine using the following PCR profile: 2 min at 94 °C, followed by 40 cycles of 15 s at 94 °C, 15 s at 60 °C, and 30 s at 72 °C. The dissociation curve analysis was conducted to verify the PCR specificity. Three biological replicates with three technical replicates were analyzed to quantify the levels of gene expression. The soybean actin and ATP synthase genes were used as internal standards to normalize the expression of YL using the 2‒ΔCt method. The primers used for quantitative real-time RT-PCR were designed using the Primer Premier 5 software (Premier, San Francisco, CA, USA) and listed in Table S3.
4.6. Subcellular Localization
To determine its subcellular localization, the coding sequence of YL lacking its stop codon was amplified by PCR in a 50 μL volume that contained 200 ng cDNA from wild type, 1 × PCR buffer, 0.4 mM dNTPs, 1 U KOD FX Neo (Toyobo, Osaka, Japan) and 0.3 μM of both forward and reverse primers of YL-GFP listed in Table S3. The sample was heated to 94 °C for 2 min, followed by 45 cycles of denaturation at 94 °C for 15 s, annealing at 61 °C for 30 s, elongation at 68 °C for 1 min. The amplified product was cloned into the pCAMBIA 1302 vector between the cauliflower mosaic virus 35S promoter and the GFP-coding sequence. The construct of YL-GFP vector was sequencing confirmed by Sangon Biotech (Shanghai, China).
For transformation of Agrobacterium, 1 μg purified plasmid DNA was added to competent cells of thawing Agrobacterium tumefaciens strain GV3101 on ice. After ice bath for 30 min, the cell/DNA mix was immersed in liquid nitrogen for 1 min and subsequently incubated at 37 °C for 5 min, then ice bath for 2 min. By adding 900 μL liquid growth medium (no antibiotics), the cell/DNA mix was shocked for at least 120 min at 28 °C. After centrifugation, the cells were resuspended and plated on an agar plate containing kanamycin (50 mg/L) for selection of transformants.
For infiltration of Nicotiana benthamiana, the Agrobacterium tumefaciens strains carrying the YL-GFP, pCAMBIA 1302 and p19 of tomato bushy stunt virus plasmids were grown at 28 °C in liquid growth medium with kanamycin (50 mg/L), rifampicin (50 mg/L) and gentamicin (25 mg/L) until OD600 reached 1.0. The Agrobacterium tumefaciens strains containing the YL-GFP and p19 or pCAMBIA 1302 and p19 plasmids were mixed. After centrifugation of the strain mixtures, the harvested cells were resuspended in 10 mM MES buffer containing 10 mM MgCl2 and 100 mM acetosyringone to a final OD600 of 1.0, followed by incubation at room temperature for 120 min. Strain mixtures were infiltrated into the abaxial surface of leaves of four-week-old Nicotiana benthamiana plants using a 1 mL syringe. The transformed epidermal cells were detected using a Zeiss LSM710 confocal microscope (Carl Zeiss Microscopy GmbH, Jena, Germany).
4.7. Chloroplast RNA Editing Analysis
For chloroplast RNA editing analysis, total DNA and RNA were isolated from wild type and yl leaves as described in Section 4.4 and Section 4.5. Next, the DNA and RNA were submitted to Berry Genomics (Beijing) for DNA resequencing and rRNA-depleted strand-specific RNA-seq, respectively. For RNA-seq analysis, four cDNA libraries (two biological replicates per genotype) were constructed. All sequencing data used in this research were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under BioProject ID PRJNA616185.
Clean reads were quality checked using FastQC (version 0.11.3) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) [48]. After quality control, reads were aligned to the soybean chloroplast genome (ncbi.nlm.nih.gov/nuccore/DQ312375.1) [49] using HISAT (version 2.0.0) [50]. SNP calling was performed by GATK combined with Samtools [51,52]. SNPs with read numbers ≥ 20 from the RNA-seq and the corresponding DNA resequencing data were compared and the different chloroplast allele bases between the two data sets were considered as the candidate RNA editing sites. Because C-to-U editing in plastid and mitochondrial mRNAs appear to be ubiquitous in land plants, the editing efficiency of each editing site was calculated using the following equation: Editing (%) = U/(C + U) × 100. U represents the read number of SNPs that are different from DNAs in plastid mRNAs. C represents the read number of SNPs that are identical as DNAs in the same plastid editing site.
To experimentally validate the RNA editing sites derived from RNA-seq, the genomic and transcript regions surrounding these sites were amplified from another wild type biological replicate and yl mutant biological replicate using KOD FX High-Fidelity DNA polymerase (Toyobo, Osaka, Japan). The PCR products were sequenced and compared to identify SNP changes resulting from RNA editing. The RNA editing extent was estimated by the relative heights of the peaks of the nucleotide in the sequence analyzed. The primer sequences are listed in Table S3.
5. Conclusions
In this study, we characterized one yellow leaf mutant and identified YL, a soybean homolog of Arabidopsis ORRM1 that is involved in RNA editing. The yl mutant displayed yellow leaves, reduced chlorophyll contents, impaired photosynthesis and an altered chloroplast ultrastructure. Through fine mapping, the yl mutation was narrowed down to a 28-kb genomic region in chromosome 20 between markers S3 and S7-3. DNA sequencing revealed that the YL mutation in the yl mutant is a C to A transition in Glyma.20G187000, which causes an amino acid alteration. Further function analysis of YL uncovered that the point mutation in yl influenced the editing extent in 42% of the chloroplast sites surveyed. However, we speculate that only the decreased level of conversion of cytidine to uridine at petB-611 and psbL-2 may affect normal photosynthesis in yl leaves. Together, our findings indicate that soybean YL protein influences photosynthesis, possibly via its function in chloroplast RNA editing.
Acknowledgments
This work is dedicated to the memory of Baoge Zhu, who devoted his life to soybean breeding. We would like to thank Jingnan Liang (Institute of Microbiology, Chinese Academy of Sciences) for assistance with ultrathin sectioning and transmission electron microscopy.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/12/4275/s1. Table S1. Comparison of the extent of RNA editing of chloroplast sites between the wild type and yl mutant by RNA-seq. Table S2. Verification of the extent of chloroplast RNA editing by Sanger sequencing. Table S3. Primers used in this study.
Author Contributions
B.Z., G.Z. and Z.T. conceived and designed the experiments. X.Z. carried out the experiments and wrote the manuscript. Y.P., Z.L., D.Z. and Z.D. assisted with doing the experiments. Y.L. participated in the statistical analysis. B.Z. and G.Z. and Z.T. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2017YFD0101401) and the “Strategic Priority Research Program” of the Chinese Academy of Sciences (Grant No. XDA24030403).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Stirbet A., Lazar D., Guo Y., Govindjee G. Photosynthesis: Basics, history, and modeling. Ann. Bot. 2019;XX:1–27. doi: 10.1093/aob/mcz171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson M.P. Photosynthesis. Essays Biochem. 2016;60:255–273. doi: 10.1042/EBC20160016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Wettstein D., Gough S., Kannangara C.G. Chlorophyll biosynthesis. Plant Cell. 1995;7:1039–1047. doi: 10.2307/3870056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandhu D., Coleman Z., Atkinson T., Rai K.M., Mendu V. Genetics and physiology of the nuclearly inherited yellow foliar mutants in soybean. Front. Plant Sci. 2018;9:471. doi: 10.3389/fpls.2018.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato K.K., Palmer R.G. Duplicate chlorophyll-deficient loci in soybean. Genome. 2014;47:190–198. doi: 10.1139/g03-092. [DOI] [PubMed] [Google Scholar]
- 6.Campbell B.W., Mani D., Curtin S.J., Slattery R.A., Michno J.M., Ort D.R., Schaus P.J., Palmer R.G., Orf J.H., Stupar R.M. Identical substitutions in magnesium chelatase paralogs result in chlorophyll-deficient soybean mutants. G3 Genes Genom. Genet. 2014;5:123–131. doi: 10.1534/g3.114.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed S., Atkinson T., Gorecki C., Espinosa K., Przybylski S., Goggi A.S., Palmer R.G., Sandhu D. Candidate gene identification for a lethal chlorophyll-deficient mutant in soybean. Agronomy. 2014;4:462–469. doi: 10.3390/agronomy4040462. [DOI] [Google Scholar]
- 8.Sandhu D., Atkinson T., Noll A., Johnson C., Espinosa K., Boelter J., Abel S., Dhatt B.K., Barta T., Singsaas E., et al. Soybean proteins GmTic110 and GmPsbP are crucial for chloroplast development and function. Plant Sci. 2016;252:76–87. doi: 10.1016/j.plantsci.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Li Q., Fang C., Duan Z.B., Liu Y.C., Qin H., Zhang J.X., Sun P., Li W.B., Wang G.D., Tian Z.X. Functional conservation and divergence of GmCHLI genes in polyploid soybean. Plant J. 2016;88:584–596. doi: 10.1111/tpj.13282. [DOI] [PubMed] [Google Scholar]
- 10.Weiss M.G. Genetic linkage in soybeans: Linkage group I. Crop Sci. 1970;10:69–72. doi: 10.2135/cropsci1970.0011183X001000010027x. [DOI] [Google Scholar]
- 11.Weiss M.G. Genetic linkage in soybeans: Linkage group VII. Crop Sci. 1970;10:627–629. doi: 10.2135/cropsci1970.0011183X001000060005x. [DOI] [Google Scholar]
- 12.Palmer R.G., Nelson R.L., Bernard R.L., Stelly D.M. Genetics and linkage of three chlorophyll-deficient mutants in soybean: y19, y22, and y23. J. Hered. 1990;81:404–406. [Google Scholar]
- 13.Zou J.J., Singh R.J., Hymowitz T. Association of the yellow leaf (y10) mutant to soybean chromosome 3. J. Hered. 2003;94:352–355. doi: 10.1093/jhered/esg065. [DOI] [PubMed] [Google Scholar]
- 14.Palmer R.G., Xu M. Positioning 3 qualitative trait loci on soybean molecular linkage group E. J. Hered. 2008;99:674–678. doi: 10.1093/jhered/esn070. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H., Zhang D., Han S., Zhang X., Yu D. Identification and gene mapping of a soybean chlorophyll-deficient mutant. Plant Breed. 2011;130:133–138. doi: 10.1111/j.1439-0523.2010.01844.x. [DOI] [Google Scholar]
- 16.Sandhu D., Ghosh J., Johnson C., Baumbach J., Baumert E., Cina T., Grant D., Palmer R.G., Bhattacharyya M.K. The endogenous transposable element Tgm9 is suitable for generating knockout mutants for functional analyses of soybean genes and genetic improvement in soybean. PLoS ONE. 2017;12:e0180732. doi: 10.1371/journal.pone.0180732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takenaka M., Zehrmann A., Verbitskiy D., Hartel B., Brennicke A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013;47:335–352. doi: 10.1146/annurev-genet-111212-133519. [DOI] [PubMed] [Google Scholar]
- 18.Ichinose M., Sugita M. RNA editing and its molecular mechanism in plant organelles. Genes. 2016;8:5. doi: 10.3390/genes8010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun T., Bentolila S., Hanson M.R. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 2016;21:962–973. doi: 10.1016/j.tplants.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Sun T., Germain A., Giloteaux L., Hammani K., Barkan A., Hanson M.R., Bentolila S. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. USA. 2013;110:E1169–E1178. doi: 10.1073/pnas.1220162110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackett J.B., Shi X., Kobylarz A.T., Lucas M.K., Wessendorf R.L., Hines K.M., Bentolila S., Hanson M.R., Lu Y. An organelle RNA recognition motif protein is required for photosystem II subunit psbF transcript editing. Plant Physiol. 2017;173:2278–2293. doi: 10.1104/pp.16.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi X., Hanson M.R., Bentolila S. Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Res. 2015;43:3814–3825. doi: 10.1093/nar/gkv245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi X., Germain A., Hanson M.R., Bentolila S. RNA recognition motif-containing protein ORRM4 broadly affects mitochondrial RNA editing and impacts plant development and flowering. Plant Physiol. 2016;170:294–309. doi: 10.1104/pp.15.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi X., Castandet B., Germain A., Hanson M.R., Bentolila S. ORRM5, an RNA recognition motif-containing protein, has a unique effect on mitochondrial RNA editing. J. Exp. Bot. 2017;68:2833–2847. doi: 10.1093/jxb/erx139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bentolila S., Heller W.P., Sun T., Babina A.M., Friso G., van Wijk K.J., Hanson M.R. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. USA. 2012;109:E1453–E1461. doi: 10.1073/pnas.1121465109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takenaka M., Zehrmann A., Verbitskiy D., Kugelmann M., Hartel B., Brennicke A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA. 2012;109:5104–5109. doi: 10.1073/pnas.1202452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barkan A., Small I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 28.Zhang F., Tang W.J., Hedtke B., Zhong L.L., Liu L., Peng L.W., Lu C.M., Grimm B., Lin R.C. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. USA. 2014;111:2023–2028. doi: 10.1073/pnas.1316183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun T., Shi X., Friso G., Van Wijk K., Bentolila S., Hanson M.R. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 2015;11:e1005028. doi: 10.1371/journal.pgen.1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X.B., Huang J.Y., Chory J. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. USA. 2019;116:10162–10167. doi: 10.1073/pnas.1820426116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phytozome. [(accessed on 14 June 2020)]; Available online: www.phytozome.net.
- 32.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 33.Small I., Peeters N., Legeai F., Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 34.Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 2001;42:264–273. doi: 10.1093/pcp/pce031. [DOI] [PubMed] [Google Scholar]
- 35.Burrows P.A., Sazanov L.A., Svab Z., Maliga P., Nixon P.J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998;17:868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voelker R., Barkan A. Nuclear genes required for post-translational steps in the biogenesis of the chloroplast cytochrome b6f complex in maize. Mol. Gen. Genet. 1995;249:507–514. doi: 10.1007/BF00290576. [DOI] [PubMed] [Google Scholar]
- 37.Zito F., Kuras R., Choquet Y., Kossel H., Wollman F.A. Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol. 1997;33:79–86. doi: 10.1023/A:1005734809834. [DOI] [PubMed] [Google Scholar]
- 38.Monde R.A., Zito F., Olive J., Wollman F.A., Stern D.B. Post-transcriptional defects in tobacco chloroplast mutants lacking the cytochrome b6/f complex. Plant J. 2000;21:61–72. doi: 10.1046/j.1365-313x.2000.00653.x. [DOI] [PubMed] [Google Scholar]
- 39.Szczepaniak A., Black M.T., Cramer W.A. Topography of the chloroplast cytochrome b6: Orientation of the cytochrome and accessibility of the lumen-side interhelix loops. Z. Naturforsch. C J. Biosci. 1989;44:453–461. doi: 10.1515/znc-1989-5-619. [DOI] [PubMed] [Google Scholar]
- 40.Hankamer B., Morris E., Nield J., Carne A., Barber J. Subunit positioning and transmembrane helix organisation in the core dimer of photosystem II. FEBS Lett. 2001;504:142–151. doi: 10.1016/S0014-5793(01)02766-1. [DOI] [PubMed] [Google Scholar]
- 41.Swiatek M., Regel R.E., Meurer J., Wanner G., Pakrasi H.B., Ohad I., Herrmann R.G. Effects of selective inactivation of individual genes for low-molecular-mass subunits on the assembly of photosystem II, as revealed by chloroplast transformation: The psbEFLJ operon in Nicotiana tabacum. Mol. Genet. Genom. 2003;268:699–710. doi: 10.1007/s00438-002-0791-1. [DOI] [PubMed] [Google Scholar]
- 42.Suorsa M., Regel R.E., Paakkarinen V., Battchikova N., Herrmann R.G., Aro E.M. Protein assembly of photosystem II and accumulation of subcomplexes in the absence of low molecular mass subunits PsbL and PsbJ. Eur. J. Biochem. 2004;271:96–107. doi: 10.1046/j.1432-1033.2003.03906.x. [DOI] [PubMed] [Google Scholar]
- 43.Krieger-Liszkay A. Singlet oxygen production in photosynthesis. J. Exp. Bot. 2005;56:337–346. doi: 10.1093/jxb/erh237. [DOI] [PubMed] [Google Scholar]
- 44.Eva-Mari A., Ivar V., Bertil A. Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- 45.Liu M., Wang Y., Nie Z., Gai J., Bhat Akhter J., Kong J., Zhao T. Double mutation of two homologous genes YL1 and YL2 results in a leaf yellowing phenotype in soybean [Glycine max (L.) Merr] Plant Mol. Biol. 2020 doi: 10.1007/s11103-020-01008-9. online. [DOI] [PubMed] [Google Scholar]
- 46.Lichtenthaler H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Method Enzymol. 1987;148:350–382. [Google Scholar]
- 47.SoyBase. [(accessed on 14 March 2014)]; Available online: http://soybase.org/
- 48.FastQC. [(accessed on 15 September 2015)]; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 49.Glycine Max Cultivar PI 437654 Chloroplast, Complete Genome. [(accessed on 12 January 2016)]; Available online: ncbi.nlm.nih.gov/nuccore/DQ312375.1.
- 50.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.