The emergence of XDR Salmonella Typhi in Pakistan has left azithromycin as the only viable oral treatment option. Here, we report the detection of an azithromycin resistance-associated mutation in one S. Typhi isolate. This finding is important because any possible spread of azithromycin resistance in S. Typhi isolates would make it nearly impossible to treat in outpatient settings due to the need of injectable antibiotics. Our findings also signify the importance of introduction of typhoid conjugate vaccine in regions of endemicity such as Pakistan.
KEYWORDS: Salmonella Typhi, typhoid fever, antimicrobial resistance, azithromycin higher MIC, Pakistan
ABSTRACT
Antimicrobial resistance is an ongoing issue in the treatment of typhoid fever. Resistance to first-line antimicrobials and extensively drug resistant (XDR) Salmonella Typhi isolates in Pakistan have left azithromycin as the only remaining effective oral treatment. Here, we report the emergence of organisms with a single point mutation in acrB gene, implicated in azithromycin resistance, in a S. Typhi isolate from Pakistan. The isolation of this organism is worrisome and highlights the significance of the introduction of typhoid conjugate vaccine in South Asia.
IMPORTANCE The emergence of XDR Salmonella Typhi in Pakistan has left azithromycin as the only viable oral treatment option. Here, we report the detection of an azithromycin resistance-associated mutation in one S. Typhi isolate. This finding is important because any possible spread of azithromycin resistance in S. Typhi isolates would make it nearly impossible to treat in outpatient settings due to the need of injectable antibiotics. Our findings also signify the importance of introduction of typhoid conjugate vaccine in regions of endemicity such as Pakistan.
OBSERVATION
Typhoid fever, the disease caused by the bacterium Salmonella Typhi, is responsible for an estimated 11.8 million infections and 128,200 deaths annually worldwide (1). S. Typhi is a human-restricted pathogen that is transmitted via the fecal-oral route. Typhoid mortality ranged from 10–30% of cases in the preantimicrobial era (2), but when treated with effective antimicrobials, typhoid has a case fatality rate of <1% (3). The rise of multidrug resistance (MDR) in the 1990s (4), followed by fluoroquinolone resistance (5), resulted in limited treatment options. The emergence and spread of an extensively drug-resistant (XDR) S. Typhi variant in Pakistan (6, 7), which is resistant to chloramphenicol, ampicillin, co-trimoxazole, streptomycin, fluoroquinolones, and third-generation cephalosporins, has left azithromycin as only realistic option for typhoid treatment in Pakistan (8). The recent report of azithromycin-resistant S. Typhi in Bangladesh highlights the issues associated with the reliance on this drug and signals the potential of untreatable typhoid (9).
Typhoid is notifiable in Pakistan, and the Aga Khan University has conducted standardized prospective facility and laboratory-based blood culture surveillance in outpatient and inpatient wards at Aga Khan University Hospital and Kharadar General Hospital between September 2016 and September 2019 through the Surveillance for Enteric fever in Asia Project (SEAP). These hospitals serve ∼30 million people, including densely populated informal urban settlements. Subjects presenting to outpatient clinics living in predefined catchment areas with three consecutive days of fever for whom a study clinician recommended a blood culture were enrolled. Inpatients with clinical suspicion of typhoid or with nontraumatic ileal perforation were also enrolled. After blood culture, serologically confirmed S. Typhi isolates were subjected to antimicrobial susceptibility testing against azithromycin, ampicillin, co-trimoxazole, chloramphenicol, ciprofloxacin, levofloxacin, ceftriaxone, cefepime, cefixime, and ceftazidime by disk diffusion; resistant organisms (according to CLSI guidelines) were confirmed by Etest (bioMérieux, France) (10).
Between the specified dates, 10,080 patients were enrolled in SEAP in Karachi; 2,104 had a positive blood culture for S. Typhi, and 139 had a positive blood culture for S. Paratyphi A. Six S. Typhi isolates exhibited potential azithromycin resistance by disc diffusion (diameter ≤ 12 mm). Upon MIC testing, one failed to revive, four isolates had azithromycin MICs ranging between 1 and 2 μg/ml and one S. Typhi isolate had an MIC of 12 μg/ml (CLSI susceptibility breakpoint ≤ 16 μg/ml) (10). This places this isolate at the upper range of the wild-type azithromycin susceptibility distribution, with additional resistance to chloramphenicol, fluoroquinolones, and co-trimoxazole, but it was susceptible to third-generation cephalosporins.
We aimed to investigate the genetic basis of the higher azithromycin MIC and place this organism into phylogenetic context with contemporaneous S. Typhi through whole-genome sequencing (WGS). Genomic DNA was extracted and subjected to WGS on a Hiseq2500 (Illumina, San Diego, CA) to generate 125-bp paired-end reads. The resulting sequence data were mapped against the CT18 reference sequence (accession no. AL513382) using the RedDog mapping pipeline to identify single-nucleotide variants (SNVs) and to confirm the S. Typhi genomes were within H58 lineage I (4.3.1.1) (7, 9, 11–19). (https://github.com/katholt/genotyphi). After removing repetitive sequences and recombination (20), we generated a final alignment 7,661 chromosomal SNVs for 664 isolates (see Table S1 in the supplemental material). Maximum-likelihood phylogenetic trees were inferred from the chromosomal SNV alignments with RAxML (v8.2.9) (21) and visualized in Microreact (22) (https://microreact.org/project/8FjPCdisk) and the ggtree package in R (23). SRST2 (24) was used with ARGannot (25) and PlasmidFinder (26) to identify antimicrobial resistance genes and plasmid replicons, respectively. Mutations in gyrA, and parC, as well as the R717Q mutation in acrB, were detected using GenoTyphi (https://github.com/katholt/genotyphi).
Details of n = 664 sequences used in the inference of a H58 lineage I (genotype 4.3.1.1) phylogenetic tree. Accession numbers, source, and year of isolation data for n = 664 S. Typhi whole-genome sequences analyzed in this study are presented. Download Table S1, XLSX file, 0.09 MB (93.2KB, xlsx) .
Copyright © 2020 Iqbal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
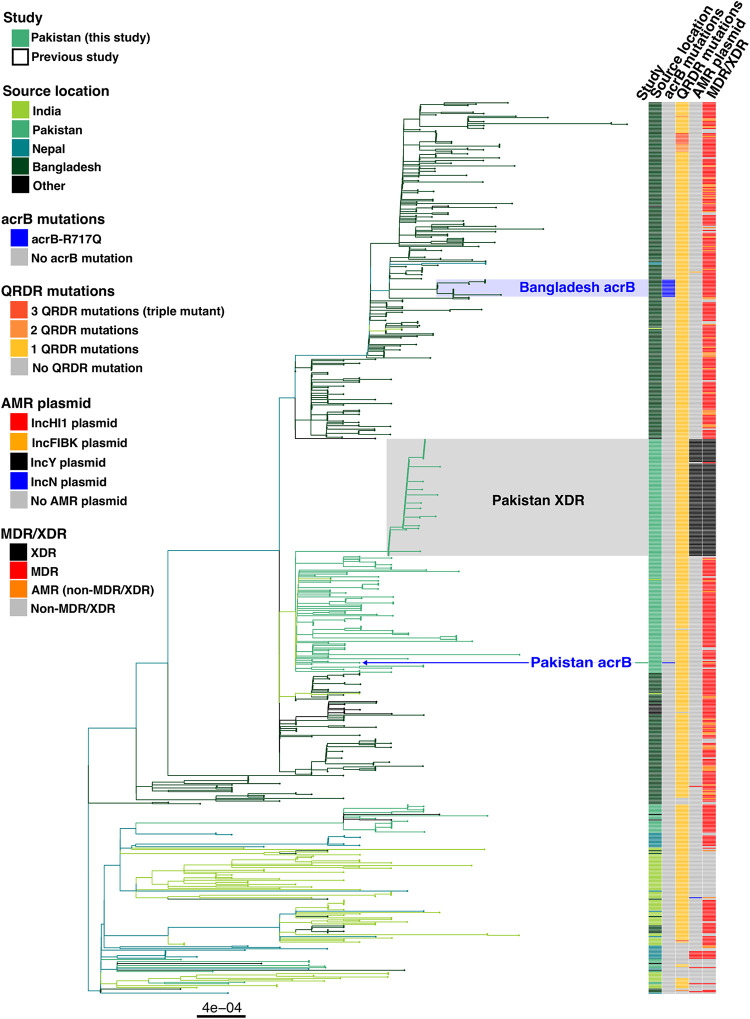
This higher azithromycin MIC S. Typhi isolate (MIC of 12 μg/ml), was typed as genotype 4.3.1.1 (H58 lineage I), which is the same sublineage at the XDR clade circulating in Pakistan. The organism additionally had single mutation in gyrA (S83F), resulting in reduced fluoroquinolone susceptibility. The apparent mechanism of higher MIC against azithromycin was an R717Q mutation in the gene encoding AcrB, a mutation identical to the recently described azithromycin resistant (MIC of ≥32 μg/ml) S. Typhi 4.3.1.1 in Bangladesh (9). The identification of this mutation in S. Typhi in Pakistan raises the possibilities that this was either a de novo mutation in the Pakistan-specific 4.3.1.1 cluster or an organism that was part of larger, internationally disseminating, azithromycin-resistant clone. To determine which was more likely, we used a collection of 663 South Asian 4.3.1.1 (H58 lineage I) sequences to contextualize S. Typhi isolate FQ2181 (7, 9, 11–19). The resulting phylogenetic tree demonstrated that this was a spontaneous mutation which emerged in Pakistan, since it was distantly related (relative within H58 lineage I) to the organisms with acrB mutations in Bangladesh, and independent of the proximal XDR sublineage (Fig. 1).
FIG 1.
South Asian H58 lineage I (genotype 4.3.1.1) phylogenetic tree (n = 664 genomes). Branches are colored by source country according to the inset legend and first color bar. The second color bar indicates genomes containing the acrB-R717Q mutation. The third color bar indicates mutations in the quinolone resistance determining region (QRDR) of genes gyrA, and parC. The final color bar indicates MDR and XDR sequences.
Typically, the isolation of a single S. Typhi exhibiting resistance to the primary treatment would not be a major cause for concern. However, this isolate demonstrates an additional, independent acquisition of the same mutation that has been observed in Bangladesh (9). Given the reliance of azithromycin for the treatment of typhoid and other bacterial infections and the “fluoroquinolone experience,” we predict that we are likely to see more of these homoplasies arising. It is too early to predict how these particular organisms may spread, and it is encouraging that these mutations have not yet been reported in XDR S. Typhi. However, given the nature of these mutations, one could arise in XDR S. Typhi, and/or the XDR plasmid may be mobilized into an azithromycin-resistant lineage.
Pakistan has initiated a nationwide typhoid conjugate vaccine (TCV) rollout program, which began with a mass vaccination in Sindh province in November 2019 (27). Now, there is in a race against time in the prevention of untreatable typhoid fever. With one World Health Organization prequalified manufacturer of TCV supplying vaccine for Gavi-eligible countries and several additional manufacturers in late-stage clinical development (28), there is reason to be optimistic about typhoid control. However, the vaccine is not yet available in all countries of endemicity, and effective treatment is still paramount for typhoid control. Consequently, we need to progress with additional intervention strategies and not overlook that antimicrobials have a substantial impact on typhoid disease control. In addition, as part of this sustained effort, we need to continue to track phenotypic and genotypic antimicrobial resistance in S. Typhi to inform best practices for antimicrobial prescribing and the impact of TCV implementation.
REFERENCES
- 1.GBD Typhoid and Paratyphoid Collaborators. 2019. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis 19:369–381. doi: 10.1016/S1473-3099(18)30685-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler T, Knight J, Nath SK, Speelman P, Roy SK, Azad MA. 1985. Typhoid fever complicated by intestinal perforation: a persisting fatal disease requiring surgical management. Rev Infect Dis 7:244–256. doi: 10.1093/clinids/7.2.244. [DOI] [PubMed] [Google Scholar]
- 3.Crump JA. 2019. Progress in typhoid fever epidemiology. Clin Infect Dis 68:S4–S9. doi: 10.1093/cid/ciy846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirza SH, Beeching NJ, Hart CA. 1996. Multidrug-resistant typhoid: a global problem. J Med Microbiol 44:317–319. doi: 10.1099/00222615-44-5-317. [DOI] [PubMed] [Google Scholar]
- 5.Britto CD, Wong VK, Dougan G, Pollard AJ. 2018. A systematic review of antimicrobial resistance in Salmonella enterica serovar Typhi, the etiological agent of typhoid. PLoS Negl Trop Dis 12:e0006779. doi: 10.1371/journal.pntd.0006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yousafzai MT, Qamar FN, Shakoor S, Saleem K, Lohana H, Karim S, Hotwani A, Qureshi S, Masood N, Rauf M, Khanzada JA, Kazi M, Hasan R. 2019. Ceftriaxone-resistant Salmonella Typhi outbreak in Hyderabad City of Sindh, Pakistan: high time for the introduction of typhoid conjugate vaccine. Clin Infect Dis 68:S16–S21. doi: 10.1093/cid/ciy877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker S, Shaheen G, Qureshi S, Yousafzai MT, Saleem MK, Hasan Z, Dougan G, Hasan R. 2018. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio 9:e00105-18. doi: 10.1128/mBio.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine MM, Simon R. 2018. The gathering storm: is untreatable typhoid fever on the way? mBio 9:e00482-18. doi: 10.1128/mBio.00482-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooda Y, Sajib MSI, Rahman H, Luby SP, Bondy-Denomy J, Santosham M, Andrews JR, Saha SK, Saha S. 2019. Molecular mechanism of azithromycin resistance among typhoidal Salmonella strains in Bangladesh identified through passive pediatric surveillance. PLoS Negl Trop Dis 13:e0007868. doi: 10.1371/journal.pntd.0007868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Wong VK, Baker S, Connor TR, Pickard D, Page AJ, Dave J, Murphy N, Holliman R, Sefton A, Millar M, Dyson ZA, Dougan G, Holt KE, International Typhoid Consortium. 2016. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun 7:12827. doi: 10.1038/ncomms12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britto CD, Dyson ZA, Duchene S, Carter MJ, Gurung M, Kelly DF, Murdoch DR, Ansari I, Thorson S, Shrestha S, Adhikari N, Dougan G, Holt KE, Pollard AJ. 2018. Laboratory and molecular surveillance of paediatric typhoidal Salmonella in Nepal: antimicrobial resistance and implications for vaccine policy. PLoS Negl Trop Dis 12:e0006408. doi: 10.1371/journal.pntd.0006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Britto CD, Dyson ZA, Mathias S, Bosco A, Dougan G, Jose S, Nagaraj S, Holt KE, Pollard AJ. 2020. Persistent circulation of a fluoroquinolone-resistant Salmonella enterica Typhi clone in the Indian subcontinent. J Antimicrob Chemother 75:337–341. doi: 10.1093/jac/dkz435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanmoy AM, Westeel E, De Bruyne K, Goris J, Rajoharison A, Sajib MSI, van Belkum A, Saha SK, Komurian-Pradel F, Endtz HP. 2018. Salmonella enterica serovar Typhi in Bangladesh: exploration of genomic diversity and antimicrobial resistance. mBio 9:e02112-18. doi: 10.1128/mBio.02112-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pham Thanh D, Karkey A, Dongol S, Ho Thi N, Thompson CN, Rabaa MA, Arjyal A, Holt KE, Wong V, Tran Vu Thieu N, Voong Vinh P, Ha Thanh T, Pradhan A, Shrestha SK, Gajurel D, Pickard D, Parry CM, Dougan G, Wolbers M, Dolecek C, Thwaites GE, Basnyat B, Baker S. 2016. A novel ciprofloxacin-resistant subclade of H58 Salmonella Typhi is associated with fluoroquinolone treatment failure. Elife 5:e14003. doi: 10.7554/eLife.14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingle DJ, Nair S, Hartman H, Ashton PM, Dyson ZA, Day M, Freedman J, Chattaway MA, Holt KE, Dallman TJ. 2019. Informal genomic surveillance of regional distribution of Salmonella Typhi genotypes and antimicrobial resistance via returning travellers. PLoS Negl Trop Dis 13:e0007620. doi: 10.1371/journal.pntd.0007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahman SIA, Dyson ZA, Klemm EJ, Khanam F, Holt KE, Chowdhury EK, Dougan G, Qadri F. 2020. Population structure and antimicrobial resistance patterns of Salmonella Typhi isolates in urban Dhaka, Bangladesh, from 2004 to 2016. PLoS Negl Trop Dis 14:e0008036. doi: 10.1371/journal.pntd.0008036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Study. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole-genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 22.Argimon S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt BG, Aanensen DM. 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 24.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.EMRO/WHO. 2019. Pakistan becomes first country to introduce new typhoid vaccine into routine immunization program, p 10–12. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 28.Sahastrabuddhe S, Saluja T. 2019. Overview of the typhoid conjugate vaccine pipeline: current status and future plans. Clin Infect Dis 68:S22–S26. doi: 10.1093/cid/ciy884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of n = 664 sequences used in the inference of a H58 lineage I (genotype 4.3.1.1) phylogenetic tree. Accession numbers, source, and year of isolation data for n = 664 S. Typhi whole-genome sequences analyzed in this study are presented. Download Table S1, XLSX file, 0.09 MB (93.2KB, xlsx) .
Copyright © 2020 Iqbal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.