Abstract
Cercospora leaf spot, caused by the fungal pathogen Cercospora beticola, is the most destructive foliar disease of sugar beet worldwide. This review discusses C. beticola genetics, genomics, and biology and summarizes our current understanding of the molecular interactions that occur between C. beticola and its sugar beet host. We highlight the known virulence arsenal of C. beticola as well as its ability to overcome currently used disease management strategies. Finally, we discuss future prospects for the study and management of C. beticola infections in the context of newly employed molecular tools to uncover additional information regarding the biology of this pathogen.
Taxonomy
Cercospora beticola Sacc.; Kingdom Fungi, Phylum Ascomycota, Class Dothideomycetes, Order Capnodiales, Family Mycosphaerellaceae, Genus Cercospora.
Host range
Well‐known pathogen of sugar beet (Beta vulgaris subsp. vulgaris) and most species of the Beta genus. Reported as pathogenic on other members of the Chenopodiaceae (e.g., lamb's quarters, spinach) as well as members of the Acanthaceae (e.g., bear's breeches), Apiaceae (e.g., Apium), Asteraceae (e.g., chrysanthemum, lettuce, safflower), Brassicaceae (e.g., wild mustard), Malvaceae (e.g., Malva), Plumbaginaceae (e.g., Limonium), and Polygonaceae (e.g., broad‐leaved dock) families.
Disease symptoms
Leaves infected with C. beticola exhibit circular lesions that are coloured tan to grey in the centre and are often delimited by tan‐brown to reddish‐purple rings. As disease progresses, spots can coalesce to form larger necrotic areas, causing severely infected leaves to wither and die. At the centre of these spots are black spore‐bearing structures (pseudostromata). Older leaves often show symptoms first and younger leaves become infected as the disease progresses.
Management
Application of a mixture of fungicides with different modes of action is currently performed although elevated resistance has been documented in most employed fungicide classes. Breeding for high‐yielding cultivars with improved host resistance is an ongoing effort and prudent cultural practices, such as crop rotation, weed host management, and cultivation to reduce infested residue levels, are widely used to manage disease.
Useful website
https://www.ncbi.nlm.nih.gov/genome/11237?genome_assembly_id=352037
Keywords: cercosporin, effector, fungicide resistance, secondary metabolite
The hemibiotrophic fungus Cercospora beticola applies various virulence strategies to infect sugar beet and is currently only managed in‐field through integrated practices.
![]()
1. DOMESTICATION HISTORY OF BETA VULGARIS AND THE IMPACT OF CERCOSPORA BETICOLA ON MODERN FARMING
The world currently relies on sugar beet (Beta vulgaris subsp. vulgaris) for approximately one‐fifth of its total sugar (ISO, 2018). While the other major source of sucrose, sugar cane, is grown in more tropical climates, sugar beet is largely grown in temperate regions with the northern tier states in the United States, France, Germany, and Russia being the top producers (FAOSTAT, 2019). All cultivated beets (fodder, chard, table beet, sugar beet) are regarded to be derived from wild Mediterranean sea beet (B. vulgaris subsp. maritima) (OECD, 2001). In the late 18th century, the German chemist Andreas Marggraf showed that sucrose in both white and red beetroot was chemically identical to the prohibitively expensive tropical cane sugar (Marggraf, 1748). Marggraf's student Franz Karl Achard noticed that conical white fodder beets had high sugar content and began to grow them for sugar production (Fischer, 1989). Achard is credited with establishing the sugar beet industry in Europe by building the first processing factory in Silesia (now Poland) in 1801. Sugar beet was subsequently commercialized in the United States in 1879 (Magnuson, 1918). Continuous breeding over the last 200 years has led to an increase in sugar content from 8% to more than 18% in cultivars grown today (Dohm et al., 2014). Additionally, the discovery of male sterile cytoplasm permitted the development of hybrid varieties for yield increase (Biancardi et al., 2010). Although breeding and improved cultivation practices yielded significant increases in sugar beet production and sugar yield in recent decades, many abiotic and biotic stresses remain that continue to affect sugar beet growth and ultimately attenuate sugar production worldwide.
The most common and destructive foliar disease of sugar beet globally is cercospora leaf spot (CLS) (Holtschulte, 2000). The disease was first described on Beta cicla in Italy by Saccardo (1876) but now has been identified across the globe wherever sugar beet is grown. CLS is most pernicious in warm, humid growing regions (Lartey et al., 2010), which make up almost one‐third of the total sugar beet production area in the United States (USDA‐NASS, 2010). The main adversity is the loss of recoverable sucrose, reaching almost 50% under uncontrolled moderate to high disease pressure (Lamey et al., 1987; Shane and Teng, 1992). Additional economic losses occur as a result of increased impurities that complicate sucrose recovery processes, leading to higher processing costs and reduced extractable sucrose (Shane and Teng, 1992). Diseased plants are also more susceptible to storage rot in winter storage piles (Smith and Ruppel, 1973). Large economic losses were seen in sugar beet crops in southern Germany in the late 1980s and early 1990s due to severe CLS epidemics (Wolf and Verreet, 2005). In the United States, Minnesota and North Dakota suffered losses from CLS in 1998 estimated at $113 million from yield reduction and fungicide application costs (Cattanach, 1999). In these areas, with conducive environmental conditions, it is crucial to appropriately apply fungicides otherwise substantial loss of foliar photosynthetic area can occur due to CLS (Figure 1a,b). The upper Midwest has seen epidemics in three out of the last four years in large part due to fungicide‐resistant Cercospora beticola populations that have resulted in several hundred million dollars in lost revenue (Mike Metzger, Minn‐Dak Farmers Cooperative, personal communication). A deeper understanding of the C. beticola–sugar beet interaction may allow for the development of innovative strategies to prevent the disease and ultimately cut much of these losses. Here, we discuss the biology of this pathogen and explore the recent advances in the molecular and genetic understanding of CLS.
Figure 1.
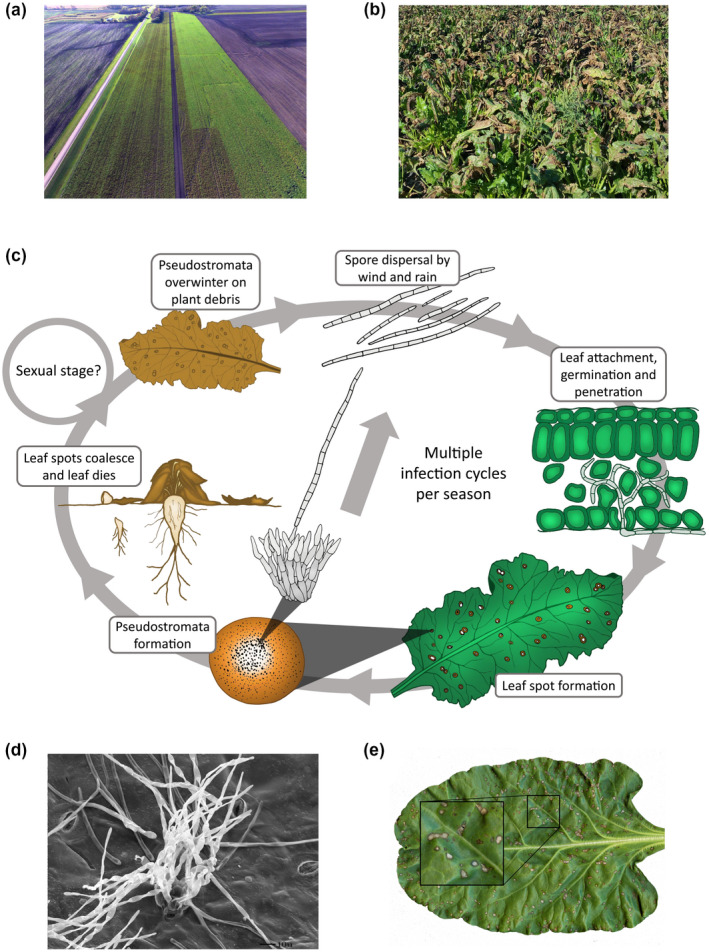
Cercospora leaf spot (CLS) of sugar beet. (a) Drone image highlighting the importance of fungicides for disease management. While the disease was well‐managed on the right side of the field, an abrupt halt in fungicide application ultimately resulted in increased CLS disease as evidenced by the brown colour noted on the left side of the field. (b) Extensive CLS disease in a sugar beet field. (c) Disease cycle of Cercospora beticola on sugar beet. Infection is initiated by airborne or splash‐dispersed conidia that penetrate the sugar beet leaf through stomata and give rise to intercellular hyphal growth. Leaf spots form on the leaves after the switch to necrotrophy, which typically occurs 7 days after infection. Pseudostromata develop in these lesions and asexually produce spores, leading to multiple infection cycles during the growing season. The pseudostromata are also the overwintering structures on plant debris at the end of the season. It is possible that C. beticola can sexually reproduce and produce ascospores, in a similar way to Zymoseptoria tritici, but this stage has not been observed. Extensive CLS disease in a sugar beet field. (d) Scanning electron micrograph exhibiting C. beticola conidiophores emerging from sugar beet. (e) CLS symptomology on sugar beet
2. INFECTION BIOLOGY
The causal agent of CLS, C. beticola, has the ability to complete several asexual cycles within a single season under conducive conditions (McKay and Pool, 1918; Nagel, 1945; Vereijssen et al., 2007). Between sugar beet growing seasons, the fungus survives primarily as desiccation‐resistant hyphal structures on infected plant residues within leaf substomatal cavities. These specialized overwintering structures are known as pseudostromata, or false stromata (conidia‐producing hyphae), because they are composed of both fungal tissue and remnants of host tissue (Eriksson, 1981). Pseudostromata can persist on plant debris for 2 years and have long been regarded as the primary inoculum sources for infection (Pool and McKay, 1916; Khan et al., 2008). However, many population studies have questioned the role of clonally reproducing primary inoculum (Groenewald et al., 2006, 2008; Knight et al., 2018, 2019). Other potential sources of initial inocula include dispersal of C. beticola‐infested plant material via tools or machinery (Knight et al., 2018, 2019), infested seed, windborne conidia, or stromata from other host plants (Khan et al., 2008; Franc, 2010; Skaracis et al., 2010; Tedford et al., 2018; Knight et al., 2020).
C. beticola is an ascomycete fungus only known to exist in the anamorphic (asexual) state (Crous et al., 2001; Groenewald et al., 2013). To reproduce, melanized conidiophores form from pseudostromata to produce conidia in the spring but this may be preceded by saprophytic, vegetative growth of fungal mycelia (Pool and McKay, 1916) (Figure 1c). Transmission electron microscopy has shown that bundles of 10–20 conidiophores (18–25 µm in diameter) often form on both upper and lower leaf surfaces and are produced from subepidermal or substomatal pseudostromata (Pons et al., 1985). Pseudostromata are three to six cells deep and up to eight to 10 cells wide. Conidiophores consist of one or two cells and are 10–25 μm long × 3–5 μm wide at the base (Figure 1d). Conidia are needle‐shaped (2–3 × 36–107 μm) and colourless with several cross‐walls (Weiland and Koch, 2004). The minimum requirements for conidial development are temperatures of at least 15°C and a relative humidity of 60% or higher (Pool and McKay, 1916; Solel and Minz, 1971a). Through wind, rain, irrigation, water splash, or insect transfer, spores are disseminated and make contact with the abaxial surface of sugar beet leaves or petioles to initiate infection (Lawrence and Meredith, 1970; Khan et al., 2007). Some studies have suggested that roots may also act as primary infection sites (Vereijssen et al., 2005), although the specific requirements for root infection remain elusive (Khan et al., 2008). Germination of conidia is optimal at high relative humidity (near 100%) and at temperatures of approximately 25‐°C (Ruppel, 1986; Khan et al., 2009). After germination, appressoria are produced, allowing hyphae to penetrate leaf tissue through stomata and spread intercellularly with no visible leaf symptoms (Rathaiah, 1977; Steinkamp et al., 1979). As the fungus switches to its necrotrophic phase, the production of phytotoxins and degradative enzyme activity leads to the necrotizing of infected cells (Steinkamp et al., 1979). Symptoms appear as circular spots 3–5 mm in size and tan to grey in colour, encircled by a tan‐brown to red‐purple border (Windels et al., 1998) (Figure 1e). Symptoms can develop on older leaves as quickly as 5 days after infection when there are favourable conditions of high humidity (>90%) and warm temperatures (day 27–32°C, night > 17°C) (Pool and McKay, 1916; Solel and Minz, 1971b). In the field, these characteristic CLS lesions are typically observed after sugar beet canopy closure (Khan et al., 2008). Pseudostromata formed within the lesions become the site for production of new conidia as early as 7 days after infection under favourable conditions (Jacobsen and Franc, 2009). Conidia are again disseminated by wind, rain splash, or insect transfer to initiate another infection cycle. Early studies show evidence for rain splash being the major factor in spore dispersal (Carlson, 1967) while studies by Khan et al. (2008) suggested that wind was the major dispersal factor for C. beticola inoculum because higher disease severity was observed on exposed plants when compared to plants in plastic cages or with ground cover. The distance that conidia can travel whilst retaining viability has not been reported, but genetic studies have provided evidence that long‐range dispersal has occurred (Groenewald et al., 2008; Vaghefi et al., 2017a; Knight et al., 2018).
3. POPULATION GENETICS
The population biology of C. beticola has been extensively explored to quantify genetic structure, gene flow, and genotypic and genetic diversity in space and time. Tools used to study C. beticola's evolutionary ecology have included random amplified polymorphic DNA, amplified fragment length polymorphisms, microsatellites, and single nucleotide polymorphisms (Groenewald et al., 2007; Turgay et al., 2010; Vaghefi et al., 2017b). A consistent theme in these studies has been to provide indirect evidence for sexual recombination, migration, and mutation. These findings have supported the development of hypotheses surrounding pathogen biology and movement with important implications for disease epidemiology and management. For example, the lack of a known teleomorph for C. beticola means a substantial gap exists in the knowledge surrounding primary inoculum sources and between‐field inoculum movement (Pethybridge et al., 2018).
C. beticola populations are generally characterized by high allelic, genetic, and genotypic diversity, typically coinciding with remarkably high phenotypic diversity (Ruppel, 1972; Moretti et al., 2004) that is often exhibited in in vitro colony morphology (Figure S1). Although C. beticola is heterothallic (self‐sterile) where any one isolate has either mating type (MAT) gene (MAT1‐1‐1 or MAT1‐2‐1) at the MAT1 locus, additionally each isolate has MAT1‐1‐1 and MAT1‐2‐1 exon fragments located at ostensibly random loci across the genome (Bolton et al., 2014). Interestingly, these MAT fragments could largely reconstitute a hypothetical fused organization of MAT genes, suggesting a homothallic (self‐fertile) ancestral state for C. beticola and other related species (Bolton et al., 2014). Ratios of isolates with different mating types (MAT1‐1 and MAT1‐2) within populations have been location‐specific. Isolates of one mating type have dominated populations from sugar beet in Iran (Bakhshi et al., 2011) and one production region within the United States (Obuya et al., 2011). In contrast, mating type ratios in equilibrium have been characterized in populations from sugar beet fields in Minnesota and North Dakota (Bolton et al., 2012c), and table beet in New York (Vaghefi et al., 2017c). Equal mating type ratios together with high genotypic diversity have supported the potential for sexual recombination within populations (Groenewald et al., 2006, 2008; Bolton et al., 2012c). Additionally, microsatellite alleles in linkage disequilibrium when combined with equal mating type ratios and high genotypic diversity in certain C. beticola populations from table beet provide evidence to suggest that a mixed reproductive mode is more likely (Vaghefi et al., 2016, 2017c; Knight et al., 2018).
Several studies have also quantified genetic homogeneity and genetic differentiation between C. beticola populations on variable spatial scales ranging from different hosts in the same field (Vaghefi et al., 2017c), fields within and between regions (Vaghefi et al., 2016, 2017b), and between continents (Groenewald et al., 2008; Vaghefi et al., 2017a; Knight et al., 2019). High levels of gene flow and low differentiation between C. beticola populations on different continents have supported evidence for genetic similarity and panmictic populations and the potential role of infested plant material dissemination in pathogen movement and initiation of epidemics (Groenewald et al., 2008; Vaghefi et al., 2017b; Knight et al., 2019). Knight et al. (2019) reported genetic diversity, differentiation, and relationships among 948 C. beticola isolates from 28 populations in eight regions and identified two major clusters. One cluster was specific to New York, with evidence of recent expansion and divergence, and another was common across North and South America, Eurasia, Hawaii, and selected states within the United States (North Dakota and Michigan) with shared origin (Knight et al., 2019). Human‐mediated dispersal of C. beticola‐infested plant material may be responsible for primary inoculum within individual seasons, while findings from studies that evaluated the temporal stability of populations within fields have challenged the role of overwintering inoculum in epidemic initiation (Knight et al., 2018, 2019).
4. POPULATION GENOMICS
Genome sequencing has provided valuable insight into the evolution and biology of diverse fungal plant pathogens (Soanes et al., 2007). Genome sequencing of C. beticola was initially aimed at identifying new polymorphic markers for genotyping of field isolates (Vaghefi et al., 2017b) and unravelling biosynthetic gene cluster evolution (de Jonge et al., 2018).
The genome size of C. beticola is comparable to other closely related Dothideomycetes, but the repeat content is considerably lower and includes less than 2% of the entire genome (de Jonge et al., 2018). In comparison, genomes of the two related species Mycosphaerella fijiensis and Cladosporium fulvum belonging to the same order Capnodiales have significantly higher repeat contents of 37% and 44%, respectively (Ohm et al., 2012). This indicates the presence of efficient genome defence mechanisms in C. beticola, such as repeat induced point mutations and DNA methylation, that may prevent the propagation of repetitive elements (Cambareri et al., 1989; Gladyshev, 2017).
Population genomic sequencing of some other fungal plant pathogens, including Leptosphaeria maculans and Zymoseptoria tritici, have demonstrated that repetitive elements can associate with the presence/absence variation of genes (Rouxel et al., 2011; Plissonneau et al., 2016). Despite the low repeat content in C. beticola (de Jonge et al., 2018), recent analyses of population genomic data have revealed a substantial number of genes showing presence/absence variation (Spanner and Bolton, unpublished data). It is possible that a dynamic repertoire of effector genes is driven by a coevolutionary arms race between plant and pathogen and facilitates the infection of C. beticola on different hosts.
We analysed North American field populations of C. beticola to assess the amount and distribution of genetic variation and infer about the recent demography of the pathogen. When analysing resequencing data from more than 100 field isolates, we identified approximately 500,000 single nucleotide polymorphisms from which we computed a mean nucleotide diversity of π = 0.0035 (Potgieter, Bolton and Stukenbrock, unpublished data). We found that π varies considerably along the C. beticola genome, including regions with a 10‐fold increase in genetic variation. Further analyses should scrutinize patterns of genetic variation along the genome to understand the functional relevance of variation “hotspots” and the underlying drivers of genetic variability. Research on the genome evolution of C. beticola could have great importance in the development of new management strategies, including the identification of new fungicide targets.
Population genomic data can also clarify the demographics within a species (Dutheil and Hobolth, 2012; Excoffier et al., 2013). Different demographic scenarios, such as population bottleneck and population expansion, affect the distribution of genetic variation in a population. A population bottleneck will significantly reduce genetic variation in the population while a population expansion will lead to an excess of newly derived mutations. Different measures, derived from the distribution of allele frequencies along the genome, reflect recent demographic events that have affected genetic variation in a population (Alcala et al., 2013). We used the population genomic data of C. beticola to compute the Tajima's D statistic (Tajima, 1989) along the genome to measure genetic variation at the DNA level via a standardized pairwise comparison of the number of nucleotide differences to the number of segregation sites. While the value varies significantly along chromosomes, we found an overall positive Tajima's D value of 1.1. Tajima's D > 0 reflects an excess of alleles of intermediate frequencies, which is expected under a scenario of population contraction. While additional analyses are required to infer the recent demography of C. beticola in North America, this positive Tajima's D value could indicate a recent loss of genetic variation in the pathogen population due to demographic changes. Recent demographic changes in populations of C. beticola may result from selection on the fungus imposed by fungicide treatment and resistant beet cultivars. It may also reflect the recent emergence of the pathogen on sugar beet as a new host. Comparative population genomic analyses can provide an opportunity to date different demographic events that have shaped genetic variation in the pathogen.
Population genomic sequencing of C. beticola from other continents and from wild hosts will furthermore provide important information on the centre of origin and the global population genetic structure of the pathogen, as well as patterns of gene flow and signatures of selection in agricultural and wild ecosystems.
5. DEVELOPMENT AND CHARACTERIZATION OF THE C. BETICOLA REFERENCE GENOME
We previously reported the C. beticola reference genome size to be 37 Mb (de Jonge et al., 2018). The final, high‐quality annotation contained 12,281 genes and 12,495 transcripts, encompassing 28,389 unique exons. On average, protein‐coding genes contained 2.3 exons and 1.3 introns that were approximately 780 and 73 bp long, respectively. A significant proportion of genes (4,384 genes or 35.7%) did not contain an intron and are referred to as single‐exon genes. We assigned both 5′ untranslated regions (UTRs) and 3′ UTRs to 7,147 genes, 5′ or 3′ UTR to 1,561 genes, and 3,573 genes did not have an assigned UTR. The average length of the 5′ UTR was 188 bp and that of the 3′ UTR 300 bp. Most intronic sequences were found in the coding regions (15,195 or 96%). We identified 446 introns (2.8% of all introns) in 5′ UTRs and 181 (1.2% of all introns) in 3′ UTRs, representing a significant under‐representation of 3′ UTR introns exemplified by the 25‐fold lower intron density (7.6 × 10−5 introns/mRNA transcripts) as compared to 5′ UTR introns (3.0 × 10−4 introns/mRNA transcripts). In Arabidopsis and some mammals, a pronounced under‐representation of 3′ UTR introns may be explained by a requirement for the nonsense‐mediated decay (NMD) pathway (Lejeune and Maquat, 2005; Chung et al., 2006). NMD is a cellular process that is involved in the detection and decay of mRNA transcripts that contain premature stop codons. Our results suggest that filamentous fungi may use a similar NMD pathway. In this regard it is interesting to note that an examination of the C. beticola proteome resulted in the identification of three important protein components of the exon junction complex, which is essential for NMD (Lejeune and Maquat, 2005). Examination of UTR characteristics in other related fungi based on available gene structures supports this view, albeit limited by the number as well as the accuracy of these gene structures.
A detailed overview of all gene model statistics, previously reported in de Jonge et al. (2018), is shown in Table 1 as well as a comparison of these statistics with five related plant pathogenic Dothideomycete fungi with well‐characterized genome sequences. These are Dothistroma septosporum (teleomorph Mycosphaerella pini), Z. tritici (previously known as Mycosphaerella graminicola), L. maculans, Parastagonospora nodorum (previously known as Stagonospora nodorum), and Pyrenophora tritici‐repentis as well as a more distantly related Eurotiomycete Aspergillus nidulans.
Table 1.
Cercospora beticola genome statistics and comparison to other related fungi
| Species a | Cbe | Cbr | Ccn | Dse | Ztr | Lma | Ptr | Pno | Ani |
|---|---|---|---|---|---|---|---|---|---|
| Assembly statistics | |||||||||
| Total assembly length (Mb) | 37.1 | 37.4 | 34.0 | 30.2 | 39.7 | 44.9 | 37.8 | 37.2 | 30.5 |
| Total length of gaps (Mb) | 3.2 | 0.3 | 0.0 | 0.1 | 0.0 | 1.1 | 0.6 | 0.2 | 0.7 |
| No. of scaffolds/contigs | 248 | 28,905 | 6,126 | 20 | 21 | 41 | 47 | 108 | 8 |
| NG50 scaffolds (no.) | 4 | 111 | 422 | 5 | 6 | 10 | 6 | 13 | 4 |
| LG50 scaffolds (Mbp) | 4.17 | 0.096 | 0.023 | 2.60 | 2.67 | 1.77 | 1.99 | 1.05 | 3.76 |
| NG95 scaffolds (no.) | 10 | 11,497 | 2,320 | 12 | 18 | 25 | 20 | 43 | 8 |
| GC content (%) | |||||||||
| Overall (excl. gaps) | 52.2 | 51.5 | 52.6 | 53.1 | 52.1 | 45.2 | 51.0 | 50.4 | 50.4 |
| Coding (CDS) | 53.8 | 53.9 | 54.3 | 54.6 | 55.6 | 54.1 | 53.6 | 54.6 | 53.4 |
| Repeat content (Mb) | 0.51 | ND | ND | 1.08 | 6.98 | 15.93 | 0.80 | 2.88 | 1.07 |
| Protein‐coding genes | |||||||||
| Protein‐coding genes (no.) | 12,281 | 11,972 | 11,556 | 12,580 | 10,951 | 12,469 | 12,169 | 12,380 | 10,680 |
| Mean gene length (bp) | 1,885 | 1,584 | 1,556 | 1,896 | 1,602 | 1,446 | 1,616 | 1,468 | 1,736 |
| Percentage coding | 68.4 | 51.1 | 52.9 | 79.3 | 44.2 | 41.2 | 52.9 | 49.1 | 62.2 |
| Mean gene density (no. genes/100 kb | 36.3 | 32.3 | 34.0 | 41.8 | 27.6 | 28.5 | 32.7 | 33.4 | 35.8 |
| Mean CDS length (bp) | 1,469 | 1,473 | 1,450 | 1,223 | 1,310 | 1,258 | 1,349 | 1,271 | 1,456 |
| Exons b | |||||||||
| No. of exons | 28,100 | 28,046 | 26,848 | 28,937 | 28,611 | 35,201 | 32,716 | 32,994 | 34,743 |
| Exons/gene | 2.3 | 2.3 | 2.3 | 2.3 | 2.6 | 2.8 | 2.7 | 2.7 | 3.3 |
| Mean exon length (bp) | 780 | 629 | 624 | 773 | 532 | 446 | 530 | 495 | 477 |
| Introns b | |||||||||
| Introns (no. introns) | 15,819 | 16,074 | 15,292 | 16,356 | 17,660 | 22,732 | 20,547 | 20,614 | 24,062 |
| Introns/gene | 1.3 | 1.3 | 1.3 | 1.3 | 1.6 | 1.8 | 1.7 | 1.7 | 2.3 |
| Mean intron length (bp) | 73 | 83 | 80 | 91 | 133 | 103 | 114 | 90 | 82 |
ND, not determined (ND).
Genomes are Cercospora beticola (Cbe), Cercospora berteroae (Cbr), Cercospora canescens (Ccn), Dothistroma septosporum (Dse), Zymoseptoria tritici (Ztr), Leptosphaeria maculans (Lma), Pyrenophora tritici‐repentis (Ptr), Parastagonospora nodorum (Pno), and Aspergillus nidulans (Ani).
Only considering the longest transcript (if alternatives exist).
5.1. Proteome characterization of C. beticola and related fungi
We used BLAST and InterProScan to obtain functional annotation for the full C. beticola proteome encompassing in total 12,495 proteins. BLASTp analyses of all C. beticola proteins against the NCBI nonredundant database identified significant similarity (E‐value ≤ 1e−10; percentage coverage of query ≥ 70%) for 9,580 proteins (77%) whereas similar analyses against a database containing all fungal protein sequences available at the Joint Genome Institute MycoCosm portal resulted in identification of 10,529 proteins (84%) with significant similarity. Similar results were obtained for the six other ascomycete species, ranging from 63% (L. maculans) to 86% (A. nidulans). InterProScan led to the identification of 12,965 Pfam domains, 5,039 SMART domains and 12,541 superfamily domains in the C. beticola proteome. Integration of the BLASTp and InterProScan results by BLAST2GO yielded gene ontology (GO) terms for 7,380 C. beticola proteins (59%).
To compare the proteomes of these seven ascomycete fungi including C. beticola, all proteins were clustered into 12,167 orthologous protein families based on all‐versus‐all BLASTp similarity via orthoMCL analysis (Li et al., 2003). From the resulting 12,167 ortholog clusters, we selected 3,554 that contain one protein sequence from each genome. We further filtered these protein clusters by retaining only those that contain single‐copy homologs in 39 ascomycete fungi using data from orthoDB (Waterhouse et al., 2012) for phylogenetic analyses, resulting in a reduced set of 850 protein families. Concatenated protein alignments were then used to build a well‐supported phylogenetic tree that clusters C. beticola most closely with D. septosporum and secondly to Z. tritici (Figure S2a). This phylogeny is in congruence with the majority of other studies using single‐ and multilocus analyses that place the genus Zymoseptoria adjacent to the Cercospora, Pseudocercospora, Dothistroma, and Sphaerulina genera (Goodwin et al., 2001; de Wit et al., 2012; Ohm et al., 2012; Arango Isaza et al., 2016; Chang et al., 2016; de Jonge et al., 2018). Orthologous protein information was also used to further enhance the functional annotation of the C. beticola proteins. More specifically, GO terms from orthologous A. nidulans proteins, for which extensive GO‐term annotation is available through AMIGO (Carbon et al., 2009), were replicated to C. beticola proteins. Using this approach, we were able to assign a GO term to a combined total of 8,608 C. beticola proteins.
5.2. Secondary metabolite cluster expansion in C. beticola
Secondary metabolites (SMs) are small bioactive molecules produced by many organisms including bacteria, plants, and fungi (Stringlis et al., 2018). They are abundant in filamentous fungi in which they play crucial roles in the establishment of specific ecological niches. Unlike primary metabolites, SMs are not essential for fungal growth, development, or reproduction but contribute to adaptation (e.g., protection against environmental stresses) or pathogenicity. Many enzymes are involved in the synthesis of a single SM. Polyketide synthases (PKSs) and nonribosomal peptide synthases (NRPSs), which catalyse the elongation of polyketides and oligopeptides, respectively, are among the most prominent and well‐studied SM biosynthetic genes. C. beticola is known to produce a number of SMs, including cercosporin (Daub and Ehrenshaft, 2000; de Jonge et al., 2018) and beticolin (Goudet et al., 1998). The genome of C. beticola possesses 15 type I PKSs, 23 NRPSs, three PKS‐NRPS hybrids, six terpene cyclases, and one demethylallyl tryptophan synthase (de Jonge et al., 2018). The closely related dothideomycetes D. septosporum and Z. tritici possess a significantly lower number of both type I PKSs (3 and 10, respectively) and NRPSs (5 and 8, respectively). The expansion of NRPSs in C. beticola is particularly notable when compared to L. maculans, P. nodorum, and P. tritici‐repentis (this study) and C. fulvum (de Wit et al., 2012). The expansion in NRPSs is further exemplified by a high number of NRPS‐related Pfam domains (PF00501, PF00550, PF00668, and PF13193). Interestingly, the predicted C. beticola SM clusters are preferentially found in subtelomeric regions (Figure 2a), a common feature of fungal SM clusters (Palmer and Keller, 2010). Probably, the observed co‐regulation of expression among genes within SM clusters is achieved by epigenetic forces such as chromatin‐level control (Palmer and Keller, 2010). Chromosomal location of gene clusters near centromeres and telomeres possibly correlates with regions that experience facultative heterochromatin, that is, large regions that can efficiently be silenced as well as activated by chromatin‐mediating factors (Palmer and Keller, 2010).
Figure 2.
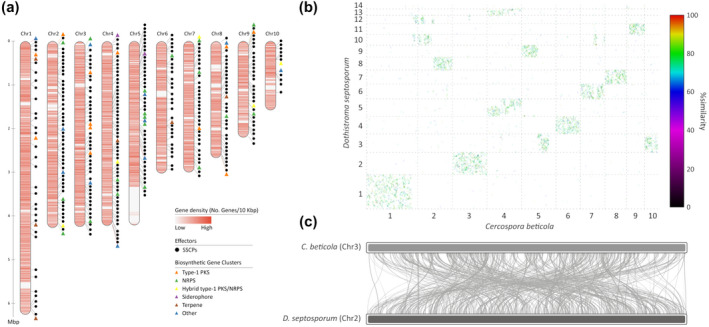
(a) Schematic representation of the 10 largest (pseudo)chromosomes and/or scaffolds of Cercospora beticola, highlighting the density of genes (genes/10 kb; ranging from 0 [white] to 10 [dark red]), the location of candidate effectors (the small, secreted cysteine‐rich proteins or SSCPs; black circles) and the type and location of biosynthetic gene clusters (coloured triangles) for secondary metabolites. (b) Whole‐genome alignment between C. beticola and Dothistroma septosporum highlighting extensive chromosome mesosynteny. Whole‐genome, protein sequence‐based alignments between C. beticola and D. septosporum reveal many short syntenic regions that are spread over each scaffold or chromosome pair, exemplary for mesosynteny. (c) Example of the shuffled homologous regions shared between chromosomes 3 and 2 from C. beticola and D. septosporum, respectively. (a) and (c) prepared using RIdeogram (Hao et al., 2019); (b) prepared by promer in MUMMER3 (Kurtz et al., 2004)
5.3. Definition and annotation of the C. beticola secretome
The arsenal of potentially secreted proteins (i.e., the secretome) among plant pathogens includes key pathogenicity molecules that are generally referred to as effectors (Kamoun, 2007). To identify candidate effectors, we scanned the proteome of C. beticola for proteins that are predicted to be secreted and lack transmembrane domains. We identified 1,087 such proteins of which 333 exhibited additional effector characteristics including small size (≤300 amino acids) and cysteine‐rich (≥2 cysteine residues) (Figure 2a; SSCPs). Analysis of genomic localization revealed an under‐representation of SSCPs on chromosome 1 (p < .0001, chi‐square test, Figure 2a) that might be an indication of genome compartmentalization, often referred to as the two‐speed genome (Raffaele et al., 2010). Of the 333 candidate effectors, 62 (approximately 19%) had no BLASTp hits in the nonredundant database from NCBI nor in the collection of 347 fungal proteomes obtained from the MycoCosm fungal genomics portal and are therefore considered C. beticola‐specific.
5.4. C. beticola synteny with related fungi
Conservation of sequence between chromosomes or scaffolds of different species or strains is typically referred to as synteny. Through gene loss, inversions, translocations, or other chromosomal rearrangements, synteny between homologous chromosomes (i.e., of common descent) can be broken. However, major chromosomal rearrangements are rare when comparing distantly related eukaryotes, and as a result large collinear regions with similar genes in similar order and orientation can be found across these species. This kind of “global” chromosome conservation is known as macrosynteny. Such macrosynteny is only rarely observed among species in the fungal kingdom. In fungi, a type of synteny known as microsynteny is common in which small segments of up to 10 conserved genes can be found. Moreover, a phenomenon termed mesosynteny in which homologous chromosomes display significant conservation of gene content but not gene order is typical in the ascomycete fungi and especially prominent in the Dothideomycete class to which C. beticola belongs (Hane et al., 2011). Conservation of gene content but not of gene order can be attributed to a large number of intrachromosomal rearrangements such as inversions and a very limited number of interchromosomal rearrangements. Hane et al. (2011) hypothesized that a high frequency of inversions might occur during meiosis, although this does not explain why Dothideomycetes in particular, and only ascomycete fungi, display such a striking conservation of chromosome gene content because many species in this class do not have a known sexual stage.
To investigate whether C. beticola chromosomes also display mesosynteny to related fungal species, we aligned C. beticola chromosomes with those of the closely related fungus, D. septosporum (Figure 2b,c). Notably, strict conservation of chromosome content but not order can be observed, the hallmark for mesosynteny. Considering a limited number of intrachromosomal rearrangements, it is expected that some collinear segments remain because insufficient rearrangements have occurred to randomize the complete gene order within a chromosome. On the other hand, some collinear segments might be the result of continuing selection pressure to keep particular genes involved in the same or similar biological processes tightly clustered on the genome, as we have previously observed is the case for biosynthetic gene clusters for cercosporin (de Jonge et al., 2018; Ebert et al., 2019) and melanin (Ebert et al., 2019).
6. C. BETICOLA EFFECTOR REPERTOIRE
Effectors can be described as microbially secreted molecules such as proteins, SMs, and small RNAs that contribute to niche colonization (Rovenich et al., 2014; Snelders et al., 2018). This definition implies that effector functions are not necessarily restricted to interaction with a host plant but also include involvement in microbial competition and nutrition acquisition (Fatima and Senthil‐Kumar, 2015; Snelders et al., 2018) that may have roles during a saprophytic growth period. Effectors that are secreted during host colonization may have the ability to modulate host biochemistry and physiology, including defence responses, to facilitate host colonization (de Jonge et al., 2011). The type of effectors produced depends on the lifestyle of the pathogen and stage of infection. Biotrophic fungi obtain their nutrients from living plant tissues and their effectors typically act to inhibit plant defence responses and facilitate nutrient acquisition. However, necrotrophic fungi secrete toxins and proteins to evoke host cell death and induce nutrient release (Koeck et al., 2011). The hemibiotrophic lifestyle of C. beticola begins with a latent symptomless (biotrophic) phase where invasive hyphae grow within the host plant followed by a fine‐tuned transition to necrotrophy. The two different phases of growth probably require different effector repertoires. Future time‐course transcriptome studies of sugar beet infection may help to elucidate how phase transition occurs and identify candidate effectors contributing to biotrophy or necrotrophy, as has been shown in other pathosystems (Gan et al., 2013; Rudd et al., 2015; Zuluaga et al., 2016). To date, both SM and proteinaceous effectors have been identified in C. beticola and are discussed below.
7. CERCOSPORIN
Cercosporin is a light‐activated, nonhost‐specific toxin produced by most Cercospora species (Daub and Ehrenshaft, 2000). Mutant lines that are unable to produce this SM experience a virulence penalty, implicating cercosporin as a virulence factor for C. beticola, C. nicotianae, C. kikuchii, C. coffeicola, and Cercospora cf. flagellaris (Callahan et al., 1999; Choquer et al., 2005; Weiland et al., 2010; Rezende et al., 2020; Souza et al., 2019). However, there are Cercospora species that naturally lack the ability to produce this toxin and yet are virulent phytopathogens, demonstrating that cercosporin is not solely necessary for pathogen success in the genus (Weiland et al., 2010; Swart et al., 2017). The metabolic pathway genes involved in cercosporin formation are organized in a cercosporin toxin biosynthesis (CTB) cluster. While cercosporin was thought to be a unique feature of fungi that belong to the Cercospora genus, it has been shown that the CTB cluster experienced duplications and multiple horizontal gene transfers across a variety of taxa, including many Colletotrichum species, and Colletotrichum fioriniae has been confirmed to also produce this potent toxin (de Jonge et al., 2018).
7.1. Toxicology
The photosensitizing nature of various perylenequinones, including cercosporin, has long been known (Brockmann et al., 1950; Weiss et al., 1957; Yamazaki et al., 1975). The essential structural feature responsible for photodynamic activity is the 3,10‐dihydroxy‐4,9‐perylenequinone chromophore (Hudson et al., 1997), which allows absorption of visible and near‐UV light that elevates the perylenequinone to an electronically excited triplet state (Foote, 1968; 1976). Once in this activated triplet state, two types of reactions are known to occur (DeRosa and Crutchley, 2002; Guedes and Eriksson, 2007): the excited perylenequinone can react with oxygen either indirectly (type I reaction) through a reducing substrate or directly (type II reaction) (Figure S3). Interaction with an electron donor leads to the formation of free radicals or radical ions that in turn react with oxygen to produce reactive oxygen species (ROS) such as H2O2 and free radical forms such as O2 •−, HO2 •, and OH•. In a direct interaction between a triplet perylenequinone and oxygen, energy can be transferred from the excited triplet state perylenequinone to oxygen, resulting in an excited singlet state of oxygen, also known as “singlet oxygen” (1O2). Both type I and II reactions yield ROS that at high concentrations are harmful to cells as they can cause lipid peroxidation as well as protein and DNA damage (Blokhina et al., 2003; Birben et al., 2012).
7.2. Biosynthesis
While extensive research on the CTB pathway has shed light on selected pathway steps, it has not yet been possible to determine a full biosynthesis scheme due to the extreme instability of most pathway intermediates and the potential occurrence of feedback inhibition (Newman and Townsend, 2016). To date, 12 genes have been found to be part of the CTB biosynthetic pathway, involved in either cercosporin production or export (Figure 3a). The iterative, nonreducing PKS CTB1 is essential for cercosporin production and acts as the keystone enzyme to initiate the biosynthetic process (Choquer et al., 2005; Crawford and Townsend, 2010; Newman et al., 2012). CTB1 harbours six functional domains that work together to form nor‐toralactone, the first intermediate in the cercosporin assembly line using one acetyl‐CoA and 6 × malonyl‐CoA units as substrate (Choquer et al., 2005; Newman et al., 2012; Newman and Townsend, 2016). Next, the nor‐toralactone intermediate is processed to toralacatone and subsequently to cercoquinone C by CTB3, a predicted O‐methyltransferase FAD‐dependent monooxygenase (Dekkers et al., 2007; Newman and Townsend, 2016; de Jonge et al., 2018). Further processing of this intermediate may be mediated by CTB2, CTB6, and CTB11/CTB12, which are hypothesized to methylate, reduce, and dimerize the molecule, respectively (Staerkel et al., 2013; Newman and Townsend, 2016). However, the corresponding intermediates have not been directly observed but are rather logically inferred. The CTB5 and/or CTB7 gene products are hypothesized to prime the cercosporin intermediate for methylenedioxy bridge formation by CTB9 and CTB10 to yield the final cercosporin molecule (Chen et al., 2007; Newman and Townsend, 2016; Swart et al., 2017; de Jonge et al., 2018). The CTB8 gene product is a Zn(II)Cys6 zinc finger transcription factor that is not directly involved in the modification of the toxin itself but is responsible for mediation of the CTB gene cluster expression (Chen et al., 2007). Lastly, the gene cluster holds two major facilitator superfamily (MFS) transmembrane transporters, CTB4 and the cercosporin facilitator protein (CFP) (Chen et al., 2007; Choquer et al., 2007; de Jonge et al., 2018). C. nicotianae CTB4 mutant strains displayed a reduction in cercosporin production by at least 35% while cercosporin accumulated in fungal mycelium and was not secreted into the medium, suggesting that CTB4 mediates cercosporin export (Choquer et al., 2007). When stimulated by high light conditions, CTB4 mutants secreted a dark brown compound of unknown nature that quickly diffused into the solid medium. Interestingly, a CTB4 homolog is also present in the putative phleichrome biosynthetic pathway, but is lacking in the predicted elsinochrome and hypocrellin clusters (Ebert et al., 2019). As elsinochrome and hypocrellin are secreted by Elsinoë fawcettii and Shiraia bambusicola, respectively, despite the lack of a CTB4 homolog, the question arises whether CTB4 is indeed solely responsible for toxin export or whether other transporter proteins can functionally substitute toxin export in the absence of CTB4 (Choquer et al., 2007). The CFP gene is incorporated in the CTB cluster of C. beticola (de Jonge et al., 2018) and is hypothesized to partially provide tolerance to cercosporin (auto‐resistance) via toxin export (Callahan et al., 1999).
Figure 3.
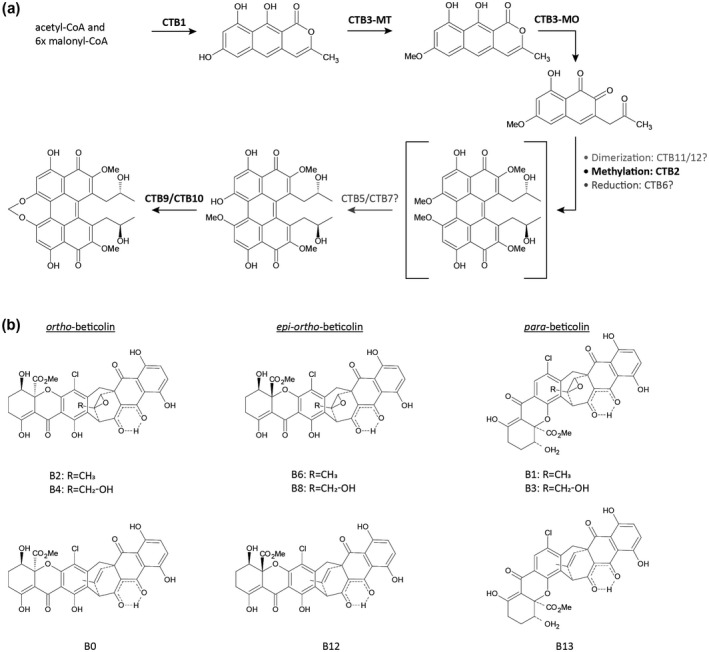
(a) Preliminary scheme of the cercosporin biosynthetic pathway consisting of 12 clustered genes. The polyketide synthase CTB1 forms nor‐toralactone, which is processed to cercoquinone C by CTB3 methyltransferase (CTB3‐MT) and monooxygenase (CTB3‐MO). Further processing of this cercosporin intermediate might be mediated by CTB2, CTB6, CTB11, and CTB12 to yield the cercosporin intermediate displayed in the square brackets, which has not been directly observed but is rather logically inferred. CTB5 and CTB7 are hypothesized to prime the cercosporin molecule for methylenedioxy bridge formation by CTB9 and CTB10. (b) Beticolin structures and isoforms. Beticolins are structurally related but can differ by residues (R) and isoforms (ortho‐, epi‐ortho‐, or para‐beticolin). Beticolins that carry the same residues are able to transform into each other by switching isomery. For example, ortho‐beticolin B2 can transform into the epi‐ortho‐beticolin B6 or para‐beticolin B1 based on the position of the oxygen and chlorine atoms
7.3. Auto‐resistance
It is essential for fungi that produce antifungal SMs to protect themselves from their own toxin. The phenomenon of avoiding self‐intoxication is known as auto‐resistance (AR). Genes with potential AR roles have been identified in highly conserved toxin biosynthesis gene clusters, with no direct involvement in toxin production. Three self‐resistance strategies have been highlighted in the literature: toxin export, detoxification, and duplication of the toxin target (Keller, 2015). However, to this date only toxin export via transporter proteins and detoxification of toxic compounds have been found in cercosporin‐producing fungi.
Protection by toxin efflux relies on the transportation of toxic substances from the inside of a cell to the outside through membrane transport proteins. Transporters involved in self‐resistance mainly belong to the MFS and ABC superfamilies (Cannon et al., 2009; Costa et al., 2014).
The MFS transporter protein, CFP, was first identified in C. kikuchii and has been shown to be involved in cercosporin AR as targeted gene disruption of CFP resulted in mutant strains with increased sensitivity to exogenous cercosporin (Callahan et al., 1999). With the recent discovery that the CTB cluster consists of additional genes (de Jonge et al., 2018), it was possible to demonstrate that the MFS transporter homolog in C. beticola, CbCFP, is incorporated in the cluster and flanked on both sides by genes necessary for cercosporin production (de Jonge et al., 2018). Mutants that lack CbCFP are more sensitive to exogenous cercosporin in vitro (Ebert and Bolton, unpublished data), suggesting that this AR mechanism is conserved in the Cercospora genus.
Besides toxin export, Cercospora species are known to have a second mechanism of AR through reductive detoxification of the cercosporin molecule (Daub et al., 1992, 2000; Leisman and Daub, 1992; Sollod et al., 1992). It has been shown that stably methylated and acetylated reduced cercosporin derivatives absorb less light and generate significantly less singlet oxygen (1O2) compared to wild‐type cercosporin (Leisman and Daub, 1992). Further investigation has revealed that the cell surface of cercosporin‐resistant strains is surrounded by a reducing environment (Leisman and Daub, 1992), showing that resistant fungi can reduce proximate cercosporin into its less reactive form and maintain this detoxification so long as the molecule is nearby. Although specific genes have been reported in other fungi that are linked to active cercosporin detoxification (Ververidis et al., 2001; Panagiotis et al., 2007), no CTB genes have yet been affiliated with AR via cercosporin reduction.
8. BETICOLINS
Beticolins are a group of nonhost‐specific phytotoxins of which 20 members (B0 to B19) have been identified to be produced by C. beticola (Milat and Blein, 1995; Goudet et al., 1998, 2000) and the hoary alyssum (Berteroa incana) pathogen Cercospora berteroae (Assante et al., 1977). Alternative names for these toxins, such as Gelbe Fration (Schlösser, 1962; Langfelder et al., 2003), Cercospora beticola toxin (CBT) (Assante et al., 1977), and cebetins (Jalal et al., 1992; Robeson and Jalal, 1993), arose due to simultaneous research efforts by different groups and limited data concerning their structure during early research. Later, analyses of their chemical structures revealed that beticolins are structurally closely related (Figure 3b). All have a chlorine atom attached to the central aromatic ring, while their octocyclic basic structure is composed of two subunits: a partially hydrogenated anthraquinone and a partially hydrogenated xanthone that are connected through a seven‐membered ring (Ducrot et al., 1994, 1996; Simon‐Plas et al., 1996; Goudet et al., 1998, 2000). Structural differences between beticolins are due to different isomeric configurations (ortho‐, para‐, or epi‐ortho‐) and variable residues (Milat et al., 2010). Interestingly, beticolins can switch isomery. For example, ortho‐beticolin B2 is able to transform into the para‐beticolin B1 or epi‐ortho‐beticolin B6 and vice versa (Ducrot et al., 1994). Early research on their biological function indicated that beticolins have antibacterial and phytotoxic properties (Schlösser, 1962). However, necrosis formation in plants upon beticolin application was only induced in the presence of light. Later it was found that due to their ability to form complexes with Mg2+, beticolins inhibit tumoral cell growth in mice (Ding et al., 1996, 2001), interfere with H+‐ATPase activity (Gomès et al., 1996a, 1996b, 1996c; Simon‐Plas et al., 1996), and are able to incorporate themselves into lipid bilayers to form ion channels with poor ion selectivity (Goudet et al., 1998, 1999, 2000). The latter property led to the classification of beticolins as ion channel‐forming toxins (Goudet et al., 2000). While chemical structures and biological activity have been evaluated in recent decades, the biosynthetic pathway of these toxins is unknown, therefore it is currently not possible to assess to what extent beticolin production and associated phytotoxic effects contribute to C. beticola virulence.
9. MELANIN
Fungal phytopathogen melanin production has been implicated in appressorial penetration of host plants and pathogenesis as well as being an integral component of the cell wall that can be useful for tolerating environmental stresses (Langfelder et al., 2003; Liu and Nizet, 2009). Recent research mining the C. beticola genome for novel PKS genes has revealed the secondary metabolite production of melanin (Ebert et al., 2019). Phylogenetic analysis of the CbPKS1 gene belonging to the DHN‐melanin clade grouped C. beticola with other well‐characterized melanin biosynthetic clusters from various ascomycetes. Additionally, whole‐cluster homology with other PKS genes that are involved in melanin production provided further evidence that these genes may be functional. Knockout mutants in melanin CbPKS1 resulted in albino phenotypes that did not affect the PKS biosynthetic cluster involved in cercosporin production (Ebert et al., 2019). Interestingly, a gene adjacent to CbPKS1 encoding for a predicted tetra‐hydroxynaphthalene reductase (Cb4HNR) involved with melanin biosynthesis has been shown to be induced in fungicide‐resistant C. beticola strains (Bolton et al., 2016). This proposes a role for the SM production of melanin that is probably an adaptation involved in survival in a challenging environment.
10. PROTEIN EFFECTORS
Effector proteins are known to be employed by a broad variety of plant pathogenic fungi to evade detection by the host during colonization. Upon pathogen recognition, the plant will initiate defence responses such as the production of chitinases that target fungal hyphae for degradation. To shield the fungal cell wall from plant‐derived chitinases, the biotrophic fungus C. fulvum, which causes leaf mould on tomato, secretes the virulence factor CfAvr4 (van Esse et al., 2007). This effector can bind to chitin in the fungal cell wall and protect fungal hyphae from hydrolysis by plant chitinases (van den Burg et al., 2003, 2004; van Esse et al., 2007; Chang and Stergiopoulos, 2015). A homologous gene was discovered in C. beticola (Stergiopoulos et al., 2010) and Cercospora cf. flagellaris (Rezende et al., 2020), and in vitro studies found that the CbAvr4 and Cfla‐Avr4 gene products could functionally bind to chitin (Mesarich et al., 2016; Rezende et al., 2020). As chitin‐binding appears to be a conserved biological trait between CfAvr4 and Avr4 homologs, it is hypothesized that Avr4 homologs including CbAvr4 also share the CfAvr4 function of shielding fungal hyphae from lysis by plant chitinases.
Effector protein identification through comparative genomics has served as a useful tool to detect another C. beticola effector named CbAve1. CbAve1 is a homolog of VdAve1 (Avirulence on Ve1 tomato), an effector secreted by the vascular wilt pathogen Verticillium dahliae. In V. dahliae the VdAve1 effector is recognized by the tomato cell surface‐localized immune receptor Ve1 and confers resistance to the pathogen. In the absence of Ve1, VdAve1 has been shown to contribute to fungal virulence but the mechanism remains elusive (de Jonge et al., 2012). In the C. beticola–B. vulgaris system, CbAve1 is expressed during infection and its product acts as a virulence factor (Boshoven et al., 2015).
Due to its hemibiotrophic lifestyle, it was hypothesized that C. beticola also secretes proteinaceous effectors that promote its necrotrophic phase. Using a phenotype‐based forward genetics approach, a proteinaceous virulence factor named CbNIP1 (Cercospora beticola necrosis‐inducing protein 1) was identified due to its ability to necrotize sugar beet and Nicotiana benthamiana leaves (Ebert et al., 2018). Interestingly, CbNIP1’s ability to induce necrosis within 48 hr was highly regulated by light. While other necrosis‐inducing proteins such as ZtNIP1 and ZtNIP2 of the wheat pathogen Z. tritici require light for full functionality (M’Barek et al., 2015), CbNIP1 was most active in complete darkness as exposure of CbNIP1‐infiltrated sugar beet leaves with a 12 hr light‐dark cycle led initially to chlorosis formation that gradually turned necrotic over time (Ebert et al., 2018). Furthermore, CbNIP1 appears to contribute to necrotic symptom development, as up‐regulated CbNIP1 expression in planta correlates with necrotic lesion appearance. At present, the mode of action of CbNIP1 and its location during infection remains unknown.
11. DISEASE MANAGEMENT
The integrated management of CLS comprises cultural practices, moderate host resistance, and the timely application of fungicides. Cultural practices aim to reduce the amount of initial inoculum for the following season through rotation with nonhost crops, tillage (burying infested debris), and avoiding planting next to fields previously sown with sugar beets. Epidemiological models regarding the disease progress of CLS have been established to predict disease severity and timing of fungicide application (Rossi and Battilani, 1991; Windels et al., 1998; Pitblado and Nichols, 2005; Racca and Jörg, 2007). Fungicides should be sprayed early in a protective manner to avoid the development of conidial populations, which can infect new unprotected foliage. Although there have been several studies regarding the potential of different bacteria and fungi as biocontrol agents for CLS, including Trichoderma and Bacillus subtilis (Collins and Jacobsen, 2003; Galletti et al., 2008), there have been no current reports of their success as a management tool in the field. Alternatively, the presence of several microbial groups has been correlated with disease incidence in sugar beet fields and these microbes may be useful as biological markers for predicting disease outbreaks (Kusstatscher et al., 2019).
11.1. Host resistance
The improvement in CLS resistance in sugar beet varieties over the last few decades has been a concerted effort by geneticists and breeders. Wild sea beet, B. vulgaris subsp. maritima, has long been a source of CLS resistance genes (Rossi, 1995). Other wild Beta relatives, such as Beta procumbens, have displayed nonhost resistance but are sexually incompatible with B. vulgaris (Panella and Frese, 2000). Inheritance of CLS resistance in sugar beet lines after introgression from wild sea beet is complex and has low heritability (Smith and Ruppel, 1974), while at the same time incorporation into high‐yielding commercial sugar beet hybrids remains a challenge (Smith and Campbell, 1996). Promisingly, recent field trials in Germany showed that several European sugar beet varieties with CLS resistance lacked a yield penalty in the absence of disease and had better economic performance than susceptible varieties (Vogel et al., 2018). CLS resistance is typically managed by at least four identifiable quantitative trait loci (QTLs) (Smith and Gaskill, 1970; Nilsson et al., 1999; Setiawan et al., 2000; Taguchi et al., 2011). Precise mapping of resistance QTLs helps marker‐assisted selection (MAS) in breeding programmes to introgress CLS resistance (Taguchi et al., 2011). The more precise the mapping, the higher the chances of breaking the potential linkage between CLS resistance and unfavourable traits. The underlying gene products can also be identified and used as molecular markers to identify alleles associated with resistance (Hunger et al., 2003). Previously, monogenic resistance was identified to race C2 of C. beticola in a sugar beet cultivar (Lewellen and Whitney, 1976) but the resistance was shown to be unstable (Koch and Jung, 2000). No other examples of monogenic resistance to CLS have been described since in sugar beet. Current and future breeding efforts for CLS resistance can exploit the reference‐quality 567 Mb genome sequence of sugar beet with 27,421 transcript‐supported genes published by Dohm et al. (2014). An additional reference‐quality genome was recently developed for a different sugar beet variety with focused annotation of nucleotide‐binding (NB‐ARC), leucine‐rich repeat (NLR) disease resistance genes, including 231 tentative NB‐ARC loci (Funk et al., 2018). When comparing these loci to validated resistance genes from monocots and eudicots, there appeared to be extensive B. vulgaris‐specific subfamily expansions. Draft genomes of Beta patula and sea beet were also recently released (del Río et al., 2019) and represent valuable resources for sugar beet breeding research.
11.2. Molecular basis of host defence
Although few studies have focused on the molecular basis of resistance, there is some published work examining plant defences upon C. beticola infection. The interaction between sugar beet and C. beticola begins with an initial defence response by the plant up‐regulating phenylalanine ammonia‐lyase (PAL) involved in the various biosynthetic pathways for many plant‐related SM compounds (i.e., lignins, flavonoids, and phytoalexins) used during biotic attacks (Liang et al., 1989). These plant defence mechanisms have been found to be suppressed by C. beticola via an interaction with a pathogen‐induced molecule on the PAL core promoter (Schmidt et al., 2004). Later it was discovered that on initial infection with the pathogen and through mid‐ to late‐stages of disease development, the plant hormone abscisic acid (ABA) was elevated (Schmidt et al., 2008). ABA was also found to reduce PAL gene expression in sugar beet through an unknown mechanism (Schmidt et al., 2008). ABA has been found to interfere with biotic stress signalling in other plants, including via suppression of PAL transcription and activity, which negatively impact disease resistance (Mauch‐Mani and Mauch, 2005).
Additional research into the defence response of sugar beet cultivars to CLS showed that three cultivars with varying resistance genotypes (susceptible, polygenic partial resistance, or monogenic resistance) differed in timing and strength of defence reactions on infection (Weltmeier et al., 2011). In all three cultivars, genes were activated in hormone production (ethylene, jasmonic acid, and gibberellin), lignin and alkaloid synthesis, signalling, and pathogenesis‐related (PR) genes by the time symptoms had appeared. The monogenic resistant genotype (resistant to C. beticola isolate C2) displayed strong defences (PR and WRKY gene expression) 1 day after inoculation and there was no significant increase in C. beticola biomass. The partial resistance genotype had a stronger defence response than the susceptible genotype and a 50% reduction in C. beticola biomass, but the pathogen was still able to infect and cause disease when there was late initiation of defence responses at 15 days after inoculation.
12. FUNGICIDE USAGE AND RESISTANCE DEVELOPMENT
The use of fungicides has been an integral part of CLS management primarily due to the lack of effective nonchemical alternatives. There are two main types of chemistries available for disease management: protectant fungicides with broad‐spectrum activity and systemic fungicides that target a specific site in the fungus. Of the former, the most commonly used are the ethylene bisdithiocarbamate (EBDC, Fungicide Resistance Action Committee or FRAC Group M03) class of fungicides, copper‐based fungicides (FRAC group M01), and, in the United States, the organotin class of compounds (FRAC Group 30) such as triphenyltin hydroxide and triphenyltin acetate. The three main classes of systemic fungicides that have been employed globally are the benzimidazoles (FRAC Group 1), triazoles (sterol demethylation inhibitors or DMIs, FRAC Group 3), and quinone outside inhibitors (QoIs, FRAC Group 11).
The continued efficacy of these fungicide classes has been marred by the emergence of resistant strains in C. beticola populations over the last few decades (Figure 4). C. beticola resistance has been noted to occur after widespread and repeated use of the same fungicide classes (Giannopolitis, 1978; Secor et al., 2010; Rosenzweig et al., 2020). Other factors contributing to the development of fungicide resistance are the polycyclic nature of this pathogen, its high rate of sporulation and common spray programmes being used over large areas for disease management (Dekker, 1986). The rotated use of different fungicide classes has been implemented to suppress selection for fungicide‐resistant C. beticola strains. Systemic fungicides (such as DMIs) are also commonly mixed with a protectant fungicide for higher efficacy, reduced costs, and as an additional step in resistance management (Ioannidis, 1994).
Figure 4.
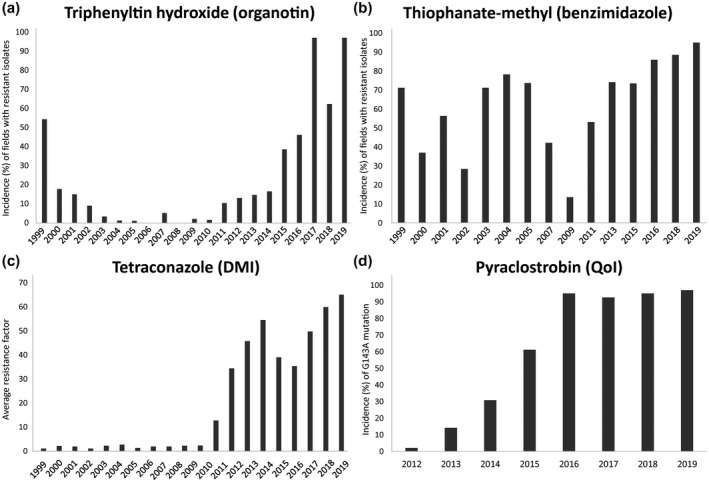
Fungicide resistance surveys for Cercospora beticola in the Red River Valley region. (a) Incidence (percentage) of fields sampled annually from 1999 to 2019 with isolates resistant to the organotin fungicide triphenyltin hydroxide at 1 µg/ml. (b) Incidence (percentage) of fields sampled from 1999 to 2019 with resistance to the benzimidazole fungicide thiophanate‐methyl at 5 µg/ml. (c) Average annual resistance factor values to the demethylation inhibitor (DMI) fungicide tetraconazole for isolates sampled annually from 1999 to 2019, where the resistance factor values are the calculated EC50 values divided by the baseline sensitivity values. (d) Incidence (percentage) of sampled isolates annually from 2012 to 2019 harbouring the G143A mutation in cytochrome b, conferring resistance to the quinone outside inhibitor (QoI) fungicide pyraclostrobin
With the emergence of resistance to most available fungicide chemistries, many recent studies have focused on characterizing the molecular basis of resistance. For fungicides with single target sites, such as the benzimidazoles, QoIs and DMIs, target genes can be sequenced and compared in both sensitive and resistant isolates to identify probable causal mutations.
12.1. FRAC group 30
Triphenyltin acetate was used in Europe in the 1970s, while triphenyltin hydroxide was used extensively in the United State throughout the 1980s (Windels et al., 1998). Tolerance to triphenyltin fungicides quickly emerged in Greece (Giannopolitis, 1978), Serbia (Marić et al., 1984), North Dakota, and Minnesota (Bugbee, 1995; Campbell et al., 1998). However, it was noted that triphenyltin‐resistant strains of C. beticola were less competitive than sensitive strains in the absence of this fungicide (Giannopolitis and Chrysayi‐Tokousbalides, 1980). Furthermore, annual surveys of triphenyltin hydroxide resistance in C. beticola isolates from central North America's Red River Valley (RRV) region have also suggested that a fitness penalty is associated with triphenyltin hydroxide resistance (Secor et al., 2010). Recent surveys of this region have shown high incidence of triphenyltin resistance (97% of isolates surveyed in 2017) but the severity of resistance was low (average spore germination rate was still <30%), perhaps suggesting that organotins may still be effective fungicides (Secor et al., 2017). Organotins act by inhibiting ATP synthase activity to stop oxidative phosphorylation in mitochondria but the genetic basis of resistance in fungi is unknown (Gadd, 2000). Although one of the most effective fungicide groups in use, organotins are no longer permitted for use within the European Union because of their associated consumer risks (Risk and Policy Analysts Limited, 2005).
12.2. FRAC group 1
Benzimidazole fungicides were implemented in the early 1970s. The first report of economic losses due to benzimidazole resistance in C. beticola populations was in Greece in 1973 (Georgopoulos and Dovas, 1973), followed by appearances in other production areas worldwide such as the United States (Ruppel and Scott, 1974; Bugbee, 1982), China (Dafang and Shuzhi, 1982), and India (Pal and Mukhopadhyay, 1983). EBDC and DMI fungicides were subsequently introduced to manage these resistant populations alongside the organotins. Benzimidazoles inhibit microtubule assembly during mitosis by binding to β‐tubulin subunits (Davidse, 1986). Sequencing of the target β‐tubulin gene identified a glutamic acid to alanine amino acid change at codon 198 (designated E198A) associated with high benzimidazole resistance in multiple populations of C. beticola in the United States and Europe (Davidson et al., 2006; Trkulja et al., 2013). An additional β‐tubulin mutation, phenylalanine to tyrosine at codon 167 (designated F167Y), has been found in low‐to‐moderate resistant isolates from Serbia (Trkulja et al., 2013), causing F167Y isolates to be more sensitive to low temperatures while E198A isolates had no detectable fitness penalty.
12.3. FRAC group 3
Another important class of fungicides used to manage C. beticola are DMIs. They have both protective and curative activity against Cercospora spp. and low levels of phytotoxicity (Brown et al., 1986; Dahmen and Staub, 1992). Although DMIs were initially thought to have a moderate risk of resistance development (Brown et al., 1986), C. beticola resistance has now been found in Europe (Karaoglanidis et al., 2001a), Morocco (El Housni et al., 2018), Canada (Trueman et al., 2017), and the United States (Secor et al., 2010; Bolton et al., 2012b; Rosenzweig et al., 2020). Resistance to DMIs is observed as a near‐continuum, ranging from high to low EC50 values (Karaoglanidis and Ioannidis, 2010). C. beticola isolates with EC50 values of greater than 1 ppm caused significantly more disease on sugar beet after application of a DMI fungicide than isolates with EC50 values below 1 ppm, implicating 1 ppm as a reasonable threshold value for DMI resistance (Bolton et al., 2012b). In the 2017 RRV region survey, 25.9% of tested C. beticola isolates were resistant (EC50 > 1 ppm) to tetraconazole while 47.1% of the same isolates were resistant to another DMI difenoconazole, which suggests that there is not strict cross‐resistance within the DMIs (Secor et al., 2017) and supports the earlier findings of Karaoglanidis and Thanassoulopoulos (2003). One study suggested that there may be some fitness penalties associated with DMI resistance, namely reduced virulence and spore production (Karaoglanidis et al., 2001b), but this has yet to be evidenced in field surveys.
The mechanism of resistance to DMIs is typically more complex than to benzimidazoles and QoIs. DMIs target the lanosterol 14α‐demethylase CYP51, which is a cytochrome P450 enzyme catalysing a key step in the fungal ergosterol biosynthesis pathway. Without the synthesis of the cell membrane sterol ergosterol, there is inhibition of fungal cell growth. Resistance can occur not only through target site modifications of CYP51, but also through overexpression of CYP51, increased active efflux of DMIs, and multiple copies of the target CYP51 gene (Leroux et al., 2007; Ziogas and Malandrakis, 2015). In the RRV region of the United States, CbCYP51 was overexpressed in several C. beticola isolates with EC50 values >1 ppm, in both quantitative PCR and RNA‐Seq studies, but the genetic mechanism underpinning this expression is unknown (Bolton et al., 2012a, 2016). No evidence has been found for alternative splicing or differential methylation of CbCYP51 between sensitive and resistant isolates (Bolton et al., 2012a). Interestingly, a silent mutation at codon 170 has been identified to be present only in highly resistant C. beticola isolates (EC50 values >50 ppm) from northern Greece (Nikou et al., 2009) and the RRV region of the United States (EC50 values >20 ppm) (Obuya et al., 2015). Recently, nonsynonymous polymorphisms in CbCYP51 have also been discovered that appear to be linked to DMI resistance (Trkulja et al., 2017; Shrestha et al., 2020). The amino acid substitutions L144F, I309T, I387M, and Y464S in isolates from the RRV region of the United States (Spanner and Bolton, unpublished data) are all associated with DMI EC50 values >1 ppm.
12.4. FRAC group 11
The QoI class of fungicides were introduced in 1996 and first used for CLS in 2002, proving to be highly effective fungicides against C. beticola (Karadimos et al., 2005; Secor et al., 2010). Pathogen surveys in Europe (Birla et al., 2012; Piszczek et al., 2018), Morocco (El Housni et al., 2018), Japan (Kayamori et al., 2020), Canada (Trueman et al., 2013), and the United States (Secor et al., 2010; Kirk et al., 2012) have indicated the rapid development of resistance to QoIs, which appears to be stable. In 2017, 89.1% of C. beticola isolates surveyed in the RRV region of the United States were resistant to the QoI pyraclostrobin and therefore its use is no longer recommended for CLS management in the region (Secor et al., 2017).
QoIs act by binding the quinol oxidation site of the cytochrome bc1 complex in the mitochondria, which disrupts ATP production (Fernández‐Ortuño et al., 2008). The membrane protein cytochrome b forms the core of the complex and is encoded by the cytochrome b (cytb) gene. Similar to other fungi (Fernández‐Ortuño et al., 2008), QoI‐resistant isolates of C. beticola found to date have the substitution of glycine by alanine at codon 143 (designated G143A) (Birla et al., 2012; Bolton et al., 2013; Trkulja et al., 2017; Piszczek et al., 2018).
The identification of mutations underlying resistance to fungicide classes enables the rapid detection of resistance via PCR methods. Real‐time PCR methods are already being employed for QoI resistance in annual surveys of C. beticola isolates (Malandrakis et al., 2011; Bolton et al., 2013) and methods have been developed to detect benzimidazole and DMI‐resistant isolates (Nikou et al., 2009; Trkulja et al., 2013; Rosenzweig et al., 2015; Shrestha et al., 2020). A method of amplifying DNA under isothermal conditions has been developed and termed loop‐mediated isothermal amplification (LAMP) (Notomi et al., 2015). This tool may eventually allow for fungicide resistance profiling of a C. beticola field population prior to spraying to best determine the appropriate chemical regime.
13. CONCLUSIONS AND FUTURE PERSPECTIVES
Future studies of CLS disease in sugar beet can exploit the wealth of genetic and genomic resources that have become available in recent years for both C. beticola (de Jonge et al., 2018) and its hosts (Dohm et al., 2014; del Río et al., 2019). The development of gene‐editing technologies such as CRISPR‐Cas9 (Doudna and Charpentier, 2014), which also revolutionized genetic editing in a broad variety of filamentous fungi as highlighted in multiple reviews (Idnurm and Meyer, 2018; Schuster and Kahmann, 2019; Song et al., 2019; Vicente et al., 2019), could be employed to investigate gene function via allele replacement in addition to gene knockouts. This would facilitate the identification of important genes and mutations for virulence in C. beticola. Breeding for effective host resistance to CLS with minimal yield penalty will continue to be important and may be expedited by gene‐editing techniques if effective gene targets are identified. Additionally, because cercosporin is a virulence factor for the fungus, transferring a cassette of fungal‐derived cercosporin AR genes to sugar beet could be a method to establish durable resistance in the host. Molecular advances will continue to help us understand additional aspects of CLS disease. The elucidation of the molecular basis of fungicide resistances is allowing us to better monitor and manage resistant populations in the field so that we can maintain the efficacy of current available fungicides. It will be imperative in the future to identify key primary sources of inoculum and establish biological mechanisms used by C. beticola to generate genetic diversity. Ultimately, improved knowledge of host–pathogen interactions will aid in successful integrated management of CLS.
Supporting information
ACKNOWLEDGEMENTS
The authors thank Dr Michael Metzger (Minn‐Dak Farmers Cooperative) for images in Figure 1a and b and Dr John Weiland (USDA – ARS) for the image in Figure 1d. The contribution by S. Pethybridge was supported by the United States Department of Agriculture National Institute of Food and Agriculture Hatch project NYG‐625424 and the Federal Capacity Funds Initiative Project 2018‐19‐113 managed by Cornell AgriTech at The New York State Agricultural Experiment Station, Cornell University, New York. The M. Bolton laboratory is supported by USDA project 3060‐21000‐044 and grants from the Sugarbeet Research & Education Board of Minnesota and North Dakota, and the Beet Sugar Development Foundation.
Rangel LI, Spanner RE, Ebert MK, et al. Cercospora beticola: The intoxicating lifestyle of the leaf spot pathogen of sugar beet. Molecular Plant Pathology. 2020;21:1020–1041. 10.1111/mpp.12962
Lorena I. Rangel, Rebecca E. Spanner, and Malaika K. Ebert contributed equally to this work.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- Alcala, N. , Streit, D. , Goudet, J. and Vuilleumier, S. (2013) Peak and persistent excess of genetic diversity following an abrupt migration increase. Genetics, 193, 953–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango Isaza, R.E. , Diaz‐Trujillo, C. , Dhillon, B. , Aerts, A. , Carlier, J. , Crane, C.F. et al (2016) Combating a global threat to a clonal crop: banana black Sigatoka pathogen Pseudocercospora fijiensis (synonym Mycosphaerella fijiensis) genomes reveal clues for disease control. PLoS Genetics, 12, e1005876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assante, G. , Locci, R. , Camarda, L. , Merlini, L. and Nasini, G. (1977) Screening of the genus Cercospora for secondary metabolites. Phytochemistry, 16, 243–247. [Google Scholar]
- Bakhshi, M. , Arzanlou, M. and Babai‐Ahari, A. (2011) Uneven distribution of mating type alleles in Iranian populations of Cercospora beticola, the causal agent of Cercospora leaf spot disease of sugar beet. Phytopathologia Mediterranea, 50, 101–109. [Google Scholar]
- Biancardi, E. , McGrath, J.M. , Panella, L.W. , Lewellen, R.T. and Stevanato, P. (2010) Sugar beet In: Bradshaw J. (Ed.) Root and Tuber Crops. 1, New York, NY, USA: Springer Science & Business Media, pp. 173–219. [Google Scholar]
- Birben, E. , Sahiner, U.M. , Sackesen, C. , Erzurum, S. and Kalayci, O. (2012) Oxidative stress and antioxidant defense. World Allergy Organization Journal, 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birla, K. , Rivera‐Varas, V. , Secor, G.A. , Khan, M.F. and Bolton, M.D. (2012) Characterization of cytochrome b from European field isolates of Cercospora beticola with quinone outside inhibitor resistance. European Journal of Plant Pathology, 134, 475–488. [Google Scholar]
- Blokhina, O. , Virolainen, E. and Fagerstedt, K.V. (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Annals of Botany, 91, 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, M.D. , Birla, K. , Rivera‐Varas, V. , Rudolph, K.D. and Secor, G.A. (2012a) Characterization of CbCyp51 from field isolates of Cercospora beticola . Phytopathology, 102, 298–305. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , Rivera‐Varas, V. , del Río Mendoza, L.E. , Khan, M.F. and Secor, G.A. (2012b) Efficacy of variable tetraconazole rates against Cercospora beticola isolates with differing in vitro sensitivities to DMI fungicides. Plant Disease, 96, 1749–1756. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , Secor, G.A. , Rivera, V. , Weiland, J.J. , Rudolph, K. , Birla, K. et al (2012c) Evaluation of the potential for sexual reproduction in field populations of Cercospora beticola from USA. Fungal Biology, 116, 511–521. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , Rivera, V. and Secor, G. (2013) Identification of the G143A mutation associated with QoI resistance in Cercospora beticola field isolates from Michigan, United States. Pest Management Science, 69, 35–39. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , de Jonge, R. , Inderbitzin, P. , Liu, Z. , Birla, K. , Van de Peer, Y. et al (2014) The heterothallic sugarbeet pathogen Cercospora beticola contains exon fragments of both MAT genes that are homogenized by concerted evolution. Fungal Genetics and Biology, 62, 43–54. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , Ebert, M.K. , Faino, L. , Rivera‐Varas, V. , de Jonge, R. , Van de Peer, Y. et al (2016) RNA‐sequencing of Cercospora beticola DMI‐sensitive and ‐resistant isolates after treatment with tetraconazole identifies common and contrasting pathway induction. Fungal Genetics and Biology, 92, 1–13. [DOI] [PubMed] [Google Scholar]
- Boshoven, J.C. , Ebert, M.K. , Song, Y. , Rovenich, H. , Rojas‐Padilla, E. , Bolton, M.D. et al (2015) Effector Biology of the Sugar Beet Pathogen Cercospora Beticola. Ch 3: Homologs of Verticillium Dahliae Effector Ave1 Contribute to Virulence of Fungal Pathogens of Diverse Plant Hosts. (PhD Dissertation), Wageningen University, Wageningen, Netherlands. [Google Scholar]
- Brockmann, H. , Weber, E. and Sander, E. (1950) Fagopyrin, ein photodynamischer Farbstoff aus Buchweizen (Fagopyrum esculentum). Naturwissenschaften, 37, 43–43. [Google Scholar]
- Brown, M. , Waller, C. , Charlet, C. and Palmieri, R. (1986) The use of flutriafol based fungicides for the control of sugar beet diseases in Europe. Presented at the 1986 British Crop Protection Conference Pests and Diseases, 3, 1055–1061. [Google Scholar]
- van den Burg, H.A. , Spronk, C.A. , Boeren, S. , Kennedy, M.A. , Vissers, J.P. , Vuister, G.W. et al (2004) Binding of the Avr4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein‐protein interactions: the chitin‐binding site of Avr3 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin‐binding domain. Journal of Biological Chemistry, 279, 16786–16796. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A. , Westerink, N. , Francoijs, K.‐J. , Roth, R. , Woestenenk, E. , Boeren, S. et al (2003) Natural disulfide bond‐disrupted mutants of Avr4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf‐4‐mediated resistance, but retain their chitin binding ability. Journal of Biological Chemistry, 278, 27340–27346. [DOI] [PubMed] [Google Scholar]
- Bugbee, W. (1982) Storage rot of sugar beet. Plant Disease, 66, 871–873. [Google Scholar]
- Bugbee, W. (1995) Cercospora beticola tolerant to triphenyltin hydroxide. Journal of Sugarbeet Research, 32, 167–174. [Google Scholar]
- Callahan, T.M. , Rose, M.S. , Meade, M.J. , Ehrenshaft, M. and Upchurch, R.G. (1999) CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Molecular Plant‐Microbe Interactions, 12, 901–910. [DOI] [PubMed] [Google Scholar]
- Cambareri, E.B. , Jensen, B.C. , Schabtach, E. and Selker, E.U. (1989) Repeat‐induced GC to AT mutations in Neurospora . Science, 244, 1571–1575. [DOI] [PubMed] [Google Scholar]
- Campbell, L. , Smith, G. , Lamey, H. and Cattanach, A. (1998) Cercospora beticola tolerant to triphenyltin hydroxide and resistant to thiophanate methyl in North Dakota and Minnesota. Journal of Sugarbeet Research, 35, 29–41. [Google Scholar]
- Cannon, R.D. , Lamping, E. , Holmes, A.R. , Niimi, K. , Baret, P.V. , Keniya, M.V. et al (2009) Efflux‐mediated antifungal drug resistance. Clinical Microbiology Reviews, 22, 291–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon, S. , Ireland, A. , Mungall, C.J. , Shu, S. , Marshall, B. , Lewis, S. et al (2009) AmiGO: online access to ontology and annotation data. Bioinformatics, 25, 288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, L. (1967) Relation of weather factors to dispersal of conidia of Cercospora beticola (Sacc). Journal of the American Society of Sugar Beet Technologists, 14, 319. [Google Scholar]
- Cattanach, A. (1999) Managing Cercospora leaf spot tolerance or resistance to fungicide. American Crystal Sugar Company, Ag. Notes, #354. [Google Scholar]
- Chang, T.‐C. , Salvucci, A. , Crous, P.W. and Stergiopoulos, I. (2016) Comparative genomics of the Sigatoka disease complex on banana suggests a link between parallel evolutionary changes in Pseudocercospora fijiensis and Pseudocercospora eumusae and increased virulence on the banana host. PLoS Genetics, 12, e1005904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, T.C. and Stergiopoulos, I. (2015) Inter‐ and intra‐domain horizontal gene transfer, gain–loss asymmetry and positive selection mark the evolutionary history of the CBM14 family. FEBS Journal, 282, 2014–2028. [DOI] [PubMed] [Google Scholar]
- Chen, H.‐Q. , Lee, M.‐H. and Chung, K.‐R. (2007) Functional characterization of three genes encoding putative oxidoreductases required for cercosporin toxin biosynthesis in the fungus Cercospora nicotianae . Microbiology, 153, 2781–2790. [DOI] [PubMed] [Google Scholar]
- Choquer, M. , Dekkers, K.L. , Chen, H.‐Q. , Cao, L. , Ueng, P.P. , Daub, M.E. et al (2005) The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae . Molecular Plant‐Microbe Interactions, 18, 468–476. [DOI] [PubMed] [Google Scholar]
- Choquer, M. , Lee, M.‐H. , Bau, H.‐J. and Chung, K.‐R. (2007) Deletion of a MFS transporter‐like gene in Cercospora nicotianae reduces cercosporin toxin accumulation and fungal virulence. FEBS Letters, 581, 489–494. [DOI] [PubMed] [Google Scholar]
- Chung, B.Y. , Simons, C. , Firth, A.E. , Brown, C.M. and Hellens, R.P. (2006) Effect of 5'UTR introns on gene expression in Arabidopsis thaliana . BMC Genomics, 7, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, D.P. and Jacobsen, B.J. (2003) Optimizing a Bacillus subtilis isolate for biological control of sugar beet Cercospora leaf spot. Biological Control, 26, 153–161. [Google Scholar]
- Costa, C. , Dias, P.J. , Sá‐Correia, I. and Teixeira, M.C. (2014) MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Frontiers in Physiology, 5, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, J.M. and Townsend, C.A. (2010) New insights into the formation of fungal aromatic polyketides. Nature Reviews Microbiology, 8, 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous, P.W. , Kang, J.‐C. and Braun, U. (2001) A phylogenetic redefinition of anamorph genera in Mycosphaerella based on ITS rDNA sequence and morphology. Mycologia, 93, 1081–1101. [Google Scholar]
- Dafang, H. and Shuzhi, W.X.Z. (1982) Studies on resistance of Cercospora beticola to benzimidazole fungicides. Journal of Plant Protection, 2, 11. [Google Scholar]
- Dahmen, H. and Staub, T. (1992) Protective, curative, and eradicant activity of difenoconazole against Venturia inaequalis, Cercospora arachidicola, and Alternaria solani . Plant Disease, 76, 774–777. [Google Scholar]
- Daub, M.E. and Ehrenshaft, M. (2000) The photoactivated Cercospora toxin cercosporin: contributions to plant disease and fundamental biology. Annual Review of Phytopathology, 38, 461–490. [DOI] [PubMed] [Google Scholar]
- Daub, M.E. , Leisman, G.B. , Clark, R.A. and Bowden, E.F. (1992) Reductive detoxification as a mechanism of fungal resistance to singlet oxygen‐generating photosensitizers. Proceedings of the National Academy of Sciences of the United States of America, 89, 9588–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub, M.E. , Li, M. , Bilski, P. and Chignell, C.F. (2000) Dihydrocercosporin singlet oxygen production and subcellular localization: a possible defense against cercosporin phototoxicity in Cercospora . Photochemistry and Photobiology, 71, 135–140. [DOI] [PubMed] [Google Scholar]
- Davidse, L.C. (1986) Benzimidazole fungicides: mechanism of action and biological impact. Annual Review of Phytopathology, 24, 43–65. [Google Scholar]
- Davidson, R. , Hanson, L. , Franc, G. and Panella, L. (2006) Analysis of β‐tubulin gene fragments from benzimidazole‐sensitive and ‐tolerant Cercospora beticola . Journal of Phytopathology, 154, 321–328. [Google Scholar]
- Dekker, J. (1986) Preventing and Managing Fungicide Resistance. US National Research Council Committee on Strategies for the Management of Pesticide‐Resistant Pest Populations, Pesticide resistance: Strategies and Tactics for Management. Washington, DC: National Academy Press, pp. 347–354. [Google Scholar]
- Dekkers, K.L. , You, B.‐J. , Gowda, V.S. , Liao, H.‐L. , Lee, M.‐H. , Bau, H.‐J. et al (2007) The Cercospora nicotianae gene encoding dual O‐methyltransferase and FAD‐dependent monooxygenase domains mediates cercosporin toxin biosynthesis. Fungal Genetics and Biology, 44, 444–454. [DOI] [PubMed] [Google Scholar]
- del Río, Á.R. , Minoche, A.E. , Zwickl, N.F. , Friedrich, A. , Liedtke, S. , Schmidt, T. et al (2019) Genomes of the wild beets Beta patula and Beta vulgaris ssp. maritima . The Plant Journal, 99, 1242–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRosa, M.C. and Crutchley, R.J. (2002) Photosensitized singlet oxygen and its applications. Coordination Chemistry Reviews, 233, 351–371. [Google Scholar]
- Ding, G. , Maume, G. , Milat, M.L. , Humbert, C. , Blein, J.P. and Maume, B.F. (1996) Inhibition of cellular growth and steroid 11β‐hydroxylation in RAS‐transformed adrenocortical cells by the fungal toxins beticolins. Cell Biology International, 20, 523–530. [DOI] [PubMed] [Google Scholar]
- Ding, G. , Maume, G. , Osman, H. , Padieu, M. , Milat, M. , Humbert, C. et al (2001) Effects of 12 beticolins, Cercospora beticola toxins, on proliferation of RAS‐transformed adrenocortical cell. Acta Pharmacologica Sinica, 22, 769–776. [PubMed] [Google Scholar]
- Dohm, J.C. , Minoche, A.E. , Holtgräwe, D. , Capella‐Gutiérrez, S. , Zakrzewski, F. , Tafer, H. et al (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature, 505, 546–549. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A. and Charpentier, E. (2014) The new frontier of genome engineering with CRISPR‐Cas9. Science, 346, 1258096. [DOI] [PubMed] [Google Scholar]
- Ducrot, P.‐H. , Einhorn, J. , Kerhoas, L. , Lallemand, J.‐Y. , Milat, M.‐L. , Blein, J.‐P. et al (1996) Cercospora beticola toxins. Part XI1: isolation and structure of beticolin 0. Tetrahedron Letters, 37, 3121–3124. [Google Scholar]
- Ducrot, P.‐H. , Lallemand, J.‐Y. , Milat, M.‐L. and Blein, J.‐P. (1994) The yellow toxins produced by Cercospora beticola. Part VIII: Chemical equilibrium between beticolins; structures of minor compounds: Beticolin 6 and beticolin 8. Tetrahedron Letters, 35, 8797–8800. [Google Scholar]
- Dutheil, J.Y. and Hobolth, A. (2012) Ancestral population genomics In: Anisimova M. (Ed.), Evolutionary Genomics. Clifton, NJ, USA: Humana Press, pp. 293–313. [Google Scholar]
- Ebert, M.K. , Spanner, R.E. , De Jonge, R. , Smith, D.J. , Holthusen, J. , Secor, G.A. et al (2019) Gene cluster conservation identifies melanin and perylenequinone biosynthesis pathways in multiple plant pathogenic fungi. Environmental Microbiology, 21, 913–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, M.K. , Wang, X. , Friesen, T.L. , de Jonge, R. , Neubauer, J.D. , Secor, G.A. , et al (2018) Effector Biology of the Sugar Beet Pathogen Cercospora beticola. Ch 4: Identification of Cercospora beticola Necrosis‐Inducing Effector CbNIP1 (PhD Dissertation), Wageningen University, Wageningen, Netherlands. [Google Scholar]
- El Housni, Z. , Ezrari, S. , Tahiri, A. , Ouijja, A. and Lahlali, R. (2018) First report of benzimidazole, DMI and QoI‐insensitive Cercospora beticola in sugar beet in Morocco. New Disease Reports, 38, 17. [Google Scholar]
- Eriksson, O. (1981) The families of bitunicate ascomycetes. Nordic Journal of Botany, 1, 800–800. [Google Scholar]
- van Esse, H.P. , Bolton, M.D. , Stergiopoulos, I. , de Wit, P.J. and Thomma, B.P. (2007) The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Molecular Plant‐Microbe Interactions, 20, 1092–1101. [DOI] [PubMed] [Google Scholar]
- Excoffier, L. , Dupanloup, I. , Huerta‐Sánchez, E. , Sousa, V.C. and Foll, M. (2013) Robust demographic inference from genomic and SNP data. PLoS Genetics, 9, e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT (2019) Food and Agriculture Organization of the United Nations. World Sugar Beet Production 1961–2018. Rome, Italy: Available at: http://www.fao.org/faostat/en/#data/QC/visualize [Accessed 19 June 2020]. [Google Scholar]
- Fatima, U. and Senthil‐Kumar, M. (2015) Plant and pathogen nutrient acquisition strategies. Frontiers in Plant Science, 6, 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Ortuño, D. , Torés, J.A. , De Vicente, A. and Pérez‐García, A. (2008) Mechanisms of resistance to QoI fungicides in phytopathogenic fungi. International Microbiology, 11, 1–9. [PubMed] [Google Scholar]
- Fischer, H.E. (1989) Origin of the ‘Weisse Schlesische Rübe’ (white Silesian beet) and resynthesis of sugar beet. Euphytica, 41, 75–80. [Google Scholar]
- Foote, C. (1976) Photosensitized oxidation and singlet oxygen: Consequences in biological systems In: Pryor W.(Ed.) Free Radicals in Biology, Vol. 2. NY: Academic Press; pp. 85–133. [Google Scholar]
- Foote, C.S. (1968) Mechanisms of photosensitized oxidation. Science, 162, 963–970. [DOI] [PubMed] [Google Scholar]
- Franc, G. (2010) Ecology and epidemiology of Cercospora beticola In: Lartey R., Weiland J., Panella L.W., Crous P.W. and Windels C.E. (Eds.) Cercospora Leaf Spot of Sugar Beet and Related Species. St. Paul, MN: American Phytopathological Society, pp. 7–19. [Google Scholar]
- Funk, A. , Galewski, P. and McGrath, J.M. (2018) Nucleotide‐binding resistance gene signatures in sugar beet, insights from a new reference genome. The Plant Journal, 95, 659–671. [DOI] [PubMed] [Google Scholar]
- Gadd, G. (2000) Microbial interactions with tributyltin compounds: detoxification, accumulation, and environmental fate. Science of the Total Environment, 258, 119–127. [DOI] [PubMed] [Google Scholar]
- Galletti, S. , Burzi, P.L. , Cerato, C. , Marinello, S. and Sala, E. (2008) Trichoderma as a potential biocontrol agent for Cercospora leaf spot of sugar beet. BioControl, 53, 917–930. [Google Scholar]
- Gan, P. , Ikeda, K. , Irieda, H. , Narusaka, M. , O'Connell, R.J. , Narusaka, Y. et al (2013) Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytologist, 197, 1236–1249. [DOI] [PubMed] [Google Scholar]
- Georgopoulos, S. and Dovas, C. (1973) A serious outbreak of strains of Cercospora beticola resistant to benzimidazole fungicides in Northern Greece. Plant Disease Report, 57, 321–324. [Google Scholar]
- Giannopolitis, C. (1978) Occurrence of strains of Cercospora beticola resistant to triphenyltin fungicides in Greece. Plant Disease Report, 62, 205–208. [Google Scholar]
- Giannopolitis, C. and Chrysayi‐Tokousbalides, M. (1980) Biology of triphenyltin‐resistant strains of Cercospora beticola from sugar beet. Plant Disease, 64, 940–942. [Google Scholar]
- Gladyshev, E. (2017) Repeat‐induced point mutation and other genome defense mechanisms in fungi. The Fungal Kingdom, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomès, E. , Gordon‐Weeks, R. , Simon‐Plas, F. , Pugin, A. , Milat, M.‐L. , Leigh, R.A. et al (1996a) Cercospora beticola toxins. Part XVII. The role of the beticolin/Mg2+ complexes in their biological activity. Study of plasma membrane H+‐ATPase, vacuolar H+‐PPase, alkaline and acid phosphatases. Biochimica et Biophysica Acta, 1285, 38–46. [DOI] [PubMed] [Google Scholar]
- Gomès, E. , Kees, V. , Francoise, S.‐P. , Marie‐Louise, M. , Gjedde, P.M. and Jean‐Pierre, B. (1996b) Activation of the plant plasma membrane H+‐ATPase. Is there a direct interaction between lysophosphatidylcholine and the C‐terminal part of the enzyme? FEBS Letters, 398, 48–52. [DOI] [PubMed] [Google Scholar]
- Gomès, E. , Simon‐Plas, F. , Milat, M.L. , Gapillout, I. , Mikès, V. , Pugin, A. et al (1996c) Cercospora beticola toxins. IX. Relationship between structure of beticolins, inhibition of plasma membrane H+‐ATPase and partition in lipid membranes. Physiologia Plantarum, 98, 133–139. [Google Scholar]
- Goodwin, S.B. , Dunkle, L.D. and Zismann, V.L. (2001) Phylogenetic analysis of Cercospora and Mycosphaerella based on the internal transcribed spacer region of ribosomal DNA. Phytopathology, 91, 648–658. [DOI] [PubMed] [Google Scholar]
- Goudet, C. , Benitah, J.‐P. , Milat, M.‐L. , Sentenac, H. and Thibaud, J.‐B. (1999) Cluster organization and pore structure of ion channels formed by beticolin 3, a nonpeptidic fungal toxin. Biophysical Journal, 77, 3052–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet, C. , Milat, M.‐L. , Sentenac, H. and Thibaud, J.‐B. (2000) Beticolins, nonpeptidic, polycyclic molecules produced by the phytopathogenic fungus Cercospora beticola, as a new family of ion channel‐forming toxins. Molecular Plant‐Microbe Interactions, 13, 203–209. [DOI] [PubMed] [Google Scholar]
- Goudet, C. , Véry, A.A. , Milat, M.L. , Ildefonse, M. , Thibaud, J.B. , Sentenac, H. et al (1998) Magnesium ions promote assembly of channel‐like structures from beticolin 0, a non‐peptide fungal toxin purified from Cercospora beticola . The Plant Journal, 14, 359–364. [DOI] [PubMed] [Google Scholar]
- Groenewald, J. , Nakashima, C. , Nishikawa, J. , Shin, H.‐D. , Park, J.‐H. , Jama, A. et al (2013) Species concepts in Cercospora: spotting the weeds among the roses. Studies in Mycology, 75, 115–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewald, M. , Groenewald, J.Z. , Harrington, T.C. , Abeln, E.C. and Crous, P.W. (2006) Mating type gene analysis in apparently asexual Cercospora species is suggestive of cryptic sex. Fungal Genetics and Biology, 43, 813–825. [DOI] [PubMed] [Google Scholar]
- Groenewald, M. , Groenewald, J.Z. , Linde, C.C. and Crous, P.W. (2007) Development of polymorphic microsatellite and single nucleotide polymorphism markers for Cercospora beticola (Mycosphaerellaceae). Molecular Ecology Notes, 7, 890–892. [Google Scholar]
- Groenewald, M. , Linde, C. , Groenewald, J.Z. and Crous, P.W. (2008) Indirect evidence for sexual reproduction in Cercospora beticola populations from sugar beet. Plant Pathology, 57, 25–32. [Google Scholar]
- Guedes, R.C. and Eriksson, L.A. (2007) Photophysics, photochemistry, and reactivity: molecular aspects of perylenequinone reactions. Photochemical & Photobiological Sciences, 6, 1089–1096. [DOI] [PubMed] [Google Scholar]
- Hane, J.K. , Williams, A.H. and Oliver, R.P. (2011) Genomic and comparative analysis of the class Dothideomycetes In: Pöggeler S. and Wöstemeyer J. (Eds.) Evolution of Fungi and Fungal‐Like Organisms. Berlin, Germany: Springer, pp. 205–229. [Google Scholar]
- Hao, Z. , Lv, D. , Ge, Y. , Shi, J. , Weijers, D. , Yu, G. et al (2019) RIdeogram: Drawing SVG Graphics to Visualize and Map Genome‐Wide Data on the Idiograms: PeerJ Preprints. R package version 0.2.0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtschulte, B. (2000) Cercospora beticola—worldwide distribution and incidence In: Asher M., Holtschulte B., Molard M., Rosso F., Steinrucken G. and Beckers R. (Eds.) IIRB Advances in Sugar Beet Research, Cercospora beticola Sacc. Biology, Agronomic Influences and Control Measures in Sugar Beet. 2, Brussels: International Institute for Beet Research, pp. 5–16. [Google Scholar]
- Hudson, J. , Imperial, V. , Haugland, R. and Diwu, Z. (1997) Antiviral activities of photoactive perylenequinones. Photochemistry and Photobiology, 65, 352–354. [DOI] [PubMed] [Google Scholar]
- Hunger, S. , Gaspero, G.D. , Möhring, S. , Bellin, D. , Schäfer‐Pregl, R. , Borchardt, D.C. et al (2003) Isolation and linkage analysis of expressed disease‐resistance gene analogues of sugar beet (Beta vulgaris L.). Genome, 46, 70–82. [DOI] [PubMed] [Google Scholar]
- Idnurm, A. and Meyer, V. (2018) The CRISPR revolution in fungal biology and biotechnology, and beyond. Fungal Biology and Biotechnology, 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis, P. (1994) Fungicides chemicals and techniques for controlling Cercospora beticola Sacc. in Greece. Presented at the Proceedings of Mediterranean Commitee Meeting of IIRB, Thessaloniki, Greece, pp. 139–151. [Google Scholar]
- ISO (2018) Sugar yearbook 2018 International Sugar Organization Sugar Yearbook. London, UK: International Sugar Organization; Available at: https://www.isosugar.org/sugarsector/sugar [Accessed 22 July 2019]. [Google Scholar]
- Jacobsen, B. and Franc, G. (2009) Cercospora leaf spot In: Harveson R., Hanson L. and Hein G. (Eds.) Compendium of Beet Diseases and Pests. 2, St. Paul, MN: American Phytopathological Society, pp. 7–10. [Google Scholar]
- Jalal, M.A. , Hossain, M.B. , Robeson, D.J. and Van der Helm, D. (1992) Cercospora beticola phytotoxins: cebetins that are photoactive, magnesium ion‐binding, chlorinated anthraquinone‐xanthone conjugates. Journal of the American Chemical Society, 114, 5967–5971. [Google Scholar]
- de Jonge, R. , Bolton, M.D. and Thomma, B.P. (2011) How filamentous pathogens co‐opt plants: the ins and outs of fungal effectors. Current Opinion in Plant Biology, 14, 400–406. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , Ebert, M.K. , Huitt‐Roehl, C.R. , Pal, P. , Suttle, J.C. , Spanner, R.E. et al (2018) Gene cluster conservation provides insight into cercosporin biosynthesis and extends production to the genus Colletotrichum . Proceedings of the National Academy of Sciences of the United States of America, 115, E5459–E5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M.K. et al (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proceedings of the National Academy of Sciences of the United States of America, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. (2007) Groovy times: filamentous pathogen effectors revealed. Current Opinion in Plant Biology, 10, 358–365. [DOI] [PubMed] [Google Scholar]
- Karadimos, D. , Karaoglanidis, G. and Tzavella‐Klonari, K. (2005) Biological activity and physical modes of action of the Qo inhibitor fungicides trifloxystrobin and pyraclostrobin against Cercospora beticola . Crop Protection, 24, 23–29. [Google Scholar]
- Karaoglanidis, G. and Ioannidis, P. (2010) Fungicide resistance of Cercospora beticola in Europe In: Lartey R., Weiland J., Panella L., Crous P. and Windels C. (Eds.) Cercospora Leaf Spot of Sugar Beet and Related Species. St. Paul, MN, USA: American Phytopathological Society, pp. 189–211. [Google Scholar]
- Karaoglanidis, G. , Ioannidis, P. and Thanassoulopoulos, C. (2001a) Influence of fungicide spray schedules on the sensitivity of Cercospora beticola to the sterol demethylation‐inhibiting fungicide flutriafol. Crop Protection, 20, 941–947. [Google Scholar]
- Karaoglanidis, G. , Thanassoulopoulos, C.C. and Ioannidis, P. (2001b) Fitness of Cercospora beticola field isolates–resistant and—sensitive to demethylation inhibitor fungicides. European Journal of Plant Pathology, 107, 337–347. [Google Scholar]
- Karaoglanidis, G. and Thanassoulopoulos, C. (2003) Cross‐resistance patterns among sterol biosynthesis inhibiting fungicides (SBIs) in Cercospora beticola . European Journal of Plant Pathology, 109, 929–934. [Google Scholar]
- Kayamori, M. , Shimizu, M. , Yamana, T. , Komatsu, T. , Minako, S. , Shinmura, A. et al (2020) First report of QoI resistance in Cercospora beticola in sugar beet in Japan. Journal of General Plant Pathology, 86, 149–153. [Google Scholar]
- Keller, N.P. (2015) Translating biosynthetic gene clusters into fungal armor and weaponry. Nature Chemical Biology, 11, 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, J. , Del Río, L. , Nelson, R. and Khan, M. (2007) Improving the Cercospora leaf spot management model for sugar beet in Minnesota and North Dakota. Plant Disease, 91, 1105–1108. [DOI] [PubMed] [Google Scholar]
- Khan, J. , Qi, A. and Khan, M. (2009) Fluctuations in number of Cercospora beticola conidia in relationship to environment and disease severity in sugar beet. Phytopathology, 99, 796–801. [DOI] [PubMed] [Google Scholar]
- Khan, J. , Rio, L.D. , Nelson, R. , Rivera‐Varas, V. , Secor, G. and Khan, M. (2008) Survival, dispersal, and primary infection site for Cercospora beticola in sugar beet. Plant Disease, 92, 741–745. [DOI] [PubMed] [Google Scholar]
- Kirk, W. , Hanson, L. , Franc, G. , Stump, W. , Gachango, E. , Clark, G. et al (2012) First report of strobilurin resistance in Cercospora beticola in sugar beet (Beta vulgaris) in Michigan and Nebraska, USA. New Disease Reports, 26, 3. [Google Scholar]
- Knight, N. , Koenick, L. , Sharma, S. and Pethybridge, S.J. (2020) Detection of Cercospora beticola and Phoma betae on table beet seed using quantitative PCR. Phytopathology, 110, 943–951. [DOI] [PubMed] [Google Scholar]
- Knight, N.L. , Vaghefi, N. , Hansen, Z.R. , Kikkert, J.R. and Pethybridge, S.J. (2018) Temporal genetic differentiation of Cercospora beticola populations in New York table beet fields. Plant Disease, 102, 2074–2082. [DOI] [PubMed] [Google Scholar]
- Knight, N.L. , Vaghefi, N. , Kikkert, J.R. , Bolton, M.D. , Secor, G.A. , Rivera, V.V. et al (2019) Genetic diversity and structure in regional Cercospora beticola populations from Beta vulgaris subsp. vulgaris suggest two clusters of separate origin. Phytopathology, 109, 1280–1292. [DOI] [PubMed] [Google Scholar]
- Koch, G. and Jung, C. (2000) Genetic localization of Cercospora resistance genes. In: Asher M., Holtschulte B., Molard M., Rosso F., Steinrucken G. and Beckers R. (Eds.) IIRB Advances in Sugar Beet Research, Cercospora beticola Sacc. Biology, Agronomic Influences and Control Measures in Sugar Beet. 2, Brussels: International Institute for Beet Research, pp. 197–209. [Google Scholar]
- Koeck, M. , Hardham, A.R. and Dodds, P.N. (2011) The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cellular Microbiology, 13, 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz, S. , Phillippy, A. , Delcher, A.L. , Smoot, M. , Shumway, M. , Antonescu, C. et al (2004) Versatile and open software for comparing large genomes. Genome Biology, 5, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusstatscher, P. , Cernava, T. , Harms, K. , Maier, J. , Eigner, H. , Berg, G. et al (2019) Disease incidence in sugar beet fields is correlated with microbial diversity and distinct biological markers. Phytobiomes Journal, 3, 22–30. [Google Scholar]
- Lamey, H.A. , Cattanach, A.W. and Bugbee, W. (1987) Cercospora leafspot of sugar beet. North Dakota State University Extension Circular, 764. [Google Scholar]
- Langfelder, K. , Streibel, M. , Jahn, B. , Haase, G. and Brakhage, A.A. (2003) Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genetics and Biology, 38, 143–158. [DOI] [PubMed] [Google Scholar]
- Lartey, R.T. , Weiland, J. , Panella, L. , Crous, P. and Windels, C. (2010) Cercospora Leaf Spot of Sugar Beet and Related Species. St. Paul, MN, USA: American Phytopathological Society. [Google Scholar]
- Lawrence, J. and Meredith, D. (1970) Wind dispersal of conidia of Cercospora beticola . Phytopathology, 60, 1076–1078. [Google Scholar]
- Leisman, G.B. and Daub, M.E. (1992) Singlet oxygen yields, optical properties, and phototoxicity of reduced derivatives of the photosensitizer cercosporin. Photochemistry and Photobiology, 55, 373–379. [Google Scholar]
- Lejeune, F. and Maquat, L.E. (2005) Mechanistic links between nonsense‐mediated mRNA decay and pre‐mRNA splicing in mammalian cells. Current Opinion in Cell Biology, 17, 309–315. [DOI] [PubMed] [Google Scholar]
- Leroux, P. , Albertini, C. , Gautier, A. , Gredt, M. and Walker, A.S. (2007) Mutations in the CYP51 gene correlated with changes in sensitivity to sterol 14α‐demethylation inhibitors in field isolates of Mycosphaerella graminicola . Pest Management Science, 63, 688–698. [DOI] [PubMed] [Google Scholar]
- Lewellen, R. and Whitney, E. (1976) Inheritance of resistance to race C2 of Cercospora beticola in sugar beet 1. Crop Science, 16, 558–561. [Google Scholar]
- Li, L. , Stoeckert, C.J. and Roos, D.S. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research, 13, 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Dron, M. , Cramer, C. , Dixon, R.A. and Lamb, C.J. (1989) Differential regulation of phenylalanine ammonia‐lyase genes during plant development and by environmental cues. Journal of Biological Chemistry, 264, 14486–14492. [PubMed] [Google Scholar]
- Liu, G.Y. and Nizet, V. (2009) Color me bad: microbial pigments as virulence factors. Trends in Microbiology, 17, 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M’Barek, S.B. , Cordewener, J.H. , Ghaffary, S.M.T. , van der Lee, T.A. , Liu, Z. , Gohari, A.M. et al (2015) FPLC and liquid‐chromatography mass spectrometry identify candidate necrosis‐inducing proteins from culture filtrates of the fungal wheat pathogen Zymoseptoria tritici . Fungal Genetics and Biology, 79, 54–62. [DOI] [PubMed] [Google Scholar]
- Magnuson, T.A. (1918) History of the beet sugar industry in California. Annual Publication of the Historical Society of Southern California, 11, 68–79. [Google Scholar]
- Malandrakis, A.A. , Markoglou, A.N. , Nikou, D.C. , Vontas, J.G. and Ziogas, B.N. (2011) Molecular diagnostic for detecting the cytochrome b G143S–QoI resistance mutation in Cercospora beticola . Pesticide Biochemistry and Physiology, 100, 87–92. [Google Scholar]
- Marggraf, A.S. (1748) Experiences chymiques faites dans le dessein de tirer un veritable sucre de diverses plantes, qui croissent dans nos contrées In: Votre M. (Ed.) Histoire de l’cadémie Royale des Sciences et Belles Lettres. Berlin, Germany: Académie des sciences, pp. 79–90. [Google Scholar]
- Marić, A. , Maširević, S. and Jerković, Z. (1984) Increase in resistance of Cercospora beticola to benomyl and first occurrence of strains tolerant of fentin acetate in Vojvodini. Zaštita Bilja, 35, 207–215. [Google Scholar]
- Mauch‐Mani, B. and Mauch, F. (2005) The role of abscisic acid in plant–pathogen interactions. Current Opinion in Plant Biology, 8, 409–414. [DOI] [PubMed] [Google Scholar]
- McKay, M.B. and Pool, V.W. (1918) Field studies of Cercospora beticola . Phytopathology, 8, 119–136. [Google Scholar]
- Mesarich, C.H. , Stergiopoulos, I. , Beenen, H.G. , Cordovez, V. , Guo, Y. , Karimi Jashni, M. et al (2016) A conserved proline residue in Dothideomycete Avr4 effector proteins is required to trigger a Cf‐4‐dependent hypersensitive response. Molecular Plant Pathology, 17, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milat, M.‐L. and Blein, J.‐P. (1995) Cercospora beticola toxins III. Purification, thin‐layer and high performance liquid chromatographic analyses. Journal of Chromatography, 699, 277–283. [Google Scholar]
- Milat, M.‐L. , Prange, T. , Wiedemann‐Merdinoglu, S. and Blein, J.‐P. (2010) Beticolins: chemistry and biological activities In: Lartey R., Weiland J., Anella L., Crous P.W. and Windels C.E. (Eds.) Cercospora Leaf Spot of Sugar Beet and Related Species. Minneapolis, MN: APS Press, pp. 119–128. [Google Scholar]
- Moretti, M. , Saracchi, M. and Farina, G. (2004) Morphological, physiological and genetic diversity within a small population of Cercospora beticola Sacc. Annals of Microbiology, 54, 129–150. [Google Scholar]
- Nagel, C.M. (1945) Epiphytology and control of sugar beet leaf spot caused by Cercospora beticola Sacc. Iowa Agriculture and Home Economics Experiment Station Research Bulletin, 27, 1. [Google Scholar]
- Newman, A.G. and Townsend, C.A. (2016) Molecular characterization of the cercosporin biosynthetic pathway in the fungal plant pathogen Cercospora nicotianae . Journal of the American Chemical Society, 138, 4219–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A.G. , Vagstad, A.L. , Belecki, K. , Scheerer, J.R. and Townsend, C.A. (2012) Analysis of the cercosporin polyketide synthase CTB1 reveals a new fungal thioesterase function. Chemical Communications, 48, 11772–11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikou, D. , Malandrakis, A. , Konstantakaki, M. , Vontas, J. , Markoglou, A. and Ziogas, B. (2009) Molecular characterization and detection of overexpressed C‐14 alpha‐demethylase‐based DMI resistance in Cercospora beticola field isolates. Pesticide Biochemistry and Physiology, 95, 18–27. [Google Scholar]
- Nilsson, N.O. , Hansen, M. , Panagopoulos, A. , Tuvesson, S. , Ehlde, M. , Christiansson, M. et al (1999) QTL analysis of Cercospora leaf spot resistance in sugar beet. Plant Breeding, 118, 327–334. [Google Scholar]
- Notomi, T. , Mori, Y. , Tomita, N. and Kanda, H. (2015) Loop‐mediated isothermal amplification (LAMP): principle, features, and future prospects. Journal of Microbiology, 53, 1–5. [DOI] [PubMed] [Google Scholar]
- Obuya, J. , Hanson, L. and Franc, G. (2011) Mating type idiomorphs distribution and their correlation to benzimidazole‐resistance in Cercospora beticola from the Central High Plains region, USA. Presented at the American Society of Sugar Beet Technologists, Proceedings from the 36th Biennial Meeting, March 2‐5, 2011, Albuquerque, NM, USA, pp. 10–114. [Google Scholar]
- Obuya, J.O. , Ananga, A. and Franc, G.D. (2015) Silent mutation: characterization of its potential as a mechanism for sterol 14 [alpha]‐demethylase resistance in Cercospora beticola field isolates from the United States. Journal of Plant Pathology & Microbiology, 6, 280. [Google Scholar]
- OECD . (2001) Consensus Document on the Biology of Beta vulgaris L. (Sugar Beet). Series on Harmonization of Regulatory Oversight in Biotechnology, No 18. OECD Environment Health and Safety Publications. Retrieved from http://www.oecd.org/ehs/. [Google Scholar]
- Ohm, R.A. , Feau, N. , Henrissat, B. , Schoch, C.L. , Horwitz, B.A. , Barry, K.W. et al (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathogens, 8, e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, V. and Mukhopadhyay, A. (1983) Occurrence of strains of Cercospora beticola resistance to carbendazim (MBC) in India. Indian Journal of Mycology and Plant Pathology, 13, 333–334. [Google Scholar]
- Palmer, J.M. and Keller, N.P. (2010) Secondary metabolism in fungi: does chromosomal location matter? Current Opinion in Microbiology, 13, 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotis, M. , Kritonas, K. , Irini, N.O. , Kiriaki, C. , Nicolaos, P. and Athanasios, T. (2007) Expression of the yeast cpd1 gene in tobacco confers resistance to the fungal toxin cercosporin. Biomolecular Engineering, 24, 245–251. [DOI] [PubMed] [Google Scholar]
- Panella, L. and Frese, L. (2000) Cercospora resistance in Beta species and the development of resistant sugar beet lines In: Asher M., Holtschulte B., Molard M., Rosso F., Steinrucken G. and Beckers R. (Eds.) IIRB Advances in Sugar Beet Research, Cercospora beticola Sacc. Biology, Agronomic Influences and Control Measures in Sugar Beet. 2, Brussels, Belgium: International Institute for Beet Research, pp. 163–176. [Google Scholar]
- Pethybridge, S. , Kikkert, J. , Hanson, L. and Nelson, S. (2018) Challenges and prospects for building resilient disease management strategies and tactics for the New York table beet industry. Agronomy, 8, 112. [Google Scholar]
- Piszczek, J. , Pieczul, K. and Kiniec, A. (2018) First report of G143A strobilurin resistance in Cercospora beticola in sugar beet (Beta vulgaris) in Poland. Journal of Plant Diseases and Protection, 125, 99–101. [Google Scholar]
- Pitblado, R. and Nichols, I. (2005) The implementation of BEETCAST‐a weather‐timed fungicide spray program for the control of Cercospora leaf spot in Ontario and Michigan. Journal of Sugarbeet Research, 42, 53–54. [Google Scholar]
- Plissonneau, C. , Stürchler, A. and Croll, D. (2016) The evolution of orphan regions in genomes of a fungal pathogen of wheat. MBio, 7, e01231‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons, N. , Sutton, B. and Gay, J. (1985) Ultrastructure of conidiogenesis in Cercospora beticola . Transactions of the British Mycological Society, 85, 405–416. [Google Scholar]
- Pool, V.W. and McKay, M. (1916) Climatic conditions as related to Cercospora beticola . Journal of Agricultural Research, 6, 21–60. [Google Scholar]
- Racca, P. and Jörg, E. (2007) CERCBET 3–a forecaster for epidemic development of Cercospora beticola . EPPO Bulletin, 37, 344–349. [Google Scholar]
- Raffaele, S. , Farrer, R.A. , Cano, L.M. , Studholme, D.J. , MacLean, D. , Thines, M. et al (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science, 330, 1540–1543. [DOI] [PubMed] [Google Scholar]
- Rathaiah, Y. (1977) Stomatal tropism of Cercospora beticola in sugar beet. Phytopathology, 67, 358–362. [Google Scholar]
- Rezende, J.S. , Zivanovic, M. , de Novaes, M.I.C. and Chen, Z.Y. (2020) The AVR4 effector is involved in cercosporin biosynthesis and likely affects the virulence of Cercospora cf. flagellaris on soybean. Molecular Plant Pathology, 21, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risk & Policy Analysts Limited (2005) Risk Assessment Studies on Targeted Consumer Applications of Certain Organotin Compounds—Final Report. Available at https://ec.europa.eu/docsroom/documents/13041/attachments/13041/translations/en/renditions/pdf [Accessed 17 December 2019]. [Google Scholar]
- Robeson, D.J. and Jalal, M.A. (1993) A Cercospora isolate from soybean roots produces cebetin B and cercosporin. Phytochemistry, 33, 1546–1548. [Google Scholar]
- Rosenzweig, N. , Hanson, L. , Clark, G. , Franc, G. , Stump, W. , Jiang, Q. et al (2015) Use of PCR‐RFLP analysis to monitor fungicide resistance in Cercospora beticola populations from sugarbeet (Beta vulgaris) in Michigan, United States. Plant Disease, 99, 355–362. [DOI] [PubMed] [Google Scholar]
- Rosenzweig, N. , Hanson, L.E. , Mambetova, S. , Jiang, Q. , Guza, C. , Stewart, J. et al (2020) Temporal population monitoring of fungicide sensitivity in Cercospora beticola from sugarbeet (Beta vulgaris) in the Upper Great Lakes. Canadian Journal of Plant Pathology. 10.1080/07060661.2019.1705914. [DOI] [Google Scholar]
- Rossi, V. (1995) Effect of host resistance in decreasing infection rate of Cercospora leaf spot epidemics on sugar beet. Phytopathologia Mediterranea, 149–156. [Google Scholar]
- Rossi, V. and Battilani, P. (1991) CERCOPRI: a forecasting model for primary infections of Cercospora leaf spot of sugarbeet 1. EPPO Bulletin, 21, 527–531. [Google Scholar]
- Rouxel, T. , Grandaubert, J. , Hane, J.K. , Hoede, C. , Van de Wouw, A.P. , Couloux, A. et al (2011) Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat‐induced point mutations. Nature Communications, 2, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovenich, H. , Boshoven, J.C. and Thomma, B.P. (2014) Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Current Opinion in Plant Biology, 20, 96–103. [DOI] [PubMed] [Google Scholar]
- Rudd, J.J. , Kanyuka, K. , Hassani‐Pak, K. , Derbyshire, M. , Andongabo, A. , Devonshire, J. et al (2015) Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiology, 167, 1158–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppel, E. (1972) Variation among isolates of Cercospora beticola from sugar beet. Phytopathology, 62, 134–136. [Google Scholar]
- Ruppel, E. (1986) Cercospora leaf spot In: Whitney E. and Duffus J. (Eds.) Compendium of Beet Diseases and Insects. St. Paul, MN: American Phytopathological Society, pp. 8–9. [Google Scholar]
- Ruppel, E. and Scott, P. (1974) Strains of Cercospora beticola resistant to benomyl in the USA. Plant Disease Report, 58, 434–436. [Google Scholar]
- Saccardo, P. (1876) Funghi Veneti novi vel critici. Series V. Nuovo Giornale Botanico Italiano, 8, 161–211. [Google Scholar]
- Schlösser, E. (1962) Über eine biologisch aktive Substanz aus Cercospora beticola . Journal of Phytopathology, 44, 295–312. [Google Scholar]
- Schmidt, K. , Heberle, B. , Kurrasch, J. , Nehls, R. and Stahl, D.J. (2004) Suppression of phenylalanine ammonia lyase expression in sugar beet by the fungal pathogen Cercospora beticola is mediated at the core promoter of the gene. Plant Molecular Biology, 55, 835–852. [DOI] [PubMed] [Google Scholar]
- Schmidt, K. , Pflugmacher, M. , Klages, S. , Maeser, A. , Mock, A. and Stahl, D.J. (2008) Accumulation of the hormone abscisic acid (ABA) at the infection site of the fungus Cercospora beticola supports the role of ABA as a repressor of plant defence in sugar beet. Molecular Plant Pathology, 9, 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, M. and Kahmann, R. (2019) CRISPR‐Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genetics and Biology, 130, 43–53. [DOI] [PubMed] [Google Scholar]
- Secor, G. , Rivera, V. and Bolton, M. (2017) Sensitivity of Cercospora beticola to Foliar Fungicides in 2017. The Sugarbeet Research and Education Board of Minnesota and North Dakota, pp. 161–168. [Google Scholar]
- Secor, G.A. , Rivera, V.V. , Khan, M. and Gudmestad, N.C. (2010) Monitoring fungicide sensitivity of Cercospora beticola of sugar beet for disease management decisions. Plant Disease, 94, 1272–1282. [DOI] [PubMed] [Google Scholar]
- Setiawan, A. , Koch, G. , Barnes, S. and Jung, C. (2000) Mapping quantitative trait loci (QTLs) for resistance to Cercospora leaf spot disease (Cercospora beticola Sacc.) in sugar beet (Beta vulgaris L.). Theoretical and Applied Genetics, 100, 1176–1182. [Google Scholar]
- Shane, W. and Teng, P. (1992) Impact of Cercospora leaf spot on root weight, sugar yield, and purity of Beta vulgaris . Plant Disease, 76, 812–820. [Google Scholar]
- Shrestha, S. , Neubauer, J. , Spanner, R. , Natwick, M. , Rios, J. , Metz, N. et al (2020) Rapid detection of Cercospora beticola in sugar beet and mutations associated with fungicide resistance using LAMP or probe‐based qPCR. Plant Disease.104, 1654–1661. [DOI] [PubMed] [Google Scholar]
- Simon‐Plas, F. , Gomes, E. , Milat, M.‐L. , Pugin, A. and Blein, J.‐P. (1996) Cercospora beticola toxins. Part X. Inhibition of plasma membrane H+‐ATPase by beticolin‐1. Plant Physiology, 111, 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaracis, G.N. , Pavli, O.I. and Biancardi, E. (2010) Cercospora leaf spot disease of sugar beet. Sugar Tech, 12, 220–228. [Google Scholar]
- Smith, G. and Campbell, L. (1996) Association between resistance to Cercospora and yield in commercial sugar beet hybrids. Plant Breeding, 115, 28–32. [Google Scholar]
- Smith, G. and Gaskill, J. (1970) Inheritance of resistance to Cercospora leaf spot in sugar beet. Journal of the American Society of Sugar Beet, 16, 172–180. [Google Scholar]
- Smith, G. and Ruppel, E. (1973) Association of Cercospora leaf spot, gross sucrose, percentage sucrose, and root weight in sugar beet. Canadian Journal of Plant Science, 53, 695–696. [Google Scholar]
- Smith, G. and Ruppel, E. (1974) Herability of resistance to Cercospora leaf spot in sugar beet 1. Crop Science, 14, 113–115. [Google Scholar]
- Snelders, N.C. , Kettles, G.J. , Rudd, J.J. and Thomma, B.P. (2018) Plant pathogen effector proteins as manipulators of host microbiomes? Molecular Plant Pathology, 19, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soanes, D.M. , Richards, T.A. and Talbot, N.J. (2007) Insights from sequencing fungal and oomycete genomes: what can we learn about plant disease and the evolution of pathogenicity? The Plant Cell, 19, 3318–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solel, Z. and Minz, G. (1971a) Infection process of Cercospora beticola in sugar beet in relation to susceptibility. Phytopathology, 61, 463–466. [Google Scholar]
- Solel, Z. and Minz, G. (1971b) Infection process of Cercospora beticola in sugarbeet in relation to susceptibility. Phytopathology, 61, 463–466. [Google Scholar]
- Sollod, C.C. , Jenns, A.E. and Daub, M.E. (1992) Cell surface redox potential as a mechanism of defense against photosensitizers in fungi. Applied and Environmental Microbiology, 58, 444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R. , Zhai, Q. , Sun, L. , Huang, E. , Zhang, Y. , Zhu, Y. et al (2019) CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Applied Microbiology and Biotechnology, 103, 6919–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, A. , Herrero, S. and Daub, M. (2019) The toxin cercosporin is a virulence factor for infection of coffee by Cercospora coffeicola . BioRxiv, 818328 [Preprint]. [Google Scholar]
- Staerkel, C. , Boenisch, M.J. , Kröger, C. , Bormann, J. , Schäfer, W. and Stahl, D. (2013) CbCTB2, an O‐methyltransferase is essential for biosynthesis of the phytotoxin cercosporin and infection of sugar beet by Cercospora beticola . BMC Plant Biology, 13, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkamp, M. , Martin, S. , Hoefert, L. and Ruppel, E. (1979) Ultrastructure of lesions produced by Cercospora beticola in leaves of Beta vulgaris . Physiological Plant Pathology, 15, 13–26. [Google Scholar]
- Stergiopoulos, I. , van den Burg, H.A. , Ökmen, B. , Beenen, H.G. , van Liere, S. , Kema, G.H. et al (2010) Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proceedings of the National Academy of Sciences of the United States of America, 107, 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis, I.A. , Zhang, H. , Pieterse, C.M. , Bolton, M.D. and de Jonge, R. (2018) Microbial small molecules—weapons of plant subversion. Natural Products Reports, 35, 410–433. [DOI] [PubMed] [Google Scholar]
- Swart, V. , Crampton, B.G. , Ridenour, J.B. , Bluhm, B.H. , Olivier, N.A. , Meyer, J.M. et al (2017) Complementation of CTB7 in the maize pathogen Cercospora zeina overcomes the lack of in vitro cercosporin production. Molecular Plant‐Microbe Interactions, 30, 710–724. [DOI] [PubMed] [Google Scholar]
- Taguchi, K. , Kubo, T. , Takahashi, H. and Abe, H. (2011) Identification and precise mapping of resistant QTLs of Cercospora leaf spot resistance in sugar beet (Beta vulgaris L.). G3: Genes, Genomes, Genetics, 1, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedford, S.L. , Burlakoti, R.R. , Schaafsma, A.W. and Trueman, C.L. (2018) Relationships among airborne Cercospora beticola conidia concentration, weather variables and cercospora leaf spot severity in sugar beet (Beta vulgaris L.). Canadian Journal of Plant Pathology, 40, 1–10. [Google Scholar]
- Trkulja, N. , Ivanović, Ž. , Pfaf‐Dolovac, E. , Dolovac, N. , Mitrović, M. , Toševski, I. et al (2013) Characterisation of benzimidazole resistance of Cercospora beticola in Serbia using PCR‐based detection of resistance‐associated mutations of the β‐tubulin gene. European Journal of Plant Pathology, 135, 889–902. [Google Scholar]
- Trkulja, N.R. , Milosavljević, A.G. , Mitrović, M.S. , Jović, J.B. , Toševski, I.T. , Khan, M.F. et al (2017) Molecular and experimental evidence of multi‐resistance of Cercospora beticola field populations to MBC, DMI and QoI fungicides. European Journal of Plant Pathology, 149, 895–910. [Google Scholar]
- Trueman, C. , Hanson, L. , Rosenzweig, N. , Jiang, Q. and Kirk, W. (2013) First report of QoI insensitive Cercospora beticola on sugar beet in Ontario, Canada. Plant Disease, 97, 1255. [DOI] [PubMed] [Google Scholar]
- Trueman, C. , Hanson, L. , Somohano, P. and Rosenzweig, N. (2017) First report of DMI‐insensitive Cercospora beticola on sugar beet in Ontario, Canada. New Disease Reports, 36, 20. [DOI] [PubMed] [Google Scholar]
- Turgay, E.B. , Bakir, M. , Ozeren, P. , Katircioglu, Y.Z. and Maden, S. (2010) Detection of pathotypes and genetic diversity of Cercospora beticola . Plant Pathology Journal, 26, 306–312. [Google Scholar]
- USDA‐NASS (2010) Field Crops: Usual Planting and Harvesting Dates. Washington, DC: USDA National Agricultural Statistics Service, Agricultural Handbook. [Google Scholar]
- Vaghefi, N. , Hay, F.S. , Kikkert, J.R. and Pethybridge, S.J. (2016) Genotypic diversity and resistance to azoxystrobin of Cercospora beticola on processing table beet in New York. Plant Disease, 100, 1466–1473. [DOI] [PubMed] [Google Scholar]
- Vaghefi, N. , Kikkert, J.R. , Bolton, M.D. , Hanson, L.E. , Secor, G.A. , Nelson, S.C. et al (2017a) Global genotype flow in Cercospora beticola populations confirmed through genotyping‐by‐sequencing. PLoS ONE, 12, e0186488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaghefi, N. , Kikkert, J.R. , Bolton, M.D. , Hanson, L.E. , Secor, G.A. and Pethybridge, S.J. (2017b) De novo genome assembly of Cercospora beticola for microsatellite marker development and validation. Fungal Ecology, 26, 125–134. [Google Scholar]
- Vaghefi, N. , Nelson, S.C. , Kikkert, J.R. and Pethybridge, S.J. (2017c) Genetic structure of Cercospora beticola populations on Beta vulgaris in New York and Hawaii. Scientific Reports, 7, 1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereijssen, J. , Schneider, J. and Jeger, M. (2007) Epidemiology of Cercospora leaf spot on sugar beet: modeling disease dynamics within and between individual plants. Phytopathology, 97, 1550–1557. [DOI] [PubMed] [Google Scholar]
- Vereijssen, J. , Schneider, J.H. and Termorshuizen, A.J. (2005) Root infection of sugar beet by Cercospora beticola in a climate chamber and in the field. European Journal of Plant Pathology, 112, 201–210. [Google Scholar]
- Ververidis, P. , Davrazou, F. , Diallinas, G. , Georgakopoulos, D. , Kanellis, A. and Panopoulos, N. (2001) A novel putative reductase (Cpd1p) and the multidrug exporter Snq2p are involved in resistance to cercosporin and other singlet oxygen‐generating photosensitizers in Saccharomyces cerevisiae . Current Genetics, 39, 127–136. [DOI] [PubMed] [Google Scholar]
- Vicente, I. , Sarrocco, S. , Malfatti, L. , Baroncelli, R. and Vannacci, G. (2019) CRISPR‐Cas for fungal genome editing: a new tool for the management of plant diseases. Frontiers in Plant Science, 10, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J. , Kenter, C. , Holst, C. and Märländer, B. (2018) New generation of resistant sugar beet varieties for advanced integrated management of Cercospora leaf spot in central Europe. Frontiers in Plant Science, 9, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, R.M. , Tegenfeldt, F. , Li, J. , Zdobnov, E.M. and Kriventseva, E.V. (2012) OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Research, 41, D358–D365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland, J. , Chung, K. and Suttle, J. (2010) The role of cercosporin in the virulence of Cercospora spp. to plant hosts In: Lartey R.T., Weiland J.J., Panella L., Crous P.W. and Windels C.E. (Eds.) Cercospora Leaf Spot of Sugar Beet and Related Species. St Paul, MN: APS Press, pp. 109–117. [Google Scholar]
- Weiland, J. and Koch, G. (2004) Sugar beet leaf spot disease (Cercospora beticola Sacc.). Molecular Plant Pathology, 5, 157–166. [DOI] [PubMed] [Google Scholar]
- Weiss, U. , Flon, H. and Burger, W.C. (1957) The photodynamic pigment of some species of Elsinoë and Sphaceloma . Archives of Biochemistry and Biophysics, 69, 311–319. [DOI] [PubMed] [Google Scholar]
- Weltmeier, F. , Mäser, A. , Menze, A. , Hennig, S. , Schad, M. , Breuer, F. et al (2011) Transcript profiles in sugar beet genotypes uncover timing and strength of defense reactions to Cercospora beticola infection. Molecular Plant‐Microbe Interactions, 24, 758–772. [DOI] [PubMed] [Google Scholar]
- Windels, C.E. , Lamey, H.A. , Hilde, D. , Widner, J. and Knudsen, T. (1998) A Cerospora leaf spot model for sugar beet: in practice by an industry. Plant Disease, 82, 716–726. [DOI] [PubMed] [Google Scholar]
- de Wit, P.J.M. , van der Burgt, A. , Ökmen, B. , Stergiopoulos, I. , Abd‐Elsalam, K.A. , Aerts, A.L. et al (2012) The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genetics, 8, e1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, P. and Verreet, J. (2005) Factors affecting the onset of Cercospora leaf spot epidemics in sugar beet and establishment of disease‐monitoring thresholds. Phytopathology, 95, 269–274. [DOI] [PubMed] [Google Scholar]
- Yamazaki, S. , Okubo, A. , Akiyama, Y. and Fuwa, K. (1975) Cercosporin, a novel photodynamic pigment isolated from Cercospora kikuchii . Agricultural and Biological Chemistry, 39, 287–288. [Google Scholar]
- Ziogas, B.N. and Malandrakis, A.A. (2015) Sterol biosynthesis inhibitors: C14 demethylation (DMIs) In: Ishii H. and Hollomon D. (Eds.) Fungicide Resistance in Plant Pathogens. Tokyo, Japan: Springer, pp. 199–216. [Google Scholar]
- Zuluaga, A.P. , Vega‐Arreguín, J.C. , Fei, Z. , Ponnala, L. , Lee, S.J. , Matas, A.J. et al (2016) Transcriptional dynamics of Phytophthora infestans during sequential stages of hemibiotrophic infection of tomato. Molecular Plant Pathology, 17, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


