Abstract
Myeloid-derived suppressor cells (MDSC) promote colorectal cancer by several mechanisms, including suppression of antitumor T cells and production of tumorigenic factors. We previously showed that an intermediate MDSC subset (I-MDSC) is expanded in an intestinal tumor model (ApcMin/+ mice), but the importance of this subset in promoting tumors is unclear. Here, we show that I-MDSCs are a distinct heterogeneous subset due to differential and reduced expression of the monocytic marker, Ly6C, and granulocytic marker, Ly6G. Besides causing necroptotic cell death, receptor-interacting protein kinase 3 (RIPK3) has an alternate function as a signaling component inducing cytokine synthesis. We evaluated whether RIPK3 regulates inflammatory cytokines in I-MDSCs to assess the nonimmunosuppressive function of I-MDSCs in promoting tumors. Inhibition of RIPK3 with the commercially available small-molecule inhibitor GSK 872 showed that RIPK3-mediated inflammation promoted intestinal tumors in two intestinal tumor models, ApcMin/+ mice and an MC38 transplantable tumor model. Mechanistically, RIPK3 signaling in I-MDSC increased tumor size by expanding IL17-producing T cells in MC38 tumors. Collectively, these data suggest RIPK3 signaling as a potential therapeutic target in colorectal cancer.
Introduction
Myeloid-derived suppressor cells (MDSC) in patients with colorectal cancer correlate with reduced survival and have emerged as a resistance mechanism to existing therapies (1). Targeting MDSC function is beneficial for improving the treatment efficacy; however, MDSCs are functionally complex. Success relies on a better understanding of functional differences among MDSC subsets because each subset appears to have a different role in tumorigenesis. In the intestinal cancer models, much of the focus has been on how monocytic MDSCs (M-MDSCs, CD11b+Ly6c+Ly6G−) and polymorphonuclear MDSCs (PMN-MDSCs, CD11b+Ly6G+Ly6Clow) promote tumor growth (2, 3). We observed a new MDSC subset termed intermediate MDSC (I-MDSC) expressing reduced Ly6C and Ly6G in tumor-bearing ApcMin/+ mice that has APC mutation, commonly found in patients with colorectal cancer (4). Phenotypic heterogeneity is a characteristic feature of MDSCs, but less is known about this variation, particularly in I-MDSCs (5). Earlier we showed that tumor reduction in ApcMin/+ mice deficient in Stat6 resulted in the reduction of all MDSC subsets (4). From previous studies, the suppressive function of M-MDSCs and PMN-MDSCs are known to promote tumors; however, the mechanism by which I-MDSCs function in ApcMin/+ mice is not known. Delineating this process would shed light on a targetable tumor promoting mechanism as the MDSC depletion or inhibition of MDSC function are an attractive therapeutic strategies for patients with colorectal cancer.
Receptor-interacting protein kinase 1 (Ripk1) and RIPK3 induce necroptotic cell death, but also signal through ERK pathway to induce cytokines (6). Upon TNFα activation, Ripk1 in complex 1 activates apoptosis via caspase 8. When caspases are inhibited, the RIP homotypic interacting motif (RHIM) of Ripk1 interacts with RIPK3 RHIM in complex 2 to activate kinase activity and phosphorylation of mixed lineage kinase domain–like (MLKL) protein, resulting in necroptosis (7). Hence, Ripk1 activation can lead to either apoptosis or necroptosis. However, deleting Ripk1 in keratinocytes resulted in RIPK3-mediated necroptosis causing excessive inflammation and Ripk1-deficient hematopoietic cells underwent increased apoptosis and necroptosis (8, 9). This was found to be due to the interaction of ZBP-1 RHIM with RIPK3 RHIM, which was prevented by Ripk1 RHIM (10). Other RHIM-containing proteins such as Toll/IL1 receptor domain containing adaptor protein–inducing IFNβ (TRIF) or DNA-dependent activator of IFN regulatory factor (DAI) can also activate RIPK3 RHIM to promote its kinase activity (11).
In addition to causing cell death, RIPK3-induced inflammatory cytokines promoted disease severity in inflammatory bowel disease (IBD) and TNFα-induced systemic inflammatory response syndrome (SIRS; refs. 12, 13). Therefore, RIPK3 is an ideal target for inhibiting inflammation. However, RIPK3 defect produced contradictory responses in murine colitis and intestinal cancer. One study showed that RIPK3 deletion did not affect colitis (12), disease was exacerbated in another study (14), and disease was ameliorated in a different study (15). Deletion of RIPK3 RHIM in CD11c+ cells reduced IL22 required for tissue repair, exacerbating colitis (16). These RIPK3 RHIM–deficient dendritic cellS (DC) produced less IL23 and IL1β due to the reduced activation of NFκBp50, suggesting that RIPK3 RHIM is required for its signaling function. Although RIPK3 RHIM hasa different function from its kinase domain, interaction of RHIM domains is essential for RIPK3 kinase activity (17). Exacerbation of colitis after deletion of RIPK3 RHIM (16) may not result in intestinal tumor development. This is possible as shown by increased colitis but reduced AOM-DSS–induced tumors after NFκBp50 deletion (18). However, total deletion of RIPK3 increased AOM-DSS–induced colonic tumors by promoting tumor cell proliferation and upregulating inflammatory cytokines (19). Conversely, deletion of RIPK3 reduced AOM-DSS–induced colon tumors promoted by Hace1 deficiency (15). Moreover, RIPK3 inhibition in bone marrow–derived macrophages (BMDM) reduced inflammatory cytokines and chemokines (20). These reports show that additional evidence is required to define how RIPK3 function impacts the intestinal tumors. Considering the importance of inflammatory cytokines in promoting intestinal tumors and the demonstrated role of RIPK3 in inducing these cytokines in myeloid cells, we reasoned that RIPK3-induced cytokines in MDSCs could promote intestinal tumors.
We targeted RIPK3 activity using a commercially available inhibitor GSK 872 that was found to be a highly specific inhibitor of RIPK3 after testing against 300 other protein tyrosine kinases (21). Given its overall tolerability among several cell types at low concentrations, we used it as a tool to elucidate the function of RIPK3 in intestinal cancer and in MDSCs. The correlation between MDSC accumulation and Th17 cells with reduced survival of patients with colorectal cancer (1) and the importance of MDSC-induced IL17 in inflammation-driven tumorigenesis (22, 23) suggest that analysis of the signaling mechanism in MDSCs driving IL17 induction would reveal therapeutic candidates. In this report, we demonstrate that RIPK3 promotes the intestinal tumors by upregulating cytokines required for expanding IL17-producing T cells through I-MDSCs, a distinct MDSC subset.
Materials and Methods
Mice
C57BL/6J and ApcMin/+ mice were purchased from Charles River Laboratories and Jackson Laboratory, respectively. Both male and female ApcMin/+ mice at 13 to 24 weeks were used in all experiments. Wild-type (WT) littermates were used as controls. ApcMin/+ mice were genotyped to identify ApcMin mutation using previously published primers (24). All mouse protocols were approved by the Yale University Institutional Animal Care and Use Committee.
RNA preparation and quantitative RT-PCR
RNA was extracted with TRIzol (Invitrogen) from size-matched ApcMin/+ polyps, WT, small intestine, ApcMin/+ MDSCs subsets (gated as shown in Supplementary Fig. S1A), or in vitro–treated MDSC subsets. The suppression assays for MDSCs are described in Supplementary Materials and Methods. Polyps and small intestine tissue were homogenized in disposable Lysing matrix tubes (MP Biomedicals) before RNA extraction. cDNA was reverse-transcribed from DNase-treated RNA using Superscript III Reverse Transcriptase (Invitrogen). Target gene was amplified by SYBR Green PCR Master Mix (Bio-Rad). Expression was normalized to housekeeping genes, RPL32 or GAPDH. The standard curve method was used to quantify the relative gene expression. Primer sequences for all target genes were obtained from the Harvard Medical School PrimerBank (25) and checked for specificity by Primer-BLAST on NCBI (26).
ApcMin/+ and MC38 tumor models
ApcMin/+ mice were injected every 3 days with GSK 872 [N-(6-(Isopropylsulfonyl)quinolin-4-yl)benzo[d]thiazol-5-amine, Millipore] at 0.25 mg/kg body weight when mice were 13 weeks old. Mice in the control group received vehicle (DMSO) dissolved in PBS. GSK 872 was well-tolerated as treated mice showed no signs of lethargy or allergic reaction at 24 hours after treatment with this inhibitor and every 3 days thereafter. At 20 weeks, tumors in the small intestine and colon of ApcMin/+ mice were counted and harvested for qRT-PCR. Splenomegaly was assessed by measuring spleen length. Splenocytes were mechanically dissociated to prepare single-cell suspensions for flow cytometry. For MC38 murine colon tumor model, 2 × 104 MC38 cells were implanted subcutaneously in WT female C57BL/6J mice. A total of 0.8 × 106 or 106 splenic MDSCs of each subset from tumor-bearing ApcMin/+ mice or BM-MDSCs (purity of sorted MDSCs is shown in Supplementary Fig. S1B) were injected directly into similar sized MC38 tumors. After I-MDSC injection, mice were treated with GSK 872 at 0.5 mg/kg every other day or once with anti-IL17A or isotype control antibody (75 μg/mouse). Tumor volume was monitored every other day, and calculated by the following formula: short diameter2 × (long diameter)/2. MC38 cells were kindly provided by Dr. Alan Sartorelli at Yale University (New Haven, CT). MC38 cells were regularly tested for Mycoplasma and found to be contamination-free.
Preparation of cells for flow cytometry and cell sorting
Tumor-infiltrating lymphocytes (TIL) from MC38 tumors were prepared by digestion of tumor fragments with collagenase (Sigma) and DNase (Roche) at 37°C for 30 minutes, live lymphocytes were separated on Percoll density gradient and stained with described antibodies. For the intracellular cytokine staining, TILs were activated with 50 ng/mL of phorbol 12-myristate 13-acetate (PMA; Sigma) and 1 μg/mL of ionomycin (Sigma) for 5 hours; brefeldin A (BD Biosciences) was added during the last 4 hours of culture before fixation. Cells were surface-stained with T-cell markers (CD45, TCRβ, CD4, CD8), permeabilized with permeabilization buffer (eBioscience), and stained with anti-IL17 or isotype control antibody (eBioscience). For sorting MDSC subsets, splenic lymphocytes were depleted of T cells, B cells, and natural killer cells using biotin-labeled CD3, CD19, and CD49d antibodies (Tonbo and eBioscience) and streptavidin-coated magnetic beads (BioLegend) to enrich CD11b+ cells. Enriched CD11b+ cells were stained for MDSC markers, including CD11b, Ly6C, Ly6G, and CD11c to sort M-MDSCs, I-MDSCs, and PMN-MDSCs after excluding DCs on BD FACSAria sorter (BD Biosciences). WT monocytes and neutrophils were sorted similarly. Sorting purity for these cells is shown in Supplementary Fig. S1B. Viability was maintained by storing the cells in RPMI containing 20% FBS until sorting was completed and assessed by trypan blue exclusion assay. Lamina propria (LP) lymphocytes were prepared from small intestines of ApcMin/+ mice by Percoll density gradient and CD11b+ cells were isolated by positive selection with biotin-labeled CD11b antibody (BD Biosciences) and streptavidin-coated magnetic beads (BioLegend). Prior to LP isolation, the intestinal epithelial cells (IEC) were collected from the EDTA-PBS wash in which the cleaned small intestine fragments were incubated at 37°C for 30 minutes. Purity of the isolated LP CD11b+ cells and IECs are shown in Supplementary Fig. S1C. Refer to the Supplementary Materials and Methods for details.
Flow cytometry data analysis
FlowJo was used to generate flow cytometry plots and mean fluorescence intensity (MFI) values from FCS files obtained by analyzing the cells on the Stratedigm instrument. Isotype control MFI was subtracted from target MFI to generate the heatmap. ApcMin/+ MDSCs were analyzed by FlowSOM tool in FlowJo to identify subpopulations within MDSC subsets (27). FlowSOM generated a minimal spanning tree (MST) consisting of 4 main clusters based on Ly6C and Ly6G expression. Each subpopulation of a MDSC subset included closely arranged nodes, where each node represents a group of similar cells and the size of the node represents the number of cells. Nodes arranged next to each other on the MST are more closely linked than nodes that are further away. Frequencies and MFI values for the sub-populations of MDSC subsets were also generated by the FlowSOM tool.
In vitro BM-MDSC generation and RIPK3 inhibition in MDSCs Bone marrow cells from WT mice were cultured at 106 per mL in MDSC medium containing RPMI, 10% FBS, 1% penicillin/streptomycin, MC38 tumor-conditioned medium (TCM), and 40 ng/mL of recombinant murine (rm) GM-CSF (Peprotech) for 3 days to generate BM-MDSCs. Single-cell suspension of MC38 tumors were cultured in RPMI with 10% FBS at 5 × 105 cells per mL for 24 hours to produce TCM. This protocol was modified from published methods (28, 29). Loosely adherent cells were separated into MDSC subsets based on Ly6C and Ly6G expression on CD11b-positive cells. A total of 106 M-MDSCs, I-MDSCs, or PMN-MDSCs sorted from in vitro–generated BM-MDSCs were stimulated with lipopolysaccharide (LPS; Invivogen) at 10 ng/mL with or without zVAD (carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone) at 50 μmol/L; GSK 872 was added at 5 μmol/L or 20 μmol/L. RNA was extracted after 2 hours and analyzed for cytokine expression by qRT-PCR. Viability of the treated cells was assessed with CellTiter-Glo assay (Promega) using a luminometer and ATP units were calculated as percent of untreated cells.
Statistical analysis
Student t test was used to determine statistical significance. Data are presented as means ± SEM. P value less than 0.05 was considered significant.
Results
RIPK3 inhibition regressed intestinal tumors
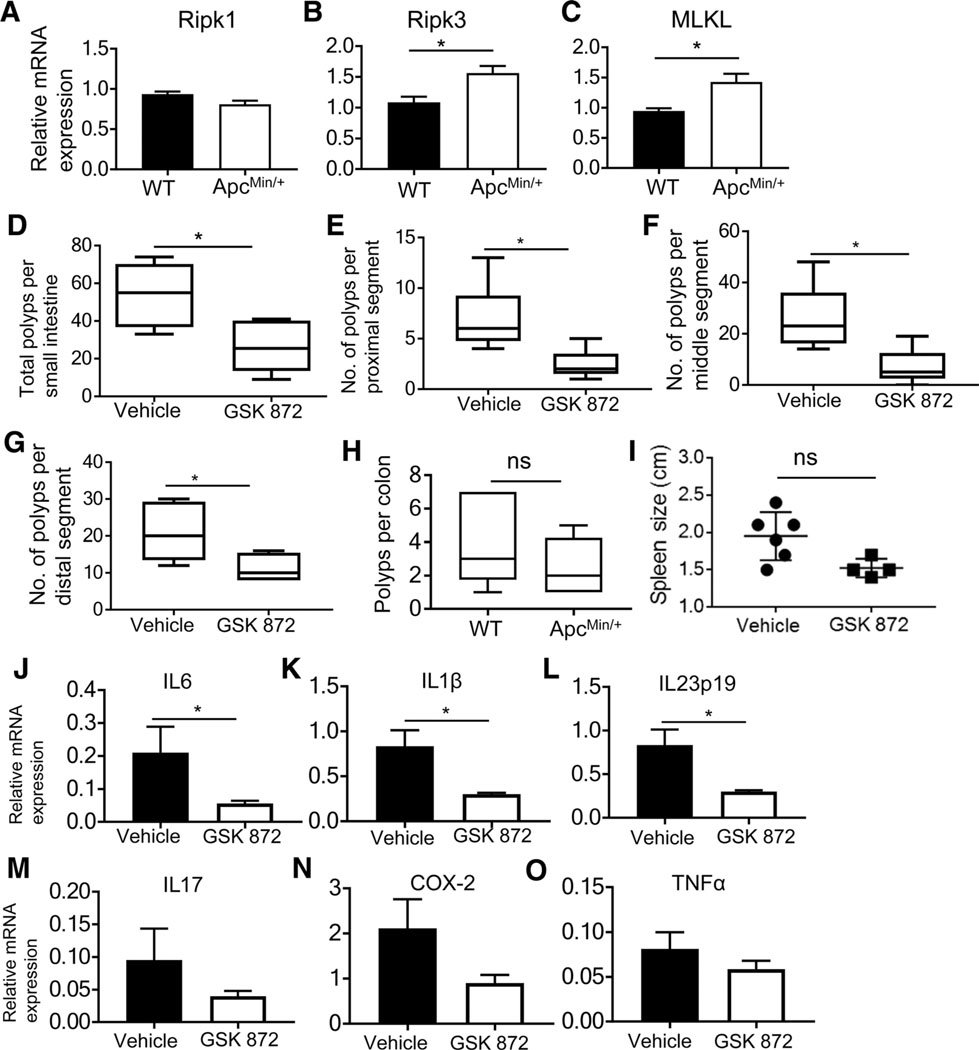
The outcome of RIPK3 function in intestinal cancer is unclear due to different results about its role in tumor formation (12, 15, 16, 19, 30). We used the ApcMin/+ model that is genetically modified to form intestinal tumors, to assess how RIPK3 affects the tumor development. RIPK3 and MLKL were elevated in ApcMin/+ intestinal polyps compared with WT intestinal tissue, but Ripk1 expression was unchanged (Fig. 1A–C), suggesting that RIPK3 mediated tumor growth. To inhibit RIPK3-induced inflammatory cytokines, we used GSK 872, a RIPK3 inhibitor with an IC50 of 1.3 nmol/L (21). A total of 10 μmol/L of GSK 872 induced apoptosis in tumor cells, but not in BMDMs that were relatively viable. This suggested that GSK 872 could inhibit RIPK3-induced cytokines without inducing excessive apoptosis if used at a low and effective concentration, which is beneficial for controlling inflammation-driven colorectal cancer. There were significantly fewer tumors in the small intestine of ApcMin/+ mice treated with GSK 872 compared with mice treated with vehicle (Fig. 1D). Tumors in the proximal, middle, and distal segments of the small intestine of GSK 872–treated ApcMin/+ mice were significantly reduced compared with control (Fig. 1E–G). Colon tumors and splenomegaly were also reduced, but did not reach statistical significance (Fig. 1H and I). IL6, IL1β, and IL23 were significantly reduced in polyps from GSK 872–treated ApcMin/+ mice (Fig. 1J–L); IL17 and COX-2 were also suppressed, but there was no change in TNFα (Fig. 1M–O). This shows that RIPK3 inhibition reduced tumor-promoting cytokines. Surprisingly, epithelial stem cell markers including Sox-9 were increased in polyps of GSK 872–treated ApcMin/+ mice (Supplementary Fig. S2A). This could be due to the prosurvival function of Ripk1 when RIPK3 is inhibited or due to other immunoregulatory mechanisms, such as PD-1 expression (4). Together, these results show that the intestinal tumors in ApcMin/+ mice are dependent on RIPK3 enzymatic activity.
Figure 1.
RIPK3 inhibition reduces intestinal tumors and inflammatory cytokines. Ripk1 (A), RIPK3 (B), and MLKL (C) mRNA expression from WT normal small intestine tissue and ApcMin/+ polyps. D, Polyps in the small intestine of 20-week-old ApcMin/+ mice treated with vehicle or GSK 872 (n = 6). Number of polyps in the proximal (E), middle (F), and distal (G) segments of the small intestine and the colon (H) of ApcMin/+ mice treated as in D. Spleen size (I) and mRNA expression (J) of IL6, IL1β (K), IL23p19 (L), IL17 (M), COX-2 (N), and TNFα (O) in polyps from ApcMin/+ mice treated as in D. *, P < 0.05; ns, nonsignificant.
Inhibition of RIPK3 in ApcMin/+ mice reduced I-MDSC accumulation
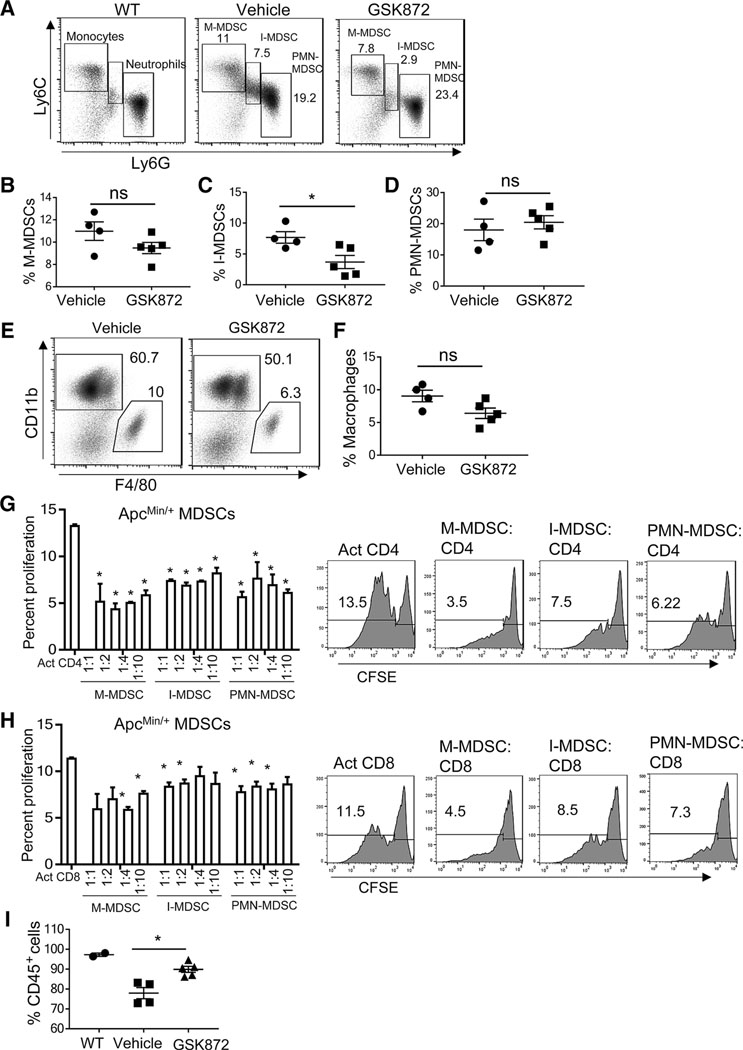
M-MDSCs and PMN-MDSCs correlate with tumor growth in a transplanted colorectal cancer model and in ApcMin/+ mice (2, 3). Tumor reduction in ApcMin/+ mice deficient in Stat6 reduced the frequency of splenic M-MDSCs and PMN-MDSCs and strikingly depleted I-MDSCs (4). In GSK 872–treated ApcMin/+ mice, the frequency of M-MDSCs and macrophages was moderately reduced, PMN-MDSCs were unchanged, and I-MDSCs were significantly reduced (Fig. 2A–F). Therefore, I-MDSC accumulation is tumor-dependent due to its consistent correlation with intestinal tumor burden (Fig. 1D; ref. 4). I-MDSCs appeared to be less suppressive than M-MDSCs in inhibiting proliferation of non-antigen specific CD4 and CD8 T cells (Fig. 2G and H). Moreover, GSK 872 treatment did not significantly alter the frequency of T cells (Supplementary Fig. S2B–S2D). We did not assess the cytotoxic quality of CD8 T cells or the suppressive function of MDSCs in GSK 872–treated ApcMin/+ mice. However, tumor reduction in GSK 872–treated ApcMin/+ mice increased CD45+ cells, indicating reversal of lymphodepletion that is typically observed in diseased ApcMin/+ mice (Fig. 2I; ref. 31). Overall, the reduction of I-MDSCs and tumors after RIPK3 inhibition suggests that I-MDSCs could be related to RIPK3-mediated tumor progression.
Figure 2.
I-MDSCs are reduced in ApcMin/+ mice treated with GSK 872. A, Representative flow cytometry plots of WT monocytes and neutrophils, and ApcMin/+ MDSC subsets; percentages of M-MDSCs (B), I-MDSCs (C), and PMN-MDSCs (D) gated on CD45+CD11bhiCD11clo live splenocytes, macrophages (CD11b+F4/80+; E and F), and CD45+ (I) cells from ApcMin/+ mice receiving vehicle control (n = 4) or GSK 872 (n = 5). G and H, ApcMin/+ MDSC subsets suppress CFSE-labeled, polyclonally activated (anti-CD3 and anti-CD28 antibodies) WT, CD4, and CD8 T-cell proliferation. Representative flow cytometry plots show the suppression of CD4 and CD8 T cells by MDSCs (1:1 MDSCs to T cells). Activated T cells without MDSC coculture were positive controls. Comparisons were made between positive control and each MDSC to T-cell coculture. *, P < 0.05; ns, nonsignificant.
I-MDSCs are a distinct subset of MDSCs dependent on GM-CSF
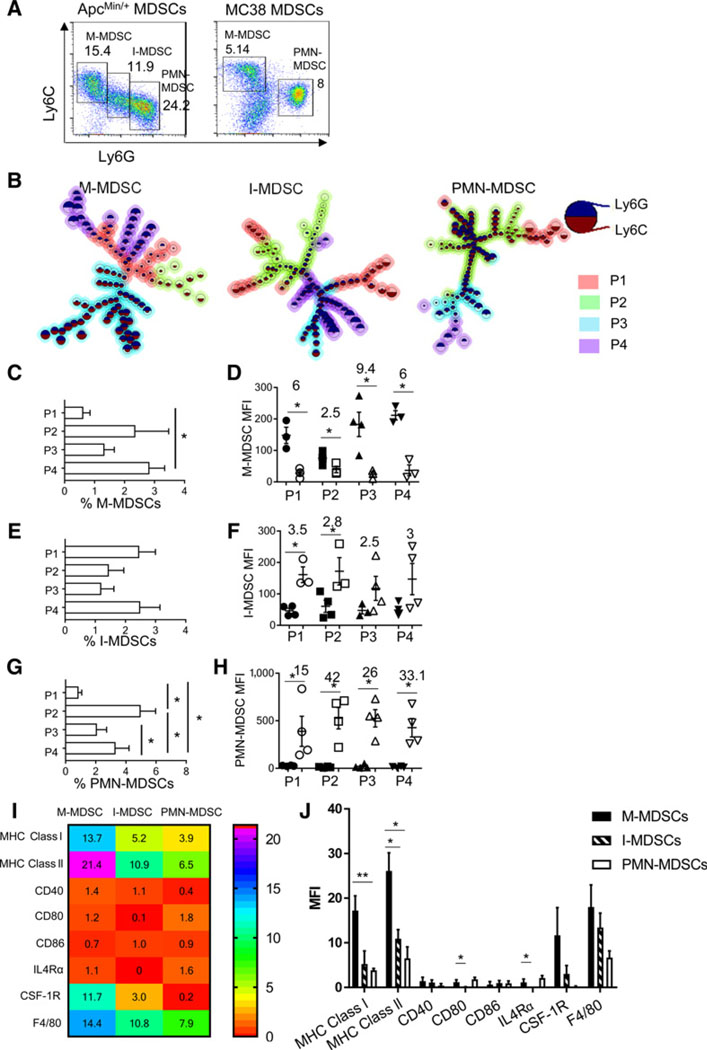
We reasoned that I-MDSCs are phenotypically different from other MDSC subsets, based on the intermediate expression of Ly6C and Ly6G. I-MDSCs accumulated in ApcMin/+ mice, but not in MC38 tumor-bearing mice (Fig. 3A). Other ApcMin/+ MDSC subsets were similar to MC38 MDSCs due to comparable expression of antigen-presenting and costimulatory molecules (Supplementary Fig. S3A and S3B). To better understand the heterogeneity of I-MDSCs, we analyzed each ApcMin/+ MDSC subset for differences in Ly6C and Ly6G expression with Flow-SOM tool. This revealed four sub populations within each MDSC subset (Fig. 3B). There are more P4 M-MDSCs than other subsets (Fig. 3C). All M-MDSC subpopulations (P1–P4) expressed Ly6C and Ly6G, which was unexpected because M-MDSCs are CD11b+Ly6C+Ly6G− (Fig. 3D); however, Ly6C was significantly higher than Ly6G. P1, P3, and P4 have similar Ly6C/Ly6G ratio and are located close to each other in the MST (Fig. 3B). P2 is phenotypically different from other subpopulations because its Ly6C/Ly6G ratio (2.5) is lower and it is located further from other subpopulations (Fig. 3B). Frequencies of all I-MDSC subpopulations were similar (Fig. 3E). The difference between Ly6G and Ly6C was significant in P1 and P2 of I-MDSCs, but not in P3 and P4 (Fig. 3F). A P1 subset and P2, and P3 and P4 are located close to each other on the MST, showing that they are related (Fig. 3B). However, other subsets within P1 are separated on the MST, suggesting that they could be different subsets. Therefore, I-MDSCs appear to be heterogeneous. Among PMN-MDSCs, P2 was most frequent with the highest Ly6G to Ly6C ratio (Fig. 3G and H). More than 10-fold increase in Ly6G in all PMN-MDSCs shows that they are all granulocytic. MHC class l and ll is reduced in I-MDSCs and PMN-MDSCs compared with M-MDSCs, indicating a shared feature between I-MDSCs and PMN-MDSCs (Fig. 3I and J). Notably, M-MDSCs and I-MDSCs express 29 and 15-fold more CSF-1R, and 3 and 2-fold more F4/80, compared with PMN-MDSCs. This suggests that I-MDSCs are similar to M-MDSCs, but lack CD80 and IL4Rα. Therefore, I-MDSCs are a distinct MDSC subset including a spectrum of cells with heterogeneous Ly6C and Ly6G expression and some similarity to other MDSC subsets.
Figure 3.
I-MDSCs accumulating in ApcMin/+ mice are a distinct subset. A, Representative flow cytometry plots of splenic MDSCs from ApcMin/+ mice and MC38 tumor-bearing mice (tumor volume:1,200 mm3. B, MST diagrams of ApcMin/+ MDSC subsets produced by FlowSOM shows subpopulations (P1, P2, P3, P4) in all MDSCs. Each node represents a group of cells with similar MFI; location of the nodes indicates interconnectedness, and size of the node indicates the number of cells. C–H, Frequency and expression level of Ly6C (closed symbols) and Ly6G (open symbols) in subpopulations of each MDSC subset. Numbers above plots in D, F, and H indicate fold, difference between Ly6C and Ly6G (D), and Ly6G versus Ly6C (F and H). Heatmap (I) and column plot (J) of MFIs of MHC class I and II, CD40, CD80, CD86, IL-4Rα, CSF-1R, and F4/80 on ApcMin/+ MDSC subsets. n = 3 to 4; *, P < 0.05.
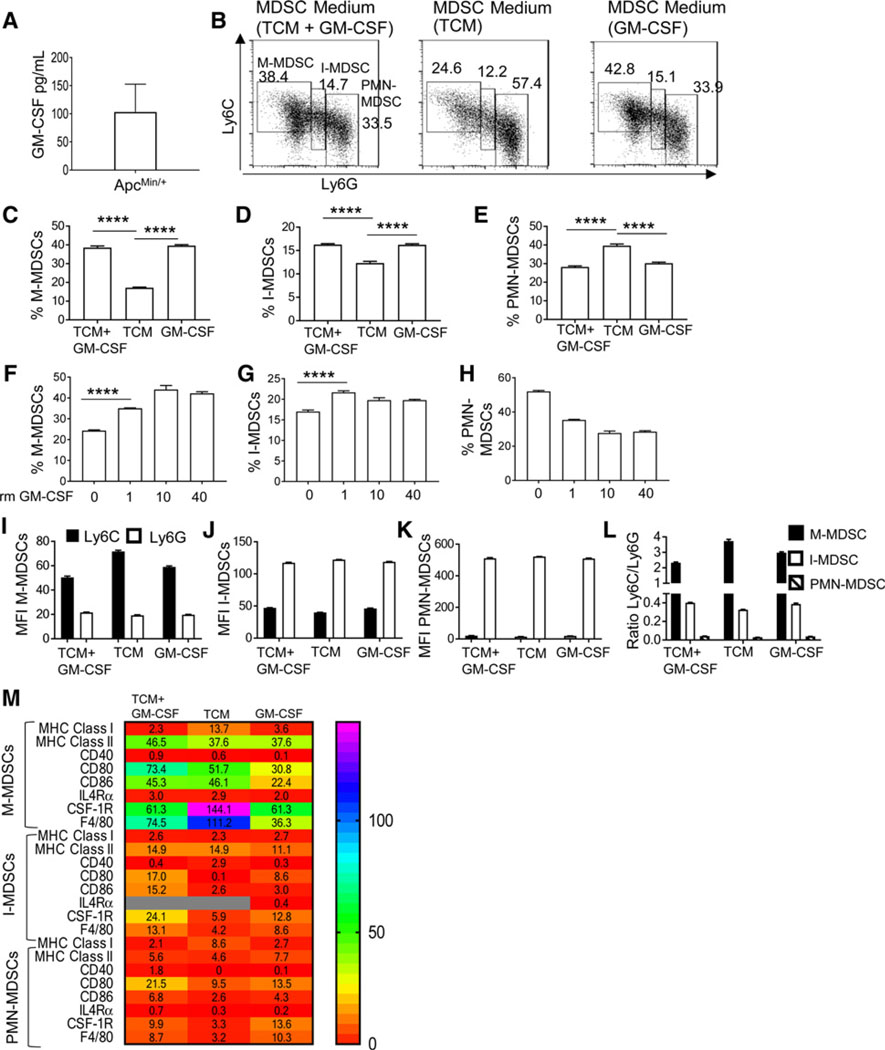
CT26 colon tumor cells overexpressing GM-CSF promoted the accumulation of I-MDSC–like cells with intermediate expression of Ly6C and Ly6G (32). The 4T1 breast cancer cells producing high GM-CSF contributed to the accumulation of I-MDSC–like cells (33). Hence, GM-CSF could selectively favor I-MDSC increase in ApcMin/+ mice, which has high circulating levels of GM-CSF (34) and polyps producing GM-CSF (Fig. 4A). To evaluate whether GM-CSF increased I-MDSC accumulation in ApcMin/+ mice, BM cells were treated with MC38 TCM and rm GM-CSF or rm GM-CSF or TCM. Both I-MDSCs and M-MDSCs accumulated more in either condition including GM-CSF (Fig. 4B–D). BM-MDSCs generated with TCM alone produced more PMN-MDSCs (Fig. 4E). Furthermore, I-MDSCs accumulated even at low concentration of GM-CSF and M-MDSCs increased with more GM-CSF (Fig. 4F–H). Thus, GM-CSF promotes the accumulation of M-MDSCs and I-MDSCs.
Figure 4.
I-MDSC accumulation is GM-CSF–dependent. A, GM-CSF production by intestinal cells including tumor cells in ApcMin/+ mice was assessed by ELISA. B, Representative plots of MDSC subsets in BM-MDSCs generated from bone marrow cells cultured with MC38 TCM and recombinant mouse GM-CSF (40 ng/mL), TCM alone, or GM-CSF alone. C–E, Percentage of MDSCs generated as described earlier (n = 4). Percentage of M-MDSCs (F), I-MDSCs (G), and PMN-MDSCs (H) in BM-MDSCs generated with increasing doses of rm GM-CSF. MFIs of Ly6C and Ly6G (I–K) and ratio of Ly6C versus Ly6G (L) of in vitro–generated MDSCs. M, Heatmap showing the expression (MFI) of MHC class I, MHC class II, CD40, CD80, CD86, IL4Ra, CSF-1R, and F4/80 in subsets of BM-MDSCs generated as described in A (n = 2). Gray boxes in heatmap show undetected values. ****, P < 0.0001.
BM-MDSCs suppress nonantigen–specific T cells, similar to ApcMin/+ MDSCs (Supplementary Fig. S4). Among BM-MDSCs generated in all conditions, M-MDSCs have the highest Ly6C expression, followed by I-MDSCs and PMN-MDSCs (Fig. 4I–L). TCM appears to promote MHC class ll, CD80, and CD86 expression on M-MDSCs and I-MDSCs (Fig. 4M; Supplementary Table S1). MHC ll and CD80 expression on M-MDSCs, and MHC l, CD80, and CD86 on I-MDSCs were dependent on GM-CSF. Both GM-CSF and TCM contributed to CSF-1R and F4/80 expression on I-MDSCs, whereas only TCM promoted F4/80 expression on M-MDSCs. Therefore, GM-CSF is required for I-MDSC accumulation and both TCM and GM-CSF are essential for their distinct phenotype.
I-MDSCs accelerate MC38 tumor growth by RIPK3-mediated IL17 production
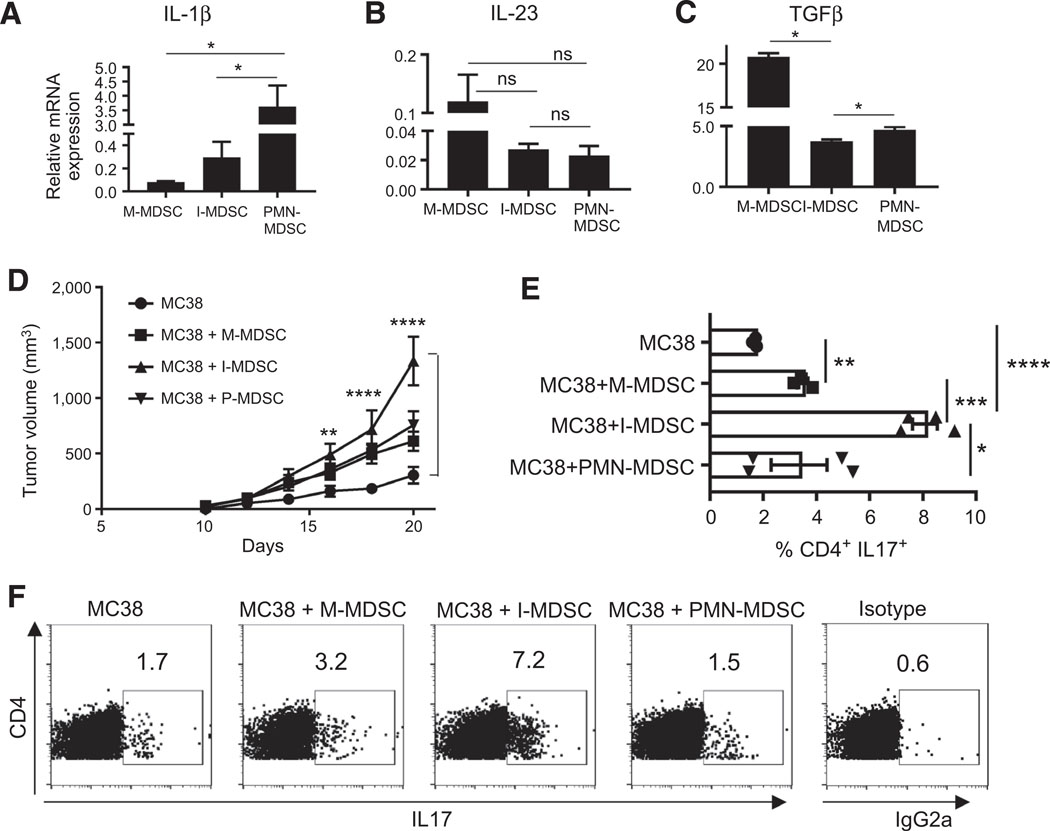
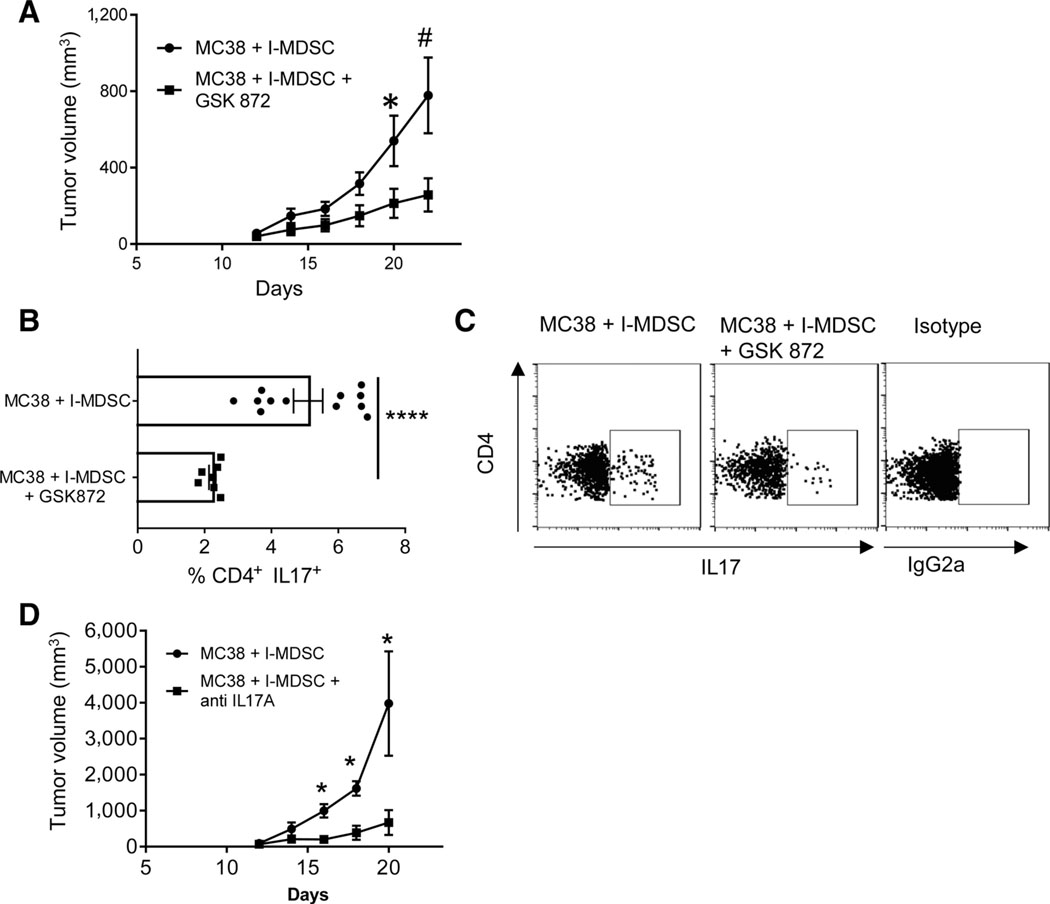
The Inhibition of RIPK3 activity reduced the intestinal tumors, I-MDSCs, and IL17 in ApcMin/+ mice. This suggests that splenic I-MDSCs could systemically promote Th17 cells by producing cytokines required for generating Th17 cells. ApcMin/+ I-MDSCs produced more IL1β than M-MDSCs, but less TGFβ than M-MDSCs and PMN-MDSCs (Fig. 5A and C). All MDSC subsets expressed IL23 (Fig. 5B). This is consistent with MDSCs from ovarian cancer, which produced IL23, IL6, or IL1β and generated Th17 cells (35). Because of different levels of cytokines produced by MDSC subsets, we evaluated whether they differ in driving the tumor growth by injecting them into MC38 tumors. The volume of tumors injected with I-MDSCs increased significantly at day 16, 18, and 20 compared with control tumors (Fig. 5D). Notably, IL17-producing CD4 T cells were significantly increased in MC38 tumors injected with I-MDSCs compared with the tumors injected with M-MDSCs or PMN-MDSCs or control tumors (Fig. 5E and F). Tumors receiving M-MDSCs had more CD4+IL17+ T cells, but were not significantly larger than control tumors. Although PMN-MDSCs expressed IL23, IL1β, and TGFβ, they did not significantly increase the tumor volume or IL17-producing T cells (Fig. 5). Fewer T cells in the tumors and spleen of MC38 tumor-bearing mice receiving I-MDSCs suggest lymphodepletion in larger tumors compared with smaller tumors in the control group (Supplementary Fig. S5A–S5D). However, there were no differences in CD44 or CD62L expression on CD4 or CD8 T cells, respectively (Supplementary Fig. S5E–S5L). To show that I-MDSCs promote tumor growth by RIPK3 signaling, MC38 tumors were injected with I-MDSCs and treated with GSK 872. I-MDSC–dependent tumor growth and frequency of CD4+IL17+ cells were significantly reduced after RIPK3 inhibition (Fig. 6A–C). Depleting IL17 after injecting MC38 tumors with I-MDSCs resulted in smaller tumors, showing that I-MDSCs promoted tumor growth by IL17(Fig. 6D). Together, these results show that RIPK3 signaling in I-MDSCs expand IL17-producing CD4 T cells in tumors to accelerate tumor growth.
Figure. 5.
I-MDSCs promote tumor growth by expanding CD4+IL17+ cells. mRNA expression of IL1β (A), IL23p19 (B; n = 10), and TGFβ (C; n = 4) in splenic ApcMin/+ MDSC subsets. D, A total of 106 M-MDSCs, I-MDSCs, and PMN-MDSCs from ApcMin/+ mice were injected into similar sized MC38 tumors (n 4). Tumor volume in these mice and WT MC38 tumor-bearing mice that were not injected with MDSCs (control group) was measured every 2 days up to day 20. E and F, Percentage (E) and representative flow cytometry plots (F) of IL17-producing CD4 T cells in TILs from MC38 tumor-bearing mice that were injected with each MDSC subset or control tumor-bearing mice. n = 4; *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001; ns, nonsignificant.
Figure 6.
RIPK3 mediates tumor growth by I-MDSCs via CD4+IL17+ cells. A, Tumor volume in MC38 tumors injected with I-MDSCs, followed by GSK 872 treatment or anti-IL17A (D) compared with control group (n = 4). B and C, Percentage (B) and representative flow cytometry plots (C) of IL17-producing CD4 T cells in TILs from MC38 tumor-bearing mice from A. *, P < 0.05; ****, P < 0.0001; #, P < 0.06.
TLR4-activated RIPK3 promotes IL23 and IL1β expression in I-MDSCs
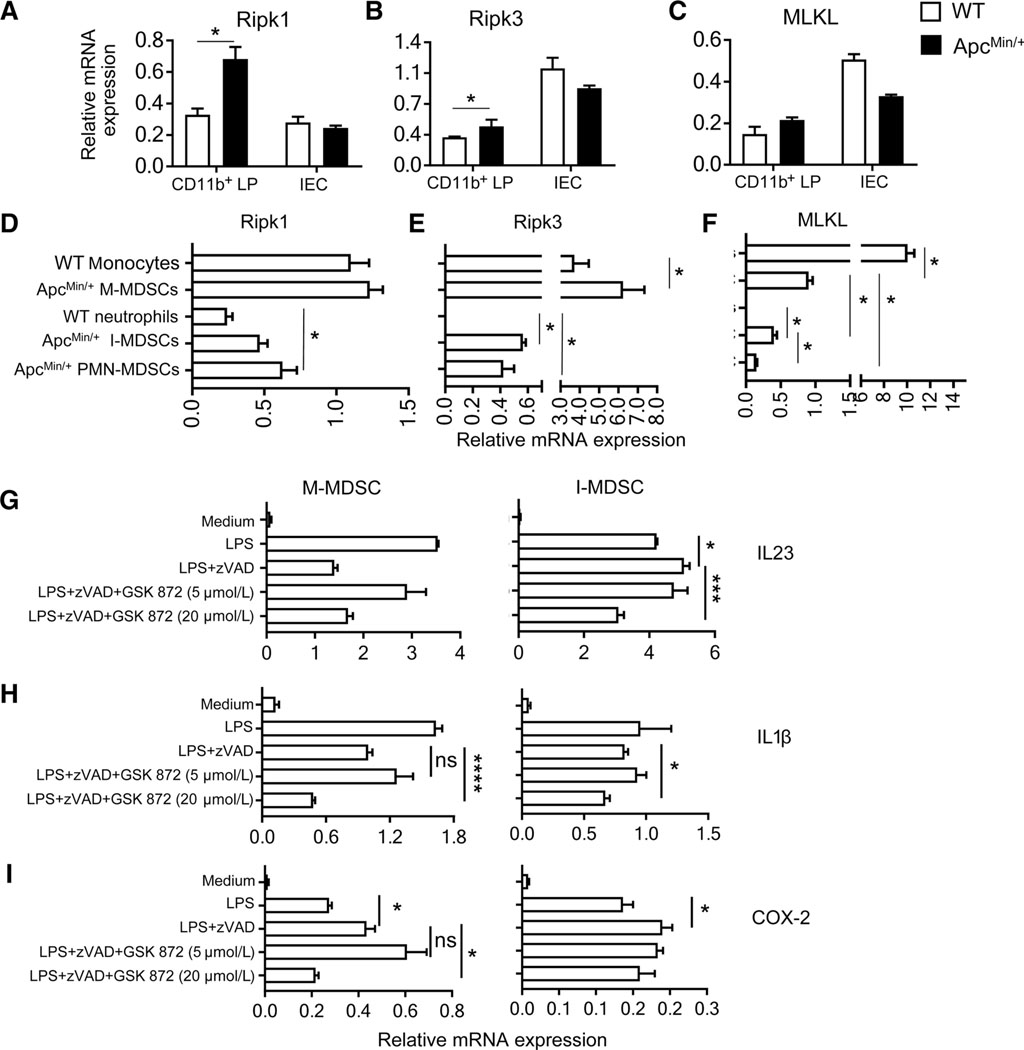
RIPK3 activity is mediated by TLR4-activated TRIF RHIM independent of Ripk1 (7, 36). This could sustain steady production of inflammatory cytokines in MDSCs. Ripk1 and RIPK3 was upregulated in myeloid cells (CD11b+) cells from ApcMin/+ LP compared with WT cells, but MLKL was unchanged (Fig. 7A–C). There was no change in Ripk1, RIPK3, or MLKL between ApcMin/+ and WT IECs. In spleen, M-MDSCs expressed more Ripk1, RIPK3, and MLKL than I-MDSCs or PMN-MDSCs (Fig. 7D–F; ref. 37). Reduced RIPK3 in I-MDSCs and in intestinal CD11b+ cells from ApcMin/+ mice is more suitable for RIPK3-induced cytokine induction because higher RIPK3 and MLKL expression results in cell death (38). The bacterial ligand LPS promotes IL23-dependent intestinal inflammation through TLR-Myd88 pathway (23). We used LPS to evaluate whether TLR4-activated RIPK3 mediated cytokine induction in MDSCs. TLR4 was similarly expressed on all MDSC subsets, but TLR2 and TLR3 were reduced on neutrophils and MDSCs (Supplementary Fig. S6A). To activate TLR4-induced RIPK3 signaling by inhibiting apoptosis, in vitro–generated M-MDSCs and I-MDSCs were treated with LPS and zVAD. These cells were treated with GSK 872 to show that LPS-induced RIPK3 is critical for cytokine induction. IL23 and COX-2 were upregulated in I-MDSCs and GSK 872 inhibited IL23 and IL1β, showing that induction of these cytokines is RIPK3-dependent (Fig. 7G–I). In M-MDSCs, COX-2 was upregulated and GSK 872 significantly inhibited both IL1β and COX-2. All the cells were viable at 2 hours after treatment, indicating that RIPK3 signaling did not cause cell death (Supplementary Fig. S6B). As M-MDSCs underwent some cell death with LPS treatment alone and GSK872 did not reverse cell death, it is unlikely to be caused by necroptosis. The specific cause of cell death in this case was not analyzed. Together, these results show that RIPK3 signaling regulates Th17-promoting cytokines in I-MDSCs.
Figure 7.
RIPK3 promotes inflammatory cytokine production in I-MDSCs. Expression of Ripk1 (A), RIPK3 (B), and MLKL (C) in WT and ApcMin/+ mice LP CD11b+ cells and IECs (n 8). Expression of Ripk1 (D), RIPK3 (E), and MLKL (F; n = 6) in WT monocytes (CD11b+Ly6C+), WT neutrophils (CD11b+Ly6G+), and MDSC subsets from ApcMin/+ mice. M-MDSCs and I-MDSCs generated in vitro from bone marrow were stimulated with LPS (10 ng/mL) with or without zVAD (50 μmol/L), or treated with GSK 872 for 2 hours and analyzed for IL23 (G), IL1β (H), and COX-2 (I) mRNA expression. n = 4; m, P < 0.05; ***, P < 0.005; ****, P < 0.0001; ns, nonsignificant.
Discussion
Intestinal microbial load is critical for the progression of intestinal tumorigenesis (39). The TLR4 ligand, LPS, is a bacterial component and activates Ripk1 and RIPK3 signaling pathway (7). Apart from regulating cell death, Ripk1 and RIPK3 mediate ERK1/2 signaling in myeloid cells, indicating their importance in cytokine production (16, 20). Demonstrating the protumor role of RIPK3 in MDSCs will identify another strategy for targeting MDSCs to improve cancer treatment. Using a RIPK3 inhibitor, GSK 872, we show that RIPK3 activity in I-MDSCs promotes tumors by expanding IL17-producing CD4 T cells.
A rare Ripk1 mutation increases RIPK3-induced inflammatory cytokines, causing IBD, which was reversed by hematopoietic stem cell transplantation (40). Moreover, RIPK3 deletion reduces tumor progression in mice with Hace1 deficiency (15). Therefore, there is a strong premise for RIPK3-induced inflammation in promoting the intestinal tumorigenesis; however, there is no evidence supporting this mechanism. In our study, IECs including tumor cells in ApcMin/+ mice expressed more RIPK3 than myeloid cells, suggesting more cell death. However, tumors continue to grow and increase in numbers. Ectopic RIPK3 expression in a lung cancer cell line induced cell death to reduce tumor growth in a xenograft model (41). In contrast, RIPK3 deletion in breast and colon cancer cell lines reduced tumor growth in xenograft models (30). These studies suggest that the role of RIPK3 in tumor cells is unclear. RIPK3-mediated cell death also activates innate immune response by releasing danger-associated molecular patterns (DAMP; ref. 42). However, we assessed whether the sustained inflammation promoted by RIPK3 signaling in MDSCs propagates tumor-promoting T cells. Low RIPK3 expression is more conducive for signaling because high RIPK3 expression generally results in necroptotic cell death (38). The Intestinal myeloid cells and splenic I-MDSCs from ApcMin/+ mice expressed less RIPK3, suggesting that RIPK3 could induce cytokine induction in these cells. Although I-MDSCs are not well defined in LP, they could be present at very low levels, and could be included in total myeloid cells from the small intestine of ApcMin/+ mice (4). By studying how splenic I-MDSCs regulate Th17 cells in MC38 tumors, we show that they could function systemically and possibly migrate to tumors to function locally by this mechanism.
Homozygous RIPK3 kinase dead mutants (D161N) undergo embryonic death due to excessive apoptosis (43). However, heterozygotes are viable, suggesting that inhibiting RIPK3 with a low concentration of GSK 872 could be less toxic. At high concentrations, GSK 872 induces apoptosis in cells, but the sensitivity to apoptosis differed among cell types (21). For example, BMDMs (macrophages) treated with 10 μmol/L of GSK 872 retained at least 75% viability up to 18 hours. As BM-MDSCs are different from macrophages, 20 μmol/L of GSK 872 was required to inhibit RIPK3 activity, but viability was maintained at this concentration. In ApcMin/+ mice, 0.25 mg/Kg of GSK 872 every 3 days inhibited intestinal tumorigenesis. We did not use a higher concentration of GSK 872 to avoid inflammation caused by excessive apoptosis. As tumor cells undergo apoptosis at concentrations greater than 10 μmol/L, GSK 872 could promote some tumor cell apoptosis in our study. The reduction in I-MDSCs directly correlates with reduction in intestinal tumors and is likely not due to apoptosis induced by GSK 872. This is supported by no change in the frequency of lymphocytes and myeloid cells. Temporal inhibition of RIPK3 could be insufficient for a dramatic change in MDSC frequency as opposed to a genetic deletion of RIPK3. In addition, RIPK3 inhibition could also modify the immunosuppressive function of MDSCs because GSK 872 inhibited both IL1β and COX-2 in ApcMin/+ polyps and M-MDSCs; these cytokines mediate suppressive function of MDSCs (44). We will evaluate this aspect of the RIPK3 function to understand the total effect of inhibiting RIPK3 in MDSCs. This suggests that MDSC subsets could regulate more than one arm of protumor T-cell response by promoting pathogenic Th17 cells and suppressing the antitumor T cells.
MDSCs from ovarian cancer promoted IL17-producing T cells (35). Moreover, in a colon tumor model of CPC-APC mice, MDSCs (CD11b+Gr-1+) and not macrophages produced IL23 to drive tumors by IL17 (23). However, the role of RIPK3 in MDSC-mediated Th17 generation is unknown. IL23 and IL1β have tumor-promoting functions in the intestinal cancer (4, 45). This is consistent with upregulation of IL23 and IL1β in ApcMin/+ polyps and MDSC subsets including I-MDSCs. GSK 872 reduced the intestinal tumors in ApcMin/+ mice and Th17-differentiating cytokines including IL6, IL23, and IL1β, suggesting that RIPK3 could be regulating Th17 cells and intestinal tumor growth in ApcMin/+ mice to promote tumors. Using a transplantable colorectal cancer tumor model, we show that ApcMin/+ I-MDSCs were more efficient in promoting tumor growth by expanding IL17-producing CD4 T cells by RIPK3 signaling.
IL23, IL1β, and IL6 generates pathogenic Th17 cells causing EAE disease progression, whereas TGFβ, IL1β, and IL6 generate conventional Th17 cells (46). T cells treated with TGFβ dramatically reduced IL17 expression, which is consistent with the role of TGFβ in suppressing IL17 production by reducing IL23R expression (46). This would explain why ApcMin/+M-MDSCs and PMN-MDSCs producing more TGFβ are less efficient in promoting Th17 cells. These Th17 cells could also be conventional Th17 cells that produce IL10 to suppress inflammation (46) and is likely the reason for reduced tumor growth. In our study, I-MDSCs produced less TGFβ than other subsets and promoted more IL17 production, resulting in increased tumor growth. This supports the possibility that low TGFβ production by I-MDSCs likely promoted the pathogenic Th17 cells. A similar mechanism could be occurring in human colorectal cancer by the generation of pathogenic Th17 cells by a MDSC subset. In addition to M-MDSCs and PMN-MDSCs, e-MDSCs lacking both CD14 and CD15 have been identified in humans (47); however, their role in Th17 induction is undefined. We will delineate the role of pathogenic versus conventional Th17 cells in intestinal tumorigenesis in future studies.
MDSCs are heterogeneous with relatively few phenotypic markers (monocytic marker Ly6C and granulocytic marker Ly6G) to adequately classify them into different subsets (5). We identified four subpopulations within each ApcMin/+ MDSC subset based on the differential expression of Ly6C and Ly6G, which is consistent with the overall heterogeneity of MDSCs. Three M-MDSC subpopulations with high Ly6C expression indicate their monocytic origin. Notably, subpopulation P2 within M-MDSC could be related to I-MDSCs due to its location on the MST and low Ly6C/Ly6G ratio. I-MDSCs with an overall reduction of Ly6C and Ly6G are distinct from M-MDSCs and PMN-MDSCs. However, they exhibit some similarity to M-MDSCs due to the similar expression of macrophage markers (F4/80 and CSF-1R) and to PMN-MDSCs due to reduction of the antigen-presenting molecules. GM-CSF promotes I-MDSC accumulation, but GM-CSF and TCM were essential for F4/80 and CSF-1R expression on I-MDSCs. Accumulation of I-MDSCs was dependent on GM-CSF, due to which, ApcMin/+ mice with high circulating levels of GM-CSF (34) have more I-MDSCs. Similarly, other studies show the expansion of I-MDSC–like cells in mice with colon tumor cells expressing GM-CSF or breast cancer cells that produce high levels of GM-CSF (32, 33). The final fate of I-MDSCs is unknown. They could convert into macrophages, DCs, or neutrophils similar to MDSCs in different tumor models (44). However, more evidence is required to delineate this process.
Collectively, we have shown that RIPK3 regulates inflammatory signaling in I-MDSCs to promote IL17 production in T cells and that it has a tumor-promoting function in intestinal cancer. Overall, modulation of colorectal cancer–promoting cytokines by GSK 872 demonstrates that RIPK3 is targetable in intestinal cancer.
Supplementary Material
Significance:
The specific role of RIPK3 in intestinal tumors and MDSC function sheds light on a key inflammatory mechanism driving tumorigenesis and allows for possible therapeutic intervention.
Acknowledgments
This work was supported by NCI diversity reentry supplement for A. Jayakumar to parent R01CA168670–01A1 awarded to A.L.M. Bothwell. We thank Dr. Nikhil Joshi for insightful discussions and expert advice.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Limagne E, Euvrard R, Thibaudin M, Rebe C, Derangere V, Chevriaux A, et al. Accumulation of MDSC and Th17 cells in patients with metastatic colorectal cancer predicts the efficacy of a FOLFOX-bevacizumab drug treatment regimen. Cancer Res 2016;76:5241–52. [DOI] [PubMed] [Google Scholar]
- 2.Garton AJ, Seibel S, Lopresti-Morrow L, Crew L, Janson N, Mandiyan S, et al. Anti-KIT monoclonal antibody treatment enhances the antitumor activity of immune checkpoint inhibitors by reversing tumor-induced immuno-suppression. Mol Cancer Ther 2017;16:671–80. [DOI] [PubMed] [Google Scholar]
- 3.Chun E, Lavoie S, Michaud M, Gallini CA, Kim J, Soucy G, et al. CCL2 Promotes colorectal carcinogenesis by enhancing polymorphonuclear myeloid-derived suppressor cell population and function. Cell Rep 2015;12:244–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayakumar A, Bothwell ALM. Stat6 promotes intestinal tumorigenesis in a mouse model of adenomatous polyposis by expansion of MDSCs and inhibition of cytotoxic CD8 Response. Neoplasia 2017;19:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016;7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature 2015;517:311–20. [DOI] [PubMed] [Google Scholar]
- 7.Wegner KW, Saleh D, Degterev A. Complex pathologic roles of RIPK1 and RIPK3: moving beyond necroptosis. Trends Pharmacol Sci 2017;38: 202–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dannappel M, Vlantis K, Kumari S, Polykratis A, Kim C, Wachsmuth L, et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature 2014;513:90–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roderick JE, Hermance N, Zelic M, Simmons MJ, Polykratis A, Pasparakis M, et al. Hematopoietic RIPK1 deficiency results in bone marrow failure caused by apoptosis and RIPK3-mediated necroptosis. Proc Natl Acad Sci U S A 2014;111:14436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton K, Wickliffe KE, Maltzman A, Dugger DL, Strasser A, Pham VC, et al. RIPK1 inhibits ZBP1-driven necroptosis during development. Nature 2016;540:129–33. [DOI] [PubMed] [Google Scholar]
- 11.Moriwaki K, Chan FK. The inflammatory signal adaptor RIPK3: functions beyond necroptosis. Int Rev Cell Mol Biol 2017;328:253–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newton K, Dugger DL, Maltzman A, Greve JM, Hedehus M, Martin-McNulty B, et al. RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ 2016;23:1565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierdomenico M, Negroni A, Stronati L, Vitali R, Prete E, Bertin J, et al. Necroptosis is active in children with inflammatory bowel disease and contributes to heighten intestinal inflammation. Am J Gastroenterol 2014; 109:279–87. [DOI] [PubMed] [Google Scholar]
- 14.Moriwaki K, Balaji S, McQuade T, Malhotra N, Kang J, Chan FK. The necroptosis adaptor RIPK3 promotes injury-induced cytokine expression and tissue repair. Immunity 2014;41:567–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tortola L, Nitsch R, Bertrand MJM, Kogler M, Redouane Y, Kozieradzki I, et al. The tumor suppressor hace1 is a critical regulator of TNFR1-mediated cell fate. Cell Rep 2016;15:1481–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriwaki K, Balaji S, Bertin J, Gough PJ, Chan FK. Distinct kinase-independent role of RIPK3 in CD11c+ mononuclear phagocytes in cytokine-induced tissue repair. Cell Rep 2017;18:2441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu XN, Yang ZH, Wang XK, Zhang Y, Wan H, Song Y, et al. Distinct roles of RIP1-RIP3 hetero- and RIP3-RIP3 homo-interaction in mediating necroptosis. Cell Death Differ 2014;21:1709–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porta C, Ippolito A, Consonni FM, Carraro L, Celesti G, Correale C, et al. Protumor steering of cancer inflammation by p50 NF-kappaB enhances colorectal cancer progression. Cancer Immunol Res 2018;6:578–93. [DOI] [PubMed] [Google Scholar]
- 19.Bozec D, Iuga AC, Roda G, Dahan S, Yeretssian G. Critical function of the necroptosis adaptor RIPK3 in protecting from intestinal tumorigenesis. Oncotarget 2016;7:46384–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Najjar M, Saleh D, Zelic M, Nogusa S, Shah S, Tai A, et al. RIPK1 and RIPK3 kinases promote cell-death-independent inflammation by toll-like receptor 4. Immunity 2016;45:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandal P, Berger SB, Pillay S, Moriwaki K, Huang C, Guo H, et al. RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol Cell 2014;56:481–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, Bothwell AL. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci U S A 2010;107:5540–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012;491:254–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luongo C, Moser AR, Gledhill S, Dove WF. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res 1994;54:5947–52. [PubMed] [Google Scholar]
- 25.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 2010;38:D792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 2012;13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 2015;87:636–45. [DOI] [PubMed] [Google Scholar]
- 28.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol 2008;181: 5791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thevenot PT, Sierra RA, Raber PL, Al-Khami AA, Trillo-Tinoco J, Zarreii P, et al. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity 2014;41: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Zhou M, Mei L, Ruan J, Hu Q, Peng J, et al. Key roles of necroptotic factors in promoting tumor growth. Oncotarget 2016;7:22219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coletta PL, Muller AM, Jones EA, Muhl B, Holwell S, Clarke D, et al. Lymphodepletion in the ApcMin/+ mouse model of intestinal tumorigenesis. Blood 2004;103:1050–8. [DOI] [PubMed] [Google Scholar]
- 32.Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity 2010;32:790–802. [DOI] [PubMed] [Google Scholar]
- 33.Dolcetti L, Peranzoni E, Ugel S, Marigo I, Fernandez Gomez A, Mesa C, et al. Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF. Eur J Immunol 2010;40: 22–35. [DOI] [PubMed] [Google Scholar]
- 34.Mohammed A, Janakiram NB, Li Q, Choi CI, Zhang Y, Steele VE, et al. Chemoprevention of colon and small intestinal tumorigenesis in APC (Min/+) mice by licofelone, a novel dual 5-LOX/COX inhibitor: potential implications for human colon cancer prevention. Cancer Prev Res 2011;4: 2015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obermajer N, Wong JL, Edwards RP, Chen K, Scott M, Khader S, et al. Induction and stability of human Th17 cells require endogenous NOS2 and cGMP-dependent NO signaling. J Exp Med 2013;210:1433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He S, Liang Y, Shao F, Wang X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc Natl Acad Sci U S A 2011;108:20054–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell 2014;54:133–46. [DOI] [PubMed] [Google Scholar]
- 38.He S, Wang L, Miao L, Wang T, Du F, Zhao L, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009;137:1100–11. [DOI] [PubMed] [Google Scholar]
- 39.Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, et al. Microbes and cancer. Annu Rev Immunol 2017;35:199–228. [DOI] [PubMed] [Google Scholar]
- 40.Cuchet-Lourenco D, Eletto D, Wu C, Plagnol V, Papapietro O, Curtis J, et al. Biallelic RIPK1 mutations in humans cause severe immunodeficiency, arthritis, and intestinal inflammation. Science 2018;361:810–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C, Li J, Yu L, Zhang Z, Xu F, Jiang L, et al. Regulation of RIP3 by the transcription factor Sp1 and the epigenetic regulator UHRF1 modulates cancer cell necroptosis. Cell Death Dis 2017;8:e3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity 2013;38:209–23. [DOI] [PubMed] [Google Scholar]
- 43.Newton K, Dugger DL, Wickliffe KE, Kapoor N, de Almagro MC, Vucic D, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 2014;343:1357–60. [DOI] [PubMed] [Google Scholar]
- 44.Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest 2015;125: 3356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol 2013;35:229–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 2010;467:967–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol 2018;19:108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.