Abstract
Abscisic acid (ABA) is an important phytohormone responsible for activating drought resistance, but the regulation mechanism of exogenous ABA on tea plants under drought stress was rarely reported. Here, we analyzed the effects of exogenous ABA on genes and metabolites of tea leaves under drought stress using transcriptomic and metabolomic analysis. The results showed that the exogenous ABA significantly induced the metabolic pathways of tea leaves under drought stress, including energy metabolism, amino acid metabolism, lipid metabolism and flavonoids biosynthesis. In which, the exogenous ABA could clearly affect the expression of genes involved in lipid metabolism and flavonoid biosynthesis. Meanwhile, it also increased the contents of flavone, anthocyanins, flavonol, isoflavone of tea leaves under drought stress, including, kaempferitrin, sakuranetin, kaempferol, and decreased the contents of glycerophospholipids, glycerolipids and fatty acids of tea leaves under drought stress. The results suggested that the exogenous ABA could alleviate the damages of tea leaves under drought stress through inducing the expression of the genes and altering the contents of metabolites in response to drought stress. This study will be helpful to understand the mechanism of resilience to abiotic stress in tea plant and provide novel insights into enhancing drought tolerance in the future.
Subject terms: Plant molecular biology, Plant stress responses
Introduction
Drought is a major abiotic stress for tea plant, it not only limits the productivity of tea plants but also affects the quality of teas1. It is essential for us to explore appropriate ways to mitigate the influences of drought stress on tea plants. At present, the research on the impacts of drought stress in tea plants mainly focused on the mechanisms underlying the stress response, which included morphological, physiological and molecular changes1–3.
The physiological changes, including the contents of chlorophyll, proline, MDA and the activities of antioxidant enzymes, have been extensively investigated in tea plants under drought stress4,5. A large number of drought-inducible genes and proteins in tea plants have also been identified by transcriptomic and proteomic analysis3,6,7. A former study in our lab showed that a large numbers of differentially expressed genes (DEGs) in response to drought stress were mainly enriched in volatile compounds, flavonoids, theanine biosynthesis pathways, and some DEGs were also involved in leaf senescence, such as lipoxygenase (LOX), salicylate/benzoate carboxyl methyltransferase (BSMT), arginine decarboxylase (ADC), glutamine synthetase (GS), glutaminase (GLS), chalcone isomerase (CHI), flavonoid 3′,5′-hydroxylase (F3′5′H), flavonol synthase (FLS), flavanone 3-hydroxylase (F3H)1. Another study in our lab showed that a large numbers of the differentially expressed proteins in response to drought stress were mainly involved in glycolysis/gluconeogenesis, starch and sucrose synthesis, or degradation metabolism, and second metabolism6. Recent research in our lab found that many lysine ubiquitination proteins (Kub proteins) related to catechins biosynthesis (e.g., PAL, CHS, CHI and F3H), carbohydrate and amino acid metabolism (e.g., FBPase, FBA and GAD1), were significantly induced by drought stress, suggesting that these Kub proteins might affect the degradation of proteins, the synthesis of catechins, and the accumulation of sucrose, fructose and GABA in tea leaves7.
Abscisic acid (ABA) is an important phytohormone responsible for activating drought resistance. Drought stress induces ABA biosynthesis, which is the signal that triggers a number of molecular and cellular responses, ultimately resulting in stomatal closure. Several studies indicated that ABA could enhance stress tolerance either as a result of its endogenous concentration or through exogenous application8,9. For instance, the exogenous ABA is beneficial for plants to enhance drought resistance, as it induces stomatal closure, osmotic adjustment, and increases activities of antioxidant enzymes in tomato, spring wheat and Populus5,10,11. Moreover, the exogenous ABA could improve protein transport, carbon metabolism and expression of resistance proteins to enhance drought tolerance of tea plants12. However, to our knowledge, the regulation mechanism of exogenous ABA on genes and metabolites of tea plants under drought stress was rarely reported. In the present study, we analyzed the effects of exogenous ABA on genes and metabolites of tea leaves under drought stress using transcriptome and metabolomics. The study will contribute to further an improved understanding of the molecular mechanism of tea plants in response to drought stress.
Results
Physiological changes of tea leaves under AT and SD
To investigate the phenotype changes of tea plants treated with exogenous ABA and drought, their stress phenotypes (displayed visible morphological changes) were photographed. The results showed that a number of wilted and curled leaves were observed under SD (Fig. 1). While the wilted and curled leaves were remarkably relieved under AT, indicating that the exogenous ABA partly relieved the damage and maintained the growth of tea plants under drought stress.
Figure 1.
The phenotype changes of tea leaves under CK, MD, AT and SD. The phenotype of CK was photographed under normal growth conditions, the phenotype of MD was photographed with drought for 24 h, the phenotypes of AT and SD were photographed at 53 h after spraying with equal ABA solution and distilled water, respectively.
To assess the effects of exogenous ABA on physiological characterization of tea leaves, we detected several physiological indexes of tea leaves under well-watered plants (CK), mild drought (MD), ABA treatment (AT) and severe drought (SD). As shown in Table 1, the content of total chlorophyll (TC) and leaf water content (LWC) decreased as the duration of the drought treatment increased under AT and SD, but it was higher under AT than that under SD. The value of maximum quantum yield of PSII (Fv/Fm) also showed similar changes. The content of MDA increased as the duration of the drought treatment increased under AT and SD, but it was lower under AT than that under SD. The results indicated that the exogenous ABA could reduce lipid peroxidation, prevent the degradation of chlorophyll and maintain photosynthesis of tea leaves under drought stress. In addition, the contents of three endogenous hormones and the activities of four antioxidant enzymes were also measured, the activity of ascorbate peroxidase was significantly lower under AT than that under SD, there was no difference in the activity of CAT, POD and GR. The content of endogenous ABA in tea leaves is significantly higher under AT than that under SD, but the content of endogenous GA3 is significantly lower, there was no difference in the content of endogenous IAA. The results suggested that the exogenous ABA could affect the changes of endogenous hormones and antioxidant enzymes of tea leaves under drought stress.
Table 1.
Physiological indexes of tea leaves during drought stress.
| Parameters | CK | MD | AT | SD |
|---|---|---|---|---|
| MDA (nmol/g) | 43.87 ± 0.88 b | 46.25 ± 0.88 b | 47.54 ± 0.92 ab | 51.85 ± 2.01 a |
| LWC (%) | 75.88 ± 1.02 | 73.23 ± 0.37 | 68.46 ± 0.7 | 65.36 ± 0.82 |
| CAT (U/g) | 98.48 ± 3.29 a | 80.21 ± 1.75 c | 82.76 ± 1.72 bc | 89.01 ± 1.80 b |
| POD (U/g) | 105.09 ± 3.09 a | 110.33 ± 2.63 a | 89.17 ± 0.92 b | 86.18 ± 0.64 b |
| APX (μmol/min/g) | 1.55 ± 0.05 a | 0.6186 ± 0.01 c | 0.68 ± 0.02 c | 0.9517 ± 0.04 b |
| GR (μmol/min/g) | 483.26 ± 2.81 bc | 433.22 ± 16.81 c | 536.16 ± 16.96 ab | 594.06 ± 25.23 a |
| ABA (μg/g) | 0.4669 ± 0.0194 b | 0.533 ± 0.024 b | 2.6333 ± 0.031 a | 0.4473 ± 0.018 b |
| IAA (μg/g) | 0.6124 ± 0.0113b | 0.8129 ± 0.0338a | 0.6303 ± 0.0158b | 0.5862 ± 0.0124b |
| GA3 (μg/g) | 0.495 ± 0.0393 d | 0.6711 ± 0.0417 c | 0.9108 ± 00,397 b | 1.1986 ± 0.0996a |
| Fv/Fm | 0.83 ± 0.21 a | 0.79 ± 0.19 ab | 0.73 ± 0.18 b | 0.65 ± 0.57 c |
| Chl content (mg/g) | 3.17 ± 0.33 a | 3.01 ± 0.27 ab | 2.88 ± 0.12 ab | 2.46 ± 0.21 b |
The date in the table are represented as the mean ± standard deviation of three biological replicates, lowercase letters indicated statistical significance—samples not sharing a letter differed significantly according to Duncan test at P < 0.05.
CK control group, MD mild drought group, AT ABA treatment group, SD severe drought group, MDA malondialdehyde, CAT catalase, POD Peroxidase, APX ascorbate peroxidase, GR glutathione reductase. The above data were determined under CK (0 h), MD (24 h), AT (77 h) and SD (77 h).
The analysis of transcriptome
To explore the transcript events of exogenous ABA on tea plants under drought, the sample leaves of CK, MD, AT and SD were conducted to RNA-seq analysis. A total of 619,817,958 clean reads were obtained from 12 RNA-Seq libraries. The Q30 percentage were over 92.54%, and the average GC content was over 44.52%. Overall, the results indicated that the RNA-Seq datasets were robust quality and could be used for further analysis (Supplementary Table S1).
A total of 2,210 DEGs (1,361 up- and 849 down-regulated) were obtained in AT/MD, 13,007 DEGs (6,465 up- and 6,542 down-regulated) were obtained in SD/MD. A total of 2,459 DEGs (1,567 up- and 892 down-regulated) were also obtained in AT/CK, 12,926 DEGs (6,447 up- and 6,479 down-regulated) were also obtained in SD/CK. The DEGs were obtained between AT/SD and MD/CK (Fig. 2). The results revealed that exogenous ABA can effectively relieve the drought stress of tea plants and decrease the expression amounts of response genes to drought.
Figure 2.
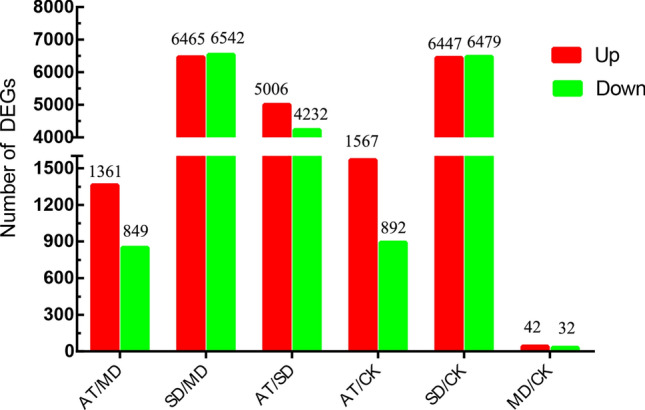
Statistics of the differentially expressed genes (DEGs) between different treatment groups.
GO analysis of the above DEGs revealed that the highly enriched terms of the biological process category were ‘cellular process’, ‘metabolic process’, ‘biological regulation’ and ‘response to stimulus’. Within the cellular component category, the highly enriched terms were ‘cell’, ‘cell part’ and ‘organelle’. Within the molecular function category, the highly represented terms were ‘binding’, ‘catalytic activity’ and ‘transporter activity’ (Supplementary Fig S1).
KEGG pathway enrichment analysis revealed that the DEGs in AT/MD were mainly enriched in starch and sucrose metabolism, lipid metabolism, plant-pathogen interaction, plant hormone signal transduction. The DEGs in AT/CK were mainly enriched in fructose and mannose metabolism, starch and sucrose metabolism, plant hormone signal transduction, glycerolipid metabolism. The DEGs in SD/MD were mainly enriched in photosynthesis, biosynthesis of amino acids, biosynthesis of secondary metabolites, starch and sucrose metabolism. The DEGs in SD/CK were mainly enriched in photosynthesis, Glycine, serine and threonine metabolism, starch and sucrose metabolism, Phenylalanine, tyrosine and tryptophan biosynthesis. The DEGs in MD/CK were mainly enriched in starch and sucrose metabolism and plant hormone signal transduction. The DEGs in AT/SD were mainly enriched in carbon metabolism, biosynthesis of amino acids, biosynthesis of secondary metabolites (Supplementary Fig S2). KEGG enrichment analyses indicated that the DEGs between CK and other three treatments were mainly enriched in energy metabolism, biosynthesis of amino acids, lipid metabolism and biosynthesis of secondary metabolites.
The effect of exogenous ABA on genes related to energy metabolism and amino acid metabolism of tea plants under drought stress
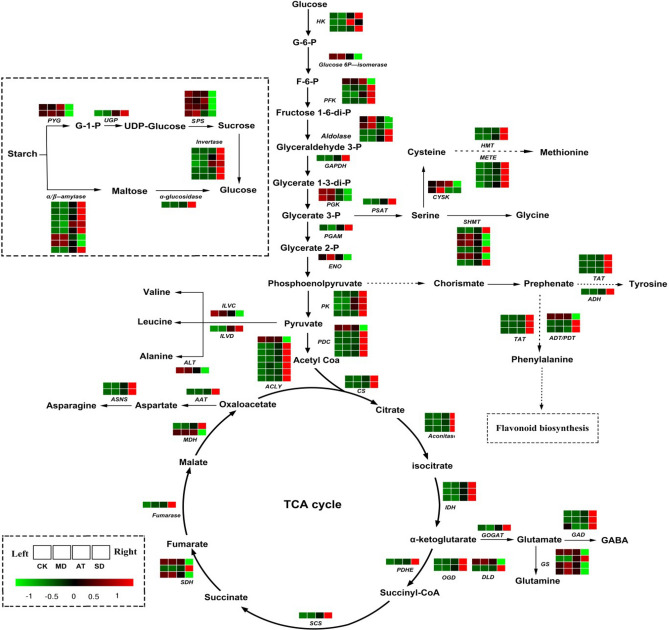
As for starch and sucrose metabolism, the genes involved in starch synthesis were significantly down-regulated in both SD/MD and SD/CK, but unchanged or slightly down-regulated in both AT/MD and AT/CK, including ADP-glucose pyrophosphorylase (AGPase), starch synthase (SS), granule-bound starch synthase (GBSS), starch-branching enzyme (SBE). While the starch degradation-related genes were up-regulated in both SD/MD and SD/CK, but unchanged or slightly up-regulated in both AT/MD and AT/CK, including AMY (α-amylase) and BAM (β-amylase). Similarly, four SPS genes (sucrose-phosphate synthase) involved in sucrose synthesis were down-regulated in both SD/MD and SD/CK, but unchanged or slightly down-regulated in both AT/MD and AT/CK. While the sucrose degradation-related genes were obviously up-regulated in both SD/MD and SD/CK, but unchanged or slightly up-regulated in both AT/MD and AT/CK, such as INV (invertase) (Fig. 3, Supplementary Table S2).
Figure 3.
The expression of genes related to starch and sucrose metabolism, glycolysis, TCA cycle and amino acids pathway under CK, MD, AT and SD. HK, hexokinase; G6PI, glucose-6-phosphate; PFK, 6-phosphofructokinase; Aldolase, fructose-bisphosphate aldolase; GAPDH, glyceraldehyde 3-phosphate; PGK, phosphoglycerate kinase; PGAM, phosphoglycerate mutase; ENO, enolase; PK, pyruvate kinase; PDC, pyruvate dehydrogenase; ACLY, ATP citrate synthase; CS, citrate synthase; Aconitase, aconitate hydratase; IDH, isocitrate dehydrogenase; PDHE, pyruvate dehydrogenase; OGD, oxoglutarate dehydrogenase complex; DLD, dihydrolipoamide dehydrogenase; SCS, succinyl-CoA synthetase; SDH, Succinate dehydrogenase; FUM, fumarate hydratase; MDH, malate dehydrogenase; PYG, glycogen phosphorylase; UGP, UTP-glucose-1-phosphate; SUS, sucrose synthase; α-glucosidase, alpha-glucosidase; Invertase, beta-fructofuranosidase; AMY, alpha-amylase; BAM, beta-amylase; GOGAT, glutamate synthase; GAD, glutamate decarboxylase; GS, glutamine synthetase; AAT, aspartate aminotransferase; ASNS, asparagine synthase; ILVC, ketol-acid reductoisomerase; ILVD, dihydroxy-acid dehydratase, ALT, branched-chain amino acid aminotransferase; TAT, tyrosine aminotransferase; ADH, arogenate dehydrogenase; ADT/PDT, arogenate/prephenate dehydratase; PSAT, phosphoserine aminotransferase; SHMT, Serine hydroxymethyltransferase; CYSK, cysteine synthase; HMT, homocysteine S-methyltransferase; METE, 5-methyltetrahydropteroyltriglutamate.
Regarding the glycolysis and TCA cycle, most of genes involved in glycolysis and TCA cycle were significantly up-regulated in both SD/MD and SD/CK, but unchanged or slightly up-regulated in both AT/MD and AT/CK, including hexokinase (HK), pyruvate kinase (PK), phosphofructokinase (PFK), phosphoglycerate mutase (PGAM) in glycolysis; ATP citrate synthase (ACLY), citrate synthase (CS), aconitate hydratase (Aconitase), isocitrate dehydrogenase (IDH), succinyl-CoA synthetase (SCS) in TCA cycle (Fig. 3, Supplementary Table S2).
Regarding the amino acid metabolism, the expression of most genes was significantly up-regulated in both SD/MD and SD/CK, but unchanged or slightly up-regulated in both AT/MD and AT/CK under drought stress, including tyrosine aminotransferase (TAT), arogenate dehydrogenase (ADH), glutamate decarboxylase (GAD), glutamate synthase (GOGAT), asparagine synthase (ASNS) (Fig. 3, Supplementary Table S2).
The effect of exogenous ABA on genes related to lipid metabolism of tea plants under drought stress
To investigate the effect of exogenous ABA on lipid metabolism of tea plants, we mainly analyzed the expression of genes involved in lipid metabolism under drought stress. A total of 81 DEGs related to lipid metabolism were selected, of which 40 DEGs were clearly up-regulated in both SD/MD and SD/CK, but unchanged or slightly up-regulated in both AT/MD and AT/CK, including diacylglycerol kinase (DGK), fatty acyl-ACP thioesterase B (FATB), ethanolamine kinase (EKI), phospholipase D1/2 (PLD1/2). In addition, 39 DEGs were significantly down-regulated in both SD/MD and SD/CK, but unchanged or slightly down-regulated in both AT/MD and AT/CK, including lipoxygenase (LOX2S), 12-oxophytodienoic acid reductase (OPR), phosphatidylserine decarboxylase (PSD), phosphatidylserine synthase (PTDSS). In addition, the lysophospholipase (LYPLA) and linoleate 9S-lipoxygenase (LOX1_5) were found to be only up-regulated in both AT/MD and AT/CK.
The effect of exogenous ABA on genes related to phenylpropanoid and flavonoid metabolism of tea plants under drought stress
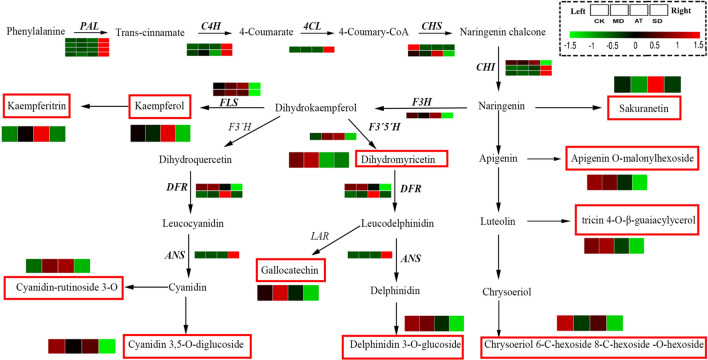
To investigate the effect of exogenous ABA on phenylpropanoid and flavonoid metabolism of tea plants, we analyzed the expression of genes involved in phenylpropanoid and flavonoid biosynthesis under drought stress (Fig. 4). The results showed that the vital genes related to phenylpropanoid biosynthesis were highly up-regulated in both SD/MD and SD/CK, but unchanged or slightly up-regulated in both AT/MD and AT/CK, such as cinnamyl-alcohol dehydrogenase (CAD), 4-coumarate-CoA ligase (4CL), ferulate-5-hydroxylase (F5′H), phenylalanine ammonia-lyase (PAL). While the key genes related to flavonoid biosynthesis were significantly down-regulated in both SD/MD and SD/CK, but unchanged or slightly down-regulated in both AT/MD and AT/CK, including F3′H, F3′5′H, FLS and DFR (dihydroflavonol 4-reductase).
Figure 4.
The expression of genes related to phenylpropanoid and flavonoid metabolism under CK, MD, AT and SD. PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, Flavonoid 3′,5′-hydroxylase; DFR, Dihydroflavonol 4-reductase; LAR, Leucoanthocyanidin reductase; ANS, leucoanthocyanidin dioxygenase; FLS, flavonol synthase.
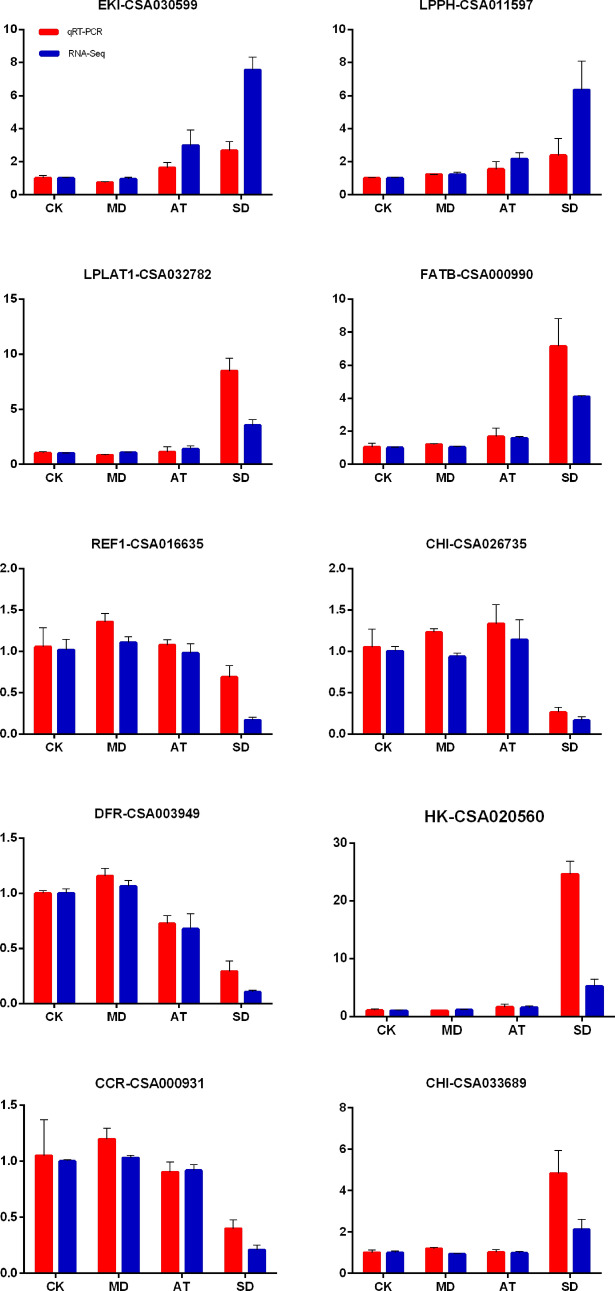
To validate the accuracy and repeatability of transcriptome sequencing data, ten DEGs were randomly selected to validate the RNA-seq data by qRT-PCR (Fig. 5). The results showed the similar expression patterns between RNA-seq and qRT-PCR, suggesting that the RNA-seq data is reliable.
Figure 5.
qRT-PCR analysis of a set of DEGs. The expression at CK was set as 1, and the relative expression level was calculated for several genes.
Metabolic differences of flavonoids and lipid metabolism under AT and SD
To investigate the effects of exogenous ABA on metabolites of tea leaves in response to drought stress, the second fully expanded leaves at the top of CK, MD, AT and SD were conducted to LC–ESI–MS/MS analysis. A total of 65 DEMs (differential metabolites) (43 up- and 22 down-regulated) were obtained in AT/MD, 90 DEMs (54 up- and 36 down-regulated) were obtained in SD/MD, and 81 DEMs (31 up- and 50 down-regulated) were obtained in AT/SD (Fig. 6). Interestingly, the abundances of most flavonoids were markedly increased, while the abundances of most lipid metabolites were markedly decreased in AT/MD compared to those in SD/MD.
Figure 6.
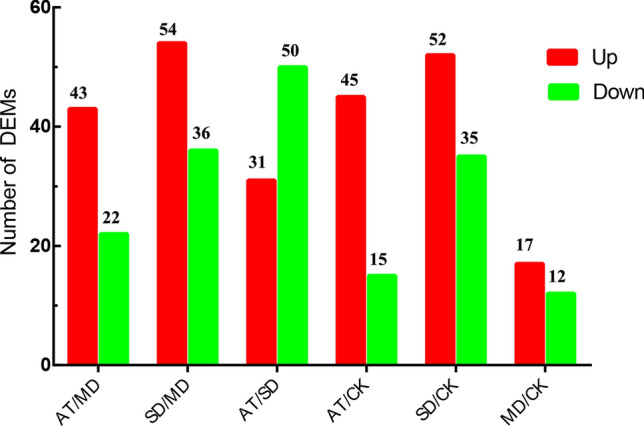
Statistics of the differential metabolites (DEMs) (VIP ≥ 1 and fold change ≥ 2 or ≤ 0.5) between different treatment groups.
For the flavonoids, the most compounds (flavone, flavanone, flavonol, isoflavone and anthocyanins) were obviously increased in AT/MD (Fig. 4). Especially, the abundances of sakuranetin in flavone showed 13.15- and 11.06-fold increase in AT/MD and SD/MD, respectively. The levels of prunetin, acacetin, laricitrin and syringetin were only increased in AT/MD. The levels of apigenin O-malonylhexoside, delphinidin 3-O-glucoside and cyanidin 3-O-rutinoside were significantly decreased in SD/MD. In addition, the levels of kaempferol, kaempferitrin, 4-methylcatechol, cyanin and fustin were higher in AT/MD than that in SD/MD.
For the lipid metabolites (Supplementary Table S3), 38 metabolites involved in fatty acids, glycerolipids and glycerophospholipids metabolisms were markedly changed under drought stress. Especially, LysoPS 22:6 showed higher abundance with 11.57- and 10-fold increase in SD/MD and AT/MD, respectively. The levels of LysoPE 14:0, LysoPE 16:0, LysoPE 18:0, LysoPE 18:1, LysoPE 18:2 (2n isomer), LysoPC 15:1, LysoPC 16:0 and LysoPC 17:0 increased significantly in SD/MD. The levels of PC 16:1/14:1 and 13-HOTrE were only increased in AT/MD. In addition, 13 metabolites were only increase in SD/MD, such as, LysoPC 18:0 (2n isomer), LysoPC 18:2 (2n isomer) and MAG (18:1) isomer2.
Interaction network analysis between genes and metabolites
Gene-metabolite interaction networks could be used to help understand functional relationship and to aid in identifying new regulatory elements13. Here, Pearson correlation tests were carried out between differentially expressed genes and metabolites related to flavonoids and lipid metabolism.
For phenylpropanoid and flavonoid biosynthesis, both 113 DEGs and 22 DEMs related to phenylpropanoid and flavonoids biosynthesis were carried out Pearson correlation analysis. The result showed that 43 DEGs had strong positive and negative correlation coefficient values (R2 > 0.8 or < − 0.8 and P value < 0.05) with 12 metabolites (Supplementary Table S4). For example, there was significantly positive correlation between the gene expression (CCR, DFR, CHI3, PER42) and metabolite abundances (gallocatechin, apigenin O-malonylhexoside, delphinidin 3-O-glucoside), but there was significantly negative correlation between the gene expression (CAD) and metabolite abundances (apigenin O-malonylhexoside, gallocatechin, dihydromyricetin and mirtillin).
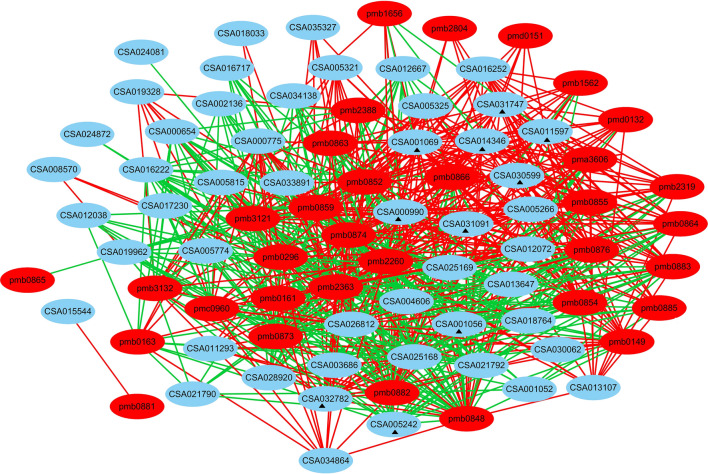
For lipid metabolism, both 81 DEGs and 38 DEMs related to lipid metabolisms were carried out Pearson correlation analysis, the result showed that 56 DEGs had strong positive and negative correlation coefficient values (R2 > 0.8 or < − 0.8 and P value < 0.05) with 34 metabolites (Fig. 7, Supplementary Table S5). Further analysis indicated that there was significantly positive correlation between the expression of eight key genes and abundances of most lipid metabolite, including diacylglycerol kinase 1 (DGK1), soluble epoxide hydrolase (EPHX2), fatty acyl-ACP thioesterase B (FATB), ethanolamine kinase (EKI), diacylglycerol kinase 2 (DGK2), acetyl-CoA acyltransferase 1 (ACAA1), glutathione peroxidase (GPX), lysophospholipid acyltransferase1 (LPT1). There was significantly negative correlation between the expression of two genes and abundances of most lipid metabolite, including lysophosphatidylcholine acyltransferase (LPCAT) and aldehyde dehydrogenase (ALDH).
Figure 7.
The connection network between genes and metabolites related to lipid metabolism. The red lines indicate positive correlation, the green line indicates negative correlation. Filled triangle indicated that the ten genes were significantly positive or negative correlation between the expression of genes and abundances of most lipid metabolite.
Discussion
ABA is a pivotal hormone in plant responses to biotic and abiotic stress, playing a key role in adapting metabolism and gene expression to help plant cope with stress conditions14. Previous research in tea plants showed that the exogenous ABA could improve protein transport, carbon metabolism and enhance the expression of resistance proteins (Hsp70) to enhance drought tolerance of tea plants12. In the present study, to gain further insight into the effects of exogenous ABA on genes and metabolites of tea plants under drought stress, we integrated transcriptomic and metabonomic analysis to explore the regulation mechanism. The results showed that exogenous ABA could regulate the expression of the genes related to energy metabolism, lipid metabolism and flavonoid biosynthesis to promote energy storages and the balance of primary metabolism, maintain membrane integrity, produce more flavonoids and derivatives in response to drought stress.
Exogenous ABA promoted energy storages and maintained the balance of primary metabolism
Starch is a key molecule in mediating plant responses to abiotic stresses, such as water deficit. Under drought stress conditions, plants generally remobilize starch to provide energy and carbon at times when photosynthesis is potentially limited15. The presence of sufficient sucrose can serve as the important energy source for the cells, and they act as an essential osmoprotectant to protect biomembranes and proteins against abiotic stress16. Previous study in tea plants showed that the expression of the genes related to starch synthesis (AGPase, SS) was inhibited and the expression of the genes related to starch degradation (AMY, BAM) was induced, whereas the expression of the genes related to sucrose synthesis (UDPGase, SPS) and degradation (INVs) was highly induced under drought stress1,2. However, in tea plants, the effect of exogenous ABA on genes involved in starch and sucrose metabolism was rarely reported.
In the present study, transcriptome analysis showed that the genes involved in starch and sucrose synthesis of tea leaves were up-regulated, and those related to starch and sucrose degradation were down-regulated in AT compared with SD (Fig. 3), suggesting that the exogenous ABA might promote a shift in metabolism towards a storage metabolism in tea leaves under drought stress. In addition, the exogenous ABA significantly decreased the expression of genes involved in TCA cycle, glycolysis and amino acid metabolism compared with severe drought. Metabolomic results showed that the exogenous ABA significantly increased the contents of the ketoglutaric acid, succinic acid and citric acid compared with severe drought. The results indicated that the exogenous ABA possible played the important role in promoting the energy storages and maintaining the balance of primary metabolism of tea plants under drought stress.
Exogenous ABA affected the expression of genes related to lipid metabolism in tea leaves under drought stress
Lipids exert multiple roles and functions in plant tissues: constituents of the cell membrane, storage molecules of metabolic energy, and as signaling factors in response to stressors17. Altered lipid biosynthesis, biomembrane rearrangement, and specific fatty acid changes reduce the impairment of cell membranes by abiotic stress18. Several studies showed that lipid metabolism in different plants was modified by drought stress19–22. In the moss Atrichum androgynum, the contents of phosphoglyceride and monogalactosyldiacylglycerol (MGDG) were significantly reduced, but the content of triacylglycerols (TAGs) was significantly increased under drought stress22. In peanut, the contents of phospholipids and galactolipids were significantly decreased under drought stress21. In addition, several studies showed that ABA could regulate lipid metabolism in different plant species23,24. For example, the exogenous ABA on mungbean leaves induced a slight increase in the content of phospholipids and a decrease in the content of free fatty acids, whereas monogalactosyldiacylglycerol (MGDG) content was significantly reduced in N-leaves after application of ABA24 . However, in tea plants, the effect of exogenous ABA on genes and metabolites related to lipid metabolism in response to drought stress was rarely reported.
In this work, transcriptome analysis showed that the expression of genes involved in lipid metabolism of tea leaves under drought stress was regulated by exogenous ABA. For example, the expression of the FATB, EKI, ACAA1, DGK and GPX related to lipid biosynthesis was significantly down-regulated and the expression of the LPCAT and ALDH related to lipid degradation (CSA005242, CSA012038) was significantly up-regulated by exogenous ABA (Supplementary Table S5). Aldehyde dehydrogenases (ALDHs) play a major role in the detoxification processes of aldehydes generated in plants when exposed to abiotic stress25. Enhancing antioxidant defense seems to be one of the approaches for plants to reduce the level of oxidative stress. Various plant ALDH genes have been reported to be activated by environmental stress including drought and salinity26,27, for example, Two ALDH genes from Arabidopsis and barley were shown to be up-regulated by drought stress for improving drought tolerance28,29. Previous studies showed that transgenic tobacco and Arabidopsis plants overexpressing ALDHs showed elevated tolerance against oxidative stress, which was correlated with decreased accumulation of reactive oxygen species and malondialdehyde derived from cellular lipid peroxidation30,31. It was reported previously that ABA treatment strong induced the expression of ALDH7B4 in Arabidopsis thaliana32. Therefore, we speculated that up-regulated expression of ALDHs induced by ABA might improve stress tolerance most likely by scavenging toxic aldehydes and reducing lipid peroxidation.
Metabolomic analysis showed that the exogenous ABA significantly altered the abundances of most lipid metabolites of tea leaves under drought stress. The abundances of LysoPE 14:0, LysoPE 16:0, LysoPE 18:0, LysoPE 18:1, LysoPC 15:1 and LysoPC 16:0 was significantly reduced and the abundances of PC 16:1/14:1, 13-HOTrE and MAG (18:3) isomer4 were slightly increased by exogenous ABA. To date, the association between the changes of these lipid metabolites and drought resistance of tea plants was rarely reported. While the correlation analysis of genes and metabolites showed that eight genes, FATB, DGK1, EKI, ACAA1, GPX, DGK7, LPT1, EPHX2, were significantly positive correlation with most lipid metabolites (including LysoPE 14:0, LysoPE 16:0, LysoPE 18:0, LysoPE 18:1, LysoPC 15:1 and LysoPC 16:0 ), and LPCAT and ALDH were significantly negative correlation with most lipid metabolites (Fig. 7, Supplementary Table S5), suggesting that the exogenous ABA could affect the expression of genes related to lipid metabolism for tea leaves to cause the reduction of lipid metabolites under drought stress. Previous study in Vigna unguiculata leaf demonstrated a role of ABA in the cell membrane protection against water stress by preventing drought-induced membrane lipid degradation33. Similar result was observed in the moss A. androgynum that the exogenous ABA might reduce the membrane damage by diminishing the lipid changes22. Previous research showed that the changes in lipid metabolism was correlated with alterations in membrane integrity under water stress34,35. In the present study of tea leaves, the results showed that exogenous ABA could mediate the expression of genes involved in lipid metabolism and affect the changes in lipid metabolism for maintaining membrane integrity and stability under drought stress. The interaction of ABA signaling with lipid signaling in tea plants under drought stress will be further elucidated in our future studies.
Exogenous ABA increased the expression of genes related to flavonoid biosynthesis in tea leaves under drought stress
Flavonoids are ubiquitous secondary metabolites with a vast array of biological functions, including defense against biotic and abiotic stresses36. The flavonoid biosynthesis of tea plants was affected by various environmental conditions, such as drought stress37. In the study, we analyzed the effects of exogenous ABA on genes and metabolites related to flavonoid metabolism of tea leaves under drought stress using transcriptomic and metabonomic analysis.
Previous researches on tea plants showed that the expression of the genes related to flavonoid biosynthesis was firstly decreased and subsequently increased in response to drought stress, such as CHS, DFR, LAR and ANR, and the expression of FLS and FNS was continuously up-regulated in response to drought stress3. The expression of the genes related to flavonoid biosynthesis, such as PAL, C4H, 4CL, CHS and DFR, was significantly induced by drought1. However, in tea plants, the effect of exogenous ABA on genes related to flavonoids biosynthetic in response to drought stress was rarely reported. In the present study, transcriptome analysis revealed that the exogenous ABA significantly induced the expression of genes related to flavonoid biosynthesis of tea leaves under drought stress. The expression of PAL, 4CL, F5′H was highly down-regulated and the expression of CHI, DRF, F3′H and FLS was significantly up-regulated by exogenous ABA.
Several researches on tea plants showed that the amounts of total polyphenols and catechins, including GC, EGC, C, EC, EGCG, GCG, ECG, significantly decreased in response to drought stress3,38–40, but the content of total flavonoids significantly increased in response to drought stress3. However, in tea plants, there was limited information about the effect of exogenous ABA on flavonoid metabolites in response to drought stress. In the present study, metabolomics analysis revealed that the exogenous ABA significantly affected the abundances of flavonoid metabolites of tea leaves under drought stress. The abundances of flavone, anthocyanins, flavonol, isoflavone classes,including apigenin O-malonylhexoside, delphinidin 3-O-glucosidewere, cyanidin 3-O-rutinoside, kaempferitrin, sakuranetin, prunetin, kaempferol, fustin, ancyanidin 3,5-O-diglucoside, significantly increased and the abundances of flavonol, flavone and flavone C-glycosides, including dihydromyricetin, acacetin O-acetyl hexoside, acacetin O-glucuronic acid, 6-C-hexosyl-hesperetin O-hexoside, were slightly decreased by exogenous ABA.
We also performed the correlation analysis between the genes and flavonoid metabolites of tea leaves under exogenous ABA. The results showed that the expression of CHI (CSA026735) and DFR (CSA003949) was significantly positive correlation with apigenin O-malonylhexoside, delphinidin 3-O-glucoside, gallocatechin and dihydromyricetin, while the expression of 4CL (CSA007753) was significantly negative correlation with apigenin O-malonylhexoside and delphinidin 3-O-glucoside, suggesting that the exogenous ABA could induce the expression of genes involved in flavonoid biosynthesis, and hence affected the biosynthesis of flavonoids and derivatives of tea leaves under drought stress.
Moreover, previous research showed that inhibition of lipid synthesis redirects the carbon flux into flavonoid metabolism41. Recently, CHS-mediated flavonoids were proven to repress embryonic fatty acid biosynthesis in the Arabidopsis seeds, implying a negative correlation between flavonoids and fatty acids biosynthesis in plants42. Our results showed that the exogenous ABA drastically decreased the abundances of lipid metabolites and increased the abundances of most flavonoids compared with severe drought. Therefore, another possible explanation was that the exogenous ABA induced the expression of genes involved in starch and sucrose metabolism to promote the accumulation of more carbon sources, and a large amount of carbon sources flux into flavonoid metabolism to increase the abundances of flavonoids and derivatives, so as to improve drought resistance of tea plants. Further studies are required to confirm such assumptions.
Conclusion
In this study, we analyzed the roles of exogenous ABA on genes and metabolites of tea leaves under drought stress using transcriptomic and metabolomic analysis. The study demonstrated that exogenous ABA significantly reduce the damage of drought to tea plants, maintain the balance of primary metabolism, promote energy storages and the formation of flavonoids and derivatives to enhance drought tolerance of tea plants under drought stress. This study will be helpful for us to understand the mechanism of resilience to abiotic stress in tea plant and provide novel insights into enhancing drought tolerance of tea plants in the future.
Material and methods
Plant materials and stress treatments
Tea plant cultivar, ‘QN3’ (Camellia sinensis cv. QN3) is an improved cultivar bred by the Tea Research Institute, Qingdao Agricultural University43–45. The 2-year-old tea seedling were cultured in growth chamber for 2 weeks with photoperiod (12 h light at 25°C and 12 h dark at 20°C, 75% of humidity) and a light intensity of 18, 000 lx.
The experiment included four treatments: well-watered plants (CK); based on previous studies and physiological response of tea plants at different time points of drought stress7, the tea seedlings under drought stress for 24 h were defined as mild drought (MD), The tea seedlings of MD were divided into two parts: one was immediately sampled and the other was again divided into two groups: one group was immediately sprayed with 100 mL of ABA solution (ABA was purchased from Sigma Chemical Company, St. Louis, MO, USA), concentration 50 mg L−1. The other was immediately sprayed with equal parts of distilled water. Subsequently, the two groups were continuous drought with all of the other environmental conditions remaining constant. The leaves of two groups displayed visible morphological difference after another 53 h of drought (total 77 h of drought), the leaves of ABA treatment were sampled and defined as AT, another group was sampled and defined as severe drought (SD). Three independent biological replicates were performed at each treatment, and each replicate contained 10 mature leaves (the second fully expanded leaves at the top). All samples were frozen immediately in liquid nitrogen and stored at − 70°C.
Measurement of physiological characterization
The contents of malondialdehyde (MDA) in the leaves were measured by the 2-thiobarbituric acid (TBA) method according to previous studies12. The leaf water content (LWC) of different treatment were determined as described previously46. The enzymatic assays of different treatment were determined as described previously47, including peroxidase (POD), catalase (CAT), ascorbate peroxidase (APX), glutathione reductase (GR). The contents of endogenous hormone of different treatment were determined as described previously48, including Ascorbic acid (ABA), indole-3-acetic acid (IAA) and Gibberellin (GA3). Leaf chlorophyll was extracted with ethanol according to previous studies44. Third leaf was sampled to measure maximum quantum yield of PSII according to previous studies1. All experiments were carried out with at least three independent repetitions.
RNA-Seq analysis
For RNA-seq analysis, the total RNA of each sample was extracted as described by Wang et al.49. RNA concentration and integrity of the total RNA were measured using NanoDrop 2000 Spectrophotometer (IMPLEN, Westlake Village, CA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), respectively. Subsequently, the library preparations were sequenced on the Illumina HiSeq platform to generate raw data. After sequencing, the clean reads were obtained by removing reads containing adaptors, more than 5% unknown bases and low-quality reads (> 20% of the bases with a quality score of ≤ 10). Gene function was annotated based on the following databases: NCBI non-redundant protein sequences (NR), Clusters of Orthologous (KOG/COG), Gene Ontology (GO), manually annotated and reviewed protein sequence database (Swiss-Prot), Kyoto Encyclopedia of Genes and Genomes (KEGG). Gene expression levels were represented using fragments per kilobase of transcript per million fragments mapped (FPKM) method. The differentially expressed genes (DEGs) were recruited based on False Discovery Rate (FDR) < 0.05 and |log2Fold Change|≥ 1. All DEGs were analyzed by GO enrichment using GOseq (1.10.0)50 and KEGG enrichment using KOBAS software51,52.
Quantitative real-time RT-PCR (qRT-PCR) analysis
Total RNA was extracted by using a plant RNA extraction kit (Tiangen, Beijing, China). First-strand cDNA was reverse transcribed according to the user manual of PrimeScript RT reagent kit (TaKaRa), and the qRT-PCR program was performed by using a Light Cycler 480 instrument (Roche). The genes were selected for RT-qPCR with specific primers designed by Primer Premier 5 software (Supplementary Table S6). The qRT-PCR reagents were the following: a total of 20 μL of reaction mixture, which included 10 μL SYBR Green PCR Master Mix (Roche), 1 μL of each primer, 2 μL cDNA and 6 μL distilled water. The qPCR program was performed as follows: 95°C, 10 min; 95°C, 10 s and 60°C, 15 s for 45 cycles; then a melting curve. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as reference gene for quantifying the expression levels of the target genes according to the method of 2−ΔΔCt53. The quantitative analysis of each RNA sample was repeated at least three times, and the representative data are expressed as the mean values ± standard error (n = 3).
Metabolite profiling analysis
Sample preparation, metabolite extraction and analysis were carried out as described in previous research54,55. In brief, the 100 mg freeze-dried samples were extracted using 1.0 mL 70% aqueous methanol (containing 0.1 mg/L lidocaine). Subsequently, 10,000g centrifugation for 10 min at 4 °C, then the extracts were absorbed and filtered before LC–MS analysis. A quality-control sample was prepared by equal blending of all samples; during the assay, the quality control sample was run every 10 injections to monitor the stability of the analytical conditions. The extracted samples (2 μL) were analyzed using a HPLC system (Shim-pack UFLC SHIMADZU CBM 30A) equipped with Waters ACQUITY UPLC HSS T3 C18 column (1.8 µm, 2.1 mm * 100 mm). LIT and triple quadrupole (QQQ) scans were acquired on a QTRAP-MS equipped with an ESI Turbo Ion-Spray interface under AB Sciex QTRAP 4,500 System, a positive ion mode and controlled by Analyst 1.6.1 software (AB Sciex). The solvent system, gradient program and ESI source operation parameters were carried out as described by previous research56.
The qualitative analysis of primary and secondary MS data was performed by searching the internal database using a self-compiled database MWDB (MetWare Biological Science and Technology Co., Ltd. Wuhan, China), Data pre-processing and metabolites identification were performed by the standard metabolic procedures, including comparing the m/z values, RT, and the fragmentation patterns with the standards. The variable importance of the projection (VIP) score of the application (O) PLS model was used to filter the best differentiated metabolites between treatments. Metabolites with significant differences in content were set with thresholds of variable importance in projection (VIP) ≥ 1 and fold change ≥ 2 or ≤ 0.5.
Statistical analysis
All statistical analyses were performed with SPSS 20.0 (Windows; SPSS Inc., Chicago, IL, USA) and significance tests were determined by Duncan’s test and ANOVA. P < 0.05 was considered the significant difference. The relationships visualization was performed by Cytoscape software (version 3.3.0, USA). Integrative analysis of metabolome and transcriptome was carried out using R software (version 3.2.4, USA).
Supplementary information
Acknowledgements
This research was subsidized by the Significant Application Projects of Agriculture Technology Innovation in Shandong Province, the Breeding Project of Shandong Province (2017LZN014), the Technology System of Modern Agricultural Industry in Shandong Province (SDAIT-19-01), the Special Foundation for Distinguished Taishan Scholar of Shandong Province (No.ts201712057), and the National Natural Science Foundation of China (No. 31800588).
Abbreviations
- ALDH
Aldehyde dehydrogenase
- AT
ABA treatment
- CAD
Cinnamyl-alcohol dehydrogenase
- CCR
Cinnamoyl-CoA reductase
- CHI3
Chalcone isomerase 3
- CK
Well-watered plants
- CHI
Chalcone isomerase
- DEMs
Differential metabolites
- DEGs
Differentially expressed genes
- DFR
Dihydroflavonol 4-reductase
- DGK
Diacylglycerol kinase
- EKI
Ethanolamine kinase
- FATB
Fatty acyl-ACP thioesterase B
- FLS
Flavonol synthase
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MDA
Malondialdehyde
- MD
Mild drought
- PAL
Phenylalanine ammonia-lyase
- PER42
Peroxidase 42
- LPCAT
Lysophosphatidylcholine acyltransferase
- SD
Severe drought
- 4CL
4-Coumarate-CoA ligase
Author contributions
Z.T.D and Z.S.G designed the study. H.X, L.T.S, Y.Q.D and L.J.W performed the experiments. Z.S.G, Y.W, W.J.Q and Q.P.M analyzed the data. C.Q prepared figures Z.T.D and Z.S.G wrote the manuscript. Z.W.J critically read the manuscript. All authors were involved in critically reading and editing the manuscript and gave the final approval of the version to be published. All authors read and approved the final manuscript.
Data availability
The datasets generated during the current study are available in the NCBI repository under the accession numbers PRJNA631326 after May. 2021.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhongshuai Gai and Yu Wang
Supplementary information
is available for this paper at 10.1038/s41598-020-69080-1.
References
- 1.Zheng C, Wang Y, Ding Z, Zhao L. Global transcriptional analysis reveals the complex relationship between tea quality, leaf senescence and the responses to cold-drought combined stress in Camellia sinensis. Front. Plant Sci. 2016;7:1858. doi: 10.3389/fpls.2016.01858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu SC, et al. Transcriptomic analysis of tea plant responding to drought stress and recovery. PLoS ONE. 2016;11:e0147306. doi: 10.1371/journal.pone.0147306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W, et al. Transcriptomic analysis reveals the molecular mechanisms of drought-stress-induced decreases in Camellia sinensis leaf quality. Front. Plant. Sci. 2016;7:385. doi: 10.3389/fpls.2016.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Upadhyaya H, Panda SK. Responses of Camellia sinensis to drought and rehydration. Biol. Plant. 2004;48:597–600. [Google Scholar]
- 5.Du Y, et al. Exogenous abscisic acid reduces water loss and improves antioxidant defence, desiccation tolerance and transpiration efficiency in two spring wheat cultivars subjected to a soil water deficit. Funct. Plant Biol. 2013;40:494–506. doi: 10.1071/FP12250. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Proteomic analysis of Camellia sinensis (L.) reveals a synergistic network in the response to drought stress and recovery. J. Plant Physiol. 2017;219:91–99. doi: 10.1016/j.jplph.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Xie H, et al. Global ubiquitome profiling revealed the roles of ubiquitinated proteins in metabolic pathways of tea leaves in responding to drought stress. Sci. Rep. 2019;9:4286. doi: 10.1038/s41598-019-41041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haisel D, Pospíšilová J, Synková H, Schnablová R, Bat’ková P. Effects of abscisic acid or benzyladenine on pigment contents, chlorophyll fluorescence, and chloroplast ultrastructure during water stress and after rehydration. Photosynthetica. 2006;44:606–614. [Google Scholar]
- 9.Trouverie J, Thevenot C, Rocher JP, Sotta B, Prioul JL. The role of abscisic acid in the response of a specific vacuolar invertase to water stress in the adult maize leaf. J. Exp. Bot. 2003;54:2177–2186. doi: 10.1093/jxb/erg234. [DOI] [PubMed] [Google Scholar]
- 10.Aroca R, Alguacil MDM, Vernieri P, Ruiz-Lozano JM. Plant responses to drought stress and exogenous ABA application are modulated differently by mycorrhization in tomato and an ABA-deficient mutant (Sitiens ) Microb. Ecol. 2008;56:704–719. doi: 10.1007/s00248-008-9390-y. [DOI] [PubMed] [Google Scholar]
- 11.Chunyang LI, Yin C, Liu S. Different responses of two contrasting Populus davidiana populations to exogenous abscisic acid application. Environ. Exp. Bot. 2004;51:237–246. [Google Scholar]
- 12.Zhou L, et al. Exogenous abscisic acid significantly affects proteome in tea plant (Camellia sinensis) exposed to drought stress. Hortic. Res. 2014;1:14029. doi: 10.1038/hortres.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu ZG, et al. Insights from the cold transcriptome and metabolome of Dendrobium officinale: global reprogramming of metabolic and gene regulation networks during cold acclimation. Front. Plant Sci. 2016;7:1653. doi: 10.3389/fpls.2016.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Rodriguez PL. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013;161:931–941. doi: 10.1104/pp.112.208678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thalmann M, Santelia D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017;214:943–951. doi: 10.1111/nph.14491. [DOI] [PubMed] [Google Scholar]
- 16.Cao YY, et al. Exogenous sucrose increases chilling tolerance in cucumber seedlings by modulating antioxidant enzyme activity and regulating proline and soluble sugar contents. Sci. Hortic. 2014;179:67–77. [Google Scholar]
- 17.Yozo O, Kazuki S. Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 2014;79:584–596. doi: 10.1111/tpj.12556. [DOI] [PubMed] [Google Scholar]
- 18.Deng S, et al. De novo transcriptome sequencing and gene expression profiling of Magnolia wufengensis in response to cold stress. BMC Plant Biol. 2019;19:321. doi: 10.1186/s12870-019-1933-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liljenberg CS. The effects of water deficit stress on plant membrane lipids. Prog. Lipid Res. 1992;31:335–343. doi: 10.1016/0163-7827(92)90012-8. [DOI] [PubMed] [Google Scholar]
- 20.Repellin A, et al. Leaf membrane lipids and drought tolerance in young coconut palms (Cocos nucifera L.) Eur. J. Agron. 1997;6:25–33. [Google Scholar]
- 21.Lauriano JA, Lidon FC, Carvalho CA, Campos PS, Matos MDC. Drought effects on membrane lipids and photosynthetic activity in different peanut cultivars. Photosynthetica. 2000;38:7–12. [Google Scholar]
- 22.Guschina IA, Harwood JL, Smith M, Beckett RP. Abscisic acid modifies the changes in lipids brought about by water stress in the moss Atrichum androgynum. New Phytol. 2002;156:255–264. doi: 10.1046/j.1469-8137.2002.00517.x. [DOI] [PubMed] [Google Scholar]
- 23.Ritchie S, Gilroy S. Abscisic acid stimulation of phospholipase D in the barley aleurone is G-protein-mediated and localized to the plasma membrane. Plant Physiol. 2000;124:693–702. doi: 10.1104/pp.124.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aghofack-Nguemezi J, Christmann A, Frosch S, Trémoliéres A, Wagner E. Contrasting photo- and thermoperiod-induced changes in abscisic acid and lipid contents in leaves of mungbean seedlings. Physiol. Plant. 2010;83:346–352. [Google Scholar]
- 25.Kotchoni SO, Kuhns C, Ditzer A, Kirch HH, Bartels D. Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 2006;29:1033–1048. doi: 10.1111/j.1365-3040.2005.01458.x. [DOI] [PubMed] [Google Scholar]
- 26.Ramanjulu S, Dorothea B, Hans-Hubert K. Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J. 2010;35:452–464. doi: 10.1046/j.1365-313x.2003.01819.x. [DOI] [PubMed] [Google Scholar]
- 27.Kirch HH, Bartels DY, Schnable PS, Wood AJ. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 2004;9:371–377. doi: 10.1016/j.tplants.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Seki M, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2010;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 29.Ozturk ZN, et al. Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol. Biol. 2002;48:551–573. doi: 10.1023/a:1014875215580. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, et al. Overexpression of ALDH2B8, an aldehyde dehydrogenase gene from grapevine, sustains Arabidopsis growth upon salt stress and protects plants against oxidative stress. Plant Cell Tissue Organ Cult. 2013;114:187–196. [Google Scholar]
- 31.Huang W, et al. Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays) Plant Mol. Biol. 2008;68:451. doi: 10.1007/s11103-008-9382-9. [DOI] [PubMed] [Google Scholar]
- 32.Kirch HH, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D. Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol. Biol. 2005;57:315. doi: 10.1007/s11103-004-7796-6. [DOI] [PubMed] [Google Scholar]
- 33.Campos PS, Thi ATP. Effects of an abscisic acid pretreatment on membrane leakage and lipid composition of Vigna unguiculata leaf discs subjected to osmotic stress. Plant Sci. 1997;130:11–18. [Google Scholar]
- 34.Taranto PA, Keenan TW, Potts M. Rehydration induces rapid onset of lipid biosynthesis in desiccated Nostoc commune (Cyanobacteria) Biochim. Biophys. Acta. 1993;1168:228–237. doi: 10.1016/0005-2760(93)90129-w. [DOI] [PubMed] [Google Scholar]
- 35.Platt KA, Oliver MJ, Thomson WW. Membranes and organelles of dehydrated Selaginella and Tortula retain their normal configuration and structural integrity. Protoplasma. 1994;178:57–65. [Google Scholar]
- 36.Broun P. Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 2005;8:272–279. doi: 10.1016/j.pbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Jaleel CA, et al. Water deficit stress effects on reactive oxygen metabolism in Catharanthus roseus; impacts on ajmalicine accumulation. Colloids Surf. B. 2008;62:105–111. doi: 10.1016/j.colsurfb.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 38.Munivenkatappa N, Sarikonda S, Rajagopal R, Balakrishnan R. Variations in quality constituents of green tea leaves in response to drought stress under south Indian condition. Sci. Hortic. 2018;233:359–369. [Google Scholar]
- 39.Chen XH, et al. Photosynthesis, yield, and chemical composition of Tieguanyin tea plants (Camellia sinensis (L.) O. Kuntze) in response to irrigation treatments. Agric. Water Manag. 2010;97:419–425. [Google Scholar]
- 40.Jeyaramraja PR, Pius PK, Kumar RR, Jayakumar D. Soil moisture stress-induced alterations in bioconstituents determining tea quality. J. Sci. Food Agric. 2003;83:1187–1191. [Google Scholar]
- 41.Leonard E, Lim KH, Saw PN, Koffas MAG. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl. Environ. Microbiol. 2007;73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xuan L, et al. TRANSPARENT TESTA 4-mediated flavonoids negatively affect embryonic fatty acid biosynthesis in Arabidopsis. Plant Cell Environ. 2018;41:2773–2790. doi: 10.1111/pce.13402. [DOI] [PubMed] [Google Scholar]
- 43.Jiang J, et al. Comprehensive proteome analyses of lysine acetylation in tea leaves by sensing nitrogen nutrition. BMC Genomics. 2018;19:840. doi: 10.1186/s12864-018-5250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gai ZS, et al. The quality evaluation of tea (Camellia sinensis) varieties based on the metabolomics. HortScience. 2019;54:409–415. doi: 10.21273/hortsci13713-18. [DOI] [Google Scholar]
- 45.Sun JH, et al. Ammonium triggered the response mechanism of lysine crotonylome in tea plants. BMC Genomics. 2019;20:1–14. doi: 10.1186/s12864-019-5716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su L, Dai Z, Li S, Xin H. A novel system for evaluating drought–cold tolerance of grapevines using chlorophyll fluorescence. BMC Plant Biol. 2015;15:1–12. doi: 10.1186/s12870-015-0459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, et al. Alleviation of cold damage by exogenous application of melatonin in vegetatively propagated tea plant (Camellia sinensis (L.) O. Kuntze) Sci. Hortic. 2018;238:356–362. [Google Scholar]
- 48.Zhao PF. Effect of exogenous abscisic acid on psbA expression at grain filling stage in two wheat cultivars under drought stress. Acta Agron. Sin. 2011;37:1372–1377. [Google Scholar]
- 49.Wang L, et al. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis. Ann. Bot. 2017;119:1195–1209. doi: 10.1093/aob/mcx011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14–R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao X, Tao CJGO, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 52.Minoru K, Yoko S, Masayuki K, Miho F, Mao T. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Cui Y, Vainstein A, Chen S, Ma H. Regulation of fig (Ficus carica L.) fruit color: metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front. Plant Sci. 2017;8:1990. doi: 10.3389/fpls.2017.01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen J, et al. Metabolite profiling of tea (Camellia sinensis L.) leaves in winter. Sci. Hortic. 2015;192:1–9. [Google Scholar]
- 56.Chen W, et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant. 2013;6:1769–1780. doi: 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the NCBI repository under the accession numbers PRJNA631326 after May. 2021.