Summary
Plant metabolism is broadly reprogrammed during acclimation to abiotic changes. Most previous studies have focused on transitions from standard to single stressful conditions. Here, we systematically analyze acclimation processes to levels of light, heat, and cold stress that subtly alter physiological parameters and assess their reversibility during de-acclimation. Metabolome and transcriptome changes were monitored at 11 different time points. Unlike transcriptome changes, most alterations in metabolite levels did not readily return to baseline values, except in the case of cold acclimation. Similar regulatory networks operate during (de-)acclimation to high light and cold, whereas heat and high-light responses exhibit similar dynamics, as determined by surprisal and conditional network analyses. In all acclimation models tested here, super-hubs in conditional transcriptome networks are enriched for components involved in translation, particularly ribosomes. Hence, we suggest that the ribosome serves as a common central hub for the control of three different (de-)acclimation responses.
Subject Areas: Biological Sciences, Metabolomics, Plant Biology, Plants, Transcriptomics
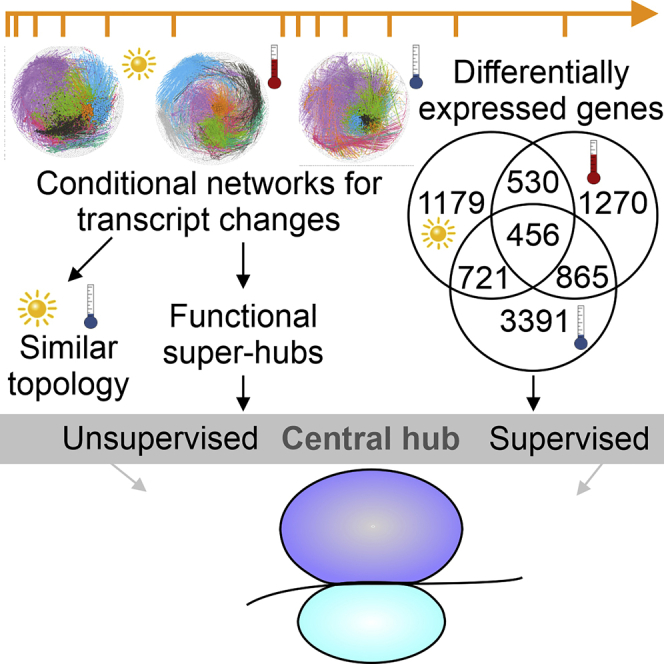
Graphical Abstract
Highlights
-
•
Time series of acclimation and de-acclimation without pleiotropic stresses
-
•
Kinetics of transcriptomes and metabolomes under three environmental conditions
-
•
Transcriptomes return to a baseline, but HL and heat metabolomes do not
-
•
All acclimation responses involve the translational machinery and photosynthesis
Biological Sciences; Metabolomics; Plant Biology; Plants; Transcriptomics
Introduction
Plants continuously adapt to abiotic environmental changes by acclimation, but even moderate deviations from the optimum can markedly limit crop yields (Ashraf et al., 2012; Buchanan et al., 2000; Lobell and Gourdji, 2012; Reddy, 2015). In the era of global warming, acclimation becomes even more relevant, as plants encounter progressive environmental changes (Anjum, 2015). Therefore, a comprehensive understanding of plant acclimation responses is required to design strategies that stabilize or enhance yields in increasingly hostile environments.
Acclimation can act on timescales that vary from minutes to days; involves the modification of gene expression, protein activity, and metabolite profiles; and ultimately affects all cellular compartments (Beine-Golovchuk et al., 2018; Caldana et al., 2011; Kaplan et al., 2007; Mueller et al., 2015; Suzuki et al., 2015; Vogel et al., 2014; Zandalinas et al., 2019; Zhao et al., 2017). Acclimation-associated physiological changes can be divided into three phases. In the first phase, environmental changes immediately affect cellular reactions (Karpinski et al., 2013; Mullineaux and Karpinski, 2002; Schmitz et al., 2014), perturbing metabolism by inhibition/activation of metabolic reactions, and leading to dearth/excess of substrates or products, increased demand for specific compounds, or a combination of these factors. Well-known examples are the acidification of the chloroplast lumen during exposure to high light (HL) levels, which activates photoprotective mechanisms such as non-photochemical quenching (NPQ) (Pinnola and Bassi, 2018; Ruban, 2016), and protein misfolding induced by heat (Guy et al., 1997; Park and Seo, 2015; Wang et al., 2004). Transient changes occur during the second phase, reconfiguring primary and energy metabolism by accumulating protective compounds and balancing levels of reactive oxygen species (Foyer et al., 2009; Nishizawa et al., 2008; Noctor et al., 2011; Panikulangara et al., 2004). Carbohydrates are re-allocated to sustain metabolic demands (Dyson et al., 2015; Nägele and Heyer, 2013; Patzke et al., 2019) and secondary metabolism is also redirected, to produce anthocyanins in response to HL or cold, for instance (Grotewold, 2006; Schulz et al., 2016; Speiser et al., 2015). Under prolonged environmental change, a new steady state is eventually established (third phase) (Huang et al., 2019; Kaplan et al., 2004; Wang et al., 2020).
Owing to the complexity of the process, enhancing acclimation by altering the activity of single components can have unexpected consequences. For instance, overexpressing the same three photoprotective proteins in two different species has opposite effects on growth under natural conditions (Garcia-Molina and Leister, 2020; Kromdijk et al., 2016), most likely due to interspecific differences in acclimation and metabolic networks. Therefore, system-wide approaches are needed to dissect acclimation responses, so as to ensure that targeted modifications occur with minimal trade-offs. Various studies of transcriptomic, proteomic, and metabolomic responses to changes in light or temperature have been done in Arabidopsis thaliana (hereafter Arabidopsis) (Caldana et al., 2011; Carrera et al., 2017; Hannah et al., 2005; Higashi et al., 2015; Huang et al., 2019; Kaplan et al., 2004, 2007; Larkindale and Vierling, 2008; Rossel et al., 2002; Wang et al., 2020), but few of them considered more than one environmental condition (Caldana et al., 2011; Carrera et al., 2017; Cerny et al., 2014; Kaplan et al., 2004; Rocco et al., 2013) or the return from stressful to control conditions (de-acclimation) (Kaplan et al., 2004; Miki et al., 2018; Nakaminami et al., 2014; Pagter et al., 2017; Vyse et al., 2019; Zuther et al., 2015, 2019).
To fill this gap, we have comprehensively monitored physiological, metabolomic, and transcriptomic changes in plants during acclimation and subsequent de-acclimation to HL, heat, and cold. Our study reveals that altered metabolic steady states are established during acclimation, which persist after de-acclimation, whereas transcriptome changes are partially (HL and heat) or fully (cold) reversible. Bioinformatic analyses indicate that components of translation, especially ribosomal proteins (RPs), play central roles in acclimation to changes in the abiotic environment.
Results
Design of Acclimation and De-acclimation Kinetics
To study how the model plant Arabidopsis acclimates to omnipresent changes in temperature and light intensity, we looked at the effects of increased light level (HL) and suboptimal temperature (heat or cold). As the standard growth condition (“control condition”), we used a long photoperiod (16 h/8 h day/night) with temperatures of 18°C at night and 22°C during the day. Plants were grown under light-emitting diode (LED) lights to avoid unwanted heat development and ensure reproducibility among independent replicates (LEDs have a longer half-life than fluorescent tubes). Plant growth and development were optimal at light intensities of 80 μmol photons m−2 s−1 (Figures S1A–S1C), and this level was used as our control condition.
Young Arabidopsis plants are reported to display 50% photoinhibition at a photosynthetically active radiation (PAR) of 535 μmol photons m−2 s−1 (Carvalho et al., 2015) and possess a basal thermotolerance ranging from −4°C to 44°C (Kaplan et al., 2004). To establish moderate acclimation conditions that do not irreversibly affect plant development, we first tested different light intensities. To this end, 14-day-old plants grown under control conditions were subjected to a PAR level of 450 or 800 μmol photons m−2 s−1 for 7 days and their physiological responses were recorded. Intensities of 450 μmol photons m−2 s−1 had a minimal impact on plant growth, whereas those of 800 μmol photons m−2 s−1 limited growth and led to anthocyanin accumulation within 4 days (Figure S1B). To evaluate photoinhibition, we monitored the maximum quantum yield of photosystem II (PSII) (Fv/Fm) during the 4 days of HL treatment, followed by 4 more days under control conditions to assess reversibility. Compared with control conditions (Fv/Fm = 0.8 during the whole time course), plants exposed to 450 μmol photons m−2 s−1 exhibited photoinhibition, which reached its maximum after 3 days (Fv/Fm = 0.57), but was fully reversible during de-acclimation (Figure S1C and Table S1). At 800 μmol photons m−2 s−1, photoinhibition was exacerbated (Fv/Fm = 0.58 after 1 day) and was only partially reversible (Figure S1C and Table S1). Therefore, 450 μmol photons m−2 s−1 was selected for the HL condition. Growth at 32°C throughout the light and dark phase resulted in the typically elongated petioles (Crawford et al., 2012; Sanchez-Bermejo et al., 2015) (Figure S1D) and control-like Fv/Fm values (Figure S1C) and served as the heat stress condition. Cold stress was imposed by reducing the growth temperature to 4°C, and decreasing the light intensity (35 μmol photons m−2 s−1), as reported previously (Fowler and Thomashow, 2002; Hoermiller et al., 2017; Juszczak et al., 2016; Zuther et al., 2019), to avoid photoinhibition (Fv/Fm of 0.67 versus 0.79 after 4 days under 80 and 35 μmol photons m−2 s−1, respectively; Figure S1C).
As 2-week-old plants grown under standard conditions returned to normal growth and development after 4 days each of acclimation and de-acclimation (Figure S1D), this design was used to systematically study reversible acclimation to HL, heat, and cold.
Physiological Changes during (De-)acclimation
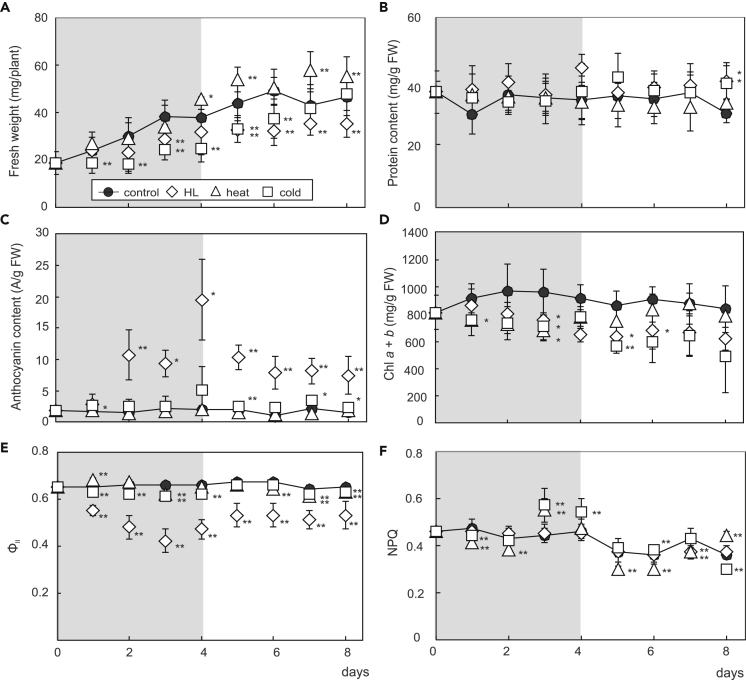
To monitor the effects of (de-)acclimation on plant development and physiology, we measured fresh weight (FW) and total proteins, anthocyanins, and chlorophylls (Figure 1 and Table S1). In all cases, FW of plants increased over the time course, e.g., from ∼19 to ∼47 mg/plant under standard conditions. In HL-exposed plants, the increase set in only on day 3 of the acclimation phase, but growth was partially restored during de-acclimation. Thus, heat-treated plants gained significantly more FW than control plants under standard conditions from day 4 of acclimation onward. Cold treatment affected FW increase most strongly, but the deficit relative to controls disappeared after 3 days of de-acclimation (Figure 1A and Table S1). HL- and cold-treated plants accumulated significantly increased levels of total proteins and anthocyanins; otherwise, these parameters were unchanged (Figures 1B and 1C). Conversely, average chlorophyll levels in treated plants were lower than in controls (Figure 1D). Photosynthetic performance was assessed in terms of the effective quantum yield of PSII (ФII) and NPQ. In general, ФII remained steady over the time course (0.62–0.67 depending on the treatment and time point), except for a progressive and significant decrease of ФII to 0.42 by day 3 of acclimation to HL, which was reversed almost completely during de-acclimation (0.53 on day 4) (Figure 1E and Table S1) and is seen in the behavior of Fv/Fm (Figure S1). This reflects the course of, and recovery from, photoinhibition (Yamori, 2016). Heat and cold each had significant effects on NPQ, in accordance with previous studies that used moderate levels of temperature stress (Harvaux and Kloppstech, 2001; Zhang et al., 2010) (Figure 1F).
Figure 1.
Physiological Performance of Arabidopsis Plants during (De-)acclimation to High Light, Heat, and Cold
(A–F) 14-day-old plants grown under standard growth conditions were exposed to high light (HL), heat, and cold for 4 days (acclimation period, highlighted by gray background), followed by a return to standard growth conditions for 4 days (de-acclimation period). Fresh weight (A), total protein content (B), anthocyanin content (C), chlorophyll (Chl a + b) level (D), effective quantum yield of photosystem II (ФII) (E), and non-photochemical quenching (NPQ) (F) were recorded daily. Values correspond to the mean ± SD of n ≥ 4 independent experiments. ∗∗p < 0.01; ∗p < 0.05 (Student’s t test). See Table S1 for standard deviations and statistics. See also Figure S1.
Thus, in agreement with the restoration of growth after de-acclimation (see Figure S1), most physiological parameters monitored are reversed during de-acclimation experiments.
Molecular Changes during (De-)acclimation: Metabolite Profiling
To obtain a comprehensive picture of molecular changes at the metabolite level, plant material was harvested from shoots at several time points during (de-)acclimation (Figure S2). Because alterations in metabolite or transcript levels can occur rapidly upon exposure to sudden changes in environmental parameters (Caldana et al., 2011; Kaplan et al., 2007; Mueller et al., 2015; Suzuki et al., 2015; Vogel et al., 2014; Zandalinas et al., 2019), measurements were made at both early (1, 5, and 15 min and 3 h) and later time points (2 and 4 days). Samples from control plants grown in parallel and harvested at the same time points were always included.
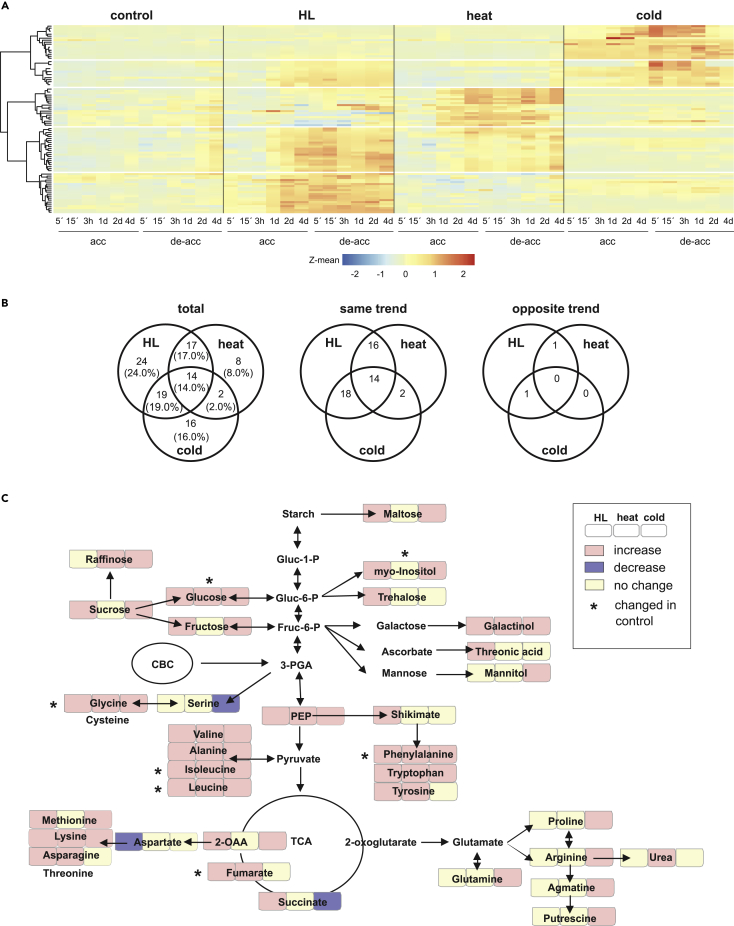
Changes in metabolite levels were identified by gas chromatography (GC)-time-of-flight mass spectrometry (MS), and a total of 98 primary metabolites and 36 unidentified compounds were detected. “Significantly altered metabolites (SAMs)” were defined as metabolites that showed at least a 2-fold change (FC ≥ 2) in concentration relative to the value at time point 0 (Student's t test with a false discovery rate [FDR] ≤ 0.05). Under control conditions, 11 metabolites changed significantly; e.g., leucine, isoleucine, fumarate, and glucose increased, as expected, as plants got older (Chia et al., 2000). Under the acclimation treatments, numbers of SAMs began to increase within 1 or 2 days, and 74, 41, and 51 SAMs were noted for HL, heat, and cold conditions, respectively (Table S2 and Figure S3A).
To infer the dynamics of metabolome changes under each treatment, hierarchical clustering was used to generate heatmaps for all metabolites that changed significantly in level at least once in any (de-)acclimation experiment (Figure 2A). The heatmaps reflected the negligible changes in metabolites under control conditions, which only become discernible toward the end of the time course (Figure 2A). Following HL or heat treatment, 70%–80% of metabolome perturbations persisted during de-acclimation, but 90% returned to the initial state after cold exposure (Figures 2A and S3B and Table S2).
Figure 2.
Changes in Metabolite Composition during (De-)acclimation to High Light, Heat, and Cold
(A) Heatmaps based on Z-means of fold changes of SAMs and generated by hierarchical clustering according to Ward d2.
(B) Venn diagrams depicting shared or unique SAMs, comprising total numbers (left panel, “total”), or only the ones that show the same (up-up or down-down, “same trend”) or opposite (up-down or down-up, “opposite trend”) regulation polarities.
(C) Scheme summarizing the changes in concentration of metabolites involved in central metabolism in Arabidopsis. acc, acclimation; de-acc, de-acclimation; CBC, Calvin-Benson cycle; TCA, tricarboxylic acid cycle; Gluc, glucose; Fruc, fructose; P, phosphate; 3-PGA, 3-phosphoglycerate; PEP, phosphoenolpyruvate; 2-OAA, 2-oxaloacetate. Metabolites whose levels also changed under control conditions are indicated by ∗. Note that glycine levels decreased under control conditions.
See also Figures S2 and S3.
Venn diagrams revealed that the fraction of metabolites altered under two different treatments was similar or significantly higher (super exact test, Wang et al., 2015, p = 0.03) than that of metabolites that were exclusively altered by one treatment (Figure 2B). In all, 31 metabolites reacted to both HL and heat and 33 to both HL and cold, whereas 24, 8, and 16 metabolites were specifically altered under HL, heat, and cold, respectively. Furthermore, the vast majority of the first set responded in the same direction (increase or decrease), indicating that metabolome responses during acclimation to the different stress conditions can target overlapping sets of compounds.
In addition, levels of 14 metabolites were increased under all three conditions. Nine of these compounds are shown in an annotated integrative map of central metabolism in Arabidopsis (Figure 2C) and four of them are unknown. Taking into account the changed levels of some of these metabolites under control conditions (Table S2), galactinol, 3-phosphoglycerate (PEP) and the amino acids glycine, tryptophan, valine, alanine, and lysine are the metabolites whose levels are increased under all treatments (Figure 2C).
Sugars like glucose, sucrose, fructose, and maltose increased during HL and cold, and trehalose increased only during HL. Central organic acids, especially fumarate, which is one of the main carbon (C) storage compounds in Arabidopsis (Chia et al., 2000), and succinate, increased during exposure to HL, and 2-oxalacetate, during HL and cold (Figure 2C and Table S2). Thus the accumulation of C blocks that can store energy or provide skeletons for larger biomolecules is characteristic for the metabolic response to HL. Notably, fumarate was reported to be important for cold acclimation of 8-week-old plants grown under short-day conditions and a light intensity of 100 μmol photons m−1 s−2 (Dyson et al., 2016). The only moderate increase of fumarate under our cold conditions could be attributable to the usage of young (2- to 3-week-old) plants exposed to long-day conditions and a lower light intensity of 35 μmol photons m−2 s−1. In addition, the accumulation of compatible solutes like galactinol under all tested conditions, myo-inositol under HL and cold, raffinose under heat and cold, and mannitol and proline under cold points to a need to mitigate osmotic stress (Nishizawa et al., 2008; Panikulangara et al., 2004) (Figure 2C).
Overall, acclimation resulted in a general increase in amino acid pools under all treatments, and accumulation of carbohydrates and organic acids particularly under HL and cold treatment. Thus the relatively subtle alterations of physiological parameters (see Figure 1) are accompanied by marked and mostly irreversible (except in the case of cold stress) changes in metabolite levels.
Molecular Changes during (De-)acclimation: Transcript Profiling
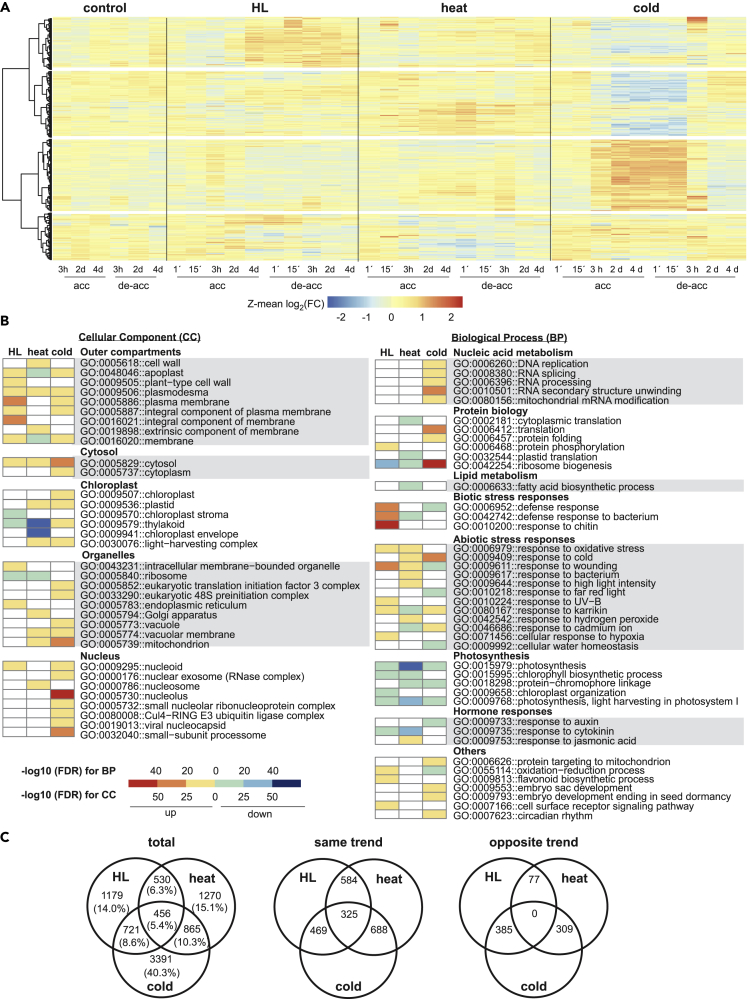
Because only a subset of metabolites in a plant cell can be detected by GC-MS, we also performed transcript profiling, using RNA sequencing to track both nuclear and organellar transcripts. First, we studied the accumulation of bona fide marker mRNAs for the circadian clock and the cell cycle. Clearly, the circadian oscillations observed under control conditions were disrupted by the HL, heat, and cold treatments (Figure S4A), and the expression pattern of cell-cycle markers was altered (Figure S4B). In light of this, we refrained from direct treatment versus control comparisons between individual time points. Instead, transcripts were defined as originating from “differentially expressed genes (DEGs)” if they displayed an absolute FC ≥ 2 with an FDR ≤0.05 (Student's t test) compared with the t = 0 min time point. We identified 1,568 DEGs under control conditions, 4,731 under HL, 4,801 under heat, and 6,426 under cold conditions (Table S3 and Figure S5A). Transcriptome changes under standard conditions were attributed to circadian oscillations (365 and 648 DEGs after 3 h acclimation and de-acclimation) and advancing plant development at the last time point of the kinetics experiment (1,053 DEGs on day 4 of de-acclimation) (Figure S5A). Notably, HL had less effect on transcripts for heat shock proteins and heat shock factors than did heat or cold treatment (Figure S5B), indicating that our HL conditions did not impose heat stress on plants (Huang et al., 2019).
The general behavior of the transcriptomes during acclimation and de-acclimation phases was visualized in heatmaps, which showed limited transcriptome changes under control conditions, whereas marked changes in mRNA expression patterns appeared after 3 h and 2 days of acclimation (Figure 3A). Large gene sets (regulons) continued to be differentially expressed during de-acclimation, although in many instances the trend of regulation was reversed (Figure 3A). Indeed, the largest numbers of DEGs were observed in the central part of the time series, from day 2 of acclimation to 15 min of de-acclimation (Figure S5A). Therefore, the corresponding 2,886 DEGs under HL, 3,121 under heat, and 5,433 observed under cold treatment (Table S4) were examined for their functional annotation into non-redundant Gene Ontology (GO) terms that were then grouped into main categories. Their protein products were assigned to a wide range of cellular compartments (extracellular, cytosol, organelles, especially chloroplasts, and nucleus) (Figure 3B). In the category “biological process,” transcripts up-regulated under all treatments were mainly associated with abiotic stress responses, whereas HL and cold treatments specifically affected biotic stress responses and nucleic acid metabolism, respectively (Figure 3B). Moreover, transcripts associated with ribosome biogenesis were repressed under HL and heat, but those for protein synthesis and folding, as well as phosphorylation, increased in cold conditions. Down-regulated transcripts encoded proteins involved in responses to cytokinins (HL and heat) and auxin (cold). Under all treatments, transcripts for photosynthesis proteins were down-regulated (Figure 3B).
Figure 3.
Changes in Transcript Accumulation during (De-)acclimation to High Light, Heat, and Cold
(A) Heatmaps based on Z-means of fold changes of DEGs. Major regulons were identified by clustering of Z-scores using Ward d2.
(B) Heatmaps illustrating non-redundant Gene Ontology (GO) term enrichment according to DAVID and REVIGO (Huang da et al., 2009a, 2009b; Supek et al., 2011). The color scale corresponds to the -log10 transformation of the FDR for the enrichment according to the Fisher's exact test. The regulatory trend of the transcripts in each bin (up or down) is indicated.
(C) Venn diagrams illustrating shared or unique DEGs, comprising total numbers (left panel, “total”), or only those that show the same (up-up or down-down, “same trend”) or opposite (up-down or down-up, “opposite trend”) polarities of regulation. Note that “same trend” and “opposite trend” sets do not always sum up to “total,” because in several instances the polarity of regulation changed during the time course.
See also Figures S4 and S5.
Moreover, global comparison of transcriptomes identified a set of 456 transcripts (Figure 3C) that changed, in the same direction, under all three conditions (Fisher's exact test, p = 1.05 × 10−34; Table S5). This indicates that all three environmental stimuli trigger a common transcriptional response, and analysis of the annotation of these proteins showed that chloroplast functions were overrepresented in this gene set (102 or 22% of genes, representing a 1.46-fold enrichment for GO term 0009507 “chloroplast”; Fisher's exact test, p = 5.89 × 10−5) (Table S6). It includes transcripts for several light-harvesting chlorophyll a/b-binding proteins (Lhcbs) and PSII subunits (Psb27, PsbO2, PsbQ1), and metabolic enzymes, along with proteins involved in redox reactions and heme and chlorophyll biosyntheses (Table S7). In general, most of these transcripts, except those involved in metabolism, were down-regulated during acclimation (Figure 3B and Table S6).
In an attempt to identify transcription factors (TFs) tentatively mediating acclimation responses toward all examined conditions, the overlap between the list of 456 common genes and genes encoding TFs cataloged in the Database for Arabidopsis Transcription Factors (DATF; http://datf.cbi.pku.edu.cn) and in TAIR (https://www.arabidopsis.org) was determined. Overall, 41 TFs were identified (Table S5), among them the C-REPEAT/DRE BINDING FACTORs CBF1 and 2 and ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 6 (ERF6), which are established factors in the response to low temperatures (Zhao et al., 2016) and, in the case of ERF6, also HL (Vogel et al., 2014). In general, several TFs have been identified that were not related to acclimation responses before; they are members of diverse families including those of the bZIP-, MYC-, zinc-finger-, AP2/ERF-, Myb-, homeobox-, and NAC-type. At least two of them, MYC2 and G-BOX BINDING FACTOR 3 (GBF3), are binding to the G-box motif CACGTG, which is associated with various pathways, among them ambient temperature and light signaling (Ezer et al., 2017). Therefore, to obtain a deeper insight into putatively over-represented cis-acting elements, the 1,000-bp promoter sequences of the 456 common genes were scanned for 8-bp elements with the expectation maximization-based program Amadeus (Linhart et al., 2008). The highest similarity between queried motifs is defined by the lowest E-value, and the three identified over-represented motifs with the lowest E-value (Figure S6) were used to search the JASPAR 2020 plant database (http://jaspar.genereg.net/) (Fornes et al., 2020) for plant transcription factors known to bind to similar sequences. The motif with the lowest E-value found by JASPAR contained indeed the G-box motif (Figure S6). The second over-represented motif identified is predicted to be bound by MYB3, which represses phenylpropanoid biosynthesis gene expression (Zhou et al., 2017), whereas no known plant-associated element could be found for the third over-represented motif.
Surprisal Analysis of Transcriptome Changes during (De-)acclimation
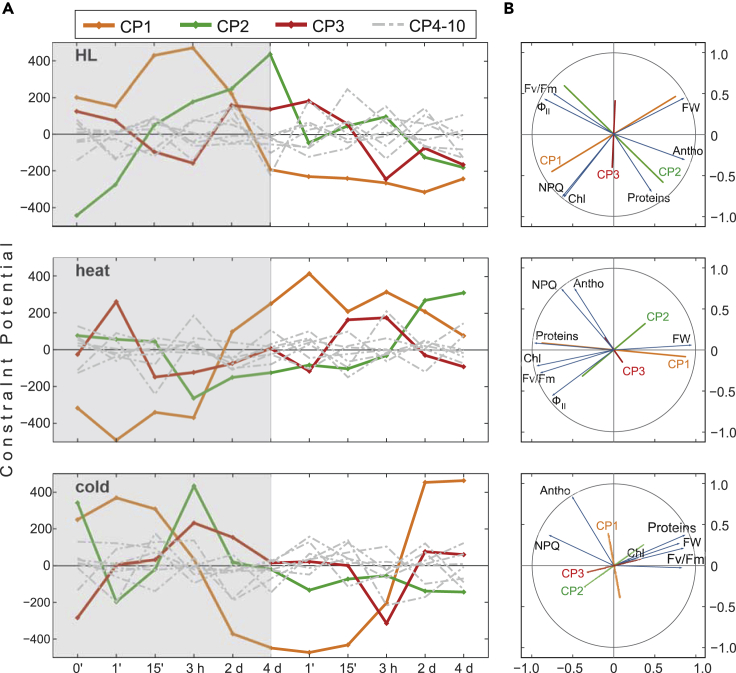
To obtain insight into the molecular dynamics of the acclimation responses, we applied an unbiased thermodynamic maximal-entropy-based approach (surprisal analysis). In biological time series data, surprisal analysis can first identify the theoretical baseline state of minimal free energy of the system and then the constraints that prevent the system from reaching that baseline state. The contribution of the individual constraints is reflected by an associated time-dependent weight, the constraint potential (CP) (Gross et al., 2013; Kravchenko-Balasha et al., 2012; Levine, 1978, 1980; Remacle et al., 2010). Changes in sign of the CPs indicate whether the contribution of the input information (here transcript profile) to the constraint is inverted. Moreover, recovery of the CPs during de-acclimation to the original values for t = 0 of acclimation implies recovery of the underlying molecular dynamics.
For our transcriptome data, a stable baseline state for each condition was indeed obtained (Figure S7) and the datasets were therefore eligible for CP identification. All identified major CPs (CP1 to CP3) were found to change their signs in all three acclimation responses, whereas this tendency was less prominent for the minor CPs (CP4–CP10). Under HL conditions, the first major CP (HL-CP1) changed its sign at 4 days of acclimation and did not return to its original sign during de-acclimation. HL-CP2 and HL-CP3 changed their sign very early, at 15 min of acclimation, with HL-CP2 showing a tendency to return to the starting value at the end of the de-acclimation phase (Figure 4A). Heat-CP1 changed its sign at 2 days of acclimation and remained steady thereafter. As in the case of HL, heat-CP2 and heat-CP3 rapidly changed their sign (at 1 min and 3 h, respectively) and returned to values similar to the starting value during de-acclimation. Cold-CP1 was clearly reversible, changing its sign at 3 h of acclimation and again at 3 h of de-acclimation, to recover to initial levels. CP2 and CP3 under cold (de-)acclimation changed their sign three times (Figure 4A).
Figure 4.
Surprisal Analysis of Transcriptome Changes during (De-)acclimation to High Light, Heat, and Cold
(A) Time course of the constraint potentials (CPs) derived from the transcriptome profiles of (de-)acclimation samples (high light [HL], heat, and cold). The minor constraints (4–10) are shown as gray lines; the acclimation phase is indicated by a gray square. Sign changes of CPs indicate reconfiguration of the underlying molecular dynamics.
(B) Circle correlation plots derived from partial least-squares regression analysis of CP1–3 of the time course shown in (A) and the physiological profiles depicted in Figure 1. Antho, anthocyanin content; Chl, chlorophyll content; FW, fresh weight; ФII, effective quantum yield of PSII; Fv/Fm, maximum quantum efficiency of PSII. Lines of CP1–CP3 in this plot were mirrored to reflect the fact that constraints contain positive and negative weights. Therefore, it is not possible to distinguish whether the effect was caused by positive or negative correlation.
See also Figure S7.
In subsets of transcripts, HL and heat provoke expression changes that are then maintained over the time span investigated here, possibly representing “memory” or “priming” effects (Crisp et al., 2017; Larkindale et al., 2005). A unique pattern was found for cold (de-)acclimation, which is the only condition under which the main acclimation state was fully reversed during the de-acclimation phase.
To investigate whether the constraints identified help to explain the physiological changes during acclimation and de-acclimation, we conducted partial least-squares (PLS) regression analysis of the three major CPs and the physiological parameters from Figure 1 (Figure 4B). A strong correlation in either direction on these plots indicates how well the physiological parameters can be explained using the CP time courses. This also allows one to rank the effects with respect to the constraint index as a proxy for energy investment, meaning primary effects are mainly explained by CP1, secondary effects by CP2, etc. For transcriptome changes under HL, primary energy investment (HL-CP1) showed strong correlations with FW, NPQ, and chlorophyll content, but was almost perpendicular to all other physiological parameters, meaning there was no correlation with them (Figure 4B). However, Fv/Fm, ФII, and contents of anthocyanin and proteins correlated well with HL-CP2. Heat-CP1 correlated with FW, contents of proteins and chlorophyll, and Fv/Fm, and heat-CP2, with ФII. The only phenotypic parameter that correlated well with cold-CP1 was the anthocyanin content and, to a much lesser extent, NPQ. Cold-CP2 and, to a greater extent, cold-CP3 showed correlations with all other physiological parameters. Interestingly, PLS analysis also revealed that the physiological parameters FW and total protein content correlated positively only under cold (de-)acclimation.
Importantly, energy investment in these cases does not necessarily mean investment in the synthesis of these compounds, but relative investment in the creation of the respective pattern. Therefore, it can be concluded that different energy sources are used to sustain growth under HL and changing temperature conditions, with FW and chlorophyll content being common to HL and heat, and anthocyanin content characteristic for cold (de-)acclimation (Figure 4B).
Comparison of Topology and Dynamics of the Three Acclimation Responses
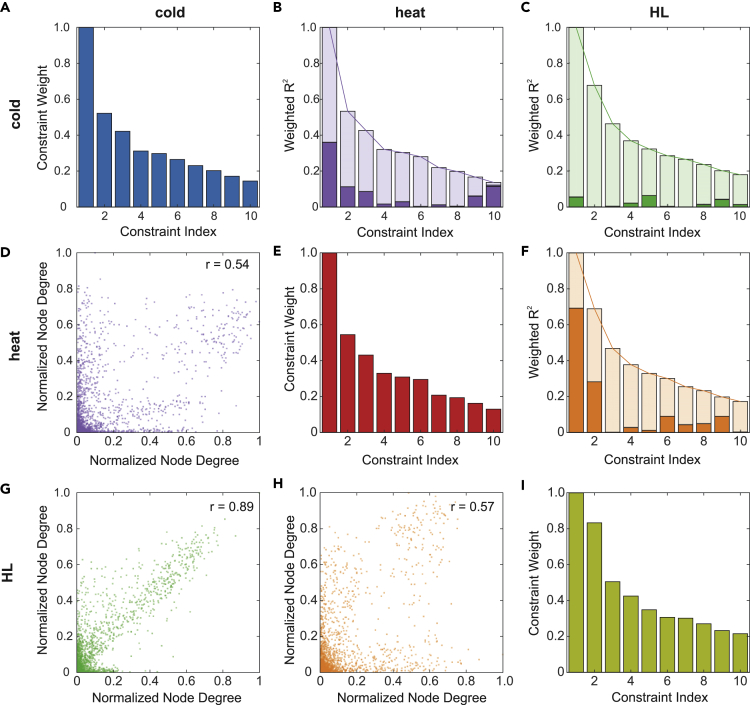
The transcriptome datasets were further analyzed in an unsupervised manner by the construction and subsequent analysis of conditional correlation networks. Briefly, co-expression matrices based on pairwise Pearson correlations among all detected transcripts for each condition were computed and mapped onto a literature-curated reference network to validate interactions and extract subnetworks that operate as conditional networks (Figure S8 and Table S8). The three transcriptional networks were similar in scale (comprising 4,600 to 5,600 nodes), although they differed in the number of connections (Table S8). The properties of the molecular topologies of these networks were then integrated with the data from CPs obtained by surprisal analysis. To this end, Pearson correlations among normalized degree distributions in each network were calculated. The networks under HL and cold were the most similar in their topology (r = 0.89), whereas more divergent patterns were obtained on comparing heat to either HL or cold networks (r = 0.57 and r = 0.54, respectively) (Figures 5D, 5G and 5H). A density plot of the Jaccard index was used as an additional measure of network similarity and showed the same pattern for the conditions, with the cold versus HL comparison yielding the highest index (Figure S8). Therefore, similar features might be involved in the response pathways seen under HL and cold (de-)acclimation conditions.
Figure 5.
Commonalities and Differences between Acclimation Responses Revealed by Comparing Results of Conditional Network and Surprisal Analyses
Matrices showing correlations between constraint indices and weights, normalized node degrees, and weighted R2 scores. Column and row headers indicate which conditions were compared with one another (blue = cold, red = heat, yellow = HL, purple = heat/cold, green = HL/cold, orange = heat/HL).
(A–I) (A, E, and I) Weights of the constraints determined by surprisal analysis for the three conditions. (B, C, and F) Weighted coefficients of determination (R2) of linear regression analyses of constraint potential (CP) time courses determined by surprisal analysis for each constraint index (1–10) and depicted as column plots. Linear regression scores were calculated pairwise for CPs with the same index. R2 was then weighted by the mean of the respective constraint weights of the two compared conditions shown on the diagonal. The open columns indicate the upper bound resulting from the weighting procedure. (D, G, and H) Correlation plots of normalized node degrees of the conditional network nodes. The Pearson correlation (r) is displayed for each plot.
See also Figures S8 and S9.
To compare the dynamics of the three different (de-)acclimation responses as reflected by the CPs obtained by surprisal analysis, we performed pairwise linear regression on all CPs with the same index (1–10) in each treatment, using the intrinsic weighting scheme of surprisal analysis (Figures 5A, 5E, and 5I) to weight the resulting coefficients of determination (R2). Interestingly, a pattern was observed that was inverted with respect to the topological comparison: although the pair HL and heat did not show strong correlation on the network level, they scored highly in the linear regression of CPs, especially for the first two CPs (weighted R2 = 0.69 and 0.28, respectively) (Figure 5F). The same was true for heat and cold, albeit to a lesser extent (weighted R2 = 0.36 and 0.11, respectively) (Figure 5B), whereas there was no notable correlation between HL and cold (Figure 5C).
Overall, therefore, HL and cold (de-)acclimation responses share similar regulatory networks, but display different dynamics. Conversely, the dynamics of the response to heat resembles that to HL (or cold), although different regulatory mechanisms seem to be at work.
Ribosomes Are Hubs of Acclimation
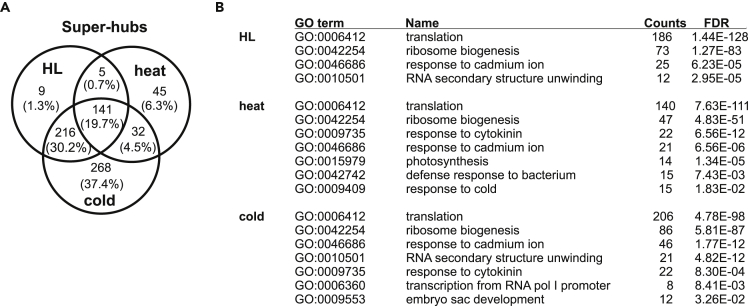
To identify the central components of acclimation responses, conditional networks were explored for hub composition. Hubs are highly connected nodes, providing information about sets of co-expressed nodes and therefore instances of coordinated transcriptional changes. If nodes with >100 edges are defined as super-hubs (Vandereyken et al., 2018), 371, 223, and 657 super-hubs were detected for the HL, heat, and cold networks, respectively (Tables S8 and S9). Interestingly, a significantly enriched fraction of 141 super-hubs (∼20%; super exact test, p = 1.04 × 10−8) was shared among the three treatments. Moreover, 357 (216 + 141) super-hubs were common to HL and cold sets (∼96% and ∼55% of super-hubs under HL and cold, respectively; p = 4.06 × 10−71) (Figure 6A).
Figure 6.
Analysis of Super-hubs in Conditional Networks for Transcripts
(A) Venn diagram depicting super-hubs (degree >100) common to the three networks.
(B) GO term enrichment of the identified super-hubs. The counts are provided, together with the FDR obtained with Fisher's exact test based on all nodes included in the networks.
To gain insight into common responses to acclimation, these super-hubs were functionally annotated. To this end, significant GO terms (Fisher's exact test; FDR ≤0.05) were filtered for the non-redundant ones by employing REVIGO (Supek et al., 2011). Strikingly, GO enrichment analysis indicated that most of the super-hubs in each of the networks were related to the biological process categories “translation” (186 super-hubs under HL, 140 under heat, and 206 under cold, respectively) and “ribosome biogenesis” (73 under HL, 47 under heat, and 86 under cold). Most of these transcripts code for RPs (Figure 6B and Table S9).
To obtain a more detailed picture of the role of translation, and in particular the ribosome, in acclimation responses, the RP members of the HL, heat, and cold super-hubs (96, 108 and 207 members, respectively) were classified according to the subcellular localizations of their gene products using MapMan v.3.5.1 (Thimm et al., 2004). The HL, heat, and cold super-hubs included 26%, 30%, and 57% of all annotated transcripts coding for RPs with bona fide subcellular localization, and RPs from all three genetic compartments, cytosol, chloroplasts, and mitochondria were covered. Subtle compartment-specific enrichments of RPs were observed: cytosolic RPs appeared as over-represented super-hubs for cold and chloroplast-localized RPs were over-represented under HL and heat, but under-represented during cold, whereas mitochondrion-located RPs showed significant depletion under all conditions (Table 1). In contrast to these subtle changes, very clear trends were observed with respect to up- or down-regulation of RP-related super-hubs on day 4 of acclimation relative to t = 0 in the (de-)acclimation time series. The vast majority of ribosome-related transcripts that act as super-hubs tended to be down-regulated at the end of the acclimation phase (day 4) under HL and heat conditions, but up-regulated at the end of cold acclimation (Table 1). Heatmap analysis indicated that transcript levels of RPs were transiently up-regulated during HL and heat acclimation, and transiently down-regulated at the beginning of cold acclimation, before being down- (HL and heat) or up- (cold) regulated at the end of the acclimation phase (Figure S10 and Table S10).
Table 1.
Subcellular Localization, Numbers, and Enrichment of Ribosomal Proteins (RPs) Regulated at the Transcript Level during Acclimation
| Subcellular Localization & Subunit | HL |
Heat |
Cold |
Total | |||
|---|---|---|---|---|---|---|---|
| # (Up/Down) | FE | # (Up/Down) | FE | # (Up/Down) | FE | ||
| Cytosol | |||||||
| 40S | 20 (0/20) | 0.83 | 28 (2/26) | 1.01 | 56 (56/0) | 1.10 | 101 |
| 60S | 41 (2/39) | 1.04 | 39 (0/39) | 0.86 | 110 (110/0) | 1.32∗ | 165 |
| All | 61 (2/59) | 0.96 | 67 (2/65) | 0.91 | 166 (166/0) | 1.24∗ | 266 |
| Chloroplast | |||||||
| 30S | 9 (0/9) | 1.44 | 10 (0/10) | 1.40 | 3 (2/1) | 0.23∗ | 26 |
| 50S | 16 (0/16) | 1.31 | 23 (0/23) | 1.64∗ | 9 (7/2) | 0.35∗ | 51 |
| All | 25 (0/25) | 1.35∗ | 33 (0/33) | 1.56∗ | 12 (9/3) | 0.31∗ | 77 |
| Mitochondrion | |||||||
| 30S | – | 0.00∗ | – | 0.00∗ | – | 0.00∗ | 13 |
| 50S | 1 (0/1) | 0.60 | – | 0.00 | 5(5/0) | 1.42 | 7 |
| All | 1 (0/1) | 0.21∗ | 0 (0/0) | 0.00∗ | 5 (5/0) | 0.50∗ | 20 |
| Total | |||||||
| 87 (2/85) | 100 (2/98) | 183 (180/3) | 363 | ||||
Transcriptional super-hubs for RPs were classified according to the subcellular localization of the corresponding ribosomal subunits based on MapMan (Thimm et al., 2004). The total number of transcripts in each category and the fold enrichment (FE) relative to the expected frequency of RPs are listed. The regulatory trend (up- or down-regulation) refers to changes in the transcript levels at 4 days of acclimation compared with the initial time point. Statistical significance of the enrichment according Fisher's two-sided exact test (p ≤ 0.05) is indicated by an asterisk.
Together, these observations indicate that modification of translation is a common and central element of acclimation responses, with very similar responses ensuing during HL and heat acclimation, and a different response occurring during cold acclimation.
Discussion
A Comprehensive Analysis of (De-)acclimation to High Light, Heat, and Cold
In this work, we exposed Arabidopsis plants to moderate levels of light, heat, and cold stress that resulted in minimal, and largely reversible, physiological perturbations (see Figure 1). In contrast to previous studies (Suzuki et al., 2015; Vogel et al., 2014), we identified low numbers of DEGs after 1 and 15 min of HL (3 and 300; see Figure S5). This is attributable to our use of lower light dosages and LED sources, which did not cause heat stress or trigger expression of heat-shock proteins. Moreover, we also avoided photoinhibition under our cold conditions by reducing light intensities (see Figure S1C). However, our acclimation treatments did lead to desynchronization of cells, as indicated by the perturbation of circadian gene expression and cell-cycle periodicity (see Figure S4).
Metabolome changes during acclimation comprised the accumulation of sugars and amino acids, especially under HL and cold treatments (see Table S2). At first glance, the accumulation of sugars appears to be in contradiction with the minimal FW losses observed under these two conditions and a transitory arrest of the photosynthetic light reactions during HL (see Figure 1). However, the increased content of central sugars such as maltose, sucrose, glucose, or fructose might result from increased starch turnover rather than increased photosynthesis. Under cold conditions, accumulation of sugars is not only aimed at providing energy but also to assure production of osmoprotectants like galactinol, raffinose, or myo-inositol. Similarly, the increased amino acid pools during the three conditions (see Figure 2) failed to correlate with changes in protein content (see Figure 1). This could imply that storage molecules, preferentially starch and/or lipids, rather than proteins are catabolized to meet the energetic demands of the acclimation response and maintain plant growth.
In general, HL and heat irreversibly altered metabolite profiles, whereas changes induced by cold treatment were readily reversible during de-acclimation (see Figure 2A). In contrast, transcriptome changes under HL and heat were only partially reversible (see Figures 3 and 4). Previous studies have observed similar patterns for both transcriptomes (Byun et al., 2014; Higashi et al., 2015; Huang et al., 2019; Zuther et al., 2015) and metabolites (Pagter et al., 2017; Vyse et al., 2019; Zuther et al., 2015). The (virtually) complete reversibility of the transcriptional response to cold may reflect the fact that the Arabidopsis ecotype Col-0 displays enhanced tolerance to low temperatures (Hannah et al., 2006; Kaplan et al., 2004; Yano et al., 2005).
As changes in metabolite profiles persisted during de-acclimation (see Figures 2 and S3), metabolites are obvious candidates for signals that prime plants to cope with future environmental challenges (Schwachtje et al., 2019; Serrano et al., 2019). We identified 7 metabolites (galactinol, phosphoenolpyruvate, glycine, tryptophan, lysine, alanine, and valine) whose concentrations increased under all conditions tested (Figure 2). Although accumulation of carbohydrates and organic acids, and their relocation between compartments, are well-known acclimation strategies (Dyson et al., 2015, 2016; Nägele and Heyer, 2013; Patzke et al., 2019; Pommerrenig et al., 2018), accumulation of amino acids during extended abiotic stress has been described only for cold-treated Arabidopsis plants (Espinoza et al., 2010; Kaplan et al., 2004; Lee et al., 2012; Pagter et al., 2017; Song et al., 2017) and deserves further investigation.
Our bioinformatic analysis suggested that HL and heat treatment induce similar temporal patterns of energy re-allocation. Conversely, topologies of conditional networks derived from co-expressed transcripts (see Figure 4) indicated that very similar regulatory pathways were triggered by HL and cold treatments. This implies that each acclimation response is characterized by a unique pattern of regulation that follows its own schedule.
Translation during (De-)acclimation
The existence of central elements that come into play under various types of adverse conditions regardless of the specific environmental input has been suggested in previous works (Kaplan et al., 2004; Kimura et al., 2003; Rocco et al., 2013). In our work, both unsupervised and supervised analyses of acclimation transcriptomes identified mRNAs for cytosolic and organellar RPs as shared and central factors in responses to diverse abiotic stresses (see Figure 6).
In mammalian cells, nucleoli and particularly ribosomes, are considered as hubs that integrate cellular responses to unfavorable conditions (Pfister, 2019; Warner and McIntosh, 2009; Yang et al., 2018). Under nucleolar stress (“ribostress”), RPs are exported to the cytosol and become pivotal components that help to trigger cell arrest or even apoptosis, depending on the level of stress (Pfister, 2019; Warner and McIntosh, 2009; Yang et al., 2018). A similar regulatory role for ribosomes during abiotic and biotic stresses is emerging in several plant species (Kim et al., 2004; Moin et al., 2016; Saez-Vasquez et al., 2000; Wang et al., 2013). A role of gene expression, in particular translation and the ribosome, in acclimation to abiotic environmental changes has been emerging in recent analyses of corresponding Arabidopsis mutants. In particular, various instances of impaired cold tolerances due to inactivation of chloroplast proteins involved in translation have been described, including subunits (Rogalski et al., 2008; Wang et al., 2017; Zhang et al., 2016), associated proteins (Pulido et al., 2018), and biogenesis factors (Paieri et al., 2018; Reiter et al., 2020) of the plastid ribosome, translation initiation or elongation factors (Liu et al., 2010; Marino et al., 2019), and RNA-binding proteins (Kupsch et al., 2012). Furthermore, the cytosolic pre-18S rRNA processing factor RH7 is involved in cold tolerance (Huang et al., 2016), indicating that impairments in cytosolic translation can also affect cold tolerance. Fewer reports are available for an interplay of translation and tolerance to heat or HL, including enhanced heat sensitivity in the absence of the cytosolic translation initiation factor 5B (Zhang et al., 2017) or the plastid elongation factor Tu (Li et al., 2018), and HL sensitivity of seedlings lacking the cytosolic translation initiation factor 2a (Lokdarshi et al., 2020).
Why are transcripts for RPs mainly down-regulated at the end of the HL and heat acclimation phase, but up-regulated after cold acclimation? These trends coincide with previous studies employing alternative setups for HL, heat, and cold in Arabidopsis and other model organisms (Beine-Golovchuk et al., 2018; Guo et al., 2002; Guy, 1990; Kim et al., 2004; Moin et al., 2016; Thomashow, 1998). Down-regulation of RPs under high temperatures was initially supposed to reflect either increased mRNA instability or the need to restrict protein production, whereas up-regulation of RPs during cold stress was thought to maintain rates of protein synthesis under thermodynamically unfavorable conditions. However, a recent report has shown that mild oxidative stress affects the numbers of ribosomes in plants, but not overall translation efficiency (Salih et al., 2020), although plant genomes possess multiple gene copies for each RP (Barakat et al., 2001). This prompted the view that changes in RP expression under adverse conditions might be related to alterations of ribosomal composition required to adjust translation rates appropriately (Carroll et al., 2008; Giavalisco et al., 2005; Wang et al., 2013). Interestingly, in our experiments, neither the total protein content nor the pool of free amino acids in leaves decreased during acclimation (see Figures 1 and 2). Therefore, translation during (de-)acclimation is selectively reconfigured during HL/heat and cold, most likely involving changes in ribosome composition and/or readjustment of protein biosynthesis rate in subcellular compartments. A promising starting point to investigate a regulatory role of ribosomal activity, or more generally translational activity, during acclimation to other conditions than cold (for which such a function is well established) could be our list of nine genes encoding RNA-binding or ribosome-associated proteins, the mRNA levels of which changed during all conditions (see Table S5). Among them are COLD, CIRCADIAN RHYTHM, and RNA BINDING 2 (CCR2)/GLYCINE RICH PROTEIN 7 (GRP7) and AT2G27710, which encodes a 60S acidic RP. Interestingly, this protein was shown before to be regulated under cold conditions in a proteome analysis using differential in-gel electrophoresis (Amme et al., 2006).
Conclusion
Our work provides a holistic picture of three acclimation responses and suggests translation as the central process involved. Common up- or down-regulation of transcripts for RPs might reflect activation or repression of translation as a whole during acclimation, whereas regulatory factors that are transiently associated with the ribosome seem more likely to mediate central aspects of acclimation. Such modifications can be refined by taking advantage of promoters containing cis-elements being either specific to certain types of (de-)acclimation or being common to several responses (see Figure S6). In fact, our transcriptome kinetics is an ideal starting point for the identification of all kinds of (de-)acclimation-relevant promoters.
Limitations of the Study
Our conclusions are based on the analyses of large-scale omics data. Although the involvement of translational processes in cold acclimation is established (see Discussion section), future work is needed to validate the predicted central function of translation during heat and HL acclimation in vivo. Furthermore, targeted experiments are required to confirm the involvement of the proposed regulatory proteins and predicted cis-acting elements in the acclimation response. We used relatively young plants for our experiments. It will be of interest whether our conclusion can be transferred to (1) older plants and also (2) other plant species. As changes of most metabolites did not return to initial levels during heat and HL de-acclimation, future kinetics experiments should consider longer de-acclimation time periods.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dario Leister (leister@lmu.de).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The datasets generated during this study have been deposited to Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/): GSE125950.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Paul Hardy and Jose Muino for critical reading of the manuscript and the Deutsche Forschungsgemeinschaft (DFG) for funding (project TR175).
Author Contributions
Conceptualization, D.L.; Methodology, K.S. and T.M.; Formal Analysis, A.G.-M., T.K., K.S., and T.M.; Investigation, A.G.-M., T.K., and M.L., Resources: D.L.; Writing: A.G.-M., T.K., and D.L. with input from all authors; Supervision: T.K. and D.L.; Funding acquisition: T.K., T.M., and D.L.
Declaration of Interests
The authors declare no competing interests.
Published: July 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101331.
Supplemental Information
Values are provided as mean ± standard deviation (n ≥ 4 independent experiments). and the mean values correspond to the values provided in Figures 1 and S1 (Fv/Fm). Statistically significant differences compared to the control condition were assessed according the Student's t test (∗, p ≤ 0.05; ∗∗, p ≤ 0.01).
Metabolites were identified by GC-MS-TOF. We considered those metabolites as SAMs, which displayed in at least one time point an absolute log2 fold-change (FC) ≥ 1 compared with the initial value (0 min) according to the Student's t test with a false discovery rate (FDR) ≤ 0.05. Raw data are normalized to an internal standard and fresh weight. Missing values were replaced by either the average of the other replicates (single missing values) or the detection threshold (multiple missing values).
Transcriptome changes under standard conditions, HL, heat, and cold according to an absolute log2 fold change (FC) ≥ 1 to time point 0 min and a false discovery rate (FDR) ≤ 0.05 for the Student's t test. The symbol and brief description of each transcript is provided.
Transcriptional changes were qualified as significant based on absolute fold change ≥1 to the initial time point (0 min) and a false discovery rate (FDR) ≤ 0.05 for the Student's t test in at least one time point from the central phase of the kinetic (2 days acclimation to 15 min de-acclimation). Transcripts are listed with gene ID, symbol, and brief description.
Transcriptome changes during the central phase of the kinetics to HL, heat, and cold (2 days acclimation to 15 min de-acclimation) were compared by means of Venn diagrams to identify the common fraction of changes to the three treatments. The selected transcripts are listed along with the gene ID, symbol, and brief description.
Transcriptome changes under HL, heat, and cold conditions according to an absolute log2 fold change (FC) ≥ 1 to time point 0 min and a false discovery rate (FDR) ≤ 0.05 for the Student's t test in at least one time point were filtered by presence in the Gene Ontology term 0009507 “chloroplast.” The symbol and brief description of each transcript is provided.
Co-expression networks were investigated for nodes with a connectivity ≥ 100 edges (super-hubs) in each condition and listed along with the gene ID, symbol, and brief description.
Ribosomal proteins were extracted from MapMan and information on their gene ID, subcellular localization, assignment to subunit and regulatory trend at the fourth day of acclimation compared with the initial time point (0 mi). DOWN/UP means down-/up-regulated; -, not changed.
References
- Amme S., Matros A., Schlesier B., Mock H.P. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J. Exp. Bot. 2006;57:1537–1546. doi: 10.1093/jxb/erj129. [DOI] [PubMed] [Google Scholar]
- Anjum N.A. Plant acclimation to environmental stress: a critical appraisal. Front. Plant Sci. 2015;6:445. [Google Scholar]
- Ashraf M., Ahmad M.S.A., Öztürk M., Aksoy A. Crop improvement through different means: challenges and prospects. In: Ashraf M., Öztürk M., Ahmad M.S.A., Aksoy A., editors. Crop Production for Agricultural Improvement. Springer Netherlands; 2012. pp. 1–15. [Google Scholar]
- Barakat A., Szick-Miranda K., Chang I.F., Guyot R., Blanc G., Cooke R., Delseny M., Bailey-Serres J. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001;127:398–415. [PMC free article] [PubMed] [Google Scholar]
- Beine-Golovchuk O., Firmino A.A.P., Dabrowska A., Schmidt S., Erban A., Walther D., Zuther E., Hincha D.K., Kopka J. Plant temperature acclimation and growth rely on cytosolic ribosome biogenesis factor homologs. Plant Physiol. 2018;176:2251–2276. doi: 10.1104/pp.17.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan B.B., Gruissem W., Jones R.L. American Society of Plant Physiologists; 2000. Biochemistry & Molecular Biology of Plants. [Google Scholar]
- Byun Y.J., Koo M.Y., Joo H.J., Ha-Lee Y.M., Lee D.H. Comparative analysis of gene expression under cold acclimation, deacclimation and reacclimation in Arabidopsis. Physiol. Plant. 2014;152:256–274. doi: 10.1111/ppl.12163. [DOI] [PubMed] [Google Scholar]
- Caldana C., Degenkolbe T., Cuadros-Inostroza A., Klie S., Sulpice R., Leisse A., Steinhauser D., Fernie A.R., Willmitzer L., Hannah M.A. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011;67:869–884. doi: 10.1111/j.1365-313X.2011.04640.x. [DOI] [PubMed] [Google Scholar]
- Carrera D.Á., Oddsson S., Grossmann J., Trachsel C., Streb S. Comparative proteomic analysis of plant acclimation to six different long-term environmental changes. Plant Cell Physiol. 2017;59:510–526. doi: 10.1093/pcp/pcx206. [DOI] [PubMed] [Google Scholar]
- Carroll A.J., Heazlewood J.L., Ito J., Millar A.H. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell Proteomics. 2008;7:347–369. doi: 10.1074/mcp.M700052-MCP200. [DOI] [PubMed] [Google Scholar]
- Carvalho F.E., Ware M.A., Ruban A.V. Quantifying the dynamics of light tolerance in Arabidopsis plants during ontogenesis. Plant Cell Environ. 2015;38:2603–2617. doi: 10.1111/pce.12574. [DOI] [PubMed] [Google Scholar]
- Cerny M., Jedelsky P.L., Novak J., Schlosser A., Brzobohaty B. Cytokinin modulates proteomic, transcriptomic and growth responses to temperature shocks in Arabidopsis. Plant Cell Environ. 2014;37:1641–1655. doi: 10.1111/pce.12270. [DOI] [PubMed] [Google Scholar]
- Chia D.W., Yoder T.J., Reiter W.D., Gibson S.I. Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species. Planta. 2000;211:743–751. doi: 10.1007/s004250000345. [DOI] [PubMed] [Google Scholar]
- Crawford A.J., McLachlan D.H., Hetherington A.M., Franklin K.A. High temperature exposure increases plant cooling capacity. Curr. Biol. 2012;22:R396–R397. doi: 10.1016/j.cub.2012.03.044. [DOI] [PubMed] [Google Scholar]
- Crisp P.A., Ganguly D.R., Smith A.B., Murray K.D., Estavillo G.M., Searle I., Ford E., Bogdanovic O., Lister R., Borevitz J.O. Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell. 2017;29:1836–1863. doi: 10.1105/tpc.16.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson B.C., Allwood J.W., Feil R., Xu Y., Miller M., Bowsher C.G., Goodacre R., Lunn J.E., Johnson G.N. Acclimation of metabolism to light in Arabidopsis thaliana: the glucose 6-phosphate/phosphate translocator GPT2 directs metabolic acclimation. Plant Cell Environ. 2015;38:1404–1417. doi: 10.1111/pce.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson B.C., Miller M.A., Feil R., Rattray N., Bowsher C.G., Goodacre R., Lunn J.E., Johnson G.N. FUM2, a cytosolic fumarase, is essential for acclimation to low temperature in Arabidopsis thaliana. Plant Physiol. 2016;172:118–127. doi: 10.1104/pp.16.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C., Degenkolbe T., Caldana C., Zuther E., Leisse A., Willmitzer L., Hincha D.K., Hannah M.A. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One. 2010;5:e14101. doi: 10.1371/journal.pone.0014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D., Shepherd S.J.K., Brestovitsky A., Dickinson P., Cortijo S., Charoensawan V., Box M.S., Biswas S., Jaeger K.E., Wigge P.A. The G-Box transcriptional regulatory code in Arabidopsis. Plant Physiol. 2017;175:628–640. doi: 10.1104/pp.17.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranasic D. Jaspar 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Thomashow M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 2009;60:455–484. doi: 10.1146/annurev.arplant.043008.091948. [DOI] [PubMed] [Google Scholar]
- Garcia-Molina A., Leister D. Accelerated relaxation of photoprotection impairs biomass accumulation in Arabidopsis. Nat. Plants. 2020;6:9–12. doi: 10.1038/s41477-019-0572-z. [DOI] [PubMed] [Google Scholar]
- Giavalisco P., Wilson D., Kreitler T., Lehrach H., Klose J., Gobom J., Fucini P. High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol. Biol. 2005;57:577–591. doi: 10.1007/s11103-005-0699-3. [DOI] [PubMed] [Google Scholar]
- Gross A., Li C.M., Remacle F., Levine R.D. Free energy rhythms in Saccharomyces cerevisiae: a dynamic perspective with implications for ribosomal biogenesis. Biochemistry. 2013;52:1641–1648. doi: 10.1021/bi3016982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Guo Y., Xiong L., Ishitani M., Zhu J.-K. An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl. Acad. Sci. U S A. 2002;99:7786. doi: 10.1073/pnas.112040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C., Haskell D., Li Q.-B., Zhang C. Molecular chaperones: do they have a role in cold stress responses of plants? In: Li P.H., Chen T.H.H., editors. Plant Cold Hardiness: Molecular Biology, Biochemistry, and Physiology. Springer US; 1997. pp. 109–129. [Google Scholar]
- Guy C.L. Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1990;41:187–223. [Google Scholar]
- Hannah M.A., Heyer A.G., Hincha D.K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005;1:e26. doi: 10.1371/journal.pgen.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah M.A., Wiese D., Freund S., Fiehn O., Heyer A.G., Hincha D.K. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 2006;142:98–112. doi: 10.1104/pp.106.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvaux M., Kloppstech K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta. 2001;213:953–966. doi: 10.1007/s004250100572. [DOI] [PubMed] [Google Scholar]
- Higashi Y., Okazaki Y., Myouga F., Shinozaki K., Saito K. Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci. Rep. 2015;5:10533. doi: 10.1038/srep10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoermiller, Naegele T., Augustin H., Stutz S., Weckwerth W., Heyer A.G. Subcellular reprogramming of metabolism during cold acclimation in Arabidopsis thaliana. Plant Cell Environ. 2017;40:602–610. doi: 10.1111/pce.12836. [DOI] [PubMed] [Google Scholar]
- Huang C.K., Shen Y.L., Huang L.F., Wu S.J., Yeh C.H., Lu C.A. The DEAD-box RNA helicase AtRH7/PRH75 participates in pre-rRNA processing, plant development and cold tolerance in Arabidopsis. Plant Cell Physiol. 2016;57:174–191. doi: 10.1093/pcp/pcv188. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhao X., Chory J. The Arabidopsis transcriptome responds specifically and dynamically to high light stress. Cell Rep. 2019;29:4186–4199.e3. doi: 10.1016/j.celrep.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczak I., Cvetkovic J., Zuther E., Hincha D.K., Baier M. Natural variation of cold deacclimation correlates with variation of cold-acclimation of the plastid antioxidant system in Arabidopsis thaliana accessions. Front. Plant Sci. 2016;7:305. doi: 10.3389/fpls.2016.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F., Kopka J., Haskell D.W., Zhao W., Schiller K.C., Gatzke N., Sung D.Y., Guy C.L. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F., Kopka J., Sung D.Y., Zhao W., Popp M., Porat R., Guy C.L. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007;50:967–981. doi: 10.1111/j.1365-313X.2007.03100.x. [DOI] [PubMed] [Google Scholar]
- Karpinski S., Szechynska-Hebda M., Wituszynska W., Burdiak P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 2013;36:736–744. doi: 10.1111/pce.12018. [DOI] [PubMed] [Google Scholar]
- Kim K.Y., Park S.W., Chung Y.S., Chung C.H., Kim J.I., Lee J.H. Molecular cloning of low-temperature-inducible ribosomal proteins from soybean. J. Exp. Bot. 2004;55:1153–1155. doi: 10.1093/jxb/erh125. [DOI] [PubMed] [Google Scholar]
- Kimura M., Yamamoto Y.Y., Seki M., Sakurai T., Sato M., Abe T., Yoshida S., Manabe K., Shinozaki K., Matsui M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 2003;77:226–233. doi: 10.1562/0031-8655(2003)077<0226:ioagrb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kravchenko-Balasha N., Levitzki A., Goldstein A., Rotter V., Gross A., Remacle F., Levine R.D. On a fundamental structure of gene networks in living cells. Proc. Natl. Acad. Sci. U S A. 2012;109:4702. doi: 10.1073/pnas.1200790109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromdijk J., Głowacka K., Leonelli L., Gabilly S.T., Iwai M., Niyogi K.K., Long S.P. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science. 2016;354:857–861. doi: 10.1126/science.aai8878. [DOI] [PubMed] [Google Scholar]
- Kupsch C., Ruwe H., Gusewski S., Tillich M., Small I., Schmitz-Linneweber C. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell. 2012;24:4266–4280. doi: 10.1105/tpc.112.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J., Hall J.D., Knight M.R., Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005;138:882. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J., Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146:748–761. doi: 10.1104/pp.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.P., Babakov A., de Boer B., Zuther E., Hincha D.K. Comparison of freezing tolerance, compatible solutes and polyamines in geographically diverse collections of Thellungiella sp. and Arabidopsis thaliana accessions. BMC Plant Biol. 2012;12:131. doi: 10.1186/1471-2229-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R.D. Information theory approach to molecular reaction dynamics. Ann. Rev. Phys. Chem. 1978;29:59–92. [Google Scholar]
- Levine R.D. An information theoretical approach to inversion problems. J. Phys. A Math. Gen. 1980;13:91–108. [Google Scholar]
- Li X., Cai C., Wang Z., Fan B., Zhu C., Chen Z. Plastid translation elongation factor Tu is prone to heat-induced aggregation despite its critical role in plant heat tolerance. Plant Physiol. 2018;176:3027–3045. doi: 10.1104/pp.17.01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart C., Halperin Y., Shamir R. Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008;18:1180–1189. doi: 10.1101/gr.076117.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Rodermel S.R., Yu F. A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol. 2010;10:287. doi: 10.1186/1471-2229-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell D.B., Gourdji S.M. The influence of climate change on global crop productivity. Plant Physiol. 2012;160:1686–1697. doi: 10.1104/pp.112.208298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokdarshi A., Guan J., Urquidi Camacho R.A., Cho S.K., Morgan P.W., Leonard M., Shimono M., Day B., von Arnim A.G. Light activates the translational regulatory kinase GCN2 via reactive oxygen species emanating from the chloroplast. Plant Cell. 2020;32:1161–1178. doi: 10.1105/tpc.19.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G., Naranjo B., Wang J., Penzler J.F., Kleine T., Leister D. Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. Plant J. 2019;99:521–535. doi: 10.1111/tpj.14342. [DOI] [PubMed] [Google Scholar]
- Miki Y., Takahashi D., Kawamura Y., Uemura M. Temporal proteomics of Arabidopsis plasma membrane during cold- and de-acclimation. J. Proteomics. 2018;197:71–81. doi: 10.1016/j.jprot.2018.11.008. [DOI] [PubMed] [Google Scholar]
- Moin M., Bakshi A., Saha A., Dutta M., Madhav S.M., Kirti P.B. Rice ribosomal protein large subunit genes and their spatio-temporal and stress regulation. Front. Plant Sci. 2016;7:1284. doi: 10.3389/fpls.2016.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.P., Krause D.M., Mueller M.J., Fekete A. Accumulation of extra-chloroplastic triacylglycerols in Arabidopsis seedlings during heat acclimation. J. Exp. Bot. 2015;66:4517–4526. doi: 10.1093/jxb/erv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux P., Karpinski S. Signal transduction in response to excess light: getting out of the chloroplast. Curr. Opin. Plant Biol. 2002;5:43–48. doi: 10.1016/s1369-5266(01)00226-6. [DOI] [PubMed] [Google Scholar]
- Nägele T., Heyer A.G. Approximating subcellular organisation of carbohydrate metabolism during cold acclimation in different natural accessions of Arabidopsis thaliana. New Phytol. 2013;198:777–787. doi: 10.1111/nph.12201. [DOI] [PubMed] [Google Scholar]
- Nakaminami K., Matsui A., Nakagami H., Minami A., Nomura Y., Tanaka M., Morosawa T., Ishida J., Takahashi S., Uemura M. Analysis of differential expression patterns of mRNA and protein during cold-acclimation and de-acclimation in Arabidopsis. Mol. Cell Proteomics. 2014;13:3602–3611. doi: 10.1074/mcp.M114.039081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Queval G., Mhamdi A., Chaouch S., Foyer C.H. Glutathione. Arabidopsis Book. 2011;9:e0142. doi: 10.1199/tab.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagter M., Alpers J., Erban A., Kopka J., Zuther E., Hincha D.K. Rapid transcriptional and metabolic regulation of the deacclimation process in cold acclimated Arabidopsis thaliana. BMC Genomics. 2017;18:731. doi: 10.1186/s12864-017-4126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paieri F., Tadini L., Manavski N., Kleine T., Ferrari R., Morandini P., Pesaresi P., Meurer J., Leister D. The DEAD-box RNA helicase RH50 is a 23S-4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol. 2018;176:634–648. doi: 10.1104/pp.17.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikulangara T.J., Eggers-Schumacher G., Wunderlich M., Stransky H., Schoffl F. Galactinol synthase1. A novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiol. 2004;136:3148–3158. doi: 10.1104/pp.104.042606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.J., Seo Y.S. Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 2015;31:323–333. doi: 10.5423/PPJ.RW.08.2015.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzke K., Prananingrum P., Klemens P.A.W., Trentmann O., Rodrigues C.M., Keller I., Fernie A.R., Geigenberger P., Bölter B., Lehmann M. The plastidic sugar transporter pSuT influences flowering and affects cold responses. Plant Physiol. 2019;179:569–587. doi: 10.1104/pp.18.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister A.S. Emerging role of the nucleolar stress response in autophagy. Front. Cell Neurosci. 2019;13:156. doi: 10.3389/fncel.2019.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnola A., Bassi R. Molecular mechanisms involved in plant photoprotection. Biochem. Soc. Trans. 2018;46:467–482. doi: 10.1042/BST20170307. [DOI] [PubMed] [Google Scholar]
- Pommerrenig B., Ludewig F., Cvetkovic J., Trentmann O., Klemens P.A.W., Neuhaus H.E. In concert: orchestrated changes in carbohydrate homeostasis are critical for plant abiotic stress tolerance. Plant Cell Physiol. 2018;59:1290–1299. doi: 10.1093/pcp/pcy037. [DOI] [PubMed] [Google Scholar]
- Pulido P., Zagari N., Manavski N., Gawronski P., Matthes A., Scharff L.B., Meurer J., Leister D. CHLOROPLAST RIBOSOME ASSOCIATED supports translation under stress and interacts with the ribosomal 30S subunit. Plant Physiol. 2018;177:1539–1554. doi: 10.1104/pp.18.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P.P. Climate Resilient Agriculture for Ensuring Food Security. Springer India; 2015. Impacts of climate change on agriculture; pp. 43–90. [Google Scholar]
- Reiter B., Vamvaka E., Marino G., Kleine T., Jahns P., Bolle C., Leister D., Ruhle T. The Arabidopsis protein CGL20 is required for plastid 50S ribosome biogenesis. Plant Physiol. 2020;182:1222–1238. doi: 10.1104/pp.19.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle F., Kravchenko-Balasha N., Levitzki A., Levine R.D. Information-theoretic analysis of phenotype changes in early stages of carcinogenesis. Proc. Natl. Acad. Sci. U S A. 2010;107:10324. doi: 10.1073/pnas.1005283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco M., Arena S., Renzone G., Scippa G.S., Lomaglio T., Verrillo F., Scaloni A., Marra M. Proteomic analysis of temperature stress-responsive proteins in Arabidopsis thaliana rosette leaves. Mol. Biosyst. 2013;9:1257–1267. doi: 10.1039/c3mb70137a. [DOI] [PubMed] [Google Scholar]
- Rogalski M., Schottler M.A., Thiele W., Schulze W.X., Bock R. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell. 2008;20:2221–2237. doi: 10.1105/tpc.108.060392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel J.B., Wilson I.W., Pogson B.J. Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 2002;130:1109–1120. doi: 10.1104/pp.005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban A.V. Nonphotochemical chlorophyll fluorescence quenching: mechanism and effectiveness in protecting plants from photodamage. Plant Physiol. 2016;170:1903–1916. doi: 10.1104/pp.15.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J., Gallois P., Delseny M. Accumulation and nuclear targeting of BnC24, a Brassica napus ribosomal protein corresponding to a mRNA accumulating in response to cold treatment. Plant Sci. 2000;156:35–46. doi: 10.1016/s0168-9452(00)00229-6. [DOI] [PubMed] [Google Scholar]
- Salih K.J., Duncan O., Li L., O'Leary B., Fenske R., Troesch J., Millar A.H. Impact of oxidative stress on the function, abundance and turnover of the Arabidopsis 80S cytosolic ribosome. Plant J. 2020 doi: 10.1111/tpj.14713. [DOI] [PubMed] [Google Scholar]
- Sanchez-Bermejo E., Zhu W., Tasset C., Eimer H., Sureshkumar S., Singh R., Sundaramoorthi V., Colling L., Balasubramanian S. Genetic architecture of natural variation in thermal responses of Arabidopsis. Plant Physiol. 2015;169:647. doi: 10.1104/pp.15.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Heinrichs L., Scossa F., Fernie A.R., Oelze M.L., Dietz K.J., Rothbart M., Grimm B., Flugge U.I., Hausler R.E. The essential role of sugar metabolism in the acclimation response of Arabidopsis thaliana to high light intensities. J. Exp. Bot. 2014;65:1619–1636. doi: 10.1093/jxb/eru027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz E., Tohge T., Zuther E., Fernie A.R., Hincha D.K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci. Rep. 2016;6:34027. doi: 10.1038/srep34027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J., Whitcomb S.J., Firmino A.A.P., Zuther E., Hincha D.K., Kopka J. Induced, imprinted, and primed responses to changing environments: does metabolism store and process information? Front. Plant Sci. 2019;10:106. doi: 10.3389/fpls.2019.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano N., Ling Y., Bahieldin A., Mahfouz M.M. Thermopriming reprograms metabolic homeostasis to confer heat tolerance. Sci. Rep. 2019;9:181. doi: 10.1038/s41598-018-36484-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Liu L., Wei Y., Li G., Yue X., An L. Metabolite profiling of adh1 mutant response to cold stress in Arabidopsis. Front. Plant Sci. 2017;7:2072. doi: 10.3389/fpls.2016.02072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speiser A., Haberland S., Watanabe M., Wirtz M., Dietz K.-J., Saito K., Hell R. The significance of cysteine synthesis for acclimation to high light conditions. Front. Plant Sci. 2015;5:776. doi: 10.3389/fpls.2014.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Devireddy A.R., Inupakutika M.A., Baxter A., Miller G., Song L., Shulaev E., Azad R.K., Shulaev V., Mittler R. Ultra-fast alterations in mRNA levels uncover multiple players in light stress acclimation in plants. Plant J. 2015;84:760–772. doi: 10.1111/tpj.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., Selbig J., Muller L.A., Rhee S.Y., Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998;118:1. doi: 10.1104/pp.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandereyken K., Van Leene J., De Coninck B., Cammue B.P.A. Hub protein controversy: taking a closer look at plant stress response hubs. Front. Plant Sci. 2018;9:694. doi: 10.3389/fpls.2018.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M.O., Moore M., Konig K., Pecher P., Alsharafa K., Lee J., Dietz K.J. Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell. 2014;26:1151–1165. doi: 10.1105/tpc.113.121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyse K., Pagter M., Zuther E., Hincha D.K. Deacclimation after cold acclimation—a crucial, but widely neglected part of plant winter survival. J. Exp. Bot. 2019;70:4595–4604. doi: 10.1093/jxb/erz229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lan P., Gao H., Zheng L., Li W., Schmidt W. Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics. 2013;14:783. doi: 10.1186/1471-2164-14-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ma K.B., Lu Z.G., Ren S.X., Jiang H.R., Cui J.W., Chen G., Teng N.J., Lam H.M., Jin B. Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 2020;20:86. doi: 10.1186/s12870-020-2292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhao Y., Zhang B. Efficient test and visualization of multi-set intersections. Sci. Rep. 2015;5:16923. doi: 10.1038/srep16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wang W.J., Zheng K.L., Gong X.D., Xu J.L., Huang J.R., Lin D.Z., Dong Y.J. The rice TCD11 encoding plastid ribosomal protein S6 is essential for chloroplast development at low temperature. Plant Sci. 2017;259:1–11. doi: 10.1016/j.plantsci.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Warner J.R., McIntosh K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W. Photosynthetic response to fluctuating environments and photoprotective strategies under abiotic stress. J. Plant Res. 2016;129:379–395. doi: 10.1007/s10265-016-0816-1. [DOI] [PubMed] [Google Scholar]
- Yang K., Yang J., Yi J. Nucleolar stress: hallmarks, sensing mechanism and diseases. Cell Stress. 2018;2:125–140. doi: 10.15698/cst2018.06.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Nakamura M., Yoneyama T., Nishida I. Starch-related α-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis. Plant Physiol. 2005;138:837–846. doi: 10.1104/pp.104.056374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandalinas S.I., Sengupta S., Burks D., Azad R.K., Mittler R. Identification and characterization of a core set of ROS wave-associated transcripts involved in the systemic acquired acclimation response of Arabidopsis to excess light. Plant J. 2019;98:126–141. doi: 10.1111/tpj.14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yuan H., Yang Y., Fish T., Lyi S.M., Thannhauser T.W., Zhang L., Li L. Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 2016;67:2731–2744. doi: 10.1093/jxb/erw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu X., Gaikwad K., Kou X., Wang F., Tian X., Xin M., Ni Z., Sun Q., Peng H. Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana. Plant Cell. 2017;29:1952–1969. doi: 10.1105/tpc.16.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wise R.R., Struck K.R., Sharkey T.D. Moderate heat stress of Arabidopsis thaliana leaves causes chloroplast swelling and plastoglobule formation. Photosynth Res. 2010;105:123–134. doi: 10.1007/s11120-010-9572-6. [DOI] [PubMed] [Google Scholar]
- Zhao C., Wang P., Si T., Hsu C.C., Wang L., Zayed O., Yu Z., Zhu Y., Dong J., Tao W.A. MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell. 2017;43:618–629.e5. doi: 10.1016/j.devcel.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J.K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016;171:2744–2759. doi: 10.1104/pp.16.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Zhang K., Sun Z., Yan M., Chen C., Zhang X., Tang Y., Wu Y. LNK1 and LNK2 corepressors interact with the MYB3 transcription factor in phenylpropanoid biosynthesis. Plant Physiol. 2017;174:1348–1358. doi: 10.1104/pp.17.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E., Juszczak I., Lee Y.P., Baier M., Hincha D.K. Time-dependent deacclimation after cold acclimation in Arabidopsis thaliana accessions. Sci. Rep. 2015;5:12199. doi: 10.1038/srep12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E., Schaarschmidt S., Fischer A., Erban A., Pagter M., Mubeen U., Giavalisco P., Kopka J., Sprenger H., Hincha D.K. Molecular signatures associated with increased freezing tolerance due to low temperature memory in Arabidopsis. Plant Cell Environ. 2019;42:854–873. doi: 10.1111/pce.13502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Values are provided as mean ± standard deviation (n ≥ 4 independent experiments). and the mean values correspond to the values provided in Figures 1 and S1 (Fv/Fm). Statistically significant differences compared to the control condition were assessed according the Student's t test (∗, p ≤ 0.05; ∗∗, p ≤ 0.01).
Metabolites were identified by GC-MS-TOF. We considered those metabolites as SAMs, which displayed in at least one time point an absolute log2 fold-change (FC) ≥ 1 compared with the initial value (0 min) according to the Student's t test with a false discovery rate (FDR) ≤ 0.05. Raw data are normalized to an internal standard and fresh weight. Missing values were replaced by either the average of the other replicates (single missing values) or the detection threshold (multiple missing values).
Transcriptome changes under standard conditions, HL, heat, and cold according to an absolute log2 fold change (FC) ≥ 1 to time point 0 min and a false discovery rate (FDR) ≤ 0.05 for the Student's t test. The symbol and brief description of each transcript is provided.
Transcriptional changes were qualified as significant based on absolute fold change ≥1 to the initial time point (0 min) and a false discovery rate (FDR) ≤ 0.05 for the Student's t test in at least one time point from the central phase of the kinetic (2 days acclimation to 15 min de-acclimation). Transcripts are listed with gene ID, symbol, and brief description.
Transcriptome changes during the central phase of the kinetics to HL, heat, and cold (2 days acclimation to 15 min de-acclimation) were compared by means of Venn diagrams to identify the common fraction of changes to the three treatments. The selected transcripts are listed along with the gene ID, symbol, and brief description.
Transcriptome changes under HL, heat, and cold conditions according to an absolute log2 fold change (FC) ≥ 1 to time point 0 min and a false discovery rate (FDR) ≤ 0.05 for the Student's t test in at least one time point were filtered by presence in the Gene Ontology term 0009507 “chloroplast.” The symbol and brief description of each transcript is provided.
Co-expression networks were investigated for nodes with a connectivity ≥ 100 edges (super-hubs) in each condition and listed along with the gene ID, symbol, and brief description.
Ribosomal proteins were extracted from MapMan and information on their gene ID, subcellular localization, assignment to subunit and regulatory trend at the fourth day of acclimation compared with the initial time point (0 mi). DOWN/UP means down-/up-regulated; -, not changed.
Data Availability Statement
The datasets generated during this study have been deposited to Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/): GSE125950.