Abstract
Glioblastoma (‘GBM’) is the most aggressive type of primary malignant adult brain tumor, with very heterogeneous radio-graphic, histologic, and molecular profiles. A growing body of advanced computational analyses are conducted towards further understanding the biology and variation in glioblastoma. To address the intrinsic heterogeneity among different computational studies, reference standards have been established to facilitate both radiographic and molecular analyses, e.g., anatomical atlas for image registration and housekeeping genes, respectively. However, there is an apparent lack of reference standards in the domain of digital pathology, where each independent study uses an arbitrarily chosen slide from their evaluation dataset for normalization purposes. In this study, we introduce a novel stain normalization approach based on a composite reference slide comprised of information from a large population of anatomically annotated hematoxylin and eosin (‘H&E’) whole-slide images from the Ivy Glioblastoma Atlas Project (‘IvyGAP’). Two board-certified neuropathologists manually reviewed and selected annotations in 509 slides, according to the World Health Organization definitions. We computed summary statistics from each of these approved annotations and weighted them based on their percent contribution to overall slide (‘PCOS’), to form a global histogram and stain vectors. Quantitative evaluation of pre- and post-normalization stain density statistics for each annotated region with PCOS > 0.05% yielded a significant (largest p = 0.001, two-sided Wilcoxon rank sum test) reduction of its intensity variation for both ‘H’ & ‘E’. Subject to further large-scale evaluation, our findings support the proposed approach as a potentially robust population-based reference for stain normalization.
Keywords: Histology, Digital pathology, Computational pathology, Stain normalization, Pre-processing, Glioblastoma, Brain tumor
1. Introduction
Glioblastoma (‘GBM’) is the most aggressive, and common, type of primary malignant adult brain tumor. GBMs are usually de novo, meaning they frequently appear without any precursor lesions. If left untreated, the tumor is quickly fatal, and even with treatment, median survival is about 16 months [1, 2]. If a GBM is suspected, multi-parametric magnetic resonance imaging (‘mpMRI’) will be done to follow up, and presumptive diagnosis can typically be given. Ideally, surgical gross total resection is performed. When extensive surgery is not possible, often needle or excisional biopsies are performed to confirm the diagnosis. GBMs, due to their serious and sudden nature, lack of effective treatment options, as well as their reported heterogeneity [3], have been the subject of research in the realm of personalized medicine and diagnostics. However, investigating their precise characterization requires large amounts of data. Fortunately, publicly available datasets with abundant information are becoming much more available.
With the advent of data collection and storage, not only are large datasets becoming available for public use, but they are also becoming more detailed in multiple scales, i.e., macro- and micro-scopic. Large comprehensive datasets publicly available in various repositories, such as The Cancer Imaging Archive (TCIA - www.cancerimagingarchive.net) [4] have shown promise on expediting discovery. One of the exemplary data collections of glioblastoma, is the TCGA-GBM [5], which since its initiation has included longitudinal radiographic scans of GBM patients, with corresponding detailed molecular characterization hosted in the National Cancer Institute’s Genomic Data Commons (gdc.cancer.gov). This dataset enabled impactful landmark studies on the discovery on integrated genomic analyses of gliomas [6, 7]. TCIA has also made possible the release of ‘Analysis Results’ from individual research groups, with the intention of avoiding study replication and allowing reproducibility analyses, but also expediting further discoveries. Examples of these ‘Analysis Results’ describe the public release of expert annotations [8–11], as well as exploratory radiogenomic and outcome prediction analyses [12–15]. The further inclusion of available histology whole-slide images (‘WSIs’) corresponding to the existing radiographic scans of the TCGA-GBM collection contributes to the advent of integrated diagnostic analyses, which in turn raises the need for normalization. Specifically, such analyses attempt to identify integrated tumor phenotypes [16, 17] and are primarily based on extracting visual descriptors, such as the tumor location [18, 19], and intensity values [20], as well as subtle sub-visual imaging features [21–28].
Intensity normalization is considered an essential step for performing such computational analysis of medical images. Specifically, for digital pathology analyses, stain normalization is an essential pre-processing step directly affecting subsequent computational analyses. Following the acquisition of tissue up until the digitization and storage of WSIs, nearly every step introduces variation into the final appearance of a slide. Prior to staining, tissue is fixed for variable amounts of time. Slide staining is a chemical process, and thus is highly prone to not only the solution preparation, but also the environmental conditions. While preparing a specimen, the final appearance can be determined by factors such as: stain duration, manufacturer, pH balance, temperature, section thickness, fixative, and numerous other biological, chemical, or environmental conditions. Additionally, the advent of digital pathology has incurred even more variation on the final appearance of WSIs, including significant differences in the process of digitization that varies between scanners (vary by manufacturers and models within a given company).
Various approaches have been developed to overcome these variations in slide appearance. Techniques such as Red-Green-Blue (‘RGB’) histogram transfers [29] and Macenko et al. [30] use the general approach of converting an image to an appropriate colorspace, and using a single example slide as a target for modifying the colors and intensities of a source image. Recently, techniques such as Reinhard [31], Vahadane [32], and Khan [33] have been developed to separate the image into optical density (‘OD’) stain vectors (S), as well as corresponding densities (W) of each stain per pixel. This process (known as ‘stain deconvolution’) has been one of the more successful and popular techniques in recent years. Additionally, a number of generative deep learning techniques, such as StainGAN [34] and StaNoSA [35], have also been developed for stain normalization. While such techniques [34, 35] have been shown to outperform many transfer-based approaches [30–33], they also have multiple downsides. First, these techniques are generative, which means that rather than modifying existing information, they attempt to generate their own information based on distributive models. These generative techniques apply a “blackbox” to input data, making it difficult to discern if the model is biased, and hence may influence all downstream processing without notice. For example, if a StainGAN model had not seen an uncommon structure during training, it would be unable to accurately model the staining of that structure, and fail in producing an accurate result. For a much more thorough review of stain normalization algorithms, see [36]. Our approach, in comparison to StainGAN or other generative methods, attempts to expand upon prior transformative stain transfer techniques. Our motivation is that an approach as the one we propose will obviate the “black box” presented by generative methods, and prevent a potential entry for insidious bias in the normalization of slides, while still maintaining a robust and accurate representation of a slide batch.
While the technical aspects of stain transfer algorithms have progressed significantly, their application remains fairly naive. In demonstration, most studies arbitrarily pick either a single WSIs, or even a single patch within a WSI, as a normalization step prior to further analysis. In this paper, we sought to build upon current stain normalization techniques by using a publicly available dataset in an effort to form a composite reference slide for stain transfer, and avoid the use of arbitrarily chosen slides from independent studies. By doing this, we have developed a standardized target for stain normalization, thus allowing the creation of more robust, accurate, and reproducible digital pathology techniques through the employment of a universal pre-processing step.
2. Materials and Methods
2.1. Data
The Ivy Glioblastoma Atlas Project (‘IvyGAP’) [37, 38] describes a comprehensive radio-patho-genomic dataset of GBM patients systematically analyzed, towards developing innovative diagnostic methods for brain cancer patients [37].
IvyGAP is a collaborative project between the Ben and Catherine Ivy Foundation, the Allen Institute for Brain Science, and the Ben and Catherine Ivy Center for Advanced Brain Tumor Treatment. The radiographic scans of IvyGAP are made available on TCIA (wiki.cancerimagingarchive.net/display/Public/Ivy+GAP). In situ hybridization (‘ISH’), RNA sequencing data, and digitized histology slides, along with corresponding anatomic annotations are available through the Allen Institute (glioblastoma.alleninstitute.org). Furthermore, the detailed clinical, genomic, and expression array data, designed to elucidate the pathways involved in GBM development and progression, are available through the Swedish Institute (ivygap.swedish.org).
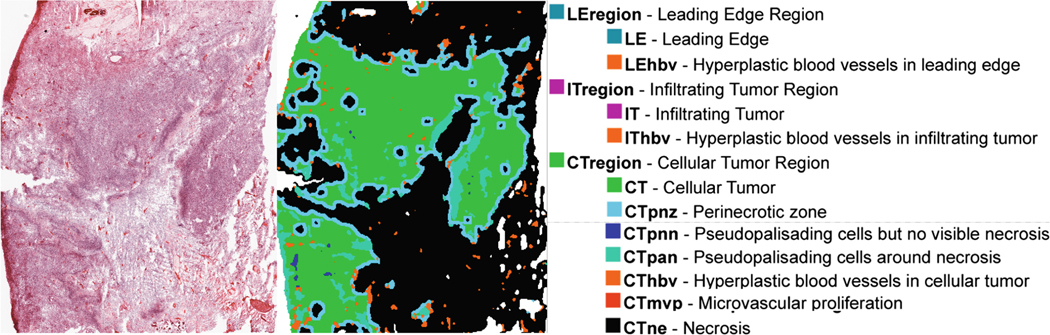
The histologic data contains approximately 11,000 digitized and annotated frozen tissue sections from 42 tumors (41 GBM patients) [37] in the form of hematoxylin and eosin (‘H&E’) stained slides along with accompanying ISH tests and molecular characterization. Tissue acquisition, processing, and staining occurred at different sites and times by different people following specific protocols [37]. Notably to this study, this resource contains a large number of H&E-stained GBM slides, each with a corresponding set of annotations corresponding to structural components of the tumor (Fig. 1).
Fig. 1.
An example slide and corresponding label map for IvyGAP is shown on the left, and on the right is the list of anatomical features annotated in the IvyGAP dataset, as well as their corresponding color in the label map.
2.2. Data Selection/Tissue Review
As also mentioned in the landmark manuscript describing the complete IvyGAP data [37], we note that the annotation labels (Fig. 1) a) do not comply with the current World Health Organization (‘WHO’) classification, and b) have as low as 60% accuracy due to their semi-supervised segmentation approach (as noted by IvyGAP’s Table S11 [37]). Therefore, for the purpose of this study, and to ensure consistency and compliance with the clinical evaluation WHO criteria during standard practice, 509 IvyGAP annotated histological images were reviewed by two board-certified neuropathologists (A.N.V. and M.P.N.). For each image, the structural features/regions that were correctly identified and labeled by IvyGAP’s semi-automated annotation application according to their published criteria [37] were marked for inclusion in this study, and all others were excluded from the analysis.
2.3. Stain Deconvolution
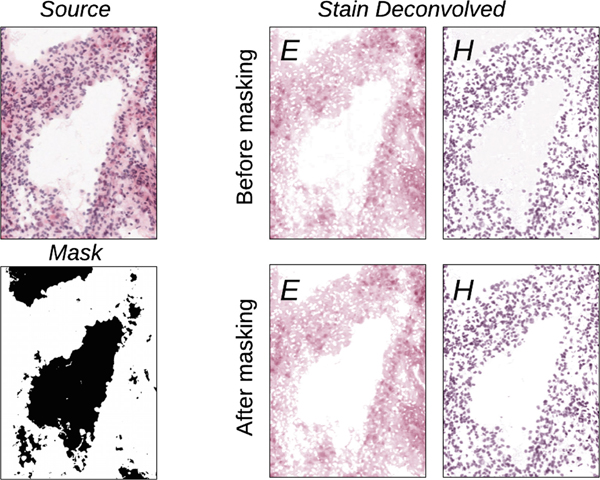
Each slide was paired with a corresponding label map of anatomical features, and then on a per-annotation basis, each region was extracted and the stains were deconvolved. A example deconvolution is illustrated in Fig. 2. Our method of stain deconvolution was based off the work of Ruifrok and Johnston [39], whose work has become the basis for many popular normalization methods. More directly, our method stems from Vahadane et al. [32], but modified so the density transformation is not a linear mapping, but a two-channel stain density histogram matching.
Fig. 2.
An example of stain deconvolution on a patch from an IvyGAP WSI. The top and bottom row illustrate the effect of deconvolution before and after applying background masking, where is a substantial artifact of background intensity.
First, a source image is flattened to a list of pixels, and represented by the matrix I. It can be stated that:
| (1) |
where m is the number of color channels in an RGB image (r = 3), and n is the number of pixels in the flattened source image. This source image can be deconvolved into the color basis for each stain, S, and the density map, W, with each matrix element representing how much one of the stains contributes to the overall color of one of the pixels. In matrix form, let:
| (2) |
where r is the number of stains in the slide (in this case, we consider r = 2 for ‘H&E’), and n is the number of pixels in the image, Also let:
| (3) |
where m is once more the number of color channels in an RGB image, and r is the number of stains present in the slide. Additionally, I0 is a matrix which represents the background of light shining through the slide, which in the case is an RGB value of (255, 255, 255). Putting it all together, we get:
| (4) |
To accomplish this, we used an open source sparse modeling library for python, namely SPAMS [40]. However, it should be noted that more robust libraries for sparse modeling exist, such as SciPy. First, we used dictionary learning to find a sparse representation of an input reach, resulting in a 2 × 3 OD representation of the stain vectors, S. Then, using these stain vectors, we used SPAMS’ lasso regression function to deconvolve the tissue into the density map W, which is a matrix showing the per-pixel contribution of each of the stains to each pixel’s final color.
The actual process of anatomical region extraction begins by pairing an ‘H&E’ with its corresponding label (Fig. 2). Then, each tissue region corresponding to each anatomical label was extracted. The pixels were converted to the CIELAB color space, and pixels over 0.8 L were thresholded out as background intensity. The remaining pixels were stain-deconvolved, and the stain vectors were saved. The ratio of red to blue was found for each stain vector, and each stain’s identity was then inferred from it being more pink (eosin) or blue (hematoxylin) when compared with the other stain vector. The stain densities were converted into a sparse histogram, and also saved. Additional region statistics (mean, standard deviation, median, and interquartile range) were also saved for each anatomical region for validation.
2.4. Composite Histograms
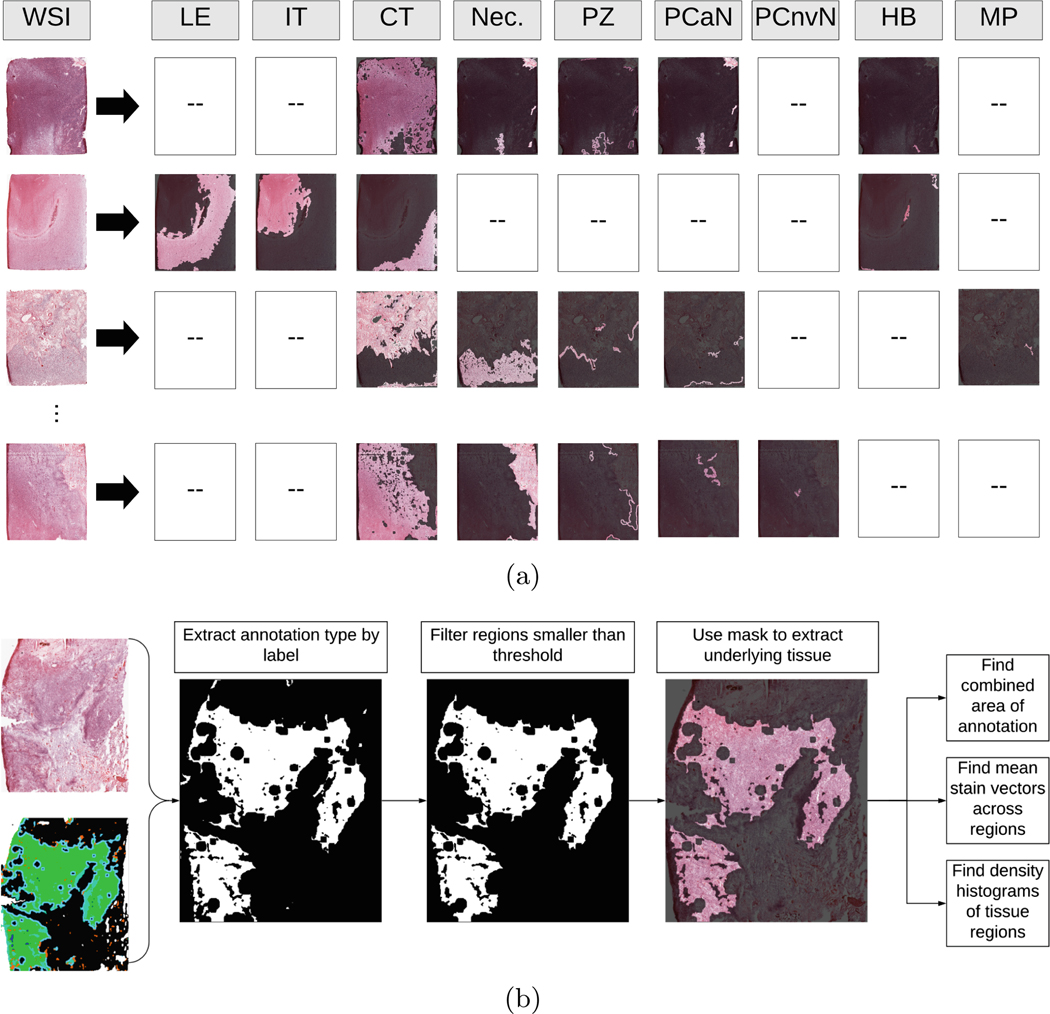
The global histogram composition of our approach is based on the assessment of each independent region (Fig. 3(a)). To create each composite histograms, first, a slide and associated label map were loaded. Then, the label map was downsampled to one tenth of its original size, and a list of colors present in the label map were found. Next, each present annotation was converted into a binary mask, and all connected components for that annotation were found. Components with an area smaller than 15,000 (0.5% of WSI size) pixels were discarded. Next, the remaining components were used as a mask to extract the underlying tissue, and flatten the region into a list of pixels. Then, stain vectors were estimated for the region, and a density map was found. Two sparse histograms were then created from the associated density values for each stain (Fig. 3(b)). This process was repeated for each annotation within an image, and then across all images. Histograms and stain vectors were kept specific to each annotation type. Each annotation type had an associated cumulative area, master histogram, and list of stain vectors.
Fig. 3.
(a) Examples of decomposing WSIs into various annotated anatomical regions. The labels correspond to the following respective components: leading edge, infiltrating tumor, cellular tumor, necrosis, perinecrotic zone, pseudopalisading cells around necrosis, pseudopalisading but no visible necrosis, hyperplastic blood, and microvascular proliferation. (b) Overview of the process used to separate and analyze regions from WSIs. First, a slide and a corresponding annotation map were loaded. Then, on a per-annotation basis, a pixel mask was created, and regions smaller than a threshold were removed. Then, each underlying region of tissue was extracted, and broken down into stain vectors and density maps via SNMF encoding [32]. Finally, a histogram was computed for the annotated tissue region. Repeat for each annotation present in the slide.
2.5. Image Transformation
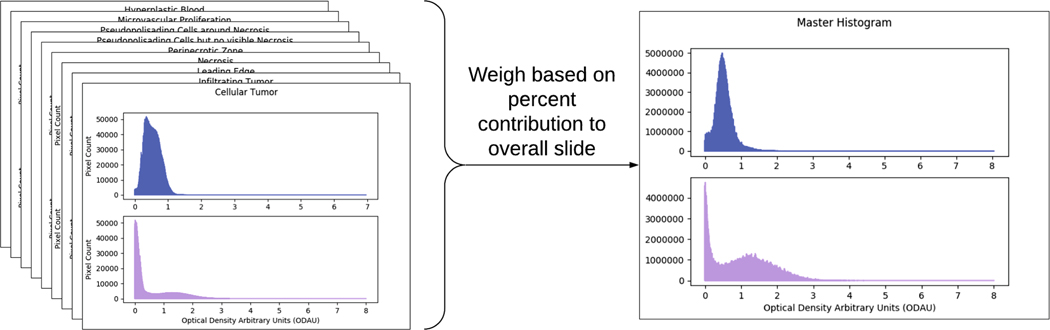
Statistics across regions were summed across all slides. To account for differences in the area representing the whole slide, each annotated anatomical structure’s overall histograms were weighed according to their percent contribution to overall slide (‘PCOS’), then merged to create a master histogram (Fig. 4). Additionally, mean stain vectors from each annotation region were computed, then weighted according to their annotation’s PCOS, and finally combined to give a master set of stain vectors.
Fig. 4.
Weighing all summary histograms based on total area, and merging to create a final summary histogram.
With the target histogram and stain vectors computed, we are able to transform a source slide in a number of ways, but we choose to use a technique derived from [32]. To do this, we first converted the slide to CIELAB color space, thresholded out background pixels, and transformed the remaining pixels using non-negative matrix factorization (‘NMF’) as proposed in [41] and extended in [32]. Then, a cumulative density function (‘CDF’) was found for each stain in both the source image and the target image, and a two-channel histogram matching function transformed each of the source’s stain density maps to approximate the distribution of the target. Finally, the stain densities and corresponding stain vectors were reconvolved, and the transformed source image closely resembles the composite of all target images in both stain color and density distributions.
3. Results
We validate our approach by quantitatively evaluating the reduction in variability of each annotated region in WSIs of our dataset. To do this, we computed the standard deviation of each anatomical annotation independently across all selected IvyGAP slides by extracting the annotated anatomical structure per slide, filtering background pixels, deconvolving, and finding the distibutions of standard deviation of each stain’s density within that tissue type. We then used the master stain vectors and histograms to batch normalize each of these slides. Following stain normalization, we recomputed the standard deviation of each transformed slide by using the same process as with the pre-convolved slides.
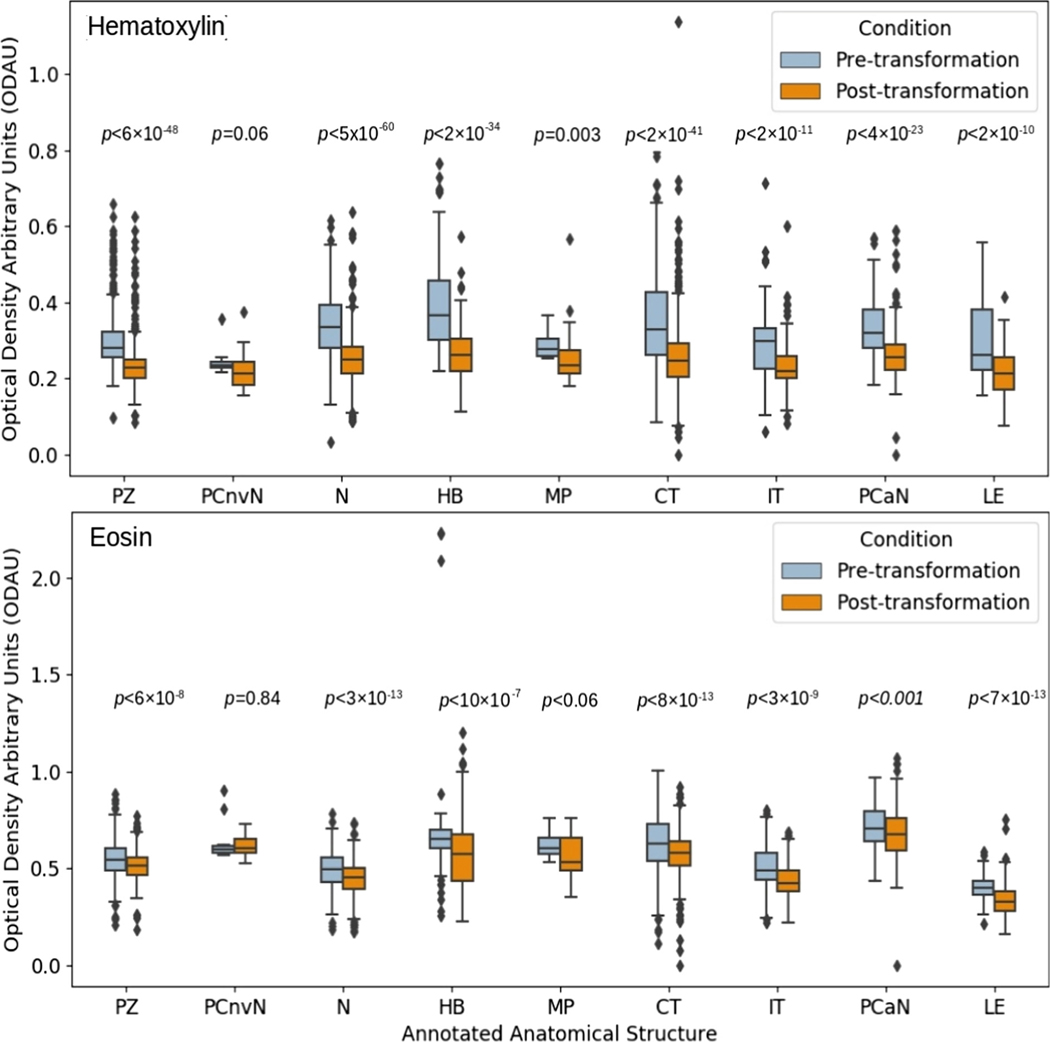
Results of each distribution of standard deviations for each stain are shown in Fig. 5. In other words, the boxes of Fig. 5 denote the spread of the spreads. Comparing pre- and post-transformation slides, through the distributions of standard deviations, shows a significant (largest p = 0.001, two-sided Wilcoxon rank sum test) decrease in standard deviation across tissue types, for all regions contributing more than 0.05% PCOS.
Fig. 5.
The overall reduction in intra-cohort variability of stain density by annotated anatomical structure. p value based on two-sided Wilcoxon rank sum test. Abbreviations for annotated regions are as follows: ‘PZ’ = Perinecrotic Zone, ‘PCnvN’ = Pseudopalisading Cells with no visible Necrosis, ‘N’ = Necrosis, ‘HB’ = Hyperplastic Blood, ‘MP’ = Microvascular Proliferation, ‘CT’ = Cellular Tumor, ‘IT’ = Infiltrating Tumor, ‘PCaN’ = Psuedopalisading Cells around Necrosis, ‘LE’ = Leading Edge
We identified the master stain vectors for hematoxylin (‘H’) and eosin (‘E’) as RGBH = [141, 116, 203] and RGBE = [148, 117, 180], respectively.
4. Discussion and Conclusion
We found that it is feasible to create composite statistics of a batch of images, to create a robust and biologically significant representation of the target GBM slide for future pre-processing. The multi-site nature of the dataset used for validating the proposed approach further emphasizes its potential for generalizability.
The technique proposed here, when compared with deep learning approaches [34, 35], obviates a “black box” entirely by nature of being a transformative technique, and not a generative one. Through the law of large numbers, we attempt to approximate the general distribution of stain densities for GBM slides from a large batch, and use it to transform slides to match said specifications. Thus, no new information in synthesized, and previously unseen structures can be transformed without issue.
Using the very specific set of slides in this study, we identified specific master stain vectors for ‘H’ and ‘E’, provided in the “Results” section above. The colors of these master stain vectors seem fairly close to each other in an RGB space, owing this partly to the “flattened color appearance” of slides fixed in frozen tissue sections, as the ones provided in the IvyGAP dataset. We expect to obtain master stain vectors of more distinct colors with formalin-fixed paraffin- embedded (FFPE) tissue slides.
While this approach has been shown to be feasible, there is still much room for improvement. For instance, the current approach is limited by the slide transformation algorithm that it implements. Stain transfer algorithms are heavily prone to artifacting [33]. While this algorithm avoids issues such as artificially staining background density, it still struggles in certain areas. Notably, NMF approaches rely on the assumption that the number of stains in a slide is already known, and that every pixel can be directly reconstructed from a certain combination of the stains. While with ‘H&E’, we can assuredly say there are only two stains, it neglects other elements such as red blood cells that introduce a third element of color into the pixel. Thus, an area for expansion is in the ability of stain transfer algorithms to cope with other variations in color.
The current application of this approach has been on a dataset containing frozen tissue, which is not representative of most slides in anatomical pathology cases or research. However, it offers the methodology to apply on a larger, more representative set of curated slides to yield a more accurate target for clinically relevant stain normalization. Furthermore, even though we note the benefit of the proposed approach by the overall reduction in the intra-cohort variability of stain density across multiple annotated anatomical structures (Fig. 5), further investigation is needed to evaluate its relevance in subsequent image analysis methods [42].
Future work includes expansion of the proposed approach through the implementation and comparison of other stain transfer techniques, and a general refinement to create a more accurate composite representation. While our proposed method is still prone to artifacts and other complications seen throughout stain transfer techniques, we believe that this study shows the feasibility of using large, detailed, publicly available multi-institutional datasets to create robust and biologically accurate reference targets for stain normalization.
Acknowledgement.
Research reported in this publication was partly supported by the National Institutes of Health (NIH) under award numbers NIH/NINDS: R01NS042645, NIH/NCI:U24CA189523, NIH/NCATS:UL1TR001878, and by the Institute for Translational Medicine and Therapeutics (ITMAT) of the University of Pennsylvania. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH, or the ITMAT of the UPenn.
References
- 1.Ostrom Q, et al. : Females have the survival advantage in glioblastoma. Neuro-Oncology 20, 576–577 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrlinger U, et al. : Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): a randomised, open-label, phase 3 trial. Lancet 393, 678–688 (2019) [DOI] [PubMed] [Google Scholar]
- 3.Sottoriva A, et al. : Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci 110, 4009–4014 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark K, et al. : The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26(6), 1045–1057 (2013). 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scarpace L, et al. : Radiology data from the cancer genome atlas glioblastoma [TCGA-GBM] collection. Cancer Imaging Arch. 11(4) (2016) [Google Scholar]
- 6.Verhaak RGW, et al. : Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1), 98–110 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network: Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med 372(26), 2481–2498 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beers A, et al. : DICOM-SEG conversions for TCGA-LGG and TCGA-GBM segmentation datasets. Cancer Imaging Arch. (2018) [Google Scholar]
- 9.Bakas S, et al. : Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat. Sci. Data 4, 170117 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakas S, et al. : Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. Cancer Imaging Arch. 286 (2017) [Google Scholar]
- 11.Bakas S, et al. : Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. Cancer Imaging Arch. (2017) [Google Scholar]
- 12.Gevaert O: Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 273(1), 168–174 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutman DA, et al. : MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 267(2), 560–569 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binder Z, et al. : Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development. Cancer Cell 34, 163–177 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain R: Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology 272(2), 484–93 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aerts HJWL: The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2(12), 1636–1642 (2016) [DOI] [PubMed] [Google Scholar]
- 17.Davatzikos C, et al. : Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging 5(1), 011018 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bilello M, et al. : Population-based MRI atlases of spatial distribution are specific to patient and tumor characteristics in glioblastoma. NeuroImage: Clin. 12, 34–40 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akbari H, et al. : In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro-Oncology 20(8), 1068–1079 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakas S, et al. : In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index. Clin. Cancer Res 23, 4724–4734 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwanenburg A, et al. : Image biomarker standardization initiative, arXiv:1612.07003 (2016) [Google Scholar]
- 22.Lambin P, et al. : Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 48(4), 441–446 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haralick RM, et al. : Textural features for image classification. IEEE Trans. Syst. Man Cybern 3, 610–621 (1973) [Google Scholar]
- 24.Galloway MM: Texture analysis using grey level run lengths. Comput. Graph. Image Process 4, 172–179 (1975) [Google Scholar]
- 25.Chu A, et al. : Use of gray value distribution of run lengths for texture analysis. Pattern Recogn. Lett 11, 415–419 (1990) [Google Scholar]
- 26.Dasarathy BV, Holder EB: Image characterizations based on joint gray level—run length distributions. Pattern Recogn. Lett 12, 497–502 (1991) [Google Scholar]
- 27.Tang X: Texture information in run-length matrices. IEEE Trans. Image Process 7, 1602–1609 (1998) [DOI] [PubMed] [Google Scholar]
- 28.Amadasun M, King R: Textural features corresponding to textural properties. IEEE Trans. Syst. Man Cybern 19, 1264–1274 (1989) [Google Scholar]
- 29.Jain A: Fundamentals of Digital Image Processing. Prentice Hall, Englewood Cliffs: (1989) [Google Scholar]
- 30.Macenko M, et al. : A method for normalizing histology slides for quantitative analysis. In: 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 1107–1110 (2009) [Google Scholar]
- 31.Reinhard E, et al. : Color transfer between images. IEEE Comput. Graphics Appl 21(5), 34–41 (2001) [Google Scholar]
- 32.Vahadane A, et al. : Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med. Imaging 35, 1962–1971 (2016) [DOI] [PubMed] [Google Scholar]
- 33.Khan A, et al. : A non-linear mapping approach to stain normalisation in digital histopathology images using image-specific colour deconvolution. IEEE Trans. Biomed. Eng 61(6), 1729–1738 (2014) [DOI] [PubMed] [Google Scholar]
- 34.Shaban MT, et al. : StainGAN: stain style transfer for digital histological images. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI), pp. 953–956 (2019) [Google Scholar]
- 35.Janowczyk A, et al. : Stain normalization using sparse AutoEncoders (StaNoSA). Comput. Med. Imaging Graph 50–61, 2017 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bianconi F, Kather JN, Reyes-Aldasoro CC: Evaluation of colour pre-processing on patch-based classification of H&E-stained images In: Reyes-Aldasoro CC, Janowczyk A, Veta M, Bankhead P, Sirinukunwattana K. (eds.) ECDP 2019. LNCS, vol. 11435, pp. 56–64. Springer, Cham; (2019). 10.1007/978-3-030-23937-47 [DOI] [Google Scholar]
- 37.Puchalski R, et al. : An anatomic transcriptional atlas of human glioblastoma. Science 360, 660–663 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah N, et al. : Data from Ivy GAP. Cancer Imaging Arch. (2016) [Google Scholar]
- 39.Ruifrok A, Johnston D: Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol 23(4), 291–299 (2001) [PubMed] [Google Scholar]
- 40.Mairal J, et al. : Online learning for matrix factorization and sparse coding. J. Mach. Learn. Res 11, 19–60 (2010) [Google Scholar]
- 41.Rabinovich A, et al. : Unsupervised color decomposition of histologically stained tissue samples. In: Advances in Neural Information Processing Systems, vol. 16, pp. 667–674 (2004) [Google Scholar]
- 42.Li X, et al. : A complete color normalization approach to histopathology images using color cues computed from saturation-weighted statistics. IEEE Trans. Biomed. Eng 62(7), 1862–1873 (2015) [DOI] [PubMed] [Google Scholar]