Abstract
Merkel cell carcinoma (MCC) is a rare primary cutaneous neoplasm of neuroendocrine carcinoma of the skin. About 80% of the MCC occurs due to Merkel cell polyomavirus (MCPyV) and 20% of the tumors usually occur due to severe UV exposure which is a more aggressive type of MCC. It tends to have an increased incidence rate among elderly and immunosuppressed individuals. On therapeutic level, sub-classification of MCC through molecular subtyping has emerged as a promising technique for MCC prognosis. In current study, two consistent distinct molecular subtypes of MCCs were identified using gene expression profiling data. Subtypes I MCCs were associated with spliceosome, DNA replication and cellular pathways. On the other hand, genes overexpressed in subtype II were found active in TNF signalling pathway and MAPK signalling pathway. We proposed different therapeutic targets based on subtype specificity, such as PTCH1, CDKN2A, AURKA in case of subtype I and MCL1, FGFR2 for subtype II. Such findings may provide fruitful knowledge to understand the intrinsic subtypes of MCCs and the pathways involved in distinct subtype oncogenesis, and will further advance the knowledge in developing a specific therapeutic strategy for these MCC subtypes.
Keywords: Merkel cell carcinoma, Molecular subtype, Gene expression, Subtype specific treatment
Highlights
-
•
Merkel cell carcinoma (MCC) a rare and highly aggressive neuroendocrine carcinoma of the skin
-
•
Sub-classification of MCC through molecular subtyping
-
•
Identification of two distinct molecular subtypes of MCCs using gene expression profiling data
-
•
Classification of different therapeutic targets based on subtype specificity
Introduction
Merkel cell carcinoma (MCC), though with a low occurrence rate is a lethal form of skin cancer. Merkel cell carcinoma resembles Merkel cells, somatosensory cells present in epidermis's basal layer. Merkel cells are believed to act as slow mechanoreceptors [1]. MCC has become the centre of attention due to the discovery of Merkel cell polyomavirus and rapid increase in occurrence rate [tripled since 1986] [2]. MCC's incident rate is higher in older people and occurs to those parts of the skin which are exposed to the sun, suggesting its occurrence related to UV exposure [3,4]. Oxygen radicals produced due to UV-A exposure cause DNA damage by chromosomal breakage resulting in loss or gain of chromosomal material [5]. MCC, though fully understood yet, is also thought to contribute towards the development of other malignancies such as malignant melanoma and chronic lymphocytic leukemia etc. [6,7] Patients with MCC developed in neck and head, and those with positive lymph nodes are initially subjected to local excision, biopsy of sentinel nodes and are later provided radiotherapy [8]. But in case of MCC metastasis, chemotherapy such as cytotoxic chemotherapy, is found to have less efficacy in most of the cases. However, the major drawback of chemotherapeutics is the high reoccurrence rate of the disease. Thus, various alternative therapies are searched to overcome the shortfalls of these traditional therapeutics. One of the alternatives to the traditional therapies is immuno-therapeutics, in which monoclonal antibodies are used to treat stage IV MCC [9,10]. Therefore, molecular target therapies are considered a viable option for the immunocompromised patients with failed track record against immunotherapies. Molecular subtyping based on gene expression patterns has allowed researchers to develop tools for the identification of molecular events associated subtypes in various tumors such as colon, lung, ovarian and prostate cancer, and provided somewhat success in prognosis and treatment [[11], [12], [13], [14], [15], [16], [17], [18]]. To date, limited data is available to correlate MCC with distinct molecular subtypes. In current study, two distinct molecular subtypes were identified through gene expression profile data. Various therapeutic genes and pathways were identified in the sample molecular subtypes which can be helpful in unfolding the novel target therapies for MCCs based on MCC molecular subtypes.
Material and methods
Determination of subtypes in MCC
Gene expression Omnibus (GEO) was used to collect the Expression profiling data of Merkel cell carcinoma cases. Dataset of GSE39612 which contains 30 MCC cases [ 19] and the second Dataset of GSE22396 having 35 MCC cases [20] were used to determine the molecular subtypes of MCC. For the identification of MCC molecular subtypes, each dataset was filtered with standard deviation and subsequently transformed on the basis of gene-based centring, then transformed dataset was separately ran on R package of Consensus Clustering Plus [21], having distance factors of 1-pearson correlation and 80% gene resampling, maximum evaluated k of 12, and agglomerative hierarchical clustering algorithm over 1000 iterations. The accuracy of Consensus Clustering Plus was checked by calculation of silhouette width (R package clustering) [22].
Reproducibility measurement of the MCC subtypes
Reproducibility of MCC molecular subtypes between the cohorts of GSE39612 and GSE22396, was determined through Subclass Mapping (SubMap) of the Gene Pattern. Subclass Mapping was carried out with parameters of (num. marker. genes = 300, num. perm = 1000 and num. perm. fisher = 1000) [23].
Gene ontology and gene set enrichment (GSEA) analysis
For the examination of Subtype specific genes, SAM [24] was performed with false discovery rate (FDR) of 0.05. The David Bioinformatics Resources online version 6.7 (https://david.ncifcrf.gov/) was preferred to emulate the GO and KEGG pathways. Whereas GSEA [25] was used to examined gene expression patterns and pathways of each subtypes. Potential therapeutic genes of each subtype of MCCs were explored through Target V2 database (http://www.broadinsitute.org/ccancer/cga/target).
Statistical analysis
Kaplan-Meier plots were plotted to calculate the survival curve. Whereas Chi-square and Fisher's exact tests were performed to evaluate the statistical significance of association between subtypes and clinical factors, and those having p value lower than 0.05 was considered as significant.
Results
Consensus clustering identifies two distinct MCC molecular subtypes
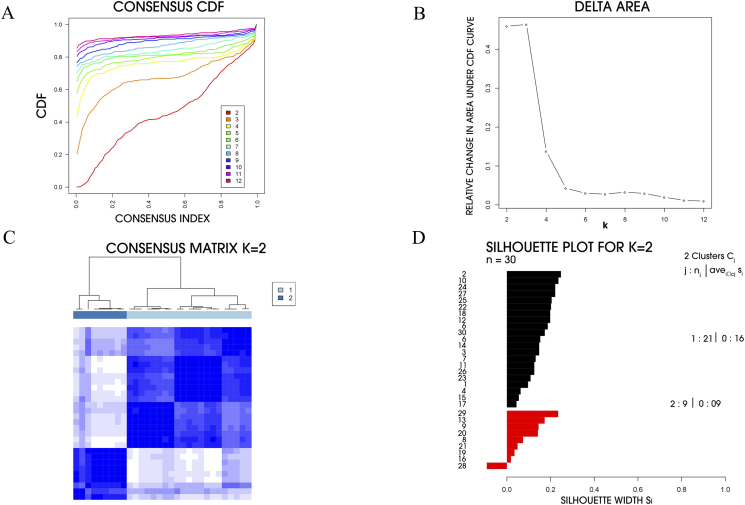
MCC molecular subtypes were determined through Consensus clustering by checking the genes with high variety of expression levels in MCC cohort. Two molecular subtypes were identified in GSE39612 by choosing the optimal number of clusters with highest increase in the area under curve of empirical cumulative distribution (CDF) (Fig. 1, A-C). Silhouette width determination was used to confirm the subtype assignment confidence and those found with positive silhouette value were selected and subsequently analyzed. Out of 30 MCC cases, 29 cases (core samples) were found to have positive silhouette values, 21 out of these 29 cases belong to subtype I and subtype II related cases were found to be 9 in number (Fig. 1D).
Fig. 1.
Identification of two molecular subtypes of MCC in GSE39612. (A) Empirical cumulative distribution plot determines the optimal number of MCC molecular subtypes. (B) Relative increase in the area under the CDF curve along with increasing assumed number of molecular subtypes. (C) Consensus clustering matrix of MCC samples using two molecular subtypes. (D) Silhouette analysis of MCC samples based on the assignment from Consensus Clustering.
Validation of subtypes in the independent dataset
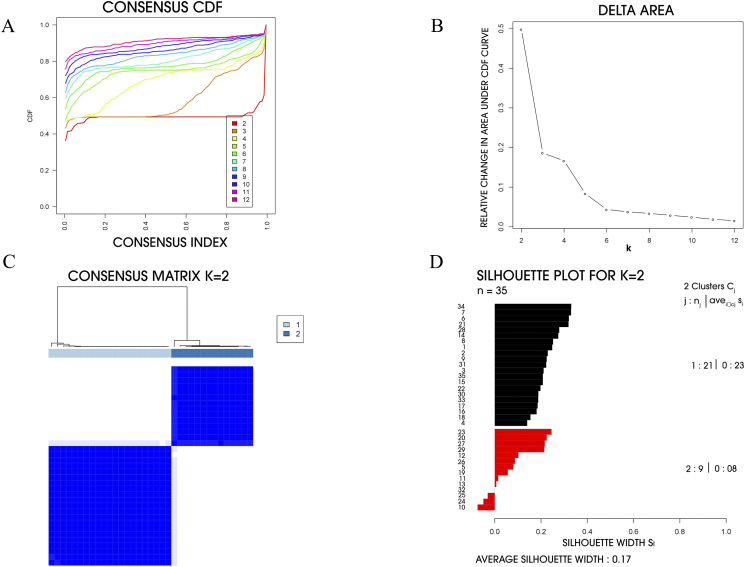
In order to confirm the validity of MCC molecular subtypes for clinic use, additional independent dataset (GSE22396) containing 35 cases was obtained from GEO as an independent validation cohort. As a result, two different molecular subtypes were observed through Consensus Clustering analysis as well (Fig. 2). Silhouette analysis revealed that 31 out of 35 samples had positive silhouette values and were defined as core samples. In 31 silhouette positive samples 21 belong to subtype I and 10 belong to subtype II.
Fig. 2.
Identification of two molecular subtypes of MCC in GSE22396. (A) Empirical cumulative distribution plot determines the optimal number of MCC molecular subtypes. (B) Relative increase in the area under the CDF curve along with increasing assumed number of molecular subtypes. (C) Consensus clustering matrix of MCC samples using two molecular subtypes. (D) Silhouette analysis of MCC samples based on the assignment from Consensus Clustering.
Reproducible molecular subtypes in the two cohorts
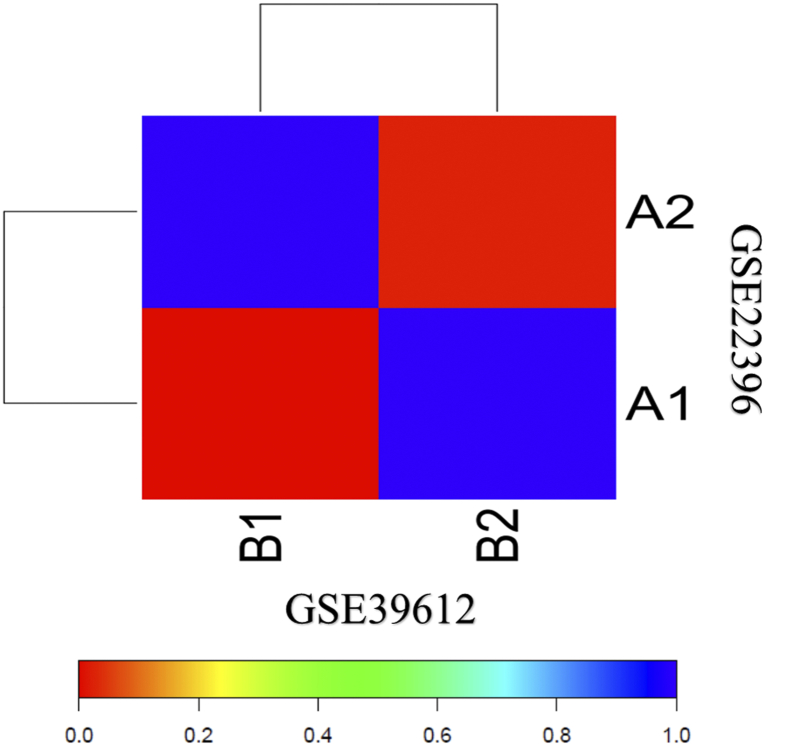
SubMap analysis was used to measure whether molecular subtypes from the two datasets can be recapitulated or not. Results suggested the significant correlation between A1-A2 subtypes and B1-B2 subtypes of GSE22396 and GSE39612 respectively, indicating the presence of two consistent different MCC molecular subtypes with distinct gene expression patterns (Fig. 3).
Fig. 3.
Association in the SubMap matrix between the subtypes of two independent dataset GSE3916 and GSE22396 showing the significant correlation. The correlation significance was denoted by FDR-corrected p-value.
Molecular subtypes and their relation with clinical symptoms
The relationship between molecular subtypes and clinical characteristics was analyzed based on GSE39612 (Supplementary Table S1). MCC molecular subtype I presented significantly high rate in upper site (Head and Neck) tumor (10/30, 33.33%) as compared to MCC subtype II (1/30, 3%) (p = 0.0468⁎). In this dataset of MCC, the number of female patients (46.6%) is higher than the male (43.3%) patients. Between the two molecular subtypes a significant difference was observed among the patients on the basis of their age group (p = 0.0273⁎). The mean age of subtype I was 79 years old which was higher than subtype II patients which was 70 years old. The median time of overall survival (OS) for subtype I MCCs was found to be 13.5 months which was lower than the patients with subtype II MCCs who had 16.5 months, although the difference is not significant (p = 0.507). In MCC subtype I, 38% of the patients were found to be MCPyV positive while the 57.1% were MCPyV negative. Whereas in subtype II of MCC 25% of the patients were MCPyV positive and 50% were MCPyV negative (p = 0.483). In addition, no remarkable difference was observed in the Kaplan-Meier plots (survival curve analysis) (Supplementary Table S1).
Functional enrichment analysis of MCC subtype-specific genes
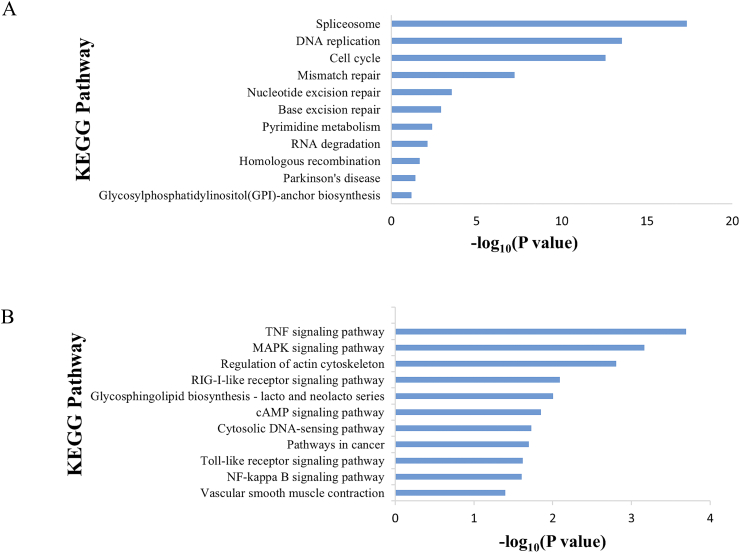
By SAM analysis of GSE39612, 28,912 genes were found to be significantly differentially expressed between the two MCC molecular subtypes. According to the results, 10,920 out of 28,912 genes were specifically overexpressed in subtype I MCCs as compared to subtype II MCCs, whereas, the remaining 17,992 genes were found overexpressed in subtype II MCCs (Supplementary Table S2). GO and KEGG analysis were performed to obtain the biological information of the subtypes. Top 1000 most overexpressed genes in each MCC subtype were selected. Out of total, 145 biological processes and 19 KEGG pathways were found to be enriched in subtype I MCCs, whereas 139 biological processes and 31 KEGG pathways were found to be enriched in subtype II MCCs. Remarkably, 43 (4.3%) genes of the subtype I MCC specific top 1000 genes were found to be related to DNA replication pathway (Supplementary Table S3). Additional pathways found enriched in subtype I MCCs were spliceosome, Mismatch repair and Cell cycle etc. (Fig. 4A). On the other hand, genes overexpressed in subtype II MCCs were related with distinct biological pathways such as TNF signalling pathway, MAPK signalling, myeloma formation and other regulatory pathways etc. (Fig. 4B).
Fig. 4.
Pathways enriched in each MCC subtypes. (A) KEGG pathways in subtype I. (B) KEGG pathways in subtype II.
Potential clinical implication of MCC subtyping
The basic aim behind the determination of MCC molecular subtypes is to develop various therapeutic routes specific towards distinct subtypes and their further use in clinical studies and discourses. For the determination of therapeutic modules related to each MCC subtype, overexpressed genes based on subtype specificity were compared with target Database (http://www.braodinsitute.org/cancer/cga/target) containing gene targets and functional inhibitors [26], for further studies to be carried out on the gene targets to convert them into potential clinical targets [[27], [28], [29]].
In total, 11 genes were found to be specific against molecular subtypes and would benefit subtype-specific MCC patients (Table 1). Subtype I MCCs contains PTCH1, SMARCA4, CDKN2A, AURKA and BRCA1, whereas subtype II MCCs contains 6 overexpressed genes namely MCL1, FGFR2, BRD4, MET, SMO and PDGFRA.
Table 1.
Gene overexpressed in each MCC molecular subtype.
| Gene overexpressed | Examples of potential therapeutic agents | |
|---|---|---|
| Subtype I | PTCH1 | Vismodegib, hedgehog inhibitors |
| SMARCA4 | HDAC | |
| CDKN2A | CDK4/6 inhibitors | |
| AURKA | AURKA inhibitors | |
| BRCA1 | PARP inhibitor | |
| MCL1 | Tubulins | |
| FGFR2 | FGFR inhibitors | |
| Subtype II | BRD4 | HDAC inhibitors Bromodomain inhibitors |
| MET | Gefitinib, erlotinib, EGFR inhibitors Crizotinib, MET inhibitors |
|
| SMO | Vismodegib, hedgehog inhibitors | |
| PDGFRA | Imatinib | |
Discussion
MCC is an aggressive type of tumor found in the skin with higher metastasis, recurrence, and mortality rate. Since last two decades, the incidences of MCC have nearly tripled. Overall, 5-year survival rate of MCC patients is almost 30–60% [30]. Advanced age, UV exposure and immunosuppression increase the risk of MCC [31]. Usual course of treatment is radiation therapy followed by wide surgical excision for most of the cases, but in-case of metastasis and positive lymph nodes, only chemotherapy is used [[32], [33], [34]].
The survival rate of MCC patients is extended if it is diagnosed early. Molecular subtyping of tumors based on gene expression profiling has guided subtype-specific diagnosis, prognosis, and aided in developing subtype targeted therapies [21]. The clinical trial of Herceptin treatment in breast cancer is an ample example of subtype targeted therapies. Wherein, HER2-negative breast cancer patients did not get any benefit from Herceptin treatment yet HER2-positive breast cancer patients responded very well and benefited from this treatment [35]. Therefore, analysing the molecular heterogeneity of MCC is posed to provide deep knowledge of genes and pathways involved in MCC. It will also provide the new opportunities to target subtype specific patients of MCC.
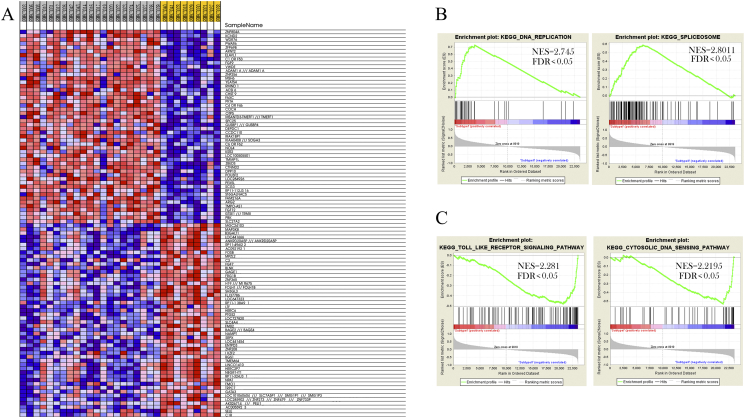
In our study, we have identified two distinct MCC molecular subtypes. Gene ontology and gene set enrichment analysis revealed distinct gene signatures in both subtypes of MCC. Spliceosome, Cell cycle, DNA replication and Mismatch repair etc., were the pathways over-expressed in subtype I MCCs (Fig. 4) along with the overexpression of DEPDC1, ELAVL and OIP5 genes in subtype I (Fig. 5). DEPDC1 is a cell cycle related gene in the bladder cancer [[36], [37], [38], [39]]. Dysregulation of DEPDC1 plays a significant role in the regulation of cell cycle, motility and progression of nasopharyngeal carcinoma [[40], [41], [42]], as well as in other human cancers (colorectal cancer, hepatocellular carcinoma and glioblastoma) [[43], [44], [45]]. Whereas, ELAVL1 gene encodes HuR protein. The important function of HuR is for mRNA stability. High levels of cytoplasmic HuR were observed in numbers of various cancers [[46], [47], [48], [49]]. For instance, in glioma, it was found involved in tumor growth and in the onset of drug resistance [[50], [51], [52], [53], [54], [55], [56], [57]]. A protein coding gene, Opa interacting protein 5 (OIP5) belongs to cancer/testis antigens (CTAs) [58]. OIP5 is found involved in different biological processes of tumor [59], Gong et al. found overexpression of OIP5 gene in clear renal cell carcinoma [60]. On the basis of above gene expression and cellular pathways found in subtype I MCCs of this cohort, potential therapies based on established roles of such determinants can be developed for subtype I MCC patients. In contrast, gene ontology and gene set enrichment analysis revealed the overexpressed pathways found in subtype II include TNF signalling pathway, MAPK signalling pathway, Pathways in cancer (Supplementary Table S4). The expression, repression and re-expression cycle of genes like H19 (Human Long Noncoding) was found to be an important oncogenic factor in several cancer tissues such as bladder cancer and hepatocellular carcinoma etc. [[61], [62], [63], [64], [65]] Whereas, expression pattern of RGS1 gene plays an important role in pathogenesis of the different malignancies and regulation of chemokine induced signalling of B cells [[66], [67], [68], [69], [70], [71]]. Overexpression of these genes in subtype II MCCs and their roles in other cancers may open new doors of opportunities to understand the patients of subtype II MCC.
Fig. 5.
GSEA reveals different gene expression signature in distinct MCC molecular subtypes. (A) Representing different gene expression patterns in subtype I and subtype II. Red, overexpressed genes; blue, down expressed genes. (B) GSEA shows the activity of DNA replication and Spliceosome pathways in subtype I. (C) GSEA demonstrated the activity of Toll like receptor signalling and cytosolic DNA sensing pathways in subtype II. Whereas NES is denoting normalized enriched score and FDR is denoting false discovery rate.
We also identified a number of known target genes and their potential therapeutic agents using TARGET V2 database in subtype I and II MCCs. These genes include PTCH1, CDKN2A, AURKA and BRCA1 in subtype I, and MCL1 in subtype II (Table 1). Cellular pathways such as Hh pathway is involved in the formation of organs, tissues and motor neurons [72]. Under biological conditions, the Hh signalling pathway is regulated through a transmembrane receptor called Patched 1 (PTCH1). Hh receptor Ptch1 is found to be overexpressed in many types of cancers such as Breast, prostate, melanoma, lung cancer and myeloid leukemia [[73], [74], [75], [76], [77]]. Interestingly, as a novel small molecule inhibitor of Hh, vismodegib was found actively involved in generation of anti-tumor response in advanced Basal cell carcinoma and got approval by the US FDA priority review program on January 30, 2012 [[78], [79], [80], [81]]. However, in another study, Carroll et al. suggested that Hh inhibition could not be an active therapy for MCC [82]. In current study, we found that more than 60% patients of both datasets GSE39612 and GSE22396 belong to subtype I of MCC (Fig. 1D, Fig. 2D and Supplementary Table S1). Our results revealed the importance of PTCH1 inhibitors in MCC subtype I (Table 1). Similar outcome was also reported in two other studies indicating the importance of Hh inhibitors in MCC. They observed the presence of amplified Hh pathway components in MCC tumors and proposed the importance of Hh pathway in pathogenesis of MCC [83]. Which further warrant the use of Hh inhibitors in the treatment of MCC [84]. We have also identified Histone deacetylase inhibitors (HDACi) in patients of subtype I MCC. Function of HDACis is to upregulate the low HLA class-I expression in MCC patients to contain the resistance of MCC towards PD-1/PD-L1 inhibition and low influx of CD8+ T which allows the tumor to avoid host immune response [85]. Thus, possible use of PTCH1 and HDACi inhibitors might be beneficial for the patients of subtype I MCC. Moreover, extension of antiapoptotic BCL2 family proteins (MCL1, BCL2, and BCL-xL) has been reported in relation to chemotherapy resistance in various types of cancers [86]. MCL1 is an influencing therapeutic target in cancer. Various gene knockdown and pharmacological inhibition models have demonstrated the potential of MCL1 as a critical pro-survival factor in tumors such as AML, multiple myeloma, B-cell lymphomas, and breast cancer [[87], [88], [89], [90], [91], [92]]. Microtubules also play a fundamental role in various cellular processes and are favourite antitubulin target chemotherapies in different cancers. Microtubule inhibitory target agents such as Vinscristine and Taxol are widely used in antitubulin chemotherapeutics. They are able to cause mitotic arrest in cells and ultimately lead towards apoptosis. MCL1 being an important regulator in the process of apoptosis is a key target of antitubulin chemotherapies [93]. In one study, 10 out of 11 MCC xenografts, ABT-263 successfully inhibited the multiple pro-survival Bcl-2 proteins and strongly support the therapeutic effect of these molecules MCC targeting [94]. Another pan-active BCL2 inhibitor, Sabutoclax was found capable of targeting all 5 anti-apoptotic proteins. A1210477 and S63845 also followed up the promise that MCL1 inhibition seems to offer as MCL1 specific inhibitors [95]. According to our gene expression analysis, we found that MCL1 inhibitor played a significant role in subtype II MCC under the influence of BCL-2 family proteins. Similarly, one more prospective inhibitor against targeted MCC called Imatinib was also identified. Imatinib inhibitor therapy is considered as an efficient therapy for patients with MCC metastasis and complications of inoperability [96]. These results suggested that patients of Subtype II MCC might benefit from these inhibitors.
In conclusion, we defined distinct molecular subtypes of MCCs. Our findings provided subtype-specific mechanisms underlying tumorigenesis and tumor progression. These outcomes might help in better targeting of the disease and developing an individualized medicine through subtype-specific manner for MCCs patients.
Author contributions statement
Xiangqian Guo: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Resources, Data Curation, Writing - Original Draft, Writing - Review & Editing, Supervision, Project administration, Funding acquisition. Umair Ali Khan Saddozai: Validation, Formal analysis, Investigation, Data Curation, Writing - Original Draft, Writing - Review & Editing, Visualization. Fengling Wang: Formal analysis. Zhang Lu: Formal analysis. Xinying Ji: Resources, Writing - Review & Editing, Supervision, Project administration. Yongqiang Li: Resources, Supervision, Project administration. Muhammad Usman Akbar: Writing - Original Draft, Writing - Review & Editing. Wan Zhu: Writing - Original Draft, Writing - Review & Editing. Yu Cheng: Writing - Review & Editing.
The following are the supplementary data related to this article.
Clinopathological characteristics of GSE39612 (N = 30).
Results of SAM analysis between different subtypes of MCC in GSE39612. Data is representing over expression of gene in two subtypes in positive and minus value.
(1). Gene were ordered according the SAM significance, gene with the positive value were overexpressed in subtype II of MCC whereas minus value genes were overexpressed in subtype I.
(2). Significance increased with decreased of absolute values of ranks from 10 to 1 or −10 to 1. Rank 1 or −1 genes are the most significant of SAM results.
Biological process enriched in each subtypes of MCC.
Enriched KEGG pathways in each subtypes of MCC.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the program for Innovative Talents of Science and Technology in Henan Province (No. 18HASTIT048). The funding bodies were not involved in the study design, data collection, analysis and interpretation of data, or writing of this manuscript.
Contributor Information
Yongqiang Li, Email: liyongqiang@vip.henu.edu.cn.
Xinying Ji, Email: 10190096@vip.henu.edu.cn.
Xiangqian Guo, Email: xqguo@henu.edu.cn.
References
- 1.Halata Z., Grim M., Bauman K.I. Friedrich Sigmund Merkel and his Merkel cell, morphology, development and physiology; review and new results. Anat. Rec. 2003;271:225–239. doi: 10.1002/ar.a.10029. [DOI] [PubMed] [Google Scholar]
- 2.Zackheim H.S., Amin S., Kashani-Sabet M., McMillan A. Prognosis in cutaneous T-cell lymphoma by skin stage; long term survival in 489 patients. J. Am. Acad. Dermatol. 1999;40:418–425. doi: 10.1016/s0190-9622(99)70491-3. [DOI] [PubMed] [Google Scholar]
- 3.Agelli C., Rollison B. The etiology and epidemiology of merkel cell carcinoma. Curr. Probl. Cancer. 2010;34:14–37. doi: 10.1016/j.currproblcancer.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Kukko H., Bohling T., Koljonen V., Tukiainen E., Haglund C., Pokhrel A., Sankila R., Pukkala E. Merkel cell carcinoma - a population-based epidemiological study in Finland with a clinical series of 181 cases. Eur.J.Cancer. 2012;48:737–742. doi: 10.1016/j.ejca.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Popp S., Waltering S., Herbst C., Moll I., Boukamp P. UV-B-type mutations and chromosomal imbalances indicate common pathways for the development of Merkel and skin squamous cell carcinomas. Int. J. Cancer. 2002;99:352–360. doi: 10.1002/ijc.10321. [DOI] [PubMed] [Google Scholar]
- 6.Howard R.A., Dores G.M., Curtis R.E., Anderson W.F., Travis L.B. Merkel cell carcinoma and multiple primary cancers. Cancer Epidemiol. Biomark. Prev. 2006;15:1545–1549. doi: 10.1158/1055-9965.EPI-05-0895. [DOI] [PubMed] [Google Scholar]
- 7.Goessling W., McKee P.H., Mayer R.J. Merkel cell carcinoma. J. Clin. Oncol. 2002;20:588–598. doi: 10.1200/JCO.2002.20.2.588. [DOI] [PubMed] [Google Scholar]
- 8.Hughes M.P., Hardee M.E., Cornelius L.A., Hutchins L.F., Becker J.C., Gao L. Merkel cell carcinoma: epidemiology, target, and therapy. Curr Dermatol Rep. 2014;3:46‐53. doi: 10.1007/s13671-014-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaufman H.L., Russell J., Hamid O., Bhatia S., Terheyden P., Angelo S.P. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016;17:1374–1385. doi: 10.1016/S1470-2045(16)30364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman H.L., Russell J.S., Hamid O., Bhatia S., Terheyden S.P., Angelo K.C., Shih C., Lebbe M., Milella I. Brownell. Updated efficacy of avelumab in patients with previously treated metastatic Merkel cell carcinoma after 1 year of follow-up: JAVELIN Merkel 200, a phase 2 clinical trial. J. Immunother.Cancer. 2018;6 doi: 10.1186/s40425-017-0310-x. 017-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins M.J., Baselga J. Targeted therapies for breast cancer. J. Clin. Invest. 2011;121:3797–3803. doi: 10.1172/JCI57152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marisa L., de Reynies A., Duval A.J., Selves J., Pierre M., Vescovo L., Etienne-Grimaldi M., Schiappa R., Guenot D., Ayadi M. Gene expression classification of colon cancer into molecular subtypes; characterization, validation and prognostic value. PLoS Med. 2013;10:1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akbani R., Akdemir K.C., Aksoy B.A. Cancer Genome Atlas Research Network. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F., Zhang Y., Parra E., Rodriguez J., Behrens C., Akbani R., Lu Y., Kurie J.M., Gibbons D.L., Mills G.B. Multiplatform-based molecular subtypes of non–small cell lung cancer. Oncogene. 2017;36:1384–1393. doi: 10.1038/onc.2016.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo X., Jo V.Y., Mills A.M., Zhu S.X., Lee C.H., Espinosa I., Nucci M.R., Forgo S. Varma E., Hastie T. Clinically relevant molecular subtypes in leiomyosarcoma. Clin. Cancer Res. 2015;21:3501. doi: 10.1158/1078-0432.CCR-14-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An Y., Wang H., Jie J., Tang Y., Zhang W., Ji S., Guo X. Identification of distinct molecular subtypes of uterine carcinosarcoma. Oncotarget. 2017;8:15878–15886. doi: 10.18632/oncotarget.15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An Y., Wang S., Li S., Zhang L., Wang D., Zhu S., Li Y., Chen W. Distinct molecular subtypes of uterine leiomyosarcoma respond differently to chemotherapy treatment. BMC Cancer. 2017;17:639. doi: 10.1186/s12885-017-3568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F., Zhongyi Y., Jiajia L., Xin J., Dang Y., Sun X., An Y., Qing Y., Zhu W. Gene expression profiling reveals distinct molecular subtypes of oesophageal squamous cell carcinoma in Asian population. Neoplasia. 2019;21:571–581. doi: 10.1016/j.neo.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms P.W., Patel R.M., Verhaegen M.E., Giordane T.J., Nash K.T., Johnson C.N., Daignault S., Thomas D.G., Gudjonsson J.E., Elder J.T. Distinct gene expression profiles of viral and non-viral associated Merkel cell carcinoma revealed by transcriptome analysis. J Invest Dermatol. 2013;133:936–945. doi: 10.1038/jid.2012.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulson K.G., Tegeder A., Willmes C., Lyer J.G., Affanasiev O.K., Schrama D., Koba S., Thibodeau R., Nagase K., Simonson W.T. Downregulation of MHC-I expression is prevalent but reversible in Merkel cell carcinoma. Cancer Immunol Res. 2014;2:1071–1079. doi: 10.1158/2326-6066.CIR-14-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkerson M.D., Hayes D.N. Consensus Cluster Plus; a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rousseeuw P.J. Silhouettes; a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. [Google Scholar]
- 23.Hoshida Y., Brunet J.P., Tamayo P., Golub T.R., Mesirov J.P. Subclass mapping; identifying common subtypes in independent disease data sets. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub R., Lander E.S. Gene set enrichment analysis; a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Allen E.M., Wagle N., Stojanov P., Perrin D.L., Cibulskis K., Marlow S., Jane-Valbuena J., Friedrich D.C., Kryukov G., Carter S.L. Whole-exome sequencing and clinical interpretation of formalin fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroni M., Veronese S., Benvenuti S., Benvenuti S., Marrapese G., Sartore-Bianchi A., Nicolantono F.D., Gambacorta M., Siena S., Bardeli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to anti-EGFR treatment in colorectal cancer; a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y., Pietenpol J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J. EGFR mutations in lung cancer; correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 30.Bichakjian C.K., Lowe L., Lao C.D., Sandler H.M., Bradford C.R., Johnson T.M. Merkel cell carcinoma; critical review with guidelines for multidisciplinary management. Cancer. 2007;110:1–12. doi: 10.1002/cncr.22765. [DOI] [PubMed] [Google Scholar]
- 31.Becker J.C. Merkel cell carcinoma. Ann. Oncol. 2010;21:81–85. doi: 10.1093/annonc/mdq366. [DOI] [PubMed] [Google Scholar]
- 32.Jabbour J., Cumming R., Scolyer R.A., Hruby G., Thompson J.F., Lee S. Merkel cell carcinoma; assessing the effect of wide local excision, lymph node dissection and radiotherapy on recurrence and survival in early-stage disease—results from a review of 82 consecutive cases diagnosed between 1992 and 2004. Ann. Surg. Oncol. 2007;14:1943–1952. doi: 10.1245/s10434-006-9327-y. [DOI] [PubMed] [Google Scholar]
- 33.Iyer J., Parvathaneni U., Gooley T., Natalie J., Miller E., Markowitz A., Blom C., Lewis W., Doumani R.F., Parvathaneni K. Single-fraction radiation therapy in patients withmetastatic Merkel cell carcinoma. Cancer Medicine. 2015;4:1161–1170. doi: 10.1002/cam4.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desch L., Kunstfeld R. Merkel cell carcinoma; chemotherapy and emerging new therapeutic options. J. Skin Cancer. 2013;9:1–9. doi: 10.1155/2013/327150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piccart-Gebhart M.J., Procter M., Leyland-Jones B. Trastuzumab after adjuvant chemotherapy in HER2- positive breast cancer. N. Engl. J. Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 36.Kanehira M., Harada Y., Takata R., Shuin T., Miki T., Fujioka T., Nakamura Y., Katagiri T. Involvement of upregulation of DEPDC1 (DEP domain containing 1) in bladder carcinogenesis. Oncogene. 2007;26:6448–6455. doi: 10.1038/sj.onc.1210466. [DOI] [PubMed] [Google Scholar]
- 37.Harada Y., Kanehira M., Fujisawa Y., Takata R., Shuin T., Miki T., Fujioka T., Nakamura Y., Katagiri T. Cell-permeable peptide DEPDC1-ZNF224 interferes with transcriptional repression and oncogenicity in bladder cancer cells. Cancer Res. 2010;70:5829–5839. doi: 10.1158/0008-5472.CAN-10-0255. [DOI] [PubMed] [Google Scholar]
- 38.Obara W., Ohsawa R., Kanehira M., Takata R., Tsunoda T., Yoshida K., Takeda K., Katagiri T., Nakamura Y., Fujioka T. Cancer peptide vaccine therapy developed from oncoantigens identified through genome-wide expression profile analysis for bladder cancer. Jpn. J. Clin. Oncol. 2012;42:591–600. doi: 10.1093/jjco/hys069. [DOI] [PubMed] [Google Scholar]
- 39.Mi Y., Zhang C., Bu Y., Zhang Y., He L., Li H., Zhu H., Li Y., Lei Y., Zhu J. DEPDC1 is a novel cell cycle related gene that regulates mitotic progression. BMB Rep. 2015;48:413–418. doi: 10.5483/BMBRep.2015.48.7.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y., Shen J.K., Hornicek F.J., Kan Q., Duan Z. The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (CDK11) in human cancer. Oncotarget. 2016;7:40846–40859. doi: 10.18632/oncotarget.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evelyn C.R., Lisabeth E.M., Wade S.M., Haak A.J., Johnson C.N., Lawlor E.R., Neubig P.R. Small-Molecule Inhibition of Rho/MKL/SRF Transcription in Prostate Cancer Cells; Modulation of Cell Cycle, ER Stress and Metastasis Gene Networks. Microarrays (Basel) 2016;5:13. doi: 10.3390/microarrays5020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng X., Zhang C., Zhu L., Zhang L., Li H., He H., Mi Y., Wang Y., Zhu J., Bu Y. DEPDC1 is required for cell cycle progression and motility in nasopharyngeal carcinoma. Oncotarget. 2017;8:63605–63619. doi: 10.18632/oncotarget.18868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan S.G., Liao W.J., Yang J.J., Huang G.J., Huang Z.Q. DEP domain containing 1 is a novel diagnostic marker and prognostic predictor for hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2014;15:10917–10922. doi: 10.7314/apjcp.2014.15.24.10917. [DOI] [PubMed] [Google Scholar]
- 44.Miyata Y., Kumagai K., Nagaoka T., Kitaura K., Kaneda G., Kanazawa H. Clinicopathological significance and prognostic value of Wilms' tumor gene expression in colorectal cancer. Cancer Biomark. 2015;15:789–797. doi: 10.3233/CBM-150521. [DOI] [PubMed] [Google Scholar]
- 45.Stangeland B., Mughal A.A., Grieg Z., Sandberg C.J., Joel M., Nygård S. Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells. Oncotarget. 2015;6:26192–26215. doi: 10.18632/oncotarget.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kakuguchi W., Nomura T., Kitamura T., Otsuguro S., Matsushita K., Sakaitani M., Maenaka K., Tei Suramin. screened from an approved drug library, inhibits HuR functions and attenuates malignant phenotype of oral cancer cells. Cancer Medicine. 2018;7:6269–6280. doi: 10.1002/cam4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koljonen V., Bohling T., Haglund C., Ristimaki A. Expression of HuR in Merkel cell carcinoma and in normal skin. J. Cutan. Pathol. 2008;35:10–14. doi: 10.1111/j.1600-0560.2007.00762.x. [DOI] [PubMed] [Google Scholar]
- 48.Kim K.Y., Li S., Cha J.D., Zhang X., Cha I.H. Significance of molecular markers in survival prediction of oral squamous cell carcinoma. Head Neck. 2012;34:929–936. doi: 10.1002/hed.21856. [DOI] [PubMed] [Google Scholar]
- 49.Denkert C., Koch I., Von Keyserlingk N., Noske A., Dietel N.M., Wechert W. Expression of the ELAV-like protein HuR in human colon cancer. Association with tumor stage and cyclooxygenase-2. Mod. Pathol. 2016;19:1261–1269. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- 50.Suswam E.A., Nabors L.B., Huang Y., Yang X., King P.H. IL-1beta induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3' untranslated region. Involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int. J. Cancer. 2005;113:911–919. doi: 10.1002/ijc.20675. [DOI] [PubMed] [Google Scholar]
- 51.Nabors L.B., Suswam E., Huang Y., Yang X., Johnson M.J., King P.H. Tumor necrosis factor alpha induces angiogenic factor up-regulation in malignant glioma cells. A role for RNA stabilization and HuR. Cancer Res. 2003;63:4181–4187. [PubMed] [Google Scholar]
- 52.Chae K.S., Kang M.J., Lee J.H., Ryu B.K., Lee M.G., Her N.G. T.K, Ha, Y.K. Kim, S.G. Chi, Opposite functions of HIF-alpha isoforms in VEGF induction by TGF-beta1 under non-hypoxic conditions. Oncogene. 2011;30:1213–1228. doi: 10.1038/onc.2010.498. [DOI] [PubMed] [Google Scholar]
- 53.Nabors L.B., Gillespie G.Y., Harkins L., King P.H. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3' untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2011;61:2154–2161. [PubMed] [Google Scholar]
- 54.Filippova N., Yang X., Wang Y., Gillespie G.Y., Langford C., King P.H., Wheeler C., Nabors L.B. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol. Cancer Res. 2011;9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urano N., Fujiwara Y., Doki Y., Kim S.J., Miyoshi Y., Noguchi S., Miyata H., Takiguchi S., Yasuda T., Yano M. Clinical significance of class III beta-tubulin expression and its predictive value for resistance to docetaxel-based chemotherapy in gastric cancer. Int. J. Oncol. 2006;28:375–381. [PubMed] [Google Scholar]
- 56.Lee K.M., Cao D., Itami A., Hruban P.M., Maitra A. M.M, Ouellette, Class III beta-tubulin, a marker of resistance to paclitaxel, is overexpressed in pancreatic ductal adenocarcinoma and intraepithelial neoplasia. Histopathology. 2007;51:539–546. doi: 10.1111/j.1365-2559.2007.02792.x. [DOI] [PubMed] [Google Scholar]
- 57.Seve P., Isaac S., Tredan O., Souquet P., Pacheco Y., Perol M., Lafanechere L., Penet A., Peiller E., Dumontet C. Expression of class III β-tubulin is predictive of patient outcome in patients with non-small cell lung cancer receiving vinorelbine-based chemotherapy. Clin. Cancer Res. 2005;11:5481–5486. doi: 10.1158/1078-0432.CCR-05-0285. [DOI] [PubMed] [Google Scholar]
- 58.Afsharpad M., Nowroozi M.R., Mobasheri M.B., Ayati M., Nekoohesh L., Saffari M., Zendehel K., Modarressi M.H. Cancer-Testis Antigens as New Candidate Diagnostic Biomarkers for Transitional Cell Carcinoma of Bladder. Pathology oncology research; POR. 2017:20. doi: 10.1007/s12253-017-0313-4. [DOI] [PubMed] [Google Scholar]
- 59.Koinuma J., Akiyama H., Fujita M., Hosokawa M., Tsuchiya E., Kondo S., Nakamura Y., Daigo Y. Characterization of an Opa interacting protein 5 involved in lung and esophageal carcinogenesis. Cancer Sci. 2012;103:577–586. doi: 10.1111/j.1349-7006.2011.02167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong M., Xu Y., Dong W., Guo G., Ni W., Wang Y., An R. Expression of Opa interacting protein 5 (OIP5) is associated with tumor stage and prognosis of clear cell renal cell carcinoma. Acta Histochem. 2013;118:810–815. doi: 10.1016/j.acthis.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 61.Luo M., Li Z., Wang W., Zeng Y., Liu Z., Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280:1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- 62.Jia J., Zhnag X., Zhan D., Li J., Li Z., Li H., Qian J. LncRna H19 interacted with miR-130a-3p and miR-17-5p to modify radio-resistance and chemo-resistance and chemo-sensitivity of cardiac carcinoma cells. Cancer Medicine. 2019;8:1604–1618. doi: 10.1002/cam4.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lottin S., Adriaenssens E., Dupressoir T., Berteaux N., Montpellier C., Coll J., Dugimont T., Curgy J.J. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002;23:1885–1895. doi: 10.1093/carcin/23.11.1885. [DOI] [PubMed] [Google Scholar]
- 64.Murphy S.K., Huang Z., Wen Y., Spillman M.A., Whitaker R.S., Simel L.R., Nichols T.D., Marks J.R., Berchuck A. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol. Cancer Res. 2006;4:283–292. doi: 10.1158/1541-7786.MCR-05-0138. [DOI] [PubMed] [Google Scholar]
- 65.Kondo M., Suzuki H., Ueda R., Osada H., Takagi K., Takahashi T. T, Takahashi, Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193–1198. [PubMed] [Google Scholar]
- 66.Reif K., Cyster J.G. RGS molecule expression in murine B lymphocytes and ability to down-regulate chemotaxis to lymphoid chemokines. J. Immunol. 2000;164:4720–4729. doi: 10.4049/jimmunol.164.9.4720. [DOI] [PubMed] [Google Scholar]
- 67.Curtin J.A., Fridlyand J., Kageshita T., Patel H.N., Busam J.K., Kutzner H., Cho K., Aiba S., Brocker E., LeBoit P.E. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 68.Mehra S., Messner H., Minden M., Chaganti R.S. Molecular cytogenetic characterization of non-Hodgkin lymphoma cell lines. Genes Chromosom. Cancer. 2002;33:225–234. doi: 10.1002/gcc.10025. [DOI] [PubMed] [Google Scholar]
- 69.Chen D., Gallie B.L., Squire J.A. Minimal regions of chromosomal imbalance in retinoblastoma detected by comparative genomic hybridization. Cancer Genet. Cytogenet. 2001;129:57–63. doi: 10.1016/s0165-4608(01)00427-7. [DOI] [PubMed] [Google Scholar]
- 70.Tirado C.A., Sandberg A.A., Stone J.F. Identification of novel amplicon at 1q31 in pancreatic cancer cell lines. Cancer Genet. Cytogenet. 1999;113:110–114. doi: 10.1016/s0165-4608(99)00012-6. [DOI] [PubMed] [Google Scholar]
- 71.Chien G., Yuen P.W., Kwong D. Comparative genomic hybridization analysis of nasopharyngeal carcinoma; consistent patterns of genetic aberrations and clinicopathological correlations. Cancer Genet. Cytogenet. 2001;126:63–67. doi: 10.1016/s0165-4608(00)00392-7. [DOI] [PubMed] [Google Scholar]
- 72.Evangelista M., Tian H., de Sauvage F.J. The hedgehog signaling pathway in cancer. Clin. Cancer Res. 2006;12:5924–5928. doi: 10.1158/1078-0432.CCR-06-1736. [DOI] [PubMed] [Google Scholar]
- 73.Queiroz K.C., Ruela-de-Sousa R.R., Fuhler G.M., Aberson H.L., Ferreira C.V., Peppelenbosch M.P., Spek C.A. Hedgehog signaling maintains chemoresistance in myeloid leukemic cells. Oncogene. 2019;29:6314–6322. doi: 10.1038/onc.2010.375. [DOI] [PubMed] [Google Scholar]
- 74.Zhao C., Chen A., Jamieson C.H., Fereshteh M., Abrahamsson A., Blum J., Kwon H.Y., Kim J., Chute J.P., Rizzieri D. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2019;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scales S., de Sauvage F. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 76.Blotta S., Jakubikova J., Calimeri T., Roccaro A.M., Amodio N., Azab A.K., Foresta U., Mitsiades C.S., Rossi M., Todoerti K. Canonical and noncanonical Hedgehog pathway in the pathogenesis of multiple myeloma. Blood. 2012;120:5002–5013. doi: 10.1182/blood-2011-07-368142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeng K.S., Sheen I.S., Jeng W.J., Yu M.C., Hsiau H., Chang F.Y. High expression of Sonic Hedgehog signaling pathway genes indicates a risk of recurrence of breast carcinoma. Onco-Targets Ther. 2013;7:79–86. doi: 10.2147/OTT.S54702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rudin C.M., Hann C.L., Laterra J., Yauch R.L., Callahan C.A., Fu L., Holcomb T., Stinson J., Gould S.E., Coleman B. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC‑0449. N Engl J Med. 2019;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vismodegib . 2012. approved by US FDA for treatment of basal cell carcinoma. Available from; http;//www.medwirenews.md/46/97315/Oncology/Vismodegib_approved_by_US_FDA_for_treatment_of_basal_cell_ carcinoma.html. [Google Scholar]
- 80.FDA . 2012. approval for vismodegib. Available from; http;//www.cancer.gov/ cancertopics/ druginfo/fda vismodegib. [Google Scholar]
- 81.Von Hoff D.D., LoRusso P.M., Rudin C.M. Inhibition of the hedgehog pathway in advanced basal‑cell carcinoma. N. Engl. J. Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 82.Carroll T., Williams J., Daily K., Rogers T., Gelb T., Coxon A., Wang S.Q., Crago A.M., Busam K.J., Brownell I. Hedgehog signaling inhibitors fail to reduce Merkel cell carcinoma viability. Invest Dermatol. 2017;137:1187–1190. doi: 10.1016/j.jid.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brunner M., Thurnher D., Pammer J., Heiduschka G., Petzelbauer P., Schmind C., Schneider S., Erovic B.M. Expression of hedgehog signaling molecules in Merkel cell carcinoma. Head Neck. 2010;32:333–340. doi: 10.1002/hed.21191. [DOI] [PubMed] [Google Scholar]
- 84.Li C., Chi S., Xie J. Hedgehog signaling in skin cancers. Cell. Signal. 2011;23:1235–1243. doi: 10.1016/j.cellsig.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ugurel S., Spassova I., Wohlfarth I.J., Drusio C., Cherouny A., Melior A., Sucker A., Zimmer L., Ritter C., Schadendorf D. MHC class-I downregulation in PD-1/PD-L1 inhibitor refractory Merkel cell carcinoma and its potential reversal by histone deacetylase inhibition: a case series. Cancer Immunol. Immunother. 2019;68:983–990. doi: 10.1007/s00262-019-02341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Czabotar P.E., Lessene G., Strasser A., Adams J.M. Control of apoptosis by the BCL-2 protein family; implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 87.Gong J.N., Khong T., Segal D., Yao Y., Riffkin C.D., Garnier J., Khaw S.L., Lessene G., Spencer A., Herold M.J. Hierarchy for targeting prosurvival BCL2 family proteins in multiple myeloma; pivotal role of MCL1. Blood. 2016;128:1834–1844. doi: 10.1182/blood-2016-03-704908. [DOI] [PubMed] [Google Scholar]
- 88.Glaser S.P., Lee E.F., Trounson E., Bouilet P., Wei A., Fairlie W.D., Izon D.J., Zuber J., Rappaport A.R., Herold M.J. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev. 2012;26:120–125. doi: 10.1101/gad.182980.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelly G., Grabow S., Glaser S.P., Flitzsimmons L., Aubery B.J., Okamato T., Valentene L.J., Robatil M. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev. 2014;28:58–70. doi: 10.1101/gad.232009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tiedemann R.E., Zhu Y.X., Schmidt J., Shi C.X., Sereduk C., Yin H., Mousses S., Stewert A.K. Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res. 2012;72:757–768. doi: 10.1158/0008-5472.CAN-11-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiao Y., Nimmer P., Sheppard G.S., Bruncklo M., Lu X., Robert-Rapp L., Pappano W.N., Elmore S.W., Souers A.J. MCL-1 Is a Key Determinant of Breast Cancer Cell Survival; Validation of MCL-1 Dependency Utilizing a Highly Selective Small Molecule Inhibitor. Mol. Cancer Ther. 2015;14:1837–1847. doi: 10.1158/1535-7163.MCT-14-0928. [DOI] [PubMed] [Google Scholar]
- 92.Merino D., Whittle J.R., Vaillant F., Serrano A., Gong J., Ginger G., Maragno A.L., Chanrion M., Schneider E., Pal B. Synergistic action of the MCL-1 inhibitor S63845 with current therapies in preclinical models of triplenegative and HER2-amplified breast cancer. Sci. Transl. Med. 2017;9:401. doi: 10.1126/scitranslmed.aam7049. [DOI] [PubMed] [Google Scholar]
- 93.Wertz I., Kusam E.S., Lam C., Okamoto T., Sandoval W., Anderson D.J., Helgason E., Ernst J.A., Eby M., Liu J. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 94.Verhaegen M., Mangelberger D., Weick J., Vozheiko T.D., Harms P.W., Nash K.T., Quintana E., Baciu P., Johnson T.M., Bichakjian C.K. Merkel Cell Carcinoma Dependence on Bcl-2 Family Members for Survival. Journal of Investigative Dermatolgy. 2014;134:2241–2250. doi: 10.1038/jid.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robert H. Whitaker and William J, Placzek. Regulating the BCL2 Family to Improve Sensitivity to Microtubule Targeting Agents. Cells. 2019;8:346. doi: 10.3390/cells8040346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frenard C., Peuvrel L., Brocard A., Saint-Jan M., Moreau A., Dreno B., Quereux G. Dramatic response of an inoperable Merkel cell carcinoma with imatinib. JAAD Case Rep. 2016;2:16–18. doi: 10.1016/j.jdcr.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinopathological characteristics of GSE39612 (N = 30).
Results of SAM analysis between different subtypes of MCC in GSE39612. Data is representing over expression of gene in two subtypes in positive and minus value.
(1). Gene were ordered according the SAM significance, gene with the positive value were overexpressed in subtype II of MCC whereas minus value genes were overexpressed in subtype I.
(2). Significance increased with decreased of absolute values of ranks from 10 to 1 or −10 to 1. Rank 1 or −1 genes are the most significant of SAM results.
Biological process enriched in each subtypes of MCC.
Enriched KEGG pathways in each subtypes of MCC.