Abstract
A novel ascomycetous genus, Elongaticollum, occurring on leaf litter of Hedychium coronarium (Zingiberaceae) in Taiwan, is described and illustrated. Elongaticollum is characterized by dark brown to black, superficial, obpyriform, pycnidial conidiomata with a distinct elongate neck, and oval to oblong, hyaline, aseptate conidia. Phylogenetic analyses (maximum likelihood, maximum parsimony and Bayesian) of combined ITS, LSU, SSU and tef1-α sequence data revealed Elongaticollum as a distinct genus within the family Phaeosphaeriaceae with high statistical support. In addition, Ophiosphaerella taiwanensis and Phaeosphaeriopsis beaucarneae are described as new species from dead leaves of Agave tequilana and Beaucarnea recurvata (Asparagaceae), respectively. Neosetophoma poaceicola is reported as a new host record from dead leaves of Musa acuminata (Musaceae). Newly described taxa are compared with other similar species and comprehensive descriptions and micrographs are provided.
Keywords: Asparagaceae, Dothideomycetes; leaf litter; new taxa; Zingiberaceae
Introduction
Plant litter is considered as one of the main contributors to net above-ground primary productivity of terrestrial ecosystems (Swift et al. 1979; Berg and McClaugherty 2008; Krishna and Mohan 2017). Since plant litter is returned back to the soil, it represents a major source of organic carbon in forest soils (Berg 2003). Plant litter can be defined as a collection of fallen leaves, twigs, seeds and other woody debris that accumulate on the ground as a natural part of the forest ecosystem (Johnson and Catley 2002; Berg and McClaugherty 2008). In particular, leaf litter is the main source of organic matter and nutrients of the soil, compared to other litter types (Robertson and Paul 1999; Berg and McClaugherty 2008; Krishna and Mohan 2017). Leaf litter decomposition is a key process contributing to biogeochemical cycles in any forest ecosystem. Microorganisms are the primary agents in this process (Purahong et al. 2016; Mlambo et al. 2019). Fungi are considered as the “key players” in leaf litter decomposition, because of their ability to produce a wide range of extracellular enzymes (Pointing et al. 2005; Berg and McClaugherty 2008; Bani et al. 2018). Many researchers have been carrying out studies of fungal species inhabiting leaf litter and have described numerous new species in Dothideomycetes (Hyde et al. 2019; Phookamsak et al. 2019; Tennakoon et al. 2019).
The family Phaeosphaeriaceae is considered to be one of the most species-rich families in Dothideomycetes and includes species that inhabit a wide range of ecosystems (i. e., marine, terrestrial, and mangroves) (Phookamsak et al. 2014, 2017; Bakhshi et al. 2019; Jones et al. 2019; Luo et al. 2019; Tennakoon et al. 2019). Phaeosphaeriaceae was established by Barr (1979), who designated Phaeosphaeria I. Miyake as the generic type of the family. Phaeosphaeriaceae species have immersed to superficial, globose to subglobose ascomata, short papilla, bitunicate asci and hyaline to pigmented, fusiform to ellipsoidal, filiform, or muriform ascospores (Bakhshi et al. 2019; Chaiwan et al. 2019; Maharachchikumbura et al. 2019; Yang et al. 2019). Members of Phaeosphaeriaceae are cosmopolitan, since they exhibit diverse lifestyles as saprobes, endophytes and pathogens of economically important plants (Barr 1992; Phookamsak et al. 2014, 2017; Yang et al. 2016; Hyde et al. 2020; Mapook et al. 2020). Apart from being cosmopolitan in nature, it appears that this family is phylogenetically highly diverse. Thus, recent studies have revealed a large number of new genera in this family. For instance, in the space of two years, eleven genera have been introduced, viz. Bhagirathimyces S.M. Singh & S.K. Singh (Hyde et al. 2020), Hydeomyces Maharachchikumbura et al. (Maharachchikumbura et al. 2019), Hydeopsis J.F. Zhang et al. (Zhang et al. 2019), Neostagonosporella C.L. Yang, et al. (Yang et al. 2019), Parastagonosporella M. Bakhshi, Arzanlou & Crous (Bakhshi et al. 2019), Pseudoophiosphaerella J.F. Zhang et al. (Zhang et al. 2019), Murichromolaenicola Mapook & K.D. Hyde (Mapook et al. 2020), Neoophiobolus Mapook & K.D. Hyde (Mapook et al. 2020), Paraleptospora Mapook & K.D. Hyde (Mapook et al. 2020), Pseudostaurosphaeria Mapook & K.D. Hyde (Mapook et al. 2020) and Vittaliana Devadatha et al. (Devadatha et al. 2019). Currently, more than 70 genera are accommodated in this family (Wanasinghe et al. 2018; Bakhshi et al. 2019; Maharachchikumbura et al. 2019; Phookamsak et al. 2019; Hongsanan et al. 2020; Hyde et al. 2020).
We are investigating the diversity of microfungi on leaf litter in the tropics with the aim of clarifying their taxonomy based on morphology coupled with multi-gene phylogeny. As a part of this study, we have collected and isolated four taxa from Taiwan, which belong to the family Phaeosphaeriaceae. We present herein comprehensive morphological descriptions and an in-depth phylogenetic investigation of the newly introduced species.
Materials and methods
Sample collection, morphological studies and isolation
Decaying leaf litter samples of Agave tequilana F.A.C. Weber (Asparagaceae), Beaucarnea recurvata Lem. (Asparagaceae), Hedychium coronarium J.Koenig (Zingiberaceae), and Musa acuminata Colla (Musaceae) were collected from Dahu Forest Area in Chiayi, Taiwan and taken to the laboratory in Zip lock plastic bags. Specimens were examined with a LEICA EZ4 stereomicroscope. Micro-morphological characters were determined using AXIOSKOP 2 PLUS compound microscope and images were captured with a Zeiss AXIOCAM 506 COLOR digital camera. Observations and photomicrographs were made from materials mounted in water. Permanent slides were preserved in lactoglycerol, sealed by applying nail-polish around the margins of cover slip. All measurements were made with ZEN2 (blue edition) and images used for figures were processed with Adobe Photoshop CS3 Extended version 10.0 software (Adobe Systems, USA).
Single ascospore and conidial isolation was carried out following the method described in Phookamsak et al. (2014). The single germinated spore was picked up and transferred to potato dextrose agar (PDA) and incubated at 25 °C in natural light. Subsequent sub-culturing was done carefully to obtain pure culture and ensure absence of contaminants. Culture characteristics were observed after three weeks. Colonies were photographed and colonial characters were noted and described. Type specimens of new taxa were deposited at the herbarium of Mae Fah Luang University (MFLU) and National Chiayi University Herbarium (NCYU). Living cultures were deposited in Mae Fah Luang University Culture Collection (MFLUCC) and National Chiayi University Culture Collection (NCYUCC). Faces of Fungi and Index Fungorum numbers were provided as in Jayasiri et al. (2015) and Index Fungorum (2020).
DNA extraction and PCR amplification
Total genomic DNA was extracted from scraped fresh fungal mycelium using the DNA extraction kit E.Z.N.A Fungal DNA Mini Kit (D3390-02, Omega Bio-Tek) following the manufacturer’s protocol. The DNA product was kept at 4 °C for DNA amplification and maintained at -20 °C for long term storage. DNA was amplified by polymerase chain reaction (PCR) for four genes, the large subunit (28S, LSU), small subunit (18S, SSU), internal transcribed spacers including the 5.8s rDNA (ITS1-5.8S-ITS2) and translation elongation factor 1 alpha (tef1-α). The partial LSU gene was amplified by using the primer combination LR0R and LR5 (Vilgalys and Hester 1990; Rehner and Samuels 1994); partial SSU was amplified with NS1 and NS4 (White et al. 1990), nuclear ITS was amplified with primers ITS5 and ITS4 (White et al. 1990), and tef1-α gene was amplified using the primers EF1-983F and EF1-2218R (Rehner et al. 2001). Amplification reactions were performed in 25 µl of total reaction that contained 9.5 µl of sterilized water, 12.5 µl of 2×Power Taq PCR MasterMix (Tri-I Biotech, Taipei, Taiwan), 1 μl of each forward and reverse primers and 1 μl of DNA template. The PCR thermal cycle program of ITS, LSU, SSU and tef1-α gene was processed initially at 94 °C for 3 minutes, followed by 35 cycles of denaturation at 94 °C for 30 seconds, annealing at 55 °C for 50 seconds, elongation at 72 °C for 1 minute and a final extension at 72 °C for 10 minutes and a holding temperature of 4 °C. The PCR products were analyzed by 1.5% agarose gels containing the Safeview DNA stain (GeneMark, Taipei, Taiwan) to confirm their expected molecular weight. PCR products were purified and sequenced with primers mentioned above by Tri-I Biotech, Taipei, Taiwan. Nucleotide sequences were deposited in GenBank (Table 1).
Table 1.
GenBank and culture collection accession numbers of species included in the present phylogenetic study. Newly generated sequences are shown in bold.
| Species | Strain/Voucher no. | GenBank accession no. | |||
|---|---|---|---|---|---|
| LSU | SSU | ITS | tef1–α | ||
| Acericola italica | MFLUCC 13-0609 | MF167429 | MF167430 | MF167428 | – |
| Allophaeosphaeria muriformia | MFLUCC 13-0277 | KX910089 | KX950400 | KX926415 | – |
| Alloneottiosporina thailandica | MFLUCC 15-0576 | – | – | – | – |
| Amarenographium ammophilicola | MFLU 17-2571 | MN017847 | MN017913 | MN047087 | MN077065 |
| Amarenomyces dactylidis | KUMCC 18-0154 | MK356345 | MK356359 | MK356371 | – |
| Arezzomyces cytisi | MFLUCC 15-0649 | KT306950 | KT306954 | KT306947 | – |
| Banksiophoma australiensis | CBS 142163 | KY979794 | – | KY979739 | KY979889 |
| Bhagirathimyc es himalayensis | AMH 10127 | MK836020 | MN121697 | MK836021 | – |
| Bhatiellae rosae | MFLUCC 17-0664 | MG828989 | MG829101 | MG828873 | – |
| Brunneomurispora lonicerae | KUMCC 18-0157 | MK356346 | MK356360 | MK356373 | MK359065 |
| Camarosporioides phragmitis | MFLUCC 13-0365 | KX572345 | KX572350 | KX572340 | KX572354 |
| Chaetosphaeronema achilleae | MFLUCC 16-0476 | KX765266 | – | KX765265 | – |
| C. hispidulum | CBS 216.75 | KF251652 | EU754045 | KF251148 | KF253108 |
| Dactylidina shoemaker | MFLUCC 14-0963 | MG829003 | MG829114 | MG828887 | MG829200 |
| Dematiopleospora cirsii | MFLUCC 13-0615 | KX274250 | – | KX274243 | KX284708 |
| D. mariae | MFLUCC 15-0612 | KJ749653 | KJ749652 | KX274244 | KJ749655 |
| Didymocyrtis xanthomendozae | CBS 129666 | – | – | KP170651 | KP170677 |
| Diederichomyces ficuzzae | CBS 128019 | JQ238616 | – | KP170647 | KP170673 |
| Dlhawksworthia clematidicola | MFLUCC 17-0693 | MG829038 | MG829144 | MG828929 | – |
| D. lonicera | MFLUCC 14-0955 | MG829012 | MG829121 | MG828902 | MG829203 |
| Edenia gomezpompae | JLCC 34533 | – | – | KC193601 | – |
| LVPEI 3225 | – | – | KU578033 | – | |
| Elongaticollum hedychii | MFLUCC 18-1638 | MT321810 | MT321803 | MT321796 | MT328753 |
| E. hedychii | MFLUCC 17-2653 | MT321811 | MT321804 | MT321797 | MT328754 |
| NCYUCC 19-0286 | MT321812 | MT321805 | MT321798 | MT328755 | |
| Embarria clematidis | MFLUCC 14-0652 | KT306953 | KT306956 | KT306949 | – |
| MFLUCC 14-0976 | MG828987 | MG829099 | MG828871 | MG829194 | |
| Equiseticola fusispora | MFLUCC 14-0522 | KU987669 | KU987670 | KU987668 | MG520895 |
| Galiicola baoshanensis | HKAS 102234 | MK356348 | MK356362 | MK356374 | MK359066 |
| G. pseudophaeosphaeria | MFLU 14-0524 | – | – | – | MG520896 |
| Hydeomyces desertipleosporoides | SQUCC 15259 | MK290839 | MK290843 | MK290841 | MK290848 |
| SQUCC 15260 | MK290840 | MK290844 | MK290842 | MK290849 | |
| Hydeopsis verrucispora | SD 2016-5 | MK522498 | MK522504 | MK522508 | MK523388 |
| Italica achilleae | MFLUCC 14-0955 | MG829012 | MG829121 | MG828902 | MG829203 |
| I. luzulae | MFLUCC 14-0932 | KT306951 | – | – | – |
| Jeremyomyces labinae | CBS 144617 | MK442529 | – | MK442589 | MK442695 |
| Juncaceicola italica | MFLUCC 13-0750 | – | – | KX500110 | MG520897 |
| J. luzulae | MFLUCC 13-0780 | KX449530 | KX449531 | KX449529 | – |
| Kwanghwaensis miscanthi | FU31017 | MK503823 | MK503829 | MK503817 | MT009126 |
| Leptosphaeria doliolum | CBS 505.75 | GU301827 | GU296159 | JF740205 | GU349069 |
| Leptospora rubella | CPC 11006 | DQ195792 | DQ195803 | DQ195780 | – |
| L. thailandica | MFLUCC 16-0385 | KX655549 | KX655554 | KX655559 | KX655564 |
| Longispora clematidis | MFLU 15–1277 | ||||
| Loratospora aestuarii | CBS 117592 | – | – | MH863024 | – |
| Mauginiella scaettae | CBS 239.58 | MH869303 | – | MH857770 | – |
| Melnikia anthoxanthii | MFLUCC 14-1011 | KU848204 | KU848205 | – | – |
| Murichromolaenicola chiangraiensis | MFLUCC 17-1488 | MN994559 | MN994605 | MN994582 | MN998163 |
| M. chromolaenae | MFLUCC 17-1489 | MN994560 | MN994606 | MN994583 | MN998164 |
| Muriphaeosphaeria galatellae | MFLUCC 14-0614 | KT438329 | KT438331 | KT438333 | MG520900 |
| MFLUCC 15-0769 | KT438330 | KT438332 | – | – | |
| Neoophiobolus chromolaenae | MFLUCC 17-1467 | MN994562 | MN994606 | MN994583 | MN998164 |
| N. chromolaenae | MFLUCC 17-1449 | MN994561 | MN994607 | MN994584 | MN998165 |
| Neosetophoma sp. | MFLUCC 17-0844 | MG829035 | MG829141 | MG828926 | MG829219 |
| N. aseptata | CBS 145363 | MK540024 | – | MK539953 | – |
| N. camporesii | MFLUCC 15-0682 | KU302778 | – | KU302779 | – |
| N. clematidis | MFLUCC 13-0734 | KP684153 | KP684154 | KP744450 | – |
| N. garethjonesii | MFLUCC 14-0528 | – | KY501126 | – | KY514402 |
| N. guiyangensis | GZ13 | MH018132 | MH018136 | MH018134 | MH051889 |
| N. italica | MFLU 14-0809 | KP711361 | KP711366 | KP711356 | – |
| N. lonicerae | KUMCC 18-0155 | MK356349 | MK356363 | MK356375 | MK359067 |
| N. lunariae | CPC 26671 | KX306789 | – | KX306763 | – |
| N. miscanthi | MFLU 18-2675 | MK503826 | MK503832 | MK503820 | – |
| N. phragmitis | CBS 145364 | MK540025 | – | MK539954 | MK540148 |
| N. poaceicola | MFLUCC 16-0886 | KY550382 | KY550383 | KY568986 | – |
| MFLUCC 18-1632 | MT321809 | MT321802 | MT321795 | – | |
| N. rosae | MFLUCC 17-0844 | MG829035 | MG829141 | MG828926 | MG829219 |
| N. rosaena | MFLUCC 17-0768 | MG829037 | MG829143 | MG828928 | – |
| N. rosarum | MFLU 17-0308 | MG829036 | MG829142 | MG828927 | – |
| N. salicis | MFLU 17-0118 | MK608026 | – | MK608025 | – |
| N. samarorum | CBS 138.96 | KF251664 | GQ387517 | MH862569 | KF253119 |
| N. sambuci | CBS 145365 | MK540026 | – | MK539955 | MK540149 |
| N. shoemakeri | MFLU 16-1606 | MG602199 | MG602201 | MG602203 | MG844352 |
| MFLUCC 17-0780 | MG844348 | MG844350 | MG844346 | MG844352 | |
| N. tienshanensis | MFLUCC 17-0844 | MG829035 | MG829141 | MG828926 | MG829219 |
| N. xingrensis | GZAAS18 0100 | MH018133 | – | MH018135 | – |
| Neosphaerellopsis thailandica | CPC 21659 | KP170721 | – | KP170652 | KP170678 |
| Neostagonospora caricis | CBS 135092 | KF251667 | – | KF251163 | – |
| N. phragmitis | MFLUCC 16-0493 | KX910090 | KX950401 | KX926416 | MG520902 |
| Neostagonosporella sichuanensis | MFLUCC 18-1228 | – | – | – | MK313854 |
| MFLUCC 18-1231 | – | – | – | MK313851 | |
| Neosulcatispora agaves | CPC 26407 | KT950867 | – | KT950853 | – |
| Nodulosphaeria multiseptata | MFLUCC 15-0078 | KY496728 | – | KY496748 | – |
| N. scabiosae | MFLUCC 14-1111 | KU708846 | KU708842 | KU708850 | KU708854 |
| Ophiobolopsis italica | MFLUCC 17-1791 | MG520959 | MG520977 | MG520939 | MG520903 |
| Ophiobolus disseminans | MFLUCC 17-1787 | MG520961 | MG520980 | MG520941 | MG520906 |
| O. rossicus | MFLU 17-1639 | MG520964 | MG520983 | MG520944 | MG520909 |
| Ophiosimulans tanaceti | MFLUCC 14-0525 | KU738891 | KU738892 | KU738890 | MG520910 |
| Ophiosphaerella agrostidis | MFLUCC 11-0152 | KM434281 | KM434290 | KM434271 | KM434299 |
| MFLUCC 12-0007 | KM434282 | KM434291 | KM434272 | KM434300 | |
| MFLUCC 16-0895 | MF197563 | MF351604 | MF351996 | – | |
| IGM35 | MF197563 | MF351604 | – | – | |
| MFLUCC 11-0152 | KM434281 | KM434290 | KM434271 | KM434299 | |
| O. aquatica | MFLUCC 14-0033 | KX767089 | KX767090 | KX767088 | MG520911 |
| MFLUCC 14-0033 | KX767089 | KX767090 | KX767088 | MG520911 | |
| O. herpotricha | k28 | – | – | KP690992 | KP691016 |
| KS29 | – | – | KP690986 | KP691015 | |
| O. korrae | ATCC 56289 | – | – | KC848509 | KC848515 |
| O. narmari | ATCC 64688 | – | – | KC848510 | KC848516 |
| ATCC 201719 | – | – | KC848508 | KC848514 | |
| O. taiwanensis | MFLU 18-2534 | MT321815 | MT321808 | MT321801 | MT328758 |
| O. taiwanica | NTUCC 17-024 | MN082419 | – | MN082417 | – |
| NTUCC 17-025 | MN082420 | – | MN082418 | – | |
| Paraleptosphaeria dryadis | CBS 643.86 | GU301828 | KC584632 | JF740213 | GU349009 |
| Paraleptospora chromolaenae | MFLUCC 17-1481 | MN994563 | MN994609 | MN994587 | MN998167 |
| P. chromolaenicola | MFLUCC 17-1450 | MN994564 | MN994610 | MN994588 | MN998168 |
| Paraophiobolus arundinis | MFLUCC 17-1789 | MG520965 | MG520984 | MG520945 | MG520912 |
| P. plantaginis | MFLUCC 17-0245 | KY815010 | KY815012 | KY797641 | MG520913 |
| Paraloratospora camporesii | MFLU 18-0915 | MN756637 | MN756635 | MN756639 | – |
| Paraphoma chrysanthemicola | CBS 522.66 | KF251670 | GQ387521 | KF251166 | KF253124 |
| P. radicina | CBS 111.79 | KF251676 | EU754092 | KF251172 | KF253130 |
| Parastagonospora dactylidis | MFLUCC 13-0375 | KU058722 | – | KU058712 | – |
| Parastagonosporella fallopiae | CBS 135981 | MH460545 | – | MH460543 | MH460549 |
| P. fallopiae | CCTU 1151-1 | MH460546 | – | MH460544 | MH460550 |
| Phaeopoacea muriformis | MFLUCC 17-0372 | MF611638 | MF611639 | MF611637 | – |
| P. festucae | MFLUCC 17-0056 | KY824767 | KY824769 | KY824766 | – |
| Phaeoseptoriella zeae | CBS 144614 | MK442547 | – | MK442611 | MK442702 |
| Phaeosphaeria musae | MFLUCC 11-0133 | KM434277 | KM434287 | KM434267 | KM434296 |
| P. oryzae | CBS 110110 | KF251689 | GQ387530 | KF251186 | – |
| P. papayae | CBS 135416 | – | – | MH866082 | – |
| Phaeosphaeriopsis agapanthi | CPC 26303 | KX228311 | – | KX228260 | – |
| P. agavacearum | CPC 29122 | KY173520 | – | KY173430 | – |
| P. agavensis | CBS 102206 | KY090669 | KY090693 | KY090635 | – |
| P. aloes | CBS 145367 | MK540030 | – | MK539959 | MK540153 |
| P. aloicola | CBS 145368 | MK540031 | – | MK539960 | MK540154 |
| P. amblyospora | CBS 110131 | – | – | MH862851 | – |
| P. beaucarneae | MFLU 18-2586 | MT321813 | MT321806 | MT321799 | MT328756 |
| MFLU 18-2587 | MT321814 | MT321807 | MT321800 | MT328757 | |
| P. dracaenicola | MFLUCC 11-0157 | KM434283 | KM434292 | KM434273 | KM434301 |
| P. glaucopunctata | MFLUCC 13-0265 | KJ522477 | KJ522481 | KJ522473 | MG520918 |
| P. grevilleae | CBS 145369 | MK540032 | – | MK539961 | MK540155 |
| P. nolinae | CBS 102205 | KY090667 | KY090694 | KY090637 | – |
| P. obtusispora | CBS 246.64 | JX681119 | – | KY090644 | – |
| P. omaniana | SQUCC:14333 | MT075849 | – | MT075840 | – |
| P. phacidiomorpha | CBS 198.35 | AF275496 | AF275515 | FJ462742 | – |
| P. pseudoagavacearum | CBS 145370 | MK540033 | – | MK539962 | – |
| MFLU 17-1800A | MN750592 | MN750607 | MN750613 | MN756837 | |
| P. triseptata | MFLUCC 13-0271 | KJ522479 | KJ522484 | KJ522475 | MG520919 |
| P. yuccae | MFLUCC 16-0558 | KY554481 | KY554480 | KY554482 | MG520920 |
| Piniphoma wesendahlina | CBS 145032 | MK442551 | – | MK442615 | MK442706 |
| Populocrescentia ammophilae | MFLUCC 17-0665 | MG829059 | MG829164 | MG828949 | MG829231 |
| P. rosacea | MFLU 17-0128 | MG829060 | MG829165 | – | MG829232 |
| Pseudoophiobolus achilleae | MFLU 17-0925 | MG520966 | – | MG520946 | – |
| P. galii | MFLUCC 17-2257 | MG520967 | MG520989 | MG520947 | MG520926 |
| Pseudoophiosphaerella huishuiensis | HS13 | MK522499 | MK522505 | MK522509 | MK523389 |
| Pseudophaeosphaeria rubi | MFLUCC 14-0259 | KX765299 | KX765300 | KX765298 | MG520934 |
| Pseudostaurosphaeria chromolaena | MFLUCC 17-1490 | MN994570 | MN994616 | MN994593 | MN998174 |
| P. chromolaenicola | MFLUCC 17-1491 | MN994571 | MN994617 | MN994594 | MN998175 |
| Poaceicola arundinis | MFLU 16-0158 | MG829057 | MG829162 | MG828947 | MG829229 |
| P. bromi | MFLUCC 13-0739 | KU058727 | – | KU058717 | – |
| Sclerostagonospora rosicola | MFLUCC 15-0129 | MG829068 | MG829172 | MG828957 | MG829237 |
| Scolicosporium minkeviciusii | MFLUCC 12-0089 | KF366382 | KF366383 | – | – |
| Septoriella phragmitis | CPC 24118 | KR873279 | – | KR873251 | – |
| S. pseudophragmitis | CBS 145417 | – | – | MK560161 | MK559452 |
| Setomelanomma holmii | CBS 110217 | GU301871 | GU296196 | KT389542 | GU349028 |
| Setophoma antiqua | LC6594 | MK511947 | – | MK511909 | MK525070 |
| S. chromolaenae | CBS 135105 | KF251747 | – | KF251244 | KF253195 |
| S. endophytica | LC3163 | MK511956 | – | MK511931 | MK525092 |
| S. longinqua | LC6593 | MK511946 | – | MK511908 | MK525069 |
| S. pseudosacchari | CBS 145373 | MK540039 | – | MK539969 | |
| S. sacchari | MFLUCC 11-0154 | KJ476146 | KJ476148 | KJ476144 | KJ461319 |
| MFLUCC 12-0241 | KJ476147 | KJ476149 | KJ476145 | KJ461318 | |
| S. terrestris | CBS 335.29 | KF251749 | GQ387526 | KF251246 | KF253196 |
| S. vernoniae | CBS 137988 | KJ869198 | – | KJ869141 | MK540162 |
| S. yingyisheniae | LC12696 | MK511950 | – | MK511914 | MK525075 |
| S. yunnanensis | LC6532 | MK511945 | – | MK511907 | MK525068 |
| Stagonospora foliicola | CBS 110111 | KF251759 | EU754118 | KF251256 | KF253206 |
| Sulcispora sp. | MFLUCC 14-0995 | KP271444 | KP271445 | KP271443 | MH665366 |
| Sulcispora pleurospora | CBS 460.84 | – | – | AF439498 | – |
| Tintelnotia destructans | CBS 127737 | KY090664 | KY090698 | KY090652 | – |
| T. opuntiae | CBS 376.91 | GU238123 | GU238226 | KY090651 | – |
| Vagicola vagans | CBS 604.86 | KU058727 | – | KF251193 | KF253149 |
| Vittaliana mangrovei | NFCCI 4251 | MG767312 | MG767313 | MG767311 | MG767314 |
| Vrystaatia aloeicola | CBS 135107 | KF251781 | – | KF251278 | – |
| Wingfieldomyces cyperi | CBS 141450 | KX228337 | – | KX228286 | MK540163 |
| Wojnowiciella eucalypti | CPC 25024 | KR476774 | – | KR476741 | LT990617 |
| W. kunmingensis | KUMCC 18-0159 | MK356354 | MK356368 | MK356380 | MK359071 |
| Xenophoma puncteliae | CBS 128022 | JQ238619 | – | – | KP170686 |
| Xenoseptoria neosaccardoi | CBS 120.43 | KF251783 | – | KF251280 | KF253227 |
| CBS 128665 | KF251784 | – | KF251281 | KF253228 | |
| Yunnanensis chromolaenae | MFLUCC 17-1486 | MN994573 | MN994619 | MN994596 | MN998177 |
| MFLUCC 17-1487 | MN994574 | MN994620 | MN994597 | MN998178 | |
| Yunnanensis phragmitis | MFLUCC 17-0315 | MF684863 | MF684867 | MF684862 | MF683624 |
| MFLUCC 17-1361 | MF684865 | MF684864 | MF684869 | – | |
Phylogenetic analysis
Phylogenetic analyses were performed using a combined LSU, SSU, ITS and tef1-α sequence dataset. Newly generated sequence data were initially subjected to blast search in NCBI to obtain the closest matches in GenBank. Sequences generated from this study were analyzed with related taxa in the family Phaeosphaeriaceae, which were obtained from GenBank and from recently published data (Bakhshi et al. 2019; Hyde et al. 2019; Maharachchikumbura et al. 2019; Yang et al. 2019; Mapook et al. 2020) (Table 1). The combined dataset consisted of 168 sequences including our newly generated sequences. Multiple alignments were automatically made with MAFFT v. 7 at the web server (http://mafft.cbrc.jp/alignment/server), using default settings (Katoh and Standley 2013). The alignment was refined manually with BioEdit v. 7.0.5.2 (Hall 1999), where necessary.
Evolutionary models for phylogenetic analyses were selected independently for each locus using MrModeltest v. 3.7 (Posada and Crandall 1998) under the Akaike Information Criterion (AIC). Phylogenetic trees were obtained from Randomized Accelerated Maximum Likelihood (RAxML), maximum parsimony analysis (MP) and Bayesian inference analyses (BI). ML trees were generated using the RAxML-HPC2 on XSEDE (8.2.8) (Stamatakis et al. 2008; Stamatakis 2014) in the CIPRES Science Gateway platform (Miller et al. 2010) using GTR+I+G model of evolution. The MP analysis was performed using PAUP (Phylogenetic Analysis Using Parsimony) version 4.0b10 (Swofford 2002), with parameters as described in Tennakoon et al. (2019). Descriptive tree statistics for parsimony, such as Tree Length (TL), Consistency Index (CI), Retention Index (RI), Relative Consistency Index (RC) and Homoplasy Index (HI) were calculated.
The BI analysis was conducted with MrBayes v. 3.1.2 (Huelsenbeck and Ronquist 2001) to evaluate posterior probabilities (PP) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002) by Markov Chain Monte Carlo sampling (MCMC). Six MCMC chains were run simultaneously, starting from random trees for 3,000,000 generations. Trees were sampled every 100th generation for a total of 30,000 trees. The first 6,000 trees were discarded as the burn-in phase of each analysis. Posterior probabilities (Rannala and Yang 1996) were determined from a majority-rule consensus tree generated with the remaining 24,000 trees. Phylograms were visualized with FigTree v1.4.0 (Rambaut 2012) and annotated in Microsoft Power Point (2010). Sequences of the new strains generated in this study are deposited in GenBank. The final alignment and trees were deposited in TreeBASE, submission ID: 26088.
Results
Phylogenetic analysis
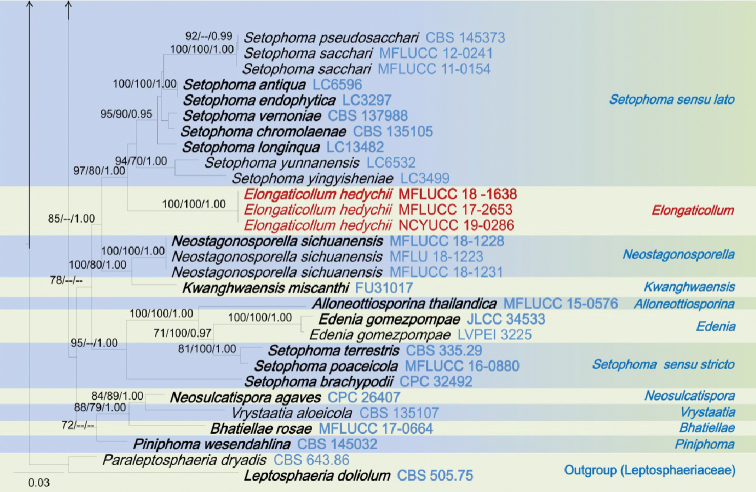
The combined dataset of ITS, LSU, SSU and tef1-α sequences comprised 3423 characters, of which 2418 characters are constant, 697 characters are parsimony-informative, while 308 variable characters are parsimony-uninformative in the maximum parsimony (MP) analysis (TL = 6364, CI = 0.250, RI = 0.657, RC = 0.164, HI = 0.750). The RAxML analysis of the combined dataset yielded a best scoring tree (Figure 1) with a final ML optimization likelihood value of – 34492.801018. The matrix had 1331 distinct alignment patterns, with 37.25% of undetermined characters or gaps. Estimated base frequencies are; A = 0.247120, C = 0.228182, G = 0.268238, T = 0.256459; substitution rates AC = 1.250439, AG = 3.526348, AT = 2.517351, CG = 0.798250, CT = 6.907432, GT = 1.000; proportion of invariable sites I = 0.596400; gamma distribution shape parameter α = 0.492378. All analyses (ML, MP and BI) gave similar results and are in agreement with previous studies based on multi-gene analyses (Hyde et al. 2019, 2020; Phookamsak et al. 2019). Phylogenetic analyses of the combined data matrix resulted in well-resolved clades, many of which had considerably high statistical support (Figure 1). Bootstrap support values for maximum likelihood, maximum parsimony ≥70%, and Bayesian posterior probabilities (BYPP) ≥0.95 are given above each branch in that order (Figure 1). Phylogenetic position and statistical support are noted in the taxonomy section.
Figure 1.
RAxML tree inferred from combined dataset of ITS, LSU, SSU and tef1-α partial sequences of 168 strains of Phaeosphaeriaceae. Bootstrap support values for maximum likelihood (ML), maximum parsimony (MP) values ≥70%, and Bayesian posterior probabilities (BYPP) ≥0.95 are given above each branch respectively. The new species are highlighted in red, and the new record in green. Ex-type strains are in bold. The tree is rooted by Leptosphaeria doliolum (CBS 505.75) and Paraleptosphaeria dryadis (CBS 643.86).
Figure 1.
Continued.
Figure 1.
Continued.
Figure 1.
Continued.
Taxonomy
Elongaticollum
Tennakoon, C.H. Kuo & K.D. Hyde gen. nov.
099B265A-B2F3-55FD-85FE-20DDD861F112
Index Fungorum number: IF 557486
Facesoffungi number: FoF07849
Etymology.
Refers to the fact that the pycnidia have elongated necks.
Diagnosis.
Saprobic on dead leaves of Hedychium coronarium J. Koenig. Sexual morph: Undetermined. Asexual morph: Coelomycetous. Conidiomata pycnidial, solitary, superficial, dark brown to black, obpyriform, papillate. Neck elongate, dark brown, usually straight, but sometimes slightly curved. Conidiomatal wall composed of 4–5 layers of light brown cells, arranged in textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, aseptate, smooth, ampulliform, arising from the inner cell wall of the apex. Conidia oval to oblong, smooth and thin-walled, hyaline, aseptate, with 1–2-minute guttules.
Type species.
Elongaticollum hedychii Tennakoon, C.H. Kuo & K.D. Hyde.
Elongaticollum hedychii
Tennakoon, C.H. Kuo & K.D. Hyde sp. nov.
934058B4-73A6-54E5-AD90-1F316FFB339C
Index Fungorum number: IF 557487
Facesoffungi number: FoF07850
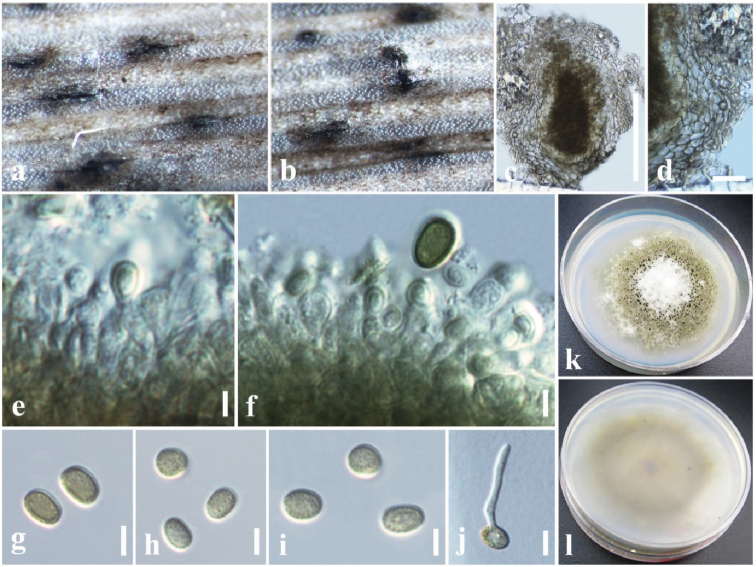
Figure 2.
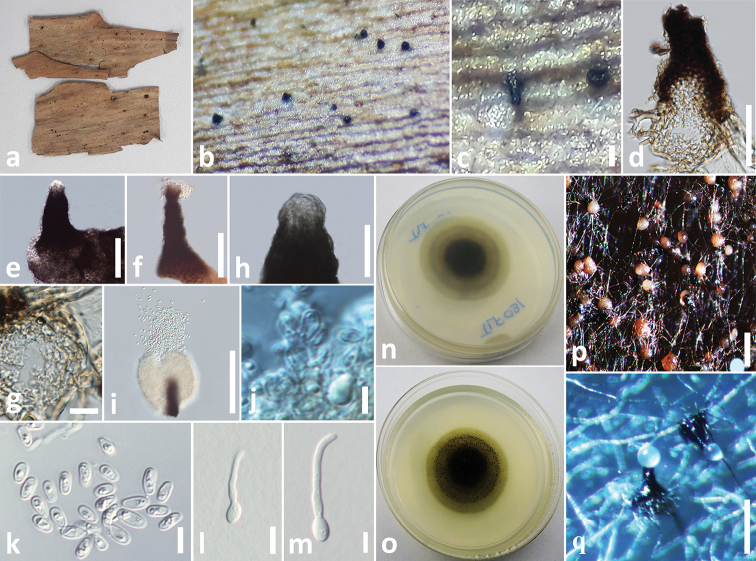
Elongaticollum hedychii (MFLU 18-2542, holotype) a specimen b appearance of conidiomata on host c close up of conidiomata on host d vertical section through conidioma e, f squash mount of conidioma g conidioma wall h, i elongated conidiomatal necks j conidiogenous cells k conidia l, m germinated conidia n colony from below o colony from above p, q pycnidia formed on PDA. Scale bars: 100 µm (c), 50 µm (d–h), 10 µm (g), 30 µm (i), 3 µm (j–m), 100 µm (p, q).
Etymology.
Name reflects the host Hedychium coronarium J. Koenig, from which the holotype was collected.
Holotype.
MFLU 18-2542.
Diagnosis.
Saprobic on dead leaves of Hedychium coronarium J. Koenig. Sexual morph: Undetermined. Asexual morph: Coelomycetous. Conidiomata 120–140 µm high, 60–70 µm diam., pycnidial, solitary, scattered, superficial, visible as small black spots on host surface, dark brown to black, obpyriform, papillate. Neck up to 80–100 μm long, 20–30 µm diam., elongated, dark brown, usually straight, but sometimes slightly curved. Conidiomatal wall 10–20 µm wide, composed of 4–5 layers of light brown, thick-walled cells, arranged in textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–4 × 3–3.5 μm (x̄ = 3.6 × 3.2 μm, n = 10), arising from the inner cell wall of the apex, hyaline, aseptate, smooth, ampulliform. Conidia 4–5 × 1.8–2.2 μm (x̄ = 4.6 × 2.1 μm, n = 30), oval to oblong, smooth, thin-walled, hyaline, aseptate, with 1–2-minute guttules.
Culture characteristics.
Colonies on PDA reaching 30 mm diameter after 3 weeks at 20–25 °C, colonies medium sparse, circular, raised, surface slightly rough with entire edge, margin entire, colony from above: light brown to grey at the margin, dark brown at middle, dark brown to black at the center; reverse, light brown to yellowish at the margin, brown at middle, dark brown to black at the center; mycelium light brown to grey with tufts; not producing pigments in PDA.
Material examined.
Taiwan, Chiayi, Fanlu Township area, Dahu Forest, dead leaves of Hedychium coronarium J. Koenig (Zingiberaceae), 15 August 2018 (23°27.514'N, 120°36.302'E), D.S. Tennakoon, TLF031-A (MFLU 18-2542, holotype), ex-type living culture (MFLUCC 18-1638 = NCYUCC 19-0163); ibid. 20 August 2018 (23°27.530'N, 120°36.314'E), TLF031-B (NCYU19-0139, paratype), living culture (NCYUCC19-0286); ibid. 25 August 2018 (23°27.512'N, 120°36.301'E), TLF031-C (NCYU19-0140, paratype), living culture (NCYUCC 19-0287).
Notes.
The genus Elongaticollum differs from other asexual morphs in Phaeosphaeriaceae in dark brown to black, superficial, obpyriform, pycnidial conidiomata with distinct elongate necks (80–100 μm) and a globose base and oval to oblong, hyaline, aseptate conidia (Figure 2). Multi-gene phylogenetic analyses (LSU, SSU, ITS, tef1-α), show Elongaticollum strains constitute a highly supported independent lineage nested between Setophomasensu lato and Neostagonosporella (97% ML, 80% MP, 1.00 BYPP, Figure 1). However, the asexual morph of Setophoma can be distinguished from Elongaticollum in having setose conidiomata without elongate necks and oblong to ellipsoidal conidia, whereas, Elongaticollum have conidiomata with distinct elongate necks and lacking setae and oval to oblong conidia (De Gruyter et al. 2010; Phookamsak et al. 2014). Despite some Setophoma species not having setae (i.e. S. antiqua, S. endophytica, and S. yunnanensis) (Liu et al. 2019), Elongaticollum species can be distinguished by its superficial conidiomata with elongate necks.
The asexual morph of Neostagonosporella differs from Elongaticollum in having multiloculate conidiomata without distinct elongate necks and two types of conidia (macroconidia: subcylindrical to cylindrical, transversely multi-septate, hyaline and microconidia oval, ellipsoidal or long ellipsoidal, aseptate, hyaline), whereas Elongaticollum has uni-loculate conidiomata with distinct elongate necks and oval to oblong conidia (Figure 2, Yang et al. 2019).
Phylogenetic investigations herein provide insights into the taxonomy of Setophoma as well (Figure 1). Two major clades of Setophoma are recovered (Setophomasensu stricto and Setophomasensu lato. The Setophomasensu stricto clade includes S. brachypodii, S. poaceicola and S. terrestris (type species). Setophomasensu lato comprises S. antiqua, S. chromolaenae, S. endophytica, S. pseudosacchari, S. sacchari, S. vernoniae, S. yingyisheniae and S. yunnanensis (Figure 1). Elongaticollum, differs from Setophomasensu lato in having distinct superficial, obpyriform, pycnidial conidiomata with a globose base and distinct elongated necks (Figure 2, Liu et al. 2019). Further work is needed to resolve relationships between Setophomasensu stricto and Setophomasensu lato.
Ophiosphaerella
Speg., Anal. Mus. nac. B. Aires, Ser. 3 12: 401 (1909)
8624C0BF-7DAC-5E5D-9E1F-4F266473DD1C
Notes.
Ophiosphaerella was introduced by Spegazzini (1909) to accommodate O. graminicola Speg. as the type species. The species of this genus are characterized by papillate ascomata bearing fissitunicate, cylindrical asci frequently narrower near the base, with a short furcate pedicel and filamentous, pale brown, multi-septate ascospores without swollen cells or separating into part spores. Barr (1987) placed Ophiosphaerella in Phaeosphaeriaceae and this was confirmed by Zhang et al. (2009, 2012) and Hyde et al. (2013) based on molecular phylogeny. Most Ophiosphaerella species are often found as pathogens or saprobes worldwide on Poaceae and Cyperaceae (Câmara et al. 2000). Currently, twelve Ophiosphaerella species are listed in Index Fungorum (2020). In this study, we introduce Ophiosphaerella taiwanensis from Agave tequilana F.A.C. Weber (Asparagaceae) as a new species.
Ophiosphaerella taiwanensis
Tennakoon, C.H. Kuo & K.D. Hyde sp. nov.
4D2BB74E-F917-5ACB-B0D0-16979236E2FF
Index Fungorum number: IF 557488
Facesoffungi number: FoF07851
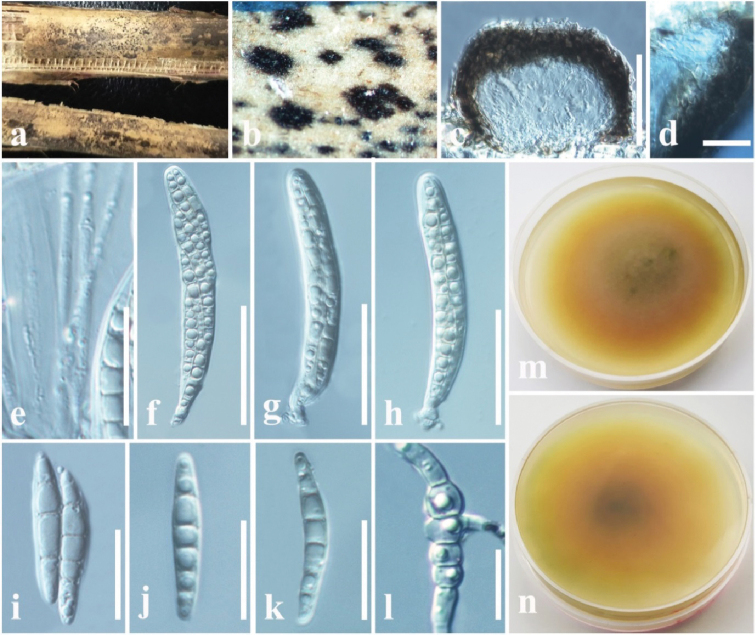
Figure 3.
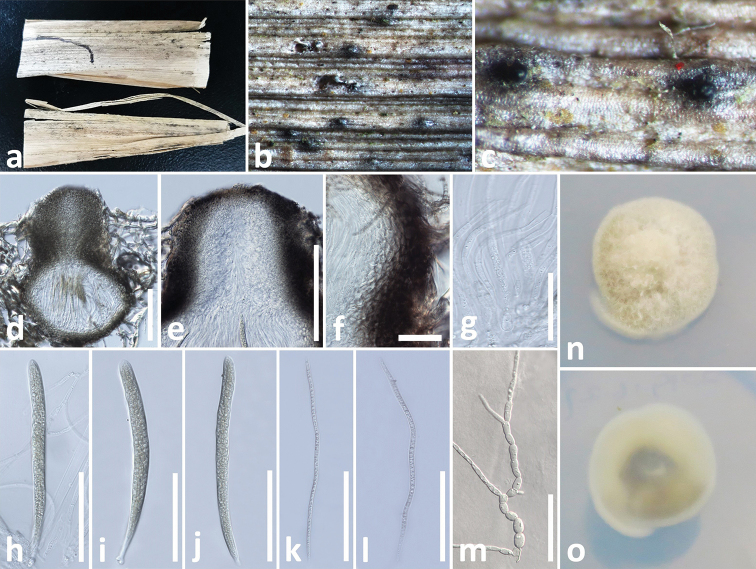
Ophiosphaerella taiwanensis (MFLU 18-2534, holotype) a, b appearance of ascomata on host c close-up of ascomata d vertical section through ascoma e apex of ascoma f peridium g pseudoparaphyses h–j asci k, l ascospores m germinated ascospore in PDAn colony from above o colony from below. Scale bars: 100 µm (d, e), 15 µm (f), 50 µm (g–m).
Etymology.
Named after Taiwan, where this fungus was collected.
Holotype.
MFLU 18-2534.
Diagnosis.
Saprobic on dead leaf of Agave tequilana F.A.C. Weber (Asparagaceae). Sexual morph: Ascomata 270–310 μm high, 220–260 μm diam., solitary, scattered, immersed to slightly erumpent through host tissue with papilla, visible as raised, small black dots in host surface, globose to subglobose, uniloculate, glabrous, dark brown to black, ostiole central, periphysate. Peridium 20–25 μm wide, thick-walled, of equal thickness, composed of 6–7 layers of small, flattened, brown to dark brown pseudoparenchymatous cells, hyaline towards the inside, arranged in a textura angularis, fusing and indistinguishable from the host tissues. Hamathecium of 1.5–2.5 µm wide, cellular, septate, rarely branching, pseudoparaphyses, anastomosing mostly above the asci and embedded in a mucilaginous matrix. Asci 115–140 × 8.5–10 μm (x̄ = 121.6 × 9.2 μm, n = 20), 8-spored, bitunicate, fissitunicate, cylindrical to cylindric-clavate, short pedicellate, apically rounded, with a well-developed ocular chamber. Ascospores 110–132 × 2.2–2.7 μm (x̄ = 117.2 × 2.4 μm, n = 20), fasciculate, parallel, scolecosporous, filiform, 12–13-septate, narrowing towards ends, pale brown to brown, smooth-walled. Asexual morph: Undetermined.
Culture characteristics.
Colonies on PDA reaching 25 mm diameter after 3 weeks at 20–25 °C, colonies medium sparse, circular, raised, surface slightly rough with entire edge, margin well-defined, colony from above: gray to light brown at the margin, gray to cream at the center; reverse, gray to light brown at the margin, dark brown to black at the center; mycelium whitish gray with tufting; not producing pigments in PDA.
Material examined.
Taiwan, Chiayi, Fanlu Township area, Dahu Forest, dead leaf of Agave tequilana F.A.C. Weber (Asparagaceae), 15 August 2018 (23°27.520'N, 120°36.310'E), D.S. Tennakoon, TLF016 (MFLU 18-2534, holotype); ibid. (NCYU19-0131, isotype), ex-type living culture, NCYUCC 19-0152.
Notes.
The scolecosporous specimen was collected from dead leaves of Agave tequilana (Asparagaceae) in Taiwan. The multi-gene phylogenetic analysis (Figure 1) shows our strain (Ophiosphaerella taiwanensis, NCYUCC 19-0152), cluster with other Ophiosphaerella species, in particular with close affinity to Ophiosphaerella agrostidis with high bootstrap support (88% ML, 70% MP, 0.99 BYPP, Figure 1). Morphological characters of our collection (NCYUCC 19-0152) differ from Ophiosphaerella agrostidis in having periphyses in the ostiole, 12–13 septate ascospores and host occurrence (Asparagaceae). Ophiosphaerella agrostidis was introduced by Câmara et al. (2000) on Agrostis palustris (Poaceae), and is lacking periphyses, comprises 15-septate ascospores (Phookamsak et al. 2014). A comparison of the 619 nucleotides across the tef1-α gene region of Ophiosphaerella taiwanensis and O. agrostidis (MFLUCC 11-0152) reveals 17 base pair differences (2.74%).
Phaeosphaeriopsis
M.P.S. Câmara, M.E. Palm & A.W. Ramaley, Mycol. Res. 107(5): 519 (2003)
06CCEE9D-81EC-5D0B-967E-99E7810387E3
Notes.
The genus Phaeosphaeriopsis was introduced by Câmara et al. (2003) to accommodate Paraphaeosphaeria-like taxa, viz. P. agavensis A.W. Ramaley, M.E. Palm & M.E. Barr, P. glaucopunctata (Grev.) Shoemaker & C.E. Babc., P. nolinae A.W. Ramaley, P. obtusispora (Speg.) O.E. Erikss, Phaeosphaeriopsis amblyspora A. W. Ramaley and Phaeosphaeriopsis amblyspora A. W. Ramaley. The genus is typified by P. glaucopunctata and characterized by having immersed, sub-epidermal, globose to subglobose to pyriform ascomata, cylindric asci and septate, punctate or verrucose ascospores (Câmara et al. 2003; Phookamsak et al. 2014; Thambugala et al. 2014; Tibpromma et al. 2017). Currently, 17 Phaeosphaeriopsis species are accepted in Index Fungorum (2020). In this paper, Phaeosphaeriopsis beaucarneae is introduced from Beaucarnea recurvata (Asparagaceae) as a new species and the sexual/asexual morph connection between strains isolated from the natural habitat was established based on molecular sequence data.
Phaeosphaeriopsis beaucarneae
Tennakoon, C.H. Kuo & K.D. Hyde sp. nov.
B91A27ED-8901-57B1-87F7-C3FF701314F0
Index Fungorum number: IF 557489
Facesoffungi number: FoF07852
Figure 4.
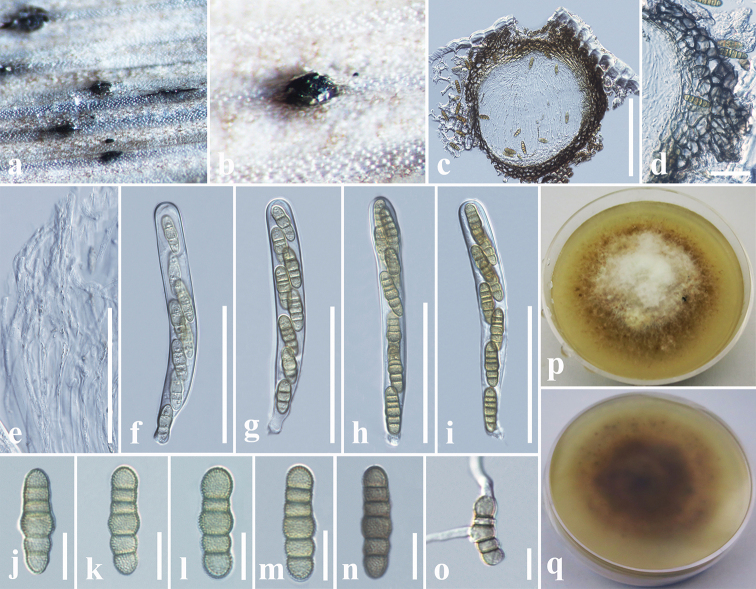
Phaeosphaeriopsis beaucarneae (MFLU 18-2586, holotype) a appearance of ascomata on host b close up of ascoma c vertical section through ascoma d peridium e pseudoparaphyses f–i asci j–n ascospores o germinated ascospore in PDAp colony from above q colony from below. Scale bars: 100 µm (c), 15 µm (d), 50 µm (e–i), 10 µm (j–o).
Figure 5.
Phaeosphaeriopsis beaucarneae (MFLU 18-2586, paratype) a appearance of conidiomata on host b close up of conidiomata c vertical section through conidioma d conidiomatal wall e, f conidiogenous cells and developing conidia g–i conidia j germinated conidium in PDAk colony from above l colony from below. Scale bars: 100 µm (c), 20 µm (d), 3 µm (e, f), 5 µm (g–j).
Etymology.
Name reflects the host Beaucarnea recurvata Lem., from which the holotype was collected.
Holotype.
MFLU 18-2586.
Diagnosis.
Saprobic on dead leaf of Beaucarnea recurvata Lem. (Asparagaceae). Sexual morph: Ascomata 160–200 μm high, 220–250 μm diam., scattered, solitary, gregarious, coriaceous, immersed to semi-immersed, slightly raised, erumpent, visible as black spots on host surface, uniloculate, dark brown to black, globose to subglobose, ostiolate. Ostiole central, papillate. Peridium 20–30 μm wide, thick-walled, of equal thickness, composed of 4–5 layers of dark brown to brown, thick-walled, pseudoparenchymatous cells of textura angularis. Hamathecium of 1.5–2.5 µm wide, cellular, septate, rarely branching, pseudoparaphyses, anastomosing mostly above the asci and embedded in a mucilaginous matrix. Asci 80–90 × 9–10 µm (x̄ = 86.5 × 9.6 µm, n = 25), 8-spored, bitunicate, fissitunicate, cylindrical to cylindric-clavate, short pedicellate, apically rounded, with a well-developed ocular chamber. Ascospores 20–25 × 5.5–7 μm (x̄ = 22.6 × 6.2 μm, n = 20), overlapping 1–2-seriate, oblong to cylindrical, yellowish to light brown, slightly narrowing towards the end cells, mostly 5-septate, constricted at the septa, enlarged at the 4th cell from above, verruculose, straight to curved, lacking a mucilaginous sheath. Asexual morph: Conidiomata 180–200 µm high, 140–160 µm diam., pycnidial, solitary, immersed to erumpent, small black spots on host surface, globose to subglobose with centrally placed ostiole. Conidiomatal wall 28–34 µm wide, composed of 6–7 layers of dark brown cells, arranged in textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–4 × 2.6–3.1 μm, holoblastic, phialidic, single, discrete, sometimes integrated, ampulliform or cylindric-clavate, hyaline, arising from basal stratum. Conidia 6.8–7.4 × 3–4 μm (x̄ = 7.1 × 3.4 μm, n = 30), 1-celled, globose to subglobose, initially hyaline, becoming brown to dark brown, aseptate, rough-walled.
Culture characteristics.
Colonies on PDA reaching 27 mm diameter after 3 weeks at 20–25 °C, colonies medium sparse, circular, raised, surface slightly rough with entire edge, margin irregular, colony from above: light brown at the margin, white to cream at the center; reverse, yellow to light brown at the margin, light brown to brown at the center; mycelium white to cream with tufting; not producing pigments in PDA.
Material examined.
Taiwan, Chiayi, Fanlu Township area, Dahu Forest, dead leaf of Beaucarnea recurvata Lem. (Asparagaceae), 21 July 2018 (23°27.514'N, 120°36.302'E), D.S. Tennakoon, SV027 (MFLU 18-2586, holotype); ibi. (NCYU19-0184, isotype), ex-type living culture, NCYUCC 19-0106; ibid., Dahu forest, dead leaf of Beaucarnea recurvata Lem. (Asparagaceae), 25 July 2018 (23°26.534'N, 120°36.220'E), D.S. Tennakoon, SV028 (MFLU 18-2587, paratype); living culture, NCYUCC 19-0107.
Notes.
Phaeosphaeriopsis beaucarneae is similar to other Phaeosphaeriopsis species in having scattered, semi-immersed to erumpent, globose to subglobose, ostiolate ascomata and cylindrical to clavate asci and light brown, verrucose ascospores (Phookamsak et al. 2014; Thambugala et al. 2014; Hyde et al. 2020). According to the present multi-gene phylogenetic analyses (Figure 1), Phaeosphaeriopsis beaucarneae is grouped with other Phaeosphaeriopsis species, in particularly closely to P. grevilleae (CBS 145369) with high statistical support (70% ML, 75% MP, 0.99 BYPP, Figure 1). The asexual morph of P. grevilleae was isolated from leaves of Grevillea sp. (Proteaceae) and introduced by Marin-Felix et al. (2019). Phaeosphaeriopsis beaucarneae differs from P. grevilleae in having larger conidia (6.8–7.4 × 3–4 μm), whereas P. grevilleae has comparatively smaller conidia (5 × 3.5 μm). A comparison of the 516 nucleotides across the ITS (+5.8S rDNA) gene region of Phaeosphaeriopsis beaucarneae and P. grevilleae (CBS 145369) revealed 16 base pair differences (3.10%). In addition, we compared our new taxon with P. grevilleae based on base pair differences in the tef1-α gene region. We found a total of 19 base pair differences (3.06%) across 619 nucleotides.
Recent studies have revealed that Phaeosphaeriopsis is a species rich genus and numerous Phaeosphaeriopsis species have been described during the last few years (Thambugala et al. 2014; Tibpromma et al. 2017; Marin-Felix et al. 2019; Al-Jaradi et al. 2020; Hyde et al. 2020). With this study, the number of Phaeosphaeriopsis species increases to 18.
Neosetophoma
Gruyter, Aveskamp & Verkley, Mycologia 102(5): 1075 (2010)
A4D85E50-AC97-57DC-B533-2418745AD589
Notes.
Neosetophoma was introduced by de Gruyter et al. (2010), typified by N. samararum (Desm.) Gruyter, Aveskamp. & Verkley. Species of Neosetophoma are characterized by globose to irregular conidiomata, with papillate ostioles, and yellowish conidia that are attenuate at one end (De Gruyter et al. 2010; Liu et al. 2015). Tibpromma et al. (2017) introduced Neosetophoma garethjonesii Tibpromma, E.B.G. Jones & K.D. Hyde as the first report of the sexual morph of Neosetophoma. Neosetophoma species have a diverse distribution as saprobes, endophytes, plant pathogens and soil fungi (Phookamsak et al. 2014; Hernandez-Restrepo et al. 2016; Karunarathna et al. 2017; Tibpromma et al. 2017; Wanasinghe et al. 2018). Currently, 19 Neosetophoma species are accepted in Index Fungorum (2020). In this study, we found Neosetophoma poaceicola Goonas., Thambug. & K.D. Hyde from dead leaves of Musa acuminata Colla in Taiwan. This is the first Neosetophoma species recorded from the plant family Musaceae.
Neosetophoma poaceicola
Goonas., Thambug. & K.D. Hyde. Mycosphere 8: 742 (2017)
4C1AA9B6-EA48-5A22-89E6-0AF230FD2C5A
Index Fungorum number: IF552974
Facesoffungi number: FoF00262
Figure 6.
Neosetophoma poaceicola (MFLU 18–2597, new host record) a appearance of ascomata on host b close up of ascomata c vertical section through ascoma d peridium e pseudoparaphyses f–h asci i–k ascospores l germinated ascospore in PDAm colony from above n colony from below. Scale bars: 50 µm (c), 20 µm (d), 30 µm (e–h), 15 µm (i–l).
Diagnosis.
Saprobic on dead leaf petioles of Musa acuminata Colla (Musaceae). Sexual morph: Ascomata 70–100 μm high, 90–130 μm diam., solitary, gregarious, coriaceous, immersed to semi-immersed, slightly raised, visible as black spots on host surface, uni-loculate, dark brown to black, globose to ovoid. Peridium 15–20 μm wide, thick-walled, of equal thickness, composed of several layers of dark brown to brown, pseudoparenchymatous cells of textura angularis. Hamathecium of 1–2 µm wide, cellular, rarely branching, pseudoparaphyses, anastomosing mostly above the asci and embedded in a mucilaginous matrix. Asci 60–80 × 7–8 μm (x̄ = 70.6 × 7.6 μm, n = 30), 8-spored, bitunicate, fissitunicate, cylindric-clavate with a short, rounded pedicel, apically rounded. Ascospores 20–30 × 3–4 μm (x̄ = 25.5 × 3.7 μm, n = 40), overlapping 1–2-seriate, hyaline, fusiform, with acute ends, 1-septate, 3–4 eu-septate, cell near the septum slightly larger, slightly constricted at the septum, straight to curved, smooth-walled, guttulate. Asexual morph: Undetermined.
Culture characteristics.
Colonies on PDA reaching 30 mm diameter after 3 weeks at 20–25 °C, colonies medium sparse, circular, flat, surface slightly rough with entire edge, margin well-defined, colony from above: yellow to light brown at the margin, brown at the center; reverse, yellow to light brown at the margin, dark brown at the center; mycelium light brown to whitish grey with tufting; not producing pigments in PDA.
Material examined.
Taiwan, Chiayi, Fanlu Township area, Dahu Forest, dead leaf petiole of Musa acuminata Colla (Musaceae), 21 July 2018 (23°27.530'N, 120°36.340'E), D.S. Tennakoon, SV049 (MFLU 18-2597, new host record), living culture, MFLUCC 18-1632, NCYUCC 19-0119.
Notes.
As morphological characters (immersed to semi-immersed ascomata, cylindric-clavate, apically rounded asci with short rounded pedicel and hyaline, fusiform, 1-septate ascospores) largely overlap with those of Neosetophoma poaceicola (MFLUCC 16–0886), we report our collection (MFLUCC 18-1632) as a new host record of N. poaceicola from dead leaves of Musa acuminata (Musaceae) in Taiwan. Combined multi-gene (LSU, SSU, ITS and tef1-α) based phylogenies also showed that our collection clustered with Neosetophoma poaceicola (MFLUCC 16-0886), with high bootstrap support (100% ML, 100% MP, 1.00 BYPP, Figure 1). Neosetophoma poaceicola was introduced by Thambugala et al. (2017) from dead leaves of grass species in Thailand. However, our collection slightly differs from Neosetophoma poaceicola (MFLUCC 16-0886) in having comparatively slightly larger ascospores (20–30 × 3–4 μm, versus 18.5–22.5 × 3.5–5 μm).
Neosetophoma species have been recorded from various host families, viz. Brassicaceae, Caprifoliaceae, Iridaceae, Malvaceae, Ranunculaceae, Salicaceae, but most are reported from Poaceae (Phookamsak et al. 2014; Karunarathna et al. 2017; Tibpromma et al. 2017, Wanasinghe et al. 2018; Marin-Felix et al. 2019). Interestingly, this is the first Neosetophoma species record (MFLU 18-2597) from the plant family Musaceae.
Discussion
The taxonomy of Phaeosphaeriaceae has been subjected to several changes in recent years. Traditionally, morphology-based identification was the main means for identifying Phaeosphaeriaceae species (Barr 1979, 1992; Tomilin 1993). However, species identification has been revolutionized by the application of molecular based approaches incorporating DNA sequence data in Phaeosphaeriaceae (Phookamsak et al. 2014, 2017; Tennakoon et al. 2016; Wanasinghe et al. 2018; Bakhshi et al. 2019; Chethana et al. 2020; Hyde et al. 2020). Phaeosphaeriaceae species are adapted to a wide range of ecological environments and are present in soils, fresh and marine habitats and cause infections in humans (Yuan 1994; Phookamsak et al. 2014, 2017; Ahmed et al. 2017; Maharachchikumbura et al. 2019; Valenzuela-Lopez et al. 2019). Members of the Phaeosphaeriaceae have also been recorded from both temperate and tropical countries (i.e. Austria, Belgium, Bulgaria, Canada, China, Germany, Italy, Japan, Norway, Poland, Thailand, Sweden, Switzerland) and from different host families (i. e. Acoraceae, Arecaceae, Cyperaceae, Asparagaceae, Brassicaceae, Fabaceae, Poaceae, Marantaceae) (Shoemaker and Babcock 1989; Phookamsak et al. 2014, 2019; Wanasinghe et al. 2018; Maharachchikumbura et al. 2019; Farr and Rossman 2020). Due to their cosmopolitan distribution, in the last few years, many researchers have paid significant attention to the Phaeosphaeriaceae (Phookamsak et al. 2014, 2019; Tennakoon et al. 2016; Wanasinghe et al. 2018; Bakhshi et al. 2019; Hyde et al. 2020).
The fungi that decay leaf litter are highly diverse and may be host-specific (Parungao et al. 2002). Several studies have examined the succession of leaf degrading communities and found unique sets of species on different types of litter (Promputtha et al. 2002, 2017; Duong et al. 2008). Additional ecological studies are therefore needed to establish whether these fungi are generalists or specialists. This study provides evidence to indicate the fungal diversity in leaf litter, even within a single family, Phaeosphaeriaceae. Additional work is necessary to identify if the newly described species are host specific.
Supplementary Material
Acknowledgments
The authors would like to thank T.K. Goh for his valuable suggestions and help. Shaun Pennycook is thanked for checking species names. This research work was partially supported by Chiang Mai University and K.D. Hyde thanks Chiang Mai University for the award of Visiting Professorship. He also thanks the Thailand Research Fund for the Grant No. RDG613001, entitled “Impact of Climate Change on Fungal Diversity and Biogeography in the Greater Mekong Subregion”. D.N. Wanasinghe would like to thank the CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2019PC0008), the National Science Foundation of China and the Chinese Academy of Sciences for financial support under the following grants: 41761144055, 41771063 and Y4ZK111B01. Wanasinghe also thanks the 64th batch of China Postdoctoral Science Foundation (grant no: Y913083271).
Citation
Tennakoon DS, Thambugala KM, Wanasinghe DN, Gentekaki E, Promputtha I, Kuo CH, Hyde KD (2020) Additions to Phaeosphaeriaceae (Pleosporales): Elongaticollum gen. nov., Ophiosphaerella taiwanensis sp. nov., Phaeosphaeriopsis beaucarneae sp. nov. and a new host record of Neosetophoma poaceicola from Musaceae. MycoKeys 70: 59–88. https://doi.org/10.3897/mycokeys.70.53674
References
- Al-Jaradi AJ, Maharachchikumbura SS, Al-Sadi AM. (2020) Phaeosphaeriopsis omaniana (Phaeosphaeriaceae, Pleosporales), a novel fungus from Oman. Phytotaxa 436: 187–192. 10.11646/phytotaxa.436.2.8 [DOI] [Google Scholar]
- Ahmed SA, Hofmueller W, Seibold M, de Hoog GS, Harak H, Tammer I, Van Diepeningen AD, Behrens‐Baumann W. (2017) Tintelnotia, a new genus in Phaeosphaeriaceae harbouring agents of cornea and nail infections in humans. Mycoses 60: 244–253. 10.1111/myc.12588 [DOI] [PubMed] [Google Scholar]
- Bakhshi M, Arzanlou M, Groenewald JZ, Quaedvlieg W, Crous PW. (2019) Parastagonosporella fallopiae gen. et sp. nov. (Phaeosphaeriaceae) on Fallopia convolvulus from Iran. Mycological Progress 18: 203–214. 10.1007/s11557-018-1428-z [DOI] [Google Scholar]
- Bani A, Pioli S, Ventura M, Panzacchi P, Borruso L, Tognetti R, Tonon G, Brusetti L. (2018) The role of microbial community in the decomposition of leaf litter and deadwood. Applied soil ecology 126: 75–84. 10.1016/j.apsoil.2018.02.017 [DOI] [Google Scholar]
- Barr ME. (1979) A classification of Loculoascomycetes. Mycologia 71: 935–957. 10.1080/00275514.1979.12021099 [DOI] [Google Scholar]
- Barr ME. (1987) New taxa and combinations in the Loculoascomycetes. Mycotaxon 29: 501–505. [Google Scholar]
- Barr ME. (1992) Additions to and notes on the Phaeosphaeriaceae (Pleosporales, Loculoascomycetes). Mycotaxon 43: 371–400. [Google Scholar]
- Berg B, McClaugherty C. (2003) Plant Litter. Decomposition, Humus Formation, Carbon Sequestration. Springer-Verlag, Berlin Heidelberg, New York.
- Berg B, McClaugherty C. (2008) Plant Litter. Decomposition, Humus Formation, Carbon Sequestration (2nd ed.). Springer. 10.1007/978-3-540-74923-3 [DOI]
- Câmara MP, O’Neill NR, Berkum PV, Dernoeden PH, Palm ME. (2000) Ophiosphaerella agrostis sp. nov. and its relationship to other species of Ophiosphaerella. Mycologia 92: 317–325. 10.1080/00275514.2000.12061162 [DOI] [Google Scholar]
- Câmara MPS, Ramaley AW, Castlebury LA, Palm ME. (2003) Neophaeosphaeria and Phaeosphaeriopsis, segregates of Paraphaeosphaeria. Mycological Research 107: 516–522. 10.1017/S0953756203007731 [DOI] [PubMed] [Google Scholar]
- Chaiwan N, Wanasinghe DN, Camporesi E, Tibpromma S, Boonmee S, Lumyong S, Hyde KD. (2019) Molecular taxonomy reveals the sexual morph of Nodulosphaeria digitalis in Phaeosphaeriaceae from Campanula trachelium in Italy. Phytotaxa 400: 1–13. 10.11646/phytotaxa.400.1.1 [DOI] [Google Scholar]
- Chethana KWT, Jayawardena RS, Hyde KD. (2020) Hurdles in fungal taxonomy: Effectiveness of recent methods in discriminating taxa. Megataxa 1: 114–122. 10.11646/megataxa.1.2.2 [DOI] [Google Scholar]
- De Gruyter J, Woudenberg JH, Aveskamp MM, Verkley GJ, Groenewald JZ, Crous PW. (2010) Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia 102: 1066–1081. 10.3852/09-240 [DOI] [PubMed] [Google Scholar]
- Devadatha B, Mehta N, Wanasinghe DN, Baghela A, Sarma VV. (2019) Vittaliana mangrovei gen. nov, sp. nov. (Phaeosphaeriaceae), from mangroves near Pondicherry (India), based on morphology and multigene phylogeny. Cryptogamie Mycologie 40: 117–132. 10.5252/cryptogamiemycologie2019v40a7 [DOI] [Google Scholar]
- Duong LM, McKenzie EHC, Lumyong S, Hyde KD. (2008) Fungal succession on senescent leaves of Castanopsis diversifolia in Doi Suthep-Pui National Park, Thailand. Fungal Diversity 30: 23–36. http://cmuir.cmu.ac.th/jspui/handle/6653943832/60073 [Google Scholar]
- Farr DF, Rossman AY. (2020) Fungal databases, Systematic mycology and microbiology laboratory, ARS, USDA. [Retrieved April 10, 2020] http://nt.ars-grin.gov/fungaldatabases
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hernandez-Restrepo M, Schumacher RK, Wingfield MJ, Ishtiaq A, Cai L, Duong TA, Edwards J, Gene J, Groenewald JZ, Sana J, Khalid AN. (2016) Fungal systematics and evolution: FUSE 2. Sydowia 68: 193–230. 10.12905/0380.sydowia68-2016-0193 [DOI] [Google Scholar]
- Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, McKenzie HCE, Sarma VV, Boonmee S, Lücking R, Pem D, Bhat JD, Liu N, Tennakoon DS, Karunarathna A, Jiang SH, Jones EBG, Phillips AJL, Manawasinghe I, Tibpromma S, Jayasiri SC, Sandamali D, Jayawardena RS, Wijayawardene NN, Ekanayaka AH, Jeewon R, Lu YZ, Dissanayake AJ, Zeng XY, Luo Z, Tian Q, Phukhamsakda C, Thambugala KM, Dai D, Chethana TKW, Ertz D, Doilom M, Liu JK, Pérez-Ortega S, Suija A, Senwanna C, Wijesinghe SN, Konta S, Niranjan M, Zhang SN, Ariyawansa HA, Jiang HB, Zhang JF, de Silva NI, Thiyagaraja V, Zhang H, Bezerra JDP, Miranda-Gonzáles R, Aptroot A, Kashiwadani H, Harishchandra D, Aluthmuhandiram JVS, Abeywickrama PD, Bao DF, Devadatha B, Wu HX, Moon KH, Gueidan C, Schumm F, Bundhun D, Mapook A, Monkai J, Chomnunti P, Samarakoon MC, Suetrong S, Chaiwan N, Dayarathne MC, Jing Y, Rathnayaka AR, Bhunjun CS, Xu J, Zheng J, Liu G, Feng Y, Xie N. (2020) Refined families of Dothideomycetes. Fungal diversity. [In press]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Jones EBG, Liu JK, Ariyawansa H, Boehm E, Boonmee S, Braun U, Chomnunti P, Crous PW, Dai DQ, Diederich P, Dissanayake A, Doilom M, Doveri F, Hongsanan S, Jayawardena R, Lawrey JD, Li YM, Liu YX, Lücking R, Monka J, Muggia L, Nelsen MP, Pang KL, Phookamsak R, Senanayake IC, Shearer CA, Suetrong S, Tanaka K, Thambugala KM, Wijayawardene NN, Wikee S, Wu HX, Zhang Y, Begoña AH, Alias SA, Aptroot A, Bahkali AH, Bezerra JL, Bhat DJ, Camporesi E, Chukea E, Gueidan C, Hawksworth DL, Hirayama K, Hoog SD, Kang JK, Knudsen K, Li WJ, Li XH, Liu ZY, Mapook A, Mckenzie EHC, Miller AN, Mortimer PE, Phillips AJL, Raja HA, Scheuer C, Schumm F, Taylor JE, Tian Q, Tibpromma S, Wanasinghe DN, Wang Y, Xu JC, Yacharoen S, Yan JY, Zang M. (2013) Families of Dothideomycetes. Fungal Diversity 63: 1–313. 10.1007/s13225-013-0263-4 [DOI] [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Dhanushka N, Wanasinghe DN, Luücking R, Aptroot A, Cáceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, de Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo ZL, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Bart B, Randrianjohany E, Hofstetter V, Gibertoni TB, da Silva Soares AM, Plautz Jr HL, Sotão HMP, Xavier WKS, Bezerra JDP, de Oliveira TGL, de Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang SN, Bao DF, Samarakoon MC, Pem D, Karunarathna A, Lin CG, Yang J, Perera RH, Kumar V, Huang SK, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao YP, Konta S, Niskanen T, Liimatainen K, Dai YC, Ji XH, Tian XM, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu YZ, Jiang HB, Zhang JF, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei DP, Réblová M, Fournier J, Nekvindová J, do Nascimento Barbosa R, dos Santos JEF, de Oliveira NT, Li GJ, Ertz D, Shang QJ, Phillips AJL, Kuo CH, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng XY, Fryar S, Tkalčec Z, Liang J, Li GS, Wen TC, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao RL, Zhao Q, Kirk PM, Liu JK, Yan JY, Mortimer PE, Xu JC. (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Jones EBG, Liu NG, Abeywickrama PD, Mapook A, Wei DP, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao DF, Li JF, Samarakoon MC, Chaiwan N, Lin CG, Phutthacharoen K, Zhang SN, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang HB, Yang J, Zeng M, Huanraluek N, Liu JK, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang SK, Thiyagaraja V, Lu YZ, Jayawardena LS, Dong W, Yang EF, Singh SK, Singh SM, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Alvarenga RLM, Gibertoni TB, Pliegler WP, Horváth E, Imre A, Alves AL, Santos ACDS, Tiago RV, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu JC, Sheng J. (2020) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 1–273. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Index Fungorum (2020) Index Fungorum. http://www.indexfungorum.org/names/Names.asp [accessed 6 April 2020]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat DJ, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu JK, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo JM, Ghobad-Nejhad M, Nilsson H, Pang KA, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen TC, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li WJ, Perera RH, Phookamsak R, de Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao RL, Zhao Q, Kang JC, Promputtha I. (2015) The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74: 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Johnson EA, Catley KM. (2002) Life in the Leaf Litter. American Museum of Natural History, New York.
- Jones EBG, Pang KL, Abdel-Wahab MA, Scholz B, Hyde KD, Boekhout T, Ebel R, Rateb ME, Henderson L, Sakayaroj J, Suetrong S, Dayarathne MC, Kumar V, Raghukumar S, Sridhar KR, Bahkali AH, Gleason FH, Norphanphoun C. (2019) An online resource for marine fungi. Fungal Diversity 96: 347–433. 10.1007/s13225-019-00426-5 [DOI] [Google Scholar]
- Karunarathna A, Papizadeh M, Senanayake IC, Jeewon R, Phookamsak R, Goonasekara ID, Wanasinghe DN, Wijayawardene NN, Amoozegar MA, Shahzadeh Fazeli SA, Camporesi E. (2017) Novel fungal species of Phaeosphaeriaceae with an asexual/sexual morph connection. Mycosphere 8: 1818–1834. 10.5943/mycosphere/8/10/8 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna MP, Mohan M. (2017) Litter decomposition in forest ecosystems: a review. Energy Ecology and Environment 2: 236–249. 10.1007/s40974-017-0064-9 [DOI] [Google Scholar]
- Liu F, Wang J, Li H, Wang W, Cai L. (2019) Setophoma spp. on Camellia sinensis. Fungal Systematics and Evolution 4: 43–57. 10.3114/fuse.2019.04.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Hyde KD, Jones EBG, Ariyawansa HA, Bhat DJ, Boonmee S, Maharachchikumbura SSN, McKenzie EHC, Phookamsak R, Phukhamsakda C, Shenoy BD, Abdel-Wahab MA, Buyck B, Chen J, Chethana KWT, Singtripop C, Dai DQ, Dai YC, Daranagama DA, Dissanayake AJ, Doilom M, D’souza MJ, Fan XL, Goonasekara ID, Hirayama K, Hongsanan S, Jayasiri SC, Jayawardena RS, Karunarathna SC, Li WJ, Mapook A, Norphanphoun C, Pang KL, Perera RH, Peršoh D, Pinruan U, Senanayake IC, Somrithipol S, Suetrong S, Tanaka K, Thambugala KM, Tian Q, Tibpromma S, Udayanga D, Wijayawardene NN, Wanasinghe DN, Wisitrassameewong K, Zeng XY, Abdel-Aziz FA, Adamčík S, Bahkali AH, Boonyuen N, Bulgakov T, Callac P, Chomnunti P, Greiner K, Hashimoto A, Hofstetter V, Kang JC, Lewis D, Li XH, Liu XZ, Liu ZY, Matsumura M, Mortimer PE, Rambold G, Randrianjohany E, Sato G, Sri-Indrasutdhi V, Tian CM, Verbeken A, von Brackel W, Wang Y, Wen TC, Xu JC, Yan JY, Zhao RL, Camporesi E. (2015) Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. 10.1007/s13225-015-0324-y [DOI] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Maharachchikumbura SSN, Jeewon R, Bao DF, Bhat DJ, Lin CG, Li WL, Yang J, Liu NG, Lu YZ, Jayawardena RS, Li JF, Su HY. (2019) Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. 10.1007/s13225-019-00438-1 [DOI] [Google Scholar]
- Maharachchikumbura SSN, Ariyawansa HA, Wanasinghe DN, Dayarathne MC, Al-Saady NA, Al-Sadi AM. (2019) Phylogenetic classification and generic delineation of Hydeomyces desertipleosporoides gen. et sp. nov., (Phaeosphaeriaceae) from Jebel Akhdar Mountain in Oman. Phytotaxa 391: 28–38. 10.11646/phytotaxa.391.1.2 [DOI] [Google Scholar]
- Mapook A, Hyde KD, McKenzie EHC, Jones EBG, Bhat DJ, Jeewon R, Stadler M, Samarakoon MC, Malaithong M, Tanunchai B. (2020) Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Diversity. [In press] 10.1007/s13225-020-00444-8 [DOI]
- Marin-Felix Y, Hernández-Restrepo M, Iturrieta-González I, García D, Carnegie AJ, Cheewangkoon R, Gramaje D, Groenewald JZ, Guarnaccia V, Halleen F, Lombard L. (2019) Genera of phytopathogenic fungi: GOPHY 3. Studies in Mycology 94: 1–124. 10.1016/j.simyco.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. SC10 Workshop on Gateway Computing Environments (GCE10). 10.1109/GCE.2010.5676129 [DOI]
- Mlambo MC, Paavola R, Fritze H, Louhi P, Muotka T. (2019) Leaf litter decomposition and decomposer communities in streams affected by intensive forest biomass removal. Ecological indicators 101: 364–372. 10.1016/j.ecolind.2019.01.035 [DOI] [Google Scholar]
- Parungao MM, Fryar SC, Hyde KD. (2002) Diversity of fungi on rainforest litter in North Queensland, Australia. Biodiversity & Conservation 11: 1185–1194. 10.1023/A:1016089220042 [DOI] [Google Scholar]
- Phookamsak R, Liu JK, McKenzie EHC, Manamgoda DS, Ariyawansa HA, Thambugala KM, Dai DQ, Camporesi E, Chukeatirote E, Wijayawardene NN, Bahkali AH, Mortimer PE, Xu JC, Hyde KD. (2014) Revision of Phaeosphaeriaceae. Fungal Diversity 68: 159–238. 10.1007/s13225-014-0308-3 [DOI] [Google Scholar]
- Phookamsak R, Wanasinghe DN, Hongsanan S, Phukhamsakda C, Huang SK, Tennakoon DS, Norphanphoun C, Camporesi E, Bulgakov TS, Promputtha I, Mortimer PE. (2017) Towards a natural classification of ophiobolus and ophiobolus-like taxa; introducing three novel genera Ophiobolopsis, Paraophiobolus and Pseudoophiobolus in Phaeosphaeriaceae (Pleosporales). Fungal Diversity 87: 299–339. 10.1007/s13225-017-0393-1 [DOI] [Google Scholar]
- Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Jones EBJ, Maharachchikumbura SSN, Raspé O, Karunarathna SC, Wanasinghe DN, Hongsanan S, Doilom M, Tennakoon DS, Machado AR, Firmino AL, Ghosh A, Karunarathna A, Mešić A, Dutta AK, Thongbai B, Devadatha B, Norphanphoun C, Senwanna C, Wei D, Pem D, Ackah FK, Wang GN, Jiang HB, Madrid H, Lee HB, Goonasekara ID, Manawasinghe IS, Kušan Cano J, Gené J, Li J, Das K, Acharya K, Raj KNA, Latha KPD, Chethana KWT, He MQ, Dueñas M, Jadan M, Martín MP, Samarakoon MC, Dayarathne MC, Raza M, Park MS, Telleria MT, Chaiwan N, Matočec N, de Silva NI, Pereira OL, Singh PN, Manimohan P, Uniyal P, Shang QJ, Bhatt RP, Perera RH, Alvarenga RLM, Nogal-Prata S, Singh SK, Vadthanarat S, Oh SY, Huang SK, Rana S, Konta S, Paloi S, Jayasiri SC, Jeon SJ, Mehmood T, Gibertoni TB, Nguyen TTT, Singh U, Thiyagaraja V, Sarma VV, Dong W, Yu XD, Lu YZ, Lim YW, Chen Y, Tkalčec Z, Zhang ZF, Luo ZL, Daranagama DA, Thambugala KM, Tibpromma S, Camporesi E, Bulgakov T, Dissanayake AJ, Senanayake IC, Dai DQ, Tang LZ, Khan S, Zhang H, Promputtha I, Cai L, Chomnunti P, Zhao RL, Lumyong S, Boonmee S, Wen TC, Mortimer PE, Xu J. (2019) Fungal diversity notes 929–1036: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 95: 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Pointing SB, Pelling AL, Smith GJD, Hyde KD. (2005) Screening of basidiomycetes and xylariaceous fungi for lignin peroxidase and laccase gene-specific sequences. Mycological Research 109: 115–124. 10.1017/S0953756204001376 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Promputtha I, Lumyong S, Lumyong P, McKenzie EC, Hyde KD. (2002) Fungal succession on senescent leaves of Manglietia garrettii in Doi Suthep-Pui National Park, northern Thailand. Fungal Diversity 10: 89–100. [Google Scholar]
- Promputtha I, Mckenzie EH, Tennakoon DS, Lumyong S, Hyde KD. (2017) Succession and natural occurrence of saprobic fungi on leaves of Magnolia liliifera in a tropical forest. Cryptogamie Mycologie 38: 213–225. 10.7872/crym/v38.iss2.2017.213 [DOI] [Google Scholar]
- Purahong W, Wubet T, Lentendu G, Schloter M, Pecyna MJ, Kapturska D, Hofrichter M, Krüger D, Buscot F. (2016) Life in leaf litter: novel insights into community dynamics of bacteria and fungi during litter decomposition. Molecular Ecology 25: 4059–4074. 10.1111/mec.13739 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2012) FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/ [accessed 10 March 2020]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Rehner SA, Samuels GJ. (1994) Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625–634. 10.1016/S0953-7562(09)80409-7 [DOI] [Google Scholar]
- Rehner S. (2001) Primers for Elongation Factor 1-α (EF1-α). http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf
- Robertson GP, Paul EA. (1999) Decomposition and soil organic matter dynamics. In: Sala OE, Jackson RB, Mooney HA, Howarth RW. (Eds) Methods of Ecosystem Science.Springer, New York, 104–116. 10.1007/978-1-4612-1224-9_8 [DOI]
- Shoemaker RA, Babcock CE. (1989) Phaeosphaeria. Canadian Journal of Botany 67: 1500–1599. 10.1139/b89-199 [DOI] [Google Scholar]
- Spegazzini C. (1909) Mycetes Argentinenses. Series IV. Anales del Museo Nacional de Historia Natural Buenos Aires. Ser. 3, 12: 257–458.
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic biology 57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift MJ, Heal OW, Anderson MM. (1979) Decomposition in Terrestrial Ecosystems. Blackwell Scientific Publications, Oxford.
- Swofford DL. (2002) PAUP: phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Associates, Sunderland.
- Tennakoon DS, Hyde KD, Phookamsak R, Wanasinghe DN, Camporesi E, Promputtha I. (2016) Taxonomy and phylogeny of Juncaceicola gen. nov.(Phaeosphaeriaceae, Pleosporinae, Pleosporales). Cryptogamie Mycologie 37: 135–156. 10.7872/crym/v37.iss2.2016.135 [DOI] [Google Scholar]
- Tennakoon DS, Jeewon R, Gentekaki E, Kuo CH, Hyde KD. (2019) Multi-gene phylogeny and morphotaxonomy of Phaeosphaeria ampeli sp. nov. from Ficus ampelas and a new record of P. musae from Roystonea regia. Phytotaxa 406: 111–128. 10.11646/phytotaxa.406.2.3 [DOI] [Google Scholar]
- Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN, Liu JK, Bhat DJ, Jones EBG, McKenzie E, Camporesi E, Bulgakov TS, Doilom M, Santiago AM, Das K, Manimohan P, Gibertoni TB, Lim YW, Ekanayaka AH, Thongbai B, Lee HB, Yang J, Kirk PM, Sysouphanthong P, Singh SK, Boonmee S, Dong W, Raj KN, Latha KP, Phookamsak R, Phukhamsakda C, Konta S, Jayasiri SC, Norphanphoun C, Tennakoon D, Li J, Dayarathne MC, Perera RH, Xiao Y, Wanasinghe DN, Senanayake IC, Goonasekara ID, Silva NI, Mapook A, Jayawardena RS, Dissanayake AJ, Manawasinghe IS, Chethana KW, Luo Z, Hapuarachchi KK, Baghela A, Soares AM, Vizzini A, Meiras-Ottoni A, Mešić A, Dutta AK, Souza CA, Richter C, Lin C, Chakrabarty D, Daranagama DA, Lima DX, Chakraborty D, Ercole E, Wu F, Simonini G, Vasquez G, Silva GA, Plautz HL, Ariyawansa HA, Lee HS, Kušan I, Song J, Sun J, Karmakar J, Hu K, Semwal KC, Thambugala KM, Voigt K, Acharya K, Rajeshkumar KC, Ryvarden L, Jadan M, Hosen MI, Mikšík M, Samarakoon MA, Wijayawardene NN, Kim NK, Matočec N, Singh PN, Tian Q, Bhatt RP, Oliveira RJ, Tulloss RE, Aamir S, Kaewchai S, Marathe SD, Khan S, Hongsanan S, Adhikari S, Mehmood T, Bandyopadhyay TK, Svetasheva TY, Nguyen TT, Antonín V, Li W, Wang Y, Indoliya Y, Tkalčec Z, Elgorban AM, Bahkali AH, Tang A, Su H, Zhang H, Promputtha I, Luangsa-Ard J, Xu J, Yan J, Kang JC, Stadler M, Mortimer PE, Chomnunti P, Zhao Q, Phillips AJ, Nontachaiyapoom S, Wen T, Karunarathna SC. (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 83: 1–261. 10.1007/s13225-017-0378-0 [DOI] [Google Scholar]
- Thambugala KM, Camporesi E, Ariyawansa HA, Phookamsak R, Liu ZY, Hyde KD. (2014) Phylogeny and morphology of Phaeosphaeriopsis triseptata sp. nov., and Phaeosphaeriopsis glaucopunctata. Phytotaxa 176: 238–250. 10.11646/phytotaxa.176.1.23 [DOI] [Google Scholar]
- Thambugala KM, Wanasinghe DN, Phillips AJL, Camporesi E, Bulgakov TS, Phukhamsakda C, Ariyawansa HA, Goonasekara ID, Phookamsak R, Dissanayake A, Tennakoon DS, Tibpromma S, Chen YY, Liu ZY, Hyde KD. (2017) Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 8: 697–796. 10.5943/mycosphere/8/4/13 [DOI] [Google Scholar]
- Tomilin BA. (1993) New species of Loculoascomycetes (fam. Phaeosphaeriaceae Barr.). Novosti Sistematiki Nizshikh Rasteniĭ 29: 69–73. [Google Scholar]
- Valenzuela-Lopez N, Sutton DA, Cano-Lira JF, Paredes K, Wiederhold N, Guarro J, Stchigel AM. (2017) Coelomycetous fungi in the clinical setting: morphological convergence and cryptic diversity. Journal of clinical microbiology 55: 552–567. 10.1128/JCM.02221-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Jones EBG, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y, Camporesi E, Bulgakov TS, Ekanayake AH, Perera RH, Samarakoon MC, Goonasekara ID, Mapook A, Li WJ, Senanayake IC, Li JF, Norphanphoun C, Doilom M, Bahkali AH, Xu JC, Mortimer PE, Tibell L, Tibell S, Karunarathna SC. (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89: 1–236. 10.1007/s13225-018-0395-7 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yang CL, Xu XL, Wanasinghe DN, Jeewon R, Phookamsak R, Ying-Gao L, Li-Juan L, Hyde KD. (2019) Neostagonosporella sichuanensis gen. et sp. nov. (Phaeosphaeriaceae, Pleosporales) on Phyllostachys heteroclada (Poaceae) from Sichuan Province, China. MycoKeys 46: 119–150. 10.3897/mycokeys.46.32458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JW, Yeh YH, Kirschner R. (2016) A new endophytic species of Neostagonospora (Pleosporales) from the coastal grass Spinifex littoreus in Taiwan. Botany 94: 593–598. 10.1139/cjb-2015-0246 [DOI] [Google Scholar]
- Yuan ZQ. (1994) Barria, a new ascomycetous genus in the Phaeosphaeriaceae. Mycotaxon 51: 313–316. [Google Scholar]
- Zhang Y, Schoch CL, Fournier J, Crous PW, De Gruyter J, Woudenberg JHC, Hirayama K, Tanaka K, Pointing SB, Spatafora JW, Hyde KD. (2009) Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Studies in Mycology 64: 85–102. 10.3114/sim.2009.64.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Crous PW, Schoch CL, Hyde KD. (2012) Pleosporales. Fungal Diversity 52: 1–225. 10.1007/s13225-011-0117-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JF, Liu JK, Jeewon R, Wanasinghe DN, Liu ZY. (2019) Fungi from Asian Karst formations III. Molecular and morphological characterization reveal new taxa in Phaeosphaeriaceae. Mycosphere 10: 202–220. 10.5943/mycosphere/10/1/3 [DOI] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. MBC genomics 3: 1–4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.