Abstract
Almost all pancreatic ductal adenocarcinomas (PDA) develop following KRAS activation which triggers epithelial transformation and recruitment of desmoplastic stroma through additional transcriptional and epigenetic regulation, but only a few of these regulatory mechanisms have been described. We profiled dysregulated microRNAs (miRNAs) starting with the earliest premalignant pancreatic intraepithelial neoplasias (PanINs) in genetically engineered mutated KRAS and P53 (KPC) mice programmed to recapitulate human PDA tumorigenesis. We identified miR-21 and miR-224 as cell-specific and compartment-specific regulators in PanINs and PDA. miR-21 is overexpressed in tumor epithelial cells of premalignant ducts, while miR-224 is overexpressed in cancer-associated fibroblasts in PDA stroma. Inhibition of miR-21 reverted pro-tumorigenic functionalities to baseline levels. Overexpression of miR-224 induced activated phenotypes in normal fibroblasts. In vivo miR-21 inhibition improved survival in established PDA. Importantly, early systemic miR-21 inhibition completely intercepted premalignant progression. Finally, an evaluation of miR-21 expression in the PDA cohort of The Cancer Genome Atlas (TCGA) identified a correlation between tumor epithelial cell content and miR-21 expression in human tumors providing further rationale for conducting human studies. Thus, miR-21 may be useful for early PanIN detection, and for intercepting developing premalignant pancreatic lesions and other KRAS-driven premalignancies.
Keywords: pancreatic ductal adenocarcinoma, miR-21, miR-224, KRAS, microRNA, pancreatic intraepithelial neoplasias
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDA) is increasing in incidence, and remains among the deadliest cancers with an overall median survival of 4–6 months[1]. Along with factors such as late diagnosis, poor prognosis results from its complex tumor microenvironment (TME) composed of transformed epithelial cells and tumor-promoting stromal cells that renders PDA refractory to radiation, chemotherapies and immunotherapies[2,3]. The stromal compartment consists of PDA-recruited cancer-associated fibroblasts (CAFs), pancreatic stellate cells, immune cells and extracellular matrix proteins that drive disease progression from the earliest premalignant pancreatic intraepithelial neoplasias (PanINs) to PDA[4].
PDA tumorigenesis is initiated, propagated and maintained by the expression of the driver mutation KRAS (mKRAS)[2,5]. mKRAS is one of the earliest catalytic events and most highly expressed mutation in over 90% of human PanIN1 lesions and PDA[6]. mKRAS persistently activates downstream signaling pathways resulting in increased cell proliferation, survival and metabolic reprogramming[7]. Concurrent to the initiation of PanIN development, mKRAS also triggers the formation of the cancer-supportive microenvironment via recruitment of fibro-inflammatory stroma[2]. These cancer-associated cell populations express additional signals that support PDA development, progression, and metastasis[8,9].
Attempts to develop therapies that directly target mKRAS have been met with challenges[10]. Most strategies have focused on inhibiting mKRAS directly or downstream effectors of mKRAS signaling[10]. However, the clinical benefit of these downstream therapeutics has been limited by compensatory mechanisms, narrow therapeutic windows with associated drug toxicities, the accumulation of additional genetic alterations in tumor cells, and the complexity of signaling by cancer-associated stromal cells[10,11]. Although it is difficult to target mKRAS therapeutically, it may be possible to develop strategies that intercept mKRAS in early premalignancies, before additional genetic and regulatory mechanisms occur[12].
MicroRNAs (miRNAs) are cellular regulators that have been associated with the etiology of many cancers at various stages of development and progression[13,14]. miRNAs are an emerging class of novel therapeutic targets being tested in clinical trials for cancer and other diseases. miRNAs are short non-coding RNAs (ncRNAs), 19–25 nucleotides in length, that post-transcriptionally regulate gene expression by binding to complementary 3’ untranslated regions (3’UTR) of target mRNAs resulting in their degradation or translational repression[13,15]. Currently, three miRNA therapeutics have advanced to clinical trials for the treatment of solid malignancies and leukemia[13,16]. Recently, interfering RNAs as a therapeutic approach to PDA have been explored using tumor-penetrating nanoparticles as a delivery modality that can penetrate the dense tumor stroma[17,18]. This technique has demonstrated efficacy in xenograft murine models with dysregulated miRNAs, specifically when both miR-21 and KRAS were simultaneously targeted[18,19]. Although miRNA expression in PDA has been reported, progressive profiling of dysregulated miRNAs with PanIN progression has been minimally studied.
We identified miR-21 and miR-224 as cell-specific and compartment-specific regulators in both PanINs and PDA. To study miRNA regulation of early PanIN development and progression, we used transgenic KrasG12D/+;Trp53R172H/+;Pdx-1-Cre (KPC) mice that exhibit stepwise progression through multiple stages of PanINs (PanIN1–3) to invasive PDA[20]. KPC mice resemble human PDA by genetic instability and histology, including desmoplastic TME[20]. Endogenous inhibition of miR-21 in primary KPC tumor cells decreased pro-tumorigenic functionalities relative to baseline levels, while overexpression of miR-224 in normal pancreatic fibroblasts conferred activated phenotypes. Inhibition of miR-21 in tumor cells simultaneously downregulated multiple tumorigenic pathways downstream of activated mKRAS. In vivo studies demonstrated that miR-21 inhibition increased survival in mice with established PDA, while early administration of a miR-21 inhibitor intercepted tumorigenesis and delayed premalignant progression. Collectively our studies identify miR-21 as a novel target for interception of mKRAS-driven PDA development.
MATERIALS & METHODS
KPC Mice
All animal studies were approved by the Institutional Animal Care and Use Committee of Johns Hopkins University. Lox-STOP-Lox-Trp53R172H/þ; Lox-STOP-Lox-KrasG12D/þ; and Pdx-1-Cre strains on a mixed C57BL/6 background, were gifted from Dr. David Tuveson (Cold Spring Harbor Laboratory, Cold Spring, NY). These mice were backcrossed to the C57BL/6 genetic background for 12 generations and interbred to obtain KPC mice. All WT C57BL/6 mice were purchased from The Jackson Laboratories. All animals were kept in pathogen-free conditions and treated in accordance with the Institutional Animal Care and Use Committee of the Johns Hopkins University and American Association of Laboratory Animal Care approved guidelines. Animals were fed Teklad Global 18% Protein Extruded Rodent Diet. The Allentown Individually Ventilated Caging System (IVC) is used to house animals with Tek-Fresh paper bedding that is also an acceptable enrichment modality. Interventions were performed during light cycle.
Laser Capture Microdissection (LCM)
Pancreata were isolated from aged KPC (4–20 weeks) or WT C57BL/6 mice and fresh-frozen in Tissue-Tek OCT (Sakura Finetek USA) or formalin-fixed and paraffin-embedded (FFPE) by the Johns Hopkins University Oncology Tissue Services. Guide slides were sectioned and stained with H&E by the Johns Hopkins University Oncology Tissue Services. Guide slides containing various grades of PanINs were identified by a pathologist. 10, 10 um frozen sections adjacent to guide sections were mounted on membrane slides (Zeiss) and stained with Cresyl Violet (Ambion). FFPE sections were stained with the Arcturus Paradise Plus Reagent System Staining kit (Thermo Fisher Scientific). Normal, low grade PanIN1, high grade PanIN2/3, and PDA tissue were microdissected using LCM (Leica LMD7000).
miRNA Profiling Using miRNA Microarray
RNA was extracted from fresh-frozen LCM samples using the RNAqueous-Micro Total RNA Isolation Kit (Thermo Fisher Scientific), quantified with a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific), and converted to cDNA using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) with Megaplex RT Primers Rodent Pool A and B (Thermo Fisher Scientific). cDNA was preamplified 18 cycles using the TaqMan PreAmp Master Mix (Thermo Fisher Scientific) and Megaplex PreAmp Primers Rodent Pool A and B (Thermo Fisher Scientific). miRNA microarray was performed by the Johns Hopkins School of Medicine Genetic Resources Core Facility using the TaqMan OpenArray Rodent MicroRNA Panel (Thermo Fisher Scientific) and TaqMan OpenArray Real-Time PCR Master Mix (Thermo Fisher Scientific). All Ct values were normalized to endogenous levels of U6 snRNA. Fold changes were quantified utilizing the ΔΔCt method in which all PanIN groups were normalized to the normal group (normal ducts isolated from WT C57BL/6 mice). P-values for differential expression statistics were generated and adjusted for multiple hypothesis testing using ExpressionSuite (Thermo Fisher Scientific).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism v7.0b (GraphPad Software). The data are presented as the means ±SEM. Comparisons between groups were made using two-tailed unpaired Student t tests and one-way or two-way ANOVA. Survival data were plotted using a Kaplan-Meier curve and differences in treatment groups were tested using the log-rank (Mantel-Cox) test. For all analyses, statistical significance is defined and shown as *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Statistical analyses for RNA-seq and TCGA are performed using R as described in the Supplementary Methods.
RESULTS
Comprehensive profiling identifies miRNAs expressed in early and late stage PanINs in KPC mice
We conducted a comprehensive miRNA profiling study of multiple PanIN stages and PDA using KPC mice. We performed microdissections on increasing grades of PanINs from sections of KPC pancreata followed by a miRNA microarray analysis utilizing the Taqman OpenArray platform to quantify the levels of 750 known murine miRNAs.
miRNA candidates were selected using specific criteria. First, candidate miRNAs had cycle threshold (Ct) values < 30 and within the detection limits of the assay. Second, candidate miRNAs had large fold changes in expression that were consistently increasing or decreasing with PanIN progression to PDA. Third, we chose miRNAs with significant fold changes (adjusted P-value < 0.05) in each PanIN stage relative to normal ducts of wildtype (WT) C56BL/6 mice. Confirmatory qPCR was completed to validate all miRNAs identified by OpenArray. Lastly, the miRNAs had to have either putative or proven roles in cancer promoting pathways.
Our selection criteria resulted in 14 miRNA candidates. Supplementary Tables 1 and 2 summarize the fold changes observed with increasing stages of PanIN development to PDA of our top 14 candidates and the primers used for subsequent validation. We found significant dysregulation of some miRNAs (miR-21, miR-16, and miR-28) as early as low-grade PanIN1 (P1), while dysregulation of others (miR-224, miR-28 and miR-216b) began at higher PanIN grades (P2/3). All 14 candidate miRNAs have significantly dysregulated expression in PDA compared to expression levels in normal ducts of WT mice.
Increased spatial-temporal expression of miR-21 and miR-224 with PanIN progression to PDA development
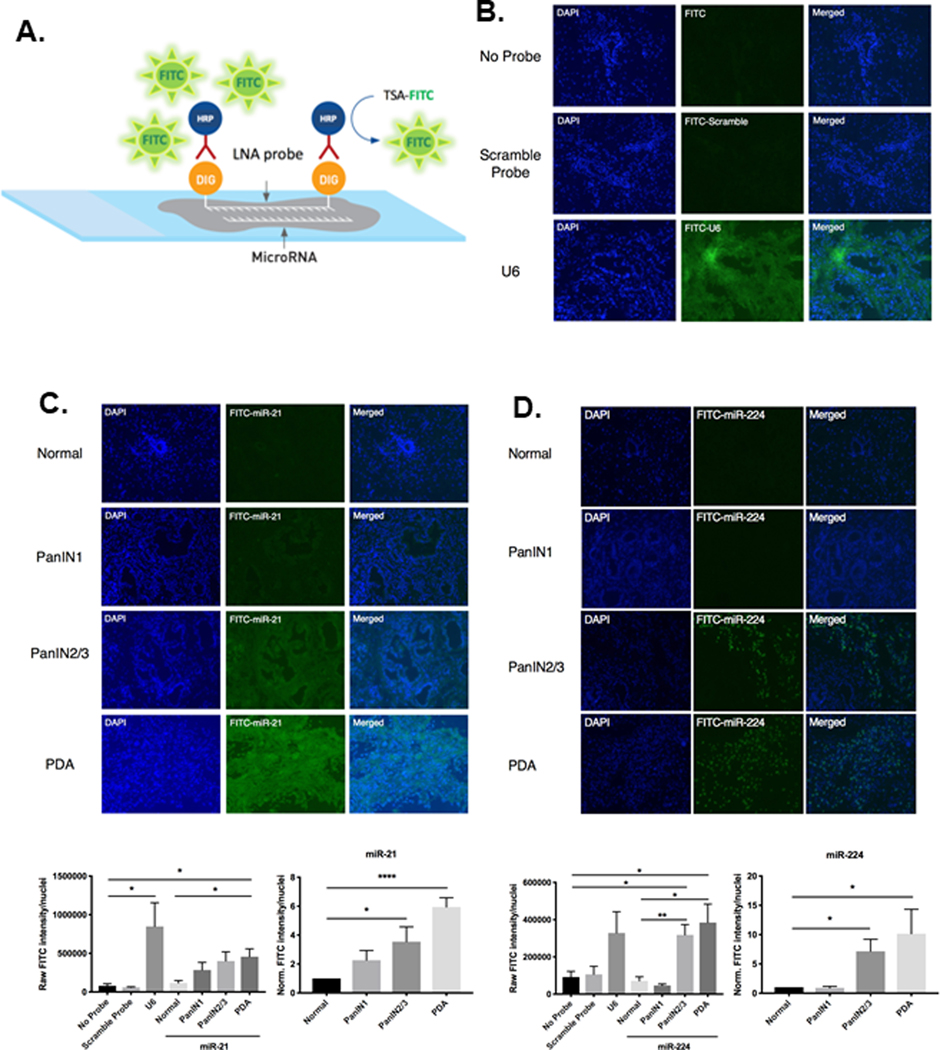
We chose to further evaluate miR-21 and miR-224 because these two miRNAs have been reported to regulate targets in cancer-promoting inflammatory pathways[21] and epithelial-mesenchymal transition[22], respectively. We performed miRNA fluorescence in situ hybridization (miR-FISH) (Figure 1A, B) to examine the endogenous expression of miR-21 and miR-224 in specific cell populations in early and late PanINs and in PDA in late PanINs and in PDA (Figure 1C, D). miR-FISH allows the fluorescent visualization of miRNAs in sections of whole tissue. The intensity of the fluorescent signal directly correlates with levels of endogenous miRNA expression. We used modified bicyclic locked-nucleic acid (LNA) probes that are complimentary in sequence and therefore will stably and accurately hybridize to miRNAs of interest at specific hybridization temperatures in situ.
Figure 1. miR-FISH analysis quantifies endogenous spatial-temporal expression of miR-21 and miR-224 throughout premalignant progression.
A. Schematic of miR-FISH, which detects endogenous miRNA in tissue sections. B. Representative fluorescent images of sections of pancreas stained with either no probe, non-specific scramble-miRNA probe, or U6 positive control probe. No fluorescent signal was observed with no probe or scramble-miRNA probe staining, however strong ubiquitous fluorescent signal was observed upon staining with U6 positive control probe. C. Representative fluorescent images of normal ducts, increasing grades of PanIN lesions, and PDA tissue stained with miR-21. Expression of miR-21 is concentrated in ductal epithelial cells. Displayed below is the quantified image analysis of miR-21 fluorescent signal. D. Representative fluorescent images of normal ducts, increasing grades of PanIN lesions, and PDA tissue stained with miR-224. Expression of miR-224 is concentrated in the stromal compartment surrounding high-grade PanIN2/3 lesions and PDA. Below is the quantified image analysis of miR-224 fluorescent signal. Raw fluorescent signals were quantified by FITC intensity/nuclei. PanIN1, PanIN2/3, and PDA raw signals were normalized to that of normal pancreatic ducts of WT C57BL/6 mice to generate relative signal intensities. (n=4–6 experiments).
miR-FISH staining of WT ducts, increasing grades of KPC PanINs, and PDA tissue demonstrates spatial-temporal expression of miR-21 is specifically concentrated in the ductal epithelial cells of PanIN lesions and increases in expression with progression to PDA (Figure 1C). The quantified raw fluorescent intensities of miR-21 in PDA tissue is significantly increased compared to normal WT ducts, while the normalized fluorescent signal indicates that high grade PanIN2/3 lesions have a significant 3.5-fold increase and PDAs have a significant 6-fold increase of miR-21 expression compared to normal ducts (Figure 1C), which correlates closely to the 7-fold increase of miR-21 expression in KPC tumor cells relative to WT pancreatic epithelial cells quantified by qPCR (Supplementary Figure 1). In contrast, miR-224 expression localized in the invading stromal compartment surrounding high grade PanIN2/3 lesions and persists with progression to PDA (Figure 1D). These data are consistent with our findings in KPC CAFs that miR-224 expression is upregulated relative to normal pancreatic associated fibroblasts (PNAFs) early in the premalignant stromal cell compartment of developing PDAs. Moreover, the normalized fluorescent signal of miR-224 produced by miR-FISH (Figure 1D) demonstrates that the significant 10-fold upregulation of miR-224 in PDA stroma equals the 10-fold increase of miR-224 expression in KPC CAFs as compared to PNAFs (Supplementary Figure 1). The miR-FISH results qualitatively and quantitatively confirm qPCR results and further informs the compartmental-specific expression of miR-21 and miR-224 throughout early PDA development.
miR-21 inhibition decreased tumor cell proliferation, migration, and invasion, whereas miR-224 inhibition increased CAF cell migration
To investigate the functional roles of miR-21 and miR-224 in regulating early PDA development, we stably transduced the primary cell lines with miRNA inhibitors and mimics. Since KPC tumor cells express higher levels of miR-21 than WT epithelial cells and KPC CAFs express higher levels of miR-224 than PNAFs, we used lentiviral vectors to stably inhibit endogenous levels of miR-21 in KPC tumor cells and miR-224 in KPC CAFs. The inhibition occurs through expression of miRNA inhibitors that bind to target miRNAs via sequence complementation to prevent the loading of target miRNAs onto the RNA-induced silencing complex (RISC). Conversely, we transduced WT pancreatic epithelial cells and PNAFs with lentivirus overexpressing miR-21 and miR-224, respectively. Successfully transduced cells were selected using antibiotics specific to each construct, and further enriched using FACs sorting (Supplementary Figure 2, Supplementary Figure 3A–F). Supplementary Figure 3G summarizes the lentiviral vectors, target cell lines, fluorescent markers, and antibiotic selection used to generate stable cell lines expressing their associated miRNA mimic or inhibitor.
The degree of miRNA inhibition was measured by qPCR. miRNA inhibitors significantly reduced the expression of miR-21 in KPC tumor cells and miR-224 in KPC CAFs about 2-fold compared to non-transduced cells. However, inhibition was incomplete due to residual miR-21 and miR-224 expression being higher than in WT epithelial cells (Figure 2A) and PNAFs, respectively.
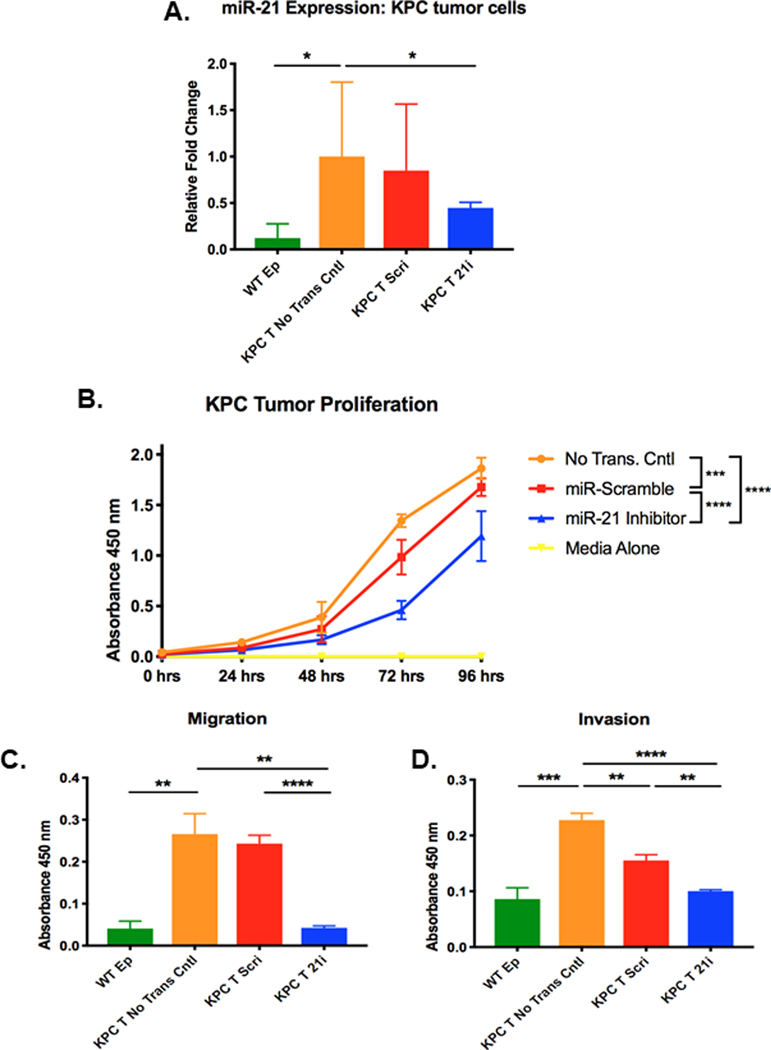
Figure 2. Inhibition of miR-21 in KPC tumor cells decreases cell proliferation, migration, and invasion.
A. Relative miR-21 expression in WT pancreatic epithelial cells (WT Ep) and KPC tumor cells transduced with either miR-scramble inhibitor (KPC T Scri) or miR-21 inhibitor (KPC T 21i) compared to expression in non-transduced KPC tumor cells. Relative fold change of miR-21 was quantified using the ΔΔCt method in which all Ct values were first normalized to endogenous control U6 snRNA and all experimental groups were normalized to non-transduced KPC tumor cells (n=3 samples/group, significance was determined by one-way ANOVA). B. Proliferation of non-transduced KPC tumor cells, KPC tumor cells transduced with either miR-scramble inhibitor or miR-21 inhibitor, and media alone (n=3 experiments, significance was determined by two-way ANOVA). C. Migration and D. Invasion of WT pancreatic epithelial cells (WT Ep), non-transduced KPC tumor cells, and KPC tumor cells transduced with either miR-scramble inhibitor or miR-21 inhibitor (n=3 experiments).
Next, we investigated functional changes in KPC tumor cells following reduced expression of miR-21. miR-21 inhibition significantly reduced proliferation by 2-fold relative to non-transduced KPC tumor cells (Figure 2B). More importantly, downregulation of miR-21 significantly reduced both migration and invasion to levels comparable to that of WT pancreatic epithelial cells (Figure 2C-D), demonstrating that reduction of miR-21 levels in KPC tumor cells can revert the cell’s migratory and invasive capacities to that of normal un-transformed epithelial cells. Interestingly, downregulation of miR-224 in KPC CAFs did not affect cell proliferation (Figure 3A, B). However, the migratory capacity of KPC CAFs was significantly increased by 1.7-fold in miR-224 inhibited CAFs compared to non-transduced CAFs (Figure 3C). Invasion was not affected by downregulation of miR-224 in KPC CAFs (Figure 3D).
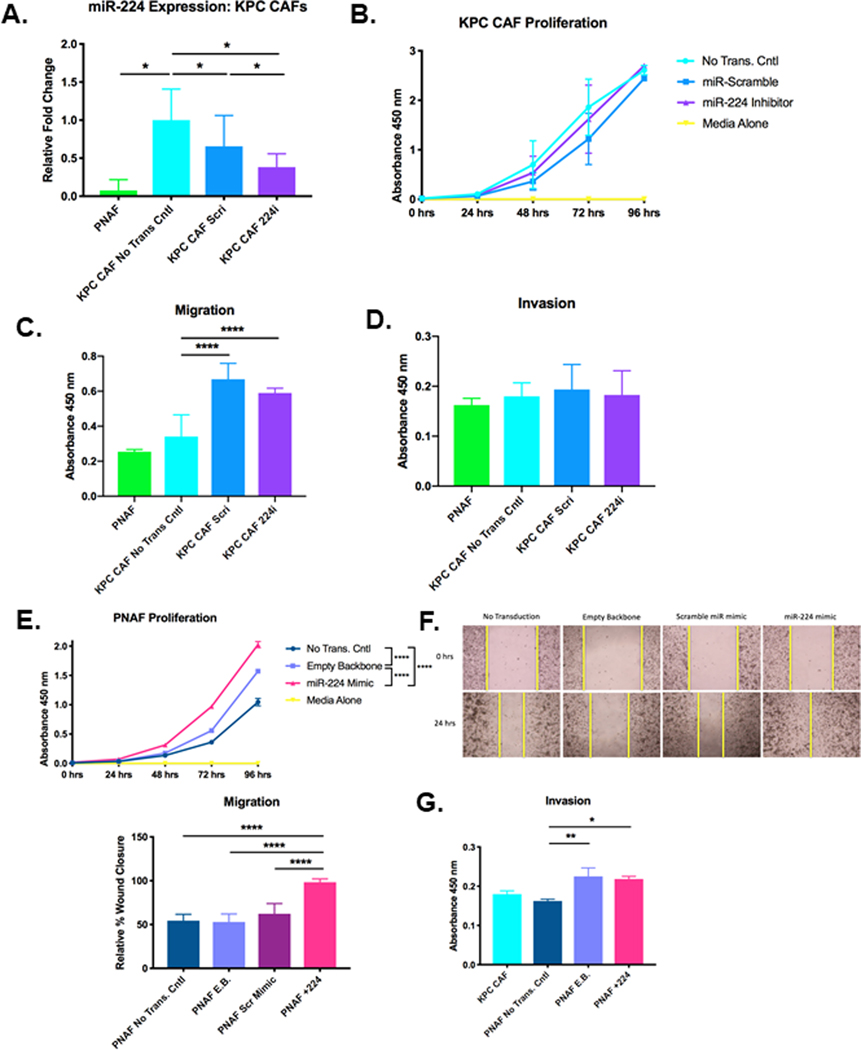
Figure 3. Inhibition of miR-224 in KPC CAFs increases migration, and overexpression of miR-224 in normal fibroblasts increases cell proliferation, migration, and invasion.
A. Relative miR-224 expression in PNAFs and CAFs transduced with either miR-scramble inhibitor (KPC CAF Scri) or miR-224 inhibitor (KPC CAF 224i) compared to expression in non-transduced CAFs. Relative fold change of miR-224 was quantified using the ΔΔCt method in which all Ct values were first normalized to endogenous control U6 snRNA and all experimental groups were normalized to non-transduced KPC CAFs (n=3 samples/group, significance was determined by one-way ANOVA). B. Proliferation assay of non-transduced KPC CAFs, KPC CAFs transduced with either miR-scramble inhibitor or miR-224 inhibitor, and media alone (n=3 experiments, significance was determined by two-way ANOVA). C. Migration and D. Invasion of PNAFs, non-transduced KPC CAFs, and KPC CAFs transduced with either miR-scramble inhibitor or miR-224 inhibitor (n=3 experiments).E. Proliferation assay of non-transduced PNAFs, PNAFs transduced with either empty backbone or miR-224 mimic, and media alone (n=3 samples/group, significance was determined by two-way ANOVA). F. Cell migration was quantified by relative % wound closure after 24 hours for all groups (n=3 experiments, significance was determined by one-way ANOVA). G. Invasion assay of KPC CAFs, non-transduced PNAFs, and PNAFs transduced with either empty backbone (PNAF E.B.) or miR-224 mimic (PNAF +224) (n=5 samples/group)
PNAF miR-224 overexpression increased cell proliferation, migration and invasion
PNAFs overexpressing miR-224 have a significant 2-fold increase in cell proliferation compared to non-transduced PNAFs (Figure 3E). Scratch assay demonstrated upregulation of miR-224 expression significantly increased PNAF migratory capacity by 2-fold compared to all other control groups to achieve 100% wound closure (Fig. 3F). This migratory capacity is comparable to KPC CAFs, indicating that miR-224 overexpression can confer activated phenotypes like migration to PNAFs. Additionally, PNAFs overexpressing miR-224 have significantly increased cell invasion compared to non-transduced PNAFs (Figure 3G). Collectively, this demonstrates increased expression of miR-224 in normal fibroblasts produces CAF-liked activated cellular functions.
Stable inhibition of miR-21 in KPC tumor cells downregulates pro-tumorigenic pathways downstream of mKRAS activation
RNA-seq was employed to investigate global miRNA-mediated modulation of key genes and pathways regulated by miR-21 in tumor cells and miR-224 in CAFs. Log fold changes for all gene transcripts were compared between (1) miR-21-inhibited and scramble-inhibited KPC tumor cells and (2) miR-224-inhibited and scramble-inhibited KPC CAFs. GAGE[23] pathway analyses on the differentially expressed gene sets produced by miR-21 and miR-224 inhibition revealed miR-21 inhibition significantly downregulated the MAPK, mTOR and actin cytoskeleton KEGG pathways in tumor cells (Supplementary Figure 4A), while miR-224 inhibition significantly downregulated the DNA replication, cell cycle and p53 signaling KEGG pathways in CAFs (Supplementary Figure 4B).
The heatmaps for the three KEGG pathways significantly downregulated by miR-21 inhibition in KPC tumor cells (highlighted top 4 rows) are shown in Supplementary Figure 4A. Although not statistically significant (P=0.06), the RAS pathway was included for analysis due to the biological importance and relevance of the pathway to both early PanIN progression and PDA development. All 4 heatmaps display downregulation in all 4 pathways (in blue) for the two miR-21-inhibited tumor samples compared to the two scramble-inhibited tumor samples which have higher gene expression. In contrast, there is less consistent downregulation in the CAF samples (bottom 4 rows), indicating that the regulation of these 4 KEGG pathways are specific to miR-21’s role in PDA cells. The significant downregulation of MAPK, mTOR and actin cytoskeleton pathways by miR-21 inhibition in tumor cells is in accordance with the reduced proliferation, migration and invasion observed with stable miR-21 inhibition. We selected 5 differentially expressed genes from each pathway for qPCR validation of fold changes (Supplementary Figure 5A–E, Supplementary Table 3).
The heatmaps for the three KEGG pathways that were significantly downregulated by miR-224 inhibition in KPC CAFs (highlighted bottom 4 rows) are shown in Supplementary Figure 4B. Despite being statistically significant, the heatmaps of all three pathways lack consistent gene expression changes between samples within each treatment group. Additionally, there is minimal clustering within the two samples of scramble-inhibited KPC CAFs. The transcriptional variability between biological replicates may indicate that there is greater heterogeneity of expressed genetic pathways following miR-224 inhibition in CAFs than can be adequately measured by this approach. We chose 5 genes (Shisa2, Cxcl12, Rasgef1b, Vav3, Dusp9), regardless of pathway classification, that were significantly regulated by miR-224 inhibition for qPCR validation of fold changes (Supplementary Figure 5F). Of the genes screened, Shisa2 was most significantly downregulated by miR-224 inhibition in CAFs, with a 255-fold reduction by qPCR (greater than 86-fold reduction by RNA-seq) when compared to the expression in CAFs transduced with scramble inhibitor. Shisa2, a regulatory endoplasmic reticulum (ER) protein, inhibits fibroblast growth factor (FGF) signaling by preventing the maturation and cell-surface expression of FGF receptors (FGFRs)[24]. FGF signaling through FGFRs have been shown to promote cell migration in many cancer types and embryogenesis[25,26]. Therefore, the significant reduction in Shisa2 levels by miR-224 inhibition and potential increase in FGFR may explain the increased migration of miR-224-inhibited CAFs in vitro (Figure 3C).
miR-21 inhibition in vivo increases overall survival in established PDA
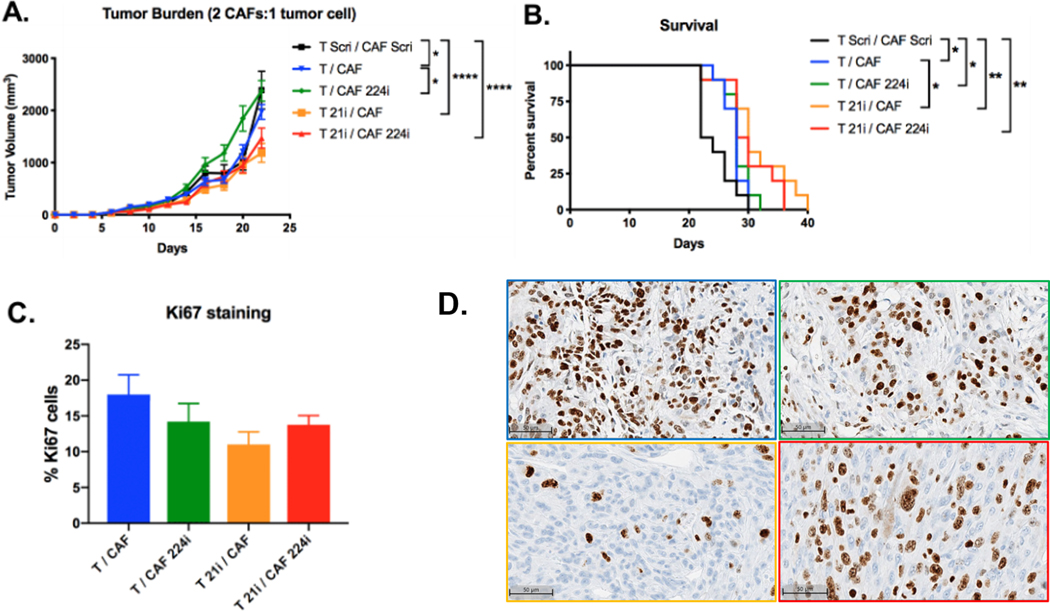
We next investigated the effects of inhibiting miR-21 in vivo. Between 60 and 90% of primary PDA tumor volume is fibrotic stroma[8,27]. Therefore, to more accurately recapitulate the major components of the PDA TME, we established subcutaneous tumors in C57BL/6 mice with a 2 CAF:1 KPC tumor cell ratio of different combinations of miRNA-inhibited or normal KPC tumor cells or CAFs. Mice with tumors composed of miR-21-inhibited KPC tumor cells and normal KPC CAFs (T 21i/CAF) had the lowest tumor burden and longest survival compared to all other groups (Figure 4A, B). IHC staining of resected tumors with the proliferation marker Ki67, demonstrated that T 21i/CAF tumors had the lowest percentage of actively proliferating Ki67+ cells (Figure 4C, D), suggesting that reduced tumor size is due to decreased proliferative capacity of miR-21-inhibited tumor cells. Tumors composed of normal KPC tumor cells and miR-224-inhibited CAFs (T/CAF 224i) were comparable in size with scramble-inhibited tumors (T Scri/CAF Scri) and larger than normal non-transduced tumors (T/CAF). Collectively, these data show that only miR-21 inhibition of tumor cells reduces tumor burden and confers survival benefit in established PDA.
Figure 4. Effects of in vivo miR-21 and miR-224 inhibition on tumor burden and survival.
Subcutaneous tumors composed of 2 CAF:1 tumor cell ratio of various combinations of scramble-inhibited, miRNA-inhibited, or normal KPC cells were implanted into the flanks of immunocompetent WT C57BL/6 mice. A. Tumor growth (n=10 mice/group, significance was determined by two-way ANOVA) and B. Percent survival of mice that received subcutaneous tumors of various combinations of KPC tumor cells and CAFs (n=10 mice/group, significance was determined by the log-rank (Mantel-Cox) test). C. %Ki67 positive cells in sections of resected tumors as determined by IHC analysis (n=5 mice/group, significance was determined by one-way ANOVA). D. Representative ki-67 IHC images from the subcutaneous xenograft model, 20x magnification. IHC images are outline color corresponds to the condition in Figure 4C. Ki67 staining was unsuccessful for the scramble-inhibited tumors (T Scri/CAF Scri) due to necrotic tissue in that treatment group.
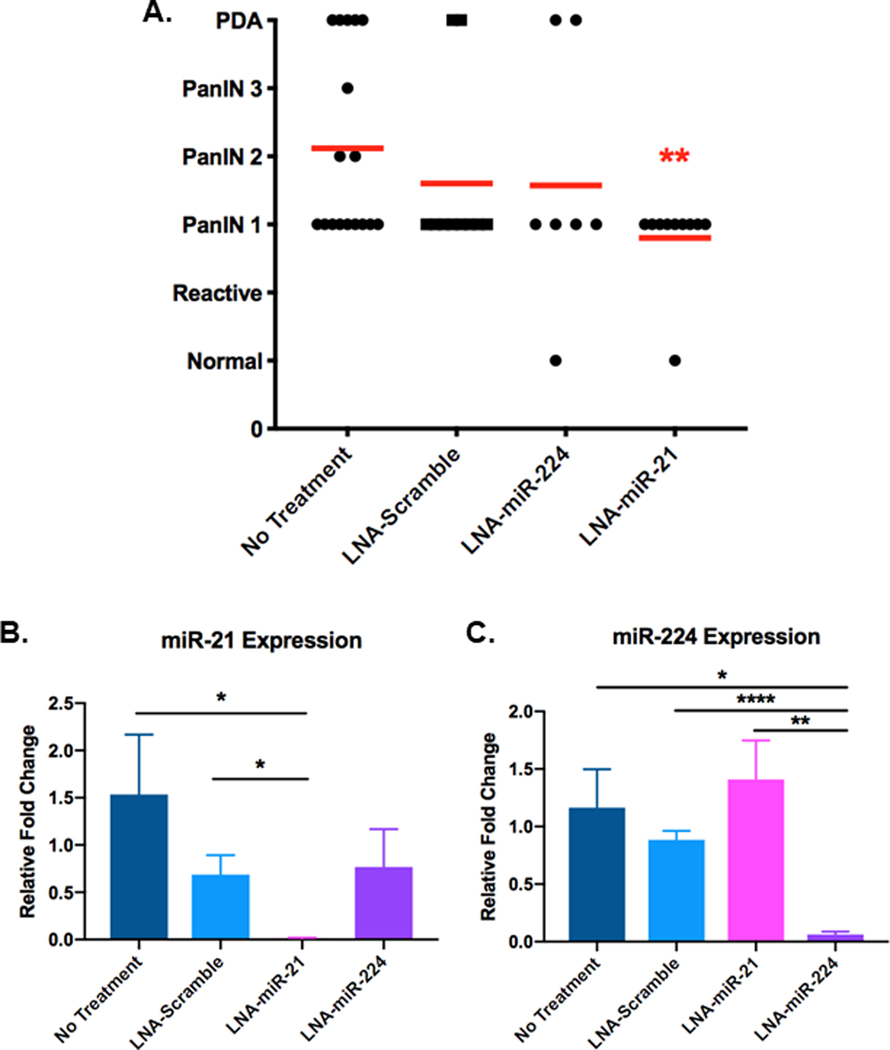
Systemic administration of LNA-miR-21 inhibitor intercepts early PanIN progression
Systemic administration of miR-21 and miR-224 inhibitors delayed progression of PanIN lesions to PDA in KPC mice. To deliver miR-21 and miR-224 inhibitors, oligonucleotides were chemically modified with a LNA ribose ring structure and phosphorothioate backbone for enhanced pharmacokinetic stability in vivo[28]. 4–5 week old KPC mice, which are expected to already have low-grade PanIN1 lesions, were dosed with negative control LNA-scramble inhibitor, LNA-miR-21 inhibitor, or LNA-miR-224 inhibitor once every 5 days for 6 weeks. At the end of the dosing regimen, the pancreas, liver, lung and kidney from treated and untreated mice were collected and analyzed for drug efficacy and toxicity. Each pancreas was pathologically evaluated for the highest-grade lesion present. KPC mice receiving early systemic administration of LNA-miR-21 inhibitor developed the fewest advanced PanINs compared to other treatment groups (Figure 5A). The average grade of premalignant lesions in the pancreas of mice dosed with LNA-miR-21 inhibitor was significantly lower than that of untreated mice. Furthermore, mice that received LNA-miR-21 inhibitor did not develop any lesions beyond the low-grade PanIN1 stage, demonstrating that early systemic miR-21 inhibition successfully delays premalignant progression. Additionally, our studies show that as a monotherapy, LNA-miR-21 inhibitor effectively intercepts PDA tumorigenesis when administered early at the time of mKRAS activation. No adverse effects of systemic miR-21 inhibition were observed in post-mortem examination (Supplementary Figure 6).
Figure 5. Effect of systemic miRNA inhibition on progression of PanIN lesions in KPC mice.
KPC mice at 4–5 weeks of age were left untreated or I.V. dosed with 25 mg/kg of LNA-scramble inhibitor, LNA-miR-224 inhibitor, or LNA-miR-21 inhibitor every 5 days for 6 weeks. RNA was isolated from whole pancreata and qPCR quantified miR-21 and miR-224 levels in each treatment group. A. Red lines indicate the average PanIN stage that was detected in the pancreata of each treatment group (n=at least 7 mice/group, significance was determined by one-way ANOVA). B. Relative fold changes of miR-21 expression and C. miR-224 expression compared to levels in untreated mice. Relative fold changes of miR-21 and miR-224 were quantified using the ΔΔCt method in which all Ct values were first normalized to endogenous control U6 snRNA and all experimental groups were normalized to untreated KPC mice (n=5 mice/group)
qPCR quantification of miRNA levels in the whole pancreas of dosed KPC mice showed that LNA-miR-21 inhibitor-treated mice had a significant 112-fold decrease and 50-fold decrease of miR-21 levels in their pancreas compared to untreated mice and LNA-scramble inhibitor-treated mice, respectively (Figure 5B). Mice that received LNA-miR-224 inhibitor had a significant 19-fold decrease and 14-fold decrease of miR-224 levels in their pancreas as compared to untreated mice and LNA-scramble inhibitor-treated mice, respectively (Figure 5C). Additionally, the data demonstrate that systemic miR-224 inhibition does not affect localized expression of miR-21 and vice versa, thereby demonstrating the specificity of each miRNA inhibitor. Collectively, these data show that systemically administered miRNA inhibitors successfully localized to the pancreas and inhibited their targets with a high degree of specificity.
Correlation of miR-21 and miR-224 Expression in TCGA Recapitulates Cellular Specificity in Human Tumors
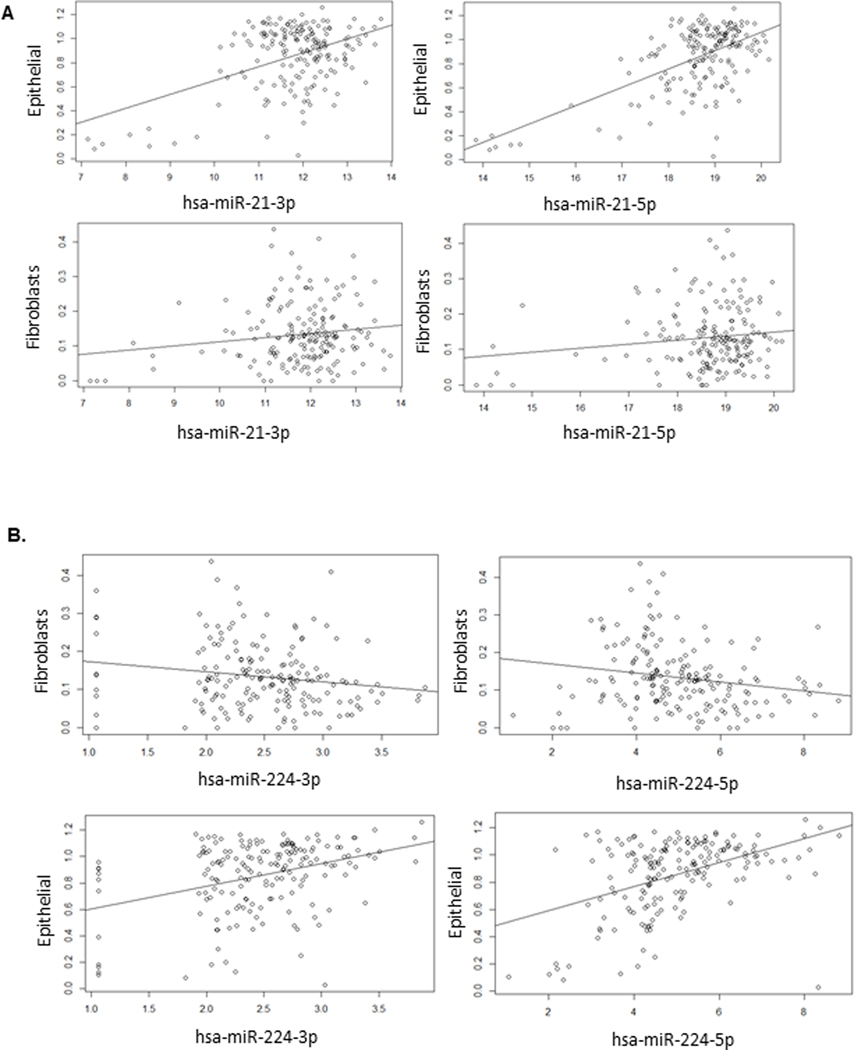
To examine translation of our findings to human PDA, we investigated miR-21 and miR-224 expression in the PDA cohort of TCGA. Specifically, we looked for a correlation between miR-21 expression and epithelial cell content as well as miR-224 expression and fibroblast content. This corresponds to our murine findings suggesting miR-21 and miR-224 cellular and compartmental specificity. Using the publicly available xCell quantification for TCGA[29], we compared epithelial and fibroblast content with miR-21 and miR-224 expression in 177 patients in the PDA cohort. Correlation analysis revealed a significant direct relationship between miR-21 expression and tumor epithelial cell content with p-values of 8.7e-7 and 2.1e-13 for miR-21–3p and miR-21–5p, respectively (Figure 6A). This relationship was not seen with comparison between miR-21 expression and tumor fibroblast content and is consistent with our findings in KPC model.
Figure 6. Analysis from TCGA examines miR expression with tumor cellular content.
The PDA cohort of TCGA was used to correlate miR expression in relation to tumor cellular composition estimated from computational microdissection with xCell. A. miR-21 expression is significantly associated with tumor epithelial cell content. P-value for epithelial cell content for hsa-miR-21–3p= 8.7e-7; hsa-miR-21–5p = 2.1e-13. There is not a significant association between miR-21 expression and tumor fibroblast content. B. miR-224 expression is inversely associated with tumor fibroblast content and associated with epithelial cell content. P-value for fibroblast content for hsa-224–3p = 0.022; hsa-224–5p = 0.00015. P-value for epithelial content for hsa-224–3p = 2.0e-5; hsa-224–5p = 0.00021. Plotted miR expression values are variance stabilized.
Similar comparison for miR-224 expression revealed an inverse relationship between miR-224 expression and tumor fibroblast content and a direct relationship between miR-224 expression and tumor epithelial cell content (Figure 6B). Though the miR-224 findings were not consistent with our in vitro and in vivo murine findings, the broad classification of fibroblasts in the dataset may not be limited to CAFs, which would impact our analysis.
DISCUSSION
MiRNAs are regulatory non-coding RNAs controlling signaling pathways in both normal and tumor cells. We identified miR-21 and miR-224 as regulators of specific cell types within developing mKRAS-driven PDAs. We demonstrate that miR-21 is a potent regulator of epithelial to mesenchymal transition (EMT), modulating transformed epithelial cell invasion and migration. We show for the first time that miR-21 can serve as a target for intercepting PDA development following mKRAS activation in early PanIN lesions. We also identified miR-224 as a key regulator of TME fibroblasts, activating normal fibroblasts to exhibit CAF activity. Finally, we extrapolated our findings to the PDA cohort of TCGA and demonstrated a correlation between miR-21 expression and tumor epithelial cell content.
Since mKRAS targeting remains a challenge in the clinical setting, inhibiting downstream effector pathways is a leading strategy for developing mKRAS therapeutics[10,11]. Our study demonstrates miR-21 inhibition simultaneously downregulates pro-tumorigenic effector pathways including MAPK, mTOR and actin cytoskeleton pathways, downstream of mKRAS in KPC tumor cells. Previous studies demonstrated miR-21 directly inhibits individual tumor suppressor gene targets such as phosphatase and tensin homolog (PTEN)[30,31], programmed cell death protein 4 (PDCD4)[32], Sprouty2 (Spry2)[33], tropomyosin (TPM1)[34] and RAS p21 protein activator 1 (RASA1)[35,36] to modulate cell proliferation, migration, invasion, and apoptosis in various cancer types. However, here we show miR-21 inhibition is capable of simultaneously downregulating multiple mKRAS-activated biologic pathways responsible for tumor cell growth, metabolism, and EMT, indicating that miR-21 is a more potent regulator of mKRAS signaling than previously realized. Additionally, miR-21 has been identified as a downstream transcriptional target of KRAS activation via transcription factor ELK1, which directly binds to miR-21’s proximal promoter region, leading to increased miR-21 expression[35]. Thus, miR-21 is a ncRNA effector of KRAS activation, providing additional rationale for its inhibition in mKRAS-driven PDA. Studies have shown that targeting individual downstream KRAS effectors such as MEK1/2, Erk1/2, or Akt has limited clinical efficacy due to compensatory mechanisms swiftly leading to acquired resistance, coupled with substantial toxicities[10,11]. We have also demonstrated that miR-21 expression is directly related to tumor epithelial cell content for the PDA cohort of TCGA. miR-21 is a promising therapeutic target because its inhibition can block oncogenic KRAS signaling through the regulation of multiple signaling proteins in multiple key pro-tumorigenic pathways, thereby potentially reducing mechanisms of acquired resistance and toxicities associated with combination drug treatments.
Our study shows for the first time that miR-224 is specifically upregulated in activated KPC CAFs compared to normal fibroblasts, and its increased spatial-temporal expression in the developing TME is specifically localized in the stromal compartment surrounding high grade PanIN2/3 lesions and PDA. The in situ localization of miR-224 contrasts that of miR-21’s, whose expression is concentrated in transformed ductal epithelial cells. Moreover, endogenous overexpression of miR-224 in normal pancreatic fibroblasts significantly increased cell proliferation, migration, and invasion, indicating that overexpression of miR-224 may generate CAFs by conferring activated phenotypes to normal fibroblasts.
We postulate that the early upregulation of miR-224 may be due to the increased expression of hypoxia-inducible factor 1 alpha (HIF-1α) and NFκB in the developing TME[8,37]. Both HIF-1α in gastric cancer and NFκB in hepatocellular carcinoma (HCC) transcriptionally regulate miR-224 by binding directly to its promoter region[38,39]. Although only demonstrated in tumor cells, this same mechanism of action may lead to increased miR-224 expression and potential miR-224-mediated activation of normal resident fibroblasts in the premalignant and PDA microenvironment. Conversely, endogenous inhibition of miR-224 in KPC CAFs did not affect cell proliferation or invasion, but rather increased cell migration, an effect possibly mediated by significant downregulation of Shisa2 in miR-224-inhibited CAFs quantified by RNA-seq. Shisa2, a negative regulator of FGF signaling[24], may promote cell migration upon downregulation in KPC CAFs. Although miR-224 inhibition has been shown to reduce pro-tumorigenic functionalities of tumor cells in many cancer types[38,40], the effects of miR-224 inhibition in CAFs have not been reported. Future studies are warranted to determine how best to regulate miR-224 activity in CAFs to revert or reduce their activated phenotype, and to determine if such reversion contributes to reduced tumorigenicity of PDA, either through reduced tumor cell growth and metastasis or through improved anticancer therapy access to the PDA TME, or both.
Our in vivo studies demonstrate that miR-21 inhibition produces therapeutic effects at multiple stages of PDA development. We report for the first time that early systemic administration of the LNA-miR-21 inhibitor significantly decreased the occurrence of high-grade PanINs and PDA in KPC mice. Our study shows that inhibiting miR-21 may be a promising strategy to prevent or slow the development of other KRAS-driven cancers. Our data are consistent with previous findings that prove miR-21 to be an oncogenic miRNA (oncomir) and that its inhibition impedes tumorigenesis in vivo. Utilizing a doxycycline-inducible mir-21LSL-Tetoff mouse model, induction of miR-21 produced pre-B-cell lymphoma, while miR-21 inactivation led to complete tumor regression[41]. Additionally, in a spontaneous transgenic mouse model of KRAS-driven NSCLC, overexpression of miR-21 enhanced tumorigenesis while knock-out of miR-21 reduced tumor formation in KRASLA2 mice[42]. In this study, we report that early systemic miR-21 inhibition can specifically impede early mKRAS-driven PDA tumorigenesis.
Our data along with those of previous groups show miR-21 as a potential diagnostic marker for the presence of PanINs and PDA due to miR-21’s increased expression throughout premalignant to PDA development[43,44]. Stable exosome encapsulated miR-21 is also elevated in the serum of patients with premalignancies and PDA, demonstrating that miR-21 is a potential circulating diagnostic marker for noninvasive early and established PDA screening reflective of increased in-tissue expression[45,46]. Therefore, elevated levels of miR-21 may indicate when miR-21 inhibition treatment should be initiated during early disease development. However, more accurate PanIN and PDA detection is necessary for the success of preventative therapies.
Our studies demonstrate that miR-21 is an important regulator of KPC tumor cell function both in vitro and in vivo. However, increasing evidence supports that miR-21 is also a regulator of fibroblast activation and CAF functionality. miR-21 expression is increased in CAFs compared to normal fibroblasts and miR-21 overexpression in normal fibroblasts enhances levels of CAF-specific markers, cell motility, and proliferation, indicating that miR-21, like miR-224, may induce CAF formation[47,48]. Moreover, inhibition of miR-21 reduces CAF migration, invasion, and proliferation[47]. miR-21 achieves this regulation in fibroblasts by directly targeting and downregulating SMAD7, an inhibitor of the TGFβ pathway, thereby sensitizing fibroblasts to TGFβ-mediated CAF formation and fibrosis[49]. In addition to cancer models, fibroblast activation in models of diabetic nephropathy and pulmonary fibrosis has been shown to occur via the same TGFβ/miR-21/SMAD7 regulatory axis[49,50]. In fact, in vivo inhibition of miR-21 attenuated fibrosis in a mouse model of fibrotic lung disease[50], a finding that is highly applicable to PDA intervention because the fibrotic stroma is an integral component of the TME that propagates PDA. Furthermore, the reported findings that miR-21 inhibition reduces fibrosis provides another mechanism of action for the PanIN-intercepting efficacy of our early systemic miR-21 inhibitor treatment reported in this study. Systemic dosing of LNA-miR-21 inhibitor exposed the entire premalignant microenvironment to miR-21 inhibition which inhibited transformed epithelial cells and possibly the activation of fibroblasts.
In addition to CAFs in the stromal compartment, miR-21 has also been shown to promote pro-tumorigenic immune populations. miR-21 has been shown to induce the polarization of tumor-supportive anti-inflammatory M2 macrophages[51], the differentiation of naïve CD4+ T cells to T regulatory cells (Tregs)[52], and the expansion of myeloid-derived suppressor cells (MDSCs) that exhibit augmented suppressive capabilities upon miR-21 overexpression[53]. Inhibition of miR-21 in the premalignant microenvironment may act to simultaneously reduce fibrosis and suppress the generation of immunosuppressive cell populations, which may cumulatively delay premalignant progression and reduce tumor growth. Therefore, miR-21 exerts its oncogenic effects on multiple cell populations, providing abundant evidence for inhibiting this miRNA in early disease progression. miR-21 inhibition shows efficacy as a monotherapy; however, future studies should test miR-21 inhibition in combination with other agents to modulate several components of the TME simultaneously to more effectively impede disease development.
In summary, we have shown that miR-21 and miR-224 are dysregulated throughout early premalignant progression to established PDA, and demonstrate cell-specific overexpression in the developing microenvironment. These data provide new opportunities for intercepting KRAS-driven premalignancy through impeding epithelial cell transformation and by reducing the PDA stromal barrier to therapeutic access.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Sidney Kimmel Comprehensive Cancer Center (SKCCC), the Skip Viragh Center for Pancreas Cancer, and the Bloomberg~Kimmel Institute for Cancer Immunotherapy at Johns Hopkins. The authors also thank the Johns Hopkins University Oncology Tissue Services for all histology and slide preparation, the SKCCC Cell Imaging Core for fluorescent and infrared imaging, the SKCCC Flow Cytometry Core for cell sorting, the SKCCC Experimental and Computational Genomics Core for RNA-seq, and the Johns Hopkins School of Medicine Genetic Resources Core Facility for miRNA microarray profiling. The authors thank L. Zheng (Johns Hopkins University School of Medicine, Baltimore, MD) for primary KPC and PNAF cell lines.
GRANT SUPPORT:
The authors acknowledge financial support from the National Institutes of Health/National Cancer Institute grant R01CA184926 to EMJ, Bloomberg Philanthropies, The Bloomberg Kimmel Institute for Cancer Immunology, The Skip Viragh Center for Pancreatic Cancer, The McGlothlin Foundation, and Lefkovsky Family Foundation grant 90058175 to EMJ. RAA and EMJ have support from NCI SPORE P50 CA062924. JWZ is supported by 5T32 CA009071.
DISCLOSURES:
EMJ and the Johns Hopkins University have the potential to receive royalties for GVAX and mesothelin through a licensing agreement between Johns Hopkins and Aduro Biotech; receives research funding from Aduro Biotech and Bristol Myers Squibb; serves as a consultant for Adaptive Biotech, Dragonfly, CSTONE and Genocea; serves on the scientific advisory board for the Parker Institute for Cancer Immunology and the Lustgarten Foundation. RAA receives research support or has consulting arrangements with Bristol Myers Squibb, Merck, Incyte, FLX biologics and adaptive biotech. EJF serves as a consultant for Champions Oncology. NJC is now employed by AstraZeneca. All other authors declare no competing interests.
ABBREVIATIONS:
- ANOVA
analysis of variance
- CAF
cancer associated fibroblast
- EMT
epithelial to mesenchymal transition
- ER
endoplasmic reticulum
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- GAGE
generally applicable gene-set enrichment
- GO
gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- KPC
KrasG12D/+;Trp53R172H/+;Pdx-1-Cre
- LNA
locked nucleic acid
- MDSC
myeloid-derived suppressor cell
- miRNA
microRNA
- miR-FISH
miRNA fluorescence in situ hybridization
- mKRAS
mutated KRAS
- PanIN
pancreatic intraepithelial neoplasias
- PDA
pancreatic ductal adenocarcinoma
- PNAF
normal pancreatic associated fibroblasts
- TCGA
the cancer genome atlas
- TME
tumor microenvironment
- Treg
T regulatory cells
Footnotes
REFERENCES
- 1.Hidalgo M Pancreatic Cancer. N Engl J Med. 2010;362:1605–22. [DOI] [PubMed] [Google Scholar]
- 2.Collins MA, Bednar F, Zhang Y, Brisset JC, Galbán S, Galbán CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.di Magliano MP, Logsdon CD. Roles for KRAS in pancreatic tumor development and progression. Gastroenterology. 2013;144:1220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lou E, Subramanian S, Steer CJ. Pancreatic cancer: modulation of KRAS, MicroRNAs, and intercellular communication in the setting of tumor heterogeneity. Pancreas [Internet]. 2013. [cited 2019 Oct 4];42:1218–26. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24152947 [DOI] [PubMed] [Google Scholar]
- 5.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. Cell Press; 2003;4:437–50. [DOI] [PubMed] [Google Scholar]
- 6.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. W.B. Saunders; 2012;142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. Nature Publishing Group; 2014;111:817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Ahrens D, Bhagat TD, Nagrath D, Maitra A, Verma A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J Hematol Oncol [Internet]. 2017. [cited 2019 Oct 4];10:76 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28351381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris JP, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer. 2010. page 683–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi M, Bien H, Mofunanya A, Powers S. Challenges in Ras therapeutics in pancreatic cancer. Semin Cancer Biol. 2019;54:101–8. [DOI] [PubMed] [Google Scholar]
- 11.Bournet B, Buscail C, Muscari F, Cordelier P, Buscail L. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities. Eur J Cancer. 2016;54:75–83. [DOI] [PubMed] [Google Scholar]
- 12.Keenan BP, Saenger Y, Kafrouni MI, Leubner A, Lauer P, Maitra A, et al. A listeria vaccine and depletion of t-regulatory cells activate immunity against early stage pancreatic intraepithelial neoplasms and prolong survival of mice. Gastroenterology. 2014;146:1784–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov [Internet]. 2017. [cited 2019 Jul 16];16:203–22. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28209991 [DOI] [PubMed] [Google Scholar]
- 14.Nana-Sinkam SP, Croce CM. Clinical Applications for microRNAs in Cancer. Clin Pharmacol Ther. 2013;93:98–104. [DOI] [PubMed] [Google Scholar]
- 15.Taucher V, Mangge H, Haybaeck J. Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application. Cell Oncol. Cellular Oncology; 2016;39:295–318. [DOI] [PubMed] [Google Scholar]
- 16.Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol. Blackwell Publishing Ltd; 2018;183:428–44. [DOI] [PubMed] [Google Scholar]
- 17.Lo JH, Hao L, Muzumdar MD, Raghavan S, Kwon EJ, Pulver EM, et al. IRGD-guided tumor-penetrating nanocomplexes for therapeutic siRNA delivery to pancreatic cancer. Mol Cancer Ther. American Association for Cancer Research Inc.; 2018;17:2377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilles ME, Hao L, Huang L, Rupaimoole R, Lopez-Casas PP, Pulver E, et al. Personalized RNA medicine for pancreatic cancer. Clin Cancer Res [Internet]. American Association for Cancer Research Inc.; 2018. [cited 2020 Mar 16];24:1734–47. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29330203 [DOI] [PubMed] [Google Scholar]
- 19.Gilles ME, Hao L, Brown K, Lim J, Bhatia SN, Slack FJ. Tumor penetrating nanomedicine targeting both an oncomiR and an oncogene in pancreatic cancer. Oncotarget. Impact Journals LLC; 2019;10:5349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83. [DOI] [PubMed] [Google Scholar]
- 21.Sheedy FJ. Turning 21: Induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. Frontiers Media S.A; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui R, Meng W, Sun H-L, Kim T, Ye Z, Fassan M, et al. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci U S A [Internet]. 2015. [cited 2019 Oct 4];112:E4288–97. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26187928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics [Internet]. 2009. [cited 2019 Oct 4];10:161 Available from: http://www.ncbi.nlm.nih.gov/pubmed/19473525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aizawa S, Yamamoto A, Takehara S, Takahashi M, Nagano T. Shisa2 promotes the maturation of somitic precursors and transition to the segmental fate in Xenopus embryos. Development. 2006;133:4643–54. [DOI] [PubMed] [Google Scholar]
- 25.Stehbens SJ, Ju RJ, Adams MN, Perry SR, Haass NK, Bryant DM, et al. FGFR2-activating mutations disrupt cell polarity to potentiate migration and invasion in endometrial cancer cell models. J Cell Sci. Company of Biologists Ltd; 2018;131. [DOI] [PubMed] [Google Scholar]
- 26.Criscitiello C, Esposito A, Curigliano G. Targeting FGFR pathway in breast cancer. Breast Cancer Innov Res Manag. 2017;37:819–22. [Google Scholar]
- 27.Rucki AA, Foley K, Zhang P, Xiao Q, Kleponis J, Wu AA, et al. Heterogeneous stromal signaling within the tumor microenvironment controls the metastasis of pancreatic cancer. Cancer Res. American Association for Cancer Research Inc.; 2017;77:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagedorn PH, Persson R, Funder ED, Albæk N, Diemer SL, Hansen DJ, et al. Locked nucleic acid: modality, diversity, and drug discovery. Drug Discov. Today. Elsevier Ltd; 2018. page 101–14. [DOI] [PubMed] [Google Scholar]
- 29.Aran D, Hu Z, Butte AJ. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. BioMed Central Ltd.; 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo M, Tan X, Mu L, Luo Y, Li R, Deng X, et al. MiRNA-21 mediates the antiangiogenic activity of metformin through targeting PTEN and SMAD7 expression and PI3K/AKT pathway. Sci Rep. Nature Publishing Group; 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue X, Liu Y, Wang Y, Meng M, Wang K, Zang X, et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget. Impact Journals LLC; 2016;7:84508–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Q, Chen S, Zhu Z, Yu L, Ren Y, Jiang M, et al. miR-21 promotes EGF-induced pancreatic cancer cell proliferation by targeting Spry2. Cell Death Dis [Internet]. 2018. [cited 2019 Jul 16];9:1157 Available from: http://www.ncbi.nlm.nih.gov/pubmed/30464258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282:14328–36. [DOI] [PubMed] [Google Scholar]
- 35.Shi L, Middleton J, Jeon YJ, Magee P, Veneziano D, Laganà A, et al. KRAS induces lung tumorigenesis through microRNAs modulation article. Cell Death Dis. Nature Publishing Group; 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong B, Liu WW, Nie WJ, Li DF, Xie ZJ, Liu C, et al. MiR-21/RASA1 axis affects malignancy of colon cancer cells via RAS pathways. World J Gastroenterol. WJG Press; 2015;21:1488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I. The hypoxic tumour microenvironment. Oncogenesis [Internet]. 2018. [cited 2019 Oct 4];7:10 Available from: http://www.ncbi.nlm.nih.gov/pubmed/29362402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He C, Wang L, Zhang J, Xu H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol Cancer. BioMed Central Ltd.; 2017;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scisciani C, Vossio S, Guerrieri F, Schinzari V, De Iaco R, D’Onorio de Meo P, et al. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J Hepatol [Internet]. 2012. [cited 2019 Oct 4];56:855–61. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22178270 [DOI] [PubMed] [Google Scholar]
- 40.Zhu G, Zhou L, Liu H, Shan Y, Zhang X. MicroRNA-224 promotes pancreatic cancer cell proliferation and migration by targeting the TXNIP-mediated HIF1α pathway. Cell Physiol Biochem. S. Karger AG; 2018;48:1735–46. [DOI] [PubMed] [Google Scholar]
- 41.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature [Internet]. 2010. [cited 2019 Jul 16];467:86–90. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20693987 [DOI] [PubMed] [Google Scholar]
- 42.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell [Internet]. 2010. [cited 2019 Jul 16];18:282–93. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20832755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J, Li A, Hong S-M, Hruban RH, Goggins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res [Internet]. 2012. [cited 2019 Jul 16];18:981–92. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22114139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem [Internet]. 2010. [cited 2019 Aug 28];56:603–12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20093556 [DOI] [PubMed] [Google Scholar]
- 45.Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu S, et al. Circulating miRNA-21–5p as a diagnostic biomarker for pancreatic cancer: Evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep. 2017;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, et al. An elevated expression of serum exosomal microRNA-191, - 21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. BMC Cancer; 2018;18:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ali S, Suresh R, Banerjee S, Bao B, Xu Z, Wilson J, et al. Contribution of microRNAs in understanding the pancreatic tumor microenvironment involving cancer associated stellate and fibroblast cells. Am J Cancer Res [Internet]. 2015. [cited 2019 Oct 4];5:1251–64. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26046003 [PMC free article] [PubMed] [Google Scholar]
- 48.Kunita A, Morita S, Irisa TU, Goto A, Niki T, Takai D, et al. MicroRNA-21 in cancer-associated fibroblasts supports lung adenocarcinoma progression. Sci Rep. Nature Publishing Group; 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang JY, Bin Gao Y, Zhang N, Zou DW, Wang P, Zhu ZY, et al. MiR-21 overexpression enhances TGF-β1-induced epithelial-to-mesenchymal transition by target smad7 and aggravates renal damage in diabetic nephropathy. Mol Cell Endocrinol. Elsevier Ireland Ltd; 2014;392:163–72. [DOI] [PubMed] [Google Scholar]
- 50.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. Rockefeller University Press; 2010;207:1589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren W, Hou J, Yang C, Wang H, Wu S, Wu Y, et al. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21–5p delivery. J Exp Clin Cancer Res. BioMed Central Ltd.; 2019;38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Li X, Wu X, Zhang T, Zhu Q, Wang X, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. American Association for Cancer Research Inc.; 2018;6:1578–92. [DOI] [PubMed] [Google Scholar]
- 53.Li L, Zhang J, Diao W, Wang D, Wei Y, Zhang C-Y, et al. MicroRNA-155 and MicroRNA-21 Promote the Expansion of Functional Myeloid-Derived Suppressor Cells. J Immunol. The American Association of Immunologists; 2014;192:1034–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.